ABSTRACT
The Corynebacterium diphtheriae hemoglobin-binding protein HbpA is critical for the acquisition of iron from the hemoglobin-haptoglobin complex (Hb-Hp). Previous studies using C. diphtheriae strain 1737 showed that large aggregates formed by HbpA are associated with iron transport activity and enhanced binding to Hb-Hp; however, specific regions within HbpA required for Hb-Hp binding or iron uptake have not been identified. In this study, we characterized two clinical isolates from Austria, designated 07-18 and 09-15, which express HbpA proteins that share only 53% and 44% sequence identity, respectively, to the strain 1737 HbpA protein. The HbpA proteins expressed by the Austrian strains had functional and structural properties similar to those of the HbpA protein in strain 1737 despite the limited sequence similarity. These shared characteristics between the HbpA proteins included similar cellular localization, aggregate formation, and Hb and Hb-Hp binding. Additionally, the Austrian strains were able to acquire iron from Hb and Hb-Hp, and deletion of the hbpA gene from these two clinical isolates reduced their ability to use Hb-Hp as an iron source. A sequence comparison between the HbpA proteins from 1737 and the Austrian strains assisted in the identification of a putative Hb-binding site that shared similar characteristics with the Hb-binding regions in Staphylococcus aureus NEAT domains. Amino acid substitutions within this conserved Hb-binding region significantly reduced Hb and Hb-Hp binding and diminished the hemin-iron uptake function of HbpA. These findings represent important advances in our understanding of the interaction of HbpA with human hemoproteins.
IMPORTANCE Hemoglobin (Hb) is the primary source of iron in humans, and the acquisition of hemin-iron from Hb is critical for many bacterial pathogens to infect and survive in the human host. In this study, we have examined the C. diphtheriae Hb-binding protein HbpA in two clinical isolates and show that these proteins, despite limited sequence similarity, are functionally equivalent to the previously described HbpA protein in strain 1737. A sequence comparison between these three strains led to the identification of a conserved Hb-binding site, which will further our understanding of how this novel protein functions in hemin-iron transport and, more generally, will expand our knowledge on how Hb interacts with proteins.
KEYWORDS: Corynebacterium, HbpA, diphtheria, hemoglobin, iron transport
INTRODUCTION
Diphtheria toxin-producing strains of Corynebacterium diphtheriae are often associated with severe respiratory disease in humans (1, 2). Nontoxigenic isolates are frequently the cause of milder cutaneous and respiratory infections, although nontoxigenic strains occasionally produce severe infections that are not prevented by current diphtheria toxoid vaccines (3). While the iron-regulated diphtheria toxin is an important virulence factor for toxin-producing strains of C. diphtheriae, additional virulence factors associated with both toxigenic and nontoxigenic clinical isolates have not been well defined. Factors important for adherence to epithelial cells and for acquisition of essential nutrients, such as iron, likely have an important role in colonization and survival in the host for both toxigenic and nontoxigenic strains (3–6).
Bacterial pathogens encounter a host environment that is limited in available iron, and this low-iron condition can serve as a signal to induce the expression of specific virulence factors such as diphtheria toxin and iron and hemin transport systems in C. diphtheriae. Many bacterial pathogens, including C. diphtheriae, have evolved a variety of mechanisms to obtain hemin-iron for growth in the iron-depleted host environment (6–10). Heme is the most abundant source of iron in humans and is primarily associated with hemoglobin (Hb), which is present in erythrocytes. Hb released from erythrocytes is rapidly bound to haptoglobin (Hp) in serum to form the Hb-Hp complex (11, 12). Two of the primary functions of the Hb-Hp complex are to limit the toxicity associated with free Hb and to remove Hb from circulation through uptake and degradation in macrophages (12).
In Staphylococcus aureus, the use of Hb as an iron source requires surface receptors known as iron-regulated surface determinants (Isd), which are covalently anchored to the cell wall by sortases and are able to bind hemin, Hb, and Hb-Hp (13, 14). The IsdB and IsdH proteins in S. aureus bind Hb at the bacterial surface through regions known as NEAT (near-iron transporter) domains where hemin is extracted from Hb and subsequently transported through the cell wall in a cascade manner by binding to a series of cell wall-anchored hemin-binding proteins (14–16). NEAT domain-containing proteins that bind hemin and/or Hb and Hb-Hp are also present in Bacillus anthracis, Listeria monocytogenes, and Streptococcus pyogenes (17–20).
Factors that are critical for the transport of hemin-iron by C. diphtheriae include the HmuTUV ABC transporter and HtaA and HtaB, which are iron-regulated hemin-binding proteins (7). HtaA binds to hemin and to various hemoproteins through a pair of domains designated conserved regions 1 and 2 (CR1 and CR2) (6, 21). The CR domains show no significant sequence similarity to the NEAT domains found in the hemin- and Hb-binding proteins in other Gram-positive bacteria (22). A C. diphtheriae htaA deletion mutant showed reduced growth when Hb or Hb-Hp was provided as the sole iron source, indicating that HtaA has a crucial role in the use of these hemoproteins as iron sources (6). The hemin-binding and surface-exposed ChtA and ChtC proteins in C. diphtheriae are also involved in the use of hemin as an iron source and have important roles in the acquisition of iron from Hb-Hp (6). The ChtA and ChtC proteins exhibit significant sequence similarity, and each contains a single CR domain. The ChtA and ChtC proteins were shown to have redundant function, since deletion of both the chtA and chtC genes was required for a reduction in the use of Hb-Hp as an iron source (6). In addition, the defect in growth with Hb-Hp observed with the chtA/C double mutant was restored by complementation with either the chtA or chtC gene (23). Most of the studies examining hemin-iron transport in C. diphtheriae were conducted in strain 1737, a clinical isolate associated with the diphtheria outbreak in the former Soviet Union in the 1990s (24). Strain 1737 is also closely related to NCTC13129, which was the first C. diphtheriae strain to have its complete genome sequenced and fully annotated in 2003 (25).
HbpA is a novel iron-regulated protein in C. diphtheriae that was recently shown to bind Hb and Hb-Hp and is involved in the use of Hb-Hp as an iron source in strain 1737 (26). HbpA shares no significant sequence similarity to other known proteins and has been identified only in a subgroup of C. diphtheriae strains, which includes clinical isolates that are closely related to strain 1737 (24–26). HbpA, like other C. diphtheriae Hb-binding proteins, is found both associated with the membrane and secreted into the culture medium. Whole-cell enzyme-linked immunosorbent assays (WC ELISAs) showed that HbpA was exposed on the surface of C. diphtheriae strain 1737 and was responsible for much of the Hb and Hb-Hp binding observed on the bacterial surface (26). Neither CR nor NEAT domains were identified in HbpA, and a region involved in Hb or Hb-Hp binding has not been reported (26). In a previous study, we showed that the presence of the C-terminal transmembrane region of HbpA is essential for hemin-iron acquisition from Hb-Hp and further demonstrated that this region is required for membrane anchoring and aggregate formation (23). The native form of HbpA is a high-molecular-weight complex that has not been structurally characterized; however, this aggregate structure of HbpA is required for optimal binding to Hb and Hb-Hp (23).
In this study, we examined two clinical isolates from Austria that express HbpA proteins that share only 53% and 44% amino acid sequence identity to strain 1737 HbpA (27). Despite the limited sequence similarity, we showed here that the HbpA proteins expressed by the Austrian strains have cellular localization and function similar to those of the HbpA protein from 1737. A sequence comparison between the HbpA proteins from 1737 and the Austrian strains facilitated the identification of a putative Hb-binding site that shares sequence characteristics with the Hb-binding region in S. aureus NEAT domains. Amino acid substitutions within this putative Hb-binding sequence of HbpA resulted in significantly diminished Hb and Hb-Hp binding and abolished the heme-iron uptake function of HbpA.
RESULTS
Sequence analysis of HbpA from Austrian clinical isolates.
A recent study described the genetic characterization of 57 C. diphtheriae clinical isolates from Austria that were obtained from either respiratory or cutaneous infections between 2011 and 2019 (27). Two of the strains, 07-18 and 09-15, which were isolated from cutaneous infections, express HbpA proteins that have only 53% and 44% amino acid sequence identity, respectively, to the previously characterized HbpA protein from strain 1737 (23, 26) (Table 1). The HbpA proteins in 07-18 and 09-15 have only 46% amino acid identity to each other. A BLAST search using the 1737 HbpA amino acid sequence identified three distinct groups of HbpA homologs based on sequence similarity, and strains 1737, 07-18, and 09-15 encode HbpA proteins that are representative members of each of these three unique sequence groups. Because strains 07-18 and 09-15 encode HbpA proteins that represent unique sequence groups, these strains were selected for additional studies to examine the function and structure of their HbpA proteins and the potential use of their HbpA sequences to identify conserved regions associated with Hb binding and hemin-iron transport.
TABLE 1.
C. diphtheriae strain comparisone
| Strain | Source |
HbpA |
% AA identity to 1737 homolog: |
% AA identity to diphtheria toxin | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Location | Infection type | % AA identity to 1737 | Predicted size (kDa) | HtaA | ChtA | ChtB | ChtC | HmuT | ||
| 1737 | FSUf | Respiratory | 100 | 38.5 | 100 | 100 | 100 | 100 | 100 | 100 |
| 06-15 | Austria | Cutaneous | 100 | 38.5 | 100 | 100 | 100 | 99 | 100 | (−)a |
| 07-18 | Austria | Cutaneous | 53b | 35.9 | 99 | (−) | 99 | 99 | 100 | (−) |
| 09-15 | Austria | Cutaneous | 44b | 36.2 | 99 | 98 | 99 | 99 | 100 | (−) |
| HC02 | Brazil | Endocarditis | 53c | 35.9 | 99 | (−) | (−) | 99 | 100 | (−) |
| VA01 | Brazil | Respiratory | 44d | 36.2 | 99 | 99 | 99 | 87 | 100 | (−) |
(−), gene not present and/or protein not expressed.
HbpA has 46% amino acid identity between strains 07-18 and 09-15.
HbpA has 100% amino acid identity between strains HC02 and 07-18.
HbpA has 100% amino acid identity between strains VA01 and 09-15.
AA, amino acid.
FSU, Former Soviet Union.
In addition to HbpA, the two Austrian strains encode other proteins associated with iron acquisition from Hb-Hp, including HtaA, HmuT, ChtA (present only in 09-15), ChtB, and ChtC; all of these proteins show at least 98% sequence identity to the homologous proteins present in 1737 (Table 1). Strain 07-18 does not encode the ChtA protein, which is functionally redundant with ChtC (26). The diphtheria toxin gene (tox) is absent in the Austrian strains, which is commonly observed for strains associated with cutaneous infections or other less severe disease.
Amino acid sequence alignment of the HbpA proteins from 1737 and the two Austrian strains showed that the highest similarity was observed at the N and C termini, which contain the signal sequence and the transmembrane region, respectively (see Fig. S1 in the supplemental material) (28). Sequences with significant similarity to known hemin- or Hb-binding regions, such as CR or NEAT domains, were not identified from this sequence alignment. The HbpA proteins encoded by the Austrian strains were slightly smaller than 1737 HbpA due to deletions in a poorly conserved but highly charged region of unknown function near the C terminus (Fig. S1 and Table 1). The hbpA genes present in the Austrian strains are carried at the same chromosomal location as that observed for 1737 hbpA (26), and the DNA sequences upstream from their hbpA coding regions contain a highly conserved DtxR-binding site and −10 promoter element, suggesting that hbpA in the Austrian strains is regulated by DtxR in an iron-dependent manner (Fig. S2). While DtxR was previously reported to control the expression of 1737 hbpA and to associate with the 1737 binding site shown in Fig. S2 (26), DtxR regulation of the hbpA genes in the Austrian strains has not been directly demonstrated.
Structural and functional characterization of the HbpA proteins from the Austrian clinical strains.
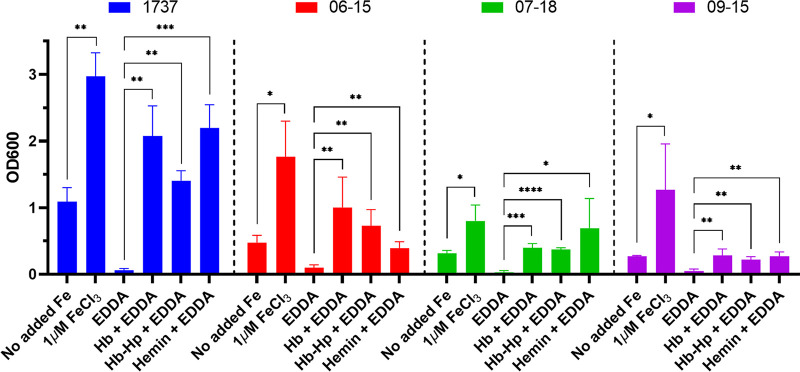
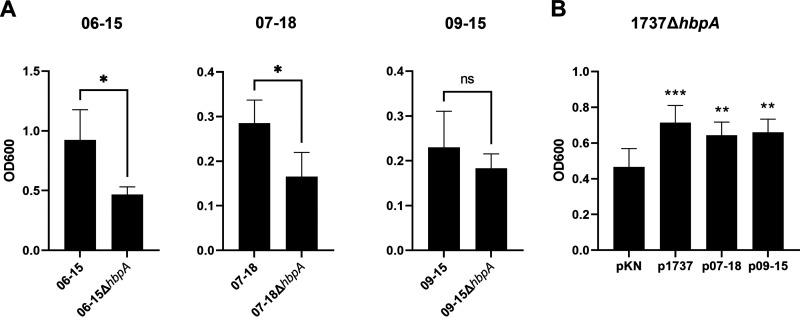
We next examined the structural and functional characteristics of the Austrian HbpA proteins. In these studies, we included a control from the Austrian strain collection, strain 06-15, which produces an HbpA protein with an amino acid sequence identical to that of 1737 HbpA (Table 1). Since the 1737 HbpA protein is involved in heme-iron transport and the subsequent use of heme as an iron source, we first determined whether the Austrian strains could use various heme compounds as iron sources. Initial studies with the Austrian strains revealed that they grew poorly in the low-iron mPGT medium previously used in iron uptake studies with strain 1737 (23, 26). Poor growth in mPGT medium relative to growth observed with strain 1737 was previously observed for other C. diphtheriae clinical isolates (6), and the reason for the reduced growth was not determined. The mPGT medium was adjusted to improve the growth of the Austrians strains under iron-depleted conditions (see Materials and Methods); these modifications to the mPGT medium also enhanced the growth of strain 1737. The three Austrian strains were able to use hemin, Hb, and Hb-Hp as iron sources when grown in the altered low-iron mPGT medium (Fig. 1). None of the Austrian strains were able to grow as well as 1737 in the presence of the various iron or heme sources, but all strains showed an increase in growth in the presence of hemin or the hemoproteins relative to the growth observed in media with no added iron source and the iron chelator EDDA [ethylenediamine di(o-hydroxyphenylacetic acid)].
FIG 1.
The Austrian clinical isolates use Hb, Hb-Hp, and hemin as iron sources for growth. Strains were grown in mPGT medium that was optimized for the growth experiments; supplemental iron sources were added as indicated below the graph (see Materials and Methods for medium details and hemoprotein concentrations). The iron chelator EDDA was added to remove nonhemin iron from the growth medium, and growth was measured by OD600 at 48 ± 1 h. Results show the mean and standard deviation from at least three experiments; analysis of significance used GraphPad Prism v9.1.2 by unpaired t tests. ****, P < 0.0001; ***, P < 0.001; **, P < 0.005; *, P < 0.05.
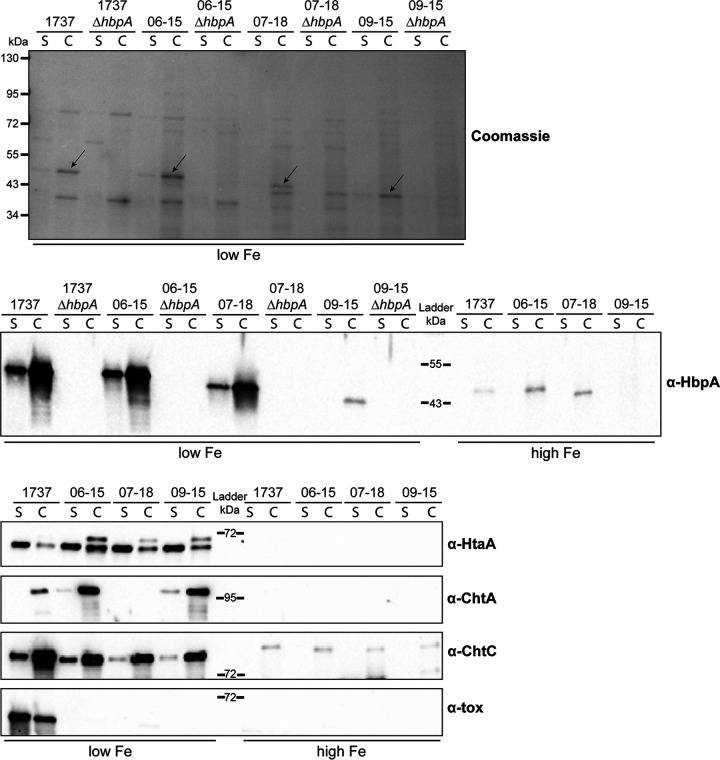
Expression of HbpA in the Austrian strains was regulated by iron levels with optimal production under low-iron conditions; similar iron-dependent expression was observed for the hemin-binding proteins HtaA, ChtA, and ChtC (Fig. 2). Diphtheria toxin was not detected from the three Austrian strains but was present in 1737, where it was optimally expressed in low-iron medium as observed previously (26). HbpA produced by the Austrian strains was observed both in the extracellular medium (S) and associated with the bacteria (C) (Fig. 2). The 09-15 HbpA protein was weakly detected in the Western blot assays using the 1737 HbpA antiserum, which was likely due to its low sequence similarity with 1737 HbpA. It was also observed that the HbpA protein from 09-15 migrated faster in SDS gels than HbpA from 07-18 even though these proteins are predicted to be similar in size (Table 1). HbpA was not detected in any of the hbpA deletion mutants, as expected.
FIG 2.
Expression of HbpA and other iron-regulated proteins in C. diphtheriae strains. Strains were grown in PGTH medium under low (2 μM EDDA)- or high (10 μM FeCl3)-iron conditions. The top panel shows supernatant (S) and cellular (C) protein fractions of each strain, separated by SDS-PAGE and stained with Coomassie blue; arrows indicate HbpA. The lower panels show Western blots probed with anti-HbpA, anti-HtaA, anti-ChtA, anti-ChtC, and anti-diphtheria toxin (α-tox) antibodies. Weak detection of HbpA in strain 09-15 is due to poor recognition of this protein by the anti-HbpA antibody. A representative experiment is shown.
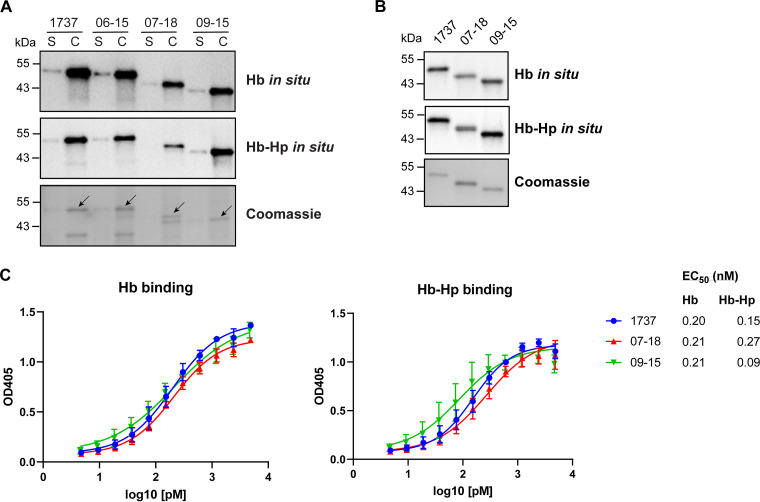
The native HbpA proteins expressed from the three Austrian strains, as well as purified recombinant HbpA proteins from strains 09-15 and 07-18, bound both Hb and Hb-Hp using a previously described in situ binding method (26) (Fig. 3A and B). The binding of the recombinant HbpA proteins to Hb and Hb-Hp was quantified by ELISA, and the findings showed that each of the HbpA proteins exhibited similar binding to the hemoproteins (Fig. 3C). HbpA expressed from a 1737 hbpA deletion mutant (1737ΔhbpA) that carried the cloned Austrian hbpA genes showed that the Austrian HbpA proteins had hemoprotein binding and cellular localization similar to those observed in their native strains (Fig. S3).
FIG 3.
The HbpA proteins from the Austrian strains bind Hb and Hb-Hp. (A) The indicated strains were grown in low-iron PGTH (2 μM EDDA) medium, and supernatant (S) and cellular (C) protein fractions are shown. Proteins were separated by SDS-PAGE and then stained with Coomassie blue (bottom panel) or assessed for binding to Hb or Hb-Hp as indicated using an in situ method. Arrows indicate HbpA protein bands in the Coomassie blue-stained panel. (B) Purified recombinant HbpA protein (0.25 μg per lane) derived from strain 1737, 07-18, or 09-15 was assessed as for panel A. Representative experiments are shown for panels A and B. (C) Binding of HbpA to Hb and Hb-Hp was measured by ELISA as described in Materials and Methods. EC50 values were calculated using GraphPad Prism v9.1.2. Results show the mean and standard deviation from three experiments.
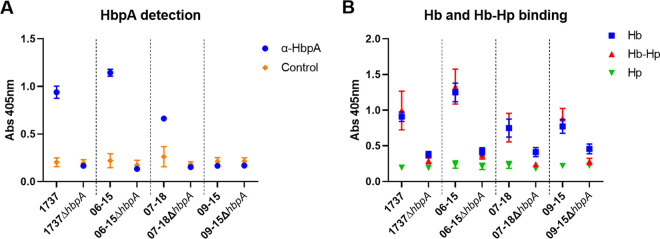
Whole-cell ELISAs (WC ELISAs), which are used to detect surface exposure of bacterial proteins, showed that HbpA was surface exposed in the Austrian strains (Fig. 4A and B). Additionally, the HbpA proteins from all three of the Austrian strains were able to bind both Hb and Hb-Hp at the bacterial surface; binding was only minimally detected in strains deleted for hbpA, which indicates that HbpA was responsible for most of the surface binding observed in these strains (Fig. 4B). The binding of HbpA to Hb and Hb-Hp at the cell surface was also observed in WC ELISAs in which the cloned Austrian hbpA genes were expressed in 1737ΔhbpA (Fig. S4).
FIG 4.
HbpA expressed from the Austrian strains binds Hb and Hb-Hp at the bacterial surface. (A) WC ELISAs were used to assess surface exposure of HbpA in the strains indicated. Bacteria were grown overnight in low-iron PGTH medium (2 μM EDDA), cell densities were normalized by OD600, and the cultures were used to coat microtiter plates. Expression and surface exposure of HbpA were measured by anti-HbpA antibody, with the secondary antibody used as a negative control. Abs, absorbance. (B) Binding to Hb and Hb-Hp was measured by incubation of cultures bound to microtiter plates with either Hb or Hb-Hp followed by detection of the bound hemoprotein with anti-Hb or anti-Hp antibody, respectively. Hp binding (with anti-Hp detection) was used as a negative control. Results show the mean and standard deviation from at least three experiments. For Hb and Hb-Hp binding (B), all comparisons between each strain and its corresponding deletion mutant were statistically significant (P < 0.0005) by unpaired t test using GraphPad Prism v9.1.2.
Deletion of the hbpA gene from the three Austrian strains resulted in reduced ability to use Hb-Hp as an iron source (Fig. 5A). Additionally, expression of the cloned Austrian hbpA genes in 1737ΔhbpA resulted in wild-type (WT) (p1737) levels of growth relative to the vector control (pKN) when Hb-Hp was the sole iron source (Fig. 5B). These findings show that the HbpA proteins in strains 07-18 and 09-15 have the ability to acquire iron from Hb-Hp, despite the low sequence similarity with the 1737 HbpA protein. Although statistical significance was not observed with strain 09-15 in the Hb-Hp iron utilization studies, likely due to variability of the assay and poor growth of the strain, the cloned hbpA gene from strain 09-15 was able to restore wild-type (p1737) levels of growth to 1737ΔhbpA.
FIG 5.
The Austrian HbpA proteins are involved in the use of Hb-Hp as an iron source. (A) The Austrian wild-type strains and their corresponding hbpA deletion mutants were grown in iron-limited medium with Hb-Hp as the sole iron source. Medium and growth conditions were optimized for each strain (see Materials and Methods). Growth of strains 07-18, 07-18ΔhbpA, 09-15, and 09-15ΔhbpA was measured at 48 ± 1 h after inoculation; 06-15 and 06-15ΔhbpA were measured at 20 to 24 h after inoculation. (B) pKN2.6Z plasmids carrying the cloned hbpA gene from strains 1737, 07-18, and 09-15 were transformed into strain 1737ΔhbpA to assess the use of Hb-Hp as a sole iron source. Strains were grown in low-iron mPGT medium containing Hb-Hp. Statistical significance in panel B is shown for each strain compared to the growth of pKN (vector-only control). Results in panels A and B show the mean and standard deviation from at least three experiments; analysis of significance used GraphPad Prism v9.1.2 by unpaired t tests. ***, P < 0.001; **, P < 0.005; *, P < 0.05; ns, no significant difference.
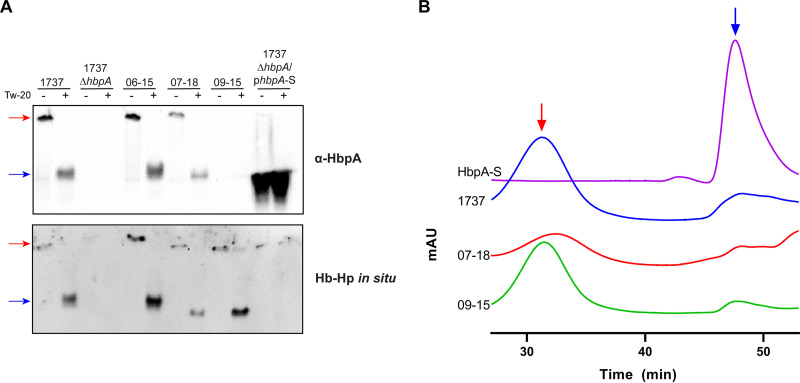
It was previously reported that 1737 HbpA formed soluble aggregates when secreted into the culture supernatant and also following purification of the recombinant HbpA protein (23). HbpA secreted from the Austrian strains exhibited migration in native gels similar to that of 1737 HbpA, in which much of the protein is unable to enter the gel in the absence of a detergent (Tween 20), suggesting that secreted HbpA from all three strains forms high-molecular-weight complexes (Fig. 6A). Similar findings were observed when the cloned Austrian hbpA genes were expressed in 1737ΔhbpA (Fig. S5). Size exclusion chromatography (SEC)-fast protein liquid chromatography (FPLC) using the purified recombinant HbpA from strains 07-18 and 09-15 also showed that both proteins form large aggregates (Fig. 6B), with estimated sizes of 988 kDa and 1,025 kDa, respectively (Fig. S6). It was previously reported that the aggregate formed by the 1737 HbpA protein is approximately 987 kDa (23); however, the HbpA complexes observed on the sizing column form broad peaks, which suggests that the HbpA aggregates are variable in size and may not form a well-defined structure (Fig. 6B). The 1737 HbpA-S protein, which contains a deletion of the C-terminal region, was used as a control in these studies, since it does not form aggregates and migrates as a monomer on native gels and in SEC (Fig. 6A and B) (23). These findings demonstrate that the HbpA proteins from 1737, 07-18, and 09-15 exhibit similar functional and structural characteristics despite significant sequence differences.
FIG 6.
The Austrian HbpA proteins form large soluble aggregates. (A) Native gels were used to compare proteins present in supernatant fractions from strains grown in low-iron PGTH medium (2 μM EDDA). Protein samples were prepared in the presence (+) or absence (−) of 0.1% Tween 20, which was used to solubilize large aggregates (23). Supernatant samples were normalized by OD600, separated by native PAGE, and then probed with anti-HbpA antibody (upper panel) or assessed for Hb-Hp binding in situ (lower panel). Plasmid phbpA-S expresses the HbpA-S protein, which runs as a monomer and was used as a control (23). A representative experiment is shown. (B) SEC was used to analyze purified recombinant HbpA from strains 07-18, 09-15, 1737, and HbpA-S. A lower elution time (minutes) indicates a higher-molecular-weight species. The red arrow indicates aggregated protein; the blue arrow indicates the monomer peak for HbpA-S. Samples were run on a Superose 6 increase 3.2/300 column for large aggregate sizing; each trace shows a representative sample from at least three replicates. mAU, milli-absorbance units.
Identification of a putative Hb- and Hb-Hp-binding site in HbpA.
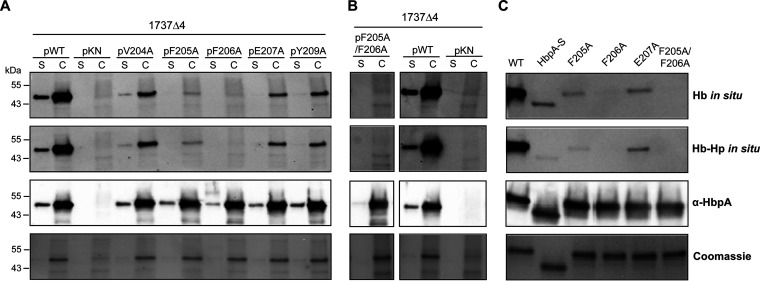
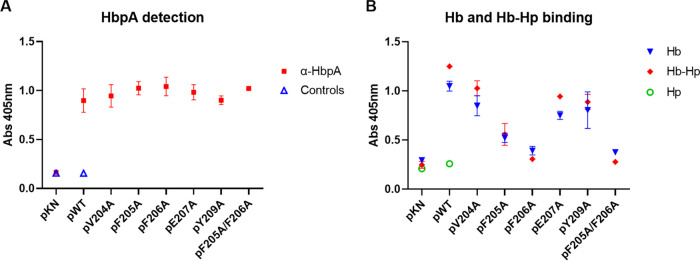
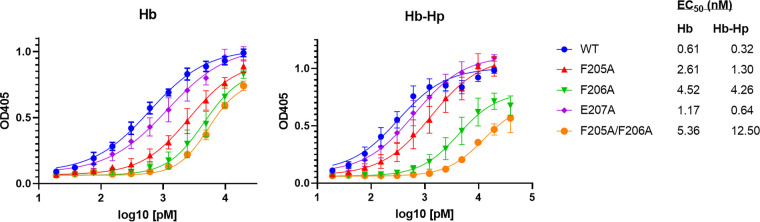
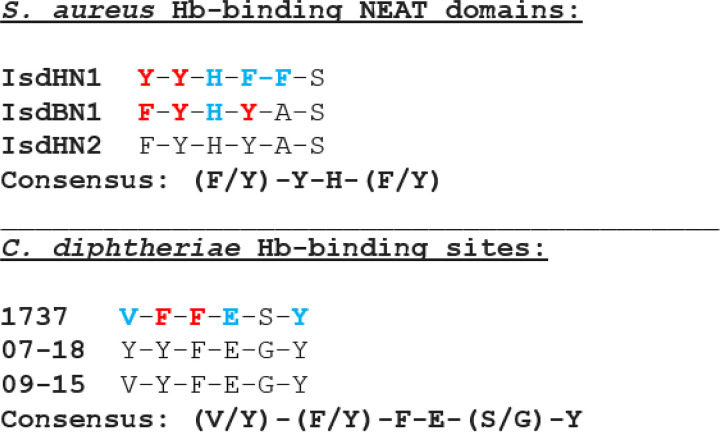
Regions associated with Hb and Hb-Hp binding or iron transport have not been previously determined for HbpA (23). Sequences required for binding to Hb were identified in the S. aureus IsdB and IsdH NEAT domains using biochemical and crystallographic analysis as well as sequence comparisons (13, 29). The Hb-binding regions on the IsdB/IsdH proteins include the aromatic amino acids Phe and Tyr and a conserved His residue, which constitute the 4-amino-acid consensus sequence (F/Y)-Y-H-(Y/F) (13, 29). Since we established that the HbpA proteins from the Austrian strains are functionally and structurally similar to 1737 HbpA, we used the sequence alignment in Fig. S1 to identify conserved regions that may be associated with HbpA function. In a search for an Hb- and Hb-Hp-binding region in HbpA, we identified a 6-amino-acid sequence that has characteristics similar to those of the S. aureus binding sites and was broadly conserved between the Austrian strains and strain 1737: this region has the consensus sequence (V/Y)-(F/Y)-F-E-(S/G)-Y (Fig. S1). To determine if this region was required for Hb-Hp binding, we made individual amino acid substitutions at five of the six residues within this sequence in 1737 HbpA and then tested the modified HbpA proteins for their ability to bind Hb and Hb-Hp. All of the substitutions resulted in reduced binding to both Hb and Hb-Hp relative to the wild-type protein; however, changes in residues F205A and F206A had the strongest impact on binding (Fig. 7A), and an F205A/F206A double mutant also showed a similar reduction in binding to both hemoproteins (Fig. 7B). These studies showed that all of the modified HbpA proteins were both cell associated and secreted into the medium, indicating that the amino acid changes did not affect the localization of the proteins (Fig. 7A and B). Binding studies using purified recombinant HbpA proteins carrying the various amino acid substitutions confirmed the crucial role that these amino acids have in binding to Hb and Hb-Hp (Fig. 7C). WC ELISA studies showed that all of the HbpA proteins containing the various amino acid substitutions were exposed on the surface at levels equivalent to that of the wild-type protein, and all had various levels of reduced binding to Hb and Hb-Hp (Fig. 8A and B).
FIG 7.
Amino acid substitutions in the HbpA Hb-binding site cause reduced binding to Hb and Hb-Hp. (A) Supernatant (S) and cellular (C) protein fractions were obtained from strains grown in low-iron mPGT medium (0.25 μM FeCl3). Protein fractions were separated by SDS-PAGE and then stained with Coomassie blue (bottom panel), probed with anti-HbpA antibody (third panel from top), or assessed in situ for binding to Hb or Hb-Hp as indicated. The various point mutations were constructed in the 1737 hbpA gene. The hbpA genes carrying the various point mutants were expressed from plasmid pKN2.6Z in strain 1737Δ4, which is deleted for hbpA, htaA, and chtA/C (26). Strain 1737Δ4 was used in these studies to reduce background bands associated with binding to hemoproteins by HtaA and ChtA/C. WT indicates the cloned wild-type 1737 hbpA gene, and pKN indicates vector control. (B) The F205A/F206A double mutant was expressed and run as in panel A, along with WT and pKN controls. All samples were run on the same gel, but the F205A/F206A sample was run in a lane that was not adjacent to the control samples. (C) Purified recombinant 1737 HbpA proteins (0.25 μg per lane) with amino acid substitutions indicated were separated by SDS-PAGE and then assessed as in panel A. Representative experiments are shown.
FIG 8.
HbpA proteins carrying the amino acid substitutions described in Fig. 7A and B were examined in WC ELISAs. (A) Cell cultures were normalized by OD600, and HbpA on the cell surface was detected using anti-HbpA antibody (“Controls” indicates use of secondary detection antibody only). (B) Binding to hemoproteins was measured by incubation with Hb or Hb-Hp, followed by detection using anti-Hb or anti-Hp antibody, respectively (Hp was used as a negative control). The hbpA genes carrying the various point mutants were expressed from plasmid pKN2.6Z in strain 1737Δ4, which is deleted for hbpA, htaA, and chtA/C (26). WT indicates the cloned wild-type 1737 hbpA gene, and pKN indicates vector control. Results show the mean and standard deviation from at least three experiments.
Plasmids encoding HbpA proteins containing the amino acid substitutions F205A, F206A, and F205A/F206A were unable to restore wild-type levels of growth to 1737ΔhbpA when Hb-Hp was the sole iron source (Fig. 9). The growth of 1737ΔhbpA carrying the cloned genes for these three HbpA proteins was not significantly different from the growth of 1737ΔhbpA carrying only the vector (pKN), but the growth of these strains was significantly different from the growth of the WT (Fig. 9). These findings indicate that these amino acid changes in the Hb-binding site significantly affect the hemin-iron transport function of HbpA.
FIG 9.
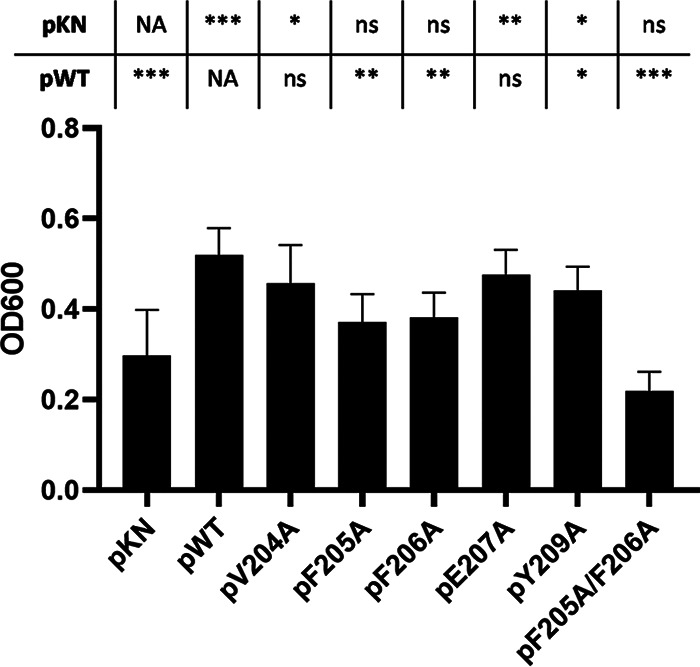
Amino acid substitutions in the HbpA Hb-binding site impact the use of Hb-Hp as an iron source. C. diphtheriae 1737ΔhbpA carrying various cloned hbpA genes on plasmid pKN2.6Z was examined for growth in low-iron mPGT medium that contained Hb-Hp as the sole iron source. WT indicates cloned wild-type hbpA gene, and pKN indicates vector control. Results show the mean and standard deviation from at least three experiments. Statistical significance was calculated in GraphPad Prism v9.1.2 using unpaired t tests; each strain was compared to the pKN and pWT controls as indicated above the graph. ***, P < 0.001; **, P < 0.005; *, P < 0.05; ns, no significant difference; NA, not applicable.
ELISAs used to quantitate the binding of the various HbpA proteins to Hb and Hb-Hp showed that the HbpA proteins with reduced hemin-iron transport function (F205A, F206A, and F205A/F206A) also showed large reductions in Hb and Hb-Hp binding as determined by 50% effective concentration (EC50) calculations (Fig. 10). SEC analysis showed that the modified HbpA proteins with the most significant reduction in Hb-Hp binding maintained the ability to form large aggregates similar to those found with wild-type HbpA (Fig. S7), indicating that the various amino acid substitution in the putative Hb-binding site did not affect aggregate formation.
FIG 10.
Binding of purified recombinant HbpA proteins to Hb and Hb-Hp was quantified by ELISA. Purified HbpA proteins with amino acid substitutions in the Hb-binding site are indicated; WT is the wild-type HbpA protein. EC50 values were calculated using GraphPad Prism v9.1.2, using upper and lower concentrations that best represent the range of performance for each protein. Results show the mean and standard deviation from at least three experiments.
The HbpA-S protein, which lacks the C-terminal transmembrane domain, exists as a monomer and is unable to form aggregates or anchor in the cytoplasmic membrane (23). HbpA-S also lacks iron transport activity and has reduced but detectable binding to Hb and Hb-Hp using the in situ binding method (23) (Fig. 7C). To provide additional support for the involvement of the consensus sequence described above in Hb binding, we constructed recombinant HbpA-S proteins that contained the F206A and the F205A/F206A amino acid substitutions. Both of these modified HbpA-S proteins showed a marked decrease in binding to both Hb and Hb-Hp in the in situ binding assay (Fig. 11), providing further evidence of the importance of this region in Hb and Hb-Hp binding.
FIG 11.
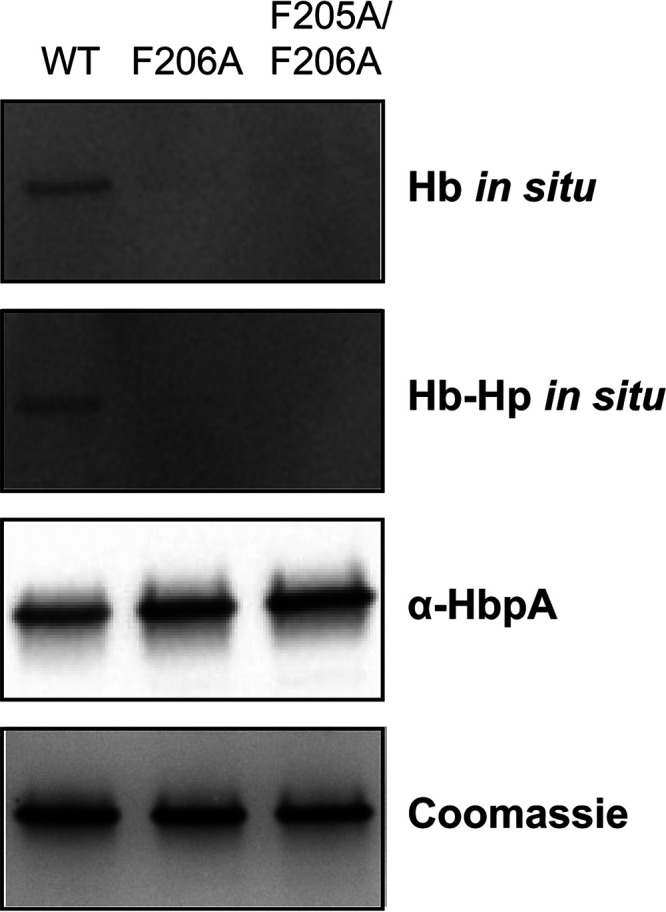
The HbpA-S monomer protein carrying amino acid substitutions in the Hb-binding site showed reduced binding to Hb and Hb-Hp. Purified recombinant proteins (2 μg per lane) were separated by SDS-PAGE, and subsequent analysis was performed as described in the legend to Fig. 7A. A representative experiment is shown.
DISCUSSION
Although Hb exists as a tetramer inside erythrocytes, following release from red blood cells, Hb forms oxidized dimers that can rapidly bind Hp to form the Hb-Hp complex (11). Hb-Hp, Hb, and free hemin all serve as iron sources for C. diphtheriae and numerous other bacterial pathogens (30). Prior to this report, the HbpA protein had been studied only in strain 1737, a toxigenic clinical isolate obtained during the diphtheria epidemic in the former Soviet Union in the 1990s (24). In this study, we have characterized the HbpA proteins from two Austrian clinical isolates, 07-18 and 09-15, which encode HbpA variants that have low sequence similarity to the 1737 HbpA protein (53% and 44% identity, respectively) and only 46% sequence identity with each other (Table 1) (27). Despite the limited sequence identity between the HbpA proteins in 07-18 and 09-15, single nucleotide polymorphism (SNP) analysis and core genome multilocus sequence typing (cgMLST) showed that these two isolates have a high degree of overall genetic similarity, and both strains were nontoxigenic and associated with cutaneous infections (27).
Proteins encoded by the Austrian strains (07-18 and 09-15) that are predicted to be involved in the use of iron from Hb-Hp include HtaA, ChtA/C, and HmuT, all of which show greater than 98% sequence identity to the homologous proteins in 1737 (Table 1). Genes that flank the hbpA gene in the chromosome (dip2331 and dip2329) show greater than 98% sequence identity between strains 1737, 07-18, and 09-15 (25). We also observed that the hbpA gene in 1737 has a GC content of 45%, whereas the average genomic GC content for 1737 and related strains is approximately 53.5% (25). The GC content of the hbpA genes from strains 07-18 and 09-15 is similarly low at 43% and 45%, respectively (27). The below-average GC content suggests that the hbpA gene was introduced into each of these strains by horizontal transfer. However, the mechanism of transfer is unclear, since transposon, phage, or insertion sequence-related genes in the regions flanking hbpA were not identified (25, 27). Surprisingly, two clinical strains from Brazil, HC02 and VA01, have an HbpA amino acid sequence identical to that of strains 07-18 and 09-15, respectively (31, 32) (Table 1).
Unusual sequence characteristics of the HbpA proteins described in this study include the high level of sequence similarity that was observed between 1737 and the two Austrian strains in the HbpA N-terminal secretion signal and in the C-terminal transmembrane region (see Fig. S1 in the supplemental material). In most proteins, these terminal regions typically maintain structural similarity, rather than a high level of specific sequence identity as seen here. Another novel feature of the HbpA sequence is a highly charged region in the C-terminal portion of the protein (Fig. S1, underlined sequence). While a function for this region is not known, an alignment of the amino acid sequences between 1737 and the two Austrian strains indicates this region is poorly conserved. Additionally, the small deletions within this region in strains 07-18 and 09-15 do not appear to significantly impact any of the functional or structural features of HbpA that were identified in this study, including hemin-iron transport, Hb-Hp binding, aggregate formation, and cellular localization (Fig. 3 to 5). The lack of sequence conservation in this C-terminal segment suggests that the presence of charged residues may be more important than the specific sequence or overall size of this region.
Because the HbpA proteins from 1737 and the two Austrian strains maintained both structural and functional similarities despite limited sequence identity, we reasoned that areas important for function and structure may be located in regions with a higher level of amino acid sequence similarity. Using this approach, we identified a region in the HbpA protein sequences that had characteristics similar to those of the Hb-binding sites present in the S. aureus NEAT domains (13, 29) (Fig. 12). The consensus sequence for the C. diphtheriae Hb-binding site, (V/Y)-(F/Y)-F-E-(S/G)-Y, was conserved in all of the HbpA sequences that were identified in a BLAST search of the NCBI database, indicating that this region is conserved even between proteins with low sequence similarity.
FIG 12.
Analysis of Hb-binding sites in S. aureus and C. diphtheriae. Amino acid substitutions were constructed in the 1737 HbpA Hb-binding site and at targeted residues in the Hb-binding N1 NEAT domains in IsdB and IsdH, as described previously (13, 29). Residues in red showed the strongest contribution to Hb binding, while residues in blue had a weaker effect on binding (all other residues were not tested for binding).
The Hb-binding NEAT domains in the IsdH and IsdB proteins include the amino acid consensus sequence (F/Y)-Y-H-(F/Y) (Fig. 12). The IsdH protein contains two Hb-binding NEAT domains with slightly different sequences: the binding sites for N1 and N2 NEAT domains are Y-Y-H-F and F-Y-H-Y, respectively, while IsdB contains only a single Hb-binding NEAT domain (N1) that is identical to the IsdH N2 domain (Fig. 12). Alanine substitutions at each of the aromatic residues and the conserved His residue in IsdH N1 and IsdB N1 resulted in decreased binding to Hb (Fig. 12) (29). However, substitutions in the Tyr and Phe residues in the N1 domain of IsdB showed the greatest decrease in Hb binding, while changes in the two Tyr residues in the N1 domain of IsdH had the greatest impact on Hb binding (Fig. 12).
In this study, we show that the amino acid substitutions with the strongest contribution to Hb binding were in two adjacent Phe residues, F205 and F206 (Fig. 12). In the Austrian strains, a Tyr residue replaces the Phe at position 205, suggesting that Phe and Tyr residues may be interchangeable at this position in the Hb-binding site. A similar situation occurs at the Hb-binding site in the NEAT domains, where critical Tyr and Phe residues are also interchangeable at specific locations within the consensus sequence (Fig. 12).
Additional studies will be needed to advance our understanding of the structural basis for the binding of Hb at the putative Hb-binding site in HbpA. These studies will confront the challenges in attempting to analyze the binding of Hb to native HbpA that exists as a complex protein aggregate.
MATERIALS AND METHODS
Bacterial strains and growth media.
Table 2 lists the strains of C. diphtheriae, Corynebacterium ulcerans, and Escherichia coli used in this study. Bacterial stocks were stored in the appropriate growth medium with 20% glycerol and maintained at −80°C. Corynebacterium strains were grown in heart infusion broth (Difco) with 0.2% Tween 80 (Millipore Sigma) (HIBTW); E. coli strains were grown in Luria-Bertani (LB) medium (Difco). A semidefined medium, mPGT, was used for iron-restricted growth of C. diphtheriae and formulated as described previously (33). Modifications to mPGT for growth of the Austrian strains are described below (see “Bacterial growth assays”). For growth in mPGT, ferric chloride (FeCl3) was added at 0.25 μM for low iron and 10 μM for high iron. Where appropriate, hemin was used at 5 μM, ethylenediamine di(o-hydroxyphenylacetic acid) (EDDA) (Millipore Sigma) at 10 μM, and kanamycin (Millipore Sigma) at 50 μg/mL.
TABLE 2.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics or usea | Reference or source |
|---|---|---|
| Strains | ||
| C. diphtheriae | ||
| 1737 | Wild type, Gravis biotype, tox+ | 24 |
| 1737ΔhbpA | Deletion of hbpA in 1737 | 26 |
| 1737Δ4 | Deletion of chtA, chtC, htaA, and hbpA in 1737 | 26 |
| 06-15 | Wild-type clinical isolate | 27 |
| 06-15ΔhbpA | Deletion of hbpA in 06-15 | This study |
| 07-18 | Wild-type clinical isolate | 27 |
| 07-18ΔhbpA | Deletion of hbpA in 07-18 | This study |
| 09-15 | Wild-type clinical isolate | 27 |
| 09-15ΔhbpA | Deletion of hbpA in 09-15 | This study |
| C. ulcerans 712 | Wild type used for cloning | 34 |
| E. coli | ||
| BL21(DE3) | Protein expression strain | New England BioLabs |
| NEBα | Cloning strain | New England BioLabs |
| S17-1 | Mating strain | 35 |
| Plasmids | ||
| pET24(a)+ | Expression vector, Knr | Millipore |
| pET-hbpA-S | pET24(a)+ carrying strep-tagged hbpA (no TM domain) from 1737 | 26 |
| pET-hbpA | pET24(a)+ carrying strep-tagged full-length hbpA from 1737 | 23 |
| pET-hbpA-07-18 | pET24(a)+ carrying strep-tagged full-length hbpA from 07-18 | This study |
| pET-hbpA-09-15 | pET24(a)+ carrying strep-tagged full-length hbpA from 09-15 | This study |
| pET-hbpA-V204A | pET24(a)+ carrying strep-tagged full-length hbpA from 1737 with V204A point mutation | This study |
| pET-hbpA-F205A | pET24(a)+ carrying strep-tagged full-length hbpA from 1737 with F205A point mutation | This study |
| pET-hbpA-F206A | pET24(a)+ carrying strep-tagged full-length hbpA from 1737 with F206A point mutation | This study |
| pET-hbpA-E207A | pET24(a)+ carrying strep-tagged full-length hbpA from 1737 with E207A point mutation | This study |
| pET-hbpA-Y209A | pET24(a)+ carrying strep-tagged full-length hbpA from 1737 with Y209A point mutation | This study |
| pET-hbpA-F205A/F206A | pET24(a)+ carrying strep-tagged full-length hbpA from 1737 with F205A and F206A point mutations | This study |
| pKN2.6Z | C. diphtheriae shuttle vector, Knr | 21 |
| pKN-hbpA-S | pKN2.6Z carrying the 1737 hbpA gene (no TM domain) | 23 |
| pKN-hbpA-FL | pKN2.6Z carrying the 1737 hbpA gene | 23 |
| pKN-hpbA-07-18 | pKN2.6Z carrying the 07-18 hbpA gene | This study |
| pKN-hpbA-09-15 | pKN2.6Z carrying the 09-15 hbpA gene | This study |
| pKN-hbpA-V204A | pKN2.6Z carrying the 1737 hbpA gene with V204A point mutation | This study |
| pKN-hbpA-F205A | pKN2.6Z carrying the 1737 hbpA gene with F205A point mutation | This study |
| pKN-hbpA-F206A | pKN2.6Z carrying the 1737 hbpA gene with F206A point mutation | This study |
| pKN-hbpA-E207A | pKN2.6Z carrying the 1737 hbpA gene with E207A point mutation | This study |
| pKN-hbpA-Y209A | pKN2.6Z carrying the 1737 hbpA gene with Y209A point mutation | This study |
| pKN-hbpA-F205A/F206A | pKN2.6Z carrying the 1737 hbpA gene with F205A and F206A point mutations | This study |
| pK18mobsacB | C. diphtheriae shuttle vector, Knr | 36 |
| pK18Δ06-15hbpA | Suicide vector for deletion of hbpA in strain 06-15 | This study |
| pK18Δ07-18hbpA | Suicide vector for deletion of hbpA in strain 07-18 | This study |
| pK18Δ09-15hbpA | Suicide vector for deletion of hbpA in strain 09-15 | This study |
Kn, kanamycin; strep, streptavidin; TM, transmembrane.
Purified human Hb was purchased from MP Biomedical, and human Hp1-1 was acquired from Millipore Sigma or Athens Research and Technology. Hemoglobin was prepared as described previously to remove free heme (23). To form the Hb-Hp complex, appropriate amounts of Hb and Hp were mixed and incubated at room temperature for 20 min prior to dilution into growth medium or buffer. For growth assays using C. diphtheriae 1737-derived strains, Hb was used at 4.37 μg/mL and Hp at 8.75 μg/mL; for the Austrian diphtheria strains, Hb was used at 6.56 μg/mL and Hp at 13.13 μg/mL. For 1737 strains expressing HbpA with point mutations, Hb was used at 2.92 μg/mL and Hp at 5.83 μg/mL.
Bacterial growth assays.
Growth assays for C. diphtheriae under iron-restricted conditions were performed as follows. On day 1, cultures were inoculated from freezer stocks into HIBTW (with antibiotics as appropriate) and grown at 37°C with shaking overnight. On day 2, an additional 1 mL of HIBTW was added to cultures and cells were incubated with shaking for an additional 1 to 2 h. Subsequently, 500 μL of culture was harvested and spun at 13,000 rpm for 1 min, and the cell pellets were resuspended in 1 mL mPGT with 1 μM FeCl3 and incubated at 37°C with shaking for 4 to 6 h. The cultures were then used to inoculate tubes containing medium with appropriate additives at a starting cell density of an optical density at 600 nm (OD600) at 0.03 and incubated at 37°C with shaking for 16 to 20 h. On day 3, the OD600 of the culture(s) was measured and cells were harvested for downstream experiments as needed.
In growth studies with the Austrian strains using Hb-Hp as the sole iron source, mPGT was supplemented with additional Casamino Acids to a final concentration of 1.5% (0.5% in standard mPGT). For other iron-restricted growth, 10% HIBTW was added to standard mPGT to make PGTH. EDDA was added to PGTH at 2 μM to create a low-iron growth condition; 10 μM FeCl3 was added to PGTH for a high-iron condition. C. diphtheriae strain 1737 was grown in mPGT medium derivations for side-by-side growth with the Austrian strains. Overnight growth was measured by OD600 following 16 to 20 h of incubation at 37°C with shaking, unless otherwise indicated.
DNA cloning and plasmid construction.
DNA for cloning was derived from PCR amplification of C. diphtheriae strain 1737, 06-15, 07-18, or 09-15 as appropriate, unless otherwise noted. Plasmids and PCR products were assembled using the NEBuilder HiFi DNA assembly cloning kit (New England Biolabs). Plasmids constructed and used in this study are listed in Table 2.
Recombinant protein expression constructs were generated in the pET24(a)+ expression vector. Briefly, coding sequence for hbpA excluding the native secretion signal was fused with an N-terminal streptavidin tag (Strep-tag II) and inserted into prepared vector. Assembly reaction mixtures were transformed into E. coli NEBα. Insertions were confirmed by sequencing, and then plasmids were moved into E. coli BL21(DE3) for expression.
Complementation plasmids for HbpA from strains 06-15, 07-18, and 09-15 were constructed in the pKN2.6Z vector using DNA synthesized by GenScript, which included the full coding region and upstream promoter and regulatory elements from each strain. These fragments were inserted into pKN2.6Z and then transformed into C. ulcerans 712. C. ulcerans was used as an intermediate in place of E. coli as the native hbpA genes are toxic to E. coli. Plasmids were then purified using a Qiagen miniprep kit and electroporated into C. diphtheriae 1737.
C. diphtheriae 1737 hbpA point mutants were created using primers designed to introduce nucleotide changes into the hbpA coding regions in pET-hbpA and pKN-hbpA. The resulting product was assembled using one-fragment assembly cloning and moved into E. coli NEBα cells [for pET24(a)+ constructs] or C. ulcerans 712 (for pKN2.6Z constructs). Plasmids were then purified and transformed into E. coli BL21(DE3) [pET24(a)+ constructs] or C. diphtheriae 1737ΔhbpA and 1737Δ4 (pKN2.6Z constructs). Double point mutants were made sequentially; sequenced plasmid DNA containing the F206A mutation was used as the PCR template material for introducing an additional mutation.
Gene knockout construction.
hbpA deletion mutants in the three Austrian strains of C. diphtheriae were constructed using the same method described previously for strain 1737, with the exception that assembly cloning was used in place of restriction digestion and ligation (26). Briefly, DNA sequence flanking the hbpA regions was generated by PCR from each strain and cloned into the suicide vector pK18mobsacB. Each construct retains 6 to 9 amino acids of the N- and C-terminal regions of the gene coding sequence. Assembly reaction mixtures were transformed into E. coli NEBα cells, purified and sequenced, and then transformed into the E. coli S-17 mating strain.
Purification of Strep-tag II-tagged proteins.
Recombinant proteins expressed with Strep-tag II were purified as described previously with modifications (23). Briefly, Strep-TactinXT Superflow resin (IBA Life Sciences) was used with the addition of 1% poly(ethylene glycol) octyl ether (octyl-POE) (Thermo Fisher Scientific) to the culture prior to lysis to facilitate recovery of HbpA from the purification process. Following dialysis into phosphate-buffered saline (PBS) and then 0.2× PBS plus 4% glycerol, a SpeedVac vacuum concentrator (Thermo Fisher Scientific) was used to reduce samples to one-fifth the initial volume. The resulting protein was in 1× PBS plus 20% glycerol and was stored at −20°C.
C. diphtheriae cell lysis.
C. diphtheriae cells grown for analysis of proteins in the supernatant and cellular fractions were grown in mPGT (with modifications as needed; see “Bacterial strains and growth media” and “Bacterial growth assays”) with limiting iron (mPGT with 0.25 μM FeCl3 or PGTH with 2 μM EDDA). Cell number was normalized by OD600 at harvest. Cells were then treated with lysozyme (from chicken egg white) and sodium lauroyl sarcosinate (Sarkosyl) as previously described (23) and used for gels as described below.
Gel electrophoresis and blotting.
Precast 4 to 15% gradient TGX (Tris-glycine extended) PAGE gels (Bio-Rad) (which are manufactured without SDS) were used for separation under both denaturing and native conditions. Samples of lysed cells, culture supernatant, or purified protein were boiled in Laemmli sample buffer for 10 min and separated using Tris-glycine-SDS running buffer for denaturing experiments; native sample buffer (with no heating step) and Tris-glycine running buffer were used for native conditions (all reagents from Bio-Rad). Tween 20 (0.1%) was added to samples as indicated to solubilize protein aggregates for analysis under native conditions. Coomassie blue staining, transfers to nitrocellulose, Western blotting, and in situ analysis of Hb and Hb-Hp binding were performed as previously described. Briefly, in situ analysis involves the separation of proteins by PAGE, transfer to nitrocellulose membranes, and incubation with Hb or Hb-Hp followed by detection with anti-Hb or anti-Hp antibody, respectively.
Direct ELISAs and EC50 calculation.
To assess the binding kinetics of purified HbpA proteins, Hb (18.8 μg/mL) or Hb-Hp (18.8:35 μg/mL) was immobilized on a microtiter plate. Wells were blocked with Tris-buffered saline plus 0.1% Tween 20 (TBST) with 5% blotting-grade blocker (Bio-Rad) and then incubated with serial dilutions of HbpA proteins. Plates were then incubated with a primary antibody against Strep-tag II, washed with TBST, incubated with an alkaline phosphatase-labeled secondary antibody, and developed by the addition of p-nitrophenyl phosphate (pNPP; Millipore Sigma). Results were measured at OD405. Details of the protocol are as described in reference 23. For analysis of the Austrian HbpA proteins, 1:2 serial dilutions were used from approximately 4.9 nM to 0.005 nM, using protein aggregate sizes derived from SEC. For the 1737 HbpA point mutants, 1:2 serial dilutions from 40 nM to 0.02 nM were used, based on the 1737 protein aggregate size previously determined (987 kDa) (23). Fifty percent effective concentration (EC50) calculations were done using GraphPad Prism v9.1.2 from a variable-slope four-parameter curve using the upper concentration that best represented a stable plateau at the upper asymptote for each protein.
WC ELISAs for protein surface exposure and Hb and Hb-Hp binding.
WC ELISAs were performed as previously described (23). Briefly, microtiter plates were coated with C. diphtheriae cultures grown under low-iron conditions (mPGT with 0.25 μg FeCl3 or PGTH with 2 μM EDDA; see “Bacterial strains and growth media” and “Bacterial growth assays”) and standardized by OD600. HbpA on the surface was detected by incubation with anti-HbpA antibody, while binding was assessed by incubation with Hb or Hb-Hp (as indicated) and detection with anti-Hb or anti-Hp antibody, respectively. An alkaline phosphatase-labeled secondary antibody was used for detection, followed by the addition of pNPP and measurement of OD405.
SEC-FPLC.
An ÄKTApur fast protein liquid chromatograph (FPLC) (Cytiva) was used to assess purified recombinant proteins by size exclusion chromatography (SEC) as previously described (23). A Superdex 200 increase 3.2/300 column was used to identify the presence of large protein aggregates in the HbpA point mutants, while a Superose 6 increase 3.2/300 column was used to determine the size of the aggregates for the HbpA proteins from the Austrian strains. Protein sizing standards, run conditions, and detection were all the same as reported previously. GraphPad Prism 9.1.2 was used to calculate the line of best fit for the sizing standards, which was then used to determine the aggregate size for the Austrian strains of HbpA.
ACKNOWLEDGMENTS
This work was supported by the intramural research program at the Center for Biologics Evaluation and Research, Food and Drug Administration.
We thank Eric Peng, Scott Stibitz, and Jessica Hastie for helpful comments on the manuscript.
Footnotes
Supplemental material is available online only.
Contributor Information
Michael P. Schmitt, Email: michael.schmitt@fda.hhs.gov.
George O’Toole, Geisel School of Medicine at Dartmouth.
REFERENCES
- 1.Zasada AA. 2015. Corynebacterium diphtheriae infections currently and in the past. Przegl Epidemiol 69:439–444, 569–574. [PubMed] [Google Scholar]
- 2.Hadfield TL, McEvoy P, Polotsky Y, Tzinserling VA, Yakovlev AA. 2000. The pathology of diphtheria. J Infect Dis 181(Suppl 1):S116–S120. 10.1086/315551. [DOI] [PubMed] [Google Scholar]
- 3.Timms VJ, Nguyen T, Crighton T, Yuen M, Sintchenko V. 2018. Genome-wide comparison of Corynebacterium diphtheriae isolates from Australia identifies differences in the pan-genomes between respiratory and cutaneous strains. BMC Genomics 19:869. 10.1186/s12864-018-5147-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gaspar AH, Ton-That H. 2006. Assembly of distinct pilus structures on the surface of Corynebacterium diphtheriae. J Bacteriol 188:1526–1533. 10.1128/JB.188.4.1526-1533.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kunkle CA, Schmitt MP. 2003. Analysis of the Corynebacterium diphtheriae DtxR regulon: identification of a putative siderophore synthesis and transport system that is similar to the Yersinia high-pathogenicity island-encoded yersiniabactin synthesis and uptake system. J Bacteriol 185:6826–6840. 10.1128/JB.185.23.6826-6840.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Allen CE, Schmitt MP. 2015. Utilization of host iron sources by Corynebacterium diphtheriae: multiple hemoglobin-binding proteins are essential for the use of iron from the hemoglobin-haptoglobin complex. J Bacteriol 197:553–562. 10.1128/JB.02413-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Allen CE, Schmitt MP. 2009. HtaA is an iron-regulated hemin binding protein involved in the utilization of heme iron in Corynebacterium diphtheriae. J Bacteriol 191:2638–2648. 10.1128/JB.01784-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crosa JH, Mey AR, Payne SM (ed). 2004. Iron transport in bacteria. ASM Press, Washington, DC. [Google Scholar]
- 9.Choby JE, Skaar EP. 2016. Heme synthesis and acquisition in bacterial pathogens. J Mol Biol 428:3408–3428. 10.1016/j.jmb.2016.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Otto BR, Verweij-van Vught AM, MacLaren DM. 1992. Transferrins and heme-compounds as iron sources for pathogenic bacteria. Crit Rev Microbiol 18:217–233. 10.3109/10408419209114559. [DOI] [PubMed] [Google Scholar]
- 11.Andersen CB, Torvund-Jensen M, Nielsen MJ, de Oliveira CL, Hersleth HP, Andersen NH, Pedersen JS, Andersen GR, Moestrup SK. 2012. Structure of the haptoglobin-haemoglobin complex. Nature 489:456–459. 10.1038/nature11369. [DOI] [PubMed] [Google Scholar]
- 12.Schaer DJ, Buehler PW, Alayash AI, Belcher JD, Vercellotti GM. 2013. Hemolysis and free hemoglobin revisited: exploring hemoglobin and hemin scavengers as a novel class of therapeutic proteins. Blood 121:1276–1284. 10.1182/blood-2012-11-451229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pishchany G, Sheldon JR, Dickson CF, Alam MT, Read TD, Gell DA, Heinrichs DE, Skaar EP. 2014. IsdB-dependent hemoglobin binding is required for acquisition of heme by Staphylococcus aureus. J Infect Dis 209:1764–1772. 10.1093/infdis/jit817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ellis-Guardiola K, Mahoney BJ, Clubb RT. 2020. NEAr transporter (NEAT) domains: unique surface displayed heme chaperones that enable Gram-positive bacteria to capture heme-iron from hemoglobin. Front Microbiol 11:607679. 10.3389/fmicb.2020.607679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bowden CFM, Chan ACK, Li EJW, Arrieta AL, Eltis LD, Murphy MEP. 2018. Structure-function analyses reveal key features in Staphylococcus aureus IsdB-associated unfolding of the heme-binding pocket of human hemoglobin. J Biol Chem 293:177–190. 10.1074/jbc.M117.806562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sjodt M, Macdonald R, Marshall JD, Clayton J, Olson JS, Phillips M, Gell DA, Wereszczynski J, Clubb RT. 2018. Energetics underlying hemin extraction from human hemoglobin by Staphylococcus aureus. J Biol Chem 293:6942–6957. 10.1074/jbc.RA117.000803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xiao Q, Jiang X, Moore KJ, Shao Y, Pi H, Dubail I, Charbit A, Newton SM, Klebba PE. 2011. Sortase independent and dependent systems for acquisition of haem and haemoglobin in Listeria monocytogenes. Mol Microbiol 80:1581–1597. 10.1111/j.1365-2958.2011.07667.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maresso AW, Garufi G, Schneewind O. 2008. Bacillus anthracis secretes proteins that mediate heme acquisition from hemoglobin. PLoS Pathog 4:e1000132. 10.1371/journal.ppat.1000132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Honsa ES, Maresso AW. 2011. Mechanisms of iron import in anthrax. Biometals 24:533–545. 10.1007/s10534-011-9413-x. [DOI] [PubMed] [Google Scholar]
- 20.Bates CS, Montanez GE, Woods CR, Vincent RM, Eichenbaum Z. 2003. Identification and characterization of a Streptococcus pyogenes operon involved in binding of hemoproteins and acquisition of iron. Infect Immun 71:1042–1055. 10.1128/IAI.71.3.1042-1055.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Allen CE, Schmitt MP. 2011. Novel hemin binding domains in the Corynebacterium diphtheriae HtaA protein interact with hemoglobin and are critical for heme iron utilization by HtaA. J Bacteriol 193:5374–5385. 10.1128/JB.05508-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Allen CE, Burgos JM, Schmitt MP. 2013. Analysis of novel iron-regulated, surface-anchored hemin-binding proteins in Corynebacterium diphtheriae. J Bacteriol 195:2852–2863. 10.1128/JB.00244-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lyman LR, Peng ED, Schmitt MP. 2021. The Corynebacterium diphtheriae HbpA hemoglobin-binding protein contains a domain that is critical for hemoprotein binding, cellular localization, and function. J Bacteriol 203:e0019621. 10.1128/JB.00196-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Popovic T, Kombarova SY, Reeves MW, Nakao H, Mazurova IK, Wharton M, Wachsmuth IK, Wenger JD. 1996. Molecular epidemiology of diphtheria in Russia, 1985–1994. J Infect Dis 174:1064–1072. 10.1093/infdis/174.5.1064. [DOI] [PubMed] [Google Scholar]
- 25.Cerdeno-Tarraga AM, Efstratiou A, Dover LG, Holden MT, Pallen M, Bentley SD, Besra GS, Churcher C, James KD, De Zoysa A, Chillingworth T, Cronin A, Dowd L, Feltwell T, Hamlin N, Holroyd S, Jagels K, Moule S, Quail MA, Rabbinowitsch E, Rutherford KM, Thomson NR, Unwin L, Whitehead S, Barrell BG, Parkhill J. 2003. The complete genome sequence and analysis of Corynebacterium diphtheriae NCTC13129. Nucleic Acids Res 31:6516–6523. 10.1093/nar/gkg874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lyman LR, Peng ED, Schmitt MP. 2018. Corynebacterium diphtheriae iron-regulated surface protein HbpA is involved in the utilization of the hemoglobin-haptoglobin complex as an iron source. J Bacteriol 200:e00676-17. 10.1128/JB.00676-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schaeffer J, Huhulescu S, Stoeger A, Allerberger F, Ruppitsch W. 2021. Assessing the genetic diversity of Austrian Corynebacterium diphtheriae clinical isolates, 2011 to 2019. J Clin Microbiol 59:e02529-20. 10.1128/JCM.02529-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Madeira F, Pearce M, Tivey ARN, Basutkar P, Lee J, Edbali O, Madhusoodanan N, Kolesnikov A, Lopez R. 2022. Search and sequence analysis tools services from EMBL-EBI in 2022. Nucleic Acids Res 10.1093/nar/gkac240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pilpa RM, Robson SA, Villareal VA, Wong ML, Phillips M, Clubb RT. 2009. Functionally distinct NEAT (NEAr Transporter) domains within the Staphylococcus aureus IsdH/HarA protein extract heme from methemoglobin. J Biol Chem 284:1166–1176. 10.1074/jbc.M806007200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Akinbosede D, Chizea R, Hare SA. 2022. Pirates of the haemoglobin. Microb Cell 9:84–102. 10.15698/mic2022.04.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trost E, Blom J, Soares SDC, Huang I-H, Al-Dilaimi A, Schröder J, Jaenicke S, Dorella FA, Rocha FS, Miyoshi A, Azevedo V, Schneider MP, Silva A, Camello TC, Sabbadini PS, Santos CS, Santos LS, Hirata R, Jr, Mattos-Guaraldi AL, Efstratiou A, Schmitt MP, Ton-That H, Tauch A. 2012. Pangenomic study of Corynebacterium diphtheriae that provides insights into the genomic diversity of pathogenic isolates from cases of classical diphtheria, endocarditis, and pneumonia. J Bacteriol 194:3199–3215. 10.1128/JB.00183-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Viguetti SZ, Pacheco LG, Santos LS, Soares SC, Bolt F, Baldwin A, Dowson CG, Rosso ML, Guiso N, Miyoshi A, Hirata R, Jr, Mattos-Guaraldi AL, Azevedo V. 2012. Multilocus sequence types of invasive Corynebacterium diphtheriae isolated in the Rio de Janeiro urban area, Brazil. Epidemiol Infect 140:617–620. 10.1017/S0950268811000963. [DOI] [PubMed] [Google Scholar]
- 33.Tai SP, Krafft AE, Nootheti P, Holmes RK. 1990. Coordinate regulation of siderophore and diphtheria toxin production by iron in Corynebacterium diphtheriae. Microb Pathog 9:267–273. 10.1016/0882-4010(90)90015-i. [DOI] [PubMed] [Google Scholar]
- 34.Serwold-Davis TM, Groman N, Rabin M. 1987. Transformation of Corynebacterium diphtheriae, Corynebacterium ulcerans, Corynebacterium glutamicum, and Escherichia coli with the C. diphtheriae plasmid pNG2. Proc Natl Acad Sci USA 84:4964–4968. 10.1073/pnas.84.14.4964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simon R, Priefer U, Puhler A. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Nat Biotechnol 1:784–791. 10.1038/nbt1183-784. [DOI] [Google Scholar]
- 36.Schafer A, Tauch A, Jager W, Kalinowski J, Thierbach G, Puhler A. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69–73. 10.1016/0378-1119(94)90324-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to S7. Download jb.00349-22-s0001.pdf, PDF file, 0.3 MB (325.7KB, pdf)