Abstract
A minitransposon mutant of Salmonella enterica serovar Typhimurium SR-11, SR-11 Fad−, is unable to utilize gluconeogenic substrates as carbon sources and is avirulent and immunogenic when administered perorally to BALB/c mice (M. J. Utley et al., FEMS Microbiol. Lett., 163:129–134, 1998). Here, evidence is presented that the mutation in SR-11 Fad− that renders the strain avirulent is in the cra gene, which encodes the Cra protein, a regulator of central carbon metabolism.
Since Salmonella enterica serovar Typhimurium is able to utilize phosphatidylserine as the sole source of carbon, nitrogen, and phosphate for growth (11) and since phosphatidylserine is a component of eucaryotic cell membranes, it was of interest to determine whether mutants unable to utilize either the glycerophosphate portion or the fatty acid portion of the phospholipid for growth are attenuated. Several mutants unable to utilize oleate as a sole carbon source were isolated after mini-Tn10::d cam transposon mutagenesis (6), and one of them, designated serovar Typhimurium SR-11 Fad− (fatty acid), was totally avirulent and immunogenic in BALB/c mice when administered perorally (21). In addition, SR-11 Fad− was found in the liver and spleen in numbers 4 to 5 orders of magnitude lower than SR-11 (21). SR-11 Fad− was also less virulent than SR-11 when administered to mice intraperitoneally (21). Furthermore, SR-11 Fad−, in addition to being unable to grow utilizing oleate as a sole carbon source, was also unable to grow utilizing citrate, isocitrate, and acetate as sole carbon sources, all of which were utilized well by SR-11 (21). SR-11 Fad− grew slowly and to only one-tenth the final yield of SR-11 utilizing pyruvate as the sole carbon source in liquid culture (21). SR-11 Fad− was, however, able to utilize glucose, glycerol, and a number of sugars sensitive to catabolite repression as the sole carbon sources (14). In the present study, we identify the gene made defective in SR-11 Fad− by mini-Tn10::d cam transposon mutagenesis and show that the defect is responsible for SR-11 Fad− avirulence in BALB/c mice.
SR-11 Fad− is a cra (fruR) mutant.
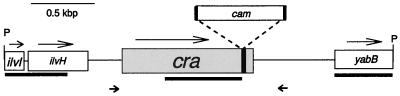
In order to identify the gene that is inactivated in SR-11 Fad−, Southern hybridization was performed using a digoxigenin (DIG)-labeled transposon-specific probe made from pJHA1 (Table 1). The transposon-specific probe was prepared with a DIG High Prime DNA Labeling and Detection Starter Kit II (Boehringer Mannheim, Indianapolis, Ind.) as described by the manufacturer. The Tn10::d cam minitransposon was located on a 4.5-kb PstI SR-11 Fad− DNA fragment which was inserted into the unique PstI site of pBluescript II SK(+) to create pJHA7 (Table 1). The first 489 bp in the 4.5-kb SR-11 Fad− insert in pJHA7 at the T3 promoter end were sequenced. The sequence was essentially identical to the last 180 bp of the serovar Typhimurium LT2 ilvI gene, the first 306 bp of the serovar Typhimurium LT2 ilvH gene, and the three intervening base pairs (GenBank accession number X55456). The first 470 bp at the opposite end of the 4.5-kb SR-11 Fad− insert in pJHA7, sequenced from the T7 promoter, were 87% identical to yabB, a gene of unknown function in Escherichia coli (GenBank accession number AE000118 U00096). In both serovar Typhimurium LT2 and E. coli, ilvI and ilvH are immediately upstream of the fruR gene, which is 601 bp upstream of yabB in E. coli (GenBank accession number AE000118 U00096). Sequencing from the chloramphenicol resistance gene in pJHA7 determined that the mini-Tn10::d cam transposon was indeed inserted 45 bp upstream of the 3′ end of the LT2 fruR gene (reference 9 and GenBank accession number X55456). The fruR gene has recently been renamed cra (catabolite repressor/activator) (18). A diagram of the genes present in the 4.5-kb PstI SR-11 Fad− DNA fragment in pJHA7 and the portions sequenced is presented in Fig. 1.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant genotype and/or phenotype | Source or reference |
|---|---|---|
| S. enterica serovar Typhimurium strains (plasmid) | ||
| SR-11 | gyr1816 | 4 |
| SR-11 Fad− | gyr1816 cra::Tn10 d cam | 21 |
| SR-11 Fad− AX-2 | gyr1816 cra::Tn10 d cam | This study |
| SR-11 Fad−(pBR322) | gyr1816 cra::Tn10 d cam, bla tet | This study |
| SR-11 Fad−(pJHA8) | gyr1816 cra::Tn10 d cam, cra+ tet | This study |
| LJ2443 | fruR51::Tn10 | 3 |
| UK-1 | Virulent avian strain | 5 |
| UK-1 Fad− AX-1 | cra::Tn10 d cam | This study |
| E. coli S-17 λ pir | Host for pLD55 and pMJN10 | V. de Lorenzo |
| Plasmids | ||
| pBR322 | bla tet | 2 |
| pJHA1 | 1.4-kb cam gene from Tn10::d cam in BamHI site of pBR322 | This study |
| pJHA7 | 4.3-kb PstI fragment containing cra::Tn10 d cam in pBR322, cam tet | This study |
| pJHA8 | 1.4-kb cra+ gene in PstI site of pBR322, tet | This study |
| pLD55 | Suicide vector for allelic exchange, bla tetAR | 15 |
| pMJN10 | 4.3-kb PstI fragment containing cra::Tn10 d cam in pLD55, cat bla tetAR | This study |
FIG. 1.
Genes present in the SR-11 Fad− 4.5-kb PstI DNA fragment that contains the chloramphenicol resistance gene (cam). Heavy lines below the genes denote regions in the fragment that were sequenced. Sequencing reactions were performed with the BigDye Terminator Cycle Sequencing Kit, and reactions were analyzed on the ABI PRISM 310 (PE Applied Biosystems, Forest City, Calif.). Nucleotide sequences were analyzed using Clone Manager and Align Plus 3 programs (Sci-Ed Software, Durham, N.C.). Arrows above the genes denote the direction of transcription. Arrows below the genes denote the positions of the forward and reverse primers used for PCR amplification of the wild-type and defective cra genes. P, PstI.
Serovar Typhimurium LJ2443, a known cra mutant (3), is unable to grow on minimal agar plates containing gluconeogenic substrates as sole carbon sources (3, 18). Its phenotype with respect to utilization of different carbon sources was compared to SR-11 Fad−. Both mutants were essentially identical with respect to growth on M9 minimal agar plates containing either sodium oleate (5 mM), sodium citrate (0.2% [wt/wt]), potassium acetate (0.4% [wt/wt]), sodium pyruvate (0.4% [wt/wt]), sodium succinate (0.6% [wt/wt]), potassium fumarate (0.4% [wt/wt]), glucose (0.2% [wt/wt]), or glycerol (0.2% [wt/wt]), i.e., they failed to grow on the gluconeogenic substrates as sole carbon sources in 48 h at 37°C but grew as well as their wild-type parents on glucose and glycerol. The wild-type SR-11 cra gene was cloned into pBR322 to create pJHA8 (Table 1). Both SR-11 Fad− and LJ2443, when complemented with pJHA8, regained the ability to grow on M9 agar plates containing the aforementioned gluconeogenic substrates as sole carbon sources. pBR322 did not functionally complement the cra mutation in either SR-11 or LJ2443. These experiments established conclusively that serovar Typhimurium SR-11 Fad− is a cra mutant.
SR-11 Fad− (pJHA8) is virulent.
It was possible that inactivation of the cra gene with the mini-Tn10::d cam insertion caused a downstream effect that resulted in avirulence. We therefore complemented SR-11 Fad− with pJHA8, which contains the wild-type SR-11 cra gene as the only nonvector gene in pBR322, and tested its virulence.
Groups of four BALB/c mice were perorally infected with either 3.1 × 108 CFU per mouse of SR-11 Fad− (pBR322) or 4.3 × 108 CFU per mouse of SR-11 Fad− (pJHA8). By 8 days postinfection, three of the four mice infected with SR-11 Fad− (pJHA8) had died. The fourth mouse infected with SR-11 Fad− (pJHA8) appeared to be very sick for several days (ruffled fur, loss of appetite, huddled) but recovered. In contrast, the four mice infected with SR-11 Fad− (pBR322) remained alive and healthy. Since complementing SR-11 Fad− with the wild-type cra gene results in renewed virulence, it is the defect in the cra gene caused by the mini-Tn10::d cam insertion that renders the strain avirulent.
A serovar Typhimurium UK-1 cra mutant constructed by allelic exchange is avirulent.
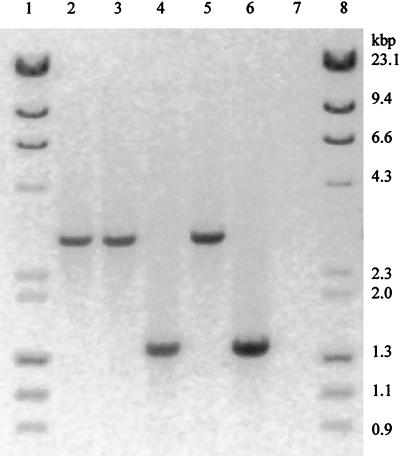
It was of interest to determine whether a cra mutant of a second serovar Typhimurium strain was also avirulent. Therefore, a cra mutant of the virulent strain serovar Typhimurium UK-1 (23), designated UK-1 Fad− AX-1 (allelic exchange), was constructed by allelic exchange of the wild-type cra gene with the mutant cra gene using pMJN10 (see Table 1). pMJN10 contains the mutant cra gene and flanking sequences as a 4.5-kb PstI DNA fragment (Fig. 1) in pLD55, an allelic exchange vector (15). That the only cra gene that UK-1 Fad− AX-1 contained was the defective cra gene was shown by four lines of evidence. First, UK-1 Fad− AX-1 was unable to grow on any of the gluconeogenic substrates. Second, complementing UK-1 Fad− AX-1 with the wild-type cra gene returned its ability to grow on the gluconeogenic substrates. Third, when PstI-cut UK-1 Fad− AX-1 DNA was probed with either the wild-type cra gene or the chloramphenicol resistance gene in Southern hybridization experiments, a single 4.5-kb band was detected, as expected if the 3.1-kb PstI DNA fragment containing the wild-type cra gene was replaced with the defective cra gene (not shown). Fourth, when UK-1 and UK-1 AX-1 DNA were amplified by PCR using the forward and reverse primers used to amplify the SR-11 wild-type cra gene (see Fig. 1 and the legend to Fig. 2), the amplified fragments were as expected (Fig. 2), i.e., in UK-1 a 1.4-kb fragment containing the wild-type cra gene and in UK-1 Fad− AX-1 a 2.8-kb fragment (the 1.4-kb cra fragment containing the inserted 1.4-kb chloramphenicol resistance gene).
FIG. 2.
PCR amplification of the defective cra genes of serovar Typhimurium SR-11 Fad−, SR-11 Fad− AX-2, and UK-1 Fad− AX-1. Genomic DNA preparations were performed by standard techniques (13) or by following the manufacturer's instructions. Genomic DNA preparations of SR-11, SR-11 Fad−, SR-11 Fad− AX-2, UK-1, and UK-1 AX-1 were amplified by PCR using 5′-ATCGACTGCAGTGCGAAATCCGTGGTAACCCGG-3′ as the forward primer and 5′-TAGCTCTGCAGCCTGTTTAACGTGTGCGGTGCC-3′ as the reverse primer. PCR reaction mixtures contained 2.0 mM magnesium chloride and 5% dimethyl sulfoxide and were subjected to 25 cycles of 94°C for 30 s, followed by 65°C for 30 s, followed by 72°C for 2.5 min. Lane 1, kilobase ladder; lane 2, SR-11 Fad− AX-2; lane 3, SR-11 Fad−; lane 4, SR-11; lane 5, UK-1 Fad− AX-1; lane 6, UK-1; lane 7, no DNA added; lane 8, kilobase ladder.
To determine whether UK-1 Fad− AX-1 is avirulent, four BALB/c mice were infected perorally with UK-1 (2.6 × 108 CFU per mouse), and four mice were identically infected with UK-1 Fad− AX-2 (1.8 × 108 CFU per mouse). By day 8 postinfection, all four UK-1 infected mice had died, whereas the four mice infected with UK-1 AX-1 were healthy and active and had never appeared to be sick. The UK-1 Fad− AX-1-infected mice were still healthy and active when the experiment was terminated at 3 weeks postinfection. Therefore, avirulence caused by a defect in cra is not restricted to SR-11.
SR-11 Fad− AX-2 was also constructed by allelic exchange of the wild-type cra gene with the mutant cra gene using pMJN10 (see Table 1). SR-11 Fad− AX-2 had the phenotype of a cra mutant with respect to utilization of carbon sources. Moreover, SR-11 Fad− AX-2 regained the wild-type phenotype, when complemented with the wild-type cra gene, and contained the chloramphenicol resistance gene on a 4.5-kb PstI fragment, and the expected 2.8-kb PCR product was amplified from SR-11 Fad− AX-2 using the forward and reverse primers specific to the SR-11 wild-type cra gene (Fig. 2). SR-11 Fad− AX-2 was also found to be avirulent. Four BALB/c mice were infected perorally with SR-11 (2.1 × 108 CFU per mouse), and five mice were identically infected with SR-11 Fad− AX-2 (2.8 × 108 CFU per mouse). By day 8 postinfection, all four SR-11 infected mice had died, whereas the five mice infected with SR-11 Fad− AX-2 were healthy and active and had never appeared to be sick. The SR-11 Fad− AX-2-infected mice were still healthy and active when the experiment was terminated at 3 weeks postinfection.
Behavior of SR-11 and SR-11 Fad− in infection models.
Serovar Typhimurium utilizes nutrients present in intestinal mucus for growth in the intestine (12, 14). Mammalian intestinal mucus contains several sugars that SR-11 and SR-11 Fad− could metabolize via glycolytic pathways, including N-acetylglucosamine, N-acetylgalactosamine, galactose, fucose, sialic acid, ribose, arabinose, mannose, gluconate, and the hexuronates glucoronate and galacturonate (1, 17, 20). Since serovar Typhimurium SR-11 Fad− is not defective in its ability to utilize sugars for growth (21), it is unlikely that it is defective in the ability to grow in the mouse intestine. In support of this view, in a previous study (8), when SR-11 and SR-11 Fad− were inoculated (4.0 × 104 CFU/ml) into mouse intestinal mucus in vitro, they grew equally well, reaching levels of about 6.0 × 108 CFU/ml in 24 h at 37°C. For this reason, we turned our attention to the ability of SR-11 Fad− to adhere to and invade intestinal epithelial cells and to survive within macrophages.
S. enterica serovar Typhimurium passes through the epithelium by penetrating and destroying the M cells of the Peyer's patches (10). Both SR-11 and SR-11 Fad− invaded BALB/c mouse M cells equally well after infection of ileal loops, made as described previously (10), and both were found in enclosed vacuoles within the M cells. In addition, differences were not found in the ability of SR-11 and SR-11 Fad− to adhere to and invade human T-84 intestinal epithelial monolayers using procedures described previously (13), i.e., 2.41 ± 0.74% (mean ± the standard deviation [SD] of triplicate samples) of the SR-11 inoculum (20 CFU per T84 cell) and 2.06 ± 0.50% of the SR-11 Fad− inoculum (20 CFU per T84 cell) became cell associated in 60 min. In the same time, 0.094 ± 0.009% of the SR-11 inoculum and 0.089 ± 0.016% of the SR-11 Fad− inoculum were internalized. Collectively, these results make it unlikely that SR-11 Fad− is avirulent because it is defective in its ability to adhere to and invade M cells and/or intestinal epithelial cells.
The ability to survive in macrophages in vitro has been correlated with S. enterica pathogenicity in mice (7, 16). Resident peritoneal macrophages were isolated from BALB/c mice (7), and survival of SR-11 and SR-11 Fad− in macrophages was determined as described previously (7). Both SR-11 and SR-11 Fad− survived equally well in resident peritoneal macrophages. That is, after 60 min 0.057 ± 0.0075% (mean ± the SD of triplicate samples) of the SR-11 inoculum and 0.056 ± 0.018% of the SR-11 Fad− inoculum survived and, in each case, both strains remained at the 1-h survival level for the next 23 h. While these data rule out the possibility that SR-11 Fad− is inherently inferior to SR-11 in surviving in BALB/c peritoneal macrophages, they do not rule out the possibility that SR-11 Fad− is avirulent because its ability to survive and grow in macrophages in vivo is decreased relative to that of SR-11. Serovar Typhimurium may, in fact, utilize gluconeogenic substrates for survival and growth in macrophages in vivo (e.g., succinate, pyruvate, etc.). If so, SR-11 Fad− would be at a distinct disadvantage. In the present experiments, the BALB/c resident peritoneal macrophages were continuously bathed in RPMI 1640 medium. RPMI 1640 medium contains 2 g of glucose per liter which SR-11 and SR-11 Fad− metabolize equally well.
In summary, a functional cra gene is necessary for serovar Typhimurium virulence in BALB/c mice. The Cra protein is a regulator of central carbon metabolism (18). When interacting with the genes it regulates, i.e., in the presence of gluconeogenic substrates (18), the Cra protein positively regulates transcription of those genes encoding biosynthetic and oxidative enzymes (e.g., key enzymes in the tricarboxylic acid cycle, the glyoxylate bypass, the gluconeogenic pathway, and electron transport) and negatively regulates transcription of genes encoding glycolytic enzymes, e.g., key enzymes in the Embden-Meyerhof and Entner-Doudoroff pathways (18). Since in a cra mutant the genes involved in glycolytic pathways are highly expressed, whereas the expression of genes involved in the gluconeogenic pathway and the glyoxylate bypass is reduced, it is possible that gluconeogenesis is required for serovar Typhimurium virulence. In support of this view, it has recently been reported that a MudJ insertion in a putative malate oxidoreductase gene, involved in gluconeogenesis, renders serovar Typhimurium avirulent and immunogenic (22). Since SR-11 Fad− grows as well as SR-11 in intestinal mucus (8), enters M cells as well as SR-11 in vivo, and adheres to epithelial cells as well as SR-11 in vitro, it is unlikely that SR-11 Fad− is avirulent because it is either defective in its ability to grow in the intestine or defective in its ability to adhere to and enter intestinal epithelial cells and M cells in vivo. If, however, the only available carbon sources for S. enterica serovar Typhimurium survival and growth in M cells and/or in macrophages in vivo are the gluconeogenic substrates, since SR-11 Fad− would be unable to grow under these conditions, it would not be surprising if this were the reason for its avirulence in mice. In situ experiments will be required to test this hypothesis.
Acknowledgments
This study was supported by a grant to P.S.C. from Intervet International, Boxmeer, The Netherlands, and from the University of Rhode Island Foundation and by NIH grant DK50989 to B.A.M.
We thank Roy R. Curtiss III, Washington University, St. Louis, Mo; V. de Lorenzo, Centro de Investigaciones Biologicas, Consejo Superior de Investigaciones, Cientificas, Madrid, Spain; and Milton Saier, University of California at San Diego, La Jolla, for their kind gifts of bacterial strains. We also thank Nafisa Ghori, Stanford University School of Medicine, Stanford, Calif., for excellent technical assistance with the transmission electron microscopy.
REFERENCES
- 1.Allen A. The structure and function of gastrointestinal mucous. In: Boedecker E C, editor. Attachment of organisms to the gut mucosa. II. Boca Raton, Fla: CRC Press, Inc.; 1984. pp. 3–11. [Google Scholar]
- 2.Balbas P, Soberon X, Merino E, Zurita M, Lomeli H, Valle F, Flores N, Bolivar F. Plasmid vector pBR322 and its special derivatives—a review. Gene. 1986;50:3–40. doi: 10.1016/0378-1119(86)90307-0. [DOI] [PubMed] [Google Scholar]
- 3.Chin A M, Feldheim D A, Saier M H., Jr Altered transcriptional patterns affecting several metabolic pathways in strains of Salmonella typhimurium which overexpress the fructose regulon. J Bacteriol. 1989;171:2424–2434. doi: 10.1128/jb.171.5.2424-2434.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Curtiss R, III, Kelly S M. Salmonella typhimurium deletion mutants lacking adenylate cyclase and cyclic AMP receptor protein are avirulent and immunogenic. Infect Immun. 1987;55:3035–3043. doi: 10.1128/iai.55.12.3035-3043.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Curtiss R, III, Porter S B, Munson M, Tinge S A, Hassan J O, Gentry-Weeks C, Kelly S M. Nonrecombinant and recombinant avirulent Salmonella live vaccines for poultry. In: Blankenship L C, Bailey J S, Cox N A, Stern N J, Meinersmann R J, editors. Colonization control of human bacterial enteropathogens in poultry. New York, N.Y: Academic Press, Inc.; 1991. pp. 169–198. [Google Scholar]
- 6.Elliot T, Roth J R. Characterization of Tn10d-Cam: a transposition-defective Tn10 specifying chloramphenicol resistance. Mol Gen Genet. 1988;213:332–338. doi: 10.1007/BF00339599. [DOI] [PubMed] [Google Scholar]
- 7.Fields P L, Swanson R V, Haidaris C G, Heffron R. Mutants of Salmonella typhimurium that cannot survive within the macrophage are avirulent. Proc Natl Acad Sci USA. 1986;83:5189–5193. doi: 10.1073/pnas.83.14.5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Franklin D P. Isolation and characterization of mouse intestinal derived phosphatidylserine that serves as a Salmonella nutrient: its role in pathogenesis and the competitive colonization of the streptomycin-treated mouse. Ph.D. dissertation. Kingston: University of Rhode Island; 1992. [Google Scholar]
- 9.Jahreis K, Postma P W, Lengeler J W. Nucleotide sequence of the ilvH-fruR gene region of Escherichia coli K12 and Salmonella typhimurium LT2. Mol Gen Genet. 1991;226:332–336. doi: 10.1007/BF00273623. [DOI] [PubMed] [Google Scholar]
- 10.Jones B D, Ghori N, Falkow S. Salmonella typhimurium initiates murine infection by penetrating and destroying the specialized epithelial M cells of the Peyer's patches. J Exp Med. 1994;180:15–23. doi: 10.1084/jem.180.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krivan H C, Franklin D P, Wang W T, Laux D C, Cohen P S. Phosphatidylserine found in intestinal mucus serves as a sole source of carbon and nitrogen for salmonellae and Escherichia coli. Infect Immun. 1992;60:3943–3946. doi: 10.1128/iai.60.9.3943-3946.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Licht T R, Tolker-Nielsen T, Holmstrøm K, Krogfelt K A, Molin S. Inhibition of Escherichia coli precursor 16S rRNA processing by mouse intestinal contents. Environ Microbiol. 1999;1:23–33. doi: 10.1046/j.1462-2920.1999.00001.x. [DOI] [PubMed] [Google Scholar]
- 13.McCormick B A, Colgan S P, Archer C D, Miller S I, Madara J L. Salmonella typhimurium attachment to human intestinal epithelial monolayers: trancellular signalling to subepithelial neutrophils. J Cell Biol. 1993;123:895–907. doi: 10.1083/jcb.123.4.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McCormick B A, Stocker B A D, Laux D C, Cohen P S. The role of motility, chemotaxis, penetration through, and growth in intestinal mucus in the ability of an avirulent strain of Salmonella typhimurium to colonize the large intestines of streptomycin-treated mice. Infect Immun. 1988;56:2209–2217. doi: 10.1128/iai.56.9.2209-2217.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Metcalf, W. W., W. Jiang, L. L. Daniels, S.-K. Kim, A. Haldimann, and B. L. Wanner. Conditionally replicative and conjugative plasmids carrying lacZα for cloning, mutagenesis, and allele replacement in bacteria. Plasmid 35:1–13. [DOI] [PubMed]
- 16.Miller S I, Mekalanos J J. Constitutive expression of the PhoP regulon attenuates Salmonella virulence and survival within macrophages. J Bacteriol. 1990;172:2485–2490. doi: 10.1128/jb.172.5.2485-2490.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peekhaus N, Conway T. What's for dinner? Entner-Doudoroff metabolism in Escherichia coli. J Bacteriol. 1998;180:3495–3502. doi: 10.1128/jb.180.14.3495-3502.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saier M H, Jr, Ramseier T M. The catabolite repressor/activator (Cra) protein of enteric bacteria. J Bacteriol. 1996;178:3411–3417. doi: 10.1128/jb.178.12.3411-3417.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 20.Sweeney N J, Klemm P, McCormick B A, Moller-Nielsen E, Utley M, Schembri M A, Laux D C, Cohen P S. The Escherichia coli gntP gene allows E. coli F-18 to occupy a distinct nutritional niche in the streptomycin-treated mouse large intestine. Infect Immun. 1996;64:3497–3503. doi: 10.1128/iai.64.9.3497-3503.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Utley M, Franklin D P, Krogfelt K A, Laux D C, Cohen P S. A Salmonella typhimurium mutant unable to utilize fatty acids and citrate is avirulent and immunogenic in mice. FEMS Microbiol Lett. 1998;163:129–134. doi: 10.1111/j.1574-6968.1998.tb13036.x. [DOI] [PubMed] [Google Scholar]
- 22.Valentine P J, Devore B P, Heffron F. Identification of three highly attenuated Salmonella typhimurium mutants that are more immunogenic and protective in mice than a prototypical aroA mutant. Infect Immun. 1998;66:3378–3383. doi: 10.1128/iai.66.7.3378-3383.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang X, Kelly S M, Bollen W S, Curtiss R., III Characterization and immunogenicity of Salmonella typhimurium SL1344 and UK-1 Δcrp and Δctd deletion mutants. Infect Immun. 1997;65:5381–5387. doi: 10.1128/iai.65.12.5381-5387.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]