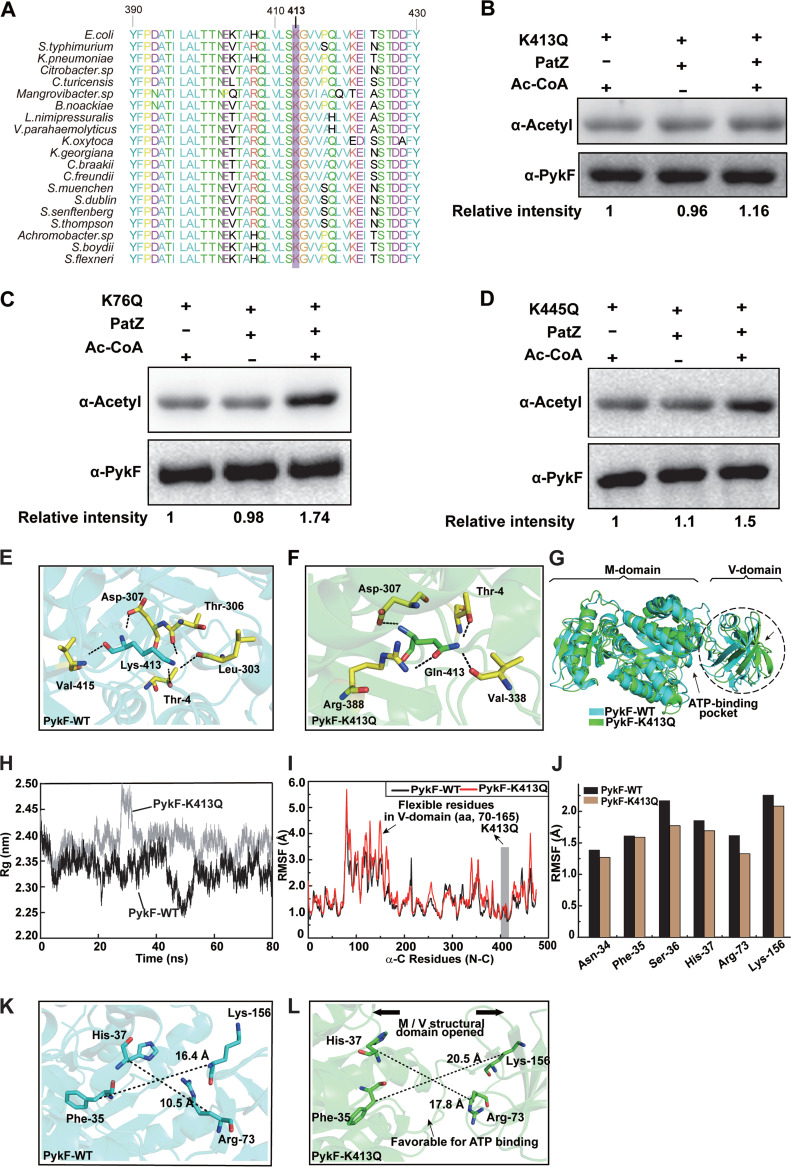
FIG 4.
K413 of PykF is the acetylation site by PatZ. (A) Conservation analysis of PykF K413 in E. coli through sequence alignment. Purple frame denotes conserved lysine residues, and the result was analyzed by BioEdit v7.0. (B to D) PatZ acetylates K413Q, K76Q, and K445Q in vitro. (E) Hydrogen bonds between Lys-413 and the surrounding residues within 3.5 Å. (F) Hydrogen bonds between Gln-413 and its surrounding residues. (G) The overall display of protein structure includes two domains (M-domain and V-domain). Comparison of the average conformations of PykF-WT and PykF-K413Q in an equilibrium state. Dashed box represents the three-dimensional structure area with the largest conformational difference. The active center of ATP is marked by a black arrow. (H) Time evolution of the C-alpha of the radius of gyration (Rg) values. (I) The root-mean-square fluctuation (RMSF) values of the C-alpha for all residues of PykF-WT and PykF-K413Q proteins were calculated over the 80-ns trajectory. The RMSF value of residues in the V-domain of PykF-K413Q is higher than that of PykF-WT (black arrow). (J) RMSF of the C-alpha profiles of residues in the ATP active binding sites of PykF-WT and PykF-K413Q MD trajectories. (K to L) The differences in the ATP-binding pocket located between the V- and M-domains in PykF-WT or PykF-K413Q were compared, and the distances between the selected residues were measured.