ABSTRACT
Bacterial restriction-modification (R-M) systems are a first-line immune defense against foreign DNA from viruses and other bacteria. While R-M systems are critical in maintaining genome integrity, R-M nucleases unfortunately present significant barriers to targeted genetic modification. Bacteria of the genus Fusobacterium are oral, Gram-negative, anaerobic, opportunistic pathogens that are implicated in the progression and severity of multiple cancers and tissue infections, yet our understanding of their direct roles in disease have been severely hindered by their genetic recalcitrance. Here, we demonstrate a path to overcome these barriers in Fusobacterium by using native DNA methylation as a host mimicry strategy to bypass R-M system cleavage of transformed plasmid DNA. We report the identification, characterization, and successful use of Fusobacterium nucleatum type II and III DNA methyltransferase (MTase) enzymes to produce a multifold increase in gene knockout efficiency in the strain Fusobacterium nucleatum subsp. nucleatum 23726, as well as the first system for efficient gene knockouts and complementations in F. nucleatum subsp. nucleatum 25586. We show plasmid protection can be accomplished in vitro with purified enzymes, as well as in vivo in an Escherichia coli host that constitutively expresses F. nucleatum subsp. nucleatum MTase enzymes. In summary, this proof-of-concept study characterizes specific MTases that are critical for bypassing R-M systems and has enhanced our understanding of enzyme combinations that could be used to genetically modify clinical isolates of Fusobacterium that have thus far been inaccessible to molecular characterization.
IMPORTANCE Fusobacterium nucleatum is an oral opportunistic pathogen associated with diseases that include cancer and preterm birth. Our understanding of how this bacterium modulates human disease has been hindered by a lack of genetic systems. Here, we show that F. nucleatum DNA methyltransferase-modified plasmid DNA overcomes the transformation barrier and has allowed the development of a genetic system in a previously inaccessible strain. We present a strategy that could potentially be expanded to enable the genetic modification of highly recalcitrant strains, thereby fostering investigational studies to uncover novel host-pathogen interactions in Fusobacterium.
KEYWORDS: Fusobacterium, Fusobacterium nucleatum, DNA methyltransferase, methylase, methylome, restriction-modification, R-M systems, transformation, bacterial genetics, bacteriophage genetics, cancer, virulence
INTRODUCTION
Bacteria have multiple mechanisms to keep out foreign DNA elements, including physical barriers in the form of membranes and innate and adaptive nucleotide recognition systems to degrade foreign DNA before costly genome integration (1–3). This ability to recognize self versus nonself DNA is critical for productive genetic exchanges through horizontal gene transfers between close species to receive adaptive advantages (4–6). The two main nucleic acid surveillance systems bacteria deploy are restriction-modification (R-M) systems and CRISPR-Cas (clustered, regularly interspaced palindromic repeat-CRISPR-associated proteins) systems. In addition, a new system known as DISARM has joined the bacterial arsenal of DNA defense systems (7). CRISPR-Cas systems are considered adaptive immune components because of their ability to chromosomally integrate foreign (i.e., viral) DNA to create memory for subsequent encounters (8–11). In addition, the rather newly characterized bacteriophage exclusion (BREX) systems exist in 10% of the sequenced bacterial genomes and block phage DNA replication and lysogeny in infected cells (12, 13). BREX differentiates itself from R-M systems in that phage DNA is not cleaved or digested, which suggests a unique bacterial defense system. While R-M systems serve bacteria well for their survival and adaptation, they present significant challenges for researchers aiming to understand these organisms through genetic manipulation in the form of gene knockouts. This genetic recalcitrance is widespread throughout the bacterial kingdom, and in many cases it has led researchers to gravitate toward using strains that have weak R-M systems that can be genetically modified, instead of using target strains that have desired phenotypes.
R-M systems consist of restriction endonucleases (REases) and DNA methyltransferases (MTase), which can either exist as a paired REase/MTase operon that can also contain additional specificity genes or as singular MTase genes (14–16). The system works when REases cleave DNA that does not have the proper MTase-induced methylation sequences, thereby signaling to the bacteria that the detected DNA is foreign and unwanted. R-M systems are classified as type I, II, III, or IV, according to their molecular structure, subunit composition, cleavage position, restriction site, and cofactor specifications (see Fig. S1 in the supplemental material). Type I systems (hsdRMS genes) cut exogenous DNA by forming protein complexes, and random cleavage usually happens at substantial distances from an asymmetric recognition sequence (400 to 7,000 bp) (17), while type II systems consist of an individual restriction endonuclease and methyltransferase that cleave DNA at symmetrical recognition sites (18). In a similar way to type I, type III forms a protein complex necessary for the restriction enzyme activity; however, the methyltransferase can function independently. DNA cleavage for type III R-M systems takes place 25 to 27 bp 3′ to an asymmetrical recognition sequence that is 5 to 6 bp in length (19, 20). Furthermore, type IV systems asymmetrically recognize DNA sequences and cleavage by REases at a defined distance from the recognition sites. In addition, some of these systems contain multiple MTases that can be adenine or cytosine specific, as well as the REase showing methyltransferase activity (17, 21–23).
Fusobacterium, especially the species Fusobacterium nucleatum, has garnered significant attention since this bacterium was reported to be overrepresented in colorectal cancer tumors more than a decade ago (24–26). Classical studies have mainly focused on the role of F. nucleatum in oral infections and diseases, including periodontitis (27, 28), severe organ infections (29–32), and preterm birth (33–35). The majority of recent studies have shifted the focus to a potential direct causal role in adverse cancer phenotypes, including heightened inflammation (36–38), production of carcinogenic metabolites (39), induced metastasis (40–42), DNA damage (43–45), increased resistance to frontline chemotherapy drugs (46, 47), and overall worse patient prognosis (36, 48, 49). Despite an increasing interest in understanding how this bacterium contributes to cancer, there have been very few mechanistic studies of specific bacterial effector proteins, due to the genetic recalcitrance in most strains. Because of this, our current molecular studies have been limited to a few Fusobacterium strains that are able to acquire “naked,” unmethylated DNA and incorporate it into their genome by recombination with homologous sequences or, in the case of multicopy plasmids, by establishing a new episome. Of these strains, F. nucleatum subsp. nucleatum 23726 (transformation by electroporation) (50–52), F. nucleatum subsp. polymorphum 10953 (transformation by electroporation) (53), F. nucleatum subsp. polymorphum 12230 (transformation by sonoporation) (54), and a recent paper highlighting the first gene interruption in Fusobacterium necrophorum using DNA conjugation from Escherichia coli (55, 56). Needless to say, these four strains do not encompass the Fusobacterium subspecies landscape and their respective infections and diseases that researchers desire to study, and they highlight the need for molecular biology and biochemical studies to achieve universal genetics.
Seminal studies have successfully used MTases to modify and protect plasmid DNA to facilitate molecular genetics in several other bacterial species (57–59). This technique has been used successfully in many studies, but it was coined plasmid artificial modification (PAM) when it was used to enhance transformation in Bifidobacterium adolescentis (60). What we currently know about the R-M systems of Fusobacterium largely exist as bioinformatic predictions based on MTase classification in the REBASE database (61). However, this bioinformatic classification in most cases did not come with experimental DNA methylation analyses to match enzymes with their target sequences. Additionally, even when an MTase is matched with its recognition and methylation sequence, this does not guarantee that these modifications will be important for effective protection of plasmid DNA to increase transformation efficiency. Therefore, the goal of this study was to biochemically characterize and utilize a broad range of F. nucleatum MTases for host mimicry by methylation to accelerate bacterial genetics in previously inaccessible strains. We successfully report the use of F. nucleatum MTase enzymes produced in E. coli to protect plasmid DNA, facilitating a significant increase in chromosomal incorporation of plasmid and transposons in multiple F. nucleatum strains, as well as the development of the first gene deletion system in F. nucleatum subsp. nucleatum 25586 after only one previous study reporting a single-crossover gene interruption of the FomA porin (62). Our study is not exhaustive, because of the number of strains (>100 F. nucleatum strains in the NCBI genome database) and R-M systems (average of seven systems per genome [Fig. 1]) that could have been tested. However, we believe our strategies will provide a flexible roadmap for the scientific community to adopt MTase-based methods for genetic manipulation in Fusobacterium.
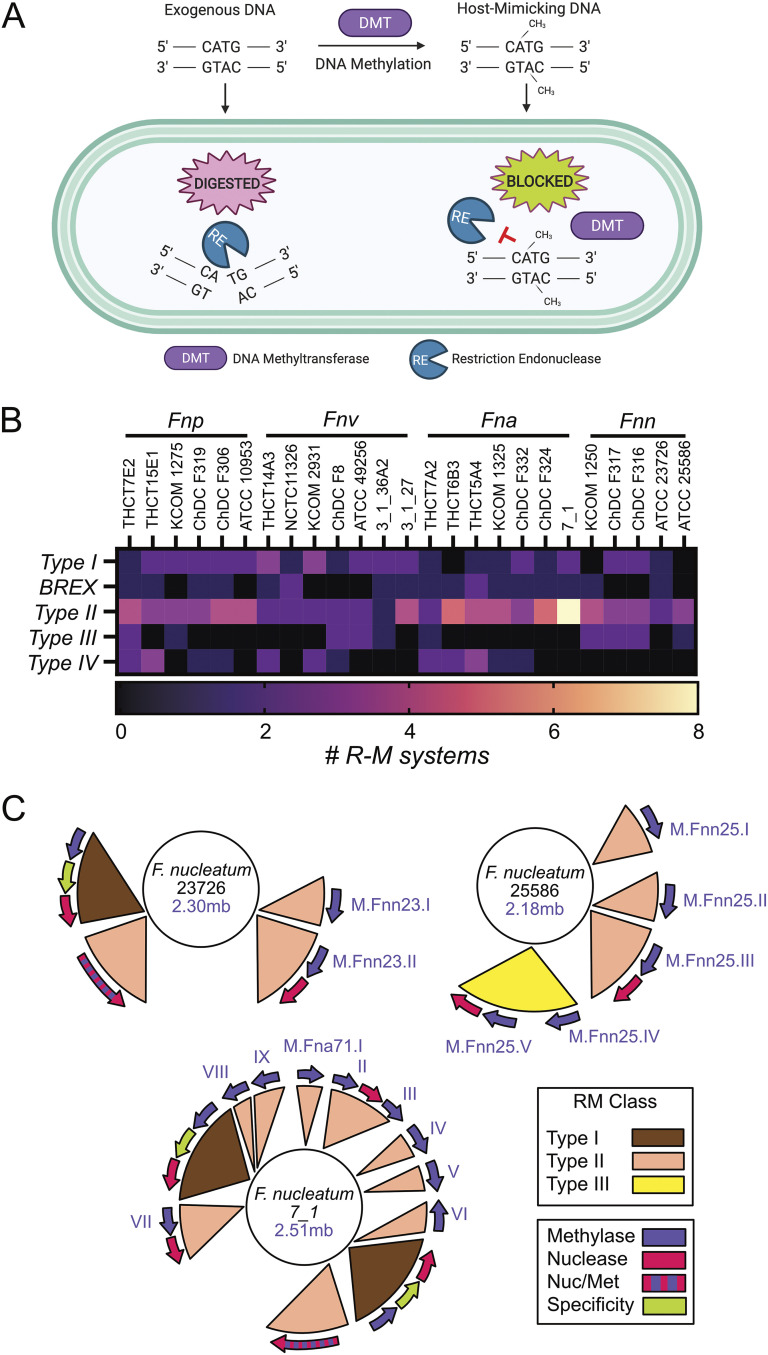
FIG 1.
Restriction-modification system classification in Fusobacterium. (A) Overview of how R-M systems utilize bacteria-specific DNA methylation to mark the chromosome as self DNA, thereby restriction digesting invading DNA that does not contain the proper methylation patterns. Adapted from Hirokazu Suzuki (80). (B) Classification and quantitation of R-M systems in 25 strains of F. nucleatum covering the four subspecies: F. nucleatum subsp. polymorphum (Fnp), F. nucleatum subsp. vincentii (Fnv), F. nucleatum subsp. animalis (Fna), and F. nucleatum subsp. nucleatum (Fnn). (C) Genome location and renaming of type II and type III MTases in three strains of F. nucleatum subsp. nucleatum used in this study. R-M systems mapped on the genomes were recreated from those on the REBASE website.
RESULTS
Bioinformatic identification and classification of R-M systems in Fusobacterium.
As shown in Fig. 1A, bacterial R-M systems act by blocking exogenous DNA from entering and being incorporated into the genome by digesting foreign, improperly methylated DNA. Scientists have exploited this defense mechanism by using strain-specific MTase enzymes to pretreat DNA before electroporation or natural competence to improve transformation efficiency (59). In this study, to identify potential Fusobacterium MTases to increase the efficiency of transformation and DNA recombination, we queried the online databases REBASE (61), FusoPortal (63), and NCBI (64) to characterize R-M systems. We analyzed 25 strains of Fusobacterium nucleatum in REBASE, covering the subspecies F. nucleatum subsp. nucleatum, F. nucleatum subsp. animalis, F. nucleatum subsp. vincentii, and F. nucleatum subsp. polymorphum, and cataloged the number and classification of their R-M systems, as shown in Fig. 1B (see also Table S1 in the supplemental material). There was an overall propensity for F. nucleatum strains to have a higher number of type II MTase genes, yet there was not a strong overall pattern for the number or class of R-M systems that differentiated the subspecies. As shown in Fig. 1C, we highlighted three strains of F. nucleatum covering subspecies F. nucleatum subsp. nucleatum and F. nucleatum subsp. animalis. The genetically tractable strain F. nucleatum subsp. nucleatum 23726 encodes 4 R-M systems, as shown in Fig. 1C: one type I, two type II, and one BREX system. F. nucleatum subsp. nucleatum 25586 lacks type I R-M systems but has three type II and one type III system containing two MTases that proved critical for enabling molecular genetics in this strain. Finally, 11 R-M systems (two type I and nine type II) were identified in F. nucleatum subsp. animalis 7_1; this was the highest quantity of R-M systems in our analyzed strains.
An orthodox type II R-M system includes two independent genes in an operon: an MTase and a REase. However, as shown in Fig. 1C, several lone type II MTases were discovered in multiple F. nucleatum subsp. nucleatum strains, and we later showed these are crucial for DNA methylation and molecular genetics. Our bioinformatic studies also confirmed the presence of the type II BREX system in several F. nucleatum subsp. nucleatum strains. The BREX system is generally composed of a 4- to 8-gene cluster (12), and in Fusobacterium it is predicted to methylate adenine residues in a mechanism similar to that in E. coli (65). However, since the restriction site for this enzyme is yet to be characterized and these systems have not been shown to be important for efficient molecular microbiology efforts, we did not focus on using these enzymes for plasmid protection. Finally, multiple type IV R-M systems were discovered in the F. nucleatum strains analyzed in this study, with type IV systems consisting of REases that will only cleave DNA that is methylated or glycosylated at either adenine or cytosine residues (21).
Recombinant production and characterization of F. nucleatum MTases.
To focus our study, we chose to utilize and characterize all type II and type III MTase enzymes in the strains F. nucleatum subsp. nucleatum 23726 and F. nucleatum subsp. nucleatum 25586. We chose to focus on these MTase classes because the strain F. nucleatum subsp. nucleatum 25586 lacks type I systems and is still highly recalcitrant to genetic manipulation. However, we do realize that type I R-M systems could play a role in other strains and could be analyzed in the future. As shown in Fig. 2, we cloned (Fig. 2A), expressed, and purified (Fig. 2B) five enzymes (M.Fnn23.I, M.Fnn23.II, M.Fnn25.I, M.Fnn25.IV, and M.Fnn25.V). M.Fnn23.I and M.Fnn23.II were used to treat the plasmid pDJSVT13 as described below, which we previously used to knock out the galKT genes in F. nucleatum subsp. nucleatum 23726 (66).
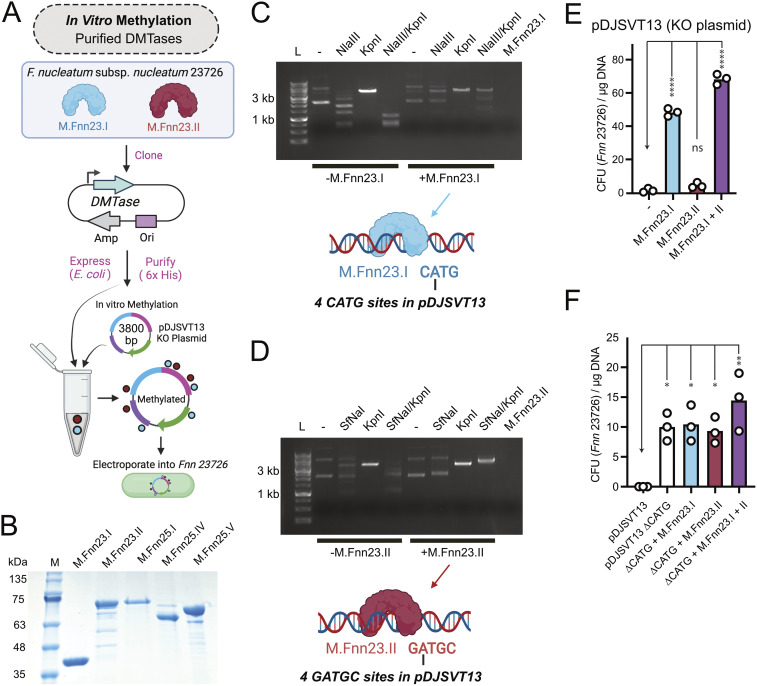
FIG 2.
F. nucleatum MTases protect plasmid DNA and allow for more efficient chromosomal plasmid incorporation in F. nucleatum subsp. nucleatum 23726. (A) Schematic of our process to produce recombinant MTases that were next used to treat plasmid DNA in vitro prior to electroporation into F. nucleatum subsp. nucleatum 23726. (B) SDS-PAGE gel of five purified MTases from F. nucleatum subsp. nucleatum 23726 and F. nucleatum subsp. nucleatum 25586. (C) Methylation of pDJSVT13 with M.Fnn23.I protects against DNA cleavage by the REase NlaIII (NEB), which cuts at CATG sites. KpnI was used as a unique single-cut enzyme in pDJSVT13. The M.Fnn23.I lane is enzyme alone and shows no contaminating DNA brought in with the pure protein. (D) Methylation of pDJSVT13 with M.Fnn23.II protected against DNA cleavage by the REase SfNaI (NEB), which cut at GATGC sites. (E) Methylation of pDJSVT13 resulted in an increased number of transformants (in CFU per microgram of DNA). (F) By changing the four CATG sequences to CACG (ΔCATG), which are the target for the MTase M.Fnn23.I, transformation efficiency was significantly increased, even in the absence of methylation. Statistical values are as follows: ns, P > 0.05; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. For panels E and F data, we used a two-way ANOVA.
Recombinant F. nucleatum MTases protect plasmid DNA from REase digestion.
To show that our recombinant enzymes from F. nucleatum subsp. nucleatum 23726 were active, we identified commercially available REases that matched the REBASE-predicted methylation sequences of M.Fnn23.I and M.Fnn23.II. By methylating the plasmid pDJVST13 with M.Fnn23.I, we showed that adenine methylation in the sequence CATG blocked cleavage by the endonuclease NlaIII, which recognized the same sequence and cleaved 3′ to the guanine (Fig. 2C). Next, we methylated pDJSVT13 with M.Fnn23.II, and we determined that methylation of the adenine in GATGC protected DNA from cleavage by SfNaI, which recognizes GCATC(N5) and cleaves 3′ to the N5 sequence (Fig. 2D). This protection of DNA from cleavage by methylation indicates that use of these enzymes in tandem would allow more efficient homologous recombination in F. nucleatum subsp. nucleatum 23726 postelectroporation.
Plasmid DNA methylated with recombinant MTases increases chromosomal integration for the galKT gene knockout plasmid pDJSVT13 in F. nucleatum subsp. nucleatum 23726.
As shown in Fig. 2E, methylation of pDJSVT13 with M.Fnn23.I resulted in significantly more colonies after transformation, indicating that protected DNA was not degraded before homologous recombination with the galKT operon in F. nucleatum subsp. nucleatum 23726. M.Fnn23.II alone did not have a drastic effect but did increase efficiency. Finally, the combination of M.Fnn23.I and M.Fnn23.II resulted in an even further increase in transformation and chromosomal incorporation, thereby greatly enhancing the efficiency of creating gene knockouts.
As M.Fnn23.I appeared to be the dominant enzyme for protecting plasmid DNA in pDJSVT13, we made a pDJSVT13 ΔCATG plasmid, now called pDJSVT21, in which the four CATG sites were eliminated with silent single-nucleotide mutations to CACG. Figure 2F shows that pDJSVT21 transformed significantly better than pDJSVT13. The addition of M.Fnn23.I or M.Fnn23.II individually did not increase transformation efficiency over that of pDJSVT21. However, the addition of both enzymes did, which could mean that these enzymes are methylating at more than their bioinformatically predicted sites.
In vivo methylation of plasmids increased transformation of gene knockout and transposon plasmids.
We next developed plasmids that placed the M.Fnn23.I and M.Fnn23.II MTase genes downstream of a strong constitutive Anderson promoter (iGEM part BBa_J23101) and before a short terminator (iGEM part BBa_00014). Plasmid pDJSVT24 contains M.Fnn23.I, pDJSVT25 contains M.Fnn23.II, and pDJSVT26 contains both M.Fnn23.I and M.Fnn23.II (Fig. 3A). TOP10 E. coli containing one of the aforementioned plasmids expressing F. nucleatum subsp. nucleatum 23726 MTases was transformed with the galKT gene knockout plasmid pDJSVT13, followed by plasmid purification from overnight cultures. Upon transformation of this mixed pool of plasmids into F. nucleatum subsp. nucleatum 23726 and selection on thiamphenicol-containing medium to select for chromosomal incorporation of pDJSVT13, we showed that this simple method of plasmid methylation was effective at significantly increasing transformation rate. We noted that since this was a mixed pool of plasmids, the efficiency of our transformations was likely higher than that shown in Fig. 3. M.Fnn23.I alone resulted in a marginal increase in efficiency, but methylation by both enzymes significantly increased transformation rates by more than 50-fold (Fig. 3B). As Top10 E. coli does possess Dam+ and Dcm+ methylation systems, we also used methylation-free E. coli ER2796 (67) and showed that plasmids purified from both strains transformed at the same rate into F. nucleatum subsp. nucleatum 23726 when pDJSVT26 was present and expressing M.Fnn23.I and M.Fnn23.II (Fig. 3C).
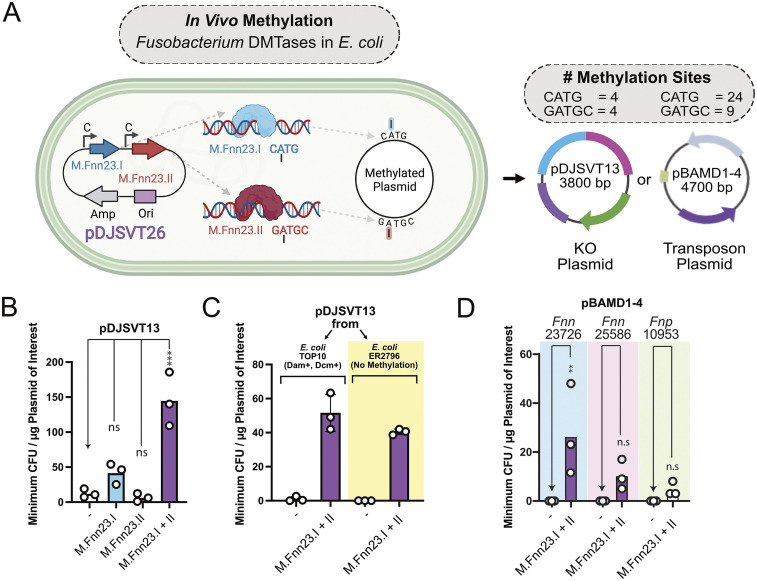
FIG 3.
In vivo methylation in E. coli expressing M.Fnn23.I and M.Fnn23.II enhanced plasmid transformation and chromosomal incorporation of plasmids and transposons. (A) Schematic of in vivo methylation of plasmids with F. nucleatum subsp. nucleatum 23726 MTases. The C above the arrow indicates a promoter that is constitutively active for constitutive expression in E. coli. (B) Transformation of pDJSVT13 was significantly increased by coexpressing M.Fnn23.I and M.Fnn23.II. (C) Comparison of methylation-positive (TOP10) and methylation-negative (ER2796) E. coli strains revealed that native E. coli methylation did not inhibit the transformation of pDJSVT13 when F. nucleatum subsp. nucleatum 23726 MTases were concurrently expressed. (D) In vivo methylation of the pBAMD1-4 transposon plasmid allowed for transformation and chromosomal transposon insertion into multiple strains of F. nucleatum. Statistical values are as follows: ns, P > 0.05; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. For panel B data we used a two-way ANOVA; for panel D data we used Student's t test analysis.
We next showed that the mini-Tn5 transposon harboring plasmid pBAMD1-4 (68) could be transformed into F. nucleatum subsp. nucleatum 23726, F. nucleatum subsp. nucleatum 25586, and F. nucleatum subsp. polymorphum 10953 after methylation with M.Fnn23.I and M.Fnn23.II. Importantly, unmethylated plasmid was unsuccessful at producing transposon insertions in these three strains (Fig. 3D). We did note that this system is not highly efficient and would benefit from use of a more complete repertoire of MTases from the respective strains. Overall, compared with in vitro plasmid treatment with recombinant MTases, creation of an E. coli strain expressing F. nucleatum MTases worked just as well and required less effort than purifying multiple proteins. However, the difficulty of creating plasmids with a significant number of MTase genes makes this method increasingly challenging.
Passaging of a plasmid in F. nucleatum allowed for transformation into additional F. nucleatum strains.
A common method of permitting a plasmid to be transformed into a genetically recalcitrant strain of interest is to first transform into a similar yet genetically competent strain, followed by repurification of the plasmid containing species-specific methylation patterns (Fig. 4A) (58). This plasmid frequently can then be transformed into the strain of interest. Here, we tested this classic method and showed that passage of the episomal, multicopy Fusobacterium plasmid pHS30 (50) in F. nucleatum subsp. nucleatum 23726 could be purified and then transformed into F. nucleatum subsp. polymorphum 10953, but not F. nucleatum subsp. nucleatum 25586, F. nucleatum subsp. animalis 7_1, or F. nucleatum subsp. animalis 4_8. When plasmid was repurified from F. nucleatum subsp. polymorphum 10953, this plasmid could only be retransformed back into F. nucleatum subsp. nucleatum 23726, revealing that the R-M systems in the other strains were not compatible with F. nucleatum subsp. nucleatum 23726 and F. nucleatum subsp. polymorphum 10953 (Fig. 4B). After methylating pHS30 with five MTases to allow transformation into F. nucleatum subsp. nucleatum 25586, repurified plasmid was only able to be transformed into F. nucleatum subsp. nucleatum 23726. And once again, repurification of the plasmid from F. nucleatum subsp. nucleatum 23726 was only able to be transformed back into F. nucleatum subsp. polymorphum 10953.
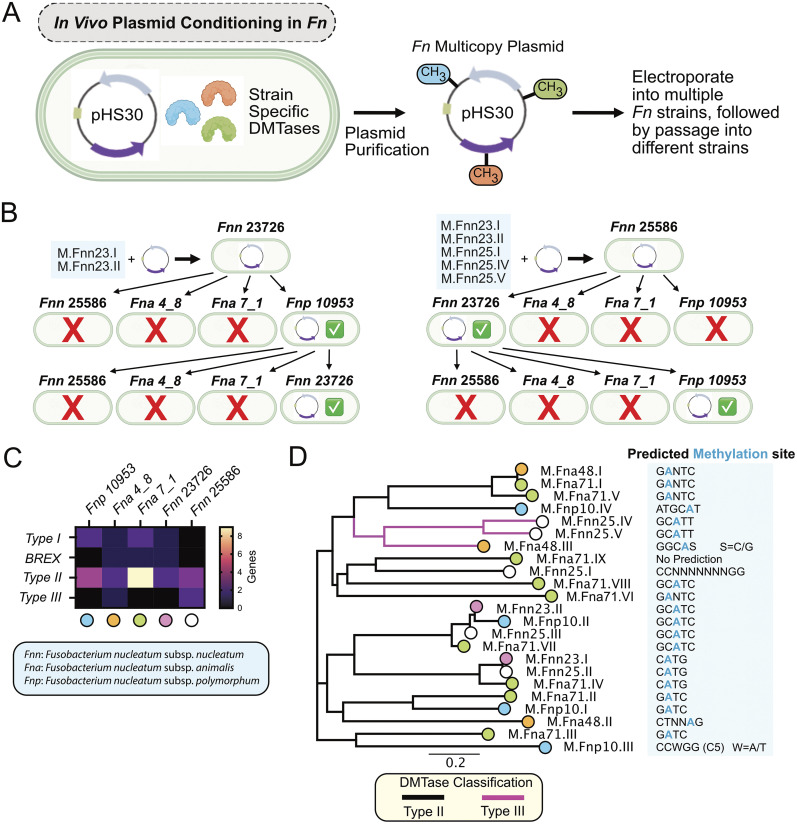
FIG 4.
Passaging of a multicopy plasmid in F. nucleatum allowed passage to additional strains. (A) Schematic of our passaging method for the Fusobacterium multicopy plasmid pHS30 and purification of this plasmid for retransformation into different F. nucleatum strains. (B) pHS30 from F. nucleatum subsp. nucleatum 23726 can be transformed into F. nucleatum subsp. polymorphum 10953, and repurification from this strain allowed transformation back into F. nucleatum subsp. nucleatum 23726. pHS30 from F. nucleatum subsp. nucleatum 25586 can be transformed into F. nucleatum subsp. nucleatum 23736, and repurification from this strain allowed transformation back into F. nucleatum subsp. polymorphum 10953. (C) Heat map of the number of R-M systems in the five F. nucleatum strains analyzed. Colored dots below the strains correlate with the strains of the enzymes found in the phylogenetic tree in panel D. (D) Phylogenetic tree of 23 type II and III MTase genes from five F. nucleatum strains. Methylation sites are those predicted by REBASE.
To better understand why there was limited plasmid passaging between F. nucleatum subsp. nucleatum strains, we analyzed the type II and type III DMTases in the five F. nucleatum strains tested above. We first compared the number of genes present in the strains for all classes of MTases and noted that all strains contained a higher number of type II genes than the other classes (Fig. 4C). However, other than strain F. nucleatum subsp. animalis 7_1 having the highest number of type II R-M systems in strains analyzed, these data did not provide an obvious answer as to why the majority of these Fusobacterium strains were so genetically recalcitrant. To take a deeper look, we assembled a phylogenetic tree of the 23 type II and type III MTases from the five strains (Fig. 4D). Utilizing REBASE, we identified the predicted DNA recognition and methylation sites for all 23 type II and type III MTases in the five strains of F. nucleatum that we used in this study: F. nucleatum subsp. nucleatum 23726, F. nucleatum subsp. nucleatum 25586, F. nucleatum subsp. animalis 4_8, F. nucleatum subsp. animalis 7_1, and F. nucleatum subsp. polymorphum 10953 (Table S2). Nearly all MTases are predicted to be adenine DNA methyltransferases, where methylation occurs at the nitrogen at position 6 in the ring (N6) of the adenine (N6-mA or m6A), which is a common theme for AT-rich bacterial genomes (approximate GC concentration of 30%). Only one enzyme from F. nucleatum subsp. polymorphum 10953 was predicted to methylate the carbon at position 5 in the ring (C5) of the cytosine (C5-mC or m5C). We found clusters of enzymes with predicted MTase recognition sites that could be exploited to produce a library of enzymes that could be used for bypassing R-M systems in multiple strains. When analyzing the 23 MTases from these five F. nucleatum strains, it stands out that the enzymes are predicted to methylate only 10 recognition sites. These data also uncovered that of the nine enzymes in F. nucleatum subsp. animalis 7_1, which covered six predicted recognition sequences, only two of these sequences were predicted to be methylated by F. nucleatum subsp. nucleatum 23726 and F. nucleatum subsp. nucleatum 25586, leaving a large number of sequences unmethylated and the likely reason why plasmid was unable to be passed from these strains to F. nucleatum subsp. animalis 7_1.
F. nucleatum subsp. nucleatum 25586 and F. nucleatum subsp. nucleatum 23726 MTases allowed for the development of the first genetic system in F. nucleatum subsp. nucleatum 25586.
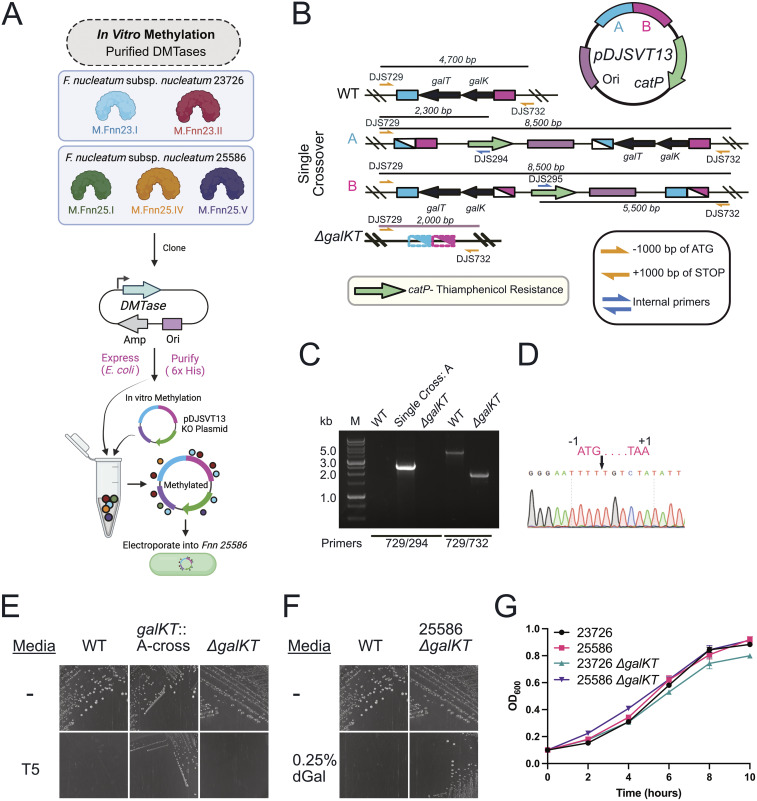
F. nucleatum subsp. nucleatum 25586 is a virulent and biomedically relevant strain that has been studied for more than 4 decades (69), yet molecular studies have not been possible because of the strain’s inability to be transformed. Our goal was to use the same system we developed previously for gene knockouts in F. nucleatum subsp. nucleatum 23726 (66). As shown in Fig. 5, we used two MTases from F. nucleatum subsp. nucleatum 23726 (M.Fnn23.I and M.Fnn23.II; the same exact enzymes as M.Fnn25.II and M.Fnn25.III) (Fig. 1C) and three from F. nucleatum subsp. nucleatum 25586 (M.Fnn25.I, M.Fnn25.IV, and M.Fnn25.V) to bypass R-M systems in F. nucleatum subsp. nucleatum 25586. Purification of these recombinant enzymes was followed by methylation of pDJSVT13 and transformation by electroporation (Fig. 5A). Colonies that grew on thiamphenicol-containing plates indicated chromosomal integration by homologous recombination before (fragment A) or after (fragment B) the galKT operon (Fig. 5B). PCR and sequencing verification of chromosomal integration (A or B single crossover) (Fig. 5C and D) was followed by double-crossover events in nonselective medium and plating on medium containing deoxygalactose, which verified excision of the galKT operon because the presence of galKT makes 2-deoxy-d-galactose toxic (Fig. 5E and F) (66). F. nucleatum subsp. nucleatum 25586 ΔgalKT grew with the same fitness as wild-type F. nucleatum subsp. nucleatum 25586, WT F. nucleatum subsp. nucleatum 23726, and F. nucleatum subsp. nucleatum 23726 ΔgalKT (Fig. 5G).
FIG 5.
Development of a galactose-selectable genetic system in F. nucleatum subsp. nucleatum 25586. (A) Schematic of the strategy to use five purified F. nucleatum MTases to methylate plasmid pDJSVT13 to transform into F. nucleatum 25586. (B) Schematic of single-crossover and galKT gene deletions using plasmid pDJSVT13, which was first homologously recombined with up- and downstream sequences of the galKT operon. Primers used for PCR verification of positive clones are denoted with yellow or blue arrows, and the labels DJSnnn and TNnnn refer to specific primers found in Table S5 in the supplemental material. (C) PCR verification of the initial chromosomal incorporation (a crossover) as well as the full operon deletion (ΔgalKT). (D) Sanger sequencing verification of a full, clean, deletion of the galKT operon. (E) Selection for A-crossover strains on thiamphenicol (T5) containing plates, and verification that the ΔgalKT strain had removed the vector and antibiotic cassette and no longer grew on thiamphenicol. (F) Proof of survival of ΔgalKT on plates containing deoxygalactose (dGal), which is toxic to wild-type F. nucleatum subsp. nucleatum 25586. (G) Growth curves showed no growth defect for F. nucleatum subsp. nucleatum 25586 ΔgalKT compared to WT F. nucleatum subsp. nucleatum 25586, WT F. nucleatum subsp. nucleatum 23726, and F. nucleatum subsp. nucleatum 23726 ΔgalKT.
Development of F. nucleatum subsp. nucleatum 25586 Δfap2 and ΔfadA strains.
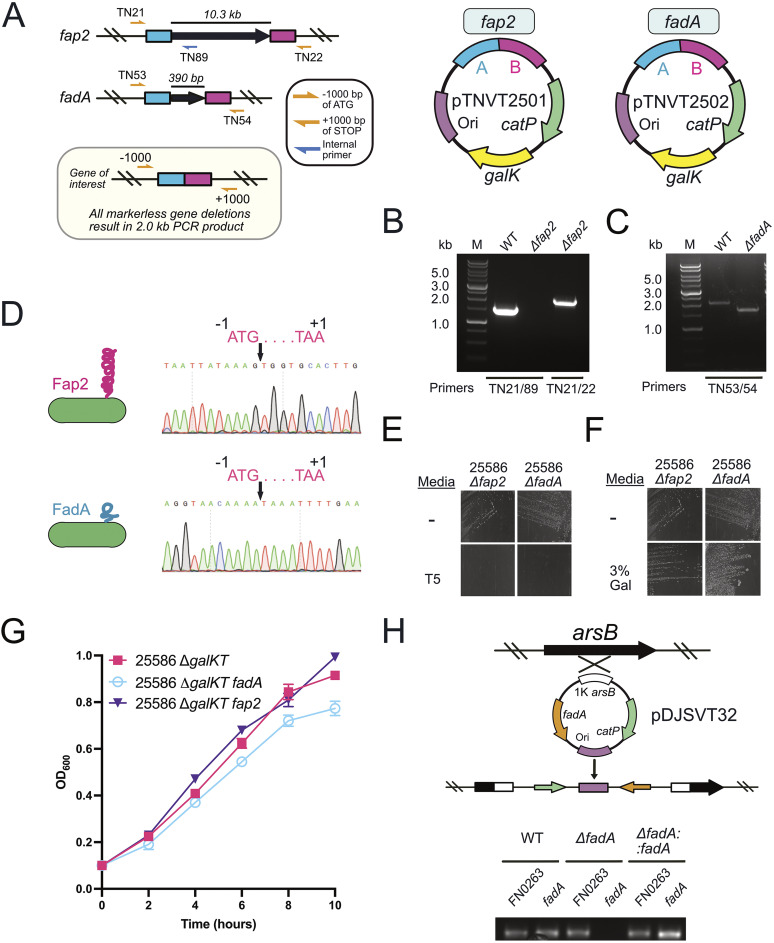
As a proof of concept, we made clean chromosomal deletions in genes fap2 and fadA in F. nucleatum subsp. nucleatum 25586 ΔgalKT (Fig. 6). This approach followed the same system that we initially used to knock out the galKT operon (Fig. 5) to make a galactose-selectable system possible. Briefly, we cloned and fused 1,000 bp up- and downstream of the target gene before ligating into our gene knockout vector that worked by double-crossover homologous recombination of gene deletion. This method resulted in clean deletions of the large, outer membrane, autotransporter adhesin fap2 (>10 kb) and the small, outer membrane adhesin fadA (390 bp), both of which have been studied extensively for their roles in F. nucleatum pathogenicity (Fig. 6A to F) (70–73). These gene deletions did not cause any adverse growth phenotypes compared to the parent strain F. nucleatum subsp. nucleatum 25586 ΔgalKT (Fig. 6G). Our final experiment was to complement the fadA gene deletion back onto the chromosome of F. nucleatum subsp. nucleatum 25586 ΔgalKT fadA at the arsB gene (66) (Fig. 6H), which confers arsenic resistance to bacteria but is not essential or necessary for F. nucleatum grown under laboratory conditions (74). In addition, we report that like the system for F. nucleatum subsp. nucleatum 23726, there appear to be no differences in efficiency when deleting large (fap2; 10 kb) or small (fadA; 390 bp) genes in F. nucleatum subsp. nucleatum 25586.
FIG 6.
Gene deletions of fap2 and fadA, as well as fadA complementation in F. nucleatum subsp. nucleatum 25586. (A) Schematic for deletion of the genes fap2 (>10 kb) and fadA (390 bp). Primers used for PCR verification of positive clones are denoted with yellow or blue arrows, and the labels TNnnn refer to specific primers found in Table S5. Plasmids pTNVT01 and pTNVT02 correspond to plasmids created to delete fap2 and fadA, respectively. (B) PCR verification of the Δfap2 mutant in F. nucleatum subsp. nucleatum 25586. (C) PCR verification of the ΔfadA mutant in F. nucleatum subsp. nucleatum 25586. (D) Sanger sequencing verification of a full, clean, deletion of the fap2 and fadA genes. (E) Streaking of F. nucleatum subsp. nucleatum 25586 Δfap2 and ΔfadA on thiamphenicol-containing plates (T5) verified the chromosomally integrated plasmid had been excised by homologous recombination. (F) Streaking of F. nucleatum subsp. nucleatum 25586 Δfap2 and ΔfadA on galactose-containing plates (T5) verified the chromosomally integrated plasmid had been excised by homologous recombination. (G) Growth curves showed no fitness effects from the fap2 and fadA gene deletions. (H) Complementation of the fadA gene (ΔfadA::fadA) onto the chromosome at the arsB gene using a single-crossover homologous recombination plasmid and confirmation by PCR.
DISCUSSION
Bacterial restriction-modification systems are important in both protection of bacteria from invading foreign DNA as well as using methylation as an epigenetic switch to control gene regulation (75). Our hypothesis was that if we could bypass Fusobacterium restriction-modification systems, it would enhance genetic efficiency in currently tractable strains, as well as leap the hurdle of developing new systems in strains that are currently inaccessible to molecular methods. Here, we showed that using strain-specific MTases from Fusobacterium nucleatum to methylate custom gene deletion plasmids led to more efficient gene deletions and gene complementations on the chromosome, as well as the introduction of a multicopy plasmid that could be used for a range of tasks, including gene complementation and protein overexpression. Our results showed a multifold increase in the efficiency of transformations and subsequent chromosomal incorporation of gene deletion plasmids in the genetically tractable strain F. nucleatum subsp. nucleatum 23726. To enhance genetics in this strain, we cloned, expressed, and characterized two type II MTase enzymes, which we renamed M.Fnn23.I and M.Fnn23.II. Using both in vitro and in vivo analyses, we verified that methylation of plasmid DNA blocked cleavage by the enzymes NlaIII and SfNaI, which cut at CATG and GATGC sites, respectively. We next showed that each enzyme individually increased the efficiency of plasmid introduction but that combining the two enzymes had a statistically significant effect. This synergistic effect could be attributed to blocking all of the potential restriction sites, where even one cut of the vector would likely stop it from being transformed. We chose to use the chromosomal integration plasmid pDJSVT13 for most of our studies, over the multicopy episomal plasmid pHS30, to focus on gene knockout improvement and not just introduction of a plasmid. Two areas that we noted that are variable between laboratories are (i) how competent bacteria are prepared and (ii) the plasmids used for transformation and subsequent generation of mutants and gene expression. Our analysis showed that the number of CATG and GATGC sites could be the differences in how other studies have shown variable transformation efficiencies, as we have not analyzed the sequences of these plasmids.
We next set our focus on creating the first gene knockouts in F. nucleatum subsp. nucleatum 25586, which had not been accomplished in over 40 years of studying the strain (20). To accomplish this, we produced five recombinantly expressed MTase enzymes to treat plasmid DNA in vitro, followed by transformation by electroporation. Through this method we were able to create the first clean gene deletions and complementations in F. nucleatum subsp. nucleatum 25586, with deletions in galKT, fap2, and fadA and complementation of fadA back onto the chromosome. This markerless gene deletion system can produce iterative gene deletions in a single strain, as the final gene deletions are free of antibiotic markers. We noted that all five enzymes were necessary to protect plasmid DNA for transformation into F. nucleatum subsp. nucleatum 25586; however, all MTases that we produced and used in these studies retained enzymatic activity after at least one freeze-thaw cycle (data not quantitated), making it a robust solution for researchers to implement.
We acknowledge that there are still major limitations to genetically modifying most strains of Fusobacterium because of their unique R-M compositions. Therefore, we understand that using a core combination of MTase enzymes for universal protection across multiple Fusobacterium species may not be possible, as each strain frequently has unique MTases that create a broad range of methylation patterns between strains. This has been reported before, as type II and type III R-M systems vary significantly even in evolutionarily similar strains of bacteria. Future studies using DNA methylation analysis of MTase deletion strains to identify exact methylation sequences with specific enzymes will lead to the experimental determination of methylation sites by specific enzymes. To support this claim, previous studies have shown that use of PacBio SMRTseq technology to determine the methylome of a bacterium results in the identification of specific methylation sites, which can then be used to guide the creation of syngenic DNA plasmids devoid of methylation and REase cleavage sites that are not degraded by the host (15). An additional study that used SMRTseq technology identified all DNA recognition sites and methylation patterns in multiple species of bacteria, followed by placing these sequences in a methylation cassette within a plasmid that was then incubated with purified enzymes to identify specific methylation patterns (76). Using this technique for highly recalcitrant strains of Fusobacterium would allow for the first true matching of methylation sites with F. nucleatum MTases outside of bioinformatic predictions. Finally, one advantage we believe that use of recombinant MTases has over this approach is that DNA methylation analysis and synthetic DNA-based plasmids do not need to be made for each strain, which can keep down costs. Ultimately, we believe these methods are complementary and can be used in combination to enhance the chances of genetic modification in highly recalcitrant strains of F. nucleatum.
Potential future studies include an investigation into the role of the type I MTase systems in plasmid methylation. In this area, we briefly tried to recombinantly express HsdM and HsdS from F. nucleatum subsp. nucleatum 23726 but had difficulty achieving a pure, soluble protein complex. In addition, we reported that we tried to use a type I restriction modification inhibitor (Lucigen) in our transformations of F. nucleatum subsp. nucleatum 23726, but this did not change the transformation efficiency (data not shown). On a final note of the potential contribution of type I R-M systems, F. nucleatum subsp. nucleatum 25586 had no type I systems and was still genetically intractable until we used type II and type III enzymes. However, many transformable strains of bacteria have been made hsdRMS negative, which should be considered in the future as a method to potentially make more efficiently transformable strains of Fusobacterium.
Two additional strategies that could be used to increase transformation efficiency would be to delete the known REases in target F. nucleatum strains and creating E. coli strains with chromosomal integrations of F. nucleatum MTases. One disadvantage of deleting REase genes in F. nucleatum is the need to first transform and create a genetic system to be able to subsequently knock out these genes. But once accomplished, an REase-free strain could potentially bypass the need to treat entering plasmid DNA with MTases. However, many of the type II MTases do not have a paired REases, as shown in Fig. 1C, and therefore it is difficult to understand what could be cleaving the unmethylated DNA sequences that correspond to specific enzymes. E. coli strains with chromosomal integrations of F. nucleatum MTases would allow for efficient in vivo methylation of target plasmids while also not introducing plasmids that could be incompatible with the origin of replication or antibiotic resistance of the target plasmid to be methylated. In addition, expanding beyond the realm of F. nucleatum to other Fusobacterium species, including F. necrophorum, could be key to understanding the pathogenicity of this species in Lemierre’s syndrome in humans (77), as well as serious organ infections in livestock (78).
In conclusion, we report that F. nucleatum MTases can be used to methylate plasmid DNA, which then allows for efficient transformation and gene deletion in the well-studied strain F. nucleatum subsp. nucleatum 23726 as well as in the previously genetically intractable strain F. nucleatum subsp. nucleatum 25586. The broader implications of this work are the enhanced ability to study the role of specific genes and corresponding virulence factors expressed by F. nucleatum during infection and disease. The methods in this study can be directly applied to target strains of interest within the scientific community and therefore provide a roadmap for discovery biology that could lead to better understanding of how to inhibit the disease-driving mechanisms of this oral, opportunistic pathogen.
MATERIALS AND METHODS
Bacterial strains and plasmids.
All E. coli strains utilized in these studies were grown aerobically overnight at 37°C on solid Luria-Bertani (LB) agar plates (10 g/liter NaCl, 5 g/liter tryptone, 10 g/liter yeast extract) or in liquid LB medium. Fusobacterium strains were grown on solid agar plates made with Columbia broth (Gibco), supplemented with hemin (5 μg/mL), menadione (0.5 μg/mL), and resazurin (1 μL/mL) under anaerobic conditions (90% N2, 5% H2, 5% CO2) at 37°C (designated CBHK medium). Liquid growths were inoculated from single F. nucleatum colonies and grown in CBHK liquid medium under anaerobic conditions. Where necessary, antibiotics were supplemented at the suggested concentrations: gentamicin, 20 μg/mL; carbenicillin, 100 μg/mL; chloramphenicol, 10 or 25 μg/mL; thiamphenicol, 5 μg/mL (CBHK plates); or streptomycin 50 μg/mL (CBHK plates). The plasmids and bacterial strains utilized in these experiments are listed in Table S3 and Table S4 in the supplemental material, respectively.
Identification and classification of F. nucleatum DNA methyltransferases.
REBASE, a curated database of restriction enzymes, was used to identify the DNA methyltransferases present in F. nucleatum subsp. nucleatum ATCC 23726 (GCA_003019875.1), F. nucleatum subsp. nucleatum ATCC 25586 (GCA_003019295.1), F. nucleatum subsp. animalis 7_1 (GCA_000158275.2), F. nucleatum subsp. animalis 4_8 (GCA_000400875.1), and F. nucleatum subsp. polymorphum 10953 (GCA_000153625.1) from the NCBI database. Type II and type III MTases were further bioinformatically characterized using NIH SMARTBLAST and pHMMER. SMARTBLAST and pHMMER provided conserved domains indicating functions of MTases. Phylogenetic analysis of F. nucleatum MTase genes identified in REBASE were downloaded from NCBI and the NCBI identification numbers are supplied in Table S2. The tree and analysis were done in Geneious Prime 2022.1.1 using the Geneious Tree Builder function.
Cloning, expression, and purification of MTases.
The MTases M.Fnn23.I, M.Fnn23.II, M.Fnn25.I, M.Fnn25.IV, and M.Fnn25.V were cloned into pET16b under the control of an isopropyl-β-d-thiogalactopyranoside (IPTG)-induced promoter for purification of the recombinant proteins using the C-terminal 6×His tag and benchtop metal affinity chromatography. In addition, M.Fnn23.I and M.Fnn23.II were cloned under the control of a constitutive promoter for continual expression in TOP10 E. coli cells to drive in vivo methylation of plasmids. All plasmids utilized and created in these studies are described in Table S3 along with the bacterial strains in Table S4 and primers in Table S5. The primers to clone the DNA methyltransferases were all ordered from Integrated DNA Technologies. For M.Fnn23.I and M.Fnn23.II, all constructs were made with E. coli, and codon-optimized synthetic DNA was used for PCR. For MTases from F. nucleatum subsp. nucleatum ATCC 25586, PCR was run with genomic DNA that was prepared with Wizard genomic DNA purification kits (Promega).
Genes were amplified by PCR, and products were purified utilizing a PCR purification kit (Biobasic) and digested for 2 h at 37°C along with pET16b, which was used as the expression vector and was obtained through EZ-10 spin column plasmid miniprep (Biobasic) with the restriction enzymes listed in Table S5 with their respective primers. The vector was then dephosphorylated with Antarctic phosphatase (FastAP; Thermo Fisher Scientific) for 1 h at 37°C. Digested products were purified utilizing a spin column and ligated by T4 DNA ligase (New England Biolabs) for 1 h at room temperature following the manufacturer's recommendations. Ligations were transformed into competent Mix&Go (Zymo Research) Top10 E. coli and plated on LB solid agar plates supplemented with 100 μg/mL carbenicillin (ampicillin). Confirmation of positive clones was performed by digestion and, if applicable, positive clones were then transformed into ARTIC(DE3) RIL or LOBSTR-BL21(DE3) RIL (79) for recombinant protein expression.
For protein expression, E. coli cells were grown in LB (15 g/liter NaCl, 15 g/liter tryptone, 10 g/liter yeast extract) medium at 37°C, 250 rpm shaking until the optical density (OD) reached 0.6. At an OD of 0.6, cells were induced with 50 μM IPTG (GoldBio). Expression was carried out at 8°C and cells were collected at 20 h after inoculation by centrifugation at 5,000 × g for 20 min at 4°C. Bacterial pellets were resuspended in a lysis buffer (20 mM Tris [pH 7.5], 400 mM NaCl, 20 mM imidazole). Bacteria were lysed with an EmulsiFlex-C3 homogenizer (Avestin) at 10,000 kPa. Unlysed cells and insoluble material were separated by centrifugation at 15,000 × g for 20 min at 4°C and then discarded. The supernatant containing the 6×His-tagged MTases was stirred with 6 mL of NiCl2-charged chelating Sepharose beads (GE Healthcare) for 30 min at 4°C. The column was washed with 400 mL of wash buffer (20 mM Tris [pH 7.5], 400 mM NaCl, 40 mM imidazole). After washing, the methyltransferases were eluted in 10 mL of elution buffer (20 mM Tris [pH 7.5], 400 mM NaCl, 250 mM imidazole). The purified protein was then directly put into dialysis in a buffer (20 mM Tris [pH 7.5], 150 mM NaCl, 10% glycerol). Protein concentrations were calculated using a Qubit fluorometer and bicinchoninic acid assays, followed by freezing at −80°C for long-term storage.
In vitro treatment of plasmid DNA with Fusobacterium DNA methyltransferases.
Plasmid DNA (35 to 40 μg), prepared from E. coli TOP10 using the EZ-10 Spin column plasmid DNA miniprep from Biobasic, was combined in a 30-μL reaction mixture with 160 μM S-adenosylmethionine (SAM; New England Biolabs), 1× Cutsmart buffer (New England Biolabs), and 1 μM of one or more MTases. The reaction mixes were incubated at 37°C for 2 h, and then plasmid was extracted by adding 1 volume of phenol:chloroform:isoamyl alcohol (25:24:1; bioWORLD) and vortexed for 20 s. Mixtures were then centrifuged at 16,000 × g for 5 min. Plasmid DNA was precipitated and washed with ethanol and dissolved in ultrapure water (bioWORLD), followed by further purification. Plasmid DNA purified from overnight expression or coexpression was isolated with an alkaline lysis and column purification technique using the EZ-10 spin column plasmid miniprep (Biobasic). Plasmid DNA was further purified for use in electroporation by precipitation overnight at −80°C in 75% ethanol with sodium acetate (pH 5.5) and 0.1 μg/mL glycogen. After a 3-h minimum incubation at −80°C, the sample was spun at 4°C for 30 min at 16,000 × g to pellet the DNA and washed five times with 70% ethanol carefully by spinning at 14,000 × g for 3 min. Pellet was then dried at room temperature for 10 to 13 min. Finally, 15 μL of ultrapure water was added and the mixtures were incubated at 37°C for 1 h to solubilize the pellet. DNA concentrations were determined using a NanoDrop spectrophotometer.
Coexpression of plasmid DNA with F. nucleatum MTases for in vivo methylation.
Using the expression vector (constitutive activity) pET16b with the DNA methyltransferase under an Anderson medium promoter (described in Table S3), we methylated pDJSVT13 in vivo. Both pET16b (gene 622 and gene 635) and pDJSVT13 were transformed into E. coli TOP10 and grown in LB (15 g/liter NaCl, 15 g/liter tryptone, 10g/liter yeast extract) at 37°C with 250 rpm shaking for 24 h.
REase protection assays.
Plasmid DNA (1 μg), prepared from E. coli TOP10 cells using the Biobasic minipreparation procedure, was combined with Cutsmart buffer (New England Biolabs; 50 mM potassium acetate, 20 mM Tris-acetate, 10 mM magnesium acetate, 100 μg/mL bovine serum albumin [pH 7.9]), 160 μM SAM (New England Biolabs), and 1 μM of the correspondent MTases. As a control, plasmid DNA (1 μg) was mock treated in reaction buffer without the methyltransferases. All samples were incubated 1 h at 37°C with the restriction enzymes, single cutters KpnI and MluI, or predicted restriction sites NlaIII and SfaNI (New England Biolabs [NEB]). For single-cut linearization, plasmid DNA was digested with restriction enzyme KpnI (NEB) following the manufacturer's instructions. After 2 h at 37°C, the ultrapure DNA underwent phenol-chloroform extraction and ethanol precipitation at −80°C, as described previously for ultrapure DNA purification. Samples were analyzed in a 1% agarose gel with ethidium bromide and imaged on a Syngene G:Box imager (Fig. 2C and D).
F. nucleatum transformation by electroporation.
All F. nucleatum strains were competently prepared by inoculating and growing a 100-mL anaerobic culture in CBHK medium to lag phase (A600 = 0.1) followed by centrifugation of bacteria at 3,200 × g for 10 min. The supernatant was removed, and the resulting pellet was washed three successive times utilizing 1 mL of ice-cold 20% glycerol in deionized H2O and 1 mM morpholinopropanesulfonic acid (MOPS) at 14,000 × g for 3 min. Bacterial pellet was then resuspended in a final volume of 80 μL of ice-cold 20% glycerol and 1 mM MOPS. Bacteria were transferred to cold, 1-mm electroporation cuvettes (Lonza), and 3 μg (concentration of >300 ng/μL) of plasmid was added before electroporating at 2.5 kV/cm, 50 μF, 360 Ω, using a BTX Electro Cell Manipulator 600 (Harvard Apparatus). The electroporated cells were promptly transferred by syringe into a sterile, anaerobic tube with 4 mL of recovery medium (CBHK; 1 mM MgCl2) and incubated at 37°C for 20 h with no shaking in an anaerobic chamber. After the recovery outgrowth, cells were centrifuged at 14,000 × g for 3 min, supernatant was removed, and pelleted cells were resuspended in 0.2 mL of recovery medium. Resuspension was plated on CBHK plates with 5 μg/mL thiamphenicol and incubated in an anaerobic 37°C incubator for 2 days for colony growth. The transformation efficiency represented the number of thiamphenicol- or streptomycin-resistant colonies per microgram of DNA. Electroporation was conducted in triplicate as independent experiments.
Utilization of plasmid methylation to enable a galactose-selectable gene deletion system in F. nucleatum subsp. nucleatum 25586.
A galactose-selectable gene deletion system for F. nucleatum subsp. nucleatum 23726 was previously developed in our lab and reported in detail by Casasanta et al. (66). As F. nucleatum subsp. nucleatum 23726 and F. nucleatum subsp. nucleatum 25586 are extremely similar at the DNA level, the plasmid pDJSVT13 that was previously used to delete the galKT operon in F. nucleatum subsp. nucleatum 23726 was also used on F. nucleatum subsp. nucleatum 25586 because of 100% nucleotide identity in the up- and downstream regions cloned for homologous recombination and gene deletion. pDJSVT13 was conditioned with five MTase enzymes (M.Fnn23.I, M.Fnn23.II, M.Fnn25.I, M.Fnn25.IV, and M.Fnn25.V) using the same conditions as describe above for the in vitro methylation protocol. Ultrapure DNA (3 μg) was electroporated (2.5 kV, 50-μF capacitance, 360-Ω resistance, 0.2-cm cuvette) into competent F. nucleatum subsp. nucleatum 25586, and single chromosomal crossovers of the pDJSVT13 plasmid were selected on thiamphenicol. Colonies were then inoculated into antibiotic-free CBHK medium overnight at 37°C to allow for a second crossover event, which effectively deleted the target gene and also the remaining plasmid that was integrated into the chromosome. Next, 100 μL from this culture was streaked on solid medium containing 0.25% 2-deoxy-d-galactose to select for galKT gene deletions, as the absence of the galT gene makes 2-deoxy-d-galactose nontoxic to F. nucleatum subsp. nucleatum. galKT gene deletions were verified by PCR and Sanger sequencing. This new strain, F. nucleatum subsp. nucleatum 25586 ΔgalKT, which we have named TNVT2501, is now the base strain used to create all future targeted gene deletions. Bacterial transformation of TNVT2501 allows for initial chromosomal integration and selection with thiamphenicol, followed by selection for double-crossover gene deletions on solid medium containing 3% galactose. We have shown that deletion of the galKT operon in F. nucleatum subsp. nucleatum 25586 does not result in altered fitness.
Creating F. nucleatum subsp. nucleatum 25586 Δfap2 and F. nucleatum subsp. nucleatum 25586 ΔfadA.
As a proof of concept, we next generated targeted gene deletions in the F. nucleatum subsp. nucleatum 25586 ΔgalKT background and in the two most well-studied F. nucleatum virulence factors: fap2 and fadA. The first step was to use the plasmid pDJSVT7, which contained a FLAG::galK gene under the control of an F. necrophorum promoter. Briefly, 750 bp directly upstream and downstream of the fap2 and fadA genes was amplified by PCR and fused by overlap-extension-PCR. PCR product was digested with KpnI/MluI ligated into pDJSVT7 digested with the same enzymes, followed by transformation into TOP10 E. coli and selection on LB plates containing chloramphenicol. Positive clones were identified by restriction digest and Sanger sequencing to verify the new gene deletion plasmids pTNVT2501 (fap2) and pTNVT2502 (fadA) (Fig. 6A). pTNVT2501 and pTNVT2502 were next electroporated (3 μg of DNA, 2.5 kV, 50-μF capacitance, 360-Ω resistance, 0.2-cm cuvette) into competent F. nucleatum subsp. nucleatum 25586 ΔgalKT, and chromosomal integration was selected for on thiamphenicol (single chromosomal crossover), followed by selection on solid medium containing 3% galactose, which produced either complete gene deletions or wild-type bacteria revertants. Gene deletions were verified by PCR and Sanger sequencing (Fig. 6). The new strain names are TNVT02 and TNVT03 for the Δfap2 and ΔfadA mutants of F. nucleatum subsp. nucleatum 25586. We showed that this system was accurate down to the single base level for creating clean genome excisions that therefore allowed for the deletion of an unlimited number of genes.
Complementation of a fadA gene deletion in F. nucleatum subsp. nucleatum 25586.
We previously created the gene complementation vector pDJSVT11 to create single-copy chromosomal complementation at a chromosomal location within the arsB gene (66). Our previously developed plasmid, pDJSVT32, was used to complement F. nucleatum subsp. nucleatum 23726 ΔgalKT fadA and was also used to complement F. nucleatum subsp. nucleatum 25586 ΔgalKT fadA (TNVT03). Briefly, this plasmid contained a 1,000-bp central region of the arsB gene, driving homologous recombination, which resulted in chromosomal insertion of the thiamphenicol resistance plasmid. Complementation was selected for on CBHK plates containing thiamphenicol, followed by inoculation into liquid CHBK containing thiamphenicol. Complementation was further verified by PCR of the fadA gene as shown in Fig. 6H.
Statistical analysis.
All statistical analyses were performed in GraphPad Prism version 8.2.1. For single analysis, an unpaired Student's t test was used. For grouped analyses, two-way analysis of variance (ANOVA) was used. In each case, the following P values correspond to symbols in figures: ns, P > 0.05, *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. To obtain statistics, all studies were performed as three independent biological experiments. For all experiments in which statistical analysis was applied, an N of 3 independent experiments was used (see details in figure legends).
Data availability.
Materials are available upon reasonable request with a material transfer agreement with Virginia Tech for bacterial strains or through the Addgene repository for plasmids. Plasmid sequences and maps can be accessed on our Open Science Framework repository at https://osf.io/yrbgj/?view_only=24a28a13c5194d22957f2fae1ce2f84b.
ACKNOWLEDGMENTS
This research was supported by the National Institutes of Health through an NCI R21 Award (grant number 1R21CA238630-01A1 to D.J.S.), the College of Agriculture and Life Sciences at Virginia Tech (to D.J.S.), the Institute for Critical Technology and Applied Science at Virginia Tech (to D.J.S.), and the USDA National Institute of Food and Agriculture (to D.J.S.). Select figures were made with a paid subscription of Biorender.com.
A.U., T.T.D.N., and B.E.S. curated and analyzed the data, designed and optimized the methodology, and wrote, reviewed, and edited the manuscript. K.J.W. and B.W. curated the data and reviewed and edited the manuscript. D.J.S. helped conceptualize, supervise, and acquire funding for the study, performed data analysis, curated and analyzed the data, designed and optimized the methodology, and wrote, reviewed, and edited the manuscript.
Footnotes
Supplemental material is available online only.
Contributor Information
Daniel J. Slade, Email: dslade@vt.edu.
Joseph Bondy-Denomy, University of California San Francisco.
REFERENCES
- 1.Forsberg KJ, Malik HS. 2018. Microbial genomics: the expanding universe of bacterial defense systems. Curr Biol 28:R361–R364. 10.1016/j.cub.2018.02.053. [DOI] [PubMed] [Google Scholar]
- 2.Puigbo P, Makarova KS, Kristensen DM, Wolf YI, Koonin EV. 2017. Reconstruction of the evolution of microbial defense systems. BMC Evol Biol 17:94. 10.1186/s12862-017-0942-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Makarova KS, Wolf YI, Koonin EV. 2013. Comparative genomics of defense systems in archaea and bacteria. Nucleic Acids Res 41:4360–4377. 10.1093/nar/gkt157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sneppen K, Semsey S, Seshasayee AS, Krishna S. 2015. Restriction modification systems as engines of diversity. Front Microbiol 6:528. 10.3389/fmicb.2015.00528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vasu K, Nagaraja V. 2013. Diverse functions of restriction-modification systems in addition to cellular defense. Microbiol Mol Biol Rev 77:53–72. 10.1128/MMBR.00044-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kobayashi I. 2001. Behavior of restriction-modification systems as selfish mobile elements and their impact on genome evolution. Nucleic Acids Res 29:3742–3756. 10.1093/nar/29.18.3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ofir G, Melamed S, Sberro H, Mukamel Z, Silverman S, Yaakov G, Doron S, Sorek R. 2018. DISARM is a widespread bacterial defence system with broad anti-phage activities. Nat Microbiol 3:90–98. 10.1038/s41564-017-0051-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Knott GJ, Doudna JA. 2018. CRISPR-Cas guides the future of genetic engineering. Science 361:866–869. 10.1126/science.aat5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jansen R, Embden JD, Gaastra W, Schouls LM. 2002. Identification of genes that are associated with DNA repeats in prokaryotes. Mol Microbiol 43:1565–1575. 10.1046/j.1365-2958.2002.02839.x. [DOI] [PubMed] [Google Scholar]
- 10.Haurwitz RE, Jinek M, Wiedenheft B, Zhou K, Doudna JA. 2010. Sequence- and structure-specific RNA processing by a CRISPR endonuclease. Science 329:1355–1358. 10.1126/science.1192272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Makarova KS, Haft DH, Barrangou R, Brouns SJJ, Charpentier E, Horvath P, Moineau S, Mojica FJM, Wolf YI, Yakunin AF, van der Oost J, Koonin EV. 2011. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol 9:467–477. 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goldfarb T, Sberro H, Weinstock E, Cohen O, Doron S, Charpak-Amikam Y, Afik S, Ofir G, Sorek R. 2015. BREX is a novel phage resistance system widespread in microbial genomes. EMBO J 34:169–183. 10.15252/embj.201489455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barrangou R, van der Oost J. 2015. Bacteriophage exclusion, a new defense system. EMBO J 34:134–135. 10.15252/embj.201490620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arber W, Hattman S, Dussoix D. 1963. On the host-controlled modification of bacteriophage lambda. Virology 21:30–35. 10.1016/0042-6822(63)90300-3. [DOI] [PubMed] [Google Scholar]
- 15.Johnston CD, Cotton SL, Rittling SR, Starr JR, Borisy GG, Dewhirst FE, Lemon KP. 2019. Systematic evasion of the restriction-modification barrier in bacteria. Proc Natl Acad Sci USA 116:11454–11459. 10.1073/pnas.1820256116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koonin EV, Makarova KS, Wolf YI. 2017. Evolutionary genomics of defense systems in Archaea and Bacteria. Annu Rev Microbiol 71:233–261. 10.1146/annurev-micro-090816-093830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Loenen WA, Dryden DT, Raleigh EA, Wilson GG. 2014. Type I restriction enzymes and their relatives. Nucleic Acids Res 42:20–44. 10.1093/nar/gkt847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilson GG. 1988. Type II restriction-modification systems. Trends Genet 4:314–318. 10.1016/0168-9525(88)90109-6. [DOI] [PubMed] [Google Scholar]
- 19.Donahue J.P, Peek RM, Jr.. 2001. Restriction and modification systems. In Mobley HLT, Mendz GL, Hazell SL (ed), Helicobacter pylori: physiology and genetics. ASM Press, Washington, DC. [PubMed] [Google Scholar]
- 20.Falkler WA, Jr, Smoot CN, Mongiello JR. 1982. Attachment of cell fragments of Fusobacterium nucleatum to oral epithelial cells, gingival fibroblasts and white blood cells. Arch Oral Biol 27:553–559. 10.1016/0003-9969(82)90069-3. [DOI] [PubMed] [Google Scholar]
- 21.Sitaraman R. 2016. The role of DNA restriction-modification systems in the biology of Bacillus anthracis. Front Microbiol 7:11. 10.3389/fmicb.2016.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loenen WA, Dryden DT, Raleigh EA, Wilson GG, Murray NE. 2014. Highlights of the DNA cutters: a short history of the restriction enzymes. Nucleic Acids Res 42:3–19. 10.1093/nar/gkt990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lepikhov K, Tchernov A, Zheleznaja L, Matvienko N, Walter J, Trautner TA. 2001. Characterization of the type IV restriction modification system BspLU11III from Bacillus sp. LU11. Nucleic Acids Res 29:4691–4698. 10.1093/nar/29.22.4691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kostic AD, Gevers D, Pedamallu CS, Michaud M, Duke F, Earl AM, Ojesina AI, Jung J, Bass AJ, Tabernero J, Baselga J, Liu C, Shivdasani RA, Ogino S, Birren BW, Huttenhower C, Garrett WS, Meyerson M. 2012. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res 22:292–298. 10.1101/gr.126573.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Castellarin M, Warren RL, Freeman JD, Dreolini L, Krzywinski M, Strauss J, Barnes R, Watson P, Allen-Vercoe E, Moore RA, Holt RA. 2012. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res 22:299–306. 10.1101/gr.126516.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kostic AD, Chun E, Robertson L, Glickman JN, Gallini CA, Michaud M, Clancy TE, Chung DC, Lochhead P, Hold GL, El-Omar EM, Brenner D, Fuchs CS, Meyerson M, Garrett WS. 2013. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe 14:207–215. 10.1016/j.chom.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Slots J. 1977. The predominant cultivable microflora of advanced periodontitis. Scand J Dent Res 85:114–121. 10.1111/j.1600-0722.1977.tb00541.x. [DOI] [PubMed] [Google Scholar]
- 28.Chen Y, Shi T, Li Y, Huang L, Yin D. 2022. Fusobacterium nucleatum: the opportunistic pathogen of periodontal and peri-implant diseases. Front Microbiol 13:860149. 10.3389/fmicb.2022.860149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kearney A, Knoll B. 2015. Myopericarditis associated with Fusobacterium nucleatum-caused liver abscess. Infect Dis (Lond) 47:187–189. 10.3109/00365548.2014.969306. [DOI] [PubMed] [Google Scholar]
- 30.Toumeh N, Mudireddy M, Smith B, Guerrero DM. 2021. Fatal case of liver and brain abscesses due to Fusobacterium nucleatum. Cureus 13:e19671. 10.7759/cureus.19671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jayasimhan D, Wu L, Huggan P. 2017. Fusobacterial liver abscess: a case report and review of the literature. BMC Infect Dis 17:440. 10.1186/s12879-017-2548-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Young EJ, Harper WK, Taylor RL. 1977. Hepatic necrobacillosis. Report of a case resembling metastatic tumor. Arch Intern Med 137:804–807. 10.1001/archinte.137.6.804. [DOI] [PubMed] [Google Scholar]
- 33.Xu B, Han YW. 2022. Oral bacteria, oral health, and adverse pregnancy outcomes. Periodontol 2000 89:181–189. 10.1111/prd.12436. [DOI] [PubMed] [Google Scholar]
- 34.Parhi L, Abed J, Shhadeh A, Alon-Maimon T, Udi S, Ben-Arye SL, Tam J, Parnas O, Padler-Karavani V, Goldman-Wohl D, Yagel S, Mandelboim O, Bachrach G. 2022. Placental colonization by Fusobacterium nucleatum is mediated by binding of the Fap2 lectin to placentally displayed Gal-GalNAc. Cell Rep 38:110537. 10.1016/j.celrep.2022.110537. [DOI] [PubMed] [Google Scholar]
- 35.Han YW, Redline RW, Li M, Yin L, Hill GB, McCormick TS. 2004. Fusobacterium nucleatum induces premature and term stillbirths in pregnant mice: implication of oral bacteria in preterm birth. Infect Immun 72:2272–2279. 10.1128/IAI.72.4.2272-2279.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Salvucci M, Crawford N, Stott K, Bullman S, Longley DB, Prehn JHM. 2022. Patients with mesenchymal tumours and high Fusobacteriales prevalence have worse prognosis in colorectal cancer (CRC). Gut 71:1600–1612. 10.1136/gutjnl-2021-325193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Queen J, Domingue JC, White JR, Stevens C, Udayasuryan B, Nguyen TTD, Wu S, Ding H, Fan H, McMann M, Corona A, Larman TC, Verbridge SS, Housseau F, Slade DJ, Drewes JL, Sears CL. 2022. Comparative analysis of colon cancer-derived Fusobacterium nucleatum subspecies: inflammation and colon tumorigenesis in murine models. mBio 13:e02991-21. 10.1128/mbio.02991-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Engevik MA, Danhof HA, Ruan W, Engevik AC, Chang-Graham AL, Engevik KA, Shi Z, Zhao Y, Brand CK, Krystofiak ES, Venable S, Liu X, Hirschi KD, Hyser JM, Spinler JK, Britton RA, Versalovic J. 2021. Fusobacterium nucleatum secretes outer membrane vesicles and promotes intestinal inflammation. mBio 12. 10.1128/mBio.02706-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ternes D, Tsenkova M, Pozdeev VI, Meyers M, Koncina E, Atatri S, Schmitz M, Karta J, Schmoetten M, Heinken A, Rodriguez F, Delbrouck C, Gaigneaux A, Ginolhac A, Nguyen TTD, Grandmougin L, Frachet-Bour A, Martin-Gallausiaux C, Pacheco M, Neuberger-Castillo L, Miranda P, Zuegel N, Ferrand J-Y, Gantenbein M, Sauter T, Slade DJ, Thiele I, Meiser J, Haan S, Wilmes P, Letellier E. 2022. The gut microbial metabolite formate exacerbates colorectal cancer progression. Nat Metab 4:458–475. 10.1038/s42255-022-00558-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yin H, Miao Z, Wang L, Su B, Liu C, Jin Y, Wu B, Han H, Yuan X. 2022. Fusobacterium nucleatum promotes liver metastasis in colorectal cancer by regulating the hepatic immune niche and altering gut microbiota. Aging (Albany NY) 14:1941–1958. 10.18632/aging.203914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li Z, Shi C, Zheng J, Guo Y, Fan T, Zhao H, Jian D, Cheng X, Tang H, Ma J. 2021. Fusobacterium nucleatum predicts a high risk of metastasis for esophageal squamous cell carcinoma. BMC Microbiol 21:301. 10.1186/s12866-021-02352-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang Y, Jobin C. 2020. Far reach of Fusobacterium nucleatum in cancer metastasis. Gut 10.1136/gutjnl-2020-323496. [DOI] [PubMed] [Google Scholar]
- 43.Geng F, Zhang Y, Lu Z, Zhang S, Pan Y. 2020. Fusobacterium nucleatum caused DNA damage and promoted cell proliferation by the Ku70/p53 pathway in oral cancer cells. DNA Cell Biol 39:144–151. 10.1089/dna.2019.5064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guo P, Tian Z, Kong X, Yang L, Shan X, Dong B, Ding X, Jing X, Jiang C, Jiang N, Yu Y. 2020. FadA promotes DNA damage and progression of Fusobacterium nucleatum-induced colorectal cancer through up-regulation of chk2. J Exp Clin Cancer Res 39:202. 10.1186/s13046-020-01677-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Okita Y, Koi M, Takeda K, Ross R, Mukherjee B, Koeppe E, Stoffel EM, Galanko JA, McCoy AN, Keku TO, Okugawa Y, Kitajima T, Toiyama Y, Martens E, Carethers JM. 2020. Fusobacterium nucleatum infection correlates with two types of microsatellite alterations in colorectal cancer and triggers DNA damage. Gut Pathog 12:46. 10.1186/s13099-020-00384-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu T, Guo F, Yu Y, Sun T, Ma D, Han J, Qian Y, Kryczek I, Sun D, Nagarsheth N, Chen Y, Chen H, Hong J, Zou W, Fang J-Y. 2017. Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell 170:548–563.e516. 10.1016/j.cell.2017.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang S, Yang Y, Weng W, Guo B, Cai G, Ma Y, Cai S. 2019. Fusobacterium nucleatum promotes chemoresistance to 5-fluorouracil by upregulation of BIRC3 expression in colorectal cancer. J Exp Clin Cancer Res 38:14. 10.1186/s13046-018-0985-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee D-W, Han S-W, Kang J-K, Bae JM, Kim H-P, Won J-K, Jeong S-Y, Park KJ, Kang GH, Kim T-Y. 2018. Association between Fusobacterium nucleatum, pathway mutation, and patient prognosis in colorectal cancer. Ann Surg Oncol 25:3389–3395. 10.1245/s10434-018-6681-5. [DOI] [PubMed] [Google Scholar]
- 49.Mima K, Nishihara R, Qian ZR, Cao Y, Sukawa Y, Nowak JA, Yang J, Dou R, Masugi Y, Song M, Kostic AD, Giannakis M, Bullman S, Milner DA, Baba H, Giovannucci EL, Garraway LA, Freeman GJ, Dranoff G, Garrett WS, Huttenhower C, Meyerson M, Meyerhardt JA, Chan AT, Fuchs CS, Ogino S. 2016. Fusobacterium nucleatum in colorectal carcinoma tissue and patient prognosis. Gut 65:1973–1980. 10.1136/gutjnl-2015-310101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kinder Haake S, Yoder S, Gerardo SH. 2006. Efficient gene transfer and targeted mutagenesis in Fusobacterium nucleatum. Plasmid 55:27–38. 10.1016/j.plasmid.2005.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu C, Al Mamun AAM, Luong TT, Hu B, Gu J, Lee JH, D’Amore M, Das A, Ton-That H. 2018. Forward genetic dissection of biofilm development by Fusobacterium nucleatum: novel functions of cell division proteins FtsX and EnvC. mBio 9. 10.1128/mBio.00360-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Peluso EA, Scheible M, Ton-That H, Wu C. 2020. Genetic manipulation and virulence assessment of Fusobacterium nucleatum. Curr Protoc Microbiol 57:e104. 10.1002/cpmc.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Haake SK, Yoder SC, Attarian G, Podkaminer K. 2000. Native plasmids of Fusobacterium nucleatum: characterization and use in development of genetic systems. J Bacteriol 182:1176–1180. 10.1128/JB.182.4.1176-1180.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Han YW, Ikegami A, Chung P, Zhang L, Deng CX. 2007. Sonoporation is an efficient tool for intracellular fluorescent dextran delivery and one-step double-crossover mutant construction in Fusobacterium nucleatum. Appl Environ Microbiol 73:3677–3683. 10.1128/AEM.00428-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.He X, Jiang K, Xiao J, Lian S, Chen Y, Wu R, Wang L, Sun D, Guo D. 2022. Interaction of 43K OMP of Fusobacterium necrophorum with fibronectin mediates adhesion to bovine epithelial cells. Vet Microbiol 266:109335. 10.1016/j.vetmic.2022.109335. [DOI] [PubMed] [Google Scholar]
- 56.Claypool BM, Yoder SC, Citron DM, Finegold SM, Goldstein EJC, Haake SK. 2010. Mobilization and prevalence of a Fusobacterial plasmid. Plasmid 63:11–19. 10.1016/j.plasmid.2009.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhao H, Xu L, Rong Q, Xu Z, Ding Y, Zhang Y, Wu Y, Li B, Ji X. 2018. Application of methylation in improving plasmid transformation into Helicobacter pylori. J Microbiol Methods 150:18–23. 10.1016/j.mimet.2018.05.016. [DOI] [PubMed] [Google Scholar]
- 58.Monk IR, Shah IM, Xu M, Tan MW, Foster TJ. 2012. Transforming the untransformable: application of direct transformation to manipulate genetically Staphylococcus aureus and Staphylococcus epidermidis. mBio 3. 10.1128/mBio.00277-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang G, Wang W, Deng A, Sun Z, Zhang Y, Liang Y, Che Y, Wen T. 2012. A mimicking-of-DNA-methylation-patterns pipeline for overcoming the restriction barrier of bacteria. PLoS Genet 8:e1002987. 10.1371/journal.pgen.1002987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yasui K, Kano Y, Tanaka K, Watanabe K, Shimizu-Kadota M, Yoshikawa H, Suzuki T. 2009. Improvement of bacterial transformation efficiency using plasmid artificial modification. Nucleic Acids Res 37:e3. 10.1093/nar/gkn884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Roberts RJ, Vincze T, Posfai J, Macelis D. 2007. REBASE–enzymes and genes for DNA restriction and modification. Nucleic Acids Res 35:D269–D270. 10.1093/nar/gkl891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nakagaki H, Sekine S, Terao Y, Toe M, Tanaka M, Ito H-O, Kawabata S, Shizukuishi S, Fujihashi K, Kataoka K. 2010. Fusobacterium nucleatum envelope protein FomA is immunogenic and binds to the salivary statherin-derived peptide. Infect Immun 78:1185–1192. 10.1128/IAI.01224-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sanders BE, Umana A, Lemkul JA, Slade DJ. 2018. FusoPortal: an interactive repository of hybrid MinION-sequenced Fusobacterium genomes improves gene identification and characterization. mSphere 3. 10.1128/mSphere.00379-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 65.Gordeeva J, Morozova N, Sierro N, Isaev A, Sinkunas T, Tsvetkova K, Matlashov M, Truncaite L, Morgan RD, Ivanov NV, Siksnys V, Zeng L, Severinov K. 2019. BREX system of Escherichia coli distinguishes self from non-self by methylation of a specific DNA site. Nucleic Acids Res 47:253–265. 10.1093/nar/gky1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Casasanta MA, et al. 2020. Fusobacterium nucleatum host-cell binding and invasion induces IL-8 and CXCL1 secretion that drives colorectal cancer cell migration. Sci Signal 13:eaba9157. 10.1126/scisignal.aba9157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Anton BP, Mongodin EF, Agrawal S, Fomenkov A, Byrd DR, Roberts RJ, Raleigh EA. 2015. Complete genome sequence of ER2796, a DNA methyltransferase-deficient strain of Escherichia coli K-12. PLoS One 10:e0127446. 10.1371/journal.pone.0127446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Martinez-Garcia E, Aparicio T, de Lorenzo V, Nikel PI. 2014. New transposon tools tailored for metabolic engineering of gram-negative microbial cell factories. Front Bioeng Biotechnol 2:46. 10.3389/fbioe.2014.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Barker HA, Kahn JM, Hedrick L. 1982. Pathway of lysine degradation in Fusobacterium nucleatum. J Bacteriol 152:201–207. 10.1128/jb.152.1.201-207.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Abed J, Emgård JEM, Zamir G, Faroja M, Almogy G, Grenov A, Sol A, Naor R, Pikarsky E, Atlan KA, Mellul A, Chaushu S, Manson AL, Earl AM, Ou N, Brennan CA, Garrett WS, Bachrach G. 2016. Fap2 mediates Fusobacterium nucleatum colorectal adenocarcinoma enrichment by binding to tumor-expressed Gal-GalNAc. Cell Host Microbe 20:215–225. 10.1016/j.chom.2016.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Coppenhagen-Glazer S, Sol A, Abed J, Naor R, Zhang X, Han YW, Bachrach G. 2015. Fap2 of Fusobacterium nucleatum is a galactose-inhibitable adhesin involved in coaggregation, cell adhesion, and preterm birth. Infect Immun 83:1104–1113. 10.1128/IAI.02838-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Meng Q, Gao Q, Mehrazarin S, Tangwanichgapong K, Wang Y, Huang Y, Pan Y, Robinson S, Liu Z, Zangiabadi A, Lux R, Papapanou PN, Guo XE, Wang H, Berchowitz LE, Han YW. 2021. Fusobacterium nucleatum secretes amyloid-like FadA to enhance pathogenicity. EMBO Rep 22:e52891. 10.15252/embr.202152891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Han YW, Ikegami A, Rajanna C, Kawsar HI, Zhou Y, Li M, Sojar HT, Genco RJ, Kuramitsu HK, Deng CX. 2005. Identification and characterization of a novel adhesin unique to oral fusobacteria. J Bacteriol 187:5330–5340. 10.1128/JB.187.15.5330-5340.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cai J, Salmon K, DuBow MS. 1998. A chromosomal ars operon homologue of Pseudomonas aeruginosa confers increased resistance to arsenic and antimony in Escherichia coli. Microbiology (Reading) 144:2705–2729. 10.1099/00221287-144-10-2705. [DOI] [PubMed] [Google Scholar]
- 75.Wion D, Casadesus J. 2006. N6-methyl-adenine: an epigenetic signal for DNA-protein interactions. Nat Rev Microbiol 4:183–192. 10.1038/nrmicro1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jensen TØ, Tellgren-Roth C, Redl S, Maury J, Jacobsen SAB, Pedersen LE, Nielsen AT. 2019. Genome-wide systematic identification of methyltransferase recognition and modification patterns. Nat Commun 10:3311. 10.1038/s41467-019-11179-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Foo EC, Tanti M, Cliffe H, Randall M. 2021. Lemierre's syndrome. Pract Neurol 21:442–444. 10.1136/practneurol-2021-002928. [DOI] [PubMed] [Google Scholar]
- 78.Pillai DK, Amachawadi RG, Baca G, Narayanan SK, Nagaraja TG. 2021. Leukotoxin production by Fusobacterium necrophorum strains in relation to severity of liver abscesses in cattle. Anaerobe 69:102344. 10.1016/j.anaerobe.2021.102344. [DOI] [PubMed] [Google Scholar]
- 79.Andersen KR, Leksa NC, Schwartz TU. 2013. Optimized E. coli expression strain LOBSTR eliminates common contaminants from His-tag purification. Proteins 81:1857–1861. 10.1002/prot.24364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Suzuki H. 2012. Host-mimicking strategies in DNA methylation for improved bacterial transformation. In Dricu A (ed), Methylation: from DNA, RNA and histones to diseases and treatment. IntechOpen, London, England. 10.5772/51691. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 and Tables S1 to S5. Download jb.00279-22-s0001.pdf, PDF file, 0.4 MB (441.3KB, pdf)
Data Availability Statement
Materials are available upon reasonable request with a material transfer agreement with Virginia Tech for bacterial strains or through the Addgene repository for plasmids. Plasmid sequences and maps can be accessed on our Open Science Framework repository at https://osf.io/yrbgj/?view_only=24a28a13c5194d22957f2fae1ce2f84b.