Abstract
The rise of antimicrobial resistant (AMR) bacteria is a global public health threat. AMR Achromobacter bacteria pose a challenging clinical problem, particularly for those with cystic fibrosis (CF) who are predisposed to chronic bacterial lung infections. Lytic bacteriophages (phages) offer a potential alternative to treat AMR infections, with the possible benefit that phage selection for resistance in target bacteria might coincide with reduced pathogenicity. The result is a genetic “trade-off,” such as increased sensitivity to chemical antibiotics, and/or decreased virulence of surviving bacteria that are phage resistant. Here, we show that two newly discovered lytic phages against Achromobacter were associated with stabilization of respiratory status when deployed to treat a chronic pulmonary infection in a CF patient using inhaled (nebulized) phage therapy. The two phages demonstrate traits that could be generally useful in their development as therapeutics, especially the possibility that the phages can select for clinically useful trade-offs if bacteria evolve phage resistance following therapy. We discuss the limitations of the current study and suggest further work that should explore whether the phages could be generally useful in targeting pulmonary or other Achromobacter infections in CF patients.
Keywords: antibiotic, antimicrobial resistance, cystic fibrosis, bacteriophage, personalized medicine, phage therapy, IL-8, trade-off, immune reaction
Introduction
Antimicrobial resistance (AMR) is a global public health threat, exacerbated by the current COVID-19 pandemic [1]. AMR bacteria are emerging due to the widespread, often inappropriate, use of antimicrobials [2], which strongly select for evolved resistance in bacterial pathogens. The stepwise evolution of resistance to each novel antibiotic has left clinicians with few, if any, options to treat highly resistant organisms. Also, many antibiotics are broad spectrum, creating collateral damage on commensal organisms within the natural human microbiomes [3]. The result is an emerging pandemic where AMR is predicted to contribute to more human deaths than all forms of cancer by 2050 [4]. Drug-resistant infections pose a particular problem to patients with chronic diseases that require elevated exposure to antibiotics, such as individuals with the genetic disease cystic fibrosis (CF). CF is characterized by lung inflammation and injury that results in bronchiectasis and persistent lung infections because these bacteria are not effectively cleared [5]. The result is that many CF patients are treated with multiple courses of antibiotics to treat these chronic infections, and consequently bacteria in the CF lung develop drug resistance.
Lytic bacteriophages (phages) – viruses of bacteria – have a long history of successful application as antibacterials in countries such as Russia and Poland, as well as prior Soviet Republics, such as Georgia [6]. More recently, Western medicine has shown interest in phage therapy to treat AMR infections [7]. The greater specificity of phages over antibiotics for target bacteria should minimize the “collateral damage” – ie, decreased species diversity and increased lability [8] – to the human microbiome. Furthermore, the co-evolution that has occurred between phages and their host bacteria for billions of years suggests that additional phage-therapy candidates can be discovered quickly if the target bacteria develop resistance to administered phage(s) [9].
Although not approved as drugs by the US Food and Drug Administration (USFDA), phages have been deployed on a compassionate-use basis at a handful of US institutions via FDA single patient investigational new drug (IND) approval, which includes the Center for Phage Biology & Therapy at Yale University, New Haven, CT. While there are current clinical trials examining safety/efficacy of phage therapy in CF patients (CYPHY; NCT04684641), these trials are currently limited to patients who are colonized with Pseudomonas aeruginosa.
The opportunistic bacterial pathogens of the genus Achromobacter are aerobic, Gram-negative rods that readily form biofilms in the airways of patients with CF [10]. Achromobacter can lead to increased inflammation, pulmonary exacerbations, pneumonia, or bacteremia [11]. Colonization is associated with advanced disease that is often difficult to eradicate. Part of the difficulty in managing these infections arises from their drug resistance, both intrinsic and acquired. It is estimated that 5% of CF patients are colonized with Achromobacter spp., many of which are drug resistant [12]. Pathogenic Achromobacter species include both A. xylosoxidans and A. denitrificans [13]. As colonization with Achromobacter is associated with increased antibiotic use in CF, and the presence of AMR Achromobacter is a relative contraindication to transplantation, alternative therapeutic strategies, such as phage therapy, are needed.
Here, we describe the treatment of a CF patient with AMR Achromobacter pulmonary infection using two newly discovered lytic phages isolated from sewage samples in New Haven, CT, USA. The phages were characterized using in vitro phenotypic assays by determining phage burst size, attachment time, biofilm inhibition capability, and stability at pH and temperatures that mimic clinically relevant conditions. Additionally, the phage genomes were completely sequenced and annotated. Further cell culture experiments were completed to determine possible trade-off effects of phage resistant Achromobacter. Results showed that phage treatment was associated with decreased sputum Achromobacter (eg, microbiological success) and the patient achieved clinical stability. In addition, data from in vitro studies showed that these phages were stable in long-term storage, highly efficient for bacterial killing, and did not select for clinically problematic changes in virulence or antibiotic resistance, when bacteria evolved phage resistance. This phage characterization and case report add to the growing body of literature supporting the successful compassionate use of phage therapy to treat Achromobacter or other AMR bacteria in Western medicine.
Methods
Strains and Culture Conditions
Achromobacter strain #YA1.3 was isolated from patient sputum and served as the host bacteria used in all phage-characterization assays, and to prepare phage lysates for in vitro studies. Bacteria were cultured at 37oC in Lysogeny Broth (LB) medium (10g tryptone, 5g yeast extract, 10g NaCl per L), and grown as colonies on 1.5% LB agar plates. Shaking incubation occurred at 250 rpm.
Preparation of Phage Stocks
To produce phage stocks, host bacteria were cultured in LB medium to exponential phase as measured by optical density at 600 nm wavelength (OD600) of 0.8, and inoculated with phages at low multiplicity of infection (MOI; ratio of particles to host cells). The mixture was incubated with shaking until the culture became visibly cleared by phage lysis (< 24 hours). The solution was then centrifuged at 4000 rcf for 10 min. The supernatant was mixed with phage precipitation buffer (30% w/v PEG-8000, 3 M NaCl), and stored overnight at 4°C. The next day, the stock was centrifuged at 4000 rcf for 15 min, and the pellet was resuspended in 20 mL phage buffer (1x PBS) followed by filter (0.22 µm) sterilization. Titer (plaque-forming units (PFU)/mL) of a phage stock was estimated via plaques formed in spot assays.
Spot Assay
Spot assays were used to estimate the titers of phage lysates. Phage lysates were serially diluted 1:10 in phage buffer and spotted on a double-layer agar plate consisting of 1.5% bottom agar and 0.75% top agar mixed with 100 µl host bacteria. The plate was then incubated at 37°C until the spots could be counted.
Phage Stability Assays
A 20-µL sample of phage lysate with known titer was placed in a sterile PCR tube, moved to a preheated Eppendorf Thermocycler for 1 hour incubation at temperature gradients between 30 to 82°C, and then used in a spot assay to estimate any change in titer. To examine effects of pH on phage stability, phages were diluted to a specific titer in pH-adjusted Phage Buffer (adjusted with NaOH and HCl to pH 1-14) and incubated at 4°C for 24 h, followed by spot assay to estimate titer changes.
Adsorption/Attachment Assay
Adsorption (attachment) assays mixed 500 µL of overnight bacterial culture with 4 mL LB medium and allowed bacteria to grow until OD600 reached 0.6. Then, 500 µL phage lysate (106 PFU/mL) was added, and the mixture was incubated at 37°C with shaking at 250 rpm. At various times, 100 µL samples were removed in triplicates and centrifuged at 12,000 rcf for 5 min. The concentration (titer) of unbound phage particles in the supernatant was determined via spot assay.
Burst Assay
Burst assays mixed 500 µL of overnight bacterial culture with 4 mL LB medium and allowed bacteria to grow until OD600 reached 0.6. Then, 500 µL phage lysate (106 PFU/mL) was added, and the mixture was incubated at 37°C with shaking. At different timepoints, 100 µL samples were removed and measured in triplicates by using spot assays to estimate changes in phage titer. Phage burst size was calculated using the method of Garbe et al. [14].
Biofilm Inhibition Assay
Bacterial cultures were preconditioned in LB medium overnight from individual colonies, then transferred to Tryptic Soy Broth in 200 µL total volume in a U-bottom 96-well plate. Cultures in each well were incubated for 24 h at 37°C. After biofilm formed, 33 µL of a phage lysate was added at different timepoints and concentrations. Crystal Violet (CV) staining was used to estimate cell density following phage infection. Medium was removed from the well, and the biofilm was washed. Then, 125 µL CV was added and incubated for 10 minutes at room temperature. Liquid was removed, and the well was washed 3 times with ddH2O. To dislodge cells, 150 µL acetic acid was applied, followed by 10-minute incubation at room temperature. The solution was transferred to a 96-well flat bottom plate and the absorbance measured at optical density at 550 nm (OD550).
Bacterial-Growth Assay
Bacterial cultures were preconditioned in LB medium overnight from individual colonies, then transferred to fresh LB in 200 µL total volumes. Cultures were incubated at 37°C with shaking and OD600 was monitored at 5-minute intervals for 24 h using a TECAN microplate reader.
Minimum Inhibitory Concentration Assay
The minimum inhibitory concentration (MIC) of antibiotics against bacteria was determined via E-TEST strip (bioMerieux) per manufacturer’s guidelines. Briefly, an overnight culture of bacteria was diluted to a McFarland standard of 0.5, and then spread on Mueller-Hinton agar (2g beef extract, 17.5g casein hydrolysate, 1.5g starch, 17g agar per L) using a sterile swab. The bacterial lawn was allowed to dry, and an E-TEST strip was placed on top of the lawn. After overnight incubation at 37°C the plate was scored by recording the lowest concentration of antibiotic that inhibited growth of the bacterial lawn.
Inflammation Assay
Bacteria cultured overnight in LB were diluted to OD600 of 0.5. A 10 mL volume of diluted bacteria was centrifuged at 4,000 rpm for 15 min, and the supernatant was filtered (0.22 µm). To prepare human-derived cells, 1 mL of 106 immortalized human bronchial epithelial (16HBE) cells were seeded in 24-well plates and grown at 37°C until confluent in growth medium minimal essential medium (MEM; Gibco) 10% fetal bovine serum (FBS; Gemini Bio-Products), supplemented with 2 mM l-glutamine, 5 U/ml penicillin, and 5 μg/ml streptomycin (Sigma) in submerged cultures. Then, cells were washed with 1 mL 1x PBS, and 700 µL of bacterial supernatant (pre- or post-phage) were added to cells and stimulated for 24 h. Identical assays were performed using an immortalized cystic fibrosis ΔF508/ΔF508 bronchial epithelial (CFBE41o-) cell line derived from a CF patient homozygous for F508del mutation. Both 16HBE and CFBE41o- cells were generously provided by Dr. J. Bomberger (University of Pittsburgh). Interleukin-8 concentration in cell culture supernatants was measured using DuoSetTM ELISA Human IL-8/CXCL8 kit (R&D Systems).
Genome Sequencing and Analysis
Phage or bacterial DNA was isolated from a phage stock or overnight culture via phenol-chloroform precipitation. DNA sequencing via Illumina was completed at Yale Center for Genome Analysis. After trimming, genome annotation was conducted using CPT Galaxy and Apollo. Structural gene prediction was completed using GLIMMER, MetaGeneAnnotator, and SixPack. Structural decisions were manually and individually confirmed based on the presence of Shine-Dalgarno Sequences, translation start/stop codons, and gene overlaps. Gene functions were predicted using BLASTp. Putative functions were manually and individually assigned upon review of the BLASTp results. The complete genomic sequences will be uploaded to GenBank.
Data and Statistical Analyses
Data were plotted using Prism GraphPad. The data were statistically analyzed by one- and two-way ANOVA, linear regression, and t-test, as described in the Results.
Isolation of Phage-Resistant Mutants
To isolate phage-resistant bacterial clones, 80 µL of bacterial overnight culture was incubated with 4 µL phage stock in 4 mL LB medium for 12-16 hours. Bacteria were plated on agar and one colony was chosen at random. A lawn of this strain was grown alongside a test phage in top agar, to ensure that the phage could not form plaques.
Results
Case Report
A 39-year-old man with cystic fibrosis (CF) was referred to the Center for Phage Biology & Therapy at Yale for consideration of bacteriophage therapy. His CF genotype is delF508/p.M1V and his disease was characterized by chronic rhinosinusitis, pancreatic insufficiency, and moderate obstructive lung disease. Referral occurred due to the isolation from sputum of increasingly multi-drug resistant (MDR) Achromobacter, and the patient continued with significant symptoms after multiple courses of both oral and intravenous antibiotics. He also showed a recent, stepwise, decline in lung function, as assessed by forced expiratory volume in 1 second (FEV1) from ~50% to 38% predicted, which corresponded with new exertional dyspnea.
The patient’s medication regimen included mucomyst, albuterol, and cromolyn. At the time of evaluation for phage therapy, the CF transmembrane conductance regulator (CFTR) modulator Elexacaftor/tezacaftor/ivacaftor (Trikafta/Kaftrio®) was not available. Sputum cultures showed evidence for three variants of Achromobacter bacteria (Table 1), two of which were multi-drug resistant (MDR), and one of which (#YA1.3) was pan-drug resistant (PDR).
Table 1. Antimicrobial Sensitivities of Three Achromobacter Clinical Isolates.
| Achromobacter #1 | Achromobacter #2 | Achromobacter #3 (strain #YA1.3) | |
| Amikacin* | R | R | R |
| Aztreonam | R | R | R |
| Cefepime | I | R | R |
| Ceftazidime | S | I | R |
| Ciprofloxacin | R | R | R |
| Gentamicin | R | R | R |
| Imipenem | R | S | R |
| Levofloxacin | R | I | R |
| Meropenem | R | R | R |
| Piperacillin-tazobactam | S | S | R |
| Tetracycline | R | R | R |
| Tobramycin | R | R | R |
| Trimethoprim/sulfamethoxazole | S | S | R |
*Broth microdilution determined whether bacteria were sensitive (S), intermediate (I), or resistant (R) to each of the listed antibiotics per CLSI breakpoints.
At the Center for Phage Biology & Therapy, two species of Achromobacter were identified in spontaneously expectorated sputum: A. denitrificans and A. xylosoxidans (via 16s sequencing; data not shown). Two lytic phages, Achr-1 and Achr-2 were isolated from sewage samples (New Haven, CT, USA) and found to be capable to infect these pathogens (see phage characterization below). As this patient was ineligible for enrollment in a phage therapy clinical trial, USFDA single-patient IND and Yale University IRB approval were obtained to administer compassionate phage therapy. The patient signed informed written consent.
Phage lysates were prepared by growing A. xylosoxidans (American Type Culture Collection #27061) in the laboratory to exponential phase in TSB (30g of TSB powder (non-animal origin)) in 1 L diH20 (per manufacturer protocol, Sigma Aldrich). A phage was then mixed with bacteria at MOI of ~0.01 and incubated at 37oC with shaking (100 rpm). After 6 hours, cultures were centrifuged and filtered (pore size: 0.22 µm) to obtain a cell-free phage lysate. Lysate was then concentrated with Centricon small pore concentrators (100 kDa MWCO) and dialyzed in 1000x volume PBS with MgSO4. Endotoxin concentration was quantitated with Hyglos EndoNext kits (bioMerieux Inc., Durham, NC). Per FDA recommendations, USP 71 testing was completed by a third-party laboratory (Accugen Laboratories, Addison, IL) on all phage preparations that were used for phage therapy. The final preparation was diluted in PBS with MgSO4 to a concentration of 1.0 x 1010 PFU/mL for individual phages. Final phage concentration was diluted into 3 ml PBS for nebulization.
Phage Achr-1 was administered once daily at a dose of 1x1010 PFU/mL via the patient’s own nebulizer. The first dose was supervised in the Adult CF Clinic without any evidence for toxicity. Subsequent daily doses were completed at home for 7 days. The patient tolerated therapy without incident. Pre- (days -7 and 0) and post-treatment (days 3, 7, 14) sputum samples showed a reduction in A. denitrificans from 5.5x106 to 1.7x104 CFU/mL but the total Achromobacter burden remained unchanged (Figure 1).
Figure 1.
Estimated densities (CFU/mL) of Achromobacter species in longitudinal samples of patient sputum. Total density of Achromobacter (A. denitrificans and A. xylosoxidans; filled circles) remained relatively constant over time, whereas the subpopulation of A. denitrificans bacteria (open circles) declined by orders of magnitude following phage treatment. Lines are results of regression analyses, where linear regressions are depicted for total Achromobacter and A. denitrificans bacteria CFU data, respectively.
Over 3 months of follow-up, the patient reported one pulmonary exacerbation that required antibiotics and his FEV1 stabilized at 39% predicted. In an attempt to reduce total Achromobacter burden, Achr-2 was used for phage therapy to target A. xylosoxidans. Again, the initial phage therapy was observed at Yale’s Adult CF Clinic. However, post-treatment sputum analysis was not available because the patient did not provide post-phage therapy sputum samples. Thus, we were unable to determine if there was a decline in total Achromobacter CFU/mL or if the patient’s isolates demonstrated evidence of phage resistance after the second phage therapy. After the second round of phage treatment, the patient’s FEV1 remained stable at 36% 2 months post-therapy before declining to 26% at 6 months post-therapy. At that point, the recently approved modulator therapy elexacaftor/tezacaftor/ivacaftor was initiated and after elexacaftor/tezacaftor/ivacaftor, the patient’s FEV1 improved to 43% predicted.
In Vitro Measures of Phage Kinetics
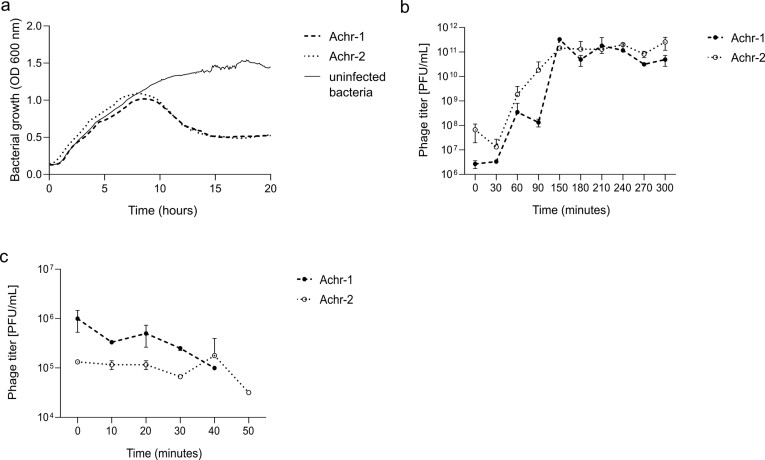
We determined the kinetics of bacterial growth (see Methods) in the presence and absence of independent infection by phages Achr-1 and Achr-2. Bacteria strain #YA1.3 (Table 1) was grown in LB medium with shaking. Results (Figure 2a) showed that the population grew exponentially until resources in the medium were exhausted and then entered stationary phase where growth ceased. By contrast, both phages Achr-1 and Achr-2 were capable of negatively impacting host bacterial growth when mixed with host cells at low MOI, as indicated by the decrease in optical density estimates around 8 hr (Figure 2a). However, at later time points the bacterial population was observed to recover and to increase in density (Figure 2a). The latter results showed that spontaneous evolution of host resistance to each phage could readily occur, suggesting that examination of changes in phenotypic traits of phage-resistant mutants were warranted (see below) as a similar outcome was considered possible during patient treatment.
Figure 2.
In vitro kinetics of bacteria and phages. (a) Uninfected bacteria (solid line) show expected growth kinetics in the absence of phage infection, achieving stationary phase within 20 hours. Whereas, incubation with either phage Achr-1 or Achr-2 (dashed and dotted lines) negatively impacted growth of host bacteria within 10 hours, followed by host recovery due to growth of phage-resistant mutant cells. (b) Populations of both phage Achr-1 (filled circles, dashed line) and phage Achr-2 (open circles, dotted line) show expected kinetics of lytic phage growth on host bacteria in one-step burst assays. Standard deviation of technical duplicates represented by error bars. (c) Attachment of phages Achr-1 (filled circles, dashed line) and Achr-2 (open circles, dotted line) are similar preceding growth of the virus population on hosts cells (Supplemental Figure S1), as both viruses irreversibly bind to susceptible host cells by 40 min and 50 min, respectively. Standard deviation of technical duplicates represented by error bars.
We then used burst assays (see Methods) to examine the kinetics of growth for each phage independently on host bacteria, to compare/contrast individual phage traits. Each phage was mixed with host bacteria #YA1.3 in duplicate, and every 30 min samples were obtained and subjected to plating assays that estimated phage titers (PFU/mL). Results (Figure 2b) showed that the kinetics of the two phages were similar. Each of the lytic phages experienced a latent (eclipse) period of roughly 30 min that preceded virus population growth, followed by exponential phage population growth on host cells as they were infected and lysed, and finally a period of non-increase because the phage population exhausted the susceptible host cells capable of supporting phage infection (Figure 2a). Using the method of Garbe et al. [14], we calculated that phage Achr-1 produced roughly 9,900 particles per infected host cell, and phage Achr-2 produced about 5,600 particles per cell.
To more closely examine the kinetics of phage adsorption (attachment) to host bacterial cells, each phage was mixed with host bacteria at low MOI and plated samples obtained every 10 min on host lawns to estimate how quickly particles bound to susceptible cells. Results (Figure 2c) showed that particles “disappeared” from solution over time, as they became irreversibly bound to susceptible host bacterial cells. The attachment kinetics of the two phages were roughly similar. The data indicated that phage Achr-1 attached to cells by around 40 min (Figure 2c), which preceded an observed increase in the phage population by 50 min (Supplemental Figure S1). Whereas, data for phage Achr-2 presented an estimated 50-min adsorption (Figure 2c) that preceded expansion of the phage population at a relatively later time point of 60 min (Supplemental Figure S1).
In Vitro Measures of Biofilm Inhibition
The ability for the two phages to reduce biofilms of host bacteria was examined by infecting cells with single phage versus together in a mixture (cocktail). Biofilm inhibition assays estimated the reduction in cell numbers within established biofilms prior to phage inoculation. Uninfected Achromobacter bacteria established biofilms (Figure 3). Fluorescence was used as a proxy for cell density, whereas growth medium (control) showed minimal background fluorescence. By contrast, after 12-hour exposure of host bacteria to the inocula of each phage alone, or together in a 1:1 cocktail, reduced fluorescence was observed (Figure 3). Assays that subjected bacteria to phage exposure for 24 hours total were similar to 12 hours, which suggests that the ability of phage(s) to inhibit biofilm growth was maximized within the first 12 hours. Interestingly, results for application of phage(s) twice in the 24-hour assay (ie, initial inoculum, followed by a second inoculum at 12 hours) did not improve biofilm inhibition, and these results for phage Achr-1 alone seemed detrimental for biofilm reduction.
Figure 3.
Ability for phage(s) to inhibit bacterial biofilms. Exposure of bacterial biofilms to phages Achr-1 and Achr-2 alone and together (cocktail) generally did not significantly reduce cell numbers (using fluorescence as proxy for cell density), relative to absence of phage infection. Longer exposure (12 vs. 24 hr) to phage(s) does not generally improve reduction of biofilm inhibition, nor do two-step exposures to phage(s). Standard deviation of biological triplicates represented by error bars.
Phage Stability
For phage therapy in humans, a phage should show stability. In addition, for convenient long-term on-demand use, ideal phages should be stable under typical storage conditions. Achr-1 and Achr-2 were stable (did not decrease in titer) when stored at 4°C for at least 3 months (data not shown). To look at generalized effects of phage thermotolerance as a proxy for stability, we incubated lysates of each phage for 1 hour at a range of temperatures between -80°C and 80°C (see Methods). Both phages were similar in thermotolerance, as both were stable at low temperatures and up to 49°C, which greatly exceeds human body temperature (Figure 4). Some differences were observed at temperatures above 49°C, especially greater sensitivity of phage Achr-1 to mortality at elevated temperatures between 52°C and 65°C, whereas phage Achr-1 showed slightly better stability than phage Achr-2 at the highest temperatures examined (Figure 4a). The effect of pH was also investigated as a proxy for phage stability because the CF lung environment is characterized as lower pH (approx. 6.5) relative to healthy lungs of non-CF individuals (approx. 7.5) [15]. Phage lysates were investigated in a range of pH-adjusted phage buffers for 24 h total (see Methods). Results (Figure 4b) showed no evidence for survival of either phage at pH 1 and 2, and at pH 13 and 14. By contrast, stability of both phages was nearly maximal between pH 3 and 11, which includes pH levels of both CF and non-CF lung environments. Our data suggested that phage Achr-1 was more sensitive to elevated pH, evidenced by its lower survival at pH 12.
Figure 4.
Stability of phages when exposed to varying temperatures and pH levels. (a) Both phages Achr-1 and Achr-2 (filled, open circles) were relatively stable in a single measurement when subjected to temperature shocks at low and high temperatures for 1-hour duration. (b) Both phages were stable in single measurements across a wide range of pH levels following 24-hour incubations.
Effects of Phage Resistance on Antibiotic Sensitivity
As previous studies show acquisition of phage resistance can alter sensitivity of AMR bacteria to antibiotics [16], Achromobacter resistant to phage infection by phage Achr-1 (ie, mutant “Achr-1 res”), and a separate mutant resistant to phage Achr-2 (ie, mutant “Achr-2 res”) were isolated. Both phage-resistant mutants showed a mucoid phenotype compared to the phage-susceptible wildtype bacteria (data not shown). MIC assays were performed with five antibiotics, to compare drug sensitivities of wildtype #YA1.3 and of phage-resistant mutants. Results (Figure 5a) for the Achr-1-resistant mutant showed increased sensitivity (ie, fold-change in MIC exceeding 1.0) for ertapenem, omadacycline, and minocycline. Similarly, we observed that the Achr-2-resistant mutant became more-sensitized to omadacycline and minocycline (Figure 5b). Taken together, the data suggested that evolution of bacterial resistance to either phage created a trade-off, whereby the mutants became more-easily inhibited by one or two classes of antibiotics: tetracyclines and a carbapenem. Whereas, the data showed that evolution of phage resistance was not associated with substantial changes in MIC for the other three chemical antibiotics examined (Figure 5a,b).
Figure 5.
Impact of evolved phage-resistance on potential trade-offs in Achromobacter. (a) Fold change in sensitivity (MIC) measured in singlets for wildtype Achromobacter ancestor relative to MIC for its phage-resistant Achr-1 res mutant was evident for Ertapenem, Omadacycline, and Minocycline antibiotics. (b) Fold change (MIC) for the Achr-2 res mutant measured in singlets was evident for Omadacycline and Minocycline antibiotics. (c) IL-8 inflammation assays in laboratory tissue culture showed that phage-resistant mutants presented values lower (Achr-1 res) or unchanged (Achr-2 res) compared to the wildtype Achr ancestor on 16HBEo cells. Standard deviation of biological duplicates measured in technical triplicates making up a total of six measurements represented by error bars. (d) IL-8 assays on CFBE cells indicated that neither mutant caused inflammation that differed from that of their Achr ancestor. Standard deviation of biological duplicates measured in technical triplicates making up a total of six measurements represented by error bars.
Effects of Phage Resistance on Bacteria-Induced Inflammation
We sought to determine whether evolution of phage-resistance affected the ability of bacteria to cause inflammation in an in vitro model. One concern for phage therapy is that bacteria that develop resistance to phage(s) to survive may develop increased or decreased virulence factors (eg, lipopolysaccharide (LPS)). Thus, in response to phage therapy surviving bacteria may produce more LPS, which increases lung inflammation, or less LPS, which results in decreased inflammation.
For example, if evolved phage resistance was due to loss or downregulation of lipopolysaccharide (LPS), or other virulence factors, which phage(s) used as receptors to infect bacteria (see below) [17], this could increase bacterial virulence, thereby causing greater inflammation (relative to wildtype bacteria) [18,19] in patient tissue as a negative consequence of administered phage therapy.
To examine the results of this type of trade-off, bacterial supernatants of wildtype (PDR strain #YA1.3) and two spontaneous phage-resistant Achromobacter bacteria (Achr-1 res, Achr-2 res) were added to confluent monolayers of human-derived cell lines for 24 h. Cystic fibrosis epithelial (CFBEo) cells, an immortalized human airway epithelial cell line isolated from a CF patient with F508del homozygous mutation (by comparison, patient genetics included one copy of F508del), and 16HBE cells, a standard laboratory immortalized human airway epithelial cell line, were used in these assays. Following incubation, IL-8 concentration in the supernatants of CFBEo- and 16HBE cells was measured after 24 h. IL-8 was selected because it is a neutrophil chemokine, and neutrophilic inflammation is one characteristic of CF lung disease.
In these experiments, Achr-1 res caused significantly lower IL-8 concentrations produced by CFBE cells, relative to wildtype (Achr) bacteria. While stimulation with the mutant Achr-2 res was measurably lower than wildtype Achr on these cells (Figure 5d), results with 16HBE cells (Figure 5d) showed lower IL-8 for the two Achr mutants relative to wildtype bacteria. However, these differences were not statistically significant. Altogether, the combined data did not suggest that evolution of phage resistance caused an adverse “trade-up” in cellular inflammation. In fact, there is evidence that resistant mutants resulted in less stimulation of IL-8 production.
Whole-Genome Sequencing and Annotation of Phages
Newly discovered phages Achr-1 and Achr-2 were fully sequenced using Illumina, and the resulting data were annotated using Galaxy CPT Public and Apollo. We then created inferred gene maps for the phages using GenomeVx and determined the GC content of each phage using GeeCee. The virus family for each phage was predicted using BLASTn search.
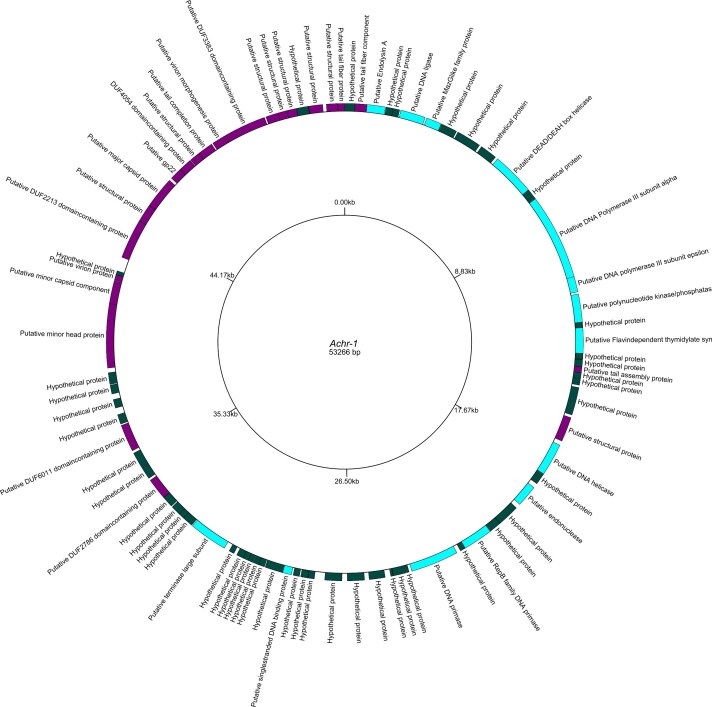
The analysis of phage Achr-1 (Figure 6) determined a genome size of roughly 53,266 bp and predicted that the virus genome contained 81 ORFs. No terminal repeats were found indicating the genome of Achr-1 was linear. Of these ORFs, the annotation identified 20 putative functional proteins, 17 putative structural proteins, and 44 hypothetical proteins. Importantly, no ORFs were predicted to have integrase functions, which suggested that phage Achr-1 was strictly lytic and thereby incapable of converting Achromobacter host bacterial cells to become lysogenic. Last, our analysis determined that the GC content of phage Achr-1 was 55%.
Figure 6.
Draft genome map of Achromobacter phage Achr-1. Genome sequencing and annotation of phage Achr-1 showed hypothetical proteins (dark green), putative structural genes (purple), and putative functional proteins (light blue).
A whole-genome BLASTn search revealed high sequence similarity (98.98%) of phage Achr-1 to viruses of the genus Pbunavirus, which belong to the virus family Myoviridae. No other known virus families showed significant sequence similarity to phage Achr-1. Comparative analysis of the genome sequence of phage-resistant mutant Achr-1 res to that of the phage-susceptible Achr ancestor bacteria revealed the core of LPS of Achromobacter as the putative attachment motif for phage Achr-1 (see Discussion).
The genome analysis of phage Achr-2 (Figure 7) showed a linear genome size of roughly 64,169 bp with no terminal repeats and the annotation of this virus suggested 94 predicted ORFs, for which 16 putative functional proteins, four putative structural proteins, and 74 hypothetical proteins were inferred. No ORFs had predicted integrase functions, indicating that phage Achr-2 was strictly lytic. The GC content of phage Achr-2 was determined to be 55%.
Figure 7.
Draft genome map of Achromobacter phage Achr-2. Genome sequencing and annotation of phage Achr-2 showed hypothetical proteins (dark green), putative structural genes (purple) and putative functional proteins (light blue).
A whole-genome BLASTn search revealed high sequence similarity (97.52%) of phage Achr-2 to viruses of the genus Pbunavirus, in the virus family Myoviridae. No other virus families showed significant sequence similarity to phage Achr-2. Sequence comparison between phage-resistant mutant Achr-2 res to that of the phage-susceptible Achr ancestor bacteria revealed LPS as the putative attachment motif for phage Achr-2 (see Discussion).
Discussion
This study shows the discovery and characterization of two lytic phages that were capable of killing Achromobacter, in particular MDR and PDR strains of A. xylosoxidans that were isolated from the sputum of a CF patient. These phages were deployed in the compassionate use treatment of this patient who had declining respiratory status. Nebulization was tolerated without toxicity, and phage therapy showed efficacy in reducing bacterial burden of the A. denitrificans subpopulation. Following therapy, the patient achieved stabilization of respiratory status. This report adds to a growing literature that shows clinical improvement when phage(s) are used to target refractory Achromobacter infections in the lungs of CF patients [20-22].
In vitro characterization of the two Achr phages revealed traits that could be generally useful for inhaled phage therapy. In particular, the high efficiency in their growth abilities on target bacteria, stability in the face of different temperatures and pH and rapid attachment (adsorption) to host bacterial cells. Equally important, in vitro assays showed few concerns over phage selection for evolved resistance in host bacteria that could coincide with “trade-ups” whereby pathogens become more dangerous following administered phage treatment. In fact, these results indicated that selection for phage-resistant mutants sometimes showed trade-offs: 1) increased sensitivity to one or more classes of antibiotics, and 2) reduced IL-8 production induced by bacterial supernatants on human-derived cells. Further characterization is warranted to study a greater number of spontaneous mutants resistant to phage infection, in particular to examine additional antibiotics that could be used in patient treatment. Furthermore, we believe that additional in vitro studies of two-phage cocktails (mixtures) should be examined alongside assays mimicking individually-administered or sequentially-delivered phages Achr-1 and Achr-2, to determine which strategy would be most effective in killing target bacteria. For example, our assays that looked at ability for each phage to suppress bacterial growth alone showed that the host-cell population became enriched for mutants capable of resisting phage infection, and it is unknown whether a cocktail of the two phages, or sequential exposure to the phages, would be superior to reducing outgrowth of these phage-resistant variants.
Our study has a number of limitations: First, the patient was unable to provide post-treatment sputum samples after the second round of phage therapy, which limited our ability to assess for a microbiological response to therapy. Second, while this study showed evidence for biofilm inhibition, these assays were based on CV staining, which measures cell density without distinguishing between live and dead cells. Therefore, an assay that indicates cell viability, such as the XTT assay [23], could be used in future experiments. Third, we did not phenotypically characterize phage-resistant isolates. Specifically, future studies could explore the mucoid phenotype that tends to occur when Achromobacter evolves resistance to either phage (data not shown). This phenotype indicates that the bacteria form a capsule surrounding the cell, presumably to shield against phage binding, rather than down-regulating or deleting the currently unknown attachment motif [24]. This change to mucoid phenotype could also explain why measurably reduced IL-8 was observed for each phage-resistant mutant supernatant. Fourth, only IL-8 was measured as an inflammatory marker. By measuring other cytokines, it may be observed that a stronger immune reaction is elicited by the supernatants of phage-resistant bacteria.
Nearly 50 years ago, phage phi-X174 yielded the first ever published genome sequence [25], whereas the first sequenced Achromobacter phages appeared in the literature only within the last 8 years [26]. Thus, the longstanding ease of phage isolation from natural sources since the early days of phage therapy now coincides with the increasing affordability of whole-genome sequencing of newly discovered viruses. Phage research in the twentieth century mainly focused on a handful of coliphages, shown to infect genotypes of the popular lab model, Escherichia coli. But the rise of antibiotic resistance in bacteria such as Achromobacter has provided strong motivation for researchers to discover promising phage candidates which could be used alongside, or in replacement of, chemical antibiotics (eg, [23,27,28]). Not surprisingly, increasing accounts of Achromobacter phages have appeared in the literature since the work of Whitman et al. [26], as biomedical researchers are motivated to address AMR infections by discovering and characterizing novel phage candidates. Thus, our study expands the work conducted on Achromobacter-specific phages, with an emphasis on possible usefulness of the Achr-1 and Achr-2 viruses in development of phage therapy.
Ideally, binding of candidate therapeutic phages to target bacteria can be determined to foster predictions of how phage selection may or may not lead to evolved trade-offs in infecting pathogens [7,29]. For the phages described in the current study, no knockout library of Achromobacter was available to identify the attachment motif. A first hint for the attachment motif was the mucoid phenotype of the resistant bacteria, indicating the bacteria has formed a capsule around itself instead of shedding-off or downregulating the phages attachment motif. This is a phenomenon mostly seen when LPS is the attachment motif. Therefore, we used the comparative sequencing results (data not shown) of the phage-resistant and susceptible wildtype bacteria, as well as the phenotypic trade-off assays, to infer whether purported receptor(s), especially LPS-related proteins were used in phage binding. The sequence analysis for phage Achr-1 revealed no change in sequences of the mutant and ancestor in the outer membrane protein H1 (100% sequence identity). But sequence differences in the genes for proteins in LPS export system of ATP-binding protein LptB (29.44% sequence identity), GDP-6-deoxy-D-mannose reductase (28.01% sequence identity), Adenosine 5’-monophosphoramisase HNT1 (33.64% sequence identity), and Beta-ketoacyl-[acyl-carrier-protein] synthase FabY (26.52% sequence identity) were detected. The LPS export system ATP-binding protein LptB is part of the ABC transporter complex LptBFG involved in the translocation of LPS from the inner membrane to the outer membrane, probably responsible for energy coupling to the transport system. GDP-mannose 4,6-dehydratase is part of the LPS biosynthesis complex. Beta-ketoacyl-[acyl-carrier-protein] synthase FabY is thought to catalyze the first elongation reaction of type-II fatty acid synthesis. These results suggested that the core of LPS could be the attachment motif of phage Achr-1.
In an identical comparative analysis for the wildtype strain and the mutant resistant to phage Achr-2, the results showed no sequence differences in purported genes for attachment proteins. However, the aforementioned sequence comparison of the Achr-2 res mutant and wildtype Achromobacter did not further suggest this possible binding target, and it is unclear how phage Achr-2 may be attaching to host cells.
Not long after Felix d’Herelle co-discovered bacteriophages in 1917, he conducted groundbreaking studies on phage ability to address bacterial infections in humans and animal models. Some of d’Herelle’s pioneering work in phage therapy appeared in the current journal, and our study continues a longstanding tradition of Yale research on phages as therapeutics [30-32], given that d’Herelle spent a brief period at Yale (1928-33) as the only academic appointment in his scientific career [33].
Acknowledgments
This work was supported by the National Institutes of Health grants T32AI007517-21A1 to to SG. PET, BKC, and JLK acknowledge generous support for this work from Yale University for the Center for Phage Biology & Therapy.
Glossary
- AMR
antimicrobial resistance
- CF
cystic fibrosis
- USFDA
U.S. Food and Drug Administration
- IND
investigational new drug
- LB
Lysogeny Broth
- MOI
multiplicity of infection
- PFU
plaque-forming units
- MIC
minimum inhibitory concentration
- CV
Crystal Violet
Appendix A.
Data Availability
Sequenced genomes will be deposited to GenBank. Accession numbers to be provided on deposit.
References
- Centers for Disease Control and Prevention, National Center for Emerging and Zoonotic Infectious Diseases, Division of Healthcare Quality Promotion. COVID-19: U.S. Impact on Antimicrobial Resistance, Special Report 2022 [Internet];June/14/2022. Available from: https://stacks.cdc.gov/view/cdc/117915
- Wise R, Hart T, Cars O, Streulens M, Helmuth R, Huovinen P, et al. Antimicrobial resistance. Is a major threat to public health. BMJ. 1998. Sep;317(7159):609–10. 10.1136/bmj.317.7159.609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchings MI, Truman AW, Wilkinson B. Antibiotics: past, present and future. Curr Opin Microbiol. 2019. Oct;51:72–80. 10.1016/j.mib.2019.10.008 [DOI] [PubMed] [Google Scholar]
- Murray CJ, Ikuta KS, Sharara F, Swetschinski L, Robles Aguilar G, Gray A, et al. Antimicrobial Resistance Collaborators. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet. 2022. Feb;399(10325):629–55. 10.1016/S0140-6736(21)02724-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyczak JB, Cannon CL, Pier GB. Lung infections associated with cystic fibrosis. Clin Microbiol Rev. 2002. Apr;15(2):194–222. 10.1128/CMR.15.2.194-222.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutateladze M, Adamia R. Phage therapy experience at the Eliava Institute. Med Mal Infect. 2008. Aug;38(8):426–30. 10.1016/j.medmal.2008.06.023 [DOI] [PubMed] [Google Scholar]
- Kortright KE, Chan BK, Koff JL, Turner PE. Phage Therapy: A Renewed Approach to Combat Antibiotic-Resistant Bacteria. Cell Host Microbe. 2019. Feb;25(2):219–32. 10.1016/j.chom.2019.01.014 [DOI] [PubMed] [Google Scholar]
- Yassour M, Vatanen T, Siljander H, Hämäläinen AM, Härkönen T, Ryhänen SJ, et al. DIABIMMUNE Study Group. Natural history of the infant gut microbiome and impact of antibiotic treatment on bacterial strain diversity and stability. Sci Transl Med. 2016. Jun;8(343):343ra81. 10.1126/scitranslmed.aad0917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Classen AY, Würstle S, Willy C, Vehreschild MJ. Clinical use of bacteriophages [Klinische Anwendung von Bakteriophagen]. Pharmakon. 2021;9(6):469–75. [Google Scholar]
- Busse HJ, Auling G. Achromobacter. In: Whitman WB, Rainey F, Kämpfer P, Trujillo M, Chun J, DeVos P, et al. Bergey’s Manual of Systematics of Archaea and Bacteria. Wiley; 2015. pp. 1–14. [Google Scholar]
- Swenson CE, Sadikot RT. Achromobacter respiratory infections. Ann Am Thorac Soc. 2015. Feb;12(2):252–8. 10.1513/AnnalsATS.201406-288FR [DOI] [PubMed] [Google Scholar]
- Barrado L, Brañas P, Orellana MÁ, Martínez MT, García G, Otero JR, et al. Molecular characterization of Achromobacter isolates from cystic fibrosis and non-cystic fibrosis patients in Madrid, Spain. J Clin Microbiol. 2013. Jun;51(6):1927–30. 10.1128/JCM.00494-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glupczynski Y, Hansen W, Freney J, Yourassowsky E. In vitro susceptibility of Alcaligenes denitrificans subsp. xylosoxidans to 24 antimicrobial agents. Antimicrob Agents Chemother. 1988. Feb;32(2):276–8. 10.1128/AAC.32.2.276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbe J, Wesche A, Bunk B, Kazmierczak M, Selezska K, Rohde C, et al. Characterization of JG024, a pseudomonas aeruginosa PB1-like broad host range phage under simulated infection conditions. BMC Microbiol. 2010. Nov;10(1):301. 10.1186/1471-2180-10-301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkebile AR, McCray PB Jr. Effects of airway surface liquid pH on host defense in cystic fibrosis. Int J Biochem Cell Biol. 2014. Jul;52:124–9. 10.1016/j.biocel.2014.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurney J, Pradier L, Griffin JS, Gougat-Barbera C, Chan BK, Turner PE, et al. Phage steering of antibiotic-resistance evolution in the bacterial pathogen, Pseudomonas aeruginosa. Evol Med Public Health. 2020. Jul;2020(1):148–57. 10.1093/emph/eoaa026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCallin S, Oechslin F. Bacterial Resistance to Phage and Its Impact on Clinical Therapy. In: Górski A, Międzybrodzki R, Borysowski J. Phage Therapy: A Practical Approach. Cham: Springer International Publishing; 2019. pp. 59–88. 10.1007/978-3-030-26736-0_3 [DOI] [Google Scholar]
- Gram A, Kowalewski MP. Molecular Mechanisms of Lipopolysaccharide (LPS) Induced Inflammation in an Immortalized Ovine Luteal Endothelial Cell Line (OLENDO). Vet Sci. 2022. Feb;9(3):99. 10.3390/vetsci9030099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira PC, Dorneles GP, Santana Filho PC, da Silva IM, Schipper LL, Postiga IA, et al. Increased LPS levels coexist with systemic inflammation and result in monocyte activation in severe COVID-19 patients. Int Immunopharmacol. 2021. Nov;100:108125. 10.1016/j.intimp.2021.108125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gainey AB, Burch AK, Brownstein MJ, Brown DE, Fackler J, Horne B, et al. Combining bacteriophages with cefiderocol and meropenem/vaborbactam to treat a pan-drug resistant Achromobacter species infection in a pediatric cystic fibrosis patient. Pediatr Pulmonol. 2020. Nov;55(11):2990–4. 10.1002/ppul.24945 [DOI] [PubMed] [Google Scholar]
- Hoyle N, Zhvaniya P, Balarjishvili N, Bolkvadze D, Nadareishvili L, Nizharadze D, et al. Phage therapy against Achromobacter xylosoxidans lung infection in a patient with cystic fibrosis: a case report. Res Microbiol. 2018. Nov;169(9):540–2. 10.1016/j.resmic.2018.05.001 [DOI] [PubMed] [Google Scholar]
- Lebeaux D, Merabishvili M, Caudron E, Lannoy D, Van Simaey L, Duyvejonck H, et al. A Case of Phage Therapy against Pandrug-Resistant Achromobacter xylosoxidans in a 12-Year-Old Lung-Transplanted Cystic Fibrosis Patient. Viruses. 2021. Jan;13(1):60. 10.3390/v13010060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Chen P, Lin Z, Wang T. Characterization of Two Pseudomonas aeruginosa Viruses vB_PaeM_SCUT-S1 and vB_PaeM_SCUT-S2. Viruses. 2019. Apr;11(4):318. 10.3390/v11040318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhry W, Lee E, Worthy A, Weiss Z, Grabowicz M, Vega N, et al. Mucoidy, a general mechanism for maintaining lytic phage in populations of bacteria. FEMS Microbiol Ecol. 2020. Oct;96(10):fiaa162. 10.1093/femsec/fiaa162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F, Air GM, Barrell BG, Brown NL, Coulson AR, Fiddes CA, et al. Nucleotide sequence of bacteriophage phi X174 DNA. Nature. 1977. Feb;265(5596):687–95. 10.1038/265687a0 [DOI] [PubMed] [Google Scholar]
- Wittmann J, Dreiseikelmann B, Rohde M, Meier-Kolthoff JP, Bunk B, Rohde C. First genome sequences of Achromobacter phages reveal new members of the N4 family. Virol J. 2014. Jan;11(1):14. 10.1186/1743-422X-11-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HC, Chen CR, Lin JW, Shen GH, Chang KM, Tseng YH, et al. Isolation and characterization of novel giant Stenotrophomonas maltophilia phage phiSMA5. Appl Environ Microbiol. 2005. Mar;71(3):1387–93. 10.1128/AEM.71.3.1387-1393.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen M, Le S, Jin X, Li G, Tan Y, Li M, et al. Characterization and Comparative Genomic Analyses of Pseudomonas aeruginosa Phage PaoP5: New Members Assigned to PAK_P1-like Viruses. Sci Rep. 2016. Sep;6(1):34067. 10.1038/srep34067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortright KE, Chan BK, Turner PE. High-throughput discovery of phage receptors using transposon insertion sequencing of bacteria. Proc Natl Acad Sci USA. 2020. Aug;117(31):18670–9. 10.1073/pnas.2001888117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Herelle F. Studies Upon Asiatic Cholera. Yale J Biol Med. 1929. Mar;1(4):195–219. [PMC free article] [PubMed] [Google Scholar]
- d’Herelle F. The Carrier Problem. Yale J Biol Med. 1930. Oct;3(1):21–38. [PMC free article] [PubMed] [Google Scholar]
- D’Herelle F. Bacterial Mutations. Yale J Biol Med. 1931. Oct;4(1):55–61. [PMC free article] [PubMed] [Google Scholar]
- Summers WC. Félix Hubert d’Herelle (1873-1949): history of a scientific mind. Bacteriophage. 2017. Jan;6(4):e1270090. 10.1080/21597081.2016.1270090 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Sequenced genomes will be deposited to GenBank. Accession numbers to be provided on deposit.