ABSTRACT
Cyclic GMP-AMP synthase (cGAS), a key DNA sensor, detects cytosolic viral DNA and activates the adaptor protein stimulator of interferon genes (STING) to initiate interferon (IFN) production and host innate antiviral responses. Duck enteritis virus (DEV) is a duck alphaherpesvirus that causes an acute and contagious disease with high mortality in waterfowl. In the present study, we found that DEV inhibits host innate immune responses during the late phase of viral infection. Furthermore, we screened DEV proteins for their ability to inhibit the cGAS-STING DNA-sensing pathway and identified multiple viral proteins, including UL41, US3, UL28, UL53, and UL24, which block IFN-β activation through this pathway. The DEV tegument protein UL41, which exhibited the strongest inhibitory effect, selectively downregulated the expression of interferon regulatory factor 7 (IRF7) by reducing its mRNA accumulation, thereby inhibiting the DNA-sensing pathway. Ectopic expression of UL41 markedly reduced viral DNA-triggered IFN-β production and promoted viral replication, whereas deficiency of UL41 in the context of DEV infection increased the IFN-β response to DEV and suppressed viral replication. In addition, ectopic expression of IRF7 inhibited the replication of the UL41-deficient virus, whereas IRF7 knockdown facilitated its replication. This study is the first report identifying multiple viral proteins encoded by a duck DNA virus, which inhibit the cGAS-STING DNA-sensing pathway. These findings expand our knowledge of DNA sensing in ducks and reveal a mechanism through which DEV antagonizes the host innate immune response.
IMPORTANCE Duck enteritis virus (DEV) is a duck alphaherpesvirus that causes an acute and contagious disease with high mortality, resulting in substantial economic losses in the commercial waterfowl industry. The evasion of DNA-sensing pathway-mediated antiviral innate immunity is essential for the persistent infection and replication of many DNA viruses. However, the mechanisms used by DEV to modulate the DNA-sensing pathway remain poorly understood. In the present study, we found that DEV encodes multiple viral proteins to inhibit the cGAS-STING DNA-sensing pathway. The DEV tegument protein UL41 selectively diminished the accumulation of interferon regulatory factor 7 (IRF7) mRNA, thereby inhibiting the DNA-sensing pathway. Loss of UL41 potently enhanced the IFN-β response to DEV and impaired viral replication in ducks. These findings provide insights into the host-virus interaction during DEV infection and help develop new live attenuated vaccines against DEV.
KEYWORDS: duck enteritis virus, cGAS-STING, DNA sensing, innate immunity
INTRODUCTION
Duck enteritis virus (DEV), also known as Anatid herpesvirus type 1, causes an acute and contagious fatal disease with high morbidity and mortality in domestic waterfowl, which results in severe economic losses to the duck industry (1). Domestic and wild ducks, geese, and swans of all ages are considered susceptible to DEV infection. The migratory waterfowl plays a crucial role in DEV transmission within and between continents, resulting in the worldwide distribution of this virus. DEV is considered a pantropic virus that replicates primarily in the mucosa of the digestive tract and then spreads to the bursa of Fabricius, thymus, spleen, and liver, leading to pathological lesions in several organs (2). DEV exhibits latent infection in trigeminal ganglia after establishing primary infection in ducks; thereafter, viral reactivation occurs that results in disease outburst (3). DEV belongs to the Herpesviridae family and Alphaherpesvirinae subfamily. The viral genome is approximately 158 kb and contains 78 open reading frames predicted to encode potential functional proteins. Despite several advances in understanding DEV pathogenesis, little is known regarding the innate immune responses and virus-host interaction during DEV infection in ducks.
Innate immunity is involved in the first line of host defense against invading pathogens and is initiated by various host pattern-recognition receptors (PRRs) that recognize conserved pathogen-associated molecular patterns and trigger the production of type I interferons (IFN-I) and the expression of multiple IFN-stimulated genes (ISGs) (4, 5). In addition to Toll-like receptors (TLRs), retinoic acid-inducible gene I-like receptors (RLRs), and Nod-like receptors, several cytosolic DNA sensors have been recently discovered (5–7). Viral DNA recognition by DNA sensors is a central host cellular defense against DNA virus infection. Among the identified DNA sensors, cyclic GMP-AMP (cGAMP) synthase (cGAS) has been demonstrated as a predominant cytosolic DNA sensor that recognizes various DNA ligands in different cell types (8). Upon binding to viral DNA, cGAS utilizes ATP and GTP to synthesize the second messenger cyclic GMP-AMP (cGAMP) to activate stimulator of interferon genes (STING). Active STING then recruits TANK-binding kinase 1 (TBK1) to phosphorylate and activate interferon regulatory factor 3 (IRF3). Phosphorylated IRF3 dimerizes and then translocates to the nucleus, ultimately leading to the production of IFN-I and several inflammatory cytokines (9, 10). Recently, the cGAS-STING DNA-sensing pathway was reported to play an important role in IFN-I responses against herpesviruses, including herpes simplex virus 1 (HSV-1), Kaposi sarcoma herpesvirus (KSHV), and Marek’s disease virus (MDV) (11–13). Evasion of host antiviral innate immunity is essential for herpesviruses to establish persistent infection and replication in the host (14). Moreover, several viral proteins that inhibit IFN-I production through modulation of the DNA-sensing signaling pathway have been identified (15–17).
Birds are natural reservoirs for many kinds of viruses. IFN-I has been characterized and assessed for its antiviral activities in various avian species. Duck IFN-I was first detected in duck embryo fibroblasts (DEFs) after infection with reovirus serotype 3 (18). Avian PRRs are reported to differ from their mammalian counterparts (18). Comparatively, chicken, duck, and goose cGAS exhibits shortened N termini with the least similarity of 22.4%, 17.4%, and 7.7% with human cGAS, respectively (19, 20). Like in mammals, the cGAS-STING axis plays a critical role in restricting DNA virus infection in chicken and duck cells (21, 22). Nevertheless, the mechanisms by which DEV modulates the cGAS-STING-mediated antiviral signaling remain obscure.
The HSV-1 virion host shutoff (VHS) protein, encoded by the UL41 gene, is an mRNA-specific RNase, which triggers host mRNA degradation to promote the sequential expression of viral proteins in the early stage of infection (23–25). Although HSV-1 VHS is known to be important for viral pathogenesis in vivo (26), the role of DEV UL41 remains unclear. In this study, we demonstrate for the first time that DEV encodes multiple viral proteins to inhibit the cGAS-STING DNA-sensing pathway. The DEV VHS protein selectively reduces the accumulation of interferon regulatory factor 7 (IRF7) mRNA, thereby inhibiting the DNA-sensing pathway. These findings expand our knowledge of the host-virus interaction during DEV infection and reveal a mechanism through which DEV antagonizes the host innate immune responses.
RESULTS
DEV inhibits IFN-β production during infection in duck embryo fibroblasts and in ducks.
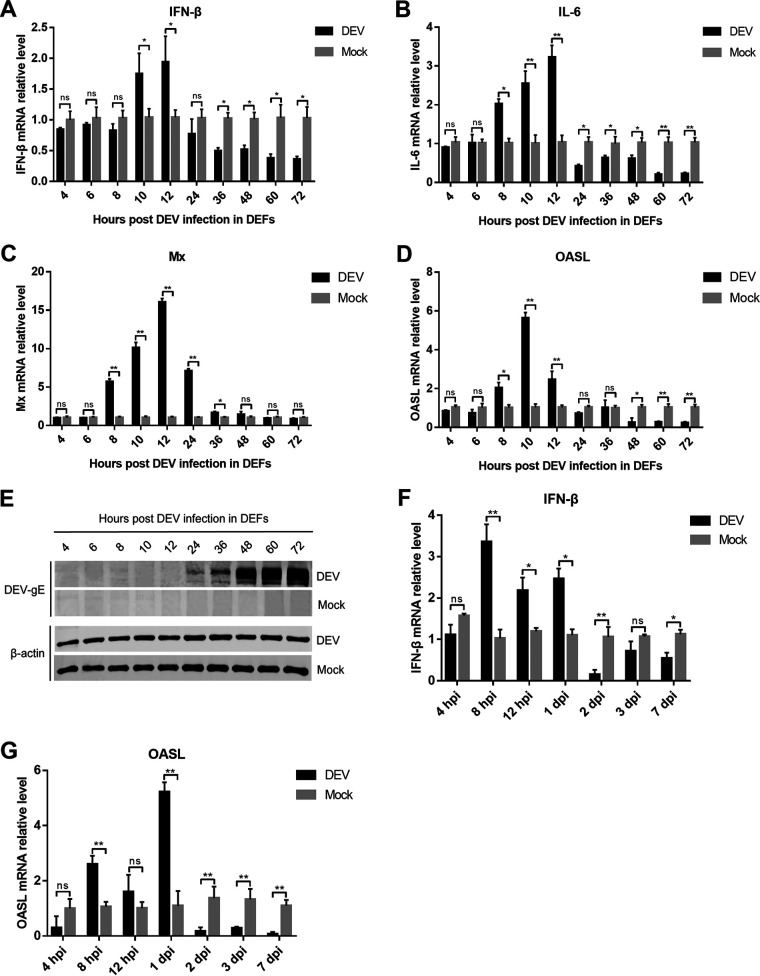
To determine whether DEV could inhibit type I IFN induction during viral infection, we infected DEFs with the virulent DEV CV strain and analyzed the mRNA expression of IFN-β and IL-6 using real-time quantitative PCR (qPCR). The results showed that IFN-β induction in DEFs upon DEV infection was prominent at 10- and 12-h postinfection (hpi), which decreased from 24 hpi and was significantly lower than that in mock cells after 36 hpi (Fig. 1A). IL-6 expression was also decreased during the late phase of DEV infection (Fig. 1B). Furthermore, DEV infection induced the expression of duck IFN-stimulated genes (ISGs), including myxovirus-resistance protein (Mx) and interferon-induced oligoadenylate synthetase-like protein (OASL), between 8 and 12 hpi, whereas ISG expression was significantly inhibited after 24 hpi (Fig. 1C and D). Consistent with IFN-β and ISGs inhibition, DEV viral proteins were expressed during the late phase of infection in DEFs (Fig. 1E), suggesting that the viral proteins might contribute to the modulation of the IFN-β response during DEV infection.
FIG 1.
DEV inhibits the production of IFN-β and IFN-stimulated genes (ISGs) during infection in DEFs and in ducks. (A to D) DEFs were infected with the virulent DEV CV strain at a multiplicity of infection (MOI) of 0.01. The mRNA levels of IFN-β (A), IL-6 (B), and duck ISGs Mx (C) and OASL (D) were measured by real-time qPCR from 4 to 72 hpi. (E) The expression of DEV protein gE during viral infection in DEFs was monitored by Western blotting. (F to G) 2-week-old specific pathogen-free ducks were inoculated intramuscularly with 10 TCID50 of DEV CV strain, and the mRNA levels of IFN-β (F) and OASL (G) in the spleen samples were measured by real-time qPCR. The relative amounts of IFN-β, IL-6, Mx, and OASL mRNA were normalized to the β-actin mRNA level in each sample, and the fold differences were compared with those in the mock samples. All controls and treated groups were performed and examined in triplicate. *, P < 0.05; **, P < 0.01; ns, no significant difference.
We also infected ducks with a virulent DEV CV strain and assessed the expression of IFN-β and OASL. The results showed that DEV infection triggered an IFN-β response during the early phase of infection (within 24 hpi). However, IFN-β induction significantly decreased in DEV-infected ducks after 2 days of infection (Fig. 1F). Consistently, OASL transcription was also greatly reduced at later time points of DEV infection in vivo (Fig. 1G). These results indicate that DEV inhibits host innate immune responses during the late phase of viral infection in ducks.
Multiple DEV proteins block cGAS-STING-mediated IFN-β activation.
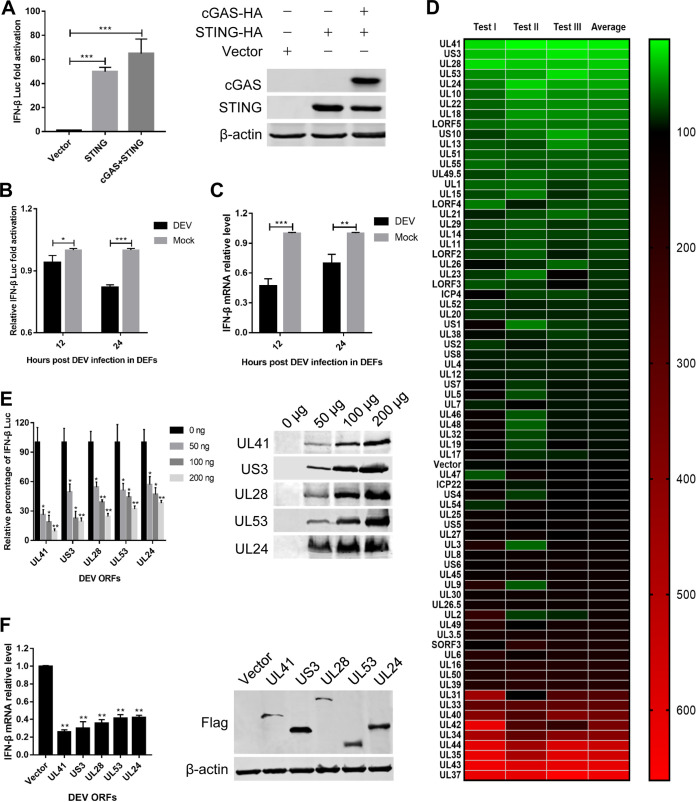
As the cGAS-STING DNA-sensing pathway plays a critical role in the induction of type I IFNs in response to herpesviruses, inhibition of IFN-β expression during DEV infection suggests that DEV may encode proteins that antagonize this pathway. In DEFs, the IFN-β promoter was highly activated when the same amounts of plasmids encoding cGAS and STING were cotransfected along with an IFN-β promoter luciferase reporter construct (Fig. 2A). However, inoculation of DEV in DEFs significantly inhibited IFN-β promoter activity and IFN-β mRNA production (Fig. 2B and C). To evaluate whether individual viral proteins could regulate the cGAS-STING pathway, DEFs were cotransfected with an empty vector or the expression plasmids encoding DEV viral proteins along with those encoding cGAS, STING, and the IFN-β promoter constructs and subjected to dual-luciferase reporter (DLR) assays to detect IFN-β promoter activity. Based on this model, we identified multiple DEV proteins, including UL41, US3, UL28, UL53, and UL24, which significantly inhibited cGAS-STING-mediated activation of the IFN-β promoter (Fig. 2D). We further found that each viral inhibitor inhibited IFN-β promoter activation in a dose-dependent manner (Fig. 2E). Next, we validated the DLR results by measuring IFN-β mRNA levels through quantitative reverse transcription-PCR (qRT-PCR) and obtained similar results (Fig. 2F).
FIG 2.
Screening of DEV viral proteins that modulate the cGAS-STING pathway. (A) DEFs were cotransfected with the IFN-β Luc reporter, pRL-TK, and the empty vector or cGAS-HA and STING-HA combined. The luciferase activity was measured at 36 h posttransfection. (B) DEFs were cotransfected with the IFN-β Luc reporter, cGAS-HA, and STING-HA expression plasmids and then inoculated with DEV (MOI = 0.01). The luciferase activity was measured at 12 h and 24 h postinfection. (C) DEFs were cotransfected with cGAS-HA and STING-HA expression plasmids and then inoculated with DEV (MOI = 0.01). The IFN-β mRNA level was measured by real-time qPCR at 12 h and 24 h postinfection. (D) DEFs were transfected with the IFN-β Luc reporter and pRL-TK, together with cGAS-HA, STING-HA, and an DEV ORF expression plasmid or the empty vector. The IFN-β luciferase activity was measured at 36 h posttransfection. The data of DEV ORFs and the empty vector from three independent tests are presented as a heat map. Higher IFN-β Luc activation levels are indicated by red, whereas lower levels are indicated by green, which corresponds to a higher degree of inhibition. (E) Various doses of the top five DEV viral protein inhibitors and the empty vector were cotransfected with cGAS-HA and STING-HA expression plasmids into DEFs, and IFN-β Luc activity was measured at 36 h posttransfection. (F) The top five DEV viral protein inhibitors and the empty vector were cotransfected with cGAS-HA and STING-HA expression plasmids into DEFs. The IFN-β mRNA levels were measured by real-time qPCR at 36 h posttransfection. The fold changes were compared with those of the empty vector controls. All controls and treated groups were performed and examined in triplicate. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
DEV UL41 inhibits IFN-β production induced by interferon-stimulatory and viral DNA.
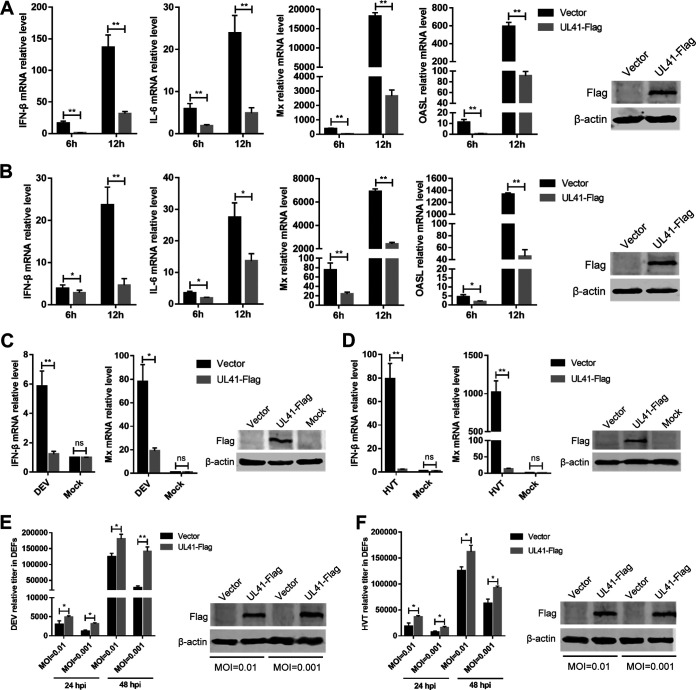
In this study, we focused on the DEV tegument protein UL41, because it showed the strongest ability to inhibit the cGAS-STING pathway. To verify the role of DEV UL41 in regulating cytosolic DNA-induced IFN-β production, DEFs were transfected with a UL41 expression plasmid or empty vector, and after 24 h, they were transfected with interferon-stimulatory DNA (ISD) fragments, which induce IFN-β expression in various cells. As shown in Fig. 3A, transfection with ISD induced high levels of IFN-β and IL-6 mRNAs in DEFs; however, ectopic expression of UL41 markedly inhibited ISD-triggered IFN-β and IL-6 mRNA expression. Compared with the control vector, ectopic expression of UL41 also significantly inhibited ISD-induced expression of Mx and OASL (Fig. 3A). Poly(dA·dT) is a synthetic analog of B-DNA that is recognized by several DNA sensors. IFN-β and IL-6 mRNA levels were significantly increased in response to poly(dA·dT) stimulation, which was attenuated in UL41-expressing DEFs (Fig. 3B). Furthermore, the expression of Mx and OASL was also reduced by the ectopic expression of UL41 in DEFs.
FIG 3.
DEV UL41 inhibits IFN-β production induced by interferon-stimulatory and viral DNA. (A) DEFs were transfected with UL41-Flag expression plasmid or empty vector for 24 h and then transfected with interferon-stimulatory DNA (ISD); the mRNA levels of IFN-β, IL-6, Mx, and OASL were measured by qPCR 6 h and 12 h later. UL41 expression was examined by Western blotting at 12 h after ISD transfection. (B) DEFs were transfected with UL41-Flag expression plasmid or empty vector for 24 h and then transfected with poly(dA·dT); the mRNA levels of IFN-β, IL-6, Mx, and OASL were measured by qPCR 6 h and 12 h later. UL41 expression was examined by Western blotting at 12 h after poly(dA·dT) transfection. (C) DEFs were transfected with UL41-Flag expression plasmid or empty vector and then left uninfected or infected with DEV (MOI = 0.01). The mRNA levels of IFN-β and Mx in these cells were measured by qPCR at 12 h postinfection, and UL41 expression was detected by Western blotting. (D) DEFs were transfected with UL41-Flag expression plasmid or empty vector and then left uninfected or infected with HVT (MOI = 0.01). The mRNA levels of IFN-β and Mx in these cells were measured by qPCR at 12 h postinfection, and UL41 expression was detected by Western blotting. (E) DEFs transfected with UL41-Flag plasmid or empty vector were infected with DEV at various MOIs (0.01 or 0.001). The viral titer was tested by qPCR at 24 and 48 hpi, and UL41 expression was examined by Western blotting at 48 hpi. (F) DEFs transfected with UL41-Flag plasmid or empty vector were infected with HVT at various MOIs (0.01 or 0.001). The viral titer was tested by qPCR at 24 and 48 hpi, and UL41 expression was examined by Western blotting at 48 hpi. The relative amount of IFN-β, IL-6, Mx, and OASL mRNA was normalized to β-actin mRNA levels in each sample, and the relative amount of DEV and HVT genomic DNA was normalized to GAPDH levels in each sample. All controls and treated groups were performed and examined in triplicate. *, P < 0.05; **, P < 0.01; ns, no significant difference.
We then examined the effects of ectopically expressed UL41 on IFN-β production triggered by DNA virus infection. The UL41-expressing cells were infected with DEV, and IFN-β and Mx mRNA levels were evaluated by quantitative PCR. The results showed that ectopic UL41 expression led to a reduced IFN-β response against DEV, compared with the empty vector-transfected control cells (Fig. 3C). Mx expression was also reduced by UL41 overexpression during DEV infection in DEFs. We also infected UL41-expressing DEFs with herpesvirus of turkey (HVT) and found that UL41 expression led to diminished production of IFN-β and Mx in HVT-infected cells, compared with that observed in control cells (Fig. 3D). Further results showed that both DEV and HVT exhibited higher replication titers in UL41-expressing cells (Fig. 3E and F). These results indicate that DEV UL41 inhibits viral DNA-triggered IFN-β activation and promotes viral replication.
UL41 deficiency enhances DEV-triggered IFN-β induction.
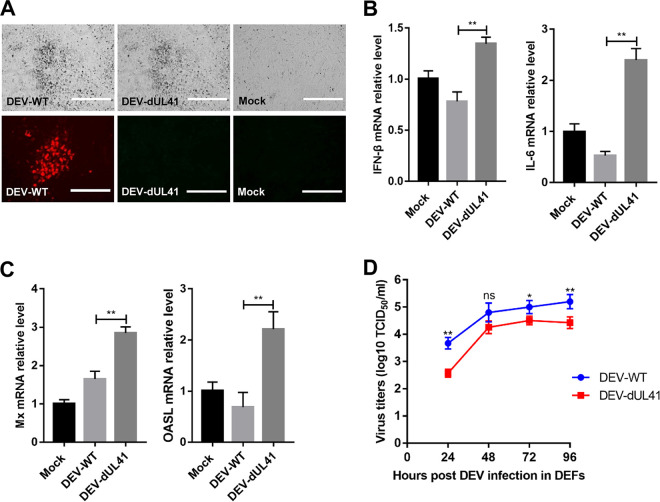
To further investigate whether UL41 acts as an IFN-β inhibitor in the context of DEV infection, we generated UL41-deficient DEV (DEV-dUL41) using overlapping fosmid clones of the virulent DEV CV strain. Deletion of the UL41 gene from the DEV genome was confirmed using an indirect immunofluorescence assay. As expected, wild-type DEV (DEV-WT) expressed the UL41 protein, whereas DEV-dUL41 did not (Fig. 4A). We next examined the expression of IFN-β, IL-6, and IFN-stimulated genes in DEFs infected with wild-type or UL41-deficient DEV. The results indicated that the mRNA levels of IFN-β, IL-6, and the IFN-stimulated genes Mx and OASL induced by DEV-dUL41 were notably higher than those induced by wild-type DEV in DEFs (Fig. 4B and C). We detected the replication curves of DEV-WT and DEV-dUL41 in DEFs, which showed that the replication ability of DEV-dUL41 was decreased in comparison with that of DEV-WT (Fig. 4D). These results demonstrate that UL41 deficiency increases IFN-β induction and reduces viral replication during DEV infection in DEFs.
FIG 4.
UL41 deficiency enhances DEV-triggered induction of IFN-β and downstream antiviral genes. (A) Cytopathic effects (bar length = 400 μm) and indirect immunofluorescence (bar length = 200 μm) analysis of the DEFs infected with DEV-WT or DEV-dUL41 using rabbit anti-UL41 and anti-rabbit IgG-TRITC antibodies. (B and C) Effects of UL41 deficiency on transcription of IFN-β, IL-6, Mx, and OASL in DEFs. DEFs were infected with DEV-WT or DEV-dUL41 (MOI = 0.01) for 24 h before analysis of IFN-β, IL-6, Mx, and OASL mRNA levels. The amounts of IFN-β, IL-6, Mx, and OASL mRNA were normalized to the β-actin mRNA level in each sample, and the fold difference relative to the mock controls was determined. (D) The growth properties of DEV-WT and DEV-dUL41 in DEFs. Data are presented as means ± SDs from three independent experiments. *, P < 0.05; **, P < 0.01; ns, no significant difference.
DEV UL41 inhibits the cGAS-STING pathway by downregulating IRF7 expression.
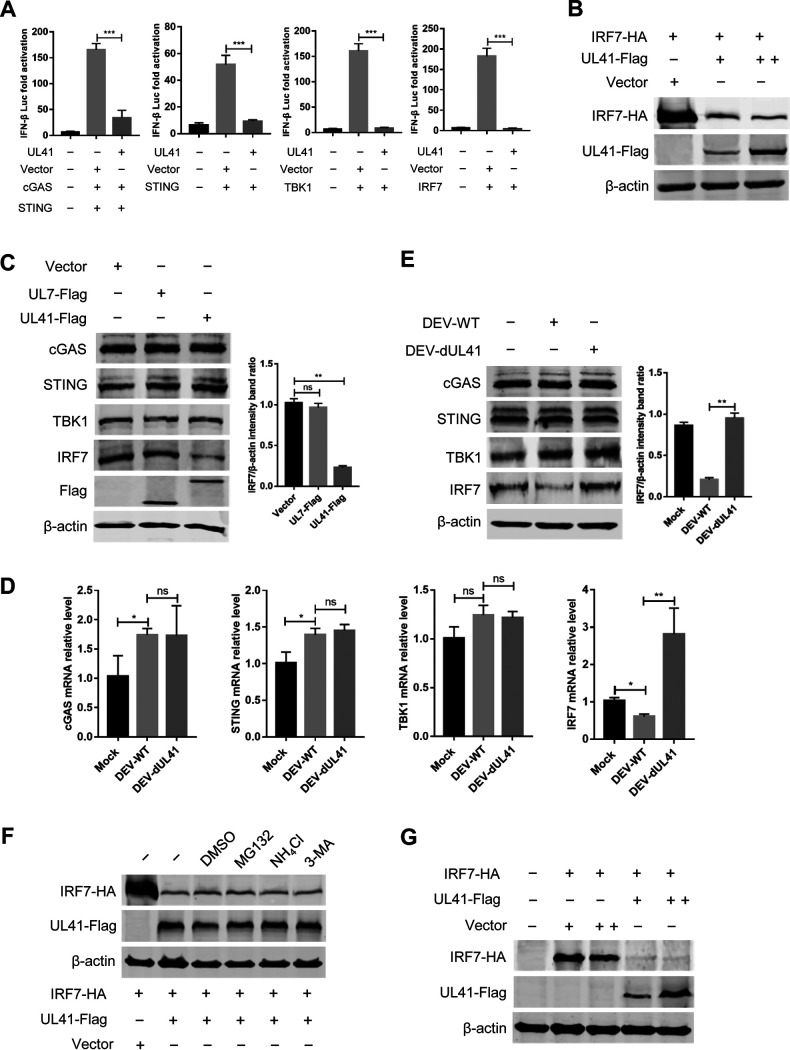
Ducks are IRF3 deficient, and IFN-β transcription in duck cells is dependent on the binding of IRF7 to distinct regulatory domains in the promoter. To clarify the components in the cGAS-STING pathway targeted by UL41, DEFs were cotransfected with UL41 plasmid, IFN-β Luc reporter plasmids, and expression plasmids encoding IRF7 signaling pathway components, including STING, TBK1, and IRF7. The results showed that transfection of STING, TBK1, or IRF7 in DEFs elicited IFN-β Luc reporter activity; however, UL41 expression inhibited the IFN-β Luc activity triggered by these constructs (Fig. 5A; Fig. S1 in the supplemental material). These findings indicate that UL41 probably suppresses the cGAS-STING pathway by targeting IRF7.
FIG 5.
DEV UL41 inhibits the cGAS-STING pathway by downregulating IRF7 expression. (A) DEFs were cotransfected with IFN-β-luc reporter, pRL-TK, and cGAS-HA, STING-HA, TBK1-HA, or IRF7-HA along with empty vector or UL41-Flag expression plasmid as indicated. Cells were harvested 36 h after transfection and subjected to dual-luciferase reporter assays. All controls and treated groups were performed and examined in triplicate. (B) 293T cells were cotransfected with IRF7-HA and UL41-Flag plasmids. Twenty-four hours after transfection, cells were harvested and subjected to Western blot analysis. (C) DEFs were transfected with UL41-Flag, UL7-Flag plasmid, or the empty vector. Twenty-four hours after transfection, cells were harvested and subjected to Western blot analysis with the indicated antibodies. (D) DEFs were infected with DEV-WT or DEV-dUL41 (MOI = 0.01). Twenty-four hours after infection, cells were harvested and analyzed with qRT-PCR. (E) DEFs were infected with DEV-WT or DEV-dUL41 (MOI = 0.01). Twenty-four hours after infection, cells were harvested and subjected to Western blot analysis with the indicated antibodies. (F) 293T cells were cotransfected with IRF7-HA and UL41-Flag plasmids. Twenty-four hours after transfection, cells were treated with dimethyl sulfoxide (DMSO), MG132 (10 μM), NH4Cl (20 mM), and 3-MA (10 mM) for 12 h. Protein expression was measured using Western blotting. (G) Vero cells were cotransfected with IRF7-HA and UL41-Flag plasmids, and cells were subjected to Western blot analysis 36 h after transfection. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, no significant difference.
The tegument protein UL41 is an endoribonuclease with substrate specificity similar to that of RNase A. Therefore, UL41 possibly abolishes IRF7 expression via its RNase activity to degrade IRF7 mRNA. To confirm this hypothesis, 293T cells were cotransfected with IRF7-HA and UL41-Flag plasmids; cells were then harvested and subjected to Western blot analysis. As shown in Fig. 5B, DEV UL41 significantly downregulated IRF7 expression in a dose-dependent manner. To confirm whether UL41 was specifically involved in the degradation of endogenous IRF7, DEFs were cotransfected with UL41-Flag or UL7-Flag expression plasmids and harvested for Western blot analysis. The results showed that DEV protein UL41 specifically reduced the expression of endogenous IRF7, whereas UL7 did not (Fig. 5C).
To investigate whether IRF7 mRNA accumulation was reduced during DEV infection, DEFs were left uninfected or infected with either wild-type DEV (DEV-WT) or UL41-deficient DEV (DEV-dUL41) for 24 h, followed by qRT-PCR and Western blot analyses. Compared with mock-infected DEFs, DEV-WT infection significantly reduced the IRF7 mRNA levels, whereas DEV-dUL41 infection did not (Fig. 5D). Moreover, DEFs infected with DEV-dUL41 produced significantly higher levels of IRF7 mRNA than those infected with DEV-WT or mock-infected cells. Western blot analysis further indicated that DEV-WT significantly inhibited endogenous IRF7 expression compared with DEV-dUL41 (Fig. 5E). To demonstrate whether UL41 specifically targets IRF7, we also examined the mRNA levels and expression of key molecules in the cGAS-STING pathway during viral infection. The results showed that DEV-WT infection failed to reduce the mRNA levels and expression of endogenous cGAS, STING, and TBK1 (Fig. 5D and E). Taken together, these data demonstrate that DEV selectively reduces IRF7 mRNA accumulation as well as protein expression via UL41.
To test whether IRF7 reduction by UL41 is via proteasomal, lysosomal, and autophagy pathway, 293T cells were cotransfected with IRF7-HA and UL41-Flag plasmids and treated with a proteasome inhibitor (MG-132), a lysosomal inhibitor (NH4Cl), or an autophagy inhibitor (3-MA). The results showed that none of the above inhibitors blocked IRF7 reduction (Fig. 5F). To rule out the reduction of IRF7 expression is due to the inhibition of IFN-β signaling by UL41 since IRF7 is an interferon-inducible gene, the IRF7-HA and UL41-Flag plasmids were cotransfected into African green monkey kidney cells (Vero), which are devoid of an IFN response (27, 28). As shown in Fig. 5G, DEV UL41 significantly reduced IRF7 expression in Vero cells, which is consistent with the results in 293T and DEF cells. These data suggested that DEV UL41 reduced IRF7 expression by decreasing the accumulation of IRF7 mRNA but not via the proteasome or lysosomal pathway or due to the inhibition of IFN-β response.
IRF7 mediates defense against replication of UL41-deficient DEV.
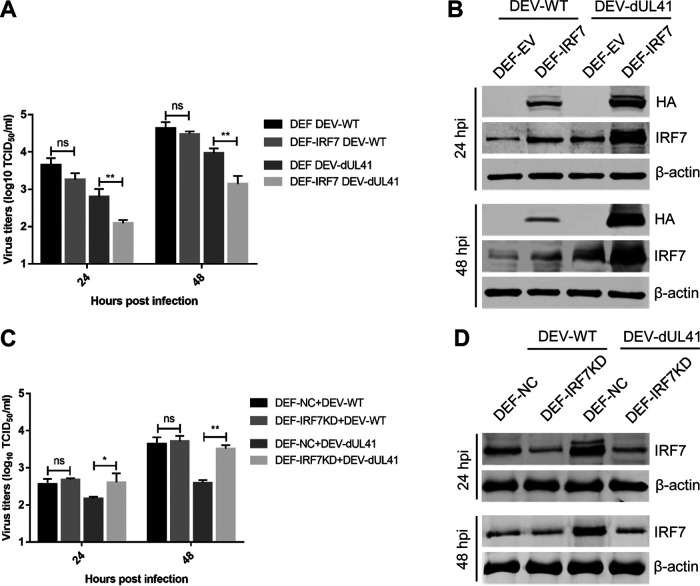
The above results suggested that DEV inhibits the IFN-β response by reducing the accumulation of IRF7 mRNA via UL41, leading us to hypothesize that IRF7 could restrict the replication of the UL41-deficient DEV. To test this assumption, DEFs with ectopic IRF7 expression were infected with DEV-WT or DEV-dUL41 and harvested at the indicated time points for viral replication analyses. As shown in Fig. 6A, ectopic IRF7 expression significantly inhibited the replication of DEV-dUL41 but not DEV-WT. Compared with cells infected with DEV-WT, the IRF7 level was notably enhanced in DEFs that ectopically expressed IRF7 (DEF-IRF7) and were infected with DEV-dUL41 (Fig. 6B). DEFs were then transfected with an IRF7-specific small interfering RNA (siRNA) before infection, and the replication of DEV-WT and DEV-dUL41 was compared with that in cells transfected with a nontargeting siRNA. IRF7 knockdown did not affect the replication of DEV-WT but did promote the replication of DEV-dUL41 (Fig. 6C). The successful knockdown of IRF7 expression in DEF-IRF7KD cells was confirmed using Western blot analyses (Fig. 6D). These data indicate that IRF7 mediates the defense against DEV-dUL41 replication.
FIG 6.
IRF7 mediates the defense against the replication of UL41-deficient DEV. (A, B) DEFs were transfected with the empty vector (DEF-EV) or with the IRF7-HA plasmid (DEF-IRF7). Twenty-four hours after transfection, the cells were infected with DEV-WT or DEV-dUL41 (MOI = 0.01), and then cells were harvested at 24 and 48 hpi for viral titration (A) and Western blot (B) analyses. (C and D) DEFs were transfected with negative-control siRNA (DEF-NC) or with siRNA specific to IRF7 (DEF-IRF7KD). Twenty-four hours after transfection, the cells were infected with DEV-WT or DEV-dUL41 (MOI = 0.01), and then cells were harvested at 24 and 48 hpi and subjected to viral titration (C) and Western blot (D) analyses. Data are presented as means ± SDs from three independent experiments. *, P < 0.05; **, P < 0.01; ns, no significant difference.
UL41 contributes to the evasion of the innate immune response against DEV.
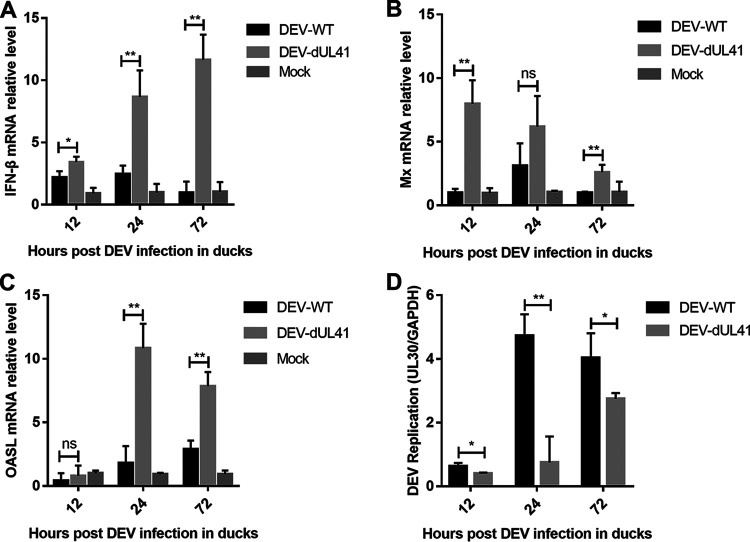
To evaluate the role of UL41 in the immune evasion of DEV, we infected ducks with wild-type or UL41-deficient DEV and examined the expression of IFN-β and its downstream antiviral genes. The results showed that the DEV-dUL41 virus induced significantly higher levels of IFN-β and IFN-stimulated genes Mx and OASL in ducks than wild-type DEV, especially during the late phase of infection (Fig. 7A to C). Furthermore, the viral DNA loads of DEV-WT and DEV-dUL41 in the infected ducks were analyzed using qPCR. As shown in Fig. 7D, the DEV-dUL41 virus replicated at a lower level than the wild-type DEV during infection. In the pathogenicity studies, 6 out of 10 ducks in the DEV-WT group exhibited gross DEV-specific lesions during the experiment; in comparison, only 2 out of 10 ducks in the DEV-dUL41-infected group showed gross DEV-specific lesions. These results suggest that deletion of DEV UL41 attenuated the replication and virulence of DEV in ducks. We speculated that the effect of UL41 on viral replication may involve its ability to inhibit the host immune response, which resulted in enhanced viral replication in vivo.
FIG 7.
UL41 deficiency facilitated IFN-β induction and attenuated DEV replication in ducks. (A to C) Ducks were infected intramuscularly with 100 TCID50 of DEV-WT or DEV-dUL41, and IFN-β (A), Mx (B), and OASL (C) production was measured by qPCR at the indicated time points. (D) Ducks were infected intramuscularly with 100 TCID50 of DEV-WT or DEV-dUL41, and virus genome copy numbers in the spleen were monitored by qPCR at the indicated time points. All samples from the mock control and DEV-infected groups were examined in triplicate. *, P < 0.05; **, P < 0.01; ns, no significant difference.
DISCUSSION
DEV replicates quickly in many cell types and tissues and establishes latent and persistent infection in ducks (1). DEV infection upregulates the expression of multiple PRRs and ISGs in ducks, indicating the activation of innate immune responses to restrict DEV infection (2). Nevertheless, DEV exhibits broad cell tropism in ducks and is difficult to be eradicated by the host immune system, which suggested that DEV might have evolved efficient strategies to evade innate antiviral responses. In the present study, we found that DEV triggers an IFN-β response during the early phase of infection in ducks; however, the production of IFN-β and ISGs was inhibited from 2 dpi. These observations suggest that DEV can efficiently modulate the antiviral innate immune responses to evade host surveillance and immunity, which appears to be critical for the successful infection and quick replication of DEV in ducks. Thus, it was considered worthwhile to determine whether DEV encodes proteins that inhibit IFN-β production along with the underlying mechanisms.
Given the key role of the cGAS-STING DNA-sensing pathway in host antiviral responses, many DNA viruses have evolved various mechanisms to block this signaling pathway (29, 30). Several viral proteins that inhibit IFN-I production through modulation of the cGAS-STING DNA-sensing pathway have been identified, including HSV-1 VP22 (16), VP24 (31), ICP27 (32), UL24 (33), and VP11/12 (34), as well as viral proteins encoded by KSHV (12), human cytomegalovirus (35, 36), and murine gammaherpesvirus 68 (37). However, in contrast to their mammalian counterparts, our current knowledge regarding avian herpesvirus proteins involved in regulating this pathway remains limited (38). The involvement of DEV proteins in antagonizing the DNA-sensing pathway remain unclear. In this study, upon screening a total of 73 DEV ORFs, we successfully identified multiple viral proteins that inhibit the cGAS-STING DNA-sensing pathway. We inferred that these viral inhibitors might function cooperatively to inhibit IFN-I induction, explaining why DEV efficiently inhibited the host antiviral innate immune response during infection in ducks.
The alphaherpesvirus UL41 gene encodes the VHS protein, which is a late tegument protein and an mRNA-specific RNase that induces host shutoff in the early stage of infection (23–25). VHS degrades viral mRNAs to facilitate the sequential expression of different classes of viral genes and degrades host mRNAs to redirect the cell from host to viral gene expression and to evade host antiviral responses (39–42). HSV-1 VHS is known to be important for viral pathogenesis in vivo, and the UL41-deleted recombinant HSV-1 strain has been evaluated as a prophylactic live attenuated vaccine against lethal HSV-1 infection in a mouse model (43). The UL41-null pseudorabies virus showed significantly smaller plaques and lower titers in Vero cells along with impaired lethality and neurological invasion in mice (44). DEV UL41 was found to be dispensable for viral replication; however, UL41 deletion affects viral DNA replication and viral release (45). In this study, DEV UL41 was shown to evade the cGAS-STING DNA-sensing pathway by reducing IRF7 mRNA accumulation, which may facilitate DEV replication. Compared with UL41-deficient DEV, DEV-WT infection significantly inhibited IFN-β and ISG expression in DEFs and in ducks. Furthermore, IRF7 knockdown facilitated the replication of DEV-dUL41 but not DEV-WT. The results also showed that DEV-WT infection did not affect the expression of cGAS, STING, or TBK1. The VHS protein UL41 was reported to selectively degrade both viral and cellular mRNAs containing AU-rich elements (ARE) in its 3′ untranslated region (UTR) (46). We found one ARE core motif (ATTTA) in the 3′ UTR of duck IRF7 but no motif in STING and TBK1, which explains why DEV UL41 decreases IRF7 expression without affecting other adaptors in the DNA-sensing pathway.
IRF7 plays a crucial role in IFN-β production in avian cells because all signals converge at this transcription activator. Thus, avian viruses are more likely to evolve strategies for counteracting IRF7 activation. We recently reported that MDV VP23 targets IRF7 to inhibit DNA-sensing signaling in chicken cells (21). The MDV oncoprotein Meq inhibits this pathway through interaction with STING and IRF7, preventing the associations of STING with TBK1 and IRF7, and suppressing IRF7 activation as well as IFN-β production (13). The results of this study indicate that UL41 inhibits IFN-β production triggered by multiple inducers of the cGAS-STING pathway, including ISD, poly(dA·dT), and DNA virus infection. The function of UL41 in antagonizing the IFN-I responses to these DNA stimuli makes the UL41-expressing cells more susceptible to other DNA viruses besides DEV. Furthermore, the IRF7 activation step is shared by many other pathways, such as the TLR and RLR pathways (4, 5); thus, UL41 could also affect other pathways induced by RNA virus infections. These findings suggest that evasion of DNA-sensing signaling by DEV might facilitate the secondary infection of other viruses.
In summary, we demonstrated for the first time that multiple viral proteins have evolved to inhibit the DNA-sensing pathway during DEV infection. DEV UL41 counteracts the cGAS-STING DNA-sensing pathway by reducing the accumulation of IRF7 mRNA. These findings lead us to better understand the mechanisms applied by DEV to evade host antiviral innate immunity and facilitate its rapid replication and persistent infection in ducks.
MATERIALS AND METHODS
Animals and ethics statement.
Specific-pathogen-free (SPF) ducks and fertilized SPF duck eggs were purchased from the National Laboratory Poultry Animal Resource Center (Harbin, China). Ten-day-old SPF duck embryos were used for preparing primary DEFs. This study was carried out following the recommendations in the Guide for the Care and Use of Laboratory Animals of the Ministry of Science and Technology of China. The use of SPF ducks and the animal experiments were approved by the Animal Ethics Committee of Harbin Veterinary Research Institute of Chinese Academy of Agricultural Sciences and were performed in accordance with animal ethics guidelines and approved protocols (SYXK [Hei] 2017-009).
Cells, viruses, and antibodies.
Primary DEFs were prepared from 10-day-old SPF duck embryos and cultured in Dulbecco’s modified Eagle’s medium (DMEM; Life Technologies, Grand Island, NY) supplemented with 5% fetal bovine serum (FBS; Sigma-Aldrich, St. Louis, MO). 293T and Vero cells were cultured in DMEM containing 10% FBS (Sigma-Aldrich). The DEV CV strain (GenBank no. JQ673560) and HVT FC126 strain (GenBank no. AF291866) were propagated in DEFs before use in this study. Commercially available antibodies, including mouse anti-Flag, rabbit anti-HA, and mouse anti-actin (Sigma-Aldrich), rabbit anti-cGAS, and rabbit anti-TBK1 (Abcam, Cambridge, UK), were used. Rabbit anti-STING, rabbit anti-IRF7, rabbit anti-UL41, and rabbit anti-gE antibodies were prepared in our laboratory. ISD and poly(dA·dT) were purchased from InvivoGen (San Diego, CA, USA).
Plasmid construction.
The DEV UL41 gene was amplified from the genome of the DEV CV strain and cloned into the pCAGGS vector for expression. Plasmids encoding duck cGAS (GenBank no. XM_021271479.1), STING (GenBank no. XM_027468120), TBK1 (GenBank no. KY963947), and IRF7 (GenBank no. MG707077) were constructed by cloning the synthesized sequence into pCAGGS with a Flag or HA tag fused to the 3′ end. The duck IFN-β promoter luciferase reporter IFN-β-luc was constructed by inserting the −96 to +1 fragment of the duck IFN-β promoter into the pGL3-basic vector.
Real-time qPCR.
Total RNA was extracted and reverse transcription was performed using ReverTra Ace qPCR RT kit (Toyobo, Osaka, Japan). The quantity of each cDNA was determined by real-time qPCR using LightCycler 480 (Roche, Basel, Switzerland). Specific primers for IFN-β, IL-6, Mx, OASL, cGAS, STING, TBK1, and IRF7 are shown in Table S1 and synthesized by Invitrogen (Shanghai, China). Fold differences between treated samples and control samples were calculated. To determine the DEV and HVT titers, total DNA was extracted using the AxyPrep BodyFluid Viral DNA/RNA Miniprep kit (Corning Life Sciences, Shanghai, China) and tested using real-time qPCR by measuring the copy numbers of DEV UL30 and HVT UL30 genes as the viral genome target and the duck GAPDH gene was used as a reference. The qPCR was performed under the following cycling conditions: 95°C for 1 min for initial denaturation, followed by 40 cycles of 95°C for 15 s for denaturation, 60°C for 1 min, and collection of PCR product signals. All controls and treated samples were examined in triplicate in the same plate.
Transfection and dual-luciferase reporter assays.
To determine duck IFN-β promoter activity, DEF cells were cotransfected with the firefly luciferase reporter plasmid (IFN-β Luc) and with the Renilla luciferase reporter pRL-TK, which served as an internal control, with or without expression plasmids, as indicated, using the TransIT-X2 dynamic delivery system (Mirus, Madison, WI, USA). At 36 h posttransfection, cells were lysed, and samples were assayed for firefly and Renilla luciferase activity using the dual-luciferase reporter assay system (Promega, Madison, WI, USA). Relative luciferase activity was normalized to Renilla luciferase activity. The reporter assays were repeated at least three times.
Western blot assays.
Expression plasmids harboring Flag or HA tags were transfected into DEFs or 293T cells using the TransIT-X2 dynamic delivery system. At 36 h posttransfection, whole-cell lysates were obtained by lysing cells in NP-40 lysis buffer (Beyotime, Beijing, China). Protein concentrations were determined using a bicinchoninic acid protein assay kit (Thermo Fisher Scientific). The proteins were separated by electrophoresis on 12% SDS-polyacrylamide gels, transferred onto nitrocellulose membranes, and incubated with the indicated primary and secondary antibodies. Images were acquired using the Odyssey infrared imaging system (LI-COR Biosciences, Lincoln, NE, USA).
Generation of UL41-deleted recombinant DEV.
Five fosmid clones containing sequences encompassing the entire genome of DEV were constructed in our preliminary studies and used here for generating the UL41-deficient DEV. Fosmid CV1, containing the DEV genome fragment from 1 to 42,401 and the coding sequence of UL41 (32,792 to 34,288 in DEV CV genome), was used for deleting UL41 with the Counter-Selection BAC Modification kit (Gene Bridges, Heidelberg, Germany). The CV1-dUL41 fosmid in which the UL41 gene was deleted was identified by PCR and sequencing. To rescue the UL41-deleted virus DEV-dUL41, 2 μg of each purified fosmid DNA (CV1-dUL41, CV2, CV3, CV4, and CV5) was used to transfect DEFs using the calcium phosphate procedure. Five days after transfection, cells were trypsinized, seeded onto a new dish, and monitored for cytopathic effects. Viral stocks were subsequently generated in DEFs for further analyses.
DEV titration.
DEV titers were estimated by the median tissue culture infective dose (TCID50). DEFs were seeded in 96-well plates at a density of 1 × 105 cells per well. Cells were infected with serial 10-fold DEV dilutions and incubated for 5 days at 37°C in a 5% CO2 atmosphere. Cell pathological changes were observed under a microscope and recorded. Viral titers were calculated according to the Reed-Muench method (47).
RNA interference.
siRNAs specifically targeting duck IRF7 (5′-GCC AAG TGG AAG ACC AAC T-3′) as well as a scrambled negative-control siRNA (5′-GTT CTC CGA ACG TGT CAC GT-3′) were synthesized by GenePharma (Shanghai, China). The siRNA transfections were performed in DEFs using the TransIT-X2 dynamic delivery system (Mirus) according to the manufacturer’s instructions. After 12 h of transfection, cells were infected with the wild-type DEV or DEV-dUL41 for further analysis. The knockdown efficiency of IRF7 was verified by real-time qPCR and Western blotting.
Animal studies.
To determine the effects of DEV infection on the induction of IFN-β and downstream antiviral genes, 35 2-week-old SPF ducks were inoculated intramuscularly with the virulent DEV CV strain, and the mock control group including 35 ducks was inoculated with DMEM. At the indicated time points shown in Fig. 1, spleen samples were collected from five ducks in each group, and the mRNA levels of IFN-β, IL-6, Mx, and OASL were measured by real-time qPCR. To characterize DEV-WT and DEV-dUL41 viruses, a total of 45 2-week-old SPF ducks were randomly divided into three groups. Two groups were inoculated intramuscularly with DEV-WT or DEV-dUL41, and the third group was mock injected with DMEM. After 12 h, 24 h, and 72 h of infection as shown in Fig. 7, five ducks from each group were humanely euthanized, and spleen samples were collected for analyzing IFN-β, Mx, and OASL expression as well as viral DNA copy numbers. For DEV pathogenicity analyses, groups of 10 2-week-old SPF ducks were inoculated with DEV-WT, DEV-dUL41, or DMEM and monitored for clinical signs and mortality for 14 days. All ducks were necropsied at the end of the experiment to evaluate the presence of DEV-specific lesions.
Statistical analysis.
The data are presented as means ± SD. Statistical significance between groups was determined using Student’s t test with GraphPad Prism 7.0 software (La Jolla, CA, USA). Statistical significance was set at P < 0.05.
ACKNOWLEDGMENTS
This research was supported by a grant from Heilongjiang Natural Science Foundation of China (YQ2020C024), National Natural Science Foundation of China (31970162, U20A2061), and Central Public-interest Scientific Institution Basal Research Fund (Y2022QC26).
Footnotes
Supplemental material is available online only.
Contributor Information
Yulong Gao, Email: gaoyulong@caas.cn.
Kai Li, Email: likai01@caas.cn.
Jae U. Jung, Lerner Research Institute, Cleveland Clinic
REFERENCES
- 1.Dhama K, Kumar N, Saminathan M, Tiwari R, Karthik K, Kumar MA, Palanivelu M, Shabbir MZ, Malik YS, Singh RK. 2017. Duck virus enteritis (duck plague)–a comprehensive update. Vet Q 37:57–80. 10.1080/01652176.2017.1298885. [DOI] [PubMed] [Google Scholar]
- 2.Li N, Hong T, Li R, Guo M, Wang Y, Zhang J, Liu J, Cai Y, Liu S, Chai T, Wei L. 2016. Pathogenicity of duck plague and innate immune responses of the Cherry Valley ducks to duck plague virus. Sci Rep 6:32183. 10.1038/srep32183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qi X, Yang X, Cheng A, Wang M, Zhu D, Jia R. 2008. The pathogenesis of duck virus enteritis in experimentally infected ducks: a quantitative time-course study using TaqMan polymerase chain reaction. Avian Pathol 37:307–310. 10.1080/03079450802043775. [DOI] [PubMed] [Google Scholar]
- 4.Wu J, Chen ZJ. 2014. Innate immune sensing and signaling of cytosolic nucleic acids. Annu Rev Immunol 32:461–488. 10.1146/annurev-immunol-032713-120156. [DOI] [PubMed] [Google Scholar]
- 5.Brubaker SW, Bonham KS, Zanoni I, Kagan JC. 2015. Innate immune pattern recognition: a cell biological perspective. Annu Rev Immunol 33:257–290. 10.1146/annurev-immunol-032414-112240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xia P, Wang S, Gao P, Gao G, Fan Z. 2016. DNA sensor cGAS-mediated immune recognition. Protein Cell 7:777–791. 10.1007/s13238-016-0320-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen Q, Sun L, Chen ZJ. 2016. Regulation and function of the cGAS-STING pathway of cytosolic DNA sensing. Nat Immunol 17:1142–1149. 10.1038/ni.3558. [DOI] [PubMed] [Google Scholar]
- 8.Sun L, Wu J, Du F, Chen X, Chen ZJ. 2013. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339:786–791. 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barber GN. 2015. STING: infection, inflammation and cancer. Nat Rev Immunol 15:760–770. 10.1038/nri3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shu C, Li X, Li P. 2014. The mechanism of double-stranded DNA sensing through the cGAS-STING pathway. Cytokine Growth Factor Rev 25:641–648. 10.1016/j.cytogfr.2014.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reinert LS, Lopušná K, Winther H, Sun C, Thomsen MK, Nandakumar R, Mogensen TH, Meyer M, Vægter C, Nyengaard JR, Fitzgerald KA, Paludan SR. 2016. Sensing of HSV-1 by the cGAS-STING pathway in microglia orchestrates antiviral defence in the CNS. Nat Commun 7:13348. 10.1038/ncomms13348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma Z, Jacobs SR, West JA, Stopford C, Zhang Z, Davis Z, Barber GN, Glaunsinger BA, Dittmer DP, Damania B. 2015. Modulation of the cGAS-STING DNA sensing pathway by gammaherpesviruses. Proc Natl Acad Sci USA 112:E4306–E4315. 10.1073/pnas.1503831112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li K, Liu Y, Xu Z, Zhang Y, Luo D, Gao Y, Qian Y, Bao C, Liu C, Zhang Y, Qi X, Cui H, Wang Y, Gao L, Wang X. 2019. Avian oncogenic herpesvirus antagonizes the cGAS-STING DNA-sensing pathway to mediate immune evasion. PLoS Pathog 15:e1007999. 10.1371/journal.ppat.1007999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zheng C. 2018. Evasion of cytosolic DNA-stimulated innate immune responses by herpes simplex virus 1. J Virol 92:e00099-17. 10.1128/JVI.00099-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.You H, Zheng S, Huang Z, Lin Y, Shen Q, Zheng C. 2019. Herpes simplex virus 1 tegument protein UL46 inhibits TANK-binding kinase 1-mediated signaling. mBio 10:e00919-19. 10.1128/mBio.00919-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang J, You H, Su C, Li Y, Chen S, Zheng C. 2018. Herpes simplex virus 1 tegument protein VP22 abrogates cGAS/STING-mediated antiviral innate immunity. J Virol 92:e00841-18. 10.1128/JVI.00841-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Su C, Zheng C. 2017. Herpes simplex virus 1 abrogates the cGAS/STING-mediated cytosolic DNA-sensing pathway via its virion host shutoff protein, UL41. J Virol 91:e02414-16. 10.1128/JVI.02414-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Santhakumar D, Rubbenstroth D, Martinez-Sobrido L, Munir M. 2017. Avian interferons and their antiviral effectors. Front Immunol 8:49. 10.3389/fimmu.2017.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu X, Wu FH, Wang X, Wang L, Siedow JN, Zhang W, Pei ZM. 2014. Molecular evolutionary and structural analysis of the cytosolic DNA sensor cGAS and STING. Nucleic Acids Res 42:8243–8257. 10.1093/nar/gku569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Neerukonda SN, Katneni U. 2020. Avian pattern recognition receptor sensing and signaling. Vet Sci 7:14. 10.3390/vetsci7010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao L, Li K, Zhang Y, Liu Y, Liu C, Zhang Y, Gao Y, Qi X, Cui H, Wang Y, Wang X. 2019. Inhibition of DNA-sensing pathway by Marek's disease virus VP23 protein through suppression of interferon regulatory factor 7 activation. J Virol 93:e01934-18. 10.1128/JVI.01934-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Y, Wang M, Cheng A, Jia R, Yang Q, Chen S, Zhu D, Liu M, Zhao X, Zhang S, Huang J, Ou X, Mao S, Yu Y, Zhang L, Liu Y, Pan L, Tian B, Rehman MU, Chen X. 2020. Duck enteritis virus VP16 antagonizes IFN-β-mediated antiviral innate immunity. J Immunol Res 2020:9630452. 10.1155/2020/9630452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taddeo B, Roizman B. 2006. The virion host shutoff protein (UL41) of herpes simplex virus 1 is an endoribonuclease with a substrate specificity similar to that of RNase A. J Virol 80:9341–9345. 10.1128/JVI.01008-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Everly DN, Jr, Feng P, Mian IS, Read GS. 2002. mRNA degradation by the virion host shutoff (Vhs) protein of herpes simplex virus: genetic and biochemical evidence that Vhs is a nuclease. J Virol 76:8560–8571. 10.1128/jvi.76.17.8560-8571.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Esclatine A, Taddeo B, Roizman B. 2004. The UL41 protein of herpes simplex virus mediates selective stabilization or degradation of cellular mRNAs. Proc Natl Acad Sci USA 101:18165–18170. 10.1073/pnas.0408272102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pasieka TJ, Lu B, Crosby SD, Wylie KM, Morrison LA, Alexander DE, Menachery VD, Leib DA. 2008. Herpes simplex virus virion host shutoff attenuates establishment of the antiviral state. J Virol 82:5527–5535. 10.1128/JVI.02047-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang S, Wang K, Lin R, Zheng C. 2013. Herpes simplex virus 1 serine/threonine kinase US3 hyperphosphorylates IRF3 and inhibits beta interferon production. J Virol 87:12814–12827. 10.1128/JVI.02355-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ye J, Chen Z, Li Y, Zhao Z, He W, Zohaib A, Song Y, Deng C, Zhang B, Chen H, Cao S. 2017. Japanese encephalitis virus NS5 inhibits type i interferon (IFN) production by blocking the nuclear translocation of IFN regulatory factor 3 and NF-κB. J Virol 91:e00039-17. 10.1128/JVI.00039-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin Y, Zheng C. 2019. A tug of war: DNA-sensing antiviral innate immunity and herpes simplex virus type I infection. Front Microbiol 10:2627. 10.3389/fmicb.2019.02627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu H, Zheng C. 2020. The race between host antiviral innate immunity and the immune evasion strategies of herpes simplex virus 1. Microbiol Mol Biol Rev 84:e00099-20. 10.1128/MMBR.00099-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang D, Su C, Zheng C. 2016. Herpes simplex virus 1 serine protease VP24 blocks the DNA-sensing signal pathway by abrogating activation of interferon regulatory factor 3. J Virol 90:5824–5829. 10.1128/JVI.00186-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Christensen MH, Jensen SB, Miettinen JJ, Luecke S, Prabakaran T, Reinert LS, Mettenleiter T, Chen ZJ, Knipe DM, Sandri-Goldin RM, Enquist LW, Hartmann R, Mogensen TH, Rice SA, Nyman TA, Matikainen S, Paludan SR. 2016. HSV-1 ICP27 targets the TBK1-activated STING signalsome to inhibit virus-induced type I IFN expression. EMBO J 35:1385–1399. 10.15252/embj.201593458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu H, Su C, Pearson A, Mody CH, Zheng C. 2017. Herpes simplex virus 1 UL24 abrogates the DNA sensing signal pathway by inhibiting NF-κB activation. J Virol 91:e00025-17. 10.1128/JVI.00025-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deschamps T, Kalamvoki M. 2017. Evasion of the STING DNA-sensing pathway by VP11/12 of herpes simplex virus type 1. J Virol 91:e00535-17. 10.1128/JVI.00535-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fu YZ, Su S, Gao YQ, Wang PP, Huang ZF, Hu MM, Luo WW, Li S, Luo MH, Wang YY, Shu HB. 2017. Human cytomegalovirus tegument protein UL82 inhibits STING-mediated signaling to evade antiviral immunity. Cell Host Microbe 21:231–243. 10.1016/j.chom.2017.01.001. [DOI] [PubMed] [Google Scholar]
- 36.Huang ZF, Zou HM, Liao BW, Zhang HY, Yang Y, Fu YZ, Wang SY, Luo MH, Wang YY. 2018. Human cytomegalovirus protein UL31 inhibits DNA sensing of cGAS to mediate immune evasion. Cell Host Microbe 24:69–80.e4. 10.1016/j.chom.2018.05.007. [DOI] [PubMed] [Google Scholar]
- 37.Kang HR, Cheong WC, Park JE, Ryu S, Cho HJ, Youn H, Ahn JH, Song MJ. 2014. Murine gammaherpesvirus 68 encoding open reading frame 11 targets TANK binding kinase 1 to negatively regulate the host type I interferon response. J Virol 88:6832–6846. 10.1128/JVI.03460-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gao L, Zheng S, Wang Y. 2021. The evasion of antiviral innate immunity by chicken DNA viruses. Front Microbiol 12:771292. 10.3389/fmicb.2021.771292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shen G, Wang K, Wang S, Cai M, Li ML, Zheng C. 2014. Herpes simplex virus 1 counteracts viperin via its virion host shutoff protein UL41. J Virol 88:12163–12166. 10.1128/JVI.01380-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Su C, Zhang J, Zheng C. 2015. Herpes simplex virus 1 UL41 protein abrogates the antiviral activity of hZAP by degrading its mRNA. Virol J 12:203. 10.1186/s12985-015-0433-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zenner HL, Mauricio R, Banting G, Crump CM. 2013. Herpes simplex virus 1 counteracts tetherin restriction via its virion host shutoff activity. J Virol 87:13115–13123. 10.1128/JVI.02167-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jiang Z, Su C, Zheng C. 2016. Herpes simplex virus 1 tegument protein UL41 counteracts IFIT3 antiviral innate immunity. J Virol 90:11056–11061. 10.1128/JVI.01672-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Page HG, Read GS. 2010. The virion host shutoff endonuclease (UL41) of herpes simplex virus interacts with the cellular cap-binding complex eIF4F. J Virol 84:6886–6890. 10.1128/JVI.00166-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ye C, Chen J, Wang T, Xu J, Zheng H, Wu J, Li G, Yu Z, Tong W, Cheng X, Zhou S, Tong G. 2018. Generation and characterization of UL41 null pseudorabies virus variant in vitro and in vivo. Virol J 15:119. 10.1186/s12985-018-1025-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.He T, Wang M, Cheng A, Yang Q, Jia R, Wu Y, Huang J, Tian B, Liu M, Chen S, Zhao XX, Zhu D, Zhang S, Ou X, Mao S, Gao Q, Sun D. 2021. DPV UL41 gene encoding protein induces host shutoff activity and affects viral replication. Vet Microbiol 255:108979. 10.1016/j.vetmic.2021.108979. [DOI] [PubMed] [Google Scholar]
- 46.Esclatine A, Taddeo B, Evans L, Roizman B. 2004. The herpes simplex virus 1 UL41 gene-dependent destabilization of cellular RNAs is selective and may be sequence-specific. Proc Natl Acad Sci USA 101:3603–3608. 10.1073/pnas.0400354101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reed LJ, Muench H. 1938. A simple method of estimation of fifty percent end points. Am J Hyg 27:493–497. 10.1093/oxfordjournals.aje.a118408. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 and Fig. S1. Download jvi.01578-22-s0001.pdf, PDF file, 0.3 MB (279.9KB, pdf)