ABSTRACT
The first pandemic of the 21st century was caused by an H1N1 influenza A virus (IAV) introduced from pigs into humans, highlighting the importance of swine as reservoirs for pandemic viruses. Two major lineages of swine H1 circulate in North America: the 1A classical swine lineage (including that of the 2009 H1N1 pandemic) and the 1B human seasonal-like lineage. Here, we investigated the evolution of these H1 IAV lineages in North American swine and their potential pandemic risk. We assessed the antigenic distance between the HA of representative swine H1 and human seasonal vaccine strains (1978 to 2015) in hemagglutination inhibition (HI) assays using a panel of monovalent antisera raised in pigs. Antigenic cross-reactivity varied by strain but was associated with genetic distance. Generally, the swine 1A lineage viruses that seeded the 2009 H1 pandemic were antigenically most similar to the H1 pandemic vaccine strains, with the exception of viruses in the genetic clade 1A.1.1.3, which had a two-amino acid deletion mutation near the receptor-binding site, which dramatically reduced antibody recognition. The swine 1B lineage strains, which arose from previously circulating (pre-2009 pandemic) human seasonal viruses, were more antigenically similar to pre-2009 human seasonal H1 vaccine viruses than post-2009 strains. Human population immunity was measured by cross-reactivity in HI assays to representative swine H1 strains. There was a broad range of titers against each swine strain that was not associated with age, sex, or location. However, there was almost no cross-reactivity in human sera to the 1A.1.1.3 and 1B.2.1 genetic clades of swine viruses, and the 1A.1.1.3 and 1B.2.1 clades were also the most antigenically distant to the human vaccine strains. Our data demonstrate that the antigenic distances of representative swine strains from human vaccine strains represent an important part of the rational assessment of swine IAV for zoonotic risk research and pandemic preparedness prioritization.
IMPORTANCE Human H1 influenza A viruses (IAV) spread to pigs in North America, resulting in a sustained circulation of two major groups of H1 viruses in swine. We quantified the genetic diversity of H1 in swine and measured antigenic phenotypes. We demonstrated that the swine H1 lineages were significantly different from the human vaccine strains and that this antigenic dissimilarity increased over time as the viruses evolved in swine. Pandemic preparedness vaccine strains for human vaccines also demonstrated a loss in similarity with contemporary swine strains. Human sera revealed a range of responses to swine IAV, including two groups of viruses with little to no immunity. The surveillance and risk assessment of IAV diversity in pig populations are essential to detect strains with reduced immunity in humans and provide critical information for pandemic preparedness.
KEYWORDS: influenza A virus, swine, antigenic cartography, H1N1, pandemic risk assessment
INTRODUCTION
The first pandemic of the 21st century was caused by a strain of influenza A virus (IAV) that was introduced from pigs into humans, highlighting the importance of swine as reservoirs for pandemic influenza A viruses. Many strains of IAV that circulate in pigs are derived from repeated introductions of human IAV into swine populations (1–4). Due to differences in population structure and movement, viruses in pigs may evolve differently from those circulating in humans (1). Instead of strain replacement being driven by targeted antibody responses to surface proteins and existing herd immunity, subpopulations of pigs with variable immunity and movement within production systems but with limited movement of pigs between populations results in an uneven antigenic change and multiple cocirculating lineages. In addition, reassortment with endemic swine IAV and further adaptation result in a diverse population of viruses (5, 6).
Three major lineages of viruses bearing the H1 hemagglutinin (HA) cocirculate in swine: these are named 1A, 1B, and 1C, and they link the evolutionary history of the genes to common ancestral lineages (7). H1-1A are derived from the “classical” 1918 human pandemic virus and have spread globally in swine. The 1A lineage also includes the H1 genetic clade, which resulted in the 2009 H1N1 pandemic. Continued transmission of the pandemic virus in humans resulted in the reintroduction of the virus into swine in multiple locations around the world, in some cases with onward transmission within pigs (8). In North America, genetic clades within the 1A lineage were previously classified into alpha (α), beta (β) and gamma (γ), and they are now referred to as 1A.1.x, 1A.2, and 1A.3.x, respectively (where “x” designates further subdivision). The viruses in 1A.1, the longest circulating lineage in swine, are classified under a single clade, following the global nomenclature system (7). However, multiple paraphyletic clades have been detected as a result of the extinction of certain clades and the sporadic surveillance practices in the earlier decades of the 20th century (7). We proposed two new clades of circulating viruses, 1A.1.1.1 and 1A.1.1.3, that are genetically and antigenically distinct from the other 1A lineage clades. The 1B viruses are derived from human seasonal H1 viruses that circulated prior to the 2009 pandemic, and they were introduced into pigs at different points in time. 1B.1 viruses were first described in the United Kingdom in the 1990s (9). In the 2000s, there were incursions of human seasonal H1 viruses into U.S. swine (the 1B.2.1 [formerly δ2] and 1B.2.2 [formerly δ1] viruses) (10, 11), as well as introductions and onward transmission in several other geographical locations, including Chile, Argentina, Brazil, Australia, and Vietnam (12). The 1C HA was derived from an avian H1N1 virus introduction that circulated in pigs in Europe in the 1970s and then spread to Asia (13, 14). A single detection of a 1C lineage HA gene was reported in Mexico (15), but no other 1C HA were identified in surveillance activities in North America. However, given that the N1 and M segments of the 2009 H1N1 pandemic virus originate from the 1C, it is likely that there was undetected circulation of this strain in this region.
Given the relatively frequent transmission of IAVs between humans and swine and the ever-present risk of another swine-origin IAV pandemic, it is critical to objectively rank the zoonotic risk of the IAVs circulating in pigs. The haemagglutinin (HA) of avian influenza viruses commonly have receptor binding site (RBS) profiles for α2,3 linkage-type sialic acids that are prevalent in bird airways and intestinal cells. However, human-derived swine IAV HAs maintain the α2,6 linkage-type sialic acids dominant in human airways after their introduction into pigs and consistently demonstrate the potential for replication and transmission in humans (16–18). As the main target of host antibody responses, HAs that deviate significantly from those to which the human population have prior immunity would pose a greater zoonotic risk if a variant virus were capable of human-to-human transmission. Further, it is plausible that cross-immunity and pandemic risk correlate with the antigenic distance between a swine virus HA and human seasonal vaccine strains. The antigenic differences between swine and human HAs can be measured in the lab by using binding assays, such as the hemagglutination inhibition (HI) assay (19). These data may subsequently be visualized and quantified using antigenic cartography to position viruses and sera in 2D or 3D “antigenic maps” such that the map distances directly correspond to HI measurements (20, 21). Consequently, characterizing the antigenic diversity of HA circulating in swine provides an objective assessment of the pandemic risk.
In this study, we used virus isolates, swine antisera, and genetic sequences generated as part of the NIH CEIRS network pipeline to characterize currently circulating swine IAVs in North America (USA., Mexico, and Canada). We measured the antigenic distances between a representative sample of currently circulating H1 swine IAVs and H1 human vaccine strains since the 1970s. We also tested representative swine strains against pandemic preparedness vaccines, termed candidate vaccine viruses (CVV), using ferret sera. We then tested a subset of these viruses with age-stratified postvaccination and postexposure human sera to assess potential immunological cross-reactivity in the human population against diverse swine IAV strains. These data showed that the antigenic distance from swine IAVs to human vaccine strains is a rational measure with which to rank swine strains for pandemic risk and that the antigenic distance should be linked with genomic epidemiology and existing risk assessment tools to inform public health pandemic preparedness measures.
RESULTS
Genetic analysis of North American swine H1 IAV strains.
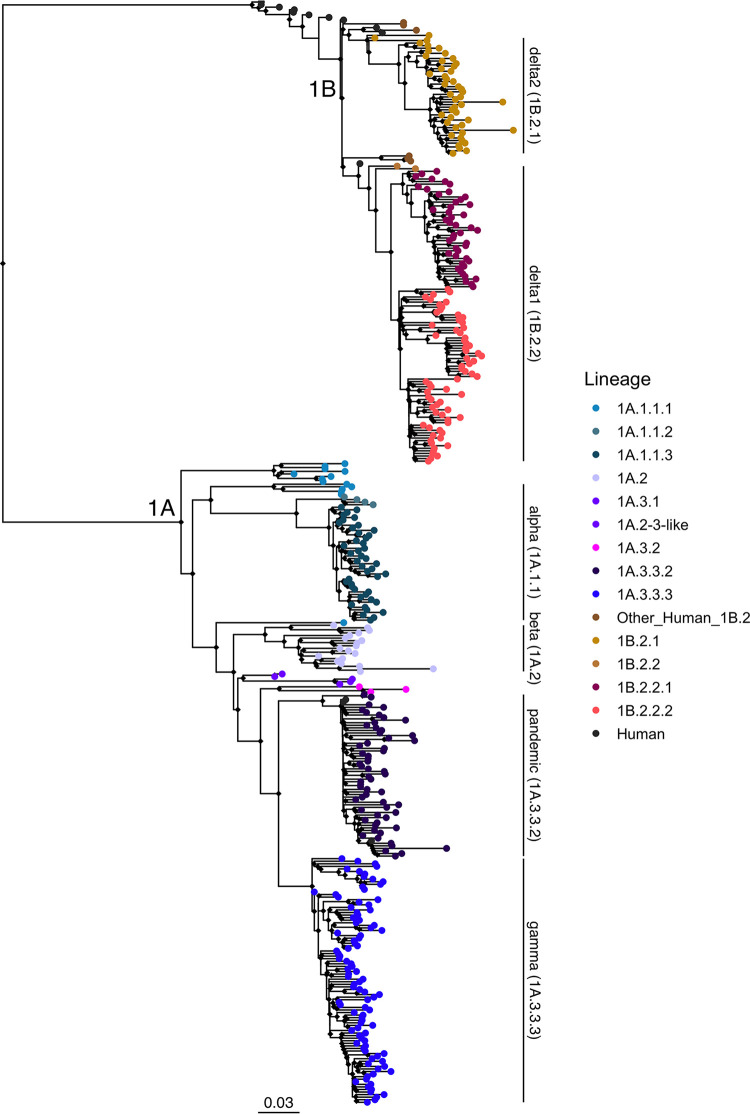
The maximum-likelihood phylogenetic tree inferred with representative HA gene sequences from human and swine hosts collected between 2012 and 2019 demonstrated the concurrent circulation of two major lineages, 1A and 1B, in North America (Fig. 1). Consistent with prior studies, the 1C (Eurasian-avian) lineage was not detected among our sequences. The majority of the HA data were collected in the contiguous USA through the USDA Influenza A Virus in Swine Surveillance System (Fig. S1A).
FIG 1.
Phylogenetic tree of H1N1 influenza A viruses from North American swine populations during the period of study (2012 to 2019). Swine IAV lineages are labeled on the right with annotated bars, according to the global H1 clade classification. Monophyletic clades containing strains characterized in the risk pipeline are colored by clade at the tip point. All of the H1N1 human vaccine strain tip points, which are colored in black, are included in the analysis but are not labeled. The tree is midpoint rooted. All branch lengths are drawn to scale. The scale bar indicates the number of nucleotide substitutions per site.
1A lineage viruses circulated in the USA, Mexico, and Canada throughout the study period. The 1A.1.1 lineage viruses were detected within swine populations in Canada and the USA, while the 1A.2 isolates were observed in Mexico and the USA. The 1A.3.3.2 (H1N1pdm09 lineage) viruses were detected in all three countries. As shown by the clustering of the contemporary swine and human strains in this clade in Fig. S1B, the majority of the 1A.3.3.2 transmission appeared to have been derived from multiple introductions of human H1 into swine in each country rather than from continued circulation within swine and transmission across countries. The 1A.3.3.3 lineage viruses were only detected in pig populations in the USA.
The swine 1B lineage viruses (1B.2.1 and 1B.2.2) were detected primarily in the USA, although a few isolates were also detected in Mexico. The 1B.2.1 viruses detected in swine were not similar to any human vaccine strain in use in the early 2000. This is likely a result of significant genetic evolution within pigs following the initial spillover. Genetically, the 1B.2.2.x clade viruses were most evolutionarily similar to a human seasonal strain, A/Michigan/2/2003 (approximate likelihood ratio test [aLRT] node support of 99.7%). A/Solomon Islands/3/2006 and A/Brisbane/59/2007 formed a distinct sister clade to the 1B.2.1 strains and were the most similar, but the low levels of public IAV sequence data in humans and swine prior to 2009 limit our ability to quantify genetic and antigenic relationships for this period.
Antigenic evolution of swine H1 IAV in North America and distance to H1 human vaccine strains.
Based on the genetic representation and the availability of virus isolates, we selected a panel of swine H1 strains from each clade, together with H1 human vaccine strains from 1977 to 2015. These strains were antigenically characterized in hemagglutinin inhibition (HI) assays against swine antisera, and antigenic maps were generated (Fig. 2). All of the swine strains displayed significant antigenic heterogeneity within and between lineages. The human H1N1pdm09 strains (CA09, MX09, MI15) were antigenically distant from the pre-2009 human vaccine strains and were more similar to the swine 1A strains (Fig. 2A and B). Previously reported swine H3N2 exhibited substantially less overall genetic diversity and reflected the antigenic drift that occurs in humans. In contrast, H1 antigenic diversity in swine is complex, as it has evolved over time in two hosts with distinct patterns of antigenic evolution within independent swine lineages (1, 21–23).
FIG 2.
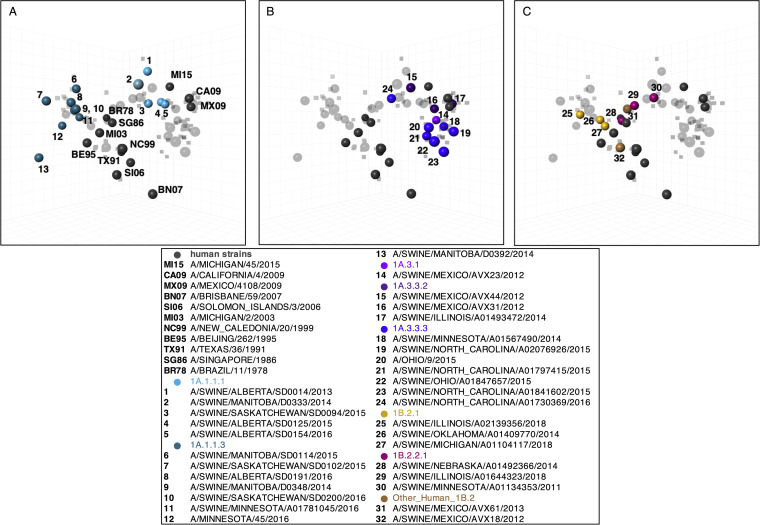
Antigenic relationships between human H1N1 vaccine strains and North American swine H1 strains. The three-dimensional maps are displayed in the same rotation in all panels. Human H1N1 vaccine strains (gray) are shown in all panels but are labeled only in panel A with abbreviated strain names. North American swine H1N1 strains are split by lineages (A) 1A.1.1.x, (B) 1A.3.3.x and (C) 1B.2.x. Each sphere in the maps represents a strain, and each is colored according to the phylogenetic clade, as described in the legend. One unit of distance represents a two-fold change in the HI assay.
The 1A and 1B clades both formed groups that were large, diffuse, and not chronologically oriented. However, there was evidence that the antigenic distances between swine strains correlated with genetic distance (Fig. S2). We also assessed the potential risk of incursion into the human population by measuring the distance from each swine strain to human vaccine strains (Fig. 3).
FIG 3.
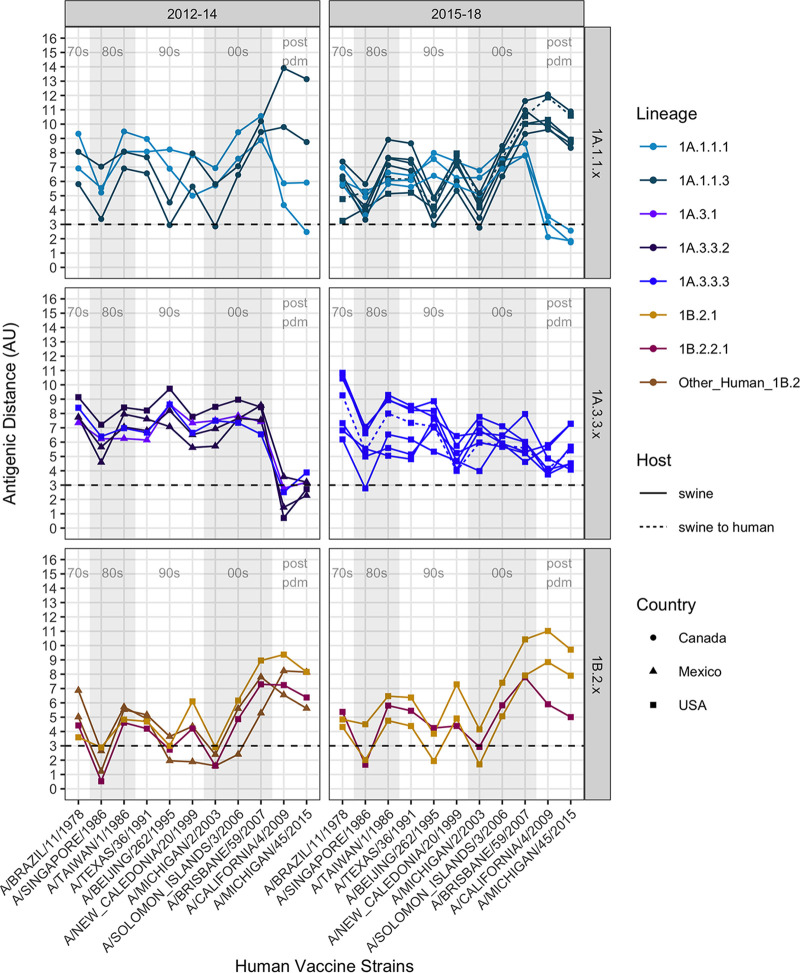
Antigenic distances between contemporary swine H1N1 strains and human seasonal H1N1 vaccine strains. Graphs are divided by swine strain lineage (1A.1.1.x, 1A.3.3.x, or 1B.2.x) and were grouped using the year of isolation to facilitate visualization. Column shades of light gray and white indicate human H1N1 vaccine strains by decade, as listed along the x axes of the lower graphs in chronological order (1978 to 2015). The antigenic distance between the swine strain and human vaccine strain is plotted on the y axis. Strains are colored by phylogenetic lineage, as in Fig. 1 and 2. The shape of the strain represents its country of origin (circle, Canada; triangle, Mexico; square, USA). The dotted line indicates a significant antigenic distance (3 AU, ~8-fold loss in HI titer) between the swine H1 strains on the y axis and the human seasonal H1N1 vaccine strains along the x axis. In general, 1A lineages that have been circulating in swine since the 1918 pandemic had variable distance to vaccine strains prior to 2009, but they were closer to the H1N1pdm09 vaccine strains. The double deletion lineages in 1A.1.1.3 deviated from 1A.1.1.1 and were antigenically highly distant from most human vaccine strains, especially the H1N1pdm09 strains. Conversely, the 1B lineages are closer to the prepandemic vaccine strains and are >3AU from the postpandemic vaccine strains.
Antigenically, the 1A.1.x and the 1A.3.x lineage viruses in pigs were distinct from the 1B lineage strains (Fig. 2A–C). The 1A.1.1.1 lineage viruses circulating in Canadian pigs were more antigenically similar (1.75 to 5.92 antigenic units [AU]) to the 2009-pdm-like vaccines than to the 1A.1.1.3 lineage viruses (8.34 to 11.86 AU) (Fig. 2A and 3). The positioning of the 1A.1.1.3 lineage viruses, with deletions at positions 129 and 130, were antigenically closer to those of the swine 1B and related human vaccine strains in the map, as the 1B lineage strains have a single deletion at residue 130 of a mature H1 peptide. The 1A.3.3.2 lineage swine viruses and the 2009 pandemic and subsequent seasonal human vaccine strains were also antigenically distant from the seasonal influenza strains circulating in humans prior to 2009 (6.19 to 9.62 AU) and from the 1B lineage swine strains seeded by the pre-2009 human strains. Unlike other genetic clades within the 1A lineage, the 1A.3.x viruses were antigenically more similar to the human H1N1pdm09 viruses, but they displayed significant strain to strain variation (0.71 to 7.29 AU) (Fig. 2B and 3). The antigenic map reflected antigenic drift that occurred in humans and resulted in the updated 2015 H1N1 pandemic vaccine strain (2.29 AU distance to the previous vaccine A/California/4/2009). However, in swine, our selected viruses were detected between 2009 and 2014, and all of these HA genes were antigenically similar to the 2009 human vaccine strains. The other dominant circulating clade within 1A.3 were the 1A.3.3.3 strains that are geographically restricted to the USA and showed significant genetic variation as well as corresponding antigenic diversity (range of 3 to 11 AU from the tested human vaccine strains), which is likely driven by changes in the putative antigenic sites of H1 (24) (Fig. S3A–D).
There was antigenic distinction between the 1B.2.2.x and 1B.2.1 viruses in the antigenic map, which is consistent with the phylogeny and accounts for the separate introductions of human seasonal influenza viruses into pigs in the early 2000s (Fig. 2C). The antigenic distances of the 1B strains against all of the human vaccine strains (Fig. 3) showed that the most antigenically similar human vaccine strains were A/Beijing/262/1995 and A/Singapore/1986, whereas the closest antigenic distance was to the seasonal field strain A/Michigan/2/2003 (1.6 to 4.4 AU). While the 1B.2.2.x and 1B.2.1 strains both showed an increased antigenic distance to the H1N1pdm09 human strains, the distance was greater in 1B.2.1, relative to those of the 1B.2.2.x strains.
Candidate vaccine virus reactivity to contemporary swine H1 strains.
To assess the potential of CVV and human seasonal H1 vaccines to protect against contemporary clades in pigs, we tested representative swine strains of each of the regularly detected H1 clades from the 1A and 1B lineages against reference ferret antisera. The sera were raised against a human seasonal H1N1 vaccine strain A/Idaho/7/2018 and CVVs A/Ohio/24/2017 (1A.1.1.3), A/Ohio/9/2015 (1A.3.3.3), A/Iowa/32/2016 (1B.2.2.1), A/Ohio/35/2017, and A/Michigan/383/2018 (1B.2.1) (Table 1 and 2). Swine viruses from clades 1A.1.1.x (including those that have the two amino acid deletion), 1A.3.3.3, and 1A.3.3.2 (H1N1pdm09 HA clade) showed moderate to significant cross-reactivity with sera raised against their respective within-clade CVV/human vaccine strains, displaying ranges of 3 to 5-fold decreases in the HI assay (Table 1). The 1A.2 and 1A.2.3-like viruses do not have a within-clade CVV or vaccine strain, and they demonstrated limited antigenic similarity to the other 1A CVVs but retained cross-reactivity with the high-titer A/Idaho/7/2018 vaccine strain. 1B.2.1 and 1B.2.2.1 viruses retained some cross-reactivity to sera from lineage-specific CVVs, even if at 2 to 3-fold decreases (Table 2). 1B.2.2.2 viruses with no associated within-clade CVV reacted poorly with the other 1B CVVs and had barely detectable titers. These represent a minor clade detected in surveillance in the United States.
TABLE 1.
Ferret antisera against human seasonal vaccine strains or candidate virus vaccines (CVVs) demonstrated variable recognition of the swine H1 1A.1.x lineagea
| Strain | Category | Clade | A/Ohio/24/2017 | IDCDC-RG59 a/Ohio/24/2017-like | A/Ohio/9/2015 RG48A | A/Idaho/7/2018 |
|---|---|---|---|---|---|---|
| IDCDC-RG59 A/Ohio/24/2017-like | CVV | 1A.1.1 | 5120 | 2560 | <10 | 1,280 |
| A/swine/Iowa/A02478635/2019 | Test | 1A.1.1 | 160 | 160 | 10 | 80 |
| A/swine/Oklahoma/A02245237/2019 | Test | 1A.2 | 40 | 20 | 320 | 2,560 |
| A/swine/Texas/A01104132/2019 | Test | 1A.2-3-like | 20 | 20 | 80 | 1,280 |
| A/Ohio/9/2015 | CVV | 1A.3.3.3 | <10 | <10 | 1,280 | 10 |
| A/swine/North Carolina/A02478571/2019 | Test | 1A.3.3.3 | <10 | 10 | 1,280 | 20 |
| A/swine/Iowa/A01731653/2016 | Test | 1A.3.3.3 | 10 | <10 | 160 | 20 |
| A/swine/Nebraska/A02214231/2017 | Test | 1A.3.3.3 | 10 | <10 | 320 | 10 |
| A/Idaho/7/2018 | Human vaccine | 1A.3.3.2 | 80 | 160 | 160 | 10,240 |
| A/swine/Utah/A02432386/2019 | Test | 1A.3.3.2 | 80 | 40 | 160 | 5,120 |
Hemagglutination inhibition (HI) titers of CVV or human seasonal vaccine antisera against the contemporary North American swine H1 1A.1.x lineage. Homologous titers are highlighted in gray, and boxes mark titers against swine viruses of the same lineage.
TABLE 2.
Ferret antisera against human seasonal vaccine strains or candidate virus vaccines (CVVs) demonstrated variable recognition of the swine H1 1B.2.x lineagea
| Strain | Category | Clade | A/Iowa/32/2016 | A/Ohio/35/2017 | A/Michigan/383/2018 |
|---|---|---|---|---|---|
| A/Iowa/32/2016 | CVV | 1B.2.2.1 | 640 | 20 | 20 |
| A/swine/South Dakota/A01481702/2014 | Test | 1B.2.2.1 | 80 | 20 | 80 |
| A/swine/Minnesota/A02478597/2019 | Test | 1B.2.2.2 | 40 | 20 | 20 |
| A/Ohio/35/2017 | CVV | 1B.2.1 | 80 | 640 | 160 |
| A/Michigan/383/2018 | CVV | 1B.2.1 | 40 | 160 | 1,280 |
| A/swine/Illinois/A02139356/2018 | Test | 1B.2.1 | 20 | 320 | 1,280 |
Hemagglutination inhibition (HI) titers of CVV or human seasonal vaccine antisera against the contemporary North American swine 1B.2.x lineage. Homologous titers are highlighted in gray, and boxes mark titers against swine viruses of the same lineage.
There were zoonotic events from the 1A.1.1.3 clade in humans in 2016 (A/Minnesota/45/2016) and 2017 (A/Ohio/24/2017), with a CVV selected based on the 2017 strain. Sera raised in ferrets against the 2017 A/Ohio/24/2017 CVV reacted to the 2019 isolate from this clade in an HI assay (Table 1), but this occurred with a >8-fold decrease in cross-reactivity. The reduction in reactivity may be associated with two amino acid substitutions, one at a putative antigenic site Sb (H1-G153D) and a second at a putative receptor binding site (H1-T222A). Viruses from lineages such as the 1A.3.3.3 resulted in human zoonotic cases, with one (A/Ohio/09/2015) being selected as a CVV. Sera raised against this virus appear to retain cross-reactivity to contemporary viruses from the same 1A.3.3.3 clade (A/swine/North Carolina/A02478571/2019), but titers were decreased by 2 to 3-fold for other viruses within the 1A.3.3.3 clade.
Human population immunity against swine H1 IAV.
To understand the implications of IAV genetic and antigenic diversity in swine for human population immunity, we tested a subset of human vaccine and swine viruses against sera from age-stratified human cohorts. One strain from each lineage was chosen according to the risk-ranking scoring system described in the Materials and Methods section (see Table S3). One set of sera was collected from people vaccinated against the pandemic lineage virus A/Michigan/45/2015 (n = 40; age, 22 to 68) at the Johns Hopkins Hospital, Baltimore, MD, USA (postvaccination cohort), and a second set of sera was collected from patients with confirmed influenza (n = 20; age, 21 to 71) in Taiwan (n = 10) or in Baltimore (n = 10) (postexposure cohort). The cohorts included adults of both sexes between the ages of 21 and 71. The sera were tested against human vaccine strains: A/Beijing/262/1995 (BE95) and A/Michigan/45/2015 (MI15) as well as the following strains: A/swine/Nebraska/A01492366/2014 (NE14 1B.2.2.1), A/swine/Illinois/A02139356/2018 (IL18 1B.2.1), A/swine/Mexico/AVX18/2012 (MX12 OtHu1B.2), A/Swine/Saskatchewan/SD0094/2015 (SK15 1A.1.1.1), A/Swine/Minnesota/A01781045/2016 (MN16 1A.1.1.3), A/swine/Iowa/A01731653/2016 (IA16 1A.3.3.3), and A/Swine/Mexico/AVX23/2012 (MX12 1A.3.1).
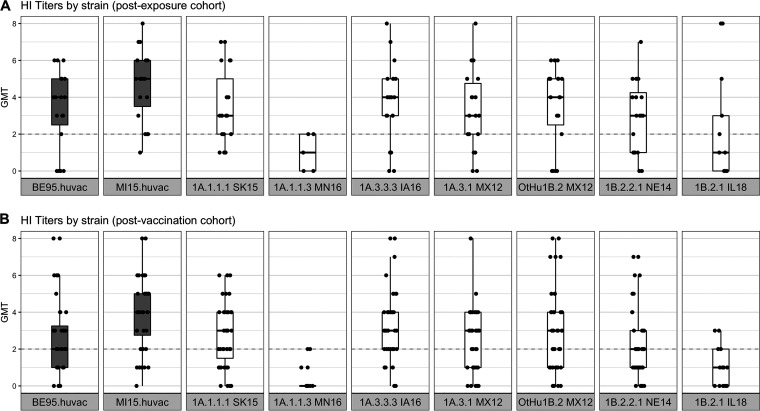
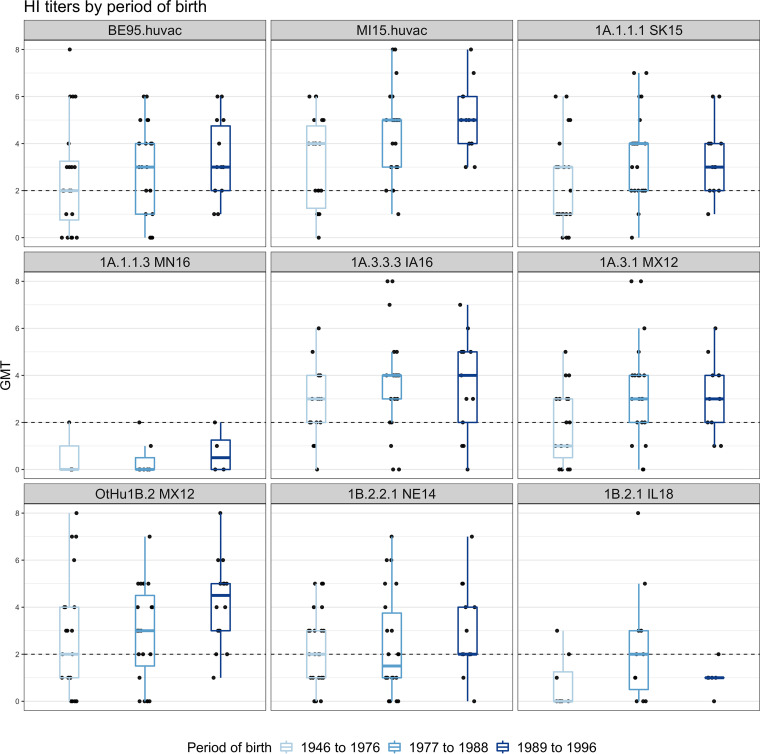
We did not detect major differences between the titers of the postexposure (Fig. 4A) and postvaccination (Fig. 4A) cohorts in their overall responses to different strains (Table S4). There were heterogenous responses to most viruses, with a broad range of titers being observed against each strain. The MN16 1A.1.1.3 and IL18 1B.2.1 strains were exceptions, as there appeared to be no cross-reactivity in almost all subjects (geometric mean titer [GMT] of <2) (Fig. 4A and B). These data mirror the swine sera HI data (Fig. 3), in which the MN16 1A.1.1.3 strain was antigenically distant to both the pandemic and the prepandemic human seasonal strains that were circulating in the 2000s with average distances of AU 9.6 and 7.07, respectively. It was also significantly drifted from human strains from prior decades (average distances of AU 4.17 and 5.57 for vaccines used in the 1970 to the 1980s and in the 1990s, respectively). The IL18 1B.2.1 strain had a similar trend, with the most similar human vaccine strain being A/Beijing/262/1995 at a distance of 3.85 AU, and was above the cutoff used when human vaccine strains are recommended for revision due to decreases in cross-reactivity to antisera (Table S3). We parsed the HI data by age to test whether the range of cross-reactive titers could be separated into high or low reactors (Fig. 5). The data showed that birth years split into categories of 1946 to 1976, 1977 to 1978, and 1989 to 1996 were not associated with the titer response to swine or human IAV strains. Regardless of the birth periods, there were both high and low reactors to each of the viruses, with the exception of MN16 1A.1.1.3, which had consistently low reactions (Fig. 5; Fig. S4). The IL18 1B.2.1 demonstrated a unique pattern, with two individual samples in the 1977 to 1988 birth year cohort exhibiting a high response to the virus (Fig. 5; Fig. S4B).
FIG 4.
Human convalescent (A) and postvaccination (B) sera geometric mean log2 hemagglutination inhibition (HI) titers against North American H1N1 swine strains. Boxplots show the median of the aggregated HI titers against H1N1 strains with the 5th and 95th percentiles and the standard deviation. Each dot represents the geometric mean titer (GMT), and the log2 (HI titer/10) of human sera is shown on the y axis against each strain (shown on the x axis). Boxplots in gray indicate the human H1N1 vaccine strains, and boxplots in white indicate the swine H1 strains. The gray dotted line indicates the minimum positive HI titer threshold (≥40 or 2). The human sera show a range of titers against most swine and human vaccine strains, with the exception of the MN16 (1A.1.1.3) and IL18 (1B.2.1) strains, to which there was little to no cross-reactivity.
FIG 5.
Hemagglutination inhibition responses of postvaccination and postexposure human sera, stratified by period of birth, against North American H1 swine strains. Shades of light to dark blue represent the periods of birth of the human participants (1946 to 1976, 1977 to 1988, and 1989 to 1996). Boxplots show the median HI titers against H1 strains with the 5th and 95th percentiles and the standard deviation. Each dot represents the geometric mean titer (GMT) of the human sera on the y axis against each strain (shown on the x axis). The gray dotted line indicates the minimum positive HI titer threshold (≥40 or 2). No major differences were observed between the different age groups.
We hypothesized that the sera that showed high cross-reactivity with the post-pandemic MI15 human vaccine strain, either by exposure or by vaccination, would also have higher responses to those swine strains which are antigenically similar to the other 1A lineage strains that were derived from pre-2009 human seasonal strains (MX12 1A.3.1 [AU 2.99], SK15 1A.1.1.1 [AU 3.05], IA16 1A.3.3.3 [AU 4.29]). To address this question, we divided the titers into three groups of high, medium, and low responders to the MI15, and we found a modest increase in the titers of the high MI15 responders against the swine 1A.x viruses (Fig. S4A). This pattern was not found for the distribution of titers for MX12 OtHu1B.2 (AU 8.15) or NE14 1B.2.2.1 (AU 6.38), nor was it found for the distribution of titers for the MN16 1A.1.1.3 (AU 8.90) strain or the IL18 1B.2.1 (AU 9.72) strain. There was no effect of age, as both high and low responders were found within each age-group (Fig. S4B).
DISCUSSION
Influenza A viruses in swine have posed a consistent challenge to food and animal security around the world. In this study, we quantified the genetic and antigenic diversity in swine influenza A H1 viruses that were isolated between 2011 and 2019 in Canada, the USA, and Mexico. We explicitly defined how evolution in swine, following the introduction of human seasonal IAV lineages into pigs, has resulted in significant drift from both human seasonal vaccines and pandemic preparedness candidate vaccine viruses. Our risk assessment pipeline integrated data derived from genomic surveillance as well as antigenic characterization incorporated into an objective strain selection process. This process identified priority swine IAV genetic clades for the assessment of zoonotic potential by using human population sera. This process quantified the public health risk of the genetically and antigenically diverse North American swine H1 influenza viruses and supported a need for the rigorous evaluation of IAV at the human-swine interface in risk assessment. Our characterization identified H1 IAVs circulating in swine to which the human population likely has minimal immunity from either prior exposure or current vaccination efforts.
Surveillance for swine IAVs revealed an expansion in the diversity of some genetic clades in North America, notably, the 1A.1.1 (α) (25). A single clade of North American 1A.1.1 viruses was defined in the current global swine H1 classification scheme (7). Additional surveillance and our characterization revealed the regional persistence and circulation of genetically and antigenically distinct viruses within different statistically supported clades in 1A.1.1. Currently circulating 1A.1.1 strains formed two major groups, both phylogenetically and antigenically. Within this study, we defined the group reported previously as a-1 as 1A.1.1.1, and the group reported as a-3 with a two amino acid deletion in the HA1 as 1A.1.1.3 from Nelson et al. (2017) (25). A third clade, the group reported as a-2 from Nelson et al. (2017) (25), was given the global name of 1A.1.1.2, but since they represent a small fraction of detections in Canada, we did not antigenically characterize these viruses.
The antigenic distances between swine IAV strains and human vaccine strains from the past ~50 years (1977 to 2015) were heterogeneous. The 1A lineage swine viruses averaged between 5.51 and 8.4 AU from the human vaccine strains. This diversity reflects decades of evolution in swine, following the human-to-swine spillover event. The 1B lineage swine viruses, originating from human seasonal strains from the 2000s, remained antigenically similar to the putative human ancestral strains. However, these contemporary swine strains had an average distance of 3.72 to 6.14 AU to more recent H1 human seasonal vaccine strains. These data are consistent with estimates of the antigenic drift of H1 1B viruses of 0.17 to 0.85 AU per year (1), and they reflect the introduction and decades of evolution in regional swine populations. Notably, some IAVs, such as the IL18 1B.2.1 virus, were antigenically distant from all of the human vaccine strains (averages of AU 7.31 and 10.37 from the post-2000 and postpandemic human strains, respectively).
This antigenic diversity in cross-reactivity characterized using swine and ferret antisera was reflected in the human serology assays, where there was also heterogenous cross-reactivity in the human population samples. Consequently, although some of the 1A and 1B viruses retain antigenic cross-reactivity with the human vaccine strains, the antigenic distance was above the threshold that is applied to guide vaccine strain updates, and human population immunity reflected this antigenic variation. In our data, we did not find associations between age, sex, or prior exposure that indicated a demographic of the human population that is more at risk to an incursion by a swine IAV into humans. Instead, our data demonstrated that certain clades of swine IAVs represent a more pressing zoonotic threat and that this dynamic is the result of evolution within swine and is unrelated to human population immunity. Although our cohorts were not comprehensive, they were representative of various ages and genders, and two distinct geographies were represented. However, we did not have human sera from a birth year period that would most likely have encountered the progenitors of the swine 1B viruses (early 2000s) in early childhood. Given that the spatial heterogeneity of the contemporary IAV lineages in pigs might influence the emergent risk at the human-animal interface, future work should focus on regional risk assessments using locally derived human serum cohorts. For example, swine surveillance efforts in Mexico and Canada suggest distinct evolutionary dynamics in those regions, with different continuously circulating lineages in swine herds (15, 25). Similarly, assessing zoonotic risk using human sera collected in the USA from occupationally exposed humans and from groups at critical swine-human interface settings (i.e., agricultural fairs) may be appropriate. Despite these caveats, our data conclusively reveal that the evolution of IAVs in swine has resulted in specific clades (1A.1.1.3 and 1B.2.1) across two major lineages to which there is almost no preexisting immunity in the human population.
A key component in human pandemic preparedness is surveillance for currently circulating and emerging IAVs in human populations around the world. These viruses are genetically and antigenically characterized for human risk assessment using polyclonal ferret antisera raised to human vaccine strains or to CVVs. When detected, cases of variant swine influenza A viruses are characterized against existing CVVs and are potentially identified for a new CVV if not recognized by existing ferret antisera (12). Our data demonstrate that CVVs address some of the gaps in cross-reactivity from human population sera. However, the antigenic evolution of influenza in swine is dynamic, and we have little understanding of the breath of diversity in contemporary global swine influenza viruses as it relates to human immunity. Given the correlation between the antigenic distance of swine strains to human vaccine strains and the measures of reduced human population immunity, our process presents an efficient means by which to evaluate swine IAVs for zoonotic threats. Linking sustainable, in-field genomic surveillance efforts with broader cross-HI assays and with more representative cohorts of human sera can establish a global pandemic preparedness protocol.
Our data revealed at least 12 genetic clades from within two major evolutionary H1 lineages that are cocirculating in North American swine. These genetic clades demonstrated considerable antigenic diversity, and the swine strains typically had significant losses in cross-reactivity to human vaccines that were in use from 1977 to 2015. Using a metric that encompassed genetic and antigenic diversity, we objectively ranked the H1 swine strains, and we tested seven prioritized viruses against two cohorts of human sera to serve as one indication of zoonotic potential. These data demonstrated significant variation in the human antibody recognition of the swine strains among and between the phylogenetic clades of swine H1. Two of the swine strains from major H1 clades (1A.1.1.3 and 1B.2.1) did not react with most of the samples of human population sera. The observed diversity of swine IAVs represents a significant challenge to pandemic risk assessment, but our process that integrates genomic surveillance, antigenic characterization, and testing against human population sera provides a template for objective risk assessment. Building on the system reported here, subsequent testing against representative human sera from the region where the swine strains circulate and from additional birth years would add valuable information to the determination of the pandemic potential of IAVs in swine. These data are fluid and will need future reassessment as IAV continues to evolve in swine, but this is the first comprehensive report to identify H1 clades in swine that represent zoonotic threats. These findings can be used to inform the design of candidate vaccine viruses for humans and identify strains that may be preemptively targeted through vaccination in the pig population.
MATERIALS AND METHODS
Genetic analysis.
All available swine H1 HA sequences from Canada, the USA, and Mexico were downloaded from the Influenza Research Database (IRD) (26, 27) on October 14, 2019 (5,517 sequences). To restrict the data set to relevant field viruses, sequences of laboratory origin were excluded. All duplicate sequences and sequences with >30% ambiguous bases (“N”) were removed (4,862 sequences). For visualization, the sequences were down-sampled by nucleotide similarity using the Cluster Database at High Identity with Tolerance (cd-hit) (28, 29), removing sequences with >98.0% sequence identity across the HA gene (375 sequences). The HA sequences of reference strains used in the HI assays were added to this data set, and all duplicates were removed to create the final data set (432 sequences). The sequences were aligned using MAFFT v7.407 (30, 31) and trimmed to a starting ATG and ending stop codon. A maximum-likelihood (ML) phylogenetic tree was inferred using IQ-TREE v1.5.5 (32), following automatic model selection with statistical support determined using the Shimodaira-Hasegawa-like approximate likelihood ratio test (aLRT, 1,000 replicates) (33). Trees were plotted in R v3.6 using the ggtree package (34).
To characterize the genetic diversity, we selected 45 swine and human influenza A viruses (IAV). The swine strains were chosen based on the genetic representation and availability of virus isolates, whereas the human seasonal vaccine strains were chosen as a representation of the dominant strain in circulation in humans and in vaccine immunity. We tested these viruses by HI against swine sera raised to 35 IAV strains, 26 against swine strains, 1 against an H1N1 variant strain, 1 against an early H1N1pdm09 human strain, and 7 against human vaccine strains. The test panel included some previously characterized swine H1N1 and H1N2 strains (4, 10). These antisera strains represented historical or contemporary clades of viruses from the USA. For some international strains, we were restricted to viruses for which an isolate was available. The 45 antigens were selected by classifying all of the H1 genes to a genetic clade, generating an HA1 consensus sequence for each clade, and identifying the best matched field-strain to the consensus. In some cases, a genetic clade was represented more than once if it reflected a statistically supported monophyletic clade in the gene tree or if detections of the clade came from different geographic locations. Strain names, subtypes, clades, and GenBank accession numbers can be found in Table S1. No strains were selected from the 1A.2 (β) clade, as it was infrequently detected in surveillance efforts, with only 47 strains sequenced after 2015 (35). Similarly, we did not select strains from within the 1B.2.2.2 clade; despite it representing a large number of HA genes in the earlier years of the data set, it was detected infrequently from 2016 to 2019 (i.e., the numerical dominance changed from 1B.2.2.2 to 1B.2.2.1 in 2015 [4, 36]).
Viruses and antisera production.
Viruses were grown in Madin-Darby canine kidney cells, with the exceptions of A/New Caledonia/20/1999 and A/Solomon Island/3/2006, which were grown in embryonated chicken eggs. The viruses used for serum production were clarified from cell culture supernatant, concentrated, and UV inactivated. Each vaccine dose was approximately 128 HAU of antigen mixed 5:1 with oil-and-water adjuvant (Emulsigen D, MVP Laboratories, Inc., Ralston, NE).
3-week-old cross-bred pigs that were free of IAV and IAV-antibodies were used for IAV antisera production. All of the pigs were treated with ceftiofur crystalline-free acid (Excede; Pfizer, New York, NY) and enrofloxacin (Baytril; Bayer Animal Health, Shawnee Mission, KS). Two pigs per virus received a prepared vaccine via intramuscular injection. Two or three doses were given two to three weeks apart. When HI titers to the homologous virus reached at least 1:160, the pigs were humanely euthanized with pentobarbital (Fatal Plus, Vortech Pharmaceuticals, Dearborn, MI) for blood collection. Sera was obtained through centrifugation and stored at − 20°C until use. Pigs were cared for in compliance with the Institutional Animal Care and Use Committee of the National Animal Disease Center, USDA-ARS.
Ferret antisera produced against CVV strains (1A.1.1 A/Ohio/24/2017, 1A.3.3.3 A/Ohio/09/2015, 1B.2.1 A/Ohio/35/2017, 1B.2.1 A/Michigan/383/2018, 1B.2.2.1 A/Iowa/32/2016) and a contemporary human pdm lineage H1N1 seasonal vaccine strain (1A.3.3.2 A/Idaho/7/2018) were provided by the Virology, Surveillance and Diagnosis Branch, Influenza Division, Centers for Disease Control and Prevention (CDC), Atlanta, Georgia.
Hemagglutination inhibition assays.
Prior to HI testing, swine sera were heat-inactivated at 56°C for 30 min and then treated with a 20% suspension of kaolin (Sigma-Aldrich, St. Louis, MO). This was followed by adsorption with 0.5% turkey red blood cells (RBCs). HI assays were performed by testing the reference antisera panel against the panel of H1 viruses according to standard techniques (37). The ferret antisera were treated and tested in a similar manner; however, 0.75% guinea pig RBCs were used for adsorption and in the HI assay. The human sera were similarly heat-inactivated and then treated with receptor destroying enzyme (RDE) and adsorbed with 0.5% turkey RBCs. HI assays were performed according to standard techniques. Two biological replicates of each virus antisera were used. Geometric mean titers were obtained via the log2 transformation of reciprocal titers divided by 10, and these were used in the analyses. Human sera with reciprocal HI titers against A/Michigan/45/2015 of 0 to 80 represent low reactors, while HI titers of 160 to 320 represent medium reactors, HI titers of 640 or greater represent high reactors.
Antigenic cartography.
Cross-HI tables generated using swine antisera were mapped and merged in three dimensions via multidimensional scaling implemented at https://acmacs-web.antigenic-cartography.org/. The antigenic distances between viruses were calculated in antigenic units (AU), where 1 AU is equivalent to a 2-fold loss in HI cross-reactivity. As defined for the human seasonal vaccine strain update, we used 3 AU or a ≥8-fold loss in cross-reactivity as the threshold of a significant loss for the risk ranking system described below. Only antigen and serum points supported by four or more titer values and antigens from strains isolated in 2012 or later were retained. Antigenic maps were exported from acmacs-web, and antigenic distances between antigens generated in the 3D map were extracted from the maps and plotted using ggplot2 (38) in R v3.6 (39). A subset of human seasonal vaccine strain antisera were used as replicates in each HI panel. The selected sera were determined by grouping vaccine strains by decade and determining the distribution of antigens/sera in the map with cross-reactivity to each sera. The merged HI table with all antigens and sera used can be found in Table S2.
Prioritizing swine H1 strains for pandemic risk assessment.
Given the large number of North American swine H1 IAV strains in our HI assays and the limited volume of human and ferret serum samples, we applied a scoring system to objectively select swine strains for additional characterization against human sera. As our goal was to identify swine H1 strains that have elevated pandemic potential, the ranking system determined whether a strain was representative of the currently circulating H1 genetic diversity in swine, with an accompanying assessment of whether the strain was antigenically unique that was similar to a previously developed method for H3 strains (40). Our assessment scored the relative risk to humans of a swine H1 virus from the panel (query strain) as , where the variables accounted for genetic sequence diversity, and the A variables accounted for antigenic diversity. The variable was the number of BLASTp hits that the query strain had with ≥97% amino acids to the data set (BLASTp search of the query strain against circulating strains) to identify the degree to which the tested strains represented all of the available swine H1 HA1. The variable was the number of BLASTp hits that the query strain had from the data set BLASTp search of circulating strains against the query strain to indicate which of the test strains best represented the HA1 of all of the available swine H1. The variable was the count of the number of times the query test strain was ≥3 AU from human vaccine strains to estimate the adult human immunity developed from seasonal H1 exposure or vaccination. The variance and outliers in the antigenic data were controlled for by Av (three times the standard deviation of all of the antigenic distance values, σ×3). The data set used for the genetic sequence diversity included 2 years of USA H1 HA data and 10 years of non-USA H1 HA data, due to the sparsity of recent data outside the USA. The highest scoring swine strains from each lineage and genetic clade were selected to be assayed against human sera (Table S3).
Human sera cohorts.
Human convalescent-phase sera representing different geographic locations and ages were tested against contemporary swine IAVs to assess human immunity. Two cohorts of human sera, a postinfection cohort with seasonal H1N1 and a postvaccination cohort, were tested against the selected swine H1 viruses. The sera were collected in a multicenter cohort study in two hospitals in the USA and three hospitals in Taiwan, coordinated by the Johns Hopkins Center of Excellence in Influenza Research and Surveillance (Johns Hopkins University IRB00091667). In both cohorts, patient information was recorded by dedicated research coordinators, following an informed consent process. The first cohort was composed of convalescent-phase sera of influenza-infected individuals (n = 10) from Taiwan and (n = 10) from the USA during the 2015 to 2016 season. The samples were collected at approximately 28 days postinfection with seasonal H1N1. Sera was collected from adult patients who presented at the hospital with an influenza-like illness. This was defined as a documented or reported fever and any of the three respiratory symptoms (cough, headache, sore throat) within the past 7 days of visiting the hospital. This cohort presented neutralizing antibody titers against the H1N1 vaccine strain (A/Michigan/45/2015). The second cohort was composed of sera collected from individuals (n = 40) vaccinated in the fall of 2017 with a quadrivalent influenza vaccine at the Johns Hopkins Hospital Employee Occupational Health Clinic (Baltimore, MD, USA). These subjects presented neutralizing antibody titers against the H1N1 vaccine strain (A/Michigan/45/2015), with samples having been taken approximately 28 days postvaccination. Females and males were grouped by decade of age: 20 to 29, 30 to 39, 40 to 49, and >50. Subsequently, a random number generator was used to select 5 per age group per gender. For the HI results analysis, the geometric mean HI titers were plotted against the different birth cohorts (1946 to 1976, 1977 to 1988, and 1989 to 1996) based on marked changes in the antigenicity of the circulating human H1 viruses (pre- and post-1977 outbreak, and the 1989 vaccine update). Spearman’s rank correlation was calculated to assess the association between the HI titers of the human seasonal H1N1 vaccine strains and the swine H1 strains. Figures were plotted using the ggplot2 package in R v3.6. The raw titer data set is provided in Table S4.
Data availability.
The data associated with this study are available as supplemental material and are posted at https://github.com/flu-crew/h1-risk-pipeline.
ACKNOWLEDGMENTS
We gratefully acknowledge pork producers, swine veterinarians, and laboratories for participating in the USDA Influenza A Virus in Swine Surveillance System and publicly sharing sequences in NCBI GenBank. We thank Michelle Harland, Gwen Nordholm, and Marcus Bolton for laboratory assistance and Jason Huegel, Keiko Sampson, and Justin Miller for animal care and handling assistance. We thank Susan Detmer from the University of Saskatchewan for the swine H1N1 viruses and the antisera as well as Adolfo García-Sastre and Ignacio Mena from the Icahn School of Medicine at Mount Sinai, NY, for providing the Mexican swine H1N1 strains. We thank Todd Davis and the members of the Virology, Surveillance, and Diagnosis Branch, Influenza Division, Centers for Disease Control and Prevention, for the provision of the reagents used in these studies. This work was supported in part by the U.S. Department of Agriculture (USDA) Agricultural Research Service (ARS project number 5030-32000-231-000D); a National Institute of Allergy and Infectious Diseases (NIAID) at the National Institutes of Health (NIH) Center of Excellence in Influenza Research and Surveillance interagency agreement associated with the Center of Research in Influenza Pathogenesis (HHSN272201400008C) and the Johns Hopkins Center of Excellence in Influenza Research and Surveillance (HHSN272201400007C); and the NIAID, NIH, Department of Health and Human Services (contract number 75N93021C00015). J.B.K, C.K.S., and J.C. were supported by an appointment to the USDA Agricultural Research Service Research Participation Program of the Oak Ridge Institute for Science and Education (ORISE) through an interagency agreement between the U.S. Department of Energy (DOE) and the USDA Agricultural Research Service (contract number DE-AC05-06OR23100) and through the SCINet project of the USDA Agricultural Research Service (ARS project number 0500-00093-001-00-D). The funders had no role in the study design, the data collection and interpretation, or the decision to submit the work for publication. The mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply a recommendation or an endorsement by the USDA, DOE, or ORISE. USDA is an equal opportunity provider and employer.
Footnotes
Supplemental material is available online only.
Contributor Information
Amy L. Vincent Baker, Email: amy.vincent@usda.gov.
Rafael A. Medina, Pontificia Universidad Católica de Chile
REFERENCES
- 1.Lewis NS, Russell CA, Langat P, Anderson TK, Berger K, Bielejec F, Burke DF, Dudas G, Fonville JM, Fouchier RA, Kellam P, Koel BF, Lemey P, Nguyen T, Nuansrichy B, Peiris JM, Saito T, Simon G, Skepner E, Takemae N, Webby RJ, Van Reeth K, Brookes SM, Larsen L, Watson SJ, Brown IH, Vincent AL, ESNIP3 consortium . 2016. The global antigenic diversity of swine influenza A viruses. Elife 5:e12217. doi: 10.7554/eLife.12217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ma W, Lager KM, Vincent AL, Janke BH, Gramer MR, Richt JA. 2009. The role of swine in the generation of novel influenza viruses. Zoonoses Public Health 56:326–337. doi: 10.1111/j.1863-2378.2008.01217.x. [DOI] [PubMed] [Google Scholar]
- 3.Nelson MI, Gramer MR, Vincent AL, Holmes EC. 2012. Global transmission of influenza viruses from humans to swine. J Gen Virol 93:2195–2203. doi: 10.1099/vir.0.044974-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rajao DS, Anderson TK, Kitikoon P, Stratton J, Lewis NS, Vincent AL. 2018. Antigenic and genetic evolution of contemporary swine H1 influenza viruses in the United States. Virology 518:45–54. doi: 10.1016/j.virol.2018.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gao S, Anderson TK, Walia RR, Dorman KS, Janas-Martindale A, Vincent AL. 2017. The genomic evolution of H1 influenza A viruses from swine detected in the United States between 2009 and 2016. J Gen Virol 98:2001–2010. doi: 10.1099/jgv.0.000885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rajao DS, Vincent AL, Perez DR. 2019. Adaptation of human influenza viruses to swine. Front Vet Sci 5. doi: 10.3389/fvets.2018.00347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anderson TK, Macken CA, Lewis NS, Scheuermann RH, Reeth KV, Brown IH, Swenson SL, Simon G, Saito T, Berhane Y, Ciacci-Zanella J, Pereda A, Davis CT, Donis RO, Webby RJ, Vincent AL. 2016. A phylogeny-based global nomenclature system and automated annotation tool for H1 hemagglutinin genes from swine influenza A viruses. mSphere 1. doi: 10.1128/mSphere.00275-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nelson MI, Viboud C, Vincent AL, Culhane MR, Detmer SE, Wentworth DE, Rambaut A, Suchard MA, Holmes EC, Lemey P. 2015. Global migration of influenza A viruses in swine. Nat Commun 6:6696. doi: 10.1038/ncomms7696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown IH, Harris PA, McCauley JW, Alexander DJ. 1998. Multiple genetic reassortment of avian and human influenza A viruses in European pigs, resulting in the emergence of an H1N2 virus of novel genotype. J General Virology 79:2947–2955. doi: 10.1099/0022-1317-79-12-2947. [DOI] [PubMed] [Google Scholar]
- 10.Lorusso A, Vincent AL, Harland ML, Alt D, Bayles DO, Swenson SL, Gramer MR, Russell CA, Smith DJ, Lager KM, Lewis NS. 2011. Genetic and antigenic characterization of H1 influenza viruses from United States swine from 2008. J Gen Virol 92:919–930. doi: 10.1099/vir.0.027557-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vincent AL, Ma W, Lager KM, Gramer MR, Richt JA, Janke BH. 2009. Characterization of a newly emerged genetic cluster of H1N1 and H1N2 swine influenza virus in the United States. Virus Genes 39:176–185. doi: 10.1007/s11262-009-0386-6. [DOI] [PubMed] [Google Scholar]
- 12.Anderson TK, Chang J, Arendsee ZW, Venkatesh D, Souza CK, Kimble JB, Lewis NS, Davis CT, Vincent AL. 2021. Swine influenza A viruses and the tangled relationship with humans. Cold Spring Harb Perspect Med 11:a038737. doi: 10.1101/cshperspect.a038737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guan Y, Shortridge KF, Krauss S, Li PH, Kawaoka Y, Webster RG. 1996. Emergence of avian H1N1 influenza viruses in pigs in China. J Virol 70:8041–8046. doi: 10.1128/JVI.70.11.8041-8046.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pensaert M, Ottis K, Vandeputte J, Kaplan MM, Bachmann PA. 1981. Evidence for the natural transmission of influenza A virus from wild ducks to swine and its potential importance for man. Bull World Health Organ 59:75–78. [PMC free article] [PubMed] [Google Scholar]
- 15.Mena I, Nelson MI, Quezada-Monroy F, Dutta J, Cortes-Fernández R, Lara-Puente JH, Castro-Peralta F, Cunha LF, Trovão NS, Lozano-Dubernard B, Rambaut A, van Bakel H, García-Sastre A. 2016. Origins of the 2009 H1N1 influenza pandemic in swine in Mexico. Elife 5:e16777. doi: 10.7554/eLife.16777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pulit-Penaloza JA, Jones J, Sun X, Jang Y, Thor S, Belser JA, Zanders N, Creager HM, Ridenour C, Wang L, Stark TJ, Garten R, Chen L-M, Barnes J, Tumpey TM, Wentworth DE, Maines TR, Davis CT. 2018. Antigenically diverse swine origin H1N1 variant influenza viruses exhibit differential ferret pathogenesis and transmission phenotypes. J Virol 92:e00095-18. doi: 10.1128/JVI.00095-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun X, Pulit-Penaloza JA, Belser JA, Pappas C, Pearce MB, Brock N, Zeng H, Creager HM, Zanders N, Jang Y, Tumpey TM, Davis CT, Maines TR. 2018. Pathogenesis and transmission of genetically diverse swine-origin H3N2 variant influenza A viruses from multiple lineages isolated in the United States, 2011–2016. J Virol 92:e00665-18. doi: 10.1128/JVI.00665-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaplan BS, Kimble JB, Chang J, Anderson TK, Gauger PC, Janas-Martindale A, Killian ML, Bowman AS, Vincent AL. 2020. Aerosol transmission from infected swine to ferrets of an H3N2 virus collected from an agricultural fair and associated with human variant infections. J Virol 94:e01009-20. doi: 10.1128/JVI.01009-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hirst GK. 1942. The quantitative determination of influenza virus and antibodies by means of red cell agglutination. J Exp Med 75:49–64. doi: 10.1084/jem.75.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lapedes A, Farber R. 2001. The geometry of shape space: application to influenza. J Theor Biol 212:57–69. doi: 10.1006/jtbi.2001.2347. [DOI] [PubMed] [Google Scholar]
- 21.Smith DJ, Lapedes AS, de Jong JC, Bestebroer TM, Rimmelzwaan GF, Osterhaus ADME, Fouchier RAM. 2004. Mapping the antigenic and genetic evolution of influenza virus. Science 305:371–376. doi: 10.1126/science.1097211. [DOI] [PubMed] [Google Scholar]
- 22.Abente EJ, Santos J, Lewis NS, Gauger PC, Stratton J, Skepner E, Anderson TK, Rajao DS, Perez DR, Vincent AL. 2016. The molecular determinants of antibody recognition and antigenic drift in the H3 hemagglutinin of swine influenza A virus. J Virol 90:8266–8280. doi: 10.1128/JVI.01002-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bedford T, Suchard MA, Lemey P, Dudas G, Gregory V, Hay AJ, McCauley JW, Russell CA, Smith DJ, Rambaut A. 2014. Integrating influenza antigenic dynamics with molecular evolution. Elife 3:e01914. doi: 10.7554/eLife.01914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Caton AJ, Brownlee GG, Yewdell JW, Gerhard W. 1982. The antigenic structure of the influenza virus A/PR/8/34 hemagglutinin (H1 subtype). Cell 31:417–427. doi: 10.1016/0092-8674(82)90135-0. [DOI] [PubMed] [Google Scholar]
- 25.Nelson MI, Culhane MR, Trovão NS, Patnayak DP, Halpin RA, Lin X, Shilts MH, Das SR, Detmer SE. 2017. The emergence and evolution of influenza A (H1α) viruses in swine in Canada and the United States. J Gen Virol 98:2663–2675. doi: 10.1099/jgv.0.000924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Squires RB, Noronha J, Hunt V, García-Sastre A, Macken C, Baumgarth N, Suarez D, Pickett BE, Zhang Y, Larsen CN, Ramsey A, Zhou L, Zaremba S, Kumar S, Deitrich J, Klem E, Scheuermann RH. 2012. Influenza Research Database: an integrated bioinformatics resource for influenza research and surveillance. Influenza Other Respir Viruses 6:404–416. doi: 10.1111/j.1750-2659.2011.00331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Y, Aevermann BD, Anderson TK, Burke DF, Dauphin G, Gu Z, He S, Kumar S, Larsen CN, Lee AJ, Li X, Macken C, Mahaffey C, Pickett BE, Reardon B, Smith T, Stewart L, Suloway C, Sun G, Tong L, Vincent AL, Walters B, Zaremba S, Zhao H, Zhou L, Zmasek C, Klem EB, Scheuermann RH. 2017. Influenza Research Database: an integrated bioinformatics resource for influenza virus research. Nucleic Acids Res 45:D466–D474. doi: 10.1093/nar/gkw857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li W, Godzik A. 2006. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 29.Fu L, Niu B, Zhu Z, Wu S, Li W. 2012. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28:3150–3152. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Katoh K, Standley DM. 2013. MAFFT Multiple Sequence Alignment Software Version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 34.Yu G, Smith DK, Zhu H, Guan Y, Lam TT-Y. 2017. ggtree: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol Evol 8:28–36. doi: 10.1111/2041-210X.12628. [DOI] [Google Scholar]
- 35.Arendsee ZW, Chang J, Hufnagel DE, Markin A, Janas-Martindale A, Vincent AL, Anderson TK. 2021. octoFLUshow: an interactive tool describing spatial and temporal trends in the genetic diversity of influenza A virus in U.S. swine. Microbiol Resour Announc 10:e01081-21. doi: 10.1128/MRA.01081-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zeller MA, Anderson TK, Walia RW, Vincent AL, Gauger PC. 2018. ISU FLUture: a veterinary diagnostic laboratory web-based platform to monitor the temporal genetic patterns of influenza A virus in swine. BMC Bioinformatics 19:397. doi: 10.1186/s12859-018-2408-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kitikoon P, Gauger PC, Vincent AL. 2014. Hemagglutinin inhibition assay with swine sera, p 295–301. In Spackman E (ed), Animal Influenza Virus. Springer, New York, NY. [DOI] [PubMed] [Google Scholar]
- 38.Wickham H. 2016. ggplot2: elegant graphics for data analysis. Springer-Verlag, New York, NY. https://ggplot2.tidyverse.org. [Google Scholar]
- 39.R Core Team. 2020. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 40.Souza CK, Anderson TK, Chang J, Venkatesh D, Lewis NS, Pekosz A, Shaw-Saliba K, Rothman RE, Chen KF, Vincent AL. 2022. Antigenic distance between North American Swine and Human Seasonal H3N2 Influenza A Viruses as an Indication of Zoonotic Risk to Humans. J Virol 96:e0137421. doi: 10.1128/JVI.01374-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download spectrum.01781-22-s0001.pdf, PDF file, 2.9 MB (2.9MB, pdf)
Data Availability Statement
The data associated with this study are available as supplemental material and are posted at https://github.com/flu-crew/h1-risk-pipeline.