ABSTRACT
The emergence of Plasmodium falciparum resistance raises an urgent need to find new antimalarial drugs. Here, we report the rational repurposing of the anti-hepatitis C virus drug, alisporivir, a nonimmunosuppressive analog of cyclosporin A, against artemisinin-resistant strains of P. falciparum. In silico docking studies and molecular dynamic simulation predicted strong interaction of alisporivir with PfCyclophilin 19B, confirmed through biophysical assays with a Kd value of 354.3 nM. Alisporivir showed potent antimalarial activity against chloroquine-resistant (PfRKL-9 with resistance index [Ri] 2.14 ± 0.23) and artemisinin-resistant (PfKelch13R539T with Ri 1.15 ± 0.04) parasites. The Ri is defined as the ratio between the IC50 values of the resistant line to that of the sensitive line. To further investigate the mechanism involved, we analyzed the expression level of PfCyclophilin 19B in artemisinin-resistant P. falciparum (PfKelch13R539T). Semiquantitative real-time transcript, Western blot, and immunofluorescence analyses confirmed the overexpression of PfCyclophilin 19B in PfKelch13R539T. A 50% inhibitory concentration in the nanomolar range, together with the targeting of PfCyclophilin 19B, suggests that alisporivir can be used in combination with artemisinin. Since artemisinin resistance slows the clearance of ring-stage parasites, we performed a ring survival assay on artemisinin-resistant strain PfKelch13R539T and found significant decrease in parasite survival with alisporivir. Alisporivir was found to act synergistically with dihydroartemisinin and increase its efficacy. Furthermore, alisporivir exhibited antimalarial activity in vivo. Altogether, with the rational target-based Repurposing of alisporivir against malaria, our results support the hypothesis that targeting resistance mechanisms is a viable approach toward dealing with drug-resistant parasite.
KEYWORDS: alisporivir, antimalarial, artemisinin resistance, PfCyclophilin 19B, cyclosporin A
INTRODUCTION
Malaria is a vector borne life-threatening disease caused by obligate intracellular Plasmodium species parasites, causing an estimated 0.627 million deaths in 2020 (1). Worryingly, Plasmodium falciparum has developed drug resistance to all known antimalarial therapeutics, leading to discontinuation of many first-line treatments, including chloroquine, proguanil, pyrimethamine, sulfadoxine-pyrimethamine, and mefloquine. More recently, reports of emerging resistance against artemisinin-based combination therapies threaten ongoing global efforts to eliminate malaria and highlight an urgent need to discover novel therapeutics with potentially new mechanisms of action (2). Artemisinin therapy causes immediate and rapid clearance of ring-stage parasites, resulting in faster clinical response. However, the short half-life of artemisinin in the blood warrants the discovery of novel partner drugs that can maintain drug pressure for a prolonged duration (3). A number of pharmacologically active compounds have been tested, in combination with artemisinin, against resistant and sensitive parasite strains to increase their efficacy in killing parasites.
Cyclophilins are a potential class of drug targets for various diseases that, along with FK506-binding proteins, belong to the immunophilins (4). Cyclophilins are ubiquitous cellular proteins that, in addition to acting as chaperones, possess peptidyl-proline isomerase (PPIase) activity (5). In the P. falciparum database, there are several cyclophilin and cyclophilin-like gene sequences, including PfCyclophilin 19A and PfCyclophilin 19B (6, 7). The role of malaria parasite encoded cyclophilins is largely unknown, but they are identified as a novel class of antimalarial drug targets. P. falciparum cyclophilins have demonstrated chaperone function that is suggested to be essential for the development and survival of P. falciparum. These cyclophilins have different subcellular localizations and are expressed in at least one stage of the P. falciparum life cycle (8). Cyclosporin A, a known inhibitor of cyclophilins, binds to cyclophilins and inhibits their PPIase activity (9, 10). This suggests both the importance of P. falciparum cyclophilins in a possible mechanism of antimalarial action of cyclosporin A and its potential as drug target.
Among P. falciparum cyclophilins, only two, PfCyclophilin 19A and PfCyclophilin 19B, possess the cyclosporin A-binding property and PPIase activity. Cyclosporin A inhibits the PPIase activity of PfCyclophilin 19A and PfCyclophilin 19B (8, 9, 11). However, all P. falciparum cyclophilins can act as chaperones (8). PfCyclophilin 19B is the major cytosolic cyclophilin protein of the P. falciparum (9). Connected to this is the observation that artemisinin resistance involves the upregulation and downregulation of several genes in protein folding and repair pathways. Previous reports describe significant upregulation of a chaperone PfCyclophilin 19B in artemisinin-resistant field parasites (12, 13). Cyclosporin A has been demonstrated to potentiate the action of artemisinin in the early ring stages and display potent antimalarial activity (14). This indicated the significance of PfCyclophilin 19B in the mode of action of cyclosporin A and in the development of artemisinin-resistant parasites. However, because of the immunosuppressive nature of cyclosporin A due to its interaction with human calcineurin, and its ability to cause eryptosis, cyclosporin A has never been clinically approved as an antimalarial drug (15, 16).
A recently developed cyclosporin A analogue, alisporivir (C63H113 N11O12, molecular weight 1,216.6, also known as Debio-025 or DEB025 [d-MeAla3–EtVal4] cyclosporin A), remains the most promising second-generation cyclophilin inhibitor (17, 18). Alisporivir is effective for the treatment of chronic hepatitis C virus, HIV, equine arteritis virus infections, as well as for treating Duchenne muscular dystrophy (19, 20). It differs slightly from the parent cyclic undecapeptide in that the sarcosine at position 3 is replaced with Me-alanine, the leucine at position 4 is replaced with valine, and the nitrogen at position X is N-ethylated instead of being N-methylated. These chemical modifications not only enhance alisporivir’s binding affinity for cyclophilins, they also abolish its binding to calcineurin, leading to the cessation of immunosuppressive activity (19). Alisporivir has a plasma half-life of 60 to 90 h; because of this, alisporivir has been chosen for a once-daily administration (20), further supporting its candidature as an artemisinin partner drug.
We report here, for the first time, the repurposing of the anti-hepatitis C virus drug alisporivir, a nonimmunosuppressive analog of cyclosporin A, against artemisinin-resistant strains of P. falciparum. Alisporivir exhibited potent antiparasitic effect against P. falciparum parasite, both in in vitro culture and in an in vivo mice model. Alisporivir potently inhibited the ring-stage survival of artemisinin-resistant P. falciparum (PfKelch13R539T). Mechanistically, the potent effect of alisporivir on artemisinin-resistant P. falciparum strain can be explained through its specific targeting of upregulated PfCyclophilin 19B in the resistant strain. Docking interaction studies further confirm specific binding of PfCyclophilin 19B with alisporivir. In addition, alisporivir treatment enhanced the efficacy of dihydroartemisinin (DHA) against parasite growth in a ring survival assay (RSA) and combination assays, suggesting that targeting the resistance mechanism of the parasite is an effective approach when dealing with the rapid emergence of drug-resistant P. falciparum.
RESULTS
Interaction study of PfCyclophilin 19B with alisporivir and cyclosporin A.
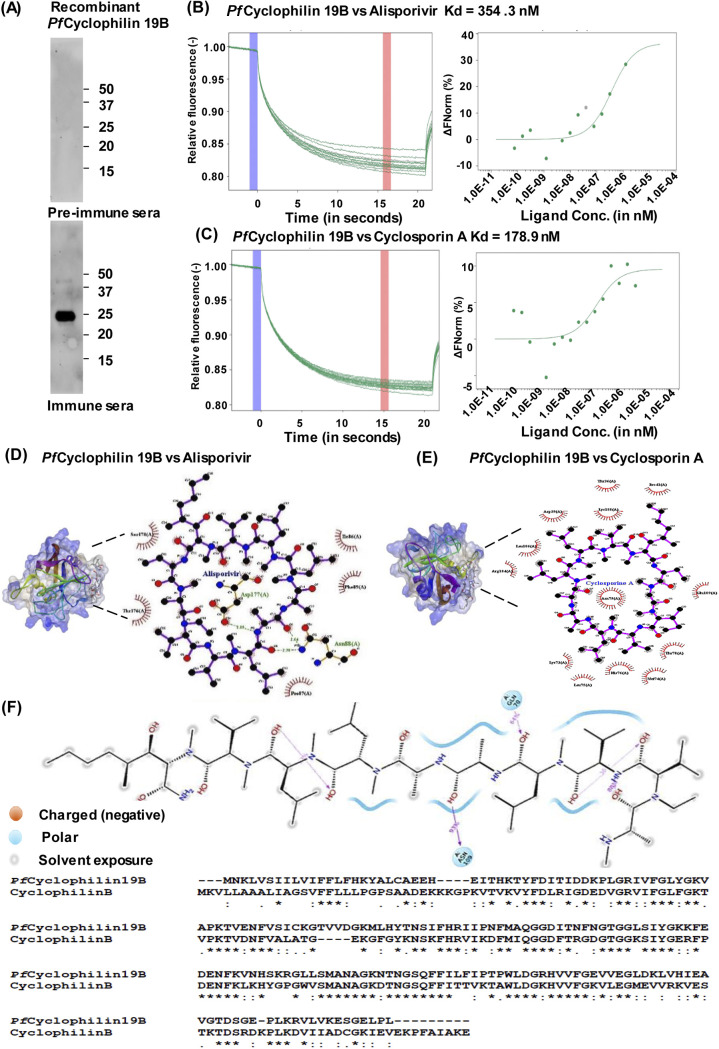
P. falciparum cyclophilins have been identified as a potential therapeutic target (9). To evaluate the binding of parasite cyclophilin with alisporivir, the expression and purification of PfCyclophilin 19B protein was performed in a bacterial system. For this, the PfCyclophilin 19B gene was cloned in a pET28a expression vector, and recombinant PfCyclophilin 19B (rPfCyclophilin 19B) protein was expressed and purified (see Fig. S1 in the supplemental material). Antibodies were generated against rPfCyclophilin 19B protein. Western blot analysis with the raised antibody detected the specific band of rPfCyclophilin 19B protein (Fig. 1A).
FIG 1.
PfCyclophilin 19B interaction study with alisporivir and cyclosporin A. (A) Western blot analysis of rPfCyclophilin 19B protein by using antibodies raised against purified protein. (B and C) MST interaction study of PfCyclophilin 19B with alisporivir (B) and cyclosporin A (C). The concentration of fluorescently labeled PfCyclophilin 19B was kept constant at 20 nM, and the drug concentrations varied. Thermographs show the binding of PfCyclophilin 19B with alisporivir and cyclosporin A (left). The cold spot at 0 s is indicated as a blue region, and the hot spot at 15 s is represented as a red region. The analysis of thermophoresis was performed by plotting the difference in normalized fluorescence. A dose-response curve shows the interaction of PfCyclophilin 19B protein with alisporivir and cyclosporin A (right). In the bound state with protein, there was a stronger decrease in fluorescence compared to the unbound state. (D) 3D surface model of the alisporivir-PfCyP19B complex with a LigPlot analysis of the drug and protein. (E) 3D surface model of the cyclosporin A-PfCyP19B complex with a LigPlot analysis of the drug and protein. (F) Analysis of the percent occupancy of the direct interaction between alisporivir and PfCyclophilin 19B residues. Sequence analysis of PfCyclophilin 19B with human Cyclophilin B.
To evaluate the interaction of alisporivir and cyclosporin A with PfCyclophilin 19B protein, microscale thermophoresis (MST) using NanoTemper Monolith was performed. MST is based on thermophoresis, which is combined with fluorescence detection in order to study molecular interactions (21). PfCyclophilin 19B protein was labeled using the prescribed NanoTemper protocol. Alisporivir concentration was varied, whereas the PfCyclophilin 19B concentration was kept constant. Thermograph showed no sign of molecule adsorption or aggregation to the capillary (Fig. 1B, C right). The Kd value was measured from the binding curve plotted between normalized fluorescence and substrate concentration. Interaction of PfCyclophilin 19B with alisporivir resulted in a calculated Kd of 354.3 nM, while cyclosporin A interacted with PfCyclophilin 19B with a Kd of 178.9 nM (Fig. 1B and C, left). Next, we performed in silico docking studies of alisporivir with PfCyclophilin 19B. The structure-refined model of PfCyclophilin 19B with an RMSD score of <2.0 was used for docking with alisporivir and cyclosporin A. The molecular docking results revealed that alisporivir could efficiently bind to PfCyclophilin 19B with a minimum binding energy of −5.72 (Fig. 1D). Moreover, alisporivir forms more hydrogen bonds with PfCyclophilin 19B compared to cyclosporin A (Fig. 1E). Interaction analysis of alisporivir with PfCyclophilin 19B protein residues demonstrated strong binding of alisporivir to the protein (see Fig. S2). Analysis of percentage occupancy of the direct interaction between alisporivir and PfCyclophilin 19B residues is shown in Fig. 1F.
Confirmation of alisporivir and cyclosporin A binding with PfCyclophilin 19B protein.
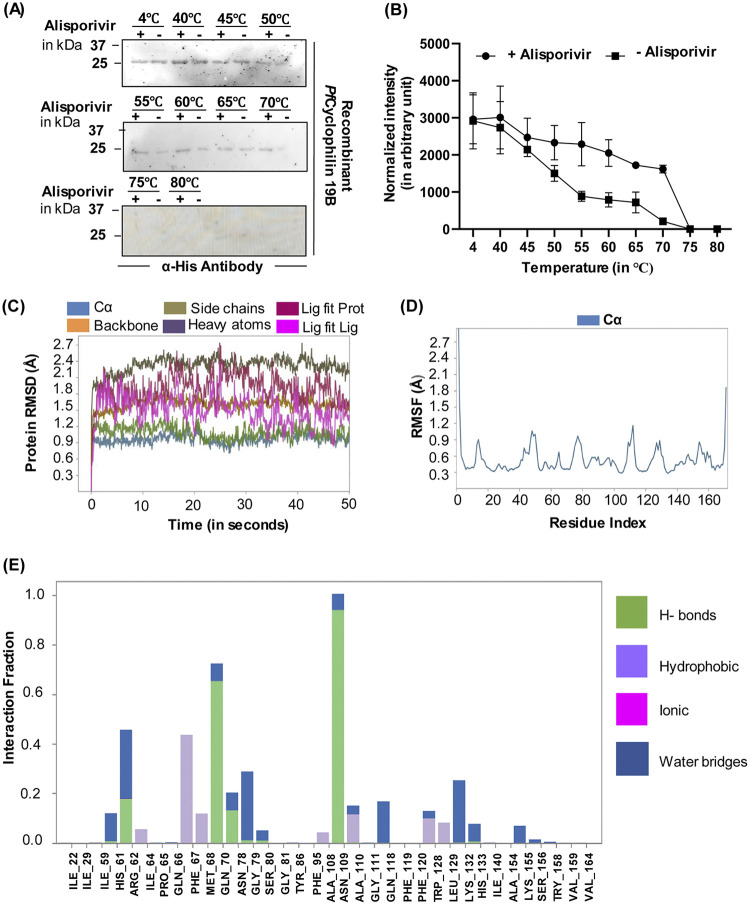
To further confirm the binding of alisporivir and cyclosporin A with PfCyclophilin 19B, protein thermal shift assay, on the same principle of cellular thermal shift assay (CETSA), was performed. In this method, thermostability of purified PfCyclophilin 19B protein was determined across a temperature gradient (40 to 80°C) in the presence or absence of alisporivir. Samples at 4°C were taken as a control. At an elevated temperature, the specific binding of ligand with target protein stabilizes the protein from denaturation and precipitation, thus increasing the stability of the target protein in the bound state. The amount of soluble protein in the samples was determined by Western blot analysis of the soluble protein fraction (Fig. 2A). An increase in band intensities in the presence of alisporivir indicates thermal protection of PfCyclophilin 19B protein at higher temperatures (Fig. 2B).
FIG 2.
Confirmation of PfCyclophilin 19B interaction with alisporivir and cyclosporin A. (A) Protein thermal shift assay. The figure depicts the immune-based thermal stability of PfCyclophilin 19B protein in the presence or absence of alisporivir across a temperature gradient between 40 and 80°C. (B) Densitometry quantification of the respective immunoblot was performed using ImageJ software, and the plotted line graph shows the increased thermostability of PfCyclophilin 19B in the presence of alisporivir (2× IC50). The intensity of the 4°C sample was used to normalize the intensity of the heated samples. The data represent the means ± the SD of three independent experiments. (C) RMSD of the simulated complex. (D) RMSF of the simulated complex. (E) Protein-ligand interactions of the MD simulation. All residues that interact with the ligand are shown in the histogram, along with the fraction of the simulation for each type of interaction.
PfCyclophilin 19B-alisporivir contacts during dynamic simulation.
Molecular dynamic simulation (MDS) was used to verify alisporivir’s binding modes and interactions. Root mean square deviations (RMSD) and root mean square fluctuations (RMSF) between proteins, ligands, and protein-ligand interactions were examined for 50 ns, while simulating the PfCyclophilin 19B and alisporivir. As shown in Fig. 2C, the docked complex’s C and backbone RMSD values were within the allowed range of 0 to 2.7 Å. Lig fit Prot and Lig fit RMSD values were both within the range of 0 to 2.5 Å. As a result, the protein chain’s shape was not altered by the docked molecules. Fig. 2D shows the portions of the protein that change the most throughout the simulation. The protein’s N- and C-terminal tails tend to vary the most. Secondary structural components, such as alpha helices and beta strands, tend to be more stable than the protein’s unstructured regions and thus change less (see Fig. S3). Throughout the simulation, the alisporivir and PfCyclophilin 19B interactions were carefully observed. Four forms of interactions between proteins and ligands (or “contacts”) have been grouped into hydrogen bonds, hydrophobic, ionic, and water bridges (Fig. 2E). Interactions and contacts (hydrogen bonds, hydrophobic, ionic, and water bridges) seen in Fig. S4 are shown in a time-lapse diagram (Fig. 2E). The number of interactions between the protein and the ligand is shown in the top panel. The residues that interact with the ligand at various places along the trajectories may be seen in the bottom panel. On the scale to the right of the graphic, residues that make more than one particular contact with the ligand are shown in a deeper orange tone (see Fig. S5 and S6). All of the candidates had interactions with the amino acids GLN70 and ASN109, which are depicted in Fig. 1C for totals of 64 and 93% of the simulation time. Important candidates have also created more than four interactions, which are responsible for the complex’s steady behavior (see Fig. S7).
In vitro antimalarial activity of alisporivir against P. falciparum.
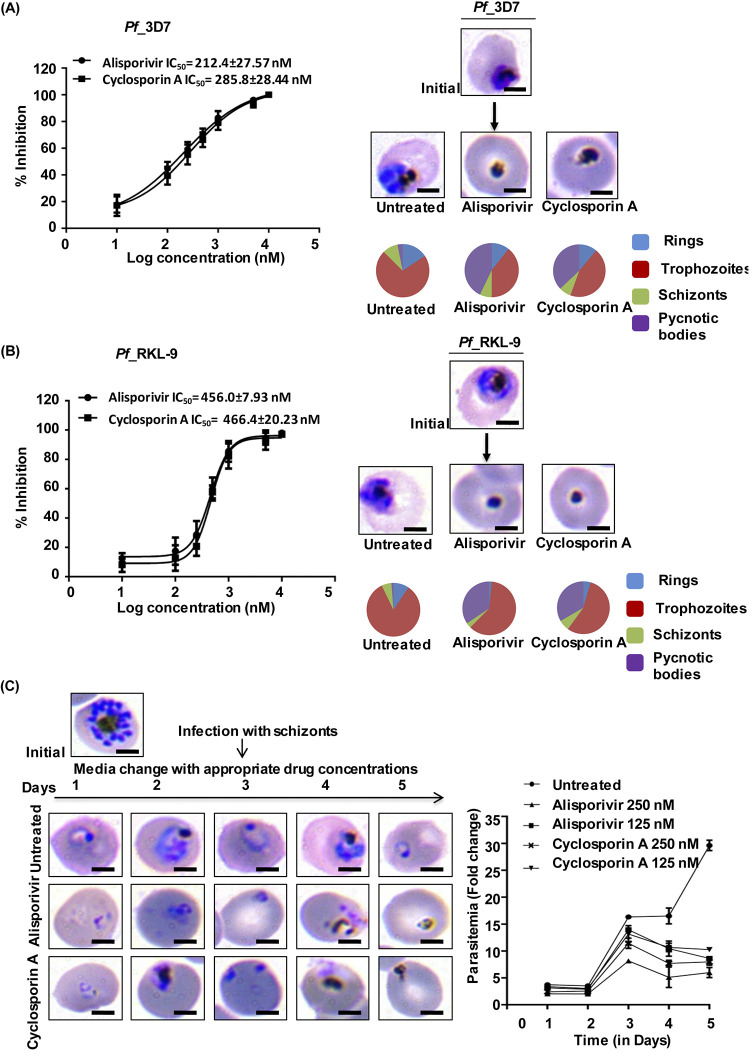
We performed growth inhibition assays (GIA) against Pf3D7 strain to investigate the antimalarial potential of alisporivir. The results, assessed by Giemsa staining, indicate that alisporivir inhibits parasite growth with a 50% inhibitory concentration (IC50) of 212.4 ± 27.57 nM compared to 285.8 ± 28.44 nM for cyclosporin A (Fig. 3A). Furthermore, alisporivir and cyclosporin A potently inhibited the growth of the chloroquine-resistant PfRKL-9 strain (Rourkela, Odisha, India) with IC50s of 456.0 ± 7.93 nM and 466.4 ± 20.23 nM, respectively (Fig. 3B) (22, 23). Morphology assessment of the Giemsa-stained parasites was done under the microscope 48 h posttreatment to evaluate the development of parasites. Alisporivir or cyclosporin A treated Pf3D7 and PfRKL-9 strains could not develop into healthy trophozoites and instead demonstrate the formation of pycnotic bodies.
FIG 3.
Effect of the alisporivir on P. falciparum growth in vitro. The dose-dependent inhibition of parasite growth was monitored in the presence of alisporivir and cyclosporin A for the Pf3D7 strain (A) and the Pf RKL-9 strain (B). IC50 values of the drugs were calculated by plotting the values of the percent growth inhibition against the log concentrations of the drug and then calculated by using nonlinear regression analysis with GraphPad Prism 6 software. The data represent the means ± the SEM of three independent experiments. Pie charts show the relative distributions of different stages of parasites and pycnotic body formations at the respective IC50s after 48 h of infection. (C) Light microscopy images of Giemsa-stained smears of alisporivir or cyclosporin A (250 nM)-treated P. falciparum parasites. The fold change in parasitemia at different time points of progression was determined. A significant reduction was observed compared to the control at day 5 (alisporivir or cyclosporin A, 125 nM [*, P < 0.05]; alisporivir or cyclosporin A, 250 nM [**, P < 0.01]). Pie diagrams show the relative proportions of different stages of parasites and pycnotic bodies. Alisporivir potentiates the antimalarial efficacy of DHA against artemisinin-resistant P. falciparum.
Effect of alisporivir on the progression of P. falciparum asexual blood stages.
To investigate the effect of alisporivir on progression of asexual parasite blood stages, the Pf3D7 parasite culture at the schizont stage was treated with alisporivir or cyclosporin A (125 or 250 nM) and evaluated for 5 days, until the ring stage of the third cycle. At each stage, parasite development was monitored by analyzing Giemsa-stained smears. The fold change in parasitemia remarkably decreased at the end of the third cycle in treated cultures compared to the control. As expected, a significant increase in the formation of pycnotic bodies was observed in treated parasite cultures (Fig. 3C).
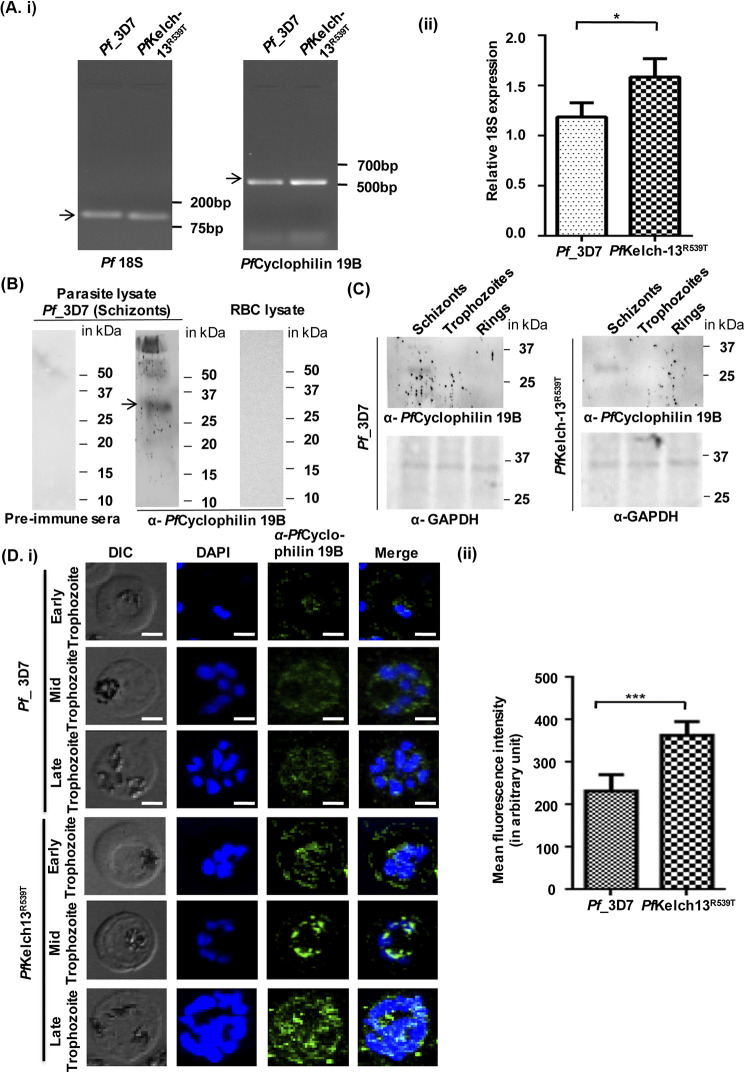
Expression analysis of PfCyclophilin 19B in P. falciparum 3D7 (artemisinin-sensitive) and PfKelch13R539T (artemisinin-resistant) strains.
Previous reports indicate upregulation of PfCyclophilin 19B in artemisinin-resistant field strains (12, 13). We investigated PfCyclophilin 19B expression levels in Pf3D7 and an artemisinin-resistant strain (PfKelch13R539T) by reverse transcription-PCR (RT-PCR) analysis (Fig. 4A, subpanels i and ii). Increased expression of PfCyclophilin 19B was observed in the PfKelch13R539T strain of malaria parasite. This is similar to transcriptomics data published earlier (12, 13). The specificity of raised PfCyclophilin 19B antibody was evaluated against parasite and red blood cell (RBC) lysates. The results identified a specific band in parasite lysate representing PfCyclophilin 19B protein, whereas no band was detected in the RBC lysate (Fig. 4B). We next assessed the expression of PfCyclophilin 19B at the protein level by Western blotting (Fig. 4C) and immunofluorescence assay (Fig. 4D). Western blot analysis showed the expression of PfCyclophilin 19B protein in the PfKelch13R539T strain and the Pf3D7 strain. Next, the expression of PfCyclophilin 19B was investigated in parasite-infected RBCs by immunofluorescence assay using PfCyclophilin 19B antibody. The expression of PfCyclophilin 19B can be detected in the Pf3D7 strain of malaria parasites, observed as green, fluorescent staining; however, the intensity of the green fluorescent staining in artemisinin-resistant parasites (PfKelch13R539T) was higher compared to the Pf3D7 strain (Fig. 4Di). Parasite nuclei were stained with DAPI (4′,6′-diamidino-2-phenylindole). This indicates elevated expression of PfCyclophilin 19B in artemisinin-resistant parasites compared to wild-type parasites, since the execution was carried out at an identical gain setting. Further, the fluorescence intensity was measured, and a significant increase in the expression of PfCyclophilin 19B was confirmed in the PfKelch13R539T strain (Fig. 4Dii). The results of RT-PCR analysis and immunofluorescence assay showed increased expression of the PfCyclophilin 19B protein in the PfKelch13R539T strain compared to the Pf3D7 strain. These results suggest that targeting the overexpression of PfCyclophilin 19B with drugs, including alisporivir, has therapeutic potential for the treatment of artemisinin-resistant malaria.
FIG 4.
Expression analysis of PfCyclophilin 19B in P. falciparum 3D7 and artemisinin-resistant PfKelch13R539T strains. (Ai) Representative image of the RNA expression level of PfCyclophilin 19B in Pf3D7 and PfKelch13R539T strains. (ii) Quantification of changes in the PfCyclophilin 19B RNA expression level was calculated relative to the RNA level of Pf18S (internal control). The data represent the means ± the SD of three independent experiments. The statistical significance of the difference in RNA levels was calculated using an unpaired t test with Welch’s correction (*, P < 0.05). (B) Western blot showing the specific band of the PfCyclophilin 19B protein in the Pf3D7 parasite lysate, while no such band was observed in the RBC lysate. Western blot analysis of Pf3D7 parasite lysate with preimmune sera was used as a negative control. The arrow indicates the band of PfCyclophilin 19B protein in the parasite lysate. (C) Expression analysis of the PfCyclophilin 19B protein at different stages (schizont, trophozoite, and ring) of parasite lysates of the Pf3D7 and PfKelch13R539T strains. Blotting with GAPDH (glyceraldehyde-3-phosphate dehydrogenase) served as a loading control. (Di) Confocal microscopic imaging to evaluate expression pattern of PfCyclophilin 19B protein in parasitized Pf3D7 and PfKelch13R539T strains. Scale bar, 5 μm. (ii) Measurement of the mean fluorescence intensity of the PfCyclophilin 19B protein in the Pf3D7 strain, as well as in the PfKelch13R539T strain. The statistical significance of difference was calculated using an unpaired t test with Welch’s correction (***, P < 0.001).
Localization of PfCyclophilin 19B in malaria parasites.
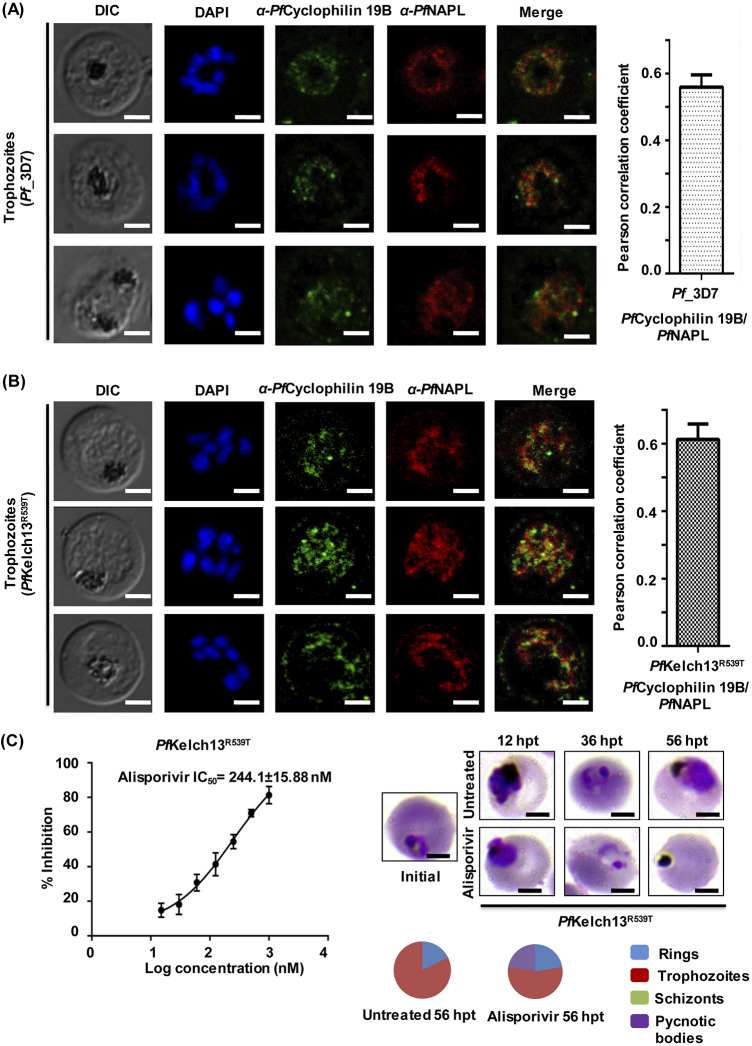
Next, we investigated the localization of PfCyclophilin 19B in the trophozoite stage of parasites by immunofluorescence assay in P. falciparum 3D7 and artemisinin-resistant strains (PfKelch13R539T). In both strains, localization of PfCyclophilin 19B was observed in the cytoplasm of the parasite. Localization of a cytoplasmic protein, PfNAPL (nucleosome assembly protein-large), was also observed in these parasites. In the asexual and sexual stages of the parasite, PfNAPL is reported to be predominantly localized in the cytoplasm (24). PfCyclophilin 19B colocalizes with PfNAPL and both were observed in the parasite cytosol (Fig. 5A and B, left). The degree of colocalization was quantified by determining the pearson correlation coefficient. The results confirm colocalization of PfCyclophilin 19B with PfNAPL in the parasite cytosol (Fig. 5A and B, right).
FIG 5.
Localization of PfCyclophilin 19B in malaria parasite. The expression and colocalization of PfCyclophilin 19B (green channel) and PfNAPL (red channel) in the trophozoite stage of the parasite (left) are evident. (A) P. falciparum 3D7 strain. (B) PfKelch13R539T strain. Nuclei were counterstained with DAPI. Scale bar, 5 μm. The Pearson correlation coefficient of PfCyclophilin 19B colocalization with PfNAPL is shown (right). (C) Determination of the inhibitory potential of alisporivir against PfKelch13R539T. The IC50 value was calculated using GraphPad Prism 6 software. The data represent the means ± the SEM of three independent experiments. Light microscopic examination of the parasite morphology was performed after treatment. A pie chart shows the relative distributions of different stages of parasites and pycnotic body formations at the respective IC50s.
The increased expression of PfCyclophilin 19B in artemisinin-resistant parasites suggests that it plays an important role in the development of drug resistance against artemisinin. We next examined the antimalarial activity of alisporivir against the artemisinin-resistant strain (PfKelch13R539T) through parasite GIA at various concentrations (15 to 1,000 nM) of alisporivir. Alisporivir was found to be potent against artemisinin-resistant parasites. We observed an IC50 of 244.1 ± 15.88 nM against artemisinin-resistant parasites, which is similar to the IC50 for wild-type parasites. Stage-specific analysis of the morphology of treated parasites at IC50 of alisporivir again demonstrated that treated parasites were reduced to pycnotic bodies, whereas control parasites progressed normally (Fig. 5C).
The ratio between the IC50 of the resistant line (Pf RKL-9 and PfKelch13R539T strain) and that of the sensitive line (Pf3D7 strain) is defined as the resistance index (Ri). A higher level of drug resistance is indicated by a higher Ri. The calculated Ri values for artemisinin-resistant and chloroquine-resistant strains were 1.15 ± 0.04 and 2.14 ± 0.23, respectively. Interestingly, a lower Ri indicates that alisporivir has strong antimalarial activity against the artemisinin-resistant strain.
Alisporivir potentiates antimalarial efficacy of DHA against artemisinin-resistant P. falciparum.
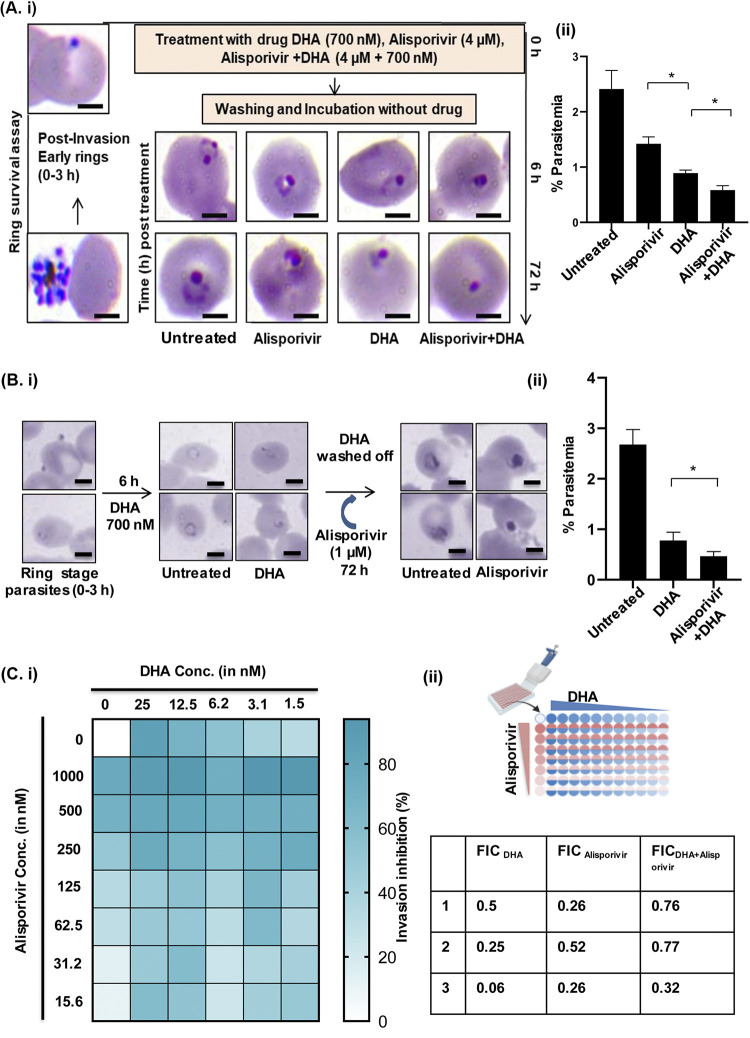
The lower rate of ring-stage parasite clearance is defined as artemisinin resistance (25). An RSA was designed to access the presence of these dormant slow-clearing parasites leading to drug resistance. To determine the potency of alisporivir against artemisinin-resistant parasites, an RSA was performed on the PfKelch13R539T strain with DHA, alisporivir, or a combination of DHA and alisporivir. A tightly synchronized ring-stage culture was treated with the drugs for 6 h. After treatment, the culture was maintained without drug pressure to determine parasite viability. Ring-stage development of parasites was observed to study the effect of drug exposure after 72 h of posttreatment. Control parasites could progress to form healthy ring-stage parasites, whereas in DHA- and alisporivir-treated cultures dead parasites could be observed (Fig. 6Ai). At 72 h posttreatment, alisporivir and DHA were associated with decreased parasite survival in the artemisinin-resistant strain. Parasitemia was decreased by ~50% after treatment with alisporivir. Interestingly, the combination of alisporivir with DHA further potentiated the efficacy of DHA by decreasing the parasitemia by 75% in the resistant strain after 72 h of treatment and decreased the parasite survival at the early ring stage (Fig. 6Aii).
FIG 6.
(A) Effect of alisporivir on the ring survival of the artemisinin-resistant parasite strain. (i) Light microscopic images of Giemsa-stained parasites treated with DHA, alisporivir, and DHA plus alisporivir. For RSA, early-ring-stage parasites (0 to 3 h) were incubated with DHA (700 nM), alisporivir (4 μM), or DHA+alisporivir (700 nM + 4 μM) for 6 h. Postincubation, the drug was removed by extensive washing, and the parasites were allowed to grow for 72 h. (ii) Bar graph representing the percent parasitemia in untreated, DHA-treated, alisporivir-treated, and DHA+alisporivir-treated samples. The combination of alisporivir with DHA enhances the antiparasitic activity of DHA against an artemisinin-resistant strain (*, P < 0.05). Scale bar, 5 μm. The data represent the means ± the SD of three independent experiments. The statistical significance was calculated using an unpaired t test. (Bi) Effect of alisporivir (1 μM) on a 700 nM DHA-treated ring-stage culture of the artemisinin-resistant strain. The culture was incubated with DHA for 6 h and then washed with culture medium; alisporivir (1 μM) was added, and the culture was allowed to grow for 72 h. (ii) Significant reduction in the parasitemia of a DHA-treated ring-stage culture of an artemisinin-resistant strain was observed after treatment with 1 μM alisporivir. The data represent the means ± the SD of three independent experiments. The statistical significance was calculated using an unpaired t test. (C) Checkerboard combination assay showing synergistic antiparasitic activity of alisporivir with DHA against an artemisinin-resistant strain at three different combinations of DHA and alisporivir with an FIC of <1.
Alisporivir targets recovered artemisinin-resistant parasites pretreated with DHA.
Next, we investigated whether alisporivir can kill the recovered remaining parasites after DHA treatment. To assess the effect of alisporivir on the ring-stage parasite survival of PfKelch13R539T strain, parasites were treated with DHA (700 nM) for 6 h. Alisporivir (1 μM) was added to the culture for 72 h, and the percent parasite survival was determined by Giemsa counting (Fig. 6Bi). We observed a significant decrease in DHA-treated parasite survival in the presence of 1 μM alisporivir (Fig. 6Bii). These results showed that alisporivir kills artemisinin-resistant parasites (PfKelch13R539T). Altogether, alisporivir can be used in combination with artemisinin.
Monotherapy for malaria treatment is not recommended because of an increased risk of drug resistance development. Combination therapy is more effective and efficient strategy to combat drug-resistant malaria. Artemisinin-based combination therapies are the most successful option for the treatment of malaria. Emerging resistance against artemisinin and partner drugs raises an urgent need to discover effective antimalarial drugs to combine with artemisinin (26). Here, to determine the combination potential of alisporivir with artemisinin, we performed a checkerboard combination assay and evaluated the effect of different combinations of alisporivir with DHA on the PfKelch13R539T strain (Fig. 6Ci). The results showed that alisporivir and DHA act synergistically at some concentration with an FIC index of <1 (Fig. 6Cii). Thus, alisporivir potentiates the antimalarial activity of artemisinin against an artemisinin-resistant strain.
In vivo antimalarial efficacy of alisporivir against P. berghei.
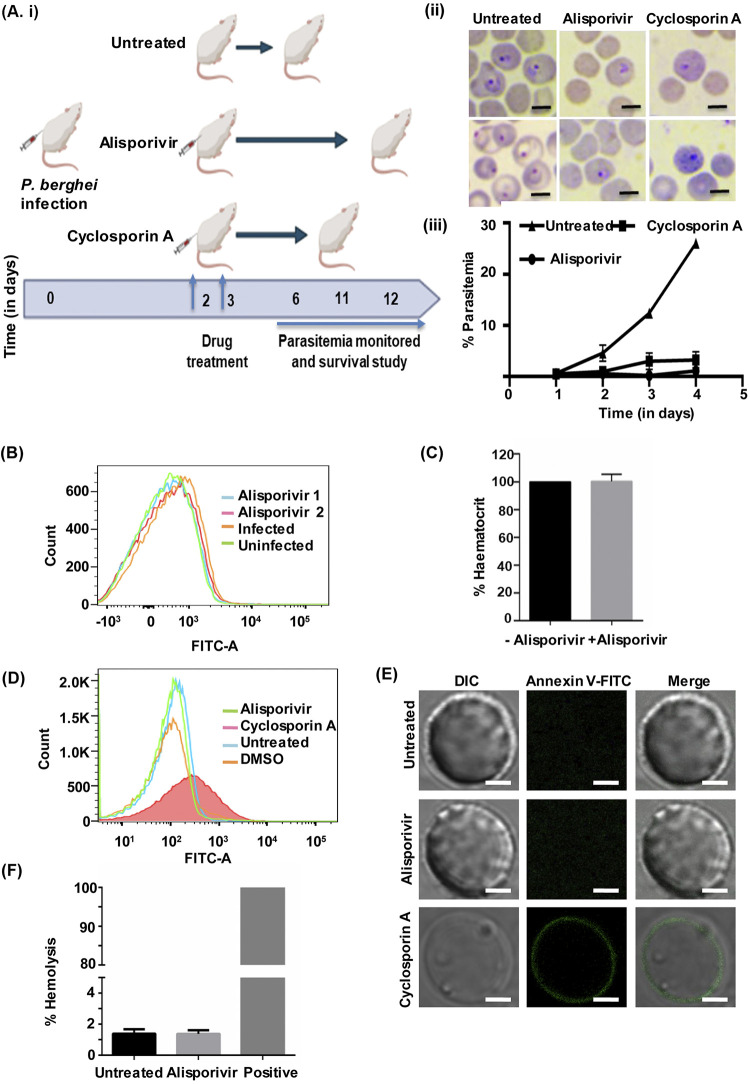
After confirming the efficacy of alisporivir in vitro, we studied the antimalarial potency of alisporivir in vivo on P. berghei mouse malaria parasites. Alisporivir and cyclosporin A were injected at 5 mg kg−1 body weight in P. berghei-infected mice for 2 days. The parasitemia in untreated mice increased with time, up to 25% within 5 days of infection. However, we did not detect a significant increase in parasitemia in alisporivir-treated mice. Cyclosporin A treatment prolonged the prepatent period, in line with previous studies (27, 28). Crucially, alisporivir treatment increased the survival of P. berghei-infected mice. We observed that alisporivir-treated mice survived the infection for more than 12 days, whereas untreated animals died within 6 days of infection (Fig. 7Ai and ii). In order to study whether alisporivir caused eryptosis in P. berghei-infected erythrocytes, the annexin V-binding property of erythrocytes was evaluated by using flow cytometry. Alisporivir-treated erythrocytes did not show any significant change in annexin V-binding property compared to uninfected erythrocytes (Fig. 7B). The percent hematocrit levels were also found to be similar in infected mice compared to alisporivir-treated infected mice (Fig. 7C).
FIG 7.
(Ai) Survival study. (ii) In vivo antimalarial effect of alisporivir and cyclosporin A in mice infected with the P. berghei malaria parasite. Parasitemia was determined in P. berghei-infected mice with or without alisporivir or cyclosporin A (5 mg/kg [body weight]) treatment. The data represent means ± the SD. (B) Representative histogram of annexin V binding in erythrocytes from a P. berghei-infected mouse with or without alisporivir treatment. (C) Hematocrit percentage in alisporivir-treated mice compared to the control. A hematocrit control value was considered as 100%. Both groups have similar packed cell volumes. Scale bar, 5 μm. (D) Evaluation of eryptosis by confocal microscopy and flow cytometry in human erythrocytes treated with 10 μM alisporivir or cyclosporin A for 48 h at 37°C. (E) Representative histogram of the annexin V binding of erythrocytes after exposure to alisporivir or cyclosporin A. (F) The percent hemolysis was calculated at 10 μM alisporivir. The hemolysis value of distilled water-diluted RBCs was taken as 100%.
The parent molecule cyclosporin A induces death of erythrocytes (15). Therefore, we next investigated the safety of alisporivir toward erythrocytes. Erythrocytes were incubated with alisporivir or cyclosporin A (10 μM) for 48 h, stained with FITC-annexin V, and assessed by flow cytometry (Fig. 7D) and confocal microscopy (Fig. 7E). The number of annexin V-binding erythrocytes was found to be extremely low upon alisporivir treatment, similar to untreated erythrocytes; however, cyclosporin A exposure significantly increased the annexin V-positive erythrocytes (Fig. 7D). Alisporivir-treated erythrocytes did not show any green fluorescence, whereas green fluorescence was observed on the surfaces of cyclosporin A-treated erythrocytes, indicating the exposure of phosphatidylserine on the surface, suggesting that alisporivir does not induce eryptosis (Fig. 7E). Next, we investigated the possible hemolysis of erythrocytes following alisporivir treatment. To measure hemolysis, hemoglobin release from erythrocytes into the media was assessed after 24 h of incubation with 10 μM alisporivir, and we found that it remains unaltered. This observation points toward the fact that alisporivir is not toxic to the erythrocytes (Fig. 7F).
Taken together, PfCyclophilin 19B appears to be involved in artemisinin resistance, and our results exploit the possible mechanism that contributes to drug resistance by targeting PfCyclophilin 19B with alisporivir.
DISCUSSION
The unavailability of new potent antimalarials and emerging resistance against artemisinin and its partner drugs are the major obstacles in malaria control (2). Therefore, there is an urgent need for identification of novel drug targets and antimalarial agents to prevent the occurrence of drug resistance. Repurposing existing drugs as novel antimalarials offers a new way for drug development (29, 30). In this study, we have repurposed the anti-hepatitis C virus drug alisporivir and demonstrate its potency against artemisinin-resistant strain (PfKelch13R539T) of P. falciparum and mouse malaria in vivo. Importantly, alisporivir does not cause eryptosis, which is a common side effect of the parent molecule cyclosporin A. Earlier studies demonstrated that cyclosporin A is an effective antimalarial against multiple strains of parasite, but due to its immunosuppressive and eryptosis-inducing nature, it could not be pursued further for malaria drug development (14, 15, 27, 28, 31–33). However, we demonstrate that alisporivir, which is a nonimmunosuppressive drug, does not induce eryptosis either in vitro (human erythrocytes) or in vivo (mouse erythrocytes). Our results and previous studies thus validate alisporivir as a safe drug which can be taken forward for antimalarial drug development.
We further demonstrate that alisporivir targets overexpressed PfCyclophilin 19B in PfKelch13R539T and strongly bind with PfCyclophilin 19B in in silico studies. Next, we performed biophysical assays, MST, and protein thermal shift assay studies to confirm the binding of alisporivir with PfCyclophilin 19B. The results of the protein thermal shift assay showed that alisporivir stabilizes PfCyclophilin 19B protein at an elevated temperature. Further, alisporivir exhibited strong interaction with the PfCyclophilin 19B protein (Kd = 354.3 nM) in an MST experiment. More importantly, in an RSA, combination of alisporivir with DHA potently inhibited the survival of the artemisinin-resistant parasites, indicating the important role of PfCyclophilin 19B in artemisinin resistance development. Interestingly, alisporivir kills survivor parasites pretreated with DHA. In addition, a synergy checkerboard assay demonstrated the synergistic interaction between alisporivir and DHA. Altogether, our data demonstrate that alisporivir can be used as an effective antimalarial or as a partner drug with artemisinin for malaria treatment.
Fast-acting antimalarial drugs represent a novel class of therapeutic agents with efficacy against all parasite strains. However, an increase in the number of cases of drug resistance development and treatment failure has been observed in certain populations. For the curative treatment of Plasmodium parasite infection, a therapeutic regimen combining the fast-acting antimalarial potency of a drug, along with a partner drug having a longer plasma life, is recommended. This will ensure the killing of residual parasites and detain the development of drug resistance (26). Alisporivir, with an estimated plasma half-life in range of 60 to 90 h, fulfills this need and hence is suitable for once-daily administration (19, 20). In addition, clinical pharmacokinetic studies have shown the rapid absorption of alisporivir after oral administration (<2 h) and high plasma accumulation of alisporivir after 4 weeks of medication. Alisporivir has demonstrated an acceptable clinical safety profile in clinical studies of hepatitis C patients thus far (19, 20).
Cyclophilins are present in all domains of life and are crucial for protein folding (34). Cyclophilins possess PPIase and chaperone activity. In Plasmodium spp., cyclophilins represent an important drug target (8). Binding of cyclosporin A with P. falciparum cyclophilins suggested its role in antimalarial action of cyclosporin A. Cyclosporin A have been shown to bind with PfCyclophilin 19B and inhibits its PPIase activity (9). The upregulation of PfCyclophilin 19B has been demonstrated in artemisinin-resistant field parasites (12, 13). Human cyclophilin B has also been suggested as a novel druggable target (33). In our investigation, we observed that the chemical alisporivir efficiently docks around the binding site in PfCyclophilin 19B with minimum free binding energy. We also discovered that the docked complex deviation values were smaller or equal to those for the reference protein, i.e., the initial simulation frame, while using molecular dynamic simulations running at 50 ns in our research. According to this, no substantial changes in the complexes’ structure were seen during molecular dynamics modeling. As a whole, the in silico data show that alisporivir docks efficiently in the binding region of the PfCyclophilin 19B protein.
Next, to confirm interaction of PfCyclophilin 19B with alisporivir and cyclosporin A, we performed an MST, which is based on principle of thermophoresis and fluorescence detection. Compared to other biophysical techniques such as surface plasmon resonance, MST requires a smaller amount of sample and is easy to perform. In addition, MST combines various aspects of other techniques to measure the binding affinity of the interaction, hence MST is very sensitive (21). MST results showed strong binding of PfCyclophilin 19B with alisporivir (Kd = 354.3 nM) and cyclosporin A (Kd = 178.9 nM). In addition, a protein thermal shift assay results demonstrated the thermal protection of PfCyclophilin 19B protein at a higher temperature (60°C) in the presence of alisporivir and cyclosporin A. Alisporivir increases the stability of the PfCyclophilin 19B protein and protects it from denaturation, which confirmed the interaction between alisporivir and this protein.
Furthermore, the observation that expression of PfCyclophilin 19B upregulates artemisinin-resistant field parasites opens new avenues for targeting it and combating drug resistance malaria (12, 13). Our data confirm the increased expression of the PfCyclophilin 19B transcript, as well as protein, in artemisinin-resistant P. falciparum parasites. The in silico and in vitro results that demonstrate the strong binding of alisporivir with PfCyclophilin 19B encouraged us to examine alisporivir in detail for its prospective antimalarial properties. Alisporivir demonstrates potent antimalarial activity against chloroquine-sensitive (212.4 ± 27.57 nM), chloroquine-resistant (456.0 ± 7.93 nM), and artemisinin-resistant (244.1 ± 15.88 nM) strains of P. falciparum. Interestingly, the chloroquine-resistant and artemisinin-resistant strains have lower Ri values (2.14 ± 0.23 and 1.15 ± 0.04, respectively), which indicates that alisporivir is a potent antimalarial against drug-resistant strains. In addition, alisporivir shows antimalarial activity against P. berghei-infected mice. Notably, two-dose alisporivir treatment reduces the P. berghei load in mice and increases the survival of the host. Understanding the mechanistic importance of PfCyclophilin 19B overexpression in artemisinin resistance and its specific targeting by alisporivir offers a novel approach for drug development.
The P. falciparum database contains gene sequences for cyclophilin and cyclophilin-like proteins (6, 7). Among these cyclophilins, only two—PfCyclophilin 19A and PfCyclophilin 19B—demonstrated the cyclosporin A-binding property and PPIase activity. Interestingly, cyclosporin A inhibited the PPIase activities of PfCyclophilin 19A and PfCyclophilin 19B (8, 9). It has also been reported that the immunosuppressive activity and cyclophilin-binding property of cyclosporin A are not linked (10, 35). Previous studies reported that intraerythrocytic schizont maturation is affected by cyclosporin A (36). The largest amount of PfCyclophilin 19B was also detected in the intraerythrocytic schizont-stage parasites (9). In addition, overexpression of PfCyclophilin 19B was suggested to be associated with artemisinin resistance (12, 13). Alisporivir, which is a nonimmunosuppressive analogue of cyclosporin A, was reported to have strong cyclophilin inhibition activity.
In our study, we demonstrated the potent antimalarial activity of alisporivir against an artemisinin-resistant strain of P. falciparum. We therefore examined whether PfCyclophilin 19B is the main target of alisporivir. The results demonstrated the strong interaction of alisporivir with PfCyclophilin 19B, suggesting it as a main target of alisporivir. To support our study, future research could be performed with PfCyclophilin 19B mutated and/or disrupted parasites or by inducing resistance that can alter PfCyclophilin 19B expression levels. However, our study could not investigate the possibility of the presence of other antimalarial targets of alisporivir. Alisporivir can bind to other targets, such as PfCyclophilin 19A, which also contributes to the PPIase activity. This could be the reason for the observed no shift in the IC50 values of artemisinin-resistant parasites in comparison to the wild-type parasites. To affect IC50 values, it is also necessary that enough free alisporivir is available to interact with other antimalarial targets after binding with PfCyclophilin 19B. Further, we investigated the role of overexpression of PfCyclophilin 19B in artemisinin resistance development. Our study indicated that targeting overexpression of PfCyclophilin 19B through alisporivir inhibits the development of artemisinin resistance; this is suggested by the results of the RSA. These findings support a significant role for PfCyclophilin 19B in artemisinin resistance development.
The secret of artemisinin-resistant parasites is their potential to remain dormant under drug pressure, but as the drug levels in plasma go down (notably, the elimination half-life of artemisinin in plasma is 2 to 5 h), the parasites recover from the artemisinin-exposed dormancy (37). Therefore, a conventional 48-h assay cannot detect the presence of drug-resistant parasites. Hence, we performed an RSA to identify the presence of resistant parasites in the culture and to demonstrate that alisporivir in combination with DHA potently reduces the ring-stage survival of artemisinin-resistant parasites. These data highlight the efficacy of alisporivir against drug-resistant parasites and fulfill the requirement of novel drugs for treating drug-resistant malaria. Moreover, we investigated the inhibitory effect of alisporivir on parasites that survived after treatment with DHA. Interestingly, alisporivir significantly killed the surviving parasites, thus supporting its candidature as a partner drug with artemisinin.
In a previous study, Tong et al. demonstrated that the cyclophilin-targeting drug cyclosporin A in combination with artesunate has a synergistic antimalarial activity against the artemisinin-resistant strain IPC 5202 in a ring-stage survival assay. The combination of cyclosporin A with artesunate displayed an FIC50 value of 0.679 ± 0.017, suggesting that cyclosporin A could be a drug partner with artesunate (14). Here, we performed a checkerboard assay to test the synergistic antiparasitic effect of alisporivir with DHA on an artemisinin-resistant strain (PfKelch13R539T). In this assay, drugs were combined in many doses, and the survival of parasites was monitored by using a Giemsa-stained smear. We observed a synergistic effect of alisporivir with DHA with an FIC of <1. These results further support our findings for alisporivir, which is a nonimmunosuppressive analog of cyclosporin A.
The management and progression of complex diseases depends on multiple molecular pathways. Despite treatment with a single agent, it is beneficial to use a combination of drugs targeting different mechanisms in order to reduce the risk of drug resistance. However, development of a successful combination regimen requires the consideration of several factors, including affordability, therapeutic efficacy, and the safety and toxicity of the proposed drug combination. To address the scientific challenges associated with drug combination therapy, extensive information in terms of the proper preclinical study of drugs, effective clinical trial design, reduced toxicity, and decreased treatment duration and cogent drug delivery options are required. Other challenges, such as legal, economic, and regulatory barriers, must also be addressed (38). Importantly, repurposing of U.S. Food and Drug Administration-approved drugs is a potential solution to address the high cost and safety challenges presented by combination regimens. In our study, we repurposed the anti-hepatitis C virus drug alisporivir, which has a known and tested safety profile, thus resolving most of these challenges.
In conclusion, the identification of the potent inhibitory effect of alisporivir on malaria parasite growth, and its functioning with minimal side effects, opens up the immediate possibility of its use, either alone or in combination with existing antimalarials, as a potent candidate drug against malarial parasites.
MATERIALS AND METHODS
Chemicals.
Alisporivir (DEB-025; Debio-025; MedChem Express, USA) and cyclosporin A (Sigma-Aldrich, USA) stocks were prepared in DMSO.
Parasite culture.
(i) In vitro culture of P. falciparum. The 3D7, RKL-9, and PfKelch13R539T strains of P. falciparum were maintained at 2% hematocrit in RPMI 1640 medium (Invitrogen, USA) containing 0.5% Albumax I and 27.2 mgL−1 hypoxanthine in a 37°C mixed-gas environment (5% O2, 5% CO2, and 90% N2) using O+ RBCs (obtained from Rotary Blood Bank, New Delhi, India), as described previously (3, 39). The culture was synchronized with 5% sorbitol and routinely monitored by Giemsa staining (Sigma, USA). P. falciparum strain RKL-9 was obtained from the Malaria Parasite Bank, National Institute of Malaria Research, New Delhi, India. Transgenic parasite isolates of the Pf3D7 strain carrying the artemisinin-resistant mutation PfKelch13R539T were used in this study, and the mutant parasite line was generated as described previously (40). An RSA of parasite line 3D7:PfKelch13-R539T, suggesting artemisinin resistance, was validated in a previous report (3).
(ii) In vitro growth inhibition assays. Synchronized P. falciparum-infected erythrocytes at 2% hematocrit and 0.8 to 1.5% initial parasitemia were treated with alisporivir or cyclosporin A at concentrations ranging from 10 nM to 10 μM (Pf 3D7 and PfRKL-9 strain) and 15 to 1,000 nM (PfKelch13R539Tstrain); a DMSO-treated culture was used as a control. After incubation, parasitemia was calculated by counting Giemsa-stained blood smears. More than 2,000 RBCs were counted per slide, and the IC50 was calculated using Prism 6.0 (GraphPad, San Diego, CA). Constraints were not used for the determination of IC50 values. Data from three independent experiments were used to calculate the IC50 values, along with the standard errors of the mean (SEM).
Th percent growth inhibition was calcualted as follows: % inhibition = (1 − % parasitemia of treatment/% parasitemia of control) × 100. To analyze the progression of the drug-treated parasite in comparison to the control, a drug-treated culture was monitored for 5 days until reaching the ring stage of the third cycle. At each stage, the parasitemia and the morphology of the parasites were examined by using Giemsa-stained blood smears.
Ring survival assay.
To assess the antimalarial potential of alisporivir with DHA (Sigma-Aldrich, USA), a ring survival assay (RSA) was performed on an artemisinin-resistant strain as described previously (3, 41). Briefly, a Percoll-purified, early-ring-stage parasite culture (0 to 3 h postinvasion) was treated with alisporivir (4 μM), DHA (700 nM), and alisporivir in combination with DHA (4 μM plus 700 nM) for 6 h, followed by washing with complete media and incubation in fresh complete medium without the drug for 66 h. After 6 h of drug treatment and after incubation for 72 h, thin blood smears of the treated samples, as well as the control, were prepared, and >10,000 RBCs were analyzed using Giemsa staining. The percent parasite survival was determined by comparing the number of viable parasites in treated samples compared to the untreated control. In parallel, to study the effect of alisporivir with DHA on ring survival, a ring-stage culture of the artemisinin-resistant strain was treated with DHA (700 nM) for 6 h (42). After a washing step with culture medium, alisporivir (1 μM) was added for 72 h, and the parasitemia was determined by counting Giemsa-stained smears.
Checkerboard combination assay.
To perform a synergy checkerboard assay, parasite-infected erythrocytes were seeded into a 96-well plate at 2% hematocrit and 1% parasitemia. Alisporivir and DHA at various concentrations were added to the culture, followed by incubation at 37°C for 48 h. In addition, the drugs DHA and alisporivir were both individually tested. Parasite growth inhibition was monitored after treatment by observing Giemsa-stained blood smears. An untreated parasite culture was used as a control. The percent parasitemia of treated samples was determined compared to the untreated control. The fractional inhibition concentrations (FICs) were calculated as described previously (43). FIC values of <1 were considered to indicate a synergistic interaction.
Infection (in vivo proliferation of P. berghei).
Animal studies were approved by the Institutional Animal Ethics Committee of Jawaharlal Nehru University (JNU), New Delhi, India, and performed according to CPCSEA guidelines. Mice were provided by the Central Laboratory Animal Resources, JNU, and maintained under standard conditions.
Mouse erythrocytes (1 × 105) infected with the ANKA strain of P. berghei (ATCC, India), were injected intraperitoneally into BALB/c mice. Three groups of mice were injected with cyclosporin A (5 mg/kg, intraperitoneally), alisporivir (5 mg/kg, intravenously [44]), and buffer control, respectively, for 2 days. Parasitemia was determined from Giemsa-stained tail blood smears.
Flow cytometric analysis of annexin V binding.
To evaluate eryptosis in erythrocytes, flow cytometric analysis was performed by measuring the annexin V fluorescence intensity. After treatment, the erythrocytes were washed in annexin V binding buffer (140 mM NaCl, 10 mM HEPES [pH 7.4], 2.5 mM CaCl2). Erythrocytes were stained with annexin V-FITC (Thermo Scientific, USA) in annexin V binding buffer (1:20) for 20 min at room temperature. Samples were then analyzed by flow cytometry (BD LSR Fortessa; BD Biosciences) using an excitation at 488 nm and an emission at 530 nm. An identical procedure was followed in order to measure eryptosis in mice samples.
Hemolysis assay and hematocrit determination.
For the hemolysis assay, RBCs (1 × 107) were treated with alisporivir (10 μM) at 37°C in a 5% CO2 incubator for 24 h. After incubation, the absorbance of the supernatant was measured at 415 nm with a microplate reader (Thermo Scientific, USA) to assess hemoglobin leakage. Erythrocytes lysed in distilled water were used as a positive control. To measure the hematocrit, blood was collected from infected and alisporivir-treated infected mice. The hematocrit was determined as the percentage of blood cells in the total volume of blood.
Reverse transcription-PCR.
Total RNA of the Pf3D7and PfKelch13R539T strains was isolated using TRIzol (Life Technologies, USA). Equal amounts of total RNA were used for cDNA synthesis using a cDNA synthesis kit (Thermo Fisher, USA). RT-PCR was performed with specific primers (see Table S1 in the supplemental material) for 20 cycles of amplification, and the expression level of PfCyclophilin 19B gene (PlasmoDB ID Pf3D7_1115600) was quantified using ImageJ software (NIH). Pf18S was used as an internal control.
Immunofluorescence assay.
Localization and the expression levels of PfCyclophilin 19B in Pf3D7 and PfKelch13R539T strains were monitored by using an immunofluorescence assay. Erythrocytes infected with ring-, trophozoite-, and schizont-stage P. falciparum were used to prepare thin smears. After fixation with chilled methanol, slides were blocked using 3% bovine serum albumin (BSA) and then stained with PfCyclophilin 19B antibody (1:200 dilution) at room temperature for 2 h. For the colocalization study, further staining was carried out with anti-PfNAPL antibodies. After a washing step, the slides were incubated with Alexa 488-conjugated anti-mouse (1:500 dilution; Invitrogen) or Alexa 568-goat anti-rabbit (1:500 dilution; Life Technologies) secondary antibody at room temperature for 1 h. Mounted slides with DAPI-antifade (Invitrogen, USA) were examined with a confocal microscope (Nikon, USA). To determine the degree of colocalization, the Pearson’s correlation coefficient was measured.
Cloning, expression, and purification of PfCyp19B protein.
For cloning, Bla1cutpET28a expression vector was prepared by digestion with the SnaB1 enzyme and then dephosphorylated. Subsequently, PfCyclophilin 19B insert digested with SnaB1 was ligated with a prepared vector. For expression of the PfCyclophilin 19B protein, a cloned construct was transformed into Escherichia coli BL21(DE3) cells. Next, mid-log-phase cultures were induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) at 25°C for 10 h to express PfCyclophilin 19B protein tagged with C-terminal hexahistidine. The induced cell pellet was sonicated in phosphate-buffered saline (PBS; 4.3 mM Na2HPO4, 1.47 mM KH2PO4, 150 mM NaCl, 2.7 mM KCl [pH 7.4]) containing 2 mM phenylmethylsulfonyl fluoride, and the resulting supernatant was bound overnight with Ni-NTA agarose beads at 4°C. The bound rPfCyp19B protein was eluted using an imidazole solution (50 to 250 mM imidazole containing PBS [pH 7.4]). Purified recombinant protein was collected and dialyzed against PBS buffer to remove the imidazole. BALB/c mice (6 weeks old, female) were immunized with purified PfCyclophilin 19B protein for antibody production, and sera were collected after the first and second booster doses.
Microscale thermophoresis.
For analysis of PfCyclophilin 19B protein and drugs interaction using microscale thermophoresis (MST), a NanoTemper Monolith (NanoTemper Technologies, Germany) instrument was used with blue/red filters. Experiments were performed in PBS with Tween. The protein sample was diluted in buffer. In all MST experiments, measurements were performed using 40% MST power at 22°C in standard treated capillaries. For the binding assay, fluorescently labeled PfCyclophilin 19B protein was diluted in buffer and centrifuged for 10 min. Further, a 16-sample serial dilution of drugs was prepared by using a 1:1 dilution in 10 μL of buffer. The labeled protein was added to a ligand solution, followed by incubation for 10 min at room temperature and then centrifugation. Data analysis was performed using MO.Affinity software (NanoTemper). The binding constant Kd was calculated by plotting dose-response Fnorm curve against the ligand concentration according to the NanoTemper protocol.
Protein thermal shift assay.
A thermal shift assay was performed to determine the interaction of PfCyclophilin 19B with alisporivir and cyclosporin A, which is based on the same principle of a cellular thermal shift assay (CETSA). Briefly, rPfCyclophilin 19B alone or incubated with drugs was heated at different temperaturea (4, 40, 45, 50, 55, 60, 65, 70, 75, or 80°C). Samples were centrifuged to isolate the soluble fraction of the protein. The soluble fraction was analyzed by Western blotting with an anti-His antibody. ImageJ software (NIH, USA) was used to determine the band intensities of the proteins.
Western blotting.
Protein samples were run on an SDS-PAGE gel and subsequently transferred onto a nitrocellulose membrane (Bio-Rad, USA). Equal amounta of protein were loaded for Western blot comparisons of the PfCyclophilin 19B expression in PfKelch13R539T strains compared to the Pf3D7 strain. In the parasite lysates, the protein levels were quantified by using a Bradford assay. After blocking with 5% BSA, blots were incubated with the respective primary antibody (anti-PfCyclophilin 19B or anti-His antibody) for 2 h at room temperature. After a washing step, secondary horseradish peroxidase-conjugated anti-mouse antibody (Thermo Scientific, USA) was added for 1 h at room temperature. The blot was washed and developed using the Luminol reagent (Clarity Western ECL Substrate; Bio-Rad).
In silico docking studies.
Protein structural files were obtained from the Protein Data Bank. The Plasmodium cyclophilin (PDB 1QNG) three-dimensional (3D) protein structure was found in the Protein Data Bank (45). The Drug Bank Database was used to extract the chemical structures of the compounds. The protein and ligand structures were improved using PDB viewer 4.1.0 and ChemBio3D Ultra-12.0. Docking instructions were run using Autodock 1.5.7rc1 and the Cygwin terminal. Around its catalytic domain residues, the ligand’s binding site was selected. In order to further analyze and visualize docking data, we used Chimera, Ligplus, Discovery Studio v19.1.0, and PyMOL v2.3.2 software (46, 47).
Molecular dynamics simulation.
Molecular dynamics simulation (MDS) was carried out using Desmond v2018.4, running on Linux (48). Using Desmond module’s protein preparation wizard and the explicit solvent model, SPC, and orthorhombic box shape, the protein-ligand complexes (such as the docked crystal structure of PfCyclophilin 19B coupled to alisporivir) were first created. The box size was established at a distance of 10 nm from the protein-ligand complex’s outermost atoms. In order to establish the ionic strength, sodium chloride with an estimated physiological value of 0.15 M was added to the simulation box. Desmond’s MD simulation software was used to run MD simulations on this model system, which had been relaxed into a local energy minimization once the minimization operations had been completed. The Simulation Event Analysis tool in Maestro was used to examine MD simulations, and the Simulation Interaction Diagram tool was used to identify ligand-receptor interactions.
Compliance with ethical standards.
Animal handling and experiments were performed as per CPCSEA guidelines and approved by the Institutional Animal Ethics Committee, JNU, New Delhi, India. All experiments were approved by the institutional IBSC committee and conducted according to the guidelines and regulations of JNU.
Statistical analysis.
The statistical significance of data was measured by using a two-tailed Student t test. Error bars in the figures represent the means ± the standard deviations (SD) of three independent experiments.
ACKNOWLEDGMENTS
We sincerely thank the Advanced Instrumentation and Research Facility (JNU) for providing a confocal microscopy facility. We also thank the Central Instrumentation Facility of the Special Centre for Molecular Medicine, JNU.
This study was supported by IRHPA IPA/2020/000007 (A.R. and S.S.), the Department of Science and Technology, SERB grant CRG/2019/00223, and grant BT/PR30534/BIC/101/1095/2018 (J.S., A.R., and S.S.). This study was also funded by the Drug and Pharmaceuticals Research Program (project P/569/2016-1/TDT [S.S.]) and Science and Engineering Research Board (EMR/2016/005644 [S.S.]). A.C. is supported by senior research fellowship from the Department of Biotechnology, JNU.
A.R. and S.S. conceptualized the idea of study and supervised and designed the experiments. A.C. and G.K. performed the experiments, validated, analyzed data, and wrote the manuscript. A.C., G.K., S.G., R.S., and J.K. set up malarial cultures and conducted experiments. A.C., S.G., and J.S. helped with imaging analysis. N.J. performed bioinformatics analysis. Z.A. wrote parts of the manuscript. Z.A., J.S., N.S., S.K., A.K.K., and N.D. helped with analysis and experimental design. M.K.M., P.S., and M.M. helped in experimental methodology. A.R., S.S., G.D., A.C., and G.K. critically analyzed the data and the manuscript. S.B. generated the artemisinin-resistant strain. This manuscript has been screened by software turnitin.
We declare that there are no conflicts of interest.
Footnotes
Supplemental material is available online only.
Contributor Information
Shailja Singh, Email: shailja.jnu@gmail.com.
Anand Ranganathan, Email: anand.icgeb@gmail.com.
REFERENCES
- 1.World Health Organization. 2022. World malaria report–2021. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 2.Dondorp AM, Smithuis FM, Woodrow C, Seidlein LV. 2017. How to contain artemisinin- and multidrug-resistant falciparum malaria. Trends Parasitol 33:353–363. 10.1016/j.pt.2017.01.004. [DOI] [PubMed] [Google Scholar]
- 3.Kannan D, Yadav N, Ahmad S, Namdev P, Bhattacharjee S, Lochab B, Singh S. 2019. Pre-clinical study of iron oxide nanoparticles fortified artesunate for efficient targeting of malarial parasite. EBioMedicine 45:261–277. 10.1016/j.ebiom.2019.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bell A, Monaghan P, Page AP. 2006. Peptidyl-prolyl cis-trans isomerases (immunophilins) and their roles in parasite biochemistry, host-parasite interaction and antiparasitic drug action. Int J Parasitol 36:261–276. 10.1016/j.ijpara.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 5.Fischer G, Wittmann-Liebold B, Lang K, Kiefhaber T, Schmid FX. 1989. Cyclophilin and peptidyl-prolyl cis-trans isomerase are probably identical proteins. Nature 337:476–478. 10.1038/337476a0. [DOI] [PubMed] [Google Scholar]
- 6.Krucken J, Greif G, von Samson-Himmelstjerna G. 2009. In silico analysis of the cyclophilin repertoire of apicomplexan parasites. Parasit Vectors 2:27. 10.1186/1756-3305-2-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marin-Menendez A, Bell A. 2011. Overexpression, purification and assessment of cyclosporin binding of a family of cyclophilins and cyclophilin-like proteins of the human malarial parasite Plasmodium falciparum. Protein Expr Purif 78:225–234. 10.1016/j.pep.2011.04.012. [DOI] [PubMed] [Google Scholar]
- 8.Marin-Menendez A, Monaghan P, Bell A. 2012. A family of cyclophilin-like molecular chaperones in Plasmodium falciparum. Mol Biochem Parasitol 184:44–47. 10.1016/j.molbiopara.2012.04.006. [DOI] [PubMed] [Google Scholar]
- 9.Gavigan CS, Kiely SP, Hirtzlin J, Bell A. 2003. Cyclosporin-binding proteins of Plasmodium falciparum. Int J Parasitol 33:987–996. 10.1016/s0020-7519(03)00125-5. [DOI] [PubMed] [Google Scholar]
- 10.Bell A, Wernli B, Franklin RM. 1994. Roles of peptidyl-prolyl cis-trans isomerase and calcineurin in the mechanisms of antimalarial action of cyclosporin, FK506, and rapamycin. Biochem Pharmacol 48:495–503. 10.1016/0006-2952(94)90279-8. [DOI] [PubMed] [Google Scholar]
- 11.Hirtzlin J, Farber PM, Franklin RM, Bell A. 1995. Molecular and biochemical characterization of a Plasmodium falciparum cyclophilin containing a cleavable signal sequence. Eur J Biochem 232:765–772. 10.1111/j.1432-1033.1995.tb20871.x. [DOI] [PubMed] [Google Scholar]
- 12.Mok S, Ashley EA, Ferreira PE, Zhu L, Lin Z, Yeo T, Chotivanich K, Imwong M, Pukrittayakamee S, Dhorda M, Nguon C, Lim P, Amaratunga C, Suon S, Hien TT, Htut Y, Faiz MA, Onyamboko MA, Mayxay M, Newton PN, Tripura R, Woodrow CJ, Miotto O, Kwiatkowski DP, Nosten F, Day NP, Preiser PR, White NJ, Dondorp AM, Fairhurst RM, Bozdech Z. 2015. Drug resistance: population transcriptomics of human malaria parasites reveals the mechanism of artemisinin resistance. Science 347:431–435. 10.1126/science.1260403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paloque L, Ramadani AP, Mercereau-Puijalon O, Augereau JM, Benoit-Vical F. 2016. Plasmodium falciparum: multifaceted resistance to artemisinins. Malar J 15:149. 10.1186/s12936-016-1206-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tong JX, Ang SEL, Tan EHN, Tan KSW. 2019. Viability screen of LOPAC(1280) reveals tyrosine kinase inhibitor tyrphostin A9 as a novel partner drug for artesunate combinations to target the Plasmodium falciparum Ring Stage. Antimicrob Agents Chemother 63:e02389-18. 10.1128/AAC.02389-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Niemoeller OM, Akel A, Lang PA, Attanasio P, Kempe DS, Hermle T, Sobiesiak M, Wieder T, Lang F. 2006. Induction of eryptosis by cyclosporin. Naunyn Schmiedebergs Arch Pharmacol 374:41–49. 10.1007/s00210-006-0099-5. [DOI] [PubMed] [Google Scholar]
- 16.Liu J, Farmer JD, Jr, Lane WS, Friedman J, Weissman I, Schreiber SL. 1991. Calcineurin is a common target of cyclophilin-cyclosporin and FKBP-FK506 complexes. Cell 66:807–815. 10.1016/0092-8674(91)90124-H. [DOI] [PubMed] [Google Scholar]
- 17.Sweeney ZK, Fu J, Wiedmann B. 2014. From chemical tools to clinical medicines: nonimmunosuppressive cyclophilin inhibitors derived from the cyclosporin and sanglifehrin scaffolds. J Med Chem 57:7145–7159. 10.1021/jm500223x. [DOI] [PubMed] [Google Scholar]
- 18.Paeshuyse J, Kaul A, De Clercq E, Rosenwirth B, Dumont JM, Scalfaro P, Bartenschlager R, Neyts J. 2006. The non-immunosuppressive cyclosporin DEBIO-025 is a potent inhibitor of hepatitis C virus replication in vitro. Hepatology 43:761–770. 10.1002/hep.21102. [DOI] [PubMed] [Google Scholar]
- 19.Gallay PA, Lin K. 2013. Profile of alisporivir and its potential in the treatment of hepatitis C. Drug Des Dev Ther 7:105–115. 10.2147/DDDT.S30946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stanciu C, Trifan A, Muzica C, Sfarti C. 2019. Efficacy and safety of alisporivir for the treatment of hepatitis C infection. Expert Opin Pharmacother 20:379–384. 10.1080/14656566.2018.1560424. [DOI] [PubMed] [Google Scholar]
- 21.Jerabek-Willemsen M, Wienken CJ, Braun D, Baaske P, Duhr S. 2011. Molecular interaction studies using microscale thermophoresis. Assay Drug Dev Technol 9:342–353. 10.1089/adt.2011.0380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wadi I, Pillai CR, Anvikar AR, Sinha A, Nath M, Valecha N. 2018. Methylene blue induced morphological deformations in Plasmodium falciparum gametocytes: implications for transmission-blocking. Malar J 17:11. 10.1186/s12936-017-2153-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jinky G, Dipak C, Mukesh KK, Mithun R. 2016. Synthesis and antimalarial activity evaluation of some mannich bases of tetraoxane-phenol conjugate. Ijper 50:591–597. 10.5530/ijper.50.4.11. [DOI] [Google Scholar]
- 24.Chandra BR, Olivieri A, Silvestrini F, Alano P, Sharma A. 2005. Biochemical characterization of the two nucleosome assembly proteins from Plasmodium falciparum. Mol Biochem Parasitol 142:237–247. 10.1016/j.molbiopara.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 25.Tilley L, Straimer J, Gnadig NF, Ralph SA, Fidock DA. 2016. Artemisinin action and resistance in Plasmodium falciparum. Trends Parasitol 32:682–696. 10.1016/j.pt.2016.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ouji M, Augereau JM, Paloque L, Benoit-Vical F. 2018. Plasmodium falciparum resistance to artemisinin-based combination therapies: a sword of Damocles in the path toward malaria elimination. Parasite 25:24. 10.1051/parasite/2018021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bobbala D, Koka S, Lang C, Boini KM, Huber SM, Lang F. 2008. Effect of cyclosporin on parasitemia and survival of Plasmodium berghei-infected mice. Biochem Biophys Res Commun 376:494–498. 10.1016/j.bbrc.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 28.Nickell SP, Scheibel LW, Cole GA. 1982. Inhibition by cyclosporin of rodent malaria in vivo and human malaria in vitro. Infect Immun 37:1093–1100. 10.1128/iai.37.3.1093-1100.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nzila A, Ma Z, Chibale K. 2011. Drug repositioning in the treatment of malaria and TB. Future Med Chem 3:1413–1426. 10.4155/fmc.11.95. [DOI] [PubMed] [Google Scholar]
- 30.Ashburn TT, Thor KB. 2004. Drug repositioning: identifying and developing new uses for existing drugs. Nat Rev Drug Discov 3:673–683. 10.1038/nrd1468. [DOI] [PubMed] [Google Scholar]
- 31.Pradhan A, Siwo GH, Singh N, Martens B, Balu B, Button-Simons KA, Tan A, Zhang M, Udenze KO, Jiang RH, Ferdig MT, Adams JH, Kyle DE. 2015. Chemogenomic profiling of Plasmodium falciparum as a tool to aid antimalarial drug discovery. Sci Rep 5:15930. 10.1038/srep15930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ch’ng JH, Mok S, Bozdech Z, Lear MJ, Boudhar A, Russell B, Nosten F, Tan KS. 2013. A whole cell pathway screen reveals seven novel chemosensitizers to combat chloroquine resistant malaria. Sci Rep 3:1734. 10.1038/srep01734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prakash P, Zeeshan M, Saini E, Muneer A, Khurana S, Kumar Chourasia B, Deshmukh A, Kaur I, Dabral S, Singh N, Anam Z, Chaurasiya A, Kaushik S, Dahiya P, Kalamuddin M, Kumar Thakur J, Mohmmed A, Ranganathan A, Malhotra P. 2017. Human cyclophilin B forms part of a multi-protein complex during erythrocyte invasion by Plasmodium falciparum. Nat Commun 8:1548. 10.1038/s41467-017-01638-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang P, Heitman J. 2005. The cyclophilins. Genome Biol 6:226. 10.1186/gb-2005-6-7-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gavigan CS, Shen M, Machado SG, Bell A. 2007. Influence of the Plasmodium falciparum P-glycoprotein homologue 1 (pfmdr1 gene product) on the antimalarial action of cyclosporin. J Antimicrob Chemother 59:197–203. 10.1093/jac/dkl461. [DOI] [PubMed] [Google Scholar]
- 36.Matsumoto Y, Perry G, Scheibel LW, Aikawa M. 1987. Role of calmodulin in Plasmodium falciparum: implications for erythrocyte invasion by the merozoite. Eur J Cell Biol 45:36–43. [PubMed] [Google Scholar]
- 37.Codd A, Teuscher F, Kyle DE, Cheng Q, Gatton ML. 2011. Artemisinin-induced parasite dormancy: a plausible mechanism for treatment failure. Malar J 10:56. 10.1186/1475-2875-10-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Humphrey RW, Brockway-Lunardi LM, Bonk DT, Dohoney KM, Doroshow JH, Meech SJ, Ratain MJ, Topalian SL, Pardoll DM. 2011. Opportunities and challenges in the development of experimental drug combinations for cancer. J Natl Cancer Inst 103:1222–1226. 10.1093/jnci/djr246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trager W, Jensen JB. 1976. Human malaria parasites in continuous culture. Science 193:673–675. 10.1126/science.781840. [DOI] [PubMed] [Google Scholar]
- 40.Bhattacharjee S, Coppens I, Mbengue A, Suresh N, Ghorbal M, Slouka Z, Safeukui I, Tang HY, Speicher DW, Stahelin RV, Mohandas N, Haldar K. 2018. Remodeling of the malaria parasite and host human red cell by vesicle amplification that induces artemisinin resistance. Blood 131:1234–1247. 10.1182/blood-2017-11-814665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Witkowski B, Amaratunga C, Khim N, Sreng S, Chim P, Kim S, Lim P, Mao S, Sopha C, Sam B, Anderson JM, Duong S, Chuor CM, Taylor WR, Suon S, Mercereau-Puijalon O, Fairhurst RM, Menard D. 2013. Novel phenotypic assays for the detection of artemisinin-resistant Plasmodium falciparum malaria in Cambodia: in-vitro and ex-vivo drug-response studies. Lancet Infect Dis 13:1043–1049. 10.1016/S1473-3099(13)70252-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Teuscher F, Gatton ML, Chen N, Peters J, Kyle DE, Cheng Q. 2010. Artemisinin-induced dormancy in plasmodium falciparum: duration, recovery rates, and implications in treatment failure. J Infect Dis 202:1362–1368. 10.1086/656476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Suberu JO, Gorka AP, Jacobs L, Roepe PD, Sullivan N, Barker GC, Lapkin AA. 2013. Anti-plasmodial polyvalent interactions in Artemisia annua L. aqueous extract: possible synergistic and resistance mechanisms. PLoS One 8:e80790. 10.1371/journal.pone.0080790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gomez L, Thibault H, Gharib A, Dumont JM, Vuagniaux G, Scalfaro P, Derumeaux G, Ovize M. 2007. Inhibition of mitochondrial permeability transition improves functional recovery and reduces mortality following acute myocardial infarction in mice. Am J Physiol Heart Circ Physiol 293:H1654. 10.1152/ajpheart.01378.2006. [DOI] [PubMed] [Google Scholar]
- 45.Peterson MR, Hall DR, Berriman M, Nunes JA, Leonard GA, Fairlamb AH, Hunter WN. 2000. The three-dimensional structure of a Plasmodium falciparum cyclophilin in complex with the potent anti-malarial cyclosporin. J Mol Biol 298:123–133. 10.1006/jmbi.2000.3633. [DOI] [PubMed] [Google Scholar]
- 46.Anam ZE, Joshi N, Gupta S, Yadav P, Chaurasiya A, Kahlon AK, Kaushik S, Munde M, Ranganathan A, Singh S. 2020. A de novo peptide from a high-throughput peptide library blocks myosin A-MTIP complex formation in Plasmodium falciparum. Int J Mol Sci 21:6158. 10.3390/ijms21176158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chaurasiya A, Garg S, Khanna A, Narayana C, Dwivedi VP, Joshi N, Z EA, Singh N, Singhal J, Kaushik S, Kaur Kahlon A, Srivastava P, Marothia M, Kumar M, Kumar S, Kumari G, Munjal A, Gupta S, Singh P, Pati S, Das G, Sagar R, Ranganathan A, Singh S. 2021. Pathogen induced subversion of NAD+ metabolism mediating host cell death: a target for development of chemotherapeutics. Cell Death Discov 7:10. 10.1038/s41420-020-00366-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bowers KJ, Chow DE, Xu H, Dror RO, Eastwood MP, Gregersen BA, Klepeis JL, Kolossvary I, Moraes MA, Sacerdoti FD. 2006. Scalable algorithms for molecular dynamics simulations on commodity clusters, p 43–43. In Proceedings of the 2006 ACM/IEEE Conference on Supercomputing. IEEE, New York, NY. 10.1109/SC.2006.54. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download aac.00392-22-s0001.pdf, PDF file, 0.8 MB (833.2KB, pdf)