ABSTRACT
Most of the HIV DNA in infected individuals is noninfectious because of deleterious mutations. However, it is unclear how much of the transcribed HIV RNA is potentially infectious or defective. To address this question, we developed and validated a novel intact viral RNA assay (IVRA) that uses droplet digital reverse transcriptase PCR (dd-RT-PCR) for the commonly mutated packaging signal (Psi) and Rev response element (RRE) regions (from the intact proviral DNA assay [IPDA]) to quantify likely intact (Psi+ RRE+), 3′ defective (Psi+ RRE−), and 5′ defective (Psi− RRE+) HIV RNA. We then applied the IPDA and IVRA to quantify intact and defective HIV DNA and RNA from peripheral CD4+ T cells from 9 antiretroviral therapy (ART)-suppressed individuals. Levels of 3′ defective HIV DNA were not significantly different from those of 5′ defective HIV DNA, and both were higher than intact HIV DNA. In contrast, 3′ defective HIV RNA (median 86 copies/106 cells; 94% of HIV RNA) was much more abundant than 5′ defective (2.1 copies/106 cells; 5.6%) or intact (0.6 copies/106 cells; <1%) HIV RNA. Likewise, the frequency of CD4+ T cells with 3′ defective HIV RNA was greater than the frequency with 5′ defective or intact HIV RNA. Intact HIV RNA was transcribed by a median of 0.018% of all proviruses and 2.2% of intact proviruses. The vast excess of 3′ defective RNA over 5′ defective or intact HIV RNA, which was not observed for HIV DNA, suggests that HIV transcription is completely blocked prior to the RRE in most cells with intact proviruses and/or that cells transcribing intact HIV RNA are cleared at very high rates.
IMPORTANCE We developed a new assay that can distinguish and quantify intact (potentially infectious) as well as defective HIV RNA. In ART-treated individuals, we found that the vast majority of all HIV RNA is defective at the 3′ end, possibly due to incomplete transcriptional processivity. Only a very small percentage of all HIV RNA is intact, and very few total or intact proviruses transcribe intact HIV RNA. Though rare, this intact HIV RNA is tremendously important because it is necessary to serve as the genome of infectious virions that allow transmission and spread, including rebound after stopping ART. Moreover, intact viral RNA may contribute disproportionately to the immune activation, inflammation, and organ damage observed with untreated and treated HIV infection. The intact viral RNA assay can be applied to many future studies aimed at better understanding HIV pathogenesis and barriers to HIV cure.
KEYWORDS: HIV DNA, HIV provirus, intact proviral DNA assay (IPDA), HIV RNA, HIV-1, defective, infectious, intact, latent infection, transcription, transcriptional regulation, viral replication
INTRODUCTION
The major barrier to curing HIV is a reservoir of cells that harbor replication-competent proviruses and are capable of producing infectious virions. Most of these cells are in a state of latent infection, in which the cell does not constitutively produce virions but can be induced by certain stimuli (such as activation of T cells) to produce infectious viruses (1–3). Low-level spontaneous reactivation from the latent reservoir is thought to be the source of the plasma virus that rebounds after stopping antiretroviral therapy (ART). In addition, the viral RNA and proteins produced by these cells likely contribute to the higher average levels of immune activation and inflammation, higher incidence of diseases in many organ systems, and lower average life expectancy observed in ART-suppressed than in HIV-uninfected individuals (4–12).
Using techniques that can sequence almost the full length of individual proviruses (near-full-length sequencing [nFL] or full-length individual provirus sequencing [FLIPS]), prior studies showed that the vast majority of HIV DNA in HIV-infected individuals is defective due to proviral mutations such as deletions and APOBEC-induced hypermutations (13–17). The intact proviral DNA assay (IPDA) was developed as a more high-throughput means to quantify those proviruses that are likely to be intact (lacking obvious sequence defects) and distinguish them from defective proviruses (18). The IPDA is a droplet digital PCR (ddPCR) assay in which cellular DNA is partitioned into oil nanodroplets expected to have at most one provirus and a duplex quantitative PCR (qPCR) is used to simultaneously test each provirus for two HIV regions that are frequently deleted (>90%) or hypermutated (>97%) in defective proviruses: the packaging signal (Psi; detected with 6-carboxyfluorescein [FAM]-labeled probe) and Rev response element (RRE; detected with VIC) (18). The assay for the RRE includes both a VIC-labeled wild-type probe designed to mismatch with hypermutated sequences and a nonlabeled, hypermutated probe (18). Droplets in which both the Psi and RRE are detected (Psi+ RRE+, or double positive) likely contain intact proviruses and can be quantified separately from single-positive droplets containing only the more 5′ Psi (Psi+ RRE−, or 3′ defective) or the more 3′ RRE (Psi− RRE+, or 5′ defective) (18). In validation studies, ~70% of proviruses classified as intact by IPDA lacked detectable defects by sequencing, and the IPDA was able to exclude 97% of defects detected by sequencing (18). Other multiplex digital PCR assays have also been designed to quantify intact and defective proviruses (19, 20).
While these assays are extremely useful for quantifying intact and defective HIV DNA, no comparable assay exists for HIV RNA, and it is unclear how much of the HIV RNA that exists in cells or plasma from HIV-infected individuals is intact or defective. In theory, HIV transcripts can be genome defective for multiple reasons. First, if proviral defects do not preclude transcription, some HIV RNA may have been transcribed from proviruses carrying sequence defects, so that the RNA may be missing deleted regions or contain hypermutations. Second, the vast majority of HIV RNA from ART-suppressed individuals is incomplete as a result of sequential transcriptional blocks at different stages of elongation (5′, mid-, and distal) or a continual loss of processivity along the genome (21–23). It is not clear how much of the incomplete or complete HIV transcripts are transcribed from proviruses that are intact or defective. Using a new technique that can simultaneously analyze the HIV RNA, integration site, and proviral sequence from single infected cells parallel HIV-1 RNA, integration site, and proviral sequencing (PRIP-seq), one recent publication showed that around 27% of all cells with intact proviruses are transcribing 5′ elongated HIV RNA (24). However, this percentage was similar for some kinds of defective proviruses, and it is not clear how many cells with intact proviruses are also transcribing more processive or completed HIV transcripts (24). Third, a small fraction of all HIV RNA in ART-suppressed individuals is incomplete as a result of single or multiple splicing (21, 22), which facilitates translation of certain HIV proteins but also prevents the RNA from serving as an infectious HIV genome.
HIV RNA that is intact (lacking proviral mutations, being processive/complete, and being unspliced) is likely to be important for several reasons. First, intact viral RNA is necessary to serve as the genome of infectious virions that allow new infections, including transmission and recrudescence after stopping ART. However, a large proportion of cells with intact or infectious proviruses may not constitutively transcribe intact HIV RNA and may not be inducible even after multiple rounds of activation ex vivo (25) or in vivo. Conversely, most cell-associated HIV RNA in ART-suppressed individuals is incomplete and therefore incapable of serving as infectious genomic RNA (21, 22), while it is unclear how much of the polyadenylated or complete HIV RNA, or even plasma HIV RNA, is defective because of proviral mutations (26). Second, intact HIV RNA can encode more HIV proteins and may be more likely to be translated into HIV proteins, which are also necessary to produce viral particles. Therefore, intact viral RNA may contribute more to the immune activation, inflammation, and organ damage observed in untreated and ART-treated infection.
In order to distinguish and quantify viral RNA that is likely to be intact, we developed a new intact viral RNA assay (IVRA) that extends the principles and validations underlying the IPDA to the study of HIV RNA. Using one-step droplet digital reverse transcriptase PCR (dd-RT-PCR), cellular RNA is first fractionated and encapsulated in oil droplets at limiting dilution, so that any given droplet is extremely unlikely to have more than a single molecule of HIV RNA. Next, reverse transcription of each single HIV RNA molecule is performed within the droplet, followed by duplex PCR using the same primers and probes from the IPDA. Since the Psi is removed by the first HIV splicing event, and RRE is removed by the second, all Psi+ droplets should contain unspliced HIV RNA. Double-positive Psi+ RRE+ droplets contain an HIV RNA molecule that does not contain deletions or hypermutations in the Psi or RRE regions and therefore is likely to have been transcribed from an intact provirus. Moreover, Psi+ RRE+ droplets contained an HIV RNA in which transcription had proceeded at least through the RRE region. Since most blocks to HIV transcription occur prior to the Env/Nef regions (21), a large proportion of these Psi+ RRE+ transcripts would be expected to be complete. Single-positive Psi+ RRE− (3′ defective) droplets may result from multiple causes, including (i) HIV RNA transcribed from a provirus with mutations in RRE, (ii) incomplete HIV RNA resulting from premature termination of transcription between the Psi and RRE regions, or (iii) RNA shearing or degradation. Conversely, single-positive Psi− RRE+ droplets (5′ defective) could result from proviral mutations in Psi, removal of the Psi from single splicing, incomplete antisense transcripts that terminate between RRE and Psi, or RNA shearing/degradation.
After validating the IVRA using HIV RNA standards, we applied the IPDA and IVRA to quantify intact and defective HIV DNA and RNA from replicates of peripheral CD4+ T cells from ART-suppressed individuals. We observed a vast excess of 3′ defective (median 94% of total) over 5′ defective (5.6%) HIV RNA, which was not observed in the HIV DNA and likely results from a block to HIV transcriptional completion. Intact HIV RNA was detected at very low levels (<1%), and only a very small proportion of intact proviruses (2.2%) transcribed intact HIV RNA. These findings suggest that HIV transcription is completely blocked prior to the RRE in most cells with intact proviruses and/or that cells transcribing intact HIV RNA are cleared at very high rates.
RESULTS
Performance of the IVRA on HIV RNA standards.
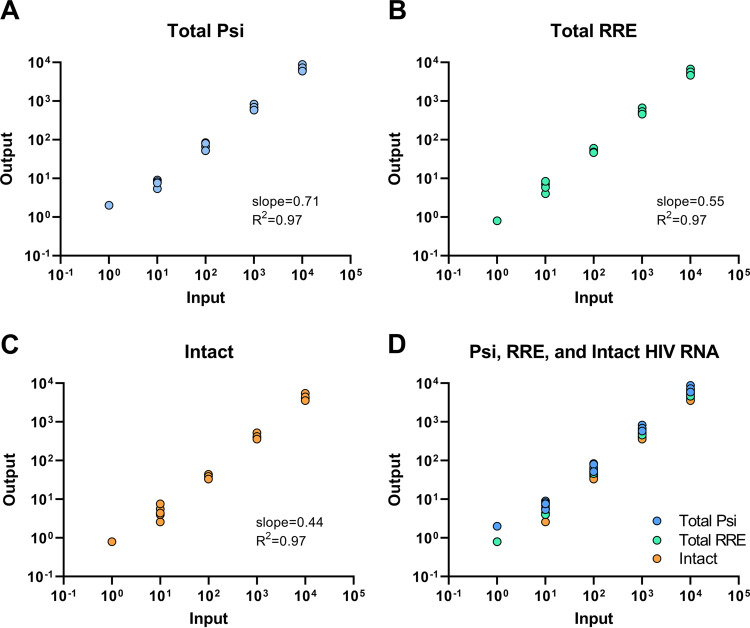
Two different supernatant HIV RNA standards were used to evaluate the performance of the IVRA. Using inputs of 100 and 1,000 copies of an older NL4-3 supernatant HIV RNA standard, double-positive droplets (Psi+ RRE+, suggesting intact HIV RNA) accounted for approximately 67% and 63% (respectively) of all positive droplets (see Fig. S1A in the supplemental material). With inputs of 101 to 104 copies of a newer supernatant HIV RNA standard, approximately 60% of all positive droplets contained intact HIV RNA (Fig. S1B). Intact droplets were detected with an input of 1 to 10 copies of the RNA standard (Fig. 1). With inputs of 100 to 104 copies of the RNA standard, we observed linear quantification (R2 ≥ 0.97) of total Psi+ RNA (Psi+ RRE− plus Psi+ RRE+), total RRE+ RNA (Psi− RRE+ plus Psi+ RRE+), and intact (Psi+ RRE+) RNA. The detection efficiencies (slope of measured versus total input copies) were 71% for total Psi+ RNA, 55% for total RRE+ RNA, and 44% for intact (Psi+ RRE+) RNA (Fig. 1). An example of the ddPCR plot is shown in Fig. S2.
FIG 1.
Linearity, efficiency, and detection limit of IVRA on supernatant HIV RNA standards. HIV RNA standards were prepared from the supernatant of in vitro infections and independently quantified using the Abbott clinical assay. The HIV RNA standards were diluted to achieve predicted inputs of 1 to 104 copies (x axis), and the copies of Psi (A and D), RRE (B and D), and Psi+ RRE+ (intact) (C and D) HIV RNA were measured by dd-RT-PCR (IVRA). Slopes and R2 values were determined by linear regression analysis.
To determine whether “background” human cellular RNA would inhibit the IVRA, we measured the outputs using 0 or 100 copies of the supernatant HIV RNA standard and a range of 0 to 1,000 ng of RNA isolated from CD4+ T cells from an HIV-negative donor. No false positives were detected in wells containing only donor RNA (Fig. S3). With HIV RNA and inputs of up to 1,000 ng of background RNA, measured levels of total Psi+ or RRE+ droplets were at least as high as those with no background (Fig. S3). However, the total HIV RNA levels and percentage of intact droplets peaked at an input of 300 ng. Therefore, in all succeeding experiments, we limited the RNA input to 300 ng per well.
HIV DNA levels in ART-suppressed individuals.
To investigate the landscape of proviral mutations in vivo, we applied the IPDA and assays for 4 other HIV DNA regions (U3-U5, 5′ long terminal repeat [LTR], Pol, and Nef) to total cellular DNA from peripheral CD4+ T cells from 9 ART-suppressed individuals. The U3-U5 LTR region was generally present at the highest level (median 485.6 copies/106 cells; P = 0.0039, 0.0078, and 0.012 for comparison to Psi, Pol, and RRE, respectively) (Fig. S4A and B), which may be expected since this target is present at both ends of an intact provirus. Of the remaining regions, which are all present at one copy per intact provirus, levels of the 5′ LTR region (median 365.9 copies/106 cells) were higher than those of the Psi (P = 0.027), Pol (P = 0.0039), or RRE (P = 0.0078) regions. We found no significant difference between levels of total Psi and RRE DNA. As expected, levels of intact HIV DNA were lower than those of any other single target. Levels of all HIV DNA regions correlated with each other, and all except Pol correlated with intact HIV DNA (Fig. S4C).
Levels of defective and intact HIV DNA and RNA.
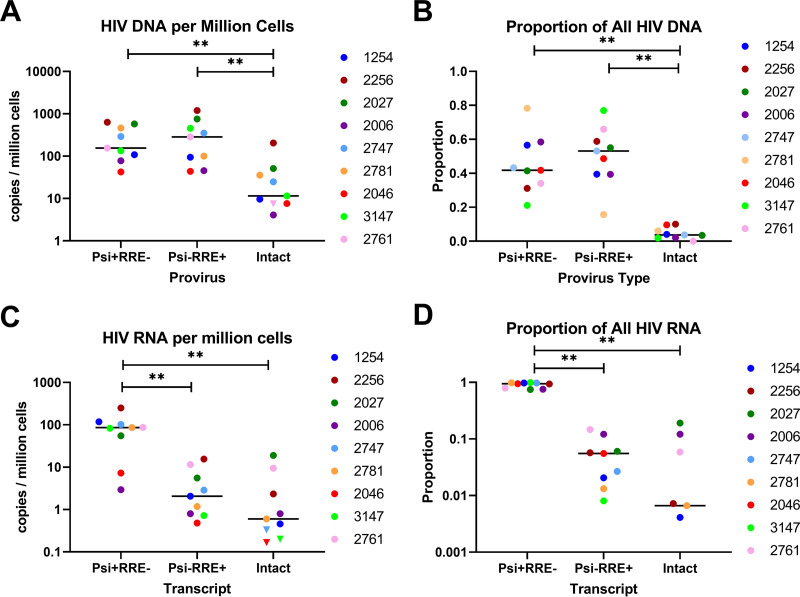
Next, we used the IPDA and IVRA to measure levels of intact and defective HIV DNA and HIV RNA in replicates of peripheral CD4+ T cells from 9 ART-suppressed individuals. Intact HIV DNA was detected in 8 of 9 individuals (Fig. 2A). Levels of intact HIV DNA (median = 11.5 copies/106 cells) were lower than those of either Psi+ RRE− HIV DNA (3′ defective; median = 155.4 copies/106 cells) or Psi− RRE+ HIV DNA (5′ defective; median = 285.3 copies106 cells) (P = 0.0039 for both). However, we detected no significant difference between levels of 3′ defective and 5′ defective HIV DNA (P = 0.30). The proportion of all HIV DNA that was intact (median 3.7%) was lower than that of Psi+ RRE− (median 42%) or Psi− RRE+ DNA (median 53%) (P = 0.002 for both), while no difference was observed between the proportions of Psi+ RRE− and Psi− RRE+ DNA (P = 0.65) (Fig. 2B).
FIG 2.
Levels and proportions of defective and intact HIV DNA and RNA. RNA and DNA were extracted in parallel from replicate dilutions of peripheral CD4+ T cells from 9 ART-suppressed individuals. Average levels (A) and proportions (B) of defective and intact HIV DNA were measured by ddPCR using the IPDA. Defective and intact HIV RNA levels (C) and proportions (D) were measured by dd-RT-PCR using the IVRA. Each color represents a different individual. Samples in which intact HIV DNA or RNA was not detected were assigned a maximum value (inverted triangles) based on 1 copy divided by the total cell equivalents analyzed across all cell replicates. Bars indicate the medians. ** indicates P < 0.005 (Wilcoxon signed-rank test).
Of the negative controls tested concurrently with RNA from HIV-infected individuals, we observed no Psi or RRE false positives in a total of at least 77 wells without RT, 29 wells with water, and 5 wells with HIV-negative donor CD4 RNA. Intact HIV RNA levels were detected in 6 of 9 HIV-infected individuals, albeit at very low levels (median 0.6 copies/106 cells), while Psi+ RRE− HIV RNA (3′ defective) and Psi− RRE+ HIV RNA (5′ defective) were detected in all 9 individuals (Fig. 2C; Fig. S5). In contrast to what was observed for HIV DNA, we observed no difference between levels of intact and Psi− RRE+ HIV RNA (median = 2.1 copies/106 cells; P = 0.20), while levels of Psi+ RRE− RNA (median = 86.2 copies/106 cells) were significantly higher than those of either Psi− RRE+ (P = 0.0039) or intact (P = 0.0039) HIV RNA. Intact HIV RNA correlated with Psi− RRE+ RNA (Spearman r = 0.77; P = 0.021) but not Psi+ RRE− RNA (P = 0.78). The proportion of all HIV RNA that was Psi+ RRE− (median 94%) was significantly higher than the proportions of Psi− RRE+ RNA (median 5.6%; P = 0.0039) or intact HIV RNA (median 0.66%; P = 0.0039), while no significant difference was observed between the proportions of Psi− RRE+ and intact HIV RNA (P = 0.20) (Fig. 2D).
Average copies per provirus of intact and defective HIV RNA.
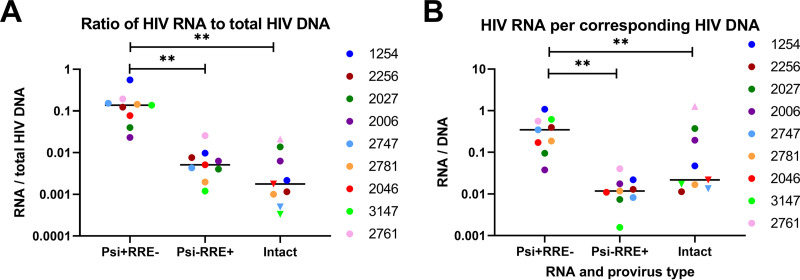
Next, we calculated the ratios of each HIV RNA type to total HIV DNA and to the corresponding HIV DNA (e.g., Psi+ RRE− RNA/Psi+ RRE− DNA) in order to express the average levels of each HIV RNA per provirus and (for ratios of <1) the maximum proportion of proviruses that transcribe each HIV RNA. The median ratio of intact HIV RNA to total HIV DNA was 0.0018 copies/provirus (Fig. 3A). The ratio of Psi+ RRE− RNA to Psi+ RRE− DNA (median 0.35 copies/provirus) was significantly higher than the ratio of Psi− RRE+ RNA to Psi− RRE+ DNA (median 0.012 copies/provirus) or of intact HIV RNA to intact DNA (median 0.022 copies/provirus) (P = 0.0039 for both) (Fig. 3B). It should be remembered that Psi+ RRE− RNA and Psi− RRE+ RNA are not necessarily the products of defective proviruses, since they can also arise from intact proviruses in which sense or antisense transcription (respectively) terminates between the two regions, while Psi− RRE+ RNA can arise from single splicing. However, even if some of the single-positive RNA is from intact proviruses, the ratios of Psi+ RRE− RNA/Psi+ RRE− DNA and Psi− RRE+ RNA/Psi− RRE+ DNA (which are both <1) still express an upper limit on the proportion of these defective proviruses that are transcribing elongated HIV RNA.
FIG 3.
Ratios of intact and defective HIV RNA to DNA. The copies/106 cells of intact and defective HIV RNA were divided by the copies/106 cells of total (containing Psi and/or RRE) HIV DNA (A) or the corresponding type of HIV DNA (B) to express the HIV RNA/DNA ratio (y axis). Each color represents a different individual. Samples in which either intact HIV RNA or DNA was not detected were assigned a maximum value (set to 1 copy divided by the total cell equivalents analyzed) in order to calculate the maximum (inverted triangle) or minimum (triangle) HIV RNA/DNA levels. Bars represent the medians. ** indicates P < 0.005 (Wilcoxon signed-rank test).
Frequency of CD4+ T cells with intact and defective HIV RNA.
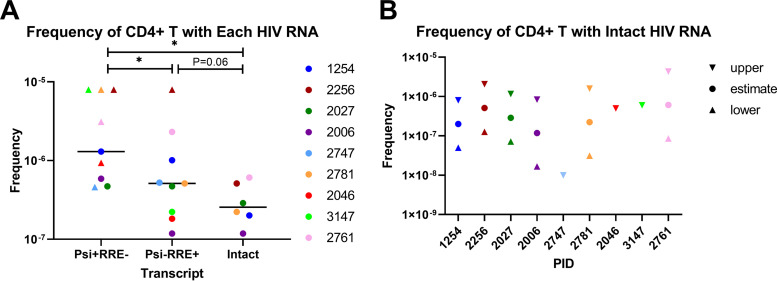
Next, we used the numbers of replicates in which each HIV RNA was or was not detected and the total number of CD4+ T cells per replicate to determine the frequency of CD4+ T cells that contain each HIV RNA, according to the method of extreme limiting dilution analysis (ELDA) (27). The frequency of CD4+ T cells with Psi+ RRE− HIV RNA was greater than the frequency with Psi− RRE+ or intact HIV RNA (P = 0.031 for both), and the frequency with Psi− RRE+ RNA tended to exceed the frequency with intact HIV RNA (P = 0.063) (Fig. 4A). The estimated median frequency of CD4+ T cells that expressed intact HIV RNA was 2.6 × 10−7 (Fig. 4A and B).
FIG 4.
Frequencies of peripheral CD4+ T cells harboring defective and intact HIV RNA. Using the proportion of cell replicates in which each type of HIV RNA was detected, along with the total number of cells per replicate (starting cell counts), we calculated the frequencies (y axes) of CD4+ T cells that contain each type of HIV RNA (A) and intact HIV RNA (B) according to the method of extreme limiting dilution analysis (ELDA). Each color represents a different individual. Circles indicate estimated frequencies; triangles indicate minimums; inverted triangles represent maximums. Bars represent medians. * indicates P < 0.05 (Wilcoxon signed-rank test). PID, patient identifier.
DISCUSSION
We developed a new intact viral RNA assay (IVRA) that uses the primer probe sets from the IPDA to classify HIV transcripts as intact, 5′ defective, or 3′ defective. After validating the assay with HIV RNA standards and in the presence of background donor RNA, we applied the IPDA and IVRA to quantify intact and defective HIV DNA and RNA in ART-suppressed individuals.
Using the supernatant HIV RNA standards, the IVRA showed sensitive, linear detection of Psi+, RRE+, and Psi+ RRE+ (intact) HIV RNA over a range of 1 to 104 copies, with minimal if any inhibition from background cellular RNA. Amplification plots showed sufficient separation between positive and negative droplets, with 4 populations that were easily distinguishable using the standards and RNA from peripheral CD4+ T cells (see Fig. S2 and S5 in the supplemental material).
Using two different supernatant HIV RNA standards, approximately 60 to 67% of all the HIV RNA was found to contain both Psi and RRE regions. One might expect that most of the HIV RNA from these standards would consist of unspliced, full-length genomic HIV RNA from virions, which should be Psi+ RRE+. Since no reverse transcription or PCR would be expected to be 100% sensitive for 1 copy, it is likely that both targets are not detected 100% of the time even when they are present in the same droplet. A similar limitation may apply to the IPDA, where we have observed double-positive rates of 70 to 80% using plasmid (pNL4-3) HIV DNA, similar to the published frequency observed after incubation of pNL4-3 for a few days at 4°C (18). However, the exact composition of the supernatant HIV RNA standards is unclear. It is possible that the RNase digestion of free HIV RNA (not in virions) is incomplete and/or results in short fragments that can still be detected by RT-PCR. Alternatively, it is possible that some incomplete HIV RNA is packaged into virions, and there could be shearing or degradation during or after RNA extraction. We attempted to prepare a different HIV RNA standard by in vitro transcription (IVT) of a plasmid, HIV-1 BH10 noninfectious molecular clone (pBKBH10S DNA), which contains T3/T7 promoters and nearly the full length of the HIV genome. However, we were unable to obtain sufficient quantity or purity of the full-size product despite multiple attempts at optimization using two different commercial kits for IVT.
To investigate the proviral landscape in ART-suppressed individuals, we compared levels of 6 different HIV DNA regions and Psi+ RRE+ (intact) HIV DNA in total peripheral CD4+ T cells. The highest total HIV DNA levels were observed for the U3-U5 region, which may reflect the fact that this region is present at 2 copies per provirus and/or that proviral mutations are less common in the LTRs. Of those regions that are present at only 1 copy per provirus, the 5′ LTR region was present at significantly higher levels than the Psi, RRE, or Pol regions, despite similar efficiencies on HIV DNA standards, likely because of the higher frequency of proviral mutations in these other regions. Therefore, assays for the U3-U5 and 5′ LTR regions may be particularly useful for quantifying total HIV DNA in individuals with very low infection frequencies. Levels of most HIV DNA regions (all but Pol) correlated with levels of intact HIV DNA, in agreement with two published studies (28, 29).
In agreement with prior studies, levels of intact HIV DNA were much lower than those of either 5′ defective or 3′ defective HIV DNA (18, 30). The median level of intact HIV DNA was 11 copies/106 cells, which is lower than reported from some prior studies using the IPDA (54 to 100 copies/106 cells) (18, 30). Some explanations could include differences in the study participants, cell type (total instead of resting CD4+ T cells), extraction method, and normalization (by DNA mass instead of RPP30, with no increase based on shearing index). Intact proviruses accounted for a median of 3.6% of all proviruses, which accords with similar measurements (2.4 to 8%) using single provirus sequencing and the IPDA (13–18). In accord with prior studies using the IPDA, we found no significant difference between levels or proportions of 5′ defective (42% of total) and 3′ defective (53% of total) HIV DNA (18, 30).
5′ defective and 3′ defective HIV RNA were detected in all individuals, but the pattern was very different from that of HIV DNA. Levels of 3′ defective HIV RNA were much higher than those of 5′ defective HIV RNA, such that 3′ defective HIV RNA accounted for most of the total (median 94%, versus 5.6%). This difference is not explained by the slightly higher efficiency for the Psi assay, which was observed for both RNA and DNA. Likewise, the difference cannot be explained by proviral mutations, since it was not observed in the HIV DNA and persisted despite normalization of each type of HIV RNA to the same HIV DNA. Instead, the excess of 3′ defective over 5′ defective or intact HIV RNA suggests incomplete processivity of HIV transcription, consistent with previously observed blocks to distal HIV transcription and completion (21), and/or a much greater clearance rate of cells transcribing the Env region.
Intact HIV RNA was detected in 6 of 9 individuals at very low levels (median 1 copy/106 cells, or 0.66% of all HIV RNA). Unlike the situation observed for HIV DNA, intact HIV RNA levels were not significantly lower than those of Psi− RRE+ (5′ defective) HIV RNA. It is likely that levels of both are limited by blocks to distal HIV transcription, and the RRE may be a surrogate for processive and/or intact HIV RNA. However, other factors may contribute to the very low observed levels of intact HIV RNA. First, cell counts can be inaccurate, and the RNA extraction was likely incomplete, although the HIV RNA levels were corrected for RNA input. Second, the efficiency of reverse transcription and of detection of both targets is not 100%. Third, it is possible that there was some shearing or degradation of the HIV RNA.
It would be ideal to have a control for RNA shearing or degradation, similar to the duplex assay for the human RPP30 gene used in the IPDA (18) (which does not work for RNA). We attempted to develop a duplex assay for a human cellular RNA that could be used to assess for shearing, but this proved much less feasible than for DNA. First, most human RNAs are small. The median size of human mRNA is about 1.4 kb, with a log normal distribution and a very small proportion of transcripts that are 9 to 10 kb (31, 32). Moreover, the situation is complicated by RNA splicing, which removes introns, reduces the distance between exons, and can remove entire exons by alternative splicing. For these reasons, the ideal assay would consist of 2 targets at opposite ends of the same long exon. In a bioinformatic analysis of total transcriptome sequencing (RNA-seq) data from peripheral CD4+ T cells from 2 individuals, we searched for transcripts that comprise a single exon, represent >95% of the mean normalized counts from a given gene, and are at least 1,000 bp long. However, we found only 3 RNA species longer than 9.5 kb (and 9 longer than 7 kb), of which the longest coding region was only 6.7 kb, and all were present at frequencies of <50 copies per million transcripts. We designed and tested multiple duplex assays targeting each of 4 transcripts, but all showed low numbers of total positive and double-positive droplets. Future experiments could try other human transcripts, such as long noncoding RNAs, or possibly transcripts from human endogenous retroviruses (if transcribed as long RNAs).
Although shearing/degradation may contribute to the low levels of intact HIV RNA, we started with recently isolated cells and extracted using methods that had been shown to give good RNA quality (by Nanodrop and Bioanalyzer), while the supernatant HIV RNA standards showed a maximum shearing/degradation rate of 34 to 40%. Moreover, one might expect that shearing or degradation would result in similar levels of single-positive Psi+ RRE− and Psi− RRE+ droplets, but the observed levels of single-positive Psi+ RNA were much higher than those of single-positive RRE+ RNA, which seems inconsistent with shearing or degradation (unless specific for the 3′ end).
To correct the measured HIV RNA levels for the frequency of different proviruses, we also calculated the ratios of defective and intact HIV RNA to total and intact proviruses. These HIV RNA/DNA ratios also indicate the average level of each HIV RNA per provirus, and (for ratios of <1) the maximum frequency of proviruses that express each HIV RNA. Using these HIV RNA/DNA ratios, we found that a median of ≤0.18% of all proviruses and ≤2.2% of intact proviruses transcribe intact HIV RNA. These findings suggest that >98% of intact proviruses have a complete block to HIV transcription prior to the RRE region and/or that intact proviruses transcribing the RRE are cleared much more rapidly than those that do not.
By performing the IVRA on replicate terminal cell dilutions, we calculated that a median of 3 in 107 CD4+ T cells harbors intact HIV RNA. This frequency is slightly lower than the frequency of latently infected cells as measured by quantitative viral outgrowth assays (QVOA) (~1 in 106), although the former is a measure of spontaneous HIV transcription in vivo, while the latter is a measure of induced viral expression ex vivo. It should also be noted that the calculated frequencies of CD4+ T cells harboring intact HIV RNA have wide confidence intervals (given the low number of replicates) and that these frequencies are likely an underestimate because they are based on the starting cell counts and do not consider RNA losses during extraction.
Other limitations of this study should be acknowledged. The ability to detect intact HIV RNA likely depends on the total number of cells and RNAs analyzed, and there can be considerable imprecision in quantifying intact HIV RNA due to the low number of Psi+ RRE+ droplets. The measured levels of intact HIV RNA may be an underestimate, since there are losses in RNA extraction (partly corrected by normalizing to RNA input), the IVRA is likely not completely sensitive for detecting both targets in the same droplet (minimum of 44% efficiency and ~60% double positives on HIV RNA standards), and the number of intact HIV transcripts can be reduced by RNA shearing or degradation. At the same time, the IVRA may overestimate the actual number of intact HIV transcripts, since there may be other defects outside the Psi or RRE regions, not all of the Psi+ RRE+ transcripts may be complete and polyadenylated, and a given Psi+ RRE+ droplet could by chance have two different defective HIV RNA molecules. However, the latter possibility seems rare given the low numbers of Psi+ droplets/well, much lower number of RRE+ droplets, and relatively similar numbers of Psi− RRE+ and Psi+ RRE+ droplets. It is unclear to what extent the under- and overestimating factors counteract each other, and most of the same limitations also apply to the IPDA. However, it is also possible that some cells transcribing intact HIV RNA have posttranscriptional blocks that prevent translation and/or production of infectious viral particles (33).
Although we used the IVRA to quantify spontaneous transcription of intact and defective HIV RNA, these measurements do not reflect the capacity of proviruses to express intact HIV RNA after activation. Future experiments should apply the IVRA to cell aliquots that are unstimulated and activated ex vivo in order to determine the degree to which activation can increase total levels of intact HIV RNA and to measure the frequency of cells that can be induced to transcribe intact HIV RNA. Finally, it should be remembered that the IVRA was not designed to measure the frequencies of intact or defective proviruses that are transcribing each HIV RNA. While some information can be gained from the ratios of IVRA/IPDA and by PRIP-seq, there is still a need for high-throughput, single-cell studies that can sequence the entire provirus and sequence or quantify all of its HIV RNA.
Nevertheless, the IVRA serves as an approximate measure of intact HIV transcripts, which are necessary to produce infectious viral particles. Moreover, the IVRA is likely much less expensive, faster, higher in throughput, and more sensitive in detecting intact HIV RNA than single-molecule RNA sequencing (26, 34), which has failed to detect intact HIV RNA in ART-suppressed individuals (34). Since plasma contains considerable amounts of defective HIV RNA (26) and assays to quantify total plasma HIV RNA do not distinguish between intact and defective forms, the IVRA could be applied to clinical or research assays designed to measure an intact viral load, especially in individuals who are not on ART or are undergoing ART interruption. The IVRA may also serve as an alternative means to quantify intact HIV RNA in the supernatant of quantitative viral outgrowth assays (QVOA), allowing for a less expensive, faster, higher-throughput, and less time-consuming version of the QVOA. In addition, the IVRA could be applied to measure reactivation from latent proviruses in plasma or cell samples from clinical trials with latency-reversing agents or other approaches aimed at cure. Moreover, intact viral RNA may serve as a surrogate for HIV protein, a biomarker for immune activation/inflammation, a harbinger of viral rebound after ART interruption, or a biomarker for HIV transmissibility. Future studies should apply the IVRA to address these questions, which may provide insight into studies aimed at HIV pathogenesis and cure.
MATERIALS AND METHODS
Supernatant HIV RNA standards.
Two HIV RNA standards were prepared from viral stocks containing the supernatant of peripheral blood mononuclear cells (PBMC) infected in vitro with the lab virus NL4-3. The first standard was prepared as described in a prior study (21, 35), and when this standard was exhausted, a second standard was prepared from a different stock of NL4-3. Briefly, these supernatants from infected cells were diluted in phosphate-buffered saline (PBS), exposed to sequential freeze-thaw cycles (to lyse cells but not virions [36, 37]), incubated with DNase and RNase (to degrade any viral RNA or DNA not protected in virions), extracted using the Qiagen viral RNA minikit with on-column DNase, and stored in aliquots at −80°C. The total HIV RNA copies per microliter in each standard were then measured using the Abbott RealTime HIV-1 assay (average from three replicate aliquots), and freshly thawed aliquots were serially diluted to obtain expected inputs of 1 to 104 HIV RNA copies.
Assay sensitivity in the presence of background RNA.
A combination of the RNA standard and isolated donor CD4+ T cell RNA was used to determine the efficiency of each assay in the presence of background cellular RNA. One hundred copies of the standard along with 0 to 1,000 ng of background RNA were added to 20-μL ddPCR mixtures and run as described below.
Participant recruitment, sample collection, and processing.
Fresh venous blood was obtained from 9 ART-suppressed HIV-infected individuals enrolled in the University of California, San Francisco (UCSF) Scope/Options cohort. The study procedures were approved by the UCSF institutional review board (IRB), and all participants provided written informed consent. PBMC were isolated using Ficoll as previously described (21). CD4+ T cells were isolated from PBMC using the EasySep human CD4+ T cell isolation kit (Stemcell Technologies), counted, aliquoted into replicates (7 replicates of 300,000 CD4+ T cells for participant 2761; 4 to 12 replicates of 1,000,000 CD4+ T cells for other individuals), and stored at −80°C.
Isolation of DNA and RNA from CD4+ T cells.
Total cellular DNA and RNA were isolated in parallel from each replicate of CD4+ T cells using the Qiagen AllPrep DNA/RNA/microRNA (miRNA) universal kit protocol, modified per the manufacturer’s protocol to include a DNase I treatment of the RNA and recovery of short RNA transcripts. DNA and RNA concentrations and quality were measured using the NanoDrop One (Thermo Scientific) UV-visible (UV-Vis) spectrophotometer.
HIV DNA region quantification.
The number of copies of the U3-U5 long terminal repeat (LTR) (U3-U5), R-U5-pre-Gag (5′ LTR or long LTR), Pol, and Nef HIV DNA regions was measured in duplicate ddPCR mixtures (each containing up to 530 ng of DNA) as described previously (21). The intact proviral DNA assay (IPDA; duplex ddPCR for the Psi and RRE regions [18]) was performed as described previously (29) with 2 to 10 cell aliquots (maximum input of 530 ng/well) to measure levels of total Psi, total RRE, Psi+ RRE−, Psi− RRE+, and Psi+ RRE+ HIV DNA. HIV DNA copies were normalized to cell equivalents using the mass of DNA added to each well (from sample volume and nanograms per microliter as measured by Nanodrop).
Intact viral RNA assay (IVRA).
Mixes were prepared for replicate 20-μL one-step droplet digital reverse transcriptase PCR (dd-RT-PCR) mixtures. Each 20-μL reaction mixture included 5 μL of the Bio-Rad 4× Advanced One-Step RT-ddPCR Supermix, 2 μL of reverse transcriptase (RT), 1 μL of 300 mM dithiothreitol (DTT), 1 μL of 20× primers/probe mix for the Psi (final 1× = 900 nM [each] Psi primer and 250 nM FAM-labeled Psi probe), and 1 μL of 20× Env primer/probe mix (final 1× = 900 nM [each] RRE primer, 250 nM VIC-labeled RRE wild-type probe, and 250 nM unlabeled hypermutated RRE probe), and up to 10,000 copies of the supernatant HIV RNA standards (with or without RNA from donor CD4+ T cells) or 300 ng of total cellular RNA from people living with HIV (PWH). Primer and probe sequences are as described previously for the IPDA, and all probes were prepared with MGB. For each experiment, RNA from one cell aliquot was tested without reverse transcriptase to detect any contaminating HIV DNA. The reaction mixtures were encapsulated in oil droplets (Bio-Rad droplet generator) and subjected to RT-PCR in a Mastercycler nexus (Eppendorf, Hamburg, Germany) thermocycler using the following conditions: 60 min at 50°C, 10 min at 95°C, 40 cycles of 30 s at 95°C and 59°C for 60 s, and a final droplet cure step of 10 min at 98°C. Droplets were read and analyzed using the Bio-Rad QX100 or QX200 system and QuantaSoft software in the absolute quantification mode. A minimum of 11,000 droplets was required for wells to be analyzed. For clinical samples, the HIV RNA levels from each cell aliquot (merged data from all replicate dd-RT-PCR wells from that aliquot) were normalized to cell equivalents using the mass of RNA added to each well (from sample volume and nanograms per microliter as measured by Nanodrop) and the observation that 1 μg RNA corresponds to approximately 106 cells (38). HIV RNA levels from all cell aliquots were then averaged to calculate the copies/106 cells.
Statistical analysis.
For HIV RNA standards, slopes and R2 values were calculated by linear regression analysis. Pairwise comparisons between different types of HIV DNA, RNA, or RNA/DNA were performed using the Wilcoxon signed-rank test. Correlations between the levels of various HIV DNA and RNA regions were performed using Spearman correlations. These statistics were calculated using GraphPad Prism v9.0. The frequencies of CD4+ T cells containing each HIV RNA transcript were calculated using the number of replicates that did and did not contain each type of HIV RNA and the total number of cells per replicate using the method of extreme limiting dilution analysis (27).
Data availability.
The data used and/or analyzed in the current study are presented in the text and supplemental material.
ACKNOWLEDGMENTS
We thank the study participants for their generous donation of samples and the SCOPE project staff at Zuckerberg San Francisco General Hospital.
This work was supported by the National Institute of Allergy and Infectious Diseases (R01AI132128 [S.Y./J.W.], P01AI169606 [S.Y.]) and the National Institute of Diabetes and Digestive and Kidney Diseases (R01DK108349 [S.Y.], R01DK120387 [S.Y.]). S.T. is supported by a CFAR Mentored Scientist in HIV Award (grant no. P30 AI027763, award no. A120163, principal investigator [PI] Paul Volberding) and the California HIV/AIDS Research Program (award no. BB19-SF-009/A135087).
We declare that we have no competing interests.
S.Y. designed the study; R.H., S.D., and J.W. provided samples; H.A.M., G.N.K., S.T., A.W., T.-H.C., and S.M.-L. designed experiments; H.A.M., G.N.K., S.T., A.W., T.-H.C., and S.M.-L. conducted experiments; H.A.M., G.N.K., D.A., and S.Y. analyzed data; S.Y. wrote the original draft; all authors reviewed and edited the manuscript; S.Y. and J.W. provided supervision and funding.
Footnotes
Supplemental material is available online only.
Contributor Information
Steven A. Yukl, Email: Steven.Yukl@ucsf.edu.
Frank Kirchhoff, Ulm University Medical Center.
REFERENCES
- 1.Chun TW, Stuyver L, Mizell SB, Ehler LA, Mican JA, Baseler M, Lloyd AL, Nowak MA, Fauci AS. 1997. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc Natl Acad Sci USA 94:13193–13197. 10.1073/pnas.94.24.13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wong JK, Hezareh M, Gunthard HF, Havlir DV, Ignacio CC, Spina CA, Richman DD. 1997. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science 278:1291–1295. 10.1126/science.278.5341.1291. [DOI] [PubMed] [Google Scholar]
- 3.Finzi D, Hermankova M, Pierson T, Carruth LM, Buck C, Chaisson RE, Quinn TC, Chadwick K, Margolick J, Brookmeyer R, Gallant J, Markowitz M, Ho DD, Richman DD, Siliciano RF. 1997. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science 278:1295–1300. 10.1126/science.278.5341.1295. [DOI] [PubMed] [Google Scholar]
- 4.Phillips AN, Neaton J, Lundgren JD. 2008. The role of HIV in serious diseases other than AIDS. AIDS 22:2409–2418. 10.1097/QAD.0b013e3283174636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kuller LH, Tracy R, Belloso W, De Wit S, Drummond F, Lane HC, Ledergerber B, Lundgren J, Neuhaus J, Nixon D, Paton NI, Neaton JD, INSIGHT SMART Study Group . 2008. Inflammatory and coagulation biomarkers and mortality in patients with HIV infection. PLoS Med 5:e203. 10.1371/journal.pmed.0050203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hunt PW, Cao HL, Muzoora C, Ssewanyana I, Bennett J, Emenyonu N, Kembabazi A, Neilands TB, Bangsberg DR, Deeks SG, Martin JN. 2011. Impact of CD8+ T-cell activation on CD4+ T-cell recovery and mortality in HIV-infected Ugandans initiating antiretroviral therapy. AIDS 25:2123–2131. 10.1097/QAD.0b013e32834c4ac1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sandler NG, Wand H, Roque A, Law M, Nason MC, Nixon DE, Pedersen C, Ruxrungtham K, Lewin SR, Emery S, Neaton JD, Brenchley JM, Deeks SG, Sereti I, Douek DC, INSIGHT SMART Study Group . 2011. Plasma levels of soluble CD14 independently predict mortality in HIV infection. J Infect Dis 203:780–790. 10.1093/infdis/jiq118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boulware DR, Hullsiek KH, Puronen CE, Rupert A, Baker JV, French MA, Bohjanen PR, Novak RM, Neaton JD, Sereti I, INSIGHT Study Group . 2011. Higher levels of CRP, D-dimer, IL-6, and hyaluronic acid before initiation of antiretroviral therapy (ART) are associated with increased risk of AIDS or death. J Infect Dis 203:1637–1646. 10.1093/infdis/jir134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ledwaba L, Tavel JA, Khabo P, Maja P, Qin J, Sangweni P, Liu X, Follmann D, Metcalf JA, Orsega S, Baseler B, Neaton JD, Lane HC, Project Phidisa Biomarkers Team . 2012. Pre-ART levels of inflammation and coagulation markers are strong predictors of death in a South African cohort with advanced HIV disease. PLoS One 7:e24243. 10.1371/journal.pone.0024243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.El-Diwany R, Breitwieser FP, Soliman M, Skaist AM, Srikrishna G, Blankson JN, Ray SC, Wheelan SJ, Thomas DL, Balagopal A. 2017. Intracellular HIV-1 RNA and CD4+ T-cell activation in patients starting antiretrovirals. AIDS 31:1405–1414. 10.1097/QAD.0000000000001480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pasternak AO, Berkhout B. 2018. What do we measure when we measure cell-associated HIV RNA. Retrovirology 15:13. 10.1186/s12977-018-0397-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Olson A, Coote C, Snyder-Cappione JE, Lin N, Sagar M. 2021. HIV-1 transcription but not intact provirus levels are associated with systemic inflammation. J Infect Dis 223:1934–1942. 10.1093/infdis/jiaa657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ho YC, Shan L, Hosmane NN, Wang J, Laskey SB, Rosenbloom DI, Lai J, Blankson JN, Siliciano JD, Siliciano RF. 2013. Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell 155:540–551. 10.1016/j.cell.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bruner KM, Murray AJ, Pollack RA, Soliman MG, Laskey SB, Capoferri AA, Lai J, Strain MC, Lada SM, Hoh R, Ho YC, Richman DD, Deeks SG, Siliciano JD, Siliciano RF. 2016. Defective proviruses rapidly accumulate during acute HIV-1 infection. Nat Med 22:1043–1049. 10.1038/nm.4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hiener B, Horsburgh BA, Eden JS, Barton K, Schlub TE, Lee E, von Stockenstrom S, Odevall L, Milush JM, Liegler T, Sinclair E, Hoh R, Boritz EA, Douek D, Fromentin R, Chomont N, Deeks SG, Hecht FM, Palmer S. 2017. Identification of genetically intact HIV-1 proviruses in specific CD4(+) T cells from effectively treated participants. Cell Rep 21:813–822. 10.1016/j.celrep.2017.09.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee GQ, Orlova-Fink N, Einkauf K, Chowdhury FZ, Sun X, Harrington S, Kuo HH, Hua S, Chen HR, Ouyang Z, Reddy K, Dong K, Ndung’u T, Walker BD, Rosenberg ES, Yu XG, Lichterfeld M. 2017. Clonal expansion of genome-intact HIV-1 in functionally polarized Th1 CD4+ T cells. J Clin Invest 127:2689–2696. 10.1172/JCI93289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sharaf R, Lee GQ, Sun X, Etemad B, Aboukhater LM, Hu Z, Brumme ZL, Aga E, Bosch RJ, Wen Y, Namazi G, Gao C, Acosta EP, Gandhi RT, Jacobson JM, Skiest D, Margolis DM, Mitsuyasu R, Volberding P, Connick E, Kuritzkes DR, Lederman MM, Yu XG, Lichterfeld M, Li JZ. 2018. HIV-1 proviral landscapes distinguish posttreatment controllers from noncontrollers. J Clin Invest 128:4074–4085. 10.1172/JCI120549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bruner KM, Wang Z, Simonetti FR, Bender AM, Kwon KJ, Sengupta S, Fray EJ, Beg SA, Antar AAR, Jenike KM, Bertagnolli LN, Capoferri AA, Kufera JT, Timmons A, Nobles C, Gregg J, Wada N, Ho YC, Zhang H, Margolick JB, Blankson JN, Deeks SG, Bushman FD, Siliciano JD, Laird GM, Siliciano RF. 2019. A quantitative approach for measuring the reservoir of latent HIV-1 proviruses. Nature 566:120–125. 10.1038/s41586-019-0898-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levy CN, Hughes SM, Roychoudhury P, Reeves DB, Amstuz C, Zhu H, Huang ML, Wei Y, Bull ME, Cassidy NAJ, McClure J, Frenkel LM, Stone M, Bakkour S, Wonderlich ER, Busch MP, Deeks SG, Schiffer JT, Coombs RW, Lehman DA, Jerome KR, Hladik F. 2021. A highly multiplexed droplet digital PCR assay to measure the intact HIV-1 proviral reservoir. Cell Rep Med 2:100243. 10.1016/j.xcrm.2021.100243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gaebler C, Falcinelli SD, Stoffel E, Read J, Murtagh R, Oliveira TY, Ramos V, Lorenzi JCC, Kirchherr J, James KS, Allard B, Baker C, Kuruc JD, Caskey M, Archin NM, Siliciano RF, Margolis DM, Nussenzweig MC. 2021. Sequence evaluation and comparative analysis of novel assays for intact proviral HIV-1 DNA. J Virol 95:e01986-20. 10.1128/JVI.01986-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yukl SA, Kaiser P, Kim P, Telwatte S, Joshi SK, Vu M, Lampiris H, Wong JK. 2018. HIV latency in isolated patient CD4(+) T cells may be due to blocks in HIV transcriptional elongation, completion, and splicing. Sci Transl Med 10:eaap9927. 10.1126/scitranslmed.aap9927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Telwatte S, Lee S, Somsouk M, Hatano H, Baker C, Kaiser P, Kim P, Chen TH, Milush J, Hunt PW, Deeks SG, Wong JK, Yukl SA. 2018. Gut and blood differ in constitutive blocks to HIV transcription, suggesting tissue-specific differences in the mechanisms that govern HIV latency. PLoS Pathog 14:e1007357. 10.1371/journal.ppat.1007357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moron-Lopez S, Xie G, Kim P, Siegel D, Lee S, Wong JK, Price J, Elnachef N, Greenblatt RM, Tien P, Roan NR, Yukl SA. 2021. Tissue-specific differences in HIV DNA levels and mechanisms that govern HIV transcription in blood, gut, genital tract, and liver in ART-treated women. J Int AIDS Soc 24:e25738. 10.1002/jia2.25738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Einkauf KB, Osborn MR, Gao C, Sun W, Sun X, Lian X, Parsons EM, Gladkov GT, Seiger KW, Blackmer JE, Jiang C, Yukl SA, Rosenberg ES, Yu XG, Lichterfeld M. 2022. Parallel analysis of transcription, integration, and sequence of single HIV-1 proviruses. Cell 185:266–282.e15. 10.1016/j.cell.2021.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kwon KJ, Timmons AE, Sengupta S, Simonetti FR, Zhang H, Hoh R, Deeks SG, Siliciano JD, Siliciano RF. 2020. Different human resting memory CD4(+) T cell subsets show similar low inducibility of latent HIV-1 proviruses. Sci Transl Med 12:eaax6795. 10.1126/scitranslmed.aax6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fisher K, Wang XQ, Lee A, Morcilla V, de Vries A, Lee E, Eden JS, Deeks SG, Kelleher AD, Palmer S. 2022. Plasma-derived HIV-1 virions contain considerable levels of defective genomes. J Virol 96:e0201121. 10.1128/jvi.02011-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hu Y, Smyth GK. 2009. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J Immunol Methods 347:70–78. 10.1016/j.jim.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 28.Papasavvas E, Azzoni L, Ross BN, Fair M, Yuan Z, Gyampoh K, Mackiewicz A, Sciorillo AC, Pagliuzza A, Lada SM, Wu G, Goh SL, Bahnck-Teets C, Holder DJ, Zuck PD, Damra M, Lynn KM, Tebas P, Mounzer K, Kostman JR, Abdel-Mohsen M, Richman D, Chomont N, Howell BJ, Montaner LJ. 2021. Intact human immunodeficiency virus (HIV) reservoir estimated by the intact proviral DNA assay correlates with levels of total and integrated dna in the blood during suppressive antiretroviral therapy. Clin Infect Dis 72:495–498. 10.1093/cid/ciaa809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moron-Lopez S, Bernal S, Wong JK, Martinez-Picado J, Yukl SA. 2022. ABX464 decreases the total human immunodeficiency virus (HIV) reservoir and HIV transcription initiation in CD4+ T cells from antiretroviral therapy-suppressed individuals living with HIV. Clin Infect Dis 74:2044–2049. 10.1093/cid/ciab733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simonetti FR, White JA, Tumiotto C, Ritter KD, Cai M, Gandhi RT, Deeks SG, Howell BJ, Montaner LJ, Blankson JN, Martin A, Laird GM, Siliciano RF, Mellors JW, Siliciano JD. 2020. Intact proviral DNA assay analysis of large cohorts of people with HIV provides a benchmark for the frequency and composition of persistent proviral DNA. Proc Natl Acad Sci USA 117:18692–18700. 10.1073/pnas.2006816117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sommer SS, Cohen JE. 1980. The size distributions of proteins, mRNA, and nuclear RNA. J Mol Evol 15:37–57. 10.1007/BF01732582. [DOI] [PubMed] [Google Scholar]
- 32.Lopes I, Altab G, Raina P, de Magalhaes JP. 2021. Gene size matters: an analysis of gene length in the human genome. Front Genet 12:559998. 10.3389/fgene.2021.559998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Verneuil A, Migraine J, Mammano F, Molina JM, Gallien S, Mouquet H, Hance AJ, Clavel F, Dutrieux J. 2018. Genetically intact but functionally impaired HIV-1 Env glycoproteins in the T-cell reservoir. J Virol 92:e01684-17. 10.1128/JVI.01684-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Imamichi H, Dewar RL, Adelsberger JW, Rehm CA, O’Doherty U, Paxinos EE, Fauci AS, Lane HC. 2016. Defective HIV-1 proviruses produce novel protein-coding RNA species in HIV-infected patients on combination antiretroviral therapy. Proc Natl Acad Sci USA 113:8783–8788. 10.1073/pnas.1609057113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaiser P, Joshi SK, Kim P, Li P, Liu H, Rice AP, Wong JK, Yukl SA. 2017. Assays for precise quantification of total (including short) and elongated HIV-1 transcripts. J Virol Methods 242:1–8. 10.1016/j.jviromet.2016.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fischer M, Wong JK, Russenberger D, Joos B, Opravil M, Hirschel B, Trkola A, Kuster H, Weber R, Gunthard HF, Swiss HIV Cohort Study . 2002. Residual cell-associated unspliced HIV-1 RNA in peripheral blood of patients on potent antiretroviral therapy represents intracellular transcripts. Antivir Ther 7:91–103. [PubMed] [Google Scholar]
- 37.Kaiser P, Niederost B, Joos B, von Wyl V, Opravil M, Weber R, Gunthard HF, Fischer M. 2006. Equal amounts of intracellular and virion-enclosed hepatitis C virus RNA are associated with peripheral-blood mononuclear cells in vivo. J Infect Dis 194:1713–1723. 10.1086/508431. [DOI] [PubMed] [Google Scholar]
- 38.Fischer M, Huber W, Kallivroussis A, Ott P, Opravil M, Luthy R, Weber R, Cone RW. 1999. Highly sensitive methods for quantitation of human immunodeficiency virus type 1 RNA from plasma, cells, and tissues. J Clin Microbiol 37:1260–1264. 10.1128/JCM.37.5.1260-1264.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to S6. Download jvi.01605-22-s0001.pdf, PDF file, 0.5 MB (540KB, pdf)
Data Availability Statement
The data used and/or analyzed in the current study are presented in the text and supplemental material.