ABSTRACT
Alginate (ALG) is known to alleviate intestinal inflammation in inflammatory bowel disease, but its mechanism of action remains elusive. In the present study, we studied the involvement of the intestinal microbiota and bile acid (BA) metabolism in ALG-mediated anti-inflammatory effects in mice. A combination of 16S rRNA gene amplicon sequencing, shotgun metagenomic sequencing, and targeted BA metabolomic profiling was employed to investigate structural and functional differences in the colonic microbiota and BA metabolism in dextran sulfate sodium (DSS)-treated mice with or without dietary supplementation of ALG. We further explored the role of the intestinal microbiota as well as a selected ALG-enriched bacterium and BA in DSS-induced colitis. Dietary ALG alleviated DSS-mediated intestinal inflammation and enriched a small set of bacteria including Bifidobacterium animalis in the colon (P < 0.05). Additionally, ALG restored several bacteria carrying secondary BA-synthesizing enzymes such as 7α-hydroxysteroid dehydrogenase and BA hydrolase to healthy levels in DSS-treated mice. Although a majority of BAs were suppressed by DSS, a few secondary BAs such as hyodeoxycholic acid (HDCA) were markedly enriched by ALG. Furthermore, ALG significantly upregulated the expression of a major BA receptor, the farnesoid X receptor, while suppressing NF-κB and c-Jun N-terminal kinase (JNK) activation. Depletion of the intestinal microbiota completely abrogated the protective effect of ALG in DSS-treated mice. Similar to ALG, B. animalis and HDCA exerted a strong anti-inflammatory effect in DSS-induced colitis by downregulating inflammatory cytokines (interleukin-1β [IL-1β], IL-6, and tumor necrosis factor alpha [TNF-α]). Taken together, these results indicated that ALG achieves its alleviating effect on intestinal inflammation through regulation of the microbiota by enriching B. animalis to promote the biosynthesis of specific secondary BAs such as HDCA. These findings have revealed intricate interactions among the intestinal microbiota, BA metabolism, and intestinal health and further provided a novel strategy to improve intestinal health through targeted manipulation of the intestinal microbiota and BA metabolism.
IMPORTANCE ALG has been shown to ameliorate inflammatory bowel disease (IBD), but little is known about the mechanism of its anti-inflammatory action. This study was the first to demonstrate that ALG provided a preventive effect against colitis in an intestinal microbiota-dependent manner. Furthermore, we confirmed that by selectively enriching intestinal B. animalis and secondary BA (HDCA), ALG contributed to the attenuation of DSS-induced colitis. These findings contribute to a better understanding of the mechanism of action of ALG on the attenuation of colitis and provide new approaches to IBD therapy by regulating gut microbial BA metabolism.
KEYWORDS: alginate, bile acid, hyodeoxycholic acid, inflammatory bowel disease, microbiome
INTRODUCTION
Inflammatory bowel disease (IBD) is a group of disorders that cause chronic intestinal inflammation (1). Accumulating evidence has linked IBD to intestinal microbiota dysbiosis (2, 3). Manipulation of the intestinal microbiota and its metabolites has shown potential in IBD therapy (4–6). However, the specific involvement of intestinal microbes and metabolites in the pathogenesis of IBD remains elusive.
Primary bile acids (BA) are produced in the liver and deconjugated and transformed by the intestinal microbiota to secondary BAs such as deoxycholic acid (DCA) and hyodeoxycholic acid (HDCA) in the intestinal tract (7, 8). Certain strains of Lactobacillus, Erysipelotrichaceae, Lachnospiraceae, Clostridium, and Bacteroides carry bile salt hydrolase (bsh), which is responsible for deconjugation of glycine or taurine from primary BAs, while other strains of Lactobacillus, Lachnospiraceae, Ruminococcaceae, Clostridiaceae, Eubacterium, and Peptostreptococcus carry the BA-inducible (bai) gene operon responsible for 7α-dehydroxylation and biotransformation to generate secondary BAs (9–11). Deconjugation and biotransformation of BAs were recently shown to be impaired in IBD patients (12). However, a mechanistic understanding of the microbiota-BA-IBD association is lacking. Thus, it is essential to elucidate the changes in the intestinal microbiome and BA metabolism in the context of intestinal inflammation in order to devise effective therapeutic strategies for IBD (13).
Alginate (ALG), a linear polymer of α-l-guluronic acid and β-d-mannuronic acid, is a natural water-soluble polysaccharide extracted from brown seaweed. ALG is known to be protective against gastric ulcers and esophageal reflux (14, 15). It is also beneficial for ameliorating experimental colitis in mice (16–18). However, poor absorption of ALG by the intestinal tract implies microbial involvement in the metabolism of ALG (19). Consistently, dietary supplementation of ALG increased the bsh enzyme activity and total BA levels and altered the BA profile in the feces of rats (20–22). However, the role of ALG in microbial BA metabolism in the context of intestinal inflammation remains largely unknown.
We hypothesized that ALG alleviates intestinal inflammation by targeting specific intestinal bacteria and BAs. To test this hypothesis, a DSS-induced colitis model was employed to investigate structural and functional differences in the intestinal microbiota and BAs in response to dietary supplementation of ALG. We further selected a bacterium and BA that were highly differentially enriched by ALG and explored their role in DSS-induced intestinal inflammation. This study has shed light on ALG-mediated regulation of the intestinal microbiota and BA metabolism and provided new approaches to the prevention and treatment of IBD.
RESULTS
Alginate alleviates DSS-induced intestinal inflammation.
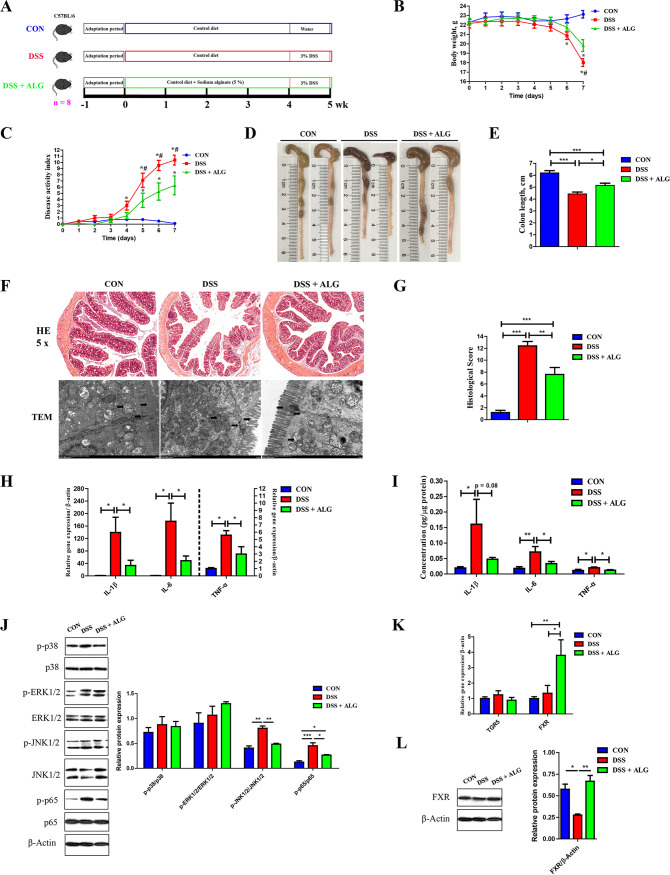
To study the influence of ALG on intestinal inflammation, we supplemented mice with or without 5% ALG in the diet and then subjected them to 3% DSS in their drinking water to induce colitis (Fig. 1A). ALG significantly ameliorated DSS-induced colitis (P < 0.05), as evidenced by an improvement in weight loss (Fig. 1B), disease activity index (DAI) score (Fig. 1C), and colonic shortening (Fig. 1D and E). Consistently, ALG supplementation showed significant alleviation of mucosal inflammatory cell infiltration and epithelial layer destruction in DSS-treated mice (P < 0.05) (Fig. 1F and G). Additionally, the mRNA and protein expression levels of proinflammatory cytokines (interleukin-1β [IL-1β], IL-6, and tumor necrosis factor alpha [TNF-α]) in the colon of DSS-treated mice were significantly suppressed by ALG (P < 0.05) (Fig. 1H and I). Moreover, DSS-induced phosphorylation of c-Jun N-terminal kinase 1/2 (JNK1/2) and NF-κB p65 in the colon was significantly decreased in response to ALG; however, phosphorylation was unchanged with p38 mitogen-activated protein kinase (MAPK) and extracellular signal-regulated kinase 1/2 (ERK1/2) (Fig. 1J).
FIG 1.
Dietary supplementation of alginate (ALG) alleviates DSS-induced colitis. (A) Experimental scheme of ALG supplementation in a mouse model of DSS-induced acute colitis. (B and C) While DSS (3%) was provided in drinking water for a week to induce colitis, body weight (B) and the disease activity index (DAI) score (C) were recorded daily in the final week of DSS treatment for three groups of mice (n = 8). The colon was collected from each mouse at the end of the experiment. (D to G) Representative images of the colon (D), the colon length (E), H&E staining (magnification of ×50), and TEM of the colon (F) and colonic histology score (G) are shown. (H and I) The mRNA (H) and protein (I) expression levels of three inflammatory cytokines in the colon were also measured. (J) Phosphorylation of p38, ERK1/2, JNK1/2, and p65 NF-κB in the colon. (K) The mRNA gene expression levels of two BA receptors in the colon. (L) Expression of FXR at the protein level. Data are presented as means ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001; #, P < 0.05; compared with the DSS plus ALG group.
G-protein-coupled receptor (TGR5) and farnesoid X receptor (FXR) are two major receptors for BAs (23). TGR5 mRNA was not affected by DSS or ALG, while FXR mRNA was significantly induced by ALG in the colon (Fig. 1K). At the protein level, DSS suppressed FXR expression, but it was largely restored by ALG (Fig. 1L). These results indicated that dietary ALG is capable of alleviating intestinal inflammation, possibly by suppressing the NF-κB and JNK signaling pathways.
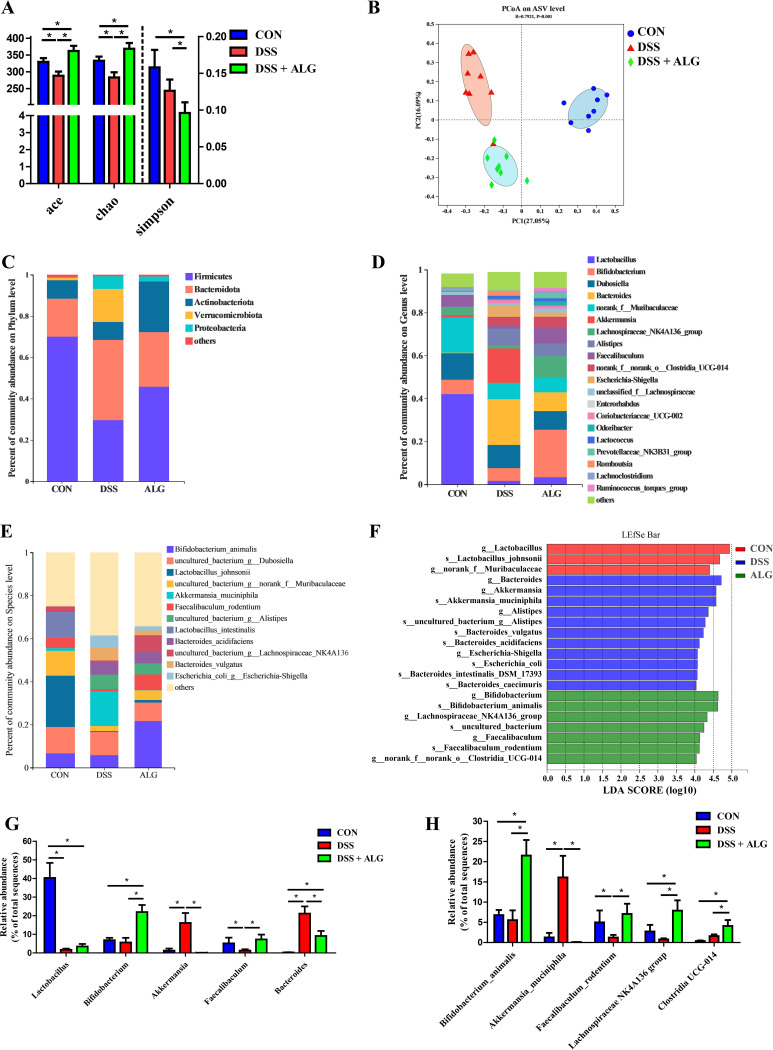
Alginate selectively promotes Bifidobacterium animalis and alters the microbial metabolic function in DSS-induced colitis. To examine the changes in the intestinal microbiota of DSS-treated mice in response to ALG, we analyzed the colonic microbiota composition using 16S rRNA gene sequencing. While ALG restored DSS-induced loss in the richness of the microbiota as indicated by ACE and Chao1, overall α-diversity was significantly increased in DSS-treated mice in response to ALG as indicated by the Simpson index (P < 0.05) (Fig. 2A). Principal-coordinate analysis (PCoA) based on the Bray-Curtis distance showed clear segregation among the three groups of mice (Fig. 2B). The colonic microbiota composition was different at the phylum (Fig. 2C), genus (Fig. 2D), and amplicon sequence variant (ASVs) levels (Fig. 2E). The linear discriminant analysis effect size (LEfSe) algorithm further identified several ASVs that were differentially abundant among three groups of mice (Fig. 2F). The relative abundance of Lactobacillus in the colon was significantly suppressed by DSS but failed to be recovered by ALG (Fig. 2G). Bacteroides, on the other hand, was markedly expanded in response to DSS but suppressed by ALG (Fig. 2G). It is important to note that Bifidobacterium animalis, a highly abundant bacterium in the colon, was drastically increased from 6.79% in healthy or DSS-treated mice to 21.5% in the DSS plus ALG mice (Fig. 2H). ALG also markedly induced Lachnospiraceae NK4A136 group and Clostridia UCG-014 in both control and DSS-treated mice and restored Faecalibacterium rodentium in DSS-treated mice to the healthy level (Fig. 2H). Interestingly, Akkermansia muciniphila was significantly increased by DSS but almost completely abolished in response to ALG supplementation (Fig. 2H).
FIG 2.
Dietary supplementation with ALG alters the composition of the intestinal microbiota in DSS-treated mice. Mice supplemented with or without 5% ALG for 4 weeks, followed by induction of colitis with 3% DSS in drinking water for another week (n = 8). The colonic digesta was collected from each mouse at the end of the experiment and subjected to DNA isolation and 16S rRNA gene sequencing. (A) The α-diversity of the colonic microbiota among three groups of mice. (B) PCoA plot depicting β-diversity of the colonic microbiota based on the Bray-Curtis distance. (A to C) The colonic microbiota composition is shown at the phylum (C), genus (D), and ASVs levels (E). (F) LEfSe analysis of differential enrichment of the colonic bacteria at the ASV level (linear discriminant analysis [LDA] > 4). (G and H) Bar graphs of the top five differentially enriched bacteria are shown at the genus (G) and ASV levels (H). Data are presented as means ± SEM. *, P < 0.05.
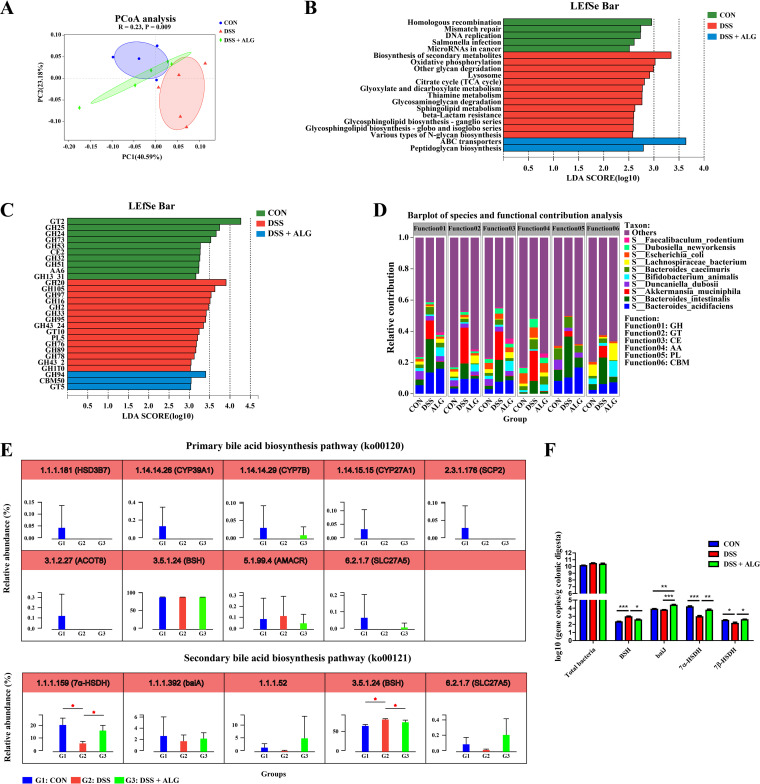
To further understand the functional changes of the colonic microbiota of mice in response to DSS and ALG, shotgun metagenome sequencing was used. The PCoA plot again revealed distinct functional profiles among the three groups based on the KEGG enzymes (analysis of similarity [ANOSIM], P < 0.05; Fig. 3A). LEfSe analysis showed differential enrichment of multiple metabolic pathways. For example, ATP-binding cassette (ABC) transporters that utilize ATP hydrolysis to provide energy for the translocation of substrates across cell membranes (24) were enriched in the DSS plus ALG group (Fig. 3B). We further identified the enrichment of several carbohydrate-active enzymes (CAZymes) such as glycoside hydrolase 94 (GH94), carbohydrate-binding module 50 (CBM50), and glycosyltransferase 5 (GT5) in the DSS plus ALG group (Fig. 3C). In fact, B. animalis was the main contributor (top 3) of these CAZymes (GH, GT, and CBM) (Fig. 3D).
FIG 3.
Dietary supplementation with ALG modulates the function of the intestinal microbiota in DSS-treated mice. Mice supplemented with or without 5% ALG for 4 weeks, followed by induction of colitis with 3% DSS in drinking water for another week (n = 8). The colonic digesta was collected from each mouse at the end of the experiment and subjected to DNA isolation and shotgun metagenomic sequencing. (A) PCoA plot based on the KEGG pathways of identified bacterial genes. (B) LEfSe analysis of differential enrichment of microbial KEGG pathways (LDA > 2.5). (C) LEfSe analysis of differential enrichment of microbial carbohydrate-active enzymes (CAZymes) (LDA > 3). (D) Functional contribution of top 10 differentially enriched CAZymes. (E) Relative abundances (%) of the major enzymes involved in the biosynthesis of primary and secondary bile acids (BAs). (F) Quantitative PCR analysis of the copy number of total bacteria and specific bacterial groups carrying BA hydrolase (bsh), BA 7-dehydroxylase (baiJ), 7α-hydroxysteroid dehydrogenase (7α-HSDH), and 7β-hydroxysteroid dehydrogenase (7β-HSDH). Data are presented as means ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Alginate alters microbial BA metabolism and increases intestinal HDCA in DSS-induced colitis.
The intestinal microbiota plays a key role in the biotransformation of primary BAs to secondary BAs by deconjugation and dihydroxylation (23). The major enzymes involved in the biosynthesis of primary and secondary BAs are bsh, baiJ, 7α-hydroxysteroid dehydrogenase (7α-HSDH), and 7β-hydroxysteroid dehydrogenase (7β-HSDH) (23). To study the impact of dietary supplementation of ALG on intestinal BA biosynthesis, we examined the changes in the major enzymes involved in the biosynthesis of primary and secondary BAs based on the shotgun metagenomic data. The host enzymes responsible for the biosynthesis of primary BAs were largely unaltered by DSS or ALG (Fig. 3E). Among those major enzymes involved in secondary BA synthesis, 7α-HSDH was significantly suppressed by DSS, while bsh was enriched; however, ALG restored relative abundances of both enzymes to healthy levels (Fig. 3E). Consistently, the quantitative PCR (qPCR) analysis revealed that the bacteria carrying 7α-HSDH, 7β-HSDH, and baiJ were significantly increased, whereas those carrying bsh were decreased in DSS-treated mice in response to dietary ALG (P < 0.05) (Fig. 3F).
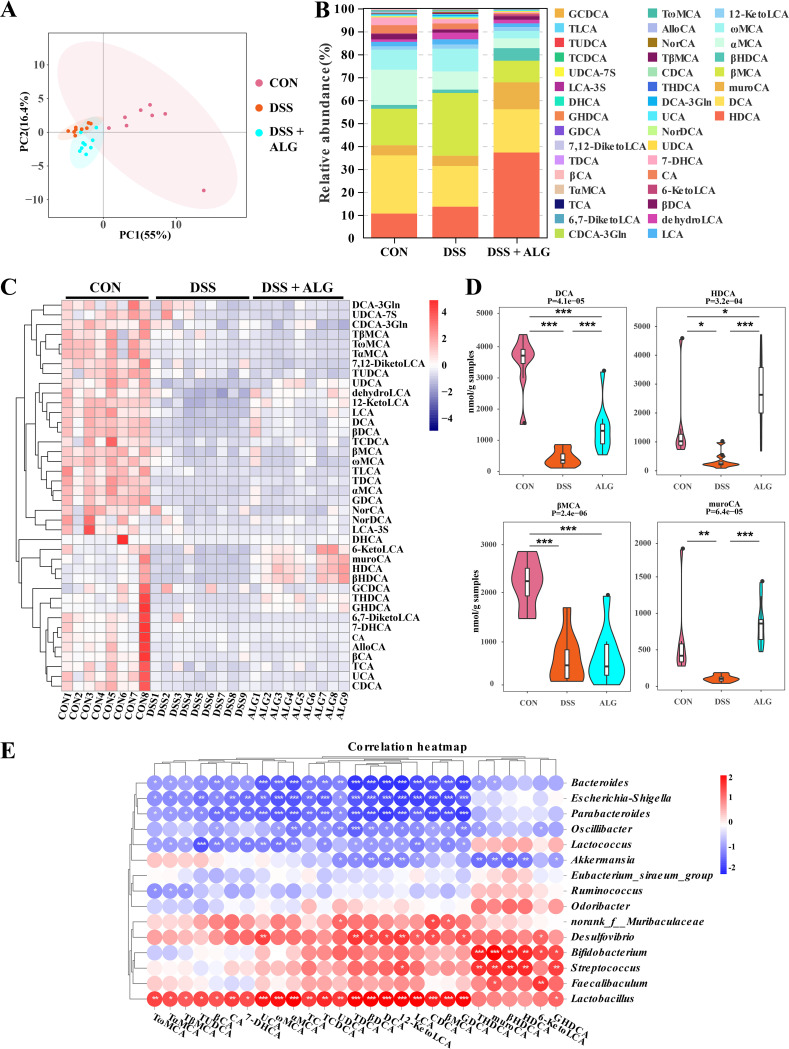
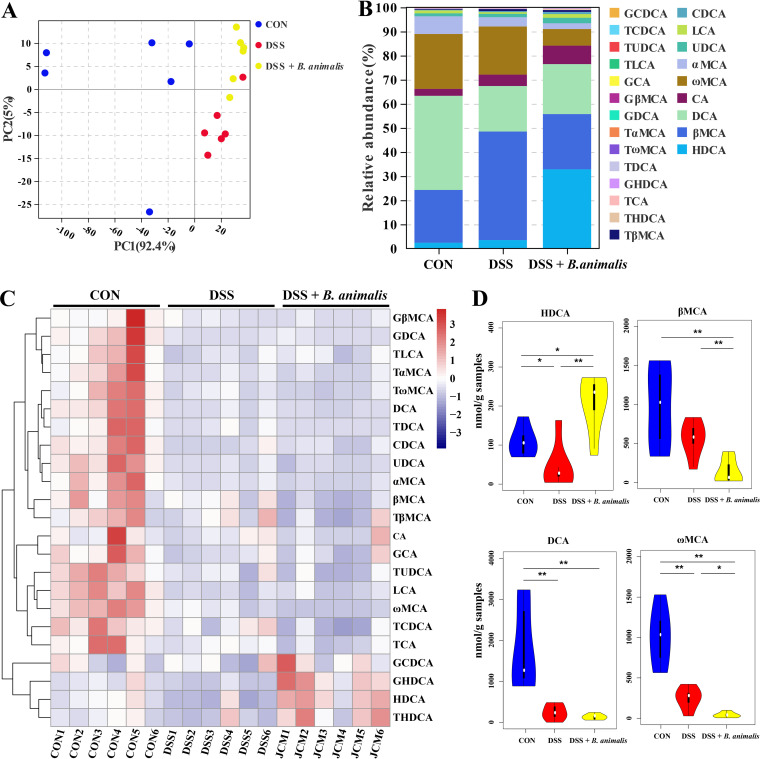
Targeted BA metabolomics was further employed to examine the effect of ALG on intestinal BAs. Clear separation in the BA profile among the three groups of mice was observed (Fig. 4A). Of 40 identified BAs, HDCA, DCA, β-muricholic acid (βMCA), murocholic acid (muroCA), and α-muricholic acid (αMCA) were the five most abundant BAs in the mouse colon (Fig. 4B). Although a majority of BAs were suppressed by DSS (Fig. 4C), a few BAs, such as DCA and muroCA, were largely restored in response to ALG (Fig. 4D). Notably, HDCA was significantly reduced by DSS but enriched drastically by ALG to become the most abundant BA in the mouse colon (Fig. 4D). Spearman’s correlation analysis further revealed strong positive and negative correlations between many of the significantly altered colonic bacteria and BAs (Fig. 4E). For example, the HDCA concentration was positively correlated with B. animalis. Collectively, these results suggested that the anti-inflammatory effect of ALG in DSS-induced colitis may be due to the alteration of intestinal microbial BA metabolism.
FIG 4.
Dietary supplementation with ALG alters the intestinal BA profile in DSS-treated mice. Mice supplemented with or without 5% ALG for 4 weeks, followed by induction of colitis with 3% DSS in drinking water for another week (n = 8). The colonic digesta was collected from each mouse at the end of the experiment and subjected to targeted BA profiling. (A) PCA plot of the BA profile among three groups of mice. (B) Relative abundances (%) of different BAs among three groups. (C) Z-score plot of all BAs identified among three groups. (D) Alterations of representative BAs among three groups. (E) Spearman correlation between the relative abundance of the intestinal microbiota and BA concentrations. Data are presented as means ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Alginate-mediated alleviation of intestinal inflammation is dependent upon the microbiota.
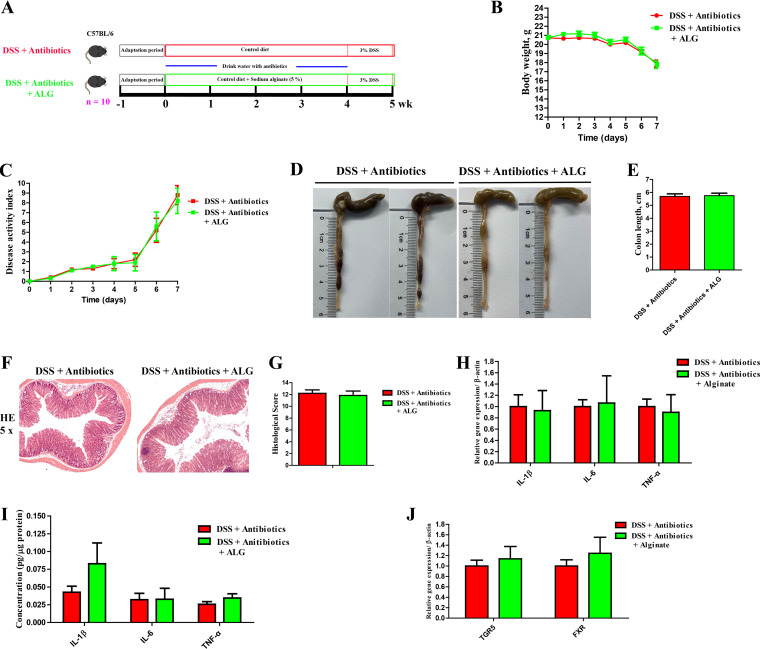
Because of drastic changes in the intestinal microbiota in DSS-treated mice in response to ALG, we hypothesized that the anti-inflammatory effect of ALG is mediated through the intestinal microbiota. To test this hypothesis, we supplemented ALG to mice with or without depletion of the intestinal microbiota with a cocktail of antibiotics followed by DSS treatment (Fig. 5A). ALG lost its protective effect on weight loss (Fig. 5B), DAI score (Fig. 5C), colonic length (Fig. 5D and E), and colonic integrity (Fig. 5F and G) in the absence of the intestinal microbiota. Consistently, similar mRNA and protein levels of proinflammatory cytokines (IL-1β, IL-6, and TNF-α) in the colon were observed in microbiota-depleted DSS-treated mice with or without ALG supplementation (Fig. 5H and I). The mRNA expression levels of BA receptors (TGR5 and FXR) were also similar between the two groups of mice (Fig. 5J). Collectively, these results suggested that the intestinal microbiota is responsible for mediating the anti-inflammatory effect of ALG in DSS-induced colitis.
FIG 5.
Microbiota depletion abrogates the anti-inflammatory effect of ALG in DSS-treated mice. (A) Experimental scheme to show depletion of the intestinal microbiota by a cocktail of antibiotics and timing of ALG and DSS treatment (n = 10). (B and C) Body weight (B) and disease activity index (DAI) score (C) were recorded daily in the final week of DSS treatment for both groups of mice. The colon was collected from each mouse at the end of the experiment. (D to F) Representative images of the colon (D), the colon length (E), H&E staining (magnification, ×50 ) of the colon (F), and colonic histology score (G) are shown. (H and I) The mRNA (H) and protein (I) expression levels of three inflammatory cytokines in the colon were also measured. (J) The mRNA gene expression levels of two BA receptors in the colon.
B. animalis ameliorates DSS-induced intestinal inflammation in parallel with altered BA metabolism.
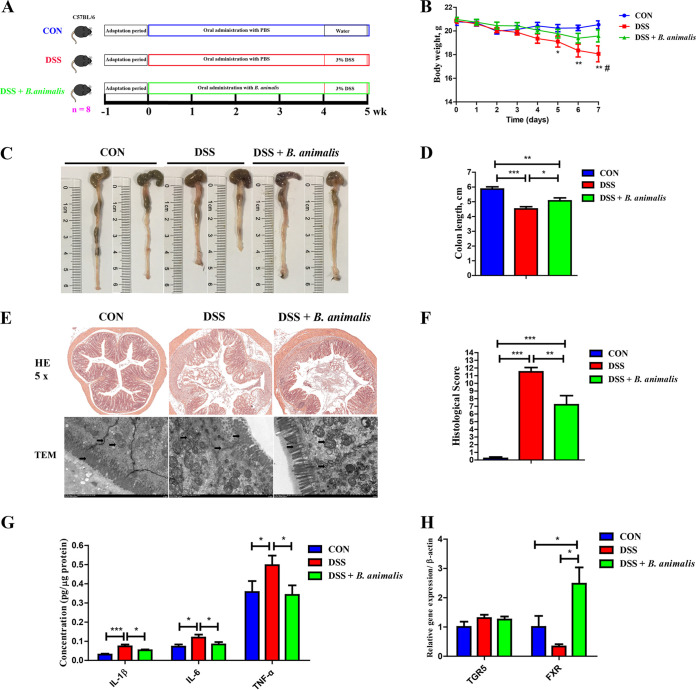
Since B. animalis was the most abundant bacterium that was differentially enriched by ALG in DSS-treated mice, B. animalis subsp. animalis JCM 1190, which is isolated from mouse feces, was chosen to directly study its anti-inflammatory effect in DSS-treated mice (Fig. 6A). Oral gavage with B. animalis significantly ameliorated DSS-induced colitis as evidenced by markedly reducing weight loss (Fig. 6B), colonic shortening (Fig. 6C and D), and epithelial destruction (Fig. 6E and F). Consistently, proinflammatory cytokines (IL-1β, IL-6, and TNF-α) were largely restored to healthy levels in the colon of DSS-treated mice in response to ALG (Fig. 6G). Furthermore, the mRNA expression of FXR in the colon was significantly induced by B. animalis (P < 0.05) (Fig. 6H). B. animalis also obviously altered the BA profile in DSS-treated mice (Fig. 7A to 7C). Among the most abundant BAs, HDCA was significantly enhanced by B. animalis, while βMCA and ωMCA were significantly reduced (Fig. 7D). Collectively, these results demonstrated that similar to ALG, B. animalis is capable of alleviating DSS-induced intestinal inflammation.
FIG 6.
Oral gavage of B. animalis alleviates DSS-induced acute colitis in mice. (A) Experimental scheme for DSS-induced colitis and B. animalis treatment (n = 8). (B) Body weight was recorded daily in the final week of DSS treatment for three groups of mice. The colon was collected from each mouse at the end of the experiment. (C to F) Representative images of the colon (C), the colon length (D), and H&E staining (magnification, ×50) and TEM image of the colon (E) and colonic histology score (F) are shown. (G and H) The mRNA gene expression levels of three inflammatory cytokines (G) and two BA receptors (H) in the colon were also measured by RT-qPCR. Data are presented as means ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001; #, P < 0.05; compared with the DSS plus B. animalis group.
FIG 7.
Oral gavage of B. animalis alters the intestinal BA profile in DSS-treated mice. Mice supplemented with or without B. animalis for 4 weeks, followed by induction of colitis with 3% DSS in drinking water for another week (n = 8). The colonic digesta was collected from each mouse at the end of the experiment and subjected to targeted BA profiling. (A) PCA plot of the BA profile among three groups of mice. (B) Relative abundances (%) of different BAs among three groups. (C) Z-score plot of all BAs identified among three groups. (D) Alterations of representative BAs among three groups. Data are presented as the mean ± SEM. *, P < 0.05; **, P < 0.01.
HDCA exerts an anti-inflammatory effect in the DSS-induced colitis mouse model.
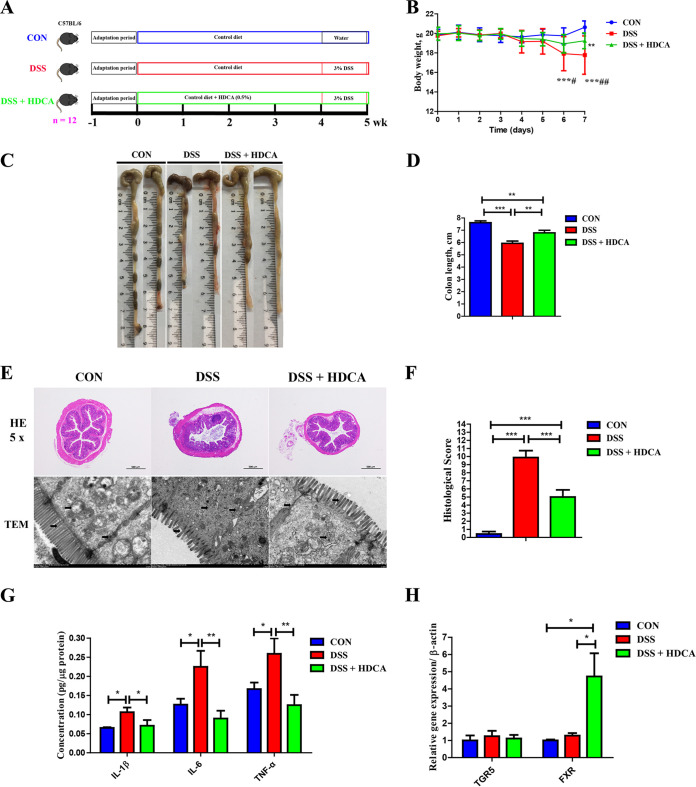
Because HDCA was drastically increased by ALG and B. animalis in DSS-treated mice, we further investigated its role in DSS-induced colitis (Fig. 8A). Dietary supplementation with HDCA significantly improved DSS-induced weight loss (Fig. 8B), colonic shortening (Fig. 8C and D), and mucosal epithelial destruction (Fig. 8E and F). Proinflammatory cytokines (IL-1β, IL-6, and TNF-α) in the colon of DSS-treated mice were restored in response to HDCA (Fig. 8G). Similar to ALG and B. animalis, HDCA also significantly increased the mRNA expression of FXR (P < 0.05) (Fig. 8H). These results suggest that HDCA might be involved in the ALG-mediated anti-inflammatory effect in DSS-induced colitis.
FIG 8.
Dietary supplementation with HDCA ameliorates DSS-induced acute colitis. (A) Experimental scheme for DSS-induced colitis and HDCA treatment (n = 12). (B) Body weight was recorded daily in the final week of DSS treatment for three groups of mice. The colon was collected from each mouse at the end of the experiment. (C to F) Representative images of the colon (C), the colon length (D), and H&E staining (magnification, ×50), TEM image of the colon (E), and colonic histology score (F) are shown. (G and H) The mRNA gene expression levels of three inflammatory cytokines (G) and two BA receptors (H) in the colon were also measured by RT-qPCR. Data are presented as the mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001; #, P < 0.05; ##, P < 0.01; compared with the DSS plus HDCA group.
DISCUSSION
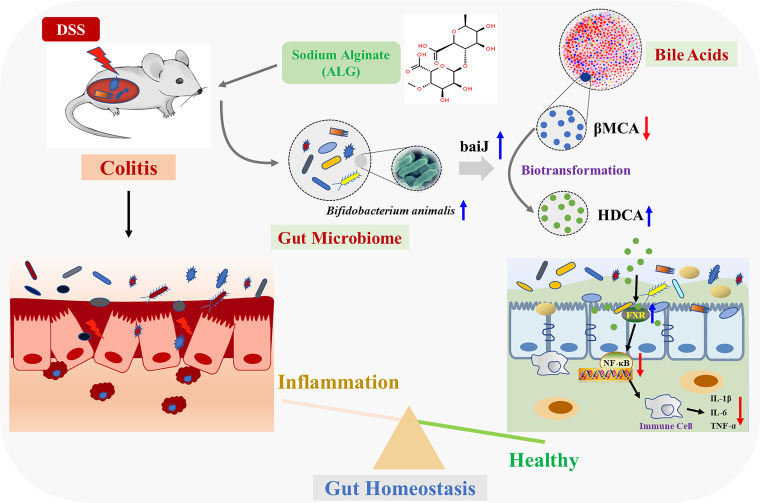
IBD is a chronic inflammatory disorder of the intestinal tract. Although the etiology of IBD remains elusive, the prevailing theory is that IBD results from an exaggerated immune response triggered by dysbiosis and altered microbial BA metabolism in a genetically prone host (12, 25, 26). However, the role of the intestinal microbiota in IBD pathogenesis is unclear. ALG is a soluble dietary fiber known to alter intestinal microbial composition and metabolism (27–29). ALG has been shown to ameliorate experimental IBD in mice (16–18), but little is known about the mechanism of its anti-inflammatory action. In the current study, using a DSS-induced colitis mouse model for IBD, we found that ALG ameliorates IBD-associated intestinal disorder by selectively enriching B. animalis and HDCA (Fig. 9).
FIG 9.
Schematic model showing the potential mechanism of feeding sodium alginate (ALG) that alleviated DSS-induced colitis. Dietary ALG induced an alteration in the gut microbiota to enrich probiotic bacteria such as Bifidobacterium animalis, which subsequently led to an increased production of hyodeoxycholic acid (HDCA), which is biotransformed from β-muricholic acid (βMCA) by BA 7-dehydroxylase (baiJ), triggering a cascade of anti-inflammatory responses by activating bile acid receptor, farnesoid X receptor (FXR), and the NF-κB signaling pathway. Ultimately, intestinal epithelial homeostasis is attenuated and colitis was attenuated.
Of note, microbiota depletion by antibiotics blocks the anti-inflammatory effect of ALG, implying a critical involvement of the intestinal microbiota. In addition, oral administration of B. animalis exerts a similar anti-inflammatory effect in DSS-treated mice. Furthermore, HDCA synthesis is also enhanced by B. animalis in DSS mice. These results, to the best of our knowledge, have demonstrated for the first time that ALG alleviates intestinal inflammation by modulating intestinal microbiota and microbial BA metabolism. The outcome provides the rationale for using ALG or its enriched bacteria (e.g., B. animalis) or bacterial metabolites (e.g., HDCA) for IBD therapy.
In this study, dietary supplementation of low-molecular-weight ALG led to an impressive, greater than a 3-fold expansion of B. animalis, which is recognized as a probiotic, making it the most abundant bacterium in the colon (21.5% ± 3.8%) in DSS-treated mice. Consistently, alginate or alginate oligosaccharides have been reported to stimulate the growth of Bifidobacterium both in vitro and in vivo (30, 31). In fact, β-D-mannuronic acid, a component of ALG, is also capable of enriching the intestinal Bifidobacterium in mice (32), suggesting that ALG is a preferred carbohydrate source for B. animalis, which is consistent with the fact that the genes involved in carbohydrate utilization account for nearly 13% of the genes present in the genome of Bifidobacterium (33). In addition, ALG enriches other polysaccharide-utilizing bacteria, including Lachnospiraceae NK4A136 group, Faecalibaculum rodentium, and Clostridia UCG-014. In agreement, metagenomic analysis has confirmed the enrichment of ABC transporters and CAZymes such as GH94, CBM50, and GT5, indicative of ALG-induced alterations of the microbial function. Moreover, it is also intriguing that in the current study, Akkermansia muciniphila a recognized probiotic, showed a significant increase in the DSS group, which is consistent with previous studies (34, 35). A. muciniphila exhibits a modulatory effect on the mucosal immune response in experiments involving both mice and humans and has the potential to be used as a next-generation beneficial microbe (36, 37). A. muciniphila, a symbiotic bacterium of the mucus layer, can utilize mucin as its sole carbon, nitrogen, and energy source (37). Thus, we speculate that the increased A. muciniphila in the colon of mice with colitis mice may cause, by the large number of intestinal mucosae, shedding induced by inflammation. Whether A. muciniphila could exert an adverse effect during DSS-induced colitis still needs further investigation.
It is noteworthy that we observed significant changes in BA metabolism in response to dietary supplementation of ALG. Although a majority of BAs are suppressed by DSS, HDCA is markedly enhanced by ALG. HDCA is believed to be converted from βMCA, a primary BA, by 7α-dehydroxylase (baiJ), which is present in a variety of intestinal bacteria, such as Lactobacillus, Lachnospiraceae, Ruminococcaceae, Clostridiaceae, Eubacterium, and Peptostreptococcus (9–11, 38, 39). We found a reciprocal change of HDCA and βMCA in response to ALG, with the former being significantly increased, while the latter remained suppressed in the colonic digesta of DSS-treated mice. This is perhaps not surprising given the significant enrichment of Lachnospiraceae NK4A136 group and Clostridia UCG-014 as well as those bacteria encoding the baiJ gene in response to ALG. A large amount of HDCA is likely converted from βMCA as a result of an increase in baiJ in ALG-supplemented mice. Consistent with our study, a strong correlation was observed earlier between HDCA and a member of Clostridium genus (40).
We further demonstrated in this study that ALG supplementation enhances intestinal HDCA synthesis, which is positively correlated with the relative abundance of B. animalis. Direct administration of B. animalis also similarly promotes the intestinal HDCA concentration, suggesting that B. animalis may be a major player in the biosynthesis of HDCA. Collectively, these findings indicated that ALG alleviates intestinal inflammation by modulating the structure and function of the intestinal microbiota. B. animalis and HDCA are perhaps major mediators of the anti-inflammatory activity of ALG. In fact, B. animalis, Faecalibaculum rodentium, and Clostridia_UCG-014 as well as several secondary BAs are known to be anti-inflammatory (41–43). In addition to BAs, short-chain fatty acids are possibly regulated by ALG and may contribute to the beneficial effect of ALG in intestinal inflammation, which warrants further investigation.
HDCA was shown to prevent gallstone formation (44), inhibit atherosclerosis formation (45), and improve glucose homeostasis (46). We found that HDCA significantly downregulates the expression of proinflammatory cytokines (IL-1β, IL-6, and TNF-α) in the colon of DSS-treated mice. Consistent with our findings, HDCA shows an anti-inflammatory effect in neurovascular and microglial cells (47, 48). In addition, dietary supplementation with 2 g/kg of HDCA improves humoral immunity by elevating serum IgA and IgG concentrations in weaned piglets (49). In the current study, HDCA appears to be a major factor in mediating the anti-inflammatory effect of ALG in the intestinal tract.
BAs function through interactions with host BA receptors such as FXR and TGR5 (50). In general, unconjugated BAs are high-affinity ligands of FXR (23). Activation of FXR strongly induces anti-inflammatory genes (51), suppresses NF-κB activation (52), and reduces inflammation in the intestine (53). Conversely, FXR deficiency exacerbates intestinal inflammation in DSS-induced colitis (53). We found in this study that FXR, but not TGR5, is induced by supplementation of ALG, B. animalis, and HDCA, suggesting that FXR might be a major receptor that mediates their anti-inflammatory action. Consistently, HDCA suppresses intestinal epithelial cell proliferation via FXR, but not TGR5, in porcine IPEC-J2 cells (54). In addition, dietary supplementation of HDCA significantly enhances intestinal FXR gene expression in weaned piglets (54).
In conclusion, ALG effectively suppresses intestinal inflammation in a DSS-induced colitis model in an intestinal microbiota-dependent manner. ALG preferentially enriches a small set of the bacteria, such as B. animalis, which subsequently facilitates the biotransformation of primary BAs to secondary BAs such as HDCA, resulting in NF-κB inhibition and suppression of intestinal inflammation. Our findings contribute to a better understanding of the mechanism of action of ALG and provide new approaches to IBD therapy.
MATERIALS AND METHODS
Characterization of sodium alginate.
Sodium alginate (>98%) was obtained from Qingdao Bright Moon Seaweed Group Co., Ltd. (Qingdao, China). The average molecular weight of ALG was determined using gel permeation chromatography (GPC). Briefly, samples were dissolved in NaNO3 at 2 mg/mL, and 200 μL was injected in an Agilent 1100 high-pressure liquid chromatography (HPLC) system equipped with organic and aqueous GPC with a flow rate of 1 mL/min and a column thermostat temperature of 35°C. The eluent was monitored by a refractive index detector with an optical unit temperature of 35°C and peak width of 0.1 min. The ratio of α-l-guluronic (G) to β-d-mannuronic acid (M) in ALG was analyzed using infrared spectroscopy with a Mattson 1000 spectrophotometer (Unicam, Cambridge, England). Measurements were recorded of between 400 and 4,000 cm−1. ALG used in the present study has a number average molecular weight (Mn) and weight average molecular weight (Mw) of 8.98 kDa and 21.51 kDa, respectively, and the G/M ratio is 0.69 (see Fig. S1 in the supplemental material).
Mouse model of DSS-induced colitis.
Female C57BL/6 mice (7 weeks old) were purchased from SPF Biotechnology Co., Ltd. (Beijing, China), and housed in an environmentally controlled room (temperature, 25 ± 2°C; relative humidity, 45 to 60%; lighting cycle, 12 h light [L]:12 h dark [D]) with ad libitum access to food and water throughout the studies. All experimental procedures were approved by the Institutional Animal Care and Use Committee of China Agricultural University, Beijing, China.
Acute colitis was induced by the addition of 3% dextran sulfate sodium (DSS, 36 to 50 kDa; MP Biomedicals, Irvine, CA, USA) to drinking water for 7 days. Body weight was measured daily. On day 8, mice were sacrificed, and the entire colon was removed (from the cecum to the anus), and its length was measured. A 2-cm segment of the descending colon was fixed in 4% formalin and processed for hematoxylin and eosin (H&E) staining. Histological scoring was blindly performed by an experienced pathologist as previously described (55). The degree of epithelial loss on the intestinal surface, crypt destruction, and inflammatory cell infiltration were assessed and included in the histopathological examination (Table S1). The severity of colitis was evaluated using a disease activity index (DAI) as described previously (Table S2) (55). The remaining colon samples were stored at −80°C for gene and protein expression analyses. Colonic digesta was also collected and frozen at −80°C for microbial DNA extraction and BA profiling.
Dietary supplementation of sodium alginate.
After 1 week of acclimation, mice were randomly divided into three groups: a control group (CON), a DSS-treated group (DSS), and a DSS-treated group supplemented with ALG (DSS plus ALG) (n = 8). Mice in the CON group were fed the standard diet alone for 5 weeks, while those in the DSS group were fed the standard diet for 5 weeks and colitis was induced with 3% DSS in drinking water in the last week of the experiment. Mice in the DSS plus ALG group were supplemented with 5% ALG for 4 weeks, followed by induction of colitis with 3% DSS in drinking water for another week. The dosage of ALG in the diet was based on previous studies (56, 57). At the end of the experiment, all mice were sacrificed, and postmortem procedures were performed as described above.
Antibiotic depletion of the intestinal microbiota in mice.
After 1 week of acclimation, mice were provided with a cocktail of antibiotics (1 g/L streptomycin, 0.5 g/L ampicillin, 1 g/L gentamicin, and 0.5 g/L vancomycin) in the drinking water to deplete the intestinal microbiota as previously described (58). All antibiotics were purchased from Meilun Bio (Dalian, China), and drinking water was replenished every other day. Mice were fed the standard diet with or without 5% ALG for 5 weeks, followed by colitis induction in week 5. Antibiotics were provided throughout the duration of the experiment. The postmortem procedures were performed as described above.
Dietary supplementation of Bifidobacterium animalis.
B. animalis subsp. animalis JCM 1190 was purchased from BeNa Culture Collection (BNCC 186305, Beijing, China) and cultured in MRS medium. After 1 week of acclimation, mice were orally gavaged with 0.2 mL phosphate-buffered saline (PBS) supplemented with or without 1 × 109 CFU B. animalis subsp. animalis JCM 1190 every other day for 5 weeks. The dosage of B. animalis subsp. animalis JCM 1190 was based on a previous study (59). DSS was added to the drinking water in week 5 to induce colitis. At the end of the trial, all animals were sacrificed, and postmortem procedures were performed as described above.
Dietary supplementation of hyodeoxycholic acid.
HDCA (>98%) was procured from Shanghai Yuanye Bio-Technology Co., Ltd. (Shanghai, China). After 1 week of acclimation, mice were randomly divided into three groups (n = 12) fed with a standard diet supplemented with or without 0.5% hyodeoxycholic acid for 5 weeks. Colitis was induced with 3% DSS in drinking water in week 5, except for those mice in the control group. At the end of the trial, all animals were sacrificed, and postmortem procedures were performed as described above.
Transmission electron microscopy (TEM) analysis.
A 5-mm segment of fresh colonic tissues was flushed with PBS, fixed in 2.5% glutaraldehyde at 4°C for 4 h, and fixed in 1% osmium tetroxide for 2 h at room temperature. Tissues were embedded in pure EMBed 812 (SPI Supplies, West Chester, PA, USA) at 37°C overnight, processed, and stained sequentially in 2% uranium acetate saturated alcohol solution to avoid light staining for 8 min, rinsed in 70% ethanol 3 times, and then rinsed in ultrapure water 3 times; 2.6% lead citrate was used to avoid CO2 staining for 8 min and then rinsed with ultrapure water 3 times. After being dried with the filer paper, the cuprum grids were put into the grid board and dried overnight at room temperature. Images were acquired using a transmission HT7700 electron microscope (Hitachi, Tokyo, Japan).
RNA extraction and Real-Time Quantitative PCR (RT-qPCR).
Total RNA was extracted from a segment of the colon using TRIzol (TaKaRa Bio, Otsu, Shiga, Japan), and 1 μg RNA was reverse-transcribed using a PrimeScript RT reagent kit with gDNA Eraser (TaKaRa Bio Inc, Dalian, China). Quantitative PCR (qPCR) was performed with gene-specific primers (Table S3) on an ABI 7300 real-time PCR system (Applied Biosystems, Foster, CA, USA) with SYBR green master mix (TaKaRa Bio) as previously described (60). Relative changes in mRNA expression were analyzed using the ΔΔCT method as previously described (60).
Enzyme-linked immunosorbent assay (ELISA).
Frozen colon samples were ground into powder in liquid nitrogen and lysed in RIPA buffer (150 mM NaCl, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris-HCl, pH 7.4) containing a cocktail of protease inhibitors (Thermo Fisher Scientific, Rockford, IL, USA). After centrifugation at 13,000 × g for 10 min at 4°C, the protein concentration of each supernatant was measured using a bicinchoninic acid (BCA) protein assay kit (Beyotime Biotechnology, Beijing, China). The concentrations of IL-1β, TNF-α, and IL-6 in the supernatant were measured the with respective ELISA kits (Thermo Fisher Scientific) according to the manufacturer’s instructions.
Western blot analysis.
An equal amount of protein (30 μg) from each colonic sample was electrophoresed on SDS polyacrylamide gels with prestained protein markers. Proteins were then transferred to polyvinylidene difluoride (PVDF) membranes (Millipore, Billerica, MA, USA) and blocked in 5% fat-free powdered milk at room temperature in Tris-Tween-buffered saline (TTBS: 20 mM Tris/150 mM NaCl, pH 7.5, and 0.1% Tween 20) for 1 h. The membranes were incubated separately with the following primary antibodies at 4°C overnight with gentle rocking (all from Cell Signaling Technology, Danvers, MA, USA): FXR (catalog [cat.] no. 72105, 1:1,000), p65 (cat. no. 6956, 1:1,000), p-p65 (cat. no. 3033, 1:1,000), p38 (cat. no. 8690, 1:1,000), p-p38 (cat. no. 4511, 1:1,000), ERK1/2 (cat. no. 4695, 1:1,000), p-ERK1/2 (cat. no. 4370, 1:2,000), JNK1/2 (cat. no. 9252, 1:1,000), and p-JNK1/2 (cat. no. 4668, 1:1,000). Membranes were then washed and incubated with a horseradish peroxidase-conjugated secondary antibody (goat anti-rabbit IgG; Thermo Fisher Scientific) at a 1:5,000 dilution for 1 h at room temperature. Signals were developed with SuperSignal West Dura extended duration substrate (Pierce, Rockford, IL, USA) and quantified on an Alpha Imager 2200 (Alpha Innotech Corporation, California, USA). The signal intensities were normalized to β-actin.
Microbial DNA extraction, 16S rRNA gene sequencing, and data analysis.
Total microbial genomic DNA in the colonic digesta was extracted using a QIAamp fast DNA stool minikit (Qiagen, Germany). The V3 to V4 regions of the 16S rRNA gene were amplified with primers 341F (5′-ACTCCTACGGGAGGCAGCAG-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′) as previously described (61). PCR products were purified, quantified, and pooled at equal amounts for PE300 sequencing on an Illumina MiSeq instrument (Illumina, San Diego, CA, USA) according to the standard protocols of Majorbio Bio-Pharm Technology Co., Ltd. (Shanghai, China). Raw sequences were analyzed using QIIME 2 (version 2020.2) (62) on the Majorbio I-Sanger cloud platform (https://cloud.majorbio.com/). Quality control and denoising were performed using DADA2 under default settings to generate ASVs (63). Only those ASVs with a minimum of two reads and present in more than one sample were retained. A phylogenetic tree was generated using the SEPP algorithm against the Silva 138 database under default settings (64). The α- and β-diversities were calculated using vegan (version 3.3.1). PCoA was performed using Bray-Curtis distance metrics. Differential taxa were identified using the LEfSe (linear discriminant analysis effect size) algorithm and further confirmed using BLAST against the NCBI 16S rRNA database (63). The functional potential of the intestinal microbiota was predicted using PICRUSt2 (65). The raw reads were deposited in the NCBI Sequence Read Archive (SRA) database (accession number SRP303220). In addition, the total bacterial and functional microbial genes, including baiJ, bsh, 7α-HSDH, and 7β-HSDH, involved in the conversion of primary to secondary BAs were quantified using qPCR and bacterial group-specific primers (Table S3) on an ABI 7300 real-time PCR system (Applied Biosystems, Foster City, CA, USA) with SYBR green master mix (TaKaRa Bio) (60).
DNA extraction, library preparation, and metagenomic sequencing.
Colonic digesta samples were also subjected to shotgun metagenomic sequencing. Briefly, microbial genomic DNA was extracted using the E.Z.N.A soil DNA kit (Omega Bio-Tek, Norcross, GA, USA). DNA concentration was measured with a TBS-380 fluorometer (Promega, Madison, WA, USA). The quality of extracted DNA was checked on 1% agarose gel. DNA was then fragmented to approximately 400 bp using a Covaris M220 ultrasonicator (Gene Company, China). Adapter ligation, cleanup, and enrichment were performed using a NEXT-FLEX rapid DNA sequencing (DNA-seq) kit (Bioo Scientific, Austin, TX, USA). The library was analyzed for size distribution, quantified using qPCR, and sequenced PE150 on an Illumina NovaSeq 6000 device at a depth of greater than 10 Gb raw reads/sample.
Metagenome assembly and functional annotation.
Shotgun sequencing reads were analyzed on the online platform of Majorbio Cloud platform (www.majorbio.com). Quality control was performed using fastp (version 0.20.0) to trim adapters and filter low-quality reads with the parameter –cut_by_quality3 -W 4 -M 20 -n 5 -c -l 50 -w 3 (63). Filtered high-quality reads were assembled using MEGAHIT (version 1.1.2) using the option –min-count 2 –min-contig-len 300 (63). Gene prediction from the assembled contig was predicted using prodigal (version 2.6.3) with -meta mode (63). A total of 5,538,849 genes were identified. A custom script was used to remove incomplete genes, and 1,507,615 complete genes were retained. A nonredundant gene catalog was constructed using CD-HIT (version 4.6.1) with 90% sequence identity and 90% coverage at the protein level (63). High-quality reads were mapped back to the constructed nonredundant gene catalog with 95% identity using SOAPaligner to calculate gene abundance within each sample (66). The clustered amino acid sequences in the gene catalog were then aligned to the GENES database using Diamond (version 0.8.35) to annotate the Kyoto Encyclopedia of Genes and Genomes (KEGG) gene profile (67). The protein sequences were also aligned to the CAZy database using HMMER (version 3.1b2) to annotate the carbohydrate-active enzyme (CAZyme) profile (68). Genes were assigned to the microbial taxa with the highest scores, and the total read count of all genes assigned to a specific taxon was used to estimate its abundance. Normalized RPKM (reads per kilobase of a gene per million mapped reads) values of taxa or KEGG terms were used for statistical analyses.
Quantitative profiling of BAs.
BAs in colonic digesta were measured as previously described (69). A Waters ACQUITY ultraperformance LC system coupled with a Waters XEVO TQ-S mass spectrometer with an electrospray ionization (ESI) source controlled by MassLynx 4.1 software (Waters, Milford, MA, USA) was used for all analyses. Chromatographic separations were performed with an ACQUITY BEH C18 column (1.7 μm, 100 mm by 2.1 mm internal dimensions) (Waters). Ultraperformance liquid chromatography mass spectroscopy (UPLC-MS) raw data obtained with the negative mode were analyzed using TargetLynx applications manager (version 4.1; Waters) to obtain calibration equations and the concentration of each BA in the samples.
Statistical analysis.
All statistical significance was assessed using a one-way analysis of variance (ANOVA) (SPSS 20 software, IBM, Armonk, NY, USA) except for the intestinal microbiome data. For intestinal microbiome data analysis, α-diversity was analyzed using the Kruskal-Wallis test. PCoA was analyzed using ANOSIM. The relative abundances of the intestinal microbiota and functional genes were analyzed using the Kruskal-Wallis test with false-discovery rate (FDR) adjustment. All results were expressed as the mean ± the standard error of the mean (SEM). Differences were considered statistically significant if P was <0.05.
Data availability.
The bacterial 16S rRNA sequencing data reported in this paper were deposited in the NCBI Sequence Read Archive (SRA) database under accession number PRJNA694703.
ACKNOWLEDGMENTS
We sincerely thank all members for assisting in conducting the experiments, technical support, and data analysis.
Yu Pi and Junjun Wang: designed and supervised the study; Yu Pi, Yujun Wu, Xiangyu Zhang, Zhenyu Wang, and Xiaoyi Liu: performed the experiments and analyzed the data; Yu Pi: wrote the manuscript; Yu Pi, Dandan Han, Jinbiao Zhao, Isabel Tobin, Jiangchao Zhao, Guolong Zhang, and Junjun Wang: contributed to the manuscript revision and discussion.
This research was financially supported by the National Natural Science Foundation of China (32002189, 32125036, 31972596, and 31902170).
We declare no competing interests.
Footnotes
Supplemental material is available online only.
Contributor Information
Junjun Wang, Email: wangjj@cau.edu.cn.
Yunhe Fu, Jilin University.
REFERENCES
- 1.Molodecky NA, Soon IS, Rabi DM, Ghali WA, Ferris M, Chernoff G, Benchimol EI, Panaccione R, Ghosh S, Barkema HW, Kaplan GG. 2012. Increasing incidence and prevalence of the inflammatory bowel diseases with time, based on systematic review. Gastroenterology 142:46–54.e42. doi: 10.1053/j.gastro.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 2.Schirmer M, Franzosa EA, Lloyd-Price J, McIver LJ, Schwager R, Poon TW, Ananthakrishnan AN, Andrews E, Barron G, Lake K, Prasad M, Sauk J, Stevens B, Wilson RG, Braun J, Denson LA, Kugathasan S, McGovern DPB, Vlamakis H, Xavier RJ, Huttenhower C. 2018. Dynamics of metatranscription in the inflammatory bowel disease gut microbiome. Nat Microbiol 3:337–346. doi: 10.1038/s41564-017-0089-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Franzosa EA, Sirota-Madi A, Avila-Pacheco J, Fornelos N, Haiser HJ, Reinker S, Vatanen T, Hall AB, Mallick H, McIver LJ, Sauk JS, Wilson RG, Stevens BW, Scott JM, Pierce K, Deik AA, Bullock K, Imhann F, Porter JA, Zhernakova A, Fu J, Weersma RK, Wijmenga C, Clish CB, Vlamakis H, Huttenhower C, Xavier RJ. 2019. Gut microbiome structure and metabolic activity in inflammatory bowel disease. Nat Microbiol 4:293–305. doi: 10.1038/s41564-018-0306-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Colman RJ, Rubin DT. 2014. Fecal microbiota transplantation as therapy for inflammatory bowel disease: a systematic review and meta-analysis. J Crohns Colitis 8:1569–1581. doi: 10.1016/j.crohns.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu WH, Winter MG, Byndloss MX, Spiga L, Duerkop BA, Hughes ER, Buttner L, Romao ED, Behrendt CL, Lopez CA, Sifuentes-Dominguez L, Huff-Hardy K, Wilson RP, Gillis CC, Tükel C, Koh AY, Burstein E, Hooper LV, Bäumler AJ, Winter SE. 2018. Precision editing of the gut microbiota ameliorates colitis. Nature 553:208–211. doi: 10.1038/nature25172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kostic AD, Xavier RJ, Gevers D. 2014. The microbiome in inflammatory bowel disease: current status and the future ahead. Gastroenterology 146:1489–1499. doi: 10.1053/j.gastro.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ridlon JM, Harris SC, Bhowmik S, Kang DJ, Hylemon PB. 2016. Consequences of bile salt biotransformations by intestinal bacteria. Gut Microbes 7:22–39. doi: 10.1080/19490976.2015.1127483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wahlström A, Sayin SI, Marschall HU, Bäckhed F. 2016. Intestinal crosstalk between bile acids and microbiota and its impact on host metabolism. Cell Metab 24:41–50. doi: 10.1016/j.cmet.2016.05.005. [DOI] [PubMed] [Google Scholar]
- 9.He YY, Mao CX, Wen H, Chen ZY, Lai T, Li LY, Lu W, Wu HD. 2017. Influence of ad libitum feeding of piglets with Bacillus subtilis fermented liquid feed on gut flora, luminal contents and health. Sci Rep 7:44553. doi: 10.1038/srep44553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Theriot CM, Bowman AA, Young VB. 2016. Antibiotic-induced alterations of the gut microbiota alter secondary bile acid production and allow for Clostridium difficile spore germination and outgrowth in the large intestine. mSphere 1:e00045-15. doi: 10.1128/mSphere.00045-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang YC, Limaye PB, Renaud HJ, Klaassen CD. 2014. Effect of various antibiotics on modulation of intestinal microbiota and bile acid profile in mice. Toxicol Appl Pharmacol 277:138–45145. doi: 10.1016/j.taap.2014.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duboc H, Rajca S, Rainteau D, Benarous D, Maubert MA, Quervain E, Thomas G, Barbu V, Humbert L, Despras G, Bridonneau C, Dumetz F, Grill JP, Masliah J, Beaugerie L, Cosnes J, Chazouillères O, Poupon R, Wolf C, Mallet JM, Langella P, Trugnan G, Sokol H, Seksik P. 2013. Connecting dysbiosis, bile-acid dysmetabolism and gut inflammation in inflammatory bowel diseases. Gut 62:531–539. doi: 10.1136/gutjnl-2012-302578. [DOI] [PubMed] [Google Scholar]
- 13.Staley C, Weingarden AR, Khoruts A, Sadowsky MJ. 2017. Interaction of gut microbiota with bile acid metabolism and its influence on disease states. Appl Microbiol Biotechnol 101:47–64. doi: 10.1007/s00253-016-8006-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poynard T, Vernisse B, Agostini H. 1998. Randomized, multicentre comparison of sodium alginate and cisapride in the symptomatic treatment of uncomplicated gastro-oesophageal reflux. Aliment Pharm Ther 12:159–165. doi: 10.1046/j.1365-2036.1998.00283.x. [DOI] [PubMed] [Google Scholar]
- 15.Dettmar PW, Sykes J, Little SL, Bryan J. 2006. Rapid onset of effect of sodium alginate on gastro-oesophageal reflux compared with ranitidine and omeprazole, and relationship between symptoms and reflux episodes. Int J Clin Pract 60:275–283. doi: 10.1111/j.1368-5031.2006.00800.x. [DOI] [PubMed] [Google Scholar]
- 16.Yamamoto A, Itoh T, Nasu R, Nishida R. 2013. Effect of sodium alginate on dextran sulfate sodium- and 2,4,6-trinitrobenzene sulfonic acid-induced experimental colitis in mice. Pharmacology 92:108–116. doi: 10.1159/000353192. [DOI] [PubMed] [Google Scholar]
- 17.Mirshafiey A, Khodadadi A, Rehm BH, Khorramizadeh MR, Eslami MB, Razavi A, Saadat F. 2005. Sodium alginate as a novel therapeutic option in experimental colitis. Scand J Immunol 61:316–321. doi: 10.1111/j.1365-3083.2005.01571.x. [DOI] [PubMed] [Google Scholar]
- 18.Razavi A, Khodadadi A, Eslami MB, Eshraghi S, Mirshafiey A. 2008. Therapeutic effect of sodium alginate in experimental chronic ulcerative colitis. Iran J Allergy Asthm 7:13–18. [PubMed] [Google Scholar]
- 19.Humphreys ED, Triffitt J. 1968. Absorption by the rat of alginate labelled with carbon-14. Nature 219:1172–1173. doi: 10.1038/2191172a0. [DOI] [PubMed] [Google Scholar]
- 20.Seal CJ, Mathers JC. 2001. Comparative gastrointestinal and plasma cholesterol responses of rats fed on cholesterol-free diets supplemented with guar gum and sodium alginate. Br J Nutr 85:317–324. doi: 10.1079/bjn2000250. [DOI] [PubMed] [Google Scholar]
- 21.Wu J, Peng SS. 1997. Comparison of hypolipidemic effect of refined konjac meal with several common dietary fibers and their mechanisms of action. Biomed Environ Sci 10:27–37. [PubMed] [Google Scholar]
- 22.Chen QC, Liu M, Zhang PY, Fan SJ, Huang JL, Yu SY, Zhang CH, Li HJ. 2019. Fucoidan and galactooligosaccharides ameliorate high-fat diet-induced dyslipidemia in rats by modulating the gut microbiota and bile acid metabolism. Nutrition 65:50–59. doi: 10.1016/j.nut.2019.03.001. [DOI] [PubMed] [Google Scholar]
- 23.Jia W, Xie GX, Jia WP. 2018. Bile acid-microbiota crosstalk in gastrointestinal inflammation and carcinogenesis. Nat Rev Gastroenterol Hepatol 15:111–128. doi: 10.1038/nrgastro.2017.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rees DC, Johnson E, Lewinson O. 2009. ABC transporters: the power to change. Nat Rev Mol Cell Biol 10:218–227. doi: 10.1038/nrm2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khan I, Ullah N, Zha L, Bai Y, Khan A, Zhao T, Che T, Zhang C. 2019. Alteration of gut microbiota in inflammatory bowel disease (IBD): Cause or consequence? IBD treatment targeting the gut microbiome. Pathogens 8:126. doi: 10.3390/pathogens8030126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Devkota S, Chang EB. 2015. Interactions between diet, bile acid metabolism, gut microbiota, and inflammatory bowel diseases. Dig Dis 33:351–356. doi: 10.1159/000371687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shang QS, Jiang H, Cai C, Hao JJ, Li GY, Yu GL. 2018. Gut microbiota fermentation of marine polysaccharides and its effects on intestinal ecology: an overview. Carbohydr Polym 179:173–185. doi: 10.1016/j.carbpol.2017.09.059. [DOI] [PubMed] [Google Scholar]
- 28.Wang XL, Wang X, Jiang H, Cai C, Li GY, Hao JJ, Yu GL. 2018. Marine polysaccharides attenuate metabolic syndrome by fermentation products and altering gut microbiota: an overview. Carbohydr Polym 195:601–612. doi: 10.1016/j.carbpol.2018.05.003. [DOI] [PubMed] [Google Scholar]
- 29.Ejima R, Akiyama M, Sato H, Tomioka S, Yakabe K, Kimizuka T, Seki N, Fujimura Y, Hirayama A, Fukuda S, Hase K, Kim YG. 2021. Seaweed dietary fiber sodium alginate suppresses the migration of colonic inflammatory monocytes and diet-induced metabolic syndrome via the gut microbiota. Nutrients 13:2812. doi: 10.3390/nu13082812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ramnani P, Chitarrari R, Tuohy K, Grant J, Hotchkiss S, Philp K, Campbell R, Gill C, Rowland I. 2012. In vitro fermentation and prebiotic potential of novel low molecular weight polysaccharides derived from agar and alginate seaweeds. Anaerobe 18:1–6. doi: 10.1016/j.anaerobe.2011.08.003. [DOI] [PubMed] [Google Scholar]
- 31.Wang Y, Han F, Hu B, Li JB, Yu WG. 2006. In vivo prebiotic properties of alginate oligosaccharides prepared through enzymatic hydrolysis of alginate. Nutr Res 26:597–603. doi: 10.1016/j.nutres.2006.09.015. [DOI] [Google Scholar]
- 32.Liu F, Wang X, Shi HJ, Wang YM, Xue CH, Tang QJ. 2017. Polymannuronic acid ameliorated obesity and inflammation associated with a high-fat and high-sucrose diet by modulating the gut microbiome in a murine model. Br J Nutr 117:1332–1342. doi: 10.1017/S0007114517000964. [DOI] [PubMed] [Google Scholar]
- 33.Milani C, Lugli GA, Duranti S, Turroni F, Bottacini F, Mangifesta M, Sanchez B, Viappiani A, Mancabelli L, Taminiau B, Delcenserie V, Barrangou R, Margolles A, van Sinderen D, Ventura M. 2014. Genomic encyclopedia of type strains of the genus Bifidobacterium. Appl Environ Microbiol 80:6290–6302. doi: 10.1128/AEM.02308-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Håkansson A, Tormo-Badia N, Baridi A, Xu J, Molin G, Hagsltt ML, Karlsson C, Jeppsson B, Cilio CM, Ahrné S. 2015. Immunological alteration and changes of gut microbiota after dextran sulfate sodium (DSS) administration in mice. Clin Exp Med 15:107–120. doi: 10.1007/s10238-013-0270-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang G, Huang S, Cai S, Yu HT, Wang YM, Zeng XF, Qiao SY. 2020. Lactobacillus reuteri ameliorates intestinal inflammation and modulates gut microbiota and metabolic disorders in dextran sulfate sodium-induced colitis in mice. Nutrients 12:2298. doi: 10.3390/nu12082298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang T, Ji XH, Lu GC, Zhang FM. 2021. The potential of Akkermansia muciniphila in inflammatory bowel disease. Appl Microbiol Biotechnol 105:5785–5794. doi: 10.1007/s00253-021-11453-1. [DOI] [PubMed] [Google Scholar]
- 37.Zhai QX, Feng SS, Arjan N, Chen W. 2019. A next generation probiotic, Akkermansia muciniphila. Crit Rev Food Sci Nutr 59:3227–3236. doi: 10.1080/10408398.2018.1517725. [DOI] [PubMed] [Google Scholar]
- 38.Fedorowski T, Salen G, Tint GS, Mosbach E. 1979. Transformation of chenodeoxycholic acid and ursodeoxycholic acid by human intestinal bacteria. Gastroenterology 77:1068–1073. doi: 10.1016/S0016-5085(79)80079-7. [DOI] [PubMed] [Google Scholar]
- 39.Ridlon JM, Hylemon PB. 2012. Identification and characterization of two bile acid coenzyme A transferases from Clostridium scindens, a bile acid 7 alpha-dehydroxylating intestinal bacterium. J Lipid Res 53:66–76. doi: 10.1194/jlr.M020313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trudeau MP, Zhou Y, Leite FL, Gomez A, Urriola PE, Shurson GC, Chen C, Isaacson RE. 2018. Fecal hyodeoxycholic acid is correlated with tylosin-induced microbiome changes in growing pigs. Front Vet Sci 5:196. doi: 10.3389/fvets.2018.00196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang CC, Du Y, Ren DY, Yang XB, Zhao Y. 2021. Gut microbiota-dependent catabolites of tryptophan play a predominant role in the protective effects of turmeric polysaccharides against DSS-induced ulcerative colitis. Food Funct 12:9793–9807. doi: 10.1039/d1fo01468d. [DOI] [PubMed] [Google Scholar]
- 42.Zagato E, Pozzi C, Bertocchi A, Schioppa T, Saccheri F, Guglietta S, Fosso B, Melocchi L, Nizzoli G, Troisi J, Marzano M, Oresta B, Spadoni I, Atarashi K, Carloni S, Arioli S, Fornasa G, Asnicar F, Segata N, Guglielmetti S, Honda K, Pesole G, Vermi W, Penna G, Rescigno M. 2020. Endogenous murine microbiota member Faecalibaculum rodentium and its human homologue protect from intestinal tumour growth. Nat Microbiol 5:511–524. doi: 10.1038/s41564-019-0649-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pang JM, Wang SL, Wang ZY, Wu YJ, Zhang XY, Pi Y, Han DD, Zhang S, Wang JJ. 2021. Xylo-oligosaccharide alleviates Salmonella induced inflammation by stimulating Bifidobacterium animalis and inhibiting Salmonella colonization. FASEB J 35:e21977. [DOI] [PubMed] [Google Scholar]
- 44.Singhal AK, Cohen BI, Mosbach EH, Une M, Stenger RJ, McSherry CK, May-Donath P, Palaia T. 1984. Prevention of cholesterol-induced gallstones by hyodeoxycholic acid in the prairie dog. J Lipid Res 25:539–549. doi: 10.1016/S0022-2275(20)37766-X. [DOI] [PubMed] [Google Scholar]
- 45.Sehayek E, Ono JG, Duncan EM, Batta AK, Salen G, Shefer S, Neguyen LB, Yang K, Lipkin M, Breslow JL. 2001. Hyodeoxycholic acid efficiently suppresses atherosclerosis formation and plasma cholesterol levels in mice. J Lipid Res 42:1250–1256. doi: 10.1016/S0022-2275(20)31575-3. [DOI] [PubMed] [Google Scholar]
- 46.Zheng XJ, Chen TL, Jiang RQ, Zhao AH, Wu Q, Kuang JL, Sun DN, Ren ZX, Li MC, Zhao ML, Wang SL, Bao YQ, Li HT, Hu C, Dong B, Li DF, Wu JY, Xia JL, Wang XM, Lan K, Rajani C, Xie GX, Lu AP, Jia WP, Jiang CT, Jia W. 2021. Hyocholic acid species improve glucose homeostasis through a distinct TGR5 and FXR signaling mechanism. Cell Metab 33:791–803.e7. doi: 10.1016/j.cmet.2020.11.017. [DOI] [PubMed] [Google Scholar]
- 47.Li CX, Wang XQ, Cheng FF, Yan X, Luo J, Wang QG. 2019. Hyodeoxycholic acid protects the neurovascular unit against oxygen-glucose deprivation and reoxygenation-induced injury in vitro. Neural Regen Res 14:1941–1949. doi: 10.4103/1673-5374.259617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bai YY, Zhu H, Huang F, Wu XJ. 2021. Hyodeoxycholic acid rescues lipopolysaccharide-induced neuroinflammation via regulating TGR5/AKT/NF-kappa B pathway. Psychoneuroendocrinology 131:105482. doi: 10.1016/j.psyneuen.2021.105482. [DOI] [Google Scholar]
- 49.Zong E, Yan S, Wang M, Yin L, Wang Q, Yin J, Li J, Li Y, Ding X, Huang P, He S, Yang H, Yin Y. 2019. The effects of dietary supplementation with hyodeoxycholic acid on the differentiation and function of enteroendocrine cells and the serum biochemical indices in weaned piglets. J Anim Sci 97:1796–1805. doi: 10.1093/jas/skz059. [DOI] [PubMed] [Google Scholar]
- 50.Chen ML, Takeda K, Sundrud MS. 2019. Emerging roles of bile acids in mucosal immunity and inflammation. Mucosal Immunol 12:851–861. doi: 10.1038/s41385-019-0162-4. [DOI] [PubMed] [Google Scholar]
- 51.Zhang SW, Liu QY, Wang J, Harnish DC. 2009. Suppression of interleukin-6-induced C-reactive protein expression by FXR agonists. Biochem Biophys Res Commun 379:476–479. doi: 10.1016/j.bbrc.2008.12.117. [DOI] [PubMed] [Google Scholar]
- 52.Wang YD, Chen WD, Wang MH, Yu DN, Forman BM, Huang WD. 2008. Farnesoid X receptor antagonizes nuclear factor kappa B in hepatic inflammatory response. Hepatology 48:1632–1643. doi: 10.1002/hep.22519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gadaleta RM, van Erpecum KJ, Oldenburg B, Willemsen ECL, Renooij W, Murzilli S, Klomp LWJ, Siersema PD, Schipper MEI, Danese S, Penna G, Laverny G, Adorini L, Moschetta A, van Mil SWC. 2011. Farnesoid X receptor activation inhibits inflammation and preserves the intestinal barrier in inflammatory bowel disease. Gut 60:463–472. doi: 10.1136/gut.2010.212159. [DOI] [PubMed] [Google Scholar]
- 54.Song M, Yang Q, Zhang FL, Chen L, Su H, Yang XH, He HW, Liu FF, Zheng JS, Ling MF, Lai XM, Zhu XT, Wang LN, Gao P, Shu G, Jiang QY, Wang SB. 2020. Hyodeoxycholic acid (HDCA) suppresses intestinal epithelial cell proliferation through FXR-PI3K/AKT pathway, accompanied by alteration of bile acids metabolism profiles induced by gut bacteria. FASEB J 34:7103–7117. doi: 10.1096/fj.201903244R. [DOI] [PubMed] [Google Scholar]
- 55.Wang SL, Zhang SY, Huang SM, Wu ZH, Pang JM, Wu YJ, Wang JJ, Han DD. 2020. Resistant maltodextrin alleviates dextran sulfate sodium-induced intestinal inflammatory injury by increasing butyric acid to inhibit proinflammatory cytokine levels. Biomed Res Int 2020:7694734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Horibe S, Tanahashi T, Kawauchi S, Mizuno S, Rikitake Y. 2016. Preventative effects of sodium alginate on indomethacin-induced small-intestinal injury in mice. Int J Med Sci 13:653–663. doi: 10.7150/ijms.16232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Miyazaki T, Shirakami Y, Kubota M, Ideta T, Kochi T, Sakai H, Tanaka T, Moriwaki H, Shimizu M. 2016. Sodium alginate prevents progression of non-alcoholic steatohepatitis and liver carcinogenesis in obese and diabetic mice. Oncotarget 7:10448–10458. doi: 10.18632/oncotarget.7249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yin J, Li YY, Han H, Chen S, Gao J, Liu G, Wu X, Deng JP, Yu QF, Huang XG, Fang RJ, Li TJ, Reiter RJ, Zhang D, Zhu CR, Zhu GQ, Ren WK, Yin YL. 2018. Melatonin reprogramming of gut microbiota improves lipid dysmetabolism in high-fat diet-fed mice. J Pineal Res 65:e12524. doi: 10.1111/jpi.12524. [DOI] [PubMed] [Google Scholar]
- 59.Zhang Q, Hu J, Feng JW, Hu XT, Wang T, Gong WX, Huang K, Guo YX, Zou Z, Lin X, Zhou R, Yuan YQ, Zhang AD, Wei H, Cao G, Liu C, Chen LL, Jin ML. 2020. Influenza infection elicits an expansion of gut population of endogenous Bifidobacterium animalis which protects mice against infection. Genome Biol 21:99. doi: 10.1186/s13059-020-02007-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang CJ, Yu M, Yang YX, Mu CL, Su Y, Zhu WY. 2017. Differential effect of early antibiotic intervention on bacterial fermentation patterns and mucosal gene expression in the colon of pigs under diets with different protein levels. Appl Microbiol Biotechnol 101:2493–2505. doi: 10.1007/s00253-016-7985-7. [DOI] [PubMed] [Google Scholar]
- 61.Hong X, Chen J, Liu L, Wu H, Tan H, Xie G, Xu Q, Zou H, Yu W, Wang L, Qin N. 2016. Metagenomic sequencing reveals the relationship between microbiota composition and quality of Chinese rice wine. Sci Rep 6:26621. doi: 10.1038/srep26621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar F, Bai Y, Bisanz JE, Bittinger K, Brejnrod A, Brislawn CJ, Brown CT, Callahan BJ, Caraballo-Rodríguez AM, Chase J, Cope EK, Da Silva R, Diener C, Dorrestein PC, Douglas GM, Durall DM, Duvallet C, Edwardson CF, Ernst M, Estaki M, Fouquier J, Gauglitz JM, Gibbons SM, Gibson DL, Gonzalez A, Gorlick K, Guo JR, Hillmann B, Holmes S, Holste H, Huttenhower C, Huttley GA, Janssen S, Jarmusch AK, Jiang LJ, Kaehler BD, Kang KB, Keefe CR, Keim P, Kelley ST, Knights D, et al. . 2019. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 37:852–857. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang ZY, Bai Y, Pi Y, Gerrits WJJ, de Vries S, Shang LJ, Tao SY, Zhang SY, Han DD, Zhu ZP, Wang JJ. 2021. Xylan alleviates dietary fiber deprivation-induced dysbiosis by selectively promoting Bifidobacterium pseudocatenulatum in pigs. Microbiome 9:227. doi: 10.1186/s40168-021-01175-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glockner FO. 2013. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Douglas GM, Maffei VJ, Zaneveld JR, Yurgel SN, Brown JR, Taylor CM, Huttenhower C, Langille MGI. 2020. PICRUSt2 for prediction of metagenome functions. Nat Biotechnol 38:685–688. doi: 10.1038/s41587-020-0548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li RQ, Yu C, Li YR, Lam TW, Yiu SM, Kristiansen K, Wang J. 2009. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics 25:1966–1967. doi: 10.1093/bioinformatics/btp336. [DOI] [PubMed] [Google Scholar]
- 67.Kanehisa M, Goto S. 2000. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lombard V, Ramulu HG, Drula E, Coutinho PM, Henrissat B. 2014. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res 42:D490–D495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ma JL, Hong Y, Zheng NN, Xie GX, Lyu YZ, Gu Y, Xi CC, Chen LL, Wu GS, Li Y, Tao X, Zhong J, Huang ZZ, Wu WB, Yuan L, Lin M, Lu X, Zhang WD, Jia W, Sheng LL, Li HK. 2020. Gut microbiota remodeling reverses aging-associated inflammation and dysregulation of systemic bile acid homeostasis in mice sex-specifically. Gut Microbes 11:1450–1474. doi: 10.1080/19490976.2020.1763770. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download spectrum.02979-22-s0001.pdf, PDF file, 0.4 MB (414.1KB, pdf)
Data Availability Statement
The bacterial 16S rRNA sequencing data reported in this paper were deposited in the NCBI Sequence Read Archive (SRA) database under accession number PRJNA694703.