ABSTRACT
Type III interferons (IFN-λ) are shown to be preferentially produced by epithelial cells, which provide front-line protection at barrier surfaces. Transmissible gastroenteritis virus (TGEV), belonging to the genus Alphacoronavirus of the family Coronaviridae, can cause severe intestinal injuries in porcine, resulting in enormous economic losses for the swine industry, worldwide. Here, we demonstrated that although IFN-λ1 had a higher basal expression, TGEV infection induced more intense IFN-λ3 production in vitro and in vivo than did IFN-λ1. We explored the underlying mechanism of IFN-λ induction by TGEV and found a distinct regulation mechanism of IFN-λ1 and IFN-λ3. The classical RIG-I-like receptor (RLR) pathway is involved in IFN-λ3 but not IFN-λ1 production. Except for the signaling pathways mediated by RIG-I and MDA5, TGEV nsp1 induces IFN-λ1 and IFN-λ3 by activating NF-κB via the unfolded protein responses (UPR) PERK-eIF2α pathway. Furthermore, functional domain analysis indicated that the induction of IFN-λ by the TGEV nsp1 protein was located at amino acids 85 to 102 and was dependent on the phosphorylation of eIF2α and the nuclear translocation of NF-κB. Moreover, the recombinant TGEV with the altered amino acid motif of nsp1 85-102 was constructed, and the nsp1 (85-102sg) mutant virus significantly reduced the production of IFN-λ, compared with the wild strain. Compared to the antiviral activities of IFN-λ1, the administration of IFN-λ3 showed greater antiviral activity against TGEV infections in IPEC-J2 cells. In summary, our data point to the significant role of IFN-λ in the host innate antiviral responses to coronavirus infections within mucosal organs and in the distinct mechanisms of IFN-λ1 and IFN-λ3 regulation.
IMPORTANCE Coronaviruses cause infectious diseases in various mammals and birds and exhibit an epithelial cell tropism in enteric and respiratory tracts. It is critical to explore how coronavirus infections modulate IFN-λ, a key innate cytokine against mucosal viral infection. Our results uncovered the different processes of IFN-λ1 and IFN-λ3 production that are involved in the classical RLR pathway and determined that TGEV nsp1 induces IFN-λ1 and IFN-λ3 production by activating NF-κB via the PERK-eIF2α pathway in UPR. These studies highlight the unique regulation of antiviral defense in the intestine during TGEV infection. We also demonstrated that IFN-λ3 induced greater antiviral activity against TGEV replication than did IFN-λ1 in IPEC-J2 cells, which is helpful in finding a novel strategy for the treatment of coronavirus infections.
KEYWORDS: coronavirus, type III IFN, RLR-mediated signaling, protein kinase R-like ER kinase, transmissible gastroenteritis virus, nonstructural protein 1 (nsp1), NF-κB
INTRODUCTION
Interferons (IFNs), as the key components of the host innate antiviral response, provide the primary defense against viral infections. Among the three types of IFNs (types I, II, and III), type III IFN-lambda (IFN-λ) is shown to predominantly act at epithelial barriers, including the gastrointestinal, respiratory, and reproductive tracts, which are major entry points for many pathogens (1–3). Furthermore, IFN-λ can modulate the functions of immune cells at mucosal sites and can thereby protect and maintain the integrity and barrier functions of the mucosae (4, 5). IFN-λ is a multigenic family of cytokines. So far, only mouse IFN-λ2 and IFN-λ3, human IFN-λ1, IFN-λ2, IFN-λ3, and IFN-λ4 (1, 6), and porcine IFN-λ1 and IFN-λ3 are reported (7, 8).
Transmissible gastroenteritis (TGE) is an acute and highly contagious disease that results in vomiting, watery diarrhea, and dehydration in neonatal piglets (9, 10). TGE virus (TGEV), as the causative agent, is an enveloped, positive-strand RNA virus that is grouped in the genus Alphacoronavirus of the family Coronaviridae in the order Nidovirale (11). The TGEV genome is 28 kb in length and codes for four structural proteins, namely, spike (S), envelope (E), membrane (M), and nucleocapsid (N), as well as an accessory protein ORF3 and the nonstructural proteins nsp1 through nsp16, which are produced by the two large polyproteins pp1a and pp1ab (12). Notably, nsp1 is produced only from alphacoronaviruses and betacoronaviruses, and it performs multiple functions, including inhibiting the expression of host genes and virulence (13, 14).
It is important to understand the pathogenesis of TGEV in depth for developing new strategies by which to treat TGEV infection. In contrast to most CoVs, TGEV infection activates nuclear factor-kappa B (NF-κB) and induces significant type I interferon (IFN-I) production (15–18). Intestinal villous epithelial cells are the primary target cells of TGEV infections. While all nucleated cells can respond to IFN-α/β, IFN-λ responses are largely concentrated at epithelial and barrier surfaces (19). However, whether and how TGEV modulates the IFN-λ response in swine intestinal epithelial cells remains unknown.
The induction and regulatory mechanisms of type III IFNs (IFN-III) are different from those of IFN-I (2, 20). Although all components of the IFN-I enhanceosome are required to induce IFN-I, NF-κB and the interferon regulatory factor (IRF) can independently mediate the production of IFN-III (21). The NF-κB family consists of five mammalian members: RelA (p65), RelB, cRel, NF-κB1 (p50), and NF-κB2 (p52) (22). A heterodimer composed of p65 and p50 is the most usual activated form of NF-κB, and it interacts with inhibitory kappa B (IκB) in resting cells (23). For RNA viruses, IFN-I can be produced following the recognition of the retinoic acid-inducible gene-I (RIG-I) and the melanoma-differentiation-associated gene 5 (MDA5) by the virus-related molecules. It has been demonstrated that TGEV infections activate NF-κB, depending on the RIG-I-like receptor (RLR) signaling pathway (24).
The endoplasmic reticulum (ER) is the largest membrane-bound compartment that is key for many cellular functions, including protein biosynthesis as well as the folding, assembly, posttranslational modification, transport, and degradation of a great quantity of membrane and secreted proteins. The accumulation of unfolded or misfolded proteins in the ER lumen rapidly induces ER stress and activates the unfolded protein response (UPR) process. Many studies have reported that the coronavirus infection of cells leads to ER stress and activates the UPR response (25–28). Our previous report showed that TGEV infection could induce ER stress and activate the PERK-eIF2α pathway. The phosphorylation of eIF2α can cause the activation of NF-κB by reducing the steady-state levels of the short-lived regulatory proteins IκBα (29). However, it is unclear whether ER stress is involved in the intense induction of IFN-λ.
Given the pivotal role of IFN-λ in TGEV-susceptible porcine intestinal epithelial cells (IEC), we investigated how IFN-λ affects viral replication and explored the ability and mechanism of TGEV to induce IFN-λ production. Our study showed that TGEV regulated the expression of IFN-λ1 and IFN-λ3 through different mechanisms. Moreover, the inhibitory effect of recombinant porcine IFN-λ3 (rpIFN-λ3) was better than that of rpIFN-λ1 against the TGEV infection of IPEC-J2 cells.
RESULTS
TGEV infection induces higher production of IFN-λ3 than IFN-λ1 in vitro and in vivo.
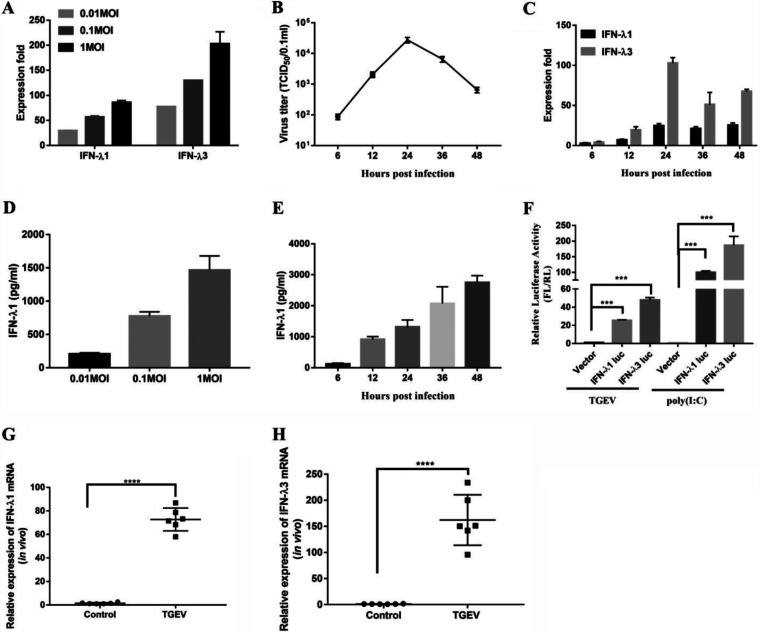
TGEV mainly infects and replicates in porcine small intestinal epithelial cells in vivo (11). To investigate whether TGEV infections induce IFN-λ in vitro, we tested the expression of IFN-λ1 and IFN-λ3 following a TGEV infection in IPEC-J2 cells. The IPEC-J2 cell line, derived from jejunum epithelium isolated from unsuckled newborn piglets, is used widely as an in vitro model system for studying host-pathogen interactions in pig intestines (30, 31). The results showed that TGEV infection triggered a dose-dependent enhancement of the levels of IFN-λ1 and IFN-λ3 at 24 h postinfection (hpi) (Fig. 1A). Although IFN-λ1 had a higher basal expression in the mock-treated control cells (data not shown), the level of IFN-λ3 induced by a TGEV infection is significantly higher than that of IFN-λ1 (Fig. 1A). The growth curve of the virus initially rose exponentially, reaching its peak at 24 hpi (Fig. 1B). To identify the kinetics and the magnitude of IFN-λ production, we determined the expression of IFN-λ1 and IFN-λ3 at different time points (6, 12, 24, 36, and 48 h) after a TGEV infection. The induction of IFN-λ1 and IFN-λ3 started at 6 hpi and then peaked at 24 hpi (Fig. 1C). The induction trend of IFN-λ is paralleled to the growth kinetics curve of TGEV. Consistent with Fig. 1A, TGEV induced higher levels of IFN-λ3 than IFN-λ1. To confirm these results, we assessed the protein levels of IFN-λ1 from IPEC-J2 cells at different multiplicities of infection (MOI) or at different time points (6, 12, 24, 36, and 48 h) after a TGEV infection. The production of IFN-λ1 appeared to be dose-dependent, beginning at 6 hpi and then gradually rising up to 48 hpi (Fig. 1D and E). To confirm IFN-λ production in the IPEC-J2 cells, we isolated the IPEC-J2 cell genomic fragments, including the pig ifnλ1 −500 to +10 and pig ifnλ3 −486 to +8, relative to the transcription initiation site, and cloned them into a firefly luciferase reporter vector (pIFN-λ1 [−500/+10] Luc and pIFN-λ3 [−486/+8] Luc). As shown in Fig. 1F, the activity of the IFN-λ3-Luc-dependent promoter increased significantly more than that of the IFN-λ1 promoter after the TGEV infection. Poly (I:C), a synthetic dsRNA, induced significant IFN-λ3 production, as previously reported (7). These results demonstrated that a TGEV infection could induce IFN-λ production in IPEC-J2 cells, and the production of IFN-λ3 was higher than that of IFN-λ1.
FIG 1.
TGEV infection induces IFN-λ production in vitro and in vivo. (A) IPEC-J2 cells were mock-infected or infected with TGEV H87 for 24 h at an MOI of 0.01, 0.1, or 1. The IFN-λ1 and IFN-λ3 expression was measured via RT-qPCR. (B) IPEC-J2 cells were infected with TGEV H87 at an MOI of 1. The samples were collected at 0, 6, 12, 24, 36, and 48 hpi. One-step growth curve of TGEV in IPEC-J2 cells. (C) A time-dependent increase of IFN-λ1 and IFN-λ3 expression was revealed via RT-qPCR in IPEC-J2 cells. (D) The IFN-λ1 expression at different MOI values was confirmed via ELISA. (E) The expression of IFN-λ1 in the supernatants in TGEV infected cells at different time points was tested via ELISA. (F) The IFN-λ1 and IFN-λ3 promoters were activated in TGEV-infected IPEC-J2 cells. Poly (I:C) (1 μg/mL) was used as a positive-control for IFN-λ activation. (G and H) TGEV infection induces IFN-λ1 and IFN-λ3 production in vivo. 12 two-day-old SPF piglets were orally inoculated with TGEV H87 strain or DMEM to serve as uninfected controls. All of the piglets were euthanized by the end of the study, which was terminated at 48 hpi. The expression of IFN-λ1 and IFN-λ3 expression in the ileum tissues was detected via RT-qPCR.
To investigate whether TGEV infection induces IFN-λ in vivo, the expression of IFN-λ1 and the expression of IFN-λ3 were surveyed in the ileum of piglets infected with TGEV at 48 hpi. TGEV infection led to a more than 60-fold rise in IFN-λ1 expression and a 100 to 350-fold rise in IFN-λ3 expression in the ileum tissues, compared to a control (Fig. 1G and H). Collectively, these results suggested that TGEV infection induces IFN-λ1 and IFN-λ3 production to different extents.
NF-κB, but not IRF3, is critical to IFN-λ production during a TGEV infection.
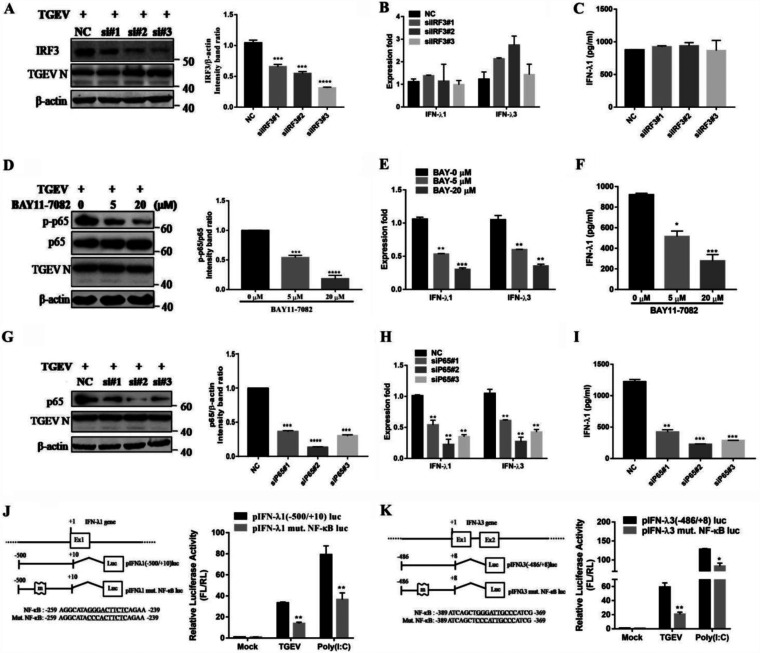
Independent actions of IRF and NF-κB can produce IFN-λ, and it is known that TGEV infection leads to IRF3 activation (24). To explore whether IRF3 is involved in IFN-λ induction, siRNA-mediated IRF3 knockdown was conducted. The silencing efficiency of IRF3 siRNA was determined via Western blotting (34.39% decrease for the no. 1, 45.37% for the no. 2, and 70.49% for the no. 3 siRNA) (Fig. 2A). Compared with a control, no significant difference was found in the IRF3-mediated induction of IFN-λ1 and IFN-λ3 (Fig. 2B and C).
FIG 2.
NF-κB, but not IRF3, is critical to producing IFN-λ during a TGEV infection. (A) The IRF3 knockdown efficiency was confirmed in IRF3-knockdown cells via Western blotting. (B) The IFN-λ1 and IFN-λ3 expression in IRF3-knockdown cells was analyzed via RT-qPCR after the TGEV infection. (C) The protein levels of IFN-λ1 were analyzed in IRF3-knockdown cells. (D) The effect of the NF-κB inhibitor BAY-11-7082 on NF-κB activation and on TGEV N expression was confirmed via Western blotting. (E) TGEV-induced IFN-λ1 and IFN-λ3 was reduced in a dose-dependent manner after treatment with BAY11-7082. (F) The protein levels of IFN-λ1 were analyzed after IPEC-J2 cells were pretreated with BAY11-7082. (G) The p65 knockdown efficiency was confirmed in the p65-knockdown cells via Western blotting. (H) The expression levels of IFN-λ1 and IFN-λ3 in the p65-knockdown cells were analyzed via RT-qPCR after the TGEV infection. (I) The protein levels of IFN-λ1 were analyzed in p65-knockdown cells. (J) Schematic representation of the mutation of the binding sites for NF-κB (underlined) in the pig IFN-λ1 promoter. IPEC-J2 cells were transfected with empty vector pIFN-λ1 (−500/+10) Luc and its mutation construct for 6 h, and this was followed by either TGEV infection or stimulation with poly (I·C) for 24 h. The cell lysates were then prepared for dual-luciferase reporter assays. (K) Schematic representation of the mutation of the binding sites for NF-κB (underlined) in the pig IFN-λ3 promoter. IPEC-J2 cells were transfected with empty vector pIFN-λ3 (−486/+8) Luc and its mutation construct for 6 h, and this was followed by TGEV infection or stimulation with poly (I·C) for 24 h. The cell lysates were then prepared for dual-luciferase reporter assays.
It has been reported that TGEV infection activates NF-κB (24). Therefore, we explored whether NF-κB is involved in the induction of IFN-λ production during a TGEV infection. As shown in Fig. 2D, treatment with the NF-κB-specific inhibitor BAY11-7082 decreased NF-κB activation in TGEV-infected cells. The production of TGEV-induced IFN-λ1 and IFN-λ3 was reduced in a dose-dependent manner after the inhibition of NF-κB (Fig. 2E and F). The results suggest that the induction of IFN-λ by TGEV infection in IPEC-J2 cells is closely connected with NF-κB. In addition, we also observed that siRNA-mediated NF-κB p65 silence significantly downregulates IFN-λ1 and IFN-λ3 mRNA expression (Fig. 2H). Consistent with these results, the knockdown of p65 by siRNA resulted in decreased IFN-λ1 production in TGEV-infected cells (Fig. 2I). The silencing efficiency of p65 siRNA was determined via Western blotting (65.47% decrease for the no. 1, 81.46% for the no. 2 and 76.49% for the no. 3 siRNA) (Fig. 2G). The mutation of the NF-κB binding site within the IFN-λ1 and IFN-λ3 promoter was prepared to confirm the role of NF-κB in IFN-λ production in IPEC-J2 cells (Fig. 2J and K). Luciferase assays showed that TGEV infection induced high levels of IFN-λ1 and IFN-λ3 promoter luciferase activity. The mutation of the binding sites for NF-κB depressed the level of TGEV-induced IFN-λ1 and IFN-λ3 promoter luciferase activity. These results suggest that NF-κB is key to IFN-λ induction.
The RLR signaling pathway is involved in IFN-λ3 production but not in IFN-λ1 production.
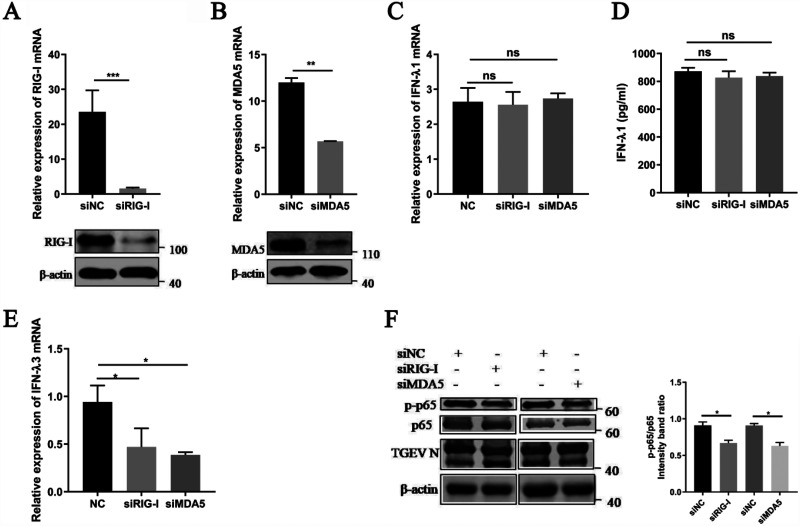
Previous studies demonstrated that TGEV infections activate NF-κB by RLR-mediated signaling (24). As NF-κB is critical to producing IFN-λ during a TGEV infection, TGEV-induced IFN-λ production potentially depends on the RLR pathway. Specific siRNAs targeting RIG-I and MDA5 were synthesized to verify this hypothesis. The mRNA and protein levels of endogenous RIG-I and MDA5 were efficiently knocked down using the synthesized siRNAs (Fig. 3A and B). After validating the successful silence of RIG-I and MDA5 expression with siRNAs, the IPEC-J2 cells were infected with TGEV (MOI = 1) for an additional 24 h. Unexpectedly, no significant change in IFN-λ1 production was detected in the siRIG-I or siMDA5 transfectants, compared with the NC siRNA transfectants (Fig. 3C and D). We then examined the effect of RIG-I and MDA5 knockdown on IFN-λ3 and found that the expression of IFN-λ3 was downregulated after RIG-I knockdown or MDA5 knockdown (Fig. 3E). These results suggested that the RLR signaling pathway is only involved in TGEV-induced IFN-λ3 production. Further, we analyzed the activation of NF-κB after the knockdown of RIG-I or MDA5. The results showed sufficient NF-κB activation, despite RIG-I or MDA5 knockdown (Fig. 3F), indicating that IFN-λ can also be induced by another yet-to-be-determined pathway.
FIG 3.
The RLR signaling pathway is involved in IFN-λ3 production but not in IFN-λ1 production. The IPEC-J2 cells were transfected with 50 nM specific siRNAs targeting RIG-I, MDA5, or negative-control siRNA for 24 h, and the cells were infected with TGEV (MOI = 1). (A) At 24 hpi, cells were collected for the analysis of the RIG-I mRNA levels via RT-qPCR assays and protein expression via Western blotting. (B) Cells transfected with specific siRNAs targeting MDA5 were collected for the analysis of the MDA5 mRNA levels via RT-qPCR assays and protein expression via Western blotting. (C) Cells transfected with specific siRNAs targeting RIG-I or MDA5 were collected for the analysis of IFN-λ1 mRNA levels via RT-qPCR assays. (D) Cell supernatants transfected with specific siRNAs targeting RIG-I or MDA5 were harvested and subjected to ELISA to assess the protein level of IFN-λ1. (E) Cells transfected with specific siRNAs targeting RIG-I or MDA5 were collected for the analysis of the IFN-λ3 mRNA levels via RT-qPCR assays. (F) Western blotting was performed to test p-p65, p65, and TGEV N in RIG-I or MDA5-knockdown cells. β-actin was used as a sample loading control. The ratio of p-p65 to p65 (p-p65/p65) was calculated.
TGEV-derived PERK of ER stress is involved in producing IFN-λ through activating NF-κB.
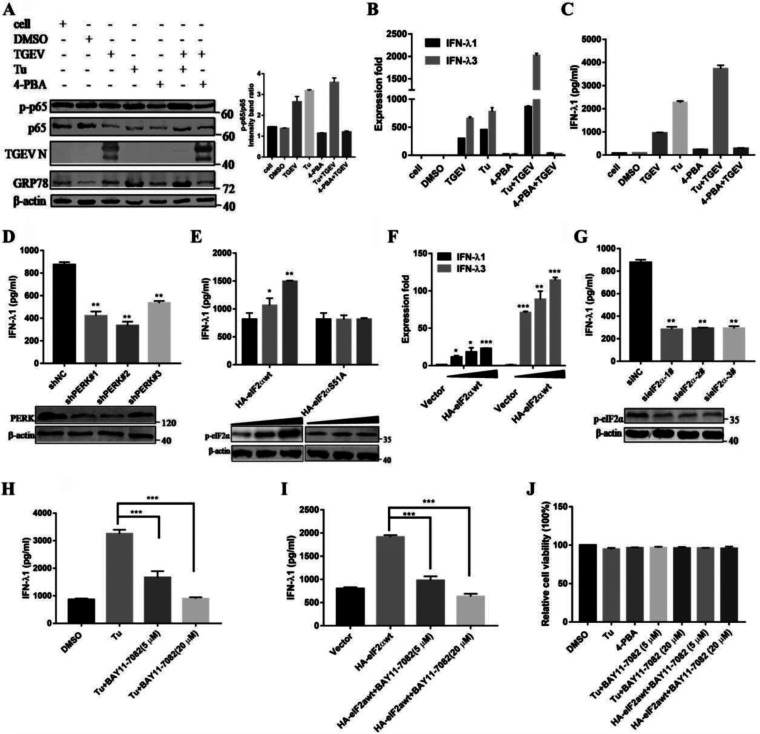
Our previous results have shown that TGEV infection in cultured cells leads to ER stress and activates the PERK signal pathway of the UPR, which is involved in the activation of NF-κB (29). To monitor whether ER stress affects the production of IFN-λ, IPEC-J2 cells were pretreated with the ER stress inducer tunicamycin (Tu) or the ER stress inhibitor 4-phenyl butyric acid (4-PBA), and this was followed either by infection with TGEV (MOI = 1) or by leaving the cells uninfected. After incubation for 2 h, the supernatants were removed and replaced with new cell culture media comprising Tu or 4-PBA for 24 h. The expression of GRP78 indicates the activation of ER stress. As shown in Fig. 4A, compared with DMSO-treated control cells, Tu treatment significantly prompted NF-κB activation in TGEV-infected cells. In contrast to the activation of ER stress, the suppression by 4-PBA treatment decreased NF-κB activation. The expression level of IFN-λ increased after Tu treatment but decreased after 4-PBA treatment. Tu treatment and TGEV infection exhibited a cumulative impact when combined, and the effect on IFN-λ3 was greater than that on IFN-λ1 (Fig. 4B). Consistent with these results, Tu treatment enhanced the protein levels of IFN-λ1 in TGEV-infected IPEC-J2 cells. In striking contrast to Tu, 4-PBA resulted in decreased IFN-λ1 production in TGEV-infected cells (Fig. 4C). These results indicate that ER stress contributes to the production of IFN-λ in response to TGEV infection.
FIG 4.
TGEV-induced, PERK-specific ER stress is critical to producing IFN-λ through activating NF-κB. (A) IPEC-J2 cells were either uninfected or pretreated with Tu (2 μg/mL), 4-PBA (2 mM), or DMSO carrier control for 2 h, and this was followed by infection with TGEV (MOI = 1) and maintainance at that concentration after infection. Western blotting was performed to test the activation of NF-κB and GRP78, as well as the viral infection. β-actin was used as a sample loading control. The ratio of p-p65 to p65 (p-p65/p65) was calculated. (B) At 24 hpi, cells were collected for the analysis of the IFN-λ1 and IFN-λ3 mRNA levels via RT-qPCR assays. (C) Cell supernatants were harvested and subjected to ELISA to assess the protein level of IFN-λ1. (D) IPEC-J2 cells were transfected with shRNA targeting PERK or control shRNA for 24 h, and then the cells were challenged with TGEV. After 24 hpi, the cell supernatants were harvested and subjected to ELISA to assess the protein level of IFN-λ1. The PERK knockdown efficiency was analyzed via Western blotting. (E) IPEC-J2 cells were transfected with HA-eIF2αwt or HA-eIF2αS51A for 24 h and were then infected with TGEV H87. At 24 hpi, the cell supernatants were harvested and subjected to ELISA to assess the protein level of IFN-λ1. The phosphorylation of eIF2α was subjected to Western blotting with antibodies against p-eIF2α. β-actin was used as a loading control. (F) Cells transfected with HA-eIF2αwt were collected for the analysis of the IFN-λ1 and IFN-λ3 mRNA levels via RT-qPCR assays. (G) IPEC-J2 cells were transfected with siRNA of eIF2α for 24 h and were then infected with TGEV H87. After 24 hpi, cell supernatants were harvested and subjected to ELISA to assess the protein level of IFN-λ1. The eIF2α knockdown efficiency was analyzed via Western blotting. (H) IPEC-J2 cells were pretreated with Tu (2 μg/mL) or DMSO carrier control 2 h before infection and maintained at that concentration after infection. Then, cells were treated or nontreated (exposed to equal amounts of DMSO) with 5 or 20 μM BAY11-7082. At 24 hpi, cell supernatants were harvested and subjected to ELISA to assess the protein level of IFN-λ. (I) IPEC-J2 cells were transfected with HA-eIF2αwt for 24 h and then infected with TGEV H87. Then, cells were treated or nontreated (exposed to equal amounts of DMSO) with 5 or 20 μM BAY11-7082. At 24 hpi, the cell supernatants were harvested and subjected to ELISA to assess the protein level of IFN-λ1. (J) Effect of Tu and 4-PBA on cell viability. IPEC-J2 cells were treated with Tu and 4-PBA or Tu plus 5 or 20 μM BAY11-7082 and HA-eIF2αwt plus 5 or 20 μM BAY11-7082. Cell cytotoxicity was analyzed with a CCK-8 system.
Our previous studies have shown that the PERK phosphorylation of eIF2α-mediated TGEV-induced NF-κB activation (29). To identify the effect of the PERK-eIF2α pathway in TGEV-induced IFN-λ production in IEC, we initially silenced the PERK signaling pathway via the shRNA knockdown of PERK. As shown in Fig. 4D, TGEV-induced IFN-λ1 was significantly decreased in PERK knockdown cells. To further elucidate the effect of eIF2α Ser-51 phosphorylation on TGEV-induced IFN-λ production, we examined IFN-λ1 expression in IPEC-J2 cells upon transfection with wild-type eIF2α (HA-eIF2αwt) or with a nonphosphorylatable mutant form of eIF2α that contained Ala instead of Ser at residue 51 (HA-eIF2αS51A). While enhanced IFN-λ1 production occurred in the eIF2αwt cells, no increase in IFN-λ1 expression was shown in the eIF2αS51A cells (Fig. 4E). The upregulation of IFN-λ3 was higher than that of IFN-λ1 in the HA-eIF2αwt overexpressed cells, which is consistent with the results of the Tu treatment in Fig. 4B (Fig. 4F). Furthermore, we monitored the importance of eIF2α phosphorylation to the production of IFN-λ in response to eIF2α siRNA knockdown, which downregulated eIF2α phosphorylation. In contrast to the overexpression of eIF2α, eIF2α silence by siRNA in IPEC-J2 cells reduced IFN-λ1 production (Fig. 4G).
To verify whether the induction of IFN-λ via the PERK signaling pathway of ER stress is dependent on NF-κB, we monitored IFN-λ production after inhibiting NF-κB by BAY11-7082 in IPEC-J2 cells treated with Tu or transfected with HA-eIF2α. The disruption of NF-κB reduced IFN-λ1 production in Tu-treated or HA-eIF2α-overexpressed cells (Fig. 4H and I). The IFN-λ production observed with Tu, 4-PBA, and BAY11-7082 was not due to cytotoxicity, as no obvious cytotoxicity was observed when measuring cell viability with a Cell Counting Kit 8 (CCK-8) (Fig. 4J). Collectively, these results suggested that the TGEV-derived PERK of ER stress is critical to producing IFN-λ in that it depends on NF-κB activation.
TGEV nsp1 induces IFN-λ production.
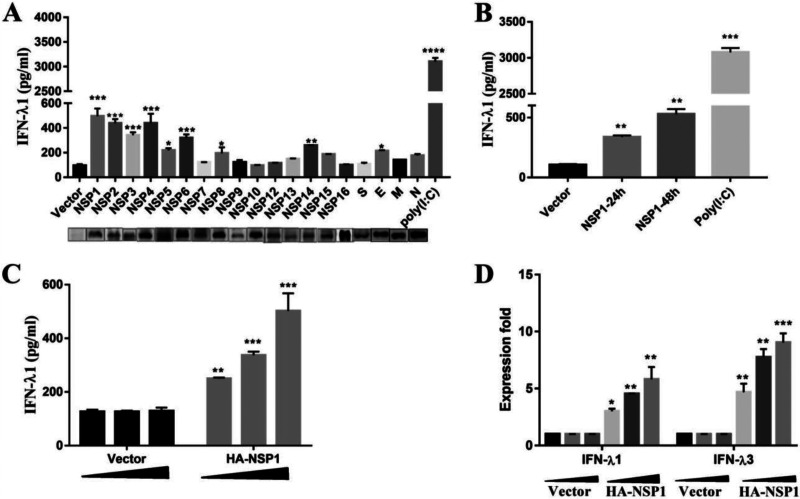
TGEV encodes 4 structural proteins (S, N, E, and M) and 16 nonstructural proteins (nsp1 to 16). To identify the pivotal viral protein(s) involved in IFN-λ induction, IFN-λ1 enzyme-linked immunosorbent assay (ELISA) was applied to screen TGEV-encoded proteins for their relative abilities to induce IFN-λ1 production. IPEC-J2 cells were transfected with different TGEV protein expression plasmids. As shown in Fig. 5A, nsp1 to 6, nsp8, nsp14, and E could induce IFN-λ1 production to various degrees. Of these, nsp1 exhibited the strongest capability for induction.
FIG 5.
TGEV nsp1 induces IFN-λ. (A) IPEC-J2 cells were transfected with various TGEV protein expression vectors. Cell supernatants were subjected to ELISA to assess the production of IFN-λ1. (B) IPEC-J2 cells were transfected with pcaggs or pcaggs-TGEV-nsp1-HA (TGEV nsp1) at 24 h and 48 h posttransfection. Cell supernatants were harvested and subjected to ELISA to assess the production of IFN-λ1. (C and D) IPEC-J2 cells were transfected with different concentrations of pcaggs or pcaggs-TGEV-nsp1-HA (TGEV nsp1). After 24 hpi the cell supernatants were subjected to ELISA to assess the production of IFN-λ1. Cells were harvested and subjected to RT-qPCR to assess the expression of IFN-λ1 and IFN-λ3.
To confirm the capacity of nsp1 to induce IFN-λ production, we detected the protein level of IFN-λ1 induced by nsp1 via ELISA and found that the protein level of IFN-λ1 also increased at different time points (Fig. 5B) and did so in a dose-dependent manner (Fig. 5C) in the IPEC-J2 cells. Then, the expression of IFN-λ1 and IFN-λ3 was tested in nsp1-transfected cells via quantitative reverse transcription polymerase chain reaction (RT-qPCR), and the results verified that TGEV nsp1 induced IFN-λ1 and IFN-λ3 production in a dose-dependent manner (Fig. 5D).
TGEV nsp1 induces IFN-λ production via eIF2α phosphorylation-mediated NF-κB activation.
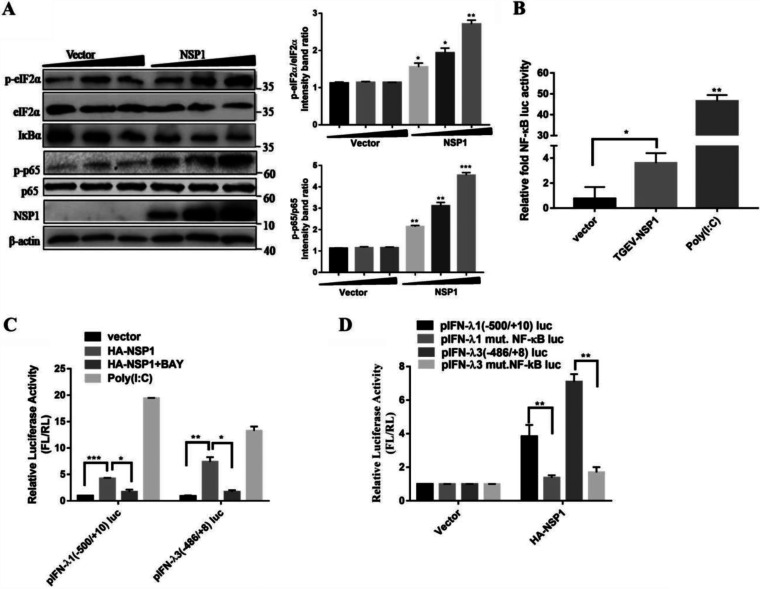
Our studies and others have shown that the phosphorylation of eIF2α by PERK inhibits protein synthesis and contributes to the activation of the NF-κB (29, 32). TGEV nsp1 was reported to inhibit host gene expression, and we explored whether nsp1 can phosphorylate eIF2α and activate NF-κB. Western blotting was conducted to explore whether TGEV nsp1 could induce eIF2α and NF-κB phosphorylation. The results showed that the phosphorylation of eIF2α by nsp1 was dose-dependent. IκBα decreased, whereas the ratio of p-p65/p65 was upregulated, as the amount of nsp1 increased (Fig. 6A). NF-κB reporter assays were carried out in nsp1-transfected cells to confirm the induction of NF-κB by nsp1. NF-κB transcription was upregulated by poly (I·C) stimulation as expected, and nsp1 was shown to upregulate NF-κB activity (Fig. 6B). Altogether, it was concluded that TGEV nsp1 could induce the phosphorylation of eIF2α and activate NF-κB.
FIG 6.
TGEV nsp1 induces IFN-λ via eIF2α phosphorylation-mediated NF-κB activation. (A) IPEC-J2 cells were transfected with different concentrations of pcaggs or pcaggs-TGEV-nsp1-HA (TGEV nsp1). Cell extracts were subjected to Western blotting using an anti-p-eIF2α antibody, anti-eIF2α antibody, anti-HA antibody, anti-IκBα antibody, anti-p-p65 antibody, anti-p65 antibody, or anti-β-actin antibody. The ratios of p-eIF2α/eIF2α and p-p65/p65 were calculated. (B) 293T cells were cotransfected with pNF-κB-Luc (NF-κB luciferase reporter plasmid) and pcaggs or pcaggs-TGEV-nsp1-HA (TGEV nsp1). Cells transfected with poly (I:C) were used as the positive control. At 24 h posttransfection, cell lysates were prepared and subjected to luciferase assays. Error bars show the standard deviations (SDs) of the results from three independent experiments. (C) 293T cells were cotransfected with pIFN-λ1 (−500/+10) Luc or pIFN-λ3 (−486/+8) Luc and pcaggs or pcaggs-TGEV-nsp1-HA (TGEV nsp1). Then, cells were treated or nontreated (exposed to equal amounts of DMSO) with 20 μM BAY11-7082. Cells transfected with poly (I:C) were used as the positive control. At 24 h posttransfection, cell lysates were prepared and subjected to luciferase assays. (D) 293T cells were transfected with pIFN-λ 1(−500/+10)Luc, pIFN-λ1 mut. NF-κB Luc, pIFN-λ3 (−486/+8)Luc, or pIFN-λ3 mut. NF-κB Luc, along with either an empty vector or pcaggs-TGEV-nsp1-HA (TGEV nsp1). At 24 h posttransfection, cell lysates were prepared and subjected to luciferase assays.
To verify whether the TGEV nsp1 induction of IFN-λ is dependent on NF-κB, we monitored the IFN-λ production after inhibiting NF-κB with BAY11-7082 in IPEC-J2 cells transfected with nsp1. The disruption of NF-κB reduced IFN-λ1 and IFN-λ3 production via nsp1 overexpression (Fig. 6C). Luciferase assays further confirmed the importance of activated NF-κB on IFN-λ production. As shown in Fig. 6D, nsp1 expression induced high levels of IFN-λ1 and IFN-λ3. The mutation of the binding sites for NF-κB decreased the level of nsp1-induced IFN-λ1 and IFN-λ3 expression. These results suggest that TGEV nsp1 induced IFN-λ production via eIF2α phosphorylation-mediated NF-κB activation.
TGEV nsp1 85-102aa causes the phosphorylation of eIF2α, activates NF-κB, and induces IFN-λ.
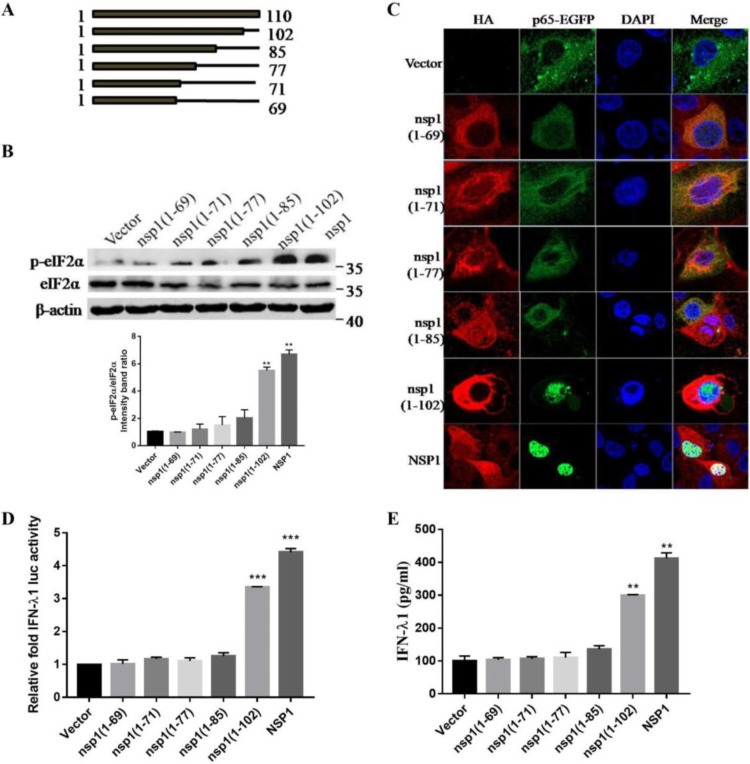
To ascertain which region in TGEV nsp1 was essential for inducing eIF2α phosphorylation, truncated nsp1 was constructed (Fig. 7A). The nsp1 and the C-terminally truncated plasmids nsp1(1-102), nsp1(1-85), nsp1(1-77), nsp1(1-71), and nsp1(1-69) were transfected into cells, and the phosphorylation of eIF2α was tested via Western blotting. The results showed that nsp1(1-102) and nsp1-transfected cells induced a higher level of eIF2α phosphorylation than did nsp1(1-85), nsp1(1-77), nsp1(1-71), and nsp1(1-69) (Fig. 7B). These results indicated that residues 85 to 102 might be involved in inducing eIF2α phosphorylation.
FIG 7.
TGEV nsp1 (amino acids 85 to 102) is important in activating NF-κB and in inducing IFN-λ production. (A) The truncations (at the C terminus) of TGEV nsp1 are shown below the bars. (B) IPEC-J2 cells were transfected with TGEV nsp1 truncation mutants. At 24 h posttransfection, cell extracts were subjected to Western blotting, using an anti-p-eIF2α antibody, anti-eIF2α antibody, or anti-β-actin antibody. The ratio of p-eIF2α to eIF2α (p-eIF2α/eIF2α) was calculated. (C) IPEC-J2 cells were cotransfected with the truncation mutants of TGEV nsp1 and pEGFP-p65. The location of NF-κB p65 was detected via immunofluorescence and was observed under laser confocal microscopy. (D) 293T cells were cotransfected with pIFN-λ1 (−500/+10) Luc and pcaggs or with the truncation mutants of TGEV nsp1. At 24 h posttransfection, the cell lysates were prepared and subjected to luciferase assays. (E) IPEC-J2 cells were transfected with the truncation mutants of TGEV nsp1. At 24 h posttransfection, cell supernatants were harvested and subjected to ELISA to assess the production of IFN-λ1.
Next, we investigated whether this motif (amino acids 85 to 102) plays a key role in activating NF-κB. To this end, nsp1 or its truncated genes were cotransfected with EGFP-p65, and the nuclear translocation of p65 was examined. The results showed that p65 was distributed diffusely throughout the cytoplasm in the vector transfected cells and in the nsp1(1-85), nsp1(1-77), nsp1(1-71), and nsp1 (1-69) transfected cells. When the cells were transfected with nsp1(1-102) and nsp1, the nuclear staining of p65 was evident, confirming that the motif comprised of amino acids 85 to 102 of TGEV nsp1 is important in inducing p65 nuclear translocation (Fig. 7C).
To further determine the specific amino acid residues for nsp1-mediated IFN-λ induction, reporter assays were performed using the IFN-λ1 luciferase constructs. HEK-293T cells were cotransfected with individual nsp1 constructs accompanied by pIFN-λ1 (−500/+10) Luc. Among the nsp1 truncation mutants, nsp1(1-102) and full-length nsp1 appeared to induce IFN-λ1 expression (Fig. 7D). Subsequently, an ELISA analysis indicated that nsp1(1-102) and nsp1 induced IFN-λ1 to a significantly higher level than did the nsp1(1-85), nsp1(1-77), nsp1(1-71) and nsp1(1-69) transfected cells (Fig. 7E), again demonstrating that the motif containing amino acids 85 to 102 was a key region for the induction of IFN-λ production. Taken together, these results indicated that the motif containing amino acids 85 to 102 of nsp1 plays vital roles in activating NF-κB and in inducing IFN-λ.
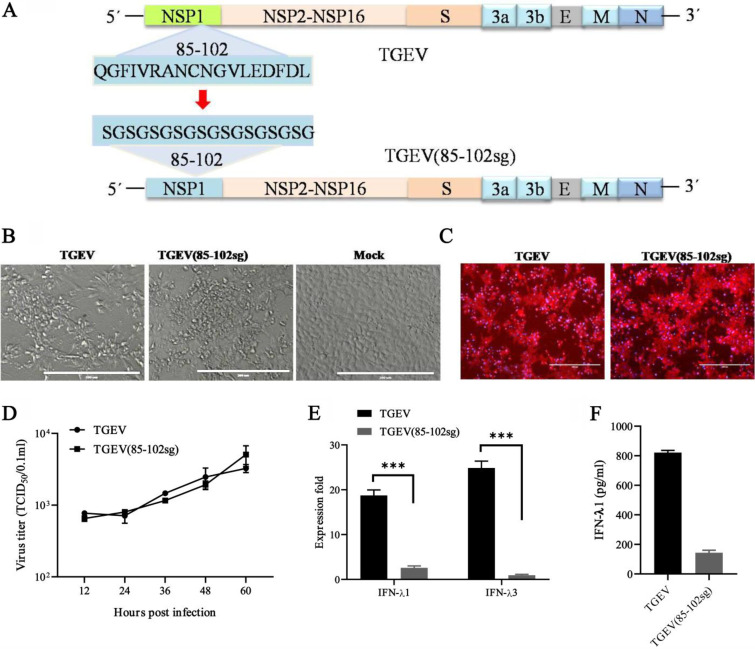
To further assess whether nsp1 (85-102) is attributed to the induction of IFN-λ, we utilized a reverse genetics system to engineer an infectious clone of recombinant TGEV with the replacement of amino acids 85 to 102 in the nsp1-coding sequence (Fig. 8A). Both the wild-type (WT) and the nsp1 (85-102sg) mutants produced live virus progeny and showed similar growth dynamics in ST cells (Fig. 8A, B, and C). However, the cells infected with WT rTGEV produced more than 8-fold higher IFN-λ1 and 20-fold higher IFN-λ3 transcripts than did the nsp1 (85-102sg) mutants (Fig. 8D). Moreover, the concentration of IFN-λ1 was also lower in the supernatants of cells infected with nsp1 (85-102sg) mutants. Altogether, nsp1 (85-102sg) mutants have the capacities for replication, presenting similar growth dynamics to those of the WT virus, and for the induction of a decreased IFN-λ response. These results support our observed correlation between nsp1 and the induction of the IFN-λ response that occurs following a TGEV infection.
FIG 8.
The nsp1 (85-102) mutants (TGEV [85-102sg]) induced lower IFN-λ responses. (A) DNA-based reverse genetics BAC system for TGEV and TGEV (85-102sg). The amino acid replacements in position 85 to 102 of NSP1 are indicated. (B and C) The recombinant virus was successfully used to infect ST cells. TGEV-BAC and TGEV (85-102sg)-BAC were transfected into ST cells with Lipo3000, and then the virus was passaged once in ST cells. The CPE was observed after 36 h (B), and the indirect immunofluorescence assay was used to detect the TGEV N protein (C). (D) Multistep growth curves for TGEV and TGEV (85-102sg) in ST cells at a MOI of 1. The virus titers at different time points, as indicated, were determined via an endpoint dilution assay. The data are represented as the mean ± the standard deviation, with n = 3. (E) TGEV (85-102sg) induced lower IFN-λ1 and IFN-λ3 expression than did WT TGEV in the IPEC-J2 cells. (F) The IFN-λ1 production of the cells infected with WT TGEV and TGEV (85-102sg) was confirmed via ELISA.
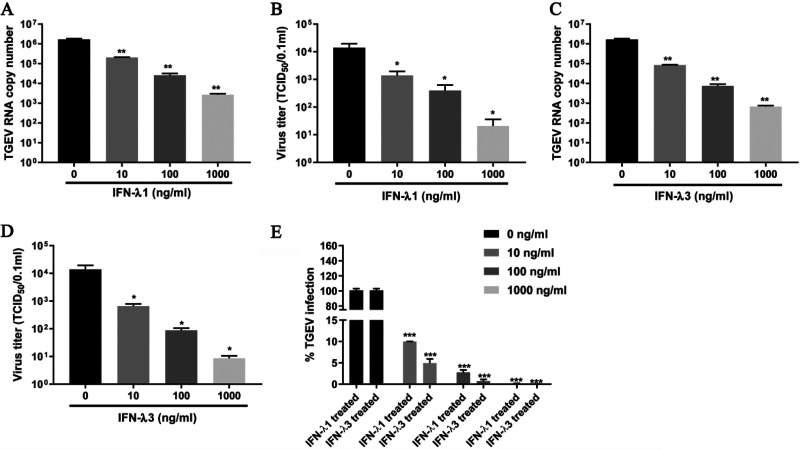
IFN-λ3 exhibits greater antiviral activity against TGEV than does IFN-λ1.
To investigate the role of IFN-λ in TGEV replication, we detected the effects of rpIFN-λ1 and rpIFN-λ3 treatment on TGEV replication. The rpIFN-λ1 treatment decreased the TGEV virus genomes and the progeny virus titers by up to 7.3 to 636-fold, compared with no treatment, as assessed by the quantification of the viral RNA and measurement of the viral titers, respectively (Fig. 9A and B). The rpIFN-λ3 treatment decreased the TGEV virus genomes and the progeny virus titers by up to 17.7 to 2,546-fold, compared with no treatment (Fig. 9C and D). The TGEV inhibition by the rpIFN-λ treatment manifested in a dose-dependent manner, and rpIFN-λ3 exhibited 2.1 to 4.6-fold greater antiviral activity against the TGEV infection of IPEC-J2 cells than that exhibited by rpIFN-λ1 (Fig. 9E). Collectively, these results demonstrate that IFN-λ3 preferentially inhibits enteropathogenic TGEV in IPEC-J2, compared with IFN-λ1.
FIG 9.
IFN-λ3 exhibits greater antiviral activity against TGEV than that of IFN-λ1. (A and B) IPEC-J2 cells were pretreated with the indicated concentration of rpIFN-λ1 2 h before infection and were maintained at that concentration after infection. TGEV infection was determined at 24 hpi via either RT-qPCR (A) or titration (B). (C and D) IPEC-J2 cells were pretreated with the indicated concentration of rpIFN-λ3 2 h before infection and were maintained at that concentration after infection. TGEV infection was determined at 24 hpi via either RT-qPCR (C) or titration (D). (E) The infection percentage of TGEV treated with rpIFN-λ1 and rpIFN-λ3 was calculated.
DISCUSSION
TGEV is an enteric coronavirus that contacts highly sensitive intestinal epithelial cells after infection, thereby triggering the immune response (33–35). Here, we describe the potential mechanisms of the antiviral host defense of IFN-λ against TGEV infections in intestinal epithelial cells. This study initially demonstrates that infection by TGEV results in the predominant production of IFN-λ3 over IFN-λ1. Notably, the classical RLR pathway is involved in IFN-λ3 production, but not in IFN-λ1 production, in IPEC-J2 cells (Fig. 3), suggesting distinct regulation mechanisms between IFN-λ1 and IFN-λ3. A previous report showed that porcine cells treated with dsRNA, the viral mimic, intensely increased IFN-λ3 expression in IPEC-J2 cells (7). It is unknown which receptor-mediated signaling pathway is mainly responsible for the induction of IFN-λ3 in porcine epithelial cells. Our results indicated that the RIG-I and MDA5 pathways are involved in the regulation of IFN-λ3, but not IFN-λ1, in IPEC-J2 cells. As the synthesized dsRNA is a ligand of some kinds of innate immune receptors, including RIG-I and MDA5, the results explain that dsRNA induces the production of high levels of IFN-λ3.
IRF3 and NF-κB are the key regulators of type I IFN expression, and it is known that TGEV infection leads to IRF3 and NF-κB activation (24). Hence, we investigated whether IRF3 and/or NF-κB were involved in IFN-λ production in TGEV-infected IPEC-J2 cells. Interestingly, IRF3 is not related to the induction of IFN-λ. Except for the signaling pathways mediated by RIG-I and MDA5, TGEV induces IFN-λ via a novel, developed mechanism of eIF2α kinase-mediated IκB degradation and consequent NF-κB activation. NF-κB induction by the ER stress pathway also occurs via the hepatitis C virus (HCV) and the human coronavirus 229E (HCoV-229E) (36, 37). For HCV, NS4B induces ER overload response-dependent NF-κB activation (36). Both classical and alternative NF-κB activation are involved in the induction of IFN-λ3, which explains the fact that TGEV infection induces higher levels of IFN-λ3 than IFN-λ1. This is the first report that TGEV regulates IFN-λ1 and IFN-λ3 expression by distinct mechanisms.
Coronavirus nsp1 is one of the most diverse viral proteins among the four coronavirus genera (13). Only Alphacoronavirus and Betacoronavirus encode nsp1, whereas Gammacoronavirus and Deltacoronavirus lack nsp1 (13). The biological functions of coronavirus nsp1 include the regulation of type I IFN production and signaling, the induction of host mRNA degradation, and the inhibition of host protein synthesis (38). PEDV nsp1 depresses the phosphorylation and subsequent degradation of IκBα and blocks the translocation of p65 to the nucleus (39). SARS-CoV nsp1 plays an important role in the induction of chemokines in the lung epithelial cells of humans through the activation of NF-κB (40). Recently, the genomic monitoring of SARS-CoV-2 uncovered an nsp1 deletion variant (Δ500-532) that showed a reduced IFN-I response and that nsp1 caused the greatest increase in IL-6 secretion among all of the SARS-CoV-2 proteins (41, 42). Similar to SARS-CoV-2, TGEV nsp1 also potently mediated IFN-λ induction through NF-κB. A previous study reported that, with the exceptions of nsp7 and ORF3b, all of the TGEV-encoded proteins could activate the NF-κB signaling pathway to various extents (43). Although they found that TGEV nsp1 could also activate NF-κB, they did not explore it further. Our study showed that TGEV nsp1 could induce the phosphorylation of eIF2α and could activate NF-κB. A previous study showed that TGEV nsp1 does not inhibit protein translation in rabbit reticulocyte lysate but does in mammalian cells and in cell-free HeLa cell extracts (44). They hypothesized that TGEV nsp1 might activate the heme-regulated eIF2α kinase and induce eIF2α phosphorylation, leading to the inhibition of translation in HeLa cell extracts. However, no significant differences were observed in eIF2α phosphorylation in HeLa cell extracts that were incubated with purified TGEV nsp1 proteins from Escherichia coli. Here, we demonstrated that the eukaryotic expressed TGEV nsp1 induces the phosphorylation of eIF2α at S51 in a dose-dependent manner in IPEC-J2 cells (Fig. 6A). The biological activities of the TGEV nsp1 protein may be influenced in prokaryotic cells or by the host cell types and species. That is, the eukaryotic expressed TGEV nsp1 protein may efficiently induce the phosphorylation of eIF2α in certain types of cells.
The nsp1 size of Alphacoronavirus is different from that of Betacoronavirus nsp1, and they share no significant sequence similarity (14). Both SARS-CoV and murine hepatitis virus (MHV) nsp1 are localized only in the cytoplasm of virus-infected cells. The subcellular distribution of transiently expressed TGEV nsp1 in IPEC-J2 cells was analyzed via confocal microscopy. It was found that TGEV nsp1 was located in both the cytoplasm and the nucleus (Fig. 7C). SARS-CoV nsp1 extensively inhibits host gene expression, including the internal reference protein β-actin. However, TGEV nsp1 could not inhibit β-actin expression, suggesting that the inhibition of protein translation by TGEV nsp1 may not be universal (Fig. 6A and 7B). In addition, SARS-CoV nsp1 suppressed the expression of its own genes, whereas TGEV nsp1 had only a slight impact on its own gene expression (Fig. 6A), suggesting that the mechanisms of the inhibition of TGEV nsp1-mediated host gene expression are different from those of the SARS CoV nsp1 protein. Unlike SARS-CoV nsp1, the nsp1 protein of TGEV could not bind 40S ribosomal subunits or facilitate the degradation of the host mRNA (44). Increased eIF2α phosphorylation at S51 results in the global inhibition of protein synthesis and improves the translation of selective mRNAs that encode proteins that regulate cell adaptation to stress. The underlying mechanism might be that TGEV nsp1 inhibits protein translation through the phosphorylation of eIF2α.
IFN-λ is a functionally nonredundant ingredient of the mucosal antiviral innate immune system (2, 3). PEG-rIFN-λ1 treatment improved survival, reduced graft-versus-host disease severity, and enhanced epithelial proliferation after allogeneic bone marrow transplantation (45). During the development and progression of herpetic stromal keratitis (SK), exogenous local rIFN-λ treatment may be a new method by which to control HSV-1-induced inflammation and the related vision impairment (46). Among the most studied IFN-λs, human IFN-λ3 has the highest antiviral activity in an in vitro model of EMCV infection (47). Similarly, we found that rpIFN-λ3 exhibited more anti-TGEV activity than did rpIFN-λ1 in IPEC-J2 cells. Since the less adverse effect-prone IFN-λs are good candidates for the management of COVID-19 (48), IFN-λ3, with its higher antiviral activity, can be screened as an effective drug against coronaviruses.
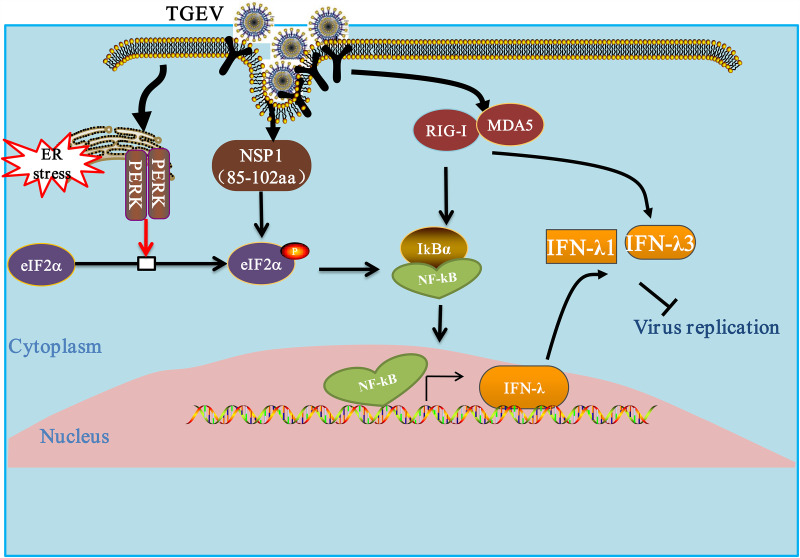
Collectively, our results are unique in that they unveil the distinct regulatory mechanisms of TGEV on IFN-λ1 and IFN-λ3 (Fig. 10). In addition to the classical NF-κB signaling pathway, an alternative NF-κB pathway plays a key role in the induction of IFN-λ. Although IFN-λ1 and IFN-λ3 are important ingredients of the innate immune defense, our data suggest that rpIFN-λ3 showed stronger antiviral activity against TGEV than did rpIFN-λ1. IFN-λ3, with its stronger antiviral ability, might be a potent therapeutic target with which to protect the intestinal epithelium from viral infection.
FIG 10.
Modulation of the IFN-λ response by TGEV. TGEV infection leads to the phosphorylation of initiation factor eIF2α through the action of the kinase PERK. Once active, eIF2α inhibits translation initiation, thereby reducing the synthesis of IκBα. The reduction in IκBα leads to the activation of NF-κB and binds DNA to promote IFN-λ1 and IFN-λ3 expression. TGEV nsp1 leads to the phosphorylation of eIF2α, which in turn suppresses IκBs. The reduced IκB synthesis reduces the inhibition of NF-κB and increases nuclear NF-κB and IFN-λ1 and IFN-λ3 expression. The RLR signaling pathway is involved in TGEV-induced IFN-λ3 production but not in IFN-λ1 production. The arrows indicate activation, and the blunt-ended lines indicate inhibition.
MATERIALS AND METHODS
Cells and viruses.
The porcine small intestinal epithelial cell line (IPEC-J2) was grown in Dulbecco’s modified Eagle’s medium: nutrient mixture F-12 (Ham) (1:1) (DMEM:F12) (Gibco) containing 10% fetal bovine serum (FBS), 5 ng/mL epidermal growth factor (Gibco), 5 μg/mL insulin-transferring-selenium supplements (Gibco), and 1% penicillin-streptomycin. Swine testicular cell line (ST cells) and HEK-293T (ATCC) were maintained in Gibco DMEM (high glucose, 4.5 g/L) containing 10% FBS at 37°C with 5% CO2. The TGEV H87 strain was propagated in ST cells and preserved in our laboratory.
Cloning and construction of plasmids.
Plasmids expressing TEGV structural and nonstructural proteins were inserted into the pcaggs vector with a hemagglutinin (HA) tag at the N terminus. The nsp1 truncated fragment was designed according to the tertiary structure of TGEV nsp1 to determine the nsp1 functional area. The C-terminally truncated constructions included nsp1(1-102), nsp1(1-85), nsp1(1-77), nsp1(1-71), and nsp1(1-69). The expression of each viral protein was validated via Western blotting.
The full-length infectious cDNA clone of TGEV was constructed based on a bacterial artificial chromosome. To construct the recombinant virus containing mutant nsp1, the clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated protein 9 (Cas9) systems were used as reported previously (49). The primers of sgRNA were synthesized as shown in Table 1, using a similar method described in a previous report (50).
TABLE 1.
Sequences of the primers used for CRISPR/Cas9
| Target | Sense strand sequence (5′–3′)a |
|---|---|
| sgnsp1 (85-102) F | GATCACTAATACGACTCACTATATcatgtttgacagcttatcatGTTTTAGAGCTAGAAA |
| sgnsp1 (85-102) R | GATCACTAATACGACTCACTATATggtgtcactttgggtgatatGTTTTAGAGCTAGAAA |
| sgNSP1R | AAAAGCACCGACTCGGTGCCACTTTTTCAAGTTGATAACGGACTAGC CTTATTTTAACTTGCTATTTCTAGCTCTAAAAC |
The T7 promoter sequence is underlined, and the guide sequence for targeting transcription template DNA is shown in lower case. The black sequence of sgnsp1 (85-102) F/sgnsp1 (85-102) R are the regions that overlap the sgNSP1R sequence. SgNSP1R is a constant primer used for transcription template DNA.
The promoter region of pig ifn-λ1 and ifn-λ3 was amplified by genomic PCR (upper primer, 5′-CGGGGTACCTATCTCAACATAATAAAGGCCAC-3′ for ifn-λ1 and 5′-GGGGTACCAGGTCAGAGACGCCTGACAAG-3′ for ifn-λ3; lower primer, 5′-CCCAAGCTTTGCGTGTTGGTTTTGTATCCTGC-3′ for ifn-λ1 and 5′-CCCAAGCTTCAGGGCCATGTCTGTGCCA-3′ for ifn-λ3), and the PCR fragment was inserted into the KpnI-HindIII site of a luciferase vector (pIFN-λ1 [−500/+10] Luc and pIFN-λ3 [−486/+8] Luc). The mutants (pIFN-λ1 mut. NF-κB Luc and pIFN-λ3 mut. NF-κB Luc) were constructed using site-directed mutagenesis to introduce the NF-κB substitution. All of the recombinant expression plasmids were verified via sequencing. A Dual-Luciferase Reporter Assay System (Promega) was used for the luciferase assays. The Renilla luciferase construct pRL-TK (Promega) was used as an internal control for the dual-luciferase assay.
Cell culture, virus infection, and treatments with chemicals.
IPEC-J2 cells were either infected with appropriately diluted viruses or mock-infected. Two hours later, virus cultures were removed, and the cells were washed with PBS three times and maintained with DMEM supplemented with 0.3% trypsin (0.25%; Gibco) and 1% DMSO at 37°C.
Tu (Sigma), 4-PBA (Sigma), and BAY11-7082 (a specific and potent inhibitor of NF-κB activation; Selleckchem) were dissolved in DMSO. The rpIFN-λ1 and rpIFN-λ3 were provided by PrositeSole Biotechnology (Beijing, China). IPEC-J2 cells were pretreated with various concentrations of chemicals or with the same volume of DMSO, and this was followed by inoculation with TGEV H87 (MOI = 1). After incubation for 2 h, the supernatant was removed and replaced with cell culture media containing various doses of the chemicals. Cells were collected at the indicated times postinfection and then subjected to SDS-PAGE and Western blotting. Supernatants were harvested at 24 hpi for viral titration or were subjected to ELISA to assess the protein level of IFN-λ1.
Experimental infection of piglets.
12 two-day-old, specific pathogen-free (SPF) piglets were divided into two groups by random and were orally inoculated with 5 mL of either viruses at 1 × 105 TCID50 or DMEM, respectively. The clinical signs and the symptoms of the piglets were recorded daily after the virus infection. At the end of the study, all of the piglets were euthanized at 48 hpi. The Animal Ethics Committee approval number is Heilongjiang-SYXK-2006-032. The intestine tissue of the piglets was collected and tested as previously described (29).
SDS-PAGE and Western blotting.
Cell samples were collected to determine the success of plasmid transfection or the effect of the chemical treatment in the cells. Cells were lysed with NP-40 lysis buffer (Beyotime Biotechnology) supplemented with a protease inhibitor cocktail (Roche Molecular Biochemicals) for 30 min. Equal amounts of total protein boiled with 5× SDS loading buffer (P0015L, Beyotime) for 10 min were separated on 10% SDS-PAGE gels and transferred onto polyvinylidene fluoride (PVDF) membranes. First, the membrane was blocked in 5% (wt/vol) skim milk for 2 h at room temperature and incubated with monoclonal or polyclonal antibodies overnight at 4°C. Antibodies against NF-κB P65 (L8F6), phospho-NF-κB P65 (P-P65, 93H1), and IκBα (L35A5) were purchased from Cell Signaling Technology. The antibody against IRF3 (A11118) was purchased from Abclonal. Antibodies against RIG-I and MDA5 were produced in rabbits immunized with purified, recombinant target protein. Antibodies against GRP78, PERK, p-eIF2α, and eIF2α were described previously (29). Subsequently, after being washed three times (10 min each) with TBST, the membrane was incubated with Alexa Fluor 680 rabbit anti-goat IgG, Alexa Fluor 680 goat anti-rabbit IgG, or IRDy800 labeled anti-mouse IgG (Invitrogen, 1:10,000-diluted) in a blocking buffer solution at room temperature for 1 h. Finally, after TBST cleaning, the membrane was scanned in an Odyssey Infrared Imaging System (LI-COR Biosciences).
RNA interference.
The siRNA targeting IRF3, RIG-I, MDA5, p65, and eIF2α were designed by Genepharma Company (Shanghai, China). The sequences of the RNA oligonucleotides are listed in Table 2. 1 × 106 IPEC-J2 cells were seeded into 6-well plates and were incubated overnight. The lipofectamine 2000 transfection reagent (Invitrogen) was used to transfect siIRF3, siRIG-I, siMDA5, sieIF2α, sip65, and nontarget control siRNA, according to the manufacturer’s instructions. Cells were then infected with TGEV at 24 h posttransfection and were harvested at the indicated time points for virus titration and protein expression analysis.
TABLE 2.
Sequences of the sense strand of siRNA used to ablate p65, eIF2α, RIG-I, and MDA5 protein expression in the IPEC-J2 cells
| Target | Sense strand sequence (5′–3′) |
|---|---|
| p65-siRNA | CCUGGAACAGGCCAUCAAUTTCCCUAUCCCUUUACGCCAUTTCCAUUGCGGACAUGGACUUTT |
| eIF2α-siRNA | GGAAUACAACAACAUCGAAGGGCAGAUAUUGAAGTGGCUUGUGCCCAAAGUGGUUACAGAUAC |
| PERK-shRNA | CGACAACCCGAAUUACAACAAAGGUCUAGGGAGCGAACCUCCCUGCAGAUUGUGGAGGCGGUA |
| IRF3-siRNA | GGAAAGAAGCAUUGCGUUUTTUCUGAUUGCCUUCAUCGAATTGCACAUUUCCAACAGCCACTT |
| RIG-I-siRNA | CCAUAACUCUUGGAGGCUUTT |
| MDA5-siRNA | GCACUUGCCCGCGAAUUAATT |
| Control-siRNA | UUCUCCGAACGUGUCACGUTT |
RNA isolation and real-time quantitative RT-PCR.
The cellular total RNA from the IPEC-J2 cells that underwent various treatments was extracted using the RNeasy Mini Kit (Qiagen Sciences). The cDNA templates were synthesized from the purified RNA using the PrimeScript II 1st Strand cDNA Synthesis Kit (TaKaRa). Quantitative real-time PCR (qPCR) was conducted in triplicate using Power SYBR Green PCR Master Mix (TaKaRa) with LightCycler 480 real-time PCR machines (Roche Molecular Biochemicals). The primers were designed using Oligo 6 software, and the sequences are shown in Table 3. The results were analyzed using the cycle threshold (ΔΔCT) method with the LightCycler 480 software 1.5. GAPDH served as the internal control.
TABLE 3.
Real-time PCR primers used in this study
| Target | Sequence |
|---|---|
| IFN-λ1 | Forward: 5′-CCACGTCGAACTTCAGGCTT-3′ |
| Reverse: 5′-ATGTGCAAGTCTCCACTGGT-3′ | |
| IFN-λ3 | Forward: 5′-AACTCATCCCTGCACAGCAGC-3′ |
| Reverse: 5′-CAGCTCTGGGGCTCCTTCTTCT-3′ | |
| RIG-I | Forward: 5′-AGAGCAGCGGCGGAATC-3′ |
| Reverse: 5′-GGCCATGTAGCTCAGGATGAA-3′ | |
| MDA5 | Forward: 5′-TCTCCGGGAAACAGGCAAC-3′ |
| Reverse: 5′-CAAAGGATGGAGAGGGCAAGT-3′ | |
| TGEV-S | Forward: 5′-GCTTGATGAATTGAGTGCTGATG-3′ |
| Reverse: 5′-CCTAACCTCGGCTTGTCTGG-3′ | |
| GAPDH | Forward: 5′-CCTTCCGTGTCCCTACTGCCAAC-3′ |
| Reverse: 5′-GACGCCTGCTTCACCACCTTCT-3′ |
The S gene of TGEV was used as the standard with which to evaluate the TGEV replication. The total viral RNA was isolated, and qPCR was conducted as described above. The primer sequences based on the TGEV S gene are listed in Table 3.
Virus titration.
IPEC-J2 cells were treated with various concentrations of IFN-λ or the DMSO control and were infected with TGEV for 24 h. At 24 hpi, the infected cell supernatants were harvested and serially diluted 10-fold from 10−1 to 10−10. The diluents were then added to confluent ST cells in a flat-bottomed 96-well plate (Costar). A cytopathic effect (CPE) and TGEV inhibition by IFN-λ were observed after 72 hpi. The titers of the virus were calculated using the Reed-Muench method and were expressed as TCID50/0.1 mL.
Indirect immunofluorescence (IFA).
At 24 h posttransfection, IPEC-J2 cells were fixed with 4% paraformaldehyde for 30 min and permeabilized with 0.2% Triton X-100 in PBS for 15 min at room temperature. After being washed with PBS three times, the cells were blocked with 5% nonfat dried milk in PBS for 30 min at 37°C. The samples were incubated with anti-NF-κB p65 antibody (Cell Signaling Technology) for 2 h at 37°C and treated with Alexa Fluor 546 goat anti-rabbit IgG (Thermo Fisher Scientific) for 1 h. Finally, the cells were treated with DAPI (4′,6-diamidino-2-phenylindole) for 10 min at room temperature. The samples were visualized after three rinses using an Evos FL Auto2 fluorescence microscope.
Monitoring IFN-λ1 production via ELISA.
IFN-λ1 produced in cells was detected using an optimized indirect ELISA method. Briefly, 96-well microtiter plates (Greiner, Germany) were coated with 0.2 μg rabbit anti-IFN-λ1 polyclonal antibody (PrositeSole Biotechnology, Beijing, China) in 100 μL/well of coating buffer (0.05 M carbonate-bicarbonate, pH 9.6). After incubating overnight at 4°C, the plates were washed with PBS containing 0.05% (vol/vol) Tween 20 (PBS-Tween) and blocked with 5% nonfat dried milk/PBST for 2 h at room temperature. The cell supernatant (diluted 1:2) and the IFN-λ1 protein (as a standard protein sample) were added to wells in duplicate and incubated for 2 h at 37°C. A blocking buffer was used as a blank. The plates were washed, and 100 μL mouse anti-IFN-λ1 monoclonal antibody (1 μg/mL in PBST; PrositeSole Biotechnology, Beijing, China) were added per well and incubated for 1 h at 37°C. The wells were washed with wash buffer and incubated with 100 μL of a 1/5,000 dilution of horseradish peroxidase (HRP)-conjugated anti-mouse IgG for 1 h (Sigma). After three additional washes, 100 μL of tetramethylbenzidine (TMB) substrate were added and incubated in the dark for 30 min. The enzyme reaction was terminated by the addition of 2 M sulfuric acid after 30 min, and the absorbance was measured at a wavelength of 450 nm.
Cell viability measurement.
Cell viability was tested using a CCK-8 assay (Beyotime, Hangzhou, China), according to the manufacturer’s instructions.
Statistical analysis.
All of the data in the figures are expressed as the mean ± the standard deviation (SD) from three independent experiments. Unpaired t tests were carried out using the GraphPad Prism software package (version 9.0). A P value of <0.05 was considered to be indicative of a statistically significant result. P values are indicated as follows: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
ACKNOWLEDGMENTS
This work was supported by the National Key Research and Development Program of China (no. 2021YFD1801103) and the Natural Science Foundation of Heilongjiang Province (no. TD2020C002).
We declare no conflict of interest.
Contributor Information
Pinghuang Liu, Email: liupinghuang@cau.edu.cn.
Li Feng, Email: fengli_h@163.com.
Tom Gallagher, Loyola University Chicago.
REFERENCES
- 1.Lazear HM, Nice TJ, Diamond MS. 2015. Interferon-lambda: immune functions at barrier surfaces and beyond. Immunity 43:15–28. 10.1016/j.immuni.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mordstein M, Neugebauer E, Ditt V, Jessen B, Rieger T, Falcone V, Sorgeloos F, Ehl S, Mayer D, Kochs G, Schwemmle M, Gunther S, Drosten C, Michiels T, Staeheli P. 2010. Lambda interferon renders epithelial cells of the respiratory and gastrointestinal tracts resistant to viral infections. J Virol 84:5670–5677. 10.1128/JVI.00272-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pott J, Mahlakoiv T, Mordstein M, Duerr CU, Michiels T, Stockinger S, Staeheli P, Hornef MW. 2011. IFN-lambda determines the intestinal epithelial antiviral host defense. Proc Natl Acad Sci USA 108:7944–7949. 10.1073/pnas.1100552108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou JH, Wang YN, Chang QY, Ma P, Hu Y, Cao X. 2018. Type III interferons in viral infection and antiviral immunity. Cell Physiol Biochem 51:173–185. 10.1159/000495172. [DOI] [PubMed] [Google Scholar]
- 5.Sommereyns C, Paul S, Staeheli P, Michiels T. 2008. IFN-lambda (IFN-lambda) is expressed in a tissue-dependent fashion and primarily acts on epithelial cells in vivo. PLoS Pathog 4:e1000017. 10.1371/journal.ppat.1000017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dellgren C, Gad HH, Hamming OJ, Melchjorsen J, Hartmann R. 2009. Human interferon-lambda3 is a potent member of the type III interferon family. Genes Immun 10:125–131. 10.1038/gene.2008.87. [DOI] [PubMed] [Google Scholar]
- 7.Sang Y, Rowland RR, Blecha F. 2010. Molecular characterization and antiviral analyses of porcine type III interferons. J Interferon Cytokine Res 30:801–807. 10.1089/jir.2010.0016. [DOI] [PubMed] [Google Scholar]
- 8.Wang D, Fang L, Zhao F, Luo R, Chen H, Xiao S. 2011. Molecular cloning, expression and antiviral activity of porcine interleukin-29 (poIL-29). Dev Comp Immunol 35:378–384. 10.1016/j.dci.2010.11.003. [DOI] [PubMed] [Google Scholar]
- 9.van Nieuwstadt AP, Zetstra T, Boonstra J. 1989. Infection with porcine respiratory coronavirus does not fully protect pigs against intestinal transmissible gastroenteritis virus. Vet Rec 125:58–60. 10.1136/vr.125.3.58. [DOI] [PubMed] [Google Scholar]
- 10.Cruz JL, Sola I, Becares M, Alberca B, Plana J, Enjuanes L, Zuniga S. 2011. Coronavirus gene 7 counteracts host defenses and modulates virus virulence. PLoS Pathog 7:e1002090. 10.1371/journal.ppat.1002090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weiss SR, Navas-Martin S. 2005. Coronavirus pathogenesis and the emerging pathogen severe acute respiratory syndrome coronavirus. Microbiol Mol Biol Rev 69:635–664. 10.1128/MMBR.69.4.635-664.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weiwei H, Qinghua Y, Liqi Z, Haofei L, Shanshan Z, Qi G, Kongwang H, Qian Y. 2014. Complete genomic sequence of the coronavirus transmissible gastroenteritis virus SHXB isolated in China. Arch Virol 159:2295–2302. 10.1007/s00705-014-2080-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Narayanan K, Ramirez SI, Lokugamage KG, Makino S. 2015. Coronavirus nonstructural protein 1: common and distinct functions in the regulation of host and viral gene expression. Virus Res 202:89–100. 10.1016/j.virusres.2014.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shen Z, Wang G, Yang Y, Shi J, Fang L, Li F, Xiao S, Fu ZF, Peng G. 2019. A conserved region of nonstructural protein 1 from alphacoronaviruses inhibits host gene expression and is critical for viral virulence. J Biol Chem 294:13606–13618. 10.1074/jbc.RA119.009713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baudoux P, Carrat C, Besnardeau L, Charley B, Laude H. 1998. Coronavirus pseudoparticles formed with recombinant M and E proteins induce alpha interferon synthesis by leukocytes. J Virol 72:8636–8643. 10.1128/JVI.72.11.8636-8643.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.La Bonnardiere C, Laude H. 1981. High interferon titer in newborn pig intestine during experimentally induced viral enteritis. Infect Immun 32:28–31. 10.1128/iai.32.1.28-31.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Riffault S, Carrat C, van Reeth K, Pensaert M, Charley B. 2001. Interferon-alpha-producing cells are localized in gut-associated lymphoid tissues in transmissible gastroenteritis virus (TGEV) infected piglets. Vet Res 32:71–79. 10.1051/vetres:2001111. [DOI] [PubMed] [Google Scholar]
- 18.Zhou Y, Wu W, Xie L, Wang D, Ke Q, Hou Z, Wu X, Fang Y, Chen H, Xiao S, Fang L. 2017. Cellular RNA helicase DDX1 is involved in transmissible gastroenteritis virus nsp14-induced interferon-beta production. Front Immunol 8:940. 10.3389/fimmu.2017.00940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mahlakoiv T, Hernandez P, Gronke K, Diefenbach A, Staeheli P. 2015. Leukocyte-derived IFN-alpha/beta and epithelial IFN-lambda constitute a compartmentalized mucosal defense system that restricts enteric virus infections. PLoS Pathog 11:e1004782. 10.1371/journal.ppat.1004782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Onoguchi K, Yoneyama M, Takemura A, Akira S, Taniguchi T, Namiki H, Fujita T. 2007. Viral infections activate types I and III interferon genes through a common mechanism. J Biol Chem 282:7576–7581. 10.1074/jbc.M608618200. [DOI] [PubMed] [Google Scholar]
- 21.Thomson SJ, Goh FG, Banks H, Krausgruber T, Kotenko SV, Foxwell BM, Udalova IA. 2009. The role of transposable elements in the regulation of IFN-lambda1 gene expression. Proc Natl Acad Sci USA 106:11564–11569. 10.1073/pnas.0904477106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen FE, Huang DB, Chen YQ, Ghosh G. 1998. Crystal structure of p50/p65 heterodimer of transcription factor NF-kappaB bound to DNA. Nature 391:410–413. 10.1038/34956. [DOI] [PubMed] [Google Scholar]
- 23.Malek S, Huxford T, Ghosh G. 1998. Ikappa Balpha functions through direct contacts with the nuclear localization signals and the DNA binding sequences of NF-kappaB. J Biol Chem 273:25427–25435. 10.1074/jbc.273.39.25427. [DOI] [PubMed] [Google Scholar]
- 24.Ding Z, An K, Xie L, Wu W, Zhang R, Wang D, Fang Y, Chen H, Xiao S, Fang L. 2017. Transmissible gastroenteritis virus infection induces NF-kappaB activation through RLR-mediated signaling. Virology 507:170–178. 10.1016/j.virol.2017.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fung TS, Huang M, Liu DX. 2014. Coronavirus-induced ER stress response and its involvement in regulation of coronavirus-host interactions. Virus Res 194:110–123. 10.1016/j.virusres.2014.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Y, Li JR, Sun MX, Ni B, Huan C, Huang L, Li C, Fan HJ, Ren XF, Mao X. 2014. Triggering unfolded protein response by 2-Deoxy-D-glucose inhibits porcine epidemic diarrhea virus propagation. Antiviral Res 106:33–41. 10.1016/j.antiviral.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fung TS, Liao Y, Liu DX. 2014. The endoplasmic reticulum stress sensor IRE1alpha protects cells from apoptosis induced by the coronavirus infectious bronchitis virus. J Virol 88:12752–12764. 10.1128/JVI.02138-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shaban MS, Muller C, Mayr-Buro C, Weiser H, Meier-Soelch J, Albert BV, Weber A, Linne U, Hain T, Babayev I, Karl N, Hofmann N, Becker S, Herold S, Schmitz ML, Ziebuhr J, Kracht M. 2021. Multi-level inhibition of coronavirus replication by chemical ER stress. Nat Commun 12:5536. 10.1038/s41467-021-25551-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xue M, Fu F, Ma Y, Zhang X, Li L, Feng L, Liu P. 2018. The PERK arm of the unfolded protein response negatively regulates transmissible gastroenteritis virus replication by suppressing protein translation and promoting type I interferon production. J Virol 92. 10.1128/JVI.00431-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu F, Li G, Wen K, Bui T, Cao D, Zhang Y, Yuan L. 2010. Porcine small intestinal epithelial cell line (IPEC-J2) of rotavirus infection as a new model for the study of innate immune responses to rotaviruses and probiotics. Viral Immunol 23:135–149. 10.1089/vim.2009.0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Geens MM, Niewold TA. 2011. Optimizing culture conditions of a porcine epithelial cell line IPEC-J2 through a histological and physiological characterization. Cytotechnology 63:415–423. 10.1007/s10616-011-9362-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiang HY, Wek SA, McGrath BC, Scheuner D, Kaufman RJ, Cavener DR, Wek RC. 2003. Phosphorylation of the alpha subunit of eukaryotic initiation factor 2 is required for activation of NF-kappaB in response to diverse cellular stresses. Mol Cell Biol 23:5651–5663. 10.1128/MCB.23.16.5651-5663.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Campbell N, Yio XY, So LP, Li Y, Mayer L. 1999. The intestinal epithelial cell: processing and presentation of antigen to the mucosal immune system. Immunol Rev 172:315–324. 10.1111/j.1600-065x.1999.tb01375.x. [DOI] [PubMed] [Google Scholar]
- 34.Allaire JM, Crowley SM, Law HT, Chang SY, Ko HJ, Vallance BA. 2018. The intestinal epithelium: central coordinator of mucosal immunity. Trends Immunol 39:677–696. 10.1016/j.it.2018.04.002. [DOI] [PubMed] [Google Scholar]
- 35.Peterson LW, Artis D. 2014. Intestinal epithelial cells: regulators of barrier function and immune homeostasis. Nat Rev Immunol 14:141–153. 10.1038/nri3608. [DOI] [PubMed] [Google Scholar]
- 36.Li S, Ye L, Yu X, Xu B, Li K, Zhu X, Liu H, Wu X, Kong L. 2009. Hepatitis C virus NS4B induces unfolded protein response and endoplasmic reticulum overload response-dependent NF-kappaB activation. Virology 391:257–264. 10.1016/j.virol.2009.06.039. [DOI] [PubMed] [Google Scholar]
- 37.Poppe M, Wittig S, Jurida L, Bartkuhn M, Wilhelm J, Muller H, Beuerlein K, Karl N, Bhuju S, Ziebuhr J, Schmitz ML, Kracht M. 2017. The NF-kappaB-dependent and -independent transcriptome and chromatin landscapes of human coronavirus 229E-infected cells. PLoS Pathog 13:e1006286. 10.1371/journal.ppat.1006286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Narayanan K, Huang C, Lokugamage K, Kamitani W, Ikegami T, Tseng CT, Makino S. 2008. Severe acute respiratory syndrome coronavirus nsp1 suppresses host gene expression, including that of type I interferon, in infected cells. J Virol 82:4471–4479. 10.1128/JVI.02472-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang X, Zhu Y, Zhu X, Shi H, Chen J, Shi D, Yuan J, Cao L, Liu J, Dong H, Jing Z, Zhang J, Wang X, Feng L. 2017. Identification of a natural recombinant transmissible gastroenteritis virus between Purdue and Miller clusters in China. Emerg Microbes Infect 6:e74. 10.1038/emi.2017.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Law AH, Lee DC, Cheung BK, Yim HC, Lau AS. 2007. Role for nonstructural protein 1 of severe acute respiratory syndrome coronavirus in chemokine dysregulation. J Virol 81:416–422. 10.1128/JVI.02336-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin JW, Tang C, Wei HC, Du B, Chen C, Wang M, Zhou Y, Yu MX, Cheng L, Kuivanen S, Ogando NS, Levanov L, Zhao Y, Li CL, Zhou R, Li Z, Zhang Y, Sun K, Wang C, Chen L, Xiao X, Zheng X, Chen SS, Zhou Z, Yang R, Zhang D, Xu M, Song J, Wang D, Li Y, Lei S, Zeng W, Yang Q, He P, Zhang Y, Zhou L, Cao L, Luo F, Liu H, Wang L, Ye F, Zhang M, Li M, Fan W, Li X, Li K, Ke B, Xu J, Yang H, He S, et al. 2021. Genomic monitoring of SARS-CoV-2 uncovers an Nsp1 deletion variant that modulates type I interferon response. Cell Host Microbe 29:489–502. 10.1016/j.chom.2021.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rauti R, Shahoha M, Leichtmann-Bardoogo Y, Nasser R, Paz E, Tamir R, Miller V, Babich T, Shaked K, Ehrlich A, Ioannidis K, Nahmias Y, Sharan R, Ashery U, Maoz BM. 2021. Effect of SARS-CoV-2 proteins on vascular permeability. Elife 10. 10.7554/eLife.69314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang L, Qiao X, Zhang S, Qin Y, Guo T, Hao Z, Sun L, Wang X, Wang Y, Jiang Y, Tang L, Xu Y, Li Y. 2018. Porcine transmissible gastroenteritis virus nonstructural protein 2 contributes to inflammation via NF-kappaB activation. Virulence 9:1685–1698. 10.1080/21505594.2018.1536632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang C, Lokugamage KG, Rozovics JM, Narayanan K, Semler BL, Makino S. 2011. Alphacoronavirus transmissible gastroenteritis virus nsp1 protein suppresses protein translation in mammalian cells and in cell-free HeLa cell extracts but not in rabbit reticulocyte lysate. J Virol 85:638–643. 10.1128/JVI.01806-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Henden AS, Koyama M, Robb RJ, Forero A, Kuns RD, Chang K, Ensbey KS, Varelias A, Kazakoff SH, Waddell N, Clouston AD, Giri R, Begun J, Blazar BR, Degli-Esposti MA, Kotenko SV, Lane SW, Bowerman KL, Savan R, Hugenholtz P, Gartlan KH, Hill GR. 2021. IFN-lambda therapy prevents severe gastrointestinal graft-versus-host disease. Blood 138:722–737. 10.1182/blood.2020006375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Antony F, Pundkar C, Sandey M, Jaiswal AK, Mishra A, Kumar A, Channappanavar R, Suryawanshi A. 2021. IFN-lambda regulates neutrophil biology to suppress inflammation in herpes simplex virus-1-induced corneal immunopathology. J Immunol 206:1866–1877. 10.4049/jimmunol.2000979. [DOI] [PubMed] [Google Scholar]
- 47.Palma-Ocampo HK, Flores-Alonso JC, Vallejo-Ruiz V, Reyes-Leyva J, Flores-Mendoza L, Herrera-Camacho I, Rosas-Murrieta NH, Santos-Lopez G. 2015. Interferon lambda inhibits dengue virus replication in epithelial cells. Virol J 12:150. 10.1186/s12985-015-0383-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Felgenhauer U, Schoen A, Gad HH, Hartmann R, Schaubmar AR, Failing K, Drosten C, Weber F. 2020. Inhibition of SARS-CoV-2 by type I and type III interferons. J Biol Chem 295:13958–13964. 10.1074/jbc.AC120.013788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang G, Liang R, Liu Z, Shen Z, Shi J, Shi Y, Deng F, Xiao S, Fu ZF, Peng G. 2019. The N-Terminal domain of spike protein is not the enteric tropism determinant for transmissible gastroenteritis virus in piglets. Viruses 11:313. 10.3390/v11040313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu Y, Tao W, Wen S, Li Z, Yang A, Deng Z, Sun Y. 2015. In vitro CRISPR/Cas9 system for efficient targeted DNA editing. mBio 6:e01714-15–e01715. 10.1128/mBio.01714-15. [DOI] [PMC free article] [PubMed] [Google Scholar]