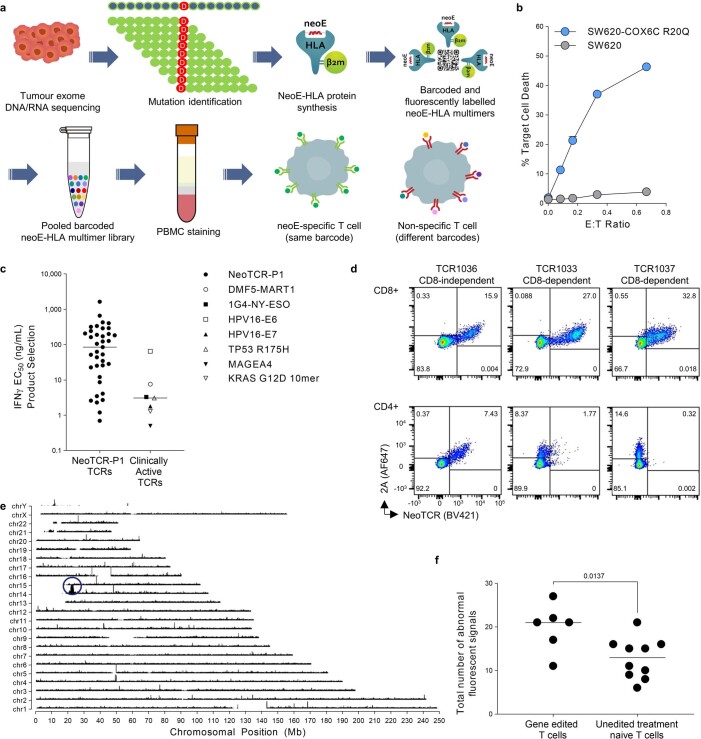
Extended Data Fig. 1. NeoTCR isolation, cytotoxicity, potency, gene editing and gene insertion.
a) Neoantigen-specific T cell capture. b) NeoTCR specific killing of an SW620 COX6C-R20Q mutant colorectal cancer cell line. Healthy donor T cells engineered to express a neoTCR from the blood of a patient with colorectal cancer targeting the COX6C-R20Q mutation, cocultured with either the parental SW620 cell line (without R20Q mutation), or with SW620-COX6C R20Q. c) Potency (IFNγ EC50) of neoTCRs isolated from the 16 patients compared with seven clinically active TCRs. d) Example gene editing (as measured by staining for 2A peptide) and neoTCR binding on CD4 and CD8 T cells for the three TCRs in a manufactured cell product. TCR1036 showed 2A expression and neoTCR binding of dextramer in CD4 and CD8 T cells, considered CD8-independent. TCR1033 and TCR1037 showed only 2A expression but no neoTCR binding by dextramer when transfected into CD4 T cells, considered CD8-dependent. e) Targeted locus amplification (TLA) was performed on 0010 TCR445 drug product. Primers specific for transgene and integrated transgene were used to amplify TLA processed genomic DNA. High coverage at the chromosome 14 integration site was observed (blue circle), indicating on-target TRAC transgene integration. A similar peak was not observed at chromosome 7, the site of TRBC knockout. f) Six clinical drug products from three patients were analysed using fluorescent in-situ hybridization (FISH) for chromosomal anomalies involving chromosome 7 and chromosome 14. All abnormal signals from each drug product tested were summed and compared to the total number of abnormal signals found in unedited cells from 10 separate donors. A p value was generated using an unpaired two-tailed t-test.