ABSTRACT
Lipo-chitooligosaccharides (LCOs) are historically known for their role as microbial-derived signaling molecules that shape plant symbiosis with beneficial rhizobia or mycorrhizal fungi. Recent studies showing that LCOs are widespread across the fungal kingdom have raised questions about the ecological function of these compounds in organisms that do not form symbiotic relationships with plants. To elucidate the ecological function of these compounds, we investigate the metabolomic response of the ubiquitous human pathogen Aspergillus fumigatus to LCOs. Our metabolomics data revealed that exogenous application of various types of LCOs to A. fumigatus resulted in significant shifts in the fungal metabolic profile, with marked changes in the production of specialized metabolites known to mediate ecological interactions. Using network analyses, we identify specific types of LCOs with the most significant effect on the abundance of known metabolites. Extracts of several LCO-induced metabolic profiles significantly impact the growth rates of diverse bacterial species. These findings suggest that LCOs may play an important role in the competitive dynamics of non-plant-symbiotic fungi and bacteria. This study identifies specific metabolomic profiles induced by these ubiquitously produced chemicals and creates a foundation for future studies into the potential roles of LCOs as modulators of interkingdom competition.
IMPORTANCE The activation of silent biosynthetic gene clusters (BGC) for the identification and characterization of novel fungal secondary metabolites is a perpetual motion in natural product discoveries. Here, we demonstrated that one of the best-studied symbiosis signaling compounds, lipo-chitooligosaccharides (LCOs), play a role in activating some of these BGCs, resulting in the production of known, putative, and unknown metabolites with biological activities. This collection of metabolites induced by LCOs differentially modulate bacterial growth, while the LCO standards do not convey the same effect. These findings create a paradigm shift showing that LCOs have a more prominent role outside of host recognition of symbiotic microbes. Importantly, our work demonstrates that fungi use LCOs to produce a variety of metabolites with biological activity, which can be a potential source of bio-stimulants, pesticides, or pharmaceuticals.
KEYWORDS: lipo-chitooligosaccharides, secondary metabolites, synergism, biosynthetic gene clusters, bacteria, Aspergillus, network analyses
INTRODUCTION
Fungal biosynthetic gene clusters (BGCs) often encode natural products with potent antimicrobial properties (1). However, since because these compounds mediate specific ecologies found in nature, many BGCs are quiescent under normal laboratory conditions. Given the ecological and pharmacological potential of these compounds, considerable effort has focused on how to activate BGCs. Abiotic stressors (e.g., temperature, light, pH) are well known to induce the production of fungal secondary metabolites that influence the growth of neighboring microorganisms (1–3). The expression of biosynthetic gene clusters encoding fungal natural products can also be activated by neighboring plant hosts or microbes. Recently, we observed that the specialized microbial signaling molecules lipo-chitooligosaccharides (LCOs), initially discovered in rhizobia and known for their role in the establishment of root nodule symbiosis with legumes (4), impact fungi at the physiological and molecular levels (5, 6).
LCOs are amphiphilic, short-chain, chitin-derived molecules that were believed to be exclusively produced by rhizobial bacteria and mycorrhizal fungi to establish symbiotic associations with host plants (4, 7–13). The molecules are now known to be ubiquitously produced across most phyla within the fungal kingdom (5). However, the role that LCOs play in nonsymbiotic fungi remains unclear. While research on the alternative roles of LCOs in fungi is still in its infancy, recent findings have opened new areas of inquiry regarding other functions of LCOs in microbes.
Foliar applications of LCOs to infected soybeans increased the disease incidence of Sclerotinia stem rot (14). In addition, legumes have been able to coordinate defense mechanisms against plant pathogens while facilitating symbiosis responses in the presence of LCOs (15). Similarly, dose-dependent phenotypic and morphological changes were observed in known virulence-associated traits when LCOs were applied to the opportunistic human pathogens Aspergillus fumigatus and Candida glabrata, including increased spore germination, pseudohypha formation, and decreased hyphal branching (5). These observations were accompanied by significant transcriptomic changes in A. fumigatus (5). Likewise, Rhodotorula mucilaginosa displayed increased cell production following exposure to LCOs (5). Finally, LCOs resulted in proteomic changes, decreased hyphal branching, and increased clamp connections in Laccaria bicolor (6). The diversity and breadth of phenotypic responses to LCOs emphasize the need to characterize and investigate the ecological function of LCO-induced metabolic profiles.
The present study investigates if LCOs affect the production of fungal secondary metabolites in ways that impact the growth of surrounding microbes, which could be likely competitors. To study this, we used the common soilborne opportunistic human pathogen Aspergillus fumigatus which has well-characterized genetic and metabolomic profiles (16–21) and has been confirmed to both produce and respond to LCOs (5). We first assessed how the exogenous application of LCOs to A. fumigatus alters the fungus’ metabolome at temperatures that are relevant to the environmental (25°C) and clinical (37°C) ecology of this organism (19, 22–25). These studies have shown that there were more secondary metabolites identified and characterized at 25°C than at 37°C. To explore the importance of various LCO treatments at inducing the production of a secondary metabolite or feature, we used network analysis as previously reported (26, 27). Observed secondary metabolites were identified using comparative fragmentation patterns, confirmation with commercial standards, and associated biosynthetic gene expressions. The biological effect of these LCO-induced metabolic profiles on bacterial development was assessed on seven strains representative of four different bacterial classes. These bacterial classes were defined by Carper et al. (28).
Our results provide the first line of evidence showing that the production of known, putative, and unknown metabolites in A. fumigatus is differentially induced by specific LCOs. Several of these LCO-specific metabolic profiles significantly impact bacterial growth. We identify several BGCs as likely candidates to explain LCO-mediated differences in bacterial growth. Taken together, our results demonstrate that LCO-dependent metabolic profiles could mediate competitive interactions of A. fumigatus through the induction of specific BGCs.
RESULTS
LCO-treated samples resulted in different metabolomic profiles at 25°C and 37°C.
(i) Nomenclature. The treatments C16:0 sulfated LCOs, C16:0 nonsulfated LCOs, C18:1 sulfated LCOs, and C18:1 nonsulfated LCOs will be abbreviated as C16:0 sLCOs, C16:0 nsLCOs, C18:1 sLCOs, and C18:1 nsLCOs, respectively. LCO structures and chemical formulas are depicted in Fig. 1.
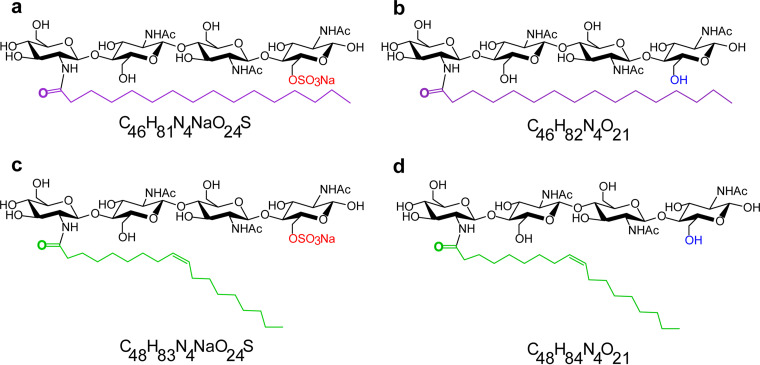
FIG 1.
Chemical structures of LCO standards. (a to d) The LCO structures are (a) C16:0 sulfated LCOs, (b) C16:0 nonsulfated LCOs, (c) C18:1 sulfated LCOs, and (d) C18:1 nonsulfated LCOs. The palmitic acid chain is in purple, the oleic acid chain is in green, the sulfated group is in red, and the nonsulfated group is in blue. The chemical formula for each LCO is shown below the structure. LCO standards were dissolved in 0.005% EtOH for use as the solvent control. The concentration of LCOs used in the experiments was 10−8 M.
(ii) Temperature conditions. Two different growth conditions were assessed; the first consisted of incubation at 25°C and 250 rpm for 6 days, whereas the second consisted of incubation at 37°C and 250 rpm for 4 days. Differences in the growth period were necessary to accommodate for the slow growth at 25°C. At 37°C, the fungal dry biomass produced under treatments C16:0 sLCOs, C16:0 nsLCOs, C18:1 sLCOs, and C18:1 nsLCOs was higher than the dry biomass of samples treated the same way at 25°C, with respective fold changes of 2.1, 2.1, 2.7, and 1.9. There were no significant differences in dry biomass between treatments grown at the same temperature (see Fig. S1 in the supplemental material).
Dry biomass weight of A. fumigatus at (a) 25°C and (b) 37°C when exposed to LCO treatments and compared to the solvent control. There were 10 biological replications. Welch’s unpaired t test was used to compare individual treatments to the solvent control for each temperature, resulting in no significant differences. Download FIG S1, TIF file, 14.3 MB (14.3MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
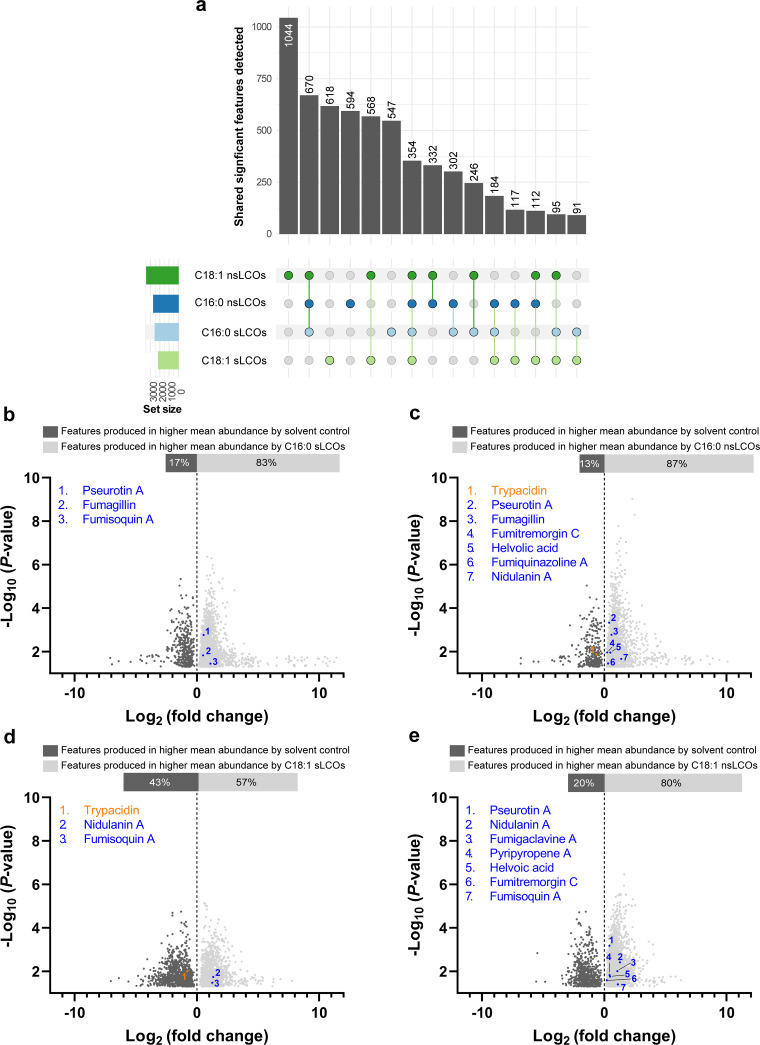
(iii) Metabolomic regulation at 25°C. We used liquid chromatography-mass spectrometry (LC/MS) to obtain a general metabolomic profile. LC/MS data revealed that all treatments using individual LCO-related compounds significantly induce the production of features compared to the solvent control (Fig. 2a). Features are defined as analytes with specific mass-to-charge (m/z) values and retention times. In total, 5,874 significant features with a P value of <0.05 were detected (Fig. 2a). Treatment with C18:1 nsLCOs resulted in the highest number of features increased in relative abundance compared to the control (17.8%), followed by C18:1 sLCOs (10.5%), C16:0 nsLCOs (10.1%), and C16:0 sLCOs (9.3%) (Fig. 2a). Comparing data collected from the different individually applied treatments revealed that some features are induced by multiple treatments. In this regard, C16:0 nsLCOs, C16:0 sLCOs, and C18:1 nsLCOs, had the highest amount of shared induced features (11.4%), followed by two treatment groups that are similar in structure, C18:1 sLCOs and C18:1 nsLCOs (9.7%) (Fig. 2a). Other features that were also induced by multiple treatments amounted to less than 5% relative abundance to the solvent control per group (Fig. 2a).
FIG 2.
LCO-induced A. fumigatus metabolomic profiles at 25°C. (a) The total significant features produced were counted and distributed by treatments that coincidentally or uniquely induced their production. Set size is the number of features coincidentally or uniquely produced by each of the treatments. The plots are separated based on features detected under the 25°C growth condition. Features showing significant differences (P < 0.05) in pairwise comparison between treatment and solvent control with intersections with at least 10 members are displayed (n = 10 per treatment). (b to e) Volcano plots representing the number of features and known secondary metabolites identified in (b) C16:0 sLCO-, (c) C16:0 nsLCO-, (d) C18:1 sLCO-, and (e) C18:1 nsLCO-treated samples. Features showing significant differences (P < 0.05) in pairwise comparison between treatment and control are displayed (n = 10 per treatment).
To identify known and putative secondary metabolites produced by A. fumigatus under the various treatments, we used the MAVEN (29) and XCMS (30) software platforms to identify analytical features above our peak area threshold (see Materials and Methods). This approach identified the secondary metabolites fumiquinazolines A and F, fumitremorgin C, helvolic acid, pseurotin A, pyripyropene A, and trypacidin, which are known to modulate microbial interactions (19). The production of these metabolites was compared across treatments (Fig. 2b to e). We also identified the production of fumagillin and fumigaclavine A, compounds known to affect disease and other interactions with the human immune system (19). A subset of the samples (4 out of 10 biological replications) was randomly selected and subjected to LC-MS/MS for confirmation of all secondary metabolites listed above by matching their fragmentation patterns to procured standards or an in silico database search if a standard was unavailable for purchase. LC-MS/MS feature confirmation data were previously shown in reference 26. Later, we evaluated whether the production of these known metabolites is regulated at the transcriptional level through quantitative PCR (qPCR) analysis of their specific backbone genes (core biosynthetic genes) or transcription factors (Fig. S2). The gene IDs and PCR primers information are provided in reference 26. The putative secondary metabolites fumigaclavine C, fumisoquin A, and nidulanin A were predicted from crude extracts based on m/z values but were not confirmed by fragmentation or comparison to a standard due to their unavailability. The production of known and putative secondary metabolites in response to the different treatments was evaluated compared to the solvent control. All LCO treatments showed a significant positive impact on the relative abundance of known or putative secondary metabolites, except for two LCO treatments: C16:0 nsLCO and C18:1 sLCO. In these treatments, trypacidin was less abundant in the treatments compared to the solvent control (Fig. 2c and d). However, no significant differences were observed in expression levels of the trypacidin backbone gene between the control and the treatments (Fig. S2). Several secondary metabolites with known antimicrobial properties—including fumisoquin A, fumiquinazolines A and F, fumitremorgin C, helvolic acid, pseurotin A, and pyripyropene A—were overproduced under specific LCO treatments (Fig. 2b to e). Fumagillin, fumigaclavine A, gliotoxin, helvolic acid, and pseurotin A backbone genes were also overexpressed in response to LCO treatments (Fig. S2).
ΔΔCT values and peak area for confirmed secondary metabolites. Welch’s unpaired t test was used to compare individual treatments to the solvent control. *, P value < 0.005; **, P value < 0.01; ***, P < 0.001; ****, P < 0.0001. There were three biological replications for the ΔΔCT data and 10 biological replications for the peak area data set. (a to h) Welch’s ANOVA P values were (a) <0.0001 for ΔΔCT data and 0.0110 for peak area, (b) <0.0001 for ΔΔCT and 0.0070 for peak area, (c) 0.0007 for ΔΔCT and not significant for peak area, (d) 0.0010 for ΔΔCT and not significant for peak area, (e) 0.0025 for ΔΔCT and not significant for peak area, (f) <0.0001 for ΔΔCT and 0.0017 for peak area, (g) not significant, and (h) not significant. The primers used for ΔΔCT analysis can be found in reference 26. Download FIG S2, TIF file, 14.3 MB (14.3MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
At 25°C, the relative abundance of fumagillin and pseurotin A were significantly increased by both C16:0 LCO treatments compared to the solvent control (Fig. 2b and c). The backbone gene implicated in pseurotin A production and the C6 transcription factor regulating fumagillin production were significantly overexpressed as a result of these treatments compared with the solvent control (Fig. S2), which is noteworthy considering that these two metabolites have biosynthetic genes within an intertwined supercluster (31).
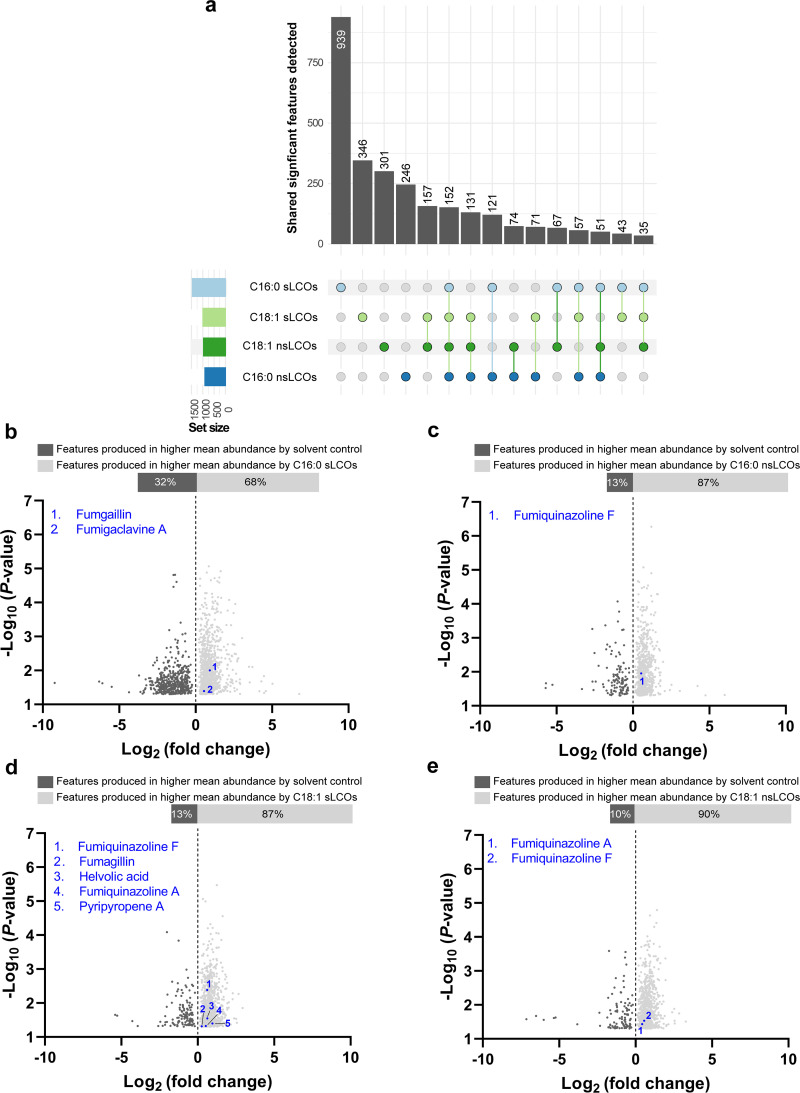
(iv) Metabolomic regulation at 37°C. LC/MS data revealed that all individually applied treatments significantly induce the production of features compared to the solvent control, like the results shown at 25°C (Fig. 3a). However, there was a 52.5% decrease in the total number of induced features compared to the 25°C data set (Fig. 2a). In total, 2,791 significant features with a P value of <0.05 were detected. (Fig. 3a). The C16:0 sLCO-treated samples had the highest number of total features induced (33.6%), followed by C18:1 sLCOs (12.4%), C18:1 nsLCOs (10.7%), and C16:0 nsLCOs (8.8%) (Fig. 2a). Comparing data collected from the different individually applied treatments revealed that some features are induced by multiple treatments. In this regard, two treatment groups that are similar in structure, C18:1 sLCOs and C18:1 nsLCOs (5.6%), followed by all treatments, shared 5.4% of features (Fig. 3a). Other features that were induced by multiple treatments had less than 5% relative abundance compared to the solvent control per group (Fig. 3a).
FIG 3.
LCO-induced A. fumigatus metabolomic profiles at 37°C. (a) The total significant features produced were counted and distributed by treatments that coincidentally or uniquely induced their production. Set size is the number of features coincidentally or uniquely produced by each of the treatments. The plots are separated based on features detected under the 37°C growth condition. Features showing significant differences (P < 0.05) in pairwise comparison between treatment and solvent control with intersections with at least 10 members are displayed (n = 10 per treatment). (b to e) Volcano plots representing the number of features and known secondary metabolites identified in (b) C16:0 sLCO-, (c) C nsLCO-, (d) C18:1 sLCO-, and (e) C18:1 nsLCO-treated samples. Features showing significant differences (P < 0.05) in pairwise comparison between treatment and control are displayed (n = 10 per treatment).
Certainly, there is a stark contrast in number of metabolites produced at 25°C compared to 37°C (Fig. S3). As under the 25°C growth condition, all LCO treatments showed a significant positive impact on the production of known secondary or putative metabolites. Again, several secondary metabolites with known antimicrobial properties—such as fumiquinazolines A and F, helvolic acid, pseurotin A, and pyripyropene A—were overproduced under various LCO-treatments (Fig. 3b to e). Previously, we observed the coregulation of pseurotin A and fumagillin in the 25°C data set by both C16:0-treated samples. However, these two metabolites do not seem to follow the same regulation at 37°C, as only one of them, fumagillin, was detected (Fig. 3b and d). Also, fumagillin was shown to be significantly overproduced under both sulfated C16:0 and C18:1 LCO treatments at 37°C, but not under nsLCOs, as shown in the 25°C data set. C16:0 sLCOs induce the production of fumagillin at both temperatures.
Principal-component analysis of treatments and temperature influence on identified features. Download FIG S3, TIF file, 14.3 MB (14.3MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
In our previous study (5) conducted at 37°C, treatment with C16:0 sLCOs caused reduced hyphal branching, whereas the other LCO treatments showed no effect on branching. The mechanisms by which C16:0 sLCOs influence branching remain unexplored. Considering that secondary metabolites can regulate sporulation and hyphal branching (32, 33), it is tempting to speculate that fumagillin, or other specific metabolites, or a combination of metabolites induced in response to the C16:0 sLCO treatment may be in part responsible for the previously observed physiological changes. For a future study, we will investigate this hypothesis by examining mutants lacking the global regulator of fungal secondary metabolism, LaeA, which was shown to regulate the expression of 20% to 40% of major classes of secondary metabolites (SM) biosynthesis genes in A. fumigatus, including the pseurotin and fumagillin supercluster (1, 16, 18). Taken together, metabolomic analyses conducted at 37°C revealed significantly fewer features and detected known secondary metabolites, regardless of the treatment (Fig. 3). These findings correlate well with a previous study showing overexpression of biosynthetic gene clusters in A. fumigatus at 30°C relative to 37°C (23, 25).
Network analysis determines which LCO treatments had the largest influence on known, putative, and unknown metabolite production.
Leveraging from our previous efforts, we used direct and auxiliary network analysis approaches (26) to determine the influence a treatment has on known or putative metabolites and unknown features. The direct network analysis approach was performed to determine which treatments impacted negatively or positively the relative abundance of a specific known metabolite produced by A. fumigatus (Fig. 4 and 5). More specifically, network theory was applied to rank the most effective treatments and receptive secondary metabolites based on node strength (size of nodes in Fig. 4a and 5a) and PageRank measures (Fig. 4b and c and Fig. 5b and c) as described in our previous work (26, 27). On the other hand, we employed the auxiliary network analysis approach illustrated in reference 26 to extract unknown features from the raw LC/MS data sets (Fig. 4d and 5d). To eliminate noise, a baseline correction was employed as previously done in reference 26. Features were identified via KEGG for both 25°C and 37°C using the online identification option within MZmine using the parameters ionization type: [M+H]+, number of results: 20, m/z tolerance: 0.001 m/z or 5.0 ppm. To illustrate the most robustly evidenced features, we filtered our network to only include features with a greater than 1 absolute value log2 fold change between the control and a treatment; however, networks of all statistically significant analyte peaks regardless of log2 fold change are also provided. We emphasize that while the auxiliary network analysis approach assesses the production of unknown features, the direct network analysis approach determines the synthesis of known and putative metabolites. Details on generating the network, definitions, and computing of the measures can be found in Materials and Methods of the current study and reference 26.
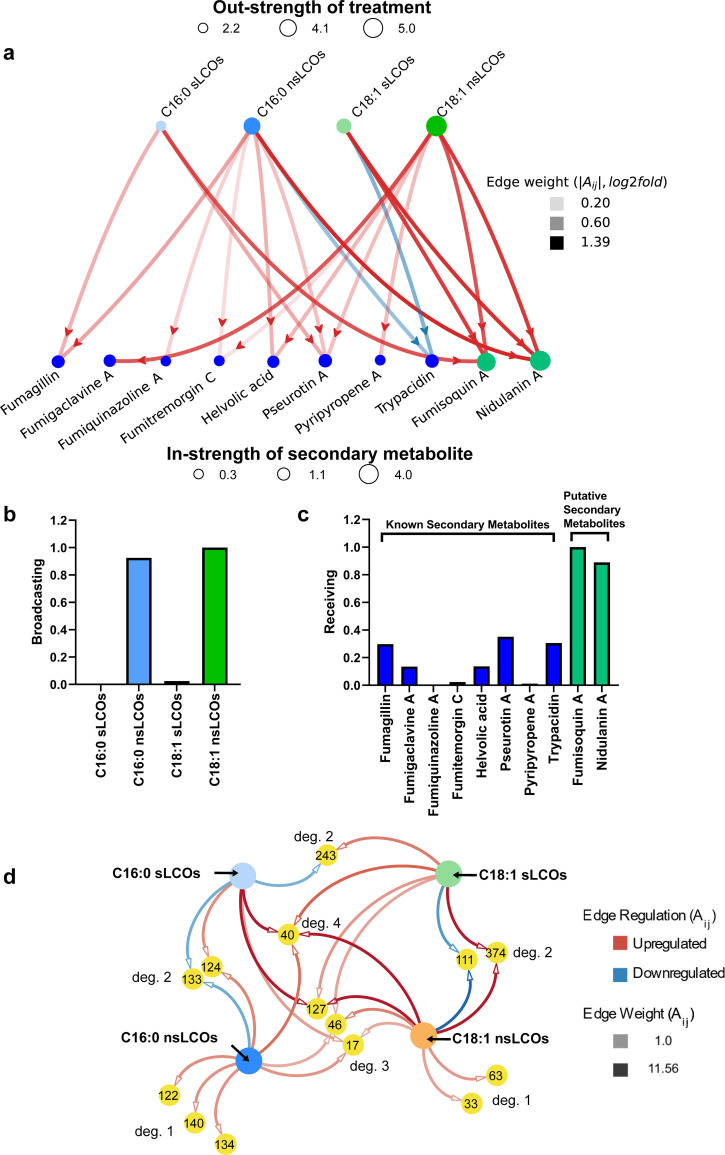
FIG 4.
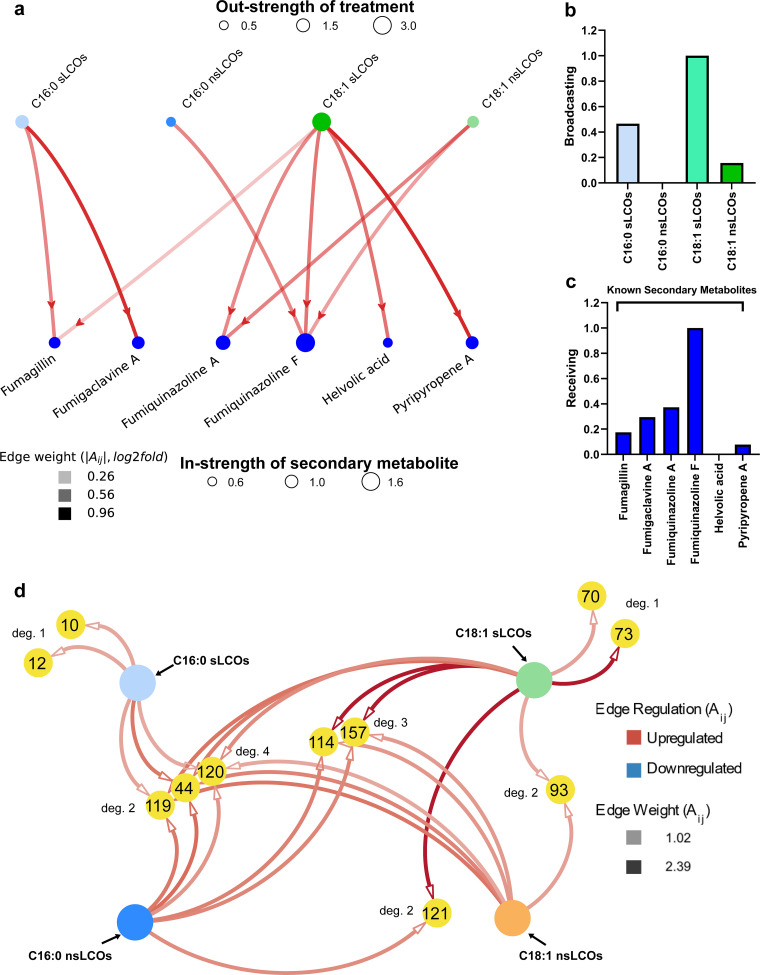
Network analysis of metabolomic changes in response to LCO treatments at 25°C. Graphical representation of the directed network analysis showing the influence of the treatments on secondary metabolite production. (a) Directed PageRank measures: (b) broadcasting for treatments and (c) receiving for secondary metabolites. (d) Auxiliary network analysis of significantly differentially regulated XICs uniquely (degree of 1 noted as “deg. 1”) or coincidentally (degrees of 2, 3, or 4 noted as “deg. 2,” “deg. 3,” or “deg. 4”) produced by all treatments. Metabolomic regulation was determined by the log2 fold change and is indicated by edge color, where red shows upregulation and blue shows downregulation.
FIG 5.
Network analysis of metabolomic changes in response to LCO treatments at 37°C. Graphical representation of the direct network analysis showing the influence of the treatments on secondary metabolite production. (a) PageRank measures: (b) broadcasting for treatments and (c) receiving for secondary metabolites. (d) Auxiliary network analysis of significantly differentially regulated XICs uniquely (degree of 1 noted as “deg. 1”) or coincidentally (degrees of 2, 3, or 4 noted as “deg. 2,” “deg. 3,” or “deg. 4”) produced by all treatments. Metabolomic regulation was determined by the log2 fold change and is indicated by edge color, where red shows upregulation and blue shows downregulation.
(i) Network analysis at 25°C. Through the directed network analysis, positive and negative regulations of both known and putative secondary metabolites by LCOs are evident (see the graph visualization in Fig. 4a). While the importance of each LCO and secondary metabolite is easily visualized using the node strength (size of the nodes), these total regulation values do not consider the relative importance of the nodes (LCOs or secondary metabolites) (see reference 26 for the shortcomings of the node strength measure). The PageRank measures (Fig. 4b and c) identify the most influential LCOs and receptive secondary metabolites (those most induced by LCOs). Through the broadcasting PageRank measure (34), it is evident that the nonsulfated LCOs (C18:1 followed by C16:0) have the largest effect on triggering the production of secondary metabolites. These results align with the inferences drawn from the UpSet plots in Fig. 2a. Moreover, the receiving PageRank measure identifies the putative metabolite fumisoquin A, followed by nidulanin A, to be the most receptive to being triggered by various LCO treatments.
Through the auxiliary network analysis, we found 14 unique extracted ion chromatograms (XIC) defined by their m/z, retention time, and area under the curve. These 14 XICs elicited 25 differentially metabolomic regulations of interest, which were extracted from the treatments with a greater than 1.0 log2 fold change (Fig. 4d, Table 1). All extracted XICs induced at 25°C (including the unfiltered XIC with log2 fold changes below 1.0) can be viewed in Table S1 and Fig. S4. None of these putative metabolites have been confirmed through fragmentation or comparison to a commercial standard, except for fumigaclavine A.
TABLE 1.
Putative metabolites produced at 25°C identified by the auxiliary method
| XIC ID | Regulation | Treatment | Log2 fold change | Putative metabolite ID | KEGG ID |
|---|---|---|---|---|---|
| 17 | Up | C16:0 nsLCOs | 1.32 | Unknown | |
| 17 | Up | C18:1 nsLCOs | 1.02 | Unknown | |
| 17 | Up | C16:0 sLCOs | 1.18 | Unknown | |
| 33 | Up | C18:1 nsLCOs | 1.13 | Fraxetin | C09265 |
| 40 | Up | C16:0 nsLCOs | 1.78 | Unknown | |
| 40 | Up | C18:1 nsLCOs | 2.08 | Unknown | |
| 40 | Up | C18:1 sLCOs | 1.96 | Unknown | |
| 40 | Up | C16:0 sLCOs | 2.08 | Unknown | |
| 46 | Up | C16:0 nsLCOs | 1.00 | Unknown | |
| 46 | Up | C18:1 nsLCOs | 1.59 | Unknown | |
| 46 | Up | C18:1 sLCOs | 1.09 | Unknown | |
| 63 | Up | C18:1 nsLCOs | 1.27 | Fumigaclavine A | C20436 |
| 111 | Down | C18:1 nsLCOs | –2.54 | Unknown | |
| 111 | Down | C18:1 sLCOs | –1.83 | Unknown | |
| 122 | Up | C16:0 nsLCOs | 1.41 | Borrerine | C09054 |
| 124 | Up | C16:0 nsLCOs | 1.52 | Fumigaclavine A | C20436 |
| 124 | Up | C16:0 sLCOs | 1.41 | Fumigaclavine A | C20436 |
| 133 | Down | C16:0 nsLCOs | –1.49 | Ergonovine | C07543 |
| 133 | Down | C16:0 sLCOs | –1.46 | Ergonovine | C07543 |
| 134 | Up | C16:0 nsLCOs | 1.34 | Unknown | |
| 140 | Up | C16:0 nsLCOs | 1.34 | 5,10-Dihydrophenazine-1-carboxylate | C20981 |
| 243 | Up | C18:1 sLCOs | 1.63 | Epithienamycin B | C17396 |
| 243 | Down | C16:0 sLCOs | –1.38 | Epithienamycin B | C17396 |
| 374 | Up | C18:1 nsLCOs | 11.56 | Stearidonic acid | C16300 |
| 374 | Up | C18:1 sLCOs | 9.43 | Stearidonic acid | C16300 |
Full auxiliary network analysis of metabolomic changes in response to LCO treatments at 25°C. The whole unfiltered network of significant log2 fold changes has 68 total nodes, 4 of which are the treatments themselves, with 130 total significant treatments as edges. Of those, 63 are downregulated and 67 are upregulated by their corresponding treatments. Of the 64 XIC analyte nodes, 29 of the nodes within the network are of degree 1, 14 of the nodes are degree 2, 11 nodes are of degree 3, and 10 are of degree 4. Of the 64 nodes, 45% of the total were of degree one, denoting that they were elicited from a single treatment. Considering summary statistics for each of the treatments within the network, C16:0 sLCOs had 19 upregulated and 13 downregulated XICs with a mean value of 0.41 and a median of 0.65. C16:0 nsLCOs had 19 upregulated and 18 downregulated XICs with a mean value of 0.33 and a median of 0.6. C18:1 sLCOs had 18 total upregulated and 18 downregulated XICs with a mean value of 0.40 and a median of 0.12. C18:1 nsLCOs had 12 upregulated and 14 downregulated XICs with a mean value of 0.83 yet with a median value of −0.1, denoting that, though there were more downregulated XICs, they were heavily outweighed by their upregulated counterparts. Two nodes of interest within the whole network are XIC IDs 243 and 374 due to having both up- and downregulated treatments. As noted in “Network Analysis at 25°C,” ID 24, identified by KEGG as an epithienamycin B-like metabolite, is upregulated by C18:1 sLCOs with a log2 fold change of 1.63, yet downregulated by C16:0 sLCOs with a log2 fold change of 1.38. XIC ID 374, when filtered for edges with log2 fold changes greater than 1, has two edges with log2 fold changes of 11.56 and 9.43, upregulated by the C18:1 nsLCOs and the C18:1 sLCOs respectively, yet when not filtered, XIC 374 has a third edge, which is downregulated by C16 nsLCOs with a log2 fold change of 0.87. All detailed XIC information is listed in Table S1. Download FIG S4, TIF file, 14.3 MB (14.3MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Full network for auxiliary route at 25°C. Download Table S1, CSV file, 0.03 MB (27.6KB, csv) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
The XIC with the highest relative abundance is ID 374 (a stearidonic acid-like metabolite). Putative stearidonic acid has two edges with log2 fold changes of 11.56 and 9.43, upregulated by sulfated and nonsulfated C18:1 LCOs, respectively. KEGG pathway analysis reveals that stearidonic acid plays a role in alpha-linolenic acid metabolism and biosynthesis of secondary metabolites. Another unknown metabolite (XIC 111) is worth noting, as it is significantly downregulated by both C18:1 nsLCOs, with a log2 fold change of 2.45, and C18:1 sLCOs, with a log2 fold change of 1.83. XIC 111 was not identified by database queries. The only other XIC to be downregulated by multiple treatments is an ergonovine-like metabolite downregulated by both C16:0 nsLCOs and C16:0 sLCOs, with log2 fold changes of 1.49 and 1.46, respectively. Erogonovine is also named ergometrine, which is an ergot alkaloid used as medication to stop postpartum hemorrhage (35). Several other ergot alkaloids have been identified from A. fumigatus, including fumigaclavine A (36). Ergometrine has not been identified in A. fumigatus before. Another XIC of interest is ID 243, an epithienamycin B-like metabolite, which is upregulated by C18:1 sLCOs with a log2 fold change of 1.63 yet downregulated by C16:0 sLCOs with a log2 fold change of 1.38. Little is known about epithienamycin B other than it is an antibiotic extracted from Streptomyces (37). Borrerine and 5,10-dihydrophenazine-1-carboxylate-like metabolites were both upregulated by C16:0 nsLCOs treatments. Borrerine is a harmala alkaloid derived from tryptophan and anthranilic acid from the plant Borreria verticillata (syn., Spermacoce verticillata) (38). KEGG pathway analysis reveals that 5,10-dihydrophenazine-1-carboxylate is involved in phenazine biosynthesis, metabolic pathways, and biosynthesis of secondary metabolites. Also, it is involved in the pyocyanine biosynthesis of the chorismate reaction into pyocyanine. Lastly, C18:1 nsLCOs induced the production of a fraxetin-like metabolite, which was recently described in A. fumigatus as an antibiotic (39) and is involved in the biosynthesis of plant secondary metabolites.
(ii) Network analysis at 37°C. The directed route (26) reveals that treatment with LCOs at 37°C has fewer secondary metabolites being regulated compared to 25°C, as evident from the sparse graph structure (comparatively smaller number of edges) in Fig. 5a. Moreover, the broadcasting PageRank measure (Fig. 5b) identifies the sulfated LCOs to be the most influential at regulating secondary metabolites at this temperature. These results align with the inferences drawn from the UpSet plots in Fig. 3a. Among the sulfated LCOs, in contrast to the UpSet plots, the sulfated C18:1 is identified to be the most influential by the network measures, as this treatment regulates a greater number of secondary metabolites with higher amplitudes compared to sulfated C16:0.
The receiving PageRank measure (Fig. 5c) identifies fumiquinazoline F as the compound most commonly regulated by various LCO treatments. Interestingly, fumiquinazoline F is the only unique secondary metabolite that is induced by LCOs at 37°C compared to the other metabolites induced at 25°C. Many more unique metabolites are regulated at 25°C than at 37°C. These include fumitremorgin C, pseurotin A, trypacidin, fumisoquin A, and nidulanin A. Interestingly, the secondary metabolite fumiquinazoline A was identified as the second most receptive secondary metabolite at 37°C, whereas it was the least receptive at 25°C.
Through the auxiliary route, we found 11 XICs and 26 differential metabolomic regulations of interest with a log2 fold change greater than 1 (Fig. 5d). The highest of these is XIC IDs 73 and 157 (alprazolam-like), 114 and 121 (clofibrate-like), all induced by the C18:1 sLCO treatment. All extracted XICs induced at 37°C (including the unfiltered XIC with log2 fold changes below 1.0) can be viewed in Table S2 and Fig. S5. None of these putative metabolites have been confirmed through fragmentation or comparison to a commercial standard, except for fumiquinazoline F.
Full auxiliary network analysis of metabolomic changes in response to LCO treatments at 37°C. The whole unfiltered network of significant log2 fold changes has 31 total nodes, 4 of which are treatments with 57 edges. Of those edges, 32 are upregulated and 25 are downregulated by their corresponding treatments. Of the 27 total nodes, 12 are of degree 1, 5 are of degree 2, 5 are of degree 3, and 5 are of degree 4. Of the 27 total analyte nodes, 44% are of degree 1, denoting that they were elicited from a single treatment. Considering summary statistics for each of the treatments within the network, treatment C16:0 nsLCOs has 8 upregulated and 8 downregulated XICs with a mean value of 0.55 and a median value of 0.37. The C18:1 nsLCO treatment has 7 upregulated and 3 downregulated XICs with a mean value of 0.74 and a median value of 1.09. C18:1 sLCO has 9 upregulated and 8 downregulated XICs with a mean value of 0.81 and a median value of 1.03. C16:0 sLCO has 8 upregulated and 6 downregulated XICs with a mean value of 0.40 and a median value of 0.83. Additional nodes of interest could be those which are downregulated with respect to their treatment, particularly given that the edges primarily tend toward upregulation. All detailed XIC information is listed in Table S2. Download FIG S5, TIF file, 14.3 MB (14.3MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Full network for auxiliary route at 37°C. Download Table S2, CSV file, 0.01 MB (14.7KB, csv) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
A 4′-O-desmethyl-3-O-acetylpapaveroxine-like compound (XIC 70) was overproduced under the C18:1 sLCO treatment. KEGG pathway analysis shows that 4′-O-Desmethyl-3-O-acetylpapaveroxine plays a role in isoquinoline alkaloid biosynthesis. Fumisoquins are isoquinoline alkaloids produced by A. fumigatus (40). An increase in production of fumisoquin A was observed with C16:0 sLCOs, C18:1 sLCOs, and C18:1 nsLCO treatments (Fig. 2). Fumisoquin A is also one of the unique putative metabolites triggered at 25°C as identified by the direct network analysis approach, but it was not detected at 37°C. With the isoquinoline alkaloid biosynthesis pathways being observed in A. fumigatus, perhaps this 4′-O-desmethyl-3-O-acetylpapaveroxine-like compound could be a novel metabolite in A. fumigatus. Alprazolam-like metabolites, XICs 73 and 157, have been putatively identified by KEGG analysis with corresponding log2 fold change intensities of 2.11 and 2.39, respectively. Alprazolam, also known as Xanax, is a neuropsychiatric agent used to reduced anxiety as a minor tranquilizer. A burseran-like compound (XIC 44) was upregulated by all LCO treatments. Burseran is a phenylpropanoid lignan first isolated from the tree Bursera microphylla and has displayed several cytotoxic and antitumor activities (41). Clofibrate-like metabolites, XICs 114 and 121, were identified by KEGG analysis and were found to be induced by most LCO treatments. KEGG pathway analysis identifies clofibrate as a chemical carcinogenesis receptor activator. Moreover, clofibrate is a lipid-lowering agent used to lower cholesterol and triglycerides. Although this compound was never identified in A. fumigatus, a different type of cholesterol-lowering drug called lovastatin was is characterized from different species of Aspergillus (42). Outside the KEGG analysis, we used Lipid Maps to identify two other putative compounds, 6′-hydroxysiphonaxanthin decanoate, an isoprenoid isolated from the chlorophyte Polyblepharides amylifera (43), and minabeolide-2, a sterol isolated from soft corals (Paraminabea) (44).
To our knowledge, there are no reports of Aspergillus fumigatus producing any of the compounds listed in Tables 1 and 2, except for fumigaclavine A and fumiquinazoline F. Therefore, further examination by comparison to a commercial standard and identification as demonstrated in reference 26 and characterization of a BGC as described in reference 27 are required. As a validation of our method, when confirmed metabolites from the direct method were cross-referenced to the predicted putative metabolites from the auxiliary method, we observed the upregulation of fumigaclavine A (XIC 63 and 124) and fumiquinazoline F (XIC 70 and 120) in most LCO-treated samples (Tables 1 and 2), which was comparable to our results in Fig. 2 and 3.
TABLE 2.
Putative metabolites produced at 37°C identified by the auxiliary method
| XIC ID | Regulation | Treatment | Log2 fold change | Putative metabolite ID | KEGG ID/Lipid Maps ID |
|---|---|---|---|---|---|
| 10 | Up | C16:0 sLCOs | 1.06 | 6′-Hydroxysiphonaxanthin decenoate | LMPR01070951 |
| 12 | Up | C16:0 sLCOs | 1.07 | Unknown | |
| 44 | Up | C16:0 nsLCOs | 1.82 | Burseran | C10547 |
| 44 | Up | C18:1 nsLCOs | 1.52 | Burseran | C10547 |
| 44 | Up | C18:1 sLCOs | 1.61 | Burseran | C10547 |
| 44 | Up | C16:0 sLCOs | 1.66 | Burseran | C10547 |
| 70 | Up | C18:1 sLCOs | 1.13 | 4′-O-desmethyl-3-O-acetylpapaveroxine | C21590 |
| 73 | Up | C18:1 sLCOs | 2.11 | Alprazolam | C06817 |
| 93 | Up | C18:1 nsLCOs | 1.15 | Unknown | |
| 93 | Up | C18:1 sLCOs | 1.03 | Unknown | |
| 114 | Up | C16:0 nsLCOs | 1.54 | Clofibrate | C06916 |
| 114 | Up | C18:1 nsLCOs | 1.27 | Clofibrate | C06916 |
| 114 | Up | C18:1 sLCOs | 2.36 | Clofibrate | C06916 |
| 119 | Up | C16:0 nsLCOs | 1.66 | Minabeolide-2 | LMST01160003 |
| 119 | Up | C18:1 nsLCOs | 1.70 | Minabeolide-2 | LMST01160003 |
| 119 | Up | C18:1 sLCOs | 1.71 | Minabeolide-2 | LMST01160003 |
| 119 | Up | C16:0 sLCOs | 1.09 | Minabeolide-2 | LMST01160003 |
| 120 | Up | C16:0 nsLCOs | 1.29 | Fumiquinazoline F | C22145 |
| 120 | Up | C18:1 nsLCOs | 1.04 | Fumiquinazoline F | C22145 |
| 120 | Up | C18:1 sLCOs | 1.42 | Fumiquinazoline F | C22145 |
| 120 | Up | C16:0 sLCOs | 1.03 | Fumiquinazoline F | C22145 |
| 121 | Up | C16:0 nsLCOs | 1.40 | Clofibrate | C06916 |
| 121 | Up | C18:1 sLCOs | 2.12 | Clofibrate | C06916 |
| 157 | Up | C16:0 nsLCOs | 1.48 | Alprazolam | C06817 |
| 157 | Up | C18:1 nsLCOs | 1.23 | Alprazolam | C06817 |
| 157 | Up | C18:1 sLCOs | 2.39 | Alprazolam | C06817 |
Finally, of all the XIC data with log2 fold changes greater than 1, only XICs 12 and 93 did not match any database or individually verified XICs. An unknown analyte (XIC 12) is induced by C16:0 sLCOs and has a log2 fold change of 1.07. Another unknown analyte (XIC 93) is induced by both C18:1 nsLCOs and C18:1 sLCOs with log2 fold changes of 1.15 and 1.03, respectively. Though no downregulation of XICs occurred with a log2 fold change greater than 1.0 in response to treatments at 37°C, the only significantly downregulated XIC within the entire set of extracted XICs is ID 155, with a log2 fold change of 0.92 elicited from the C16:0 sLCO treatment (Table S2).
LCO-induced fungal metabolites influence bacterial log-phase growth.
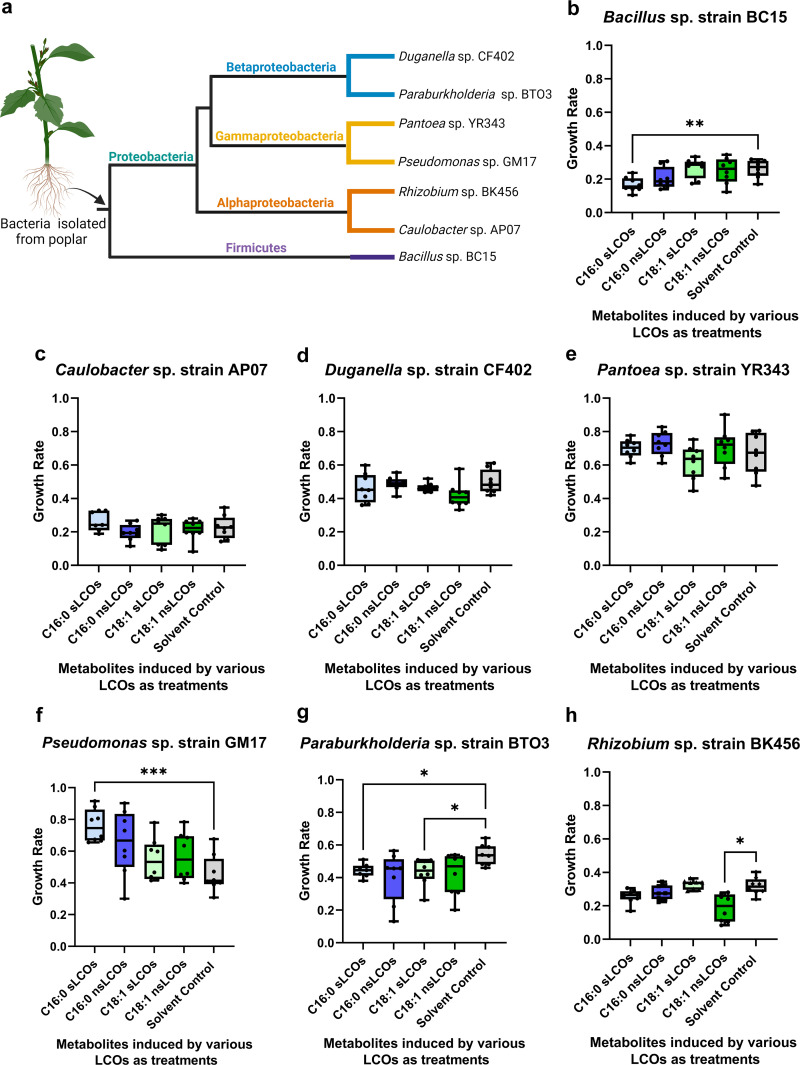
Recently, a molecular microbiome study showed that A. fumigatus and several plant growth-promoting bacteria were found in abundance on poplar wood chips (45). Since these organisms are found together on poplar and are known to be soilborne organisms, we speculate they might have interactions below ground, in soil or root systems. Therefore, we were interested in investigating if A. fumigatus exudates induced by LCOs would have an impact on bacteria isolated from poplar root systems. We used the crude exudates extracted at 25°C because they are better representatives of a forest-soil environment than those extracted at 37°C. Additionally, most of the known metabolites secreted at 25°C are known to be involved in the microbe-microbe competition (19). Bacterial strains isolated from the Populus native microbiome are known to be plant growth promoters and include the Gram-positive bacterium, Bacillus sp. strain BC15 and the Gram-negative bacteria Caulobacter sp. strain AP07, Duganella sp. strain CF402, Pantoea sp. strain YR343, Paraburkholderia sp. strain BTO3, and Pseudomonas sp. strain GM17 (28, 46, 47). Moreover, related bacterial strains are known to be affected by A. fumigatus metabolites (48–50). Additionally, since none of these genera are reported to produce LCOs, we included the soil bacterium Rhizobium sp. strain BK456, which harbors nif and nod genes (28) that are critical for LCO production, the initiation of rhizobia infection thread, and organogenesis in legumes (51). These bacteria represent four different taxonomic classes (Fig. 6a), which gives insight into how these fungal secreted metabolites might influence bacterial growth for future microcosm studies.
FIG 6.
Log-phase growth rates of bacterial species following exposure to LCO-induced secreted metabolites. (a) Poplar-promoting bacteria that were examined in this study and their taxonomic placement. Data are mean quantiles based on the inflection point of growth for each bacterial species. Statistical analysis was conducted with a Welch one-way ANOVA test and a Dunnett T3. (b to h) Welch’s ANOVA P values were (b) 0.0050, (c) not significant, (d) not significant, (e) not significant, (f) 0.0266, (g) 0.0011, and (f) 0.0013. *, P < 0.05; **, P < 0.01; ***, P < 0.001. The point of inflection for each bacterial strain can be found in Table S3. There were eight biological replications for each bacterial species. (Figure 6a created with www.BioRender.com.)
Bacterial point of inflection. Download Table S3, CSV file, 0.00 MB (280B, csv) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Bacterial growth rates varied significantly between the different LCO-induced fungal metabolic profile treatments. Crude extracts obtained from cultures of A. fumigatus in the presence of exogenous C16:0 sLCOs reduced the Bacillus sp. strain BC15 growth rate by 44.7% (Fig. 6b) and increased the Pseudomonas sp. strain GM17 growth rate by 49.5% (Fig. 6g). Crude extracts obtained from cultures with exogenous C16:0 nsLCOs reduced the growth rate of Paraburkholderia sp. strain BTO3 by 29.3% (Fig. 6f). In contrast, extracts obtained from cultures in the presence of C18:1 nsLCOs reduced the growth rate of Rhizobium sp. strain BK456 by 51.4% (Fig. 6h). Exogenous application of individual LCOs (Fig. 1a to d) did not influence bacterial growth rates (Fig. S6a to g), which indicates that the differential bacterial growth rates reported earlier were caused by metabolomic changes observed in the fungus as a response to LCOs rather than the LCO treatments themselves. The commercially available standard of helvolic acid decreased the growth rate of Bacillus sp. strain BC15 by 32.2%, whereas other commercially available secondary metabolite standards did not show any influence on the bacteria used in this study (Fig. S7). These results suggest that the crude extract’s effects on bacterial growth are caused either by synergistic interactions between specific metabolites or by uncharacterized metabolites.
Log phase growth rates of bacterial species following exposure to LCO standards. Data are mean quantiles based on the inflection point of growth for each bacterial species. Statistical analysis was conducted using Welch’s one-way ANOVA test and Dunnett’s T3. (a to g) LCO standards had no significant impact on bacterial growth rates compared to the solvent control. The point of inflection for each bacterial strain can be found in Table S3. There were eight biological replications for each bacterial species. Download FIG S6, TIF file, 14.3 MB (14.3MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Log-phase growth rates of bacterial species following exposure to secondary metabolite standards. Secondary metabolite standards were commercially purchased and dissolved in either solvent control 1 (100% ethanol), shown in gray, or solvent control 2 (100% methanol), shown in orange, according to the manufacturer’s instructions. Data are mean quantiles based on the inflection point of growth for each bacterial species. Statistical analysis was conducted with Welch’s one-way ANOVA test and Dunnett’s T3. The points of inflection can found in Table S3. *, P < 0.05; **, P < 0.01; ***, P < 0.001. N values represents the biological replications as follows: Bacillus sp. strain BC15 is eight, Caulobacter sp. strain APO7 is six, Duganella sp. strain CF402 is six, Pantoea sp. strain YR343 is four, Paraburkholderia sp. strain BTO3 is six, Pseudomonas sp. strain GM17 is eight, and Rhizobium sp. strain BK456 is four. (a) Helvolic acid has a negative significant impact on Bacillus sp. strain BC15 log-phase growth, the Welch’s ANOVA test P value is 0.0387. (b to g) Secondary metabolite standards had no significant impact (Welch’s ANOVA test P > 0.05) on bacterial growth compared to the solvent control. Caulobacter sp. strain AP07 could not grow in the presence of ethanol. Download FIG S7, TIF file, 14.3 MB (14.3MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
DISCUSSION
Research on LCOs has largely focused on the role of these compounds as signaling molecules that orchestrate associations between host plants and symbiotic microbes (11). Here, we present the first evidence of an alternative function for LCOs that involves transkingdom signaling between a fungus and multiple soilborne bacteria through the regulation of fungus-secreted metabolites with potential bioactivities. Pairing metabolomics with network and transcriptomic analyses highlights that several known and putative secondary metabolites have increased production by A. fumigatus in the presence of LCOs. We demonstrated that A. fumigatus had a higher production of known, putative, and unknown metabolites at 25°C than at 37°C despite having lower dry biomass, with fold changes ranging between 1.9 to 2.7 at this temperature (Fig. S1). Moreover, the induction of specific metabolites by various LCO treatments was temperature dependent (Fig. 2 and 5, Fig. S2 and S3). For example, the largest impact on feature production at 25°C was observed in response to C18:1 sLCO treatment, whereas at 37°C, the C16:0 sLCO treatment had the largest influence. In our previous study, C16:0 sLCOs were reported to also have an impact on physiology and transcriptomics at 37°C (5). Future investigations are needed to determine the influence C18:1 sLCOs have on the fungus at 25°C. Taken together, these findings show that sulfated LCOs have the largest impact on the metabolomics of A. fumigatus at both temperatures, but they might play pivotal roles in different biological systems. For example, C16:0 sLCOs seem to have a critical role in a clinical setting, as their effects were more pronounced at 37°C, whereas C18:1 sLCOs might potentially have an impact on the organization of a soil microbiome. Although our study focuses on the roles of LCOs in a poplar rhizosphere-soil microbiome setting, this experimental design can be applied to gain a better understanding of the role of these molecules in a clinical setting when A. fumigatus coinfects with other opportunistic human pathogens such as Pseudomonas aeruginosa or Staphylococcus aureus. Along the same line, Rush et al. (5) showed that the human pathogen Candida albicans produces LCOs, whereas Candida glabrata does not. Both pathogens were shown to coinfect patients, causing oropharyngeal candidiasis (52). Also, C. glabrata was once believed to be the only species of Candida that does not form pseudohyphae, which are required for infection (53). Therefore, it will be engaging in future studies to explore how LCOs might be involved in the interactions between these two organisms.
Although we were not able to pinpoint a specific metabolite responsible for regulating bacterial growth rate in our study, we did observe the consistent induction of fumagillin, fumigaclavine A, fumiquinazoline A, helvolic acid, and pyripyropene A production in our treated samples. The commercially available standards of those metabolites did not influence bacterial growth rate, except for helvolic acid, which decreased the growth rate of Bacillus sp. strain BC15 (Fig. S7). The metabolites induced by the C16:0 sLCO treatment had the same effect on Bacillus sp. strain BC15. However, they were not shown to induce the production of helvolic acid (Fig. 2 and 3). Therefore, we believe the reduction of Bacillus growth rate may reflect the synergistic activity of more than one secondary metabolite. While synergistic interactions are rarely studied (54), there is one example where (55) showed that aflatoxin is more toxic to insects in the presence of kojic acid. Another key observation is that the fungal extracts impacted growth across the Bacteria kingdom and did not show any specificity to a bacterial taxon. This result indicates that the bacterial response is not conserved within a class. These findings could be used to understand how LCOs organize a plant microbiome, which is critical, as it was shown that the microbiome has an influence on the host fitness (56). Another aspect we are considering for future experiments is to identify and knock out genes responsible for LCO production in A. fumigatus. Although we observed that different types of LCOs resulted in different metabolomic profiles, without a mutant lacking LCO production, we cannot exclude the effect of LCOs produced by A. fumigatus in our system.
Another critical point of this study was the elucidation of unknown XICs with potential biological activities using various LCO treatments. Here, we identified multiple unknown XICs, through network analysis, that were differentially induced by LCO treatments at different temperatures. Interestingly, there were more highly expressed XICs produced at 25°C than at 37°C (Tables 1 and 2). In terms of validation of the identity of the potential metabolites identified through the auxiliary route, the KEGG results showed that fumigaclavine A and fumiquinazoline F were upregulated in all treatments between the network analysis, which aligns well with our LC/MS/MS data. This indicates that the auxiliary route can be a good approach to predict uncharacterized metabolites in A. fumigatus, such as alprazolam, burseran, clofibrate, ergometrine, epithienamycin B, fraxetin, and stearidonic acid. This list includes several other metabolites as well that were potentially identified but with a log2 fold change (<1.0) that did not meet our cutoff threshold (Tables S1 and 2). These results are exciting considering how challenging it can be to activate BGC expression under standard laboratory conditions. Therefore, our future efforts will be directed toward characterizing some of these unknown XICs through RNA sequencing experiments and overexpression of their putative backbone genes.
Finally, LCOs with concentrations ranging from micromolar to picomolar have been shown to have multiple effects on fungi, plants, rhizobial bacteria, and mammalian cells. LCOs were associated with transcriptomic and physiologic changes in plants and fungi (5, 6, 11, 57). Moreover, these molecules allowed rhizobia to start the infection thread process leading to organogenesis (11) or an increased number of nodules with a limited population (58). LCOs have also been shown to help mycorrhizal fungi colonize their host (7, 13, 59). Lastly, LCOs have been shown to modulate the development of mammalian endothelial cells in rat aorta biological assays and affect the endothelial cells’ adhesion to extracellular matrix components (60). Given the wide-ranging impacts of LCO on diverse organisms, we were surprised that LCO treatments had no effect on the growth of the bacterial species examined here. Possibly, LCOs are signaling molecules exclusively perceived by eukaryotes, although more evidence is needed to address this speculation, as we only investigated LCOs’ impact on bacterial growth and no other phenotypes.
MATERIALS AND METHODS
LCO standards.
Standard LCO compounds (C16:0 sLCOs, C16:0 nsLCOs, sC18:1 LCOs, and nsC18:1 LCOs) were synthesized at CERMAV (Grenoble, France) and used at 10−8 M in 0.005% ethanol (EtOH)/water as previously described (5, 6). The chemical structures are shown in Fig. 1a to d. The LCOs used in these experiments are the commonly found LCO types from fungi (5).
Experiments conducted with Aspergillus fumigatus.
(i) Organism and inoculum. Aspergillus fumigatus strain Af293 (61) was used in this study. The growth medium used was liquid glucose minimal medium (GMM) without 1% thiamine (62). GMM was supplemented with LCOs dissolved in EtOH for a final LCO concentration of 10−8 M, which has been shown to be biologically functional for changes in plants or fungal physiology (5). The negative control was supplemented with EtOH for a final concentration of 0.005%. Streak GMM plates of A. fumigatus were grown at 37°C for 4 days. Afterward, 0.01% Tween 80 solution was added to the plates, spores were harvested with a sterile L-shaped hockey-stick cell spreader, and spore suspension was collected with a 25-mL serological pipette. The spore concentration was adjusted to 106 spores in the GMM broth medium. Inoculated Aspergillus fumigatus spores were grown at 25°C for 6 days and at 37°C for 4 days at 250 rpm in a New Brunswick Excella E25 incubator shaker. Fungal exudates were filtered through a 0.45-μm Durapore polyvinylidene fluoride membrane filter (Millipore Sigma, item number SE1M003M00). After the incubation periods, fungal biomass was collected and lyophilized to estimate the dry biomass (Fig. S1). Statistical analyses were performed using Prism software version 9.0.0 (GraphPad, San Diego, CA). A one-way analysis of variance (ANOVA) was performed per treatment. Then Dunnett’s test was performed per species to test treatment group responses against solvent control responses. The error bars in all figures indicate the standard error of the mean. To determine the relationship of metabolites exuded between 25°C and 37°C treated samples, significant analytes were used for principal-component analysis (PCA) using the R package ggfortify (Fig. S3).
Liquid chromatography-mass spectrometry (LC/MS) of culture filtered fungal exudates identified and quantified metabolites produced in response to different treatments were as provided in the materials and methods of reference 26.
(ii) Profiling putative metabolites using ultra-high-pressure liquid chromatography-mass spectrometry (UHPLC-MS). The effect of LCOs on secondary metabolite production by A. fumigatus strain Af293 was assessed by UHPLC-MS analysis. About 106 fresh spores were grown in 125-mL flasks containing 50 mL of liquid GMM supplemented with the LCO treatments mentioned above. Two different growth conditions were assessed; the first consisted of incubation at 25°C and 250 rpm for 6 days, whereas the second consisted of incubation at 37°C and 250 rpm for 4 days. Differences in growth period were necessary to standardize the total amount of growth. For secondary metabolite analysis, 3 mL of supernatant was homogenized with 3 mL of ethyl acetate (Millipore Sigma, item number 270989). Organic and aqueous layers were separated by centrifugation at 3,000 rpm for 5 min, and the organic layer was collected and air dried. Samples were later resuspended in acetonitrile:water (50:50 vol/vol) and filtered through an Acrodisc syringe filter with a nylon membrane (0.45 μm, Pall Corporation) into 1-mL HPLC vials. Samples were subjected to high-resolution UHPLC-MS analysis performed on a Thermo Scientific Vanquish UHPLC system connected to a Thermo Scientific Q Exactive Orbitrap operated in electrospray positive-ionization mode (63). Data acquisition and processing for the UHPLC-MS were done using Thermo Scientific Xcalibur software version 4.2.47. Files were converted to .mzXML using MassMatrix MS data file conversion grouped by condition and run in MAVEN, an open-source software program (29), and the XCMS open-source package (https://xcmsonline.scripps.edu/) using a pairwise comparison between treatments and controls with the parameter UPLC/Q-Exactive 3110 (30). Volcano plots were based on features similar or uniquely produced by each treatment as determined by XCMS and illustrated in Prism software version 9.4.1 (681) (GraphPad, San Diego, CA). Metabolic profiling is in reference to the fungus, and not the bacteria examined in this study.
(iii) Confirmation of putative metabolites by targeted LC-MS/MS analysis. To confirm the identities of several putative metabolites detected by UHPLC-MS analysis as described above, commercial standards (see “Group 3,” below) were procured and analyzed by UHPLC-MS/MS both independently to glean confirmatory fragmentation spectra and when spiked into fungal extracts to confirm retention times through putative metabolite chromatographic peak augmentation. Commercial standards/putative metabolites were targeted by high-resolution parallel reaction monitoring (PRM) using a Vanquish UHPLC plumbed directly to a Q Exactive Plus mass spectrometer (Thermo Scientific) outfitted with a 75-μm-inside diameter (i.d.) nanospray emitter packed with 15-cm Kinetex C18 resin (1.7 μm particle size; Phenomenex). Mobile phases included solvent A (95% H2O, 5% acetonitrile, 0.1% formic acid) and solvent B (5% H2O, 90% acetonitrile, 5% isopropanol, 0.1% formic acid). Nanoliter flow rates were achieved by UHPLC split-flow and measured at 300 nL/min at the nanospray emitter. Then, 10 μL of each sample was auto-injected prior to the split, leading to the separation and analysis of 20 nL of extract over a 40- min chromatographic method: 17% to 100% B over 20 min; hold at 100% B for 2 min; 100% to 17% B over 3 min; hold at 17% B for 15 min to reequilibrate the column. Putative metabolites/commercial standards were targeted for PRM analysis in positive-ion mode with a duty cycle that included a full scan (270 to 600 m/z range; resolution 70,000; 3 microscan spectrum averaging), followed by a PRM scan targeting each metabolite/standard (resolution, 17,500; isolation window, 1.0 m/z; normalized collision energy in the higher-energy collisional dissociation (HCD) cell was stepped: 30, 35, and 40). The resulting data were analyzed with Skyline (64) to confirm the identities of each putative analyte (26) identified in the preceding UHPLC-MS analysis.
Assessment of LCO-induced fungal metabolic profile on bacterial species.
(i) Organisms and inoculum. The described Gram-positive and Gram-negative bacterial members of the Populus spp. microbial communities were used and previously identified and characterized by Henning et al. (46), Carper et al. (28), and Wang et al. (47). The strains used were the Gram-positive bacterium Bacillus sp. strain BC15 and the Gram-negative bacteria Caulobacter sp. strain AP07, Duganella sp. strain CF402, Pantoea sp. strain YR343, Paraburkholderia sp. strain BTO3, Pseudomonas sp. strain GM17, and Rhizobium sp. strain BK456. The growth medium used was Reasoner’s 2A (R2A) broth mix from Teknova, Inc., (Hollister, CA, USA), prepared according to the manufacturer’s instructions. The cell concentration was adjusted to a 0.01 optical density at 600 nm (OD600).
(ii) Metabolite extractions. Crude fungal extracts. A 6-mL sample of filtered fungal extracts was vortexed with an equal volume of ethyl acetate for secondary metabolite analysis for 10 min. Organic and aqueous layers were separated by centrifugation at 3,000 rpm for 5 min, and the organic layer was collected, dried, and stored at 4°C. Samples were resuspended in acetonitrile:water (50:50 vol/vol) and filtered through a 0.45-μm Durapore polyvinylidene fluoride membrane filter (Millipore Sigma, item number SE1M003M00) into sterile 10-mL glass scintillation vials and stored at 4°C.
(iii) Treatment groups and application on individual bacterial species. Three groups of treatments were assessed for their effect on bacterial growth.
Group 1 was the effect of crude extracts on bacterial growth (as mentioned in “Crude Fungal Extracts,” above).
Group 2 was the effect of exogenous LCO on bacterial growth (as mentioned in the “LCO Standards,” above).
Group 3 was the effect of exogenous secondary metabolite standards on bacterial growth in response. Fumagillin (CAS number 23110-15-8), fumitremorgin C (CAS number 118974-02-0), helvolic acid (CAS registry number 29400-42-8), pseurotin A (CAS number 58523-30-1), and pyripyropene A (CAS number 147444-03-9) were purchased from Cayman Chemicals, Inc. (Ann Arbor, MI, USA). Fumigaclavine A (CAS number 6879-59-0) and trypacidin (CAS number 1900-29-4) were purchased from AdipoGen Life Sciences (Waltham, MA, USA). Gliotoxin (CAS number 67–99-2) was purchased from Enzo Life Sciences, Inc. (Farmingdale, NY, USA). To obtain solubility, the following metabolites were dissolved in the corresponding solvents as described by the manufacturer’s instructions, except for gliotoxin: fumagillin in ethanol at a concentration of 0.25 mg/mL, fumigaclavine A in methanol at a concentration of 1 mg/mL, fumitremorgin C in methanol at a concentration of 1 mg/mL, gliotoxin in ethanol at a concentration of 10 mg/mL, helvolic acid in ethanol at a concentration of 20 mg/mL, pseurotin A in methanol at a concentration of 1 mg/mL, pyripyropene A in methanol at a concentration of 1 mg/mL, and trypacidin in acetone at a concentration 1 mg/mL. Our gliotoxin sample did not appear to be soluble in methanol; however, it was soluble in ethanol following sonication. Trypacidin was not used in the assay due to its insolubility in R2A broth. Fumitremorgin C was not used in the bacterial assay due to the limited amount of compound available. However, trypacidin and fumitremorgin C were used in the confirmation of metabolites in our LC/MS samples (26). All solutions were placed in a sterile 25-mL or 50-mL class A glass medium bottle sonicated in a water bath for 15 min at 25°C before use. Once in solution, fumagillin, fumigaclavine A, gliotoxin, helvolic acid, pseurotin A, pyripyropene A, and trypacidin were stored at −20°C, and fumitremorgin C was kept at 4°C. The bottles were wrapped with aluminum to provide long-term protection from light.
(iv) 96-Well plate experiments with oCelloScope and bacterial communities. To assess the growth of bacterial species, oCelloScope version 9.0 (BioSense Solutions, Inc.) was used. In preparation for the experiment, bacterial species were grown overnight at 25°C at 250 rpm in 125-mL flasks with 50 mL of liquid R2A. Bacterial growth was measured with a spectrometer to obtain an initial absorbance reading and then was used to calculate an optical density (OD) of 0.01. Bacterial samples were mixed with their treatments and added to a flat-bottom 96-well plate (Corning, Inc., item number 351172). The total volume in each well with R2A, treatment, and bacterial suspension was 250 μL. Bacteria were treated with 10−8 M LCOs (final concentration), 30 μg/mL of secondary metabolite standards, or 150 μg of crude fungal extracts. Antimicrobial or insecticidal MICs have been reported for fumagillin as low as 5 μg/mL (65–67), fumigaclavines at 7.81 μg/mL (68), gliotoxin up to 8 μg/mL (69), helvolic acid up to 16 μg/mL (70), pseurotin A up to 64 μg/mL (49), and pyripyropene A up to 100 ppm (71). Pseurotin A was not used up to the suggested 64 μg/mL because of the limited amount of compound available. Bacteria were grown under treatment conditions at 25°C for 24 h. Bacterial growth was determined with four biological replications and two technical replications each time, and experiments were repeated twice. Experiments where bacteria were treated with secondary metabolite standards consisted of only three biological replications per trial. Bacterial growth was determined by normalized segmentation and extraction of surface area (SESA), which is an algorithm that measures microbial growth with high sensitivity based on segmentation and contrast-based identification of all objects that are in a scan area yet not affected by changes in illumination or condensation (oCelloScope User Manual version 9.0, BioSense, Inc.). The SESA-normalized log phase of bacteria species was used for the analysis.
(v) Calculating the inflection point of growth and determining the log phase. All bacterial strains examined were grown for 24 h to find the inflection point of growth with the SESA-normalized algorithm with the oCelloScope. There were eight biological replications. The inflection point of growth was calculated in R version 4.02 (72) with the package Growthcurver version 0.3.1 (73). Table S3 lists the point of inflection for each bacterial strain.
Statistical analysis.
Statistical analyses were performed using Prism software version 9.4.1 (681) (GraphPad, San Diego, CA). Outliers were removed by the robust regression and outlier removal (ROUT) method, which is based on robust nonlinear regression and false-discovery rate, where Q is the maximum false-discovery rate, set at 1% (74). Additional outliers were removed if a biological replication had a technical replication with all negative values caused by analysis with an out-of-focus camera. Individual negative values within a technical data set were eliminated from the analysis. A Bartlett test was performed for the equality of variances across treatments per bacterial species. Next, a one-way ANOVA was performed per species, implementing either (i) a simple F test for the equality of means in a one-way ANOVA or (ii) an approximate method of Welch, which generalizes the two-sample Welch test to the case of many samples. Implementation of either option depended on whether Bartlett test results suggested equal variance (ii) or not (ii) among treatments. Finally, Dunnett’s test was performed per species to test treatment group responses against solvent control responses. The error bars in all figures indicate the standard error of the mean. R version 4.0.2 (2020-06-22) with library(ggplot2) and library(dplyr) was used.
Network analysis.
A more formal and elaborate description of the network analysis is available in reference 26. The following discussion is a high-level explanation of the methodology. The interactions between elements in a system can be represented by a network (or graph), mathematically denoted by G(V, E, W) The network comprises vertices (or nodes), V, which represent the elements in the system, and in the current study, they are the treatments and analytes (known or putative metabolites and unknown features) at a particular temperature. The nodes are connected to each other using edges, E, which represent the interactions between the nodes. In the current study, these edges denote the up- or downregulations triggered by a treatment on an analyte. Thus, the edges of the network in the current study are weighted with weights, W, rather than binary connections. We use the magnitude of the log2 fold value change caused by a treatment on an analyte to represent the edges (weighted edges).
The resulting network represents the influence of the treatments on triggering the production of known or putative metabolites and unknown features in A. fumigatus. Building a network representation of a system primarily allows us to rank the nodes of the network based on their ability to influence each other. We mainly use two measures to rank the nodes in the current network: (i) node strength and (ii) PageRank. The network ranking measures of the treatments quantify the ability of the treatments to influence the production of the analytes, whereas for the analytes, the measures quantify their ability to be receptive to various treatments.
Node strength is simply the sum of all the edges from (or to) a particular node. In the current analysis, the node strength of a treatment is the total amount of regulation triggered by the treatment, which is formally called the out-strength of the node, as the edges are from the node to the others. The node strength of the analytes is the total amount by which the analyte is regulated by all the treatments, formally called the in-strength of a node.
While node strength can provide a ranking of the treatments and analytes, the number of edges from/to a node (treatment or analyte) is significant to understand the importance of the node, which is not considered for computing the node strength. The number of analytes influenced by a treatment can show how diverse the effect of a treatment is. Similarly, the number of treatments affecting an analyte denotes how easily it is influenced by the chemical signals. Another criterion to understand the importance of a node is to quantify the relative importance of one node to another node to which the first one is connected. If a treatment is connected to an analyte that is influenced by many other treatments, this emphasizes the ability of the analyte to be influenced and not the unique ability of the treatment. Such factors are not highlighted by the node strength measure.
The PageRank considers the above-mentioned factors to rank the nodes in a network. PageRank is the underlying method used by the Google search engine to rank a webpage based on the number of directed links among the webpages, also considering the popularity of the webpages being linked to and from the webpage. For the treatments, the ability to be influential is measured by the broadcasting version of the PageRank measure, whereas the ability to be receptive to treatments for the analytes is denoted by the receiving version of the PageRank measure.
Furthermore, we classify the network analysis into two based on the analytes being considered. When the network is made using the treatments and the known and putative metabolites, we call the analysis the direct network analysis. The analysis is called the auxiliary network analysis when the nodes comprise the treatments and the unknown features. The auxiliary network analysis requires further data processing involving KEGG, MZmine, and Lipid Maps to recover the unknown features, which are elaborated in reference 26.
Code availability.
The algorithms and source codes used for the network analyses are available through reference 26.
Data availability.
LC/MS data are available through the MassIVE repository (https://massive.ucsd.edu/ProteoSAFe/static/massive.jsp; Data set ID MSV000090566).
ACKNOWLEDGMENTS
We thank Olivia Shafer for suggestions and manuscript proofreading.
This research was funded by the Genomic System Sciences Program, U.S. Department of Energy, Office of Science, Biological and Environmental Research, as part of the Plant-Microbe Interfaces Scientific Focus Area at Oak Ridge National Laboratory (ORNL; http://pmi.ornl.gov; ORNL is managed by UT-Battelle, LLC, for the U.S. Department of Energy under contract DEAC05-00OR22725). To perform the network-theoretic analysis, M.G.M. used the resources of the Oak Ridge Leadership Computing Facility at ORNL, which is supported by the DOE Office of Science under contract number DE-AC05-00OR22725.
S.F. and S.C. acknowledge the French National Research Agency (ANR) through the Labex ARCANE and CBH-EUR-GS (ANR-17-EURE-0003), Glyco@Alps (ANR-15-IDEX-02), and Carnot PolyNat (CARN-025-01) programs for its support and NanoBio ICMG (UAR 2607) for providing facilities for mass spectrometry (A. Durand, L. Fort, R. Gueret) and NMR analyses (I. Jeacomine).
T.A.R. and J.T. initiated and designed the project. T.A.R., J.T., A.A.C., J.J.G., N.P.K., D.A.P., and J.L.L. designed and implemented the experiments with Aspergillus fumigatus and bacterial species. S.C., S.F., and J.-M.A. provided LCO molecules. M.G.M., M.J.L., D.K., and D.A.J. conducted the network analysis. P.E.A., R.J.G., J.T., and T.A.R. conducted and analyzed the mass spectrometry data sets. T.A.R., J.T., M.J.L., M.G.M., A.A.C., J.J.G., M.T.D., P.E.A., R.J.G., and J.L.L. wrote the manuscript with feedback from all the coauthors.
We declare no competing interests.
Contributor Information
Tomás A. Rush, Email: rushta@ornl.gov.
Jesse L. Labbé, Email: jlabbe@invaio.com.
Christopher R. Anderton, Pacific Northwest National Laboratory
REFERENCES
- 1.Keller NP. 2019. Fungal secondary metabolism: regulation, function and drug discovery. Nat Rev Microbiol 17:167–180. doi: 10.1038/s41579-018-0121-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atanasov AG, Zotchev SB, Dirsch VM, Supuran CT, International Natural Product Sciences Taskforce . 2021. Natural products in drug discovery: advances and opportunities. Nature Rev Drug Discovery 20:200–216. doi: 10.1038/s41573-020-00114-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Netzker T, Fischer J, Weber J, Mattern DJ, König CC, Valiante V, Schroeckh V, Brakhage AA. 2015. Microbial communication leading to the activation of silent fungal secondary metabolite gene clusters. Front Microbiol 6:299. doi: 10.3389/fmicb.2015.00299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lerouge P, Roche P, Faucher C, Maillet F, Truchet G, Promé JC, Dénarié J. 1990. Symbiotic host-specificity of Rhizobium meliloti is determined by a sulphated and acylated glucosamine oligosaccharide signal. Nature 344:781–784. doi: 10.1038/344781a0. [DOI] [PubMed] [Google Scholar]
- 5.Rush TA, Puech-Pagès V, Bascaules A, Jargeat P, Maillet F, Haouy A, Maës AQ, Carriel CC, Khokhani D, Keller-Pearson M, Tannous J, Cope KR, Garcia K, Maeda J, Johnson C, Kleven B, Choudhury QJ, Labbé J, Swift C, O’Malley MA, Bok JW, Cottaz S, Fort S, Poinsot V, Sussman MR, Lefort C, Nett J, Keller NP, Bécard G, Ané J-M. 2020. Lipo-chitooligosaccharides as regulatory signals of fungal growth and development. Nat Commun 11:3897. doi: 10.1038/s41467-020-17615-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Villalobos Solis MI, Engle NL, Spangler MK, Cottaz S, Fort S, Maeda J, Ané J-M, Tschaplinski TJ, Labbé JL, Hettich RL, Abraham PE, Rush TA. 2022. Expanding the biological role of lipo-chitooligosaccharides and chitooligosaccharides in Laccaria bicolor growth and development. Front Fungal Biol 3. doi: 10.3389/ffunb.2022.808578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cope KR, Bascaules A, Irving TB, Venkateshwaran M, Maeda J, Garcia K, Rush TA, Ma C, Labbé J, Jawdy S, Steigerwald E, Setzke J, Fung E, Schnell KG, Wang Y, Schlief N, Bücking H, Strauss SH, Maillet F, Jargeat P, Bécard G, Puech-Pagès V, Ané J-M. 2019. The ectomycorrhizal fungus Laccaria bicolor produces lipochitooligosaccharides and uses the common symbiosis pathway to colonize populus roots. Plant Cell 31:2386–2410. doi: 10.1105/tpc.18.00676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dénarié J, Debellé F, Promé JC. 1996. Rhizobium lipo-chitooligosaccharide nodulation factors: Signaling molecules mediating recognition and morphogenesis. Annu Rev Biochem 65:503–535. doi: 10.1146/annurev.bi.65.070196.002443. [DOI] [PubMed] [Google Scholar]
- 9.Genre A, Chabaud M, Balzergue C, Puech-Pagès V, Novero M, Rey T, Fournier J, Rochange S, Bécard G, Bonfante P, Barker DG. 2013. Short-chain chitin oligomers from arbuscular mycorrhizal fungi trigger nuclear Ca2+ spiking in Medicago truncatula roots and their production is enhanced by strigolactone. New Phytol 198:190–202. doi: 10.1111/nph.12146. [DOI] [PubMed] [Google Scholar]
- 10.Girardin A, Wang T, Ding Y, Keller J, Buendia L, Gaston M, Ribeyre C, Gasciolli V, Auriac M-C, Vernié T, Bendahmane A, Ried MK, Parniske M, Morel P, Vandenbussche M, Schorderet M, Reinhardt D, Delaux P-M, Bono J-J, Lefebvre B. 2019. LCO receptors involved in arbuscular mycorrhiza are functional for rhizobia perception in legumes. Curr Biol 29:4249–4259.e5. doi: 10.1016/j.cub.2019.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khokhani D, Carrera Carriel C, Vayla S, Irving TB, Stonoha-Arther C, Keller NP, Ané J-M. 2021. Deciphering the chitin code in plant symbiosis, defense, and microbial networks. Annu Rev Microbiol 75:583–607. doi: 10.1146/annurev-micro-051921-114809. [DOI] [PubMed] [Google Scholar]
- 12.Liang Y, Tóth K, Cao Y, Tanaka K, Espinoza C, Stacey G. 2014. Lipochitooligosaccharide recognition: an ancient story. New Phytol 204:289–296. doi: 10.1111/nph.12898. [DOI] [PubMed] [Google Scholar]
- 13.Maillet F, Poinsot V, André O, Puech-Pagès V, Haouy A, Gueunier M, Cromer L, Giraudet D, Formey D, Niebel A, Martinez EA, Driguez H, Bécard G, Dénarié J. 2011. Fungal lipochitooligosaccharide symbiotic signals in arbuscular mycorrhiza. Nature 469:58–63. doi: 10.1038/nature09622. [DOI] [PubMed] [Google Scholar]
- 14.Marburger DA, Willbur JF, Weber ME, Ané J-M, Kabbage M, Conley SP, Smith DL. 2018. Characterizing the effect of foliar lipo-chitooligosaccharide application on sudden death syndrome and sclerotinia stem rot in soybean. Plant Heal Prog 19:46–53. doi: 10.1094/PHP-10-17-0058-RS. [DOI] [Google Scholar]
- 15.Rey T, André O, Nars A, Dumas B, Gough C, Bottin A, Jacquet C. 2019. Lipo-chitooligosaccharide signalling blocks a rapid pathogen-induced ROS burst without impeding immunity. New Phytol 221:743–749. doi: 10.1111/nph.15574. [DOI] [PubMed] [Google Scholar]
- 16.Bok JW, Keller NP. 2004. LaeA, a regulator of secondary metabolism in Aspergillus spp. Eukaryot Cell 3:527–535. doi: 10.1128/EC.3.2.527-535.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frisvad JC, Rank C, Nielsen KF, Larsen TO. 2009. Metabolomics of Aspergillus fumigatus. Med Mycol 47:S53–S71. doi: 10.1080/13693780802307720. [DOI] [PubMed] [Google Scholar]
- 18.Keller N, Bok J, Chung D, Perrin RM, Keats Shwab E. 2006. LaeA, a global regulator of Aspergillus toxins. Med Mycol 44:83–85. doi: 10.1080/13693780600835773. [DOI] [PubMed] [Google Scholar]
- 19.Raffa N, Keller NP. 2019. A call to arms: mustering secondary metabolites for success and survival of an opportunistic pathogen. PLoS Pathog 15:e1007606. doi: 10.1371/journal.ppat.1007606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Romsdahl J, Wang CCC. 2019. Recent advances in the genome mining of Aspergillus secondary metabolites (covering 2012–2018). Medchemcomm 10:840–866. doi: 10.1039/c9md00054b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Van De Veerdonk FL, Gresnigt MS, Romani L, Netea MG, Latgé JP. 2017. Aspergillus fumigatus morphology and dynamic host interactions. Nat Rev Microbiol 15:661–674. doi: 10.1038/nrmicro.2017.90. [DOI] [PubMed] [Google Scholar]
- 22.Berthier E, Lim FY, Deng Q, Guo C-J, Kontoyiannis DP, Wang CCC, Rindy J, Beebe DJ, Huttenlocher A, Keller NP. 2013. Low-volume toolbox for the discovery of immunosuppressive fungal secondary metabolites. PLoS Pathog 9:e1003289. doi: 10.1371/journal.ppat.1003289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hagiwara D, Sakai K, Suzuki S, Umemura M, Nogawa T, Kato N, Osada H, Watanabe A, Kawamoto S, Gonoi T, Kamei K. 2017. Temperature during conidiation affects stress tolerance, pigmentation, and trypacidin accumulation in the conidia of the airborne pathogen Aspergillus fumigatus. PLoS One 12:e0177050. doi: 10.1371/journal.pone.0177050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kosalec I, Pepljnjak S, Jandrlić M. 2005. Influence of media and temperature on gliotoxin production in Aspergillus fumigatus strains. Arh Hig Rada Toksikol 3:269–273. [PubMed] [Google Scholar]
- 25.Lind AL, Smith TD, Saterlee T, Calvo AM, Rokas A. 2016. Regulation of secondary metabolism by the velvet complex is temperature-responsive in Aspergillus. G3 (Bethesda) 6:4023–4033. doi: 10.1534/g3.116.033084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gopalakrishnan Meena M, Lane MJ, Tannous J, Carrell AA, Abraham PE, Giannone RJ, Ané J-M, Keller NP, Labbé JL, Kainer D, Jacobson DA, Rush TA. 2022. A guidance into the fungal metabolomic abyss: network analysis for revealing relationships between exogenous compounds and their outputs. bioRxiv. doi: 10.1101/2022.08.11.503656. [DOI] [PMC free article] [PubMed]
- 27.Rush TA, Shrestha HK, Gopalakrishnan Meena M, Spangler MK, Ellis JC, Labbé JL, Abraham PE. 2021. Bioprospecting Trichoderma: a systematic roadmap to screen genomes and natural products for biocontrol applications. Front Fungal Biol 2. doi: 10.3389/ffunb.2021.716511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carper DL, Weston DJ, Barde A, Timm CM, Lu T-Y, Burdick LH, Jawdy SS, Klingeman DM, Robeson MS, Veach AM, Cregger MA, Kalluri UC, Schadt CW, Podar M, Doktycz MJ, Pelletier DA. 2021. Cultivating the bacterial microbiota of Populus roots. mSystems 6:e01306-20. doi: 10.1128/mSystems.01306-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clasquin MF, Melamud E, Rabinowitz JD. 2012. LC-MS data processing with MAVEN: A metabolomic analysis and visualization engine. Curr Protoc Bioinforma 37:14.11. doi: 10.1002/0471250953.bi1411s37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tautenhahn R, Patti GJ, Rinehart D, Siuzdak G. 2012. XCMS online: a web-based platform to process untargeted metabolomic data. Anal Chem 84:5035–5039. doi: 10.1021/ac300698c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wiemann P, Guo C-J, Palmer JM, Sekonyela R, Wang CCC, Keller NP. 2013. Prototype of an intertwined secondary-metabolite supercluster. Proc Natl Acad Sci USA 110:17065–17070. doi: 10.1073/pnas.1313258110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Calvo AM, Wilson RA, Bok JW, Keller NP. 2002. Relationship between secondary metabolism and fungal development. Microbiol Mol Biol Rev 66:447–459. doi: 10.1128/MMBR.66.3.447-459.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schimmel TG, Coffman AD, Parsons SJ. 1998. Effect of butyrolactone I on the producing fungus, Aspergillus terreus. Appl Environ Microbiol 64:3707–3712. doi: 10.1128/AEM.64.10.3707-3712.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Page L, Brin S. 1998. The anatomy of a large-scale hypertextual Web search engine. Comput Networks 30:107–117. doi: 10.1016/S0169-7552(98)00110-X. [DOI] [Google Scholar]
- 35.Spencer SPE, Lowe SA. 2019. Ergometrine for postpartum hemorrhage and associated myocardial ischemia: two case reports and a review of the literature. Clin Case Rep 7:2433–2442. doi: 10.1002/ccr3.2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Panaccione DG, Coyle CM. 2005. Abundant respirable ergot alkaloids from the common airborne fungus Aspergillus fumigatus. Appl Environ Microbiol 71:3106–3111. doi: 10.1128/AEM.71.6.3106-3111.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Box SJ, Hood JD, Spear SR. 1979. Four further antibiotics related to olivanic acid produced by Streptomyces olivaceus: fermentation, isolation, characterisation and biosynthetic studies. J Antibiot (Tokyo) 32:1239–1247. doi: 10.7164/antibiotics.32.1239. [DOI] [PubMed] [Google Scholar]
- 38.Pousset J-L, Kerharo J, Maynart G, Monseur X, Cavé A, Goutarel R. 1973. La borrerine: nouvel alcaloïde isolé du Borreria verticillata. Phytochemistry 12:2308–2310. doi: 10.1016/0031-9422(73)85148-9. [DOI] [Google Scholar]
- 39.Zihad SMNK, Hasan MT, Sultana MS, Nath S, Nahar L, Rashid MA, Uddin SJ, Sarker SD, Shilpi JA. 2022. Isolation and characterization of antibacterial compounds from Aspergillus fumigatus: an endophytic fungus from a mangrove plant of the Sundarbans. Evid Based Complement Alternat Med 2022:9600079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baccile JA, Spraker JE, Le HH, Brandenburger E, Gomez C, Bok JW, Macheleidt J, Brakhage AA, Hoffmeister D, Keller NP, Schroeder FC. 2016. Plant-like biosynthesis of isoquinoline alkaloids in Aspergillus fumigatus. Nat Chem Biol 12:419–424. doi: 10.1038/nchembio.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marcotullio MC, Curini M, Becerra JX. 2018. An ethnopharmacological, phytochemical and pharmacological review on lignans from Mexican Bursera spp. Molecules 23:1976. doi: 10.3390/molecules23081976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Seenivasan A, Subhagar S, Aravindan R, Viruthagiri T. 2008. Microbial production and biomedical applications of lovastatin. Indian J Pharm Sci 70:701–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Egeland ES, Guillard RRL, Liaaen-Jensen S. 1997. Additional carotenoid prototype respresentatives and a general chemosystematic evaluation of carotenoids in prasinophyceae (chlorophyta). Phytochemistry 44:1087–1097. doi: 10.1016/S0031-9422(96)00650-4. [DOI] [Google Scholar]
- 44.Ksebati MB, Schmitz FJ. 1988. Minabeolides: a group of with anolides from soft coral, Minabea sp. J Org Chem 53:3926–3929. doi: 10.1021/jo00252a008. [DOI] [Google Scholar]
- 45.Zöhrer J, Probst M, Dumfort S, Lenz H, Pecenka R, Insam H, Ascher-Jenull J. 2021. Molecular monitoring of the poplar wood chip microbiome as a function of storage strategy. International Biodeterioration & Biodegradation 156:105133. doi: 10.1016/j.ibiod.2020.105133. [DOI] [Google Scholar]
- 46.Henning JA, Weston DJ, Pelletier DA, Timm CM, Jawdy SS, Classen AT. 2019. Relatively rare root endophytic bacteria drive plant resource allocation patterns and tissue nutrient concentration in unpredictable ways. Am J Bot 106:1423–1434. doi: 10.1002/ajb2.1373. [DOI] [PubMed] [Google Scholar]
- 47.Wang J, Carper DL, Burdick LH, Shrestha HK, Appidi MR, Abraham PE, Timm CM, Hettich RL, Pelletier DA, Doktycz MJ. 2021. Formation, characterization and modeling of emergent synthetic microbial communities. Comput Struct Biotechnol J 19:1917–1927. doi: 10.1016/j.csbj.2021.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Margalit A, Carolan JC, Sheehan D, Kavanagh K. 2020. The Aspergillus fumigatus secretome alters the proteome of Pseudomonas aeruginosa to stimulate bacterial growth: implications for co-infection. Mol Cell Proteomics 19:1346–1359. doi: 10.1074/mcp.RA120.002059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mehedi MAU, Molla AH, Khondkar P, Sultana S, Islam MA, Rashid MA, Chowdhury R. 2010. Pseurotin a: an antibacterial secondary metabolite from aspergillus fumigatus. Asian J Chem 22:2611–2614. [Google Scholar]
- 50.Reece E, Doyle S, Greally P, Renwick J, McClean S. 2018. Aspergillus fumigatus inhibits Pseudomonas aeruginosa in co-culture: implications of a mutually antagonistic relationship on virulence and inflammation in the CF airway. Front Microbiol 9:1205. doi: 10.3389/fmicb.2018.01205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.D’Haeze W, Holsters M. 2002. Nod factor structures, responses, and perception during initiation of nodule development. Glycobiology 12:79R–105R. doi: 10.1093/glycob/12.6.79r. [DOI] [PubMed] [Google Scholar]
- 52.Tati S, Davidow P, McCall A, Hwang-Wong E, Rojas IG, Cormack B, Edgerton M. 2016. Candida glabrata binding to Candida albicans hyphae enables its development in oropharyngeal candidiasis. PLoS Pathog 12:e1005522. doi: 10.1371/journal.ppat.1005522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fidel PL, Jr, Vazquez JA, Sobel JD. 1999. Candida glabrata: Review of epidemiology, pathogenesis, and clinical disease with comparison to C. albicans. Clin Microbiol Rev 12:80–96. doi: 10.1128/CMR.12.1.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bignell E, Cairns TC, Throckmorton K, Nierman WC, Keller NP. 2016. Secondary metabolite arsenal of an opportunistic pathogenic fungus. Philos Trans R Soc B 371:20160023. doi: 10.1098/rstb.2016.0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dowd PF. 1988. Synergism of aflatoxin B1 toxicity with the co-occurring fungal metabolite kojic acid to two caterpillars. Entomol. Exp. Appl 47:69–71. [Google Scholar]
- 56.Gould AL, Zhang V, Lamberti L, Jones EW, Obadia B, Korasidis N, Gavryushkin A, Carlson JM, Beerenwinkel N, Ludington WB. 2018. Microbiome interactions shape host fitness. Proc Natl Acad Sci U S A 115:E11951–E11960. doi: 10.1073/pnas.1809349115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jaiswal SK, Mohammed M, Ibny FYI, Dakora FD. 2021. Rhizobia as a source of plant growth-promoting molecules: potential applications and possible operational mechanisms. Front Sustain Food Syst 4:619676. doi: 10.3389/fsufs.2020.619676. [DOI] [Google Scholar]
- 58.Macchiavelli RE, Brelles-Mariño G. 2004. Nod factor-treated Medicago truncatula roots and seeds show an increased number of nodules when inoculated with a limiting population of Sinorhizobium meliloti. J Exp Bot 55:2635–2640. doi: 10.1093/jxb/erh261. [DOI] [PubMed] [Google Scholar]
- 59.Oláh B, Brière C, Bécard G, Dénarié J, Gough C. 2005. Nod factors and a diffusible factor from arbuscular mycorrhizal fungi stimulate lateral root formation in Medicago truncatula via the DMI1/DMI2 signalling pathway. Plant J 44:195–207. doi: 10.1111/j.1365-313X.2005.02522.x. [DOI] [PubMed] [Google Scholar]
- 60.Djordjevic MA, Bezos A, Marmuse L, Driguez H, Samain E, Vauzeilles B, Beau J-M, Kordbacheh F, Rolfe BG, Schwörer R, Daines AM, Gresshoff PM, Parish CR. 2014. Lipo-chitin oligosaccharides, plant symbiosis signalling molecules that modulate mammalian angiogenesis in vitro. PLoS One 9:e112635. doi: 10.1371/journal.pone.0112635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brookman JL, Denning DW. 2000. Molecular genetics in Aspergillus fumigatus. Curr Opin Microbiol 3:468–474. doi: 10.1016/S1369-5274(00)00124-7. [DOI] [PubMed] [Google Scholar]
- 62.Kaminskyj SGW. 2001. Fundamentals of growth, storage, genetics and microscopy of Aspergillus nidulans. Fungal Genetics Rep 48:25–31. doi: 10.4148/1941-4765.1175. [DOI] [Google Scholar]
- 63.Pfannenstiel BT, Zhao X, Wortman J, Wiemann P, Throckmorton K, Spraker JE, Soukup AA, Luo X, Lindner DL, Lim FY, Knox BP, Haas B, Fischer GJ, Choera T, Butchko RAE, Bok J-W, Affeldt KJ, Keller NP, Palmer JM. 2017. Revitalization of a forward genetic screen identifies three new regulators of fungal secondary metabolism in the genus Aspergillus. mBio 8:e01246-17. doi: 10.1128/mBio.01246-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, Kern R, Tabb DL, Liebler DC, MacCoss MJ. 2010. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26:966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mills RF. 1955. The action of fumagillin on a bacteriophage of Staphylococcus aureus. J Gen Microbiol 13:39–44. doi: 10.1099/00221287-13-1-39. [DOI] [PubMed] [Google Scholar]
- 66.Shadduck JA. 1980. Effect of fumagillin on in vitro multiplication of Encephalitozoon cuniculi. J Protozool 27:202–208. doi: 10.1111/j.1550-7408.1980.tb04681.x. [DOI] [PubMed] [Google Scholar]
- 67.Zhang P, Nicholson DE, Bujnicki JM, Su X, Brendle JJ, Ferdig M, Kyle DE, Milhous WK, Chiang PK. 2002. Angiogenesis inhibitors specific for methionine aminopeptidase 2 as drugs for malaria and leishmaniasis. J Biomed Sci 9:34–40. doi: 10.1007/BF02256576. [DOI] [PubMed] [Google Scholar]
- 68.Pinheiro EAA, Carvalho JM, dos Santos DCP, Feitosa AdO, Marinho PSB, Guilhon GMSP, de Souza ADL, da Silva FMA, Marinho AMDR. 2013. Antibacterial activity of alkaloids produced by endophytic fungus Aspergillus sp. EJC08 isolated from medical plant Bauhinia guianensis. Nat Prod Res 27:1633–1638. doi: 10.1080/14786419.2012.750316. [DOI] [PubMed] [Google Scholar]
- 69.Esteban P, Redrado S, Comas L, Domingo MP, Millán-Lou MI, Seral C, Algarate S, Lopez C, Rezusta A, Pardo J, Arias M, Galvez EM. 2021. In vitro and in vivo antibacterial activity of gliotoxin alone and in combination with antibiotics against Staphylococcus aureus. Toxins (Basel) 13:85. doi: 10.3390/toxins13020085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Qin L, Li B, Guan J, Zhang G. 2009. In vitro synergistic antibacterial activities of helvolic acid on multi-drug resistant Staphylococcus aureus. Nat Prod Res 23:309–318. doi: 10.1080/14786410801972813. [DOI] [PubMed] [Google Scholar]
- 71.Horikoshi R, Goto K, Mitomi M, Oyama K, Sunazuka T, Ōmura S. 2017. Identification of pyripyropene A as a promising insecticidal compound in a microbial metabolite screening. J Antibiot (Tokyo) 70:272–276. doi: 10.1038/ja.2016.155. [DOI] [PubMed] [Google Scholar]
- 72.R Core Team . 2020. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/. [Google Scholar]
- 73.Sprouffske K, Wagner A. 2016. Growthcurver: an R package for obtaining interpretable metrics from microbial growth curves. BMC Bioinformatics 17:172. doi: 10.1186/s12859-016-1016-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Motulsky HJ, Brown RE. 2006. Detecting outliers when fitting data with nonlinear regression: a new method based on robust nonlinear regression and the false discovery rate. BMC Bioinformatics 7:123. doi: 10.1186/1471-2105-7-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Dry biomass weight of A. fumigatus at (a) 25°C and (b) 37°C when exposed to LCO treatments and compared to the solvent control. There were 10 biological replications. Welch’s unpaired t test was used to compare individual treatments to the solvent control for each temperature, resulting in no significant differences. Download FIG S1, TIF file, 14.3 MB (14.3MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
ΔΔCT values and peak area for confirmed secondary metabolites. Welch’s unpaired t test was used to compare individual treatments to the solvent control. *, P value < 0.005; **, P value < 0.01; ***, P < 0.001; ****, P < 0.0001. There were three biological replications for the ΔΔCT data and 10 biological replications for the peak area data set. (a to h) Welch’s ANOVA P values were (a) <0.0001 for ΔΔCT data and 0.0110 for peak area, (b) <0.0001 for ΔΔCT and 0.0070 for peak area, (c) 0.0007 for ΔΔCT and not significant for peak area, (d) 0.0010 for ΔΔCT and not significant for peak area, (e) 0.0025 for ΔΔCT and not significant for peak area, (f) <0.0001 for ΔΔCT and 0.0017 for peak area, (g) not significant, and (h) not significant. The primers used for ΔΔCT analysis can be found in reference 26. Download FIG S2, TIF file, 14.3 MB (14.3MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Principal-component analysis of treatments and temperature influence on identified features. Download FIG S3, TIF file, 14.3 MB (14.3MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Full auxiliary network analysis of metabolomic changes in response to LCO treatments at 25°C. The whole unfiltered network of significant log2 fold changes has 68 total nodes, 4 of which are the treatments themselves, with 130 total significant treatments as edges. Of those, 63 are downregulated and 67 are upregulated by their corresponding treatments. Of the 64 XIC analyte nodes, 29 of the nodes within the network are of degree 1, 14 of the nodes are degree 2, 11 nodes are of degree 3, and 10 are of degree 4. Of the 64 nodes, 45% of the total were of degree one, denoting that they were elicited from a single treatment. Considering summary statistics for each of the treatments within the network, C16:0 sLCOs had 19 upregulated and 13 downregulated XICs with a mean value of 0.41 and a median of 0.65. C16:0 nsLCOs had 19 upregulated and 18 downregulated XICs with a mean value of 0.33 and a median of 0.6. C18:1 sLCOs had 18 total upregulated and 18 downregulated XICs with a mean value of 0.40 and a median of 0.12. C18:1 nsLCOs had 12 upregulated and 14 downregulated XICs with a mean value of 0.83 yet with a median value of −0.1, denoting that, though there were more downregulated XICs, they were heavily outweighed by their upregulated counterparts. Two nodes of interest within the whole network are XIC IDs 243 and 374 due to having both up- and downregulated treatments. As noted in “Network Analysis at 25°C,” ID 24, identified by KEGG as an epithienamycin B-like metabolite, is upregulated by C18:1 sLCOs with a log2 fold change of 1.63, yet downregulated by C16:0 sLCOs with a log2 fold change of 1.38. XIC ID 374, when filtered for edges with log2 fold changes greater than 1, has two edges with log2 fold changes of 11.56 and 9.43, upregulated by the C18:1 nsLCOs and the C18:1 sLCOs respectively, yet when not filtered, XIC 374 has a third edge, which is downregulated by C16 nsLCOs with a log2 fold change of 0.87. All detailed XIC information is listed in Table S1. Download FIG S4, TIF file, 14.3 MB (14.3MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Full network for auxiliary route at 25°C. Download Table S1, CSV file, 0.03 MB (27.6KB, csv) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Full auxiliary network analysis of metabolomic changes in response to LCO treatments at 37°C. The whole unfiltered network of significant log2 fold changes has 31 total nodes, 4 of which are treatments with 57 edges. Of those edges, 32 are upregulated and 25 are downregulated by their corresponding treatments. Of the 27 total nodes, 12 are of degree 1, 5 are of degree 2, 5 are of degree 3, and 5 are of degree 4. Of the 27 total analyte nodes, 44% are of degree 1, denoting that they were elicited from a single treatment. Considering summary statistics for each of the treatments within the network, treatment C16:0 nsLCOs has 8 upregulated and 8 downregulated XICs with a mean value of 0.55 and a median value of 0.37. The C18:1 nsLCO treatment has 7 upregulated and 3 downregulated XICs with a mean value of 0.74 and a median value of 1.09. C18:1 sLCO has 9 upregulated and 8 downregulated XICs with a mean value of 0.81 and a median value of 1.03. C16:0 sLCO has 8 upregulated and 6 downregulated XICs with a mean value of 0.40 and a median value of 0.83. Additional nodes of interest could be those which are downregulated with respect to their treatment, particularly given that the edges primarily tend toward upregulation. All detailed XIC information is listed in Table S2. Download FIG S5, TIF file, 14.3 MB (14.3MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Full network for auxiliary route at 37°C. Download Table S2, CSV file, 0.01 MB (14.7KB, csv) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Bacterial point of inflection. Download Table S3, CSV file, 0.00 MB (280B, csv) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Log phase growth rates of bacterial species following exposure to LCO standards. Data are mean quantiles based on the inflection point of growth for each bacterial species. Statistical analysis was conducted using Welch’s one-way ANOVA test and Dunnett’s T3. (a to g) LCO standards had no significant impact on bacterial growth rates compared to the solvent control. The point of inflection for each bacterial strain can be found in Table S3. There were eight biological replications for each bacterial species. Download FIG S6, TIF file, 14.3 MB (14.3MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Log-phase growth rates of bacterial species following exposure to secondary metabolite standards. Secondary metabolite standards were commercially purchased and dissolved in either solvent control 1 (100% ethanol), shown in gray, or solvent control 2 (100% methanol), shown in orange, according to the manufacturer’s instructions. Data are mean quantiles based on the inflection point of growth for each bacterial species. Statistical analysis was conducted with Welch’s one-way ANOVA test and Dunnett’s T3. The points of inflection can found in Table S3. *, P < 0.05; **, P < 0.01; ***, P < 0.001. N values represents the biological replications as follows: Bacillus sp. strain BC15 is eight, Caulobacter sp. strain APO7 is six, Duganella sp. strain CF402 is six, Pantoea sp. strain YR343 is four, Paraburkholderia sp. strain BTO3 is six, Pseudomonas sp. strain GM17 is eight, and Rhizobium sp. strain BK456 is four. (a) Helvolic acid has a negative significant impact on Bacillus sp. strain BC15 log-phase growth, the Welch’s ANOVA test P value is 0.0387. (b to g) Secondary metabolite standards had no significant impact (Welch’s ANOVA test P > 0.05) on bacterial growth compared to the solvent control. Caulobacter sp. strain AP07 could not grow in the presence of ethanol. Download FIG S7, TIF file, 14.3 MB (14.3MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Data Availability Statement
LC/MS data are available through the MassIVE repository (https://massive.ucsd.edu/ProteoSAFe/static/massive.jsp; Data set ID MSV000090566).