Summary
Epigenetic reprogramming to pluripotency requires extensive remodeling of chromatin landscapes to silence existing cell-type-specific genes and activate pluripotency genes. ATP-dependent chromatin remodeling complexes are important regulators of chromatin structure and gene expression; however, the role of recently identified Bromodomain-containing protein 9 (BRD9) and the associated non-canonical BRG1-associated factors (ncBAF) complex in reprogramming remains unknown. Here, we show that genetic or chemical inhibition of BRD9, as well as ncBAF complex subunit GLTSCR1, but not the closely related BRD7, increase human somatic cell reprogramming efficiency and can replace KLF4 and c-MYC. We find that BRD9 is dispensable for human induced pluripotent stem cells under primed but not under naive conditions. Mechanistically, BRD9 inhibition downregulates fibroblast-related genes and decreases chromatin accessibility at somatic enhancers. BRD9 maintains the expression of transcriptional regulators MN1 and ZBTB38, both of which impede reprogramming. Collectively, these results establish BRD9 as an important safeguarding factor for somatic cell identity whose inhibition lowers chromatin-based barriers to reprogramming.
Keywords: BRD9, reprogramming, iPSC, chromatin remodeling, naive and primed pluripotency
Graphical abstract

Highlights
-
•
CRISPR-Cas9 screen identified ncBAF subunits as barriers to reprogramming
-
•
BRD9 inhibition enhances reprogramming efficiency and induces pluripotency by OS
-
•
BRD9 maintains the accessible chromatin at putative fibroblast enhancers
-
•
BRD9 inhibition downregulates fibroblast-related genes which impede reprogramming
In this article, Onder and colleagues revealed ncBAF as a barrier to human somatic cell reprogramming to pluripotency. They found that ncBAF-specific subunit BRD9 is necessary for maintaining accessible chromatin at putative fibroblast enhancers and its inhibition downregulates expression of somatic cell-type-specific genes before and during reprogramming.
Introduction
Expression of transcription factors such as OCT4, SOX2, KLF4, and MYC (OSKM) can erase somatic cell identity and reprogram the cells to pluripotency (Takahashi and Yamanaka, 2006; Takahashi et al., 2007). In doing so, reprogramming factors reset the entire epigenetic landscape established throughout development (Papp and Plath, 2013). The varying and low efficiencies of this process point to the presence of intrinsic barriers to cell fate conversions. Several chromatin factors such as DOT1L methyltransferase (Onder et al., 2012), histone chaperone CAF-1 (Cheloufi et al., 2015), BET family proteins (Shao et al., 2016), RNA Pol II regulator RPAP1 (Lynch et al., 2018), SUMO modification (Cossec et al., 2018), chromatin regulator FACT (Kolundzic et al., 2018), and CBP/EP300 bromodomains (Ebrahimi et al., 2019) have emerged as potent barriers to reprogramming acting mainly by safeguarding pre-existing gene expression programs. Inhibition of these factors greatly facilitates reprogramming of a wide range of cell types (Brumbaugh et al., 2019). Discovery of additional safeguarding factors will likely yield important insights into chromatin-based mechanisms that maintain cell identity and restrict cell plasticity.
ATP-dependent chromatin remodeling complexes evict, exchange, and space nucleosomes driven by the hydrolysis of ATP (Hota and Bruneau, 2016). Chromatin remodelers can facilitate transcriptional activation or repression based on the genomic location they bind to and additional chromatin factors they recruit (Ho et al., 2011; Hodges et al., 2016; Kadoch et al., 2017). Among these, NuRD, INO80, and SWI/SNF complexes have been shown to modulate reprogramming in a variety of contexts (dos Santos et al., 2014; Singhal et al., 2010; Wang et al., 2014; Zhuang et al., 2018). For example, overexpression of BAF complex subunits Smarca4 and Smarcc1 enhances murine somatic cell reprogramming by facilitating binding of Oct4 to its gene targets (Singhal et al., 2010). In contrast, BAF complex subunits, Smarca2 and Smarcc2, have shown to be barriers in this context through upregulation of Stat3 and its target genes (Jiang et al., 2015). Suppressing these somatic BAF subunits has been shown to activate the pluripotency circuit (Jiang et al., 2015). These studies point to regulatory roles for different ATP-dependent chromatin remodeling complexes in various reprogramming frameworks.
Non-canonical BAF (ncBAF) complex is a recently identified SWI/SNF complex that includes specific subunits such as BRD9 and Glioma tumor suppressor candidate region gene 1 (GLTSCR1) (Alpsoy and Dykhuizen, 2018; Mashtalir et al., 2018; Michel et al., 2018; Pan et al., 2018; Wang et al., 2019). BRD9 binds to enhancers in a cell-type-specific manner and inhibition of its bromodomain leads to apoptosis in acute myeloid leukemia cells (Del Gaudio et al., 2019). Similarly, BRD9 inhibition leads to decreased cell proliferation, G1-arrest, and apoptosis in rhabdoid tumor cells (Wang et al., 2019). In mouse embryonic stem cells (mESCs), ncBAF has been shown to co-localize with key regulators of naive pluripotency (Gatchalian et al., 2018). These studies suggest that BRD9 is important for regulating cell identity and survival. However, the role of BRD9 and the ncBAF complex in somatic cell reprogramming remains unknown. In this study, we addressed this question using a combination of chemical and genetic tools in somatic cells and revealed an important role for BRD9 in safeguarding cell identity in the context of human reprogramming.
Results
Genetic suppression of ncBAF-specific subunits increases reprogramming efficiency
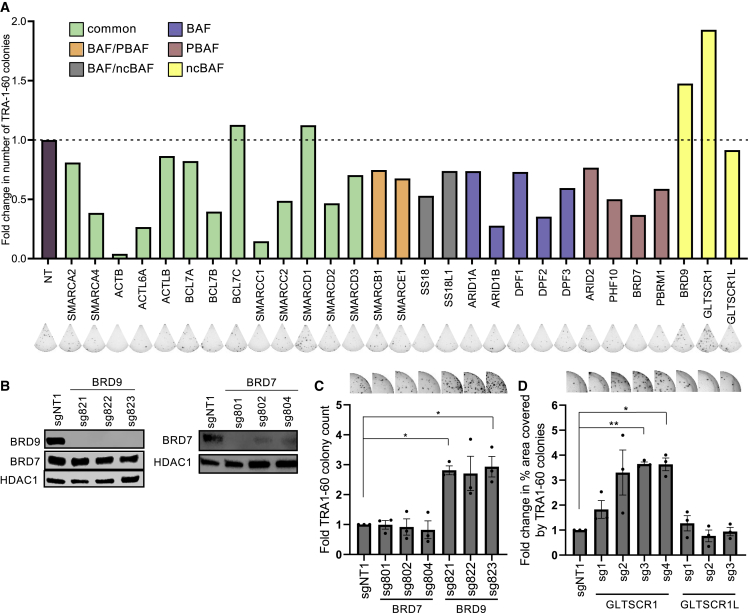
To investigate the role of BAF complexes in human somatic cell reprogramming, we depleted individual complex members in fibroblasts using guide RNMAs (gRNAs) and Cas9 and quantified the resulting reprogramming efficiency. gRNAs targeting common BAF complex members such as SMARCC1 (BAF155) and SMARCA4 (BRG1) inhibited reprogramming. In contrast, depletion of BRD9 and GLTCSR1 increased human induced pluripotent stem cell (iPSC) generation efficiency (Figure 1A). BRD9 and GLTCSR1 are specific members of the ncBAF complexes, which led us to hypothesize that among the various BAF complexes in somatic cells, BRD9-containing ncBAF complex is a specific barrier for reprogramming. To test this, we used two genetic loss-of-function approaches. First, knockdown of BRD9 using two independent short hairpin RNAs (shRNAs) increased reprogramming efficiency 2-fold (Figures S1A and S1B). In contrast, suppression of BRD9 paralog and PBAF-specific subunit, BRD7, had no effect on reprogramming (Figures S1A and S1C). In the second approach, knockout of BRD9, but not BRD7, boosted reprograming efficiency up to 3-fold compared with control gRNA expression (Figures 1B and 1C). Similarly, knockout of ncBAF-specific subunit GLTSCR1 increased reprogramming efficiency up to 3.5-fold (Figure 1D). Targeting GLTSCR1 paralog, GLTSCR1L, had no effect on reprogramming (Figures 1D and S1D). BRD9 is expressed at similar levels in fibroblasts and on day 6 of reprogramming, modestly decreasing in fully reprogrammed human iPSCs (Figure S1E). Exogenous overexpression of BRD9 does not block iPSC generation (Figures S1F and S1G). Collectively, these results suggest that endogenous ncBAF complex members act as barriers to human somatic cell reprogramming.
Figure 1.
Genetic suppression of ncBAF-specific subunits increases reprogramming efficiency
(A) Fold change in TRA-1-60 positive colonies for average of two sgRNAs targeting each gene compared with control sgRNA (NT). n = 2, independent experiments in technical duplicates. Representative well images are shown below the graph.
(B) Western blots for BRD9 and BRD7 in control (sgNT1), BRD9-targeted (top) and BRD7-targeted cells (bottom). HDAC1 serves as loading control for nuclear lysates.
(C) Fold change in the number of TRA-1-60-positive colonies with indicated sgRNAs compared with control (sgNT1). Representative well images are shown above the graph. Bar graphs show the mean, and error bars represent SEM. n = 3 independent experiments. p values are calculated by one-sample t test for mu = 1.
(D) Fold change in the number of TRA-1-60-positive colonies with indicated sgRNAs compared with control (sgNT1). Representative well images are above relative bars. Bar graphs show the mean and error bars represent SEM. n = 3, independent experiments. p values are calculated by one-sample t test for mu = 1.
Bromodomain inhibition and degradation of BRD9 facilitate reprogramming
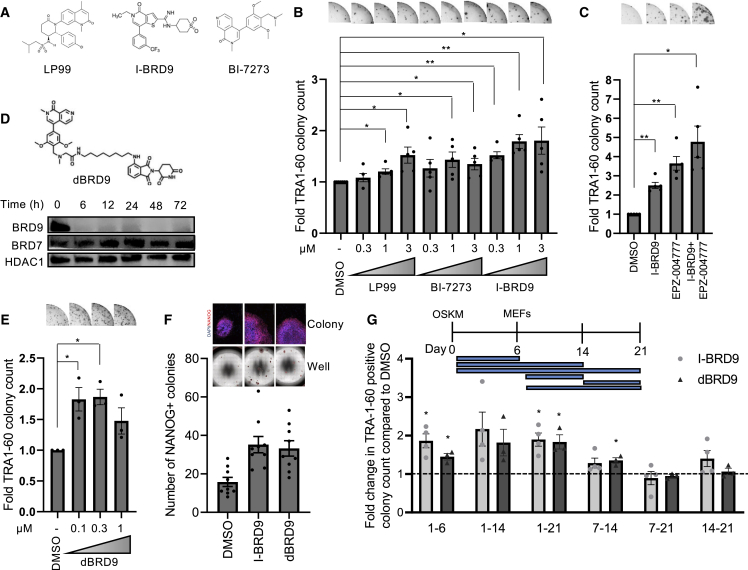
To confirm the role of BRD9 in somatic cell reprogramming, we used three structurally different inhibitors, LP99, BI-7273, and I-BRD9, all of which selectively target the bromodomain of BRD9 (Clark et al., 2015; Martin et al., 2016; Theodoulou et al., 2016) (Figure 2A). All three BRD9 inhibitors significantly increased reprogramming efficiency up to 2-fold (Figure 2B). Importantly, I-BRD9 had an additive effect with inhibition of DOT1L, a potent reprogramming enhancer that we had previously identified (Onder et al., 2012). Combined inhibition of BRD9 and DOT1L led to a 4.5-fold increase in the number of iPSCs generated from human fibroblasts (Figure 2C). Next, we took advantage of a recently described PROteolysis Targeting Chimera (PROTAC) targeting BRD9, dBRD9, to acutely deplete this protein (Remillard et al., 2017). Time-course treatment with dBRD9 dramatically decreased BRD9 protein levels for up to 72 h without any effects on the closely related BRD7 (Figure 2D). Reprogramming efficiency increased to 2-fold compared with control treatment even at the lowest concentration of 0.1 μM dBRD9 tested (Figures 2E and 2F). iPSCs generated from both control and BRD9 inhibitor-treated cells exhibited canonical characteristics of pluripotency, such as expression of OCT4, SSEA4, and NANOG, silencing of retroviral transgenes and generation of teratomas containing differentiated cells from all three germ layers (Figures S2A–S2C). BRD9 inhibition or degradation did not increase the expression levels of exogenous OSKM during reprogramming (Figure S2D). Episomal plasmid-based reprogramming of an additional human dermal fibroblast line was increased with I-BRD9 or dBRD9, indicating that BRD9 inhibition enhances iPSC generation independently of reprogramming strategy or cell line (Figure S2E). Upon I-BRD9 or dBRD9 treatment, OCT4-GFP reporter fibroblasts (Balboa et al., 2017) generated an increased number of GFP-positive colonies that activated the endogenous POU5F1 loci (Figure S2F). Taken together, these results show that bromodomain inhibition or acute degradation of BRD9 increases human somatic cell reprogramming efficiency.
Figure 2.
Bromodomain inhibition and degradation of BRD9 facilitate reprogramming
(A) Chemical structures of LP99, BI-7273, and I-BRD9.
(B) Fold change in TRA-1-60-positive colonies with compounds compared with DMSO. Representative well images are shown above. Bar graphs show the mean and error bars represent SEM. n = 5, independent experiments with technical triplicates. p values are calculated by one-sample t test for mu = 1.
(C) Fold change in TRA-1-60-positive colonies upon I-BRD9, EPZ00477 (DOT1L inhibitor), and their combination treatment to DMSO. Representative well images are above the graph. Bar graphs show the mean and error bars represent SEM. n = 5, independent experiments. p values for DMSO comparisons are calculated by one-sample t test for mu = 1.
(D) Chemical structure of dBRD9 (top), western blots for BRD9 and BRD7 at indicated time points after treatment with 0.3 μM dBRD9 (bottom). HDAC1 serves as loading control for nuclear lysates.
(E) Fold change in TRA-1-60-positive colonies with dBRD9 compared with DMSO. Representative well images are above the graph. Bar graphs show the mean and error bars represent SEM. n = 3, independent experiments. p values are calculated by one-sample t test for mu = 1.
(F) NANOG-positive colonies generated with indicated chemicals. Representative well and colony images are shown above the graph. DAPI was used to stain the nuclei. Scale bars denote 200 μm. Bar graphs show the mean and error bars represent SEM. n = 3, independent experiments with technical triplicates.
(G) Schematic depicting the time intervals for treatment of compounds during reprogramming (top). Fold change in the number of TRA-1-60-positive colonies upon inhibitor treatment between indicated days of reprogramming (bottom). Bar graphs show the mean and error bars represent SE of mean. n = 4, independent experiments for I-BRD9 treatment. n = 3, independent experiments for dBRD9 treatment. p values are calculated by one-sample t test for mu = 1. Dotted line indicates a fold change of 1 compared with respective controls.
To understand how BRD9 blocks iPSC generation, we determined when in the reprogramming process its inhibition has the maximal effect. I-BRD9 and dBRD9 had the most effect on reprogramming efficiency when applied during the first 6 days after OSKM expression (Figures 2G and S2G). As there were no further increases in the number of iPSCs with longer periods of treatments, we concluded that BRD9 is a barrier for the initial stage of reprogramming. In addition, the percentage of emerging TRA-1-60-positive cells on day 6, as assessed by flow cytometry, was significantly higher in BRD9 inhibitor-treated cultures compared with controls (Figure S2H). Taken together, these results indicate that the initial stage of reprogramming is most sensitive to BRD9 inhibition and that even a transient BRD9 inhibition is sufficient to increase the efficiency of human iPSC generation.
BRD9 inhibition and degradation enable iPSC generation without KLF4 and c-MYC
To investigate if BRD9 inhibition can enable human iPSC generation with fewer Yamanaka factors, we carried out reprogramming with only OSK or OS. In both cases, reprogramming efficiency was increased with BRD9 inhibition (Figures 3A and 3B). PCR with vector-specific primers validated the absence of KLF4 and MYC transgenes in genomic DNAs of iPSCs derived from OS-transduced fibroblasts (Figure 3C). OS-iPSCs could be stably propagated and exhibited pluripotency characteristics such as silencing of retroviral transgenes, expression of pluripotency markers such as OCT4, SSEA4, and NANOG, and ability to form teratomas containing differentiated cells from all three germ layers (Figures 3D–3F). These results show that BRD9 inhibition can enable iPSC generation with fewer exogenous factors.
Figure 3.
BRD9 inhibition and degradation enable iPSC generation without KLF4 and c-MYC
(A) Fold change in TRA-1-60-positive colonies generated by OSK in the presence of DMSO, I-BRD9, or dBRD9. Representative well images are above the graph. Bar graphs show the mean and error bars represent SE of mean. n = 5, independent experiments. p values are calculated by one-sample t test for mu = 1.
(B) TRA-1-60 positive colonies generated by OCT4 and SOX2 (OS) in the presence of DMSO, I-BRD9, or dBRD9. Representative well images are above the graph. n = 5, independent experiments with at least technical triplicates. p values are calculated by two sample t test.
(C) Agarose gel images of PCR products for exogenous OCT4, SOX2, KLF4, and c-MYC transgenes in iPSCs generated from fibroblasts.
(D) Phase contrast and GFP fluorescence images of colonies (marked by dotted lines) derived by OS and dBRD9 (upper) or I-BRD9 (lower) treatments showing typical iPSC morphology and silencing of retroviral GFP transgene. Scale bars, 100 μm.
(E) OCT4, SSEA4 (left), and NANOG (right) immunofluorescence of iPSCs derived from OS-expressing fibroblasts. Hoechst 33324 was used to stain the nuclei. Scale bars, 300 μm.
(F) Hematoxylin and eosin-stained sections of teratomas of iPSCs derived from OS-expressing fibroblasts treated with dBRD9 or I-BRD9 show tissues from endoderm, ectoderm, and mesoderm lineages. Scale bars, 100 μm.
BRD9 has state-specific roles in pluripotency induction
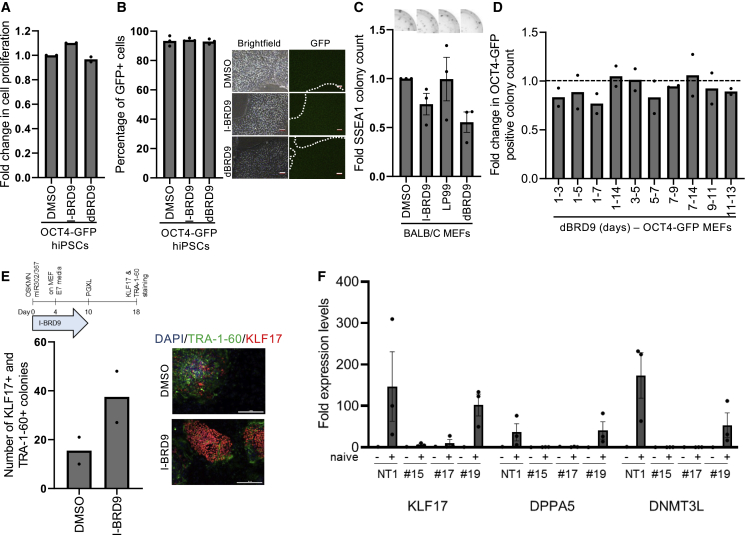
While small molecules allow for transient inhibition or degradation of BRD9 during reprogramming, single guide RNAs (sgRNAs) may result in a permanent knockout in the resulting iPSCs. iPSCs generated from fibroblasts expressing BRD9 sgRNAs were expanded in culture (Figure S3A). Nine iPSC lines out of 14 did not express BRD9 at all, suggesting a homozygous knockout. Four iPSC lines had reduced protein expression suggestive of a heterozygous knockout and one iPSC colony retained wild-type levels of BRD9 (Figure S3B). Complete knockout clones robustly expressed OCT4, NANOG, and SSEA4 similar to control iPSC lines (Figure S3C). To further characterize the pluripotency of BRD9 knockout human iPSCs, we investigated their differentiation capacity. BRD9-knockout iPSCs were able to form teratomas containing cells from all three germ layers (Figure S3D). These results suggest that BRD9 is dispensable for human primed pluripotency acquisition.
Naive mouse ESCs are sensitive to BRD9 bromodomain inhibition (Gatchalian et al., 2018), therefore we investigated the role of BRD9 in human pluripotent stem cells. BRD9 inhibitor or degrader treatment of primed human iPSCs did not change the proliferation rate nor the percentage of OCT4-positive cells compared with controls (Figures 4A and 4B). These results, in combination with the knockout iPSC lines, suggest that BRD9 inhibition does not abrogate human primed pluripotency despite negatively affecting naive pluripotency in the mouse. To specifically examine if the role of BRD9 in pluripotency acquisition is state-specific, we reprogrammed mouse embryonic fibroblasts in the presence of BRD9 inhibitors. BRD9 inhibitors did not increase murine somatic cell reprogramming; in fact, we observed a modest decrease in efficiency with the degrader (Figure 4C). A time-course experiment using the dBRD9 on Oct4-GFP MEFs also indicated no difference in the generation of Oct4-positive mouse iPSCs (Figure 4D).
Figure 4.
BRD9 has species-specific roles in acquisition of pluripotency
(A) Cell proliferation analysis upon 2-day treatment of OCT4-GFP reporter iPSCs with indicated small molecules. Bar graphs show the mean. n = 2, independent experiments with technical triplicates.
(B) Percentage of GFP-positive cells in OCT4-GFP reporter hiPSCs upon indicated treatments (left). Bar graphs show the mean. n = 3, independent experiments. Brightfield and GFP fluorescence images of OCT4-GFP reporter hiPSCs (marked by dotted lines) after 2-day treatment (right). Scale bars, 100 μm.
(C) Fold change in the number of SSEA1-positive colonies generated from MEFs. Representative well images are above the graph. Bar graphs show the mean and error bars represent SEM. n = 3, independent experiments.
(D) Fold change in the number of Oct4-GFP-positive colonies generated with dBRD9 at indicated time intervals compared with DMSO. Dotted line indicates a fold change of 1 compared with respective controls. Bar graphs show the mean. n = 2, independent experiments.
(E) Schematic depicting the timeline for naive reprogramming (left). Number of TRA-1-60 and KLF17-positive colonies generated with DMSO and I-BRD9 treatment (right). Bar graphs show the mean. n = 2, independent experiments. Representative colony images stained with TRA-1-60 (green) and KLF17 (red) are shown above the graph. DAPI was used to stain the nuclei. Scale bars, 200 μm.
(F) Expression levels of naive pluripotency markers before and after naive conversion of BRD9 KO primed iPSCs. Bar graphs show the mean and error bars indicate the SEM. n = 3, independent experiments.
To investigate the effects of transient BRD9 inhibition during reprogramming of human somatic cells to a naive-like state, we transduced fibroblasts with lentiviruses expressing OSKM, mir302/367 and NANOG and subsequently cultured them in PGXL medium (Bredenkamp et al., 2019). Transient BRD9 inhibition during reprogramming did not block the generation of TRA-1-60 and KLF17-positive naive-like human iPSCs (Figure 4E). To assess if BRD9 is required for maintenance of the naive-like state in human cells, we cultured control and BRD9 knockout primed iPSCs under naive conversion conditions and monitored the induction of naive markers KLF17, DNMT3L, and DPPA5. While control primed iPSCs robustly activated the naive marker genes, three independent BRD9 KO lines failed to do so (Figure 4F). This result suggests that, in contrast to primed cells, naive human PSCs require BRD9 expression.
BRD9 maintains somatic-specific gene expression and enhancer accessibility
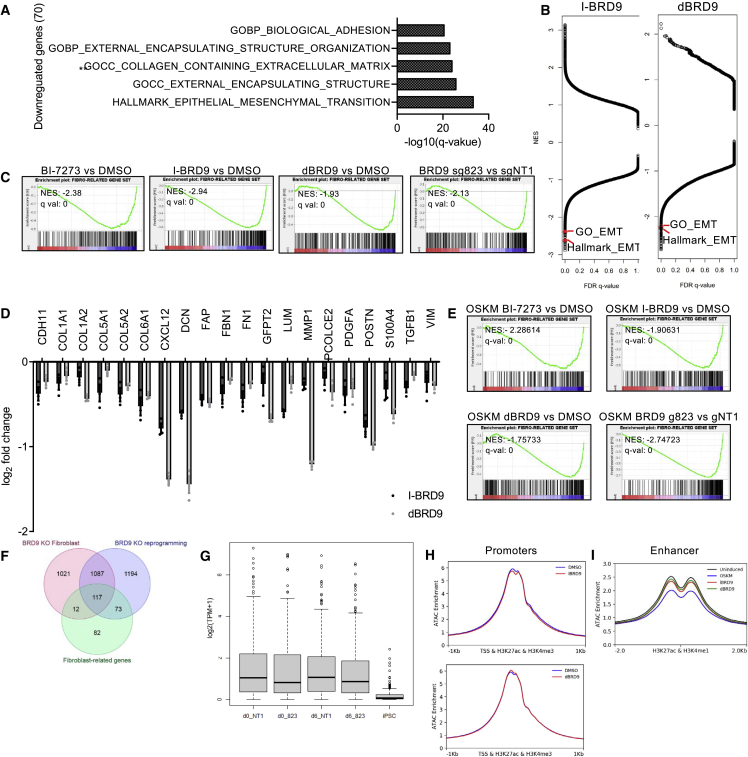
Given that BRD9 inhibition is most effective in early reprogramming, we next sought to identify the transcriptional effects elicited by different modes of BRD9 inhibition. We performed mRNA-sequencing from fibroblasts treated with BI-7273, I-BRD9, and dBRD9, as well as those expressing Cas9 and BRD9 sgRNA. BRD9 inhibitors differentially downregulated 928, 170, and 577 genes (dBRD9, I-BRD9, and BI-7273, respectively), of which 70 were common to all treatments (Figures S4A and S4B). In contrast, 22 genes were commonly upregulated with BRD9 inhibitors (Figure S4B). Cas9 and sgBRD9 expression resulted in greater numbers of differentially expressed genes (2,115 up, 2,237 down), but a majority of the commonly downregulated genes with inhibitors were also downregulated by the knockout (48 of 70 genes). Gene ontology (GO) analysis of this common set of downregulated genes revealed that they were highly enriched in cellular process linked to fibroblast identity and function, such as epithelial-to-mesenchymal transition (EMT), extracellular matrix components, and adhesion (Figure 5A). More broadly, Gene Set Enrichment Analysis (GSEA) indicated EMT gene sets were among the topmost negatively regulated gene sets upon I-BRD9 and dBRD9 treatments (Figure 5B). This finding led us to ask specifically whether the fibroblast expression program as a whole was downregulated upon BRD9 inhibition. GSEA revealed that all BRD9 perturbations resulted in a highly significant downregulation of the fibroblast-related gene set (Ebrahimi et al., 2019) (Figure 5C). A total of 117 out of the 307 fibroblast-related genes were significantly downregulated by at least one BRD9 inhibitor and the commonly downregulated genes had significant overlap with the fibroblast-related gene set (26 of 70; Hypergeometric p value = 4.1e-31). Single-cell RNA-sequencing (RNA-seq) studies have identified genes such as DCN and POSTN, which are commonly expressed by fibroblasts of various origin (Deng et al., 2021; Muhl et al., 2020; Vorstandlechner et al., 2020). Examination of our RNA-seq data revealed that a majority of such fibroblast-related genes were downregulated by BRD9 inhibition (Figures 5D and S4C). In contrast, we did not observe positive enrichment of pluripotency-associated gene sets with any BRD9 perturbation, indicating that BRD9 inhibition on its own does not activate the pluripotency network (Figure S4D). To understand if BRD9 inhibition suppresses fibroblast-related genes during reprogramming, we performed RNA-seq on day 6 post-OSKM. Inhibition of BRD9 during reprogramming resulted in significant negative enrichment of fibroblast-related gene set (Figures 5E–5G). Long-term treatment of fibroblasts with inhibitors did not affect fibroblast cell morphology but slightly decreased cell proliferation (Figures S4E and S4F). In addition, reprogramming of human iPSC-derived motor neural progenitors with dBRD9 resulted in 2.5-fold greater efficiency, suggesting that BRD9’s barrier role in pluripotency induction is independent of the starting somatic cell type (Figure S4G). Taken together, these analyses suggest that BRD9 acts as a barrier to reprogramming at a transcriptional level by sustaining starting cell-type-specific gene expression.
Figure 5.
BRD9 maintains fibroblast-related gene expression and enhancer accessibility
(A) Top five GO and Hallmark gene sets enriched in common downregulated genes with dBRD9, BI-7273, and I-BRD9. Number of genes (n) in comparison were 70. p values were calculated by hypergeometric distribution.
(B) GSEA on pre-ranked gene lists according to log2FC value for comparisons of fibroblasts treated with I-BRD9 and DMSO (left) and dBRD9 and DMSO (right) for all genesets available at The Molecular Signatures Database (MSigDB). Red circles indicate Hallmark_EMT and GO_EMT gene sets.
(C) GSEA of indicated treatments for the fibroblast-related gene set. NES: normalized enrichment score; q val: False discovery rate q-value.
(D) log2 fold change in expression levels of fibroblast marker genes. Bar graphs show the mean and n = 3, independent experiments.
(E) GSEA of reprogramming cells at day 6 for the fibroblast-related gene set.
(F) Venn diagram showing intersection of fibroblast-related genes with downregulated genes upon BRD9 knockout (KO) in both fibroblasts and reprogramming cells (day 6).
(G) Average expression levels of fibroblast-related genes upon BRD9 KO in fibroblasts and reprogramming cells (day 6).
(H) Aggregate ATAC-seq plots from fibroblasts treated with I-BRD9 (top), dBRD9 (bottom), and DMSO on the +/−1 kb of transcription start sites of all fibroblast H3K27ac and H3K4me3 summits. n = 3, independent experiments.
(I) Aggregate ATAC-seq plots from fibroblasts expressing OSKM (day 6) or treated with I-BRD9, dBRD9 and DMSO around +/−2kb of all fibroblast H3K27ac and H3K4me1 summits. n = 3, independent experiments.
To gain insight into how BRD9 functions to maintain expression of somatic-specific genes, we performed Assay for Transposase-Accessible Chromatin using sequencing (ATAC-seq) in fibroblasts treated with I-BRD9 and dBRD9. Both inhibitors had no effect on accessible chromatin around promoters marked by overlap of H3K27ac and H3K4me3 (Figure 5H). However, BRD9 inhibitors reduced the chromatin accessibility around putative active enhancers as marked by overlap of H3K27ac with H3K4me1 (Figure 5I). Importantly, fibroblast-related active enhancers start to lose accessibility upon OSKM expression, suggesting that BRD9 inhibition augments this process (Figure 5I) (Li et al., 2017). The loss of accessibility was more pronounced at enhancers, but not promoters, of genes downregulated by BRD9 inhibition (Figure S4H). These results indicate that BRD9 constitutes a barrier to reprogramming by maintaining accessibility of active enhancers in the starting cell populations.
MN1 and ZBTB38 are putative BRD9 target genes that suppress reprogramming
Among the most consistently downregulated genes upon BRD9 inhibition were transcriptional regulators MN1 and ZBTB38 (Figure 6A). MN1 has not been implicated in reprogramming, but regulates palate development (Mak et al., 2020) and can act as co-factor for various transcription factors, such as retinoic acid receptor/retinoic X receptor (RAR/RXR) (Meester-Smoor et al., 2008). It is also implicated in transcriptional control of leukemic transformation in collaboration with DOT1L (Riedel et al., 2016). ZBTB38 is predicted to be a master regulator in fibroblasts and is controlled by a fibroblast-specific super-enhancer (Hnisz et al., 2013). We hypothesized that downregulation of these two factors soon after OSKM expression is necessary for efficient reprogramming. To test this notion, we overexpressed MN1 or ZBTB38 at a relatively low level along with OSKM, which resulted in a 40% decrease in reprogramming efficiency (Figures 6B and 6C). This result suggested that these two factors may themselves obstruct reprogramming. Interestingly, experimental downregulation of ZBTB38 has recently been shown to increase reprogramming (Mellis et al., 2021). To corroborate this finding and investigate whether the same holds true for MN1, we lowered the expression levels of these two factors in fibroblasts using shRNAs and assessed reprogramming efficiency. Downregulation of MN1 or ZBTB38 resulted in 30% to 40% more iPSCs compared with controls (Figures 6D and 6E). These results suggest that BRD9 may obstruct reprogramming in part by sustaining the expression these two transcriptional regulators (Figure 6F).
Figure 6.
BRD9-regulated MN1 and ZBTB38 act as barrier to reprogramming
(A) Expression of MN1 and ZBTB38 in TPM across different treatments in fibroblasts (left) and for BRD9 sgRNA and control sgRNA expression (right). Bar graphs show the mean and error bars represent SE of mean. n = 3, independent experiments. p values were calculated by two-sample t test.
(B) Fold change in relative MN1 and ZBTB38 mRNA levels upon their overexpression in fibroblasts compared with empty vector controls. n = 2, technical replicates.
(C) TRA-1-60-positive colonies upon overexpression of MN1 and ZBTB38. Representative well images are above the graph. Bar graphs show the mean. n = 2, independent experiments with technical duplicates for MN1 and ZBTB38 and quadruplicate for empty vector.
(D) Fold change in relative MN1 and ZBTB38 mRNA levels upon shRNA transduction compared with non-targeting shFF controls. n = 2, technical replicates.
(E) TRA-1-60-positive colonies generated upon shRNA expression targeting MN1 and ZBTB38. Representative well images are above the graph. Bar graphs show the mean. n = 2, independent experiments with technical duplicates.
(F) Model for BRD9-containing BAF complex’s role in human somatic cell reprogramming.
Discussion
In this study, we investigated the role of BRD9 in human somatic cell reprogramming via a combination of genetic and chemical perturbation approaches. Knockdown or knockout BRD9 increased human reprogramming efficiency. Importantly, the contrasting effects of BRD9 and BRD7 inhibition on reprograming efficiency indicate that the recently identified BRD9-containing ncBAF, but not BRD7-containing PBAF, is a barrier to reprogramming. This notion is supported by our finding that loss of an additional specific member of ncBAF complex, GLTSCR1, has a similar positive effect on iPSC formation. To acutely block BRD9 function, we took advantage of selective bromodomain inhibitors and a PROTAC degrader, all of which significantly increased reprogramming efficiency and enabled iPSC generation in the absence of KLF4 and cMYC. Taken together, these findings demonstrate that BRD9-containing ncBAF complexes act as a barrier to reprogramming.
Notably, we observed that BRD9 is dispensable for induction of human primed pluripotency. BRD9 knockout fibroblast could efficiently generate iPSCs that could be propagated and expanded while retaining canonical properties of primed pluripotent stem cells. This is in contrast to the non-BRD9 containing ES-specific BAF complexes that are required for pluripotency and self-renewal of mouse ESCs (Ho et al., 2009). Naive mESCs have been shown to lose self-renewal capacity and enter into a primed, epiblast-like transcriptional state upon BRD9 inhibition (Gatchalian et al., 2018). Human iPSCs are considered to be in a primed state, and our data indicate that BRD9 is dispensable for their maintenance. It is therefore likely that BAF complexes other than ncBAF support human primed pluripotency (Huang et al., 2021). Murine reprogramming was not enhanced upon BRD9 inhibition nor degradation in two different MEF reprogramming systems. This is consistent with a recent study in which I-BRD9 was found not to increase reprogramming efficiency (Janiszewski et al., 2019). Collectively, these observations point to distinct roles for BRD9 in somatic cell reprogramming and pluripotency maintenance.
BRD9 inhibition leads to a broad downregulation of fibroblast-enriched genes accompanied by decreased chromatin accessibility across putative active enhancers. Previous studies show BRD9 occupies distal enhancers (Bell et al., 2019; Del Gaudio et al., 2019) and co-localizes with CTCF (Inoue et al., 2019; Michel et al., 2018) and is important for maintaining cell-type-specific transcription programs (Loo et al., 2020). Genes downregulated by BRD9 inhibition in fibroblasts are specific to this cell type, such as collagens and ECM components. Downregulation of such genes does not cause major defects in cell viability, suggesting that interfering with ncBAF can downregulate cell-type-specific genes to an extent that will facilitate reprogramming while avoiding cell death that may result from a more extensive shut down of cell identity. Despite our extensive efforts to perform BRD9 chromatin immunoprecipitation sequencing (ChIP-Seq) to understand whether regulatory elements of target genes are occupied by BRD9 and ncBAF, we were unable to do so. As there are no publicly available data regarding BRD9 ChIP-Seq in human fibroblasts, it remains to be investigated whether the genes identified herein are directly bound by BRD9. Nonetheless, we transcriptionally identified several key transcription factors such as MN1 and ZBTB38 as putative BRD9 targets. Our functional data collectively establish these putative BRD9-regulated genes as important barriers to reprogramming.
The present findings add BRD9 inhibitors to the arsenal of small molecule inhibitors that can be used to regulate and direct cell fate changes. We also show that BRD9 inhibition can be combined with other modulators such as DOT1L inhibitors to boost reprogramming efficiency. Importantly, inhibition of DOT1L-mediated H3K79 methylation facilitates the generation of chemically induced pluripotent stem cells (ciPSCs) from both mouse and human cells (Guan et al., 2022; Zhao et al., 2015) and results in a permissive epigenome state that enables reprogramming by alternative transcription factors (Kim et al., 2020). Interestingly, DOT1L and BRD9 seem to regulate largely non-overlapping sets of genes (Onder et al., 2012). Identification of BRD9 as a mechanism that sustains cell identity-related expression programs suggests that combinatorial perturbations, which include BRD9 inhibitors, can enhance various reprogramming methods.
Experimental procedures
Resource availability
Corresponding author
Further information and requests for resources and reagents should be directed to and will be fulfilled by the corresponding author, Tamer Onder (tonder@ku.edu.tr).
Materials availability
Unique materials generated in this study are available from the lead contact with a completed Materials Transfer Agreement. Stable reagents generated in this study are available from the lead contact without restriction.
Data and code availability
RNA-seq and ATAC-seq data are deposited to the NCBI GEO database with the accession number GSE161640. This study did not generate any novel codes but used previously published methods.
Reprogramming assays
Reprogramming assays were performed as described previously (Ebrahimi et al., 2019; Park et al., 2008). Unless stated otherwise, I-BRD9, LP99, BI-7273, and dBRD9 were used at final concentrations of 1, 3, 1, and 0.3 μM, respectively, for the first 2 weeks of reprogramming. EPZ-004777 was used at final concentration of 3 μM.
Cloning
sgRNA and shRNA oligonucleotides (Table S1) were cloned into lentiCRISPR v2 (Addgene #52961) and into pSMP vector (Addgene #36394), respectively, as previously described (Onder et al., 2012). ZBTB38 expression vector in pLenti6.2/V5-DEST was obtained from DNASU Plasmid Repository (Clone IDs: HsCD00436332). MSCV-MN1 plasmid was a gift from Kathrin Bernt (University of Pennsylvania).
Virus production and transduction
A total of 2.5 × 106 293T cells were transfected with 2.5 μg viral vector, 0.25 μg pCMV-VSV-G (Addgene #8454), 2.25 μg psPAX2 (Addgene #12260) for lentivirus or pUMVC (Addgene #8449) for retroviruses using 20 μL FuGENE 6 (Promega) in 400 μL DMEM per plate. Supernatants were collected 48 h and 72 h post-transfection and filtered. Fibroblasts were doubly transduced in consecutive days in the presence of 8 μg/mL protamine sulfate (Sigma) and selected by puromycin at 1 μg/mL.
Western blot
Cell pellets were resuspended in cytosolic lysis buffer (10 mM HEPES pH 7.9, 10 mM KCl, 0.1 mM EDTA, 0.4% NP-40, Protease Inhibitor [1X] [Roche]) and shaken on ice for 15 min. After centrifugation at 3,000g for 3 min, pellets were washed once with cytosolic buffer and resuspended in nuclear lysis buffer (20 mM HEPES pH 7.9, 0.4M NaCl, 1 mM EDTA, 10% Glycerol, Protease Inhibitor [1X] [Roche]) and sonicated. Membranes were incubated with BRD9 antibody (Active Motif, Catalog: 61,537) at 1:1,000, BRD7 antibody (Cell Signaling, D9K2T) at 1:1,000, and HDAC1 antibody (Santa Cruz, sc-7872) at 1:500 overnight at 4°C.
Quantitative RT-PCR analyses
qPCR assays were performed as described previously (Ebrahimi et al., 2019) with the indicated primers (Table S1).
Immunostaining
TRA-1-60 staining was performed as previously described (Onder et al., 2012) and anti-SSEA1 antibody (Biolegend, 125604) was used to quantify MEF reprogramming. For immunofluorescence-based characterization of iPSCs, cells were passaged onto mitomycin-C-treated MEFs in hESC medium as previously described (Ebrahimi et al., 2019). The antibodies used were KLF17 (Atlas, HPA024629), OCT4 (Abcam, ab19857), SSEA4/A647 (BD, 560218), and NANOG (Abcam, ab21624).
RNA-seq and analysis
Fibroblasts and OSKM transduced cells were treated for 5 days with DMSO, BI-7273 (1 μM), I-BRD9 (1 μM), and dBRD9 (0.3 μM). Total RNA was prepared using Direct-zol kit according to manufacturer’s instructions (Zymo Research). Libraries were prepared using NEBNext Poly(A) mRNA Magnetic Isolation Module from the NEBNext ultra-directional RNA kit to create a first stranded library. Reads were mapped to hg19 built-in genome by HISAT2 after assessing their quality by FastQC. DESeq2 package was used to identify differentially expressed genes between samples. Genes were considered as differentially regulated based on adjusted p value <0.05. Rank-ordered gene lists were used for GSEA (Subramanian et al., 2005). A list of genes that are differentially downregulated upon at least 2 BRD9 compound treatment is provided (Table S2).
ATAC-seq and analysis
ATAC-seq was performed using 100,000 cells as described previously (Buenrostro et al., 2013) using in-house-produced Tn5 transposase. Samples were purified using the GeneJET PCR purification kit (Thermo). PCR amplification was performed using the following protocol: 3 min at 72°C, 30 s at 98°C, and 11 cycles of 10 s at 98°C, 30 s at 63°C, and 3 min at 72°C. Samples were run on a Tapestation (Agilent) to determine library size and quantification before paired-end (2 × 41 base pair) sequencing on a NextSeq 500 (Illumina). Sequence reads were quality controlled with FastQC and mapped to the most recent human reference genome (hg38) using bowtie (Langmead et al., 2009), allowing a maximum of two mismatches and only uniquely mapped reads. Reads with mapping quality score below 30 and blacklisted regions were filtered. DeepTools (Ramírez et al., 2016) ComputeMatrix and plotProfile commands were used to generate aggregate ATAC plots. For this, BigWig files were generated using deepTools bamCoverage command, eliminating duplicates and normalizing by sequencing depth and effective genome size. Intersecting regions among fibroblast ChIP peaks for H3K27ac, H3K4me1, and H3K4me3 were identified using previously published fibroblast ChIP-seq data (Ebrahimi et al., 2019), calling the peaks using macs2 (Zhang et al., 2008) with the parameters --keep-dup 1 and –broad, and finding the intersecting regions using bedtools (Quinlan and Hall, 2010) intersect.
Statistical analysis
Statistical analysis was performed using GraphPad Prism 8 for t tests or R 4.0.2 for Wilcoxon-signed rank test. The test details, number of independent experiments, and exact p values were provided in related figure legends.
Author contributions
Conceptualization: K.S., G.G.S., U.O., T.T.O. Software: A.P.C. Formal analysis: K.S., G.G.S., A.D.C., A.P.C., T.T.O. Investigation: K.S., G.G.S., M.P., S.K., A.P.C., H.C., A.Y., D.A., A.B.Y., E.S.A., D.H.A., J.E.D. Resources: A.P.C., D.A., L.H.S., J.Q. Data Curation: K.S., A.P.C. Writing—original draft: K.S., G.G.S., T.T.O. Writing—review & editing: K.S., G.G.S., U.O., T.T.O. Supervision: J.Q., U.O., T.T.O. Funding acquisition: U.O., T.T.O.
Acknowledgments
We thank Ahmet Kocabay and Ali Cihan Taşkın for help with mouse experiments, and Arzu Ruacan (Koç University, School of Medicine, Department of Pathology) for examination of histological sections. The authors gratefully acknowledge use of the services and facilities of the Koç University Research Center for Translational Medicine (KUTTAM), funded by the Republic of Turkey Ministry of Development. K.S. and D.H.A. were supported by TUBITAK BIDEB Scholarship. Funding: Newton Advanced Fellowship (T.T.O.), TUBITAK Projects 231S182 and 219Z209 (T.T.O.), Arthritis Research UK, program grant 20522 (U.O.), Cancer Research UK (U.O.), LEAN project of the Leducq Foundation (U.O.), People Program Marie Curie Actions of the European Union’s Seventh Framework Program FP7/2007–2013 under REA grant agreement no. 609305 (U.O.).
CONFLICT OF INTERESTS
The authors declare no competing interests.
Published: November 3, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stemcr.2022.10.005.
Supplemental information
References
- Alpsoy A., Dykhuizen E.C. Glioma tumor suppressor candidate region gene 1 (GLTSCR1) and its paralog GLTSCR1-like form SWI/SNF chromatin remodeling subcomplexes. J. Biol. Chem. 2018;293:3892–3903. doi: 10.1074/jbc.RA117.001065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balboa D., Weltner J., Novik Y., Eurola S., Wartiovaara K., Otonkoski T. Generation of an OCT4 reporter human induced pluripotent stem cell line using CRISPR/SpCas9. Stem Cell Res. 2017;23:105–108. doi: 10.1016/j.scr.2017.07.006. [DOI] [PubMed] [Google Scholar]
- Bell C.M., Raffeiner P., Hart J.R., Vogt P.K. PIK3CA cooperates with KRAS to promote MYC activity and tumorigenesis via the bromodomain protein BRD9. Cancers. 2019;11:E1634. doi: 10.3390/cancers11111634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredenkamp N., Yang J., Clarke J., Stirparo G.G., von Meyenn F., Dietmann S., Baker D., Drummond R., Ren Y., Li D., et al. Wnt inhibition facilitates RNA-mediated reprogramming of human somatic cells to naive pluripotency. Stem Cell Rep. 2019;13:1083–1098. doi: 10.1016/j.stemcr.2019.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumbaugh J., Di Stefano B., Hochedlinger K. Reprogramming: identifying the mechanisms that safeguard cell identity. Development. 2019;146:dev182170. doi: 10.1242/dev.182170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro J.D., Giresi P.G., Zaba L.C., Chang H.Y., Greenleaf W.J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheloufi S., Elling U., Hopfgartner B., Jung Y.L., Murn J., Ninova M., Hubmann M., Badeaux A.I., Euong Ang C., Tenen D., et al. The histone chaperone CAF-1 safeguards somatic cell identity. Nature. 2015;528:218–224. doi: 10.1038/nature15749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark P.G.K., Vieira L.C.C., Tallant C., Fedorov O., Singleton D.C., Rogers C.M., Monteiro O.P., Bennett J.M., Baronio R., Müller S., et al. LP99: discovery and synthesis of the first selective BRD7/9 bromodomain inhibitor. Angew. Chem. Int. Ed. Engl. 2015;54:6217–6221. doi: 10.1002/anie.201501394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cossec J.-C., Theurillat I., Chica C., Búa Aguín S., Gaume X., Andrieux A., Iturbide A., Jouvion G., Li H., Bossis G., et al. SUMO safeguards somatic and pluripotent cell identities by enforcing distinct chromatin states. Cell Stem Cell. 2018;23:742–757.e8. doi: 10.1016/j.stem.2018.10.001. [DOI] [PubMed] [Google Scholar]
- Deng C.C., Hu Y.F., Zhu D.H., Cheng Q., Gu J.J., Feng Q.L., Zhang L.X., Xu Y.P., Wang D., Rong Z., Yang B. Single-cell RNA-seq reveals fibroblast heterogeneity and increased mesenchymal fibroblasts in human fibrotic skin diseases. Nat. Commun. 2021;12:3709–3716. doi: 10.1038/s41467-021-24110-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- dos Santos R.L., Tosti L., Radzisheuskaya A., Caballero I.M., Kaji K., Hendrich B., Silva J.C.R. MBD3/NuRD facilitates induction of pluripotency in a context-dependent manner. Cell Stem Cell. 2014;15:102–110. doi: 10.1016/j.stem.2014.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebrahimi A., Sevinç K., Gürhan Sevinç G., Cribbs A.P., Philpott M., Uyulur F., Morova T., Dunford J.E., Göklemez S., Arı Ş., et al. Bromodomain inhibition of the coactivators CBP/EP300 facilitate cellular reprogramming. Nat. Chem. Biol. 2019;15:519–528. doi: 10.1038/s41589-019-0264-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatchalian J., Malik S., Ho J., Lee D.-S., Kelso T.W.R., Shokhirev M.N., Dixon J.R., Hargreaves D.C. A non-canonical BRD9-containing BAF chromatin remodeling complex regulates naive pluripotency in mouse embryonic stem cells. Nat. Commun. 2018;9:5139. doi: 10.1038/s41467-018-07528-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Gaudio N., Di Costanzo A., Liu N.Q., Conte L., Migliaccio A., Vermeulen M., Martens J.H.A., Stunnenberg H.G., Nebbioso A., Altucci L. BRD9 binds cell type-specific chromatin regions regulating leukemic cell survival via STAT5 inhibition. Cell Death Dis. 2019;10:338. doi: 10.1038/s41419-019-1570-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan J., Wang G., Wang J., Zhang Z., Fu Y., Cheng L., Meng G., Lyu Y., Zhu J., Li Y., et al. Chemical reprogramming of human somatic cells to pluripotent stem cells. Nature. 2022;605:325–331. doi: 10.1038/s41586-022-04593-5. [DOI] [PubMed] [Google Scholar]
- Hnisz D., Abraham B.J., Lee T.I., Lau A., Saint-André V., Sigova A.A., Hoke H.A., Young R.A., HniszBrian D. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L., Ronan J.L., Wu J., Staahl B.T., Chen L., Kuo A., Lessard J., Nesvizhskii A.I., Ranish J., Crabtree G.R. An embryonic stem cell chromatin remodeling complex, esBAF, is essential for embryonic stem cell self-renewal and pluripotency. Proc. Natl. Acad. Sci. USA. 2009;106:5181–5186. doi: 10.1073/pnas.0812889106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L., Miller E.L., Ronan J.L., Ho W.Q., Jothi R., Crabtree G.R. EsBAF facilitates pluripotency by conditioning the genome for LIF/STAT3 signalling and by regulating polycomb function. Nat. Cell Biol. 2011;13:903–913. doi: 10.1038/ncb2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges C., Kirkland J.G., Crabtree G.R. The many roles of BAF (mSWI/SNF) and PBAF complexes in cancer. Cold Spring Harb. Perspect. Med. 2016;6:a026930. doi: 10.1101/cshperspect.a026930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hota S.K., Bruneau B.G. ATP-dependent chromatin remodeling during mammalian development. Development. 2016;143:2882–2897. doi: 10.1242/dev.128892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Park K.-M., Gontarz P., Zhang B., Pan J., McKenzie Z., Fischer L.A., Dong C., Dietmann S., Xing X., et al. OCT4 cooperates with distinct ATP-dependent chromatin remodelers in naïve and primed pluripotent states in human. Nat. Commun. 2021;12:5123. doi: 10.1038/s41467-021-25107-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue D., Chew G.-L., Liu B., Michel B.C., Pangallo J., D’Avino A.R., Hitchman T., North K., Lee S.C.-W., Bitner L., et al. Spliceosomal disruption of the non-canonical BAF complex in cancer. Nature. 2019;574:432–436. doi: 10.1038/s41586-019-1646-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janiszewski A., Talon I., Chappell J., Collombet S., Song J., De Geest N., To S.K., Bervoets G., Marin-Bejar O., Provenzano C., et al. Dynamic reversal of random X-Chromosome inactivation during iPSC reprogramming. Genome Res. 2019;29:1659–1672. doi: 10.1101/gr.249706.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Z., Tang Y., Zhao X., Zhang M., Donovan D.M., Tian X.C. Knockdown of Brm and Baf170, components of chromatin remodeling complex, facilitates reprogramming of somatic cells. Stem Cells Dev. 2015;24:2328–2336. doi: 10.1089/scd.2015.0069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadoch C., Williams R.T., Calarco J.P., Miller E.L., Weber C.M., Braun S.M.G., Pulice J.L., Chory E.J., Crabtree G.R. Dynamics of BAF–Polycomb complex opposition on heterochromatin in normal and oncogenic states. Nat. Genet. 2017;49:213–222. doi: 10.1038/ng.3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K.P., Choi J., Yoon J., Bruder J.M., Shin B., Kim J., Arauzo-Bravo M.J., Han D., Wu G., Han D.W., et al. Permissive epigenomes endow reprogramming competence to transcriptional regulators. Nat. Chem. Biol. 2020;17:47–56. doi: 10.1038/s41589-020-0618-6. [DOI] [PubMed] [Google Scholar]
- Kolundzic E., Ofenbauer A., Bulut S.I., Uyar B., Baytek G., Sommermeier A., Seelk S., He M., Hirsekorn A., Vucicevic D., et al. FACT sets a barrier for cell fate reprogramming in Caenorhabditis elegans and human cells. Dev. Cell. 2018;46:611–626.e12. doi: 10.1016/j.devcel.2018.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Liu J., Yang X., Zhou C., Guo J., Wu C., Qin Y., Guo L., He J., Yu S., et al. Chromatin accessibility dynamics during iPSC reprogramming. Cell Stem Cell. 2017;21:819–833.e6. doi: 10.1016/j.stem.2017.10.012. [DOI] [PubMed] [Google Scholar]
- Loo C.-S., Gatchalian J., Liang Y., Leblanc M., Xie M., Ho J., Venkatraghavan B., Hargreaves D.C., Zheng Y. A genome-wide CRISPR screen reveals a role for the non-canonical nucleosome-remodeling BAF complex in Foxp3 expression and regulatory T cell function. Immunity. 2020;53:143–157.e8. doi: 10.1016/j.immuni.2020.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch C.J., Bernad R., Calvo I., Nóbrega-Pereira S., Ruiz S., Ibarz N., Martinez-Val A., Graña-Castro O., Gómez-López G., Andrés-León E., et al. The RNA polymerase II factor RPAP1 is critical for mediator-driven transcription and cell identity. Cell Rep. 2018;22:396–410. doi: 10.1016/j.celrep.2017.12.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak C.C.Y., Doherty D., Lin A.E., Vegas N., Cho M.T., Viot G., Dimartino C., Weisfeld-Adams J.D., Lessel D., Joss S., et al. MN1 C-terminal truncation syndrome is a novel neurodevelopmental and craniofacial disorder with partial rhombencephalosynapsis. Brain. 2020;143:55–68. doi: 10.1093/brain/awz379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin L.J., Koegl M., Bader G., Cockcroft X.-L., Fedorov O., Fiegen D., Gerstberger T., Hofmann M.H., Hohmann A.F., Kessler D., et al. Structure-based design of an in vivo active selective BRD9 inhibitor. J. Med. Chem. 2016;59:4462–4475. doi: 10.1021/acs.jmedchem.5b01865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashtalir N., D’Avino A.R., Michel B.C., Luo J., Pan J., Otto J.E., Zullow H.J., McKenzie Z.M., Kubiak R.L., St Pierre R., et al. Modular organization and assembly of SWI/SNF family chromatin remodeling complexes. Cell. 2018;175:1272–1288.e20. doi: 10.1016/j.cell.2018.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meester-Smoor M.A., Janssen M.J.F.W., Grosveld G.C., de Klein A., van IJcken W.F.J., Douben H., Zwarthoff E.C. MN1 affects expression of genes involved in hematopoiesis and can enhance as well as inhibit RAR/RXR-induced gene expression. Carcinogenesis. 2008;29:2025–2034. doi: 10.1093/carcin/bgn168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellis I.A., Edelstein H.I., Truitt R., Goyal Y., Beck L.E., Symmons O., Dunagin M.C., Linares Saldana R.A., Shah P.P., Pérez-Bermejo J.A., et al. Responsiveness to perturbations is a hallmark of transcription factors that maintain cell identity in vitro. Cell Syst. 2021;12:885–899.e8. doi: 10.1016/j.cels.2021.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel B.C., D’Avino A.R., Cassel S.H., Mashtalir N., McKenzie Z.M., McBride M.J., Valencia A.M., Zhou Q., Bocker M., Soares L.M.M., et al. A non-canonical SWI/SNF complex is a synthetic lethal target in cancers driven by BAF complex perturbation. Nat. Cell Biol. 2018;20:1410–1420. doi: 10.1038/s41556-018-0221-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhl L., Genové G., Leptidis S., Liu J., He L., Mocci G., Sun Y., Gustafsson S., Buyandelger B., Chivukula I.V., et al. Single-cell analysis uncovers fibroblast heterogeneity and criteria for fibroblast and mural cell identification and discrimination. Nat. Commun. 2020;11:3953. doi: 10.1038/s41467-020-17740-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onder T.T., Kara N., Cherry A., Sinha A.U., Zhu N., Bernt K.M., Cahan P., Marcarci B.O., Unternaehrer J., Gupta P.B., et al. Chromatin-modifying enzymes as modulators of reprogramming. Nature. 2012;483:598–602. doi: 10.1038/nature10953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J., Meyers R.M., Michel B.C., Mashtalir N., Sizemore A.E., Wells J.N., Cassel S.H., Vazquez F., Weir B.A., Hahn W.C., et al. Interrogation of mammalian protein complex structure, function, and membership using genome-scale fitness screens. Cell Syst. 2018;6:555–568.e7. doi: 10.1016/j.cels.2018.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp B., Plath K. Epigenetics of reprogramming to induced pluripotency. Cell. 2013;152:1324–1343. doi: 10.1016/j.cell.2013.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park I.H., Zhao R., West J.A., Yabuuchi A., Huo H., Ince T.A., Lerou P.H., Lensch M.W., Daley G.Q. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F., Ryan D.P., Grüning B., Bhardwaj V., Kilpert F., Richter A.S., Heyne S., Dündar F., Manke T. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016;44:W160–W165. doi: 10.1093/nar/gkw257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remillard D., Buckley D.L., Paulk J., Brien G.L., Sonnett M., Seo H.S., Dastjerdi S., Wühr M., Dhe-Paganon S., Armstrong S.A., Bradner J.E. Degradation of the BAF complex factor BRD9 by heterobifunctional ligands. Angew. Chem. Int. Ed. Engl. 2017;56:5738–5743. doi: 10.1002/anie.201611281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedel S.S., Haladyna J.N., Bezzant M., Stevens B., Pollyea D.A., Sinha A.U., Armstrong S.A., Wei Q., Pollock R.M., Daigle S.R., et al. MLL1 and DOT1L cooperate with meningioma-1 to induce acute myeloid leukemia. J. Clin. Invest. 2016;126:1438–1450. doi: 10.1172/JCI80825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Z., Yao C., Khodadadi-Jamayran A., Xu W., Townes T.M., Crowley M.R., Hu K. Reprogramming by de-bookmarking the somatic transcriptional program through targeting of BET bromodomains. Cell Rep. 2016;16:3138–3145. doi: 10.1016/j.celrep.2016.08.060. [DOI] [PubMed] [Google Scholar]
- Singhal N., Graumann J., Wu G., Araúzo-Bravo M.J., Han D.W., Greber B., Gentile L., Mann M., Schöler H.R. Chromatin-remodeling components of the BAF complex facilitate reprogramming. Cell. 2010;141:943–955. doi: 10.1016/j.cell.2010.04.037. [DOI] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mesirov J.P. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Theodoulou N.H., Bamborough P., Bannister A.J., Becher I., Bit R.A., Che K.H., Chung C.W., Dittmann A., Drewes G., Drewry D.H., et al. Discovery of I-BRD9, a selective cell active chemical probe for bromodomain containing protein 9 inhibition. J. Med. Chem. 2016;59:1425–1439. doi: 10.1021/acs.jmedchem.5b00256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorstandlechner V., Laggner M., Kalinina P., Haslik W., Radtke C., Shaw L., Lichtenberger B.M., Tschachler E., Ankersmit H.J., Mildner M. Deciphering the functional heterogeneity of skin fibroblasts using single-cell RNA sequencing. FASEB J. 2020;34:3677–3692. doi: 10.1096/fj.201902001RR. [DOI] [PubMed] [Google Scholar]
- Wang L., Du Y., Ward J.M., Shimbo T., Lackford B., Zheng X., Miao Y.l., Zhou B., Han L., Fargo D.C., et al. INO80 facilitates pluripotency gene activation in embryonic stem cell self-renewal, reprogramming, and blastocyst development. Cell Stem Cell. 2014;14:575–591. doi: 10.1016/j.stem.2014.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Wang S., Troisi E.C., Howard T.P., Haswell J.R., Wolf B.K., Hawk W.H., Ramos P., Oberlick E.M., Tzvetkov E.P., et al. BRD9 defines a SWI/SNF sub-complex and constitutes a specific vulnerability in malignant rhabdoid tumors. Nat. Commun. 2019;10:4445. doi: 10.1038/s41467-019-12524-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W., Liu X.S. Model-based analysis of ChIP-seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Zhao T., Guan J., Zhang X., Fu Y., Ye J., Zhu J., Meng G., Ge J., Yang S., et al. A XEN-like state bridges somatic cells to pluripotency during chemical reprogramming. Cell. 2015;163:1678–1691. doi: 10.1016/j.cell.2015.11.017. [DOI] [PubMed] [Google Scholar]
- Zhuang Q., Li W., Benda C., Huang Z., Ahmed T., Liu P., Guo X., Ibañez D.P., Luo Z., Zhang M., et al. NCoR/SMRT co-repressors cooperate with c-MYC to create an epigenetic barrier to somatic cell reprogramming. Nat. Cell Biol. 2018;20:400–412. doi: 10.1038/s41556-018-0047-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq and ATAC-seq data are deposited to the NCBI GEO database with the accession number GSE161640. This study did not generate any novel codes but used previously published methods.