DNA-damaging cancer therapies induce delayed activation of cGAS–STING-dependent type I interferon signaling in the heart. DNA damage–induced cGAS–STING signaling drives cardiac inflammation and late pathologic remodeling of the heart, progressive cardiac functional impairment, and late lethal cardiac toxicity.
Abstract
Late cardiac toxicity is a potentially lethal complication of cancer therapy, yet the pathogenic mechanism remains largely unknown, and few treatment options exist. Here we report DNA-damaging agents such as radiation and anthracycline chemotherapies inducing delayed cardiac inflammation following therapy due to activation of cGAS- and STING-dependent type I interferon signaling. Genetic ablation of cGAS–STING signaling in mice inhibits DNA damage–induced cardiac inflammation, rescues late cardiac functional decline, and prevents death from cardiac events. Treatment with a STING antagonist suppresses cardiac interferon signaling following DNA-damaging therapies and effectively mitigates cardiac toxicity. These results identify a therapeutically targetable, pathogenic mechanism for one of the most vexing treatment-related toxicities in cancer survivors.
Introduction
Radiation therapy (RT) and anthracycline chemotherapeutics such as doxorubicin are central to the curative treatment regimens of many cancers. These treatments elicit cytotoxic effects in cancer cells through creation of DNA double-stranded breaks and are often used individually or in combination to treat cancer of the breast, thorax, Hodgkin’s, and non-Hodgkin’s lymphomas (Dühmke et al., 2001; Darby et al., 2011; Rodrigues et al., 2015). As cancer long-term survival continues to increase, late normal tissue toxicities, especially late cardiac toxicity, are a major source of long-term morbidity and mortality in cancer survivors (Aleman et al., 2003; Swain et al., 2003). As such, there is a growing need to identify mechanisms of late normal tissue toxicity and develop therapeutic strategies to mitigate these late effects. RT-induced cardiac toxicity doubles with every gray of mean heart dose following treatment for breast cancer (Darby et al., 2013) and its lifetime occurrence was found to be at 8% in long-term survivors of Hodgkin’s lymphoma, fivefold higher than the general population (Aleman et al., 2007). Similarly, the risk of late anthracycline-induced cardiac toxicity exceeds 35% with cumulative doxorubicin doses over 600 mg/m2 (Mitry and Edwards, 2016).
While the acute effects of DNA double-stranded breaks from RT and anthracyclines are well known, the mechanisms by which DNA damage leads to normal tissue remodeling and late tissue toxicity years after cancer treatment are poorly understood (De Ruysscher et al., 2019). The generation of reactive oxygen species and topoisomerase-2β–mediated mitochondrial dysfunction as well as apoptosis of cardiomyocytes following treatment have been implicated in the pathogenesis of cardiac toxicity following RT and anthracyclines (Cappetta et al., 2017; Zhang et al., 2012). Concurrent administration of dexrazoxane, an iron chelator that reduces superoxide radical generation and inhibits topoisomerase-2β, is an FDA-approved therapy for prevention of anthracycline-associated cardiac toxicity (Curigliano et al., 2020). However, the intervening mechanisms connecting acute tissue stress to delayed and chronic tissue damage are unclear. Experimental data have shown that DNA damage induces sterile inflammation in tumors and in tissue (Chen and Nuñez, 2010). However, studies of inflammatory signaling following DNA-damaging therapies have predominantly focused on TGF-β and downstream pro-inflammatory cytokines and have yet to translate into effective strategies to mitigate late normal tissue toxicity.
Results
An experimental model of DNA damage–induced cardiac toxicity
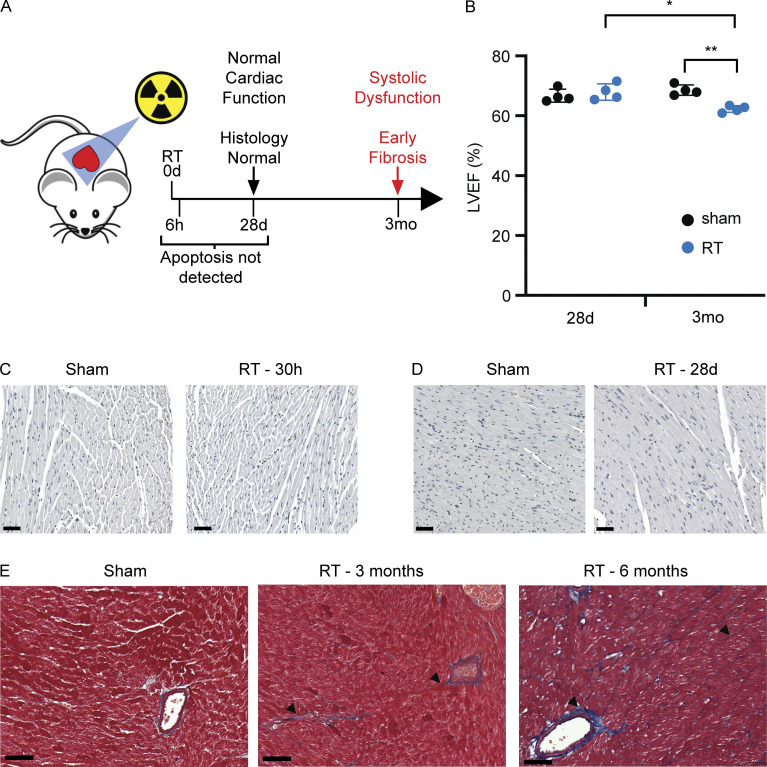
Since late normal tissue toxicity likely stems from a combination of cell intrinsic and microenvironmental responses to cardiac DNA damage, we developed an animal model of DNA damage–induced cardiac toxicity to examine the temporal response to cardiac DNA damage. Mice were treated with a single dose of cardiac RT using an image-guided radiotherapy platform to isolate high-dose exposure to the heart and minimize dose to other organs. In this model, similar to prior studies in rodent models of radiation exposure, cardiac systolic dysfunction develops within 3–6 mo of cardiac radiation, as measured by echocardiography (Fig. 1, A and B; Dreyfuss et al., 2021; Mezzaroma et al., 2012; Schlaak et al., 2019). We observed no evidence of radiation-induced apoptosis of cardiomyocytes at 30 h after RT and 28 d after RT (Fig. 1, C and D), markedly contrasting with extensive apoptosis evident in the spleen (Fig. S1 A). Cardiac systolic function remains normal 1 mo after RT but systolic dysfunction becomes apparent in this model beginning 3 mo after RT exposure and is associated with increased interfiber and perivascular collagen deposition (fibrosis) that continues to progress from the period of 3 to 6 mo after RT (Fig. 1 E). These features are similar to histopathologic and cardiophysiologic effects observed in late cardiac toxicity in cancer patients following exposure of the heart to high doses of RT, confirming that this experimental system accurately models crucial aspects of the pathophysiologic process observed in cancer patients (Burns et al., 1983; Gomez et al., 1983; Gustavsson et al., 1990; Savage et al., 1990; Stewart et al., 1995). Since late cardiac toxicity occurs in a significantly delayed manner relative to the time of DNA damage exposure, we reasoned that a delayed DNA damage–induced pathogenic signal is likely at work driving cardiac pathogenesis 4 wk after RT. Crucially, identification of a pathogenic signaling process prior to overt cardiac dysfunction would provide an opportunity to therapeutically intervene to prevent toxicity.
Figure 1.
A mouse model of cardiac toxicity following DNA damage. (A) Schematic representation of experimental model of radiation-induced cardiac toxicity and timeline cardiophysiologic, cellular, and histopathologic changes after RT. (B) LVEF in four individual irradiated and four individual sham-treated wildtype mice 1 and 3 mo after treatment. While LVEF was similar between irradiated and sham-treated mice 1 mo after RT, all irradiated mice had declines in LVEF by 3 mo after RT. N = 4 for each treatment condition, repeated twice. *P = 0.008, **P = 0.001, two-tailed Student’s T test. (C and D) TUNEL stained left ventricle myocardium 30 h (C) and 28 d (D) after RT or sham treatment. (E) Left ventricle myocardium stained with Masson’s trichrome in mice 3 and 6 mo after cardiac radiation shows progressive accumulation of perivascular and interfiber collagen (blue) with fibrosis (arrowheads) in comparison with a sham-treated Sting+/+ mouse. Scale bar = 50 μm. Experiment performed with N = 4 mice for each treatment condition and timepoint; representative images shown, repeated once.
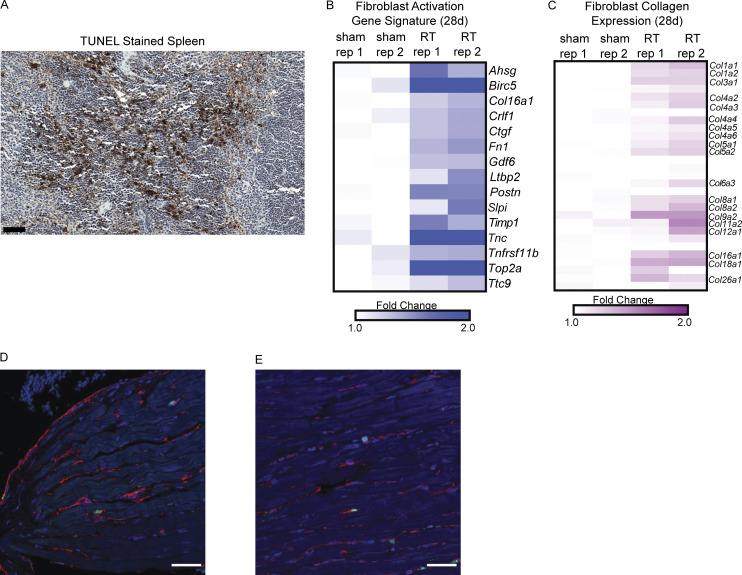
Figure S1.
A mouse model of radiation-induced cardiac toxicity. (A) TUNEL staining in mouse spleen as positive control for cellular apoptosis in tissue. Scale bar = 50 μm. (B) Expression of genes associated with fibroblast activation in cardiac fibroblasts from C57BL/6 as defined in Park. et al. (2018) in fibroblasts isolated from hearts 28 d after sham treatment or RT. (C) Expression of collagen genes in fibroblasts isolated from hearts 28 d after sham treatment or RT. (D and E) Low magnification (40×) immunofluorescence images of hearts sections of Sting+/+ mice 28 d after sham treatment (D) and cardiac RT (E). cGAS, green; Vimentin, red; DAPI, blue. Experiment performed with N = 4 mice for each treatment condition and timepoint; representative image shown.
Delayed activation of type I IFN signaling in fibroblasts after DNA damage
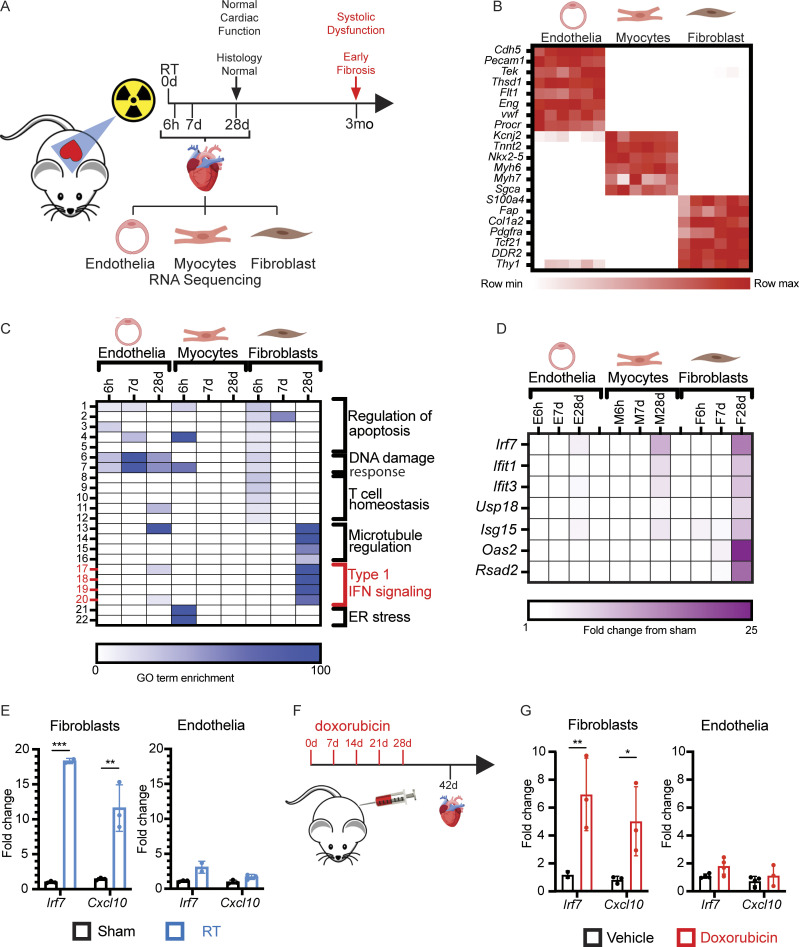
To distinguish between sub-acute to chronic signals that may drive cardiac toxicity from acute, transient DNA damage responses, we examined the transcriptional response in the principle cardiac cell populations of the heart from 6 h to 4 wk following cardiac irradiation. Mice were euthanized at timepoints from 6 h to 28 d following treatment with a single dose of 12 Gy RT, at which time hearts tissues were minced and dissociated to single cells suspensions, and cardiomyocytes, endothelia, and fibroblasts were isolated and RNA extracted for gene expression analyses by RNA-seq (Fig. 2 A). Comparison of tissue-specific gene expression across specimens confirmed successful enrichment for each tissue population (Fig. 2 B). The acute response to cardiac DNA damage at 6 h was similar across all three cell populations, driven by expression of p53-regulated genes involved in the DNA damage response, apoptosis regulation, and cell cycle regulation, all of which subsequently waned by 28 d following DNA damage (Fig. 2 A and Table S1). While radiation induced acute, transient expression of apoptotic regulators, we found no evidence of apoptosis 30 h or 28 d after RT (Fig. 1, B and C), consistent with prior studies demonstrating consistent activation of p53-dependent apoptosis regulators across cell types, including in tissues highly resistant to acute toxicity from radiation, including the heart (Tanikawa et al., 2017). In contrast, 28 d after RT exposure, cardiac fibroblasts have increased expression of the vast majority of genes that were previously associated with an activated, pro-fibrotic cardiac fibroblast phenotype in C57BL/6 mice (Fig. S1 B) and demonstrate increased expression of many collagen genes (Fig. S1 C), consistent with the radiation-induced, pro-fibrotic phenotype observed in this model (Fig 1 E; Park et al., 2018).
Figure 2.
Delayed activation of type I IFN signaling after cardiac DNA damage. (A) Schematic representation of timeline of tissue collection relative to RT and histopathologic changes and adverse physiologic effects on the heart. (B) Heatmap of gene expression of cell-specific markers in endothelia, myocytes, and fibroblasts quantified by RNA-seq. (C) Heatmap of GO term fold enrichment in endothelia, myocytes, and fibroblasts at 6 h, 7 d, and 28 d after cardiac RT. (D) Expression heatmap of type I IFN–related genes at 6 h, 7 d, and 28 d after RT in endothelial cells, myocytes, and fibroblasts. (B–D) N = 2 biologic replicates for each timepoint and each treatment condition with each biologic replicate containing pooled cell populations from four individual hearts. (E) Gene expression of Irf7 and Cxcl10 measured by qRT-PCR in endothelial cells and fibroblasts for 28 d after 12 Gy cardiac RT or sham treatment. N = 2–3 for presented data from one representative experiment; four independent experiments were performed. (F) Schematic representation of mouse model of doxorubicin-induced cardiac toxicity. (G) qRT-PCR of endothelial cells and fibroblasts for Irf7 and Cxcl10 14 d after completing 5 wk doses of doxorubicin or vehicle only. N = 2–3 for presented data from one representative experiment; four independent experiments were performed. *P < 0.05, **P < 0.001, ***P < 0.0001, two-tailed T test.
Gene ontology (GO) enrichment analysis also revealed substantial enrichment of genes associated with IFN responses and anti-viral signaling, such as Irf7, amongst genes induced 28 d after RT, a response that was largely restricted to cardiac fibroblasts (Fig. 2, C–E). We next examined whether DNA damage–induced IFN signaling in hearts also occurred in response to DNA damage by anthracyclines. Cardiac endothelial cells and fibroblasts were isolated from mice 2 wk after completion of treatment with 5 wk doses of doxorubicin (Fig. 2 F), a regimen previously shown to induce cardiac toxicity (Zhang et al., 2012). Similar to mice after cardiac RT, cardiac fibroblasts, but not cardiac endothelial cells, responded to DNA damage with delayed activation of IFN signaling and upregulation of Irf7 (Fig. 2 G). Therefore, delayed activation of IFN stimulated genes (ISGs) specifically in cardiac fibroblasts is a common response to heart exposure to DNA-damaging agents.
Cytosolic DNA and cGAS–STING drive type I IFN signaling
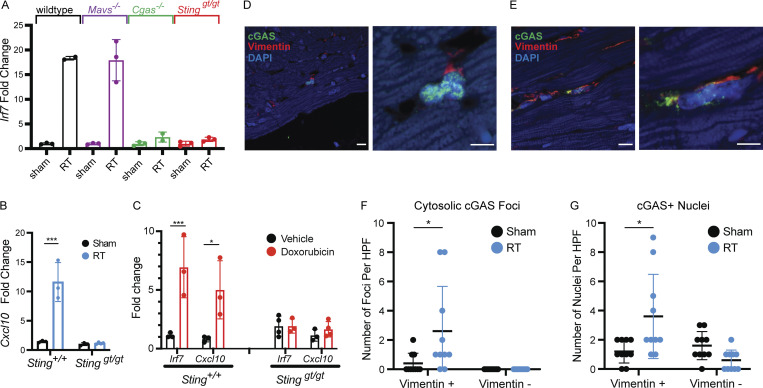
Given the strong activation of a pro-inflammatory, type I IFN response following RT and doxorubicin, we sought to determine the mechanism of this activation. Multiple endogenous molecules, collectively referred to as damage-associated molecular patterns, have been shown to drive a robust IFN activation (Gong et al., 2020). Given the DNA damaging action of both radiation and doxorubicin, we reasoned that this type I IFN response may be driven by intracellular recognition of damaged nucleic acid through intracellular pattern recognition receptors (Roers et al., 2016; Zhang et al., 2020). Mice deficient in cytosolic double-stranded RNA signaling (Mavs−/−) still exhibited strong Irf7 induction following RT, suggesting that detection of cytosolic double-stranded RNA is not the driver of response (Fig. 3 A). However, ablation of the cytosolic dsDNA recognition pathway in Cgas−/− and Stinggt/gt mice completely prevented Irf7 and Cxcl10 induction following RT (Fig. 3, A and B). Similarly, absence of Irf7 and Cxcl10 induction was noted in Stinggt/gt mice following doxorubicin administration (Fig. 3 C). Pathogenic cytosolic DNA is detected by cyclic GMP-AMP synthase (cGAS), which signals to stimulator of IFN response cGAMP interactor 1 (STING) through production of the second messenger cyclic guanisine monophosphate-adenosine monophosphate (cyclic GMP-AMP; Kwon and Bakhoum, 2020). Prior reports have revealed that recruitment of cGAS to cytosolic DNA generates cGAS foci (Li et al., 2021; Motwani et al., 2019). Since cGAS–STING-dependent ISG induction following DNA damage was restricted largely to cardiac fibroblasts, we next examined cardiac tissue for cytosolic cGAS foci following RT. Using immunofluorescence imaging of heart tissue 28 d following RT, both nuclear and cytosolic cGAS foci were increased in vimentin-positive cells, a marker of cardiac fibroblasts, but not in vimentin-negative cells (Fig. 3, D–G; and Fig. S1, D and E).
Figure 3.
Type I IFN signaling after DNA damage requires cGAS and STING. (A) Irf7 expression measured by qRT-PCR in fibroblasts of mice genetically deficient in different components of pattern recognition receptor signaling, 28 d after cardiac RT. N = 2–3 for presented data from one representative experiment, three independent experiments were performed. (B) Cxcl10 expression measured by qRT-PCR in fibroblasts of Sting+/+ vs. Stinggt/gt 28 d after cardiac RT. N = 3 for presented data from one representative experiment; three independent experiments were performed. (C) Irf7 and Cxcl10 expression measured by qRT-PCR in fibroblasts of Sting+/+ vs. Stinggt/gt 14 d after completing 5 wk doses of doxorubicin. N = 3–4 for presented data from one representative experiment; four independent experiments were performed. (D) Immunofluorescence for cGAS (green) and Vimentin (red) in heart sections of Sting+/+ mice 28 d after sham treatment. N = 4 hearts imaged with immunofluorescence. (E) Immunofluorescence for cGAS (green) and Vimentin (red) in heart sections Sting+/+ mice 28 d after 20 Gy cardiac RT. N = 4 hearts imaged with immunofluorescence. (F) Quantification of the number of cells with cytosolic cGAS-positive foci in sham-treated and RT-treated Sting+/+ mice. Each measurement represents number of cells in one high-powered field of view. (G) Quantification of the number of cells with cGAS-positive nuclei in sham-treated and RT-treated Sting+/+ mice. Each measurement represents number of cells in one high-powered field of view. (F and G) N = 4 for sham and N = 4 for RT. Quantification represents the mean number of foci per high-power field (HPF) from four HPF images each of the four hearts imaged. *P < 0.05, ***P < 0.001, two-tailed T test.
DNA damage–induced, STING-dependent type I IFN signaling remodels the heart microenvironment
Expression of ISGs after DNA damage raised the possibility that IFN signaling could remodel the inflammatory infiltrate in hearts in response to DNA-damaging cancer therapy. We performed single-cell RNA sequencing (scRNA-seq) on CD45+ leukocytes isolated from the mouse heart to determine whether cardiac DNA damage and ensuing cGAS–STING signaling cooperate to remodel the leukocyte repertoire in the heart (Fig. 4 A). At 28 d after cardiac RT, when cGAS foci and ISG induction is apparent in cardiac fibroblasts, IFN-activated monocytes and macrophages, dendritic cells, and B cells are recruited to the hearts of Sting+/+ mice, but this response is attenuated in the hearts of Stinggt/gt mice (Fig. 4, B–D; Fig. S2, A and B; and Table S2). These same cell populations are recruited to the hearts of mice following myocardial infarction and have been associated with pathologic cardiac remodeling and fibrosis leading to loss of cardiac function and worsened survival (Cao et al., 2018; King et al., 2017; Lee et al., 2018; Zouggari et al., 2013). In addition to recruiting IFN-activated leukocytes to the heart, RT-induced STING signaling increased the expression of multiples ISGs in several cardiac leukocyte populations (Fig. 4 E). Therefore, cGAS–STING-dependent type I IFN signaling induced by cardiac DNA damage drives inflammation in the cardiac microenvironment, which is associated with cardiac pathologic processes and death from cardiac events.
Figure 4.
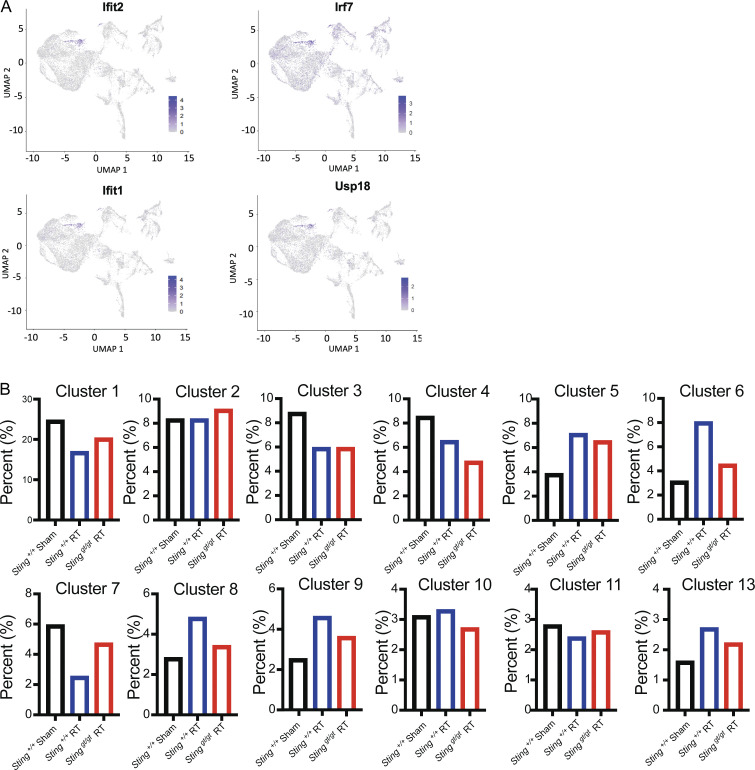
Inflammatory remodeling of the heart microenvironment by STING signaling following DNA damage. (A) UMAP defined clusters of cells from scRNA-seq of CD45-positive leukocytes isolated from the myocardium of mice 28 d after cardiac RT. (B) Dot plot of expression of select IFN associated genes in each cell cluster. Cluster 12 identified IFN-induced (IFNIC) monocytes and macrophages. NK, natural killer. (C) Histogram of the expression of ISGs in each scRNA-seq cluster. (D) Cluster 12 (IFNIC macrophages and monocytes) as a percentage of the total cell population in Sting+/+ after 28 d after sham treatment, Sting+/+ mice 28 d after cardiac RT, and Stinggt/gt mice 28 d after cardiac RT. Data represent two independent biologic replicates each with CD45+ cell isolated from four mice for each condition; scRNA-seq performed once. (E) Histogram of the expression of ISGs in each scRNA-seq cluster in mice of indicated genotype and treatment condition. IFNIC macrophages highlighted. Data in each biologic condition represent two independent scRNA-seq libraries, each composed of CD45+ cells isolated from four mice.
Figure S2.
Characteristics of DNA damage and STING recruited leukocyte population in the heart. (A) Expression of ISGs is especially associated with cells in cluster 12, the IFNIC monocytes and macrophages, with expression of each cell overlaid on the UMAP projections. (B) Extending from Fig. 4 D, with data from each additional cluster identified in UMAP. Plotting the percentage of each cluster of the total cell population of CD45+ cells in Sting+/+ 28 d after sham treatment, Sting+/+ mice 28 d after cardiac RT, and Stinggt/gt mice 28 d after cardiac RT. In addition to IFNIC monocytes and macrophages, we also observed increased STING-dependent recruitment of B cells (clusters 6) and dendritic cells (cluster 8) 1 mo after DNA damage. Data in each biologic condition represent two independent scRNA-seq libraries, each composed of CD45+ cells isolated from four mice.
STING signaling drives cardiac toxicity after DNA damage
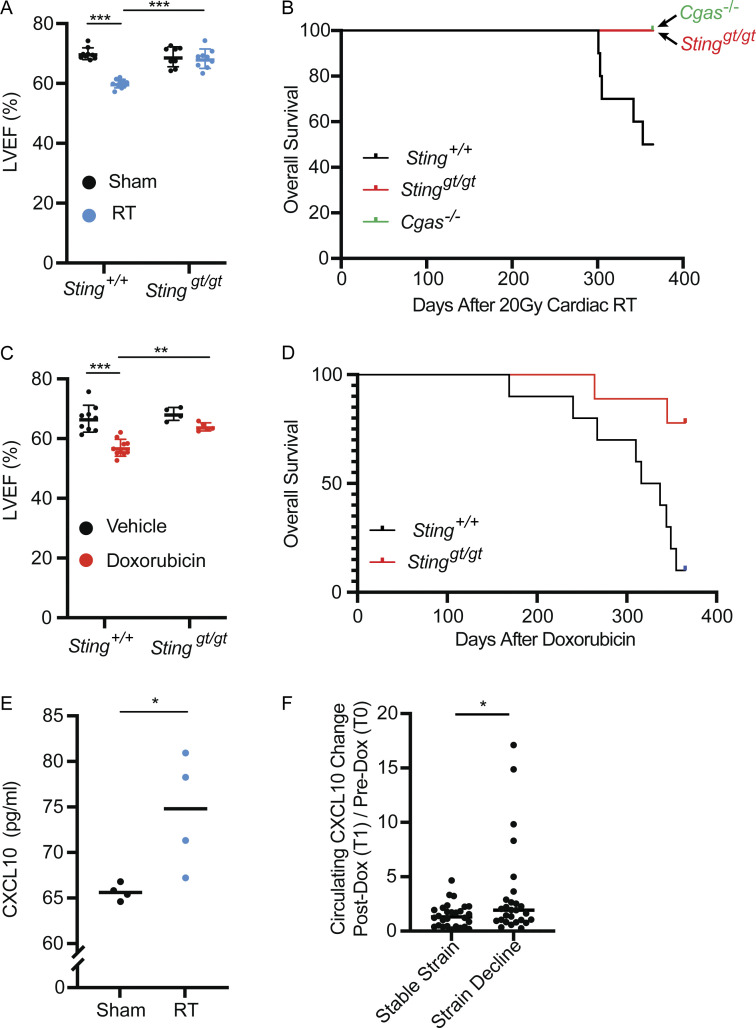
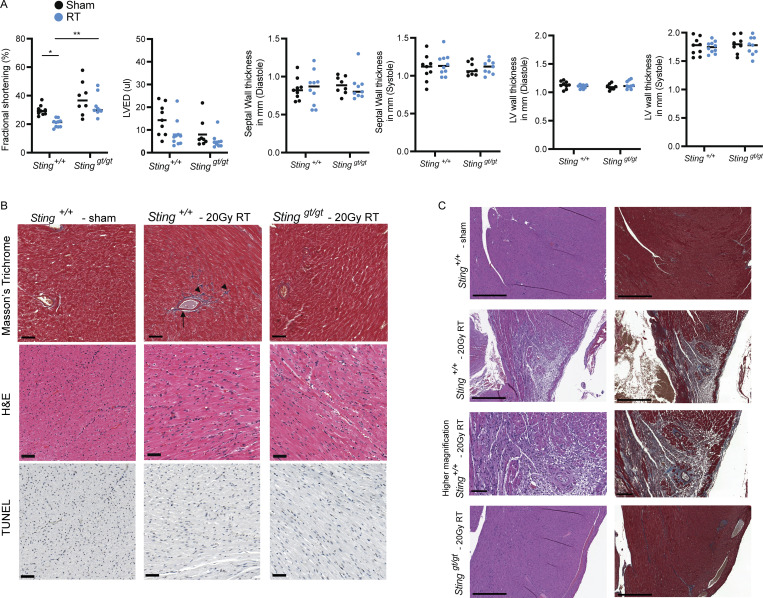
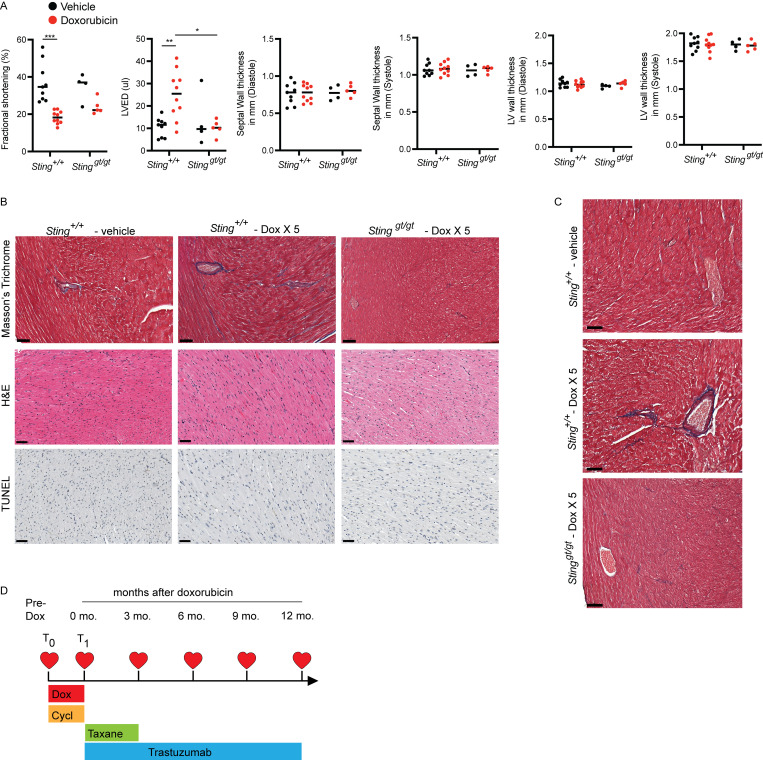
Cardiac functional decline as measured by echocardiography is an indication of cardiac toxicity from cancer therapy and is associated with subsequent risk of cardiac events. Indeed, in cancer survivors and animal models, cardiotoxic doses of anthracyclines lead to progressive myocardial fibrosis and worsening left ventricular ejection fraction (LVEF) a measure of left ventricular systolic function (Lee Chuy and Yu, 2019; Zhang et al., 2012). Similarly, exposure of large volumes of the heart to high-radiation doses results in myocardial fibrosis, declines in LVEF, and diminished fractional shortening (FS) in experimental animal models of radiation-induced cardiac toxicity and survivors of Hodgkin’s lymphoma who received mantle field radiation with substantial cardiac radiation exposure (Burns et al., 1983; Dreyfuss et al., 2021; Gomez et al., 1983; Gustavsson et al., 1990; Mezzaroma et al., 2012; Savage et al., 1990; Schlaak et al., 2019; Stewart et al., 1995). We next evaluated whether STING-dependent inflammatory signaling also contributed to worsening cardiac function following DNA-damaging cancer therapies. Initially, wildtype mice treated with cardiac RT to 20 Gy had no discernable change in cardiac function 1 mo following RT; however, by 3 mo, irradiated animals began to show signs of cardiac dysfunction with declines in LVEF (Fig. 5 A) and diminished FS in comparison with sham irradiated wildtype mice of the same age (Fig. S3 A). In contrast, identical cardiac irradiation of Stinggt/gt mice demonstrated no sign of cardiac functional decline compared to age-matched sham-irradiated mice (Fig. 5 A and Fig. S3 A). Cardiac histopathology 3 mo after cardiac irradiation revealed mild radiation-induced interfiber and perivascular fibrosis in Sting+/+ mice, compared to irradiated Stinggt/gt mice and sham-treated Sting+/+ mice, but no evidence of cardiomyocyte apoptosis at this time (Fig. S3 B). Furthermore, cardiac function at 3 mo following cardiac RT was predictive of survival by 1 yr after irradiation. Death from cardiac events in wildtype mice (n = 10) occurred beginning 300 d after RT with 50% incidence of death from cardiac events by 1 yr (Fig. 5 B). Cardiac events were associated uniformly with signs of severe respiratory distress prior to death and pleural effusions at the time of necropsy. Consistent with cardiac toxicity, histopathology of hearts from Sting+/+ mice at the time of death revealed extensive cardiomyocyte degeneration and loss, myocardial and perivascular fibrosis, and mural and luminal ventricular thrombi, while irradiated Stinggt/gt had no histopathologic abnormalities, similar to unirradiated Sting+/+ mice of the same age (Fig. S3 C). In marked contrast, 100% of Cgas−/− (n = 12) and Stinggt/gt (n = 12) mice survived at 365 d following cardiac RT with no signs of cardiac toxicity (Fig. 5 B and Fig. S3 C). Similar to mice treated with cardiac radiation, doxorubicin-treated wildtype mice experienced reduced LVEF, decreased FS, and increased left ventricular end-diastolic volume compared to sham-treated mice 2 wk after completing a five-dose regimen of doxorubicin, while loss of cGAS–STING signaling in Stinggt/gt mice rescued cardiac function, similar to results in mice treated with cardiac RT (Fig. 5 C and Fig. S4 A). Similar to prior reports, Sting+/+ mice treated with doxorubicin had increased interfiber and perivascular fibrosis, in contrast to doxorubicin-treated Stinggt/gt mice and vehicle-treated Sting+/+ mice, but no evidence of cardiomyocyte apoptosis (Fig. S4 B). 90% of doxorubicin treated Sting+/+ mice died of cardiac events within 1 yr of completing doxorubicin, with evidence of cardiac perivascular and interfiber fibrosis at the time of death, in contrast to 11% of Stinggt/gt that died within 1 yr of doxorubicin therapy (Fig. 5 D and Fig. S4 C). Therefore, cGAS–STING signaling initiated in the weeks following cardiac DNA damage drives functional cardiac decline associated with interfiber and perivascular fibrosis and death from late cardiac toxicity in the majority of Sting+/+ mice within 1 yr of treatment.
Figure 5.
cGAS–STING signaling drives cardiac toxicity after DNA-damaging cancer therapy. (A) LVEF as measured by 2D echocardiography in Sting+/+ vs. Stinggt/gt mice 3 mo after cardiac RT; N = 8–11 for each condition; two independent experiments were performed. (B) Kaplan–Meier plot of overall survival of wildtype (n = 10), Cgas−/− (n = 12), and Stinggt/gt (n = 12) mice after cardiac RT; day 0 is denoted as day of cardiac RT; tick marks indicate that 12 of 12 Cgas−/− and 12 of 12 Stinggt/gt remain alive 365 d after cardiac RT; P < 0.01, log-rank test. (C) LVEF as measured by 2D echocardiography Sting+/+ vs. Stinggt/gt mice 14 d after completing 5 wk doses of doxorubicin; N = 9–10 for Sting+/+ mice and N = 4–5 for Stinggt/gt mice; two independent experiments were performed. (D) Kaplan–Meier plot of overall survival of wildtype (n = 10) and Stinggt/gt (n = 9) mice after 5 wk doses of doxorubicin; day 0 is denoted the last dose of doxorubicinl; tick marks indicate that 7 of 9 Stinggt/gt remain alive 365 d after the last dose of doxorubicin; P < 0.01, log-rank test. (E) Circulating CXCL10 chemokine levels in mice 28 d after cardiac RT or sham treatment, as measured by ELISA; N = 4 mice for each condition; two independent experiments were performed. (F) The change in circulating CXCL10 levels from T0 to T1 levels in patients who developed >15% decline in GLS (N = 27), an early imaging biomarker of cardiac toxicity, compared to those who did not (N = 29). CXCL10 independently measured for each patient sample twice. *P < 0.05, **P < 0.01, ***P < 0.0001, two-tailed T test.
Figure S3.
Functional and histologic characterization of cardiac toxicity after radiation. (A) Echocardiogram in sham and RT-treated mice of indicated genotypes 3 mo after treatment. N = 8–11 for each condition, repeated once. *P < 0.05, **P < 0.01, two tailed T test. (B) Masson’s trichrome, H&E, and TUNEL staining of the left ventricle of indicated genotype 3 mo after cardiac irradiation or sham-treatment only; scale bars = 50 μm. Mild interfiber fibrosis (arrowhead) and perivascular fibrosis (arrow) is present in irradiated Sting+/+. No apoptosis was evident. Experiment performed with N = 4 mice for each treatment condition and timepoint; representative image shown. (C) H&E (left) and Masson’s trichrome (right) stains from the left ventricular wall of a Sting+/+ mouse 1 yr after sham treatment or cardiac radiation with 20 Gy, demonstrating cardiac fibrosis and cardiomyocyte degeneration and loss, and in a Stinggt/gt mouse 1 yr after cardiac radiation with 20 Gy. Scale bars = 500 μm, except for the high magnification image, 100 μm. N = 3–4 for each condition; representative image shown.
Figure S4.
Functional and histologic characterization of cardiac toxicity after doxorubicin. (A) Echocardiogram results from mice of indicated genotypes 14 d after completing 5 wk doses of doxorubicin or vehicle. N = 9–10 for Sting+/+ mice and N = 4–5 for Stinggt/gt mice; two independent experiments were performed. *P < 0.05, **P < 0.01, ***P < 0.0001, two tailed T test. (B). Masson’s trichrome, H&E, and TUNEL stains of the left ventricle of indicated genotype 2 wk after completing five doses of doxorubicin or vehicle treatment only; scale bars = 50 μm. Fibrosis was evident in doxorubicin treated Sting+/+ mice, but no evidence of apoptosis was apparent. Experiment performed with N = 4 mice for each treatment condition and timepoint; representative image shown. (C) Masson’s trichrome stain of the left ventricle in mice of indicated genotype 10 mo after completing five doses of doxorubicin or vehicle treatment only; scale bars = 50 μm. N = 3–4 for each condition; representative image shown. (D) Study schema for MSKCC IRB 14-099, a clinical trial to identify biomarkers of cardiac toxicity in HER2+ breast cancer patients receiving doxorubicin-based polychemotherapy and trastuzumab. Timepoints of echocardiograms and blood biospecimens obtained on the trial in relation to the treatment schedule. T0 denotes pre-treatment timepoint, and T1 denotes a post-doxorubicin, pre-trastuzumab timepoint.
Since cardiac DNA damage induces the expression of Cxcl10 in fibroblasts in a cGAS–STING-dependent manner and circulating levels of CXCL10 have previously been associated with cardiovascular disease (Rothenbacher et al., 2006), we next examined whether cardiac DNA damage increases the circulating levels of CXCL10 in mice. CXCL10 is significantly increased in the blood of mice receiving cardiac RT 28 d after treatment relative to sham-treated mice of the same age (Fig. 5 E), corresponding to the time at which Cxcl10 is induced in a cGAS–STING-dependent manner after RT (Fig. 3 A). Since circulating CXCL10 is induced by cardiac DNA damage, we next queried whether CXCL10 dynamics following cardiac DNA damage was associated with clinical correlates of cardiac toxicity. In cancer patients treated with cardiotoxic therapies such as RT or anthracyclines, a decline in global longitudinal strain (GLS) is considered as an early imaging biomarker of cardiac toxicity that precedes overt cardiac functional decline. Therefore, we next examined the association of CXCL10 with changes in GLS in cancer patients receiving cardiotoxic therapy. Memorial Sloan Kettering Cancer Center (MSKCC) Institutional Review Board (IRB) 14–099 enrolled 80 HER2+ breast cancer patients receiving anthracycline-based polychemotherapy followed by trastuzumab to monitor for cardiac toxicity using serial echocardiograms and to obtain blood specimens for identification of predictive biomarkers of cardiac toxicity. In this trial, 45% of patients developed a GLS decline of >15% within 1 yr of completing doxorubicin. Using these biospecimens, we quantified the circulating CXCL10 levels at pre-treatment (T0) and post-anthracycline, pre-trastuzumab (T1) and examined their relationship to GLS decline (Fig. S4 D). Specimens were available at T0 and T1 for 56 of these patients, of whom 27 (48%) had >15% decline in GLS within 1 yr of completing doxorubicin while 29 (52%) did not. The change in circulating CXCL10 levels from T0 to T1 was significantly higher in patients who subsequently experience a decline in GLS compared to those who did not (Fig. 5 F). Since these data points precede exposure to paclitaxel or trastuzumab, they are thus solely a metric of a patient’s inflammatory response to a 2-mo regimen of DNA-damaging chemotherapy including doxorubicin and cyclophosphamide and are consistent with our results in mouse models that activation of type I IFN signaling after cardiac DNA damaging therapy drives late cardiac toxicity.
STING inhibitor therapy prevents cardiac toxicity after DNA damage
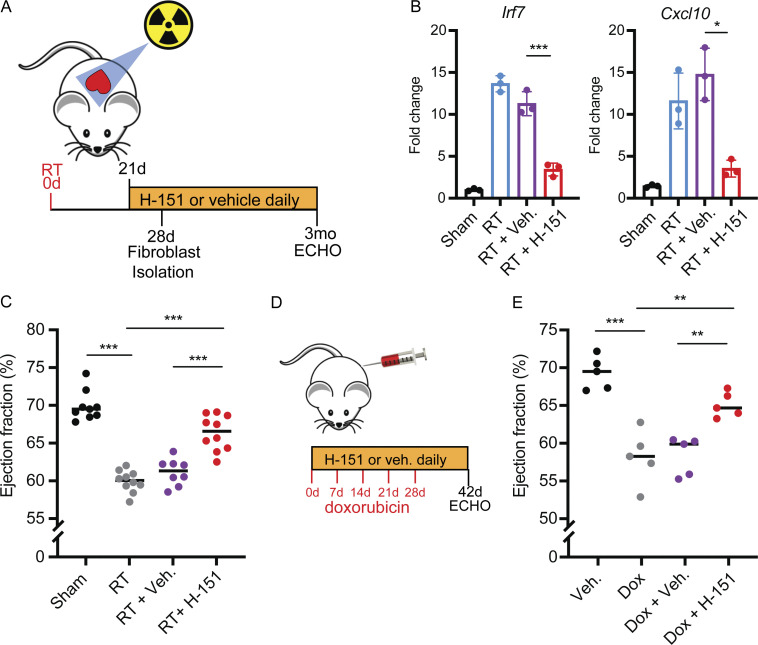
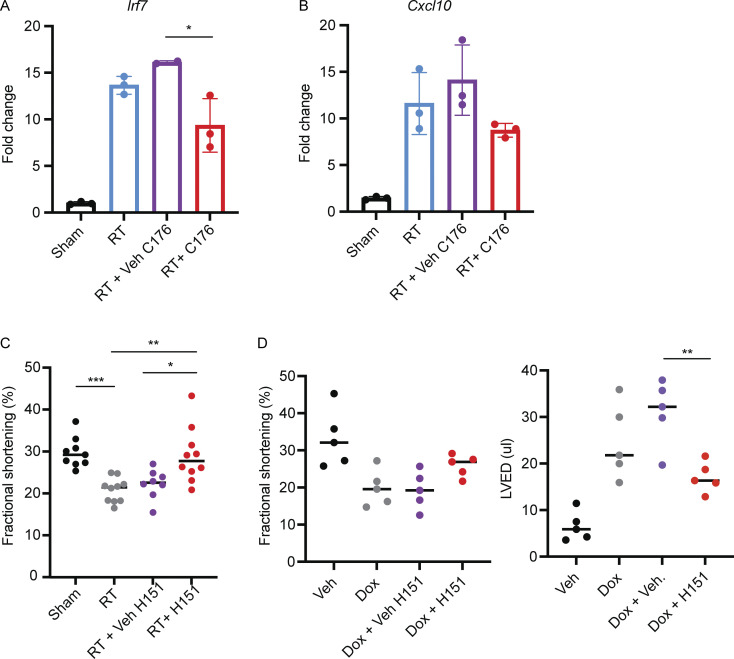
The cGAS–STING pathway is the subject of intensive pre-clinical drug development in oncology and autoimmune diseases and several small molecule antagonists have recently been developed. We next wanted to test whether pharmacologic inhibition of the cGAS–STING pathway is an effective therapeutic approach for the prevention of cardiac toxicity following DNA-damaging cancer therapy. The recent characterization of small molecule covalent inhibitors of STING, such as H-151 and C-176, offered an attractive tool (Haag et al., 2018). Since we observed that DNA damage–induced, cGAS–STING-dependent type I IFN signaling was detectable by 28 d following RT, we began daily treatments with H-151 21 d after RT and continued until the end point of the study (Fig. 6 A). H-151 treatments effectively attenuated Irf7 and Cxcl-10 induction 28 d after RT in cardiac fibroblasts of wildtype animals, reducing levels similar to those in Stinggt/gt mice (Fig. 6 B). C-176 treatments, on the other hand, had a less robust effect on Irf7 and Cxcl-10 following RT (Fig. S5, A and B). Having confirmed the activity of H-151 to inhibit cGAS–STING signaling in vivo in our model of cardiac toxicity, we next examined whether H-151 could prevent cardiac functional decline following cardiac DNA damage. Echocardiography of wildtype mice treated with H-151 or vehicle alone showed that H-151 prevented a decline in LVEF and FS in wildtype animals following RT (Fig. 6 C and Fig. S5 C). Similarly, in doxorubicin-treated mice, H-151 treatments prevented LVEF decline, as well as left ventricular dilation manifested through increase in left ventricular end-diastolic volume (Fig. 6, D and E; and Fig. S5 D). Therefore, cGAS–STING pathway antagonists are promising therapies for prevention of cardiac toxicity following cancer therapies that cause cardiac DNA damage.
Figure 6.
A covalent STING inhibitor effectively prevents cardiac toxicity from RT and anthracyclines. (A) Schematic representation of experimental design for STING antagonist H-151 treatments following cardiac RT relative to study endpoints, echocardiogram (ECHO). (B) Irf7 and Cxcl10 gene expression in cardiac fibroblasts isolated from Sting+/+ after cardiac RT or sham treatment and subsequent treatment with H-151 or vehicle, measured by qRT-PCR; N = 3 mice for each condition; three independent experiments were performed. (C) LVEF as measured by echocardiography in Sting+/+ mice 3 mo after cardiac RT or shame treatment and subsequent treatment with either STING antagonist H-151 or vehicle until the time of echocardiography; N = 8–10 mice per condition. (D) Schematic representation of experimental design for STING antagonist H-151 treatments in mice treated with doxorubicin to induce cardiac toxicity. (E) LVEF of Sting+/+ mice 14 d after completing 5 wk doses of doxorubicin or vehicle and concurrent H-151 of vehicle until the time of echocardiography; N = 5 mice per condition. *P < 0.05, **P < 0.01, ***P < 0.0001, two-tailed T test.
Figure S5.
Cardiac outcomes following treatment with STING inhibitors. (A and B) qRT-PCR for expression of Irf7 (A) and Cxcl10 (B) in cardiac fibroblasts 28 d after cardiac RT or sham and 7 d of treatment with STING antagonist C-176 or vehicle; N = 2–3 mice per condition; two independent experiments were performed. (C) FS as measured by echocardiography in Sting+/+ mice 3 mo after cardiac RT or sham treatment and subsequent treatment with either STING antagonist H-151 or vehicle until the time of echocardiography. (D and E) FS (D) and LVED (E) of Sting+/+ mice 14 d after completing 5 wk doses of doxorubicin or vehicle and concurrent H-151 of vehicle until the time of echocardiography. In D, N = 8–10 mice per condition, experiment performed once. In E, N = 5 mice per condition. *P < 0.05, **P < 0.01, ***P < 0.001, two tailed T test.
Discussion
Our findings provide new mechanistic understanding of cardiac toxicity following DNA-damaging cancer therapy. These results suggest that unrepaired double-stranded DNA damage following RT or anthracyclines drives cytosolic mis-localization of DNA in fibroblasts weeks after DNA damage, which in turn is detected by cGAS and leads to the activation of the type I IFN response. The lag in between the DNA-damage event and the induction of cGAS–STING-dependent signaling is likely related to a requirement for fibroblasts to undergo cell cycling prior to cytosolic mis-localization of genomic DNA as previously reported (Harding et al., 2017). Ultimately, the cardiac inflammation that results from this activation leads to cardiac mortality months after the initial insult in our model.
Early clinical observations of radiation-induced cardiac toxicity, including systolic heart disease and cardiomyopathy, were observed in cancer survivors who had been treated with mantle field radiation for Hodgkin’s lymphoma without blocking of the heart (Burns et al., 1983; Gomez et al., 1983; Gustavsson et al., 1990; Savage et al., 1990). Indeed, in one series evaluating cardiac outcomes of 58 survivors of Hodgkin’s lymphoma who received mantle radiation using echocardiography, 20 patients (34.5%) had echocardiographic abnormalities, including 5 (8.6%) demonstrating systolic dysfunction with LVEF <50% (Greenfield et al., 2006). Notably, left ventricular systolic dysfunction is also observed in rodent models of radiation-induced cardiac toxicity in which the entire heart is exposed to radiation (Dreyfuss et al., 2021; Mezzaroma et al., 2012; Schlaak et al., 2019). That modern clinical series of radiation induced cardiac toxicity infrequently observe systolic dysfunction as a feature is likely due to routinely employed strategies to block as much of the heart as possible, resulting in partial heart irradiation. Nevertheless, the DNA damage–induced, cGAS–STING-dependent signals that sustain cardiac inflammation and cardiac injury following doxorubicin or radiation in our animal models are expected to be at work in that pathogenesis of cardiac toxicity following partial irradiation of the heart. Furthermore, these data delineate a mechanism of late cardiac toxicity that develops over months to years that is sustained by cGAS–STING-dependent sub-acute to chronic inflammation arising from cardiac fibroblasts. This contrasts markedly with a recent report that STING signaling mediates rapid, acute cardiomyocyte apoptosis, and acute heart dysfunction following exposure to high doses of DNA crosslinking agent cisplatin, a class of therapeutics rarely associated with cardiac toxicity and over a timeline that does not recapitulate the disease process observed in cancer patients (Wang et al., 2021).
We have shown that DNA damage and STING induce type I IFN signaling in cardiac fibroblasts in a delayed manner, 1 mo after DNA damage. This results in cardiac inflammation and recruits cell populations that have previously been associated with pathologic remodeling of the myocardium in other heart diseases, including IFN-induced macrophages and monocytes, B cells, and dendritic cells (Cao et al., 2018; King et al., 2017; Lee et al., 2018; Zouggari et al., 2013). While each of these cell populations has been associated with pathologic remodeling of the myocardium and in myocardial fibrosis, this study does not ascertain whether one or more of these cell populations is required for the pathogenesis of DNA damage–induced cardiac fibrosis. Future studies are needed to rigorously dissect the mechanistic contribution of each cell population that is recruited to the heart in a STING-dependent manner after DNA damage, in order to clarify the cell populations driving STING-dependent cardiac fibrosis.
These data revealed that targeting of the cGAS–STING pathway holds great potential as a treatment to prevent normal tissue toxicity and improve the therapeutic ratio of DNA damaging cancer treatments. Further studies are needed to determine whether the delayed cGAS–STING signaling that drives cardiac toxicity is also a pathogenic part of a more general mechanism of delayed normal tissue injury following DNA-damaging cancer therapies. Indeed, our data indicate that clinical trials utilizing cGAS–STING antagonists to prevent late cardiac toxicity following DNA-damaging cancer therapy are warranted. Finally, our results demonstrate the consequences of pathogenic cGAS–STING signaling in normal tissues, which may have implications for drugs seeking to exploit cGAS–STING activation to treat cancer.
Materials and methods
Mice, reagents, and injections
Animal housing, handling, and all procedures were approved by MSKCC ethical committees and were performed in accordance to Institutional Animal Care and Use Committee guidelines. All mice strains were purchased from The Jackson Laboratory, namely, C57BL/6J (Cat#: 000664) were used as controls in experiments, and Stinggt (Cat#: 017537) were generated in the C57BL/6J and backcrossed at Jackson Laboratory to Jackson’s C57BL/6J (000664) twice, Cgastm1d(EUCOMM)Hmgu (Cat#: 026554, referred to as Cgas−/−) were generated at C57BL/6J mice and backcrossed at Jackson Laboratory to Jackson’s C57BL/6J (000664) twice, and Mavstm1Zjc (Cat#: 008634, referred to as Mavs−/−) are in a mixed C57BL/6 and 129 background after 129 embryonic stem cells were used to generate the mutation and chimera mice were bred to C57BL/6 mice. In all experiments, wildtype or Sting+/+ mice refer to Jackson Laboratory’s C57BL/6J stock (000664). Littermate mice were not used for genetic comparisons due to the large number of genetic strains used and the large number of mice required for experiments for this study precluded using littermates for the experiments.
Pharmaceutical grade doxorubicin was purchased from MSKCC pharmacy at a concentration of 2 mg/ml in PBS. For experiments, mice were weighted prior to each injection and subsequently injected with 5 mg/kg doxorubicin or vehicle (PBS) weekly for 5 wk to a total dose of 25 mg/kg. For inhibitor treatments, mice were injected intraperitoneally with daily 750 nmol of H151 (Cat#: 6675; Tocris) in 200 μl PBS 0.1% Tween-80 or vehicle and 750 nmol of C176 STING (Cat#: 7040; Tocris) in 200 μl corn oil or vehicle starting at 7 d prior to radiation or doxorubicin treatment and continuing until experimental endpoint.
Radiation treatments
Mice aged 8–10 wk were anesthetized using 3% isoflurane during induction and 2% during maintenance. X-Rad225Cx small animal irradiator (Precision X-ray) was used to administer either 12 or 20 Gy of radiation to the heart using 225 kVp x rays, a 0.9-mm half value layer Cu beam, and a 10-mm circular collimator. The isocenter was placed at the center of the heart as determined by a cone beam computed tomomgraphy scan. The 12 Gy treatment was delivered using parallel opposed obliques while the 20 Gy treatment was delivered using a 4-field box. Both treatments were delivered at a dose rate of 400 cGy/min. After analysis of dosimetry using a standard radiation protocol on multiple animals, total dose varied <5% across animals despite sex and age differences; therefore, beam angles and radiation plan were not adjusted for sex or weight. Sham irradiation involved all steps including sedation, imaging, and recovery, except actual radiation delivery.
Survival analyses
Mice were euthanized when showing evidence of overt heart failure including hunching, rapid breathing, lethargy, and abdominal distention. Kaplan–Meier curves were plotted using GraphPad prism. Hearts were paraffin-embedded and examined by pathologist to confirm histopathologic evidence of radiation-induced cardiac toxicity.
Heart cell separation
Mice were euthanized at experimental timepoints per Institutional Animal Care and Use Committee guidelines. To reduce variability and red blood cell contamination as well as RNA degradation, upon entering the thoracic cavity, cold PBS was perfused immediately before further cardiac extraction. The great vessels were cut at their interface to the heart, and pericardium and surrounding adipose were discarded.
Endothelia and fibroblasts
Hearts were harvested and digested with gentleMACS C Tube (Cat#: 130-093-237; Miltenyi) and the neonatal heart dissociation kit (Cat#: 130-098-373; Miltenyi) in a gentleMACS Octo Dissociator with heaters according to the manufacturer’s protocol. Following dissociation, debris removal and red blood cell lysis were performed using debris removal solution (Cat#: 130-109-398; Miltenyi) and red blood cell lysis solution (Cat#: 130-094-183; Miltenyi), respectively, according to the manufacturer’s protocol. Lysates were subsequently sequentially immunoprecipitated on an OctoMACS magnetic separator in MS columns (Cat#: 130-042-201; Miltenyi) using mouse CD45 (Cat#: 130-052-301; Miltenyi) to deplete leukocytes, CD31 (Cat#: 130-097-418; Miltenyi), and CD90.2 (Cat#: 130-121-278; Miltenyi) microbeads according to the manufacturer’s protocol.
Myocytes
Following acute heart extraction, myocytes were isolated utilizing an enzymatic isolation protocol through manufacturer instructions (Adumyt Non-perfusion Adult Cardiomyocyte Isolation, Cellutron Life Technologies). Because cells were not intended for culture, a Langendorf perfusion system was not utilized.
RNA isolation, purification, and cDNA generation
RNA isolation was performed using RNeasy Mini Kit (Cat#: 74106; Qiagen) according to the manufacturer’s protocol. Following RNA isolation, RNA was purified using RNAClean XP (Cat#: A63987; Beckman Coulter) per the manufacturer’s instruction. Genomic DNA was digested using ezDNAse Enzyme (Cat#: 11756050; Thermo Fisher Scientific ) for 2 min at 37°C, and cDNA was generated using SuperScript IV Vilo Master Mix (Cat#: 11756050; Thermo Fisher Scientific) according to the manufacturer’s protocol.
Quantitative RT-PCR (qRT-PCR)
qRT-PCR was performed using TaqMan Fast Advanced Master Mix (Cat#: 4444556; Thermo Fisher Scientific) on an Applied Biosytems QuantStudio 6 Flex Real-Time PCR system using the indicated primers: Irf7 (Cat#: Mm00516793_g1; Thermo Fisher Scientific), Cxcl10 (Cat#: Mm00445235; Thermo Fisher Scientific, _m1), and GAPDH (Cat#: Mm99999915_g1; Thermo Fisher Scientific). Expression was normalized to GAPDH (mice).
RNA-seq and analysis
RNA-seq on poly-A selected RNA was performed by Novogene on the Illumina NovaSeq 6000 Sequencing System. Samples were quantitated for total RNA and subsequent steps were carried out by Novogene, including library preparation and paired-end 150-bp sequencing. Bioinformatics analyses including quality control (QC) and DEseq were performed by Basepair with false discovery rate corrections for multiple tests. False discovery rate <0.05 was considered a statistically significant difference. Additional analysis for GO enrichment was performed using the web-based software Panther (http://geneontology.org).
Single-cell transcriptome sequencing
Dissociated, sorted cells were stained with Trypan blue and the Countess II Automated Cell Counter (Thermo Fisher Scientific) was used to assess both cell number and viability (74–82%). Following QC, the single-cell suspension was loaded onto Chromium Chip B (10x Genomics PN 2000060) and Gel Beads in emulasion generation, cDNA synthesis, cDNA amplification, and library preparation of 4,500–9,300 cells proceeded using the Chromium Single Cell 3′ Reagent Kit v3 (10x Genomics PN 1000075) according to the manufacturer’s protocol. cDNA amplification included 11 cycles, and 44–87 ng of the material was used to prepare sequencing libraries with 12 cycles of PCR. Indexed libraries were pooled equimolar and sequenced on a NovaSeq 6000 in a PE28/91 paired end run using the NovaSeq 6000 SP, S1, or S2 Reagent Kit (100 cycles; Illumina). An average of 201 million paired reads was generated per sample.
Single-cell transcriptome analysis
The raw sequence data (FASTQ files) were first processed with 10x Genomics CellRanger (ver. 5) software to compute the cell/barcode by gene count matrices. The command used was: cellranger count and the reference database was refdata-gex-mm10-2020-A. The count matrix was then processed with a series of R scripts (R version 3.6.1) using the Seurat (version 3.2.2) R library. The workflow used included standard processing with the SCTransform method for normalization and sample integration. First after reading in the 10x data, the cells in each sample were filtered to remove those that had any of the following QC measures: <1,500 detected genes per cell or <5,000 unique molecular identifiers per cell or >10% of the cells reads mapping to mitochondrial genes. Cells that failed any of these QC tests were removed from the analysis. Next each cell was scored for its cell-cycle phase using Seurat’s CellCycleScoring function. Note the Seurat library only has cell-cycle genes for human so we obtained a mouse version using R’s biomaRt packaged to convert between human and mouse homologues.
At this stage, all samples were processed independently. The next step was to integrate all of them into one unified dataset using Seurat’s integration workflow with SCTransform to normalize the data before integration. This first step both normalizes and regresses out the cell-cycle component. We then followed the rest of the integration workflow from the Seurat vignette.
At this point, we made two passes through the following pipeline. In the first pass, the data were clustered and cell types were identified using custom gene lists and Seurat’s AddModuleScore function. Clusters that contained irrelevant biological cell types were then filtered out and we repeated the entire processing pipeline with the reduced set of cells. The cell type filtering was done after the integration step. We ran the principal component analysis (PCA) projection (RunPCA) and computed the Uniform Manifold Approximation and Projection embedding (UMAP; RunUMAP) using the first 20 PCA components. The data were then clustered using the FindNeighbors/FindClusters functions with a resolution of 0.2 in the clustering step.
We then found cluster-specific marker genes and re-computed the cell type identifications as described above.
To compute differential gene expression, we use the Muscat package (ver. 1.5.0, for muscat ver. 4.0 of R was used for compatibility reasons). Specifically, we used the pseudo-bulk method with edgeR.
Cardiac 2D echocardiography
Mice were anesthetized using 3% isoflurane during induction and 2% during maintenance. Mice were placed on a heated stage to 37℃ and 2D echocardiograms were obtained using FujiFilm-Vevo 2100 micro-Ultrasound in B mode. Parasternal long axis views were utilized for LVEF and FS analysis using the Vivo lab LV trace tool. Investigators analyzing echocardiograms were blinded to animal genotype and/or intervention at the time of analysis.
Sample preparation for immunofluorescence
The immunofluorescence detection of cGAS was performed at Molecular Cytology Core Facility of MSKCC, using Discovery XT processor (Ventana Medical Systems). After 32 min of heat and CC1 (Cat#: 950-500; Cell Conditioning 1, Ventana) retrieval, the tissue sections were blocked first for 30 min in Background Blocking reagent (Cat#: NB306; Innovent). A rabbit monoclonal antibody to cGAS (Cat#: 31659; Cell Signaling) was used at a concentration of 0.5 μg/ml. The incubation with the primary antibody was done for 5 h, followed by 60 min incubation with biotinylated goat anti-rabbit IgG (Cat#: PK6101; Vector labs) at a concentration of 5.75 mg/ml, followed by Blocker D, Streptavidin-HRP and TSA Alexa 488 (Cat#: B40932; Life Tech) for 16 min.
The next day, a rabbit monoclonal Vimentin antibody (Cat#: 5741; Cell Signaling) was used in a 0.045 μg/ml concentration. The incubation with the primary antibody was done for 5 h, followed by a 60 min incubation with a goat anti-rabbit IgG (Cat#: PK6101; Vector labs) to a 5.75 mg/ml concentration. Blocker D, Streptavidin-HRP, and Tyramide-CF594 (Cat#: 92174; Biotium) were prepared according to manufacturer instructions in a 1:2,000 dilution and incubated for 16 min.
All slides were counterstained with 5 μg/ml DAPI (Cat#: D9542; Sigma-Aldrich) for 5 min at room temperature, mounted with anti-fade mounting medium Mowiol (Mowiol 4-88 CalbioChem code: 475904), and coverslip was added.
Immunofluorescence image acquisition
The images were acquired with an inverted Zeiss LSM880 (Carl Zeiss Microscopy GmbH) equipped with an Airy Scan detector (gain 850, digital gain 1) in super resolution mode. The objectives used for acquisition were a 20× air immersion (Plan-Apochromat 20×/0.8 M27) and a 63× oil immersion (Plan-Apochromat 63×/1.4 Oil DIC M27). With the 63× magnification, we used a pixel size of 32 nm and dwell time of 19.8 μs for the zoomed images and a pixel size and dwell time of 106 nm and 39.6 μs, respectively, for the bigger field of view. With the 20× objective, we used a pixel dwell time of 39.6 μs and a pixel size of 334 nm. We used the same optical configuration, splitting the three laser lines (405 nm at 0.7% power, 488 nm at 0.4%, and 561 nm at 0.5%) with a 488/561/633 Mirror Beam Splitter and an invisible light beam splitter at 405. In detection, we applied a band pass filter 495–550 nm and a long pass 570 nm.
For quantification, we acquired 10 images for each sample with the same optical configuration and 63× objective, but with pixel size of 265 nm and pixel dwell time of 1.24 μs. Z-stacks were acquired when the thickness of the sample required it, with a spacing of 500 nm between the stacks, and a maximum intensity projection was performed before quantification using the Zeiss ZEN black software.
All the images were processed using the feature AiryScan processing in the Zeiss ZEN software for image acquisition and analysis, using default parameters. Further image processing was performed in ImageJ (Rasband, W.S., U.S. National Institutes of Health, https://imagej.nih.gov).
MSKCC IRB 14-099 clinical trial and biospecimens
A prospective cohort of 80 women with non-metastatic breast cancer patients who were planned for treatment with anthracycline-based polychemotherapy followed by trastuzumab were enrolled. All patients underwent routine 2D and speckle tracking echocardiography at baseline (pre-anthracycline) and, after the completion of anthracyclines (2 mo) and thereafter at 3-mo intervals until 1 yr. Blood was collected at the pre-anthracycline and the 2-mo timepoint and analyzed for biomarkers. The study was conducted with approval of the MSKCC IRB, and all patients consented to blood collection.
Cytokine analyses
Mice were euthanized at indicated timepoint and intracardiac puncture was performed using a 21 gauge needle. Blood was collected at room temperature and held upright before for 30 min. Blood was spun at 1500 g for 15 min, and serum was collected by collecting supernatant. For ELISA, 100 μl of serum was run in duplicate using the mouse CXCL10 sandwich ELISA (Cat#: DY466; R&D Systems) according to the manufacturer’s protocol. For Luminex, 25 μl of serum was run in duplicate using the CXCL10 mouse Procarta simplex kit (Cat# EPX01A-26018-901; Thermo Fisher Scientific) according to the manufacturer’s protocol. Samples were red on a Flexmap 3D machine.
Online supplemental material
Fig. S1 provides additional characterization of the mouse model of RT-induced cardiac toxicity with control histology images for apoptosis detection by TUNEL and IHC staining, and expression of gene associated with fibrosis in cardiac fibroblasts. Fig. S2 includes additional analyses of the proportion of cell types and their ISG expression from scRNA-seq of CD45+ cells isolated from the heart. Fig. S3 includes detailed echocardiographic data and histology for mice with cardiac toxicity after RT. Fig. S4 includes detailed echocardiographic data and histology for mice with cardiac toxicity after doxorubicin. Fig. S5 includes comparison of STING antagonists H-151 and C-176 in suppressing RT-induced cardiac IFN signaling and additional echocardiographic data for cardiac outcomes in mice treated with H-151 after RT or doxorubicin. Table S1 provides a complete list of GO terms associated with rows in Fig. 2 C. Table S2 includes genes most significantly associated with each of the clusters identified in scRNA-seq of CD45+ leukocytes in Fig. 4 and Fig. S2.
Supplementary Material
shows specific GO terms corresponding to rows in Fig. 2 C.
shows genes most significantly associated with each scRNA-seq cluster in Fig. 4 A and Fig. S2.
Acknowledgments
We thank Simon Powell and Azusa Tanaka for careful reviews of the manuscript and helpful discussions. We thank Christina Marney for able assistance.
The study was supported by Department of Defense Discovery Award W81XWH1910002 (S.H. Patel, S.F. Bakhoum, A.M. Schmitt), National Institutes of Health R35 GM 124909 (A.M. Schmitt), a Memorial Sloan Kettering Cancer Center (MSKCC) Imaging and Radiation Sciences grant (S.H. Patel, S.F. Bakhoum, A.M. Schmitt), a MSKCC Chanel Endowment for Survivorship Research Grant (A.F. Yu), and MSKCC Cancer Center Support Grant P30 CA 008748.
Author contributions: Conceptualization: S.H. Patel and A.M. Schmitt. Methodology: A. Shamseddine, S.H. Patel, Z.R. Moore, A.T. Chan, A.F. Yu, A. Piersigilli, N.D. Socci, S.F. Bakhoum, A.M. Schmitt. Investigation: A. Shamseddine, S.H. Patel, V. Chavez, Z.R. Moore, M. Adnan, M.D. Bona, J. Li, N.D. Socci, C.T. Dang, L.V. Ramanathan, K.C. Oeffinger, J.E. Liu, R.M. Steingart, A.F. Yu, A.M. Schmitt. Visualization: A. Shamseddine, S.H. Patel, Z.R. Moore, M.D. Bona, N.D. Socci. Funding acquisition: S.H. Patel, S.F. Bakhoum, A.F. Yu, A.M. Schmitt. Project administration: A.M. Schmitt. Supervision: S.F. Bakhoum and A.M. Schmitt. Writing – original draft: A. Shamseddine and A.M. Schmitt. Writing – review & editing: All authors.
Data availability
Sequencing data have been deposited under GEO SuperSeries GSE218449 including bulk RNA-seq for specific cardiac cell populations (GSE218447) and scRNA-seq of CD45+ cells isolated from the heart (GSE218448). All other data are available in the main text or supplementary material.
References
- Aleman, B.M., van den Belt-Dusebout A.W., Klokman W.J., Van’t Veer M.B., Bartelink H., and van Leeuwen F.E.. 2003. Long-term cause-specific mortality of patients treated for Hodgkin’s disease. J. Clin. Oncol. 21:3431–3439. 10.1200/JCO.2003.07.131 [DOI] [PubMed] [Google Scholar]
- Aleman, B.M., van den Belt-Dusebout A.W., De Bruin M.L., van ’t Veer M.B., Baaijens M.H., de Boer J.P., Hart A.A., Klokman W.J., Kuenen M.A., Ouwens G.M., et al. 2007. Late cardiotoxicity after treatment for Hodgkin lymphoma. Blood. 109:1878–1886. 10.1182/blood-2006-07-034405 [DOI] [PubMed] [Google Scholar]
- Burns, R.J., Bar-Shlomo B.Z., Druck M.N., Herman J.G., Gilbert B.W., Perrault D.J., and McLaughlin P.R.. 1983. Detection of radiation cardiomyopathy by gated radionuclide angiography. Am. J. Med. 74:297–302. 10.1016/0002-9343(83)90631-9 [DOI] [PubMed] [Google Scholar]
- Cao, D.J., Schiattarella G.G., Villalobos E., Jiang N., May H.I., Li T., Chen Z.J., Gillette T.G., and Hill J.A.. 2018. Cytosolic DNA sensing promotes macrophage transformation and governs myocardial ischemic injury. Circulation. 137:2613–2634. 10.1161/CIRCULATIONAHA.117.031046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappetta, D., De Angelis A., Sapio L., Prezioso L., Illiano M., Quaini F., Rossi F., Berrino L., Naviglio S., and Urbanek K.. 2017. Oxidative stress and cellular response to doxorubicin: A common factor in the complex milieu of anthracycline cardiotoxicity. Oxid. Med. Cell. Longev. 2017:1521020. 10.1155/2017/1521020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, G.Y., and Nuñez G.. 2010. Sterile inflammation: Sensing and reacting to damage. Nat. Rev. Immunol. 10:826–837. 10.1038/nri2873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curigliano, G., Lenihan D., Fradley M., Ganatra S., Barac A., Blaes A., Herrmann J., Porter C., Lyon A.R., Lancellotti P., et al. 2020. Management of cardiac disease in cancer patients throughout oncological treatment: ESMO consensus recommendations. Ann. Oncol. 31:171–190. 10.1016/j.annonc.2019.10.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darby, S.C., Ewertz M., McGale P., Bennet A.M., Blom-Goldman U., Brønnum D., Correa C., Cutter D., Gagliardi G., Gigante B., et al. 2013. Risk of ischemic heart disease in women after radiotherapy for breast cancer. N. Engl. J. Med. 368:987–998. 10.1056/NEJMoa1209825 [DOI] [PubMed] [Google Scholar]
- De Ruysscher, D., Niedermann G., Burnet N.G., Siva S., Lee A.W.M., and Hegi-Johnson F.. 2019. Radiotherapy toxicity. Nat. Rev. Dis. Prim. 5:13. 10.1038/s41572-019-0064-5 [DOI] [PubMed] [Google Scholar]
- Dreyfuss, A.D., Goia D., Shoniyozov K., Shewale S.V., Velalopoulou A., Mazzoni S., Avgousti H., Metzler S.D., Bravo P.E., Feigenberg S.J., et al. 2021. A novel mouse model of radiation-induced cardiac injury reveals biological and radiological biomarkers of cardiac dysfunction with potential clinical relevance. Clin. Cancer Res. 27:2266–2276. 10.1158/1078-0432.CCR-20-3882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dühmke, E., Franklin J., Pfreundschuh M., Sehlen S., Willich N., Rühl U., Müller R.P., Lukas P., Atzinger A., Paulus U., et al. 2001. Low-dose radiation is sufficient for the noninvolved extended-field treatment in favorable early-stage Hodgkin’s disease: Long-term results of a randomized trial of radiotherapy alone. J. Clin. Oncol. 19:2905–2914. 10.1200/JCO.2001.19.11.2905 [DOI] [PubMed] [Google Scholar]
- Gomez, G.A., Park J.J., Panahon A.M., Parthasarathy K.L., Pearce J., Reese P., Bakshi S., and Henderson E.S.. 1983. Heart size and function after radiation therapy to the mediastinum in patients with Hodgkin’s disease. Cancer Treat. Rep. 67:1099–1103 [PubMed] [Google Scholar]
- Gong, T., Liu L., Jiang W., and Zhou R.. 2020. DAMP-sensing receptors in sterile inflammation and inflammatory diseases. Nat. Rev. Immunol. 20:95–112. 10.1038/s41577-019-0215-7 [DOI] [PubMed] [Google Scholar]
- Greenfield, D.M., Wright J., Brown J.E., Hancock B.W., Davies H.A., O’Toole L., Eiser C., Coleman R.E., and Ross R.J.. 2006. High incidence of late effects found in Hodgkin’s lymphoma survivors, following recall for breast cancer screening. Br. J. Cancer. 94:469–472. 10.1038/sj.bjc.6602974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustavsson, A., Eskilsson J., Landberg T., Svahn-Tapper G., White T., Wollmer P., and Akerman M.. 1990. Late cardiac effects after mantle radiotherapy in patients with Hodgkin’s disease. Ann. Oncol. 1:355–363. 10.1093/oxfordjournals.annonc.a057774 [DOI] [PubMed] [Google Scholar]
- Haag, S.M., Gulen M.F., Reymond L., Gibelin A., Abrami L., Decout A., Heymann M., van der Goot F.G., Turcatti G., Behrendt R., and Ablasser A.. 2018. Targeting STING with covalent small-molecule inhibitors. Nature. 559:269–273. 10.1038/s41586-018-0287-8 [DOI] [PubMed] [Google Scholar]
- Harding, S.M., Benci J.L., Irianto J., Discher D.E., Minn A.J., and Greenberg R.A.. 2017. Mitotic progression following DNA damage enables pattern recognition within micronuclei. Nature. 548:466–470. 10.1038/nature23470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, K.R., Aguirre A.D., Ye Y.X., Sun Y., Roh J.D., Ng R.P. Jr, Kohler R.H., Arlauckas S.P., Iwamoto Y., Savol A., et al. 2017. IRF3 and type I interferons fuel a fatal response to myocardial infarction. Nat. Med. 23:1481–1487. 10.1038/nm.4428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon, J., and Bakhoum S.F.. 2020. The cytosolic DNA-sensing cGAS-STING pathway in cancer. Cancer Discov. 10:26–39. 10.1158/2159-8290.CD-19-0761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Chuy, K., and Yu A.F.. 2019. Cardiotoxicity of contemporary breast cancer treatments. Curr. Treat. Options Oncol. 20:51. 10.1007/s11864-019-0646-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J.S., Jeong S.J., Kim S., Chalifour L., Yun T.J., Miah M.A., Li B., Majdoubi A., Sabourin A., Keler T., et al. 2018. Conventional dendritic cells impair recovery after myocardial infarction. J. Immunol. 201:1784–1798. 10.4049/jimmunol.1800322 [DOI] [PubMed] [Google Scholar]
- Li, J., Duran M.A., Dhanota N., Chatila W.K., Bettigole S.E., Kwon J., Sriram R.K., Humphries M.P., Salto-Tellez M., James J.A., et al. 2021. Metastasis and immune evasion from extracellular cGAMP hydrolysis. Cancer Discov. 11:1212–1227. 10.1158/2159-8290.CD-20-0387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mezzaroma, E., Di X., Graves P., Toldo S., Van Tassell B.W., Kannan H., Baumgarten C., Voelkel N., Gewirtz D.A., and Abbate A.. 2012. A mouse model of radiation-induced cardiomyopathy. Int. J. Cardiol. 156:231–233. 10.1016/j.ijcard.2012.01.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitry, M.A., and Edwards J.G.. 2016. Doxorubicin induced heart failure: Phenotype and molecular mechanisms. Int. J. Cardiol. Heart Vasc. 10:17–24. 10.1016/j.ijcha.2015.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motwani, M., Pesiridis S., and Fitzgerald K.A.. 2019. DNA sensing by the cGAS-STING pathway in health and disease. Nat. Rev. Genet. 20:657–674. 10.1038/s41576-019-0151-1 [DOI] [PubMed] [Google Scholar]
- Park, S., Ranjbarvaziri S., Lay F.D., Zhao P., Miller M.J., Dhaliwal J.S., Huertas-Vazquez A., Wu X., Qiao R., Soffer J.M., et al. 2018. Genetic regulation of fibroblast activation and proliferation in cardiac fibrosis. Circulation. 138:1224–1235. 10.1161/CIRCULATIONAHA.118.035420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues, G., Choy H., Bradley J., Rosenzweig K.E., Bogart J., Curran W.J. Jr, Gore E., Langer C., Louie A.V., Lutz S., et al. 2015. Definitive radiation therapy in locally advanced non-small cell lung cancer: Executive summary of an American Society for Radiation Oncology (ASTRO) evidence-based clinical practice guideline. Pract. Radiat. Oncol. 5:141–148. 10.1016/j.prro.2015.02.012 [DOI] [PubMed] [Google Scholar]
- Roers, A., Hiller B., and Hornung V.. 2016. Recognition of endogenous nucleic acids by the innate immune system. Immunity. 44:739–754. 10.1016/j.immuni.2016.04.002 [DOI] [PubMed] [Google Scholar]
- Rothenbacher, D., Müller-Scholze S., Herder C., Koenig W., and Kolb H.. 2006. Differential expression of chemokines, risk of stable coronary heart disease, and correlation with established cardiovascular risk markers. Arterioscler. Thromb. Vasc. Biol. 26:194–199. 10.1161/01.ATV.0000191633.52585.14 [DOI] [PubMed] [Google Scholar]
- Savage, D.E., Constine L.S., Schwartz R.G., and Rubin P.. 1990. Radiation effects on left ventricular function and myocardial perfusion in long term survivors of Hodgkin’s disease. Int. J. Radiat. Oncol. Biol. Phys. 19:721–727. 10.1016/0360-3016(90)90502-B [DOI] [PubMed] [Google Scholar]
- Schlaak, R.A., Frei A., Schottstaedt A.M., Tsaih S.W., Fish B.L., Harmann L., Liu Q., Gasperetti T., Medhora M., North P.E., et al. 2019. Mapping genetic modifiers of radiation-induced cardiotoxicity to rat chromosome 3. Am. J. Physiol. Heart Circ. Physiol. 316:H1267–H1280. 10.1152/ajpheart.00482.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart, J.R., Fajardo L.F., Gillette S.M., and Constine L.S.. 1995. Radiation injury to the heart. Int. J. Radiat. Oncol. Biol. Phys. 31:1205–1211. 10.1016/0360-3016(94)00656-6 [DOI] [PubMed] [Google Scholar]
- Swain, S.M., Whaley F.S., and Ewer M.S.. 2003. Congestive heart failure in patients treated with doxorubicin: A retrospective analysis of three trials. Cancer. 97:2869–2879. 10.1002/cncr.11407 [DOI] [PubMed] [Google Scholar]
- Tanikawa, C., Zhang Y.Z., Yamamoto R., Tsuda Y., Tanaka M., Funauchi Y., Mori J., Imoto S., Yamaguchi R., Nakamura Y., et al. 2017. The transcriptional landscape of p53 signalling pathway. EBioMedicine. 20:109–119. 10.1016/j.ebiom.2017.05.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darby, S., McGale P., Correa C., Taylor C., Arriagada R., Clarke M., Cutter D., Davies C., Ewertz M., Godwin J., et al. ; Early Breast Cancer Trialists’ Collaborative Group . 2011. Effect of radiotherapy after breast-conserving surgery on 10-year recurrence and 15-year breast cancer death: Meta-analysis of individual patient data for 10,801 women in 17 randomised trials. Lancet. 378:1707–1716. 10.1016/S0140-6736(11)61629-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L., Zhang S., Han J., Nie X., Qi Y., Han Y., Chen X., and He C.. 2021. Activation of STING pathway contributed to cisplatin-induced cardiac dysfunction promoting the activation of TNF-α-AP-1 signal pathway. Front. Pharmacol. 12:711238. 10.3389/fphar.2021.711238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, S., Liu X., Bawa-Khalfe T., Lu L.S., Lyu Y.L., Liu L.F., and Yeh E.T.. 2012. Identification of the molecular basis of doxorubicin-induced cardiotoxicity. Nat. Med. 18:1639–1642. 10.1038/nm.2919 [DOI] [PubMed] [Google Scholar]
- Zhang, X., Bai X.C., and Chen Z.J.. 2020. Structures and mechanisms in the cGAS-STING innate immunity pathway. Immunity. 53:43–53. 10.1016/j.immuni.2020.05.013 [DOI] [PubMed] [Google Scholar]
- Zouggari, Y., Ait-Oufella H., Bonnin P., Simon T., Sage A.P., Guérin C., Vilar J., Caligiuri G., Tsiantoulas D., Laurans L., et al. 2013. B lymphocytes trigger monocyte mobilization and impair heart function after acute myocardial infarction. Nat. Med. 19:1273–1280. 10.1038/nm.3284 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
shows specific GO terms corresponding to rows in Fig. 2 C.
shows genes most significantly associated with each scRNA-seq cluster in Fig. 4 A and Fig. S2.
Data Availability Statement
Sequencing data have been deposited under GEO SuperSeries GSE218449 including bulk RNA-seq for specific cardiac cell populations (GSE218447) and scRNA-seq of CD45+ cells isolated from the heart (GSE218448). All other data are available in the main text or supplementary material.