Abstract
The scope of proteins accessible to total chemical synthesis via native chemical ligation (NCL) is often limited by slow ligation kinetics. Here we describe Click-Assisted NCL (CAN), in which peptides are incorporated with traceless “helping hand” lysine linkers that enable addition of dibenzocyclooctyne (DBCO) and azide handles. The resulting strain-promoted alkyne-azide cycloaddition (SPAAC) increases their effective concentration to greatly accelerate ligations. We demonstrate that copper(I) protects DBCO from acid-mediated rearrangement during acidic peptide cleavage, enabling direct production of DBCO synthetic peptides. Excitingly, triazole-linked model peptides ligated rapidly and accumulated little side product due to the fast reaction time. Using the E. coli ribosomal subunit L32 as a model protein, we further demonstrate that SPAAC, ligation, desulfurization, and linker cleavage steps can be performed in one pot. CAN is a useful method for overcoming challenging ligations involving sterically hindered junctions. Additionally, CAN is anticipated to be an important stepping stone towards a multi-segment, one-pot, templated ligation system.
Keywords: Chemical Protein Synthesis, Native Chemical Ligation, NCL, Click-Assisted NCL, CAN, Traceless Templated Peptide Ligation, Strain-Promoted Alkyne-Azide Cycloadditions, SPAAC, Copper-free Click Chemistry, DBCO, Tetrakis(acetonitrile)copper(I) Tetrafluoroborate, Helping Hand Lysine Linker, Ddap, Ribosome
Graphical Abstract
INTRODUCTION
The combination of solid-phase peptide synthesis (SPPS)1 with chemoselective ligation reactions enables the chemical synthesis of uniquely modified proteins. Of the chemoselective ligation strategies available, the Kent group’s native chemical ligation (NCL) has proven to be highly robust and is widely used in the chemical synthesis of proteins.2, 3 With NCL, peptide segments bearing an N-terminal Cys and C-terminal thioester are chemoselectively ligated to generate synthetic proteins. NCL has been extended beyond Cys junctions via free-radical desulfurization, a robust technique that allows the more common Ala, as well as other amino acids amenable to thiol surrogates, to be used as ligation junctions.4, 5 NCL has produced many useful synthetic proteins,6, 7 including targets for mirror-image drug discovery,8-12 functional native and mirror-image enzymes,13-19 mirror-image proteins for racemic crystallography,20-23 and proteins containing complex post-translational modifications.24-27
However, NCL has several challenges that limit its scope in chemical protein synthesis (CPS).7, 28 First, NCL requires high peptide concentrations, typically ≥ 1 mM, at neutral pH for efficient ligation. Unfortunately, peptide segments are often too insoluble to reach these ideal concentrations, even in denaturing conditions.29 While many groups have developed strategies to increase peptide solubility (e.g., incorporating temporary solubilizing tags29-32 and using buffers containing hexafluoro-2-propanol33, 34 or ionic liquids35), these do not overcome the inherent issue of requiring high concentrations. Additionally, the availability of suitable Cys or Ala ligation junctions is limited for many synthesis projects, forcing the selection of suboptimal segments for NCL.11, 36 Although alternative thiol-containing amino acids, such as penicillamine (thiol derivative of Val), can be used as junctions to increase access to alternative ligation strategies, these often suffer from slow ligation rates.37-39 In cases where limited suitable junctions exist, the use of sterically hindered thioesters may be required (e.g., Thr, Ile, and Val).39-41 Even under ideal NCL conditions, such thioesters suffer from long reaction times that often result in loss of peptide because of competing side reactions, creating complex HPLC purifications that further lower yields. The NCL rate is also dependent on the type of thioester used.42 Most commonly, 4-mercaptophenylacetic acid (MPAA) is used for NCL due to its favorable transthioesterification kinetics.43 However, MPAA has several disadvantages including strong near-UV absorbance, incompatibility with one-pot desulfurization, and potential co-elution with products in RP-HPLC.44-46 Several alternatives can be used in place of MPAA (e.g., trifluoroethanethiol,44 methyl thioglycolate (MTG),45 imidazole,46 and 3-mercaptopropionic acid47), though at the cost of slower NCL kinetics.
Templated ligation strategies can overcome these major barriers by increasing the proximity of the reactive partners, the C-terminal thioester and the N-terminal thiol, thereby increasing their effective concentration (Fig. 1A). Templated ligations occur in an intramolecular fashion, converting reaction kinetics from 2nd to 1st order. An early example of such a proximity-driven reaction was demonstrated with Kemp’s thiol capture.48, 49 Effectively, templated ligations are macrocyclization reactions, a strategy commonly used in the synthesis of cyclic natural products and other compounds.50-53 Importantly, application of this strategy can improve the efficiency of slow thioesters and enable ligations at dramatically lower peptide concentrations, especially helpful for poorly soluble peptides.
Figure 1:
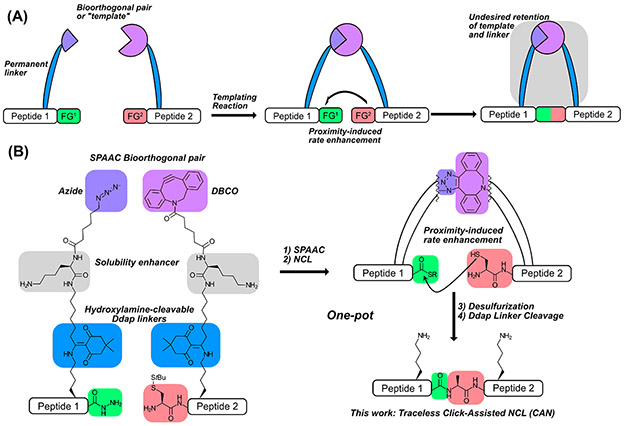
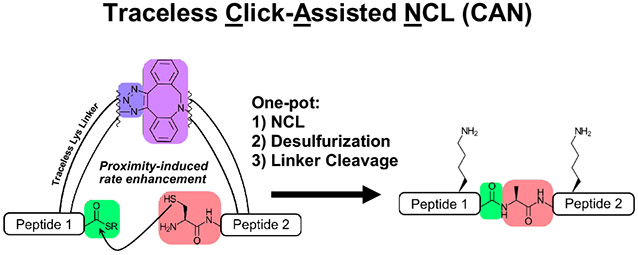
Overview of templated peptide ligation strategies. (A) Peptides functionalized with bioorthogonal groups are linked together, increasing the effective concentration of two functional groups (FG1 and FG2). While this strategy increases reaction efficiency, the bioorthogonal groups remain bound to the ligated peptide, leaving “scars” that are unacceptable for most CPS applications. (B) With Click-Assisted NCL (CAN), peptides functionalized with traceless Lys linkers (Ddap) and either DBCO or azide are first linked via SPAAC. These templated peptides then undergo a proximity-enhanced NCL, desulfurization (if desired), and gentle hydroxylamine treatment to cleave the Ddap linker and tracelessly afford native peptide. Importantly, all steps can be performed in one pot.
Several templated ligation strategies currently exist, but specific caveats limit the general utility of each strategy for CPS. Protein-templated ligations were the first demonstration of templated NCL, as peptides containing specific secondary structures were brought into close proximity via their non-covalent interactions.54-61 While protein-templated ligation works well in a variety of contexts (e.g., self-replicating peptides, conformationally assisted protein ligations, and protein labeling), the method is infeasible for most CPS applications, as peptides do not usually self-associate, and denaturants are typically required for peptide segment solubilization. Nucleic acids and peptide nucleic acids (PNAs) have also been used to template ligations, as peptides conjugated with DNA or PNA can be brought into proximity via specific base-pairing interactions.62-67 Nucleic acid-templated ligations complete quickly with only nM-pM template, but DNA/PNA linkers cannot be removed from ligated product. In addition, this method has only been used in non-denaturing buffers, predominantly with short peptide sequences (<20 residues). Recently, several traceless templated ligation strategies have been developed. The first reported method uses peptides functionalized with UV-cleavable linkers attached to nucleic acid handles to enable templated ligation at low peptide concentrations. The Diederichsen group initially developed this strategy by incorporating PNA into the linkers,68 and the Okamoto group later created a similar approach by using DNA in their linkers.69 This traceless nucleic acid-templated ligation is not yet robust enough for routine CPS applications, however, as the required UV irradiation and copper-catalyzed click reactions may cause oxidative damage.70-72 The Bode group has also designed a traceless templated ligation using streptavidin-binding linkers, which cleave upon amide bond formation.73 However, complex linker synthesis is currently needed, and the methodology has not yet been demonstrated on peptides.
A New Strategy for Traceless Templated Ligations
We propose that the following fundamental elements are required for a traceless templated ligation strategy to be broadly applicable in CPS:
Simple and robust bioorthogonal linker synthesis
Bioorthogonal chemistry can be performed in typical peptide-solubilizing denaturants (e.g., 6 M guanidine)
Bioorthogonal linkers are stable to commonly encountered CPS conditions (e.g., resin cleavage, ligation, and desulfurization)
Removal of bioorthogonal linkers is chemoselective, traceless, and efficient
Flexible positioning of bioorthogonal linkers within peptide sequences (e.g., placement not restricted to termini or infrequent amino acids)
Here, we describe Click-Assisted NCL (CAN), which meets all these criteria by combining a traceless “helping hand” Lys linker (Ddap)74 with the bioorthogonal strained-click pair dibenzocyclooctyne (DBCO) and azide (Fig. 1B). In the CAN strategy, peptide segments are first coupled with a Ddap linker on a Lys side chain on-resin during SPPS before functionalization with either DBCO or azide. After resin cleavage and HPLC purification, the DBCO/azide-bearing peptides are combined for strain-promoted alkyne-azide cycloaddition (SPAAC). The resultant triazole linkage effectively templates the C-terminal thioester and N-terminal thiol for ligation, allowing NCL to proceed with high efficiency. After ligation, mild hydroxylamine treatment chemoselectively removes the templating linkers,74 affording the native ligated peptide. Using model peptides, we show that the CAN strategy is compatible with all canonical amino acids. CAN greatly accelerates ligation in the context of sterically hindered thioesters. Furthermore, these demonstrations use methyl thioglycolate (MTG) as a slower, relative to MPAA, but desulfurization-compatible thiol additive for NCL.45 Finally, we illustrate the utility of CAN for CPS by performing a strain-promoted click reaction, NCL, desulfurization, and linker cleavage to produce the E. coli 50S ribosomal subunit L32 in one pot.
RESULTS AND DISCUSSION
Synthesis of DBCO Peptides via Fmoc-SPPS
In order for CAN to be a robust CPS technique, peptides need to be templated together through a bioorthogonal reaction that will not cause side reactions during conjugation. For this purpose, we chose SPAAC75, 76 over copper-catalyzed azide-alkyne cycloaddition (CuAAC)77, 78 because SPAAC avoids potential copper-catalyzed oxidation during the click reaction.71, 72 Additionally, we wanted to perform one-pot templating with NCL and anticipated that the presence of CuAAC compounds within NCL conditions may cause side-product formation (e.g., during in situ thioester formation).79
The first obstacle to developing CAN was generating a robust, SPPS-compatible method for installing azide and DBCO cycloaddition partners. While azide-containing peptides can routinely be synthesized via SPPS,80 DBCO peptides have only been synthesized using post-cleavage couplings, such as thiol-maleimide81-83 or amine-NHS ester.84 These post-cleavage workarounds are necessary because DBCO undergoes an inactivating rearrangement under the strongly acidic conditions, 95% trifluoroacetic acid (TFA), used for peptide deprotection and cleavage from the resin.82, 83, 85, 86 This rearrangement has been demonstrated in DBCO and related biarylazacyclooctynones and is thought to result from an acid-catalyzed 5-endo-dig cycloisomerization.86, 87 Longer peptides are unlikely to have a single primary amine or thiol for post-cleavage couplings, so DBCO needs to be incorporated during Fmoc-SPPS. While DBCO can withstand lower TFA concentrations (<30%),85, 88, 89 most commonly used protecting groups and resins require higher TFA for efficient removal.
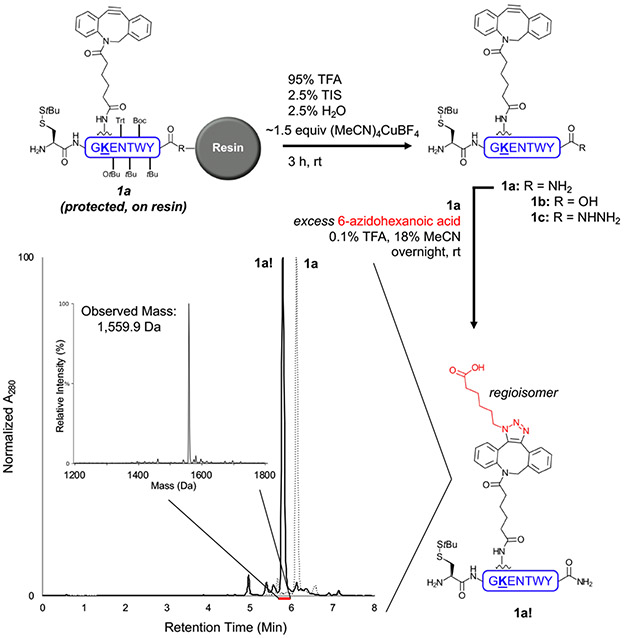
We set out to investigate the stability of DBCO in standard TFA cleavage conditions using the model peptides H-C(StBu)GK(DBCO)ENTWY-R (R = NH2 1a, OH 1b, or NHNH2 1c; see Table S1 for all peptide sequences) and Ac-RRRYSTEVEK(DBCO)NV-NHNH2 2a (DBCO coupled to Lys side chains and Cys protected with StBu to prevent potential thiol-yne reactions).90 Our initial attempts to synthesize DBCO-containing model peptides via Fmoc-SPPS and a standard TFA peptide cleavage cocktail failed due to 5-endo-dig cycloisomerization and associated side products (Fig. S1-S4).
The Hosoya group previously reported that a copper(I) salt, tetrakis(acetonitrile)copper(I) tetrafluoroborate or (MeCN)4CuBF4, could reversibly protect cyclooctynes from reacting with azides.91, 92 This strategy builds on the finding that various metals can form complexes with cycloalkyne rings,93 which led us to examine copper as an additive to prevent the acid-mediated 5-endo-dig cycloisomerization of DBCO. Guided by this methodology, initial attempts at protecting peptides 1a and 2a were performed by treating peptide resins with ~3 equiv (MeCN)4CuBF4 in dimethylformamide (DMF) for 1 h. The resin was then washed with DMF and dichloromethane (DCM) before cleavage in the standard cocktail at rt for 3 h. Standard ether precipitation and air drying produced the crude DBCO peptides. As envisioned, a transient DBCO-Cu complex was protected from acid-promoted ring rearrangement, and the resultant DBCO peptide product 1a was able to react fully with excess 6-azidohexanoic acid (Fig. S5). However, we discovered that (MeCN)4CuBF4 caused premature cleavage of peptides assembled on 2-chlorotrityl chloride resin. Additionally, the Cu(I) salt-DMF mixture reacted with the C-terminal hydrazide-containing peptide 2a to form undesired side products (Fig. S6).
To address these issues, we next tested the direct addition of ~1.5 equiv (MeCN)4CuBF4 to the peptide resin as a dry powder, immediately followed by addition of the TFA cleavage cocktail for 3 h at rt (Fig. 2). Again, standard ether precipitation and air drying afforded the crude DBCO peptide 1a. Incubation of 1a with 6-azidohexanoic acid demonstrated a successful SPAAC (Fig. 2 and Fig. S7; see Table S2 for symbols used to denote common modifications such as triazole). This simplified protection method was also successful in producing peptide 1b with reactive DBCO (Fig. S8). However, side reactions occurred when attempting to react crude, hydrazide-containing 2a or 1c with 6-azidohexanoic acid. Copper test strips indicated that Cu was only partially removed during ether precipitation and was likely causing the undesired side reactions during SPAAC. Gratifyingly, HPLC purification removed the remaining Cu, as pure 1c did not contain Cu (copper test strips), and the peptide successfully coupled with 6-azidohexanoic acid to afford the triazole with minimal side products (Fig. S9-10). Inductively coupled plasma mass spectrometry (ICP-MS) confirmed removal of Cu, as several HPLC-purified DBCO peptides contained the same background Cu levels as peptides that were never exposed to the Cu(I) salt (Table S3).
Figure 2:
Protection of DBCO peptide 1a from acid-mediated degradation using (MeCN)4CuBF4 in standard peptide cleavage conditions. This protection strategy generated a relatively clean crude peptide (dashed line in LC/MS chromatogram) corresponding to the expected mass of 1a (observed mass: 1,401.9 Da, calculated average mass: 1,402.6 Da, Fig. S7). This crude peptide successfully underwent SPAAC when incubated with excess 6-azidohexanoic acid, as the expected triazole formed (solid line in LC/MS chromatogram, 1a!; observed mass: 1,559.9 Da, calculated average mass: 1,559.8 Da). LC/MS method C was used.
Robustness of the DBCO Protection Method
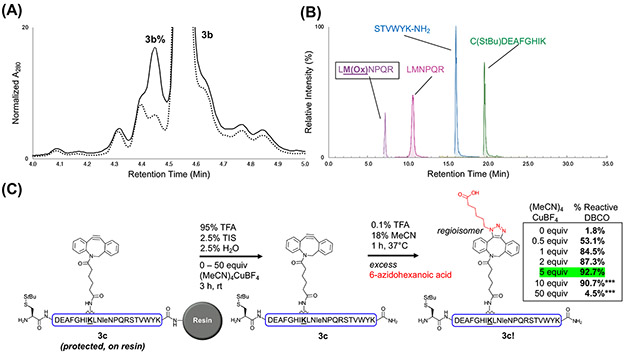
Concerned with the potential of Cu(I) salts to oxidize certain amino acids, particularly Met and His,71 we examined the compatibility of the DBCO protection strategy using the more diverse model peptide H-C(StBu)DEAFGHIK(DBCO)LMNPQRSTVWYK-NH2 3a (and without DBCO, 3b), which contains every canonical amino acid. When 3b was cleaved in the presence or absence of ~3 equiv (MeCN)4CuBF4, the analytical HPLC and LC/MS chromatograms were similar (Fig. 3A and Fig. S11-12). However, the presumed oxidation side-product (+16 Da) increased from 6% to 12.5% in Cu(I)-treated 3b (Fig. 3A). Using trypsin digestion and high-resolution LC/MS/MS on 3b ± Cu(I) during cleavage, Met was identified as the oxidation-prone residue (Fig. 3B). Furthermore, when the Cu(I) salt was used in the cleavage of model peptides H-C(StBu)GKENTWYX-NH2 4a (X=H) and 4b (X=M), oxidation was only observed in the Met-containing peptide 4b (Fig. S13-14). While increased Met oxidation is a potential concern for the DBCO protection method, all other canonical amino acids are fully compatible. In addition, Met is one of the least abundant amino acids and can typically be substituted with an isosteric norleucine (Nle) residue without affecting protein activity and folding.47, 94, 95 In cases where Met is required, any oxidized Met peptide can also be removed via standard HPLC purification.
Figure 3:
Assessment of the (MeCN)4CuBF4 DBCO protection strategy’s robustness. Peptide 3b (all canonical residues represented) was used to determine the strategy’s oxidation potential, and peptide 3c (Met-to-Nle variant of 3a) was used to establish effective amounts of Cu(I) salt to use for DBCO protection. (A) Overlaid LC/MS chromatogram showing 3b cleaved with (solid line) or without (dashed line) ~3 equiv (MeCN)4CuBF4. Integration of the labeled peak areas suggested oxidation (3b%) increased from 6% to 12.5% with Cu(I) salt addition to the standard cleavage cocktail. LC/MS method C was used, and the individual chromatograms are shown in Fig. S11. (B) Trypsin digestion and LC/MS/MS analysis of 3b revealed Met as the oxidation-susceptible residue. The 3b sequence is color-coded by trypsin-digested fragments and matches the appropriate ion signal identified through MS/MS analysis. LM(Ox)NPQR (far left peak, purple) indicates peptide containing oxidized Met. (C) Cleavage of 3c with different equiv of (MeCN)4CuBF4 indicated that 5 equiv provides the most effective DBCO protection (green). After performing SPAAC on crude 3c, A280 peak areas of reacted (3c!) and unreacted peptide were integrated to calculate % reactive DBCO. ***Indicates higher equiv of (MeCN)4CuBF4 that were found to complex with the DBCO peptide, preventing reaction with azide after cleavage. Although the % reactive DBCO is lower, this decrease is not due to 5-endo-dig-cycloisomerization. Representative LC/MS chromatograms of crude 3c (before and after SPAAC) are shown in Fig. S15-17.
We next investigated the ideal concentration of the (MeCN)4CuBF4 additive. Considering the objective of these experiments was to determine the amount of active DBCO following cleavage across a range of (MeCN)4CuBF4 equivalents, the analysis was simplified by synthesizing 3a with a Met-to-Nle substitution to give oxidation-resistant 3c (Fig. 3C). 3c was incubated with varying amounts of (MeCN)4CuBF4 through standard cleavage conditions for 3 h before ether precipitation and air drying. Note that the (MeCN)4CuBF4 equivalents are relative to the theoretical yield of 3c (as described in the supplementary methods). Crude 3c was then dissolved in HPLC buffer (0.1% TFA, 18% MeCN) and coupled with excess 6-azidohexanoic acid for 1 h at 37°C to determine the amount of reactive DBCO present (Fig. 3C and Fig. S15-17). DBCO protection increased until 5 equiv (MeCN)4CuBF4. Surprisingly, 50 equiv increased the amount of DBCO that failed to click with azide. This loss of reactivity may be attributed to excessive Cu(I) salts that are incompletely removed following ether precipitation, which could form a complex with the DBCO peptide, thus precluding efficient cycloaddition reactivity (Fig. S17). Based on these optimizations, the use of 5 equiv of (MeCN)4CuBF4 was determined to be ideal for preventing the rearrangement of DBCO-containing peptides through standard cleavage conditions.
Click-Assisted NCL with Model Peptides
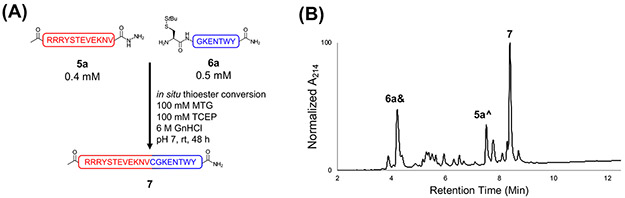
With a DBCO protection strategy in hand, we next investigated Click-Assisted NCL (CAN). We substituted the commonly used 4-mercaptophenylacetic acid (MPAA) thiol for methyl thioglycolate (MTG) during NCL, as MTG elutes in the void during RP-HPLC, avoiding co-elution with peptide. Additionally, MTG is compatible with one-pot desulfurizations.45 Although MTG has ~2-fold slower ligation kinetics compared to MPAA,45 we hypothesized that the effective concentration enhancement of the CAN strategy would render the choice of thiol additive less impactful. The non-templated control peptides STEVE (5a) and KENT (6a) were synthesized without SPAAC linkers to determine their in-solution NCL rate under conditions representative of poorly soluble peptides (0.4 mM 5a and 0.5 mM 6a; Fig. 4A). 5a was also designed with a C-terminal Val thioester, among the slowest in NCL due to steric hindrance.40, 41 After 48 h, the ligation reaction was incomplete with several side products and only 29% of the total peak area corresponding to ligated product 7 (Fig. 4B and Fig. S18-21).
Figure 4:
Non-templated native chemical ligation between purified 5a and 6a. (A) Simplified schematic of the ligation. Note that MTG was allowed to form the thioester for 10 min prior to TCEP addition, as shown in the more detailed scheme (Scheme S1). (B) HPLC chromatogram of the ligation endpoint (48 h). 6a& refers to StBu-deprotected 6a, and 5a^ refers to 5a containing a C-terminal MTG thioester. After 48 h (from MTG addition), the reaction was incomplete, and numerous side products were observed. Analytical method B was used. HPLC and LC/MS chromatograms of more time points for this ligation are shown in Fig. S20-21.
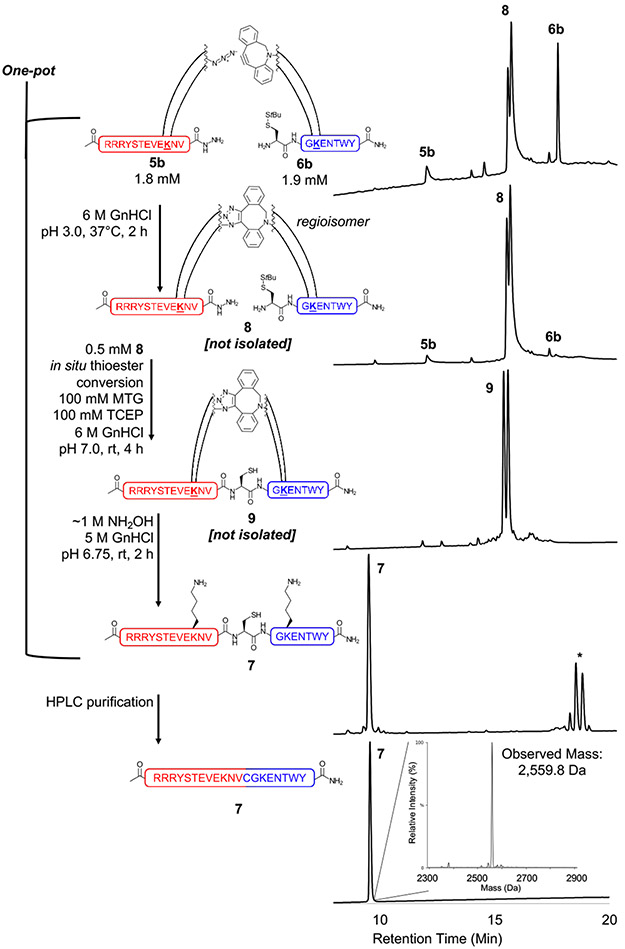
As a proof-of-concept for CAN, we then synthesized model peptides 5b and 6b, each functionalized with the traceless Lys linker Ddap,74 a single Lys (to enhance solubility), and azide or DBCO as shown in Fig. 1B. Approximately equal amounts of purified 5b and 6b were dissolved together in 6 M GnHCl at pH 3 to undergo SPAAC (Fig. 5 and Fig. S22-25). Importantly, to the best of our knowledge, this is the first demonstration of SPAAC in denaturing conditions using guanidine. HPLC analysis revealed the presence of two triazole cycloadduct peptides (both referred to as 8). The two distinguishable isobaric peaks are attributed to the two anticipated triazole regioisomers (Fig. 5 and Fig. S24). Notably, the SPAAC reaction still finished within 2 h when the 5b and 6b concentrations were reduced by ~4-fold (matching the concentrations used in the non-templated NCL reaction: 0.4 mM 5b and 0.5 mM 6b, which mimic poorly soluble peptide concentrations, Fig. S24-25). Using comparable conditions to the non-templated NCL, the C-terminal hydrazide was converted to an MTG thioester in situ prior to performing CAN (0.5 mM 8, Fig. 5). Excitingly, the ligation to form 9 was complete at 4 h (Fig. 5 and Fig. S26-27). The relative peak area of 9 from this CAN reaction was much higher (85%) than the non-templated control NCL with significantly diminished side products (Fig. 5). We then cleaved the Ddap linkers of 9 in one pot via the addition of hydroxylamine to ~1 M at pH 6.75 for 2 h (Fig. 5 and Fig. S28). The native, ligated product without Ddap linkers was easily purified by HPLC to afford 7 (Fig. 5 and Fig. S29). CAN with 8 was then repeated for a more precise kinetic analysis (Fig. 6 and Fig. S30-32).
Figure 5:
One-pot SPAAC, CAN, and linker cleavage. 5b and 6b were first templated together via SPAAC to produce triazole-linked 8. CAN was then performed on 8 to efficiently generate 9 after only 4 h. Linker cleavage of 9 resulted in native 7, which was easily HPLC purified. The HPLC chromatograms show, in descending order, 0 and 2 h SPAAC, 4 h CAN (after MTG addition), 2 h linker cleavage, and purified 7 (each y-axis is normalized A214). The double peaks observed after SPAAC and CAN are expected, as the triazole forms two regioisomers. * indicates the four cleaved Ddap linker peaks (triazole regioisomer and a second nucleophilic addition via hydroxylamine on the dimedone ring). Note that unreacted STEVE and KENT peptides observed after linker cleavage do not bind to the column with the HPLC gradient used here (Fig. S28 shows these unreacted peptides). The mass spectrum of purified 7 shows the correct full-length STEVEKENT peptide was generated (observed mass: 2,559.8 Da, calculated average mass: 2,559.9 Da). A detailed one-pot reaction schematic is shown in Scheme S2. Analytical method C and LC/MS method C were used. HPLC and LC/MS chromatograms of time points are shown in Fig. S24-29.
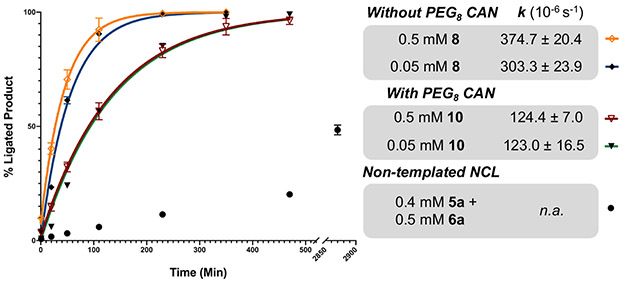
Figure 6:
Comparison of non-templated NCL and CAN reaction rates. NCL or CAN was performed using the STEVE and KENT model peptides (5a, 6a, 8, and 10). The relative integrated peak area of ligated products are shown at various time points relative to TCEP addition (1, 20, 50, 110, 230, 350, 470, 1430, and 2870 min). CAN ligations were fit to first-order rate constants. Importantly, the 8 CAN is complete after 4 h at both peptide concentrations. Each data point represents the average of two independent experiments, with the error bars corresponding to standard deviation.
To investigate the impact of linker length on the CAN reaction rate, peptides 5c and 6c were each functionalized with Ddap, a single Lys, and a PEG8 linker before addition of their respective SPAAC partners. The PEG8 spacers double the theoretical fully extended distance between the triazole-linked peptides. Using the same conditions as with 5b and 6b, the PEG8 peptides first underwent SPAAC to give triazole-linked 10 (2.5 mM 5c and 2.6 mM 6c at 37°C for 2 h, Fig. S33-36) before performing CAN at 0.5 mM to yield ligated product 11 (Fig. S37-38). CAN with PEG8 spacers was ~3-fold slower than non-PEGylated CAN (Fig. 6).
One of the more attractive uses of CAN, and templated ligations in general, is the ability to perform ligations at lower peptide concentrations (i.e., the triazole-linkage converts NCL to an intramolecular reaction). To validate this property, we repeated CAN with 8 and 10 (±PEG8) at a 10-fold lower concentration (0.05 mM). The MTG and TCEP concentrations were also both reduced to 10 mM, as 100 mM MTG and 100 mM TCEP caused undesired N-terminal Cys peptide capping (Fig. S39-42), and 10 mM MTG with 100 mM TCEP caused desulfurization. Using these optimized conditions, CAN kinetics were similar for both the 0.05 and 0.5 mM reactions (Fig. 6 and Fig. S43-46).
To directly compare the non-templated NCL and CAN reaction kinetics, we estimated the initial reaction rates (Table S4). At 0.5 mM, CAN without the PEG8 spacers was ~30-fold faster than the control NCL, while the PEG8 CAN was ~11-fold faster. The 0.05 mM CAN initial rates are slightly slower than the 0.5 mM reactions due to StBu removal occurring more slowly from the lower TCEP concentrations used. Importantly, the CAN rates should follow a 1st-order rate law, whereas the control non-templated NCL follows a 2nd-order rate law. Therefore, a 10-fold reduction in concentration of both peptides would slow the solution NCL reaction 100-fold, whereas a reduction in triazole-linked peptide concentration does not significantly alter ligation rate.
Click-Assisted NCL of the E. coli 50S ribosomal subunit L32
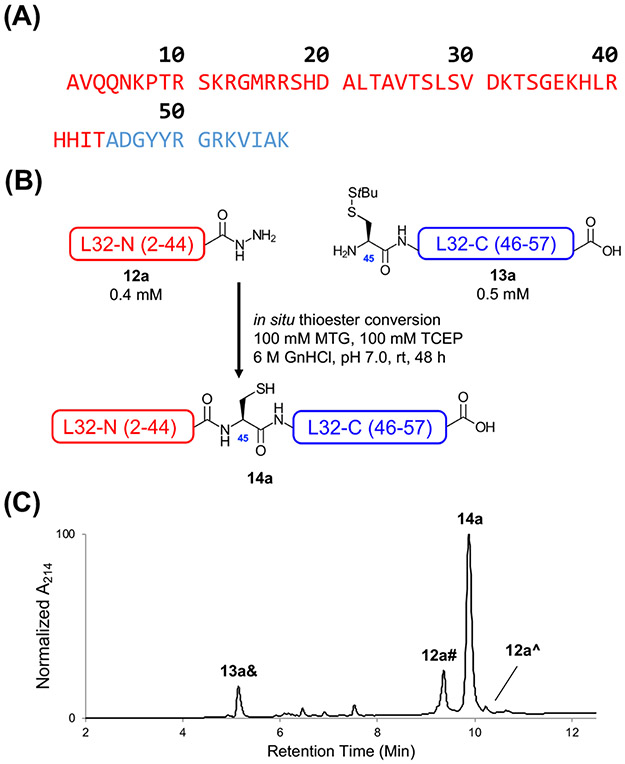
To demonstrate the general utility of CAN in CPS, we pursued a representative protein target – the E. coli 50S ribosomal subunit L32 (Fig. 7A). L32 is an ideal CAN choice for several reasons. First, the 56-residue L32 only contains challenging ligation junctions (e.g., Thr-Ala) with slow NCL kinetics.40, 41 Second, L32 lacks any native Cys residues, so desulfurization must be performed after NCL to produce native L32, allowing us to test four reactions (SPAAC, CAN, desulfurization, and Ddap linker cleavage) in a one-pot fashion (Fig. 8). Finally, L32-C (45-57) conveniently bears a Lys residue near its N-terminus, and L32-N (2-44, removed initiator Met) contains Lys residues both proximal and distal to the C-terminal thioester, allowing us to investigate the impact of Ddap linker placement on CAN reaction rate (Fig. 8). Following the same procedure for the model peptide control reactions, we first performed a representative control NCL between peptide 12a (L32-N 2-44; 0.4 mM) and 13a (L32-C 45-57; 0.5 mM) that was nearly complete after 48 h (Fig. 7B, Fig. 7C, and Fig. S47-50), with 70% of the peak area corresponding to ligated 14a (Fig. 7C). Notably, a significant amount of 12a was lost due to thioester hydrolysis.
Figure 7:
Non-templated native chemical ligation of E. coli 50S ribosomal subunit L32 (14a). (A) Sequence of L32 with L32-N (2-44, red) and L32-C (45-57, blue). The cleaved initiator Met was excluded. (B) Simplified schematic of the non-templated NCL between L32-N (12a) and L32-C (13a). Note that MTG was allowed to form the thioester for 10 min prior to TCEP addition, as shown in the more detailed scheme (Scheme S3). (C) HPLC chromatogram of the ligation endpoint. The ligation was considered complete after 48 h (after MTG addition), as nearly all of the reactive L32-N peptide (12a^) was depleted. However, a significant amount of L32-N peptide underwent hydrolysis (12a#) instead of ligation. 13a& refers to StBu-deprotected 13a. Analytical method B was used. HPLC and LC/MS chromatograms of more time points are shown in Fig. S49-50.
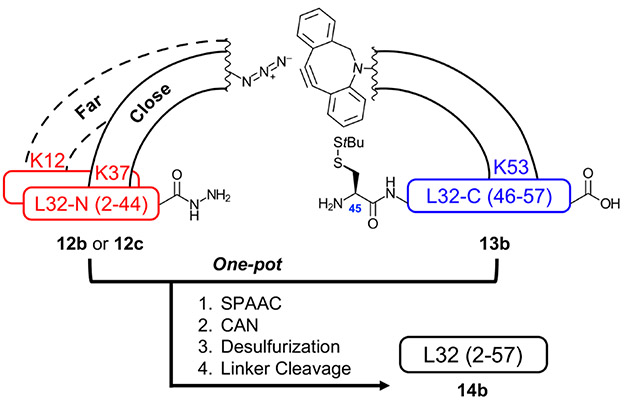
Figure 8:
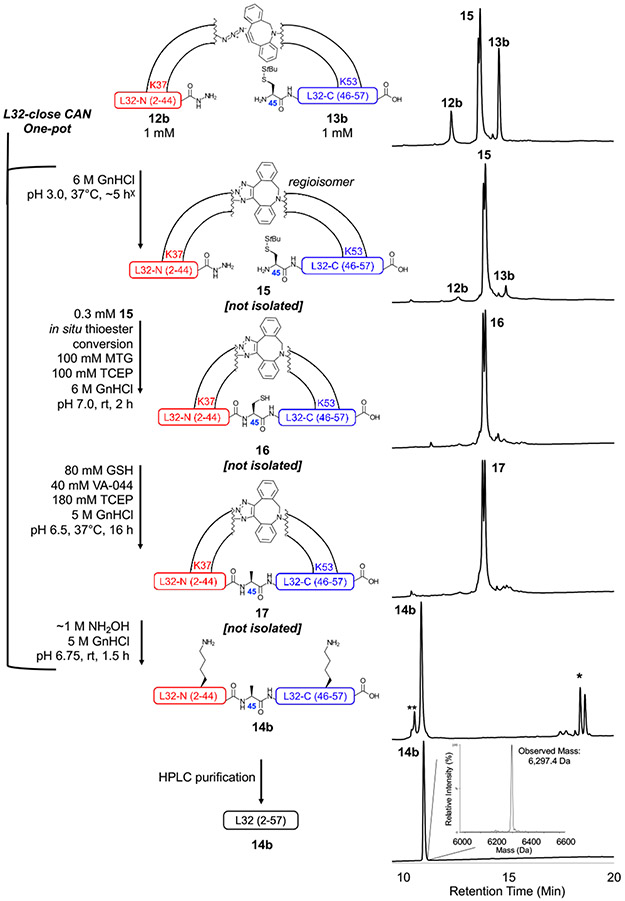
One-pot SPAAC, CAN, desulfurization, and linker cleavage assembly strategy for L32-far (dotted line) and L32-close (solid line), showing location of Ddap attachment points at specified Lys residues.
To investigate CAN for L32, L32-N (2-44) was functionalized with Ddap followed by a single Lys and 6-azidohexanoic acid at Lys37 (7 residues from the thioester, 12b, L32-close) or Lys12 (32 residues from the thioester, 12c, L32-far) (Fig. 8). Nle was substituted for the sole Met, as cleavage cocktails containing NH4I additive were found to cause azide degradation in peptide 2b, and we desired to avoid Met oxidation during peptide cleavage. L32-C (45-57) was functionalized at Lys53 with Ddap followed by a single Lys and DBCO-C6 to give 13b. We first investigated CAN of L32-close by clicking 12b with 13b in 6 M GnHCl at pH 3 to produce 15 (Fig. 9 and Fig. S51-54), which has a distance of 15 AA between Lys Ddap linkers. CAN was then performed using the same ligation conditions as the non-templated L32 ligation (0.3 mM of triazole-linked 15, Fig. 9). 15 ligated to completion within 2 h, much faster than the 48 h needed for the non-templated ligation. Furthermore, 88% of the total peak area corresponded to ligated product 16 (Fig. 9 and Fig. S55-56). Finally, we performed desulfurization to generate 17 and Ddap linker cleavage to produce full-length L32 14b in a one-pot synthesis (Fig. 9 and Fig. S57-59).
Figure 9:
One-pot SPAAC, CAN, desulfurization, and linker cleavage of 12b and 13b to generate full-length L32-Nle (14b). 12b and 13b were first templated together via ~5 h SPAAC to produce triazole-linked 15. The x in the reaction scheme indicates that additional 12b was added to the reaction after 4 h, as a significant amount of unreacted 13b was observed (see Scheme S4). CAN was then performed on 15 to generate 16 after only 2 h. Desulfurization was then performed for 16 h to produce 17. A 1.5 h linker cleavage of 17 resulted in 14b, which was easily HPLC purified. The HPLC chromatograms show, in descending order, 0 h and ~5 h SPAAC, 2 h CAN (after MTG addition), 16 h desulfurization, 1.5 h linker cleavage, and purified 14b (each y-axis is normalized A214). The double peaks within the SPAAC, CAN, and desulfurization chromatograms are expected, as the triazole forms two regioisomers. * indicates cleaved Ddap linker peaks. ** indicates hydrolyzed L32-N and desulfurized L32-C. The mass spectrum of purified 14b indicates that the correct full-length L32-Nle was generated (observed mass: 6,297.4 Da, calculated average mass: 6,297.1 Da). Analytical method C and LC/MS method C were used. HPLC and LC/MS chromatograms of time points taken for SPAAC, CAN, desulfurization, linker cleavage, and purification are shown in Fig. S53-59.
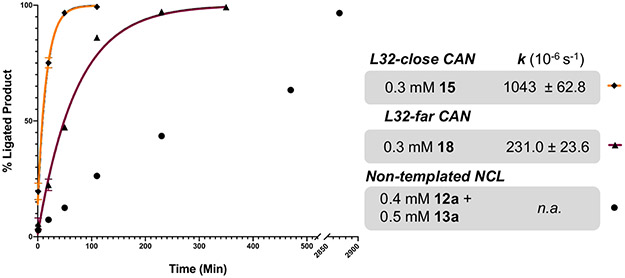
To determine how placing Ddap linkers further from ligation junctions affects CAN rate, we investigated CAN of L32-far by performing SPAAC with 12c and 13b in 6 M GnHCl at pH 3 (1.6 mM 12c and 1.7 mM 13b at 37°C for 4 h, Fig. S60-62). L32-far CAN was then tested with 0.3 mM of the triazole-linked 18 (distance of 40 AA between Ddap linkers) under the same ligation conditions as L32-close. The L32-far CAN was complete after 6 h to produce peptide 19, compared to the 48 h non-templated ligation (Fig. 10 and Fig. S63-64). The 1st order rate constant for L32-close (98-membered macrocycle) was 4.5-fold faster than for L32-far (173-membered macrocycle) (Fig. 10). This modest reduction in CAN rate is similar to that observed for PEG8 incorporation into the model peptides (~3-fold slower, Fig. 6). These results suggest that linker length and placement is easily adaptable for the CAN technique, especially since CAN with increased distances was still more efficient than non-templated NCL.
Figure 10:
Comparison of non-templated NCL and CAN reaction rates using L32 peptides (12a, 13a, 15, and 18). Relative peak area of ligated product is shown at various time points (relative to TCEP addition). CAN ligations were fit to first-order rate constants. For non-templated NCL, peak areas of both product (14a) and L32-N reactant (12a^) were calculated using estimated extinction coefficients, as described in the supplementary methods. Each data point represents the average of two independent experiments, with error bars showing standard deviation.
The synthesis of L32 using CAN highlights some of the key features of this technique. First, implementing CAN does not add extra intermediate HPLC purifications. Second, the distance between linker sites on the two peptides is quite flexible – the L32 ligation was separated by 40 AA yet still achieved a significant improvement in ligation rate relative to non-templated NCL. Finally, CAN dramatically increases the throughput of in-solution CPS steps, as CAN enabled full-length, L32 to be easily HPLC purified in ~25 h (including SPAAC, ligation, desulfurization, and linker removal), whereas the non-templated L32 NCL requires 48 h to reach completion plus a subsequent desulfurization step.
CONCLUSIONS
For the first time, DBCO peptides were directly synthesized via Fmoc-SPPS by using (MeCN)4CuBF4 to protect DBCO from acid-mediated rearrangement during standard acid cleavage. This DBCO protection strategy was shown to be robust, as DBCO peptides with various C-terminal functional groups were successfully prepared on different resins. Furthermore, all canonical amino acids were compatible, although Met is slightly oxidation-prone. This DBCO protection method enabled development of Click-Assisted NCL (CAN), which is a traceless templated ligation strategy that can be used to dramatically increase NCL efficiency. Using the STEVE and KENT model peptides, CAN required only 4 h to completely ligate a bulky Val thioester junction at 0.05 mM templated peptide concentration, while the non-templated NCL was incomplete after 48 h (0.4 mM 5a with 0.5 mM 6a). CAN also improved yield and reduced side products compared to non-templated NCL. The utility of CAN in CPS was also demonstrated through the preparation of the E. coli 50S ribosomal subunit L32. Compared to non-templated NCL, CAN enabled a faster ligation with one-pot SPAAC, ligation, desulfurization, and linker removal. Finally, the CAN rate was shown to modestly drop with template distance (i.e., increased macrocycle size), maintaining significant NCL rate enhancements even with distant attachment points – up to 40 AA separation demonstrated.
This new method to directly synthesize DBCO peptides via Fmoc-SPPS will be a useful tool for many peptide chemists. For example, peptide and protein nanoparticle conjugates have exciting biomedical applications (e.g., drug delivery and in vivo imaging), but there is a need to develop additional methods enabling site-specific incorporation of peptides onto nanoparticles.96 DBCO peptides can be easily conjugated to azide-functionalized nanoparticles,85 and this method will enable larger and more sequence-diverse DBCO peptides to be incorporated. Additionally, several methods exist to incorporate azides into proteins via engineered tRNA synthetases or Met replacement with azide-containing analogs,97-101 enabling DBCO peptides to tag recombinant azido-proteins for various applications (e.g., proteome labeling and imaging). Although cyclooctynes can be genetically encoded into proteins,99, 101-105 these methods are more challenging to implement compared to azide.99 The finding that (MeCN)4CuBF4 prevents acid-catalyzed 5-endo-dig cycloisomerization in DBCO, and perhaps other alkyne-derived isomerization reactions,106 is an important observation. For example, several groups have reported synthetic limitations when working with various strained alkynes due to the 5-endo-dig cycloisomerization,85-89 and protecting these strained alkynes with (MeCN)4CuBF4 should increase the number of synthetic strategies available. This (MeCN)4CuBF4 protection may also be effective in protecting DBCO peptides after resin cleavage and ether precipitation, as suggested by the 50 equiv (MeCN)4CuBF4 data shown in Fig. 3C. The larger quantity of (MeCN)4CuBF4 in the cleavage solution appear to allow DBCO to remain protected after resin cleavage, as 6-azidohexanoic acid did not efficiently react with DBCO in aqueous conditions. Liberation of copper from the DBCO peptides could be accomplished with aqueous ammonia, EDTA, or metal scavengers, as reported by the Hosoya group.91, 92 As other metal salts have been demonstrated to complex with cycloalkyne rings,93, 107 these may also be able to protect against acid-mediated degradation.
We expect CAN to find use as a tool in CPS for efficiently ligating peptides suffering from sluggish thioesters and/or poor solubility. Although DBCO can present SPPS coupling difficulties and reduce in-solution peptide solubility, these issues can be circumvented by incorporating short spacers (e.g., PEG) and solubilizing residues (e.g., Lys and Arg) onto the Ddap linker prior to DBCO coupling. If DBCO on a particular peptide still proves to be problematic, then that peptide could be coupled with the less sterically demanding azide moiety instead. Interestingly, since lower-concentration CAN is expected to maintain its ligation rate, the relative benefit of CAN over solution NCL increases with lower triazole-linked peptide concentrations. The current CAN methodology is expected to be applicable to many CPS strategies, especially since the reaction rate enhancement is relatively insensitive to linker attachment points. For example, there is a 49% likelihood that a Lys is within 20 residues on both sides of a ligation junction, and this probability increases to 69% for Lys being within 30 residues (based on the Lys abundance published on UniProt’s database).108 The L32-far CAN suggests that linkers can be placed at least 40 residues away from each other while still achieving enhanced ligation rates. While we only used Lys as attachment sites for the click handles, we expect that other amine-containing amino acids (e.g., ornithine) or the N-terminal amine could also be used to attach these handles. We expect that CAN may be particularly useful for sluggish ligations using unnaturally thiolated amino acids that enable NCL outside of Cys, such as thiolated valine.37, 38 CAN should also be compatible with other peptide linker strategies,31, 32 due to the simplicity of preparing both azide and DBCO peptides via SPPS. Notably, the Payne group recently developed reductive diselenide-selenoester ligation (rDSL) that can also overcome difficult junctions and poorly soluble peptides. The rDSL strategy has remarkably fast kinetics (nM peptides were efficiently ligated) and therefore does not require templating.109 However, selenocysteine is much more expensive relative to cysteine building blocks, and alternative selenylated amino acids are not yet commercially available. Both CAN and rDSL have the potential to make ligations at difficult junctions with hydrophobic peptides more routine in CPS, and combination of these strategies could provide an even more broadly useful method.
Future improvements to the CAN methodology have the potential to dramatically streamline CPS projects. For example, CAN currently requires relatively high peptide concentrations to efficiently perform SPAAC (typically >50 μM)110 before NCL. Although we limited our demonstration to aqueous conditions with denaturant, it is worth noting that SPAAC is much more versatile than NCL since SPAAC can be performed in solvent mixtures more conducive for solubilizing peptides. For example, MeCN/H2O is compatible with being directly lyophilized.111, 112 Additionally, solvents containing hexafluoro-2-propanol or ionic liquids have been shown to increase hydrophobic peptide solubility for NCL,33-35 and we expect SPAAC and CAN to be compatible with such solvents. Furthermore, unlike NCL, SPAAC does not suffer from competition with thioester hydrolysis. Future iterations of CAN may employ faster bioorthogonal chemistries for linking the peptides, as long as the chemistries are SPPS compatible. In fact, such bioorthogonal reactions that are also mutually orthogonal to DBCO and azide SPAAC would potentially enable multiple ligations to be performed in one pot via a CAN strategy. Such a combination would greatly increase yields in CPS by eliminating the need for intermediate HPLC purifications after each ligation. Finally, CAN parameters can be incorporated into an improved version of our “Automated Ligator” (Aligator) program,113 to predict segments that will benefit most from CAN.
Supplementary Material
ACKNOWLEDGMENT
The authors would like to thank Aman Makaju, Anna Bakhtina, and Dr. Sarah Franklin for mass spectrometry analysis of trypsin-digested samples, and Dr. Diego Fernandez for ICP-MS analysis of peptides (ICP-MS Metals and Strontium Isotope Facility, University of Utah). We also thank the Synthetic and Medicinal Chemistry Core (University of Utah and USTAR) for large-scale synthesis of Fmoc-Ddap-OH. The authors also thank Adedeji Aderounmu, Dr. Weiliang Xu, Jonah Holbrook, Dr. Sarah Apple, Zachary Cruz, Judah Evangelista, Dr. Riley Giesler, and Dr. Steven Draper for experimental/material assistance. We also thank Dr. Michael Jacobsen for experimental/material assistance, helpful discussions, and thoughtful advice. Finally, we thank Dr. Debra Eckert, Dr. Andrew Roberts, and Dr. Riley Giesler for critical review of this manuscript.
Funding Sources
Funding for this work was provided by a University of Utah Graduate Research Fellowship (to P.W.E.), the Marjorie Riches Gunn Award for Graduate Student Excellence (to J.M.F.), and NIH grants T32-GM122740 (to P.S.) and P50-AI150464 and R01-AI076168 (to M.S.K.).
Footnotes
ASSOCIATED CONTENT
Materials, methods, supplemental tables, supplemental schemes, and supplemental figures are available in the supplementary information document.
The authors declare no competing financial interests.
REFERENCES
- 1.Merrifield RB Solid phase peptide synthesis. I. The synthesis of a tetrapeptide. J. Am. Chem. Soc 1963, 85 (14), 2149–2154. [Google Scholar]
- 2.Dawson PE; Muir TW; Clark-Lewis I; Kent SB Synthesis of proteins by native chemical ligation. Science 1994, 266 (5186), 776–779. [DOI] [PubMed] [Google Scholar]
- 3.Dawson PE; Kent SB Synthesis of native proteins by chemical ligation. Annu. Rev. Biochem 2000, 69, 923–960. [DOI] [PubMed] [Google Scholar]
- 4.Wan Q; Danishefsky SJ Free-radical-based, specific desulfurization of cysteine: A powerful advance in the synthesis of polypeptides and glycopolypeptides. Angew. Chem., Int. Ed. Engl 2007, 46 (48), 9248–9252. [DOI] [PubMed] [Google Scholar]
- 5.Noisier AFM; Albericio F Advance in ligation techniques for peptide and protein synthesis. In Amino acids, peptides and proteins, Ryadnov M; Farkas E, Eds. The Royal Society of Chemistry: 2014; Vol. 39, pp 1–20. [Google Scholar]
- 6.Kent SBH Novel protein science enabled by total chemical synthesis. Protein Sci. 2019, 28 (2), 313–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Agouridas V; El Mahdi O; Diemer V; Cargoet M; Monbaliu JM; Melnyk O Native chemical ligation and extended methods: Mechanisms, catalysis, scope, and limitations. Chem. Rev 2019, 119 (12), 7328–7443. [DOI] [PubMed] [Google Scholar]
- 8.Chang HN; Liu BY; Qi YK; Zhou Y; Chen YP; Pan KM; Li WW; Zhou XM; Ma WW; Fu CY, et al. Blocking of the pd-1/pd-11 interaction by a d-peptide antagonist for cancer immunotherapy. Angew. Chem., Int. Ed. Engl 2015, 54 (40), 11760–11764. [DOI] [PubMed] [Google Scholar]
- 9.Clinton TR; Weinstock MT; Jacobsen MT; Szabo-Fresnais N; Pandya MJ; Whitby FG; Herbert AS; Prugar LI; McKinnon R; Hill CP, et al. Design and characterization of ebolavirus gp prehairpin intermediate mimics as drug targets. Protein Sci. 2015, 24 (4), 446–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Petersen ME; Jacobsen MT; Kay MS Synthesis of tumor necrosis factor alpha for use as a mirror-image phage display target. Org. Biomol. Chem 2016, 14 (23), 5298–5303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levinson AM; McGee JH; Roberts AG; Creech GS; Wang T; Peterson MT; Hendrickson RC; Verdine GL; Danishefsky SJ Total chemical synthesis and folding of all-l and all-d variants of oncogenic kras(g12v). J. Am. Chem. Soc 2017, 139 (22), 7632–7639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Rosa L; Di Stasi R; D'Andrea LD Total chemical synthesis by native chemical ligation of the all-d immunoglobulin-like domain 2 of axl. Tetrahedron 2019, 75 (7), 894–905. [Google Scholar]
- 13.Weinstock MT; Jacobsen MT; Kay MS Synthesis and folding of a mirror-image enzyme reveals ambidextrous chaperone activity. Proc. Natl. Acad. Sci. U. S. A 2014, 111 (32), 11679–11684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vinogradov AA; Evans ED; Pentelute BL Total synthesis and biochemical characterization of mirror image barnase. Chem. Sci 2015, 6 (5), 2997–3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Z; Xu W; Liu L; Zhu TF A synthetic molecular system capable of mirror-image genetic replication and transcription. Nat. Chem 2016, 8 (7), 698–704. [DOI] [PubMed] [Google Scholar]
- 16.Jiang W; Zhang B; Fan C; Wang M; Wang J; Deng Q; Liu X; Chen J; Zheng J; Liu L, et al. Mirror-image polymerase chain reaction. Cell Discovery 2017, 3, 17037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pech A; Achenbach J; Jahnz M; Schulzchen S; Jarosch F; Bordusa F; Klussmann S A thermostable d-polymerase for mirror-image pcr. Nucleic Acids Res. 2017, 45 (7), 3997–4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu W; Jiang W; Wang J; Yu L; Chen J; Liu X; Liu L; Zhu TF Total chemical synthesis of a thermostable enzyme capable of polymerase chain reaction. Cell Discovery 2017, 3, 17008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fan C; Deng Q; Zhu TF Bioorthogonal information storage in l-DNA with a high-fidelity mirror-image pfu DNA polymerase. Nat Biotechnol 2021, 10.1038/s41587-021-00969-6. [DOI] [PubMed] [Google Scholar]
- 20.Mandal K; Uppalapati M; Ault-Riche D; Kenney J; Lowitz J; Sidhu SS; Kent SB Chemical synthesis and x-ray structure of a heterochiral {d-protein antagonist plus vascular endothelial growth factor} protein complex by racemic crystallography. Proc. Natl. Acad. Sci. U. S. A 2012, 109 (37), 14779–14784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yan B; Ye L; Xu W; Liu L Recent advances in racemic protein crystallography. Bioorg. Med. Chem 2017, 25 (18), 4953–4965. [DOI] [PubMed] [Google Scholar]
- 22.Kent SB Racemic & quasi-racemic protein crystallography enabled by chemical protein synthesis. Curr. Opin. Chem. Biol 2018, 46, 1–9. [DOI] [PubMed] [Google Scholar]
- 23.Chen CC; Gao S; Ai HS; Qu Q; Tian CL; Li YM Racemic x-ray structure of l-type calcium channel antagonist calciseptine prepared by total chemical synthesis. Science China Chemistry 2018, 61 (6), 702–707. [Google Scholar]
- 24.Jbara M; Laps S; Morgan M; Kamnesky G; Mann G; Wolberger C; Brik A Palladium prompted on-demand cysteine chemistry for the synthesis of challenging and uniquely modified proteins. Nat. Commun 2018, 9 (1), 3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lewis YE; Galesic A; Levine PM; De Leon CA; Lamiri N; Brennan CK; Pratt MR O-glcnacylation of alpha-synuclein at serine 87 reduces aggregation without affecting membrane binding. ACS Chem. Biol 2017, 12 (4), 1020–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Araman C; Thompson RE; Wang S; Hackl S; Payne RJ; Becker CFW Semisynthetic prion protein (prp) variants carrying glycan mimics at position 181 and 197 do not form fibrils. Chem. Sci 2017, 8 (9), 6626–6632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lechner CC; Agashe ND; Fierz B Traceless synthesis of asymmetrically modified bivalent nucleosomes. Angew. Chem., Int. Ed. Engl 2016, 55 (8), 2903–2906. [DOI] [PubMed] [Google Scholar]
- 28.Sun H; Brik A The journey for the total chemical synthesis of a 53 kda protein. Acc. Chem. Res 2019, 52 (12), 3361–3371. [DOI] [PubMed] [Google Scholar]
- 29.Giesler RJ; Fulcher JM; Jacobsen MT; Kay MS Controlling segment solubility in large protein synthesis. In Total chemical synthesis of proteins, Brik A; Dawson PE; Liu L Eds. WILEY-VCH GmbH: 2021; pp 185–209. [Google Scholar]
- 30.Jacobsen MT; Petersen ME; Ye X; Galibert M; Lorimer GH; Aucagne V; Kay MS A helping hand to overcome solubility challenges in chemical protein synthesis. J. Am. Chem. Soc 2016, 138 (36), 11775–11782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao DD; Fan XW; Hao H; Zhang HL; Guo Y Temporary solubilizing tags method for the chemical synthesis of hydrophobic proteins. Curr. Org. Chem 2019, 23 (1), 2–13. [Google Scholar]
- 32.Yoshiya T; Tsuda S; Masuda S Development of trityl group anchored solubilizing tags for peptide and protein synthesis. ChemBioChem 2019, 20 (15), 1906–1913. [DOI] [PubMed] [Google Scholar]
- 33.Shen F; Tang S; Liu L Hexafluoro-2-propanol as a potent cosolvent for chemical ligation of membrane proteins. Sci China Chem 2011, 54 (1), 110–116. [Google Scholar]
- 34.Baumruck AC; Tietze D; Steinacker LK; Tietze AA Chemical synthesis of membrane proteins: A model study on the influenza virus b proton channel. Chem Sci 2018, 9 (8), 2365–2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baumruck AC; Yang J; Thomas GF; Beyer LI; Tietze D; Tietze AA Native chemical ligation of highly hydrophobic peptides in ionic liquid-containing media. J Org Chem 2021, 86 (2), 1659–1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roberts AG; Johnston EV; Shieh JH; Sondey JP; Hendrickson RC; Moore MA; Danishefsky SJ Fully synthetic granulocyte colony-stimulating factor enabled by isonitrile-mediated coupling of large, side-chain-unprotected peptides. J. Am. Chem. Soc 2015, 137 (40), 13167–13175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen J; Wan Q; Yuan Y; Zhu J; Danishefsky SJ Native chemical ligation at valine: A contribution to peptide and glycopeptide synthesis. Angew. Chem., Int. Ed. Engl 2008, 47 (44), 8521–8524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haase C; Rohde H; Seitz O Native chemical ligation at valine. Angew. Chem., Int. Ed. Engl 2008, 47 (36), 6807–6810. [DOI] [PubMed] [Google Scholar]
- 39.Chen H; Xiao Y; Yuan N; Weng J; Gao P; Breindel L; Shekhtman A; Zhang Q Coupling of sterically demanding peptides by β-thiolactone-mediated native chemical ligation. Chem. Sci 2018, 9 (7), 1982–1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hackeng TM; Griffin JH; Dawson PE Protein synthesis by native chemical ligation: Expanded scope by using straightforward methodology. Proc. Natl. Acad. Sci. U. S. A 1999, 96 (18), 10068–10073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Conibear AC; Watson EE; Payne RJ; Becker CFW Native chemical ligation in protein synthesis and semi-synthesis. Chem. Soc. Rev 2018, 47 (24), 9046–9068. [DOI] [PubMed] [Google Scholar]
- 42.Dawson PE; Churchill MJ; Ghadiri MR; Kent SBH Modulation of reactivity in native chemical ligation through the use of thiol additives. J. Am. Chem. Soc 1997, 119 (19), 4325–4329. [Google Scholar]
- 43.Johnson ECB; Kent SBH Insights into the mechanism and catalysis of the native chemical ligation reaction. J. Am. Chem. Soc 2006, 128 (20), 6640–6646. [DOI] [PubMed] [Google Scholar]
- 44.Thompson RE; Liu X; Alonso-Garcia N; Pereira PJ; Jolliffe KA; Payne RJ Trifluoroethanethiol: An additive for efficient one-pot peptide ligation-desulfurization chemistry. J. Am. Chem. Soc 2014, 136 (23), 8161–8164. [DOI] [PubMed] [Google Scholar]
- 45.Huang YC; Chen CC; Gao S; Wang YH; Xiao H; Wang F; Tian CL; Li YM Synthesis of l- and d-ubiquitin by one-pot ligation and metal-free desulfurization. Chem. - Eur. J 2016, 22 (22), 7623–7628. [DOI] [PubMed] [Google Scholar]
- 46.Sakamoto K; Tsuda S; Mochizuki M; Nohara Y; Nishio H; Yoshiya T Imidazole-aided native chemical ligation: Imidazole as a one-pot desulfurization-amenable non-thiol-type alternative to 4-mercaptophenylacetic acid. Chem. - Eur. J 2016, 22 (50), 17940–17944. [DOI] [PubMed] [Google Scholar]
- 47.Kumar KS; Bavikar SN; Spasser L; Moyal T; Ohayon S; Brik A Total chemical synthesis of a 304 amino acid k48-linked tetraubiquitin protein. Angew. Chem., Int. Ed. Engl 2011, 50 (27), 6137–6141. [DOI] [PubMed] [Google Scholar]
- 48.Kemp DS; Leung SL; Kerkman DJ Models that demonstrate peptide bond formation by prior thiol capture i. Capture by disulfide formation. Tetrahedron Lett. 1981, 22 (3), 181–184. [Google Scholar]
- 49.Kemp DS; Galakatos NG; Bowen B; Tan K Peptide synthesis by prior thiol capture. 2. Design of templates for intramolecular o,n-acyl transfer. 4,6-disubstituted dibenzofurans as optimal spacing elements. J. Org. Chem 1986, 51 (10), 1829–1838. [Google Scholar]
- 50.Tam JP; Wong CTT Chemical synthesis of circular proteins. J. Biol. Chem 2012, 287 (32), 27020–27025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lin H; Thayer DA; Wong CH; Walsh CT Macrolactamization of glycosylated peptide thioesters by the thioesterase domain of tyrocidine synthetase. Chemistry & Biology 2004, 11 (12), 1635–1642. [DOI] [PubMed] [Google Scholar]
- 52.Meutermans WDF; Bourne GT; Golding SW; Horton DA; Campitelli MR; Craik D; Scanlon M; Smythe ML Difficult macrocyclizations: New strategies for synthesizing highly strained cyclic tetrapeptides. Org. Lett 2003, 5 (15), 2711–2714. [DOI] [PubMed] [Google Scholar]
- 53.Sengupta S; Mehta G Macrocyclization via c–h functionalization: A new paradigm in macrocycle synthesis. Org. Biomol. Chem 2020, 18 (10), 1851–1876. [DOI] [PubMed] [Google Scholar]
- 54.Lee DH; Granja JR; Martinez JA; Severin K; Ghadiri MR A self-replicating peptide. Nature 1996, 382 (6591), 525–528. [DOI] [PubMed] [Google Scholar]
- 55.Yao S; Ghosh I; Zutshi R; Chmielewski J A ph-modulated, self-replicating peptide. J. Am. Chem. Soc 1997, 119 (43), 10559–10560. [Google Scholar]
- 56.Beligere GS; Dawson PE Conformationally assisted protein ligation using c-terminal thioester peptides. J. Am. Chem. Soc 1999, 121 (26), 6332–6333. [Google Scholar]
- 57.Rubinov B; Wagner N; Rapaport H; Ashkenasy G Self-replicating amphiphilic beta-sheet peptides. Angew. Chem., Int. Ed. Engl 2009, 48 (36), 6683–6686. [DOI] [PubMed] [Google Scholar]
- 58.Reinhardt U; Lotze J; Zernia S; Morl K; Beck-Sickinger AG; Seitz O Peptide-templated acyl transfer: A chemical method for the labeling of membrane proteins on live cells. Angew. Chem., Int. Ed. Engl 2014, 53 (38), 10237–10241. [DOI] [PubMed] [Google Scholar]
- 59.Reinhardt U; Lotze J; Morl K; Beck-Sickinger AG; Seitz O Rapid covalent fluorescence labeling of membrane proteins on live cells via coiled-coil templated acyl transfer. Bioconjugate Chem. 2015, 26 (10), 2106–2117. [DOI] [PubMed] [Google Scholar]
- 60.Lotze J; Reinhardt U; Seitz O; Beck-Sickinger AG Peptide-tags for site-specific protein labelling in vitro and in vivo. Mol. BioSyst 2016, 12 (6), 1731–1745. [DOI] [PubMed] [Google Scholar]
- 61.Petszulat H; Seitz O A fluorogenic native chemical ligation for assessing the role of distance in peptide-templated peptide ligation. Bioorg. Med. Chem 2017, 25 (18), 5022–5030. [DOI] [PubMed] [Google Scholar]
- 62.Bruick RK; Dawson PE; Kent SB; Usman N; Joyce GF Template-directed ligation of peptides to oligonucleotides. Chemistry & Biology 1996, 3 (1), 49–56. [DOI] [PubMed] [Google Scholar]
- 63.Vazquez O; Seitz O Templated native chemical ligation: Peptide chemistry beyond protein synthesis. J. Pept. Sci 2014, 20 (2), 78–86. [DOI] [PubMed] [Google Scholar]
- 64.Michaelis J; Roloff A; Seitz O Amplification by nucleic acid-templated reactions. Org. Biomol. Chem 2014, 12 (18), 2821–2833. [DOI] [PubMed] [Google Scholar]
- 65.Di Pisa M; Seitz O Nucleic acid templated reactions for chemical biology. ChemMedChem 2017, 12 (12), 872–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lores Lareo P; Linscheid MW; Seitz O Nucleic acid and snp detection via template-directed native chemical ligation and inductively coupled plasma mass spectrometry. J. Mass Spectrom 2019, 54 (8), 676–683. [DOI] [PubMed] [Google Scholar]
- 67.Sayers J; Payne RJ; Winssinger N Peptide nucleic acid-templated selenocystine-selenoester ligation enables rapid mirna detection. Chem. Sci 2018, 9 (4), 896–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Middel S; Panse CH; Nawratil S; Diederichsen U Native chemical ligation directed by photocleavable peptide nucleic acid (pna) templates. ChemBioChem 2017, 18 (23), 2328–2332. [DOI] [PubMed] [Google Scholar]
- 69.Hayashi G; Yanase M; Nakatsuka Y; Okamoto A Simultaneous and traceless ligation of peptide fragments on DNA scaffold. Biomacromolecules 2019, 20 (3), 1246–1253. [DOI] [PubMed] [Google Scholar]
- 70.Pattison DI; Rahmanto AS; Davies MJ Photo-oxidation of proteins. Photochem. Photobiol. Sci 2012, 11 (1), 38–53. [DOI] [PubMed] [Google Scholar]
- 71.Li S; Cai H; He J; Chen H; Lam S; Cai T; Zhu Z; Bark SJ; Cai C Extent of the oxidative side reactions to peptides and proteins during the cuaac reaction. Bioconjugate Chem. 2016, 27 (10), 2315–2322. [DOI] [PubMed] [Google Scholar]
- 72.Conibear AC; Farbiarz K; Mayer RL; Matveenko M; Kahlig H; Becker CF Arginine side-chain modification that occurs during copper-catalysed azide-alkyne click reactions resembles an advanced glycation end product. Org. Biomol. Chem 2016, 14 (26), 6205–6211. [DOI] [PubMed] [Google Scholar]
- 73.Osuna Galvez A; Bode JW Traceless templated amide-forming ligations. J. Am. Chem. Soc 2019, 141 (22), 8721–8726. [DOI] [PubMed] [Google Scholar]
- 74.Fulcher JM; Petersen ME; Giesler RJ; Cruz ZS; Eckert DM; Francis JN; Kawamoto EM; Jacobsen MT; Kay MS Chemical synthesis of shiga toxin subunit b using a next-generation traceless "helping hand" solubilizing tag. Org. Biomol. Chem 2019, 17 (48), 10237–10244. [DOI] [PubMed] [Google Scholar]
- 75.Jewett JC; Bertozzi CR Cu-free click cycloaddition reactions in chemical biology. Chem. Soc. Rev 2010, 39 (4), 1272–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jewett JC; Sletten EM; Bertozzi CR Rapid cu-free click chemistry with readily synthesized biarylazacyclooctynones. J. Am. Chem. Soc 2010, 132 (11), 3688–3690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang Q; Chan TR; Hilgraf R; Fokin VV; Sharpless KB; Finn MG Bioconjugation by copper(i)-catalyzed azide-alkyne [3 + 2] cycloaddition. J. Am. Chem. Soc 2003, 125 (11), 3192–3193. [DOI] [PubMed] [Google Scholar]
- 78.Lee DJ; Mandal K; Harris PW; Brimble MA; Kent SB A one-pot approach to neoglycopeptides using orthogonal native chemical ligation and click chemistry. Org. Lett 2009, 11 (22), 5270–5273. [DOI] [PubMed] [Google Scholar]
- 79.Zheng JS; Tang S; Qi YK; Wang ZP; Liu L Chemical synthesis of proteins using peptide hydrazides as thioester surrogates. Nat. Protoc 2013, 8 (12), 2483–2495. [DOI] [PubMed] [Google Scholar]
- 80.Schneggenburger PE; Worbs B; Diederichsen U Azide reduction during peptide cleavage from solid support-the choice of thioscavenger? J. Pept. Sci 2010, 16 (1), 10–14. [DOI] [PubMed] [Google Scholar]
- 81.Kizil C; Iltzsche A; Thomas AK; Bhattarai P; Zhang Y; Brand M Efficient cargo delivery into adult brain tissue using short cell-penetrating peptides. Plos One 2015, 10 (4), e0124073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Perols A; Arcos Famme M; Eriksson Karlstrom A Site-specific antibody labeling by covalent photoconjugation of z domains functionalized for alkyne-azide cycloaddition reactions. ChemBioChem 2015, 16 (17), 2522–2529. [DOI] [PubMed] [Google Scholar]
- 83.Witte MD; Theile CS; Wu T; Guimaraes CP; Blom AE; Ploegh HL Production of unnaturally linked chimeric proteins using a combination of sortase-catalyzed transpeptidation and click chemistry. Nat. Protoc 2013, 8 (9), 1808–1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Huey L Development of a monodisperse oligomeric hemoglobin-based oxygen carrier for acute blood replacement therapy. WWU Honors Program Senior Projects 2018, 83. [Google Scholar]
- 85.Wang X; Gobbo P; Suchy M; Workentin MS; Hudson RHE Peptide-decorated gold nanoparticles via strain-promoted azide-alkyne cycloaddition and post assembly deprotection. RSC Adv. 2014, 4 (81), 43087–43091. [Google Scholar]
- 86.Chigrinova M; McKay CS; Beaulieu LP; Udachin KA; Beauchemin AM; Pezacki JP Rearrangements and addition reactions of biarylazacyclooctynones and the implications to copper-free click chemistry. Org. Biomol. Chem 2013, 11 (21), 3436–3441. [DOI] [PubMed] [Google Scholar]
- 87.Debets MF; van Berkel SS; Schoffelen S; Rutjes FP; van Hest JC; van Delft FL Aza-dibenzocyclooctynes for fast and efficient enzyme pegylation via copper-free (3+2) cycloaddition. ChemComm 2010, 46 (1), 97–99. [DOI] [PubMed] [Google Scholar]
- 88.Loy DM; Klein PM; Krzysztoń R; Lächelt U; Rädler JO; Wagner E A microfluidic approach for sequential assembly of sirna polyplexes with a defined structure – activity relationship. PeerJ Materials Science 2019, 1, e1. [Google Scholar]
- 89.Klein PM; Wagner E Click-shielded and targeted lipopolyplexes. Methods Mol. Biol 2019, 2036, 141–164. [DOI] [PubMed] [Google Scholar]
- 90.van Geel R; Pruijn GJ; van Delft FL; Boelens WC Preventing thiol-yne addition improves the specificity of strain-promoted azide-alkyne cycloaddition. Bioconjugate Chem. 2012, 23 (3), 392–398. [DOI] [PubMed] [Google Scholar]
- 91.Yoshida S; Hatakeyama Y; Johmoto K; Uekusa H; Hosoya T Transient protection of strained alkynes from click reaction via complexation with copper. J. Am. Chem. Soc 2014, 136 (39), 13590–13593. [DOI] [PubMed] [Google Scholar]
- 92.Yoshida S; Kuribara T; Ito H; Meguro T; Nishiyama Y; Karaki F; Hatakeyama Y; Koike Y; Kii I; Hosoya T A facile preparation of functional cycloalkynes via an azide-to-cycloalkyne switching approach. ChemComm 2019, 55 (24), 3556–3559. [DOI] [PubMed] [Google Scholar]
- 93.Bennett MA; Schwemlein HP Metal complexes of small cycloalkynes and arynes. Angew. Chem., Int. Ed. Engl 1989, 28 (10), 1296–1320. [Google Scholar]
- 94.Li J; Li Y; He Q; Li Y; Li H; Liu L One-pot native chemical ligation of peptide hydrazides enables total synthesis of modified histones. Org. Biomol. Chem 2014, 12 (29), 5435–5441. [DOI] [PubMed] [Google Scholar]
- 95.Ohayon S; Spasser L; Aharoni A; Brik A Targeting deubiquitinases enabled by chemical synthesis of proteins. J. Am. Chem. Soc 2012, 134 (6), 3281–3289. [DOI] [PubMed] [Google Scholar]
- 96.Spicer CD; Jumeaux C; Gupta B; Stevens MM Peptide and protein nanoparticle conjugates: Versatile platforms for biomedical applications. Chem. Soc. Rev 2018, 47 (10), 3574–3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Liu Y; Conboy MJ; Mehdipour M; Liu Y; Tran TP; Blotnick A; Rajan P; Santos TC; Conboy IM Application of bio-orthogonal proteome labeling to cell transplantation and heterochronic parabiosis. Nat. Commun 2017, 8 (1), 643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yang AC; du Bois H; Olsson N; Gate D; Lehallier B; Berdnik D; Brewer KD; Bertozzi CR; Elias JE; Wyss-Coray T Multiple click-selective trna synthetases expand mammalian cell-specific proteomics. J. Am. Chem. Soc 2018, 140 (23), 7046–7051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Merten H; Schaefer JV; Brandl F; Zangemeister-Wittke U; Pluckthun A Facile site-specific multiconjugation strategies in recombinant proteins produced in bacteria. Methods Mol. Biol 2019, 2033, 253–273. [DOI] [PubMed] [Google Scholar]
- 100.Saleh AM; Wilding KM; Calve S; Bundy BC; Kinzer-Ursem TL Non-canonical amino acid labeling in proteomics and biotechnology. J. Biol. Eng 2019, 13, 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lee KJ; Kang D; Park HS Site-specific labeling of proteins using unnatural amino acids. Mol. Cells 2019, 42 (5), 386–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Plass T; Milles S; Koehler C; Schultz C; Lemke EA Genetically encoded copper-free click chemistry. Angew. Chem., Int. Ed. Engl 2011, 50 (17), 3878–3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Borrmann A; Milles S; Plass T; Dommerholt J; Verkade JM; Wiessler M; Schultz C; van Hest JC; van Delft FL; Lemke EA Genetic encoding of a bicyclo[6.1.0]nonyne-charged amino acid enables fast cellular protein imaging by metal-free ligation. ChemBioChem 2012, 13 (14), 2094–2099. [DOI] [PubMed] [Google Scholar]
- 104.Lang K; Davis L; Wallace S; Mahesh M; Cox DJ; Blackman ML; Fox JM; Chin JW Genetic encoding of bicyclononynes and trans-cyclooctenes for site-specific protein labeling in vitro and in live mammalian cells via rapid fluorogenic diels-alder reactions. J. Am. Chem. Soc 2012, 134 (25), 10317–10320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Alamudi SH; Satapathy R; Kim J; Su D; Ren H; Das R; Hu L; Alvarado-Martinez E; Lee JY; Hoppmann C, et al. Development of background-free tame fluorescent probes for intracellular live cell imaging. Nat. Commun 2016, 7, 11964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Baldwin JE Rules for ring closure. J. Chem. Soc., Chem. Commun 1976, 734–736. [Google Scholar]
- 107.Gobbo P; Romagnoli T; Barbon SM; Price JT; Keir J; Gilroy JB; Workentin MS Expanding the scope of strained-alkyne chemistry: A protection-deprotection strategy via the formation of a dicobalt-hexacarbonyl complex. ChemComm 2015, 51 (30), 6647–6650. [DOI] [PubMed] [Google Scholar]
- 108.The UniProt Consortium Uniprot: A worldwide hub of protein knowledge. Nucleic Acids Res. 2019, 47 (D1), D506–D515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Chisholm TS; Kulkarni SS; Hossain KR; Cornelius F; Clarke RJ; Payne RJ Peptide ligation at high dilution via reductive diselenide-selenoester ligation. J. Am. Chem. Soc 2020, 142 (2), 1090–1100. [DOI] [PubMed] [Google Scholar]
- 110.Saito F; Noda H; Bode JW Critical evaluation and rate constants of chemoselective ligation reactions for stoichiometric conjugations in water. ACS Chem. Biol 2015, 10 (4), 1026–1033. [DOI] [PubMed] [Google Scholar]
- 111.Zayas J; Annoual M; Das JK; Felty Q; Gonzalez WG; Miksovska J; Sharifai N; Chiba A; Wnuk SF Strain promoted click chemistry of 2- or 8-azidopurine and 5-azidopyrimidine nucleosides and 8-azidoadenosine triphosphate with cyclooctynes. Application to living cell fluorescent imaging. Bioconjugate Chem. 2015, 26 (8), 1519–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Dommerholt J; Rutjes FPJT; van Delft FL Strain-promoted 1,3-dipolar cycloaddition of cycloalkynes and organic azides. Top. Curr. Chem 2016, 374 (2), 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Jacobsen MT; Erickson PW; Kay MS Aligator: A computational tool for optimizing total chemical synthesis of large proteins. Bioorg. Med. Chem 2017, 25 (18), 4946–4952. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.