Abstract
Background: The importance of gut microbiota in human health is being increasingly studied. Imbalances in gut microbiota have been associated with infection, inflammation, and obesity. Antibiotic use is the most common and significant cause of major alterations in the composition and function of the gut microbiota and can result in colonization with multidrug-resistant bacteria. Methods: The purpose of this review is to present existing evidence on how microbiota modulation and prevention of gut dysbiosis can serve as tools to combat antimicrobial resistance. Results: While the spread of antibiotic-resistant pathogens requires antibiotics with novel mechanisms of action, the number of newly discovered antimicrobial classes remains very low. For this reason, the application of alternative modalities to combat antimicrobial resistance is necessary. Diet, probiotics/prebiotics, selective oropharyngeal or digestive decontamination, and especially fecal microbiota transplantation (FMT) are under investigation with FMT being the most studied. But, as prevention is better than cure, the implementation of antimicrobial stewardship programs and strict infection control measures along with newly developed chelating agents could also play a crucial role in decreasing colonization with multidrug resistant organisms. Conclusion: New alternative tools to fight antimicrobial resistance via gut microbiota modulation, seem to be effective and should remain the focus of further research and development.
Keywords: Microbiome, resistome, gut microbiota, antimicrobial resistance, fecal bacteriotherapy, fecal microbiota transplantation, antimicrobial stewardship, dysbiosis, multidrug-resistant organisms, prebiotics, probiotics, beta-lactamases, charcoal agent
Introduction
Humans are colonized with a very large number of microorganisms (bacteria, archaea, viruses, and unicellular eukaryotes) immediately after birth. These commensal and symbiotic microorganisms, which were formerly believed to outnumber human cells 10-fold, but now their ratio is estimated closer to 1:1, comprise the host microbiota. [1,2]. The term microbiota was first described by Joshua Lederberg referring to the community of all microorganisms residing in the human body and their collective genome [3]. The majority of colonizing bacteria are found in the gastrointestinal tract (GIT), with approximately two-thirds of all microorganisms, residing in the colon [4]. The gut microbiota includes several hundred to more than 1,000 species [5]. The predominant organisms are anaerobes, followed by facultative anaerobes and aerobic bacteria and predominant phyla are Firmicutes and Bacteroidetes, followed by Proteobacteria, Fusobacteria, Cyanobacteria, Verrucomicrobia, and Actinobacteria [6].
It is known that the microbial populations residing in the GIT change throughout life in terms of both content and function [7,8]. Each individual has a unique GIT microbiota at genus and species level, influenced by host genetics, ethnicity, diet, early microbial exposure, environmental conditions, lifestyle, and immune and overall health status. However, the composition of the human microbiota is fairly stable at the phylum level. The major phyla that dominate the human intestine are conserved between all individuals, although the proportions of these groups can vary [9]. Other factors that likely physiologically contribute to microbiota variations between individuals include type of delivery, feeding pattern, diet and age-related changes in the GIT, namely low-grade inflammation [10]. Although the microbiota composition differs between individuals, certain functions encoded in the gut microbiota (core microbiota) are shared between individuals [11]. Some of these functions that are most important to the host are digestion of polysaccharides, vitamin production, lipid metabolism, regulation of the host immune response, and protection against pathogenic organisms [12].
Alterations in the composition of gut microbiota, known as dysbiosis, can be induced by several exogenous factors, with antimicrobial use probably being the most important one. Dysbiosis can promote disease, impair immune responses, but also facilitate colonization resistance imbalance and a shift to predominance of resistant pathogens [13]. The problem of increasing antimicrobial resistance worldwide is an imminent public health threat. The discovery pace of new antimicrobials cannot catch up with the development and spread of novel resistance mechanisms. Hence, there is need for novel effective preventive or therapeutic approaches, at individual or population level, against these difficult to eradicate resistant pathogens.
The purpose of this review is to describe how antimicrobial use affects the human microbiota towards the development of antimicrobial resistance and to present existing evidence on the role microbiota modulation strategies in reducing antimicrobial resistance potential.
Methods/Data Search
Literature search included articles published in English, until April 2022, belonging to journals indexed in PubMed. We also searched the reference lists of the initial papers for further relevant articles.
The Importance of Microbiota in Health and Disease
The importance of microbiota in human health is being increasingly recognized. The role of gut microbiota in several diseases has been well studied (inflammatory bowel disease, obesity, diabetes mellitus, irritable bowel syndrome, colorectal cancer) and its association with many others is currently being investigated [6,14]. Gut microbiota plays a fundamental role in the development of both local and systemic immunity. Specifically, it provides its host with a physical barrier to invading pathogens by competitive exclusion, and production of antimicrobial products and it also stimulates the host to produce various antimicrobial compounds [12,15-17]. Some of its other beneficial functions include digestion of plant polysaccharides and host glycans in the colon, production of essential vitamins, functional and structural maturation of the GIT and development of the intestinal surface area and microvasculature [18]. Moreover, a healthy gut microbiota influences the gut-brain axis and shapes stress related symptoms such as anxiety and pain [19] and is also implicated in appetite control [20].
Gastrointestinal microbiota contribute in the regulation of gut homeostasis by maintaining epithelial barrier integrity, stimulating angiogenesis, inducing T regulatory cells, and by their anti-inflammatory and immunostimulatory properties [21]. Gut barrier function is also regulated by the brush border enzyme, intestinal alkaline phosphatase (IAP), whose absence has been associated in the pathophysiology of certain diseases such as inflammatory bowel disease, necrotizing enterocolitis, metabolic syndrome, and type 2 diabetes mellitus [22,23].
A quantitative, qualitative, metabolic, or locational imbalance of gut commensals, called dysbiosis [24], may be associated with diseases like dental plaque, bacterial vaginosis, psoriasis, atopic dermatitis, asthma, inflammatory bowel disease, diabetes, obesity, colon cancer, and recurrent Clostridium difficile infection (RCDI) [25]. Critical illness has also been associated with the loss of normal, “health promoting” bacteria [26].
Effect of Antimicrobial Use on the Microbiota and Resistome
Disturbance in Composition and Function
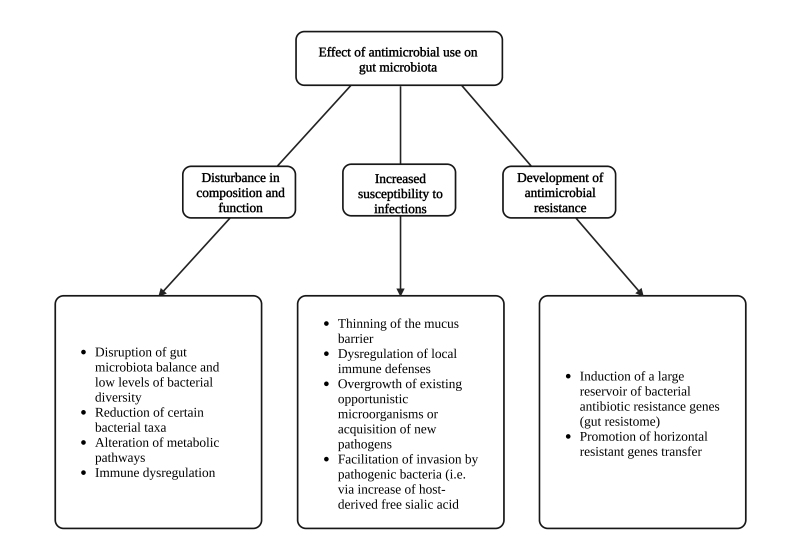
Many factors can harm the beneficial GIT microbiota, including antibiotic use, psychological and physical stress, radiation, altered GIT peristalsis, gastrointestinal infections, and dietary changes [27]. Antibiotic use is the most common and significant cause of major alterations in the composition and function of the normal gut microbiota [28] (Figure 1). Antibiotics cause serious alterations in gut microbiota which result in low levels of bacterial diversity, and expansions of the presence of certain taxa [29]. The potential for an antimicrobial agent to influence gut microbiota is related to its spectrum of activity, mode of action, potency, pharmacokinetics, dosage and length of administration [30], but is also associated with the existing microbiota of the host and the presence of antimicrobial resistance genes in this community [31].
Figure 1.
Effect of antimicrobial use on gut microbiota.
Altered diversity of gut bacteria can lead to irritable bowel syndrome or infections by gut pathogens such as C. difficile, inherently resistant to many antimicrobials [32]. Additional unintended consequences of antibiotic use on gut microbiota include the selection for a reservoir of bacterial antibiotic resistance (AR) genes, promotion of horizontal gene transfer between bacterial strains, increased populations of enteric bacteria through altered carbohydrate composition, depletion of vitamin-producing bacteria, changes in metabolic activities that contribute to nutrition and immune dysregulation [29].
Increased Susceptibility to Infections
Besides alteration of microbiota composition, antibiotics also interfere with local gut immune defenses [33]. For example, decreased IL-17 and INF-γ production, was observed in the small intestine and decreased numbers of Treg cells in the colon post antimicrobials [34]. Hence, dysbiosis is likely an effect not only of the bactericidal properties of antibiotics, but also of the altered host-microbiota interactions [35]. Additionally, the gut microbiota induces mucin production, while antibiotics result in thinning of the mucus barrier thereby increasing susceptibility to bacterial invasion [36]. Alterations of the bacterial populations, which normally colonize the gut lumen, may result in intestinal infections either from newly acquired pathogens or from the overgrowth and pathogenic potential of opportunistic microorganisms.
It is of interest that antibiotic exposure, by altering the gut microbiota and by changing the balance between species, as well as their interactions, can lead to an increase in host-derived free sialic acid, which may result in easier invasion by pathogens such as Salmonella typhymorium [9,37]. Several studies have been performed in infants treated with antibiotics, especially preterm ones. Treatment with various antibiotics such as cephalexin, gentamicin, vancomycin, and erythromycin altered the normal bacterial microbiota of infants increasing the percentage of potentially pathogenic Enterobacteriaceae and lowering the amount of bacteria like Bifidobacteriaceae, Bacilli, and Lactobacillus which are part of the healthy microbiota [38].
Development of Antimicrobial Resistance
Abuse of antibiotics has led to the development of multidrug resistant organisms (MDROs). Infections with MDROs are a major cause of morbidity and mortality worldwide [39].
Microbes can develop defensive mechanisms and employ resistance mechanisms against the agents used for their elimination. The human gut microbiota harbors a large reservoir of resistance genes, named as the gut resistome. Using metagenomic sequencing, Forslund et al. were able to detect resistance genes for 50 of 68 classes of antibiotics in 252 fecal metagenomes from individuals from different continents with an average of 21 AR genes per sample [40]. Additionally, Hu et al., again in an international cohort of 162 persons, identified a total of 1093 AR genes [41].
The gut resistance reservoir encompasses naturally occurring bacteria, bacteria with acquired resistance genes and acquired bacteria, harboring resistance genes, which do not normally colonize the gut [42,43]. The latter may survive and dominate in the gut microbiota for a long period of time. Transfer of resistance genes or virulence traits between non-pathogenic and pathogenic isolates is possible although not common. One example is the vanB-type vancomycin resistance transposon, which is commonly carried by anaerobic gut commensals of the phylum Firmicutes [44], and can be transferred to Enterococcus faecium, rendering it resistant to vancomycin [45]. As it has been shown from experimental studies but also in human cohorts, decreased gut microbiota variability can decrease colonization resistance and facilitate colonization of pathogenic and MDROs [33,46].
Gut Microbiota Modulation as a Tool Against Antimicrobial Resistance
While the spread of antibiotic-resistant pathogens requires antibiotics with novel mechanisms of action, the number of new antimicrobials approved for therapy remains very low [39]. For this reason, an intriguing alternative modality to combat antimicrobial resistance could be the modulation of the gut microbiota. We describe the most important interventions that have been employed for targeted as well as non-specific microbiota modulation (Table 1).
Table 1. Gut Microbiota Modulation as a Tool Against Antimicrobial Resistance.
| Intervention | Mechanism of action | Advantages | Disadvantages | References |
| Diet and dietary supplements | -Wide variety, low fat and plant polysaccharide diet, low protein or
high in fiber diets preserve the bacterial diversity of the gut
microbiome and the colonization resistance. -Addition to the diet of substances such as konjac glucomannan, HMOs or some Chinese remedies protect the gut microbiome and promote its restoration. -Oral administration of IAP which maintains or even restores gut microbiota. |
-Simple to apply. -Accessible to everyone. -Naturally derived components so fewer side effects. |
-Lack of evidence in humans. | [53-57,59-61] |
| Prebiotics and probiotics | -Prebiotics stimulate, while probiotics serve as, lactobacilli or
bifidobacterial that reduce the growth or interfere with the survival of
pathogenic microorganisms in the gut. -Targeted eradication of pathogens by newly developed engineered probiotics. |
-Easily accessible and administered. | -Difficult to find the most suitable probiotic for each dysbiosis
condition. -Data mainly on ICU patients. -Conflicting results in protecting gut microbiota from MDROs, especially gram(-) pathogens. -Reports of bacteremias in ICU patients. |
[65,66,71,73] |
| Fecal Microbiota Transplantation (FMT) | -Infusion of donor feces into patient’s gut (administered mainly orally)
in order to repopulate it with a healthy and balanced microbiota as a
weapon against C. difficile infections (especially
recurrent), as a “barrier” to colonization by multi-drug resistant
bacteria, and as a method of reducing the load of antibiotic resistance
genes. -Enhancing host responses. |
-Successful against difficult-to-treat situations. -High rates of effectiveness. |
-Lack of large randomized clinical trials. -Incidents of serious adverse events. |
[9,88-90,92,93,95-97,103,108,111-113] |
| Antimicrobial compounds | -Use of bacteriocins to inhibit the growth of pathogenic bacteria and preservation of gut microbiota. | -Targeted therapy. -Avoidance of using broad-spectrum antibiotics. |
-Lack of scientific data. -No available clinical trials. |
[119] |
| Selective Digestive Decontamination (SDD) and Selective Oropharyngeal Decontamination (SOD) | -Prophylactic use of antibiotics to reduce the gut colonization with MDROs. | -Successful into wards with low rates of resistant bacteria. | -Lack of data in centers with high rates of resistance. -Need for rigorous surveillance of patients. |
[122-124,124,126,127] |
HMOs: human milk oligosaccharides, IAP: intestinal alkaline phosphatase, ICU: intensive care unit, MDRO: multi-drug resistant organism
Diet and Dietary Supplements
Dietary composition affects the makeup and genetic diversity of gut microbiota. The role of individual dietary components, predominantly the ratio and type of protein, carbohydrates, and fat intake in gut microbiota variation is increasingly being studied [47]. It was shown that the feces of omnivores contained more species of the Clostridial clusters IV and XIVa, bacteria which are able to convert fiber to short chain fatty acids (SCFAs), compared with those of vegetarians and lactovegetarians [48,49]. SCFAs can regulate the expression of virulence genes of Salmonella spp. or E. coli in vitro [50]. Moreover, in a study where mice were fed a “Western” high-fat/simple carbohydrate or a low-fat/complex plant polysaccharide diet, the former had less bacterial diversity, a lower proportion of Bacteroidetes and an increased proportion of Firmicutes compared to the group which received a low-fat diet [51].
We know that limited bacterial diversity is considered an “unhealthy” microbiota [52], which is a risk factor for decreased colonization resistance. Elderly individuals who resided in long-term facilities and had limited variety in their diet, also had decreased gut bacterial diversity, compared to their counterparts that resided in the community; this was associated with worse health status [53]. In another experimental model, a high-protein diet disrupted gut microbiota, suggesting that avoiding a high-protein diet could help preserve colonization resistance [54]. On the other hand, a diet high in fibers was associated with a quicker restoration of the gut microbiota after antibiotic exposure when compared to a high-protein diet [55]. There are also reports of specific substances such as konjac glucomannan (glucomannan derived from Amorphophallus konjac, a plant with edible tubers) with protective effects on the gut microbiota [56], and reports of Chinese dietary remedies that have a beneficial result on the gut restoration [57,58]. Other studies found that chemically created human milk oligosaccharides (HMOs), if given as a dietary supplement, could restore human gut microbiota by promoting the development of beneficial commensal bacteria (bifidobacteria) [59,60]. In addition, oral supplementation of IAP has been linked with the maintenance or even restoration of normal gut microbiota after its disruption [61]. In two experimental mouse models, oral supplementation of IAP was effective in preventing infections from Salmonella enterica (serovar Typhimurium), C. difficile, and possibly other pathogens, by restoring commensal gut microbiota [62]. An ongoing cross-sectional study, named the Wisconsin microbiota study, will provide us with fruitful information about the relation between diet, gut microbiota, and MDROs [63]. Although, relevant studies in humans are still lacking, modulation of diet in order to increase bacterial diversity could serve as an adjuvant strategy in order to decrease antimicrobial resistance.
Prebiotics and Probiotics
Prebiotics are defined as “selectively fermented ingredients that allow specific changes, both in the composition and/or activity in the GI microbiota that confer benefits upon host well-being and health” [64]. Prebiotics are considered to stimulate lactobacilli or bifidobacteria growth and have been associated with beneficial effects in human metabolism through modulation of the gut microbiota [65,66]. In a murine study, diet supplementation with SCFAs or fructooligosaccharides caused a shift in microbiota composition [67]. Similarly, fructooligosaccharides were found to result in a reinstitution of the gut microbiota [68], while other inulin-type probiotics were found to inhibit the disruption of gut microbiota by preserving the commensal bacteria [69].
Probiotics are “live microorganisms which when administered in adequate amounts confer a health benefit on the host” as defined by the World Health Organization [70]. The most commonly used probiotics are strains from the genera Lactobacillus and Bifidobacterium. According to Rijkers et al., probiotics exert their beneficial role in three ways, namely by interfering with the growth or survival of pathogenic microorganisms in the gut lumen, by improving mucosal barrier function or mucosal immune system and by having an effect on the systemic immune system and other organs [71]. Lactobacillus plantarum and L. acidophilus were shown to reduce enteric counts of multidrug resistant enteroaggregative E. coli in an experimental murine study [72]. However, because of the great interindividual variability of gut microbiota, and the different mechanisms with which dysbiosis promotes disease, more research is needed to determine the most suitable probiotic for each dysbiosis-related condition.
More recently, engineered probiotics have been employed for targeted P. aeruginosa eradication in two studies. These probiotics are programmed to detect quorum sensing molecules and upon detection of the pathogen, they express antimicrobial compounds or activate other previously engineered mechanisms in order to eradicate their target [73]. Apart from probiotics, phages have also been the focus of genetic engineering to enable targeted killing of bacteria with AR or specific virulence traits [74]. Although there are many systematic reviews with a recent umbrella review focusing on the role of probiotics on reducing infections among critically ill patients, there are no large studies examining the effect of probiotics on colonization resistance [75]. There are many studies showing their effectiveness on gram-positive bacteria, such as MRSA or VRE [76-78], while the outcome in gram-negative pathogens is disappointing [79,80].
The current need for the development of future probiotics is to determine which bacteria could enhance colonization resistance, as well as to design a more customized probiotic administration [81]. Finally, as there are reports of clinically significant bacteremias with bacterial strains contained in probiotic supplements in ICU patients, it is advisable to use them with caution in this population [82,83], and always in the context of a clinical trial.
Fecal Microbiota Transplantation
Fecal microbiota transplantation (FMT), the transplantation of stool from a healthy donor into the GIT of a patient, was first used 1,700 years ago, in China, to treat food poisoning and severe persistent diarrhea [84]. The first case of application of this method in the treatment of pseudomembranous colitis was described by Eiseman in 1958 [85]. In recent years, and most importantly after the epidemic of the hypervirulent BI/NAP1/027 strain in North America, donor feces infusion is used for the treatment of RCDI, showing excellent results with high cure rate, up to 90%, and minimal adverse effects [86,87].
FMT has shown important clinical success in eliminating C. difficile, in decreasing C. difficile relapses and in resolving C. difficile infection (CDI) associated symptoms. Patients suffering from RCDI have decreased diversity of bacterial species and reduced number of Bacteroidetes and Firmicutes phyla in their feces compared with patients experiencing the first episode of CDI or antibiotic-associated diarrhea (AAD) [88]. The rationale of using FMT is to repopulate the colon with a healthy and balanced microbiota, characterized by wide diversity and to displace harmful bacteria, which colonize the gut [89].
It is believed that the transplanted microbiota may hinder colonization by pathogenic bacteria through changes in the luminal microenvironment and antagonism for nutrients and binding sites. Also, pathogenic bacteria can detect microbiota derived signals or host derived signals that have been modified by the microbiota, which alter their virulence or colonization potential [9]. Such mechanisms may also come into play for the inhibition of colonization by multi-drug resistant pathogens, although strong data to support such an argument is still lacking.
Also, there is evidence that changes in gut microbial metabolites may enhance host responses. Enrichment in secondary bile acids after FMT is associated with alterations in regulatory T cells [90]. FMT has also been associated with restoration of IgA mediated interactions, and T cell populations, even reversing a CDI-related immunosenescent phenotype [91]. Murine models also suggest a beneficial immune response associated with FMT [92,93]. In one study, ceftriaxone-induced dysbiosis leading to intestinal membrane compromise and increased inflammatory cytokine release, was restored and cytokines decreased, three weeks post-FMT [94]. Similarly, FMT reversed intestinal lymphocyte and dendritic cell depletion induced by broad spectrum antibiotic use in an experimental model [95].
Considering the favorable outcome in C. difficile infection and relapse other applications of FMT were explored, especially in diseases where dysbiosis is thought to play an important role [96]. Moreover, researchers observed that persons who underwent FMT for RCDI also had more favorable outcome regarding gut decolonization from resistant pathogens [96]. In a case report by Crum-Cianflone et al., a critically ill patient received FMT to treat C. difficile colitis. After FMT the investigators observed that except for the resolution of symptoms related to CDI, the patient was also decolonized from MDROs and had reduced episodes of sepsis, health care associated infections. and antibiotic use [97]. The rationale for using FMT to eradicate MDROs is that especially for gram-negative pathogens, the gut is their main reservoir and thus elimination from the GIT may lessen the risk for systemic infection due to bacterial translocation and may eradicate them from other body sites [97]. Evidence on the role of FMT for MDRO decolonization or treatment is increasing [98-106]. However, findings are often contradictory and due to the lack of large randomized trials (RCTs), assessment of long-term effectiveness is limited.
Recent studies have revealed that the normalization of gut microbiota following successful FMT resulted in reduction of the load of AR genes and this favorable outcome was maintained during the follow up period [91,105]. Furthermore, in a single center prospective study, FMT was associated with a decrease in the number as well as downregulation of the expression of antibiotic resistant genes (Van A, blaKPC, blaNDM, blaOXA). Data from metagenomic sequencing showed that after FMT, there was depletion of 95 resistance genes, including important quinolone, β-lactamase, ESBL, and vancomycin resistance genes [107]. There is also evidence from a murine study, suggesting that transplantation of a healthy microbiota displaced both VRE and Klebsiella pneumoniae from the intestinal lumen despite an increased colonization burden [108]. In a small study by Wei et al., FMT resulted in cure of MRSA enteritis in five patients and in eradication of gut MRSA colonization [109].
However, RCTs have yet to prove such beneficial effect for FMT. The only RCT directly addressing MDRO decolonization found only a small, non-significant reduction in ESBL and CRE colonization after the combination of antibiotics and FMT [110]. FMT appears to be a safe and potentially effective intervention in eradicating Carbapenem-resistant Enterobacteriaceae (CRE) colonization. In a recent systematic review, which included ten studies (one RCT) CRE decolonization rate was estimated 61.1% and 78.7% at 1 month and 6-12 months after FMT, respectively [99].
FMT appears to have few, often mild to moderate and self-limiting side effects including nausea, fever, abdominal tenderness, constipation or diarrhea, cramping, and abdominal distension [111]. However, there have been reports of serious adverse events such as death, aspiration pneumonia, viral and bacterial infections, transient relapse of irritable bowel disease, and adverse effects related to the procedure such as sore throat and bowel perforation [112]. Furthermore, a possible transmission of Van B resistance gene after FMT has been described [113]. Last, concerns about long-term outcomes of FMT are not negligible. Remarkably, FMT was safe even when used in severely immunocompromised patients, such as those with hematological malignancies receiving intensive chemotherapy and immunosuppressive drugs and in patients with allogeneic HSCT; bacteremia due to pre-FMT colonizing bacteria was potentially prevented [114-116]. In summary, FMT may protect against intestinal translocation of MDROs preventing bloodstream infections [100] independently of gut decolonization, as several studies showed a reduction in the incidence of clinical infection post-FMT in MDRO-colonized patients [109,114,116,117]. Currently, there are several ongoing studies designed to evaluate the role of FMT in decolonization from MDROs (NCT03479710, NCT04181112, NCT04759001, NCT04431934, NCT04593368, NCT02922816, NCT04583098, NCT04759001, NCT02543866, NCT04146337, NCT04746222, NCT0418874, www.clinicaltrials.gov).
Antimicrobial Compounds for Targeted Therapy
Several gut microbial strains produce bacteriocins, which are antimicrobial compounds of high-potency and low toxicity [118]. The role of a gut derived bacteriocin, namely thiuricin CD, has been used in a CDI mouse model and showed that it was able to inhibit the growth of C. difficile, without a major shift in the gut microbiota [119]. Such molecules could represent future targeted therapeutic interventions in order to minimize unnecessary use of broad-spectrum antimicrobials [52].
Selective Digestive Decontamination (SDD) and Selective Oropharyngeal Decontamination (SOD)
SOD was introduced as a theoretical concept of preventing bacterial pneumonia by altering the pharyngeal flora and averting the aspiration of these pathogens [120] while the idea of SDD was first introduced as a means to reduce the load of resistant pathogens colonizing the digestive tract of ICU patients [121]. The first study which showed a reduction of MDRO gut colonization was a single-center study [122] followed by two larger randomized cross-over studies supporting that SOD/SDD resulted in a significant decrease of resistant pathogens on the gut [123,124]. However, a more recent randomized multicenter study revealed that the use of SOD/SDD did not lead to a change in the gut resistant pathogens [125]. An explanation of the different outcomes between the first three and the last study could lie in the rates of antimicrobial resistance in the participating ICUs. Therefore, SOD/SDD could be used in patients hospitalized in ICUs with low rates of resistant bacteria while more research should be done in centers with high prevalence of antimicrobial resistance [126]. However, a substantial concern raised from the use of SDD was the emergence of colistin-resistant Enterobacteriaceae in the gut of SDD-treated patients [127].
Prevention of Dysbiosis
We describe here the main approaches of preventing gut dysbiosis which are also presented briefly in Table 2.
Table 2. Prevention of Dysbiosis.
| Intervention | Mechanism of action/Application | Advantages | Disadvantages | References |
| Control measures | -Screening via nasal or rectal swabs for multidrug-resistant
organisms. -Good hygiene, environmental cleaning, contact precautions etc. |
-Easy to apply. | -Difficult to measure its direct effectiveness. | [103,128-130] |
| Antimicrobial stewardship programs | -Wise choice of antibiotics (e.g. avoidance of anti-anaerobic
antimicrobials) and use of narrow-spectrum agents help to preserve gut
microbiota. -Shorter courses of antibiotics lead to fewer microbiota disruptions and easier restoration. -Lower doses of antibiotics result in a lower risk of resistant genes. -Use of antibiotics that are not faecally and/or biliary excreted prevents the development of gut resistance. |
-Good rates of preventing infections with multidrug-resistant
organisms. -Applicable in every healthcare facility. -No need for adjuvant equipment/substances. |
-No many directly focused studies on the colonization of the gut
microbiota. -In some cases, it is inevitable to avoid some antibiotics. -Needs coordinated action between many specialties. |
[102,103,129,131,136,137,139,140,143,144] |
| Chelating/degradating agents | -Use of substances such as beta-lactamase enzymes and charcoal-based substances that absorb the remaining amount of antibiotic before reaching the colon protecting this way the gut microbiota without affecting the serum levels of the antibiotic. | -Promising preliminary results. -No serious adverse events. |
-Need for more clinical trials. -Difficult to measure their long-term clinical benefit. |
[102,145,147,150-153,156,158,159,161,163] |
Control Measures
A first step to prevent gut colonization with MDROs in hospitalized patients is the application of effective infection control measures [103,128,129]. Measures to prevent in-hospital transmission of MDROs include primarily hand hygiene followed by environmental cleaning, contact precautions, or even topical decolonization processes for some pathogens (ie, MRSA) [130].
Antimicrobial Stewardship and the Gut Antimicrobial Resistome
A significant approach in order to avoid major disruptions in gut microbiota is rationalizing antimicrobial use by implementing effective antimicrobial stewardship strategies [102,103,129,131].
Several large meta-analyses have supported that antimicrobial stewardship programs (ASPs) lead to a decrease in infections with MDROs [129,132,133] while others found the correlation inconclusive [134]. One large meta-analysis concluded that ASPs are successful in reducing MDRO colonization, independently from infection, and this success was higher when ASPs were combined with good infection control protocols (ie, hand hygiene) [129].
Choice of antibiotics: The type and spectrum of the antimicrobial used, is crucial for the development of resistance. For example, the unnecessary and prolonged use of anti-anaerobic antimicrobials has been related to a higher possibility of colonization with MDROs [135]. Avoiding anti-anaerobic antimicrobials and using narrow-spectrum agents whenever possible is beneficial to the human’s gut microbiota as fewer commensals will be affected [131,136].
Duration of antimicrobial therapy: Duration of antimicrobial therapy has been linked to greater alterations of gut microbiota. Shorter courses of antibiotics result in fewer microbiota disruptions and easier restoration of gut microbiota, which is supported by many studies in neonates [131]. Also, the use of ceftriaxone for more than 14 days was correlated with higher number of resistance genes in gut microbiota [137]. However, the duration of treatment with fluoroquinolones was not associated with the emergence of resistant E. coli strains in another study [138]. Similarly, in patients receiving antibiotic therapy for gram-negative bacteremia, shorter antibiotic treatment (7 days) was not associated with reduced resistant genes in the gut microbiota or with better preservation or restoration of the gut microbiota when compared with longer treatment courses (14 days) [139].
Dose: Lower doses of antibiotics have been associated with a lower or slower risk of acquiring resistant genes [140]. However, appropriate dosing is very important since antimicrobial underdosing can also lead to resistance.
Route of administration: Several animal studies showed that oral antibiotics disturb the gut microbiota more prominently than parenterally administered ones (IV or IM) [140,141]. However, recently published evidence had contradictory findings, showing that oral or parenteral route of administration has the same disrupting effect on gut microbiota [142]. Besides, the key components contributing to the preservation of gut microbiota after antibiotic exposure relies on the latter’s properties, like bile excretion, intestinal absorption, and presence in the feces [143,144]. Antibiotics that are not or only partially fecally and/or biliary excreted, may have fewer repercussions on the gut microbiota and therefore prevent the augmentation of gut resistome [131,136]. Furthermore, alternative modes of administration, like local application of antimicrobial agents, nebulized agents, or even transdermal administration lack strong data from clinical trials to support their non-inferior efficacy and beneficial profile for the gut microbiota [131,136]. Another interesting approach is to limit the use of oral antibiotics on discharge, granted there is clinical amelioration, after the completion of an inpatient intravenous antibiotic treatment course [136].
Chelating/degradating Agents
Another interesting approach for maintaining gut microbiota, is the use of newly developed substances such as beta-lactamase enzymes and charcoal-based substances, that absorb the remaining amount of a fecally or biliary excreted antibiotic before reaching the colon, thus protecting the gut microbiota without affecting antibiotic serum levels [102,145].
Beta-lactamase enzymes: The idea of using a beta-lactamase was derived by the observation that the simultaneous existence of cephalosporins and b-lactamase-producing pathogens resulted in less bacterial colonization [146]. P1A was the first beta-lactamase used to degrade the residual penicillin, aminopenicillins, and ureidopenicillins in the gut [147]. Except for its effectiveness in animal models [148,149], P1A was found to prevent the disruption of gut microbiota and the emergence of antibiotic resistant genes in colonizing bacteria [147,150].
A new agent was further developed to include the class of cephalosporins. Ribaxamase is an oral b-lactamase, and when administered concurrently with an IV b-lactam antibiotic, like ceftriaxone, or a β-lactam/β-lactamase inhibitor combination, can hydrolyze the excess of antibiotic at the small intestine, potentially enhancing colonization resistance [151-153]. This agent was well-tolerated and effective in both animal models [149,154] and humans [151-153]. Another ribaxamase-based agent, SYN-007, was evaluated in animal models to expand the use of oral ribaxamase. SYN-007 resulted in the protection of the microbiota and prevention of emergence and proliferation of resistant genes (ie, encoding ESBLs) while simultaneously preserving the concentration of antibiotics in serum [155,156]. However, ribaxamase is not effective against all b-lactam antibiotics and especially against carbapenems, which are associated with a greater risk of gut dysbiosis and antimicrobial resistance [157]. Therefore, a novel carbapenemase, SYN-006, was developed and tested in a rigorous animal model to expand the protection of gut microbiota from the detrimental effects of all beta-lactam classes [158,159]. Clinical trials in humans are needed to investigate the safety and efficacy of such complementary therapeutic strategies. A more recent discovery is the development of a beta-lactamase containing engineered live biotherapeutic product (eLBP), which introduces a more effective delivery of the active enzyme, also combining easier and cheaper manufacturing [160].
Charcoal-based agents: DaV-132 is a novel, orally administered, absorbent charcoal targeted-agent aiming to degrade the remaining antibiotic from the GIT by acting on the late ileum or proximal colon [161,162]. Although in clinical trials it was only administered with fluoroquinolones [161-166], the agent showed effectiveness in protecting gut microbiota and preventing colonization of resistant strains, without compromising therapeutic efficacy and safety [161,163].
Conclusion
Modulating the gut microbiota through diet, probiotics, prebiotics, antimicrobial molecules, phage therapy, or FMT could be a promising intervention in the fight against antimicrobial resistance. Strategies that will selectively inhibit pathogens without causing major shifts in the gut microbiota are most desirable. As our knowledge of the host-microbiota interactions, microbial quorum sensing and the drivers of colonization resistance will expand, a more targeted modulation of the microbial communities may be possible, allowing for a selective elimination of pathogenic bacteria and a less disruptive approach to gut microbiota homeostasis.
Glossary
- FMT
Fecal Microbiota Transplantation
- MDRO
Multidrug-Resistant Organism
- GIT
Gastrointestinal Tract
- ICU
Intensive Care Unit
- IL-17
Interleukin-17
- INF-γ
Interferon-γ
- AAD
Antibiotic-Associated Diarrhea
- CDI
Clostridium difficile Infection
- ESBL
Extended-Spectrum Beta-Lactamase
- CRE
Carbapenem-Resistant Enterobacteriaceae
- VRE
Vancomycin-Resistant Enterococci
- MRSA
Methicillin-Resistant Staphylococcus aureus
- HMO
Human Milk Oligosaccharide
- RCDI
Recurrent Clostridium difficile Infection
- SCFAs
Short-Chain Fatty Acids
- AR
Antibiotic Resistance
- RCT
Randomized Clinical Trial
- HSCT
Hematopoietic Stem Cell Transplantation
- SDD
Selective Digestive Decontamination
- SOD
Selective Oropharyngeal Decontamination
- IAP
Intestinal Alkaline Phosphatase
- ASP
Antimicrobial Stewardship Program
- eLBP
engineered Live Biotherapeutic Product
Funding
This work was not funded.
Author Contributions
EC and AN formulated the idea and outline, RM and AN did literature review, EC, RM, AN, and EP edited, reviewed and approved the manuscript.
References
- Peterson J, Garges S, Giovanni M, McInnes P, Wang L, Schloss JA, et al. NIH HMP Working Group. The NIH Human Microbiome Project. Genome Res. 2009. Dec;19(12):2317–23. 10.1101/gr.096651.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sender R, Fuchs S, Milo R. Are We Really Vastly Outnumbered? Revisiting the Ratio of Bacterial to Host Cells in Humans. Cell. 2016. Jan;164(3):337–40. 10.1016/j.cell.2016.01.013 [DOI] [PubMed] [Google Scholar]
- Lederberg J. Infectious history. Science. 2000. Apr;288(5464):287–93. 10.1126/science.288.5464.287 [DOI] [PubMed] [Google Scholar]
- Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006. Feb;124(4):837–48. 10.1016/j.cell.2006.02.017 [DOI] [PubMed] [Google Scholar]
- Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature. 2012. Jun;486(7402):207–14. 10.1038/nature11234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemente JC, Ursell LK, Parfrey LW, Knight R. The impact of the gut microbiota on human health: an integrative view. Cell. 2012. Mar;148(6):1258–70. 10.1016/j.cell.2012.01.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claesson MJ, Cusack S, O’Sullivan O, Greene-Diniz R, de Weerd H, Flannery E, et al. Composition, variability, and temporal stability of the intestinal microbiota of the elderly. Proc Natl Acad Sci USA. 2011. Mar;108(Suppl 1 Suppl 1):4586–91. 10.1073/pnas.1000097107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adlerberth I, Wold AE. Establishment of the gut microbiota in Western infants. Vol. 98, Acta Paediatrica, International Journal of Paediatrics. 2009. p. 229–38. 10.1111/j.1651-2227.2008.01060.x [DOI] [PubMed]
- Bäumler AJ, Sperandio V. Interactions between the microbiota and pathogenic bacteria in the gut. Nature. 2016. Jul;535(7610):85–93. 10.1038/nature18849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschi C. Inflammaging as a Major Characteristic of Old People: Can It Be Prevented or Cured? Nutr Rev. 2007;65(SUPPL.3). [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Gordon JI. The core gut microbiome, energy balance and obesity. J Physiol. 2009. Sep;587(Pt 17):4153–8. 10.1113/jphysiol.2009.174136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekirov I, Russell SL, Antunes LC, Finlay BB. Gut microbiota in health and disease. Physiol Rev. 2010. Jul;90(3):859–904. 10.1152/physrev.00045.2009 [DOI] [PubMed] [Google Scholar]
- Pilmis B, Le Monnier A, Zahar JR. Gut Microbiota, Antibiotic Therapy and Antimicrobial Resistance: A Narrative Review. Microorganisms. 2020. Feb;8(2):269. 10.3390/microorganisms8020269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarner F. Decade in review-gut microbiota: the gut microbiota era marches on. Nat Rev Gastroenterol Hepatol. 2014. Nov;11(11):647–9. 10.1038/nrgastro.2014.156 [DOI] [PubMed] [Google Scholar]
- Macpherson AJ, Harris NL. Interactions between commensal intestinal bacteria and the immune system. Nat Rev Immunol. 2004. Jun;4(6):478–85. 10.1038/nri1373 [DOI] [PubMed] [Google Scholar]
- O’Hara AM, Shanahan F. The gut flora as a forgotten organ. EMBO Rep. 2006. Jul;7(7):688–93. 10.1038/sj.embor.7400731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neish AS. Microbes in gastrointestinal health and disease. Gastroenterology. 2009. Jan;136(1):65–80. 10.1053/j.gastro.2008.10.080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper LV, Wong MH, Thelin A, Hansson L, Falk PG, Gordon JI. Molecular analysis of commensal host-microbial relationships in the intestine. Science. 2001. Feb;291(5505):881–4. 10.1126/science.291.5505.881 [DOI] [PubMed] [Google Scholar]
- Cryan JF, O’Mahony SM. The microbiome-gut-brain axis: from bowel to behavior. Neurogastroenterol Motil. 2011. Mar;23(3):187–92. 10.1111/j.1365-2982.2010.01664.x [DOI] [PubMed] [Google Scholar]
- Fetissov SO, Hamze Sinno M, Coëffier M, Bole-Feysot C, Ducrotté P, Hökfelt T, et al. Autoantibodies against appetite-regulating peptide hormones and neuropeptides: putative modulation by gut microflora. Nutrition. 2008. Apr;24(4):348–59. 10.1016/j.nut.2007.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L, Zhang J. Role of intestinal microbiota and metabolites on gut homeostasis and human diseases. BMC Immunol. 2017. Jan;18(1):2. 10.1186/s12865-016-0187-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos GM, Ismael S, Morais J, Araújo JR, Faria A, Calhau C, et al. Intestinal Alkaline Phosphatase: A Review of This Enzyme Role in the Intestinal Barrier Function. Microorganisms. 2022. Mar;10(4):746. 10.3390/microorganisms10040746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malo J, Alam MJ, Islam S, Mottalib MA, Rocki MM, Barmon G, et al. Intestinal alkaline phosphatase deficiency increases the risk of diabetes. BMJ Open Diabetes Res Care. 2022. Jan;10(1):e002643. 10.1136/bmjdrc-2021-002643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill DA, Hoffmann C, Abt MC, Du Y, Kobuley D, Kirn TJ, et al. Metagenomic analyses reveal antibiotic-induced temporal and spatial changes in intestinal microbiota with associated alterations in immune cell homeostasis. Mucosal Immunol. 2010. Mar;3(2):148–58. 10.1038/mi.2009.132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paliy O, Agans R. Application of phylogenetic microarrays to interrogation of human microbiota. FEMS Microbiol Ecol. 2012. Jan;79(1):2–11. 10.1111/j.1574-6941.2011.01222.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latorre M, Krishnareddy S, Freedberg DE. Microbiome as mediator: do systemic infections start in the gut? World J Gastroenterol. 2015. Oct;21(37):10487–92. 10.3748/wjg.v21.i37.10487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawrelak JA, Myers SP. The causes of intestinal dysbiosis: a review. Altern Med Rev. 2004. Jun;9(2):180–97. [PubMed] [Google Scholar]
- Francino MP. Antibiotics and the human gut microbiome: dysbioses and accumulation of resistances. Vol. 6. Front Microbiol. 2016;6. 10.3389/fmicb.2015.01543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langdon A, Crook N, Dantas G. The effects of antibiotics on the microbiome throughout development and alternative approaches for therapeutic modulation. Genome Med. 2016. Apr;8(1):39. 10.1186/s13073-016-0294-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nord CE, Edlund C. Impact of antimicrobial agents on human intestinal microflora. J Chemother. 1990. Aug;2(4):218–37. 10.1080/1120009X.1990.11739021 [DOI] [PubMed] [Google Scholar]
- Pérez-Cobas AE, Artacho A, Knecht H, Ferrús ML, Friedrichs A, Ott SJ, et al. Differential effects of antibiotic therapy on the structure and function of human gut microbiota. PLoS One. 2013. Nov;8(11):e80201. 10.1371/journal.pone.0080201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashid MU, Weintraub A, Nord CE. Effect of new antimicrobial agents on the ecological balance of human microflora. Anaerobe. 2012. Apr;18(2):249–53. 10.1016/j.anaerobe.2011.11.005 [DOI] [PubMed] [Google Scholar]
- Ubeda C, Pamer EG. Antibiotics, microbiota, and immune defense. Trends Immunol. 2012. Sep;33(9):459–66. 10.1016/j.it.2012.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candon S, Perez-Arroyo A, Marquet C, Valette F, Foray AP, Pelletier B, et al. Antibiotics in early life alter the gut microbiome and increase disease incidence in a spontaneous mouse model of autoimmune insulin-dependent diabetes. PLoS One. 2015. May;10(5):e0125448. 10.1371/journal.pone.0125448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov II, Frutos RL, Manel N, Yoshinaga K, Rifkin DB, Sartor RB, et al. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe. 2008. Oct;4(4):337–49. 10.1016/j.chom.2008.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wlodarska M, Willing B, Keeney KM, Menendez A, Bergstrom KS, Gill N, et al. Antibiotic treatment alters the colonic mucus layer and predisposes the host to exacerbated Citrobacter rodentium-induced colitis. Infect Immun. 2011. Apr;79(4):1536–45. 10.1128/IAI.01104-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pamer EG. Resurrecting the intestinal microbiota to combat antibiotic-resistant pathogens. Science. 2016. Apr;352(6285):535–8. 10.1126/science.aad9382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwood C, Morrow AL, Lagomarcino AJ, Altaye M, Taft DH, Yu Z, et al. Early empiric antibiotic use in preterm infants is associated with lower bacterial diversity and higher relative abundance of Enterobacter. J Pediatr. 2014. Jul;165(1):23–9. 10.1016/j.jpeds.2014.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, Rice LB, et al. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin Infect Dis. 2009. Jan;48(1):1–12. 10.1086/595011 [DOI] [PubMed] [Google Scholar]
- Forslund K, Sunagawa S, Kultima JR, Mende DR, Arumugam M, Typas A, et al. Country-specific antibiotic use practices impact the human gut resistome. Genome Res. 2013. Jul;23(7):1163–9. 10.1101/gr.155465.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Yang X, Qin J, Lu N, Cheng G, Wu N, et al. Metagenome-wide analysis of antibiotic resistance genes in a large cohort of human gut microbiota. Nat Commun. 2013;4(1):2151. 10.1038/ncomms3151 [DOI] [PubMed] [Google Scholar]
- Wellington EM, Boxall AB, Cross P, Feil EJ, Gaze WH, Hawkey PM, et al. The role of the natural environment in the emergence of antibiotic resistance in gram-negative bacteria. Lancet Infect Dis. 2013. Feb;13(2):155–65. 10.1016/S1473-3099(12)70317-1 [DOI] [PubMed] [Google Scholar]
- Sommer MO, Church GM, Dantas G. The human microbiome harbors a diverse reservoir of antibiotic resistance genes. Virulence. 2010;1(4):299–303. 10.4161/viru.1.4.12010 [DOI] [PubMed] [Google Scholar]
- Stinear TP, Olden DC, Johnson PD, Davies JK, Grayson ML. Enterococcal vanB resistance locus in anaerobic bacteria in human faeces. Lancet. 2001. Mar;357(9259):855–6. 10.1016/S0140-6736(00)04206-9 [DOI] [PubMed] [Google Scholar]
- van Schaik W. The human gut resistome. Philos Trans R Soc Lond B Biol Sci. 2015. Jun;370(1670):20140087. 10.1098/rstb.2014.0087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donskey CJ, Chowdhry TK, Hecker MT, Hoyen CK, Hanrahan JA, Hujer AM, et al. Effect of antibiotic therapy on the density of vancomycin-resistant enterococci in the stool of colonized patients. N Engl J Med. 2000. Dec;343(26):1925–32. 10.1056/NEJM200012283432604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kau AL, Ahern PP, Griffin NW, Goodman AL, Gordon JI. Human nutrition, the gut microbiome and the immune system. Nature. 2011. Jun;474(7351):327–36. 10.1038/nature10213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeerdoss J, Devi RS, Mary RR, Ramakrishna BS. Faecal microbiota composition in vegetarians: comparison with omnivores in a cohort of young women in southern India. Br J Nutr. 2012. Sep;108(6):953–7. 10.1017/S0007114511006362 [DOI] [PubMed] [Google Scholar]
- Matijašić BB, Obermajer T, Lipoglavšek L, Grabnar I, Avguštin G, Rogelj I. Association of dietary type with fecal microbiota in vegetarians and omnivores in Slovenia. Eur J Nutr. 2014. Jun;53(4):1051–64. 10.1007/s00394-013-0607-6 [DOI] [PubMed] [Google Scholar]
- Sun Y, O’Riordan MX. Regulation of bacterial pathogenesis by intestinal short-chain Fatty acids. Adv Appl Microbiol. 2013;85:93–118. 10.1016/B978-0-12-407672-3.00003-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Bäckhed F, Fulton L, Gordon JI. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe. 2008. Apr;3(4):213–23. 10.1016/j.chom.2008.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh CJ, Guinane CM, O’Toole PW, Cotter PD. Beneficial modulation of the gut microbiota. FEBS Lett. 2014. Nov;588(22):4120–30. 10.1016/j.febslet.2014.03.035 [DOI] [PubMed] [Google Scholar]
- Claesson MJ, Jeffery IB, Conde S, Power SE, O’Connor EM, Cusack S, et al. Gut microbiota composition correlates with diet and health in the elderly. Nature. 2012. Aug;488(7410):178–84. 10.1038/nature11319 [DOI] [PubMed] [Google Scholar]
- Caballero-Flores G, Pickard JM, Fukuda S, Inohara N, Núñez G. An Enteric Pathogen Subverts Colonization Resistance by Evading Competition for Amino Acids in the Gut. Cell Host Microbe. 2020. Oct;28(4):526–533.e5. 10.1016/j.chom.2020.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzorati M, Vilchez-Vargas R, Bussche JV, Truchado P, Jauregui R, El Hage RA, et al. High-fiber and high-protein diets shape different gut microbial communities, which ecologically behave similarly under stress conditions, as shown in a gastrointestinal simulator. Mol Nutr Food Res. 2017. Jan;61(1):1600150. 10.1002/mnfr.201600150 [DOI] [PubMed] [Google Scholar]
- Mao YH, Song AX, Yao ZP, Wu JY. Protective effects of natural and partially degraded konjac glucomannan on Bifidobacteria against antibiotic damage. Carbohydr Polym. 2018. Feb;181:368–75. 10.1016/j.carbpol.2017.10.083 [DOI] [PubMed] [Google Scholar]
- Wu G, Zhang C, Wang J, Zhang F, Wang R, Shen J, et al. Diminution of the gut resistome after a gut microbiota-targeted dietary intervention in obese children. Sci Rep. 2016. Apr;6(1):24030. 10.1038/srep24030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv W, Liu C, Ye C, Sun J, Tan X, Zhang C, et al. Structural modulation of gut microbiota during alleviation of antibiotic-associated diarrhea with herbal formula. Int J Biol Macromol. 2017. Dec;105(Pt 3):1622–9. 10.1016/j.ijbiomac.2017.02.060 [DOI] [PubMed] [Google Scholar]
- Elison E, Vigsnaes LK, Rindom Krogsgaard L, Rasmussen J, Sørensen N, McConnell B, et al. Oral supplementation of healthy adults with 2′-O-fucosyllactose and lacto-N-neotetraose is well tolerated and shifts the intestinal microbiota. Br J Nutr. 2016. Oct;116(8):1356–68. 10.1017/S0007114516003354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šuligoj T, Vigsnæs LK, Abbeele PV, Apostolou A, Karalis K, Savva GM, et al. Effects of Human Milk Oligosaccharides on the Adult Gut Microbiota and Barrier Function. Nutrients. 2020. Sep;12(9):2808. 10.3390/nu12092808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malo MS, Alam SN, Mostafa G, Zeller SJ, Johnson PV, Mohammad N, et al. Intestinal alkaline phosphatase preserves the normal homeostasis of gut microbiota. Gut. 2010. Nov;59(11):1476–84. 10.1136/gut.2010.211706 [DOI] [PubMed] [Google Scholar]
- Alam SN, Yammine H, Moaven O, Ahmed R, Moss AK, Biswas B, et al. Intestinal alkaline phosphatase prevents antibiotic-induced susceptibility to enteric pathogens. Ann Surg. 2014. Apr;259(4):715–22. 10.1097/SLA.0b013e31828fae14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggers S, Malecki KM, Peppard P, Mares J, Shirley D, Shukla SK, et al. Wisconsin microbiome study, a cross-sectional investigation of dietary fibre, microbiome composition and antibiotic-resistant organisms: rationale and methods. BMJ Open. 2018. Mar;8(3):e019450. 10.1136/bmjopen-2017-019450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson GR, Probert HM, Loo JV, Rastall RA, Roberfroid MB. Dietary modulation of the human colonic microbiota: updating the concept of prebiotics. Nutr Res Rev. 2004. Dec;17(2):259–75. 10.1079/NRR200479 [DOI] [PubMed] [Google Scholar]
- Roberfroid M, Gibson GR, Hoyles L, McCartney AL, Rastall R, Rowland I, et al. Prebiotic effects: metabolic and health benefits. Br J Nutr. 2010. Aug;104(November Suppl 2):S1–63. 10.1017/S0007114510003363 [DOI] [PubMed] [Google Scholar]
- Vieira AT, Teixeira MM, Martins FS. The role of probiotics and prebiotics in inducing gut immunity. Front Immunol. 2013. Dec;4:445. 10.3389/fimmu.2013.00445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vadder F, Kovatcheva-Datchary P, Goncalves D, Vinera J, Zitoun C, Duchampt A, et al. Microbiota-generated metabolites promote metabolic benefits via gut-brain neural circuits. Cell. 2014. Jan;156(1-2):84–96. 10.1016/j.cell.2013.12.016 [DOI] [PubMed] [Google Scholar]
- Shi Y, Zhai Q, Li D, Mao B, Liu X, Zhao J, et al. Restoration of cefixime-induced gut microbiota changes by Lactobacillus cocktails and fructooligosaccharides in a mouse model. Microbiol Res. 2017. Jul;200:14–24. 10.1016/j.micres.2017.04.001 [DOI] [PubMed] [Google Scholar]
- Soldi S, Vasileiadis S, Lohner S, Uggeri F, Puglisi E, Molinari P, et al. Prebiotic supplementation over a cold season and during antibiotic treatment specifically modulates the gut microbiota composition of 3-6 year-old children. Benef Microbes. 2019. Apr;10(3):253–63. 10.3920/BM2018.0116 [DOI] [PubMed] [Google Scholar]
- Pineiro M, Stanton C. Probiotic bacteria: legislative framework— requirements to evidence basis. J Nutr. 2007. Mar;137(3 Suppl 2):850S–3S. 10.1093/jn/137.3.850S [DOI] [PubMed] [Google Scholar]
- Rijkers GT, Bengmark S, Enck P, Haller D, Herz U, Kalliomaki M, et al. Guidance for substantiating the evidence for beneficial effects of probiotics: current status and recommendations for future research. J Nutr. 2010. Mar;140(3):671S–6S. 10.3945/jn.109.113779 [DOI] [PubMed] [Google Scholar]
- Kumar M, Dhaka P, Vijay D, Vergis J, Mohan V, Kumar A, et al. Antimicrobial effects of Lactobacillus plantarum and Lactobacillus acidophilus against multidrug-resistant enteroaggregative Escherichia coli. Int J Antimicrob Agents. 2016. Sep;48(3):265–70. 10.1016/j.ijantimicag.2016.05.014 [DOI] [PubMed] [Google Scholar]
- Hwang IY, Tan MH, Koh E, Ho CL, Poh CL, Chang MW. Reprogramming microbes to be pathogen-seeking killers. ACS Synth Biol. 2014. Apr;3(4):228–37. 10.1021/sb400077j [DOI] [PubMed] [Google Scholar]
- Yosef I, Manor M, Kiro R, Qimron U. Temperate and lytic bacteriophages programmed to sensitize and kill antibiotic-resistant bacteria. Proc Natl Acad Sci USA. 2015. Jun;112(23):7267–72. 10.1073/pnas.1500107112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naseri A, Seyedi-Sahebari S, Mahmoodpoor A, Sanaie S. Probiotics in Critically Ill Patients: An Umbrella Review. Indian J Crit Care Med. 2022. Mar;26(3):339–60. 10.5005/jp-journals-10071-24129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggers S, Barker AK, Valentine S, Hess T, Duster M, Safdar N. Effect of Lactobacillus rhamnosus HN001 on carriage of Staphylococcus aureus: results of the impact of probiotics for reducing infections in veterans (IMPROVE) study. BMC Infect Dis. 2018. Mar;18(1):129. 10.1186/s12879-018-3028-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szachta P, Ignyś I, Cichy W. An evaluation of the ability of the probiotic strain Lactobacillus rhamnosus GG to eliminate the gastrointestinal carrier state of vancomycin-resistant enterococci in colonized children. J Clin Gastroenterol. 2011;45(10):872–7. 10.1097/MCG.0b013e318227439f [DOI] [PubMed] [Google Scholar]
- Manley KJ, Fraenkel MB, Mayall BC, Power DA. Probiotic treatment of vancomycin-resistant enterococci: a randomised controlled trial. Med J Aust. 2007. May;186(9):454–7. 10.5694/j.1326-5377.2007.tb00995.x [DOI] [PubMed] [Google Scholar]
- Salomão MC, Heluany-Filho MA, Menegueti MG, Kraker ME, Martinez R, Bellissimo-Rodrigues F. A randomized clinical trial on the effectiveness of a symbiotic product to decolonize patients harboring multidrug-resistant Gram-negative bacilli. Rev Soc Bras Med Trop. 2016;49(5):559–66. 10.1590/0037-8682-0233-2016 [DOI] [PubMed] [Google Scholar]
- Ljungquist O, Kampmann C, Resman F, Riesbeck K, Tham J. Probiotics for intestinal decolonization of ESBL-producing Enterobacteriaceae: a randomized, placebo-controlled clinical trial. Clin Microbiol Infect. 2020. Apr;26(4):456–62. 10.1016/j.cmi.2019.08.019 [DOI] [PubMed] [Google Scholar]
- Duan H, Yu L, Tian F, Zhai Q, Fan L, Chen W. Antibiotic-induced gut dysbiosis and barrier disruption and the potential protective strategies. Crit Rev Food Sci Nutr. 2022;62(6):1427–52. 10.1080/10408398.2020.1843396 [DOI] [PubMed] [Google Scholar]
- Salminen MK, Rautelin H, Tynkkynen S, Poussa T, Saxelin M, Valtonen V, et al. Lactobacillus bacteremia, clinical significance, and patient outcome, with special focus on probiotic L. rhamnosus GG. Clin Infect Dis. 2004. Jan;38(1):62–9. 10.1086/380455 [DOI] [PubMed] [Google Scholar]
- Yelin I, Flett KB, Merakou C, Mehrotra P, Stam J, Snesrud E, et al. Genomic and epidemiological evidence of bacterial transmission from probiotic capsule to blood in ICU patients. Nat Med. 2019. Nov;25(11):1728–32. 10.1038/s41591-019-0626-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Luo W, Shi Y, Fan Z, Ji G. Should we standardize the 1,700-year-old fecal microbiota transplantation? Am J Gastroenterol. 2012. Nov;107(11):1755. 10.1038/ajg.2012.251 [DOI] [PubMed] [Google Scholar]
- Eiseman B, Silen W, Bascom GS, Kauvar AJ. Fecal enema as an adjunct in the treatment of pseudomembranous enterocolitis. Surgery. 1958. Nov;44(5):854–9. [PubMed] [Google Scholar]
- Gough E, Shaikh H, Manges AR. Systematic review of intestinal microbiota transplantation (fecal bacteriotherapy) for recurrent Clostridium difficile infection. Clin Infect Dis. 2011. Nov;53(10):994–1002. 10.1093/cid/cir632 [DOI] [PubMed] [Google Scholar]
- Floch MH. Fecal bacteriotherapy, fecal transplant, and the microbiome. J Clin Gastroenterol. 2010. Sep;44(8):529–30. 10.1097/MCG.0b013e3181e1d6e2 [DOI] [PubMed] [Google Scholar]
- Chang JY, Antonopoulos DA, Kalra A, Tonelli A, Khalife WT, Schmidt TM, et al. Decreased diversity of the fecal Microbiome in recurrent Clostridium difficile-associated diarrhea. J Infect Dis. 2008. Feb;197(3):435–8. 10.1086/525047 [DOI] [PubMed] [Google Scholar]
- Kyne L, Farrell RJ, Kelly CP. Clostridium difficile [ix–x.]. Gastroenterol Clin North Am. 2001. Sep;30(3):753–77. 10.1016/S0889-8553(05)70209-0 [DOI] [PubMed] [Google Scholar]
- Campbell C, McKenney PT, Konstantinovsky D, Isaeva OI, Schizas M, Verter J, et al. Bacterial metabolism of bile acids promotes generation of peripheral regulatory T cells. Nature. 2020. May;581(7809):475–9. 10.1038/s41586-020-2193-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huus KE, Frankowski M, Pučić-Baković M, Vučković F, Lauc G, Mullish BH, et al. Changes in IgA-targeted microbiota following fecal transplantation for recurrent Clostridioides difficile infection. Gut Microbes. 2021;13(1):1–12. 10.1080/19490976.2020.1862027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrello C, Garavaglia F, Cribiù FM, Ercoli G, Lopez G, Troisi J, et al. Therapeutic faecal microbiota transplantation controls intestinal inflammation through IL10 secretion by immune cells. Nat Commun. 2018. Dec;9(1):5184. 10.1038/s41467-018-07359-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal JP, Mullish BH, Quraishi MN, Iqbal T, Marchesi JR, Sokol H. Mechanisms underpinning the efficacy of faecal microbiota transplantation in treating gastrointestinal disease. Therap Adv Gastroenterol. 2020. Sep;13:1756284820946904. 10.1177/1756284820946904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Liang P, Li Z, Wang Y, Zhang G, Gao H, et al. Fecal microbiota transplantation and bacterial consortium transplantation have comparable effects on the re-establishment of mucosal barrier function in mice with intestinal dysbiosis [Internet]. Front Microbiol. 2015. Jul;6:692. [cited 2022 Jun 15] Available from: https://www.frontiersin.org/article/10.3389/fmicb.2015.00692 10.3389/fmicb.2015.00692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekmekciu I, von Klitzing E, Fiebiger U, Escher U, Neumann C, Bacher P, et al. Immune Responses to Broad-Spectrum Antibiotic Treatment and Fecal Microbiota Transplantation in Mice [Internet]. Front Immunol. 2017. Apr;8:397. [cited 2022 Jun 15] Available from: https://www.frontiersin.org/article/10.3389/fimmu.2017.00397 10.3389/fimmu.2017.00397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoruts A, Weingarden AR. Emergence of fecal microbiota transplantation as an approach to repair disrupted microbial gut ecology. Immunol Lett. 2014. Dec;162(2 2 Pt A):77–81. 10.1016/j.imlet.2014.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crum-Cianflone NF, Sullivan E, Ballon-Landa G. Fecal microbiota transplantation and successful resolution of multidrug-resistant-organism colonization. J Clin Microbiol. 2015. Jun;53(6):1986–9. 10.1128/JCM.00820-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghani R, Mullish BH, Roberts LA, Davies FJ, Marchesi JR. The potential utility of fecal (or intestinal) microbiota transplantation in controlling infectious diseases. Gut Microbes. 2022;14(1):2038856. 10.1080/19490976.2022.2038856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macareño-Castro J, Solano-Salazar A, Dong LT, Mohiuddin M, Espinoza JL. Fecal microbiota transplantation for Carbapenem-Resistant Enterobacteriaceae: A systematic review. J Infect. 2022. Jun;84(6):749–59. 10.1016/j.jinf.2022.04.028 [DOI] [PubMed] [Google Scholar]
- Ghani R, Mullish BH, Davies FJ, Marchesi JR. How to adapt an intestinal microbiota transplantation programme to reduce the risk of invasive multidrug-resistant infection. Clin Microbiol Infect. 2022. Apr;28(4):502–12. 10.1016/j.cmi.2021.11.006 [DOI] [PubMed] [Google Scholar]
- Bilsen MP, Lambregts MM, van Prehn J, Kuijper EJ. Faecal microbiota replacement to eradicate antimicrobial resistant bacteria in the intestinal tract - a systematic review. Curr Opin Gastroenterol. 2022. Jan;38(1):15–25. 10.1097/MOG.0000000000000792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuethrich I, W Pelzer B, Khodamoradi Y, Vehreschild MJ. The role of the human gut microbiota in colonization and infection with multidrug-resistant bacteria. Gut Microbes. 2021;13(1):1–13. 10.1080/19490976.2021.1911279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gargiullo L, Del Chierico F, D’Argenio P, Putignani L. Gut Microbiota Modulation for Multidrug-Resistant Organism Decolonization: Present and Future Perspectives [Internet]. Front Microbiol. 2019. Jul;10:1704. [cited 2022 Jun 10] Available from: https://www.frontiersin.org/article/10.3389/fmicb.2019.01704 10.3389/fmicb.2019.01704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalsamy SN, Woodworth MH, Wang T, Carpentieri CT, Mehta N, Friedman-Moraco RJ, et al. The Use of Microbiome Restoration Therapeutics to Eliminate Intestinal Colonization With Multidrug-Resistant Organisms. Am J Med Sci. 2018. Nov;356(5):433–40. 10.1016/j.amjms.2018.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha S, Tariq R, Tosh PK, Pardi DS, Khanna S. Faecal microbiota transplantation for eradicating carriage of multidrug-resistant organisms: a systematic review. Clin Microbiol Infect. 2019. Aug;25(8):958–63. 10.1016/j.cmi.2019.04.006 [DOI] [PubMed] [Google Scholar]
- Woodworth MH, Hayden MK, Young VB, Kwon JH. The Role of Fecal Microbiota Transplantation in Reducing Intestinal Colonization With Antibiotic-Resistant Organisms: The Current Landscape and Future Directions. Open Forum Infect Dis. 2019. Jul;6(7):ofz288. 10.1093/ofid/ofz288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung V, Vincent C, Edens TJ, Miller M, Manges AR. Antimicrobial Resistance Gene Acquisition and Depletion Following Fecal Microbiota Transplantation for Recurrent Clostridium difficile Infection. Clin Infect Dis. 2018. Jan;66(3):456–7. 10.1093/cid/cix821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caballero S, Carter R, Ke X, Sušac B, Leiner IM, Kim GJ, et al. Distinct but Spatially Overlapping Intestinal Niches for Vancomycin-Resistant Enterococcus faecium and Carbapenem-Resistant Klebsiella pneumoniae. PLoS Pathog. 2015. Sep;11(9):e1005132. 10.1371/journal.ppat.1005132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Gong J, Zhu W, Guo D, Gu L, Li N, et al. Fecal microbiota transplantation restores dysbiosis in patients with methicillin resistant Staphylococcus aureus enterocolitis. BMC Infect Dis. 2015. Jul;15(1):265. 10.1186/s12879-015-0973-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttner BD, de Lastours V, Wassenberg M, Maharshak N, Mauris A, Galperine T, et al. R-Gnosis WP3 study group. A 5-day course of oral antibiotics followed by faecal transplantation to eradicate carriage of multidrug-resistant Enterobacteriaceae: a randomized clinical trial. Clin Microbiol Infect. 2019. Jul;25(7):830–8. 10.1016/j.cmi.2018.12.009 [DOI] [PubMed] [Google Scholar]
- Baxter M, Colville A. Adverse events in faecal microbiota transplant: a review of the literature. J Hosp Infect. 2016. Feb;92(2):117–27. 10.1016/j.jhin.2015.10.024 [DOI] [PubMed] [Google Scholar]
- Wang S, Xu M, Wang W, Cao X, Piao M, Khan S, et al. Systematic review: adverse events of fecal Microbiota transplantation. Vol. 11. PLoS One. 2016;11(8):e0161174. 10.1371/journal.pone.0161174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouhten H, Mattila E, Arkkila P Sr, Satokari R. Reduction of Antibiotic Resistance Genes in Intestinal Microbiota of Patients With Recurrent Clostridium difficile Infection After Fecal Microbiota Transplantation. Clin Infect Dis. 2016. Sep;63(5):710–1. 10.1093/cid/ciw390 [DOI] [PubMed] [Google Scholar]
- Battipaglia G, Malard F, Rubio MT, Ruggeri A, Mamez AC, Brissot E, et al. Fecal microbiota transplantation before or after allogeneic hematopoietic transplantation in patients with hematologic malignancies carrying multidrug-resistance bacteria. Haematologica. 2019. Aug;104(8):1682–8. 10.3324/haematol.2018.198549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilinski J, Grzesiowski P, Sorensen N, Madry K, Muszynski J, Robak K, et al. Fecal Microbiota Transplantation in Patients With Blood Disorders Inhibits Gut Colonization With Antibiotic-Resistant Bacteria: Results of a Prospective, Single-Center Study. Clin Infect Dis. 2017. Aug;65(3):364–70. 10.1093/cid/cix252 [DOI] [PubMed] [Google Scholar]
- Ghani R, Mullish BH, McDonald JA, Ghazy A, Williams HR, Brannigan ET, et al. Disease Prevention Not Decolonization: A Model for Fecal Microbiota Transplantation in Patients Colonized With Multidrug-resistant Organisms. Clin Infect Dis. 2021. Apr;72(8):1444–7. 10.1093/cid/ciaa948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Yoseph H, Carasso S, Shklar S, Korytny A, Even Dar R, Daoud H, et al. Oral Capsulized Fecal Microbiota Transplantation for Eradication of Carbapenemase-producing Enterobacteriaceae Colonization With a Metagenomic Perspective. Clin Infect Dis. 2021. Jul;73(1):e166–75. 10.1093/cid/ciaa737 [DOI] [PubMed] [Google Scholar]
- Lakshminarayanan B, Guinane CM, O’Connor PM, Coakley M, Hill C, Stanton C, et al. Isolation and characterization of bacteriocin-producing bacteria from the intestinal microbiota of elderly Irish subjects. J Appl Microbiol. 2013. Mar;114(3):886–98. 10.1111/jam.12085 [DOI] [PubMed] [Google Scholar]
- Rea MC, Dobson A, O’Sullivan O, Crispie F, Fouhy F, Cotter PD, et al. Effect of broad- and narrow-spectrum antimicrobials on Clostridium difficile and microbial diversity in a model of the distal colon. Proc Natl Acad Sci USA. 2011. Mar;108(Suppl 1 Suppl 1):4639–44. 10.1073/pnas.1001224107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johanson WG, Pierce AK, Sanford JP. Changing pharyngeal bacterial flora of hospitalized patients. Emergence of gram-negative bacilli. N Engl J Med. 1969. Nov;281(21):1137–40. 10.1056/NEJM196911202812101 [DOI] [PubMed] [Google Scholar]
- Stoutenbeek CP, van Saene HK, Miranda DR, Zandstra DF. The effect of selective decontamination of the digestive tract on colonisation and infection rate in multiple trauma patients. Intensive Care Med. 1984;10(4):185–92. 10.1007/BF00259435 [DOI] [PubMed] [Google Scholar]
- de Jonge E, Schultz MJ, Spanjaard L, Bossuyt PM, Vroom MB, Dankert J, et al. Effects of selective decontamination of digestive tract on mortality and acquisition of resistant bacteria in intensive care: a randomised controlled trial. Lancet. 2003. Sep;362(9389):1011–6. 10.1016/S0140-6736(03)14409-1 [DOI] [PubMed] [Google Scholar]
- de Smet AM, Kluytmans JA, Cooper BS, Mascini EM, Benus RF, van der Werf TS, et al. Decontamination of the digestive tract and oropharynx in ICU patients. N Engl J Med. 2009. Jan;360(1):20–31. 10.1056/NEJMoa0800394 [DOI] [PubMed] [Google Scholar]
- Oostdijk EA, Kesecioglu J, Schultz MJ, Visser CE, de Jonge E, van Essen EH, et al. Effects of decontamination of the oropharynx and intestinal tract on antibiotic resistance in ICUs: a randomized clinical trial. JAMA. 2014. Oct;312(14):1429–37. 10.1001/jama.2014.7247 [DOI] [PubMed] [Google Scholar]
- Wittekamp BH, Plantinga NL, Cooper BS, Lopez-Contreras J, Coll P, Mancebo J, et al. Decontamination Strategies and Bloodstream Infections With Antibiotic-Resistant Microorganisms in Ventilated Patients: A Randomized Clinical Trial. JAMA. 2018. Nov;320(20):2087–98. 10.1001/jama.2018.13765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittekamp BH, Oostdijk EA, Cuthbertson BH, Brun-Buisson C, Bonten MJ. Selective decontamination of the digestive tract (SDD) in critically ill patients: a narrative review. Intensive Care Med. 2020. Feb;46(2):343–9. 10.1007/s00134-019-05883-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen AB, van Hout D, Bonten MJ, Willems RJ, van Schaik W. Microevolution of acquired colistin resistance in Enterobacteriaceae from ICU patients receiving selective decontamination of the digestive tract. J Antimicrob Chemother. 2020. Nov;75(11):3135–43. 10.1093/jac/dkaa305 [DOI] [PubMed] [Google Scholar]
- Barnes SL, Morgan DJ, Harris AD, Carling PC, Thom KA. Preventing the transmission of multidrug-resistant organisms: modeling the relative importance of hand hygiene and environmental cleaning interventions. Infect Control Hosp Epidemiol. 2014. Sep;35(9):1156–62. 10.1086/677632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baur D, Gladstone BP, Burkert F, Carrara E, Foschi F, Döbele S, et al. Effect of antibiotic stewardship on the incidence of infection and colonisation with antibiotic-resistant bacteria and Clostridium difficile infection: a systematic review and meta-analysis. Lancet Infect Dis. 2017. Sep;17(9):990–1001. 10.1016/S1473-3099(17)30325-0 [DOI] [PubMed] [Google Scholar]
- Mitchell BG, Hall L, White N, Barnett AG, Halton K, Paterson DL, et al. An environmental cleaning bundle and health-care-associated infections in hospitals (REACH): a multicentre, randomised trial. Lancet Infect Dis. 2019. Apr;19(4):410–8. 10.1016/S1473-3099(18)30714-X [DOI] [PubMed] [Google Scholar]
- Tan GS, Tay HL, Tan SH, Lee TH, Ng TM, Lye DC. Gut Microbiota Modulation: Implications for Infection Control and Antimicrobial Stewardship. Adv Ther. 2020. Oct;37(10):4054–67. 10.1007/s12325-020-01458-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karanika S, Paudel S, Grigoras C, Kalbasi A, Mylonakis E. Systematic Review and Meta-analysis of Clinical and Economic Outcomes from the Implementation of Hospital-Based Antimicrobial Stewardship Programs. Antimicrob Agents Chemother. 2016. Jul;60(8):4840–52. 10.1128/AAC.00825-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuts EC, Hulscher ME, Mouton JW, Verduin CM, Stuart JW, Overdiek HW, et al. Current evidence on hospital antimicrobial stewardship objectives: a systematic review and meta-analysis. Lancet Infect Dis. 2016. Jul;16(7):847–56. 10.1016/S1473-3099(16)00065-7 [DOI] [PubMed] [Google Scholar]
- Davey P, Marwick CA, Scott CL, Charani E, McNeil K, Brown E, et al. Interventions to improve antibiotic prescribing practices for hospital inpatients. Cochrane Database Syst Rev [Internet]. 2017. [cited 2022 Jun 10];(2). Available from: https://www.cochranelibrary.com/cdsr/doi/10.1002/14651858.CD003543.pub4/full 10.1002/14651858.CD003543.pub4 [DOI] [PMC free article] [PubMed]
- Bhalla A, Pultz NJ, Ray AJ, Hoyen CK, Eckstein EC, Donskey CJ. Antianaerobic antibiotic therapy promotes overgrowth of antibiotic-resistant, gram-negative bacilli and vancomycin-resistant enterococci in the stool of colonized patients. Infect Control Hosp Epidemiol. 2003. Sep;24(9):644–9. 10.1086/502267 [DOI] [PubMed] [Google Scholar]
- Shahi F, Redeker K, Chong J. Rethinking antimicrobial stewardship paradigms in the context of the gut microbiome. JAC Antimicrob Resist. 2019. May;1(1):dlz015. 10.1093/jacamr/dlz015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meletiadis J, Turlej-Rogacka A, Lerner A, Adler A, Tacconelli E, Mouton JW, the SATURN Diagnostic Study Group. Amplification of Antimicrobial Resistance in Gut Flora of Patients Treated with Ceftriaxone. Antimicrob Agents Chemother. 2017. Oct;61(11):e00473–17. 10.1128/AAC.00473-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lastours V, Chau F, Roy C, Larroque B, Fantin B. Emergence of quinolone resistance in the microbiota of hospitalized patients treated or not with a fluoroquinolone. J Antimicrob Chemother. 2014. Dec;69(12):3393–400. 10.1093/jac/dku283 [DOI] [PubMed] [Google Scholar]
- Leo S, Lazarevic V, von Dach E, Kaiser L, Prendki V, Schrenzel J, et al. Effects of antibiotic duration on the intestinal microbiota and resistome: the PIRATE RESISTANCE project, a cohort study nested within a randomized trial. EBioMedicine. 2021. Sep;71:103566. 10.1016/j.ebiom.2021.103566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Huang Y, Zhou Y, Buckley T, Wang HH. Antibiotic administration routes significantly influence the levels of antibiotic resistance in gut microbiota. Antimicrob Agents Chemother. 2013. Aug;57(8):3659–66. 10.1128/AAC.00670-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Surathu A, Raplee I, Chockalingam A, Stewart S, Walker L, et al. The effect of antibiotics on the gut microbiome: a metagenomics analysis of microbial shift and gut antibiotic resistance in antibiotic treated mice. BMC Genomics. 2020. Mar;21(1):263. 10.1186/s12864-020-6665-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly SA, Nzakizwanayo J, Rodgers AM, Zhao L, Weiser R, Tekko IA, et al. Antibiotic Therapy and the Gut Microbiome: Investigating the Effect of Delivery Route on Gut Pathogens. ACS Infect Dis. 2021. May;7(5):1283–96. 10.1021/acsinfecdis.1c00081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan A, Edlund C, Nord CE. Effect of antimicrobial agents on the ecological balance of human microflora. Lancet Infect Dis. 2001. Sep;1(2):101–14. 10.1016/S1473-3099(01)00066-4 [DOI] [PubMed] [Google Scholar]
- Kelly SA, Rodgers AM, O’Brien SC, Donnelly RF, Gilmore BF. Gut Check Time: Antibiotic Delivery Strategies to Reduce Antimicrobial Resistance. Trends Biotechnol. 2020. Apr;38(4):447–62. 10.1016/j.tibtech.2019.10.008 [DOI] [PubMed] [Google Scholar]
- Szychowiak P, Villageois-Tran K, Patrier J, Timsit JF, Ruppé É. The role of the microbiota in the management of intensive care patients. Ann Intensive Care. 2022. Jan;12(1):3. 10.1186/s13613-021-00976-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Léonard F, Andremont A, Leclerq B, Labia R, Tancrède C. Use of β-lactamase-producing anaerobes to prevent ceftriaxone from degrading intestinal resistance to colonization. J Infect Dis. 1989. Aug;160(2):274–80. 10.1093/infdis/160.2.274 [DOI] [PubMed] [Google Scholar]
- Tarkkanen AM, Heinonen T, Jõgi R, Mentula S, van der Rest ME, Donskey CJ, et al. P1A recombinant β-lactamase prevents emergence of antimicrobial resistance in gut microflora of healthy subjects during intravenous administration of ampicillin. Antimicrob Agents Chemother. 2009. Jun;53(6):2455–62. 10.1128/AAC.00853-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmoinen J, Mentula S, Heikkilä M, van der Rest M, Rajala-Schultz PJ, Donskey CJ, et al. Orally administered targeted recombinant Beta-lactamase prevents ampicillin-induced selective pressure on the gut microbiota: a novel approach to reducing antimicrobial resistance. Antimicrob Agents Chemother. 2004. Jan;48(1):75–9. 10.1128/AAC.48.1.75-79.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokai-Kun JF, Bristol JA, Setser J, Schlosser M. Nonclinical Safety Assessment of SYN-004: An Oral β-lactamase for the Protection of the Gut Microbiome From Disruption by Biliary-Excreted, Intravenously Administered Antibiotics. Int J Toxicol. 2016. May;35(3):309–16. 10.1177/1091581815623236 [DOI] [PubMed] [Google Scholar]
- Pitout JDD. IPSAT P1A, a class A beta-lactamase therapy for the prevention of penicillin-induced disruption to the intestinal microflora. Curr Opin Investig Drugs Lond Engl. 2009. Aug;10(8):838–44. [PubMed] [Google Scholar]
- Kokai-Kun JF, Roberts T, Coughlin O, Le C, Whalen H, Stevenson R, et al. Use of ribaxamase (SYN-004), a β-lactamase, to prevent Clostridium difficile infection in β-lactam-treated patients: a double-blind, phase 2b, randomised placebo-controlled trial. Lancet Infect Dis. 2019. May;19(5):487–96. 10.1016/S1473-3099(18)30731-X [DOI] [PubMed] [Google Scholar]
- Kokai-Kun JF, Le C, Trout K, Cope JL, Ajami NJ, Degar AJ, et al. Ribaxamase, an Orally Administered β-Lactamase, Diminishes Changes to Acquired Antimicrobial Resistance of the Gut Resistome in Patients Treated with Ceftriaxone. Infect Drug Resist. 2020. Jul;13:2521–35. 10.2147/IDR.S260258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokai-Kun JF, Roberts T, Coughlin O, Sicard E, Rufiange M, Fedorak R, et al. The Oral β-Lactamase SYN-004 (Ribaxamase) Degrades Ceftriaxone Excreted into the Intestine in Phase 2a Clinical Studies. Antimicrob Agents Chemother. 2017. Feb;61(3):e02197–16. 10.1128/AAC.02197-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connelly S, Bristol JA, Hubert S, Subramanian P, Hasan NA, Colwell RR, et al. SYN-004 (ribaxamase), an oral beta-lactamase, mitigates antibiotic-mediated dysbiosis in a porcine gut microbiome model. J Appl Microbiol. 2017. Jul;123(1):66–79. 10.1111/jam.13432 [DOI] [PubMed] [Google Scholar]
- Connelly S, Fanelli B, Hasan NA, Colwell RR, Kaleko M. SYN-007, an Orally Administered Beta-Lactamase Enzyme, Protects the Gut Microbiome from Oral Amoxicillin/Clavulanate without Adversely Affecting Antibiotic Systemic Absorption in Dogs. Microorganisms. 2020. Jan;8(2):152. 10.3390/microorganisms8020152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connelly S, Fanelli B, Hasan NA, Colwell RR, Kaleko M. Oral Beta-Lactamase Protects the Canine Gut Microbiome from Oral Amoxicillin-Mediated Damage. Microorganisms. 2019. May;7(5):150. 10.3390/microorganisms7050150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P, Curtis N. The effect of antibiotics on the composition of the intestinal microbiota - a systematic review. J Infect. 2019. Dec;79(6):471–89. 10.1016/j.jinf.2019.10.008 [DOI] [PubMed] [Google Scholar]
- Connelly S, Parsley T, Ge H, Kaleko M. Identification, Characterization, and Formulation of a Novel Carbapenemase Intended to Prevent Antibiotic-Mediated Gut Dysbiosis. Microorganisms. 2019. Jan;7(1):22. 10.3390/microorganisms7010022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connelly S, Fanelli B, Hasan NA, Colwell RR, Kaleko M. Oral Metallo-Beta-Lactamase Protects the Gut Microbiome From Carbapenem-Mediated Damage and Reduces Propagation of Antibiotic Resistance in Pigs [Internet]. Front Microbiol. 2019. Feb;10:101. [cited 2022 May 26] Available from: https://www.frontiersin.org/article/10.3389/fmicb.2019.00101 10.3389/fmicb.2019.00101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubillos-Ruiz A, Alcantar MA, Donghia NM, Cárdenas P, Avila-Pacheco J, Collins JJ. An engineered live biotherapeutic for the prevention of antibiotic-induced dysbiosis. Nat Biomed Eng. 2022. Jul;6(7):910–21. 10.1038/s41551-022-00871-9 [DOI] [PubMed] [Google Scholar]
- de Gunzburg J, Ghozlane A, Ducher A, Le Chatelier E, Duval X, Ruppé E, et al. Protection of the Human Gut Microbiome From Antibiotics. J Infect Dis. 2018. Jan;217(4):628–36. 10.1093/infdis/jix604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vehreschild MJ, Ducher A, Louie T, Cornely OA, Feger C, Dane A, et al. An open randomized multicentre Phase 2 trial to assess the safety of DAV132 and its efficacy to protect gut microbiota diversity in hospitalized patients treated with fluoroquinolones. J Antimicrob Chemother. 2022. Mar;77(4):1155–65. 10.1093/jac/dkab474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guk J, Guedj J, Burdet C, Andremont A, de Gunzburg J, Ducher A, et al. Modeling the Effect of DAV132, a Novel Colon-Targeted Adsorbent, on Fecal Concentrations of Moxifloxacin and Gut Microbiota Diversity in Healthy Volunteers. Clin Pharmacol Ther. 2021. Apr;109(4):1045–54. 10.1002/cpt.1977 [DOI] [PubMed] [Google Scholar]
- Ducher A, Vehreschild MJ, Vehreschild MJ, Louie TJ, Cornely OA, Féger C, et al. LB-5. DAV132 Protects Intestinal Microbiota of Patients Treated with Quinolones, a European Phase II Randomized Controlled Trial (SHIELD). Open Forum Infect Dis. 2020. Oct;7 Supplement_1:S845–6. 10.1093/ofid/ofaa515.1902 [DOI] [Google Scholar]
- Grall N, Massias L, Nguyen TT, Sayah-Jeanne S, Ducrot N, Chachaty E, et al. Oral DAV131, a charcoal-based adsorbent, inhibits intestinal colonization by β-lactam-resistant Klebsiella pneumoniae in cefotaxime-treated mice. Antimicrob Agents Chemother. 2013. Nov;57(11):5423–5. 10.1128/AAC.00039-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuzuriha K, Yakabe K, Nagai H, Li S, Zendo T, Zai K, et al. Protection of gut microbiome from antibiotics: development of a vancomycin-specific adsorbent with high adsorption capacity. Biosci Microbiota Food Health. 2020;39(3):128–36. 10.12938/bmfh.2020-002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouhten H, Mattila E, Arkkila P, Satokari R. Reduction of Antibiotic Resistance Genes in Intestinal Microbiota of Patients With Recurrent Clostridium difficile Infection After Fecal Microbiota Transplantation. Clin Infect Dis. 2016. Sep;63(5):710–1. 10.1093/cid/ciw390 [DOI] [PubMed] [Google Scholar]