Abstract
The microbiome contributes to the development and maturation of the immune system. In response to commensal bacteria, intestinal CD4+ T lymphocytes differentiate into functional subtypes with regulatory or effector functions. The development of small intestine intraepithelial lymphocytes that co-express CD4 and CD8αα homodimers (CD4IELs) depends on the microbiota. However, the identity of the microbial antigens recognized by CD4+ T cells that can differentiate into CD4IELs remains unknown. We identified β-hexosaminidase, a conserved enzyme across commensals of the Bacteroidetes phylum, as a driver of CD4IEL differentiation. In a mouse model of colitis, β-hexosaminidase–specific lymphocytes protected against intestinal inflammation. Thus, T cells of a single specificity can recognize a variety of abundant commensals and elicit a regulatory immune response at the intestinal mucosa.
One-Sentence Summary:
A commensal-derived antigen drives regulatory immune responses in the intestine.
The microbiota contributes to functional specification of adaptive immunity, both through direct interactions and via soluble mediators released into the environment (1–3). Colonic bacteria such as Helicobacter hepaticus promote differentiation of antigen-specific CD4+ T cells into Foxp3+ regulatory T cells (Tregs) in the colon, whereas segmented filamentous bacteria (SFB) induce quasi-clonal pro-inflammatory T helper 17 (TH17) cells in the ileum (4–10). Such interactions are not only specific for the bacterial species concerned, but depending on location and context, can also influence T cell fates (4, 11). Because fate decisions are made at the clonal level, different T cell receptor (TCR) specificities ought to drive distinct developmental and functional outcomes.
Two subsets of CD4+ T lymphocytes are known to regulate adaptive immunity at the intestinal mucosa: peripherally induced regulatory T cells (pTregs) and CD8αα-expressing intraepithelial lymphocytes (CD4IELs) (12–16). Depletion of CD4IELs aggravates intestinal inflammation in mice that lack expression of Foxp3 (14). CD4IELs and pTregs thus cooperate in the regulation of local intestinal inflammation. In specific-pathogen-free (SPF) mice, CD4IELs are present mostly in the small intestinal epithelium (SIE). Their abundance varies with age, diet, and microbiota (17, 18) (fig. S1, A and B and, data S1, A and B). CD4IELs can develop in a microbiota-dependent manner, either from conventional CD4+ T cells or from Treg precursors (11, 14). Germ-free (GF) mice have few if any CD4IELs (14, 19) (fig. S1C). Mice treated with antibiotics show a similar decline in CD4IELs (fig. S1, D and E) (14, 19), whereas fecal transplants from SPF mice restore the CD4IEL population in GF mice to varying extents (fig. S1F) (17, 18). The microbiota therefore contributes not only to the development but also to the maintenance of CD4IELs. However, the microbial antigens recognized by CD4IELs are unknown. Most IEL populations, including CD4IELs, display a restricted TCR repertoire (17, 20, 21). A limited array of antigens might thus suffice to shape this compartment. We used a microbiota-specific transnuclear (TN) monoclonal T cell model (11) to identify naturally occurring TCR ligands of CD4IELs in SPF mice (fig. S2A). T cells from the TN mouse carry a monoclonal TCR, cloned from a pTreg obtained from a mesenteric lymph node (mLN) of a SPF mouse (11). Naïve monoclonal T cells isolated from TN mice populate not only the recipient’s mLNs where they differentiate into pTregs, but also the small intestinal epithelium, where they develop into CD4IELs in a microbiota-dependent fashion (11).
We thus sought to identify the commensal member(s) recognized by TN T cells by selectively growing fecal bacteria derived from Taconic “excluded flora” (EF) mice, which carry a microbiota enriched for the antigen recognized by the TN TCR (11). TN T cells proliferated strongly in the presence of bacterial extracts derived from Bacteroides bile esculin (BBE) agar plates or aerotolerant bacteria recovered from Schaedler blood agar plates (SBA). 16S rRNA sequencing showed enrichment for operational taxonomic units (OTUs) corresponding to the Parabacteroides genus in activating extracts (Fig. 1, A to C, and data S1C). TN T cells proliferated robustly in the presence of Parabacteroides goldsteinii extracts (Fig. 1D), the predominant member of the altered Schaedler flora (ASF) (22). This proliferation was dependent on antigen presentation by dendritic cells (fig. S2B). Neither the related Parabacteroides distasonis nor any other species tested induced proliferation of TN T cells in vitro (Fig. 1D, and fig. S2C). Next, we transferred CD4+ TN T cells into congenic recipients, which were then immunized with bacterial extracts. Proliferation of TN cells occurred in the draining lymph nodes in response to P. goldsteinii extract but not P. distasonis extract or PBS (Fig. 1E). Thus, P. goldsteinii, a commensal abundant in Taconic mice but less so in Jackson (Jax) (fig. S3, A to C, and data S1, D and E), can engage the TN TCR in vivo. TN T cells failed to proliferate and differentiate into CD4IELs in Rag1−/− recipients treated with antibiotics or when colonized with unrelated bacteria (Clostridium tertium) but expanded and differentiated into CD4IELs in mice colonized with P. goldsteinii (Fig. 1F, and fig. S3D). Thus, P. goldsteinii promotes the in vivo development of CD4IELs from naïve T cells carrying the TN TCR.
Fig. 1. P. goldsteinii induces CD4IELs in both monoclonal and polyclonal SPF mice.
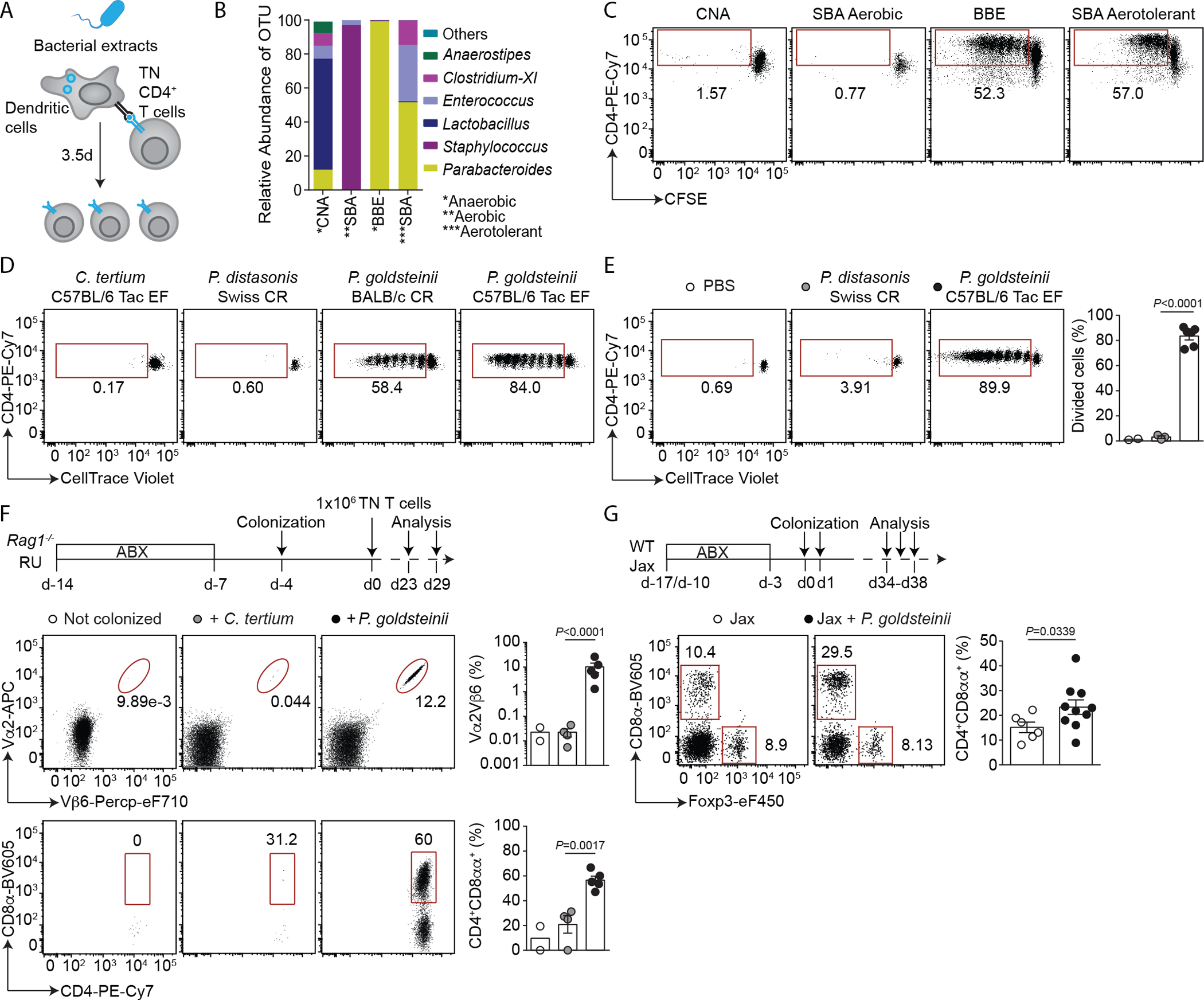
(A) Schematic of the in vitro proliferation assay. (B) 16S rRNA sequencing of isolated bacterial extracts used in C. (C) CFSE dilution among TN cells in response to the indicated bacterial extracts derived from Taconic EF fecal bacteria, isolated using the indicated growth conditions. (D) CellTrace Violet dilution among CD4+ TN T cells in response to the indicated bacteria. (C, D) Representative of flow cytometry dot plots of three experiments. (E) Representative flow cytometry dot plots showing CellTrace Violet dilution (left) and frequency of divided cells (right) among CD4+ TN T cells harvested from the draining inguinal lymph nodes (LNs) of mice immunized with the indicated bacterial extracts (n=2–6 mice per group). (F) Rag1−/− hosts were treated with antibiotics (ABX), then either colonized or not colonized with the indicated bacteria prior to receiving CD4+ TN T cells. Cells from the small intestinal epithelium (SIE) were analyzed by flow cytometry (n=2–5). (top) Experimental design. (middle) Representative flow cytometry dot plots of Vα2 and Vβ6 expression among CD45+ cells and CD4 and CD8α expression among TN cells (Vα2+Vβ6+) in SIE (bottom). (right) Frequency of TN cells among CD45+ cells (top) and frequency of CD4IELs (CD4+CD8αα+) in the SIE among TN CD4+ T cells (bottom) in all mice analyzed in (F). (G) WT Jax mice were treated with ABX and colonized with P. goldsteinii. Cells from the SIE were analyzed by flow cytometry (n=6–10). (top) Experimental design. (bottom) Representative dot plots showing Foxp3 and CD8α expression (left) and frequency of CD4IELs (right) among CD4+ T cells. The graphs in (F, G) show the means ± SEM and each symbol represents a mouse from two to three experiments. P-values were calculated using unpaired two-tailed Student’s t test (E and F) and two-tailed Student’s t test with Welch correction (G). (Tac: Taconic; CR: Charles River; SBA: Schaedler blood agar; BBE: Bacteroides bile esculin; CNA: colistin–nalidixic acid).
We next tested whether P. goldsteinii could induce CD4IELs in wild-type (WT) mice with a polyclonal T cell repertoire. SPF mice obtained from Jax, which naturally harbor few CD4IELs (fig. S1A), developed more CD4IELs upon colonization with P. goldsteinii (Fig. 1G). By contrast, colonization with the unrelated commensal SFB, also absent in Jax mice (10), did not boost CD4IEL frequencies in these mice (fig. S3, E and F). Monocolonization of GF animals with P. goldsteinii was not sufficient to induce CD4IELs however (fig. S4A). One possible explanation is that monocolonized mice have low tissue expression of IFNγ and lack class II MHC (MHCII) on intestinal epithelial cells (fig. S4, B to D), both required for CD4IEL development (20, 23). Thus, P. goldsteinii promotes the accumulation of CD4IELs from both TN and WT precursors in SPF mice.
We next fractionated P. goldsteinii lysates to identify potential TCR ligands derived from this species (fig. S5, A to D). This procedure yielded fractions that activated TN T cells in vitro (fig. S5E). Liquid chromatography with tandem mass spectrometry (LC-MS/MS) analysis of fraction 17 identified 33 proteins, the top 15 of which were expressed recombinantly in Escherichia coli (table S1). Their sonicates served as a source of candidate antigens tested in the proliferation assay. We identified β-N-acetylhexosaminidase (β-hex) as the protein recognized by the TN TCR (Fig. 2A). TN T cells proliferated in mice immunized with protein extract of E. coli expressing P. goldsteinii β-hex, but not when immunized with β-hex derived from closely related species (Fig. 2B). By analyzing the activity of truncated versions of the protein, we defined a ~70-residue stretch that contained the cognate epitope of the TN TCR. Using overlapping peptides, we identified YKGSRVWLN as the minimal epitope (fig. S6, A to D). The corresponding β-hex peptide from Parabacteroides merdae failed to stimulate TN T cells, even though it bound to I-Ab (Fig. 2C; fig. S6, E to G; and table S2). Thus, the P. goldsteinii YKGSRVWLN β-hex epitope is a natural ligand of the TN TCR. Homologous β-hex peptides derived from Bacteroides vulgatus and Bacteroides luti also induced proliferation of TN cells in vitro, but only at higher concentrations (Fig. 2D). Using a combination of BlastP and Jackhmmer analyses (24), we found putative β-hex immunostimulatory sequences in many Bacteroidetes members —the most abundant phylum found exclusively in the gastrointestinal tract (25, 26)—as well as in more distantly related species, such as Spirosoma panaciterrae (fig. S7). Thus, the TN TCR likely recognizes an even broader collection of bacterial species that belong to the Bacteroidetes phylum.
Fig. 2. The TN TCR recognizes epitopes from Bacteroidetes β-N-acetylhexosaminidase in complex with I-Ab.

(A) Candidate polypeptides identified by mass spectrometry were recombinantly expressed in E. coli and bacterial extracts were tested in vitro for their ability to activate TN T cells. Representative dot plots (left) and frequency of divided TN T cells in the presence of tested extracts (right). Graph shows means of two technical replicates. (B and C) In vivo proliferation of CD4+ TN T cells in response to 25 μg of bacterial extracts (B, n=3–7) or 2 μg of peptide (C, n=5). Mutations in the core epitope are indicated in red. (D) Same as (A), using peptide concentrations of 500 nM-50 pM in serial 10-fold dilutions (left). Alignment of sequences homologous to the TN epitope (right). “Activity” is the ability of each peptide to induce proliferation of TN T cells in vitro (see left). (E and F) CD45.1+ recipients were treated with antibiotics (ABX) and received CD45.2+ CD4+ TN T cells. The mice were then either colonized or not colonized with the indicated bacteria (n=7–8). Four weeks post-colonization, small intestine intraepithelium lymphocytes (E) and mesenteric lymph nodes (F) were analyzed. Dot plots show CD8α and Foxp3 expression and graphs show the frequency of CD4IELs (CD4+CD8αα+) in the SIE (E) and mLN Tregs (Foxp3+) (F) among TN CD4+ T cells. In vitro experiments are representative of three experiments (A and D). Dot plots for in vivo experiments show one representative mouse and the graphs all mice analyzed in two experiments. Each symbol represents a mouse and graphs show means ± SEM (B, C, and E and F). P-values were calculated using one-way ANOVA with Dunnett’s post-hoc test in (B, E and F) or unpaired two-tailed Student’s t test in (C). (P.g.: P. goldsteinii; P.d.: P. distasonis; P.m.: P. merdae).
Colonization of SPF mice with P. goldsteinii promoted the expansion and development of CD4IELs and pTregs from TN CD4+ T cells. Likewise, B. vulgatus, which also encodes a TN β-hex epitope (Fig. 2D), supported the expansion and development of TN CD4IELs (Fig. 2, E and F, and fig. S8, A to C). By contrast, colonization of mice with a β-hex–deficient strain of B. vulgatus failed to do so (fig. S8, D to H). Despite similar expression of β-hex along the intestinal tract (fig. S9, A and B), TN cells transferred into SPF mice proliferated predominantly in the jejunum/ileum–draining mLN (fig. S9, C to E). Thus, the β-hex epitope is presented in the distal mLN and is necessary for the differentiation of CD4IELs from TN precursors.
To determine whether this prevalent β-hex epitope is also recognized by intestinal CD4+ lymphocytes of WT SPF mice, we designed P. goldsteinii β-hex–MHCII tetramers spanning the β-hex epitope (table S3) and enumerated β-hex specific CD4+ T cells by flow cytometry (Fig. 3, A to C, and fig. S10, A to L). Splenic CD4+ T cells stained equally poorly with control and β-hex tetramers, whereas β-hex–specific cells were readily identified in both gut-draining mLNs and small intestinal epithelium (Fig. 3, A to C, and fig. S10, A to F). The frequency of β-hex–specific cells was significantly higher among mLNs Tregs of mice housed at both Boston Children’s Hospital and Rockefeller University animal facilities (Fig. 3B and S10G). β-hex–specific cells accounted for up to 3.5% of all CD25+ CD4+ T cells (enriched in Tregs (27)) in mLNs (Fig. 3B). Despite the intrinsically reduced ability of CD4IELs to bind MHCII tetramers (fig. S10H), 40% of β-hex tetramer+ cells in the epithelium were CD4IELs (fig. S10, I and J). The frequencies of β-hex–specific CD4+ T cells varied by mouse, with age, and even between rooms within the same facility, and ranged from 1 to 20% in the epithelium and 0.2 to 2% in the mLN (fig. S10, C to F, I, and K). We then examined the extent to which the commensal–derived β-hex antigen shapes the epithelial lymphocyte compartment (Fig. 3C and fig. S10L). In Jax mice, which carry low frequencies of CD4IELs (fig. S1A) and are virtually devoid of P. goldsteinii (fig. S3, A and B), the proportion of β-hex–specific IELs was lower than in Taconic mice (Fig. 3, C and D). Taconic mice have high frequencies of CD4IELs and are natural carriers of intestinal bacteria encoding the β-hex antigen, including P. goldsteinii and other Bacteroidetes (See fig. S1E, and fig. S3, A to C). Similarly, Jax mice co-housed with Taconic mice were efficiently colonized with P. goldsteinii and showed a higher proportion of β-hex–specific IELs, close to that observed in the Taconic mice themselves (Fig. 3, C and D). Treatment of Jax mice with β-hex peptide slightly increased the frequencies of β-hex–specific IELs, although this difference did not reach statistical significance (Fig. 3C). Thus, the P. goldsteinii β-hex epitope is a prominent natural T cell receptor ligand for mLN Tregs and intraepithelial CD4+ T cells. We conclude that a microbiota rich in P. goldsteinii supports the expansion of commensal-specific IELs using as example the β-hex antigen derived from abundant intestinal commensals such as Parabacteroides and Bacteroides spp.
Fig. 3.
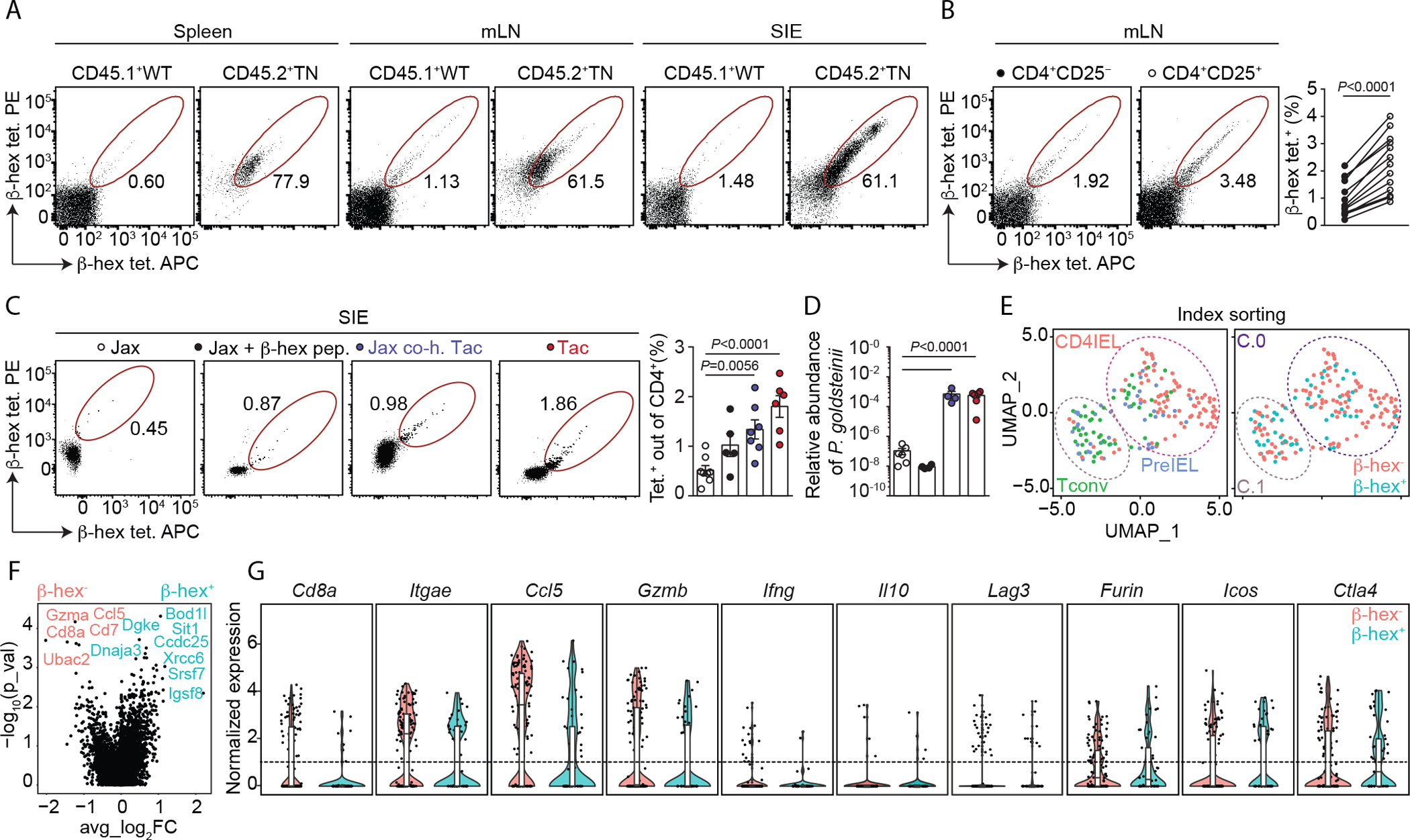
The β-hexosaminidase epitope is recognized by intraepithelial lymphocytes and Tregs. (A) Cells from spleen, mesenteric lymph nodes (mLNs), and the small intestinal epithelium (SIE) were harvested from SPF mice housed at BCH. Cells from CD45.1+ WT mice and CD45.2+ TN mice were mixed at 10:1 (WT:TN) ratio and stained with β-hex tetramers. Gates show the frequency of tetramer+ CD45.1+ or CD45.2+ cells among CD4+ T cells. (B) Representative dot plots of mLNs from Tac SPF mice (n=13) stained with the indicated tetramers (left). Frequencies of tetramer+ cells in the indicated populations among CD4+ T cells isolated from mLNs (right). (C) Tetramer analysis among SIE CD4+ T cells of WT mice sourced from the indicated facilities, co-housed or treated orally with β-hex peptide as indicated (n=5–8 per group). (D) Quantification of P. goldsteinii abundance by qPCR in fecal samples derived from the mice shown in (C). (E-G) scRNAseq analysis of β-hex–specific (β-hex+) and β-hex− CD4+ T cells sorted from SIE of three Taconic SPF mice. (E) Uniform manifold approximation and projection (UMAP) plots according to gene expression analysis showing the distribution of conventional T cells (Tconv, CD103− CD8α−), PreIEL (CD103+CD8α−) and CD4IELs (CD103+CD8α+) according to the index sorting (left) and clustering of β-hex+ and β-hex− cells (right). (F) Volcano plot comparing the gene expression of β-hex+ and β-hex− cells. (G) Expression of selected genes by β-hex+ and β-hex− cells. All mice shown of two (E to G), three (A), five (B), or six (C, D) experiments. Error bars represent means ± SEM. P-values were calculated using paired two-tailed Student’s t tests in (B), one-way ANOVA with Dunnett’s post-hoc test (C, D) and Wilcoxon rank sum test (F, G). (Jax: Jackson; Tac: Taconic; co-h: co-housed).
To define the transcriptional signature of β-hex–specific IELs, we index-sorted β-hex+ and β-hex− IELs from Taconic mice (fig. S11A), and profiled them by single-cell RNA sequencing (scRNAseq) using the SMART-Seq2 methodology (28). This approach enabled us to analyze the gene expression profile of commensal-induced β-hex–specific IELs at homeostasis. We identified two major clusters visualized by Uniform Manifold Approximation and Projection (UMAP) (Fig. 3E and fig. S11B). Cluster 0 contained mostly Cd8a and Itgae (encoding CD103)–expressing cells, and a gene signature typical of CD4IELs, which includes both cytotoxic and regulatory profiles as defined by expression of granzymes and Ctla4 (20, 29). Cluster 1 was composed predominantly of Cd8a− IELs, including preIELs (20, 29) (fig. S11C). β-hex+ and β-hex− IELs were largely indistinguishable from each other and were evenly distributed among the two clusters (Fig. 3E, and fig. S11, D to F). There were no differentially expressed genes between β-hex+ and β-hex− IELs, and both subsets expressed typical IEL markers (Fig. 3, F and G, and fig. S11, E to G), associated with both regulatory (i.e., Ctla4) and cytotoxic (i.e., Gzmb) profiles. We observed a slight enrichment of β-hex+ IELs in cluster 1 compared to cluster 0 (Fig. 3, F and G, and fig. S11D). This enrichment may be related to the reduced ability of CD4IELs to bind MHCII-tetramers (fig. S10H), presumably because of the inhibitory interactions between the thymus leukemia (TL) antigen on epithelial cells and CD8αα on CD4IELs (30). Thus, the gene expression profile of β-hex–specific IELs is representative of all IELs. Consequently, β-hex likely represents one of many possible specificities that lead CD4+ T cells to acquire an IEL phenotype after initial priming by a commensal antigen.
Although CD4IELs express IFNγ and granzyme B, they display a regulatory phenotype—characterized by the expression of IL-10 and Lag3 (11, 20, 29) (Fig. 3G)—and can protect against intestinal inflammation (14–16). To test the anti-inflammatory potential of β-hex–specific T cells, we transferred CD4+ TN T cells into immunodeficient Rag2−/− mice and then induced colitis by WT CD4+ T cells (fig. S12A) (31). WT cells expanded in the intestine, irrespective of whether TN cells were present or not (fig. S12B). TN cells efficiently differentiated into CD4IELs (fig. S12C). In contrast, they poorly differentiated into pTregs (fig. S12D), similar to what has been observed for other commensal-specific T cells transferred into Rag1−/− recipients (32). TN cells are found predominantly in the small intestine and mLNs (11), but in this colitis model, TN cells were also present in the large intestine, where they acquired expression of CD8αα (fig. S12, B and C). Mice that received TN cells were partially protected from colitis induced by colitogenic WT cells. We observed reduced weight loss, improved histopathological scores, and lower levels of fecal lipocalin-2 (LCN-2), a marker of intestinal inflammation (33) (Fig. 4, A to D). Oral administration of β-hex peptide—a procedure that expands TN cells and leads to their differentiation into CD4IELs (fig. S12E)—further reduced intestinal inflammation in recipients of TN and WT CD4+ T cells (Fig. 4, B to D). Similarly, mice that received only colitogenic WT cells and oral β-hex peptide also exhibited decreased intestinal inflammation and increased frequency of total CD4IELs in the small intestine (Fig. 4B and fig. S12C). At an earlier time point, we also observed an increased frequency of total CD4IELs as well as of β-hex–specific IELs, which was accompanied by a decreased frequency of Tregs in the epithelium (fig. S12, F to H). The frequency of WT-derived Tregs was low in all groups, suggesting little contribution by pTregs to such protection (fig. S12D), as has been observed previously (34). To definitively exclude a role for WT Tregs, we induced colitis by transferring T cells from Foxp3DTR mice and depleted Tregs with diphtheria toxin (DT) (Fig. 4, E to H, and fig. S12, I to J). Depletion of Tregs in this colitis model leads to severe weight loss, intestinal inflammation, and death (35). Despite this depletion, TN T cells still protected mice against intestinal inflammation. Although mice lost weight due to toxicity associated with repeated injections of DT (36, 37), there was no difference between the two groups (recipients of Foxp3DTR vs Foxp3WT naïve T cells) (Fig. 4, F to H). Similar to WT Tregs, the proportion of TN T cells that differentiated into Tregs was low but not null in this model (fig. S12D). To evaluate a possible contribution of TN-derived Tregs to the anti-inflammatory response induced by β-hex–specific TN T cells, we ablated Foxp3 directly in pTreg TN/Rag1−/−/Foxp3eGFP zygotes using CRISPR/Cas 9 genome editing. We generated mice with either a 1-bp deletion or a 1-bp insertion frameshift in exon 8 of the Foxp3 gene (fig. S13, A to C). We adoptively transferred splenic CD4+ T cells from these Foxp3-deficient TN mice into Rag2−/− recipients and then induced colitis using WT naïve CD4+ T cells as outlined above (fig. S13D). Foxp3-deficient TN T cells (fig. S13E) conferred partial protection against intestinal inflammation (Fig. 4, I to L). Thus, the anti-inflammatory effect exerted by TN T cells in this model can occur in the absence of Foxp3 expression. Notably, Foxp3-deficient and -sufficient TN T cells were equally able to expand in all intestinal sites surveyed (fig. S13F). In this colitis model, TN T cells differentiated into CD4IELs (up to 80%) and secreted IFNγ and granzyme B, which are hallmarks of IELs, irrespective of the presence of a functional Foxp3 gene in the TN donors (fig. S13, G to I). Thus, β-hex–specific T cells that migrate to, expand within, and differentiate into CD4IELs in response to commensal antigens can protect against intestinal inflammation.
Fig. 4. β-hexosaminidase–specific T cells confer protection against intestinal inflammation.

(A to D) Taconic SPF Rag2−/− either received or did not receive CD45.2+CD4+ TN T cells (day 0) and naïve CD45.1+ CD4+ T cells from WT mice (day 12). On days 2, 4, and 13, some mice received β-hex peptide orally (n=8–10). (A) Percent of initial weight over time. (B) Fecal lipocalin (LCN-2) levels throughout the experiment (n=5–8). (C) Representative photomicrographs of H&E-stained colonic sections from indicated groups. (D) Histology score of H&E-stained sections of colon. (E to H) Same as (A to D) but using either WT (n=7) or Foxp3DTR (n=10) CD4+ T cells (on day 12 post-transfer of TN T cells). These mice received diphtheria toxin (DT) intraperitoneally once per week throughout the experiment. (E) Percent of initial weight over time. (F) Fecal LCN-2 levels throughout the experiment. (G) Representative photomicrographs of H&E-stained colonic sections from indicated groups. (H) Histology score of H&E-stained sections of colon. (I to L) Same as (A to D) but using either WT (n=5) or Foxp3−/− TN CD4+ cells (n=5, day 0). (I) Percent of initial weight over time. (J) Fecal LCN-2 levels. (K) Representative photomicrographs of H&E-stained sections of colon of indicated groups. (L) Histology scores of H&E-stained sections of colon. (A to L). All mice analyzed in two experiments. Graphs show means ± SEM. P-values were calculated using two-way ANOVA for repeated measures with Tukey post-hoc test in (A, B, E, F, I, and J), Mann-Whitney test (H) and Kruskal–Wallis test followed by Dunn’s multiple comparisons post-hoc test (D, L). (C and G) Scale bar: 200 μm (top), 60 μm (bottom). (K) Scale bar: 200 μm (top), 100 μm (bottom). Low- and high-magnification photomicrographs show two representative mice in each group (C and G and K). ns, not significant.
Commensal bacteria shape the differentiation and function of intestinal T cells (1), including CD4IELs. We show that P. goldsteinii and other Bacteroidetes can induce antigen-specific differentiation of CD4IELs. It is possible that a fraction of β-hex–specific pTregs generated in the mLNs migrate to the epithelium. There, they can further differentiate into preIELs or CD4IELs after re-encountering their cognate antigen (14, 20). Because CD4+ T cells require local engagement with MHCII to differentiate into CD4IELs (20), recognition of an antigen such as β-hex, present across a range of abundant commensals, may provide CD4IEL precursors with a competitive advantage to populate the intestinal epithelium. Indeed, the same β-hex–specific TCR rearrangement found in the TN strain is present among CD4IELs and their epithelial precursors in mice that carry only the Vβ6 chain of the TN TCR and a polyclonal TCRα chain (20). Although we have identified β-hex as a natural ligand of intraepithelial CD4+ T cells, it is likely that other commensal-derived antigens are also recognized by these cells. Because activation of CD4IELs is attenuated by interactions between TL antigen expressed by intestinal epithelial cells and CD8αα on IELs (38), antigen abundance and TCR affinity may play a role in the development of CD4IELs. In addition, commensals such as P. goldsteinii and B. vulgatus may not only provide TCR ligands, but also metabolites essential for the creation of an environment that supports the development and maintenance of regulatory IELs. Here we showed that β-hex–specific T cells that reside in the epithelium can protect against intestinal inflammation. The TCR-titratable regulation imposed by the interaction between the TL antigen and CD8αα (38) may control the anti-inflammatory and cytotoxic functions of CD4IELs. Deciphering the rules that govern the mutualism between commensals, pathobionts, and immune cells is essential for a better understanding of homeostasis and inflammation at the intestinal mucosa.
Supplementary Material
Acknowledgments:
We thank J. Leube for helping with culture experiments, P. Silver for access to anaerobic chambers, C. McClune for assistance with bioinformatic analyses, S. Jain, J. Bhagat and M. Suarez-Farinas for Statistical consultation, and O. Venezia from the Moon lab for helping with tetramer production. We are grateful to M. Fischbach for supporting D.B. in performing experiments during revision of this manuscript, M. Laub and D. Irvine for guidance throughout the project. We thank S. Kolifrath, A. Rogoz, and S. Gonzalez for mouse husbandry, R. Mathieu from the flow cytometry facilities of BCH and K. Gordon from RU, E. Spooner (WIBR) for mass spectrometry, R. Bronson (Harvard) and Y. Alvarez (RU) and the NYU Histology core facility for histology and colitis scoring, and the NIH tetramer core facility for providing tetramers. We thank members of the Ploegh, Victora, and Mucida laboratories for discussions and suggestions.
Funding:
H.L.P. was supported by CMIT and D.M. by FASI/FARE. D.B. and H.L.P. were supported by NIH R01AI087879 and DP1AI150593-01. A.M.B., M.L., and D.M. by NIH R01DK093674-07, R01DK113375 and R01DK093674. T.B.R.C., J.B. and G.D.V. by NIH DP1AI144248. J.J.M was supported by NIH R01DK126910 and P30DK043351, and MGH ECOR. D.B. is currently funded by an Early Postdoc Mobility Fellowship from the Swiss National Science Foundation.
Footnotes
Competing interests: E.J.A. is a consultant and has equities at Finch therapeutics. The other authors declare no competing interests. T.K.L. is a co-founder of Senti Biosciences, Synlogic, Engine Biosciences, Tango Therapeutics, Corvium, BiomX, Eligo Biosciences, Bota.Bio, Avendesora, and NE47Bio. T.K.L. also holds financial interests in nest.bio, Armata, IndieBio, CognitoHealth, Quark Biosciences, Personal Genomics, Thryve, Lexent Bio, MitoLab, Vulcan, Serotiny, Avendesora, Pulmobiotics, Provectus Algae, Invaio, and NSG Biolabs.
Data and materials availability:
All data are available in the main text or the supplementary materials. Raw and processed scRNAseq data can be accessed at the Gene Expression Omnibus (GEO) under accession number GSE198037.
References and Notes
- 1.Belkaid Y, Harrison OJ, Homeostatic Immunity and the Microbiota. Immunity 46, 562–576 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ivanov II, Honda K, Intestinal commensal microbes as immune modulators. Cell Host Microbe 12, 496–508 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Geuking MB, Burkhard R, Microbial modulation of intestinal T helper cell responses and implications for disease and therapy. Mucosal Immunology 13, 855–866 (2020). [DOI] [PubMed] [Google Scholar]
- 4.Ansaldo E et al. , Akkermansia muciniphila induces intestinal adaptive immune responses during homeostasis. Science 364, 1179–1184 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sefik E et al. , MUCOSAL IMMUNOLOGY. Individual intestinal symbionts induce a distinct population of RORgamma(+) regulatory T cells. Science 349, 993–997 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lathrop SK et al. , Peripheral education of the immune system by colonic commensal microbiota. Nature 478, 250–254 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang Y et al. , Focused specificity of intestinal TH17 cells towards commensal bacterial antigens. Nature 510, 152–156 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu M et al. , c-MAF-dependent regulatory T cells mediate immunological tolerance to a gut pathobiont. Nature 554, 373–377 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chai JN et al. , Helicobacter species are potent drivers of colonic T cell responses in homeostasis and inflammation. Sci Immunol 2, eaal5068 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ivanov II et al. , Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139, 485–498 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bilate AM et al. , Tissue-specific emergence of regulatory and intraepithelial T cells from a clonal T cell precursor. Sci Immunol 1, eaaf7471 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bilate AM, Lafaille JJ, Induced CD4+Foxp3+ regulatory T cells in immune tolerance. Annu Rev Immunol 30, 733–758 (2012). [DOI] [PubMed] [Google Scholar]
- 13.Shale M, Schiering C, Powrie F, CD4(+) T-cell subsets in intestinal inflammation. Immunol Rev 252, 164–182 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sujino T et al. , Tissue adaptation of regulatory and intraepithelial CD4+ T cells controls gut inflammation. Science 352, 1581–1586 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reis BS, Rogoz A, Costa-Pinto FA, Taniuchi I, Mucida D, Mutual expression of the transcription factors Runx3 and ThPOK regulates intestinal CD4(+) T cell immunity. Nat Immunol 14, 271–280 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Basu J et al. , Essential role of a ThPOK autoregulatory loop in the maintenance of mature CD4(+) T cell identity and function. Nat Immunol 22, 969–982 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cervantes-Barragan L et al. , Lactobacillus reuteri induces gut intraepithelial CD4(+)CD8alphaalpha(+) T cells. Science 357, 806–810 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Umesaki Y, Setoyama H, Matsumoto S, Okada Y, Expansion of alpha beta T-cell receptor-bearing intestinal intraepithelial lymphocytes after microbial colonization in germ-free mice and its independence from thymus. Immunology 79, 32–37 (1993). [PMC free article] [PubMed] [Google Scholar]
- 19.Mucida D et al. , Transcriptional reprogramming of mature CD4(+) helper T cells generates distinct MHC class II-restricted cytotoxic T lymphocytes. Nat Immunol 14, 281–289 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bilate AM et al. , T Cell Receptor Is Required for Differentiation, but Not Maintenance, of Intestinal CD4(+) Intraepithelial Lymphocytes. Immunity 53, 1001–1014 e1020 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wojciech L et al. , Non-canonicaly recruited TCRalphabetaCD8alphaalpha IELs recognize microbial antigens. Sci Rep 8, 10848 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dewhirst FE et al. , Phylogeny of the defined murine microbiota: altered Schaedler flora. Appl Environ Microbiol 65, 3287–3292 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reis BS, Hoytema van Konijnenburg DP, Grivennikov SI, Mucida D, Transcription factor T-bet regulates intraepithelial lymphocyte functional maturation. Immunity 41, 244–256 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eddy SR, Accelerated Profile HMM Searches. PLoS Comput Biol 7, e1002195 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wexler AG, Goodman AL, An insider’s perspective: Bacteroides as a window into the microbiome. Nat Microbiol 2, 17026 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.C. Human Microbiome Project, Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fontenot JD, Gavin MA, Rudensky AY, Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat Immunol 4, 330–336 (2003). [DOI] [PubMed] [Google Scholar]
- 28.Trombetta JJ et al. , Preparation of Single-Cell RNA-Seq Libraries for Next Generation Sequencing. Curr Protoc Mol Biol 107, 4 22 21–24 22 17 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.London M, Bilate AM, Castro TBR, Sujino T, Mucida D, Stepwise chromatin and transcriptional acquisition of an intraepithelial lymphocyte program. Nat Immunol 22, 449–459 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Olivares-Villagomez D et al. , Thymus leukemia antigen controls intraepithelial lymphocyte function and inflammatory bowel disease. Proc Natl Acad Sci U S A 105, 17931–17936 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Powrie F, Leach MW, Mauze S, Caddle LB, Coffman RL, Phenotypically distinct subsets of CD4+ T cells induce or protect from chronic intestinal inflammation in C. B-17 scid mice. Int Immunol 5, 1461–1471 (1993). [DOI] [PubMed] [Google Scholar]
- 32.Nutsch K et al. , Rapid and Efficient Generation of Regulatory T Cells to Commensal Antigens in the Periphery. Cell Rep 17, 206–220 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chassaing B et al. , Fecal lipocalin 2, a sensitive and broadly dynamic non-invasive biomarker for intestinal inflammation. PLoS One 7, e44328 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Izcue A et al. , Interleukin-23 restrains regulatory T cell activity to drive T cell-dependent colitis. Immunity 28, 559–570 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boehm F et al. , Deletion of Foxp3+ regulatory T cells in genetically targeted mice supports development of intestinal inflammation. BMC Gastroenterol 12, 97 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dombrowski Y et al. , Regulatory T cells promote myelin regeneration in the central nervous system. Nat Neurosci 20, 674–680 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goldwich A, Steinkasserer A, Gessner A, Amann K, Impairment of podocyte function by diphtheria toxin--a new reversible proteinuria model in mice. Lab Invest 92, 1674–1685 (2012). [DOI] [PubMed] [Google Scholar]
- 38.Olivares-Villagomez D, Van Kaer L, TL and CD8alphaalpha: Enigmatic partners in mucosal immunity. Immunol Lett 134, 1–6 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miura H, Quadros RM, Gurumurthy CB, Ohtsuka M, Easi-CRISPR for creating knock-in and conditional knockout mouse models using long ssDNA donors. Nat Protoc 13, 195–215 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brunkow ME et al. , Disruption of a new forkhead/winged-helix protein, scurfin, results in the fatal lymphoproliferative disorder of the scurfy mouse. Nat Genet 27, 68–73 (2001). [DOI] [PubMed] [Google Scholar]
- 41.Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R, Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell 118, 229–241 (2004). [DOI] [PubMed] [Google Scholar]
- 42.Kino Y et al. , Counterselection employing mutated pheS for markerless genetic deletion in Bacteroides species. Anaerobe 42, 81–88 (2016). [DOI] [PubMed] [Google Scholar]
- 43.Mimee M, Tucker AC, Voigt CA, Lu TK, Programming a Human Commensal Bacterium, Bacteroides thetaiotaomicron, to Sense and Respond to Stimuli in the Murine Gut Microbiota. Cell Syst 1, 62–71 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Simon MD et al. , Rapid flow-based peptide synthesis. Chembiochem 15, 713–720 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sidney J et al. , Measurement of MHC/peptide interactions by gel filtration or monoclonal antibody capture. Curr Protoc Immunol Chapter 18, Unit 18 13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cheng Y, Prusoff WH, Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem Pharmacol 22, 3099–3108 (1973). [DOI] [PubMed] [Google Scholar]
- 47.Gulukota K, Sidney J, Sette A, DeLisi C, Two complementary methods for predicting peptides binding major histocompatibility complex molecules. J Mol Biol 267, 1258–1267 (1997). [DOI] [PubMed] [Google Scholar]
- 48.Valdez Y et al. , Nramp1 drives an accelerated inflammatory response during Salmonella-induced colitis in mice. Cell Microbiol 11, 351–362 (2009). [DOI] [PubMed] [Google Scholar]
- 49.Moon JJ et al. , Quantitative impact of thymic selection on Foxp3+ and Foxp3− subsets of self-peptide/MHC class II-specific CD4+ T cells. Proc Natl Acad Sci U S A 108, 14602–14607 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dobin A et al. , STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li B, Dewey CN, RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stuart T et al. , Comprehensive Integration of Single-Cell Data. Cell 177, 1888–1902 e1821 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Maeda H et al. , Quantitative real-time PCR using TaqMan and SYBR Green for Actinobacillus actinomycetemcomitans, Porphyromonas gingivalis, Prevotella intermedia, tetQ gene and total bacteria. FEMS Immunol Med Microbiol 39, 81–86 (2003). [DOI] [PubMed] [Google Scholar]
- 54.Gomes-Neto JC et al. , A real-time PCR assay for accurate quantification of the individual members of the Altered Schaedler Flora microbiota in gnotobiotic mice. J Microbiol Methods 135, 52–62 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Matsuki T et al. , Development of 16S rRNA-gene-targeted group-specific primers for the detection and identification of predominant bacteria in human feces. Appl Environ Microbiol 68, 5445–5451 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available in the main text or the supplementary materials. Raw and processed scRNAseq data can be accessed at the Gene Expression Omnibus (GEO) under accession number GSE198037.


