Abstract
Bacteroides fragilis produces a capsular polysaccharide complex (CPC) that is directly involved in its ability to induce abscesses. Two distinct capsular polysaccharides, polysaccharide A (PS A) and PS B, have been shown to be synthesized by the prototype strain for the study of abscesses, NCTC9343. Both of these polysaccharides in purified form induce abscesses in animal models. In this study, we demonstrate that the CPC of NCTC9343 is composed of at least three distinct capsular polysaccharides: PS A, PS B, and PS C. A previously described locus contains genes whose products are involved in the biosynthesis of PS C rather than PS B as was originally suggested. The actual PS B biosynthesis locus was cloned, sequenced, and found to contain 22 genes in an operon-type structure. A mutant with a large chromosomal deletion of the PS B biosynthesis locus was created so that the contribution of PS B to the formation of abscesses could be assessed in a rodent model. Although purified PS B can induce abscesses, removal of this polysaccharide does not attenuate the organism's ability to induce abscesses.
Bacteroides fragilis is the anaerobic species most frequently isolated from intra-abdominal abscesses. The capsular polysaccharide complex (CPC) of the prototype strain 9343 is the major virulence factor for abscess formation, and purified CPC stimulates the formation of abscesses in animal models (10). Two distinct capsular polysaccharides have been identified from strain NCTC9343: polysaccharide A (PS A) and PS B (12). The repeating units of PS A and PS B contain both positively and negatively charged groups (19). The presence of both types of charged groups is required for the induction of abscesses (18).
We characterized a locus of B. fragilis NCTC9343 that is involved in the synthesis of a high-molecular-weight capsular polysaccharide (4). This locus contains 16 genes, all tightly clustered and transcribed from the same DNA strand, whose products are similar to other products involved in polysaccharide biosynthesis. No PS B was recovered from a mutant, 9343ΔwcfD-L, in which 10 genes of this locus were removed (4). This finding, coupled with the putative functions of the gene products encoded by the region and the belief that B. fragilis NCTC9343 produced only two capsular polysaccharides, led us to conclude that this locus was involved in the synthesis of PS B. A second mutant was created, 9343ΔwcfF, in which only a single gene of the region was mutated. Using whole-organism antiserum adsorbed with the 9343ΔwcfD-L mutant, we demonstrated that the single-gene mutant was also unable to synthesize the high-molecular-weight polysaccharide encoded by this locus.
A second locus of strain NCTC9343, shown to be the biosynthesis locus of PS A, has been cloned and sequenced (unpublished data). In this report, we describe the characterization of a third capsular polysaccharide biosynthesis locus of NCTC9343. Current data demonstrate that this locus, rather than the previously characterized region (4), is the actual PS B biosynthesis locus. We demonstrate that the previously described region encodes gene products involved in the synthesis of a third, previously uncharacterized capsular polysaccharide, referred to here as PS C.
MATERIALS AND METHODS
Bacteria, plasmids, and media.
The bacterial strains and plasmids used in this study are described in Table 1. Escherichia coli strains were grown in Luria (L) broth or on L agar plates. B. fragilis strains were grown anaerobically in basal medium (12) or on brain heart infusion (BHI) agar plates supplemented with 50 μg of hemin and 0.5 μg of menadione (BHIS) per ml. The following concentrations of antibiotic supplements were added when appropriate: ampicillin, 100 μg/ml; kanamycin, 20 μg/ml; erythromycin, 5 μg/ml; trimethoprim, 100 μg/ml; and gentamicin, 200 μg/ml.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| Strains | ||
| E. coli HB101 | F−hsdS20 (rB− mB−) recA13 leuB6 ara-14 proA2 lacY1 galK2 rpsL20 (Smr) xyl-5 mtl-1 supE44 λ− | 14 |
| E. coli DH5α | F− Φ80dlacZΔM15 Δ(lacZYA-argF) U169 endA1 recA1 hsdR17 (rK− mK+) deoR thi-1 supE44 gyrA96 relA1 λ− | 2 |
| B. fragilis US52540 | Clinical isolate | A. B. Onderdonk |
| B. fragilis mutant 1-45 | Strain US52540 with pNJR6Ω4 insertion in wcfW | This study |
| B. fragilis NCTC9343 | Type strain, appendix abscess | 8; NCTC |
| B. fragilis 9343ΔPS B | NCTC9343 mutant with a chromosomal deletion removing wcfV–wcgQ | This study |
| Plasmid | ||
| pHC79 | Cosmid vector; Apr | 7 |
| pBluescript II SK | Phagemid cloning vector; Apr | Stratagene |
| RK231 | Mobilizable plasmid used to move plasmids from E. coli to B. fragilis, RK2 derivative; Tra+ Tetr Kmr | 6 |
| R751 | Mobilizable plasmid used to move plasmids from E. coli to B. fragilis; Tra+ Tpr | 16 |
| pJST55 | Bacteroides suicide vector (Emr, Bacteroides); mob+ Tra− Apr | 17 |
| pNJR6Ω4 | Plasmid pNJR6 containing duplicate copies of Tn4351 (Emr, Bacteroides); mob+, Tra− Kmr | 4 |
| pLEC29 | Junctional clone of B. fragilis US52540 mutant 1-45 containing a portion of pNJR6Ω4 and ∼8 kb of B. fragilis DNA; Kmr | This study |
| pLEC29.2 | Subclone of pLEC29 containing 3.5-kb EcoRI fragment (B. fragilis DNA) in pBluescript; Apr | This study |
| pLEC29.4 | Subclone of pLEC29 containing 3.2-kb EcoRI fragment (B. fragilis-pNJR6Ω4 junction) in pBluescript; Apr | This study |
| pLEC32 | B. fragilis NCTC9343 gene bank clone in pHC79; Apr | This study |
| pLEC33 | B. fragilis NCTC9343 gene bank clone in pHC79; Apr | This study |
| pLEC32.1 | 5.5-kb EcoRI subclone of pLEC32 in pBluescript; Apr | This study |
| pLEC32.2 | 3.5-kb EcoRI subclone of pLEC32 in pBluescript; Apr | This study |
| pLEC32.4 | 1.4-kb EcoRI subclone of pLEC32 in pBluescript; Apr | This study |
| pLEC32.5 | 1.2-kb EcoRI subclone of pLEC32 in pBluescript; Apr | This study |
| pLEC33.1 | 7.2-kb EcoRI subclone of pLEC33 in pBluescript; Apr | This study |
| pLEC35 | bp 7415 through bp 18022 of PS B region, with bp 9836 to 15576 deleted; in pJST55; Apr | This study |
Transposon mutagenesis.
Transposon mutagenesis was performed as previously described (4). E. coli DH5α containing pNJR6Ω4 and the conjugal helper plasmid R751 was mated with B. fragilis US52540 overnight aerobically on BHIS plates, plated onto BHIS containing erythromycin and gentamicin, and incubated anaerobically. The resulting colonies were screened by colony immunoblot with MAb G9 (11).
Cloning of DNA at the transposon junction of mutant 1-45.
The chromosome of mutant 1-45 (MAb G9-negative) yielded an SstI fragment containing the origin of replication and the Kmr gene of pNJR6, as well as ∼8 kb of B. fragilis US52540 DNA. This fragment was cloned by ligation of SstI-digested 1-45 chromosomal DNA (favoring intramolecular ligations), transformation of E. coli DH5α, and selection with kanamycin. EcoRI fragments of the resulting plasmid (pLEC29) were subcloned into pBluescript II SK. Subclone pLEC29.4 contains a portion of the transposon and ∼2 kb of US52540 DNA immediately flanking the transposon. Plasmid pLEC29.2 contains a 3.7-kb EcoRI insert that represents the US52540 DNA adjacent to pLEC29.4. The 3.7-kb insert of pLEC29.2 was used as a probe to select cosmid clones from a B. fragilis NCTC9343 cosmid genebank constructed as previously described (4). Two cosmid clones were selected: pLEC32 and pLEC33.
Construction of pLEC32 and pLEC33 subclones.
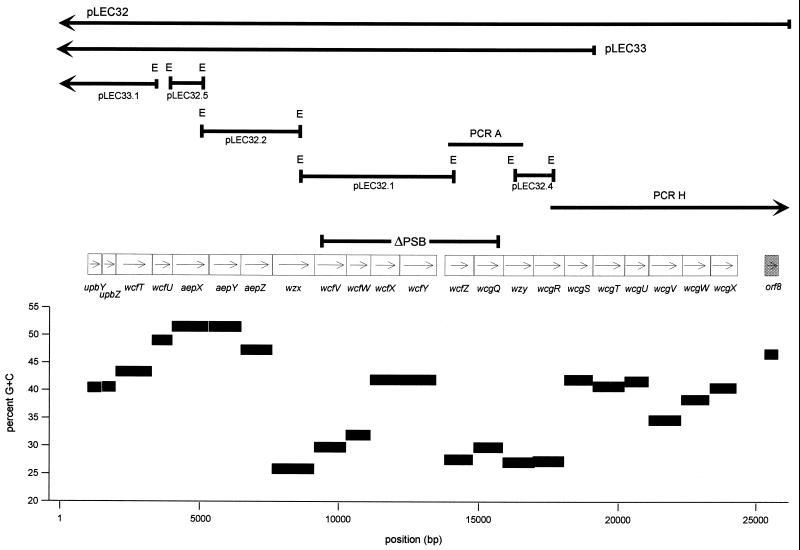
EcoRI fragments of pLEC32 and pLEC33 were subcloned into pBluescript II SK. The plasmids used for sequencing template and their locations are diagrammed in Fig. 2 and described in Table 1.
FIG. 2.
ORF map of the B. fragilis NCTC9343 PS B biosynthesis locus. The direction of transcription of each ORF is designated with an arrow. The region deleted in mutant 9343ΔPS B is indicated above the ORFs. Cosmid clones, subclones, and PCR products used as sequencing templates are shown. The average G+C content of each ORF is presented below.
Creation of deletion mutant 9343ΔPS B.
Mutant 9343ΔPS B is devoid of six genes of the PS B locus beginning with wcfV and continuing into wcgQ (see Fig. 2). To create this mutant, a primer internal to and oriented upstream of wcfV (primer 2; XhoI, 5′-GGGACTCGAGGACACGACTATCGCAGCCATTCAAATAGGG) was used in a PCR with primer 1 (BamHI, 5′-GCATGGATCCATACCACGCACCATGAAACGGGTACG) located approximately 2.4 kb upstream of primer 2 (restriction sites are underlined). A second PCR used a primer within wcgQ oriented downstream (primer 3; XhoI, 5′-GAGACTCGAGAAGATAATCGGGGGGTGTCAATAGCCAGAG) with a primer located approximately 2.4 kb downstream of primer 3 (primer 4; BamHI, 5′-CAGAGGATCCGCTTTCGCCTCAACACCATTAGGTACAGAC) (restriction sites are underlined). PCRs with primers 1 and 2 and primers 3 and 4 were performed with high-fidelity DNA polymerase (Life Technologies, Gaithersburg, Md.). The PCR products were digested with BamHI and XhoI and were gel purified. The products were cloned by three-way ligation into the Bacteroides suicide vector pJST55 (17) at the BglII site. A plasmid containing the correct orientation of the PCR products was selected by PCR and designated pLEC35. Plasmid pLEC35 was introduced into NCTC9343 by mobilization from E. coli using the conjugal helper plasmid RK231. Cointegrates that were formed by integration of the suicide plasmid into the chromosome of NCTC9343 were detected by selection for stable Emr colonies. The cointegrate was passaged three times in basal medium to allow for resolution of the cointegrate and plated onto BHIS. The resulting colonies were replica plated onto BHIS containing erythromycin, and the Ems colonies were tested for the mutant genotype by PCR. The mutant with the wcfV-wcgQ region deletion was designated 9343ΔPS B.
DNA sequencing and analyses.
DNA sequencing and analyses were performed as described elsewhere (4).
SDS-PAGE and Western blotting (immunoblotting).
Bacterial cell lysates were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on discontinuous SDS-polyacrylamide gels (4 to 20% gradient; ESA, Inc., Chelmsford, Mass.), and transferred to polyvinylidene difluoride (Immobilon) by standard techniques. Blots were blocked in Tris-buffered saline (TBS) containing 5% nonfat dry milk (TBS-milk) and then incubated for 1 h in TBS-milk containing primary antibody. The blots were washed with TBS and incubated with alkaline phosphatase-conjugated goat anti-mouse immunoglobulin G (Sigma Chemical Co.) at 1:1,000 in TBS for 1 h. After being washed again with TBS, the blots were developed with a colorimetric detection solution. Culture supernatants of monoclonal antibody (MAb) G9 (B. fragilis NCTC9343 PS B specific) and QUBF7 (B. fragilis NCTC9343 PS C specific [9]) were used at 1:50.
Mouse model of intra-abdominal abscess formation.
A previously described animal model of intra-abdominal infection (15) was modified for these studies. Briefly, male C57BL/6 mice (4 to 6 weeks old; Charles River Laboratories, Wilmington, Mass.) were challenged via the intraperitoneal route with 0.1 ml of inoculum containing 10-fold dilutions of NCTC9343 or 9343ΔPS B organisms mixed 1:1 (vol/vol) with sterilized rat fecal solution (SFC). SFC is used as an adjuvant for abscess formation and does not induce abscesses when implanted alone into the peritonea of mice. Six days after challenge, the animals were necropsied and examined for intraabdominal abscesses. The presence of one or more abscesses in an animal was scored as a positive result. To compare the abscess-inducing potential of these strains, a previously described mathematical model was used to calculate the dose of bacteria required to induce abscesses in 50% of animals (termed the AD50) (18).
RESULTS
The 9343ΔwcfF mutant strain, previously demonstrated by Western blot to be devoid of a high-molecular-weight polysaccharide believed at the time to be PS B, was used in a large-scale fermentation for the isolation of PS A. Nuclear magnetic resonance analysis of the capsular polysaccharides isolated from this strain clearly demonstrated that PS B, in addition to PS A, was synthesized by 9343ΔwcfF. This unexpected finding led to a reevaluation of the designation of the PS B locus and a search for the actual locus involved in the synthesis of PS B.
Cloning of the PS B biosynthesis locus of NCTC9343.
Pantosti et al. described an MAb, G9, that reacted with the PS B of B. fragilis NCTC9343 (11). We previously demonstrated that the 9343ΔwcfD–L and 9343ΔwcfF mutants also reacted with MAb G9; however, we attributed this to cross-reactivity with PS A, which we had previously demonstrated using purified polysaccharide in Western blots (4). Recent data suggest that the reactivity of MAb G9 with purified PS A by Western blot is an artifact. We have determined that MAb G9 does not react with purified PS A by enzyme-linked immunosorbent assay or on the surface of the bacteria (data not shown). These data demonstrate that MAb G9 is PS B specific, as originally described. Therefore, it became clear that the previously cloned region, a mutant of which retains MAb G9 reactivity, is not the biosynthesis locus of PS B. In order to clone the PS B biosynthesis locus of NCTC9343, we selected transposon mutants that no longer reacted with MAb G9. Since NCTC9343 is somewhat resistant to the introduction of foreign DNA, transposon mutagenesis of this strain is difficult. B. fragilis US52540, which also produces a high-molecular-weight capsular polysaccharide that reacts specifically with MAb G9, was used for transposon mutagenesis.
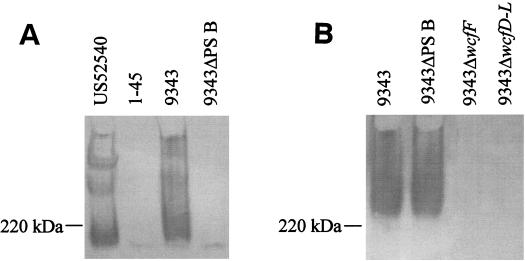
One transposon mutant, 1-45, was demonstrated to be MAb G9 negative by Western blot (Fig. 1A). The DNA at the junction of the transposon insertion of US52540 mutant 1-45 was cloned, creating plasmid pLEC29. The DNA sequence surrounding the transposon showed that the transposon had inserted into an open reading frame (ORF) with similarity to α1,2-fucosyltransferases. Since the repeating unit of PS B contains a terminal fucose with an α1-2 linkage to galacturonic acid, this finding provided evidence that this transposon had inserted into the PS B biosynthesis locus. An EcoRI subclone of pLEC29 (pLEC29.4) was used as a probe to select NCTC9343 gene bank clones pLEC32 and pLEC33. All subsequent analyses and DNA sequences were performed only with DNA from strain NCTC9343. The cosmid clones were subcloned as indicated in Fig. 2 and used as sequencing template. In total, 26,041 bp were sequenced. Analysis of this sequence revealed 22 ORFs, most of which encode putative products with various degrees of similarity to proteins implicated in polysaccharide biosynthesis in other organisms (Table 2). These 22 ORFs are all oriented in the same direction and are tightly clustered. These genetic characteristics are similar to those of the other capsular polysaccharide biosynthesis locus sequenced from NCTC9343 and suggest that this region is an operon. The genes were named in accordance with the nomenclature of polysaccharide biosynthesis genes (13).
FIG. 1.
Western immunoblot analysis of B. fragilis wild-type and mutant strains. Bacterial cultures were grown to an optical density at 600 nm of 0.8, and the bacteria were pelleted by centrifugation and lysed with 1× loading buffer. Bacteria (equivalent to 40 μl of culture) were added to each well of the 4 to 20% gradient SDS-polyacrylamide gel, transferred to PVDF, and probed with MAb G9 (PS B specific) (A) or MAb QUBF7 (PS C specific) (B).
TABLE 2.
Ability of B. fragilis 9343 and 9343ΔPS B mutant to induce abscessesa
| Challenge dose | No. of animals with abscesses when challenged with:
|
|
|---|---|---|
| 9343 (n = 10) | 9343ΔPS B (n = 10) | |
| 108 | 9 | 9 |
| 107 | 8 | 8 |
| 106 | 7 | 8 |
| 105 | 6 | 6 |
| 104 | 3 | 3 |
Of four mice challenged with sterile fecal contents alone, none developed abscesses.
The G+C content of the B. fragilis chromosome has been reported to be 41 to 44% (Fig. 2). The G+C content of this locus is highly variable, ranging from 25.9% (wzx) to 51.5% (aepX). This disparity suggests that this locus was acquired as a result of several ancestral horizontal transfer events.
NCTC9343 PS B mutant and analysis of phenotype.
Mutant 1-45 (MAb G9 negative) was created using strain US52540. To determine whether a mutation in the homologous region of the NCTC9343 chromosome would yield a similar phenotype, a deletion mutant was created in which six genes (wcfV to wcgQ) of the NCTC9343 chromosome were removed (Fig. 2). This mutant was demonstrated by Western blot to be MAb G9 negative (Fig. 1A) and was therefore designated 9343ΔPS B. We have determined that a previously described MAb, designated QUBF7 (9), does not react with the 9343ΔwcfF or 9343ΔwcfD–L mutants (Fig. 1B). Therefore, this MAb is specific for the polysaccharide encoded by the polysaccharide biosynthesis locus originally believed to be PS B. MAb QUBF7 reacts with 9343ΔPS B; this reaction demonstrates that the previously identified genetic region and the PS B locus encode separate polysaccharides identifiable with distinct MAbs. Several previous findings indicate that the QUBF7-reactive polysaccharide encoded by the previously described genetic locus is also a capsular polysaccharide. Lutton et al. showed by electron microscopic analysis of colloidal gold-labeled bacteria that MAb QUBF7 reacts with the bacterial cell surface. Additionally, these authors demonstrated that MAb QUBF7 reacts with a high-molecular-weight polysaccharide. We have also shown by immunoadsorption that the previously cloned locus encodes a high-molecular-weight surface polysaccharide (4). On the basis of these data, the polysaccharide encoded by the previously described locus, which is MAb QUBF7 reactive, is designated PS C.
Abscess induction by 9343ΔPS B.
We showed that a PS C mutant (9343ΔwcfF) was still fully virulent in an animal model of abscess formation compared with wild-type NCTC9343 (4). Since the structure of PS C has still not been determined, it is possible that this polysaccharide does not contain the necessary charge motif for abscess induction. If so, its removal would not be expected to diminish abscess formation. PS B, however, does contain the necessary charge motif for the induction of abscesses, and purified PS B elicits abscesses in rodent models (18). To assess the contribution of PS B to abscess formation in the context of intact bacteria, where other capsular polysaccharides are expressed, both NCTC9343 and 9343ΔPS B were tested in the rodent model. The results showed that the PS B mutant strain retains the full virulence of the wild-type, yielding AD50s of 105.1 and 105.2, respectively (Table 2).
DISCUSSION
The CPC of B. fragilis has been demonstrated to be composed of two capsular polysaccharides: PS A and PS B. We provide data demonstrating that the prototype organism for the study of abscess formation, B. fragilis NCTC9343, synthesizes at least three capsular polysaccharides: the previously described PS A and PS B, as well as the newly recognized PS C. Genetic and MAb analyses have revealed that a previously described capsular polysaccharide biosynthesis locus sequenced from B. fragilis NCTC9343 is involved in the synthesis of PS C and not PS B. The data clearly demonstrate that the locus described herein is the actual PS B locus. In addition to the nonreactivity of a deletion mutant lacking this region with the PS B-specific MAb G9, the presence of genes whose putative products are similar to those necessary for the synthesis of PS B constitutes further evidence that this region is the PS B biosynthesis locus. In light of these data, the previously described PS B2 region of B. fragilis 638R (3) will similarly be renamed the PS C2 biosynthesis locus.
The PS B of NCTC9343 is composed of the repeating unit [→4)α-l-QuipNAc(1→3)β-d-QuipNAc(1→4)[α-l-Fucp(1→2) β-d-GalpA(1→3)β-d-GlcpNAc(1→3)]α-d-Galp(1→], with a 2-aminoethylphosphonate (AEP) substituent on O-4 of the N-acetyl-β-d-glucopyranosyl residue (19). The PS B locus contains three genes, designated aepX, aepY, and aepZ, that encode putative products highly similar to three products involved in the formation of AEP (Table 2). The presence of these genes in this locus was not anticipated since they had not previously been found within a bacterial polysaccharide biosynthesis locus; however, their presence further identifies this region as the PS B locus.
This locus also contains other genes whose presence was anticipated given the structure of PS B; wcfX and wcfY are adjacent genes with nearly identical G+C contents which significantly differ from that of the surrounding genes (Fig. 1), suggesting that products of these genes may function in a common pathway. On the basis of the PS B structure and the proposed functions of the homologs of these gene products, we propose that WcfY is a UDP-glucose dehydrogenase that converts UDP-glucose to UDP-glucuronic acid, which is then epimerized by WcfX to UDP-galacturonic acid, one of the nucleotide sugar precursors of PS B.
The product of wcfW is similar to a family of α1,2-fucosyltransferases. The presence of this gene in the PS B locus was expected since its product is necessary for formation of the α1,2 linkage of the terminal l-fucose to d-galacturonic acid. Genes that encode products with putative functions similar to those of WcfY and WcfW are also found in the PS C biosynthesis locus and contributed to the misidentification of the region.
In addition to WcfY, the putative α1,2-fucosyltransferase, five other genes (wcfZ, wcgQ, wcgR, wcgV, and wcgX) encode products with similarity to glycosyltransferases. Unlike the PS C region, which contains seven genes whose products are similar to glycosyltransferases, the PS B region contains the expected number of glycosyltransferases for the transfer of the six residues in the assembly of the repeating unit of PS B.
Three genes of the PS B locus encode products with nine or more predicted membrane-spanning domains: wzy, wzx, and wcfT. Wzy and Wzx were assigned as the putative polymerase and flippase, respectively, based on their size, low G+C content, and similarity to other products assigned to these functions (Table 2). The third highly hydrophobic protein encoded within this region, WcfT, has 12 potential membrane-spanning domains. Homologs of this protein have not been described in other bacterial polysaccharide biosynthesis loci. However, in our description of the PS C biosynthesis locus of NCTC9343 (4), we reported the sequence of a partial gene, designated orf1, that is approximately 1.8 kb upstream of the PS C biosynthesis region and is transcribed in the opposite direction from the PS C biosynthesis genes. The product of orf1 is also predicted to span the membrane multiple times and is 62% similar to WcfT.
The repeating unit of PS B contains both l-N-acetylquinovosamine (l-QuipNAc) and d-N-acetylquinovosamine (d-QuipNAc). The biosynthesis pathways for the nucleotide precursors of these two residues have not been reported. Given the structures of the polysaccharides whose loci contain homologs of these genes, it is likely that wcgW encodes a 6-dehydratase involved in the direct conversion of UDP–d-N-acetylglucosamine to UDP–d-QuiNAc. Similarly, we predict that the products of three clustered genes, wcgS, wcgT, and wcgU, which are similar to a 4,6-dehydratase, a 3,5-epimerase, and a 4-reductase, respectively, are involved in the formation of UDP–l-QuiNAc from UDP–d-N-acetylmannosamine.
The only sugar residue of the PS B repeating unit whose nucleotide sugar precursor would not be synthesized by products encoded by the PS B locus is l-fucose. In the synthesis of colanic acid of E. coli, the products of two genes, gmd and fcl, have been shown to convert GDP–d-mannose to GDP–l-fucose in a three-step reaction (1). No homologs of gmd or fcl were found in the PS B biosynthesis locus and are likely present elsewhere on the chromosome. The PS C locus, which also contains a gene whose product is similar to α1,2 fucosyltransferases, likewise does not contain gmd or fcl homologs.
Comparison of the PS B locus to the PS C locus of strain NCTC9343 has revealed that not only are the terminal two genes of each locus highly similar at the protein level (Table 3), but also that the DNA regions comprising these genes are 76.1% identical between loci. We predict that the products of wcgW and wcgX of the PS B locus are involved in the synthesis and transfer of UDP–d-QuiNAc to undecaprenylphosphate as the first step in the synthesis of the PS B repeating unit. Based on the similarity of these gene products, we predict that the PS C of NCTC9343 will also contain d-QuiNAc.
TABLE 3.
Identity of the products encoded by the NCTC9343 PS B region with sequences in the databasea
| ORF | % G+C | Size (aa) | Organism and/or gene | Gene product | % Identical/ % similar (aa)b | Accession no.c |
|---|---|---|---|---|---|---|
| upbY | 40.6 | 174 | Bacteroides fragilis upcY | Putative involvement in transcriptional antitermination | 29/52 (2–172) | AF048749 |
| upbZ | 40.7 | 161 | Bacteroides fragilis upcZ | Unknown | 37/57 (6–128) | AF048749 |
| wcfT | 43.2 | 427 | Bacteroides fragilis orf1 | Unknown | 34/62 (12–314) | AF048749 |
| Danio rerio frizzled 8b protein | Involved in regulation of developmental genes | 27/45 (310–407) | AF060696 | |||
| wcfU | 49.0 | 239 | Streptococcus pneumoniae cps23fM | CpsfM | 30/51 (5–233) | AF057294 |
| Archaeoglobus fulgidus rfbF | Putative glucose 1-phosphate cytidyltransferase | 26/46 (5–235) | AE001025 | |||
| aepX | 51.5 | 433 | Streptomyces wedmorensis fom1 | Phosphoenolpyruvate phosphomutase | 59/75 (5–430) | AB016934 |
| Tetrahymena pyriformis | Phosphoenolpyruvate phosphomutase | 37/53 (149–428) | P33182 | |||
| Bacillus subtilis | Glycerol-3-phosphate cytidylyltransferase | 29/53 (3–138) | P27623 | |||
| aepY | 51.4 | 378 | Streptomyces wedmorensis fom2 | Phosphonopyruvate decarboxylase | 50/67 (15–327) | S60212 |
| aepZ | 47.2 | 366 | Salmonella enterica serovar Typhimurium phnW | 2-aminoethylphosphonate aminotransferase | 35/54 (1–363) | U69493 |
| wzx | 25.9 | 497 | Salmonella enterica group C1 wzx | Putative flippase | 23/39 (11–429) | G47677 |
| Bacteroides fragilis 638R PS C region wzx | Putative flippase | 20/42 (11–437) | AF125164 | |||
| wcfV | 29.9 | 381 | None | |||
| wcfW | 31.9 | 289 | Vibrio cholerae O22 wblA | Probable α1,2-fucosyltransferase | 31/52 (1–283) | AB012957 |
| Helicobacter pylori fucT | α1,2-fucosyltransferase | 28/51 (3–286) | AF093828 | |||
| wcfX | 42.0 | 350 | Vibrio cholerae O139 | Putative nucleotide sugar epimerase | 56/72 (3–335) | U47057 |
| Sinorhizobium meliloti | UDP-glucuronic acid epimerase | 47/62 (1–348) | Q58455 | |||
| wcfY | 42.1 | 440 | Burkholderia pseudomallei udg | Putative UDP-glucose dehydrogenase | 51/70 (1–436) | AF159428 |
| wcfZ | 27.7 | 340 | Vibrio cholerae O139 wbfO | Putative glycosyltransferase | 39/56 (8–229) | Y07786 |
| wcgQ | 29.7 | 348 | Streptococcus agalactiae type III cpsK | Putative glycosyltransferase | 30/49 (15–287) | AF163833 |
| Streptococcus pneumoniae type 14 cps14J | β1,4-galactosyltransferase | 32/53 (15–223) | X85787 | |||
| wzy | 27.1 | 376 | Streptococcus suis cps1H | Putative capsular polysaccharide polymerase | 23/45 (33–371) | AF155804 |
| Escherichia coli K30 wzy | Repeat unit polymerase | 21/37 (15–348) | AF104912 | |||
| wcgR | 27.3 | 371 | Escherichia coli O113 wbnE | Putative glycosyltransferase | 24/43 (77–369) | AF172324 |
| wcgS | 42.1 | 338 | Bacteroides fragilis 638R wcgJ | Putative epimerase/dehydratase | 84/90 (1–338) | AF125164 |
| Pseudomonas aeruginosa O11 wbjB | WbjB | 71/84 (1–332) | AF147795 | |||
| wcgT | 40.8 | 376 | Pseudomonas aeruginosa wbjD | WbjD | 60/78 (2–374) | AF147795 |
| Staphylococcus aureus cap5G | Cap5G | 53/72 (2–374) | U81973 | |||
| wcgU | 41.8 | 287 | Actinobacillus actinomycetemcomitans rmlD | dTDP–4-keto-l-rhamnose reductase | 33/50 (5–155) | AB030032 |
| Escherichia coli K-12 rmlD | dTDP–4-keto-l-rhamnose reductase | 35/47 (6–143) | P37760 | |||
| wcgV | 34.5 | 403 | Acinetobacter lwoffii weeG | Putative glycosyltransferase | 25/44 (1–394) | AJ243431 |
| Escherichia coli wcaI | Putative glycosyltransferase | 26/47 (1–325) | P32057 | |||
| wcgW | 38.4 | 335 | Bacteroides fragilis 9343 PS C locus wcfK | Putative epimerase or dehydratase | 81/86 (1–335) | AF048749 |
| Pseudomonas aeruginosa wbpV | WbpV | 28/44 (62–331) | AF035937 | |||
| wcgX | 40.4 | 316 | Bacteroides fragilis 9343 PS C locus wcfL | Putative UndP–GlcNAc-phosphotransferase | 80/87 (1–313) | AF048749 |
| Yersinia enterocolitica trsF | Probable N-acetylgalactosaminyltransferase | 28/46 (23–303) | S51265 | |||
| orf8 | 46.6 | 157 | Rhizobium leguminosarum | DNA-binding protein HLR53 | 30/50 (32–98) | P02348 |
aa, amino acids.
The identity and similarity values are based on the indicated region of the B. fragilis gene product.
Numbers are either GenBank accession numbers or cross-referenced in GenBank.
The overall genetic composition of the PS B biosynthesis locus has similarities to the biosynthesis loci of other bacteria. The exceptions are the aminoethylphosphonate biosynthesis genes (aepX, aepY, and aepZ), wcfT (predicted to encode a transmembrane product), and wcfV, for which no homologs are present in the database. Comparison of the NCTC9343 PS B locus with the only other reported polysaccharide biosynthesis loci of B. fragilis, PS C1 of NCTC9343 and PS C2 of strain 638R, reveals that the genes of all three loci are organized in an operon type structure in that all genes are oriented in the same direction and tightly clustered. The genetic complement of these three loci, however, differ as the repeating units of each of these capsular polysaccharides are structurally and serologically distinct. Products similar to sugar dehydrogenases are encoded by all three loci. These products likely confer a negative charge to the repeating unit of each of the respective polysaccharides. This negative charge is part of the motif necessary for abscess induction.
The genetic arrangement of the PS C biosynthesis locus has been analyzed for 50 B. fragilis strains. Two small genes, previously designated orf3 and orf4, were found just upstream of the PS C biosynthesis genes in all 50 strains analyzed, regardless of the variability of the downstream PS C biosynthesis genes (5). We have redesignated orf3 and orf4 as upcY and upcZ (designation derived from upstream PS C), respectively. The PS B biosynthesis locus similarly contains homologs of these two genes just upstream of the PS B biosynthesis genes. These genes have therefore been designated upbY and upbZ (designation derived from upstream PS B). Future studies will examine the prevalence of upbY and upbZ to determine whether these genes are upstream of the PS B biosynthesis genes in all B. fragilis strains.
Purified PS A and PS B have each been shown to be potent abscess-inducing polysaccharides. In the rat intra-abdominal abscess model, the AD50 of purified PS A is 0.67 μg, whereas the AD50 of PS B is 25 μg (18). We found the PS C mutant (9343ΔwcfF) to be fully virulent in the rodent abscess model. Since the structure of the PS C of NCTC9343 has not been determined, it is possible that it does not have the necessary charge motif for the induction of abscesses. Moreover, as PS A and PS B continue to be expressed by this mutant, it would be expected to retain some abscess-inducing ability. The present study indicates that the PS B mutant is fully virulent in the rodent abscess model. Although purified PS B does induce abscesses, the more potent PS A is probably responsible for the abscess-inducing ability of the PS B mutant. Mutants deficient in the synthesis of two or more of these capsular polysaccharides may be necessary for the production of an attenuated strain.
ACKNOWLEDGMENTS
We are grateful to Vincent Carey for aid in statistical analysis of animal data, to Sheila Patrick for supplying MAb QUBF7, and to Michael Malamy for plasmid pJST55.
This work was supported by the Edward and Amalie Kass Fellowship and by Public Health Service grants AI44193 and AI39576 from the National Institute of Allergy and Infectious Diseases.
REFERENCES
- 1.Andrianipoulos K, Wang L, Reeves P R. Identification of the fucose synthetase gene in the colanic acid gene cluster of Escherichia coli K-12. J Bacteriol. 1998;180:998–1001. doi: 10.1128/jb.180.4.998-1001.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bethesda Research Laboratories. BRL pUC host: E. coli DH5 α competent cells. Focus. 1986;8:9. [Google Scholar]
- 3.Comstock L E, Coyne M J, Tzianabos A O, Kasper D L. Interstrain variation of the polysaccharide B biosynthesis locus of Bacteroides fragilis: characterization of the region from strain 638R. J Bacteriol. 1999;181:6192–6196. doi: 10.1128/jb.181.19.6192-6196.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Comstock L E, Coyne M J, Tzianabos A O, Pantosti A, Onderdonk A B, Kasper D L. Analysis of a capsular polysaccharide biosynthesis locus of Bacteroides fragilis. Infect Immun. 1999;67:3525–3532. doi: 10.1128/iai.67.7.3525-3532.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Comstock L E, Pantosti A, Kasper D L. Genetic diversity of the capsular polysaccharide C biosynthesis region of Bacteroides fragilis. Infect Immun. 2000;68:6182–6188. doi: 10.1128/iai.68.11.6182-6188.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guiney D, Hasegawa P, Davis C. Plasmid transfer from Escherichia coli to Bacteroides fragilis: differential expression of antibiotic resistance phenotypes. Proc Natl Acad Sci USA. 1984;83:7203–7206. doi: 10.1073/pnas.81.22.7203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hohn B, Collins J. A small cosmid for efficient cloning of large DNA fragments. Gene. 1980;11:291–298. doi: 10.1016/0378-1119(80)90069-4. [DOI] [PubMed] [Google Scholar]
- 8.Johnson J L. Taxonomy of the Bacteroides. I. Deoxyribonucleic acid homologies among Bacteroides fragilis and other saccharolytic Bacteroides species. Int J Syst Bacteriol. 1978;28:245–268. [Google Scholar]
- 9.Lutton D, Patrick S, Crockard A, Stewart L, Larkin M, Dermott E, McNeill T. Flow cytometric analysis of within-strain variation in polysaccharide expresion by Bacteroides fragilis by use of murine monoclonal antibodies. J Med Microbiol. 1991;35:229–237. doi: 10.1099/00222615-35-4-229. [DOI] [PubMed] [Google Scholar]
- 10.Onderdonk A B, Kasper D L, Cisneros R L, Bartlett J G. The capsular polysaccharide of Bacteroides fragilis as a virulence factor: comparison of the pathogenic potential of encapsulated and unencapsulated strains. J Infect Dis. 1977;136:82–89. doi: 10.1093/infdis/136.1.82. [DOI] [PubMed] [Google Scholar]
- 11.Pantosti A, Colangeli R, Tzianabos A O, Kasper D L. Monoclonal antibodies to detect capsular diversity among Bacteroides fragilis isolates. J Clin Microbiol. 1995;33:2647–2652. doi: 10.1128/jcm.33.10.2647-2652.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pantosti A, Tzianabos A O, Onderdonk A B, Kasper D L. Immunochemical characterization of two surface polysaccharides of Bacteroides fragilis. Infect Immun. 1991;59:2075–2082. doi: 10.1128/iai.59.6.2075-2082.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reeves P R, Hobbs M, Valvano M A, Skurnik M, Whitfield C, Coplin D, Kido N, Klena J, Maskell D, Raetz C R H, Rick P D. Bacterial polysaccharide synthesis and gene nomenclature. Trends Microbiol. 1996;4:495–503. doi: 10.1016/s0966-842x(97)82912-5. [DOI] [PubMed] [Google Scholar]
- 14.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Press; 1989. [Google Scholar]
- 15.Shapiro M E, Kasper D L, Zaleznik D F, Spriggs S, Onderdonk A B, Finberg R W. Cellular control of abscess formation: role of T cells in the regulation of abscesses formed in response to Bacteroides fragilis. J Immunol. 1986;137:341–346. [PubMed] [Google Scholar]
- 16.Shoemaker N B, Getty C E, Salyers A A, Gardner J F. Evidence that the clindamycin-erythromycin resistance gene of Bacteroides plasmid pBF4 is on a transposable element. J Bacteriol. 1985;162:626–632. doi: 10.1128/jb.162.2.626-632.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thompson J S, Malamy M H. Sequencing the gene for an imipenem-cefoxitin-hydrolyzing enzyme (CfiA) from Bacteroides fragilis. J Bacteriol. 1990;172:2584–2593. doi: 10.1128/jb.172.5.2584-2593.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tzianabos A O, Onderdonk A B, Rosner B, Cisneros R L, Kasper D L. Structural features of polysaccharides that induce intra-abdominal abscesses. Science. 1993;262:416–419. doi: 10.1126/science.8211161. [DOI] [PubMed] [Google Scholar]
- 19.Tzianabos A O, Pantosti A, Baumann H, Brisson J R, Jennings H J, Kasper D L. The capsular polysaccharide of Bacteroides fragilis comprises two ionically linked polysaccharides. J Biol Chem. 1992;267:18230–18235. [PubMed] [Google Scholar]