ABSTRACT
Mitochondria are critical cellular organelles that perform a wide variety of functions, including energy production and immune regulation. To perform these functions, mitochondria contain approximately 1,500 proteins, the majority of which are encoded in the nuclear genome, translated in the cytoplasm, and translocated to the mitochondria using distinct mitochondrial targeting sequences (MTS). Bacterial proteins can also contain MTS and localize to the mitochondria. For the obligate intracellular human pathogen Chlamydia trachomatis, interaction with various host cell organelles promotes intracellular replication. However, the extent and mechanisms through which Chlamydia cells interact directly with mitochondria remain unclear. We investigated the presence of MTS in the C. trachomatis genome and discovered 30 genes encoding proteins with around 70% or greater probability of mitochondrial localization. Five are translocated to the mitochondria upon ectopic expression in HeLa cells. Mass spectrometry of isolated mitochondria from infected cells revealed that two of these proteins localize to the mitochondria during infection. Comparison of mitochondria from infected and uninfected cells suggests that chlamydial infection affects the mitochondrial protein composition. Around 125 host proteins were significantly decreased or absent in mitochondria from infected cells. Among these were proapoptotic factors and those related to mitochondrial fission/fusion dynamics. Conversely, 82 host proteins were increased in or specific to mitochondria of infected cells, many of which act as antiapoptotic factors and upregulators of cellular metabolism. These data support the notion that C. trachomatis specifically targets host mitochondria to manipulate cell fate decisions and metabolic function to support pathogen survival and replication.
IMPORTANCE Obligate intracellular bacteria have evolved multiple means to promote their intracellular survival and replication within the otherwise harsh environment of the eukaryotic cell. Nutrient acquisition and avoidance of cellular defense mechanisms are critical to an intracellular lifestyle. Mitochondria are critical organelles that produce energy in the form of ATP and regulate programmed cell death responses to invasive pathogenic microbes. Cell death prior to completion of replication would be detrimental to the pathogen. C. trachomatis produces at least two and possibly more proteins that target the mitochondria. Collectively, C. trachomatis infection modulates the mitochondrial protein composition, favoring a profile suggestive of downregulation of apoptosis.
KEYWORDS: Chlamydia, apoptosis, mitochondria, secreted effector
INTRODUCTION
The obligate intracellular pathogen Chlamydia trachomatis is the etiological agent of the most commonly reported bacterial infection, causing both blinding trachoma and sexually transmitted disease, affecting over 1.8 million people and 100 million people, respectively (1, 2). Chlamydia bacteria have a unique biphasic developmental cycle where the bacteria alternate between two morphologically and functionally distinct forms, the infectious elementary body (EB) and the replicative reticulate body (RB). Critical to the developmental cycle is the establishment and maintenance of the chlamydia-containing vacuole, or inclusion. The inclusion serves as a protective membrane allowing survival of the replicative body and, as such, is a key interaction point between bacteria and host (3).
For C. trachomatis, inclusion interaction with host cell organelles has been demonstrated to be critical for nutrient uptake, inclusion localization, and regulation of escape mechanisms (4). These known interactions occur through recruitment of organelles to the inclusion membrane through protein-protein interactions between inclusion membrane proteins (Incs) and organelle proteins. An example of these interactions includes the recruitment of Golgi-derived vesicles as a strategy for lipid acquisition from the host (5, 6) and interactions of the inclusion membrane with the endoplasmic reticulum (7, 8), endocytic vesicles (9), lipid droplets (10), peroxisomes (11), and cytoskeleton (12). The ability to manipulate mitochondria is critical for pathogens relying on the host for nutrients and survival (13), as is expected for the obligate intracellular pathogen C. trachomatis.
Mitochondria are critical cellular organelles with roles in diverse pathways, including energy and metabolite production, as well as regulation of cell survival. Often referred to as “the powerhouses of the cell,” these double-membrane-bound structures are responsible for the production of most of the cell’s ATP through the tricarboxylic acid (TCA) cycle and oxidative phosphorylation (14). Along with the production of ATP, these pathways are responsible for producing precursors for biosynthetic pathways, particularly for fatty acid, nucleotide, and amino acid metabolism (15). Inherently linked with the role of mitochondria in energy production is the maintenance of homeostasis within the cell and, therefore, the promotion of cell death in dysregulated states, such as during stress or infection. Intrinsic apoptosis is induced when an internal stimulus leads to the activation of proapoptotic factors, which results in the depolarization and permeabilization of the outer mitochondrial membrane, ultimately leading to cell death (16, 17).
Due to their diverse functions, mitochondria contain many proteins functioning in structural and enzymatic roles. Mitochondrial DNA encodes only about 1% of the mitochondrial proteome, and most of the proteins encoded by mitochondrial DNA are important in oxidative phosphorylation or translational machinery (18). However, hundreds of proteins found within the mitochondria are translated in the cytoplasm and translocated to the mitochondria using distinct mitochondrial targeting sequences (MTS). These MTS are typically amphiphilic cleavable signals between 20 and 40 amino acids in length, located on the N terminus, containing multiple positively charged residues and no acidic residues (19). It has been shown that bacterial proteins can also contain MTS and localize to the mitochondria (20, 21). For example, EspF of enteropathogenic Escherichia coli is injected into the host via a type III secretion system (T3SS) and localizes to the mitochondria, causing cytochrome c release and apoptosis (22).
While it has been shown that Chlamydia bacteria modulate host processes associated with mitochondria, such as inhibition of apoptosis and promotion of mitochondrial fusion (23), the extent and mechanisms through which Chlamydia bacteria interact directly with mitochondria remain unclear. Interestingly, mitochondria are not recruited to the C. trachomatis inclusion membrane, like other organelles (24). We hypothesized that rather than recruiting mitochondria to the inclusion, secreted effectors might instead be trafficked to the mitochondria. In this study, we show that C. trachomatis encodes proteins that contain MTS, are secreted by a type III secretion system, and localize to the mitochondria during infection. Additionally, mitochondria from infected cells have unique proteomes that reflect global changes caused by the presence of C. trachomatis.
(Preliminary accounts of this work were presented at the Gordon Conference on Microbial Toxins and Pathogenesis in 2010 and the Chlamydia Basic Research Society Biannual Meetings in 2011 and 2013.)
RESULTS
C. trachomatis proteins contain predicted and functional MTS.
To investigate the presence of mitochondrial targeting sequences (MTS) within the chlamydial genome, a bioinformatic screen was used to analyze the primary sequence for C. trachomatis D/UW-3 proteins. The computational MTS predictor, MitoProt (25), calculates a mitochondrial export probability based on N-terminal MTS parameters. Thirty C. trachomatis proteins with predicted MTS and mitochondrial export probabilities near or above 0.7 were identified (Table 1). The majority of candidate proteins were Chlamydia-specific hypothetical proteins, but five putative or demonstrated inclusion membrane proteins, two known type III secreted proteins, and two of the putative cytotoxin pseudogenes were represented as well.
TABLE 1.
C. trachomatis proteins with mitochondrial targeting sequences by MitoProt prediction
| Gene | Annotated function | MTSa | Mitochondrial export probability | Mitochondrial localization? |
|---|---|---|---|---|
| CT011 | Hypothetical protein | 1:47 | 0.979 | No |
| CT037 | Hypothetical protein | 1:70 | 0.742 | NTb |
| CT058 | Hypothetical protein | 1:87 | 0.985 | No |
| CT060 | FHIPEP family type III secretion protein | 1:154 | 0.969 | NT |
| CT080 | Late transcription unit protein B (ltuB) | 1:24 | 0.976 | No |
| CT087 | 4-Alpha glucanotransferase (malQ) | 1:41 | 0.941 | No |
| CT101 | Hypothetical protein | 1:70 | 0.858 | NT |
| CT132 | Hypothetical protein | 1:34 | 0.962 | Yes |
| CT133 | Class I SAM-dependent methyltransferase | 1:17 | 0.831 | No |
| CT166 | Putative cytotoxin pseudogene | 1:35 | 0.945 | NT |
| CT168 | Putative cytotoxin pseudogene | 1:29 | 0.810 | NT |
| CT195 | Putative inclusion membrane protein | 1:127 | 0.960 | No |
| CT229 | Inclusion membrane protein, CpoS | 1:97 | 0.712 | No |
| CT244 | Hypothetical protein | 1:8 | 0.686 | No |
| CT256 | Hypothetical protein | 1:155 | 0.757 | No |
| CT300 | Hypothetical protein | 1:112 | 0.952 | No |
| CT339 | DNA internalization-related competence protein, ComEC | 1:135 | 0.997 | No |
| CT351 | Hypothetical protein | 1:45 | 0.676 | NT |
| CT352 | Hypothetical protein | 1:48 | 0.962 | NT |
| CT385 | Hypothetical protein | 1:5 | 0.7054 | No |
| CT484 | Hypothetical protein | 1:85 | 0.846 | No |
| CT529 | Putative inclusion membrane protein | 1:37 | 0.985 | Yes |
| CT550 | Hypothetical protein | 1:3 | 0.887 | No |
| CT552 | Hypothetical protein | 1:47 | 0.996 | No |
| CT616 | Hypothetical protein | 1:86 | 0.968 | No |
| CT618 | Putative inclusion membrane protein | 1:37 | 0.896 | Yes |
| CT642 | Putative inclusion membrane protein | 1:8 | 0.808 | Yes |
| CT647 | Hypothetical protein | 1:32 | 0.753 | Yes |
| CT700 | Hypothetical Protein | 1:40 | 0.717 | No |
| CT862 | Type III secretion low calcium response chaperone, lcrH2 | 1:47 | 0.735 | No |
Analyzed region (amino acid sequence number).
NT, not tested.
Eukaryotic expression vectors were generated using the pEGFP-N1 plasmid to express the product of each candidate gene as an enhanced green fluorescent protein (EGFP) fusion protein that could be expressed ectopically in HeLa cells. We utilized HeLa cells for this screening due to their uniformity, ease of transfection, and permissiveness to chlamydial infection. We utilized C. trachomatis L2 coding regions for the ability to perform future genetic experiments, and therefore were unable to generate vectors for CT166, CT168, and CT352, which are absent in the C. trachomatis L2 genome (26). Genes with known functions (CT060 [27], CT101 [28], and pseudogenes CT037 and CT300) were excluded from construct generation, while CT616 proved challenging to clone. The remaining twenty-two candidate genes from the MTS screen were examined for localization of the encoded proteins in HeLa cells (Table 1).
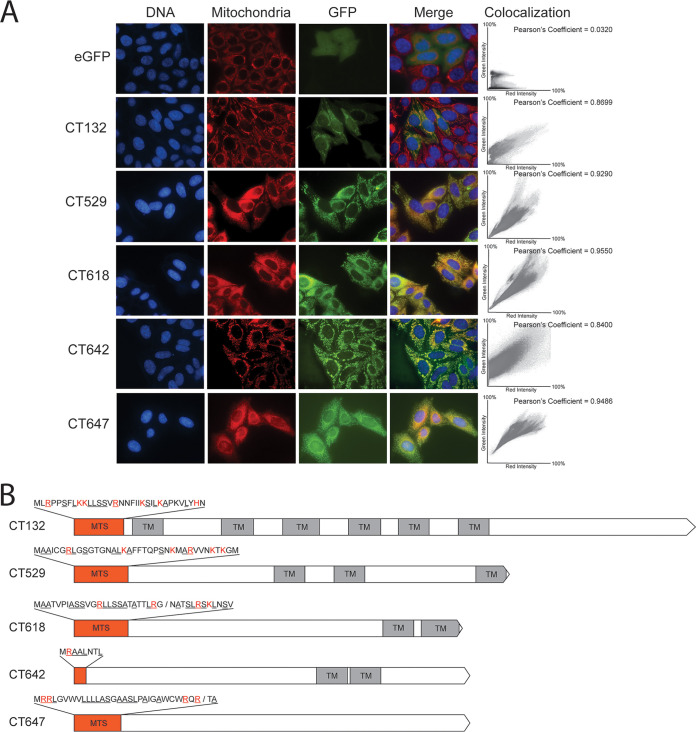
Five candidates, CT132, CT529, CT618, CT642, and CT647, showed colocalization of their encoded proteins with mitochondria upon transfection of their vectors compared with the diffuse cytoplasmic staining of the vector control (Fig. 1A). Colocalization was quantified with Pearson correlation coefficients. Closer examination of the MTS for each of these proteins revealed that each was enriched for the canonical amino acids alanine, arginine, leucine, and serine, as well as for positive residues (Fig. 1B). CT529, CT618, and CT642 are all predicted inclusion membrane proteins with characteristic bilobed transmembrane domains, although CT529 has a possible third transmembrane domain at the C terminus of the protein (29). CT132 is a hypothetical protein with six putative transmembrane domains, which may be characteristic of a multipass integral membrane protein (30). CT132 also shows homology, using the Basic Local Alignment Search Tool (BLAST), to transporter protein BrkB of Bordetella pertussis which similarly has six membrane-spanning domains (31). CT647 is a hypothetical protein with no known or predicted functional domains.
FIG 1.
(A) Microscopy shows GFP localization of transfected C. trachomatis fusion proteins with functional MTS. HeLa cells on coverslips were transfected with either the empty EGFP-N1 vector (eGFP) or GFP-tagged C. trachomatis proteins (CT###) and incubated for 48 h before staining with 100 nM MitoTracker (red) and NucBlue (blue). Bar = 10 μm. Coverslips were imaged under ×60 magnification. Colocalization scatterplots were determined for the regions of interest in each representative image, with green intensity on the y axis and red intensity on the x axis, and Pearson coefficients are reported. (B) Gene maps for the five candidate MTS sequences. Orange boxes represent MTS sequences, which are shown above the maps. Red amino acids indicate positive residues, and underlined residues denote canonically enriched amino acids. Predicted cleavage sites are indicated with a slash. Transmembrane domains (TM) are indicated.
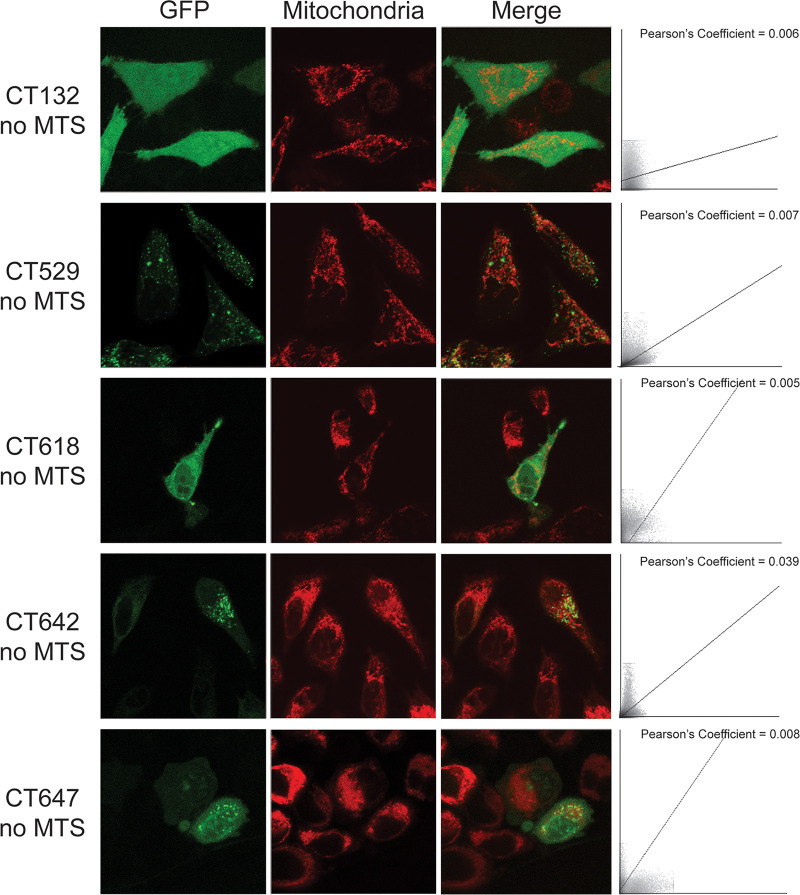
To verify that the mitochondrial localization for these five proteins was due to recognition of the MTS, constructs were designed where the N-terminal predicted MTS was removed, leaving the methionine start codon intact. For all five mitochondrially localized constructs, removal of the MTS resulted in an inability to colocalize with mitochondria (Fig. 2). Both the CT132-GFP and CT618-GFP signals were cytoplasmic and appeared similar to the majority of nonfunctional MTS proteins (Fig. S1 in the supplemental material). CT529-GFP, CT618-GFP, and CT647-GFP signals had punctate staining that was no longer correlated with mitochondria. These data suggest that the bioinformatically predicted MTS of CT132, CT529, CT618, and CT647 are functionally recognized in HeLa cells, resulting in translocation of these proteins to the mitochondria.
FIG 2.
Immunofluorescent staining shows GFP localization of mitochondrially targeted C. trachomatis proteins after removal of the MTS. HeLa cells on coverslips were transfected with GFP-tagged C. trachomatis proteins with the MTS removed (CT### no MTS) and incubated for 24 h before staining with 100 nM MitoTracker (red). Bar = 10 μm. Colocalization scatterplots were determined for each representative image, and Pearson coefficients are reported.
(A) Immunofluorescent staining shows GFP localization of transfected C. trachomatis proteins that did not have a functional MTS. HeLa cells on coverslips were transfected with either the empty EGFP-N1 vector (eGFP) or GFP-tagged C. trachomatis proteins (CT###) and incubated for 24 h before staining with 100 nM MitoTracker (red). (B) Colocalizations with other organelles demonstrated with anti-calnexin antibodies or anti-Golgi apparatus antibodies. Bar = 10 μm. Download FIG S1, TIF file, 2.6 MB (2.7MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Seventeen proteins did not localize with mitochondria after transfection (Fig. S1A). The majority of these candidates had diffuse cytoplasmic localization, similar to the EGFP vector control. Three fusion proteins appeared to localize to discrete sites within the cell other than the mitochondria. CT229 is a known inclusion membrane protein, named CpoS, and inserts into the inclusion membrane, where it interacts with Rab GTPases to regulate vesicular trafficking (32). The CT229-GFP fusion protein in this study appeared to be maintaining interaction with clathrin-coated vesicles of the trans-Golgi network, as previously described (33). Hypothetical protein CT700-GFP fusion protein staining was similar to that of CT229, and we demonstrated that this protein also localized to the Golgi apparatus during infection (Fig. S1B). Additionally, putative inclusion membrane protein CT195 has previously been demonstrated to colocalize with endoplasmic reticulum (ER) structures, and our construct had similar colocalization with calnexin-stained ER membranes (Fig. S1B) (34).
CT529, CT618, and CT642 are type III secreted proteins.
In order for the MTS to be recognized by cognate chaperones and transporters, the chlamydial MTS would need to be present in host cytosol; therefore, we examined whether the five mitochondrial-targeting candidate proteins could be secreted. In C. trachomatis, the type III secretion system (T3SS) is critical for the delivery of effectors into the host cell, as well as Incs into the inclusion membrane (35). The bioinformatic prediction pipeline T3SEpp was used to predict the likelihood of being type III secreted for each protein-coding sequence (36). Both CT132 and CT647 were identified as non-type III secreted, while CT529, CT618, and CT642 were predicted to be type III secreted, which matches their identification as putative Incs (Fig. 3A).
FIG 3.
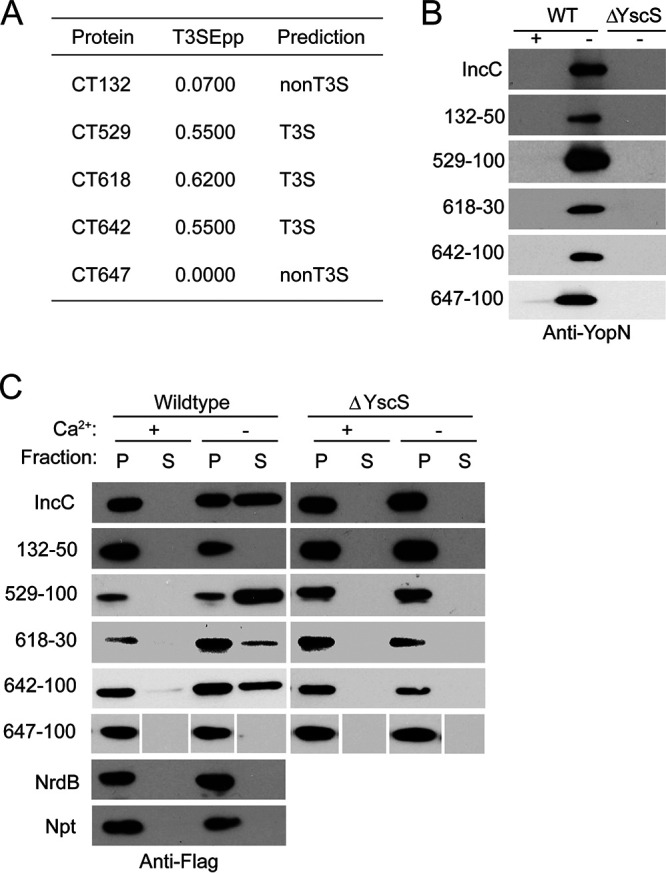
Select mitochondrial targeting Chlamydia proteins are secreted by type III secretion. (A) Results from computational integrated prediction pipeline T3SEpp for predicting bacterial type III effectors. (B) Y. pseudotuberculosis cells expressing the N-terminal sequences (30 to 100 bp, as indicated) of the indicated chlamydial proteins (132 is CT132, etc.) or the vector control were grown in the presence or absence of 5 mM Ca2+, where its absence promotes type III secretion. Cultures were induced with 0.01 mM IPTG and the temperature shifted from 26 to 37°C, and supernatants were probed for YopN (Yersinia T3SS control). (C) Flag-tagged Npt fusion constructs were expressed as described for panel B, and samples were fractionated into pelleted cells (P) for nonsecreted fractions and supernatant (S) for secreted fractions. The samples of CT647 were run in a different order, and certain panels were spliced for consistency in the presentation of this figure, as indicated by gaps.
The chlamydial T3SS is heterologous to that of Yersinia pseudotuberculosis, which can be utilized as a model to demonstrate type III secretion (T3S) of C. trachomatis proteins (37, 38). The N-terminal sequences of the five candidate effectors were fused to the type III secreted carrier protein neomycin phosphotransferase (Npt) of either wild-type Y. pseudotuberculosis or the T3SS-deficient ΔYscS strain. As a control, we probed for the Yersinia secreted effector YopN in the supernatants for each of the samples to ensure that expression of the fusion construct did not disrupt normal yersinial T3S (Fig. 3B). In wild-type Y. pseudotuberculosis, fusion of the N-terminal regions of CT132 and CT647 to Npt did not result in secretion in the absence of calcium, which activates T3S, and the results were not different from the results for the NrdB negative control, which is a chlamydial gene product with no T3S signal. In contrast, the results for CT529, CT618, and CT642 fusion constructs were comparable to the results for the IncC positive control, which is a known T3S effector in C. trachomatis and has been previously demonstrated to be type III secreted in Y. pseudotuberculosis (Fig. 3C) (38). In the ΔYscS strain, these constructs were not detected in the supernatant, demonstrating that these are in fact type III secreted effectors.
CT529 and CT618 proteins are identified in mitochondria of infected cells.
To confirm the localization of the candidate effectors during infection, a proteomics approach was taken. One benefit to using a proteomics screen was that proteins that localized with mitochondria through alternative mechanisms to canonical MTS could also be identified, thus providing the most comprehensive way to identify mitochondrial targeting chlamydial effectors. Mitochondria from HeLa cells either uninfected or infected for 24 h with C. trachomatis L2 were isolated using fluorescence-activated mitochondrial sorting (39). Isolated mitochondria were lysed, and total protein content was identified using mass spectrometry. Triplicate experiments were performed for each condition with and without disuccinimidyl suberate (DSS) cross-linking, which allowed the identification of proteins with potential transient or noncovalent mitochondrial interactions.
Mitochondrial proteins were either cross-linked or not; however, there was no significant difference between cross-linked and non–cross-linked conditions. No C. trachomatis proteins were present only under cross-linked conditions. Forty-nine chlamydial proteins with more than two unique peptides in at least two replicates were detected in the mitochondria of infected cells after elimination of those detected under the uninfected condition. Of the original candidate proteins, both CT529 and CT618 were detected in at least four replicates (Table 2). CT132, CT642, and CT647 were not identified in any replicate. Of the remaining chlamydial proteins, 12 (24.5%) were ribosomal proteins and five (10.2%) were hypothetical proteins.
TABLE 2.
C. trachomatis proteins identified in mitochondria from infected cells
| Gene (plasmid) or plasmid | Annotated function, gene name | No. of unique peptides | Coverage (%) | MitoProt MTS predictiona |
|---|---|---|---|---|
| CTL0259 (CT004) | Aspartyl/glutamyl-tRNA amidotransferase subunit B, gatB | 2 | 5 | 0.012 |
| CTL0277 (CT022) | Type B 50S ribosomal protein L31, rpmE2 | 2 | 24 | 0.047 |
| CTL0283 (CT028) | 50S ribosomal protein L19, rplS | 3 | 26 | 0.146 |
| CTL0301 (CT045) | Leucyl aminopeptidase, pepA | 4 | 8 | 0.015 |
| CTL0321 (CT065) | ADP, ATP carrier protein, ntp1 | 3 | 4 | 0.229 |
| CTL0322 (CT066) | Hypothetical protein | 2 | 23 | 0.342 |
| CTL0323 (CT067) | ABC transporter substrate-binding protein, ytgA | 4 | 16 | 0.578 |
| CTL0365 (CT110) | Chaperonin, groL | 33 | 79 | 0.168 |
| CTL0381 (CT126) | 30S ribosomal protein S9, rpsI | 5 | 47 | 0.716 |
| CTL0423 (CT170) | Tryptophan synthase subunit B, trpB | 13 | 38 | 0.161 |
| CTL0519 (CT267) | DNA binding protein, ihfA | 3 | 46 | 0.530 |
| CTL0544 (CT292) | Deoxyuridine 5′-triphosphate nucleotidohydrolase, dut | 2 | 28 | 0.042 |
| CTL0567 (CT315) | DNA-directed RNA polymerase subunit beta, rpoB | 9 | 11 | 0.047 |
| CTL0568 (CT316) | 50S ribosomal protein L7/L12, rplL | 2 | 20 | 0.209 |
| CTL0570 (CT318) | 50S ribosomal protein L1, rplA | 4 | 18 | 0.049 |
| CTL0595 (CT341) | Chaperone protein, dnaJ | 3 | 11 | 0.003 |
| CTL0655 (CT398) | Hypothetical protein | 3 | 19 | 0.013 |
| CTL0679 (CT421.1) | Hypothetical protein | 3 | 30 | 0.699 |
| CTL0696 (CT437) | Elongation factor G, fusA | 19 | 32 | 0.132 |
| CTL0752 (CT491) | Transcription termination factor, rho | 4 | 11 | 0.040 |
| CTL0769 (CT507) | DNA-directed RNA polymerase subunit alpha, rpoA | 4 | 18 | 0.052 |
| CTL0770 (CT508) | 30S ribosomal protein S11, rpsK | 3 | 31 | 0.907 |
| CTL0771 (CT509) | 30S ribosomal protein S13, rpsM | 4 | 35 | 0.325 |
| CTL0774 (CT512) | 30S ribosomal protein S5, rpsE | 2 | 10 | 0.156 |
| CTL0776 (CT514) | 50S ribosomal protein L6, rplF | 3 | 25 | 0.629 |
| CTL0786 (CT524) | 30S ribosomal protein S19, rpsS | 2 | 14 | 0.759 |
| CTL0788 (CT526) | 50S ribosomal protein L23, rplW | 3 | 45 | 0.300 |
| CTL0789 (CT527) | 50S ribosomal protein L4, rplD | 4 | 24 | 0.018 |
| CTL0791 (CT529) | Putative inclusion membrane protein | 3 | 20 | 0.970 |
| CTL0863 (CT600) | Peptidoglycan-associated lipoprotein, pal | 2 | 15 | 0.509 |
| CTL0866 (CT603) | Thioredoxin peroxidase, ahpC | 11 | 74 | 0.280 |
| CTL0872 (CT608) | ATP-dependent DNA helicase, uvrD | 2 | 6 | 0.203 |
| CTL0874 (CT610) | Virulence protein, CADD | 5 | 44 | 0.259 |
| CTL0882 (CT618) | Putative inclusion membrane protein | 4 | 29 | 0.817 |
| CTL0018 (CT650) | Recombinase RecA | 3 | 10 | 0.021 |
| CTL0024 (CT655) | 2-dehydro-3-deoxyphosphooctonate aldolase, kdsA | 3 | 25 | 0.012 |
| CTL0046 (CT677) | Ribosome recycling factor, frr | 2 | 20 | 0.043 |
| CTL0060 (CT691) | Hypothetical protein | 2 | 8 | 0.072 |
| CTL0112 (CT743) | Histone H1-like protein, hctA | 2 | 14 | 0.332 |
| CTL0137 (CT768) | Hypothetical protein | 4 | 8 | 0.006 |
| CTL0140 (CT771) | Hydrolase | 3 | 25 | 0.080 |
| CTL0142 (CT773) | Leucine dehydrogenase, ldh | 4 | 18 | 0.009 |
| CTL0168 (CT799) | 50S ribosomal protein L25, rplY | 5 | 21 | 0.287 |
| CTL0183 (CT812) | Polymorphic membrane protein, pmpD | 6 | 7 | 0.009 |
| CTL0213 (CT841) | ATP-dependent zinc metalloprotease, ftsH | 3 | 9 | 0.0081 |
| CTL0233 (CT858) | Protease-like activity factor, CPAF | 2 | 5 | 0.914 |
| CTL0250 (CT871) | Polymorphic membrane protein, pmpG | 4 | 7 | 0.412 |
| CTL0254 (CT874) | Polymorphic membrane protein, pmpI | 3 | 4 | 0.011 |
| pL2-03 (pGP1) | Putative helicase | 2 | 7 | 0.038 |
See reference 25.
Infection with C. trachomatis alters the mitochondrial proteome.
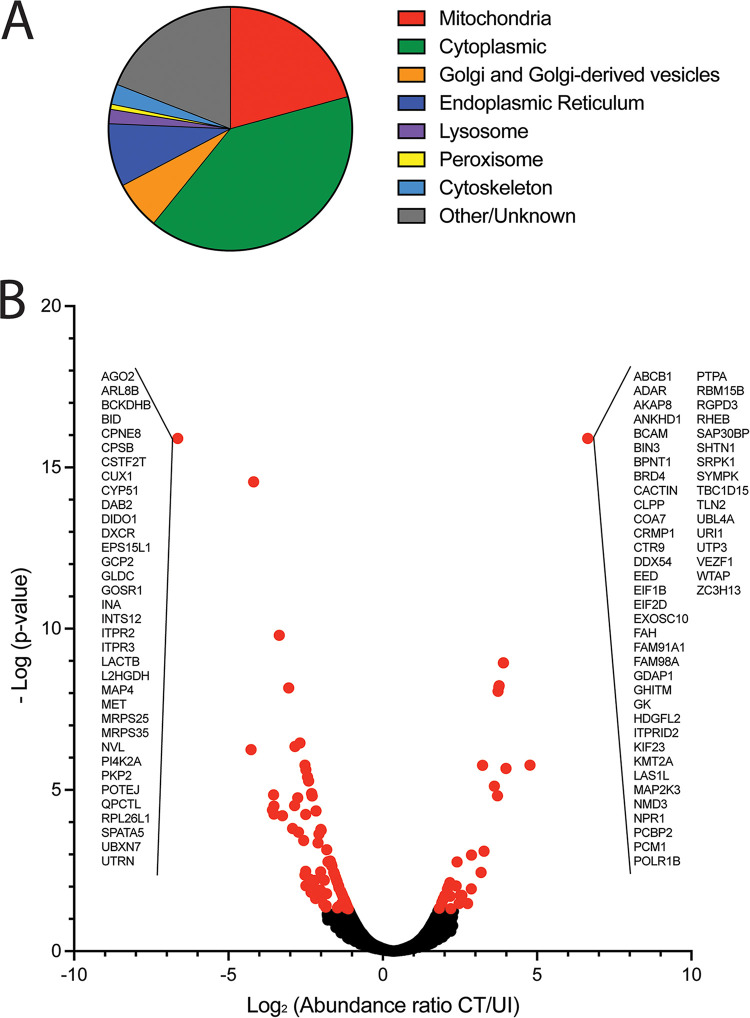
Performing mass spectrometry on total protein content allowed the comparison of host protein changes between mitochondria isolated from uninfected and infected cells. There was an average of 2,306 proteins identified per replicate, with no significant difference between experimental replicates or between treatment conditions. When comparing cross-linked to non–cross-linked conditions, there were few differences in protein content, with only 75 (2.8%) total proteins identified only under non–cross-linked conditions and 57 (2.1%) proteins only identified under cross-linked conditions out of a total of 2,685 proteins in all replicates. Of all proteins identified, over 20% were known mitochondrial proteins (Fig. 4A). Forty percent of proteins identified were cytoplasmic proteins and likely interacted with mitochondrial proteins. One concern with isolating mitochondria is the close association with other organelles, particularly the endoplasmic reticulum and Golgi-derived vesicles. Within the proteomes, around 8% and 5% of proteins were known to interact with the ER or Golgi apparatus/Golgi-derived vesicles respectively, indicating that there was minimal pulldown of these organelles with the isolated mitochondria. Less than 2% of the proteins identified are predicted to be localized to the lysosomes, peroxisomes, or cytoskeletal elements. Proteins known to localize with these nonmitochondrial organelles may indicate direct interactions between mitochondria and nonmitochondrial organelles.
FIG 4.
(A) Gene Ontology (GO) Cellular Compartments for proteins identified in mass spectrometry screen of mitochondria from uninfected and C. trachomatis-infected HeLa cells (GO terms GO:0005739, GO:0005759, GO:0005743, GO:0005741, GO:0005758, GO:0097679, GO:0000139, GO:0044233, GO:0005783, GO:0032865, GO:0015629, GO:0005764, and GO:0005777). (B) Host protein peak abundances for each identified peptide were compared between groups and reported as the log2 of the ratio of C. trachomatis-infected HeLa cells (CT) to uninfected cells (UI), where significant changes (red) were determined at a P value of <0.05. All peptides identified under only one condition have the same value on the plot and are listed in insets.
To determine mitochondrial-proteome differences between C. trachomatis-infected and uninfected conditions, an abundance ratio was calculated by dividing the abundance of protein under the C. trachomatis-infected condition by the abundance under the uninfected condition (CT/UI). There were 83 proteins identified with significantly (P > 0.05) increased abundance under the infected condition (Table S1) and 125 that were significantly decreased under the infected condition (Table S2), totaling 208 (7.8%) proteins with differential abundances (Fig. 4A). Of the proteins with differential abundances, there were subsets of proteins identified that were unique to one condition. Thirty-five proteins were present only in mitochondria of uninfected cells, and 50 proteins were present only in mitochondria of infected cells (Fig. 5). Among these proteins, there was no apparent bias toward or against mitochondrial-DNA-encoded proteins compared with nuclear-DNA-encoded proteins.
FIG 5.
Mitochondria from uninfected (UI) or C. trachomatis L2-infected (CT) HeLa cells have unique protein contents. Proteins unique to one condition (uninfected or C. trachomatis infected) are shown. Known or estimated subcellular locations based on Gene Ontology Cellular Compartments (GO terms GO:0005739, GO:0005759, GO:0005743, GO:0005741, GO:0005758, GO:0097679, GO:0000139, GO:0044233, GO:0005783, and GO:0032865) and functions based on Gene Ontology Biological Process or Molecular Function (GO:0006915, GO:0043065, GO:0043066, GO:0045087, GO:0019722, GO:0050848, GO:0007165, GO:0005975, GO:0008610, GO:0006633, GO:0046112, GO:0009058, GO:0015031, GO:0016197, GO:0030036, GO:0005200, GO:0003723, GO:1903108, GO:0003677, GO:0006306, GO:0000266, and GO:0008053) are indicated by filled squares.
Proteins with decreased abundance in mitochondria of infected cells. Download Table S1, DOCX file, 0.03 MB (28.9KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Proteins with increased abundance in mitochondria of infected cells. Download Table S2, DOCX file, 0.02 MB (23.3KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Proteins present at the mitochondria only of uninfected cells may be recruited elsewhere during infection. Specifically, infection restricted the presence of several Golgi apparatus and ER proteins. Functional analysis of proteins unique to the uninfected condition showed that they were representative of diverse functions, including RNA binding (26%), metabolism (23%), calcium signaling (20%), and apoptosis (17%). The proteins involved in apoptosis that were present only in the uninfected cell mitochondria were all proapoptotic effectors, including the well-characterized BH3 domain Bid, which localizes to the mitochondrial membrane during the initiation of intrinsic apoptosis (40). Additionally, three positive regulators of mitochondrial fission were present only in uninfected cells.
Of the proteins present only in infected replicates, 37 were known cytoplasmic proteins, suggesting that chlamydial infection resulted in the relocation of these proteins to the mitochondria, potentially by interacting with proteins on the mitochondrial membrane. There was a functional symmetry to those proteins unique to the infected condition and those absent during infection. Of the unique mitochondrial proteins in the infected cells, the majority were involved in apoptosis (34%), RNA binding (26%), and metabolism (14%) (Fig. 5). However, under this condition, proteins involved in apoptosis acted largely as antiapoptotic factors, and those involved in metabolism were oxidative phosphorylation and glycolysis activators.
DISCUSSION
In this study, we describe the identification of mitochondrial targeting sequences (MTS) in the C. trachomatis genome and demonstrate that at least five of these signals are recognized and result in the translocation of an EGFP reporter fusion protein to the mitochondria in HeLa cells. Previous work has shown that knockdown of components of the Tom complex is essential for the replication of Chlamydia caviae either in Drosophila cells or mammalian cells (41). The Tom complex is a multiprotein complex involved in the recognition and import of nuclear-DNA-encoded mitochondrial proteins to mitochondria. This requirement for components of the Tom complex, Tom40 and Tom22, was specific for C. caviae, however, as knockdown of Tom40 and Tom22 was not inhibitory to C. trachomatis (41). It is unclear which, if any, C. caviae proteins might be trafficked to mitochondria or whether mitochondrially targeted C. trachomatis proteins might utilize different mechanisms. The chlamydial effectors that colocalized with mitochondria are the first C. trachomatis proteins reported that may directly interact with host mitochondria. From the initial bioinformatic screen, eight proteins were not tested in the ectopic expression analysis. While those with C. trachomatis L2 orthologs were not later identified in the mitochondria of infected cells, it remains possible that the three proteins unique to serovar D, CT166, CT168, and CT352, may also be mitochondrial targeting effectors.
With the apparent differences in effects on mitochondria between C. trachomatis and C. caviae and the possibility of differences between serovars, we investigated the presence of homologs of the MTS-containing proteins that localized to mitochondria in other chlamydial species (Table S3). All five genes appeared to be conserved among the chlamydial species analyzed, with the exception of CT618, which did not have an apparent homolog in Chlamydia pneumoniae. The direct homologs for CT132 and CT529 appeared to have conserved MTS as well, even when protein identity was low. CT642 and CT647 MTS appeared to be less conserved. The conservation of CT529’s mitochondrial export probability suggests that its role in mitochondria may be important for chlamydial infection across species, while the loss of CT618 or CT618 mitochondrial export suggests that CT618 may have a role in only specific host species or tissues.
Chlamydia homologs to mitochondrial targeting candidate effectors. Download Table S3, DOCX file, 0.01 MB (15.6KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Examination of the MTS for the five positive colocalizing proteins after ectopic expression reveals that there may be a preference for an MTS of around 35 amino acids in length. Only CT642 had an MTS that was outside the 32- to 37-amino-acid range, and interestingly, this MTS was one of the shortest detected by MitoProt, at 8 amino acids. Despite being about a quarter the length of the other sequences, the CT642 MTS had an equal ratio of conserved residues (Fig. 1B, underlined) that likely contribute to the ability to be recognized by cognate machinery. Cleavage sequences were identified for only two of the five proteins, CT618 and CT647. It remains unclear whether these signals are cleaved or if there are alternate cleavage mechanisms for the other MTS. Due to the use of trypsin in the mass spectrometry protein identification, we were not able to determine whether CT618 or CT529 MTS were cleaved during infection. Adding evidence to the functionality of the MTS signal, deleting the sequence from the GFP fusion proteins led to loss of mitochondrial localization. Both CT529 and CT642 localized to discrete subcellular compartments rather than producing a diffuse cytoplasmic staining. This phenotype may be due to their Inc-like structure, as it has been shown previously that ectopically expressed Incs cause the formation of or associate with distinct vesicles within HeLa cells (42).
Because CT529, CT618, and CT642 have been previously identified as putative Incs (34), it was not surprising that each was confirmed as type III secreted by a heterologous Y. pseudotuberculosis system (Fig. 3). Within Chlamydia species, both CT618 and CT642 appear to be conserved (43), so it is possible that the MTS is conserved as well. CT132 and CT647 were not predicted to be type III secreted; however, it is possible that these proteins could be secreted through the general secretory system.
The proteomics analysis of isolated mitochondria was critical in demonstrating the validity of the mitochondrial localization for CT529 and CT618 from the initial screen, as both were included in the 49 chlamydial proteins identified (Table 2). Both CT529 and CT618 were previously identified in a T3S component screen as interacting with the same unique T3S chaperone, CT260 (Mcsc, multiple cargo secretion chaperone) (27). This chaperone may be important for proper secretion of CT529 and CT618, as they are translocated through the T3SS and recognized by host chaperone proteins. CT529 has been previously demonstrated to colocalize with lipid droplets; however, this study artificially stimulated lipid droplet formation, which may not reflect the localization of this protein during a normal infection of HeLa cells (10). Additionally, when CT529 was expressed with a C-terminal GFP, there did appear to be protein predominately localized outside lipid droplets, likely mitochondria considering the data presented herein. This localization was absent when CT529 was expressed with an N-terminal mCherry tag that likely disrupted the MTS, inhibiting the normal translocation of this effector. The function of CT529 and CT618 at the mitochondria remains unknown, and additional studies will be needed to determine the role these proteins play during chlamydial infection.
In the initial MitoProt MTS screen, we were only able to identify N-terminal MTS sequences. While many mitochondrial proteins rely on an N-terminal MTS signal, there are both internal MTS (44) and unique non-N-terminal MTS in β-barrel proteins (45), which are recognized and translocated by distinct mechanisms. There is no current bioinformatic mechanism to recognize these MTS within bacterial proteins. Therefore, the proteomics screen enabled us to identify additional chlamydial candidate effectors that may localize to mitochondria during infection (Table 2). It is possible that these chlamydial effectors may traffic to the mitochondria through alternate MTS or through direct or indirect proteins that associate with the mitochondrial outer membrane.
While the majority of chlamydial proteins identified at the mitochondria of infected cells have not been verified, two proteins have previously reported functions that support interaction with mitochondrial proteins: CT858 (CPAF) and CT610 (CADD). CPAF (chlamydial protease-like activity factor) has been hypothesized and demonstrated to have a variety of functions, particularly in cleaving a set of host and chlamydial proteins (46). Pirbhai et al. provided evidence that CPAF may cleave BH3 family proteins (like Bid) during chlamydial infection to prevent apoptosis (47), and in a study in Protochlamydia, a CPAF homolog was shown to disrupt mitochondrial membrane potential (48). These studies suggest that CPAF may target proteins of the mitochondria, which is supported by its identification in the mitochondrial proteome here. In fact, the absence of Bid at the mitochondria of infected cells (Fig. 5B) could be explained by its cleavage by CPAF. While the protein targets of CPAF have been disputed, Bid cleavage has not been specifically examined in vivo (49). CADD (Chlamydia protein associating with death domains) has shown to be involved in the apoptotic pathway, and although the initial analysis of CADD did not detect CADD localization at mitochondria with anti-CADD antiserum (50), our data suggest that CADD may interact with mitochondrially localized apoptotic effectors. Additionally, our initial bioinformatic screen did not identify CT421.1 as a candidate MTS, as the MitoProt probability fell just below the cutoff of 0.7. However, this protein was identified in all replicates of mitochondria from infected cells, and it is possible that CT421.1 does have a functional MTS. Further studies will be needed to investigate the validity of this promising candidate. Some caution will need to be exercised in evaluating the functions of the many chlamydial proteins identified by mass spectrometry. We cannot rule out the possibility that certain chlamydial proteins identified in mitochondria of infected cells were the result of lysed bacteria, particularly the ribosomal proteins and chaperones that have been shown in other proteomics studies to be common contaminants (51).
Through the proteomic screen, we were able to demonstrate changes in the protein content of mitochondria during infection (Fig. 4A). For this initial screen, we elected to use HeLa cells due to their ubiquitous utilization in chlamydial research, including for many protein-protein interaction screens within the field (52–56). Subsequent investigations of the specific pathways that may be altered by chlamydial infection and chlamydial effector function will likely incorporate primary cell models to validate the findings presented here.
Infection with C. trachomatis resulted in the absence of several proapoptotic factors and the recruitment of antiapoptotic factors. One regulatory pathway of apoptosis that appeared to be disproportionately affected was the mTORC1 (molecular target of rapamycin complex 1) pathway. The master activator of this pathway, RheB, was present only in infected mitochondria, suggesting activation of this pathway by Chlamydia (57). Along with RheB, the downstream mTORC1 effector, URI1, which maintains the phosphorylation and subsequent inactivation of Bad, a proapoptotic Bcl-2 family protein, was present at mitochondria only in infected cells (58). Several putative negative regulators of mTORC1 were also absent from or decreased in abundance in mitochondria of infected cells, including NRDC (nardilysin) (59). Another apoptotic pathway affected by infection involved the calcium influx into the mitochondria by the endoplasmic reticulum. Inositol 1,4,5-triphosphate receptor interacts with the mitochondrial outer membrane porin VDAC1 (voltage-dependent anion channel type 1), resulting in Ca2+ efflux into the mitochondria (60). The inositol 1,4,5-triphosphate receptor(s) are known to be recruited to the inclusion membrane by a microdomain Inc, MrcA, which may explain their absence in the infected proteome (61, 62). This report contributes to a growing body of knowledge within the field detailing the variety of mechanisms Chlamydia spp. utilize to inhibit apoptosis (56, 63–65).
In addition to changes to apoptotic pathways, C. trachomatis infection resulted in a shift of mitochondrial dynamics toward promotion of mitochondrial fusion, which corroborates similar findings (23, 64, 66, 67). These changes were identified by the absence of profission regulator UTRN (utrophin) (68) and the presence of pro-fusion regulators ANKHD1 (ankyrin repeat and KH domain-containing protein 1) (69) and UBL4A (ubiquitin-like protein 4A) (70) only in infected cells (Fig. 5). Mitochondrial dynamics play a critical role in metabolism, and fusion has been demonstrated to support increased oxidative phosphorylation and the production of ATP (71). Additionally, there were changes in the mitochondrial proteome to reflect this shift in increased metabolism. In particular, glycerol kinase (GK), which has been suggested to bind to the mitochondrial porin during increased metabolic activity (72), was bound to mitochondria only in infected cells. The proteomics data presented here suggest shifts in mitochondrial function during infection and can serve as a resource for further research into the specific changes induced by C. trachomatis.
In summary, we demonstrate that C. trachomatis secretes at least two proteins that associate with host mitochondria. The individual and collective roles of these proteins remain to be resolved, but it is clear that chlamydial infection induces changes in the mitochondrial proteome composition that provide multiple targets for detailed studies of pathogen subversion of host cells to promote the pathogen’s survival. An improved understanding of how Chlamydia bacteria manipulate mitochondria and other cellular organelles should provide new insights into how intracellular pathogens create favorable intracellular niches for replication.
MATERIALS AND METHODS
Strains and cell culture.
C. trachomatis serovar L2 (LGV 434/Bu) was propagated in HeLa 229 cells in RPMI 1640 medium containing 5% fetal bovine serum (FBS) at 37°C with 5% CO2. Elementary bodies (EBs) were purified by renografin density gradient as previously described (73), and infectious EBs were quantified by titration and manual counting as previously described (74) using indirect immunofluorescence with a rabbit polyclonal anti-C. trachomatis L2 EB antibody followed by an anti-rabbit IgG Alexa Fluor 488-conjugated secondary antibody (Invitrogen). Chlamydial genomic DNA was purified using the DNeasy blood & tissue kit (Qiagen). Briefly, purified EBs were boiled for 10 min and then incubated at room temperature in dithiothreitol (DTT) at a final concentration of 20% (vol/vol) for 15 min. The DNeasy blood & tissue DNA isolation kit (Qiagen) was used to extract genomic DNA, following the manufacturer’s instructions for Gram-negative bacteria.
Generation of C. trachomatis protein-GFP fusion constructs.
For each candidate effector, the open reading frame was amplified from C. trachomatis L2 genomic DNA by PCR with primers that added restriction sites on each end, using AccuPrime Pfx DNA polymerase (Invitrogen). DNA was extracted by gel purification using the QIAquick gel extraction kit (Qiagen). The pEGFP-N1 plasmid was purified using the maxiprep kit (Qiagen). Plasmid and insert products were digested using appropriate restriction enzymes (New England Biolabs) and ligated using T4 ligase (New England Biolabs). Constructs were transformed into chemically competent Turbo E. coli cells (New England Biolabs) according to the manufacturer’s instructions, and transformants verified by colony PCR and Sanger sequencing. Plasmids were extracted using a maxiprep kit. For specific effectors, the MTS was removed by generating additional primers inside the predicted MTS (or the first 33 amino acids for CT642) and adding the start codon back into the forward primer sequences.
Transfections and microscopy.
Purified plasmids were transfected into HeLa 229 cells using the Xfect transfection reagent (TaKaRa Biosciences) according to the manufacturer’s instructions. Briefly, 105 cells were plated onto 24-well plates with glass coverslips and allowed to adhere overnight. Plasmid DNA (1 μg per well) was mixed with reaction buffer (up to 25 μL per well). Polymer (0.3 μL per well) was mixed with reaction buffer (up to 25 μL per well), added to the plasmid-reaction buffer solution, and incubated at room temperature for 10 min. Fifty microliters of solution was added to each well and incubated for 4 h before the medium was replaced with fresh RPMI medium plus 5% FBS. Transfected cells were incubated for 48 h before staining with 100 nM MitoTracker red (Invitrogen) and DAPI (4′,6-diamidino-2-phenylindole) (NucBlue live ReadyProbes; Invitrogen) for 20 min at 37°C. Coverslips were washed twice in phosphate-buffered saline (PBS) and imaged under ×60 magnification on a Nikon Ti2e microscope using DAPI, fluorescein isothiocyanate (FITC) (GFP), and tetramethylrhodamine 5-isothiocyanate (TRITC) (MitoTracker) filters. Colocalization was determined quantitatively by NIS-Elements 64-bit software (version 5.11.02; Nikon). For images with less than 100% transfection (Fig. 1, eGFP and CT132), boundaries were drawn around the transfected cells, determined by positive GFP staining, and the plots were determined only for transfected cells. All microscopy was performed in at least duplicate experiments, with representative images presented.
T3S analysis using Y. pseudotuberculosis.
The first 50 or 100 amino acids of C. trachomatis genes were fused to Npt (neomycin phosphotransferase) in pFlag-CTC as previously described (37, 38). The constructs were transformed into wild-type Yersinia pseudotuberculosis (pIB102) cells or the ΔyscS strain (pIB68), and cultures were grown at 26°C for 2 h either in the presence or absence of 5 mM calcium. Cultures were shifted to 37°C, and fusion protein expression was induced with 0.01 mM IPTG (isopropyl-β-d-thiogalactopyranoside). Cultures were harvested after 4 h postinduction and separated into pellet or supernatant fractions. Samples were analyzed by immunoblotting with anti-Flag M2 antibodies or anti-YopN antibodies.
Isolation of mitochondria.
In triplicate experiments, four T-75 flasks were seeded with 1 × 106 HeLa 229 cells and allowed to adhere overnight. Two flasks were infected with C. trachomatis L2 at a multiplicity of infection of 1 to initiate a uniform infection. The cultures were centrifuged at 550 × g for 20 min at room temperature, fed with RPMI medium plus 5% FBS, and incubated at 37°C for 20 h to capture a middle stage of infection. The remaining flasks were mock infected. After incubation, cells were trypsinized and resuspended in PBS. One infected and one uninfected flask were treated with DSS Crosslinker (Thermo Scientific), following the manufacturer’s instructions for intracellular cross-linking. All samples were then resuspended in fluorescence-activated cell sorting (FACS) buffer (PBS with 1% bovine serum albumin [BSA] and 2.5 mM EDTA) (39) and stained with 100 nM MitoView 405 (Invitrogen) for 15 min at 37°C. Cells were pelleted and resuspended in ice-cold cell lysis buffer (200 mM sucrose, 10 mM Tris, pH 7.4, 0.5 mM EDTA, and 1× Halt protease inhibitor cocktail [Invitrogen] in PBS).
Mitochondria were sorted on a MACSQuant Tyto cell sorter (Miltenyi Biotech). The MACSQuant Tyto HS cartridge (Miltenyi Biotec) was primed using 0.4 mL of MACSQuant Tyto running buffer (Miltenyi Biotec) according to the manufacturer’s instructions. Fluorescently labeled beads were used to accurately identify the gating threshold for removal of debris and instrument noise. Cell lysates were loaded to a MACSQuant Tyto HS cartridge (product number 130-121-549; Miltenyi Biotec) and sorted according to the instrument instructions until 1 × 107 positive events had been collected. Purified mitochondria were resuspended with 2× Laemmli buffer with β-mercaptoethanol and incubated at 100°C for 10 min to denature proteins.
Sample preparation and LC-MS/MS.
Equal quantities of mitochondrial proteins were denatured using 10 mM TCEP [tris (2-carboxyethyl) phosphine] for 45 min at 56°C and alkylated using 20 mM iodoacetamide for 1 h at room temperature in the dark. The SP3 protocol was applied for protein cleanup (75). On-bead trypsin digestion was performed at 37°C overnight. The resulting peptides were desalted with C18 ZipTip pipette tips (Millipore, Bedford, MA, USA) for liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis.
A Q Exactive plus mass spectrometer (Thermo Fisher Scientific, San Jose, CA, USA) coupled with an Easy-nLC 1200 high-performance liquid chromatography (HPLC) system was utilized and was controlled by Xcalibur software (Thermo Fisher Scientific). Peptide samples were loaded onto an Acclaim PepMap 100 C18 trap column (75 μm by 20 mm, 3-μm particle size, 100 Å) in 0.1% formic acid and further separated on an Acclaim PepMap rapid-separation liquid chromatography (RSLC) C18 analytical column (75 μm by 250 mm, 2-μm particle size, 100 Å) using an acetonitrile-based gradient (solvent A was 0% acetonitrile, 0.1% formic acid, and solvent B was 80% acetonitrile, 0.1% formic acid) at a flow rate of 300 nL/min. The gradient was 2 to 25% B from 0 to 90 min, 25 to 40% B from 90 to 120 min, 40 to 100% B from 120 to 125 min, and 100% B from 125 to 127 min, followed by column wash and reequilibration to 2% solvent B. Electrospray ionization was carried out with an EASY-Spray source at a 275°C capillary temperature, 50°C column temperature, and 1.9 kV spray voltage. The mass spectrometer was operated in data-dependent acquisition mode with a mass range of 350 to 2,000 m/z. The full scan resolution was set to 70,000, with the automatic gain control (AGC) target at 1e6 and a maximum fill time of 30 ms. The fragment scan resolution was set to 17,500, with the AGC target at 5e4 and a maximum fill time of 50 ms. The normalized collision energy was set to 27. The dynamic exclusion was set for a 60-s duration and a repeat count of 1.
MS data analysis.
Raw data were acquired by the Xcalibur 4.2 system (Thermo Scientific, Bremen, Germany) and analyzed using Proteome Discoverer 2.4. The raw files were searched against Homo sapiens (Swiss-Prot version 2017-10-25), Chlamydia trachomatis (UniProtKB UP000000795), and common contaminants provided by MaxQuant. The search parameters were as follows: full trypsin digestion with two missed cleavages. Carbamidomethylation on cysteine residues was set as the fixed modification. A total of five variable modifications were allowed per peptide from the following list: oxidation on methionine, acetylation on the protein N terminus, pyro-glutamate conversion on N-terminal glutamate, methionine loss on the protein N terminus, and methionine loss and acetylation on the protein’s N-terminal methionine. The precursor peptide mass tolerance was set to 5 ppm. The fragment ion mass tolerance was set to 0.02 Da. The Percolator algorithm was used for peptide-spectrum match (PSM) validation. Proteins reported in the result have at least two distinct peptides detected in the analysis. Proteins detected fewer than 2 times in 3 replicates were excluded from statistical analysis. The statistical analysis of these data was completed using the unpaired, two-tailed Student’s t test. The full LCMS data set is shown in Table S4.
Proteome data from LC-MS. Download Table S4, XLSX file, 0.6 MB (651.1KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Bioinformatic analyses.
To predict MTS, open reading frames were translated from C. trachomatis serovar D (D/UW-3) and analyzed with MitoProt II version 1.101 (25). Genes encoding proteins with export-to-mitochondria probability scores of more than 0.7 were selected as candidate genes. C. trachomatis L2 proteins from the proteomics data set were also analyzed for MTS. Type III secretion prediction was performed using T3Sepp (accessible from http://www.szu-bioinf.org/T3SEpp/) (36). Functional analysis of proteomics hits was performed using DAVID (Database for Annotation, Visualization and Integrated Discovery) Bioinformatics Resources (https://david.ncifcrf.gov/home.jsp) (76, 77). Graphs were generated using GraphPad Prism (version 9.3.1). Homologs to C. trachomatis effectors were identified using the Protein Basic Local Alignment Search Tool (BLAST) against the specific chlamydial genome of interest, as follows: Chlamydia caviae strain GPIC (NC_003361), Chlamydia muridarum strain Nigg (NC_002620), Chlamydia suis strain R19 (NZ_CP035278.1), Chlamydia pneumoniae strain CWL029 (NC_000922.1), and Chlamydia pecorum (NC_015408.1). The percent identity was determined through MAFFT alignment of the amino acid sequence of the homolog to the C. trachomatis serovar D gene.
ACKNOWLEDGMENTS
This work was supported by the Intramural Research Program of the NIAID, NIH.
We thank Rebecca Miller for technical assistance and Adam Nock for review of this manuscript.
Contributor Information
Ted Hackstadt, Email: Ted_Hackstadt@nih.gov.
Sarah E. F. D’Orazio, University of Kentucky
REFERENCES
- 1.Solomon AW, Burton MJ, Gower EW, Harding-Esch EM, Oldenburg CE, Taylor HR, Traore L. 2022. Trachoma. Nat Rev Dis Primers 8:32. doi: 10.1038/s41572-022-00359-5. [DOI] [PubMed] [Google Scholar]
- 2.Senior K. 2012. Chlamydia: a much underestimated STI. Lancet Infect Dis 12:517–518. doi: 10.1016/S1473-3099(12)70161-5. [DOI] [PubMed] [Google Scholar]
- 3.Cocchiaro JL, Valdivia RH. 2009. New insights into Chlamydia intracellular survival mechanisms. Cell Microbiol 11:1571–1578. doi: 10.1111/j.1462-5822.2009.01364.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gitsels A, Sanders N, Vanrompay D. 2019. Chlamydial infection from outside to inside. Front Microbiol 10:2329. doi: 10.3389/fmicb.2019.02329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carabeo RA, Mead DJ, Hackstadt T. 2003. Golgi-dependent transport of cholesterol to the Chlamydia trachomatis inclusion. Proc Natl Acad Sci USA 100:6771–6776. doi: 10.1073/pnas.1131289100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hackstadt T, Rockey DD, Heinzen RA, Scidmore MA. 1996. Chlamydia trachomatis interrupts an exocytic pathway to acquire endogenously synthesized sphingomyelin in transit from the Golgi apparatus to the plasma membrane. EMBO J 15:964–977. doi: 10.1002/j.1460-2075.1996.tb00433.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Derre I. 2015. Chlamydiae interaction with the endoplasmic reticulum: contact, function and consequences. Cell Microbiol 17:959–966. doi: 10.1111/cmi.12455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dumoux M, Clare DK, Saibil HR, Hayward RD. 2012. Chlamydiae assemble a pathogen synapse to hijack the host endoplasmic reticulum. Traffic 13:1612–1627. doi: 10.1111/tra.12002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scidmore MA, Fischer ER, Hackstadt T. 2003. Restricted fusion of Chlamydia trachomatis vesicles with endocytic compartments during the initial stages of infection. Infect Immun 71:973–984. doi: 10.1128/IAI.71.2.973-984.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saka HA, Thompson JW, Chen YS, Dubois LG, Haas JT, Moseley A, Valdivia RH. 2015. Chlamydia trachomatis infection leads to defined alterations to the lipid droplet proteome in epithelial cells. PLoS One 10:e0124630. doi: 10.1371/journal.pone.0124630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boncompain G, Muller C, Meas-Yedid V, Schmitt-Kopplin P, Lazarow PB, Subtil A. 2014. The intracellular bacteria Chlamydia hijack peroxisomes and utilize their enzymatic capacity to produce bacteria-specific phospholipids. PLoS One 9:e86196. doi: 10.1371/journal.pone.0086196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scidmore MA. 2011. Recent advances in Chlamydia subversion of host cytoskeletal and membrane trafficking pathways. Microbes Infect 13:527–535. doi: 10.1016/j.micinf.2011.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marchi S, Morroni G, Pinton P, Galluzzi L. 2022. Control of host mitochondria by bacterial pathogens. Trends Microbiol 30:452–465. doi: 10.1016/j.tim.2021.09.010. [DOI] [PubMed] [Google Scholar]
- 14.Vercellino I, Sazanov LA. 2022. The assembly, regulation and function of the mitochondrial respiratory chain. Nat Rev Mol Cell Biol 23:141–161. doi: 10.1038/s41580-021-00415-0. [DOI] [PubMed] [Google Scholar]
- 15.Spinelli JB, Haigis MC. 2018. The multifaceted contributions of mitochondria to cellular metabolism. Nat Cell Biol 20:745–754. doi: 10.1038/s41556-018-0124-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bock FJ, Tait SWG. 2020. Mitochondria as multifaceted regulators of cell death. Nat Rev Mol Cell Biol 21:85–100. doi: 10.1038/s41580-019-0173-8. [DOI] [PubMed] [Google Scholar]
- 17.Estaquier J, Vallette F, Vayssiere JL, Mignotte B. 2012. The mitochondrial pathways of apoptosis. Adv Exp Med Biol 942:157–183. doi: 10.1007/978-94-007-2869-1_7. [DOI] [PubMed] [Google Scholar]
- 18.Chacinska A, Koehler CM, Milenkovic D, Lithgow T, Pfanner N. 2009. Importing mitochondrial proteins: machineries and mechanisms. Cell 138:628–644. doi: 10.1016/j.cell.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Omura T. 1998. Mitochondria-targeting sequence, a multi-role sorting sequence recognized at all steps of protein import into mitochondria. J Biochem 123:1010–1016. doi: 10.1093/oxfordjournals.jbchem.a022036. [DOI] [PubMed] [Google Scholar]
- 20.Lobet E, Letesson JJ, Arnould T. 2015. Mitochondria: a target for bacteria. Biochem Pharmacol 94:173–185. doi: 10.1016/j.bcp.2015.02.007. [DOI] [PubMed] [Google Scholar]
- 21.Kozjak-Pavlovic V, Ross K, Rudel T. 2008. Import of bacterial pathogenicity factors into mitochondria. Curr Opin Microbiol 11:9–14. doi: 10.1016/j.mib.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 22.Holmes A, Muhlen S, Roe AJ, Dean P. 2010. The EspF effector, a bacterial pathogen’s Swiss army knife. Infect Immun 78:4445–4453. doi: 10.1128/IAI.00635-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chowdhury SR, Reimer A, Sharan M, Kozjak-Pavlovic V, Eulalio A, Prusty BK, Fraunholz M, Karunakaran K, Rudel T. 2017. Chlamydia preserves the mitochondrial network necessary for replication via microRNA-dependent inhibition of fission. J Cell Biol 216:1071–1089. doi: 10.1083/jcb.201608063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsumoto A, Bessho H, Uehira K, Suda T. 1991. Morphological studies of the association of mitochondria with chlamydial inclusions and the fusion of chlamydial inclusions. J Electron Microsc (Tokyo) 40:356–363. [PubMed] [Google Scholar]
- 25.Claros MG, Vincens P. 1996. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur J Biochem 241:779–786. doi: 10.1111/j.1432-1033.1996.00779.x. [DOI] [PubMed] [Google Scholar]
- 26.Thomson NR, Holden MT, Carder C, Lennard N, Lockey SJ, Marsh P, Skipp P, O’Connor CD, Goodhead I, Norbertzcak H, Harris B, Ormond D, Rance R, Quail MA, Parkhill J, Stephens RS, Clarke IN. 2008. Chlamydia trachomatis: genome sequence analysis of lymphogranuloma venereum isolates. Genome Res 18:161–171. doi: 10.1101/gr.7020108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spaeth KE, Chen YS, Valdivia RH. 2009. The Chlamydia type III secretion system C-ring engages a chaperone-effector protein complex. PLoS Pathog 5:e1000579. doi: 10.1371/journal.ppat.1000579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mital J, Miller NJ, Fischer ER, Hackstadt T. 2010. Specific chlamydial inclusion membrane proteins associate with active Src family kinases in microdomains that interact with the host microtubule network. Cell Microbiol 12:1235–1249. doi: 10.1111/j.1462-5822.2010.01465.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bannantine JP, Griffiths RS, Viratyosin W, Brown WJ, Rockey DD. 2000. A secondary structure motif predictive of protein localization to the chlamydial inclusion membrane. Cell Microbiol 2:35–47. doi: 10.1046/j.1462-5822.2000.00029.x. [DOI] [PubMed] [Google Scholar]
- 30.Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. 2002. Molecular biology of the cell, 4th ed, Garland Science, New York, NY. [Google Scholar]
- 31.Fernandez RC, Weiss AA. 1994. Cloning and sequencing of a Bordetella pertussis serum resistance locus. Infect Immun 62:4727–4738. doi: 10.1128/iai.62.11.4727-4738.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rzomp KA, Moorhead AR, Scidmore MA. 2006. The GTPase Rab4 interacts with Chlamydia trachomatis inclusion membrane protein CT229. Infect Immun 74:5362–5373. doi: 10.1128/IAI.00539-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Faris R, Merling M, Andersen SE, Dooley CA, Hackstadt T, Weber MM. 2019. Chlamydia trachomatis CT229 subverts Rab GTPase-dependent CCV trafficking pathways to promote chlamydial infection. Cell Rep 26:3380–3390.e5. doi: 10.1016/j.celrep.2019.02.079. [DOI] [PubMed] [Google Scholar]
- 34.Li Z, Chen C, Chen D, Wu Y, Zhong Y, Zhong G. 2008. Characterization of fifty putative inclusion membrane proteins encoded in the Chlamydia trachomatis genome. Infect Immun 76:2746–2757. doi: 10.1128/IAI.00010-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ferrell JC, Fields KA. 2016. A working model for the type III secretion mechanism in Chlamydia. Microbes Infect 18:84–92. doi: 10.1016/j.micinf.2015.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hui X, Chen Z, Lin M, Zhang J, Hu Y, Zeng Y, Cheng X, Ou-Yang L, Sun MA, White AP, Wang Y. 2020. T3SEpp: an integrated prediction pipeline for bacterial type III secreted effectors. mSystems 5:e00288-20. doi: 10.1128/mSystems.00288-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fields KA, Fischer ER, Mead DJ, Hackstadt T. 2005. Analysis of putative Chlamydia trachomatis chaperones Scc2 and Scc3 and their use in the identification of type III secretion substrates. J Bacteriol 187:6466–6478. doi: 10.1128/JB.187.18.6466-6478.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bauler LD, Hackstadt T. 2014. Expression and targeting of secreted proteins from Chlamydia trachomatis. J Bacteriol 196:1325–1334. doi: 10.1128/JB.01290-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.MacDonald JA, Bothun AM, Annis SN, Sheehan H, Ray S, Gao Y, Ivanov AR, Khrapko K, Tilly JL, Woods DC. 2019. A nanoscale, multi-parametric flow cytometry-based platform to study mitochondrial heterogeneity and mitochondrial DNA dynamics. Commun Biol 2:258. doi: 10.1038/s42003-019-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Korsmeyer SJ, Wei MC, Saito M, Weiler S, Oh KJ, Schlesinger PH. 2000. Pro-apoptotic cascade activates BID, which oligomerizes BAK or BAX into pores that result in the release of cytochrome c. Cell Death Differ 7:1166–1173. doi: 10.1038/sj.cdd.4400783. [DOI] [PubMed] [Google Scholar]
- 41.Derre I, Pypaert M, Dautry-Varsat A, Agaisse H. 2007. RNAi screen in Drosophila cells reveals the involvement of the Tom complex in Chlamydia infection. PLoS Pathog 3:1446–1458. doi: 10.1371/journal.ppat.0030155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mital J, Miller NJ, Dorward DW, Dooley CA, Hackstadt T. 2013. Role for chlamydial inclusion membrane proteins in inclusion membrane structure and biogenesis. PLoS One 8:e63426. doi: 10.1371/journal.pone.0063426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lutter EI, Martens C, Hackstadt T. 2012. Evolution and conservation of predicted inclusion membrane proteins in chlamydiae. Comp Funct Genomics 2012:362104. doi: 10.1155/2012/362104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Diekert K, Kispal G, Guiard B, Lill R. 1999. An internal targeting signal directing proteins into the mitochondrial intermembrane space. Proc Natl Acad Sci USA 96:11752–11757. doi: 10.1073/pnas.96.21.11752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jores T, Klinger A, Groß LE, Kawano S, Flinner N, Duchardt-Ferner E, Wöhnert J, Kalbacher H, Endo T, Schleiff E, Rapaport D. 2016. Characterization of the targeting signal in mitochondrial beta-barrel proteins. Nat Commun 7:12036. doi: 10.1038/ncomms12036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Snavely EA, Kokes M, Dunn JD, Saka HA, Nguyen BD, Bastidas RJ, McCafferty DG, Valdivia RH. 2014. Reassessing the role of the secreted protease CPAF in Chlamydia trachomatis infection through genetic approaches. Pathog Dis 71:336–351. doi: 10.1111/2049-632X.12179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pirbhai M, Dong F, Zhong Y, Pan KZ, Zhong G. 2006. The secreted protease factor CPAF is responsible for degrading pro-apoptotic BH3-only proteins in Chlamydia trachomatis-infected cells. J Biol Chem 281:31495–31501. doi: 10.1074/jbc.M602796200. [DOI] [PubMed] [Google Scholar]
- 48.Matsuo J, Nakamura S, Ito A, Yamazaki T, Ishida K, Hayashi Y, Yoshida M, Takahashi K, Sekizuka T, Takeuchi F, Kuroda M, Nagai H, Hayashida K, Sugimoto C, Yamaguchi H. 2013. Protochlamydia induces apoptosis of human HEp-2 cells through mitochondrial dysfunction mediated by chlamydial protease-like activity factor. PLoS One 8:e56005. doi: 10.1371/journal.pone.0056005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen AL, Johnson KA, Lee JK, Sutterlin C, Tan M. 2012. CPAF: a chlamydial protease in search of an authentic substrate. PLoS Pathog 8:e1002842. doi: 10.1371/journal.ppat.1002842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stenner-Liewen F, Liewen H, Zapata JM, Pawlowski K, Godzik A, Reed JC. 2002. CADD, a Chlamydia protein that interacts with death receptors. J Biol Chem 277:9633–9636. doi: 10.1074/jbc.C100693200. [DOI] [PubMed] [Google Scholar]
- 51.Mellacheruvu D, Wright Z, Couzens AL, Lambert JP, St-Denis NA, Li T, Miteva YV, Hauri S, Sardiu ME, Low TY, Halim VA, Bagshaw RD, Hubner NC, Al-Hakim A, Bouchard A, Faubert D, Fermin D, Dunham WH, Goudreault M, Lin ZY, Badillo BG, Pawson T, Durocher D, Coulombe B, Aebersold R, Superti-Furga G, Colinge J, Heck AJ, Choi H, Gstaiger M, Mohammed S, Cristea IM, Bennett KL, Washburn MP, Raught B, Ewing RM, Gingras AC, Nesvizhskii AI. 2013. The CRAPome: a contaminant repository for affinity purification-mass spectrometry data. Nat Methods 10:730–736. doi: 10.1038/nmeth.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mirrashidi KM, Elwell CA, Verschueren E, Johnson JR, Frando A, Von Dollen J, Rosenberg O, Gulbahce N, Jang G, Johnson T, Jager S, Gopalakrishnan AM, Sherry J, Dunn JD, Olive A, Penn B, Shales M, Cox JS, Starnbach MN, Derre I, Valdivia R, Krogan NJ, Engel J. 2015. Global mapping of the inc-human interactome reveals that retromer restricts chlamydia infection. Cell Host Microbe 18:109–121. doi: 10.1016/j.chom.2015.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bugalhao JN, Luis MP, Pereira IS, da Cunha M, Pais SV, Mota LJ. 2022. The Chlamydia trachomatis inclusion membrane protein CT006 associates with lipid droplets in eukaryotic cells. PLoS One 17:e0264292. doi: 10.1371/journal.pone.0264292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dickinson MS, Anderson LN, Webb-Robertson BM, Hansen JR, Smith RD, Wright AT, Hybiske K. 2019. Proximity-dependent proteomics of the Chlamydia trachomatis inclusion membrane reveals functional interactions with endoplasmic reticulum exit sites. PLoS Pathog 15:e1007698. doi: 10.1371/journal.ppat.1007698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ohmer M, Tzivelekidis T, Niedenfuhr N, Volceanov-Hahn L, Barth S, Vier J, Borries M, Busch H, Kook L, Biniossek ML, Schilling O, Kirschnek S, Hacker G. 2019. Infection of HeLa cells with Chlamydia trachomatis inhibits protein synthesis and causes multiple changes to host cell pathways. Cell Microbiol 21:e12993. [DOI] [PubMed] [Google Scholar]
- 56.Tan GM, Lim HJ, Yeow TC, Movahed E, Looi CY, Gupta R, Arulanandam BP, Abu Bakar S, Sabet NS, Chang LY, Wong WF. 2016. Temporal proteomic profiling of Chlamydia trachomatis-infected HeLa-229 human cervical epithelial cells. Proteomics 16:1347–1360. doi: 10.1002/pmic.201500219. [DOI] [PubMed] [Google Scholar]
- 57.Groenewoud MJ, Zwartkruis FJ. 2013. Rheb and mammalian target of rapamycin in mitochondrial homoeostasis. Open Biol 3:130185. doi: 10.1098/rsob.130185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Elmore S. 2007. Apoptosis: a review of programmed cell death. Toxicol Pathol 35:495–516. doi: 10.1080/01926230701320337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yoon WH, Sandoval H, Nagarkar-Jaiswal S, Jaiswal M, Yamamoto S, Haelterman NA, Putluri N, Putluri V, Sreekumar A, Tos T, Aksoy A, Donti T, Graham BH, Ohno M, Nishi E, Hunter J, Muzny DM, Carmichael J, Shen J, Arboleda VA, Nelson SF, Wangler MF, Karaca E, Lupski JR, Bellen HJ. 2017. Loss of nardilysin, a mitochondrial co-chaperone for alpha-ketoglutarate dehydrogenase, promotes mTORC1 activation and neurodegeneration. Neuron 93:115–131. doi: 10.1016/j.neuron.2016.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kania E, Roest G, Vervliet T, Parys JB, Bultynck G. 2017. IP3 receptor-mediated calcium signaling and its role in autophagy in cancer. Front Oncol 7:140. doi: 10.3389/fonc.2017.00140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nguyen PH, Lutter EI, Hackstadt T. 2018. Chlamydia trachomatis inclusion membrane protein MrcA interacts with the inositol 1,4,5-trisphosphate receptor type 3 (ITPR3) to regulate extrusion formation. PLoS Pathog 14:e1006911. doi: 10.1371/journal.ppat.1006911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chamberlain NB, Hackstadt T. 2022. Chlamydia trachomatis suppresses host cell store-operated Ca2+ entry and inhibits NFAT/calcineurin signaling. bioRxiv. doi: 10.1101/2022.03.15.484523. [DOI] [PMC free article] [PubMed]
- 63.Verbeke P, Welter-Stahl L, Ying S, Hansen J, Hacker G, Darville T, Ojcius DM. 2006. Recruitment of BAD by the Chlamydia trachomatis vacuole correlates with host-cell survival. PLoS Pathog 2:e45. doi: 10.1371/journal.ppat.0020045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang Y, Lei W, Zhao L, Wen Y, Li Z. 2022. Insights into mitochondrial dynamics in chlamydial infection. Front Cell Infect Microbiol 12:835181. doi: 10.3389/fcimb.2022.835181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wong WF, Chambers JP, Gupta R, Arulanandam BP. 2019. Chlamydia and its many ways of escaping the host immune system. J Pathog 2019:8604958. doi: 10.1155/2019/8604958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kurihara Y, Itoh R, Shimizu A, Walenna NF, Chou B, Ishii K, Soejima T, Fujikane A, Hiromatsu K. 2019. Chlamydia trachomatis targets mitochondrial dynamics to promote intracellular survival and proliferation. Cell Microbiol 21:e12962. doi: 10.1111/cmi.12962. [DOI] [PubMed] [Google Scholar]
- 67.Chowdhury SR, Rudel T. 2017. Chlamydia and mitochondria—an unfragmented relationship. Microb Cell 4:233–235. doi: 10.15698/mic2017.07.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pant M, Sopariwala DH, Bal NC, Lowe J, Delfin DA, Rafael-Fortney J, Periasamy M. 2015. Metabolic dysfunction and altered mitochondrial dynamics in the utrophin-dystrophin deficient mouse model of Duchenne muscular dystrophy. PLoS One 10:e0123875. doi: 10.1371/journal.pone.0123875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kitamata M, Hanawa-Suetsugu K, Maruyama K, Suetsugu S. 2019. Membrane-deformation ability of ANKHD1 is involved in the early endosome enlargement. iScience 17:101–118. doi: 10.1016/j.isci.2019.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang H, Zhao Y, Yao Q, Ye Z, Manas A, Xiang J. 2020. Ubl4A is critical for mitochondrial fusion process under nutrient deprivation stress. PLoS One 15:e0242700. doi: 10.1371/journal.pone.0242700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yao CH, Wang R, Wang Y, Kung CP, Weber JD, Patti GJ. 2019. Mitochondrial fusion supports increased oxidative phosphorylation during cell proliferation. Elife 8:e41351. doi: 10.7554/eLife.41351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Östlund A-K, Göhring U, Krause J, Brdiczka D. 1983. The binding of glycerol kinase to the outer membrane of rat liver mitochondria: its importance in metabolic regulation. Biochem Med 30:231–245. doi: 10.1016/0006-2944(83)90089-3. [DOI] [PubMed] [Google Scholar]
- 73.Caldwell HD, Kromhout J, Schachter J. 1981. Purification and partial characterization of the major outer membrane protein of Chlamydia trachomatis. Infect Immun 31:1161–1176. doi: 10.1128/iai.31.3.1161-1176.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Furness G, Graham DM, Reeve P. 1960. The titration of trachoma and inclusion blennorrhoea viruses in cell cultures. J Gen Microbiol 23:613–619. doi: 10.1099/00221287-23-3-613. [DOI] [PubMed] [Google Scholar]
- 75.Hughes CS, Moggridge S, Muller T, Sorensen PH, Morin GB, Krijgsveld J. 2019. Single-pot, solid-phase-enhanced sample preparation for proteomics experiments. Nat Protoc 14:68–85. doi: 10.1038/s41596-018-0082-x. [DOI] [PubMed] [Google Scholar]
- 76.Huang da W, Sherman BT, Lempicki RA. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 77.Sherman BT, Hao M, Qiu J, Jiao X, Baseler MW, Lane HC, Imamichi T, Chang W. 2022. DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res 50(W1):W216–W221. doi: 10.1093/nar/gkac194. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Immunofluorescent staining shows GFP localization of transfected C. trachomatis proteins that did not have a functional MTS. HeLa cells on coverslips were transfected with either the empty EGFP-N1 vector (eGFP) or GFP-tagged C. trachomatis proteins (CT###) and incubated for 24 h before staining with 100 nM MitoTracker (red). (B) Colocalizations with other organelles demonstrated with anti-calnexin antibodies or anti-Golgi apparatus antibodies. Bar = 10 μm. Download FIG S1, TIF file, 2.6 MB (2.7MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Proteins with decreased abundance in mitochondria of infected cells. Download Table S1, DOCX file, 0.03 MB (28.9KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Proteins with increased abundance in mitochondria of infected cells. Download Table S2, DOCX file, 0.02 MB (23.3KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Chlamydia homologs to mitochondrial targeting candidate effectors. Download Table S3, DOCX file, 0.01 MB (15.6KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Proteome data from LC-MS. Download Table S4, XLSX file, 0.6 MB (651.1KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.