Abstract
Diabetic kidney disease (DKD) is the leading cause of kidney failure and is associated with substantial risk of cardiovascular disease, morbidity, and mortality. Traditionally, DKD prevention and management have focused on addressing hyperglycemia, hypertension, obesity, and renin-angiotensin system activation as important risk factors for disease. Over the last decade, sodium-glucose cotransporter-2 inhibitors and glucagon-like peptide-1 receptor agonists have been shown to meaningfully reduce risk of diabetes-related kidney and cardiovascular complications. Additional agents demonstrating benefit in DKD such as non-steroidal mineralocorticoid receptor antagonists and endothelin A receptor antagonists are further contributing to the growing arsenal of DKD therapies. With the availability of greater therapeutic options comes the opportunity to individually optimize DKD prevention and management. Novel applications of transcriptomic, proteomic, and metabolomic/lipidomic technologies, as well as use of artificial intelligence and reinforced learning methods through consortia such as the Kidney Precision Medicine Project and focused studies in established cohorts hold tremendous promise for advancing our understanding and treatment of DKD. Specifically, enhanced understanding of the molecular mechanisms underlying DKD pathophysiology may allow for the identification of new mechanism-based DKD subtypes and the development and implementation of targeted therapies. Implementation of personalized care approaches has the potential to revolutionize DKD care.
PROBLEM
Diabetes affects approximately 10% of the overall United States population, amounting to roughly 32 million people.1 Diabetic kidney disease (DKD), defined as persistent albuminuria and/or decreased estimated glomerular filtration rate (eGFR) attributed to diabetes, occurs in approximately 30% of people with type 1 and type 2 diabetes (T1D and T2D, respectively).2,3 DKD is the leading cause of kidney failure in the United States, accounting for 47% of incident cases of kidney failure in 2019.4 Based on a 2021 report from the International Diabetes Federation, the prevalence of diabetes is projected to increase both in the United States and worldwide through 2045, ultimately affecting 780 million people globally.1 With this, the burden of DKD and diabetes-related kidney failure is expected to proportionally increase over time.5,6 Further contributing to the growing prevalence of DKD is population aging, compounded with decreasing diabetes-related mortality.6,7
DKD is associated with substantially increased risk of cardiovascular disease and related morbidity and mortality. One study using data from the third National Health and Nutrition Examination Survey (NHANES III) found that the absolute risk of cardiovascular mortality over 10 years was 13% higher among those with T2D and kidney disease compared to those with T2D alone, adjusting for age, sex, race, smoking status, blood pressure, and cholesterol.8 Similarly, increased mortality has been reported in adults with T1D and kidney disease compared to adults with T1D and normal kidney function in longitudinal cohort studies based in the United States and Finland.9,10 Furthermore, cardiovascular risk and mortality risk increase progressively with lower GFR and higher albuminuria.
While coronary heart disease and diastolic dysfunction are the most commonly observed cardiovascular complications among people with chronic kidney disease (CKD), increased risk of peripheral vascular disease and stroke has also been observed. Accordingly, the American Heart Association has recognized those with CKD as belonging to the “highest-risk group” for the prevention, identification, and treatment of cardiovascular risk factors.11 The increased risk of cardiovascular disease seen with CKD is believed to result from the presence of both traditional cardiovascular risk factors (hypertension, endothelial dysfunction, chronic inflammation) and CKD-specific metabolic and hormonal derangements (mineral metabolism disorders, anemia, renin-angiotensin system [RAS] activation).12 However, the specific mechanisms by which kidney dysfunction contributes to cardiovascular risk have yet to be fully elucidated.
DKD also meaningfully contributes to population-based health care disparities in the United States. The prevalence of diagnosed T2D is higher among American Indian, Black, Hispanic, and Asian adults compared to White adults, placing these populations at greater risk of related complications, including DKD.13 Additionally, members of minority racial and ethnic groups with diabetes are more likely to develop DKD and progress to kidney failure compared to their White counterparts.4,14 Heightened risk of kidney disease in these populations is in part due to low socioeconomic status and reduced access to health care resulting from the perpetuation of systemic social and environmental barriers.
The substantial, growing burden of DKD has been and will continue to be an important driver of increased morbidity and mortality in the United States and worldwide. Here, we detail advances made in recent decades in the care and prevention of DKD and highlight promising innovations in DKD research that are primed to yield meaningful progress in the field. Specifically, we discuss how novel applications of transcriptomic, proteomic, and lipidomic technologies combined with improved care implementation will culminate an enhanced understanding of DKD pathophysiology and the development of targeted diagnostic, prognostic, and therapeutic strategies. Ultimately, these efforts represent the ushering of a new era of precision medicine for DKD.
BACKGROUND
Until recently, DKD prevention and management have focused primarily on addressing hyperglycemia as an important risk factor for disease. Early, intensive glycemic control was demonstrated to reduce the risk of DKD and other microvascular complications in both T1D (as amply supported by the Diabetes Control and Complications Trial and Epidemiology of Diabetes Interventions and Complications [DCCT/EDIC]) and T2D (in the UK Prospective Diabetes Study [UKPDS] and ADVANCE).15–17 In the ADVANCE trial of T2D, tight glucose control also reduced the risk of kidney failure.17 In accordance with these recognized benefits, guidelines from the American Diabetes Association, European Society for the Study of Diabetes (ADA/EASD), and other professional organizations emphasize maintenance of tight glycemic control, with a recommended individualized hemoglobin A1c goal of less than approximately 7%.18 However, this must be balanced with risk of severe hypoglycemia, which may occur with specific glucose-lowering regimens in an effort to achieve intensive glycemic control and may contribute to poor outcomes.19 Additionally, along with management of blood glucose, treatment of hypertension and obesity have been recognized as important for reducing risk of DKD incidence and progression.16,20
Since the time RAS inhibition was discovered to slow decline in kidney function in diabetes, angiotensin converting enzyme inhibitors (ACEi) and angiotensin receptor blockers (ARB) have been a mainstay of DKD management. In randomized, controlled trials, RAS inhibition slowed eGFR decline and reduced albuminuria progression.21 These benefits are independent of blood-pressure lowering. While effective for preventing the progression of established DKD (with albuminuria), RAS inhibition has not been effective at reducing the incidence of DKD.22
Over the last decade, novel, glucose-lowering agents have been shown to meaningfully reduce the risk of kidney and cardiovascular complications of T2D, revolutionizing diabetes care (Figure 1). Sodium-glucose cotransporter-2 inhibitors (SGLT2i) have been demonstrated to significantly reduce the risk of progressive kidney function decline, incident kidney failure, cardiovascular death, and heart failure hospitalizations in adults with T2D with and without kidney disease.23 The profound renoprotective effect of SGLT2i has been observed in both primary cardiovascular and kidney outcome trials and is consistent across baseline eGFR and albuminuria subgroups.24 Trials of glucagon-like peptide-1 receptor agonists (GLP-1 RA) in T2D populations have also demonstrated significant kidney and cardiovascular benefits. Specifically, studies of GLP-1 RA have reported a reduction in major adverse cardiovascular events as primary outcomes, and a reduction in eGFR decline or albuminuria progression as secondary outcomes.25 These findings have transformed the management of diabetes and related complications from a “glucose-centric” to a “cardio-renal” risk reduction focused approach.26 The most recent ADA guidelines emphasize consideration of DKD when deciding on additional glucose-lowering therapies.27 For the management of DKD, 2022 Kidney Disease Improving Global Outcomes (KDIGO) guidelines consider SGLT2i along with metformin as first-line drug therapy, after which GLP-1 RA are recommended if needed to achieve glycemic target or if albuminuria is persistent.28
Figure 1. Timeline of current and future randomized controlled trials of sodium glucose cotransporter 2 inhibitors (SGLT2i) and glucagon-like receptor agonists (GLP-1 RA).
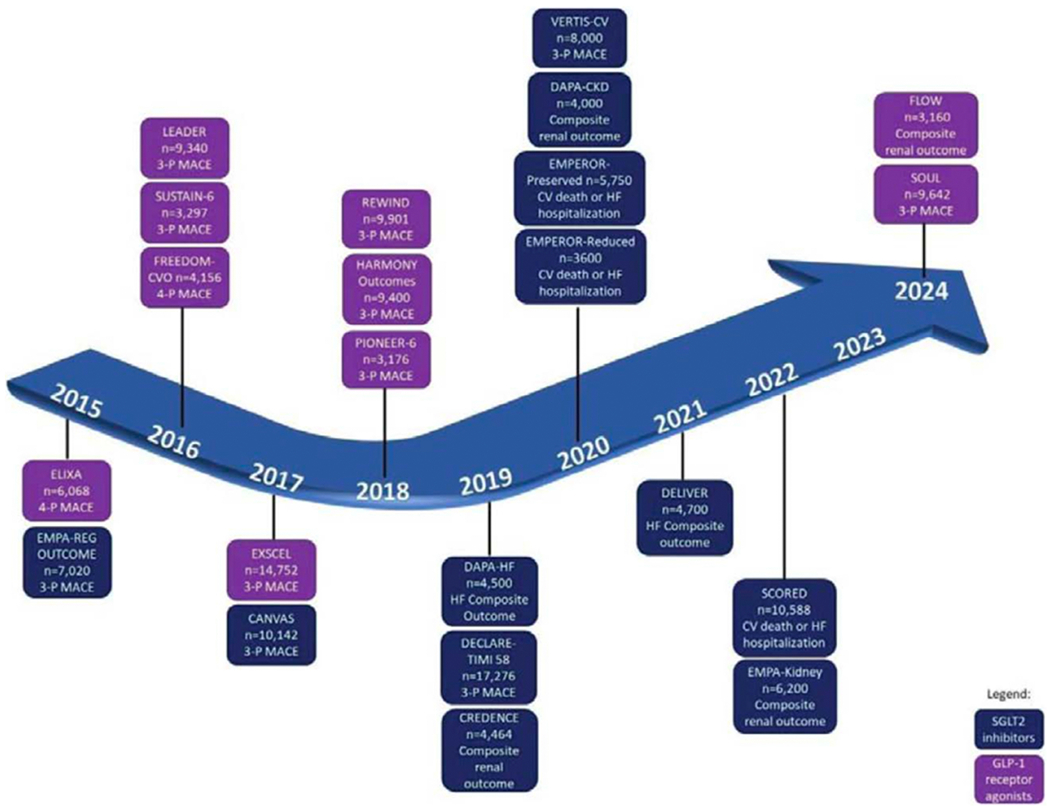
Reprinted with permission. Circulation.2020;142:e265-e286. ©2020 American Heart Association, Inc.
Further expanding the arsenal of DKD therapies is the non-steroidal mineralocorticoid receptor antagonist, finerenone. In randomized control trials of adults with T2D and albuminuria despite maximum-tolerated RAS inhibition, finerenone significantly reduced the risk of sustained eGFR decline and kidney failure as well as that of cardiovascular events.29 This agent has also been recommended for use in ADA and upcoming KDIGO 2022 guidelines.27,28 Other novel drug classes, such as endothelin A receptor antagonists, have demonstrated promise in slowing DKD progression and are undergoing further study.30
Despite recent advances in treatments for DKD, important questions remain. First, substantial residual risks of CKD progression and cardiovascular events are evident for patients with DKD even when optimally treated with currently-available drugs. Second, while some interventions (e.g. lifestyle modification, SGLT2i) are likely to have substantial benefits with low risks across wide subgroups of patients, benefits and risks may vary with novel drugs. Third, with multiple drugs now available for treatment of people with T2D and CKD, combinations of drugs need to be considered and optimized. Therefore, “one-size-fits-all” approach is unlikely to be optimal for the prevention and management of DKD. Knowledge of the mechanisms underlying DKD pathology and differences in disease severity and prognosis in individuals is lacking. Enhanced insight into the molecular drivers of DKD is essential for targeting the right drugs to the right patients and for the development of new therapies.
INNOVATIVE METHODS
The use of novel, cutting-edge approaches for molecular interrogation of kidney pathology holds tremendous promise for advancing our understanding and treatment of DKD. These tools are paving the way for the discovery and application of precision medicine methodology in DKD, directly addressing the limitations of the current, broad treatment approaches. Specifically, precision medicine refers to the concept of personally curating medical prevention and treatment strategies, accounting for individual variability.31 A number of new consortia aim to advance precision medicine in DKD, including the Kidney Precision Medicine Project (KPMP), TRIDENT, and BEAT-DKD.32–34 These and focused studies in established cohorts are applying novel multiomic data to better understand mechanism, risk factors, and subtypes of DKD.
Consortia-based Approaches: The Kidney Precision Medicine Project
The KPMP is a national consortium funded by the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) which aims to redefine common forms of CKD and acute kidney injury (AKI) by integrating molecular kidney data procured from protocol kidney biopsies with standardized clinical phenotypic and outcome data.32 The KPMP is enrolling adults with DKD, hypertensive kidney disease, or AKI, and excluding those with glomerular diseases, kidney transplant, malignancy, or pregnancy. In obtaining kidney biopsies from participants with common forms of CKD (such as DKD) who are not usually biopsied for clinical purposes, the KPMP is by design ideally positioned to elucidate the mechanisms underlying the most prevalent kidney disease phenotypes. Overall, the KPMP proposes a model for molecularly defining disease and targeted treatments beginning with cohort-based, kidney tissue-centered clinical and molecular phenotyping, followed by multiscalar integration of clinical, histopathologic, transcriptomic, proteomic, and metabolomic data, and resulting in identification of disease pathways at the patient level. Specifically, the goals of the KPMP are as follows: (1) to ethically collect kidney biopsies from participants with AKI and CKD; (2) to create a reference kidney tissue atlas; (3) to define molecular disease subgroups; (4) to identify new therapeutic targets for kidney disease.
Kidney tissue within the KPMP undergoes comprehensive structural and molecular phenotyping using a wide breadth of multimodal technologies spanning transcriptomics, proteomics, metabolomics, epigenetics, and spatial imaging (Figure 2). The tissue interrogation pipeline is subjected to rigorous quality control measures across all phases of tissue handling including collection, processing, and preservation.35 This approach facilitates harmonization of the resulting data. Following processing, tissue samples are distributed across Tissue Interrogation Sites (TIS) for application of molecular technologies. The data ascertained from these molecular platforms are at once complementary and redundant, yielding comprehensive results that are amenable to validation across technologies. For example, transcriptomic data is obtained across different TIS via single-cell RNA sequencing (RNAseq), single-nucleus RNAseq, spatial transcriptomic profiling, and in bulk from laser microdissected biopsy samples.
Figure 2.
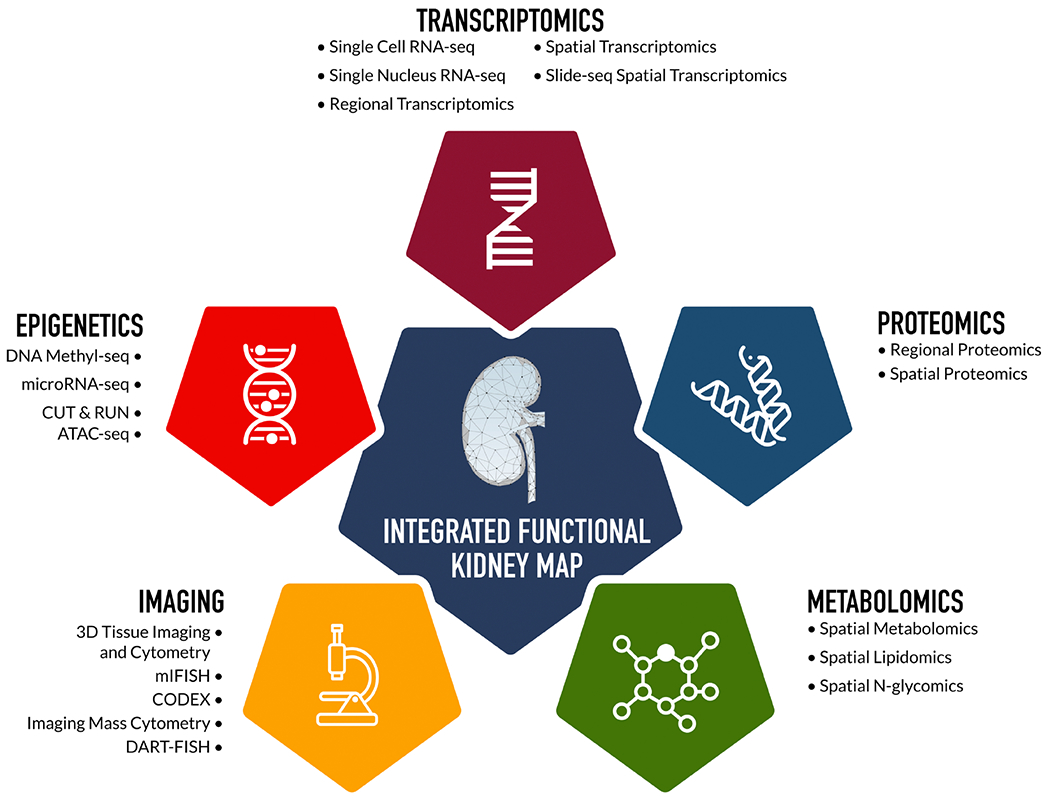
Summary of KPMP technologies.
Technological advancements implemented within the KPMP allow for the capture and integration of high-dimensional molecular and spatial data, holding the potential for deeper phenotyping of kidney disease than ever before possible. With single-cell RNAseq, individual transcriptomic profiles from thousands of kidney cells can be compared and contrasted such that cell-type-specific changes in gene expression that occur with disease can be thoroughly characterized.36 Compartmental and near-single-cell profiling of proteins provide complementary information on the functional activation and regulation of molecular pathways.37,38 Manual kidney histopathology assessment is further enhanced by computer-based image interpretation technologies with the capacity to automatically and rapidly detect, segment, and quantify high-volume structural features.39 Additionally, blood and urine metabolomic and proteomic assessments are being performed. Once this wealth of data is collected, computational data reduction using machine learning and other approaches allows for identification of latent molecular patterns and networks which may have biological significance.40 Ultimately, integration of omics, histopathology, and clinical data has the potential to provide insight into the molecular mechanisms underlying DKD and other kidney pathologies.
Alongside the application of pioneering technologies and integrative data approaches, patient engagement is central to the mission of the KPMP.41 As key stakeholders, patient partners have actively been involved in the KPMP since its inception, serving on the NIDDK’s Advisory Council which reviewed the KPMP grant application and as standing members of key committees within the KPMP. Patient partners with CKD and AKI, in their role as part of the KPMP’s Community Engagement Committee and other committees, contribute perspectives which guide scientific research priorities and communication of research findings, as well as essential study components such as the informed consent process. This degree of involvement of patient partners within the KPMP is novel in large-scale precision medicine research and serves as a benchmark for future studies.
In addition to the KPMP, a number of other consortia have been working to define the molecular bases underlying DKD and other forms of kidney disease. These include the Biomarker Enterprise to Attack DKD (BEAt DKD), the Transformative Research in Diabetic Nephropathy cohort (TRIDENT), the Nephrotic Syndrome Study Network (NEPTUNE), the Cure Glomerulonephropathy Network (CureGN), and the Clinical Phenotyping and Resource Biobank Core (C-PROBE).33,34,42–44
Focused Approaches: Metabolomics and Lipidomics
Metabolomics attempts to systematically identify and quantitate small molecule metabolites from biological systems. The recent rapid development of a variety of analytical platforms based on mass spectrometry coupled with sophisticated chromatography platforms have enabled identification of complex metabolic phenotypes.45 Given the metabolic underpinning of diabetes and its complications, the technology is ideally suited to deep phenotype to understand molecular mechanisms of metabolic dysregulation beyond glucose.
Lipids, a class of metabolites, comprise the majority of biologic molecules in human plasma and are important sources of energy for the kidney and other tissues.46 Both diabetes and CKD are accompanied by substantial metabolic derangements and alterations in lipid profiles which augment risk of cardiovascular disease.47,48 Specifically, diabetes is associated with increased serum concentrations of triglyceride-rich lipoprotein, small dense low-density lipoprotein (sdLDL), and very low density lipoprotein (VLDL), as well as decreased serum concentrations of high-density lipoprotein (HDL). These derangements are both exacerbated by and contribute to progressive kidney function decline.49 Historically, studies investigating lipid associations with kidney function have utilized traditional lipid panels that measure total cholesterol, lipoproteins, and triglycerides. With the advent of omic technologies, combined with growing appreciation for the role of lipid abnormalities in kidney disease pathology, meaningful developments in the field of lipidomics, a subset of metabolomic techniques geared toward identifying lipids, have been made.
The kidney is a highly metabolically active organ, with different nephron segments exhibiting distinct preferences in energy substrates – for example, fatty acids are the preferred substrate for ATP production in the tubule, whereas glucose is used more readily in the glomerulus.50 In diabetes, fatty acid metabolism (β-oxidation and lipid synthesis) and glycolysis in the kidney are enriched in the proximal tubule and other nephron segments.51 Notably, these metabolic changes are specific to the kidney, and are distinct from those observed in the retina and nerve in diabetic mouse models, suggesting tissue-specific metabolic reprogramming. Despite increased flux through glycolytic and β-oxidation pathways, ATP production is not increased, suggesting an uncoupling between mitochondrial metabolism and oxidative phosphorylation. The resulting accumulation of tricarboxylic acid (TCA) cycle metabolites is proposed to contribute to DKD pathophysiology. Results from studies conducted in adults with T1D and T2D and CKD have also identified distinct patterns of urinary metabolites derived from mitochondrial oxidative phosphorylation, with higher concentrations in those with preserved kidney function and lower levels in those with advanced CKD (eGFR < 40ml/min/1.73m2).51,52 It is likely that samples from subjects with advanced DKD reflected tubular damage, whereas samples from those in earlier disease stages reflected increased metabolic activity at the proximal tubule.
Dysregulation of lipid transport and metabolism may lead to lipid accumulation within the kidney, with visible tubular and glomerular lipid deposits reported in human DKD biopsy samples.53 Advancements in mass spectrometry and bioinformatics approaches have allowed for enhanced lipid profiling both at the plasma and tissue level (Figure 3).54 These approaches have facilitated the distinction and quantification of lipid species and their corresponding chemical characteristics, including class, chain length, and degree of saturation. High-resolution lipid characterization is especially valuable when assessing alterations related to kidney dysfunction because in this setting changes in lipid profiles occur primarily at the class level and may otherwise not be captured using traditional methods.48 Analysis of the resulting lipidomic data using dimensionality reduction methods can identify subgroups of highly correlated lipids which may bear relevance to specific disease processes.
Figure 3. Identification of lipid classes using mass spectrometry.
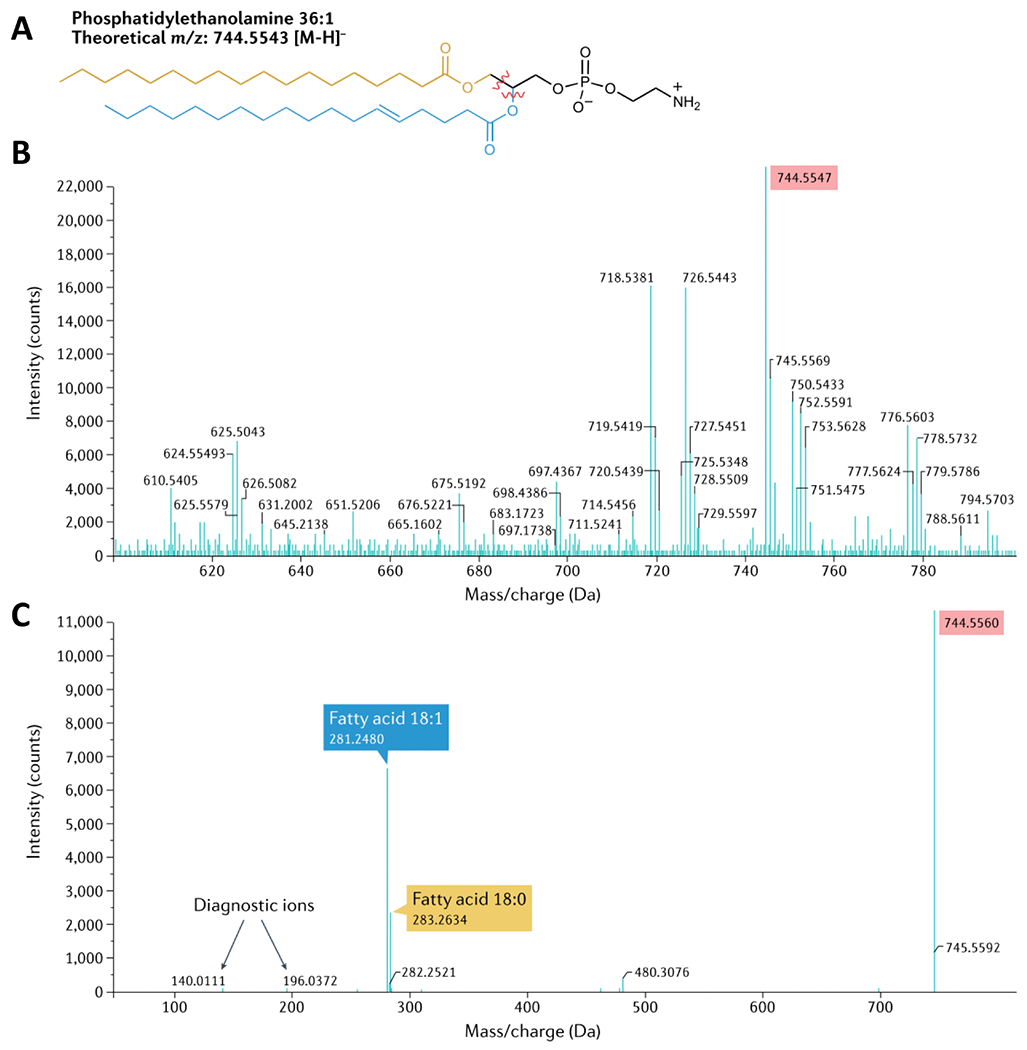
(a) Chemical structure of phosphatidylethanolamine (PE) 36:1. The fragments identified using tandem mass spectrometry (MS–MS) are highlighted. (b) MS1 spectra, identifying the [M–H] precursor ion with its expected m/z (highlighted in pink on the spectra). (c) MS–MS (or MS2) spectra, which identify the fragments of PE 36:1 derived from the precursor ions. Reprinted with permission from Springer Nature: Elsevier, from Nature Reviews Nephrology, “Lipidomic approaches to dissect dysregulated lipid metabolism in kidney disease,” Baek et al., 18:1, 2022; permission conveyed through Copyright Clearance Center, Inc.
Furthermore, application of differential network enrichment analyses can provide insight into how correlations between lipids differ across disease scenarios. Ultimately, integration of lipidomic data with tissue transcriptomic and proteomic data can enrich understandings of how the various biological processes underscoring these molecular components are intertwined. Essential for the success of these approaches is the incorporation of detailed lipidomic data into broader metabolomic datasets and linkage of these with related gene and protein pathways. Overall, application of lipidomic technologies is enhancing understandings of metabolic reprogramming in DKD. These approaches are instrumental for deciphering how alterations in lipid handling in DKD contribute to disease pathology.
PROVOCATIVE NEW DIRECTIONS
The foundation of current clinical practice is centered on optimizing glycemic control managing blood pressure and dyslipidemia, and maximizing RAS inhibition reduce diabetes-related kidney complications. Molecular characterization of DKD has the potential to transform both our understanding of DKD pathophysiology and our approaches to therapy. Specifically, combining molecular with histopathologic and clinical data may allow for the distinction of disease subtypes and the development of targeted therapies. Additionally, integrating metabolomic/lipidomic profiles to identify metabolic subtypes of patients, termed as “metabotype” will aid molecular understanding and risk stratification beyond glycemic control.
Integration of transcriptomic, proteomic, and metabolomic/lipidomic kidney tissue data can yield novel insights into subcellular pathways within the kidney and their functional relationships. Research along these lines has already resulted in a growing appreciation of the kidney as an organ that is both influenced by and capable of influencing systemic metabolic processes. For example, in a study performed using nephrectomy samples from 56 healthy adults within the KPMP, dynamic enrichment analysis of top differentially expressed genes and protein markers was able to reveal the dominant physiological processes within proximal tubule cell types, of which energy generation processes accounted for a substantial proportion (Figure 4A).55 Additionally, by mapping genes and proteins to aerobic and anaerobic energy metabolic pathways, energy generation profiles could be derived for each tubular segment (Figure 4B). Other work conducted in adults with T2D demonstrated an association between expression of genes involved in fatty acid synthesis in the glomerulus and serum free fatty acid concentrations, implying that kidney-specific changes in lipid metabolism may account for changes in serum lipid profiles.56
Figure 4.
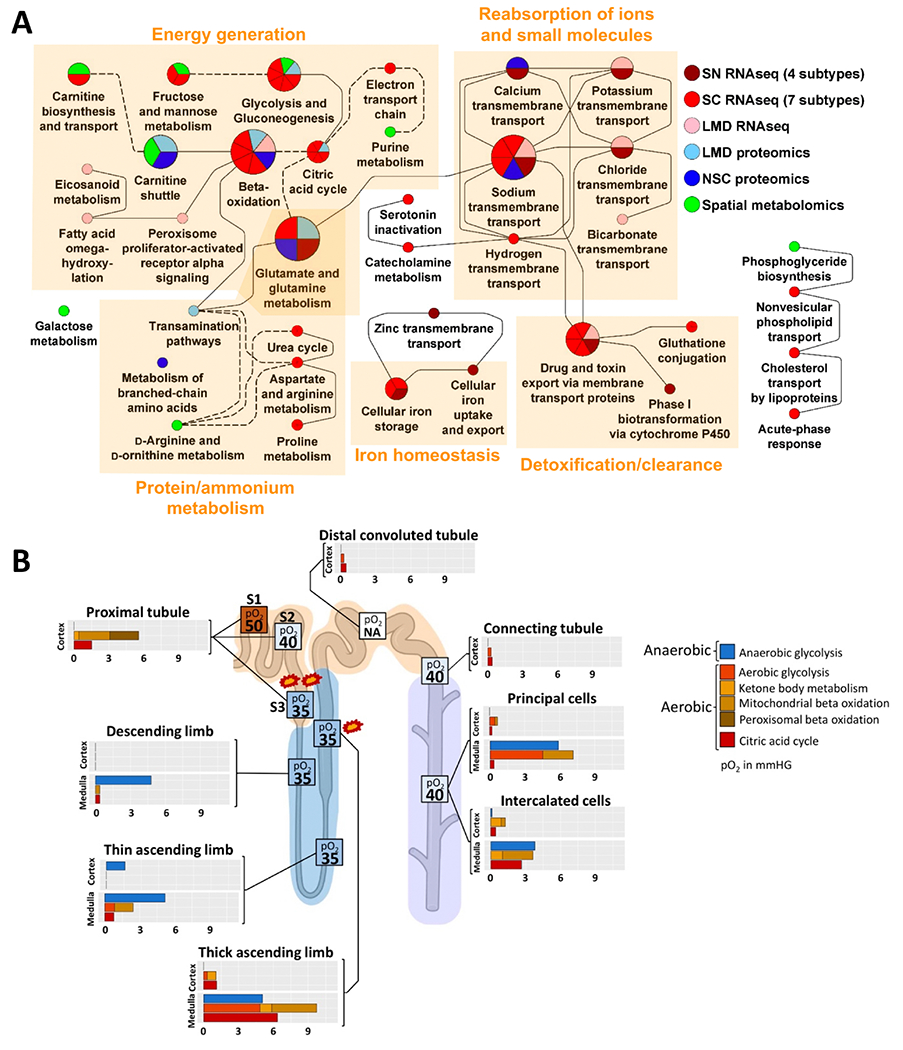
(A) Enrichment analysis of proximal tubular marker genes, proteins, and metabolites. Dynamic enrichment analysis of marker genes, proteins, and metabolites of each proximal tubule subtype/subsegment reveals corresponding top subcellular processes. (B) Tubular aerobic and anaerobic energy generation profiles by cell type. Energy profiles were generated by combining a novel ontology allowing for distinction of aerobic and anaerobic energy pathways with cell marker genes. Reprinted from Hansen J et al., Science Advances 2022. From Hansen et al., “A reference tissue atlas for the human kidney” Sci Adv. 2022 Jun 10;8(23):eabn4965. doi: 10.1126/sciadv.abn4965. Epub 2022 Jun 8. PMID: 35675394; PMCID: PMC9176741. © The Authors, some rights reserved; exclusive licensee AAAS. Distributed under a CC BY-NC 4.0 license http://creativecommons.org/licenses/by-nc/4.0/[creativecommons.org]”. Reprinted with permission from AAAS.
Kidney tissue transcriptomic studies have also begun to help clarify alterations in molecular pathways that occur with diabetes across specific cell types.57 Single-cell RNAseq performed in isolated glomerular cells from mouse models showed dynamic changes in endothelial and mesangial cell transcription with diabetes induction.58 Specifically, among diabetic mice compared to controls, genes involved in regulation of angiogenesis and protein stabilization were enriched in endothelial and mesangial cells, respectively. In a human nephrectomy study where single nuclear RNAseq was performed in laser microdissected interstitial samples, genes involved in extracellular matrix organization and small-molecule catabolism were upregulated in DKD versus controls.59 Identification of molecular changes consistent with DKD early in the course of disease may enable early diagnosis of DKD, even before typical histopathological changes are apparent.60
Molecular tissue data can additionally be used to discern mechanism-based DKD subtypes which may explain some of the heterogeneity in the clinical presentation and trajectory of DKD. This process involves identifying differences in gene expression patterns or cell processes that exist within diabetes populations, then correlating these with kidney function and related outcomes. In one study conducted in adults with T2D, expression of a set of genes associated with mitochondrial dysfunction, inflammation, and tubular metabolism was associated with concomitant tubulointerstitial injury and subsequent 10-year kidney function decline.61 As another example, integration of transcriptomic and spatial imaging data has revealed the existence of adaptive and degenerative cellular states within the kidney, and enabled the identification of the specific expression signatures and environmental interactions of these distinct cell conditions.62 Interestingly, there was overlap in the molecular signatures of epithelial cells undergoing maladaptive and reparative processes, such as upregulation of pathways involved in cytokine signaling and fibrosis. Notably, in a longitudinal cohort, increased proportions of thick ascending limb and proximal tubule cells expressing adaptive phenotypes were associated with kidney function decline. Greater proportions of these adaptive cell types were also seen with DKD compared to healthy controls.
Lipidomic profiles have also demonstrated the capacity to draw important kidney prognostic distinctions among adults with diabetes. In a population of adults with T2D, a greater abundance of longer triacylglycerols and a lower abundance of longer acylcarnitines were associated with kidney disease progression (Figure 5). Additionally, greater abundance of serum unsaturated phosphatidylethanolamines and lower unsaturated free fatty acid concentrations were associated with kidney function decline.56 Collectively, these findings reveal a previously unrecognized link between lipid markers of impaired mitochondrial β-oxidation and enhanced de novo lipogenesis with DKD progression among individuals with preserved kidney function. In contrast, in type 1 diabetes lower concentrations of unsaturated free fatty acids may be protective against rapid eGFR decline, suggesting the presence of insulin resistance versus reduced insulin availability may underlie differential mechanisms for lipid handling and kidney dysfunction in these groups.63 Analyses focusing on changes in lipid networks have also demonstrated that differences in correlations between lipids of the same class may predict progression to kidney failure.64
Figure 5. Differences in carbon chain length and number of double bonds in complex lipids and acylcarnitines between adults with type 2 diabetes with and without progressive kidney function decline.
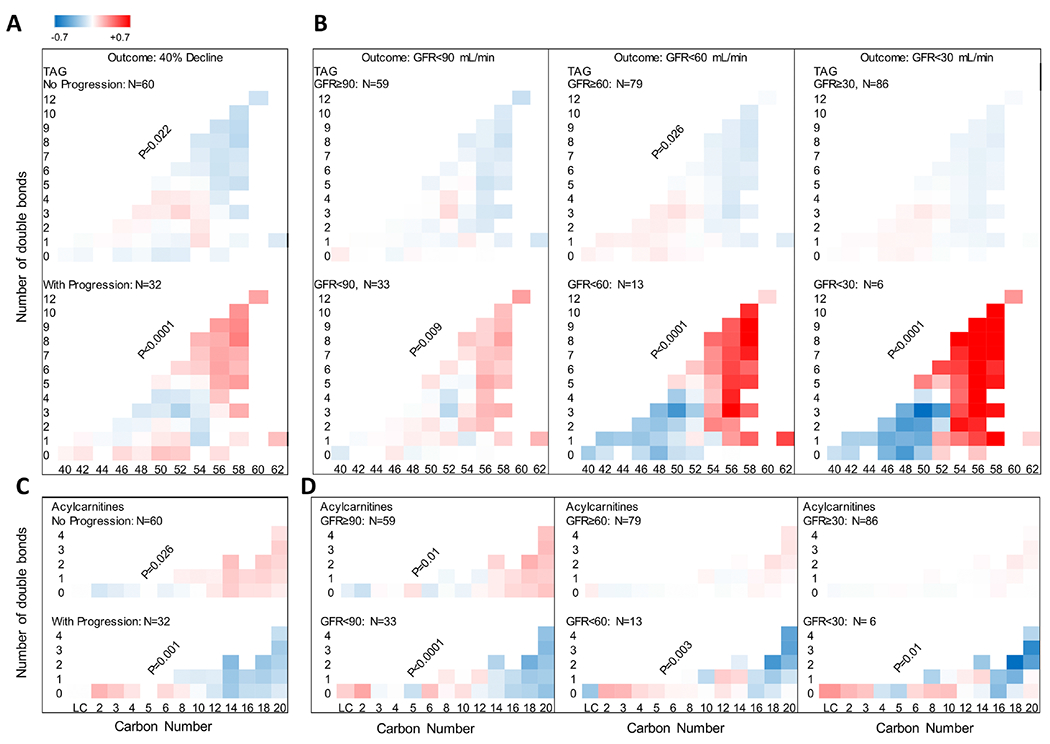
(A) Greater relative abundance of longer unsaturated triacylglycerols (TAGs) in the serum of adults with progressive (n = 32) versus non-progressive (n = 60) eGFR decline. (B) A similar pattern was found when all participants were grouped based on whether they achieved a sustained GFR <90 (n = 33), <60 (n = 13), and <30 mL/min (n = 6). (C) Lower relative abundance of longer unsaturated acylcarnitines (ACs) in adults with progressive versus non-progressive eGFR decline. (D) A similar trend was noted in abundance of ACs by carbon number, when all participants were grouped based on whether they achieved a sustained GFR <90 (n = 33), <60 (n = 13), or <30 mL/min (n = 6). Reprinted from Afshinnia F et al., JCI Insight 2019. Reprinted with permission of American Society for Clinical Investigation, from JCI Insight, “Increased lipogenesis and impaired β-oxidation predict type 2 diabetic kidney disease progression in American Indians,” Afshinnia et al., 21:4, 2019; permission conveyed through Copyright Clearance Center, Inc.
Understanding molecular changes in DKD and their response to various treatment regimens may facilitate the curation of therapies to specific disease mechanisms. Studies conducted in mouse models of DKD have demonstrated different patterns of transcriptional changes across distinct kidney cell types in response to treatment with ACEi, SGLT2i, and/or rosiglitizone.65 Different treatment regimens also differentially normalized intracellular signaling patterns across cell types. Notably, combination therapy with ACEi and SGLT2i offered the greatest degree of proximal tubular protection after two weeks. Similarly, renin-angiotensin system inhibition ameliorated diabetes-related remodeling of triacylglycerol lipid network connections in mouse models.66
The identification of altered metabolic and molecular pathways in DKD (primarily from laboratory models) has already resulted in the development of novel, mechanism-targeted therapies.67 Drugs combating oxidative stress, including antioxidants such as coenzyme Q10 and reservatrol, NADPH oxidase inhibitors, and NF-κB inhibitors, have been tested in animal and human DKD models with mixed success.68,69 The glycosaminoglycan sulodexide, which failed to show benefit in clinical trials, purportedly may also protect against oxidative stress injury via increased klotho expression.70,71 Agents targeting MAPK and JAK-STAT pathways (the ASK1 inhibitor GS-444,217 and the JAK1/JAK2 inhibitor baracitinib, respectively) have been shown to reduce albuminuria and slow kidney disease progression in early studies and merit further research.72,73 Pathways involved in autophagy, inflammation, and fibrosis additionally represent promising drug targets in DKD.67 Moving forward, human molecular kidney data will be to validate laboratory model data, identify new drug targets, and guide the implementation of mechanism-based therapies.
Essential for amplifying the use and clinical incorporation of multiomic kidney tissue data is the development and maintenance of a comprehensive, high-quality kidney tissue reference database.55 A central aim of the KPMP is the development of a reference kidney atlas. This atlas is to serve as an integrated source for clinical, histopathologic, blood and urine biomarker, and kidney tissue transcriptomic, proteomic, metabolomic data across the spectrums of health, AKI, and CKD. The development of detailed ontologies which more precisely classify genes, proteins, metabolites, and related processes in relation to kidney cell types is key to defining disease pathways and to integration with existing ontologies.74,75 In housing and promoting access to molecular kidney tissue data, the atlas will provide a space for advancing knowledge pertaining to the cellular and subcellular pathways involved in kidney disease progression. Additionally, it will serve as a communal environment for the testing and development of new scientific hypotheses. Ultimately, complex, multifaceted datasets comprised of multiomic, clinical, and histopathologic data, along with data on environmental factors and historical treatment responses can be integrated using artificial intelligence to develop personalized care algorithms.76
FINAL RECOMMENDATIONS
The national and worldwide prevalence of diabetes is high and growing. DKD affects a substantial proportion of people with diabetes and is associated with significant morbidity and mortality. The last decade has borne witness to the advent of a number of new therapies for people with diabetes which substantially reduce the risk of DKD incidence and progression. The application of novel technologies which allow for deep molecular and structural phenotyping of kidney tissue holds great promise for the advancement of knowledge on the mechanisms underlying DKD pathology. Consortia such as the KPMP are actively working to integrate kidney transcriptomic, proteomic, and metabolomic/lipidomic data with clinical and histopathologic data to generate novel, mechanism-based understandings of DKD. Ultimately, these methods are paving the way for the implementation of precision diagnostics, as well as targeted therapeutic and prognostic approaches in DKD.
Highlights for “Present and future directions in diabetic kidney disease” by Limonte et al.
Diabetic kidney disease is a leading cause of kidney failure and is associated with substantial risk of cardiovascular disease, morbidity, and mortality.
Use of novel technologies enabling deep molecular and structural phenotyping of kidney tissue is expanding our knowledge of the mechanisms underlying diabetic kidney disease pathology.
Integration of molecular and clinical data may reveal mechanism-based diabetic kidney disease subtypes and pave the way for the implementation of targeted diagnostic, prognostic, and therapeutic approaches.
ACKNOWLEDGMENTS/FUNDING
This manuscript is a summary of the data presented during lectures and workshops included in the NIDDK/DiaComp funded “Frontiers in Diabetic Complications- From Biology to Technology” Conference, held in May 2022 Financial support for this work was provided by the NIDDK Diabetic Complications Consortium (RRID:SCR_001415, [www.diacomp.org]www.diacomp.org), grants DK076169 and DK115255. CPL receives funding from the American Kidney Fund’s Clinical Scientist in Nephrology program. MK and SP acknowledge support from P30DK089503, R24DK082841, P30DK081943 and the JDRF Center of Excellence (5-COE-2019-861-S-B). RPB is supported by R01DK107956; U01DK119083; 1U01 DK0945157; R01DK116723, and the JDRF Center of Excellence. IHdB receives funding from R01DK126373, R01DK125084, R01DK132399, U2CDK114886.
DISCLOSURES
CPL has nothing to disclose. M.K. reports grants from NIH/NIDDK and JDRF in support of this manuscript. Grants and contracts outside the submitted work through the University of Michigan with NIH, Chan Zuckerberg Initiative, AstraZeneca, NovoNordisk, Eli Lilly, Gilead, Goldfinch Bio, Janssen, Boehringer-Ingelheim, Moderna, European Union Innovative Medicine Initiative, Certa, Chinook, amfAR, Angion, RenalytixAI, Travere, Regeneron, IONIS, consulting fees through the University of Michigan from Astellas, Poxel, Janssen and UCB. In addition, M.K. has a patent PCT/EP2014/073413 “Biomarkers and methods for progression prediction for chronic kidney disease” licensed. SP has nothing to disclose. RPB consults for Novo Nordisk and Roche. IHdB consults for AstraZeneca, Bayer, Boehringer-Ingelheim, Cyclerion Therapeutics, George Clinical, Goldfinch Bio, Ironwood, Lilly, Otsuka and receives research equipment and supplies from DexCom.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.International Diabetes Federation. IDF Diabetes Atlas, 10th Edn. Vol 102. (International Diabetes Federation, ed.).; 2021. doi: 10.1016/j.diabres.2013.10.013 [DOI] [Google Scholar]
- 2.de Boer IH, Rue TC, Cleary PA, et al. Long-term renal outcomes of patients with type 1 diabetes mellitus and microalbuminuria. Arch Intern Med. 2011;171(5):412–420. doi: 10.1001/archinternmed.2011.413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zelnick LR, Weiss NS, Kestenbaum BR, et al. Diabetes and CKD in the United States population, 2009–2014. Clin J Am Soc Nephrol. 2017;12(12):1984–1990. doi: 10.2215/CJN.03700417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ríos Burrows N, Koyama A, Pavkov ME. Reported cases of end-stage kidney disease—United States, 2000–2019. Am J Transplant. 2022;22(5):1483–1486. doi: 10.1111/ajt.16657 [DOI] [PubMed] [Google Scholar]
- 5.Afkarian M, Zelnick LR, Hall YN, et al. Clinical manifestations of kidney disease among US adults with diabetes, 1988-2014. JAMA. 2016;316(6):602–610. doi: 10.1001/jama.2016.10924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McCullough KP, Morgenstern H, Saran R, Herman WH, Robinson BM. Projecting ESRD incidence and prevalence in the United States through 2030. J Am Soc Nephrol. 2019;30(1):127–135. doi: 10.1681/ASN.2018050531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harding JL, Pavkov ME, Magliano DJ, Shaw JE, Gregg EW. Global trends in diabetes complications: a review of current evidence. Diabetologia. 2019;62(1):3–16. doi: 10.1007/s00125-018-4711-2 [DOI] [PubMed] [Google Scholar]
- 8.Afkarian M, Sachs MC, Kestenbaum B, et al. Kidney disease and increased mortality risk in type 2 diabetes. J Am Soc Nephrol. 2013;24(2):302–308. doi: 10.1681/ASN.2012070718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Groop PH, Thomas MC, Moran JL, et al. The presence and severity of chronic kidney disease predicts all-cause mortality in type 1 diabetes. Diabetes. 2009;58(7):1651–1658. doi: 10.2337/db08-1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Orchard TJ, Secrest AM, Miller RG, Costacou T. In the absence of renal disease, 20 year mortality risk in type 1 diabetes is comparable to that of the general population: A report from the Pittsburgh Epidemiology of Diabetes Complications Study. Diabetologia. 2010;53(11):2312–2319. doi: 10.1007/s00125-010-1860-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sarnak M, Levey A, Schoolwerth A, et al. Kidney Disease as a Risk Factor for Development of Cardiovascular Disease. Circulation. 2003;108:2154–2169. [DOI] [PubMed] [Google Scholar]
- 12.Pálsson R, Patel UD. Cardiovascular complications of diabetic kidney disease. Adv Chronic Kidney Dis. 2014;21(3):273–280. doi: 10.1053/j.ackd.2014.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Centers for Disease Control and Prevention. National Diabetes Statistics Report website. Accessed July 12, 2022. https://www.cdc.gov/diabetes/data/statistics-report/index.html
- 14.Nicholas SB, Kalantar-Zadeh K, Norris KC. Socioeconomic disparities in chronic kidney disease. Adv Chronic Kidney Dis. 2015;22(1):6–15. doi: 10.1053/j.ackd.2014.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Diabetes Control and Complications Trial Research Group, Nathan D, Genuth S, et al. The Effect of Intensive Treatment of Diabetes on the Development and Progression of Long-term Complications in Insulin-dependent Diabetes Mellitus. N Engl J Med. 1993;329(14):977–986. http://www.nejm.org/doi/pdf/10.1056/NEJM199309303291401 [DOI] [PubMed] [Google Scholar]
- 16.Retnakaran R, Cull CA, Thorne KI, Adler AI, Holman RR. Risk factors for renal dysfunction in type 2 diabetes: U.K. Prospective Diabetes Study 74. Diabetes. 2006;55(6):1832–1839. doi: 10.2337/db05-1620 [DOI] [PubMed] [Google Scholar]
- 17.Wong MG, Perkovic V, Chalmers J, et al. Long-term benefits of intensive glucose Control for preventing end-stage kidney disease: ADVANCE-ON. Diabetes Care. 2016;39(5):694–700. doi: 10.2337/dc15-2322 [DOI] [PubMed] [Google Scholar]
- 18.Classification I. Standards of medical care in diabetes-2009. Diabetes Care. 2009;32(SUPPL. 1). doi: 10.2337/dc09-S013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.The Action to Control Cardiovascular Risk in Diabetes Study Group. Effects of Intensive Glucose Lowering in Type 2 Diabetes. N Engl J Med. 2008;358(24):2545–2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bakris GL, Weir MR, Shanifar S, et al. Effects of blood pressure level on progression of diabetic nephropathy: results from the RENAAL study. Arch Intern Med. 2003;163(13):1555–1565. doi: 10.1001/archinte.163.13.1555 [DOI] [PubMed] [Google Scholar]
- 21.Strippoli GFM, Bonifati C, Craig M, Navaneethan SD, Craig JC. Angiotensin converting enzyme inhibitors and angiotensin II receptor antagonists for preventing the progression of diabetic kidney disease. Cochrane Database Syst Rev. 2006;(4). doi: 10.1002/14651858.CD006257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mauer M, Zinman B, Gardiner R, et al. Renal and Retinal Effects of Enalapril and Losartan in Type 1 Diabetes (RASS). N Engl J Med. 2009;361(1):40–51. doi: 10.1056/nejmoa0808400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Staplin N, Roddick AJ, Emberson J, et al. Net effects of sodium-glucose co-transporter-2 inhibition in different patient groups: a meta-analysis of large placebo-controlled randomized trials. eClinicalMedicine. 2021;41:1–11. doi: 10.1016/j.eclinm.2021.101163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neuen BL, Young T, Heerspink HJL, et al. SGLT2 inhibitors for the prevention of kidney failure in patients with type 2 diabetes: a systematic review and meta-analysis. Lancet Diabetes Endocrinol. 2019;7(11):845–854. doi: 10.1016/S2213-8587(19)30256-6 [DOI] [PubMed] [Google Scholar]
- 25.Giugliano D, Scappaticcio L, Longo M, et al. GLP-1 receptor agonists and cardiorenal outcomes in type 2 diabetes: an updated meta-analysis of eight CVOTs. Cardiovasc Diabetol. 2021;20(1):1–11. doi: 10.1186/s12933-021-01366-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jacob S, Krentz AJ, Deanfield J, Rydén L. Evolution of Type 2 Diabetes Management from a Glucocentric Approach to Cardio-Renal Risk Reduction: The New Paradigm of Care. Drugs. 2021;81(12):1373–1379. doi: 10.1007/s40265-021-01554-6 [DOI] [PubMed] [Google Scholar]
- 27.Care D, Suppl SS. Summary of Revisions: Standards of Medical Care in Diabetes—2022. Diabetes Care. 2022;45(January):S4–S7. doi: 10.2337/dc22-SREV [DOI] [PubMed] [Google Scholar]
- 28.Kidney Disease: Improving Global Outcomes (KDIGO) Diabetes Work Group. KDIGO 2022 Clinical Practice Guideline for Diabetes Management in Chronic Kidney Disease. [in Press. [Google Scholar]
- 29.Bakris GL, Agarwal R, Anker SD, et al. Effect of Finerenone on Chronic Kidney Disease Outcomes in Type 2 Diabetes. N Engl J Med. 2020;383(23):2219–2229. doi: 10.1056/nejmoa2025845 [DOI] [PubMed] [Google Scholar]
- 30.Heerspink HJL, Parving HH, Andress DL, et al. Atrasentan and renal events in patients with type 2 diabetes and chronic kidney disease (SONAR): a double-blind, randomised, placebo-controlled trial. Lancet. 2019;393(10184):1937–1947. doi: 10.1016/S0140-6736(19)30772-X [DOI] [PubMed] [Google Scholar]
- 31.Collins FS, Varmus H. A new initiative on precision medicine. N Eng J Med. 2015;372(9):793–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Boer IH, Alpers CE, Azeloglu EU, et al. Rationale and design of the Kidney Precision Medicine Project. Kidney Int. 2021;99(3):498–510. doi: 10.1016/j.kint.2020.08.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kammer M, Heinzel A, Willency JA, et al. Integrative analysis of prognostic biomarkers derived from multiomics panels helps discrimination of chronic kidney disease trajectories in people with type 2 diabetes. Kidney Int. 2019;96(6):1381–1388. doi: 10.1016/j.kint.2019.07.025 [DOI] [PubMed] [Google Scholar]
- 34.Townsend RR, Guarnieri P, Argyropoulos C, et al. Rationale and design of the Transformative Research in Diabetic Nephropathy (TRIDENT) Study. Kidney Int. 2020;97(1):10–13. doi: 10.1016/j.kint.2019.09.020 [DOI] [PubMed] [Google Scholar]
- 35.El-Achkar TM, Eadon MT, Menon R, et al. A multimodal and integrated approach to interrogate human kidney biopsies with rigor and reproducibility: Guidelines from the kidney precision medicine project. Physiol Genomics. 2021;53(1):1–11. doi: 10.1152/physiolgenomics.00104.2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Menon R, Otto EA, Hoover P, et al. Single cell transcriptomics identifies focal segmental glomerulosclerosis remission endothelial biomarker. JCI Insight. 2020;5(6):1–21. doi: 10.1172/jci.insight.133267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sigdel TK, Piehowski PD, Roy S, et al. Near-Single-Cell Proteomics Profiling of the Proximal Tubular and Glomerulus of the Normal Human Kidney. Front Med. 2020;7(September):1–11. doi: 10.3389/fmed.2020.00499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ayoub I, Shapiro JP, Song H, et al. Establishing a Case for Anti-complement Therapy in Membranous Nephropathy. Kidney Int Reports. 2021;6(2):484–492. doi: 10.1016/j.ekir.2020.11.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barisoni L, Lafata KJ, Hewitt SM, Madabhushi A, Balis UGJ. Digital pathology and computational image analysis in nephropathology. Nat Rev Nephrol. 2020;16(11):669–685. doi: 10.1038/s41581-020-0321-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eddy S, Mariani LH, Kretzler M. Integrated multi-omics approaches to improve classification of chronic kidney disease. Nat Rev Nephrol. 2020;16(11):657–668. doi: 10.1038/s41581-020-0286-5 [DOI] [PubMed] [Google Scholar]
- 41.Tuttle KR, Knight R, Appelbaum PS, et al. Integrating patient priorities with science by community engagement in the kidney precision medicine project. Clin J Am Soc Nephrol. 2021;16(4):660–668. doi: 10.2215/CJN.10270620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hodgin JB, Mariani LH, Zee J, et al. Quantification of Glomerular Structural Lesions: Associations With Clinical Outcomes and Transcriptomic Profiles in Nephrotic Syndrome. Am J Kidney Dis. 2022;79(6):807–819.e1. doi: 10.1053/j.ajkd.2021.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mariani L, Bomback A, Canetta P, et al. CureGN Study Rationale, Design, and Methods: Establishing a Large Prospective Observational Study of Glomerular Disease. Am J Kidney Dis. 2019;73(2):218–229. doi: 10.1053/j.ajkd.2018.07.020.Corresponding [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Afshinnia F, Rajendiran TM, Soni T, et al. Impaired B-oxidation and altered complex lipid fatty acid partitioning with advancing CKD. J Am Soc Nephrol. 2018;29(1):295–306. doi: 10.1681/ASN.2017030350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sas KM, Karnovsky A, Michailidis G, Pennathur S. Metabolomics and diabetes: Analytical and computational approaches. Diabetes. 2015;64(3):718–732. doi: 10.2337/db14-0509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Quehenberger O, Dennis EA. The Human Plasma Lipidome. N Engl J Med. 2011;365(19):1812–1823. doi: 10.1056/nejmra1104901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bahiru E, Hsiao R, Phillipson D, Watson KE. Mechanisms and Treatment of Dyslipidemia in Diabetes. Curr Cardiol Rep. 2021;23(4):26. doi: 10.1007/s11886-021-01455-w [DOI] [PubMed] [Google Scholar]
- 48.Baek J, He C, Afshinnia F, Michailidis G, Pennathur S. Lipidomic approaches to dissect dysregulated lipid metabolism in kidney disease. Nat Rev Nephrol. 2022;18(1):38–55. doi: 10.1038/s41581-021-00488-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Russo GT, De Cosmo S, Viazzi F, et al. Plasma triglycerides and HDL-C levels predict the development of diabetic kidney disease in subjects with type 2 diabetes: The AMD annals initiative. Diabetes Care. 2016;39(12):2278–2287. doi: 10.2337/dc16-1246 [DOI] [PubMed] [Google Scholar]
- 50.Forbes JM, Thorburn DR. Mitochondrial dysfunction in diabetic kidney disease. Nat Rev Nephrol. 2018;14(5):291–312. doi: 10.1038/nrneph.2018.9 [DOI] [PubMed] [Google Scholar]
- 51.Sas KM, Kayampilly P, Byun J, et al. Tissue-specific metabolic reprogramming drives nutrient flux in diabetic complications. JCI Insight. 2016;1(15). doi: 10.1172/jci.insight.86976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sharma K, Karl B, Mathew AV, et al. Metabolomics reveals signature of mitochondrial dysfunction in diabetic kidney disease. J Am Soc Nephrol. 2013;24(11):1901–1912. doi: 10.1681/ASN.2013020126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Herman-Edelstein M, Scherzer P, Tobar A, Levi M, Gafter U. Altered renal lipid metabolism and renal lipid accumulation in human diabetic nephropathy. J Lipid Res. 2014;55(3):561–572. doi: 10.1194/jlr.P040501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhao YY, Wu SP, Liu S, Zhang Y, Lin RC. Ultra-performance liquid chromatography-mass spectrometry as a sensitive and powerful technology in lipidomic applications. Chem Biol Interact. 2014;220:181–192. doi: 10.1016/j.cbi.2014.06.029 [DOI] [PubMed] [Google Scholar]
- 55.Hansen J, Sealfon R, Menon R, et al. A reference tissue atlas for the human kidney. Sci Adv. 2022;8(23). doi: 10.1126/sciadv.abn4965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Afshinnia F, Nair V, Lin J, et al. Increased lipogenesis and impaired B-oxidation predict type 2 diabetic kidney disease progression in American Indians. JCI Insight. 2019;4(21). doi: 10.1172/jci.insight.130317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wilson PC, Wu H, Kirita Y, et al. The single cell transcriptomic landscape of early human diabetic nephropathy. Proc Natl Acad Sci. Published online 2019. doi: 10.1017/CBO9781107415324.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fu J, Akat KM, Sun Z, et al. Single-cell RNA profiling of glomerular cells shows dynamic changes in experimental diabetic kidney disease. J Am Soc Nephrol. 2019;30(4):533–545. doi: 10.1681/ASN.2018090896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Barwinska D, El-Achkar TM, Ferreira RM, et al. Molecular characterization of the human kidney interstitium in health and disease. Sci Adv. 2021;7(7):1–14. doi: 10.1126/sciadv.abd3359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Patel J, Torrealba J, Poggio E, et al. Molecular Signatures of Diabetic Kidney Disease Hiding in a Patient with Hypertension-Related Kidney Disease. Clin J Am Soc Nephrol. Published online 2021:CJN.10350721. doi: 10.2215/cjn.10350721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nair V, Komorowsky CV., Weil EJ, et al. A molecular morphometric approach to diabetic kidney disease can link structure to function and outcome. Kidney Int. 2018;93(2):439–449. doi: 10.1016/j.kint.2017.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lake BB, Menon R, Winfree S, et al. An atlas of healthy and injured cell states and niches in the human kidney. bioRxiv. Published online 2021:2021.07.28.454201. https://www.biorxiv.org/content/10.1101/2021.07.28.454201v1%0Ahttps://www.biorxiv.org/content/10.1101/2021.07.28.454201v1.abstract [DOI] [PMC free article] [PubMed]
- 63.Afshinnia F, Rajendiran TM, He C, et al. Circulating Free Fatty Acid and Phospholipid Signature Predicts Early Rapid Kidney Function Decline in Patients With Type 1 Diabetes. Diabetes Care. Published online July 2021. doi: 10.2337/dc21-0737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ma J, Karnovsky A, Afshinnia F, et al. Differential network enrichment analysis reveals novel lipid pathways in chronic kidney disease. Bioinformatics. 2019;35(18):3441–3452. doi: 10.1093/bioinformatics/btz114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wu H, Villalobos RG, Yao X, et al. Mapping the single-cell transcriptomic response of murine diabetic kidney disease to therapies. Cell Metab. 2022;34:1–15. doi: 10.1016/j.cmet.2022.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sas K, Lin J, Wang CH, et al. Renin-angiotensin system inhibition reverses the altered triacylglycerol metabolic network in diabetic kidney disease. Metabolomics. 2022;17(7). doi: 10.1007/s11306-021-01816-0.Renin-angiotensin [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Warren AM, Knudsen ST, Cooper ME. Diabetic nephropathy: an insight into molecular mechanisms and emerging therapies. Expert Opin Ther Targets. 2019;23(7):579–591. doi: 10.1080/14728222.2019.1624721 [DOI] [PubMed] [Google Scholar]
- 68.Tanase DM, Gosav EM, Anton MI, et al. Oxidative Stress and NRF2/KEAP1/ARE Pathway in Diabetic Kidney Disease (DKD): New Perspectives. Biomolecules. 2022;12(9):1–25. doi: 10.3390/biom12091227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fontecha-Barriuso M, Lopez-Diaz AM, Guerrero-Mauvecin J, et al. Tubular Mitochondrial Dysfunction, Oxidative Stress, and Progression of Chronic Kidney Disease. Antioxidants. 2022;11(7):1–21. doi: 10.3390/antiox11071356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lewis EJ, Lewis JB, Greene T, et al. Sulodexide for kidney protection in type 2 diabetes patients with microalbuminuria: A randomized controlled trial. Am J Kidney Dis. 2011;58(5):729–736. doi: 10.1053/j.ajkd.2011.06.020 [DOI] [PubMed] [Google Scholar]
- 71.Liu YN, Zhou J, Li T, et al. Sulodexide Protects Renal Tubular Epithelial Cells from Oxidative Stress-Induced Injury via Upregulating Klotho Expression at an Early Stage of Diabetic Kidney Disease. J Diabetes Res. 2017;2017. doi: 10.1155/2017/4989847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tesch GH, Ma FY, Han Y, Liles JT, Breckenridge DG, Nikolic-Paterson DJ. ASK1 inhibitor halts progression of diabetic nephropathy in Nos3-deficient mice. Diabetes. 2015;64(11):3903–3913. doi: 10.2337/db15-0384 [DOI] [PubMed] [Google Scholar]
- 73.Tuttle KR, Brosius FC, Adler SG, et al. JAK1/JAK2 inhibition by baricitinib in diabetic kidney disease: Results from a Phase 2 randomized controlled clinical trial. Nephrol Dial Transplant. 2018;33(11):1950–1959. doi: 10.1093/ndt/gfx377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ong E, Wang LL, Schaub J, et al. Modelling kidney disease using ontology: insights from the Kidney Precision Medicine Project. Nat Rev Nephrol. 2020;16(November). doi: 10.1038/s41581-020-00335-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.He Y, Steck B, Ong E, et al. KTAO: A kidney tissue atlas ontology to support community-based kidney knowledge base development and data integration. CEUR Workshop Proc. 2018;2285(Icbo):1–6. [Google Scholar]
- 76.Makino M, Yoshimoto R, Ono M, et al. Artificial intelligence predicts the progression of diabetic kidney disease using big data machine learning. Sci Rep. 2019;9(1):1–9. doi: 10.1038/s41598-019-48263-5 [DOI] [PMC free article] [PubMed] [Google Scholar]


