Abstract
Extracellular vesicles (EVs) are tissue-specific particles containing valuable diagnostic information. However, single EV analysis in blood is challenging due to their physical properties, the molecular complexity of plasma, and a lack of robust data interpretation methods. We assess the applicability of our recently-developed calibrated Imaging Flow Cytometry (IFCM)-based methodology to detect/characterize circulating tissue-specific EV subsets in the clinical setting of kidney transplantation. Platelet-poor plasma was generated from 36 HLA-A3 mismatched donor (HLA-A3 +) and kidney transplant recipients (KTRs; HLA-A3-). Samples taken before transplantation, 3 days, 7 days, and 6 months after transplantation as well as before ‘for-cause’ kidney transplant biopsies were stained with anti-CD9 (plasma EV-marker) and anti-HLA-A3. Before transplantation, no significant differences in total CD9 + EV concentrations were detected between donor and KTR samples. Tissue-specific EVs were identified as CD9 + HLA-A3 + . Serial dilution experiments of HLA-A3 + in HLA-A3- PPP showed that single CD9 + HLA-A3 + EVs were detectable down to ~ 1% above the recipient ‘self-signal’. After transplantation, CD9 + HLA-A3 + EVs were detected above pre-transplantation concentrations in individuals with stable allograft function, but not in individuals with allograft dysfunction. These results demonstrate the applicability of our calibrated IFCM-based methodology in the direct detection of tissue-specific EV subsets in clinical samples. We believe that this EV methodology is applicable in a variety of clinical contexts.
Subject terms: Nanoparticles, Flow cytometry, High-throughput screening
Introduction
Extracellular vesicles (EVs) are lipid bilayer-delimited membrane particles (30–8000 nm in diameter1) excreted by all cell types, which act as signaling intermediaries during normal homeostasis and during pathologic processes2–6. EVs carry proteins on their surface and/or a variety of macromolecules as cargo (e.g. DNA, RNA, lipids and proteins7), which are thought to reflect the status of their cell of origin. Indeed, EVs are regarded as “snapshots” of the status of their cell of origin and are examined to assess the presence of various diseases, e.g., cancer or viral infection8,9. As EVs are present in body fluids (e.g. blood1/saliva10/urine11,12), they are considered to be minimally-invasive biomarkers and so-called "liquid biopsies"2,13–16.
The quantification and characterization of EVs is hampered by their physical characteristics such as their small size, low epitope copy number17, and the variety of protein markers depending on the cell source18,19, all of which contribute to the well-documented EV heterogeneity. The identification of EVs in plasma is further hindered by the molecular complexity of the plasma, which contains multiple elements (e.g., lipoproteins, cell debris and soluble proteins), that interfere with EV analysis18,20. Moreover, a lack of robust methods and ambiguities in how data should be interpreted for EV analysis, makes comparison between studies challenging21,22.
To overcome the selection biases or introduction of artefacts that are introduced by performing commonly used EV isolation methods18,20, our group recently developed a standardized Imaging Flow Cytometry (IFCM)-based methodology for the direct measurement of single EVs ≤ 400 nm in diameter in diluted plasma samples, without prior isolation of EVs23. By omitting the need for sample isolation, this method has the potential to directly show the status of an individual by measuring distinct EV subsets, which is greatly beneficial for the monitoring of EVs in health and disease21,22.
Expanding upon the previously reported ability of our methodology to directly detect and discriminate human- and mouse-derived EVs in mixed human/mouse plasma samples23, we here aimed to assess the ability of our protocol to identify, discriminate and analyze EV subsets within human patient plasma samples. To this end, the setting of clinical organ transplantation offers a unique scenario in which tissue-specific EVs originating from the allograft are released into circulation after transplantation14,24,25. We present an application of our methodology that allows for the direct detection of donor-derived EVs (dd-EVs) in plasma samples from kidney transplant recipients (KTRs) on the basis of donor-recipient Human Leukocyte Antigen (HLA) mismatching by analyzing samples obtained before (control, no donor EVs present) and after (signal validation, detection of dd-EVs originating from the allograft) transplantation. Dd-EVs were measured against a background of recipient EVs in longitudinally collected KTR samples, and the dynamics of dd-EVs was monitored over time, as well as at time of ‘for-cause’ biopsy samples.
Results
Direct detection of single EVs ≤ 400 nm in donor and KTR plasma samples
Compared to donor samples, the plasma composition of KTRs contains multiple elements (e.g., elevated creatinine and urea, (medicinal) waste products) which may interfere with single EV detection. Therefore, we first tested the ability of our protocol to directly detect single EVs ≤ 400 nm in diameter in plasma samples obtained from KTRs before transplantation. To this end, PPP samples from living donors (used as healthy controls) and KTRs were diluted in fPBS, and stained with either anti-CD9 (as common plasma EV marker) or isotype control (to infer labelling specificity) before analysis with IFCM. After initial acquisition, detergent treatment was applied on each anti-CD9 labeled sample to discriminate between vesicular and non-vesicular events.
Total concentrations of CD9 + objects/mL (Fig. 1a) as measured in donor and KTR plasma samples were compared (Fig. 1b). In the donor group, we detected 1.26E8 ± 6E7 objects/mL before, and 2.74E6 ± 6.7E6 objects/mL after detergent treatment. This represented an approximate 98% reduction—which implies that the majority of CD9 + events analyzed represent EVs. In the plasma samples obtained from KTRs, we detected 1.07E8 ± 5.3E7 objects/mL before, and 1.6E6 ± 5.1E6 objects/mL after detergent treatment (~ 97% reduction). No significant differences in total CD9 + EV concentrations were observed between both groups. Additionally, isotype staining resulted in 4.16E5 ± 3.85E5 objects/mL and 5.27E5 ± 1.22E5 objects/mL for donor and KTR samples, respectively, illustrating the high specificity of our labelling strategy (~ 300-fold difference with mAb labelling). Buffer only controls (measured each day of sample acquisition) showed no detectable events – indicating that no signals derived from antibody aggregates were detected.
Figure 1.
Detection of CD9 + EVs in both (living) donor and kidney transplant recipient (KTR) samples obtained before transplantation. (a) left to right: representative histogram plots of a PPP sample stained with anti-CD9, the same sample after detergent treatment (to discriminate between vesicular and non-vesicular events), and isotype staining. (b) For each of the controls applied: quantification of total CD9 + EV concentrations as detected in PPP samples obtained from (living) donors, KTRs, nd buffer only controls.
These data suggest that our IFCM protocol is able to directly detect single CD9 + EVs in diluted plasma samples obtained from donors and KTRs before transplantation, despite the differences in molecular composition of the plasma between both groups.
Identification and discrimination of single EVs on the basis of HLA phenotype
In order to identify dd-EVs in the circulation of KTRs after kidney transplantation, we assessed whether IFCM is capable of discriminating EVs based on HLA phenotype. To this end, we labelled the same PPP samples (donors—HLA-A3 + , and recipients (before KTx)—HLA-A3 − ) with anti-CD9 and anti-HLA-A3, and performed the same control experiments as described above.
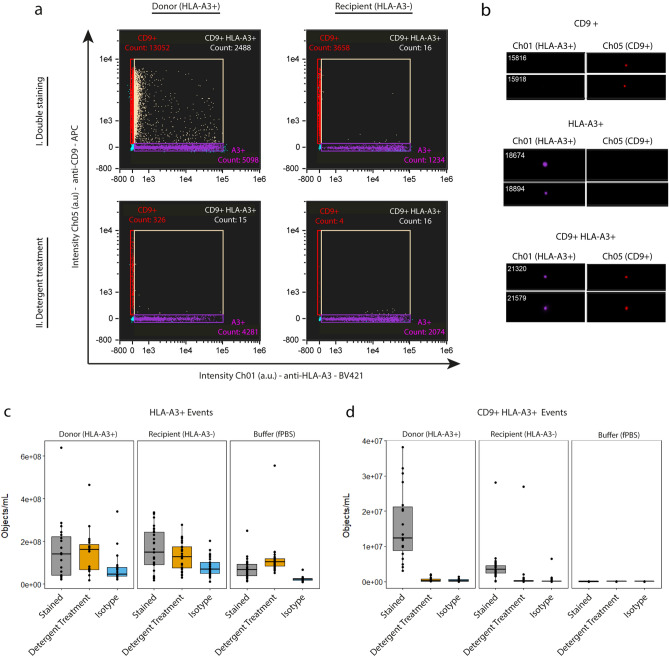
Figure 2a shows representative scatter plots for both a donor and recipient PPP sample analyzed with IFCM, before and after detergent treatment. Visual examination of events representative for each fluorescent population (before detergent treatment) demonstrated the selection and analysis of particles showing single spot fluorescence (no coincidence events), indicating the selection and analysis of single EVs23 (Fig. 2b).
Figure 2.
Discrimination of EVs based on HLA phenotype. (a) Representative scatter plots of an HLA-A3 + PPP sample (left) and an HLA-A3- PPP sample (right) stained with anti-HLA-A3 (X-axis) and anti-CD9-APC (Y-axis) before and after detergent treatment (top and bottom row, respectively). (b) Visual examination of events representative for each fluorescent population demonstrated the selection and subsequent analysis of particles showing single spot fluorescence (no coincidence events), indicating the selection and analysis of single EVs. (c,d) Concentrations of HLA-A3 + and CD9 + HLA-A3 + fluorescent events in all pre-transplantation donor, recipient, and buffer only (control) samples before and after detergent treatment, and after isotype staining (grey, orange, and blue boxes, respectively). HLA-A3 + events were found to not be indicative of EV signals and were excluded from analysis. Double-positive events were found to be indicative of EV signals, as well as to be discriminative between HLA-A3 + and HLA-A3- PPP samples.
For HLA-A3 + single-positive events, we observed only marginal reduction by detergent treatment, showing that these events are not representative of biological particles. Moreover, buffer only controls showed a high degree of fluorescent events, indicating that these signals are most likely representative of antibody aggregates (Fig. 2c). Therefore, HLA-A3 single-positive fluorescent events were interpreted to not be representative of EVs, and were excluded from further analysis.
Analysis of CD9 + HLA-A3 + double-positive fluorescent event concentrations in the KTR group showed that our assay produced background signals that varied among the different recipients (recipient-specific background) despite recipients all being HLA-A3 negative. Luminex single antigen assay for the anti-HLA-A3 antibody showed no cross reactivity of the antibody with other HLA epitopes (Supplementary Data S1 online), thus validating the specificity of the antibody. Comparison of CD9 + HLA-A3 + concentrations between the donor and recipient groups (1.60E7 ± 1.03E7 objects/mL vs 4.29E6 ± 4.48E6 objects/mL, respectively) showed that these double-positive fluorescent events can be used to discriminate samples on the basis of HLA phenotype. Concentrations of CD9 + HLA-A3 + double-positive events as measured in donor PPP samples showed an approximately 96% reduction after detergent treatment, with concentrations after detergent treatment residing in the range of the isotype and buffer controls (Fig. 2d). Consequently, CD9 + HLA-A3 + double-positive fluorescent events were identified to represent EVs.
It should be noted that some samples showed low/minimal reduction in concentrations of fluorescent events detected after detergent treatment (as illustrated by 1 recipient sample in Fig. 2d). These events were interpreted to not represent membrane-delimited vesicular events, but rather be indicative for protein complexes (which are unaffected by detergent treatment). To identify and exclude such samples from future analysis, we established a threshold of ≥ 95% reduction (in concentration) after detergent treatment for samples to be included in future analysis. This cut-off value was established to ensure that only samples are included that are representative (≥ 95%) of membrane-delimited particles (as opposed to protein complexes).
These observations show that our IFCM protocol is able to identify and discriminate single EVs in diluted PPP samples on the basis of their HLA phenotype by analyzing events that are both CD9 + (plasma EV marker) and HLA-A3 + (to discriminate between HLA-A3 + and HLA-A3 − individuals).
IFCM discriminates single CD9 + HLA-A3 + EVs down to 1% above recipient-specific background
To determine the discriminative power of our IFCM protocol to detect CD9 + HLA-A3 + EVs above (recipient-self) background signals in KTR samples, we serially diluted (twofold dilutions) HLA-A3 + PPP into HLA-A3 − PPP (5 matched donor-recipient couples), labelled these samples with anti-CD9 and anti-HLA-A3, and analyzed these with IFCM.
For each of the 5 donor-recipient couples, we quantified the concentrations of CD9 + HLA-A3 + EVs as detected for each dilution step (Fig. 3a). Similar to the previous observation, we observed that each of the 5 HLA-A3 − PPP recipient samples (without any spiked-in HLA-A3 + PPP) yielded varying background concentrations. These were interpreted to represent the recipient-self sample-specific background (indicated with dashed green lines, Fig. 3a). We were able to detect CD9 + HLA-A3 + EVs above their respective sample-specific background concentrations down to ~ 100-fold dilution, thus with CD9 + HLA-A3 + EVs being detectable ~ 1% above the recipient ‘self’ signal.
Figure 3.
Quantification of CD9 + HLA-A3 + events in mixed PPP samples. (a) Serial dilutions (twofold) of 5 double-stained (anti-CD9 and anti-HLA-A3) matched donor-recipient samples shows that our setup is able to discriminate CD9 + HLA-A3 + EVs above recipient ‘self’ background (HLA-A3- recipient signals, indicated by the green dashed lines) down to ~ 1%. (b) Coincidence testing through serial dilution. A linear correlation between the concentrations of CD9 + HLA-A3 + EVs and dilution factor (range: 100% to 1.56% HLA-A3 + PPP) for each serial dilution experiment confirmed single particle analysis in mixed PPP samples. (c) Stable (standardized) fluorescent signals for both detection channels (Ch01 – HLA-A3 / Ch05 – CD9) were observed irrespective of the dilution factor.
Next, we examined the accuracy of our IFCM protocol to detect single CD9 + HLA-A3 + EVs in mixed plasma samples – as opposed to the coincidence detection of multiple particles recorded as a single event. For each individual (donor-in-recipient) dilution experiment, the concentrations of CD9 + HLA-A3 + EVs as detected for each dilution step (range: 100–1.56% HLA-A3 + PPP) were analyzed using a linear regression model (Fig. 3b). We observed that the concentrations of CD9 + HLA-A3 + EVs were linearly proportional to the dilution factor, as indicated by the R2 values. Additionally, analysis of the (standardized) fluorescent intensities as detected in both detection channels revealed that the fluorescent intensities were unaffected by serial dilution; mean: 1315 (range 1085–1604) ERF for BV421 (HLA-A3), and 29.5 (range 25.7–35.8) ERF for APC (CD9) (Fig. 3c).
Taken together, these data indicate that IFCM is able to detect single dd-EVs directly in recipients’ circulation if their fraction exceeds ~ 1% above their recipient specific background.
dd-EVs are detected in KTRs with stable allograft function
Next, we examined whether dd-EVs could be directly detected in PPP of KTRs after transplantation. As stated previously, samples not passing the threshold of ≥ 95% reduction after detergent treatment were excluded from analysis; excluded samples were not associated with any specific time-point. Sample exclusion on the basis of detergent treatment resulted in the exclusion of 10 KTR sample series, leaving 26 KTR sample series in the analysis. Table 1 shows the patient characteristics corresponding to the samples included in our analysis, stratified into the four groups as described in the methods section.
Table 1.
Patient characteristics corresponding to the samples included in our analysis after exclusion of samples not passing the 95% reduction after detergent treatment.
| Control | Other diagnosis | Presumed rejection | Rejection | p-test | |
|---|---|---|---|---|---|
| n = | 11 | 3 | 5 | 7 | |
| Recipient age (Years) | 58.64 (10.06) | 62.33 (10.21) | 61.20 (14.55) | 58.29 (16.43) | 0.95 |
| Recipient gender (Male) | 9 (81.8) | 3 (100.0) | 1 (20.0) | 3 (42.9) | 0.04 |
| Recipient BMI | 27.45 (5.13) | 28.03 (11.94) | 26.62 (2.27) | 27.44 (4.92) | 0.99 |
| Donor type | 0.60 | ||||
| DBD | 3 (27.3) | 1 (33.3) | 0 (0.0) | 3 (42.9) | |
| DCD | 3 (27.3) | 2 (66.7) | 1 (20.0) | 2 (28.6) | |
| Living donor-related | 1 (9.1) | 0 (0.0) | 1 (20.0) | 0 (0.0) | |
| Living donor-unrelated | 4 (36.4) | 0 (0.0) | 3 (60.0) | 2 (28.6) | |
| Donor age (Years) | 54.09 (17.68) | 66.00 (2.65) | 61.20 (13.55) | 64.00 (6.90) | 0.38 |
| Donor gender (Male) | 4 (36.4) | 1 (33.3) | 4 (80.0) | 3 (42.9) | 0.40 |
| Missmatch HLA-A | 0.217 | ||||
| 0 | 0 (0.0) | 0 (0.0) | 1 (20.0) | 0 (0.0) | |
| 1 | 9 (81.8) | 2 (66.7) | 1 (20.0) | 4 (57.1) | |
| 2 | 2 (18.2) | 1 (33.3) | 3 (60.0) | 3 (42.9) | |
| Missmatch HLA-B | 0.56 | ||||
| 0 | 2 (18.2) | 0 (0.0) | 0 (0.0) | 0 (0.0) | |
| 1 | 4 (36.4) | 1 (33.3) | 1 (20.0) | 4 (57.1) | |
| 2 | 5 (45.5) | 2 (66.7) | 4 (80.0) | 3 (42.9) | |
| Missmatch HLA-DR | 0.02 | ||||
| 0 | 4 (36.4) | 0 (0.0) | 0 (0.0) | 3 (42.9) | |
| 1 | 5 (45.5) | 0 (0.0) | 5 (100.0) | 2 (28.6) | |
| 2 | 2 (18.2) | 3 (100.0) | 0 (0.0) | 2 (28.6) | |
| Induction therapy | |||||
| Basiliximab | 11 (100.0) | 3 (100.0) | 5 (100.0) | 5 (71.4) | 0.12 |
| Alemtuzumab | 0 (0.0) | 0 (0.0) | 0 (0.0) | 2 (28.6) | 0.12 |
| Maintenance immunosuppression | |||||
| TAC / MMF / Prednisolone | 11 (100.0) | 3 (100.0) | 5 (100.0) | 7 (100.0) | NA |
For the dd-EV analysis, samples from a total of 26 KTRs were included in the analysis (Fig. 4B). Continued variables are described as mean (SD). Categorical variables as number of cases (%).
ATG Antithymocyte globulin, BMI Body mass index, DBD Donation after brain death, DCD Donation after circulatory death, MMF Mycophenolate mofetil, MPA Mycophenolic Acid, TAC Tacrolimus.
First, we compared the total concentrations of double-positive events measured in PPP samples taken before (n = 13) and 2–4 days (n = 17) after transplantation (Fig. 4a). We observed a statistically significant difference between these time points which confirms the release of dd-EVs into KTR's circulation.
Figure 4.
Detection of dd-EVs after transplantation. (a) Analysis of the concentrations of CD9 + HLA-A3 + events before and 2–4 days after kidney transplantation revealed a significant increase after transplantation. (b) longitudinal analysis of dd-EV concentrations in KTR blood samples subdivided into either (1) the control group (representing all samples from patients whom did not experience any complications after transplantation) or (2) the biopted group (encompassing all samples from patients whom underwent a biopsy), showing that dd-EVs are detectable above baseline (before transplantation) only in patients whom did not experience complications after transplantation. (c) Representative CD9 vs HLA-A3 scatter plots of PPP samples taken 6 months after KTx from an individual in the control (left) and biopted (right) group. (d) dd-EV concentrations as detected in ‘for-cause’ biopsy samples compared to time-matched control samples.
We then performed a longitudinal analysis of dd-EV concentrations comparing KTRs in the control group with KTRs who underwent a ‘for-cause’ kidney biopsy (‘Biopted’ group) (Fig. 4b). In the control group, we observed significant increases in dd-EV concentrations at 2–4 days, 6–8 days and 6 months after transplantation compared to the concentrations detected before transplantation. These dd-EV concentrations were observed to be stable throughout follow-up, suggesting that stable allograft function (without a biopsy) leads to a detectable dd-EV signal in KTR plasma. This notion was further strengthened by the observation that dd-EV concentrations did not increase (compared to levels detected before transplantation) in individuals who did experience allograft complications (‘Biopted’ group); this effect was observed up to at least 6 months after transplantation (Fig. 4c).
To examine a potential diagnostic value of dd-EVs, we next compared the concentrations of CD9 + HLA-A3 + EVs measured in ‘for-cause’ biopsy samples taken at 6 days after transplantation (median, range: 2–60) with the concentrations as detected in samples taken 6–8 days after transplantation from patients in the ‘control’ group.
The concentration of dd-EVs was higher in time-matched KTR samples without a biopsy compared to concentrations detected at the moment of a for-cause biopsy (irrespective of their pathological classification, 5.36E6 ± 2.05E6 objects/mL vs 3.24E6 ± 8.55E5 objects/mL, respectively, p = 0.05). However, no statistical differences were observed when comparing dd-EV concentrations between the control samples and biopsies indicative for ATN (N = 2), Presumed Rejection (N = 3), or Biopsy Proven Acute Rejection (BPAR—3 × aABMR and 3 × aTCMR2a) , demonstrating that dd-EVs were unable to discriminate between the type of complication (Fig. 4d).
Discussion
EVs have great potential value as (minimally invasive) biomarkers26, but sensitive and reproducible methods for single EV analysis are essential to understand the role of EVs in human health and disease22. In the current work, we assessed the applicability of our recently developed IFCM-based methodology23 to directly measure EV subsets in human patient plasma samples – as exemplified by the detection of single dd-EVs in KTR plasma samples after kidney transplantation.
In addition to their physical properties and current technological challenges, the detection of EV subsets during health and disease may – depending on the disease – be hampered by the presence of ‘contaminating’ agents in the sample matrix. In the current study, components such as urea and creatinine are elevated as a consequence of kidney failure and may interfere with single EV detection. We show that our IFCM protocol is able to detect single CD9 + EVs in plasma samples obtained from both donors (healthy controls) and KTRs before transplantation, thereby suggesting that single EV detection by our IFCM protocol is uninfluenced by such contaminating components. The concentration of EVs in urine is related to nephron mass, explaining a lower concentration of total CD9 + EVs in KTRs compared to donors27. Here, we did not observe a difference in total CD9 + EVs between both groups in blood plasma, which is most likely due to the contribution of multiple organs to the total plasma CD9 + EV pool.
The concept of plasma circulating dd-EV detection and characterization with IFCM has been presented previously4. Mastoridis et al. showed that by analyzing CFSE + events in combination with an exosome-specific marker (CD63) and an origin-specific marker (donor-HLA), circulating dd-EVs could be detected in the circulation of a liver transplantation recipient4. However, our protocol provides several improvements over the approach presented in previous work: (1) our IFCM platform is both SSC (size) and fluorescence calibrated—which enhances reproducibility, (2) no EV isolation was performed—thus the ‘full spectrum’ of detectable EVs was analyzed, and (3) by analyzing CD9 + EVs (shown to be highly prevalent in plasma samples)4,23 in combination with donor-HLA, we were able to analyze a broad spectrum of circulating dd-EVs – as opposed to the analysis of donor-derived CFSE + CD63 + exosomes (a donor EV subset). Additionally, we prove the detection of single dd-EVs by our IFCM protocol and determined its sensitivity through serial dilution of HLA-A3 + plasma into HLA-A3 − plasma demonstrating a linear correlation with the dilution factor and stable ERF22.
Currently, it is consensus that CD9 + EVs detected in PPP are most likely platelet-derived. As platelets express HLA Class I antigens on their surface, one might expect a high degree of co-localization of HLA Class I antigens with CD9 on these EVs. However, we found that CD9 + HLA-A3 + particles are only a fraction (~ 16%) of the total CD9 + particles in HLA-A3 + donors. This observation may be explained by (1) different modes of EV biogenesis influencing whether HLA Class I antigens are present on EVs and/or whether their epitope topology is directed to the vesicular surface (and thus detectable with our setup), and (2) CD9 (but not HLA Class I antigens) may also be found on lipoproteins18.
The identification and quantification of single dd-EVs with IFCM as presented here is also subject to limitations. First, IFCM needs a minimum of 3 pixels before an event is recorded23. Consequently, EVs with a low HLA-A3 epitope-density might be missed by our assay. Second, although we calibrated the arbitrary fluorescent SSC intensities of our platform to reflect particle-size, we obtained a goodness-of-fit measure (R2) of 91%, which implies a 9% error when selecting particles with SSC intensities corresponding to EVs ≤ 400 nm23. For reference, (conventional) clinical-grade Flow Cytometers typically result in R2 values > 0.99—although lacking the resolution to detect EVs < 300 nm18,28,29. Additionally, we excluded samples not passing the threshold of ≥ 95% reduction after detergent treatment from analysis, which imposes an analytical bias towards samples containing ‘high’ concentrations of fluorescent events. However, by applying this selection criteria on our samples we ensured the analysis of fluorescent events of biological origin well above the (fluorescent) background of our assay. An alternative to the detergent treatment threshold could be the selective analysis of samples with a minimum number of events in the gate of interest (exceeding the number of events acquired in negative control samples e.g., buffer + reagents, unstained plasma, plasma + isotype staining) after acquisition.
In transplantation, the potential of EVs as biomarker for the detection of allograft rejection has been reported by a few groups30–36. Most notably, animal models of heterotopic heart transplantation (mouse into mouse)24, islet xenotransplantation (human into mouse)14, and a lung transplantation model between rats (Wistar into Lewis)25, have provided evidence that concentrations of allograft-derived EVs diminish during rejection well before alterations in classical biomarkers occur or histologic manifestations of injury were observed. In the current study, we observed a stable release of dd-EVs in KTRs who did not experience allograft dysfunction after transplantation but were unable to detect dd-EVs above pre-transplantation signals in KTRs who underwent a ‘for-cause’ biopsy.
Mechanistically, it is currently unknown whether the lower concentrations of dd-EVs during allograft dysfunction is a consequence of decreased production of EVs by the allograft, increased consumption of dd-EVs by recipient immune cells, or a combination of both24. Donor exosomes have been shown to be involved in donor antigen presentation to recipient alloreactive T cells in lymphoid organs by the recipient dendritic cells in a phenomenon known as cross-dressing35,37–39. As Habertheuer et al. suggested, this mechanism of T cell activation may suppress production of exosomes by the transplanted tissue even before there is targeted injury to the allograft24. At any rate, the detection of dd-EVs can be associated with stable allograft function – which is in contrast to the detection of e.g. donor-derived cell-free DNA, which has been observed to increase in concentration as a consequence of allograft damage40.
In conclusion, our calibrated IFCM-based methodology to directly detect and characterize plasma-derived EV subsets is applicable in patient human plasma samples. We believe that this methodology—after validation of markers of interest—could boost the EV biomarker research in a variety of clinical contexts, of which monitoring of kidney transplant integrity appears especially promising.
Materials and methods
Clinical sample selection
To analyze distinct donor-derived EVs (dd-EVs) in KTR plasma samples, 36 donor-KTR couples were selected on the basis of an HLA-A3 mismatch between donors (HLA-A3 +) and KTRs (HLA-A3 − ). KTRs had not received a previous HLA-A3 + graft. All donor-KTR couples participated in an observational study which aimed to identify minimally invasive biomarkers for the diagnosis of acute kidney transplant rejection, and was approved by the institutional review board of the Erasmus MC (Medical Ethical Review Board number 2018-035); details of this study are described elsewhere40. All patients provided written informed consent. Blood samples from living donors were obtained before donation and collected as part of our ongoing Biobank program (Medical Ethical Review Board number 2010-022). These donors served as healthy controls. The study was conducted in accordance with the principles of the Declaration of Helsinki.
Sample collection and processing
From both donors and KTRs, whole blood samples (EDTA) were collected before transplantation, and, for KTRs, 3 days, 7 days, and 6 months after transplantation. Additionally, blood samples were collected on the morning of (or the day preceding) a ‘for-cause’ kidney transplant biopsy.
Blood was drawn from each individual into two BD Vacutainer K3-EDTA-coated collection tubes (BD Biosciences, San Jose, USA). Whole blood was centrifuged at 1910 × g for 10 min at room temperature and the plasma layer was then collected at 16,000 × g for 10 min at room temperature in 1 mL aliquots using Safe-Lock Eppendorf tubes (Eppendorf AG, Hamburg, Germany). The resulting platelet-poor plasma (PPP) was first pooled before being divided into 700-µL aliquots in cryovials containing 28 µL of a 25 × concentrated protease inhibitor cocktail solution (4% v/v) (cOmplete Protease inhibitor cocktail tablets, Roche, Mannheim, Germany) according to the manufacturers’ instructions and stored at − 80 °C.
Stratification of KTRs based on biopsy scores
Biopsies were scored by an experienced renal pathologist, and KTRs were divided into 4 groups based on (1) no ‘for-cause’ biopsy and no anti-rejection therapy (‘Control’ group), (2) acute tubular necrosis (ATN) as the main finding in the biopsy and no anti-rejection therapy (‘ATN’ group), (3) no histopathological signs of rejection but treated with anti-rejection therapy on the basis of clinical suspicion of rejection (‘Presumed Rejection’ group), and (4) biopsy-proven acute rejection (BPAR—aABMR or aTCMR2A) in combination with anti-rejection therapy (‘Rejection’ group). In case patients underwent multiple biopsies over the course of the study, only the first biopsy was used in the analysis.
Sample labelling
PPP samples were stained with monoclonal antibodies (mAbs) and isotype controls as extensively described in our previous work23. In the absence of a specific marker, EVs are identified by their expression of common markers such as CD9, CD63 and CD8141. In this study, we used CD9 (which has been shown to be highly prevalent on plasma-derived EVs) as common EV marker23. Additionally, following the characterization of kidney-derived EVs released during normothermic machine perfusion, we determined that CD9 co-localizes to a higher degree with MHC Class I molecules compared to CD63 (~ threefold difference) (Supplementary Figure S1online). mAbs used in this study were CD9–APC, clone HI9a (6 µg/mL, Biolegend, San Diego, USA), and HLA-A3-BV421, clone GAP-A3 (200 µg/mL, BD Biosciences, New York, USA). Matched isotype controls were IgG1, k-APC, clone MOPC-21 (200 µg/mL, BioLegend), and IgG2a, k-BV421, clone G155-178 (200 µg/mL, BD Biosciences). Specificity of the anti-HLA-A3 mAbs was confirmed by Luminex single antigen assay (Supplementary Data S1 online). Prior to staining, mAbs and isotypes were centrifuged at 16,000 × g for 10 min at room temperature to remove potential mAb clumps (to reduce false-positive signals from analysis), and were diluted in 0.20 µm filtered PBS (fPBS) before staining (final concentrations: 200 ng/mL).
Staining was performed by addition of the diluted mAbs/isotypes to 30 µL of PPP followed by a pre-determined volume of fPBS (Vtot = fPBS + sample + mAbs = 130 µL) followed by O/N incubation at 4 °C. All samples were brought to a total volume of 380 µL using fPBS before IFCM measurements.
Controls
To ascertain EV measurements the following controls were applied, as recommended by the MIFlowCyt-EV framework22: buffer only, buffer with reagents, unstained samples, isotype controls, and detergent treatment, which aims to disrupt the membranous structure of EVs thereby allowing discrimination between membrane-enclosed vesicles (which lyse upon detergent treatment) and other protein complexes (which are unaffected by detergent treatment). Detergent treatment was performed by adding 20 µL of 10% (V/V) TritonX-100 to the samples followed by 30 min of incubation at room temperature prior to acquisition.
Data acquisition
All samples were acquired on an ImageStreamX MkII instrument (ISx; Luminex). Settings as extensively described elsewhere were used23. In brief, lasers were turned on as applicable per fluorophore and set to their maximum power (405 nm : 200 mW, 642 nm :150 mW) with the exception of the 785 nm SSC laser (1.25 mW). High Gain mode—an upgrade of the IFCM that increases the photonic sensitivity and object detection of the system – was activated. Data was acquired over fixed time periods – to standardize among samples – of 180 s using the 60 × objective with fluidics set to ‘low speed / high sensitivity’. This resulted in a flow speed of 43.59 ± 0.07 mm/sec (mean ± standard deviation). Core size was set at 6 µm, autofocus was activated and the ‘Remove Speedbead’ option was checked. BV421 fluorescence signals were collected in channel 1 (435–505-nm filter), APC signals in channel 5 (642–745 nm filter), and SSC signals in channel 6 (745–785 nm filter). Particle enumeration was achieved through the advanced fluidic control of the ISx coupled with continuously running speed beads, resulting in the “objects/mL” feature within the ISx Data Exploration and Analysis Software (IDEAS).
Data analysis
Data analysis was performed using Amnis IDEAS software (version 6.2). Image display mapping was linearly adjusted for all fluorescent events for each channel and then applied to all files of the respective experiment. To ensure the analysis of EVs we (1) selected all particles with SSC intensities ≤ 900 a.u., and (2) identified and excluded coincidence detection by counting the number of fluorescent spots within the pixel grid for each event acquired; events showing multiple spots were excluded from analysis23. This gating strategy ensures the selection and analysis of single spot fluorescent particles ≤ 400 nm. Gating areas and cut-offs were established through identification of (fluorescent) populations in unstained and single stained samples, and arbitrary fluorescent intensities were converted into Equivalent number of Reference Fluorophores (ERF) values based on previously published calibration data23. Lower and upper gating area cut-offs were defined as 677 – 112,201 ERF for BV421, and 6.40 – 123 ERF for APC.
Statistical analysis
Statistical analysis was performed using R version 4.0.2 and RStudio (RStudio Team (2016). RStudio: Integrated Development for R. RStudio, Inc., Boston, MA URL http://www.rstudio.com/.) version 1.1.463. All concentrations reported in this work were corrected for sample dilution (before acquisition − 380 µL total volume per test containing 30 µL sample = ~ 12.33-fold dilution factor) and are shown as the mean ± standard deviation unless specified otherwise. Statistical significance between EV concentrations and groups was determined through two-sided t-tests, 95% CI with unpaired data.
Supplementary Information
Acknowledgements
The authors would like to thank Manou van Alphen and Wenda Verschoor for their assistance in sample measurements with IFCM, Marian C. Clahsen – van Groningen for the pathological scoring of the biopsy samples, and the Immunohaematology and Blood Transfusion lab of the Leiden University Medical Center (LUMC) for performing the Luminex single antigen assay for the HLA-A3 mAb.
Abbreviations
- EVs
Extracellular vesicles
- dd-EVs
donor-derived extracellular vesicles
- IFCM
Imaging flow cytometry
- KTR
Kidney transplant recipient
- HLA
Human leukocyte antigen
- PPP
Platelet-poor plasma
- SSC
Side scatter
Author contributions
W.W.W. participated in the research design, performance of the research, data analysis, article drafting and is the corresponding author. D.A.H., C.C.B., M.J.H., K.B. participated in the research design and writing of the manuscript. All authors reviewed the manuscript and approved its final version.
Data availability
All source data underlying the figures presented in this work are provided as ‘Supplementary Data Figs. 1–4’. Any other relevant data are available from the corresponding author upon reasonable request.
Competing interests
In the last three years, D.A. Hesselink has received lecture fees and consulting fees from Astellas Pharma, Chiesi Pharma, Medincell, Novartis Pharma, Sangamo Therapeutics and Vifor Pharma. He has received grant support from Astellas Pharma and Chiesi Pharma [paid to his institution]. D.A. Hesselink does not have employment or stock ownership at any of these companies, and neither does he have patents or patent applications. All other authors declare to not have competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-26580-6.
References
- 1.Arraud N, Linares R, Tan S, Gounou C, Pasquet JM, Mornet S, et al. Extracellular vesicles from blood plasma: Determination of their morphology, size, phenotype and concentration. J. Thromb. Haemost. 2014;12(5):614–627. doi: 10.1111/jth.12554. [DOI] [PubMed] [Google Scholar]
- 2.Karpman D, Stahl AL, Arvidsson I. Extracellular vesicles in renal disease. Nat. Rev. Nephrol. 2017;13(9):545–562. doi: 10.1038/nrneph.2017.98. [DOI] [PubMed] [Google Scholar]
- 3.Marar C, Starich B, Wirtz D. Extracellular vesicles in immunomodulation and tumor progression. Nat. Immunol. 2021;22(5):560–570. doi: 10.1038/s41590-021-00899-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mastoridis S, Bertolino GM, Whitehouse G, Dazzi F, Sanchez-Fueyo A, Martinez-Llordella M. Multiparametric analysis of circulating exosomes and other small extracellular vesicles by advanced imaging flow cytometry. Front. Immunol. 2018;9:1583. doi: 10.3389/fimmu.2018.01583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pitt JM, Kroemer G, Zitvogel L. Extracellular vesicles: Masters of intercellular communication and potential clinical interventions. J. Clin. Invest. 2016;126(4):1139–1143. doi: 10.1172/JCI87316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ashcroft J, Leighton P, Elliott TR, Hosgood SA, Nicholson ML, Kosmoliaptsis V. Extracellular vesicles in kidney transplantation: A state-of-the-art review. Kidney Int. 2022;101(3):485–497. doi: 10.1016/j.kint.2021.10.038. [DOI] [PubMed] [Google Scholar]
- 7.Zaborowski MP, Balaj L, Breakefield XO, Lai CP. Extracellular vesicles: Composition, biological relevance, and methods of study. Bioscience. 2015;65(8):783–797. doi: 10.1093/biosci/biv084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Han L, Lam EW, Sun Y. Extracellular vesicles in the tumor microenvironment: Old stories, but new tales. Mol Cancer. 2019;18(1):59. doi: 10.1186/s12943-019-0980-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Urbanelli L, Buratta S, Tancini B, Sagini K, Delo F, Porcellati S, et al. The role of extracellular vesicles in viral infection and transmission. Vaccines. 2019;7(3):102. doi: 10.3390/vaccines7030102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Han Y, Jia L, Zheng Y, Li W. Salivary exosomes: Emerging roles in systemic disease. Int. J. Biol. Sci. 2018;14(6):633–643. doi: 10.7150/ijbs.25018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salih M, Zietse R, Hoorn EJ. Urinary extracellular vesicles and the kidney: Biomarkers and beyond. Am. J. Physiol. Renal Physiol. 2014;306(11):F1251–F1259. doi: 10.1152/ajprenal.00128.2014. [DOI] [PubMed] [Google Scholar]
- 12.Wang S, Kojima K, Mobley JA, West AB. Proteomic analysis of urinary extracellular vesicles reveal biomarkers for neurologic disease. EBioMedicine. 2019;45:351–361. doi: 10.1016/j.ebiom.2019.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McBride JD, Rodriguez-Menocal L, Badiavas EV. Extracellular vesicles as biomarkers and therapeutics in dermatology: A focus on exosomes. J. Invest. Dermatol. 2017;137(8):1622–1629. doi: 10.1016/j.jid.2017.04.021. [DOI] [PubMed] [Google Scholar]
- 14.Vallabhajosyula P, Korutla L, Habertheuer A, Yu M, Rostami S, Yuan CX, et al. Tissue-specific exosome biomarkers for noninvasively monitoring immunologic rejection of transplanted tissue. J. Clin. Invest. 2017;127(4):1375–1391. doi: 10.1172/JCI87993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Verhoeven JGHP, Boer K, Van Schaik RHN, Manintveld OC, Huibers MMH, Baan CC, et al. Liquid biopsies to monitor solid organ transplant function: A review of new biomarkers. Ther. Drug Monit. 2018;40(5):515–525. doi: 10.1097/FTD.0000000000000549. [DOI] [PubMed] [Google Scholar]
- 16.Wu L, Boer K, Woud WW, Udomkarnjananun S, Hesselink DA, Baan CC. Urinary extracellular vesicles are a novel tool to monitor allograft function in kidney transplantation: A systematic review. Int. J. Mol. Sci. 2021;22(19):10499. doi: 10.3390/ijms221910499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Welsh JA, van der Pol E, Bettin BA, Carter DRF, Hendrix A, Lenassi M, et al. Towards defining reference materials for measuring extracellular vesicle refractive index, epitope abundance, size and concentration. J. Extracell. Vesicles. 2020;9(1):1816641. doi: 10.1080/20013078.2020.1816641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Simonsen JB. What are we looking at? Extracellular vesicles, lipoproteins, or both? Circ. Res. 2017;121(8):920–922. doi: 10.1161/CIRCRESAHA.117.311767. [DOI] [PubMed] [Google Scholar]
- 19.Blijdorp CJ, Tutakhel OAZ, Hartjes TA, van den Bosch TPP, van Heugten MH, Rigalli JP, et al. Comparing approaches to normalize, quantify, and characterize urinary extracellular vesicles. J. Am. Soc. Nephrol. 2021;32(5):1210. doi: 10.1681/ASN.2020081142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karimi N, Cvjetkovic A, Jang SC, Crescitelli R, Hosseinpour Feizi MA, Nieuwland R, et al. Detailed analysis of the plasma extracellular vesicle proteome after separation from lipoproteins. Cell Mol. Life Sci. 2018;75(15):2873–2886. doi: 10.1007/s00018-018-2773-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Konoshenko MY, Lekchnov EA, Vlassov AV, Laktionov PP. Isolation of extracellular vesicles: General methodologies and latest trends. Biomed. Res. Int. 2018;2018:8545347. doi: 10.1155/2018/8545347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Welsh JA, Van Der Pol E, Arkesteijn GJA, Bremer M, Brisson A, Coumans F, et al. MIFlowCyt-EV: A framework for standardized reporting of extracellular vesicle flow cytometry experiments. J. Extracell. Vesicles. 2020;9(1):1713526. doi: 10.1080/20013078.2020.1713526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Woud WW, van der Pol E, Mul E, Hoogduijn MJ, Baan CC, Boer K, et al. An imaging flow cytometry-based methodology for the analysis of single extracellular vesicles in unprocessed human plasma. Commun. Biol. 2022;5(1):633. doi: 10.1038/s42003-022-03569-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Habertheuer A, Korutla L, Rostami S, Reddy S, Lal P, Naji A, et al. Donor tissue-specific exosome profiling enables noninvasive monitoring of acute rejection in mouse allogeneic heart transplantation. J. Thorac. Cardiovasc. Surg. 2018;155(6):2479–2489. doi: 10.1016/j.jtcvs.2017.12.125. [DOI] [PubMed] [Google Scholar]
- 25.Habertheuer A, Ram C, Schmierer M, Chatterjee S, Hu R, Freas A, et al. Circulating donor lung-specific exosome profiles enable noninvasive monitoring of acute rejection in a rodent orthotopic lung transplantation model. Transplantation. 2021;106:754. doi: 10.1097/TP.0000000000003820. [DOI] [PubMed] [Google Scholar]
- 26.Yanez-Mo M, Siljander PR, Andreu Z, Zavec AB, Borras FE, Buzas EI, et al. Biological properties of extracellular vesicles and their physiological functions. J. Extracell. Vesicles. 2015;4:27066. doi: 10.3402/jev.v4.27066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blijdorp CJ, Hartjes TA, Wei KY, van Heugten MH, Bovee DM, Budde RPJ, et al. Nephron mass determines the excretion rate of urinary extracellular vesicles. J. Extracell. Vesicles. 2022;11(1):e12181. doi: 10.1002/jev2.12181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Larson MC, Luthi MR, Hogg N, Hillery CA. Calcium-phosphate microprecipitates mimic microparticles when examined with flow cytometry. Cytometry A. 2013;83(2):242–250. doi: 10.1002/cyto.a.22222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gyorgy B, Modos K, Pallinger E, Paloczi K, Pasztoi M, Misjak P, et al. Detection and isolation of cell-derived microparticles are compromised by protein complexes resulting from shared biophysical parameters. Blood. 2011;117(4):e39–48. doi: 10.1182/blood-2010-09-307595. [DOI] [PubMed] [Google Scholar]
- 30.Zhang H, Huang E, Kahwaji J, Nast CC, Li P, Mirocha J, et al. Plasma exosomes from HLA-sensitized kidney transplant recipients contain mRNA transcripts which predict development of antibody-mediated rejection. Transplantation. 2017;101(10):2419–2428. doi: 10.1097/TP.0000000000001834. [DOI] [PubMed] [Google Scholar]
- 31.Morelli AE. Exosomes: From cell debris to potential biomarkers in transplantation. Transplantation. 2017;101(10):2275–2276. doi: 10.1097/TP.0000000000001856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tower CM, Reyes M, Nelson K, Leca N, Kieran N, Muczynski K, et al. Plasma C4d+ endothelial microvesicles increase in acute antibody-mediated rejection. Transplantation. 2017;101(9):2235–2243. doi: 10.1097/TP.0000000000001572. [DOI] [PubMed] [Google Scholar]
- 33.Kennel PJ, Saha A, Maldonado DA, Givens R, Brunjes DL, Castillero E, et al. Serum exosomal protein profiling for the non-invasive detection of cardiac allograft rejection. J. Heart Lung Transplant. 2018;37(3):409–417. doi: 10.1016/j.healun.2017.07.012. [DOI] [PubMed] [Google Scholar]
- 34.Gunasekaran M, Xu Z, Nayak DK, Sharma M, Hachem R, Walia R, et al. Donor-derived exosomes with lung self-antigens in human lung allograft rejection. Am. J. Transplant. 2017;17(2):474–484. doi: 10.1111/ajt.13915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Campana S, De Pasquale C, Carrega P, Ferlazzo G, Bonaccorsi I. Cross-dressing: An alternative mechanism for antigen presentation. Immunol. Lett. 2015;168(2):349–354. doi: 10.1016/j.imlet.2015.11.002. [DOI] [PubMed] [Google Scholar]
- 36.Golebiewska JE, Wardowska A, Pietrowska M, Wojakowska A, Debska-Slizien A. Small extracellular vesicles in transplant rejection. Cells. 2021;10(11):2989. doi: 10.3390/cells10112989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Burlingham WJ. "Cross-dressing" becomes fashionable among transplant recipients. Am. J. Transplant. 2017;17(1):5–6. doi: 10.1111/ajt.14032. [DOI] [PubMed] [Google Scholar]
- 38.Macedo C, Tran LM, Zahorchak AF, Dai H, Gu X, Ravichandran R, et al. Donor-derived regulatory dendritic cell infusion results in host cell cross-dressing and T cell subset changes in prospective living donor liver transplant recipients. Am. J. Transplant. 2021;21(7):2372–2386. doi: 10.1111/ajt.16393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mastoridis S, Londono MC, Kurt A, Kodela E, Crespo E, Mason J, et al. Impact of donor extracellular vesicle release on recipient cell "cross-dressing" following clinical liver and kidney transplantation. Am. J. Transplant. 2021;21(7):2387–2398. doi: 10.1111/ajt.16123. [DOI] [PubMed] [Google Scholar]
- 40.Verhoeven J, Boer K, Peeters AMA, Clahsen-van Groningen MC, Roodnat JI, van de Wetering J, et al. A novel high-throughput droplet digital pcr-based indel quantification method for the detection of circulating donor-derived cell-free dna after kidney transplantation. Transplantation. 2022;106:1777. doi: 10.1097/TP.0000000000004078. [DOI] [PubMed] [Google Scholar]
- 41.Andreu Z, Yanez-Mo M. Tetraspanins in extracellular vesicle formation and function. Front. Immunol. 2014;5:442. doi: 10.3389/fimmu.2014.00442. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All source data underlying the figures presented in this work are provided as ‘Supplementary Data Figs. 1–4’. Any other relevant data are available from the corresponding author upon reasonable request.