Abstract
Regulation of virulence gene expression in enteropathogenic Escherichia coli (EPEC) and enterohemorrhagic E. coli (EHEC) is incompletely understood. In EPEC, the plasmid-encoded regulator Per is required for maximal expression of proteins encoded on the locus of enterocyte effacement (LEE), and a LEE-encoded regulator (Ler) is part of the Per-mediated regulatory cascade upregulating the LEE2, LEE3, and LEE4 promoters. We now report that Ler is essential for the expression of multiple LEE-located genes in both EPEC and EHEC, including those encoding the type III secretion pathway, the secreted Esp proteins, Tir, and intimin. Ler is therefore central to the process of attaching and effacing (AE) lesion formation. Ler also regulates the expression of LEE-located genes not required for AE-lesion formation, including rorf2, orf10, rorf10, orf19, and espF, indicating that Ler regulates additional virulence properties. In addition, Ler regulates the expression of proteins encoded outside the LEE that are not essential for AE lesion formation, including TagA in EHEC and EspC in EPEC. Δler mutants of both EPEC and EHEC show altered adherence to epithelial cells and express novel fimbriae. Ler is therefore a global regulator of virulence gene expression in EPEC and EHEC.
Enteropathogenic Escherichia coli (EPEC) and enterohemorrhagic E. coli (EHEC) are important enteric pathogens for humans. EPEC is the most common bacterial cause of diarrhea in infants (35), while EHEC, especially those of serotype O157:H7, are important emerging pathogens causing diarrhea, hemorrhagic colitis, and hemolytic-uremic syndrome (35). Central to the pathogenesis of both EPEC and EHEC infections is the formation of attaching and effacing (AE) lesions on infected host intestinal epithelial cells. The AE lesion is characterized by the loss of microvilli (effacement) and the induction of a pedestal of polymerized actin and other cytoskeletal elements that forms underneath and around the infecting bacterium (13, 24, 35). In EPEC strain E2348/69, the AE phenotype is encoded by a 35.6-kb pathogenicity island, the locus of enterocyte effacement (LEE) (31, 32). The LEE contains genes encoding an outer membrane protein (intimin), a type III secretion system (Esc, Sep, and Ces proteins), secreted proteins (Esp), and the translocated intimin receptor (Tir), as well as a number of open reading frames of undetermined function (8). These genes are also found in the same organization on the LEE of EHEC (36) and are necessary but not sufficient for AE lesion formation by EHEC in vitro (11).
In addition to the LEE pathogenicity island and the AE phenotype, other parts of the genome in both EPEC and EHEC encode additional virulence factors and pathogenic mechanisms. The EPEC virulence plasmid encodes the regulator Per (18) and the type IV bundle-forming pili (BFP) (16), which are necessary both for in vitro EPEC adherence to HEp-2 cells in the characteristic localized-adherence pattern and for full virulence in humans (2). The EHEC virulence plasmid encodes a large number of known or potential virulence factors (4) including an RTX cytotoxin-hemolysin, Hly, and the autotransporter toxin, EspP (3), and contains tagA, which encodes a lipoprotein homologous to the cryptic ToxR-activated TagA of Vibrio cholerae. The EHEC, but not EPEC, chromosome contains phages encoding Shiga toxins 1 and/or 2, which are central to the pathogenesis of both hemorrhagic colitis and hemolytic-uremic syndrome (35). The EPEC chromosome contains an additional small pathogenicity island encoding the autotransporter toxin EspC (43; J. L. Mellies, F. Navarro-Garcia, J. P. Nataro, and J. B. Kaper, submitted for publication).
The way in which EPEC and EHEC regulate the expression of these multiple virulence genes is not well understood, and regulation studies have been largely confined to BFP and the LEE-encoded genes. It has been shown that BFP (16) and EspC (26) (in EPEC) and the LEE-encoded Esps (26) are maximally secreted when bacteria are grown in tissue culture media. The genetic basis for regulation has focused, in EPEC, on the role of the plasmid-encoded regulator, Per (18), which upregulates the expression of BFP and the LEE-encoded genes in EPEC (18, 33, 44). An analogous specific regulator has not been described for EHEC. It has recently been demonstrated that quorum sensing is also involved in the regulation of EHEC and EPEC LEE genes (42).
In contrast to EPEC and EHEC, much more is known about regulation in related pathogens and other type III secretory systems (reviewed in references 7 and 21). A common theme is gene activation by an AraC-like protein and repression with a second DNA binding protein such as YmoA (in Yersinia) or H-NS. H-NS, the “histone-like nonstructural protein,” binds strongly to curved (i.e., usually AT-rich) DNA, causing changes in supercoiling and packing and influencing gene expression (7, 21). In Shigella, H-NS counters the upregulatory effect of VirF and represses transcription of virB, while in enterotoxigenic E. coli, H-NS represses CfaD-mediated activation of cfa (7). H-NS is important for flagellar synthesis, F1-fimbrial phase switching, and regulation of proU and csgA among other genes (1, 5, 7, 21). H-NS can also negatively regulate its own expression (6).
A large family of H-NS-like proteins has been described which includes orthologs such as BpH3 in Bordetella (19) and the paralogous E. coli protein StpA (40). These proteins diverge significantly from H-NS and may have a different spectrum of activity but can functionally substitute for H-NS in several assays (1).
A gene whose predicted protein product has similarity to the H-NS family of DNA binding proteins was recently found in the LEE of EPEC (8). Originally termed orf1, this open reading frame is shown here to encode a protein able to regulate virulence gene expression but to be functionally distinct from H-NS. We recently reported that orf1 in EPEC is part of a regulatory cascade involving the AraC homolog Per and renamed this gene ler (for “LEE-encoded regulator”) (33). ler was found to activate the transcription of several LEE operons. Another group has also recently found that ler can activate transcription from LEE operons and is required for expression of LEE-encoded proteins (14). They also demonstrated that integration host factor binds upstream of ler and is required for Ler expression.
We report here that ler also regulates LEE genes in EHEC O157:H7 and, more surprisingly, affects the expression of phenotypes encoded elsewhere in the genome of both EPEC and EHEC O157:H7. These results expand the role of Ler as a global regulator of virulence gene expression in both EPEC and EHEC.
MATERIALS AND METHODS
Bacterial strains, plasmids, and PCR primers.
Bacterial strains and plasmids used in this study are listed in Table 1, and PCR primers are listed in Table 2. Unless otherwise stated, bacteria were grown at 37°C in Luria broth. In experiments where minimal essential medium (MEM; Life Technologies, Bethesda, Md.) was used, bacteria were grown overnight at 37°C in MEM prior to inoculation into fresh MEM and grown until an optical density at 600 nm of 1.0 (late log phase) was reached. The growth medium was supplemented with ampicillin (200 μg/ml), chloramphenicol (25 μg/ml), kanamycin (25 μg/ml), or nalidixic acid (100 μg/ml) as needed.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Details | Reference or sourcea |
|---|---|---|
| Strains | ||
| E2348/69 | Wild-type EPEC O127:H6 | 29 |
| EDL933 | Wild-type EHEC O157:H7 | 37 |
| 85-170 | EHEC O157:H7 Δstx | 45 |
| 86-24 | Wild-type EHEC O157:H7 | 20 |
| SE941 | O55:H7 strain | CVD diarrheal isolate |
| SE796 | EPEC E2348/69 Δler::kan | This study |
| SE1099 | EHEC 85-170 Δler::kan | This study |
| SE1101 | EHEC 86-24 Δler::kan | This study |
| SE860 | SE796(pCVD456) | This study |
| SE1104 | SE796(pSE1100) | This study |
| SE1110 | SE796(pSE1092) | This study |
| SE1140 | SE1099(pSE1092) | This study |
| TE2680 | MC4100 recD::Tn10 | 12 |
| Plasmids | ||
| pCVD456 | pMOB::ler orf2345EPEC | 31 |
| pSE1092 | pACYC184::lerEHEC | This study |
| pSE1093 | pBR322::lerEHEC | This study |
| pSE1100 | pBR322::lerEPEC | This study |
| pTHK113 | hns | 25 |
| pJG9 | oritscat sacB | J. Galen, unpublished data |
| pRS551 | lacZ reporter gene fusion vector | 41 |
CVD, Center for Vaccine Development.
TABLE 2.
Primers used in this study
| Name | Sequence (5′ to 3′) |
|---|---|
| K590 | AAG ACA TTC TAC CCC GGG AAA ATA TTT AAC |
| K591 | CTG GCT TTC AGG ATC CTT ATT TTG GC |
| K592 | GTG AAT TAG TTT CCC GGG TCA TAA TAA ATA |
| K593 | CTT CAC ATT TTG GGA TCC TAT CTC TC |
| K803 | GAC ATA TCA TCA TGG ATC CTG AAT AAT GC |
| K848 | GCG TTA ATT GCT GAG ATT C |
| K874 | TGC GAT CCT TCA TAA TCA T |
| K893 | CGT ACC TAG CGT AGG TT |
| K1226 | CGG GAT CCG CGG TTA CTT GTT CAG CTA |
| K1229 | CGG GAT CCT TAT CCT CTG GTA TGA TAT C |
| K1230 | CGG GAT CCA AAG CGA CTG CGA CAG CAG GA |
| K1370 | AGA GGA TTC CTC TTC ACC ATA TGT GTA CCC CTC AA |
| K1371 | TAT TTA TTA CCC GGG CCG CTG AAA AAT ATT TAA CAT GAA AT |
| K1372 | CCG GAA TTC CTG TAA CTC GAA TTA AGT AGA GT |
| K1373 | TTT CAG CGG CCC GGG TAA TAA ATA ATC TCC GCA TGC TTT |
| K1420 | CGC GGA TCC AGC TCA CGT TAT CGT TAT CAT T |
| K1502 | CGC GGA TCC AGC TAC AGG AAG CTC ATC CTT |
| K1503 | ACA TGA ATT CAG CGA TGC TGC CCA TGA A |
| K1547 | ACT TTC TCC GAC AGC ACC |
| K1548 | TCA GAC GCA GAC TGG TAG |
| K1938 | GAG GGA TCC AGT TCG GAT ACG CAA TCA |
| K1939 | GCA GAA TTC ATC ATG GCT CCG GGA GAG AGA |
| K1942 | GCAGAATTCTGACTCGTATGACAACGCGA |
| K1943 | AGA GGA TCC GGT AGT TTC AGG GTA GGA GCC A |
| K1944 | AGC GGA TCC AAC GAA CCC TGC AGA TCA T |
| K1945 | AGA GGA ATT CAG GAT AAT GAG CTT ACC CAG CA |
| K1964 | AGC GAT TAA CCC TCC TGT A |
| K1965 | AAC ACA TTG GCG GAC TCG AT |
| K1966 | TCC AAT GTT ATC CCA AAC GTA |
| K1967 | GAT TCT CCT ATC TGG TTT GTA |
Molecular techniques.
Where cloning required PCR amplification, the proofreading polymerase Pwo (Boehringer-Mannheim) was used and the resultant clones were examined for fidelity by sequencing. All other PCR amplifications were performed using Taq polymerase (Life Technologies). Automated sequencing was performed at the University of Maryland Biopolymer Core Facility. All other molecular techniques were performed by standard methods. DNA analysis was performed with DNAsis v5 (Hitachi) and with the suite of programs provided by the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov). Homology searches were performed using PSI-BLAST (http://www.ncbi.nlm.gov/blast/psiblast.cgi) with the filter off and gap function activated. Protein localization was predicted using the PSORT algorithm (http://www.psort.nibb.ac.jp).
Cloning and mutagenesis.
The ler regions from EPEC and EHEC were cloned into a variety of vectors. pCVD456 (31) is a 3.1-kb EcoRI fragment from the LEE of EPEC E2348/69 cloned into pMOB and containing ler orf2345. Primers K1372 and K1420 were used to amplify ler from the EPEC chromosome, which was cloned into pBR322 digested with EcoRI and BamHI, creating pSE1100. pSE1092 was constructed by amplification of the ler region from EHEC 85-170 using primers K1372 and K1370 and cloning as an EcoRI-blunt fragment into an EcoRI-PvuII fragment of pACYC184. In all cases the ler region was transcriptionally isolated from plasmid promoters, since we observed that clones containing ler under the control of a strong plasmid promoter grew poorly and the plasmid was unstable.
A nonpolar in-frame deletion mutation of ler was constructed in both EPEC E2348/69 and EHEC O157:H7 strains by allelic exchange. For EPEC, primers K590 through K593 were used to amplify ca. 500-bp regions flanking ler which contained a SmaI site at the deletion junctions into which was cloned the promoterless kanamycin resistance cassette, aphA3 (hereafter referred to as kan). This fragment, containing the EPEC ler promoter upstream and the resistance cassette and orf2 downstream, was initially cloned as an EcoRI-BamHI fragment in pBluescript and was then moved as a PvuII fragment into the suicide vector pJG9. pJG9 contains a temperature-sensitive replicon and sacB gene for counter selection. The resultant plasmid, pSE774, was electrotransformed into E2348/69, and, via allelic exchange, the wild-type chromosomal ler gene was replaced with an in-frame, nonpolar kan cassette, generating strain SE796. The site of insertion was confirmed by PCR and Southern hybridization.
To construct a ler mutation in the LEE of EHEC, primers K1370 through K1373 were used to PCR amplify and ligate regions flanking ler from the chromosome of EHEC strain 85-170, using a strategy similar to that employed for pSE774. The ultimate construct of pSE1096 was used to mutate the ler gene on the EHEC chromosome, as was previously done with EPEC. The mutation was confirmed with PCR and by sequencing the flanking regions. pSE1096 was used to mutate ler in EHEC O157:H7 strain 85-170, generating SE1099, and strain 86-24, generating SE1101. The ler regions in both these strains are identical to that of EDL933. The mutant phenotypes exhibited by SE1099 and SE1105 were complemented by cloned ler.
Primer extension.
Primer extension was performed as described previously (33). Briefly, primers hybridizing to the sense strand between 20 and 50 nucleotides downstream of the ATG start codon were end labeled using T4 DNA kinase and [γ-32P]ATP. Labeled primers were hybridized with 35 μg of DNA-free total bacterial RNA and reverse transcribed for 1 h at 42°C using Life Technologies Superscript II reverse transcriptase and the recommended buffers, reagents, and methods. The resultant cDNA mix was treated with RNase H, precipitated, and resolved through a 6% acrylamide–urea sequencing gel, and the bands were visualized by autoradiography.
Assays for virulence-associated phenotypes.
The fluorescent actin stain (FAS) test (27) utilizes fluorescein isothiocyanate-phalloidin to visualize the accumulation of actin beneath and around bacteria attached to HEp-2 cells. A weak or unfocused accumulation of actin underneath bacteria appears as a faint halo of fluorescence known as the shadow phenotype (27). The assay for Tir translocation has been previously described (38). EspA filaments were visualized using anti-EspA antibodies and immunofluorescence microscopy, as described previously (28). Expression of bacterial proteins was examined in supernatants and purified membrane and cytoplasmic fractions prepared as outlined previously (9, 23). Following separation by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, proteins were either stained with Coomassie blue or blotted to polyvinylidene difluoride and Western blotted with monospecific polyclonal rabbit antibodies against Tir, intimin, and all EPEC secreted proteins as previously described (9, 23).
Adherence was examined by the modified method of Scaletsky et al. (40) as previously described (10). Shiga toxin production was assessed by quantitative killing of Vero cells as previously described (15). Bacterial motility was examined in Craigie tubes containing 0.25% agar in Luria broth. Fimbriae were examined by electron microscopy on negatively stained bacteria as previously described (17).
Assessing promoter activity with lacZ fusions.
To assay the effect of ler on gene expression, regions containing the promoter and at least 200 bp of flanking DNA were amplified with Pwo polymerase and cloned into plasmid pRS551, which contains a promoterless lac operon (41). To generate single-copy lacZ fusions, these plasmids were linearized with XhoI and the linear DNA was transformed into E. coli K-12 strain TE2680 and integrated into the chromosome as previously described (12). Promoter activity (in Miller units) was assayed by quantification of β-galactosidase activity in bacterial cultures as previously described (34). For promoters from the EHEC LEE, bla and stx, we used previously constructed fusions (33, 42). New promoter fusions were constructed with the following primers: bfp (K1546 and K1548), espC (K1948 and K1939), tagA (K1944 and K1945), espP (K1942 and K1943), and hly (K1502 and K1503).
Examination of Ler for properties similar to those of H-NS.
To examine if ler is functionally related to hns, we examined the ability of ler to rescue and/or interfere with hns function by using lacZ reporter strains as described by Donato et al. (5). pCVD456, containing ler orf234, or pTHK113, containing hns, was transformed into THK60 (hns+ proU::lacZYA), THK62 (Δhns::tet proU::lacZYA), THK88 (hns+ fimB::lacZYA), and THK90 (Δhns::tet fimB::lacZYA) and assayed for β-galactosidase activity (in Miller units) as described above.
RESULTS
Characterization of ler and the ler gene product.
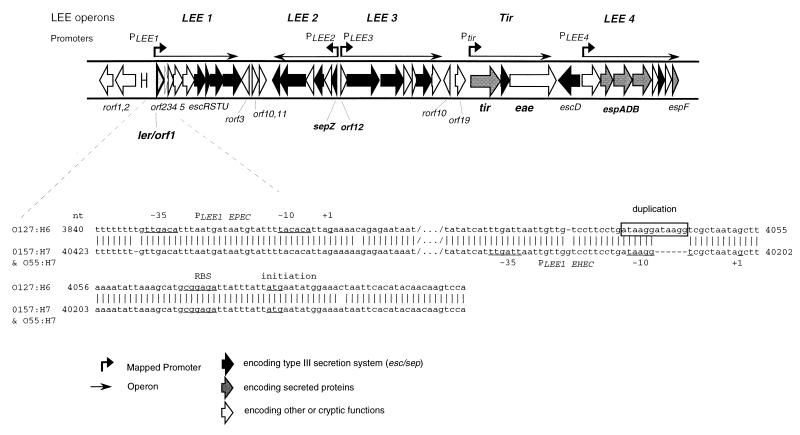
ler, previously known as orf1, is the first in a series of codirectional genes in the LEE (Fig. 1). It has been recently demonstrated by reverse transcription-PCR that these genes form a polycistronic operon denoted LEE1 in EPEC O127:H6 strain E2348/69 (33). The LEE1 operon contains nine genes, ler orf2345 escRSTU, and is highly conserved with respect to EHEC O157:H7 (36). Primer extension has demonstrated that the promoter for LEE1 in EPEC is different from that in EHEC O157:H7 and is 169 nucleotides upstream (33, 36), although the region upstream of ler is conserved. Alignment of these two regions demonstrated the duplication of a 6-nucleotide (ATAAGG) sequence in EPEC O127:H7 compared to the same region in O157:H7 strain EDL933 (Fig. 1). This duplication in EPEC falls in the region predicted as the −10 region for the LEE1 promoter in O157:H7, disrupting this corresponding area in EPEC. This duplication may be responsible for the inactivity of the downstream promoter in EPEC.
FIG. 1.
The LEE, showing the structures of the LEE1 through LEE4 and tir operons and the mapped promoters. The promoter driving ler expression (PLEE1) has been expanded to show the differences between the promoter of EPEC O127:H6 and those of EHEC O157:H7 and EPEC O55:H7. EPEC contains an ATAAGG duplication that disrupts the region corresponding to the −10 sequence found in EHEC (33, 42). Open reading frames have been shaded to distinguish those encoding secreted proteins from those involved in type III secretion or other functions. In the alignment of the sequences, /…/ represents an area deleted from the figure for presentation and − represents a nucleotide missing in that sequence when compared with the other sequence.
Sequencing of the ler region from two other EHEC O157:H7 strains and an EPEC O55:H7 isolate (Table 1) demonstrated that the ler region in these strains was 100% identical to that of EHEC O157:H7 strain EDL933 and presumably contained identical promoters. Unlike EPEC O127:H6, which belongs to the EPEC1 evolutionary group, these four strains are all members of the EHEC1 evolutionary group, with the O55:H7 serotype believed to be a progenitor of the O157:H7 lineage (48).
To demonstrate the ability of orf1/ler to encode a protein product, ler was cloned into and expressed from the T7 expression vector pET21. A 14-kDa protein was detected, which is consistent with the 15.1-kDa mass predicted from the amino acid sequence (results not shown).
Comparison of the predicted Ler sequence with previously described proteins by using PSI-BLAST demonstrated that Ler is distantly related to proteins that are members of the H-NS family of DNA binding proteins, including BpH3 from Bordetella (29% identical, 47% similar over the homologous region, and 41% similar over the entire length), StpA from E. coli (23% identical, 48% similar over the homologous region, and 40% similar over the entire length) and, more distantly, H-NS (36% identical, 55% similar over the homologous region, but only 20% similar over the entire length) (Fig. 2). The highest similarity between Ler and its homologs is found in the DNA binding C-terminal domain, and Ler possessed the conserved DNA binding motif of H-NS family proteins (TWTGXGRXP) (5, 46). No homology was seen within the N-terminal oligomerization domain (6). Both Ler and H-NS have similar predicted pI values (5.86 and 5.29, respectively), and both were predicted by the PSORT algorithm to be localized in the cytoplasm. Together, these predictions are consistent with the classification of Ler as a member of the H-NS-family of DNA binding proteins, although somewhat distant from E. coli H-NS itself.
FIG. 2.
Comparison of Ler and its homologs. Ler homologs are aligned to show identical (marked by their letter) and similar but nonidentical (+) amino acids. Ler from EHEC and EPEC are highly conserved and are related to the H-NS family of DNA binding proteins, including BbH3 and BpH3 of Bordetella, StpA, other H-NS orthologs and paralogs, and, more distantly, E. coli H-NS. Similarity is highest at the C-terminal region, which mediates DNA binding. The conserved DNA binding motif (TWTGXGRXP) contained in Ler and all members of the H-NS family is underlined (6). The percent identity (%ID) and similarity (%Sm) over the homologous region are listed to the right of each alignment, as is the number of gap initiations in the alignment (#gaps, gaps not shown).
Mutation of ler disrupts functions associated with the LEE.
An in-frame nonpolar deletion mutation of ler was constructed in EPEC O127:H6 and both stx+ and stx strains of EHEC O157:H7, as described in Materials and Methods. For biosafety reasons, the stx EHEC strain was used for all experiments unless otherwise stated.
EPEC and EHEC Δler mutants were defective in the formation of AE lesions on HEp-2 cells as determined by the FAS test (Table 3). EHEC Δler mutants were negative in the FAS assay, and a shadow FAS phenotype was observed after 6-h incubations of EPEC Δler on HEp-2 cells (data not shown) which suggests that the ability to form AE lesions was not completely abolished in this strain but was, rather, strongly diminished. The FAS phenotype in both EPEC and EHEC mutants was restored by complementation with ler from their respective parent strains cloned on a multicopy plasmid vector (Table 3), and complemented strains exhibited a FAS reaction that was visibly enhanced over that of the wild type. Interestingly, the cloned EHEC ler was also able to restore FAS in the EPEC Δler mutant (Table 3), indicating that EHEC ler is able to functionally substitute for EPEC ler.
TABLE 3.
Virulence phenotypes affected by Ler
| Strain | Genotype | FASa | Production of EspABD, Tir EspC/P, and intimin | Adherence pattern | Production of LFF |
|---|---|---|---|---|---|
| E2348/69 | EPEC wild type | ++ | + | LA | − |
| SE796 | EPEC Δler::kan | − | − | LA-DA-AA | + |
| SE860 | EPEC Δler::kan(pLER ORF2345EPEC) | ++++ | + | LA | − |
| SE1104 | EPEC Δler::kan(pLEREPEC) | ++++ | + | LA | − |
| SE1110 | EPEC Δler::kan(pLEREHEC) | ++++ | NTb | LA | − |
| 85-170 | EHEC wild type | + | + | LA-DA | − |
| SE1099 | EHEC Δler::kan | − | − | LA-DA-AA | + |
| SE1105 | EHEC Δler::kan(pLEREHEC) | ++ | + | LA-DA | − |
+ to ++++; qualitative assessment of activity in the assay.
NT, not tested.
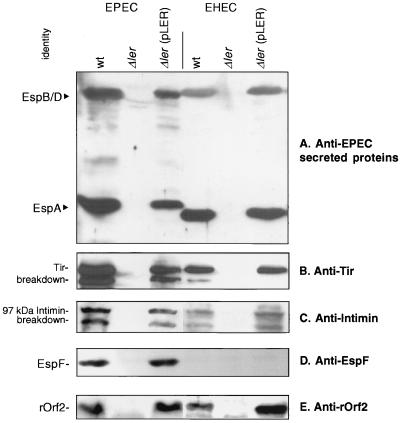
The coordinated expression of multiple virulence factors, including the Esp proteins, Tir, and intimin, is necessary for AE lesion formation. We investigated which of these factors is regulated by Ler. A comparison of the wild-type, Δler mutant, and complemented strains revealed that ler is necessary for EPEC and EHEC to secrete Esp proteins into the supernatant (Fig. 3A) and for production of the EspA filament by EPEC, since the filament was not observed in mutant strains (Stuart Knutton, personal communication). Similarly, Tir was not secreted by the ler mutants (Fig. 3B) and was not translocated by EPEC Δler into HEp-2 cells as determined by staining HEp-2 cell lysates with antiphosphotyrosine antibodies (data not shown). Immunoblotting of membrane preparations demonstrated that intimin levels were markedly reduced in the absence of ler (Fig. 3C). In addition to the lack of Esp secretion observed in the Δler mutants, Esp proteins were not found at detectable levels in the whole cell as determined by Western blot analyses performed on lysates, probed with antibodies against secreted Esp proteins (data not shown).
FIG. 3.
Mutation of ler affects the production of LEE-encoded proteins. Western blots of secreted proteins probed with antibodies against all-EPEC secreted proteins (A) or Tir (B), membrane preparations stained with antibodies against the outer membrane protein intimin (C), and whole-cell lysates stained with antisera against EspF (D) or rOrf2 (E) indicate that multiple LEE-encoded virulence factors are regulated by Ler. wt, wild type.
The LEE also encodes a number of proteins unnecessary for AE lesion formation, including EspF and rOrf2. espF is cotranscribed with espADB on LEE4, while rorf2 is transcribed on an operon divergent from LEE1 and separate from the main LEE operons (8, 33, 36). Production of these proteins was demonstrated to be dependent on Ler since Δler mutants did not produce them, as determined by Western blot analyses using antisera specific for EspF (Fig. 3D) (a gift of M. Donnenberg) or rOrf2 (Fig. 3E). The antiserum raised against EPEC EspF did not react with EHEC EspF, presumably reflecting the high sequence divergence in EspF between EPEC and EHEC (36).
Ler affects the level of proteins encoded outside the LEE.
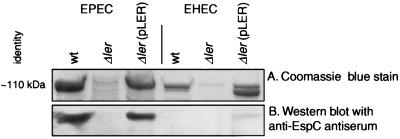
The ler gene product was found to control the expression of genes encoded outside the LEE in both EPEC and EHEC. We observed high-molecular-mass (∼110 kDa) proteins in the supernatants of wild-type and complemented strains grown in MEM but not in supernatants from Δler mutants (Fig. 4A). In EPEC, this band has been identified as EspC, an autotransported toxin encoded by a separate chromosomal pathogenicity island (43; Mellies et al., submitted). Western blotting with an antiserum that recognizes EspC (22) supported this identification (Fig. 4B).
FIG. 4.
Regulation of high-molecular-mass secreted proteins by Ler. (A) Coomassie blue-stained gels of secreted protein preparations from EPEC and EHEC demonstrated that ∼110-kDa proteins were not secreted from Δler mutants. (B) Antibodies identify this protein in EPEC supernatants as EspC. wt, wild type.
Similarly, Ler regulated a high-molecular-mass protein(s) in EHEC, observed as a doublet in supernatants (Fig. 4A). These bands may correspond either to one full-length protein and a breakdown product of the same protein or two distinct secreted proteins coregulated by Ler. An EspC-analogous high-molecular-mass autotransporter, EspP, has been identified in EHEC supernatants (3), but we lack a sufficiently specific antiserum to confirm this identity.
The major virulence factor produced by EHEC but not EPEC is Shiga toxin (Stx). Ler does not affect the production of Stx since the wild-type EHEC 86-24 (Stx1+ Stx2+) produced toxin in amounts equivalent to those produced by its Δler derivative, SE1101, when assayed on Vero cells (A. O'Brien, personal communication). Similarly, Ler did not appear to affect the production of enterohemolysin from EHEC, as assayed on 5% washed sheep erythrocyte agar (data not shown).
Mutation of ler affects the pattern of EPEC adherence and causes hyperadherence in EHEC.
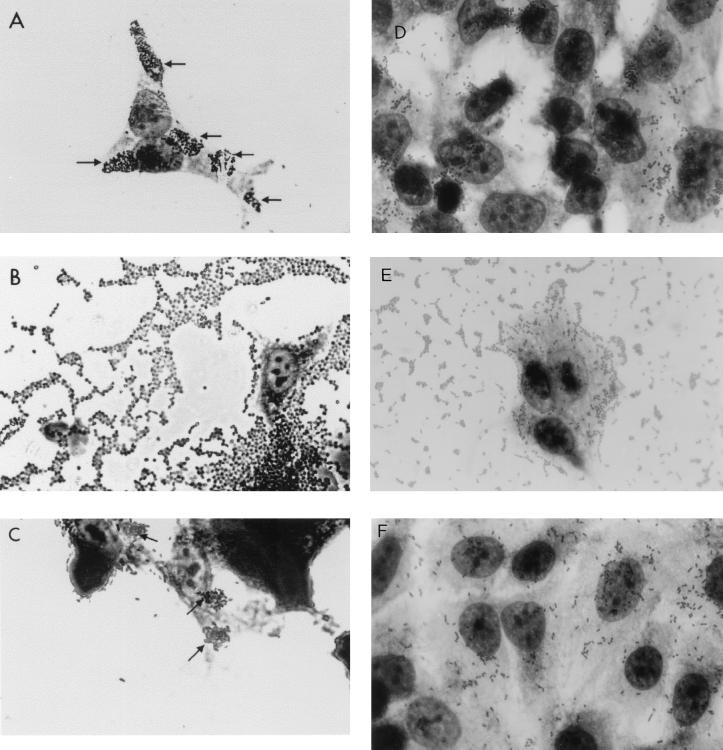
Mutation of ler in EPEC and especially in EHEC was associated with changes in the pattern of adherence (Fig. 5, Table 3). Wild-type EPEC normally exhibits localized adherence (LA) to HEp-2 cells, with the formation of microcolonies (Fig. 5A). Mutation of ler in EPEC was associated with the appearance of a complex-aggregative (AA)–diffuse-adherence (DA) phenotype and proportionally decreased localized adherence (LA), although some microcolonies were observed (Fig. 5B). The differences between wild-type and Δler mutant strains were more clearly visible after a 6-h incubation with HEp-2 cells rather than the more common 3-h incubation period (data not shown). Complementation of the Δler mutant with cloned ler abolished the DA-AA pattern and restored the LA pattern (Fig. 5C).
FIG. 5.
Altered adherence phenotypes of Δler mutants. (A to C) EPEC (A) and the complemented Δler mutant (C) exhibited LA and formed large microcolonies on the surface of the HEp-2 cells; the EPEC Δler mutant (B) displayed a complex mixture of DA and AA patterns with some LA microcolony formation. (D to F) With EHEC 85-170, the wild type adhered to HEp-2 cells in a DA-LA pattern (D) while the Δler mutant displayed increased adherence, especially to glass, and an AA pattern (E); complementation of the Δler mutation with plasmid-encoded ler restored the wild-type adherence phenotype (F). Magnification, ×2,500.
EHEC 85-170 normally displays moderate LA-DA patterns of adherence (Fig. 5D), while its corresponding Δler mutant, SE1099, was more adherent than the wild type and adhered in an AA pattern (Fig. 5E). Transformation of SE1099 with pSE1092 (pLEREHEC) restored the wild-type adherence pattern and abolished the AA pattern (Fig. 5F).
Mutation of ler causes the expression of novel fimbriae in EPEC and EHEC.
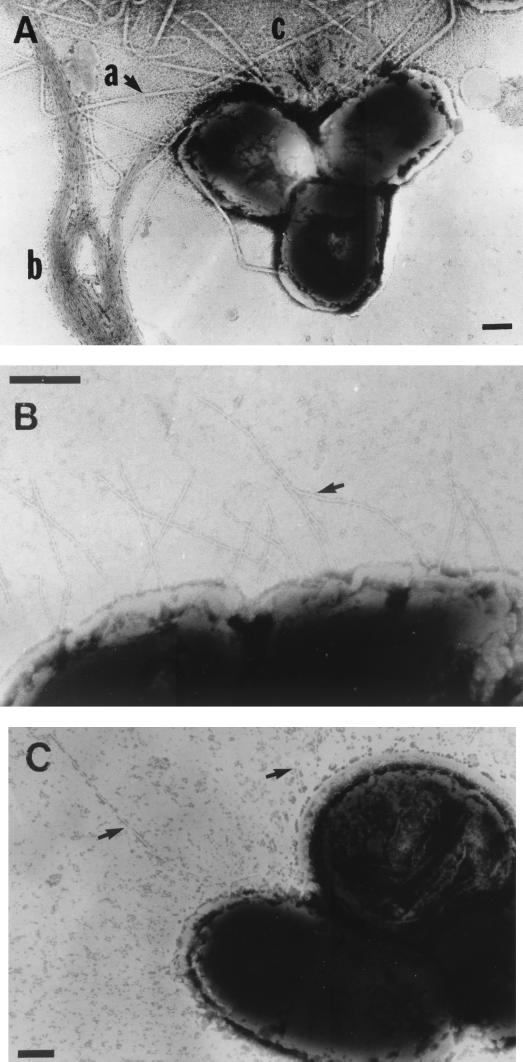
Electron microscopy studies of the Δler mutants demonstrated that the alterations in adherence observed in these strains were accompanied by changes in fimbrial expression. In addition to the BFP normally produced by EPEC, the EPEC Δler strain produced additional fimbriae with novel morphologies (Fig. 6A). We clearly distinguished several morphologically distinct fimbrial types including long fine fimbriae, more rigid bent fimbriae, and short fine fimbriae (Fig. 6A and B). The complemented mutant, E2348/69 Δler(pLER), did not express these novel fimbriae (data not shown) and displayed the wild-type phenotype.
FIG. 6.
Novel fimbriae expressed by Δler mutants. (A and B) The EPEC Δler mutant expressed a number of novel fimbriae (A, a and c) that are clearly distinguishable from BFP (b) and include numerous LFF (c) as well as “bent” fimbriae (b) and shorter fine fimbriae (B, arrow). (C) EHEC 85-170 does not express fimbriae (not shown), but the Δler mutant exhibited long fine fimbriae (arrow). Bar = 200 μm.
Immunoelectron microscopy using antibodies conjugated to gold particles was used to confirm that the novel fimbriae were not derived from BFP. Antibodies directed against BfpA reacted to BFP but not to fimbriae with novel morphologies (data not shown). Further, no differences were found in the levels of BfpA between wild-type and Δler strains in Western blot analyses using anti-BfpA antiserum against whole-cell lysates of bacteria grown both in MEM and on agar (data not shown).
When we examined EHEC, we also observed that mutation of ler was associated with enhanced fimbrial expression. Wild-type 85-170 typically expressed few fimbriae (data not shown). The Δler mutant SE1099, however, produced long fine fimbriae (Fig. 6C) that were not observed in the wild type or the complemented strain (data not shown). It is not yet known if these fimbriae are identical to the morphologically similar long fine fimbriae observed in the EPEC Δler mutant. We are currently characterizing these novel Ler-regulated fimbriae from EPEC and EHEC.
Ler activates promoters in the absence of other EPEC- or EHEC-specific genes.
Ler is able to activate promoters from the LEE and elsewhere in the genome in the absence of other EPEC- or EHEC-specific genes. This was demonstrated by fusing promoters to a lacZ reporter gene and introducing them as single copies into the chromosome of E. coli K-12. These strains were then transformed with pSE1093 (pBR322 containing ler from EHEC) when the promoter was derived from EHEC or pSE1100 (pBR322 containing ler from EPEC) when the promoter was EPEC derived. Control strains containing only the pBR322 vector were also constructed and tested.
The genes within the LEE are arranged in at least five large polycistronic operons designated LEE1 through LEE4 and tir (Fig. 1), and the transcription start sites have been determined by primer extension (9, 33, 42). Using previously constructed reporter fusions (9, 42), we demonstrated that Ler upregulated transcription from EHEC LEE promoters for the tir, LEE2, and LEE3 operons in the range of about eightfold (Table 4). These data demonstrate that Ler upregulates the expression of several virulence-associated proteins and demonstrate that Ler acts directly as an activator of transcription of LEE operons from EHEC and EPEC, including those containing esc/sep and tir. Ler did not regulate PLEE1 (i.e., the ler promoter) or eae, indicating that intimin expression is controlled via the tir promoter, which regulates the tir cesT eae polycistronic operon. Similar results have been previously reported by us for EPEC LEE operons LEE1, LEE2, and LEE3 (33). Interestingly, Ler did not regulate the LEE4 operon in EHEC and caused only a twofold upregulation of the LEE4 operon in EPEC (33). The observation that Ler is, at best, a weak activator of the LEE4 promoter contrasts with the dramatic increase in EspABD protein levels in the presence of Ler (Fig. 3).
TABLE 4.
Activation of gene expression by Ler
| Operon | Source of promoter | β-Galactosidase activitya
|
Fold activationb | |
|---|---|---|---|---|
| pBR322 | pLER | |||
| LEE1 | EHEC LEE | 111 ± 6 | 100 ± 6 | 0.9 |
| LEE2 | EHEC LEE | 27 ± 4 | 207 ± 14 | 7.6c |
| LEE3 | EHEC LEE | 22 ± 5 | 175 ± 22 | 8.1c |
| tir | EHEC LEE | 10 ± 1 | 80 ± 16 | 7.6c |
| eae | EHEC LEE | 34 ± 9 | 33 ± 2 | 0.9 |
| LEE4 | EHEC LEE | 861 ± 51 | 942 ± 38 | 1.1 |
| stx | EHEC phage | 15 ± 4 | 20 ± 3 | 1.3 |
| espP | EHEC plasmid | 230 ± 11 | 218 ± 8 | 0.9 |
| hly | EHEC plasmid | 1,169 ± 54 | 951 ± 62 | 0.8 |
| tagA | EHEC plasmid | 12 ± 1 | 245.3 ± 20 | 19.5c |
| espC | EPEC chromosome | 33 ± 3 | 1,027 ± 70 | 31.0c |
| bfp | EPEC plasmid | 85 ± 1 | 61 ± 0.2 | 0.7 |
| bfpd | EPEC plasmid | 2,836 ± 81 | 2,476 ± 170 | 0.9 |
| bla | control | 160 | 170 | 1.1 |
Mean β-galactosidase activity (in Miller units) present in bacteria containing indicated promoter-reporter fusion ± standard error. EHEC promoters were fused with a promoterless lacZ reporter and introduced as single-copy fusions into the chromosome of E. coli K-12 strain TE2680. These reporter strains were transformed with pBR322 or pSE1093, except for espC and bfp fusions, which were transformed with pSE1100 containing EPEC ler.
β-Galactosidase activity of pLER divided by activity of pBR322.
Significant at P = 0.05 compared to vector-only control.
Plasmid pRS551 containing bfp::lacZ fusion in EPEC Δler or EPEC wild-type backgrounds, respectively.
A number of genes found outside the LEE also were regulated by Ler in a K-12 background (Table 4). Ler strongly activated transcription from promoters for the EHEC plasmid-located tagA gene (20-fold) and the EPEC chromosomally located espC gene (30-fold). In contrast, promoters for EHEC genes stx (on a chromosomally integrated phage) and plasmid gene hly were not activated by Ler in a K-12 host background. These results agree with phenotypic observations, Western blotting results, or preliminary data from a (not shown) DNA array. Interestingly, the espP promoter was not regulated by Ler. This suggests that the high-molecular-mass Ler-regulated secreted protein in EHEC is not EspP or that espP is regulated in EHEC via a second regulator.
Finally, the promoter for the EPEC plasmid operon bfp was not regulated by Ler in either K-12 or EPEC backgrounds (Table 4). This result confirms Western blot data and supports the identity of novel, Ler-regulated EPEC fimbriae as distinct from BFP.
Primer extension identifies additional genes regulated by Ler in wild type EPEC and EHEC.
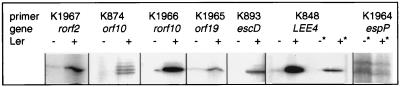
Primer extension was used to find whether particular mRNA transcripts were synthesized in EPEC and EHEC in either the presence or absence of Ler. We have previously (33) used primer extension to demonstrate that Per upregulates the LEE1 and (via Ler) LEE2 transcripts. We now report that Ler is absolutely necessary for full transcription of the LEE4 operon in both EPEC and EHEC (Fig. 7), supporting our observations that levels of EspADB and EspF are markedly reduced in Δler mutants. This finding is in contrast with data from LEE4::lacZ reporters in an E. coli K-12 background and strongly implies the presence of a specific LEE4 regulator that is present in EPEC and EHEC but not K-12.
FIG. 7.
Regulation of multiple EPEC and EHEC genes by Ler. Primer extension was performed on mRNA extracted from Δler mutants (−) and Ler+ complements (+) of EHEC (∗) or EPEC (no ∗) by using primers K848 through K1967, which are directed against transcripts from genes rorf2 through espP, as shown. The resultant cDNA transcripts were visualized by autoradiography.
We also found that mRNA transcripts for rorf2, orf10, rorf10, orf19, and escD (rorf11) were absent or strongly reduced in the absence of Ler. Therefore, Ler regulates all major operons within the LEE. Finally, we examined espP transcription and could not find evidence of Ler regulation. This agrees with our espP::lacZ fusion data and suggests that the high-molecular-mass Ler-regulated protein from EHEC is not EspP but another, as yet unidentified protein.
Ler is distinct from H-NS.
Alleles of H-NS may have divergent amino acid sequences but nonetheless be able to functionally substitute for H-NS in hns mutant strains, as has been observed with BpH3 (19), StpA (40), and others (1). At the same time, some H-NS homologs, such as StpA, form heterodimers with H-NS that may affect the normal DNA binding properties of H-NS, and so StpA action is more fully understood in an hns+ background (6). We examined to what extent Ler might be functionally analogous to H-NS or whether Ler could alter H-NS activity by using reporter fusions of lacZ to either proU or fim (5). hns+ and hns fusion strains were transformed with pCVD456 (ler) or pTHK113 (hns). We found that while cloned H-NS affected the expression of proU::lacZ or fim::lacZ up to 18-fold (Table 5), Ler had less than a 2-fold effect, which we do not consider significant. Therefore, Ler is neither functionally equivalent to H-NS nor able to have dominant negative effects on H-NS function.
TABLE 5.
Inability of Ler to complement hns mutation
| Strain | Genotype | β-Galactosidase activitya
|
||
|---|---|---|---|---|
| No plasmid | pHNSb | pLERc | ||
| THK60 | hns+ proU::lacZ | 200 ± 16 | 27 ± 2 | 137 ± 9 |
| THK62 | hns proU::lacZ | 553 ± 16 | 30 ± 6 | 932 ± 17 |
| THK88 | hns+ fimB::lacZ | 74 ± 2 | 26 ± 2 | 54 ± 1 |
| THK90 | hns fimB::lacZ | 417 ± 8 | 30 ± 3 | 475 ± 7 |
β-Galactosidase activity (in Miller units) present in bacteria containing indicated promoter-reporter fusion ± standard error.
pTHK113 encoding H-NS.
pCVD456 encoding Ler.
Mutation in hns has been reported to repress flagellar synthesis and therefore motility (1, 7, 19). In contrast, Ler did not affect motility, since the motility of EPEC Δler mutants was not diminished from that of the wild type as assessed in the Craigie tube, semisolid agar, and hanging-drop methods (data not shown). The results of all of these experiments indicate that Ler is functionally distinct from H-NS.
DISCUSSION
The ler locus encodes a regulator of virulence gene expression in both EPEC and EHEC O157:H7, directly regulating genes within the LEE elements and elsewhere in the genome. ler was originally described as orf1 and was proposed as a DNA binding protein on the basis of homology to the H-NS family of regulators (8, 30). Once it was demonstrated that the locus fulfills the functions of a regulator by activating transcription of LEE operons, it was renamed ler, the LEE-encoded regulator (33).
Ler is essential for the formation of AE lesions, since all the genes known to be important for AE lesion formation are regulated by ler or, in the case of the LEE1 operon, coregulated with ler. Both EPEC O127:H6 and EHEC O157:H7, with nonpolar deletions of ler, were unable to form AE lesions on HEp-2 cells, form the EspA filament, express intimin, translocate Tir, or secrete into the supernatant the type III secreted proteins EspA, EspB, EspD, or Tir. In addition, Δler mutants failed to express the type III secreted proteins, in contrast to mutants with mutations in escN or some other genes involved in type III secretion, which produce Esp and Tir proteins normally but are unable to secrete or translocate these proteins (9, 22, 23, 47). Therefore, the Δler phenotype is consistent with Ler as a regulator of LEE-located genes. Ler was then directly demonstrated to act as a regulator able to activate the transcription of LEE promoters in the absence of other EPEC- or EHEC-specific genomic elements with the use of lacZ reporter fusions in an E. coli K-12 background. This has been demonstrated for several EPEC promoters (33), and we have now demonstrated it for EHEC LEE promoters, including that of tir.
The level of induction observed in a K-12 background for LEE2,3 and tir promoters, from both EPEC and EHEC, was ca. eightfold. It is possible that induction of these operons is more dramatic in the wild-type strain due to contributions of accessory factors, modifications of Ler, or loss of topological features in the single-copy fusions. This is clearly the case with the promoter for LEE4. In a K-12 background, LEE4 is strongly expressed in the absence of Ler and is induced at most twofold by Ler. In a wild-type background, by contrast, LEE4 is not expressed in Δler mutants and is strongly induced by Ler, as assessed from the levels of EspADBF proteins and from primer extension experiments. This implies both the presence of EPEC- and EHEC-specific accessory factors and the normal repression of LEE4 in the wild type. Further, it suggests that an accessory factor normally represses LEE4 and that Ler may function as a derepressor of expression from this operon. For example, lack of type III secretion may feed back to inhibit transcription.
While Ler is clearly responsible for regulating the expression of the elements involved in the AE phenotype, it also regulates other LEE-encoded factors not involved in AE lesion formation. Based on primer extension, Ler increased transcription from rorf2, orf10, rorf10, orf19, escD, and LEE4 operons. From the operon structure of the LEE (8, 33), this also predicts increased transcription of rorf1, orf11, and orf27 through espF. We observed increased levels of rOrf2 and EspF proteins in strains containing Ler, consistent with the results predicted from primer extension. Ler therefore is potentially able to activate the entire LEE.
Our findings compare with those recently published by Friedberg et al. (14) who screened, in an EPEC background, a multicopy plasmid library containing random fragments of the EPEC LEE fused to a gfp reporter. They demonstrated that Ler was necessary for production of EspADB, Tir, intimin, and EspF and could activate promoters for LEE2, LEE3, and eae in the range of 5- to 44-fold. They found that Ler did not activate rorf2, nor did they find evidence that LEE4 or other LEE genes were activated by Ler. While the use of reporter fusions in a wild-type background has certain advantages, our laboratory has found that the use of multicopy plasmids containing strong promoters fused to a toxic protein such as green fluorescent protein in a RecA+ background may mislead. For example, highly expressed genes may be toxic in this system and cannot be cloned, and so they are missed in a genetic screen. Furthermore, DNA topology is an important factor in the function of H-NS-like proteins, and studying regulation of a chromosomal gene cloned into a multicopy plasmid may result in misleading conclusions (such as the level of activation) due to differences in DNA topology between supercoiled plasmids and chromosomal genes. Consequently, we continue to use stable, chromosomally integrated fusions to the nontoxic lacZ reporter and support or extend our findings with Western blots, primer extensions, and other assays of activity in the wild-type host. We believe that the differences observed in the levels of activation between the two studies and the failure of Friedberg et al. (14) to observe Ler activation of rorf2, orf10, rorf10, orf19, escD, and LEE4 reflect differences in methodology.
In addition to the effects of Ler on LEE-located genes, we found that Ler regulates the expression of phenotypes and proteins encoded outside the LEE. We observed that Ler strongly activates (31-fold) the espC promoter and increases the levels of EspC secreted from EPEC. The 110-kDa secreted protein EspC is encoded on a second chromosomal pathogenicity island in EPEC and has recently been demonstrated to be an enterotoxin in vitro (Mellies et al., submitted). The homologous EHEC O157:H7 protein EspP has been identified in EHEC culture supernatants and is encoded on the EHEC virulence plasmid. We observed that Ler regulated the levels of a large secreted protein(s) from EHEC but could not demonstrate Ler activation of an espP::lacZ fusion in K-12 or activation of espP transcription in the wild type. This suggests that Ler regulates another, as yet unidentified autotransporter and that the protein observed in EHEC 85-170 supernatants is not EspP. Another gene carried on the EHEC plasmid, tagA (4), was, however, regulated by ler, as judged by tagA::lacZ fusions in K-12. tagA has no known function in EHEC or in V. cholerae, where it was first described as a ToxR-activated protein. It is interesting that the function of TagA remains cryptic yet it is activated by major virulence regulons in two unrelated enteric pathogens.
Ler also regulates fimbrial expression and adherence phenotypes. In EHEC, mutation of ler was associated with enhanced adherence to tissue culture monolayers, altered adherence patterns, and expression of long fine fimbriae (LFF). These parallels suggest that Ler is a repressor of LFF (or perhaps an activator of another repressor) and that these fimbriae mediate the DA-AA pattern of adherence observed in vitro. No fimbrial adhesin has yet been clearly defined for EHEC O157:H7, and we are conducting further investigation into the identity and properties of these fimbriae. It should be noted that it is difficult to determine the roles of particular Ler-regulated fimbriae in altered adherence phenotypes since other factors involved in adherence, such as intimin and the type III secretion system, are not expressed in Δler strains.
The EPEC Δler mutant exhibited a mixture of both DA-AA and LA to HEp-2 cells, while the wild type and the complemented strain exhibited LA. Electron microscopy demonstrated that the EPEC Δler mutant expressed a number of fimbriae of morphologic types not observed in the wild type and not previously described in EPEC or EHEC. These fimbriae in EPEC were unrelated to BFP since they were expressed under conditions normally nonpermissive for BFP production, they were demonstrated by immunoelectron microscopy to be distinct from BFP, and both Western blots and bfp::lacZ fusions demonstrated that Ler does not regulate BFP production. We are currently characterizing these novel fimbriae, and it is possible that the LFF observed in the EPEC Δler are identical to those observed in EHEC Δler and may represent a common EPEC and EHEC adhesin mediating DA-AA adherence to HEp-2 cells.
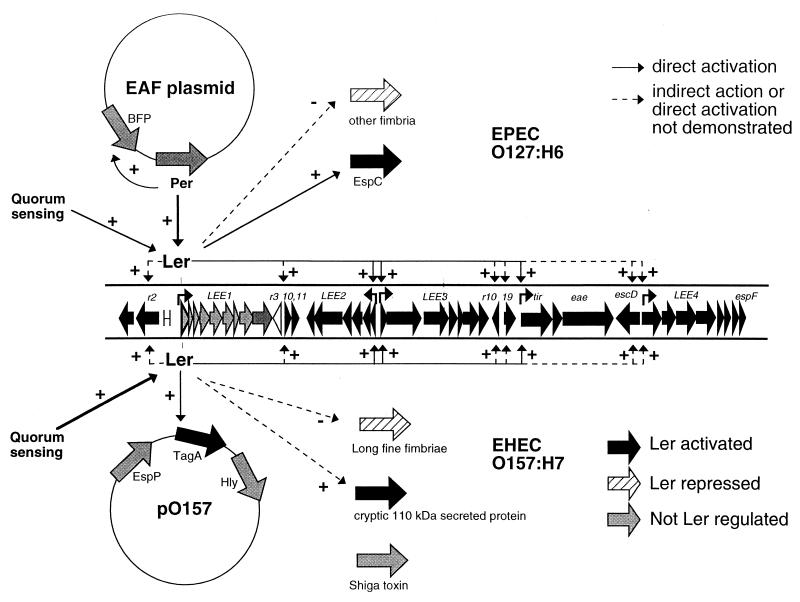
Ler therefore can have both negative and positive effects and thereby can play a central role in regulating many virulence and virulence-related phenotypes including AE lesion formation, adherence, and toxin production in both EPEC and EHEC. The fact that Ler coregulates so many genes suggests that a number of cryptic genes may play important roles in pathogenesis. We propose an entire ler regulon (Fig. 8), which suggests a pattern of gene expression in pathogenesis. Low levels of Ler expression are associated with enhanced production of fimbriae and/or adhesins which could be involved in initial colonization. High levels of Ler expression are associated with upregulation of genes involved in AE lesion formation, intimate adherence, and toxin production. This would imply at least two distinct phases in the regulation of virulence genes involved in pathogenesis.
FIG. 8.
The ler regulon. Ler regulates the expression of many genes both within the LEE and elsewhere on the genome. Some genes are directly regulated by Ler, as shown in gene fusion studies in E. coli K-12 (indicated by solid lines), while other genes are indirectly regulated or direct regulation has not been demonstrated (dashed lines). Expression of Ler is activated by quorum sensing in both EPEC and EHEC and additionally by Per in EPEC. Genes in the top half of the figure apply to EPEC O127:H6, and genes in the bottom half apply to EHEC O157:H7.
Since Ler expression is central to regulation of virulence genes, it follows that the regulation of Ler expression is important to pathogenesis. Ler expression in EPEC is activated by Per (33) and IHF (14). Per is not present in EHEC, but there is at least one shared regulatory pathway, since we have recently shown that quorum sensing activates the LEE1 promoter in both EPEC and EHEC (42). Differences in the pathways of Ler activation reflect differences in EPEC and EHEC pathogenesis. EHEC infects the large intestine and so could potentially use autoinducer secreted by the large number of resident bacteria to signal activation of virulence gene expression. EPEC normally infects the small intestine, where the concentration of bacteria is low, and may overcome this deficit by the presence of Per, which may respond to other environmental signals.
The structure of the Ler regulon also reflects the evolutionary history of EPEC and EHEC as proposed by Whittam and McGraw (48), in which the LEE elements were inherited first and other virulence factors added to the genome in later evolution. It would appear that many of the later elements have come under the control of Ler as they have been acquired, including other virulence loci and the large virulence plasmid (in EHEC). In contrast, it would appear that in EPEC the Per regulator may have evolved on the BFP plasmid first to regulate BFP and subsequently to regulate Ler.
The mechanism by which Ler regulates the expression of these genes remains to be determined. Ler is distantly related to the H-NS family of proteins but is functionally distinct from H-NS, as demonstrated by several assays (Table 5), and neither protein can functionally substitute for the other. Therefore, while H-NS is an important global regulator of housekeeping genes, Ler appears to be specific for virulence-associated genes and is encoded in a pathogenicity island. Furthermore, Ler also appears to be different from other H-NS homologs that appear to act as antagonists or modifiers or H-NS action, since Ler could not suppress H-NS-mediated phenotypes. This suggests that Ler may represent a new member of the H-NS family of DNA binding proteins but one that is neither analogous nor antagonistic to H-NS.
ACKNOWLEDGMENTS
We thank the staff of the University of Maryland Biopolymer Laboratory for sequencing, Stuart Knutton for examination of EspA filament production, and Maria S. Dubois for assistance with protein techniques. We especially thank Gina Donato and Tom Kawula, University of North Carolina, for assistance with H-NS experiments and the laboratory of Alison O'Brien, Uniformed Services University of the Health Sciences, for Shiga toxin assays.
This research was supported by NIH grants AI21657 and AI41325.
REFERENCES
- 1.Bertin P, Benhabiles N, Krin E, Laurent-Winter C, Tendeng C, Turlin E, Thomas A, Danchin A, Brasseur R. The structural and functional organization of H-NS-like proteins is evolutionarily conserved in gram-negative bacteria. Mol Microbiol. 1999;31:319–329. doi: 10.1046/j.1365-2958.1999.01176.x. [DOI] [PubMed] [Google Scholar]
- 2.Bieber D, Ramer S W, Wu C Y, Murray W J, Tobe T, Fernandez R, Schoolnik G K. Type IV pili, transient bacterial aggregates, and virulence of enteropathogenic Escherichia coli. Science. 1998;280:2114–2118. doi: 10.1126/science.280.5372.2114. [DOI] [PubMed] [Google Scholar]
- 3.Brunder W, Schmidt H, Karch H. EspP, a novel extracellular serine protease of enterohaemorrhagic Escherichia coli O157:H7 cleaves human coagulation factor V. Mol Microbiol. 1997;24:767–778. doi: 10.1046/j.1365-2958.1997.3871751.x. [DOI] [PubMed] [Google Scholar]
- 4.Burland V, Shao Y, Perna N T, Plunkett G, Sofia H J, Blattner F R. The complete DNA sequence and analysis of the large virulence plasmid of Escherichia coli O157:H7. Nucleic Acids Res. 1998;26:4196–4204. doi: 10.1093/nar/26.18.4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Donato G M, Lelivelt M J, Kawula T H. Promoter-specific repression of fimB expression by the Escherichia coli nucleoid-associated protein H-NS. J Bacteriol. 1997;179:6618–6625. doi: 10.1128/jb.179.21.6618-6625.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dorman C J, Hinton J C D, Free A. Domain organization and oligomerization among H-NS-like nucleoid-associated proteins in bacteria. Trends Microbiol. 1999;7:124–129. doi: 10.1016/s0966-842x(99)01455-9. [DOI] [PubMed] [Google Scholar]
- 7.Dorman C J, Bhriain N N. Co-ordinate regulation of virulence gene expression in Escherichia coli. In: Sussman M, editor. Escherichia coli: mechanisms of virulence. Cambridge, United Kingdom: Cambridge University Press; 1999. pp. 373–399. [Google Scholar]
- 8.Elliott S, Wainwright L A, McDaniel T, MacNamara B, Donnenberg M, Kaper J B. The complete sequence of the locus of enterocyte effacement (LEE) from enteropathogenic Escherichia coli E2348/69. Mol Microbiol. 1998;28:1–4. doi: 10.1046/j.1365-2958.1998.00783.x. [DOI] [PubMed] [Google Scholar]
- 9.Elliott S J, Dubois M S, Hutcheson S W, Wainwright L A, Batchelor M, Frankel G, Knutton S, Kaper J B. Identification of CesT, a chaperone for the type III secretion of Tir in enteropathogenic Escherichia coli. Mol Microbiol. 1999;33:1176–1189. doi: 10.1046/j.1365-2958.1999.01559.x. [DOI] [PubMed] [Google Scholar]
- 10.Elliott S J, Kaper J B. Role of type 1 fimbriae in EPEC infections. Microb Pathog. 1997;23:113–118. doi: 10.1006/mpat.1997.0135. [DOI] [PubMed] [Google Scholar]
- 11.Elliott S J, Yu J, Kaper J B. The cloned locus of enterocyte effacement (LEE) from enterohemorrhagic Escherichia coli O157:H7 is unable to confer the attaching and effacing phenotype upon E. coli K-12. Infect Immun. 1999;67:4260–4263. doi: 10.1128/iai.67.8.4260-4263.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elliott T. A method for constructing single-copy lac fusions in Salmonella typhimurium and its application to the hemA-prfA operon. J Bacteriol. 1992;174:245–253. doi: 10.1128/jb.174.1.245-253.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frankel G, Phillips A D, Rosenshine I, Dougan G, Kaper J B, Knutton S. Enteropathogenic and enterohaemorrhagic E. coli: more subversive elements. Mol Microbiol. 1998;30:911–921. doi: 10.1046/j.1365-2958.1998.01144.x. [DOI] [PubMed] [Google Scholar]
- 14.Friedberg D, Umanski T, Fang Y, Rosenshine I. Hierarchy in the expression of the locus of enterocyte effacement genes of enteropathogenic Escherichia coli. Mol Microbiol. 1999;34:941–952. doi: 10.1046/j.1365-2958.1999.01655.x. [DOI] [PubMed] [Google Scholar]
- 15.Gentry M K, Dalrymple J M. Quantitative microtiter cytotoxicity assay for Shigella toxin. J Clin Microbiol. 1980;12:361–366. doi: 10.1128/jcm.12.3.361-366.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Girón J A, Ho A S, Schoolnik G K. An inducible bundle-forming pilus of enteropathogenic Escherichia coli. Science. 1991;254:710–713. doi: 10.1126/science.1683004. [DOI] [PubMed] [Google Scholar]
- 17.Girón J A, Ho A S, Schoolnik G K. Characterization of fimbriae produced by enteropathogenic Escherichia coli. J Bacteriol. 1993;175:7391–7403. doi: 10.1128/jb.175.22.7391-7403.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gómez-Duarte O G, Kaper J B. A plasmid-encoded regulatory region activates chromosomal eaeA expression in enteropathogenic Escherichia coli. Infect Immun. 1995;63:1767–1776. doi: 10.1128/iai.63.5.1767-1776.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goyard S, Bertin P. Characterization of BpH3, an H-NS-like protein in Bordetella pertussis. Mol Microbiol. 1997;24:815–823. doi: 10.1046/j.1365-2958.1997.3891753.x. [DOI] [PubMed] [Google Scholar]
- 20.Griffin P M, Ostroff S M, Tauxe R V, Greene K D, Wells J G, Lewis J H, Blake P A. Illnesses associated with Escherichia coli O157:H7 infections. Ann Intern Med. 1988;109:705–712. doi: 10.7326/0003-4819-109-9-705. [DOI] [PubMed] [Google Scholar]
- 21.Hueck C J. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol Mol Biol Rev. 1998;62:379–433. doi: 10.1128/mmbr.62.2.379-433.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jarvis K G, Girón J A, Jerse A E, McDaniel T K, Donnenberg M S, Kaper J B. Enteropathogenic Escherichia coli contains a specialized secretion system necessary for the export of proteins involved in attaching and effacing lesion formation. Proc Natl Acad Sci USA. 1995;92:7996–8000. doi: 10.1073/pnas.92.17.7996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jarvis K G, Kaper J B. Secretion of extracellular proteins by enterohemorrhagic Escherichia coli via a putative type III secretion system. Infect Immun. 1996;64:4826–4829. doi: 10.1128/iai.64.11.4826-4829.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaper J B, Elliott S J, Sperandio V, Perna N T, Mayhew G F, Blattner F R. Attaching-and-effacing intestinal histopathology and the locus of enterocyte effacement. In: Kaper J B, O'Brien A D, editors. Escherichia coli O157:H7 and other Shiga toxin-producing E. coli strains. Washington, D.C.: ASM Press; 1998. pp. 163–182. [Google Scholar]
- 25.Kawula T H, Orndorff P E. Rapid site-specific DNA inversion in Escherichia coli mutants lacking the histonelike protein H-NS. J Bacteriol. 1991;173:4116–4123. doi: 10.1128/jb.173.13.4116-4123.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kenny B, Abe A, Stein M, Finlay B B. Enteropathogenic Escherichia coli protein secretion is induced in response to conditions similar to those in the gastrointestinal tract. Infect Immun. 1997;65:2606–2612. doi: 10.1128/iai.65.7.2606-2612.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Knutton S, Baldwin T, Williams P H, McNeish A S. Actin accumulation at sites of bacterial adhesion to tissue culture cells: basis of a new diagnostic test for enteropathogenic and enterohemorrhagic Escherichia coli. Infect Immun. 1989;57:1290–1298. doi: 10.1128/iai.57.4.1290-1298.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Knutton S, Rosenshine I, Pallen M J, Nisan I, Neves B C, Bain C, Wolff C, Dougan G, Frankel G. A novel EspA-associated surface organelle of enteropathogenic Escherichia coli involved in protein translocation into epithelial cells. EMBO J. 1998;17:2166–2176. doi: 10.1093/emboj/17.8.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Levine M M, Bergquist E J, Nalin D R, Waterman D H, Hornick R B, Young C R, Sotman S, Rowe B. Escherichia coli strains that cause diarrhoea but do not produce heat-labile or heat-stable enterotoxins and are non-invasive. Lancet. 1978;i:1119–1122. doi: 10.1016/s0140-6736(78)90299-4. [DOI] [PubMed] [Google Scholar]
- 30.McDaniel T K. Ph.D. thesis. Baltimore: University of Maryland; 1996. [Google Scholar]
- 31.McDaniel T K, Jarvis K G, Donnenberg M S, Kaper J B. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc Natl Acad Sci USA. 1995;92:1664–1668. doi: 10.1073/pnas.92.5.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McDaniel T K, Kaper J B. A cloned pathogenicity island from enteropathogenic Escherichia coli confers the attaching and effacing phenotype on E. coli K-12. Mol Microbiol. 1997;23:399–407. doi: 10.1046/j.1365-2958.1997.2311591.x. [DOI] [PubMed] [Google Scholar]
- 33.Mellies J, Elliott S J, Sperandio V, Donnenberg M S, Kaper J. The Per regulon of enteropathogenic Escherichia coli: identification of a regulatory cascade and a novel transcriptional activator, the locus of enterocyte effacement (LEE)-encoded regulator (Ler) Mol Microbiol. 1999;33:296–306. doi: 10.1046/j.1365-2958.1999.01473.x. [DOI] [PubMed] [Google Scholar]
- 34.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 35.Nataro J P, Kaper J B. Diarrheagenic Escherichia coli. Clin Microbiol Rev. 1998;11:142–201. doi: 10.1128/cmr.11.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perna N T, Mayhew G F, Pósfal G, Elliott S, Donnenberg M S, Kaper J B, Blattner F R. Molecular evolution of a pathogenicity island from enterohemorrhagic Escherichia coli O157:H7. Infect Immun. 1998;66:3810–3817. doi: 10.1128/iai.66.8.3810-3817.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Riley L W, Remis R S, Helgerson S D, McGee H B, Wells J G, Davis B R, Hebert R J, Olcott E S, Johnson L M, Hargrett N T, Blake P A, Cohen M L. Hemorrhagic colitis associated with a rare Escherichia coli serotype. N Engl J Med. 1983;308:681–685. doi: 10.1056/NEJM198303243081203. [DOI] [PubMed] [Google Scholar]
- 38.Rosenshine I, Donnenberg M S, Kaper J B, Finlay B B. Signal transduction between enteropathogenic Escherichia coli (EPEC) and epithelial cells: EPEC induces tyrosine phosphorylation of host cell proteins to initiate cytoskeletal rearrangement and bacterial uptake. EMBO J. 1992;11:3551–3560. doi: 10.1002/j.1460-2075.1992.tb05438.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scaletsky I C A, Silva M L M, Trabulsi L R. Distinctive patterns of adherence of enteropathogenic Escherichia coli to HeLa cells. Infect Immun. 1984;45:534–536. doi: 10.1128/iai.45.2.534-536.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shi X, Bennett G N. Plasmids bearing hfq and the hns-like gene stpA complement hns mutants in modulating arginine decarboxylase gene expression in Escherichia coli. J Bacteriol. 1994;176:6769–6775. doi: 10.1128/jb.176.21.6769-6775.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Simons R W, Houman F, Kleckner N. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene. 1987;53:85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- 42.Sperandio V, Mellies J, Nguyen W, Kaper J B. Quorum sensing controls expression of the type III secretion gene transcription and protein secretion in enterohemorrhagic and enteropathogenic Escherichia coli. Proc Natl Acad Sci USA. 1999;96:15196–15201. doi: 10.1073/pnas.96.26.15196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stein M, Kenny B, Stein M A, Finlay B B. Characterization of EspC, a 110-kilodalton protein secreted by enteropathogenic Escherichia coli which is homologous to members of the immunoglobulin A protease-like family of secreted proteins. J Bacteriol. 1996;178:6546–6554. doi: 10.1128/jb.178.22.6546-6554.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tobe T, Schoolnik G K, Sohel I, Bustamente V H, Puente J L. Cloning and characterization of bfpTVW, genes required for the transcriptional activation of bfpA in enteropathogenic Escherichia coli. Mol Microbiol. 1996;55:3117–3125. doi: 10.1046/j.1365-2958.1996.531415.x. [DOI] [PubMed] [Google Scholar]
- 45.Tzipori S, Karch H, Wachsmuth I K, Robins-Browne R M, O'Brien A D, Lior H, Cohen M L, Smithers J, Levine M M. Role of a 60-megadalton plasmid and Shiga-like toxins in the pathogenesis of infection caused by enterohemorrhagic Escherichia coli O157:H7 in gnotobiotic piglets. Infect Immun. 1987;55:3117–3125. doi: 10.1128/iai.55.12.3117-3125.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ueguchi C, Suzuki T, Yoshida T, Tanaka K, Mizuno T. Systematic mutational analysis revealing the functional domain organization of Escherichia coli nucleoid protein H-NS. J Mol Biol. 1996;263:149–162. doi: 10.1006/jmbi.1996.0566. [DOI] [PubMed] [Google Scholar]
- 47.Wainwright L A, Kaper J B. EspB and EspD require a specific chaperone for proper secretion from enteropathogenic Escherichia coli. Mol Microbiol. 1998;27:1247–1260. doi: 10.1046/j.1365-2958.1998.00771.x. [DOI] [PubMed] [Google Scholar]
- 48.Whittam T S, McGraw E A. Clonal analysis of EPEC serogroups. Rev Microbiol Sao Paulo. 1996;27(Suppl. 1):7–16. [Google Scholar]