Abstract
The COVID-19 pandemic has been affecting the world since December 2019, and nowadays, the number of infected is increasing rapidly. Chest X-ray images are clinical adjuncts that can be used in the diagnosis of COVID-19 disease. Because of the rapid spread of COVID-19 disease worldwide and the limited number of expert radiologists, the proposed method uses the automatic diagnosis method rather than a manual diagnosis method. In the paper, COVID-19 Positive/Negative (2275 Positive, 4626 Negative) and Normal/Pneumonia (2313 Normal, 2313 Pneumonia) are diagnosed using chest X-ray images. Herein, 80 % and 20 % of the images are used in the training and validation set, respectively. In the proposed method, six different classifiers are trained using chest X-ray images, and the five most successful classifiers are used in both phases. In Phase-1 and Phase-2, image features are extracted using the Bag of Features method for Cosine K-Nearest Neighbor (KNN), Linear Discriminant, Logistic Regression, Bagged Trees Ensemble, Medium Gaussian Support Vector Machine (SVM), excluding SqueezeNet Deep Learning (K = 2000 and K = 1500 for Phase-1 and Phase-2, respectively). In both phases, the five most successful classifiers are determined, and images classify with the help of the Majority Voting (Mathematical Evaluation) method. The application of the proposed method is designed for users to diagnose COVID-19 Positive, Normal, and Pneumonia. The results show that accuracy values obtained by Majority Voting (Mathematical Evaluation) method for Phase-1 and Phase-2 are equal to 99.86 % and 99.28 %, respectively. Thus, it indicates that the accuracy of the whole system is 99.63 %. When we analyze the classification performance metrics for Phase-1 and Phase-2, Specificity (%), Precision (%), Recall (%), F1 Score (%), Area Under Curve (AUC), and Matthews Correlation Coefficient (MCC) are equal to 99.98–99.83–99.07–99.51–0.9974–0.9855 and 99.73–99.69–98.63–99.23–0.9928–0.9518, respectively. Moreover, if the classification performance metrics of the whole system are examined, it is seen that Specificity (%), Precision (%), Recall (%), F1 Score (%), AUC, and MCC are 99.88, 99.78, 98.90, 99.40, 0.9956, and 0.9720, respectively. When the studies in the literature are examined, the results show that the proposed model is better than its counterparts. Because the best performance metrics for the dataset used were obtained in this study. In addition, since the biphasic majority voting technique is used in the study, it is seen that the proposed model is more reliable. On the other hand, although there are tens of thousands of studies on this subject, the usability of these models is debatable since most of them do not have graphical user interface applications. Already, in artificial intelligence technologies, besides the performance of the developed models, their usability is also important. Because the developed models can generally be used by people who are less knowledgeable about artificial intelligence.
Keywords: COVID-19, Machine learning, Deep learning, Bag of features, Majority voting
1. Introduction
The coronavirus pandemic has broken out on December 1st, 2019, in Wuhan, China. World Health Organization (WHO) has announced that this disease caused by the new virus has named COVID-19 on February 11th, 2020 (Wu et al., 2020). To date (as of May 27th, 2021), 169,404,850 coronavirus cases have been detected in the world, and 3,518,899 people died due to the COVID-19 disease. Additionally, the number of people recovered is equal to 151,144,574 from December 1st, 2019, to May 27th, 2021 (COVID-19 Pandemic @ https://www.worldometers.info). Herein, it seems that the COVID-19 disease detection phase is the most important for the successful treatment of COVID-19 patients. While there are so many cases of COVID-19 all over the world, and the number of cases is increasing day by day, it seems that the number of expert radiologists who diagnose disease by examining chest X-ray images is insufficient. To reduce the workload of expert radiologists, save time, and successfully diagnose by obtaining high classification performance metrics, COVID-19 disease is diagnosed automatically using machine learning and deep learning methods. It seems that machine learning and deep learning techniques, which can apply to all areas of life, provide advantages like COVID-19 disease detection thanks to the developing technologies (Selcuk and Alkan, 2019, Alkan et al., 2014).
Herein, the fact that the COVID-19 disease can easily be transmitted from person to person and the total number of cases around the world is still increasing rapidly affects all humanity indirectly and directly. There are different methodologies developed to solve this problem relatively or to reduce its effect, but the developed artificial intelligence-based models have a high potential to be used because they can diagnose COVID-19 quickly, successfully and automatically. As mentioned earlier, it is significant that these models have both high performance metrics and usability. Therefore, it is aimed to develop models that have high performance metrics and can reliably diagnose COVID-19 in this study. On the other hand, it is seen that the models developed in the studies in the literature generally do not have a graphical user interface application. Therefore, we have designed a user-friendly graphical user interface application that can diagnose COVID-19 in about 15 s. Unfortunately, such applications are widely used today. Therefore, it is aimed to reduce the workload of experts, reduce costs and save time in this study.
In this context, there are many studies conducted recently. In one of these studies, medical imaging and the use of artificial intelligence have been investigated for COVID-19 prediction, and the multi-center cohort is collected there. Also, a data-driven consensus has been established for predicting disease using deep learning (Chassagnon et al., 2021). Signoroni et al. (2021) has proposed a deep learning architecture using chest X-ray images for predicting the multi-regional score conveying the degree of lung compromise in the patients. This architecture is called BS-Net, and it has performed a high degree of accuracy in all phases. A system, which includes techniques such as Recurrent Neural Networks (RNN), Long-Short Term Memory (LSTM), Generative Adversarial Networks (GANs), and Extreme Learning Machine (ELM), have been developed for the diagnosis of COVID-19 in (Jamshidi et al., 2020). In this way, the COVID-19 virus can be fought faster and more effectively. In (Oh et al., 2020), a patch-based Convolutional Neural Networks (CNN) approach with fewer parameters has been proposed for COVID-19 diagnosis, and segmentation network, majority voting, and images consisting of five different classes have been used. The results show that the accuracy values of the proposed method and COVID-19-Net are equal to 91.9 % and 92.4 %, respectively. In (Ismael and Şengür, 2021), deep feature extraction, fine-tuning of pre-trained CNN, and developed CNN architecture have been used to classify chest X-ray images from COVID-19 Positive and Normal subjects. From the results, it seems that the accuracy of the designed system using fine-tuned ResNet50 is equal to 92.6 %. By examining ethnic and genetic differences, a feature group based on laboratory findings has been formed for interpreting blood data, and a new deep learning classifier architecture that detects COVID-19 has been suggested using this feature group. From the results, it appears that its accuracy is equal to 94.95 % (Göreke et al., 2021). In one of the other studies, the novel tree component method is suggested in Li et al. (2021). While the first is the deep multiple instance learning, the second is the bag-level data augmentation component. Third, the self-supervised pretext component is used to aid the learning process. The results show that the average accuracy of the proposed method designed to determine COVID-19 severity is equal to 95.8 %. (Ozyurt et al., 2021) suggests a system that can diagnose COVID-19 using Computed Tomography (CT) images, and that system uses a new feature generation technique and a hybrid feature selector. From the results, it seems that its accuracy of the proposed method is equal to 95.84 %. In (Serte and Demirel, 2021), an artificial intelligence approach is recommended to classify each CT image obtained from the 3D CT scan as COVID-19 Positive or Normal. The results render that the proposed method provides 96 % of the AUC value for COVID-19 detection on CT scans. Fifteen images have been obtained from one chest image using Fuzzy Tree Transformation (3-level F-tree). After the exemplar division is applied to these images, the multi-kernel local binary pattern is applied to each exemplar division and image to extract features. Here, the features are determined according to the iterative neighborhood component feature selector. The best classifier used in the study is Cubic SVM, and its accuracy is 97.01 % (Tuncer et al., 2021). It seems that the deep feature extracted using chest X-ray images from COVID-19 and Pneumonia subjects classifies by ResNet152, and the results show that the accuracy of the method is equal to 97.7 % (Kumar et al., 2020). Panwar et al. (2020) proposes a system that detects COVID-19 based on deep learning using visual indicators found in the chest radiography imaging for COVID-19 patients, and the name of the system is nCOVnet. Here, it seems that the training accuracy and confidence of the proposed method are equal to 97 % and 97.97 %, respectively. Chandra et al. (2021) suggests a system that consists of two phases and examines chest X-ray images from Normal/Abnormal and COVID-19 Positive/Pneumonia subjects using majority voting. The results seem that the accuracy values of the proposed method are equal to 98.062 % and 91.329 % for Phase-1 and Phase-2, respectively. Karakanis and Leontidis (2021) proposes an approach to augment the limited amount of data and to generate synthetic images that can be used to detect COVID-19. When we analyze the accuracy values of the proposed method for two classes (COVID-19 Positive/Normal) and three classes (COVID-19 Positive/Bacteria/Normal), they are 98.7 % and 98.3 %, respectively. Sedik et al. (2020) presents two data augmentation models to improve the performance metric of CNN-based and Convolutional LSTM (ConvLSTM)-based deep learning architectures. The results render that the accuracy of the system created using the proposed method can reach 99 %. Sheykhivand et al. (2021) uses GANs, without involving feature extraction/selection to classify pneumonia. It seems that the accuracy of the system can achieve 99 % when COVID-19 separates from the healthy group.
In the paper, we propose a new and novel method for diagnosing COVID-19 disease. Primarily, we divide the chest X-ray images used in the study into COVID-19 Positive/Negative and Normal/Pneumonia. From these images, we extract their features using the Bag of Features method, and we train six different classifiers, which are Cosine KNN, Linear Discriminant, Logistic Regression, Bagged Trees Ensemble, Medium Gaussian SVM, and SqueezeNet Deep Learning, using 80 % of the features. Afterward, we select the five most successful classifiers for Phase-1 and Phase-2, and classify chest X-ray images as COVID-19 Positive or Negative and Normal or Pneumonia, respectively, using the Majority Voting (Mathematical Evaluation) method or the probability values of independent events. We create the theoretical framework so that we can explain the Majority Voting (Mathematical Evaluation) method. In addition, we design a user-friendly application for users to diagnose the COVID-19 disease.
In light of this information, this study is innovative and competitive in terms of:
-
•
The number of images in the data set used in the study enabled us to obtain reliable models. In this context, the best performance metrics for the dataset used were obtained in this study.
-
•
In addition, the biphasic majority voting technique used in the study has been designed specifically for this problem. Thus, it will be easier to make comparisons between different studies.
-
•
Indeed, although there are many studies in the literature that can diagnose COVID-19 disease, it seems that most of them do not have user-friendly software. Therefore, we have developed user-friendly software so that the proposed models can be easily used by users. Hereabouts, it is thought that this study will contribute significantly to the literature.
The paper is organized as follows. Section 2 presents the statistic of the number of chest X-ray images used in the study. Section 3 expresses the feature extraction method, classification algorithms, Majority Voting (Mathematical Evaluation) method, and Graphical User Interface (GUI) application of the proposed method. Section 4 renders the performance metrics obtained for the experimental study, and Section 5 interprets the results and discussion of the study. Lastly, Section 6 examines the conclusion of the proposed method.
2. Materials
In the study, we used chest X-ray images found in (COVID19, Pneumonia, and Normal Chest X Ray PA Dataset https://www.kaggle.com). The file types of these images are MATLAB-aided PNG, JPG, and JPEG. To better understand the dataset used in the proposed method, Table 1 and Fig. 1 can be given as follows:
Table 1.
Statistic of the number of chest X-ray images in training and validation set.
| Dataset Size | Training Set (80 %) | Validation Set (20 %) | |||
|---|---|---|---|---|---|
| Phase-1 | COVID-19 Positive | 2275 | 5521 | 1380 | |
| COVID-19 Negative | 4626 | ||||
| Phase-2 | COVID-19 Negative | Pneumonia | 2313 | 3701 | 925 |
| Normal | 2313 | ||||
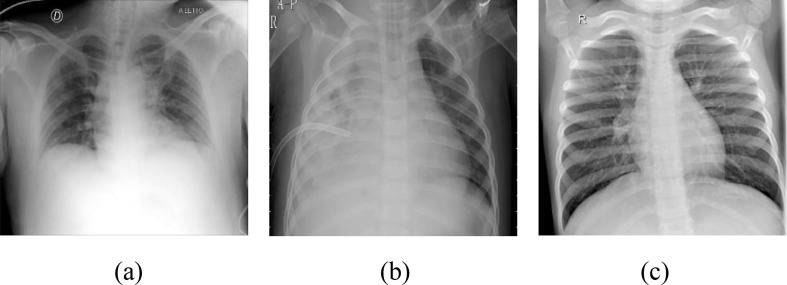
Fig. 1.
Chest X-ray images from subjects a) COVID-19 Positive b) Pneumonia c) Normal.
Table 1 shows the statistic of the number of chest X-ray images used for the training and validation set. The dataset includes chest X-ray images from 2275 COVID-19 Positive (we do not use 38 images because MATLAB does not support these images), 2313 Pneumonia, and 2313 Normal subjects. The classifiers used in Phase-1 examine COVID-19 Positive and Negative images, where the number of chest X-ray images from COVID-19 Negative subjects is equal to 4626. For Phase-1 and Phase-2, 80 % and 20 % of the images are used in the training and validation set, respectively. Additionally, the chest X-ray images included in the dataset are shown in Fig. 1, where Fig. 1.a, 1.b, and 1.c show the chest X-ray images from COVID-19 Positive, Pneumonia, and Normal subjects, respectively.
3. Methods
In this study, six different classifiers are trained using chest X-ray images from COVID-19 Positive/Negative and Normal/Pneumonia subjects. Classifiers used in the study are Cosine KNN, Linear Discriminant, Logistic Regression, Bagged Trees Ensemble, Medium Gaussian SVM, and SqueezeNet Deep Learning. Image features are extracted using the Bag of Features method for other classifiers, excluding SqueezeNet Deep Learning. The proposed method consists of two steps that are Phase-1 and Phase-2 where the majority voting is used separately. Thus, the images can classify as COVID-19 Positive, Normal, Pneumonia. In the study, the theoretical framework and tables are created to examine the classification performance metrics by the majority voting. Also, the application of the proposed COVID-19 decision support system is developed using the MATLAB GUI.
3.1. Feature extraction
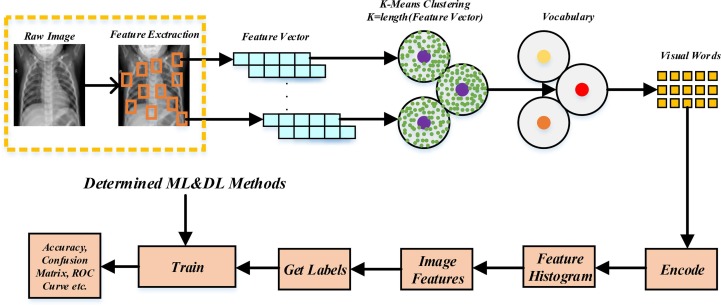
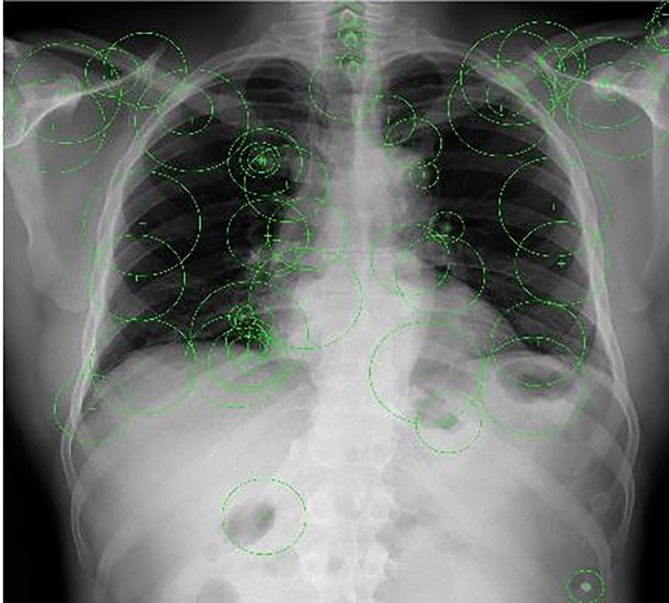
The block diagram of the feature extraction method that is used for the proposed method is shown in Fig. 2 . The bag of features method is used for feature extraction (O’Hara and Draper, 2011). Features in the raw images obtained from the dataset are selected by using the Detector Method. The extracted features are obtained like Fig. 3 using Speed-up Robust Feature (SURF) from selected feature points location (Bay et al., 2006).
Fig. 2.
The block diagram of the feature extraction method using Bag of Features.
Fig. 3.
Feature extraction according to SURF.
The obtained feature vector includes features such as scale, sign of laplacian, orientation, location, and metric. In the proposed method, features are derived from Phase-1 (COVID-19 Positive-COVID-19 Negative) and Phase-2 (Normal-Pneumonia) separately. Afterward, 80 % of the strongest features are kept from each category. It is determined that the image category with the least number of strongest features. The other image category uses the same number of the strongest features, either. So, visual word vocabulary is obtained by using the K-Means clustering method (Alkan and Akben, 2011). The K for Phase-1 and Phase-2 is equal to 2000 and 1500, respectively. Each cluster center represents a feature or a visual word. Obtained visual words are encoded so that a feature histogram can be created for each image. Then, image features are determined using the feature histogram. Labels get for images that are used in the training set. In this way, output parameters can be calculated with the help of machine learning (ML) or deep learning (DL) methods.
3.2. Classification algorithms
In this section, six different classifiers used in the study are explained separately. They are Cosine KNN, Linear Discriminant, Logistic Regression, Bagged Trees Ensemble, Medium Gaussian SVM, and SqueezeNet Deep Learning. These classifiers can be explained as follows:
3.2.1. Cosine KNN
The KNN classifier is one of the most used classification algorithms, and it is one of the simplest classifiers. The KNN classifier calculates distances between testing and training data samples. Here, k is the number of the nearest neighbor. When the KNN uses the cosine similarity measure, the testing set is classified using Cosine KNN (Ali et al., 2019).
3.2.2. Linear discriminant
The Linear Discriminant analysis is one of the widely used data analysis methods. The purpose of the Linear Discriminant is to define a low-dimensional linear subspace for two or more classes in which data points can be separated optimally. It can be used for dimensionality reduction, recognition, and supervised classification. The Linear Discriminant analysis has advantages such as low-cost implementation, correspondence to Bayes’s optimal classification, and easy adaptation for discriminating non-linearly separable classes (Markopoulos, 2017).
3.2.3. Logistic regression
The Logistic Regression is a supervised classification algorithm used because of the correlation between discrete variables, and it works well on numerical datasets. Here, the Logistic Regression consists of two dependent variables (0,1), where they express as false and true, respectively. Thus, it aims to find the most suitable model among the independent variables (Mary Gladence et al., 2015).
3.2.4. Bagged trees ensemble
The ensemble method is a supervised classification technique that combines different machine learning algorithms, and it aims to obtain more accurate predictions. In particular, the ensemble of decision trees is one of the most widely used successful classification algorithms among the classification algorithms. These techniques can be used for classification, regression, and ranking. Here, bagging can also be expressed as bootstrap aggregating, and each classifier is individually trained, and combined by getting an average (González et al., 2020).
3.2.5. Medium Gaussian SVM
The SVM is one of the supervised machine learning algorithms, and one of the most robust prediction methods. The SVM creates a model by aiming to maximize the width of the margin between the two categories. Although the SVM is an efficient classifier algorithm for memory and high-dimensional space, it may not be an efficient classifier for noisy datasets. Besides, the SVM classifier is helpful in text, hypertext, image, and signal classification. If Medium Gaussian is determined as the kernel function for the SVM, the Medium Gaussian SVM classifier can be used in the training phase (Bhati and Rai, 2020-Alkan, 2011).
3.2.6. SqueezeNet deep learning
SqueezeNet is one of the most widely used CNN, and it has 18 deep layers. Additionally, SqueezeNet Convolutional Neural Network has an overall of 68 layers, and this network can be called the pre-trained network. Herein, the pre-trained network can classify images into 1000 categories, such as COVID-19 Positive, Normal, and Pneumonia, etc. The input size of the images used in this network is 227*227*3 (Iandola et al., 2016).
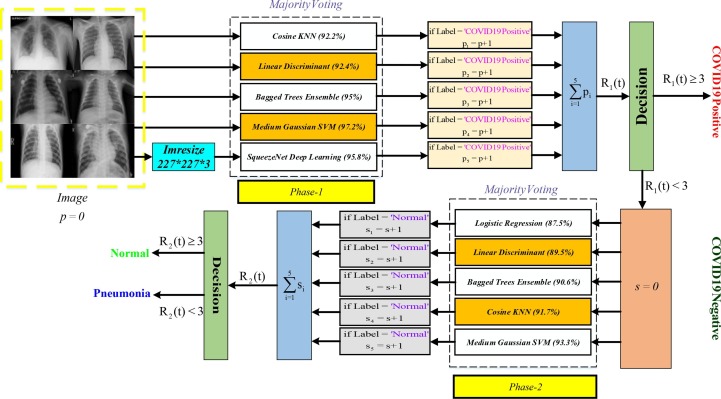
In Fig. 4 , the block diagram of the proposed method is given for COVID-19 disease detection. Here, systems are trained using six different classifiers. The systems used in the study are Cosine KNN, Linear Discriminant, Bagged Trees Ensemble, Medium Gaussian SVM, Logistic Regression, and SqueezeNet Deep Learning. The five most successful classifiers are identified in both phases. In phase-1, the person is determined to be COVID-19 Positive or Negative. For phase-1, the five most successful systems are Cosine KNN (92.2 %), Linear Discriminant (92.4 %), Bagged Trees Ensemble (95 %), SqueezeNet Deep Learning (95.86 %), and Medium Gaussian SVM (97.2 %), respectively. Besides, for SqueezeNet Deep Learning, the image size is set as 227*227*3. The accuracy values (%) are calculated according to test sets. Then, the images in the test set apply to the trained classifiers used in Phase-1. In the process, the COVID-19 Positive label is examined for each classifier. If the output is ‘COVID-19 Positive’, the pi-value is increased by 1. The obtained pi values are summed for the five most successful systems. Thus, the function R1(t) is obtained from the summation symbol. In the decision process, it is determined that the image is COVID-19 Positive or Negative by using majority voting. If the output is COVID-19 Negative, s is set to 0, and the image is examined for phase-2.
Fig. 4.
The detailed block diagram of the proposed method developed for COVID-19 disease detection.
In phase-2, the five most successful systems are Logistic Regression (87.5 %), Linear Discriminant (89.5 %), Bagged Trees Ensemble (90.6 %), Cosine KNN (91.7 %), and Medium Gaussian SVM (93.3 %), respectively. These classifiers have been trained using chest X-ray images from Normal and Pneumonia subjects. Afterward, the same image is evaluated for the Normal label (Fig. 4). If the output is Normal for phase-2, the value of si is increased by 1. So, R2(t) function is obtained by summing si values for the five most successful systems. If the person is not COVID-19 Positive, the image classifies as Normal or Pneumonia according to the R2(t) function. Hence, Rx(t) and Ry(t) functions can be given as follows:
| (1) |
| (2) |
where Rx(t) and Ry(t) functions represent the outputs of the majority voting for Phase-1 and Phase-2, respectively. Since n (nЄ{2 t-1 | tЄZ+}) classifiers are used in the proposed method, the threshold value (Th) is determined as (n + 1)/2 (R1(t),R2(t) Th / R1(t),R2(t) < Th / n = 5,Th = 3). According to Eq. (1), the images classify as COVID-19 Positive or Negative. If the output of Phase-1 is COVID-19 Negative, Eq. (2) is examined, and the images are determined as Normal or Pneumonia. To better understand the majority voting used in the study, Majority Voting (Mathematical Evaluation) can be given as follows:
3.3. Majority voting (mathematical evaluation)
The majority Voting system is a decision system that determines the winner for a decision according to more than half of the votes cast (Randhawa et al., 2018). This section indicates the detailed theoretical framework for generalized majority voting systems. The majority voting method can be expressed using the probability of the independent events (Maity, 2018). If we analyze these probability values, A(i) and B(i) functions can be written by,
| (3) |
| (4) |
where A(i) and B(i) functions can be explained as the cluster of the probabilities of correct (P) and incorrect (P'=1-P) estimation of the image, respectively. The number of elements of the clusters is imax for both functions, where imax is equal to C(n,m). Additionally, for A(i) and B(i) functions, the number of elements each i-th subset is k + 1 and n-(k + 1), respectively, where k is equal to m-1. For example, if n = 5 and k = 2, A(1) and B(1) are equal to {P 1, P 2, P 3} and {P 4′, P5′}, respectively. Thus, we can be obtained f(t) and g(t) functions using Eq. (3), (4).
| (5) |
| (6) |
where f(t) and g(t) functions denote the product of the subset elements of A(i) and B(i) functions, respectively. Here, Pr and Pr' show each element of subsets of these functions. For example, if n = 5, m = 3 and i = 4, f(t) and g(t) functions are set to P 1.P 3.P 4 and P 2′.P5′, respectively. Using the f(t) and g(t) functions, Eq. (7) can be given as follows:
| (7) |
Eq. (7) gives the accuracy expression used in the designed system, and the accuracy can be found for Phase-1 or Phase-2. fi(t) and gi(t) functions are multiplied from i = 1 to i = imax to obtain the hi(t) functions (hi(t) = fi(t).gi(t)). The obtained hi(t) functions are summed from m = Th to m = n. Hence, the accuracy of the system is determined using the majority voting. To better understand this situation, we can create Table 2 .
Table 2.
Calculation of the accuracy using majority voting a) for m = 3 b) for m = 4 c) for m = 5.
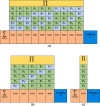 |
Table 2 shows the majority voting for Th m n used in the proposed method where m values for Table 2.a, 2.b, and 2.c are 3, 4, and 5, respectively. Each row in the tables consists of knowledge of the accuracy or error rates of different classifiers. Given that 5 different classifiers are used in the proposed method, it is determined as Th = 3, n = 5, and m 3. Primarily, each element in the column is multiplied among themselves, and hi(t) functions are obtained. The obtained hi(t) functions are summed from i = 1 to i = imax, so that Probability A, Probability B, and Probability C values can be calculated for Table 2.a, 2.b, and 2.c, respectively. These probabilities can be calculated directly using Eq. (7), hence accuracy of the system is as follows:
| (8) |
where the accuracy expressions for Phase-1 or Phase-2 can be calculated by summing the values of the Probability A, Probability B, and Probability C. Probability A, B, and C values are expressed according to C(5,3), C(5,4), and C(5,5), respectively. The training parameters and evaluation metrics obtained for Phase-1 and Phase-2 in the proposed method are given in Table 3 and Table 4 .
Table 3.
Training parameters and evaluation metrics of successful classifiers used for Phase-1.
|
Phase-1 |
Accuracy (100*Pi) | Total Misclassification Cost | Prediction Speed ( obs/sec) | Training Time | Model Type | Feature Selection |
|---|---|---|---|---|---|---|
| Cosine KNN | 92.2 | 107 | 53 | 877.97 sec | K = 10, Distance weight = Equal | 2000 features |
| Linear Discriminant | 92.4 | 105 | 1500 | 75.487 sec | Covariance structure = Full | 2000 features |
| Bagged Trees Ensemble | 95 | 69 | 5200 | 1092.3 sec | Maximum number of splits = 6901, Number of learners = 30 | 2000 features |
| Medium Gaussian SVM | 97.2 | 39 | 250 | 214.64 sec | Kernel scale = 45, Multi class method = One-vs-One | 2000 features |
| SqueezeNet Deep Learning | 95.86 | 59 | – | 950 min 20 sec | Iteration = 1000, Learning rate = 10-4, Image size = 227*227*3 | – |
Table 4.
Training parameters and evaluation metrics of successful classifiers used for Phase-2.
|
Phase-2 |
Accuracy (100* Pi) | Total Misclassification Cost | Prediction Speed ( obs/sec) | Training Time (sec) |
Model Type | Feature Selection |
|---|---|---|---|---|---|---|
| Logistic Regression | 87.5 | 116 | 2400 | 307.04 | – | 1500 features |
| Linear Discriminant | 89.5 | 97 | 1300 | 52.806 | Covariance structure = Full | 1500 features |
| Bagged Trees Ensemble | 90.6 | 87 | 3800 | 354.2 | Maximum number of splits = 4625, Number of learners = 30 | 1500 features |
| Cosine KNN | 91.7 | 77 | 79 | 331.2 | K = 10, Distance weight = Equal | 1500 features |
| Medium Gaussian SVM | 93.3 | 62 | 170 | 226.07 | Kernel scale = 39, Multi class method = One-vs-One | 1500 features |
Table 3 shows the training parameters and evaluation metrics of the five most successful classifiers used for Phase-1. For the systems used in Phase-1, it is seen that Medium Gaussian SVM is the most successful system in terms of accuracy, total misclassification cost, prediction speed, and training time. The accuracy, total misclassification cost, prediction speed, and training time of the Medium Gaussian SVM are equal to 97.2 %, 39, 250 obs/sec, 214.68 sec, respectively. Moreover, the computer that is used in the study has Intel(R) Core(TM) i5-6400 CPU @2.70 GHz 8.00 GB RAM (x64). It is seen that SqueezeNet Deep Learning is the slowest system in terms of time between the classifiers. In Phase-1, 2000 features are extracted using the Bag of Features method in other classifiers, except for SqueezeNet Deep Learning. The deep learning neural network is trained using 227*227*3 COVID-19 Positive/Negative images. Principal Component Analysis that predicts the correlation structure of the variables is not used for any classifier in Phase-1.
Table 4 indicates the training parameters and evaluation metrics of the five most successful classifiers (Logistic Regression 87.5 %, Linear Discriminant 89.5 %, Bagged Trees Ensemble 90.6 %, Cosine KNN 91.7 %, Medium Gaussian SVM 93.3 %) used for Phase-2. It is seen that the most successful system in these classifiers is Medium Gaussian SVM. The accuracy, total misclassification cost, prediction speed, and training time of the Medium Gaussian SVM are equal to 93.3 %, 62, 170 obs/sec, 226.07 sec, respectively. For each classifier used in Phase-2, 1500 features are extracted using the Bag of Features method. Principal Component Analysis methods are not also used in Phase-2 as in Phase-1. By using Eqs. (3)–(7) or Table 2, and Eq. (8) with the help of the required values in Table 1, Table 2, Table 3, Table 4, Eq. (9) can be calculated as follows:
| (9) |
where the overall accuracy of the designed system is given by a generalized formula. Accuracy for Phase-1 and Phase-2 can be calculated using Eq. (8). Since the dataset sizes that are given in Table 1 for Phase-1 and Phase-2 are different in the proposed method, the accuracy of the whole system is obtained by using the weighted arithmetic mean. Additionally, the application of the proposed method, which is designed using the MATLAB GUI, is given as follows:
3.4. GUI application of the proposed method
Since it is easily understood and used by users who are not interested in the subject, the GUI is developed in the study. For this purpose, the designed graphical user interface program provides advantages in terms of both accessibility and time-saving due to the limited number of expert radiologists. Here, the untagged image can apply to the program by the user with the help of a button. Primarily, it is evaluated according to Phase-1, where Positive and Negative values are set to 0. These variables are increased one by one according to the decisions of the five most successful classifiers used in Phase-1. Afterward, the images classify as COVID-19 Positive or COVID-19 Negative using the Majority Voting (Mathematical Evaluation) method. If Decision-1 is COVID-19 Positive, then the image is not examined for Phase-2. When Decision-1 is COVID-19 Negative, Normal and Pneumonia values are set to 0. The Normal and Pneumonia variables are also increased one by one according to the decisions of the five most successful classifiers determined in Phase-2. Likewise, Decision-2 indicates as Normal (Normal > Pneumonia) or Pneumonia (Pneumonia > Normal) using the Majority Voting (Mathematical Evaluation) method for Decision-1: COVID-19 Negative.
4. Experimental study
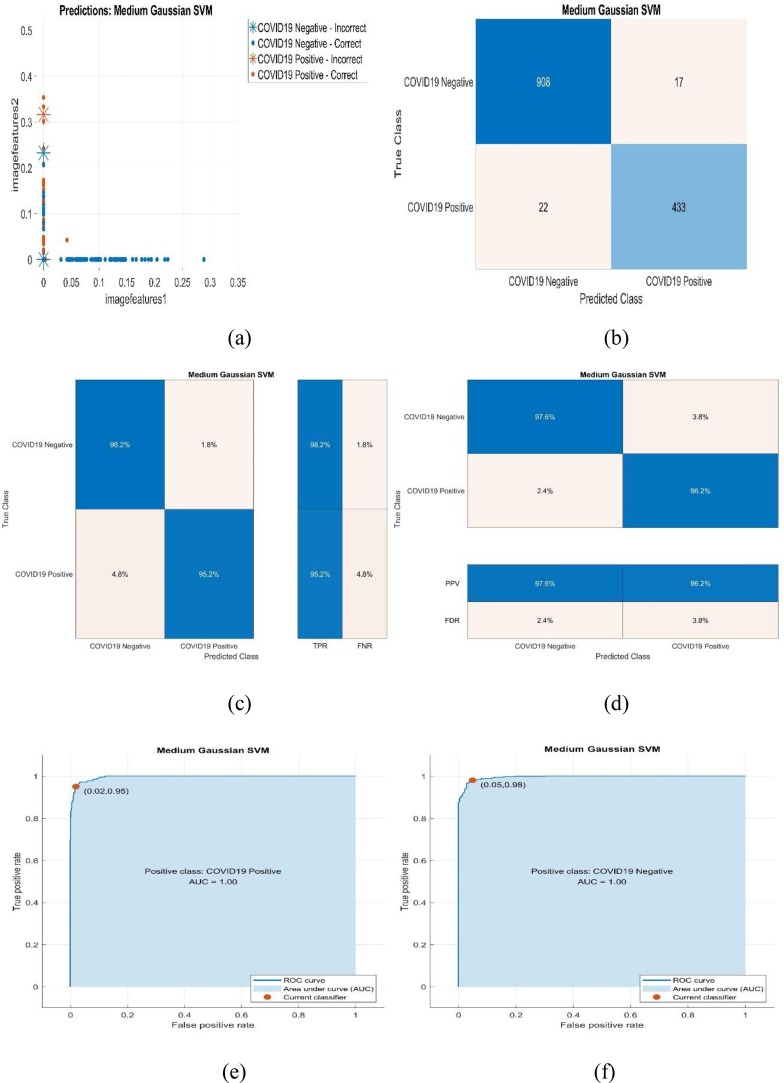
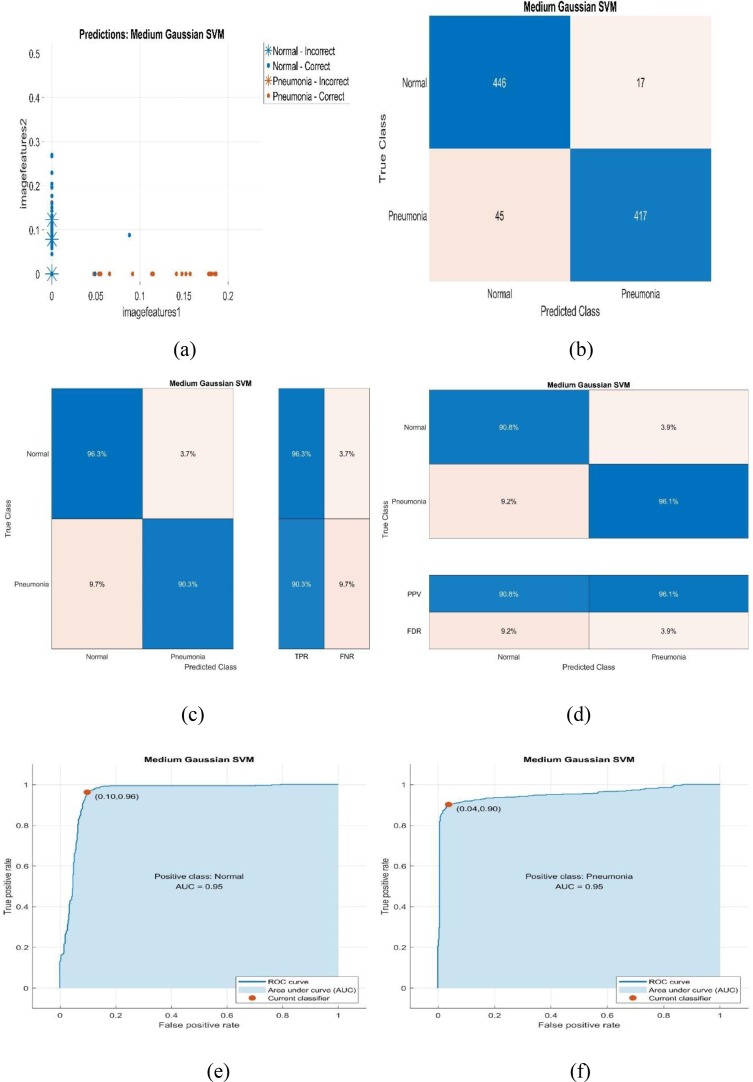
In this section, scatter plots and evaluation metrics of Medium Gaussian SVM, which is the most successful classifier for Phase-1 and Phase-2 in the proposed study, are given. Additionally, it is shown that overall accuracy is calculated using Majority Voting (Mathematical Evaluation) method. Classification performance metrics of the classifiers used by the proposed method are given for Phase-1 and Phase-2. Herein, Majority Voting-1 and Majority Voting-2 performance metrics are denoted according to Phase-1 and Phase-2, respectively. Hence, the classification performance metrics and evaluation metrics of the whole system can be analyzed as Fig. 5, Fig. 6 and Tables 5 :
Fig. 5.
Scatter plot and evaluation metrics of Medium Gaussian SVM that is the most successful classifier for Phase-1.
Fig. 6.
Scatter plot and evaluation metrics of Medium Gaussian SVM that is the most successful classifier for Phase-2.
Table 5.
Classification performance metrics of the proposed method.
| Specificity (%) | Precision (%) | Recall (%) | F1 Score (%) | AUC | MCC | ||
|---|---|---|---|---|---|---|---|
| Phase-1 | Cosine KNN | 95.35 | 90.09 | 85.93 | 87.96 | 0.9064 | 0.8230 |
| Linear Discriminant | 94.70 | 89.06 | 87.69 | 88.37 | 0.9120 | 0.8272 | |
| Bagged Trees Ensemble | 97.19 | 94.06 | 90.55 | 92.27 | 0.9387 | 0.8861 | |
| SqueezeNet Deep Learning | 99.67 | 99.25 | 88.13 | 93.36 | 0.9390 | 0.9072 | |
| Medium Gaussian SVM | 98.16 | 96.22 | 95.16 | 95.69 | 0.9666 | 0.9359 | |
| Majority Voting-1 | 99.98 | 99.83 | 99.07 | 99.51 | 0.9974 | 0.9855 | |
| Phase-2 | Logistic Regression | 88.12 | 87.94 | 86.79 | 87.36 | 0.8746 | 0.7492 |
| Linear Discriminant | 91.14 | 90.82 | 87.88 | 89.33 | 0.8951 | 0.7907 | |
| Bagged Trees Ensemble | 91.57 | 91.39 | 89.61 | 90.49 | 0.9059 | 0.8120 | |
| Cosine KNN | 97.19 | 96.83 | 86.14 | 91.18 | 0.9167 | 0.8386 | |
| Medium Gaussian SVM | 96.33 | 96.08 | 90.26 | 93.08 | 0.9329 | 0.8675 | |
| Majority Voting-2 | 99.73 | 99.69 | 98.63 | 99.23 | 0.9928 | 0.9518 | |
| Overall Majority Voting | 99.88 | 99.78 | 98.90 | 99.40 | 0.9956 | 0.9720 |
Fig. 5 shows the scatter plot and evaluation metrics of the Medium Gaussian SVM that is the most successful classifier for Phase-1, where the Fig. 5.a-5.f show scatter plot, the number of observation, True Positive Rate (TPR)-False Negative Rate (FNR), Positive Predictive Value (PPV)-False Discovery Rate (FDR) for confusion matrix, Receiver Operating Characteristic (ROC) curves for Positive Class: COVID-19 Positive-Negative, respectively. When Fig. 5.c is examined, it is seen that TPR and FNR are equal to 98.2 %-1.8 % and 95.2 %-4.8 % for COVID-19 Negative and Positive classes, respectively. When Fig. 5.d is examined, it is concluded that PPV and FDR are equal to 97.6 %-2.4 % and 96.2 %-3.8 % for COVID-19 Negative and Positive classes, respectively. Fig. 5.e and 5.f show that AUC is 1 for Positive Class: COVID-19 Positive-Negative, respectively.
Fig. 6.a, 6.b, 6.c, 6.d, 6.e, and 6.f show scatter plot, number of observations, TPR-FNR, PPV- FDR for confusion matrix, ROC curves for Positive Class: Normal-Pneumonia, respectively. Here, the most successful classifier used for Phase-2 is Medium Gaussian SVM. When Fig. 6.c and Fig. 6.d are examined, it is seen that TPR, FNR, PPV, and FDR are equal to 96.3 %-90.3 %, 3.7 %-9.7 %, 90.8 %-96.1 %, and 9.2 %-3.9 % for COVID-19 Normal and Pneumonia classes, respectively. From Fig. 6.e and Fig. 6.f, it can be seen that the AUC for Positive Class: Normal-Pneumonia is equal to 0.95. Table 5 is given as follows to examine the classification performance metrics of the proposed method.
Table 5 indicates that the classification performance metrics of the proposed method for Phase-1 and Phase-2. These metrics can be listed as Specificity (%), Precision (%), Recall (%), F1 Score (%), AUC, and MCC. Furthermore, classification performance metrics for Majority Voting 1–2 that can be calculated by using the classification performance metrics of the most successful systems are shown in the table. The table shows that for Majority Voting-1, Specificity (%), Precision (%), Recall (%), F1 Score, AUC, and MCC are equal to 99.98, 99.83, 99.07, 99.51, 0.9974, and 0.9855, respectively. Likewise, the table denotes that they, which are given for Majority Voting-2, are 99.73, 99.69, 98.63, 99.23, 0.9928, and 0.9518, respectively. The classification performance metrics of the whole system can be found by using Majority Voting-1,2 classification performance metrics, Table 1, and Eq. (9). The results show that the classification performance metrics, which are Specificity (%), Precision (%), Recall (%), F1 Score, AUC, and MCC for Overall Majority Voting, are equal to 99.88, 99.78, 98.90, 99.40, 0.9956, and 0.9720, respectively. For Overall Majority Voting, the results show that the classification performance metrics, which are Specificity (%), Precision (%), Recall (%), F1 Score, AUC, and MCC are equal to 99.88, 99.78, 98.90, 99.40, 0.9956, and 0.9720, respectively.
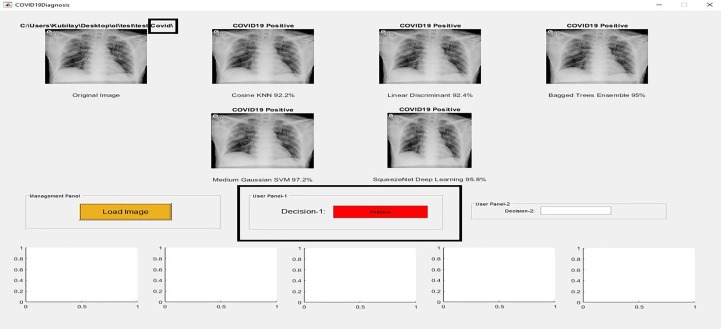
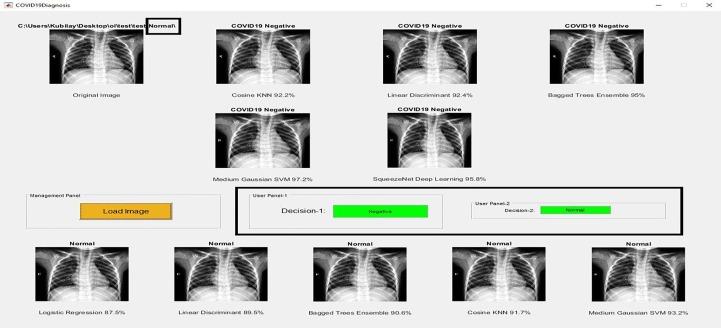
The application of the proposed method is developed in the MATLAB GUI page, which can be considered a decision support system that users can utilize to diagnose COVID-19 disease. Screenshots of the application designed for users to diagnose the COVID-19 disease are given in Fig. 7, Fig. 8, Fig. 9 , where COVID-19 Positive, Normal, and Pneumonia images are shown in Fig. 7, Fig. 8, Fig. 9, respectively.
Fig. 7.
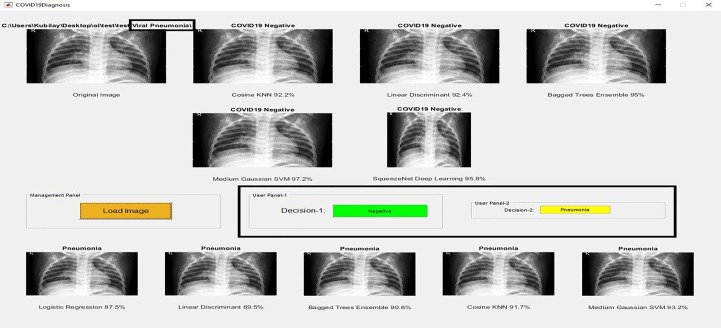
Screenshot of the application designed for users to diagnose the COVID-19 disease, Decision: COVID-19 Positive.
Fig. 8.
Screenshot of the application designed for users to diagnose the COVID-19 disease, Decision: Normal.
Fig. 9.
Screenshot of the application designed for users to diagnose the COVID-19 disease, Decision: Pneumonia.
Original images used in the application do not include in the training or validation set. Here, the image can be transferred to the application by using the 'Load Image' button that is found in 'Management Panel'. This image is applied to the classifiers trained in Phase-1 and Phase-2, and the class of the image is estimated. In application, Cosine KNN, Linear Discriminant, Bagged Trees Ensemble, Medium Gaussian SVM, and SqueezeNet Deep Learning are used for Phase-1. In these classifiers, 'Decision-1′ that is found in 'User Panel-1′ is determined by majority voting. If the image classifies as COVID-19 Positive, 'Decision-1′ will be Positive.
Otherwise, 'Decision-1′ would be Negative. If the image classifies as COVID-19 Negative, it is examined for Phase-2. The classifiers used for Phase-2 are Logistic Regression, Linear Discriminant, Bagged Trees Ensemble, Cosine KNN, and Medium Gaussian SVM. After the image classifies as COVID-19 Negative in Phase-1, it is examined for Normal and Pneumonia. If the image classifies as Normal, 'Decision-2′ that is found in 'User Panel-2′ will be Normal. Otherwise, 'Decision-2′ would be Pneumonia.
5. Results and discussion
In the paper, COVID-19 disease is detected using chest X-ray images from COVID-19 Positive, Normal, and Pneumonia subjects. In the proposed method, six different classifiers are trained using these images. These classifiers are listed as Cosine KNN, Linear Discriminant, Logistic Regression, Bagged Trees Ensemble, Medium Gaussian SVM, and SqueezeNet Deep Learning. The proposed method consists of two steps that are Phase-1 and Phase-2. In Phase-1 and Phase-2, images classify as COVID-19 Positive/Negative and Normal/Pneumonia, respectively. Image features are extracted using the Bag of Features method for other classifiers, excluding SqueezeNet Deep Learning. Here, the five most successful classifiers in Phase-1 and Phase-2 are determined, and images classify using the Majority Voting (Mathematical Evaluation) method. Therefore, a detailed theoretical infrastructure is created for the Majority Voting (Mathematical Evaluation) method used in the study. For Phase-1, the accuracy values of Cosine KNN, Linear Discriminant, Bagged Trees Ensemble, Medium Gaussian SVM, and SqueezeNet Deep Learning classifiers are equal to 92.2 %, 92.4 %, 95 %, 97.2 %, and 95.86 %, respectively. For Phase-2, the accuracy values of Logistic Regression, Linear Discriminant, Bagged Trees Ensemble, Cosine KNN, and Medium Gaussian SVM classifiers are equal to 87.5 %, 89.5 %, 90.6 %, 91.7 %, and 93.3 %, respectively. So, the accuracy values for both phases can be combined with the help of Majority Voting (Mathematical Evaluation). Results show that the overall accuracy values of Phase-1 and Phase-2 are equal to 99.86 % and 99.28 %, respectively. Thus, the accuracy of the whole system is calculated using Majority Voting (Mathematical Evaluation), and it is equal to 99.63 %. When the classification performance metrics are examined for Phase-1,2, the results show that Specificity (%), Precision (%), Recall (%), F1 Score (%), AUC, and MCC are 99.98–99.83–99.07–99.51–0.9974–0.9855 and 99.73–99.69–98.63–99.23–0.9928–0.9518, respectively. Specificity (%), Precision (%), Recall (%), F1 Score (%), AUC, and MCC for the whole system is equal to 99.88, 99.78, 98.90, 99.40, 0.9956, and 0.9720, respectively. Furthermore, the application of the proposed method is designed using the MATLAB GUI for users to diagnose the COVID-19 disease.
Table 6 shows the accuracy values of the proposed method and (Chandra et al., 2021), which is one of the most successful studies. From the table, it is seen that both systems are designed according to Phase-1 and Phase-2. While the proposed method uses chest X-ray images from COVID-19 Positive and Negative (Normal-Pneumonia) subjects for Phase-1, (Chandra et al., 2021) uses chest X-ray images obtained from Normal and Abnormal (COVID-19 Positive-Pneumonia) subjects. If the output of Phase-1 is Abnormal for (Chandra et al., 2021), COVID-19 Positive and Pneumonia images are examined for Phase-2.
Table 6.
Three classes performance comparison of the proposed method and (Chandra et al., 2021) that is using majority voting (ground truth and mathematical evaluation).
| Chandra et al. (2020) | Proposed Method | Accuracy (%) | ||
|---|---|---|---|---|
| Phase-1 | Naïve Bayes | Cosine KNN | 88.372 | 92.2 |
| Decision Tree | Linear Discriminant | 90.698 | 92.4 | |
| KNN | Bagged Trees Ensemble | 95.736 | 95 | |
| SVM (Poly Kernel) | SqueezeNet Deep Learning | 96.124 | 95.86 | |
| ANN | Medium Gaussian SVM | 96.512 | 97.2 | |
| Majority Voting (Ground Truth) | 98.062 | – | ||
| Majority Voting (Mathematical Evaluation) | 99.79 | 99.86 | ||
| Phase-2 | KNN | Logistic Regression | 72.093 | 87.5 |
| ANN | Linear Discriminant | 73.256 | 89.5 | |
| Decision Tree | Bagged Trees Ensemble | 79.070 | 90.6 | |
| Naïve Bayes | Cosine KNN | 80.814 | 91.7 | |
| SVM (RBF Kernel) | Medium Gaussian SVM | 86.628 | 93.3 | |
| Majority Voting (Ground Truth) | 91.329 | – | ||
| Majority Voting (Mathematical Evaluation) | 93.08 | 99.28 | ||
| Overall | Majority Voting (Ground Truth) | 93.41 | – | |
| Majority Voting (Mathematical Evaluation) | 97.11 | 99.63 | ||
If the output of Phase-1 is COVID-19 Negative for the proposed method, Normal and Pneumonia images are examined for Phase-2. Whereas the classifiers used by (Chandra et al., 2021) in Phase-1 are Naïve Bayes, Decision Trees, KNN, SVM (Poly Kernel), and Artificial Neural Network (ANN), the proposed method uses Cosine KNN, Linear Discriminant, Bagged Trees Ensemble, SqueezeNet Deep Learning, and Medium Gaussian SVM classifiers. For Phase-1, the accuracy of (Chandra et al., 2021) that can be calculated the performance evaluation using the Majority Voting (Ground Truth) method is equal to 98.062 %. Here, the accuracy of both systems can be calculated according to the Majority Voting (Mathematical Evaluation). From Table 6, it is seen that the accuracy of the proposed method and (Chandra et al., 2021) is equal to 99.86 % and 99.79 %, respectively.
Whereas the classifiers used by (Chandra et al., 2021) in Phase-2 are KNN, ANN, Decision Trees, Naïve Bayes, and SVM (RBF Kernel), the proposed method uses Logistic Regression, Linear Discriminant, Bagged Trees Ensemble, Cosine KNN, and Medium Gaussian SVM classifiers. In Phase-2, the accuracy of (Chandra et al., 2021) obtained using the Majority Voting (Ground Truth) method is 91.329 %. When the accuracy of the proposed method and (Chandra et al., 2021) determined using the Majority Voting (Mathematical Evaluation) method is analyzed, it is seen that they are equal to 99.28 % and 93.08 %, respectively. The overall accuracy of (Chandra et al., 2021) calculated using Majority Voting (Ground Truth) for Phase-1 and Phase-2 is equal to 93.41 %. As the accuracy of the systems determined according to the Majority Voting (Mathematical Evaluation) method is analyzed, it can be concluded that the accuracy of the proposed method and (Chandra et al., 2021) is 99.63 % and 97.11 %, respectively. It is seen that the accuracy of the proposed method is better than (Chandra et al., 2021).
In the light of this information, it is one of the current and innovative studies published in the Expert Systems with Applications journal (Chandra et al., 2021) that can diagnose COVID-19. It is seen that the proposed system is more advantageous because it has better performance metrics and a user-friendly GUI application. In the study of (Chandra et al., 2021), Normal-Abnormal and COVID-19 Positive-Pneumonia is detected in Phase-1 and Phase-2, respectively. Herein, it seems that our proposed system also gives better results as Normal and Pneumonia are evaluated as COVID-19 Negative. Because, as can be understood from both studies, it can be seen from the numerical results that pneumonia is more difficult to detect. Therefore, the proposed method is grouped as such. In this context, as can be seen in Table 6, the common evaluation metrics of these two parallel studies can be determined. It is seen that the Majority Voting (Mathematical Evaluation) technique, which gives final results based on the model accuracies used in both studies, can facilitate the comparison considerably. Thus, the originality of the study can be revealed. In addition, although there are many studies in the literature that can detect COVID-19 disease (please see Introduction), the usability of these models is unfortunately low. Because the proposed models generally do not have user-friendly software. At this point, we are designing a GUI application that experts can use easily. Thus, we aim to ensure that the reliable models developed for the solution of the identified problem can be easily used by experts, etc. In addition, we predict that the proposed models with high-performance metrics and their GUI applications can contribute significantly to the literature.
6. Conclusions
We propose a hybrid method that can classify COVID-19 Positive, Normal, and Pneumonia images using machine learning and deep learning methods in the paper. We analyze the chest X-ray images used in the study for two steps, which are Phase-1 and Phase-2, where Cosine KNN, Linear Discriminant, Logistic Regression, Bagged Trees Ensemble, Medium Gaussian SVM, and SqueezeNet Deep Learning is used. Additionally, we extract the image features using the Bag of Features for other classifiers, excluding SqueezeNet Deep Learning. In both phases, we train six different classifiers using chest X-ray images from COVID-19 Positive, Normal, and Pneumonia subjects, where COVID-19 Positive/Negative and Normal/Pneumonia images classify for Phase-1 and Phase-2, respectively. The five most successful classifiers found in Phase-1 and Phase-2 are determined, and these classifiers are selected. Then, chest X-ray images classify using the Majority Voting (Mathematical Evaluation) method, where it is found the most successful classifiers for both phases. Besides, we have designed the application of the proposed method using MATLAB GUI, because it is easy to use, provides time-saving, and the number of expert radiologists is limited.
As mentioned before, image features for all classifiers except SqueezeNet architecture are extracted using the Bag of Features method. With the help of deep learning techniques, it is now seen that the image features can be extracted from the models themselves. Thus, deep learning models can be considered more practical. However, it can be seen in this study that classical machine learning techniques can also give more successful results for this problem. Therefore, we have not select the 5 classifiers from deep learning models in this study. We also seen that classical machine learning techniques can be trained more quickly than pre-trained deep learning techniques. On the other hand, when the proposed systems in the study were examined, we chose the five most successful classifiers from 6 different classifiers. If we had used 6 classifiers, any 3 different classifiers would have made the same decision. In this case, the proposed model would not be able to decide. In order to avoid this situation, we preferred to use 5 different classifiers. Thus, it is seen that the biphasic majority voting technique used for this study can work successfully. Another prominent issue in the study is the finding of total metrics using the weighted average technique. This is because the number of images of the classes in the dataset is different. Herein, if the number of images in different classes were equal, we could use the arithmetic mean technique. Therefore, this study, which was constructed in accordance with the problem, is innovative in terms of the methods it uses. Also, Normal and COVID-19 Positive/Pneumonia are detected in the first phase of (Chandra et al., 2021). However, for this problem, it can be seen that these two classes have more similar properties than the normal class. Therefore, in this study, we first classified the COVID-19 Positive and COVID-19 Negative images. In addition, the results show that the best performance metrics for the dataset used were obtained in this study. (Chandra et al., 2021) and the proposed system are relatively parallel studies, but it can be clearly seen that the proposed system is better than it. The proposed system is innovative and competitive as it is more successful than one of the most recent studies published in Expert Systems with Applications (Chandra et al., 2021) and has a user-friendly GUI application. Already, thanks to the GUI application developed, it is possible to diagnose whether the subject has COVID-19 in 15 s, and whether the same subject is Normal or Pneumonia in about 40 s. In future studies, it is planned to work on reducing the running time of the proposed model.
CRediT authorship contribution statement
Kubilay Muhammed Sunnetci: Methodology, Software, Investigation, Writing – original draft, Conceptualization, Resources, Data curation. Ahmet Alkan: Methodology, Writing – original draft, Conceptualization, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors would like to thank the anonymous referees for their helpful comments and suggestions.
Footnotes
Peer review under responsibility of Submissions with the production note ‘Please add the Reproducibility Badge for this item’ the Badge and the following footnote to be added:The code (and data) in this article has been certified as Reproducible by the CodeOcean: https://codeocean.com. More information on the Reproducibility Badge Initiative is available at https://www.elsevier.com/physicalsciencesandengineering/computerscience/journals.
References
- Ali N., Neagu D., Trundle P. Evaluation of k-nearest neighbour classifier performance for heterogeneous data sets. SN Applied Sciences. 2019;1(12):1–15. doi: 10.1007/s42452-019-1356-9. [DOI] [Google Scholar]
- Alkan A. Analysis of knee osteoarthritis by using fuzzy c-means clustering and SVM classification. Scientific Research and Essays. 2011;6(20):4213–4219. doi: 10.5897/sre11.068. [DOI] [Google Scholar]
- Alkan A., Akben S.B. Use of K-means clustering in migraine detection by using EEG records under flash stimulation. International Journal of Physical Sciences. 2011;6(4):641–650. doi: 10.5897/IJPS11.174. [DOI] [Google Scholar]
- Alkan A., Tuncer S.A., Gunay M. Comparative MR image analysis for thyroid nodule detection and quantification. Measurement: Journal of the International Measurement Confederation. 2014;47(1):861–868. doi: 10.1016/j.measurement.2013.10.009. [DOI] [Google Scholar]
- COVID-19 Pandemic (2021). Retrieved May 27, 2021, from https://www.worldometers.info/coronavirus/.
- Bay H., Tuytelaars T., Van Gool L. Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics) 2006. SURF: Speeded up robust features. [DOI] [Google Scholar]
- Bhati B.S., Rai C.S. Analysis of Support Vector Machine-based Intrusion Detection Techniques. Arabian Journal for Science and Engineering. 2020;45(4):2371–2383. doi: 10.1007/s13369-019-03970-z. [DOI] [Google Scholar]
- Chandra T.B., Verma K., Singh B.K., Jain D., Netam S.S. Coronavirus disease (COVID-19) detection in Chest X-Ray images using majority voting based classifier ensemble. Expert Systems with Applications. 2021;165 doi: 10.1016/j.eswa.2020.113909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chassagnon G., Vakalopoulou M., Battistella E., Christodoulidis S., Hoang-Thi T.N., Dangeard S.…Paragios N. AI-driven quantification, staging and outcome prediction of COVID-19 pneumonia. Medical Image Analysis. 2021;67 doi: 10.1016/j.media.2020.101860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [Dataset] COVID19, Pneumonia, and Normal Chest X Ray PA Dataset (2021). Retrieved May 27, 2021, from https://www.kaggle.com/amanullahasraf/covid19-pneumonia-normal-chest-xray-pa-dataset.
- González S., García S., Del Ser J., Rokach L., Herrera F. A practical tutorial on bagging and boosting based ensembles for machine learning: Algorithms, software tools, performance study, practical perspectives and opportunities. Information Fusion. 2020;64:205–237. doi: 10.1016/j.inffus.2020.07.007. [DOI] [Google Scholar]
- Göreke V., Sarı V., Kockanat S. A novel classifier architecture based on deep neural network for COVID-19 detection using laboratory findings. Applied Soft Computing. 2021;106 doi: 10.1016/j.asoc.2021.107329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iandola, F. N., Han, S., Moskewicz, M. W., Ashraf, K., Dally, W. J., & Keutzer, K. (2016). SqueezeNet: AlexNet-level accuracy with 50x fewer parameters and <0.5MB model size, (February), 0–13. http://arxiv.org/abs/1602.07360.
- Ismael A.M., Şengür A. Deep learning approaches for COVID-19 detection based on chest X-ray images. Expert Systems with Applications. 2021;164 doi: 10.1016/j.eswa.2020.114054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamshidi M., Lalbakhsh A., Talla J., Peroutka Z., Hadjilooei F., Lalbakhsh P.…Mohyuddin W. Artificial Intelligence and COVID-19: Deep Learning Approaches for Diagnosis and Treatment. IEEE Access. 2020;8:109581–109595. doi: 10.1109/ACCESS.2020.3001973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karakanis S., Leontidis G. Lightweight deep learning models for detecting COVID-19 from chest X-ray images. Computers in Biology and Medicine. 2021;130:1–9. doi: 10.1016/j.compbiomed.2020.104181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R., Arora R., Bansal V., Sahayasheela V.J., Buckchash H., Imran J.…Raman B. Accurate Prediction of COVID-19 using Chest X-Ray Images through Deep Feature Learning model with SMOTE and Machine Learning Classifiers. 2020;medRxiv:1–10. doi: 10.1101/2020.04.13.20063461. [DOI] [Google Scholar]
- Li Z., Zhao W., Shi F., Qi L., Xie X., Wei Y.…Shen D. A novel multiple instance learning framework for COVID-19 severity assessment via data augmentation and self-supervised learning. Medical Image Analysis. 2021;69:101978. doi: 10.1016/j.media.2021.101978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markopoulos, P. (2017). Linear Discriminant Analysis with Few Training Data. Panos P. Markopoulos Department of Electrical and Mircoelectronic Engineering Rochester Institute of Technology. IEEE International Conference on Acoustics, Speech, and Signal Processing (ICASSP) 2017, 4626–4630.
- Maity R. Statistical Methods in Hydrology and Hydroclimatology. Springer Transactions in Civil and Environmental Engineering. Springer; Singapore: 2018. Basic Concepts of Probability and Statistics. [DOI] [Google Scholar]
- Mary Gladence L., Karthi M., Maria Anu V. A statistical comparison of logistic regression and different bayes classification methods for machine learning. ARPN Journal of Engineering and Applied Sciences. 2015;10(14):5947–5953. [Google Scholar]
- O’Hara, S., & Draper, B. A. (2011). Introduction to the Bag of Features Paradigm for Image Classification and Retrieval, (June 2014). http://arxiv.org/abs/1101.3354.
- Oh Y., Park S., Ye J.C. Deep learning COVID-19 features on CXR using limited training data sets. arXiv. 2020;39(8):2688–2700. doi: 10.1109/TMI.2020.2993291. [DOI] [PubMed] [Google Scholar]
- Ozyurt F., Tuncer T., Subasi A. An automated COVID-19 detection based on fused dynamic exemplar pyramid feature extraction and hybrid feature selection using deep learning. Computers in Biology and Medicine. 2021;132 doi: 10.1016/j.compbiomed.2021.104356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panwar H., Gupta P.K., Siddiqui M.K., Morales-Menendez R., Singh V. Application of deep learning for fast detection of COVID-19 in X-Rays using nCOVnet. Chaos, Solitons and Fractals. 2020;138 doi: 10.1016/j.chaos.2020.109944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randhawa K., Loo C.K., Seera M., Lim C.P., Nandi A.K. Credit Card Fraud Detection Using AdaBoost and Majority Voting. IEEE Access. 2018;6:14277–14284. doi: 10.1109/ACCESS.2018.2806420. [DOI] [Google Scholar]
- Sedik A., Iliyasu A.M., El-Rahiem B.A., Abdel Samea M.E., Abdel-Raheem A., Hammad M.…Abd El-Latif A.A. Deploying machine and deep learning models for efficient data-augmented detection of COVID-19 infections. Viruses. 2020;12(7) doi: 10.3390/v12070769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selcuk T., Alkan A. Detection of microaneurysms using ant colony algorithm in the early diagnosis of diabetic retinopathy. Medical Hypotheses. 2019;129 doi: 10.1016/j.mehy.2019.109242. [DOI] [PubMed] [Google Scholar]
- Serte S., Demirel H. Deep learning for diagnosis of COVID-19 using 3D CT scans. Computers in Biology and Medicine. 2021;132 doi: 10.1016/j.compbiomed.2021.104306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheykhivand S., Mousavi Z., Mojtahedi S., Yousefi Rezaii T., Farzamnia A., Meshgini S., Saad I. Developing an efficient deep neural network for automatic detection of COVID-19 using chest X-ray images. Alexandria Engineering Journal. 2021;60(3):2885–2903. doi: 10.1016/j.aej.2021.01.011. [DOI] [Google Scholar]
- Signoroni A., Savardi M., Benini S., Adami N., Leonardi R., Gibellini P.…Farina D. BS-Net: Learning COVID-19 pneumonia severity on a large chest X-ray dataset. Medical Image Analysis. 2021;71 doi: 10.1016/j.media.2021.102046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuncer T., Ozyurt F., Dogan S., Subasi A. Chemometrics and Intelligent Laboratory Systems A novel Covid-19 and pneumonia classi fi cation method based on. Chemometrics and Intelligent Laboratory Systems. 2021;210 doi: 10.1016/j.chemolab.2021.104256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y.C., Chen C.S., Chan Y.J. The outbreak of COVID-19: An overview. Journal of the Chinese Medical Association. 2020;83(3):217–220. doi: 10.1097/JCMA.0000000000000270. [DOI] [PMC free article] [PubMed] [Google Scholar]