Abstract
The two-component regulatory system PmrA-PmrB confers resistance of Salmonella spp. to cationic antimicrobial peptides (AP) such as polymyxin (PM), bactericidal/permeability-increasing protein, and azurocidin. This resistance occurs by transcriptional activation of two loci termed pmrE and pmrHFIJKLM. Both pmrE and pmrHFIJKLM produce products required for the biosynthesis of lipid A with 4-aminoarabinose (Ara4N). Ara4N addition creates a more positively charged lipopolysaccharide (LPS) and thus reduces cationic AP binding. Experiments were conducted to further analyze the regulation of the pmrHFIJKLM operon and the role of this operon and the surrounding genomic region in LPS modification and antimicrobial peptide resistance. The pmrHFIJKLM genes are cotranscribed and over 3,000-fold regulated by PmrA-PmrB. The pmrHFIJKLM promoter bound PmrA, as determined by gel shift analysis, as did a 40-bp region of the PmrA-PmrB-regulated pmrCAB promoter. Construction of nonpolar mutations in the pmrHFIJKLM genes showed that all except pmrM were necessary for the Ara4N addition to lipid A and PM resistance. The flanking genes of the operon (pmrG and pmrD) were not necessary for PM resistance, but pmrD was shown to be regulated by the PhoP-PhoQ regulatory system. BALB/c mice inoculated with pmrA and pmrHFIJKLM mutant strains demonstrated virulence attenuation when the strains were administered orally but not when they were administered intraperitoneally, indicating that Ara4N addition may be important for resistance to host innate defenses within intestinal tissues.
The key to success for many bacteria in causing infection is colonization of host tissues. Enteric bacteria, such as Salmonella spp., have to survive in harsh host microenvironments including the intestinal mucosa. At the intestinal mucosa, these bacteria encounter host defense mechanisms including antimicrobial peptides (AP), which are cationic, amphipathic molecules that kill bacteria by membrane permeabilization. Within the intestine, AP are secreted into the lumen by Paneth cells located in the base of intestinal crypts. AP are also found within phagocytic cells located in the intestinal submucosa. Following oral ingestion, typhoid fever-causing strains of Salmonella can transcytose through M cells and intestinal epithelial cells and are then taken up by and survive within resident phagocytes. The ability of salmonellae to survive within the host intestine and within professional phagocytes is likely to depend, at least in part, on mechanisms of resistance to AP.
Lipopolysaccharade (LPS) is the major surface component of gram-negative bacteria. Lipid A is the bioactive component of LPS that comprises the outer leaflet of the gram-negative bacterial outer membrane. Phosphate groups on lipid A and LPS core components result in the bacterial surface having a net negative charge. This charge plays a significant role in the electrostatic interaction of cationic AP with the bacterial surface. Gram-negative organisms can synthesize LPS with specific lipid A and core modifications in response to environmental conditions that include those found in host tissues. Lipid A modifications are best characterized for Salmonella spp. in which the addition of palmitate or 4-aminoarabinose (Ara4N) to lipid A is coordinately regulated by the two-component system PhoP-PhoQ (9–11), which is necessary for bacterial survival within macrophages and within the host (6, 12, 15). Ara4N-containing lipid A results in a less negatively charged bacterial surface, which reduces AP binding and promotes resistance to the cationic AP polymyxin B (PM), bactericidal/permeability-increasing protein, and azurocidin (17, 18).
PhoP-PhoQ-mediated Ara4N addition to lipid A occurs through transcriptional activation of the genes encoding another two-component system, PmrA-PmrB, which can itself be activated independent of PhoP-PhoQ by low-pH or high-iron conditions (9, 14, 20). Two loci, pmrE and pmrHFIJKLM, are regulated by PmrA-PmrB and are essential for both biosynthesis of lipid A containing Ara4N and for resistance to PM (1, 8). This study was conducted to examine the individual and collective roles of the genes of the pmrHFIJKLM operon, as well as flanking genes, in AP resistance, modification of lipid A, and Salmonella enterica serovar Typhimurium virulence.
MATERIALS AND METHODS
Bacterial strains, culture conditions, and reagents.
Bacterial strains used in this study are listed in Table 1. Cultures were grown on Luria-Bertani (LB) agar plates or broth at 37°C with aeration. Antibiotics were used at the following concentrations: chloramphenicol, 25 μg/ml; ampicillin, 50 μg/ml; kanamycin, 45 μg/ml; tetracycline, 15 μl/ml; streptomycin, 1,000 μg/ml. The chromogenic substrate for β-galactosidase, 5-bromo-4-chloro-indolyl-β-d-galactopyranoside (X-Gal), was used at a concentration of 40 μg/ml.
TABLE 1.
Bacterial strains and plasmids and relevant properties
| Strain or plasmid | Genotype or relevant phenotype | Source or reference |
|---|---|---|
| Strainsa | ||
| SM10λpir | thi-1 thr-1 leuB6 supE44 tonA21 lacY1 recA::RP4-2-Tc::Mu | |
| JSG210 | ATCC 14028s (WT) | ATCC |
| JSG430 | pho24 pmrA::Tn10d (PhoPc PmrA−) | 9 |
| JSG437 | phoP::Tn10d-tet pmrA505 zjd::Tn10d-cam (PhoP− PmrAc) | 9 |
| JSG435 | ATCC 14028s pmrA 505 zjd::Tn10d-cam (PmrAc) | 9 |
| JSG421 | pmrA::Tn10d (PmrA−) | 9 |
| JSG485 | JSG435 pmrF1::Tn10d | 8 |
| JM109 | F′traD36 lacI Δ(lacZ)M15 proA+B+/e14− (mcrA) Δ(lac-proAB) thi gyrA96 (Nalr) endA1 hsdR17 (rK− mK−) relA1 supE44 recA1 | Promega |
| JSG224 | phoN2 zxx::6251 Tn10d-cam (CS019) | 15 |
| JSG844 | JSG435 rpsL (PmrAc Smr) | This study |
| JSG1097 | JSG844; ΔpmrH | This study |
| JSG911 | JSG844; ΔpmrF | This study |
| JSG868 | JSG844; ΔpmrI | This study |
| JSG1098 | JSG844; ΔpmrJ | This study |
| JSG883 | JSG844; ΔpmrK | This study |
| JSG884 | JSG844; ΔpmrL | This study |
| JSG1046 | JSG421 pmrM::luc | This study |
| JSG1047 | JSG435 pmrM::luc | This study |
| JSG1048 | JSG485 pmrM::luc | This study |
| JSG989 | JSG224 pmrD::luc | This study |
| Plasmids | ||
| pGPL01 | Firefly luciferase recorder, suicide vector | 9 |
| pKAS32 | rpsL suicide vector | 19 |
| pSR01 | pKAS32; ΔpmrH | This study |
| pSR02 | pKAS32; ΔpmrI | This study |
| pSR03 | pKAS32; ΔpmrK | This study |
| pSR04 | pKAS32; ΔpmrL | This study |
| pSR05 | pKAS32; ΔpmrF | This study |
| pSR06 | pKAS32; ΔpmrJ | This study |
| pLB02 | Firefly luciferase recorder, suicide vector | 7 |
| pGPLM02 | pGPL01 with pmrM EcoRI-KpnI PCR fragment amplified with JG252 and JG253 | This study |
| pMRD1 | pGPL01 with pmrD EcoRI-KpnI PCR fragment amplified with JG177 and JG178 | This study |
| pLB2223 | 118-bp pmrHFIJKLM operon promoter-fragment (JG122 and JG123) in pLB02 (EcoRI-KpnI) | This study |
| pQPA01 | pQE30 containing the pmrA (WT) gene amplified with primers JG106 and JG107 (BamHI-PstI) | This study |
| pQP505 | pQE30 containing the pmrA-constitutive (JSG435) gene amplified with primers JG106 and JG107 (BamHI-PstI) | This study |
| pQE30 | Amino-terminal His tag vector | Qiagen |
Strain SMI0λpir is E. coli; the other strains are S. enterica serovar Typhimurium.
Construction of luciferase fusions and transcription assays.
pmrD was amplified from plasmid pKK01 (8) by PCR using primers JG177 (5′ GGG AAT TCT GCC ATG TTC TGG TGC TGT GC 3′) and JG178 (5′ GGG GTA CCG CGC GTC AAC CGC TGC CAT TC 3′). This fragment was digested with the restriction enzymes EcoRI and KpnI and ligated into pGPL01, which is a Pir-dependent suicide vector containing a promoterless firefly luciferase gene downstream of the multiple cloning site (9). This clone, transformed into Escherichia coli SM10λPir, was mated with CS019, and a single recombinant was identified. P22HTint phage transduction was used to transduce the pmrD∷luc fusion into various strains. Lucifierase activity was determined as previously described (9) after growth of strains to log phase (optical density at 600 nm [OD600] ∼0.6). A pmrM luciferase fusion was constructed in a manner identical to that for the pmrD fusion with primers JG252 (5′ GGAATTCGGACTGATAAGCGTTGCG 3′) and JG253 (5′ GGGGTACCTGATGCACGCTGTTATTCC 3′). The luciferase fusion to the pmrHFIJKLM promoter was also constructed similarly to the pmrD and pmrM fusions, except that the promoter fragment was cloned into pLB02 (7), which contains DNA homologous to a region downstream of pagC, where the vector is recombined. The primers used to amplify the pmrHFIJKLM promoter were JG122 (5′ GGGGTACCTGAAAGCCGCTTT TC 3′) and JG123 (5′ GGAATTCTTTTTACTTCACCT 3′).
Analysis of LPS modification.
LPS was isolated by Mg2+-ethanol precipitation as described by Darveau and Hancock (4), and lipid A was isolated by hydrolysis in 1% sodium dodecyl sulfate at pH 4.5 (3). Negative-ion matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry was performed as described previously (5). Lyophilized lipid A was dissolved with 5 μl of a 5-chloro-2-mercaptobenzothiazole (CMBT) MALDI matrix in chloroform-methanol (1:1 [vol/vol]) and then applied (1 μl) to the sample plate. All MALDI-TOF experiments were performed using a BiflexIII mass spectrometer (Bruker Daltonics, Inc., Billerica, Mass.). Mass spectroscopy tracings were analyzed for the presence or absence of Ara4N based on known peak locations at m/z 1,929 (hexa-acylated lipid A with Ara4N) and 2,167 (hepta-acylated lipid A with Ara4N).
Assays of DNA binding.
PCR fragments of the pmrHFIJKLM and pmrCAB operons were digested with EcoRI (located at one end of each fragment) and labeled with [α-32P]dATP (10 μCi/reaction) with the Klenow fragment of DNA polymerase I. Binding reactions were carried out by the incubation of the purified PmrA or PmrA-constitutive (PmrAc) His-tagged proteins with binding buffer (10mM Tris-HCl [pH 7.5], 1mM EDTA, 100 mM KCl, 0.1 mM dithiothreitol, 5% glycerol, 75 μg of sonicated salmon sperm DNA/ml, 10 mg of bovine serum albumin/ml for 5 min at room temperature. Labeled probe (1 to 5 ng) was added, and the reaction mixture was incubated for 10 min at room temperature. Samples were electophoresed at 4°C on a 5% acrylamide gel at 200 V. Gels were subsequently dried and autoradiographed.
Animal studies.
Survival assays were accomplished as follows. Female BALB/c mice were inoculated orally with 20 μl of an overnight culture (washed and diluted to 106 organisms/20 μl in phosphate-buffered saline (PBS), which is 1 log unit above the 50% lethal dose [LD50]) using the animal's swallowing reflex. Mice were prefed 20 μl of 10% sodium bicarbonate 30 min prior to bacterial inoculation. Alternatively, after dilution in PBS, animals were inoculated via the intraperitoneal route with various numbers of organisms in a 100-μl volume. Overnight cultures were plated to enumerate the numbers of organisms inoculated. The average days of survival and the numbers of surviving mice were recorded. In competition assays, female BALB/c mice were inoculated orally as described above. Mice were inoculated with a mixture of 10 μl of two different strains (each containing 106 organisms/10 μl). Mice were sacrificed when clearly moribund, and livers and spleens were removed. The organs were homogenized in 1 × PBS, diluted, and plated onto the appropriate antibiotic-containing agar plates, which selected for one of the two competing strains.
MIC assays of AP resistance.
PM (U.S. Biochemicals; 8,040 U mg−1) was used at concentrations of 0.0084 to 12 μg/ml in both plate and broth assays. Standard MIC testing of susceptibility to PM was accomplished as described by Steinberg et al. (21).
Deletion analysis of the pmrHFIJKLM operon.
Flanking regions of the genes of the operon were amplified using PCR and digested with the restriction enzyme NcoI. Primers that were used are listed in Table 2. The flanking regions were then ligated together and digested with the restriction enzymes EcoRI (3′) and XbaI (5′). This fragment was then ligated into plasmid pKAS32, a suicide vector containing a dominant streptomycin sensitivity allele (19). Deletion constructs in SM10λPir were mated with JSG844, and single recombinants were selected by plasmid-encoded resistance to ampicillin and chromosome-encoded resistance to chloramphenicol and sensitivity to streptomycin. Losses of the plasmid (secondary recombination events) were selected following overnight incubation by growth on 1,000-μg/ml streptomycin-containing agar. Colonies were further screened by PCR to confirm the incorporation of the deletion. Mutation of the pmrM gene was accomplished not by the construction of a nonpolar deletion but by the construction of the pmrM::luc fusion described above, which essentially inserts plasmid pGPL01 into the pmrM coding sequence.
TABLE 2.
Primers used for construction of each nonpolar deletion
| Deleted gene | Primer | Primer sequence (5′ to 3′) |
|---|---|---|
| pmrH | JG234 | CAT GCC ATG GCA TTG AAA GCC GCT TTT C |
| JG235 | GCT CTA GAC CCG TGC TGT CTG ACA GG | |
| JG236 | CAT GCC ATG GGA TAG CAG GAC AAT AAG CAT G | |
| JG237 | CGG AAT TCG TGA TGC ACC GGG ATC TC | |
| pmrF | JG183 | CAT GCC ATG GGT CAA ACA TGC TTA TTG TCC TGC TAT C |
| JG181 | GCT CTA GAC CCT TGA CCG TAA TCC CCG TG | |
| JG184 | CAT GCC ATG GGC CGT TTA CTG AGG AAA GCC ACC AAT G | |
| JG202 | CGG AAT TCG CCG TTA TAG CTG AAC GCG CC | |
| pmrI | JG185 | CAT GCC ATG GTG GTG GCT TTC CTC AGT AAA C |
| JG187 | GCT CTA GAC AGG ACA GCC TGT TTC GCA AAT CG | |
| JG186 | CAT GCC ATG GCG ATA TCG CGG AAC GCG CAT C | |
| JG188 | CGG AAT TCC CAG GTA TTG AAC GAC TGC GE | |
| pmrJ | JG245 | CAT GCC ATG GGC GTA AAC CGA CTT TCG TCA TGA TGC G |
| JG246 | GCT CTA GAC AGC GGT AAA CGT CGC ATT CG | |
| JG247 | CAT GCC ATG GGG CAG CGA GCG CCT CAT GAT G | |
| JG248 | CGG AAT TCG AAA GAA CAT AGG TGG GCA GTT TC | |
| pmrK | JG191 | CAT GCC ATG GGC GTA TCG ATT TCA TCA TGA G |
| JG193 | GCT CTA GAC ACC ATG CGT GGC AGA CTC AC | |
| JG192 | CAT GCC ATG GGG TGT TAA TTC AGT ATC GGC C | |
| JG196 | CGG AAT TCG CAA GCC ATC ATG ATC TGG CG | |
| pmrL | JG194 | CAT GCC ATG GCA GAA CGA CGC CGA TCA TTT AGG C |
| JG193 | GCT CTA GAC ACC ATG CGT GGC AGA CTC AC | |
| JG195 | CAT GCC ATG GGG CAT AAT GGG CGT AAT GTG G | |
| JG196 | CGG AAT TCG CAA GCC ATC ATG ATC TGG CG |
Confirmation of in-frame deletions and complementation experiments.
PCR fragments overlapping the deletion sites were sequenced to confirm correct incorporation of the in-frame deletion. Deletions in pmrH and pmrF were complemented with plasmid pKK013 (carrying only the intact pmrH and pmrF genes). The pmrJ, pmrK, pmrL, and pmrM genes were complemented by pKK012-1 (carrying only the intact pmrJKL and pmrM genes). To further confirm nonpolarity, the pmrM::luc fusion was mated into each deletion strain and luciferase activity was monitored.
Isolation of the PmrA protein.
The pmrA gene or the mutant, constitutive allele of this gene (pmrA505) was amplified by PCR with primers JG106 (5′ GCGGATCCAAGATACTGATTGTTGAAG 3′) and JG107 (5′ AACTGCAGTTAGCTTTCCTCAGTGGC 3′) from strains ATCC 14028s (wild type [WT]) and JSG435, respectively. These fragments were cloned into the N-terminal His tag-encoding vector pQE30 and transformed into strain XL1-Blue-MRF′. Transcription of the genes was induced with IPTG (isopropyl-β-d-thiogalactopyranoside) upon movement of the plasmids into strain M15/pREP4, and the His-tagged proteins were purified as previously described (7).
RESULTS
Definition of the pmrHFIJKLM transcriptional unit and promoter.
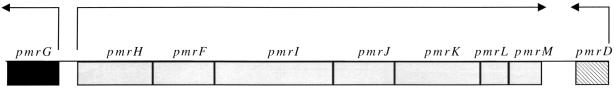
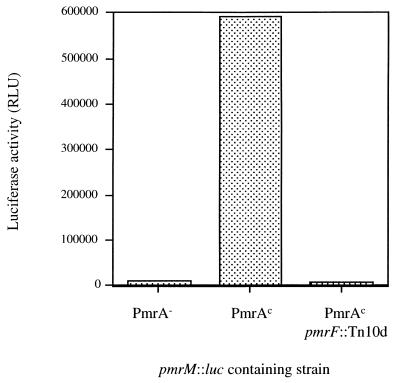
The pmrHFIJKLM genes are unidirectionally transcribed, and the individual open reading frames are separated by no more than 5 bp, as shown in the diagram of this chromosomal region (Fig. 1). This arrangement suggested that these genes formed a typical prokaryotic operon. To demonstrate that these genes were indeed cotranscribed, a luciferase fusion to pmrM, the last gene of the operon, was constructed in single copy on the chromosome. Upon the movement of this fusion into PmrA-null (PmrA−), PmrAc, and PmrAc pmrF::Tn10d backgrounds by P22-mediated transduction, luciferase activity was monitored. As shown in Fig. 2, this fusion was highly regulated by PmrA-PmrB, and the reporter activity in a PmrA− strain was similar to that in a strain containing the pmrF::Tn10d insertion, demonstrating that the insertion in pmrF had a polar effect on downstream gene transcription. Therefore, these data strongly suggest that the pmrHFIJKLM genes are cotranscribed and that the promoter is upstream of this operon.
FIG. 1.
Diagram of the chromosomal region containing the pmrG, pmrD, and pmrHFIJKLM genes. Arrows, direction of transcription.
FIG. 2.
The pmrHFIJKLM genes are expressed as an operon. PmrA−, PmrAc, and PmrAc pmrF::Tn10d strains were examined. Each strain contained a single-copy pmrM::luc fusion and was assayed in logarithmic phase (OD600, 0.6) for firefly luciferase-activity (expressed as relative light units [RLU]). The data are from a single experiment of three independent assays that gave similar results.
To define the DNA region of the pmrHFIJKLM operon necessary for PmrA-mediated activation, a 118-bp region of the predicted promoter was examined for regulation in a single-copy luciferase assay system and also by gel mobility shift experiments for DNA binding. To examine promoter regulation, the fragment was cloned into the luciferase fusion/suicide vector pLB02 (7) and subsequently recombined (into an innocuous site downstream of pagC) onto the chromosomes of PmrA− and PmrAc strains. As shown in Fig. 3, analysis of luciferase activity showed this promoter to be 3,442-fold activated by PmrA. Furthermore, little activity of the pmrHFIJKLM promoter-luciferase gene fusion was observed in a PmrA− or PhoPc PmrA− strain. This demonstrates that there is little transcription of the pmrHFIJKLM genes in the absence of PmrA and that direct activation of the promoter is mediated by PmrA and not PhoP.
FIG. 3.
The pmrHFIJKLM promoter is highly PmrA-PmrB activated and tightly regulated. PmrA−, PmrAc, PhoPc PmrA−, and PhoP− PmrAc strains containing a single-copy pmrHFIJKLM promoter::luc fusion were assayed in logarithmic phase (OD600, 0.6) for firefly luciferase activity (expressed as relative light units [RLU]). Note that the y-axis scale is logarithmic. The data are from a single experiment of three independent assays that gave similar results.
To demonstrate PmrA binding to this promoter fragment, gel mobility shift experiments were conducted with the purified PmrA-His protein, as well as a mutant version of this protein (17) previously shown to result in constitutive activation of PmrA-PmrB regulated genes (PmrAc-His). As shown in Fig. 4, the pmrHFIJKLM promoter fragment bound to both PmrA-His and PmrAc-His. Densitometry studies suggest that the PmrAc-His protein resulted in a threefold increase in the amount of shifted promoter fragment, suggesting that constitutive activation of PmrA-regulated genes by this protein may be due to increased affinity for promoter binding.
FIG. 4.
Binding of the PmrA-His and PmrAc-His proteins to the pmrHFIJKLM promoter (called pmrF) and the pmrCAB promoter (called pmrC). The sizes of the fragments used and the primers used to amplify the fragments are noted. Arrows, shifted fragments. N, no protein added; A, PmrA-His added; Ac, PmrAc-His added.
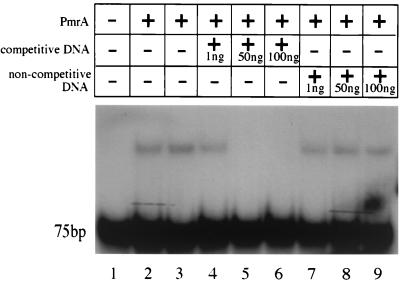
To extend the results discussed above to the definition of a consensus DNA binding site for PmrA, we performed gel shift analysis on promoter fragments of another PmrA-regulated locus, pmrCAB. The pmrCAB operon is positively autoregulated by PmrA (9, 20). PCR fragments extending 75, 100, 125, and 200 bp upstream from the ATG start codon of the pmrC gene were examined by gel mobility shift experiments, and all fragments were shown to bind PmrA (Fig. 4; data not shown for the 100- and 125-bp fragments). Based on these studies, a 40-bp fragment corresponding to a region directly upstream of the predicted −35 sequence was amplified by PCR and analyzed in the gel shift assay. As shown in Fig. 5, this fragment also bound PmrA. In addition, the binding of PmrA to the pmrCAB promoter was shown to be specific. Binding could be completely abolished with 50 ng of unlabeled promoter fragment DNA; however, 100 ng of unlabeled, noncompetitive (pUC19) DNA did not produce any reduction of PmrA-pmrCAB promoter binding. Therefore, the PmrA binding site is contained within this 40-bp sequence.
FIG. 5.
PmrA-His protein binding to the pmrCAB promoter is specific. The presence or absence of the PmrA-His protein, competitive DNA, and noncompetitive DNA is noted at the top. The numbers at the bottom are the lane numbers.
Regulation and role in PM resistance of the flanking genes of the operon.
It has been shown that the gene upstream of and divergently transcribed from the pmrHFIJKLM operon is the PmrA-PmrB-regulated gene, pmrG (8). The gene directly downstream and divergently transcribed from the pmrHFIJKLM operon is pmrD, which imparts increased PM resistance to S. enterica serovar Typhimurium when expressed from a multicopy plasmid (16). To further the studies of this chromosomal region, experiments were designed to determine the potential regulation of pmrD and the role of pmrD and pmrG in resistance to PM.
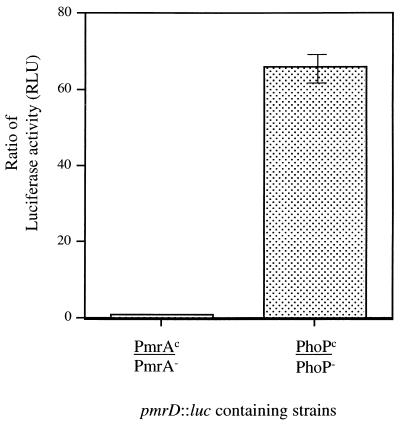
To examine both the regulation and role in AP resistance of the pmrD gene, a fragment internal to the gene was cloned into the luciferase fusion/suicide vector pGPL01, and this fusion was recombined onto the chromosome. This resulted in both a gene disruption and a transcriptional fusion of pmrD to the luc gene. Regulation of this locus was examined by moving this region into PhoP−, PhoPc, PmrA−, and PmrAc backgrounds by P22-mediated transduction, followed by assay of firefly luciferase activity. We chose to examine regulation by PhoP and PmrA as these two regulators are directly or indirectly involved in the regulation of pmrG and the pmrHFIJKLM operon. As shown in Fig. 6, this locus is activated 66-fold by PhoP-PhoQ but is not regulated by PmrA-PmrB. This pmrD mutant (in a high-level-PM-resistant strain background [PmrAc]) was also examined on PM-containing plates and by MIC analysis for an effect on resistance to PM. Both plate and MIC assays showed that the (single-copy) loss of pmrD had no effect on resistance to PM under the growth conditions employed (data not shown).
FIG. 6.
PhoP-PhoQ but not PmrA-PmrB regulates the pmrD gene. PmrA−, PmrAc, PhoPc, and Phop− strains containing a single-copy pmrD::luc fusion were assayed in logarithmic phase (OD600, 0.6) for firefly luciferase activity (expressed as relative light units [RLU]). Results are expressed as ratios of firefly luciferase activity in PmrAc versus PmrA− backgrounds and in PhoPc versus PhoP− backgrounds.
To examine the effect of the loss of the PmrA-PmrB-regulated pmrG gene on PM resistance, pmrG::TnphoA was transduced into a PmrAc background and the resulting strains were examined for resistance to PM on plates and by MIC analysis. Like the loss of pmrD, the loss of pmrG had no effect on resistance to PM under standard growth conditions. Therefore, this chromosomal region contains three regulated loci, one by PhoP-PhoQ (pmrD) and two by PmrA-PmrB (pmrG and pmrHFIJKLM), while of these three loci only mutations in pmrHFIJKLM affect PM resistance when strains are grown on standard laboratory media.
Deletion analysis of the pmrHFIJKLM operon: roles of individual loci in LPS modification and PM resistance.
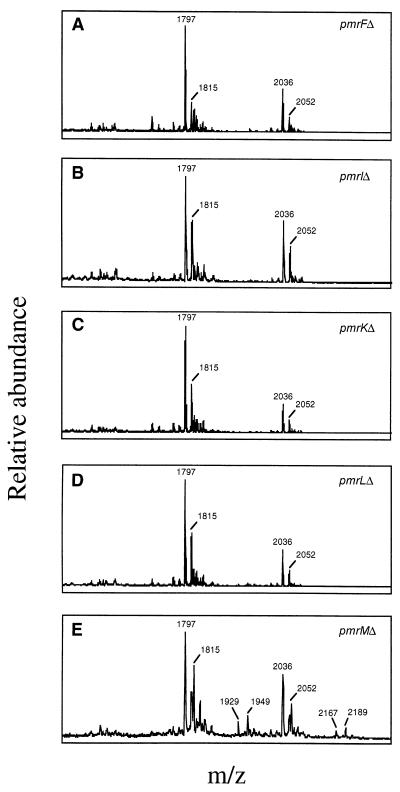
To examine the roles of individual genes of the pmrHFIJKLM operon in resistance to PM and in Ara4N modification of lipid A, nonpolar deletions were constructed in pmrH, pmrF, pmrI, pmrJ, pmrK, and pmrL. An insertion mutation was created in pmrM by recombination of a suicide plasmid within this locus. The chromosomally incorporated deletions were confirmed to be nonpolar by sequencing the deletion junctions and by demonstrating the lack of an effect on luciferase activity of a pmrM::luc fusion mated into each deletion background. These nonpolar deletion or mutant strains (constructed in a PmrAc background) were then examined for resistance to PM. All mutations except that in pmrM had a major effect on resistance to PM (MIC of PmrAc and ΔpmrM strains, 4 μg/ml; MIC of strains with pmrHFIJKL deletions, 0.1 μg/ml). To determine if the loss in resistance to PM was due to the loss of Ara4N in lipid A, MALDI-TOF mass spectroscopy was performed on purified lipid A. This analysis confirmed the absence of Ara4N in deletion strains with reduced resistance to PM (Fig. 7). This can be seen by the absence of peaks at m/z 1,929 and 2,167, which correspond to hexa- and hepta-acyl lipid A with Ara4N, respectively. Loss of the peaks at m/z 1,949 and 2,189 is also evident in the mutants, as these peaks correspond to hexa- and hepta-acyl lipid A, respectively, with Ara4N and the PhoP-PhoQ-mediated hydroxyl addition to myristate. The profile of the pmrM mutant was identical to that of the PmrAc strain (the parental strain of all mutants examined [8]), further demonstrating that this gene plays no role in Ara4N addition to lipid A or PM resistance.
FIG. 7.
Mass spectroscopy of the lipid A from pmrF, pmrI, pmrK, pmrL, and pmrM nonpolar deletion strains. The m/z ratios and related structures are as follows: 1,797, unmodified hexa-acyl lipid A; 1,815, hexa-acyl lipid A with 2-OH myristate; 1,929, hexa-acyl lipid A with Ara4N; 1,949, hexa-acyl lipid A with 2-OH myristate and Ara4N; 2,036, unmodified hepta-acyl lipid A; 2,052, hepta-acyl lipid A with 2-OH myristate; 2,167, hepta-acyl lipid A with Ara4N; 2,189, hepta-acyl lipid A with 2-OH myristate and Ara4N. The lipid A from strains with pmrH and pmrJ nonpolar deletions was also examined, and the results were identical to those shown in panels A through D but were not included due to differences in the scale of m/z values examined and the difficulty in comparing these tracings to those for the other samples. The PmrAc strain that is the parental background of the strains with the examined deletions has been examined previously (8), and results for it are identical to those shown in panel E.
LPS modification affects oral virulence.
It has been shown previously that a strain containing a pmrF::Tn10d insertion was unable to add Ara4N to lipid A, which resulted in a significant decrease in resistance to PM (8). To determine the effect on S. enterica serovar Typhimurium virulence of the lipid A modification with Ara4N, the pmrF::Tn10d mutant was examined in the mouse model of typhoid fever.
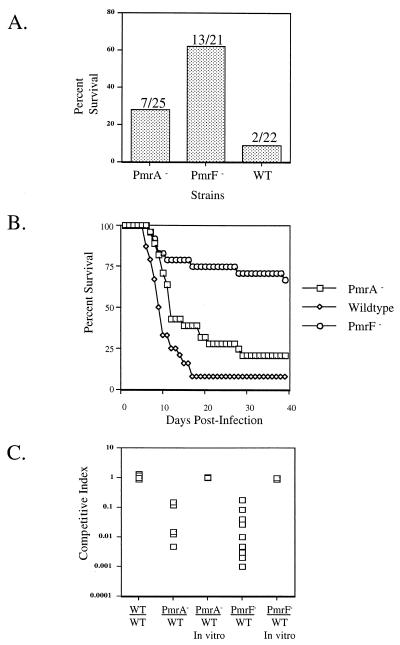
Mice inoculated orally with the pmrF::Tn10d mutant at a dose approximately 1 log unit above the LD50 had a sevenfold increase in survival versus mice inoculated with the WT strain (62 versus 9%) (Fig. 8A). Furthermore, of those mice inoculated with the pmrF::Tn10d mutant, the time to death of some of the mice was greater than 20 days, while the mice inoculated with the WT consistently averaged about 12 days (Fig. 8B). To confirm the observed virulence defect, competition assays were performed with the pmrF::Tn10d mutant and the WT strain. These data show that the pmrF::Tn10d mutant was dramatically impaired (10- to 1,000-fold) in its ability to compete with the WT strain (Fig. 8C). In vitro competition assays demonstrated that the defect was not due to a growth deficiency (Fig. 8C). A PmrA− strain was also examined in all of the above-mentioned virulence assays. Although it was expected that a PmrA− strain should show the same, if not a greater, defect than the pmrF::Tn10d mutant, the virulence defect that was observed was not as dramatic as that seen with the pmrF::Tn10d mutant (Fig. 8).
FIG. 8.
Mutations in pmrA or the pmrHFIJKLM operon affect virulence by the oral route. Mice were inoculated orally with a dose of 106 CFU. BALB/c mice were infected with ATCC 14028s (WT), WT with a pmrA::Tn10d insertion (PmrA−), WT with a pmrF::Tn10d insertion (PmrF−), or a combination of two of these strains. (A) Percent survival of BALB/c mice receiving the WT, PmrA−, or PmrF− strain. The numbers above the bars represent the number of surviving mice per the number of mice tested. (B) Percent survival of mice over 40 days postinfection. (C) Competition assays of WT versus PmrA− and WT versus PmrF− strains. The competitive index is the ratio of CFU in the livers of mice infected with a PmrA−, WT, or PmrF− strain to CFU in livers of mice infected with the WT strain. The WT-versus-WT experiment involved two WT strains marked with different antibiotic resistances. The competitions labeled in vitro were performed by growth overnight in LB medium to ensure that any in vivo effects were not due to a growth disadvantage. Both liver and spleen were examined in the competition assays, but only the data for the liver are shown.
Intraperitoneal inoculation of JSG485 (pmrF::Tn10d) and PmrA− strains into mice had no effect on the LD50 (<20 organisms) or the survival time of the mice compared to inoculation with the WT (data not shown). Therefore, loss of Ara4N affects S. enterica serovar Typhimurium virulence during early events in a natural infection but not at later stages of macrophage survival and growth within the liver and spleen.
DISCUSSION
Activation of the PmrA-PmrB two-component regulatory system by environmental signals or by mutations within pmrA that result in a constitutive phenotype activates the transcription of genes whose products covalently modify LPS. These modifications (Ara4N and phosphoethanolamine) result in increased resistance to AP, which are membrane-active pore formers present at mucosal surfaces and within macrophages and neutrophils. Increased resistance to these peptides provides the bacterium with a survival advantage within the host. In this work, we presented a detailed characterization of an island of PmrA-PmrB- and PhoP-PhoQ-regulated genes. Experiments were conducted to examine the roles of the individual genes in this region in resistance to AP and what effect the loss of genes in this region has on S. enterica serovar Typhimurium virulence.
The pmrHFIJKLM locus is regulated by PmrA-PmrB, and a pmrF::Tn10d insertion was shown to eliminate the ability to modify lipid A with Ara4N and markedly reduced resistance to PM (8). These genes were shown to be cotranscribed, as a pmrM::luc fusion was activated by PmrA-PmrB and luciferase activity was dramatically reduced when the reporter fusion was recombined downstream of the pmrF::Tn10d insertion. Fusion of the promoter of this operon to the gene encoding firefly luciferase showed it to be over 3,000-fold activated by PmrA-PmrB. It is intriguing to propose that this highly expressed in vivo-activated promoter may be useful in a live-attenuated Salmonella vaccine for the expression of heterologous antigens. This approach has several advantages including high-level expression in host antigen-presenting cells and the ability to construct stable, single-copy chromosomal constructs. In fact, this approach has been successfully tested using phoP-regulated gene promoters (2, 13).
Studies of the regulators of two-component systems have shown that they bind to consensus sequences in the promoters of regulated genes. Therefore, experiments were designed to examine the interaction of PmrA with pmr promoters. The PmrA protein was purified (both WT and constitutive forms) and used in promoter binding studies. A 118-bp region of the pmrHFIJKLM promoter and a 40-bp region of the pmrCAB operon promoter were found to shift with each protein. The 40-bp region of the pmrCAB promoter is located just upstream of the −35 region, which places the putative binding site in a position comparable to those of other transcriptional activators of the OmpR family. Recently, Wøsten and Groisman (22) demonstrated similar binding of the PmrA protein to the promoters of activated genes and defined a consensus binding site of 5′ TTAAKTTCTTAAKGTT 3′ for PmrA. This site is located within both the pmrCAB and pmrHFIJKLM operon promoter fragments that bound PmrA (16 of 16 match in each). The PmrAc protein bound approximately threefold more promoter fragments than the WT PmrA protein, suggesting that the constitutive activity of this protein may be due to increased promoter affinity.
Nonpolar deletions were constructed in each gene of the pmrHFIJKLM operon (except pmrM, which was disrupted by insertion), and the effect of these mutations on PM resistance was measured. All of the genes except pmrM had an effect on PM resistance, which resulted in an MIC level similar to that of a PmrA− strain (MIC of PmrAc and ΔpmrM strains, 4 μg/ml; MIC of pmrHFIJKL deletion strains, 0.1 μg/ml). Based on protein homologies and known sugar biosynthesis pathways, a model involving the pmrHFIJKLM operon and pmrE has been proposed for Ara4N biosynthesis and addition to lipid A (1). This pathway begins with the conversion of UDP-glucose to UDP-glucuronic acid by the pmrE gene product, followed by the conversion to UDP-4-amino-l-deoxyarabinose and attachment of this moiety to lipid A by the products of the pmrHFIJKLM operon. This model, however, was unable to identify functions for PmrL and PmrM, as neither of these small proteins has any strong similarities to proteins in GenBank. From this study, it is clear that PmrL does belong at some point in the pathway but PmrM does not. Therefore, the function of PmrM remains unknown.
Previous studies showed that the gene upstream of the pmrHFIJKLM operon, pmrG, was regulated by PmrA-PmrB (8). Because the effect of this gene on the PM resistance phenotype had not been previously examined, a pmrG mutant was assayed for PM resistance in a PmrAc background and the pmrG mutation was shown to have no effect. The gene downstream of the operon, pmrD, however, had previously been shown to have an effect on PM resistance only in high copy number. To see if it too was regulated by PmrA-PmrB, a firefly luciferase fusion was created and expression was measured when strains were grown in various backgrounds. This data surprisingly demonstrated that pmrD was regulated by PhoP-PhoQ and not PmrA-PmrB. Interestingly, work by others accomplished concurrently with our studies also showed that pmrD was regulated by PhoP-PhoQ and that the pmrD gene product mediates interaction of the PhoP-PhoQ system with the PmrA-PmrB system, as PmrA-regulated genes are not expressed in a pmrD mutant grown under PhoP-PhoQ-inducing conditions (low magnesium) (14). PmrD is thought to exert its effect on PmrA-PmrB through a posttranscriptional mechanism, possibly involving its effect on PmrA phosphorylation. These results likely explain why pmrD had an effect on PM resistance only when in multicopy and when strains were grown in LB medium (non-phoP-PhoQ-inducing conditions).
Loss of expression of the pmrHFIJKLM operon eliminates Ara4A addition to lipid A and PM resistance. To determine if this loss played a role in S. enterica serovar Typhimurium pathogenesis, a pmrF mutant was examined in mice and was shown to have markedly reduced virulence by the oral route but not by the intraperitoneal route. In addition, for most of the mice that did eventually die, the time from inoculation to death was much longer than for mice infected with a WT strain. A pmrA mutant showed similar results, but did not display as severe of a virulence defect as the pmrF mutant strain. This was surprising, as PmrA activates the pmrF-containing operon and therefore was expected to give similar results. These data suggest that the pmrHFIJKLM operon may be expressed in vivo by a mechanism independent of PmrA-PmrB.
At this time, it is unclear at which stage of infection the pmrHFIJKLM locus plays a role. The PmrA-PmrB system (and the PhoP-PhoQ system, which can activate transcription of pmrAB) is thought to be induced within macrophages, and if the role of the PmrA-PmrB regulatory system is to promote survival within macrophages, then a defect should have been observed in mice by both the intraperitoneal and oral routes. Therefore, the data suggest that the defect is prior to the interaction with macrophages. In vitro experiments show no defect in invasion or type III secretion with pmrA or pmrF mutants (J. S. Gunn, unpublished results). Therefore, it is intriguing to speculate that PmrA-PmrB may be activated by unknown environmental signals within the small intestine and that the PmrA-PmrB-induced LPS modifications play a role in resistance to intestinal AP or other intraintestinal antimicrobial factors.
The results of this work further characterize a highly regulated island of genes necessary for LPS modification, AP resistance, survival in mice, and two-component system interactions. Furthermore, these data demonstrate that in vivo-regulated modifications of LPS are an important part of Salmonella pathogenesis and suggest that the PhoP-PhoQ and PmrA-PmrB regulatory systems may be important at locations other than within host cells.
ACKNOWLEDGMENTS
This work was supported by grants AI30479 (S.I.M.), AI43521 (J.S.G.), and T32AI07271-15 (S.S.R.) from the National Institutes of Health.
REFERENCES
- 1.Baker S J, Daniels C, Morona R. PhoP/Q regulated genes in Salmonella typhi: identification of melittin sensitive mutants. Microb Pathog. 1997;22:165–179. doi: 10.1006/mpat.1996.0099. [DOI] [PubMed] [Google Scholar]
- 2.Bullifent H L, Griffin K F, Jones S M, Yates A, Harrington L, Titball R W. Antibody responses to Yersinia pestis F1-antigen expressed in Salmonella typhimurium aroA from in vivo-inducible promoters. Vaccine. 2000;18:2668–2676. doi: 10.1016/s0264-410x(00)00053-0. [DOI] [PubMed] [Google Scholar]
- 3.Caroff M, Tacken A, Szabo L. Detergent-accelerated hydrolysis of bacterial endotoxins and determination of the anomeric configuration of the glycosyl phosphate present in the “isolated lipid A” fragment of the Bordetella pertussis endotoxin. Carbohydr Res. 1988;175:273–282. doi: 10.1016/0008-6215(88)84149-1. [DOI] [PubMed] [Google Scholar]
- 4.Darveau R P, Hancock R E. Procedure for isolation of bacterial lipopolysaccharides from both smooth and rough Pseudomonas aeruginosa and Salmonella typhimurium strains. J Bacteriol. 1983;155:831–838. doi: 10.1128/jb.155.2.831-838.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ernst R K, Yi E C, Guo L, Lim K B, Burns J L, Hackett M, Miller S I. Specific lipopolysaccharide found in cystic fibrosis airway Pseudomonas aeruginosa. Science. 1999;286:1561–1565. doi: 10.1126/science.286.5444.1561. [DOI] [PubMed] [Google Scholar]
- 6.Fields P I, Swanson R V, Haidaris C G, Heffron F. Mutants of Salmonella typhimurium that cannot survive within the macrophage are avirulent. Proc Natl Acad Sci USA. 1986;83:5189–5193. doi: 10.1073/pnas.83.14.5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gunn J S, Hohmann E L, Miller S I. Transcriptional regulation of Salmonella virulence: a PhoQ periplasmic domain mutation results in increased net phosphotransfer to PhoP. J Bacteriol. 1996;178:6369–6373. doi: 10.1128/jb.178.21.6369-6373.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gunn J S, Lim K B, Krueger J, Kim K, Guo L, Hackett M, Miller S I. PmrA-PmrB regulated genes necessary for 4-aminoarabinose lipid A modification and polymyxin resistance. Mol Microbiol. 1998;27:1171–1182. doi: 10.1046/j.1365-2958.1998.00757.x. [DOI] [PubMed] [Google Scholar]
- 9.Gunn J S, Miller S I. PhoP/PhoQ activates transcription of pmrA/B, encoding a two-component system involved in Salmonella typhimurium antimicrobial peptide resistance. J Bacteriol. 1996;178:6857–6864. doi: 10.1128/jb.178.23.6857-6864.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guo L, Lim K, Gunn J S, Bainbridge B, Darveau R, Hackett M, Miller S I. Regulation of lipid A modifications by Salmonella typhimurium virulence genes phoP-phoQ. Science. 1997;276:250–253. doi: 10.1126/science.276.5310.250. [DOI] [PubMed] [Google Scholar]
- 11.Guo L, Lim K B, Poduje C M, Daniel M, Gunn J S, Hackett M, Miller S I. Lipid A acylation and bacterial resistance against vertebrate antimicrobial peptides. Cell. 1998;95:189–198. doi: 10.1016/s0092-8674(00)81750-x. [DOI] [PubMed] [Google Scholar]
- 12.Hohmann E L, Oletta C A, Killeen K P, Miller S I. phoP/phoQ-deleted Salmonella typhi (TY800) is a safe and immunogenic single dose typhoid fever vaccine in volunteers. J Infect Dis. 1996;173:1408–1414. doi: 10.1093/infdis/173.6.1408. [DOI] [PubMed] [Google Scholar]
- 13.Hohmann E L, Oletta C A, Loomis W P, Miller S I. Macrophage-inducible expression of a model antigen in Salmonella typhimurium enhances immunogenicity. Proc Natl Acad Sci USA. 1995;92:2904–2908. doi: 10.1073/pnas.92.7.2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kox L F, Wosten M M, Groisman E A. A small protein that mediates the activation of a two-component system by another two-component system. EMBO J. 2000;19:1861–1872. doi: 10.1093/emboj/19.8.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller S I, Kukral A M, Mekalanos J J. A two-component regulatory system (phoP phoQ) controls Salmonella typhimurium virulence. Proc Natl Acad Sci USA. 1989;86:5054–5058. doi: 10.1073/pnas.86.13.5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roland K L, Esther C R, Spitznagel J K. Isolation and characterization of a gene, pmrD, from Salmonella typhimurium that confers resistance to polymyxin when expressed in multiple copies. J Bacteriol. 1994;176:3589–3597. doi: 10.1128/jb.176.12.3589-3597.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roland K L, Martin L E, Esther C R, Spitznagel J K. Spontaneous pmrA mutants of Salmonella typhimurium LT2 define a new two-component regulatory system with a possible role in virulence. J Bacteriol. 1993;175:4154–4164. doi: 10.1128/jb.175.13.4154-4164.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shafer W M, Casey S G, Spitznagel J K. Lipid A and resistance of Salmonella typhimurium to antimicrobial granule proteins of human neutrophils. Infect Immun. 1984;43:834–838. doi: 10.1128/iai.43.3.834-838.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Skorupski K, Taylor R K. Positive selection vectors for allelic exchange. Gene. 1996;169:47–52. doi: 10.1016/0378-1119(95)00793-8. [DOI] [PubMed] [Google Scholar]
- 20.Soncini F C, Groisman E A. Two-component regulatory systems can interact to process multiple environmental signals. J Bacteriol. 1996;178:6796–6801. doi: 10.1128/jb.178.23.6796-6801.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Steinberg D A, Hurst M A, Fujii C A, Kung A H, Ho J F, Cheng F C, Loury D J, Fiddes J C. Protegrin-1: a broad-spectrum, rapidly microbicidal peptide with in vivo activity. Antimicrob Agents Chemother. 1997;41:1738–1742. doi: 10.1128/aac.41.8.1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wøsten M M, Groisman E A. Molecular characterization of the PmrA regulon. J Biol Chem. 1999;274:27185–27190. doi: 10.1074/jbc.274.38.27185. [DOI] [PubMed] [Google Scholar]