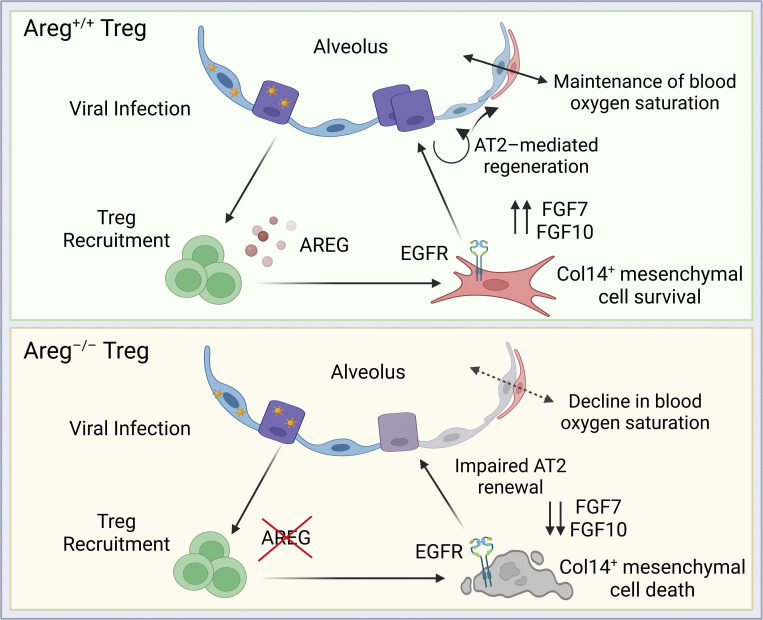
During influenza virus infection, Treg cell–derived amphiregulin is sensed by EGFR+Collagen-14+ lung mesenchymal cells, stimulating their activation and survival and supporting their role as signaling relay intermediates to promote effective alveolar regeneration.
Abstract
Following respiratory viral infection, regeneration of the epithelial barrier is required to preserve lung function and prevent secondary infections. Lung regulatory T (Treg) cells are critical for maintaining blood oxygenation following influenza virus infection through production of the EGFR ligand amphiregulin (Areg); however, how Treg cells engage with progenitors within the alveolar niche is unknown. Here, we describe local interactions between Treg cells and an Areg-responsive population of Col14a1+EGFR+ lung mesenchymal cells that mediate type II alveolar epithelial (AT2) cell-mediated regeneration following influenza virus infection. We propose a mechanism whereby Treg cells are deployed to sites of damage and provide pro-survival cues that support mesenchymal programming of the alveolar niche. In the absence of fibroblast EGFR signaling, we observe impaired AT2 proliferation and disrupted lung remodeling following viral clearance, uncovering a crucial immune/mesenchymal/epithelial network that guides alveolar regeneration.
Graphical Abstract
Introduction
Host resilience against respiratory viral infections requires a calibrated immune response that effectively clears pathogens while limiting excessive inflammation. As seen in patients with COVID-19 and other severe respiratory infections, inappropriate inflammatory and tissue protective responses generated early during infection can lead to acute respiratory distress syndrome and mortality (Levy and Serhan, 2014; Matthay et al., 2019; Rolfes et al., 2018; Tay et al., 2020). Although infrequent within non-lymphoid tissues such as the lung, Foxp3+CD4+ regulatory T (Treg) cells have a profound effect on mitigating tissue damage and promoting repair across multiple organ systems through their production of tissue-specific growth factors and cytokines (Zaiss et al., 2019; Zhang et al., 2017). Among these factors is amphiregulin (Areg), an epidermal growth factor receptor (EGFR) ligand involved in many developmental, regenerative, and proliferative processes (Berasain and Avila, 2014; Burzyn et al., 2013; Green et al., 2017; Hui et al., 2017; Ito et al., 2019; Meulenbroeks et al., 2015), and a critical modulator of lung repair (Arpaia et al., 2015; Ding et al., 2016; Manzo et al., 2012; Minutti et al., 2019; Monticelli et al., 2011; Morimoto et al., 2018; Nordgren et al., 2018). Although EGFR ligands including Areg are abundantly produced by the epithelium and other immune cells in the lung, we previously demonstrated that Treg cell–derived Areg is uniquely important for maintaining lung function during influenza virus infection (Arpaia et al., 2015). Mice conditionally lacking Treg cell sources of Areg suffer a severe reduction in blood oxygen saturation (SpO2) without alteration in viral load, immune cell infiltrate, or viral antigen–specific conventional T cell responses—suggesting that Treg cell–derived Areg promotes a tissue-protective process within the alveolar epithelium that is distinct from the canonical role of Treg cells in immunosuppression. While considerable research has correlated EGFR-family signaling with lung diseases such as fibrosis and cancer (Green et al., 2017; Vallath et al., 2014), studies have only recently begun to uncover the distinct cell types that respond to immune-derived EGFR ligands in vivo and the spatial relationships that promote non-hematopoietic regeneration following acute injury.
In the distal lung, type II alveolar epithelial cells (AT2) are a progenitor population capable of self-renewal and differentiation into type I alveolar epithelial cells (AT1), elongated squamous cells that line the alveolus and are responsible for gas exchange (Barkauskas et al., 2013). AT2 also support barrier defense and alveolar function through their production of surfactants and antimicrobial modulators (Hogan et al., 2014). Upon infection by PR8/H1N1 influenza virus, AT2 and AT1 are lost as a result of virally induced apoptosis and local inflammatory responses, prompting the expansion and differentiation of a subpopulation of WNT-responsive alveolar epithelial progenitors (AEP) that aid in the reconstruction of lung alveoli (Ibricevic et al., 2006; Nabhan et al., 2018; Zacharias et al., 2018). Many studies have described the cell-intrinsic molecular signaling that drives AT2 activation and differentiation, including the engagement of the WNT, YAP/TAZ, STAT3, BMP/SMAD, TP53, and NOTCH pathways (Basil et al., 2020; Chung et al., 2018; Finn et al., 2019; Frank et al., 2016; Jiang et al., 2020; Kobayashi et al., 2020; Paris et al., 2020; Strunz et al., 2020). AT2 maintenance and activation is modulated by signals from the lung stroma both at steady state and after insult (Adamson et al., 1990; Barkauskas et al., 2013; Nabhan et al., 2018). Distinct subtypes of lung mesenchymal cells respond to different sources of tissue injury and regulate epithelial cell differentiation (Lee et al., 2017; Xie et al., 2018; Zepp et al., 2017). However, in the context of significant immune infiltration, the extent to which hematopoietic cells—and specifically Treg cell–derived growth factors—influence epithelial/mesenchymal cell signaling is poorly understood.
Here, we investigate a mechanism by which Treg cells stimulate the alveolar regenerative niche during influenza virus–induced injury. We observe the rapid mobilization of lung Treg cells to virally infected regions and their distinct colocalization with the lung stroma. Additionally, we identify a population of Collagen-14+ (Col14+) mesenchymal cells that express high levels of EGFR, are uniquely AREG-responsive, and are effective in supporting alveolar differentiation. In the absence of Treg cell–derived Areg, we observe an increase in Col14+ cell death and dysregulated signaling that leads to dysplastic alveolar regeneration. Mechanistically, ineffectual mesenchymal programming leads to impaired AT2 differentiation—thereby resulting in a delayed repair response and reduced lung function during infection.
Results
Treg cells uniquely infiltrate to sites of active viral infection
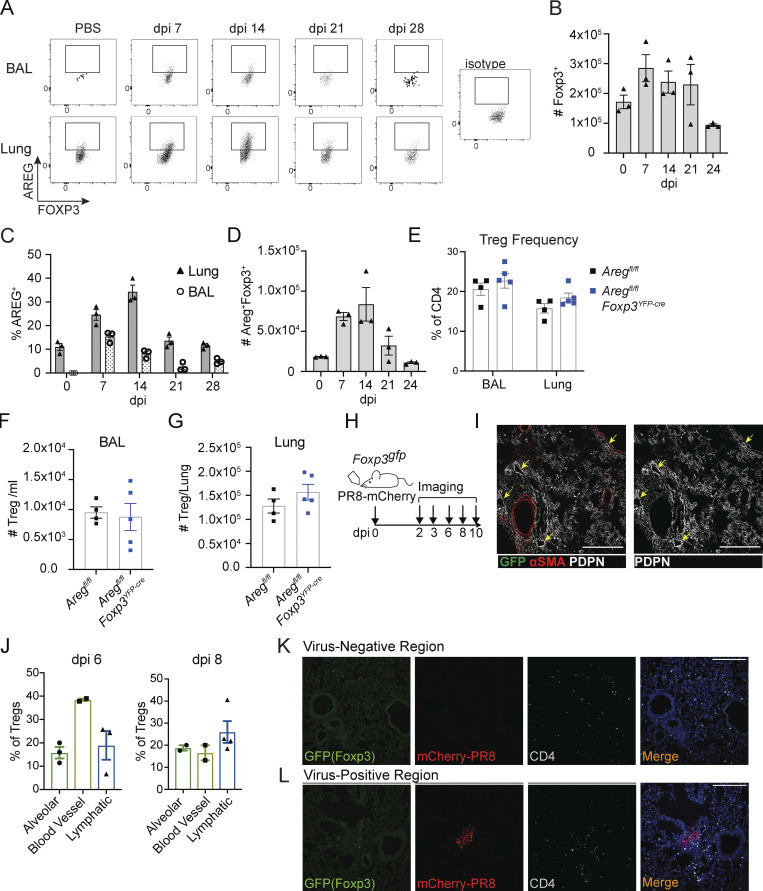
During influenza virus infection, Areg-producing Treg cells help to maintain blood oxygenation and protect against tissue damage through an unknown mechanism (Arpaia et al., 2015). To understand how Treg cells may be acting within the infected lung, we initially characterized Treg infiltration and Areg production in the lung and bronchoalveolar lavage (BAL) by flow cytometry during the peak anti-viral T cell response and inflammatory resolution phases. Treg cells produce Areg soon after infection, with the maximal number of Areg+ Tregs appearing between day post-infection (dpi) 7 and dpi 14, and gradually lose expression to baseline ∼3 wk after viral challenge (Fig. S1, A–D). As previously reported, the frequency or number of Treg cells in the lung and BAL is not altered in Aregfl/flFoxp3YFP-cre mice that lack Areg from Treg cells, indicating that Areg expression does not directly influence their recruitment or expansion within the lung tissue (Fig. S1, E–G).
Figure S1.
Treg cells produce Areg and localize to sites of active influenza virus infection. (A) Kinetic flow cytometric analysis of Areg protein expression in Treg cells at dpi 0, 7, 14, 21, and 28 in the BAL and lung tissue of wildtype mice; n = 3 mice per time point. (B–D) Kinetic flow cytometric analysis of influenza virus infection, with quantification of (B) total Treg cells/lung, (C) frequency of Areg+ Treg cells among CD4+ T cells in the lung and BAL, and (D) total number of Areg+ Treg cells in the lung. (E–G) Flow cytometric analysis of the frequency of Treg cells among all CD4+ T cells in the lung and BAL (E) and the number of Treg cells in the BAL (F) and lung (G) of influenza virus–infected Aregfl/flFoxp3YFP-cre and Aregfl/fl littermate control mice at dpi 6. Representative data from one of two independent experiments shown; n = 4–5 mice/group. (H) Foxp3gfp mice intranasally infected with PR8/H1N1-mCherry or PBS (mock) and analyzed by immunofluorescence imaging at dpi 2, 3, 6, 8, and 10. Treg cells identified by staining with anti-GFP, quantifications performed from multiple images of different regions of the lung. (I and J) Foxp3gfp mice intranasally infected with non-fluorescent PR8/H1N1 analyzed at dpi 6 and dpi 8; n = 3 animals per time point. (I) Treg cell (GFP+) distribution analyzed in relation to vasculature (αSMA+, red) and AT1/lymphatic vessels (PDPN, white) at dpi 8. Yellow arrows indicate lymphatics. Scale bar = 200 µm. (J) Quantification of the percentage of Treg cells found in alveolar regions, within 50 μm of a blood vessel, or in lymphatic vessels at dpi 6 (left) and dpi 8 (right). (K and L) Foxp3gfp mice intranasally infected with PR8/H1N1-mCherry analyzed at dpi 4. Representative images of Treg cells (green) and total CD4+ cells (white) in relation to mCherry-PR8/H1N1 (red) at dpi 3 influenza infection. Presence of each cell type in virus-negative lung regions (K) and virus-positive lung regions (L). Scale bar = 200 µm.
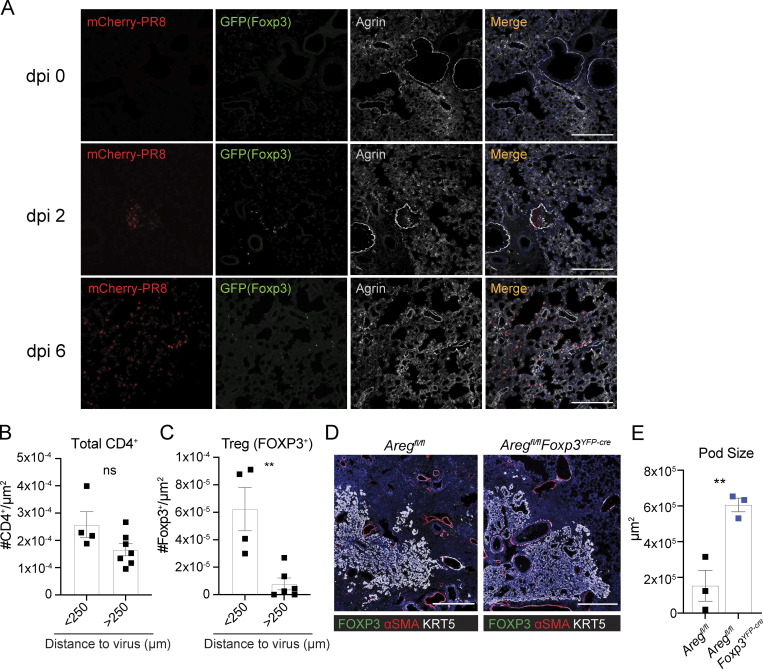
Given that Treg cells are a minor immune cell population within the anti-viral inflammatory response and that many other lung cells produce Areg, we hypothesized that Treg localization may guide their unique function. We first determined the spatial distribution of Treg cells in relation to virally infected cells by infecting Foxp3gfp mice with an mCherry-fluorescent strain of PR8/H1N1 influenza (Fukuyama et al., 2015) and monitoring Treg cell localization over time by immunofluorescence imaging (Fig. S1 H). Lung sections were stained with anti-GFP to identify Treg cells and anti-Agrin, a secreted proteoglycan and component of the extracellular matrix (ECM) that demarcates lung structures (Fig. 1 A). In the absence of infection, Treg cells are extremely infrequent and can occasionally be found near bronchioalveolar duct junctions. Shortly after infection with influenza, at dpi 2, we observe a patchy distribution of Treg cells surrounding pockets of infected mCherry+ upper airway cells. As the virus spreads to distal airways (around dpi 6), Treg cells disperse throughout alveolar spaces. Additional staining for α-smooth-muscle actin (αSMA) expressed on cells lining the endothelium, and podoplanin (PDPN), a protein expressed on lymphatic endothelial cells, AT1, and mesenchymal cells, reveals Treg cells in close proximity to major blood vessels during early stages of infection and more broadly distributed throughout alveolar regions and within/near lymphatic vessels at later time points (Fig. S1, I and J). Notably, Treg localization within alveolar compartments is restricted to regions of active viral replication or areas that were previously infected as identified by regions containing viral debris not encapsulated within a cell. The distribution pattern of Treg cells is distinct from total CD4+ T cells, which are found in both virally infected and uninfected regions, while Treg cells are found nearly exclusively near virus (Fig. 1, B and C; and Fig. S1, K and L). Together, these observations illustrate that Treg cells are among the earliest immune cell types to act at sites of barrier perturbation.
Figure 1.
Treg cells infiltrate to sites of active viral infection and influence epithelial repair. (A) Foxp3gfp mice intranasally infected with PR8/H1N1-mCherry or PBS (mock) were analyzed by immunofluorescence imaging to assess the proximity of Treg cells (green) to PR8-mCherry virus (red) in relation to the secreted ECM proteoglycan, Agrin, at the indicated dpi. Scale bar = 200 µm. (B and C) Quantification of total CD4+ T cells (B) or Foxp3+ Treg cells (C) located within less than or greater than 250 μm from PR8/H1N1-mCherry virus infected regions at dpi 4. (D) Immunofluorescence imaging of KRT5+ pods (white), αSMA+ cells (red), and FOXP3+ Treg cells in Aregfl/fl (control) or Aregfl/flFoxp3YFP-cre mice at 17 dpi with PR8/H1N1. Scale bar = 400 μm. (E) Quantification of the total area of KRT5+ pods in whole mouse lung sections; n = 3 mice/group. Representative data from one of two independent experiments shown. On vertical bar plots, each point represents a biologic replicate and bars show mean ± SEM. Data were analyzed by two-tailed unpaired Student’s t test (** P < 0.01).
While it is surprising that a relatively small population of Areg-producing Treg cells has such a considerable impact on lung protection (Arpaia et al., 2015), Areg signaling is thought to be highly spatially regulated by its heparin-binding domain that allows interaction with heparin sulfate proteoglycans (HSPGs) of the ECM (Berasain and Avila, 2014; Sarrazin et al., 2011). In fact, HSPGs are required for AREG activation of EGFR, as treatment with heparinase in vitro abrogates AREG signaling (Johnson and Wong, 1994). Therefore, Areg activity within tissues may be uniquely shaped by cell–cell contacts, short-range activity, and surrounding ECM.
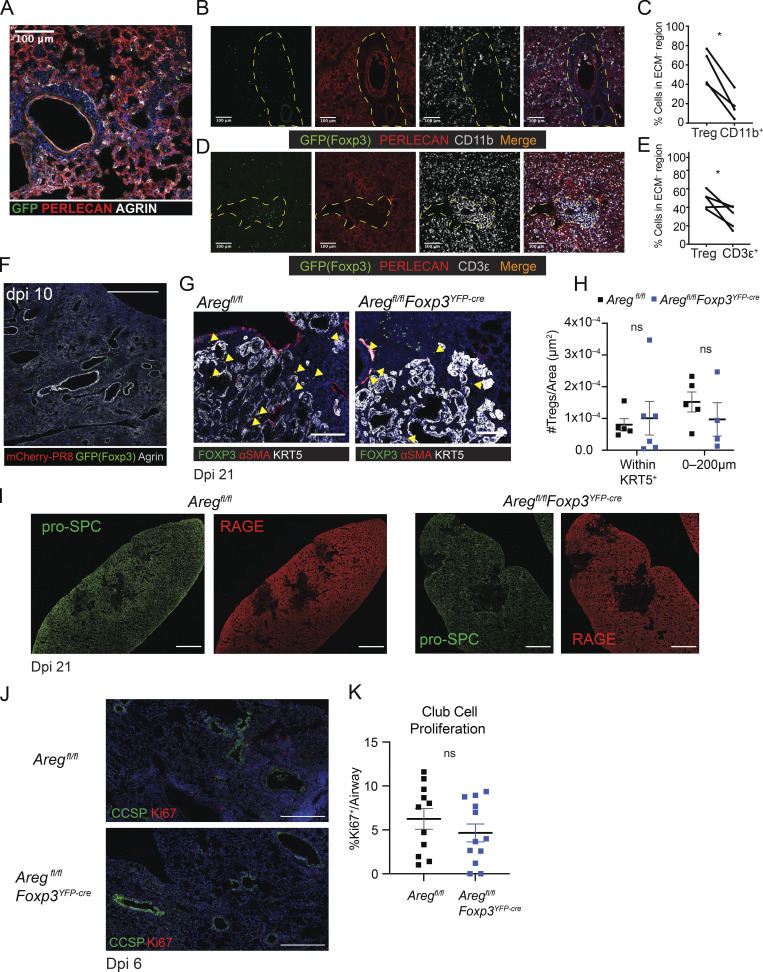
To further explore this connection, we characterized the spatial relationship of Treg cells and HSPG-containing ECM during influenza infection by immunofluorescence imaging. Notably, Treg cells preferentially localize to regions devoid of the extracellularly deposited ECM HSPGs perlecan and agrin (Fig. S2, A–E). In contrast to other immune cell populations, Treg exclusion from HSPG+ areas appears to be cell type specific. CD11b+ myeloid cells are predominantly found within HSPG+ areas (Fig. S2, B and C) and total CD3ε+ T cells are observed more uniformly distributed throughout the tissue (Fig. S2, D and E). Furthermore, Treg cells persist in lung sites lacking HSPGs following complete viral clearance at dpi 10 (Fig. S2 F). Given the observation that Treg cells exist in HSPG– regions yet Areg requires HSPGs to signal, we believe that this spatial segregation of Treg cells within HSPG– regions may be a key to regulating Areg signaling to only the adjacent HSPG+ regions to which Areg can diffuse.
Figure S2.
Treg cells are uniquely distributed in infected lung and loss of Areg from Treg cells does not impact alveolar or club cell regeneration at late repair stages. (A) Treg cell exclusion from perlecan (red) and agrin (white) containing areas. (B–E) Lung tissue distribution of Treg cells as compared to CD11b+ (B and C) and total CD3e+ T cells (D and E) within tissue sections taken from the same lungs at dpi 8 influenza challenge. HSPG+ ECM identified by co-staining with anti-perlecan (red). Treg cells are found in proteoglycan-devoid regions (channels outlined by yellow dotted lines). Scale bar = 200 µm. Statistical significance evaluated by two-tailed paired Student’s t test comparing Treg:CD11b+ or Treg:CD3ε+ within one large image of each biologic replicate (* P < 0.05). (F) Treg cell (green) persistence in the lung after viral clearance at dpi 10 following PR8/H1N1 challenge. Scale bar = 400 µm. (G–I) Aregfl/flFoxp3YFP-cre and Aregfl/fl mice infected with PR8/H1N1; lung tissue was harvested at dpi 21 for immunofluorescence imaging analysis. (G) Presence of Foxp3+ Treg cells (green) within KRT5+ pods (white) in Aregfl/flFoxp3YFP-cre and Aregfl/fl lungs at dpi 21 following influenza infection. Yellow arrowheads indicate Treg cells within KRT5+ pods. Scale bar = 100 µm. (H) Quantification of the number of Treg cells/μm2 within a KRT5+ pod area or within 200 μm from a KRT+ pod. Statistical significance evaluated by two-way ANOVA. (I) Representative imaging of AT2 (pro-SPC, green) and AT1 (RAGE, red) regeneration in Aregfl/fl (left) and Aregfl/flFoxp3YFP-cre (right) mice. Scale bar = 1,000 µm. (J) Aregfl/flFoxp3YFP-cre and Aregfl/fl mice infected with PR8/H1N1 and analyzed at dpi 6 for proliferation of club cells (CCSP+, green) as indicated by Ki67 staining (red). Scale bar = 450 µm. (K) Quantification of the percentage of Ki67+ club cells per airway quantified in at least three airways per mouse, with four mouse replicates per group. Data represented as mean ± SEM. Statistical significance evaluated by two-tailed unpaired Student’s t test.
Disrupted alveolar repair in mice lacking Treg cell–derived Areg
Infection with influenza virus results in the acute loss of alveolar epithelial cells (AECs), in turn requiring the rapid activation and differentiation of remaining AT2 to re-establish the barrier epithelium (Basil et al., 2020; Zacharias et al., 2018). Defects in rapid AT2-mediated regeneration engages an alternate repair pathway that leads to the formation of long-lasting keratin 5-expressing (Krt5+) pod structures with reduced capacity to support normal blood oxygenation (Fernanda de Mello Costa et al., 2020; Kumar et al., 2011; Ray et al., 2016; Vaughan et al., 2015; Zuo et al., 2015). Inappropriate AT2 repair signaling has been shown to expand KRT5+ pods (Paris et al., 2020). Although the dramatic decline in lung function observed in Aregfl/flFoxp3YFP-cre mice suggests a deficiency in alveolar repair, a direct role for Treg cell–derived Areg in the restitution of alveolar epithelial barrier integrity has not been examined. To evaluate potential lasting impacts on lung remodeling from conditional loss of Areg during influenza virus infection, Aregfl/flFoxp3YFP-cre and littermate control Aregfl/fl mice were infected with PR8/H1N1 and alveolar structures were assessed at dpi 17. Strikingly, Aregfl/flFoxp3YFP-cre lungs exhibited a significant expansion in Krt5+ pods (Fig. 1, D and E), indicative of dysplastic alveolar regeneration and suggesting that Treg cell–derived Areg promotes normal alveolar epithelial programming during an acute viral infection. While there is a downward trend in density of Treg cells within Krt5+ pods, their overall number and localization is unchanged in Areg-deficient animals at dpi 21 (Fig. S2, G and H). Further, Treg cells cease producing Areg after inflammatory resolution and AT1 and AT2 cells show similar reconstitution in both Aregfl/fl and Aregfl/flFoxp3YFP-cre animals (Fig. S1, A–D, and Fig. S2 I). Taken together, these data suggest that Treg cell–derived Areg acts in earlier phases of alveolar regeneration to promote AT2 activation and differentiation.
T cell–derived Areg promotes AEC regeneration during influenza virus infection
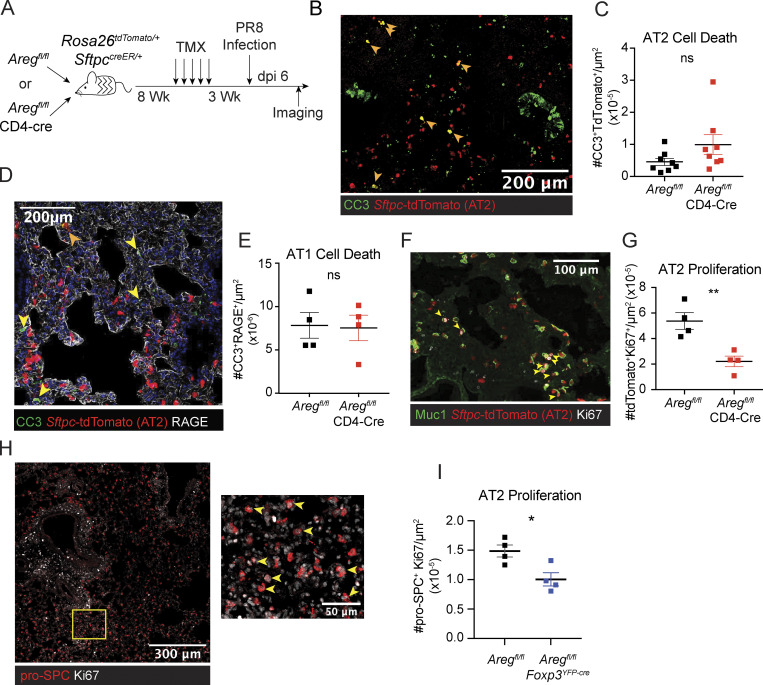
To determine the cellular effects of T cell–derived Areg on the alveolar niche, we created a tractable system for evaluating AEC dynamics at the cellular level using lineage-tracing bone marrow chimeras. SftpccreER/+Rosa26tdTomato/+ hosts were lethally irradiated and engrafted with T cell–depleted donor bone marrow from Aregfl/flCD4-Cre (targeting all T cells) or littermate control Aregfl/fl mice (Fig. 2 A). Following reconstitution of the hematopoietic compartment, mice were treated with tamoxifen to induce recombination and permanent lineage tracing of host-derived AT2. After a 3-wk chase, animals were infected with PR8/H1N1 influenza virus and lung tissue sections were processed for immunofluorescence imaging analysis. First, we directly quantified epithelial cell death in our bone marrow chimera system by immunofluorescence imaging of cleaved caspase 3 (CC3), a marker for apoptotic cell death. No differences in cell death were observed in Sftpc-tdTomato+ AT2 or RAGE+ AT1 (Fig. 2, B–E). We next examined early AT2 regeneration by measuring proliferation of tdTomato+ cells at the borders of damaged lung regions. Here, we found a significant reduction in Ki67 staining of AT2-derived cells in Aregfl/flCD4-Cre bone marrow chimeras as compared to those reconstituted with bone marrow from control Aregfl/fl mice (Fig. 2, F and G). A similar reduction in Ki67+ pro-SPC+ AT2 was also observed through direct assessment of AT2 proliferation in Treg cell–specific conditional knockout mice (Aregfl/flFoxp3YFP-cre; Fig. 2, H and I). CCSP+ club cell proliferation was not impaired in Aregfl/flFoxp3YFP-cre mice, suggesting that loss of Treg cell–derived Areg results in specific dysregulation of AT2 activating signals needed to stimulate alveolar regeneration at early post-infection time points (Fig. S2, J and K).
Figure 2.
Treg cell– and T cell–specific ablation of Areg leads to defects in AT2 proliferation. (A–E) (A) Schematic of bone marrow chimeras generated from lethally irradiated SPCcreRosa2tdTomato/+ hosts reconstituted with bone marrow from Aregfl/fl or Aregfl/flCD4-Cre littermate animals. Following reconstitution, AT2 labeling was induced by oral tamoxifen (TMX) administration and animals were subsequently infected with PR8/H1N1. (B–E) Imaging at dpi 6 of influenza-infected lungs described in A. Each point on image quantification graphs represents one biologic replicate. Two independent experiments were performed using this bone marrow chimera system. (B and C) Representative image and quantification of apoptotic cell death analyzed by CC3 staining of tdTomato+ AT2. Filled orange arrowheads indicate apoptotic AT2. Pooled data from two independent experiments are shown. (D and E) AT1 apoptosis quantified by CC3 staining of RAGE+ cells (AT1) indicated by filled yellow arrowheads. (F) Immunofluorescent staining of lungs from bone marrow chimeras harvested at dpi 6, with AT2 proliferation measured by Ki67 immunostaining (white) of Sftpc-tdTomato+ cells, with Muc1 staining also used to guide determination of AT2. (G) Quantification of tdTomato+Ki67+ cells per μm2. (H) Imaging of AT2 proliferation in lungs of Aregfl/fl (wildtype) at dpi 6 by Ki67 (white) of pro-SPC+ cells. Zoomed image of yellow box in left image is shown in right image highlighting Ki67+ pro-SPC+ proliferating AT2. (I) Quantification of AT2 proliferation in Aregfl/flFoxp3YFP-cre mice and Aregfl/fl littermate controls as in shown in H at dpi 6. Representative data from one of two independent experiments shown. On vertical bar plots, each point represents a biologic replicate and bars show mean ± SEM. Data were analyzed by two-tailed unpaired Student’s t test (* P < 0.05, ** P < 0.01).
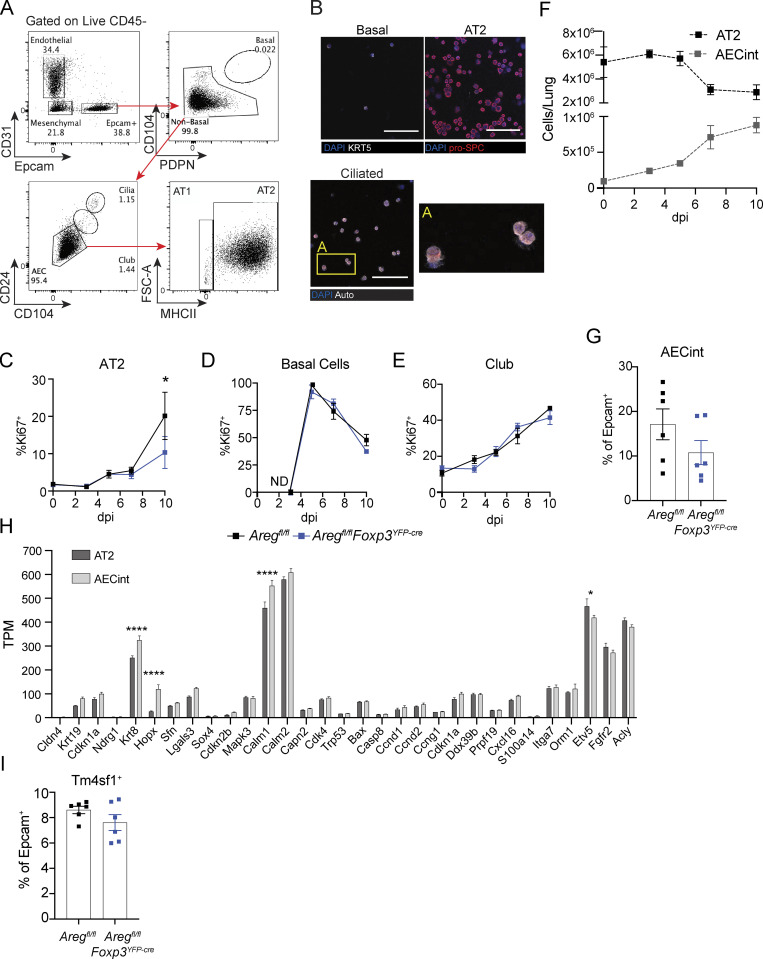
To quantitatively assess dynamics of lung epithelial cell regeneration, we developed an antibody staining panel to specifically identify AT2 and other lung epithelial cell populations by flow cytometry throughout influenza infection (Fig. S3 A). Cell identity was confirmed through cytospin and immunofluorescence imaging with pro-SPC, KRT5, and by ciliated cell morphology (Fig. S3 B). To broadly assess the effects of Treg cell–derived Areg on epithelial progenitors, we quantified proliferation of basal, club, and AT2 cells by flow cytometric analysis following intracellular staining for Ki67. Confirming our imaging observations, bulk AT2 exhibit significantly reduced proliferation (Fig. S3 C). In contrast, proliferation of basal cells and club cells was unaltered in Areg-deficient animals (Fig. S3, D and E), further supporting the conclusion that AT2 are the primary progenitor cell type influenced by Treg cell–derived Areg.
Figure S3.
AT2 have reduced proliferation in the absence of Treg cell–derived Areg, and arising AECint are comprised of transitional AEC populations that share features of AT2 and AT1. (A) Gating strategy of live lung epithelial cells following agarose-prep isolation from lungs of mice at dpi 3 with PR8/H1N1-mCherry. (B) Cytospin analysis of sorted lung basal cell, AT2, and ciliated cells stained for pro-SPC (red) or KRT5. Box A shows a zoomed image of ciliated cells. Scale bar = 100 μm. (C–E) Proliferation of lung progenitor epithelial cells in Aregfl/flFoxp3YFP-cre and Aregfl/fl controls during influenza infection at dpi 0, 3, 5, 7, and 10 assessed by flow cytometry for Ki67 intracellular staining; n = 4 mice per group per time point, representative data from one of at least two independent experiments per time point shown; statistical significance evaluated by two-tailed unpaired Student’s t test at each time point (* P <0.05). (F) Kinetic assessment of the number of wildtype AT2 and AECint during influenza A virus infection determined by flow cytometry. (G) Flow cytometric analysis of AECs isolated from Aregfl/flFoxp3YFP-cre and Aregfl/fl littermate control mice at dpi 6 influenza A virus quantifying the frequency of AECint of total Epcam+ cells; n = 6 animals per group, representative data from one of two independent experiments shown. (H) Bulk RNA-Seq of AT2 and AECint at dpi 7 of influenza infection isolated from four wildtype animals. Gene expression shown in transcripts per million (TPM) of genes associated with damage-associated transitional progenitor cells. Data represented as mean ± SEM; statistical significance evaluated by two-way ANOVA (* P < 0.05, **** P < 0.0001). (I) Flow cytometric analysis of AECs at dpi 6 of influenza infection as in G. Frequency of TM4SF1+ AECs within total Epcam+ cells.
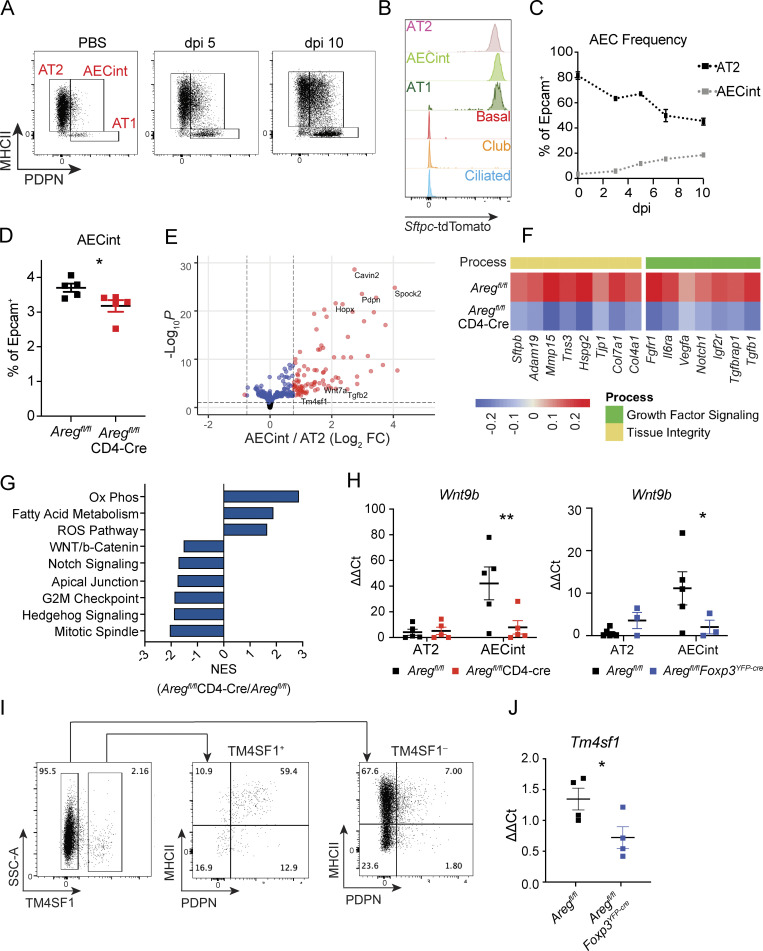
Through further flow cytometric analysis of AT2, we identified a transitional AEC population that co-expresses markers for both AT2 (MHCII) and AT1 (PDPN), which we termed “intermediate AEC” (AECint; Fig. 3 A). Using lineage tracing in SftpccreER/+Rosa26tdTomato/+ mice, we determined that AECint arise entirely from Sftpc-expressing AT2 (Fig. 3 B). By frequency and number, AECint expand over the course of infection (Fig. 3 C and Fig. S3 F). Notably, there is a significant reduction in AECint in the absence of T cell–derived Areg, a trend that is matched with Treg cell–specific deletion of Areg (Fig. 3 D and Fig. S3 G).
Figure 3.
AECint emerge during influenza-induced lung damage. (A) Flow cytometry gating strategy of AECs isolated from lungs of naive mice and following mock treatment or PR8/H1N1 infection at dpi 5 and 10. (B) AT2 lineage tracing based on expression of tdTomato in SPCcreER/+Rosa26tdTomato mice treated with tamoxifen 3 wk prior to infection with PR8/H1N1. Epithelial cells analyzed at dpi 6, populations identified as in Fig. S3, AECint gated as in Fig. 2 F. (C) Frequency of AT2 and AECint in relation to total epithelial cells isolated at dpi 0 3, 5, 7, and 10. (D) Frequency of AECint within total Epcam+ cells isolated from lungs of Aregfl/flCD4-Cre and Aregfl/fl control mice infected with PR8/H1N1 at dpi 4 by flow cytometry. Representative data from one of four independent experiments performed within “early” post-infection time window shown, with early defined as dpi 3–4. Data were analyzed by two-tailed unpaired Student’s t test (* P < 0.05). (E–G) RNA-Seq analysis of AT2 and AECint isolated from Aregfl/flCD4-Cre and Aregfl/fl littermate control mice at dpi 5 following PR8/H1N1 infection (n = 4 per group). (E) Volcano plot showing genes significantly upregulated in AECint, as compared to AT2, isolated from infected Aregfl/fl control mice. (F) Heatmap representation of average gene expression in AECint isolated from control (Aregfl/fl) and conditional knock-out mice (Aregfl/fl CD4-Cre) for genes involved in growth factor signaling and tissue homeostasis; n = 4 mice per group. (G) GSEA of AECint isolated from PR8/H1N1-infected Aregfl/fl vs. Aregfl/flCD4-Cre mice. Bar graph depicts normalized enrichment scores (NES) for statistically significant (false discovery rate < 0.05) gene sets. (H) AECint sorted by FACs from Aregfl/flFoxp3YFP-cre (left) and Aregfl/flCD4-Cre (right) mice and Aregfl/fl littermate controls for qPCR of Wnt9b at dpi 3. Representative data from one of three independent experiments per genotype shown. Statistical significance evaluated by two-way ANOVA (* P < 0.05, ** P < 0.01). (I) Staining of TM4SF1 on alveolar epithelial cells by flow cytometry at dpi 6. Pre-gated on CD45–CD31–Epcam+CD104–CD24–. Representative staining data for one mouse, from one of two independent experiments, are shown. (J) qPCR analysis of Tm4sf1 expression normalized to Tbp in sorted AECint from Aregfl/fl littermate controls and Aregfl/flFoxp3YFP-cre mice at dpi 5 following PR8/H1N1 infection. Representative data from one of two independent experiments shown. Statistical significance evaluated by two-tailed unpaired Student’s t test ( * P < 0.05).
Given that AECint are induced in the context of epithelial damage, we hypothesized that these transitional cells are critical to support lung re-epithelialization. We therefore took advantage of our newly developed gating strategy to isolate AT2 and AECint from influenza virus–infected wildtype mice for transcriptional profiling by bulk RNA-sequencing (RNA-Seq). Confirming our FACS analysis, AECint display increased expression of canonical AT1 marker genes such as Hopx and Pdpn compared to AT2 (Fig. 3 E). Spock2, an interferon-induced gene implicated in protection against viral infection and induced during AT2 to AT1 differentiation, is also upregulated in AECint, supporting the conclusion that AECint are a transitional population between AT2 and AT1 (Fig. 3 E; Ahn et al., 2019; Choi et al., 2020). Of note, the cells observed through this gating strategy are likely a mixture of cells in transition and not a uniform cell state.
To characterize the molecular changes that occur within AECint in the absence of T cell–derived Areg, we isolated AECint from infected Aregfl/flCD4-Cre and littermate control Aregfl/fl mice at dpi 5 for profiling by RNA-Seq. AECint from Aregfl/flCD4-Cre mice exhibit a distinct transcriptional profile that reflects a failure to upregulate key receptors and ligands involved in growth factor signaling that promote AT2 cell differentiation to AT1. These include Il6ra, Notch1, and Fgfr1, receptors upstream of signaling pathways that all have been shown promote transdifferentiation (Fig. 3 F; Zepp et al., 2017). AECint from Aregfl/flCD4-Cre also show reduced production of structural and tissue remodeling elements, including collagens, perlecan (Hspg2; an ECM-deposited heparan sulfate proteoglycan), metalloproteinases, junction proteins, and surfactant protein B (Sftpb; Fig. 3 F).
Several single-cell RNA-Seq (scRNA-Seq) studies of bleomycin-induced alveolar injury have identified damage-associated transient progenitor AECs that express Krt8+—also described as alveolar differentiation intermediates and the pre-alveolar type-1 transitional cell state (Choi et al., 2020; Jiang et al., 2020; Kobayashi et al., 2020; Strunz et al., 2020). We observe increased Krt8 expression in AECint as compared to AT2, consistent with their transitional identity (Fig. S3 H). However, they lack gene expression signatures associated with p53 signaling, cellular senescence, and dysfunction that have been attributed to cells with stalled differentiation. We therefore conclude that AECint are transitional KRT8+ cells undergoing normal differentiation following injury and not dysfunctional or stalled progenitors as observed in the case of bleomycin-induced injury.
At the transcriptomic level, gene set enrichment analysis (GSEA) reveals a reduction in WNT, NOTCH, and cell cycle signaling in the mixed Aregfl/flCD4-Cre AECint population (Fig. 3 G), consistent with the reduction in cellular activation and proliferation observed by immunofluorescence imaging and flow cytometry (Fig. 2, F–I and Fig. S3 C). In comparison with AT2, AECint from influenza virus–infected wildtype mice express greater levels of Wnt7a and Tm4sf1 (Fig. 3 E), markers of the recently described WNT-responsive AEPs that drive early epithelial repair following influenza virus infection (Nabhan et al., 2018; Zacharias et al., 2018). We also see a robust induction of autocrine Wnt9b in the emerging AECint population specifically at early time points (dpi 3) in comparison with AT2 (Fig. 3 H). Critically, AECint sorted from lungs of infected Aregfl/flFoxp3YFP-cre or Aregfl/flCD4-Cre mice fail to upregulate Wnt9b (Fig. 3 H), in accordance with the negative enrichment of WNT signaling pathway-associated genes inferred by RNA-Seq analysis of AECint in Aregfl/flCD4-Cre animals (Fig. 3 G). Together, these data suggest that Treg cell–derived Areg contributes to the induction of WNT signaling in earliest-responding transitioning alveolar epithelial cells. This in turn leads to the observed delay in differentiation and a failure to upregulate pro-differentiation genes including Il6ra, Notch1, and Fgfr (Fig. 3 B).
To further understand the composition of the AECint population, we stained for the AEP marker, TM4SF1, by flow cytometry (Zacharias et al., 2018). Consistent with our RNA-Seq data demonstrating higher Tm4sf1 expression in AECint in comparison with AT2 (Fig. 3 E), TM4SF1+ AECs primarily fall within the AECint gate by flow cytometry. Of note, not all AECint are TM4SF1+, reinforcing that they are a mixed population of activated AT2 participating in the regenerative response (Fig. 3 I). Along with a trending reduction in the frequency of TM4SF1+ AECs within Epcam+ cells in the absence of Treg cell–derived Areg (Fig. S3 I), expression of Tm4sf1 by AECint isolated from infected Aregfl/flFoxp3YFP-cre mice is significantly reduced compared to littermate Aregfl/fl controls (Fig. 3 J). Cumulatively, these data highlight that Treg cell–derived Areg is a factor that supports the proliferation and differentiation of AT2 at early time points following acute viral injury.
Lung mesenchymal cells express EGFR and co-localize with Treg cells
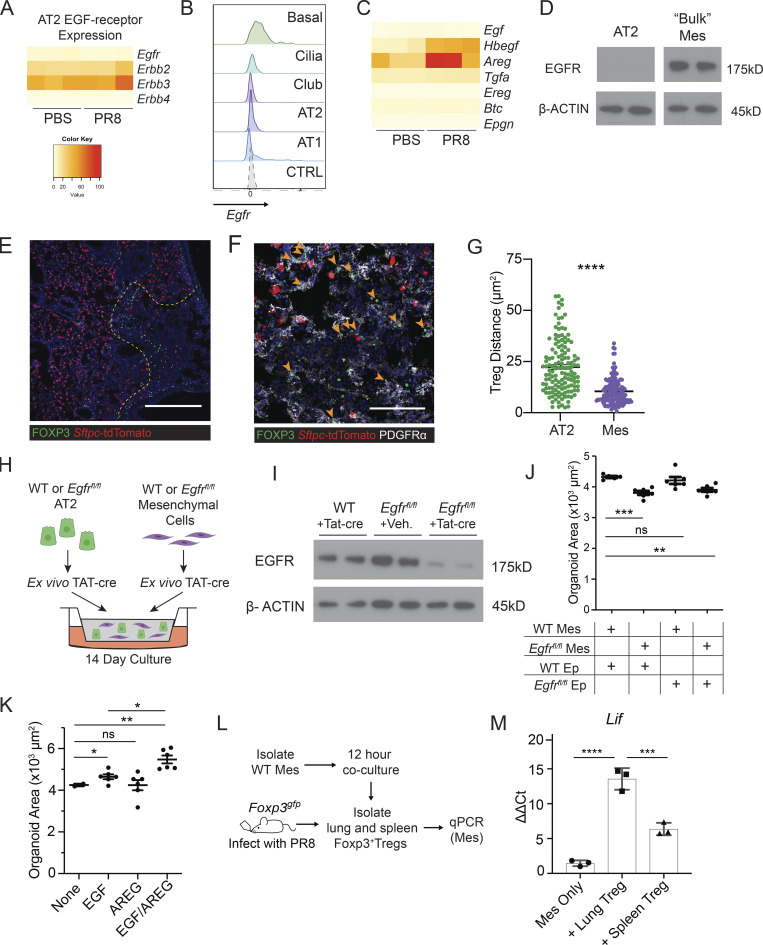
Given the literature describing the role of EGFR—the only receptor through which Areg has been reported to signal (Freed et al., 2017)—in lung epithelial cells, our first hypothesis was that Treg cell–derived Areg signals directly to AT2 to initiate alveolar regeneration (Finigan et al., 2012; Vallath et al., 2014; Yamaoka et al., 2019). To our surprise, extremely low levels of Egfr were detected in sorted AT2 from mock- or PR8/H1N1-infected lungs by bulk RNA-Seq (Fig. 4 A). This result was further confirmed by RNA flow cytometry probing for Egfr, wherein we found no Egfr expression on AT2 and higher levels of Egfr transcripts in basal cells and AT1 (Fig. 4 B). This contrasts with other EGFR-family members, Erbb2 and Erbb3, which are abundantly expressed by AT2 but do not directly engage in Areg signaling (Inoue et al., 2020; Ohmori et al., 2021). Moreover, Areg is highly expressed by AT2 themselves, leading us to consider that Treg cell–derived Areg does not directly signal to AT2, but may instead be indirectly sensed by an intermediate cell type that then provides factors that support AT2 activation and differentiation (Fig. 4 C). Thus, to examine Egfr-expressing cell types that could be responsible for sensing Treg cell–derived Areg within damaged alveoli, we probed publicly available mouse scRNA-Seq datasets (Han et al., 2018; Schaum et al., 2018) and identified lung mesenchymal cells as one of the highest Egfr-expressing cell types in the lung. In support of this observation, we detected high levels of EGFR protein in lung mesenchymal cells, but not AT2, sorted from uninfected wildtype mice by immunoblot (Fig. 4 D).
Figure 4.
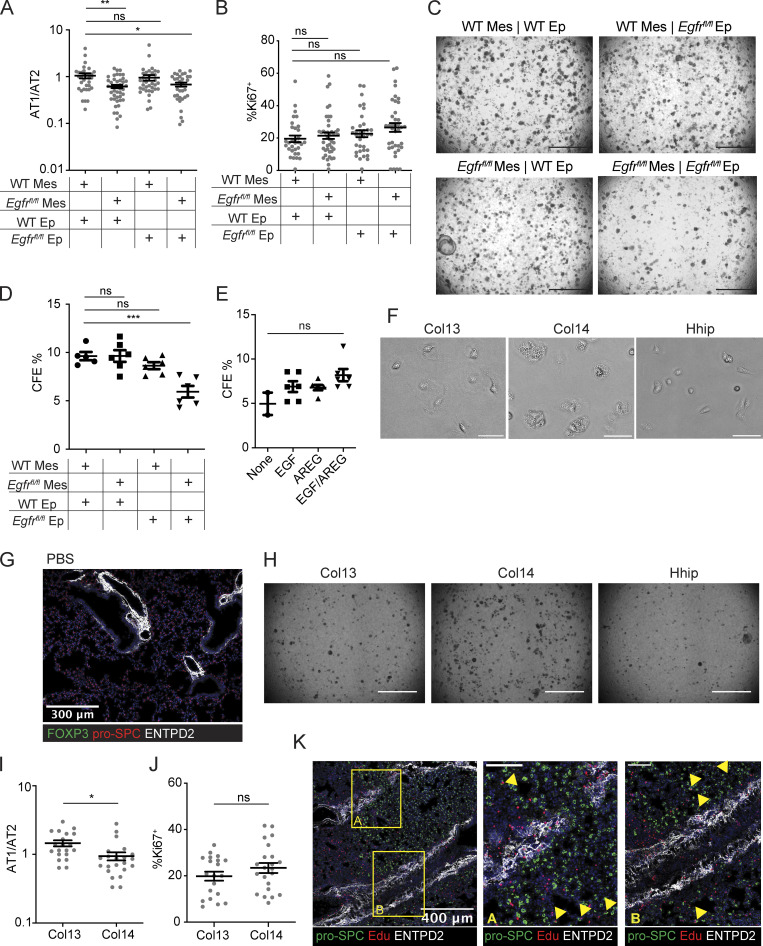
EGFR activation on mesenchymal cells stimulates alveolar growth in vitro. (A) Heatmap of EGFR-family receptor expression by RNA-Seq of FACS-sorted AT2 from PBS-treated and PR8/H1N1-infected mice at dpi 7; n = 4 animals per group. (B) RNA flow cytometry of Egfr expression by epithelial cell type analyzed by RNA PrimeFlow. Control probe = Dapb. (C) Heatmap of EGFR ligand expression determined by RNA sequencing of AT2 from mock-treated (PBS) and PR8/H1N1-infected wildtype animals; n = 4 animals per group. (D) Immunoblot for EGFR protein expression in wildtype mesenchymal and epithelial cells isolated from C57BL/6 mice and cultured overnight prior to protein extraction. Protein input normalized by Bradford assay and validated by staining for β-actin (bottom panels). Representative data from one of two independent experiments shown. (E and F) Immunofluorescence imaging of lung tissue sections from tamoxifen-treated SPCcreER/+Rosa26tdTomato mice infected with PR8/H1N1 influenza at dpi 6. (E) Treg cell (FOXP3+, green) distribution in relation to AT2 (red) or (F) PDGFRα+ mesenchymal cells (Mes, white). Scale bar in E = 400 μm; scale bar in F = 100 μm; filled orange arrowheads indicate Treg cells. (G) Quantification of Treg:AT2 and Treg:Mes distances calculated by K-nearest neighbor analysis. At least four damaged areas were analyzed in each of n = 3 mice. Data represented as mean ± SEM; statistical significance evaluated by two-tailed unpaired Student’s t test (**** P < 0.0001). (H) Alveolar organoid culture setup of lung epithelial and mesenchymal cells isolated from C57BL/6 or Egfrfl/fl mice cultured for 14 d following treatment with TAT-cre to induce Egfr deletion. (I) Immunoblot for deletion of EGFR following in vitro treatment with TAT-cre in Egfrfl/fl lung mesenchymal cells with wildtype mesenchyme and vehicle controls. Protein input normalized by Bradford assay and validated by staining for β-actin (bottom panels). Representative data from one of three independent experiments shown. (J) Quantification of the average area of all organoids ≥1000 µm2 with mesenchymal EGFR deletion, epithelial EGFR deletion, both, or neither (control). Data represented as mean ± SEM. Experiment performed five times (mesenchymal deletion conditions alone: three experiments; epithelial deletion conditions included alongside mesenchymal conditions: two experiments), with normalized data from the latter two experiments represented on graphs. Statistical significance evaluated by one-way ANOVA (** P < 0.01, *** P < 0.001). (K) Organoids generated from co-culture of AT2 and lung mesenchymal cells isolated from C57BL/6 mice grown with or without the addition of rmAREG (100 ng/ml), rmEGF (25 ng/ml), or both, for 14–16 d. Quantification of the average area of all organoids ≥1000 µm2. Data represented as mean ± SEM; statistical significance evaluated by one-way ANOVA (* P < 0.05, ** P < 0.01). Experiment performed three times, with normalized data from all experiments shown. (L and M) In vitro setup of Treg cells co-cultured with isolated lung mesenchymal cells for 12 h at a 1:2 (Treg:Mes) ratio (L) and transcriptional analysis of adherent mesenchymal cells for EGFR activation (M) assessed by qPCR for Lif expression normalized to housekeeping (Tbp). Data represented as mean ± SEM; n = 3 wells/group. Representative data from one of three independent experiments shown; statistical significance evaluated by one-way ANOVA (*** P < 0.001, **** P < 0.0001). Source data are available for this figure: SourceData F4.
In light of this observation, we examined the spatial relationship between Treg cells, AT2/AT2-derived cells, and EGFR-expressing lung mesenchymal cells utilizing the AT2 lineage-trace mouse (SftpccreER/+Rosa26tdTomato/+) to readily identify AT2 and AT2-derived cells by orally dosing adult animals with tamoxifen 3 wk prior to infection. Imaging of lung tissue sections from influenza virus–infected animals revealed large swaths of alveolar regions devoid of AT2 or AT2-derived cells, reflective of virally induced AT2 apoptosis and widespread disruption of normal lung architecture following infection (Fig. 4 E). Intriguingly, Foxp3+ Treg cells were detected at the borders of AT2-depleted regions in areas spatially segregated from intact AT2, where they instead appeared to predominantly colocalize with adjacent PDGFRα+ mesenchymal cells (Fig. 4, E–G). Given the distinct association of Treg cells with the lung stroma, we hypothesized that Treg cell–derived Areg may be sensed by adjacent mesenchymal cells—in turn activating these cells to signal to the epithelium and facilitate alveolar regeneration.
EGFR activation on lung mesenchymal cells supports growth of alveolar organoids
To determine whether the activation of EGFR in lung mesenchymal cells promotes alveolar regeneration, we employed an alveolar organoid culture system (Barkauskas et al., 2013; Lee et al., 2017; McQualter et al., 2010; Zepp et al., 2017). AT2 and lung mesenchymal cells isolated from wildtype C57BL/6 or Egfrfl/fl mice were treated ex vivo with a recombinant, cell-permeable fusion of cre-recombinase (TAT-cre) to induce Egfr deletion in either AT2, mesenchymal cells, or both cell types (Fig. 4 H; Peitz et al., 2002). By immunoblot, we confirmed partial deletion of EGFR in mesenchymal cells (Fig. 4 I). Surprisingly, deletion of Egfr in mesenchymal cells—but not AT2—resulted in significantly smaller alveolar organoids, a size reduction comparable to that observed following Egfr deletion in both epithelial and mesenchymal cell types (Fig. 4 J). Organoids grown in the presence of Egfr-deficient mesenchyme also had lower AT1/AT2 ratios, suggesting a defect in AT2 to AT1 conversion, although no difference in Ki67 staining was observed (Fig. S4, A and B). The size of organoids increases as more AT1 create spaces within the center of the structure; as a result, the lower AT1/AT2 ratio observed in Egfr-null conditions could explain smaller organoids. A reduction in total colony forming efficiency was also detected when Egfr was deleted on both epithelial and mesenchymal cell types (Fig. S4, C and D). From these data, we conclude that Egfr signaling on mesenchymal cells promotes factors that drive AT2 differentiation in vitro.
Figure S4.
Col14+ lung mesenchymal cells express EGFR and promote alveolar organoid growth. (A and B) Quantification of organoids grown in experimental setup described in Fig. 4 H. (A) Ratio of RAGE + AT1 to pro-SPC + AT2 per organoid. Statistical significance evaluated by one-way ANOVA (* P < 0.05, ** P < 0.01). (B) Percentage of Ki67+ cells per organoid. (C) 2.5× images of organoid cultures following Egfr deletion with TAT-cre. (D) Colony forming efficiency of organoids following EGFR deletion, calculated by total number of organoids/10,000 cell input. Statistical significance evaluated by one-way ANOVA (*** P < 0.001). (E) Organoids generated from co-culture of AT2 and lung mesenchymal cells isolated from C57BL/6 mice grown with or without rmAREG (100 ng/ml), rmEGF (25 ng/ml), or both, for 14 d. Quantification of colony forming efficiency calculated by the total number of organoids/10,000 cell input. Statistical significance evaluated by one-way ANOVA. (F) Brightfield images of FACS-isolated live mesenchymal cell subsets grown overnight in culture. Scale bar = 50 μm. (G) Immunofluorescence image of Col14+ (ENTPD2+), Treg (absent), and AT2 (pro-SPC+) in PBS-treated, wildtype lungs. (H) 2.5× magnification images of organoid culture wells grown with Col13+, Col14+, or Hhip+ lung mesenchymal cells. Scale bar = 1,000 μm. (I) Ratio of the number of AT1 (RAGE+)/AT2 (pro-SPC+) per organoid when grown with Col13+ or Col14+ support cells. (J) Percentage of Ki67+ cells per organoid grown with Col13+ or Col14+ cells. For I and J, data represented as mean ± SEM. Statistical significance evaluated by two-tailed unpaired Student’s t test (* P < 0.05). (K) Immunofluorescence imaging of proliferating AT2 analyzed by Edu incorporation injected 6 h before animal harvest. Box A and B are two zoomed regions demonstrating localization of AT2 in proximity to Col14+ stromal cells (ENTPD2+, white). Yellow arrowheads indicate Edu+ AT2.
We also tested the effect of adding recombinant mouse (rm) AREG, EGF, or both EGFR ligands on the growth of alveolar organoids. While a modest increase in the size of alveolar organoids can be seen following addition of rmEGF alone, organoids are larger, but not more numerous, following addition of both rmAREG and rmEGF (Fig. 4 K and Fig. S4 E). These data illustrate that AREG/EGFR signaling on mesenchymal cells contributes to alveolar organoid growth and that AREG and EGF may cooperate to promote alveolar regeneration.
To test the hypothesis that Treg cell–derived Areg can activate EGFR on lung mesenchymal cells, Treg cells from lungs or spleens of influenza virus–infected Foxp3gfp animals were co-cultured with mesenchymal cells isolated from lungs of naïve adult mice (Fig. 4 L). Expression of Lif, a downstream target of EGFR signaling (Kerpedjieva et al., 2012), was upregulated in mesenchymal cells following co-culture with lung Treg cells in comparison with spleen Treg cells from influenza-infected mice (Fig. 4 M). These results indicate that signals derived from Treg cells isolated from the lungs of influenza-infected mice are capable of activating lung mesenchymal cells.
EGFR+Col14+ mesenchymal cells promote alveolar regeneration
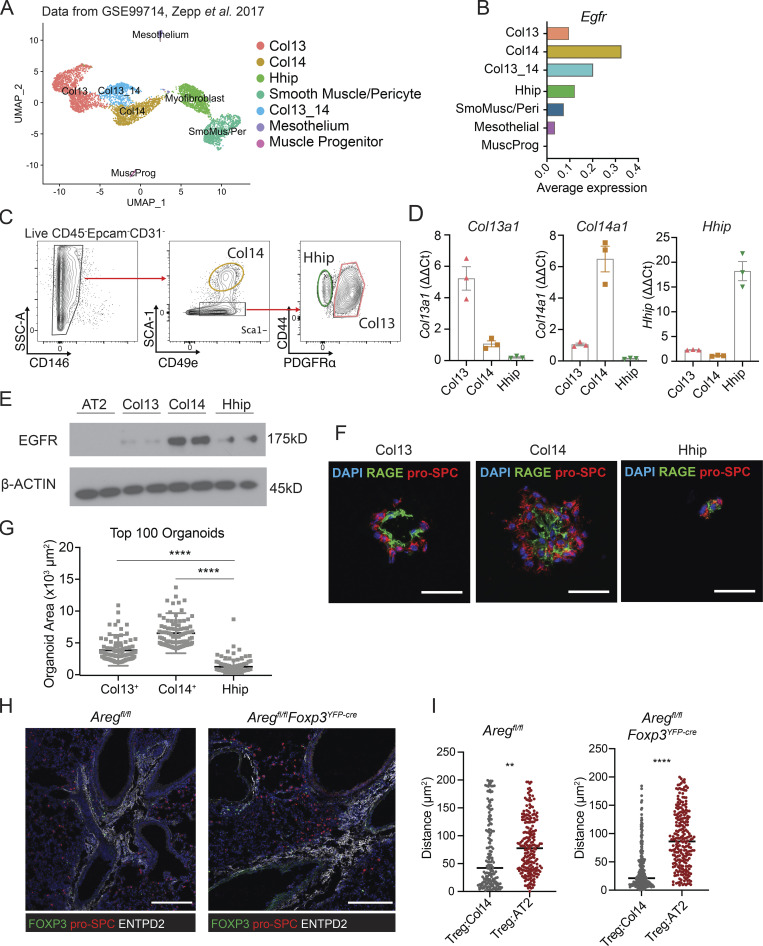
Several recent studies have used scRNA-Seq to describe the heterogeneity of lung stromal cells in terms of their expression of unique receptors and ligands, tissue distribution relative to epithelial and endothelial cells, and unique functions during homeostasis and disease (Dahlgren et al., 2019; Lee et al., 2017; Tsukui et al., 2020; Xie et al., 2018; Zepp et al., 2017). Based on our findings that EGFR signaling on mesenchymal cells influences alveolar organoid development (Fig. 4, H and I), we sought to identify which lung mesenchymal subtypes may be responding to Treg cell–derived Areg. We initially evaluated Egfr expression levels within different lung mesenchymal populations at steady state by reanalyzing scRNA-Seq datasets from published sources (Fig. 5 A; Zepp et al., 2017). We noted that while Egfr is expressed in multiple clusters, its expression is highest in the Col14 (Col14a1)-expressing population, an adventitial lung mesenchymal subset identified in mice and humans based on expression of Pi16, Entpd2, Serpinf1, and Il33 (Fig. 5 B and Table S1; Dahlgren et al., 2019; Stenmark et al., 2011; Travaglini et al., 2020; Tsukui et al., 2020; Xie et al., 2018). Several studies have implicated Col14+ mesenchymal cells—in some cases referred to as mesenchymal alveolar niche cells—in alveolar regeneration and immune cell modulation (Benabid and Peduto, 2020; Paris et al., 2020; Zepp et al., 2017).
Figure 5.
Treg cells co-localize with Col14+ lung mesenchymal cells that express EGFR and support alveolar organoid growth. (A) UMAP projection of reanalyzed scRNA-Seq dataset from Zepp et al. 2017 (GSE99714) of lung mesenchyme from 6-wk-old C57BL/6 mice. (B) Average Egfr expression across lung mesenchymal cell subsets from A. (C) Gating strategy for lung mesenchymal cells isolated from C57BL/6 mice informed by dataset in A. Pre-gated on live (Sytox–) CD45–Epcam–CD31– cells. (D) Confirmation of marker gene expression of FACS-sorted Col13+, Col14+, and Hhip+ by qPCR, normalized to Hprt. Data represented as mean ± SEM. (E) Immunoblot for EGFR expression in sorted AT2 and mesenchymal subsets. Protein input normalized by Bradford assay and validated by staining for β-actin (bottom panel). Representative data from one of two independent experiments shown. (F and G) Alveolar organoids grown in the presence of individually sorted lung mesenchymal populations assessed by (F) immunofluorescence imaging of organoids stained for pro-SPC (AT2) and RAGE (AT1), with DAPI nuclear staining (blue), and (G) size quantification of the top 100 organoids in each culture condition. Scale bar in F = 50 μm; data in G represented as mean ± SEM, statistical significance evaluated by one-way ANOVA (**** P < 0.0001). Shown are representative data from two independent experiments for Col13+ and Col14+ mesenchymal cells, and one experiment for Hhip+. (H) Immunofluorescence imaging of Treg cells (green) in proximity to pro-SPC+ AT2 (red) and ENTPD2+ Col14+ cells in Aregfl/fl (left) and Aregfl/flFoxp3YFP-cre (right). Scale bar = 200 μm. (I) Quantification of H, with the distance of Treg to AT2 or Treg to Col14 calculated by k-nearest neighbor analysis in Aregfl/fl (left) and Aregfl/flFoxp3YFP-cre (right). Data represented as mean ± SEM; statistical significance evaluated by two-tailed unpaired Student’s t test (** P < 0.01, **** P < 0.0001). Source data are available for this figure: SourceData F5.
Using these scRNA-Seq datasets, we developed a panel of surface markers to identify and isolate several major lung mesenchymal cell populations by flow cytometry (Fig. 5 C). We confirmed the identities of the three most abundant stromal subsets through FACS purification and quantitative PCR (qPCR) of subset-defining genes based on the designations of previous publications: Col13+ (Col13a1); Col14+ (Col14a1); and Hhip+ (Hhip; Fig. 5 D; Xie et al., 2018). High EGFR expression was observed specifically on the Col14+ subset by immunoblot (Fig. 5 E). Of note, these cell types exhibit unique morphologies after ex vivo isolation and adherence in overnight culture, with Col14+ being distinctly vesicularized in comparison to the other subsets (Fig. S4 F). We also validated the adventitial fibroblast designation of Col14+ by immunofluorescence staining of ectonucleoside triphosphate diphosphohydrolase 2 (ENTPD2), an adenosine-processing ectoenzyme that is highly expressed by Col14+ cells. In accordance with previous reports, Col14+ cells surround major blood vessels in the adventitia in wildtype uninfected lungs (Fig. S4 G; Dahlgren et al., 2019; Travaglini et al., 2020; Tsukui et al., 2020).
We next assessed if Col14+ mesenchymal cells are functionally distinct in their ability to promote alveolar epithelial regeneration. CD45–CD31–Epcam–CD146– cells that are Sca1+—which we established to be a defining surface marker for Col14+ mesenchymal cells (Fig. 5 C)—have been previously found to support lung epithelial cell growth in vitro (McQualter et al., 2010). To determine the ability of different mesenchymal subsets to promote alveolar regeneration, we isolated Col13+, Col14+, and Hhip+ cells for alveolar organoid culture with lung AT2 (Barkauskas et al., 2013). Organoids grown with Col14+ support cells were significantly larger and contained more pro-SPC+ AT2 surrounding a robust interior of RAGE+ AT1 (Fig. 5, F and G; and Fig. S4 H). The AT1/AT2 ratio for organoids grown in the presence of Col14+ mesenchymal cells is modestly lower than those grown with Col13+ mesenchyme (Fig. S4 I), which can be attributed to the overall greater number of AT2 cells per organoid observed in Col14+ cultures. Notably, organoids grown with Col13+ cells were smaller, and those grown with Hhip+ cells were unable to form multicellular structures, demonstrating that Col14+ mesenchymal cells are highly proficient in driving alveolar cell growth in vitro. No differences were observed in Ki67 expression between cultures (Fig. S4 J).
Finally, we examined the localization of Treg cells in relation to ENTPD2+Col14+ cells in wildtype mice in the absence of Treg cell–derived Areg. In both settings, Treg cells are found closer in proximity to Col14+ cells than to AT2 (Fig. 5, H and I). Of note, proliferating AT2 are not found directly near adventitia, indicating that AT2:Col14+ cell signaling likely occurs through secreted factors (Fig. S4 K).
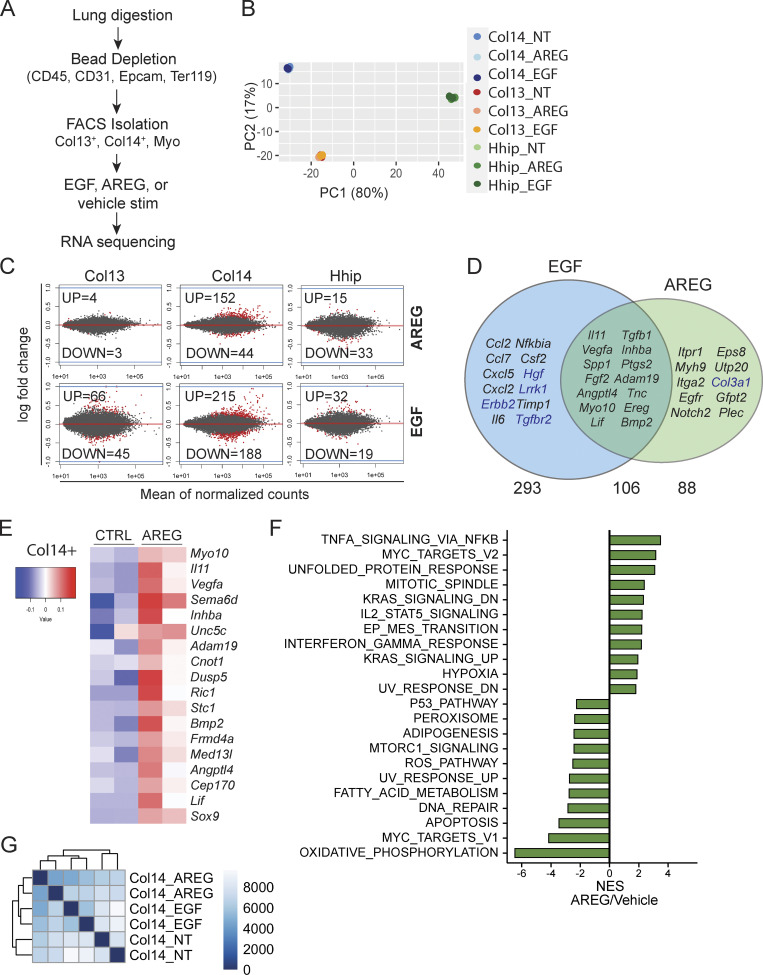
Col14+ stromal cells are uniquely AREG responsive
To determine the transcriptional responses induced by EGFR activation within each subset, we performed bulk RNA-Seq on FACS-sorted Col13+, Col14+, and Hhip+ lung mesenchymal cells stimulated with rmEGF, rmAREG, or vehicle control (Fig. 6 A). In accordance with our sort strategy, each population clusters based on cell identity by principle component analysis (Fig. 6 B). Importantly, Col14+ cells are most responsive to AREG treatment in vitro compared to Col13+ and Hhip+ subsets (Fig. 6 C)—evidenced by nearly 200 significantly differentially expressed genes in Col14+ cells vs. only 7 and 48 differentially expressed genes in AREG-treated Col13+ and Hhip+ populations, respectively (Fig. 6 C, top). While many of the same genes were induced in Col14+ cells in response to both AREG and EGF stimulation, many cytokines and chemokines including Il6, Ccl2, Ccl7, Cxcl5, and Cxcl2 were selectively induced by EGF treatment (Fig. 6 D). Of the genes most highly induced in Col14+ cells by AREG treatment, several have been reported for their roles in pulmonary function and disease, including osteopontin (Spp1), Il11, Vegfa, and Tenascin (Tnc; Fig. 6, D and E; Gremlich et al., 2020; Morimoto et al., 2018; Ng et al., 2019; Vila Ellis et al., 2020). Furthermore, GSEA of genes induced by AREG treatment revealed an enrichment in proliferative and activation pathways and a reduction in pro-apoptotic signaling (Fig. 6 F). The distinct transcriptional changes elicited by treatment with EGF or AREG are also demonstrated through hierarchical clustering of Col14+ samples, which clearly segregate by treatment group (Fig. 6 G). Taken together, these findings support the notion that the Col14+ stromal cells are an EGFR ligand–responsive subpopulation of lung mesenchymal cells that respond to signals provided by lung Treg cells and promote the expansion and differentiation of alveolar epithelial cells.
Figure 6.
Col14+ lung mesenchymal cells selectively respond to Areg. (A) Schematic of RNA-Seq analysis of lung mesenchymal cell subsets treated with rmEGF (50 ng/ml) or rmAREG (200 ng/ml) following isolation from naive C57BL/6 mice. Cells were plated overnight prior to stimulation. (B) Principal component analysis of RNA-Seq data from individually sorted lung mesenchymal cell populations left untreated or stimulated in vitro with rmEGF or rmAREG. (C) RNA-Seq analysis of the number of statistically significant (Padj < 0.05) differentially expressed genes (red) after AREG (top) or EGF (bottom) stimulation in each population. (D) Venn diagram showing statistically significant (Padj < 0.05) differentially expressed genes induced by rmAREG or rmEGF treatment of Col14+ lung mesenchymal cells. Subset of genes selected based on relevance to lung disease or tissue repair. Upregulated genes shown in black text, downregulated genes shown in blue text. (E) Heatmap of top genes induced in Col14+ lung mesenchymal cells following rmAREG treatment. (F) GSEA of Col14+ lung mesenchymal cells following in vitro stimulation with rmAREG vs. vehicle. Bar graph depicts normalized enrichment scores (NES) for statistically significant (false discovery rate < 0.05) gene sets. (G) Poisson-based clustering of Col14+ cells left untreated (“Col14_NT”) or stimulated with EGF (“Col14_EGF”) or AREG (“Col14_AREG”).
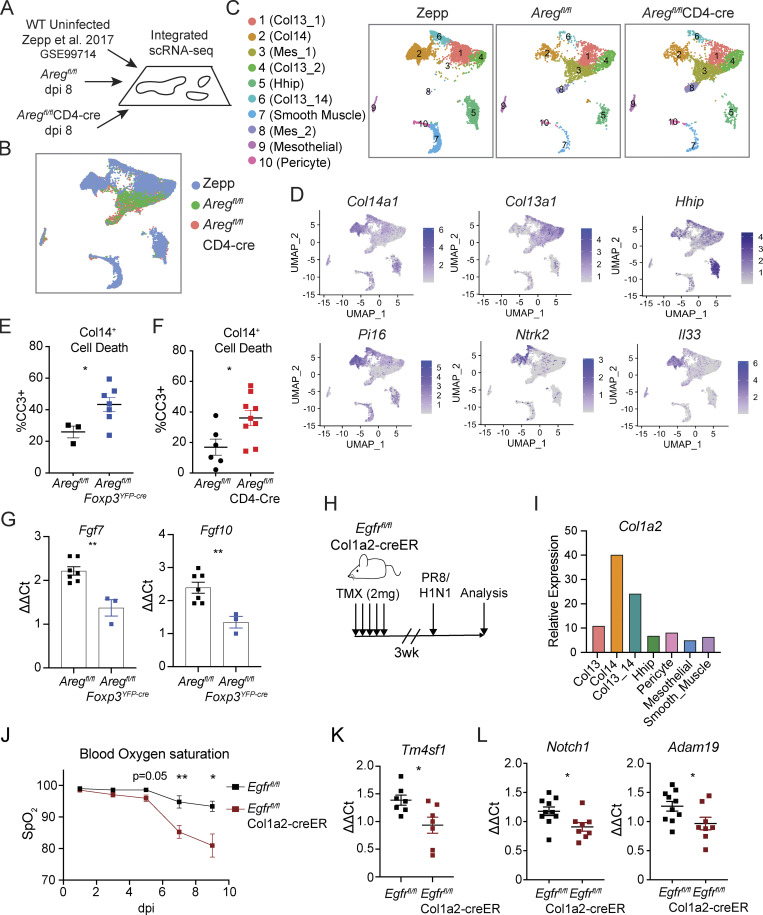
Dysregulated activation and increased apoptosis of Col14+ mesenchymal cells in the absence of Treg cell–derived Areg
Given the immense heterogeneity within lung stromal cells and dynamic changes in surface marker expression during infection, we performed scRNA-Seq to assess transcriptional differences in specific mesenchymal subsets in the absence of T cell–derived Areg. We isolated mesenchymal cells from Aregfl/flCD4-Cre and littermate control mice for analysis at day 8 following influenza virus infection (Fig. 7 A). To compare the mesenchymal populations identified during infection to those observed at steady-state, we integrated our scRNA-Seq results with a previously published dataset generated for naive lung mesenchyme (Fig. 7, A and B; Xie et al., 2018; Zepp et al., 2017). Although many mesenchymal clusters were readily identified in untreated and flu-infected lungs by principal components analysis–based clustering and Uniform Manifold Approximation and Projection (UMAP) visualization (Seurat v3), additional activated or differentiating stromal subsets (“Mes” clusters) greatly expand upon influenza infection (Fig. 7, B and C). We subsequently identified a cluster corresponding to Col14+ cells based high expression of Col14a1, Il33, Pi16, and Ntrk2 and low expression of Col13a1 and Hhip (Fig. 7 D).
Figure 7.
Treg-derived Areg promotes Col14+ cell survival and growth factor production during infection. (A–D) Integration of previously published scRNA-Seq data from C57BL/6 lung mesenchymal cells (GSE99714) with data from lung mesenchymal cells isolated from Aregfl/flCD4-Cre and Aregfl/fl littermate control mice at dpi 8 following influenza virus infection. (B) UMAP rendering of integrated datasets described in A. (C) Overlay of cell clusters within UMAP projection comparing mesenchymal subsets at steady state (Zepp), and at 8 dpi of Aregfl/flCD4-Cre and Aregfl/fl littermate control mice with PR8/H1N1. (D) Feature plots of identity genes for Col14+ mesenchymal cells. (E and F) Apoptosis of Col14+ lung mesenchymal cells, as quantified by flow cytometric analysis of CC3 staining at dpi 6 in (E) Aregfl/flFoxp3YFP-cre and corresponding Aregfl/fl littermate control and (F) Aregfl/flCD4-Cre and corresponding Aregfl/fl littermate control mice. Representative data from one of three independent experiments shown, depicted as mean ± SEM, statistical significance evaluated by two-tailed unpaired Student’s t test (* P <0.05). (G) Expression of Fgf7 and Fgf10 in Col14+ mesenchymal cells isolated from Aregfl/flFoxp3YFP-cre in comparison to Aregfl/fl littermate controls at dpi 6 by qPCR. Data are normalized to Tbp and represented as mean ± SEM, statistical significance evaluated by two-tailed unpaired Student’s t test (** P <0.01). Representative data from one of two independent experiments shown. (H) 8-wk-old Egfrfl/flCol1a2-creER and Egfrfl/fl littermate control mice were treated with five consecutive doses of tamoxifen (TMX) followed by 3-wk chase period prior to infection with PR8/H1N1. (I) Col1a2 expression by mesenchymal cell clusters identified within Zepp naive lung scRNA-Seq dataset. (J) Blood oxygen saturation measured by MouseOx pulse oximeter during influenza infection of Egfrfl/flCol1a2-creER mice and Egfrfl/fl controls at dpi 1, 3, 5, 7, and 9. Representative data from one of two independent experiments shown, statistical significance evaluated at each time point by two-tailed unpaired Student’s t test (* P <0.05, ** P <0.01). (K) AECint from Egfrfl/flCol1a2-creER and Egfrfl/fl littermate control mice isolated by FACS at dpi 5 for transcriptional analysis of Tm4sf1 normalized to Tbp. Representative data from one of two independent experiments shown, depicted as mean ± SEM. (L) Expression of Notch1 and Adam19 in AECint isolated from Egfrfl/flCol1a2-creER and Egfrfl/fl normalized to Tbp assessed by qPCR at dpi 4. Representative data from one of two independent experiments shown, depicted as mean ± SEM. Statistical significance for K and L evaluated by two-tailed unpaired Student’s t test (* P <0.05).
Given that type-I interferons induced during influenza virus infection are known to drive programmed cell death (Fujikura and Miyazaki, 2018; Malireddi and Kanneganti, 2013) and because we observed a negative enrichment for apoptosis following Areg treatment of Col14+ cells (Fig. 6 F), we hypothesized that Areg may be an important pro-survival signal that enables mesenchymal cells to endure tissue damage and inflammation. In support of this concept, we found that Col14+ cells undergo significantly higher rates of apoptotic cell death during influenza virus infection in the absence of total T cell– and Treg cell–derived Areg (Aregfl/flCD4-Cre and Aregfl/flFoxp3YFP-cre mice, respectively) in comparison to Aregfl/fl littermate controls, as determined by flow cytometric analysis of CC3 staining (Fig. 7, E and F). Furthermore, Col14+ cells sorted from infected Aregfl/flFoxp3YFP-cre mouse lungs express reduced levels of growth factors, Fgf7 and Fgf10, which have found to promote AT2 activation following acute injury (Fig. 7 G; Paris et al., 2020). Taken together, these data suggest that increased Col14+ mesenchymal cell apoptosis in the absence of Treg cell–derived Areg leads to a dysregulation in signaling networks that promote alveolar regeneration following virus-induced damage.
Treg cells signal through mesenchymal cell intermediates to support alveolar regeneration
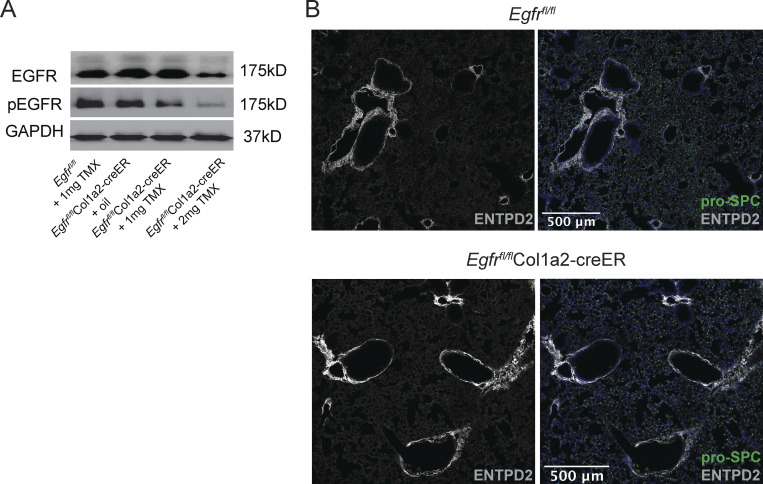
To directly test if Treg cell–derived Areg signals through EGFR+ mesenchymal cells, we crossed mesenchymal–specific Col1a2-creER transgenic mice with a conditional Egfrfl/fl allele to enable inducible deletion of Egfr on mesenchymal cells in vivo (Fig. 7 H). Although Col1a2-creER is broadly considered a pan-mesenchymal inducible Cre driver, our scRNA-Seq analyses demonstrate that Col1a2 is most highly expressed on lung Col14+ mesenchymal cells at steady state (Fig. 7 I). Following oral administration of tamoxifen, partial deletion of EGFR on bulk lung mesenchymal cells was confirmed by immunoblot (Fig. S5 A), with partial deletion likely due to incomplete recombination efficiency and the presence of non-targeted Col1a2– stromal cells contained within the bulk mesenchymal cell enrichment. No baseline defects in Col14+ cells were observed in Egfrfl/flCol1a2-creER mice following tamoxifen administration (Fig. S5 B). Strikingly, loss of EGFR on Col1a2-expressing mesenchymal cells results in a severe reduction in blood oxygen saturation following influenza virus infection, a decline that phenocopies loss of Areg from Treg cells (Fig. 7 J; Arpaia et al., 2015). Similar to the loss of Treg cell–derived Areg, qPCR analysis of AECint isolated from influenza-infected Egfrfl/flCol1a2-creER and Egfrfl/fl littermate control mice reveals reduced expression of Tm4sf1 in the absence of EGFR signaling within the Col1a2+ population (Fig. 7 K). Further, AECint gene expression from Egfrfl/flCol1a2-creER conditional knockout mice mirrors the transcriptional changes observed by RNA-Seq of Aregfl/flCD4-Cre mice, with significantly reduced expression of genes including Notch1 and Adam19 (Fig. 7 L and Fig. 3 F). From these data, we conclude that loss of Treg cell–derived Areg signaling in EGFR+ Col14+ lung mesenchymal cells impairs AT2-mediated repair—resulting in defective alveolar regeneration and increased disease severity following virally induced injury.
Figure S5.
Loss of Egfr on mesenchymal cells phenocopies loss of Treg cell–derived Areg, related to Fig. 7. (A) Mesenchymal cells isolated by negative bead enrichment from Egfrfl/flCol1a2-creER mice 3 wk after tamoxifen delivery. Mesenchymal cells plated overnight and prior to stimulation with 25 ng/ml rmEGF. Egfr deletion was confirmed by Western blot of EGFR and phospho-EGFR (pEGFR). Protein input normalized by Bradford assay and validated by immunoblotting for GAPDH or β-actin. Representative data from one of two independent experiments shown. (B) Representative immunostaining of ENTPD2+Col14+ cells (white) and pro-SPC+AT2 (green) in uninfected Egfrfl/flCol1a2-creER and Egfrfl/fl controls 3 wk after tamoxifen treatment. Source data are available for this figure: SourceData FS5.
Discussion
As a mobile organ, the immune system is uniquely capable of trafficking throughout the body to execute distinct responses to context-specific stimulatory cues. Here, we demonstrate that lung Treg cells are rapidly recruited to sites of active viral infection and provide key tissue-protective factors that drive early tissue repair programs in non-hematopoietic cells. This unique pro-repair Treg cell pool seems to operate outside of classical MHC/TCR interactions and may rely on innate signals such as cytokines, alarmins, TLR ligands, or lipid mediators for activation (Arpaia et al., 2015). We discovered that the exquisite localization of Treg cells to sites of tissue injury is distinct from conventional CD4+ T cells. While the cues that recruit Treg cells to the site of damage are not well understood, we believe that this unique patterning enables highly spatially regulated signaling. It is well appreciated that heparin sulfate proteoglycans are required for Areg signaling. We hypothesize that Areg diffuses within the area immediately surrounding the Treg cells to Col14+ cells in matrix-rich areas. Although untested in these studies, we suspect that Treg cell migration within the tissue may also rely on interactions with these components of the extracellular matrix. Future studies are needed to understand if these cells acquire memory features or tissue residency properties that enable tissue retention and rapid response to rechallenge.
A growing number of studies have revealed circumstances in which the immune system interacts with tissue stem cells to influence their activation and differentiation following injury (Naik et al., 2018). Recently, several reports have also shown the direct influence of inflammatory cytokines on alveolar epithelial cell differentiation (Broggi et al., 2020; Choi et al., 2020; Major et al., 2020). Our work elucidates an indirect mechanism by which pro-survival signals from Treg cells mediate stromal cell-guided activation of alveolar progenitors. We identify a Col14a1-expressing mesenchymal cell subtype, referred to as Col14+ mesenchymal cells, that is uniquely sensitive to Treg cell–derived signals and also highly expresses IL-33, an alarmin known to induce Areg production by Treg cells and also support ILC2, another Areg-producing immune cell type found within the adventitia (Monticelli et al., 2011). While not directly examined in this study, this positive feedback loop may serve to strengthen Treg–stromal interactions or to polarize recently extravasated lung Treg cells toward a pro-repair phenotype. Ultimately, we established Treg cell–derived Areg as a critical factor for Col14+ resilience to cell death within highly inflammatory tissue environments during influenza viral challenge. This Treg–Col14+ mesenchymal cell axis supports alveolar epithelial progenitor cell programming that rapidly regenerates the barrier epithelium following virus-induced damage.
The downstream implications of this dramatic cell death phenotype are observed in the regenerating intermediate AECs, here termed AECint, which are less proliferative and found in reduced numbers in the absence of Treg cell–derived Areg. In the absence of scRNA-Seq, we have a limited ability to describe the cell identities within this broad collection of transitioning cells; however, AECint lack key features of dysfunction described for stalled transitioning cells found after bleomycin-induced lung injury (Choi et al., 2020; Jiang et al., 2020; Kobayashi et al., 2020; Strunz et al., 2020). Over the course of infection, we expect that this transitioning population will differentiate and be transcriptionally distinct as they receive anti-inflammatory signals following viral clearance. While not investigated here, it would be interesting to understand if repair defects observed in Aregfl/flFoxp3YFP-cre mice results in increased susceptibility to subsequent bacterial or viral infections.
These studies also highlight an intriguing aspect of EGFR signaling, in that treatment with EGF or AREG has profoundly different downstream consequences depending on cell type. This may be due to signaling through heterodimerization of EGFR with other EGFR family members (Erbb2, Erbb3, or Erbb4, which have not been shown to bind Areg themselves), competition for receptor engagement by other ligands, and/or other co-receptors and downstream signaling modules that have yet to be identified. It is clear that the functional outcomes of AREG signaling are highly cell specific. Elucidating the distinct transcriptional changes elicited by EGFR activation in discrete lung mesenchymal cell subsets has important implications, not only for tissue repair, but in regenerative medicine, fibrosis, and cancer.
Materials and methods
Mice
Aregfl/flCD4-Cre and Aregfl/flFoxp3YFP-cre have been previously described (Arpaia et al., 2015). EgfrFl/Fl mice (Lee and Threadgill, 2009) were generously provided by Dr. David Threadgill (Texas A&M University, College Station, TX, USA) and crossed to Col1a2-creER (Stock# 029567; Jackson Laboratories; Zheng et al., 2002). SftpcCreER mice were obtained from Jackson Laboratories (Stock# 028054; Rock et al., 2011) and subsequently crossed to Ai14 tomato lineage tracing mice (Stock# 007914; Jackson Laboratories; Madisen et al., 2010). All animal procedures and experimental treatments were performed under protocol AC-AABD8554 approved by the Institutional Animal Care and Use Committee at Columbia University Irving Medical Center. All mouse strains were maintained in Columbia University’s animal facilities in accordance with institutional guidelines.
Pulse oximetry
Blood oxygen saturation (SpO2) during infection was measured using a MouseOx Plus pulse oximeter (Starr Life Sciences). Fur was removed from the neck at the time of viral inoculation using Nair hair removal product. Beginning on the first day post-infection, SpO2 was assessed every other day. Oximeter collar clips were secured to the neck of conscious animals and SpO2 was recorded for approximately 3–5 min per mouse. Results are shown as the mean SpO2 recorded during this interval.
Tamoxifen treatment
SftpcCreER animals received daily doses of tamoxifen (200 mg/kg) resuspended in corn oil by oral gavage for 5 consecutive days. Col1a2-creER animals received daily intraperitoneal doses of tamoxifen (100 mg/kg) for 5 consecutive days.
Bone marrow chimeras
Recipient animals were irradiated with 2 doses of 500 rad from a Cesium-137 irradiator. Donor bone marrow was depleted of T cells and RBCs using the Dynabeads FlowComp Mouse Pan T CD90.2 kit (Thermo Fisher Scientific), with addition of biotinylated anti-Ter119 antibody (BioLegend). 5–10 × 106 cells were delivered retro-orbitally under isoflurane anesthesia. Mice were kept on neomycin for 3 wk following generation of chimeras.
PR8/H1N1 viral strains
PR8/H1N1 was harvested in the laboratory of Dr. Donna Farber (Columbia University, New York, NY, USA) and kindly provided for these studies. PR8/H1N1-mCherry (Fukuyama et al., 2015) was generously donated by Dr. Yoshihiro Kawaoka (University of Wisconsin, Madison, Madison, WI, USA). For propagating PR8/H1N1-mCherry influenza virus, 200 μl of 1 × 103 TCID50 viral stock was injected into the air sac of 10-d-old embryonated chicken eggs and harvested 72 h after incubation. To measure viral titer, 10-fold serial dilutions of harvest were used to infect Madin–Darby canine kidney epithelial cells in the presence of 1 μg/ml tosyl phenylalanyl chloromethyl ketone–treated trypsin (Worthington Biochemical Corporation). Following 3-d culture, supernatants from infected Madin–Darby canine kidney cells were collected to determine lung viral titers by relative performance in a hemagglutination assay using chicken RBC. Mice were infected intranasally with 250–450 TCID50 PR8/H1N1 or 1.5 TCID50 PR8/H1N1-mCherry under ketamine anesthesia.
Immunofluorescence imaging
Lungs were perfused with periodate-lysine-paraformaldehyde buffer followed by fixation in 4% PFA for 1 h. Lungs were dehydrated in 30% sucrose overnight prior to embedding in OCT freezing medium (Fisher Healthcare), and 10-μm sections were prepared using an Avantik cryostat. Sections were then blocked with 10% serum matching the species of secondary antibodies in 0.3% Triton X-100 for 1 h. Sections were stained with antibodies in 1% serum in 0.3% Triton X-100 in a humidified chamber overnight at 4°C and with secondary antibodies for 1 h at room temperature. Primary antibodies were used against the following targets: pro-SPC (1:1,000; Sigma-Aldrich); Foxp3 (1:200; Invitrogen); Muc1 (1:250; Thermo Fisher Scientific); Ki67 (1:100; Thermo Fisher Scientific); PDPN (1:300; Developmental Studies Hybridoma Bank); GFP (1:200; Thermo Fisher Scientific); PDGFRα (1:200; R&D); αSMA (1:200; Sigma-Aldrich); RAGE (1:500; R&D); Agrin (1:200; R&D); CC3 (1:100; Cell Signaling). Secondary antibodies purchased from Jackson ImmunoResearch raised in donkey or goat. Slides were mounted using ProLong Diamond Antifade Mountant (Thermo Fisher Scientific). Confocal images were acquired on a Nikon Ti Eclipse inverted microscope. All image quantifications were performed in Fiji. For all analyses, multi-image swaths containing >20 20× images that encompassed both damaged and normal adjacent tissue were acquired and stitched together for global evaluation of the lung. At least 3 mice per group were quantified (exact numbers indicated in figure legends). Identification of cell types was performed by automatic thresholding of positive signal and particle counting. For each cell type, individual cell coordinates were exported and distances were calculated by k-nearest neighbor in R.
For determining cells in proximity to lung structures (i.e., 250 μm from pockets of virally infected cells, as in Fig. 1, B and C; 50 μm from blood vessels or lymphatics, as in Fig. S1 J), lung regions of interest were manually demarcated and a 250 or 50 μm band was automatically drawn surrounding the initial region of interest designation. Cells were then automatically counted within or outside of the band. The final quantification is represented as number of cells/area of region of interest. When assessing alveolar proliferation, regions of viral infection were determined similarly to “zones” described in Zacharias et al. (2018), where regions closest to alveolar damage were evaluated for proliferation of AT2 per unit area.
Isolation of epithelial cells for sequencing and flow cytometry
Distal lung epithelial cells were isolated by agarose prep as previously described (Gereke et al., 2012). Briefly, mice were perfused with 10 ml PBS prior to performing BAL three times. 25 U/ml dispase (Gibco) dissolved in HBSS (Gibco) was delivered intratracheally in 1 ml to the lungs and a plug was created by adding 200 μl 1% low melt agarose prepared in PBS. After agarose hardening, whole lung lobes were separated, then digested for 45 min at room temperature in 5 ml 5 U/ml dispase. Following digestion, lungs were then shredded in 10 ml DMEM with 50 U/ml DNAse (Roche) followed by incubation at 37°C for 10 min. Cells were then sequentially passed through 100-, 70-, and 40-μm filters to create a single-cell suspension for antibody staining and downstream analyses.
Flow cytometric analysis and cell sorting
For extraction of immune and mesenchymal cells, a single-cell lung suspension was prepared by digesting minced lung tissue in 1 mg/ml collagenase (Worthington Biochemical Corporation), 1 mg/ml dispase (Gibco), and 5 U/ml DNAase (Roche), then passing cells through a 100-μm filter. Dead cells were excluded by staining cells with fixable viability dye for prior to fixation for fixed samples (Ghost Dye Red 780, Tonbo Biosciences), or Sytox Blue for live cells (Thermo Fisher Scientific). Antibodies were prepared in staining buffer containing 1% BSA and 2.5 mM EDTA in PBS and stained for 20 min at 4°C. Cells were fixed using the Foxp3 Transcription Factor Staining Buffer Kit (Tonbo Biosciences) for 1 h at 4°C, per manufacturer’s instructions. For staining of intracellular targets, fixation and permeabilization was performed according to the Foxp3 Transcription Factor Staining Kit protocol (Tonbo Biosciences) and cell were stained with antibody mixes for 45 min at 4°C. All panels included blocking with anti-CD16/32 (2.4G2; Tonbo) and staining with anti-CD45 (30-F11; BD). Myeloid cells were stained with anti-CD11b (M1/70; BioLegend), -Ly6G (1A8; BioLegend), -Ly6C (HK1.4; BioLegend), -Siglec-F (E50-2440; BD), -CD11c (HL3; BD), -MHCII (M5/114.15.2; Tonbo). Lymphocytes were stained using anti-NK1.1 (PK136; BioLegend), -TCRβ (H57-597; BioLegend), -CD4 (RM4-5; BD), -CD8α (53–6.7;Tonbo), and -Foxp3 (FJK-16s; eBioscience). Mesenchymal cells were stained with anti-EpCAM (G8.8; BioLegend), -CD31 (390; BioLegend), -CD44 (IM7; Tonbo), -Ly-6A (D7; Sca 1 BioLegend), -CD49e (5H10-27[MFR5]; BioLegend), -CD146 (ME-9F1; BioLegend), and -CD140α (APA5; Thermo Fisher Scientific). Egfr RNA expression assessed using PrimeFlow RNA assay as per manufacturers protocol.
Bulk RNA-Seq
In vitro AREG and EGF stimulation of sorted mesenchymal cells
Lungs were digested with collagenase (1 mg/ml), dispase (1 mg/ml), and DNAase (5 U/ml). Lung mesenchyme was isolated by negative selection using streptavidin beads (BD Streptavidin Particles Plus) using biotinylated anti-CD31, -Epcam, -CD45, and -Ter119 (Biolegend) antibodies. Bead-attached cells were removed using DynaMag-2 Magnet (Thermo Fisher Scientific). Populations were sorted and plated at 50,000 cells/well in a 48-well tissue culture–treated plate overnight at 37°C prior to washing away any dead cells and debris. 8 h later, cells were stimulated with 50 ng/ml rmEGF (BioLegend), 200 ng/ml rmAREG (R&D), or vehicle control for 4 h. Cells were lysed with Trizol for RNA extraction by the Columbia Molecular Pathology Core Facility and sequencing was performed by the Columbia Genome Center.
AECs
Aregfl/fl and Aregfl/flCD4-Cre mice were infected with 400 TCID50 PR8/H1N1 prior to cell isolation by epithelial tissue prep protocol. Epcam+ cells were further enriched by negative selection using biotinylated anti-CD31, -CD45, and -Ter119 (Biolegend) antibodies. Live AT2 and AECint were sorted and pelleted for RNA isolation and sequencing by GENEWIZ low input RNA-Seq. All gene assignments were generated from Gencode (GRCm38 vM24) annotated genome. Differential expression analysis was performed in R with DESeq2 (Apeglm).
scRNA-Seq
Aregfl/fl and Aregfl/flCD4-Cre mice were infected with 440 TCID50 PR8/H1N1. Lung mesenchymal cells were harvested at dpi 8. Lung tissue was digested with collagenase (1 mg/ml), dispase (1 mg/ml), and DNAase (5 U/ml), and lung mesenchymal cells were isolated by negative selection using streptavidin beads (BD Streptavidin Particles Plus) using biotinylated anti-CD31, -Epcam, -CD45, and -Ter119 (BioLegend) antibodies. 10× scRNA-Seq was performed by the Columbia University Genome Center and scRNA-Seq reads were processed with Cell Ranger (10X Genomics). Further analysis was computed according to the Satija Lab methods. Cells with <500 or >2,500 features and >5% mitochondrial DNA were excluded. Minor clusters of CD45+, Epcam+, or CD31+ cells were removed from our analysis. Single cells were visualized by UMAP projection. Pathway analysis was computed by GSEA and Metascape.
In vitro co-culture
Mesenchymal cells were isolated and plated as for RNA-Seq. Briefly, Foxp3gfp mice were infected with 400 TCID50 PR8/H1N1 7 d prior to mesenchymal cell isolation. After plating mesenchymal cells overnight at 50,000 cells/well, Treg cells were FACS-sorted from Foxp3gfp mice at dpi 8 and co-cultured with mesenchymal cells for 12 h at a 1:2 ratio (Treg:Mes) in the presence of anti-CD3/anti-CD28 T cell activation beads (Thermo Fisher Scientific, Dynabeads Mouse T-cell Activator). For stimulation experiments and controls, rmEGF was used at 100 ng/ml and rmAREG was used at 200 ng/ml. At the end of each incubation, wells were washed once with EDTA to remove Treg cells, then twice with PBS, followed by lysis of mesenchymal cells in Trizol for RNA isolation and qPCR.
Western blot
Lung mesenchymal cells and AT2 were isolated as described above then treated with TAT-CRE recombinase as described in for Lung alveolar organoids (below). After TAT-CRE treatment, 100,000 cells/well were plated overnight in 24-well tissue culture–treated plates (Corning) in DMEM supplemented with 10% FBS (Corning), and 1× penicillin, streptomycin, Glutamax, sodium pyruvate, and nonessential amino acids (Gibco). The following morning, cells were lysed for protein extraction: for mesenchymal cells, nonadherent dead cells were washed away with 2 washes of ice-cold PBS, and cells were lysed by scraping wells with TNT Lysis Buffer, pH 8.0 (20 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, and 1% Triton X-100) with freshly added Halt Protease and Phosphatase Inhibitor Cocktail (Pierce). For AT2, the same procedure was followed, except PBS washes were centrifuged to lyse loosely adherent cells and combined with lysates taken from the same well. Lysates were shaken on ice for 30 min, then centrifuged at full speed for 15 min, with the supernatant taken and used for Western blotting. Lysate supernatants were mixed with SDS (Thermo Fisher Scientific) with bromophenol blue (Amresco), placed at 95°C for 5 min, allowed to cool to room temperature, and loaded into Bolt 4–12% Bis-Tris Plus Gels (Thermo Fisher Scientific). The Mini Blot Module (Thermo Fisher Scientific) was used both for SDS-PAGE (in Bolt MOPS SDS Running Buffer [Thermo Fisher Scientific]), and protein transfer (to Immobilon-P PVDF [Millipore] in Bolt Transfer Buffer [Thermo Fisher Scientific]). Membranes were washed with TBS; blocked with 5% nonfat dry milk in TBST (TBS with 0.1% Tween-20 [Thermo Fisher Scientific]); washed three times in TBST; stained with primary antibodies in 5% milk or BSA in TBST, overnight at 4°C (β-Actin Mouse mAb [clone 8H10D10; Cell Signaling], EGF Receptor Rabbit mAb [clone C74B9; Cell Signaling]); washed three times in TBST; stained with secondary antibodies in 5% milk in TBST for 1 h at room temperature (anti-mouse IgG HRP-linked antibody [Cell Signaling]) or anti-rabbit IgG HRP-linked antibody [Cell Signaling]); washed three times; and incubated with chemiluminescent substrate (Immobilon Classico Western HRP Substrate [Millipore] or Immobilon Forte Western HRP Substrate [Millipore]). Membranes were exposed to film, and films were developed on a Kodak X-OMAT.
Lung alveolar organoids
For AT2, alveolar epithelial cells were isolated from lungs of naive 8–10-wk-old C57BL/6 male mice (as described above) then FACS-sorted using a BD FACS Aria system, with gating strategy as shown in Fig. S3 A. Cells were handled as follows for three separate lines of experimentation: (1) For EGFR ligand treatment experiments, cells were sorted directly into “Pre-Organoid Media” (Airway Epithelial Cell Basal Medium [Promocell] with 5% FBS and 1× penicillin, streptomycin, and Glutamax [Gibco]). (2) For TAT-CRE treatment of AT2 isolated from C57BL/6 or Egfrfl/fl mice, after FACS-sorting, cells were washed and resuspended in ADCF-mAb serum-free media (Hyclone) at a concentration of 1 × 106 cells/ml, and incubated at 37°C for 10 min. Simultaneously, TAT-CRE recombinase was diluted in ADCF-mAb media to a final concentration of 100 μg/ml, pre-incubated at 37°C for 10 min, then combined 1:1 will cell suspensions, gently mixed, and incubated at 37°C for 45 min, followed by dilution into Pre-Organoid Media. (3) For co-incubation of AT2 with mesenchymal subtypes, cells were sorted directly into Pre-Organoid Media.
Mesenchymal cells for organoid experiments were isolated from lungs of naive 8–10-wk-old C57BL/6 male mice using either negative selection with streptavidin beads or sorting (as described above). Cells were handled as follows for three separate lines of experimentation: (1) For EGFR ligand treatment experiments, following negative selection, cells were washed into Pre-Organoid Media. (2) TAT-CRE recombinase treatment of lung mesenchymal cells isolated from C57BL/6 or Egfrfl/fl mice was performed as described for AT2. (3) For co-incubation of individual mesenchymal subsets with AT2, mesenchymal cells were sorted directly into Pre-Organoid Media.
Alveolar organoid cultures were performed as previously described (Barkauskas et al., 2013; Lee et al., 2017; Zepp et al., 2017), with the following modifications. At the time of organoid plating and each subsequent media change, fresh rmEGF (25 ng/ml; BioLegend) and/or rmAREG (100 ng/ml; R&D Systems) were added to media. At day 14–16 of culture, brightfield (2.5× magnification) images were acquired using a Zeiss AxioObserver.Z1 inverted microscope. Images were quantified using Fiji’s “Analyze Particles” function after cleaning images using thresholding, the “Fill Holes” function, and the “Watershed” function, with a cutoff for particle size identification of >1000 µm2.
For immunofluorescence imaging, matrigel/organoid plugs from transwell inserts were flash-frozen in Tissue-Plus OCT Compound (Fisher) and 10-μm sections were prepared using an Avantik cryostat. Frozen sections were then thawed, fixed with paraformaldehyde, permeabilized with 0.3% Triton X-100 (Sigma-Aldrich) in PBS, blocked with 5% horse serum (Gibco) and 0.05% Triton X-100 (Sigma-Aldrich) in PBS, and stained overnight with primary antibodies (rat anti-mouse RAGE [clone 175410; R&D Systems], rabbit anti human/mouse-pro-SPC [AB3786; Sigma-Aldrich]). Secondary antibodies (Cy3 donkey anti-rabbit IgG, AlexaFluor 488 donkey anti-rat IgG [Jackson ImmunoResearch]) were stained in 1% horse serum. Slides were then stained with DAPI (Life Technologies) and sealed with ProLong Diamond Antifade Mountant (Life Technologies). Slides were imaged at 40–60× on a Nikon A1R Confocal Laser Scanning microscope.
Statistical analysis
Statistical analyses were performed using GraphPad Prism or R. Two-tailed Student’s t test was used for the statistical analysis of differences between two groups and one-way ANOVA with Tukey’s post-test was used for the statistical analysis of multiple groups. Pooled experiments are represented as mean ± SEM, as indicated. For depicting statistical analysis values on graphical representations, * P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.0001, and ns (not statistically significant). All experiments were performed ≥2 times, as indicated within the figure legend for each corresponding dataset.
Online supplemental material
Fig. S1 describes the kinetics of Treg cell infiltration, Areg production, and spatial distribution during acute influenza infection. Fig. S2 evaluates Treg distribution during early infection and upon resolution and determines that Treg cell–derived Areg minimally impacts club cells. Fig. S3 describes different populations of lung epithelial cells by flow cytometry. Fig. S4 shows that EGFR signaling in lung mesenchymal supports robust growth of alveolar organoids and that organoids grow most proficiently with support from the Col14+ subset of lung cells. Fig. S5 establishes that the Egfrfl/flCol1a2-creER mouse model has reduced mesenchymal cell EGFR signaling but is morphologically equivalent to wildtype animals. Table S1 contains the marker gene lists of each mesenchymal cell cluster identified by scRNA-Seq.
Supplementary Material
is a list of each mesenchymal cell.
contains original blots for Fig. 4.
contains original blots for Fig. 5.
contains original blots for Fig. S5.
Acknowledgments
We are extremely grateful for intellectual support from L. Rankin and T. Savage. We also thank Dr. D. Farber and R. Guyer for providing PR8/H1N1 and for help with growing and harvesting PR8/H1N1-mCherry. We appreciate the generous donation of Egfrfl/fl mice from Dr. D. Threadgill and PR8/H1N1-mCherry from Dr. Y. Kawaoka. We thank L. Munteanu and T. Swayne for assistance with confocal imaging and quantification methods, M. Kissner and D. Troast for help with cell sorting, and Dr. P.M. Treuting (University of Washington) for advice and consultation with imaging studies and associated analysis pipelines. Graphical abstract created with BioRender.com.
This work was supported by National Institutes of Health (NIH)/National Institute of Allergy and Infectious Diseases (NIAID) K22AI127847 (N. Arpaia), NIH/NIAID R21AI149657 (N. Arpaia), NIH/National Heart, Lung, and Blood Institute R01HL148718 (N. Arpaia), Searle Scholars Program SSP-2017-2179 (N. Arpaia), and NIH/NIAID T32AI106711 (K.A. Kaiser). Research in this publication utilized the Genomics and High Throughput Screening Shared Resource and the Confocal and Specialized Microscopy Shared Resource of the Herbert Irving Comprehensive Cancer Center at Columbia University, funded in part through the NIH/National Cancer Institute Cancer Center Support Grant P30CA013696.
Author contributions: This study was conceived and designed by K.A. Kaiser and N. Arpaia; K.A. Kaiser performed experiments and analyzed data jointly with N. Arpaia; K. de los Santos-Alexis assisted with animal experiments; L.F. Loffredo assisted with alveolar organoid cultures and development and optimization of lung mesenchymal cell sorting strategies; O.R. Ringham assisted with immunofluorescence imaging of Col14+ mesenchymal cells; K.A. Kaiser and N. Arpaia wrote the manuscript.
Data availability
This paper does not report original code. All raw sequencing data have been deposited in the Gene Expression Omnibus under accession no. GSE169127. Any additional information required to reanalyze the data reported in this paper are available from the lead contact upon request.
References
- Adamson, I.Y., Hedgecock C., and Bowden D.H.. 1990. Epithelial cell-fibroblast interactions in lung injury and repair. Am. J. Pathol. 137:385–392. [PMC free article] [PubMed] [Google Scholar]
- Ahn, N., Kim W.J., Kim N., Park H.W., Lee S.W., and Yoo J.Y.. 2019. The interferon-inducible proteoglycan testican-2/SPOCK2 functions as a protective barrier against virus infection of lung epithelial cells. J. Virol. 93:93. 10.1128/JVI.00662-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arpaia, N., Green J.A., Moltedo B., Arvey A., Hemmers S., Yuan S., Treuting P.M., and Rudensky A.Y.. 2015. A distinct function of regulatory T cells in tissue protection. Cell. 162:1078–1089. 10.1016/j.cell.2015.08.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkauskas, C.E., Cronce M.J., Rackley C.R., Bowie E.J., Keene D.R., Stripp B.R., Randell S.H., Noble P.W., and Hogan B.L.. 2013. Type 2 alveolar cells are stem cells in adult lung. J. Clin. Invest. 123:3025–3036. 10.1172/JCI68782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basil, M.C., Katzen J., Engler A.E., Guo M., Herriges M.J., Kathiriya J.J., Windmueller R., Ysasi A.B., Zacharias W.J., Chapman H.A., et al. 2020. The cellular and physiological basis for lung repair and regeneration: Past, present, and future. Cell Stem Cell. 26:482–502. 10.1016/j.stem.2020.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benabid, A., and Peduto L.. 2020. Mesenchymal perivascular cells in immunity and disease. Curr. Opin. Immunol. 64:50–55. 10.1016/j.coi.2020.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berasain, C., and Avila M.A.. 2014. Amphiregulin. Semin. Cell Dev. Biol. 28:31–41. 10.1016/j.semcdb.2014.01.005 [DOI] [PubMed] [Google Scholar]
- Broggi, A., Ghosh S., Sposito B., Spreafico R., Balzarini F., Lo Cascio A., Clementi N., De Santis M., Mancini N., Granucci F., and Zanoni I.. 2020. Type III interferons disrupt the lung epithelial barrier upon viral recognition. Science. 369:706–712. 10.1126/science.abc3545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burzyn, D., Kuswanto W., Kolodin D., Shadrach J.L., Cerletti M., Jang Y., Sefik E., Tan T.G., Wagers A.J., Benoist C., and Mathis D.. 2013. A special population of regulatory T cells potentiates muscle repair. Cell. 155:1282–1295. 10.1016/j.cell.2013.10.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, J., Park J.E., Tsagkogeorga G., Yanagita M., Koo B.K., Han N., and Lee J.H.. 2020. Inflammatory signals induce AT2 cell-derived damage-associated transient progenitors that mediate alveolar regeneration. Cell Stem Cell. 27:366–382.e7. 10.1016/j.stem.2020.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung, M.I., Bujnis M., Barkauskas C.E., Kobayashi Y., and Hogan B.L.M.. 2018. Niche-mediated BMP/SMAD signaling regulates lung alveolar stem cell proliferation and differentiation. Development. 145:dev163014. 10.1242/dev.163014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlgren, M.W., Jones S.W., Cautivo K.M., Dubinin A., Ortiz-Carpena J.F., Farhat S., Yu K.S., Lee K., Wang C., Molofsky A.V., et al. 2019. Adventitial stromal cells define group 2 innate lymphoid cell tissue niches. Immunity. 50:707–722.e6. 10.1016/j.immuni.2019.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, L., Liu T., Wu Z., Hu B., Nakashima T., Ullenbruch M., Gonzalez De Los Santos F., and Phan S.H.. 2016. Bone marrow CD11c+ cell-derived amphiregulin promotes pulmonary fibrosis. J. Immunol. 197:303–312. 10.4049/jimmunol.1502479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernanda de Mello Costa, M., Weiner A.I., and Vaughan A.E.. 2020. Basal-like progenitor cells: A review of dysplastic alveolar regeneration and remodeling in lung repair. Stem Cell Rep. 15:1015–1025. 10.1016/j.stemcr.2020.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finigan, J.H., Downey G.P., and Kern J.A.. 2012. Human epidermal growth factor receptor signaling in acute lung injury. Am. J. Respir. Cell Mol. Biol. 47:395–404. 10.1165/rcmb.2012-0100TR [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn, J., Sottoriva K., Pajcini K.V., Kitajewski J.K., Chen C., Zhang W., Malik A.B., and Liu Y.. 2019. Dlk1-Mediated temporal regulation of Notch signaling is required for differentiation of alveolar type II to type I cells during repair. Cell Rep. 26:2942–2954.e5. 10.1016/j.celrep.2019.02.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank, D.B., Peng T., Zepp J.A., Snitow M., Vincent T.L., Penkala I.J., Cui Z., Herriges M.J., Morley M.P., Zhou S., et al. 2016. Emergence of a wave of Wnt signaling that regulates lung alveologenesis by controlling epithelial self-renewal and differentiation. Cell Rep. 17:2312–2325. 10.1016/j.celrep.2016.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freed, D.M., Bessman N.J., Kiyatkin A., Salazar-Cavazos E., Byrne P.O., Moore J.O., Valley C.C., Ferguson K.M., Leahy D.J., Lidke D.S., and Lemmon M.A.. 2017. EGFR ligands differentially stabilize receptor dimers to specify signaling kinetics. Cell. 171:683–695.e18. 10.1016/j.cell.2017.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujikura, D., and Miyazaki T.. 2018. Programmed cell death in the pathogenesis of influenza. Int. J. Mol. Sci. 19:2065. 10.3390/ijms19072065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuyama, S., Katsura H., Zhao D., Ozawa M., Ando T., Shoemaker J.E., Ishikawa I., Yamada S., Neumann G., Watanabe S., et al. 2015. Multi-spectral fluorescent reporter influenza viruses (Color-flu) as powerful tools for in vivo studies. Nat. Commun. 6:6600. 10.1038/ncomms7600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gereke, M., Autengruber A., Gröbe L., Jeron A., Bruder D., and Stegemann-Koniszewski S.. 2012. Flow cytometric isolation of primary murine type II alveolar epithelial cells for functional and molecular studies. J. Vis. Exp. 26:4322. 10.3791/4322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green, J.A., Arpaia N., Schizas M., Dobrin A., and Rudensky A.Y.. 2017. A nonimmune function of T cells in promoting lung tumor progression. J. Exp. Med. 214:3565–3575. 10.1084/jem.20170356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gremlich, S., Roth-Kleiner M., Equey L., Fytianos K., Schittny J.C., and Cremona T.P.. 2020. Tenascin-C inactivation impacts lung structure and function beyond lung development. Sci. Rep. 10:5118. 10.1038/s41598-020-61919-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, X., Wang R., Zhou Y., Fei L., Sun H., Lai S., Saadatpour A., Zhou Z., Chen H., Ye F., et al. 2018. Mapping the mouse cell atlas by microwell-seq. Cell. 172:1091–1107.e17. 10.1016/j.cell.2018.02.001 [DOI] [PubMed] [Google Scholar]
- Hogan, B.L., Barkauskas C.E., Chapman H.A., Epstein J.A., Jain R., Hsia C.C., Niklason L., Calle E., Le A., Randell S.H., et al. 2014. Repair and regeneration of the respiratory system: Complexity, plasticity, and mechanisms of lung stem cell function. Cell Stem Cell. 15:123–138. 10.1016/j.stem.2014.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui, S.P., Sheng D.Z., Sugimoto K., Gonzalez-Rajal A., Nakagawa S., Hesselson D., and Kikuchi K.. 2017. Zebrafish regulatory T cells mediate organ-specific regenerative programs. Dev. Cell. 43:659–672.e5. 10.1016/j.devcel.2017.11.010 [DOI] [PubMed] [Google Scholar]
- Ibricevic, A., Pekosz A., Walter M.J., Newby C., Battaile J.T., Brown E.G., Holtzman M.J., and Brody S.L.. 2006. Influenza virus receptor specificity and cell tropism in mouse and human airway epithelial cells. J. Virol. 80:7469–7480. 10.1128/JVI.02677-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue, H., Akimoto K., Homma T., Tanaka A., and Sagara H.. 2020. Airway epithelial dysfunction in asthma: Relevant to epidermal growth factor receptors and airway epithelial cells. J. Clin. Med. 9:3698. 10.3390/jcm9113698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito, M., Komai K., Mise-Omata S., Iizuka-Koga M., Noguchi Y., Kondo T., Sakai R., Matsuo K., Nakayama T., Yoshie O., et al. 2019. Brain regulatory T cells suppress astrogliosis and potentiate neurological recovery. Nature. 565:246–250. 10.1038/s41586-018-0824-5 [DOI] [PubMed] [Google Scholar]
- Jiang, P., Gil de Rubio R., Hrycaj S.M., Gurczynski S.J., Riemondy K.A., Moore B.B., Omary M.B., Ridge K.M., and Zemans R.L.. 2020. Ineffectual type 2-to-Type 1 alveolar epithelial cell differentiation in idiopathic pulmonary fibrosis: Persistence of the KRT8hi transitional state. Am. J. Respir. Crit. Care Med. 201:1443–1447. 10.1164/rccm.201909-1726LE [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, G.R., and Wong L.. 1994. Heparan sulfate is essential to amphiregulin-induced mitogenic signaling by the epidermal growth factor receptor. J. Biol. Chem. 269:27149–27154. 10.1016/S0021-9258(18)47137-2 [DOI] [PubMed] [Google Scholar]
- Kerpedjieva, S.S., Kim D.S., Barbeau D.J., and Tamama K.. 2012. EGFR ligands drive multipotential stromal cells to produce multiple growth factors and cytokines via early growth response-1. Stem Cells Dev. 21:2541–2551. 10.1089/scd.2011.0711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi, Y., Tata A., Konkimalla A., Katsura H., Lee R.F., Ou J., Banovich N.E., Kropski J.A., and Tata P.R.. 2020. Persistence of a regeneration-associated, transitional alveolar epithelial cell state in pulmonary fibrosis. Nat. Cell Biol. 22:934–946. 10.1038/s41556-020-0542-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, P.A., Hu Y., Yamamoto Y., Hoe N.B., Wei T.S., Mu D., Sun Y., Joo L.S., Dagher R., Zielonka E.M., et al. 2011. Distal airway stem cells yield alveoli in vitro and during lung regeneration following H1N1 influenza infection. Cell. 147:525–538. 10.1016/j.cell.2011.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J.H., Tammela T., Hofree M., Choi J., Marjanovic N.D., Han S., Canner D., Wu K., Paschini M., Bhang D.H., et al. 2017. Anatomically and functionally distinct lung mesenchymal populations marked by Lgr5 and Lgr6. Cell. 170:1149–1163.e12. 10.1016/j.cell.2017.07.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, T.C., and Threadgill D.W.. 2009. Generation and validation of mice carrying a conditional allele of the epidermal growth factor receptor. Genesis. 47:85–92. 10.1002/dvg.20464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy, B.D., and Serhan C.N.. 2014. Resolution of acute inflammation in the lung. Annu. Rev. Physiol. 76:467–492. 10.1146/annurev-physiol-021113-170408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen, L., Zwingman T.A., Sunkin S.M., Oh S.W., Zariwala H.A., Gu H., Ng L.L., Palmiter R.D., Hawrylycz M.J., Jones A.R., et al. 2010. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 13:133–140. 10.1038/nn.2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Major, J., Crotta S., Llorian M., McCabe T.M., Gad H.H., Priestnall S.L., Hartmann R., and Wack A.. 2020. Type I and III interferons disrupt lung epithelial repair during recovery from viral infection. Science. 369:712–717. 10.1126/science.abc2061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malireddi, R.K., and Kanneganti T.D.. 2013. Role of type I interferons in inflammasome activation, cell death, and disease during microbial infection. Front. Cell. Infect. Microbiol. 3:77. 10.3389/fcimb.2013.00077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzo, N.D., Foster W.M., and Stripp B.R.. 2012. Amphiregulin-dependent mucous cell metaplasia in a model of nonallergic lung injury. Am. J. Respir. Cell Mol. Biol. 47:349–357. 10.1165/rcmb.2011-0257OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthay, M.A., Zemans R.L., Zimmerman G.A., Arabi Y.M., Beitler J.R., Mercat A., Herridge M., Randolph A.G., and Calfee C.S.. 2019. Acute respiratory distress syndrome. Nat. Rev. Dis. Primers. 5:18. 10.1038/s41572-019-0069-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQualter, J.L., Yuen K., Williams B., and Bertoncello I.. 2010. Evidence of an epithelial stem/progenitor cell hierarchy in the adult mouse lung. Proc. Natl. Acad. Sci. USA. 107:1414–1419. 10.1073/pnas.0909207107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meulenbroeks, C., van Weelden H., Schwartz C., Voehringer D., Redegeld F.A.M., Rutten V.P.M.G., Willemse T., Sijts A.J.A.M., and Zaiss D.M.W.. 2015. Basophil-derived amphiregulin is essential for UVB irradiation-induced immune suppression. J. Invest. Dermatol. 135:222–228. 10.1038/jid.2014.329 [DOI] [PubMed] [Google Scholar]
- Minutti, C.M., Modak R.V., Macdonald F., Li F., Smyth D.J., Dorward D.A., Blair N., Husovsky C., Muir A., Giampazolias E., et al. 2019. A macrophage-pericyte Axis directs tissue restoration via amphiregulin-induced transforming growth factor beta activation. Immunity. 50:645–654.e6. 10.1016/j.immuni.2019.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monticelli, L.A., Sonnenberg G.F., Abt M.C., Alenghat T., Ziegler C.G., Doering T.A., Angelosanto J.M., Laidlaw B.J., Yang C.Y., Sathaliyawala T., et al. 2011. Innate lymphoid cells promote lung-tissue homeostasis after infection with influenza virus. Nat. Immunol. 12:1045–1054. 10.1038/ni.2131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto, Y., Hirahara K., Kiuchi M., Wada T., Ichikawa T., Kanno T., Okano M., Kokubo K., Onodera A., Sakurai D., et al. 2018. Amphiregulin-producing pathogenic memory T helper 2 cells instruct eosinophils to secrete osteopontin and facilitate airway fibrosis. Immunity. 49:134–150.e6. 10.1016/j.immuni.2018.04.023 [DOI] [PubMed] [Google Scholar]
- Nabhan, A.N., Brownfield D.G., Harbury P.B., Krasnow M.A., and Desai T.J.. 2018. Single-cell Wnt signaling niches maintain stemness of alveolar type 2 cells. Science. 359:1118–1123. 10.1126/science.aam6603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naik, S., Larsen S.B., Cowley C.J., and Fuchs E.. 2018. Two to tango: Dialog between immunity and stem cells in health and disease. Cell. 175:908–920. 10.1016/j.cell.2018.08.071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng, B., Dong J., D’Agostino G., Viswanathan S., Widjaja A.A., Lim W.W., Ko N.S.J., Tan J., Chothani S.P., Huang B., et al. 2019. Interleukin-11 is a therapeutic target in idiopathic pulmonary fibrosis. Sci. Transl. Med. 11:eaaw1237. 10.1126/scitranslmed.aaw1237 [DOI] [PubMed] [Google Scholar]
- Nordgren, T.M., Heires A.J., Bailey K.L., Katafiasz D.M., Toews M.L., Wichman C.S., and Romberger D.J.. 2018. Docosahexaenoic acid enhances amphiregulin-mediated bronchial epithelial cell repair processes following organic dust exposure. Am. J. Physiol. Lung Cell. Mol. Physiol. 314:L421–L431. 10.1152/ajplung.00273.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohmori, T., Yamaoka T., Ando K., Kusumoto S., Kishino Y., Manabe R., and Sagara H.. 2021. Molecular and clinical features of EGFR-TKI-associated lung injury. Int. J. Mol. Sci. 22:792. 10.3390/ijms22020792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paris, A.J., Hayer K.E., Oved J.H., Avgousti D.C., Toulmin S.A., Zepp J.A., Zacharias W.J., Katzen J.B., Basil M.C., Kremp M.M., et al. 2020. STAT3-BDNF-TrkB signalling promotes alveolar epithelial regeneration after lung injury. Nat. Cell Biol. 22:1197–1210. 10.1038/s41556-020-0569-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peitz, M., Pfannkuche K., Rajewsky K., and Edenhofer F.. 2002. Ability of the hydrophobic FGF and basic TAT peptides to promote cellular uptake of recombinant cre recombinase: A tool for efficient genetic engineering of mammalian genomes. Proc. Natl. Acad. Sci. USA. 99:4489–4494. 10.1073/pnas.032068699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray, S., Chiba N., Yao C., Guan X., McConnell A.M., Brockway B., Que L., McQualter J.L., and Stripp B.R.. 2016. Rare SOX2+ airway progenitor cells generate KRT5+ cells that repopulate damaged alveolar parenchyma following influenza virus infection. Stem Cell Rep. 7:817–825. 10.1016/j.stemcr.2016.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock, J.R., Barkauskas C.E., Cronce M.J., Xue Y., Harris J.R., Liang J., Noble P.W., and Hogan B.L.. 2011. Multiple stromal populations contribute to pulmonary fibrosis without evidence for epithelial to mesenchymal transition. Proc. Natl. Acad. Sci. USA. 108:E1475–E1483. 10.1073/pnas.1117988108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolfes, M.A., Foppa I.M., Garg S., Flannery B., Brammer L., Singleton J.A., Burns E., Jernigan D., Olsen S.J., Bresee J., and Reed C.. 2018. Annual estimates of the burden of seasonal influenza in the United States: A tool for strengthening influenza surveillance and preparedness. Influenza Other Respir. Viruses. 12:132–137. 10.1111/irv.12486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarrazin, S., Lamanna W.C., and Esko J.D.. 2011. Heparan sulfate proteoglycans. Cold Spring Harb. Perspect. Biol. 3:a004952. 10.1101/cshperspect.a004952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaum, N., Karkanias J., Neff N.F., May A.P., Quake S.R., Wyss-Coray T., Darmanis S., Batson J., Botvinnik O., Chen M.B., et al. 2018. Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature. 562:367–372. 10.1038/s41586-018-0590-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenmark, K.R., Nozik-Grayck E., Gerasimovskaya E., Anwar A., Li M., Riddle S., and Frid M.. 2011. The adventitia: Essential role in pulmonary vascular remodeling. Compr. Physiol. 1:141–161. 10.1002/cphy.c090017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strunz, M., Simon L.M., Ansari M., Kathiriya J.J., Angelidis I., Mayr C.H., Tsidiridis G., Lange M., Mattner L.F., Yee M., et al. 2020. Alveolar regeneration through a Krt8+ transitional stem cell state that persists in human lung fibrosis. Nat. Commun. 11:3559. 10.1038/s41467-020-17358-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay, M.Z., Poh C.M., Rénia L., MacAry P.A., and Ng L.F.P.. 2020. The trinity of COVID-19: Immunity, inflammation and intervention. Nat. Rev. Immunol. 20:363–374 10.1038/s41577-020-0311-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travaglini, K.J., Nabhan A.N., Penland L., Sinha R., Gillich A., Sit R.V., Chang S., Conley S.D., Mori Y., Seita J., et al. 2020. A molecular cell atlas of the human lung from single-cell RNA sequencing. Nature. 587:619–625. 10.1038/s41586-020-2922-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukui, T., Sun K.H., Wetter J.B., Wilson-Kanamori J.R., Hazelwood L.A., Henderson N.C., Adams T.S., Schupp J.C., Poli S.D., Rosas I.O., et al. 2020. Collagen-producing lung cell atlas identifies multiple subsets with distinct localization and relevance to fibrosis. Nat. Commun. 11:1920. 10.1038/s41467-020-15647-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallath, S., Hynds R.E., Succony L., Janes S.M., and Giangreco A.. 2014. Targeting EGFR signalling in chronic lung disease: Therapeutic challenges and opportunities. Eur. Respir. J. 44:513–522. 10.1183/09031936.00146413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan, A.E., Brumwell A.N., Xi Y., Gotts J.E., Brownfield D.G., Treutlein B., Tan K., Tan V., Liu F.C., Looney M.R., et al. 2015. Lineage-negative progenitors mobilize to regenerate lung epithelium after major injury. Nature. 517:621–625. 10.1038/nature14112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vila Ellis, L., Cain M.P., Hutchison V., Flodby P., Crandall E.D., Borok Z., Zhou B., Ostrin E.J., Wythe J.D., and Chen J.. 2020. Epithelial Vegfa specifies a distinct endothelial population in the mouse lung. Dev. Cell. 52:617–630.e6. 10.1016/j.devcel.2020.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, T., Wang Y., Deng N., Huang G., Taghavifar F., Geng Y., Liu N., Kulur V., Yao C., Chen P., et al. 2018. Single-cell deconvolution of fibroblast heterogeneity in mouse pulmonary fibrosis. Cell Rep. 22:3625–3640. 10.1016/j.celrep.2018.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaoka, T., Arata S., Homma M., Homma T., Kusumoto S., Ando K., Manabe R., Kishino Y., Ohba M., Tsurutani J., et al. 2019. Blockade of EGFR activation promotes TNF-induced lung epithelial cell apoptosis and pulmonary injury. Int. J. Mol. Sci. 20:4021. 10.3390/ijms20164021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacharias, W.J., Frank D.B., Zepp J.A., Morley M.P., Alkhaleel F.A., Kong J., Zhou S., Cantu E., and Morrisey E.E.. 2018. Regeneration of the lung alveolus by an evolutionarily conserved epithelial progenitor. Nature. 555:251–255. 10.1038/nature25786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaiss, D.M., Minutti C.M., and Knipper J.A.. 2019. Immune- and non-immune-mediated roles of regulatory T-cells during wound healing. Immunology. 157:190–197. 10.1111/imm.13057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zepp, J.A., Zacharias W.J., Frank D.B., Cavanaugh C.A., Zhou S., Morley M.P., and Morrisey E.E.. 2017. Distinct mesenchymal lineages and niches promote epithelial self-renewal and myofibrogenesis in the lung. Cell. 170:1134–1148.e10. 10.1016/j.cell.2017.07.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, C., Li L., Feng K., Fan D., Xue W., and Lu J.. 2017. “Repair” Treg cells in tissue injury. Cell. Physiol. Biochem. 43:2155–2169. 10.1159/000484295 [DOI] [PubMed] [Google Scholar]
- Zheng, B., Zhang Z., Black C.M., de Crombrugghe B., and Denton C.P.. 2002. Ligand-dependent genetic recombination in fibroblasts: A potentially powerful technique for investigating gene function in fibrosis. Am. J. Pathol. 160:1609–1617. 10.1016/S0002-9440(10)61108-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo, W., Zhang T., Wu D.Z., Guan S.P., Liew A.A., Yamamoto Y., Wang X., Lim S.J., Vincent M., Lessard M., et al. 2015. p63(+)Krt5(+) distal airway stem cells are essential for lung regeneration. Nature. 517:616–620. 10.1038/nature13903 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
is a list of each mesenchymal cell.
contains original blots for Fig. 4.
contains original blots for Fig. 5.
contains original blots for Fig. S5.
Data Availability Statement
This paper does not report original code. All raw sequencing data have been deposited in the Gene Expression Omnibus under accession no. GSE169127. Any additional information required to reanalyze the data reported in this paper are available from the lead contact upon request.