ABSTRACT
The trypanosomatid protozoan parasite Leishmania has a significant impact on human health globally. Understanding the pathways associated with virulence within this significant pathogen is critical for identifying novel vaccination and chemotherapy targets. Within this study we leverage an ultradeep proteomic approach to improve our understanding of two virulence-associated genes in Leishmania, encoding the Golgi mannose/arabinopyranose/fucose nucleotide-sugar transporter (LPG2) and the mitochondrial fucosyltransferase (FUT1). Using deep peptide fractionation followed by complementary fragmentation approaches with higher-energy collisional dissociation (HCD) and electron transfer dissociation (ETD) allowed the identification of over 6,500 proteins, nearly doubling the experimentally known Leishmania major proteome. This deep proteomic analysis revealed significant quantitative differences in both Δlpg2– and Δfut1s mutants with FUT1-dependent changes linked to marked alterations within mitochondrion-associated proteins, while LPG2-dependent changes impacted many pathways, including the secretory pathway. While the FUT1 enzyme has been shown to fucosylate peptides in vitro, no evidence for protein fucosylation was identified within our ultradeep analysis, nor did we observe fucosylated glycans within Leishmania glycopeptides isolated using hydrophilic interaction liquid chromatography (HILIC) enrichment. This work provides a critical resource for the community on the observable Leishmania proteome as well as highlighting phenotypic changes associated with LPG2 or FUT1, ablation of which may guide the development of future therapeutics.
IMPORTANCE Leishmania is a widespread trypanosomatid protozoan parasite of humans, with ~12 million cases currently, ranging from mild to fatal, and hundreds of millions asymptomatically infected. This work advances knowledge of the experimental proteome by nearly 2-fold, to more than 6,500 proteins and thus provides a great resource to investigators seeking to decode how this parasite is transmitted and causes disease and to identify new targets for therapeutic intervention. The ultradeep proteomics approach identified potential proteins underlying the “persistence-without-pathology” phenotype of mutants with deletion of the Golgi nucleotide transporter LPG2, showing many alterations and several candidates. Studies of a rare mutant with deletion of the mitochondrial fucosyltransferase FUT1 revealed changes underlying its strong mitochondrial dysfunction but did not reveal examples of fucosylation of either peptides or N-glycans. This suggests that this vital protein’s elusive target(s) may be more complex than the methods used could detect or that this target may not be a protein but perhaps another glycoconjugate or glycolipid.
KEYWORDS: N-linked glycans, N-linked glycoconjugates, trypanosomatid protozoan parasite, fucose, glycoproteome, kinetoplast, mitochondria, ultradeep proteomics
INTRODUCTION
Leishmaniasis is a devastating parasitic disease caused by species of the trypanosomatid protozoan parasite genus Leishmania. Currently, there are over 12 million cases, with 1.7 billion people at risk of infection worldwide, and estimates of asymptomatic infections ranging as high as >300 million (1–4). Clinical manifestations include self-healing, localized or diffuse cutaneous lesions (cutaneous leishmaniasis), destruction of the nasopharyngeal mucosa (mucocutaneous leishmaniasis), and enlargement of the spleen or liver, which can lead to death (visceral leishmaniasis) (3). Leishmania parasites are transmitted to mammalian hosts via the bite of phlebotomine sand flies and undergo a series of transformations throughout their life cycle, predominantly as promastigotes (insect stage) and amastigotes (intracellular in mammalian host) (3, 5). Powerful genetic tools have uncovered a number of loci that are essential to the completion of the parasite’s life cycle and/or potentially suitable as targets for chemotherapy (6–10). Here, we focus on two mutants relevant to such efforts, whose properties are depicted in Fig. 1.
FIG 1.
Overview of glycosylation pathways relevant to LPG2 or FUT1 biology and deletion mutants. LPG2 encodes is a Golgi GDP-sugar transporter, essential for LPG synthesis. Mutant Δlpg2– parasites grow comparably to WT parasites in culture but do not produce pathology. FUT1 encodes a fucosyltransferase located in the parasite mitochondria, whose substrates remain unknown. Null-mutant Δfut1s parasites exhibit severe defects, including significantly decreased growth rate, abnormal mitochondrial morphology, decreased mitochondrial membrane potential (Ψ), and loss of kDNA compactness or the kDNA network altogether. Arap, arabinopyranose (green star); Man, mannose (green circle); Fuc, fucose (red triangle); LPG, lipophosphoglycan. This figure was created with BioRender.com.
Leishmania parasites are completely coated in a dense glycocalyx largely containing glycoconjugate lipophosphoglycan (LPG), composed of (i) a 1-O-alkyl-2-lyso-phosohotidylinositol lipid anchor, (ii) a heptasaccharide core, (iii) a phosphoglycan polymer consisting of 15 to 30 Galβ1,4Manα1-PO4 repeating units (phosphoglycan [PG] repeats), often bearing side chain sugars such as galactose in Leishmania major, and (iv) a small oligosaccharide cap (11–13). Biochemical and genetic studies of L. major Δlpg1– (this nomenclature indicates homozygous deletion and precise replacement of the LPG1 ORF) mutants specifically lacking LPG have implicated it in many key steps of the parasite infectious cycle: in the sand fly vector, as well in the establishment of infection within the mammalian host following the sand fly bite (14–18). However, the amastigote stage lacks significant levels of LPG, and the few Δlpg1– parasites able to survive the initial host response are thereafter highly virulent (17). In contrast, deletion of LPG2, encoding a Golgi nucleotide sugar transporter (NST; described below), resulted in loss of LPG along with related glycoconjugates, including proteophosphoglycans, which are normally expressed in amastigotes (19). While many Δlpg2– phenotypes in both sand flies and establishment of mammalian infection mirrored those of Δlpg1– mutants, infections of susceptible mice with Δlpg2– mutants showed no overt disease and long-term persistence of a small number of parasites typical of long-term asymptomatic infections (19). This “persistence-without-pathology” phenotype induced protective immunity in a manner similar to that seen in natural healed infections (20).
Although the attenuated Δlpg2– phenotype had been attributed solely to the loss of LPG and related glycoconjugates (19), this view was challenged by studies of an analogous LPG-deficient mutant obtained by genetic deletion of the partially redundant LPG5A and LPG5B genes, encoding the Golgi UDP-galactose transporters (21). These parasites resemble the Δlpg2– mutant in lacking LPG and related PG-bearing glycoconjugates but, unlike Δlpg2– parasites, retained pathology following inoculation into susceptible mice, similar to that seen with Δlpg1– mutants lacking only LPG (22). This suggested the possibility of LPG2-dependent “off-LPG/phosphoglycan” effects, identification of which could shed new insight on both the persistence-without-pathology phenotype of the Δlpg2– strain, its ability to serve as a live vaccine line, and its tendency to revert toward virulence via second-site events (20, 23, 24).
The second mutant studied arises from finding that LPG2 was the first example of a multispecific NST, able to transport GDP-l-fucose as well as GDP-d-arapyranose (25). While the roles of GDP-Man in phosphoglycan repeat synthesis and d-Arap as a “capping” sugar able to block LPG interaction with the sand fly epithelium had been thoroughly studied (26, 27), GDP-Fuc transport was enigmatic. Although L. major expresses low levels of GDP-fucose mediated by the closely related bifunctional salvage enzymes AFKP80 and FKP40 (28, 29), convincing evidence for fucoconjugates has been hard to find. However, deletion of the two salvage enzymes could not be achieved unless “metabolic complementation” of GDP-fucose was engineered through expression of the trypanosome de novo GDP-fucose-synthetic pathway (29). Reasoning that this implied the existence of an essential fucosyltransferase (FUT), we identified 5 candidate fucosyltransferases, four of which appeared to be targeted as expected to the parasite secretory pathway. SCA1 and SCA2 function in d-Arap modifications of LPG (26), while null mutants of SCAL and FUT2 showed little phenotype in culture (30).
The fifth candidate, FUT1, however, was important for parasite survival and potentially the key player in the GDP-fucose requirement. Unlike the other candidates, FUT1 was found throughout all trypanosomatid species but not in other organisms (30, 31). Unexpectedly, FUT1 was found to target the parasite mitochondrion, as does its homolog TbFUT1 from Trypanosoma brucei, and both the mitochondrial localization and fucosyltransferase activity were essential (30, 31). While the gene was impossible to knock out by conventional approaches, through plasmid shuffling and examination of more than 1,000 events, a single, rare Leishmania Δfut1 deletion segregant (Δfut1s mutant) was obtained (30). This mutant line displayed severe growth and mitochondrial defects, which were rescued by L. major FUT1 (LmjFUT1) or TbFUT1 re-expression (30). While FUT1 proteins described in other organisms are able to fucosylate a variety of glycan substrates (32, 33), recombinant L. major FUT1 was unexpectedly able to fucosylate both glycan and peptide substrates in vitro (30). Thus far, neither the native acceptor in Leishmania nor that in trypanosomes has been identified, despite considerable effort in both species (30, 31).
Here, we generated high-coverage proteomes of wild-type (WT), Δlpg2–, and Δfut1s Leishmania major, identifying over 6,500 proteins representing nearly 80% of the predicted proteome, expanding the previously known experimental L. major proteome by nearly 2-fold. Differential proteomic analysis showed numerous differences in the Δlpg2– and Δfut1s parasites, including some impacted in both. Despite the depth of this analysis, we failed to identify any fucose-bearing proteins across these proteomes. Furthermore, while glycopeptide enrichment analysis of WT samples also identified numerous instances of N-linked glycopeptides, no O-fucosylation or fucosylated N-linked glycans were observed. Taken together, this work highlights the challenges of identifying FUT1 targets and indicates that alternative approaches will be required in future studies.
RESULTS
Acquisition of a high coverage of the L. major proteome.
We examined the WT Fn strain of L. major, whose genome is one of the best-characterized references for Leishmania spp., as well as L. major Fn Δfut1s (30) and a newly created Fn Δlpg2– mutant. CRISPR/Cas9 mutagenesis was used to readily generate homozygous open reading frame (ORF) replacements (see Fig. S1 in the supplemental material), and one Fn Δlpg2– clonal line (c14.2) was chosen for further study. Preliminary studies showed that it lacks LPG by agglutination tests and does not exhibit lesion pathology when inoculated into susceptible mice but persists at low levels, similar to the previously described L major LV39 clone 5 Δlpg2– mutant (19). Previous studies showed that restoration of LPG2 expression restores all WT phenotypes tested in both the Fn and LV39 clone 5 backgrounds (19, 34).
For each line, four replicate cultures were initiated and harvested in logarithmic growth phase. Parasite lysates were digested with trypsin and separated into 12 concatenated fractions by basic reverse-phase C18 chromatography (35) and then individually separated and analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS). To ensure the ability to localize any potential glycosylation events, precursors were subjected to both higher-energy collisional dissociation (HCD) and electron transfer dissociation (ETD) fragmentation with spectra searched against a database consisting of the predicted L. major proteome. Protein matches meeting a 0.01 false discovery rate (FDR) cutoff, excluding contaminants, reverse decoys, and those identified only by site, were retained for an initial analysis of the coverage (Table S1).
The numbers of proteins detected in WT, Δlpg2–, and Δfut1s strains were similar (6,208, 6,102, and 5,934, respectively), totaling 6,744 (Table S1) and representing nearly 80% of the predicted L. major proteome (8,307 and 8,038 in TriTrypDB and UniProt, respectively). In comparisons with two previous proteomic studies of L. major (36, 37), 3,484 proteins identified had no prior MS-based evidence, while 3,260 had prior MS-based evidence, and 308 detected previously did not appear in our data set (Fig. 2A). Thus, this work increased experimental coverage of the Leishmania proteome by nearly 2-fold.
FIG 2.
(A) Comparison of proteins identified in this work with previous studies. The combined data set (Table S1) from all lines and replicas was compared to L. major proteins with MS-based evidence annotated in TriTrypDB (https://tritrypdb.org/tritrypdb/app). Of the proteins identified here, 3,484 showed previous MS evidence and 3,260 had no prior MS-based reports; 308 proteins with previous MS-based evidence did not appear in our data sets. (B) Distribution of identified proteins among WT, Δfut1s, and Δlpg2– L. major. The total proteome (Table S1) was further parsed by considering only proteins identified in at least two of the four biological replicates, in one or more parasite lines, yielding a total of 5,841 (Table S3). The Venn diagram displays the overlap of proteins among the three lines.
No predicted mitochondrion-encoded proteins were identified, despite the inclusion of many of them in our predicted proteome database (described in Materials and Methods). Often these are omitted from proteomic computational analysis completely, and due to their hydrophobicity, they are often not detected by standard or general approaches similar to those used here (38–40).
Protein glycosylation and absence of detectable fucosylation.
We showed previously that recombinant FUT1 is able to modify both glycan and peptide substrates in vitro (30), and the expanded proteome provided an opportunity to search for evidence of in vivo fucosylation. To allow the localization of fucosylation events, ETD MS fragmentation was used, as O-fucosylated peptides are poorly localized with collision-based approaches (41). We searched first for peptide-O-fucose by considering deoxyhexose (dHex) modifications of serine or threonine residues, yet despite our proteome depth and the ability of our group to identify glycosylation events from deep proteome data sets (42, 43), no high-confidence O-fucosylation events were observed. Moreover, we were specifically unable to detect HSP70 or HSP60 peptide O-fucose, inferred previously in Leishmania donovani (44), peptides of which were fucosylated by recombinant FUT1 in vitro (30). Open searching (45) was also undertaken using MSFragger (46), which also failed to reveal any putative fucose-containing glycopeptides from our deep proteome analysis.
To further explore the diversity of glycans which may exist within L. major, we surveyed glycopeptides derived from L. major using zwitterionic-hydrophilic interaction liquid chromatography (ZIC-HILIC) (47) coupled to LC-MS analysis. Using open searching with MSFragger (46), multiple unique glycoforms were observed, the majority of which corresponded to N-linked glycoforms. While short O-linked glycopeptides can be enriched with this approach (48), our observation of predominately N-linked glycopeptide supports the idea that these escaped detection in L. major using ZIC-HILIC-based enrichment. We identified 65 glycopeptides bearing a variety of high-mannose N-linked glycans, as reported previously for Leishmania (49–52). The glycopeptides mapped to 49 different proteins, with 14 showing multiple sites and/or glyco-heterogeneity; 24 were predicted as hypothetical proteins of unknown function. The annotated glycosylated proteins are summarized in Table 1 and include many described previously, such as gp63/leishmanialysin, nucleotidases, and phosphatases (50), or ones likely to be so due to the presence of targeting motifs for the secretory pathway such as FUT2 (30). While these findings highlight the idea that multiple N-linked glycopeptides could be recovered from L. major, no fucosylated N-linked glycans were observed (Table S2). Within this data set, three additional proteins absent from the total proteome were identified, bringing the collective total identified to 6,747. These three were all hypothetical proteins, two without prior MS-based evidence (LmjF.08.0350 and LmjF.09.1330) and one previously reported (LmjF.33.1035) (36). Both LmjF.08.0350 and LmjF.09.1330 have a detected mass shift of approximately 1,380.5 Da, while LmjF.33.1035 presented a mass shift of 2,803.3 Da. Future work will be required to confirm these preliminary assignments and the potential identity of the attached modifications.
TABLE 1.
First-pass L. major glycoproteome from MS analysis of HILIC-enriched glycopeptidesa
| Protein annotation | ID |
|---|---|
| ATP pyrophosphate-lyase | LmjF.17.0200 |
| β-Galactofuranosyl transferase (LPG1) | LmjF.25.0010 |
| C8 sterol isomerase-like protein | LmjF.29.2140 |
| Cathepsin L-like protease | LmjF.08.1060 |
| Chitinase | LmjF.16.0790 |
| d-Alanyl-glycyl endopeptidase-like protein | LmjF.33.2850 |
| Dolichyl-diphosphooligosaccharide–protein glycotransferase | LmjF.35.1130 |
| GPI ethanolamine phosphate transferase 3 | LmjF.24.0340 |
| Hypothetical predicted multipass transmembrane protein | LmjF.24.0700 |
| Leishmanolysin (GP63) | LmjF.10.0465 |
| Phosphoglycan β1,3-galactosyltransferase 3 (SCG3) | LmjF.02.0010 |
| Putative 3′-nucleotidase/nuclease | LmjF.12.0400 |
| Putative 3′-nucleotidase/nuclease | LmjF.31.2310 |
| Putative amastin-like surface protein | LmjF.34.1080 |
| Putative β-fructofuranosidase | LmjF.04.0310 |
| Putative DNAJ domain protein | LmjF.24.0520 |
| Putative extracellular receptor | LmjF.19.0640 |
| Putative glycosyl transferase (FUT2) | LmjF.02.0330 |
| Putative phospholipid:diacylglycerol acyltransferase | LmjF.09.1040 |
| Putative surface protein amastin | LmjF.30.0860 |
| Signal recognition particle receptor subunit β | LmjF.33.2620 |
| Signal sequence receptor subunit α | LmjF.22.0260 |
| Stealth CR3 domain-containing protein | LmjF.16.1010 |
| Transmembrane 9 superfamily member | LmjF.34.3660 |
| Uncharacterized protein L7845.03 (thioredoxin) | LmjF.35.1330 |
Annotated proteins identified in the glycoproteomic analysis in this work. Gene IDs and protein annotations are taken from TriTrypDB (www.tritrypdb.org). Another 24 hypothetical proteins lacking annotation were also identified. GPI, glycosylphosphatidylinositol.
WT and mutant cell lines display qualitatively similar proteomes.
To compare the impacts of LPG2 and FUT1 loss, we first created a high-confidence data set by parsing the total proteome (Table S1) for proteins detected in two or more biological replicates, in at least one cell line. This retained 5,841 proteins (Table S3), of which 95% (5,576) occurred in all three lines (Fig. 2B), evidence of high congruency. Of the remainder, 16 to 21 were unique to one line, and 62 to 97 were shared by two lines (Fig. 2B). As expected, the LPG2 protein was detected in all WT and Δfut1s lysates but not in Δlpg2– replicas. FUT1 peptides, on the other hand, were detected in only one WT biological replicate, perhaps due to low abundance (53).
Mitochondrial proteome and gene ontology.
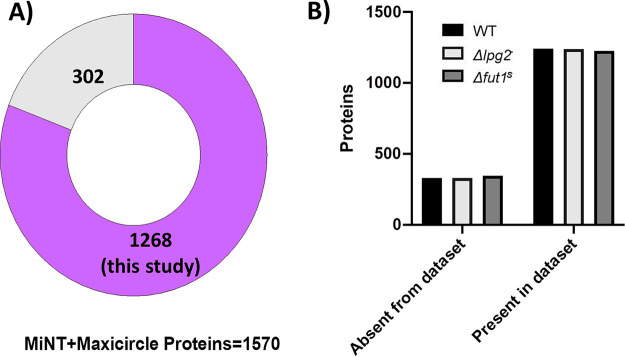
As FUT1 is targeted to the mitochondrion, where its loss results in numerous mitochondrial abnormalities (30), we queried the high-confidence proteomic data set for nuclearly encoded mitochondrial proteins present in the trypanosomatid predicted mitochondrial protein database MiNT (54). Of 1,559 mitochondrial L. major proteins in the MiNT database (approximately 19% of the total proteome), 1,268 (81%) were identified (Fig. 3A; Table S3), with the WT having a slightly higher number of identified proteins (1,241), followed by the Δlpg2– (1,239) and Δfut1s (1,226) mutants (Fig. 3B). Quantitative changes were apparent (discussed below), and no mitochondrially (maxicircle) encoded proteins were identified, as discussed above.
FIG 3.
Representation of the predicted L. major mitochondrial proteome. Proteins from WT or mutant parasite lines (Table S3) were compared to proteins in MiNT (predicted nuclearly encoded mitochondrial protein), supplemented with maxicircle (mitochondrion)-encoded proteins as described in Materials and Methods. (A) A total of 1,268 proteins, representing 81% of the mitochondrial proteome, were identified in this study. (B) Distribution of mitochondrial proteins by cell line.
A gene ontology (GO) analysis was performed using the PANTHER classification system (Fig. S2; Table S4) (55). As seen previously (36), much of the L. major proteome consists of hypothetical proteins lacking annotation (over 60%) (Fig. S2A), and GO analysis of annotated proteins showed considerable similarity among all three lines. For biological processes (Fig. S2B), the categories with the highest number of genes were cellular process (GO:0009987), metabolic process (GO:0008152), localization (GO:0051179), and biological regulation (GO:0065007). For cellular components (Fig. S2C), the most commonly assigned categories were cellular anatomical entity (GO:0110165), intracellular (GO:0005622) and protein-containing complex (GO:0032991). For molecular functions (Fig. S2D), the categories were catalytic activity (GO:0003824) and binding (GO:0005488).
Knockouts of FUT1 and LPG2 have significant differential impacts on L. major protein abundance.
Quantitative differences among WT, Δfut1s, and Δlpg2– parasites were assessed by analysis of variance (ANOVA) with an FDR of 0.05 applied to the protein abundance values (label-free quantitation [LFQ]) (Table S3), which were then normalized by Z-scoring to construct a heat map (Fig. 4). Clustering showed clear grouping of replicates within each line, as well as blocks encompassing 465 proteins differing significantly (Fig. 4). Of these, 107 increased and 142 decreased in Δfut1s parasites only, 173 decreased and 13 increased in Δlpg2– parasites only, and 30 decreased in Δfut1s parasites but increased in 2 of 4 Δlpg2– biological replicates (Tables S3, S5, and S6; Fig. 4). Again, the majority of differentially expressed proteins were hypothetical proteins (254/465 total) (Table S3).
FIG 4.
Patterns of variation of significantly differentially expressed proteins among WT and FUT1 and LPG2 knockout L. major lines. ANOVA using an FDR of 0.05 and an S0 of 1 was performed on LFQ values for the high-confidence proteome (Table S3), to identify proteins that are significantly different between WT, Δfut1s, and Δlpg2– clones. From these data, a heat map was generated, clustering the parasite lines, each with four replicas (top dendrogram), and the differentially expressed genes (left dendrogram). The cluster properties are summarized on the right, and the specific proteins included in each cluster can be found in Table S5 and S6.
SHERP, a stationary-phase/metacyclic parasite marker (56), was not seen in any Δlpg2– replicates but was unexpectedly detected in log-phase WT and Δfut1s parasites at similar levels (Tables S1 and S3). These results were corroborated by Western blot analysis with anti-SHERP antisera (Fig. S3).
Significantly different proteins accompanying the severe mitochondrial dysfunction of Δfut1s.
The Δfut1s mutant shows considerably slower growth and numerous mitochondrial abnormalities, including lowered mitochondrial membrane potential and complete loss or loss in compactness of the kinetoplast DNA (kDNA) network (30). From the cluster of 142 significantly less abundant proteins in Δfut1s parasites (Fig. 4; Table S5), 26 (18%) were predicted as mitochondrial, of which 13 were annotated (Table 2). Of 107 proteins that were significantly more abundant in Δfut1s parasites, 15 were mitochondrial, of which 2 were annotated (Table 2). The differentially expressed mitochondrial proteins are depicted in Fig. 5, many of which are positioned to contribute to Δfut1s mitochondrial dysfunction (see Discussion).
TABLE 2.
Mitochondrial proteins significantly changed in Δfut1s parasitesa
| Cluster pattern | Protein name | TriTrypDB ID | Difference in log2(LFQ intensity) |
Corrected ANOVA P value | ||
|---|---|---|---|---|---|---|
| Δlpg2– vs WT | Δfut1s vs WT | Δfut1s vs Δlpg2– | ||||
| Down in Δfut1s | 3-HMG-CoA reductase) (EC 1.1.1.34) | LmjF.30.3190 | 0.87 | −1.38 | −2.26 | 0.01 |
| Aldehyde dehydrogenase, mitochondrial (EC 1.2.1.3) | LmjF.25.1120 | 0.12 | −2.00 | −2.13 | 0.01 | |
| Chaperonin HSP60, mitochondrial | LmjF.36.2030 | 0.80 | −0.81 | −1.61 | 0.04 | |
| Kinetoplast-associated protein-like protein | LmjF.27.0240 | 1.60 | −3.80 | −5.40 | 0.00 | |
| Metallopeptidase, clan ME, family M16 (EC 1.10.2.2) | LmjF.35.1380 | 0.26 | −1.14 | −1.40 | 0.04 | |
| Proline dehydrogenase (EC 1.5.5.2) | LmjF.26.1610 | 1.34 | −0.07 | −1.40 | 0.02 | |
| Putative asparaginyl-tRNA synthetase (EC 6.1.1.22) | LmjF.34.2340 | 0.42 | −1.06 | −1.48 | 0.03 | |
| Putative carbamoyl-phosphate synthase (EC 6.3.4.16) | LmjF.16.0590 | 0.56 | −0.69 | −1.25 | 0.05 | |
| Putative cytochrome c oxidase VIII (EC 1.9.3.1) | LmjF.31.1570 | 0.63 | −2.16 | −2.79 | 0.02 | |
| Putative cytochrome c oxidase subunit 10 (EC 1.9.3.1) | LmjF.23.0370 | −1.34 | −2.49 | −1.15 | 0.04 | |
| Putative DNA-directed RNA polymerase I largest subunit (EC 2.7.7.6) | LmjF.32.0650 | 0.72 | −0.69 | −1.42 | 0.04 | |
| Putative heat shock protein | LmjF.18.1370 | 0.56 | −0.83 | −1.38 | 0.04 | |
| Putative NADH dehydrogenase (EC 1.6.99.3) | LmjF.36.5380 | 0.47 | −1.20 | −1.67 | 0.04 | |
| Putative translation initiation factor | LmjF.17.1290 | 0.78 | −0.59 | −1.36 | 0.05 | |
| Up in Δfut1s | Inositol phosphosphingolipid phospholipase C like | LmjF.08.0200 | 0.09 | 2.00 | 1.91 | 0.01 |
| Putative 3,2-trans-enoyl-CoA isomerase mitochondrial (EC 1.1.1.35) (EC 5.3.3.8) | LmjF.31.2330 | −0.79 | 2.14 | 2.93 | 0.04 | |
Annotated mitochondrial proteins differing significantly in abundance, as determined by ANOVA, between the Δfut1s and WT or Δlpg2– parasites (Fig. 4; Tables S3 and S5). Overall, there were 279 proteins in clusters where proteins decreased in Δfut1s parasites (142 total; 26 mitochondrial, of which 13 are unannotated), increased in Δfut1s parasites (107 total; 17 mitochondrial, of which 15 are unannotated), or decreased in Δfut1s and varied in Δlpg2– parasites (30 total; 13 mitochondrial, of which 12 are unannotated). CoA, coenzyme A.
FIG 5.
Schematic overview of a Leishmania mitochondrion showing proteins with significantly altered expression in Δfut1s relative to WT L. major. The specific proteins are listed in Table 1. Only mitochondrial proteins showing differential expression (Table 2) are shown. In the panel depicting the electron transport chain, the proteins decreased in the Δfut1s parasites are shown in blue, those not detected in our data set are shown in white, and those that were unchanged are shown in gray. It should be noted that trypanosomatids possess a single mitochondrion per cell whose structure differs from the typical metazoan mitochondrion shape depicted here. This figure was created with BioRender.com.
Protein changes seen in the secretory pathway mutant Δlpg2.
Overall, 174 proteins were significantly decreased in Δlpg2– parasites, of which 89 are of unknown function, while 13 increased in Δlpg2– parasites, of which 6 are of unknown function (Fig. 4; Tables S3 and S6). The predominance of proteins downregulated in Δlpg2– relative to Δfut1s organisms could be related to the relative health of the Δlpg2– mutant (this work and reference 19), as many of the changes seen in Δfut1s parasites involved stress responses arising from mitochondrial dysfunction. Notably, the proteins altered in Δlpg2– parasites mapped to a variety of cellular compartments and metabolic pathways (Tables S4 and S6), suggesting that despite the normal growth of this mutant in culture, the impact of loss of LPG2 was nonetheless profound. The broad impact of LPG2-dependent effects thus hinders efforts to pinpoint those most important to the biological alterations of most interest, such as persistence without pathology.
LPG2 loss affected several proteins involved in the glycosylation pathways. Two examples are decreases of phosphomannomutase (PMM) and phosphoacetylglucosamine mutase (PAGM), which catalyze the reversible transfer of phosphate between C-6 and C-1 hydroxyl groups of mannose and N-acetylglucosamine (57). PMM in particular is necessary for establishing infection in macrophages (27).
Potential interactions between LPG2- and FUT1-dependent pathways.
While the proteins differentially altered in Δlpg2– and Δfut1s parasites were mostly quite distinct (Fig. 4; Tables S5 and S6), in 2 of 4 replicate Δlpg2– lines a small group of 30 proteins showed inverse regulation to changes observed in Δfut1s parasites (Fig. 4; Table S5). Of these, 12 were unannotated predicted mitochondrial proteins. As the replicates were grown at the same time from the same inoculation under the same conditions, we cannot account for the variability. Changes in Δfut1s parasites were also observed in proteins outside the mitochondrion (Table S5). Interestingly, these included increase of other glycosyltransferases, including α-1,2-mannosyltransferase, phosphoglycan β1,2-arabinosyltransferase, and phosphoglycan β1,3-galactosyltransferase 3 (Table S5). We speculate that the cross-mutant effects arise from competition for GDP-fucose synthesized in the cytosol (28, 29), for transport and use by secretory pathway fucosyltransferases (dependent on LPG2) or the mitochondrion (FUT1).
DISCUSSION
This work presents a high-coverage L. major proteomic data set consisting of new mass spectrometry-based evidence for nearly 3,500 proteins beyond the 3,600 proteins typically identified experimentally in several Leishmania species, including L. major, by others (36). For quantitative analysis, the data were parsed to yield a high-confidence data set of 5,841 proteins. Comparison showed that the proteomes of WT, Δlpg2–, and Δfut1s parasites were highly congruent, but with significant quantitative variation (58). Over half of the significantly changing proteins were uncharacterized hypothetical proteins.
FUT1 and mitochondrial dysfunction.
In addition to severely delayed growth, the Δfut1s mutant exhibits profound mitochondrial dysfunction, including loss of membrane potential, bloated cristae, presence of large aggregates, loss of kDNA compactness, and complete loss of the kDNA network in some parasites (30). Benefiting from the high coverage of mitochondrial proteins in our data set (>80%) (Fig. 3), we were able to survey the impact of FUT1 deletion on these (Table 2; Table S5; Fig. 4). The Δfut1s mutant had significantly decreased levels of key components of mitochondrial respiratory chain complexes III (MMP-β) and IV (cytochrome c oxidases COX8 and COX10), (59) as well as the alternative NADH dehydrogenase (NDH2) (58, 60). Δfut1s alterations in several maxicircle (mitochondrion)-encoded components of the respiratory chain could not be assessed, as they were absent in our data sets. Collectively, downregulation of this pathway matches the changes in membrane potential and growth seen in Δfut1s parasites (30).
Δfut1s parasites showed downregulation of kinetoplast-associated proteins (KAPs), histone-like proteins attributed to packaging mitochondrial DNA (kDNA) in trypanosomatids (61). Downregulation of these has been associated with rearrangement of the kinetoplast structure, parasite growth defects, and shrinkage and complete loss of the kDNA network (62, 63), all phenotypes seen in Δfut1s parasites (30).
Enzymes impacted in Δfut1s parasites include 3-hydroxy-3-methylglutaryl coenzyme A reductase (HMGR) and proline dehydrogenase (ProDH) (Table 2; Table S5; Fig. 5). HMGR is a key enzyme in the mevalonate pathway, yielding isoprenoids important for viability and proliferation (64). ProDH mediates the first step in a pathway that leads to production of α-ketoglutarate entering the tricarboxylic acid cycle (65) and generates FADH2, which transfers electrons to coenzyme Q and the electron transport pathways (65). ProDH transcripts have been found to be present in higher levels in drug-resistant Leishmania (66).
Several proteins associated with stress responses were also impacted in Δfut1s parasites, including mitochondrial heat shock proteins HSP60 and HSP110 (67). HSP60 acts as one of the principal mediators of protein homeostasis in the mitochondrial matrix and in response to damaged or misfolded mitochondrial proteins after oxidative stress (67, 68). The inositol phosphosphingolipid phospholipase C-like (ISCL) protein, which contributes to parasite survival in acidic environments, was increased in Δfut1s parasites (69).
A first-pass L. major glycoproteome, lacking detectable fucosylation.
The FUT1 protein shows motifs characteristic of fucosyltransferases within the GT11 family, and recombinant Trypanosoma brucei and L. major FUT1 proteins were able to fucosylate various glycans in vitro (30, 31). Moreover, LmjFUT1 was shown to fucosylate several peptides in vitro (30 [TbrFUT1 was not tested]). The in vivo substrate of FUT1 has proven elusive in both parasite species, and the broad specificity of LmjFUT1 prompted us to search for protein O-fucosylation as part of this work. While glycopeptides were not detected with high confidence in the total proteomic studies, potentially reflecting technical considerations (70, 71), following HILIC enrichment we were able to identify 58 glycopeptides representing 49 Leishmania proteins, many of which were known or anticipated to be glycosylated but many of which were novel (Table 1). Several other potential modifications were identified computationally, such as pyrophosphorylation (72), which will require experimental verification. In Trypanosoma cruzi, HILIC enrichment similarly successfully identified a variety of N- and O-glycosylated proteins (73). Comprehensive studies of the Leishmania glycoproteome were lacking (50), with previous studies being mostly limited to a few glycoproteins such as GP63 (leishmanialysin). Despite the utility of the HILIC enrichment, many known glycosylated proteins of Leishmania were not detected, and thus, the proteins in Table 1 should be regarded as a first-pass (preliminary) glycoproteome, confirmation of which will be needed in any event.
Notably, we did not find evidence for peptide O-fucose or fucosylated glycans. Although glycopeptide enrichment using HILIC can be utilized for O-glycoproteome characterization, it may bias for peptides with large glycans or multiple glycosylations (74). It is possible that any potential FUT1-dependent protein fucosylation may be in a form recalcitrant to the methods used, or that other parasite stages could yield different results. It is also possible that further proteomic searches extending our ultradeep effort might eventually yield proteins or modifications (including fucosylation) not identified here. Alternatively, while it is difficult to prove a negative, the possibility that the in vivo substrate of FUT1 may be glycans or glycolipids rather than proteins or glycoproteins seems at least as likely from the absence of detectable peptide fucosylation. Resolution of this question ultimately will depend on identification of the relevant FUT1-dependent glycoconjugate in the future.
Summary and perspective.
Proteins showing altered expression in Δlpg2– parasites could contribute to the persistence-without-pathology phenotype seen in this mutant; however, as these parasites grow normally in culture, we were challenged to compose a predictive query of the 186 differentially expressed candidates for those most likely to contribute to the phenotype. These data also raised the possibility that changes in the expression of one or more of these proteins could contribute to the recovery of amastigote virulence in Δlpg2– revertants occasionally found in infected animals (23). Experimental tests of the role(s) of the LPG2-dependent proteome in the loss or recovery of amastigote virulence will be required to resolve this. Similarly, Δfut1s parasites showed 249 differentially expressed proteins, including many known or potential mitochondrial proteins, as expected from its strong impact on mitochondrial function, as well as a number of nonmitochondrial proteins. While our studies raise many hypotheses for the roles of both FUT1 and LPG2 in parasite biology, genetic confirmation will be required, as well as studies to establish whether the effects seen are direct or indirect consequences of gene ablation. Fortunately, with the advent of high-throughput knockout screening via CRISPR technology (75), probing the importance of this panoply of genes will be increasingly feasible.
MATERIALS AND METHODS
Cell culture.
All parasites were derivatives of L. major strain Fn (MHOM/IL/80/Fn). Parasites were grown as the promastigote form in vitro in complete medium 199 (M199) supplemented with 10% heat-inactivated fetal bovine serum (FBS), 40 mM HEPES (pH 7.4), 100 nM adenine, 1 μg/mL biotin, 5 μg/mL hemin, penicillin-streptomycin, and 2 μg/mL biopterin. Parasites were seeded in 200 mL medium at a density of 105/mL in roller bottles and allowed to grow until mid-log phase, corresponding to a density of 1 × 106 to 4 × 106/mL. Culture vessels were rotated at approximately 2 rpm using a cell production roller apparatus (Bellco Biotechnology). The Δfut1s segregant was described previously (30).
Generation of a Δlpg2– mutant line by CRISPR/Cas9 mutagenesis.
Parasites were engineered to express a human codon-optimized Streptococcus pyogenes Cas9 and an LPG2 sgRNA (plasmid p63Phleo-HspCas9 or B7521; single guide RNA [sgRNA] synthesized from template B7617) expressed from a U6 promoter as described previously (76). To replace the LPG2 ORF, homology-directed repair (HDR) templates were designed containing a hygromycin B (HYG) resistance gene flanked by sequences matching the 30 nucleotides flanking the LPG2 ORF. A total of 10 μg of HDR DNA was transfected into the L. major HSpCas9-B7617 parasites, which were then plated on semisolid M199 medium containing 50 μg/mL hygromycin B. Numerous hygromycin B-resistant (HYGr) colonies were obtained, of which 26 were tested and appeared to be WT/Δlpg2::HYG heterozygotes. Subsequently, parasites were transfected with a blasticidin (BSD) resistance gene HDR template, bearing the 30 nucleotides (nt) of LPG2 flanking sequence as before. The HSpCas9-B7617-HYGr parasites were transfected and plated onto semisolid medium containing 50 μg/mL hygromycin B and 10 μg/mL blasticidin. A total of 22 colonies were tested and confirmed to be Δlpg2::HYG/Δlpg2::BSD knockouts by PCR tests (Fig. S1), and preliminary data showed them to be LPG deficient, as judged by their failure to agglutinate with monoclonal anti-PG antibody WIC79.3. One clone was selected for further study (SpCas9-B7617 B+H-c14.2), referred to here as the Δlpg2– clone.
Generation of parasite lysates for proteomic analysis.
Whole-parasite lysates were prepared as described previously (77) with some modifications. After 3 washes with ice-cold phosphate-buffered saline (PBS), parasites were resuspended in 1 mL of ice-cold lysis buffer [6 M guanidinium chloride, 100 mM Tris (pH 8.5), 10 mM Tris(2-carboxyethyl)phosphine (TCEP), 40 mM 2-chloroacetamide (CAA), supplemented with a protease inhibitor tablet (Roche) and 0.2 mg/mL 1,10-phenanthroline]. Samples were immediately boiled at 95°C in a ThermoMixer at 1,500 rpm. After cooling samples on ice for 10 min, an additional boiling step was done at 95°C. One milligram of protein was collected from four biological replicates. The samples were acetone precipitated with 4 volumes of ice-cold acetone at −20°C overnight and then a second time under the same conditions for at least 4 h. After centrifugation and supernatant removal, samples were air dried while covered with a Kimwipe and stored at −80°C until ready for analysis.
Digestion of L. major proteome samples.
Precipitated protein pellets were resuspended in 50% trifluoroethanol and then heated at 50°C for 10 min with shaking at 1,000 rpm. Resuspended samples were then reduced/alkylated in a single step with 20 mM Tris(2-carboxyethyl)phosphine and 40 mM chloroacetamide for 1 h in the dark. Samples were then diluted 10-fold with 100 mM triethylammonium bicarbonate and digested with trypsin (1/100 [wt/wt]) overnight with shaking at 800 rpm. Digested samples were acidified to a final concentration of 0.5% formic acid and desalted with 50-mg tC18 Sep-Pak columns (Waters Corporation, Milford, MA, USA) according to the manufacturer’s instructions. Briefly, tC18 Sep-Pak columns were conditioned with buffer B (0.1% formic acid [FA], 80% acetonitrile [ACN]) and washed with 10 volumes of buffer A* (0.1% trifluoroacetic acid [TFA], 2% ACN), samples were loaded, columns were washed with 10 volumes of buffer A*, and bound peptides were eluted with buffer B and then dried by vacuum centrifugation.
High-pH fractionation.
Proteome samples were fractionated by basic reverse-phase chromatography according to the protocol of Batth and Olsen (78) with minor modifications. Briefly, peptides were resuspended in 1 mL of buffer A (5 mM ammonium formate, pH 10.5) and separated using a 1100 series high-performance liquid chromatograph (HPLC) (Agilent Technologies, CA) using a Gemini NX C18 column (4.6 by 250 mm, 5 μm; Phenomenex, CA) at a flow rate of 1 mL/min. Separation was accomplished using a 90-min gradient with samples loaded on the column at 2% buffer B (5 mM ammonium formate, 90% ACN [pH 10.5]) for 3 min. The concentration of buffer B was then ramped from 2% to 28% over 45 min, from 28% to 40% over 5 min, and from 40% to 80% over 5 min. The gradient was held at 80% B for 2 min, and then the column was regenerated by being returned to 2% B over 10 min and held at 2% B for 10 min. Sixty 1-min fractions were collected. Every group of five fractions was combined to generate a total of 12 pooled fractions, which were concentrated by vacuum centrifugation, desalted using C18 stage tips, and then subjected to mass-spectrometric analysis.
Proteomics analysis using reverse-phase LC-MS.
Pooled basic reverse-phase fractions were resuspended in buffer A* (2% ACN, 0.1% TFA) and separated using a two-column chromatography setup composed of a PepMap100 C18 20-mm by 75-μm trap and a PepMap C18 500-mm by 75-μm analytical column (Thermo Fisher Scientific). Samples were concentrated onto the trap column at 5 μL/min for 5 min and infused into an Orbitrap Elite mass spectrometer (Thermo Fisher Scientific). Then, 125-min gradients were run, altering the buffer composition from 2% buffer B (80% ACN, 0.1% FA) to 28% B over 95 min, from 28% B to 40% B over 10 min, and then from 40% B to 80% B over 5 min; the composition was held at 80% B for 3 min, decreased to 2% B over 2 min, and held at 2% B for another 10 min. The Elite Orbitrap mass spectrometer was operated in a data-dependent mode, automatically switching between the acquisition of a single Orbitrap MS scan (60,000 resolution) and a maximum of 10 MS-MS scans, with each ion subjected to both an HCD scan (15,000 resolution; normalized collision energy [NCE], 40; maximum fill time, 200 ms; automatic gain control [AGC], 5 × 104) and an ETD scan (ion trap analyzed; ETD reaction time, 100 ms with supplementary activation enabled; AGC, 1 × 104).
Proteomic analysis.
Proteome samples were processed using MaxQuant (v1.6.3.4) (79) and searched against the Leishmania major MHOM (UniProt accession no. UP000000542, containing 8,038 proteins) and TriTrypDB Leishmania major Fn (TrypDB 37) databases. The reference proteome database was supplemented with predicted proteins from the kinetoplast maxicircle (mitochondrion) of L. major (LmjF00.0040, NADH dehydrogenase 7 [MURF3, ND7]; LmjF00.0050, cytochrome oxidase 3; LmjF00.0060, CYb; LmjF00.0070, MURF4 [A6]; LmjF00.0080, MURF1; LmjF00.0100, NADH dehydrogenase subunit 1 [ND1]; NADH dehydrogenase subunit 4, NADH dehydrogenase subunit 5, LmjF00.0110 cytochrome oxidase 2 [CO2]; LmjF00.0120, MURF2; LmjF00.0130, cytochrome oxidase 1 [CO1]; LmjF00.0150, NADH dehydrogenase 4 [ND4]; LmjF00.0180, NADH dehydrogenase 5 [ND5]); due to extensive pan-RNA editing, other protein products that could not be reliably predicted were not included. Searches were undertaken using trypsin enzyme specificity with carbamidomethylation of cysteine as a fixed modification. Oxidation of methionine and dHex modifications of serine/threonine residues were included as variable modifications, and a maximum of 2 missed cleavages was allowed. To attempt to identify any potential complex fucosylation events, dependent peptide searching was enabled. To ensure the inclusion of only high-quality peptide spectral matches (PSMs), a PSM FDR of 0.1% was set, while an FDR of 1% was allowed at the protein level. To enhance the identification of peptides between samples, the “match between runs” option was enabled, with a precursor match window set to 2 min and an alignment window of 20 min with the LFQ option enabled (80).
The result file was then uploaded to Perseus (V1.6.0.7) (81) for statistical analysis. Potential contaminants, proteins only identified by site, and reverse decoys were removed. For LFQ comparisons, values were log2(x) transformed and biological replicates were grouped. Data were parsed further by removing proteins not identified in at least two biological replicates in at least one cell line. Missing values were inferred based on the observed total peptide intensities with a range of 0.3σ and a downshift of 1.8σ using Perseus. A multiple-sample (ANOVA) test with a permutation-based FDR set at 0.05 and the artificial within groups parameter S0 set at 1 was performed to identify significantly differentially abundant proteins between Δfut1s, WT, and Δlpg2– lines. A heat map of significantly different proteins was constructed after Z-score-based normalization and Euclidean clustering of the transformed LFQ values (82).
Glycopeptide enrichment by ZIC-HILIC.
Two hundred fifty micrograms of whole-cell lysates that had been digested, C18 Sep-Pak cleaned, and dried was resuspended in 80% acetonitrile–1% TFA, and glycopeptides were enriched using homemade ZIC-HILIC stage tips as previously described (48). Briefly, ZIC-HILIC columns were first conditioned with 80% acetonitrile–1% TFA; then, samples were loaded onto columns before being washed with 80% acetonitrile–1% TFA, and glycopeptides were eluted with Milli-Q water. Samples were dried and stored at −20°C until undergoing LC-MS.
LC-MS analysis of ZIC-HILIC-enriched samples.
ZIC-HILIC-enriched samples were resuspended in buffer A* and separated using a two-column chromatography setup composed of a PepMap100 C18 20-mm by 75-μm trap and a PepMap C18 500-mm by 75-μm analytical column (Thermo Fisher Scientific) coupled to an Orbitrap Fusion Lumos Tribrid mass spectrometer (Thermo Fisher Scientific). ZIC-HILIC-enriched samples were analyzed using 185-min gradients. Separation gradients were run for each sample, altering the buffer composition from 2% buffer B to 28% B over 106 or 166 min depending on the run length, from 28% B to 40% B over 9 min, and then from 40% B to 80% B over 3 min; the composition was held at 80% B for 2 min, dropped to 2% B over 2 min, and held at 2% B for another 3 min. The Lumos mass spectrometer was operated in a data-dependent mode with a single Orbitrap MS scan (350 to 1,800 m/z, maximal injection time of 50 ms, an AGC maximum of 4 × 105 ions, and a resolution of 120,000) acquired every 3 s followed by Orbitrap MS/MS HCD scans of precursors (NCE, 30%; maximal injection time of 100 ms; AGC set to a maximum of 1.0 × 105 ions; and a resolution of 15,000). HCD scans containing the oxonium ions (204.0867; 138.0545 and 366.1396 m/z) triggered two additional product-dependent MS/MS scans of potential glycopeptides; an Orbitrap electron transfer–higher-energy collision dissociation [EThcD] scan (NCE, 15%; maximal injection time of 250 ms; AGC set to a maximum of 2 × 105 ions with a resolution of 30,000) and an ion trap collision-induced dissociation (CID) scan (NCE, 35%; maximal injection time of 40 ms; AGC set to a maximum of 5 × 104 ions). Data files were searched using MSFragger (v15) (46) using the Leishmania major MHOM proteome (UniProt accession no. UP000000542). Open database searches were performed, allowing modifications between −150 and 1,000 Da on the deep proteome analysis, with global.modsummary.tsv used to assess the presence of fucosylated modifications. For the detection of glycoforms within ZIC-HILIC enrichments, open database searches were performed, allowing modifications between −150 and 2,000 Da. The results from the ZIC-HILIC open searches were combined within R, and only assignments with an MSFragger expectation value of <0.001 and a delta mass of >140 Da were retained for analysis.
Data availability.
All MS data, search results, and R scripts have been deposited in the PRIDE ProteomeXchange Consortium repository (83, 84) with identifiers PXD015966 and PXD035738.
ACKNOWLEDGMENTS
We acknowledge Igor Almeida for discussions and Deborah Dobson and Suzanne Hickerson for sharing preliminary data on the LPG status and mouse infectivity of L. major Fn Δlpg2–. We thank the Melbourne Mass Spectrometry and Proteomics Facility of The Bio21 Molecular Science and Biotechnology Institute at The University of Melbourne for the support of mass spectrometry analysis. We thank Greg Matlashewski for providing plasmids used in CRISPR/Cas9 editing and the members of our group for discussions.
Funding was provided by R01 AI031078 as well as the NIAID R01 research supplement to promote diversity in health-related research (G.P.). This work was supported by National Health and Medical Research Council of Australia (NHMRC) project grants awarded to N.E.S. (APP1100164).
G.P. – investigation, data analysis, writing manuscript. N.E.S. – investigation, data analysis, resources, funding. L.F.L. – investigation. S.M.B. – data analysis, writing, resources, funding.
Footnotes
Supplemental material is available online only.
Contributor Information
Stephen M. Beverley, Email: stephen.beverley@wustl.edu.
Sumiti Vinayak, University of Illinois at Urbana Champaign.
REFERENCES
- 1.Alvar J, Vélez ID, Bern C, Herrero M, Desjeux P, Cano J, Jannin J, Boer M, the WHO Leishmaniasis Control Team . 2012. Leishmaniasis worldwide and global estimates of its incidence. PLoS One 7:e35671. doi: 10.1371/journal.pone.0035671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pigott DM, Bhatt S, Golding N, Duda KA, Battle KE, Brady OJ, Messina JP, Balard Y, Bastien P, Pratlong F, Brownstein JS, Freifeld CC, Mekaru SR, Gething PW, George DB, Myers MF, Reithinger R, Hay SI. 2014. Global distribution maps of the leishmaniases. Elife 3:e28051. doi: 10.7554/eLife.02851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banuls AL, Bastien P, Pomares C, Arevalo J, Fisa R, Hide M. 2011. Clinical pleiomorphism in human leishmaniases, with special mention of asymptomatic infection. Clin Microbiol Infect 17:1451–1461. doi: 10.1111/j.1469-0691.2011.03640.x. [DOI] [PubMed] [Google Scholar]
- 4.Mannan SB, Elhadad H, Loc TTH, Sadik M, Mohamed MYF, Nam NH, Thuong ND, Hoang-Trong BL, Duc NTM, Hoang AN, Elhusseiny KM, Minh LHN, Quynh TTH, Nghia TLB, Mai Nhu Y, Tieu TM, Hirayama K, Huy NT, Hamano S. 2021. Prevalence and associated factors of asymptomatic leishmaniasis: a systematic review and meta-analysis. Parasitol Int 81:102229. doi: 10.1016/j.parint.2020.102229. [DOI] [PubMed] [Google Scholar]
- 5.Dostalova A, Volf P. 2012. Leishmania development in sand flies: parasite-vector interactions overview. Parasit Vectors 5:276. doi: 10.1186/1756-3305-5-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paape D, Prendergast CT, Price HP, Doehl JSP, Smith DF. 2020. Genetic validation of Leishmania genes essential for amastigote survival in vivo using N-myristoyltransferase as a model. Parasit Vectors 13:132. doi: 10.1186/s13071-020-3999-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duncan SM, Jones NG, Mottram JC. 2017. Recent advances in Leishmania reverse genetics: manipulating a manipulative parasite. Mol Biochem Parasitol 216:30–38. doi: 10.1016/j.molbiopara.2017.06.005. [DOI] [PubMed] [Google Scholar]
- 8.Jones NG, Catta-Preta CMC, Lima A, Mottram JC. 2018. Genetically validated drug targets in Leishmania: current knowledge and future prospects. ACS Infect Dis 4:467–477. doi: 10.1021/acsinfecdis.7b00244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Turco SJ, Späth GF, Beverley SM. 2001. Is lipophosphoglycan a virulence factor? A surprising diversity between Leishmania species. Trends Parasitol 17:223–226. doi: 10.1016/s1471-4922(01)01895-5. [DOI] [PubMed] [Google Scholar]
- 10.Beverley SM, Turco SJ. 1995. Identification of genes mediating lipophosphoglycan biosynthesis by functional complementation of Leishmania donovani mutants. Ann Trop Med Parasitol 89:11–17. doi: 10.1080/00034983.1995.11813009. [DOI] [PubMed] [Google Scholar]
- 11.Ma D, Russell DG, Beverley SM, Turco SJ. 1997. Golgi GDP-mannose uptake requires Leishmania LPG2. A member of a eukaryotic family of putative nucleotide-sugar transporters. J Biol Chem 272:3799–3805. doi: 10.1074/jbc.272.6.3799. [DOI] [PubMed] [Google Scholar]
- 12.Segawa H, Soares RP, Kawakita M, Beverley SM, Turco SJ. 2005. Reconstitution of GDP-mannose transport activity with purified Leishmania LPG2 protein in liposomes. J Biol Chem 280:2028–2035. doi: 10.1074/jbc.M404915200. [DOI] [PubMed] [Google Scholar]
- 13.Turco SJ, Descoteaux A. 1992. The lipophosphoglycan of Leishmania parasites. Annu Rev Microbiol 46:65–94. doi: 10.1146/annurev.mi.46.100192.000433. [DOI] [PubMed] [Google Scholar]
- 14.Spath GF, Epstein L, Leader B, Singer SM, Avila HA, Turco SJ, Beverley SM. 2000. Lipophosphoglycan is a virulence factor distinct from related glycoconjugates in the protozoan parasite Leishmania major. Proc Natl Acad Sci USA 97:9258–9263. doi: 10.1073/pnas.160257897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Svárovská A, Ant TH, Seblová V, Jecná L, Beverley SM, Volf P. 2010. Leishmania major glycosylation mutants require phosphoglycans (lpg2-) but not lipophosphoglycan (lpg1-) for survival in permissive sand fly vectors. PLoS Negl Trop Dis 4:e580. doi: 10.1371/journal.pntd.0000580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sacks DL, Modi G, Rowton E, Späth G, Epstein L, Turco SJ, Beverley SM. 2000. The role of phosphoglycans in Leishmania–sand fly interactions. Proc Natl Acad Sci USA 97:406–411. doi: 10.1073/pnas.97.1.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Späth GF, Garraway LA, Turco SJ, Beverley SM. 2003. The role(s) of lipophosphoglycan (LPG) in the establishment of Leishmania major infections in mammalian hosts. Proc Natl Acad Sci USA 100:9536–9541. doi: 10.1073/pnas.1530604100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang K, Barron T, Turco SJ, Beverley SM. 2004. The LPG1 gene family of Leishmania major. Mol Biochem Parasitol 136:11–23. doi: 10.1016/j.molbiopara.2004.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spath GF, Lye LF, Segawa H, Sacks DL, Turco SJ, Beverley SM. 2003. Persistence without pathology in phosphoglycan-deficient Leishmania major. Science 301:1241–1243. doi: 10.1126/science.1087499. [DOI] [PubMed] [Google Scholar]
- 20.Uzonna JE, Spath GF, Beverley SM, Scott P. 2004. Vaccination with phosphoglycan-deficient Leishmania major protects highly susceptible mice from virulent challenge without inducing a strong Th1 response. J Immunol 172:3793–3797. doi: 10.4049/jimmunol.172.6.3793. [DOI] [PubMed] [Google Scholar]
- 21.Capul AA, Barron T, Dobson DE, Turco SJ, Beverley SM. 2007. Two functionally divergent UDP-Gal nucleotide sugar transporters participate in phosphoglycan synthesis in Leishmania major. J Biol Chem 282:14006–14017. doi: 10.1074/jbc.M610869200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Capul AA, Hickerson S, Barron T, Turco SJ, Beverley SM. 2007. Comparisons of mutants lacking the Golgi UDP-galactose or GDP-mannose transporters establish that phosphoglycans are important for promastigote but not amastigote virulence in Leishmania major. Infect Immun 75:4629–4637. doi: 10.1128/IAI.00735-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spath GF, Lye LF, Segawa H, Turco SJ, Beverley SM. 2004. Identification of a compensatory mutant (Δlpg2– REV) of Leishmania major able to survive as amastigotes within macrophages without LPG2-dependent glycoconjugates and its significance to virulence and immunization strategies. Infect Immun 72:3622–3627. doi: 10.1128/IAI.72.6.3622-3627.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kébaïer C, Uzonna JE, Beverley SM, Scott P. 2006. Immunization with persistent attenuated Δlpg2 Leishmania major parasites requires adjuvant to provide protective immunity in C57BL/6 mice. Infect Immun 74:777–780. doi: 10.1128/IAI.74.1.777-780.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hong K, Ma D, Beverley SM, Turco SJ. 2000. The Leishmania GDP-mannose transporter is an autonomous, nulti-specific, hexameric complex of LPG2 subunits. Biochemistry 39:2013–2022. doi: 10.1021/bi992363l. [DOI] [PubMed] [Google Scholar]
- 26.Dobson DE, Mengeling BJ, Cilmi S, Hickerson S, Turco SJ, Beverley SM. 2003. Identification of genes encoding arabinosyltransferases (SCA) mediating developmental modifications of lipophosphoglycan required for sand fly transmission of Leishmania major. J Biol Chem 278:28840–28848. doi: 10.1074/jbc.M302728200. [DOI] [PubMed] [Google Scholar]
- 27.Garami A, Mehlert A, Ilg T. 2001. Glycosylation defects and virulence phenotypes of Leishmania mexicana phosphomannomutase and dolicholphosphate-mannose synthase gene deletion mutants. Mol Cell Biol 21:8168–8183. doi: 10.1128/MCB.21.23.8168-8183.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Turnock DC, Ferguson MAJ. 2007. Sugar nucleotide pools of Trypanosoma brucei, Trypanosoma cruzi, and Leishmania major. Eukaryot Cell 6:1450–1463. doi: 10.1128/EC.00175-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo H, Novozhilova NM, Bandini G, Turco SJ, Ferguson MAJ, Beverley SM. 2017. Genetic metabolic complementation establishes a requirement for GDP-fucose in Leishmania. J Biol Chem 292:10696–10708. doi: 10.1074/jbc.M117.778480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo H, Damerow S, Penha L, Menzies S, Polanco G, Zegzouti H, Ferguson MAJ, Beverley SM. 2021. A broadly active fucosyltransferase LmjFUT1 whose mitochondrial localization and activity are essential in parasitic Leishmania. Proc Natl Acad Sci USA 118:e2108963118. doi: 10.1073/pnas.2108963118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bandini G, Damerow S, Sempaio Guther ML, Guo H, Mehlert A, Paredes Franco JC, Beverley S, Ferguson MA. 2021. An essential, kinetoplastid-specificGDP-Fuc:β-D-Gal α-1,2-fucosyltransferase is located in the mitochondrion of Trypanosoma brucei. Elife 10:70272. doi: 10.7554/eLife.70272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rocha J, Cicéron F, de Sanctis D, Lelimousin M, Chazalet V, Lerouxel O, Breton C. 2016. Structure of Arabidopsis thaliana FUT1 reveals a variant of the GT-B class fold and provides insight into xyloglucan fucosylation. Plant Cell 28:2352–2364. doi: 10.1105/tpc.16.00519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Holgersson J, Löfling J. 2006. Glycosyltransferases involved in type 1 chain and Lewis antigen biosynthesis exhibit glycan and core chain specificity. Glycobiology 16:584–593. doi: 10.1093/glycob/cwj090. [DOI] [PubMed] [Google Scholar]
- 34.Favila MA, Geraci NS, Jayakumar A, Hickerson S, Mostrom J, Turco SJ, Beverley SM, McDowell MA. 2015. Differential impact of LPG- and PG-deficient Leishmania major mutants on the immune response of human dendritic cells. PLoS Negl Trop Dis 9:e0004238. doi: 10.1371/journal.pntd.0004238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Batth TS, Francavilla C, Olsen JV. 2014. Off-line high-pH reversed-phase fractionation for in-depth phosphoproteomics. J Proteome Res 13:6176–6186. doi: 10.1021/pr500893m. [DOI] [PubMed] [Google Scholar]
- 36.Pawar H, Renuse S, Khobragade SN, Chavan S, Sathe G, Kumar P, Mahale KN, Gore K, Kulkarni A, Dixit T, Raju R, Prasad TS, Harsha HC, Patole MS, Pandey A. 2014. Neglected tropical diseases and omics science: proteogenomics analysis of the promastigote stage of Leishmania major parasite. OMICS 18:499–512. doi: 10.1089/omi.2013.0159. [DOI] [PubMed] [Google Scholar]
- 37.Silverman JM, Clos J, de’Oliveira CC, Shirvani O, Fang Y, Wang C, Foster LJ, Reiner NE. 2010. An exosome-based secretion pathway is responsible for protein export from Leishmania and communication with macrophages. J Cell Sci 123:842–852. doi: 10.1242/jcs.056465. [DOI] [PubMed] [Google Scholar]
- 38.Skodova-Sverakova I, Horvath A, Maslov DA. 2015. Identification of the mitochondrially encoded subunit 6 of F1FO ATPase in Trypanosoma brucei. Mol Biochem Parasitol 201:135–138. doi: 10.1016/j.molbiopara.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sanchiz A, Morato E, Rastrojo A, Camacho E, Gonzalez-de la Fuente SG, Marina A, Aguado B, Requena JM. 2020. The experimental proteome of Leishmania infantum promastigote and its usefulness for improving gene annotations. Genes (Basel) 11:1036. doi: 10.3390/genes11091036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Horvath A, Nebohacova M, Lukes J, Maslov DA. 2002. Unusual polypeptide synthesis in the kinetoplast-mitochondria from Leishmania tarentolae. Identification of individual de novo translation products. J Biol Chem 277:7222–7230. doi: 10.1074/jbc.M109715200. [DOI] [PubMed] [Google Scholar]
- 41.Riley NM, Malaker SA, Driessen MD, Bertozzi CR. 2020. Optimal dissociation methods differ for N- and O-glycopeptides. J Proteome Res 19:3286–3301. doi: 10.1021/acs.jproteome.0c00218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hahne H, Gholami AM, Kuster B. 2012. Discovery of O-GlcNAc-modified proteins in published large-scale proteome data. Mol Cell Proteomics 11:843–850. doi: 10.1074/mcp.M112.019463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Swearingen KE, Eng JK, Shteynberg D, Vigdorovich V, Springer TA, Mendoza L, Sather DN, Deutsch EW, Kappe SHI, Moritz RL. 2019. A tandem mass spectrometry sequence database search method for identification of O-fucosylated proteins by mass spectrometry. J Proteome Res 18:652–663. doi: 10.1021/acs.jproteome.8b00638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rosenzweig D, Smith D, Myler PJ, Olafson RW, Zilberstein D. 2008. Post-translational modification of cellular proteins during Leishmania donovani differentiation. Proteomics 8:1843–1850. doi: 10.1002/pmic.200701043. [DOI] [PubMed] [Google Scholar]
- 45.Chick JM, Kolippakkam D, Nusinow DP, Zhai B, Rad R, Huttlin EL, Gygi SP. 2015. A mass-tolerant database search identifies a large proportion of unassigned spectra in shotgun proteomics as modified peptides. Nat Biotechnol 33:743–749. doi: 10.1038/nbt.3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kong AT, Leprevost FV, Avtonomov DM, Mellacheruvu D, Nesvizhskii AI. 2017. MSFragger: ultrafast and comprehensive peptide identification in mass spectrometry-based proteomics. Nat Methods 14:513–520. doi: 10.1038/nmeth.4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scott NE, Cordwell SJ. 2015. Enrichment and identification of bacterial glycopeptides by mass spectrometry. Methods Mol Biol 1295:355–368. doi: 10.1007/978-1-4939-2550-6_25. [DOI] [PubMed] [Google Scholar]
- 48.Mysling S, Palmisano G, Hojrup P, Thaysen-Andersen M. 2010. Utilizing ion-pairing hydrophilic interaction chromatography solid phase extraction for efficient glycopeptide enrichment in glycoproteomics. Anal Chem 82:5598–5609. doi: 10.1021/ac100530w. [DOI] [PubMed] [Google Scholar]
- 49.Olafson RW, Thomas JR, Ferguson MA, Dwek RA, Chaudhuri M, Chang KP, Rademacher TW. 1990. Structures of the N-linked oligosaccharides of Gp63, the major surface glycoprotein, from Leishmania mexicana amazonensis. J Biol Chem 265:12240–12247. doi: 10.1016/S0021-9258(19)38336-X. [DOI] [PubMed] [Google Scholar]
- 50.Mule SN, Saad JS, Fernandes LR, Stolf BS, Cortez M, Palmisano G. 2020. Protein glycosylation in Leishmania spp. Mol Omics 16:407–424. doi: 10.1039/d0mo00043d. [DOI] [PubMed] [Google Scholar]
- 51.Funk VA, Thomas-Oates JE, Kielland SL, Bates PA, Olafson RW. 1997. A unique, terminally glucosylated oligosaccharide is a common feature on Leishmania cell surfaces. Mol Biochem Parasitol 84:33–48. doi: 10.1016/s0166-6851(96)02780-6. [DOI] [PubMed] [Google Scholar]
- 52.Parodi AJ, Martin-Barrientos J, Engel JC. 1984. Glycoprotein assembly in Leishmania mexicana. Biochem Biophys Res Commun 118:1–7. doi: 10.1016/0006-291x(84)91058-1. [DOI] [PubMed] [Google Scholar]
- 53.Zubarev RA. 2013. The challenge of the proteome dynamic range and its implications for in-depth proteomics. Proteomics 13:723–726. doi: 10.1002/pmic.201200451. [DOI] [PubMed] [Google Scholar]
- 54.Becco L, Smircich P, Garat B. 2019. Conserved motifs in nuclear genes encoding predicted mitochondrial proteins in Trypanosoma cruzi. PLoS One 14:e0215160. doi: 10.1371/journal.pone.0215160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mi H, Muruganujan A, Huang X, Ebert D, Mills C, Guo X, Thomas PD. 2019. Protocol update for large-scale genome and gene function analysis with the PANTHER classification system (v.14.0). Nat Protoc 14:703–721. doi: 10.1038/s41596-019-0128-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sadlova J, Price HP, Smith BA, Votypka J, Volf P, Smith DF. 2010. The stage-regulated HASPB and SHERP proteins are essential for differentiation of the protozoan parasite Leishmania major in its sand fly vector, Phlebotomus paptasi. Cell Microbiol 12:1765–1779. doi: 10.1111/j.1462-5822.2010.01507.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bandini G, Mariño K, Güther MLS, Wernimont AK, Kuettel S, Qiu W, Afzal S, Kelner A, Hui R, Ferguson MAJ. 2012. Phosphoglucomutase is absent in Trypanosoma brucei and redundantly substituted by phosphomannomutase and phospho-N-acetylglucosamine mutase. Mol Microbiol 85:513–534. doi: 10.1111/j.1365-2958.2012.08124.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fang J, Beattie DS. 2002. Novel FMN-containing rotenone-insensitive NADH dehydrogenase from Trypanosoma brucei mitochondria: isolation and characterization. Biochemistry 41:3065–3072. doi: 10.1021/bi015989w. [DOI] [PubMed] [Google Scholar]
- 59.Zorova LD, Popkov VA, Plotnikov EY, Silachev DN, Pevzner IB, Jankauskas SS, Babenko VA, Zorov SD, Balakireva AV, Juhaszova M, Sollott SJ, Zorov DB. 2018. Mitochondrial membrane potential. Anal Biochem 552:50–59. doi: 10.1016/j.ab.2017.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Duarte M, Ferreira C, Khandpur GK, Flohr T, Zimmermann J, Castro H, Herrmann JM, Morgan B, Tomas AM. 2021. Leishmania type II dehydrogenase is essential for parasite viability irrespective of the presence of an active complex I. Proc Natl Acad Sci USA 118:e2103803118. doi: 10.1073/pnas.2103803118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.de Souza SSA, Catta-Preta CM, Alves JMP, Cavalcanti DP, Teixeira MMG, Camargo EP, De Souza W, Silva R, Motta MCM. 2017. Expanded repertoire of kinetoplast associated proteins and unique mitochondrial DNA arrangement of symbiont-bearing trypanosomatids. PLoS One 12:e0187516. doi: 10.1371/journal.pone.0187516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lukes J, Hines JC, Evans CJ, Avliyakulov NK, Prabhu VP, Chen J, Ray DS. 2001. Disruption of the Crithidia fasciculata KAP1 gene results in structural rearrangement of the kinetoplast disc. Mol Biochem Parasitol 117:179–186. doi: 10.1016/s0166-6851(01)00348-6. [DOI] [PubMed] [Google Scholar]
- 63.Beck K, Acestor N, Schulfer A, Anupama A, Carnes J, Panigrahi AK, Stuart K. 2013. Trypanosoma brucei Tb927.2.6100 is an essential protein associated with kinetoplast DNA. Eukaryot Cell 12:970–978. doi: 10.1128/EC.00352-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Peña-Diaz P, Mach J, Kriegová E, Poliak P, Tachezy J, Lukeš J. 2018. Trypanosomal mitochondrial intermediate peptidase does not behave as a classical mitochondrial processing peptidase. PLoS One 13:e0196474. doi: 10.1371/journal.pone.0196474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Paes LS, Suárez Mantilla B, Zimbres FM, Pral EMF, Diogo de Melo P, Tahara EB, Kowaltowski AJ, Elias MC, Silber AM. 2013. Proline dehydrogenase regulates redox state and respiratory metabolism in Trypanosoma cruzi. PLoS One 8:e69419. doi: 10.1371/journal.pone.0069419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Patino LH, Muskus C, Ramírez JD. 2019. Transcriptional responses of Leishmania (Leishmania) amazonensis in the presence of trivalent sodium stibogluconate. Parasit Vectors 12:348. doi: 10.1186/s13071-019-3603-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Padmanabhan PK, Zghidi-Abouzid O, Samant M, Dumas C, Aguiar BG, Estaquier J, Papadopoulou B. 2016. DDX3 DEAD-box RNA helicase plays a central role in mitochondrial protein quality control in Leishmania. Cell Death Dis 7:e2406. doi: 10.1038/cddis.2016.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Voos W. 2013. Chaperone-protease networks in mitochondrial protein homeostasis. Biochim Biophys Acta 1833:388–399. doi: 10.1016/j.bbamcr.2012.06.005. [DOI] [PubMed] [Google Scholar]
- 69.Xu W, Xin L, Soong L, Zhang K. 2011. Sphingolipid degradation by Leishmania major is required for its resistance to acidic pH in the mammalian host. Infect Immun 79:3377–3387. doi: 10.1128/IAI.00037-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zacharias LG, Hartmann AK, Song E, Zhao J, Zhu R, Mirzaei P, Mechref Y. 2016. HILIC and ERLIC enrichment of glycopeptides derived from breast and brain cancer cells. J Proteome Res 15:3624–3634. doi: 10.1021/acs.jproteome.6b00429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Illiano A, Pinto G, Melchiorre C, Carpentieri A, Faraco V, Amoresano A. 2020. Protein glycosylation investigated by mass spectrometry: an overview. Cells 9:1986. doi: 10.3390/cells9091986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Azevedo C, Livermore T, Saiardi A. 2015. Protein polyphosphorylation of lysine residues by inorganic polyphosphate. Mol Cell 58:71–82. doi: 10.1016/j.molcel.2015.02.010. [DOI] [PubMed] [Google Scholar]
- 73.Alves MJM, Kawahara R, Viner R, Colli W, Mattos EC, Thaysen-Andersen M, Larsen MR, Palmisano G. 2017. Comprehensive glycoprofiling of the epimastigote and trypomastigote stages of Trypanosoma cruzi. J Proteomics 151:182–192. doi: 10.1016/j.jprot.2016.05.034. [DOI] [PubMed] [Google Scholar]
- 74.Riley NM, Bertozzi CR, Pitteri SJ. 2021. A pragmatic guide to enrichment strategies for mass spectrometry-based glycoproteomics. Mol Cell Proteomics 20:100029. doi: 10.1074/mcp.R120.002277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Beneke T, Demay F, Hookway E, Ashman N, Jeffery H, Smith J, Valli J, Becvar T, Myskova J, Lestinova T, Shafiq S, Sadlova J, Volf P, Wheeler RJ, Gluenz E. 2019. Genetic dissection of a Leishmania flagellar proteome demonstrates requirement for directional motility in sand fly infections. PLoS Pathog 15:e1007828. doi: 10.1371/journal.ppat.1007828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang W-W, Matlashewski G. 2015. CRISPR-Cas9-mediated genome editing in Leishmania donovani. mBio 6:e00861-15. doi: 10.1128/mBio.00861-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Humphrey SJ, Azimifar SB, Mann M. 2015. High-throughput phosphoproteomics reveals in vivo insulin signaling dynamics. Nat Biotechnol 33:990–995. doi: 10.1038/nbt.3327. [DOI] [PubMed] [Google Scholar]
- 78.Batth TS, Olsen JV. 2016. Offline high pH reversed-phase peptide fractionation for deep phosphoproteome coverage. Methods Mol Biol 1355:179–192. doi: 10.1007/978-1-4939-3049-4_12. [DOI] [PubMed] [Google Scholar]
- 79.Cox J, Mann M. 2008. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 80.Cox J, Hein MY, Luber CA, Paron I, Nagaraj N, Mann M. 2014. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol Cell Proteomics 13:2513–2526. doi: 10.1074/mcp.M113.031591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tyanova S, Temu T, Sinitcyn P, Carlson A, Hein MY, Geiger T, Mann M, Cox J. 2016. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat Methods 13:731–740. doi: 10.1038/nmeth.3901. [DOI] [PubMed] [Google Scholar]
- 82.Tyanova S, Cox J. 2018. Perseus: a bioinformatics platform for integrative analysis of proteomics data in cancer research. Methods Mol Biol 1711:133–148. doi: 10.1007/978-1-4939-7493-1_7. [DOI] [PubMed] [Google Scholar]
- 83.Perez-Riverol Y, Csordas A, Bai J, Bernal-Llinares M, Hewapathirana S, Kundu DJ, Inuganti A, Griss J, Mayer G, Eisenacher M, Perez E, Uszkoreit J, Pfeuffer J, Sachsenberg T, Yilmaz S, Tiwary S, Cox J, Audain E, Walzer M, Jarnuczak AF, Ternent T, Brazma A, Vizcaino JA. 2019. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res 47:D442–D450. doi: 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Vizcaino JA, Csordas A, del-Toro N, Dianes JA, Griss J, Lavidas I, Mayer G, Perez-Riverol Y, Reisinger F, Ternent T, Xu QW, Wang R, Hermjakob H. 2016. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res 44:D447–D456. doi: 10.1093/nar/gkv1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Download spectrum.03052-22-s0001.xlsx, XLSX file, 1.6 MB (1.6MB, xlsx)
Table S2. Download spectrum.03052-22-s0002.xlsx, XLSX file, 0.2 MB (254.4KB, xlsx)
Table S3. Download spectrum.03052-22-s0003.xlsx, XLSX file, 1.4 MB (1.4MB, xlsx)
Table S4. Download spectrum.03052-22-s0004.xlsx, XLSX file, 0.5 MB (476.2KB, xlsx)
Supplemental material. Download spectrum.03052-22-s0005.pdf, PDF file, 0.4 MB (404KB, pdf)
Data Availability Statement
All MS data, search results, and R scripts have been deposited in the PRIDE ProteomeXchange Consortium repository (83, 84) with identifiers PXD015966 and PXD035738.