ABSTRACT
The molecular detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is key for clinical management and surveillance. Funded by the European Centre for Disease Prevention and Control, we conducted an external quality assessment (EQA) on the molecular detection and variant typing of SARS-CoV-2 that included 59 European laboratories in 34 countries. The EQA panel consisted of 12 lyophilized inactivated samples, 10 of which were SARS-CoV-2 variants (Alpha, Beta, Gamma, Delta, Epsilon, Eta, parental B.1 strain) ranging from 2.5 to 290.0 copies/μL or pooled respiratory viruses (adenovirus, enterovirus, influenza virus A, respiratory syncytial virus, or human coronaviruses 229E and OC43). Of all participants, 72.9% identified the presence of SARS-CoV-2 RNA correctly. In samples containing 25.0 or more genome copies/μL, SARS-CoV-2 was detected by 98.3% of the participating laboratories. Laboratories applying commercial tests scored significantly better (P < 0.0001, Kruskal-Wallis test) than those using in-house assays. Both the molecular detection and the typing of the SARS-CoV-2 variants were associated with the RNA concentrations (P < 0.0001, Kruskal-Wallis test). On average, only 5 out of the 10 samples containing different SARS-CoV-2 variants at different concentrations were correctly typed. The identification of SARS-CoV-2 variants was significantly more successful among EQA participants who combined real-time reverse transcription polymerase chain reaction (RT-PCR)-based assays for mutation detection and high-throughput genomic sequencing than among those who used a single methodological approach (P = 0.0345, Kruskal-Wallis test). Our data highlight the high sensitivity of SARS-CoV-2 detection in expert laboratories as well as the importance of continuous assay development and the benefits of combining different methodologies for accurate SARS-CoV-2 variant typing.
KEYWORDS: SARS-CoV-2, variants, COVID-19, external quality assessment, molecular diagnostics, Europe
INTRODUCTION
As of mid-August of 2022, severe acute respiratory syndrome-coronavirus 2 (SARS-CoV-2) has infected over 594 million people and has caused more than 6.4 million deaths, globally (1). The diagnostic gold standard for the detection of SARS-CoV-2 is semiquantitative real-time reverse transcription-PCR (qRT-PCR), which is widely applied in clinical management and surveillance (2). Beyond the detection of infected individuals, the diagnostic identification of specific mutations that are correlated with specific SARS-CoV-2 variants of concern or interest (VOC/VOI) and the genomic surveillance of SARS-CoV-2 has become increasingly important, as several VOCs have emerged during the pandemic (3). This was recently illustrated by the emergence and rapid spread of the Omicron variants (4). Conducting external quality assessments (EQA) is an established means by which to improve and support diagnostic accuracy (5). In 2020, an EQA on molecular SARS-CoV-2 detection among European laboratories organized by us showed limited sensitivity at low concentrations (5). Here, we conducted a follow-up EQA to assess whether laboratories maintained or improved their diagnostic testing performance and to assess the capability to type SARS-CoV-2 variants in samples containing low RNA concentrations.
MATERIALS AND METHODS
In June of 2021, laboratories involved in the coronavirus disease (COVID-19) response and/or laboratories which were part of the Emerging Viral Diseases-Expert Laboratory Network (EVD-LabNet) (6) and/or the national reference laboratories involved in the COVID-19 response were invited to participate in the 2nd SARS-CoV-2 EQA. The full list of participating laboratories is shown in the Acknowledgments.
Panel composition.
The EQA panel contained 12 samples. 10 samples contained inactivated material of different SARS-CoV-2 lineages (Alpha, Beta, Gamma, Delta, Epsilon, Eta variants, and the early B.1 isolate) (7), and 2 samples contained inactivated material of other respiratory viruses to evaluate specificity (Table 1). Of the 10 SARS-CoV-2 samples, 4 contained the Alpha variant (B.1.1.7) in various concentrations to evaluate sensitivity. The Alpha variant was chosen for the assessments of sensitivity because it was the predominant variant in Europe at the time of panel production. The panels were produced at Charité-Universitätsmedizin Berlin at low SARS-CoV-2 viral concentrations to capture molecular detection in patients with low virus concentrations, such as those in the late stages of infection or in specimens that had deteriorated (e.g., because they were taken and stored in resource-limited settings). An optional SARS-CoV-2 variant identification activity was offered during the EQA in addition to the mandatory identification of SARS-CoV-2 positive and SARS-CoV-2 negative samples.
TABLE 1.
EQA panel composition and performance of 59 participating laboratoriesa
| Correct molecular results |
Correct variant results |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Sample ID | Virus | Lineage | WHO label | Mutations | Copies/μL | % | No./total | % | No./total |
| 1 | Adenovirus | n.a. | n.a. | 5,000.0 | 98.3 | 58/59 | n.a. | n.a. | |
| Enterovirus B (Echovirus 6) | Arslan | 5,000.0 | |||||||
| Influenzavirus A | H1N1 pdm09 | 5,000.0 | |||||||
| Respiratory syncytial virus | 2,000.0 | ||||||||
| 2 | Human coronavirus 229E | n.a. | 5,000.0 | 98.3 | 58/59 | n.a. | n.a. | ||
| Human coronavirus OC43 | 5,000.0 | ||||||||
| 12 | SARS-CoV-2 | B.1.1.7 | Alpha | H69/V70 del Y144 del N501Y A570D P681H T716I S982A D1118H |
290.0 | 100.0 | 59/59 | 83.3 | 30/36 |
| 6 | SARS-CoV-2 | 11.0 | 91.5 | 54/59 | 52.8 | 19/36 | |||
| 11 | SARS-CoV-2 | 3.0 | 96.6 | 57/59 | 61.1 | 22/36 | |||
| 3 | SARS-CoV-2 | 2.5 | 72.9 | 43/59 | 27.8 | 10/36 | |||
| 10 | SARS-CoV-2 | B.1.351 | Beta | L18F D80A D215G L241/L242/A243 del K417N E484K N501Y A701V |
25.0 | 100.0 | 59/59 | 58.3 | 21/36 |
| 7 | SARS-CoV-2 | P.1 | Gamma | L18F T20N P26S D138Y R190S K417T E484K N501Y H655Y T1027I V1176F |
25.0 | 100.0 | 59/59 | 41.7 | 15/36 |
| 9 | SARS-CoV-2 | B.1.617 | Delta | T19R E156/F157 del R158G L452R T478K P681R D950N |
50.0 | 98.3 | 58/59 | 52.8 | 19/36 |
| 5 | SARS-CoV-2 | B.1.429 | Epsilon | S13I W152C L452R |
200.0 | 100.0 | 59/59 | 33.3 | 12/36 |
| 4 | SARS-CoV-2 | B.1.525 | Eta | Q52R A67V H69/V70 del Y144 del E484K Q677H F888L |
200.0 | 98.3 | 58/59 | 44.4 | 16/36 |
| 8 | SARS-CoV-2 | B.1 | Early isolateb | n.a. | 100.0 | 100.0 | 59/59 | 66.7 | 24/36 |
n.a., not applicable.
B.1 lineage isolate in circulation in the beginning of 2020 and containing none of the later, lineage-defining single nucleotide polymorphisms or deletions.
Panel preparation, validation, and dispatch.
All EQA samples were isolated from clinical specimens and cultured in Dulbecco's Modified Eagle’s Medium (DMEM) supplemented with 5.0% fetal calf serum (FCS), 1.0% nonessential amino acids, and 1.0% penicillin/streptomycin (100.0 U/mL). The samples were diluted to the desired concentrations in phosphate-buffered saline (PBS), and 500 μL of each sample were freeze-dried in screw cap tubes after heat inactivation (65.0°C, 4 h for SARS-CoV-2 and adenovirus, 2 h for all other viruses). The freeze-dried samples were shipped at ambient temperature.
To confirm the successful inactivation of SARS-CoV-2, the resuspended samples were passaged three times consecutively in cell culture. For this purpose, Vero cells were seeded to 80.0% confluence in 6-well plates. The cell culture medium was discarded, and 500 μL of the resuspended sample were added per well. The cells were inoculated for 1 h at 37.0°C and 5.0% CO2. Afterwards, the inoculum was removed, and the cells were washed twice using 1.0 mL PBS and incubated for 7 days with 1.0 mL of fresh DMEM supplemented with 5.0% FCS, 1.0% nonessential amino acids, and 1.0% penicillin/streptomycin (100.0 U/mL) added per well. The cells were checked daily for cytopathic effects. 7 days post-infection, 500 μL of the supernatant were used for the inoculation of new cells, and the remainder was tested for the presence of SARS-CoV-2 RNA via real-time RT-PCR.
The SARS-CoV-2 samples were quantified using an E-gene-based, in-house assay that was recommended by the World Health Organization (WHO) (8), relying on the EURM-019 single-stranded-RNA Joint Research Centre (JRC) standard for the molecular quantification of SARS-CoV-2 (9). The EQA panel was validated independently by two reference laboratories before distribution. These laboratories had no prior knowledge of the composition of the EQA panels and tested one EQA panel each.
Data collection.
Together with the EQA panels, participants received reconstitution and testing instructions and were asked to report on SARS-CoV-2 detection in an obligatory first step, and they were asked to perform variant typing voluntarily as an additional step. Result reporting was done via a submission form that participants received via email. They were asked to categorize their samples (as positive, negative, or inconclusive) and to provide additional information on cycle threshold (Ct) values, extraction methods and the types of RT-PCR(s) that were used. Additionally, the participants could report the determined SARS-CoV-2 variant (by WHO label or by Pango lineage), signature mutation(s), or both, along with the details of the method used. The latter was an optional activity and was evaluated separately from the results of the molecular detection. Sequences were neither collected nor evaluated; only the participants’ conclusions, as reported, were analyzed.
Evaluation of results.
The following scoring system was used for the molecular detection of SARS-CoV-2 RNA: correctly reported samples (presence or absence of SARS-CoV-2 RNA) were assessed with 0 points, inconclusive results were scored with 1 point, and false results were scored with 2 points. For the analysis of VOC/mutation testing, we differentiated between correctly determined variants/signature mutations, identification as VOC/VOI, and false typing. The determination of the signature mutation(s), the naming of the variant under any classification (for example as “Alpha” or “B.1.1.7”), or both was considered to be correct. Generalized responses (e.g., “variant of concern”) or the identification of mutations shared by VOCs/VOIs, were considered to indicate an identification as a VOC/VOI. Classifications that were considered to be false included either incorrectly identified variants or a failure to type.
Statistical analysis.
Statistical analyses were conducted using R version 4.0.2. All variables were tested for normality using the Shapiro-Wilk test, and further statistical testing was done using the appropriate tests. Each component of the diagnostic workflow can affect the diagnostic performance, including the extraction of nucleic acids (NA) (10) and the molecular test, such as real-time RT-PCR itself (5, 11). To analyze the effect of specific components of the diagnostic workflow, and because the entire workflows were highly diverse among the participating laboratories, further analyses were conducted for the individual steps of the testing process, such as RNA extraction and PCR targets. To analyze the concentration changes of NAs during extraction, we considered the reported volumes for sample reconstitution, input volumes and elution volumes, and eluate volumes used for the assays.
RESULTS
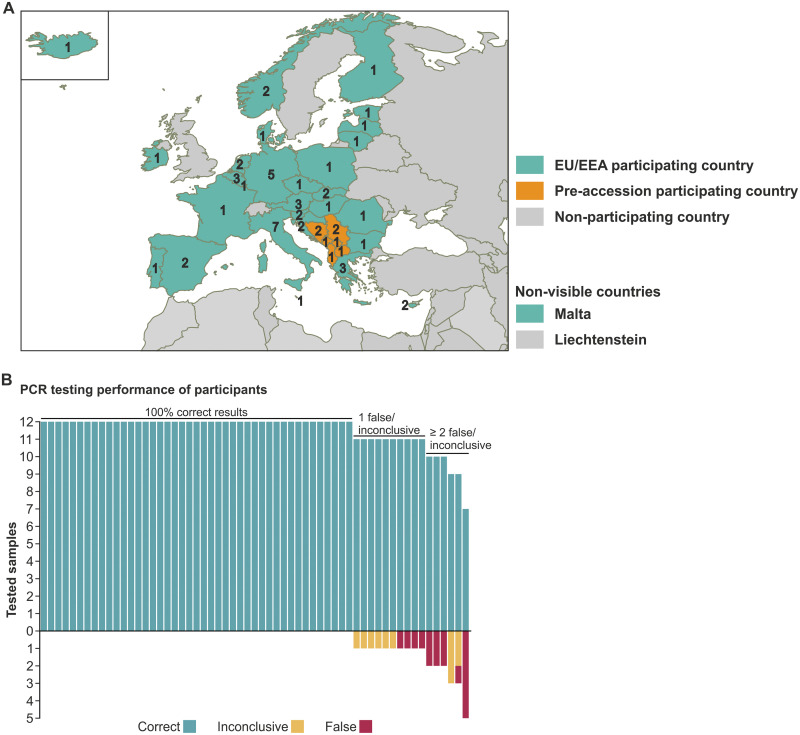
59 expert laboratories from 34 countries, covering 28 of the 30 EU/EEA countries and 6 of the 7 EU pre-accession countries, reported EQA results and technical details on testing within the scheduled time frame of 1 month (Fig. 1A).
FIG 1.
Participating laboratories and overview of laboratory performance. (A) Map of participating laboratories by country. (B) Overview of molecular SARS-CoV-2 testing by laboratory.
Molecular detection of SARS-CoV-2.
The majority of the participating laboratories, 72.9% (43/59), identified all 12 samples correctly as positive or negative for SARS-CoV-2 (Fig. 1B). 16.9% (10/59) reported correct results for 11 samples, and 10.2% (6/59) reported correct results for 10 samples or fewer. From all of the results reported, 2.3% (16/708) were incorrect. These incorrect results were exclusively reported for the SARS-CoV-2 containing samples (16/590, 2.7%), while both of the specificity control samples were either reported correctly (116/118, 98.3%) or inconclusive (2/118, 1.7%). Among the samples containing SARS-CoV-2, 1.5% (9/590) of the results were reported as inconclusive, yielding a total percentage of inconclusive results of 1.6% (11/708) (Table 1).
The risk for both false-negative SARS-CoV-2 detection and, later on, to inability to type increased significantly with lower SARS-CoV-2 RNA concentrations (P < 0.0001, Kruskal-Wallis test). Accordingly, the Ct values of positively tested samples were higher with decreasing SARS-CoV-2 RNA concentrations (Fig. 2).
FIG 2.
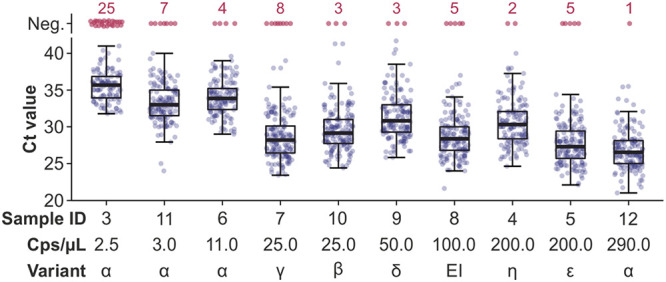
Performance of SARS-CoV-2 detection among different RNA concentrations and SARS-CoV-2 lineages. Reported Ct values by SARS-CoV-2-positive sample, considering all results reported and including multiple tests conducted by a given participant. The median Ct values are indicated by bars, quartiles by boxes, and interquartile ranges by whiskers. EI, early isolate (B.1).
Compared to an EQA on the molecular detection of SARS-CoV-2 in June and July of 2020 (5), which applied the same scoring system to evaluate the EQA performance of participants and was also composed of 12 samples, the capability to detect SARS-CoV-2 has significantly improved in this EQA, based on the overall performance score per laboratory (P = 0.0002, chi-square test). On average, the laboratories that participated in the first ECDC EQA performed slightly better in this EQA compared to the laboratories that did not (mean score 0.65 versus 0.91), although that difference was not statistically significant (P = 0.2413, Mann-Whitney U test).
Similar to the previous SARS-CoV-2 EQA done on the European level (5), the overall score-based performance was not significantly different between laboratories conducting automated NA extraction and laboratories conducting manual RNA extraction (P = 0.3033, Mann-Whitney U test). During NA extraction, the NA concentration is commonly increased due to the smaller elution volume. However, the diagnostic sensitivity among participating laboratories was not significantly correlated with the extent of NA concentration change during the extraction process (P value = 0.2677, Spearman correlation test). Beyond NA extraction, NA detection is another crucial step in molecular diagnostics. Transcription in coronaviruses typically varies among genomic and subgenomic regions, which may cause different sensitivities among assays that target different genomic sites (12). The diagnostic targets among the applied tests in this EQA were E (37 laboratories), N (35 laboratories), RdRp (29 laboratories), ORF1 (25 laboratories), S (22 laboratories), ORF8 (3 laboratories), ns2 (2 laboratories), and M genes (1 laboratory). On average, the participants applied two (multiplex) assays, in total conducting four PCRs and targeting three genes for each sample. 10 laboratories tested for targets on 1 gene, 14 on two genes, 17 on three genes, 11 on four genes, 3 on 5 genes, and 1 on 6 genes. Statistically, the diagnostic performance did not differ significantly among assays targeting different regions (P = 0.5714, Kruskal-Wallis test). Additionally, the performance of participants was not correlated with the number of different targets or RT-PCR assays they applied (P = 0.6517, Spearman correlation test). A wide range of commercial and in-house real-time SARS-CoV-2 RT-PCR assays is available. In this EQA, the score-based performance of laboratories applying commercial tests was significantly better (P < 0.0001, Kruskal-Wallis test) that that of laboratories applying in-house tests, resulting in a mean score per sample of 0.06 with commercial tests and 0.17 with in-house tests.
SARS-CoV-2 variant typing.
Of all 59 laboratories participating in the EQA, 36 provided information on variant typing as an optional activity. Only one laboratory reported fully correct results for all 10 samples, whereas 36.1% (13/36) reported correct results for at least 7 of the 10 SARS-CoV-2 positive samples and 61.1% (22/36) reported correct results for 6 or fewer of the SARS-CoV-2 positive samples (Table 1). On average, the laboratories typed 5 of the 10 samples correctly.
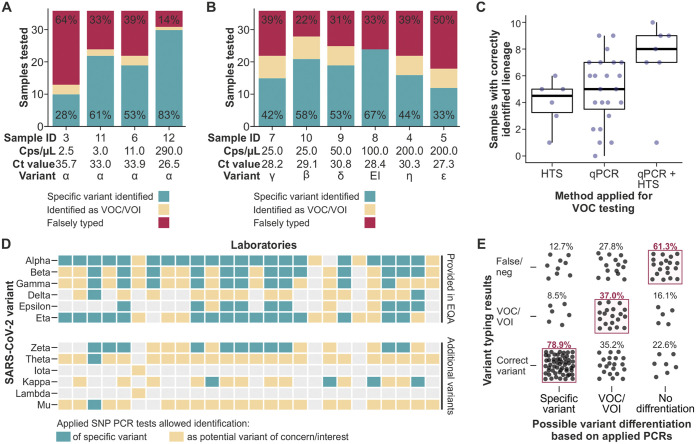
In general, the VOC/mutation testing was more accurate for samples with higher concentrations than for those with lower concentrations (Table 1; Fig. 3A and 3B). Among samples from the Alpha variant, the only variant which was represented at different concentrations, the VOC/mutation testing performance was significantly correlated with the RNA concentration (P < 0.0001, Spearman correlation test) (Fig. 3A). The sample which had the highest concentration of 290.0 copies/μL was tested correctly for VOC/mutations by 83.3% of the participants. Multiple labs submitted no answer, as expected for low-concentration samples that are not easily typed.
FIG 3.
Performance of optional SARS-CoV-2 variant/mutation identification among different RNA concentrations and SARS-CoV-2 lineages. (A) Reported VOC/mutation testing results for different SARS-CoV-2 RNA concentrations of the Alpha (α) variant. The percentage of correctly determined variants is indicated in green. EI, early isolate. (B) Reported VOC/mutation testing results for different SARS-CoV-2 RNA concentrations and variants. The percentage of correctly determined variants is indicated in green. (C) Correctly determined SARS-CoV-2 variants among laboratories using different methods for VOC/mutation typing. The percentage of correctly determined variants is indicated. (D) Conducted assays for the identification of specific VOCs/VOIs. (E) Variant typing results, stratified by the suitability of the conducted typing assays. Suitability was defined as the application of PCR(s) that were able to identify the variants present in the EQA panel. SNP, single-nucleotide polymorphism. neg, negative.
For the VOC/mutation testing, real-time RT-PCR, high-throughput sequencing (HTS), and conventional PCR with subsequent Sanger sequencing were applied (Table 2). 7 of the 36 laboratories providing details on their testing processes applied at least two of those methods, with real-time RT-PCR being the most frequently applied (31/36). The number of correctly typed variants was significantly associated with the applied methods (P = 0.0345, Kruskal-Wallis test) (Fig. 3C). The average number of correctly typed variants was 4.9 among laboratories applying real-time RT-PCR tests, 4.0 among laboratories applying HTS, and 7.3 among laboratories applying both real-time RT-PCR and HTS (Table 2). In total, 54% of the samples that were neither typed correctly nor identified as a VOC/VOI were tested negative, likely due to the low RNA concentrations of the provided samples.
TABLE 2.
Performance of different methods for variant typing among 36 laboratories participating in an optional EQA typing activity
| Applied method for VOC/mutation testinga | Mean correctly typed samples (95% CI) | Participants |
|---|---|---|
| Real-time RT-PCR | 4.9 (3.9 to 5.9) | 23 |
| HTS | 4.0 (2.7 to 5.3) | 6 |
| Real-time RT-PCR + HTS | 7.3 (5.3 to 9.3) | 7 |
aVOC, variant of concern; RT-PCR, reverse transcription PCR; HTS, high-throughput sequencing.
Among those laboratories applying real-time RT-PCR assays for VOC testing, 18 different mutations were targeted, with S:N501 and S:E484 being the most frequently tested and targeted (by 63.3% of the reporting laboratories) (Table 3). The number of tested mutations did not correlate with the number of correctly determined variants (P value = 0.9316, Spearman correlation test). Notably, the combination of real-time RT-PCR assays applied for VOC/VOI testing varied greatly, and most of the laboratories were not able to differentiate all of the provided variants, based on the available real-time RT-PCR assays (Fig. 3D). The variants that could be identified exactly using real-time RT-PCR by most laboratories were Alpha (80.8%), Eta (61.5%), and Beta (50.0%). The identification of EQA samples as the correct variant or as a potential VOC/VOI was significantly correlated with the choice of a suitable set of tested mutations (P value = 0.0084, Spearman correlation test) (Fig. 3E). Among the laboratories that applied real-time RT-PCR assays which allowed the correct identification of a given variant, 78.9% typed the variant correctly. In contrast, 61.3% of the laboratories that applied real-time RT-PCR assays that were suitable neither to determine the correct variant of a given sample nor to identify it as potential VOC/VOI typed the samples incorrectly.
TABLE 3.
Single nucleotide polymorphisms and deletions detected by 30 laboratoriesa reporting details on SARS-CoV-2 mutation testing
| Tested SNPb | Variants detectable |
Frequency of use |
||
|---|---|---|---|---|
| Pango lineage | WHO label | % | No./total | |
| S:N501 | B.1.1.7 B.1.351 P.1 |
Alpha Beta Gamma |
63.3 | 19/30 |
| S:E484 | B.1.351 B.1.525 P.1 |
Beta Eta Gamma |
63.3 | 19/30 |
| S:ΔH69-V70 | B.1.1.7 B.1.525 |
Alpha Eta |
60.0 | 18/30 |
| S:L452 | B.1.617 B.1.429 |
Delta Epsilon |
40.0 | 12/30 |
| S:K417 | B.1.351 P.1 |
Beta Gamma |
36.7 | 11/30 |
| S:W152 | B.1.429 | Epsilon | 23.3 | 7/30 |
| S:P681 | B.1.1.7 B.1.617 |
Alpha Delta |
20.0 | 6/30 |
| S:A570 | B.1.1.7 | Alpha | 10.0 | 3/30 |
| ORF1a:ΔS3675-3677 | B.1.1.7 B.1.351 |
Alpha Beta |
6.7 | 2/30 |
| S:V1176 | P.1 | Gamma | 6.7 | 2/30 |
| S:Δ242-244 | B.1.351 | Beta | 3.3 | 1/30 |
| S:ΔY144 | B.1.1.7 B.1.525 |
Alpha Eta |
3.3 | 1/30 |
| S:H655 | P.1 | Gamma | 3.3 | 1/30 |
| S:A701 | B.1.351 | Beta | 3.3 | 1/30 |
| S:Q677 | B.1.525 | Eta | 3.3 | 1/30 |
| S:L452 | B.1.617 B.1.429 |
Delta Epsilon |
3.3 | 1/30 |
| S:T478 | B.1.617 | Delta | 3.3 | 1/30 |
| S:N679 | BA.x | Omicron | 3.3 | 1/30 |
aSix laboratories performed HTS and did not provide details on variant SNPs and deletions.
bSNP, single-nucleotide polymorphism.
DISCUSSION
This SARS-CoV-2 EQA was conducted to evaluate the abilities of European expert laboratories to detect SARS-CoV-2 via molecular methods and to type SARS-CoV-2 variants via WHO labels and the Pango lineage. The overall results of this EQA demonstrated that the majority of laboratories were able to correctly detect the presence or absence of SARS-CoV-2 RNA in all samples. Reduced molecular detection levels were observed for SARS-CoV-2 samples with low RNA concentrations, which was not unexpected and is of limited clinical importance. Of all analyzed variables, only the use of commercial or in-house real-time RT-PCR tests was associated with a significant difference in molecular detection performance, with in-house assays showing reduced sensitivity. This highlights the potential lack of optimization of in-house protocols and the need for the sound harmonization of workflows and the sound validation of in-house tests, as the diagnostic performance of in-house tests that are optimized and are properly validated is not generally considered to be lower than that displayed by commercial tests (13).
In contrast to SARS-CoV-2 detection, most laboratories participating in the optional activity of variant typing reported false results for variants and mutations, which was likely also a consequence of low sample concentrations and was not correlated with specific variants. Low RNA concentrations have been previously reported to limit the robustness of VOC/mutation detection (14). As low RNA concentrations not only increase the risk for general test failure but also increase the risk for sequencing errors (15), RNA concentrations should be considered when selecting samples for VOC/mutation surveillance. Several studies and the WHO recommend the selection of samples with Ct ≤ 30.0 for VOC/mutation surveillance (16–18). However, proper sample storage and quality may be limited in remote areas, raising the demand for robust typing techniques for representative variant surveillance. Judging from the performance in this EQA, the quality of VOC/mutation testing can benefit from combining real-time RT-PCR and HTS techniques. The real-time RT-PCR-based detection of mutations requires the careful selection of assays to allow for the differentiation of VOCs/VOIs. Otherwise, it can suffer from limited specificity and may be insufficient to determine specific SARS-CoV-2 lineages (7, 19). In contrast, HTS has the advantage of allowing for untargeted, full-genome surveillance, whereas real-time RT-PCR-based VOC testing is limited to specific mutations and needs to be updated with the emergence of new variants. In contrast, HTS is laborious, more expensive, and time-consuming. Thus, it will not replace real-time RT-PCR-based approaches. As VOC/mutation testing will remain important during the course of the COVID-19 pandemic, as is best illustrated by the emergence of the Omicron sublineages (20), the central distribution of quality controls, surveillance guidelines that consider RNA concentrations, and follow-up EQAs on SARS-CoV-2 VOC/mutation testing that also consider remote and resource-limited areas is advisable (10).
This study was limited by the lack of samples with different RNA concentrations for all selected variants, which hampers the systematic comparison of different methods among variants. Moreover, the panel was composed to evaluate the molecular detection of SARS-CoV-2 rather than for variant typing. Thus, it contained samples of low RNA concentrations. Therefore, the analysis of variant typing cannot be translated into the expected performance of laboratories with samples commonly selected for variant typing and must not be interpreted as poor capabilities of variant typing during clinical routines. Nevertheless, it provides important insights into how variant typing can be optimized for samples with low RNA concentrations, which may be important in many regions of the world that lack the infrastructure for timely sample collection and storage.
Our findings highlight the benefit of continuous EQAs and the need for future EQAs on SARS-CoV-2 variant typing to strengthen genomic surveillance, both in affluent settings and in resource-limited settings, as well as the benefits of combining different methodologies for accurate SARS-CoV-2 variant detection.
ACKNOWLEDGMENTS
We thank all of the EQA participants: Center for Virology, Medical University of Vienna, Austria; Department for Pathobiology, University of Veterinary Medicine Vienna, Austria; Clinical Reference Laboratory, Institute of Tropical Medicine, Belgium; National Influenza Centre, Sciensano, Belgium; Department of Virology, National Centre of Infectious and Parasitic Diseases, Bulgaria; Molecular Department, Croatian Institute of Public Health, Croatia; Research Unit, University Hospital for Infectious Diseases, Fran Mihaljević, Croatia; Department of Biological Sciences, University of Cyprus, Cyprus; Department of Molecular Virology, Cyprus Institute of Neurology and Genetics, Cyprus; Microbiology Department, Nicosia General Hospital, Cyprus; NRL for Influenza and other Respiratory Viruses, National Institute of Public Health, Czech Republic; Diagnostic Department, SA Pärnu Hospital, Estonia; Laboratory of Communicable Diseases, Health Board, Estonia; Microbiologia Laboratory of Rakvere Hospital, Estonia; Clinical Microbiology, Helsinki University Hospital, Finland; Expert Microbiology, National Institute for Health and Welfare, Finland; Biology Department, University of Corsica Pasquale Paoli, France; Unité des virus émergents, Aix-Marseille University, France; Central Diagnostics Unit, Bundeswehr Institute of Microbiology, Germany; FG17 Influenza and Other Respiratory Viruses, Robert Koch Institute, Germany; Institute of Microbiology and Virology, Brandenburg Medical School Theodor Fontane, Germany; Institute of Novel and Emerging Infectious Diseases, Friedrich-Loeffler-Institute, Germany; ZBS1 Highly Pathogenic Viruses, Robert Koch Institute, Germany; Molecular Diagnostics Laboratory, Central Public Health Laboratory, Greece; National Influenza Reference Laboratory for N. Greece, Aristotle University of Thessaloniki, Greece; National Influenza Reference Laboratory of Southern Greece, Hellenic Pasteur Institute, Greece; National Reference Laboratory for Respiratory Viruses, Nemzeti Népegészségügyi Központ, Hungary; Medical Virology, Landspitali University Hospital, Iceland; National Virus Reference Laboratory, University College Dublin, Ireland; Department of Infectious Diseases, Istituto Superiore di Sanità, Italy; Infectious Diseases, Amedeo di Savoia Hospital, Turin, Italy; Microbiology and Virology Unit, Padova University Hospital, Italy; Molecular Virology, Fondazione IRCCS Policlinico San Matteo, Italy; Scientific Department, Army Medical Center, Italy; U.O. Microbiology, Az. Ospedaliero-Universitaria di Bologna, Italy; Virology Laboratory, National Institute for Infectious Diseases, Lazzaro Spallanzani, IRCCS, Italy; Department of Microbiology, National Institute of Public Health of Kosovo, Kosovo; Microbiology and Virology Department, National Microbiology Reference Laboratory, Latvia; Clinical testing Department, National Public Health Surveillance Laboratory, Lithuania; Microbiology Department, Laboratoire National de Santé, Luxembourg; Pathology Department, Mater Dei Hospital, Malta; Department for Molecular Diagnostics, Institute of Public Health of Montenegro, Montenegro; Laboratory for Virology and Molecular Diagnostics, Institute of Public Health, North Macedonia; Department of Medical Microbiology, St. Olavs University Hospital, Norway; Department of Microbiology, Oslo University Hospital, Norway; Department of Virology, Norwegian Institute of Public Health, Norway; Virology Department, National Institute of Public Health, Poland; Infectious Diseases, National Institute for Health, Portugal; National Influenza and Other Respiratory Viruses Reference Laboratory, National Institutes of Health, Portugal; Molecular Diagnostics Laboratory, National Institute for Infectious Diseases, Romania; Viral Respiratory Infections Laboratory, Cantacuzino National Military-Medical Institute for Research and Development, Romania; Department for Microbiology, Clinical Center of Serbia, Serbia; Department of Medical Microbiology, Public Health Authority of the Slovak Republic, Slovak Republic; Department of Virus Ecology, Biomedical Research Center of the Slovak Academy of Sciences, Slovak Republic; Laboratory for Public Health Virology, National Laboratory of Health, Environment and Food, Slovenia; Laboratory for Zoonoses, Institute of Microbiology and Immunology, Slovenia; Laboratorio Microbiología, Hospital Clinic de Barcelona, Spain; National Influenza Centre, Valladolid, National Influenza Centre, Spain; Respiratory Virus and Influenza Laboratory National Center of Microbiology-Instituto de Salud Carlos III, Spain; Servicio de Microbiología, Hospital Universitario Virgen de las Nieves, Spain; Department of Microbiology, The Public Health Agency of Sweden, Sweden; Department Viroscience, Erasmus MC, The Netherlands; Diagnostics and laboratory Surveillance (IDS), National Institute for Public Health and the Environment (RIVM), The Netherlands; National Virology Reference Laboratory, Public Health General Directorate of Turkey.
We thank the Joint Research Centre (JRC) for their assistance regarding SARS-CoV-2 reference materials. In addition, we thank the staff members who provided laboratory, administrative, and other technical support at Robert Koch Institute (RKI), Charité, and RIVM.
This work was supported by the European Centre for Disease Prevention and Control (ECDC) under the Specific Contract no. 5 ECD.11818, implementing Framework Contract no. ECDC/2017/002.
R.M. contributed to the data curation, formal analysis, investigation, methodology, project administration, validation, visualization, writing of the original draft, and review and editing of the writing. C.F. contributed to the formal analysis, investigation, methodology, software, visualization, sample preparation, writing of the original draft, and review and editing of the writing. K.R.S. contributed to the data curation, formal analysis, investigation, methodology, project administration, software, validation, visualization, writing of the original draft, and review and editing of the writing. A.Mel. contributed to the conceptualization, funding acquisition, validation, and review and editing of the writing. A.C.A.C. contributed to the sample preparation, formal analysis, investigation, validation, and review and editing of the writing. C.D., B.B., A.Mei., and A.K. contributed to the conceptualization, resources, and review and editing of the writing. J.F.D. and C.B.E.M.R. contributed to the conceptualization, funding acquisition, project administration, resources, supervision, and review and editing of the writing.
Contributor Information
Chantal B. E. M. Reusken, Email: chantal.reusken@rivm.nl.
Jan Felix Drexler, Email: felix.drexler@charite.de.
Elitza S. Theel, Mayo Clinic
REFERENCES
- 1.WHO Coronavirus (COVID-19) Dashboard [Internet]. World Health Organization. 2022. Available from: https://covid19.who.int/. [Google Scholar]
- 2.Hellewell J, Abbott S, Gimma A, Bosse NI, Jarvis CI, Russell TW, Munday JD, Kucharski AJ, Edmunds WJ, Funk S, Eggo RM, Centre for the Mathematical Modelling of Infectious Diseases COVID-19 Working Group . 2020. Feasibility of controlling COVID-19 outbreaks by isolation of cases and contacts. Lancet Glob Health 8:e488–e96. 10.1016/S2214-109X(20)30074-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mishra S, Mindermann S, Sharma M, Whittaker C, Mellan TA, Wilton T, Klapsa D, Mate R, Fritzsche M, Zambon M, Ahuja J, Howes A, Miscouridou X, Nason GP, Ratmann O, Semenova E, Leech G, Sandkühler JF, Rogers-Smith C, Vollmer M, Unwin HJT, Gal Y, Chand M, Gandy A, Martin J, Volz E, Ferguson NM, Bhatt S, Brauner JM, Flaxman S, COVID-19 Genomics UK (COG-UK) Consortium . 2021. Changing composition of SARS-CoV-2 lineages and rise of Delta variant in England. EClinicalMedicine 39:101064. 10.1016/j.eclinm.2021.101064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Karim SSA, Karim QA. 2021. Omicron SARS-CoV-2 variant: a new chapter in the COVID-19 pandemic. Lancet 398:2126–2128. 10.1016/S0140-6736(21)02758-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fischer C, Mögling R, Melidou A, Kühne A, Oliveira-Filho EF, Wolff T, Reiche J, Broberg E, Drosten C, Meijer A, Leitmeyer K, Drexler JF, Reusken CBEM. 2021. Variable sensitivity of SARS-CoV-2 molecular detection in European expert laboratories: external quality assessment, June and July 2020. J Clin Microbiol 59. 10.1128/JCM.02676-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Emerging Viral Diseases-Expert Laboratory Network (EVD-LabNet) [Available from: https://www.ecdc.europa.eu/en/about-us/partnerships-and-networks/disease-and-laboratory-networks/evd-labnet.
- 7.Sander A-L, Yadouleton A, de Oliveira Filho EF, Tchibozo C, Hounkanrin G, Badou Y, Adewumi P, René KK, Ange D, Sourakatou S, Sedjro E, Aïssi MAJ, Fidelia H, Djingarey MH, Nagel M, Jo WK, Moreira-Soto A, Drosten C, Landt O, Corman VM, Hounkpatin B, Drexler JF. 2021. Mutations associated with SARS-CoV-2 variants of concern, Benin, early 2021. Emerg Infect Dis 27:2889–2903. 10.3201/eid2711.211353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DK, et al. 2020. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.JRC. EURM-019 single stranded RNA (ssRNA) fragments of SARS-CoV-2 Available from: https://crm.jrc.ec.europa.eu/p/EURM-019.
- 10.Fischer C, Pedroso C, Mendrone A, Jr, Bispo de Filippis AM, Vallinoto ACR, Ribeiro BM, et al. 2018. External quality assessment for Zika virus molecular diagnostic testing. Brazil Emerg Infect Dis 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iglói Z, Leven M, Abdel-Karem Abou-Nouar Z, Weller B, Matheeussen V, Coppens J, Koopmans M, Molenkamp R. 2020. Comparison of commercial realtime reverse transcription PCR assays for the detection of SARS-CoV-2. J Clin Virol 129:104510. 10.1016/j.jcv.2020.104510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thi Nhu Thao T, Labroussaa F, Ebert N, V'kovski P, Stalder H, Portmann J, Kelly J, Steiner S, Holwerda M, Kratzel A, Gultom M, Schmied K, Laloli L, Hüsser L, Wider M, Pfaender S, Hirt D, Cippà V, Crespo-Pomar S, Schröder S, Muth D, Niemeyer D, Corman VM, Müller MA, Drosten C, Dijkman R, Jores J, Thiel V. 2020. Rapid reconstruction of SARS-CoV-2 using a synthetic genomics platform. Nature 582:561–565. 10.1038/s41586-020-2294-9. [DOI] [PubMed] [Google Scholar]
- 13.Oladimeji P, Pickford J. 2020. Letter of concern re: “Comparison of seven commercial RT-PCR diagnostic kits for COVID-19” van Kasteren et al., Journal of Clinical Virology. J Clin Virol 130:104536. 10.1016/j.jcv.2020.104536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wegner F, Roloff T, Huber M, Cordey S, Ramette A, Gerth Y, Bertelli C, Stange M, Seth-Smith HMB, Mari A, Leuzinger K, Cerutti L, Harshman K, Xenarios I, Le Mercier P, Bittel P, Neuenschwander S, Opota O, Fuchs J, Panning M, Michel C, Hallin M, Demuyser T, De Mendonca R, Savelkoul P, Dingemans J, van der Veer B, Boers SA, Claas ECJ, Coolen JPM, Melchers WJG, Gunell M, Kallonen T, Vuorinen T, Hakanen AJ, Bernhoff E, Hetland MAK, Golan Berman H, Adar S, Moran-Gilad J, Wolf DG, Leib SL, Nolte O, Kaiser L, Schmutz S, Kufner V, Zaheri M, Trkola A, Aamot HV, Hirsch HH, et al. 2022. External quality assessment of SARS-CoV-2-sequencing: an ESGMD-SSM pilot trial across 15 European laboratories. J Clin Microbiol 60JCM0169821. 10.1128/JCM.01698-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jacot D, Pillonel T, Greub G, Bertelli C. 2021. Assessment of SARS-CoV-2 genome sequencing: quality criteria and low-frequency variants. J Clin Microbiol 59:e0094421. 10.1128/JCM.00944-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kubik S, Marques AC, Xing X, Silvery J, Bertelli C, De Maio F, et al. 2021. Recommendations for accurate genotyping of SARS-CoV-2 using amplicon-based sequencing of clinical samples. Clin Microbiol Infect 27:1036 e1–e8. 10.1016/j.cmi.2021.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.PAHO. Guidance for SARS-CoV-2 samples selection for genomic characterization and surveillance. Pan American Health Organization; 2021. Feburary 1st. [Google Scholar]
- 18.Rockett RJ, Arnott A, Lam C, Sadsad R, Timms V, Gray K-A, Eden J-S, Chang S, Gall M, Draper J, Sim EM, Bachmann NL, Carter I, Basile K, Byun R, O'Sullivan MV, Chen SC-A, Maddocks S, Sorrell TC, Dwyer DE, Holmes EC, Kok J, Prokopenko M, Sintchenko V. 2020. Revealing COVID-19 transmission in Australia by SARS-CoV-2 genome sequencing and agent-based modeling. Nat Med 26:1398–1404. 10.1038/s41591-020-1000-7. [DOI] [PubMed] [Google Scholar]
- 19.Harvey WT, Carabelli AM, Jackson B, Gupta RK, Thomson EC, Harrison EM, Ludden C, Reeve R, Rambaut A, Peacock SJ, Robertson DL, COVID-19 Genomics UK (COG-UK) Consortium . 2021. SARS-CoV-2 variants, spike mutations and immune escape. Nat Rev Microbiol 19:409–424. 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dyson L, Hill EM, Moore S, Curran-Sebastian J, Tildesley MJ, Lythgoe KA, House T, Pellis L, Keeling MJ. 2021. Possible future waves of SARS-CoV-2 infection generated by variants of concern with a range of characteristics. Nat Commun 12:5730. 10.1038/s41467-021-25915-7. [DOI] [PMC free article] [PubMed] [Google Scholar]