SUMMARY
This review serves as an update to the previous Nocardia review by Brown-Elliott et al. published in 2006 (B. A. Brown-Elliott, J. M. Brown, P. S. Conville, and R. J. Wallace. Jr., Clin Microbiol Rev 19:259–282, 2006, https://doi.org/10.1128/CMR.19.2.259-282.2006). Included is a discussion on the taxonomic expansion of the genus, current identification methods, and the impact of new technology (including matrix-assisted laser desorption ionization-time of flight [MALDI-TOF] and whole genome sequencing) on diagnosis and treatment. Clinical manifestations, the epidemiology, and geographic distribution are briefly discussed. An additional section on actinomycotic mycetoma is added to address this often-neglected disease.
KEYWORDS: Nocardia, aerobic actinomycetes, mycetoma, nocardiosis
INTRODUCTION
Population-based descriptions of the disease nocardiosis and its etiologic agent, the bacterial genus Nocardia, in the United States are scarce, despite a substantial body of research (1–3). Nocardiosis is a medically important disease that more frequently affects immunocompromised patients (4, 5). There are varied clinical presentations that can make clinical recognition challenging (6), and as a soilborne opportunistic pathogen, prevention methods are limited (4). The disease is difficult to treat, requiring months or years of antimicrobial therapy (7), and contributes to mortality in patients with underlying conditions (8).
Analysis of Nocardia by species is also limited, and due to major taxonomic changes over time (9), retrospective analyses of Nocardia species may need to be retested using modern molecular methods because of historical misidentification and misclassification (9, 10).
HISTORY AND TAXONOMY OF NOCARDIA
Nocardiae belong to the class Actinobacteria in the order Corynebacteriales (https://lpsn.dsmz.de/order/corynebacteriales). Other notable pathogenic genera in this group include Mycobacterium and Corynebacterium. The genus was first described in 1889 by Trevisan (Sneath, 1980), and reports on the clinical relevance of Nocardia began to appear in the literature in the early 1900s. In the 1940s, case reports appear implicating Nocardia species in invasive pulmonary infections. A review of nocardiosis cases published at that time notes that with the introduction of sulfonamide therapy in the late 1930s, fatalities from pulmonary nocardiosis decreased. However, a high mortality rate remained in patients with disseminated disease (11). The taxonomic history has been significantly impacted by conflicting descriptions of the first reported isolates and reassignments of many of the earliest validated species (9). There are currently 109 validly named species with roughly half of these considered to be clinically relevant, with some first isolated from human sources. Previous reports and reviews on Nocardia and nocardiosis have relayed the complicated history of the taxonomy so an in-depth discussion will not be presented in this current review. In 2018, Conville et al. published a detailed description of the history of the genus (9).
Bacteria that belong to the genus Nocardia are high GC, aerobic, Gram-positive, partially acid-fast, lysozyme resistant, and catalase positive with a characteristic beaded branching cell morphology. On blood agar, nocardiae form distinctive colonies with white aerial mycelium giving a chalky appearance to mature growth. A few species, most notably Nocardia farcinica, can appear as raised and wet (or muccoid) when young and then begin producing the characteristic aerial hyphae with age. Chemotaxonomic hallmarks for assignment to the genus include: meso-2,6-diaminopimelic acid, arabinose and galactose as the diagnostic sugars, and mycolic acids with a chain length of 46 to 58 carbons (12). The most common sources of human clinical material include bronchial washings, bronchial lavage fluids, sputum, abscess/wound drainage, and blood. More rarely reported sources involve the central nervous system including cerebral spinal fluid and brain tissue/abscess material. Ocular infections are also reported in the literature (13–15).
Differentiation of Nocardia species has previously been based on decomposition of substrates and acid production from or utilization of carbohydrates. Currently, molecular identification methods have become the gold standard. With an improved ability to resolve the heterogeneity of the genus, 48 new Nocardia species have been validated since the last Nocardia review (6). Of these species, 14 were isolated from human sources and 30 were from non-human sources (26 from soil, 4 from plants, and the other from animals). The demographics of the type strains are given in Table 1.
TABLE 1.
New Nocardia species 2006 to 2021
| Species of Nocardia (ref) | yr | Source | Country of origin | Type strain in culture collection | GenBank accession no. of 16S rRNA gene sequence |
|---|---|---|---|---|---|
| Nocardia aciditolerans (252) | 2013 | Non-human | UKa | KACC 17155 DSM 45801 |
JX484797 |
| Nocardia altamirensis | 2008 | Non-human | Spain | CIP 109315 DSM 45049 JCM 14670 |
EU006090 |
| Nocardia amamiensis (253) | 2007 | Non- human | Japan | DSM 45066 JCM 14877 KCTC 19208 NBRC 102102 |
AB275164 |
| Nocardia amikacinitolerans (254) | 2013 | Human | USAa | CCUG 59655 DSM 45539 |
GU985442 |
| Nocardia arizonensis (93) | 2015 | Human | USAa | DSM 45748 CCUG 62754 NBRC 08935 |
JN678715 |
| Nocardia artemisiae (255) | 2011 | Non-human | China |
CTCCAA 209038 DSM 45379100526 |
GU367157 |
| Nocardia aurantia (256) | 2020 | Non-human | Germany | NRRL B65542 VKM Ac-2842 |
KY558730 |
| Nocardia aurantiaca (257) | 2020 | Non-human | Thailand | JCM 33775 TISTR 2838 |
LC495742 |
| Nocardia aurea (258) | 2019 | Non-human | China | DSM 103986 KCTC 39849 |
MH091575 |
| Nocardia barduliensis (259) | 2021 | Human | Spain | CECT 9924 DSM 109819 |
MT472102 |
| Nocardia bhagyanesis (260) | 2014 | Non-human | India | ATCC BAA-2548 KCTC 29269 |
JX076851 |
| Nocardia blacklockiae (28) | 2009 | Human | USAa | ATCC 70035 DSM 45135 JCM 16005 |
EU099360 |
| “Nocardia boironii” (261) | 2016 | Human | France | EML 1451 DSM 101696 |
KU131666 |
| Nocardia bovistercoris (262) | 2021 | Non-human | China | DSM 110681 CCTCC AA 2019090 |
MW250206 |
| Nocardia callitridis (263) | 2010 | Non-human | Australia | DSM 45353 ACM 5287 |
FJ805428 |
| Nocardia camponoti (264) | 2016 | Non-human | China | DSM 100526 CGMCC 4.7278 |
KP784782 |
| Nocardia casuarinae (265) | 2014 | Non-human | Switzerland | DSM 45978 CECT 8469 |
KF924767 |
| Nocardia cavernae (266) | 2017 | Non-human | China | KCTC 39595 YIM A1135 |
KY285257 |
| Nocardia colli (267) | 2020 | Human | China | CICC 11023 KCTC 39837 |
KJ659849 |
| Nocardia coubleae (268) | 2007 | Non-human | Kuwait | DSM 44960 CIP 108996 |
JN041456 |
| Nocardia donostiensis (269) | 2016 | Human | Spain | DSM 46814 CECT 8839 |
KM233637 |
| Nocardia endophytica (270) | 2011 | Non-human | China | KCTC 19777 CCTCC AA 2010004 |
HM153801 |
| Nocardia exalbida (271) | 2006 | Human | Japan | NBRC 100660 JCM 12667 DSM 44883 |
AB187522 |
| Nocardia gamkensis (272) | 2006 | Non-human | South Africa | DSM 44956 NRRL B-24450 |
DQ235272 |
| Nocardia gipuzkoensis (259) | 2021 | Human | Spain | CECT 30129 DSM 111366 |
MT704612 |
| Nocardia goodfellowii (273) | 2012 | Non-human | Turkey | DSM 45517 NRRL B-24834 KCTC 19985 |
HQ157183 |
| Nocardia grenadensis (274) | 2012 | Non-human | Caribbean Sea | CCUG 60970 CIP 110294 |
FR729900 |
| Nocardia halotolerans (275) | 2015 | Non-human | Iran | IBRC-M 10490 LMG 28544 |
KM577163 |
| Nocardia harenae (276) | 2006 | Non-human | Republic of Korea | KCCM 42317 NRRL B-24459 |
DQ282122 |
| Nocardia heshunensis (277) | 2017 | Non-human | China | DSM 46764 JCM 39985 |
KY039322 |
| Nocardia iowensis (278) | 2009 | Non-human | USAa | UI 122540 NRRL B-24671 DSM 45197 |
DO925490 |
| Nocardia jejuensis (279) | 2006 | Non-human | Republic of Korea | JCM13281 NRRL B-24430 |
AY964666 |
| Nocardia jiangxiensis (280) | 2005 | Non-human | China | CGMCC 4.1905 JCM 12861 | AY639902 |
| Nocardia jinanensis (281) | 2009 | Non-human | China | CCMCC 4.3508 DSM 45048 |
DQ462650 |
| Nocardia lijiangensis (282) | 2005 | Non-human | China | CCTCC AA 204005 KCTC 19028 |
AY779043 |
| Nocardia kroppenstedtii (283) | 2014 | Human | UKa | DSM 45810 NCTC 13617 |
DQ157924 |
| Nocardia lasii (284) | 2017 | Non-human | China | CGMCC 4.7279 DSM 100525 |
KP784803 |
| Nocardia macrotermitis (256) | 2020 | Non-human | NRRL B65541 VKM Ac-2841 |
KY558706 | |
| Nocardia miyunensis (280) | 2006 | Non-human | China | CGMCC 4.1904 NRRL 12860 |
AY639901 |
| Nocardia mikamii (285) | 2010 | Human | USAa | DSM 45174 JCM 15508 |
EY484388 |
| Nocardia ninae (286) | 2007 | Human | France | CIP 108950 DSM 44978 |
JF797312 |
| Nocardia niwae (287) | 2011 | Human | USAa | DSM 45340 CCUG 57756 |
FJ765056 |
| Nocardia polyresistens (288) | 2005 | Non-human | China |
CCTCCAA 204004 KCTC 19027 |
AY626158 |
| Nocardia rhamnosiphila (289) | 2010 | Non-human | South Africa | DSM 45147 NRRL 24637 |
EF418604 |
| Nocardia rhizoaphaerae (290) | 2015 | Non-human | China | CGMCC 47204 KCTC 29678 |
KP972639 |
| Nocardia shinanonensis (291) | 2016 | Human | Japan | IFM 11456 NBRC 109590 TBRC 5149 |
LC103184 |
| Nocardia speluncae (292) | 2007 | Non-human | Korea | JBRI 2006 KCTC 19223 DSM 45078 |
AM422449 |
| Nocardia sungurluensis (293) | 2014 | Non-human | Turkey | DSM 45714 KCTC 29094 |
JN989289 |
| Nocardia terpenica (294) | 2007 | Human | Japan | JCM 13033 DSM 44935 NBRC 100888 |
AB201298 |
| Nocardia thraciensis (273) | 2012 | Non-human | Turkey | DSM 45517 NRRL B-24838 KCTC 19985 |
HQ157183 |
| Nocardia vulneris (94) | 2014 | Human | USA | DSM 45737 CCUG 62683 NBRC 108936 |
JN705252 |
| Nocardia wallacei (28) | 2009 | Human | USA | ATCC 49873 DSM 45136 |
EU099357 |
UK, United Kingdom; USA, USA.
LABORATORY EVALUATION
Even though nocardiosis is rare and often underreported, the disease is of high consequence and characterized by high levels of morbidity and mortality. Subsequently, an accurate, reliable, rapid, and simple method for the identification of the etiologic agent in clinical samples is of paramount importance in order to reduce mortality and expedite appropriate antimicrobial therapy. Previously, identification of Nocardia clinical isolates was based on microscopic observation and macroscopic visualization of colony morphology and color complemented with a battery of conventional phenotypic and chemotaxonomic tests. Few of these tests are highly discriminatory and all require expertise or extensive training. Additionally, Nocardia species may grow slowly, requiring - in some cases - weeks to form colonies. The primary specimen source is often from a non-sterile site which makes them difficult to cultivate. Overgrowth by faster growing bacteria is a common delay in identification.
Accurate identification is critical in providing clinicians information regarding the infectious agent and its susceptibility profile to determine appropriate antimicrobial treatment. When aggregated, identification will inform the epidemiology of the agent, geographic and species distribution, understanding its clinical relevance and spectrum of disease, pathogenicity, and determining risk factors. In the past few decades, molecular identification has been aided by technical advances in molecular genetics and DNA/RNA sequencing technologies including whole genome sequencing. At present, molecular identification methods have increasingly supplanted phenotypic methods for definitive identification and determining the phylogenetic relationships for an increasing number of new species and taxonomic emendation.
There are many previously published descriptions of the evolution of early identification techniques for Nocardia species. The most comprehensive is the review preceding this one by Brown-Elliott et al., 2006 (6). Therefore, this review will not cover species identification by biochemical methods, serological methods, DNA probes and ribotyping, RFLP (restriction fragment length polymorphisms) analysis, and RAPD (randomly amplified polymorphic DNA) analysis. Each of these techniques have severe limitations and are currently not advised for identification of Nocardia species. As of the time of this publication, molecular identification to the species level of human clinical isolates of Nocardia is either by 16S rRNA gene sequence analysis, DNA gyrase subunit B gene (gyrB) sequence analysis, or secA1 gene sequence analysis, according to CLSI guidelines (16). Other genes widely used are hsp65 and rpoB. Multilocus sequence typing (MLST), whole genome analysis, and proteomics, such as MALDI-TOF analysis, are currently becoming more popular and with promising results. A brief description of each gene target and/or method can be found below.
Gene Sequencing
16S rRNA gene.
Of all the available gene targets, 16S rRNA gene sequences are the most frequently used and are considered the gold standard for routine identification and phylogenetic analysis of Nocardia species isolates (6, 17). Advantages of 16S rRNA gene sequencing over phenotypic methods for identification of isolates includes higher reliability and accuracy, rapid identification in 1 to 2 days versus several weeks, providing useful phylogenetic information. The wide use and utility of analysis of 16S rRNA gene sequences is due to their presence in all bacterial species, the gene has a known function and structure with a relatively slow rate of evolution, and harbors both conserved and variable regions enabling both genus-species and species-specific motifs to be used for species identification.
Using near full length 16S rRNA gene sequences for phylogenetic analysis, Ruimy et al. (1994) showed Nocardia species formed a monophyletic clade most closely related to Gordonia, Rhodococcus, Mycobacterium, Tsukamurella, and Corynebacterium, the closest related high G+C genome, mycolic acid-containing taxa (17). Likewise, partial 16S rRNA gene sequencing by Chun and Goodfellow (1995) (18) resolved the type strains of 9 Nocardia species into homogeneous taxon distinct from other taxon of aerobic actinomycetes with the genus Rhodococcus as the nearest taxon. Nocardia nucleotide signature sequences were identified and were found to be concentrated within 16S rRNA helix 37-1 (18, 19).
A phylogenetic investigation of the genus v using near full length 16S rRNA gene sequences acquired from 74 strains, comprising 25 known Nocardia species was examined to provide a more accurate method for species identification and taxonomic criteria, especially for strains of Nocardiaasteriodes (20). Their results showed sequence microheterogeneity within the 16S rRNA gene (≤5 bp difference) in 8 of 11 isolates. A low level of sequence interspecies heterogeneity was also observed among several closely related Nocardia species. For instance, 16S rRNA gene similarity was 99.5% between Nocardia paucivorans and Nocardia brevicatena, whereas the DNA-DNA relatedness was <70% (21). Of interest was the considerable sequence variation detected among 10 taxa suggesting they should be investigated further as new species of Nocardia. Strains previously classified as a single species by phenotypic methods, such as N. asteroides, were found to be a heterogenous collection of taxa, likely composed of 5 distinct species (9, 22).
Two studies evaluated the use of partial DNA sequences of approximately 500-bp from the 5′-end of the 16S rRNA gene for the identification of Nocardia species (23, 24). The MicroSeq 500 system supplemented with an expanded MicroSeq library of reference sequences was used to sequence a 529-bp fragment of the 16S rRNA gene and used to compare with phenotypic testing, REA analysis of 16S rRNA and 65-kDA genes, and sequencing a 999-bp DNA fragment of the 16S gene (23). The results showed sequencing the 16S rRNA gene to be more rapid and accurate than achieved by phenotypic methods for species identification and was able to correctly identify 6 common pathogenic Nocardia species (N. brasiliensis N. cyriacigeorgica, N. farcinica, N. nova, N. otitidiscaviarum, and N. veterana). For 94 strains representing 19 species, the MicroSeq system showed 72% agreement with phenotypic methods for species identification. While useful, the MicroSeq System was determined to be inadequate for reliable identification of many Nocardia species due to nearly indistinguishable sequence homogeneity within the first 500-bp of the 16S rRNA gene. Patel et al. (2004) similarly reported the ability of 16S rRNA gene sequences to be able to distinguish Nocardia taxa from other aerobic actinomycetes and observed many Nocardia species where composed of multiple species or complexes, including the N. nova, N. otitidiscaviarum, and N. transvalenis complexes (24). Use of near full length or full length 16S rRNA gene sequences was suggested for obtaining more accurate and reliable identification of isolates and for phylogenetic analysis (23, 24).
Molecular identification is based on the premise that 2 strains are the same species if they have identical sequences or the highest degree of sequence similarity in the available database. Therefore, the reliability and accuracy of molecular identification is heavily dependent on the accuracy and reliability of DNA sequences present in a sequence database. Unfortunately, existing DNA sequence databases may not always provide unambiguous identification. Misidentifications may occur due to the lack of curation of DNA sequence data in publicly available databases. This leads to the inclusion of misidentified or inaccurate entries, inadequate updates of newly described species, obsolete or inappropriate data entries, sequences submitted with inadequate sequence length or quality containing either incomplete or fragmented gene sequences, sequence data from only a single representative of the species or a small number of strains for a given species, and entries that do not adequately represent the extent of intraspecies gene heterogeneity (25–27). Consequently, results obtained from gene databases should be carefully scrutinized for gene sequences from non-validated species (valid species may be found at https://www.bacterio.net). The choice of database is therefore crucial for accurate identification.
Mellmann et al. (2003) evaluated the performance of 3 DNA sequence databases-GenBank, RIDOM, and MicroSeq 500-for species identification (27). Test isolates were identified by sequencing the 5′-end 429-bp of the rRNA gene from 64 culture collection strains (including 30 type strains) and 91 clinical isolates. Species breakpoints were ≥99.12% sequence similarity. All Nocardia type strains, except N. soli and N. cummidelens, were well demarcated and distinguishable. Overall, the RIDOM database provided the highest number of correctly identified species, followed by GenBank, and lastly by the MicroSeq 500 database, however, the RIDOM database is presently outdated and has been discontinued. DNA sequencing produced higher correct identifications than was obtained by phenotypic methods (27). In a similar study, the accuracy of 3 sequence databases, GenBank, Bioinformatics Bacteria Identification (BIBI), and Ribosomal Database Project (RDP-II), were evaluated for species identification against a collection of 96 Nocardia isolates by sequencing 606-bp of the 5′-end (25). All 96 isolates were correctly identified at ≥99% sequence similarity, and 86.5% of the isolates were correctly identified at 100% sequence similarity. DNA polymorphisms in the 16S rRNA gene sequences were detected with the highest degree of sequence intraspecies polymorphisms detected in isolates of N. nova, followed by isolates of N. brasiliensis. In an investigation by Conville et al. (2010), the Integrated Database Network System (IDNS) SmartGene database was used to identify a total of 102 Nocardia isolates (28). The IDNS database identifies queries for sequence comparisons based on using a DNA sequence of the most representative sequence of a species, denoted as the centroid sequence. The IDNS database correctly identified 76% of the test isolates. IDNS misidentifications were due to sequence heterogeneity within certain species, inaccurate database entries or due to inadequate size of the centroid sequence in the database.
To evaluate the performance of clustering algorithms, 364 16S rRNA gene sequences were attained from GenBank consisting of 77 different Nocardia species that were used to determine the most optimal method for alignment and distance measurements (26). Linear mapping of the alignment distance matrix identified 80 taxon using comparisons to centroid sequences. An additional 110 16S rRNA gene sequences obtained from gene databases were accurately identified by comparison to centroid sequences.
Three 16S rRNA operons were initially identified following the analysis of the whole genome sequences of N. farcinica IFM 10152 by Ishikawa et al. (29). Direct sequencing of 16S rRNA genes also suggested multiple gene copies. Multiple gene copies are generally observed as unresolved ambiguous multiple overlapping peaks present in sequencing chromatographs that cannot be resolved even after repeated sequencing (30, 31). The presence of unresolved mixed bases from multiple 16S rRNA gene copies is challenging since they may hamper identification resulting in misidentifications or altered RFLP profiles. The variable numbers of 16S rRNA gene copies that may be composed of heterogeneous gene sequences may complicate identification and phylogenetic analysis of Nocardia species isolates. Multiple 16S rRNA genes copies have been detected by cloning and sequencing individual gene copies from single clone isolates or by analysis of whole genome sequences. Conville et al. (2007) reported 2, 3 and 5 copies in the Nocardia type strains of N. concava, N. yamanashiensis, and N. ignorata, respectively, after sequencing cloned 16S rRNA genes or hybridization studies (31). Analysis of whole genome sequences for Nocardia type strains N. brasiliensis HUJEG-1, N. nova SH22a, and N. seriolae UTF1 identified 3, 3 and 4 rRNA gene copies, respectively (32–34). Mixed bases may be analyzed and resolved in some cases using RipSeq software (35).
Questions remain among researchers regarding the reliability of assignment of unknown isolates to a species based on the highest similarity score, especially when sequence similarity is below 100%. At present, there is no standard or consensus for interpretation of species with near identical 16S sequences. Sequence length, data quality, methods for alignment, and measurement of similarity or distance need to be addressed. CLSI document MM18-A recommended >99.6% sequence similarity for the identification of isolates to the species level and 99 to 99.5% sequence similarity for genus level identification (36). Some investigators suggest ≥99 sequence similarity, or a centroid percent similarity of ≥99.8% as criteria for species identification (25, 37). These criteria may be difficult to implement in all cases since several Nocardia type strains have been shown to be clearly distinct by DNA-DNA hybridization studies but have nearly indistinguishable 16S rRNA gene sequences. For instance, the 16S rRNA gene sequences of the N. kruczakiae and N. veterana type strains have 99.8% sequence similarity, the N. paucivorans and N. brevicatena type strains showed 99.6% sequence similarity, and there is only a 2 out of 1415-bp difference between N. sienata and N. testacea 16S rRNA genes (21, 38, 39).
ATPase secretory protein (secA1).
One alternative to 16S rRNA gene analysis is sequencing the single copy gene for the preprotein translocase ATPase secretory protein, secA1 (40). A 468-bp gene fragment of the secA1 gene was amplified and sequenced from 30 Nocardia type or reference strains and compared to analysis of a 1,285-bp rRNA gene fragment. Interspecies secA1 gene sequence variation was higher than obtained from 16S rRNA gene sequences; 85 to 98.7% sequence similarity compared to 94.4 to 99.8% sequence similarity, respectively. Analysis of 156 amino acid residues from 40 clinical isolates belonging to 12 Nocardia species detected intraspecies differences ranging from 0 to 3 amino acids. Analysis of secA1 amino acid sequences provided more reliable species demarcation than obtainable by DNA nucleotide sequences. Species identification was compared using a 470-bp secA1 gene fragment against the 5′ end, 606-bp 16S rRNA gene fragment for 10 reference strains and 110 clinical isolates representing 15 Nocardia species (41). Species identification by both secA1 and 16S rRNA gene sequencing was concordant for 94 of 110 clinical isolates. Discrepant species identification was suggested to be due to lateral gene transfer of secA1 gene sequences. Due to its faster molecular clock, the secA1 gene are more variable and showed a significant higher degree of interspecies and intraspecies diversity than obtained when using16S rRNA gene sequences which may be useful for epidemiologic investigations. Together, these studies show the usefulness of secA1 as an important adjunct method to 16S rRNA gene sequencing and the importance of evaluating a large number of isolates of the same species to create a robust sequence database for identification and taxonomy.
DNA gyrase subunit B (gyrB).
The DNA gyrase subunit B (gyrB) gene has been used for identification and phylogenetic studies of Nocardia species isolates (42–44). An approximate 1,200-bp gyrB fragment was amplified from 56 Nocardia type strains. Interspecies sequence similarity ranged from 82.4 to 99.9%, corresponding to a 3.6-fold higher sequence divergence than obtainable by 16S gene sequences. As expected, the majority (70%) of gyrB single nucleotide polymorphisms were silent (42). gyrB gene sequences were found to be able to distinguish Nocardia species from other mycolic acid-containing genera (44). Improved identification and enhanced discrimination has been achieved when using a combination of 16S and gyrB gene sequences (42). However, whole genome sequencing detected 2 gyrB gene copies, gyrB and gyrA, in N. farcinica (45). Currently, there is no consensus breakpoint for species demarcation.
B-subunit of RNA polymerase (rpoB).
The rpoB gene encodes the B-subunit of RNA polymerase and has provided a useful tool for identifying Nocardia isolates (43). Sequence analysis of a 354-bp rpoB gene fragment of 119 Spanish clinical isolates showed rpoB gene sequences to have a higher degree of DNA sequence polymorphisms than obtained using an approximate 500-bp 16S gene fragment. Both rpoB and gyrB gene sequences produced nearly the same degree of interspecies discrimination. Utilization of this rpoB fragment was hampered by its relatively short size and lack of comprehensive database. As with gyrB, whole genome sequencing has identified 2 rpoB genes, rpoB and rpoB2 (45).
Heat shock protein gene (hsp65).
Using PCR primers TB11 and TB12, a 441-bp DNA fragment of the heat shock protein gene, hsp65, from 44 Nocardia type and/or reference strains was amplified by PCR and compared to RFLP analysis and 16S gene sequencing (46). The investigation showed the hsp65 gene sequence to have a higher number of variable sites than detected for the 606-bp 16S rRNA gene fragment. The average dissimilarity for hsp65 gene sequence ranged from 12% to 0% and 9.5% to 0% for rRNA gene sequences for the same isolates. The hsp65 sequence variation was primarily localized to 2 hot spots located between bp 624 to 664 and bp 683 to 725. hsp65 gene microheterogeneity allowed for a more discriminatory and reliable phylogenetic tree than obtained by using 16S gene sequences. The majority of base substitutions in hsp65 occurred at the third codon position. Gene sequences of the hsp65 were found to be more variable than 16S rRNA gene sequences but were not as variable as gyrB or rpoB gene sequences (44). Sequencing of the hsp65 gene has been used for the identification of 11 ocular nocardiosis isolates from patients in China to the species level and showed the diversity of the etiologic agents responsible for Nocardia keratitis (47, 48) used both 605-bp 16S and hsp65 to identify 30 clinical isolates from patients in India.
Multilocus Sequence Typing
No single gene sequence alone has been found to provide sufficient discrimination between all species of Nocardia. Identification to the species level based on the analysis of a single gene is often unreliable due to the inability of a single gene to provide clear and unequivocal discrimination due to either low sequence variation between species, or the presence of foreign sequences from recombination or horizontal gene transfer. Multilocus sequence typing (MLST) has been reported to obtain a higher degree of discrimination and phylogenetic information than obtainable using a single gene (49, 50). MLST analysis is based on the concatenation of multiple loci for the detection of genetic relationships (49, 50). McTaggart et al. (2010) developed a MLST scheme for Nocardia and examined 11 reference strains, 36 type strains, and 190 clinical isolates by sequencing gyrB-16S-secA1-hsp65-rpoB gene fragments from each isolate, generating a 2,190-bp concatenated sequence for species identification (50). MLST detected 30 sequence clusters, denoted as species clusters; 167 of 237 isolates (71.3%) were assigned into the same species cluster as the corresponding type strain or were assigned as potential novel species. MLST showed 95% concordance between identification using phenotypic methods (50). Of interest is that MLST revealed that in 22.1% of the isolates, one or more alleles were in part comprised of foreign alleles. The presence of foreign alleles suggests interspecies recombination often leading to fuzzy species clusters. The presence of numerous insertion sequences in Nocardia genomes is consistent with the potential for frequent acquisition of foreign DNA. The authors suggest concatenation of multiple loci buffer against distortions by horizontal transfer within a single locus, which may distort their true taxonomic relationship (50). MLST analysis found that levels of genetic diversity vary widely among Nocardia species, thereby preventing the establishment of exact cutoff values for species identification. N. cyriacigeorgica and N. farcinica were the most prevalent species identified. MLST was not able to distinguish the Nocardia type strains of N. arthritidis, N. exalbida, N. gamkensis, N. ignorata, or N. coubleae, which suggests a reevaluation of their species status (50). Due to the expense and labor involved in sequencing 5 gene fragments, a 3 gene (gyrG-16S-secA1) or 4 gene (gyrB-16S-secA1-hsp65) typing system was proposed and identified correctly 98.5% and 99.5% of the isolates, respectively. A MLST scheme using 7 housekeeping genes (gyrB-hsp65-secA1-rpoB-rpoA-recA-trpB) generating a 3,639-bp concatenated sequence, detected 44 sequence types from a collection of 59 N. farcinica clinical and zoonotic isolates obtained in China (51). Population analysis identified 6 major clonal types from 46 isolates belonging to 32 sequence types. A modified MLST (16S-gyrB-hsp65-secA1) was used to identify 7 Nocardia clinical and zoonotic isolates obtained from Brazil (52). Overall, the high degree of genetic variation detected by MLST analysis has been useful for species identification, phylogenetic analysis, and epidemiologic investigations and is considered superior analysis using single gene sequences (53, 54). However, interpretation of MLST may be hampered by differences in mutation rates of different genes, differences in sequence length, gene duplications, and cost of supplies and equipment for analysis to sequence individual gene fragments.
Whole Genome Sequencing
Completed or draft quality bacterial whole genome sequences have been useful for detection of potential novel secondary metabolites, virulence genes, mobile elements or repeat motifs, antimicrobial resistance markers, bioactive and metabolic activities (55). Overall, analysis of whole genome sequences provides more reproducible, precise, and reliable phylogenetic analysis than obtained by traditional methods. The phylogenetic relationships among 26 Nocardia type strains was determined using shotgun sequencing and Roche 454 technology. Results showed Nocardia species have circular chromosomes with genomes ranging in size from 5.99 to 10.43 Mbp, an average genome size of 7.88 Mbp and a G+C ratio ranging from 65.5 to 71.7% (45). Phylogenetic trees were constructed from draft genome nucleotide or amino acid sequences by MLST analysis using 12 concatenated housekeeping genes and Bidirectional best hit (BBH) with orthologous genes. Twenty-two of twenty-five branches were common by both methods. Evolutionary relationships were subsequently calculated using the genome-to-genome distance calculator (GGDC), Average nucleotide identity (ANI), and the DNA maximal unique matches index (MuMi). Phylogenetic trees constructed using GGDC, ANI, and MUMi showed similar tree topologies and were all found to be superior to trees constructed using only the 16S rRNA gene sequences (45).
The number of Nocardia species with completed or genome sequencing projects in progress listed in the NCBI/Bioproject webpage accessed May 8, 2020 (https://www.ncbi.nlm.nih.gov/data-hub/taxonomy/1817/) is presently 84, growing rapidly, and probably is not inclusive of all known sequencing projects. In 2004, N. farcinica IFM 10152 was the first completed Nocardia genome. At present, there are 5 complete published Nocardia genomes including; N. brasiliensis HUJEG-1 (33), N. cyriacigeorgica GUH-2 (56), N. farcinica IFM 10152 (29), N. nova SH22a (32), and N. seriolae UTF1 (34). The number of predicted genes ranged from 5,674 for N. farcinica IFM 10152 to 8,414 for N. brasiliensis HUJEG-1. Comparative genome analysis detected 2,745 orthologous genes among the 5 Nocardia genomes. Whole genome sequencing confirmed the presence of multiple copies of the 16S rRNA genes. Three rrn operons were identified in N. brasiliensis, N. cyriacigeorgica, N. farcinica, and N. seriolae. Komaki et al. (2014) performed a genome-based analysis of 5 Nocardia species comparing type-1 polyketide synthases (PKS-1) and nonribosomal peptide synthase (NPRS) gene clusters (55). The number of secondary metabolite clusters varied substantially by species. PKS-1 clusters ranged from 4 to 11 copies and NRRS clusters ranged from 7 to 13. Seven PKS-1 and or NRPS clusters were detected in all the Nocardia strains examined. Many of these gene clusters were comprised of unique gene motifs not previously detected indicating the potential of Nocardia species as important sources for natural secondary metabolites. Two biosynthesis gene clusters consisting of 10 genes involved for nocobactin NA production have been identified following whole genome sequencing (57). Despite the abundance of genome projects, there are concerns about size, completeness, and accuracy of the data set (58). Likewise, in addition to the availability of whole genome sequences, it is easy to predict the use of RNA-Seq technology in the future to identify essential genes required for pathogenesis, growth, cell division, the production of bioreactive molecules, metabolism, and other important biological functions.
Vectors and Genetic Tools and Metabolic Engineering
Investigations on the molecular biology of Nocardia species has been facilitated by the development of cloning vectors, mutagenesis, and by whole genome sequencing. The first cloning vector, plasmid pCY104, was constructed by Yao (1994) by joining N. asteriodes IF03927 (mexicana) cryptic plasmid pCY101 with Escherichia coli plasmid pIJ4625, thereby supplying 3 antibiotic resistance gene markers capable of expression in N. asteriodes (59). The resulting vector, plasmid pCY104, was 8.9 kb with a transformation efficiency of 8 × 104 transformants per μg pCY104 by electroporation. The Nocardia-Escherichia shuttle vectors, pNV118 and pNV119, were constructed by combining a 1,777-bp DNA fragment carrying the Mycobacterium fortuitum plasmid pAL5000 origin of replication into the E. coli cloning vectors pK18 or pK19 (60). Plasmids NV118/NV119 have several advantages over plasmid pCY104, including a smaller size, 4.4 versus 8.9 kb, multiple cloning sites, and blue/white screening of transformants in E. coli by lacZ selection. A method for efficient transformation of Nocardia lactamdurans LC411 was developed using the pULVK series of plasmid vectors (61). Plasmid pULVK1 was originally derived from a 10.4 kb endogenous plasmid obtained from Amycolatopsis DSM 43387 by a natural deletion of 4.4 kb within plasmid pRL1. Two improved E. coli-Nocardia shuttle vectors, plasmids pULVK2 and pULVK3, were derived from plasmid pULVK1 by addition of a synthetic linker encompassing multiple cloning sites or by subcloning the Bluescript KS (+) multiple cloning sites. An apramycin gene resistance gene was subcloned into pULVK 2 to produce plasmid pULVK2A. Transformation efficiency was 7 × 105 transformants per μg of DNA and has been used to elucidate the pathway for nocardicin A biosynthesis in Nocardia uniformis (62).
Degradation of natural rubber and gutta-percha was investigated in N. nova SH22a (63). Mutants defective in rubber or gutta-percha degradation were obtained after optimization of electroporation conditions resulting in 4.3 × 107 transformants per μg DNA of vector pNC9503. A transposon insertion library composed of 12,000 insertion mutants was constructed by electroporation of plasmid MA5096 whose construct contains transposon Tn5096 and ampicillin and apramycin resistance markers. Of interest was the integration of the entire pMA5096 plasmid into the N. nova genome. Integration of pMA5096 into the genome facilitated identification of the site of integration, isolation, and sequencing of mutated loci therefore allowing for easier identification of mutant loci defective in gutta-percha degradation.
Plasmid shuttle vectors have been used on a variety of investigations to examine Nocardia metabolism and/or pathogenicity. The source of N. farcinica strains with high-level aminoglycoside resistance obtained from a clinical bovine mastitis epizootic in Canada was investigated by Kogure et al. (64). DNA from an amikacin-resistant mastitis isolate was shotgun cloned in plasmid pNV19 and used to transform the amikacin-susceptible Nocardia. farcinica IFM10152 host strain. DNA sequencing was performed on amikacin moderately resistant transformants detecting an A-to-G single point mutation at position 1408, located in the aminoacyl site of the 16S rRNA gene. Homozygous mutations at all three 16S rRNA gene loci were responsible for conferring high resistance to amikacin (64). In order to determine the contribution of the rpoB2 gene to rifampin resistance, N. asteriodes IFM 0319T, a rifampin sensitive strain, was transformed by electroporation with plasmids pNV1.2 or pNVrpoB2 a construct containing the RNA polymerase β-subunit (65). Only plasmid pNVrpoB2 transformants grew in the presence of 100 μg/mL rifampin showing the contribution of the rpoB2 gene to rifampin resistance. Whole genome sequencing of N. farcinica IFM 10152 identified a biosynthetic gene cluster consisting of 10 genes present in 2 clusters for biosynthesis of the siderophore notobactin NA (29). Construction of in-frame deletion mutants in the nbtA and nbtE genes showed highly reduced nocobactin NA production (57). The nbtS gene was shown to confer salicylate production in Streptomyces avermitilis which lacks a salicylate synthase gene.
Yields of nargenicin A1, a polyketide macrolide, was achieved by the application of DNA technologies and metabolic engineering using Nocardia sp. CS682 (66, 67). Yields of nargenicin A1 were increased 2.8- fold by the overexpression of heterologous S-adenosylmethionine synthetase (metK18) and acetyl-CoA carboxylase due to transcriptional activation of gene in the nargenicin A1 pathway. In cultures expressing the heterologous metK18 gene supplementation of cultures with methyl oleate increased yields of nargenicin A1 by 5.57-fold due to increasing the intracellular biosynthetic precursor pool.
Proteomics
Nocardia species are of biological interest due to their potential for production of novel bioactive metabolites and compounds by its members including antimicrobials, rubber, and petroleum degradation, waste management, bioconversion, steroid conversion, and biodegradation of alkanes and aromatic hydrocarbons (63). Despite the production of important biomolecules, there are relatively few investigations of the Nocardia proteome. Fourteen signature proteins were detected only in Mycobacterium and Nocardia species suggesting their close phylogenetic relationship which is in agreement with 16S rRNA gene analysis (68). Koenig et al. used comparative two-dimensional gel electrophoresis to examine the abundant soluble proteins for 5 Nocardia species isolates grown in Glucose Yeast Extract medium (69). Protein mixtures were resolved by two-dimensional gel electrophoresis, abundant spots excised, and digested with trypsin. The tryptic digests were then analyzed by matrix-assisted laser desorption/ionization mass spectrometry (MALDI-MS). Several chaperones and housekeeping enzymes were identified. Arylamine N-acetyltransferase (NAT), an important enzyme involved with inactivation of isoniazid in Mycobacterium, was the first Nocardia protein to be characterized functionally and structurally (70). The substrate profile of the 293-amino acid NAT gene suggests NAT may contribute to isoniazid resistance but not to sulfamethoxazole resistance in Nocardia. Highly expressed genes in N. farcinica have been predicted using the codon adaptation index (71). Although most of the predicted highly expressed genes are involved with housekeeping functions, 25 putative virulence or genes required for intracellular survival in a host as well as genes involved for protection from reactive oxygen produced by phagocytes were identified.
MALDI-TOF mass spectrometry.
Matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) is an increasingly important method for the accurate and rapid identification of bacterial and fungal samples, particularly when given the breadth of Nocardia taxonomy and limitations of biochemical methods (6). While the upfront cost of a MALDI-TOF instrument is significant, the per sample cost of running samples is relatively inexpensive and the quick turnaround time coupled with its ease of use have made MALDI-TOF economically viable for many clinical microbiology labs (72–74). There are numerous manufacturers of MALDI-TOF instruments worldwide, but 2 of the most prominent MALDI-TOF instruments in the United States and Europe are the Bruker MALDI Biotyper (Bruker Daltonics GmbH & Co. KG) and bioMérieux's Vitek MS (bioMérieux). Both instruments have been thoroughly examined in the literature (73, 75–77) and both instruments operate in a similar manner. Briefly, a sample is applied to a target plate, co-crystallized with a matrix solution (α-Cyano-4-hydroxycinnamic acid) and then the target plate is placed in the instrument. A vacuum is applied and samples are bombarded with short laser pulses that vaporize the sample leading to desorption and ionization of matrix polypeptides and proteins by charge transfer. The ionized materials are accelerated in an electric field and enter the flight tube. The time of flight of the ions to reach the detector is precisely measured to produce characteristic spectra and is dependent on the degree of ionization and mass (ranging from 100 Da to 100 kDa) (78, 79). Species identification is based on comparison of sample spectra with spectra in a database. The quality of spectra has been found to be dependent on variables including the instrument, sample preparation, plate cleanliness, extraction method, age of culture, culture conditions including amount of sample and length of incubation, and method of disruption (80–82).
Many of the variables impacting MALDI-TOF identification are user determined (e.g., growth conditions of cultures, type and quality of extraction procedures), but it is the contents of the reference database that can ultimately determine the success of an identification (73). As of the publication of this article, the Bruker MBT Compass Library Revision H contained 89 Nocardia species, while the Vitek MS version 3.2.0 contained only 16 Nocardia species. It is important to note that 59 of the Nocardia species in the Bruker library have only a single representative for the respective species. Carrasco et al. reported that less prevalent Nocardia species were difficult to correctly identify with MALDI-TOF and suggested that databases needed to be improved to include more of these species (53). Other investigators have also highlighted the importance of obtaining spectra from multiple strains of the same species to provide a range of spectral profiles obtainable from a given species (54, 83–85). Lack of Nocardia representation in commercial databases has led many investigators to create their own MALDI-TOF databases (54, 72, 74, 81, 86, 87), a distinct disadvantage for owners of the Vitek MS as bioMérieux does not allow for the creation of user databases with their MALDI-TOF instruments.
Not surprisingly, studies examining the efficacy of user created databases have found that the percentage of successful identifications increased when custom made databases where filled with Nocardia strains not in commercially available databases (72, 74, 81, 86, 87). When Verroken et al. (84) used the Bruker database, only 19 of 43 (44%) Nocardia test isolates were correctly identified. However, when spectra from an additional 110 Nocardia isolates confirmed by 16S rRNA gene analysis and phenotypic tests were used to supplement the spectra database supplied with the Bruker Biotyper, the number of correct identifications improved to 88%. Many investigators agree that a higher level of correct identifications can be achieved when the manufacturer’s spectra database is augmented with more diverse reference spectra, particularly from unusual and newly described species. Investigators who wish to create custom databases are advised to obtain secondary identification either by 16S rRNA gene sequences and/or MLST to confirm the correct species identification before entering reference isolates into custom made MALDI-TOF spectra databases (53, 54).
Unfortunately, the absence of a species in a MALDI-TOF database does not always result in a “no identification” score. Several studies have reported that when commercial databases are challenged with Nocardia species that are not in the manufacturer databases, the samples can be misidentified as an incorrect Nocardia species (85, 88, 89). Thus, it is imperative for users to be aware of the contents of the databases they are searching. Both the Bruker and bioMérieux MALDI-TOF instruments appear to be equally capable of identifying Nocardia species if the respective species are in their databases (74, 85, 88, 90–92), and both companies update their databases fairly regularly with additional species and better-quality spectra. However, it is important to recognize that studies examining how well the Bruker Biotyper and the bioMérieux Vitek MS identify Nocardia species are dependent on the version of the database that the authors tested at the time they conducted their study. As a result, a literature review of which manufacturer can best detect Nocardia is only evaluating the contents and quality of the respective databases at a specific point in time.
Many bacterial species can rapidly be examined via MALDI-TOF by simply spotting a colony onto a target plate and applying matrix (the direct transfer or direct smear method). While some investigators have reported good results using this method with Nocardia (92), multiple studies report that Nocardia species require a much more rigorous preparation (85, 88); bioMérieux offers an extraction kit (Vitek MS Mycobacterium/Nocardia Kit) to assist in the preparation of Nocardia samples for MALDI-TOF analysis. The procedure utilizes 0.5 mm diameter glass beads in a bead beating step, but is otherwise very similar to the Bruker extraction method that utilizes ethanol, formic acid, and acetonitrile that many authors have employed when working with Nocardia (86, 89, 91, 92). Some studies have noted that bead beating is not necessary for Nocardia MALDI-TOF extractions (89, 92).
Several authors have highlighted the importance of culture conditions when identifying Nocardia via MALDI-TOF and a wide variety of media and culture conditions have been employed with varying levels of success. Mycobacterial growth indicator tube broth, Trypticase soy broth, brain heart infusion agar with 5% rabbit or sheep blood, bromocresol purple agar, buffered charcoal yeast extract media, chocolate agar, Columbia blood agar, horse blood agar, Lowenstein-Jensen medium, Middlebrook and Cohn 7H11 agar with Oleic Albumin Dextrose Catalase, Sabouraud dextrose agar, and tryptic soy agar with and without 5% sheep blood have all been utilized to culture Nocardia for MALDI studies (53, 54, 72, 80, 81, 84–96). Cultures have been reported to be incubated at temperatures ranging from 30°C to 37°C from 18 h to 7 days, with and without 5% CO2 (53, 54, 72, 80, 81, 84–96). It is important to ensure that Nocardia are cultured with the correct media, but for use in MALDI-TOF investigators specifically examining culture conditions have reported that Columbia blood agar (or other media that can promote quick growth) incubated for less than 48 h produced the best results (81, 85, 89).
Unlike phenotypic testing and DNA sequencing, MALDI-TOF does not appear to require mature colonies for successful identification of Nocardia (81). Khot et al. reported that 52% of Nocardia isolates could not be identified (Bruker score <1.7) when cultures were allowed to grow for more than 48 h (81). But when re-grown for less than 48 h only 12% could not be identified. McTaggart et al. found that Nocardia cultures grown for 3 days on Sabouraud dextrose agar resulted in only 36% of species being identified to the species level. But, when cultures were grown on Columbia blood agar as soon as growth was observed (18 to 72 h), 81% of the cultures were correctly identified to the species level (89). Similarly, Toyokawa et al. stressed the importance of using colonies between 18 and 48 h when using MALDI-TOF to identify Nocardia (91).
Since multiple factors can lead to a failure to identify a sample, either repeating an extraction or spotting samples multiple times on a target plate can improve results (80, 85, 88). Particularly with the Bruker Biotyper, some investigators have adopted much less stringent criteria for their identifications than those recommended by the manufacturer. Specifically, for Nocardia species, Bruker scores as low 1.9 (81), 1.8 (92), and 1.7 (74) have been used and/or suggested for Nocardia species level identifications, and as low as 1.5 for genus level identifications (92). In instances where the Bruker Biotyper reports multiple species with scores above 2.0 some investigators have employed a “10% rule difference” to determine if an identification can be made (74, 97), but this appears to only be necessary with <1% of Nocardia species (74).
MALDI-TOF analysis has been shown to be a rapid method with a relatively high degree of accuracy for the identification of the majority of Nocardia isolates, but investigators are cautioned that not all Nocardia species are resolvable with MALDI-TOF (9, 81, 88, 90). MALDI-TOF alone cannot be used for identification of all organisms (73) and, therefore, it is sometimes necessary to employ other methods of identification such as DNA sequencing, which is still currently the gold standard for identification.
Direct Detection in Formalin-Fixed, Paraffin-Embedded Tissue, and Clinical Samples
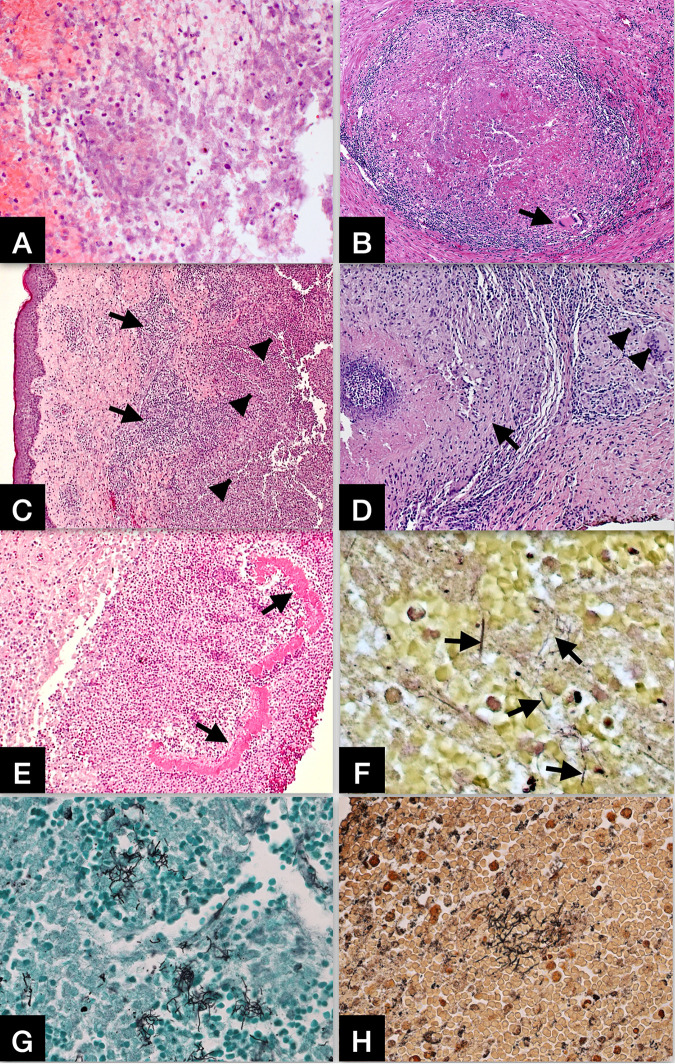
Formalin/paraformaldehyde-fixed paraffin-embedded (FFPE) tissues from biopsy or autopsy are important sources of clinical materials. Identification of Nocardia in FFPE tissue samples by direct examination of stained histologic sections is difficult because many other bacteria, especially mycobacterial organisms, are indistinguishable by morphology alone using histochemical stains. However, histopathologic features with a panel of ancillary histochemical stains can still provide insightful information for differential diagnosis and the extent of tissue damage caused by nocardial infection (98, 99). Nocardial infection frequently causes abscess formation (Fig. 1A) and granulomatous response (Fig. 1B) in the tissues involved, such as lung (Fig. 1B), skin (Fig. 1C, D, and E), and brain (Fig. 1A). Routine hematoxylin & eosin (H&E) stain shows necrosis, karyorrhectic debris, and neutrophilic infiltrate in the abscess, and lymphohistiocytic infiltrate with multinucleated cells in granulomatous inflammation. In rare instances, dense aggregated granules (grains) with a peripheral radial deposition of intensely eosinophilic material – a Splendore-Hoeppli phenomenon - may be observed in cutaneous infection (Fig. 1E). The bacilli cannot be demonstrated directly by the H&E stain. Ancillary special stains, including Gram stain, Grocott methenamine silver (GMS) stain, Steiner silver stain, and acid-fast bacilli (AFB) stain can help highlight the bacilli in tissue samples. However, the sensitivity of these special stains varies depending on the quality of the stains, the number of organisms in the tissues examined, the species of nocardia, and subjective interpretation bias. When the bacilli are observed, these stains are not specific and can only raise the index of suspicion for nocardial infection. Gram stain usually shows thin, delicate, beaded filaments with variable gram-positivity (Fig. 1F). GMS stain (Fig. 1G) and Steiner silver stain (Fig. 1H) both can readily illustrate the bacilli, and the GMS is the most sensitive screening stain for nocardiosis. AFB stain, such as Ziehl-Neelsen stain may partially highlight the bacilli but the result is often inconsistent and cannot be interpreted alone without other stains. Unlike many bacterial and fungal organisms, Nocardia species are not visualized with Periodic acid-Schiff (PAS) stain.
FIG 1.
(A) Nocardial brain abscess. Routine hematoxylin & eosin (H&E) stain shows necrosis, karyorrhectic debris, and neutrophilic infiltrate in the abscess. Original magnifications: X200. (B) Nocardial lung infection with granulomatous inflammation. H&E stain shows a granuloma with central necrosis, lymphohistiocytic infiltrate at the periphery, and rare multinucleated cells (arrow). Original magnifications: X100. (C) Nocardial skin infection. H&E stain shows a zone of mixed inflammatory infiltrate (arrows) and large area of abscess formation (arrowheads) in the dermis. Original magnifications: X50. (D) Nocardial skin infection with granulomatous inflammation. H&E stain shows a granuloma (arrow) with central necrosis and lymphohistiocytic infiltrate at the periphery, as well as scattered multinucleated cells in adjacent area (arrowheads). Original magnifications: X100. (E) Nocardial skin infection. H&E stain shows dense aggregated granules (grains) with a peripheral radial deposition of intensely eosinophilic material (arrows) – a Splendore-Hoeppli phenomenon. Original magnifications: X100. (F) Gram stain showing scattered Gram-positive and gram-variable nocardial organisms in the abscess. Original magnifications: X400. (G) Grocott methenamine silver (GMS) stain showing clusters of filamentous nocardial organisms in the abscess. Original magnifications: X400. (H) Steiner silver stain showing clusters of filamentous nocardial organisms in the abscess. Original magnifications: X400.
Identification from FFPE tissue has been accomplished by in situ hybridization with fluorescently labeled oligonucleotide probes specific to the variable regions of 16S rRNA gene sequences or by using a PCR-based assay. Due to the presence of high molecular weight mycolic acids in Nocardia species cell walls, permeabilization of N. asteroides cell walls in fixed cells was discovered to require mild acid hydrolysis (1 M HCl for 30 min) in order to allow the entry of fluorescently labeled oligonucleotide probes inside cells (100). An investigation by Carr et al. (2005) detected an enhanced fluorescent signal when using a combination of acid hydrolysis with enzyme treatments (101). Enzyme treatments included combinations of lipase/proteinase K, acid/mutanolysin/lysozyme or acid/lipase/proteinase K. Fluorescent labeled oligonucleotide probes specific to 16S rRNA gene sequences were used to identify Nocardia in 10 of 13 tissue samples (102). Whereas in situ hybridization provided a rapid and specific method for identifying Nocardia in tissues while maintaining tissue morphology using standard formalin-fixed tissues, no significant increase in sensitivity was noted after comparison to histologic staining. Negative results following in situ hybridization may signify either low sensitivity of the probe or the microorganism of interest was not present.
To detect Nocardia species directly from clinical samples (sputa, bronchoalveolar liquid [BAL], pus and skin biopsy), Couble et al. (2005) developed a PCR-based assay amplifying a 590-bp 16S rRNA gene fragment using Nocardia-specific primers, NG1 and NG2 (103). Following agarose gel electrophoresis, the PCR product was transferred to a solid support by Southern blotting and then hybridized with a chemiluminescent 16S probe. The method successfully identified 5 different Nocardia species in 18 samples. In comparison, 20 culture positive Mycobacterium tuberculosis samples were negative by the assay. The sensitivity of the assay was determined using spiked samples of BAL. Stained agarose gels detected 1,000 CFU/reaction whereas 1 CFU/reaction were detected by Southern blotting. The etiologic agent present in a brain abscess was reported (99); FFPE brain tissue from the abscess was deparaffinized and DNA purified using a commercial DNA isolation kit. DNA sequencing of a 330-bp rRNA gene target fragment showed the highest degree of sequence homology to either N.farcinica or N. otitidiscaviarum confirming histological examination showing Gram-positive, branching mycelium. The isolate was identified as N. farcinica using a species-specific PCR assay (104).
Antimicrobial Susceptibility Testing
Antimicrobial susceptibility testing of Nocardia isolates has been used both as a method of identification and a way to guide therapy. There are previous reports that delve deeply into this topic so that history will not be retraced here (6, 9, 23).
Currently, the Clinical and Laboratory Standards Institute (CLSI) broth micro dilution method is the recommended method for performing AST on Nocardia isolates (16). The most current methods are described in detail in the standard M24 3rd edition that replaced the M24-A2 in 2018 (16). First line recommended drugs for testing are: amikacin, amoxicillin-clavulanate, ceftriaxone, ciprofloxacin, clarithromycin, imipenem, linezolid, minocycline, moxifloxacin, trimethoprim-sulfamethoxazole, and tobramycin. Second line drugs are cefepime, cefotaxime, and doxycycline (16). For many of the frequently reported species such as N. brasiliensis, N. farcinica, N. nova, N. cyriacigeorgica, and the N. transvalensis complex, susceptibility and resistance to antimicrobial agents can be predicted with accurate identification (105Conville, 2012 #3606). However, it is highly recommended that AST be performed on all Nocardia isolates of clinical significance for the best clinical outcome.
Sulfonamides have historically been the drugs of choice for treatment of nocardial infections (6). When performing in vitro AST against these drugs, endpoint determination can be difficult due to the growth characteristics of Nocardia species. This issue has also been reported when testing against several other drugs of choice such as ceftriaxone and imipenem where false resistance has been documented (105). In 2012, a multisite reproducibility study by Conville et al. traced many of the difficulties with reproducibility across, and within, testing laboratories back to the physiological and growth characteristics of nocardiae (105). Cells of nocardiae are hydrophobic due to the presence of mycolic acids in their cell walls (6). Even distribution of cell mass in liquid inoculum is difficult because the culture tends to become clumpy and float. Conville et al. also found that because of differences in growth characteristics between species within the genus, susceptibility endpoint determination can be inconsistent between testing personnel (105). Their findings, therefore, presented the need for a reference strain of Nocardia for growth characteristics to be proposed, N. nova strain ATCC BAA-2227. Further, as a means of confirmation of resistance to sulfonamides, the use of a disk diffusion test for sulfisoxazole was also proposed. CLSI now has incorporated this requirement in the most recent edition of the M24 (16).
CLINICAL DISEASE: NOCARDIOSIS
Epidemiology
In 1976, Beaman and colleagues published a report with the aim of measuring the incidence of nocardiosis in North America (1). They found that because there is no national reporting system for Nocardia infections, accurately measuring the incidence of the disease was not possible. This still holds true today.
Demographics.
Many studies report that males outnumber females at a ratio of 2 to 3:1 (3, 106–109), but other studies show a more equitable distribution with slightly fewer females (5, 110–112). The differences in distribution may be associated with an underlying association of sex with the risk factor of interest in the study. Two studies among solid organ transplant recipients reported a ratio of 1.7:1 (8) and 2:1 (107) males to females, but the latter study reported a ratio of 3:2 among all transplant patients at their facility (107). Other transplant (113–115) and older HIV studies (109, 116, 117) report a disparate ratio by sex. Malignancy studies reported ratios of 1:1 (110, 111) and 1.6:1 (118), non-specialty facilities reported ratios of 1.1:1 (119) - 1.3:1 (5), while national reference laboratory reports vary widely (1.4:1 [112] to 2.4:1 [120]).
McNeil et al., in the seminal summary of medically important actinomycetes, stated that cases typically are in their “third to fourth decade” (4), although some recent studies have average ages in the 50’s (5, 121) and 60’s (112). Reports range widely in average age reported, which also may have an association with risk factors. The average age among patients for whom isolates were sent to CDC between 2008 and 2018 with age information (n = 1,894) was 58.0 (Standard deviation [SD]: 21.5, range 7 weeks-104) (unpublished CDC data).
Sources of disease.
As saprophytic bacteria, Nocardia spp. decompose organic matter in the soil (122) and have been found widely in soil and water (123). Generally, pulmonary exposure occurs when aerosolized spores or mycelia are inhaled (4); the bacteria are frequently found in dust and bioaerosols (123–126). Primary pulmonary infection more often occurs in immunocompromised individuals (6) or those with structural lung disease (127, 128).
The second exposure route is direct inoculation into the body, which is the source of most primary cutaneous infections (3, 129, 130). Inoculation can be traumatic, such as an injury sustained during a car accident (131) or mildly traumatic as a prick from a bush (132), through a nosocomial exposure (124, 133–136), or through dust or dirt entering into open wounds (3, 130). In an extreme case, a patient developed nocardial meningitis following a traumatic skull fracture (137).
Nosocomial.
As previously stated, nosocomial exposures are infrequently reported in the literature; source confirmation varies (124, 138). Exmelin et al. reported an outbreak among 3 immunocompromised heart transplant patients in the same ward with highly similar strains; no potential source was identified (135). Two instances of disseminated disease occurred in immunocompetent cases following insertion of prostheses (136, 139), but again, the source was not identified.
Houang et al. reported the source of a nocardiosis outbreak in a renal unit was contaminated air ducts, as they were able to recover a small number of Nocardia spp. colonies from air, dust, and settle plates placed in the ward (124). Because molecular typing was not yet available to confirm the relationship of the isolates, there is not confirmatory evidence of the source of the infections. A more recent investigation used molecular typing methods to determine that the source of an outbreak among 5 open heart surgery patients was traced to an anesthesiologist who was present at each surgery and was found to be colonized with the organism (140).
One study suggested that exposure to medical equipment was a possible source of infection (141). There was extensive Nocardia spp. biofilm formation found on central venous catheters used at a cancer facility where 10 patients had central line-associated bloodstream Nocardia spp. infections and another 7 were bacteremic (141). The authors did not assert that the infections were definitively from the catheters, although they recommended antimicrobial treatment of central venous catheters to reduce biofilm growth. Beyond such prophylaxis, no specific precautions are recommended to prevent Nocardia spp. nosocomial transmission due to its rarity and limited evidence for communicability. However, the clinical implications of environmental contamination with Nocardia spp. in health care settings may need to be reconsidered. For example, Rahdar et al. found evidence of 25 Nocardia spp. isolates from 63% of Iranian hospitals that were sampled (126).
Incidence.
Current knowledge of nocardiosis incidence in the United States is based on an historical survey of 171 infectious disease physicians from 1974 and isolates received at the CDC reference lab (1). This extrapolated estimate was 500-1,000 new cases annually, which has been referenced frequently and recently (142, 143) despite changing demographics of the population (144), increasing numbers of immunocompromised adults (145), longer survival (146), and greater occurrence of higher risk conditions (based on Organ Procurement and Transplant Network data as of January 17, 2019), and improved laboratory methods (10).
Although there are no current national-level estimates of nocardiosis incidence or prevalence for the United States, there are many prevalence estimates among special patients at specific facilities. A retrospective study from a transplant facility calculated an overall prevalence rate of 0.6% among their transplant patients; by organ type, rates ranged from 0.1% among liver transplant patients to 3.5% among lung transplant patients (114).
Globally, reports are in conflict whether nocardiosis incidence is increasing or remains stable (128). In Japan, a population-based analysis of isolates from 1992–2001 found the raw count to be trending upward (39), but they did not account for population growth. A study from Quebec reported increasing incidence (147), while 2 studies from Spain reported a stable incidence (120) and a non-significant positive change (121).
Risk factors among the immunocompetent.
An estimated 60% of infections occur in immunocompromised individuals, and 40% are reportedly immunocompetent (148, 149). However, it is estimated that 10% or fewer infections occur in immunocompetent individuals without any risk factors (150, 151), such as chronic lung disease, on long-term corticosteroids, or have other underlying conditions that may predispose them to a lung infection (106, 128, 143). More specifically, these include chronic obstructive pulmonary disease (127), bronchiectasis (128), and cystic fibrosis (143, 152).
In one review of 59 cases of pulmonary disease, 88% had some sort of underlying pulmonary condition, including structural changes to the lung, but most were considered immunocompetent (106). Structural changes to the lungs (127, 128) may impact the respiratory immune response (153), as can aging (153), which may increase the risk of pulmonary nocardiosis among adults 65 and older. A recent study has found an association between disseminated nocardiosis among immunocompetent individuals with granulocyte macrophage colony-stimulating factor autoantibodies (154). This association requires additional research but may help explain the ability of nocardiae to overtake an apparently healthy immune system. The remainder of immunocompetent nocardiosis cases are likely exposed to the bacteria by inoculation (132) leading to cutaneous infection, or nosocomial exposure leading to arthritis (133, 134, 155) or eye infections (14).
Corticosteroids.
Nocardiosis has been associated with the use of oral and inhaled corticosteroids (5, 121, 128). This association is not restricted to Nocardia spp.; a similar correlation has been found with non-tuberculosis Mycobacterium pulmonary infections (156). It is not clear the extent of the impact of lung structural changes compared to prolonged corticosteroid use, since many patients with structural changes are on extended corticosteroid use (127). One study of 31 pulmonary nocardiosis cases found 7 (23%) cases had COPD, 4 of whom were on prolonged steroids, and 20 cases overall (64.5%) had prolonged steroid use (157). In another study, immunocompromised patients with prolonged corticosteroid use had a much higher mortality compared to both immunocompetent and immunocompromised patients not taking corticosteroids (85%, 15%, and 20% respectively) (158). Improvements in targeted immunosuppressive medications for transplant patients and others have reduced the use of broad corticosteroids (114), which may be a factor in reports of decreasing infections among transplant recipients (107, 113).
Risk factors among the immunocompromised.
Conditions that affect cell-mediated immunity dominate the risk factors for nocardiosis among the immunocompromised (159). The immune response to nocardiae begins with innate immunity; first, monophils and neutrophils phagocytize most nocardiae and inhibit their growth, although the nocardiae are not destroyed (160, 161). Adequately functioning T-lymphocytes are then required to directly contact the nocardiae, causing subsequent lysis and killing of the bacterium (162), which prevents pulmonary or systemic nocardial infection. However, adequately functioning cell-mediated immunity may aid in the development of mycetoma granulomatous inflammation (163).
Transplant.
Solid organ and hematopoietic stem cell transplant (HSCT) recipients are at greater risk of bacterial infection due to induced immunosuppression required to prevent rejection (5, 164, 165). Transplant recipients remain at risk despite more precise effects of anti-rejection drugs on the cell-mediated immune system compared to older medications such as azathioprine (113, 142, 165).
The prevalence among heart transplant patients ranges from 0.65% (149, 166)−2.5% (114), although historical reports show rates of 13% when patients received azathioprine for immune suppression (113, 115). Peleg et al. also calculated nocardiosis rates by organ transplant types: kidney (0.2%), liver (0.1%), small bowel/multi-visceral (1.3%), and lung (3.5%) (114). Rates reported in Spain are similar, although lower among lung transplant patients (renal [0.26%], hepatic [0.18%], and lung [1.78%]) (149). Yet, multiple reports support higher rates of infections among lung transplant patients compared to other organ transplants (149, 167, 168).
Nocardia spp. infections have occurred months to years after receipt of the transplant (149, 164, 169). Although Peleg et al. found that 63% of transplant recipients developed nocardiosis within 1 year of transplant (114), a multisite study found that only 41% of recipients had onset during the same time (8). The median time from transplant to nocardiosis onset was 17.5 months (range 2–244 months), although this varied significantly by organ transplanted (8). Onset more than 3 years after transplant occurred in 31.6% of patients (8), and 14% had onset more than 5 years post-transplant (114).
Most infections are pulmonary, and nodules are a common finding (8, 114, 168). Extrapulmonary dissemination ranged widely but was more common in transplant patients than others (5). Dissemination ranged from 20% to 47% (8, 114, 168, 170). Mortality also varied widely, from 14% (114) to almost 40% (128).
A number of risk factors have been identified beyond immunosuppression. Risk factors found from 2 studies include high dose steroid use and high calcineurin inhibitor levels within the previous month (114, 169). Additional risk factors include recipient age, use of the immunosuppressive drug tacrolimus, intensive care unit length of stay following the transplant (169), and cytomegalovirus (CMV) infection or disease (8, 114). CMV reactivation occurs in patients with hemopoietic stem cell transplant (171).
Malignancy.
Patients with solid tumors and hematologic cancers are at elevated risk of nocardiosis, likely due to cell-mediated immunosuppression (111). A surveillance study in southern France found 22% of cases had a history of malignancy (172). The majority of infections occur in patients with hematologic cancers (54.5–64%), followed by solid tumors (36–43.9%) patients have frequently received stem cell transplants (31%–35.6%) (111, 118). In a cancer population, nocardiosis frequency appears to be increasing over time—infections averaged 3.3/year from 1988 to 2001 (111), 4.6/year from 2002 to 2005, and 16.4/year from 2006 to 2012 at a cancer facility (118). Incidence during the first period was 60/100,000 patients with the highest incidence among bone marrow transplant recipients (701/100,000 patients) (111); incidence was not published for the latter time periods (118). The increasing frequency has been ascribed to improved testing, recognition, and survival among cancer patients (118), especially as the toxicities from some therapeutics, such as the monoclonal antibody alemtuzumab or purine analogs, can last for months to a few years after use (173).
Monoclonal antibodies are now common, effective therapies for malignancies (174, 175), but their mechanism of action leads to defects in cell-mediated immune response. Alemtuzumab causes neutropenia lymphopenia, while rituximab causes B-cell lymphopenia; both have the potential for increased risk of infection, although viral infections or reactivation appear more frequent than bacterial infections (173, 175–177). CMV infection or reactivation is also a concern for hematologic cancer patients treated with alemtuzumab and occasionally with other monoclonal antibodies (171, 178), which in turn is associated with nocardiosis (8).
There are a small number of reported nocardiosis cases in patients receiving monoclonal antibodies used for cancer therapy (174, 179), which raises the question of the effect of the B-cell associated lymphopenia from rituximab on T-cell immune response (164, 175).
Cancer patients may also be at risk because nocardiae can compose biofilms on central venous catheters (CVC) (141). Such growth can lead to central line-associated bloodstream infections (CLABSI) (141), and may be related to disseminated bacteremia in patients with CVCs (108, 141). Although these infections appear to be rare, those with CLABSI had better outcomes than those with disseminated infection, including shorter hospital stay and lower mortality (141).
Nocardiosis symptoms, such as fever, may not be present due to immunosuppression (118). Additionally, abscesses can be difficult to differentiate from malignancies (7), and may be confused as metastasis of an existing cancer (180). This is particularly true for central nervous system (CNS) infections in patients with cancers that frequently metastasize to the brain, which can delay diagnosis and may affect prognosis (180).
HIV.
HIV is a risk factor for nocardiosis owing to its impact on cell-mediated immunity (159). Although HIV may be the primary risk factor in many cases, in one study, half of the patients also had chronic lung disease (120). Reported all-cause mortality from older studies was 63% (109) and 67% (116). More recent reports indicate that nocardiosis is less frequent in HIV-positive persons possibly as a result of prophylactic trimethoprim-sulfamethoxazole to prevent Pneumocystis infections (142). However, this prophylaxis may not provide adequate protection against nocardiosis (5, 170).
There are a few estimates of nocardiosis prevalence among HIV-infected individuals. A few articles that are frequently referenced report cases from the 1980s (109, 116, 181) when the demographics, care, and prognosis was vastly different for HIV-positive patients compared to today (146). These estimates range from approximately 0.3% (116, 181) to 1.8% (109). In Spain, a report found an incidence rate of 0.38% (120), while one study in Côte d’Ivoire found a nocardiosis prevalence of 4% among patients who died of AIDS (117).
Injection drug use is commonly reported among cases with HIV and nocardiosis. In Uttamdani et al., 53% of HIV-positive nocardiosis cases were injection drug users (IDU), compared to 30% among the HIV-positive patients who received treatment at the same time (109). Cases also frequently had onset of AIDS concurrent with the nocardiosis infection or within the previous 6 months (60%) (109).
Pulmonary nocardiosis can be mistaken for tuberculosis (7), and tuberculosis and HIV are frequent co-infections (182). Among 10 patients who were diagnosed postmortem with nocardiosis, 40% had been incorrectly diagnosed with tuberculosis prior to death (117). Of HIV-positive patients presenting to a chest clinic for suspect pulmonary tuberculosis in Sudan, 1.2% were diagnosed with nocardiosis (183); 2.9% of HIV-negative patients at the same chest clinic also had nocardiosis, but 94% of patients had a risk factor for nocardiosis (183). Another study that also evaluated patients with suspect pulmonary tuberculosis in Ghana found 16.7% were co-infected with HIV and Nocardia spp. while 8.3% were co-infected with HIV and tuberculosis (184).
Global Distribution and Ecology
Nocardia have been isolated from many sources including soil, seawater, caves, sugar cane fields, in association with ants, and humans. Infections are considered to be opportunistic and may be localized or disseminated and are widely distributed globally and found in a variety of ecological systems A few studies have attempted to evaluate the geographic distribution of Nocardia species, primarily using clinical isolates (3, 128, 185). The genus Nocardia appears to be ubiquitous in many locations around the world (2, 14, 117, 121, 126, 157, 174, 183, 185–187).
There may be geographic variation by infecting species. The previously named N. asteroides, which has been separated into multiple diverse species (40), and N. farcinica have been reported to have an even distribution across the United States (6, 151). One report claims that N. nova is less frequently identified in the southwest (151). Of N. brasiliensis case reports in the United States up to 1984, 63% were published in 5 states (Texas, California, Florida, North Carolina, and Oklahoma) (3), although these results are perhaps limited by publication bias. Another author based in the southwestern U.S. has stated that N. brasiliensis is most common in the southeast (based on the previously mentioned study) and the southwest, based on receipt of 40 N. brasiliensis isolates out of 455 Nocardia isolates in 5 years (151). Uhde et al. report that 59 of 106 (56%) of N. brasiliensis isolates originated from Tennessee and Florida over a 10-year period (2), which supports the claim of a higher prevalence in the southeast.
Ecological characteristics likely influence the presence of Nocardia spp. organisms and geographic variation of Nocardia spp. in the soil. It is generally accepted that nocardiosis is endemic in tropical and subtropical climates, while infections are less frequent in temperate climates (6). Warmth and humidity have been associated with nocardial keratitis (188) and actinomycetoma (189). This association may be intensified by greater soil exposure due to the types of clothing and shoes worn (or not worn) in hot and humid environments (187–189). These results contradict a statement of a regionally-based author that most U.S. cases occur in the hot, dry, and windy climate of the American southwest (151).
Nocardia species diversity, but not the presence of Nocardia spp., may be influenced by soil characteristics. A soil study from Iran found that 60% of “N. asteroides complex” were found in soil with pH 7.1 to 8.0, and 63.6% of N. brasiliensis isolates were from soil pH 8.01 to 9 (190). The greatest frequency of “N. asteroides complex” were recovered from a desert climate (190), contrary to associations of nocardiosis with humidity (188, 189). Despite the findings that soil characteristic preferences may differ by Nocardia species, this was not borne out in a study of human infections at a local level (128). The evaluation did not find evidence of an association of soil characteristics at case residences or geospatial clustering of case residences regardless of infecting species (128). Host susceptibility and marginal ecological variation at the local level may affect the influence of species soil preference.
Clinical Manifestations
There are 3 main forms of nocardiosis: primary cutaneous, pulmonary, and disseminated infections (6). Less frequent sources or presentations of disease include bacteremia, ocular, and other extra-pulmonary infections (6). Other authors describe additional forms of disease: CNS, extrapulmonary, and mycetoma (163). The most common forms are described below, as well as a few forms of extrapulmonary disease.
Cutaneous disease.
Primary cutaneous nocardiosis is caused by the direct inoculation with soil contaminated with Nocardia spp. (3, 5, 130). Some cases report no trauma, such as innocuous events including direct contact of open cuts with soil when gardening (3, 130) or a thorn prick (132). Traumatic inoculation has been reported, such as injuries from a car accident (131) and through nosocomial exposure (135). N. brasiliensis may account for up to 80% of cutaneous infections (6, 163). Interestingly, one study in Houston found N. farcinica to most commonly cause skin infections, although this was in a severely immunocompromised population (118).
Primary cutaneous infection can disseminate hematogenously to cause systemic, or disseminated, nocardiosis (191). Primary cutaneous infections can present as superficial, lymphocutaneous, subcutaneous/actinomycetoma (159). Meanwhile, it is estimated that 8 to 10% of cutaneous infections are secondary to a primary pulmonary infection that disseminated to the skin (159, 163).
The least severe form, patients with superficial skin infections may have ulcers (192), cellulitis (193), abscesses (194), granulomas (195), as well as pustules, plaques, or papules (129). The superficial form was found to be the most common form of cutaneous nocardiosis in the United States in one literature review of 75 cases (129). The authors also found an average time to diagnosis of 12.7 weeks (range: 0.5 to 52) from 43 patients with superficial cutaneous nocardiosis, and only 32.6% were immunocompromised (129). Only infrequently do superficial infections progress into disseminated infections (196). Differential diagnoses include other pyogenic bacterial infections, including Staphylococci and group A streptococci (3, 130), sporotrichosis (6, 191), tularemia (7, 191), or Erysipelothrix spp. infections (7).
An estimated 1/3 of cutaneous infections progress to the lymphocutaneous form (197), which involves the lymphatic system; it is also called sporotrichoid nocardiosis because of its similarity to sporotrichosis (198). In one series, patients frequently presented with a nodule, as well as local pain, edema, erythema, warmth, and induration (130). Some patients presented with lymphangitis and lymphadenopathy (130), as well as cellulitis; the lesions may progress to subcutaneous abscesses (129, 191). Average time to diagnosis of 26 patients was 20.6 weeks (range: 0.5 to 208) (129), slightly longer than that for superficial infections; 46.2% were immunocompromised (129). Differential diagnoses include sporotrichosis (198) and Mycobacterium marinum infection (7, 199).
Both cutaneous and lymphocutaneous forms often present on the extremities (118, 129, 130, 191), which may point to the ability for trauma to occur on uncovered skin. Specimens for diagnosis include fluid drained from abscesses or lesions for culture, or biopsies of the lesions to identify histopathologic evidence of Nocardia spp. bacilli (129, 130, 191). Co-infections may confuse the diagnosis (3, 199), and contamination of the wound with multiple organisms can outcompete or conflate the causative agent of the infection (3). Finally, a full examination must be done to rule out secondary cutaneous infection due to dissemination (4).
Pulmonary disease.
Primary pulmonary nocardiosis is thought to be caused from inhalation of aerosolized spores or mycelia (4). Pulmonary infection is the most common form of disease in the United States (6, 159), and more often affects patients with structural lung disease (127, 128), immunocompromising conditions (6), or those taking corticosteroids for more than 6 months (127, 128).
Respiratory tract colonization is reported in many reviews of pulmonary infections (106, 119, 143, 200). Some lung diseases, such as cystic fibrosis, may predispose patients to respiratory tract colonization (143). Georghiou et al. found 20% of Nocardia spp. isolates were not associated with clinical symptoms and the patients were assumed to be colonized (197), while Fujita et al. found 40% of immunocompetent patients were colonized (200). If a patient has symptoms or signs of lung infection and a positive Nocardia spp. culture from a respiratory specimen, the result should be assumed to be clinically important (119, 200).
The disease is characterized by an acute onset with inflammatory response, which progresses to granulomatous inflammation and necrotic abscess development (150, 151). Symptoms are usually nonspecific, including cough, dyspnea, and fever (106, 118, 150, 200, 201); pleuritic chest pain has also been reported (201). One study found that symptoms did not differ significantly by immune status (200).
Clinical signs include presence of leukocytosis and elevated C-reactive protein (118, 150, 200). Frequent radiographic findings include the presence of one or more lung nodules, lobar consolidation, and pleural effusion (106, 118, 150, 200); pulmonary infiltrates and necrotizing granulomas may be present (118), and lung findings are frequently bilateral (8). Cavitation is reported more frequently among immunocompromised patients (5, 200).
Time from onset to diagnosis varies widely. An older literature review found the average time to diagnosis was 11.7 weeks (SD: 16.5, range: 2 days to 29 months) while a more recent study found an average of 42 days (SD: 40) (157). Although reasons for the delays were not explained, more rapid diagnostic results and improved clinical recognition may be factors in the differences between periods. When differentiated by immune status, mean delay was 45.8 days (SD: 45) for immunocompetent patients who presented with subacute infections, and 7.4 days (SD: 12) for immunosuppressed patients with acute infections (200). Delays in diagnosis (202) and acute disease (150) are associated with poor outcomes and higher mortality.
The proportion of patients that progress to disseminated infection also varies and is associated with immune status. Dissemination to an extrapulmonary site has been reported in 0 to 4% of immunocompetent patients and 22 to 28% of immunosuppressed (128, 200). When immune status is not differentiated, the proportion ranges from 8.5% to 38% (118, 158, 201).
Symptoms, signs, and radiographic findings are not sufficient for diagnosis of nocardiosis due to their lack of specificity. Differential diagnoses for pulmonary nocardiosis include tuberculosis and non-tuberculosis Mycobacterium infections, various fungal (e.g., Aspergillus spp.) and bacterial infections (e.g., Rhodococcus equi in HIV-positive patients), and malignancy (4, 7, 203).
Early specimen collection, particularly prior to antimicrobial therapy, will improve the ability to recover organism for microbiological or histopathological diagnosis (127). Noninvasive collection methods for respiratory specimens produces good recovery of organism (118, 157); sputum and bronchoalveolar lavage are the most common specimens reported (106, 118, 127, 157, 200).
Disseminated disease.
Disseminated, or systemic, infection is due to the hematogenous spread of the infection to a noncontiguous organ or system (6, 163). It can result from primary cutaneous or pulmonary infection; it can cause infection anywhere in the body, but predominately affects the skin, lungs, and CNS (5, 163). Other relatively common locations of disseminated infection include the kidney (204), joints (205), retina (206), and heart (207).
Dissemination appears to occur more commonly in those with immunosuppression (5, 200). A study of 4 medical centers in Taiwan found only 6% of nocardiosis cases had dissemination (208), and a surveillance study in Spain found 13.5% had disseminated disease (121). When separated by immune status, dissemination was 0 to 9% among immunocompetent and 22 to 27% among immunocompromised patients (5, 200). Transplant recipients are at greatest risk of dissemination (5); 42.7% of patients experienced disseminated disease in a large multi-site study (8). Radiological imaging is important to locate abscesses using computed tomography (CT) or magnetic resonance imaging (MRI) (186).
Central nervous system.
In a study of 1,050 cases, 22.7% of cases (n = 238) had CNS infections, of which 42% were immunocompetent (163). Meanwhile, 44% of disseminated infections had CNS involvement (163). Similarly, 25.6% of transplant recipients had CNS involvement (8). An estimated 38% of all CNS nocardiosis infections are primary infections rather than disseminated infections (163).
Symptoms and signs of CNS infection may include fever, headaches, meningismus (209), seizures (5, 209), and neurologic deficits (5, 8). However, the absence of signs does not exclude CNS involvement; 43.3% of transplant recipients had no neurological signs or symptoms despite presence of CNS infection on imaging (8). Thus, radiological imaging (e.g., CT, MRI) and collection of cerebrospinal fluid is important for any patient with suspect nocardiosis, particularly immunocompromised patients (118, 209). In this population, disease progression may be rapid (163); the abscesses can spread by extending Nocardia spp. filaments (163). However, progression and onset of neurologic signs can take years in immunocompetent patients (148). Differential diagnoses may include malignancies (7, 209), vascular infarction, or other bacterial or fungal infections (7).
Extrapulmonary disease.
Other extrapulmonary forms are reported in the literature, which occur either via dissemination from primary cutaneous or pulmonary infection, or direct inoculation (158).
Ocular infections.
The eye can be affected with either a primary or disseminated infection (163). Corneal lesions or keratitis can result from traumatic inoculation of the eye, eye surgery, steroid use (13), or contamination of contact lenses (13, 210). Retinal involvement is more often associated with disseminated disease (163). Corticosteroids are frequently used as a treatment for bacterial keratitis and corneal ulcers but may actually produce worse outcomes for nocardial infections (14).
Osteomyelitis and septic arthritis.
Nocardial osteomyelitis has been recognized as an unusual presentation since 1963, with the first culture-proven infection of the vertebral column (211). Osteomyelitis has since been described in the vertebra and appendicular skeleton and are predominately disseminated from a primary infection site (211–213). Most primary osteomyelitis cases are described in immunocompetent individuals following traumatic inoculation (194, 214–216), although 2 cases have been reported in patients with HIV (217, 218).
Septic arthritis infected with Nocardia sp. has been reported infrequently, although the first case was reported in the English literature in 1954 (219). Infections predominately affect the knee and are described shortly following total knee replacement (220, 221) or periprosthetic infection of the knee (155, 222). Two reports have described nocardial septic arthritis of the knee joint following surgical repair of the anterior cruciate ligament in immunocompetent patients (133, 134).
Colonization
Nocardia spp. may be isolated from the respiratory tract of patients that are asymptomatic in pulmonary infections (6, 5, 7, 46, 152). In a retrospective study of 102 patient isolates in Australia, 20% of isolates were not considered clinical infections but most likely airway colonization (197). Nocardia spp. can be encountered in patients with underlying structural diseases or functional abnormalities such as bronchiectasis and cystic fibrosis (CF) (6, 5, 7, 46, 152). Some pathogenic bacterial species recovered from the airways of CF patients are indisputably associated with lung infection; however, the clinical relevance of unusual species such as nocardial infection has not been demonstrated conclusively and remains uncertain (223). In a recent study, 6 isolates of Nocardia spp. were the sole pathogen recovered and 7 isolates presented worsening of pulmonary clinical signs (46). Treatment of some of these patients with cotrimoxazole was started. Although the instigation of treatment remains uncertain at first isolation, the potential pathogenicity of Nocardia spp. in CF patients with worsening signs of infection should be considered carefully (46).
Mortality
The mortality rate ranges widely based on the form of disease, the immune status of the patient, and the era of the publication (159). Most reviews focus on all-cause mortality rather than deaths definitively due to nocardiosis. Factors that are associated with mortality include acute disease (108, 150, 158, 200), involvement of 2 or more organs (108, 158, 163), severe immunosuppression (158), and greater disease severity (150).
Primary cutaneous disease without dissemination has the lowest risk of death (4). Most individuals with primary cutaneous nocardiosis recover fully (3, 129, 130), although there have been some cases in which primary cutaneous infection disseminates and results in death (3). Patients with bacteremia have a mortality rate of ~50% (108), and the rate among patients with disseminated infections range from 44% (163) to 85% (158). CNS involvement, whether primary or secondary infection, has a poor prognosis and mortality of almost 50% (6).
Underlying conditions are also associated with higher mortality rates. Patients with malignancies have a mortality rate greater than 60% (111). Transplant recipients reportedly have the worst outcomes compared to other immunocompromised and immunocompetent patients (128). Of 47 transplant patients, fewer than 60% survived at 12 months, while immunocompetent patients had a survival rate of greater than 90% (128). Another study reported a similarly high all-cause mortality rate of 37% (149). Other studies report better survival outcomes of 82% (168) and 6-month survival at 86% (114). Although transplant recipients are severely immunosuppressed and at greater risk of mortality (224), patients with nocardiosis have poorer outcomes compared to other transplant recipients, with a comparative mortality rate of 16.2% versus 1.3% (169). Finally, mortality may have decreased over time. Cases in the literature before 1950 had a mortality rate of 70%, which dropped to 44% between 1950 and 1979, and to 26% between 1980 and 1994 (163).
Treatment
There are no standard recommendations for nocardiosis treatment. Treatment selection and duration must be customized to the patient based on the form and severity of disease, underlying conditions, and antimicrobial susceptibility testing (6, 7). Antimicrobial susceptibility varies by Nocardia species, which were historically divided into groups based on their susceptibility patterns to various antimicrobial classes (40, 225, 226). In vitro and in vivo antimicrobial susceptibilities can be inconsistent (6, 7), and the organism can be fastidious and outgrown by the presence of co-infecting bacteria (159). These factors can make treatment selection challenging. Additionally, there has been some dispute regarding resistance of trimethoprim sulfamethoxazole (TMP-SMX) (2, 147, 198), which is a first line and widely used treatment for nocardiosis (7, 227). A large multi-center study from 6 laboratories in the United States did not find evidence of substantial resistance to TMP-SMX (198).
Often only reference laboratories will perform susceptibility testing, which can further delay appropriate treatment (147). However, if susceptibility testing is not available, the group antimicrobial susceptibility profiles could be used as a rough gauge to determine treatment regimens (9, 225), although some of the groups have inconsistent patterns for some drug classes (6, 225, 228). Empiric multidrug therapy is recommended in patients with severe disease while awaiting susceptibility results, usually consisting of broad spectrum antibiotics including carbapenems (imipenem or meropenem), TMP-SMX, amikacin, linezolid, or parenteral cephalosporins (i.e., ceftriaxone, cefotaxime) (228, 229). Agents with low resistance profiles to Nocardia spp. (228, 230–232) and high bioavailability are preferred, especially agents with good bioavailability in the CNS if CNS involvement is suspected (7, 225, 228). TMP-SMX and linezolid are both orally bioavailable and along with ceftriaxone are effective at treating CNS infections (228, 233); however, adverse events may be common especially at higher doses or in vulnerable patients (114, 234–236).
Once the susceptibility results are known, the final treatment regimen should account for antagonistic effects of antibiotics (e.g., linezolid and aminoglycosides) (236), concomitant medications and potential drug-drug interactions, and adverse events (225, 228, 229). Monotherapy may be sufficient for non-severe cases while multidrug therapy remains recommended for severe pulmonary, disseminated, and CNS infections (228, 229), with an emphasis on highly bactericidal agents with synergistic effects (233, 236).
Extended duration of treatment is recommended to prevent relapse (7), although there is no set recommendation and depends on the patient’s comorbid conditions, speed of clinical and radiographic improvements, and location and severity of disease (229). Immunocompetent patients with uncomplicated infections will likely be cured with shorter courses of antibiotics of 8 weeks or more (229); for example, one study found an average treatment duration among patients with superficial cutaneous infections of 4 months (range 1 to 12 months) (129). Patients with pulmonary infections should receive 6 to 12 months of treatment, and patients with complicated or disseminated disease should receive 9 to 12 months of antimicrobial treatment (7, 227, 229). A suggestion for patients with CNS involvement is to treat with intravenous antimicrobials for 3 to 6 weeks or more, with an additional year or more of oral antimicrobial treatment (229, 237).
Studies administering intravenous bactericidal antimicrobials followed by oral treatment may suggest treatment of <6 months of primarily pulmonary infections could be effective. A subset of 17 solid organ transplant patients that received <6 months of treatment had a median treatment duration of 56 days (range: 24 to 120) with 15 (88.2%) cured at 1 year (234). A study among heart transplant patients found a similar effect of 91.7% cured (11 of 12 patients) when intravenously administering 3 to 4 weeks of bactericidal antimicrobials followed by 1 to 3 months of oral treatment with radiographic evidence of improvement (236).
Antimicrobials alone may be insufficient for many patients. Surgical excision, incision, and drainage or debridement may be required for cutaneous lesions (3, 130, 191, 238), which may be performed serially (130, 238). Skin grafts may also be needed for wound closure, particularly for necrotic lesions (3, 238). When a prosthetic joint is involved, revision, or one-stage replacement of the joint has been performed to remove infected tissue and repair the prosthetic (155, 222).
Challenges to prompt and appropriate treatment include a limited availability of rapid diagnostics to differentiate between Nocardia species, Mycobacterium species, and Actinomyces species; Nocardia spp. can have fastidious growth (4), may be overgrown by other infecting organisms (159), and clinicians may not consider an infectious etiology given similarity to some malignancies (5, 239). Additionally, treatment guidelines have not been developed because of limited reporting on treatment with new drugs, treatment failure and resistance, and lack of surveillance and reporting. Controlled trials are needed to evaluate the effectiveness of quantity, dosing, and duration of antibiotics to improve cure rates and reduce adverse events.
Prevention
Nocardia spp. are ubiquitous in the soil, so there are few techniques to prevent infection (4). Since a majority of primary cutaneous infections reported a traumatic injury to the skin (3, 129), covering skin or open wounds to avoid direct soil contact may prevent cutaneous nocardiosis. Wearing shoes may prevent actinomycetoma by preventing small inoculation injuries to the feet (4); however this has not been systematically examined (240).
TMP-SMX is given to immunocompromised individuals to prevent Pneumocystis infection (5). Some authors have suggested that this use may provide ancillary protection against nocardiosis (108, 142, 241); however, infections concurrent with prophylactic use (5, 168, 170) have led to dispute over its effectiveness for this purpose (169).
Disinfection of a ward following a nosocomial outbreak may be warranted to prevent future infections (124, 140). Antimicrobial treatment of central venous catheters may prevent the introduction of Nocardia spp. to an immunocompromised person via biofilm growth (141). There are no other specific recommendations to prevent Nocardia spp. nosocomial transmission. However, this may need further evaluation to determine the clinical importance when Nocardia spp. are found in health care settings (126).
FUTURE RESEARCH NEEDS
There are several limitations in the existing literature that preclude providing an accurate estimate of nocardiosis incidence, a generalizable description of persons at risk and risk factors, or evaluation of geospatial associations of nocardiosis. Because nocardiosis is rare, prospective cohort studies are usually not feasible, and the logistics and expense of multi-site studies can be challenging. These reasons likely drive the popularity of facility-based retrospective studies using medical chart or record review (5, 114, 118). Medical records could not be evaluated previously on a large scale for nocardiosis, such as using national hospitalization discharge databases (242), because nocardiosis was combined with other etiologic agents in the same code within the International Classification of Diseases 9th revision-clinical modification (ICD-9-CM). Nocardiosis was given a distinct diagnosis code in the 10th revision (ICD-10-CM) (243), which went into effect on October 1, 2015 for all medical billing in the United States.
The use of population-level data, such as electronic medical records, may provide generalizable descriptions of infections and risk factors. However, these data sources will likely limit analyses to patients with more severe infections, to locales with national or regional electronic medical records, and assume that the sensitivity of the clinical diagnosis is high. Additional research is needed to develop more rapid diagnostic assays and treatment recommendations for nocardiosis, since the timeliness of current diagnostics can delay appropriate treatment.
CLINICAL DISEASE: ACTINOMYCETOMA
Mycetoma is a debilitating chronic granulomatous subcutaneous inflammatory disease. Characteristics include large painless tumor-like swellings and the formation of sinuses with discharge that contains grains (187). The general description of the disease is mycetoma, however, the causative agents can be of bacterial or fungal origin and include many different genera. Specific forms of the disease are referred to as eumycetoma (fungal form) or actinomycetoma (bacterial form) (244). N. brasiliensis is reportedly the most frequent nocardial etiology of mycetoma (3, 5); other commonly attributed bacterial genera include Actonomadura and Streptomyces (245).
Treatment of actinomycetoma cases may be improved by an emphasis on species identification; however, there is still substantial need for at least a high-level differentiation of mycetoma between fungal and bacterial etiology to focus treatment. A lack of differentiation between eumycetoma and actinomycetoma can lead to unnecessary amputations when antibiotic treatment can be effective, though amputation may be warranted to remove appendages severely affected with actinomycetoma (3, 246).
Because much of the mycetoma literature is syndrome-focused and outside the narrow scope of this review, we have summarized key references by topic in Table 2. As a relatively newly recognized neglected tropical disease (NTD) that primarily affects poor and marginalized people, (247), international experts have convened to identify priorities to address areas of neglect (248–250). Mycetoma has also been included in the World Health Organization’s 2021 to 2030 road map for neglected tropical diseases (251). We hope these actions will garner support and propel progress on identified research priorities needed to combat mycetoma, including improved diagnostics and access to care by those most affected by mycetoma.
TABLE 2.
Key references for mycetoma
| Topics | First author and title | Reference |
|---|---|---|
| Establish as a neglected tropical disease; priorities and research needs | Fahal AH. Mycetoma: the journey from neglect to recognition as a neglected tropical disease. | 295 |
| Fahal AH. The Khartoum call for action Khartoum, Sudan −2019. | 248 | |
| van de Sande WWJ. Closing the mycetoma knowledge gap. | 249 | |
| Ziljstra EE. Mycetoma: A Long Journey from Neglect. | 250 | |
| Case burden estimate | Emery D. The global distribution of actinomycetoma and eumycetoma. | 296 |
| van de Sande WWJ. Global burden of human mycetoma: a systematic review and meta-analysis. | 297 | |
| Review | Nenoff P. Eumycetoma and actinomycetoma-an update on causative agents, epidemiology, pathogenesis, diagnostics and therapy. | 187 |
| Organizations and collaborations | Davey G. Research capacity building with mycetoma as the central research challenge. | 298 |
| Traxler RM. Development of the Global Mycetoma Working Group. | 299 | |
| Diagnostics | Ahmed AA. Mycetoma laboratory diagnosis: Review article. | 300 |
| Bahar ME. Mycetoma imaging: the best practice. | 301 | |
| Siddig EE. Actinomycetoma laboratory-based diagnosis: a mini-review 2021. | 245 | |
| Interventions and control programs | Bakhiet SM. A holistic approach to the mycetoma management. | 302 |
| Mitjà O. Integrated Control and Management of Neglected Tropical Skin Diseases. | 303 | |
| Roberto E. Mycetoma and the Community Dermatology Program, Mexico. | 304 | |
| Prevention and control goals | Fahal AH. The Khartoum call for action Khartoum, Sudan −2019. | 248 |
| WHO. Skin NTD road map. | 251 |
ACKNOWLEDGMENTS
In memorial to June M. Brown, in recognition of her contributions to the advancement of the microbiological understanding of Actinomycetes and the clinical implications of these infections.
In acknowledgment of Richard Rothenberg and Henry Walke for their critical review and constructive criticism of the literature review.
REFERENCES
- 1.Beaman BL, Burnside J, Edwards B, Causey W. 1976. Nocardial infections in the United States, 1972–1974. J Infect Dis 134:286–289. 10.1093/infdis/134.3.286. [DOI] [PubMed] [Google Scholar]
- 2.Uhde KB, Pathak S, McCullum I, Jr, Jannat-Khah DP, Shadomy SV, Dykewicz CA, Clark TA, Smith TL, Brown JM. 2010. Antimicrobial-resistant Nocardia isolates, United States, 1995–2004. Clin Infect Dis 51:1445–1448. 10.1086/657399. [DOI] [PubMed] [Google Scholar]
- 3.Smego RA, Jr, Gallis HA. 1984. The clinical spectrum of Nocardia brasiliensis infection in the United States. Rev Infect Dis 6:164–180. 10.1093/clinids/6.2.164. [DOI] [PubMed] [Google Scholar]
- 4.McNeil MM, Brown JM. 1994. The medically important aerobic actinomycetes: epidemiology and microbiology. Clin Microbiol Rev 7:357–417. 10.1128/CMR.7.3.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Steinbrink J, Leavens J, Kauffman CA, Miceli MH. 2018. Manifestations and outcomes of Nocardia infections: comparison of immunocompromised and nonimmunocompromised adult patients. Medicine 97:e12436. 10.1097/MD.0000000000012436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown-Elliott BA, Brown JM, Conville PS, Wallace RJ, Jr, 2006. Clinical and laboratory features of the Nocardia spp. based on current molecular taxonomy. Clin Microbiol Rev 19:259–282. 10.1128/CMR.19.2.259-282.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilson JW. 2012. Nocardiosis: updates and clinical overview. Mayo Clin Proc 87:403–407. 10.1016/j.mayocp.2011.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coussement J, Lebeaux D, van Delden C, Guillot H, Freund R, Marbus S, Melica G, Van Wijngaerden E, Douvry B, Van Laecke S, Vuotto F, Tricot L, Fernandez-Ruiz M, Dantal J, Hirzel C, Jais JP, Rodriguez-Nava V, Lortholary O, Jacobs F, European Study Group for Nocardia in Solid Organ T . 2016. Nocardia infection in solid organ transplant recipients: a multicenter European case-control study. Clin Infect Dis 63:338–345. 10.1093/cid/ciw241. [DOI] [PubMed] [Google Scholar]
- 9.Conville PS, Brown-Elliott BA, Smith T, Zelazny AM. 2018. The complexities of Nocardia taxonomy and identification. J Clin Microbiol 56. 10.1128/JCM.01419-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Salinas-Carmona MC, Rosas-Taraco AG, Welsh O. 2012. Systemic increased immune response to Nocardia brasiliensis co-exists with local immunosuppressive microenvironment. Antonie Van Leeuwenhoek 102:473–480. 10.1007/s10482-012-9779-y. [DOI] [PubMed] [Google Scholar]
- 11.Tucker FC, Hirsch EF. 1949. Nocardiosis, with a report of three cases of actinomycosis due to Nocardia asteroides. J Infect Dis 85:72–86. 10.1093/infdis/85.1.72. [DOI] [PubMed] [Google Scholar]
- 12.Lechevalier MP, Lechevalier H. 1970. Chemical Composition as a criterion in the classification of aerobic actinomycetes. Int J Systematic Bacteriology 20:435–443. 10.1099/00207713-20-4-435. [DOI] [Google Scholar]
- 13.Lalitha P. 2009. Nocardia keratitis. Curr Opin Ophthalmol 20:318–323. 10.1097/ICU.0b013e32832c3bcc. [DOI] [PubMed] [Google Scholar]
- 14.Ni N, Srinivasan M, McLeod SD, Acharya NR, Lietman TM, Rose-Nussbaumer J. 2016. Use of adjunctive topical corticosteroids in bacterial keratitis. Curr Opin Ophthalmol 27:353–357. 10.1097/ICU.0000000000000273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ifantides C, Batlle OR, Mushatt D, Ayyala RS. 2015. Nocardia exalbida blebitis: a case report. J Glaucoma 24:e19–e21. 10.1097/IJG.0b013e3182a07574. [DOI] [PubMed] [Google Scholar]
- 16.CLSI. 2018. Susceptibility testing of mycobacteria, Nocardia spp., and other aerobic actinomycetes. 3rd ed Clinical and Laboratory Standards Institute, Wayne, PA. [PubMed] [Google Scholar]
- 17.Kong F, Chen SC, Chen X, Sintchenko V, Halliday C, Cai L, Tong Z, Lee OC, Sorrell TC. 2009. Assignment of reference 5'-end 16S rDNA sequences and species-specific sequence polymorphisms improves species identification of Nocardia. Open Microbiol J 3:97–105. 10.2174/1874285800903010097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ruimy R, Boiron P, Boivin V, Christen R. 1994. A phylogeny of the genus Nocardia deduced from the analysis of small-subunit ribosomal DNA sequences, including transfer of Nocardia amarae to the genus Gordona as Gordona amarae comb. nov. FEMS Microbiol Lett 123:261–267. 10.1111/j.1574-6968.1994.tb07234.x. [DOI] [PubMed] [Google Scholar]
- 19.Chun J, Goodfellow M. 1995. A phylogenetic analysis of the genus Nocardia with 16S rRNA gene sequences. Int J Syst Bacteriol 45:240–245. 10.1099/00207713-45-2-240. [DOI] [PubMed] [Google Scholar]
- 20.Neefs JM, Van de Peer Y, De Rijk P, Chapelle S, De Wachter R. 1993. Compilation of small ribosomal subunit RNA structures. Nucleic Acids Res 21:3025–3049. 10.1093/nar/21.13.3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roth A, Andrees S, Kroppenstedt RM, Harmsen D, Mauch H. 2003. Phylogeny of the genus Nocardia based on reassessed 16S rRNA gene sequences reveals underspeciation and division of strains classified as Nocardia asteroides into three established species and two unnamed taxons. J Clin Microbiol 41:851–856. 10.1128/JCM.41.2.851-856.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yassin AF, Rainey FA, Burghardt J, Brzezinka H, Mauch M, Schaal KP. 2000. Nocardia paucivorans sp. nov. Int J Syst Evol Microbiol 50 Pt 2:803–809. 10.1099/00207713-50-2-803. [DOI] [PubMed] [Google Scholar]
- 23.Wallace RJ, Jr, Steele LC, Sumter G, Smith JM. 1988. Antimicrobial susceptibility patterns of Nocardia asteroides. Antimicrob Agents Chemother 32:1776–1779. 10.1128/AAC.32.12.1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cloud JL, Conville PS, Croft A, Harmsen D, Witebsky FG, Carroll KC. 2004. Evaluation of partial 16S ribosomal DNA sequencing for identification of Nocardia species by using the MicroSeq 500 system with an expanded database. J Clin Microbiol 42:578–584. 10.1128/JCM.42.2.578-584.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patel JB, Wallace RJ, Jr, Brown-Elliott BA, Taylor T, Imperatrice C, Leonard DG, Wilson RW, Mann L, Jost KC, Nachamkin I. 2004. Sequence-based identification of aerobic actinomycetes. J Clin Microbiol 42:2530–2540. 10.1128/JCM.42.6.2530-2540.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Helal M, Kong F, Chen SC, Bain M, Christen R, Sintchenko V. 2011. Defining reference sequences for Nocardia species by similarity and clustering analyses of 16S rRNA gene sequence data. PLoS One 6:e19517. 10.1371/journal.pone.0019517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mellmann A, Cloud JL, Andrees S, Blackwood K, Carroll KC, Kabani A, Roth A, Harmsen D. 2003. Evaluation of RIDOM, MicroSeq, and Genbank services in the molecular identification of Nocardia species. Int J Med Microbiol 293:359–370. 10.1078/1438-4221-00271. [DOI] [PubMed] [Google Scholar]
- 28.Conville PS, Brown JM, Steigerwalt AG, Brown-Elliott BA, Witebsky FG. 2008. Nocardia wallacei sp. nov. and Nocardia blacklockiae sp. nov., human pathogens and members of the “Nocardia transvalensis Complex. J Clin Microbiol 46:1178–1184. 10.1128/JCM.02011-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ishikawa J, Yamashita A, Mikami Y, Hoshino Y, Kurita H, Hotta K, Shiba T, Hattori M. 2004. The complete genomic sequence of Nocardia farcinica IFM 10152. Proc Natl Acad Sci USA 101:14925–14930. 10.1073/pnas.0406410101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Conville PS, Witebsky FG. 2005. Multiple copies of the 16S rRNA gene in Nocardia nova isolates and implications for sequence-based identification procedures. J Clin Microbiol 43:2881–2885. 10.1128/JCM.43.6.2881-2885.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Conville PS, Witebsky FG. 2007. Analysis of multiple differing copies of the 16S rRNA gene in five clinical isolates and three type strains of Nocardia species and implications for species assignment. J Clin Microbiol 45:1146–1151. 10.1128/JCM.02482-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luo Q, Hiessl S, Poehlein A, Daniel R, Steinbuchel A. 2014. Insights into the microbial degradation of rubber and gutta-percha by analysis of the complete genome of Nocardia nova SH22a. Appl Environ Microbiol 80:3895–3907. 10.1128/AEM.00473-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vera-Cabrera L, Ortiz-Lopez R, Elizondo-Gonzalez R, Ocampo-Candiani J. 2013. Complete genome sequence analysis of Nocardia brasiliensis HUJEG-1 reveals a saprobic lifestyle and the genes needed for human pathogenesis. PLoS One 8:e65425. 10.1371/journal.pone.0065425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yasuike M, Nishiki I, Iwasaki Y, Nakamura Y, Fujiwara A, Shimahara Y, Kamaishi T, Yoshida T, Nagai S, Kobayashi T, Katoh M. 2017. Analysis of the complete genome sequence of Nocardia seriolae UTF1, the causative agent of fish nocardiosis: the first reference genome sequence of the fish pathogenic Nocardia species. PLoS One 12:e0173198. 10.1371/journal.pone.0173198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wolff TY, Eickhardt S, Bjornsdottir MK, Moser C, Bjarnsholt T, Hoiby N, Thomsen TR. 2013. Direct sequencing and RipSeq interpretation as a tool for identification of polymicrobial infections. J Clin Microbiol 51:1281–1284. 10.1128/JCM.00190-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.CLSI. 2018. Interpretive Criteria for Identification of Bacteria and Fungi by Targeted DNA Sequencing, 2nd ed Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 37.Conville PS, Murray PR, Zelazny AM. 2010. Evaluation of the integrated database network system (IDNS) SmartGene software for analysis of 16S rRNA gene sequences for identification of Nocardia species. J Clin Microbiol 48:2995–2998. 10.1128/JCM.00681-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Conville PS, Brown JM, Steigerwalt AG, Lee JW, Anderson VL, Fishbain JT, Holland SM, Witebsky FG. 2004. Nocardia kruczakiae sp. nov., a pathogen in immunocompromised patients and a member of the “N. nova complex. J Clin Microbiol 42:5139–5145. 10.1128/JCM.42.11.5139-5145.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kageyama A, Yazawa K, Ishikawa J, Hotta K, Nishimura K, Kageyama A. 2003. Nocardial infections in Japan from 1992 to 2001, including the first report of infection by Nocardia transvalensis. Eur J Epidemiol 19:383–389. 10.1023/B:EJEP.0000024706.02325.c0. [DOI] [PubMed] [Google Scholar]
- 40.Conville PS, Witebsky FG. 2007. Organisms designated as Nocardia asteroides drug pattern type VI are members of the species Nocardia cyriacigeorgica. J Clin Microbiol 45:2257–2259. 10.1128/JCM.00133-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kong F, Wang H, Zhang E, Sintchenko V, Xiao M, Sorrell TC, Chen X, Chen SC. 2010. secA1 gene sequence polymorphisms for species identification of Nocardia species and recognition of intraspecies genetic diversity. J Clin Microbiol 48:3928–3934. 10.1128/JCM.01113-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carrasco G, Valdezate S, Garrido N, Medina-Pascual MJ, Villalon P, Saez-Nieto JA. 2015. gyrB analysis as a tool for identifying Nocardia species and exploring their phylogeny. J Clin Microbiol 53:997–1001. 10.1128/JCM.03072-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carrasco G, Valdezate S, Garrido N, Villalon P, Medina-Pascual MJ, Saez-Nieto JA. 2013. Identification, typing, and phylogenetic relationships of the main clinical Nocardia species in spain according to their gyrB and rpoB genes. J Clin Microbiol 51:3602–3608. 10.1128/JCM.00515-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takeda K, Kang Y, Yazawa K, Gonoi T, Mikami Y. 2010. Phylogenetic studies of Nocardia species based on gyrB gene analyses. J Med Microbiol 59:165–171. 10.1099/jmm.0.011346-0. [DOI] [PubMed] [Google Scholar]
- 45.Tamura T, Matsuzawa T, Oji S, Ichikawa N, Hosoyama A, Katsumata H, Yamazoe A, Hamada M, Suzuki K, Gonoi T, Fujita N. 2012. A genome sequence-based approach to taxonomy of the genus Nocardia. Antonie Van Leeuwenhoek 102:481–491. 10.1007/s10482-012-9780-5. [DOI] [PubMed] [Google Scholar]
- 46.Rodriguez-Nava V, Durupt S, Chyderiotis S, Freydiere AM, Karsenty J, de Montclos M, Reix P, Durieu I, Nove-Josserand R, Chiron R, Bremont F, Tetu L, Murris M, Terru D, Godreuil S, Bergeron E, Freney J, Boiron P, Vandenesch F, Marchandin H, Segonds C, Doleans-Jordheim A. 2015. A French multicentric study and review of pulmonary Nocardia spp. in cystic fibrosis patients. Med Microbiol Immunol 204:493–504. 10.1007/s00430-014-0360-3. [DOI] [PubMed] [Google Scholar]
- 47.Rudramurthy SM, Honnavar P, Kaur H, Samanta P, Ray P, Ghosh A, Chakrabarti A. 2015. Molecular identification of clinical Nocardia isolates from India. J Med Microbiol 64:1216–1225. 10.1099/jmm.0.000143. [DOI] [PubMed] [Google Scholar]
- 48.Yin X, Liang S, Sun X, Luo S, Wang Z, Li R. 2007. Ocular nocardiosis: HSP65 gene sequencing for species identification of Nocardia spp. Am J Ophthalmol 144:570–573. 10.1016/j.ajo.2007.06.031. [DOI] [PubMed] [Google Scholar]
- 49.Maiden MC, Bygraves JA, Feil E, Morelli G, Russell JE, Urwin R, Zhang Q, Zhou J, Zurth K, Caugant DA, Feavers IM, Achtman M, Spratt BG. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci USA 95:3140–3145. 10.1073/pnas.95.6.3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McTaggart LR, Richardson SE, Witkowska M, Zhang SX. 2010. Phylogeny and identification of Nocardia species on the basis of multilocus sequence analysis. J Clin Microbiol 48:4525–4533. 10.1128/JCM.00883-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Du P, Hou X, Xie Y, Xu S, Li L, Zhang J, Wan K, Lou Y, Li Z. 2016. Genotyping of Nocardia farcinica with multilocus sequence typing. Eur J Clin Microbiol Infect Dis 35:771–778. 10.1007/s10096-016-2596-x. [DOI] [PubMed] [Google Scholar]
- 52.Baio PV, Ramos JN, dos Santos LS, Soriano MF, Ladeira EM, Souza MC, Camello TC, Ribeiro MG, Hirata Junior R, Vieira VV, Mattos-Guaraldi AL. 2013. Molecular identification of Nocardia isolates from clinical samples and an overview of human nocardiosis in Brazil. PLoS Negl Trop Dis 7:e2573. 10.1371/journal.pntd.0002573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Carrasco G, de Dios Caballero J, Garrido N, Valdezate S, Canton R, Saez-Nieto JA. 2016. Shortcomings of the commercial MALDI-TOF MS Database and Use of MLSA as an arbiter in the identification of Nocardia species. Front Microbiol 7:542. 10.3389/fmicb.2016.00542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xiao M, Pang L, Chen SC, Fan X, Zhang L, Li HX, Hou X, Cheng JW, Kong F, Zhao YP, Xu YC. 2016. Accurate identification of common pathogenic Nocardia species: evaluation of a multilocus sequence analysis platform and matrix-assisted laser desorption ionization-time of flight mass spectrometry. PLoS One 11:e0147487. 10.1371/journal.pone.0147487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Komaki H, Ichikawa N, Hosoyama A, Takahashi-Nakaguchi A, Matsuzawa T, Suzuki K, Fujita N, Gonoi T. 2014. Genome based analysis of type-I polyketide synthase and nonribosomal peptide synthetase gene clusters in seven strains of five representative Nocardia species. BMC Genomics 15:323. 10.1186/1471-2164-15-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zoropogui A, Pujic P, Normand P, Barbe V, Belli P, Graindorge A, Roche D, Vallenet D, Mangenot S, Boiron P, Rodriguez-Nava V, Ribun S, Richard Y, Cournoyer B, Blaha D. 2013. The Nocardia cyriacigeorgica GUH-2 genome shows ongoing adaptation of an environmental Actinobacteria to a pathogen's lifestyle. BMC Genomics 14:286. 10.1186/1471-2164-14-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hoshino Y, Chiba K, Ishino K, Fukai T, Igarashi Y, Yazawa K, Mikami Y, Ishikawa J. 2011. Identification of nocobactin NA biosynthetic gene clusters in Nocardia farcinica. J Bacteriol 193:441–448. 10.1128/JB.00897-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Studholme DJ. 2016. Genome update. Let the consumer beware: Streptomyces genome sequence quality. Microb Biotechnol 9:3–7. 10.1111/1751-7915.12344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yao W, Yang Y, Chiao J. 1994. IS204: an insertion sequence from Nocardia asteroides (mexicana) YP21. Plasmid 32:262–269. 10.1006/plas.1994.1065. [DOI] [PubMed] [Google Scholar]
- 60.Chiba K, Hoshino Y, Ishino K, Kogure T, Mikami Y, Uehara Y, Ishikawa J. 2007. Construction of a pair of practical Nocardia-Escherichia coli shuttle vectors. Jpn J Infect Dis 60:45–47. [PubMed] [Google Scholar]
- 61.Kumar CV, Coque JJ, Martin JF. 1994. Efficient transformation of the Cephamycin C producer Nocardia lactamdurans and development of shuttle and promoter-probe cloning vectors. Appl Environ Microbiol 60:4086–4093. 10.1128/aem.60.11.4086-4093.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kelly WL, Townsend CA. 2005. Mutational analysis of nocK and nocL in the nocardicin a producer Nocardia uniformis. J Bacteriol 187:739–746. 10.1128/JB.187.2.739-746.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Luo Q, Hiessl S, Poehlein A, Steinbuchel A. 2013. Microbial gutta-percha degradation shares common steps with rubber degradation by Nocardia nova SH22a. Appl Environ Microbiol 79:1140–1149. 10.1128/AEM.03016-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kogure T, Shimada R, Ishikawa J, Yazawa K, Brown JM, Mikami Y, Gonoi T. 2010. Homozygous triplicate mutations in three 16S rRNA genes responsible for high-level aminoglycoside resistance in Nocardia farcinica clinical isolates from a Canada-wide bovine mastitis epizootic. Antimicrob Agents Chemother 54:2385–2390. 10.1128/AAC.00021-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ishikawa J, Chiba K, Kurita H, Satoh H. 2006. Contribution of rpoB2 RNA polymerase beta subunit gene to rifampin resistance in Nocardia species. Antimicrob Agents Chemother 50:1342–1346. 10.1128/AAC.50.4.1342-1346.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Maharjan S, Aryal N, Bhattarai S, Koju D, Lamichhane J, Sohng JK. 2012. Biosynthesis of the nargenicin A1 pyrrole moiety from Nocardia sp. CS682. Appl Microbiol Biotechnol 93:687–696. 10.1007/s00253-011-3567-x. [DOI] [PubMed] [Google Scholar]
- 67.Maharjan S, Koju D, Lee HC, Yoo JC, Sohng JK. 2012. Metabolic engineering of Nocardia sp. CS682 for enhanced production of nargenicin A(1). Appl Biochem Biotechnol 166:805–817. 10.1007/s12010-011-9470-1. [DOI] [PubMed] [Google Scholar]
- 68.Gao B, Paramanathan R, Gupta RS. 2006. Signature proteins that are distinctive characteristics of Actinobacteria and their subgroups. Antonie Van Leeuwenhoek 90:69–91. 10.1007/s10482-006-9061-2. [DOI] [PubMed] [Google Scholar]
- 69.Koenig S, Mehlich AM, Ackermann D, Hamid ME, Saeed NS, Mukhtar M, El Hassan M. 2008. Homologous housekeeping proteins in Nocardia–the NoDaMS proteomic database. Front Biosci 13:842–855. 10.2741/2725. [DOI] [PubMed] [Google Scholar]
- 70.Martins M, Pluvinage B, Li de la Sierra-Gallay I, Barbault F, Dairou J, Dupret JM, Rodrigues-Lima F. 2008. Functional and structural characterization of the arylamine N-acetyltransferase from the opportunistic pathogen Nocardia farcinica. J Mol Biol 383:549–560. 10.1016/j.jmb.2008.08.035. [DOI] [PubMed] [Google Scholar]
- 71.Wu G, Nie L, Zhang W. 2006. Predicted highly expressed genes in Nocardia farcinica and the implication for its primary metabolism and nocardial virulence. Antonie Van Leeuwenhoek 89:135–146. 10.1007/s10482-005-9016-z. [DOI] [PubMed] [Google Scholar]
- 72.Forero Morales MP, Hopprich R, Wise R, Shephard L, Weldhagen GF. 2018. Cost-effective implementation of a custom MALDI-TOF library for the identification of South Australian Nocardia isolates. Pathology 50:753–757. 10.1016/j.pathol.2018.08.006. [DOI] [PubMed] [Google Scholar]
- 73.Jamal W, Albert MJ, Rotimi VO. 2014. Real-time comparative evaluation of bioMerieux VITEK MS versus Bruker Microflex MS, two matrix-assisted laser desorption-ionization time-of-flight mass spectrometry systems, for identification of clinically significant bacteria. BMC Microbiol 14:289. 10.1186/s12866-014-0289-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Buckwalter SP, Olson SL, Connelly BJ, Lucas BC, Rodning AA, Walchak RC, Deml SM, Wohlfiel SL, Wengenack NL. 2016. Evaluation of matrix-assisted laser desorption ionization-time of flight mass spectrometry for identification of Mycobacterium species, Nocardia species, and other aerobic Actinomycetes. J Clin Microbiol 54:376–384. 10.1128/JCM.02128-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ferrand J, Bonnet I, Alauzet C, Lozniewski A. 2018. Evaluation of the Vitek MS and the MALDI Biotyper systems for the identification of less commonly isolated but clinically relevant anaerobes and facultative anaerobes. Anaerobe 54:210–216. 10.1016/j.anaerobe.2018.05.014. [DOI] [PubMed] [Google Scholar]
- 76.Porte L, Garcia P, Braun S, Ulloa MT, Lafourcade M, Montana A, Miranda C, Acosta-Jamett G, Weitzel T. 2017. Head-to-head comparison of Microflex LT and Vitek MS systems for routine identification of microorganisms by MALDI-TOF mass spectrometry in Chile. PLoS One 12:e0177929. 10.1371/journal.pone.0177929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wilen CB, McMullen AR, Burnham CA. 2015. Comparison of sample preparation methods, instrumentation platforms, and contemporary commercial databases for identification of clinically relevant Mycobacteria by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol 53:2308–2315. 10.1128/JCM.00567-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Marvin LF, Roberts MA, Fay LB. 2003. Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry in clinical chemistry. Clin Chim Acta 337:11–21. 10.1016/j.cccn.2003.08.008. [DOI] [PubMed] [Google Scholar]
- 79.Wieser A, Schneider L, Jung J, Schubert S. 2012. MALDI-TOF MS in microbiological diagnostics-identification of microorganisms and beyond (mini review). Appl Microbiol Biotechnol 93:965–974. 10.1007/s00253-011-3783-4. [DOI] [PubMed] [Google Scholar]
- 80.Blosser SJ, Drake SK, Andrasko JL, Henderson CM, Kamboj K, Antonara S, Mijares L, Conville P, Frank KM, Harrington SM, Balada-Llasat JM, Zelazny AM. 2016. Multicenter matrix-assisted laser desorption ionization-time of flight mass spectrometry study for identification of clinically relevant Nocardia spp. J Clin Microbiol 54:1251–1258. 10.1128/JCM.02942-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Khot PD, Bird BA, Durrant RJ, Fisher MA. 2015. Identification of Nocardia species by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol 53:3366–3369. 10.1128/JCM.00780-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Totty H, Miller E, Moreno E, Dunne WM, Jr, Deol P. 2016. Comparison of mechanical disruption techniques for rapid inactivation of Mycobacterium and Nocardia species before identification using matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry. J Clin Microbiol 54:2626–2627. 10.1128/JCM.01096-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Girard V, Mailler S, Polsinelli S, Jacob D, Saccomani MC, Celliere B, Monnin V, van Belkum A, Hagen F, Meis JF, Durand G. 2017. Routine identification of Nocardia species by MALDI-TOF mass spectrometry. Diagn Microbiol Infect Dis 87:7–10. 10.1016/j.diagmicrobio.2016.09.024. [DOI] [PubMed] [Google Scholar]
- 84.Verroken A, Janssens M, Berhin C, Bogaerts P, Huang TD, Wauters G, Glupczynski Y. 2010. Evaluation of matrix-assisted laser desorption ionization-time of flight mass spectrometry for identification of Nocardia species. J Clin Microbiol 48:4015–4021. 10.1128/JCM.01234-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Durand T, Vautrin F, Bergeron E, Girard V, Polsinelli S, Monnin V, Durand G, Dauwalder O, Dumitrescu O, Laurent F, Rodriguez-Nava V. 2020. Assessment of VITEK(R) MS IVD database V3.0 for identification of Nocardia spp. using two culture media and comparing direct smear and protein extraction procedures. Eur J Clin Microbiol Infect Dis 39:559–567. 10.1007/s10096-019-03758-x. [DOI] [PubMed] [Google Scholar]
- 86.Hasçelik G, Kostrzewa M, Müştak HK, Uner C, Diker KS. 2019. Evaluation of matrix-assisted laser desorption ionization-time of flight mass spectrometry in diagnosis of clinical Nocardia species. J Laboratory Medicine 43:157–162. 10.1515/labmed-2018-0180. [DOI] [Google Scholar]
- 87.Marin M, Ruiz A, Iglesias C, Quiroga L, Cercenado E, Martin-Rabadan P, Bouza E, Rodriguez-Sanchez B. 2018. Identification of Nocardia species from clinical isolates using MALDI-TOF mass spectrometry. Clin Microbiol Infect 24:1342.e5–1342.e8. 10.1016/j.cmi.2018.06.014. [DOI] [PubMed] [Google Scholar]
- 88.Body BA, Beard MA, Slechta ES, Hanson KE, Barker AP, Babady NE, McMillen T, Tang YW, Brown-Elliott BA, Iakhiaeva E, Vasireddy R, Vasireddy S, Smith T, Wallace RJ, Jr, Turner S, Curtis L, Butler-Wu S, Rychert J. 2018. Evaluation of the Vitek MS v3.0 matrix-assisted laser desorption ionization-time of flight mass spectrometry system for identification of mycobacterium and Nocardia Species. J Clin Microbiol 56. 10.1128/JCM.00237-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.McTaggart LR, Chen Y, Poopalarajah R, Kus JV. 2018. Incubation time and culture media impact success of identification of Nocardia spp. by MALDI-ToF mass spectrometry. Diagn Microbiol Infect Dis 92:270–274. 10.1016/j.diagmicrobio.2018.06.016. [DOI] [PubMed] [Google Scholar]
- 90.Girard V, Mailler S, Welker M, Arsac M, Celliere B, Cotte-Pattat PJ, Chatellier S, Durand G, Beni AM, Schrenzel J, Miller E, Dussoulier R, Dunne WM, Jr, Butler-Wu S, Saubolle MA, Sussland D, Bell M, van Belkum A, Deol P. 2016. Identification of Mycobacterium spp. and Nocardia spp. from solid and liquid cultures by matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS). Diagn Microbiol Infect Dis 86:277–283. 10.1016/j.diagmicrobio.2016.07.027. [DOI] [PubMed] [Google Scholar]
- 91.Toyokawa M, Ohana N, Ueda A, Imai M, Tanno D, Honda M, Takano Y, Ohashi K, Saito K, Shimura H. 2021. Identification and antimicrobial susceptibility profiles of Nocardia species clinically isolated in Japan. Sci Rep 11:16742. 10.1038/s41598-021-95870-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yarbrough ML, Lainhart W, Burnham CD. 2017. Identification of Nocardia, Streptomyces, and Tsukamurella using MALDI-TOF MS with the Bruker Biotyper. Diagn Microbiol Infect Dis 89:92–97. 10.1016/j.diagmicrobio.2017.06.019. [DOI] [PubMed] [Google Scholar]
- 93.Lasker BA, Bell M, Klenk HP, Schumann P, Brown JM. 2015. Nocardia arizonensis sp. nov., obtained from human respiratory specimens. Antonie Van Leeuwenhoek 108:1129–1137. 10.1007/s10482-015-0566-4. [DOI] [PubMed] [Google Scholar]
- 94.Lasker BA, Bell M, Klenk HP, Sproer C, Schumann C, Schumann P, Brown JM. 2014. Nocardia vulneris sp. nov., isolated from wounds of human patients in North America. Antonie Van Leeuwenhoek 106:543–553. 10.1007/s10482-014-0226-0. [DOI] [PubMed] [Google Scholar]
- 95.Yonetani S, Ohnishi H. 2022. A case of multiple abscesses caused by Nocardia farcinica rapidly diagnosed by MALDI-TOF MS. IDCases 28:e01497. 10.1016/j.idcr.2022.e01497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yoon JS, So H, Joo B, Park J, Jeong IS, Lee GJ, Park J. 2022. A canine case of Nocardia africana infection detected by matrix-assisted laser desorption ionization-time-of-flight mass spectrometry. Vet Sci 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Saffert RT, Cunningham SA, Ihde SM, Jobe KE, Mandrekar J, Patel R. 2011. Comparison of Bruker Biotyper matrix-assisted laser desorption ionization-time of flight mass spectrometer to BD Phoenix automated microbiology system for identification of gram-negative bacilli. J Clin Microbiol 49:887–892. 10.1128/JCM.01890-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sagar R, Challa S. 2017. Pathology and diagnosis of Nocardiosis. JSM Foot and Ankle 2. [Google Scholar]
- 99.Tatti KM, Shieh WJ, Phillips S, Augenbraun M, Rao C, Zaki SR. 2006. Molecular diagnosis of Nocardia farcinica from a cerebral abscess. Hum Pathol 37:1117–1121. 10.1016/j.humpath.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 100.Macnaughton SJ, O'Donnell AG, Embley TM. 1994. Permeabilization of mycolic-acid-containing actinomycetes for in situ hybridization with fluorescently labelled oligonucleotide probes. Microbiology 140:2859–2865. 10.1099/00221287-140-10-2859. [DOI] [PubMed] [Google Scholar]
- 101.Carr EL, Eales K, Soddell J, Seviour RJ. 2005. Improved permeabilization protocols for fluorescence in situ hybridization (FISH) of mycolic-acid-containing bacteria found in foams. J Microbiol Methods 61:47–54. 10.1016/j.mimet.2004.10.023. [DOI] [PubMed] [Google Scholar]
- 102.Isotalo PA, Qian X, Hayden RT, Roberts GD, Lloyd RV. 2009. In situ hybridization for the differentiation of Actinomyces and Nocardia in tissue sections. Diagn Mol Pathol 18:183–188. 10.1097/PDM.0b013e31818b3716. [DOI] [PubMed] [Google Scholar]
- 103.Couble A, Rodriguez-Nava V, de Montclos MP, Boiron P, Laurent F. 2005. Direct detection of Nocardia spp. in clinical samples by a rapid molecular method. J Clin Microbiol 43:1921–1924. 10.1128/JCM.43.4.1921-1924.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Brown JM, Pham KN, McNeil MM, Lasker BA. 2004. Rapid identification of Nocardia farcinica clinical isolates by a PCR assay targeting a 314-base-pair species-specific DNA fragment. J Clin Microbiol 42:3655–3660. 10.1128/JCM.42.8.3655-3660.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Conville PS, Brown-Elliott BA, Wallace RJ, Jr, Witebsky FG, Koziol D, Hall GS, Killian SB, Knapp CC, Warshauer D, Van T, Wengenack NL, Deml S, Woods GL. 2012. Multisite reproducibility of the broth microdilution method for susceptibility testing of Nocardia species. J Clin Microbiol 50:1270–1280. 10.1128/JCM.00994-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kurahara Y, Tachibana K, Tsuyuguchi K, Akira M, Suzuki K, Hayashi S. 2014. Pulmonary nocardiosis: a clinical analysis of 59 cases. Respir Invest 52:160–166. 10.1016/j.resinv.2013.09.004. [DOI] [PubMed] [Google Scholar]
- 107.Arduino RC, Johnson PC, Miranda AG. 1993. Nocardiosis in renal transplant recipients undergoing immunosuppression with cyclosporine. Clin Infect Dis 16:505–512. 10.1093/clind/16.4.505. [DOI] [PubMed] [Google Scholar]
- 108.Kontoyiannis DP, Ruoff K, Hooper DC. 1998. Nocardia bacteremia. Report of 4 cases and review of the literature. Medicine (Baltimore, MD)) 77:255–267. 10.1097/00005792-199807000-00004. [DOI] [PubMed] [Google Scholar]
- 109.Uttamchandani RB, Daikos GL, Reyes RR, Fischl MA, Dickinson GM, Yamaguchi E, Kramer MR. 1994. Nocardiosis in 30 patients with advanced human immunodeficiency virus infection: clinical features and outcome. Clin Infect Dis 18:348–353. 10.1093/clinids/18.3.348. [DOI] [PubMed] [Google Scholar]
- 110.Berkey P, Bodey GP. 1989. Nocardial infection in patients with neoplastic disease. Rev Infect Dis 11:407–412. 10.1093/clinids/11.3.407. [DOI] [PubMed] [Google Scholar]
- 111.Torres HA, Reddy BT, Raad II, Tarrand J, Bodey GP, Hanna HA, Rolston KV, Kontoyiannis DP. 2002. Nocardiosis in cancer patients. Medicine (Baltimore, MD) 81:388–397. 10.1097/00005792-200209000-00004. [DOI] [PubMed] [Google Scholar]
- 112.Lebeaux D, Bergeron E, Berthet J, Djadi-Prat J, Mouniee D, Boiron P, Lortholary O, Rodriguez-Nava V. 2018. Antibiotic susceptibility testing and species identification of Nocardia isolates: a retrospective analysis of data from a French expert laboratory, 2010–2015. Clin Microbiol Infect 25:489–495. 10.1016/j.cmi.2018.06.013]. [DOI] [PubMed] [Google Scholar]
- 113.Hofflin JM, Potasman I, Baldwin JC, Oyer PE, Stinson EB, Remington JS. 1987. Infectious complications in heart transplant recipients receiving cyclosporine and corticosteroids. Ann Intern Med 106:209–216. 10.7326/0003-4819-106-2-209. [DOI] [PubMed] [Google Scholar]
- 114.Peleg AY, Husain S, Qureshi ZA, Silveira FP, Sarumi M, Shutt KA, Kwak EJ, Paterson DL. 2007. Risk factors, clinical characteristics, and outcome of Nocardia infection in organ transplant recipients: a matched case-control study. Clin Infect Dis 44:1307–1314. 10.1086/514340. [DOI] [PubMed] [Google Scholar]
- 115.Simpson GL, Stinson EB, Egger MJ, Remington JS. 1981. Nocardial infections in the immunocompromised host: a detailed study in a defined population. Rev Infect Dis 3:492–507. 10.1093/clinids/3.3.492. [DOI] [PubMed] [Google Scholar]
- 116.Kim J, Minamoto GY, Grieco MH. 1991. Nocardial infection as a complication of AIDS: report of six cases and review. Rev Infect Dis 13:624–629. 10.1093/clinids/13.4.624. [DOI] [PubMed] [Google Scholar]
- 117.Lucas SB, Hounnou A, Peacock C, Beaumel A, Kadio A, De Cock KM. 1994. Nocardiosis in HIV-positive patients: an autopsy study in West Africa. Tuber Lung Dis 75:301–307. 10.1016/0962-8479(94)90137-6. [DOI] [PubMed] [Google Scholar]
- 118.Wang HL, Seo YH, LaSala PR, Tarrand JJ, Han XY. 2014. Nocardiosis in 132 patients with cancer: microbiological and clinical analyses. Am J Clin Pathol 142:513–523. 10.1309/AJCPW84AFTUWMHYU. [DOI] [PubMed] [Google Scholar]
- 119.Rosett W, Hodges GR. 1978. Recent experiences with nocardial infections. Am J Med Sci 276:279–285. 10.1097/00000441-197811000-00004. [DOI] [PubMed] [Google Scholar]
- 120.Pintado V, Gomez-Mampaso E, Fortun J, Meseguer MA, Cobo J, Navas E, Quereda C, Martin-Davila P, Moreno S. 2002. Infection with Nocardia species: clinical spectrum of disease and species distribution in Madrid, Spain, 1978-2001. Infection 30:338–340. 10.1007/s15010-002-2127-9. [DOI] [PubMed] [Google Scholar]
- 121.Minero MV, Marin M, Cercenado E, Rabadan PM, Bouza E, Munoz P. 2009. Nocardiosis at the turn of the century. Medicine (Baltimore, MD) 88:250–261. 10.1097/MD.0b013e3181afa1c8. [DOI] [PubMed] [Google Scholar]
- 122.Goodfellow M, Williams ST. 1983. Ecology of actinomycetes. Annu Rev Microbiol 37:189–216. 10.1146/annurev.mi.37.100183.001201. [DOI] [PubMed] [Google Scholar]
- 123.Orchard VA, Goodfellow M. 1980. Numerical classification of some named strains of Nocardia asteroides and related isolates from soil. J Gen Microbiol 118:295–312. 10.1099/00221287-118-2-295. [DOI] [PubMed] [Google Scholar]
- 124.Houang ET, Lovett IS, Thompson FD, Harrison AR, Joekes AM, Goodfellow M. 1980. Nocardia asteroides infection–a transmissible disease. J Hosp Infect 1:31–40. 10.1016/0195-6701(80)90029-8. [DOI] [PubMed] [Google Scholar]
- 125.Predicala BZ, Urban JE, Maghirang RG, Jerez SB, Goodband RD. 2002. Assessment of bioaerosols in swine barns by filtration and impaction. Curr Microbiol 44:136–140. 10.1007/s00284-001-0064-y. [DOI] [PubMed] [Google Scholar]
- 126.Rahdar HA, Azadi D, Shojaei H, Daei-Naser A. 2017. Molecular analysis and species diversity of Nocardia in the hospital environment in a developing country, a potential health hazard. J Med Microbiol 66:334–341. 10.1099/jmm.0.000436. [DOI] [PubMed] [Google Scholar]
- 127.Maggiorelli C, Di Pierro I, Manta C, Maccari U, Galanti I, Scala R. 2015. Nocardia and lungs in COPD: beyond immuno-deficiencies. COPD 12:315–319. 10.3109/15412555.2014.933951. [DOI] [PubMed] [Google Scholar]
- 128.Woodworth MH, Saullo JL, Lantos PM, Cox GM, Stout JE. 2017. Increasing Nocardia incidence associated with bronchiectasis at a tertiary care center. Ann Am Thorac Soc 14:347–354. 10.1513/AnnalsATS.201611-907OC. [DOI] [PubMed] [Google Scholar]
- 129.Parvu M, Schleiter G, Stratidis JG. 2012. Skin Infections caused by Nocardia species a case report and review of the literature of primary cutaneous nocardiosis reported in the United States. Infect Dis Clin Pract 20:237–241. 10.1097/IPC.0b013e31824fc95e. [DOI] [Google Scholar]
- 130.Tarchini G, Ross FS. 2013. Primary lymphocutaneous nocardiosis associated with gardening: a case series. World J Clin Infect Dis 3:86–89. 10.5495/wjcid.v3.i4.86. [DOI] [Google Scholar]
- 131.Welsh O, Morales-Toquero A, Vera-Cabrera L, Cabrera-Vera L, Vazquez-Martinez O, Gómez-Flores M, Ocampo-Candiani J. 2011. Actinomycetoma of the scalp after a car accident. Int J Dermatol 50:854–857. 10.1111/j.1365-4632.2011.04874.x. [DOI] [PubMed] [Google Scholar]
- 132.Angelika J, Hans-Jurgen G, Uwe-Frithjof H. 1999. Primary cutaneous nocardiosis in a husband and wife. J Am Acad Dermatol 41:338–340. 10.1016/S0190-9622(99)70381-6. [DOI] [PubMed] [Google Scholar]
- 133.Gupta AK, Moorman CT. 3rd, 2010. Nocardia nova infection after primary anterior cruciate ligament reconstruction with tibialis anterior allograft. Am J Sports Med 38:1483–1486. 10.1177/0363546510361217. [DOI] [PubMed] [Google Scholar]
- 134.Yong EX, Cheong EY, Boutlis CS, Chen DB, Liu EY, McKew GL. 2015. Nocardia septic arthritis complicating an anterior cruciate ligament repair. J Clin Microbiol 53:2760–2762. 10.1128/JCM.00754-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Exmelin L, Malbruny B, Vergnaud M, Prosvost F, Boiron P, Morel C. 1996. Molecular study of nosocomial nocardiosis outbreak involving heart transplant recipients. J Clin Microbiol 34:1014–1016. 10.1128/jcm.34.4.1014-1016.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Vuotto F, Faure K, Queyre V, Dessein R, Pasquet A, Lambert M, Haulon S, Beregi J, Guery B, Hatron P. 2011. Vascular nosocomial Nocardia farcinica infection after arterial stenting in an immunocompetent patient. Can J Infect Dis Med Microbiol 22:e10-–e11. 10.1155/2011/216873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Bross JE, Gordon G. 1991. Nocardial meningitis: case reports and review. Rev Infect Dis 13:160–165. 10.1093/clinids/12.5.160. [DOI] [PubMed] [Google Scholar]
- 138.Kachi S, Okazaki M, Takeda H, Igarashi H, Kobayashi O, Watanabe H, Nakata K, Kawai S, Aoshima M, Watanabe T, Goto H. 2006. Outbreak of Nocardia farcinica infection with the same pattern in randomly amplified polymorphic DNA analysis. J Hosp Infect 62:502–506. 10.1016/j.jhin.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 139.Mrozek N, Hamizi S, Gourdon F, Laurichesse H, Beytout J, Lesens O. 2008. [Potential nosocomial disseminated infection due to Nocardia asteroides after a prosthesis insertion in an immunocompetent patient]. Rev Med Interne 29:1034–1037. 10.1016/j.revmed.2008.02.016. [DOI] [PubMed] [Google Scholar]
- 140.Wenger PN, Brown JM, McNeil MM, Jarvis WR. 1998. Nocardia farcinica sternotomy site infections in patients following open heart surgery. J Infect Dis 178:1539–1543. 10.1086/314450. [DOI] [PubMed] [Google Scholar]
- 141.Al Akhrass F, Hachem R, Mohamed JA, Tarrand J, Kontoyiannis DP, Chandra J, Ghannoum M, Haydoura S, Chaftari AM, Raad I. 2011. Central venous catheter-associated Nocardia bacteremia in cancer patients. Emerg Infect Dis 17:1651–1658. 10.3201/eid1709.101810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Lederman ER, Crum NF. 2004. A case series and focused review of nocardiosis: clinical and microbiologic aspects. Medicine (Baltimore, MD))83:300–313. 10.1097/01.md.0000141100.30871.39. [DOI] [PubMed] [Google Scholar]
- 143.Schoen L, Santoro JD, Milla C, Bhargava S. 2015. Pulmonary nocardiosis in an immunocompetent patient with cystic fibrosis. Case Rep Pulmonol 2015:984171. 10.1155/2015/984171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Ortman JM, Velkoff VA, Hogan H. 2014. An aging nation: the older population in the United States. Current Population Reports, U.S. Census Bureau. Washington, DC. https://www.census.gov/library/publications/2014/demo/p25-1140.html. Accessed 20 March 2020. [Google Scholar]
- 145.Harpaz R, Dahl RM, Dooling KL. 2016. Prevalence of immunosuppression among US adults, 2013. JAMA 316:2547–2548. 10.1001/jama.2016.16477. [DOI] [PubMed] [Google Scholar]
- 146.Samji H, Cescon A, Hogg RS, Modur SP, Althoff KN, Buchacz K, Burchell AN, Cohen M, Gebo KA, Gill MJ, Justice A, Kirk G, Klein MB, Korthuis PT, Martin J, Napravnik S, Rourke SB, Sterling TR, Silverberg MJ, Deeks S, Jacobson LP, Bosch RJ, Kitahata MM, Goedert JJ, Moore R, Gange SJ, North American AIDS Cohort Collaboration on Research and Design (NA-ACCORD) of IeDEA . 2013. Closing the gap: increases in life expectancy among treated HIV-positive individuals in the United States and Canada. PLoS One 8:e81355. 10.1371/journal.pone.0081355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Tremblay J, Thibert L, Alarie I, Valiquette L, Pepin J. 2011. Nocardiosis in Quebec, Canada, 1988-2008. Clin Microbiol Infect 17:690–696. 10.1111/j.1469-0691.2010.03306.x. [DOI] [PubMed] [Google Scholar]
- 148.Beaman BL, Boiron P, Beaman L, Brownell GH, Schaal K, Gombert ME. 1992. Nocardia and nocardiosis. J Med Vet Mycol 30 Suppl 1:317–331. 10.1080/02681219280001001. [DOI] [PubMed] [Google Scholar]
- 149.Santos M, Gil-Brusola A, Morales P. 2011. Infection by Nocardia in solid organ transplantation: thirty years of experience. Transplant Proc 43:2141–2144. 10.1016/j.transproceed.2011.06.065. [DOI] [PubMed] [Google Scholar]
- 150.Chen YC, Lee CH, Chien CC, Chao TL, Lin WC, Liu JW. 2013. Pulmonary nocardiosis in southern Taiwan. J Microbiol Immunol Infect 46:441–447. 10.1016/j.jmii.2012.07.017. [DOI] [PubMed] [Google Scholar]
- 151.Saubolle MA, Sussland D. 2003. Nocardiosis: review of clinical and laboratory experience. J Clin Microbiol 41:4497–4501. 10.1128/JCM.41.10.4497-4501.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Betran A, Villuendas MC, Rezusta A, Pereira J, Revillo MJ, Rodriguez-Nava V. 2016. Clinical significance, antimicrobial susceptibility and molecular identification of Nocardia species isolated from children with cystic fibrosis. Braz J Microbiol 47:531–535. 10.1016/j.bjm.2016.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Iwasaki A, Foxman EF, Molony RD. 2017. Early local immune defences in the respiratory tract. Nat Rev Immunol 17:7–20. 10.1038/nri.2016.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Rosen LB, Rocha Pereira N, Figueiredo C, Fiske LC, Ressner RA, Hong JC, Gregg KS, Henry TL, Pak KJ, Baumgarten KL, Seoane L, Garcia-Diaz J, Olivier KN, Zelazny AM, Holland SM, Browne SK. 2015. Nocardia-induced granulocyte macrophage colony-stimulating factor is neutralized by autoantibodies in disseminated/extrapulmonary nocardiosis. Clin Infect Dis 60:1017–1025. 10.1093/cid/ciu968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Hadeed MM, MacDonell JR, Dempsey IJ, Moore CC, Browne JA. 2017. Chronic Nocardia cyriacigeorgica periprosthetic knee infection successfully treated with a two-stage revision: a case report. JBJS Case Connect 7:e74. 10.2106/JBJS.CC.16.00250. [DOI] [PubMed] [Google Scholar]
- 156.Andrejak C, Nielsen R, Thomsen VO, Duhaut P, Sorensen HT, Thomsen RW. 2013. Chronic respiratory disease, inhaled corticosteroids and risk of non-tuberculous mycobacteriosis. Thorax 68:256–262. 10.1136/thoraxjnl-2012-201772. [DOI] [PubMed] [Google Scholar]
- 157.Martinez Tomas R, Menendez Villanueva R, Reyes Calzada S, Santos Durantez M, Valles Tarazona JM, Modesto Alapont M, Gobernado Serrano M. 2007. Pulmonary nocardiosis: risk factors and outcomes. Respirology 12:394–400. 10.1111/j.1440-1843.2007.01078.x. [DOI] [PubMed] [Google Scholar]
- 158.Presant CA, Wiernik PH, Serpick AA. 1973. Factors affecting survival in nocardiosis. Am Rev Respir Dis 108:1444–1448. 10.1164/arrd.1973.108.6.1444. [DOI] [PubMed] [Google Scholar]
- 159.Lerner PI. 1996. Nocardiosis. Clin Infect Dis 22:891–903. 10.1093/clinids/22.6.891. [DOI] [PubMed] [Google Scholar]
- 160.Filice GA. 1985. Inhibition of Nocardia asteroides by neutrophils. J Infect Dis 151:47–56. 10.1093/infdis/151.1.47. [DOI] [PubMed] [Google Scholar]
- 161.Filice GA, Beaman BL, Krick JA, Remington JS. 1980. Effects of human neutrophils and monocytes on Nocardia asteroides: failure of killing despite occurrence of the oxidative metabolic burst. J Infect Dis 142:432–438. 10.1093/infdis/142.3.432. [DOI] [PubMed] [Google Scholar]
- 162.Deem RL, Doughty FA, Beaman BL. 1983. Immunologically specific direct T lymphocyte-mediated killing of Nocardia asteroides. J Immunol 130:2401–2406. [PubMed] [Google Scholar]
- 163.Beaman BL, Beaman L. 1994. Nocardia species: host-parasite relationships. Clin Microbiol Rev 7:213–264. 10.1128/CMR.7.2.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Clark NM, Reid GE, Practice ASTIDCo . 2013. Nocardia infections in solid organ transplantation. Am J Transplant 13 Suppl 4:83–92. 10.1111/ajt.12102. [DOI] [PubMed] [Google Scholar]
- 165.Filice GA. 2005. Nocardiosis in persons with human immunodeficiency virus infection, transplant recipients, and large, geographically defined populations. J Lab Clin Med 145:156–162. 10.1016/j.lab.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 166.Peraira JR, Segovia J, Fuentes R, Jimenez-Mazuecos J, Arroyo R, Fuertes B, Mendaza P, Pulpon LA. 2003. Pulmonary nocardiosis in heart transplant recipients: treatment and outcome. Transplant Proc 35:2006–2008. 10.1016/s0041-1345(03)00651-1. [DOI] [PubMed] [Google Scholar]
- 167.Husain S, McCurry K, Dauber J, Singh N, Kusne S. 2002. Nocardia infection in lung transplant recipients. J Heart Lung Transplant 21:354–359. 10.1016/s1053-2498(01)00394-1. [DOI] [PubMed] [Google Scholar]
- 168.Poonyagariyagorn HK, Gershman A, Avery R, Minai O, Blazey H, Asamoto K, Alster J, Murthy S, Mehta A, Pettersson G, Mason DP, Budev M. 2008. Challenges in the diagnosis and management of Nocardia infections in lung transplant recipients. Transpl Infect Dis 10:403–408. 10.1111/j.1399-3062.2008.00338.x. [DOI] [PubMed] [Google Scholar]
- 169.Coussement J, De Greef J, Dureault A, Lebeaux D. 2018. Can we kill two birds with this stone? Anti-pneumocystis prophylaxis to prevent Nocardia infection in hematopoietic stem cell transplant recipients. Biol Blood Marrow Transplant 24:1952–1953. 10.1016/j.bbmt.2018.05.028. [DOI] [PubMed] [Google Scholar]
- 170.Shannon K, Pasikhova Y, Ibekweh Q, Ludlow S, Baluch A. 2016. Nocardiosis following hematopoietic stem cell transplantation. Transpl Infect Dis 18:169–175. 10.1111/tid.12499. [DOI] [PubMed] [Google Scholar]
- 171.Ljungman P, de la Camara R, Robin C, Crocchiolo R, Einsele H, Hill JA, Hubacek P, Navarro D, Cordonnier C, Ward KN, European Conference on Infections in Leukaemia group . 2019. Guidelines for the management of cytomegalovirus infection in patients with haematological malignancies and after stem cell transplantation from the 2017 European Conference on Infections in Leukaemia (ECIL 7). Lancet Infect Dis 19:e260–e272. 10.1016/S1473-3099(19)30107-0. [DOI] [PubMed] [Google Scholar]
- 172.Haussaire D, Fournier PE, Djiguiba K, Moal V, Legris T, Purgus R, Bismuth J, Elharrar X, Reynaud-Gaubert M, Vacher-Coponat H. 2017. Nocardiosis in the south of France over a 10-years period, 2004-2014. Int J Infect Dis 57:13–20. 10.1016/j.ijid.2017.01.005. [DOI] [PubMed] [Google Scholar]
- 173.Morrison VA. 2014. Immunosuppression associated with novel chemotherapy agents and monoclonal antibodies. Clin Infect Dis 59 Suppl 5:S360–S364. 10.1093/cid/ciu592. [DOI] [PubMed] [Google Scholar]
- 174.Cattaneo C, Antoniazzi F, Caira M, Castagnola C, Delia M, Tumbarello M, Rossi G, Pagano L. 2013. Nocardia spp infections among hematological patients: results of a retrospective multicenter study. Int J Infect Dis 17:e610–e614. 10.1016/j.ijid.2013.01.013. [DOI] [PubMed] [Google Scholar]
- 175.Rafailidis PI, Kakisi OK, Vardakas K, Falagas ME. 2007. Infectious complications of monoclonal antibodies used in cancer therapy: a systematic review of the evidence from randomized controlled trials. Cancer 109:2182–2189. 10.1002/cncr.22666. [DOI] [PubMed] [Google Scholar]
- 176.Grim SA, Pham T, Thielke J, Sankary H, Oberholzer J, Benedetti E, Clark NM. 2007. Infectious complications associated with the use of rituximab for ABO-incompatible and positive cross-match renal transplant recipients. Clin Transplant 21:628–632. 10.1111/j.1399-0012.2007.00700.x. [DOI] [PubMed] [Google Scholar]
- 177.Martin SI, Marty FM, Fiumara K, Treon SP, Gribben JG, Baden LR. 2006. Infectious complications associated with alemtuzumab use for lymphoproliferative disorders. Clin Infect Dis 43:16–24. 10.1086/504811. [DOI] [PubMed] [Google Scholar]
- 178.O'Brien SM, Keating MJ, Mocarski ES. 2006. Updated guidelines on the management of cytomegalovirus reactivation in patients with chronic lymphocytic leukemia treated with alemtuzumab. Clin Lymphoma Myeloma 7:125–130. 10.3816/CLM.2006.n.049. [DOI] [PubMed] [Google Scholar]
- 179.Kundranda MN, Spiro TP, Muslimani A, Gopalakrishna KV, Melaragno MJ, Daw HA. 2007. Cerebral nocardiosis in a patient with NHL treated with rituximab. Am J Hematol 82:1033–1034. 10.1002/ajh.20921. [DOI] [PubMed] [Google Scholar]
- 180.Abel S, Hasan S, Kujawski B, Talwar A, Betler J, Wegner R, Colonias A, Aziz K. 2016. Cryptic Nocardia nova brain abscess postradiation treatment and neurosurgery in a patient with small cell lung cancer: a case report and review of the literature. Adv Radiat Oncol 1:290–293. 10.1016/j.adro.2016.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Holtz HA, Lavery DP, Kapila R. 1985. Actinomycetales infection in the acquired immunodeficiency syndrome. Ann Intern Med 102:203–205. 10.7326/0003-4819-102-2-203. [DOI] [PubMed] [Google Scholar]
- 182.Mendez-Samperio P. 2017. Diagnosis of tuberculosis in HIV co-infected individuals: Current Status, challenges and opportunities for the future. Scand J Immunol 86:76–82. 10.1111/sji.12567. [DOI] [PubMed] [Google Scholar]
- 183.Alnaum HM, Elhassan MM, Mustafa FY, Hamid ME. 2011. Prevalence of Nocardia species among HIV-positive patients with suspected tuberculosis. Trop Doct 41:224–226. 10.1258/td.2011.110107. [DOI] [PubMed] [Google Scholar]
- 184.Sakyi SA, Danquah KO, Ephraim RD, Enimil A, Frimpong V, Ahenkorah Fondjo L, Darkoh EL. 2018. Evaluating the contribution of Nocardia spp. and Mycobacterium tuberculosis to pulmonary infections among HIV and non-HIV patients at the Komfo Anokye Teaching Hospital, Ghana. Can J Infect Dis Med Microbiol 2018:2910198. 10.1155/2018/2910198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Hashemi-Shahraki A, Heidarieh P, Bostanabad SZ, Hashemzadeh M, Feizabadi MM, Schraufnagel D, Mirsaeidi M. 2015. Genetic diversity and antimicrobial susceptibility of Nocardia species among patients with nocardiosis. Sci Rep 5:17862. 10.1038/srep17862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Jiang Y, Huang A, Fang Q. 2016. Disseminated nocardiosis caused by Nocardia otitidiscaviarum in an immunocompetent host: a case report and literature review. Exp Ther Med 12:3339–3346. 10.3892/etm.2016.3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Nenoff P, van de Sande WW, Fahal AH, Reinel D, Schofer H. 2015. Eumycetoma and actinomycetoma–an update on causative agents, epidemiology, pathogenesis, diagnostics and therapy. J Eur Acad Dermatol Venereol 29:1873–1883. 10.1111/jdv.13008. [DOI] [PubMed] [Google Scholar]
- 188.Stapleton F, Keay LJ, Sanfilippo PG, Katiyar S, Edwards KP, Naduvilath T. 2007. Relationship between climate, disease severity, and causative organism for contact lens-associated microbial keratitis in Australia. Am J Ophthalmol 144:690–698.e1. 10.1016/j.ajo.2007.06.037. [DOI] [PubMed] [Google Scholar]
- 189.Bonifaz A, Tirado-Sanchez A, Calderon L, Saul A, Araiza J, Hernandez M, Gonzalez GM, Ponce RM. 2014. Mycetoma: experience of 482 cases in a single center in Mexico. PLoS Negl Trop Dis 8:e3102. 10.1371/journal.pntd.0003102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Kachuei R, Emami M, Mirnejad R, Khoobdel M. 2012. Diversity and frequency of Nocardia spp. in the soil of Isfahan province, Iran. Asian Pac J Trop Biomed 2:474–478. 10.1016/S2221-1691(12)60079-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Chen B, Tang J, Lu Z, Wang N, Gao X, Wang F. 2016. Primary cutaneous nocardiosis in a patient with nephrotic syndrome: a case report and review of the literature. Medicine (Baltimore, MD) 95:e2490. 10.1097/MD.0000000000002490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Sherber NS, Olivere JW, Martins CR. 2006. An 80-year-old man with a nonhealing glabellar lesion. Primary cutaneous nocardiosis. Arch Pathol Lab Med 130:e100–e101. 10.5858/2006-130-e100-AYMWAN. [DOI] [PubMed] [Google Scholar]
- 193.Sinnott J, Holt DA, Alverez C, Greene J, Sweeney MS. 1990. Nocardia brasiliensis cellulitis in a heart transplant patient. Tex Heart Inst J 17:133–135. [PMC free article] [PubMed] [Google Scholar]
- 194.Schiff TA, McNeil MM, Brown JM. 1993. Cutaneous Nocardia farcinica infection in a nonimmunocompromised patient: case report and review. Clin Infect Dis 16:756–760. 10.1093/clind/16.6.756. [DOI] [PubMed] [Google Scholar]
- 195.Bhalodia AM, Lertzman BH, Kantor GR, Granick MS. 1998. Localized cutaneous Nocardia brasiliensis mimicking foreign body granuloma. Cutis 61:161–163. [PubMed] [Google Scholar]
- 196.Ng CS, Hellinger WC. 1998. Superficial cutaneous abscess and multiple brain abscesses from Nocardia asteroides in an immunocompetent patient. J Am Acad Dermatol 39:793–794. 10.1016/S0190-9622(98)70054-4. [DOI] [PubMed] [Google Scholar]
- 197.Georghiou PR, Blacklock ZM. 1992. Infection with Nocardia species in Queensland. A review of 102 clinical isolates. Med J Aust 156:692–697. 10.5694/j.1326-5377.1992.tb121509.x. [DOI] [PubMed] [Google Scholar]
- 198.Brown-Elliott BA, Biehle J, Conville PS, Cohen S, Saubolle M, Sussland D, Wengenack N, Kriel K, Bridge L, McNulty S, Vasireddy R, Wallace RJ. Jr, 2012. Sulfonamide resistance in isolates of Nocardia spp. from a US multicenter survey. J Clin Microbiol 50:670–672. 10.1128/JCM.06243-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Hawrot AC, Carter EL. 2005. Simultaneous chronic cutaneous infection with Mycobacterium marinum and Nocardia asteroides. J Am Acad Dermatol 52:703–704. 10.1016/j.jaad.2004.09.039. [DOI] [PubMed] [Google Scholar]
- 200.Fujita T, Ikari J, Watanabe A, Tatsumi K. 2016. Clinical characteristics of pulmonary nocardiosis in immunocompetent patients. J Infect Chemother 22:738–743. 10.1016/j.jiac.2016.08.004. [DOI] [PubMed] [Google Scholar]
- 201.Hui CH, Au VW, Rowland K, Slavotinek JP, Gordon DL. 2003. Pulmonary nocardiosis re-visited: experience of 35 patients at diagnosis. Respir Med 97:709–717. 10.1053/rmed.2003.1505. [DOI] [PubMed] [Google Scholar]
- 202.Martinez R, Reyes S, Menendez R. 2008. Pulmonary nocardiosis: risk factors, clinical features, diagnosis and prognosis. Curr Opin Pulm Med 14:219–227. 10.1097/MCP.0b013e3282f85dd3. [DOI] [PubMed] [Google Scholar]
- 203.Olson ES, Simpson AJ, Norton AJ, Das SS. 1998. Not everything acid fast is Mycobacterium tuberculosis–a case report. J Clin Pathol 51:535–536. 10.1136/jcp.51.7.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Benes J, Viechova J, Picha D, Horova B, Zatloukal P. 2003. Disseminated Nocardia asteroides infection in an immunocompetent woman following an arm injury. Infection 31:112–114. 10.1007/s15010-003-3073-x. [DOI] [PubMed] [Google Scholar]
- 205.Koll BS, Brown AE, Kiehn TE, Armstrong D. 1992. Disseminated Nocardia brasiliensis infection with septic arthritis. Clin Infect Dis 15:469–472. 10.1093/clind/15.3.469. [DOI] [PubMed] [Google Scholar]
- 206.Puri S, Hadayer A, Breaux A, Barr CC. 2018. Disseminated nocardiosis with retinal abscess in a patient treated for bullous pemphigoid. Am J Ophthalmol Case Rep 10:145–147. 10.1016/j.ajoc.2018.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Tabrizi SJ. 1994. Nocardia pericarditis. BMJ 309:1495–1497. 10.1136/bmj.309.6967.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Lai CC, Liu WL, Ko WC, Chen YH, Tan HR, Huang YT, Hsueh PR. 2011. Multicenter study in Taiwan of the in vitro activities of nemonoxacin, tigecycline, doripenem, and other antimicrobial agents against clinical isolates of various Nocardia species. Antimicrob Agents Chemother 55:2084–2091. 10.1128/AAC.01808-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Khan M, Adnan MM, Shahbaz N, Hamza M, Mujeeb SA. 2014. Nocardia mikamii a novel species causing disseminated nocardiosis: a literature review of disseminated nocardiosis. Int Sch Res Notices 2014:869153. 10.1155/2014/869153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Sharma N, O'Hagan S. 2016. The role of oral co-trimoxazole in treating Nocardia farcinica keratitis: a case report. J Ophthalmic Inflamm Infect 6:21. 10.1186/s12348-016-0087-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Epstein S, Holden M, Feldshuh J, Singer JM. 1963. Unusual cause of spinal cord compression: nocardiosis. NY State. J Med 63:3422–3427. [PubMed] [Google Scholar]
- 212.Raszka D, Popelka S, Heřt J, Jahoda D, Landor I, Vavřík P. 2018. Rare case of osteomyelitis of tibial shaft caused by Nocardia cyriacigeorgica. Folia Microbiol (Praha) 63:525–532. 10.1007/s12223-018-0589-0. [DOI] [PubMed] [Google Scholar]
- 213.Tokumoto JI, Jacobs RA. 1994. Case report: Nocardia osteomyelitis. Am J Med Sci 307:428–433. 10.1097/00000441-199406000-00009. [DOI] [PubMed] [Google Scholar]
- 214.Vander Heiden T, Stahel PF, Clutter S, Price C, Peterson SL, Morgan SJ. 2009. Nocardia osteomyelitis: a rare complication after intramedullary nailing of a closed tibial shaft fracture. J Orthop Trauma 23:232–236. 10.1097/BOT.0b013e318199e8d9. [DOI] [PubMed] [Google Scholar]
- 215.Vanegas S, Franco-Cendejas R, Cicero A, Lopez-Jacome E, Colin C, Hernandez M. 2014. Nocardia brasiliensis-associated femorotibial osteomyelitis. Int J Infect Dis 20:63–65. 10.1016/j.ijid.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 216.Baraboutis IG, Argyropoulou A, Papastamopoulos V, Psaroudaki Z, Paniara O, Skoutelis AT. 2008. Primary sternal osteomyelitis caused by Nocardia nova: case report and literature review. Braz J Infect Dis 12:257–259. 10.1590/s1413-86702008000300018. [DOI] [PubMed] [Google Scholar]
- 217.Talpada M, Rauf SJ, Walling DM. 2002. Primary Nocardia osteomyelitis as a presentation of AIDS. AIDS Read 12:75–78. [PubMed] [Google Scholar]
- 218.Moore SL, Jones S, Lee JL. 2005. Nocardia osteomyelitis in the setting of previously unknown HIV infection. Skeletal Radiol 34:58–60. 10.1007/s00256-004-0784-8. [DOI] [PubMed] [Google Scholar]
- 219.Moore M, Lane CW, Gaul LE. 1954. Nocardiosis of the knee caused by Nocardia brasiliensis; report of first case in a native of the United States. AMA Arch Derm Syphilol 70:302–310. 10.1001/archderm.1954.01540210042008. [DOI] [PubMed] [Google Scholar]
- 220.Nizam I, Kohan L, Kerr D. 2007. Nocardia nova septic arthritis following total knee replacement: a case report. J Orthop Surg (Hong Kong) 15:390–392. 10.1177/230949900701500332. [DOI] [PubMed] [Google Scholar]
- 221.Ozan F, Koyuncu S, Kizilay C, Ozgenc O. 2013. The Nocardia farcinica infection developing after total knee arthroplasty surgery. Acta Orthop Traumatol Turc 47:212–217. 10.3944/aott.2013.2536. [DOI] [PubMed] [Google Scholar]
- 222.Laurent F, Rodriguez-Villalobos H, Cornu O, Vandercam B, Yombi JC. 2015. Nocardia prosthetic knee infection successfully treated by one-stage exchange: case report and review. Acta Clin Belg 70:287–290. 10.1179/2295333714Y.0000000109. [DOI] [PubMed] [Google Scholar]
- 223.Lipuma JJ. 2010. The changing microbial epidemiology in cystic fibrosis. Clin Microbiol Rev 23:299–323. 10.1128/CMR.00068-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Gondos A, Brenner H. 2011. Relative survival of transplant patients: quantifying surplus mortality among renal transplant recipients compared with the general population. Transplantation 92:913–917. 10.1097/TP.0b013e31822e0c08. [DOI] [PubMed] [Google Scholar]
- 225.Bell M, McNeil MM, Brown JM. 2017. Nocardia species (Nocardiosis), on E-Sun Technologies, Inc. http://www.antimicrobe.org/b117.asp#top. Accessed September 23, 2022. [Google Scholar]
- 226.Wallace RJ, Brown BA, Brown JM, McNeil M. 1994. Taxonomy of Nocardia species. Clin Infect Dis 18:476–477. 10.1093/clinids/18.3.476. [DOI] [PubMed] [Google Scholar]
- 227.Welsh O, Vera-Cabrera L, Salinas-Carmona MC. 2013. Current treatment for Nocardia infections. Expert Opin Pharmacother 14:2387–2398. 10.1517/14656566.2013.842553. [DOI] [PubMed] [Google Scholar]
- 228.Lafont E, Conan PL, Rodriguez-Nava V, Lebeaux D. 2020. Invasive nocardiosis: disease presentation, diagnosis and treatment - old questions, new answers? IDR 13:4601–4613. 10.2147/IDR.S249761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Restrepo A, Clark NM, Infectious Diseases Community of Practice of the American Society of T . 2019. Nocardia infections in solid organ transplantation: guidelines from the infectious diseases community of practice of the American Society of Transplantation. Clin Transplant 33:e13509. 10.1111/ctr.13509. [DOI] [PubMed] [Google Scholar]
- 230.Brown-Elliott BA, Killingley J, Vasireddy S, Bridge L, Wallace RJ. Jr, 2016. In vitro comparison of ertapenem, meropenem, and imipenem against isolates of rapidly growing Mycobacteria and Nocardia by use of broth microdilution and etest. J Clin Microbiol 54:1586–1592. 10.1128/JCM.00298-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Hamdi AM, Fida M, Deml SM, Abu Saleh OM, Wengenack NL. 2020. Retrospective analysis of antimicrobial susceptibility profiles of Nocardia species from a tertiary hospital and reference laboratory, 2011 to 2017. Antimicrob Agents Chemother 64:e01868-19. 10.1128/AAC.01868-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Schlaberg R, Fisher MA, Hanson KE. 2014. Susceptibility profiles of Nocardia isolates based on current taxonomy. Antimicrob Agents Chemother 58:795–800. 10.1128/AAC.01531-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Nau R, Sorgel F, Eiffert H. 2010. Penetration of drugs through the blood-cerebrospinal fluid/blood-brain barrier for treatment of central nervous system infections. Clin Microbiol Rev 23:858–883. 10.1128/CMR.00007-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Lebeaux D, Freund R, van Delden C, Guillot H, Marbus SD, Matignon M, Van Wijngaerden E, Douvry B, De Greef J, Vuotto F, Tricot L, Fernández-Ruiz M, Dantal J, Hirzel C, Jais J-P, Rodriguez-Nava V, Jacobs F, Lortholary O, Coussement J, Anstey JR, Antoine M, Ausselet N, Belhaj A, Boelens J, de Beenhouwer H, Denis C, Ho E, Ieven M, Jonckheere S, Knoop C, le Moine A, Rodriguez-Villalobos H, Racapé J, Roisin S, Vandercam B, Vander Zwalmen M-L, Vanfraechem G, Van Laecke S, Verhaegen J, Barrou B, Battistella P, Bergeron E, Bouvier N, Caillard S, Caumes E, Chaussade H, Chauvet C, Crochette R, Epailly E, Essig M, European Study Group for Nocardia in Solid Organ Transplantation, European Study Group for Nocardia in Solid Organ Transplantation ., et al. 2017. Outcome and treatment of nocardiosis after solid organ transplantation: new insights from a European study. Clin Infect Dis 64:1396–1405. 10.1093/cid/cix124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Davidson N, Grigg MJ, McGuinness SL, Baird RJ, Anstey NM. 2020. Safety and outcomes of linezolid use for nocardiosis. Open Forum Infect Dis 7:ofaa090. 10.1093/ofid/ofaa090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Tripodi MF, Durante-Mangoni E, Fortunato R, Cuccurullo S, Mikami Y, Farina C, Utili R. 2011. In vitro activity of multiple antibiotic combinations against Nocardia: relationship with a short-term treatment strategy in heart transplant recipients with pulmonary nocardiosis. Transpl Infect Dis 13:335–343. 10.1111/j.1399-3062.2010.00588.x. [DOI] [PubMed] [Google Scholar]
- 237.Mamelak AN, Obana WG, Flaherty JF, Rosenblum ML. 1994. Nocardial brain abscess: treatment strategies and factors influencing outcome. Neurosurgery 35:622–631. 10.1097/00006123-199410000-00007. [DOI] [PubMed] [Google Scholar]
- 238.Ricci JA, Weil AA, Eberlin KR. 2015. Necrotizing cutaneous nocardiosis of the hand: a case report and review of the literature. J Hand Microsurg 7:224–227. 10.1007/s12593-015-0173-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Singh A, Chhina D, Soni RK, Kakkar C, Sidhu US. 2016. Clinical spectrum and outcome of pulmonary nocardiosis: 5-year experience. Lung India 33:398–403. 10.4103/0970-2113.184873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Tomczyk S, Deribe K, Brooker SJ, Clark H, Rafique K, Knopp S, Utzinger J, Davey G. 2014. Association between footwear use and neglected tropical diseases: a systematic review and meta-analysis. PLoS Negl Trop Dis 8:e3285. 10.1371/journal.pntd.0003285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Molina A, Winston DJ, Pan D, Schiller GJ. 2018. Increased incidence of nocardial infections in an era of atovaquone prophylaxis in allogeneic hematopoietic stem cell transplant recipients. Biol Blood Marrow Transplant 24:1715–1720. 10.1016/j.bbmt.2018.03.010. [DOI] [PubMed] [Google Scholar]
- 242.Healthcare Cost and Utilization Project (HCUP). HCUP State Inpatient Databases (SID). 2001–2015. Rockville, MD: Agency for Healthcare Research and Quality. https://www.hcup-us.ahrq.gov/sidoverview.jsp. Accessed 29 July 2021. [PubMed] [Google Scholar]
- 243.National Center for Health Statistics. 2018. 2018 release of International Classification of Diseases, Tenth Revision. https://www.cdc.gov/nchs/icd/icd10.htm. Accessed 2 December 2018. [Google Scholar]
- 244.Beer KD, Blaney DD, Kadzik M, Asiedu KB, Shieh W, Bower W, Jackson BR, Walke H, Chiller T. 2018. A call to action for mycetoma. Curr Fungal Infect Rep 12:99–104. 10.1007/s12281-018-0317-x. [DOI] [Google Scholar]
- 245.Siddig EE, van de Sande WWJ, Fahal AH. 2021. Actinomycetoma laboratory-based diagnosis: a mini-review. Trans R Soc Trop Med Hyg 115:355–363. 10.1093/trstmh/traa176. [DOI] [PubMed] [Google Scholar]
- 246.Suleiman SH, Wadaella el S, Fahal AH. 2016. The surgical treatment of mycetoma. PLoS Negl Trop Dis 10:e0004690. 10.1371/journal.pntd.0004690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.World Health Assembly, 69. 2016. Addressing the burden of mycetoma. World Health Organization. https://apps.who.int/iris/handle/10665/252801. [Google Scholar]
- 248.Fahal AH. 2021. The Khartoum call for action Khartoum, Sudan - 2019. Trans R Soc Trop Med Hyg 115:295–296. 10.1093/trstmh/trab043. [DOI] [Google Scholar]
- 249.van de Sande W, Fahal A, Ahmed SA, Serrano JA, Bonifaz A, Zijlstra E, Eumycetoma Working Group . 2018. Closing the mycetoma knowledge gap. Med Mycol 56:153–164. [DOI] [PubMed] [Google Scholar]
- 250.Zijlstra EE, van de Sande WW, Fahal AH. 2016. Mycetoma: a long journey from neglect. PLoS Negl Trop Dis 10:e0004244. 10.1371/journal.pntd.0004244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.World Health Organization. 2020. Ending the neglect to attain the sustainable development goals: a road map for neglected tropical diseases 2021–2030. World Health Organization. https://apps.who.int/iris/handle/10665/338565. Acessed 29 July 2021. [Google Scholar]
- 252.Golinska P, Wang D, Goodfellow M. 2013. Nocardia aciditolerans sp. nov., isolated from a spruce forest soil. Antonie Van Leeuwenhoek 103:1079–1088. 10.1007/s10482-013-9887-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Yamamura H, Tamura T, Sakiyama Y, Harayama S. 2007. Nocardia amamiensis sp. nov., isolated from a sugar-cane field in Japan. Int J Syst Evol Microbiol 57:1599–1602. 10.1099/ijs.0.64829-0. [DOI] [PubMed] [Google Scholar]
- 254.Ezeoke I, Klenk HP, Potter G, Schumann P, Moser BD, Lasker BA, Nicholson A, Brown JM. 2013. Nocardia amikacinitolerans sp. nov., an amikacin-resistant human pathogen. Int J Syst Evol Microbiol 63:1056–1061. 10.1099/ijs.0.039990-0. [DOI] [PubMed] [Google Scholar]
- 255.Zhao GZ, Li J, Zhu WY, Klenk HP, Xu LH, Li WJ. 2011. Nocardia artemisiae sp. nov., an endophytic actinobacterium isolated from a surface-sterilized stem of Artemisia annua L. Int J Syst Evol Microbiol 61:2933–2937. 10.1099/ijs.0.029306-0. [DOI] [PubMed] [Google Scholar]
- 256.Benndorf R, Schwitalla JW, Martin K, de Beer ZW, Vollmers J, Kaster AK, Poulsen M, Beemelmanns C. 2020. Nocardia macrotermitis sp. nov. and Nocardia aurantia sp. nov., isolated from the gut of the fungus-growing termite Macrotermes natalensis. Int J Syst Evol Microbiol 70:5226–5234. 10.1099/ijsem.0.004398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Kanchanasin P, Yuki M, Kudo T, Ohkuma M, Phongsopitanun W, Tanasupawat S. 2020. Nocardia aurantiaca sp. nov., isolated from soil in Thailand. Int J Syst Evol Microbiol 70:5432–5438. 10.1099/ijsem.0.004432. [DOI] [PubMed] [Google Scholar]
- 258.Fang BZ, Han MX, Zhang LY, Jiao JY, Zhang XT, Zhang ZT, Wang Y, Nie GX, Li WJ. 2019. Nocardia aurea sp. nov., a novel actinobacterium isolated from a karstic subterranean environment. Int J Syst Evol Microbiol 69:159–164. 10.1099/ijsem.0.003122. [DOI] [PubMed] [Google Scholar]
- 259.Nouioui I, Cortes-Albayay C, Neumann-Schaal M, Vicente D, Cilla G, Klenk HP, Marimon JM, Ercibengoa M. 2020. Genomic virulence features of two novel species Nocardia barduliensis sp. nov. and Nocardia gipuzkoensis sp. nov., isolated from patients with chronic pulmonary diseases. Microorganisms 8:1517. 10.3390/microorganisms8101517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Vaddavalli R, Peddi S, Kothagauni SY, Linga VR. 2014. Nocardia bhagyanesis sp. nov., a novel actinomycete isolated from the rhizosphere of Callistemon citrinus (Curtis), India. Antonie Van Leeuwenhoek 105:443–450. 10.1007/s10482-013-0093-0. [DOI] [PubMed] [Google Scholar]
- 261.Gilquin JM, Riviere B, Jurado V, Audouy B, Kouatche JB, Bergeron E, Mouniee D, Molina T, Faure P, Saiz-Jimenez C, Rodriguez-Nava V. 2016. First case of actinomycetoma in France due to a novel Nocardia species, Nocardia boironii sp. Nov mSphere 1:e00309-16. 10.1128/mSphere.00309-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Zhang X, Zhang L, Yu X, Zhang J, Jiao Y, Ju H, Wang X, Zhao J, Xiang W. 2019. Nocardia bovistercoris sp. nov., an actinobacterium isolated from cow dung. Int J Syst Evol Microbiol 71. [DOI] [PubMed] [Google Scholar]
- 263.Kaewkla O, Franco CMM. 2010. Nocardia callitridis sp. nov., an endophytic actinobacterium isolated from a surface-sterilized root of an Australian native pine tree. Int J Syst Evol Microbiol 60:1532–1536. 10.1099/ijs.0.016337-0. [DOI] [PubMed] [Google Scholar]
- 264.Liu C, Guan X, Li Y, Li W, Ye L, Kong X, Song J, Wang X, Xiang W. 2016. Nocardia camponoti sp. nov., an actinomycete isolated from the head of an ant (Camponotus japonicas Mayr). Int J Syst Evol Microbiol 66:1900–1905. 10.1099/ijsem.0.000963. [DOI] [PubMed] [Google Scholar]
- 265.Ghodhbane-Gtari F, Nouioui I, Salem K, Ktari A, Montero-Calasanz MdC, Tisa LS, Klenk H-P, Gtari M. 2014. Nocardia casuarinae sp. nov., an actinobacterial endophyte isolated from root nodules of Casuarina glauca. Antonie Van Leeuwenhoek 105:1099–1106. 10.1007/s10482-014-0168-6. [DOI] [PubMed] [Google Scholar]
- 266.Li QQ, Han MX, Fang BZ, Jiao JY, Liu L, Yang ZW, Zhang WQ, Wei DQ, Li WJ. 2017. Nocardia cavernae sp. nov., a novel actinobacterium isolated from a karst cave sample. Int J Syst Evol Microbiol 67:2998–3003. 10.1099/ijsem.0.002072. [DOI] [PubMed] [Google Scholar]
- 267.Zhou T, Wang XY, Deng DQ, Xu LH, Li XL, Guo Y, Li WH, Xie H, Zhang PL, Zhou XH. 2020. Nocardia colli sp. nov., a new pathogen isolated from a patient with primary cutaneous nocardiosis. Int J Syst Evol Microbiol 70:2981–2987. 10.1099/ijsem.0.003856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Rodriguez-Nava V, Khan ZU, Potter G, Kroppenstedt RM, Boiron P, Laurent F. 2007. Nocardia coubleae sp. nov., isolated from oil-contaminated Kuwaiti soil. Int J Syst Evol Microbiol 57:1482–1486. 10.1099/ijs.0.64815-0. [DOI] [PubMed] [Google Scholar]
- 269.Ercibengoa M, Bell M, Marimon JM, Humrighouse B, Klenk HP, Potter G, Perez-Trallero E. 2016. Nocardia donostiensis sp. nov., isolated from human respiratory specimens. Antonie Van Leeuwenhoek 109:653–660. 10.1007/s10482-016-0667-8. [DOI] [PubMed] [Google Scholar]
- 270.Xing K, Qin S, Fei SM, Lin Q, Bian GK, Miao Q, Wang Y, Cao CL, Tang SK, Jiang JH, Li WJ. 2011. Nocardia endophytica sp. nov., an endophytic actinomycete isolated from the oil-seed plant Jatropha curcas L. Int J Syst Evol Microbiol 61:1854–1858. 10.1099/ijs.0.027391-0. [DOI] [PubMed] [Google Scholar]
- 271.Iida S, Kageyama A, Yazawa K, Uchiyama N, Toyohara T, Chohnabayashi N, Suzuki SI, Nomura F, Kroppenstedt RM, Mikami Y. 2006. Nocardia exalbida sp. nov., isolated from Japanese patients with nocardiosis. Int J Syst Evol Microbiol 56:1193–1196. 10.1099/ijs.0.63850-0. [DOI] [PubMed] [Google Scholar]
- 272.le Roes M, Meyers PR. 2006. Nocardia gamkensis sp. nov. Antonie Van Leeuwenhoek 90:291–298. 10.1007/s10482-006-9083-9. [DOI] [PubMed] [Google Scholar]
- 273.Sazak A, Sahin N, Camas M. 2012. Nocardia goodfellowii sp. nov. and Nocardia thraciensis sp. nov., isolated from soil. Int J Syst Evol Microbiol 62:1228–1234. 10.1099/ijs.0.031559-0. [DOI] [PubMed] [Google Scholar]
- 274.Kampfer P, Lodders N, Grun-Wollny I, Martin K, Busse HJ. 2012. Nocardia grenadensis sp. nov., isolated from sand of the Caribbean Sea. Int J Syst Evol Microbiol 62:693–697. 10.1099/ijs.0.030684-0. [DOI] [PubMed] [Google Scholar]
- 275.Moshtaghi Nikou M, Ramezani M, Ali Amoozegar M, Rasooli M, Harirchi S, Shahzadeh Fazeli SA, Schumann P, Sproer C, Ventosa A. 2015. Nocardia halotolerans sp. nov., a halotolerant actinomycete isolated from saline soil. Int J Syst Evol Microbiol 65:3148–3154. 10.1099/ijsem.0.000394. [DOI] [PubMed] [Google Scholar]
- 276.Seo JP, Lee SD. 2006. Nocardia harenae sp. nov., an actinomycete isolated from beach sand. Int J Syst Evol Microbiol 56:2203–2207. 10.1099/ijs.0.64187-0. [DOI] [PubMed] [Google Scholar]
- 277.Huang JR, Ming H, Duan YY, Li S, Zhang LY, Ji WL, Zhao ZL, Meng XL, Li WJ, Nie GX. 2017. Nocardia heshunensis sp. nov., an actinomycete isolated from soil. Int J Syst Evol Microbiol 67:3467–3473. 10.1099/ijsem.0.002143. [DOI] [PubMed] [Google Scholar]
- 278.Lamm AS, Khare A, Conville P, Lau PC, Bergeron H, Rosazza JP. 2009. Nocardia iowensis sp. nov., an organism rich in biocatalytically important enzymes and nitric oxide synthase. Int J Syst Evol Microbiol 59:2408–2414. 10.1099/ijs.0.007427-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Lee SD. 2006. Nocardia jejuensis sp. nov., a novel actinomycete isolated from a natural cave on Jeju Island, Republic of Korea. Int J Syst Evol Microbiol 56:559–562. 10.1099/ijs.0.63866-0. [DOI] [PubMed] [Google Scholar]
- 280.Cui Q, Wang L, Huang Y, Liu Z, Goodfellow M. 2005. Nocardia jiangxiensis sp. nov. and Nocardia miyunensis sp. nov., isolated from acidic soils. Int J Syst Evol Microbiol 55:1921–1925. 10.1099/ijs.0.63644-0. [DOI] [PubMed] [Google Scholar]
- 281.Sun W, Zhang YQ, Huang Y, Zhang YQ, Yang ZY, Liu ZH. 2009. Nocardia jinanensis sp. nov., an amicoumacin B-producing actinomycete. Int J Syst Evol Microbiol 59:417–420. 10.1099/ijs.0.002899-0. [DOI] [PubMed] [Google Scholar]
- 282.Xu P, Li WJ, Tang SK, Jiang Y, Gao HY, Xu LH, Jiang CL. 2006. Nocardia lijiangensis sp. nov., a novel actinomycete strain isolated from soil in China. Syst Appl Microbiol 29:308–314. 10.1016/j.syapm.2005.11.009. [DOI] [PubMed] [Google Scholar]
- 283.Jones AL, Fisher AJ, Mahida R, Gould K, Perry JD, Hannan MM, Judge EP, Brown R, Boagey K, Goodfellow M. 2014. Nocardia kroppenstedtii sp. nov., an actinomycete isolated from a lung transplant patient with a pulmonary infection. Int J Syst Evol Microbiol 64:751–754. 10.1099/ijs.0.048330-0. [DOI] [PubMed] [Google Scholar]
- 284.Liu C, Bai L, Ye L, Zhao J, Yan K, Xiang W, Wang X. 2016. Nocardia lasii sp. nov., a novel actinomycete isolated from the cuticle of an ant (Lasius fuliginosus L). Antonie Van Leeuwenhoek 109:1513–1520. 10.1007/s10482-016-0753-y. [DOI] [PubMed] [Google Scholar]
- 285.Jannat-Khah D, Kroppenstedt RM, Klenk HP, Sproer C, Schumann P, Lasker BA, Steigerwalt AG, Hinrikson HP, Brown JM. 2010. Nocardia mikamii sp. nov., isolated from human pulmonary infections in the USA. Int J Syst Evol Microbiol 60:2272–2276. 10.1099/ijs.0.015594-0. [DOI] [PubMed] [Google Scholar]
- 286.Laurent F, Rodriguez-Nava V, Noussair L, Couble A, Nicolas-Chanoine MH, Boiron P. 2007. Nocardia ninae sp. nov., isolated from a bronchial aspirate. Int J Syst Evol Microbiol 57:661–665. 10.1099/ijs.0.64476-0. [DOI] [PubMed] [Google Scholar]
- 287.Moser BD, Klenk HP, Schumann P, Potter G, Lasker BA, Steigerwalt AG, Hinrikson HP, Brown JM. 2011. Nocardia niwae sp. nov., isolated from human pulmonary sources. Int J Syst Evol Microbiol 61:438–442. 10.1099/ijs.0.020370-0. [DOI] [PubMed] [Google Scholar]
- 288.Xu P, Li WJ, Tang SK, Jiang Y, Chen HH, Xu LH, Jiang CL. 2005. Nocardia polyresistens sp. nov. Int J Syst Evol Microbiol 55:1465–1470. 10.1099/ijs.0.63352-0. [DOI] [PubMed] [Google Scholar]
- 289.Everest GJ, Cook AE, le Roes-Hill M, Meyers PR. 2011. Nocardia rhamnosiphila sp. nov., isolated from soil. Syst Appl Microbiol 34:508–512. 10.1016/j.syapm.2011.03.006. [DOI] [PubMed] [Google Scholar]
- 290.Wang Y, Liu W, Feng WW, Zhou XQ, Bai JL, Yuan B, Ju XY, Cao CL, Huang Y, Jiang JH, Lv AJ, Qin S. 2015. Nocardia rhizosphaerae sp. nov., a novel actinomycete isolated from the coastal rhizosphere of Artemisia Linn., China. Antonie Van Leeuwenhoek 108:31–39. 10.1007/s10482-015-0460-0. [DOI] [PubMed] [Google Scholar]
- 291.Matsumoto T, Negishi T, Hamada M, Komaki H, Gonoi T, Yaguchi T. 2016. Nocardia shinanonensis sp. nov., isolated from a patient with endophthalmitis. Int J Syst Evol Microbiol 66:3324–3328. 10.1099/ijsem.0.001197. [DOI] [PubMed] [Google Scholar]
- 292.Seo JP, Yun YW, Lee SD. 2007. Nocardia speluncae sp. nov., isolated from a cave. Int J Syst Evol Microbiol 57:2932–2935. 10.1099/ijs.0.65085-0. [DOI] [PubMed] [Google Scholar]
- 293.Camas M, Veyisoglu A, Sahin N. 2014. Nocardia sungurluensis sp. nov., isolated from soil. Int J Syst Evol Microbiol 64:1629–1634. 10.1099/ijs.0.051334-0. [DOI] [PubMed] [Google Scholar]
- 294.Hoshino Y, Watanabe K, Iida S, Suzuki SI, Kudo T, Kogure T, Yazawa K, Ishikawa J, Kroppenstedt RM, Mikami Y. 2007. Nocardia terpenica sp. nov., isolated from Japanese patients with nocardiosis. Int J Syst Evol Microbiol 57:1456–1460. 10.1099/ijs.0.64695-0. [DOI] [PubMed] [Google Scholar]
- 295.Fahal AH. 2021. Mycetoma: the journey from neglect to recognition as a neglected tropical disease. Trans R Soc Trop Med Hyg 115:292–294. 10.1093/trstmh/traa195. [DOI] [PubMed] [Google Scholar]
- 296.Emery D, Denning DW. 2020. The global distribution of actinomycetoma and eumycetoma. PLoS Negl Trop Dis 14:e0008397. 10.1371/journal.pntd.0008397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.van de Sande WW. 2013. Global burden of human mycetoma: a systematic review and meta-analysis. PLoS Negl Trop Dis 7:e2550. 10.1371/journal.pntd.0002550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Davey G, Callow C, Newport MJ. 2021. Research capacity building with mycetoma as the central research challenge. Trans R Soc Trop Med Hyg 115:436. 10.1093/trstmh/traa136. [DOI] [PubMed] [Google Scholar]
- 299.Traxler RM, Beer KD, Blaney DD, van de Sande WWJ, Fahal AH, Asiedu KB, Bower WA, Chiller T. 2021. Development of the global mycetoma working group. Transactions of the Royal Society of Tropical Medicine and Hygiene 115:437–440. 10.1093/trstmh/traa163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Ahmed AA, van de Sande W, Fahal AH. 2017. Mycetoma laboratory diagnosis: Review article. PLoS Negl Trop Dis 11:e0005638. 10.1371/journal.pntd.0005638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Bahar ME, Bakheet OELH, Fahal AH. 2021. Mycetoma imaging: the best practice. Trans R Soc Trop Med Hyg 115:387–396. 10.1093/trstmh/traa178. [DOI] [PubMed] [Google Scholar]
- 302.Bakhiet SM, Fahal AH, Musa AM, Mohamed ESW, Omer RF, Ahmed ES, El Nour M, Mustafa ERM, Sheikh ARME, Suliman SH, El Mamoun MAG, El Amin HM. 2018. A holistic approach to the mycetoma management. PLoS Negl Trop Dis 12:e0006391. 10.1371/journal.pntd.0006391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Mitja O, Marks M, Bertran L, Kollie K, Argaw D, Fahal AH, Fitzpatrick C, Fuller LC, Garcia Izquierdo B, Hay R, Ishii N, Johnson C, Lazarus JV, Meka A, Murdoch M, Ohene SA, Small P, Steer A, Tabah EN, Tiendrebeogo A, Waller L, Yotsu R, Walker SL, Asiedu K. 2017. Integrated control and management of neglected tropical skin diseases. PLoS Negl Trop Dis 11:e0005136. 10.1371/journal.pntd.0005136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Roberto E, Guadalupe C-L, Guadalupe E-C, Hay R. 2021. Mycetoma and the community dermatology program, Mexico. Trans R Soc Trop Med Hyg 115:383–386. 10.1093/trstmh/traa199. [DOI] [PubMed] [Google Scholar]