ABSTRACT
The global dissemination of the mobile colistin resistance (mcr) gene illustrates how the use of colistin in veterinary medicine can affect human health, exemplifying the concept of One Health. This study screened for the existence of mcr variants (from mcr-1 to mcr-10) in a 5-year collection of clinical Klebsiella short-read whole-genome sequencing (WGS) data from a tertiary hospital in China (2013 to 2018) and aimed to identify the mechanisms of mcr spread. MICs were measured for the mcr-positive isolates, and long-read sequencing was performed to complete the mcr-positive genome sequences. Six variants (mcr-1.1, mcr-8.1, mcr-8.2, mcr-9.1, mcr-9.2, and mcr-10.1) were identified in 20 genomes, with plasmids from the IncFIIK, IncHI2, IncI2, and IncX4 groups. Highly similar plasmids (coverage, >75%; nucleotide identity, >98.5%) isolated from silver gulls, chickens, pigs, wastewater treatment plants, and hospital sewage were identified in GenBank. The MICs of the mcr-1- and mcr-8-carrying isolates were ≥4 μg/mL; however, the MICs of the mcr-9- and mcr-10-carrying isolates ranged from 0.5 μg/mL to 1 μg/mL (colistin susceptible). The variants mcr-1 to mcr-9 were found only in Klebsiella pneumoniae, while mcr-10.1 was found in K. pneumoniae, Klebsiella quasipneumoniae subsp. quasipneumoniae, and Klebsiella variicola. A pair of inverted repeats (IRs) was identified for hsdSMR-ISEc36-mcr-10.1-xerC; IR-1 (5′-TCAAACGTA) was inside the attL site of xerC, indicating that mcr-10.1 was originally integrated by xerC and mobilized by ISEc36 afterwards. In conclusion, this is the first report of mcr-10.1 susceptible to colistin in three species of Klebsiella. This study shows the genetic events that happened to mcr-10.1 in a stepwise manner, with the first step being XerC integration and the second being ISEc36 mobilization. Finally, this study also highlights mcr transmission between humans and nature.
IMPORTANCE Reports of mcr-1 and mcr-8 are common in China; however, few studies have reported mcr-9 and mcr-10. One reason is that the newly described variants can be phenotypically colistin susceptible and thus may not be identified. This study identified the mcr-positive clinical isolates by investigating WGS data for 2,855 Klebsiella isolates (including K. pneumoniae, K. quasipneumoniae subsp. quasipneumoniae, and K. variicola) and found three mcr-9 and three mcr-10 cases (MICs, 0.5 μg/mL to 1 μg/mL; colistin susceptible). This study also reveals a pair of perfect 9-bp IRs of ISEc36 and the precise mcr-10.1 integration and insertion events that happened to the IncFIIK plasmids. A One Health analysis of highly similar plasmid structures from human and nonhuman sources emphasizes the plasmid transmission and evolution process.
KEYWORDS: mobile colistin resistance, mcr-10, Klebsiella pneumoniae, China, One Health, colistin resistance, mcr
INTRODUCTION
Colistin (polymyxin E) is extensively used in treating food-producing animals but was banned from human use in the 1970s due to its nephrotoxicity and neurotoxicity (1). However, with the evolution and increase in multidrug-resistant (MDR) bacteria, particularly carbapenem-resistant Enterobacteriaceae (CRE), in hospitals around the world, colistin has been reintroduced to clinical medicine. Unfortunately, resistance to colistin subsequently appeared in both human and nonhuman cases, indicating a zoonotic disseminating issue which should be addressed using a One Health perspective (2–4).
Colistin resistance can be caused by either chromosomal mutations or mobile colistin resistance (mcr) gene acquisition (5). In Enterobacteriaceae strains, mcr acquisition is more concerning, as the mcr gene can rapidly disseminate resistance via horizontal gene transfer (HGT). The mcr gene encodes a phosphoethanolamine transferase that can modify the lipid A component of lipopolysaccharide. The common mobile genetic elements (MGEs) assisting mcr mobilization include insertion sequences (ISs; e.g., ISApl1, IS903B, ISEcl1, ISKpn26, etc.), recombinases, integrases, and plasmids from the IncI, IncH, IncX, and IncF incompatibility groups (3, 6).
As of 2021, a total of 10 mcr homologs have been identified (6). The first to be reported was mcr-1 from pigs and chickens in China (7). Since then, mcr-1 has been widely described, especially in Escherichia coli (8, 9). In contrast, the other mcr variants have been reported in a limited number of species (10). In Klebsiella, mcr-8 and mcr-9 have been reported. The mcr-8 gene was initially identified in the K. pneumoniae plasmid pKP91 of swine origin (11), and mcr-9 was detected in silico from a clinical colistin-susceptible Salmonella enterica serotype Typhimurium strain isolated in 2010 in the United States (12). The expression of mcr-9 normally requires exposure to colistin and a qseBC system next to mcr-9 for expression induction (3). Notably, the IncHI2 plasmid is the predominant plasmid type disseminating mcr-9 (13); this plasmid is widely known for its carriage of multiple antimicrobial-resistant genes (ARGs), including the carbapenemase genes (3). The latest identified mcr variant is mcr-10, which was originally detected in the clinical Enterobacter roggenkampii strain 090065 in China, showing colistin MIC values ranging from 2 μg/mL (susceptible) to 4 μg/mL (resistant) (6). High-level colistin resistance (MIC, ≥16 μg/mL) of mcr-10 could be screened out using high concentrations of colistin (4 μg/mL to 8 μg/mL) (2). High mRNA levels of phoPQ were observed for the high-level resistance induced by the high colistin pressures. The mcr-10 variants have been reported in IncF plasmids, including the IncFIA, IncFIB, and IncFII groups, in six genera (Enterobacter, Klebsiella, Escherichia, Citrobacter, Kluyvera, and Raoultella) (6).
In this study, we (i) conducted a genome surveillance study of clinical mcr variants in K. pneumoniae over 5 years, (ii) determined the clinical features and phenotypic impacts of the mcr-carrying isolates, (iii) characterized the mcr-carrying isolates from species level to the mcr-surrounding genetic context, and (iv) linked the mcr-carrying plasmids from human sources to plasmids from other sources in nature.
RESULTS
Overview of the 20 mcr-carrying K. pneumoniae genomes.
Of the 2,855 Klebsiella genomes, 20 (0.70%) were positive for mcr. The median age of the infected patients was 52 years (ranging from 1 month to 78 years), and 17 (85%) were male (Table 1). Eleven patients were hospitalized, and nine were outpatients. The isolates were isolated from sputum (n = 9), blood (n = 7), urine (n = 2), drainage fluid (n = 1), and wound secretions (n = 1). Notably, 18 were MDR with phenotypic resistance to more than three antibiotic categories, and 17 were extended-spectrum beta-lactamase (ESBL) producers. Only four were carbapenem resistant and carried blaNDM-5. Comorbidities included cerebral injury (n = 4), malignant spinal tumor (n = 1), severe acute pancreatitis (n = 1), gastric cancer (n = 1), coronary artery disease (n = 1), aplastic anemia (n = 1), and acute lymphoblastic leukemia (ALL; n = 1; died with isolates carrying both mcr-8.1 and blaNDM-5).
TABLE 1.
Clinical information and genomic characterization of the 20 mcr-carrying plasmids
| Strain | Yr | Age (yr) | Gendera | Specimen type | Hospitalized | MDR | ESBL | CRE | mcr variant | Colistin MIC | Sp.b | MLST | Inc groupc | Plasmid size (kb) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 29-72 | 2014 | 49 | M | Sputum | + | − | − | − | 1.1 | 4 | KpI | ST43 | X4 | 34 |
| 15-81 | 2016 | 78 | M | Sputum | + | + | + | − | 1.1 | 4 | KpI | ST1 | X4 | 34 |
| 17-6 | 2016 | 66 | F | Blood | + | + | + | − | 1.1 | 16 | KpI | ST268 | I2 | 66 |
| 17-66 | 2016 | 19 | M | Sputum | + | + | + | − | 1.1 | 16 | KpI | ST268 | I2 | 66 |
| 1-17 | 2017 | 38 | M | Abdominal drainage | − | + | + | + | 1.1 | 16 | KpI | ST5258 | FIIK | 66 |
| 5-30d | 2017 | 8 | M | Sputum | − | + | + | − | 1.1 | 8 | KpI | ST39 | HI2 | 266 |
| 19-40 | 2016 | 37 | M | Sputum | + | + | + | + | 8.1 | 8 | KpI | ST685 | FIIK | 95 |
| 38-73 | 2016 | 37 | M | Blood | + | + | + | + | 8.1 | 8 | KpI | ST685 | FIIK | 95 |
| 38-72 | 2016 | 37 | M | Blood | + | + | + | + | 8.1 | 8 | KpI | ST685 | FIIK | 104 |
| 1-36 | 2017 | 52 | M | Urine | − | + | + | − | 8.1 | >64 | KpI | ST395 | FIIK | 110 |
| 1-44 | 2017 | 70 | M | Blood | − | + | + | − | 8.1 | >64 | KpI | ST395 | FIIK | 110 |
| 4-35 | 2017 | 70 | M | Sputum | − | + | + | − | 8.1 | >64 | KpI | ST395 | FIIK | 110 |
| 29-21 | 2015 | 63 | M | Blood | + | + | − | − | 8.2 | 4 | KpI | ST39 | FIIK | 86 |
| 11-62 | 2016 | 58 | M | Sputum | − | + | + | − | 8.2 | 4 | KpI | ST1 | FIIK | 97 |
| 36-80d | 2018 | 68 | M | Sputum | − | + | + | − | 9.1 | 1 | KpI | ST412 | HI2 | 242 |
| 28-33 | 2015 | 44 | F | Urine | − | + | + | − | 9.2 | 1 | KpI | ST3687 | HI2 | 248 |
| 28-1-0 | 2015 | 66 | M | Wound | − | + | + | − | 9.2 | 1 | KpI | ST3687 | HI2 | 251 |
| 31-62d | 2014 | 66 | M | Blood | + | − | − | − | 10.1 | 1 | KpI | ST1087 | FIIK | 153 |
| 25-52 | 2015 | 16 | F | Blood | + | + | + | − | 10.1 | 1 | KpII-A | ST5281 | FIIK | 91 |
| 15-76 | 2016 | 0.1 | M | Sputum | + | + | + | − | 10.1 | 0.5 | KpIII | ST5266 | FIIK | 128 |
Gender: M, male; F, female.
Sp., species complex; KpI, K. pneumoniae; KpII-A, K. quasipneumoniae subsp. quasipneumoniae; KpIII, K. variicola.
Inc group, incompatibility group.
Completed by hybrid assembly using both short and long reads.
A total of six mcr variants were identified in the 20 genomes, including mcr-1.1 (n = 6), mcr-8.1 (n = 6), mcr-8.2 (n = 2), mcr-9.1 (n = 1), mcr-9.2 (n = 2), and mcr-10.1 (n = 3). All but two mcr-positive genomes were K. pneumoniae. The remaining two were Klebsiella quasipneumoniae subsp. quasipneumoniae and Klebsiella variicola, both of which carried mcr-10.1. MLST revealed that the mcr-carrying genomes were from a variety of clones, indicating that the mcr dissemination was not due to clonal spread. Both the mcr-9- and mcr-10-carrying isolates were colistin susceptible (n = 6; MIC, 0.5 μg/mL to 1 μg/mL), while the MIC values of the remaining 14 were ≥4 μg/mL. No qseBC gene was found in the genomic contigs of the mcr-carrying isolates. Chromosomal genes (phoP/phoQ, pmrA/pmrB, and mgrB) known to be associated with colistin susceptibility and mcr expression regulation were investigated by amino acid sequences aligned to the reference gene sequences of K. pneumoniae MGH 78578 (GenBank accession number CP000647.1) (see Fig. S1 in the supplemental material). phoQ and pmrAB sequences in mcr-10.1-carrying K. quasipneumoniae subsp. quasipneumoniae and K. variicola genomes had more nonsynonymous mutations (n = 36) than the sequences in the mcr-10.1-carrying K. pneumoniae genome (n = 1).
Plasmid typing revealed that the most common mcr-carrying plasmid type was IncFIIK (n = 12), followed by IncHI2 (n = 4), IncI2 (n = 2), and IncX4 (n = 2). Further investigation of the mcr genetic contexts was performed, and relatively consistent genetic patterns were found, i.e., ISApl1 upstream of mcr-1.1 and pap2 (ISApl1-mcr-1.1-pap2; n = 4); IS903B downstream of mcr-8.1 with no flanking transposons upstream (dgkA-beaS-copR-mcr-8.1-//-IS903B; n = 6); ISEcl1 upstream of mcr-8.2 (ISEcl1-mcr-8.2-//-ISKpn26; n = 2); IS903B upstream of mcr-9.1 and mcr-9.2 (IS903B-mcr-9; n = 3); and xerC upstream of mcr-10.1 (xerC-mcr-10.1; n = 3) (Fig. S2).
Genetic characterization of the mcr-carrying plasmids.
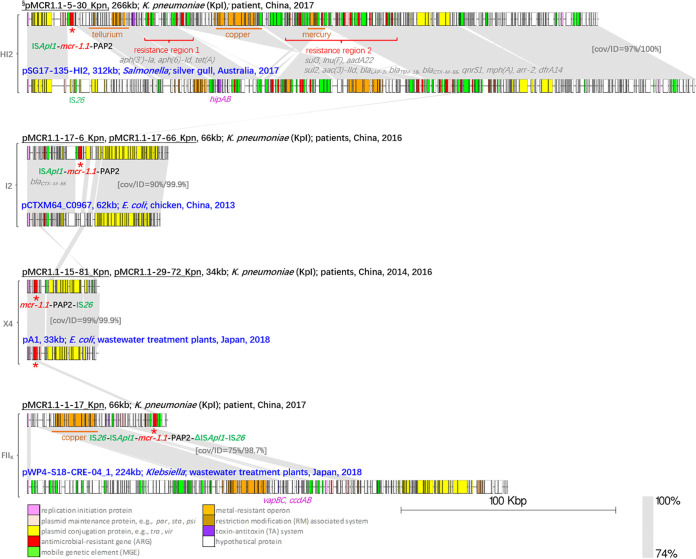
Of the six mcr-1.1-positive plasmids, one was carried by an IncHI2 plasmid, two by IncI2 plasmids, two by IncX4 plasmids, and one by an IncFIIK plasmid. It is interesting to note that pMCR1.1-5-30_Kpn (completed by hybrid assembly using both short and long reads), identified in this study, shared a similar IncHI2 backbone with pSG17-135-HI2 (GenBank accession number CP048776.1), a plasmid isolated from a gull in Australia (Fig. 1). Both pMCR1.1-5-30_Kpn and the mcr-free plasmid pSG17-135-HI2 carried resistance region 1 (three ARGs) and resistance region 2 (12 ARGs), conferring resistance to aminoglycosides [aph(3′)-Ia, aph(6)-Id, aadA22, and aac(3)-IId], tetracycline [tet(A)], sulfonamides (sul3 and sul2), lincomycin [lnu(F)], beta-lactams (blaLAP-2, blaTEM-1B, and blaCTX-M-55), quinolone (qnrS1), macrolides [mph(A)], rifamycin (arr-2), and trimethoprim (dfrA14), in addition to colistin (mcr-1.1). In addition, three heavy metal resistance operons (terABCDWXY, cusABCDRS-copABCDEFG-pcoERS, and a truncated merCDEPTR) and one toxin-antitoxin (TA) system (hipAB) were identified in pMCR1.1-5-30_Kpn. Shorter mcr-1.1-carrying plasmids were found in the other three plasmid incompatibility groups. The closest reference of the IncI2 mcr-1.1-carrying plasmids was pCTXM64_C0967 (KP091735.1), a blaCTX-M-64-carrying and mcr-free plasmid from a chicken source. An average nucleotide identity (ANI) of 99.9% was found between them. The linear map indicated that the colocation of mcr-1.1 and blaCTX-M evolved from the blaCTX-M-carrying plasmid. A 99.9% ANI and 99% coverage were found between the mcr-1.1-carrying IncX4 plasmids identified in this study and pA1 (LC477138.1), a mcr-1.1-carrying plasmid from a Japanese E. coli strain from a wastewater treatment plant. Another plasmid isolated from a wastewater treatment plant in Japan, pWP4-S18-CRE-04_1 (AP022079.1), was a close match of pMCR1.1-1-17_Kpn. The copper resistance operon and the TA systems of vapBC and ccdAB were shared between them; however, no IncFIIK tra region was identified in the genomic contigs of pMCR1.1-1-17_Kpn.
FIG 1.
Comparison of plasmids harboring mcr-1.1 obtained in this study with those reported from nonhuman sources. The complete plasmids are grouped by the incompatibility groups IncHI2, IncI2, IncX4, and IncFIIK. The plasmid name, length in kilobases, species, and isolation information (source, location, and year) are shown above each sequence. Plasmids from this study are labeled in black, and plasmids from nonhuman sources are labeled in blue. §, plasmid sequences that were completed by hybrid assembly; Δ, truncated insertion sequences. The mcr genes are indicated by a red asterisk (*), and the mcr genetic context arrays are shown next to the asterisk. Heavy metal resistance operons and resistance regions are labeled in orange and red, respectively. Gray shading connects highly similar sequences. cov, coverage; ID, identity.
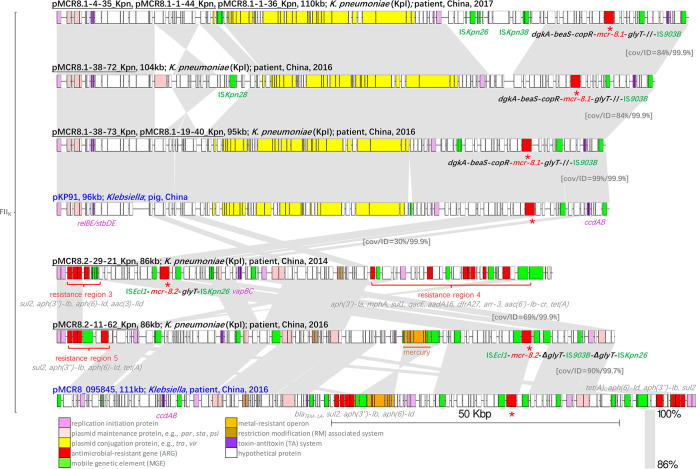
All eight mcr-8-carrying plasmids were from the IncFIIK group. However, the IncFIIK plasmid backbone of the mcr-8.1-carrying plasmids differed from that of the mcr-8.2-carrying plasmids, indicating that the transmission dynamics of the dominant mcr variants in this study were diversified (Fig. 2). The mcr-8.1-carrying plasmids shared a more conserved backbone with a long tra region, while the mcr-8.2-carrying plasmids shared a less conserved backbone, showing higher genetic plasticity with multiple resistance regions. The clinical mcr-8.1-carrying plasmids in this study showed 99.9% ANI with pKP91 (GenBank accession number MG736312.1), the initial report of mcr-8.1 in K. pneumoniae. The longest mcr-8.1-carrying plasmids (110 kb) were all isolated in 2017 and harbored an additional fragment containing heat shock proteins mobilized by multiple insertion sequences, mainly ISKpn26 and ISKpn38, compared to the shorter mcr-8.1-carrying plasmids isolated in 2016. Plasmid pMCR8.1-38-72_Kpn possessed an ISKpn28 inserted region composed of the plasmid partition protein ParB, the single-stranded DNA binding protein Ssb, the reverse transcriptase protein LtrA, and the SOS inhibitors PsiA and PsiB. By comparison, the mcr-8.2-carrying plasmids were less conserved, with multiple regions being inserted, and no close match was found in nonhuman sources. The clinical plasmid pMCR8_095845 (CP031883.1) was similar to pMCR8.2-11-62_Kpn, with resistance region 5 (four ARGs), ccdAB, antirestriction systems, and one mercury resistance operon (merCDEPTR). In pMCR8.2-29-21_Kpn, resistance region 3 differed from resistance region 5 (four ARGs) by only one gene [aac(3)-IId in region 3 versus tet(A) in region 5]. Moreover, a larger resistance region 4, constituting nine ARGs, was identified in pMCR8.2-29-21_Kpn.
FIG 2.
Comparison of IncFIIK plasmids harboring mcr-8 obtained in this study and those reported from nonhuman sources. The plasmid name, length in kilobases, species, and isolation information (source, location, and year) are shown above each sequence. Plasmids from this study are labeled in black, and plasmids from previous studies are labeled in blue. The mcr genes are indicated by a red asterisk (*), and the mcr genetic context arrays are shown next to the asterisk. Heavy metal resistance operons and resistance regions are labeled in orange and red, respectively. Gray shading connects highly similar sequences. cov, coverage; ID, identity.
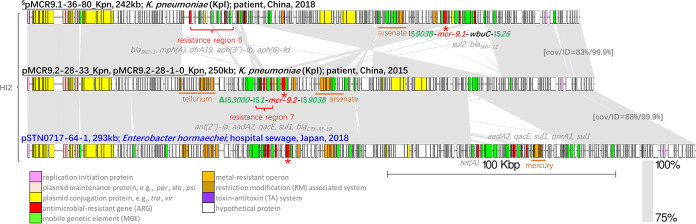
Three large IncHI2 plasmids were obtained from the mcr-9-carrying genomes. Despite a large fragment reversed between the mcr-9.1-carrying pMCR9.1-36-80_Kpn (completed by hybrid assembly) and the mcr-9.2-carrying pMCR9.2-28-33_Kpn and pMCR9.2-28-1-0_Kpn, all plasmids shared a conserved IncHI2 plasmid backbone (coverage, >83%; ANI, >99.9%) (Fig. 3). Insertion of resistance regions is the main reason for the large plasmid sizes. pMCR9.1-36-80_Kpn possessed resistance region 6, consisting of five ARGs, and the mcr-9.2-carrying plasmids harbored resistance region 7, also composed of five ARGs. Their closest match, pSTN0717-64-1 (GenBank accession number AP022511.1) from hospital sewage in Japan, also contained resistance region 7 and an additional ARG array, ranging from tet(A) to sul1. No qseBC gene was identified in the contigs of the mcr-9-carrying genomes.
FIG 3.
Comparison of IncHI2 plasmids harboring mcr-9 obtained in this study and a plasmid from a nonhuman source. The plasmid name, length in kilobases, species, and isolation information (source, location, and year) are shown above each sequence. Plasmids from this study are labeled in black, and pSTN0717-64-1 (GenBank accession number AP022511.1) is labeled in blue. §, plasmid sequences that were completed by hybrid assembly. The mcr genes are indicated by a red asterisk (*), and the mcr genetic context arrays are shown next to the asterisk. Heavy metal resistance operons and resistance regions are labeled in orange and red, respectively. Gray shading connects highly similar sequences. cov, coverage; ID, identity.
Diversified genetic contexts of mcr-10.1 due to insertions.
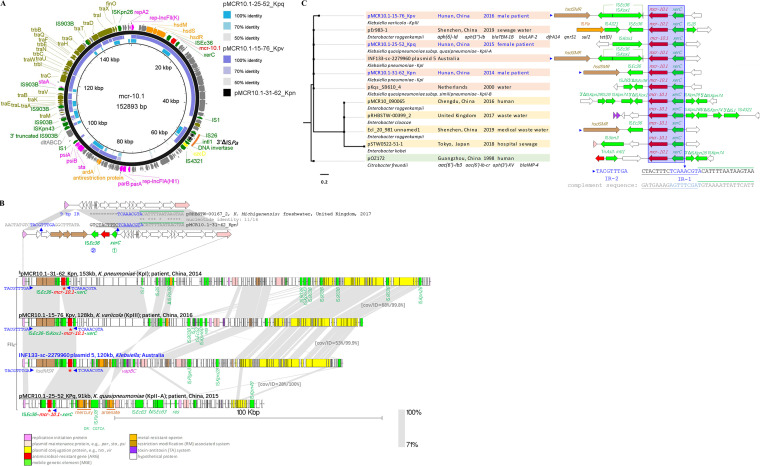
Three mcr-10.1-carrying IncFIIK plasmids were identified in three Klebsiella species. The genetic contexts of mcr-10.1 were relatively conserved, with an immediately upstream xerC and a downstream ISEc36 (Fig. S2d). Although the genetic context in pMCR10.1-15-76_Kpv was interrupted by an ISKox1, the flanking sequences downstream of ISEc36 remained unchanged.
The complete sequence of pMCR10.1-31-62_Kpn was obtained by hybrid assembly. Plasmid pMCR10.1-15-76_Kpv from K. variicola showed 53% coverage with pMCR10.1-31-62_Kpn from K. pneumoniae, while pMCR10.1-25-52_KPq from K. quasipneumoniae subsp. quasipneumoniae only shared 19% (Fig. 4A). All three plasmids contained the main genetic elements of the replication-associated genes of IncFIIK, the ISEc36-mcr-10.1-xerC genetic context, a 3′ truncated ISPa38 from the Tn3 family, and parts of the IncFIIK tra region. The mcr-10.1-carrying plasmids obtained in 2014 and 2016 during this study showed a relatively high diversity compared to the other mcr-carrying plasmids in this study.
FIG 4.
Comparison of the mcr-10.1-carrying IncF plasmids. (A) Circular map showing the three mcr-10.1-carrying IncFIIK plasmids identified in this study. All plasmids were aligned to pMCR10.1-31-62_Kpn (completed by hybrid assembly; 152,983 bp). Functional genes are labeled in different colors. (B) Schematic representation showing the hsdSMR-ISEc36-mcr-10.1-xerC insertion event and comparison of the mcr-10.1-carrying IncFIIK plasmids. The 9-bp inverted repeats (IRs) are indicated in blue. The two 16-bp parts of the entire 32-bp xerC attL sequence are indicated by underlining and a green overline. Identical bases are indicated by asterisks (*). (C) Phylogenetic tree of the mcr-10.1-carrying IncF plasmids. The isolation information (source, location, and year), mcr-10.1 genetic contexts, and additional antimicrobial-resistant genes are shown. The 9-bp IRs are indicated with arrows and highlighted in blue text. The 32-bp xerC attL site is indicated by underlining and overlines. The complement sequence of attL is also shown. Δ, truncated insertion sequences.
To fully understand the integration and insertion events of mcr-10.1 in our ISEc36-mcr-10.1-xerC-carrying plasmids, a BLAST search for highly similar sequences was performed, and the mcr-free IncFIIK plasmid pRHBSTW-00167_2 (GenBank accession number CP058119.1) was noted. By carefully comparing pRHBSTW-00167_2 and pMCR10.1-31-62_Kpn, a pair of 9-bp perfect IRs were identified (Fig. 4B). IR-1 (5′-TCAAACGTA) was 95 bp upstream of xerC and located inside the 32-bp left attachment site (attL; 5′-CTACTTTCTCAAACGTACATTTTAATAAGTAA), and IR-2 (5′-TACGTTTGA) was located downstream of mcr-10.1 and a type I restriction-modification system encoded by hsdSMR. Two 16-bp attL parts contained 11 identical nucleotides. No attR was identified in our three mcr-10.1-carrying plasmids. The presence of perfect IRs and the absence of xerC attR indicated that the ISEc36 insertion event happened after xerC integration and destroyed the complete xerC attR site. The plasmid linear map further revealed that the mcr-10.1-carrying plasmids were diversified by a variety of ISs. However, no additional ARG was found in the mcr-10.1-carrying IncFIIK plasmids. The mercury-resistant and arsenate-resistant operons were found in the shortest mcr-10.1-carrying plasmid, pMCR10.1-25-52_KPq from K. quasipneumoniae subsp. quasipneumoniae. In addition, a complete ISPa38 (also called TnPa38) was identified in pMCR10.1-25-52_KPq, with a pair of 5-bp direct repeats (DRs) of 5′-CGTCA. The closest sequence from a public database was the IncFIIK plasmid 5 in the K. pneumoniae genome of INF133-sc-2279960 (LR890193.1) from Australia. This plasmid also contained the hsdSMR-ISEc36-mcr-10.1-xerC genetic context, with the 9-bp IRs present on both sides.
In addition, the phylogenetic structure of mcr-10.1-carrying IncF plasmids and additional mcr-10.1 genetic contexts were inspected to understand mcr-10.1 evolution. The maximum likelihood trees divided 11 highly related mcr-10.1-carrying plasmids into two main groups (Fig. 4C). The mcr-10.1-carrying IncFIIK plasmids from this study were grouped into one with homologous IncFII plasmids from water. The mcr-10.1-carrying IncFIA and IncFIB plasmids formed another group. Noticeably, xerC was the only conserved MGE adjacent to mcr-10.1. The downstream of mcr-10.1 was divergent. IR-1 located exactly in xerC attL was present in all mcr-10.1-carrying plasmids, while IR-2 only existed in the hsdSMR-ISEc36-mcr-10.1-xerC-carrying sequences.
DISCUSSION
This study investigated the genetic relatedness of mcr-carrying plasmids in Klebsiella isolates recovered from humans in China with publicly available plasmids from nonhuman sources using a One Health perspective and aimed to establish potential links to understand the ways in which the plasmids evolved and were disseminated between animals, humans, plants, and the environment. This study also characterized the prevalence, clinical features, and phenotypic characteristics of the clinical mcr-positive Klebsiella genomes. It is the first worldwide report of mcr-10.1 susceptible to colistin in clinically uncommon species of K. quasipneumoniae subsp. quasipneumoniae and K. variicola. The mcr-10.1 was found embedded in a genetic structure (hsdSMR-ISEc36-mcr-10.1-xerC) with a pair of IRs.
This study demonstrates that approximately 0.70% of clinical Klebsiella genomes were mcr positive in a 5-year period before the implementation of the colistin withdrawal policy in China in 2018 (14). The overall mcr-positive rate is comparable with other reports (4, 7). The mcr-8 homolog is predominant in our collection, with mcr-1 being the second most common. Unlike the situation in Australia (3), cocarriage of the carbapenemase gene and mcr was rare in this study, occurring in only one isolate; however, the infected patient was dead in this study (15).
The identification of mcr in both human and nonhuman sources, along with the fact that colistin is heavily used in industry rather than in humans, together propose the transmission of colistin resistance in a One Health manner (4). By including humans, animals, and environmental sectors in mcr-carrying plasmid structure analysis, we found highly similar plasmid backbones from a silver gull, food animals, wastewater treatment plants, and hospital sewage. The nonhuman plasmids shared a wide range of genetic elements, including large antibiotic resistance regions and heavy metal-resistant operons, with our mcr-carrying plasmids from patients. The shared ARGs, conferring resistance to a broad spectrum of antibiotics not limited to the drugs regularly used clinically, suggest that the presence of mcr in our collection is not due to colistin pressure in the hospital but is more likely to be acquired from food animals and waste sources (9). The carriage of multiple large resistance regions in the mcr-positive IncHI2 plasmids with tra regions indicates that the mcr-carrying IncHI2 plasmids pose a greater threat to public health (3).
Our shotgun WGS study demonstrated that WGS is an effective screen for important genes, especially the silent mcr-9.1, mcr-9.2, and mcr-10.1 variants (9). Both mcr-9 and mcr-10 have been reported as mediating low-level resistance to colistin, although the mechanism remains unclear (3). The activity of mcr-10 against colistin could be even lower than that of mcr-9, as indicated by the colistin killing assay (2). Our susceptibility test results were similar to those of the previous study, with lower colistin MIC values for mcr-9 and mcr-10 (0.5 μg/mL to 1 μg/mL in K. pneumoniae, K. quasipneumoniae subsp. quasipneumoniae, and K. variicola versus 2 μg/mL to 4 μg/mL in Enterobacter roggenkampii and E. coli) (6). The WGS data provide all gene and promoter sequences of the colistin resistance associated genes and allow for comparative genomic analysis (Fig. 4; see Fig. S1 in the supplemental material). However, due to the lack of mcr-10-carrying Klebsiella genomes with colistin-resistant phenotypes, further investigation is needed to understand the key reason for the colistin susceptibility of the mcr-10-positive Klebsiella isolates.
The complementary long-read sequencing technology also helped to elucidate the mcr-carrying plasmid structures. The plasmid structures of mcr-10.1 were divergent (less than 70% coverage) (2), which may be due to the mcr carriage on the hyperdivergent IncF-type plasmids (13) and in different species of Klebsiella. However, the mcr-10.1-adjacent MGE was conserved; an XerC-type recombinase was consistently identified directly upstream of mcr-10.1 (6). Interestingly, only xerC attL was identified in our mcr-10.1-carrying plasmids, indicating that the acquisition of mcr-10.1 might have occurred after the xerC integration of mcr-10.1. The identification of the perfect 9-bp IR pair further confirmed that mcr-10.1 in our collection was acquired by ISEc36 insertion of the mcr-10.1-xerC fragment. The insertion of ISEc36-mcr-10.1-xerC may also bring an additional type I restriction modification (RM) system encoded by hsdSMR. ISEc36 was initially identified in E. coli strain W635, carrying the carbapenemase gene blaIMI-2 (16).
Conclusion.
This study characterized 20 mcr-carrying plasmids from a One Health perspective. This study reports the colistin-susceptible mcr-10.1 variant in K. quasipneumoniae subsp. quasipneumoniae and K. variicola for the first time and shows a pair of perfect IRs next to mcr-10.1, illustrating ISEc36 mobilizing mcr-10.1-xerC and hsdSMR in a stepwise manner.
MATERIALS AND METHODS
Bacterial genomes and study design.
A total of 2,855 unique Klebsiella genomes from a surveillance study were included (17). Isolates were recovered from patient specimens at a hospital in the central part of China between 5 January 2013 and 24 July 2018. Multilocus sequence typing (MLST) was done using the Klebsiella Pasteur MLST database (https://bigsdb.pasteur.fr/klebsiella/). ARGs (including the mcr variants) and plasmid incompatibility groups were assigned using ResFinder version 4.0 (18) and PlasmidFinder version 2.1 (19), respectively, via the ABRicate pipeline (https://github.com/tseemann/abricate).
Ethics.
Written informed consent was obtained from all participants. This study was approved by the ethics committees under the tracking numbers 201806861 and BGI-IRB 18061-T2.
Scaffolding of the mcr-carrying plasmids.
The sizes of the mcr-carrying contigs ranged from 1,948 bp to 59,684 bp (20). The contigs were searched using BLAST against the nonredundant/nucleotide (nr/nt) database to find suitable plasmid references. Scaffolding of the mcr-carrying plasmid contigs was completed using assembly_improvement version 1.7.0 (21) and was manually checked by mapping reads to the scaffolds using Geneious 9.1.8. In addition, three genomes carrying mcr-1.1 (strain 5-30), mcr-9.1 (strain 36-80), and mcr-10.1 (strain 31-62) were sequenced on a MinION device (Oxford Nanopore Technologies, UK). Unicycler version 0.4 was deployed to achieve the hybrid assembly of both the short reads (BGISEQ-500 [MGI, China]) and long reads (MinION) (https://github.com/rrwick/Unicycler). The mcr-carrying scaffolds were also checked and assessed by mapping the raw reads back to the assembled scaffolds.
Antimicrobial susceptibility assays.
Antimicrobial susceptibility tests (ASTs), with the exception of colistin, were performed using the Vitek 2 Compact system (BioMérieux, Marcy-l’Etoile, France) according to the manufacturer’s instructions. The susceptibility results were interpreted according to the Clinical and Laboratory Standards Institute guidelines (22). The MIC values for colistin were determined using the broth microdilution method in cation-adjusted Mueller-Hinton broth, and the results were interpreted following the European Committee on Antimicrobial Susceptibility Testing (EUCAST) breakpoints (susceptible, ≤2 μg/mL; resistant, >2 μg/mL; https://www.eucast.org/clinical_breakpoints). No induction assays were performed, as qseBC was not identified in the 20 mcr-carrying genomes.
Data availability.
The data that support the findings of this study have been deposited at the China National GeneBank DataBase (CNGBdb) in the CNGB Sequence Archive (CNSA) under accession number CNP0001198 (23, 24).
ACKNOWLEDGMENTS
This work was supported by the China National GeneBank (CNGB). We declare no competing interests.
Footnotes
Supplemental material is available online only.
Contributor Information
Wenen Liu, Email: wenenliu@163.com.
Junhua Li, Email: lijunhua@genomics.cn.
Na Pei, Email: peina@genomics.cn.
Katharina Schaufler, University of Greifswald.
REFERENCES
- 1.Poirel L, Jayol A, Nordmann P. 2017. Polymyxins: antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes. Clin Microbiol Rev 30:557–596. doi: 10.1128/CMR.00064-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xu T, Zhang C, Ji Y, Song J, Liu Y, Guo Y, Zhou K. 2021. Identification of mcr-10 carried by self-transmissible plasmids and chromosome in Enterobacter roggenkampii strains isolated from hospital sewage water. Environ Pollut 268:115706. doi: 10.1016/j.envpol.2020.115706. [DOI] [PubMed] [Google Scholar]
- 3.Macesic N, Blakeway LV, Stewart JD, Hawkey J, Wyres KL, Judd LM, Wick RR, Jenney AW, Holt KE, Peleg AY. 2021. Silent spread of mobile colistin resistance gene mcr-9.1 on IncHI2 “superplasmids” in clinical carbapenem-resistant Enterobacterales. Clin Microbiol Infect 27:1856.e7–1856.e13. doi: 10.1016/j.cmi.2021.04.020. [DOI] [PubMed] [Google Scholar]
- 4.Shi X, Li Y, Yang Y, Shen Z, Wu Y, Wang S. 2020. Global impact of mcr-1-positive Enterobacteriaceae bacteria on “one health.” Crit Rev Microbiol 46:565–577. doi: 10.1080/1040841X.2020.1812510. [DOI] [PubMed] [Google Scholar]
- 5.Wang C-H, Siu LK, Chang F-Y, Chiu S-K, Lin J-C. 2021. A resistance mechanism in non-mcr colistin-resistant Escherichia coli in Taiwan: R81H substitution in PmrA is an independent factor contributing to colistin resistance. Microbiol Spectr 9:e0002221. doi: 10.1128/Spectrum.00022-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang C, Feng Y, Liu L, Wei L, Kang M, Zong Z. 2020. Identification of novel mobile colistin resistance gene mcr-10. Emerg Microbes Infect 9:508–516. doi: 10.1080/22221751.2020.1732231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu Y-Y, Wang Y, Walsh TR, Yi L-X, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, Yu L-F, Gu D, Ren H, Chen X, Lv L, He D, Zhou H, Liang Z, Liu J-H, Shen J. 2016. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 8.Shen Y, Zhou H, Xu J, Wang Y, Zhang Q, Walsh TR, Shao B, Wu C, Hu Y, Yang L, Shen Z, Wu Z, Sun Q, Ou Y, Wang Y, Wang S, Wu Y, Cai C, Li J, Shen J, Zhang R, Wang Y. 2018. Anthropogenic and environmental factors associated with high incidence of mcr-1 carriage in humans across China. Nat Microbiol 3:1054–1062. doi: 10.1038/s41564-018-0205-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sia CM, Greig DR, Day M, Hartman H, Painset A, Doumith M, Meunier D, Jenkins C, Chattaway MA, Hopkins KL, Woodford N, Godbole G, Dallman TJ. 2020. The characterization of mobile colistin resistance (mcr) genes among 33000 Salmonella enterica genomes from routine public health surveillance in England. Microb Genom 6:e000331. doi: 10.1099/mgen.0.000331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Phetburom N, Boueroy P, Chopjitt P, Hatrongjit R, Akeda Y, Hamada S, Nuanualsuwan S, Kerdsin A. 2021. Klebsiella pneumoniae complex harboring mcr-1, mcr-7, and mcr-8 isolates from slaughtered pigs in Thailand. Microorganisms 9:2436. doi: 10.3390/microorganisms9122436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu B, Wang Y, Ling Z, Yu Z, Shen Z, Zhang S, Wang X. 2020. Heterogeneity and diversity of mcr-8 genetic context in chicken-associated Klebsiella pneumoniae. Antimicrob Agents Chemother 65:e01872-20. doi: 10.1128/AAC.01872-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carroll LM, Gaballa A, Guldimann C, Sullivan G, Henderson LO, Wiedmann M. 2019. Identification of novel mobilized colistin resistance gene mcr-9 in a multidrug-resistant, colistin-susceptible Salmonella enterica serotype Typhimurium isolate. mBio 10:e00853-19. doi: 10.1128/mBio.00853-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Y, Dai X, Zeng J, Gao Y, Zhang Z, Zhang L. 2020. Characterization of the global distribution and diversified plasmid reservoirs of the colistin resistance gene mcr-9. Sci Rep 10:8113. doi: 10.1038/s41598-020-65106-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Y, Xu C, Zhang R, Chen Y, Shen Y, Hu F, Liu D, Lu J, Guo Y, Xia X, Jiang J, Wang X, Fu Y, Yang L, Wang J, Li J, Cai C, Yin D, Che J, Fan R, Wang Y, Qing Y, Li Y, Liao K, Chen H, Zou M, Liang L, Tang J, Shen Z, Wang S, Yang X, Wu C, Xu S, Walsh TR, Shen J. 2020. Changes in colistin resistance and mcr-1 abundance in Escherichia coli of animal and human origins following the ban of colistin-positive additives in China: an epidemiological comparative study. Lancet Infect Dis 20:1161–1171. doi: 10.1016/S1473-3099(20)30149-3. [DOI] [PubMed] [Google Scholar]
- 15.Pei N, Jian Z, Liu Y, Liang T, Liu W, Li J. 2021. Draft genome sequence of a polymyxin-resistant Klebsiella pneumoniae clinical strain carrying mcr-8.1 and blaNDM-5. Microbiol Resour Announc 10:e01224-20. doi: 10.1128/MRA.01224-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rojo-Bezares B, Martin C, Lopez M, Torres C, Saenz Y. 2012. First detection of blaIMI-2 gene in a clinical Escherichia coli strain. Antimicrob Agents Chemother 56:1146–1147. doi: 10.1128/AAC.05478-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pei N, Liu Q, Cheng X, Liang T, Jian Z, Wang S, Zhong Y, He J, Zhou M, Kristiansen K, Chen W, Liu W, Li J. 2021. Longitudinal study of the drug resistance in Klebsiella pneumoniae of a tertiary hospital, China: phenotypic epidemiology analysis (2013–2018). Infect Drug Resist 14:613–626. doi: 10.2147/IDR.S294989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bortolaia V, Kaas RS, Ruppe E, Roberts MC, Schwarz S, Cattoir V, Philippon A, Allesoe RL, Rebelo AR, Florensa AF, Fagelhauer L, Chakraborty T, Neumann B, Werner G, Bender JK, Stingl K, Nguyen M, Coppens J, Xavier BB, Malhotra-Kumar S, Westh H, Pinholt M, Anjum MF, Duggett NA, Kempf I, Nykasenoja S, Olkkola S, Wieczorek K, Amaro A, Clemente L, Mossong J, Losch S, Ragimbeau C, Lund O, Aarestrup FM. 2020. ResFinder 4.0 for predictions of phenotypes from genotypes. J Antimicrob Chemother 75:3491–3500. doi: 10.1093/jac/dkaa345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carattoli A, Hasman H. 2020. PlasmidFinder and in silico pMLST: identification and typing of plasmid replicons in whole-genome sequencing (WGS). Methods Mol Biol 2075:285–294. doi: 10.1007/978-1-4939-9877-7_20. [DOI] [PubMed] [Google Scholar]
- 20.Pei N, Li Y, Liu C, Jian Z, Liang T, Zhong Y, Sun W, He J, Cheng X, Li H. 2022. Within-ward transmissions of Klebsiella pneumoniae low-endemic-risk clones resolved by a genomic analysis: highlighting the role of whole-genome sequencing in infection surveillance. Lancet doi: 10.2139/ssrn.3831167. [DOI] [Google Scholar]
- 21.Page AJ, De Silva N, Hunt M, Quail MA, Parkhill J, Harris SR, Otto TD, Keane JA. 2016. Robust high-throughput prokaryote de novo assembly and improvement pipeline for Illumina data. Microb Genom 2:e000083. doi: 10.1099/mgen.0.000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.CLSI. 2018. Performance standards for antimicrobial susceptibility testing. CLSI, Wayne, PA. [Google Scholar]
- 23.Guo X, Chen F, Gao F, Li L, Liu K, You L, Hua C, Yang F, Liu W, Peng C, Wang L, Yang X, Zhou F, Tong J, Cai J, Li Z, Wan B, Zhang L, Yang T, Zhang M, Yang L, Yang Y, Zeng W, Wang B, Wei X, Xu X. 2020. CNSA: a data repository for archiving omics data. Database (Oxford) 2020:baaa055. doi: 10.1093/database/baaa055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen FZ, You LJ, Yang F, Wang LN, Guo XQ, Gao F, Hua C, Tan C, Fang L, Shan RQ, Zeng WJ, Wang B, Wang R, Xu X, Wei XF. 2020. CNGBdb: China National GeneBank DataBase. Yi Chuan 42:799–809. doi: 10.16288/j.yczz.20-080. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
. Download spectrum.02306-22-s0001.pdf, PDF file, 1.2 MB (1.2MB, pdf)
Data Availability Statement
The data that support the findings of this study have been deposited at the China National GeneBank DataBase (CNGBdb) in the CNGB Sequence Archive (CNSA) under accession number CNP0001198 (23, 24).