ABSTRACT
Delamanid (DLM) and pretomanid (PTM) are recent additions to the anti-tuberculosis (TB) drug armamentarium, and they offer more effective options for drug-resistant TB treatment. In particular, DLM is included in Group C, which is recommended for use in longer multidrug-resistant (MDR)-TB regimens. Previous studies have shown that resistance to DLM/PTM is caused by mutations in the ddn, fgd1, fbiA, fbiB, fbiC, and fbiD genes, which are related to the F420-dependent bioactivation pathway. Herein, we conduct in vitro selection of DLM-resistant strains using clinical Mycobacterium tuberculosis (MTB) isolates with various drug resistance profiles. The spontaneous resistance frequency of drug-susceptible (DS) MTB (1.14 × 10−6 to 1.04 × 10−4) to DLM was similar to that of H37Rv (8.88 × 10−6 to 9.96 × 10−6) but higher than those of multidrug-resistant MTB (2.03 × 10−7 to 3.18 × 10−6) and extensively drug-resistant (XDR) MTB (4.67 × 10−8 to 3.60 × 10−6). Of the 100 independently selected DLM-resistant MTB mutants, 65% harbored mutations in genes associated with either DLM prodrug activation (ddn, 39.73%; fgd1, 16.44%) or the F420 biosynthetic pathway (fbiA, 16.44%; fbiB, 5.48%; fbiC, 21.92%). Of the 45 mutations we identified, 38 were not previously reported. A structure analysis revealed that several point mutations affected the ligand binding or structural stability of enzymes related to DLM resistance, which would block the enzyme activity required for prodrug activation. Our results elucidate the in vitro spontaneous DLM-resistance patterns of different clinical strains, which could improve the understanding of the causes of DLM resistance in clinical strains and of the effects on drug resistance of different mutations in genes that are related to the DLM activation pathway.
KEYWORDS: MTB, delamanid, pretomanid, resistance, F420
INTRODUCTION
Tuberculosis (TB) is one of the global public health problems that seriously threatens public health (1). The epidemic of drug-resistant TB, especially multidrug-resistant tuberculosis (MDR-TB) and extensively drug-resistant tuberculosis (XDR-TB) (referring to the old definition) (2), worsens this situation and hinders efforts for tuberculosis care and control. Due to preexisting resistance to rifampin (RIF) and isoniazid (INH), MDR-TB treatment requires a prolonged treatment duration and a high drug toxicity; however, only half of patients achieve favorable outcomes via the completion of an anti-TB treatment (3). In 2019, there were about 465,000 MDR-/rifampicin-resistant TB (MDR/RR-TB) patients worldwide, 50% of which came from India, the Russian Federation, and China (1). China has the second largest burden of MDR/RR-TB in the world, and the proportions of MDR/RR-TB patients were about 7.1% (5.6% to 8.7%) in new cases and 23% (23% to 24%) in retreated cases (1). Despite the significant decrease in TB incidence, the spread of MDR-TB constitutes a major threat to TB control and prevention in China. This serious situation highlights the urgent need for new antimicrobial development.
Delamanid (DLM), a dihydro-nitroimidazooxazole derivative, inhibits the synthesis of mycobacterial cell wall components, methoxy mycolic acid, and ketomycolic acid (4). It is a prodrug that requires bioreductive activation by the mycobacterial 8-hydroxy-5-deazaflavin (coenzyme F420)-dependent nitroreductase Ddn (5). As an important agent with a novel mechanism of action, DLM exhibited potent antibacterial activity against drug-susceptible and drug-resistant Mycobacterium tuberculosis (MTB) isolates (4). Further clinical trials demonstrated that DLM-containing regimens could significantly improve sputum culture conversion rates in comparison to a placebo group (6). In 2020, the World Health Organization (WHO) endorsed DLM as a member of the Group C drugs for the treatment of MDR/RR-TB (7). Multiple mutations within F420 biosynthesis-associated genes, such as ddn, fgd1, and fbiA-D, were associated with a decreased susceptibility to DLM (5, 8). Despite attracting more attention, the knowledge on the mechanisms conferring DLM resistance remains limited. Furthermore, there has been no comprehensive interpretation of DLM resistance at an atomic level. Pretomanid (PTM), which is often cross-resistant with DLM, is also a promising new nitroimidazole drug that has been recommended by the WHO for the treatment of MDR-TB with additional resistance to fluroquinolone (in combination with bedaquiline and linezolid) under operational research conditions (7). A variety of loss-of-function mutations in ddn, fgd, and fbiA-D that cause DLM and PTM resistance have been reported both in vitro and in vivo, and cross-resistance between them is inevitable due to their shared drug resistance mechanism (5, 8–10). In view of the increasing clinical use of DLM and PTM, the paucity of data on this issue is concerning.
To address this concern, we conducted in vitro selection of DLM-resistant strains using clinical MTB isolates with various drug resistance profiles. Our objectives were: (i) to determine the drug resistance-associated mutation rates in vitro; (ii) to investigate the molecular mechanisms that confer DLM resistance; and (iii) to establish the correlation between genetic mutations and DLM/PTM minimum inhibitory concentrations (MICs) in MTB.
RESULTS
Characterization and spoligotyping of parent strains.
We first measured the MICs of DLM against 60 clinical isolates (20 DS, 20 MDR, and 20 XDR strains) to select the strains to be used for resistance induction. Due to contamination or initial resistance to DLM, 18 strains were excluded. For further resistance induction experimentation, we finally selected 42 strains (14 DS, 14 MDR, and 14 XDR strains) with MIC values of DLM that were less than the critical MICs of resistance as the parent strains. The MIC values of DLM in the 42 parent strains are shown in Table 1. We also detected the MIC values of PTM in the parent strains, which were all lower than 0.125 μg/mL. By spoligotyping analysis, 39 (92.86%) of the 42 parent strains belonged to the Beijing family. All of the MDR and XDR strains belonged to the Beijing family. Among the DS strains, 11 (78.57%) strains belonged to the Beijing family, and 3 (21.43%) strains (DS-6, DS-8, and DS-14) belonged to the T1 family.
TABLE 1.
MIC distribution of parent strains
| Strains | No. of strains with different MIC (μg/mL) |
Critical MIC (μg/mL) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ≤0.016 | 0.031 | 0.063 | 0.125 | 0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 | ||
| DSa | 14 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.125 |
| MDRb | 14 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| XDRc | 13 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| H37Rv | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
DS, drug-susceptible.
MDR, multidrug-resistant.
XDR, extensively drug-resistant.
Drug resistance induction.
Drug-resistant progeny strains were obtained after 4 weeks of incubation. There were no detectable colony-forming units (CFU) of DLM-resistant progeny strains on plates of 8 parent strains, including 3 DS strains (DS-5, DS-8, and DS-13), 3 MDR strains (MDR-2, MDR-6, and MDR-11), and 2 XDR strains (XDR-11 and XDR-12). The spontaneous resistance frequencies to DLM of the different clinical strains are shown in Table 2. The frequencies of spontaneous resistance to DLM of all strains appeared to be similar on the two concentrations tested. In the DS strains, the resistance frequencies were around 10−5, ranging from 1.14 × 10−6 to 1.04 × 10−4. In the MDR strains, the resistance frequencies were around 10−6, ranging from 2.03 × 10−7 to 3.18 × 10−6. In the XDR strains, the resistance frequencies were around 10−7, ranging from 4.67 × 10−8 to 3.60 × 10−6.
TABLE 2.
Spontaneous resistance frequencies to DLM of clinical strains
| Strain | DLM (μg/mL) |
|
|---|---|---|
| 0.063 | 0.125 | |
| DSa -1 | 1.71 × 10−5 | 1.25 × 10−5 |
| DS-2 | 3.42 × 10−6 | 3.42 × 10−6 |
| DS-3 | 9.12 × 10−6 | 7.98 × 10−6 |
| DS-4 | 1.04 × 10−4 | 8.21 × 10−5 |
| DS-6 | 1.14 × 10−6 | 1.14 × 10−6 |
| DS-7 | 4.56 × 10−6 | 2.28 × 10−6 |
| DS-9 | 3.42 × 10−5 | 1.82 × 10−5 |
| DS-10 | 1.71 × 10−5 | 5.70 × 10−6 |
| DS-11 | 1.71 × 10−5 | 9.12 × 10−6 |
| DS-12 | 1.48 × 10−5 | 1.71 × 10−5 |
| DS-14 | 7.98 × 10−6 | 1.25 × 10−5 |
| MDRb-1 | 4.06 × 10−7 | 1.42 × 10−6 |
| MDR-3 | 2.03 × 10−6 | 1.69 × 10−6 |
| MDR-4 | 1.02 × 10−6 | 1.08 × 10−6 |
| MDR-5 | 5.41 × 10−7 | 4.06 × 10−7 |
| MDR-7 | 1.35 × 10−7 | 2.03 × 10−7 |
| MDR-8 | 2.10 × 10−6 | 1.69 × 10−6 |
| MDR-9 | 4.06 × 10−7 | 1.22 × 10−6 |
| MDR-10 | 1.08 × 10−6 | 1.15 × 10−6 |
| MDR-12 | 1.29 × 10−6 | 5.41 × 10−7 |
| MDR-13 | 2.44 × 10−6 | 3.18 × 10−6 |
| MDR-14 | 2.50 × 10−6 | 2.03 × 10−6 |
| XDRc-1 | 1.87 × 10−7 | 1.87 × 10−7 |
| XDR-2 | 3.27 × 10−7 | 2.34 × 10−7 |
| XDR-3 | 1.87 × 10−7 | 9.35 × 10−8 |
| XDR-4 | 4.21 × 10−7 | 3.27 × 10−7 |
| XDR-5 | 4.67 × 10−7 | 5.14 × 10−7 |
| XDR-6 | 8.88 × 10−7 | 3.74 × 10−7 |
| XDR-7 | 3.60 × 10−6 | 3.34 × 10−6 |
| XDR-8 | 1.03 × 10−6 | 8.41 × 10−7 |
| XDR-9 | 9.35 × 10−8 | 4.67 × 10−8 |
| XDR-10 | 4.21 × 10−7 | 4.67 × 10−8 |
| XDR-13 | 3.41 × 10−6 | 2.52 × 10−6 |
| XDR-14 | 5.61 × 10−7 | 3.27 × 10−7 |
| H37Rv | 9.96 × 10−6 | 8.88 × 10−6 |
DS, drug-susceptible.
MDR, multidrug-resistant.
XDR, extensively drug-resistant.
Mutations in resistant genes and MICs of progeny strains.
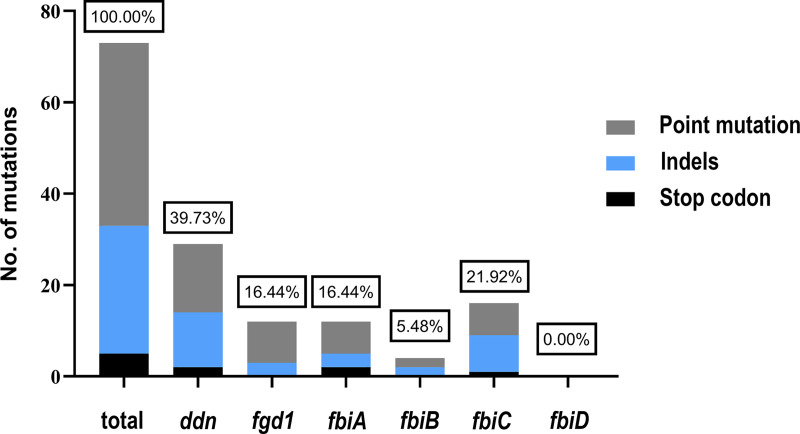
We finally obtained 1,170 drug-resistant progeny strains in all plates, and 100 strains were randomly selected for the measurement of their MICs and the sequencing of their drug-resistant genes with a ratio of about 1:10. DNA sequencing identified a total of 73 mutations, including 29 in ddn (including 15 point mutations, 12 frameshift mutations, and 2 stop codons), 12 in fgd1 (including 9 point mutations and 3 frameshift mutations), 12 in fbiA (including 7 point mutations, 3 frameshift mutations, and 2 stop codons), 4 in fbiB (including 2 point mutations and 2 frameshift mutations), and 16 in fbiC (including 8 point mutations, 7 frameshift mutations, and 1 stop codon) (Fig. 1). No mutation was found in fbiD. There were 3 synonymous mutations in the above-mentioned point mutations that appeared in four strains, namely, Ddn P63P (two strains), Fgd1 Y155Y (one strain), and FbiA E249E (one strain).
FIG 1.
Distribution of mutation frequencies among the six target genes. The sequencing of 6 resistance-related genes was performed in 100 DLM-resistant colonies. 73 unique mutations in these 6 genes were identified from progeny colonies. The relative number of genes encoding early stop codons is shown in black, whereas that of out-of-frame insertions and deletions is shown in blue and that of point mutations is shown in gray.
Out of the 100 DLM-resistant strains, 65 (65.00%) had nonsynonymous mutations in the above-mentioned resistant genes, and 35 (35.00%) had no mutations in any of the six genes. There were 26 (26.00%) strains with mutations in ddn, 10 (10.00%) with mutations in fgd1, 8 (8.00%) with mutations in fbiA, 2 (2.00%) with mutations in fbiB, 15 (15.00%) with mutations in fbiC, and 4 (4.00%) strains with mutations in two different genes (Table 3).
TABLE 3.
Nonsynonymous mutations in DLM resistance-conferring genes
| Gene | Nucleotide change | Amino acid change | MIC range (μg/mL) |
No. of strains | |
|---|---|---|---|---|---|
| DLM | PTM | ||||
| ddn | 68G>C | R23P | 0.25 | 16 | 1 |
| ddn | 113dela | G38fsd | 16 | 16 | 1 |
| ddn | 116_117insGb | G39fs | 16 | 16 | 1 |
| ddn | 117_118insC | T40fs | 0.5 to 1 | 0.5 to 16 | 2 |
| ddn | 134del | P45fs | >16 | 16 | 1 |
| ddn | 157G>T | G53C | 0.5 | 8 | 1 |
| ddn | 161del | R54fs | 16 | 16 | 1 |
| ddn | 188C>A | P63Q | >16 | >16 | 2 |
| ddn | 189G>T, 195C>G | Y65*e | 16 | 8 to 16 | 2 |
| ddn | 233C>A | S78Y | >16 | >16 | 1 |
| ddn | 237_238insGCG | K79_G80insG | 1 | 16 | 1 |
| ddn | 352G>A | E118K | >16 | 4 to 16 | 4 |
| ddn | 424_425delinsTTc | R142F | 0.125 | 8 to 16 | 4 |
| ddn | 434_435insC | P145fs | >16 | >16 | 1 |
| ddn | 447_448insC | E150fs | 0.5 to 16 | 1 to 16 | 3 |
| fgd1 | 209T>G | L70R | 16 | 2 to 16 | 4 |
| fgd1 | 278T>G | M93R | 16 | 16 | 1 |
| fgd1 | 603_604insG | L202fs | >16 | >16 | 1 |
| fgd1 | 851G>C | W284S | >16 | >16 | 1 |
| fgd1 | 870del | P290fs | >16 | 16 | 1 |
| fgd1 | 911G>T | G304V | >16 | 16 | 1 |
| fgd1 | 962T>C | L321P | 0.25 | 0.25 | 1 |
| fbiA | 22G>A | G8S | 1 | 8 | 1 |
| fbiA | 193T>C | C65R | 0.25 | 0.25 | 1 |
| fbiA | 309del | E103fs | 16 | >16 | 1 |
| fbiA | 428T>C | L143P | >16 | 16 | 1 |
| fbiA | 596C>A | A199E | 1 | 0.5 | 1 |
| fbiA | 747A>G, 748A>T | K250* | 16 | 16 | 1 |
| fbiA | 745_809del | E249fs | >16 | >16 | 1 |
| fbiA | 864G>A | W288* | >16 | >16 | 1 |
| fbiB | 201del | P67fs | 4 | 16 | 1 |
| fbiB | 735_736insC | A246fs | 8 | 8 | 1 |
| fbiC | 177C>A | C59* | >16 | >16 | 1 |
| fbiC | 343_344insC | R115fs | 2 to 16 | 2 to 16 | 6 |
| fbiC | 765del | F255fs | 8 | 8 | 1 |
| fbiC | 773G>A | G258D | >16 | >16 | 1 |
| fbiC | 1130T>G | L377R | 8 to 16 | 2 to 16 | 2 |
| fbiC | 1792G>C | E598Q | 16 | >16 | 1 |
| fbiC | 1810G>T | G604C | 16 | 16 | 1 |
| fbiC | 2003_2007del | A668fs | >16 | >16 | 1 |
| fbiC | 2092C>G | H698D | >16 | >16 | 1 |
| fbiA-ddn | 595G>A, 117_118insC | A199T, T40fs | 0.5 | 1 | 1 |
| fgd1-fbiA | 834_835insA, 954_955insG | Q279fs, M319fs | >16 | >16 | 1 |
| fbiA-fbiB | 776G>A, 991A>T | C259Y, I331L | 16 | 8 | 1 |
| fbiC-fbiB | 2155T>G, 991A>T | W719G, I331L | >16 | 16 | 1 |
del, deletion.
ins, insertion.
delins, deletion and insertion.
fs, frameshift.
*, stop codon.
All of the 65 progeny strains with mutations in their resistant genes were resistant to DLM and displayed MIC values of ≥0.125 μg/mL. Considering that the MIC values of the parent strains were between 0.016 and 0.031 μg/mL, the MIC values of the progeny strains to DLM were at least 4 times higher than those of their parent strains. We also detected the resistance of the progeny strains to PTM and found that all of the progeny strains had MIC values of ≥8 times those of the parent strains (Table 3).
The effects of point mutations on structure stability and ligand binding.
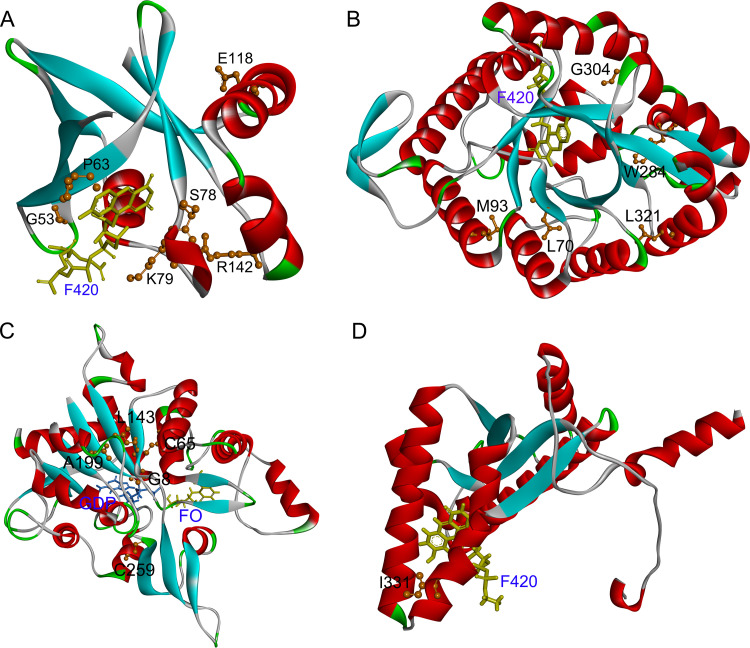
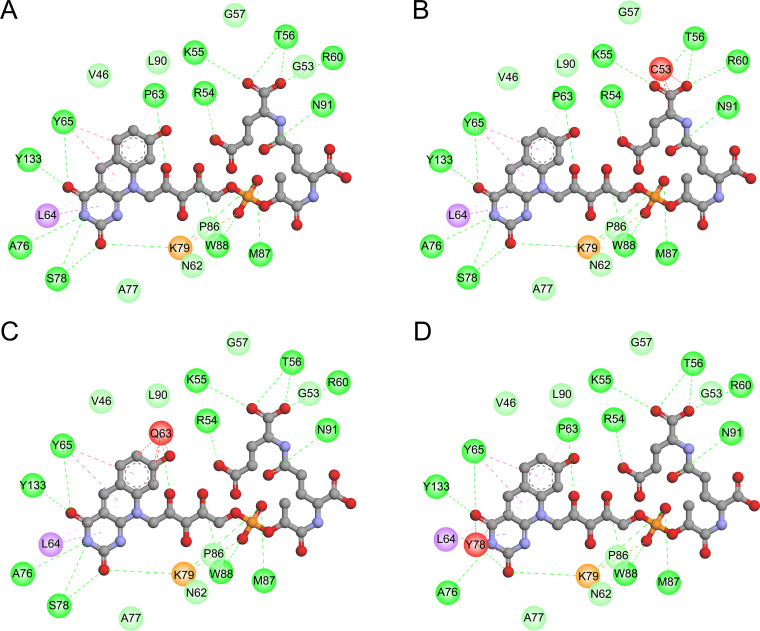
In the activation process of DLM, Ddn catalyzes the reduction of DLM using the cofactor F420. Therefore, we used the structure of the Ddn-F420 complex (PDB 3R5R) as the templates for a structure analysis of Ddn. As shown in Fig. 2A, residues G53, P63, S78, and K79 are located close to the F420 binding site. A detailed analysis of the interaction between Ddn and F420 revealed that the G53C, P63Q, and S78Y mutations introduced unfavorable bumps for receptor-ligand binding (Fig. 3). The K79_G80insG mutation extended the loop from S78 to P87, leading to the loss of local interaction between this loop and the other part of Ddn as well as a change of the binding pocket of F420. According to the structure predicted by AlphaFold (AF-P9WP15-F1), R23 is located on an α-helix at the N terminus of Ddn, and the R23P mutation probably disrupted this α-helix, leading to structural instability. The E118K mutation introduced a positive charge, resulting in charge repulsion between K118 and R72. The side chain of R142 had several hydrogen bonds with residues around the F420 binding site, such as A77, S78, A82, and E83, and the R142F mutation resulted in the loss of the above interaction and might possibly have influenced the binding of F420.
FIG 2.
Structure of the MTB Ddn (A), Fgd1 (B), FbiA (C), and FbiB (D) complexed with cofactors. (A) Ribbon representation of Ddn with cofactor F420 (PDB code 3R5R). The F420 (yellow) is shown with a display style of stick. (B) Ribbon representation of Fgd1 complexed with F420 (PDB code 3B4Y). The F420 (yellow) was displayed in stick. (C) Ribbon representations of the predicted FbiA-FO-GDP complex. The cofactors FO (yellow) and GDP (blue) were displayed in stick. (D) Ribbon representation of FbiB complexed with F420 (PDB code 4XOQ). The F420 (yellow) was displayed in stick. The identified mutated residues (orange) were represented with a display style of ball and stick. The images were obtained using the Discovery Studio Visualizer v.4.5 software.
FIG 3.
The 2D diagram showing the interactions between F420 and wild-type Ddn (A), mutant G53C (B), mutant P63Q (C), and mutant S78Y (D). The F420 molecule is shown in the middle with a display style of ball and stick. The colored balls around F420 indicate the residues involved in the direct interactions between Ddn and F420. The green, purple, and red dashed lines connecting F420 and the corresponding residues indicate intermolecular hydrogen bonds, hydrophobic interactions, and steric hindrance, respectively. Residues involved in hydrogen bonds, van der Waals interactions, or polar interactions are represented by green balls. Residues involved in hydrophobic interactions, attractive charges, and unfavorable bumps are represented by purple, orange, and red balls, respectively.
F420-dependent glucose-6-phosphate dehydrogenase (Fgd1) could provide the reduced cofactor F420-H2 for DLM activation. In our study, all identified mutations in Fgd1 are farther than 8 Å away from the cofactor F420 binding site; thus, these mutations might lead to changes in drug resistance by affecting the structural stability, not by influencing the cofactor binding (Fig. 2B). The side chain of L70 participated in the formation of a hydrophobic core and had hydrophobic interactions with several hydrophobic residues around it. The L70R mutation not only destroyed hydrophobic interactions but also introduced a positive charge, which resulted in a charge repulsion between R70 and R100. M93 and W284 are also located in the hydrophobic cores. Thus, the M93R and W284S mutations also destroyed hydrophobic interactions. Residue 304, located on the surface of the protein, and the hydrophobic side chain of V304 pointed to the hydrophilic solution in the G304V mutation, which might cause structural instability. The L321 mutation, located on an α-helix at the C terminus of Fgd1, as well as the L321P mutation probably disrupted this α-helix, leading to structural instability.
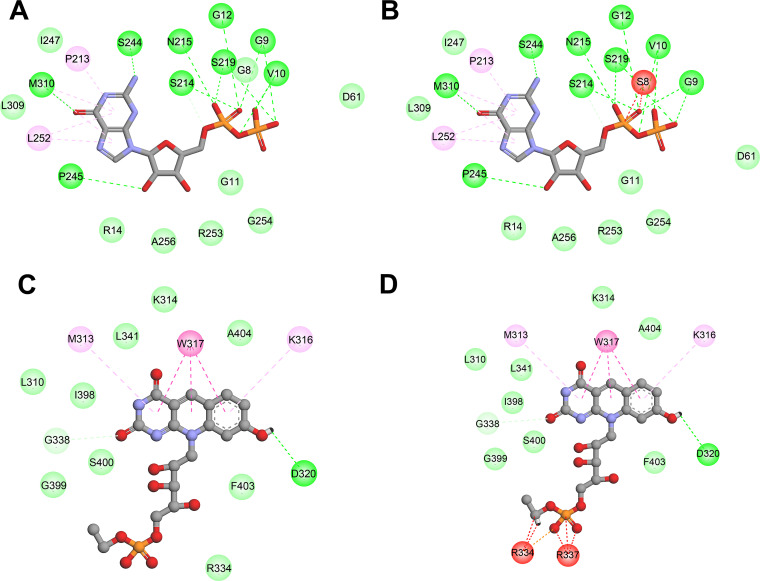
FbiA catalyzes the synthesis of dehydro-F420, using 7,8-didemethyl-8-hydroxy-5-deazariboflavin (FO) and enoylpyruvoyl-2-diphospho-5′-guanosine (EPPG) as the substrates. Thus, we modeled the structure of the FbiA-FO-GDP complex by using the crystal structure of FbiA from M. smegmatis in complex with FO and GDP (PDB 6UW5) as the templates. Among our identified mutations in FbiA, only G8S was located close to the substrate binding site (Fig. 2C), and the G8S mutation introduced unfavorable bumps between S8 and GDP (Fig. 4A and B). The C65R, L143P, A199E, and C259Y mutations might influence the structural stability of FbiA (Table 4). Due to the large side chain of the mutation residue, the C65R, A199E, and C259Y mutations introduced unfavorable bumps between the mutation residues and the residues around them, leading to structural instability. The side chain of L143 participated in the formation of a hydrophobic core and had hydrophobic interactions with several hydrophobic residues around it. The L143P mutation not only destroyed the hydrophobic interactions but also introduced structure bumps between P143 and A43. The calculated free energy for A199T was low, and the A199T mutation did not introduce any unfavorable interactions. Considering that A199T was only found in a double mutant with FbiA A199T and Ddn T40fs, the effect of FbiA A199T on drug resistance was unclear.
FIG 4.
The 2D diagram showing the interactions between GDP and wild-type FbiA(A), mutant G8S (B), and the interactions between F420 and wild-type FbiB (C), mutant I331L (D). The GDP (A and B) and F420 (C and D) molecules are shown in the middle with a display style of ball and stick. The colored balls around GDP or F420 indicate the residues involved in the direct interactions between the ligand and the protein. The green, magenta, and red dashed lines connecting the ligand and the corresponding residues indicate intermolecular hydrogen bonds, hydrophobic interactions, and steric hindrance, respectively. Residues involved in hydrogen bonds, van der Waals interactions, or polar interactions are represented by green balls. Residues involved hydrophobic interactions and unfavorable bumps are represented by magenta and red balls, respectively.
TABLE 4.
The effect of mutations on the protein stability and MIC range of mutants
| Protein | Amino acid change | ΔΔG (kcal/mol)c |
MIC range (μg/mL) |
||
|---|---|---|---|---|---|
| Eris | PremPS | DLM | PTM | ||
| Ddn | R23P | >10 | 0.09 | 0.25 | 16 |
| Ddn | G53C | −5.53 | 0.88 | 0.5 | 8 |
| Ddn | P63Q | 2.14 | 1.08 | >16 | >16 |
| Ddn | S78Y | −1.1 | 0.53 | >16 | >16 |
| Ddn | K79_G80insGa | NA | NA | 1 | 16 |
| Ddn | E118K | 0.43 | 1.39 | >16 | 4 to 16 |
| Ddn | R142F | >10 | −0.18 | 0.125 | 8 to 16 |
| Fgd1 | L70R | >10 | 1.85 | 16 | 2 to 16 |
| Fgd1 | M93R | NA | NA | 16 | 16 |
| Fgd1 | W284S | −0.3 | 1.42 | >16 | >16 |
| Fgd1 | G304V | 0.63 | 1.02 | >16 | 16 |
| Fgd1 | L321P | 3.19 | 2.73 | 0.25 | 0.25 |
| FbiA | G8S | 0.21 | 1.49 | 1 | 8 |
| FbiA | C65R | >10 | 2.25 | 0.25 | 0.25 |
| FbiA | L143P | 4.26 | 3.3 | >16 | 16 |
| FbiA | A199E | 8.18 | 1.88 | 1 | 0.5 |
| FbiA | A199T (& Ddn T40fsb) | −0.05 | 0.62 | 0.5 | 1 |
| FbiA | C259Y (& FbiB I331L) | 2.02 | 1.07 | 16 | 8 |
| FbiB | I331L (& FbiA C259Y/FbiC W719G) | −1.73 | 0.7 | 16 | 8 to 16 |
| FbiC | G258D | 1.64 | 1.26 | >16 | >16 |
| FbiC | L377R | 0.66 | 2.94 | 8 to 16 | 2 to 16 |
| FbiC | E598Q | 2.63 | 0.53 | 16 | >16 |
| FbiC | G604C | 0.12 | 0.49 | 16 | 16 |
| FbiC | H698D | 0.11 | 1.74 | >16 | >16 |
| FbiC | W719G (& FbiB I331L) | 5.29 | 2.19 | >16 | 16 |
ins, insertion but not frameshift.
fs, frameshift mutation.
The free energy (ΔΔG) was calculated for the point mutations in the available protein structures by using two endpoint methods, namely, Eris and PremPS.
FbiB catalyzes the synthesis of F420 from using dehydro-F420 and glutamate as the substrates. Therefore, the structure of the FbiB-F420 complex (PDB 4XOQ) was used as the template for the structure analysis. For FbiB, only I331L was detected in our study, which was close to the ligand F420 (Fig. 2D). I331L mutation introduced unfavorable bumps not only with the ligand F420 (Fig. 4C and D) but also with R334 and R337 of the FbiB protein, thereby influencing both protein stability and ligand binding.
FbiC catalyzes the synthesis of FO. Because no crystal structure was available for FbiC, the structure predicted by AlphaFold (AF-P9WP77-F1) was used for the structure analysis. According to the calculated results of free energy (Table 4), all of the the mutations in FbiC might lead to structural instability; thus, we only analyzed the possible interactions affecting structural stability. The G258D, L377R, and G604C mutations introduced unfavorable bumps. The E598Q mutation lost the negative charge of residue 598, leading to the loss of the electrostatic attractive interaction between E598 and R847. Residue 698, located in a hydrophobic core composed of H698, F736, L702, and V694, as well as the H698D mutation led to the loss of the hydrophobic interaction of H698 with other residues. Residue 719, located in another hydrophobic core, as well as the W719G mutation led to the loss of the hydrophobic interactions of W719 with P716 and V769.
DISCUSSION
In this study, we obtained DLM-resistant progeny strains from DS, MDR, and XDR-MTB clinical isolates and compared their frequencies of spontaneous resistance to DLM in different types of MTB clinical strains for the first time. We also discovered novel mutations in drug resistance genes. Our study provided useful information to improve the understanding of the molecular mechanism and molecular detection of DLM resistance.
First, we examined the spontaneous frequencies of resistance to DLM in DS, MDR, and XDR-MTB strains. The spontaneous resistance frequencies to DLM in the DS-MTB strains and H37Rv that we observed were similar to the findings of previous reports of the spontaneous frequencies of DLM-resistant mutants in MTB H37Rv and in M. bovis BCG Tokyo (5), and they were higher than those of PTM-resistant mutants in MTB H37Rv (9, 11). In addition, the frequencies in the DS-MTB strains were higher than those observed in the MDR and XDR-MTB strains, indicating that the different types of clinical strains had different spontaneous resistance frequencies to DLM. To our knowledge, this is the first study to demonstrate that DS, MDR, and XDR-MTB strains have different spontaneous frequencies of resistance to DLM. In a recent study, Gomez-Gonzalez et al. analyzed 8,622 clinical isolates and found that the average number of mutations in 6 DLM resistance genes per sample among the DS-MTB isolates was higher than those observed in MDR-MTB or XDR-MTB isolates, which suggested that the 3 types of strains have different mutation frequencies in the 6 DLM resistance genes (12). Several studies have found that the presence of drug resistance could impair the fitness of MTB and that the drug-resistant strains may have an increased generation time, compared with drug-susceptible strains (13–15). Consequently, the increased generation time of drug-resistant tubercle bacilli would negatively affect the accumulation of mutations that confer drug resistance and would thereby lead to relatively low mutation rates being noted in drug-resistant bacteria.
In this study, mutations of the known drug-resistant genes ddn, fgd1, fbiA, fbiB, fbiC, and fbiD were detected in 65 (65.00%) DLM-resistant progeny strains. However, 35 (35.00%) DLM-resistant progeny strains had no mutations in the aforementioned 6 genes. Pang et al. reported 4 XDR-MTB clinical isolates that were resistant to DLM, and 2 of them harbored no mutations in these resistance-related genes (16). Kardan-Yamchi et al. found 9 DLM-resistant clinical strains, and only 4 strains had known resistance-related mutations (17). In a previous study by Haver et al., 17% of the PTM-resistant strains were found to have no mutations of known drug-resistant genes (9).The above-mentioned evidence suggests that other genes associated with drug resistance have not been discovered.
Among the mutations induced by DLM in this study, the mutations in ddn accounted for the highest proportion (39.73%), followed by fbiC (21.92%), fgd1 (16.44%), fbiA (16.44%), and fbiB (5.48%). The mutations in ddn comprised a higher proportion of those selected in our study of MTB clinical isolates compared to the proportion selected in a previous study of M. bovis BCG Tokyo (20.00%). On the other hand, the mutations in fgd1 (30.00%) were more frequent in M. bovis BCG Tokyo than in our MTB clinical isolates. The mutation frequencies for fbiC, fbiA, and fbiB (23.33%, 13.33%, and 6.67%, respectively) in M. bovis BCG Tokyo were similar to our findings in the MTB clinical isolates (5). It is worth noting that no mutation in fbiD was found in our study. Whole-genome sequencing data on 8,622 MTB clinical isolates revealed that the fbiD gene accounted for the lowest number of different mutations (n = 66, 7.19% of the total mutations identified) among the 6 DLM-resistant genes (12). However, a PTM treatment study in a mouse model showed that 9.00% of the PTM-resistant isolates were fbiD mutants, and this proportion was higher than those of the fgd1 and fbiB mutants (8), implying that the fbiD mutants may have superior fitness in vivo, relative to other mutants. The mutation frequencies for resistance-related genes were also different between in vitro and in vivo studies on PTM (8, 9), and the drug resistance mutation frequencies reported in clinical strains from different studies were also different (12, 18). The different mutation distributions mentioned above may be due to the different experimental systems used in different studies or the different strains included. Overall, there were more drug resistance mutations in the ddn, fgd1, and fbiC genes, which are directly related to the F420 cycle, and there were relatively few mutations in fbiA, fbiB, and fbiD.
Among the 45 mutations detected, only 7 had related reports. Feuerriegel et al. reported that the MIC of the Ddn R23W mutant to PTM was 0.25 μg/mL (19), whereas the MICs of the Ddn R23P mutant to DLM and PTM found in our study were 0.25 and 16 μg/mL, respectively. R23 is located on the N-terminal α-helix of Ddn, which is not close to the F420 binding site. Considering that proline is a special residue which could disrupt the α-helix, R23P might play a greater influence on the structural stability of Ddn, which is a possible explanation for the different effects of R23P and R23W on the level of drug resistance. The MIC of the Ddn G53C mutant in this study was 0.5 μg/mL, which was close to that reported by Polsfuss et al. (20). Lee et al. (21) observed that the Ddn S78Y mutant was resistant to PTM with a MIC that was 64 times higher than that of H37Rv but remained susceptible to DLM. Further functional experiments confirmed that the S78Y mutant could not activate PTM but did retain the ability to activate DLM. In this study, the S78Y mutant of Ddn was significantly resistant to both DLM and PTM. Considering that only one strain in this study had the S78Y mutation and that the influence of other drug-resistance genes could not be ruled out, further research may be needed to elucidate the effect of this mutation on the MIC. The ddn 434_435insC found in this study and the 434Cdel mutation found by Fujiwara et al. (5) were both frameshift mutations at residue 434, which would lead to high levels of drug resistance. The MIC of the Fgd1 M93R mutant in this study was 16 μg/mL, but the Fgd1 M93T mutant reported by Battaglia et al. was sensitive to DLM, again suggesting that different amino acid substitutions have different effects on drug resistance (18). Our study showed that both the FbiA K250*and the FbiC L377R mutations could cause high-level resistance to DLM and PTM, which was similar to the results of previous studies (8, 22).
The remaining 38 mutations were novel mutant types that were reported for the first time, which greatly enriched the mutation spectrum associated with DLM resistance. Among the 38 new mutation types, 17 frameshift mutations and 3 stop codon mutations seriously affected the structure and integrity of the protein, and almost all of them showed high levels of resistance to DLM/PTM. The other 18 mutations were point mutations, which showed different degrees of drug resistance to DLM/PTM. Among them, only the R142F mutant of Ddn was low-level resistant to DLM (MIC = 0.125 μg/mL) but significantly resistant to PTM (MIC ≥ 8 μg/mL).
Both DLM and PTM are nitroimidazole derivatives that share the same activation pathway, which is related to F420 redox cycling. At the present time, the research on the cross-resistance of the two prodrugs is attracting more attention. Previous studies showed that mutations in the resistance genes related to DLM resistance could also cause resistance to PTM and vice versa (8, 12, 23). We found that the progeny strains induced by DLM treatment were not only resistant to DLM but also generally resistant to PTM. However, the MICs of some mutants to DLM and PTM were not completely consistent. In our study, the MICs of the Ddn R23P, G53C, and R142F mutants to PTM were more than 16 times higher than those to DLM. Lee et al. also found that the Ddn S78Y and Y133C mutants did not activate PTM but retained the ability to activate DLM (21). A possible explanation for this difference is that the binding sites of DLM and PTM on the Ddn protein are not exactly the same. In addition, Rifat et al. also reported that the resistance levels of some FbiB and FbiD mutants to PTM were significantly higher than those to DLM (8), suggesting that the six genes in the activation pathway of the nitroimidazoles might have different effects on the drug resistance of the two drugs.
The most important limitation of this study is that laboratory-induced mutations may not ultimately be reflected in clinical drug-resistant isolates, which may lead to differences between drug-induced mutations and clinical diagnoses. Although the parent strains that we used were clinical isolates that were closer to clinical facts than to laboratory standard strains, these mutations will still need to be identified in clinical isolates to confirm their clinical relevance. Another limitation was that we currently only elucidate the effects of individual mutations on drug resistance by analyzing the correlations between the MICs of the strains and the modifications of the protein structure. In the future, functional studies should be performed to explore the exact mechanism by which each mutation causes resistance.
In conclusion, using DLM for in vitro drug induction and screening, we characterized the mutation spectrum of DLM/PTM resistance that is emerging in clinical strains of MTB. Meanwhile, we also established the association between these candidate variants and their phenotypic resistance profiles. Our findings not only provide evidence for the elucidation of the causative mechanisms of DLM/PTM drug resistance and for the development of molecular detection but also have broad implications for the continued clinical development and usage of nitroimidazole antitubercular agents.
MATERIALS AND METHODS
Ethics statement.
The clinical strains and research protocols applied in this study were approved by the Ethics Committee of Beijing Chest Hospital, Capital Medical University (No. 2016KY05).
Clinical strains.
The strains in this study were classified as DS-TB, MDR-TB, or XDR-TB, using the pre-2021 WHO definition (2). 20 drug-susceptible (DS), 20 MDR, and 20 XDR clinical isolates (collected between March and September of 2016) were randomly selected from the Biobank of Beijing Chest Hospital in China. The phenotypic resistance for rifampicin, isoniazid, fluoroquinolones (including levofloxacin, moxifloxacin, and gatifloxacin), and second-line injectable agents (including amikacin, capreomycin, kanamycin, and streptomycin) was determined via the absolute concentration method on Löwenstein-Jensen (L-J) solid media that contained the anti-TB drugs, according to the recommendation of the WHO.
MIC determination of DLM and PTM.
The MICs of clinical isolates against DLM/PTM were determined via resazurin microtiter assay (REMA) (18, 22, 24, 25). The fresh subcultured MTB clones were harvested from Middlebrook 7H10 agar, and the turbidity of the cultures was adjusted to a 1.0 McFarland standard. For the inoculum, the 1.0 McFarland cell suspension was diluted to 1:20 in Middlebrook 7H9 broth supplemented with 10% OADC. Serial 2-fold dilutions of drugs in 100 μL of 7H9 broth were directly prepared in a 96-well plate. The concentration range for DLM/PTM was 0.016 to 16 μg/mL. 100 μL of the inoculum was added into each well of the prepared 96-well plate. The 96-well plate was incubated at 37°C for 7 days, and then 40 μL of 0.01% fresh resazurin solution were pipetted into each well. After additional 24 h of incubation at 37°C, the MIC was determined as the lowest concentration of drugs that prevented a color change from blue (indicating that no viable MTB cells exist) to pink (indicating that viable MTB cells exist). The MTB H37Rv strain (ATCC 27249) was conducted in all runs as a quality control. Due to the lack of an approved critical concentration for DLM in REMA by the WHO, we used 0.125 μg/mL as the critical MIC for DLM, according to previous studies (16, 18, 22). Clinical isolates with a MIC of DLM lower than 0.125 μg/mL were considered to be drug-susceptible and were chosen as the parent strains for drug resistance induction.
Drug resistance induction.
The clinical strains were inoculated on drug-free L-J media and were cultured to the logarithmic growth phase. To induce drug resistance, the fresh subcultured clones were harvested, and the turbidity of the cultures was adjusted to a 1.0 McFarland standard to obtain the inoculum. 100 μL of the inoculum were inoculated onto 6 7H10 agar plates that contained anti-TB drugs with 2 drug concentrations, namely, 0.063 μg/mL and 0.125 μg/mL (0.5 times and 1 times the critical MIC of DLM), using 2 plates for each concentration. The progeny strains growing on the drug-containing plates were obtained after being cultured for 4 weeks. Due to the large number of progeny strains, one-tenth strains were randomly harvested from each plate for the measurement of their MICs, performance of amplification, and sequencing of drug-resistant genes.
DNA extraction.
Genomic DNA extraction was performed as described previously (16). The freshly cultured bacteria were harvested from the surface of the L-J medium and were transferred into 500 μL Tris-EDTA (TE) buffer. This was followed by centrifugation at 13,000 rpm for 2 min, after which the supernatant was discarded. The pellet was resuspended in 500 μL TE buffer and then heated in a 95°C water bath for 1 h. After the centrifugation of the cellular debris, the crude DNA in the supernatant was used as the template for polymerase chain reaction (PCR).
Spoligotyping.
The genotyping of MTB was performed using a MTB McSpoligotyping Kit (Xiamen Zeesan Biotech, Xiamen, People’s Republic of China) as described previously (26–28). The amplification system was 25 μL/tube, including 19.75 μL McSpoligotyping PCR Mix (A/B/C), 0.25 μL McSpoligotyping enzyme, and 5 μL DNA. The PCR was performed on a Zeesan SLAN96 real-time fluorescent PCR instrument. The PCR cycle program was set according to the instructions: predenaturation at 50°C for 5 min; denaturation at 95°C for 10 min; denaturation at 95°C for 15 s, annealing at 57°C for 15 s, extension at 72°C for 15 s, 50 cycles; supplemental extension at 72°C for 15s. A melt curve analysis was performed as follows: 95°C for 3 min, 35°C for 1 min, and 35 to 90°C with a heating rate of 0.04°C/s to acquire the fluorescence signals for the FAM, HEX, ROX, and CY5 channels. The results were submitted to the SITVIT2 database (http://www.pasteur-guadeloupe.fr:8081/SITVIT2/submit.jsp) for the assignment of their spoligotype international types and corresponding clades.
Amplification and sequencing of resistant genes.
In this study, full-length coding regions for resistance-related genes from different strains were amplified via PCR and analyzed via Sanger sequencing. The genes studied included ddn, fgd1, fbiA, fbiB, fbiC, and fbiD. The primers that were used for the fbiD drug resistance gene amplification and sequencing are 5′-TTATATCGGGTGGTATTGATCTGA-3′ and 5′-AACTCAGCTCCCGGTTCAGGTA-3′. The primers for the other 5 genes used in this study were as reported previously (16). The PCR was performed with a final volume of 50 μL that contained 5 μL 10×PCR buffer, 200 μM each dNTP, 0.2 μM each primer set, and 1 U HotStar Taq polymerase (Qiagen). The amplification products were sent to Tianyihuiyuan Company (Beijing, China) for Sanger sequencing. The DNA sequences were aligned with the sequences of H37Rv using BioEdit Sequence Alignment Editor Version 7.1.3 (https://bioedit.software.informer.com/7.1/).
Structure analysis.
The structure files that were used as the templates for the structure analysis were obtained from the Protein Data Bank (PDB) (https://www.rcsb.org/) and the AlphaFold Protein Structure Database (http://alphafold.ebi.ac.uk/). Because only the core structure of Ddn (from R31 to P151) had been determined via experimentation, the structure predicted by AlphaFold (AF-P9WP15-F1) was used as the template for mutant R23P, and the crystal structure of Ddn with cofactor F420 (PDB file 3R5R) was used as the templates for the other Ddn mutants. The templates for the mutants of Fgd1 and FbiB were PDB files 3B4Y (crystal structure of Fgd1 complexed with F420) and 4XOQ (crystal structure of the C-terminal domain of FbiB with F420), respectively. The wild structure of FbiA was modeled via SWISS-MODEL (http://swissmodel.expasy.org/), using the crystal structure of FbiA from M. smegmatis in complex with FO and GDP (PDB file 6UW5) as the template. The structures predicted by AlphaFold (AF-P9WP77-F1) were used as the templates for the mutants of FbiC.
The structures of the mutants were also modeled via SWISS-MODEL. To predict the impact on protein stability, the free energy changes induced by the point mutations were calculated via Eris (29) and PremPS (30). The detailed intramolecular and protein-ligand interactions were analyzed with the Discovery Studio Visualizer v.4.5 software package (BIOVIA, Dassault Systèmes, San Diego, CA, USA), using the “Structure Monitor” and “Receptor-Ligand Interactions” modules.
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (81401739) and the Beijing Key Clinical Specialty Project (20201214).
We declare that there are no conflicts of interest.
J.L. and M.G. developed the concept and designed the experiments. J.S. and Y.P. assisted with the design. Y.L., L.L., and T.W. conducted the in vitro MIC experiment. Y.L., L.L., and J.S. carried out the selection of spontaneous-resistant mutants. Y.L., J.S., and L.L sequenced and analyzed the spontaneous mutants. J.L. and Y.L. performed the structure analysis. Y.P., P.C., M.G., and Y.G. analyzed the results. Y.L. and J.L. wrote the manuscript with critical input from M.G.
Contributor Information
Mengqiu Gao, Email: gaomqwdm@aliyun.com.
Jie Lu, Email: lujiebch@163.com.
REFERENCES
- 1.WHO. 2020. Global tuberculosis report. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 2.WHO. 2020. Meeting report of the WHO expert consultation on the definition of extensively drug-resistant tuberculosis.
- 3.Alene KA, Yi H, Viney K, McBryde ES, Yang K, Bai L, Gray DJ, Clements ACA, Xu Z. 2017. Treatment outcomes of patients with multidrug-resistant and extensively drug resistant tuberculosis in Hunan Province, China. BMC Infect Dis 17:573. 10.1186/s12879-017-2662-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matsumoto M, Hashizume H, Tomishige T, Kawasaki M, Tsubouchi H, Sasaki H, Shimokawa Y, Komatsu M. 2006. OPC-67683, a nitro-dihydro-imidazooxazole derivative with promising action against tuberculosis in vitro and in mice. PLoS Med 3:e466. 10.1371/journal.pmed.0030466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fujiwara M, Kawasaki M, Hariguchi N, Liu Y, Matsumoto M. 2018. Mechanisms of resistance to delamanid, a drug for Mycobacterium tuberculosis. Tuberculosis (Edinb) 108:186–194. 10.1016/j.tube.2017.12.006. [DOI] [PubMed] [Google Scholar]
- 6.Gler MT, Skripconoka V, Sanchez-Garavito E, Xiao H, Cabrera-Rivero JL, Vargas-Vasquez DE, Gao M, Awad M, Park SK, Shim TS, Suh GY, Danilovits M, Ogata H, Kurve A, Chang J, Suzuki K, Tupasi T, Koh WJ, Seaworth B, Geiter LJ, Wells CD. 2012. Delamanid for multidrug-resistant pulmonary tuberculosis. N Engl J Med 366:2151–2160. 10.1056/NEJMoa1112433. [DOI] [PubMed] [Google Scholar]
- 7.WHO. 2020. WHO consolidated guidelines on tuberculosis, module 4: treatment – drug-resistant tuberculosis treatment. World Health Organization, Geneva, Switzerland. [PubMed] [Google Scholar]
- 8.Rifat D, Li SY, Ioerger T, Shah K, Lanoix JP, Lee J, Bashiri G, Sacchettini J, Nuermberger E. 2020. Mutations in fbiD (Rv2983) as a novel determinant of resistance to pretomanid and delamanid in Mycobacterium tuberculosis. Antimicrob Agents Chemother 65. 10.1128/AAC.01948-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haver HL, Chua A, Ghode P, Lakshminarayana SB, Singhal A, Mathema B, Wintjens R, Bifani P. 2015. Mutations in genes for the F420 biosynthetic pathway and a nitroreductase enzyme are the primary resistance determinants in spontaneous in vitro-selected PA-824-resistant mutants of Mycobacterium tuberculosis. Antimicrob Agents Chemother 59:5316–5323. 10.1128/AAC.00308-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stover CK, Warrener P, VanDevanter DR, Sherman DR, Arain TM, Langhorne MH, Anderson SW, Towell JA, Yuan Y, McMurray DN, Kreiswirth BN, Barry CE, Baker WR. 2000. A small-molecule nitroimidazopyran drug candidate for the treatment of tuberculosis. Nature 405:962–966. 10.1038/35016103. [DOI] [PubMed] [Google Scholar]
- 11.Manjunatha UH, Boshoff H, Dowd CS, Zhang L, Albert TJ, Norton JE, Daniels L, Dick T, Pang SS, Barry CE 3rd. 2006. Identification of a nitroimidazo-oxazine-specific protein involved in PA-824 resistance in Mycobacterium tuberculosis. Proc Natl Acad Sci USA 103:431–436. 10.1073/pnas.0508392103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gomez-Gonzalez PJ, Perdigao J, Gomes P, Puyen ZM, Santos-Lazaro D, Napier G, Hibberd ML, Viveiros M, Portugal I, Campino S, Phelan JE, Clark TG. 2021. Genetic diversity of candidate loci linked to Mycobacterium tuberculosis resistance to bedaquiline, delamanid and pretomanid. Sci Rep 11:19431. 10.1038/s41598-021-98862-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Andersson DI, Levin BR. 1999. The biological cost of antibiotic resistance. Curr Opin Microbiol 2:489–493. 10.1016/s1369-5274(99)00005-3. [DOI] [PubMed] [Google Scholar]
- 14.Kodio O, Georges Togo AC, Sadio Sarro YD, Fane B, Diallo F, Somboro A, Degoga B, Kone M, Coulibaly G, Tolofoudje M, Bane S, Sanogo M, Kone B, Coulibaly N, Dabitao D, Baya B, Maiga M, Bougoudogo F, Samake F, Dao S, Doumbia S, Diallo S, Diarra B. 2019. Competitive fitness of Mycobacterium tuberculosis in vitro. Int J Mycobacteriol 8:287–291. 10.4103/ijmy.ijmy_97_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bhatter P, Chatterjee A, D'Souza D, Tolani M, Mistry N. 2012. Estimating fitness by competition assays between drug susceptible and resistant Mycobacterium tuberculosis of predominant lineages in Mumbai, India. PLoS One 7:e33507. 10.1371/journal.pone.0033507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pang Y, Zong Z, Huo F, Jing W, Ma Y, Dong L, Li Y, Zhao L, Fu Y, Huang H. 2017. In vitro drug susceptibility of bedaquiline, delamanid, linezolid, clofazimine, moxifloxacin, and gatifloxacin against extensively drug-resistant tuberculosis in Beijing, China. Antimicrob Agents Chemother 61. 10.1128/AAC.00900-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kardan-Yamchi J, Kazemian H, Battaglia S, Abtahi H, Foroushani AR, Hamzelou G, Cirillo DM, Ghodousi A, Feizabadi MM. 2020. Whole genome sequencing results associated with minimum inhibitory concentrations of 14 anti-tuberculosis drugs among rifampicin-resistant isolates of Mycobacterium tuberculosis from Iran. J Clin Med 9. 10.3390/jcm9020465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Battaglia S, Spitaleri A, Cabibbe AM, Meehan CJ, Utpatel C, Ismail N, Tahseen S, Skrahina A, Alikhanova N, Mostofa Kamal SM, Barbova A, Niemann S, Groenheit R, Dean AS, Zignol M, Rigouts L, Cirillo DM. 2020. Characterization of genomic variants associated with resistance to bedaquiline and delamanid in naive Mycobacterium tuberculosis clinical strains. J Clin Microbiol 58. 10.1128/JCM.01304-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feuerriegel S, Koser CU, Bau D, Rusch-Gerdes S, Summers DK, Archer JA, Marti-Renom MA, Niemann S. 2011. Impact of Fgd1 and ddn diversity in Mycobacterium tuberculosis complex on in vitro susceptibility to PA-824. Antimicrob Agents Chemother 55:5718–5722. 10.1128/AAC.05500-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Polsfuss S, Hofmann-Thiel S, Merker M, Krieger D, Niemann S, Russmann H, Schonfeld N, Hoffmann H, Kranzer K. 2019. Emergence of low-level delamanid and bedaquiline resistance during extremely drug-resistant tuberculosis treatment. Clin Infect Dis 69:1229–1231. 10.1093/cid/ciz074. [DOI] [PubMed] [Google Scholar]
- 21.Lee BM, Harold LK, Almeida DV, Afriat-Jurnou L, Aung HL, Forde BM, Hards K, Pidot SJ, Ahmed FH, Mohamed AE, Taylor MC, West NP, Stinear TP, Greening C, Beatson SA, Nuermberger EL, Cook GM, Jackson CJ. 2020. Predicting nitroimidazole antibiotic resistance mutations in Mycobacterium tuberculosis with protein engineering. PLoS Pathog 16:e1008287. 10.1371/journal.ppat.1008287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schena E, Nedialkova L, Borroni E, Battaglia S, Cabibbe AM, Niemann S, Utpatel C, Merker M, Trovato A, Hofmann-Thiel S, Hoffmann H, Cirillo DM. 2016. Delamanid susceptibility testing of Mycobacterium tuberculosis using the resazurin microtitre assay and the BACTEC MGIT 960 system. J Antimicrob Chemother 71:1532–1539. 10.1093/jac/dkw044. [DOI] [PubMed] [Google Scholar]
- 23.Wen S, Jing W, Zhang T, Zong Z, Xue Y, Shang Y, Wang F, Huang H, Chu N, Pang Y. 2019. Comparison of in vitro activity of the nitroimidazoles delamanid and pretomanid against multidrug-resistant and extensively drug-resistant tuberculosis. Eur J Clin Microbiol Infect Dis 38:1293–1296. 10.1007/s10096-019-03551-w. [DOI] [PubMed] [Google Scholar]
- 24.Lopez B, Siqueira de Oliveira R, Pinhata JMW, Chimara E, Pacheco Ascencio E, Puyen Guerra ZM, Wainmayer I, Simboli N, Del Granado M, Palomino JC, Ritacco V, Martin A. 2019. Bedaquiline and linezolid MIC distributions and epidemiological cut-off values for Mycobacterium tuberculosis in the Latin American region. J Antimicrob Chemother 74:373–379. 10.1093/jac/dky414. [DOI] [PubMed] [Google Scholar]
- 25.Palomino JC, Martin A, Camacho M, Guerra H, Swings J, Portaels F. 2002. Resazurin microtiter assay plate: simple and inexpensive method for detection of drug resistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother 46:2720–2722. 10.1128/AAC.46.8.2720-2722.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zeng X, Xu Y, Zhou Y, Li H, Zheng R, Tan Y, Sun B, Zhao Y, Li Q. 2018. McSpoligotyping, a one-step melting curve analysis-based protocol for spoligotyping of Mycobacterium tuberculosis. J Clin Microbiol 56. 10.1128/JCM.00539-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang J, Pang Y, Zhang T, Xian X, Li Y, Wang R, Wang P, Zhang M, Wang J. 2020. Molecular characteristics and in vitro susceptibility to bedaquiline of Mycobacterium tuberculosis isolates circulating in Shaanxi, China. Int J Infect Dis 99:163–170. 10.1016/j.ijid.2020.07.044. [DOI] [PubMed] [Google Scholar]
- 28.Pan J, Ye H, Wu Z, Xiao J, Liu J. 2021. One-step melting curve analysis-based McSpoligotyping reveals genotypes of Mycobacterium tuberculosis in a coastal city, China. Arch Microbiol 203:4579–4585. 10.1007/s00203-021-02441-0. [DOI] [PubMed] [Google Scholar]
- 29.Yin S, Ding F, Dokholyan NV. 2007. Eris: an automated estimator of protein stability. Nat Methods 4:466–467. [DOI] [PubMed] [Google Scholar]
- 30.Chen Y, Lu H, Zhang N, Zhu Z, Wang S, Li M. 2020. PremPS: predicting the impact of missense mutations on protein stability. PLoS Comput Biol 16:e1008543. 10.1371/journal.pcbi.1008543. [DOI] [PMC free article] [PubMed] [Google Scholar]