Abstract
Protein based therapeutics have the potential to treat a variety of diseases, however, safe and effective methods for delivering them into cells need to be developed before their clinical potential can be realized. Peptide fusions have great potential for improving intracellular delivery of proteins. However, very few peptides have been identified that can increase the intracellular delivery of proteins, and new peptides that can enhance intracellular protein delivery are greatly needed. In this report we demonstrate that the coiled-coil forming peptide (KVSALKE)5 (termed K5) can function as a cell penetrating peptide (CPP), and can also complex other proteins that contain its partner peptide E5. We show here that GFP and Cas9 fused to the K5 peptide had dramatically enhanced cell uptake in a variety of cell lines, and was able to edit neurons and astrocytes in the striatum and hippocampus of mice after a direct intracranial injection. Collectively, these studies demonstrate that the coiled-coil forming peptide (KVSALKE)5 is a new class of multifunctional CPPs that has great potential for improving the delivery of proteins into cells and in vivo.
Keywords: Cas9, gene editing, neuroscience, protein fusions, cell penetrating peptides, non-viral delivery
Graphical Abstract
Coiled-coil forming peptide (KVSALKE)5 can function as a cell penetrating peptide (CPP), and can also complex other proteins that contain its partner peptide E5. K5 peptide fused GFP and Cas9 dramatically enhance cell uptake in a variety of cell lines, and enables in vivo genome editing in the mouse brain after a direct intracranial injection.
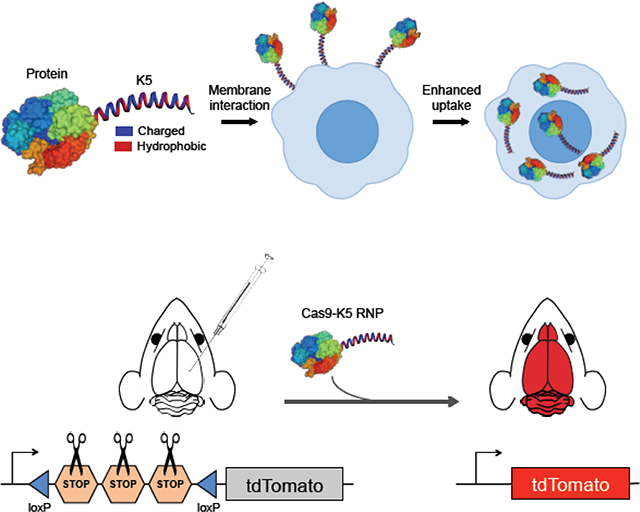
1. Introduction
Protein based therapeutics have the potential to treat a variety of diseases, such as neurodegenerative diseases, cancer, genetic diseases and numerous others1–5. However, before the tremendous potential of protein therapeutics can be realized, safe and effective methods for delivering them into cells need to be developed. Developing effective intracellular protein delivery vehicles has been challenging. Proteins are in general intolerant to chemical modifications, easily denature, and also do not complex cationic materials as readily as nucleic acids6. A wide variety of protein delivery strategies are currently being considered, based upon gold nanoparticles, block copolymers, cationic lipids and cell penetrating peptides, which have shown promising results in cell culture and animal studies7–9. However, it is unclear if these delivery vectors will have the properties needed to succeed in human clinical trials, given their ill-defined nature, large size and toxicity. The development of safe and effective protein delivery vectors is still an unsolved problem.
Intracellular protein delivery strategies based upon fusing short peptides (termed cell penetrating peptides) to proteins have tremendous potential for enhancing the intracellular delivery of proteins, and also have the properties needed to succeed in human clinical trials10, 11. In particular, protein fusions are a single molecular species and can be easily synthesized and characterized on a large scale. Protein fusions also have a much smaller molecular weight than nanoparticles and will be able to access tissues and cells that nanoparticles cannot. Finally, protein fusions should have lower toxicity than cationic nanoparticles because of their lower charge density, and will also not activate toll-like receptor (TLR) ligands, as cationic lipids do12, 13.
Pioneering studies from the Dowdy laboratory demonstrated that short cationic peptide sequences 10–20 amino acids in length derived from the TAT protein were able to deliver proteins such as CRE, beta-galactosidase and GFP into a variety of cell types and even in vivo14–18. TAT appears to enhance cell uptake via its cationic charge density, which causes binding with cellular proteoglycans. The efficiency of TAT was further improved by fusing it to an endosomal disruptive peptide from the Influenza virus, HA, to generate a fusion peptide that could stimulate both endocytosis and endosomal disruption15, 16. Based upon these studies a variety of other cell penetrating peptides have been explored. For example, the gene editing enzymes, Cas9 and zinc finger nucleases, fused to the SV40 NLS signals, were able to directly edit cells in vitro and vivo17, 19. In addition to TAT, a variety of other peptides for enhancing intracellular protein delivery have been developed, of which the peptide GALA is perhaps the most well studied20, 21. The peptide GALA is a pH sensitive membrane disruptive peptide, which forms amphiphilic helices at pH 5.0, and can enhance the endosomal release of proteins. GALA has been fused to antibodies and the ribosome inactivating protein HBP, and was able to dramatically improve the intracellular delivery of these proteins.
Despite these encouraging results, there is still a great need for improving the efficiency of cell penetrating peptides (CPPs) fused to proteins. Current delivery peptides used in protein fusions are frequently positively charged peptides and appear to function by increasing the positive charge density of their target protein22–24. However, making cell delivery dependent upon charge density is challenging because every protein has its own unique charge density, and will requires its own unique peptide fused to it. Related to this issue is the fact that CPPs have only been examined on a moderate number of proteins, and it is unclear if they have the versatility needed to act as universal protein delivery reagents. For example, GFP has been the most widely studied protein fused to CPPs, and here large enhancements in cell delivery are observed after fusion with the positively charged peptides such as the TAT peptide25. However, fusing the TAT peptide to a larger protein such as Cas9 results in negligible enhancements in uptake26. Developing CPPs that can enhance the delivery of larger functional proteins has been a major challenge because they will most likely require fusion to 40–50 amino acid peptide segments, to compensate for their large size and charge density. However, current CPPs are unstructured peptides and most proteins will not tolerate fusion to large unstructured peptides27. Thus, expanding the arsenal of CPPs will consequently have numerous applications in biotechnology.
Peptides that form coiled-coil motifs are potential candidates for cell penetrating peptides that can enhance the delivery of proteins. Coiled-coil peptides form amphiphatic alpha helices that self-assemble with partner peptides due to the complementary nature of their hydrophobic and charged domains28. Coiled-coil forming peptides also have the potential to interact strongly with cell membranes because they generate amphiphatic alpha helices, with hydrophobic and charged surfaces, which in general bind membranes with much higher affinity than un-structured peptides with a similar hydrophobic-hydrophilic balance29–31. For example, Rabe et al. demonstrated that the coiled-coil peptide (KIAALKE)3 could bind lipid bilayers with a Kd of 10−9M, and could also enhance lipid bilayer fusion and facilitate drug release from liposomal vectors intracellularly32–34. Coiled-coil forming peptides have the potential to act as cell penetrating peptides (CPPs), given their ability to bind cell membranes tightly.
Using coiled-coil peptides as CPPs could have many potential benefits over existing CPPs commonly fused to proteins. For example, coiled-coil peptides have a well-defined structure in contrast to CPPs such as TAT and SV40 NLS, which appear to be random coil peptides35. Fusing structureless peptides to proteins can be problematic because it increases the chances of generating an unfoldable protein. In addition, current CPPs are primarily composed of cationic peptides such as Argn, which bind membranes through electrostatic interactions, and do not insert into the hydrophobic core of the membrane36, 37. In contrast, coiled-coil peptides contain hydrophobic surfaces and can bind and insert into the hydrophobic core of membranes31, potentially leading to efficient endosomal disruption. Finally, proteins fused to coiled-coil peptides will also be able to complex other proteins or particles, which could enable cell targeting and endosomal disruption functions38, 39. However, despite their potential, the ability of coiled-coil peptides to enhance the delivery of proteins has never been investigated.
In this report, we demonstrate that that the coiled-coil forming peptide (KVSALKE)5 (termed K5) can function as a cell penetrating peptide, and can also complex proteins that contain its partner peptide E5 (see Figure 1). We show here that GFP and Cas9 fused to the K5 peptide had dramatically enhanced cell uptake in a variety of cell lines, and that Cas9-K5 was able to edit brain tissue in neurons and astrocytes of striatum and hippocampus after a direct intracranial injection into adult mouse brains. Collectively, these studies demonstrate that the coiled-coil forming peptide (KVSALKE)5 is a new class of multifunctional CPPs that has great potential for improving the delivery of proteins into cells and in vivo.
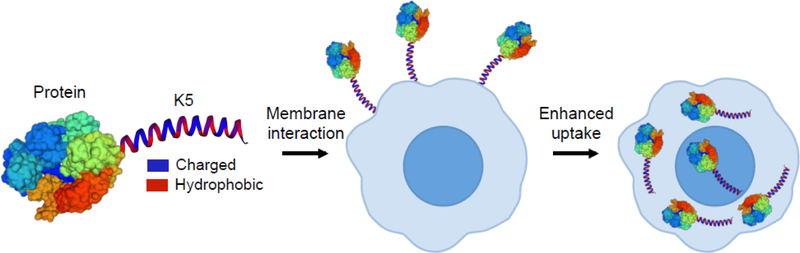
Figure 1. The coiled-coil forming peptide (KVSALKE)5 (K5) is a cell penetrating peptide and can enhance the delivery of proteins into cells.
The peptide (KVSALKE)5 (K5) is fused to proteins and enhances cell uptake via its binding with cell membranes.
2. Results and discussion
The K5 peptide enhances the delivery of EGFP into various cell types
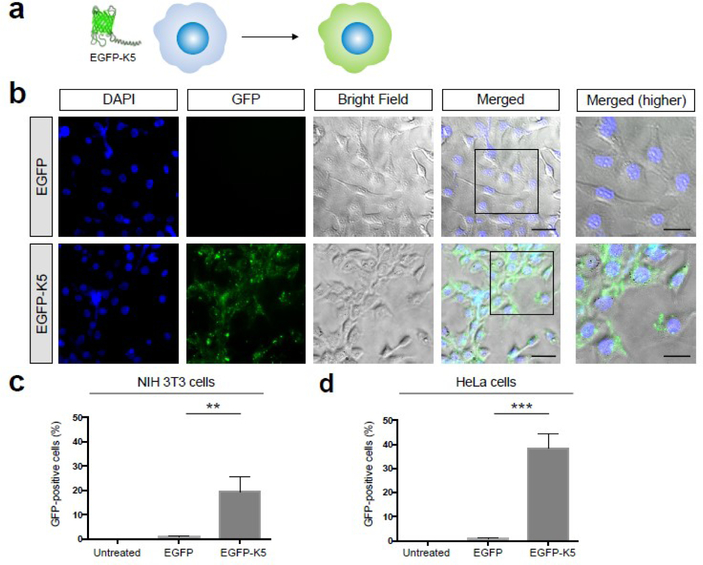
We performed experiments to determine if the coiled-coil forming peptide K5 could act as a CPP and enhance the delivery of EGFP into cells (see Figure 2a). We selected EGFP as a model protein because of its straightforward fluorescent readout, and because it is widely used as a reporter protein for investigating the efficacy of CPPs, and would allow us to compare K5 with other CPPs reported in the literature. K5 was fused to EGFP at its N-terminal and expressed in high yields in E.coli (10 mg/L). K5 was fused to the N-terminus of Cas9 because this site tolerates fusion proteins well, and several Cas9 fusions at the N-terminus have already been made, which were enzymatically active. K5-EGFP was incubated with NIH 3T3 and Hela cells at 166 μg/mL for 4 hours in the presence of 10% FBS, the cells were washed and then imaged via fluorescent microscopy. As a control, EGFP only was incubated with NIH 3T3 and Hela cells and imaged. Figure 2b demonstrates that K5 significantly enhances the delivery of EGFP into NIH 3T3 cells. Notably, cells treated with EGFP had minimal levels of fluorescence, whereas cells treated with EGFP-K5 had high levels of green fluorescence, which appeared to have distributed throughout the cell. We performed flow cytometry experiments to further quantify the increase in fluorescence observed with K5. Figures 2c and 2d demonstrates that K5 dramatically increased the number of GFP positive cells in both NIH 3T3 and HeLa cells. NIH 3T3 and HeLa cells treated with GFP were only 1–2% GFP positive, whereas cells treated with GFP-K5 were 20 and 40% GFP positive respectively (see Figures S1 and S2 for detailed flow cytometry histograms). In addition, there was no sign of toxicity in cells treated with GFP-K5 and these experiments demonstrate that K5 can dramatically increase the uptake of small proteins into cells.
Figure 2. K5 enhances the delivery of EGFP into various cell types.
(a) Schematic illustration of enhanced cellular uptake of EGFP by K5. (b) K5 facilitates the internalization of EGFP into NIH 3T3 cells. Higher magnifications of the merged images (black boxes) are shown in the right panel. Scale bars represent 50 μm (left panels) and 30 μm (right panels), respectively. (c-d) Quantification of flow cytometry data of EGFP or EGFP-K5 uptake in (c) NIH 3T3 cells and (d) HeLa cells. Mean ± SEM. ** P < 0.01, *** P < 0.001 by permutation t-test. P values were calculated between EGFP and EGFP-K5.
Cas9 fused to K5 is enzymatically active
Although CPPs have been intensely investigated for enhancing the delivery of GFP, there are very few studies investigating their ability to deliver functional proteins, and their results with proteins outside of GFP have been mixed. For example fusion of TAT to the Cas9 RNP results in only moderate increases in cell delivery, and requires co-formulation with additional TAT peptides for delivery into cells26. Fusion of 7 SV40-NLS sequences to Cas9 was able to enhance delivery into the brain19, and these results demonstrate that CPPs can enhance the delivery of even large proteins. However, outside of the SV40-NLS and TAT, the ability of other CPPs to enhance the delivery of the Cas9 RNP into cells and in vivo has not been investigated, and could improve numerous aspects of this delivery strategy, in particular, protein yield, editing efficiency and cell tropism. There is consequently great in exploring other types of CPPs that can enhance the delivery of the Cas9 RNP.
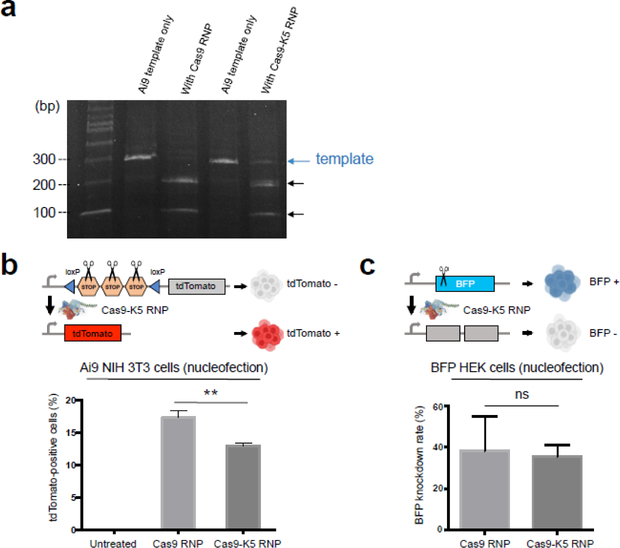
In our study, we investigated if the K5 peptide could enhance the delivery of the Cas9 RNP into cells and further in vivo. The K5 peptide was fused to the N-terminus of the Cas9 protein, and expressed in E.coli, with a yield of 10 mg/L (see Figure S3 for gel characterization). The nuclease activity of Cas9-K5 RNP was compared against Cas9 RNP (native Cas9 RNP), with regards to their ability to cleave a PCR amplicon. A total of 500 ng of Cas9-K5 RNP or Cas9 RNP were mixed with 500 ng of template DNA (1:1 mass ratio), and the cutting efficiency was determined by gel electrophoresis. Figure 3a demonstrates that although the Cas9-K5 RNP does have enzymatic activity, its ability to cleave DNA is lower than Cas9 RNP. For example, under these conditions, the Cas9 RNP cleaved 100% of the template DNA, whereas the Cas9-K5 RNP cleaved only 71%. The Cas9 RNP also had a higher level of enzymatic activity then the Cas9-K5 RNP, when mixed with template DNA at a 0.5:1 mass ratio and a 2:1 mass ratio (see Figure S4). The K5 peptide could be interfering with the activity of Cas9 in multiple ways, given the numerous conformational changes that the Cas9 protein undergoes during its catalytic cycle.
Figure 3. Cas9 fused to K5 is enzymatically active.
(a) In vitro Ai9 template DNA cleavage assay demonstrates that the Cas9-K5 RNP has 71% of the nuclease activity of the native Cas9 RNP (Cas9 RNP). (b-c) Quantification of flow cytometry data of (b) tdTomato-positive cells in Ai9 NIH 3T3 cells and (c) BFP knockdown rate in BFP HEK cells by Cas9 RNP or Cas9-K5 RNP delivered by nucleofection. Mean ± SEM. ** P < 0.01, ns P > 0.05 by permutation t-test. P values were calculated between Cas9 RNP and Cas9-K5 RNP.
Next, the ability of the Cas9-K5 RNP to edit cells after nucleofection was investigated and compared against the Cas9 RNP, using either Ai9 NIH 3T3 cells or BFP-HEK cells. Ai9 NIH 3T3 cells have a loxP flanked STOP cassette (i.e. three repeated SV40 polyA signal sequences) upstream of a tdTomato transgene that is integrated into their genome, and will generate red fluorescence after gene editing and removal of the STOP cassette40. The Cas9-K5 RNP and Cas9 RNP had similar levels of editing in cells after electroporation. The Cas9-K5 RNP edited 14% of Ai9 3T3 fibroblasts which was 77% of the editing rate of the Cas9 RNP (see Figure 3b; see Figure S5 for detailed flow cytometry histograms). We further tested the Cas9-K5 RNP activity in BFP HEK cells. Gene editing efficiency was calculated by subtracting the percentage of BFP-negative cells in the untreated cells from the percentage of BFP-negative cells in the treated cells. BFP HEK cells treated with Cas9-K5 RNP had a similar BFP knockdown rate to cells treated with Cas9 RNP targeting the BFP gene (see Figure 3c; see Figure S6 for detailed flow cytometry histograms). These results demonstrate that the Cas9-K5 RNP has sufficient enzymatic activity for gene editing in cells.
K5 peptide enhances the delivery of Cas9 RNP into various cell type
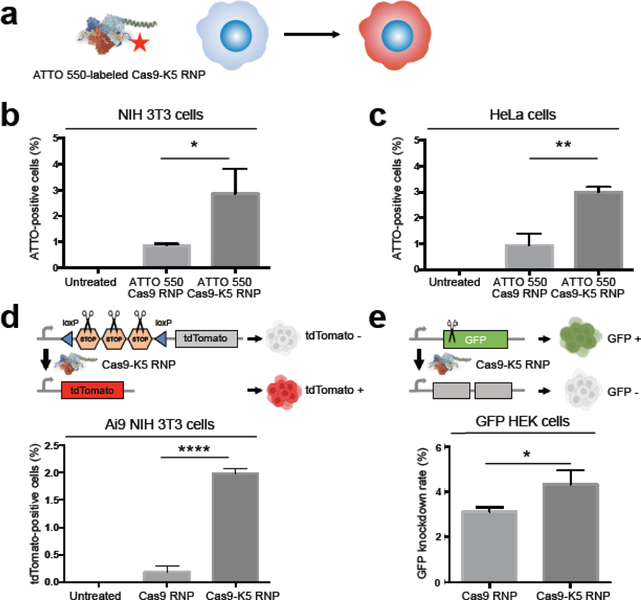
To further explore whether K5-fused Cas9 could enhance gene editing efficiency, we first explored the internalization of the Cas9-K5 RNP in various cell types. ATTO 550-labeled sgRNA was assembled with Cas9 or Cas9-K5 to form fluorescent Cas9 RNP or Cas9-K5 RNP and their uptake efficiency was measured by flow cytometry (see Figure 4a). The K-5 peptide causes a significant enhancement of cell internalization of the Cas9 RNP in NIH 3T3 cells, even in the presence of serum proteins. For example, incubating NIH 3T3 cells with Cas9-K5 RNP in DMEM medium with 10% FBS resulted in more than 3% of cells being ATTO 550-positive, while NIH 3T3 cells incubated with Cas9 RNP exhibited less than a 1% positive rate, leading to approximately a 3 fold increase in uptake (see Figure 4b; see Figure S7 for detailed flow cytometry histograms). A similar trend was confirmed in HeLa cells, where cells treated with Cas9-K5 RNP also showed more than a 3% ATTO 550-positive rate, while Cas9 RNP-treated HeLa cells were less than 1% ATTO 550-positive (see Figure 4c; see Figure S8 for detailed flow cytometry histograms).
Figure 4. The K5 peptide enhances the delivery of the Cas9 RNP into various cell types.
(a) Schematic illustration of enhanced uptake of ATTO 550-labeled Cas9 RNP by the K5 peptide in cells. (b-c) K5 enhanced Cas9 RNP uptake of ATTO 550-labeled Cas9 RNP: Quantification of flow cytometry data of ATTO 550-positive cells in (b) NIH 3T3 cells and (c) HeLa cells using ATTO 550-labeled Cas9 RNP or ATTO 550-labeled Cas9-K5 RNP. (d-e) The K5 peptide enhanced the gene editing efficiency of the Cas9 RNP: Quantification of flow cytometry data of (d) tdTomato-positive cells in Ai9 NIH 3T3 cells and (e) GFP knockdown rate in GFP HEK cells by Cas9 RNP or Cas9-K5 RNP. Mean ± SEM. * P < 0.05, ** P < 0.01, **** P < 0.0001 by permutation t-test. P values were calculated between Cas9 RNP and Cas9-K5 RNP.
The K5 peptide enhances the gene editing efficiency in Ai9 3T3 cells and GFP HEK cells
We then investigated if the K5 peptide could enhance Cas9 RNP editing in vitro. Ai9 NIH 3T3 cells were used as model cells to measure the tdTomato-positive rate to determine the Cas9 RNP editing efficiency. We found that Ai9 NIH 3T3 cells treated with Cas9 RNP (166 μg/mL) were merely 0.183% tdTomato-positive, whereas Cas9-K5 RNP-treated cells (166 μg/mL) were 1.98% tdTomato-positive, which represents approximately a 10 fold increase in editing efficiency (see Figure 4d; see Figure S9 for detailed flow cytometry histograms). We further confirmed the enhanced editing in GFP-expressing HEK cells, using a sgRNA designed to knock out the GFP gene via indel mutations. Gene editing efficiency was calculated by subtracting the percentage of GFP-negative cells in the untreated cells from the percentage of GFP-negative cells in the treated cells. Figure 4e demonstrates that the K5 peptide also improved the delivery of the Cas9 RNP in GFP HEK cells. GFP HEK cells treated with the Cas9 RNP (166 μg/mL) were 3.12% GFP-negative, whereas GFP HEK cells treated with the Cas9-K5 RNP (166 μg/mL) were 4.33% GFP-negative, a 39% increase in editing efficiency (see Figure S10 for detailed flow cytometry histograms). These results show that the K5 peptide can enhance the intracellular delivery of gene editing enzymes.
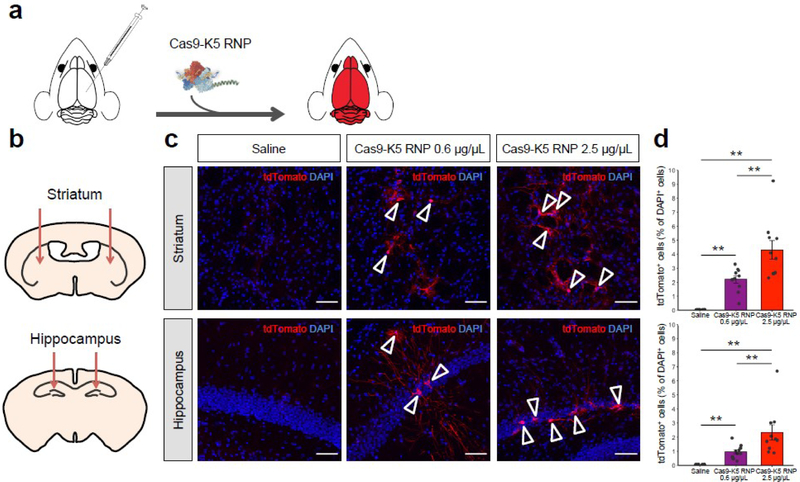
The Cas9-K5 RNP can edit genes in the striatum and hippocampus of Ai9 mice
Given that our results demonstrate that K5 fusion to Cas9 RNP leads to enhanced gene editing in various cell types (see Figures 4d and 4e), we further tested the gene editing efficiency of the Cas9-K5 RNP in vivo. To test our idea of whether Cas9-K5 RNP could deliver and edit genes in vivo, we stereotaxically injected Cas9-K5 RNP into the brains of adult Ai9 mice (see Figure 5a). The Ai9 mouse is a genetically engineered mouse model, which has a fluorescent tdTomato gene containing loxP flanked STOP cassettes upstream of it41. As mentioned earlier, the deletion of the STOP sequences allows transcription of the tdTomato gene, resulting in fluorescence expression (see Figure 5a). The sgRNA used for these experiments was designed to remove the STOP sequences, and was verified in Ai9 NIH 3T3 cells (see Figure 4d). Cas9 RNP or Cas9-K5 RNP or saline were stereotaxically injected into two brain regions, the hippocampus and the striatum, of 2–4 month old adult Ai9 mice as shown in Figure 5b, and the expression of tdTomato was measured via fluorescence histology. The gene editing efficiency was determined by detecting tdTomato-positive (tdTomato+) cells normalized by total cell number (DAPI+). Figures 5c and 5d demonstrate that Cas9-K5 RNP can efficiently edit cell genomes after stereotaxic intracerebral injection in both striatum and hippocampus, while control (saline-treated brain hemisphere) did not show edited cells. We treated two dosages (0.6 and 2.5 μg/uL) of Cas9-K5 RNP; The efficiency was significantly increased by higher dosage injection (2.5 μg/uL) compared to lower dosage injection (0.6 μg/uL), approximately 2 fold both in striatum (~2% to ~4%) and hippocampus (~1% to ~2.5%) respectively. These results indicate that Cas9-K5 RNP edited brain cells and that the gene editing efficiency was dose-dependently increased. Notably, when we counted the total number of cells (DAPI+) by DAPI nuclear staining, we did not see a significant difference in total DAPI-positive cell numbers, in comparison to the saline-treated group, in either the low or high dosages, demonstrating the Cas9-K5 has no significant impact on cell numbers (see Figure S11).
Figure 5. The Cas9-K5 RNP can edit the brains of Ai9 mice after an intracranial injection.
(a) Cas9 is fused to K5 and injected into the brains of Ai9 mice, and was able to edit neurons and astrocytes in the brain after an intracranial injection. (b) Schematic showing the injection site for the Cas9-K5 RNP, in either the striatum or hippocampus of adult Ai9 mice. Saline or Cas9-K5 RNP (0.6 μg/μL or 2.5 μg/μL) was injected into the striatum (AP: 0.50 mm, ML: ±1.87 mm, DV: −3.47 mm) or the hippocampus (AP: −2.56 mm, ML: ±1.55 mm, DV: −2.00 mm) of Ai9 mice. (c) tdTomato (red) immunostaining and DAPI (blue) nucelear staining were performed 21 days after the saline or Cas9-K5 RNP (0.6 μg/μL or 2.5 μg/μL) injection. Scale bar represents 50 μm. (d) Quantification of the percentage of tdTomato+ cells among DAPI+ cells in the striatum (permutation one-way ANOVA: F(2,27) = 27.03, P < 0.001) and hippocampus (permutation one-way ANOVA: F(2,27) = 12.93, P < 0.001). Mean ± SEM, n = 10, ** P < 0.01 by post hoc permutation t-test. Post hoc P values were calculated between control (saline) and Cas9-K5 RNP 0.6 μg/μL and 2.5 μg/μL, respectively, or Cas9-K5 RNP 0.6 μg/μL and Cas9-K5 RNP 2.5 μg/μL.
The K5 peptide edits neurons and astrocytes in the striatum and hippocampus of Ai9 mice in vivo
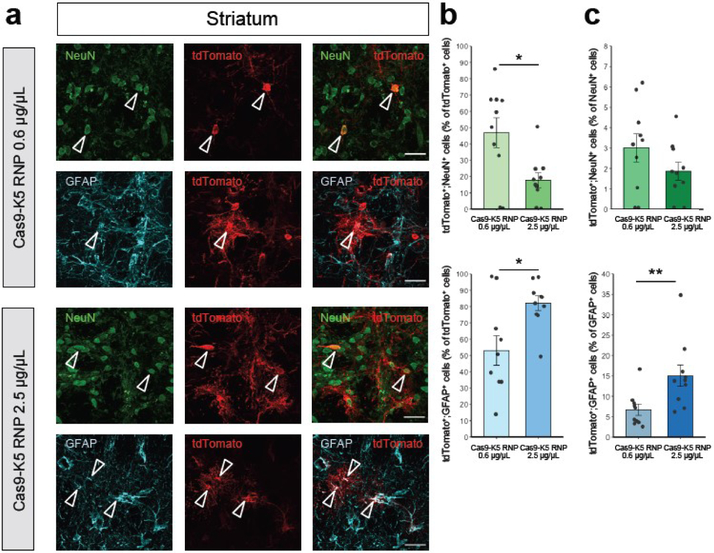
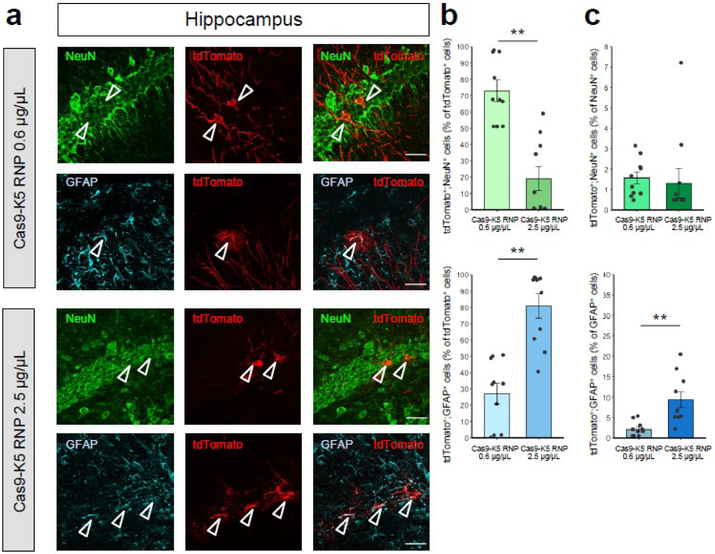
Whereas neurons are the primary working units of the brain, non-neuronal cells also play important roles in brain function by maintaining, supporting, and regulating neuronal functions. There is great interest in editing the genes of both neuronal and glial cells, since along with neurons, glial cell dysfunction causes multiple brain disorders42. We further identified the brain cell types edited by Cas9-K5 RNP to determine the editing efficiency in both neurons and astrocytes in Ai9 mouse brains. Cas9-K5 RNP-injected Ai9 brain sections were stained with the following cell markers: neuronal nuclear protein (NeuN) and glial fibrillary acidic protein (GFAP) which identify the neuronal populations and astrocytes respectively in the histology sections. The co-localization of tdTomato-positive (tdTomato+) cells with NeuN-positive (NeuN+) or GFAP-positive (GFAP+) cells demonstrates that approximately 47% and 53% among all edited cell populations (% of tdTomato+ cell) are neurons and astrocytes in the striatum (see Figure 6b; see Figure 6a for representative images) and approximately 72% and 28% in the hippocampus (see Figure 7b; see Figure 7a for representative images) when we used lower dosage injection (0.6 μg/uL) of Cas9-K5 RNP. Notably, the % astrocytes among edited cells increased by a higher dosage (2.5 μg/uL) injection of Cas9-K5 RNP compared to lower dosage (0.6 μg/uL) approximately 1.5 fold in striatum (~53% to ~82%) and approximately 2.5 fold in hippocampus (~28% to ~80%), while the % of neurons among edited cells decreased in striatum (~47% to ~18%) and in hippocampus (~72% to ~20%) (see Figure 6b for striatum and Figure 7b for hippocampus).
Figure 6. The cell specificity of Cas9-K5 RNP edited cells in the striatum of Ai9 mice.
(a) Immunostaining of tdTomato+ (red) and either NeuN+ (green) or GFAP+ (cyan) cells 21 days after stereotaxic injection of Cas9 K5 RNP. Scale bar represents 30 μm. (b-c) Quantification of (b) tdTomato+;NeuN+ or tdTomato+;GFAP+ cells among total tdTomatol+ cells (%); (c) tdTomat°+;NeuN+ or tdTomat°+;GFAP+ cells among total NeuN+ or GFAP+ cells (%) respectively in the Cas9-K5 RNP-injected area, mean ± SEM, n = 10. * P < 0.05, ** P < 0.01 by permutation t-test. P values were calculated between Cas9-K5 RNP 0.6 μg/μL and Cas9-K5 RNP 2.5 μg/μL.
Figure 7. The cell specificity of Cas9-K5 RNP edited cells in the hippocampus of Ai9 mice.
(a) Immunostaining of tdTomato+ (red) and either NeuN+ (green) or GFAP+ (cyan) cells 21 days after stereotaxic injection of Cas9 K5 RNP. Scale car represents 30 μm. (b-c) Quantification of (b) tdTomato+;NeuN+ or tdTomato+;GFAP+ cells among total tdTomato+ cells (%); (c) tdTomato+;NeuN+ or tdTomato+;GFAP+ cells among total NeuN+ or GFAP+ cells (%) respectively in the Cas9-K5 RNP-injected area, mean ± SEM, n = 10. ** P < 0.01 by permutation t-test. P values were calculated between Cas9-K5 RNP 0.6 μg/μL and Cas9-K5 RNP 2.5 μg/μL.
Next, we further determined the gene editing efficiency in each cell type in neuron and astrocyte by analyzing the edited neurons (tdTomato+;NeuN+) among neuronal populations (NeuN+) and edited astrocytes (tdTomato+;GFAP+) among astrocytes populations (GFAP+). As a result, we found that gene editing efficiency in neurons ranges from ~1.3% to ~3% in striatum and hippocampus, and did not show any significant increased gene editing efficiency by higher dosage (see the upper graphs of Figure 6c for striatum and Figure 7c for hippocampus; see Figure S12 for non-normalized raw values). Notably, the gene editing efficiency in astrocytes increased significantly approximately 2.1 fold in striatum (~7% to ~15%) and approximately 4.5 fold in hippocampus (~2% to ~9%) by a higher dosage injection of Cas9-K5 RNP compared to lower dosage (see the lower graphs of Figure 6c for striatum and Figure 7c for hippocampus; see Figure S12 for non-normalized raw values). There was a minor increase (less than 1.5 fold) of total astrocyte number by higher dosage treatment of Cas9-K5 RNP both in striatum and hippocampus while no significant total neuron number change was observed (see Figure S13).
Altogether, our results implicate that higher dosage of Cas9-K5 RNP may provide advantages when targeting astrocytes. On the other hand, neuronal gene editing efficiency was capped or saturated and did not significantly benefit by a higher dosage of Cas9-K5 RNP. This information may enable strategies for targeting brain cell types in the future with Cas9-K5. Gene editing in the brain with Cas9-K5 RNP was significantly lower in efficiency than gene editing with AAV, which usually generates 20–30% cell infection rates in the brain43. However, the Cas9-K5 RNP can be treated into patients multiple times, whereas AAV based therapies cannot, and this may compensate for their lower efficiencies.
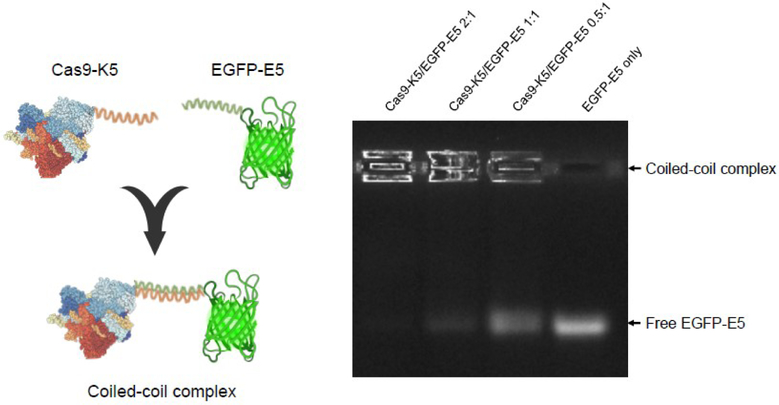
Cas9-K5 can complex proteins that contain its E5 counterpart
Hetero- or homo-oligomerization of proteins by coiled-coil interactions has shown great potential for use in biotechnology, bioengineering, and cell-based therapeutic applications44. We therefore further investigated if our K5-fused protein is capable of complexing proteins that contained its E5 counterpart (EVSALEK)5. We fused the E5 peptide to EGFP and mixed it with Cas9-K5, to investigate if Cas9-K5 could complex other proteins (see Figure 8). As shown by agarose gel electrophoresis in Figure 8, free EGFP-E5 migrates through the agarose gel, due to its negative charge, and can be directly visualized via its fluorescence. In contrast, EGFP-E5 mixed with Cas9-K5 has a very different gel migration pattern, and the mobility of the EGFP-E5 was retarded after complexation with Cas9-K5. In addition, the inhibitory effect of the Cas9-K5 was dose-dependent and high Cas9-K5 to EGFP-E5 molar ratios, completely inhibited EGFP-E5 from migrating into the gel. Additional control experiments were also performed to investigate if the E5 and K5 sequences were essential for mediating complexation between Cas9 and EGFP. Figure S14 demonstrates that complexation between Cas9 and EGFP was dependent upon the E5 and K5 sequences. These results demonstrate the great potential of K5/E5 interactions to direct the formation of multicomponent complexes of precise defined stoichiometry and will allow the introduction of different delivery functionalities into protein complexes.
Figure 8. Cas9-K5 can complex EGFP-E5.
Complexation of EGFP-E5 with Cas9-K5 at different molar ratios. Cas9-K5 is able to complex with EGFP-E5 as shown by agarose gel electrophoresis.
3. Conclusions
The development of highly efficient CPPs that can be fused to proteins remains a central problem in the field of drug delivery. Although a variety of peptides have been used to enhance the delivery of GFP, these peptides have not been investigated for more challenging proteins such as Cas9 and have also rarely been investigated in vivo. Consequently, there is still a great need for developing new CPPs. The peptide (KVSALKE)x and its analogues has been used by numerous laboratories to assemble proteins together via coiled-coil interactions. However, the ability of coiled-coil forming peptides to enhance the delivery of proteins into cells has never been investigated. In this report, we demonstrate that the peptide (KVSALKE)5 could enhance the delivery of either GFP or Cas9 RNP into cells and in vivo. Using coiled-coil forming peptides as CPPs has several advantages. In particular, the K5 peptide forms an amphiphilic alpha-helix, which generates a hydrophobic surface and a cationic surface, which should enable efficient membrane binding and potentially membrane partitioning and endosomal disruption. The K5 peptide was able to enhance the delivery of the Cas9 RNP into Ai9 fibroblasts and also was effective at editing a variety of cells in the brain after a direct intracranial injection. The K5 peptide did lower the activity of the Cas9 RNP, and appears to be the largest repeat tolerated by the Cas9 RNP. For example, Cas9 fused to (KVSALKE)7 and (KVSALKE)9 both had minimal amounts of enzymatic activity (see Figure S15). We anticipate numerous applications of the K5 peptide given its ability to enhance cell uptake, broad cell tropism and ability to complex other proteins.
4. Experimental Section/Methods
Materials.
Oligonucleotides were purchased from Integrated DNA Technologies (IDT, Coralville, IA). Phusion High-Fidelity DNA Polymerase was purchased from NEB (Ipswich, MA). Mini-PROTEAN TGX Gels (4–20%) were purchased from Bio-Rad (Hercules, CA). Cas9 proteins were purchased from Macrolab UC Berkeley. BFP HEK cells and GFP HEK cells were a generous gift from GenEdit45. NIH 3T3 cells or HeLa cells were obtained from UC Berkeley Cell Culture Facility. Ai9 NIH 3T3 cells were a generous gift from Craig Duvall's lab40. Paraform aldehyde (PFA, cat. #: 41678–0010) and D(+)-Sucrose (cat. #: 177140010) were purchased from Acros Organics; phosphate buffered saline (PBS, cat. #: BP 399–4), sodium citrate (cat. #: BP 327–500), and Tween® 20 (cat. #: 9005–64-5) from Fisher BioReagents; Superfrost Plus Microsope Slides (cat. #: 12–550-15) and Tissue-Plus O.C.T. Compound from Fisher Healthcare; Isoflurane from Vetone; ProLong Gold Antifade Reagent from Molecular Probes; Embedding Molds from Thermo Scientific; Goat Serum from Gibco; DAPI (4’, 6-diamidino-2-phenylindole), DMEM media, non-essential amino acids, penicillin-streptomycin, dPBS, and accutase were purchased from Invitrogen (Carlsbad, CA).
Antibodies.
The rabbit polyclonal RFP antibody was purchased from Rockland (cat. #: 600–401-379); the mouse monoclonal NeuN antibody (cat. #: MAB377) from Millipore (Burlington, MA), and the chicken polyclonal GFAP antibody from EnCor (cat. #: CPCA-GFAP) The donkey anti-rabbit IgG-Cy™3 (cat. #: 711–165-152), donkey anti-mouse IgG-AlexaFluor®647(cat. #: 715–605-151), and donkey anti-chicken IgY-Cy™2 (cat. #: 703–225-155) were purchased from Jackson ImmunoResearch Laboratories, Inc. (West Grove, PA).
Animal care and use.
Ai9 mice (in C57BL/6J background) were obtained from Jackson Laboratory (stock #007909). The use and care of animals in this study follow the guidelines of the UTHSCSA and UC Berkeley Institutional Animal Care and Use Committee.
Protein expression.
(KVSALKE)5 (K5), (KVSALKE)7 (K7) or (KVSALKE)9 (K9) fused Streptococcus pyogenes Cas9 (Cas9-K5, Cas9-K7 or Cas9-K9) was expressed from an expression vector that was previously published in the manuscript by Jinek et al46. It was composed of a custom pET-based expression vector encoding an N-terminal 6His-tag followed by the maltose-binding protein (MBP) and a TEV protease cleavage site followed by K5 sequence, as well as two SV40 nuclear localization signal (NLS) peptides at its C-terminus. Recombinant Cas9-K5, Cas9-K7 or Cas9-K9 protein was expressed in the Escherichia coli strain BL21 (DE3)46 (UC Berkeley MacroLab) and further purified to homogeneity as previously described. Purified Cas9 protein was stored in 50 mM HEPES at pH 7.5 with 300 mM NaCl, 10% glycerol, and 100 μM TCEP at −80°C. Cas9-K5, Cas9-K7 or Cas9-K9 protein concentration was determined by a NanoDrop 2000 (Thermo Scientific) from the absorbance at 280 nm. K5 or E5 (EVSALEK)5 fused EGFP (EGFP-K5 or EGFP-E5) was expressed from pET based vector encoding an N-terminal 6His-tag followed by EGFP and K5 or E5 sequence on the C terminus. Recombinant EGFP-K5 and EGFP-E5 protein was expressed in Escherichia coli strain BL21 (DE3) (UC Berkeley MacroLab) and further purified by HisTrap HP (Cytiva). Cas9 RNP formation was conducted by mixing Cas9, Cas9-K5, Cas9-K7 or Cas9-K9 protein with sgRNA at a molar ratio of 1:1.2 and incubated on ice for 15 min before use. sgRNA sequences used in this paper are as below:
BFP and GFP sgRNA targeting sequence: GCTGAAGCACTGCACGCCAT
Ai9 sgRNA targeting sequence: AAGTAAAACCTCTACAAATG
For coiled-coil complexation studies, a 1 mg/mL solution of Cas9-K5 was mixed with a 1 mg/mL solution of EGFP-E5 at different molar ratios (2:1, 1:1, 1:2) and incubated at room temperature for 20 min. The resulting complexes were run on an agarose gel for coiled-coil interaction analysis.
Cell culture.
Ai9 NIH 3T3 cells, NIH 3T3 cells, HeLa cells, BFP HEK cells and GFP HEK cells were cultured in DMEM supplemented with 10% fetal bovine serum (Gibco), and 1% penicillin-streptomycin and maintained at 37 °C in a humidified incubator (Thermo Electron Corporation) containing 5% CO2. When the cells reached 80% confluency, they were passaged using accutase solution; media was changed every 2 – 3 days.
Cell internalization studies.
Studies of intracellular delivery with EGFP and Cas9 RNP were achieved using fluorescence microscopic technique and fluorescence-activated cell sorting (FACS). NIH 3T3 cells or HeLa cells were seeded in 96 well plates at a density of 10K per well the day before exposing to samples. EGFP-K5 and ATTO 550-labeled Cas9-K5 RNP or their native forms were added into the cell cultures. After incubation at 37 °C for 4 hr, the cells were washed three times with PBS and either visualized with a fluorescent microscope EVOS M5000 (Invitrogen) or analyzed via Attune NxT flow cytometer (Invitrogen).
Cas9 RNP nucleofection and Cas9 RNP transfection.
For nucleofection, cells were detached using Accutase, spun down at 600 g for 3 min and washed with PBS. Nucleofection was conducted using a Lonza 4D-Nucleofector system with SE Cell Line 4D-Nucleofector™ X Kit following the manufacturer’s protocol. Briefly, 200K Ai9 NIH 3T3 cells or BFP HEK cells were suspended in 20 μL SE buffer containing Cas9 RNPs (31.25 pmole of Cas9, 37.5 pmole of sgRNAs) and transfected using the EN-158 or CM-130 Lonza program. After nucleofection, cells were spun down at 600g and resuspended in 500 μL fresh DMEM medium supplemented with 10% FBS and 1% penicillin-streptomycin and incubated at 37 °C in tissue culture plates for 5 days before flow cytometry analysis.
For direct transfection studies, 100 μL of Opti-MEM (Gibco) containing Cas9 RNP or Cas9-K5 RNP (16.6 μg Cas9 or Cas9-K5) was added to Ai9 NIH 3T3 cells or GFP HEK cells. The cells were transfected for 48 hr at 37 °C in a humidified incubator containing 5% CO2 and replaced with fresh DMEM medium supplemented with 10% FBS and 1% penicillin-streptomycin. Td-tomato signals or the GFP signals were analyzed by Attune NxT flow cytometer (Invitrogen) after another 24 hr or 72 hr. Gene editing efficiency was determined by the percentage of Td-tomato positive cells or GFP negative cells.
In vitro cleavage assays.
The Ai9 template was PCR amplified (forward: TGCTATACGAAGTTATTCGC reverse: GACAAACCACAACTAGAATG) from synthesized Ai9 DNA (ATGTATGCTATACGAAGTTATTCGCGATGAATAAATGAAAGCTTGCAGAT CTGCGACTCTAGAGGATCTGCGACTCTAGAGGATCATAATCAGCCATACCACATTTGTAG AGGTTTTACTTGCTTTAAAAAACCTCCCACACCTCCCCCTGAACCTGAAACATAAAATGA ATGCAATTGTTGTTGTTAACTTGTTTATTGCAGCTTATAATGGTTACAAATAAAGCAATA GCATCACAAATTTCACAAATAAAGCATTTTTTTCACTGCATTCTAGTTGTGGTTTGTCCAA ACTCATCAATGTATCTTATCATGTCT). For in vitro cleavage assays, 500 ng of template DNA was incubated with 500 ng (or as indicated in each Figure) of Cas9 RNP, Cas9-K5 RNP, Cas9-K7 RNP or Cas9-K9 RNP with sgRNA targeting the Ai9 sequence in 10 μL 1X Cutsmart buffer (NEB) and incubated at 37 °C for 4 hr. Gel electrophoresis was performed to verify cleavage of the template.
Stereotaxic injection.
Male and female Ai9 (Cre-dependent tdTomato reporter) mice aged 2–3 months were used in this study. Mice were anaesthetized using isoflurane (4% for induction and 2% for maintaining) and bilaterally injected with saline (left hemisphere) and Cas9-K5 RNP (right hemisphere) into the striatum (AP: 0.50 mm, ML: ±1.87 mm, DV: −3.47 mm) and hippocampus (AP: −2.56 mm, ML: ±1.55 mm, DV: −2.00 mm). Either saline or Cas9-K5 RNP (final concentrations per injection: 0.6 μg/μL or 2.5 μg/μL) were infused with 1.5 μL per injected site (0.5 μL/min) using a Hamilton Neuros syringe. After the infusion, the injector was left at the injection site for 5 min and then slowly withdrawn. A period of 21 days was given before the perfusion and immunostaining procedures. The use and care of animals in this study followed the guidelines of the UTHSCSA.
Immunostaining.
21 days after stereotaxic injection, the mice were anaesthetized by isoflurane and were perfused with ice-cold PBS followed by 4% PFA in PBS. The brains were post-fixed for 4 hr in 4% PFA, washed once with PBS, and then cryoprotected by 30% sucrose in PBS at 4°C. After cryoprotection, the brains were embedded in O.C.T. compound, frozen, and then stored at −80°C until next processing. Mouse brain sections were obtained by cryostat (CM3050S; Leica Microsystems). Slices with striatum and hippocampus were cryosectioned on the coronal plane at 20 μm, mounted on glass slides, and stored at −20°C. The sections to be immunostained were washed three times in PBS. Antigen retrieval was performed by steaming in a citrate buffer (0.294% sodium citrate, 0.05% Tween 20 in distilled water, pH 6.0) for 15 min with subsequent cooling over ice for 10 min. The sections were rinsed in PBS and blocked (5% goat serum, 0.2% Triton X-100 in PBS) at room temperature for 1 hr. The sections were next incubated in the same blocking solution with primary antibodies (1:500 rabbit anti-RFP, 1:500 mouse anti-NeuN, 1:1000 chicken anti-GFAP) overnight at 4°C. The sections were washed four times in PBS before incubation with secondary antibodies (1:500 Cy™3-conjugated donkey anti-rabbit, 1:500 AlexaFluor®647-conjugated donkey anti-mouse, 1:500 Cy™2-conjugated donkey anti-chicken) for 2 hr. After the incubation with secondary antibodies the sections were washed four times in PBS, and than incubated with DAPI diluted in PBS (1:1500 of 5 μg/μL stock solution) for 10 min. Next the sections were washed three times in PBS and mounted with Prolong Gold Antifade Reagent and imaged using a Zeiss Axio Observer 7 microscope with 20x and 40x objective, respectively and captured with camera Axiocam 503 mono (Carl Zeiss Microscopy GmbH).
Image analysis.
Quantification of gene editing efficiency in Ai9 mice was performed in a defined ROI, which was the same for all animals in striatum and hippocampus, respectively. Each injected brain area was analyzed from 5 images taken as a z-stack (10 μm) with the same magnification (40x) and same exposure times. The total number of tdTomato+, DAPI+, NeuN+, and GFAP+ cells was counted using the Cell counter plugin for ImageJ software (NIH). To determine the percentage of edited cells (tdTomato+) or to present the number of DAPI+ cells in Ai9 mice, tdTomato+ cells were counted and normalized against the number of DAPI+ cells or the number of DAPI+ cells were presented itself (analyzed from a defined ROI, which was the same size for all images analyzed for comparison). To determine the percentage of cell types among edited cells, NeuN+ or GFAP+ cells were counted in only tdTomato+ cells. The percentage of the tdTomato+ cells among the cell types was also analyzed by counting NeuN+ or GFAP+ cells co-stained with tdTomato among the total NeuN+ or GFAP+ cells. Each cell marker was stained with tdTomato and analyzed independently. The percentage of tdTomato+ and total number of DAPI+ cells among saline and Cas9-K5 RNP (0.6 μg/μL and 2.5 μg/μL)-injected brain areas were evaluated. The effect of Cas9-K5 RNP concentration (0.6 μg/μL vs. 2.5 μg/μL) on the cell specificity, total number of edited (tdTomato+) NeuN+ or GFAP+ cells, as well as total number of all edited (tdTomato+) cells in the striatum and hippocampus was also analyzed.
Statistical Analysis.
All in vitro experimental raw data were acquired in at least 3 independent experiments and presented as mean ± SEM. An unpaired two-tailed t-test was performed to evaluate statistical significance between two groups. P < 0.05 was considered significant. The statistical analyses were performed using GraphPad Prism 6. For in vivo experiment two animals (male and female) per group were used. The statistical analyses were conducted with data acquired from a total of 5 images per animal, a total of 10 images per group (n = 10). All data are presented as mean ± SEM. The statistical analysis of multiple-group comparison was performed using one-way permutation ANOVA with maximum of 5,000 permutations. The post-hoc between-group comparisons were evaluated using two-tailed permutation t-test with 10,000 permutations. Significance level was defined as P < 0.05. Statistical analyses were conducted using lmPerm and RVAideMemoire packages for R version 3.6.0.
Supplementary Material
Acknowledgements
This work was supported by the National Institute of Mental Health R01 award (R01MH125979-01) funded to H.Y.L. and N.M. This work was also supported by the UT Rising STARs award, the Simons Foundation Autism Research Initiative (SFARI) pilot award (#574967) funded to H.Y.L, and R61DA048444-01, U24HG010423, 1UM1AI164559, NIH RO1EB023776 and UG3NS115599 funded to N.M.
Footnotes
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
References
- 1.Kintzing JR, Interrante MVF, Cochrane JR, Trends. Pharmacol. Sci. 2016, 37, 993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Usmani SS, Bedi G, Samuel JS, Singh S, Kalra S, Kumar P, Ahuja AA, Sharma M, Gautam A, Raghava GPS, Plos One 2017, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lagasse HA, Alexaki A, Simhadri VL, Katagiri NH, Jankowski W, Sauna ZE, Kimchi-Sarfaty C, F1000Res. 2017, 6, 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caravella J, Lugovskoy A, Curr. Opin. Chem. Biol. 2010, 14, 520. [DOI] [PubMed] [Google Scholar]
- 5.Leader B, Baca QJ, Golan DE, Nat. Rev. Drug Discov. 2008, 7, 21. [DOI] [PubMed] [Google Scholar]
- 6.Di Marco M, Shamsuddin S, Razak KA, Aziz AA, Devaux C, Borghi E, Levy L, Sadun C, Int. J. Nanomed. 2010, 5, 37. [PMC free article] [PubMed] [Google Scholar]
- 7.Ray M, Lee YW, Scaletti F, Yu R, Rotello VM, Nanomedicine (Lond) 2017, 12, 941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stewart MP, Langer R, Jensen KF, Chem. Rev. 2018, 118, 7409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang YM, Roise JJ, Lee K, Li J, Murthy N, Curr. Opin. Biotech. 2018, 52, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gupta B, Levchenko TS, Torchilin VP, Adv. Drug Deliver. Rev. 2005, 57, 637. [DOI] [PubMed] [Google Scholar]
- 11.Chiper M, Niederreither K, Zuber G, Adv. Healthc. Mater. 2018, 7. [DOI] [PubMed] [Google Scholar]
- 12.Lonez C, Vandenbranden M, Ruysschaert JM. Adv. Drug Deliver. Rev. 2012, 64, 1749. [DOI] [PubMed] [Google Scholar]
- 13.Ruysschaert JM, Lonez C, Bba-Biomembranes 2015, 1848, 1860. [DOI] [PubMed] [Google Scholar]
- 14.Fawell S, Seery J, Daikh Y, Moore C, Chen LL, Pepinsky B, Barsoum J, P. Natl. Acad. Sci. USA 1994, 91, 664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barka T, Gresik EW, van der Noen H, J. Histochem. Cytochem. 2000, 48, 1453. [DOI] [PubMed] [Google Scholar]
- 16.Wadia JS, Stan RV, Dowdy SF, Nat. Med. 2004, 10, 310. [DOI] [PubMed] [Google Scholar]
- 17.Gaj T, Guo J, Kato Y, Sirk SJ, Barbas CF 3rd, Nat. Methods 2012, 9, 805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Patel SG, Sayers EJ, He L, Narayan R, Williams TL, Mills EM, Allemann RK, Luk LYP, Jones AT, Tsai YH, Sci Rep 2019, 9, 6298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Staahl BT, Benekareddy M, Coulon-Bainier C, Banfal AA, Floor SN, Sabo JK, Urnes C, Munares GA, Ghosh A, Doudna JA, Nat. Biotechnol. 2017, 35, 431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen YJ, Deng QW, Wang L, Guo XC, Yang JY, Li T, Xu Z, Lee HC, Zhao YJ, Chem Commun (Camb) 2021, 57, 1434. [DOI] [PubMed] [Google Scholar]
- 21.Li C, Cao XW, Zhao J, Wang FJ, J. Membr. Biol. 2020, 253, 139. [DOI] [PubMed] [Google Scholar]
- 22.Lonn P, Dowdy SF, Expert Opin. Drug Del. 2015, 12, 1627. [DOI] [PubMed] [Google Scholar]
- 23.Bemkop-Schnurch A, Adv. Drug Deliver. Rev. 2018, 136, 62. [Google Scholar]
- 24.Cronican JJ, Thompson DB, Beier KT, McNaughton BR, Cepko CL, Liu DR, Acs. Chem. Biol. 2010, 5, 747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nischan N, Herce HD, Natale F, Bohlke N, Budisa N, Cardoso MC, Hackenberger CPR, Angew. Chem. Int. Edit. 2015, 54, 1950. [DOI] [PubMed] [Google Scholar]
- 26.Ramakrishna S, Kwaku Dad AB, Beloor J, Gopalappa R, Lee SK, Kim H, Genome Res. 2014, 24, 1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rosano GL, Ceccarelli EA, Front. Microbiol. 2014, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.De Crescenzo G, Litowski JR, Hodges RS, O'Connor-McCourt MD, Biochemistry-US 2003, 42, 1754. [DOI] [PubMed] [Google Scholar]
- 29.BenTal N, BenShaul A, Nicholls A, Honig B, Biophys. J. 1996, 70, 1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seelig J, Bba-Biomembranes 2004, 1666, 40.15519307 [Google Scholar]
- 31.Nakayama N, Hagiwara K, Ito Y, Ijiro K, Osada Y, Sano K, Langmuir 2015, 31, 8218. [DOI] [PubMed] [Google Scholar]
- 32.Rabe M, Aisenbrey C, Pluhackova K, de Wert V, Boyle AL, Bruggeman DF, Kirsch SA, Bockmann RA, Kros A, Raap J, Bechinger B, Biophys. J. 2016, 111, 2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pahler G, Panse C, Diederichsen U, Janshoff A, Biophys. J. 2012, 103, 2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gillingham AK, Munro S, Bba-Mol. Cell. Res. 2003, 1641, 71. [DOI] [PubMed] [Google Scholar]
- 35.Eiriksdottir E, Konate K, Langel U, Divita G, Deshayes S, Biochim. Biophys. Acta 2010, 1798, 1119. [DOI] [PubMed] [Google Scholar]
- 36.Sauder R, Seelig J, Ziegler A, in Cell-Penetrating Peptides: Methods and Protocols, (ed. Langel U) Humana Press, 2011, [Google Scholar]
- 37.Folska A, Prevette L, Abstr. Pap. Am. Chem. S. 2017, 253. [Google Scholar]
- 38.Bode SA, Kruis IC, Adams HPJHM, Boelens WC, Pruijn GJM, van Hest JCM, Lowik DWPM, Chembiochem 2017, 18, 185. [DOI] [PubMed] [Google Scholar]
- 39.Assal Y, Mizuguchi Y, Mie M, Kobatake E, Bioconjugate Chem. 2015, 26, 1672. [DOI] [PubMed] [Google Scholar]
- 40.Evans BC, Fletcher RB, Kilchrist KV, Dailing EA, Mukalel AJ, Colazo JM, Oliver M, Cheung-Flynn J, Brophy CM, Tierney JW, Isenberg JS, Hankenson KD, Ghimire K, Lander C, Gersbach CA, Duvall CL, Nat. Commun. 2019, 10, 5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, Lein ES, Zeng H, Nat. Neurosci. 2010, 13, 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Almad AA, Maragakis NJ, Stem. Cell Res. Ther. 2012, 3, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hudry E, Vandenberghe LH, Neuron 2019, 101, 839. [DOI] [PubMed] [Google Scholar]
- 44.Lebar T, Lainscek D, Merljak E, Aupic J, Jerala R, Nat. Chem. Biol. 2020, 16, 513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee K, Conboy M, Park HM, Jiang F, Kim HJ, Dewitt MA, Mackley VA, Chang K, Rao A, Skinner C, Shobha T, Mehdipour M, Liu H, Huang WC, Lan F, Bray NL, Li S, Com JE, Kataoka K, Doudna JA, Conboy I, Murthy N, Nat. Biomed. Eng. 2017, 1, 889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E, Science 2012, 337, 816. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.