Abstract
Bacterial overgrowth in the stomach may occur under conditions of diminished or absent acid secretion. Under these conditions, secretion of the hormone gastrin is elevated. Alternatively, bacterial factors may directly stimulate gastrin. Consistent with this hypothesis, we found that mice colonized for 2 months with a mixed bacterial culture of opportunistic pathogens showed an increase in serum gastrin. To examine regulation of gene expression by bacterial proteins, stable transformants of AGS cells expressing gastrin or interleukin-8 (IL-8) promoters were cocultured with live organisms. Both whole-cell sonicates and a heat-stable fraction were also coincubated with the cells. A level of 108 organisms per ml stimulated both the gastrin and IL-8 promoters. Heat-stable proteins prepared from these bacterial sonicates stimulated the promoter significantly more than the live organism or unheated sonicates. A 38-kDa heat-stable protein stimulating the gastrin and IL-8 promoters was cloned and found to be an OmpA-related protein. Immunoblotting using antibody to the OmpA-like protein identified an Acinetobacter sp. as the bacterial species that expressed this protein and colonized the mouse stomach. Moreover, reintubation of mice with a pure culture of the Acinetobacter sp. caused gastritis. We conclude that bacterial colonization of the stomach may increase serum gastrin levels in part through the ability of the bacteria to produce OmpA-like proteins that directly stimulate gastrin and IL-8 gene expression. These results implicate OmpA-secreting bacteria in the activation of gastrin gene expression and raise the possibility that a variety of organisms may contribute to the increase in serum gastrin and subsequent epithelial cell proliferation in the hypochlorhydric stomach.
Colonization by aerobic and anerobic flora occurs in the stomach with increasing pH (45) and may be the result of increasing age, malnutrition, or iatrogenically induced achlorhydria, e.g., H2 receptor blockade or proton pump inhibitor administration. With the exception of stress ulcer treatment, the bacterial flora present under conditions of hypochlorhydria has been poorly studied. More importantly, chronic achlorhydria is a risk factor for gastric cancer (42). Patients in intensive care units often have a gastric pH of >3 due to the routine use of antacids, proton pump inhibitors, or H2 receptor antagonists to prevent stress ulcers. An increase in gastric pH permits colonization of the stomach with opportunistic pathogens that contribute to the development of nosocomial pneumonia (19). Acinetobacter baumannii, Klebsiella pneumoniae, and Pseudomonas spp. are the predominant strains cultured from the relatively alkaline stomachs of ventilated patients (19) and are implicated in nosocomial respiratory infections (10). In a previous study, which examined oropharyngeal and gastric colonization using DNA genomic analysis, gastric colonization occurred regardless of the pH, which ranged from 2.8 to 5.7. Antacids were not used, and H2 receptor antagonists were used occasionally. Interestingly, A. baumannii was responsible for 30 nosocomial pneumonias in these ventilated patients despite the use of broad-spectrum antibiotics, e.g., amoxicillin and aminoglycosides.
Gastrin regulates acid secretion and is a growth factor for the oxyntic mucosa (11). Elevated serum gastrin levels stimulate parietal cell proliferation and acid secretion. Due to the normal feedback regulation of gastrin by acid, achlorhydria is a potent activator of gastrin gene expression (7). Therefore, overexpression of gastrin has also been implicated as a risk factor for gastric cancer (47). Since achlorhydria predisposes the stomach to both colonization by a variety of bacteria and hypergastrinemia, we queried whether the two conditions might be related. Thus, the goal of the present study was to identify candidate virulence factors in gastric bacterial flora that might both stimulate gastrin promoter activity and increase serum gastrin levels.
A microaerophilic culture consisting of Acinetobacter, Pseudomonas, and Corynebacterium spp. was inoculated into mouse stomachs for 2 to 6 months, after which an increase in serum gastrin levels was observed within 2 months. In addition to the effect on gastrin, the effect of these organisms on interleukin-8 (IL-8) gene expression was studied in a cell culture model, since it has been shown that Helicobacter pylori colonization stimulates production of CXC cytokines (e.g., IL-8) from the gastric mucosa (24). This cytokine class exhibits potent chemotactic effects that contribute to the inflammatory infiltrate (31). We used a human gastric cell line stably transfected with the human gastrin or IL-8 promoter to identify putative virulence factors produced by these organisms. We observed that live cultures of the bacteria were able to stimulate gastrin and IL-8 promoter activity in a dose-dependent manner and correlated the activity with the major protein in the heat-stable fraction. This protein was N-terminally sequenced and then cloned and found to be related to the major outer membrane protein found in most gram-negative pathogens, called OmpA. Of the organisms cultured from the mouse stomach, the OmpA-like protein was expressed exclusively in the Acinetobacter strain. Therefore, the OmpA-like protein appears to be an important bacterial factor capable of stimulating gastrin and IL-8 gene expression.
MATERIALS AND METHODS
Reagents.
The IL-8 luciferase reporter construct, a gift from H. C. Reinecker (Massachusetts General Hospital, Boston), contains the first 497 bp of the human IL-8 promoter as described by Mukaida et al. (32) in pGL3 basic (Promega).
A mixed stock of Acinetobacter, Pseudomonas, and Corynebacterium spp. was used to inoculate blood agar plates consisting of 5% sterile horse blood in Campylobacter-selective agar (Difco) supplemented with 5% (vol/vol) sterile horse blood (Rockland). The plates were incubated for 5 days in a humidified microaerophilic chamber to mimic the stomach environment (BBL Gas System, with CampyPak Plus packs [Fisher]). The bacteria recovered from the mice and cultured under these conditions were urease, catalase, and oxidase positive. To document the absence of any H. pylori species, the culture was plated onto blood agar plates supplemented with multiple antibiotics (amphotericin B [Fungizone], 1.5 ml/liter; vancomycin, 6.67 mg/ml; polymyxin B sulfate, 0.22 mg/ml; bacitracin, 13.33 mg/ml; and nalidixic acid, 2.14 mg/ml). No growth was observed. Whole-cell sonicates (WCS) were prepared by removing bacterial growth with a cotton swab, suspending it in ice-cold phosphate-buffered saline (PBS) (10 ml per 150-mm plate), and then pelleting the bacteria collected in 15-ml conical tubes at 3,000 rpm for 10 min at 4°C. The washed bacterial pellet was resuspended in PBS at a final concentration of 108 organisms/ml. Five 15-s bursts at 30% power output were used to disrupt the bacterial wall at 4°C. Heated supernatants (HS) were prepared from the WCS by incubating the sonicate for 15 min at 80°C and then pelleting denatured protein at 10,000 rpm for 15 min. Fifty micrograms of either WCS or HS was added to each 35-mm well of a six-well plate. Luciferase activity normalized to protein was analyzed 6 h later after incubating cells with live organisms or 3 h later after incubation with bacterial extracts. Protein concentrations were determined by the method of Bradford (6). The results were expressed as induction relative to the untreated controls. The proteins in the HS were size fractionated in a 2-ml-volume Centricon filter (Millipore) with a membrane size limit of 10 kDa according to the manufacturer's instructions.
Bacterial colonization in mice.
C57BL/6 (Charles River) mice underwent gastric intubation with a catheter four times over four consecutive days with 100 μl of bacterial suspension containing 108 organisms in PBS. Control animals were treated with a suspension of 108 Escherichia coli DH5α organisms. The animals were subjected to fasting overnight and then sacrificed 2 or 6 months later. The stomach was fixed in 4% paraformaldehyde diluted in PBS and paraffin embedded. Three-micrometer sections were prepared, deparaffinized, and stained with hematoxylin and eosin. Acinetobacter colonization was verified by reculturing stomach homogenates. Reintubation of mice with a pure subculture of the Acinetobacter sp. was accomplished as described above after twice-daily oral administration of streptomycin (0.2 ml of a 5-mg/ml stock solution per intubation) over 5 days to reduce the endogenous microbial flora in the stomach. Serum gastrin levels were determined by radioimmunoassay performed by the University of Michigan Peptide Center.
Cell culture.
AGS cells (derived from a human gastric adenocarcinoma) (1) were purchased from the American Type Culture Collection and cultured in Dulbecco's modified Eagle's medium (Gibco-BRL) containing 10% fetal calf serum, 100 μg of penicillin per ml, and 100 μg of streptomycin per ml in a humidified atmosphere of 5% CO2 and 95% air in 35-mm six-well culture dishes at 37°C. The cells were stably transfected with the 240 gastrin luciferase reporter construct (240 GasLuc), selected in G418 (Gibco-BRL), and pooled as described previously (17). The 240 GasLuc construct contains 240 bp of the human gastrin promoter and the first exon ligated upstream of the luciferase reporter in pGL2 basic (Promega). Calcium phosphate coprecipitation (5 Prime-3 Prime) was used to perform stable transfections into AGS cells. The cells were incubated for 48 h in Ham's F-12 nutrient mixture containing 100 μg of penicillin per ml and 100 μg of streptomycin (Gibco-BRL) per ml without serum prior to treatment with 10 nM epidermal growth factor (EGF), 20 ng of IL-1β per ml, or 50 μg of bacterial extracts.
Protease digestion.
Staphylococcus aureus V8 and trypsin proteases were purchased from Sigma. Three hundred micrograms of the HS was incubated at 37°C with 7,500 U of each protease. A second aliquot of the proteases was added after 3 h, and the incubation was allowed to proceed overnight. Protease activity was inactivated by heating the proteins to 80°C for 15 min and pelleting the denatured protein. Digestion of the protein mixture in the HS was confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Coomassie blue staining. Fifty micrograms of the digest was added to the luciferase-expressing stable transformants to determine if the activity in the HS was protease sensitive. There was no morphologic change in the cells after incubation with the protein digest. Protein concentrations were determined by the method of Bradford (6).
Protein elution.
Bacterial proteins from the HS were resolved on a 4 to 12% gradient gel (Novex) and visualized by Sypro Orange (Molecular Probes). The 38-, 130-, and 15-kDa proteins were excised, minced, and then electroeluted overnight at 10 mA per sample in a Bio-Rad Electro-Eluter (Model 422) in the presence of 25 mM Tris–192 mM glycine–0.1% SDS. The SDS was removed by continuing the elution for an additional hour in the absence of SDS. Fifty microliters of the eluted protein was added per 35-mm well of a six-well plate.
Peptide analysis.
SDS-PAGE was carried out on a 4 to 12% gradient gel (Novex), and the gel was stained with Coomassie blue dye. Edman degradation analysis of the N-terminal sequence was carried out by the University of Michigan Protein Core after transferring resolved proteins to polyvinylidene difluoride (PVDF) paper. Thirty N-terminal amino acids were sequenced from the 38-kDa protein isolated from the HS and bacterial wash fraction (GVTVTPLMLGYTFQDTQHNNNGNDGELTSS). In addition, a cyanogen bromide (CNBr) digest of the 38-kDa protein was resolved on an SDS–10% polyacrylamide gel and transferred to PVDF for amino acid sequencing by Edman degradation. N-terminal sequencing of the 9-kDa fragment (ELRVFFDTNKSNIKDOYKPEIAKVAEKLVE) was used to design the reverse primer. The forward primer [5′ ATG (TC)T(CAGT) GGA TAT ACA TTT CAG GAC AC(CAGT) CA(AG) CA(TC) AA(TC) AA(TC) AAT] was a degenerate oligonucleotide sequence based upon the underlined N-terminal amino acid sequence from the full-length 38-kDa protein. The reverse primer was a degenerate oligonucleotide sequence based upon amino acid sequence from the 9-kDa peptide [5′ (AG)CT (TC)TT (AG)TT (CAGT)GT (AG)TC (AG)AA (AG)AA (CAGT)AC]. The primers produced a 600-bp fragment from genomic DNA. This fragment was labeled using the Rediprime II kit (Stratagene) and used to screen a genomic bacterial library.
Genomic library screen.
The library in lambda ZAP II (Stratagene) was prepared from genomic DNA extracted from the bacterial culture mixture using a genomic DNA extraction kit (Qiagen). A total of 107 plaques of the unamplified library were screened. The Bluescript plasmid containing genomic inserts were excised with helper phage and sequenced. Of the six positive clones identified as containing 0.4- to 1.6-kb inserts, all six were partially sequenced, and it was confirmed that they contained the same overlapping sequence. Two of the overlapping clones containing the most distal 5′ and 3′ sequences were completely sequenced to produce a 1.7-kb genomic contig for the ompA locus.
Antibody production and immunoblotting.
A synthetic peptide (250 micromol) that corresponded to the C terminus (CGSRTVLAEQPVAQ) of the OmpA-like protein was prepared by the University of Michigan Peptide Core and conjugated to keyhole limpet hemocyanin. Rabbits were injected monthly with 250 to 500 μg of the peptide (Rockland) and analyzed by Western blotting using the HS as the antigen. An immunoglobulin fraction was prepared from both the immune and the preimmune sera using protein A-agarose (Santa Cruz) prior to use. To detect the OmpA on immunoblots, protein transferred to PVDF membrane was blocked for 1 h with 0.5× Uni-Block (Analytical Genetic Testing Center, Inc.) and then incubated with a 1:10,000 dilution of the immunoglobulin fraction for 1 h at 25°C. A 1:1,000 dilution of the secondary antibody was used, and the complexes were detected by enhanced chemiluminescence (SuperSignal; Pierce).
Recombinant protein.
To produce recombinant OmpA protein, the cDNA was amplified from the Bluescript plasmid using a forward primer (5′ GGCGTAACTGTTACTCCGTTGATG [GVTV form minus putative signal peptide] or 5′ ATGGCCTATTGCGGGCTTGAGCTT [full-length form with putative signal peptide]) and a reverse primer (5′ TTGAGCAACTGGTTGTTCAGCTAAAACAG). The amplimer was then inserted into the EcoRI site of the TA cloning vector (Invitrogen) and sequenced to determine the orientation. The EcoRI fragment was excised from the TA cloning vector and directionally subcloned into the EcoRI site of the pET vector (Novagen). The orientation was verified by restriction analysis and sequencing. The resulting expression vector was used to transform BL21(DE) cells and produce recombinant protein. The recombinant protein was induced at 37°C with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 3 to 4 h and then purified from the bacterial pellet (inclusion bodies) according to the method described by Yang et al. (49). Briefly, the bacterial pellet obtained after sonication was suspended in 50 mM Tris-HCl, pH 8.0, in 0.5% Triton X-100–10 mM EGTA. It was then centrifuged at 20,000 × g for 30 min at 4°C. The pellet was washed in 50 mM Tris-HCl, pH 8.0, containing 0.1% SDS and resuspended in 50 mM Tris-HCl, pH 8.0, in 0.5% Triton X-100–10 mM EGTA. To this suspension, a final concentration of 6 M urea was added; the suspension was heated to 60°C for 30 min and then dialyzed at 4°C against 50 mM Tris-HCl, pH 8.0, overnight. Due to low solubility, this suspension was resolved on an SDS-polyacrylamide gel as described for the eluted protein above prior to addition of the AGS cell cultures.
Nucleotide sequence accession number.
The GenBank accession no. for the Acinetobacter ompA genomic sequence is AF132598.
RESULTS
Increase in serum gastrin stimulated by gastric flora.
Mice were infected with the mixture of opportunistic pathogens and then sacrificed after 2 months. The stomachs were examined by immunohistochemistry and revealed mucosal and submucosal infiltrates in the corpus and antrum. Serum gastrin levels were elevated in infected mice compared with control mice by approximately fourfold (14.2 ± 4.0 [standard error of the mean] fmol/ml, n = 3, versus 3.2 ± 1.1 [standard error of the mean] fmol/ml, n = 3, respectively).
Regulation of the gastrin promoter by bacterial coculture.
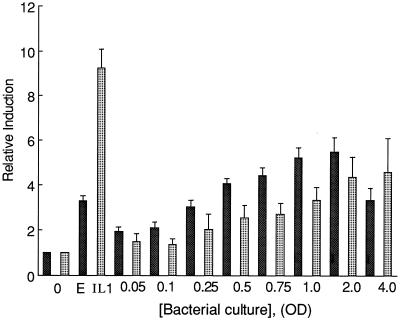
To examine whether bacterial contact directly stimulates gastrin gene expression, increasing concentrations of the bacterial mixture used to infect the mice were cocultured with stable transformants of AGS cells that express the human gastrin reporter construct. The effect of the organisms on IL-8 promoter activity was also examined. The results demonstrated a dose-dependent increase in both gastrin and IL-8 promoter activity with increasing concentrations of bacteria (Fig. 1) and demonstrated that live organisms were capable of stimulating gastrin as well as IL-8 promoter activity to an extent that correlated with their ability to increase serum gastrin levels in infected mice.
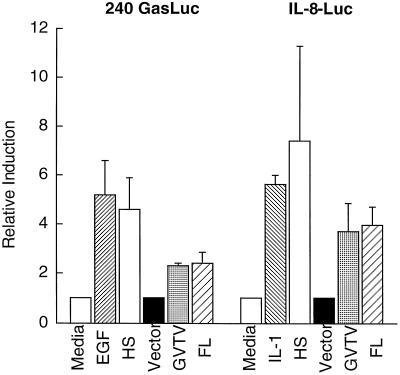
FIG. 1.
Coculture of the bacterial culture with AGS transformants stimulates gastrin and IL-8 promoters. Increasing amounts of the organisms were cocultured with stable transformants of AGS cells expressing gastrin (solid bars) or IL-8 (stippled bars) promoter constructs. One optical density (OD) unit equals 2.4 × 108 organisms. EGF (E) and IL-1β were used as positive inducers of the gastrin and IL-8 promoters, respectively. The basal luciferase activity for these constructs was ∼7,000 light units per μg of protein constructs for the gastrin promoter and ∼50,000 light units per μg of protein for the IL-8 promoter. The mean of the relative induction ± standard error for at least four experiments is shown.
Identifying a protein fraction that stimulates promoter activity.
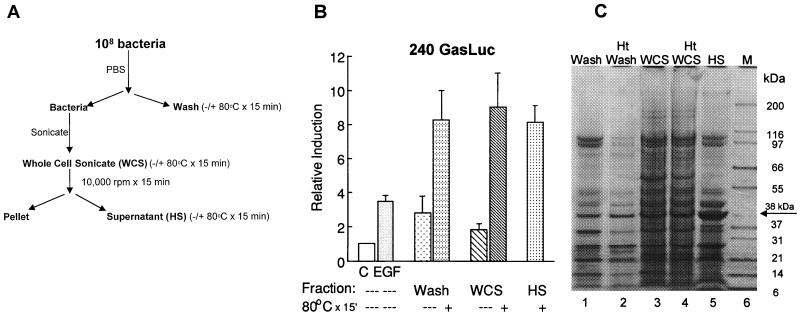
To isolate a bacterial subfraction containing the activity, the proteins recovered from the bacterial wash as well as the WCS were heated and then separated into soluble and insoluble fractions (Fig. 2A). Surprisingly, the substance stimulating an increase in promoter activity partitioned with the heat-soluble fraction (Fig. 2B). This result raised several possibilities—that the activating substance was not protein or that heating removed heat-labile inhibitory substances. A third consideration was that heating induced a conformational change in the protein such that it was more active. To address the first two possibilities, the heat-stable supernatant was resolved on a denaturing gel (Fig. 2C). A striking finding was the similarity between the heat-stable bacterial wash and the heat-stable fraction isolated from the WCS (Fig. 2C, lanes 2 and 4). The most abundant protein in the heat-stable supernatant prepared from the WCS was a 38-kDa protein which also appeared in the heated wash. Much of the protein was heat labile and was rendered insoluble. Thus, the higher specific activity may be related to the removal of an inhibitor.
FIG. 2.
The bacterial fraction stimulating the gastrin promoter is heat stable. (A) Fractionation scheme for the bacterial mixture. The PBS wash (Wash), WCS, and supernatant from the WCS were heated to 80°C for 15 min. The HS represents the activity after pelleting insoluble protein (Pellet) at 10,000 rpm for 15 min. (B) Fifty micrograms of the various fractions was used to stimulate stable transformants that express the human gastrin reporter construct. The relative induction ± standard error for at least four experiments is shown. C, control. (C) The indicated fractions were resolved on a 4 to 12% gradient gel and then stained with Coomassie blue dye. Lane M shows molecular mass markers. Ht, heated.
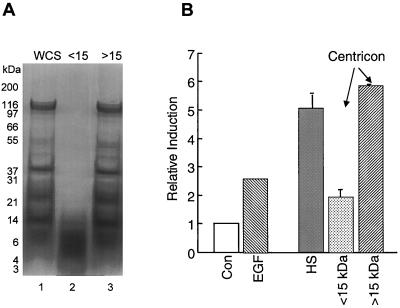
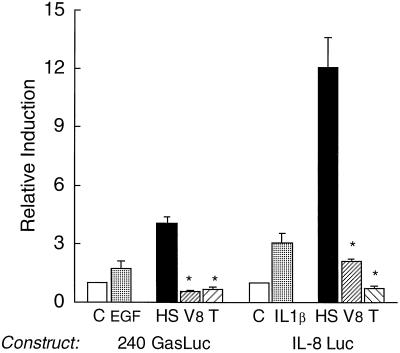
To examine whether the activating material was of protein origin or was a small organic molecule or peptide, size fractionation was performed using Centricon spin columns set to exclude masses of approximately 10 to 15 kDa (Fig. 3A). The results shown in Fig. 3B demonstrate that most of the activity segregated with the higher-molecular-mass proteins (>15 kDa), supporting the argument that the activating factor was not a small molecule. We found that a minimum amount of activity segregated with the low-molecular-weight material and concluded that this may represent either peptides or proteolytic fragments of the higher-molecular-weight proteins. To demonstrate directly that the factor was a protein, the HS was treated with S. aureus V8 or trypsin proteases (Fig. 4). Indeed, both proteases abolished the expected induction of both promoters by the heat-stable fraction. Therefore, we concluded that the gram-negative organisms produce soluble, heat-stable proteins capable of stimulating gene expression.
FIG. 3.
The bacterial activity segregates with a protein of >15 kDa. (A) The Centricon fractions greater or less than 15 kDa were prepared from the HS and then resolved on an SDS–4 to 12% polyacrylamide gradient gel. Lane 1, WCS; lane 2, protein filtrate with molecular mass less than 15 kDa (<15); lane 3, protein retained on the Centricon filter, primarily >15 kDa (>15). (B) Activity of the gastrin promoter after stimulation of the stable transformants with 10 nM EGF or with 50 μg of the HS fraction or the >15-kDa or <15-kDa protein fractions from the Centricon filter. Shown is the mean of two experiments. Con, control.
FIG. 4.
The activity in the HS is protease sensitive. The HS was digested with either V8 protease (V8) or trypsin (T) overnight. Fifty micrograms of the HS before and after protease digestion was incubated with the stable transformants expressing the human gastrin (240 GasLuc) or IL-8 (IL-8 Luc) promoter for 3 h prior to the assay for luciferase activity, and values were then expressed as relative induction. Treatment of the gastrin and IL-8 promoters with 10 nM EGF and 4 ng of IL-1β per ml, respectively, is shown. The mean ± standard error is shown for three independent experiments. ∗, P < 0.05. C, control.
Transcriptional activity attributed to 38-kDa protein.
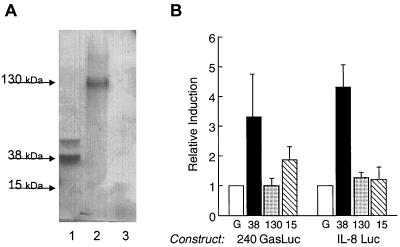
To determine if the activity resided with one or more of the abundant heat-stable proteins, three protein bands including the abundant 38-kDa protein were eluted from the SDS gel. Equivalent amounts of eluted protein verified by SDS-PAGE were tested on the stable transformants expressing the gastrin and IL-8 promoters. The results shown in Fig. 5 demonstrate that much of the induction was mediated by the 38-kDa heat-stable protein in contrast to the 130- and 15-kDa protein bands. However, since other proteins in the heat-stable fraction were not evaluated, we could not eliminate the possibility that other proteins in the HS fraction may also be capable of stimulating the promoters.
FIG. 5.
Eluted 38-kDa protein isolated from the HS stimulates the gastrin and IL-8 promoters. (A) Coomassie blue-stained SDS-PAGE of protein bands (lane 1, 38 kDa; lane 2, 130 kDa; lane 3, 15 kDa) eluted from the gel and used in reporter gene assays shown in panel B. (B) Relative induction of the gastrin (240 GasLuc) and IL-8 (IL-8 Luc) promoter constructs stably expressed in AGS cells after treatment with the eluted 38 (solid bars)-, 130 (stippled bars)-, and 15 (hatched bars)-kDa proteins. Eluted gel (G; open bars) without protein was used as the control for the eluted protein bands. Shown are results representative of three experiments performed in triplicate.
Cloning of an ompA-like gene.
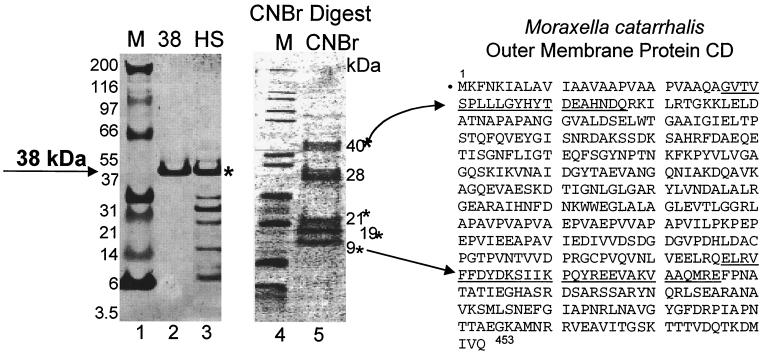
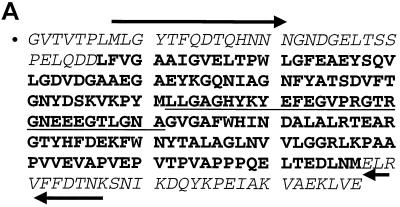
Since a significant amount of the promoter activity was attributed to the 38-kDa protein, the eluted band was submitted for peptide sequencing. N-terminal sequencing of the first 30 amino acids showed 80% similarity to outer membrane protein CD from a respiratory pathogen, Moraxella catarrhalis (Fig. 6). CNBr cleavage produced several internal peptides that were subsequently sequenced. The smaller, 9-kDa fragment was 80% identical to a sequence within the C terminus of the same Moraxella protein (Fig. 6). Thus, we concluded that the bacterial factor may also be an outer membrane protein. Degenerate PCR primers were designed from the two N- and C-terminal peptides and produced a 600-bp amplimer with genomic DNA isolated from the bacterial culture. The resulting amplimer was subcloned, sequenced, and translated to confirm overlap with the N-terminal and C-terminal peptides and one additional 21-kDa internal CNBr peptide fragment (Fig. 7A).
FIG. 6.
N-terminal sequence of 38-kDa protein related to Moraxella outer membrane protein CD. Lanes 1 to 3, Coomassie blue-stained gel; lanes 4 and 5, Coomassie blue-stained PVDF membrane. Lanes 1 and 4, molecular mass markers; lane 2, excised 38-kDa protein; lane 3, HS; lane 5, CNBr digest resolved on a 4 to 12% SDS gradient gel. The N-terminal sequences from the full-length 38-kDa protein and the 9-kDa CNBr fragment were compared to the M. catarrhalis CD protein (underlined). Asterisks indicate the CNBr fragments N-terminally sequenced.
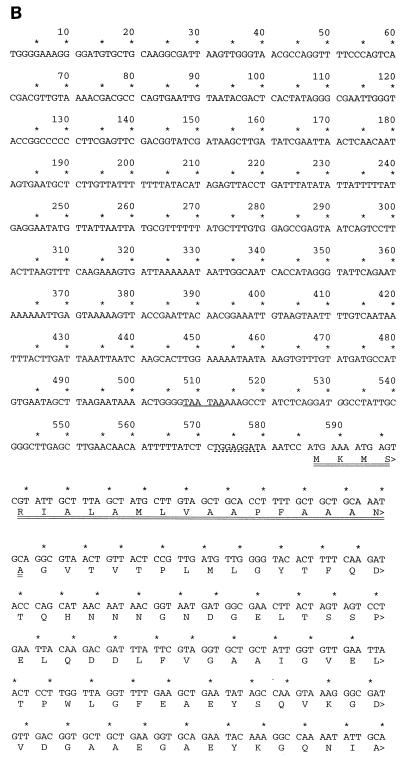
FIG. 7.
Sequence of a novel OmpA-like protein. (A) PCR primers were designed from the N-terminal peptide sequences of the full-length protein and 9-kDa CNBr fragment and used to amplify genomic DNA isolated from bacterial strains. The forward (→) and reverse (←) primers are indicated. The italicized sequences are the residues sequenced from the full-length 38-kDa peptide and 9-kDa CNBr fragments. The underlined residues were sequenced from the N terminus of the 21-kDa CNBr fragment. (B) ompA-like gene locus with translation. The in-frame upstream TAA stop codons are underlined. The dotted underline indicates the Shine-Dalgarno sequence. The putative signal sequence is double underlined. (C) Comparison of M. catarrhalis CD protein (M. cat) with the OmpA-like protein (OmpA). Plus signs indicate amino acid similarity; dashes indicate sequence gaps.
This 600-bp fragment was then used to screen a genomic library created from genomic DNA. Two of the six overlapping clones were sequenced completely, creating a 1.7-kb locus containing the 1.1-kb coding sequence (Fig. 7B). Translation of the open reading frame based upon the most N-terminal peptide revealed two TAA stop codons 78 bp upstream of a putative ATG start codon. Another in-frame ATG was identified 57 bp further upstream. However, this second ATG was not preceded by a Shine-Dalgarno sequence. Thus, the predicted ATG was preceded by a perfect Shine-Dalgarno sequence (TGGAGGAT) 6 bp upstream of the translational start codon (18). The additional 21-amino-acid sequence upstream of the original peptide beginning with GVTV was 45% identical to the E. coli OmpA signal sequence and the leader sequence of other secreted bacterial proteins (38). Amino acids 22 to 262 were similar to transmembrane domains of OmpA (29). Thus, overall the translated protein contained 349 amino acids corresponding to a molecular mass of 38 kDa (35 kDa without the N-terminal leader sequence) and a pI of 4.76. There were no cysteine residues present within the entire open reading frame. These features are consistent with the abundant heat-modifiable class of outer membrane proteins, called OmpA, found in most gram-negative bacteria. Figure 7C shows the homology between the Moraxella outer membrane CD OmpA-like protein and the newly cloned OmpA-like protein.
Recombinant OmpA-like protein stimulates gastrin and IL-8 promoter activity.
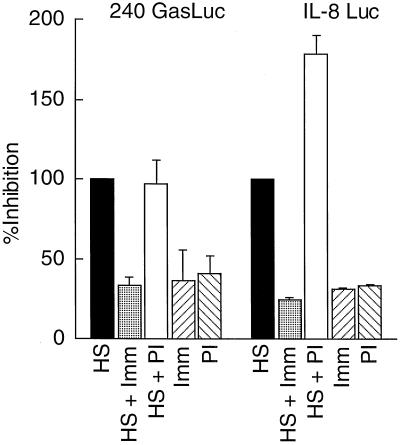
To demonstrate that the recombinant OmpA-like protein stimulated gastrin and IL-8 gene expression, the recombinant protein was prepared from E. coli protein-overexpressing transformants and tested on the stably transfected AGS cells. The results demonstrate that the recombinant OmpA-like protein stimulated the gastrin and IL-8 promoters (Fig. 8). The difference between the activity of the HS and that of the recombinant protein was due to the lower concentration of protein eluted from the gel. To demonstrate that the OmpA-like protein was the major protein responsible for the activity in the HS, antibody neutralization experiments were performed. Antibody raised against the C terminus of the OmpA-like protein specifically blocked the activation of both the gastrin and IL-8 promoters mediated by the HS, whereas the preimmune antibodies did not inhibit activity in the HS (Fig. 9).
FIG. 8.
Recombinant OmpA-like protein stimulates gastrin and IL-8 promoters. HS or two forms (minus signal sequence [GVTV] and full-length protein [FL]) of the recombinant OmpA-like protein eluted from SDS-polyacrylamide gels were incubated with AGS cells stably transfected with the 240 GasLuc or IL-8 Luc reporter construct. “Vector” indicates bacterial extract prepared as described for the recombinant protein except that the bacteria were transformed with the empty pET vector. “Media” indicates the AGS culture medium described in Materials and Methods. The results are the means and ranges of two independent experiments performed in duplicate and plotted as relative induction.
FIG. 9.
Neutralization of HS activity with OmpA-like protein antiserum. HS alone (20 μg) or in the presence of 2 μg of antibody raised against the OmpA-like protein (HS + Imm) was incubated for 4 h prior to precipitating the complexes with protein A/G. The resulting supernatant was then added to the stable AGS transformants. The same concentration of immunoglobulin G prepared from the preimmune serum was used as a negative control (HS + PI). In addition, the immune (Imm) or preimmune (PI) immunoglobulin G fractions alone were added to the stable transformants in the absence of the HS. The results are the means and ranges of two independent experiments performed in duplicate and plotted as percent inhibition relative to the HS.
The OmpA-like protein is expressed only in Acinetobacter species.
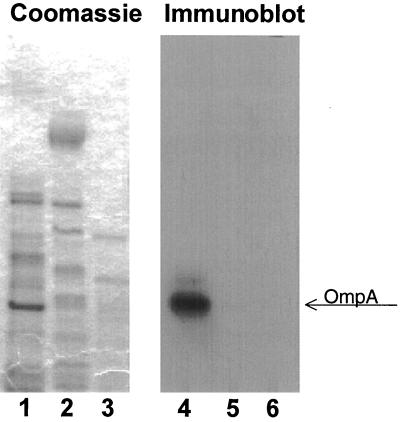
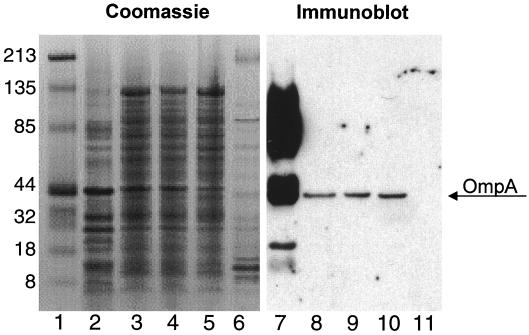
Since the original bacterial suspension contained at least two other bacterial species, the mixture was subcultured and typed. Typing of the bacterial species was performed using 16S ribosome primers, and the sequences were compared to database entries (28). Further characterization was performed using morphology, culture characteristics, and enzymatic analysis. Antibody raised to the cloned OmpA-like protein was used on an immunoblot to identify the bacterial strain expressing the cloned factor. The results shown in Fig. 10 demonstrate that the Acinetobacter sp. was the only bacterial strain isolated that expressed the cloned OmpA-like protein. To verify that this species was present in the inoculated mice, Western blotting was performed on the bacteria cultured from the mice. The results shown in Fig. 11 demonstrate that the same OmpA-like protein cloned was expressed in a subculture of Acinetobacter from infected mouse stomachs. The antibody did not cross-react with HS from a Pseudomonas sp.
FIG. 10.
The OmpA-like protein is expressed in Acinetobacter spp. HS from subcultures of Acinetobacter, Pseudomonas, and a Corynebacterium sp. were resolved on an SDS–4 to 12% polyacrylamide gel, transferred to PVDF membrane, and probed with a 1:2,000 dilution of the antibody to OmpA-like protein. Enhanced chemiluminescence was used to detect the antigen-antibody complexes. Lanes 1 to 3 contain the Coomassie blue-stained HS protein from Acinetobacter, Pseudomonas, and a Corynebacterium sp.; lanes 4 to 6 contain the corresponding immunoblot for the Acinetobacter, Pseudomonas, and Corynebacterium protein. The OmpA band is indicated.
FIG. 11.
Organisms isolated from mouse stomachs express the OmpA-like protein. HS were prepared from subcultures of bacteria isolated from two different mice infected with the gram-negative organisms 6 months previously. The Coomassie blue-stained gel (lanes 2 to 6) is shown compared with the immunoblot (lanes 7 to 11) prepared from the same samples and resolved in parallel. Antibody to the OmpA-like protein was used at a 1:10,000 dilution. Lane 1 is a Coomassie blue stain of the markers. Lanes 2 and 7 contain HS from the mixed bacterial culture used to intubate the mice; lanes 3, 4, 8, and 9 contain HS prepared from the Acinetobacter subcultured from the mice after 6 months; lanes 5 and 10 contain HS from a pure subculture of the Acinetobacter spp.; lanes 6 and 11 contains HS from a pure subculture of the Pseudomonas spp. The OmpA band is indicated. Numbers at left indicate molecular masses in kilodaltons.
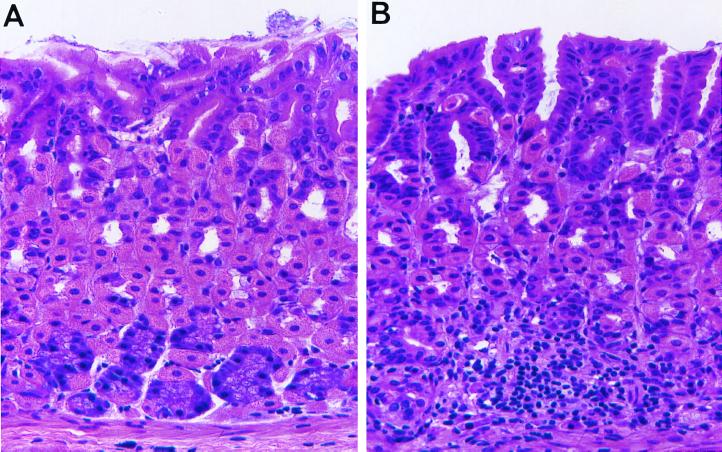
Acinetobacter species causes gastritis.
To prove that Acinetobacter alone causes gastritis, endogenous microbial flora was reduced with oral streptomycin prior to instilling 108 Acinetobacter organisms. Figure 12 shows the presence of a mucosal mononuclear infiltrate in the mouse corpus that extends into the submucosa (Fig. 12B) not present in the sham-intubated mouse (Fig. 12A). The infiltrate disrupted the normal architecture of the basally located chief and midglandular parietal cells. Thus, we concluded that Acinetobacter spp. are capable of causing gastritis.
FIG. 12.
Acinetobacter spp. cause mouse gastritis. Mice were treated orally for 5 days with streptomycin to sterilize the mouse stomach prior to instilling Acinetobacter spp. Stomachs from sham-intubated mice (A) were compared to stomachs from mice intubated with 108 Acinetobacter organisms for 2 months (B). Shown are representative sections from a paraffin-embedded stomach stained with hemotoxylin and eosin. (A) Normal (sham-intubated) mouse corpus (magnification, ×400); (B) Acinetobacter-infected mouse corpus (magnification, ×400).
DISCUSSION
We hypothesize that one of the events occuring during bacterial colonization of the stomach is the production of putative virulence factors. We report here that the OmpA-like protein from an Acinetobacter sp. stimulates the gastrin and IL-8 promoters and therefore would be capable of stimulating the production of this hormone and this cytokine under conditions that would favor its colonization in the stomach. This result has relevance to atrophic gastritis, during which the pH of the stomach rises due to atrophy of the parietal cell population, favoring colonization by aerobic and anaerobic flora (22). Atrophic gastritis is a late feature of chronic H. pylori infection but also occurs in pernicious anemia and malnutrition and with advancing age. More importantly, the loss of stomach acid is a risk factor for gastric cancer, presumably due to an increase in N-nitroso compounds produced by colonizing bacterial flora (2).
The antral G-cell response to bacterial infection of the gastric mucosa has been most intensively studied for H. pylori infection. A major consequence of H. pylori colonization is the increase in antral G cells and serum gastrin levels (21, 44). The studies reported here clearly demonstrate that bacterial species other than H. pylori may colonize the stomach and cause inflammation. Colonization by H. pylori does not occur efficiently in the hypochlorhydric stomach due to competition by other bacterial species (22). Thus, the ability of H. pylori to successfully compete with other bacteria for this rather unique niche is primarily due to its ability to survive in the low-pH environment of the stomach, e.g., at a pH of <3. Chronic H. pylori infection eventually results in atrophic gastritis, resulting in achlorhydria or severe hypochlorhydria. The bacterial overgrowth that follows is usually comprised of aerobic and anerobic flora. Moreover, where malignant mucosal transformation has occurred, H. pylori is absent (30, 41, 50). Thus, while H. pylori infection may initiate the development of atrophic gastritis, it may not mediate the final transforming event. Moreover, it may not be the only bacterial species initiating these events.
The use of the gastrin reporter construct to identify specific virulence factors was based on the proposal by Blaser and Parsonnet in 1994 that the hormone gastrin may play a role in ulcer and cancer pathogenesis triggered by H. pylori infection (5). Sustained levels of this hormone are associated with duodenal ulcers (46) as well as neoplastic transformation (8, 16, 43). Patients with chronic H. pylori infection exhibit increases in serum gastrin and G-cell populations concomitant with a decrease in D cells (21, 44). Yet, there is no evidence that H. pylori has a direct effect on the G cell. With respect to changes in G-cell numbers, several regulatory mechanisms may be operating. First, bacterial proteins may have a direct effect on the G cell. Second, there may be a loss of negative regulation due to a direct effect on the D cell. Third, parietal cell atrophy and subsequent achlorhydria may develop after longstanding H. pylori infection, resulting in elevated serum gastrin through the normal feedback regulatory mechanisms. Since atrophic gastritis is accompanied by a decrease in H. pylori colonization (15, 27), we examined whether organisms that might colonize the hypochlorhydric stomach are capable of stimulating gastrin. Gastrin activation by these organisms would provide a mechanism by which growth factors may be produced in the preneoplastic stomach.
To address the role of bacterial proteins, we investigated whether bacterial colonization activates gastrin promoter activity. We show in this report that infection by bacterial flora that includes Acinetobacter strains raises serum gastrin levels coincident with inflammation of the mucosa. Indeed, coculturing with the organisms stimulated both the gastrin and IL-8 promoters in a dose-dependent manner. The production and secretion of IL-8 or other CXC cytokines are important mechanisms to recruit inflammatory mediators to the site of infection (9). Therefore, we evaluated the ability of Acinetobacter to stimulate IL-8 gene expression. The novel finding reported here is that stimulation of both IL-8 and gastrin gene expression by a bacterial protein is due to a member of the OmpA protein family.
The OmpA-like protein has a low isoelectric point and no cysteines, suggesting that it may be acid as well as heat stable. This may be expected of proteins that are exposed to a low stomach pH. Residues 1 to 21 are hydrophobic and contain a signal peptide cleavage site (AlaXAla) (38). The putative leader sequence is 38% identical (76% similar) to the signal peptide for E. coli OmpA, which predicts protein translocation, export, and cleavage by signal peptidase I (38). The putative cleavage site for the Acinetobacter spp. OmpA follows the −3,−1 rule for signal peptides (13). Thus, the Acinetobacter spp. OmpA leader sequence is consistent with the protein being present in the outer membrane or periplasm.
The protein is closely related to the OmpA-related heat-modifiable proteins from Moraxella and Pseudomonas as well as the OmpA homologs from Salmonella, Shigella, and Klebsiella spp. Antibody raised against the Acinetobacter OmpA blocked IL-8 and gastrin promoter activation. The novel Acinetobacter spp. protein was most homologous to outer membrane protein CD precursor from Moraxella (45% identical; 60% positive over 341 amino acids). It also has ∼25% identity and 40% similarity within the C-terminal domain (268 to 333) to OmpA from E. coli and Serratia, Salmonella, Shigella, and Klebsiella bacterial species (23, 25, 26, 34, 36, 48). Apparently, OmpA proteins usually represent the major outer membrane protein, and all gram-negative bacteria have some functional equivalent of OmpA (3). The Acinetobacter spp. OmpA protein was 30% identical (47% similar) over 321 amino acids to outer membrane protein porin F (OprF) from Pseudomonas, which is also a member of the OmpA protein family. Thus, Acinetobacter spp. OmpA was also closely related to the Pseudomonas porin proteins.
OmpA-related proteins including the Pseudomonas porin F protein (OprF) appear to play a variety of roles depending on the bacterial species. In Pseudomonas, these proteins form pores in mammalian cells that permit the penetration of solutes, support survival in low-osmosis conditions, maintain cell shape, and participate in antimicrobial resistance (40). OmpA from E. coli and Shigella, Salmonella, Serratia, and Klebsiella spp. is the prevalent outer membrane protein and is highly immunogenic. It is also a receptor for bacterial phages, mediates F′-pilus-dependent conjugation, and maintains cell shape (4, 20, 35, 37, 39). Acinetobacter spp. OmpA is quite heat stable, and this property may be related to the heat-modifiable characteristic of most OmpA proteins including the M. catarrhalis CD (23, 33). This property is also a characteristic of E. coli OmpA, OmpA from enteropathogens, and H. pylori porin proteins (12, 14, 25, 26, 36, 48). Heat-modifiable proteins migrate in a heterogeneous pattern on SDS gels after heating due to a conformational change in the protein (3).
In summary, this is the first report documenting regulation of the human gastrin promoter by a bacterial protein, raising the strong possibility that bacterial proteins alone, exclusive of inflammation, may be sufficient to mediate elevated serum gastrin levels. This is also the first report of an effect of an OmpA-like protein on mammalian gene expression. IL-8 promoter activity was also activated, consistent with prior reports showing general activation of IL-8 production when epithelial cells are cocultured with a variety of bacterial strains. The results also suggest that opportunistic pathogens may regulate both gastrin and IL-8 production in part by producing OmpA-like proteins. The results do not rule out the possibility that other bacterial proteins affect cell function, but it is clear that the 38-kDa OmpA is the major protein within the heat-stable fraction of Acinetobacter spp. that is capable of activating these two promoters.
ACKNOWLEDGMENTS
J.L.M. is an investigator of the Howard Hughes Medical Institute. The work was supported in part by Public Health Service grant DK-45729.
We thank Charles Mitchell of the University of Michigan Protein and Carbohydrate Structure Facility Core and H. C. Reinecker of the Massachusetts General Hospital for the gift of the IL-8 luciferase promoter construct. Also, Susan Finnis of the University of Michigan Peptide Center (DK-34933) performed the gastrin radioimmunoassays, and the University of Michigan Clinical Microbiology Laboratory performed the bacterial subtyping.
REFERENCES
- 1.Barranco S C, Townsend C M, Jr, Casartelli C, Macik B G, Burger N L, Boerwinkle W R, Gourley W K. Establishment and characterization of an in vitro model system for human adenocarcinoma of the stomach. Cancer Res. 1983;43:1703–1709. [PubMed] [Google Scholar]
- 2.Bartsch H, Ohshima H, Pignatelli B, Calmels S. Endogenously formed N-nitroso compounds and nitrosating agents in human cancer etiology. Pharmacogenetics. 1992;2:272–277. doi: 10.1097/00008571-199212000-00005. [DOI] [PubMed] [Google Scholar]
- 3.Beher M G, Schnaitman C A, Pugsley A P. Major heat-modifiable outer membrane protein in gram-negative bacteria: comparison with the OmpA protein of Escherichia coli. J Bacteriol. 1980;143:906–913. doi: 10.1128/jb.143.2.906-913.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Behrmann M, Koch H G, Hengelage T, Wieseler B, Hoffschulte H K, Muller M. Requirements for the translocation of elongation-arrested, ribosome-associated OmpA across the plasma membrane of Escherichia coli. J Biol Chem. 1998;273:13898–13904. doi: 10.1074/jbc.273.22.13898. [DOI] [PubMed] [Google Scholar]
- 5.Blaser M J, Parsonnet J. Parasitism by the “slow” bacterium Helicobacter pylori leads to altered gastric homeostasis and neoplasia. J Clin Investig. 1994;94:4–8. doi: 10.1172/JCI117336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 7.Brand S J, Stone D. Reciprocal regulation of antral gastrin and somatostatin gene expression by omeprazole-induced achlorhydria. J Clin Investig. 1988;82:1059–1066. doi: 10.1172/JCI113662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conteas C N, Desai T K, Arlow F A. Relationship of hormones and growth factors to colon cancer. Gastroenterol Clin N Am. 1988;17:761–772. [PubMed] [Google Scholar]
- 9.Crabtree J. Role of cytokines in pathogenesis of Helicobacter pylori-induced mucosal damage. Dig Dis Sci. 1998;43(Suppl.):46S–53S. [PubMed] [Google Scholar]
- 10.Craven D E, Barber T W, Steger K A, Montecalvo M A. Nosocomial pneumonia in the 1990s: update of epidemiology and risk factors. Semin Respir Infect. 1990;5:157–172. [PubMed] [Google Scholar]
- 11.Dockray G J. Topical review. Gastrin and gastric epithelial physiology. J Physiol (London) 1999;518:315–324. doi: 10.1111/j.1469-7793.1999.0315p.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doig P, Trust T J. Identification of surface-exposed outer membrane antigens of Helicobacter pylori. Infect Immun. 1994;62:4526–4533. doi: 10.1128/iai.62.10.4526-4533.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duffaud G, Inouye M. Signal peptidases recognize a structural feature at the cleavage site of secretory proteins. J Biol Chem. 1988;263:10224–10228. [PubMed] [Google Scholar]
- 14.Exner M M, Doig P, Trust T J, Hancock R E. Isolation and characterization of a family of porin proteins from Helicobacter pylori. Infect Immun. 1995;63:1567–1572. doi: 10.1128/iai.63.4.1567-1572.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Farinati F, Valiante F, Germana B, Della Libera G, Baffa R, Rugge M, Plebani M, Vianello F, Di Mario F, Naccarato R. Prevalence of Helicobacter pylori infection in patients with precancerous changes and gastric cancer. Eur J Cancer Prev. 1993;2:321–326. doi: 10.1097/00008469-199307000-00006. [DOI] [PubMed] [Google Scholar]
- 16.Finley G G, Koski R A, Melhem M F, Pipas J M, Meisler A I. Expression of the gastrin gene in the normal human colon and colorectal adenocarcinoma. Cancer Res. 1993;53:2919–2926. [PubMed] [Google Scholar]
- 17.Ford M G, Valle J D, Soroka C J, Merchant J L. EGF receptor activation stimulates endogenous gastrin gene expression in canine G cells and human gastric cell cultures. J Clin Investig. 1997;99:2762–2771. doi: 10.1172/JCI119466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Freifelder D. Translation I: the information problem. In: Freifelder D, editor. Molecular biology. 2nd ed. Portola Valley, Calif: Jones and Bartlett; 1983. pp. 367–413. [Google Scholar]
- 19.Garrouste-Orgeas M, Chevret S, Arlet G, Marie O, Rouveau M, Popoff N, Schlemmer B. Oropharyngeal or gastric colonization and nosocomial pneumonia in adult intensive care unit patients. A prospective study based on genomic DNA analysis. Am J Respir Crit Care Med. 1997;156:1647–1655. doi: 10.1164/ajrccm.156.5.96-04076. [DOI] [PubMed] [Google Scholar]
- 20.Ghiara P, Rossi M, Marchetti M, Di Tommaso A, Vindigni C, Ciampolini F, Covacci A, Telford J L, De Magistris M T, Pizza M, Rappuoli R, Del Giudice G. Therapeutic intragastric vaccination against Helicobacter pylori in mice eradicates an otherwise chronic infection and confers protection against reinfection. Infect Immun. 1997;65:4996–5002. doi: 10.1128/iai.65.12.4996-5002.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Graham D Y, Lew G M, Lechago J. Antral G-cell and D-cell numbers in Helicobacter pylori infection: effect of H. pylori eradication. Gastroenterology. 1993;104:1655–1660. doi: 10.1016/0016-5085(93)90642-p. [DOI] [PubMed] [Google Scholar]
- 22.Houben G M, Stockbrugger R W. Bacteria in the aetio-pathogenesis of gastric cancer: a review. Scand J Gastroenterol Suppl. 1995;212:13–18. doi: 10.3109/00365529509090296. [DOI] [PubMed] [Google Scholar]
- 23.Hsiao C B, Sethi S, Murphy T F. Outer membrane protein CD of Branhamella catarrhalis: sequence conservation in strains recovered from the human respiratory tract. Microb Pathog. 1995;19:215–225. doi: 10.1016/s0882-4010(95)90272-4. [DOI] [PubMed] [Google Scholar]
- 24.Keates S, Hitti Y S, Upton M, Kelly C P. Helicobacter pylori infection activates NF-κB in gastric epithelial cells. Gastroenterology. 1997;113:1099–1109. doi: 10.1053/gast.1997.v113.pm9322504. [DOI] [PubMed] [Google Scholar]
- 25.Kennell W L, Egli C, Hancock R E, Holt S C. Pore-forming ability of major outer membrane proteins from Wolinella recta ATCC 33238. Infect Immun. 1992;60:380–384. doi: 10.1128/iai.60.2.380-384.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Komatsuzawa H, Kawai T, Wilson M E, Taubman M A, Sugai M, Suginaka H. Cloning of the gene encoding the Actinobacillus actinomycetemcomitans serotype b OmpA-like outer membrane protein. Infect Immun. 1999;67:942–945. doi: 10.1128/iai.67.2.942-945.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuipers E J. Review article: exploring the link between Helicobacter pylori and gastric cancer. Aliment Pharmacol Ther. 1999;13(Suppl. 1):3–11. doi: 10.1046/j.1365-2036.1999.00002.x. [DOI] [PubMed] [Google Scholar]
- 28.Lane D J, Pace B, Olsen G J, Stahl D A, Sogin M L, Pace N R. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc Natl Acad Sci USA. 1985;82:6955–6959. doi: 10.1073/pnas.82.20.6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lawrence J G, Ochman H, Hartl D L. Molecular and evolutionary relationships among enteric bacteria. J Gen Microbiol. 1991;137:1911–1921. doi: 10.1099/00221287-137-8-1911. [DOI] [PubMed] [Google Scholar]
- 30.McCloy R F, Arnold R, Bardhan K D, Cattan D, Klinkenberg-Knol E, Maton P N, Riddell R H, Sipponen P, Walan A. Pathophysiological effects of long-term acid suppression in man. Dig Dis Sci. 1995;40:96S–120S. doi: 10.1007/BF02214874. [DOI] [PubMed] [Google Scholar]
- 31.Moss S F, Legon S, Bishop A E, Polak J M, Calam J. Effect of Helicobacter pylori on gastric somatostatin in duodenal ulcer disease. Lancet. 1992;340:930–932. doi: 10.1016/0140-6736(92)92816-x. [DOI] [PubMed] [Google Scholar]
- 32.Mukaida N, Shiroo M, Matsushima K. Genomic structure of the human monocyte-derived neutrophil chemotactic factor IL-8. J Immunol. 1989;143:1366–1371. [PubMed] [Google Scholar]
- 33.Murphy T F, Kirkham C, DeNardin E, Sethi S. Analysis of antigenic structure and human immune response to outer membrane protein CD of Moraxella catarrhalis. Infect Immun. 1999;67:4578–4585. doi: 10.1128/iai.67.9.4578-4585.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murphy T F, Kirkham C, Lesse A J. The major heat-modifiable outer membrane protein CD is highly conserved among strains of Branhamella catarrhalis. Mol Microbiol. 1993;10:87–97. doi: 10.1111/j.1365-2958.1993.tb00906.x. [DOI] [PubMed] [Google Scholar]
- 35.Nguyen T N, Samuelson P, Sterky F, Merle-Poitte C, Robert A, Baussant T, Haeuw J F, Uhlen M, Binz H, Stahl S. Chromosomal sequencing using a PCR-based biotin-capture method allowed isolation of the complete gene for the outer membrane protein A of Klebsiella pneumoniae. Gene. 1998;210:93–101. doi: 10.1016/s0378-1119(98)00060-2. [DOI] [PubMed] [Google Scholar]
- 36.Ohnishi S, Kameyama K, Takagi T. Characterization of a heat modifiable protein, Escherichia coli outer membrane protein OmpA, in binary surfactant system of sodium dodecyl sulfate and octylglucoside. Biochim Biophys Acta. 1998;1375:101–109. doi: 10.1016/s0005-2736(98)00145-x. [DOI] [PubMed] [Google Scholar]
- 37.Ortiz V, Isibasi A, Garcia-Ortigoza E, Kumate J. Immunoblot detection of class-specific humoral immune response to outer membrane proteins isolated from Salmonella typhi in humans with typhoid fever. J Clin Microbiol. 1989;27:1640–1645. doi: 10.1128/jcm.27.7.1640-1645.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pratap J, Dikshit K L. Effect of signal peptide changes on the extracellular processing of streptokinase from Escherichia coli: requirement for secondary structure at the cleavage junction. Mol Gen Genet. 1998;258:326–333. doi: 10.1007/s004380050738. [DOI] [PubMed] [Google Scholar]
- 39.Puohiniemi R, Karvonen M, Vuopio-Varkila J, Muotiala A, Helander I M, Sarvas M. A strong antibody response to the periplasmic C-terminal domain of the OmpA protein of Escherichia coli is produced by immunization with purified OmpA or with whole E. coli or Salmonella typhimurium bacteria. Infect Immun. 1990;58:1691–1696. doi: 10.1128/iai.58.6.1691-1696.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rawling E G, Brinkman F S, Hancock R E. Roles of the carboxy-terminal half of Pseudomonas aeruginosa major outer membrane protein OprF in cell shape, growth in low-osmolarity medium, and peptidoglycan association. J Bacteriol. 1998;180:3556–3562. doi: 10.1128/jb.180.14.3556-3562.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Safatle-Ribeiro A V, Ribeiro U, Jr, Clarke M R, Sakai P, Ishioka S, Garrido A B, Jr, Gama-Rodrigues J, Safatle N F, Reynolds J C. Relationship between persistence of Helicobacter pylori and dysplasia, intestinal metaplasia, atrophy, inflammation, and cell proliferation following partial gastrectomy. Dig Dis Sci. 1999;44:243–252. doi: 10.1023/a:1026681829083. [DOI] [PubMed] [Google Scholar]
- 42.Seery J P. Achlorhydria and gastric carcinogenesis. Lancet. 1991;338:1508–1509. doi: 10.1016/0140-6736(91)92312-p. [DOI] [PubMed] [Google Scholar]
- 43.Singh P, Owlia A, Varro A, Dai B, Rajaraman S, Wood T. Gastrin gene expression is required for the proliferation and tumorigenicity of human colon cancer cells. Cancer Res. 1996;56:4111–4115. [PubMed] [Google Scholar]
- 44.Sumii M, Sumii K, Tari A, Kawaguchi H, Yamamoto G, Takehara Y, Fukino Y, Kamiyasu T, Hamada M, Tsuda T, Yoshihara M, Haruma K, Kajiyama G. Expression of antral gastrin and somatostatin mRNA in Helicobacter pylori-infected subjects. Am J Gastroenterol. 1994;89:1515–1519. [PubMed] [Google Scholar]
- 45.Torres A, El-Ebiary M, Soler N, Monton C, Fabregas N, Hernandez C. Stomach as a source of colonization of the respiratory tract during mechanical ventilation: association with ventilator-associated pneumonia. Eur Respir J. 1996;9:1729–1735. doi: 10.1183/09031936.96.09081729. [DOI] [PubMed] [Google Scholar]
- 46.Walsh J H, Grossman M I. Gastrin. N Engl J Med. 1975;292(Part 2):1377–1384. doi: 10.1056/NEJM197506262922605. [DOI] [PubMed] [Google Scholar]
- 47.Wang T C, Dangler C A, Chen D, Goldenring J R, Koh T, Raychowdhury R, Coffey R J, Ito S, Varro A, Dockray G J, Fox J G. Synergistic interaction between hypergastrinemia and Helicobacter infection in a mouse model of gastric cancer. Gastroenterology. 2000;118:36–47. doi: 10.1016/s0016-5085(00)70412-4. [DOI] [PubMed] [Google Scholar]
- 48.Wexler H M, Getty C, Fisher G. The isolation and characterisation of a major outer-membrane protein from Bacteroides distasonis. J Med Microbiol. 1992;37:165–175. doi: 10.1099/00222615-37-3-165. [DOI] [PubMed] [Google Scholar]
- 49.Yang Y P, Myers L E, McGuinness U, Chong P, Kwok Y, Klein M H, Harkness R E. The major outer membrane protein, CD, extracted from Moraxella (Branhamella) catarrhalis is a potential vaccine antigen that induces bactericidal antibodies. FEMS Immunol Med Microbiol. 1997;17:187–199. doi: 10.1111/j.1574-695X.1997.tb01012.x. [DOI] [PubMed] [Google Scholar]
- 50.Zhang Z F, Kurtz R C, Klimstra D S, Yu G P, Sun M, Harlap S, Marshall J R. Helicobacter pylori infection on the risk of stomach cancer and chronic atrophic gastritis. Cancer Detect Prev. 1999;23:357–367. doi: 10.1046/j.1525-1500.1999.99041.x. [DOI] [PubMed] [Google Scholar]