ABSTRACT
Pseudomonas aeruginosa is a Gram-negative opportunistic pathogen, the leading cause of acute and chronic infections in immunocompromised patients, frequently with high morbidity and mortality rates. The xenobiotic response element (XRE) family proteins are the second most common transcriptional regulators (TRs) in P. aeruginosa. However, only a few XRE-like TRs have been reported to regulate multiple bacterial cellular processes, encompassing virulence, metabolism, antibiotic synthesis or resistance, stress responses, and phage infection, etc. Our understanding of what roles these XRE-like small regulatory proteins play in P. aeruginosa remains limited. Here, we aimed to decipher the role of a putative XRE-type transcriptional regulator (designated LfsT) from a prophage region on the chromosome of a clinical P. aeruginosa isolate, P8W. Southern blot and reverse transcription quantitative PCR (RT-qPCR) assays demonstrated that LfsT controlled host sensitivity to the phage PP9W2 and was essential for efficient phage replication. In addition, electrophoretic mobility shift assays (EMSAs) and transcriptional lacZ fusion analyses indicated that LfsT repressed the lysogenic development and promoted the lytic cycle of phage PP9W2 by binding to the promoter regions of the gp71 gene (encoding a CI-like repressor) and several vital phage genes. Combined with RNA-seq and a series of phenotypic validation tests, our results showed that LfsT bound to the flexible palindromic sites within the promoters upstream of several genes in the bacterial genome, regulating fatty acid (FA) metabolism, spermidine (SPD) transport, as well as the type III secretion system (T3SS). Overall, this study reveals novel regulatory roles of LfsT in P. aeruginosa, improving our understanding of the molecular mechanisms behind bacterium-phage interactions.
IMPORTANCE This work elucidates the novel roles of a putative XRE family TR, LfsT, in the intricate regulatory systems of P. aeruginosa. We found that LfsT bound directly to the core promoter regions upstream of the start codons of numerous genes involved in various processes, including phage infection, FA metabolism, SPD transport, and the T3SS, regulating as the repressor or activator. The identified partial palindromic motif NAACN(5,8)GTTN recognized by LfsT suggests extensive effects of LfsT on gene expression by maintaining preferential binding to nucleotide sites under evolutionary pressure. In summary, these findings indicate that LfsT enhances metabolic activity in P. aeruginosa, while it reduces host resistance to the phage. This study helps us better understand the coevolution of bacteria and phages (e.g., survival comes at a cost) and provides clues for designing novel antimicrobials against P. aeruginosa infections.
KEYWORDS: Pseudomonas aeruginosa, XRE family transcriptional regulator, LfsT, phage infection, FA metabolism, SPD transport, T3SS, fatty acid metabolism, spermidine transport
INTRODUCTION
Phages have long been considered the predators of bacteria since 100 years ago. However, recent studies have indicated that bacterium-phage interactions are far more complex than initially thought (1). Bacterium-phage coevolution is a vital driver of many ecological and evolutionary processes such as population dynamics, the evolution of diversity, mutation rates, and evolutionary pathogen virulence (2–4). The phage-host arms race is maintained during phage adsorption, injection into bacteria, replication inside host cells, and prophage lysogeny (5). Bacterial defense mechanisms against phage predation can be broadly classified into two categories. The first category includes surface modifications that directly impair phage adsorption. The second category involves interference in DNA replication, transcription, and translation processes after the phage genome is injected into the bacteria, e.g., restriction-modification (RM) endonucleases, inhibitory proteins (Abi and Sie), bacteriophage exclusion (Brex), and CRISPR (6). Despite the many new types of defense systems that have been identified recently, including RNA editing, retron satellite DNA synthesis, chemical antiphage defense systems, and quorum sensing (7–9), our understanding of the underlying molecular mechanisms of bacterium-phage interactions remains limited. In addition, temperate phages decide between lytic (where they replicate and lyse their host) and lysogenic (where they integrate and keep the host viable) cycles during infections (10). Apart from the many intracellular signaling pathways and genetic circuits that contribute to this decision, communication between phages using small molecules also profoundly influences it (11). These interplays between bacteria and phages result in mutual benefits and progress rather than unilateral extinction.
Pseudomonas aeruginosa is the primary agent causing nosocomial infections of the lungs, wounds, blood, and urinary tract, often accompanied by high morbidity and mortality rates (12). Acute and chronic infections caused by this bacterium in critically ill patients are difficult to treat due to its adaptability and resistance to multiple antimicrobials (13). Moreover, P. aeruginosa is a versatile metabolic bacterium with intricate regulatory systems that help it survive in changing environments (14). Under different conditions, transcriptional regulation is instrumental in controlling gene expression for enduring diverse stresses (15). The xenobiotic response element (XRE) family transcriptional regulators (TRs) are the second most common TRs in P. aeruginosa, regulating the expression of various genes, encompassing virulence factor genes, metabolic genes, antibiotic synthesis and resistance genes, stress response genes, and phenotypic switching genes (16–21). Although the regulatory mechanism of XRE TRs induced by many xenobiotics (e.g., heavy metals, oxidants, toxins, and antibiotics) is well known in eukaryotes (22, 23), their performance in prokaryotes is still unclear. Furthermore, only a fraction of bacterial studies have focused on the XRE family TRs (24–27), so the functions of the XRE TRs in P. aeruginosa require further investigation.
Here, we analyze the mode of action of LfsT in phage PP9W2 infection and multiple metabolic processes of the bacterial strain P8W. Our results indicate that the putative XRE-type TR LfsT functions as a global regulator by binding directly to the core promoter regions of the gp71 gene (encoding a CI-like repressor protein) and several essential phage genes (encoding helicases or structural proteins) during infection. Besides, it also binds to the promoters of individual genes and divergent operons in the host genome, playing a role in diverse metabolic pathways, including fatty acid (FA) degradation, spermidine (SPD) transport, and the type III secretion system (T3SS). Taken together, these results show that LfsT mediates efficient phage replication and various bacterial metabolic pathways, helping the host fine-tune gene regulation during adaptation to different environments.
RESULTS
A putative regulator, LfsT, controls the phage sensitivity of the host.
Phage PP9W2 (GenBank accession number OM141125) is a linear double-stranded DNA phage with a 54-kb genome (see Fig. S1A in the supplemental material), which was isolated in our previous study (28). Its major capsid protein is highly homologous to those of seven related D3-like Pseudomonas phages (using lipopolysaccharide [LPS] as a phage receptor) according to phylogenetic tree analysis (Fig. S1B). Growth inhibition experiments indicated that the best MOI (multiplicity of infection) for phage PP9W2 was 1 (Fig. S1C). Furthermore, a one-step growth experiment demonstrated that the latency time and the burst size of phage PP9W2 were about 1.5 to 2 h and 99, respectively (Fig. S1D). We constructed a Tn5G transposon (Table 1) insertion library of the clinical P. aeruginosa strain P8W (GenBank accession number NZ_CP081477.2) (sensitive to PP9W2) to screen for phage-resistant (PR) mutants. A total of five PR mutants (Fig. S2) were found, and four insertion sites (I1 to I4) were located at a cluster of genes associated with LPS synthesis in P. aeruginosa (29–31). Meanwhile, only one (I5) was inserted immediately behind bp 81 of a putative transcription factor gene (GenBank accession number WP_006379425.1) from a predicted prophage region of P8W (32), comprising the XRE family helix-turn-helix (XRE-HTH) DNA binding domain (Fig. S3A). For convenience, we use the gene name lfsT (designated by its functions in the lytic development of phage PP9W2 and in the fatty acid metabolism, spermidine transport, and the T3SS of the bacterium P8W) throughout this article. PR1 to -4 exhibited severely truncated structures containing few core components and lost the O-chain part of the LPS profile compared to PR5 and the parent strain P8W (Fig. S3B). Moreover, we found significantly decreased LPS contents and adsorption rates of PR1 to -4 compared to PR5 and P8W (Fig. S3C and D). The phage resistance of PR1 to -4 may be attributed to the ineffective arrest of the infection process. However, the underlying molecular mechanism of the phage resistance of PR5 has not yet been determined as it displayed an LPS profile and an adsorption rate similar to those of the wild type.
TABLE 1.
Bacterial strains, phages, and plasmids used in this work
| Strain, phage, or plasmid | Genotype and/or description | GenBank accession no., reference, or source |
|---|---|---|
| Strains | ||
| P. aeruginosa | ||
| P8W | Wild-type P. aeruginosa clinical isolate | NZ_CP081477.2 |
| P8D | lfsT deletion derivative of P8W | This work |
| P8D/pCTX | Complemented (lfsT) strain of P8D | This work |
| PR1 | Tn5G insertion mutant (wapR) of P8W | This work |
| PR2 | Tn5G insertion mutant (wapP) of P8W | This work |
| PR3 | Tn5G insertion mutant (waaC) of P8W | This work |
| PR4 | Tn5G insertion mutant (waaC) of P8W | This work |
| PR5 | Tn5G insertion mutant (lfsT) of P8W | This work |
| P8D/pUCP18 | P8D carrying pUCP18 | This work |
| P8W/pUCP18 | P8W carrying pUCP18 | This work |
| P8D/pCT72N | P8D carrying pUCP18::gp72 | This work |
| P8D/pCT71N | P8D carrying pUCP18::gp71 | This work |
| P8W/pCT72N | P8W carrying pUCP18::gp72 | This work |
| P8W/pCT71N | P8W carrying pUCP18::gp71 | This work |
| E. coli | ||
| DH5α | F− ϕ80 lacZΔM15 Δ(lacZYA-argF)U169 endA1 recA1 endA1 hsdR17(rK− mK+) supE44 λ− thi-1 gyrA96 relA1 phoA | TianGen |
| S17-1 | RP4-2 Tc::Mu Km::Tn7 Tpr Smr Pro Res− Mod+ | TransGen |
| JM109 | endA1 recA1 gyrA96 thi hsdR17(rK− mK+) relA1 supE44D(lac-proAB) [F′ traD36 proAB laqIqZΔM15] | Biomed |
| BL21(DE3) | F− ompT hsdSB(rB− mB−) gal dcm(DE3) | Biomed |
| DH5α/pET32a(+) | DH5α carrying pET32a(+) | This work |
| DH5α/pOEX | DH5α carrying pET32a(+)::lfsT | This work |
| Phages | ||
| PP9W | Excised phage isolated previously | This work |
| PP9W2 | Lyses P8W or is lysogenic in it | OM141125 |
| PP27 | Excised phage isolated previously | This work |
| Plasmids | ||
| pRK2013Tn5G | Tn5G-carrying plasmid; Kmr Gmr | 69 |
| pEX18Tc | Gene replacement vector; Tcr oriT+ sacB+ | Weihui Wu |
| pEX18Tc::UD | lfsT gene knockout vector | This work |
| pUCP18 | Broad-host-range shuttle vector; Apr | Weihui Wu |
| pCTX | lfsT gene complementation vector | This work |
| pCT72N | pUCP18::gp72 | This work |
| pCT71N | pUCP18::gp71 | This work |
| pMD19 (Simple) | Linearized T vector for cloning promoter fragments; Apr | TaKaRa |
| pCP75 | gp75 gene promoter cloned into pMD19 | This work |
| pCP72 | gp72 gene promoter cloned into pMD19 | This work |
| pCP71 | gp71 gene promoter cloned into pMD19 | This work |
| pCP13 | gp13 gene promoter cloned into pMD19 | This work |
| pCP09 | gp09 gene promoter cloned into pMD19 | This work |
| pCP04 | gp04 gene promoter cloned in pMD19 | This work |
| pCP01 | gp01 gene promoter cloned into pMD19 | This work |
| pCPP | potA gene promoter cloned into pMD19 | This work |
| pCPF | faoA gene promoter cloned into pMD19 | This work |
| pCPN | popN gene promoter cloned into pMD19 | This work |
| pCP2 | PA2550 gene promoter cloned into pMD19 | This work |
| pDN19lacΩ | Promoterless lacZ fusion vector; Spr Smr Tcr | Weihui Wu |
| pDN75 | gp75 gene promoter cloned into pDN19lacΩ | This work |
| pDN71 | gp71 gene promoter cloned into pDN19lacΩ | This work |
| pDN13 | gp13 gene promoter cloned into pDN19lacΩ | This work |
| pDN09 | gp09 gene promoter cloned into pDN19lacΩ | This work |
| pDN04 | gp04 gene promoter cloned into pDN19lacΩ | This work |
| pDN01 | gp01 gene promoter cloned into pDN19lacΩ | This work |
| pDNP | potA gene promoter cloned into pDN19lacΩ | This work |
| pDNF | faoA gene promoter cloned into pDN19lacΩ | This work |
| pDNN | popN gene promoter cloned into pDN19lacΩ | This work |
| pDN2 | PA2550 gene promoter cloned into pDN19lacΩ | This work |
| pET32a(+) | Fusion vector for an N-terminal His tag; Apr | Novagen |
| pOEX | his-lfsT fusion in the pET32a(+) vector; Apr | This work |
| pEASY-T1 | Cloning vector for plotting the standard curves; Apr Kanr | TransGen |
| pEASY-L | Partially integrated fragment in pEASY-T1 | This work |
| pEASY-1789 | Partial PA1789 fragment in pEASY-T1 | This work |
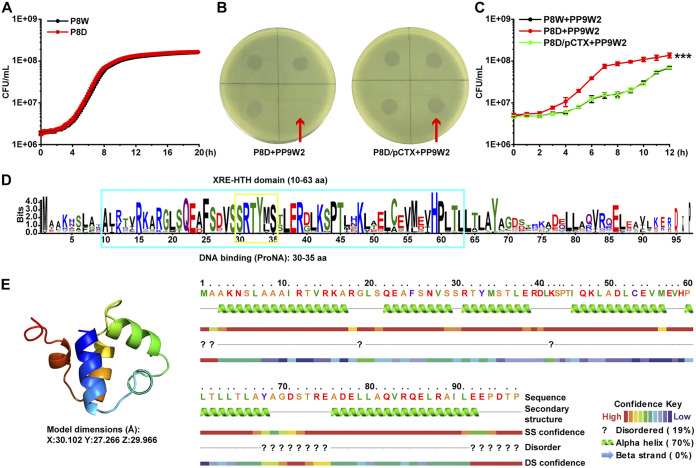
The regulation of the sensitivity of bacteria to the phage is an evolved mechanism during continual bacterium-phage interactions (33). Research in this area will improve our understanding of their relationships. We focused on the lfsT gene since it seems irrelevant to phage receptor modification. To better explore the functions of lfsT, we constructed a ΔlfsT derivate (P8D) (Table 1) of strain P8W by homologous recombination (for details, see Materials and Methods) to exclude other interferences (e.g., polar effects), rather than using the PR5 mutant directly in subsequent experiments. Growth curve assays indicated that the lfsT gene is not essential for host cell growth since no growth retardation was found in the P8D group (Fig. 1A). The introduction of the lfsT gene (using the recombinant plasmid pUCP18::lfsT [designated pCTX]) (Table 1) into P8D restored its sensitivity to phage PP9W2 (Fig. 1B). Further growth inhibition experiments (MOI = 1) demonstrated that P8W and P8D/pCTX (complementation group) displayed distinct growth delays compared to P8D when incubated with PP9W2 after 12 h (P < 0.001) (Fig. 1C). The LfsT protein (97 amino acids [aa]) contains an XRE-HTH domain (aa 10 to 63) with a predicted DNA binding site (aa 30 to 35) (Fig. 1D; Data Set S1), and the structural model of LfsT mainly included four α-helices with a λ repressor-like DNA binding domain belonging to the SinR family (Fig. 1E; Data Set S2). These data suggested that LfsT is a putative regulator of the clinical P. aeruginosa strain P8W, controlling the host’s sensitivity to the phage PP9W2.
FIG 1.
A hypothetical XRE-type transcriptional regulator, LfsT, impacts the sensitivity of bacterial strain P8W to phage PP9W2. (A) Growth experiment in LB medium for 12 h. The differently colored lines represent P8W and P8D (ΔlfsT). (B) Complementation (plasmid pCTX carrying the lfsT gene) restored host phage sensitivity in a spotting assay. (C) Growth inhibition of PP9W2 on the indicated strains (LB medium supplemented with 100 mg/L ampicillin) at an MOI (multiplicity of infection) of 1. The differently colored lines represent P8W, P8D, and P8D/pCTX. (D) Protein domain prediction for LfsT. The blue box represents an XRE-HTH domain from aa 10 to 63. The yellow box represents a DNA binding site from aa 30 to 35. More highly conserved amino acid residues are indicated by larger letters. (E, left) Protein structural model of LfsT built using SWISS-MODEL. The image is rainbow colored from the N to the C termini. (Right) Protein secondary structure of LfsT. The experiments were independently replicated three times, and each sample was tested in triplicate (A to C). Data were analyzed by one-way analysis of variance (ANOVA) with Tukey’s multiple-comparison test (α < 0.05) to examine the mean differences between the data groups. ***, P < 0.001. Error bars show standard deviations. SS, secondary structure; DS, disorder structure.
LfsT is essential for phage replication.
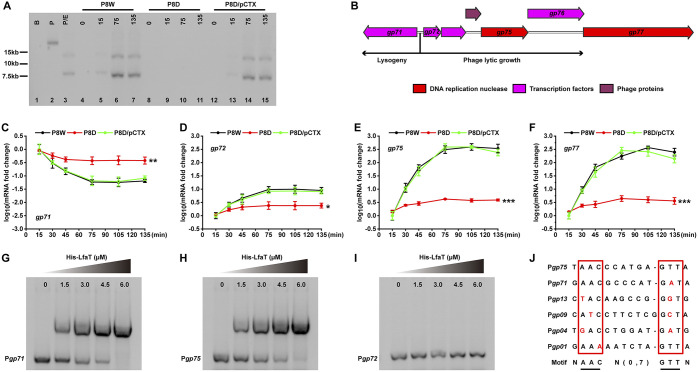
We used restriction endonuclease digestion (Table S1) to investigate the packaging mode of PP9W2 genomic DNA (gDNA) (34). As shown in Fig. S4A (lane 4), purified PP9W2 genomic DNA digested with FseI displayed two significant bands. After heat treatment, band A was replaced by two new positive bands (lane 6, bands B and C). This result was consistent with the illustration in Fig. S4B. The 13.2-kb fragment (A) was from the digested concatemeric DNA, while the 7.7-kb fragment (B) and the 5.5-kb fragment (C) were from the digested monomeric DNA. These data indicated that PP9W2 genomic DNA uses site-specific cos-type-like packaging. To identify whether the genome replication of PP9W2 was disrupted in P8D, Southern blot analysis was performed. Only fragments with the designed DNA probe (Fig. S4B) could be detected. No positive bands were detected in P8D, while P8W and P8D/pCTX displayed similar band profiles (Fig. 2A). These results demonstrated that LfsT is required for the genome replication of phage PP9W2 during infection.
FIG 2.
LfsT binds directly to the promoters of the gp71 gene and other crucial phage genes to influence phage replication. (A) Southern blotting (detailed in Materials and Methods). The genomic DNA in all lanes was digested with FseI, except for lane 2. Lane 1 (B), P8W genomic DNA; lanes 2 and 3 (P and P/E), phage PP9W2 genomic DNA; lanes 4 to 15, total DNA of different bacterial cells infected with phage PP9W2 at the indicated time intervals (0, 15, 75, and 135 min); lanes 4 to 7, P8W cells; lanes 8 to 11, P8D cells; lanes 12 to 15, P8D/pCTX cells. (B) Information on genes flanking the gp71/gp72 switch. The differently colored squares represent various proteins as indicated above. (C to F) RT-qPCR assay of the genes using cDNA of different strains infected with phage PP9W2 at the indicated time intervals. gp71 encodes a CI-like repressor protein, gp72 encodes a Cro-like repressor protein, gp75 encodes the putative helicase DnaK, and gp77 encodes the putative helicase DnaB. (G to I) EMSAs of different promoter fragments using the purified His-tagged LfsT protein. (G and H) LfsT binding to the gp71 or gp75 promoter; (I) Negative control. The purity of the His-tagged LfsT protein is about 97.9%. (J) Predicted LfsT binding sites of different promoters. gp13 encodes the putative tail component, gp09 encodes the phage gp6-like head-tail connector protein, gp04 encodes the phage portal protein, and gp01 encodes the terminase small subunit. A partial palindromic motif was speculated to be AACN(0,7)GTT. Red letters represent probable mutation sites. Each sample was tested in triplicate, and the experiments were independently replicated three times (C to F). Data were analyzed by one-way ANOVA with Tukey’s multiple-comparison test (α < 0.05) to examine the mean differences between the endpoints of data groups. Error bars show standard deviations. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Bacteriophage λ encodes two proteins that play opposing roles in phage growth. The λ repressor CI is required for lysogeny, while the Cro repressor turns off early gene transcription (35). We found a similar CI/Cro switch in the genome of PP9W2 (Fig. 2B; Fig. S5), which might control phage lysis-lysogeny decisions. To further investigate how LfsT impacts phage replication, reverse transcription quantitative PCR (RT-qPCR) assays using different primer pairs (Table 2) were conducted for the indicated strains. After 75 min of infection, the expression levels of gp71 (encoding a CI-like protein), gp72 (encoding a Cro-like protein), gp75 (encoding the putative helicase DnaK), and gp77 (encoding the putative helicase DnaB) reached the plateau phase in all of the tested strains (Fig. 2C to F). After another hour, the transcription levels of gp72, gp75, and gp77 in P8D were significantly reduced compared to those in P8W and P8D/pCTX (P < 0.05 and P < 0.001) (Fig. 2D to F). Meanwhile, the transcription level of gp71 in P8D displayed a contrasting increase (P < 0.01) (Fig. 2C). These results suggested that LfsT may be a critical factor that influences the transcription of these phage genes.
TABLE 2.
Primers used in this work
| Primer name | Sequence (5′–3′)a | Function or description |
|---|---|---|
| OTn1 | GATCCTGGAAAACGGGAAAG | Identification of Tn5G insertion sites |
| OTn2 | CCATCTCATCAGAGGGTAGT | |
| lfsTU-F | CCGGAATTCCGGTCAGTCATGGCGAAAC | 1-kb fragment upstream of lfsT |
| lfsTU-R | CGCGGATCCAACCAAGATGACAGCCCATTG | |
| lfsTD-F | CGCGGATCCGTTGGCGTGTGGTACTGAAT | 1-kb fragment downstream of lfsT |
| lfsTD-R | CCCAAGCTTGCGTCCTTGTCTTGTTGTGA | |
| lfsT-F | CCGGAATTCGTGGCAGCGAAGAACTCATTG | Cloning of the lfsT gene into pUCP18 |
| lfsT-R | CGCGGATCCCTAAGGCGTGTCGGGCTCTT | |
| 72N-F | CCGGAATTCGGAAGCTATGACCACCATCTAC | Cloning of the gp72 gene into pUCP18 |
| 72N-R | CGCGGATCCGCTGTCATGTCAGGCAACC | |
| 71N-F | CCGGAATTCTTCGGATGGAACTCAAAGAC | Cloning of the gp71 gene into pUCP18 |
| 71N-R | CGCGGATCCGCGATCTCCTATCCAATGC | |
| OEX-F | CGCGGATCCGTGGCAGCGAAGAACTCATTG | Expression of lfsT in pET32a(+) |
| OEX-R | CCCAAGCTTCTAAGGCGTGTCGGGCTCTT | |
| gp02-Fb | AATTACGGAGTGTCGGTCTTC | DNA probe synthesis for Southern blotting |
| gp02-R | GGATGGATGTGTTGGAGAGTC | |
| gp71-F | AGCATGGAGCCTTACATATTCG | RT-qPCR assay for gp71 |
| gp71-R | ATAAGCGGACTTGTCTGGATTG | |
| gp72-F | TTTGGGACTCAAGACGAGACC | RT-qPCR assay for gp72 |
| gp72-R | CAACCGCCGACAGCATTTC | |
| gp75-F | GCGATTATTACCTGTCCGTAGA | RT-qPCR assay for gp75 |
| gp75-R | GGTGTTCAGTCGGCAGATG | |
| gp77-F | GCGTTCCTGTGATTCTCCTAAG | RT-qPCR assay for gp77 |
| gp77-R | ACTCGGTATGCTCGTTGTAGA | |
| gp58-F | AGCAGCGTAGTGATGAATGGT | RT-qPCR assay for gp58 |
| gp58-R | AATCGGCTCCAGGTCGGTA | |
| gp36-F | CCACACTAAGGCAGGCAAG | RT-qPCR assay for gp36 |
| gp36-R | CGCAGGTCGTGAATCGTAA | |
| faoA-F | GCAAGCCGAAGAAGGTCAC | RT-qPCR assay for faoA |
| faoA-R | GGAAGCCGATGCCGTAGAT | |
| 2550-F | GCGGCAACATAGACCACATC | RT-qPCR assay for PA2550 |
| 2550-R | TGCATGGCGTACCAGTAGG | |
| potA-F | CAGCCTGACGATCAACACC | RT-qPCR assay for potA |
| potA-R | GCTCTGGAACACCGTATGC | |
| popN-F | GGACATCCTCCAGAGTTCCTC | RT-qPCR assay for popN |
| popN-R | AAGGCGAAGGTCAGCTCTT | |
| rpoD-F | CGTCCTCAGCGGCTATATCG | Reference gene for RT-qPCR assay |
| rpoD-R | TCTTCCTCGTCGTCCTTCTCT | |
| E71-F | ATGGGAACGCCCATGATAA | Promoter region of gp71 for EMSA |
| E71-R | CTGAGCAATATACGCCGATC | |
| E72-F | CCCTGCTGTACCGTATGAGT | Promoter region of gp72 for EMSA |
| E72-R | GACACAGGTCCTCTTTCTTGAA | |
| E75-F | GCGATTACTACAGGGCTTTGT | Promoter region of gp75 for EMSA |
| E75-R | GTTCTGTCATGCCGATCTTGT | |
| E13-F | GGACAGCCTGGAGCACATT | Promoter region of gp13 for EMSA |
| E13-R | CGCACCATCCGATCAAACC | |
| E09-F | TCTGCGGTGAGCTTCTGAC | Promoter region of gp09 for EMSA |
| E09-R | ACCATGCCAGTGAGACATCC | |
| E04-F | TCGTGGACTTCAACCAGACA | Promoter region of gp04 for EMSA |
| E04-R | TTTCCTTGGCGTCGATCTTTG | |
| E01-F | GAAATAGTCGGGTTCCATCAGC | Promoter region of gp01 for EMSA |
| E01-R | GGTGTCCTAGCGAAAGGTTCT | |
| Epot-F | GGCGAAGGAACATCGAAGAC | Promoter region of potA for EMSA |
| Epot-R | CGCATCCCGCTCTAACTAGA | |
| Efao-F | GGCGTATGAATCGAGCGTTT | Promoter region of faoA for EMSA |
| Efao-R | CAAGAGGCTTAACCGTGATGG | |
| Epop-F | GCGACGAATTTCAGTGCCA | Promoter region of popN for EMSA |
| Epop-R | CGGAGGAACTCTGGAGGATG | |
| E255-F | TCAGGTTGGCTTCGGTATAGAT | Promoter region of PA2550 for EMSA |
| E255-R | TGGTAACGATGCCGGAACA | |
| M13-47 | CGCCAGGGTTTTCCCAGTCACGAC | Amplification of the promoter regions cloned into pMD19 |
| RV-M | GAGCGGATAACAATTTCACACAGG | |
| L71-F | CCGGAATTCATGGGAACGCCCATGATAA | Promoter region of gp71 for lacZ fusion |
| L71-R | CGCGGATCCCTGAGCAATATACGCCGATC | |
| L75-F | CCGGAATTCGCGATTACTACAGGGCTTTGT | Promoter region of gp75 for lacZ fusion |
| L75-R | CGCGGATCCGTTCTGTCATGCCGATCTTGT | |
| L13-F | CCGGAATTCGGACAGCCTGGAGCACATT | Promoter region of gp13 for lacZ fusion |
| L13-R | CGCGGATCCCGCACCATCCGATCAAACC | |
| L09-F | CCGGAATTCTCTGCGGTGAGCTTCTGAC | Promoter region of gp09 for lacZ fusion |
| L09-R | CGCGGATCCACCATGCCAGTGAGACATCC | |
| L04-F | CCGGAATTCTCGTGGACTTCAACCAGACA | Promoter region of gp04 for lacZ fusion |
| L04-R | CGCGGATCCTTTCCTTGGCGTCGATCTTTG | |
| L01-F | CCGGAATTCGAAATAGTCGGGTTCCATCAGC | Promoter region of gp01 for lacZ fusion |
| L01-R | CGCGGATCCGGTGTCCTAGCGAAAGGTTCT | |
| Lpot-F | CCGGAATTCGGCGAAGGAACATCGAAGAC | Promoter region of potA for lacZ fusion |
| Lpot-R | CGCGGATCCCGCATCCCGCTCTAACTAGA | |
| Lfao-F | CCGGAATTCGGCGTATGAATCGAGCGTTT | Promoter region of faoA for lacZ fusion |
| Lfao-R | CGCGGATCCCAAGAGGCTTAACCGTGATGG | |
| Lpop-F | CCGGAATTCGCGACGAATTTCAGTGCCA | Promoter region of popN for lacZ fusion |
| Lpop-R | CGCGGATCCCGGAGGAACTCTGGAGGATG | |
| L255-F | CCGGAATTCTCAGGTTGGCTTCGGTATAGAT | Promoter region of PA2550 for lacZ fusion |
| L255-R | CGCGGATCCTGGTAACGATGCCGGAACA | |
| LC-F | AGCCACAATCCTGTGCTCTAC | Quantification of lysogenic copies |
| LC-R | AAAGGAATTTCACGATTGGCAC | |
| TC-Fc | AGGAAGGCTACAGCGTCTC | Quantification of total copies |
| TC-R | GGCGGTCTTGGTCATCAGT | |
The underlined sequences represent the sites of recognition of different restriction enzymes.
Primers gp02-F and gp02-R were used to amplify the designed probe region of gp02 (502 bp).
Primers TC-F and TC-R were used to amplify the specific fragment of phage PP9W2 integrated into the genome of P8W.
Considering the performance of LfsT during phage infection and its predicted structure, we hypothesized that it might bind to the promoters of some phage genes and verified this by an electrophoretic mobility shift assay (EMSA). As shown in Fig. 2G and H, the purified His-tagged LfsT protein bound specifically to the promoters of gp71 and gp75, and the promoter of gp72 was used as the control group (Fig. 2H; Fig. S5). Combined with the other four promoter sequences bound by LfsT (Table S2), we predicted a flexible palindromic structure, NAACN(0,7)GTTN (Fig. 2J), in these potential binding sequences using an online tool, MEME Suite (36). These data indicated that LfsT probably regulates the expression of some related phage genes by binding directly to their promoters during infection.
LfsT promotes phage lytic growth rather than lysogenic development.
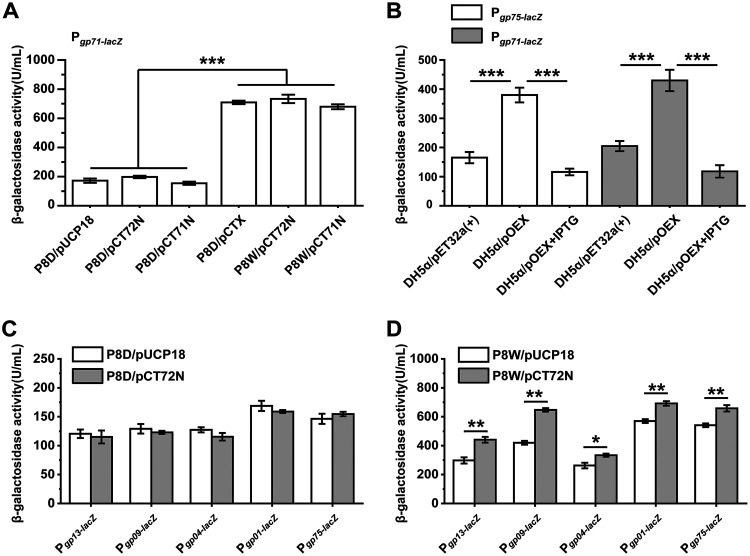
To further explore the underlying regulatory mechanism of LfsT, we measured the β-galactosidase activities of various double transformants (15). Using a transcriptional lacZ reporter fusion, we found that the gp71 promoter was more active in P8D/pCTX, P8W/pCT72N (pUCP18::gp72) (Table 1), and P8W/pCT71N (with the lfsT gene) than in P8D/pUCP18 (empty vector), P8D/pCT72N, and P8D/pCT71N (without the lfsT gene) (P < 0.001) (Fig. 3A). Whether in the presence or the absence of lfsT, the complementation of the Gp71 or Gp72 protein did not affect LacZ activity in the corresponding cell extracts. These results hinted that LfsT probably binds directly to the promoter region of gp71 to regulate its expression in P8W. Moreover, the CI/Cro-like Gp71/Gp72 switch in PP9W2 may be defective (Fig. S5). To further study the dose-effect patterns of LfsT (37), we overexpressed the lfsT gene in the heterologous host Escherichia coli DH5α treated with isopropyl β-d-1-thiogalactopyranoside (IPTG). Both the gp75 and gp72 promoters were significantly less active in response to LfsT (P < 0.001) (Fig. 3B). These data suggested the stimulatory (i.e., low dose) and inhibitory (i.e., high dose) roles of LfsT in regulating the transcription of these phage genes, which is analogous to hormetic dose-response mechanisms. Besides, the expression levels of several phage genes (encoding structural proteins) bound by LfsT (Fig. 2J) were analyzed, including the putative tail component gene gp13, the phage gp6-like head-tail connector protein gene gp09, the phage portal protein gene gp04, the terminase small-subunit gene gp01, and the putative helicase DnaK gene gp75. The expression of all of these genes is unaffected by the Gp72 protein since no difference between the P8D/pUCP18 and P8D/pCT72N groups was observed (Fig. 3C). However, it was significantly enhanced to different extents in the presence of LfsT, by 48.1% (P < 0.01), 54.2% (P < 0.01), 27.2% (P < 0.05), 21.3% (P < 0.01), and 21.6% (P < 0.01), respectively (Fig. 3D). These results indicated that the Gp72 protein is likely incapable of binding to the defective OR3-like operator and that the efficient transcription of these phage genes is dependent on LfsT.
FIG 3.
Function of LfsT in regulating vital phage genes. (A) LacZ activity in the indicated double transformants carrying pDN19lacΩ::Pgp71 (Pgp71-lacZ) evaluated by the corresponding β-galactosidase activities. pUCP18, empty vector; pCT72N, pUCP18::gp72; pCT71N, pUCP18::gp71; pCTX, pUCP18::lfsT. (B) LacZ activity in the indicated E. coli DH5α double transformants carrying pDN19lacΩ::Pgp75 (Pgp75-lacZ) or pDN19lacΩ::Pgp71 (Pgp71-lacZ). pET32a(+) is an empty expression vector. pOEX, pET32a(+)::lfsT. The overexpression of lfsT was started by the addition of 1.5 mM isopropyl β-d-1-thiogalactopyranoside (IPTG). (C) LacZ activity of different phage gene promoter-lacZ fusions in P8D carrying pUCP18 or pCT72N. (D) LacZ activity corresponding to the results in panel C for P8W carrying pUCP18 or pCT72N. All of the double transformants were grown in LB medium supplemented with 100 mg/L ampicillin, streptomycin, and tetracycline. The experiments were independently replicated three times, and each sample was tested in triplicate. Data were analyzed by ANOVA with Tukey’s multiple-comparison test (α < 0.05) to examine the mean differences between the data groups. *, P < 0.05; **, P < 0.01; ***, P < 0.001. Error bars show standard deviations.
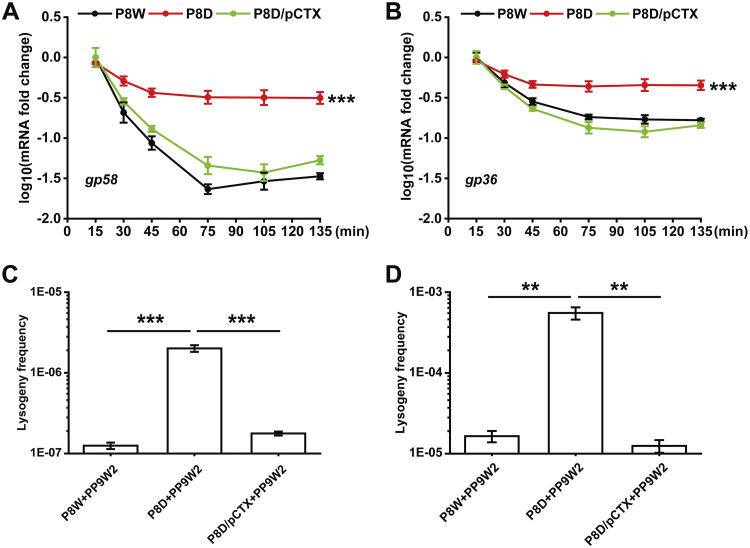
Considering that LfsT might bind to a truncated OR3-like position adjacent to the −10 promoter region (Fig. S5), we then asked whether LfsT would interfere with phage lysogenic development. First, we tested the expression of several lysogeny-related genes in the indicated strains by RT-qPCR analysis. The expression levels of the gp58 (encoding a YqaJ viral recombinase family protein) and gp36 (encoding a phage integrase) genes in P8D were significantly increased in comparison to the levels in P8W or P8D/pCTX after 135 min of infection (P < 0.001) (Fig. 4A and B). We also designed specific primer pairs (Table 2) to further calculate the related phage lysogeny frequencies by a quantitative PCR method (for details, see Materials and Methods). Although the lysogeny frequency of PP9W2 in all of the indicated strains increased with time (from 2 h to 12 h), it was significantly higher in P8D than in P8W or P8D/pCTX after 2 h (P < 0.001) (Fig. 4C) or 12 h (P < 0.01) (Fig. 4D) of growth. These data demonstrated that LfsT inhibits phage lysogenic development to a specific extent.
FIG 4.
Negative effects of LfsT on lysogenic phage development. (A and B) RT-qPCR assay for gp58 (encoding a YqaJ viral recombinase family protein) and gp36 (encoding a phage integrase) in different strains (colored lines) infected with phage PP9W2 at the indicated time intervals. (C and D) Lysogeny frequencies of phage PP9W2 in the indicated strains after 2 h (C) or 12 h (D) of growth. The experiments were independently replicated three times, and each sample was tested in triplicate. Data were analyzed by one-way ANOVA with Tukey’s multiple-comparison test (α < 0.05) to examine the mean differences between the indicated groups. **, P < 0.01; ***, P < 0.001. Error bars show standard deviations.
Identification of LfsT binding sites in the genome of P8W.
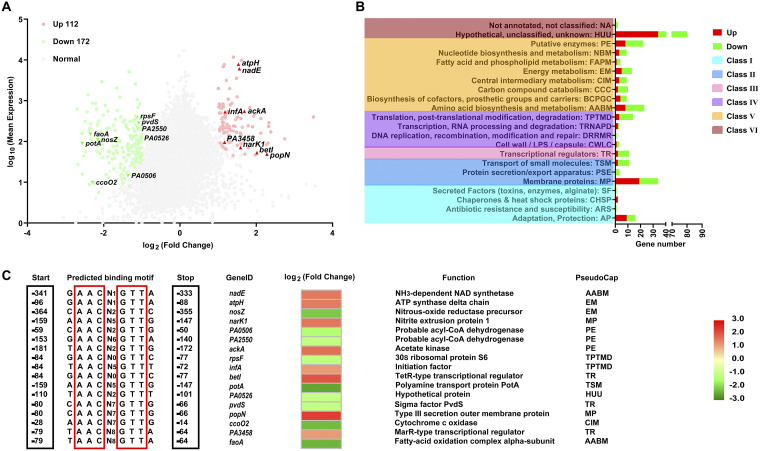
Numerous transcriptional regulators with uncharacterized XRE DNA binding domains were found to play critical roles in both secondary metabolism and bacterial adaptation to new environments (20). We conducted a transcriptome analysis to further explore the regulatory role of LfsT in host metabolic processes. As shown in Fig. 5A, the expression levels of a total of 284 genes were significantly altered between P8D and P8W (Data Set S3) (fold change [FC] of ≥2; false discovery rate [FDR] of ≤0.001), including 112 upregulated and 172 downregulated genes. These differentially expressed genes (DEGs) were grouped into 6 major classes (Fig. 5B) based on PseudoCAP (38), where most DEGs belonged to the PE (putative enzyme), AABM (amino acid biosynthesis and metabolism), MP (membrane protein), and AP (adaptation and protection) classes.
FIG 5.
Predicted LfsT-dependent genes and binding sites in the genome of P. aeruginosa P8W after RNA-seq. (A) Volcano plot of the transcriptome data (P8D versus P8W). Significantly differentially expressed genes (DEGs) (log2 fold change of ≥1; FDR of ≤0.001) are indicated in red (upregulated) and green (downregulated), while others are represented by gray dots, and genes with potential LfsT binding sites upstream of the start codons are indicated (red triangles and inverted green triangles). (B) Classification of DEGs expressed in response to LfsT deficiency based on PseudoCAP categories. When a gene was assigned to different categories, the first category was selected (see Data Set S3 in the supplemental material). Red and green bars represent the numbers of upregulated and downregulated DEGs, respectively. The PseudoCAP categories were further grouped into six classes (differently colored rectangles on the left). For clarity, some categories with no assigned DEGs are not displayed. (C) Search for the motif NAACN(0,9)GTTN across promoter regions within 500 bp upstream of 113 DEGs. Seventeen genes with predicted sites in the intergenic regions are shown.
Subsequently, we predicted the promoters of these DEGs and found 113 promoters (Data Set S4) within 500 bp upstream of their start codons (39). According to the results in Fig. 2J, we searched for derivative palindromic sequences (5′NAACN(0,9) GTTN-3′) across these potential promoter regions by MAST (Motif Alignment & Search Tool) analysis. Seventeen DEG promoters were identified to contain the putative LfsT binding sites in the intergenic regions, including 8 upregulated and 9 downregulated genes (Fig. 5A). The predicted sites were adjacent to the DEGs, ranging from 14 to 364 bp upstream, and the functions of these 17 DEGs are shown in Fig. 5C. To investigate which genes might be directly regulated by LfsT, we performed EMSAs with the 17 amplified promoter fragments using the indicated primer pairs (Table 2). As shown in Fig. S6, LfsT specifically bound to 4 promoters with different affinities, including potA (encoding a polyamine transport protein), faoA (encoding a fatty acid oxidation complex alpha subunit), popN (encoding a type III secretion outer membrane protein), and PA2550 (encoding a probable acyl-CoA dehydrogenase). Combined with the results in Fig. 2J, we speculated a more precise binding motif for LfsT, NAACN(5,8)GTTN. These results implied that LfsT possibly functions as a pleiotropic regulator with versatile roles in host metabolism.
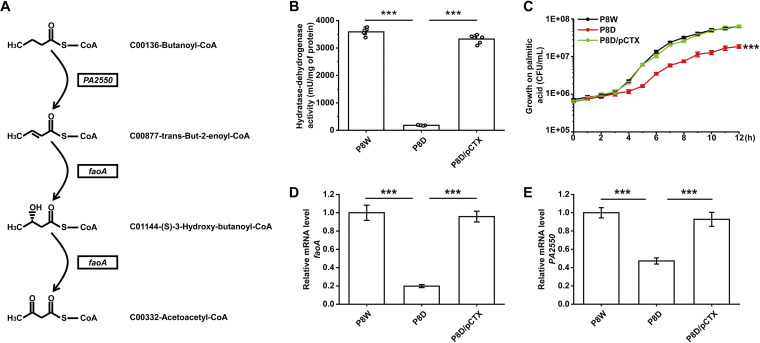
LfsT positively regulates fatty acid degradation.
P. aeruginosa grows better aerobically than E. coli on short-, medium-, and long-chain FAs as the sole carbon and energy sources (40). RNA-seq data indicated that the transcription of several FA metabolism-related genes was significantly altered in P8D, including enhanced expression of fabD, fabZ, and PA4389 but weakened expression of adhA, faoA, PA2550, and speA. Notably, faoA (encoding a fatty acid oxidation complex α-subunit) and PA2550 (encoding a probable acyl-CoA dehydrogenase) are two crucial genes in FA degradation. They function by dehydrogenation-oxidation in the gradual transformation of FAs from long-chain FAs to short-chain FAs, as shown in the schematic in Fig. 6A. A spectrophotometry assay (41) was used to measure HDT (the total multienzyme hydratase-dehydrogenase complex for FA β-oxidation) activity. We found much lower HDT activity in P8D than in P8W and P8D/pCTX, only 5.3% and 5.8%, respectively (P < 0.001) (Fig. 6B). Furthermore, we used the indicated strains to perform growth experiments on palmitic acid as the sole carbon and energy source. After 12 h of growth, significant growth retardation with less biomass of P8D than of P8W and P8D/pCTX was observed (P < 0.001) (Fig. 6C). RT-qPCR data for faoA and PA2550 were consistent with the above-described results (P < 0.001) (Fig. 6D and E). To further identify the role of LfsT in the regulation of faoA and PA2550, we constructed transcriptional lacZ reporter fusions. The faoA and PA2550 promoters were more active in P8D when complemented with the lfsT gene than with the empty vector (P < 0.001) (Fig. 7H). All of these results indicated that LfsT plays a positive role in FA degradation in strain P8W.
FIG 6.
Role of LfsT in fatty acid metabolism. (A) Schematic of the faoA (encoding the fatty acid oxidation complex α-subunit) and PA2550 (encoding a probable acyl-CoA dehydrogenase) genes engaging in the partial dehydrogenation-oxidation of fatty acids. (B) HDT activity assays of the indicated strains. (C) Growth curves of the related strains using 0.1% (wt/vol) palmitic acid as the sole carbon and energy source (supplemented with 100 mg/L ampicillin) for 12 h. (D and E) RT-qPCR assay of faoA (encoding the fatty acid oxidation complex α-subunit) and PA2550 (encoding a probable acyl-CoA dehydrogenase) in the indicated strains at mid-log phase (8 h in LB medium supplemented with 100 mg/L ampicillin). Each sample was tested in triplicate (C to E) or sextuplicate (B), and the experiments were independently replicated three times. Data were analyzed by one-way ANOVA with Tukey’s multiple-comparison test (α < 0.05) to compare the mean differences between the data groups. Error bars show standard deviations. ***, P < 0.001.
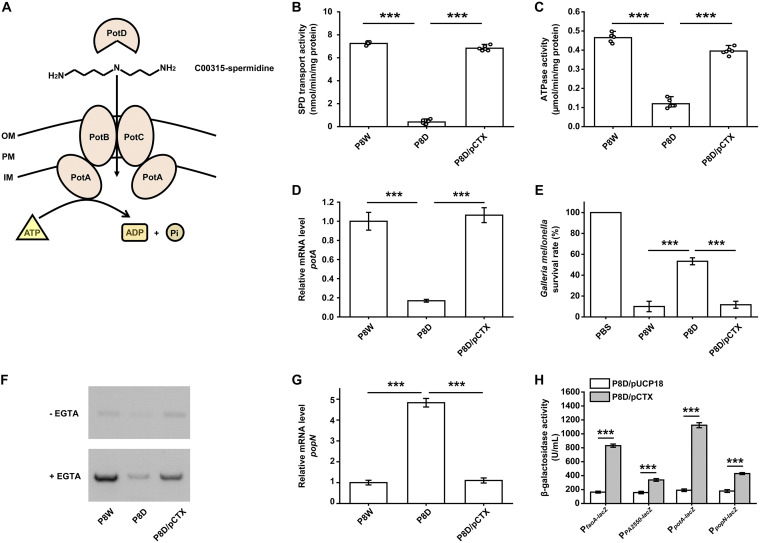
FIG 7.
Regulatory functions of LfsT in SPD (spermidine) transport and T3SS activity. (A) Schematic of the SPD transport system comprised of the PotA–PotD proteins in P. aeruginosa. OM, outer membrane; PM, plasma membrane; IM inner membrane. (B) SPD transport activity analysis of the indicated strains. (C) Corresponding ATPase activity assays of the related strains. (D) RT-qPCR assay of the potA gene (encoding a polyamine transport protein) in the indicated strains. Details are described above. (E) Galleria mellonella-killing assay using the indicated strains for 24 h. The group treated with phosphate-buffered saline (PBS) is the control group. (F) Detection of secreted ExoS (exoenzyme S) in different bacterial supernatants by Western blotting. (G) RT-qPCR assay for popN (encoding a type III secretion outer membrane protein). (H) LacZ activity of the indicated DEG promoter-lacZ fusions in P8D carrying pUCP18 or pCTX. The experiments were independently replicated three times, and each sample was tested in triplicate (D to H) or sextuplicate (B and C). Data were analyzed by one-way ANOVA with Tukey’s multiple-comparison test (α < 0.05) to compare the mean differences between the related groups. ***, P < 0.001. Error bars show standard deviations.
LfsT increases SPD transport and T3SS activity.
Polyamines (putrescine, spermidine, and spermine) are crucial for normal cell growth and viability, having been reported to modulate the functions of RNA, DNA, nucleotide triphosphates, proteins, ion channels, and other acidic substances (42). The polyamine content in cells is dependent on its biosynthesis, degradation, and transport (43). For P. aeruginosa, a spermidine-preferential uptake system comprised of PotA, PotB, PotC, and PotD is schematically depicted in Fig. 7A. The expression levels of potA, potB, potC, potD, and PA5376 were significantly downregulated in P8D, all of which belong to the ATP binding cassette (ABC) transporters in the transcriptome data. The SPD transport activity was determined by measuring [13C]spermidine uptake into ornithine-loaded inside-out membrane vesicles. We found that the SPD transport activity in P8D under specific growth conditions (for details, see Materials and Methods) was significantly downregulated compared to those in P8W and P8D/pCTX (P < 0.001) (Fig. 7B). Besides, we evaluated the ATPase activity of the indicated strains using an ATPase assay kit under the same conditions, and a similar distinct downregulation of the corresponding activity in P8D compared to P8W and P8D/pCTX was observed (P < 0.001) (Fig. 7C). Data from RT-qPCR and transcriptional lacZ reporter fusion analyses of potA further supported the above-described results (P < 0.001) (Fig. 7D and H). These data confirmed the positive impact of LfsT on SPD transport in P8W.
A critical factor in the pathogenesis of acute P. aeruginosa infections is the T3SS, by which the effector proteins are injected directly into host cells (44). Considering the significantly changed expression of T3SS-related genes in P8D (popN, pcr3, pcrD, pscO, and PA1697), we hypothesized that LfsT might influence P8W virulence in vivo. A Galleria mellonella-killing assay using the indicated strains for acute infection (24 h) was conducted. The relative rate of survival of the P8D-infected group was 5.3-fold higher than that of the P8W group, while it 4.6-fold higher than that of the P8D/pCTX group (P < 0.001) (Fig. 7E). This result was also identified by a Western blot assay. As shown in Fig. 7F, P8D exhibited less ExoS (exoenzyme S) secretion than P8W and P8D/pCTX induced by EGTA. The popN gene encodes a T3SS outer membrane protein precursor, which specifically interacts with Pcr1 and forms a T3SS repressor (45). We further determined the negative role of LfsT in regulating popN gene expression by RT-qPCR and lacZ reporter fusion assays (P < 0.001) (Fig. 7G and H). These results indicated that LfsT positively regulates T3SS activity in the host bacterium P8W.
DISCUSSION
Our understanding of bacterium-phage interplays and coevolution is limited, particularly concerning the small regulatory proteins (e.g., XRE-type TRs) in P. aeruginosa due to the lack of a deep understanding of its intricate regulatory systems. In the present study, we report a systematic functional analysis of a putative XRE-type transcriptional regulator, LfsT, in a clinical P. aeruginosa strain isolated from a burn patient, engaging in the control of phage infection and multiple host metabolic processes. These findings highlight an evolutionary mechanism behind bacterium-phage interactions and unveil a full-scale understanding of the regulatory network of P. aeruginosa metabolism and virulence, providing potential clues for novel drug targets designed to handle P. aeruginosa infections.
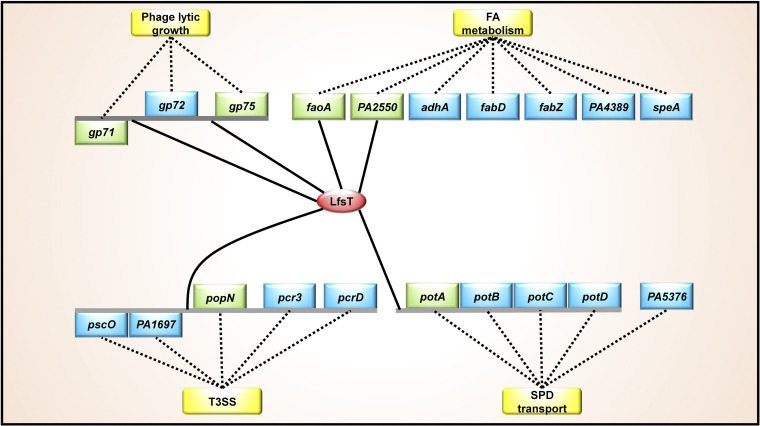
All of the identified functions of LfsT as a novel transcriptional regulator are summarized as a regulatory network in the schematic diagram in Fig. 8. Here, the global regulatory roles of LfsT are involved in several direct targets, including gp71, gp75 (phage infection), faoA, PA2550 (FA degradation), potA (SPD transport), and popN (T3SS) in P. aeruginosa. LfsT controls the phage sensitivity of a clinical P. aeruginosa strain, similar to the XRE-type regulator SrpA in P. aeruginosa reference strain PAK described previously (46). However, the underlying molecular mechanism of SrpA-mediated phage infection is coregulation with the RNA polymerase of the lytic phage K5 (46), and analogous to the SinR family λ phage repressor, LfsT regulates the transcription of numerous vital phage genes via binding specifically to the promoter regions. The gp71/gp72 gene pair resembles the ci/cro switch in phage λ, which binds to the same three right operons to perform autoregulation (47). Briefly, at low CI repressor concentrations, OR1 and OR2 are occupied to ensure that Cro synthesis is repressed while CI synthesis is activated. At high concentrations, OR3 is bound, which turns off ci gene transcription. The Cro binding pattern is just the opposite (35, 48–50). Interestingly, we found a truncated OR3-like operator in the PP9W2 genome by multiple alignments with the corresponding sequences of phage λ, which might be defective and disrupt the binding of the Gp71 and Gp72 proteins at this site. This may partially explain why no difference was found for the transcriptional Pgp71-lacZ fusion when complemented with the Gp71 or Gp72 protein. No hysteretic positive band was observed in EMSAs using the gp72 promoter fragment, indicating the specificity of the binding of LfsT to the predicted motif (Fig. 2J). The binding of LfsT to this site (in green in Fig. S5 in the supplemental material) of the core gp71 promoter sequences (in yellow) may interfere with RNA polymerase action since it is adjacent to the −10 promoter region (Fig. S5). This is probably why the transcription level of gp71 was significantly enhanced in P8D (without lfsT) during the latent period. Besides, the OR3-like operator overlaps the putative LfsT binding site, which further indicates the negative impact of LfsT on gp71 gene transcription.
FIG 8.
Scheme of the LfsT regulatory network in the host bacterium P. aeruginosa P8W. Solid lines represent LfsT-targeted sites in the promoter regions of individual genes or divergent operons. Dotted lines indicate links (direct or indirect) between the DEGs and various processes (in yellow rectangles) reported previously. The LfsT protein is depicted in the red oval. The genes directly subject to LfsT mediation are depicted in green rectangles, while others involved in the regulation of LfsT are depicted in blue rectangles. FA, fatty acid; SPD, spermidine; T3SS, type III secretion system.
In addition, the negative regulatory role of excess LfsT in some phage gene promoter activities (Fig. 3A and B) may be ascribed to DNA bending caused by the created multimers responsible for the repression state of TRs (51). Moreover, LfsT significantly represses the expression of some lysogeny-related genes and the lysogeny frequency of PP9W2 (Fig. 4). We speculate that the gp71/gp72 switch in phage PP9W2 is probably defective and disabled to fine-tune phage lysis-lysogeny decisions. The binding of LfsT to the predicted site in the gp71 promoter may remediate the defective regulation, directly limiting the predisposition to lysogeny and indirectly promoting the lytic development of the phage. The lfsT gene was possibly acquired via horizontal gene transfer (HGT) since its genomic context is between two prophage regions comprising numerous conjugal transfer-related genes (Fig. S3A). The role of LfsT in phage infection may develop to be an autoregulatory tool for specific phages for better adaption to undesirable genetic mutations under evolutionary pressure.
A more specific motif preferentially recognized by LfsT was identified as NAACN(5,8)GTTN using the other four predicted DEG promoter binding sites. It resembles a partial palindromic structure with a flexible center characterized by AT-rich regions at both ends, which is consistent with most transcriptional regulator binding sites (52). Furthermore, we searched for the motif across the genome of P8W in the Pseudomonas Genome Database, and more than 300 potential LfsT binding sites were identified (data not shown), indicating the possible broad LfsT protein interactions with DNA. Our data suggested that LfsT binding is not limited to one target. It may be speculated that LfsT maintains preferred nucleotide positions with a variable center in recognized sequences under evolutionary stress to exert a global regulation effect on host gene expression.
On the other hand, LfsT also significantly impacts versatile host metabolic processes to different extents. We found that the ΔlfsT mutant displayed worse HDT activity and adaptation to the growth of cells on palmitic acid as the sole carbon and energy source. This may be attributed to the downregulation of faoA and PA2550, which were directly subjected to LfsT-mediated regulation. Transcriptome analysis indicated another factor responsible for regulating FA metabolism, probably involving LfsT-dependent changes in DEGs such as adhA (alcohol dehydrogenase), fabD (malonyl-CoA-acyl carrier protein transacylase), fabZ [(3R)-hydroxymyristoyl-acyl carrier protein dehydratase], PA4389 (3-oxacyl-acyl carrier protein reductase), and speA (arginine decarboxylase), which have been reported to be associated with the biosynthesis, metabolism, and degradation of FA (53–57). It is worth mentioning that LfsT may regulate the transcription of divergent operons rather than individual genes (Fig. 8). The potA–potD operon is subject to LfsT regulation since the transcription levels of all of these genes were significantly diminished in the absence of LfsT. The expression of the PotA–PotD proteins is required for high SPD transport activity (43, 58–60). The lfsT-deficient mutant exhibited decreased SPD and ATPase activity compared to the parent strain, indicating stimulation by LfsT binding to the potA–potD promoter. We speculate that the interaction of LfsT with DNA may encompass core promoter sequences, positively influencing RNA polymerase activity and resulting in the promotion of potA–potD gene expression. Besides, lfsT is not an essential gene for normal cell growth in LB medium (Fig. 1A). However, it impacts SPD uptake and ATPase activity tested in medium A (see details in Materials and Methods), indicating that the regulation of the potA–potD operon by LfsT may depend on specific growth conditions. Traditionally, the development of antimicrobials for anti-P. aeruginosa therapeutics is based on interference with essential gene targets (61). Our findings expand the useful weapons of “nonessential genes” (e.g., the lfsT gene acquired by HGT) for potential targets for the development of novel drugs against P. aeruginosa infections.
In P. aeruginosa, FA and SPD have been reported to regulate T3SS expression and virulence in vivo by modulating intercellular signal molecules (62–64). Particularly, significantly increased expression levels of the pscO-PA1697 and popN-pcrD genes were found in response to LfsT deficiency. LfsT binding to the predicted site from bp 66 to 88 upstream of popN was identified, which might reduce the access of RNA polymerase to −10 promoter sequences. We observed an increased rate of Galleria mellonella survival in the absence of LfsT. This might be manifested by the decreased secretion of ExoS (Fig. 7E and F). Our results suggested the role of LfsT in popN-pcrD operon inhibition, and we speculate that it is an energy-efficient way for these genes to be repressed in a tightly controlled manner in the cell to save resources for gene expression. Taken together, these results show that the acquired LfsT, probably by HGT, has evolved into an important functional regulatory protein for host bacterial metabolism and virulence.
In light of the global regulatory effects of LfsT, we performed a DIAMOND BLASTP search of the Pseudomonas Genome Database with default parameters. A total of 134 complete genomes contained LfsT homologs (Data Set S5), about 20% of the total complete genomes online, indicating that LfsT-like proteins are widely distributed among Pseudomonas strains. However, its performance in various strains may differ due to the discrepant genomic contexts. It is not surprising that some previously described genes exert different regulatory effects on various bacterial subspecies (15, 65, 66). We speculate that this may be due to many factors, e.g., the existence of an alternative pathway in the complicated transcriptional regulatory web, which allows the complex and precise response of bacterial cells to environmental stress. This study adds a new player (LfsT) to the existing network of multiple metabolic pathways in P. aeruginosa, providing a deeper understanding of phage-host interactions and indicating the role of these XRE-type TRs in the adaptation of host cells to rapidly changing environments. However, the precise role of LfsT in P. aeruginosa awaits elucidation; e.g., (i) analogous to other XRE-type TRs, LfsT regulation probably requires other specific signaling molecules or growth conditions apart from binding sequences; (ii) LfsT improves the metabolic activity of the bacteria, while it also increases the phage sensitivity of the host, which may be speculated by adaptive costs under continual coevolution between bacteria and phages; (iii) some LfsT-dependent targets are also involved in another intricate regulatory network that may be coregulated by other TRs; and (iv) how these LfsT targets are expressed in response to excess LfsT remains to be determined. Further studies are required to determine the underlying molecular mechanisms engaged in these ways.
MATERIALS AND METHODS
Bacterial strains, phages, plasmids, and culture conditions.
The bacterial strains, phages, and plasmids used in this study are detailed in Table 1. We routinely cultured the strains in LB medium supplemented with the appropriate antibiotics at 37°C unless stated otherwise. The primers utilized in this work are detailed in Table 2.
Characterization of phage PP9W2.
We performed a phylogenetic analysis based on the major capsid proteins of the selected D3-like Pseudomonas phages. The neighbor-joining tree (topology only) was constructed using MEGA 7, and the branch lengths are shown in Fig. S1B in the supplemental material. A growth inhibition experiment was conducted as previously described (67). Briefly, the indicator strain P8W was incubated with phage PP9W2 at different MOIs (0.1, 1, and 10). Next, the mixtures were added to fresh LB medium for 12 h of growth. The optical densities of the samples were measured in triplicate at 1-h intervals. A one-step growth experiment was performed as previously described (68). In brief, the medium of a P8W culture grown overnight was mixed with a purified phage PP9W2 solution (MOI = 1) supplemented with 10 mM CaCl2. The culture was continuously incubated at 37°C for 6 h. We obtained samples (100 μL) in triplicate every 30 min and determined the phage titer by an overlay agar plate method.
Construction of a Tn5G mutant library.
We used transposon Tn5G to generate a mutant bank to screen for phage PP9W2-resistant mutants of strain P8W as described previously, with minor modifications (69). Briefly, the bank was cultivated at 37°C for 2 h and then incubated with the purified phage PP9W2 stock solution. The mixture was plated onto Luria-Bertan (LB) medium-agar plates with 100 μg/mL ampicillin and gentamicin. A spotting assay was used to identify the phage-resistant mutants.
Identification of insertion sites.
Inverse PCR was performed to identify the insertion sites in the PR mutants as described previously (70). The PCR fragments amplified by using the primer pairs listed in Table 2 were cloned into the pGEM T-Easy vector for further sequencing. The genome of P8W was used to analyze sequence homology.
LPS profile and content analyses.
LPS profile analysis of the related strains and mutants was performed as previously described (71). We used an LPS extraction kit (catalog number ab239718; Abcam) to extract LPS. The total carbohydrate content (grams per CFU) was quantified according to a total carbohydrate quantification assay (catalog number ab155891; Abcam).
Adsorption rate assay.
Briefly, the related cells cultured overnight were resuspended in fresh LB medium supplemented with 10 mM CaCl2; next, the culture was mixed with the purified phage stock solution (MOI = 1) and incubated without shaking at 37°C for 30 min. The phage adsorption rate was quantified as previously described (72).
Construction of the lfsT gene deletion derivative P8D.
We used a homologous-recombination method to knock out the lfsT gene in the P8W genome. Briefly, we first amplified the upstream and downstream fragments of the lfsT gene and ligated the molecules. Next, the fragment was ligated with the enzyme-digested plasmid pEX18Tc. The resulting plasmid, pEX18Tc::UD, was first introduced into E. coli DH5α, then transformed into E. coli S17-1, and finally introduced into P8W by conjugation. The lfsT gene mutants were screened using L-agar plates supplemented with 100 mg/L tetracycline, 50 mg/L kanamycin, and 7.5% sucrose. The positive clones were identified by PCR and DNA sequencing.
Complementation experiment.
The amplicon of the lfsT gene was digested with the corresponding restriction enzymes and cloned into plasmid pUCP18. The recombinant plasmid (pCTX) was transformed into competent cells of P8D by electroporation. The complemented clone was screened using L-agar plates supplemented with 100 mg/L ampicillin and further identified by PCR and DNA sequencing.
Growth curve experiment.
Strains cultured overnight were inoculated into fresh LB medium on 96-well plates. The optical density at 600 nm (OD600) value was quantified by using an automatic microplate reader (Bioscreen, Finland) at 5-min intervals for 20 h of growth. The test was independently replicated three times, and each sample was tested in quintuplicate.
LfsT protein structure prediction.
Domain prediction was performed using an online tool (http://smart.embl-heidelberg.de/). We searched the amino acid sequence of LfsT across the nonredundant (nr) database by PSI-BLAST with an expected threshold of 1E−10. A total of 1,000 LfsT-homologous sequences (Data Set S1) were used to generate a WebLogo figure using an online tool (http://weblogo.threeplusone.com/create.cgi). SWISS-MODEL was used to conduct homology modeling of the LfsT-like protein structure as previously described (73). Another 1,000 LfsT homologs (Data Set S2) were involved in the analysis. Sixty-four residues (66% of the LfsT sequence) were modeled with 99.5% confidence by the single highest-scoring template, d1b0na2 (SinR domain-like). Ninety residues (93%) could be modeled at >90% confidence using multiple templates.
Analysis of the packaging process for phage PP9W2 genomic DNA.
The manipulation suite (Table S1) of the phage PP9W2 genomic DNA sequence was analyzed as described previously (74). Purified phage genomic DNA was digested with the restriction enzyme FseI and then, immediately or after heating (75°C for 10 min), separated on a 0.8% agarose gel by electrophoresis.
Southern blot analysis.
Briefly, the related strains were incubated with phage PP9W2 at an MOI of 1 according to the growth inhibition experiment results. We sampled the strains at 0 min (control), 15 min (after adsorption), 75 min (1 h of infection), and 135 min (2 h of infection). Total DNA was extracted from the collected cells and then digested with the restriction endonuclease FseI. The primer pairs listed in Table 2 were used to synthesize the designed probe region of gp02 from bp 1304 to 1805 of the PP9W2 genome to detect the genomic DNA of phage PP9W2 (Fig. S4B). The genomic DNAs of P8W and phage PP9W2 were selected as negative and positive controls, respectively. Probe synthesis and DNA hybridization were performed using detection starter kit II (Roche), according to the manufacturer’s instructions.
Reverse transcription-quantitative real-time PCR.
We incubated strain P8W, P8D, or P8D/pCTX with phage PP9W2 at an MOI of 1. We sampled the culture every 15 min or 30 min and immediately performed total RNA extraction using a total RNA extraction kit (Promega). Next, 1 μg total RNA was subjected to reverse transcription using the FastKing gDNA dispelling RT supermix (TianGen). The cDNA was finally subjected to RT-qPCR analysis with the primers indicated in Table 2. A total volume of 20 μL comprising 10 μL of TB green premix ExTaq II (TaKaRa Bio), 8.2 μL of deionized water, 0.4 μL each of the primers (10 mM), and 1 μL of the template was used for analysis in triplicate. A typical 2−ΔΔCT method was used to perform relative quantification (75).
Electrophoretic mobility shift assay.
We first purified the LfsT protein. Plasmid pET32a(+) was used to generate a recombinant plasmid, pOEX, for the expression of the lfsT gene in E. coli BL21(DE3).
A total of 1.5 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) was added to induce the overexpression of lfsT. We used a one-step bacterial active protein extraction kit (Sangon Biotech) and a Ni-nitrilotriacetic acid (NTA)-Sefinose column (Bio Basic, Canada) to extract the His-tagged LfsT protein according to the manufacturers’ protocols. A desalting gravity column (Sephadex) was used to purify the target protein.
For electrophoretic mobility shift assays (EMSAs), we amplified the predicted promoter sequences (Table S2 and Data Set S4) of the tested genes with the primers indicated in Table 2. The PCR fragments (150 to 250 bp) were first cloned into pMD19 (Simple; TaKaRa), and the ligated fragments were then further amplified using the primer pair M13-47 and RV-M. We used an EMSA kit (Molecular Probes) to perform the analysis according to the manufacturer’s instructions. The DNA-protein complexes were separated by 5% nondenaturing polyacrylamide gel electrophoresis. The gel was visualized with an automatic digital gel image analysis system (Tanon 1600R).
Construction of lacZ transcriptional reporters.
The predicted promoter fragments of the tested genes were amplified with the primers indicated in Table 2. The related PCR products were cloned into the digested pDN19lacΩ plasmid (76). The recombinant plasmids were then transformed into different strains by electroporation. The activities of the corresponding promoters were represented by the β-galactosidase activities of the double transformants.
Lysogeny frequency assay.
The lysogeny frequency of phage PP9W2 in different strains was quantified by a modified quantitative PCR method as previously described (77). Briefly, the number of total P8W copies was quantified based on the single-copy gene PA1789. The number of lysogen copies was quantified using the primers indicated in Table 2, which amplified the fragment only when the phage was integrated. We sampled different strains after 2 h or 12 h of growth to quantify the lysogeny frequency.
Transcriptome analysis of P8D compared to P8W.
The related bacterial cells were grown to an OD600 of 0.8, and the harvested cells were immediately subjected to freezing in liquid nitrogen and delivered to Novogene (Beijing, China) on dry ice. Briefly, total RNA was extracted and subjected to rRNA removal; purified mRNA was fragmented before cDNA synthesis. The cDNA libraries were sequenced in triplicate on an Illumina HiSeq platform. Alignment of the clean reads to the reference genome (P. aeruginosa P8W [GenBank accession number NZ_CP081477.2]) was conducted using Bowtie (78). A total of 17,737,358 (97.37%) reads were mapped to the reference genome in the sample of P8W, and 19,416,320 (95.67%) reads were mapped to the reference genome in the sample of P8D. Genes with an adjusted P value of <0.05 by DESeq were assigned as differentially expressed, of which the DEGs with an FDR of ≤0.001 and a fold change (FC) of ≥2 were regarded as significantly changed. The Pseudomonas Genome Database (https://www.pseudomonas.com/) was used to assign the DEGs based on PseudoCAP. P. aeruginosa PAO1 was selected as the reference genome to convert gene identifiers (http://kobas.cbi.pku.edu.cn/kobas3/annotate/) for comparison.
HDT activity assay.
We measured the overall activities of enoyl-CoA hydratase and 3-hydroxyacyl-CoA dehydrogenase (HDT) as described previously (41). In brief, the related bacterial cells cultured overnight were harvested and resuspended in 1 mL of Tris-EDTA (TE) containing 0.5% (wt/vol) Triton X-100, 50 μg/mL of DNase I and RNase A, and 1 mg/mL of lysozyme. After 15 min at 37°C, the supernatants were collected and used as the cell extracts. The protein concentrations in the cell extracts were determined by BSA (bovine serum albumin) quantitative method. Next, we mixed 100-μL cell extracts with an equal volume of a reaction mixture comprised of 80 mM Tris-HCl (pH 8.0), 4 mM CoASH (coenzyme A, SH: sulfydryl), 2.5 mM 4-hydroxybenzoic acid and 4-aminoantipyrine, 0.6 mM 3-oxohexadecanoyl-CoA, 0.125% (wt/vol) Triton X-100, 0.5 mg/mL bovine serum albumin (BSA), acyl-CoA oxidase (12 U/mL), and peroxidase (12.5 U/mL). After 5 min at 37°C, 0.5% (wt/vol) SDS was added to terminate the reaction, and the corresponding absorption at 500 nm was then measured. HDT activity was calculated as described in reference (41).
Growth experiments on palmitic acid.
We cultured the related strains using 0.1% (wt/vol) palmitic acid as the sole carbon and energy source for 12 h. We sampled the cultures every hour and measured the OD600 value. The experiment was independently replicated three times, and each sample was tested in triplicate.
SPD transport activity analysis.
We measured spermidine uptake activity as previously described (43), with minor modifications. Briefly, the related bacterial cells were cultured in medium A containing 0.4% (wt/vol) glucose, 0.7% (wt/vol) K2HPO4, 0.3% (wt/vol) KH2PO4, 0.05% (wt/vol) sodium citrate, 0.1% (wt/vol) (NH4)2SO4, 0.01% (wt/vol) MgSO4 · 7H2O, 0.0002% (wt/vol) thiamine, 0.001% (wt/vol) biotin, and 0.01% (wt/vol) each of leucine, threonine, methionine, serine, glycine, and ornithine supplemented with 100 mg/L ampicillin at 37°C until the OD600 reached 0.3. Next, cells were harvested and resuspended in buffer I [0.4% glucose, 62 mM KH2PO4 (pH 7.0), 1.7 mM sodium citrate, 7.6 mM (NH4)2SO4, and 0.41 mM MgSO4] to generate a final protein concentration of 0.1 mg/mL. Next, the suspension was mixed with 0.25 mM [13C]spermidine and incubated at 30°C for 5 min. The cells were collected on cellulose acetate membrane filters, and the filters were washed three times with buffer I. δ13C values were measured using an isotope ratio mass spectrometer (Picarro analyzer).
ATPase activity assay.
We cultured the related strains under the same conditions as the ones described above for the SPD transport experiment. An ATPase assay kit (colorimetric, catalog number ab234055; Abcam) was used to monitor ATPase activity in the cell extracts.
Galleria mellonella-killing assay.
A Galleria mellonella-killing assay was performed as previously described (79). Briefly, strains cultured overnight were diluted (1:100) into fresh LB medium at 37°C until the OD600 reached 0.5. Next, the cells were collected, resuspended to a final concentration of 1 × 105 CFU/mL (plate count), and supplemented with 10 mM MgSO4. Ten microliters of the diluted strain solution or MgSO4 (aqueous) was injected into Galleria mellonella larvae and incubated at 37°C for 24 h. Each group comprised 20 larvae and 3 replicates. The experiments were independently replicated three times.
Western blot analysis.
We performed Western blotting as previously described (80). Strains cultured overnight were resuspended (1:100) in fresh LB medium supplemented with 5 mM EGTA or double-distilled water (ddH2O) and further incubated at 37°C for 4 h. The supernatants were harvested and mixed with 15% trichloroacetic acid (TCA). The samples (equal bacteria loads, with 500 μL of NuPAGE antioxidant) were run on 12% NuPAGE Bis-Tris gels (Thermo Fisher Scientific). A nitrocellulose membrane was used to transfer the protein. Rabbit antiserum (1:1,000) was used to detect ExoS. Hybridization was performed using 1 mg/mL secondary antibody (1:2,000), and the membrane was then incubated with Amersham ECL Prime Western blotting detection reagent (Thermo Fisher Scientific).
Data availability.
The whole-genome sequencing data of P8W were deposited at the Sequence Read Archive (SRA) of the National Center for Biotechnology Information (NCBI) under BioProject accession number PRJNA753640. The whole-genome sequencing data of PP9W2 have been made available at the NCBI under GenBank accession number OM141125. The RNA-seq data have been deposited at the Sequence Read Archive of the NCBI under BioProject accession number PRJNA857324. Other related data are available in this article and the supplemental material or from the corresponding authors upon request.
ACKNOWLEDGMENTS
We thank Weihui Wu for kindly providing the plasmids here.
This work was supported by Key Projects of the National Natural Science Foundation of China (41831287), the National Key R&D Program of China (2020YFC1806904), and the National Natural Science Foundation of China (31870351).
Footnotes
Supplemental material is available online only.
Contributor Information
Weihui Wu, Email: wuweihui@nankai.edu.cn.
Yi Luo, Email: luoy@nankai.edu.cn.
Lianrong Wang, Wuhan University.
REFERENCES
- 1.Fernandez L, Rodriguez A, Garcia P. 2018. Phage or foe: an insight into the impact of viral predation on microbial communities. ISME J 12:1171–1179. doi: 10.1038/s41396-018-0049-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gomez P, Buckling A. 2011. Bacteria-phage antagonistic coevolution in soil. Science 332:106–109. doi: 10.1126/science.1198767. [DOI] [PubMed] [Google Scholar]
- 3.Koskella B, Brockhurst MA. 2014. Bacteria-phage coevolution as a driver of ecological and evolutionary processes in microbial communities. FEMS Microbiol Rev 38:916–931. doi: 10.1111/1574-6976.12072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang J, Gao Q, Zhang Q, Wang T, Yue H, Wu L, Shi J, Qin Z, Zhou J, Zuo J, Yang Y. 2017. Bacteriophage-prokaryote dynamics and interaction within anaerobic digestion processes across time and space. Microbiome 5:57. doi: 10.1186/s40168-017-0272-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Golais F, Holly J, Vitkovska J. 2013. Coevolution of bacteria and their viruses. Folia Microbiol (Praha) 58:177–186. doi: 10.1007/s12223-012-0195-5. [DOI] [PubMed] [Google Scholar]
- 6.De Sordi L, Lourenco M, Debarbieux L. 2019. The battle within: interactions of bacteriophages and bacteria in the gastrointestinal tract. Cell Host Microbe 25:210–218. doi: 10.1016/j.chom.2019.01.018. [DOI] [PubMed] [Google Scholar]
- 7.Kronheim S, Daniel-Ivad M, Duan Z, Hwang S, Wong AI, Mantel I, Nodwell JR, Maxwell KL. 2018. A chemical defence against phage infection. Nature 564:283–286. doi: 10.1038/s41586-018-0767-x. [DOI] [PubMed] [Google Scholar]
- 8.Gao L, Altae-Tran H, Bohning F, Makarova KS, Segel M, Schmid-Burgk JL, Koob J, Wolf YI, Koonin EV, Zhang F. 2020. Diverse enzymatic activities mediate antiviral immunity in prokaryotes. Science 369:1077–1084. doi: 10.1126/science.aba0372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoyland-Kroghsbo NM, Maerkedahl RB, Svenningsen SL. 2013. A quorum-sensing-induced bacteriophage defense mechanism. mBio 4:e00362-12. doi: 10.1128/mBio.00362-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Erez Z, Steinberger-Levy I, Shamir M, Doron S, Stokar-Avihail A, Peleg Y, Melamed S, Leavitt A, Savidor A, Albeck S, Amitai G, Sorek R. 2017. Communication between viruses guides lysis-lysogeny decisions. Nature 541:488–493. doi: 10.1038/nature21049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davidson AR. 2017. Virology: phages make a group decision. Nature 541:466–467. doi: 10.1038/nature21118. [DOI] [PubMed] [Google Scholar]
- 12.Sherertz RJ, Sarubbi FA. 1983. A three-year study of nosocomial infections associated with Pseudomonas aeruginosa. J Clin Microbiol 18:160–164. doi: 10.1128/jcm.18.1.160-164.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Obritsch MD, Fish DN, MacLaren R, Jung R. 2004. National surveillance of antimicrobial resistance in Pseudomonas aeruginosa isolates obtained from intensive care unit patients from 1993 to 2002. Antimicrob Agents Chemother 48:4606–4610. doi: 10.1128/AAC.48.12.4606-4610.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kotecka K, Kawalek A, Kobylecki K, Bartosik AA. 2021. The MarR-type regulator PA3458 is involved in osmoadaptation control in Pseudomonas aeruginosa. Int J Mol Sci 22:3982. doi: 10.3390/ijms22083982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Modrzejewska M, Kawalek A, Bartosik AA. 2021. The LysR-type transcriptional regulator BsrA (PA2121) controls vital metabolic pathways in Pseudomonas aeruginosa. mSystems 6:e00015-21. doi: 10.1128/mSystems.00015-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Novichkov PS, Kazakov AE, Ravcheev DA, Leyn SA, Kovaleva GY, Sutormin RA, Kazanov MD, Riehl W, Arkin AP, Dubchak I, Rodionov DA. 2013. RegPrecise 3.0—a resource for genome-scale exploration of transcriptional regulation in bacteria. BMC Genomics 14:745. doi: 10.1186/1471-2164-14-745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu Y, Lu T, Zhang J, Zhang P, Tao M, Pang X. 2020. A novel XRE family regulator that controls antibiotic production and development in Streptomyces coelicolor. Appl Microbiol Biotechnol 104:10075–10089. doi: 10.1007/s00253-020-10950-z. [DOI] [PubMed] [Google Scholar]
- 18.Si M, Chen C, Zhong J, Li X, Liu Y, Su T, Yang G. 2020. MsrR is a thiol-based oxidation-sensing regulator of the XRE family that modulates C. glutamicum oxidative stress resistance. Microb Cell Fact 19:189. doi: 10.1186/s12934-020-01444-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu Y, Hu Q, Wei R, Li R, Zhao D, Ge M, Yao Q, Yu X. 2018. The XRE family transcriptional regulator SrtR in Streptococcus suis is involved in oxidant tolerance and virulence. Front Cell Infect Microbiol 8:452. doi: 10.3389/fcimb.2018.00452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trouillon J, Ragno M, Simon V, Attree I, Elsen S. 2021. Transcription inhibitors with XRE DNA-binding and cupin signal-sensing domains drive metabolic diversification in Pseudomonas. mSystems 6:e00753-20. doi: 10.1128/mSystems.00753-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eckstein S, Brehm J, Seidel M, Lechtenfeld M, Heermann R. 2021. Two novel XRE-like transcriptional regulators control phenotypic heterogeneity in Photorhabdus luminescens cell populations. BMC Microbiol 21:63. doi: 10.1186/s12866-021-02116-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoffman EC, Reyes H, Chu FF, Sander F, Conley LH, Brooks BA, Hankinson O. 1991. Cloning of a factor required for activity of the Ah (dioxin) receptor. Science 252:954–958. doi: 10.1126/science.1852076. [DOI] [PubMed] [Google Scholar]
- 23.Reyes H, Reisz-Porszasz S, Hankinson O. 1992. Identification of the Ah receptor nuclear translocator protein (Arnt) as a component of the DNA binding form of the Ah receptor. Science 256:1193–1195. doi: 10.1126/science.256.5060.1193. [DOI] [PubMed] [Google Scholar]
- 24.Lu H, Wang L, Li S, Pan C, Cheng K, Luo Y, Xu H, Tian B, Zhao Y, Hua Y. 2019. Structure and DNA damage-dependent derepression mechanism for the XRE family member DG-DdrO. Nucleic Acids Res 47:9925–9933. doi: 10.1093/nar/gkz720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Y, Liang S, Pan Z, Yu Y, Yao H, Liu Y, Liu G. 2022. XRE family transcriptional regulator XtrSs modulates Streptococcus suis fitness under hydrogen peroxide stress. Arch Microbiol 204:244. doi: 10.1007/s00203-022-02854-5. [DOI] [PubMed] [Google Scholar]
- 26.McCallum N, Hinds J, Ender M, Berger-Bächi B, Stutzmann Meier P. 2010. Transcriptional profiling of XdrA, a new regulator of spa transcription in Staphylococcus aureus. J Bacteriol 192:5151–5164. doi: 10.1128/JB.00491-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martinez-Salazar JM, Salazar E, Encarnacion S, Ramirez-Romero MA, Rivera J. 2009. Role of the extracytoplasmic function sigma factor RpoE4 in oxidative and osmotic stress responses in Rhizobium etli. J Bacteriol 191:4122–4132. doi: 10.1128/JB.01626-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Long X, Zhang H, Wang X, Mao D, Wu W, Luo Y. 2022. RecT affects prophage lifestyle and host core cellular processes in Pseudomonas aeruginosa. Appl Environ Microbiol 88:e01068-22. doi: 10.1128/aem.01068-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klein G, Muller-Loennies S, Lindner B, Kobylak N, Brade H, Raina S. 2013. Molecular and structural basis of inner core lipopolysaccharide alterations in Escherichia coli: incorporation of glucuronic acid and phosphoethanolamine in the heptose region. J Biol Chem 288:8111–8127. doi: 10.1074/jbc.M112.445981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Poon KK, Westman EL, Vinogradov E, Jin S, Lam JS. 2008. Functional characterization of MigA and WapR: putative rhamnosyltransferases involved in outer core oligosaccharide biosynthesis of Pseudomonas aeruginosa. J Bacteriol 190:1857–1865. doi: 10.1128/JB.01546-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Delucia AM, Six DA, Caughlan RE, Gee P, Hunt I, Lam JS, Dean CR. 2011. Lipopolysaccharide (LPS) inner-core phosphates are required for complete LPS synthesis and transport to the outer membrane in Pseudomonas aeruginosa PAO1. mBio 2:e00142-11. doi: 10.1128/mBio.00142-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Song W, Sun HX, Zhang C, Cheng L, Peng Y, Deng Z, Wang D, Wang Y, Hu M, Liu W, Yang H, Shen Y, Li J, You L, Xiao M. 2019. Prophage Hunter: an integrative hunting tool for active prophages. Nucleic Acids Res 47:W74–W80. doi: 10.1093/nar/gkz380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Minot S, Bryson A, Chehoud C, Wu GD, Lewis JD, Bushman FD. 2013. Rapid evolution of the human gut virome. Proc Natl Acad Sci USA 110:12450–12455. doi: 10.1073/pnas.1300833110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Catalano CE. 2000. The terminase enzyme from bacteriophage lambda: a DNA-packaging machine. Cell Mol Life Sci 57:128–148. doi: 10.1007/s000180050503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ptashne M, Jeffrey A, Johnson AD, Maurer R, Meyer BJ, Pabo CO, Roberts TM, Sauer RT. 1980. How the lambda repressor and cro work. Cell 19:1–11. doi: 10.1016/0092-8674(80)90383-9. [DOI] [PubMed] [Google Scholar]
- 36.Bailey TL, Johnson J, Grant CE, Noble WS. 2015. The MEME Suite. Nucleic Acids Res 43:W39–W49. doi: 10.1093/nar/gkv416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang D, Calabrese EJ, Lian B, Lin Z, Calabrese V. 2018. Hormesis as a mechanistic approach to understanding herbal treatments in traditional Chinese medicine. Pharmacol Ther 184:42–50. doi: 10.1016/j.pharmthera.2017.10.013. [DOI] [PubMed] [Google Scholar]
- 38.Winsor GL, Griffiths EJ, Lo R, Dhillon BK, Shay JA, Brinkman FSL. 2016. Enhanced annotations and features for comparing thousands of Pseudomonas genomes in the Pseudomonas Genome Database. Nucleic Acids Res 44:D646–D653. doi: 10.1093/nar/gkv1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reese MG. 2001. Application of a time-delay neural network to promoter annotation in the Drosophila melanogaster genome. Comput Chem 26:51–56. doi: 10.1016/s0097-8485(01)00099-7. [DOI] [PubMed] [Google Scholar]
- 40.Zarzycki-Siek J, Norris MH, Kang Y, Sun Z, Bluhm AP, McMillan IA, Hoang TT. 2013. Elucidating the Pseudomonas aeruginosa fatty acid degradation pathway: identification of additional fatty acyl-CoA synthetase homologues. PLoS One 8:e64554. doi: 10.1371/journal.pone.0064554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sato S, Hayashi M, Imamura S, Ozeki Y, Kawaguchi A. 1992. Primary structures of the genes, faoA and faoB, from Pseudomonas fragi B-0771 which encode the two subunits of the HDT multienzyme complex involved in fatty acid beta-oxidation. J Biochem 111:8–15. doi: 10.1093/oxfordjournals.jbchem.a123722. [DOI] [PubMed] [Google Scholar]
- 42.Igarashi K, Kashiwagi K. 2000. Polyamines: mysterious modulators of cellular functions. Biochem Biophys Res Commun 271:559–564. doi: 10.1006/bbrc.2000.2601. [DOI] [PubMed] [Google Scholar]
- 43.Kashiwagi K, Igarashi K. 2011. Identification and assays of polyamine transport systems in Escherichia coli and Saccharomyces cerevisiae. Methods Mol Biol 720:295–308. doi: 10.1007/978-1-61779-034-8_18. [DOI] [PubMed] [Google Scholar]
- 44.Hauser AR. 2009. The type III secretion system of Pseudomonas aeruginosa: infection by injection. Nat Rev Microbiol 7:654–665. doi: 10.1038/nrmicro2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang H, Shan Z, Kim J, Wu W, Lian W, Zeng L, Xing L, Jin S. 2007. Regulatory role of PopN and its interacting partners in type III secretion of Pseudomonas aeruginosa. J Bacteriol 189:2599–2609. doi: 10.1128/JB.01680-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.You J, Sun L, Yang X, Pan X, Huang Z, Zhang X, Gong M, Fan Z, Li L, Cui X, Jing Z, Jin S, Rao Z, Wu W, Yang H. 2018. Regulatory protein SrpA controls phage infection and core cellular processes in Pseudomonas aeruginosa. Nat Commun 9:1846. doi: 10.1038/s41467-018-04232-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Johnson AD, Poteete AR, Lauer G, Sauer RT, Ackers GK, Ptashne M. 1981. λ repressor and cro—components of an efficient molecular switch. Nature 294:217–223. doi: 10.1038/294217a0. [DOI] [PubMed] [Google Scholar]
- 48.Johnson A, Meyer BJ, Ptashne M. 1978. Mechanism of action of the cro protein of bacteriophage lambda. Proc Natl Acad Sci USA 75:1783–1787. doi: 10.1073/pnas.75.4.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aggarwal AK, Rodgers DW, Drottar M, Ptashne M, Harrison SC. 1988. Recognition of a DNA operator by the repressor of phage 434: a view at high resolution. Science 242:899–907. doi: 10.1126/science.3187531. [DOI] [PubMed] [Google Scholar]
- 50.Ohlendorf DH, Tronrud DE, Matthews BW. 1998. Refined structure of Cro repressor protein from bacteriophage lambda suggests both flexibility and plasticity. J Mol Biol 280:129–136. doi: 10.1006/jmbi.1998.1849. [DOI] [PubMed] [Google Scholar]
- 51.Monferrer D, Tralau T, Kertesz MA, Dix I, Sola M, Uson I. 2010. Structural studies on the full-length LysR-type regulator TsaR from Comamonas testosteroni T-2 reveal a novel open conformation of the tetrameric LTTR fold. Mol Microbiol 75:1199–1214. doi: 10.1111/j.1365-2958.2010.07043.x. [DOI] [PubMed] [Google Scholar]
- 52.Wang T, Sun W, Fan L, Hua C, Wu N, Fan S, Zhang J, Deng X, Yan J. 2021. An atlas of the binding specificities of transcription factors in Pseudomonas aeruginosa directs prediction of novel regulators in virulence. Elife 10:e61885. doi: 10.7554/eLife.61885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kutchma AJ, Hoang TT, Schweizer HP. 1999. Characterization of a Pseudomonas aeruginosa fatty acid biosynthetic gene cluster: purification of acyl carrier protein (ACP) and malonyl-coenzyme A:ACP transacylase (FabD). J Bacteriol 181:5498–5504. doi: 10.1128/JB.181.17.5498-5504.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Leesong M, Henderson BS, Gillig JR, Schwab JM, Smith JL. 1996. Structure of a dehydratase-isomerase from the bacterial pathway for biosynthesis of unsaturated fatty acids: two catalytic activities in one active site. Structure 4:253–264. doi: 10.1016/S0969-2126(96)00030-5. [DOI] [PubMed] [Google Scholar]
- 55.Mohan S, Kelly TM, Eveland SS, Raetz CR, Anderson MS. 1994. An Escherichia coli gene (FabZ) encoding (3R)-hydroxymyristoyl acyl carrier protein dehydrase. Relation to fabA and suppression of mutations in lipid A biosynthesis. J Biol Chem 269:32896–32903. doi: 10.1016/S0021-9258(20)30075-2. [DOI] [PubMed] [Google Scholar]
- 56.Guo QQ, Zhang WB, Zhang C, Song YL, Liao YL, Ma JC, Yu YH, Wang HH. 2019. Characterization of 3-oxacyl-acyl carrier protein reductase homolog genes in Pseudomonas aeruginosa PAO1. Front Microbiol 10:1028. doi: 10.3389/fmicb.2019.01028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Moore RC, Boyle SM. 1990. Nucleotide sequence and analysis of the speA gene encoding biosynthetic arginine decarboxylase in Escherichia coli. J Bacteriol 172:4631–4640. doi: 10.1128/jb.172.8.4631-4640.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Furuchi T, Kashiwagi K, Kobayashi H, Igarashi K. 1991. Characteristics of the gene for a spermidine and putrescine transport system that maps at 15 min on the Escherichia coli chromosome. J Biol Chem 266:20928–20933. doi: 10.1016/S0021-9258(18)54799-2. [DOI] [PubMed] [Google Scholar]
- 59.Pistocchi R, Kashiwagi K, Miyamoto S, Nukui E, Sadakata Y, Kobayashi H, Igarashi K. 1993. Characteristics of the operon for a putrescine transport system that maps at 19 minutes on the Escherichia coli chromosome. J Biol Chem 268:146–152. doi: 10.1016/S0021-9258(18)54126-0. [DOI] [PubMed] [Google Scholar]
- 60.Kashiwagi K, Innami A, Zenda R, Tomitori H, Igarashi K. 2002. The ATPase activity and the functional domain of PotA, a component of the spermidine-preferential uptake system in Escherichia coli. J Biol Chem 277:24212–24219. doi: 10.1074/jbc.M202849200. [DOI] [PubMed] [Google Scholar]
- 61.Juhas M. 2015. Pseudomonas aeruginosa essentials: an update on investigation of essential genes. Microbiology (Reading) 161:2053–2060. doi: 10.1099/mic.0.000161. [DOI] [PubMed] [Google Scholar]
- 62.Kang Y, Zarzycki-Siek J, Walton CB, Norris MH, Hoang TT. 2010. Multiple FadD acyl-CoA synthetases contribute to differential fatty acid degradation and virulence in Pseudomonas aeruginosa. PLoS One 5:e13557. doi: 10.1371/journal.pone.0013557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kang Y, Lunin VV, Skarina T, Savchenko A, Schurr MJ, Hoang TT. 2009. The long-chain fatty acid sensor, PsrA, modulates the expression of rpoS and the type III secretion exsCEBA operon in Pseudomonas aeruginosa. Mol Microbiol 73:120–136. doi: 10.1111/j.1365-2958.2009.06757.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lin Q, Wang H, Huang J, Liu Z, Chen Q, Yu G, Xu Z, Cheng P, Liang Z, Zhang L-H. 2022. Spermidine is an intercellular signal modulating T3SS expression in Pseudomonas aeruginosa. Microbiol Spectr 10:e00644-22. doi: 10.1128/spectrum.00644-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yang X, Zhang Z, Huang Z, Zhang X, Li D, Sun L, You J, Pan X, Yang H. 2019. A putative LysR-type transcriptional regulator inhibits biofilm synthesis in Pseudomonas aeruginosa. Biofouling 35:541–550. doi: 10.1080/08927014.2019.1627337. [DOI] [PubMed] [Google Scholar]
- 66.Turner KH, Vallet-Gely I, Dove SL. 2009. Epigenetic control of virulence gene expression in Pseudomonas aeruginosa by a LysR-type transcription regulator. PLoS Genet 5:e1000779. doi: 10.1371/journal.pgen.1000779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chow JJ, Batt CA, Sinskey AJ. 1988. Characterization of Lactobacillus bulgaricus bacteriophage ch2. Appl Environ Microbiol 54:1138–1142. doi: 10.1128/aem.54.5.1138-1142.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lu N, Kim C, Chen Z, Wen Y, Wei Q, Qiu Y, Wang S, Song Y. 2019. Characterization and genome analysis of the temperate bacteriophage phiSAJS1 from Streptomyces avermitilis. Virus Res 265:34–42. doi: 10.1016/j.virusres.2019.03.006. [DOI] [PubMed] [Google Scholar]
- 69.Li L, Pan X, Cui X, Sun Q, Yang X, Yang H. 2016. Characterization of Pseudomonas aeruginosa phage K5 genome and identification of its receptor related genes. J Basic Microbiol 56:1344–1353. doi: 10.1002/jobm.201600116. [DOI] [PubMed] [Google Scholar]
- 70.Vallet I, Olson JW, Lory S, Lazdunski A, Filloux A. 2001. The chaperone/usher pathways of Pseudomonas aeruginosa: identification of fimbrial gene clusters (cup) and their involvement in biofilm formation. Proc Natl Acad Sci USA 98:6911–6916. doi: 10.1073/pnas.111551898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chang PC, Wang CJ, You CK, Kao MC. 2011. Effects of a HP0859 (rfaD) knockout mutation on lipopolysaccharide structure of Helicobacter pylori 26695 and the bacterial adhesion on AGS cells. Biochem Biophys Res Commun 405:497–502. doi: 10.1016/j.bbrc.2011.01.060. [DOI] [PubMed] [Google Scholar]
- 72.Uddin MJ, Dawan J, Ahn J. 2019. Assessment of the alteration in phage adsorption rates of antibiotic-resistant Salmonella typhimurium. Arch Microbiol 201:983–989. doi: 10.1007/s00203-019-01667-3. [DOI] [PubMed] [Google Scholar]
- 73.Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Heer FT, de Beer TAP, Rempfer C, Bordoli L, Lepore R, Schwede T. 2018. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res 46:W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Stothard P. 2000. The sequence manipulation suite: JavaScript programs for analyzing and formatting protein and DNA sequences. Biotechniques 28:1102–1104. doi: 10.2144/00286ir01. [DOI] [PubMed] [Google Scholar]
- 75.Savli H, Karadenizli A, Kolayli F, Gundes S, Ozbek U, Vahaboglu H. 2003. Expression stability of six housekeeping genes: a proposal for resistance gene quantification studies of Pseudomonas aeruginosa by real-time quantitative RT-PCR. J Med Microbiol 52:403–408. doi: 10.1099/jmm.0.05132-0. [DOI] [PubMed] [Google Scholar]
- 76.Totten PA, Lory S. 1990. Characterization of the type a flagellin gene from Pseudomonas aeruginosa PAK. J Bacteriol 172:7188–7199. doi: 10.1128/jb.172.12.7188-7199.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rabinovich L, Sigal N, Borovok I, Nir-Paz R, Herskovits AA. 2012. Prophage excision activates Listeria competence genes that promote phagosomal escape and virulence. Cell 150:792–802. doi: 10.1016/j.cell.2012.06.036. [DOI] [PubMed] [Google Scholar]
- 78.de Ruijter A, Guldenmund F. 2016. The bowtie method: a review. Saf Sci 88:211–218. doi: 10.1016/j.ssci.2016.03.001. [DOI] [Google Scholar]
- 79.Andrejko M, Zdybicka-Barabas A, Cytryńska M. 2014. Diverse effects of Galleria mellonella infection with entomopathogenic and clinical strains of Pseudomonas aeruginosa. J Invertebr Pathol 115:14–25. doi: 10.1016/j.jip.2013.10.006. [DOI] [PubMed] [Google Scholar]
- 80.Springhorn A, Hoppe T. 2019. Western blot analysis of the autophagosomal membrane protein LGG-1/LC3 in Caenorhabditis elegans. Methods Enzymol 619:319–336. doi: 10.1016/bs.mie.2018.12.034. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download spectrum.03511-22-s0001.pdf, PDF file, 0.8 MB (800.2KB, pdf)
Data Sets S1 to S5. Download spectrum.03511-22-s0002.xls, XLS file, 0.9 MB (904KB, xls)
Data Availability Statement
The whole-genome sequencing data of P8W were deposited at the Sequence Read Archive (SRA) of the National Center for Biotechnology Information (NCBI) under BioProject accession number PRJNA753640. The whole-genome sequencing data of PP9W2 have been made available at the NCBI under GenBank accession number OM141125. The RNA-seq data have been deposited at the Sequence Read Archive of the NCBI under BioProject accession number PRJNA857324. Other related data are available in this article and the supplemental material or from the corresponding authors upon request.