Abstract
Strengthened DNA repair pathways in tumor cells contribute to the development of resistance to DNA-damaging agents. Consequently, targeting proteins in these pathways is a promising strategy for tumor chemosensitization. Here, we show that the expression of a subset of Fanconi anemia (FA) genes is attenuated in glioblastoma tumor cells deficient in Methylthioadenosine Phosphorylase (MTAP), a common genetic alteration in a variety of cancers. Subsequent experiments in cell line models of different cancer types illustrate that this reduced transcription of FA genes can be recapitulated by blockage of Protein Arginine Methyltransferase 5 (PRMT5), a promising therapeutically targetable epigenetic regulator whose enzymatic activity is compromised in MTAP-deficient cells. Further analyses provide evidence to support that PRMT5 can function as an epigenetic regulator that contributes to the increased expression of FA genes in cancer cells. Most notably and consistent with the essential roles of FA proteins in resolving DNA damage elicited by interstrand crosslinking (ICL) agents, PRMT5 blockage, as well as MTAP loss, sensitizes tumor cells to ICL agents both in vitro and in xenografts. Collectively, these findings reveal a novel epigenetic mechanism underlying the upregulated expression of FA genes in cancer cells and suggest that therapeutically targeting PRMT5 can have an additional benefit of chemosensitizing tumor cells to ICL agents.
Implications:
PRMT5 positively regulates the expression of FA genes. Inhibition of PRMT5 attenuates FA-dependent DNA repair pathway and sensitizes tumor cells to ICL agents.
Keywords: PRMT5, Fanconi anemia, MTAP
Introduction
The Fanconi anemia (FA) family of genes are essential components in the maintenance of genomic stability and repair of DNA damage, as highlighted by the well-established roles of their germline mutations (loss of function) in causing FA, a hereditary recessive disease characterized by predisposition to malignancies 1,2. Paradoxically, FA genes, along with their functional partners, also play important roles in promoting tumor progression and conferring chemoresistance, making them promising targets for cancer chemosensitization 3,4. Curiously, despite their well-established roles in tumorigenesis and tumor progression, aside from the findings that their transcription display characteristics of house-keeping genes 5 and is associated with the Rb/E2F pathway 6, the transcriptional regulation of FA genes, and how their expression is affected by genetic alterations common in cancer cells, remain mostly unclear.
Homozygous deletion of the MTAP occurs in a variety of human cancer types, including about 45% of all glioblastomas (GBM) 7,8. Recently, multiple lines of evidence have emerged elucidating the pathogenic effects of MTAP loss in tumorigenesis and suggest strategies for exploiting it for cancer treatments9-14. MTAP normally acts as a metabolic enzyme in the purine/methionine salvage pathway. It metabolizes methylthioadenosine (MTA), a metabolite generated in the polyamine biosynthesis pathway which produces adenine and methionine as a way for salvaging these metabolites. Consequently, therapeutic strategies have been developed to take advantage of the two consequences of MTAP loss: the compromised capacity of cells to produce these essential cellular building blocks and the accumulation of its direct substrate, MTA. For example, it has been shown that MTAP-deficient tumor cells were more susceptible to inhibitors of purine synthesis and to methionine deprivation14-16. In addition, recent studies revealed the potent activity of MTA in inhibiting several protein arginine methyltransferases (PRMTs), with the highest potency in inhibiting PRMT517-22. PRMTs have increasingly been recognized as playing critical roles in both normal development and in pathogenesis 23,24. This notion has been underscored by recent findings that PRMT genes are altered frequently in a range of cancers, and their upregulated expression is correlated with poor prognosis 23. Consequently, rapid progress has been made in developing therapeutic strategies targeting PRMTs, as highlighted by a number of PRMT-targeting compounds in preclinical tests and in clinical trials for various forms of blood and solid tumors 24. Among the PRMTs, PRMT5 has attracted particular interest: As a key post-translational modification enzyme that functions in regulating splicing of a wide range of mRNA 25-27 and in epigenetic control of gene transcription 13,28-30, PRMT5 has been shown to be involved in multiple cellular processes, such as DNA damage response (DDR) 13,27,31 and cancer cell stemness 14,32.
Mechanistically, PRMT5 modifies non-histone and histone proteins alike. In the latter case, PRMT5 possesses dual epigenetic regulatory roles. It can suppress gene expression via adding suppressive histone methylation marks and modifying DNA methylation 30,33, while in other contexts it activates gene transcription through catalyzing activating histone methylation marks, notably mono-methylation on arginine 2 and symmetric di-methylation on arginine 8 of histone 3 (H3R2me1 and H3R8me2s, respectively) 28,34-36. As a result of its involvement in multiple essential cellular processes, PRMT5 is believed to be a promising therapeutically targetable epigenetic enzyme in multiple cancer types 32,36-39. Notably, MTAP loss has been shown to sensitize tumor cells to PRMT5 inhibition due to the already weakened PRMT5 activity, providing a potential avenue for targeted therapy against MTAP-deleted tumors 21,22,39.
In this study, we demonstrate that MTAP deficiency in tumor cells leads to reduced expression of Fanconi anemia (FA) family genes, essential components in repairing inter-strand crosslink (ICL)-induced DNA damage, and this reduced FA gene expression is associated with increased vulnerability to ICL agents. We show that the effect of MTAP loss is recapitulated by PRMT5 blockage, an observation that led to the identification of PRMT5 as an epigenetic regulator in maintaining the transcription of these FA genes, at least partially via its control of the H3R2me1 in the promoter regions of the regulated FA genes. Further highlighting the epigenetic regulatory role of PRMT5, we demonstrate that tumor cells treated with a small molecular inhibitor of PRMT5 similarly displayed reduced H3R2me1, attenuated expression of these FA genes, and higher susceptibility to ICL agents. Collectively, our findings identify a new vulnerability of MTAP-deficient cells, gain insight into the transcriptional control of the FA family of genes, and provide novel rationale for improving PRMT5 blockage-based therapeutic designs.
Materials and Methods
Cell Lines
Glioblastoma cell lines including U251MG, T98G, and U118MG, previously described 13, were cultured in Dulbecco's Modified Eagle's medium (Gibco, Cat# 11995-065) with 10% (v/v) fetal bovine serum (FBS; Corning, Cat # 35-010-CV). Colorectal cancer cell lines, including HCT116 and SW48 (from ATCC and obtained via the Duke Cell Culture Facility (CCF)), were cultured in McCoy's 5A (ThermoFisher, Cat# 16600108) and Leibovitz's L-15 (ThermoFisher, Cat# 11415114) medium, respectively, supplemented with 10% FBS. Breast cancer cell line MCF-7 (from ATCC via Duke CCF) was cultured with Eagle's Minimum Essential Medium (EMEM, Sigma, Cat# M5650) with 10% FBS. The glioblastoma cell lines have not been authenticated for this study, and the remaining three cell lines were authenticated via STR profiling by the Duke CCF. The U251MG cell line (but not T98G and U118G) and the three non-glioblastoma cell lines were tested to confirm mycoplasma-free by the Duke CCF. The CRISPR-mediated gene knockout cell lines were described previously13. All cell lines were maintained in a humidified atmosphere at 37°C and with 5% CO2 except for SW48, which was cultured without 5% CO2. Cell lines were used for experiments within ~20-25 passages from the time of thawing.
Plasmids, Chemicals and Antibodies
All chemicals, inhibitors and antibodies used in the article are summarized in Supplementary Table for Reagents. The PRMT5 and MTAP overexpression plasmids were previously described (Du et al., 2019). Briefly, overexpression of PRMT5 was obtained using lentiviral vector pLX304 (Addgene #25890), and restoration of MTAP in U118MG cell line was obtained using MSCV (Addgene #24828). Exogenous expression of FANCD2 and FANCI was achieved by using lentiviral vector pLX304 (for FANCD2) and pLenti-CMV-puro-DEST (for FANCI). Cell lines with stable knockdown of PRMT5 (note the attempt to knockout PRMT5 was not successful, likely due to its essential roles in cell survival) and WDR77 were obtained using lentiviral expression construct pLKO.1 (Addgene #10878) (shRNA sequences were listed in Supplementary Table for Reagents) and selected using puromycin (1 μg/ml). All plasmids used in this article were verified by Sanger sequencing. All lentivirus and retrovirus was packaged using 293FT cells (Invitrogen, Cat# R700-07); and the tumor cells were transduced with a MOI value of 3-5.
Cell Proliferation and Viability Assays
To evaluate cellular response to inter-strand crosslinking agents, cells were seeded at 1×103 cells per well in 96-well plates. 24 hours later, cells were treated with different doses of drugs for 72 hours, and numbers of live cells were determined using Cell Counting kit-8 (CCK-8, Cat# CK04, Dojin Laboratories, Kumamoto, Japan). Briefly, CCK8 solution was added to each well to a final concentration of 10% (v/v), and the plates were incubated at 37°C for 1-4 hour. Absorbance in individual wells was determined at 450 nm using microplate reader infinite M200 PRO (TECAN, Männedorf, Switzerland). The dose response curves were simulated using “dose-response simulation” function in the GraphPad Prism 5.0 software (GraphPad Sofeware Inc.). Five replicates were included for each dose (technical repeats), and each assay was independently repeated two or three times (biological repeats). The relative cell numbers were calculated and presented as mean +/− s.d. Clonogenic formation assay was used to evaluate proliferation. Briefly, cells plated at a confluence of 70%-80% were exposed to reagents of different doses for one day, then seeded on 6-well plates at a density of 500 cells per well with fresh medium, and allowed to grow further for 12–18 days to form colonies. The colonies were fixed with 4% (v/v) paraformaldehyde and stained with 0.1% (w/v) crystal violet. The quantifications were done manually. Student’s t-test was used to calculate P value.
Reverse transcription and quantitative real-time PCR (RT-qPCR)
Total RNA was extracted using the RNA extraction kit from Qiagen (Cat# 80204). 1 μg of total RNA was reverse-transcribed using a RNA to cDNA EcoDry Premix Kit (TaKaRa, Cat# 639549) following the manufacturer’s protocols. The cDNA was amplified using the SYBR Green PCR Kit (KAPA, Cat# KK4602) with gene-specific primers listed in Supplementary Table for Reagents. RT-qPCR was performed using CFX96TM Real-Time System (BIO-RAD, Hercules, USA), and the results were analyzed using CFX Maestro Software (BIO-RAD, Hercules, USA). Internal control genes used for gene expression normalization included GAPDH or ACTB. All RT-qPCR were performed in at least two independent experiments and representative results were shown.
Protein Extraction, and Immunoblots
Total cellular protein extracts were prepared by using lysis buffer composed of 1% SDS (w/v), 1 mM DTT and protease inhibitor cocktail (ROCHE, Cat# 04693132001) in PBS buffer (pH7.4), as previously described 13. The lysis solution was mixed with the equal volume of 2X Laemmli loading buffer (BIO-RAD, Cat# 161-0737) with 5% 2-mercaptoethanol and heated at 100°C for 10 min before being used for immunoblot analysis.
Immunoblotting was performed as previously described 52. In brief, protein lysates were resolved using Bis-Tris gels (12% or 4-12% of SDS-PAGE gel, Novex, Cat# NP0341BOX and NP0335BOX) in MOPS running buffer (Novex, Cat# NP0341BOX). Proteins were transferred onto PVDF membranes (Immobilon, Millipore) and incubated overnight with primary antibodies (see Supplementary Table S1 for detailed information on antibodies) at 4°C. Then blots were incubated with horseradish peroxidase-conjugated secondary antibody (Cell Signaling Tech, Anti-mouse IgG: Cat# 7076; Anti-rabbit IgG: Cat# 7074) followed by detection with enhanced chemiluminescence by the Gel Doc XR+ System (BIO-RAD, Hercules, USA). All immunoblot experiments shown were repeated in at least two independent experiments. Whole gel images for all immunoblots are available upon request.
Chromatin Immunoprecipitation (ChIP) and Quantitative PCR (ChIP-qPCR)
Formaldehyde cross-linking and chromatin immunoprecipitations (ChIP) was performed as previously described 13,28. Briefly, cells were harvested at 70%-80% confluence, washed twice with PBS, and cross-linked with 1% (v/v) formaldehyde for 10 minutes in room temperature. Then the cells were lysed on ice using 1×107 cells / ml RIPA buffer (Santa Cruz, Cat# sc-24948) for 30 minutes, followed by sonication using Bioruptor (Diagenode SA, Belgium) for 30min at high power (1 min on, 1 min off). Ten percent of total lysate was set aside as the input for later quantification. Immunoprecipitation was carried out using antibodies specific for H3R2me1, H3R2me2s or H3K4me3, or using an IgG control antibody, with 2 ug of each antibody in 1 ml of lysate, incubating with A/G agarose beads (ThermoFisher, Cat# 20421) (20 μl per tube) overnight in 4°C. The thermocycler used for qPCR was the same as the one used for aforementioned RT-qPCR. Primers for the FA gene’s promoter regions are listed in the Supplemental Table for Reagents. The enrichment of each histone mark in the promoter region was evaluated by the comparison of the percentage of pulled-down DNA to the input (% input) between the specific antibody ChIP group and the control IgG group. All ChIP-qPCR presented in this study have been repeated in two independent experiments. Statistical analysis was performed using Student’s t-test.
Immunofluorescent Staining
Immunofluorescence was performed as previously described 13,53, with modifications. Briefly, cells cultured in chambered coverslip (Ibidi, Cat# 80826) were fixed in 4% (v/v) paraformaldehyde (20 minutes, room temperature), permeabilized in 0.25% (v/v) TritonX-100 in PBS for (5 minutes, room temperature), washed with PBS, rinsed and then blocked with 5% (v/v) bovine serum albumin (BSA) in PBS for 30 minutes. Cells were stained with the anti-FANCD2 antibody (1:400 with 5% BSA in PBS) for 1 hour in room temperature, and washed three times with PBS, followed by incubation with fluorescein coupled secondary antibody (Alexa Fluor Plus 488, ThermoFisher, Cat# A32731, 1:500 in PBS) for 30 minutes in room temperature. Finally, DAPI (0.1 μg per ml in PBS) was used to stain the cells for DNA detection (5 minutes in room temperature). Slides were scanned using a Zeiss 880 confocal microscope (Light Microscopy Core Facility, Duke Cancer Institute). Images were acquired under identical settings including fluorescence signal intensity and exposure time, saved as czi files, and converted to JPEG using LSM ZEN software. FANCD2 foci were analyzed using ImageJ (version 1.8.0, National Institutes of Health, Bethesda, U.S.). Briefly, foci with a diameter greater than 0.8 μm were detected and quantified as previously described 13. The number of foci was counted in 50 randomly selected nuclei for each experimental group. The mean values of foci between different groups were compared using a t-test.
Alkaline Comet Assay
Alkaline comet assay was performed as previously described 13,54. Briefly, cells were treated with DMSO or 1 μM of MMC for 24 hours, and harvested and suspended in 0.5% (w/v) low-melting-point agarose in PBS at a density of 1×104 per ml. The cells were plated on the glass slides provided by the OxiSelect Comet Assay Kit (MyBioSource, Cat# MBS168757) with 80ul of agarose per well, and at the agarose were allowed to solidify at 4°C for 30 min. Comet formation was induced by electrophoresis of cells (constant voltage of 1 volt per cm) in alkaline electrophoresis solution (300 mM NaOH, 1mM EDTA, pH>13), provided by the same Kit. For each sample, duplicate slides were processed. The tail moment was defined as the product of the percentage of DNA in the tail and the displacement between the head and the tail of the comet. At least 50 nuclei from each slide were quantified. All measurements were carried out using the software ImageJ OpenComet as previously described 55. Tail-moments between different groups were compared using nonparametric test (Mann–Whitney U test).
Mouse Xenografts and Treatments
Six- to eight-week-old male and female nude mice were obtained from and maintained in the Duke Cancer Center Isolation Facility (Jackson stock # 007850). Briefly, 2×106 of the indicated cancer cells were injected into the subcutaneous layer on the right flank. Tumors were measured twice a week by caliper, and mice were euthanized when individual tumor volume reached 2.0 cm3. When cohort mean tumor size reached 0.3-65 cm3, mice were intraperitoneally treated with vehicle control or with MMC (Sigma, Cat# M7949, 1 mg/kg in PBS) once a week for five weeks. For the treatment involving MTDIA, mice were treated with MDTIA (Medkoo Cat# 407244, 10 mg/kg in PBS) four times a week, with or without MMC (1 mg/kg) once a week for up to five weeks. The animal protocol was approved by the Duke University Institutional Animal Care and Use Committee (Protocol Registry Approval Number A133-19-06).
Gene Expression Correlation Analysis
Gene expression data used for correlation analyses was provided by TCGA and the analysis was performed via the GEPIA portal (http://gepia.cancer-pku.cn/detail.php)56. Human cancer cell lines’ gene expression data was provided by the Cancer Cell Line Encyclopedia (CCLE) project and downloaded from cBioportal 46-48, and gene expression z-scores were used for expression correlation analysis.
Statistical Analysis
All cell biology and biochemical experiments in this study were repeated in at least two independent experiments. Data obtained are presented as mean ± s.d. Mean values between two groups were compared using Student’s t-test if they follow normal distributions, otherwise they were compared using a nonparametric test. Multiple groups of mean values were compared using ANOVA. All tests were two-sided, deemed statistically significant if P<0.05.
Results
MTAP loss leads to reduced expression of a subset of FA genes
Previous analysis of gene expression correlation has revealed MTAP expression is positively correlated with the expression of a large fraction of DNA damage response (DDR) genes13. Among all DDR genes, we focused specifically on the FA family genes for the following three reasons: their obvious importance in human cancer genetics and hereditary diseases 1,2, our limited knowledge on the mechanism underlying their transcriptional regulation 5, 6, and their roles in resolving DNA damage, a basis on which some chemotherapeutic drugs exert their tumor killing/suppression activity 40. We postulated that understanding the regulation of the transcription of the FA genes can provide a new avenue for devising more effective treatments.
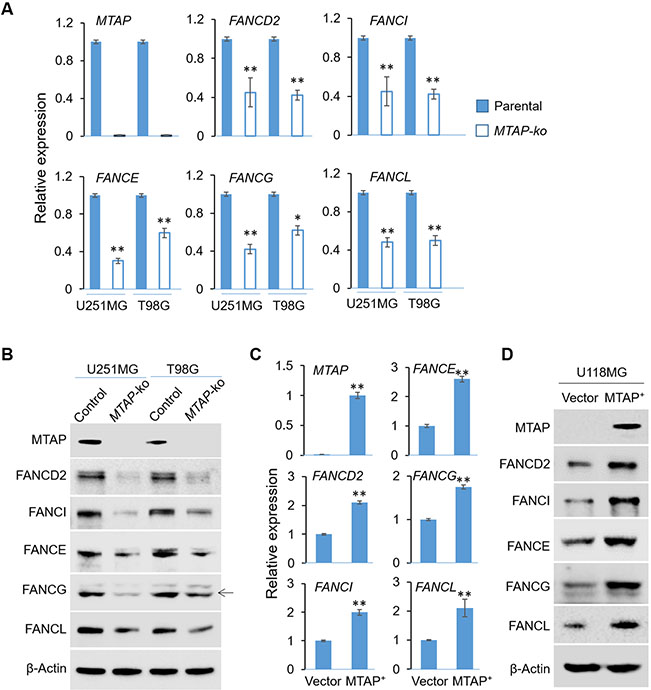
We used two endogenously expressing MTAP wild type cell lines (U251MG and T98G) and generated CRISPR-mediated MTAP knockout isogenic pairs that differ only in their MTAP status 13 to determine the effect of MTAP loss on FA gene expression. Quantitative RT-PCR analysis revealed five FA genes (FANCE, FANCG, FANCL, FANCD2 and FANCI, out of 13 FA genes selected to be examined based on their unique roles in forming the core FA complex and the subsequent ubiquitination of the ID2 complex and their correlated expression with the MTAP expression in GBMs 13) that displayed reduced expression in the MTAP-null cell lines when compared to their MTAP-intact counterparts (Fig. 1A, B). In addition, MTAP deficiency-induced suppression of FA gene expression was also observed when GBM cells were treated with an MTAP inhibitor, MTDIA 41 (Supplementary Fig. 1A), suggesting the down-regulation was due to short term effects of MTAP loss 13, instead of a long term outcome of DNA methylome reprogramming 14. Further experiments showed that this effect of MTAP loss was not limited to these GBM cell lines, as MTAP-intact HCT116 and MCF-7 cell lines treated with an MTAP inhibitor, MTDIA41, also displayed reduced expression in the FA genes (with the exception of FANCI) (Supplementary Fig. 1B, C). In complementary experiments, restoration of MTAP expression in a naturally MTAP-null GBM cell line, U118MG 13, enhanced the expression of these FA genes (Fig. 1C, D).
Fig. 1. MTAP deficient leads to attenuated FA gene expression.
(A) Total RNA was prepared from isogenic control or MTAP-ko cell lines derived from two GBM cell lines (U251MG and T98G) and RT-qPCR was performed to assess the expression of indicated FA genes. (B) Cell lines used in (C) were used for total protein extraction followed by immunoblots to assess the levels of indicated proteins. (C) Isogenic U118MG cell lines with different MTAP status, including the MTAP-null (vector control) and the MTAP-restored (MTAP+) lines, were used for RNA preparation and RT-qPCR to assess the expression of indicated FA genes. (D) Cell lines used in (e) were used for total protein extraction followed by immunoblots to assess the levels of indicated proteins. *p<0.05; ** p<0.01.
MTAP deficiency sensitizes tumor cells to ICL agents
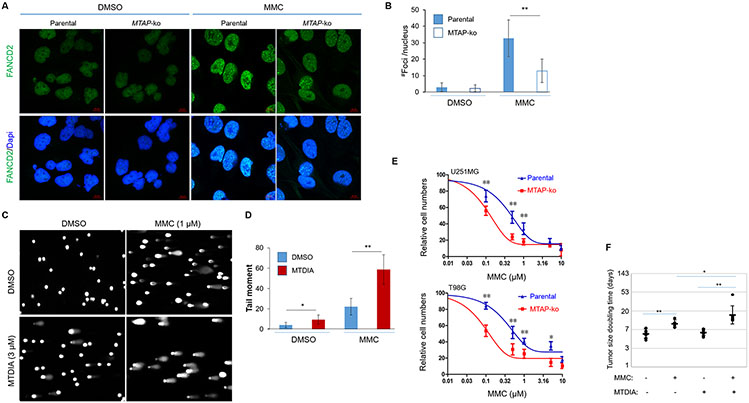
FA proteins form foci of FA complex in response to ICL-inflicted DNA damage 1. Corroborating the above results and confirming the functional consequence of the MTAP loss-induced deficiency in FA gene expression, MTAP-null cells were defective in forming FA protein foci in response to Mitomycin C (MMC), an inter-strand DNA crosslinking agent (Fig. 2A, B). Furthermore, MTAP blockage with MTDIA led to higher levels of DNA damage spontaneously and in response to MMC treatment (Fig. 2C, D). These led us to postulate that MTAP deficiency can sensitize tumor cells to treatments with ICL agents. Further experiments confirmed that indeed, compared to their matched, MTAP-intact counterparts, MTAP-deficient GBM cells (U251MG and T98G) displayed higher susceptibility to treatments of ICL agents (MMC and Cisplatin), as measured by cell propagation and colony formation assays (Fig. 2E and Supplementary Fig. 2A-C). While the poor blood brain barrier permeability of the ICL agents42,43 precluded testing the effect in orthotopic brain tumor models, treatments in U251MG-derived subcutaneous (s.c.) xenografts confirmed that this MTAP deficiency-enhanced susceptibility to MMC also occurred in vivo, as demonstrated by the measurably stronger tumor suppression of the combined treatment with MMC plus MTDIA compared with either agent alone (Fig. 2F and Supplementary Fig. 2D). Collectively, these results are in agreement with the aforementioned differential FA gene expression in MTAP-deficient cells and suggest MTAP deficiency sensitizes tumor cells’ response to ICL agents.
Fig. 2. MTAP deficiency sensitizes tumor cells to MMC.
(A) U251MG cell lines (Control or MTAP-ko) were treated with vehicle control or with MMC (0.3 uM) for 24 hours, and IF staining was performed. (B) Quantifications of foci for the IF staining experiments shown in (A). (C) U251MG cell line was treated with vehicle control or with MTAP inhibitor (MTDIA, 3 μM) for 24 hours, and alkaline comet assays were performed to assess DNA damage. (D) Quantifications of the tail moment for the alkaline comet assays shown in (C). (E) Isogenic cell lines derived from U251MG and T98G were treated with indicated doses of MMC for 3 days, and cell numbers were determined by CCK8 assay. (F) Subcutaneous xenografts derived from U251MG were treated with vehicle control (n=8), MMC alone (n=5), MTDIA alone (n=8), or MMC plus MTDIA (n=7), and the doubling times of tumor size were determined and compared.*p<0.05; ** p<0.01.
Loss-of-function of PRMT5 recapitulates MTAP deficiency-induced FA gene downregulation
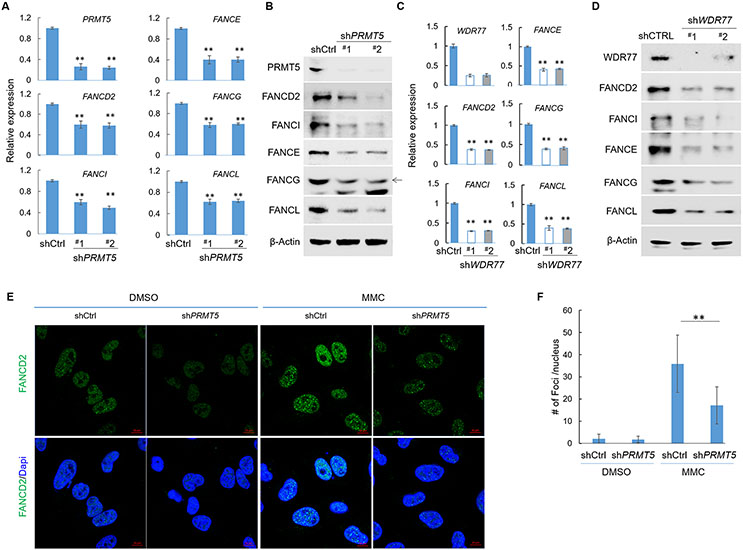
A recent study demonstrated the compromised PRMT5 functionality is a mechanism underlying the reduced transcription of RNF168 in MTAP-null GBM cells 13. Consequently, we speculated that PRMT5’s loss of function can similarly lead to the attenuated expression of the subset of FA genes. We therefore determined the expression of FA genes in two GBM cell lines with PRMT5 knockdown (shPRMT5) and found that among all FA genes tested, the same set of five FA genes, including FANCD2, FANCI, FANCE, FANCG, and FANCL, indeed displayed reduced expression in the shPRMT5 lines when compared to the control lines (Fig. 3A, B and Supplementary Fig. 3A). The reduced expression of these genes was also observed when U251MG cells were treated with a PRMT5 inhibitor, EPZ015666 38 (Supplementary Fig. 3B). Furthermore, in cells with knockdown of WDR77, the co-activator essential for PRMT5, 44 a similar down-regulation of these FA genes was also observed (Fig. 3C, D). In corroborating the observed differential gene expression, knockdown of PRMT5 in U251MG cells led to compromised formation of FA protein foci in response to MMC (Fig. 3E, F). To further determine whether this PRMT5 blockage-induced attenuated expression of FA genes was unique to the aforementioned GBM cell lines, we similarly tested the effect of PRMT5 loss of function, via a PRMT5 inhibitor or knockdown of PRMT5 in a breast cancer cell line (MCF-7) and in two colorectal cancer cell lines (HCT116 and SW48). These experiments revealed that the effect of PRMT5 loss-of-function on FA gene’s expression was not unique to the GBM cell lines, as a similar phenotype was also observed in MCF-7 (Supplementary Fig. 4A), HCT116 (Supplementary Fig. 4B), and SW48 (Supplementary Fig. 4C, D). Finally, in agreement with these differential gene expression findings, in comparison to the control SW48 cell line (shCtrl), SW48 cell line with PRMT5 knockdown displayed defective formation of FA foci in response to MMC treatment (Supplementary Fig. 4E, F).
Fig. 3. PRMT5 activates FA gene transcription.
(A) Total RNA from U251MG cell lines (shCtrl and shPRMT5) were used for reverse-transcription and quantitative PCR analysis (RT-qPCR). (B) Total protein lysates from U251MG cell lines (shCtrl and shPRMT5) were used for immunoblots using indicated antibodies. (C) Derivatives of U251MG cell line, shCtrl or shWDR77 (two different shRNA sequences) were used for RT-qPCR analysis to determine the relative expression levels of indicated genes. (D) Cell lines used in (c) were used for immunoblots to determine the levels of indicated proteins. (E) U251MG cell lines (Control or shPRMT5) were treated with vehicle control or with MMC (0.3 μM) for 24 hours, and IF staining was performed. (F) Quantification of foci for the anti-FANCD2 IF staining experiments shown in (E).
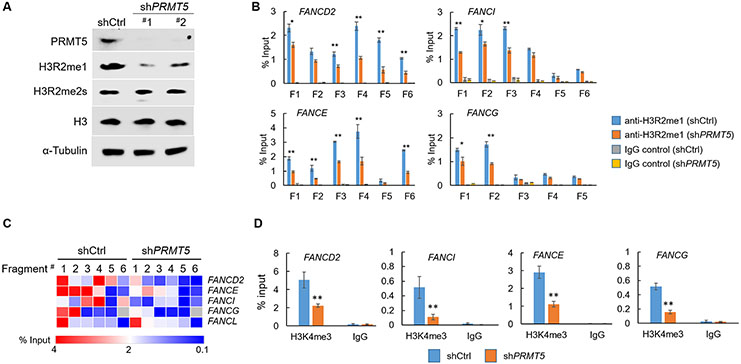
PRMT5 can catalyze H3R2me2s or H3R2me1 in activating or suppressing gene transcription28,34-36,45. In particular, PRMT5-mediated H3R2me1 has been shown to contribute to the active transcription of a DDR gene, RNF168, in GBM cells 13. To gain further insight into the mechanism underlying the effects of PRMT5’s loss of function on gene expression of FA genes, we investigated the contribution of these histone marks on the activation of FA gene’s transcription. In agreement with the previous findings13, knockdown of PRMT5 led to decreased global levels of H3R2me1, but not H3R2me2s (Fig. 4A). We then performed ChIP-qPCR to determine the level of these two histone marks in the regions spanning the promoters of the five PRMT5-regulated FA genes (Supplementary Fig. 5A). These experiments revealed positive enrichment for H3R2me1 in the promoter regions of all five genes examined (Fig. 4B, C), where RNF168 was used as a positive control for PRMT5/H3R2me1 regulation 13 (Supplementary Fig. 5B). Furthermore, with the exception of the FANCL gene, in the promoters of four other genes (FANCD2, FANCI, FANCE and FANCG), the levels of H3R2me1 was attenuated in the PRMT5-kd cells (Fig. 4B, C). In contrast, parallel ChIP-qPCR analysis in the FANCD2 and FANCI promoters revealed the absence of such a PRMT5-dependent enrichment for the other histone mark, H3R2me2s, where SP2 was used as a positive control for PRMT5/H3R2me2s regulation 29 (Supplementary Fig. 5C, D). In further supporting the effect of MTAP loss on the activity of PRMT5 and the roles of PRMT5-mediated H3R2me1 in activating the FA genes, similar ChIP-qPCR analysis similarly revealed reduced levels of H3R2m1 in the FANCD2 and FANCI promoters (Supplementary Fig. 6A, B). Finally, in agreement with the reduced transcription of the FA genes, the promoter regions of these genes also displayed decreased levels of H3K4me3, a featured histone mark for active gene transcription (Fig. 4D). Collectively, these results illustrate that loss of function of PRMT5 recapitulates MTAP deficiency-induced FA gene downregulation. They also support that PRMT5 functions as an epigenetic regulator in maintaining the transcription of a subset of FA genes in cancer cells.
Fig. 4. PRMT5 knockdown leads to reduced H3R2me1 levels in the promoter regions of four FA genes.
(A) U251MG cell line with control (shCtrl) or PRMT5 kd (shPRMT5), were used for immunoblots with indicated antibodies. (B) U251MG cell lines (shCtrl and shPRMT5) were used for anti-H3R2me1 chromatin immunoprecipitation (ChIP) followed by qPCR analyses. For each gene six amplicons covering ~1.2-1.5 kb in the promoter region were designed and their relative enrichment were quantified. (C) The heat-map shows the quantification results presented in (B). (D) Derivatives of U251MG cell line were used for anti-H3K4me3 ChIP-qPCR analysis, and the relative enrichment of the promoter regions of indicated FA genes were shown. *p<0.05; ** p<0.01.
PRMT5 contributes to the upregulated expression of a subset of FA family genes in cancer cells
The essential roles of FA family genes in maintaining genomic stability have been highlighted by the predisposition to malignancies resultant from their germline mutations 1,2. Two FA genes that are key components of the FA pathway, FANCD2 and FANCI, were analyzed among human cancers (from TCGA). Interestingly, it was revealed that in an overwhelming majority of cancer types, 17/18 (for FANCD2) and 22/23 (for FANCI), these genes were expressed at a higher level in cancer than in the matched normal tissues (Supplementary Fig. 7A, B). This observation, together with the role of PRMT5 in activating FA gene’s transcription, led us to postulate that PRMT5 contributes to the upregulated expression of at least a subset of FA genes in cancer cells.
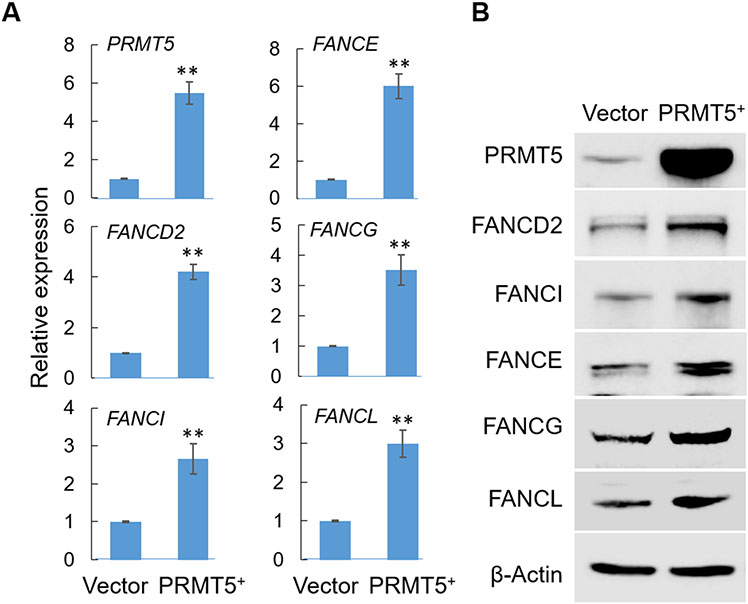
Taking advantage of the recently published Cancer Cell Line Encyclopedia project 46-48, we analyzed gene expression data from human cancer cell lines. As expected, a significant positive correlation between the expression of PRMT5 and its activated gene RNF168 13 was detected in human cancer cell lines, serving as a positive control (Supplementary Fig. 8A). Further analysis revealed that this positive correlation occurred only in cell lines originated from solid cancers but not in those from blood cancers (Supplementary Fig. 8B, C), suggesting tissue-dependent target genes of PRMT5. Expanding the same analysis to the 18 (out of 19) FA genes for which gene expression data was available, we found that 17 of them displayed moderate yet significant positive correlations with PRMT5 expression (Supplementary Fig. 8D). Similarly, the positive correlation between FA genes and PRMT5 was more prominent in solid cancer-derived cell lines than blood cancer-derived lines (Supplementary Fig. 8E). In agreement with these correlations and with the above experimental results from PRMT5 loss of function assays, overexpression of exogenous PRMT5 in the U251MG cell line led to upregulated expression of the five FA genes examined as measured by both RT-qPCR and by immunoblots (Fig. 5A, B). While the transcriptional regulation of the FA family genes is certainly a complicated, cellular context-dependent process, collectively these results led us to speculate that expression of PRMT5 likely contributes to the activated expression of at least a subset of the FA family genes in certain cellular contexts.
Fig. 5. PRMT5 overexpression leads to upregulated expression of FA genes.
(A) Total RNA from U251MG cell lines (vector control or PRMT5+) were used for RT-qPCR analysis. (B) Total protein lysates from U251MG cell lines (vector control or PRMT5+) were used for immunoblots using indicated antibodies. ** p<0.01.
PRMT5 loss-of-function sensitizes cancer cells to ICL agents
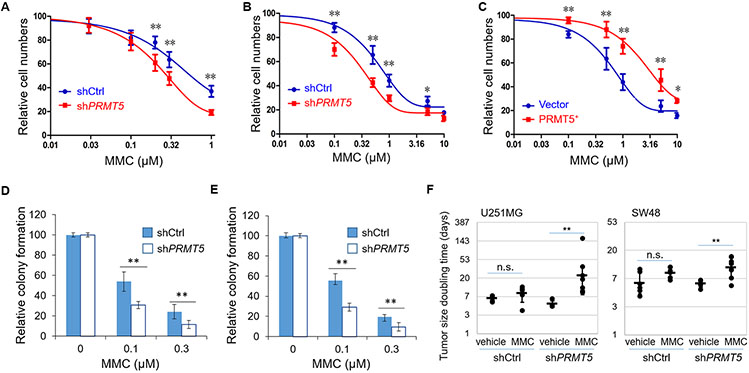
PRMT5 has been established as a promising therapeutic target in a variety of cancer types 26,37,38,49. While the essential role of PRMT5 in coordinating mRNA splicing provides a solid rationale for targeting PRMT5 in glioma initially26, its multifaceted roles in various cellular processes most likely contribute to the therapeutic efficacy. We speculate that the positive roles of PRMT5 in activating FA gene expression can be exploited for maximizing the therapeutic benefit through combining its inhibition with an ICL drug. We therefore tested the combinatory effects of PRMT5’s loss-of-function with an ICL agent. Indeed, we found that in two cell lines tested (SW48 and U251MG), in comparison to the matched controls, PRMT5-kd or PRMT5 overexpression conferred an increased or decreased susceptibility, respectively, to treatments with an ICL agent (MMC or Cisplatin), as measured by cell propagation (Fig. 6A-C and Supplementary Fig. 9A, B) and by colony formation (Fig. 6D, E and Supplementary Fig. 9C, D). Notably, this PRMT5-kd conferred susceptibility to ICL agents was mitigated by the exogenous expression of two FA genes (FANCD2 and FANCI), in agreement with the scenario in which the downregulated expression of these FA genes contributes to the susceptibility (Supplementary Fig. 10). Furthermore, this PRMT5 blockage-conferred susceptibility to MMC was also observed in subcutaneous (s.c.) xenograft models derived from U251MG or from SW48, as in each model the cell line with PRMT5 knockdown displayed a stronger response the MMC treatment in comparison to the matched control cell line (Fig. 6F and Supplementary Fig. 11). While it is unlikely that this effect was simply due to the reduced expression of FA genes given the essential roles of PRMT5 in tumor cells, collectively these results are consistent with the positive effects of PRMT5 on FA gene expression and support the idea that blocking PRMT5’s function can render tumor cells more susceptible to ICL agents.
Fig. 6. PRMT5 blockage sensitizes tumor cells to ICL agents.
(A) SW48 derivative cell lines were treated with indicated doses of MMC for 3 days and cell numbers were determined by CCK8 assay. (B - C) U251MG derivative cell lines were treated with indicated doses of MMC for 3 days, and cell numbers were determined by CCK8 assay. (D-E) Derivatives of (D) SW48 and (E) U251MG cell lines were treated with indicated doses of MMC for 24 hours, and clonogenic formation assays were performed to determine the relative colony formation. (F) Derivative U251MG and SW48 cell lines (shCtrl or shPRMT5) were subcutaneously implanted in nude mice and tumors were treated with MMC (1 mg/kg via intraperitoneal injection once per week for 3-5 weeks), and tumor sizes were measured twice per week starting from the first treatment, and tumor size’s doubling times were determined and calculated (from the left to the right group, n=5, 6, 4, 7 (for U251MG) and n=6, 7, 6, and 8 (for SW48), respectively). n.s. not significant; *p<0.05; ** p<0.01.
Discussion
In this study, we identify an epigenetic mechanism underlying the upregulated expression of a subset of FA family genes in cancer cells, including two key components in the FA complex (FANCD2 and FANCI) 1,50. We reveal that in MTAP-deficient cells, the expression of five FA genes, including FANCD2 and FANCI, was attenuated, consistent with the positively correlated expression between MTAP and a subset of FA genes and adding further evidence to support the previously established notion that MTAP loss leads to compromised DDR pathways in cancer cells 13. In accordance with the previously described effect of MTAP loss on PRMT5’s activity 13,21,22,39, we further demonstrated that loss of PRMT5 activity recapitulated this differential expression of FA genes and similarly leads to increased susceptibility of cancer cells to ICL agents.
Besides being essential genome guardians in resolving inter-strand DNA crosslinks and stalled replication forks 51, the importance of FA genes has been highlighted by their link to FA, a heritable recessive disorder featuring predisposition to malignancies 1,50. In tumor cells the same genome stability maintenance of FA proteins, as part of the DNA repair pathways, also contribute to tumor progression and development of resistance to DNA damage-based chemotherapeutic drugs, thus therapeutically targeting them have been a promising strategy for cancer’s chemosensitization 3,4. Despite their obvious importance in human cancers, our understanding of FA gene transcriptional regulation is limited by the fact that their regulatory elements display characteristics of housekeeping genes and because their expression is related to the cell cycle 5,6. Notably, their expression was elevated in cancer cells compared to normal cells of matched tissues in a majority of cancer types examined, which is consistent with their tumor-promoting roles. This suggests that illuminating the mechanism underlying their overexpression in cancer cells can potentially provide another avenue to chemosensitizing tumor cells. Combined with the fact that PRMT5 is overexpressed in a range of cancers 23, our findings suggest that the elevated expression of at least some of the FA genes in cancer cells can likely be attributed to heightened PRMT5 activity in cancer cells, thus providing a complementary epigenetic mechanism that can serve as a foundation for further illuminating the regulatory mechanisms of these genes.
Several limitations are noted in this regard. First, we note that experiments in this study primarily were limited to several cell line models with a focus on a small subset of FA genes, among numerous genes functioning in the DDR pathways. In addition, the detailed epigenetic mechanism underlying the gene expression regulation remains to be fully illuminated, including the potential involvement of other PRMTs and the downstream co-activators. For example, it has been proposed that PRMT5 partners with the WDR5/MLL complex in activating the transcription of a set of epithelial to mesenchymal genes in response to TGFβ, raising the intriguing possibility that the WDR5/MLL complex is involved in the transcriptional activation of the FA genes 28. Finally, PRMT5 inhibition has been shown to affect splicing of a large number of genes, primarily those in the cell cycle progression, in GBM cells 52. Thus it is possibly that altered splicing contributes to the differential transcript levels of FA genes upon PRMT5 blockage in addition to the epigenetic regulation at the gene transcription step. Future studies will be needed to further address these issues. Nevertheless, we speculate that besides providing opportunities for advancing our understanding of epigenetic regulation of FA gene transcription in cancer cells, findings described in this study also inform therapeutic considerations in the following ways. First, PRMT5 is considered a potential cancer therapeutic target 26,38,49, and its candidacy has been shown to be particularly promising in combination with inhibitors against other PRMTs or in the context of loss of MTAP, a common genetic alteration in numerous cancer types 21,22,37. While the essential role of PRMT5 in coordinating mRNA splicing provides a solid rationale initially for targeting PRMT5 in glioma 26, we speculate that the effects of PRMT5 blockage on gene transcription, such as the transcription of FA genes described here, can also contribute to therapeutic benefits. Consequently, we suggest that additional insight into the efficacy of PRMT5 inhibition in distinct types of cancer can be gained by further illuminating PRMT5-mediated alteration in gene transcription in tumor cells and the effects of a PRMT5 blockage on such a program. Second, it is feasible to exploit the vulnerability resulting from PRMT5 inhibition-induced differential gene expression in tumor cells and devise combinatory therapeutic strategies accordingly, as exemplified by the proof-of-principle results from the ICL agent treatment described in this study. Lastly, tumor cells bearing MTAP deficiency have been shown to be particularly susceptible to PRMT5 blockage 21,22,37. In this regard, our findings in MTAP-deficient cells suggest that therapeutic considerations based on the PRMT5-controlled gene transcription are likely also applicable to tumors with MTAP loss and warrant further investigation.
Supplementary Material
Table 1.
Thermodynamic parameters for docking between WDR5’s WIN domain and unmodified, H3R2me1, and H3R2me2s-containing histone H3 tail (NH2-terminal tail of 15 amino acids were used for the docking simulation).
| H3R2me1- WDR5 docking |
H3R2me2s- WDR5 docking |
Unmodified H3- WDR5 docking |
|
|---|---|---|---|
| Binding energy (kcal/mol) | 1.03 | 10.4 | 47.7 |
| Internal energy (kcal/mol) | 1.01 | 10.4 | 47.7 |
| van der Waals and hydrogen bond energy (kcal/mol) | 1.01 | 10.4 | 47.7 |
| Electrostatic energy (kcal/mol) | 3.29 | 3.14 | 2.58 |
| Torsional energy (kcal/mol) | 19.39 | 19.69 | 19.09 |
Acknowledgement
We thank the Functional Genomics Core facility and Light Microscopy Core Facility (Biology / DCI), and The Cancer Center Isolation Facility (CCIF/DCI) for support to this study. We thank Xudong Yao for assistance in molecular cloning and Paula K Greer, Heng Liu and Hai Yan for technical and administrative assistance. JL was supported by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences. This work was supported by The Department of Pathology and the Preston Robert Tisch Brain Tumor Center at Duke, and the National Institute of Neurological Disorders and Stroke of the National Institutes of Health, Award Number R01NS101074 (YH).
Footnotes
Conflict of interest The authors declare no conflict of interest.
References
- 1.D'Andrea AD The Fanconi road to cancer. Genes & development 17, 1933–1936, doi: 10.1101/gad.1128303 (2003). [DOI] [PubMed] [Google Scholar]
- 2.Dong H et al. Update of the human and mouse Fanconi anemia genes. Human genomics 9, 32, doi: 10.1186/s40246-015-0054-y (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andreassen PR & Ren K Fanconi anemia proteins, DNA interstrand crosslink repair pathways, and cancer therapy. Current cancer drug targets 9, 101–117, doi: 10.2174/156800909787314011 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chirnomas D et al. Chemosensitization to cisplatin by inhibitors of the Fanconi anemia/BRCA pathway. Molecular cancer therapeutics 5, 952–961, doi: 10.1158/1535-7163.MCT-05-0493 (2006). [DOI] [PubMed] [Google Scholar]
- 5.Meier D & Schindler D Fanconi anemia core complex gene promoters harbor conserved transcription regulatory elements. PloS one 6, e22911, doi: 10.1371/journal.pone.0022911 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoskins EE et al. Coordinate regulation of Fanconi anemia gene expression occurs through the Rb/E2F pathway. Oncogene 27, 4798–4808, doi: 10.1038/onc.2008.121 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brennan CW et al. The somatic genomic landscape of glioblastoma. Cell 155, 462–477, doi: 10.1016/j.cell.2013.09.034 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nobori T et al. Absence of methylthioadenosine phosphorylase in human gliomas. Cancer research 51, 3193–3197 (1991). [PubMed] [Google Scholar]
- 9.Li CF et al. Downregulated MTAP expression in myxofibrosarcoma: A characterization of inactivating mechanisms, tumor suppressive function, and therapeutic relevance. Oncotarget, doi:2552 [pii] (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhi L et al. Association of common variants in MTAP with susceptibility and overall survival of osteosarcoma: a two-stage population-based study in Han Chinese. Journal of Cancer 7, 2179–2186, doi: 10.7150/jca.16609 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marce S et al. Lack of methylthioadenosine phosphorylase expression in mantle cell lymphoma is associated with shorter survival: implications for a potential targeted therapy. Clinical cancer research : an official journal of the American Association for Cancer Research 12, 3754–3761, doi: 10.1158/1078-0432.CCR-05-2780 (2006). [DOI] [PubMed] [Google Scholar]
- 12.Lin X et al. Genetic variants at 9p21.3 are associated with risk of esophageal squamous cell carcinoma in a Chinese population. Cancer science, doi: 10.1111/cas.13130 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Du C et al. A PRMT5-RNF168-SMURF2 Axis Controls H2AX Proteostasis. Cell reports 28, 3199–3211 e3195, doi: 10.1016/j.celrep.2019.08.031 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hansen LJ et al. MTAP Loss Promotes Stemness in Glioblastoma and Confers Unique Susceptibility to Purine Starvation. Cancer Res 79, 3383–3394, doi: 10.1158/0008-5472.CAN-18-1010 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hori H et al. Methylthioadenosine phosphorylase cDNA transfection alters sensitivity to depletion of purine and methionine in A549 lung cancer cells. Cancer Res 56, 5653–5658 (1996). [PubMed] [Google Scholar]
- 16.Kamatani N, Nelson-Rees WA & Carson DA Selective killing of human malignant cell lines deficient in methylthioadenosine phosphorylase, a purine metabolic enzyme. Proc Natl Acad Sci U S A 78, 1219–1223 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bigaud E & Corrales FJ Methylthioadenosine (MTA) Regulates Liver Cells Proteome and Methylproteome: Implications in Liver Biology and Disease. Molecular & cellular proteomics : MCP 15, 1498–1510, doi: 10.1074/mcp.M115.055772 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang S Histone methyltransferases, diet nutrients and tumour suppressors. Nature reviews. Cancer 2, 469–476, doi: 10.1038/nrc819 (2002). [DOI] [PubMed] [Google Scholar]
- 19.Lee SH & Cho YD Induction of apoptosis in leukemia U937 cells by 5'-deoxy-5'-methylthioadenosine, a potent inhibitor of protein carboxylmethyltransferase. Experimental cell research 240, 282–292, doi: 10.1006/excr.1998.4000 (1998). [DOI] [PubMed] [Google Scholar]
- 20.Oliva A, Galletti P, Zappia V, Paik WK & Kim S Studies on substrate specificity of S-adenosylmethionine: protein-carboxyl methyltransferase from calf brain. European journal of biochemistry / FEBS 104, 595–602 (1980). [DOI] [PubMed] [Google Scholar]
- 21.Mavrakis KJ et al. Disordered methionine metabolism in MTAP/CDKN2A-deleted cancers leads to dependence on PRMT5. Science 351, 1208–1213, doi: 10.1126/science.aad5944 (2016). [DOI] [PubMed] [Google Scholar]
- 22.Kryukov GV et al. MTAP deletion confers enhanced dependency on the PRMT5 arginine methyltransferase in cancer cells. Science 351, 1214–1218, doi: 10.1126/science.aad5214 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stopa N, Krebs JE & Shechter D The PRMT5 arginine methyltransferase: many roles in development, cancer and beyond. Cellular and molecular life sciences : CMLS 72, 2041–2059, doi: 10.1007/s00018-015-1847-9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jarrold J & Davies CC PRMTs and Arginine Methylation: Cancer's Best-Kept Secret? Trends Mol Med 25, 993–1009, doi: 10.1016/j.molmed.2019.05.007 (2019). [DOI] [PubMed] [Google Scholar]
- 25.Radzisheuskaya A et al. PRMT5 methylome profiling uncovers a direct link to splicing regulation in acute myeloid leukemia. Nature structural & molecular biology 26, 999–1012, doi: 10.1038/s41594-019-0313-z (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Braun CJ et al. Coordinated Splicing of Regulatory Detained Introns within Oncogenic Transcripts Creates an Exploitable Vulnerability in Malignant Glioma. Cancer cell 32, 411–426 e411, doi: 10.1016/j.ccell.2017.08.018 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hamard PJ et al. PRMT5 Regulates DNA Repair by Controlling the Alternative Splicing of Histone-Modifying Enzymes. Cell reports 24, 2643–2657, doi: 10.1016/j.celrep.2018.08.002 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen H, Lorton B, Gupta V & Shechter D A TGFbeta-PRMT5-MEP50 axis regulates cancer cell invasion through histone H3 and H4 arginine methylation coupled transcriptional activation and repression. Oncogene 36, 373–386, doi: 10.1038/onc.2016.205 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Migliori V et al. Symmetric dimethylation of H3R2 is a newly identified histone mark that supports euchromatin maintenance. Nature structural & molecular biology 19, 136–144, doi: 10.1038/nsmb.2209 (2012). [DOI] [PubMed] [Google Scholar]
- 30.Zhao Q et al. PRMT5-mediated methylation of histone H4R3 recruits DNMT3A, coupling histone and DNA methylation in gene silencing. Nature structural & molecular biology 16, 304–311, doi: 10.1038/nsmb.1568 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clarke TL et al. PRMT5-Dependent Methylation of the TIP60 Coactivator RUVBL1 Is a Key Regulator of Homologous Recombination. Molecular cell 65, 900–916 e907, doi: 10.1016/j.molcel.2017.01.019 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chiang K et al. PRMT5 Is a Critical Regulator of Breast Cancer Stem Cell Function via Histone Methylation and FOXP1 Expression. Cell reports 21, 3498–3513, doi: 10.1016/j.celrep.2017.11.096 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rank G et al. Identification of a PRMT5-dependent repressor complex linked to silencing of human fetal globin gene expression. Blood 116, 1585–1592, doi: 10.1182/blood-2009-10-251116 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deng X et al. Protein arginine methyltransferase 5 functions as an epigenetic activator of the androgen receptor to promote prostate cancer cell growth. Oncogene 36, 1223–1231, doi: 10.1038/onc.2016.287 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.LeBlanc SE et al. Protein arginine methyltransferase 5 (Prmt5) promotes gene expression of peroxisome proliferator-activated receptor gamma2 (PPARgamma2) and its target genes during adipogenesis. Molecular endocrinology 26, 583–597, doi: 10.1210/me.2011-1162 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tarighat SS et al. The dual epigenetic role of PRMT5 in acute myeloid leukemia: gene activation and repression via histone arginine methylation. Leukemia 30, 789–799, doi: 10.1038/leu.2015.308 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fedoriw A et al. Anti-tumor Activity of the Type I PRMT Inhibitor, GSK3368715, Synergizes with PRMT5 Inhibition through MTAP Loss. Cancer cell 36, 100–114 e125, doi: 10.1016/j.ccell.2019.05.014 (2019). [DOI] [PubMed] [Google Scholar]
- 38.Chan-Penebre E et al. A selective inhibitor of PRMT5 with in vivo and in vitro potency in MCL models. Nature chemical biology 11, 432–437, doi: 10.1038/nchembio.1810 (2015). [DOI] [PubMed] [Google Scholar]
- 39.Marjon K et al. MTAP Deletions in Cancer Create Vulnerability to Targeting of the MAT2A/PRMT5/RIOK1 Axis. Cell reports 15, 574–587, doi: 10.1016/j.celrep.2016.03.043 (2016). [DOI] [PubMed] [Google Scholar]
- 40.Deans AJ & West SC DNA interstrand crosslink repair and cancer. Nature reviews. Cancer 11, 467–480, doi: 10.1038/nrc3088 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Basu I et al. Growth and metastases of human lung cancer are inhibited in mouse xenografts by a transition state analogue of 5'-methylthioadenosine phosphorylase. The Journal of biological chemistry 286, 4902–4911, doi: 10.1074/jbc.M110.198374 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gomes ML, de Mattos DM, de Souza Freitas R, Bezerra RJ & Bernardo-Filho M Study of the toxicological effect of mitomycin C in mice: alteration on the biodistribution of radiopharmaceuticals used for renal evaluations. Hum Exp Toxicol 20, 193–197, doi: 10.1191/096032701678766840 (2001). [DOI] [PubMed] [Google Scholar]
- 43.Jacobs SS et al. Plasma and cerebrospinal fluid pharmacokinetics of intravenous oxaliplatin, cisplatin, and carboplatin in nonhuman primates. Clinical cancer research : an official journal of the American Association for Cancer Research 11, 1669–1674, doi: 10.1158/1078-0432.CCR-04-1807 (2005). [DOI] [PubMed] [Google Scholar]
- 44.Burgos ES et al. Histone H2A and H4 N-terminal tails are positioned by the MEP50 WD repeat protein for efficient methylation by the PRMT5 arginine methyltransferase. The Journal of biological chemistry 290, 9674–9689, doi: 10.1074/jbc.M115.636894 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pal S, Vishwanath SN, Erdjument-Bromage H, Tempst P & Sif S Human SWI/SNF-associated PRMT5 methylates histone H3 arginine 8 and negatively regulates expression of ST7 and NM23 tumor suppressor genes. Molecular and cellular biology 24, 9630–9645, doi: 10.1128/MCB.24.21.9630-9645.2004 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cerami E et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer discovery 2, 401–404, doi: 10.1158/2159-8290.CD-12-0095 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gao J et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Science signaling 6, pl1, doi: 10.1126/scisignal.2004088 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ghandi M et al. Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature 569, 503–508, doi: 10.1038/s41586-019-1186-3 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yan F et al. Genetic validation of the protein arginine methyltransferase PRMT5 as a candidate therapeutic target in glioblastoma. Cancer Res 74, 1752–1765, doi: 10.1158/0008-5472.CAN-13-0884 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mathew CG Fanconi anaemia genes and susceptibility to cancer. Oncogene 25, 5875–5884, doi: 10.1038/sj.onc.1209878 (2006). [DOI] [PubMed] [Google Scholar]
- 51.Kee Y & D'Andrea AD Expanded roles of the Fanconi anemia pathway in preserving genomic stability. Genes & development 24, 1680–1694, doi: 10.1101/gad.1955310 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Du C et al. 5-Fluorouracil targets histone acetyltransferases p300/CBP in the treatment of colorectal cancer. Cancer Lett 400, 183–193, doi: 10.1016/j.canlet.2017.04.033 (2017). [DOI] [PubMed] [Google Scholar]
- 53.Gruosso T et al. Chronic oxidative stress promotes H2AX protein degradation and enhances chemosensitivity in breast cancer patients. EMBO molecular medicine 8, 527–549, doi: 10.15252/emmm.201505891 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Langie SA, Azqueta A & Collins AR The comet assay: past, present, and future. Frontiers in genetics 6, 266, doi: 10.3389/fgene.2015.00266 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gyori BM, Venkatachalam G, Thiagarajan PS, Hsu D & Clement MV OpenComet: an automated tool for comet assay image analysis. Redox biology 2, 457–465, doi: 10.1016/j.redox.2013.12.020 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tang Z et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic acids research 45, W98–W102, doi: 10.1093/nar/gkx247 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.