Abstract
Pre-existing cross-reactive immune responses to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) proteins in infection-naive subjects have been described by several studies. In particular, regions of high homology between SARS-CoV-2 and common cold coronaviruses have been highlighted as a likely source of this cross-reactivity. However, the role of such cross-reactive responses in the outcome of SARS-CoV-2 infection and vaccination is currently unclear. Here, we review evidence regarding the impact of pre-existing humoral and T cell immune responses to outcomes of SARS-CoV-2 infection and vaccination. Furthermore, we discuss the importance of conserved coronavirus epitopes for the rational design of pan-coronavirus vaccines and consider cross-reactivity of immune responses to ancestral SARS-CoV-2 and SARS-CoV-2 variants, as well as their impact on COVID-19 vaccination.
Subject terms: Immunological memory, Immunopathogenesis
This Review discusses the evidence for pre-existing cross-reactive immune responses to SARS-CoV-2, which are mainly due to infections with common cold coronaviruses, and how such cross-reactivity affects adaptive immune responses. Furthermore, it explores cross-reactivity in the context of SARS-CoV-2 variants of concern and its implications for vaccine development.
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causal agent for COVID-19, is a member of a large family of positive-sense RNA human coronaviruses (HCoVs), which include the Alphacoronaviruses (α-CoVs) 299E and NL63 and the Betacoronaviruses (β-CoVs) HKU1, OC43, Middle East respiratory syndrome coronavirus (MERS-CoV), SARS-CoV and SARS-CoV-2 (ref. 1). All HCoVs express the structural proteins spike, envelope, membrane and nucleocapsid, accessory proteins in open reading frames (ORFs) 3, 6, 7, 8, 9, 10 and 14, the much larger group of non-structural proteins (NSPs) in ORF1 (ref. 2). There is a high degree of homology among HCoVs in both structural proteins and NSPs3,4. The so-called common cold coronaviruses (CCCs) HKU1, OC43, NL63 and 299E are seasonal, are common in human populations globally and were responsible for an estimated 10–20% of viral respiratory infections in 2019, causing predominantly mild disease5. By contrast, MERS-CoV, SARS-CoV and SARS-CoV-2 have all emerged in the past 20 years and can cause severe, potentially fatal disease6. Although SARS-CoV-2 is broadly classified as pathogenic owing to its relatively high case–fatality ratio7, COVID-19 shows a diverse range of symptoms from asymptomatic or mild disease in the majority of patients, to critical disease including acute respiratory distress syndrome, pneumonia, cardiac arrythmia, encephalopathy, acute kidney injury and, in some cases, death8. Several risk factors for COVID-19 disease severity have been identified. These include demographic factors such as age9, Black and Asian ethnicity10, male gender11 and comorbidities such as diabetes12, kidney disease, cerebrovascular disease, cardiovascular disease and respiratory disease13,14. Moreover, various immunological correlates have been associated with COVID-19 severity and disease outcomes. These include autoantibodies to type I interferons15,16; altered myeloid cell populations including increased numbers of immature neutrophils and loss of non-classical monocytes17 as well as increased hyperinflammatory and aberrant CD163+ monocytes18. Elevated serum cytokine levels are associated with severe COVID-19 infections and are a strong predictor of adverse disease outcomes19. Adaptive immune responses, including the development of a coordinated SARS-CoV-2-specific CD4+ and CD8+ T cell response and neutralizing antibodies, are associated with a reduction in COVID-19 disease severity20–22; however, certain T cell responses including unconventional CD16+ T cells23, mucosal-associated invariant T (MAIT) cells24 and γδ T cells25 may be increased in severe COVID-19 and are potentially related to immunopathology.
Cross-reactive immune responses that exist in individuals before SARS-CoV-2 exposure may also affect susceptibility to infection and disease severity4,26–33. Given the high homology of SARS-CoV-2 with other HCoVs, particularly those of the β-CoV genus, and the very high prevalence of CCCs, it is probable that these viruses are the source of cross-reactive immune responses. However, the exact source and the subsequent role of pre-existing SARS-CoV-2 cross-reactivity remain a question of considerable research interest. Evidence from the study of other infectious diseases suggests that pre-existing, cross-reactive immune response may impact disease outcomes (Fig. 1), in some cases promoting protection against infections — such as in influenza A34 and Japanese encephalitis virus infections — in which beneficial cross-reactive T cell responses are found35. But in other cases, such as dengue virus and zika virus infections36–38, disease severity is potentially exacerbated (Box 1). Understanding cross-reactive immune responses to SARS-CoV-2 may contribute to our understanding of the heterogeneity of clinical outcomes in COVID-19 disease and vaccination and allows us to identify immune epitopes that are conserved between genetically diverse coronaviruses and are most likely to be functionally constrained. These may provide insights for the development of pan-coronavirus vaccines that can protect from emerging SARS-CoV-2 variants of concern (VoCs) and future novel pandemic coronaviruses.
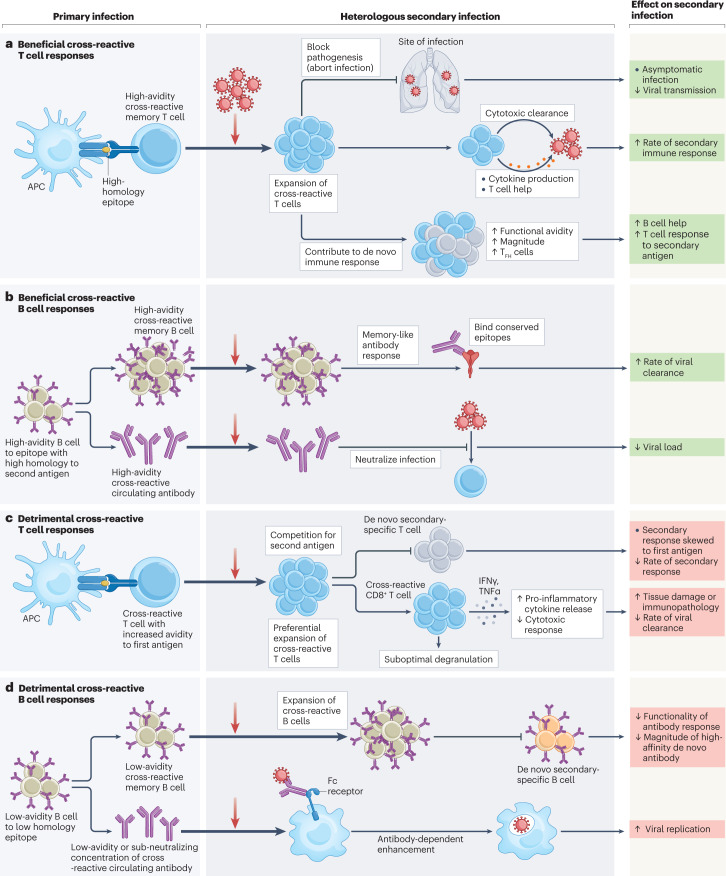
Fig. 1. Mechanisms of beneficial or detrimental cross-reactive immune responses identified in heterologous secondary viral infections.
Cross-reactive T and B cells may be generated in a primary infection with a virus that has sequence similarity to a heterologous secondary infectious agent. Cross-reactive immune cells may have beneficial or detrimental effects, and the mechanisms for such effects have been determined for many different virus families. a, Beneficial cross-reactive T cells may be generated through the priming of T cells by high-homology epitopes. These T cells then cross-react with high avidity during a heterologous secondary infection and may block pathogenicity by preventing invasive infection (aborting infection) or may expedite the rate of viral clearance by forming a ‘secondary-like’ memory immune response with an increased magnitude of B cell help and T cell responses. Evidence for such cross-reactive mechanisms was reported in studies of coronavirus infections, including severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)39,62,90,100. b, Beneficial cross-reactive memory B cells and antibodies can be generated to epitopes of high similarity between a primary and a heterologous secondary infection. Cross-reactive B cells specific to highly conserved epitopes may then go on to produce an expedited and high-functionality memory-like response to heterologous infection, including the production of cross-neutralizing antibodies that can prevent viral entry to cells. Evidence for these cross-reactive mechanisms was reported in coronavirus infections, including Betacoronaviruses and SARS-CoV-2 (refs. 49,108), influenza infection109 and influenza vaccination110 studies. c, Cross-reactive T cells generated to epitopes of relatively low homology may detrimentally impact a secondary infection with heterologous agents by dominating the response to the secondary infection, competing for presented antigen and disrupting the production of high-avidity de novo T cell responses. Cross-reactive T cells may bind with lower affinity to epitopes from the secondary infectious agent compared with the first. Low-affinity epitope binding by cross-reactive CD8+ T cells may then lead to the production of pro-inflammatory cytokines (interferon-γ (IFNγ), tumour necrosis factor-α (TNFα)) without the induction of cytolytic effector functions (degranulation) — leading to immunopathology and reducing viral clearance. Evidence for these cross-reactive mechanisms has been reported in studies of flavivirus infection36,111,112, reviewed elsewhere113,114. d, Cross-reactive B cells generated to epitopes of low homology may interfere with responses to a secondary heterologous infection by skewing the response to antibodies with higher avidity for the initial infectious agent, reducing the generation of de novo antibody responses. Evidence for such cross-reactive mechanisms was reported in coronavirus (common cold coronaviruses and SARS-CoV-2)85 and influenza virus studies, reviewed elsewhere115. Low-avidity cross-reactive antibodies or low levels of high-avidity cross-reactive antibodies that cannot neutralize a secondary heterologous infection may bind to virus and aid viral entry into cells through Fc-receptor-mediated endocytosis, a mechanism known as antibody-dependent enhancement. Evidence for antibody-dependent enhancement was reported in studies of flavivirus infection116,117. APC, antigen-presenting cell; TFH cell, follicular helper T cell.
Here, we bring together the evidence for pre-existing cross-reactive immune responses against SARS-CoV-2. We discuss theoretical evidence for a potential role of immune cross-reactivity in SARS-CoV-2 infection, experimental evidence for cross-reactivity between SARS-CoV-2 and other viruses, evidence for an impact of cross-reactivity on clinical outcomes following SARS-CoV-2 infection, and we examine the impact of cross-reactivity on response to SARS-CoV-2 vaccination. Furthermore, we reflect on the implications of cross-reactivity for future vaccine development and in the context of SARS-CoV-2 VoCs.
Box 1 Sequence homology among closely related viruses impacts on immune response to heterologous infection.
Closely related viruses can induce cross-reactive immune responses against each other, as exemplified in virus families in which high sequence homology among members impacts on the immune response to related viruses. For example, it is well known that cross-reactivity impacts on clinical outcomes in infections with members of the Flaviviridae family: this includes dengue virus (DENV) and zika virus (ZIKV), in which immune response to infection by one DENV serotype can have deleterious consequences for subsequent infection by another DENV serotype or ZIKV36–38. This phenomenon aligns with the concept of ‘original antigenic sin’. This was proposed in 1953 (ref. 118) and describes a scenario in which immunity to a primary infection alters the effectiveness of the immune response to a secondary infection. Detrimental original antigenic sin in flaviviruses is mostly antibody-mediated and due to antibody-dependent enhancement (ADE) in a secondary infection37,119,120 (Fig. 1). In the example of DENV, ADE is a phenomenon whereby low avidity or a low concentration of antibodies generated in response to a primary infection of one DENV serotype is unable to neutralize a secondary infection with another DENV serotype, but instead binds to and enhances the capacity of the virus causing the secondary infection to enter the cells through Fc-receptor-mediated endocytosis119.
Although there is substantial evidence for detrimental ADE in flavivirus infection, the role of cross-reactive T cell-mediated immune responses in flavivirus outcomes is still debated114. Initial evidence suggested that in repeat infection with DENV of different serotypes, T cells recruited in the secondary infection are of a higher affinity to the primary infectious serotype than the secondary serotype36. Furthermore, several studies have demonstrated that the magnitude of T cells in DENV infections positively correlates with disease severity112,121 and suggested that cross-reactive T cells in secondary DENV infections may play a role in the increased pathogenesis observed during secondary infections111 (Fig. 1). However, there is also evidence that cross-reactive T cells may instead be beneficial to the clinical outcomes of flavivirus infection. A recent publication identified 93 cross-reactive ZIKV T cell epitopes in 90% of the DENV-exposed, ZIKV-unexposed donors, and in vitro testing of some of these cross-reactive epitopes showed that they induced strong cross-reactive T cell response122, which, in theory, may have a protective role in subsequent ZIKV infection. In several other studies, T cell responses to cross-reactive peptides that were generated in previous DENV infections corresponded to a more rapid and higher magnitude of T cell responses to secondary infection with ZIKV or the similarly highly conserved flavivirus, Japanese encephalitis virus122–124. Epitopes of cross-reactive T cells had a high degree of sequence homology between DENV and ZIKV (>67%). These were associated with an increase in the magnitude of the secondary immune response to ZIKV122, experimentally demonstrating that the degree of sequence homology impacts on the strength of cross-reactive T cell responses. The threshold of 67% sequence homology between epitopes has subsequently also been shown to be relevant to T cell cross-reactivity between common cold coronaviruses and SARS-CoV-2 (ref. 28).
In addition to examples of beneficial cross-reactivity in flaviviruses, there are further examples of beneficial cross-reactivity in the literature. Perhaps the most famous example is the discovery by Edward Jenner in 1796 that inoculation with cowpox protected individuals from the related, but deadly smallpox virus — leading to the eventual eradication of smallpox and the foundation of the field of vaccinology125,126. More recently, detailed studies of influenza A infection during the 2009 H1N1 pandemic found that pre-existing T cells in H1N1-naive donors were cross-reactive with conserved epitopes of H1N1. Furthermore, the magnitude of these pre-existing H1N1 cross-reactive T cells was inversely correlated with the disease score in a subsequent infection, such that study participants with higher pre-existing cross-reactive interferon-γ-secreting CD8+ T cells had a reduced risk of symptomatic infection with H1N1 (ref. 34). Studies such as these demonstrate that cross-reactivity can substantially alter subsequent immune responses and disease outcomes in a heterologous infection.
SARS-CoV-2 sequence homology with other viruses
SARS-CoV-2 shows an overall sequence homology of 69% with OC43, 68% with HKU1 and 65% with NL63 and 229E (ref. 3), suggesting that cross-reactive immune responses induced by previous CCC infections may play an important role in SARS-CoV-2 infections or vaccination outcomes. Genomic and protein alignment has shown that the NSPs, the S2 domain of the spike protein and the nucleocapsid protein show the highest homology between the CoVs3,4,39 (Fig. 2). The homology between the SARS-CoV-2 and SARS-CoV nucleocapsid protein is 90%, and for the β-CoVs HKU1 and OC43, it is 34% and 35%, respectively. The S2 region of the spike protein is considerably more conserved between SARS-CoV-2 and CCCs than the S1 region40. In particular, regions such as the S2 fusion protein domain and heptapeptide repeats 2 are highly conserved across coronaviruses41,42. Immunodominant domains found on the S1 portion of the spike protein, such as the receptor-binding domain (RBD) and N-terminal domain, are relatively more variable between CoVs (for the RBD, SARS-CoV-2 has 73–76% homology to SARS-CoV, 24% to HKU1 and 23% to OC43)3,43–45. The variable conservation of SARS-CoV-2 with HCoVs means that cross-reactivity is probably primarily targeted to certain regions of the SARS-CoV-2 proteome. For instance, the relatively low sequence homology of the SARS-CoV-2 RBD region makes this a less likely target for cross-reactive antibodies. Considering that the RBD region is known to contain the majority of neutralizing antibody-binding sites46, this potentially limits the ability for cross-reactive antibodies to neutralize SARS-CoV-2. However, binding antibodies to the highly conserved S2 fusion protein region of the SARS-CoV-2 spike protein have also been shown to neutralize SARS-CoV-2 cell entry47. In the context of COVID-19 vaccines, the majority of which use the SARS-CoV-2 spike protein as immunogen, cross-reactivity of CCC is limited to the spike protein. It is therefore likely that there is a complex and dynamic interplay of cross-reactivity of CCC with COVID-19 vaccines and SARS-CoV-2 infection, potentially with enhanced cross-reactivity to conserved regions of the SARS-CoV-2 spike protein in vaccinated populations.
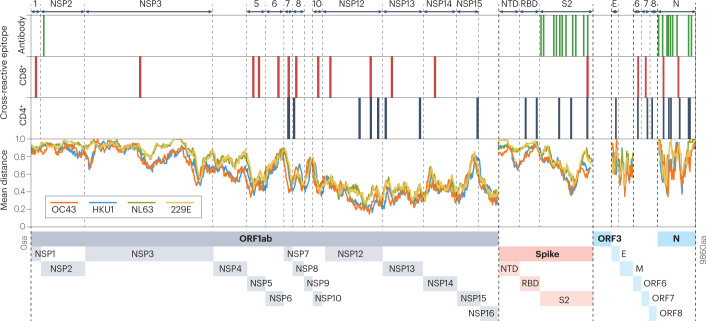
Fig. 2. Severe acute respiratory syndrome coronavirus 2 cross-reactive epitopes and common cold coronavirus homology.
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) proteome with regions of experimentally defined cross-reactive epitopes in humans, as listed in Supplementary Table 1. SARS-CoV-2 proteins containing cross-reactive epitopes are labelled along the top and depicted along the bottom of the figure. The amino acid sequences for the ancestral SARS-CoV-2 and the reference sequences [Accession numbers: 229E: NC_002645.1, OC43: NC_006213.1, NL63: NC_005831.2, HKU1: NC_006577.2] for each of the four common cold coronaviruses were aligned against each other using MATLAB multialign function and the PAM250 scoring matrix. A moving window was used to calculate the mean number of amino acid (aa) differences between the ancestral SARS-CoV-2 sequence and each common cold coronavirus (mean number of mismatches per base (mean distance)). A window size of 100 was used for ORF1ab and spike proteins, and a window size of 30 was used for other proteins. E, envelope protein; M, membrane protein; N, nucleocapsid protein; NSP, non-structural protein; NTD, N-terminal domain; ORF, open reading frame; RBD, receptor-binding domain; S2, spike subunit 2.
Evolutionary conservation of the SARS-CoV-2 spike protein with other single-stranded RNA viruses has also been described, and computational analysis has shown genetic sequence similarities between regions of SARS-CoV-2 and paramyxoviruses including mumps, measles and Nipah viruses48. Interestingly, within the spike protein, the region of highest sequence similarity between SARS-CoV-2 and paramyxoviruses corresponds with the SARS-CoV-2 S2 fusion protein domain, which is also highly conserved among HCoVs48. The high sequence homology between these viruses and SARS-CoV-2 suggests that viruses other than HCoV may also contribute to SARS-CoV-2 immune cross-reactivity and impact on infection and vaccine outcomes.
Evidence for cross-reactive immunity
Several studies have shown immune reactivity to SARS-CoV-2 epitopes or antigen in samples from individuals that were collected before the COVID-19 pandemic4,26–28,49, providing definitive evidence that SARS-CoV-2 cross-reactive immune responses may be derived from non-SARS-CoV-2 antigens. Moreover, epitopes of cross-reactive T and B cells have been predicted with computational tools and identified using conventional laboratory assays28,50–52. These are located in the regions of high homology between SARS-CoV-2 and HCoVs such as the nucleocapsid protein4, the spike protein S2 region27,49,53 and NSPs found in ORF14,27,28,39,49,53–55 (Fig. 2 and Supplementary Table 1). However, it is feasible that cross-reactivity, facilitated by receptor-binding degeneracy or epitope structural similarity53, is not limited to CCCs and that other common infectious agents may provide a source of epitopes with high homology to SARS-CoV-2 epitopes.
Cross-reactive antibodies and B cells
A number of studies have detected cross-reactive antibodies in samples taken from SARS-CoV-2-unexposed individuals before or early in the pandemic45,49,56–59. In unexposed children, adolescents and adults, a high seroprevalence was found against SARS-CoV-2 total spike and nucleocapsid proteins but not against the RBD or the S1 subunits of the spike protein, suggesting that responses were mainly targeting S2 in these cohorts49,59. Cross-reactive antibodies binding within the final 743 amino acids of the spike protein were mapped to multiple epitopes within the S2 region of the spike protein, which were largely well conserved between SARS-CoV-2 and other HCoVs49 (Table 1). Similar findings were made in an analysis of 196 overlapping peptides covering the SARS-CoV-2 spike protein, demonstrating that antibodies from serum samples taken before the pandemic can bind SARS-CoV-2 S2 regions with high sequence homology to other HCoVs.
Table 1.
Experimental evidence for beneficial, detrimental or neutral effects of cross-reactivity on T cell and B cell responses and COVID-19 clinical outcomes
| Observation | Beneficial | Neutral | Detrimental |
|---|---|---|---|
| B cells | |||
| A study of 48 SARS-CoV-2-unexposed individuals identified SARS-CoV-2-binding antibodies in 33; 29 out of 33 neutralized SARS-CoV-2 in vitro, indicating the existence of pre-existing cross-reactive protective antibodies49 | X | ||
| Increased (in vitro) neutralizing capacity of CCP from 126 donors with high titres of antibodies targeted at the coronavirus NL63 (ref. 108) | X | ||
| No correlation between the levels of anti-CCC antibodies before SARS-CoV-2 infection and subsequent SARS-CoV-2-targeted antibody response, or protection against infection in two case–control studies of 121 (ref. 84) and 502 individuals (ref. 59) | X | ||
| An increase in class-switched antibodies and reduced SARS-CoV-2 spike-protein-specific antibody functionality in the serum from 12 elderly SARS-CoV-2 convalescent donors, compared with 12 young convalescent donors. This suggests the presence of antibodies with reduced functional capacity in individuals who have experienced more CCC exposure77 | X | ||
| A positive correlation between titres of β-CoV-specific antibodies and SARS-CoV-2-specific antibodies in 21 individuals with fatal COVID-19 infection, as well as an increased ratio of β-CoV spike-protein-specific antibodies compared with SARS-CoV-2 spike-protein-specific antibodies in 21 individuals with fatal COVID-19 compared with 21 individuals with non-fatal COVID-19 who required admission to an intensive care unit85 | X | ||
| HCoV-specific antibody titres were higher in 32 patients with severe COVID-19 compared with 888 samples taken before the pandemic and from 64 patients with mild or severe COVID-19. Longitudinal assessment of HCoV-specific antibody titres in 28 patients with COVID-19 showed that HCoV antibody titres were not boosted after SARS-CoV-2 infection. Together, these data suggest that a higher HCoV-specific antibody titre before COVID-19 is associated with more severe COVID-19 (ref. 86) | X | ||
| T cells | |||
| Pre-existing cross-reactive T cells appeared to abort SARS-CoV-2 infection before seroconversion in 10 individuals following close-contacts with SARS-CoV-2-infected individuals and in 19 highly exposed health-care workers39 | X | ||
| Increased frequency of cross-reactive IL-2-producing T cells in 26 individuals who did not become PCR-positive after close-contacts with SARS-CoV-2-infected individuals, compared with 26 close-contacts who did become PCR-positive90 | X | ||
| Pre-existing SARS-CoV-2 cross-reactive T cells associated with an increase in magnitude of high-avidity T cells in 17 patients with acute SARS-CoV-2 infection. The same study also showed an association between cross-reactive T cells and an expedited ‘secondary-like’ memory T cell response to COVID-19 vaccine, increasing the rate of response to the vaccine in 31 donors62 | X | ||
| Pre-existing cross-reactive T cell responses were associated with an increased frequency of SARS-CoV-2 spike-protein-specific multicytokine producing CD4+ T cells and TFH cells in 17 individuals after COVID-19 vaccination, compared with 18 individuals without pre-existing cross-reactive T cells100 | X | ||
| Increased CD4+ and CD8+ T cell proliferation in response to COVID-19 vaccine in 48 donors with pre-existing SARS-CoV-2 cross-reactive responses, leading to an increased functional T cell response to the vaccine101 | X | ||
| Pre-existing cross-reactive T cells to SARS-CoV-2 from 17 donors had low functional avidity and T cells in 21 patients with COVID-19 were only minimally cross-reactive with CCCs. This suggests that cross-reactive T cells do not significantly contribute to the SARS-CoV-2 immune response26 | X | ||
Summary of the studies and mechanisms of CCC-SARS-CoV-2 cross-reactive immune responses in SARS-CoV-2 infection and vaccination. Clinical/biological outcomes that are beneficial, neutral or detrimental to COVID-19 clinical course are marked. β-CoV, Betacoronavirus; CCC, common cold coronavirus; CCP, COVID-19 convalescent plasma; HCoV, human coronavirus; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; TFH cells; follicular helper T cells.
Similarly, SARS-CoV-2-reactive memory B cell populations have been investigated for cross-reactivity with HCoVs57. In COVID-19 convalescent donors, up to 4% of immunoglobulin G (IgG)+ memory B cells bound to both the SARS-CoV-2 spike protein and either HKU1 or NL63, demonstrating cross-reactivity of the B cell receptor to epitopes of both SARS-CoV-2 and HCoVs57. Monoclonal antibodies generated from these cross-reactive B cells also showed cross-reactive binding to several other HCoVs, including OC43, MERS-CoV-2 and SARS-CoV57. Pre-existing cross-reactive memory B cells have also been shown to expand in the early response to SARS-CoV-2 infection60. These cross-reactive memory B cells had already undergone significant hypermutation before the initial SARS-CoV-2 infection, but their frequency decreased over time (between 3 and 6 months after SARS-CoV-2 infection). This may suggest that, although cross-reactive memory B cells form part of the initial response to SARS-CoV-2 infection, they are not selected for among the overall memory B cell pool, which forms after infection60. Notably, however, some cross-reactive memory B cells that are available for rechallenge may not be detectable by conventional flow cytometry staining in peripheral blood, and techniques such as tetramer enrichment may be required to detect rare cross-reactive memory B cell populations61.
Cross-reactive T cells
Several studies have also identified cross-reactive T cells to SARS-CoV-2 in blood samples from SARS-CoV-2-unexposed individuals4,27,28,30,31,49,58,62. Compared with antibody epitopes, and of relevance to vaccine design, a greater number of epitopes for cross-reactive T cells were identified in SARS-CoV-2 proteins other than spike (Supplementary Table 1). For example, responses to the structural nucleocapsid protein and non-structural ORF1 proteins (targeting mainly NSP7 and NSP13) were detected in 19 of 37 SARS-CoV-2-unexposed donors using an interferon-γ ELISpot assay4. These T cell epitopes mapped to a region of the nucleocapsid protein (N101–120) that was highly conserved between SARS-CoV-2 and HCoVs and contained a T cell epitope that had been identified in individuals who had recovered from SARS-CoV infection 17 years earlier (Supplementary Table 1). This epitope has undergone considerable subsequent analysis and is suggested to be an important target for cross-reactive CD8+ T cells63,64. A study by Swadling et al.39, using samples taken before the COVID-19 pandemic and samples from SARS-CoV-2-exposed but uninfected health-care workers, identified epitopes of cross-reactive T cells in the non-structural components NSP7, NSP12 and NSP13 of the replication transcription complex of SARS-CoV-2. Bioinformatic analysis in this study found significantly higher sequence homology between SARS-CoV-2 and all other HCoVs in the replication transcription complex compared with structural proteins39. Cross-reactive T cells from three out of five seronegative health-care workers showed a higher response to the corresponding HKU1 epitope when compared with the homologous SARS-CoV-2 epitope, suggesting that SARS-CoV-2 cross-reactivity is a consequence of previous HKU1 infection in these individuals39.
Short-term T cell lines derived from SARS-CoV-2-unexposed subjects and peptides representing SARS-CoV-2-immunodominant epitopes have also been used to evaluate cross-reactive T cells28. Many of these short-term T cell lines cross-reacted with HCoV epitopes, with some reacting more strongly with HCoV epitopes than the SARS-CoV-2 epitope that was used to generate them, again suggesting that they originated from HCoVs other than SARS-CoV-2 (ref. 28). As not all these epitopes have high homology with epitopes from other HCoVs, it is possible that short-term T cell lines that were not cross-reactive with HCoVs might have originated from a non-HCoV source. Substantiation of the phenotype of cross-reactive T cells shows that they were not generated de novo but had a bona fide memory phenotype (CD45RA−CCR7−/+)28. Evaluation of the homology of cross-reactive peptides showed that those with the highest homology between SARS-CoV-2 and CCCs (≥67%) were more likely to be cross-reactive than those with lower homology, suggesting, as expected, that epitope similarity is key to cross-reactivity28.
However, functional T cell responses to epitopes of low homology between HCoVs, including NSP736–50 and NSP726–40, have also been identified in other in vitro experiments4. Peripheral blood mononuclear cells (PBMCs) from a SARS-CoV-2-unexposed donor who had SARS-CoV-2 NSP736–50-reactive T cells failed to respond when stimulated with the corresponding HCoV-derived amino acid sequence4. This finding is interesting as it provides evidence for alternative sources of T cell cross-reactivity beyond HCoVs. A recent study that similarly investigated sources for SARS-CoV-2 cross-reactive T cells found that SARS-CoV-2-specific T cells identified in samples taken from individuals before the COVID-19 pandemic could cross-react with predicted, naturally processed, defined microbial peptides from common commensal bacteria65, suggesting that microbial peptides from commensal bacteria are potentially an important source of pre-existing SARS-CoV-2 cross-reactive T cells.
In addition to the studies of cross-reactive CD4+ T cells to SARS-CoV-2, other studies have identified pre-existing cross-reactive CD8+ T cells30,31,55,63,66. Human leukocyte antigen A (HLA-A)-restricted and HLA-B-restricted shared epitopes have been described in several regions of SARS-CoV-2, including the spike protein, ORF1ab-derived proteins and ORF7a4,28,30,31 (Supplementary Table 1). In particular, a high degree of pre-existing cross-reactive CD8+ T cell responses to SARS-CoV-2 was found in unexposed donors who carry the HLA-B*7:02 allele66.
Impact of cross-reactive immune responses
The impact of cross-reactive immune responses on the outcome of SARS-CoV-2 infection can be defined in two main ways: at an epidemiological level, in which the prevalence of CCC infection can be correlated with the clinical outcome of COVID-19 and prevalence of SARS-CoV-2, and in the setting of cohort studies that directly assess the impact of cross-reactive antigen on SARS-CoV-2 immune responses (Table 1). Moreover, a number of studies have aimed to decipher the mechanisms by which cross-reactive antibodies, memory B cells and T cells can affect immunity to SARS-CoV-2.
Epidemiological studies can explore the variation in CCC prevalence by geography, seasonality and across age groups and associate this prevalence with the outcome or incidence of SARS-CoV-2 infection, thus giving new insights into the role of CCCs in modulating COVID-19 severity or SARS-CoV-2 infection prevalence.
Geographical and seasonal variation in CCCs
Although CCCs are globally distributed, the prevalence of these infections is likely to vary both by geographical region and by season. One meta-analysis of 128 studies showed an overall CCC infection rate of 5%, but this ranged from 0.73% in the Philippines to a high of 21.51% in Tunisia67. In general, these studies contained small numbers of people and included those with symptomatic disease only, so the true prevalence of infections may be grossly under-reported.
The seasonality of CCCs is also well documented with an increased prevalence of CCC infections in winter months globally67–69. In Scotland, it has been shown that HCoV strains co-circulated reciprocally across seasons, with high prevalence of one coronavirus strain coinciding with low prevalence of another5. Similar findings were demonstrated in a cohort in MI, USA70. The seasonality of CCCs is potentially explained by social behaviour and waning immunity between waves. Notably, asynchronous seasonality and low levels of co-infection71 were observed between the α-CoVs HCoV-NL63 and HCoV-229E, suggesting that a potentially immune-mediated interaction (cross-reactivity) between closely related CCCs could impact the population prevalence of a different CCC5. However, the effect of the seasonality of CCCs on cross-reactivity with SARS-CoV-2 is unclear as yet.
Considering the high but variable prevalence of CCC infection globally, is there any evidence of an impact of CCC prevalence on SARS-CoV-2 infection outcome? In one large study looking to identify the relationship between the previous infection with a CCC and the clinical outcome of SARS-CoV-2 infection, 875 out of 15,928 individuals tested PCR-positive for CCC infection72. Individuals who tested positive for CCCs and subsequently became infected with SARS-CoV-2 had milder COVID-19 when compared with those with no evidence of CCC infection in the study time frame72, suggesting that there is some cross-protection from the previous CCC infection. Another study showed that participants with a history of positive tests for CCCs had less severe COVID-19 on infection with SARS-CoV-2, including a reduced likelihood for intensive care unit admission and death, which remained significant after accounting for age, sex, body mass index and diabetes mellitus status73.
Age and CCC cross-reactivity
Exploring the relationship between the prevalence of CCCs and SARS-CoV-2 infection outcomes, which is known to vary between age groups, may also give insights into CCC-SARS-CoV-2 cross-reactivity. Although children had the highest prevalence of CCCs before the COVID-19 pandemic, in the early ‘waves’ of the SARS-CoV-2 pandemic, they were the least likely to be infected with SARS-CoV-2, accounting for only 1–3% of confirmed cases in early 2020 (refs. 74,75). Although this inverse correlation may be confounded by a variation in rates of asymptomatic infection and social isolation in different age groups, it is interesting to observe that in three independent studies that were carried out before the COVID-19 pandemic, children aged under 5 years, compared with other age groups, were found to have the highest prevalence of infection with the β-CoV HCoV-OC43, the CCC that is most closely related to SARS-CoV-2 (refs. 5,70,71). By contrast, people over the age of 65 years had the highest prevalence of the α-CoV 229E and had lower overall CCC prevalence compared with children5,70,71. It is therefore possible that increased pre-existing β-CoV immune responses that cross-react with SARS-CoV-2, or niche competition between β-CoVs and SARS-CoV-2, are causally related to a decrease in infection rates and disease severity in younger age groups, whereas elderly people are less protected by exposure to α-CoV HCoV-229E, which has low homology to SARS-CoV-2.
Cross-reactive immune responses have been directly measured in different age cohorts and can be used to support or refute these epidemiological associations. In pre-pandemic samples, it has been shown that the magnitude of pre-existing SARS-CoV-2 spike protein cross-reactive IgG is higher in children compared with that in adults and that cross-reactive antibodies generated against SARS-CoV-2 spike protein domains may neutralize SARS-CoV-2 infection in vitro5,49, potentially explaining the increased protection from SARS-CoV-2 infection and COVID-19 severity in children42,49,76. In exploring Ig subsets and functionality in specific age groups, it has been shown that there are higher levels of class-switched (immunoglobulin A (IgA) and IgG) antibody responses to SARS-CoV-2–CCC-conserved regions (S2 and nucleocapsid protein) in COVID-19 convalescent older adults, compared with COVID-19 convalescent children. However, children had more functional responses (Fc-mediated phagocytosis) targeting SARS-CoV-2 RBD and S1 (ref. 77). This suggests that repeat or recurring infection with CCCs over the course of a lifetime may skew CCC-generated antibodies to a more class-switched state, but with less functional cross-reactivity than found in children. The frequency of high-avidity and functional CD4+ T cells, cross-reactive with SARS-CoV-2 S2, has also been shown to decrease significantly with age62. It is possible that this reflects both an age-related decline of immune activity, through thymic involution78, immunosenesence79 and reduced T cell receptor (TCR) repertoire diversity80, and the age-related distribution of CCC prevalence5. A similar phenomenon of decreasing CD4+ T cell functionality with age is observed in mice, in which thymic involution and reduced TCR repertoire diversity in aged mice were associated with a reduced capacity to develop de novo responses and immunity to influenza A virus81. In this case, it is plausible that the increased frequency of cross-reactive CD4+ T cells in young children and adults may contribute to their increased protection against severe COVID-19, compared with elderly adults in the early phase of the pandemic.
One model investigating the relationship between CCC cross-reactivity and the age distribution of SARS-CoV-2 incidence rates suggests that cross-protection from previous infection with CCCs is not sufficient to explain age differences in the incidence of SARS-CoV-2 infections82, but may account for some reduced susceptibility in children. Overall, these data suggest a possible correlation between previous infection with CCCs and reduced SARS-CoV-2 severity. However, identifying a causal relationship between endemic CCC prevalence and COVID-19 disease outcomes is challenging owing to the impact of substantial confounding factors that vary between age populations including immunosenescence, comorbidities, physiological changes such as endothelial damage and angiotensin-converting enzyme 2 (ACE2) receptor density83, variation in COVID-19 vaccination rates and SARS-CoV-2 variant prevalence.
Cohort studies of antibody and B cell cross-reactivity
Several studies have explored the potential effect of cross-reactive antibodies and T cells on COVID-19 severity. Although cross-reactive antibodies against SARS-CoV-2 spike protein domains can be detected in pre-pandemic samples and may neutralize SARS-CoV-2 infection in vitro (SARS-CoV-2 spike pseudotypes and authentic SARS-CoV-2)5,49, there is an ongoing debate as to whether these influence COVID-19 clinical outcomes. In a large case–control study, levels of CCC-specific antibodies were found to be elevated in response to SARS-CoV-2 infection; however, the baseline CCC antibody titre (measured before SARS-CoV-2 infection) was not associated with protection against infection or predictive of disease severity84. This finding was supported by a second study, which similarly showed that SARS-CoV-2 cross-reactive antibodies were non-neutralizing and did not protect against SARS-CoV-2 infection or hospitalization59. The in vitro and clinical studies taken together suggest that some CCC–SARS-CoV-2 cross-reactive antibodies may have neutralizing capacity when assessed in vitro, but that the clinical effect of this may be marginal as overall there is a low frequency of broadly cross-reactive memory B cell clones that target cross-neutralizing epitopes42.
One study has suggested that CCC cross-reactive antibodies may have a detrimental effect on COVID-19 severity85. In this retrospective cross-sectional study, it was shown that antibody responses to the HCoV-OC43 spike protein correlated with SARS-CoV-2 spike-protein-reactive antibodies in fatal COVID-19 and that levels of SARS-CoV-2 spike-protein-targeted antibodies were reduced in patients who died from COVID-19, compared with patients with COVID-19 who required admission to an intensive care unit but survived. This is supported by another study that suggested that patients with critical COVID-19 had higher levels of anti-HCoV antibodies compared with other individuals assessed before the pandemic or with individuals with mild-to-severe COVID-19 (ref. 86). One possible explanation is that de novo responses to SARS-CoV-2 are decreased in patients with fatal COVID-19 compared with those with non-fatal severe COVID-19 and that the de novo generation of new responses is inhibited by pre-existing HCoV-OC43 cross-reactive responses. However, these studies do not directly investigate the effect of pre-existing cross-reactive antibodies on COVID-19 severity, and the causal relationship between CCC cross-reactivity and fatal outcomes remains unclear. Furthermore, the median age of patients with fatal COVID-19 was not controlled for compared with those with non-fatal disease, which may confound the observations. To ascertain a detrimental role for cross-reactive antibodies in SARS-CoV-2 infection, further work must be carried out to explain a mechanism for this phenomenon (such as antibody-dependent enhancement or reduced de novo SARS-CoV-2 response). As yet, there is little evidence for antibody-dependent enhancement in SARS-CoV-2 infection, with or without pre-existing SARS-CoV-2 cross-reactive antibodies (reviewed elsewhere87).
Overall, there is no evidence from longitudinal cohort studies or population-level studies to suggest that pre-existing SARS-CoV-2 cross-reactive antibodies exacerbate COVID-19. Rather, current evidence suggests that cross-reactive antibodies form part of the immune response to SARS-CoV-2, alongside the de novo response. However, as the levels of pre-existing antibodies are low, they are largely inconsequential in determining the clinical outcomes of COVID-19. These conclusions do not provide any convincing evidence for detrimental antibody-mediated cross-reactivity, which would mean that vaccine strategies that use CCC cross-reactive antigens should not be pursued. Consistent with this, the development of pan-coronavirus vaccines that are designed to target different SARS-CoV-2 VoC is currently an intense area of research in academic and industry laboratories88,89.
Cohort studies of T cell cross-reactivity
Several studies have investigated the impact of SARS-CoV-2 cross-reactive T cells on COVID-19 severity and SARS-CoV-2 infection, with a growing body of evidence showing that these have a beneficial role31,39,62. Cross-reactive T cells, which are potentially capable of preventing SARS-CoV-2 infection before seroconversion, have been detected in two independent studies. The first demonstrated that SARS-CoV-2 cross-reactive T cells found in health-care workers who were highly exposed to SARS-CoV-2 but found to be both seronegative and PCR-negative for COVID-19 were associated with levels of an innate immune marker (IFNα-inducible protein 27; IFI27) that can be used as a surrogate marker for early SARS-CoV-2 infection39. The authors propose that T cell responses to NSP12, a protein with high sequence homology to CCCs, were linked with ‘abortive’ infection, whereby SARS-CoV-2 infection was cleared before seroconversion or PCR positivity. Similarly, CD4+ T cells that produce IL-2 in response to a pool of SARS-CoV-2 peptides with high CCC homology (located primarily in regions outside the spike protein, including ORF1, 4, 6, 7 and 8, nucleocapsid protein and envelope protein) have been associated with protection from PCR-positive SARS-CoV-2 infection in people who were close-contacts with SARS-CoV-2-infected individuals90. In this study, several antigenic peptides were identified that were analogous to the NSP12 epitopes associated with abortive T cell infections in the health-care worker study reported by Swadling et al.39,90, together suggesting that cross-reactivity to NSP12 may play an important role in protecting against infection.
Indirect evidence for early activation of CD8+ T cells in mild SARS-CoV-2 infection91 supports the concept that cross-reactive memory T cells are recruited and activated early in response to SARS-CoV-2 infection. Several studies have found that a delayed onset and weak induction of SARS-CoV-2 specific T cells are found in patients with severe COVID-19 (ref. 92) and that the early induction of SARS-CoV-2 reactive T cells is associated with mild COVID-19 (ref. 93) — potentially as a consequence of recruitment of cross-reactive memory T cell responses (although this was not demonstrated in these studies). More direct evidence was provided by a study that showed that patients hospitalized with COVID-19 had a lower frequency of CD4+ T cells that bind HLA-DR-restricted cross-reactive epitopes compared with individuals with mild COVID-19 (ref. 31) and that CD8+ T cell responses in individuals with severe COVID-19 were directed at epitopes that were, on average, less similar to HCoVs than to epitopes targeted by CD8+ T cell responses in individuals with mild COVID-19 (ref. 94). Recently, the important contribution of an immunodominant CD4+ T cell epitope (S816–830) in the fusion protein domain of SARS-CoV-2 S2 was reported. High-avidity responses to this epitope were generated in the majority of infected donors, and the frequency of these pre-infection cross-reactive T cells showed a positive correlation with titres of S1-targeted IgG and neutralizing antibodies after infection62. Responses to this epitope were further highlighted in a single patient, with cross-reactive CD4+ T cells specific to the S816–830 epitope detected 6 years before SARS-CoV-2 infection and subsequently boosted following SARS-CoV-2 infection53.
The role of T cell avidity in clinical outcomes in patients with COVID-19 has also been considered elsewhere, showing that SARS-CoV-2-specific T cells of low avidity are enriched in patients who are hospitalized with severe COVID-19 compared with patients with mild disease26. Low-avidity responses in hospitalized patients have not been directly or causally linked to CCC cross-reactive responses, although pre-infection cross-reactive SARS-CoV-2 responses have been shown to be of lower avidity relative to T cells that are specific for SARS-CoV-2 peptides26,95. To fully address causality, longitudinal studies that assess CCC T cell reactivity and avidity before and after SARS-CoV-2 infection, correlated with COVID-19 severity, are required.
The impact of cross-reactivity beyond CCCs
The current data support the hypothesis that beneficial immune cross-reactivity in SARS-CoV-2 infection may also be derived from sources other than HCoVs, and cross-reactive immune responses from memory T cells for unrelated pathogens have been proposed96. In one study, self-reported measles, mumps, rubella (MMR) but not Bacillus Calmette–Guérin or hepatitis B vaccination was associated with less severe COVID-19 (ref. 97), and another study showed that the presence of antibodies induced by MMR vaccination was associated with better outcomes in COVID-19 (ref. 98). More recent data suggest that recipients of MMR and tetanus/diphtheria/pertussis (Tdap) vaccine share TCR clonotypes with individuals who are convalescent or vaccinated against COVID-19 (ref. 99). This may explain why vaccination with either MMR or Tdap is associated with reduced hospitalization, intensive therapy unit (ITU) care and/or death in patients with COVID-19, particularly in women when adjusted for other confounds99. In these studies, the time interval between the MMR and Tdap vaccine dose and SARS-CoV-2-positive test was not significantly associated with outcomes99. Given the widespread deployment of MMR vaccines in the face of a high prevalence of severe COVID-19, it is unlikely that MMR cross-reactive responses are a major factor in the stratification of disease severity, although it is plausible that COVID-19 severity may be substantially worse in a global context without widespread exposure to cross-reactive antigen such as MMR. Further study of COVID-19 severity/mortality in populations with poor MMR vaccine coverage is required to fully understand the role of this cross-reactivity in COVID-19.
Heterologous humoral responses mediated by previous responses to respiratory pathogens may also contribute to some protection in COVID-19. This was suggested by a study that found weaker antibody responses to rhinoviruses, influenza viruses and enteroviruses detected in patients who were hospitalized with COVID-19, compared with those with mild COVID-19 (ref. 58).
Effect of cross-reactive immune responses on COVID-19 vaccination
The effect of pre-existing cross-reactive SARS-CoV-2 immune responses on the outcomes of COVID-19 vaccination is of great interest but has been investigated in few studies to date. One of these was a longitudinal study of 35 individuals with pre-existing cross-reactive spike-protein-specific CD4+ T cells that investigated responses to low-dose mRNA-1273 vaccination100. Here, participants had a significantly higher frequency of SARS-CoV-2 spike-protein-specific and polyfunctional CD4+ T cells (producing multiple cytokines) after the first dose and the second dose of vaccine compared with individuals without pre-existing cross-reactive CD4+ T cells, and these frequencies remained higher up to 6 months following vaccination100. Follicular helper T cells (TFH cells) were also significantly increased in vaccinees with pre-existing cross-reactive CD4+ T cells compared with those without, corresponding with increased spike-protein-specific IgG and RBD-specific IgG 15 days after the second vaccine dose in vaccinees with pre-existing SARS-CoV-2 S-specific cross-reactive CD4+ T cells100. This indicates that pre-existing SARS-CoV-2 spike-protein-specific cross-reactive CD4+ T cells facilitate a coordinated boost to adaptive immune responses after COVID-19 vaccination.
Vaccinees with pre-existing cross-reactive CD4+ T cell responses to SARS-CoV-2 S2 have also been shown to generate higher functional avidity after a first dose of BNT162b2 vaccination compared with individuals without pre-existing cross-reactive T cell responses62. Interestingly, of the early (days 0–7) T cell responses to SARS-CoV-2 S2 following vaccination, up to 100% of the CD4+ T cell response was directed at the S816–830 epitope, suggesting that this cross-reactive epitope is a major contributor to the early CD4+ T cell response to COVID-19 vaccination. Furthermore, an investigation of the role of T cells to the vaccine ChAdOx1 nCov-19 in people living with HIV found that SARS-CoV-2-seronegative individuals with baseline (before vaccination) SARS-CoV-2-specific CD4+ T cell proliferative responses had a significantly higher proportion of proliferating T cells in response to SARS-CoV-2 peptide following vaccination compared with those who had no proliferating SARS-CoV-2-specific T cells at baseline101. Together, these data suggest that SARS-CoV-2-specific CD4+ T cells in response to both BNT162b2 and ChAdOx1 nCoV-19 vaccines are boosted by the presence of pre-existing cross-reactive immune responses to SARS-CoV-2.
In evaluating humoral responses to COVID-19 vaccines, it has been shown that cross-reactive antibody responses at baseline do not correlate with post-vaccination SARS-CoV-2 antibody responses84. A follow-up study in mice showed that animals immunized with spike protein from OC43, HKU1, 229E or NL63 4 weeks before immunization with recombinant SARS-CoV-2 spike protein showed significantly lower levels of SARS-CoV-2 RBD-targeted and SARS-CoV-2 pseudoneutralizing antibodies than mice that were naive to CCCs84. Overall, data on cross-reactivity in vaccine responsiveness are analogous to that described in SARS-CoV-2 infection, in which the clinical impact of cross-reactive immune responses to vaccines is largely mediated by T cells, rather than antibodies.
Cross-reactivity between SARS-CoV-2 variants
In the context of widespread SARS-CoV-2 vaccination and infection with emerging variants of SARS-CoV-2, the landscape of coronavirus cross-reactivity has changed. Although the phenomenon of CCC cross-reactivity with SARS-CoV-2 emerged on a background of zero population exposure to the novel SARS-CoV-2 virus, cross-reactivity with subsequent variants of SARS-CoV-2 now exists on a heterogeneous background of populations exposed to various SARS-CoV-2 vaccines and variants. It is likely that the precise combination of these exposures defines the composition of the cross-reactive memory immune repertoire present in an individual and that the outcome of a subsequent SARS-CoV-2 infection is defined by these earlier combinatorial exposures. This is highlighted in a recent small, but detailed study showing that triple-vaccinated health-care workers infected first with Wuhan-Hu-1 SARS-CoV-2 in the first ‘wave’ of SARS-CoV-2 infections in the UK and then subsequently with Omicron (B.1.1.529, BA.1) had a lower magnitude of antibody and T cell responses to the Omicron spike protein than individuals who had not been initially infected with Wuhan-Hu-1 SARS-CoV-2102. The study suggests that immunological imprinting through exposure to the ancestral strain of SARS-CoV-2 impairs the cross-reactivity of the response to divergent variants of SARS-CoV-2. Given that cross-reactive T cells derived from CCCs appear to boost functional T cell responses to SARS-CoV-2 infection or vaccination62, it is surprising that immune imprinting with SARS-CoV-2 variants impairs immune responses to subsequent heterologous infection — especially considering the relatively high homology between the SARS-CoV-2 variants. Furthermore, recent epidemiological evidence suggests that unvaccinated individuals primed with both a non-Omicron variant and an Omicron variant (‘double-primed’) were almost half as likely to be reinfected with Omicron BA.2/4/5 135 days after the start of follow-up compared with those primed with only an earlier Omicron infection (‘Omicron-primed’), with an adjusted hazard ratio of reinfection of 0.52 (95% CI 0.40–0.68)103. Detailed characterization of the immune repertoire in the context of different historical variant exposures will be needed to better understand the effects of immune imprinting on subsequent immune responses to SARS-CoV-2 infections and vaccines. The contradictory evidence from the immunological and epidemiological studies presented here suggests that although measurable immune responses may be negatively impacted by immune imprinting, this does not translate into diminished protection against infection.
In the context of vaccines, these complex histories of multiple SARS-CoV-2 variant exposures should also be considered. In assessing COVID-19 vaccine efficacy in 363,646 individuals tested for SARS-CoV-2 by PCR after 10 April 2022, previous PCR-positive infection with Omicron BA.1 or BA.2, in addition to triple mRNA COVID-19 vaccination, was found to reduce Omicron BA.5 PCR-positive reinfection, compared with triple mRNA COVID-19 vaccination alone. Previous infection with either Delta or Alpha VoCs in addition to triple vaccination provided additional protection (Delta 46.9% (95% CI 27.0–61.3) and Alpha 65.4% (95% CI 49.8–76.2)) against BA.5 compared with vaccination alone, although the additional protection from infection with Delta and Alpha VoCs was lower than that from Omicron infection104. As Alpha and Delta waves occurred earlier compared with BA.1/2, the additional protection afforded by previous infection with these variants could reasonably be associated with the time between reinfections. However, the fact that Alpha infection occurred earlier than Delta and offers more protection against reinfection with BA.5 suggests that this is not the case.
To date, widely deployed COVID-19 vaccines have used only the spike protein as the vaccine antigen. In a widely vaccinated environment, SARS-CoV-2 variants have evolved with mutations primarily located in immunodominant epitopes in the spike protein RBD to evade immunity105,106. Comparatively, the identified regions of cross-reactivity between CCCs and SARS-CoV-2 are in regions located outside the RBD, which remain largely unaltered between SARS-CoV-2 variants (Supplementary Table 1). It is likely that these regions of CCC cross-reactivity have not mutated, either because they have a key role in SARS-CoV-2 viral replication or fitness, or because they are not under selection pressure to do so105. Either way, the preservation of cross-reactive epitopes between SARS-CoV-2 variants and CCCs highlights these regions as important targets for pan-coronavirus vaccine design. Multiple vaccine candidates are now being developed that use these conserved regions as immunogens, with recent studies showing that antibodies targeting a conserved region of the S2 domain of the SARS-CoV-2 spike protein are able to neutralize hACE2 receptor binding and protect against SARS-CoV-2 symptomatic infection in preclinical small animal challenge models88,89 and that antibodies generated to conserved sequences of the S2 stem helix are broadly neutralizing against β-CoVs42,76,107.
In the absence of pan-coronavirus vaccines, the lifting of government mandates to reduce SARS-CoV-2 transmission means that it is likely there will be an increase in the spread of other viruses, including CCCs. The cross-reactivity between SARS-CoV-2 and these viruses will therefore remain relevant for outcomes of COVID-19. Considering a potential back-boosting effect, whereby future CCC infection boosts immune responses to SARS-CoV-2, repeated seasonal infections with CCCs may provide an innocuous booster for SARS-CoV-2 immune responses. Furthermore, this recurring exposure may preferentially boost cross-reactive responses to epitopes that are conserved between coronaviruses, potentially enhancing pan-coronavirus immune responses. The reciprocal effect of widespread SARS-CoV-2 immunity on the seasonality and prevalence of CCCs will also give us important information on cross-reactivity between these viruses.
Conclusion
We evaluated the available evidence assessing the role of CCC and SARS-CoV-2 immune cross-reactivity in SARS-CoV-2 infections and vaccine outcomes and provide a detailed table of experimentally verified cross-reactive T and B cell epitopes of SARS-CoV-2 (Supplementary Table 1). Our analysis suggests that pre-existing cross-reactive T cells may provide some protection from COVID-19, whereas there are no data that convincingly show a role of cross-reactive antibodies in COVID-19 clinical outcomes. Similarly, CCC cross-reactive CD4+ T cells that are detectable before vaccination may enhance responses to both mRNA-based and viral vector-based vaccines62,100,101, whereas pre-existing cross-reactive antibody responses are enhanced by vaccination but do not themselves enhance overall vaccine antibodies titres. Despite evidence showing that cross-reactive T cells may be detrimental to the outcome of secondary infections in other virus families and that some unconventional23–25 or low-avidity conventional T cell26 populations may be associated with severe disease, to date there is little evidence suggesting a role for pre-existing cross-reactive T cells in exacerbating COVID-19.
Investigating the effects of CCC infections on clinical outcomes in COVID-19 in the future will be complex. High levels of SARS-CoV-2 and COVID-19 vaccine exposure globally mean that it will be almost impossible to design studies that investigate cross-reactive responses in individuals who have not been exposed to SARS-CoV-2 antigen. However, multiple factors are likely to contribute to the capacity of an individual to generate cross-reactivity, including the infectivity and genus of the virus (HCoV or otherwise), the temporal relationship between different viral infections, the HLA allotype that restricts antigen presentation to T cells31,63, age (immunological and biological)77 and history of SARS-CoV-2 exposure82, each of which may contribute to the quality and quantity of immune responses84.
In the context of repeated infections with different SARS-CoV-2 variants, it is likely that cross-reactive memory responses to CCCs, COVID-19 vaccines and SARS-CoV-2 variants will form a pool of cross-reactive responses that offer some protection from future variants. Here, it is possible that the exact order and composition of different infections may play a role in determining the efficacy of the immune response to prevent symptomatic SARS-CoV-2 infection. New vaccines that exploit CCC and SARS-CoV-2 variant immune cross-reactivity by targeting regions that are conserved across coronaviruses are currently being investigated and may offer a way to protect the population from future coronaviruses pandemics.
Supplementary information
Acknowledgements
S.M.M. is supported by the Medical Research Council. E.B. is funded by the Oxford NIHR Biomedical Research Centre and is an NIHR Senior Investigator. M.A.A. is supported by a Sir Henry Dale Fellowship jointly funded by the Royal Society and Wellcome Trust (220171/Z/20/Z). The views expressed in this article are those of the author and not necessarily those of the NHS, the NIHR or the Department of Health.
Glossary
- Antibody-dependent enhancement
Antibodies of low concentration or low avidity that are unable to neutralize a virus may instead opsonize it, thereby enhancing viral uptake into cells through antibody to Fc-receptor binding.
- Back-boosting
Enhancement of immune responses to a primary infection via cross-reactivity with a subsequent secondary infection.
- Functional avidity
The measure of how well a T cell receptor binds to its cognate antigen; that is, a T cell with high functional avidity has a higher or stronger response to a given concentration of antigen compared with a T cell with low functional avidity.
- Receptor-binding degeneracy
The ability of T cell receptors to bind with varying levels of avidity to more than one epitope presented by a particular major histocompatability complex (MHC).
- SARS-CoV-2 pseudoneutralizing antibodies
Antibodies capable of neutralizing cell entry of non-SARS-CoV-2 viral vectors that have been engineered to express the SARS-CoV-2 spike protein.
- SARS-CoV-2 spike pseudotypes
Viral vectors that have been engineered to express the SARS-CoV-2 spike protein.
- Short-term T cell lines
The in vitro expansion of T cells. Often, short-term T cell lines are generated from single-sorted T cell clones that bind a specific epitope.
- Tetramer enrichment
A technique used to detect rare antigen-specific B and T cell populations using tetramers (antigen-specific complexes that can be used to stimulate/detect specific T or B cell receptors) and antibody-bound magnetic beads to increase the relative concentration of T and B cells that are specific to an epitope of interest.
Author contributions
S.M.M., A.O., E.B. and M.A.A. researched data for the article. All authors contributed substantially to discussion of the content. S.M.M., A.O. and E.B. wrote the article. All authors reviewed and/or edited the manuscript before submission.
Peer review
Peer review information
Nature Reviews Immunology thanks Andreas Thiel, Christoph Neumann-Haefelin, Alessandro Sette and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Eleanor Barnes, Email: ellie.barnes@ndm.ox.ac.uk.
Ane Ogbe, Email: ane.ogbe@ndm.ox.ac.uk.
Supplementary information
The online version contains supplementary material available at 10.1038/s41577-022-00809-x.
References
- 1.Cui J, Li F, Shi Z-L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yadav R, et al. Role of structural and non-structural proteins and therapeutic targets of SARS-CoV-2 for COVID-19. Cells. 2021 doi: 10.3390/cells10040821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaur N, et al. Genetic comparison among various coronavirus strains for the identification of potential vaccine targets of SARS-CoV2. Infect. Genet. Evol. 2021;89:104490. doi: 10.1016/j.meegid.2020.104490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Le Bert N, et al. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature. 2020;584:457–462. doi: 10.1038/s41586-020-2550-z. [DOI] [PubMed] [Google Scholar]
- 5.Nickbakhsh S, et al. Epidemiology of seasonal coronaviruses: establishing the context for the emergence of coronavirus disease 2019. J. Infect. Dis. 2020;222:17–25. doi: 10.1093/infdis/jiaa185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu B, Guo H, Zhou P, Shi Z-L. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 2021;19:141–154. doi: 10.1038/s41579-020-00459-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Global-Change-Data-Lab. Our World Data. https://ourworldindata.org/grapher/deaths-covid-19-vs-case-fatality-rate?tab=table (2021).
- 8.Yuki K, Fujiogi M, Koutsogiannaki S. COVID-19 pathophysiology: a review. Clin. Immunol. 2020;215:108427. doi: 10.1016/j.clim.2020.108427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Romero Starke K, et al. The isolated effect of age on the risk of COVID-19 severe outcomes: a systematic review with meta-analysis. BMJ Glob. Health. 2021;6:e006434. doi: 10.1136/bmjgh-2021-006434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sze S, et al. Ethnicity and clinical outcomes in COVID-19: a systematic review and meta-analysis. EclinicalMedicine. 2020 doi: 10.1016/j.eclinm.2020.100630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peckham H, et al. Male sex identified by global COVID-19 meta-analysis as a risk factor for death and ITU admission. Nat. Commun. 2020;11:6317. doi: 10.1038/s41467-020-19741-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McGurnaghan SJ, et al. Risks of and risk factors for COVID-19 disease in people with diabetes: a cohort study of the total population of Scotland. Lancet Diabetes Endocrinol. 2021;9:82–93. doi: 10.1016/S2213-8587(20)30405-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Biswas M, Rahaman S, Biswas TK, Haque Z, Ibrahim B. Association of sex, age, and comorbidities with mortality in COVID-19 patients: a systematic review and meta-analysis. Intervirology. 2021;64:36–47. doi: 10.1159/000512592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Williamson EJ, et al. Factors associated with COVID-19-related death using OpenSAFELY. Nature. 2020;584:430–436. doi: 10.1038/s41586-020-2521-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bastard P, et al. Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science. 2020;370:eabd4585. doi: 10.1126/science.abd4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bastard P, et al. Autoantibodies neutralizing type I IFNs are present in ~4% of uninfected individuals over 70 years old and account for ~20% of COVID-19 deaths. Sci. Immunol. 2021;6:eabl4340. doi: 10.1126/sciimmunol.abl4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Silvin A, et al. Elevated calprotectin and abnormal myeloid cell subsets discriminate severe from mild COVID-19. Cell. 2020;182:1401–1418. doi: 10.1016/j.cell.2020.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Szabo PA, et al. Longitudinal profiling of respiratory and systemic immune responses reveals myeloid cell-driven lung inflammation in severe COVID-19. Immunity. 2021;54:797–814. doi: 10.1016/j.immuni.2021.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Del Valle DM, et al. An inflammatory cytokine signature predicts COVID-19 severity and survival. Nat. Med. 2020;26:1636–1643. doi: 10.1038/s41591-020-1051-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rydyznski Moderbacher C, et al. Antigen-specific adaptive immunity to SARS-CoV-2 in acute COVID-19 and associations with age and disease severity. Cell. 2020;183:996–1012. doi: 10.1016/j.cell.2020.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Unterman A, et al. Single-cell multi-omics reveals dyssynchrony of the innate and adaptive immune system in progressive COVID-19. Nat. Commun. 2022;13:440. doi: 10.1038/s41467-021-27716-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scurr MJ, et al. Magnitude of venous or capillary blood-derived SARS-CoV-2-specific T cell response determines COVID-19 immunity. Nat. Commun. 2022;13:5422. doi: 10.1038/s41467-022-32985-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Georg P, et al. Complement activation induces excessive T cell cytotoxicity in severe COVID-19. Cell. 2022;185:493–512. doi: 10.1016/j.cell.2021.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ahern DJ, et al. A blood atlas of COVID-19 defines hallmarks of disease severity and specificity. Cell. 2022;185:916–938. doi: 10.1016/j.cell.2022.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jouan Y, et al. Phenotypical and functional alteration of unconventional T cells in severe COVID-19 patients. J. Exp. Med. 2020 doi: 10.1084/jem.20200872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bacher P, et al. Low-avidity CD4+ T cell responses to SARS-CoV-2 in unexposed individuals and humans with severe COVID-19. Immunity. 2020;53:1258–1271. doi: 10.1016/j.immuni.2020.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Braun J, et al. SARS-CoV-2-reactive T cells in healthy donors and patients with COVID-19. Nature. 2020;587:270–274. doi: 10.1038/s41586-020-2598-9. [DOI] [PubMed] [Google Scholar]
- 28.Mateus J, et al. Selective and cross-reactive SARS-CoV-2 T cell epitopes in unexposed humans. Science. 2020;370:89. doi: 10.1126/science.abd3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ogbe A, et al. T cell assays differentiate clinical and subclinical SARS-CoV-2 infections from cross-reactive antiviral responses. Nat. Commun. 2021;12:2055. doi: 10.1038/s41467-021-21856-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schulien I, et al. Characterization of pre-existing and induced SARS-CoV-2-specific CD8+ T cells. Nat. Med. 2021;27:78–85. doi: 10.1038/s41591-020-01143-2. [DOI] [PubMed] [Google Scholar]
- 31.Nelde A, et al. SARS-CoV-2-derived peptides define heterologous and COVID-19-induced T cell recognition. Nat. Immunol. 2021;22:74–85. doi: 10.1038/s41590-020-00808-x. [DOI] [PubMed] [Google Scholar]
- 32.Nguyen-Contant P, et al. S protein-reactive IgG and memory B cell production after human SARS-CoV-2 infection includes broad reactivity to the S2 subunit. mBio. 2020;11:e01991–20. doi: 10.1128/mBio.01991-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meckiff BJ, et al. Imbalance of regulatory and cytotoxic SARS-CoV-2-reactive CD4+ T cells in COVID-19. Cell. 2020;183:1340–1353. doi: 10.1016/j.cell.2020.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sridhar S, et al. Cellular immune correlates of protection against symptomatic pandemic influenza. Nat. Med. 2013;19:1305–1312. doi: 10.1038/nm.3350. [DOI] [PubMed] [Google Scholar]
- 35.Turtle L, et al. Human T cell responses to Japanese encephalitis virus in health and disease. J. Exp. Med. 2016;213:1331–1352. doi: 10.1084/jem.20151517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mongkolsapaya J, et al. Original antigenic sin and apoptosis in the pathogenesis of dengue hemorrhagic fever. Nat. Med. 2003;9:921–927. doi: 10.1038/nm887. [DOI] [PubMed] [Google Scholar]
- 37.Dejnirattisai W, et al. Cross-reacting antibodies enhance dengue virus infection in humans. Science. 2010;328:745. doi: 10.1126/science.1185181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Midgley CM, et al. An in-depth analysis of original antigenic sin in dengue virus infection. J. Virol. 2011;85:410. doi: 10.1128/JVI.01826-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Swadling L, et al. Pre-existing polymerase-specific T cells expand in abortive seronegative SARS-CoV-2. Nature. 2021 doi: 10.1038/s41586-021-04186-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ng KW, Faulkner N, Wrobel AG, Gamblin SJ, Kassiotis G. Heterologous humoral immunity to human and zoonotic coronaviruses: aiming for the achilles heel. Semin. Immunol. 2021;55:101507. doi: 10.1016/j.smim.2021.101507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jaimes JA, André NM, Chappie JS, Millet JK, Whittaker GR. Phylogenetic analysis and structural modeling of SARS-CoV-2 spike protein reveals an evolutionary distinct and proteolytically sensitive activation loop. J. Mol. Biol. 2020;432:3309–3325. doi: 10.1016/j.jmb.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Low JS, et al. ACE2-binding exposes the SARS-CoV-2 fusion peptide to broadly neutralizing coronavirus antibodies. Science. 2022;377:735–742. doi: 10.1126/science.abq2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang Y, Yang C, Xu X-F, Xu W, Liu S-W. Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacol. Sin. 2020;41:1141–1149. doi: 10.1038/s41401-020-0485-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu L, et al. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature. 2020;584:450–456. doi: 10.1038/s41586-020-2571-7. [DOI] [PubMed] [Google Scholar]
- 45.Hicks J, et al. Serologic cross-reactivity of SARS-CoV-2 with endemic and seasonal betacoronaviruses. J. Clin. Immunol. 2021 doi: 10.1007/s10875-021-00997-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Piccoli L, et al. Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology. Cell. 2020;183:1024–1042. doi: 10.1016/j.cell.2020.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Poh CM, et al. Two linear epitopes on the SARS-CoV-2 spike protein that elicit neutralising antibodies in COVID-19 patients. Nat. Commun. 2020;11:2806. doi: 10.1038/s41467-020-16638-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ahmadi E, Zabihi MR, Hosseinzadeh R, Mohamed Khosroshahi L, Noorbakhsh F. SARS-CoV-2 spike protein displays sequence similarities with paramyxovirus surface proteins: a bioinformatics study. PLoS ONE. 2021;16:e0260360. doi: 10.1371/journal.pone.0260360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ng KW, et al. Preexisting and de novo humoral immunity to SARS-CoV-2 in humans. Science. 2020;370:1339. doi: 10.1126/science.abe1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Grifoni A, et al. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181:1489–1501. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tilocca B, et al. Comparative computational analysis of SARS-CoV-2 nucleocapsid protein epitopes in taxonomically related coronaviruses. Microbes Infect. 2020;22:188–194. doi: 10.1016/j.micinf.2020.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Grifoni A, et al. A sequence homology and bioinformatic approach can predict candidate targets for immune responses to SARS-CoV-2. Cell Host Microbe. 2020;27:671–680. doi: 10.1016/j.chom.2020.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Low JS, et al. Clonal analysis of immunodominance and cross-reactivity of the CD4 T cell response to SARS-CoV-2. Science. 2021;372:1336. doi: 10.1126/science.abg8985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Premkumar L, et al. The receptor-binding domain of the viral spike protein is an immunodominant and highly specific target of antibodies in SARS-CoV-2 patients. Sci. Immunol. 2020;5:eabc8413. doi: 10.1126/sciimmunol.abc8413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Saini SK, et al. SARS-CoV-2 genome-wide T cell epitope mapping reveals immunodominance and substantial CD8+ T cell activation in COVID-19 patients. Sci. Immunol. 2021;6:eabf7550. doi: 10.1126/sciimmunol.abf7550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ladner JT, et al. Epitope-resolved profiling of the SARS-CoV-2 antibody response identifies cross-reactivity with endemic human coronaviruses. Cell Rep. Med. 2021 doi: 10.1016/j.xcrm.2020.100189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Song G, et al. Cross-reactive serum and memory B-cell responses to spike protein in SARS-CoV-2 and endemic coronavirus infection. Nat. Commun. 2021;12:2938. doi: 10.1038/s41467-021-23074-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shrock E, et al. Viral epitope profiling of COVID-19 patients reveals cross-reactivity and correlates of severity. Science. 2020;370:eabd4250. doi: 10.1126/science.abd4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Anderson EM, et al. Seasonal human coronavirus antibodies are boosted upon SARS-CoV-2 infection but not associated with protection. Cell. 2021;184:1858–1864. doi: 10.1016/j.cell.2021.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sokal A, et al. Maturation and persistence of the anti-SARS-CoV-2 memory B cell response. Cell. 2021;184:1201–1213. doi: 10.1016/j.cell.2021.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Krishnamurty AT, et al. Somatically hypermutated Plasmodium-specific IgM+ memory B cells are rapid, plastic, early responders upon malaria rechallenge. Immunity. 2016;45:402–414. doi: 10.1016/j.immuni.2016.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Loyal L, et al. Cross-reactive CD4+ T cells enhance SARS-CoV-2 immune responses upon infection and vaccination. Science. 2021;374:eabh1823. doi: 10.1126/science.abh1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lineburg KE, et al. CD8+ T cells specific for an immunodominant SARS-CoV-2 nucleocapsid epitope cross-react with selective seasonal coronaviruses. Immunity. 2021;54:1055–1065. doi: 10.1016/j.immuni.2021.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nguyen THO, et al. CD8+ cells specific for an immunodominant SARS-CoV-2 nucleocapsid epitope display high naive precursor frequency and TCR promiscuity. Immunity. 2021;54:1066–1082. doi: 10.1016/j.immuni.2021.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bartolo L, et al. SARS-CoV-2-specific T cells in unexposed adults display broad trafficking potential and cross-react with commensal antigens. Sci. Immunol. 2022;7:eabn3127. doi: 10.1126/sciimmunol.abn3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Francis Joshua M, et al. Allelic variation in class I HLA determines CD8+ T cell repertoire shape and cross-reactive memory responses to SARS-CoV-2. Sci. Immunol. 2021;0:eabk3070. doi: 10.1126/sciimmunol.abk3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li P, et al. Estimating global epidemiology of low-pathogenic human coronaviruses in relation to the COVID-19 context. J. Infect. Dis. 2020;222:695–696. doi: 10.1093/infdis/jiaa321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li Y, Wang X, Nair H. Global seasonality of human seasonal coronaviruses: a clue for postpandemic circulating season of severe acute respiratory syndrome coronavirus 2. J. Infect. Dis. 2020;222:1090–1097. doi: 10.1093/infdis/jiaa436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Park S, Lee Y, Michelow IC, Choe YJ. Global seasonality of human coronaviruses: a systematic review. Open Forum Infect. Dis. 2020 doi: 10.1093/ofid/ofaa443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Monto AS, et al. Coronavirus occurrence and transmission over 8 years in the HIVE cohort of households in Michigan. J. Infect. Dis. 2020;222:9–16. doi: 10.1093/infdis/jiaa161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gaunt ER, Hardie A, Claas ECJ, Simmonds P, Templeton KE. Epidemiology and clinical presentations of the four human coronaviruses 229E, HKU1, NL63, and OC43 detected over 3 years using a novel multiplex real-time PCR method. J. Clin. Microbiol. 2010;48:2940–2947. doi: 10.1128/JCM.00636-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sagar M, et al. Recent endemic coronavirus infection is associated with less-severe COVID-19. J. Clin. Invest. 2021 doi: 10.1172/JCI143380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Aran D, Beachler DC, Lanes S, Overhage JM. Prior presumed coronavirus infection reduces COVID-19 risk: a cohort study. J. Infect. 2020;81:923–930. doi: 10.1016/j.jinf.2020.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wu Z, McGoogan JM. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72 314 cases from the Chinese Center for Disease Control and Prevention. JAMA. 2020;323:1239–1242. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- 75.Ladhani SN, et al. COVID-19 in children: analysis of the first pandemic peak in England. Arch. Dis. Child. 2020;105:1180. doi: 10.1136/archdischild-2020-320042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dacon C, et al. Broadly neutralizing antibodies target the coronavirus fusion peptide. Science. 2022;0:eabq3773. doi: 10.1126/science.abq3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Selva KJ, et al. Systems serology detects functionally distinct coronavirus antibody features in children and elderly. Nat. Commun. 2021;12:2037. doi: 10.1038/s41467-021-22236-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Thomas R, Wang W, Su D-M. Contributions of age-related thymic involution to immunosenescence and inflammaging. Immun. Ageing. 2020;17:2. doi: 10.1186/s12979-020-0173-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Aiello A, et al. Immunosenescence and its hallmarks: how to oppose aging strategically? A review of potential options for therapeutic intervention. Front. Immunol. 2019 doi: 10.3389/fimmu.2019.02247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Britanova OV, et al. Age-related decrease in TCR repertoire diversity measured with deep and normalized sequence profiling. J. Immunol. 2014;192:2689. doi: 10.4049/jimmunol.1302064. [DOI] [PubMed] [Google Scholar]
- 81.Yager EJ, et al. Age-associated decline in T cell repertoire diversity leads to holes in the repertoire and impaired immunity to influenza virus. J. Exp. Med. 2008;205:711–723. doi: 10.1084/jem.20071140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Waterlow NR, Leeuwen EV, Davies NG, Flasche S, Eggo RM. How immunity from and interaction with seasonal coronaviruses can shape SARS-CoV-2 epidemiology. Proc. Natl Acad. Sci. USA. 2021;118:e2108395118. doi: 10.1073/pnas.2108395118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zimmermann P, Curtis N. Why is COVID-19 less severe in children? A review of the proposed mechanisms underlying the age-related difference in severity of SARS-CoV-2 infections. Arch. Dis. Child. 2021;106:429. doi: 10.1136/archdischild-2020-320338. [DOI] [PubMed] [Google Scholar]
- 84.Lin C-Y, et al. Pre-existing humoral immunity to human common cold coronaviruses negatively impacts the protective SARS-CoV-2 antibody response. Cell Host Microbe. 2021 doi: 10.1016/j.chom.2021.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.McNaughton AL, et al. Fatal COVID-19 outcomes are associated with an antibody response targeting epitopes shared with endemic coronaviruses. JCI Insight. 2022 doi: 10.1172/jci.insight.156372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wratil PR, et al. Evidence for increased SARS-CoV-2 susceptibility and COVID-19 severity related to pre-existing immunity to seasonal coronaviruses. Cell Rep. 2021;37:110169. doi: 10.1016/j.celrep.2021.110169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gartlan C, et al. Vaccine-associated enhanced disease and pathogenic human coronaviruses. Front. Immunol. 2022 doi: 10.3389/fimmu.2022.882972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ng KW, et al. SARS-CoV-2 S2-targeted vaccination elicits broadly neutralizing antibodies. Sci. Transl Med. 2022;14:eabn3715. doi: 10.1126/scitranslmed.abn3715. [DOI] [PubMed] [Google Scholar]
- 89.Zhou P, et al. A human antibody reveals a conserved site on beta-coronavirus spike proteins and confers protection against SARS-CoV-2 infection. Sci. Transl Med. 2022;14:eabi9215. doi: 10.1126/scitranslmed.abi9215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kundu R, et al. Cross-reactive memory T cells associate with protection against SARS-CoV-2 infection in COVID-19 contacts. Nat. Commun. 2022;13:80. doi: 10.1038/s41467-021-27674-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chandran, A. et al. Non-severe SARS-CoV-2 infection is characterised by very early T cell proliferation independent of type 1 interferon responses and distinct from other acute respiratory viruses. Preprint at medRxiv10.1101/2021.03.30.21254540 (2021).
- 92.Bertoletti A, Tan AT, Le Bert N. The T-cell response to SARS-CoV-2: kinetic and quantitative aspects and the case for their protective role. Oxf. Open Immunol. 2021 doi: 10.1093/oxfimm/iqab006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tan AT, et al. Early induction of functional SARS-CoV-2-specific T cells associates with rapid viral clearance and mild disease in COVID-19 patients. Cell Rep. 2021;34:108728. doi: 10.1016/j.celrep.2021.108728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mallajosyula V, et al. CD8+ T cells specific for conserved coronavirus epitopes correlate with milder disease in patients with COVID-19. Sci. Immunol. 2021;6:eabg5669. doi: 10.1126/sciimmunol.abg5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Dykema AG, et al. Functional characterization of CD4+ T cell receptors crossreactive for SARS-CoV-2 and endemic coronaviruses. J. Clin. Invest. 2021 doi: 10.1172/JCI146922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mahajan S, et al. Immunodominant T-cell epitopes from the SARS-CoV-2 spike antigen reveal robust pre-existing T-cell immunity in unexposed individuals. Sci. Rep. 2021;11:13164. doi: 10.1038/s41598-021-92521-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.López-Martin I, Andrés Esteban E, García-Martínez FJ. Relationship between MMR vaccination and severity of Covid-19 infection. Survey among primary care physicians. Med. Clin. 2021;156:140–141. doi: 10.1016/j.medcli.2020.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gold Jeffrey E, et al. Analysis of measles–mumps–rubella (MMR) titers of recovered COVID-19 patients. mBio. 2020;11:e02628–02620. doi: 10.1128/mBio.02628-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mysore V, et al. Protective heterologous T cell immunity in COVID-19 induced by the trivalent measles–mumps–rubella and tetanus–diptheria–pertussis vaccine antigens. Med. 2021 doi: 10.1016/j.medj.2021.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mateus J, et al. Low-dose mRNA-1273 COVID-19 vaccine generates durable memory enhanced by cross-reactive T cells. Science. 2021;374:eabj9853. doi: 10.1126/science.abj9853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ogbe A, et al. Durability of ChAdOx1 nCov-19 vaccination in people living with HIV. JCI Insight. 2022 doi: 10.1172/jci.insight.157031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Reynolds CJ, et al. Immune boosting by B.1.1.529 (Omicron) depends on previous SARS-CoV-2 exposure. Science. 2022;377:eabq1841. doi: 10.1126/science.abq1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Chemaitelly H, et al. Immune imprinting and protection against repeat reinfection 00with SARS-CoV-2. N. Engl. J. Med. 2022 doi: 10.1056/NEJMc2211055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hansen CH, et al. Risk of reinfection, vaccine protection, and severity of infection with the BA.5 omicron subvariant: a Danish nation-wide population-based study. SSRN Electron. J. 2022 doi: 10.2139/SSRN.4165630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Harvey WT, et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021;19:409–424. doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Singh D, Yi SV. On the origin and evolution of SARS-CoV-2. Exp. Mol. Med. 2021;53:537–547. doi: 10.1038/s12276-021-00604-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Pinto D, et al. Broad betacoronavirus neutralization by a stem helix-specific human antibody. Science. 2021;373:1109–1116. doi: 10.1126/science.abj3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Morgenlander WR, et al. Antibody responses to endemic coronaviruses modulate COVID-19 convalescent plasma functionality. J. Clin. Invest. 2021 doi: 10.1172/JCI146927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.McCarthy KR, et al. Memory B cells that cross-react with group 1 and group 2 influenza A viruses are abundant in adult human repertoires. Immunity. 2018;48:174–184. doi: 10.1016/j.immuni.2017.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Galli G, et al. Fast rise of broadly cross-reactive antibodies after boosting long-lived human memory B cells primed by an MF59 adjuvanted prepandemic vaccine. Proc. Natl Acad. Sci. USA. 2009;106:7962–7967. doi: 10.1073/pnas.0903181106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Mongkolsapaya J, et al. T cell responses in dengue hemorrhagic fever: are cross-reactive T cells suboptimal. J. Immunol. 2006;176:3821. doi: 10.4049/jimmunol.176.6.3821. [DOI] [PubMed] [Google Scholar]
- 112.Duangchinda T, et al. Immunodominant T-cell responses to dengue virus NS3 are associated with DHF. Proc. Natl Acad. Sci. USA. 2010;107:16922–16927. doi: 10.1073/pnas.1010867107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Welsh RM, Che JW, Brehm MA, Selin LK. Heterologous immunity between viruses. Immunol. Rev. 2010;235:244–266. doi: 10.1111/j.0105-2896.2010.00897.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Screaton G, Mongkolsapaya J, Yacoub S, Roberts C. New insights into the immunopathology and control of dengue virus infection. Nat. Rev. Immunol. 2015;15:745–759. doi: 10.1038/nri3916. [DOI] [PubMed] [Google Scholar]
- 115.Zhang A, Stacey HD, Mullarkey CE, Miller MS. Original antigenic sin: how first exposure shapes lifelong anti-influenza virus immune responses. J. Immunol. 2019;202:335. doi: 10.4049/jimmunol.1801149. [DOI] [PubMed] [Google Scholar]
- 116.Pierson TC, et al. The stoichiometry of antibody-mediated neutralization and enhancement of West Nile virus infection. Cell Host Microbe. 2007;1:135–145. doi: 10.1016/j.chom.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Goncalvez AP, Engle RE, St. Claire M, Purcell RH, Lai C-J. Monoclonal antibody-mediated enhancement of dengue virus infection in vitro and in vivo and strategies for prevention. Proc. Natl Acad. Sci. USA. 2007;104:9422–9427. doi: 10.1073/pnas.0703498104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Francis T. Influenza: the new acquayantance. Ann. Intern. Med. 1953;39:203–221. doi: 10.7326/0003-4819-39-2-203. [DOI] [PubMed] [Google Scholar]
- 119.Dejnirattisai W, et al. Dengue virus sero-cross-reactivity drives antibody-dependent enhancement of infection with zika virus. Nat. Immunol. 2016;17:1102–1108. doi: 10.1038/ni.3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Stettler K, et al. Specificity, cross-reactivity, and function of antibodies elicited by Zika virus infection. Science. 2016;353:823–826. doi: 10.1126/science.aaf8505. [DOI] [PubMed] [Google Scholar]
- 121.Zivna I, et al. T cell responses to an HLA-B*07-restricted epitope on the dengue NS3 protein correlate with disease severity. J. Immunol. 2002;168:5959. doi: 10.4049/jimmunol.168.11.5959. [DOI] [PubMed] [Google Scholar]
- 122.Schouest B, et al. Pre-existing T cell memory against Zika virus. J. Virol. 2021;95:e00132–00121. doi: 10.1128/JVI.00132-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Grifoni A, et al. Prior dengue virus exposure shapes T cell immunity to Zika virus in humans. J. Virol. 2017;91:e01469–01417. doi: 10.1128/JVI.01469-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Saron WAA, et al. Flavivirus serocomplex cross-reactive immunity is protective by activating heterologous memory CD4 T cells. Sci. Adv. 2018;4:eaar4297. doi: 10.1126/sciadv.aar4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Vieira GF, Chies JAB. Immunodominant viral peptides as determinants of cross-reactivity in the immune system — Can we develop wide spectrum viral vaccines? Med. Hypotheses. 2005;65:873–879. doi: 10.1016/j.mehy.2005.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Moss B. Smallpox vaccines: targets of protective immunity. Immunol. Rev. 2011;239:8–26. doi: 10.1111/j.1600-065X.2010.00975.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.