ABSTRACT
BxpB (also known as ExsFA) and ExsFB are an exosporium basal layer structural protein and a putative interspace protein of Bacillus anthracis that are known to be required for proper incorporation of the BclA collagen-like glycoprotein on the spore surface. Despite extensive similarity of the two proteins, their distribution in the spore is markedly different. We utilized a fluorescent fusion approach to examine features of the two genes that affect spore localization. The timing of expression of the bxpB and exsFB genes and their distinct N-terminal sequences were both found to be important for proper assembly into the exosporium basal layer. Results of this study provided evidence that the BclA nap glycoprotein is not covalently attached to BxpB protein despite the key role that the latter plays in BclA incorporation. Assembly of the BxpB- and ExsFB-containing outer basal layer appears not to be completely abolished in mutants lacking the ExsY and CotY basal layer structural proteins despite these spores lacking a visible exosporium. The BxpB and, to a lesser extent, the ExsFB proteins, were found to be capable of self-assembly in vitro into higher-molecular-weight forms that are stable to boiling in SDS under reducing conditions.
IMPORTANCE The genus Bacillus consists of spore-forming bacteria. Some species of this genus, especially those that are pathogens of animals or insects, contain an outermost spore layer called the exosporium. The zoonotic pathogen B. anthracis is an example of this group. The exosporium likely contributes to virulence and environmental persistence of these pathogens. This work provides important new insights into the exosporium assembly process and the interplay between BclA and BxpB in this process.
KEYWORDS: Bacillus anthracis, BclA, exosporium, immunofluorescence, promoters, protein assembly, spore
INTRODUCTION
The bacterial sporulation process is a mechanism utilized by many soil-dwelling bacteria to survive the harsh environmental conditions faced (1). When growth conditions are unfavorable, the bacteria alter their physiology, switching from vegetative growth to production of an endospore. This is a survival strategy rather than a replicative process, as a single spore is produced by a sporulating cell. The metabolically inert spore produced is refractive to a variety of environmental insults, including desiccation, heat, and UV irradiation. Spores can persist in soil environments for long periods of time. Bacillus anthracis is a zoonotic animal pathogen that is of concern as a potential biothreat agent (2). For B. anthracis, the spore is the infectious form. Their spores contain an outer spore layer called an exosporium, which is separated from the spore coat by the interspace (3). The exosporium surface provides the sites of initial interactions with the infected host. The exosporium is thought to be a semipermeable barrier that excludes potentially harmful large molecules such as antibodies and hydrolytic enzymes (4, 5). The exosporium also confers hydrophobic properties on the spore, likely playing a role in the persistence of spores in soil environments (6). However, the exosporium’s most important role may be in the initial interactions between spores and macrophages and dendritic cells of the host innate immune system (7–9).
The B. anthracis exosporium layer consists of a basal layer surrounded by an external nap of hairlike projections (10–12). The collagen-like glycoprotein BclA is the principal component of this hairlike nap (13–15). The exosporium basal layer is composed of a number of different proteins (14, 16, 17). Known exosporium-associated proteins include the collagen-like proteins BclA (13–15), BclB (18–21), and BetA (22), as well as BxpB (also called ExsFA) and ExsFB, which are needed for assembly of these collagen-like proteins (23, 24). CotY is important in the formation of one pole of the exosporium, referred to as the “bottle cap,” which appears to be important for emergence of the bacterial cell when the spore undergoes germination (25). The cap region of the exosporium represents approximately 25% of the exosporium and corresponds to the mother cell-central pole of the developing spore. The protein composition of the cap region of the exosporium differs from that of the noncap region. Cap-enriched exosporium proteins include CotY and ExsFB, whereas non-cap-enriched proteins include BclB, BxpB, and alanine racemase (Alr) (21, 25, 26). Furthermore, the CotE and CotO spore coat proteins and ExsB are required for proper attachment of the exosporium layer to the developing spore (27–29). Enzymes are also associated with the exosporium, including alanine racemase, arginase, inosine-preferring nucleoside hydrolases, and superoxide dismutase (17, 30–33).
Incorporation of BclA onto the spore surface is dependent on the exosporium basal layer protein BxpB (ExsFA), as there is a dramatic reduction of BclA assembly in spores of bxpB mutants (23, 24). Although an exsFB-null single mutant displays only a modest reduction of BclA incorporation onto spores, total loss of BclA requires inactivation of both the bxpB and exsFB determinants (24). BxpB and BclA are expressed at the same time in sporulating cells and appear in the mother cell cytoplasm in similarly sized complexes (26, 34). It has been hypothesized that BclA and BxpB become associated through the formation of covalent bonds, with the release of the N-terminal (NT) 19 amino acids from BclA. However, there has been only one report indicating covalent attachment of BclA to BxpB (35), but this paper was later retracted (36). The N-terminal 35 amino acids of BclA (the N-terminal domain [NTD]), which include the localization and attachment domains of the protein, are sufficient to position the protein at the site of exosporium assembly and subsequent stable attachment to the exosporium surface (20, 34). Furthermore, we demonstrated that although there may be an interaction between BclA and BxpB in the mother cell cytoplasm prior to actual assembly into the exosporium, the proteolytic cleavage of BclA after the 19th residue does not occur until late in the assembly process, when the BclA protein has been positioned around the majority of the spore (20). When BclA is in place around 75% of the spore, a wave of proteolytic cleavage of the BclA begins at the site of initiation of exosporium synthesis, namely, the mother cell-central pole of the spore. Positioning of the protein and cleavage do not require BclA sequences beyond the N-terminal domain, indicating that trimerization of this glycoprotein is not required for the assembly process.
The Bacillus exosporium basal layer’s principle structural proteins are CotY, ExsY, BxpB, ExsFB, and BclA (3, 16, 37–39). Mutants of B. anthracis deleted for the cotY and exsY genes produce spores lacking the exosporium layer (40). Mutants deleted for bxpB and exsFB produce spores that contain an exosporium layer but lack the BclA nap. This suggests that the ExsY and CotY proteins are assembled first and constitute an inner basal sublayer, and BxpB and ExsFB are assembled later and comprise the outer basal layer. Evidence for this layered arrangement has been provided by electron microscopic studies. The basal layer of the B. anthracis exosporium is approximately 12 to 16 nm thick and appears to be comprised of two, approximately 5-nm-thick sublayers (41). Although the assembly process for BclA during sporulation has been studied in some detail, the pathways for incorporation of BxpB and ExsFB are less well understood. In this study, we examined factors that contribute to the assembly of the outer basal layer.
RESULTS
Trafficking of BxpB and ExsFB to assembly sites of the exosporium basal layer.
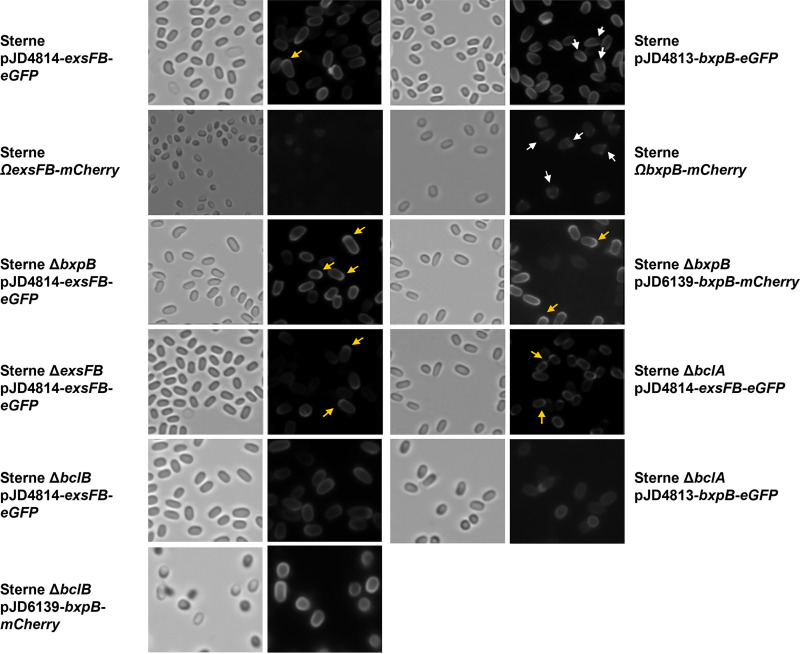
It has been shown that BxpB is found predominantly in the noncap region of the exosporium, whereas ExsFB was distributed around the spore circumference but enriched at the cap (17, 26). We examined the distribution of BxpB and ExsFB in spores using mCherry and enhanced green fluorescent protein (eGFP) fusions. The BxpB fusions exhibited fluorescence around the Sterne spores but with lesser intensity at one pole, which was previously shown to be the mother cell central spore bottlecap pole (Fig. 1) (25). When the fusion protein was expressed in a bxpB deletion mutant, fluorescence was evident over the entire spore, but there was enhanced fluorescence evident at one spore pole, consistent with the results reported by Giorno et al. (17). This indicates that the distribution of the fusion protein was altered in the absence of the wild-type BxpB protein. The pattern of BxpB spore incorporation was not altered in mutants lacking BclA or BclB, although the amount incorporated in the former was reduced. BclB assembly into spores was shown to be dependent on BxpB, but the reverse is not true (21). To reduce the potential effects of gene dosage, the bxpB-mcherry determinant was integrated into the B. anthracis genome in a single copy at the amyS (bas3291) locus. The spore-associated fluorescence was weaker with these spores, but the paucity of fluorescence at the cap pole of the spore is evident. Because of the weaker signals associated with the single-copy fusion gene integrations, the more intense mCherry fusion was utilized with ExsFB rather than the eGFP reporter used with the plasmid-containing host cells. The fluorescence intensity of the spores was quantified and is shown in Fig. S1 in the supplemental material.
FIG 1.
Bright-field and epifluorescence micrographs of spores from Sterne, ΔbxpB, ΔexsFB, and ΔbclA cells producing the BxpB-mCherry, BxpB-eGFP, or the ExsFB-eGFP fusion proteins expressed from plasmids or in a single copy at the amyS locus (denoted by Ω). Yellow arrows indicate examples of enhanced fluorescence at a spore pole, and white arrows indicate examples of reduced fluorescence at a spore pole.
ExsFB-eGFP in Sterne spores exhibited fluorescence around the periphery of the spore that was enhanced at one pole (Fig. 1). Overall fluorescence was enhanced in the absence of BxpB, but the spore distribution pattern of fluorescence was not substantially affected by the loss of the exsFB or bxpB alleles or in bclA- or bclB-null mutants. Single-copy expression of exsFB-mcherry at the amyS locus produced faint fluorescence in spores, but the overall pattern of the fluorescence was similar to that with the plasmid-borne fusions. Because of a weaker signal, the single-copy exsFB-egfp fusion gives a faint but consistent signal which does not show as well as the mCherry fusion. As a control for background fluorescence, spores were prepared from Sterne expressing unfused mcherry under the control of the promoter of the bclA spore gene. Despite an abundance of mCherry in the cytoplasm of sporulating cells bearing pGS4260, the resulting spores did not exhibit fluorescence, indicative of the mCherry protein not detectably adsorbing onto spore surfaces following mother cell lysis (data not shown). We noted in these studies that single-copy versus plasmid-borne expression of the fusion proteins was consistent in pattern but that plasmid-borne expression gives rise to a subpopulation of nonfluorescent spores, which we have seen with studies involving the exosporium basal layer protein-encoding genes cotY and exsY (42). Nonetheless, the patterns of spores actually exhibiting fluorescence are reproducible.
Complementation experiments in bxpB- and/or exsFB-null mutant strains of B. anthracis.
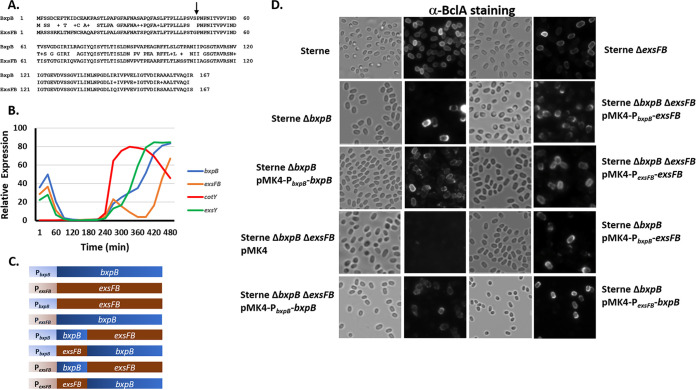
The BxpB and ExsFB proteins are highly similar in amino acid sequence (78% identical, 86% conserved) (Fig. 2A), but their distribution patterns in spores differ. The phenotype known to be associated with the bxpB and exsFB mutants is BclA incorporation. The wild-type BclA phenotype, distribution of BclA around the entire surface of the spore, was not appreciably affected by loss of the ExsFB protein (Fig. 2D). However, in the absence of BxpB, anti-BclA staining became more pronounced at one pole of the spore. Addition of the plasmid pJD4730, bearing the bxpB gene, restored a more uniform distribution of BclA, indicating that the plasmid-borne gene complemented the bxpB deletion (17) (Fig. 2D). The spores displayed less of a bias toward incorporation of BclA at one pole. However, the appearance of the antibody-labeled BclA around the spore circumference was more mottled than that observed with the Sterne parent strain. Spores lacking both BxpB and ExsFB had no detectable BclA on their surfaces. Introduction of the plasmid-borne determinants restored BclA to the spore surface, indicating that the recombinant proteins were produced in these strains and functioned in facilitating BclA assembly. Expression of bxpB (pJD4730) in the ΔbxpB ΔexsFB double mutant resulted in spores that expressed BclA in a mottled appearance around the spore, whereas expression of exsFB (pJD4732) resulted in spores with a more uniform surface distribution of BclA on the spore surface. Western blotting of spore extracts from the pMK4-bxpB and pMK4-exsFB hosts indicated that there was only a modest increase in the amount of these spore proteins in the plasmid-bearing Sterne cells (Fig. S2).
FIG 2.
Effect of expression of BxpB and ExsFB on BclA spore surface distribution. (A) Amino acid sequence alignment of BxpB and ExsFB with sequence identities and similarities (denoted by plus sign) indicated. The arrow denotes the position where the chimeric proteins were fused. (B) Microarray-based RNA expression profiles of the exosporium determinants bxpB, cotY, exsY, and exsFB during the in vitro growth and sporulation cycle. Microarray data were from Bergman et al. (43). (C) Representations of the full-length and chimeric bxpB and exsFB determinants expressed from either the bxpB or exsFB promoters. (D) Bright-field and anti-BclA fluorescence of spores of the Sterne parent strain and the ΔbxpB-, ΔexsFB-, and ΔbxpB ΔexsFB-null mutants bearing the plasmid-borne bxpB or exsFB determinants expressed from their native or swapped promoter sequences where indicated. Spores were reacted with anti-BclA polyclonal serum and goat anti-rabbit-Alexa Fluor 568 secondary antibodies.
When the exsFB determinant was expressed from the bxpB promoter in the double null mutant cells, however, BclA localized more strongly to one pole of the spore. This suggests that the timing of ExsFB expression, in the absence of BxpB, strongly influences the spore surface distribution of the BclA glycoprotein. Expression of ExsFB from its native promoter in the double mutant gave rise to spores with a BclA distribution comparable to that of the wild-type Sterne strain rather than the expected pattern of expression of BclA over the entire surface of the spore but predominantly at one pole (a BxpB-negative [BxpB−] and ExsFB-positive [ExsFB+] phenotype).
Effects of timing of expression of the bxpB and exsFB determinants on spore localization of BclA.
In a microarray analysis of gene expression throughout the B. anthracis in vitro life cycle, Bergman et al. (43) discovered that the expression profile of the bxpB and exsFB determinants differed, with bxpB and exsFB mRNA appearing at a time coincident with expression of spore coat and exosporium determinants. However, the bxpB message continued to increase, whereas the exsFB mRNA declined after this initial expression and reemerged at a time point 2.5 h after the initial expression (Fig. 2B). To evaluate if the timing of expression plays a role in the distribution of the BxpB and ExsFB proteins in the outer spore layers, we constructed genes that were driven by the paralog’s promoter and ribosome binding site sequences (Fig. 2C). BxpB and ExsFB are not functionally equivalent as evidenced by the expression of exsFB off the bxpB promoter (pJD4693), failing to complement the bxpB-null mutation (Fig. 2D). Expression of bxpB from the exsFB promoter also failed to complement the ΔbxpB mutation, suggesting that timing of expression is important for the proper assembly of the outer basal layer. The BclA phenotypes were also examined in ΔbxpB ΔexsFB cells bearing the pMK4 shuttle plasmid carrying the bxpB or exsFB determinants driven by the promoters for each of these two genes (Fig. 2D). The expression of these constructs restored BclA to the spore surface, again indicating that the BxpB or ExsFB proteins were expressed. However, the localization patterns were unexpected. Expression of bxpB from the bxpB promoter (pJD4730) resulted in a BclA surface pattern resembling that of a bxpB mutant host (enhanced pole localization) rather than the expected BxpB+ ExsFB− phenotype. Expression of bxpB off the exsFB promoter (pJD4695) or exsFB off the bxpB promoter (pJD4693) gave the same bxpB-negative BclA pole-dominant distribution pattern. Expression of exsFB from its native promoter in the double-mutant host gave a wild-type BclA distribution pattern (presence of BclA uniformly around the spore). This suggests that the individual genes can restore BclA to the spore surface in the ΔbxpB ΔexsFB spores but cannot complement the double mutant to give the correct single-mutant phenotypes.
N-terminal sequences of BxpB and ExsFB are important in the context of promoter-specific expression.
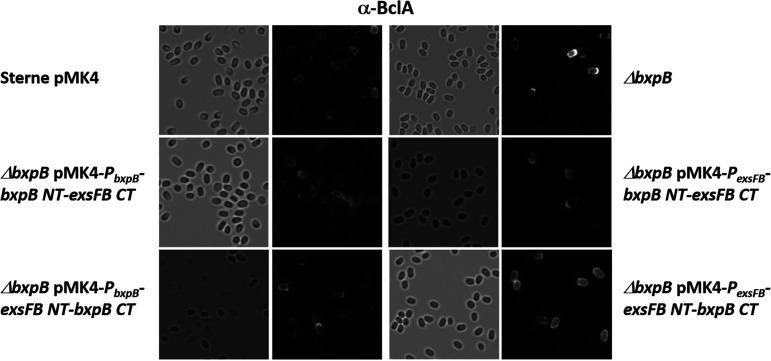
The BxpB and ExsFB proteins are similar in amino acid sequence, but there is more sequence divergence at their N terminus than with the rest of the protein (Fig. 2A). We therefore created chimeric proteins with the N-terminal sequences of one substituting for the paralog’s sequence (junction point at the arrow in Fig. 2A and depicted in Fig. 2C). These chimeric constructs were expressed in B. anthracis hosts, and the spore BclA distribution pattern was examined. The results are shown in Fig. 3. To complement the bxpB-null phenotype, results depended on the nature of chimera and the promoter element used. With the bxpB promoter, complementation (more uniform BclA distribution over the entire spore surface) was better achieved with the BxpB NT-ExsFB C-terminal (CT) construct (pJD4696), and there was no complementation with the ExsFB NT-BxpB CT chimera (pJD4698). When the chimeras were expressed using the exsFB promoter, the opposite chimera, ExsFB NT-BxpB CT (pJD4697), was more effective at complementing the bxpB-null phenotype, while the BxpB NT-ExsFB CT construct (pJD4699) failed to complement. The results indicate that the N-terminal sequences of the two proteins are important for proper positioning of the two proteins in the context of an absence of wild-type BxpB, and the N-terminal sequences were more effective with their homologous promoter element.
FIG 3.
Bright-field and anti-BclA fluorescence of spores of the Sterne parent strain and the ΔbxpB mutant bearing the plasmid-borne bxpB or exsFB chimeric determinants expressed from the bxpB or exsFB promoter elements. Spores were reacted with anti-BclA polyclonal serum and goat anti-rabbit-Alexa Fluor 568 secondary antibodies.
Is BclA anchored to BxpB?
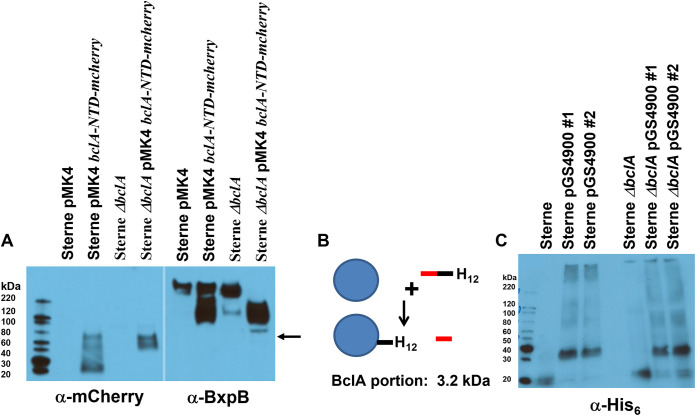
The BclA-N-terminal 35 residues (the NTD) have been shown to be the site of a proteolytic cleavage event (between residues 19 and 20) that is associated with stable attachment of BclA to the basal layer (13, 14, 20). The NTD is sufficient for localization and attachment of the protein to the spore surface. We fused the BclA NTD to the mCherry protein (32.7 kDa uncleaved and 30.6 kDa if the BclA sequence is cleaved) and expressed it on plasmid pGS4260 in sporulating cells of B. anthracis Sterne. The fusion protein was incorporated onto the spore as expected (20). The anti-mCherry-reactive species extracted from the Sterne spores existed in the expected fusion protein monomer size plus two additional major species with approximate molecular weights (MWs) of approximately 70,000 and 55,000 (Fig. 4A, lane 2, left). None of these species reacted with the anti-BxpB antiserum (lane 2, right), suggesting that BxpB was not present (i.e., covalently attached) in this mCherry-associated species. In BclA-lacking mutant spores, the BclA-NTD-mCherry fusion appeared in three species of ~90,000 (faint band), 70,000, and 55,000 molecular weights. The mass of mCherry is 28.8 kDa. With no competing wild-type BclA, all of the fusion proteins appeared in higher-molecular-weight complexes (no monomer species detected). Again, no correspondingly sized complexes were identified with the polyclonal anti-BxpB antibodies. Although not shown in Fig. 4A, expression of mCherry (lacking the BclA residues) in sporulating cells (driven by the bclA promoter) gives rise to only the expected 29-kDa species in sporulating cells, and no detectable mCherry is found in mature spores (by either Western blotting of spore extracts or by flow cytometry). These results suggest that BclA may be stably attached to an exosporium basal layer protein, but that protein is not BxpB.
FIG 4.
(A) Western blotting of spore extracts probed with antiserum against mCherry (left) or BxpB (right). The blot was probed with anti-mCherry and then stripped and reprobed with anti-BxpB. The arrow indicates the position of the larger anti-mCherry-reactive species in the Sterne ΔbclA pMK4 bclA-NTD-mcherry lane. (B) Strategy for bclA-NTD-his12 labeling of the BclA partner protein. The BclA NTD sequence (red bar represents residues 1 to 19, and black bar represents residues 20 to 35) with His12 residues at the C terminus was expressed in B. anthracis. During assembly around the spore (20), a proteolytic cleavage event removes the N-terminal 19 residues, with the remaining NTD sequences being stably attached to a partner protein in the exosporium (represented by the blue circle). (C) Western blotting of spore extracts probed with antiserum against the His tag. The pGS4900 clones 1 and 2 refer to two independent isolates bearing the plasmid. The size of the BclA-NTD-His12 protein is 5,287 daltons (Da) (3,290 Da when cleaved). A 35-kDa species was detected with the anti-His tag antiserum, and higher-MW SDS-resistant complexes are evident as well.
However, this result does not imply that there is no interaction between BclA and BxpB. Noncovalent but stable complexes form in the mother cell cytoplasm prior to attachment of the complexes to the developing exosporium (26). The expression of the BclA-NTD-mCherry fusion results in different sizes of BxpB-containing complexes extracted from the spores, or from sporulating cells, than cells expressing full-length BclA (compare Fig. 4A [anti-BxpB panel], lane 1 versus lane 2 and lane 3 versus lane 4). This supports there being common complexes containing both BxpB and BclA in the mother cell cytoplasm that subsequently get incorporated into the exosporium. Whether this interaction between the BclA-NTD and BxpB is direct or indirect and involves another exosporium protein is not known.
The smallest protein complex visible in the Western blotting shown in Fig. 4A was a 55,000-molecular-weight species, suggesting a molecular weight of approximately 26,000 for the BclA-NTD partner protein. To further support the existence of such a BclA-interacting protein, the BclA-NTD-encoding sequence was modified by the addition of a His12 tag coding sequence, and this construct, including the bclA promoter (pGS4900), was introduced into the Sterne and Sterne ΔbclA strains and spores produced. During sporulation, the BclA-NTD domain would interact with the partner protein and would be cleaved, and the C-terminal sequence (including the His12 tag) would be covalently attached to the partnering protein (Fig. 4B). Western blots of spore extracts were probed with antibodies against the His tag (Fig. 4C). In both the wild-type Sterne and the bclA-null strains, the antibodies recognized a protein migrating as a thick band between the 30- and 40-kDa standards, giving an approximately 31,000-molecular-weight value for the targeted protein (with the attached sequence that adds 3,290 in molecular weight). In the absence of the BclA-NTD-His12-encoding plasmid, only a small species, ~20 kDa, was evident as an artifact (present in Sterne spore lysates lacking the pGS4900 plasmid [lanes 1 and 4]). With both the Sterne and the Sterne ΔbclA hosts, the His-tagged protein was present as SDS-resistant high-molecular-weight complexes, a property that has been observed with certain other exosporium proteins. The mCherry and His tag fusion approaches identified a 26- to 32-kDa protein as the putative BclA-NTD attachment protein.
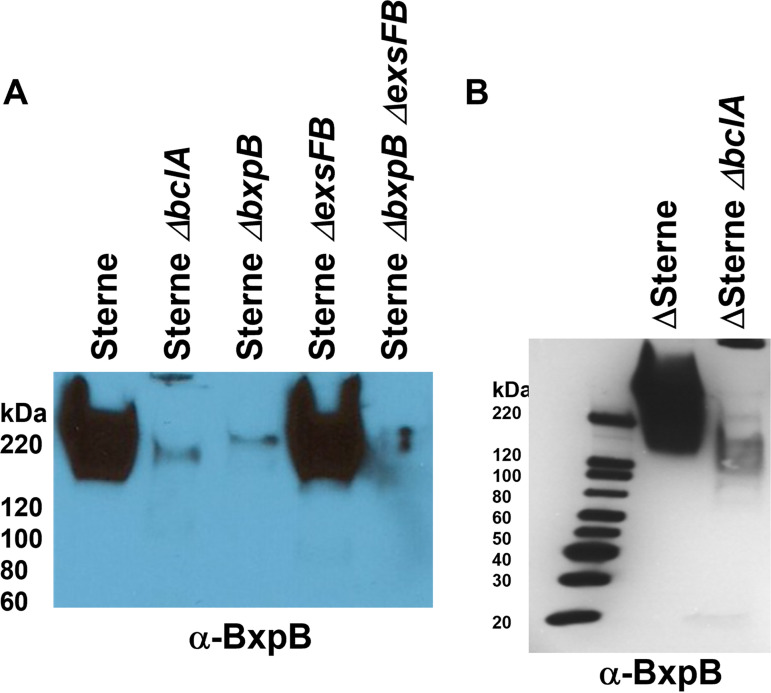
BxpB incorporation into the exosporium basal layer is largely dependent on BclA.
BxpB and BclA appear in sporulating cells with similar kinetics and are found in similarly sized high-molecular-weight complexes (26). BclA incorporation is dependent on the localization and attachment domain located at the N terminus of this protein (20, 26, 34). BxpB lacks such an exosporium-targeting sequence, which begs the question of how BxpB is positioned and assembled in the developing exosporium. Western blots of spore extracts were probed with anti-BxpB antiserum, and the results are shown in Fig. 5. The bclA deletion mutant showed a marked reduction in extractable BxpB content with both the Sterne and ΔSterne strains. These results, together with the observation that complexes between BclA and BxpB are found in the mother cell cytoplasm prior to assembly of BclA onto the spore surface (26), suggest that BclA, with its localization domain, is responsible for directing the exosporium complexes, including BxpB, from the mother cell cytoplasm to the site of assembly of the nascent exosporium. Without the BclA spore-targeting domain, the incorporation of BxpB is very inefficient.
FIG 5.
Impact of BclA on BxpB incorporation into the exosporium basal layer. (A) Western blot of Sterne spore extracts probed with antiserum against BxpB. (B) Western blot of Delta Sterne spore extracts probed with antiserum against BxpB. The faint band in the Delta bxpB lanes is due to cross reactivity with ExsFB.
Are CotY and ExsY required for the assembly of the outermost exosporium basal layer?
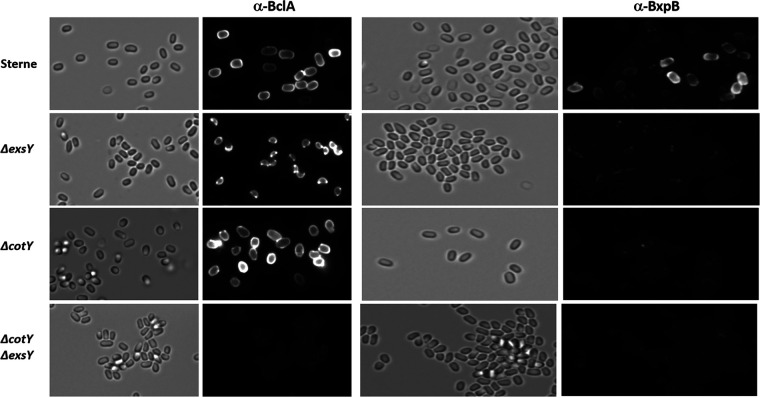
Mutation studies suggest that the B. anthracis exosporium basal layer is assembled with the inner layer (principally ExsY and CotY) incorporated and with the BxpB- and ExsFB-containing outer layer (including BclA) assembled with a slight delay onto the inner basal layer. To determine if loss of CotY and/or ExsY affects deposition of outer basal layer proteins, spores from mutants lacking the CotY and/or ExsY proteins were examined for BclA and BxpB content (Fig. 6). Sterne wild-type spores react to both the anti-BclA and anti-BxpB antisera. The anti-BxpB antiserum, which is a highly reactive antiserum, exhibited weaker binding than the relatively weaker anti-BclA serum, consistent with the BxpB epitopes lying beneath the exosporium nap layer. The BxpB epitopes are less available for binding, likely due to the shielding by the nap layer containing BclA and other collagen-like proteins such as BclB and BetA (19, 21, 22). Spores from the exsY-null strain spores possess only the cap portion of the exosporium, and both the anti-BclA and the anti-BxpB antibodies recognize this structure. The anti-BxpB antiserum exhibited greater binding along the margin of the cap than that of the pole, which may result from epitope availability along the exposed margin of the cap structure. The exosporium-containing cotY-null spores reacted with the anti-BclA antibodies but exhibited only sparse and patchy binding to the surface of these spores with the anti-BxpB serum. The failure of the anti-BxpB antibodies to bind to the CotY-deficient spores was due to the absence of surface exposure of the BxpB protein because BxpB was readily extracted from the ΔcotY spores, as evident in the Western blot analysis shown in Fig. 7.
FIG 6.
Immunofluorescence of spores from Sterne, Sterne ΔexsY, Sterne ΔcotY, and Sterne ΔcotY ΔexsY treated with antiserum against BclA or BxpB. Left-hand images are bright-field images, and the right-hand images of each pair are fluorescent images of the same field.
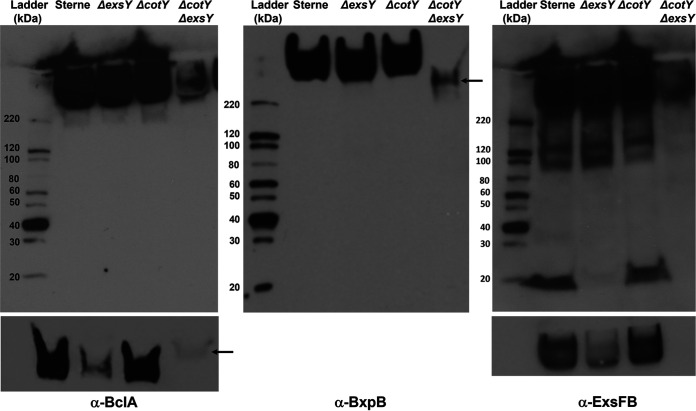
FIG 7.
Western blotting of spore extracts probed with rabbit polyclonal antiserum against rBclA (left), rBxpB (middle), and rExsFB (right). Strain designations of the spore extracts are indicated above the image of the blot. The bottom panels are from the high-molecular-weight region of the blots from a shorter exposure time of a comparable gel. The arrows in the bottom anti-BclA and the anti-BxpB panels indicate the position of the reactive species in the ΔcotY ΔexsY spore extract lane, which migrate differently from the high-MW complexes seen in the extracts from the single-mutant spores.
With the ΔcotY ΔexsY spores, which lack an exosporium, only faint and spotty surface labeling of the spores occurred with either the anti-BclA or the anti-BxpB antibodies. These results indicate that in the double mutant, BclA and BxpB are produced, and the proteins can, perhaps nonspecifically, adhere to the exosporium-less spores.
To validate the results of the spore immunofluorescence experiments, the spores from the wild-type and mutant strains were extracted by boiling in the presence of SDS and urea, the extracts were resolved by SDS-PAGE, and the blots were probed with antisera against BclA, BxpB, and ExsFB (Fig. 7). Each of the three exosporium proteins was present in the wild-type, ExsY-negative, CotY-negative, and ExsY-/CotY-negative strains, with the latter spores possessing markedly reduced levels of these proteins.
The wild-type and exsY mutant spore extracts showed abundant levels of the extracted proteins despite the ExsY-negative spores possessing only the cap portion of the exosporium. Because the cap-only exosporium structure is less stably attached to the spore, it is likely to be more efficiently extracted from the spores, giving the impression of comparable levels. The bottom panels of Fig. 7 are shorter exposures of the high-molecular-weight region of an identically loaded gel and better illustrate the differences in levels of BclA and ExsFB in the spore extracts. The results strongly indicate that production and spore incorporation of the exosporium proteins (BclA, BxpB, and ExsFB) can occur in the absence of CotY or ExsY. When both are absent, however, the exosporium does not form, but these proteins remain in spores, albeit at greatly reduced levels. The high-molecular-weight complexes recognized by the anti-rBclA or anti-ExsFB antibodies migrate differently in extracts from the cotY exsY double mutant than from extracts from the single mutants (Fig. 7, arrows) suggesting that the composition of the SDS-resistant complexes from the double mutant differ in composition. The amount of BclA, BxpB, and ExsFB extracted from the cotY mutant spores was comparable to that extracted from the Sterne parent strain spores. This result suggests that the reduced immunofluorescence with the anti-BxpB antiserum with the cotY mutant spores (Fig. 6) was likely due to reduced surface exposure of the protein rather than a reduced amount of the BxpB protein.
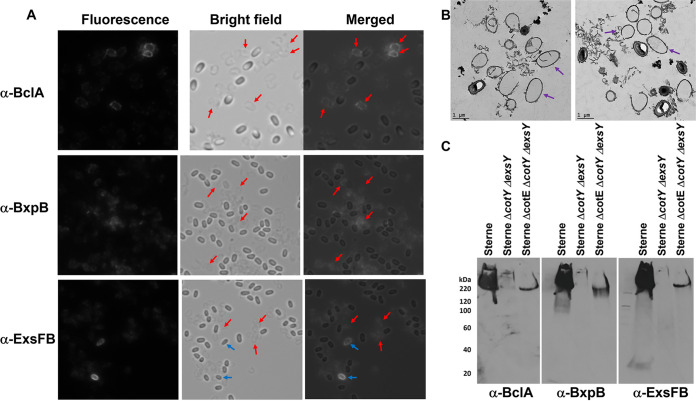
When exosporium-less spores from a triple mutant lacking CotE, CotY, and ExsY were tested for reactivity to anti-BclA, anti-BxpB, and anti-ExsFB antisera, stronger reactivities were seen in the fields that did not correspond to positions of spores (Fig. 8A). However, spore-shaped fluorescent objects were visible. The spore preparation contained what appeared to be “ghost” structures that reacted with the BclA antibodies (positions denoted by the red arrows in Fig. 8A). Transmission electron microscopy examination of the spore preparations from the triple mutant revealed the presence of ring structures which likely represent sections through these non-spore-associated outer exosporium basal layer sacculi (Fig. 8B).
FIG 8.
Assembly of the outer basal layer in the absence of the ExsY-CotY layer. (A) Spores from a triple null mutant of B. anthracis that lack CotE, CotY, and ExsY were reacted with anti-BclA, anti-BxpB, and anti-ExsFB antiserum. Fluorescent, bright-field, and merged images are shown. Red arrows in the bright-field image indicate positions of the putative basal layer casings that reacted with the antiserum. Blue arrows denote spores that reacted with the antiserum. (B) Transmission electron micrographs of the spore preparations from the cotE cotY exsY triple deletion mutant illustrating the presence of the empty vesicles (examples indicated with purple arrows). (C) Western blotting of spore extracts probed sequentially with antiserum directed against rBclA, rBxpB, and rExsFB.
Spores from the wild-type Sterne strain and the ΔexsY ΔcotY and ΔexsY ΔcotY ΔcotE deletion mutant strains were extracted by boiling in SDS and urea and the extracts analyzed by PAGE and Western blotting with rabbit polyclonal anti-BclA, anti-BxpB, and anti-ExsFB antisera. The results are shown in Fig. 8C. As expected, the ΔexsY ΔcotY mutant spores that lack the exosporium layer had reduced content of these exosporium proteins. However, the triple deletion mutant, lacking the CotE exosporium-anchoring protein, reproducibly released more high-molecular-weight complexes containing BxpB and ExsFB than did the double mutant.
The results suggest that the outer exosporium basal layer proteins BxpB, ExsFB, and, to a lesser extent, BclA, are anchored to the spore surface despite the lack of a TEM-demonstrable exosporium layer. Furthermore, its stable spore association is dependent on the CotE-associated exosporium-anchoring system. In the absence of this anchoring protein, the structure is released from the spores and is found in the spore preparations. Because we did not observe these ghost objects in the spore preparations from the cotY exsY double null mutant, it is likely that the CotE-associated exosporium-anchoring system retains some functionality in the absence of the CotY and ExsY proteins. The presence of these released shell-like structures in the spore preparations may account for the improved solubilization of BxpB- and ExsFB-associated high-MW complexes in the triple mutant extracts.
Protein-protein interactions involving BxpB and ExsFB.
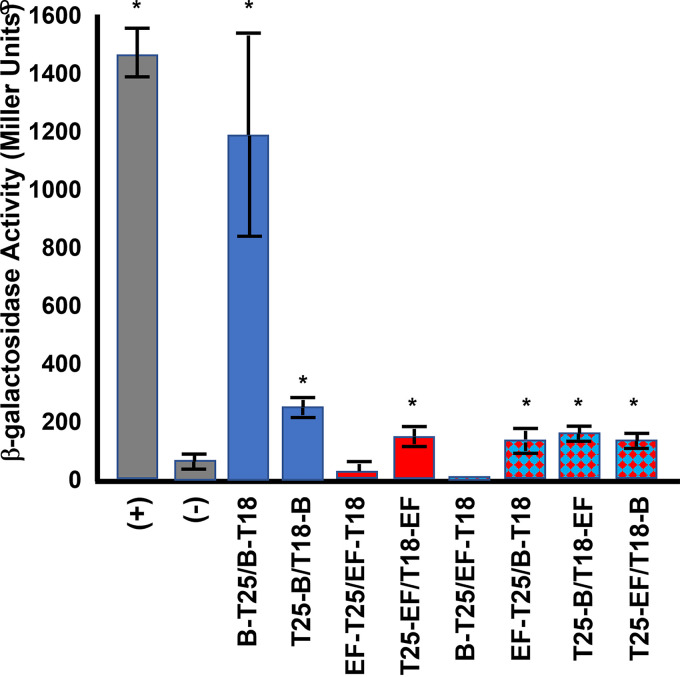
BxpB and ExsFB are structural proteins in the exosporium basal layer. The CotY and ExsY proteins, cysteine-rich inner basal layer structural proteins, have been demonstrated to be capable of self-assembly when expressed in Escherichia coli (37). As part of the assembly process of the exosporium protein shell, there may be self-interactions or heterologous interactions with other basal layer proteins. To test whether BxpB and ExsFB are capable of these interactions, a bacterial two-hybrid approach was taken (44). We examined potential self-interactions with BxpB and ExsFB and also possible interactions between BxpB and ExsFB. We tested both N-terminal and C-terminal fusions to adenylate cyclase. The results of this study are presented in Fig. 9. BxpB was shown to be capable of self-interactions in this assay, with the adenylate cyclase domain fusions at the C terminus of the protein being substantially more active than the N-terminal fusions. ExsFB self-interactions were much weaker than those observed with BxpB. With ExsFB, no detectable β-galactosidase activity above the background was obtained with the fusions of the T18 and T25 domains at the C terminus, but positive results were obtained with the fusions at the N terminus of ExsFB. Except for the BxpB-T25/ExsFB-T18 pairing, positive interactions were observed with the BxpB-ExsFB combinations, including with both C-terminal or N-terminal fusions to ExsFB. The resulting enzymatic activities were, however, substantially lower than those observed with the BxpB-BxpB C-terminal fusion interactions.
FIG 9.
Bacterial two-hybrid analysis of BxpB and ExsFB interactions. β-Galactosidase assays were conducted in E. coli bearing compatible plasmids harboring the T18 and T25 adenylate cyclase domains (44). The fusions were to BxpB (designated B) and ExsFB (designated EF). B-T25 signifies a BxpB fusion with the T25 domain fused at its C terminus, whereas T18-EF indicates ExsFB with the T18 domain fused at its N terminus. The positive control (+) was a strain containing plasmids with the zip sequence fused to each of the adenylate cyclase domains (44), and the negative control (−) was a strain bearing two fusions with B. anthracis proteins that we have shown not to be interactive in this assay (the exosporium protein ExsY and a putative nucleotide sugar dehydrogenase subfamily protein encoded on the pXO1 plasmid; locus tag AW20_5669). β-Galactosidase activity is expressed in Miller units. Controls are shown as gray bars, BxpB-BxpB interactions as blue bars, ExsFB-ExsFB as red bars, and BxpB-ExsFB as blue-and-red checkerboard bars. The error bars signify the standard deviation of ≥3 independent assays. An asterisk denotes the statistical significance (P < 0.05) for the sample relative to the negative control.
Self-assembly of BxpB and ExsFB into SDS-resistant higher-molecular-weight forms.
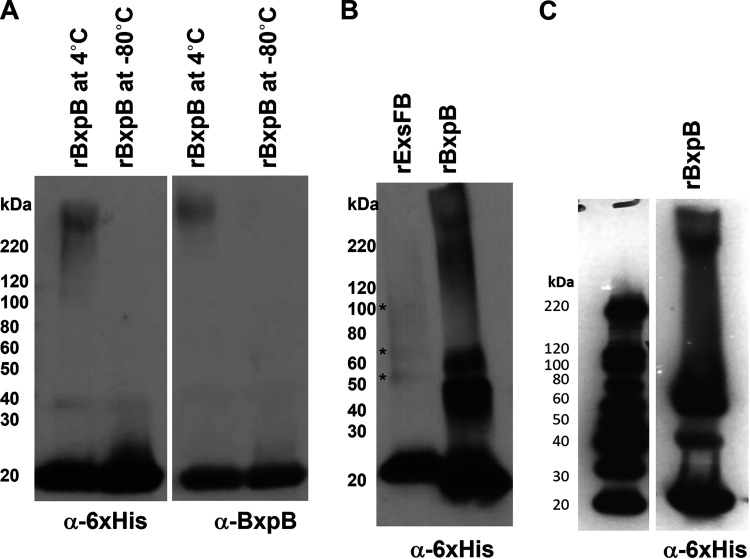
Bacillus spore coat and crust proteins, as well as the exosporium ExsY and CotY proteins, have been shown to be able to organize into higher-MW complexes through formation of disulfide bonds (37, 45, 46). To determine if the outer basal layer proteins were similarly capable of this type of assembly, we expressed recombinant BxpB and ExsFB proteins bearing an N-terminal hexameric histidine tag in E. coli, purified the proteins, and kept them in phosphate-buffered saline (PBS) at 4°C. Both proteins were found to form higher-molecular-weight complexes over time, with BxpB being substantially more proficient at it than ExsFB (Fig. 10). These higher-molecular-weight forms were resistant to denaturation by boiling in the presence of SDS and a reducing agent (Fig. 10) (47).
FIG 10.
Western blotting of recombinant BxpB and ExsFB isolated from E. coli and stored in PBS. (A) rBxpB incubated at 4°C (lane 1) and −80°C (lane 2) overnight was subjected to SDS-PAGE, transferred to immobilon filters, and probed sequentially with anti-6×His tag and anti-rBxpB antisera. (B) His-tagged BxpB and ExsFB were purified from E. coli and incubated in PBS at 4°C for 2 weeks. The samples were subjected to SDS-PAGE and blotted with anti-His tag antiserum. Positions of faint bands are denoted by an asterisk. (C) BxpB sample prepared by a different author in a different year, indicative of the reproducibility of this process.
DISCUSSION
BxpB and ExsFB are outer spore proteins of B. anthracis with distinct sites of localization. The evidence for BxpB being an outer exosporium basal layer protein includes its impact on incorporation of the BclA nap glycoprotein and its apparent partnering with BclA in the mother cell cytoplasm during the exosporium assembly process. The spore location for ExsFB is somewhat less obvious, with one report suggesting it lies in the interspace region between the exosporium basal layer and the outer spore coat layer (17) and another suggesting it is enriched in the cap portion of the basal layer (26). The two spore proteins are similar in sequence, raising the question as to what determines their site of incorporation in the developing spore. The results of this study suggest that timing of expression, in conjunction with the less similar N-terminal sequences, is important for proper positioning of the two proteins. This suggests that there may be proteins that specifically recognize the N terminus of BxpB and ExsFB that guide the proteins to their assembly sites. The dependence on BclA for BxpB incorporation suggests that BclA may be the guide protein for BxpB assembly into the exosporium.
BxpB and ExsFB have been shown to impact BclA content of the spore, with mutants lacking BxpB showing a marked reduction in BclA, while loss of ExsFB has a modest reduction in BclA (23, 24). BxpB has also been shown to be critical for the incorporation into the exosporium of other collagen-like glycoproteins, including BclB and BetA (also named BclF) (21, 22, 48). During assembly of BclA onto the spore surface, there is a cleavage event that is thought to covalently attach the glycoprotein to the basal layer (26, 34). This cleavage event occurs late in the assembly process, after BclA has been positioned approximately 75% around the spore (20). Our Western blotting results indicate that BclA may be attached to another protein, but the protein to which BclA attaches is not the predicted BxpB. However, the BxpB- and BclA-containing high-MW complexes form in the mother cell cytoplasm prior to their assembly around the spore (26). Therefore, strong noncovalent interactions between BclA and BxpB occur in the mother cell cytoplasm.
Although it is well established that BxpB plays an important role in BclA incorporation onto the exosporium surface, the reciprocal situation is also true, with the absence of BclA substantially impacting BxpB incorporation levels. There have been two published observations of this from other laboratories, but in each case, the study was focused on other issues, and no conclusions regarding the role of BclA in BxpB incorporation into spores were made. Evidence that loss of BclA negatively impacts BxpB incorporation can be found in Fig. 5 of the paper by Tan and Turnbough (34) and Fig. 1B of the paper by Cybulski et al. (49). The N terminus of BclA, containing the exosporium localization and attachment sequences, is responsible for this interaction, and site-directed mutations in this BclA N-terminal domain result in a decreased incorporation of BxpB (20, 26). Further evidence in support of BxpB residing in the same complexes with BclA is the coimmunoprecipitation of BxpB from sporulating cells expressing the BclA-NTD-eGFP fusion protein using anti-GFP antiserum (26). Assembly of the outer basal layer and nap layers would be predicted to occur through the BclA N-terminal exosporium-targeting domain delivering the BxpB- and BclA-containing complexes to the developing spore. Efficient incorporation of BclA onto the exosporium surface requires not only the BclA NTD-targeting sequences but also the presence of BxpB in these complexes. The protein(s) responsible for recognizing the BclA-targeting domain to position the complexes on the exosporium surface and for the proteolytic cleavage of BclA has not yet been identified. Whether the cleavage of BclA is associated with attachment of the protein to an exosporium basal layer protein remains speculative. We were unable to detect a candidate protein by mass spectrometry.
Spores from mutants lacking the CotY and ExsY proteins do not possess an exosporium layer. Despite this, there are residual levels of the outer basal layer proteins, BxpB, ExsFB, and BclA, in the spores. These proteins are thought to assemble onto the ExsY- and CotY-containing, self-assembling inner basal layer (37, 38). The presence of the spore-shaped shell structures in the spore preps of the cotE cotY exsY triple mutant suggests that BxpB and perhaps other outer basal layer proteins can organize into a spore-shaped sac in the absence of the inner basal layer proteins. ExsY and, to a lesser extent, CotY, have been shown to be capable of self-assembly into two-dimensional sheets, with disulfide linkages of these cysteine-rich proteins being critical to this process (37). Here, we demonstrate that BxpB and, to a lesser extent ExsFB, are also capable of self-assembly into higher oligomeric forms that remain associated during SDS-PAGE analysis. The formation of dimers, trimers, and higher-MW forms that are resistant to boiling in SDS in the presence of reducing agents is remarkable and is different from results previously seen with ExsY and CotY in that the proteins are not cysteine rich. ExsFB possesses only a single cysteine residue, and BxpB has 2, whereas CotY and ExsY contain 14 and 12 cysteine residues, respectively. If disulfide linkages are involved in the BxpB multimers, the linkages would have to be markedly resistant to reduction with β-mercaptoethanol (Fig. 10) or dithiothreitol (data not shown). Therefore, the mechanism for self-assembly is likely different than the mechanisms operative with CotY and ExsY.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains and plasmids are listed in Table S1 in the supplemental material. All Escherichia coli strains were cultivated using Luria broth (LB). Bacillus anthracis was grown using brain heart infusion broth (Difco). Agar plates were made by the addition of agar at a concentration of 1.5% (wt/vol). Nutrient broth and agar (Oxoid) were used for sporulation. Antibiotics, where needed, were added at the following final concentrations: 100 μg/mL ampicillin, 10 μg/mL chloramphenicol, 25 μg/mL kanamycin, and 100 μg/mL spectinomycin.
DNA purification.
The Wizard SV miniprep kit (Promega) was used to isolate plasmid DNA. For B. anthracis, the pellets from 5-mL cultures were frozen at −80°C overnight and thawed at 37°C prior to DNA extraction. Genomic DNA was isolated using the Wizard genomic DNA purification kit (Promega).
Construction of complementation and expression plasmids.
Expression of Bacillus proteins was accomplished by introduction of the gene with its native or heterologous promoter into the shuttle plasmids pMK4 (50) or pHPS2 (26). The bxpB and exsFB chimeras, as well as the promoter-swapped constructs, were constructed by splicing by overlapping extension PCR (51). All constructs were nucleotide sequence verified prior to transformation into B. anthracis strains.
Construction of the gene fusions to mcherry or egfp.
The fluorescent reporter fusions were generated by PCR amplifying the B. anthracis genes using primers with a 5′ SacI or PstI restriction site upstream of the native promoter element and a 3′ NheI site immediately prior to the termination codon of the open reading frame. SacI/PstI- and NheI-digested DNA fragments were then cloned into identically digested pDG4100 (for mCherry fusions) or pDG4099 (for eGFP fusions) and transformed into E. coli strain DH5α. Clones with plasmids of the correct size and restriction endonuclease pattern were identified and their insertion sequence verified by nucleotide sequence analysis. The plasmids were then transformed into E. coli GM48, and plasmid DNA was isolated and electroporated into B. anthracis Sterne.
Electroporation of B. anthracis.
To prepare B. anthracis electrocompetent cells, bacteria from an overnight BHI agar plate were inoculated into 50 mL of prewarmed BHI with 0.5% glycerol and incubated at 37°C with shaking until the optical density at 600 nm (OD600) reached ~0.6 to 0.8. The culture was then passed through a disposable analytical test filter funnel apparatus with a pore size of 0.4 μm (Thermo Scientific). The filtered cells were then washed twice with 10 mL of ice-cold electroporation buffer (1 mM HEPES, 10% glycerol, pH 7.0). The filter was then placed in a 50-mL polypropylene conical tube, and the cells were washed off the filter by vortex mixing with 5 mL electroporation buffer. Cells were aliquoted into amounts of 400 μL and used immediately or stored at −80°C. For electrotransformation, 10 μL (~1 μg) of plasmid DNA was mixed with 200 μL of electrocompetent B. anthracis cells. All plasmids used for electroporation were passed through E. coli GM48 to produce DNA lacking the Dam methylation pattern. The samples were incubated for 10 min on ice, transferred to a chilled 1-mm electroporation cuvette (Midwest Scientific), and shocked with a pulse of 2.25 kV, 25 μF, and 100 Ω with a Bio-Rad gene pulser apparatus. The bacteria were transferred to a 15-mL polypropylene conical tube with 1 mL of BGGM (BHI with 10% glycerol, 0.4% glucose, and 10 mM MgCl2) and incubated at 37°C (or 30°C for temperature-sensitive plasmids). Samples were plated onto BHI agar plates with appropriate antibiotics and incubated at 37°C or 30°C.
Generation of B. anthracis deletion mutants.
The gene deletion mutants were generated by PCR amplification (Phusion high-fidelity DNA polymerase; New England Biolabs) of 1-kb sequences upstream and downstream of the bxpB, exsFB, cotY, or exsY open reading frame of interest. The upstream and downstream fragments were fused by splicing overlap exchange PCR. The resulting 2-kb fragment was cloned into SalI-digested and alkaline phosphatase-treated pGS4294. A spectinomycin resistance cassette flanked by lox66/lox71 sites (52) was inserted into the BamHI site at the position of the deleted gene. Sequence-verified plasmids were isolated from E. coli GM48 and electroporated into B. anthracis Sterne and incubated at 30°C. Colonies exhibiting spectinomycin (Spec) resistance were inoculated onto spectinomycin and erythromycin (Ery) (the pGS4294 vector-encoded resistance) plates to ensure no spontaneous spectinomycin-resistant colonies arose and that cells from the colony harbored the allele replacement plasmid. Following confirmation of both Specr and Eryr of the transformants, the resulting B. anthracis clones were inoculated into 10 mL of BHI broth containing spectinomycin and grown overnight at 42°C with shaking. Thirty microliters of the culture was then spot inoculated onto a BHI agar with spectinomycin plate, streaked for isolation of single colonies, and incubated overnight at 37°C. Larger colonies were then selected with this semiselective incubation temperature. This process of growing an overnight liquid culture at 42°C and plating was repeated until PCR analysis of DNA from the clone using primers flanking the gene to be deleted gave only the DNA fragment size corresponding to the deletion allele. Sequence analysis of the PCR fragment confirmed the deletion.
To create double or triple mutants, the deletion mutant strain was transformed with plasmid pGS4080, a segregationally unstable plasmid encoding the cre recombinase expressed from the spac promoter. Chloramphenicol-resistant transformants were selected and then subcultured on tryptic soy agar (TSA) plates lacking chloramphenicol at 37°C. The resulting colonies were screened for sensitivity to chloramphenicol and spectinomycin. Clones with the correct antibiotic resistance profile were selected and genomic DNA extracted, and they were screened by PCR and DNA sequencing to confirm the deletion of the lox-flanked spectinomycin resistance cassette. The deletion clone was then utilized to create a deletion at another locus.
Production of spores.
Cells from a BHI broth culture of the B. anthracis strain were swab inoculated onto the surface of 150- by 15-mm Oxoid nutrient agar plates with the appropriate antibiotics. The cultures were incubated at 30°C for 5 days. The surface layer of bacterial growth was harvested with a sterile cotton swab, and the spores were dispersed into 1.5-mL microcentrifuge tubes containing 1 mL PBS. The spores were harvested by centrifugation at 15,000 rpm, and the top pellet layer containing lysed cell debris was removed by flushing and aspiration and then discarded. The process was repeated until there was no evidence of vegetative cells or cell debris present. Spores were then resuspended in PBS and stored at 4°C.
Western blot analysis.
For Western blot analysis, 10 mg of spores were pelleted in an Eppendorf 5425 microcentrifuge at 15,000 rpm for 2 min, and the resulting pellets were then resuspended in 100 μL urea-SDS-PAGE buffer (8 M Urea, 50 mM Tris-HCl [pH 10], 2% SDS, 0.002% bromophenol blue, 0.71 M β-mercaptoethanol, and 10% glycerol) and boiled for 10 min. Spores were pelleted by centrifugation for 10 min, and 15 μL of the extract was loaded onto a 15-well Mini-Protean TGX gel (Bio-Rad) and electrophoresed at 190 V in SDS buffer (25 mM Tris, 192 mM glycine, 0.1% SDS, pH 8.3). The proteins were electro-transferred to Immobilon membranes (MilliporeSigma) for 1 h on ice at 250 mA in 25 mM Tris, 192 mM glycine, and 10% methanol. Membranes were blocked overnight at 4°C using SuperBlock (Thermo Fisher) and stained with a 1:20,000 dilution of anti-mCherry rabbit polyclonal antibody (Invitrogen), anti-his tag HRP conjugate (Abcam), or rabbit polyclonal antisera against rBclA, rBxpB, or rExsFB, at room temperature for 1 h. The membranes were then washed six times with 0.1% Tween 20 in PBS for 5 min each with agitation. Except with the anti-His tag antibody conjugate, secondary staining was done with 1:20,000 anti-rabbit IgG-horseradish peroxidase (HRP) conjugate (Invitrogen) at room temperature for 1 h, and the membrane was washed eight times with 0.1% Tween 20 in PBS for 5 min with agitation. The membrane was processed using Pierce ECL Western blotting substrate and then exposed to autoradiography film (Midwest Scientific).
Immunolabeling of spores.
Ten milligrams of spores were resuspended in 750 μL SuperBlock blocking buffer and incubated for at least 20 min at room temperature. The spores were then harvested by centrifugation, and the spore pellet was resuspended in 250 μL SuperBlock blocking buffer with 1 μL primary antibody and incubated at room temperature for 20 min (with short vortex mixing every 5 min). Rabbit polyclonal antibodies used were against BclA (19), BxpB (19), and ExsFB at 1:250 dilution. The exsFB open reading frame was PCR amplified and cloned into the pQE30 plasmid (Qiagen). His-tagged proteins were expressed in E. coli and purified using the His Spin protein purification kit (Zymo Research). Anti-ExsFB antiserum was prepared in rabbits using Ribi adjuvant (Corixa). Following incubation with the primary antibody, the spores were harvested by centrifugation and washed with 750 μL of SuperBlock blocking buffer. The pellet was then resuspended in 250 μL of SuperBlock blocking buffer with secondary antibody conjugate (1:250 goat anti-rabbit IgG-Alexa Fluor 568; Invitrogen). The spores were incubated at room temperature for 20 min, pelleted, and washed with 750 μL SuperBlock blocking buffer, followed by three washes with 750 μL PBS, and finally resuspended in 250 μL PBS. The spores were examined by epifluorescence microscopy using a Nikon E600 epifluorescence microscope using the mCherry filter set. Fluorescence quantification was accomplished using the NIH ImageJ program (https://imagej.nih.gov/nih-image/).
Protein interaction analysis by the bacterial two-hybrid method.
The procedure of Karimova et al. was used (44). Plasmids pKT25, pKNT25, pUT18, and pUT18C were used, thus obtaining hybrid proteins with the T18 or T25 domains of adenylate cyclase on their N or C termini. All plasmid constructs (Table S1) were verified by restriction analysis and DNA sequencing. The compatible recombinant plasmid pairs were cotransformed into E. coli BTH101-competent cells. Transformants were selected on LB plates supplemented with ampicillin (100 μg/mL) plus kanamycin (50 μg/mL) and were cultivated at 30°C for 48 h. The plasmids pKT25-zip and pUT18C-zip served as positive control, and a pairing of ExsY with the pXO1-encoded AW20_5669 proteins was used as the negative control. β-Galactosidase assays were performed as described by Schaefer et al. (53) utilizing PopCulture reagent as the lysis reagent (MilliporeSigma) and a BioTek Synergy 96-well microplate reader.
Statistical analysis.
Statistical differences (P < 0.05) were assessed between bacterial two-hybrid sample combinations and the negative control using Welch’s t test.
Transmission electron microscopy.
Spores were fixed in 1 mL 2% glutaraldehyde-100 mM sodium cacodylate solution containing 0.1% ruthenium red (Sigma-Aldrich) for 1 h at 37°C (54). Each spore pellet was then washed in 100 mM sodium cacodylate buffer, embedded in HistoGel, and washed with 100 mM sodium cacodylate buffer containing 0.01 M 2-mercaptoethanol and 0.13 M sucrose (2-ME buffer). Samples underwent secondary fixation in 1% osmium tetroxide (Electron Microscopy Sciences). Sterne spores were used as a wild-type control and were treated identically to the mutant spores. Spores were washed in 2-ME buffer and dehydrated with sequential treatments of 20, 50, 75, 90, and 100% acetone. Polymerization occurred at 60°C in Epon-Spurrs resin after extended resin infiltration. Samples were cut into 85-nm-thick sections, and the sections were mounted on 200-mesh nickel grids and stained with 2% uranyl acetate (Electron Microscopy Sciences) for 20 min at room temperature. The samples were treated with lead for 5 min at room temperature. Grids were washed in ultrapure water and observed by TEM using a Jeol 1400 electron microscope at the University of Missouri Electron Microscopy Core Facility.
ACKNOWLEDGMENTS
We thank Bill Picking (University of Missouri) for the BTH101 strain.
This work was supported by NIAID grant R21AI101093 and the University of Missouri McKee Microbial Pathogenesis endowment fund to G.C.S.
Footnotes
Supplemental material is available online only.
Contributor Information
George C. Stewart, Email: stewartgc@missouri.edu.
Tina M. Henkin, Ohio State University
REFERENCES
- 1.Tan IS, Ramamurthi KS. 2014. Spore formation in Bacillus subtilis. Environ Microbiol Rep 6:212–225. 10.1111/1758-2229.12130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ehling-Schulz M, Lereclus D, Koehler TM. 2019. The Bacillus cereus group: Bacillus species with pathogenic potential. Microbiol Spectr 7. 10.1128/microbiolspec.GPP3-0032-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stewart GC. 2015. The exosporium layer of bacterial spores: a connection to the environment and the infected host. Microbiol Mol Biol Rev 79:437–457. 10.1128/MMBR.00050-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gerhardt P. 1967. Cytology of Bacillus anthracis. Fed Proc 26:1504–1517. [PubMed] [Google Scholar]
- 5.Gerhardt P, Black SH. 1961. Permeability of bacterial spores. II. Molecular variables affecting solute permeation. J Bacteriol 82:750–760. 10.1128/jb.82.5.750-760.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Williams G, Linley E, Nicholas R, Baillie L. 2013. The role of the exosporium in the environmental distribution of anthrax. J Appl Microbiol 114:396–403. 10.1111/jam.12034. [DOI] [PubMed] [Google Scholar]
- 7.Oliva CR, Swiecki MK, Griguer CE, Lisanby MW, Bullard DC, Turnbough CL, Jr, Kearney JF. 2008. The integrin Mac-1 (CR3) mediates internalization and directs Bacillus anthracis spores into professional phagocytes. Proc Natl Acad Sci USA 105:1261–1266. 10.1073/pnas.0709321105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bozue J, Moody KL, Cote CK, Stiles BG, Friedlander AM, Welkos SL, Hale ML. 2007. Bacillus anthracis spores of the bclA mutant exhibit increased adherence to epithelial cells, fibroblasts, and endothelial cells but not to macrophages. Infect Immun 75:4498–4505. 10.1128/IAI.00434-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brahmbhatt TN, Janes BK, Stibitz ES, Darnell SC, Sanz P, Rasmussen SB, O'Brien AD. 2007. Bacillus anthracis exosporium protein BclA affects spore germination, interaction with extracellular matrix proteins, and hydrophobicity. Infect Immun 75:5233–5239. 10.1128/IAI.00660-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hachisuka Y, Kojima K, Sato T. 1966. Fine filaments on the outside of the exosporium of Bacillus anthracis spores. J Bacteriol 91:2382–2384. 10.1128/jb.91.6.2382-2384.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kramer MJ, Roth IL. 1968. Ultrastructural differences in the exosporium of the Sterne and Vollum strains of Bacillus anthracis. Can J Microbiol 14:1297–1299. 10.1139/m68-217. [DOI] [PubMed] [Google Scholar]
- 12.Driks A. 2002. Maximum shields: the assembly and function of the bacterial spore coat. Trends Microbiol 10:251–254. 10.1016/s0966-842x(02)02373-9. [DOI] [PubMed] [Google Scholar]
- 13.Sylvestre P, Couture-Tosi E, Mock M. 2002. A collagen-like surface glycoprotein is a structural component of the Bacillus anthracis exosporium. Mol Microbiol 45:169–178. 10.1046/j.1365-2958.2000.03000.x. [DOI] [PubMed] [Google Scholar]
- 14.Steichen C, Chen P, Kearney JF, Turnbough CL, Jr.. 2003. Identification of the immunodominant protein and other proteins of the Bacillus anthracis exosporium. J Bacteriol 185:1903–1910. 10.1128/JB.185.6.1903-1910.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sylvestre P, Couture-Tosi E, Mock M. 2003. Polymorphism in the collagen-like region of the Bacillus anthracis BclA protein leads to variation in exosporium filament length. J Bacteriol 185:1555–1563. 10.1128/JB.185.5.1555-1563.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Redmond C, Baillie LW, Hibbs S, Moir AJ, Moir A. 2004. Identification of proteins in the exosporium of Bacillus anthracis. Microbiology (Reading) 150:355–363. 10.1099/mic.0.26681-0. [DOI] [PubMed] [Google Scholar]
- 17.Giorno R, Mallozzi M, Bozue J, Moody KS, Slack A, Qiu D, Wang R, Friedlander A, Welkos S, Driks A. 2009. Localization and assembly of proteins comprising the outer structures of the Bacillus anthracis spore. Microbiology (Reading) 155:1133–1145. 10.1099/mic.0.023333-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Waller LN, Stump MJ, Fox KF, Harley WM, Fox A, Stewart GC, Shahgholi M. 2005. Identification of a second collagen-like glycoprotein produced by Bacillus anthracis and demonstration of associated spore-specific sugars. J Bacteriol 187:4592–4597. 10.1128/JB.187.13.4592-4597.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thompson BM, Waller LN, Fox KF, Fox A, Stewart GC. 2007. The BclB glycoprotein of Bacillus anthracis is involved in exosporium integrity. J Bacteriol 189:6704–6713. 10.1128/JB.00762-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thompson BM, Stewart GC. 2008. Targeting of the BclA and BclB proteins to the Bacillus anthracis spore surface. Mol Microbiol 70:421–434. 10.1111/j.1365-2958.2008.06420.x. [DOI] [PubMed] [Google Scholar]
- 21.Thompson BM, Hoelscher BC, Driks A, Stewart GC. 2012. Assembly of the BclB glycoprotein into the exosporium and evidence for its role in the formation of the exosporium 'cap' structure in Bacillus anthracis. Mol Microbiol 86:1073–1084. 10.1111/mmi.12042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thompson BM, Hoelscher BC, Driks A, Stewart GC. 2011. Localization and assembly of the novel exosporium protein BetA of Bacillus anthracis. J Bacteriol 193:5098–5104. 10.1128/JB.05658-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Steichen CT, Kearney JF, Turnbough CL, Jr.. 2005. Characterization of the exosporium basal layer protein BxpB of Bacillus anthracis. J Bacteriol 187:5868–5876. 10.1128/JB.187.17.5868-5876.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sylvestre P, Couture-Tosi E, Mock M. 2005. Contribution of ExsFA and ExsFB proteins to the localization of BclA on the spore surface and to the stability of the Bacillus anthracis exosporium. J Bacteriol 187:5122–5128. 10.1128/JB.187.15.5122-5128.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Steichen CT, Kearney JF, Turnbough CL, Jr.. 2007. Non-uniform assembly of the Bacillus anthracis exosporium and a bottle cap model for spore germination and outgrowth. Mol Microbiol 64:359–367. 10.1111/j.1365-2958.2007.05658.x. [DOI] [PubMed] [Google Scholar]
- 26.Thompson BM, Hsieh HY, Spreng KA, Stewart GC. 2011. The co-dependence of BxpB/ExsFA and BclA for proper incorporation into the exosporium of Bacillus anthracis. Mol Microbiol 79:799–813. 10.1111/j.1365-2958.2010.07488.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Giorno R, Bozue J, Cote C, Wenzel T, Moody KS, Mallozzi M, Ryan M, Wang R, Zielke R, Maddock JR, Friedlander A, Welkos S, Driks A. 2007. Morphogenesis of the Bacillus anthracis spore. J Bacteriol 189:691–705. 10.1128/JB.00921-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McPherson SA, Li M, Kearney JF, Turnbough CL, Jr.. 2010. ExsB, an unusually highly phosphorylated protein required for the stable attachment of the exosporium of Bacillus anthracis. Mol Microbiol 76:1527–1538. 10.1111/j.1365-2958.2010.07182.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boone TJ, Mallozzi M, Nelson A, Thompson B, Khemmani M, Lehmann D, Dunkle A, Hoeprich P, Rasley A, Stewart G, Driks A. 2018. Coordinated assembly of the Bacillus anthracis coat and exosporium during bacterial spore outer layer formation. mBio 9:e01166-18. 10.1128/mBio.01166-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raines KW, Kang TJ, Hibbs S, Cao GL, Weaver J, Tsai P, Baillie L, Cross AS, Rosen GM. 2006. Importance of nitric oxide synthase in the control of infection by Bacillus anthracis. Infect Immun 74:2268–2276. 10.1128/IAI.74.4.2268-2276.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weaver J, Kang TJ, Raines KW, Cao GL, Hibbs S, Tsai P, Baillie L, Rosen GM, Cross AS. 2007. Protective role of Bacillus anthracis exosporium in macrophage-mediated killing by nitric oxide. Infect Immun 75:3894–3901. 10.1128/IAI.00283-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chesnokova ON, McPherson SA, Steichen CT, Turnbough CL, Jr.. 2009. The spore-specific alanine racemase of Bacillus anthracis and its role in suppressing germination during spore development. J Bacteriol 191:1303–1310. 10.1128/JB.01098-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cybulski RJ, Jr, Sanz P, Alem F, Stibitz S, Bull RL, O'Brien AD. 2009. Four superoxide dismutases contribute to Bacillus anthracis virulence and provide spores with redundant protection from oxidative stress. Infect Immun 77:274–285. 10.1128/IAI.00515-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tan L, Turnbough CL, Jr.. 2010. Sequence motifs and proteolytic cleavage of the collagen-like glycoprotein BclA required for its attachment to the exosporium of Bacillus anthracis. J Bacteriol 192:1259–1268. 10.1128/JB.01003-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tan L, Li M, Turnbough CL, Jr.. 2011. An unusual mechanism of isopeptide bond formation attaches the collagenlike glycoprotein BclA to the exosporium of Bacillus anthracis. mBio 2:e00084-11. 10.1128/mBio.00084-11. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 36.Tan L, Li M, Turnbough CL, Jr.. 2013. Retraction: an unusual mechanism of isopeptide bond formation attaches the collagenlike glycoprotein BclA to the exosporium of Bacillus anthracis. mBio 4:e00923-13. 10.1128/mBio.00923-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Terry C, Jiang S, Radford DS, Wan Q, Tzokov S, Moir A, Bullough PA. 2017. Molecular tiling on the surface of a bacterial spore–the exosporium of the Bacillus anthracis/cereus/thuringiensis group. Mol Microbiol 104:539–552. 10.1111/mmi.13650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lablaine A, Serrano M, Bressuire-Isoard C, Chamot S, Bornard I, Carlin F, Henriques AO, Broussolle V. 2021. The morphogenetic protein CotE positions exosporium proteins CotY and ExsY during sporulation of Bacillus cereus. mSphere 6:e00007-21. 10.1128/mSphere.00007-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boydston JA, Yue L, Kearney JF, Turnbough CL, Jr.. 2006. The ExsY protein is required for complete formation of the exosporium of Bacillus anthracis. J Bacteriol 188:7440–7448. 10.1128/JB.00639-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Durand-Heredia J, Hsieh H-Y, Thompson BM, Stewart GC. 2022. ExsY, CotY, and CotE effects on Bacillus anthracis outer spore layer architecture. J Bacteriol e0029122. 10.1128/jb.00291-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rodenburg CM, McPherson SA, Turnbough CL, Jr, Dokland T. 2014. Cryo-EM analysis of the organization of BclA and BxpB in the Bacillus anthracis exosporium. J Struct Biol 186:181–187. 10.1016/j.jsb.2014.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Durand-Heredia J, Stewart GC. 2022. Localization of the CotY and ExsY proteins to the exosporium basal layer of Bacillus anthracis. MicrobiologyOpen 11:e1327. 10.1002/mbo3.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bergman NH, Anderson EC, Swenson EE, Niemeyer MM, Miyoshi AD, Hanna PC. 2006. Transcriptional profiling of the Bacillus anthracis life cycle in vitro and an implied model for regulation of spore formation. J Bacteriol 188:6092–6100. 10.1128/JB.00723-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Karimova G, Gauliard E, Davi M, Ouellette SP, Ladant D. 2017. Protein-protein interaction: bacterial two-hybrid. Meth Mol Biol 1615:159–176. 10.1007/978-1-4939-7033-9_13. [DOI] [PubMed] [Google Scholar]
- 45.Jiang S, Wan Q, Krajcikova D, Tang J, Tzokov SB, Barak I, Bullough PA. 2015. Diverse supramolecular structures formed by self-assembling proteins of the Bacillus subtilis spore coat. Mol Microbiol 97:347–359. 10.1111/mmi.13030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ursem R, Swarge B, Abhyankar WR, Buncherd H, de Koning LJ, Setlow P, Brul S, Kramer G. 2021. Identification of native cross-links in Bacillus subtilis spore coat proteins. J Proteome Res 20:1809–1816. 10.1021/acs.jproteome.1c00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Spreng KA. 2012. Identification and characterization of Bacillus anthracis spore-associated proteins. PhD thesis. University of Missouri, Columbia, MO. [Google Scholar]
- 48.Leski TA, Caswell CC, Pawlowski M, Klinke DJ, Bujnicki JM, Hart SJ, Lukomski S. 2009. Identification and classification of bcl genes and proteins of Bacillus cereus group organisms and their application in Bacillus anthracis detection and fingerprinting. Appl Environ Microbiol 75:7163–7172. 10.1128/AEM.01069-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cybulski RJ, Jr, Sanz P, McDaniel D, Darnell S, Bull RL, O'Brien AD. 2008. Recombinant Bacillus anthracis spore proteins enhance protection of mice primed with suboptimal amounts of protective antigen. Vaccine 26:4927–4939. 10.1016/j.vaccine.2008.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sullivan MA, Yasbin RE, Young FE. 1984. New shuttle vectors for Bacillus subtilis and Escherichia coli which allow rapid detection of inserted fragments. Gene 29:21–26. 10.1016/0378-1119(84)90161-6. [DOI] [PubMed] [Google Scholar]
- 51.Horton RM, Cai ZL, Ho SN, Pease LR. 1990. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. Biotechniques 8:528–535. [PubMed] [Google Scholar]
- 52.Lambert JM, Bongers RS, Kleerebezem M. 2007. Cre-lox-based system for multiple gene deletions and selectable-marker removal in Lactobacillus plantarum. Appl Environ Microbiol 73:1126–1135. 10.1128/AEM.01473-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schaefer J, Jovanovic G, Kotta-Loizou I, Buck M. 2016. Single-step method for β-galactosidase assays in Escherichia coli using a 96-well microplate reader. Anal Biochem 503:56–57. 10.1016/j.ab.2016.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Waller LN, Fox N, Fox KF, Fox A, Price RL. 2004. Ruthenium red staining for ultrastructural visualization of a glycoprotein layer surrounding the spore of Bacillus anthracis and Bacillus subtilis. J Microbiol Methods 58:23–30. 10.1016/j.mimet.2004.02.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 and Fig. S1 and S2. Download jb.00290-22-s0001.pdf, PDF file, 0.4 MB (413.9KB, pdf)