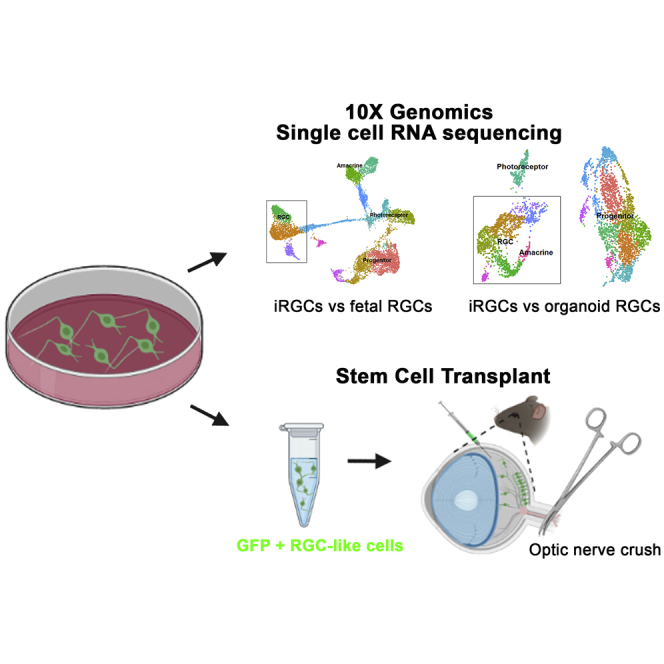
Summary
Retinal ganglion cell (RGC) replacement therapy could restore vision in glaucoma and other optic neuropathies. We developed a rapid protocol for directly induced RGC (iRGC) differentiation from human stem cells, leveraging overexpression of NGN2. Neuronal morphology and neurite growth were observed within 1 week of induction; characteristic RGC-specific gene expression confirmed identity. Calcium imaging demonstrated γ-aminobutyric acid (GABA)-induced excitation characteristic of immature RGCs. Single-cell RNA sequencing showed more similarities between iRGCs and early-stage fetal human RGCs than retinal organoid-derived RGCs. Intravitreally transplanted iRGCs survived and migrated into host retinas independent of prior optic nerve trauma, but iRGCs protected host RGCs from neurodegeneration. These data demonstrate rapid iRGC generation in vitro into an immature cell with high similarity to human fetal RGCs and capacity for retinal integration after transplantation and neuroprotective function after optic nerve injury. The simplicity of this system may benefit translational studies on human RGCs.
Keywords: induced retinal ganglion cell, stem cell, transplant, neuroprotective
Graphical abstract
Highlights
-
•
A rapid protocol to directly induce RGCs from stem cells
-
•
iRGCs were generated in about 1 week
-
•
iRGCs had relatively high similarity to human RGC in transcriptional profiles
-
•
iRGC survived and migrated for up to 1 month after transplantation
Luo et al. developed a rapid protocol to directly induce RGCs from stem cells, which only took about 1 week. Transcriptome analyses showed closer alignment between the iRGC approach and human fetal retinal development than between fetal and organoid RGCs. iRGCs survived and migrated for up to 1 month after transplantation and showed a strong neuroprotective effect on host RGCs.
Introduction
In glaucoma and optic neuropathies, retinal ganglion cell (RGC) loss is irreversible and leads to permanent visual impairment. Cell replacement therapy is a promising approach for treating advanced optic neuropathies. Primary RGC transplantation in animal models has been demonstrated successfully in prior studies (Hertz et al., 2014a; Venugopalan et al., 2016), but a supply of primary donor cells would be very limited. Thus, there is an unmet need to develop a stem cell-derived RGC therapy not only to provide protection to remaining neurons but also to restore vision loss.
Existing protocols generally include the following steps during stem cell-derived RGC differentiation. Stem cells are first directed to an anterior neural fate with Dickkopf-1 (Dkk-1; a Wnt inhibitor), Noggin (a bone morphogenetic protein inhibitor), and insulin growth factor 1 (IGF-1) (Lamba et al., 2006). Subsequently, the presence of basic fibroblast growth factor (bFGF) pushes the stem cells to an eye field fate, an essential step during retinogenesis (Furukawa et al., 1997; Mathers et al., 1997). Then progeny are differentiated into retinal progenitor cells (CHX10-/PAX6-positive) and finally into RGC-like cells (HUC/D-positive). In addition to supplying several signaling pathway regulators during neural induction, RGC fate can also be directed by overexpressing several essential transcription factors. For example, overexpressing ATOH7 allows generation of RGC-like cells from mouse and human induced pluripotent stem cells (iPSCs; including human iPSCs [hiPSCs] reprogrammed from human Tenon’s capsule fibroblasts) (Chen et al., 2010; Deng et al., 2016; Xie et al., 2014). Exogenous SOX4 expression can also greatly potentiate differentiation of RGCs from hiPSCs and shows a synergistic effect with ATOH7 on potentiating RGC differentiation (Chang et al., 2017).
However, these previous protocols usually take 2–4 weeks for RGC differentiation, and most such differentiation protocols have not explored electrophysiologic differentiation or suitability for transplantation and integration into the retina in vivo. Therefore, we aimed to establish a rapid differentiation method in the present study. Here we developed a rapid protocol for induced RGC (iRGC) differentiation from human embryonic stem cells (hESCs) and hiPSCs, driven by expression of Neurogenin-2 (NGN2), that requires less than 10 days. Physiological examination using calcium imaging in response to GABA stimulation was used to stage iRGC maturation compared with primary mouse RGCs. Single-cell transcriptional profiling confirmed iRGC fate compared with human fetal RGCs. Finally, we showed that iRGCs could survive in vivo on rodent retinas after intravitreal injection. These data support cell replacement therapy as a potential treatment for RGC loss in optic neuropathies.
Results
Rapidly inducing stem cell-derived RGCs by NGN2 overexpression
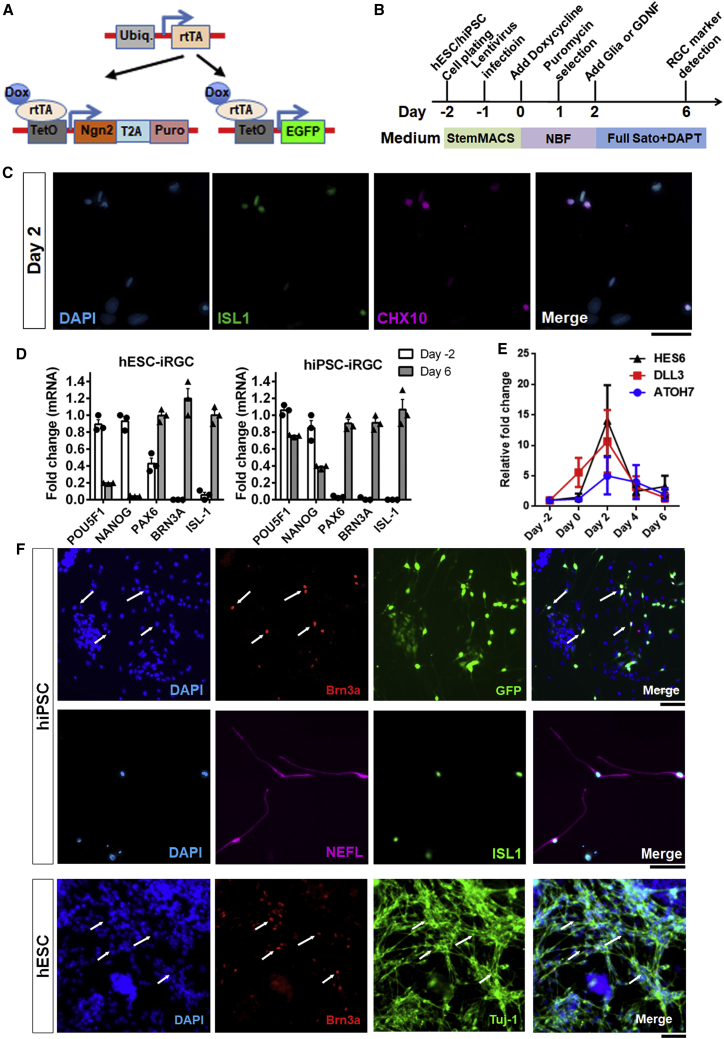
We extended the application of a previous rapid induced-neuron protocol that leverages NGN2 expression (Zhang et al., 2013) to accelerate RGC differentiation from human stem cells. We transduced stem cells with three lentiviruses carrying constitutive expression of rtTA for tetracycline-inducible expression of NGN2 and GFP (Figure 1A). To directly differentiate stem cells into RGC-like cells, we slightly modified the previously published culture approach by replacing glial cells with glial cell-derived neurotrophic factor (GDNF) on day 2 and changed the medium to a commonly used RGC SATO growth medium (Wu et al., 2018) plus the Notch inhibitor DAPT for another 4 days (Figure 1B). On day 2, after NGN2 overexpression and before switching to the RGC culture medium, some cells were already expressing CHX10, a retinal progenitor marker (Figure 1C). To investigate the status of cell differentiation, we utilized qRT-PCR to measure gene expression of the stem cell markers POU5F1 and NANOG, the neural progenitor cell marker PAX6, and the RGC markers BRN3A and ISL-1 on days 2 and 6 for hiPSCs and hESCs. We observed reductions of POU5F1 and NANOG and elevation of PAX6, BRN3A, and ISL-1 on day 6 compared with day −2, indicating loss of stem cell pluripotency and induction of neuronal fate, consistent with RGC marker expression (Figure 1D). Because previous studies have shown that NGN2 drove neuronal differentiation by activating ATOH7, DLL3, and HES6 (Hufnagel et al., 2010; Smith et al., 2016), we performed qRT-PCR through the first week of differentiation to evaluate the expression of these genes. The results demonstrated that, after turning on NGN2 by doxycycline, ATOH7, DLL3, and HES6 were transiently upregulated before returning to baseline expression (Figure 1E). By 1 week, 14% and 20% of cells were Brn3a positive in hiPSC-iRGC and hESC-iRGC cultures, respectively. These cells exhibited typical RGC morphology, extending neurites labeled by the axonal skeleton markers NEFL and Tuj1 (Figure 1F).
Figure 1.
Overexpression of NGN2 drives induced RGC (iRGC) differentiation
(A) Design of lentiviral vectors for NGN2-mediated differentiation of hESCs and hiPSCs to iRGCs (modified from Zhang et al., 2013). Cells were transduced with rtTA, which complexes with doxycycline to bind to the TetOn promoter, and either Tet promoter NGN2 or EGFP gene expression vectors.
(B) Flow diagram of iRGC cell generation (adapted from Zhang et al., 2013).
(C) After NGN2 expression was activated by doxycycline, human stem cells were directed to a retinal progenitor cell fate expressing ISL1 and CHX10. Scale bar, 200 μm.
(D) Expression of the pluripotent markers POU5F1 and NANOG decreased and the neural progenitor marker PAX6 increased in the first week of differentiation. The RGC markers Brn3a and ISL1 were significantly increased on day 6 (N = 3).
(E) Transient expression of ATOH7, DLL3, and HES6 after NGN2 overexpression (N = 3). Scale bar, 50 μm).
(F) hiPSCs and hESCs were differentiated into RGC-like cells and harvested on day 6. Brn3a was used as an RGC marker in both cultures, whereas NEFL and Tuj-1 illuminate the extending axons.
iRGCS have similar electrophysiological responses as immature primary RGCs
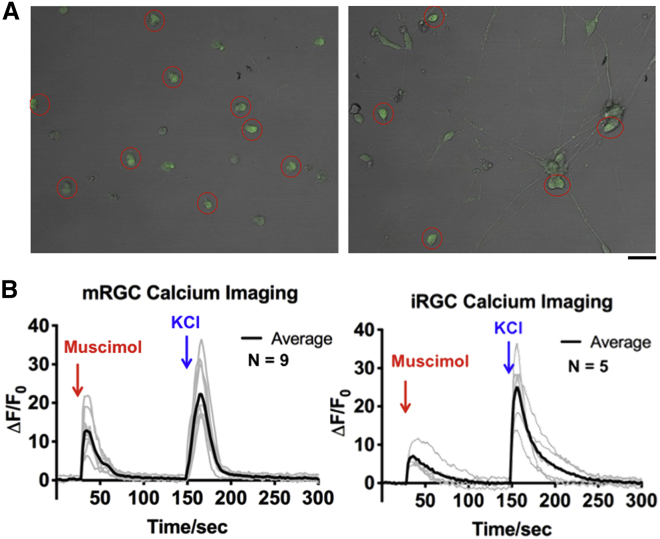
Previous studies have shown that GABAA receptor activation is excitatory in immature RGCs (Venugopalan et al., 2020). When stimulated by a selective GABAA agonist such as muscimol (Wang et al., 2007), immature RGCs depolarize and demonstrate rapid calcium entry, visualized by calcium imaging with Fluo 4-AM. We compared iRGCs after 6 days of differentiation with primary RGCs from post-natal day 2 (P2), which are developmentally immature. iRGCs and primary RGCs showed Ca2+ influx when exposed to muscimol (Figure 2), which resembles the effect of KCl-induced depolarization (Figure 2) and is in contrast to the lack of depolarization-induced calcium entry in mature RGCs (Venugopalan et al., 2020). These data show that rapidly induced RGCs predominantly exhibit immature electrophysiologic maturation, as measured by response to GABAA receptor activation.
Figure 2.
iRGCs demonstrate electrophysiological properties similar to immature primary mouse RGCs
(A) iRGCs were observed under a bright field to identify a region of interest (ROI; red circles).
(B) Calcium imaging of iRGCs shows calcium influx consistent with an excitatory GABA response to muscimol, similar to immature P2 mouse RGCs. Scale bar, 50 μm.
Single-cell transcriptional profiling demonstrates iRGC differentiation
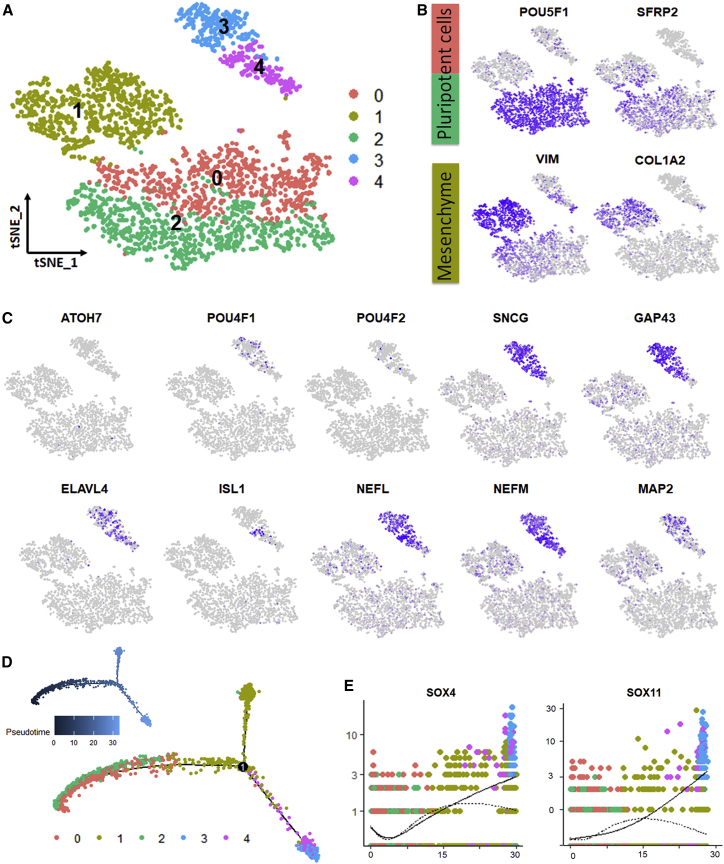
To gain insight into cell fate determination during iRGC differentiation, we performed single-cell RNA sequencing (scRNA-seq) analysis on differentiated cells collected on day 6. In total, 2,552 cells were included in the computational analysis, and the data were visualized by t-distributed stochastic neighbor embedding (tSNE) dimensional reduction (Figure 3A). We identified three different cell types generated in this differentiation, including pluripotent cells (POU5F1 and SFRP2 positive), mesenchymal cells (VIM, DES, and COL1A2 positive), and iRGCs (Figure 3B). The iRGCs expressed several RGC markers, including SNCG, NEFL, GAP43, ELAVL4 (HuD), and ISL1 (clusters 3 and 4) (Figure 3C). ATOH7, which is thought to be a gene signature of the transitioning cell population during retinal development (Sridhar et al., 2020), was barely detectable in all cell clusters. These results were consistent with our previous qRT-PCR data showing that ATOH7 expression went back to baseline after the first few days of differentiation. Similar to human fetal RGCs (Luo et al., 2019), BRN3A (POU4F1) was dominant in the iRGCs rather than BRN3B (POU4F2). iRGCs highly expressed axon skeleton markers, including NEFL, NEFM, and MAP2.
Figure 3.
Single-cell transcriptional profiling of differentiated cells
(A) tSNE plot for hiPSC-induced iRGCs shows 3 major groups statistically separated into 5 clusters.
(B) Expression of representative cell-type-specific genes. Purple indicates high expression, and gray indicates low expression. Cells in clusters 0 and 2 express POU5F1 and SFRP2, consistent with pluripotent cell identity. Cluster 1 cells express the extracellular matrix markers VIM and COL1A2, indicating mesenchymal identity.
(C) Expression of the RGC-specific markers SNCG, GAP43, ELAVL4, ISL1, POU4F1, and POU4F2 and the axon-associated genes NEFL, NEFM, and MAP2 is almost completely preferentially localized to clusters 3 and 4, except for ATOH7, which is not expressed significantly in any clusters, and RBPMS, which is expressed throughout all clusters.
(D) Monocle 2 reveals lineage bifurcation in the differentiated cells. The pluripotent cells (clusters 0 and 2 from A) were distributed at the beginning of the trajectory (pseudo-time 0), and then the cells went into two different cell fates: mesenchymal cells (cluster 1 from A) or RGCs (clusters 3 and 4 from A).
(E) Expression of the RGC specification transcription factors SOX4 and SOX11 over pseudo-time. Dashed lines and solid lines show expression in mesenchymal and iRGC lineages, respectively, and suggest SOX4 and SOX11 gene expression preferentially rise in iRGCs.
To delineate the cell fate determination process in the present rapid protocol, we used Monocle 2 to reorder the cells based on their stage-specific gene expression pattern and reconstruct their cell fate determination timeline. This algorithm arranged cells expressing mitotic cell cycle markers as the beginning of differentiation, labeled pseudo-time 0 (Figure 3D). Then developmental lineages were estimated by ranking all cell clusters in a temporal manner. The bifurcating trajectory led to two main lineages derived from the initiating stem cells, terminating in one lineage as iRGCs and in the other as mesenchymal cells (Figure 3D). Several genes involved in directing RGC fate determination were analyzed across lineages; for example, SOX4 and SOX11 have been shown to be key factors regulating RGC specification (Chang et al., 2017) and were significantly upregulated in cells in the iRGC lineage (Figure 3E). These results suggest that, although there was a cell mixture in the differentiation cultures, iRGCs were still effectively generated by overexpressing NGN2.
iRGCs show similarity to fetal RGCs more than retinal organoid-derived RGCs
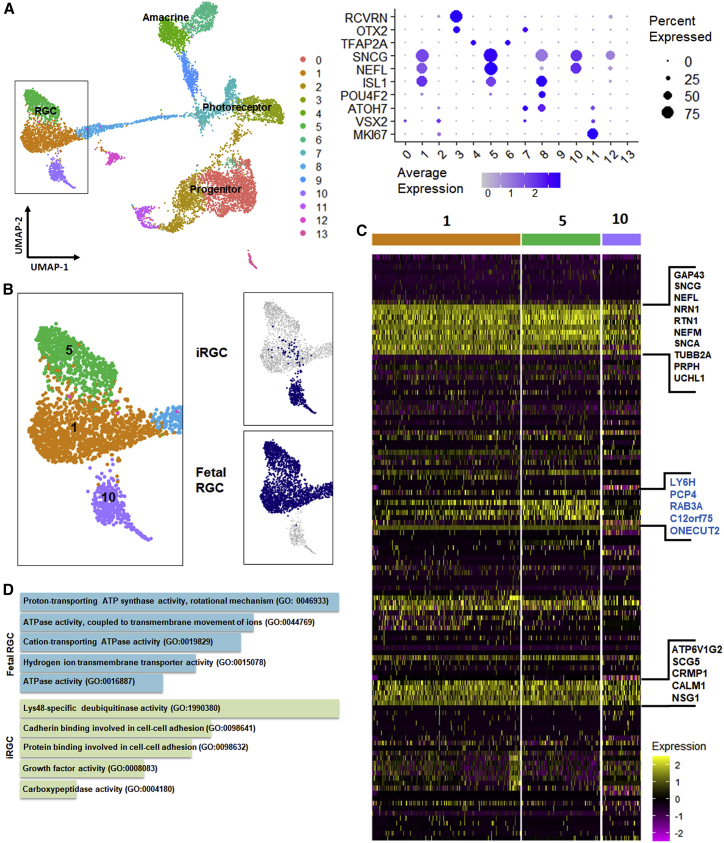
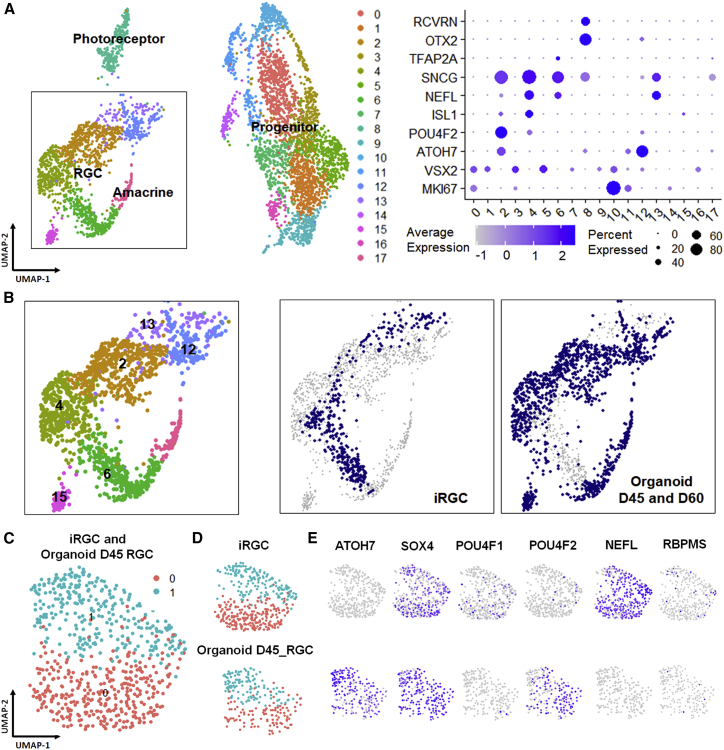
To evaluate how well the iRGCs in the present system mimicked RGCs in the developing human retina, we performed unbiased clustering to map our iRGC subsets (clusters 3 and 4 in Figure 3) to the combined dataset of day 59 and day 82 fetal human retinas. RGCs, amacrine cells, retinal progenitors, and photoreceptor cells were identified by typical marker gene expression (Figure 4A). Visualized by uniform manifold approximation and projection (UMAP) dimensional reduction, iRGCs were distributed near the fetal RGC clusters, indicating their similarities. However, the majority of these iRGCs did not overlap with fetal RGCs, which suggested that they were not identical (Figure 4B). A heatmap of variable genes showed that these three RGC clusters shared similar RGC marker genes, but cluster 10, which mostly consisted of iRGCs, also showed some differences compared with the other two clusters (Figure 4C). Gene Ontology (GO) enrichment found that fetal RGCs highly expressed genes involved in ATP synthesis and ATP utilization, whereas in iRGCs, genes involved in ubiquitination and cell-cell adhesion were predominant as differentially enriched ontologies (Figure 4D).
Figure 4.
iRGCs show transcriptional similarities to a subset of RGCs from fetal retinas
(A) Combined UMAP plot of day 59 fetal retina, day 82 fetal retina, and iRGCs shows 14 clusters, representing at least four main retinal cell types identified by typical markers shown in dot plots.
(B) Focusing on the RGC segment (boxed in A), UMAP plotted by tissue of origin suggests that iRGCs form a unique subtype closest to cluster 1- and cluster 5-type RGCs found in fetal retinas of both ages.
(C) Heatmap of genes (black) driving cluster separation among clusters 1, 5, and 10, demonstrating the few differences (blue) between fetal RGCs and iRGCs.
(D) Functional enrichment of genes that are significantly expressed in fetal RGCs and iRGCs.
Retinal organoids, stem cell-derived tissues that recapitulate fetal retinal development, are another promising source of donor RGCs. We compared iRGCs with whole retinal organoids and organoid-derived RGCs. We mapped the iRGCs to integrated day 45 and day 60 retinal organoid data. After identifying the cell types (Figure 5A), we found that iRGCs fall into similar clusters as organoid RGCs but without a substantial overlap (Figure 5B). To more closely compare organoid-derived RGCs and iRGCs, we selected day 45 retinal organoid-derived RGCs based on belonging to clusters highly expressing the BRN3 or GAP43 gene and combined them with iRGCs, followed by re-clustering (Figure 5C). The datasets largely overlapped, and the contributions of each subgroup were almost equal from both samples in the resulting clusters (Figure 5D). Although the UMAP plot indicates that they shared high similarities among samples and sub-clusters, several genes were differentially expressed in iRGCs compared with organoid RGCs (Figure 5E). For example, RGCs from retinal organoids still expressed ATOH7 but not NEFL on day 45. In contrast, iRGCs from our rapid protocol highly expressed NEFL, consistent with axon growth observed in culture. The two samples showed distinct preferences of BRN3 subtype expression. In iRGCs, cells mainly expressed BRN3A (POU4F1), similar to fetal human RGCs (Luo et al., 2019), whereas most organoid RGCs were positive for BRN3B (POU4F2). These data show that, although iRGCs only went through less than 2 weeks of differentiation, they already gained similar transcriptomic maturity as 2-month-old human fetal RGCs and retinal organoid-derived RGCs.
Figure 5.
iRGCs show transcriptional overlap with RGCs from retinal organoids
(A) Combined UMAP plots of RGCs from day 45 and day 60 retinal organoids and iRGCs. Four main retinal cell types were identified by typical markers shown in dot plots.
(B) Focusing on the RGC segment (boxed in A), UMAP plotted by tissue of origin demonstrates fully overlapping clustering between iRGCs and organoid-derived RGCs.
(C–E) Datasets of iRGCs (clusters 3 and 4 from Figure 3) and of day 45 retinal organoid-derived RGCs (defined by independent cluster analysis of those data) were combined and re-clustered in UMAP (C). Plotting by tissue of origin showed largely overlapping distributions of the iRGCs and organoid RGCs between the clusters (D), and expression patterns of representative RGC-specific genes in the two samples showed some shared genes but also some differences between the differentiation conditions (E).
iRGCs are transcriptionally distinct from NGN-2 induced neurons (iNs) and brain organoids
To determine whether the iRGCs generated in this modified protocol are transcriptionally different from those in the original iN protocol (Lin et al., 2021), we compared scRNA-seq profiles among day 5 iNs and day 59 fetal RGCs and iRGCs (Figures S1A–S1C). The iRGCs essentially all overlapped with subsets of fetal RGCs and shared a diversity of some typical RGC markers like POU4F2 and RBPMS, whereas the iNs contained some progeny that overlapped with these fetal RGCs, but the two major iN progeny clusters did not overlap with fetal RGCs and expressed non-RGC markers like NEUROD2 and PHOX2B (Figures S1B and S1C). These data indicate that iRGCs are transcriptionally more similar to fetal RGCs than the majority of iNs.
Next we compared the mixture of cells in our system with brain organoid transcriptomics profiles to confirm the modified system was deriving RGCs rather than generic neurons similar to brain-directed cultures. The whole dataset of our differentiated cells was combined with the published day 93 cortical organoid dataset (Figure S1D). Grouped by samples, the UMAP plot demonstrated that, although the two cell populations interlaced in UMAP space, iRGC progeny segregated as distinct clusters from adjacent clusters of organoid cells with almost no overlap (Figure S1E). By examining several markers typically expressed in cortical neurons, we found that only a neural crest marker was slightly expressed in our sample. VIM-positive mesenchymal cells were observed in both samples. In comparison, most of the RGC markers were distinctly expressed in iRGCs (Figure S1F). These results indicated that the rapid protocol is reliably deriving RGC-specific progeny with these cell culture modifications.
In vivo iRGC transplantation showed a neuroprotective effect
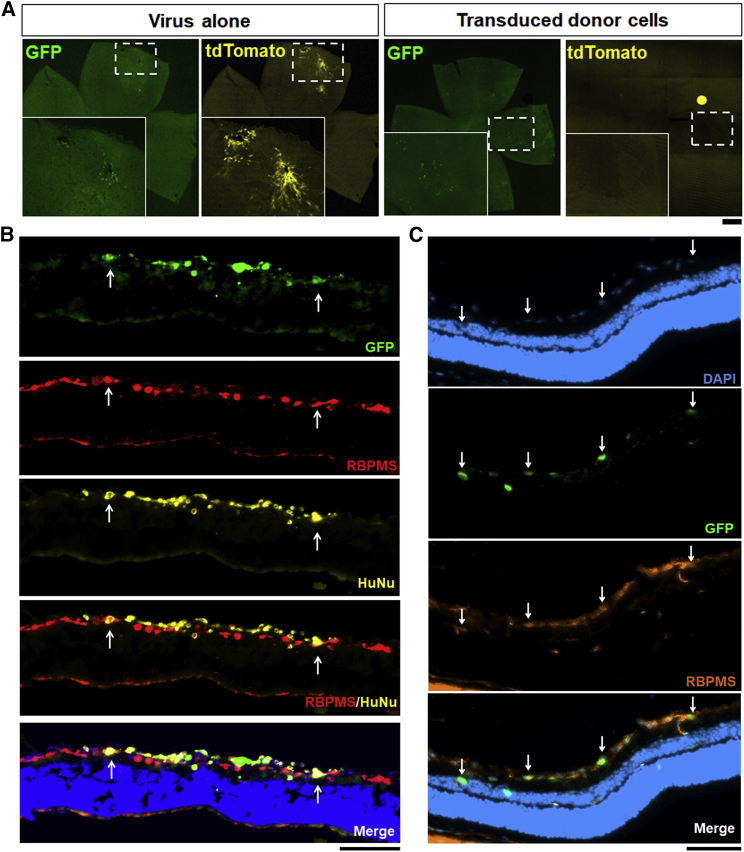
We next wanted to determine whether the differentiated cells survive and migrate into a host retina after transplantation. To track transplanted cells, we labeled differentiated cells with lentivirus (LV)-green fluorescent protein (LV-GFP), but we needed to rule out two critical sources of confounding. To exclude false positive identification of transplanted cells caused by virus contamination leading to host RGC transduction, we injected Cre-GFP-LV versus Cre-GFP-transduced cells into floxed-tdTomato mice. Any viral carryover in the donor cells or material transfer of Cre protein would be expected to turn on tdTomato in host retinal cells. Flat-mount imaging showed that intravitreal injection of Cre-GFP-LV transduced endogenous RGCs and turned on Cre-mediated expression of tdTomato (Figure 6A, left panels). However, after transplantation of donor cells that were transduced previously by Cre-GFP-LV, we did not observe any tdTomato signal (Figure 6A, right panels). Thus, transducing donor cells ex vivo does not lead to viral or protein transfer after cell transplantation in vivo.
Figure 6.
Donor cells survive and integrate into adult rodent retina 1 week and 1 month after transplantation, respectively
(A) To distinguish donor cells from host RGCs and rule out viral contamination and cell fusion with host cells, Cre-GFP-LV alone or Cre-GFP-LV-transduced donor cells were injected intravitreally (as labeled) into floxed-tdTomato mice. After direct viral injection (left panels), GFP-transduced host cells expressed tdTomato, but no tdTomato signal was observed in the retinas that were transplanted by Cre-GFP-LV-transduced donor cells (right panels).
(B and C) Differentiated cells were injected intravitreally into rat eyes, and retinas were dissected, cryosectioned, and immunostained against the RGC-specific marker RBPMS 7 days (B) or 1 month (C) after transplantation. Arrows in (B) indicate donor cells co-labeled by RBPMS and human nuclei (HuNus). Arrows in (C) indicate the migration and integration of iRGCs into the ganglion cell layer of the host retina.
Scale bars, 500 μm (A) and 100 μm (B and C).
To explore transplantation and integration of differentiated cells, we intravitreally injected cells into adult rat eyes and observed that GFP-positive donor cells survive in the host retina 1 week after transplantation. Cross-sections of the retina revealed that donor cells resided on the ganglion cell layer, rarely expressing RBPMS as host RGCs do (Figure 6B) but consistent with the lack of RBPMS detection in the iRGC scRNA-seq. The origin of GFP-labeled cells was confirmed by human nucleus staining (Figure 6B), again providing data against the possibility of material transfer (Pearson et al., 2016). By 4 weeks after transplantation in vivo, donor iRGCs survived and migrated into and, in some cases, past the ganglion cell layer in the mouse retina and, by this time point, were essentially all RBPMS positive (Figure 6C). Thus, iRGCs are competent to integrate into the normal adult retina after intravitreal transplantation.
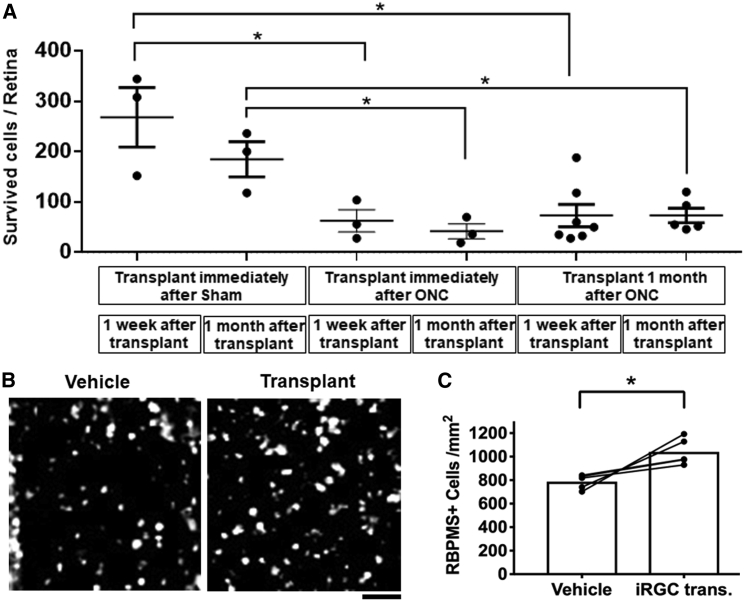
Because the long-term goal of RGC differentiation is to treat RGC loss, we next transplanted the differentiated cells to adult mouse retinas after optic nerve crush (ONC) injury. We first performed the transplants in sham or immediately after ONC. We observed donor cell survival 1 week and 1 month after transplantation. Surprisingly, fewer GFP-positive donor cells were observed in ONC eyes compared with sham eyes (Figure 7A). The reduction of donor cell survival in the ONC group might be due to trauma-induced inflammation in the eye. There was no significant difference in cell numbers between 1 week and 1 month after transplantation in the sham or ONC groups, suggesting that donor cell survival remains stable after 1 week of transplantation. To understand whether the ONC-induced inflammatory response influences donor cell survival, we transplanted differentiated cells 1 month after ONC. Similar to immediate transplantation, we found no increase in GFP-positive cells 1 week or 1 month after transplantation (Figure 7A). Putting together the data across the sham and acute versus chronic ONC experiments, these data suggest that ONC inhibits the survival and/or integration of donor cells after transplantation.
Figure 7.
Survival of donor and host RGCs in ONC models
(A) Survival of donor cells was decreased in “immediate ONC” mice. Data show similar numbers of persistent donor cells transplanted into “1 month after ONC mice” (n = 3).
(B) Example immunostaining against RBPMS of retinal RGCs with and without cell transplantation.
(C) One week after transplantation, the number of RBPMS-positive RGCs was significantly greater in the cell-transplanted group than in the vehicle group (n = 4).
Finally, we wanted to determine whether donor cells influence host RGC survival after ONC. Host RGCs were identified by RBPMS staining (Figure 7B). RBPMS-positive cells in mouse retinas were significantly increased after cell transplantation (around 200 cells/mm2) (Figure 7B). Because donor cells are much fewer in number (less than 100 cells per retina; i.e., less than 10 cells/mm2), and because, in the acute phase after transplantation, donor iRGCs express a very low level of RBPMS (Figure 6B), the detected increase in RBPMS-positive cells is most likely due to enhanced survival of endogenous RGCs. These results indicate that cells differentiated in the iRGC protocol are neuroprotective to endogenous RGCs after optic nerve injury.
Discussion
The present study describes a new, effective method that more rapidly generates RGC-like neurons from human stem cells by overexpressing a single transcription factor and leveraging a specific growth medium. The entire iRGC generation procedure takes less than 2 weeks, and the resulting iRGCs exhibit high similarity in transcriptomic profiles to 2-month-old fetal or retinal organoid-derived RGCs, although there is still room to optimize reprogramming efficiency. These iRGCs can be transplanted intravitreally, showing survival and integration into rat and mouse host retinas. Thus, the approach described here can enable highly efficient and reproducible iRGC generation, and the simplicity of the system will benefit translational studies on human RGCs.
NGN2 is one of the transcription factors in the basic-helix-loop-helix (bHLH) network that controls proliferation (Britz et al., 2006), cell fate identity (Cai et al., 2000), axon guidance (Seibt et al., 2003), and survival in the developing nervous system. NGN2 also plays a key role in regulating the initial progression of early retinal neurogenesis and RGC specification. A study on mouse retinal development revealed that NGN2 expression demarcates the leading edge of neurogenesis, in which progenitor cells exit the cell cycle to gain retinal neuronal fate (Hufnagel et al., 2010). Because CHX10 is ubiquitously expressed in retinal progenitors prior to initiation of neurogenesis (Burmeister et al., 1996; Hufnagel et al., 2010), the role of NGN2 in retinal neurogenesis is consistent with our results in the present study; before switching to RGC medium, the transfected cells were already expressing CHX10 protein and stood to accelerate RGC fate induction through exogenous NGN2 expression.
In addition, it has been well established that the Notch signaling pathway is essential in RGC fate decisions (Austin et al., 1995; Bao and Cepko, 1997). Notch activity is downregulated just before RGC differentiation (Nelson et al., 2006), and reducing Notch signaling can reinitiate RGC production after it has normally ceased during development (Silva et al., 2003), consistent with our application of the Notch inhibitor DAPT to enhance the induction of RGC fate. The Notch pathway interacts with the bHLH network by suppressing endogenous ATOH7 and NGN2 (Maurer et al., 2014; Miesfeld et al., 2018). Thus, in the present system, hiPSCs/hESCs may be directed to an RGC fate by endogenous (through Notch inhibition) and exogenous (through gene transfer) NGN2 upregulation.
Although previous studies have indicated that ATOH7 is directly activated by NGN2 (Hufnagel et al., 2010; Skowronska-Krawczyk et al., 2009), ATOH7 was barely expressed in our scRNA-seq. A recent scRNA-seq study on fetal retinas identified a transitional cell population that is ATOH7/DLL3/HES6 positive (Sridhar et al., 2020). They appear to be at critical post-mitotic junctures on their way to specifying RGC fate. Our qPCR along the first week of differentiation in vitro showed that ATOH7 was transiently upregulated on day 2 and back to baseline on day 6, consistent with no longer detecting it by scRNA-seq also collected on day 6. This result is consistent with another recent study (Smith et al., 2016), and such a transient expression pattern during RGC development has also been observed in mice (Brzezinski et al., 2012; Prasov et al., 2012). Thus, the absence of the transitional marker ATOH7 by day 6 may indicate synchronized induction of RGCs in the present system.
Comparing differentiation across approaches using deep sequencing and advanced transcriptome analyses pointed to unexpectedly closer alignment between our rapid, 2-dimensional culture approach and slower human fetal retinal development than between fetal development and the similarly slower differentiation evidenced in 3-dimensional organoids. The ganglion cell layer in human fetal retinas is not clearly defined until gestational weeks 9–10 (Luo et al., 2019; Provis et al., 1985). In stem cell-derived retinal organoids, which highly recapitulate the timing of fetal retinal development, RGCs are observed as early as differentiation week 7 (Luo et al., 2018; Zhong et al., 2014). In comparison with fetal retina and retinal organoid datasets using UMAP reduction, which preserves global data structure and demonstrates similarities among clusters (Becht et al., 2018), the similarity of our iRGCs to fetal RGCs or organoid RGCs can be clearly visualized. Further analysis shows shared marker genes between iRGCs and fetal or organoid RGCs. However, our iRGCs are more identical to fetal RGCs than organoid RGCs. For example, like fetal RGCs, the majority of RGC-like cells express POU4F1 (BRN3a), whereas most organoid RGCs are POU4F2 (BRN 3b) positive. The axon skeleton markers NEFL and NEFM are expressed at significantly higher levels in the fetal retina and iRGCs compared with retinal organoids (Sridhar et al., 2020), consistent with the RGC transcriptional maturation seen in another scRNA-seq study on human fetal retinas (Lu et al., 2020). Even so, we acknowledge that iRGC specification, electrophysiological maturation, and transcriptome consistency to human RGCs may still be improved. For example, our previous studies show that GDF15 (Chang et al., 2019) and SOX4 (Chang et al., 2017) facilitate RGC specification in stem cell-to-RGC differentiation; in future studies, we explore to what degree these factors promote RGC maturation.
Because our protocol was modified from a cortical neuron differentiation protocol, it was essential to determine how much the RGC-like cells in our protocol differ from cortical neurons, acknowledging that, because the neural retina is developmentally an outgrowth of the forebrain, RGCs share many marker genes with other forebrain neurons (Gabriel et al., 2021). Thus, we compared iRGC and iN transcriptomes, including fetal RGCs. iRGCs were much more similar to fetal RGCs than iN progeny, arguing that the modifications in our iRGC protocol are meaningful in generating RGC-like progeny. A consistent result could also be seen when comparing iRGCs and cortical organoids, where iRGCs made up distinct but non-overlapping clusters from cortical organoid progeny. Of course, from a translational standpoint, donor cells need not necessarily be identical to the host cells they are replacing, and it is still unknown to what extent donor cell transcriptional identity to host cells is needed to achieve optimal cell replacement outcomes. It is interesting that our donor cells between 1 week and 1 month showed continued migration into the retina and increased expression of the RGC marker RBPMS, suggesting that their further maturation and integration could be achieved after in vivo transplantation.
One of the primary goals in translating the basic science of stem cell differentiation into RGCs is to obtain donor cells for the purpose of transplantation and cell replacement therapy in optic neuropathies. Here we modeled this approach by intravitreal injection of iRGCs in rat and mouse eyes to evaluate the efficacy of cell transplantation. We previously reported that RGCs derived from hESCs successfully migrated into the ganglion cell layer 1 week after transplantation to the adult rat eye (Zhang et al., 2019). We now observed that iRGCs from the rapid protocol also successfully engraft into host retinas by 1 week, although at a low survival rate, consistent with our previous studies (Hertz et al., 2014b; Venugopalan et al., 2016). We also observe further migration to the inner nuclear layer 1 month after transplantation. A number of factors could improve the integration we reported here and should be explored in future studies. For example, the internal limiting membrane (ILM) is a known barrier to donor RGC survival and synaptic integration into the host retina (Zhang et al., 2021). Digestion of the ILM by Pronase E (Zhang et al., 2021) could increase iRGC survival and integration. Expressing or co-injecting survival and growth factors could also be leveraged to promote integration, as has been shown in cell transfer to the spinal cord, for example. (Lu et al., 2012) Our previous study showed that stem cell transplantation enhances donor RGC survival in the host retina (Wu et al., 2018). Donor cell type and transplant location may also be critical. For example, subretinal transplantation with human retinal organoid pieces or cells dissociated from human retinal organoids can survive for months to years in non-human primate models (Tu et al., 2019). It is still uncertain whether differences in survival or integration are due to donor cell sources, the transplantation method, or the species of the recipients. With the higher expression level of axon-related genes in our rapid protocol-induced RGCs and fetal RGCs, we hypothesize that allowing the iRGCs to extend axons before transplantation may also be a promising way to pre-stimulate the donor cells and promote their survival and axon growth in vivo. A recent study of isolation of donor RGCs 1 week after transplantation also demonstrated the importance of axon growth and guidance in donor cell survival (Oswald et al., 2021).
Finally, it will be critical to understand to what degree the injured or degenerating host retina aids or inhibits donor cell integration. To understand whether iRGCs can survive after transplantation into the injured retina, we transplanted donor cells into host eyes immediately or 1 month after optic nerve injury. Compared with transplantation into normal host eyes, survival of donor iRGCs was attenuated in the immediate group or 1 month after ONC. Inflammatory responses in the retina are elevated after optic nerve injury (Ahmad et al., 2014), and optic nerve injury-induced pro-inflammatory cytokines could contribute to donor iRGC death. Phagocytic microglial infiltration peaks around 14 days after crush and is still above basal levels at 2 months (Nadal-Nicolas et al., 2017). In addition, the donor cells could induce an inflammatory response, especially as a xenograft in the present study. Future studies should examine more explicitly the effects of donor cell transplantation on glial and immune response and examine integration at even later time points after injury to allow neuro-inflammation to subside fully and/or include immune-regulating drugs to improve donor cell survival. Conversely, cell transplantation may affect host RGCs after optic nerve injury. We found that iRGC transplantation significantly promotes endogenous RGC survival 1 week after optic nerve injury, although this significant improvement was lost by 2 weeks after ONC. This suggests that iRGC transplantation can transiently delay endogenous RGC death.
These data comprehensively assay iRGCs for gene expression, electrophysiological maturation, and competency to integrate into a host retina after transplantation. Future studies comparing donor cell differentiation methods, stages of maturation, and accompanying factors applied during transplantation may facilitate a translational pipeline toward cell replacement therapy in optic neuropathies.
Materials and methods
Corresponding Author
Further information and requests for resources and reagents should be directed to and will be fulfilled by the corresponding author, J.L.G. (luozm@stanford.edu).
Materials availability
This study did not generate new unique reagents. Details about methods, reagents, scRNA-seq analyses, and transplantation can be found in the supplemental information.
Cell lines and animal strains used in this study
The Brn3b-driven mCherry H7-A81 ESC line (Sluch et al., 2015) was a kind gift from Dr. Donald Zack (Co-Director, Johns Hopkins Center for Stem Cell and Ocular Regenerative Medicine). 297-1 hiPSCs derived from peripheral blood mononuclear cells of a healthy male donor in his 50s was obtained from the Stanford Stem Cell Core.
All animal research was conducted in compliance with the ARVO statement for use of animals in ophthalmic and vision research and was approved by the Institutional Animal Care and Use Committee at Stanford University. C57BL/6 wild-type (WT) male and female mice 4–6 weeks old and Sprague-Dawley female rats 6 weeks old were obtained from Charles River Laboratories. Cre-driven tdTomato mice were obtained from The Jackson Laboratory.
Author contributions
Conceptualization, K.-C.C., Z.L., M.W., and J.L.G.; methodology, K.-C.C. and Z. L.; investigation, K.-C.C., Z. L., S.W., C.S., X.X., M.N., M.B., R.R.W., Y.Z., and S.S.; visualization, Z.L. and B.T.; funding acquisition, K.-C.C. and J.L.G.; supervision, M.W. and J.L.G.; writing – original draft, K.-C.C., Z.L., and M.N.; writing – review & editing, J.L.G.
Acknowledgments
We thank Dr. Donald Zack for kindly providing the H7 hESC line used in this study. We thank Dr. Thomas A. Reh for intellectual contributions. Library preparation and sequencing were carried out at the DNA Technologies and Expression Analysis Cores at the UC Davis Genome Center, supported by NIH Shared Instrumentation Grant 1S10OD010786-01. This study was supported by the National Eye Institute (P30-EY026877 and F32-EY029137), a Veteran Affairs Merit Award (I01-BX002950 to J.L.G.), the BrightFocus Foundation, the Gilbert Family Foundation, Research to Prevent Blindness, Inc, and the Foundation Optic Atrophy 1.
Conflict of interests
The authors declare no competing interests.
Published: November 10, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stemcr.2022.10.011.
Supplemental information
Data and code availability
The datasets generated during this study are available on GEO: GSE200452. Additional datasets used for comparison can be found at GEO: GSE142526, GSE157019, and ArrayExpress: E-MTAB-10632.
References
- Ahmad S., Elsherbiny N.M., Bhatia K., Elsherbini A.M., Fulzele S., Liou G.I. Inhibition of adenosine kinase attenuates inflammation and neurotoxicity in traumatic optic neuropathy. J. Neuroimmunol. 2014;277:96–104. doi: 10.1016/j.jneuroim.2014.10.006. [DOI] [PubMed] [Google Scholar]
- Austin C.P., Feldman D.E., Ida J.A., Jr., Cepko C.L. Vertebrate retinal ganglion cells are selected from competent progenitors by the action of Notch. Development. 1995;121:3637–3650. doi: 10.1242/dev.121.11.3637. [DOI] [PubMed] [Google Scholar]
- Bao Z.Z., Cepko C.L. The expression and function of Notch pathway genes in the developing rat eye. J. Neurosci. 1997;17:1425–1434. doi: 10.1523/JNEUROSCI.17-04-01425.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becht E., McInnes L., Healy J., Dutertre C.A., Kwok I.W.H., Ng L.G., Ginhoux F., Newell E.W. Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol. 2018;37:38–44. doi: 10.1038/nbt.4314. [DOI] [PubMed] [Google Scholar]
- Britz O., Mattar P., Nguyen L., Langevin L.M., Zimmer C., Alam S., Guillemot F., Schuurmans C. A role for proneural genes in the maturation of cortical progenitor cells. Cereb. Cortex. 2006;16(Suppl 1):i138–i151. doi: 10.1093/cercor/bhj168. [DOI] [PubMed] [Google Scholar]
- Brzezinski J.A., 4th, Prasov L., Glaser T. Math5 defines the ganglion cell competence state in a subpopulation of retinal progenitor cells exiting the cell cycle. Dev. Biol. 2012;365:395–413. doi: 10.1016/j.ydbio.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burmeister M., Novak J., Liang M.Y., Basu S., Ploder L., Hawes N.L., Vidgen D., Hoover F., Goldman D., Kalnins V.I., et al. Ocular retardation mouse caused by Chx10 homeobox null allele: impaired retinal progenitor proliferation and bipolar cell differentiation. Nat. Genet. 1996;12:376–384. doi: 10.1038/ng0496-376. [DOI] [PubMed] [Google Scholar]
- Cai L., Morrow E.M., Cepko C.L. Misexpression of basic helix-loop-helix genes in the murine cerebral cortex affects cell fate choices and neuronal survival. Development. 2000;127:3021–3030. doi: 10.1242/dev.127.14.3021. [DOI] [PubMed] [Google Scholar]
- Chang K.C., Hertz J., Zhang X., Jin X.L., Shaw P., Derosa B.A., Li J.Y., Venugopalan P., Valenzuela D.A., Patel R.D., et al. Novel regulatory mechanisms for the SoxC transcriptional network required for visual pathway development. J. Neurosci. 2017;37:4967–4981. doi: 10.1523/JNEUROSCI.3430-13.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang K.C., Sun C., Cameron E.G., Madaan A., Wu S., Xia X., Zhang X., Tenerelli K., Nahmou M., Knasel C.M., et al. Opposing effects of growth and differentiation factors in cell-fate specification. Curr. Biol. 2019;29:1963–1975.e5. doi: 10.1016/j.cub.2019.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Chen Q., Sun X., Shen W., Liu B., Zhong X., Leng Y., Li C., Zhang W., Chai F., et al. Generation of retinal ganglion-like cells from reprogrammed mouse fibroblasts. Invest. Ophthalmol. Vis. Sci. 2010;51:5970–5978. doi: 10.1167/iovs.09-4504. [DOI] [PubMed] [Google Scholar]
- Deng F., Chen M., Liu Y., Hu H., Xiong Y., Xu C., Liu Y., Li K., Zhuang J., Ge J. Stage-specific differentiation of iPSCs toward retinal ganglion cell lineage. Mol. Vis. 2016;22:536–547. [PMC free article] [PubMed] [Google Scholar]
- Furukawa T., Kozak C.A., Cepko C.L. rax, a novel paired-type homeobox gene, shows expression in the anterior neural fold and developing retina. Proc. Natl. Acad. Sci. USA. 1997;94:3088–3093. doi: 10.1073/pnas.94.7.3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel E., Albanna W., Pasquini G., Ramani A., Josipovic N., Mariappan A., Schinzel F., Karch C.M., Bao G., Gottardo M., et al. Human brain organoids assemble functionally integrated bilateral optic vesicles. Cell Stem Cell. 2021;28:1740–1757.e8. doi: 10.1016/j.stem.2021.07.010. [DOI] [PubMed] [Google Scholar]
- Hertz J., Qu B., Hu Y., Patel R.D., Valenzuela D.A., Goldberg J.L. Survival and integration of developing and progenitor-derived retinal ganglion cells following transplantation. Cell Transplant. 2014;23:855–872. doi: 10.3727/096368913X667024. [DOI] [PubMed] [Google Scholar]
- Hertz J., Qu B., Hu Y., Patel R.D., Valenzuela D.A., Goldberg J.L. Survival and integration of developing and progenitor-derived retinal ganglion cells following transplantation. Cell Transplant. 2014;23:855–872. doi: 10.3727/096368913x667024. [DOI] [PubMed] [Google Scholar]
- Hufnagel R.B., Le T.T., Riesenberg A.L., Brown N.L. Neurog2 controls the leading edge of neurogenesis in the mammalian retina. Dev. Biol. 2010;340:490–503. doi: 10.1016/j.ydbio.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamba D.A., Karl M.O., Ware C.B., Reh T.A. Efficient generation of retinal progenitor cells from human embryonic stem cells. Proc. Natl. Acad. Sci. USA. 2006;103:12769–12774. doi: 10.1073/pnas.0601990103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H.C., He Z., Ebert S., Schörnig M., Santel M., Nikolova M.T., Weigert A., Hevers W., Kasri N.N., Taverna E., et al. NGN2 induces diverse neuron types from human pluripotency. Stem Cell Rep. 2021;16:2118–2127. doi: 10.1016/j.stemcr.2021.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu P., Wang Y., Graham L., McHale K., Gao M., Wu D., Brock J., Blesch A., Rosenzweig E.S., Havton L.A., et al. Long-distance growth and connectivity of neural stem cells after severe spinal cord injury. Cell. 2012;150:1264–1273. doi: 10.1016/j.cell.2012.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y., Shiau F., Yi W., Lu S., Wu Q., Pearson J.D., Kallman A., Zhong S., Hoang T., Zuo Z., et al. Single-cell analysis of human retina identifies evolutionarily conserved and species-specific mechanisms controlling development. Dev. Cell. 2020;53:473–491.e9. doi: 10.1016/j.devcel.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Z., Xu C., Li K., Xian B., Liu Y., Li K., Liu Y., Rong H., Tang M., Hu D., et al. Islet1 and Brn3 expression pattern study in human retina and hiPSC-derived retinal organoid. Stem Cells Int. 2019;2019 doi: 10.1155/2019/8786396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Z., Zhong X., Li K., Xie B., Liu Y., Ye M., Li K., Xu C., Ge J. An optimized system for effective derivation of three-dimensional retinal tissue via Wnt signaling regulation. Stem cells. 2018;36:1709–1722. doi: 10.1002/stem.2890. [DOI] [PubMed] [Google Scholar]
- Mathers P.H., Grinberg A., Mahon K.A., Jamrich M. The Rx homeobox gene is essential for vertebrate eye development. Nature. 1997;387:603–607. doi: 10.1038/42475. [DOI] [PubMed] [Google Scholar]
- Maurer K.A., Riesenberg A.N., Brown N.L. Notch signaling differentially regulates Atoh7 and Neurog2 in the distal mouse retina. Development. 2014;141:3243–3254. doi: 10.1242/dev.106245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miesfeld J.B., Moon M.S., Riesenberg A.N., Contreras A.N., Kovall R.A., Brown N.L. Rbpj direct regulation of Atoh7 transcription in the embryonic mouse retina. Sci. Rep. 2018;8 doi: 10.1038/s41598-018-28420-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadal-Nicolás F.M., Jiménez-López M., Salinas-Navarro M., Sobrado-Calvo P., Vidal-Sanz M., Agudo-Barriuso M. Microglial dynamics after axotomy-induced retinal ganglion cell death. J. Neuroinflammation. 2017;14:218. doi: 10.1186/s12974-017-0982-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson B.R., Gumuscu B., Hartman B.H., Reh T.A. Notch activity is downregulated just prior to retinal ganglion cell differentiation. Dev. Neurosci. 2006;28:128–141. doi: 10.1159/000090759. [DOI] [PubMed] [Google Scholar]
- Oswald J., Kegeles E., Minelli T., Volchkov P., Baranov P. Transplantation of miPSC/mESC-derived retinal ganglion cells into healthy and glaucomatous retinas. Mol. Ther. Methods Clin. Dev. 2021;21:180–198. doi: 10.1016/j.omtm.2021.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson R.A., Gonzalez-Cordero A., West E.L., Ribeiro J.R., Aghaizu N., Goh D., Sampson R.D., Georgiadis A., Waldron P.V., Duran Y., et al. Donor and host photoreceptors engage in material transfer following transplantation of post-mitotic photoreceptor precursors. Nat. Commun. 2016;7 doi: 10.1038/ncomms13029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasov L., Nagy M., Rudolph D.D., Glaser T. Math5 (Atoh7) gene dosage limits retinal ganglion cell genesis. Neuroreport. 2012;23:631–634. doi: 10.1097/WNR.0b013e328355f260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provis J.M., van Driel D., Billson F.A., Russell P. Development of the human retina: patterns of cell distribution and redistribution in the ganglion cell layer. J. Comp. Neurol. 1985;233:429–451. doi: 10.1002/cne.902330403. [DOI] [PubMed] [Google Scholar]
- Seibt J., Schuurmans C., Gradwhol G., Dehay C., Vanderhaeghen P., Guillemot F., Polleux F. Neurogenin2 specifies the connectivity of thalamic neurons by controlling axon responsiveness to intermediate target cues. Neuron. 2003;39:439–452. doi: 10.1016/s0896-6273(03)00435-5. [DOI] [PubMed] [Google Scholar]
- Silva A.O., Ercole C.E., McLoon S.C. Regulation of ganglion cell production by Notch signaling during retinal development. J. Neurobiol. 2003;54:511–524. doi: 10.1002/neu.10156. [DOI] [PubMed] [Google Scholar]
- Skowronska-Krawczyk D., Chiodini F., Ebeling M., Alliod C., Kundzewicz A., Castro D., Ballivet M., Guillemot F., Matter-Sadzinski L., Matter J.M. Conserved regulatory sequences in Atoh7 mediate non-conserved regulatory responses in retina ontogenesis. Development. 2009;136:3767–3777. doi: 10.1242/dev.033449. [DOI] [PubMed] [Google Scholar]
- Sluch V.M., Davis C.h.O., Ranganathan V., Kerr J.M., Krick K., Martin R., Berlinicke C.A., Marsh-Armstrong N., Diamond J.S., Mao H.Q., Zack D.J. Differentiation of human ESCs to retinal ganglion cells using a CRISPR engineered reporter cell line. Sci. Rep. 2015;5 doi: 10.1038/srep16595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D.K., Yang J., Liu M.L., Zhang C.L. Small molecules modulate chromatin accessibility to promote NEUROG2-mediated fibroblast-to-neuron reprogramming. Stem Cell Rep. 2016;7:955–969. doi: 10.1016/j.stemcr.2016.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridhar A., Hoshino A., Finkbeiner C.R., Chitsazan A., Dai L., Haugan A.K., Eschenbacher K.M., Jackson D.L., Trapnell C., Bermingham-McDonogh O., et al. Single-cell transcriptomic comparison of human fetal retina, hPSC-derived retinal organoids, and long-term retinal cultures. Cell Rep. 2020;30:1644–1659.e4. doi: 10.1016/j.celrep.2020.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu H.Y., Watanabe T., Shirai H., Yamasaki S., Kinoshita M., Matsushita K., Hashiguchi T., Onoe H., Matsuyama T., Kuwahara A., et al. Medium- to long-term survival and functional examination of human iPSC-derived retinas in rat and primate models of retinal degeneration. EBioMedicine. 2019;39:562–574. doi: 10.1016/j.ebiom.2018.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venugopalan P., Cameron E.G., Zhang X., Nahmou M., Muller K.J., Goldberg J.L. Physiologic maturation is both extrinsically and intrinsically regulated in progenitor-derived neurons. Sci. Rep. 2020;10:2337. doi: 10.1038/s41598-020-58120-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venugopalan P., Wang Y., Nguyen T., Huang A., Muller K.J., Goldberg J.L. Transplanted neurons integrate into adult retinas and respond to light. Nat. Commun. 2016;7 doi: 10.1038/ncomms10472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C.T., Blankenship A.G., Anishchenko A., Elstrott J., Fikhman M., Nakanishi S., Feller M.B. GABA(A) receptor-mediated signaling alters the structure of spontaneous activity in the developing retina. J. Neurosci. 2007;27:9130–9140. doi: 10.1523/JNEUROSCI.1293-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S., Chang K.C., Nahmou M., Goldberg J.L. Induced pluripotent stem cells promote retinal ganglion cell survival after transplant. Invest. Ophthalmol. Vis. Sci. 2018;59:1571–1576. doi: 10.1167/iovs.17-23648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie B.B., Zhang X.M., Hashimoto T., Tien A.H., Chen A., Ge J., Yang X.J. Differentiation of retinal ganglion cells and photoreceptor precursors from mouse induced pluripotent stem cells carrying an Atoh7/Math5 lineage reporter. PLoS One. 2014;9 doi: 10.1371/journal.pone.0112175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K.Y., Tuffy C., Mertz J.L., Quillen S., Wechsler L., Quigley H.A., Zack D.J., Johnson T.V. Role of the internal limiting membrane in structural engraftment and topographic spacing of transplanted human stem cell-derived retinal ganglion cells. Stem Cell Rep. 2021;16:149–167. doi: 10.1016/j.stemcr.2020.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Tenerelli K., Wu S., Xia X., Yokota S., Sun C., Galvao J., Venugopalan P., Li C., Madaan A., et al. Cell transplantation of retinal ganglion cells derived from hESCs. Restor. Neurol. Neurosci. 2019;38:131–140. doi: 10.3233/RNN-190941. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Pak C., Han Y., Ahlenius H., Zhang Z., Chanda S., Marro S., Patzke C., Acuna C., Covy J., et al. Rapid single-step induction of functional neurons from human pluripotent stem cells. Neuron. 2013;78:785–798. doi: 10.1016/j.neuron.2013.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong X., Gutierrez C., Xue T., Hampton C., Vergara M.N., Cao L.H., Peters A., Park T.S., Zambidis E.T., Meyer J.S., et al. Generation of three-dimensional retinal tissue with functional photoreceptors from human iPSCs. Nat. Commun. 2014;5:4047. doi: 10.1038/ncomms5047. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during this study are available on GEO: GSE200452. Additional datasets used for comparison can be found at GEO: GSE142526, GSE157019, and ArrayExpress: E-MTAB-10632.