ABSTRACT
Carbapenem-resistant hypervirulent Klebsiella pneumoniae (CR-hvKP) is recognized as a threat worldwide, but the mechanisms underlying its emergence remain unclear. As most CR-hvKP isolates are not hypermucoviscous, we speculated that the evolution of the capsule might result in the convergence of carbapenem resistance and hypervirulence. Here, 2,096 K. pneumoniae isolates were retrospectively collected to screen the ST23-K1 clone, and hypervirulence was roughly defined as being highly resistant to serum killing. The effect of wcaJ on the capsule, virulence, fitness, and resistance acquisition was further analyzed. The capsule gene wcaJ, inserted by ISKpn26/ISKpn74, was identified via whole-genome sequencing in four hvKP, but not hypermucoviscous, isolates. Uronic acid quantitation results revealed that these isolates produced significantly less capsular polysaccharides than NTUH-K2044. A significant increase in capsular production was observed in wcaJ-complemented isolates and confirmed by transmission electron microscopy. Further, all wcaJ-complemented isolates acquired greater resistance to macrophage phagocytosis, and one representative isolate resulted in a significantly higher mortality rate than the parental isolate in mice, indicating that wcaJ inactivation might compromise virulence. However, isolates with wcaJ interruption demonstrated a lower fitness cost and a high conjugation frequency of the blaKPC-2 plasmid, raising concerns about the emergence of carbapenem resistance in hvKP.
IMPORTANCE Klebsiella pneumoniae is one of the most common nosocomial pathogens worldwide, and we speculated that the evolution of the capsule might result in the convergence of carbapenem resistance and hypervirulence of K. pneumoniae. The wcaJ gene was first reported to be interrupted by insertion sequence elements in ST23-K1 hypervirulent Klebsiella pneumoniae, resulting in little capsule synthesis, which plays an important role in virulence. We examined the effect of wcaJ on the capsule, virulence, and fitness. Isolates with wcaJ interruption might compromise virulence and demonstrated a lower fitness cost and a high conjugation frequency of the blaKPC-2 plasmid, highlighting its role as a potential factor facilitating hypervirulence and carbapenem resistance.
KEYWORDS: carbapenem-resistant hypervirulent Klebsiella pneumoniae, CR-hvKP, capsule, conjugation, insertion sequences, IS, virulence, wcaJ
INTRODUCTION
Klebsiella pneumoniae is one of the most common nosocomial pathogens worldwide, posing a serious threat to public health (1). In recent years, two variants of K. pneumoniae, namely, hypervirulent K. pneumoniae (hvKP) and carbapenem-resistant K. pneumoniae (CRKP), have become particularly great concerns, with hvKP causing serious and metastatic infections, such as liver abscess, endophthalmitis, and necrotizing fasciitis, which are associated with high rates of mortality and morbidity in healthy individuals (2). In China, the majority of pyogenic liver abscesses are due to hvKP (3), and most hvKP isolates are susceptible to all available antimicrobials (4). However, the convergence of hypervirulence and carbapenem resistance was identified in K. pneumoniae, with a significant increase in its reported prevalence (5). Carbapenem-resistant hypervirulent K. pneumoniae (CR-hvKP) demonstrates a limited fitness cost compared with the classic CRKP and hvKP strains (6–8), raising concerns about its further dissemination in clinical settings.
Numerous studies have investigated the epidemiology of CR-hvKP, whereas others have dissected the characteristics underlying its antibiotic resistance and virulence (4, 6, 9–11). However, there is still limited knowledge of the molecular mechanisms driving the emergence of CR-hvKP, particularly the element that decreases the fitness cost associated with the cooccurrence of virulence and resistance.
Herein, an analysis of four K. pneumoniae genomes, with a focus on the prevalent clone ST23-K1 hvKP, along with virulence and fitness assays revealed that the wcaJ gene is a potential factor that facilitates the combination of hypervirulence and multidrug resistance. The wcaJ gene in K. pneumoniae encodes a glycosyltransferase, which is the initiating enzyme of the colanic acid synthesis pathway, and it plays an important role in polysaccharide synthesis (12). Insertion in the wcaJ gene via insertion sequence (IS) elements in ST23-K1 hvKP was found to be associated with a low fitness cost and a high conjugation capacity with respect to the blaKPC gene, increasing the potential risk of CR-hvKP emergence.
RESULTS
Microbiological characteristics of K. pneumoniae isolates.
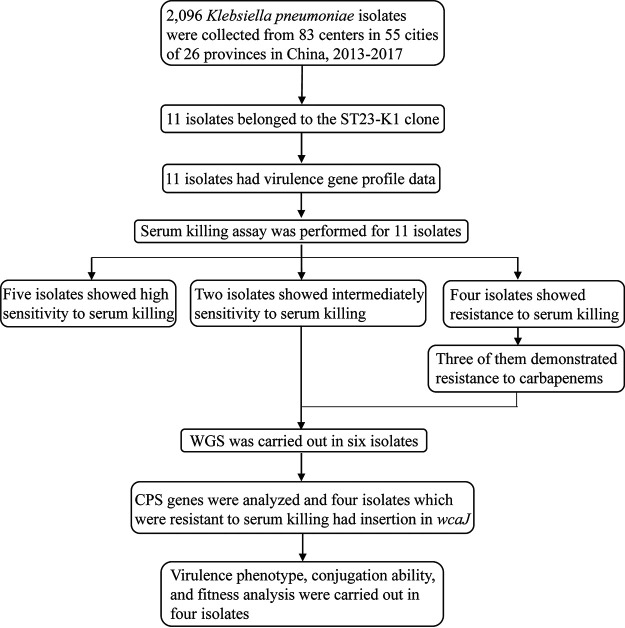
In total, 2,096 K. pneumoniae isolates were collected from the China Carbapenem-Resistant Enterobacteriaceae (CRE) Network (13) between 2013 and 2017, with 11 isolates belonging to the ST23-K1 clone (Table 1). Among these, 10 isolates were positive for all critical virulence genes tested, including iucA, iroN, rmpA, and rmpA2, indicating that they harbor the pLVPK-like virulence plasmid. Hypermucoviscosity and the ability to defend against serum killing provide valuable information regarding the characterization of hvKP (14). Interestingly, we found that six isolates (designated C228, C400, C1356, C2768, C4120, and C4599) were not sensitive to serum killing and that four of them (C400, C1356, C2768, and C4599) were highly resistant to serum killing (grade 6) but negative for the string test. In addition, four isolates (C228, C400, C1356, and C4599) harbored the blaKPC-2 carbapenemase gene. Furthermore, the O locus types of these six isolates belonged to the O1/O2v2 serotype, the same as that for NTUH-K2044. The reasons for such phenotypic differences in serum killing and the string test were further investigated in this study. The study design is summarized in Fig. 1.
TABLE 1.
Microbiological information for ST23-K1 isolatesa
| Isolate ID | Collection date (yr/mo/day) | City | K locus | ST | KPC | Serum killing | String testb | IS element in wcaJ | ISKpn26 | ISKpn74 | rmpA | rmpA2 | O-antigen serotype | O type | rmpD type |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NTUH-K2044 | Taiwan | KL1 | ST23 | Resistant (grade 6) | + | − | + | + | + | O1/O2v2 | O1 | 2 | |||
| C1356 | 2015/2/7 | Beijing | KL1 | ST23 | KPC-2 | Resistant (grade 6) | − | ISKpn26 | + | + | + | + | O1/O2v2 | O1 | 3 |
| C2768 | 2017/2/20 | Guangzhou | KL1 | ST23 | Resistant (grade 6) | − | ISKpn74 | − | + | + | + | O1/O2v2 | O1 | 3 | |
| C400 | 2015/2/10 | Beijing | KL1 | ST23 | KPC-2 | Resistant (grade 6) | − | ISKpn26 | + | + | + | + | O1/O2v2 | O1 | 3 |
| C4599 | 2017/9/12 | Kunming | KL1 | ST23 | KPC-2 | Resistant (grade 6) | − | ISKpn26 | + | + | + | + | O1/O2v2 | O1 | |
| C228 | 2014/7/4 | Nanjing | KL1 | ST23 | KPC-2 | Intermediate (grade 4) | − | − | + | − | + | O1/O2v2 | O1 | 26 | |
| C4120 | 2017/6/9 | Zhengzhou | KL1 | ST23 | Intermediate (grade 4) | + | − | + | + | + | O1/O2v2 | O1 | 2 | ||
| C1908 | 2016/12/19 | Kunming | KL1 | ST23 | Sensitive (grade 2) | + | − | + | + | + | |||||
| C2362 | 2017/2/19 | Beijing | KL1 | ST23 | Sensitive (grade 2) | + | − | + | + | + | |||||
| C787 | 2016/3/27 | Shenzhen | KL1 | ST23 | KPC-2 | Sensitive (grade 2) | + | − | + | + | + | ||||
| C1922 | 2016/2/10 | Tianjin | KL1 | ST23 | Sensitive (grade 1) | − | − | + | + | + | |||||
| C1928 | 2016/3/1 | Tianjin | KL1 | ST23 | Sensitive (grade 1) | − | − | + | + | + |
FIG 1.
Design and experimental procedures of this study.
Identification of a glycosyltransferase-encoding gene, wcaJ, as a potential reason for resistance to serum killing but negative string test results.
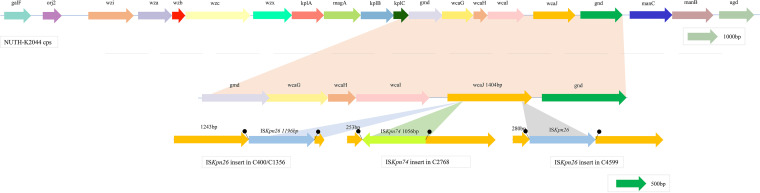
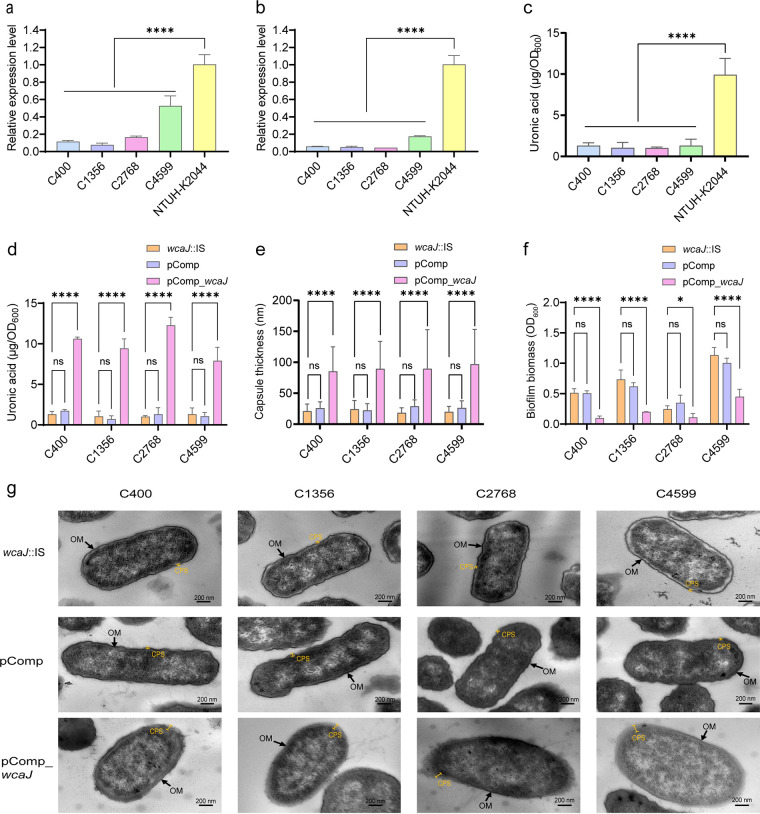
Since capsular polysaccharide synthesis (CPS) is an important factor affecting the hypermucoviscosity phenotype and serum killing (14), we speculated that genes encoding CPS could be altered, giving rise to the phenotypic diversity observed in serum killing and string tests. Four isolates (C400, C1360, C2768, and C4599) were found to have an insertion in the wcaJ gene, which was the only mutated CPS gene according to Kleborate, BLAST, and mapping results (15–18) (see Table S1 in the supplemental material). C228 contained an intact wcaJ gene but was negative in the string test, which might be due to the absence of the rmpA gene (Table 1), despite possessing an intact hypermucoviscosity-related gene, rmpD (19). In addition, four isolates with wcaJ inserted via an IS element, but not C4599, possessed an intact rmpD gene, which did not result in a hypermucoviscosity phenotype. Among these four isolates showing wcaJ inactivation, three were CR-hvKP, and ISKpn26 was inserted in wcaJ in all of these isolates (1,196 bp, IS5 group, IS5 family) (5), whereas the other was a hvKP isolate, in which wcaJ was found to contain a newly reported IS element, ISKpn74 (1,056 bp, IS903 group, IS5 family) (20). Figure 2 shows the genomic environment of the wild-type wcaJ gene in the reference isolate NTUH-K2044 (21). The wcaJ gene was determined to be located in the first contig of the NTUH-K2044 genome (GCF_000009885.1), with a length of 1,404 bp. The ISKpn26 insertion site in wcaJ was the same in C400 and C1356 (nucleotide [nt] position 1243) but was different in C4599 (nt position 280). Furthermore, ISKpn74 was inserted in the wcaJ gene of C2768 at nt position 253 (Fig. 2). To confirm the inactivation of the wcaJ gene, we designed two pairs of primers to detect wcaJ gene expression, and the positions of these are shown in Fig. S1. As expected, compared with that in the reference isolate NTUH-K2044, both fragments showed low expression in all four isolates (Fig. 3a and b) (P < 0.0001).
FIG 2.
Disruption of wcaJ in this study. The Klebsiella pneumoniae K1 capsular polysaccharide biosynthesis gene cluster is shown. The wcaJ gene of C400/C1356 was disrupted by the insertion of ISKpn26 at nt position 1243; in C4599, ISKpn26 was inserted at nt position 280, whereas in the wcaJ gene of C2768, ISKpn74 was inserted at nt position 253.
FIG 3.
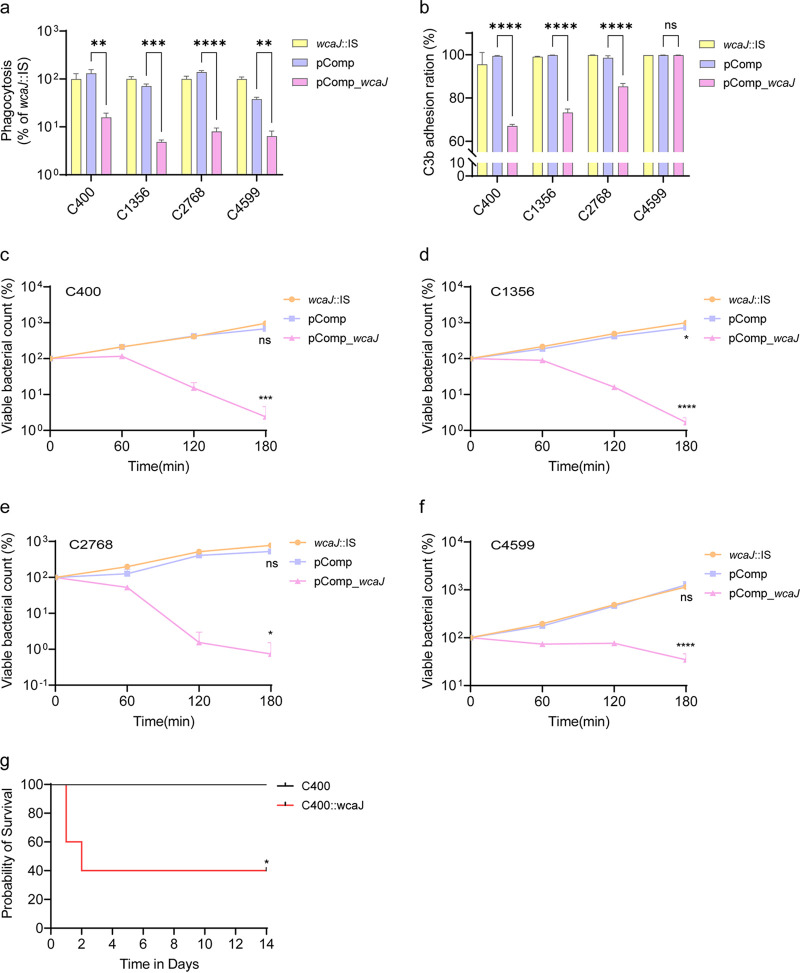
Capsule-related phenotype analysis. (a and b) RT-qPCR analysis of upstream and downstream fragments, respectively, of IS insertion in the wcaJ gene region. The NTUH-K2044 isolate was used as a reference. (c) The uronic acid contents of C400, C1356, C2768, and C4599 isolates were compared to that of NTUH-K2044. (d) Uronic acid production was significantly increased in wcaJ-complemented isolates compared to that in the parent isolates with IS insertion in wcaJ (wcaJ::IS). (e) Capsule thickness was determined by measuring five different sites for 10 cells per isolate. The data represent the average of 50 replicates ± standard deviations (SD). (f) Biofilm formation comparison between wcaJ::IS and wcaJ-complemented counterparts. Six replicates were performed per isolate. (g) Representative TEM images of wcaJ::IS, pComp, and wcaJ-complemented counterparts. Arrows indicate the outer membrane (OM). Capped lines mark the capsule (CPS). Each data point represents three replicates (n = 3), except for those in panels e and f. Data are presented as the mean ± SD. *, P < 0.05; ****, P < 0.0001; ns, not significant.
Clinical features of patients infected by isolates with wcaJ inactivation.
The medical records of four patients infected by isolates with wcaJ inactivation are summarized in Table 2. All isolates were recovered from sputum samples. Notably, patients infected with C400 and C1356 were hospitalized in the same department. All patients suffered from underlying diseases, such as hypertension and heart disease. Except for the patient infected with C4599, most (75%, 3/4) had received antimicrobials 30 days before infection. All of these four patients had pulmonary infections and recovered, except for the patient infected with C2768, who developed a fungal infection and sepsis and finally died.
TABLE 2.
Clinical characteristics of the four patients infected with wcaJ-interrupted isolates
| Parameter | Patient 1 (C400) | Patient 2 (C1356) | Patient 3 (C2768) | Patient 4 (C4599) |
|---|---|---|---|---|
| Age (yr) | 52 | 42 | 59 | 62 |
| Gender | Female | Male | Male | Male |
| City | Beijing | Beijing | Guangdong | Kunming |
| Type of specimen | Sputum | Sputum | Sputum | Sputum |
| Ward | Internal medicine | Internal medicine | Internal medicine, ICUa | Rehabilitation department |
| Underlying disease(s) | Hypertension | Cerebral hemorrhage, diabetes, hypertension, hemiplegia | Hypertension, coronary heart disease | Hypertension, heart disease, nervous system disease, hemiplegia |
| Infection type(s) | Pneumonia | Pneumonia | Pulmonary infection, sepsis, fungal infection | Pulmonary infection |
| Prior antibiotic usage within 30 days | Yes | Yes | Yes | No |
| Empirical antimicrobial usage | Cefoxitin, levofloxacin | Moxifloxacin, amikacin | Piperacillin/tazobactam, imipenem, teicoplanin | No |
| Maximum temp (°C) | Unknown | 37.4 | 37 | Unknown |
| WBCa (109/L) | Unknown | 4.46 | 16.3 | 14.79 |
| Therapeutic antimicrobial usage | Unknown | Unknown | Imipenem | Unknown |
| Outcome | Recovered | Recovered | Died | Recovered |
ICU, intensive care unit; WBC, leukocytes.
IS element insertion into the wcaJ gene among isolates in the public database.
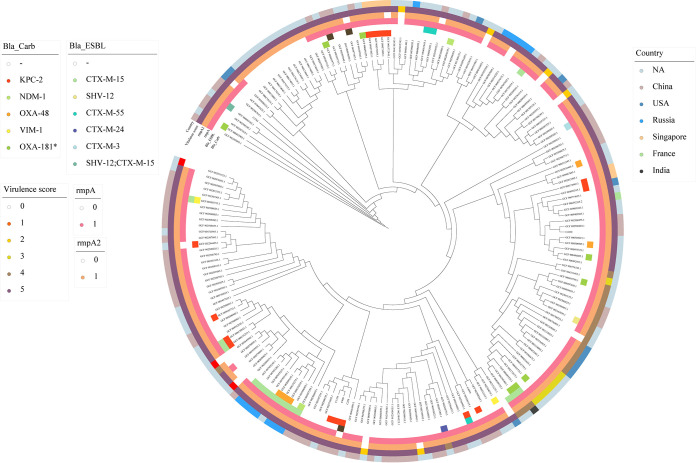
To investigate insertions in the wcaJ gene among ST23-K1 isolates in public databases, we downloaded 185 K. pneumoniae genomes of the ST23-K1 clone from the NCBI database (Table S1). Among 185 ST23-K1 isolates in the public database, 30 harbored rmpA or rmpA2 deletions and 16 had virulence scores of <4 (Table S1) (15). Isolates with an rmpA deletion and those with a virulence score of 4 discernably clustered into different groups in the evolutionary analysis. At the same time, isolates from the same country clustered together (Fig. 4). Furthermore, most ST23-K1 isolates demonstrated the possession of ISKpn74 (91/191) (Table 1; Table S2), indicating the potential risk of the insertion of such mobile elements into the wcaJ gene. In addition to the four ST23-K1 isolates in this study, we confirmed that another three isolates from the database also had insertions in the wcaJ gene (Table S3).
FIG 4.
Phylogenetic tree of ST23-K1 isolates. The core genome phylogenetic tree of 191 isolates, including 6 isolates in this study and 185 isolates downloaded from public databases, is shown. On the outside of the tree, the inner two panels provide information about the resistance genes carried by each isolate, including the β-lactamase (Bla), carbapenemase (Carb), and extended-spectrum β-lactamase (ESBL) genes; the three outer subpanels indicate the presence of rmpA and rmpA2 and the virulence score. The outermost panels are colored according to the country of isolates.
The wcaJ gene plays a critical role in capsule synthesis.
Uronic acid quantitation results revealed that these four isolates produced significantly less capsular polysaccharide than NTUH-K2044 (P < 0.0001), which was used as a high-capsule-producing reference isolate (Fig. 3c). We hypothesized that the poor capsule synthesis ability of C400, C1356, C2768, and C4599 was due to wcaJ inactivation by the IS family. To confirm this, complementation of the wild-type wcaJ gene in C400, C1356, C2768, and C4599 isolates was performed. The wcaJ-complemented isolates (pComp_wcaJ) were positive in the string test (Fig. S2). However, after low-speed centrifugation, the supernatants of two pComp_wcaJ isolates (C400 and C1356) remained clear, whereas those of the other two (C2768 and C4599) became significantly turbid with pComp_wcaJ plasmid complementation (Fig. S3), indicating hypermucoviscosity. Regarding capsular production, a significant increase was observed in all pComp_wcaJ isolates (Fig. 3d) (P < 0.001). Furthermore, a thicker capsule was detected by transmission electron microscopy (TEM) analysis of the wcaJ-complemented isolates (Fig. 3e and g) (P < 0.0001). Moreover, less biofilm was observed in wcaJ-complemented isolates than in clinical isolates with wcaJ inactivation (Fig. 3f) (P < 0.05), suggesting a greater colonization ability among wcaJ-interrupted isolates.
Effect of the wcaJ gene on virulence in vitro and in vivo.
To elucidate the effect of the wcaJ gene on virulence in vitro, serum killing and macrophage phagocytosis assays were performed. All pComp_wcaJ isolates acquired greater resistance to macrophage phagocytosis than the parent isolates (Fig. 5a) (P < 0.01). Intriguingly, in contrast to that observed in the parent isolates that were highly resistant to serum killing (grade 6), pComp_wcaJ isolates exhibited increased susceptibility (grade 2 for C400 and C2768 and grade 3 for C1356 and C4599) (Fig. 5c to f). To explore the reason for the inconsistent results between serum killing and macrophage phagocytosis assays, a flow cytometric analysis of C3b deposition on the surface of isolates was conducted. The wcaJ-inactivated isolates exhibited high C3b complement adhesion when incubated with serum and fluorescein isothiocyanate (FITC) anti-C3b antibodies, whereas pComp_wcaJ isolates showed low C3b complement adhesion, except for C4599 (Fig. 5b). The controversial phenomenon in which wcaJ-interrupted isolates with fewer capsules have greater tolerance to serum killing requires further investigation.
FIG 5.
Virulence-related phenotype analysis. (a) Phagocytosis by RAW 264.7 cells. The values shown were calculated as a ratio of the intracellular bacteria to total bacteria and normalized to that of wcaJ::IS isolates (n = 3). Data were analyzed by one-way ANOVA. Data are presented as the mean ± SD. (b) Flow cytometric analysis of C3b deposition on the surfaces of isolates (n = 3). Data were analyzed by one-way ANOVA. Data are presented as the mean ± SD. (c to f) Serum killing results for C400, C1356, C2768, C4599, and their counterparts. Samples were taken at 0, 60, 120, and 180 min, and each isolate was tested three times. Isolates were compared by two-way ANOVA (overall P < 0.0001), and Tukey’s multiple-comparison test at 180 min showed significant differences. (g) Survival of BALB/c mice. The mice were challenged with 1.96 × 106 CFU of the C400 isolate and 1.97 × 106 CFU of its wcaJ-complemented isolate. The mortality of mice was observed over 14 days. The significance was calculated using the log rank (Mantel-Cox) test. *, P < 0.05; ****, P < 0.0001; ns, not significant.
A murine infection model was also used to assess virulence in vivo, and C400 was selected as a representative isolate. The mortality rate of mice challenged with the C400 wcaJ-complemented isolate was significantly higher than that of mice challenged with the parent isolate (Fig. 5g) (P = 0.049), which indicated that insertion in wcaJ might compromise virulence.
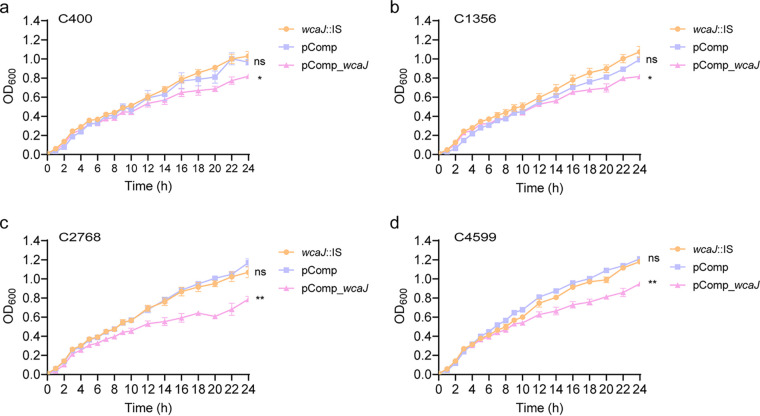
Fitness and conjugation ability of blaKPC-2 are enhanced by wcaJ gene inactivation in hvKP.
Compared to that in isolates with wcaJ interrupted by an IS, a significant fitness cost was observed in wcaJ-complemented isolates (Fig. 6) (P < 0.05). The impact of capsular changes caused by wcaJ inactivation on the process of blaKPC-2 plasmid conjugation was further considered. Since few ST23-K1 isolates with IS elements inserted into the wcaJ gene were identified in our database, the KPC plasmids of C400 and C1356 were first eliminated to construct plasmid-curing isogenic derivatives, and conjugation efficiency was then compared between ΔKPC isogenic derivatives and their wcaJ-complemented strains. Deletion of the blaKPC-2 plasmid was confirmed using PCR, and antimicrobial susceptibility testing showed that the susceptibility of carbapenems was restored (Table S4). A higher conjugation frequency (10−5) was observed in ΔKPC::pComp isolates than in ΔKPC::pComp_wcaJ isolates (conjugation frequency < 10−8, P < 0.01), which showed that complementation of wcaJ might prevent KPC conjugation by increasing capsule thickness. In this study, the other two isolates with a wcaJ truncation (C4599 and C2768) were not subjected to conjugation assays due to failure in the elimination of the KPC plasmid and the lack of appropriate antibiotic selection markers, respectively.
FIG 6.
Growth curve of C400, C1356, C2768, and C4599 isolates and their corresponding isogenic derivative strains pComp and pComp_wcaJ. (a) C400; (b) C1356; (c) C2768; (d) C4599. Data were analyzed by two-way ANOVA (overall P < 0.05), and Tukey’s multiple-comparison test at 24 h showed significant differences. *, P < 0.05; **, P < 0.01; ns, not significant.
DISCUSSION
The wcaJ gene encodes a glycosyltransferase; this enzyme loads the first glucose moiety onto the lipid carrier undecaprenyl phosphate, which is responsible for initiating capsule synthesis (22). Previous studies reported that the inactivation of wcaJ caused by ISKpn26 in the K64 isolate and IS1 in a K. pneumoniae isolate interrupts capsular polysaccharide synthesis but that wcaJ-complemented counterparts show restored CPS production (23, 24), which is consistent with our results. We observed that ISKpn26 and ISKpn74 were inserted into wcaJ in ST23-K1 isolates. Notably, ISKpn74 is widespread among ST23-K1 isolates, and ISKpn26 is one of the most common mobile elements in blaKPC-2 plasmids, indicating the potential risks of ISKpn26 and ISKpn74 transfer.
The capsule is considered an important in vitro component related to virulence, and inactivation of the wcaJ gene decreases capsule synthesis, thus affecting virulence. Previous studies have proven that ΔwcaJ mutants exhibit impaired capsule production and are susceptible to uptake by RAW264.7 macrophages (25), which was also observed for wcaJ-interrupted isolates in our study. In addition, respective wcaJ-complemented isolates showed restored CPS production and resistance to phagocytosis.
Although the CPS is the main factor protecting K. pneumoniae from the complement system (26), our study showed that wcaJ inactivation could increase serum resistance despite producing little CPS. There are two main mechanisms underlying K. pneumoniae tolerance to the bactericidal effect of the complement system (27). One is to avoid complement activation via its thick capsule, whereas the other is to prevent membrane attack complex assembly on the bacterial surface via the long O-polysaccharide side chains of lipopolysaccharide, which prevent C5b-9 formation. Our current findings confirmed an inconsistency between serum resistance and the C3b sedimentation rate, in which the wcaJ-inactivated isolates exhibited high C3b complement adhesion and high serum resistance simultaneously. This contradictory phenomenon might reflect failed complement activation, but the exact underlying reason requires further investigation. Furthermore, the bacterial capsule harbors receptors for bacteriophages. A recent study reported that mutations in genes essential for capsule biosynthesis in Acinetobacter baumannii confer resistance to phage adsorption through capsule loss (28). Similarly, the wcaJ gene is related to phage sensitivity (29), and its absence might protect isolates, particularly CR-hvKP, from lytic phage predation in vitro and in vivo (24, 30), thus enhancing their viability.
Consistent with the previous view that the conjugation rate of the resistance plasmid is higher in isolates lacking capsule barriers (31, 32), isolates with lower capsule production owing to wcaJ inactivation in this study exhibited greater conjugation efficiency with respect to the blaKPC-2 plasmid. More alarmingly, isolates with wcaJ interrupted by IS elements demonstrated a lower fitness cost than wcaJ-complemented isolates. This raises concerns about the emergence and dissemination of carbapenem resistance among ST23-K1 clones of hvKP.
This research, however, is subject to several limitations. First, the complementation experiment was used to evaluate the effects of IS element insertion in wcaJ. In fact, the in situ replacement approach could be better for demonstrating these effects, as some toxic effects might be caused by wcaJ overexpression in Escherichia coli TOP10 (33). Second, as the result in the mouse infection model barely meets statistical significance (five mice per group, P = 0.049), the virulence assessment of strains in vivo can likely be improved by increasing the number of mice per group. In addition, the inclusion of the three other isolates could also contribute to a more comprehensive understanding. Third, the inclusion of more ST23 CR-hvKP isolates in future research will contribute to an expansion of this work.
In conclusion, we report the clinical characteristics and molecular features of four clinical ST23-K1 hvKP isolates with IS elements inserted into the capsule gene wcaJ. Further, we dissected the effect of wcaJ on capsule synthesis, virulence, and fitness. Our study highlights the role of wcaJ as a potential factor that facilitates the combination of hypervirulence and carbapenem resistance, suggesting the urgent need to increase clinical awareness with respect to this matter.
MATERIALS AND METHODS
Study design and procedures.
To elucidate the mechanism of carbapenem resistance emergence in ST23-K1 hvKP, we carried out a systemic in-depth study. First, K. pneumoniae isolates, with a focus on CRKP, were retrospectively collected from our national multicenter surveillance network to screen ST23-K1 clones and the hypervirulence phenotype, which was defined as high resistance to serum killing in vitro. The capsule is one of the most critical virulence-associated factors of hvKP (34), and our previous work demonstrated that the majority of CR-hvKP strains are negative for the string test, suggesting capsule alterations (data not shown). Thus, we speculated that the evolution of capsules might be a reason for the acquisition of carbapenem resistance in hvKP. For the ST23-K1 isolates with hypervirulence, being intermediate or resistant to serum killing, whole-genome sequencing (WGS) was performed, and genes encoding the capsule were further investigated. We found that wcaJ was inserted in the four isolates that were highly resistant to serum killing. To explore the role of wcaJ alterations, the effects on capsule, virulence, fitness, and resistance transmission were analyzed.
Microbiological features.
In total, 2,096 K. pneumoniae isolates were collected from 83 centers in 55 cities in 26 provinces in China between 2013 and 2017. Multilocus sequence typing (MLST) analysis was performed according to the instructions on the MLST website (https://bigsdb.pasteur.fr/klebsiella/), and PCR was used to detect virulence genes (iucA, iroN, rmpA, and rmpA2), K serotypes (K1, K2, K5, K20, K54, K57, K47, and K64), and the carbapenemase gene (blaKPC-2), as previously described (7).
WGS and analysis.
Six isolates with intermediate or full resistance to serum killing were sequenced using an Illumina NovaSeq system and a HiSeq 2500 system to investigate capsular genes. Reads were assembled and annotated using SPAdes (version 3.13.0) (35) and Prokka (version 1.13.7) (36). The K serotype, MLST, O locus classification, antibiotic resistance genes, and virulence genes were analyzed using Kleborate v2.2.0 and Kaptive v2.0.3 (https://github.com/katholt/Kleborate, https://github.com/katholt/Kaptive) (15, 16, 37). Potential insertion and deletion sites were examined with blastn (version 2.9.0) (18), BWA-MEM (version 0.7.12) (17), and Integrative Genomics Viewer (IGV; version 2.11.4) (38) (see Fig. S4 in the supplemental material) and were further confirmed through PCR and Sanger sequencing. Meanwhile, ISfinder (39) (https://www-is.biotoul.fr/) and blastn were utilized for the identification of IS types.
Phylogenetic analysis.
To study the distribution of and phylogenetic relationships associated with antibiotic resistance and virulence genes among our isolates and other isolates in the public database, phylogenetic analysis was performed using 191 isolates, including data for 185 isolates downloaded from public databases and six isolates in this study. Kleborate and blastn were used to identify the wcaJ gene integrity in the 185 isolates from public databases, and isolates with incomplete whole-genome sequencing data for which the presence of an IS could not be discerned were not included.
RT-qPCR.
For real-time quantitative PCR (RT-qPCR), total RNA was extracted according to the manufacturer’s instructions (Qiagen, Germany), and reverse transcription was then carried out with 500 ng of RNA using the PrimeScript RT master mix kit (TaKaRa, Japan). Since the wcaJ gene was cut into two fragments via IS insertion, two pairs of primers (Table S5) were designed to amplify these fragments, namely, the upstream (156-bp) and downstream (100-bp) regions. The location of these probes is shown in Fig. S1. The reaction mixture was kept at 95°C for 30 s, followed by 40 cycles of 95°C for 5 s and 60°C for 34 s. The ΔΔCT method was used to compare expression differences among isolates. The rrsE gene encoding 6-phosphogluconate dehydrogenase and the NTUH-K2044 isolate were used as the internal reference (40) and control isolate, respectively.
Construction of the wcaJ-complemented isolates.
To evaluate the effects of wcaJ inactivation caused by IS insertion, we constructed wcaJ-complemented isolates. The isolates, primers, and plasmids used are listed in Tables S5 and S6. The ampicillin resistance gene of the pEASY plasmid (TransGen Biotech) was replaced with the tigecycline resistance gene (tetX4), resulting in the pEASY-tgc plasmid. The intact wcaJ gene from NTUH-K2044 and the rpsL promoter from the pCasKP-hph plasmid were then amplified and integrated into the pEASY-tgc plasmid by Gibson assembly, yielding a continuous-expression plasmid (pComp_wcaJ). Meanwhile, the pComp plasmid without the wcaJ gene was used as a blank control to offset the effect of a foreign plasmid on phenotypic changes.
Mucoviscosity.
The mucoviscosity of isolates was determined via the string test as previously described (25). Briefly, the string test was performed by stretching the bacterial colony on agar plates by use of a loop. Colonies that stretched at least 5 mm were positive. All experiments were conducted in triplicate. Further, the sedimentation assay was also used to evaluate mucoviscosity (41). After 8 h of incubation, cultures were normalized to 1 OD600/mL (where OD600 is optical density at 600 nm) and centrifuged at 1,000 × g for 5 min. The values of the supernatant were then detected.
Extraction and quantification of capsule.
Capsular polysaccharides were extracted and quantified as previously described (41, 42). Briefly, 500 μL of the overnight culture was mixed with 100 μL of 1% Zwittergent 3-14 detergent in 100 mM citric acid (pH 2.0) and heated at 50°C for 30 min. Samples were centrifuged for 5 min at 14,000 rpm, and 250 μL of the supernatant was transferred into a new tube and precipitated with 1 mL of absolute ethanol at 4°C for 30 min. After centrifugation for 5 min at 14,000 rpm, the pellets were dried and dissolved in 100 μL of distilled water, and then 600 μL of 12.5 mM sodium tetraborate (Sinopharm Chemical Reagent Co., Ltd) in H2SO4 was added. Samples were boiled for 5 min and cooled to room temperature for 10 min before the addition of 10 μL of 0.15% 3-phenylphenol (Aladdin). After incubation for 15 min to generate the chromophore, the absorbance at 520 nm was measured. The amounts of CPS were determined from a standard curve of glucuronic acid (Aladdin), and the results were expressed in micrograms per OD600 for each culture. All experiments were carried out in triplicate.
TEM.
To determine capsule thickness, transmission electron microscopy (TEM) (FEI Tecnai Spirit) was performed for isolates C400, C1356, C2768, and C4599 and their corresponding isogenic derivative strains. Overnight cultures were transferred to 1.5-mL polypropylene tubes and centrifuged at 15,000 rpm for 3 min. The pellets were washed with phosphate-buffered saline (PBS) three times and further fixed in 1.5% glutaraldehyde. Image analysis was conducted using ImageJ software.
Biofilm assay.
Biofilm assays were performed as previously described (43), with some modifications. Briefly, an exponential-phase culture was added to 2.5 mL of LB broth in a polystyrene tube (1:100 dilution) and incubated at 37°C for 48 h. The tubes were washed with PBS, and 4 mL of 0.02% crystal violet was then added. After 15 min of incubation, crystal violet was removed, and each tube was washed three times with PBS. Then, 3 mL of 30% acetic acid was added, and the tube was incubated at room temperature for 30 min. The OD of solubilized crystal violet was measured at 600 nm. All isolates were tested based on six replicates.
Serum killing assay.
According to our previous study (44), human serum was obtained from 10 healthy volunteers. An inoculum of 25 μL of exponential-phase culture (OD600, 0.6) was added to 75 μL of pooled human serum in a 2-mL microcentrifuge tube. Viable counts were checked after incubation for 0, 60, 120, and 180 min at 37°C with 200 rpm of shaking. Each isolate was tested in triplicate. Results were analyzed by comparing the numbers of CFU at each time point and are expressed as highly sensitive (grade 1 or 2), intermediate (grade 3 or 4), or resistant (grade 5 or 6).
Flow cytometry.
Bacterial flow cytometry was performed as previously described (45). An inoculum of log-phase bacteria (1 × 107 CFU) was incubated with 10% healthy volunteer serum at 37°C for 1 h, and the mixture was incubated with FITC anti-complement C3b antibody (Biolegend) after washing three times with PBS. Samples were analyzed on a FACSCanto system using Diva software (BD Biosciences, San Jose, CA, USA), and at least 200,000 events per sample were acquired for analysis.
Macrophage phagocytosis assay.
Raw 264.7 murine macrophages were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum. Bacterial phagocytosis assays were performed as previously described (46, 47). Macrophages were seeded into 24-well plates and infected at a multiplicity of infection (MOI) of 100 (bacteria/cell). To synchronize the infection, the plates were centrifuged at 200 × g for 5 min and incubated at 37°C with 5% CO2 for 1 h. Cells were rinsed three times with PBS, followed by the addition of 1 mL of DMEM containing 300 μg/mL gentamicin (except for C2768 and C4599 isolates, which were treated with 300 μg/mL of hygromycin due to gentamicin resistance) to eliminate extracellular bacteria. After 1 h of incubation, cells were rinsed again three times with PBS and lysed with 0.2% Triton X-100 (20-min incubation at room temperature). Tenfold serial dilutions were plated on LB agar plates to determine total CFU counts. The percentage of phagocytosed bacteria per inoculum was calculated and normalized to that of wacJ::IS isolates. Three biological replicates were carried out per isolate.
In vivo mouse lethality assay.
The mouse lethality assay was performed as previously described (48). Five 6- to 8-week-old male BALB/c mice, purchased from the Peking University Animal Center, were used for each bacterial concentration in this study. Bacterial isolates were grown overnight in LB broth and subsequently serially diluted to 107 CFU/mL in PBS. A 100-μL bacterial suspension was injected intraperitoneally using a 1-mL sterile syringe. The survival of mice was observed daily for 14 days.
Growth curve.
Overnight cultures of the wcaJ-inactivated and wcaJ-complemented K. pneumoniae isolates were diluted to an OD600 of 0.01 and grown with shaking (200 rpm) at 37°C for 24 h (49). The OD600 was measured every 2 h to obtain the growth curve.
Generation of the isogenic blaKPC-2 plasmid deletion mutant.
KPC plasmid curing in CR-hvKP was performed based on clustered regularly interspaced short palindromic repeats (CRISPR), as previously described (50). According to the detailed protocol of the two-plasmid system, pCasKP-pSGKP, for genome editing, we designed a 20-bp spacer using sgRNAcas9 software (51). The spacer was then ligated into the pSGKP-tgc plasmid. Two plasmids, pCasKP-hph and pSGKP-tgc, were electrotransformed into the target isolate one after another for cleavage of the blaKPC-2 plasmid. Primers used in this step are listed in Table S5. Antibiotics were added at the following concentrations: 100 μg/mL hygromycin and 8 μg/mL tigecycline for both the E. coli and K. pneumoniae isolates. The MICs of the parent strain and the blaKPC-cured mutants were determined using the Vitek2 automated system (bioMérieux, Marcy l’Etoile, France).
Conjugation assay of the blaKPC-2-bearing plasmid.
Conjugation assays were conducted according to previous studies (52). The clinical isolate C1925, which was found to be KPC-2 positive and ciprofloxacin susceptible, was used as a donor, whereas C400ΔKPC::pComp, C400ΔKPC::pComp_wcaJ, C1356ΔKPC::pComp, and C1356ΔKPC::pComp_wcaJ were used as recipients. The donor and recipient isolates were mixed at a ratio of 1:1 on a 0.22-μm microporous membrane for 24 h. The transconjugants were first selected on LB plates supplemented with meropenem (1 μg/mL) and ciprofloxacin (32 μg/mL) and then confirmed by PCR.
Statistics.
GraphPad Prism version 9 was used for statistical analysis, including Student’s t tests, one-way analysis of variance (ANOVA), two-way ANOVA, and log rank (Mantel-Cox) tests. A P of <0.05 was considered statistically significant.
Ethics.
The Peking University People’s Hospital Institutional Review Board approved this study (no. 2019PHB194-01). Informed consent was obtained from the healthy participants for blood sample collection, whereas patient consent was waived owing to the retrospective nature of this research, and patient data were anonymized. Animal studies were reviewed and approved by the Medical Ethics Committee of Peking University People's Hospital (no. 2019PHE001).
Data availability.
Data in this study were deposited into NCBI under BioProject accession no. PRJNA791169.
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (grant no. 81991533, 32141001 and 81961130396) and the Newton Advanced Fellowship award (grant no. NAF009\1002).
The funding sources had no role in the study design, isolate collection, data analysis, or any relevant aspect of this study.
H.W. conceived, designed, and supervised the study. S.W., Q.D., Y.Z., A.Z., Q.W., R.W., X.W., and L.J. collected and interpreted the data. S.W., Q.D., A.Z., and S.M. conducted the analysis. S.W., Q.D., and H.W. wrote the manuscript. All authors approved the final version of the manuscript. The corresponding author attests that all listed authors meet the authorship criteria and that no others meeting the criteria have been omitted.
We declare that we have no conflicts of interest.
Footnotes
Supplemental material is available online only.
Contributor Information
Hui Wang, Email: whuibj@163.com.
Monica Adriana Garcia-Solache, Brown University.
REFERENCES
- 1.Wyres KL, Lam MMC, Holt KE. 2020. Population genomics of Klebsiella pneumoniae. Nat Rev Microbiol 18:344–359. doi: 10.1038/s41579-019-0315-1. [DOI] [PubMed] [Google Scholar]
- 2.Siu LK, Yeh K-M, Lin J-C, Fung C-P, Chang F-Y. 2012. Klebsiella pneumoniae liver abscess: a new invasive syndrome. Lancet Infect Dis 12:881–887. doi: 10.1016/S1473-3099(12)70205-0. [DOI] [PubMed] [Google Scholar]
- 3.Lee C-R, Lee JH, Park KS, Jeon JH, Kim YB, Cha C-J, Jeong BC, Lee SH. 2017. Antimicrobial resistance of hypervirulent Klebsiella pneumoniae: epidemiology, hypervirulence-associated determinants, and resistance mechanisms. Front Cell Infect Microbiol 7:483. doi: 10.3389/fcimb.2017.00483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang R, Lin D, Chan EW-C, Gu D, Chen G-X, Chen S. 2016. Emergence of carbapenem-resistant serotype K1 hypervirulent Klebsiella pneumoniae strains in China. Antimicrob Agents Chemother 60:709–711. doi: 10.1128/AAC.02173-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang Y, Yang Y, Chen G, Lin M, Chen Y, He R, Galvão KN, El-Gawad El-Sayed Ahmed MA, Roberts AP, Wu Y, Zhong L-L, Liang X, Qin M, Ding X, Deng W, Huang S, Li H-Y, Dai M, Chen D-Q, Zhang L, Liao K, Xia Y, Tian G-B. 2021. Molecular characterization of carbapenem-resistant and virulent plasmids in Klebsiella pneumoniae from patients with bloodstream infections in China. Emerg Microbes Infect 10:700–709. doi: 10.1080/22221751.2021.1906163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang Y, Wang X, Wang Q, Chen H, Li H, Wang S, Wang R, Wang H. 2021. Emergence of tigecycline nonsusceptible and IMP-4 carbapenemase-producing K2-ST65 hypervirulent Klebsiella pneumoniae in China. Microbiol Spectr 9:e0130521. doi: 10.1128/Spectrum.01305-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Y, Jin L, Ouyang P, Wang Q, Wang R, Wang J, Gao H, Wang X, Wang H, China Carbapenem-Resistant Enterobacteriaceae (CRE) Network . 2020. Evolution of hypervirulence in carbapenem-resistant Klebsiella pneumoniae in China: a multicentre, molecular epidemiological analysis. J Antimicrob Chemother 75:327–336. doi: 10.1093/jac/dkz446. [DOI] [PubMed] [Google Scholar]
- 8.Long D, Zhu L-L, Du F-L, Xiang T-X, Wan L-G, Wei D-D, Zhang W, Liu Y. 2019. Phenotypical profile and global transcriptomic profile of hypervirulent Klebsiella pneumoniae due to carbapenemase-encoding plasmid acquisition. BMC Genomics 20:480. doi: 10.1186/s12864-019-5705-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gu D, Dong N, Zheng Z, Lin D, Huang M, Wang L, Chan EW-C, Shu L, Yu J, Zhang R, Chen S. 2018. A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. Lancet Infect Dis 18:37–46. doi: 10.1016/S1473-3099(17)30489-9. [DOI] [PubMed] [Google Scholar]
- 10.Parrott AM, Shi J, Aaron J, Green DA, Whittier S, Wu F. 2021. Detection of multiple hypervirulent Klebsiella pneumoniae strains in a New York City hospital through screening of virulence genes. Clin Microbiol Infect 27:583–589. doi: 10.1016/j.cmi.2020.05.012. [DOI] [PubMed] [Google Scholar]
- 11.Lipworth S, Vihta K-D, Chau K, Barker L, George S, Kavanagh J, Davies T, Vaughan A, Andersson M, Jeffery K, Oakley S, Morgan M, Hopkins S, Peto TEA, Crook DW, Walker AS, Stoesser N. 2021. Ten-year longitudinal molecular epidemiology study of Escherichia coli and Klebsiella species bloodstream infections in Oxfordshire, UK. Genome Med 13:144–144. doi: 10.1186/s13073-021-00947-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Devanga Ragupathi NK, et al. 2020. The influence of biofilms on carbapenem susceptibility and patient outcome in device associated K. pneumoniae infections: insights into phenotype vs genome-wide analysis and correlation. Front Microbiol 11:591679. doi: 10.3389/fmicb.2020.591679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Q, Wang X, Wang J, Ouyang P, Jin C, Wang R, Zhang Y, Jin L, Chen H, Wang Z, Zhang F, Cao B, Xie L, Liao K, Gu B, Yang C, Liu Z, Ma X, Jin L, Zhang X, Man S, Li W, Pei F, Xu X, Jin Y, Ji P, Wang H. 2018. Phenotypic and genotypic characterization of carbapenem-resistant Enterobacteriaceae: data from a longitudinal large-scale CRE study in China (2012–2016). Clin Infect Dis 67:S196–S205. doi: 10.1093/cid/ciy660. [DOI] [PubMed] [Google Scholar]
- 14.Guo Y, Wang S, Zhan L, Jin Y, Duan J, Hao Z, Lv J, Qi X, Chen L, Kreiswirth BN, Wang L, Yu F. 2017. Microbiological and clinical characteristics of hypermucoviscous Klebsiella pneumoniae isolates associated with invasive infections in China. Front Cell Infect Microbiol 7:24. doi: 10.3389/fcimb.2017.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lam MMC, Wick RR, Watts SC, Cerdeira LT, Wyres KL, Holt KE. 2021. A genomic surveillance framework and genotyping tool for Klebsiella pneumoniae and its related species complex. Nat Commun 12:4188. doi: 10.1038/s41467-021-24448-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wyres KL, Wick RR, Gorrie C, Jenney A, Follador R, Thomson NR, Holt KE. 2016. Identification of Klebsiella capsule synthesis loci from whole genome data. Microb Genom 2:e000102. doi: 10.1099/mgen.0.000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vasimuddin M, Misra S, Li H, Aluru S. 2019. Efficient architecture-aware acceleration of BWA-MEM for multicore systems, pp 314–324. In IEEE International Parallel and Distributed Processing Symposium (IPDPS) . IEEE, New York, NY. doi: 10.1109/IPDPS.2019.00041. [DOI] [Google Scholar]
- 18.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walker KA, Treat LP, Sepúlveda VE, Miller VL. 2020. The small protein RmpD drives hypermucoviscosity in Klebsiella pneumoniae. mBio 11:e01750-20. doi: 10.1128/mBio.01750-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eger E, Heiden SE, Becker K, Rau A, Geisenhainer K, Idelevich EA, Schaufler K. 2021. Hypervirulent Klebsiella pneumoniae sequence type 420 with a chromosomally inserted virulence plasmid. Int J Mol Sci 22:9196. doi: 10.3390/ijms22179196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pan Y-J, Lin T-L, Chen C-T, Chen Y-Y, Hsieh P-F, Hsu C-R, Wu M-C, Wang J-T. 2015. Genetic analysis of capsular polysaccharide synthesis gene clusters in 79 capsular types of Klebsiella spp. Sci Rep 5:15573. doi: 10.1038/srep15573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dorman MJ, Feltwell T, Goulding DA, Parkhill J, Short FL. 2018. The capsule regulatory network of Klebsiella pneumoniae defined by density-TraDISort. mBio 9:e01863-18. doi: 10.1128/mBio.01863-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li J, Sheng Y, Ma R, Xu M, Liu F, Qin R, Zhu M, Zhu X, He P. 2021. Identification of a depolymerase specific for K64-serotype Klebsiella pneumoniae: potential applications in capsular typing and treatment. Antibiotics (Basel) 10:144. doi: 10.3390/antibiotics10020144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tan D, Zhang Y, Qin J, Le S, Gu J, Chen L-k, Guo X, Zhu T. 2020. A frameshift mutation in wcaJ associated with phage resistance in Klebsiella pneumoniae. Microorganisms 8:378. doi: 10.3390/microorganisms8030378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tan YH, Chen Y, Chu WHW, Sham L-T, Gan Y-H. 2020. Cell envelope defects of different capsule-null mutants in K1 hypervirulent Klebsiella pneumoniae can affect bacterial pathogenesis. Mol Microbiol 113:889–905. doi: 10.1111/mmi.14447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bengoechea JA, Sa Pessoa J. 2019. Klebsiella pneumoniae infection biology: living to counteract host defences. FEMS Microbiol Rev 43:123–144. doi: 10.1093/femsre/fuy043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Merino S, Camprubí S, Albertí S, Benedí VJ, Tomás JM. 1992. Mechanisms of Klebsiella pneumoniae resistance to complement-mediated killing. Infect Immun 60:2529–2535. doi: 10.1128/iai.60.6.2529-2535.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gordillo Altamirano F, Forsyth JH, Patwa R, Kostoulias X, Trim M, Subedi D, Archer SK, Morris FC, Oliveira C, Kielty L, Korneev D, O'Bryan MK, Lithgow TJ, Peleg AY, Barr JJ. 2021. Bacteriophage-resistant Acinetobacter baumannii are resensitized to antimicrobials. Nat Microbiol 6:157–161. doi: 10.1038/s41564-020-00830-7. [DOI] [PubMed] [Google Scholar]
- 29.Song L, Yang X, Huang J, Zhu X, Han G, Wan Y, Xu Y, Luan G, Jia X. 2021. Phage selective pressure reduces virulence of hypervirulent through mutation of the gene. Front Microbiol 12:739319. doi: 10.3389/fmicb.2021.739319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cai R, Wang G, Le S, Wu M, Cheng M, Guo Z, Ji Y, Xi H, Zhao C, Wang X, Xue Y, Wang Z, Zhang H, Fu Y, Sun C, Feng X, Lei L, Yang Y, Ur Rahman S, Liu X, Han W, Gu J. 2019. Three capsular polysaccharide synthesis-related glucosyltransferases, GT-1, GT-2 and WcaJ, are associated with virulence and phage sensitivity of Klebsiella pneumoniae. Front Microbiol 10:1189. doi: 10.3389/fmicb.2019.01189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haudiquet M, Buffet A, Rendueles O, Rocha EPC. 2021. Interplay between the cell envelope and mobile genetic elements shapes gene flow in populations of the nosocomial pathogen Klebsiella pneumoniae. PLoS Biol 19:e3001276. doi: 10.1371/journal.pbio.3001276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Santos-López A, Rodríguez-Beltrán J, San Millán Á. 2021. The bacterial capsule is a gatekeeper for mobile DNA. PLoS Biol 19:e3001308. doi: 10.1371/journal.pbio.3001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chaput C, Spindler E, Gill RT, Zychlinsky A. 2013. O-antigen protects gram-negative bacteria from histone killing. PLoS One 8:e71097. doi: 10.1371/journal.pone.0071097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paczosa MK, Mecsas J. 2016. Klebsiella pneumoniae: going on the offense with a strong defense. Microbiol Mol Biol Rev 80:629–661. doi: 10.1128/MMBR.00078-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 37.Lam MMC, Wick RR, Judd LM, Holt KE, Wyres KL. 2022. Kaptive 2.0: updated capsule and lipopolysaccharide locus typing for the species complex. Microb Genom 8:000800. doi: 10.1099/mgen.0.000800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thorvaldsdottir H, Robinson JT, Mesirov JP. 2013. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform 14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Siguier P, Perochon J, Lestrade L, Mahillon J, Chandler M. 2006. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res 34:D32–D36. doi: 10.1093/nar/gkj014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang X, Chen H, Zhang Y, Wang Q, Zhao C, Li H, He W, Zhang F, Wang Z, Li S, Wang H. 2015. Genetic characterisation of clinical Klebsiella pneumoniae isolates with reduced susceptibility to tigecycline: role of the global regulator RamA and its local repressor RamR. Int J Antimicrob Agents 45:635–640. doi: 10.1016/j.ijantimicag.2014.12.022. [DOI] [PubMed] [Google Scholar]
- 41.Walker KA, Miner TA, Palacios M, Trzilova D, Frederick DR, Broberg CA, Sepúlveda VE, Quinn JD, Miller VL. 2019. A Klebsiella pneumoniae regulatory mutant has reduced capsule expression but retains hypermucoviscosity. mBio 10:00089-19. doi: 10.1128/mBio.00089-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Choi M-J, Ko KS. 2015. Loss of hypermucoviscosity and increased fitness cost in colistin-resistant Klebsiella pneumoniae sequence type 23 strains. Antimicrob Agents Chemother 59:6763–6773. doi: 10.1128/AAC.00952-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Biswas I, Mettlach J. 2019. A simple static biofilm assay for Acinetobacter baumannii. Methods Mol Biol 1946:159–165. doi: 10.1007/978-1-4939-9118-1_15. [DOI] [PubMed] [Google Scholar]
- 44.Zhang Y, Zeng J, Liu W, Zhao F, Hu Z, Zhao C, Wang Q, Wang X, Chen H, Li H, Zhang F, Li S, Cao B, Wang H. 2015. Emergence of a hypervirulent carbapenem-resistant Klebsiella pneumoniae isolate from clinical infections in China. J Infect 71:553–560. doi: 10.1016/j.jinf.2015.07.010. [DOI] [PubMed] [Google Scholar]
- 45.Talyansky Y, Nielsen TB, Yan J, Carlino-Macdonald U, Di Venanzio G, Chakravorty S, Ulhaq A, Feldman MF, Russo TA, Vinogradov E, Luna B, Wright MS, Adams MD, Spellberg B. 2021. Capsule carbohydrate structure determines virulence in Acinetobacter baumannii. PLoS Pathog 17:e1009291. doi: 10.1371/journal.ppat.1009291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu Q, Yang X, Chan EWC, Chen S. 2021. The hypermucoviscosity of hypervirulent K. pneumoniae confers the ability to evade neutrophil-mediated phagocytosis. Virulence 12:2050–2059. doi: 10.1080/21505594.2021.1960101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ares MA, Fernández-Vázquez JL, Rosales-Reyes R, Jarillo-Quijada MD, von Bargen K, Torres J, González-y-Merchand JA, Alcántar-Curiel MD, De la Cruz MA. 2016. H-NS nucleoid protein controls virulence features of Klebsiella pneumoniae by regulating the expression of type 3 pili and the capsule polysaccharide. Front Cell Infect Microbiol 6:13. doi: 10.3389/fcimb.2016.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Russo TA, MacDonald U. 2020. The Galleria mellonella infection model does not accurately differentiate between hypervirulent and classical Klebsiella pneumoniae. mSphere 5:e00850-19. doi: 10.1128/mSphere.00850-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang J, Wang H-H, Lu Y, Yi L-X, Deng Y, Lv L, Burrus V, Liu J-H. 2021. A ProQ/FinO family protein involved in plasmid copy number control favours fitness of bacteria carrying mcr-1-bearing IncI2 plasmids. Nucleic Acids Res 49:3981–3996. doi: 10.1093/nar/gkab149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang Y, Wang S, Chen W, Song L, Zhang Y, Shen Z, Yu F, Li M, Ji Q. 2018. CRISPR-Cas9 and CRISPR-assisted cytidine deaminase enable precise and efficient genome editing in Klebsiella pneumoniae. Appl Environ Microbiol 84:e01834-18. doi: 10.1128/AEM.01834-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xie S, Shen B, Zhang C, Huang X, Zhang Y. 2014. sgRNAcas9: a software package for designing CRISPR sgRNA and evaluating potential off-target cleavage sites. PLoS One 9:e100448. doi: 10.1371/journal.pone.0100448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.He T, Wang R, Liu D, Walsh TR, Zhang R, Lv Y, Ke Y, Ji Q, Wei R, Liu Z, Shen Y, Wang G, Sun L, Lei L, Lv Z, Li Y, Pang M, Wang L, Sun Q, Fu Y, Song H, Hao Y, Shen Z, Wang S, Chen G, Wu C, Shen J, Wang Y. 2019. Emergence of plasmid-mediated high-level tigecycline resistance genes in animals and humans. Nat Microbiol 4:1450–1456. doi: 10.1038/s41564-019-0445-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1 to S3. Download spectrum.02400-22-s0001.xlsx, XLSX file, 0.1 MB (124.4KB, xlsx)
Fig. S1-4. Download spectrum.02400-22-s0002.pdf, PDF file, 0.6 MB (640KB, pdf)
Data Availability Statement
Data in this study were deposited into NCBI under BioProject accession no. PRJNA791169.