ABSTRACT
Microsporidia, as unicellular eukaryotes, also have an endomembrane system for transporting proteins, which is essentially similar to those of other eukaryotes. In eukaryotes, coat protein complex II (COPII) consists of Sar1, Sec23, Sec24, Sec13, and Sec31 and mediates protein transport from the endoplasmic reticulum (ER) to the Golgi apparatus. Sar1 is the central player in the regulation of coat protein complex II vesicle formation in the endoplasmic reticulum. In this study, we successfully cloned the NbSar1, NbSec23-1, NbSec23-2, NbSec24-1, NbSec24-2, NbSec13, NbSec31-1, and NbSec31-2 genes and prepared NbSar1 polyclonal antibody. We found that NbSar1 was localized mainly in the perinuclear cytoplasm of Nosema bombycis by immunofluorescence analysis (IFA). Yeast two-hybrid assays demonstrated that NbSar1 interacts with NbSec23-2, NbSec23-2 interacts with NbSec24-1 or NbSec24-2, NbSec23-1 interacts with NbSec31, and NbSec31 interacts with NbSec13. Moreover, the silencing of NbSar1 by RNA interference resulted in the aberrant expression of NbSar1, NbSec23-1, NbSec24-1, NbSec24-2, NbSec13, NbSec31-1, and NbSec31-2 and significantly inhibited the proliferation of N. bombycis. Altogether, these findings indicated that the subunits of coat protein complex II work together to perform functions in the proliferation of N. bombycis and that NbSar1 may play a crucial role in coat protein complex II vesicle formation.
IMPORTANCE As eukaryotes, microsporidia have retained the endomembrane system for transporting and sorting proteins throughout their evolution. Whether the microsporidia form coat protein complex II (COPII) vesicles to transport cargo proteins and whether they play other roles besides cargo transport are not fully explained at present. Our results showed that NbSar1, NbSec23-1/NbSec23-2, NbSec24-1/NbSec24-2, NbSec13, and NbSec31 might be assembled to form COPII in the ER of N. bombycis, and the functions of COPII are also closely related to the proliferation of N. bombycis, this may be a new target for the prevention of pébrine disease of the silkworm.
KEYWORDS: microsporidia, Nosema bombycis, endoplasmic reticulum, coat protein complex II, Sar1
INTRODUCTION
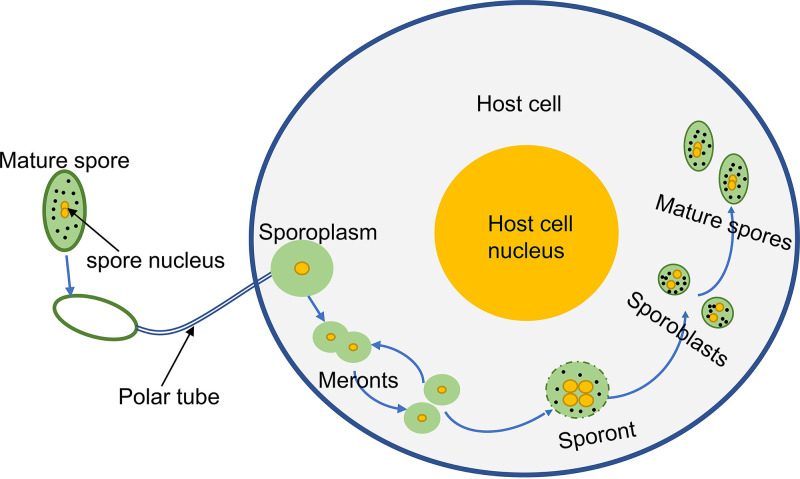
Microsporidia are obligate intracellular parasites that can infect a wide range of vertebrates and invertebrates, including humans (1). Microsporidia are classified into 200 genera including more than 1,400 species (2). Nosema bombycis, the pathogen causing pébrine disease in the silkworm, was discovered in the silkworm by Naegeli in 1857. Pébrine is transmitted both horizontally and vertically to the silkworm, resulting in great economic losses to the silkworm industry (3, 4). The life cycle of N. bombycis is divided into three phases: the infective phase, the proliferative phase, and the sporogonic phase (Fig. 1).
FIG 1.
Diagrammatic illustration of the developmental cycle of N. bombycis. The life cycle of N. bombycis is divided into an infective phase, a proliferative phase, and a sporogonic phase. In the infective phase, the mature spore germinates and injects the sporoplasm into the host cell. In the proliferative phase, the meront proliferates by binary fission or multiple fissions in the host cell. In the sporogonic phase, the sporont with four nuclei divides into two sporoblasts, and the sporoblasts further develop into mature spores with thick walls.
Eukaryotic cells maintain life activities by synthesizing proteins and lipids for secretion and transport to specific organelles and plasma membranes. Secretory and membrane proteins are synthesized in the ribosome and transferred to the compartment of the endoplasmic reticulum (ER) or integrated into the ER membrane. The transport of prefolded proteins from the ER to the Golgi apparatus is performed by coat protein complex II (COPII)-mediated vesicular transport (5). COPII consists of five proteins that are highly conserved in eukaryotes: the small G protein Sar1, the inner coat proteins Sec23 and Sec24, and the outer coat proteins Sec13 and Sec31 (6). Sar1 is inserted into the ER membranes in a Sar1-GTP-activated form to recruit the Sec23-Sec24 complex through binding Sec23, eventually forming the inner layer of COPII vesicles (7, 8). The Sec23-Sec24 complex is bent and positively charged on one side, which facilitates the binding of the complex to the ER membrane (8), while Sec23 recruits the Sec13-Sec31 complex to form the caged outer layer of COPII vesicles by binding to the proline-rich domain at the C terminus of Sec31 and driving the deformation of the ER membrane (9). The role of Sar1, a small GTPase, in vesicle budding has been extensively studied in Saccharomyces cerevisiae. Sar1 is involved in vesicle formation only when it is activated by the ER membrane protein Sec12 (10, 11). Therefore, Sar1 is a central player in the regulation of COPII vesicle formation in the ER.
Microsporidia exhibit extreme compaction in genome size due to their intracellular parasitic lifestyle, resulting in the loss of genes associated with their metabolic pathways and a high degree of organelle simplification (12–17). As eukaryotes, microsporidia have retained the endomembrane system for transporting and sorting proteins. Membrane polymers of the Golgi apparatus were observed in the early sporont and aggregated around flattened vesicles of the ER near the perinuclear lumen (18, 19). The tubular structures of the Golgi apparatus have been found in sporont and mature spores of microsporidia (20, 21), and the tubular structure is connected to the perinuclear lumen, ER, plasma membrane, and polar tube (22). Expectedly, the five subunits of COPII, Sar1, Sec23, Sec24, Sec13, and Sec31, are present in N. bombycis according to genomic analyses. Previously, it was reported that the spore wall and polar tube proteins were transported from the ER to the plasma membrane through tubular networks (TNs) in microsporidia, which does not require the involvement of vesicles produced by COPI or COPII (22). But studies on COPII of microsporidia have been rare until now. Whether the microsporidia form COPII vesicles to transport cargo proteins and whether COPII of microsporidia plays various roles besides cargo transport are not fully explained at present. In order to understand the mechanism of COPII in the proliferation of N. bombycis, we studied the regulatory and interaction relationships among N. bombycis Sar1 (NbSar1), NbSec23, NbSec24, NbSec13, and NbSec31 in the microsporidian N. bombycis for the first time. We have found that NbSar1 is localized in the ER of N. bombycis and interacts with NbSec23-2 and that NbSec23-2 interacts with NbSec24-1/NbSec24-2, NbSec23-1 interacts with NbSec31, and NbSec31 interacts with NbSec13. Moreover, RNAi (RNA interference) on NbSar1 resulted in the abnormal expression of NbSec23-1, NbSec24-1/NbSec24-2, NbSec31-1/NbSec31-2, and NbSec13. These findings indicated that NbSar1, NbSec23-1/NbSec23-2, NbSec24-1/NbSec24-2, NbSec13, and NbSec31 might be assembled to form COPII in the ER of N. bombycis.
RESULTS
Cloning and expression of the NbSar1 gene and sequence analysis.
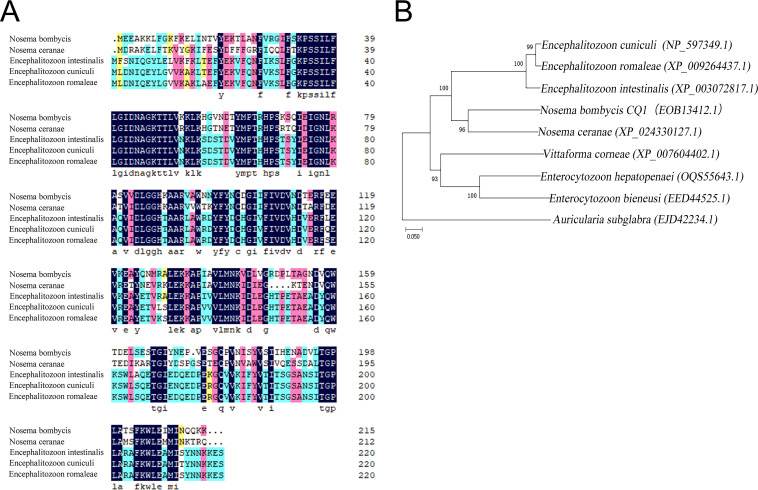
PCR amplification and sequencing results showed that the NbSar1 gene contains a complete open reading frame (ORF) of 648 bp in length that encodes 215 amino acids. Homologous multiple-sequence alignment revealed that NbSar1 is highly homologous to other microsporidian Sar1 proteins, with a homology of >35%, and that the amino acid similarities between NbSar1 and the Sar1 proteins of Nosema ceranae (GenBank accession number XP_024330127.1), Encephalitozoon intestinalis (accession number XP_003072817.1), Encephalitozoon cuniculi (accession number NP_597349.1), and Encephalitozoon romaleae (accession number XP_009264437.1) were 63.89%, 57.01%, 56.56%, and 56.56%, respectively (Fig. 2A). Phylogenetic analysis results showed that N. bombycis and N. ceranae were clustered into the same branch (Fig. 2B), indicating that they were closely related to each other.
FIG 2.
Bioinformatics analysis of Sar1. (A) Amino acid alignment of Sar1 proteins from N. bombycis and other microsporidian species. (B) Phylogenetic tree of Sar1 proteins from N. bombycis and other microsporidian species. Protein sequences were aligned using ClustalW. The maximum likelihood (ML) tree was constructed using MEGA 6.0 software, and bootstrap analyses were performed by employing Jones-Taylor-Thornton (JTT) model-based distance matrices generated from 1,000 resamplings of the alignments. Values at the branches indicate bootstrap support.
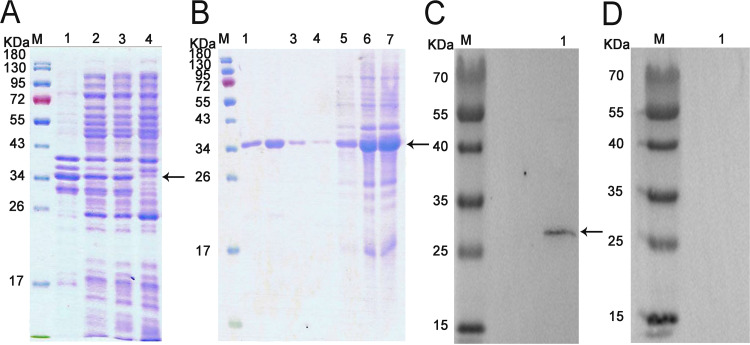
SDS-PAGE results showed that the molecular weight of the NbSar1 recombinant protein was about 34 kDa, which included the NbSar1 protein and the His tag. Ultrasonic fragmentation showed that the NbSar1 protein was present mainly in the form of inclusion bodies in isopropyl-β-d-thiogalactopyranoside (IPTG)-induced bacteria (Fig. 3A). Purified recombinant proteins were obtained by elution with imidazole at different concentrations (Fig. 3B). Western blotting indicated that a 28-kDa protein band was detected in the total protein of N. bombycis, which corresponds to the predicted size of the NbSar1 amino acid sequence (Fig. 3C). Preimmune serum, which served as the negative control, did not detect the target protein (Fig. 3D).
FIG 3.
Expression, purification, and Western blot analysis of recombinant NbSar1. (A) SDS-PAGE analysis of the expressed recombinant protein. Lane M, protein marker; lane 1, the precipitate of induced recombinant bacteria; lane 2, the supernatant of induced recombinant bacteria; lane 3, recombinant bacteria induced with 0.5 mM IPTG at 37°C for 4 h; lane 4, recombinant bacteria without induction. (B) SDS-PAGE analysis of the purified fusion protein. Lane M, protein marker; lane 1, elution with 500 mmol/L imidazole; lane 2, elution with 200 mmol/L imidazole; lane 3, elution with 50 mmol/L imidazole; lane 4, elution with 20 mmol/L imidazole; lane 5, elution with 10 mmol/L imidazole; lane 6, flowthrough; lane 7, unpurified recombinant NbSar1 protein. (C) Specificity analysis of NbSar1 antibody. Lane M, protein marker; lane 1, total protein of an N. bombycis mature spore. (D) Preimmune serum control. Lane M, protein marker; lane 1, total protein of an N. bombycis mature spore. The arrows indicate NbSar1 protein.
Subcellular localization of NbSar1 in different developmental phases of N. bombycis.
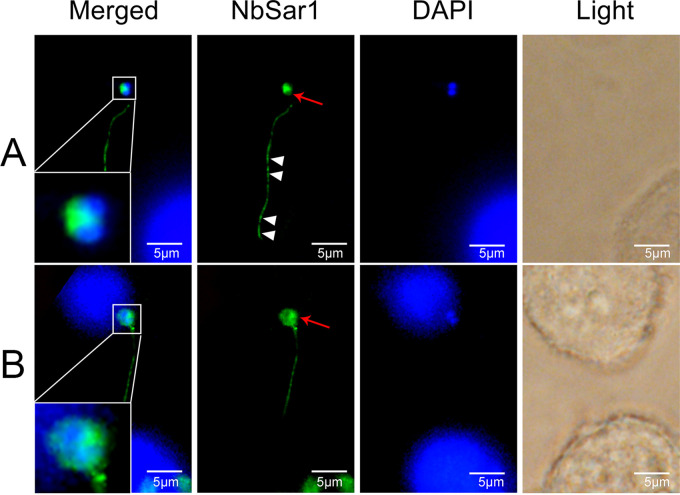
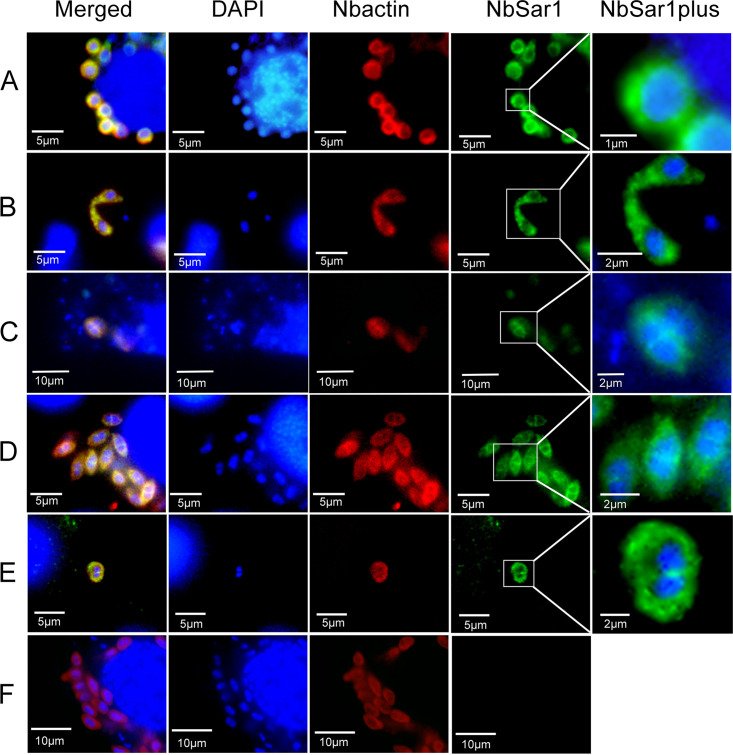
To investigate the localization of NbSar1 in the intracellular phase of N. bombycis, NbSar1 antibodies, marked with green fluorescence, were used to perform immunofluorescence analysis (IFA). In the infective phase, when the polar tube was everted from the germinating spore and the two unfused nuclei were expelled outside the spore, the green fluorescent signals were distributed mainly in the perinuclear cytoplasm of the sporoplasm and in the polar tube (Fig. 4A). After the contents of the N. bombycis spore were completely expelled from the germinated spore and injected into the new host cell, the green fluorescent signals were distributed mainly in the perinuclear cytoplasm of the sporoplasm (Fig. 4B). When the two nuclei of the invaded sporoplasm fused into one nucleus, the green and red fluorescent signals (N. bombycis actin antibodies were marked with red fluorescence as a reference label) were distributed in the perinuclear cytoplasm of the sporoplasm (Fig. 5A). In the proliferative phase, when N. bombycis was undergoing rapid proliferation, the green and red fluorescent signals were distributed mainly in the cytoplasm (Fig. 5B). In the sporogonic phase, green and red fluorescent signals were distributed in the perinuclear region of sporont with four nuclei (Fig. 5C). Sporoblasts are cells derived from the final division of the sporont. In the sporoblasts, the green and the red fluorescent signals were distributed mainly in the perinuclear region (Fig. 5D). When the N. bombycis cell neared maturation, the green and the red fluorescent signals were distributed mainly in the cytoplasm (Fig. 5E). No green fluorescent signals were observed in the preimmune serum group (Fig. 5F). These results suggested that NbSar1 exists mainly in the perinuclear region of N. bombycis throughout its life cycle.
FIG 4.
Subcellular localization of NbSar1 during intracellular infection. (A) Germinating spore. The white arrowheads indicate that the green fluorescent signals are distributed in the polar tube, and the red arrows indicate that the green fluorescent signals are distributed in the cytoplasm of the sporoplasm. (B) After spore germination. The red arrows indicate that the green fluorescent signals are distributed mainly in the perinuclear cytoplasm of the sporoplasm. DAPI (blue) was used to stain the nuclei of host cells and N. bombycis. NbSar1 antibody was coupled with Alexa Fluor 488 (green). BmN cells were infected with N. bombycis for 7 days.
FIG 5.
Subcellular colocalization of NbSar1 and N. bombycis actin in N. bombycis. (A) Sporoplasm. (B) Meront. (C) Sporont. (D) Sporoblasts. (E) Sporoblast nearing maturation into a spore. (F) Preimmune serum. DAPI (blue) was used to stain the nuclei of host cells and N. bombycis. NbSar1 antibody was coupled with Alexa Fluor 488 (green), and N. bombycis actin (Nbactin) antibody was coupled with Cy5 (red). BmN cells were infected with N. bombycis for 7 days.
Interactions between NbSar1 and NbSec23-2, NbSec23-2 and NbSec24, NbSec23-1 and NbSec31-1, and NbSec31-1 and NbSec13.
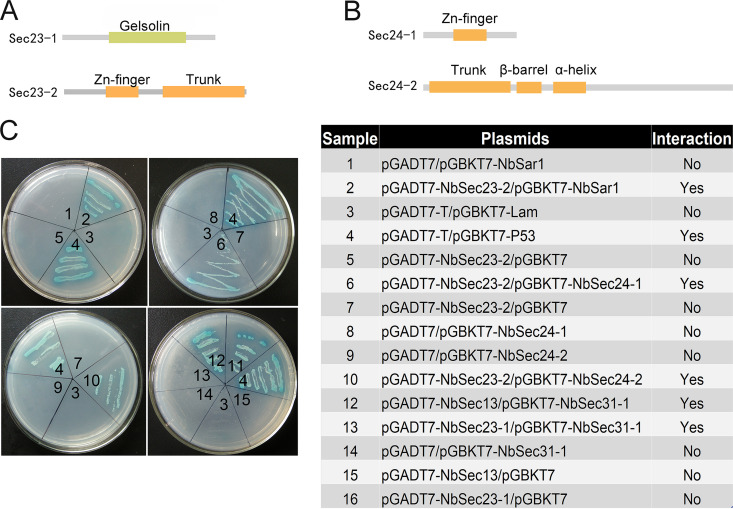
In this study, cloning results showed that NbSec23-1 (GenBank accession number EOB11408.1), NbSec23-2 (accession number EOB12942.1), NbSec24-1 (accession number EOB13378.1), NbSec24-2 (accession number EOB11737.1), NbSec13 (accession number EOB15402.1), NbSec31-1 (accession number EOB13833.1), and NbSec31-2 (accession number EOB13782.1) contain complete ORFs of 540 bp, 648 bp, 648 bp, 1,905 bp, 825 bp, 2,463 bp, and 2,721 bp, respectively. The domain prediction results showed that NbSec23-1 contains only the gelsolin domain; NbSec23-2 contains the trunk domain and the Zn finger domain (Fig. 6A); NbSec24-1 contains only the Zn finger domain; and NbSec24-2 contains the trunk domain, the β-barrel domain, and the α-helix domain (Fig. 6B). Sequence alignment results showed that NbSec31-2 includes the complete sequence of NbSec31-1. The interactions between NbSar1 and NbSec23-2, NbSec23-2 and NbSec24-1/NbSec24-2, NbSec23-1 and NbSec31-1, and NbSec31-1 and NbSec13 were examined by a yeast two-hybrid assay. The yeast two-hybrid assay showed that activating domain (AD)–NbSec23-2 and BK-NbSar1, AD–NbSec23-2 and Binding Domain (BK)–NbSec24-1/BK–NbSec24-2, AD–NbSec23-1 and BK–NbSec31-1, and AD-NbSec13 and BK–NbSec31-1 grew successfully on synthetic dropout (SD)−Ade/−His/−Leu/−Trp selective medium with 5-Bromo-4-chloro-3-indoxyl-α-d-galactopyranoside (X-α-gal) (Fig. 6C). The above results proved that NbSar1 interacts with NbSec23-2, NbSec23-2 interacts with NbSec24-1/NbSec24-2, NbSec23-1 interacts with NbSec31-1, and NbSec31-1 interacts with NbSec13, which is consistent with previous studies showing that the assembly of COPII vesicles depends on the interactions among Sar1, Sec23, Sec24, Sec13, and Sec31 and that the trunk domain of Sec23 interacts with Sec24 to form the Sec23-Sec24 complex, Sec13 interacts with Sec31 to form the Sec13-Sec13 complex, Sar1 recruits the Sec23-Sec24 complex through interaction with the trunk domain of Sec23, and the gelsolin domain of Sec23 recruits the Sec13-Sec31 complex through interaction with the proline-rich domain at the C terminus of Sec31 (7, 23–26). Our findings suggested that NbSar1, NbSec23, NbSec24, NbSec13, and NbSec31 could assemble a complex through interaction.
FIG 6.
Domains and interactions between subunits of COPII. (A) Domains of NbSec23-1 and NbSec23-2. (B) Domains of NbSec24-1 and NbSec24-2. (C) Yeast two-hybrid analysis of the interactions between NbSar1 and NbSec23-2, NbSec23-2 and NbSec24-1/NbSec24-2, NbSec23-1 and NbSec31-1, and NbSec13 and NbSec31-1. The interaction of pGADT7-T and pGBKT7-P53 was used as the positive control, and the interaction of pGADT7-T and pGBKT7-lam was used as the negative control.
RNAi on NbSar1 downregulated the expression of NbSec23-1, NbSec31-1, and NbSec31-2 but upregulated the expression of NbSec24-1, NbSec24-2, and NbSec13.
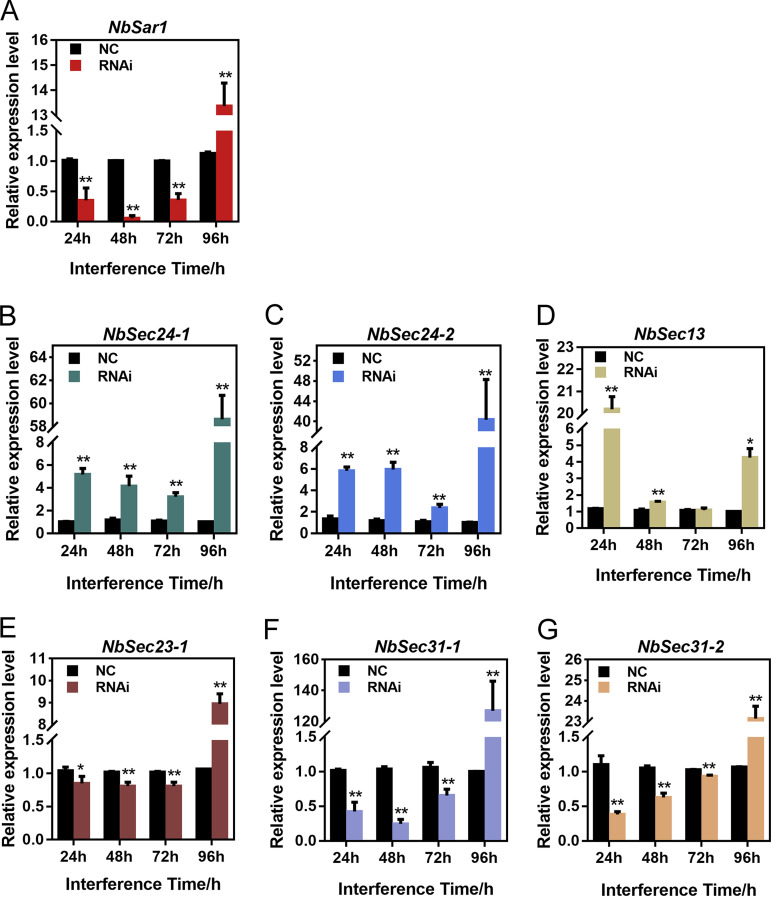
To assess the interference effects of small interfering RNA (siRNA) on NbSar1 expression, silkworms infected with N. bombycis were injected with Sar1 siRNA, while the control group was injected with nonsense siRNA. Quantitative PCR (qPCR) was performed to analyze the expression pattern of NbSar1. As a result, the relative expression level of NbSar1 was significantly decreased following siRNA treatment for 24 h, 48 h, and 72 h, whereas it was significantly increased at 96 h (Fig. 7A). This proved that RNAi on NbSar1 can effectively depress the expression of NbSar1 at 24 h, 48 h, and 72 h.
FIG 7.
Effect of the knockdown of NbSar1 on the expression of subunits of COPII. (A) Relative expression level of the NbSar1 gene after RNAi. (B) Relative expression level of the NbSec24-1 gene after RNAi. (C) Relative expression level of the NbSec24-2 gene after RNAi. (D) Relative expression level of the NbSec13 gene after RNAi. (E) Relative expression level of the NbSec23-1 gene after RNAi. (F) Relative expression level of the NbSec31-1 gene after RNAi. (G) Relative expression level of the NbSec31-2 gene after RNAi. NC, control group; RNAi, RNAi-NbSar1 group. Asterisks represent statistically significant differences (*, P < 0.05; **, P < 0.01). The error bars represent the standard deviations from three independent repeats.
To determine the effect of knocking down NbSar1 on the expression of COPII subunits, the N. bombycis β-tubulin gene served as the reference gene to analyze the expression patterns of NbSec23-1, NbSec24-1, NbSec24-2, NbSec13, NbSec31-1, and NbSec31-2 by qPCR. The results showed that NbSec23-1, NbSec31-1, and NbSec31-2 were significantly downregulated (Fig. 7B, E, and F) whereas NbSec24-1 and NbSec24-2 were significantly upregulated following siRNA treatment for 24 h, 48 h, and 72 h (Fig. 7C), NbSec13 was significantly upregulated following siRNA treatment for 24 h and 48 h (Fig. 7D). However, NbSec23-1, NbSec24-1, NbSec24-2, NbSec13, NbSec31-1, and NbSec31-2 were all significantly upregulated 96 h after siRNA treatment (Fig. 7B to F). These results suggested that NbSar1, NbSec23-1, NbSec24-1, NbSec24-2, NbSec13, NbSec31-1, and NbSec31-2 may work together as a complex for the proliferation of N. bombycis.
Knockdown of NbSar1 suppressed the proliferation of N. bombycis.
To further explore the effect of knocking down the NbSar1 gene on the proliferation of N. bombycis, the expression patterns of the small-subunit rRNA (ssrRNA) of N. bombycis were used to reflect the proliferation of N. bombycis. The effect of RNAi was analyzed by qPCR, with the BmGAPDH (Bombyx mori glyceraldehyde-3-phosphate dehydrogenase) gene serving as a reference gene. After the knockdown of NbSar1, the transcription level of N. bombycis ssrRNA was significantly downregulated in the RNAi-NbSar1 groups from 24 h to 72 h and then significantly upregulated at 96 h (Fig. 8). These results suggested that NbSar1 plays an important role in the proliferation of N. bombycis.
FIG 8.
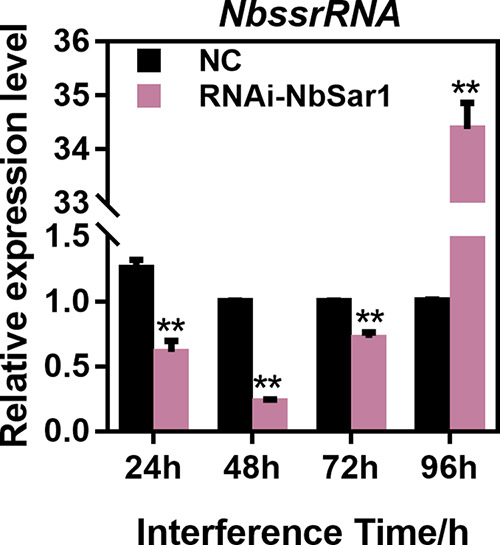
Knockdown of NbSar1 inhibited the proliferation of N. bombycis. Shown are the relative expression levels of the N. bombycis ssrRNA gene after RNAi. NC, control group; RNAi, RNAi-NbSar1 group. Asterisks represent statistically significant differences (*, P < 0.05; **, P < 0.01). The error bars represent the standard deviations from three independent repeats.
DISCUSSION
Sar1, a small G protein, acts as a molecular switch for vesicular transport and is highly conserved in fungi, mammals, as well as plants. In this study, IFA was performed to determine the localization of Sar1 at different developmental stages of N. bombycis. The results showed that NbSar1 is distributed mainly in the perinuclear region of the cytoplasm and is clearly excluded from the nucleus in the intracellular phase of N. bombycis. These findings are consistent with those of previous studies showing that Sar1 was distributed along the perinuclear contour and in ER membranes in CHO-K1 cells (27) and that Sar1 of S. cerevisiae was located mainly around the periphery of the ER and in the perinuclear region (28, 29). In microsporidia, ultrastructure analysis showed the presence of ER tubular structures in the perinuclear region of Paranosema grylli (22), and the sporont of Berwaldia singularis contains more vesicles and a rough ER surrounding the nucleus (30). After spore germination, the newly ejected sporoplasm contains the outer membrane, vesicles, cytoplasm, and ER to which ribosomes are attached (18). These results indicated that the Sar1 may be localized to the ER of N. bombycis.
Interactions between NbSar1 and NbSec23-2, NbSec23-2 and NbSec24-1/NbSec24-2, NbSec23-1 and NbSec31-1, and NbSec31-1 and NbSec13 were successfully verified by yeast two-hybrid assays in this study. In yeast, Sec23 and Sec24 contain five distinct domains: the zinc finger domain, the trunk domain or α/β vWA (von Willebrand factor [vWF] type A domain), the β-barrel domain, the α-helix domain, and the gelsolin domain (7). Although the Sec23 proteins of E. cuniculi, Enterocytozoon hepatopenaei, Mitosporidium daphniae, and N. ceranae and the Sec24 proteins of E. intestinalis and Nosema granulosis contain all five distinct domains, the Sec23 and Sec24 proteins of other microsporidia such as Dictyocoela roeselum, Hamiltosporidium magnivora, Amphiamblys sp., and Enterospora canceri contain 1 to 4 domains, respectively. In N. bombycis, NbSec23-1 has only the gelsolin domain, and NbSec23-2 has the zinc finger domain and the trunk domain, while NbSec24-1 has only the zinc finger domain, and NbSec24-2 has the trunk domain, the β-barrel domain, and the α-helix domain. The multiple-sequence alignment showed that NbSec23/NbSec24 of N. bombycis has high similarity to those of other microsporidia (see Fig. S1 and S2 in the supplemental material). Moreover, the specific and efficient Sar1 recruitment to the ER membrane is accomplished by the guanine exchange factor (GEF) Sec12, whose role in the biogenesis of COPII is crucial (8). Sec12, which shows a high degree of sequence divergence in different species, is a type II transmembrane protein, and only the K-loop motif (GGGGxxxxGϕxN, where x indicates any amino acid, the ϕ indicates a hydrophobic residue) in Sec12 is highly conserved (31, 32). In Aspergillus nidulans, a protein with the Sec12 K-loop motif and a single-pass type II transmembrane is located on the ER and promotes specific nucleotide exchange on Sar1 as a GEF (GDP to GTP) of Sar1 (33). Although we have not found proteins containing the K-loop motif in N. bombycis by using PSI-BLAST, related proteins have been screened in some microsporidia, including N. ceranae, Nosema apis, Enterospora canceri, and so on. Previous research showed that the uniparental inheritance of microsporidia led to the evolution of two unusual traits: male killing and feminization of the host under the long-term coevolution of microsporidia and the host (34). Furthermore, alleles may be lost and novel genotypes could arise during the adaptation of microsporidia to a new host (35–37). These results indicated that the categories and numbers of domains of Sec23, Sec24, and relevant proteins in microsporidia displayed diversity, which may have resulted from the coevolution relationship between the microsporidium and its host.
It has been reported that the kinase inhibitor H89 affected the early assembly of COPII and ultimately inhibited vesicular stomatitis virus glycoprotein microsome export from the ER by blocking the membrane recruitment and activation of Sar1 (38). The silencing of mammalian Sar1A and Sar1B by siRNA caused a disruption of COPII assembly (39). In order to understand the regulatory role of NbSar1 in COPII assembly by microsporidia, knockdown of NbSar1 was performed to assess the interference effect on the expression of other subunits of COPII. Interestingly, the expression levels of NbSec23-1, NbSec31-1, and NbSec31-2 were significantly downregulated (Fig. 7C, E, and F), whereas the expression levels of NbSec24-1, NbSec24-2, and NbSec13 were significantly upregulated (Fig. 7B and D). Nevertheless, whether a compensatory mechanism exists between Sec23 and Sec24 or between Sec13 and Sec31 is unknown. These findings shed new light: the knockdown of NbSar1 suppresses the normal expression of NbSec23, NbSec24, NbSec13, and NbSec31. Previous studies found that the addition of the protein kinase inhibitor H89 leads to the inactivation of Sar1 and the accumulation of Sec23, Sar1, and GalT2 at the ER exit sites (ERESs) (27). The Sar1 temperature-sensitive allele has been successfully employed in some fungal models such as S. cerevisiae and A. nidulans (40–42). In the genetic model A. nidulans, the temperature inactivation of allele mutations of SarA leads to markedly reduced SarA levels and the aggregation of Sec23-green fluorescent protein (GFP) into very few spots per cell at high temperature, suggesting that these alleles cause temperature-dependent SarA misfolding. Furthermore, the SarA6 Ser186Pro substitution leads to the formation of apical balloons resembling specialized fungal structures at 37°C, which underscores the importance of the C-terminal α-helix for SarA function/stability (40). These studies exhibited results similar to those of our research: the inactivation or silencing of Sar1 blocks the normal assembly of COPII at ERESs.
GTPases are involved in vesicular transport associated with a variety of signaling pathways that affect the regulation of cell proliferation and differentiation (43, 44). To further explore the role of Sar1 in the proliferation of N. bombycis in the present study, silkworms infected with N. bombycis were injected with Sar1 siRNA. After the knockdown of NbSar1, we found that the proliferation of N. bombycis was significantly inhibited (Fig. 8), which is consistent with previous research showing that the knockdown of Sar1B suppressed cell proliferation and induced significant apoptosis of RKO (colorectal cancer) cells (45). In Colaphellus bowringi, the knockdown of each gene of COPII considerably inhibited yolk deposition and ovarian growth. Furthermore, the silencing of Sar1, Sec23, and Sec24 suppressed feeding and increased mortality (46). The knockdown of Sar1b in zebrafish embryos also caused abnormal craniofacial skeletal development and the retention of intracellular collagen (47).
It was unexpected that the expression of NbSar1, NbSec23/NbSec24, and NbSec13/NbSec31 was significantly upregulated after interference on NbSar1 at 96 h (Fig. 6B to F). A previous study showed that the expression of the Rab11 gene was downregulated at 2 days whereas it exhibited upregulation at 7 days under continuous feeding of double-stranded RNA (dsRNA)-expressing bacteria or dsRNA for 14 days, which may be due to the stress response or the resistance of insects to RNAi (48). Moreover, similar results revealed that virus- or viroid-derived siRNAs are involved in the silencing of host genes and lead to the development of typical symptoms associated with the respective diseases (49–52). The host possesses RNAi mechanisms specific to the virus; accordingly, the virus could resist the RNAi of the host cell through mutation of its target gene, the generation of some viral proteins, and resistance to Dicer activity (53–55). In light of the function of Sar1 being similar to that of GTPase, we suspect that the N. bombycis also has a stress response or resistance to RNAi, but the mechanism remains unknown.
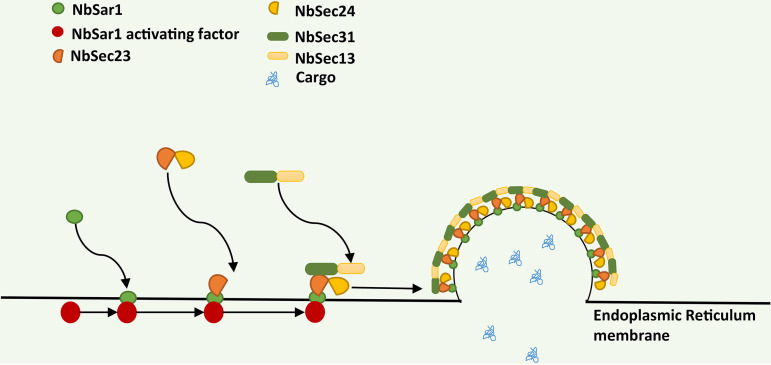
In summary, the results of this study indicate that NbSar1 is distributed mainly in the perinuclear region; that NbSar1 interacts with NbSec23-2, Nbsec23-2 interacts with NbSec24-1/NbSec24-2, NbSec23-1 interacts with NbSec31, and NbSec31 interacts with NbSec13; and that RNAi on NbSar1 resulted in the aberrant expression of NbSec23, NbSec24, NbSec31, and NbSec13 and inhibited the proliferation of N. bombycis. Based on the results of this study and previous reports, we propose a model for COPII assembly in N. bombycis (Fig. 9). According to this model, NbSar1 is activated under the action of an NbSar1-activating factor (unknown at present, but we suspect that it exists), and the activated NbSar1 then recruits the NbSec23-NbSec24 complex through binding NbSec23, thus forming the COPII inner layer. The NbSec13–NbSec31-1 complex is recruited to the ER through interactions between NbSec23 and NbSec31, thus forming the COPII outer layer. Eventually, NbSar1, NbSec23, NbSec24, NbSec13, and NbSec31 are assembled to form COPII at the ER membrane of N. bombycis. The data in this study support that NbSec23, NbSec24, NbSec31, and NbSec13 can assemble to form COPII mediated by NbSar1 of N. bombycis and that the functions of COPII are also closely related to the proliferation of N. bombycis; this may be a new target for the prevention of pébrine disease of the silkworm.
FIG 9.
Model of COPII assembly in N. bombycis. NbSar1 is activated under the action of an NbSar1-activating factor (unknown at present, but we suspect that it exists), and activated NbSar1 then binds with NbSec23-2 to recruit the NbSec23-1/NbSec23-2–NbSec24 complex, thus forming the COPII inner layer. In combination with NbSec31-1, NbSec23-1 is recruited to the NbSec13–NbSec31-1 complex, thus forming the COPII outer layer.
MATERIALS AND METHODS
Parasites, hosts, and genomes.
N. bombycis, silkworm ovarian cell line (BmN cells), and silkworms were provided by the Department of Silkworm Physiology and Pathology of the Institute of Sericulture of the Chinese Academy of Agricultural Sciences (Zhenjiang, China). The cells were cultured in TC-100 medium (AppliChem, Germany) supplemented with 10% fetal bovine serum (FBS) (Gibco) at 26°C. N. bombycis genomic data (GenBank accession number ACJZ00000000.1) were obtained from MicrosporidiaDB (https://microsporidiadb.org) or GenBank.
Cloning and expression of the NbSar1 gene and sequence analysis.
Purified mature spores of N. bombycis (108 spores/mL) were crushed in a tube containing 0.7 g of acid-washed glass beads for 1 min and then cooled on ice; this was repeated 10 times. The broken-spore suspension was used to extract genomic DNA according to the instructions of the fungal genomic DNA rapid extraction kit (TaKaRa, Japan). The purified genomic DNA was stored at −20°C after measuring the concentration. The forward primer 5′-GAATTCATGGAAGAAGCCAAAAAACTCTTCGG-3′, containing an EcoRI restriction site (GAATTC), and the reverse primer 5′-CGAGCTCTTATTTCTTTTGTTGGTTTATCATAATCTC-3′, containing a SacI restriction site (GAGCTC), were designed according to the sequences of the NbSar1 gene (GenBank accession number EOB13412.1). The 50-μL PCR amplification reaction mixture contained 2 μL (200 ng) genomic DNA of N. bombycis, 2 μL (20 μM) each forward and reverse primers, and 25 μL PrimeSTAR HS DNA polymerase (TaKaRa, Japan), with the addition of double-distilled water (ddH2O) to bring the total volume to 50 μL. The PCR amplification products were separated by 1% agarose gel electrophoresis and recovered using the AxyPrep DNA gel extraction kit (TaKaRa, Japan). The recovered products were cloned into the pMD19-T vector (Axygen, USA). The recombinant vector pMD19-T-NbSar1 was transformed into Escherichia coli Top10 competent cells (Sangon Biotech, China) and then cultured on LB plates containing ampicillin. The recombinants were identified by PCR and sequenced by Sangon Biotech (Shanghai, China). pMD19-T-NbSar1 was digested by EcoRI and SacI, and the NbSar1 gene was then ligated into the expression vector pET-28a. The pET-28a-NbSar1 recombinant plasmid was transformed into E. coli BL21(DE3) strain cells (Sangon Biotech, Shanghai, China) and cultured on LB plates containing kanamycin. Single colonies were selected for PCR identification and sequencing (Sangon Biotech, Shanghai, China). Recombinant pET-28a-NbSar1 was induced with 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 4 h. After cultivation, bacterial cells were collected and destroyed by sonication. SDS-PAGE was performed to confirm the expression of the recombinant protein, and the recombinant protein was tested by Western blotting using His tag antibody (Sangon Biotech, Shanghai, China). The expressed recombinant proteins were purified with the HisTrap FF purification kit (GE Healthcare, USA) according to the manufacturer’s instructions.
The isoelectric point (pI) and molecular weight of NbSar1 were predicted by the ExPASy Compute pI/Mw tool (https://web.expasy.org/compute_pi/). The homologous sequences were compared by using DNAMAN.
Preparation of NbSar1 polyclonal antibody and Western blot analysis.
New Zealand White rabbits were injected with 300 μg of purified recombinant protein emulsified with Freund’s complete adjuvant one time (Sigma, Germany) and then injected subcutaneously with 100 μg of purified recombinant protein emulsified with Freund’s incomplete adjuvant four times within 2 months. A small amount of rabbit antiserum was collected after immunization every time, and the antibody titer of the rabbit serum was assessed more than 1:8 0000 using enzyme-linked immunosorbent assay (ELISA). The antiserum was collected after the final immunization and purified by antigen affinity purification. Preimmune serum of rabbits was screened to confirm the absence of endogenous antibody that reacted with NbSar1. The animal experiment was carried out in Shanghai Youke Biotechnology Co., LTD. The animal Use license (SYXK(Shanghai)2018-0020) was approved by Shanghai Commission of Science and Technology (SCST).
The spore suspension broken by acid-washed glass beads was used to extract the total protein of N. bombycis. NbSar1 in the total protein was tested by Western blotting using the NbSar1 antibody.
Immunolocalization of NbSar1 in N. bombycis.
The germination of mature spores of N. bombycis was performed in 0.2 M KOH at 27°C for 1 h. BmN cells were cultured to a density of 80% in six-well plates. The cells were inoculated with the germinating spores in six-well plates and then cultured for 7 days. The cells infected with N. bombycis were fixed with 4% paraformaldehyde for 1 h, washed 3 times in phosphate-buffered saline–Tween (PBST) for 5 min, and subjected to permeabilization with 0.1% Triton X-100 for 1 h at room temperature. After being washed 3 times in PBST for 5 min, the cell samples were blocked with 5% bovine serum albumin (BSA) at 4°C overnight. The cell samples were incubated with 1 μg/mL NbSar1 antibody for 1 h at room temperature, washed 3 times in PBST for 10 min, and incubated in 1 μg/mL Alexa Fluor 488-conjugated goat anti-rabbit IgG (Sangon Biotech, Shanghai, China) at room temperature for 1 h successively. After being washed 3 times in PBST for 10 min, the cell samples were incubated with 2 μg/mL N. bombycis actin antibody (provided by the Department of Silkworm Physiology and Pathology of the Institute of Sericulture of the Chinese Academy of Agricultural Sciences, China) for 1 h, washed 3 times in PBST for 10 min, incubated with 5 μg/mL Cy5-conjugated goat anti-rabbit IgG (Sangon Biotech, Shanghai, China) for 1 h, and washed 3 times in PBST for 5 min. The nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI) for 20 min at room temperature. Negative controls were treated with preimmune serum as the primary antibody. Immunolocalization was observed by using an inverted fluorescence microscope (Olympus IX-71).
Yeast two-hybrid assay.
NbSar1, NbSec23-1, NbSec23-2, NbSec24-1, NbSec24-2, NbSec13, NbSec31-1, and NbSec31-2 were amplified from genomic DNA of N. bombycis using specific primers (Table 1), and NbSar1 digested by EcoRI and NotI was ligated into the bait vector pGBKT7-BK, while NbSec31-1 and NbSec24-2 were ligated into the yeast two-hybrid bait vector pGBKT7-BK using an In-Fusion HD cloning kit (TaKaRa, Japan). NbSec13 and NbSec23-1 digested by EcoRI and XhoI were ligated into prey vector pGADT7-AD, while NbSec24-1 digested by EcoRI and NotI was ligated into prey vector pGBKT7-BK, and NbSec23-2 was ligated into prey vector pGADT7-AD using an In-Fusion HD cloning kit (TaKaRa, Japan). The bait recombinant vector and prey recombinant vector were cotransformed into Y2H Gold yeast cells (Weidi Biotech, Shanghai, China) and then cultured on SD−Leu/−Trp plates (Clontech, USA) at 28°C. A single colony on SD−Leu/−Trp plates was selected and cultured on SD−Ade/−His/−Leu/−Trp plates containing X-α-gal (Clontech, USA) at 28°C.
TABLE 1.
Primer sequences for PCR
| Gene | Primer direction | Primer sequence (5′–3′) |
|---|---|---|
| NbSar1 | Forward | CCGGAATTCATGGAAGAAGCCAAAAAA |
| Reverse | TTGCGGCCGCTTATTTCTTTTGTTGGTTTAT | |
| NbSec23-1 | Forward | CCGGAATTCATGGCTTACTATCCAAATTTTATG |
| Reverse | CCGCTCGAGTTATTCATCACTACTCACAACAA | |
| NbSec23-2 | Forward | GGAGGCCAGTGAATTCATGGAAGAAGCAATCAGAGAAATCG |
| Reverse | TCATCTGCAGCTCGAGCTGCAGGTCATATTCCTCATAAG | |
| NbSec24-1 | Forward | CCGGAATTCATGACAGACGATTCTTCTCAAGTTTA |
| Reverse | TTGCGGCCGCCTACCACCACACTTCCCTTACATTA | |
| NbSec24-2 | Forward | CATGGAGGCCGAATTCATGTTTAAGATTGTCCTTAAAAGT |
| Reverse | GCTAGTTATGCGGCCGCTTATGTTGATGATCCTCGTACAAA | |
| NbSec13 | Forward | CCGGAATTCATGGAGTCCCATAAAATCCAC |
| Reverse | CCGCTCGAGTTATTCACCACATTTACTAAG | |
| NbSec31-1 | Forward | CGCCATATGATGATAAGTCTTTATGAACCCG |
| Reverse | TTGCGGCCGCTTAATAAACTAATTGAACCAGTGT | |
| NbSec31-2 | Forward | CGCCATATGATGAAAATCAACAAAAGGTG |
| Reverse | TTGCGGCCGCTTAATAAACTAATTGGACCAGTG | |
RNAi of NbSar1.
Small interfering RNA (siRNA) was designed according to the sequence of the NbSar1 gene and synthesized by Sangon Biotech (Shanghai, China). Fresh mulberry leaves were smeared with a 108-spore/mL suspension of N. bombycis, and P50 silkworms of the fifth instar were fed on the mulberry leaves smeared with N. bombycis. After feeding for 6 h, each silkworm was injected with 3 μL of siRNA (Table 2). After the injection of siRNA at 24 h, 48 h, 72 h, and 96 h, the midguts of the silkworms were collected and kept at −80°C. The midgut was lysed with 1 mL of RNAiso plus lysate (TaKaRa, Japan), the total RNA was then extracted with a Mini Best universal RNA extraction kit (TaKaRa, Japan), and cDNA was synthesized with PrimeScript RT master mix (TaKaRa, Japan). cDNA was used as the template to perform qPCR using the TB green premix Ex Taq II (Tli RNase H Plus) kit (TaKaRa, Japan) according to the manufacturer’s instructions, and the primer sequences are shown in Table 2. The transcription levels were calculated by the 2−ΔΔCT method with three replicates. GraphPad Prism 7.0 (GraphPad Software, San Diego, CA, USA) was used to conduct the multiple t tests.
TABLE 2.
Primer sequences for RNAi and qPCR
| Gene (purpose of primer) | Primer direction | Primer sequence (5′–3′) |
|---|---|---|
| NbSar1 (RNAi) | Sense | CCAAGUCCCAAAUUGAAAUTT |
| Antisense | AUUUCAAUUUGGGACUUGGTT | |
| Negative control (RNAi) | Sense | UUCUCCGAACGUGUCACGUTT |
| Antisense | ACGUGACACGUUCGGAGAATT | |
| NbSar1 (qPCR) | Forward | AATGCGGGTAAGACTACTCTGG |
| Reverse | CAAGCAACTCGAGCTGCTTTGTG | |
| N. bombycis β-tubulin (qPCR) | Forward | TTCCCTTCCCTAGACTTCACTTC |
| Reverse | CAGCAGCCACAGTCAAATACC | |
| NbSec23-1 (qPCR) | Forward | GCGAAAGAAATCCAACTCCTC |
| Reverse | CGTAATCACTGTCCCACTAGAA | |
| NbSec24-1 (qPCR) | Forward | GACAGACGATTCTTCTCAAGTTTATTC |
| Reverse | TCTTCATTATAATTCTCGGGTTGGA | |
| NbSec24-2 (qPCR) | Forward | CCTTTCCTAATTTCTATCCTCCTCAT |
| Reverse | CCTTATCATTCTCTGCCCTTTCT | |
| NbSec13 (qPCR) | Forward | GTGGAGGTGTGTCCTGATAATG |
| Reverse | AGAGAGACTATAGACGGGTTCAC | |
| NbSec31-1 (qPCR) | Forward | TCCACGAGTAGTTCCGAGTATG |
| Reverse | CGGGTACAGAATCCACTTGATG | |
| NbSec31-2 (qPCR) | Forward | CACGTGTAGTTCCGAGTATGAG |
| Reverse | TACAGGAGGCCTAGTACAGAAT | |
| BmGAPDH (qPCR) | Forward | TTCATGCCACAACTGCTACA |
| Reverse | AGTCAGCTTGCCATTAAGAG | |
ACKNOWLEDGMENTS
This work was supported by the China Agriculture Research System of MOF and MARA and the National Natural Science Foundation of China (31960684).
We are also grateful to all who provided the means for us to access free software, which we have used and cited in this article. We also thank all partners and laboratory staff for kind help and criticism.
We declare no conflict of interest.
Footnotes
Supplemental material is available online only.
Contributor Information
Zhongyuan Shen, Email: szysri@163.com.
Miguel A. Penalva, Centro de Investigaciones Biologicas CSIC
REFERENCES
- 1.Guo R, Cao G, Lu Y, Xue R, Kumar D, Hu X, Gong C. 2016. Exogenous gene can be integrated into Nosema bombycis genome by mediating with a non-transposon vector. Parasitol Res 115:3093–3098. doi: 10.1007/s00436-016-5064-8. [DOI] [PubMed] [Google Scholar]
- 2.Han B, Ma Y, Tu V, Tomita T, Mayoral J, Williams T, Horta A, Huang H, Weiss LM. 2019. Microsporidia interact with host cell mitochondria via voltage-dependent anion channels using sporoplasm surface protein 1. mBio 10(4):e01944-19. doi: 10.1128/mBio.01944-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pan G, Xu J, Li T, Xia Q, Liu SL, Zhang G, Li S, Li C, Liu H, Yang L, Liu T, Zhang X, Wu Z, Fan W, Dang X, Xiang H, Tao M, Li Y, Hu J, Li Z, Lin L, Luo J, Geng L, Wang L, Long M, Wan Y, He N, Zhang Z, Lu C, Keeling PJ, Wang J, Xiang Z, Zhou Z. 2013. Comparative genomics of parasitic silkworm microsporidia reveal an association between genome expansion and host adaptation. BMC Genomics 14:186. doi: 10.1186/1471-2164-14-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li Z, Wang Y, Wang L, Zhou Z. 2018. Molecular and biochemical responses in the midgut of the silkworm, Bombyx mori, infected with Nosema bombycis. Parasit Vectors 11:147. doi: 10.1186/s13071-018-2755-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schekman R, Orci L. 1996. Coat proteins and vesicle budding. Science 271:1526–1533. doi: 10.1126/science.271.5255.1526. [DOI] [PubMed] [Google Scholar]
- 6.Bonifacino JS, Glick BS. 2004. The mechanisms of vesicle budding and fusion. Cell 116:153–166. doi: 10.1016/S0092-8674(03)01079-1. [DOI] [PubMed] [Google Scholar]
- 7.Bi X, Corpina RA, Goldberg J. 2002. Structure of the Sec23/24-Sar1 pre-budding complex of the COPII vesicle coat. Nature 419:271–277. doi: 10.1038/nature01040. [DOI] [PubMed] [Google Scholar]
- 8.Yoshihisa T, Barlowe C, Schekman R. 1993. Requirement for a GTPase-activating protein in vesicle budding from the endoplasmic reticulum. Science 259:1466–1468. doi: 10.1126/science.8451644. [DOI] [PubMed] [Google Scholar]
- 9.Bi X, Mancias JD, Goldberg J. 2007. Insights into COPII coat nucleation from the structure of Sec23.Sar1 complexed with the active fragment of Sec31. Dev Cell 13:635–645. doi: 10.1016/j.devcel.2007.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Militello R, Colombo MI. 2013. Small GTPases as regulators of cell division. Commun Integr Biol 6:e25460. doi: 10.4161/cib.25460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barlowe C, Schekman R. 1993. SEC12 encodes a guanine-nucleotide-exchange factor essential for transport vesicle budding from the ER. Nature 365:347–349. doi: 10.1038/365347a0. [DOI] [PubMed] [Google Scholar]
- 12.Akiyoshi DE, Morrison HG, Lei S, Feng X, Zhang Q, Corradi N, Mayanja H, Tumwine JK, Keeling PJ, Weiss LM, Tzipori S. 2009. Genomic survey of the non-cultivatable opportunistic human pathogen, Enterocytozoon bieneusi. PLoS Pathog 5:e1000261. doi: 10.1371/journal.ppat.1000261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Campbell SE, Williams TA, Yousuf A, Soanes DM, Paszkiewicz KH, Williams BA. 2013. The genome of Spraguea lophii and the basis of host-microsporidian interactions. PLoS Genet 9:e1003676. doi: 10.1371/journal.pgen.1003676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cornman RS, Chen YP, Schatz MC, Street C, Zhao Y, Desany B, Egholm M, Hutchison S, Pettis JS, Lipkin WI, Evans JD. 2009. Genomic analyses of the microsporidian Nosema ceranae, an emergent pathogen of honey bees. PLoS Pathog 5:e1000466. doi: 10.1371/journal.ppat.1000466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Corradi N, Slamovits CH. 2011. The intriguing nature of microsporidian genomes. Brief Funct Genomics 10:115–124. doi: 10.1093/bfgp/elq032. [DOI] [PubMed] [Google Scholar]
- 16.Cuomo CA, Desjardins CA, Bakowski MA, Goldberg J, Ma AT, Becnel JJ, Didier ES, Fan L, Heiman DI, Levin JZ, Young S, Zeng Q, Troemel ER. 2012. Microsporidian genome analysis reveals evolutionary strategies for obligate intracellular growth. Genome Res 22:2478–2488. doi: 10.1101/gr.142802.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Méténier G, Vivarès CP. 2001. Molecular characteristics and physiology of microsporidia. Microbes Infect 3:407–415. doi: 10.1016/S1286-4579(01)01398-3. [DOI] [PubMed] [Google Scholar]
- 18.Vavra J, Larsson JIR. 1999. Structure of the microsporidia, p 7–84. In Wittner M, Weiss LM (ed), The microsporidia and microsporidiosis. ASM Press, Washington, DC. [Google Scholar]
- 19.Sokolova Y, Snigirevskaya E, Morzhina E, Skarlato S, Mironov A, Komissarchik Y. 2001. Visualization of early Golgi compartments at proliferate and sporogenic stages of a microsporidian Nosema grylli. J Eukaryot Microbiol 48:86S–87S. doi: 10.1111/j.1550-7408.2001.tb00464.x. [DOI] [PubMed] [Google Scholar]
- 20.Sokolova YY, Mironov AA. 2008. Structure and function of the Golgi organelle in parasitic protists, p 647–674. In Mironov A, Pavelka M (ed), The Golgi apparatus: state of the art 110 years after Camillo Golgi’s discovery. Springer, Vienna, Austria. [Google Scholar]
- 21.Takvorian PM, Cali A. 1994. Enzyme histochemical identification of the Golgi apparatus in the microsporidian, Glugea stephani. J Eukaryot Microbiol 41:63S–64S. [PubMed] [Google Scholar]
- 22.Beznoussenko GV, Dolgikh VV, Seliverstova EV, Semenov PB, Tokarev YS, Trucco A, Micaroni M, Di Giandomenico D, Auinger P, Senderskiy IV, Skarlato SO, Snigirevskaya ES, Komissarchik YY, Pavelka M, De Matteis MA, Luini A, Sokolova YY, Mironov AA. 2007. Analogs of the Golgi complex in microsporidia: structure and avesicular mechanisms of function. J Cell Sci 120(Part 7):1288–1298. doi: 10.1242/jcs.03402. [DOI] [PubMed] [Google Scholar]
- 23.Bhattacharya N, O’Donnell J, Stagg SM. 2012. The structure of the Sec13/31 COPII cage bound to Sec23. J Mol Biol 420:324–334. doi: 10.1016/j.jmb.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peotter J, Kasberg W, Pustova I, Audhya A. 2019. COPII-mediated trafficking at the ER/ERGIC interface. Traffic 20:491–503. doi: 10.1111/tra.12654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Béthune J, Wieland FT. 2018. Assembly of COPI and COPII vesicular coat proteins on membranes. Annu Rev Biophys 47:63–83. doi: 10.1146/annurev-biophys-070317-033259. [DOI] [PubMed] [Google Scholar]
- 26.Lord C, Ferro-Novick S, Miller EA. 2013. The highly conserved COPII coat complex sorts cargo from the endoplasmic reticulum and targets it to the Golgi. Cold Spring Harb Perspect Biol 5:a013367. doi: 10.1101/cshperspect.a013367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Quintero CA, Giraudo CG, Villarreal M, Montich G, Maccioni HJ. 2010. Identification of a site in Sar1 involved in the interaction with the cytoplasmic tail of glycolipid glycosyltransferases. J Biol Chem 285:30340–30346. doi: 10.1074/jbc.M110.128868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nishikawa S, Nakano A. 1991. The GTP-binding Sar1 protein is localized to the early compartment of the yeast secretory pathway. Biochim Biophys Acta 1093:135–143. doi: 10.1016/0167-4889(91)90114-D. [DOI] [PubMed] [Google Scholar]
- 29.Horikawa K, Yorimitsu T, Kodera C, Sato K. 2019. Implication of a novel function of Sar1 in the nucleus. Cell Struct Funct 44:105–112. doi: 10.1247/csf.19019. [DOI] [PubMed] [Google Scholar]
- 30.Yazar S, Koru O, Hamamcı B, Cetinkaya U, Karaman U, Kuk S. 2013. Microsporidia and microsporidiosis. Turkiye Parazitol Derg 37:123–134. doi: 10.5152/tpd.2013.28. (In Turkish.) [DOI] [PubMed] [Google Scholar]
- 31.d’Enfert C, Barlowe C, Nishikawa S, Nakano A, Schekman R. 1991. Structural and functional dissection of a membrane glycoprotein required for vesicle budding from the endoplasmic reticulum. Mol Cell Biol 11:5727–5734. doi: 10.1128/mcb.11.11.5727-5734.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McMahon C, Studer SM, Clendinen C, Dann GP, Jeffrey PD, Hughson FM. 2012. The structure of Sec12 implicates potassium ion coordination in Sar1 activation. J Biol Chem 287:43599–43606. doi: 10.1074/jbc.M112.420141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bravo-Plaza I, Hernández-González M, Pinar M, Díaz JF, Peñalva MA. 2019. Identification of the guanine nucleotide exchange factor for SAR1 in the filamentous fungal model Aspergillus nidulans. Biochim Biophys Acta 1866:118551. doi: 10.1016/j.bbamcr.2019.118551. [DOI] [PubMed] [Google Scholar]
- 34.Dunn AM, Smith JE. 2001. Microsporidian life cycles and diversity: the relationship between virulence and transmission. Microbes Infect 3:381–388. doi: 10.1016/S1286-4579(01)01394-6. [DOI] [PubMed] [Google Scholar]
- 35.Gómez-Moracho T, Bartolomé C, Bello X, Martín-Hernández R, Higes M, Maside X. 2015. Recent worldwide expansion of Nosema ceranae (Microsporidia) in Apis mellifera populations inferred from multilocus patterns of genetic variation. Infect Genet Evol 31:87–94. doi: 10.1016/j.meegid.2015.01.002. [DOI] [PubMed] [Google Scholar]
- 36.Gómez-Moracho T, Bartolomé C, Martín-Hernández R, Higes M, Maside X. 2015. Evidence for weak genetic recombination at the PTP2 locus of Nosema ceranae. Environ Microbiol 17:1300–1309. doi: 10.1111/1462-2920.12574. [DOI] [PubMed] [Google Scholar]
- 37.Pelin A, Selman M, Aris-Brosou S, Farinelli L, Corradi N. 2015. Genome analyses suggest the presence of polyploidy and recent human-driven expansions in eight global populations of the honeybee pathogen Nosema ceranae. Environ Microbiol 17:4443–4458. doi: 10.1111/1462-2920.12883. [DOI] [PubMed] [Google Scholar]
- 38.Aridor M, Balch WE. 2000. Kinase signaling initiates coat complex II (COPII) recruitment and export from the mammalian endoplasmic reticulum. J Biol Chem 275:35673–35676. doi: 10.1074/jbc.C000449200. [DOI] [PubMed] [Google Scholar]
- 39.Cutrona MB, Beznoussenko GV, Fusella A, Martella O, Moral P, Mironov AA. 2013. Silencing of mammalian Sar1 isoforms reveals COPII-independent protein sorting and transport. Traffic 14:691–708. doi: 10.1111/tra.12060. [DOI] [PubMed] [Google Scholar]
- 40.Hernández-González M, Peñalva MA, Pantazopoulou A. 2015. Conditional inactivation of Aspergillus nidulans sarASAR1 uncovers the morphogenetic potential of regulating endoplasmic reticulum (ER) exit. Mol Microbiol 95:491–508. doi: 10.1111/mmi.12880. [DOI] [PubMed] [Google Scholar]
- 41.Nakańo A, Muramatsu M. 1989. A novel GTP-binding protein, Sar1p, is involved in transport from the endoplasmic reticulum to the Golgi apparatus. J Cell Biol 109(6 Part 1):2677–2691. doi: 10.1083/jcb.109.6.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamanushi T, Hirata A, Oka T, Nakano A. 1996. Characterization of yeast sar1 temperature-sensitive mutants, which are defective in protein transport from the endoplasmic reticulum. J Biochem 120:452–458. doi: 10.1093/oxfordjournals.jbchem.a021432. [DOI] [PubMed] [Google Scholar]
- 43.Fryer LG, Jones B, Duncan EJ, Hutchison CE, Ozkan T, Williams PA, Alder O, Nieuwdorp M, Townley AK, Mensenkamp AR, Stephens DJ, Dallinga-Thie GM, Shoulders CC. 2014. The endoplasmic reticulum coat protein II transport machinery coordinates cellular lipid secretion and cholesterol biosynthesis. J Biol Chem 289:4244–4261. doi: 10.1074/jbc.M113.479980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Loftus AF, Hsieh VL, Parthasarathy R. 2012. Modulation of membrane rigidity by the human vesicle trafficking proteins Sar1A and Sar1B. Biochem Biophys Res Commun 426:585–589. doi: 10.1016/j.bbrc.2012.08.131. [DOI] [PubMed] [Google Scholar]
- 45.Lu Y, Zhou S-K, Chen R, Jiang L-X, Yang L-L, Bi T-N. 2020. Knockdown of SAR1B suppresses proliferation and induces apoptosis of RKO colorectal cancer cells. Oncol Lett 20:186. doi: 10.3892/ol.2020.12048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tian Z, Guo S, Zhu F, Liu W, Wang X-P. 2022. Targeting coat protein II complex genes via RNA interference inhibits female adult feeding and reproductive development in the cabbage beetle Colaphellus bowringi. Pest Manag Sci 78:2141–2150. doi: 10.1002/ps.6836. [DOI] [PubMed] [Google Scholar]
- 47.Levic DS, Minkel JR, Wang W-D, Rybski WM, Melville DB, Knapik EW. 2015. Animal model of Sar1b deficiency presents lipid absorption deficits similar to Anderson disease. J Mol Med (Berl) 93:165–176. doi: 10.1007/s00109-014-1247-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li X, Zhang M, Zhang H. 2011. RNA interference of four genes in adult Bactrocera dorsalis by feeding their dsRNAs. PLoS One 6:e17788. doi: 10.1371/journal.pone.0017788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shimura H, Pantaleo V, Ishihara T, Myojo N, Inaba J, Sueda K, Burgyán J, Masuta C. 2011. A viral satellite RNA induces yellow symptoms on tobacco by targeting a gene involved in chlorophyll biosynthesis using the RNA silencing machinery. PLoS Pathog 7:e1002021. doi: 10.1371/journal.ppat.1002021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smith NA, Eamens AL, Wang MB. 2011. Viral small interfering RNAs target host genes to mediate disease symptoms in plants. PLoS Pathog 7:e1002022. doi: 10.1371/journal.ppat.1002022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Catalano D, Cillo F, Finetti-Sialer M. 2012. In silico prediction of virus-derived small interfering RNAs and their putative host messenger targets in Solanum lycopersicum infected by different potato virus Y isolates. EMBnet J 18:83–84. doi: 10.14806/ej.18.A.425. [DOI] [Google Scholar]
- 52.Navarro B, Gisel A, Rodio ME, Delgado S, Flores R, Di Serio F. 2012. Small RNAs containing the pathogenic determinant of a chloroplast-replicating viroid guide the degradation of a host mRNA as predicted by RNA silencing. Plant J 70:991–1003. doi: 10.1111/j.1365-313X.2012.04940.x. [DOI] [PubMed] [Google Scholar]
- 53.Zheng ZM, Tang S, Tao M. 2005. Development of resistance to RNAi in mammalian cells. Ann N Y Acad Sci 1058:105–118. doi: 10.1196/annals.1359.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gitlin L, Karelsky S, Andino R. 2002. Short interfering RNA confers intracellular antiviral immunity in human cells. Nature 418:430–434. doi: 10.1038/nature00873. [DOI] [PubMed] [Google Scholar]
- 55.Jacque JM, Triques K, Stevenson M. 2002. Modulation of HIV-1 replication by RNA interference. Nature 418:435–438. doi: 10.1038/nature00896. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download spectrum.00719-22-s0001.pdf, PDF file, 1.1 MB (1.1MB, pdf)