SUMMARY
Stony corals build the framework of coral reefs, ecosystems of immense ecological and economic importance. The existence of these ecosystems is threatened by climate change and other anthropogenic stressors that manifest in microbial dysbiosis such as coral bleaching and disease, often leading to coral mortality. Despite a significant amount of research, the mechanisms ultimately underlying these destructive phenomena, and what could prevent or mitigate them, remain to be resolved. This is mostly due to practical challenges in experimentation on corals and the highly complex nature of the coral holobiont that also includes bacteria, archaea, protists, and viruses. While the overall importance of these partners is well recognized, their specific contributions to holobiont functioning and their interspecific dynamics remain largely unexplored. Here, we review the potential of adopting model organisms as more tractable systems to address these knowledge gaps. We draw on parallels from the broader biological and biomedical fields to guide the establishment, implementation, and integration of new and emerging model organisms with the aim of addressing the specific needs of coral research. We evaluate the cnidarian models Hydra, Aiptasia, Cassiopea, and Astrangia poculata; review the fast-evolving field of coral tissue and cell cultures; and propose a framework for the establishment of “true” tropical reef-building coral models. Based on this assessment, we also suggest future research to address key aspects limiting our ability to understand and hence improve the response of reef-building corals to future ocean conditions.
KEYWORDS: model organisms, metaorganism, reef-building corals, microbial functions
INTRODUCTION
Scleractinian or stony corals build the framework of coral reefs, which are the most biodiverse and productive marine ecosystems (1). Coral reefs provide important ecosystem services and a livelihood to over 500 million people globally (2). Climate change, together with the local stressors of pollution and overexploitation have heavily impacted coral reefs around the world, causing major habitat loss and threatening the survival of these ecosystems (3–5) (Fig. 1). Preserving the biological and ecological functions of coral reefs requires drastic reductions of global and local stressors (6) together with active conservation and restoration interventions (7, 8). The effectiveness of such interventions depends upon a deep, accurate, and comprehensive understanding of coral biology (9, 10). However, while speed is imperative, research progress is challenged by the difficulties associated with working with corals.
FIG 1.
Coral bleaching and coral diseases as major threats to coral reefs. (A) Aerial view of coral bleaching in the Great Barrier Reef (Australia) during the 2017 mass bleaching event. (B) Acropora cytherea affected by white syndrome (WS), a tissue loss disease of unknown etiology. (C) Orbicella annularis suffering from stony coral tissue loss disease (SCTLD), a new lethal disease alarmingly spreading through the Caribbean. (D) Goniopora sp. infected with black band disease (BBD) during a bleaching event (visible loss of pigmentation). BBD is caused by a microbial consortium dominated by filamentous cyanobacteria. Image credits: A, Ed Roberts/ARC Centre of Excellence for Coral Reef Studies; B, C, and D, Dr. Greta Aeby.
Holobiont Diversity and Complexity
Corals are complex metaorganisms. The coral animal hosts a vast array of microorganisms encompassing unicellular algae of the family Symbiodiniaceae, bacteria, archaea, fungi, and other protists, as well as viruses, which collectively constitute the so-called coral holobiont (see Box 1) (11–13). Each coral colony represents a rich and diverse microecosystem often hosting several Symbiodiniaceae species (14, 15), hundreds to tens of thousands of bacterial taxa (16, 17), and probably at least as many archaea, viruses, and protists (18–20). This high diversity of microorganisms can be partially explained by coral colony morphology consisting of a dynamic surface mucus layer, the coral gastrodermal and epidermal tissues, the mesoglea, and the skeleton. Each of these represents a microhabitat or niche populated by distinct microbial communities (21–23).
BOX 1: CORAL HOLOBIONT MEMBERS AND FUNCTIONS—AN OVERVIEW OF CURRENT KNOWLEDGE
Symbiodiniaceae and coral bleaching. The endosymbiotic dinoflagellate algae (Symbiodiniaceae) are the most extensively studied and better characterized members of the coral holobiont (13). The coral-algal symbiosis is obligate and based around nutritional exchange, where the metabolic contribution of the photosynthetic algae supports high productivity under oligotrophic conditions—far beyond the capacity of the coral animal alone (24, 25). This symbiosis supports the building of the structural foundation of coral reefs and represents the engine of these ecosystems (26). However, this symbiosis is under threat, primarily due to global warming and other anthropogenic stressors on both a local and a global scale. The loss of the algae from coral tissue—coral bleaching—weakens the coral host and often leads to its death (27). Increasing frequency of and decreasing recovery time between bleaching events (28) make coral bleaching the largest challenge for the persistence of reef ecosystems, which has received much attention over the last 3 decades (29). Nonetheless, a complete and detailed understanding of the underlying cellular mechanisms is still lacking.
Bacteria and coral disease. Bacteria are the best studied of the microbial coral holobiont members and are known to play an important role in holobiont health (30). They are known to exist in highly diverse communities, which appear to vary in composition depending on coral and Symbiodiniaceae genotype (31, 32), environmental conditions (33), anatomical compartments (21, 34), and even colony age (35). They have been accredited as controlling or governing key functions for the coral host including, nutrient cycling (36, 37), and immunity (38, 39), and they have even been hypothesized to facilitate rapid environmental adaptation (40).
Imbalances in the microbiome, or dysbioses, compromise coral health and can lead to the emergence of disease (30). Due to climate change and anthropogenic activities, coral diseases are increasing in frequency and number, e.g., black band disease, now occurring in coral reefs around the world (41), and gray patch disease in the Indo-Pacific (42). Some researchers are now arguing that disease rivals coral bleaching as a major cause of coral reef decline on a global scale (43, 44).
Manipulating the coral microbiome has recently been shown to increase the tolerance of corals to a number of stressors, for example, by enriching the holobiont in members with beneficial functions and traits (45–48). Research on coral probiotics aims at understanding how to perform such manipulations to accelerate the rate of coral adaptation to global change (45, 47, 49). However, although our understanding of the likely functional role many bacteria play in coral health is rapidly advancing, we still lack a mechanistic understanding of the dynamics and functions of the majority of the coral associates (reviewed in reference 50). While there are limitations inherent to culture-based methods, a recent study has shown that diverse members across many phyla can and have already been cultured and highlighted how these could be further expanded in the coming years from the adoption of more diverse culturing approaches (51).
Understudied microbial partners. The remainder of the coral’s microbiome—i.e., the “other” (endolithic) microalgae, protists, archaea, fungi, and viruses—is comparatively less well understood. However, these microbes constitute a nonnegligible proportion of the coral microbiome. Archaea were found to constitute up to half of the prokaryotic fraction in absolute abundance, fungi were the most abundant microorganism in metagenomes of Porites astreoides, for example (18, 19), and viruses have been shown to be present in abundances of upwards of ~107 viruses per mL of mucus (52). Fungi and endolithic algae specifically appear to at least spatially dominate in the coral aragonite skeleton, where they have been shown to be directly involved in carbon and nitrogen cycling and may metabolically interact with each other and the coral host (53, 54). Archaea also appear to be involved in nutrient metabolism, in particular ammonia oxidation, carbon metabolism, and the synthesis of essential vitamins (55). Viruses, however, remain the most elusive members of the coral holobiont, and both their pathogenic and their beneficial roles are currently being investigated (reviewed in reference 56).
Many of the associated microorganisms are likely to be involved in holobiont metabolism, immunity, and environmental adaptation and may therefore contribute to the health and performance of the metaorganism (reviewed in references 30 and 50). The holobiont phenotype thus results from the combination of the long-term stable host genotype and the more flexible genotypes of the associated microbes (57–60). In addition, the environment (e.g., temperature, light, and salinity) and host characteristics (e.g., trophic state and age) modulate the cross-kingdom interactions between holobiont members (i.e., host-microbe and microbe-microbe) in a complex and underexplored framework (35, 61, 62).
Our understanding of what makes a coral “tick” has recently expanded exponentially and now the majority of researchers acknowledge the importance of the holobiont as a whole rather than focus on any one aspect (30). However, we still struggle to disentangle holobiont complexity and fall short in our understanding of coral functioning from a holistic perspective (Fig. 2 and Table 1). Several fundamental questions, such as those listed below, therefore remain either fully or partially unanswered.
FIG 2.
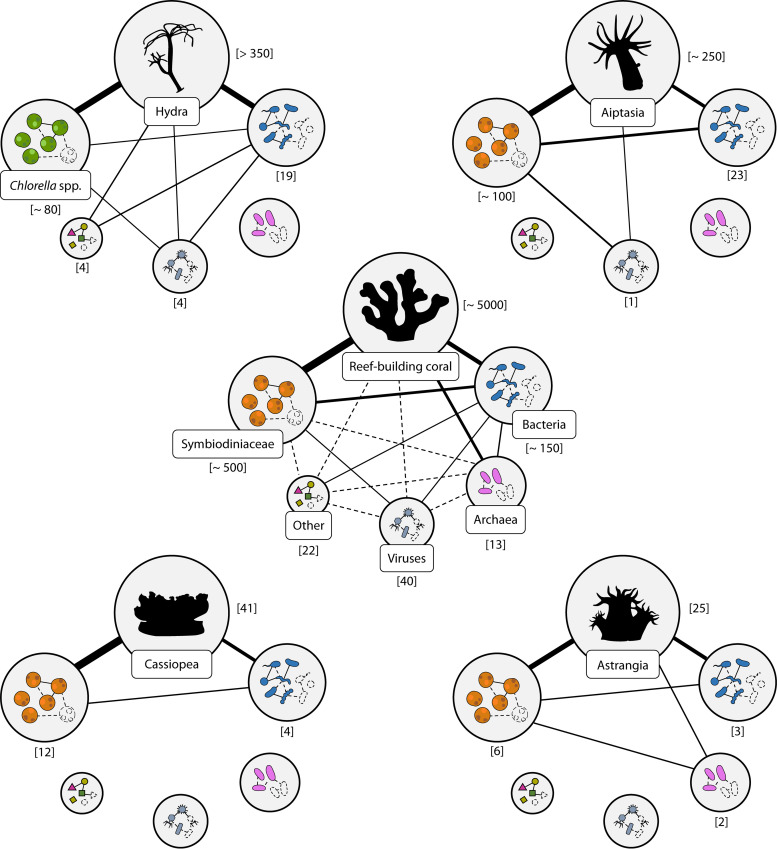
Overview of the state of knowledge on cnidarian holobionts regarding composition and functional interactions among their members. Cnidarian holobionts are disassembled into their major taxonomic compartments (gray circles). Within each taxonomic compartment, dashed-outlined microbes represent hypothesized, yet currently unidentified, taxa. Lines connecting compartments indicate known relationships, while absence of connecting lines indicate lack of information. Lines connecting individual microbes indicate known (solid), hypothesized (dashed), and presumed lack of functional relationships (no lines). The size of the taxonomic compartments and the thickness of the connecting lines approximate the assumed importance of each holobiont member or relationship. “Other” includes fungi and other protists (e.g., unicellular algae other than Symbiodiniaceae and Chlorella spp.). Numbers in square brackets report the numbers of peer-reviewed publications on the model organisms and their compartments (see the supplemental material for additional details).
TABLE 1.
Summary of the most relevant literature on functional interactions between members of the cnidarian model organisms
| Interaction | Observation(s)a |
|||
|---|---|---|---|---|
| Hydra | Aiptasia | Cassiopea | Astrangia spp. | |
| Host-symbiotic algae | ↔ Facultative endosymbiosis with Chlorella spp. (only H. viridissima); metabolic complementarity. The host provides CO2, N, P, and S. The algae provide photosynthates and amino acids (reviewed in references 84, 85, and 296). → Host controls Chlorella population size through algal cell cycle modulation, expulsion, or digestion (reviewed in references 84, 85, and 296). |
↔ Bleaching is associated with cellular and molecular responses in both partners (127, 297–300). ↔Onset of symbiosis as a modulation of the host immune response (137, 301–304). ↔ Species specific; association with non-native algal types results in altered expression patterns for metabolic exchanges, oxidative stress response, and immunity processes (119, 122, 136). ↔ Metabolic complementarity; Transfer of org. and inorg. nutrients between partners (135, 140, 305–307). ← Species-specific; association with non-native algal types is less stable and results in lower host growth and reproduction rates (132, 133). |
↔ Partner specificity: Association with non-native thermotolerant algal strain produces more heat-sensitive holobionts (164) ↔ Coupling of host and symbiont metabolism through translocation and recycling of C and N compounds (308, 309). → Host can restrict N (nitrate) availability to symbiont (170, 173). ← Symbiosis-driven development (161, 169, 171). ← Unable to heterotrophically compensate lack of alga-derived nutrients under low illumination (310) or in aposymbiotic state (162). ← Alga-fixed and translocated C satisfies >100% of host metabolic demand (311). |
↔ Facultative association; the symbiont provides mild advantages (nutrition, host growth and healing) or nondetectable effects (calcification rate) (180, 190, 312–314). → Host regulates symbiont population density through expulsion (315). → Host trophic conditions drive mutualistic/parasitic shift (181). |
| Host-bacteria | → Host influences bacterial microbiome through antimicrobial compounds and by altering bacterial quorum-sensing signaling (95, 97–99, 106). ← Bacteria protect the Hydra host against fungal infections (100). ← Bacterial involvement in host body developmental regulation (101). ← Bacterial role in spontaneous body contraction regulation (105). |
↔ Species specificity in the host-bacterium association (111). → Host state linked to bacterial microbiome structure and composition (141, 145). ← Potential role of bacteria in host defensive tissues (316). |
← Bacteria as larvae settlement cue (161, 317). | |
| Host-virus | ↔ Viromes are species specific; heat stress linked to altered viromes and altered predicted cellular functions involved in (DNA and C) metabolism and defense (318). |
↔ Diverse but stable viral assemblage (319). | ||
| Host-fungi | ← Lethal fungal pathogen (100). |
|||
| Symbiotic algae-bacteria | → Symbiotic algae provide host with colonization resistance and community-immunity against an invasive bacterium (320). | ↔ Symbiotic state linked to bacterial community composition; potential bacterial involvement in modulating N availability (170). | → Recovery of prokaryotic microbiota following antibiotic treatment is more consistent among symbiotic (than aposymbiotic) individuals (195). × Prokaryotic microbiome unaffected by seasonality or symbiotic state (77, 321). |
|
| Symbiotic algae-archaea | → Recovery of prokaryotic microbiota following antibiotic treatment is more consistent among symbiotic thann aposymbiotic individuals (195). |
|||
| Symbiotic algae-virus | ←? Consistent presence of virus in Chlorella suggests functional role (322). |
→ Relative abundance of viral taxa linked to symbiotic state (319). | ||
| Bacterium-virus | ← Bacteriophages dominate Hydra virome (318). ← Bacteriophage role in bacterial population dynamics (323, 324). |
|||
References are indicated parenthetically where applicable.
Who is there and where?
The coral microbiome has certainly not been fully characterized yet. The current knowledge of the coral microbiome is highly skewed toward Symbiodiniaceae and bacterial members, and less is known about the other microbial partners that constitute a large proportion of the microbiome in both biomass and absolute abundance (18, 19, 53). Thousands of taxa are likely yet to even be described and characterized.
Who does what?
While it is well recognized that the microbiome plays an important role in fundamental physiological functions such as nutrition, development, and immunity, the exact contribution and involvement of each microbial taxon remains to be resolved.
Who interacts with whom?
Besides coral-Symbiodiniaceae dynamics, very little is known about interactions between other members of the holobiont (63, 64). For instance, do bacteria interact with each other, the coral host, the Symbiodiniaceae, archaea, fungi, and/or viruses? Furthermore, can microbial communities in different anatomical compartments interact with each other? If so, then our next question would be as follows.
How do they interact with each other?
Individuals may affect or impact others within the community via positive (e.g., mutualistic symbiosis and facilitation), negative (e.g., competition, predation, and parasitism), or neutral (e.g., commensalism) interactions. Resolving the network of interactions between individual members of the microbiome and across Kingdoms is extremely challenging, yet it will yield very valuable information on ecological and coevolutionary processes (42, 65).
Model Organisms Unravel Complex Biological Principles
Model organisms by their very nature facilitate research because they are practically and/or ethically “more convenient to study” than the organisms of interest, and at the same time they are similar enough so that discoveries can be meaningfully transferred. Interestingly, there is no apparent fixed set of rules to define a model organism or its validity (66). Instead, the models are usually chosen based on the suitability of the organism to investigate a specific phenomenon or set of questions needing to be addressed, namely, its tractability and its informative power (67, 68). For example, tractability is typically associated with small size, fast growth rate, short reproductive cycle, broad availability, ease of maintenance under laboratory settings, and simplicity of some traits (e.g., small number of genes or simple body plan) (68). In addition to tractability, many models possess what could be perceived as “odd” or “unusual” features, which make them stand out among similar organisms. Typically, these oddities have a great informative power if harnessed for research purposes (67, 68). For example, the high regenerative capacity of Hydra or the ability to survive extreme conditions of tardigrades helped shed light on the mechanisms of aging, transdifferentiation, and de novo generation of biological patterns (69, 70) and on protection against damage of biological structures (71, 72).
Simplicity is also a very important feature for biological investigation that traditionally follows a reductionist approach. Compared to more complex or derived systems, simpler systems possess all of the fundamental features but lack much of the “extra noise”, such as additional biochemical pathways or regulatory processes, and therefore facilitate understanding of fundamental biological mechanisms. Thus, researchers exploring the mechanisms of gene expression and regulation found it more convenient to study the yeast Saccharomyces cerevisiae, a single-celled eukaryote with a simple genomic structure and comparably little non-coding DNA (73), rather than something as complex as a human cell. The same fundamental principles apply to both, to the point that essential yeast genes can be replaced by their human orthologs (74).
As scientific knowledge grows and new questions arise, new model organisms are being added to a growing list (75). Technological progress is increasing the insight from each model organism rather than replacing them. Easy access to high-throughput sequencing technologies expands the number of sequenced genomes available and facilitates the development of new and customized molecular tools (67, 68). Model organisms are not only growing in number, but they are also used in new and more varied applications. While model organisms have been extensively used to investigate fundamental biological principles (e.g., organismal development, behavior, and evolution [69]), phenomena directly affecting human health (i.e., to understand and treat diseases) or to generate economic benefits (agricultural crops and livestock), more recently, nature and biodiversity conservation represents a new niche for model organism-based research (76).
Coral research is challenged by a set of important questions that could benefit from the use of model organisms (76–79) (Fig. 2). Disentangling holobiont complexity to shed light on the mechanisms underlying coral responses to global change (e.g., coral bleaching and diseases; see Box 1 and Fig. 1) and how these can be prevented or mitigated, is one of the most pressing challenges faced today (8). To reverse the decline of coral reefs (and maintain the services they provide), scientists have been embracing efforts to increase corals resilience through approaches that span many levels of intervention, from the microscopic level (cellular and molecular, i.e., assisted evolution) to the macroscopic level (ecosystem scale, i.e., assisted gene flow), and from laboratory-based to field deployment (7). Here, we provide a comprehensive overview of the state-of-play of model organisms and systems that can be utilized to move coral holobiont research into the next stage. Our aim for this review is to (i) highlight each model system’s advantages and disadvantages and (ii) synthesize open research questions and how establishment of new model systems could address them. We review established and emerging cnidarian model systems, including the freshwater hydroid Hydra, the anemone Aiptasia, the jellyfish Cassiopea, and the temperate coral Astrangia poculata (Fig. 3). Moreover, we provide a comparative overview of their attributes, including distribution, ease of rearing in aquaria, life cycle, amenability to manipulate symbiotic states, existing knowledge base, and resources. We further introduce the different approaches that can facilitate direct experimentation on tropical stony corals, a necessary step to validate discoveries made on laboratory model systems (10, 80). These approaches include the establishment of tropical stony coral species as model organisms, as well as simplified systems such as cell and tissue cultures.
FIG 3.
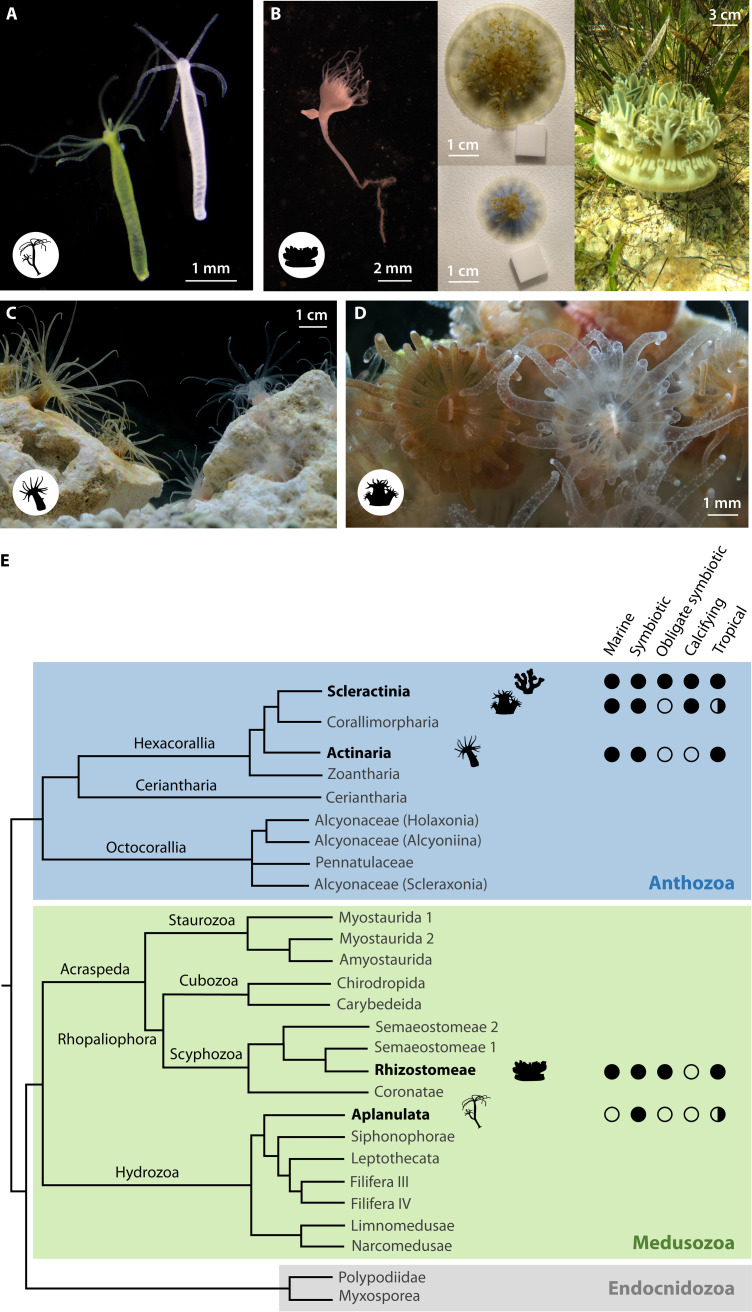
Cnidarian model organisms and systems for reef-building corals. (A) Freshwater hydroid Hydra, symbiotic with the chlorophyte Chlorella spp. (left) and aposymbiotic (right). (B) Cassiopea xamachana scyphistoma (polyp, early life stage, left), young symbiotic medusa (center top), young aposymbiotic medusa (center-bottom), and adult symbiotic medusa (right). (C) Sea anemone Aiptasia, symbiotic (left) and aposymbiotic (right) polyps. (D) Astrangia poculata naturally occurring in symbiotic (left) and aposymbiotic (right) state. (E) Phylogeny showing the relative phylogenetic distance within cnidarians and between tropical scleractinians (including reef-building corals) and the cnidarian model organisms discussed in this review. Specifically, from top to bottom are shown: tropical stony corals (Scleractinia), Astrangia spp. (Scleractinia), Exaiptasia spp. (Actinaria), Cassiopea xamachana (Rhizostomeae), and Hydra spp. (Aplanulata). Phylogeny was modified from reference 159. Image credits: A, Jay Bathia; B, Victoria Sharp, Claudia Tatiana Galindo, and Andre Morandini; C, Samuel Begood; D, Alicia Schickle.
CNIDARIAN MODEL ORGANISMS AS CORAL HOLOBIONT MODELS
Freshwater Hydroid Hydra
Hydra is the oldest cnidarian model organism and arguably the best known. Hydra species belong to the class Hydrozoa, in the anthozoan sister subphylum Medusozoa (Fig. 3E). They inhabit freshwater ecosystems worldwide (81–83) and while most do not associate with microalgae, one species (H. viridissima) can establish facultative endosymbiosis with the chlorophyte Chlorella spp. (84, 85). Hydra has a simple life cycle that can be easily completed in the lab. Under normal laboratory conditions, solitary polyps reproduce asexually by budding, while environmental shifts in temperature or food induce sexual reproduction (86, 87). This ease of rearing combined with its exceptional regenerative capacity, was already appreciated in the 1700s (88) and contributed to the birth of experimental zoology (89). This is evidenced by early fundamental discoveries using the Hydra model, such as Ethel Browne’s discovery of induced formation of a secondary axis by transplanting a head onto a polyp (90), 15 years before Mangold and Spemann published their observation of the organizer activity of the dorsal lip of the amphibian embryo (91). Hydra then grew into a model for developmental biology that helped answer questions of pattern formation at the theoretical (92) and molecular levels (93). Later, the adoption of H. viridissima as a model organism contributed to the understanding of fundamental processes in cnidarian-algal nutrient exchange and symbiosis regulation (see reference 85 and references therein).
Over the last 15 years, Hydra has become an important model for host-bacterium research. Initial analyses of the bacterial microbiome of different Hydra species revealed a high degree of species specificity reflecting the phylogenetic relationships of its Hydra host species (94, 95). Subsequent research identified that the epithelial cells produce specific antimicrobial peptides (96–99) which act as key innate immune factors responsible for shaping the species-specific bacterial associations (95).
The Hydra model has shed light on the involvement of the bacterial microbiome in shaping the holobiont phenotype. For example, the removal of the intact microbiome revealed that bacteria protect the Hydra host against fungal infections (100). Other recent results indicate that bacteria associated with Hydra are able to modify the Wnt-signaling pathway, a central signaling cascade in development (101). This pathway is involved in several developmental processes in Hydra, such as head (93) and bud (102, 103) formation, and the differentiation of stem cells (104). Functional analyses of bacterium-regulated genes revealed that the corresponding peptides have an antagonistic function to Wnt-signaling and influence stem cell differentiation (101). Studies have also shown that specific bacterial species are involved in regulating the frequency of spontaneous body contractions, which is reduced by ~50% in germ-free animals (105). Although the mechanisms underlying these responses are still unclear, studying the host-microbe signaling in detail revealed a direct interaction between Hydra and its associated bacteria based on quorum-sensing signaling molecules (106). This investigation identified a fundamental mechanism whereby the host-modified bacterial signal molecule promotes symbiosis establishment, while the non-modified signal molecule represses it (106). This demonstrates that Hydra is able to alter quorum-sensing controlled behavior of its bacterial symbionts to promote metaorganism assembly and resilience (106).
Within the bacterial communities of Hydra, one particular bacterium (a Curvibacter sp.) stands out as it is the most abundant member (95). Curvibacter sp. appears to populate and accumulate in a mucus-like layer of the ectodermis (100) and can easily be cultivated and reproduced in the lab (100). Genomic and transcriptomic data are available for both Curvibacter and its Hydra host (106–108), which facilitates functional studies in both symbiosis partners (109–111). Importantly, due to the transparent appearance of the animals, the host’s cells and those of the Curvibacter can be transgenically labeled (109, 110). This means microscopic analysis can be achieved in an in vivo context on single cells, as well as whole tissue levels across space and time. It is now even possible to generate germ-free Hydra polyps via antibiotic treatment and repopulate them with single or multiple bacterial strains (95, 100, 112). Despite differences in the surface topography between Hydra and stony corals (113), their relatively distant phylogenetic relationships (Fig. 3E), and differences in life history (Table 2), the Hydra model can help to understand mucosal host-microbe interactions in corals by providing a roadmap to controlled symbiosis-reestablishment experiments in cnidarians.
TABLE 2.
Summary of relevant features of cnidarian model organismsa
| Feature | Observation(s) |
|||||
|---|---|---|---|---|---|---|
| Hydra | Aiptasia | Cassiopea (polyp) | Cassiopea (medusa) | Astrangia | Tropical scleractinians | |
| Species | Hydra spp. | Exaiptasia diaphana (Rapp, 1829) | C. xamachana (Bigelow, 1892) | A. poculata (Ellis & Solander, 1786) | ~800 species (symbiotic) | |
| Body features | ||||||
| Form | Sessile | Sessile | Benthic | Benthic but not sessile | Benthic | Sessile |
| Motility | Limited | Limited | No | Yes | No | No |
| Calcifying | No | No | No | No | Yes | Yes |
| Colonial | No | No | No | No | Yes | Yes |
| Polyp size | 5 to 15 mm | ~1 mm to 5 cm | ~0.1 to 2 mm | ~2 mm to 20 cm | <10 mm | ~1 mm to 20 cm |
| Environment and availability | ||||||
| Water/medium | Freshwater | Seawater | Seawater | Seawater | Seawater | |
| Latitudinal distribution | Temperate | Tropical and subtropical | Tropical and subtropical | Temperate to tropical | Tropical and subtropical | |
| Trophic environment | Medium nutrients (mesotrophic) | Low nutrients (oligotrophic) | High nutrients (eutrophic) | Medium nutrients (mesotrophic) | Low nutrients (oligotrophic) | |
| Habitat | Streams and lakes | Mangroves, coral reefs | Mangroves | Mangroves, coral reefs, seagrass beds | Shallow to deep hard substrates | Coral reefs |
| Geographic area | Circumglobal | Shallow seas in the intertropical belt | Caribbean and Gulf of Mexico | Atlantic | Shallow seas in the intertropical belt | |
| Life cycle and propagation | ||||||
| Full/closed life cycle in lab (ex situ) | Yes | No | Yes | Yes | Yes | |
| Mode of clonal propagation | Budding | Pedal laceration | Budding | Regeneration | Fragmentation | Budding and fragmentation |
| Time to maturity or (sexual) generation time | Wks | NA | Mos | Mos | Yrs | |
| Rearing and maintenance in artificial settings | Easy | Easy | Easy | Moderate | Moderate/demanding | Demanding |
| Algal symbiont | ||||||
| Algal symbiont: identity | Chlorella | Symbiodiniaceae | Symbiodiniaceae | Symbiodiniaceae | Symbiodiniaceae | |
| Algal symbiont: culturable | No | Yes | Yes | Yes | Yes | |
| Needs algal symbiont to complete development | No | NA | Yes | No | Yes | |
| Obligate symbiosis at adult stage | No | Yes | Yes | No | Yes | |
| Survival in aposymbiotic state | Yrs | Indefinitely | Indefinitely | >3 wks | Indefinitely | Wks |
| Algal symbiont acquisition | Mixed | Horizontal (env) | Horizontal (env) | Horizontal (env) | Depends on species | |
| Practical aspects | ||||||
| Consortium | OpenHydra | Aiptasia Symbiosis Resource | CassiopeaBase | Temperate Coral Research Working Group | No | |
| Online open access resources | Hydra 2.0 Genome Project Portal | protocols.io, Reefgenomics | Medina Lab | Coral Microbiome Portal | Reefgenomics, SymPortal | |
| Established strain(s) | Yes | CC7, H2, EM5, JK, JKA2, VW9, VWA12, PLF3, PLF5, and PLF8 | 12 strains: T1–A, B, C, D, E, and F and T2–A, B, C, D, E, and F | No | No | |
| Genome(s) available | Assembled | 3 | Casxa1 | Draft | >45 (05.10.2022) | |
| Cites regulation | No | No | No | No | Appendix II | |
This summary can help evaluate the suitability of a system of interest based on similarities and dissimilarities with tropical scleractinian corals. NA, not applicable; mos, months; wks, weeks; yrs, years.
Sea Anemone Aiptasia
The sea anemone Aiptasia is also among the original models utilized for the study of coral-dinoflagellate symbiosis, with some publications dating back to the 1960s and early 70s (e.g., reference [114]). The name Aiptasia is a common name for Exaiptasia diaphana (Rapp, 1829), which was previously named Exaiptasia pallida (115). Similar to Hydra, Aiptasia is fast growing, hardy, clonal, and it is extremely amenable to laboratory culture. Indeed, it is considered an aquarium pest by the hobby industry due to its rapid growth and propagation, often quickly overgrowing corals and other sessile reef fauna. Animals can be purchased from animal supply companies, collected from nature with relative ease, and acquired from the growing global network of Aiptasia laboratories that commonly share strains. Its strengths and limitations as a model have been extensively documented elsewhere (e.g., see references 76, 116, and 117), so we will only summarize them here.
With Aiptasia’s popularity growing, the model has been adopted by an increasing number of laboratories around the world (117, 118). Historically, studies were primarily performed on animals of unknown or mixed genetic background, with different laboratories using different populations, strains, and possibly even different species (115). Since the early 2000s, however, the Aiptasia community has rallied and increasingly turned to clonal populations, allowing for the control (to some degree) of genetic background noise (119, 120). Two specific clonal lines, CC7 and H2 (originating from Florida and Hawaii, respectively), are now shared widely among research groups in the United States, Europe, and the Middle East (118, 121). However, different clonal populations are in laboratory culture in both Australia (117) and New Zealand (122) since these countries have strict limitations on importing non-native organisms. The genomes of CC7 (123), H2, and a clonal line from the Red Sea are available, as are numerous transcriptomes from other clonal lines (see Table 2).
The Aiptasia-Symbiodiniaceae model is a powerful system to unravel the cellular and molecular mechanism underlying coral bleaching. In contrast to tropical corals, Aiptasia can be easily bleached experimentally in a standardized and controlled manner, maintained in this aposymbiotic state indefinitely (typically under dark conditions to prevent repopulation from accidentally introduced algae) and subsequently repopulated with Symbiodiniaceae algae. Aposymbiotic animals will grow and continue to undergo pedal laceration to reproduce asexually if fed frequently (124), suggesting that the health of the animal is not impeded to any major degree. Bleaching can be achieved via a number of methods, including heat stress (125), cold shock (often in combination with the herbicide DCMU [126]), or by incubation in menthol (126). Examining aspects of the heat stress response and accompanying loss of symbionts over time in Aiptasia provides a powerful analog to coral bleaching responses (e.g., references 125, 127, and 128). However, for rendering animals aposymbiotic for use in experiments on symbiosis reestablishment, menthol bleaching is the most rapid and the most effective at eliminating essentially all symbionts from host tissues (126).
Depending on host origin, Aiptasia harbors different Symbiodiniaceae species. Most laboratory animals contain Breviolum minutum (including host strain H2), or Symbiodinium linuchae (including host strain CC7). Both of these symbiont species have been successfully brought into culture and the genomes sequenced (129, 130). In addition, Aiptasia is tolerant of a variety of non-native symbiont species, including Symbiodinium microadriaticum and Durusdinium trenchii, two species with differing susceptibility to environmental perturbation (131–134). These species can enter hosts but are less successful than native species at populating them, and the metabolic exchange and interpartner homeostasis is perturbed (122, 135, 136). Similar to polyps, Aiptasia larvae can take up a variety of algal species and subsequent persistence and proliferation appears to depend on the ability of the microalgae to suppress host innate immune response and escape expulsion (vomocytosis) (137). This diversity in specificity allows for the study of comparative Symbiodiniaceae repopulation dynamics, mechanisms of recognition, specificity and regulation, and differential susceptibility to heat stress or other perturbation (e.g., see references 133 and 138–140).
The Aiptasia microbiome has now been described in a variety of strains from around the world (141–144). Overall, there is congruence between the different studies, showing a similar microbiome makeup among animals in culture and similarity in the taxonomic diversity of microbiomes between Aiptasia and corals. These baseline descriptions of the Aiptasia microbiome set the stage for future studies on the effect of heat stress and other environmental perturbation on the Aiptasia holobiont, the interaction of the algal symbionts with the microbiome, and the possibility of manipulating the microbiome to aid in building coral resilience. Indeed, some of this work has already begun (113, 145).
The life history characteristics of Aiptasia contain both key strengths, and at present, significant limitations to its value as a model system. For example, asexual reproduction (pedal laceration) facilitates clonal propagation, but sexual reproduction has not yet been achieved in captivity. Aiptasia is gonochoristic, although there is some evidence that animals can switch sex (146). Animals spawn non-symbiotic gametes and therefore onset of symbiosis must occur anew with each host generation. Further, researchers have now developed culturing conditions that result in predictable and repeated spawning of gametes, successful fertilization and subsequent rearing of F1 larvae (147–150). These larvae can establish symbiosis with algae from culture, which again provides a powerful system to examine mechanisms of recognition and specificity. To date however, despite considerable effort by several research groups, there has been no success in achieving larval settlement and metamorphosis into juvenile polyps (unpublished data). This presents a major barrier to conventional genetics, gene editing, or other gene knockdown techniques in Aiptasia (151) that would revolutionize our ability to discern host gene function in the symbiosis.
There are other aspects of Aiptasia biology that warrant further development to cement this organism as a key model. The host processes of pedal laceration and subsequent patterning that results in development of clonal juveniles has been described (152–154). However, the role of symbiosis in these processes is just beginning to be described (155) and is a topic ripe for future work. Finally, although we believe animals harboring B. minutum occur pan-tropically, whereas those containing S. linuchae appear restricted to the Florida Keys (156, 157), there is a lack of global sampling studies describing natural symbiosis states. Such surveys could be further expanded to include other symbiotic anemone species within the Aiptasiidae. This would extend the relevance of the model system into a comparative genomic and ecological framework and explore conservation in the molecular evolution of marine endosymbioses.
Jellyfish Cassiopea
The Upside-down Jellyfish Cassiopea xamachana (Bigelow, 1892) has been a powerful model to study developmental symbiosis for more than four decades (reviewed in reference 158). In recent years, the “Cassiopea” model has gained increasing attention as a system to study cnidarian symbiosis (78, 158), with a complete genome now available (159, 160). All nine species in the genus are found in tropical and subtropical waters around the world, although the distribution range of C. xamachana is limited to the Caribbean and Gulf of Mexico (summarized in reference 158), and therefore the adoption of this species as a model organism might need to overcome transport restrictions (as previously discussed for Aiptasia). Like corals, Cassiopea establishes an obligatory association with Symbiodiniaceae algae (161, 162) and is therefore employed to study the bleaching response of cnidarian-algal symbiosis to heat stress (163, 164). Cassiopea is suited to laboratory rearing and investigations since it can tolerate a broad range of environmental conditions (163, 165, 166), is noncalcifying, and has a short life cycle that can be completed in the laboratory in four to six months. Embryos can be collected daily from the brooding region of female medusae, a trait which facilitates genetic (e.g., microinjection) and developmental studies (e.g., embryogenesis) (reviewed in reference 158).
The Cassiopea life cycle allows easy access to different life-stages that also differ in their dependence on Symbiodiniaceae. Female medusae constantly release brooded swimming larvae that settle and metamorphose into polyps upon encountering microbial settlement cues (161). These polyps (scyphistomae) reproduce by budding, and large clonal aposymbiotic populations can easily be maintained under laboratory conditions with regular feeding as “immortal lines” (158). The establishment of symbiosis with S. microadriaticum triggers strobilation, a metamorphic transition into sexual ephyra (i.e., free-living juvenile medusae), termed symbiosis-driven development (167). Interestingly, polyps can establish symbiosis with a broad range of Symbiodiniaceae species (168), but only some elicit metamorphosis (169). This system enables developmental, genetic, and physiological comparisons of the onset of symbiosis in the same host genetic background. On the contrary, adult medusae depend on their Symbiodiniaceae partner (162, 170) toward which they show a high degree of selectivity and specificity (164, 169, 171), not unlike differences in symbiotic specificity between juvenile and adult corals (172).
These characteristics make Cassiopea particularly suitable for symbiosis manipulations. Both polyps and medusae can be bleached through temperature stress (162, 171) or menthol treatment (170) and, although lack of the algal symbionts eventually leads to death (162), aposymbiotic medusae can survive for more than 3 weeks (170). Comparison between symbiotic states can help unravel each partner’s contribution to holobiont functioning, such as nutrient uptake and dynamics, and the effect of Symbiodiniaceae presence or absence on the bacterial microbiome (170). Further, aposymbiotic polyps can reestablish symbiosis with native or non-native Symbiodiniaceae strains (164, 169, 171). The ability to obtain different polyp-Symbiodiniaceae associations will in turn allow the production of clonal polyps that harbor different microbiomes. This, together with efforts to develop axenic and gnotobiotic animals, will also open doors to systematically explore host-microbiome interactions with the Cassiopea model system (158).
Another outstanding feature of Cassiopea is its ability to maintain a functional symbiosis across a wide range of environmental stressors (163, 165, 166). This trait can therefore help identify mechanisms that confer tolerance to changing environmental conditions in reef-building corals (170, 173). For example, Cassiopea can withstand high temperatures, showing onset of bleaching between 37 and 40°C (163). Also, high nutrient loads, that typically destabilize the coral-algal symbiosis and lower their bleaching threshold (174), are well tolerated by Cassiopea (175). Tracking of uptake and translocation of isotopically labeled nutrients suggest that Cassiopea is able to exert control over its algal symbionts’ capacity to access N, specifically by restricting nitrate (170, 173), the N species linked to decreased heat tolerance in corals (176).
The Cassiopea system is marked by peculiarities that further distinguish it from stony corals and that contribute to its large environmental tolerance. While corals host Symbiodiniaceae in their gastrodermal cells, in Cassiopea these are predominantly located inside amoebocytes (177). Amoebocytes are motile cells found in the mesoglea, which can be actively redistributed to meet energetic demands across different body parts (173). In addition, Cassiopea has a greater capacity to enrich its nutrient environment compared to corals (178). Rather than relying on currents to transport particle and solutes, Cassiopea uses bell pulsation to generate flows that draw particles (e.g., zooplankton) from the surrounding seawater to its feeding appendages and mobilize nutrients from the underneath sediments (178). Discoveries made on Cassiopea therefore need to be contextualized considering these aspects.
Temperate Coral Astrangia poculata
The temperate coral Astrangia poculata (Ellis & Solander, 1786) is one of the very few calcifying cnidarian model organisms currently available. Since this species is more amenable to rearing in aquaria (compared to the majority of tropical scleractinians), it represents an attractive, and increasingly popular, model system. Colonies are easy to collect since they are abundant in coastal, easily accessible locations across the western Atlantic. On average, colonies carry about 20 to 100 polyps, can grow to ~10 cm in diameter, and are gonochoric (carry separate sexes). Spawning is synchronous and inducible in the laboratory throughout the period of late July to early October, mirroring patterns of gametogenesis (179). In addition, A. poculata are gaining attention not only as an emerging model system but also as an emblem for coral and climate change research, demonstrated by its designation in 2021 as the Official State Coral of RI, USA.
Above all, two aspects (and their implications) are particularly remarkable about A. poculata: the nature of its photosymbiosis and its outstanding thermotolerance. A. poculata facultatively engages in symbiosis with the photosymbiont Breviolum psygmophilum. Sympatric colonies can be found in different symbiotic states (symbiotic, aposymbiotic, and patchy/mixed) across all seasons (77, 180), and photosymbiont density can be artificially manipulated (increased with high light intensity; decreased with low light intensity) (R. Rotjan, unpublished data). Because tropical corals often cannot be decoupled from Symbiodiniaceae without imposing stress, many critical, basal questions regarding this symbiosis are difficult to address in corals directly. Here, the A. poculata model can be particularly advantageous. Tracking nitrogen uptake and translocation in both symbiotic and aposymbiotic A. poculata, for example, helped elucidate how nutrient availability modulates the coral-algal relationship, specifically suggesting that nitrogen and carbon limitation shift the coral-photosymbiont mutualism toward parasitism (181). Similar to Cassiopea, A. poculata can tolerate extreme temperatures—withstanding what is among the largest temperature ranges that any hard coral has been documented to experience in its natural habitat. In the species’ northernmost distribution (southern New England), seawater temperature seasonally fluctuates over a range exceeding 20°C, with average temperatures spanning from 4 to 29°C (77). This annual temperature range compares to that of the Persian Gulf, the region with the most extreme environmental conditions where tropical reef-building corals persist (with recorded extremes spanning from ~11 to 36°C [182, 183]). Although the thermal environment of A. poculata is much colder than that of coral reefs, this ability to cope with such a large temperature range makes A. poculata an excellent experimental system for identification of genes and critical mechanisms of thermal tolerance. For example, comparison of gene expression between symbiotic states of A. poculata under thermal stress demonstrated that many stress-response genes previously identified in tropical corals likely belong to the host, as these were also present in aposymbiotic specimens (184), while transcriptional profiles of Symbiodiniaceae remain relatively unaffected by heat stress in corals (185). These experiments mirror physiological and metabolic patterns of the coral holobiont under stress (186, 187) and underline the potential to directly transfer insights gained from studying A. poculata to tropical reef-building corals. Furthermore, explorations of A. poculata in its natural environment across seasons and along latitudinal gradients can be used to test the influence of symbiosis and seasonality on microbe-microbe interactions within the holobiont. Across the year, A. poculata experiences shifts in photosymbiont density similar to those described for stony corals (188, 189), and the onset of a state of cold-induced quiescence (dormancy) during the winter months (190–192). Recent research efforts have utilized individuals from these naturally occurring gradients to identify microbes and multipartner (Symbiodiniaceae-bacterial) interactions important in the cnidarian response to environmental changes. Recent work on wild A. poculata colonies showed that the influence of photosymbiont density on the taxonomic structure and activity of the bacterial and archaeal community was smaller than that of seasonality (77). These findings largely agree with the stable bacterial communities found in cold shock bleached Aiptasia (141) and heat-stress bleached Porites lobata and Pocillopora acuta corals (193, 194) and support the generalization that external (environmental) factors have a stronger effect on microbiome structuring than photosymbiont density alone. Interestingly however, the presence of B. psygmophilum appears to facilitate consistent recovery of the bacterial and archaeal communities in A. poculata after antibiotics treatment (195). In these studies, the A. poculata bacterial community shares similarities in taxonomic structure with those of tropical corals but is remarkably less species-rich and more predictable (51, 77). As next steps, development of protocols is under way for spawning, embryonic development, larval rearing, larval settlement, and postsettlement growth to enable experimental examination of processes governing multipartner symbioses, including symbiont recruitment, establishment, and succession.
Increasing the Power of Cnidarian Model Systems
The traits that make the discussed cnidarian model organisms convenient study systems (and ecologically successful species) also set them apart from tropical reef-building corals. For example, features that greatly facilitate experimental investigation such as the lack of a carbonate skeleton, facultative photosymbiosis, and broad environmental tolerance (or “hardiness” of a species) have relevant physiological implications, and ignoring them might leave important biological mechanisms unaddressed. Therefore, to be informative for coral reef conservation, discoveries made from model systems should not be viewed as standalones but contextualized within a broader framework. To increase the power of these experimental model organisms, it is therefore necessary to adopt combined research approaches that use multiple models chosen for the complementarity of their features and that rely on multi-institutional collaborations (see “A Trait-Based Approach To Identify Suitable Coral Species” below and Table 2 for a summary of similarities and dissimilarities between the discussed organisms).
DIRECT TESTING AND EXPERIMENTATION ON CORALS THROUGH HOLOBIONT SIMPLIFICATION
Tissue Cultures as Structural Simplification
Structural simplifications offered by tissue cultures and cell lines allow for direct miniaturization of tropical stony coral systems. They eliminate skeletal components, which increases optical transparency aiding visualization, and liberates sample processing from the interferences of the aragonite particles and Ca2+ ions. As in the examples of the cnidarian model systems, a small sample size here aids high replication, translates into faster and less expensive workflows, and uses available live material efficiently.
The first step of structural simplification of the coral host involves the isolation or explantation of tissues or cells and maintenance of these as “primary cultures” or “tissue explants.” This can be undertaken in a number of ways (Table 3). So-called “destructive approaches” affect tissue organization by breaking down cell-cell or cell-ECM (extracellular matrix) adhesion, through the removal of divalent cations (Ca2+, Mg2+), enzymatic digestion of ECM components, and single-cell isolation through gravimetric fractionation or sieving of digested tissues (reviewed in reference 196). Isolated coral cells then reaggregate into multicellular structures. Nondestructive approaches preserve the original tissue organization and can be achieved mechanically by cutting (197, 198) or peeling off coral tissues (199, 200), or physiologically by inducing a stress response mechanism called polyp bail-out (201–203). Destructive approaches remain prone to microbial contamination that hinders long-term survival (196) (Table 3). Nondestructive approaches only require minimal treatment to control contamination and have, on average, longer viability (Table 3).
TABLE 3.
Chronological overview of studies on coral cell and tissue cultures with synthesis of culture origin and type, viability, and use of antimicrobials
| Tropical scleractinian coral species group(s) | Other cnidarian species | Life stage | Suspended and/or adherenta | Viabilityb | Proliferationc | Isolation approachd | Antibiotic and/or antimycotice | Year of publication | Reference(s) |
|---|---|---|---|---|---|---|---|---|---|
| Fungia scutaria, Pocillopora damicornis | Boloceroides sp., Cassiopea xamachana, Exaiptasia diaphana (formerly Aiptasia pulchella), Zoanthus sociatus | Adult | S | 5 h | NO | Des | 1992 | 252 | |
| Acropora microphthalma, Montipora digitata, Pocillopora damicornis, Porites sp., Seriatopora hystrix, Stylophora pistillata | Adult | S | 12 days | NO | Des | AB, AM | 1999 | 204 | |
| Pocillopora damicornis | Adult | A, S | 7 to 12 days | NO | Des | AB, AM | 2001 | 207 | |
| Pocillopora damicornis, Stylophora pistillata | Adult | A | 7 days | NO | Des | AB, AM | 2004 | 202, 211 | |
| Montipora digitata | Xenia elongata | Adult | A | 21 days | NO | Des | AB, AM | 2007 | 208 |
| Acropora millepora | Planula | S | 10 wks | CP | Des | AB, AM | 2009 | 214 | |
| Fungia sp.,Pavona divaricata | Adult | S | 4 days | NO | Des | 2009 | 205 | ||
| Pocillopora damicornis | Adult | S | 9 days to 2 mos | NO | Des | 2010 | 325 | ||
| Favia favus, Fungia granulosa | Oculina patagonica | Adult | A, S | 3 mos to >3 yrs | After | NDes | 2011 | 199 | |
| Stylophora pistillata | Adult | A | 6 to 8 wks | NA | Des | AB, AM | 2012 | 209 | |
| Pocillopora damicornis | Adult | S | >2 days | NS | Des | AB, AM | 2013 | 206 | |
| Pocillopora damicornis | Adult | S | 5 to 11 days | NA | NDes | 2014 | 197 | ||
| Fungia granulosa | Adult | S | >2 mos | NA | NDes | AB | 2015 | 200 | |
| Pocillopora damicornis, Seriatopora hystrix, Stylophora pistillata | Adult | A, S | Wks to mos | After | NDes | 2016 | 203 | ||
| Stylophora pistillata | Adult | A | >12 days | NA | Des | AB, AM | 2017 | 210 | |
| Goniopora lobata | Adult | S | NA | NA | NDes | 2017 | 198 | ||
| Pocillopora damicornis | Nematostella vectensis | Adult | S | 12 to 13 days | Reported | Des | AB | 2021 | 216 |
| Pocillopora acuta | Adult | S | 7 to 10 days | NO | Des | AB, AM | 2021 | 213 | |
| Acropora tenuis | Planula | S | >8 mos | CP | Des | AB, AM | 2021 | 212 |
S, suspended; A, adherent.
wks, weeks; mos, months; yrs, years.
NA, not applicable; CP, clear proliferation; NS, not significant; NO, not observed; After, after settlement and differentiation; Reported, reported but time limited.
Des, destructive; NDes, nondestructive.
AB, antibiotic; AM, antimycotic.
Coral tissue explants can be maintained in either suspended (204–206) or adherent cultures (202, 207–209). Suspended aggregates are ball-shaped and present a tissue organization similar to coral colonies (204–206). These also maintain a photophysiology comparable to that of the parental colony (200). They have been used to investigate the involvement of light and oxidative stress in coral bleaching (205) and to study ecotoxicology of cryoprotectants (197). Interestingly, the method developed by Vizel et al. (199) produced explants that can be kept for several months in an undeveloped state or induced to develop into a polyp that calcifies and ultimately regrows into a colony. In addition, Symbiodiniaceae-free tissue balls potentially provide a tool to complement investigation on the coral-algal symbiosis (198).
Adherent aggregates are generally flat, but spatial relationships between the diverse cell types are not well resolved between isolation protocols (202, 207–209). Only these adherent aggregates (as opposed to tissue balls) calcify in vitro, likely owing to the presence of the ECM and skeletal organic matrix which are secreted de novo (207, 208). The easy access to the site of calcification in adherent cultures has allowed researchers to reveal important details of the calcification process, such as its conditional independence from photosynthetic activity of the Symbiodiniaceae partner (209), the intracellular commencement of the biomineralization process (209), and its dependence on multicellularity (e.g., see references 207, 208, and 210). The use of adherent cultures also produced the first evidence of interactions between coral cells and an associated endolithic fungus (211). Although it appears to be limited to pocilloporid corals, polyp bail-out induced through controlled salinity stress is arguably the most successful approach for obtaining adherent coral micropropagates (201, 203). The resulting micropropagated polyps fit inside a microfluidic platform, the “coral-on-a-chip”, representing one of the most advanced tools for live study of coral physiology (203). This made it possible to observe microscopic phenomena in real time, such as calcification, coral-pathogen interactions, and coral bleaching (203).
Untapped Potential of Coral Cell Cultures
The next stage in structural simplification of the coral host can be achieved with secondary cultures and cell lines (cultures that can be propagated indefinitely). These are less-representative versions of the original biological system because they filter out some cell types, but they overcome the time limitation of primary cultures and the need to continuously source from living organisms. To date, most coral-derived cell cultures have only short viability and coral cell lines have been established only very recently (212, 213) (Table 3). The most remarkable achievements in terms of culture longevity rely on cells originating from larvae (212, 214, 215). This approach resulted in mixed cell cultures visibly proliferating and remaining viable for between 10 weeks (214) and upwards of 8 months, with the possibility of restarting the culture after cryopreservation and rewarming (212, 215). To this point proliferating cultures either contained mixed, often unidentified, cell types (214, 216), or homogeneous, but not a priori specifically selected for, cell populations (212). Nevertheless, cell lines with specific properties could be selected a posteriori from the pool of available established cell lines, each of which reportedly expressed specific and consistent sets of genes (over time) reminiscent of different cell types (e.g., gastrodermis, epidermis, and secretory, undifferentiated, and neuronal cells) (212). This was demonstrated (in principle) after cells from a line showing endoderm-like properties successfully established endosymbiosis (in vitro) when exposed to cultured Symbiodiniaceae (215).
Among the various cell types, stem-cell like cells are particularly sought after because of their potential to initiate persistent cell lines. Currently all immortal animal cell lines originate from tumors or from experimentally reprogrammed cells (217). These behave substantially differently than physiological cell populations (218). For that matter, tropical corals (and other cnidarians) are an attractive subject for cell cultures, owing to their longevity (219) and regenerative capacity, which suggests that stem-cell like cell populations remain abundant throughout the life of these organisms (220). While adult somatic cells of hydrozoans have been described to dedifferentiate (i.e., return to a pluri- or totipotent state) and transdifferentiate (i.e., differentiate into different cell types) (220–222), a mechanistic understanding of coral self-renewal is still lacking (223). Stem-cell like cell lineages have been identified and characterized in cnidarians (class Hydrozoa) as interstitial cell lines (or I cells) (220, 224). In corals (class Anthozoa), protease-treated larval-derived cells assumed amorphous shapes with extended pseudopodia capable of proliferating in vitro indefinitely. These appeared morphologically similar to endoderm precursor cells (212) and resemble amoebocytes, which are hypothesized to play a similar role to I cells in non-hydrozoan cnidarians (223). Gene expression patterns, enzymatic activity assays such as applied to human hematopoietic stem cells based on aldehyde dehydrogenase, and fluorescence-activated cell sorting (FACS) could help locate stem-cell like cells in corals (225, 226), and some coral cell lines have indeed shown properties of progenitor cells (212). However, even single-cell RNA sequencing of FACS-sorted cells from swimming larvae, primary polyps, and adult colonies of Stylophora pistillata could not identify any stem-cell like cell populations (227). The cellular basis underlying coral regenerative capacity thus remains elusive.
Another limitation in establishing coral cell cultures is the lack of coral-specific culturing formulations (213). The culture media used presently seem to favor growth of contaminants rather than of coral cells, and dilution leads to better results (196). In addition, for many coral cell types, proliferation likely requires initial adhesion to a substrate (196, 213). Therefore, lack of knowledge about the structure and function of regenerative systems and of appropriate culturing conditions hinder the culturability of coral tissues and cells (196, 213). This field might benefit from a change of perspective that focuses on the peculiarity of corals rather than on their similarity with terrestrial metazoans. Culturing techniques could be improved through reverse-engineered approaches that use (meta)genomic, transcriptomic, and proteomic information to guide the design of culturing media and protocols, as pioneered in bacterial culturing (228, 229).
Simplifying the Coral Holobiont by Disassembling Its Members
In the intact coral holobiont, the interdependence and complementarity of processes underlying fundamental functions hamper the understanding of contributions of individual members (80). Hence, as a complementary strategy to structural simplification, the complexity of the coral holobiont can also be simplified by disassembling and isolating its members (host and microorganisms, respectively). Such approaches can guide studies aimed at clarifying partner dynamics, and generate predictions that will then ultimately need validation through the controlled reassembly of metaorganism components (80, 100).
Possibility of gnotobiotic coral hosts.
Axenic (germ-free) or gnotobiotic hosts (where all associated microbes are known), are powerful tools to study the role of the microbiome in health and fitness (as reviewed in reference 80). Comparison between symbiotic and axenic individuals allows us to explore the contribution of the microbiome to host physiology, and targeted inoculation with selected microorganisms can help identify causative links to their function (100, 101, 105). By combining established Symbiodiniaceae and bacterial depletion protocols, the generation of gnotobiotic coral hosts can be broken down into sequential steps.
The removal of Symbiodiniaceae from the gastrodermal tissue by bleaching produces aposymbiotic hosts and can be seen as a first step in holobiont simplification. Ideal methods maximize bleaching efficacy while they minimize the impact on the host and the remaining microbiota. Temperature stress was among the first methods utilized to bleach corals in artificial systems (27); however, it may result in high host mortality, not be fully effective, and potentially influence thermotolerance in subsequent studies (126, 230). The herbicide Diuron or DCMU [N′-(3,4-dichlorophenyl,-N,N-dimethylurea)] overcomes most of these limitations, but it also does not lead to complete bleaching and it is a hazardous substance (231, 232). Menthol is considered a more “gentle,” yet effective, bleaching agent that has now been applied to many cnidarians (170, 232, 233). However, the exact mechanism triggering the bleaching response remains to be fully elucidated (232, 234). Menthol-bleached hosts remain aposymbiotic (for at least 15 weeks, in Aiptasia) after the cessation of the treatment (126) and can subsequently be employed in experiments and reestablish symbiosis with Symbiodiniaceae (126, 232). Of note, while a fully aposymbiotic state can be difficult to achieve (and prove), removing ~98% of the symbiont population appears sufficient to allow inoculated non-native symbionts to establish and repopulate the host (235). How these chemical bleaching agents affect other microorganisms in the coral holobiont, however, remains unknown.
Completing a fully sterile life cycle is the gold standard of developing true axenic animals (236). Similarly, protocols for creating axenic Hydra polyps make use of sterile rearing techniques and its closed life cycle in the laboratory (112), a prospect that may become attainable in research on corals given advancements in artificially producing larvae from brooding species (237, 238) and ex situ techniques for spawning coral species (239, 240). Among cnidarians, Hydra, Nematostella, and Aiptasia adult polyps can be rendered gnotobiotic through antimicrobial treatment (100, 113, 241). This represents a complementary or alternative approach, which has been successful in eliminating more than 99% of the microbial load of corals (242), when a closed life cycle is not (yet) available (100, 236, 241). However, microbial load can recover in as little as 96 h once dosing stops (242). Although no protocol for long-term maintenance of gnotobiotic corals after antimicrobial treatment is available yet, it could be argued that organisms that are allowed to develop naturally (i.e., with their native microbiome), and only later undergo microbial depletion, represent more realistic models of wild-type organisms (100, 236).
The efficacy of the holobiont disassembly process will vary between coral life histories. First, coral species have different strategies of acquiring their microbiome and vary in microbiome flexibility at early life stages (243) and as adults (33). The main division occurs between broadcast spawners and brooders (244), with the former acquiring Symbiodiniaceae and likely bacteria horizontally from the environment (244, 245), while the latter mostly inherit them vertically from the parental colony (172). Second, in contrast to axenic mice and Hydra, raising axenic corals may be challenging as it remains unclear whether corals require the presence of specific microorganisms to complete development (246). Finally, antimicrobial treatment might be detrimental as it was shown to cause disaggregation of tissue in coral larvae (214) and adults in some cases (216). Since this might be a feature of the specific antimicrobials employed, testing substances with different mechanisms of action (e.g., azoles) is warranted. Nevertheless, antimicrobial treatment may not be effective in the long-term due to difficulties of antibiotics reaching the inner skeleton (21, 242). Early-life stages after settlement may therefore be particularly suitable for manipulation, as they have more dynamic microbial communities than do adult corals (45, 243).
Symbiodiniaceae cultures.
Symbiodiniaceae cultures represent the first established and most advanced cell culturing technique in coral research. Symbiodiniaceae are routinely studied for their properties in culture (247, 248) and are used to study early symbiont acquisition by larvae, symbiosis establishment and reestablishment (e.g., following bleaching) dynamics, and comparative physiology in the host (133, 149, 169). While historically only a small proportion of coral-associated Symbiodiniaceae have been considered culturable (249), new approaches such as isolation and culturing from single cells promise innovation in the field (250).
Symbiodiniaceae in culture experience substantially different conditions compared to those in hospite (within the host) and therefore do not perfectly replicate endosymbiotic dynamics (215, 251, 252). Many cultures utilize antibiotics to keep bacterial contamination to a minimum, but such practices could affect Symbiodiniaceae, either through side effects of the antibiotics or through the induced loss of their bacterial associates (253, 254).
Bacterial cultures.
The majority of our knowledge on coral-associated bacteria is based on 16S rRNA gene amplicon sequencing, while metabolic pathways and interactions in the holobiont are less well-explored (55). The use of culture-based approaches may be one option which will likely provide additional insight into microbial functions (e.g., reference 255), but which has largely been forgotten about in favor of next generation culture-independent methods. Sweet et al. (51) recently curated data of the diversity and function of cultured bacteria (both published and unpublished) from tropical, temperate, and cold-water corals. This resulted in a catalog (isolates.reefgenomics.org) of 3,055 unique isolates that spanned 138 species and 12 putatively novel bacterial genera across the Pseudomonadota (Proteobacteria), Firmicutes, Bacteroidetes, and Actinobacteria phyla. Available genomes from these bacteria were considerably sparse, but those available (74 at the time of writing) allowed the researchers to analyze biosynthetic gene clusters underlying the production of secondary metabolites important in host health and symbiosis (51, 256). Despite this promising start, most bacteria have yet to be cultured, and some have even been deemed unculturable (257). Indeed, a metadata analysis of SSU rRNA gene sequences from bacteria and archaea associated with corals found that only 6.5% of these were generated from cultured isolates (258). Unlocking at least part of this additional diversity is likely to be achieved with alternative isolation and cultivation procedures, inspired by advancements in the broader microbiological field (259, 260). For example, the gradients of physicochemical growth conditions could be widened within the ‘culturomics’ framework (238), and implementing microfluidics systems (261–263). The growth of obligate symbiotic or syntrophic bacteria could be achieved through co-culturing (264), and growth media and sorting methods could be developed through omics-guided approaches (228, 229).
Microbial contaminants in cultures: friends or foes?
Invasion of cell cultures by microbial contaminants is a universal issue. Antimicrobial agents routinely employed in terrestrial animal cell cultures are largely ineffective against coral microbial contaminants, which are known to overgrow and cause the termination of coral cell cultures (200, 214, 216, 265). While there is clearly a need for coral-specific antimicrobial treatments, corals are exceptional in hosting microbes not only on external surfaces and mucosa but also in all other tissue compartments and in the skeleton (21, 53). In addition, although cultured Symbiodiniaceae strains are often treated with antimicrobials (249) and can in some cases be maintained axenically (118), Symbiodiniaceae cultures also harbor abundant and characteristic bacterial microbiomes (266). Recently, bacteria have also been reported to associate with Symbiodiniaceae intracellularly as well as extracellularly (267). This suggests a tight involvement of the coral microbiome in holobiont functioning and regulation. Indeed, the longest viability of coral explants was achieved from protocols that did not use antimicrobials (199, 203) (Table 3). In contrast, the first coral cell lines were grown in media containing antimicrobials (212, 215). This indicates a potential connection between the presence of associated bacteria and the formation of complex structures (tissues), and represents an incentive to investigate whether so-called microbial contaminants might comprise key coral associates.
Reassembling the Metaorganism for Hypothesis Testing
The study of isolated holobiont members is necessary to generate hypotheses on their functions and the dynamics of their interactions (100, 241); however, these hypotheses then need to be tested in a metaorganism context (80). For this purpose, reassembling metaorganisms represents the ultimate testing ground. Practically speaking, the inoculation of axenic or gnotobiotic hosts with cultured or “transplanted” (46, 268) microbial isolates will elucidate the intra- and interkingdom interactions underpinning holobiont functioning. This approach borrows from the field of human gut microbiome, where studies on animal models could demonstrate causative links between the presence of specific bacteria and the host phenotype. For example, the introduction of a single gut-residing bacteria in axenic mice led to the development of autoimmune arthritis (269) and the presence or absence of a bacterial consortium modulated food allergy in the host (270). More recently, several authors have proposed to adopt this type of approach based on success in other cnidarian models (100, 105, 110, 113, 241, 271, 272) and on promising first applications in some reef-building coral species (46).
TROPICAL STONY CORALS AS CANDIDATE MODEL SPECIES
The validation of laboratory results from model systems on true corals remains irreplaceable in the transition from controlled experiments to practical implementation of conservation activities. Non-coral model organisms lack important features such as the aragonite skeleton, obligate nature of the symbiosis with Symbiodiniaceae, and adaptation to oligotrophic conditions. Although the term “coral model” is found in the literature and attributed to a number of species (see reference 213 and references therein), to date there is no formally established or universally agreed-upon true coral model organism. Establishment of such models should start by identifying a group of promising species which possess characteristics that maximize amenability to experimentation (tractability) and informative power (transferability of knowledge to ecologically relevant contexts). Because tractability and transferability often can be antithetic, we discuss the most relevant factors that affect these two properties in selected candidate coral model species (see, e.g., Table 4).
TABLE 4.
Relevant features to identify and evaluate the suitability of candidate coral species to the establishment of true coral model organismsa
| Characteristics | Observation(s)b |
||||
|---|---|---|---|---|---|
| Pocillopora damicornis | Stylophora pistillata | Acropora millepora | Galaxea fascicularis | Orbicella faveolata | |
| Tractability | |||||
| Distribution (% of global ecoregions according to CotW) | Very broad (85.3) | Broad (68.7) | Broad (56.7) | Broad (67.3) | Localized (5.3) |
| Suitability to aquarium rearing | High, aquarium culture >10 yrs (326) | High, aquarium culture >30 yrs (273) | High, completed life cycle ex situ (240) | High (327, 328) | High (329) |
| Amenability to experimental bleaching | Unknown | High, effective with menthol (232) | Unknown | High, effective with menthol (328) | Unknown |
| Polyp size range in mm (corallite diam) | 0.8–1 (WoRMS, CTD) | 0.9–1.4 (CTD) | 0.4–1.6 (CTD) | 5–10 (CotW) | 2–3 (CotW) |
| Others | Polyp-bailout stress response (203, 330, 331) | Polyp-bailout stress response (203) | First broadcast spawning coral to produce F2 fully ex situ (240) | Protruding corallites allow easy isolation of individual polyps; polyps remain extended during daytime; large egg size (332) | Major reef builder in the Caribbean (333) |
| Transferability | |||||
| Natural occurrence: oceanic basin | IndoPacific (CotW) | IndoPacific (CotW) | IndoPacific (CotW) | IndoPacific (CotW) | Atlantic (CotW) |
| Taxonomy: host clade | Robusta (334) | Robusta (335) | Complexa (334) | Complexa (334) | Robusta (336) |
| Taxonomy: host family | Pocilloporidae (WoRMS) | Pocilloporidae (WoRMS) | Acroporidae (WoRMS) | Euphylliidae (WoRMS) | Merulinidae (WoRMS) |
| Taxonomy: Symbiodiniaceae genus | Cladocopium spp. most common, Symbidinium and Durusdinium spp. also found (CTD) | Cladocopium spp. (CTD) | Cladocopium spp. (CTD) | Cladocopium and/or Durusdinium spp. (337–341) | Brevioulum spp. predominant; Symbidinium, Cladocopium, and Durusdinium spp. also found (CTD) |
| Colony morphology | Branched, usually <30 cm tall (WoRMS) | Branching to submassive (CofW) | Corymbose cushions or clumps (CotW) | Massive (often dome-shaped) or columnar (CotW) | Massive, sizes up to 10 m (CotW) |
| Trophic strategy | Relatively autotrophic (280) | Mixotrophic but with great variability (342) | |||
| Habitat preference depth | 0 to >40 m (particularly abundant at 5 to 20 m) (WoRMS) | 1 to 65 m (343) | 2 to 30 m (CTD) (344) | 2 to 20 m (CotW, WoRMS) | 0.5 to 40 m (CTD) |
| Reproductive mode | Brooder (predominantly) and broadcast spawner (244) | Brooder (peculiarity: protandrous simultaneous hermaphrodites) (345) | Broadcast spawner (244, 346) | Broadcast spawner (peculiarity: pseudogynodioecious) (347) | Broadcast spawner (348) |
Tractability refers to traits that facilitate experimental work, while transferability indicates the most relevant aspects to consider to address the broad variation encompassed by the scleractinian taxon. This list can be considered a template or guide to be applied beyond the species listed here.
Abbreviations: CotW, Corals of the World (www.coralsoftheworld.org); CTD, Coral Trait Database (https://www.coraltraits.org/); WoRMS, World Register of Marine Species (https://www.marinespecies.org). Reference sources are indicated parenthetically where applicable.
Trait-Based Approach To Identify Suitable Coral Species
Tractability.
Tractability is the sum of a set of characteristics that make a coral species a practically more convenient study subject (Table 4). These characteristics include: (i) broad availability in geographic distribution and abundance in the reef (however, this aspect may only be relevant in the initial exploratory stage: once the species becomes established as a model organism, research will focus on a few selected strains/clonal lines shared between research groups, overcoming the need to source colonies from the wild); (ii) compatibility with growth under aquarium conditions to maintain long-term cultures or living collections (this includes for example colonies of Stylophora pistillata that have been kept in aquarium culture for more than 3 decades) (273); (iii) amenability to bleaching, maintenance in a bleached state, and symbiosis reestablishment to disentangle host and Symbiodiniaceae contribution to holobiont functioning (85); and (iv) larger polyps (e.g., in Galaxea fascicularis) offer the option to isolate a single polyp, reducing complexity from the colony to the individual level (274, 275). Corals with larger polyps are also easier to visually examine (276, 277), not to mention separating the tissue from the skeletal matrix (197–200). (v) Finally, further case-specific advantageous traits could also include the continuous release of larvae (237, 238) with the long-term prospect of achieving a closed axenic life cycle or a consistent budding and polyp bailout response to reliably produce tissue cultures (203, 278, 279). Some of these traits are correlated and tend to co-occur. For example, survival in the bleached state is linked to polyp size because it generally correlates with heterotrophic feeding capacity (280).
Transferability.
Coral model species with high tractability will be representative of only a subsample of the scleractinian taxon. Therefore, their representativeness and transferability should be considered targeting a diverse suite of model systems. We suggest identifying coral model species that aim at collectively covering the broadest range possible of the following aspects (Table 4): (i) geographic distribution, where at both larger and smaller scales, different regions are dominated by different species of both coral host and associated Symbiodiniaceae (15, 281), and (ii) phylogeny, where metabolism and microbiome composition differ between the two major coral clades (282, 283) and thermal tolerance varies between coral families (284). Of the hundreds of extant tropical reef-building coral species, only a few have been extensively investigated, with the majority of studies focusing on members of just a few families (29). Although this might seem like an underrepresentation, it is noteworthy that the three most abundant families (Acroporidae, Pocilloporidae, and Poritidae) make up ~70% of corals on the coral reefs worldwide (see reference 285 and references therein). In addition, Symbiodiniaceae identity should also be considered, since it is known to affect metabolism and thermotolerance (286, 287). The following aspects should also be taken into account: (iii) colony morphology, which plays an important role in thermotolerance (284, 288) and is influenced by external conditions (289); (iv) trophic strategy, which can range from more autotrophic to more heterotrophic and also affects holobiont thermal tolerance (280); (v) habitat preference, which determines the exposure and acclimatization potential to stressors (290, 291) and can vary with depth, distance from shore, and reef type and topography, among others; (vi) reproductive mode, which determines the mode of microbiome transmission between generations and affects the stability of the microbiome and the potential for transgenerational adaptation (23, 292, 293); and (vii) host-symbiont flexibility, since the ability of the coral host to associate with different Symbiodiniaceae strains and species has been linked to adaptive capacity (281, 294). However, to allow for comparisons between host species, a framework for the quantification of this trait is needed. Namely, it is necessary to define a methodology for symbiont characterization together with a metric to quantify flexibility (or specificity), as well as to explore within-species variation through balanced sampling efforts that account for temporal, spatial, and environmental variability (295). Further, this approach could be expanded to the other components of the microbiome (e.g., bacteria [33]).
JOINING FORCES: EXPANDING COMMUNITY-BASED APPROACHES TO CNIDARIAN MODEL ORGANISMS
While it is necessary to establish multiple and diverse model systems (67, 68), strategic focus on a limited number of species will increase efficiency of resource use. A clearly stated or universally agreed-upon selection of “best candidate” species for the establishment of true coral model organisms seems to be lacking; however, the coral field clearly has its favorites. For example, over the last 30 years the three species Pocillopora damicornis, Stylophora pistillata, and Acropora millepora were preferentially used in coral heat stress experiments (29). Although these may be excellent candidates for a true coral model, preexisting knowledge is not per se a requirement nor an indicator of the validity of species as model organisms (68), but a widespread appreciation might be a good indicator of their practical advantages that should be taken into account.
Cnidarian model systems can accelerate the rate of discovery necessary to develop solutions for coral reef preservation, and these tools can become particularly powerful when part of a structured framework with defined goals and strategies (10, 76, 80). The idea of a community-based coordinated effort to develop and establish cnidarian model systems was put forward with a “call to arms” (10, 76). The main recommendations included (i) a reframing of researchers’ attitude to put collective achievements above individual accomplishments, together with (ii) cooperation between working groups to improve efficiency, through active communication and resource sharing, and (iii) the adoption of a targeted approach that prioritizes the most pressing issues pertinent to corals and coral reef adaptation to future ocean conditions.
Examples that show encouraging progress in this direction can be found among the existing cnidarian model organism working groups and open resource networks. Researchers and educators interested in working with Hydra can find a comprehensive collection of best practices and resources through the OpenHydra (http://openhydra.org/) platform, and the Hydra 2.0 Genome Project Portal (https://research.nhgri.nih.gov/hydra/) provides easy access to the data generated from the Hydra genome sequencing projects. The Aiptasia community has developed a web site (https://aiptasia-resource.org/) and a Protocols.io group (https://www.protocols.io/workspaces/aiptasiasymbiodiniaceae-model-system) for researchers and educators who are interested in using Aiptasia in the laboratory and classroom (Table 2). Current resources for the Cassiopea model include a publicly available draft genome (https://mycocosm.jgi.doe.gov/Casxa1/Casxa1.home.html) and 10 clonal lines available upon request (http://medinalab.org/new/). The genome line T1-A is kept by multiple labs around the world (https://cassiopeabase.org/). The bacterial species, Pseudoalteromonas sp., that is an active inducer of settlement is available upon request as well as other Cassiopea spp. bacterial isolates from different developmental stages (medinalab.org [A. H. Kerwin et al., unpublished data]). Research on A. poculata has accelerated rapidly in recent years, thanks to the development of a large research collaborative focused on the temperate coral genera Astrangia and Oculina. The Temperate Coral Research Working Group (https://sites.bu.edu/astrangia/), which now exceeds 100 researchers, has met annually since 2016 (with the exception of 2020) and consists of researchers, educators, and journalists who work with Astrangia and Oculina, all of whom are addressing long-standing questions of coral-microbe symbiosis, as well as climate change education and outreach. A draft genome of the host has been assembled and is currently under annotation. Breviolum psygmophilum, the intracellular photosymbiont thought to be present in all A. poculata populations, is in culture, and draft transcriptomes of B. psygmophilum and other congeners exist. 16S rRNA sequences from A. poculata specimens from two populations in the United States (Jamestown, RI, and Woods Hole, MA) have been deposited and are publicly available on the Coral Microbiome Portal (https://www2.whoi.edu/site/amy-apprill/coral-microbiome-portal/). Protocols for spawning, embryonic development, larval rearing, larval settlement, and CRISPR protocols have been developed and will be shared so that they are publicly available for researchers. Regarding the establishment of true coral models, however, there is still a general lack of coordination, and we can draw inspiration from the work done on the other cnidarian model systems.
Community-based and coordinated efforts may further include the integrated use of the different model systems, for example, in a validation sequence where first explorations are conducted on more tractable systems and are subsequently validated on systems with higher transferability, improving both speed and efficiency. In this context, the protocols for drug development and approval provide a valuable example of integrated approaches that combine the use of several model systems selected for the complementarity of their features. Similar to preclinical trials in human medical applications, test series are performed on model systems of increasing complexity starting from in vitro assays, and proceeding on to animals that possess anatomical and physiological features comparable to humans regarding the effects of a particular drug. Only drugs that pass all of the sequential validations are considered for testing on humans in clinical trials. In analogy, coral-related biological features can first be assayed in tissue or cell cultures and/or non-coral models, to be later corroborated by direct testing on corals (80). This process has already been successfully implemented to explore the involvement of cellular and immunity responses in coral bleaching, by first using Aiptasia and subsequently verifying on stony corals (reviewed in reference 10). Therefore, joining forces and coordinating efforts among groups working on different cnidarian model systems represents a promising approach to accelerate the development of solutions for coral reef conservation.
ACKNOWLEDGMENTS
This study is part of the “Ocean2100” global change simulation project of the Colombian-German Center of Excellence in Marine Sciences (CEMarin) funded by the German Academic Exchange Service. S.F. was funded by DFG grant CRC 1182 “Origin and Function of metaorganisms,” project B1. M.M. was funded by NSF grants OCE 1442206 and OCE 1642311. V.M.W. was funded by NSF grants IOS 1645164, IOS 2124119, and EF-URoL 2025476. K.S. was supported in part by the Institutional Development Award (IDeA) Network for Biomedical Research Excellence from the National Institute of General Medical Sciences of the National Institutes of Health under grant number P20GM103430.
The cnidarian and microbe pictograms were created by Giulia Puntin (licensed under CC BY 4.0, https://github.com/sPuntinG/Coral_stuff) and modified for use here.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Reaka-Kudla M, Wilson DE, Wilson EO. 1997. The global biodiversity of coral reefs: a comparison with rain forests, p 83–108. In Biodiversity II: understanding and protecting our biological resources. Joseph Henry/National Academy Press, Washington, DC. [Google Scholar]
- 2.Costanza R, de Groot R, Sutton P, van der Ploeg S, Anderson SJ, Kubiszewski I, Farber S, Turner RK. 2014. Changes in the global value of ecosystem services. Glob Environ Chang 26:152–158. 10.1016/j.gloenvcha.2014.04.002. [DOI] [Google Scholar]
- 3.van Hooidonk R, Maynard J, Tamelander J, Gove J, Ahmadia G, Raymundo L, Williams G, Heron SF, Planes S. 2016. Local-scale projections of coral reef futures and implications of the Paris Agreement. Sci Rep 6:39666. 10.1038/srep39666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hughes TP, Kerry JT, Álvarez-Noriega M, Álvarez-Romero JG, Anderson KD, Baird AH, Babcock RC, Beger M, Bellwood DR, Berkelmans R, Bridge TC, Butler IR, Byrne M, Cantin NE, Comeau S, Connolly SR, Cumming GS, Dalton SJ, Diaz-Pulido G, Eakin CM, Figueira WF, Gilmour JP, Harrison HB, Heron SF, Hoey AS, Hobbs J-PA, Hoogenboom MO, Kennedy EV, Kuo C, Lough JM, Lowe RJ, Liu G, McCulloch MT, Malcolm HA, McWilliam MJ, Pandolfi JM, Pears RJ, Pratchett MS, Schoepf V, Simpson T, Skirving WJ, Sommer B, Torda G, Wachenfeld DR, Willis BL, Wilson SK. 2017. Global warming and recurrent mass bleaching of corals. Nature 543:373–377. 10.1038/nature21707. [DOI] [PubMed] [Google Scholar]
- 5.Castro-Sanguino C, Ortiz JC, Thompson A, Wolff NH, Ferrari R, Robson B, Magno-Canto MM, Puotinen M, Fabricius KE, Uthicke S. 2021. Reef state and performance as indicators of cumulative impacts on coral reefs. Ecol Indic 123:107335. 10.1016/j.ecolind.2020.107335. [DOI] [Google Scholar]
- 6.Kennedy EV, Perry CT, Halloran PR, Iglesias-Prieto R, Schönberg CHL, Wisshak M, Form AU, Carricart-Ganivet JP, Fine M, Eakin CM, Mumby PJ. 2013. Avoiding coral reef functional collapse requires local and global action. Curr Biol 23:912–918. 10.1016/j.cub.2013.04.020. [DOI] [PubMed] [Google Scholar]
- 7.Anthony K, Bay LK, Costanza R, Firn J, Gunn J, Harrison P, Heyward A, Lundgren P, Mead D, Moore T, Mumby PJ, van Oppen MJH, Robertson J, Runge MC, Suggett DJ, Schaffelke B, Wachenfeld D, Walshe T. 2017. New interventions are needed to save coral reefs. Nat Ecol Evol 1:1420–1422. 10.1038/s41559-017-0313-5. [DOI] [PubMed] [Google Scholar]
- 8.Duarte CM, Agusti S, Barbier E, Britten GL, Castilla JC, Gattuso JP, Fulweiler RW, Hughes TP, Knowlton N, Lovelock CE, Lotze HK, Predragovic M, Poloczanska E, Roberts C, Worm B. 2020. Rebuilding marine life. Nature 580:39–51. 10.1038/s41586-020-2146-7. [DOI] [PubMed] [Google Scholar]
- 9.Hoegh-Guldberg O, Kennedy EV, Beyer HL, McClennen C, Possingham HP. 2018. Securing a long-term future for coral reefs. Trends Ecol Evol 33:936–944. 10.1016/j.tree.2018.09.006. [DOI] [PubMed] [Google Scholar]
- 10.Weis VM. 2019. Cell biology of coral symbiosis: foundational study can inform solutions to the coral reef crisis. Integr Comp Biol 59:845–855. 10.1093/icb/icz067. [DOI] [PubMed] [Google Scholar]
- 11.Rohwer F, Seguritan V, Azam F, Knowlton N. 2002. Diversity and distribution of coral-associated bacteria. Mar Ecol Prog Ser 243:1–10. 10.3354/meps243001. [DOI] [Google Scholar]
- 12.Blackall LL, Wilson B, Van Oppen MJH. 2015. Coral: the world’s most diverse symbiotic ecosystem. Mol Ecol 24:5330–5347. 10.1111/mec.13400. [DOI] [PubMed] [Google Scholar]
- 13.LaJeunesse TC, Parkinson JE, Gabrielson PW, Jeong HJ, Reimer JD, Voolstra CR, Santos SR. 2018. Systematic revision of Symbiodiniaceae highlights the antiquity and diversity of coral endosymbionts. Curr Biol 28:2570–2580.e6. 10.1016/j.cub.2018.07.008. [DOI] [PubMed] [Google Scholar]
- 14.Quigley KM, Davies SW, Kenkel CD, Willis BL, Matz MV, Bay LK. 2014. Deep-sequencing method for quantifying background abundances of Symbiodinium types: exploring the rare Symbiodinium biosphere in reef-building corals. PLoS One 9:e94297. 10.1371/journal.pone.0094297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ziegler M, Stone E, Colman D, Takacs-Vesbach C, Shepherd U. 2018. Patterns of Symbiodinium (Dinophyceae) diversity and assemblages among diverse hosts and the coral reef environment of Lizard Island, Australia. J Phycol 54:447–460. 10.1111/jpy.12749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McDevitt-Irwin JM, Baum JK, Garren M, Vega TR. 2017. Responses of coral-associated bacterial communities to local and global stressors. Front Mar Sci 4:1–16. [Google Scholar]
- 17.Hernandez-Agreda A, Leggat W, Bongaerts P, Herrera C, Ainsworth TD. 2018. Rethinking the coral microbiome: simplicity exists within a diverse microbial biosphere. mBio 9:1–14. 10.1128/mBio.00812-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wegley L, Yu Y, Breitbart M, Casas V, Kline DI, Rohwer F. 2004. Coral-associated archaea. Mar Ecol Prog Ser 273:89–96. 10.3354/meps273089. [DOI] [Google Scholar]
- 19.Wegley L, Edwards R, Rodriguez-Brito B, Liu H, Rohwer F. 2007. Metagenomic analysis of the microbial community associated with the coral Porites astreoides. Environ Microbiol 9:2707–2719. 10.1111/j.1462-2920.2007.01383.x. [DOI] [PubMed] [Google Scholar]
- 20.Kwong WK, del Campo J, Mathur V, Vermeij MJA, Keeling PJ. 2019. A widespread coral-infecting apicomplexan with chlorophyll biosynthesis genes. Nature 568:103–107. 10.1038/s41586-019-1072-z. [DOI] [PubMed] [Google Scholar]
- 21.Sweet MJ, Croquer A, Bythell JC. 2011. Bacterial assemblages differ between compartments within the coral holobiont. Coral Reefs 30:39–52. 10.1007/s00338-010-0695-1. [DOI] [Google Scholar]
- 22.Apprill A, Weber LG, Santoro AE. 2016. Distinguishing between microbial habitats unravels ecological complexity in coral microbiomes. mSystems 1:e00143-16. 10.1128/mSystems.00143-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Oppen MJH, Blackall LL. 2019. Coral microbiome dynamics, functions and design in a changing world. Nat Rev Microbiol 17:557–567. 10.1038/s41579-019-0223-4. [DOI] [PubMed] [Google Scholar]
- 24.Muscatine L, Cernichiari E. 1969. Assimilation of photosynthetic products of zooxanthellae by reef coral. Ecol Stud 137:506–523. 10.2307/1540172. [DOI] [PubMed] [Google Scholar]
- 25.Roth MS. 2014. The engine of the reef: photobiology of the coral-algal symbiosis. Front Microbiol 5:422. 10.3389/fmicb.2014.00422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muscatine L, Porter JW. 1977. Reef corals: mutualistic symbioses adapted to nutrient-poor environments. Bioscience 27:454–460. 10.2307/1297526. [DOI] [Google Scholar]
- 27.Gates RD, Baghdasarian G, Muscatine L. 1992. Temperature stress causes host cell detachment in symbiotic cnidarians: implications for coral bleaching. Biol Bull 182:324–332. 10.2307/1542252. [DOI] [PubMed] [Google Scholar]
- 28.Hughes TP, Anderson KD, Connolly SR, Heron SF, Kerry JT, Lough JM, Baird AH, Baum JK, Berumen ML, Bridge TC, Claar DC, Eakin CM, Gilmour JP, Graham NAJ, Harrison H, Hobbs J-PA, Hoey AS, Hoogenboom M, Lowe RJ, McCulloch MT, Pandolfi JM, Pratchett M, Schoepf V, Torda G, Wilson SK. 2018. Spatial and temporal patterns of mass bleaching of corals in the Anthropocene. Science 359:80–83. 10.1126/science.aan8048. [DOI] [PubMed] [Google Scholar]
- 29.McLachlan RH, Price JT, Solomon SL, Grottoli AG. 2020. Thirty years of coral heat-stress experiments: a review of methods. Coral Reefs 39:885–902. 10.1007/s00338-020-01931-9. [DOI] [Google Scholar]
- 30.Sweet MJ, Bulling MT. 2017. On the importance of the microbiome and pathobiome in coral health and disease. Front Mar Sci 4. 10.3389/fmars.2017.00009. [DOI] [Google Scholar]
- 31.Bernasconi R, Stat M, Koenders A, Huggett MJ. 2019. Global networks of Symbiodinium-Bacteria within the coral holobiont. Microb Ecol 77:794–807. 10.1007/s00248-018-1255-4. [DOI] [PubMed] [Google Scholar]
- 32.Garrido AG, Machado LF, Zilberberg C, de Assis Leite DC. 2021. Insights into ‘Symbiodiniaceae phycosphere’ in a coral holobiont. Symbiosis 83:25–39. 10.1007/s13199-020-00735-3. [DOI] [Google Scholar]
- 33.Ziegler M, Grupstra CGB, Barreto MM, Eaton M, BaOmar J, Zubier K, Al-Sofyani A, Turki AJ, Ormond R, Voolstra CR. 2019. Coral bacterial community structure responds to environmental change in a host-specific manner. Nat Commun 10:3092. 10.1038/s41467-019-10969-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pollock FJ, McMinds R, Smith S, Bourne DG, Willis BL, Medina M, Thurber RV, Zaneveld JR. 2018. Coral-associated bacteria demonstrate phylosymbiosis and cophylogeny. Nat Commun 9:4921. 10.1038/s41467-018-07275-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Williams AD, Brown BE, Putchim L, Sweet MJ. 2015. Age-related shifts in bacterial diversity in a reef coral. PLoS One 10:e0144902-16. 10.1371/journal.pone.0144902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raina J-B, Dinsdale EA, Willis BL, Bourne DG. 2010. Do the organic sulfur compounds DMSP and DMS drive coral microbial associations? Trends Microbiol 18:101–108. 10.1016/j.tim.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 37.Pogoreutz C, Rädecker N, Cárdenas A, Gärdes A, Voolstra CR, Wild C. 2017. Sugar enrichment provides evidence for a role of nitrogen fixation in coral bleaching. Glob Chang Biol 23:3838–3848. 10.1111/gcb.13695. [DOI] [PubMed] [Google Scholar]
- 38.Ritchie K. 2006. Regulation of microbial populations by coral surface mucus and mucus-associated bacteria. Mar Ecol Prog Ser 322:1–14. 10.3354/meps322001. [DOI] [Google Scholar]
- 39.Zaneveld JR, McMinds R, Vega Thurber R. 2017. Stress and stability: applying the Anna Karenina principle to animal microbiomes. Nat Microbiol 2:17121. 10.1038/nmicrobiol.2017.121. [DOI] [PubMed] [Google Scholar]
- 40.Torda G, Donelson JM, Aranda M, Barshis DJ, Bay L, Berumen ML, Bourne DG, Cantin N, Foret S, Matz M, Miller DJ, Moya A, Putnam HM, Ravasi T, van Oppen MJH, Thurber RV, Vidal-Dupiol J, Voolstra CR, Watson S-A, Whitelaw E, Willis BL, Munday PL. 2017. Rapid adaptive responses to climate change in corals. Nat Clim Chang 7:627–636. 10.1038/nclimate3374. [DOI] [Google Scholar]
- 41.Sato Y, Ling EYS, Turaev D, Laffy P, Weynberg KD, Rattei T, Willis BL, Bourne DG. 2017. Unraveling the microbial processes of black band disease in corals through integrated genomics. Sci Rep 7:40455. 10.1038/srep40455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sweet M, Burian A, Fifer J, Bulling M, Elliott D, Raymundo L. 2019. Compositional homogeneity in the pathobiome of a new, slow-spreading coral disease. Microbiome 7:1–14. 10.1186/s40168-019-0759-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maynard J, van Hooidonk R, Eakin CM, Puotinen M, Garren M, Williams G, Heron SF, Lamb J, Weil E, Willis B, Harvell CD. 2015. Projections of climate conditions that increase coral disease susceptibility and pathogen abundance and virulence. Nat Clim Chang 5:688–694. 10.1038/nclimate2625. [DOI] [Google Scholar]
- 44.Tracy AM, Pielmeier ML, Yoshioka RM, Heron SF, Harvell CD. 2019. Increases and decreases in marine disease reports in an era of global change. Proc Biol Sci 286:20191718. 10.1098/rspb.2019.1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peixoto RS, Sweet M, Villela HDM, Cardoso P, Thomas T, Voolstra CR, Høj L, Bourne DG. 2021. Coral probiotics: premise, promise, prospects. Annu Rev Anim Biosci 9:265–288. 10.1146/annurev-animal-090120-115444. [DOI] [PubMed] [Google Scholar]
- 46.Doering T, Wall M, Putchim L, Rattanawongwan T, Schroeder R, Hentschel U, Roik A. 2021. Towards enhancing coral heat tolerance: a “microbiome transplantation” treatment using inoculations of homogenized coral tissues. Microbiome 9:102. 10.1186/s40168-021-01053-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peixoto RS, Rosado PM, de Assis Leite DC, Rosado AS, Bourne DG. 2017. Beneficial microorganisms for corals (BMC): proposed mechanisms for coral health and resilience. Front Microbiol 8:341. 10.3389/fmicb.2017.00341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rosado PM, Leite DCA, Duarte GAS, Chaloub RM, Jospin G, Nunes da Rocha U, Saraiva JP, Dini-Andreote F, Eisen JA, Bourne DG, Peixoto RS. 2019. Marine probiotics: increasing coral resistance to bleaching through microbiome manipulation. ISME J 13:921–936. 10.1038/s41396-018-0323-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Santoro EP, Borges RM, Espinoza JL, Freire M, Messias CSMA, Villela HDM, Pereira LM, Vilela CLS, Rosado JG, Cardoso PM, Rosado PM, Assis JM, Duarte GAS, Perna G, Rosado AS, Macrae A, Dupont CL, Nelson KE, Sweet MJ, Voolstra CR, Peixoto RS. 2021. Coral microbiome manipulation elicits metabolic and genetic restructuring to mitigate heat stress and evade mortality. Sci Adv 7:eabg3088. 10.1126/sciadv.abg3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pogoreutz C, Voolstra CR, Rädecker N, Weis V. 2020. The coral holobiont highlights the dependence of cnidarian animal hosts on their associated microbes, p 91–118. In Cellular dialogues in the holobiont. CRC Press, Boca Raton, FL. [Google Scholar]
- 51.Sweet M, Villela H, Keller-Costa T, Costa R, Romano S, Bourne DG, Cárdenas A, Huggett MJ, Kerwin AH, Kuek F, Medina M, Meyer JL, Müller M, Pollock FJ, Rappé MS, Sere M, Sharp KH, Voolstra CR, Zaccardi N, Ziegler M, Peixoto R. 2021. Insights into the cultured bacterial fraction of corals. mSystems 6:e01249-20. 10.1128/mSystems.01249-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nguyen-Kim H, Bouvier T, Bouvier C, Doan-Nhu H, Nguyen-Ngoc L, Rochelle-Newall E, Baudoux A-C, Desnues C, Reynaud S, Ferrier-Pages C, Bettarel Y. 2014. High occurrence of viruses in the mucus layer of scleractinian corals. Environ Microbiol Rep 6:675–682. 10.1111/1758-2229.12185. [DOI] [PubMed] [Google Scholar]
- 53.Pernice M, Raina J-B, Rädecker N, Cárdenas A, Pogoreutz C, Voolstra CR. 2020. Down to the bone: the role of overlooked endolithic microbiomes in reef coral health. ISME J 14:325–334. 10.1038/s41396-019-0548-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fine M, Loya Y. 2002. Endolithic algae: an alternative source of photoassimilates during coral bleaching. Proc Biol Sci 269:1205–1210. 10.1098/rspb.2002.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Robbins SJ, Singleton CM, Chan CX, Messer LF, Geers AU, Ying H, Baker A, Bell SC, Morrow KM, Ragan MA, Miller DJ, Forêt S, Ball E, Beeden R, Berumen M, Aranda M, Ravasi T, Bongaerts P, Hoegh-Guldberg O, Cooke I, Leggat B, Sprungala S, Fitzgerald A, Shang C, Lundgren P, Fyffe T, Rubino F, van Oppen M, Weynberg K, Robbins SJ, Singleton CM, Xin CC, Messer LF, Geers AU, Ying H, Baker A, Bell SC, Morrow KM, Ragan MA, Miller DJ, Forêt S, Voolstra CR, Tyson GW, Bourne DG, Voolstra CR, Tyson GW, Bourne DG, ReFuGe2020 Consortium. 2019. A genomic view of the reef-building coral Porites lutea and its microbial symbionts. Nat Microbiol 4:2090–2100. 10.1038/s41564-019-0532-4. [DOI] [PubMed] [Google Scholar]
- 56.Vega Thurber R, Payet JP, Thurber AR, Correa AMS. 2017. Virus–host interactions and their roles in coral reef health and disease. Nat Rev Microbiol 15:205–216. 10.1038/nrmicro.2016.176. [DOI] [PubMed] [Google Scholar]
- 57.Zilber-Rosenberg I, Rosenberg E. 2008. Role of microorganisms in the evolution of animals and plants: the hologenome theory of evolution. FEMS Microbiol Rev 32:723–735. 10.1111/j.1574-6976.2008.00123.x. [DOI] [PubMed] [Google Scholar]
- 58.Bordenstein SR, Theis KR. 2015. Host biology in light of the microbiome: ten principles of holobionts and hologenomes. PLoS Biol 13:e1002226-23. 10.1371/journal.pbio.1002226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Voolstra CR, Ziegler M. 2020. Adapting with microbial help: microbiome flexibility facilitates rapid responses to environmental change. Bioessays 42:e2000004. 10.1002/bies.202000004. [DOI] [PubMed] [Google Scholar]
- 60.Liew YJ, Howells EJ, Wang X, Michell CT, Burt JA, Idaghdour Y, Aranda M. 2020. Intergenerational epigenetic inheritance in reef-building corals. Nat Clim Chang 10:254–259. 10.1038/s41558-019-0687-2. [DOI] [Google Scholar]
- 61.Rosenberg E, Koren O, Reshef L, Efrony R, Zilber-Rosenberg I. 2007. The role of microorganisms in coral health, disease and evolution. Nat Rev Microbiol 5:355–362. 10.1038/nrmicro1635. [DOI] [PubMed] [Google Scholar]
- 62.Bourne DG, Morrow KM, Webster NS. 2016. Insights into the coral microbiome: underpinning the health and resilience of reef ecosystems. Annu Rev Microbiol 70:317–340. 10.1146/annurev-micro-102215-095440. [DOI] [PubMed] [Google Scholar]
- 63.Layeghifard M, Hwang DM, Guttman DS. 2017. Disentangling interactions in the microbiome: a network perspective. Trends Microbiol 25:217–228. 10.1016/j.tim.2016.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hirano H, Takemoto K. 2019. Difficulty in inferring microbial community structure based on co-occurrence network approaches. BMC Bioinformatics 20:1–14. 10.1186/s12859-019-2915-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Medina M, Baker DM, Baltrus DA, Bennett GM, Cardini U, Correa AMS, Degnan SM, Christa G, Kim E, Li J, Nash DR, Marzinelli E, Nishiguchi M, Prada C, Roth MS, Saha M, Smith CI, Theis KR, Zaneveld J. 2022. Grand challenges in coevolution. Front Ecol Evol 9:1–11. [Google Scholar]
- 66.Leonelli S, Ankeny RA. 2013. What makes a model organism? Endeavour 37:209–212. 10.1016/j.endeavour.2013.06.001. [DOI] [PubMed] [Google Scholar]
- 67.Goldstein B, King N. 2016. The future of cell biology: emerging model organisms. Trends Cell Biol 26:818–824. 10.1016/j.tcb.2016.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Russell JJ, Theriot JA, Sood P, Marshall WF, Landweber LF, Fritz-Laylin L, Polka JK, Oliferenko S, Gerbich T, Gladfelter A, Umen J, Bezanilla M, Lancaster MA, He S, Gibson MC, Goldstein B, Tanaka EM, Hu CK, Brunet A. 2017. Non-model model organisms. BMC Biol 15:1–31. 10.1186/s12915-017-0391-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Galliot B. 2012. Hydra, a fruitful model system for 270 years. Int J Dev Biol 56:411–423. 10.1387/ijdb.120086bg. [DOI] [PubMed] [Google Scholar]
- 70.Gierer A. 2012. The Hydra model: a model for what? Int J Dev Biol 56:437–445. 10.1387/ijdb.113458ag. [DOI] [PubMed] [Google Scholar]
- 71.Hashimoto T, Horikawa DD, Saito Y, Kuwahara H, Kozuka-Hata H, Shin-I T, Minakuchi Y, Ohishi K, Motoyama A, Aizu T, Enomoto A, Kondo K, Tanaka S, Hara Y, Koshikawa S, Sagara H, Miura T, Yokobori SI, Miyagawa K, Suzuki Y, Kubo T, Oyama M, Kohara Y, Fujiyama A, Arakawa K, Katayama T, Toyoda A, Kunieda T. 2016. Extremotolerant tardigrade genome and improved radiotolerance of human cultured cells by tardigrade-unique protein. Nat Commun 7:12808. 10.1038/ncomms12808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Boothby TC, Tapia H, Brozena AH, Piszkiewicz S, Smith AE, Giovannini I, Rebecchi L, Pielak GJ, Koshland D, Goldstein B. 2017. Tardigrades use intrinsically disordered proteins to survive desiccation. Mol Cell 65:975–984.e5. 10.1016/j.molcel.2017.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Goffeau A, Barrell BG, Bussey H, Davis RW, Dujon B, Feldmann H, Galibert F, Hoheisel JD, Jacq C, Johnston M, Louis EJ, Mewes HW, Murakami Y, Philippsen P, Tettelin H, Oliver SG. 1996. Life with 6000 genes. Science 274:546–567. 10.1126/science.274.5287.546. [DOI] [PubMed] [Google Scholar]
- 74.Kachroo AH, Laurent JM, Yellman CM, Meyer AG, Wilke CO, Marcotte EM. 2015. Systematic humanization of yeast genes reveals conserved functions and genetic modularity. Science 348:921–5925. 10.1126/science.aaa0769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fields S, Johnston M. 2005. Whither model organism research? Science 307:1885–1886. 10.1126/science.1108872. [DOI] [PubMed] [Google Scholar]
- 76.Weis VM, Davy SK, Hoegh-Guldberg O, Rodriguez-Lanetty M, Pringle JR. 2008. Cell biology in model systems as the key to understanding corals. Trends Ecol Evol 23:369–376. 10.1016/j.tree.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 77.Sharp KH, Pratte ZA, Kerwin AH, Rotjan RD, Stewart FJ. 2017. Season, but not symbiont state, drives microbiome structure in the temperate coral Astrangia poculata Microbiome 5:120. 10.1186/s40168-017-0329-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ohdera AH, Abrams MJ, Ames CL, Baker DM, Suescún-Bolívar LP, Collins AG, Freeman CJ, Gamero-Mora E, Goulet TL, Hofmann DK, Jaimes-Becerra A, Long PF, Marques AC, Miller LA, Mydlarz LD, Morandini AC, Newkirk CR, Putri SP, Samson JE, Stampar SN, Steinworth B, Templeman M, Thomé PE, Vlok M, Woodley CM, Wong JCY, Martindale MQ, Fitt WK, Medina M. 2018. Upside-down but headed in the right direction: review of the highly versatile Cassiopea xamachana system. Front Ecol Evol 6:35. 10.3389/fevo.2018.00035. [DOI] [Google Scholar]
- 79.Neff EP. 2020. The quest for an animal model of coral health and disease. Lab Anim (NY) 49:37–41. 10.1038/s41684-019-0467-7. [DOI] [PubMed] [Google Scholar]
- 80.Jaspers C, Fraune S, Arnold AE, Miller DJ, Bosch TCG, Voolstra CR, Consortium of Australian Academy of Science Boden Research Conference Participants. 2019. Resolving structure and function of metaorganisms through a holistic framework combining reductionist and integrative approaches. Zoology (Jena) 133:81–87. 10.1016/j.zool.2019.02.007. [DOI] [PubMed] [Google Scholar]
- 81.Welch PS, Loomis HA. 1924. A limnological study of Hydra oligactis in Douglas Lake, Michigan. Trans Am Microsc Soc 43:203–235. 10.2307/3221738. [DOI] [Google Scholar]
- 82.Bryden RR. 1952. Ecology of Pelmatohydra oligactis in Kirkpatricks Lake. Tennessee Ecol Soc Am Ecol Monogr 22:45–68. 10.2307/1948528. [DOI] [Google Scholar]
- 83.Ribi G, Tardent R, Tardent P, Scascighini C. 1985. Dynamics of hydra populations in Lake Zürich, Switzerland, and Lake Maggiore, Italy. Schweiz Z Hydrol 47:45–56. 10.1007/BF02538183. [DOI] [Google Scholar]
- 84.Muscatine L, Lenhoff HM. 1963. Symbiosis: on the role of algae symbiotic with hydra. Science 142:956–958. 10.1126/science.142.3594.956. [DOI] [PubMed] [Google Scholar]
- 85.Davy SK, Allemand D, Weis VM. 2012. Cell biology of cnidarian-dinoflagellate symbiosis. Microbiol Mol Biol Rev 76:229–261. 10.1128/MMBR.05014-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tardent P. 1974. Gametogenesis in the genus hydra. Am Zool 14:447–456. 10.1093/icb/14.2.447. [DOI] [Google Scholar]
- 87.Sugiyama T, Fujisawa T. 1977. Genetic analysis of developmental mechanisms in Hydra I. sexual reproduction of Hydra magnipapillata and isolation of mutants. Dev Growth Differ 19:187–200. 10.1111/j.1440-169X.1977.00187.x. [DOI] [PubMed] [Google Scholar]
- 88.Trembley A. 1744. Mémoires pour servir à l’histoire d’un genre de polypes d’eau douce, à bras en forme de cornes. Jean & Herman Verbeek, Leiden, Netherlands. https://books.google.de/books/about/Memoires_pour_servir_a_l_histoire_d_un.html?id=uR4OAAAAQAAJ&redir_esc=y. Accessed 22 November 2021. [Google Scholar]
- 89.Ratcliff MJ. 2012. The Trembley Effect or the birth of marine zoology. Int J Dev Biol 56:425–436. 10.1387/ijdb.123520mr. [DOI] [PubMed] [Google Scholar]
- 90.Browne EN. 1909. The production of new hydranths in Hydra by the insertion of small grafts. J Exp Zool 7:1–23. 10.1002/jez.1400070102. [DOI] [Google Scholar]
- 91.Spemann H, Mangold H. 1924. Über Induktion von Embryonalanlagen durch Implantation artfremder Organisatoren. Archiv f Mikr Anat u Entwicklungsmechanik 100:599–638. 10.1007/BF02108133. [DOI] [Google Scholar]
- 92.Turing AM. 1952. The chemical basis of morphogenesis. Philos Trans R Soc Lond B Biol Sci 237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hobmayer B, Rentzsch F, Kuhn K, Happel CM, Von Laue CC, Snyder P, Rothbächer U, Holstein TW. 2000. WNT signalling molecules act in axis formation in the diploblastic metazoan Hydra. Nature 407:186–189. 10.1038/35025063. [DOI] [PubMed] [Google Scholar]
- 94.Fraune S, Bosch TCG. 2007. Long-term maintenance of species-specific bacterial microbiota in the basal metazoan Hydra. Proc Natl Acad Sci USA 104:13146–13151. 10.1073/pnas.0703375104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Franzenburg S, Walter J, Künzel S, Wang J, Baines JF, Bosch TCG, Fraune S. 2013. Distinct antimicrobial peptide expression determines host species-specific bacterial associations. Proc Natl Acad Sci USA 110:E3730–E3738. 10.1073/pnas.1304960110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Augustin R, Anton-Erxleben F, Jungnickel S, Hemmrich G, Spudy B, Podschun R, Bosch TCG. 2009. Activity of the novel peptide arminin against multiresistant human pathogens shows the considerable potential of phylogenetically ancient organisms as drug sources. Antimicrob Agents Chemother 53:5245–5250. 10.1128/AAC.00826-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Augustin R, Schröder K, Rincón APM, Fraune S, Anton-Erxleben F, Herbst EM, Wittlieb J, Schwentner M, Grötzinger J, Wassenaar TM, Bosch TCG. 2017. A secreted antibacterial neuropeptide shapes the microbiome of Hydra. Nat Commun 8:1–8. 10.1038/s41467-017-00625-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bosch TCG, Augustin R, Anton-Erxleben F, Fraune S, Hemmrich G, Zill H, Rosenstiel P, Jacobs G, Schreiber S, Leippe M, Stanisak M, Grötzinger J, Jung S, Podschun R, Bartels J, Harder J, Schröder JM. 2009. Uncovering the evolutionary history of innate immunity: the simple metazoan Hydra uses epithelial cells for host defence. Dev Comp Immunol 33:559–569. 10.1016/j.dci.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 99.Fraune S, Augustin R, Anton-Erxleben F, Wittlieb J, Gelhaus C, Klimovich VB, Samoilovich MP, Bosch TCG. 2010. In an early branching metazoan, bacterial colonization of the embryo is controlled by maternal antimicrobial peptides. Proc Natl Acad Sci USA 107:18067–18072. 10.1073/pnas.1008573107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Fraune S, Anton-Erxleben F, Augustin R, Franzenburg S, Knop M, Schröder K, Willoweit-Ohl D, Bosch TCG. 2015. Bacterium-bacterium interactions within the microbiota of the ancestral metazoan Hydra contribute to fungal resistance. ISME J 9:1543–1556. 10.1038/ismej.2014.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Taubenheim J, Willoweit-Ohl D, Knop M, Franzenburg S, He J, Bosch TCG, Fraune S. 2020. Bacterium- and temperature-regulated peptides modulate β-catenin signaling in Hydra. Proc Natl Acad Sci USA 117:21459–21468. 10.1073/pnas.2010945117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lengfeld T, Watanabe H, Simakov O, Lindgens D, Gee L, Law L, Schmidt HA, Özbek S, Bode H, Holstein TW. 2009. Multiple Wnts are involved in Hydra organizer formation and regeneration. Dev Biol 330:186–199. 10.1016/j.ydbio.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 103.Watanabe H, Schmidt HA, Kuhn A, Höger SK, Kocagöz Y, Laumann-Lipp N, Özbek S, Holstein TW. 2014. Nodal signalling determines biradial asymmetry in Hydra. Nature 515:112–115. 10.1038/nature13666. [DOI] [PubMed] [Google Scholar]
- 104.Khalturin K, Anton-Erxleben F, Milde S, Plötz C, Wittlieb J, Hemmrich G, Bosch TCG. 2007. Transgenic stem cells in Hydra reveal an early evolutionary origin for key elements controlling self-renewal and differentiation. Dev Biol 309:32–44. 10.1016/j.ydbio.2007.06.013. [DOI] [PubMed] [Google Scholar]
- 105.Murillo-Rincon AP, Klimovich A, Pemöller E, Taubenheim J, Mortzfeld B, Augustin R, Bosch TCG. 2017. Spontaneous body contractions are modulated by the microbiome of Hydra. Sci Rep 7:1–9. 10.1038/s41598-017-16191-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Pietschke C, Treitz C, Forêt S, Schultze A, Künzel S, Tholey A, Bosch TCG, Fraune S. 2017. Host modification of a bacterial quorum-sensing signal induces a phenotypic switch in bacterial symbionts. Proc Natl Acad Sci USA 114:E8488–E8497. 10.1073/pnas.1706879114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Chapman JA, Kirkness EF, Simakov O, Hampson SE, Mitros T, Weinmaier T, Rattei T, Balasubramanian PG, Borman J, Busam D, Disbennett K, Pfannkoch C, Sumin N, Sutton GG, Viswanathan LD, Walenz B, Goodstein DM, Hellsten U, Kawashima T, Prochnik SE, Putnam NH, et al. 2010. The dynamic genome of Hydra. Nature 464:592–596. 10.1038/nature08830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hemmrich G, Khalturin K, Boehm AM, Puchert M, Anton-Erxleben F, Wittlieb J, Klostermeier UC, Rosenstiel P, Oberg HH, Domazet-Lošo T, Sugimoto T, Niwa H, Bosch TCG. 2012. Molecular signatures of the three stem cell lineages in Hydra and the emergence of stem cell function at the base of multicellularity. Mol Biol Evol 29:3267–3280. 10.1093/molbev/mss134. [DOI] [PubMed] [Google Scholar]
- 109.Wittlieb J, Khalturin K, Lohmann JU, Anton-Erxleben F, Bosch TCG. 2006. Transgenic Hydra allow in vivo tracking of individual stem cells during morphogenesis. Proc Natl Acad Sci USA 103:6208–6211. 10.1073/pnas.0510163103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wein T, Dagan T, Fraune S, Bosch TCG, Reusch TBH, Hülter NF. 2018. Carrying capacity and colonization dynamics of Curvibacter in the Hydra host habitat. Front Microbiol 9:443. 10.3389/fmicb.2018.00443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Klimovich A, Wittlieb J, Bosch TCG. 2019. Transgenesis in Hydra to characterize gene function and visualize cell behavior. Nat Protoc 14:2069–2090. 10.1038/s41596-019-0173-3. [DOI] [PubMed] [Google Scholar]
- 112.Rahat M, Dimentman C. 1982. Cultivation of bacteria-free Hydra viridis: missing budding factor in nonsymbiotic Hydra. Science 216:67–68. 10.1126/science.7063873. [DOI] [PubMed] [Google Scholar]
- 113.Costa RM, Cárdenas A, Loussert-Fonta C, Toullec G, Meibom A, Voolstra CR. 2021. Surface topography, bacterial carrying capacity, and the prospect of microbiome manipulation in the sea anemone coral model Aiptasia. Front Microbiol 12:637834. 10.3389/fmicb.2021.637834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Cook CB. 1971. Transfer of 35S-labeled material from food ingested by Aiptasia sp. to its endosymbiotic zooxanthellae, p 218–224. In Lenhoff H, Muscatine L, Davis LV (ed), Experimental coelenterate biology. University of Hawaii Press, Honolulu, HI. [Google Scholar]
- 115.Grajales A, Rodriguez E. 2014. Morphological revision of the genus Aiptasia and the family Aiptasiidae (Cnidaria, Actiniaria, Metridioidea). Zootaxa 3826:55–100. 10.11646/zootaxa.3826.1.2. [DOI] [PubMed] [Google Scholar]
- 116.Rädecker N, Raina JB, Pernice M, Perna G, Guagliardo P, Kilburn MR, Aranda M, Voolstra CR. 2018. Using Aiptasia as a model to study metabolic interactions in cnidarian-Symbiodinium symbioses. Front Physiol 9:214. 10.3389/fphys.2018.00214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Dungan AM, Hartman LM, Tortorelli G, Belderok R, Lamb AM, Pisan L, McFadden GI, Blackall LL, van Oppen MJH. 2020. Exaiptasia diaphana from the Great Barrier Reef: a valuable resource for coral symbiosis research. Symbiosis 80:195–206. 10.1007/s13199-020-00665-0. [DOI] [Google Scholar]
- 118.Xiang T, Hambleton EA, Denofrio JC, Pringle JR, Grossman AR. 2013. Isolation of clonal axenic strains of the symbiotic dinoflagellate Symbiodinium and their growth and host specificity. J Phycol 49:447–458. 10.1111/jpy.12055. [DOI] [PubMed] [Google Scholar]
- 119.Cziesielski MJ, Liew YJ, Cui G, Schmidt-Roach S, Campana S, Marondedze C, Aranda M. 2018. Multi-omics analysis of thermal stress response in a zooxanthellate cnidarian reveals the importance of associating with thermotolerant symbionts. Proc R Soc B 285:20172654. 10.1098/rspb.2017.2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Jinkerson RE, Russo JA, Newkirk CR, Kirk AL, Chi RJ, Martindale MQ, Grossman AR, Hatta M, Xiang T. 2022. Cnidarian-Symbiodiniaceae symbiosis establishment is independent of photosynthesis. Curr Biol 32:2402–2415. 10.1016/j.cub.2022.04.021. [DOI] [PubMed] [Google Scholar]
- 121.Sunagawa S, Wilson EC, Thaler M, Smith ML, Caruso C, Pringle JR, Weis VM, Medina M, Schwarz JA. 2009. Generation and analysis of transcriptomic resources for a model system on the rise: the sea anemone Aiptasia pallida and its dinoflagellate endosymbiont. BMC Genomics 10:258. 10.1186/1471-2164-10-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Matthews JL, Crowder CM, Oakley CA, Lutz A, Roessner U, Meyer E, Grossman AR, Weis VM, Davy SK. 2017. Optimal nutrient exchange and immune responses operate in partner specificity in the cnidarian-dinoflagellate symbiosis. Proc Natl Acad Sci USA 114:13194–13199. 10.1073/pnas.1710733114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Baumgarten S, Simakov O, Esherick LY, Liew YJ, Lehnert EM, Michell CT, Li Y, Hambleton EA, Guse A, Oates ME, Gough J, Weis VM, Aranda M, Pringle JR, Voolstra CR, Knowlton N. 2015. The genome of Aiptasia, a sea anemone model for coral symbiosis. Proc Natl Acad Sci USA 112:11893–11898. 10.1073/pnas.1513318112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Bedgood SA, Bracken MES, Ryan WH, Levell ST, Wulff J. 2020. Nutritional drivers of adult locomotion and asexual reproduction in a symbiont-hosting sea anemone Exaiptasia diaphana. Mar Biol 167. 10.1007/s00227-020-3649-3. [DOI] [Google Scholar]
- 125.Cleves PA, Krediet CJ, Lehnert EM, Onishi M, Pringle JR. 2020. Insights into coral bleaching under heat stress from analysis of gene expression in a sea anemone model system. Proc Natl Acad Sci USA 117:28906–28917. 10.1073/pnas.2015737117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Matthews JL, Sproles AE, Oakley CA, Grossman AR, Weis VM, Davy SK. 2016. Menthol-induced bleaching rapidly and effectively provides experimental aposymbiotic sea anemones (Aiptasia sp.) for symbiosis investigations. J Exp Biol 219:306–310. 10.1242/jeb.128934. [DOI] [PubMed] [Google Scholar]
- 127.Dunn SR, Thomason JC, Le Tissier MDA, Bythell JC. 2004. Heat stress induces different forms of cell death in sea anemones and their endosymbiotic algae depending on temperature and duration. Cell Death Differ 11:1213–1222. 10.1038/sj.cdd.4401484. [DOI] [PubMed] [Google Scholar]
- 128.Perez S, Weis VM. 2006. Nitric oxide and cnidarian bleaching: an eviction notice mediates breakdown of a symbiosis. J Exp Biol 209:2804–2810. 10.1242/jeb.02309. [DOI] [PubMed] [Google Scholar]
- 129.Shoguchi E, Shinzato C, Kawashima T, Gyoja F, Mungpakdee S, Koyanagi R, Takeuchi T, Hisata K, Tanaka M, Fujiwara M, Hamada M, Seidi A, Fujie M, Usami T, Goto H, Yamasaki S, Arakaki N, Suzuki Y, Sugano S, Toyoda A, Kuroki Y, Fujiyama A, Medina M, Coffroth MA, Bhattacharya D, Satoh N. 2013. Draft assembly of the Symbiodinium minutum nuclear genome reveals dinoflagellate gene structure. Curr Biol 23:1399–1408. 10.1016/j.cub.2013.05.062. [DOI] [PubMed] [Google Scholar]
- 130.González-Pech RA, Stephens TG, Chen Y, Mohamed AR, Cheng Y, Shah S, Dougan KE, Fortuin MDA, Lagorce R, Burt DW, Bhattacharya D, Ragan MA, Chan CX. 2021. Comparison of 15 dinoflagellate genomes reveals extensive sequence and structural divergence in family Symbiodiniaceae and genus Symbiodinium. BMC Biol 19. 10.1186/s12915-021-00994-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Schoenberg DA, Trench RK, Smith DC. 1980. Genetic variation in Symbiodinium (= Gymnodinium) microadriaticum Freudenthal, and specificity in its symbiosis with marine invertebrates. III. Specificity and infectivity of Symbiodinium microadriaticum. Proc R Soc London Ser B Biol Sci 207:445–460. [Google Scholar]
- 132.Belda-Baillie CA, Baillie BK, Maruyama T. 2002. Specificity of a model cnidarian-dinoflagellate symbiosis. Biol Bull 202:74–85. 10.2307/1543224. [DOI] [PubMed] [Google Scholar]
- 133.Gabay Y, Weis VM, Davy SK. 2018. Symbiont identity influences patterns of symbiosis establishment, host growth, and asexual reproduction in a model cnidarian-dinoflagellate symbiosis. Biol Bull 234:1–10. 10.1086/696365. [DOI] [PubMed] [Google Scholar]
- 134.Gabay Y, Parkinson JE, Wilkinson SP, Weis VM, Davy SK. 2019. Inter-partner specificity limits the acquisition of thermotolerant symbionts in a model cnidarian-dinoflagellate symbiosis. ISME J 13:2489–2499. 10.1038/s41396-019-0429-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Hambleton EA, Jones VAS, Maegele I, Kvaskoff D, Sachsenheimer T, Guse A. 2019. Sterol transfer by atypical cholesterol-binding NPC2 proteins in coral-algal symbiosis. Elife 8. 10.7554/eLife.43923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Sproles AE, Oakley CA, Matthews JL, Peng L, Owen JG, Grossman AR, Weis VM, Davy SK. 2019. Proteomics quantifies protein expression changes in a model cnidarian colonized by a thermally tolerant but suboptimal symbiont. ISME J 13:2334–2345. 10.1038/s41396-019-0437-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Jacobovitz MR, Rupp S, Voss PA, Maegele I, Gornik SG, Guse A. 2021. Dinoflagellate symbionts escape vomocytosis by host cell immune suppression. Nat Microbiol 6:769–782. 10.1038/s41564-021-00897-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Baumgarten S, Cziesielski MJ, Thomas L, Michell CT, Esherick LY, Pringle JR, Aranda M, Voolstra CR. 2018. Evidence for miRNA-mediated modulation of the host transcriptome in cnidarian-dinoflagellate symbiosis. Mol Ecol 27:403–418. 10.1111/mec.14452. [DOI] [PubMed] [Google Scholar]
- 139.Tivey TR, Parkinson JE, Weis VM. 2020. Host and symbiont cell cycle coordination is mediated by symbiotic state, nutrition, and partner identity in a model cnidarian-dinoflagellate symbiosis. mBio 11:e02626-19. 10.1128/mBio.02626-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Sproles AE, Oakley CA, Krueger T, Grossman AR, Weis VM, Meibom A, Davy SK. 2020. Sub-cellular imaging shows reduced photosynthetic carbon and increased nitrogen assimilation by the non-native endosymbiont Durusdinium trenchii in the model cnidarian Aiptasia. Environ Microbiol 22:3741–3753. 10.1111/1462-2920.15142. [DOI] [PubMed] [Google Scholar]
- 141.Röthig T, Costa RM, Simona F, Baumgarten S, Torres AF, Radhakrishnan A, Aranda M, Voolstra CR. 2016. Distinct bacterial communities associated with the coral model Aiptasia in aposymbiotic and symbiotic states with Symbiodinium. Front Mar Sci 3. 10.3389/fmars.2016.00234. [DOI] [Google Scholar]
- 142.Brown T, Otero C, Grajales A, Rodriguez E, Rodriguez-Lanetty M. 2017. Worldwide exploration of the microbiome harbored by the cnidarian model, Exaiptasia pallida (Agassiz in Verrill, 1864) indicates a lack of bacterial association specificity at a lower taxonomic rank. PeerJ 5:e3235. 10.7717/peerj.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Herrera M, Ziegler M, Voolstra CR, Aranda M. 2017. Laboratory-cultured strains of the sea anemone Exaiptasia reveal distinct bacterial communities. Front Mar Sci 4:115. 10.3389/fmars.2017.00115. [DOI] [Google Scholar]
- 144.Hartman LM, van Oppen MJH, Blackall LL. 2020. Microbiota characterization of Exaiptasia diaphana from the Great Barrier Reef. Anim Microbiome 2:10. 10.1186/s42523-020-00029-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ahmed HI, Herrera M, Liew YJ, Aranda M. 2019. Long-term temperature stress in the coral model Aiptasia supports the “Anna Karenina Principle” for bacterial microbiomes. Front Microbiol 10:975. 10.3389/fmicb.2019.00975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Schlesinger A, Kramarsky-Winter E, Rosenfeld H, Armoza-Zvoloni R, Loya Y. 2010. Sexual plasticity and self-fertilization in the sea anemone Aiptasia diaphana. PLoS One 5:e11874. 10.1371/journal.pone.0011874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Hambleton EA, Guse A, Pringle JR. 2014. Similar specificities of symbiont uptake by adults and larvae in an anemone model system for coral biology. J Exp Biol 217(Pt 9):1613–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Grawunder D, Hambleton EA, Bucher M, Wolfowicz I, Bechtoldt N, Guse A. 2015. Induction of gametogenesis in the cnidarian endosymbiosis model Aiptasia sp. Sci Rep 5:15677. 10.1038/srep15677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Bucher M, Wolfowicz I, Voss PA, Hambleton EA, Guse A. 2016. Development and symbiosis establishment in the cnidarian endosymbiosis model Aiptasia sp. Sci Rep 6:19867. 10.1038/srep19867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Wolfowicz I, Baumgarten S, Voss PA, Hambleton EA, Voolstra CR, Hatta M, Guse A. 2016. Aiptasia sp. larvae as a model to reveal mechanisms of symbiont selection in cnidarians. Sci Rep 6:32366. 10.1038/srep32366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Jones VAS, Bucher M, Hambleton EA, Guse A. 2018. Microinjection to deliver protein, mRNA, and DNA into zygotes of the cnidarian endosymbiosis model Aiptasia sp. Sci Rep 8:1–11. 10.1038/s41598-018-34773-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Cary LR. 1911. A study of pedal laceration in actinians. Biol Bull 20:81–107. 10.2307/1536038. [DOI] [Google Scholar]
- 153.Clayton WSJ. 1985. Pedal laceration by the anemone Aiptasia pallida. Mar Ecol Prog Ser 21:75–80. 10.3354/meps021075. [DOI] [Google Scholar]
- 154.Lin J, Tsai CC, Lai WK, Chen CP. 1992. Pedal laceration in the sea anemone Aiptasia sp. (Anthozoa: Actiniaria). Chin Biosci 35:33–41. [Google Scholar]
- 155.Presnell JS, Wirsching E, Weis VM. 2022. Tentacle patterning during Exaiptasia diaphana pedal lacerate development differs between symbiotic and aposymbiotic animals. PeerJ 10:e12770. 10.7717/peerj.12770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Thornhill DJ, Xiang Y, Pettay DT, Zhong M, Santos SR. 2013. Population genetic data of a model symbiotic cnidarian system reveal remarkable symbiotic specificity and vectored introductions across ocean basins. Mol Ecol 22:4499–4515. 10.1111/mec.12416. [DOI] [PubMed] [Google Scholar]
- 157.Bellis ES, Edlund RB, Berrios HK, Lessios HA, Denver DR. 2018. Molecular signatures of host specificity linked to habitat specialization in Exaiptasia sea anemones. Ecol Evol 8:5413–5426. 10.1002/ece3.4058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Medina M, Sharp V, Ohdera A, Bellantuono A, Dalrymple J, Gamero-mora E, Steinworth B, Hofmann DK, Martindale MQ, Morandini AC, Degennaro M, Fitt WK. 2021. The upside-down jellyfish Cassiopea xamachana as an emerging model system to study cnidarian-algal symbiosis, p 149–171. In Boutet A, Schierwater B (ed), Handbook of marine model organisms in experimental biology. CRC Press, Boca Raton, FL. [Google Scholar]
- 159.Kayal E, Bentlage B, Sabrina Pankey M, Ohdera AH, Medina M, Plachetzki DC, Collins AG, Ryan JF. 2018. Phylogenomics provides a robust topology of the major cnidarian lineages and insights on the origins of key organismal traits. BMC Evol Biol 18:1–18. 10.1186/s12862-018-1142-0.29368592 [DOI] [Google Scholar]
- 160.Ohdera A, Ames CL, Dikow RB, Kayal E, Chiodin M, Busby B, La S, Pirro S, Collins AG, Medina M, Ryan JF. 2019. Box, stalked, and upside-down? Draft genomes from diverse jellyfish (Cnidaria, Acraspeda) lineages: Alatina alata (cubozoa), Calvadosia cruxmelitensis (staurozoa), and Cassiopea xamachana (scyphozoa). Gigascience 8:1–15. 10.1093/gigascience/giz069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Hofmann DK, Fitt WK, Fleck J. 1996. Checkpoints in the life-cycle of Cassiopea spp.: control of metagenesis and metamorphosis in a tropical jellyfish. Int J Dev Biol 40:331–338. [PubMed] [Google Scholar]
- 162.McGill CJ, Pomory CM. 2008. Effects of bleaching and nutrient supplementation on wet weight in the jellyfish Cassiopea xamachana (Bigelow) (Cnidaria: Scyphozoa). Mar Freshw Behav Physiol 41:179–189. 10.1080/10236240802369899. [DOI] [Google Scholar]
- 163.Klein SG, Pitt KA, Lucas CH, Hung SH, Schmidt-Roac S, Aranda M, Duarte CM. 2019. Night-time temperature reprieves enhance the thermal tolerance of a symbiotic cnidarian. Front Mar Sci 6:1–16. [Google Scholar]
- 164.Newkirk CR, Frazer TK, Martindale MQ, Schnitzler CE. 2020. Adaptation to bleaching: are thermotolerant Symbiodiniaceae strains more successful than other strains under elevated temperatures in a model symbiotic cnidarian? Front Microbiol 11:822. 10.3389/fmicb.2020.00822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Stoner EW, Sebilian SS, Layman CA. 2016. Comparison of zooxanthellae densities from upside-down jellyfish, Cassiopea xamachana, across coastal habitats of The Bahamas. Rev Biol Mar Oceanogr 51:203–208. 10.4067/S0718-19572016000100022. [DOI] [Google Scholar]
- 166.Al-jbour SM, Zimmer M, Kunzmann A. 2017. Cellular respiration, oxygen consumption, and trade-offs of the jellyfish Cassiopea sp. in response to temperature change. J Sea Res 128:92–97. 10.1016/j.seares.2017.08.006. [DOI] [Google Scholar]
- 167.Gilbert SF. 2014. Symbiosis as the way of eukaryotic life: the dependent co-origination of the body. J Biosci 39:201–209. 10.1007/s12038-013-9343-6. [DOI] [PubMed] [Google Scholar]
- 168.Thornhill DJ, Daniel MW, LaJeunesse TC, Schmidt GW, Fitt WK. 2006. Natural infections of aposymbiotic Cassiopea xamachana scyphistomae from environmental pools of Symbiodinium. J Exp Mar Bio Ecol 338:50–56. 10.1016/j.jembe.2006.06.032. [DOI] [Google Scholar]
- 169.Mellas RE, McIlroy SE, Fitt WK, Coffroth MA. 2014. Variation in symbiont uptake in the early ontogeny of the upside-down jellyfish, Cassiopea spp. J Exp Mar Bio Ecol 459:38–44. 10.1016/j.jembe.2014.04.026. [DOI] [Google Scholar]
- 170.Röthig T, Puntin G, Wong JCY, Burian A, McLeod W, Baker DM. 2021. Holobiont nitrogen control and its potential for eutrophication resistance in an obligate photosymbiotic jellyfish. Microbiome 9:127. 10.1186/s40168-021-01075-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Newkirk CR, Frazer TK, Martindale MQ. 2018. Acquisition and proliferation of algal symbionts in bleached polyps of the upside-down jellyfish, Cassiopea xamachana. J Exp Mar Bio Ecol 508:44–51. 10.1016/j.jembe.2018.08.010. [DOI] [Google Scholar]
- 172.Quigley KM, Willis BL, Bay LK. 2017. Heritability of the Symbiodinium community in vertically- and horizontally-transmitting broadcast spawning corals. Sci Rep 7:8219. 10.1038/s41598-017-08179-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Lyndby NH, Rädecker N, Bessette S, Søgaard JL, Escrig S, Trampe E, Kühl M, Meibom A. 2020. Amoebocytes facilitate efficient carbon and nitrogen assimilation in the Cassiopea-Symbiodiniaceae symbiosis. Proc R Soc Lond B Biol Sci 287:20202393. 10.1098/rspb.2020.2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Vega Thurber RL, Burkepile DE, Fuchs C, Shantz AA, Mcminds R, Zaneveld JR. 2014. Chronic nutrient enrichment increases prevalence and severity of coral disease and bleaching. Glob Chang Biol 20:544–554. 10.1111/gcb.12450. [DOI] [PubMed] [Google Scholar]
- 175.Stoner EW, Layman CA, Yeager LA, Hassett HM. 2011. Effects of anthropogenic disturbance on the abundance and size of epibenthic jellyfish Cassiopea spp. Mar Pollut Bull 62:1109–1114. 10.1016/j.marpolbul.2011.03.023. [DOI] [PubMed] [Google Scholar]
- 176.Fernandes de Barros Marangoni L, Ferrier-Pagès C, Rottier C, Bianchini A, Grover R. 2020. Unravelling the different causes of nitrate and ammonium effects on coral bleaching. Sci Rep 10:1–14. 10.1038/s41598-020-68916-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Colley NJ, Trench RK. 1985. Cellular events in the reestablishment of a symbiosis between a marine dinoflagellate and a coelenterate. Cell Tissue Res 239:93–103. 10.1007/BF00214908. [DOI] [PubMed] [Google Scholar]
- 178.Jantzen C, Wild C, Rasheed M, El-Zibdah M, Richter C. 2010. Enhanced pore-water nutrient fluxes by the upside-down jellyfish Cassiopea sp. in a Red Sea coral reef. Mar Ecol Prog Ser 411:117–125. 10.3354/meps08623. [DOI] [Google Scholar]
- 179.Szmant A, Yevich P, Pilson M. 1980. Gametogenesis and early development of the temperate coral Astrangia danae (Anthozoa: Scleractinia). Biol Bull 158:257–269. 10.2307/1540935. [DOI] [Google Scholar]
- 180.Szmant-Froelich A, Pilson MEQ. 1984. Effects of feeding frequency and symbiosis with zooxanthellae on nitrogen metabolism and respiration of the coral Astrangia danae. Mar Biol 81:153–162. 10.1007/BF00393114. [DOI] [Google Scholar]
- 181.DiRoberts L, Dudek A, Ray N, Fulweiler R, Rotjan R. 2021. Testing assumptions of nitrogen cycling between a temperate, model coral host and its facultative symbiont: symbiotic contributions to dissolved inorganic nitrogen assimilation. Mar Ecol Prog Ser 670:61–74. 10.3354/meps13731. [DOI] [Google Scholar]
- 182.Coles SL, Fadlallah YH. 1991. Reef coral survival and mortality at low temperatures in the Arabian Gulf: new species-specific lower temperature limits. Coral Reefs 9:231–237. 10.1007/BF00290427. [DOI] [Google Scholar]
- 183.Paparella F, Xu C, Vaughan GO, Burt JA. 2019. Coral bleaching in the Persian/Arabian Gulf is modulated by summer winds. Front Mar Sci 6. 10.3389/fmars.2019.00205. [DOI] [Google Scholar]
- 184.Wuitchik DM, Almanzar A, Benson BE, Brennan S, Chavez JD, Liesegang MB, Reavis JL, Reyes CL, Schniedewind MK, Trumble IF, Davies SW. 2021. Characterizing environmental stress responses of aposymbiotic Astrangia poculata to divergent thermal challenges. Mol Ecol 30:5064–5079. 10.1111/mec.16108. [DOI] [PubMed] [Google Scholar]
- 185.Barshis DJ, Ladner JT, Oliver TA, Palumbi SR. 2014. Lineage-specific transcriptional profiles of Symbiodinium spp. unaltered by heat stress in a coral host. Mol Biol Evol 31:1343–1352. 10.1093/molbev/msu107. [DOI] [PubMed] [Google Scholar]
- 186.Hawkins TD, Krueger T, Wilkinson SP, Fisher PL, Davy SK. 2015. Antioxidant responses to heat and light stress differ with habitat in a common reef coral. Coral Reefs 34:1229–1241. 10.1007/s00338-015-1345-4. [DOI] [Google Scholar]
- 187.Krueger T, Hawkins TD, Becker S, Pontasch S, Dove S, Hoegh-Guldberg O, Leggat W, Fisher PL, Davy SK. 2015. Differential coral bleaching: contrasting the activity and response of enzymatic antioxidants in symbiotic partners under thermal stress. Comp Biochem Physiol A Mol Integr Physiol 190:15–25. 10.1016/j.cbpa.2015.08.012. [DOI] [PubMed] [Google Scholar]
- 188.Stimson J. 1997. The annual cycle of density of zooxanthellae in the tissues of field and laboratory-held Pocillopora damicornis (Linnaeus). J Exp Mar Bio Ecol 214:35–48. 10.1016/S0022-0981(96)02753-0. [DOI] [Google Scholar]
- 189.Fagoonee I, Wilson HB, Hassell MP, Turner JR. 1999. The dynamics of zooxanthellae populations: a long-term study in the field. Science 283:843–845. 10.1126/science.283.5403.843. [DOI] [PubMed] [Google Scholar]
- 190.Dimond J, Carrington E. 2007. Temporal variation in the symbiosis and growth of the temperate scleractinian coral Astrangia poculata. Mar Ecol Prog Ser 348:161–172. 10.3354/meps07050. [DOI] [Google Scholar]
- 191.Thornhill DJ, Kemp DW, Bruns BU, Fitt WK, Schmidt GW. 2008. Correspondence between cold tolerance and temperate biogeography in a western Atlantic Symbiodinium (Dinophyta) lineage. J Phycol 44:1126–1135. 10.1111/j.1529-8817.2008.00567.x. [DOI] [PubMed] [Google Scholar]
- 192.Grace S. 2017. Winter quiescence, growth rate, and the release from competition in the temperate scleractinian coral Astrangia poculata (Ellis & Solander, 1786). Northeast Nat 24:B119–B134. 10.1656/045.024.s715. [DOI] [Google Scholar]
- 193.Hadaidi G, Röthig T, Yum LK, Ziegler M, Arif C, Roder C, Burt J, Voolstra CR. 2017. Stable mucus-associated bacterial communities in bleached and healthy corals of Porites lobata from the Arabian Seas. Sci Rep 7:45362. 10.1038/srep45362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Botté ES, Cantin NE, Mocellin VJL, O’Brien PA, Rocker MM, Frade PR, Webster NS. 2022. Reef location has a greater impact than coral bleaching severity on the microbiome of Pocillopora acuta. Coral Reefs 41:63–79. 10.1007/s00338-021-02201-y. [DOI] [Google Scholar]
- 195.Bent SM, Miller CA, Sharp KH, Hansel CM, Apprill A. 2021. Differential patterns of microbiota recovery in symbiotic and aposymbiotic corals following antibiotic disturbance. mSystems 6:e01086-20. 10.1128/mSystems.01086-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Domart-Coulon I, Ostrander GK. 2015. Coral cell and tissue culture methods, p 489–505. In Diseases of coral. John Wiley & Sons, Inc, Hoboken, NJ. [Google Scholar]
- 197.Feuillassier L, Martinez L, Romans P, Engelmann-Sylvestre I, Masanet P, Barthélémy D, Engelmann F. 2014. Survival of tissue balls from the coral Pocillopora damicornis L. exposed to cryoprotectant solutions. Cryobiology 69:376–385. 10.1016/j.cryobiol.2014.08.009. [DOI] [PubMed] [Google Scholar]
- 198.Lu Q, Liu T, Tang X, Dong B, Guo H. 2017. Reformation of tissue balls from tentacle explants of coral Goniopora lobata: self-organization process and response to environmental stresses. In Vitro Cell Dev Biol Anim 53:111–122. 10.1007/s11626-016-0095-0. [DOI] [PubMed] [Google Scholar]
- 199.Vizel M, Loya Y, Downs CA, Kramarsky-Winter E. 2011. A novel method for coral explant culture and micropropagation. Mar Biotechnol (NY) 13:423–432. 10.1007/s10126-010-9313-z. [DOI] [PubMed] [Google Scholar]
- 200.Gardner SG, Nielsen DA, Petrou K, Larkum AWD, Ralph PJ. 2015. Characterization of coral explants: a model organism for cnidarian–dinoflagellate studies. Coral Reefs 34:133–142. 10.1007/s00338-014-1240-4. [DOI] [Google Scholar]
- 201.Sammarco P. 1982. Polyp bail-out: an escape response to environmental stress and a new means of reproduction in corals. Mar Ecol Prog Ser 10:57–65. 10.3354/meps010057. [DOI] [Google Scholar]
- 202.Domart-Coulon I, Tambutté S, Tambutté E, Allemand D. 2004. Short term viability of soft tissue detached from the skeleton of reef-building corals. J Exp Mar Bio Ecol 309:199–217. 10.1016/j.jembe.2004.03.021. [DOI] [Google Scholar]
- 203.Shapiro OH, Kramarsky-Winter E, Gavish AR, Stocker R, Vardi A. 2016. A coral-on-a-chip microfluidic platform enabling live-imaging microscopy of reef-building corals. Nat Commun 7:10860. 10.1038/ncomms10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Kopecky EJ, Ostrander GK. 1999. Isolation and primary culture of viable multicellular endothelial isolates from hard corals. In Vitro Cell Dev Biol Anim 35:616–624. 10.1007/s11626-999-0101-x. [DOI] [PubMed] [Google Scholar]
- 205.Nesa B, Hidaka M. 2009. High zooxanthella density shortens the survival time of coral cell aggregates under thermal stress. J Exp Mar Bio Ecol 368:81–87. 10.1016/j.jembe.2008.10.018. [DOI] [Google Scholar]
- 206.Lecointe A, Cohen S, Gèze M, Djediat C, Meibom A, Domart-Coulon I. 2013. Scleractinian coral cell proliferation is reduced in primary culture of suspended multicellular aggregates compared to polyps. Cytotechnology 65:705–724. 10.1007/s10616-013-9562-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Domart-Coulon IJ, Elbert DC, Scully EP, Calimlim PS, Ostrander GK. 2001. Aragonite crystallization in primary cell cultures of multicellular isolates from a hard coral, Pocillopora damicornis. Proc Natl Acad Sci USA 98:11885–11890. 10.1073/pnas.211439698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Helman Y, Natale F, Sherrell RM, LaVigne M, Starovoytov V, Gorbunov MY, Falkowski PG. 2008. Extracellular matrix production and calcium carbonate precipitation by coral cells in vitro. Proc Natl Acad Sci USA 105:54–58. 10.1073/pnas.0710604105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Mass T, Drake JL, Haramaty L, Rosenthal Y, Schofield OME, Sherrell RM, Falkowski PG. 2012. Aragonite precipitation by “proto-polyps” in coral cell cultures. PLoS One 7:e35049. 10.1371/journal.pone.0035049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Mass T, Drake JL, Heddleston JM, Falkowski PG. 2017. Nanoscale visualization of biomineral formation in coral proto-polyps. Curr Biol 27:3191–3196. 10.1016/j.cub.2017.09.012. [DOI] [PubMed] [Google Scholar]
- 211.Domart-Coulon IJ, Sinclair CS, Hill RT, Tambutt S, Puverel S, Ostrander GK. 2004. A basidiomycete isolated from the skeleton of Pocillopora damicornis (Scleractinia) selectively stimulates short-term survival of coral skeletogenic cells. Mar Biol 144:583–592. 10.1007/s00227-003-1227-0. [DOI] [Google Scholar]
- 212.Kawamura K, Nishitsuji K, Shoguchi E, Fujiwara S, Satoh N. 2021. Establishing sustainable cell lines of a coral, Acropora tenuis. Mar Biotechnol (NY) 23:373–388. 10.1007/s10126-021-10031-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Roger LM, Reich HG, Lawrence E, Li S, Vizgaudis W, Brenner N, Kumar L, Klein-Seetharaman J, Yang J, Putnam HM, Lewinski NA. 2021. Applying model approaches in non-model systems: a review and case study on coral cell culture. PLoS One 16:e0248953. 10.1371/journal.pone.0248953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Reyes-Bermudez A, Miller DJ. 2009. In vitro culture of cells derived from larvae of the staghorn coral Acropora millepora. Coral Reefs 28:859–864. 10.1007/s00338-009-0527-3. [DOI] [Google Scholar]
- 215.Kawamura K, Sekida S, Nishitsuji K, Shoguchi E, Hisata K, Fujiwara S, Satoh N. 2021. In vitro symbiosis of reef-building coral cells with photosynthetic dinoflagellates. Front Mar Sci 8:1–11.35685121 [Google Scholar]
- 216.Nowotny JD, Connelly MT, Traylor-Knowles N. 2021. Novel methods to establish whole-body primary cell cultures for the cnidarians Nematostella vectensis and Pocillopora damicornis. Sci Rep 11:4086. 10.1038/s41598-021-83549-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Ballarin L, Rinkevich B, Bartscherer K, Burzynski A, Cambier S, Cammarata M, Domart-Coulon I, Drobne D, Encinas J, Frank U, Geneviere A-M, Hobmayer B, Löhelaid H, Lyons D, Martinez P, Oliveri P, Peric L, Piraino S, Ramšak A, Rakers S, Rentzsch F, Rosner A, da Silva TH, Somorjai I, Suleiman S, Coelho AV. 2018. Maristem—stem cells of marine/aquatic invertebrates: from basic research to innovative applications. Sustainability 10:526. 10.3390/su10020526. [DOI] [Google Scholar]
- 218.Kaur G, Dufour JM. 2012. Cell lines. Spermatogenesis 2:1–5. 10.4161/spmg.19885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Devlin‐Durante MK, Miller MW, Precht WF, Baums IB, Carne L, Smith TB, Banaszak AT, Greer L, Irwin A, Fogarty ND, Williams DE, Caribbean Acropora Research Group. 2016. How old are you? Genet age estimates in a clonal animal. Mol Ecol 25:5628–5646. 10.1111/mec.13865. [DOI] [PubMed] [Google Scholar]
- 220.Rinkevich B. 2011. Cell cultures from marine invertebrates: new insights for capturing endless stemness. Mar Biotechnol (NY) 13:345–354. 10.1007/s10126-010-9354-3. [DOI] [PubMed] [Google Scholar]
- 221.Schmid V, Ono S-I, Reber-Müller S. 1999. Cell-substrate interactions in Cnidaria. Microsc Res Tech 44:254–268. . [DOI] [PubMed] [Google Scholar]
- 222.Reber-Muller S, Streitwolf-Engel R, Yanze N, Schmid V, Stierwald M, Erb M, Seipel K. 2006. BMP2/4 and BMP5-8 in jellyfish development and transdifferentiation. Int J Dev Biol 50:377–384. 10.1387/ijdb.052085sr. [DOI] [PubMed] [Google Scholar]
- 223.Gold DA, Jacobs DK. 2013. Stem cell dynamics in Cnidaria: are there unifying principles? Dev Genes Evol 223:53–66. 10.1007/s00427-012-0429-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Bosch TCG. 2007. Why polyps regenerate and we don’t: towards a cellular and molecular framework for Hydra regeneration. Dev Biol 303:421–433. 10.1016/j.ydbio.2006.12.012. [DOI] [PubMed] [Google Scholar]
- 225.Storms RW, Trujillo AP, Springer JB, Shah L, Colvin OM, Ludeman SM, Smith C. 1999. Isolation of primitive human hematopoietic progenitors on the basis of aldehyde dehydrogenase activity. Proc Natl Acad Sci USA 96:9118–9123. 10.1073/pnas.96.16.9118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Rosental B, Kozhekbaeva Z, Fernhoff N, Tsai JM, Traylor-Knowles N. 2017. Coral cell separation and isolation by fluorescence-activated cell sorting (FACS). BMC Cell Biol 18:30. 10.1186/s12860-017-0146-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Levy S, Elek A, Grau-Bové X, Menéndez-Bravo S, Iglesias M, Tanay A, Mass T, Sebé-Pedrós A. 2021. A stony coral cell atlas illuminates the molecular and cellular basis of coral symbiosis, calcification, and immunity. Cell 184:2973–2987.e18. 10.1016/j.cell.2021.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Gutleben J, Chaib De Mares M, van Elsas JD, Smidt H, Overmann J, Sipkema D. 2018. The multi-omics promise in context: from sequence to microbial isolate. Crit Rev Microbiol 44:212–229. 10.1080/1040841X.2017.1332003. [DOI] [PubMed] [Google Scholar]
- 229.Cross KL, Campbell JH, Balachandran M, Campbell AG, Cooper CJ, Griffen A, Heaton M, Joshi S, Klingeman D, Leys E, Yang Z, Parks JM, Podar M. 2019. Targeted isolation and cultivation of uncultivated bacteria by reverse genomics. Nat Biotechnol 37:1314–1321. 10.1038/s41587-019-0260-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Silverstein RN, Cunning R, Baker AC. 2015. Change in algal symbiont communities after bleaching, not prior heat exposure, increases heat tolerance of reef corals. Glob Change Biol 21:236–249. 10.1111/gcb.12706. [DOI] [PubMed] [Google Scholar]
- 231.Jones R. 2004. Testing the “photoinhibition” model of coral bleaching using chemical inhibitors. Mar Ecol Prog Ser 284:133–145. 10.3354/meps284133. [DOI] [Google Scholar]
- 232.Wang J-T, Chen Y-Y, Tew KS, Meng P-J, Chen CA. 2012. Physiological and biochemical performances of menthol-induced aposymbiotic corals. PLoS One 7:e46406. 10.1371/journal.pone.0046406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Dani V, Priouzeau F, Pagnotta S, Carette D, Laugier JP, Sabourault C. 2016. Thermal and menthol stress induce different cellular events during sea anemone bleaching. Symbiosis 69:175–192. 10.1007/s13199-016-0406-y. [DOI] [Google Scholar]
- 234.Clowez S, Renicke C, Pringle JR, Grossman AR. 2021. Impact of menthol on growth and photosynthetic function of Breviolum minutum (Dinoflagellata, Dinophyceae, Symbiodiniaceae) and interactions with its Aiptasia host. J Phycol 57:245–257. 10.1111/jpy.13081. [DOI] [PubMed] [Google Scholar]
- 235.Scharfenstein HJ, Chan WY, Buerger P, Humphrey C, van Oppen MJH. 2022. Evidence for de novo acquisition of microalgal symbionts by bleached adult corals. ISME J 16:1676–1679. 10.1038/s41396-022-01203-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Greer R, Dong X, Morgun A, Shulzhenko N. 2016. Investigating a holobiont: microbiota perturbations and transkingdom networks. Gut Microbes 7:126–135. 10.1080/19490976.2015.1128625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Nietzer S, Moeller M, Kitamura M, Schupp PJ. 2018. Coral larvae every day: Leptastrea purpurea, a brooding species that could accelerate coral research. Front Mar Sci 5:1–12.29552559 [Google Scholar]
- 238.Eyal-Shaham L, Eyal G, Ben-Zvi O, Sakai K, Harii S, Sinniger F, Hirose M, Cabaitan P, Bronstein O, Feldman B, Shlesinger T, Levy O, Loya Y. 2020. A unique reproductive strategy in the mushroom coral Fungia fungites. Coral Reefs 39:1793–1804. 10.1007/s00338-020-02004-7. [DOI] [Google Scholar]
- 239.Craggs J, Guest JR, Davis M, Simmons J, Dashti E, Sweet M. 2017. Inducing broadcast coral spawning ex situ: closed system mesocosm design and husbandry protocol. Ecol Evol 7:11066–11078. 10.1002/ece3.3538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Craggs J, Guest JR, Davis M, Sweet M. 2020. Completing the life cycle of a broadcast spawning coral in a closed mesocosm. Invertebr Reprod Dev 64:244–247. 10.1080/07924259.2020.1759704. [DOI] [Google Scholar]
- 241.Domin H, Zurita-Gutiérrez YH, Scotti M, Buttlar J, Hentschel HU, Fraune S. 2018. Predicted bacterial interactions affect in vivo microbial colonization dynamics in Nematostella. Front Microbiol 9:728. 10.3389/fmicb.2018.00728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Sweet MJ, Croquer A, Bythell JC. 2011. Dynamics of bacterial community development in the reef coral Acropora muricata following experimental antibiotic treatment. Coral Reefs 30:1121–1133. 10.1007/s00338-011-0800-0. [DOI] [Google Scholar]
- 243.Damjanovic K, van Oppen MJH, Menéndez P, Blackall LL. 2019. Experimental inoculation of coral recruits with marine bacteria indicates scope for microbiome manipulation in Acropora tenuis and Platygyra daedalea. Front Microbiol 10:1702. 10.3389/fmicb.2019.01702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Baird AH, Guest JR, Willis BL. 2009. Systematic and biogeographical patterns in the reproductive biology of scleractinian corals. Annu Rev Ecol Evol Syst 40:551–571. 10.1146/annurev.ecolsys.110308.120220. [DOI] [Google Scholar]
- 245.Sharp KH, Ritchie KB, Schupp PJ, Ritson-Williams R, Paul VJ. 2010. Bacterial acquisition in juveniles of several broadcast spawning coral species. PLoS One 5:e10898. 10.1371/journal.pone.0010898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Tebben J, Motti CA, Siboni N, Tapiolas DM, Negri AP, Schupp PJ, Kitamura M, Hatta M, Steinberg PD, Harder T. 2015. Chemical mediation of coral larval settlement by crustose coralline algae. Sci Rep 5:10803. 10.1038/srep10803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Wang J-T, Keshavmurthy S, Chu T-Y, Chen CA. 2017. Diverse responses of Symbiodinium types to menthol and DCMU treatment. PeerJ 5:e3843. 10.7717/peerj.3843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Wong JCY, Enríquez S, Baker DM. 2021. Towards a trait-based understanding of Symbiodiniaceae nutrient acquisition strategies. Coral Reefs 40:625–639. 10.1007/s00338-020-02034-1. [DOI] [Google Scholar]
- 249.Krueger T, Gates RD. 2012. Cultivating endosymbionts: host environmental mimics support the survival of Symbiodinium C15 ex hospite. J Exp Mar Bio Ecol 413:169–176. 10.1016/j.jembe.2011.12.002. [DOI] [Google Scholar]
- 250.Nitschke MR, Craveiro SC, Brandão C, Fidalgo C, Serôdio J, Calado AJ, Frommlet JC. 2020. Description of Freudenthalidium gen. nov. and Halluxium gen. nov. to formally recognize clades Fr3 and H as genera in the family Symbiodiniaceae (Dinophyceae). J Phycol 56:923–940. 10.1111/jpy.12999. [DOI] [PubMed] [Google Scholar]
- 251.Maruyama S, Weis VM. 2021. Limitations of using cultured algae to study cnidarian-algal symbioses and suggestions for future studies. J Phycol 57:30–38. 10.1111/jpy.13102. [DOI] [PubMed] [Google Scholar]
- 252.Gates RD, Muscatine L. 1992. Three methods for isolating viable anthozoan endoderm cells with their intracellular symbiotic dinoflagellates. Coral Reefs 11:143–145. 10.1007/BF00255468. [DOI] [Google Scholar]
- 253.Frommlet JC, Sousa ML, Alves A, Vieira SI, Suggett DJ, Serôdio J. 2015. Coral symbiotic algae calcify ex hospite in partnership with bacteria. Proc Natl Acad Sci USA 112:6158–6163. 10.1073/pnas.1420991112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Frommlet JC, Wangpraseurt D, Sousa ML, Guimarães B, Medeiros da Silva M, Kühl M, Serôdio J. 2018. Symbiodinium-induced formation of microbialites: mechanistic insights from in vitro experiments and the prospect of its occurrence in nature. Front Microbiol 9:998. 10.3389/fmicb.2018.00998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Rypien KL, Ward JR, Azam F. 2010. Antagonistic interactions among coral-associated bacteria. Environ Microbiol 12:28–39. 10.1111/j.1462-2920.2009.02027.x. [DOI] [PubMed] [Google Scholar]
- 256.Silva SG, Blom J, Keller‐Costa T, Costa R. 2019. Comparative genomics reveals complex natural product biosynthesis capacities and carbon metabolism across host‐associated and free‐living Aquimarina (Bacteroidetes, Flavobacteriaceae) species. Environ Microbiol 21:4002–4019. 10.1111/1462-2920.14747. [DOI] [PubMed] [Google Scholar]
- 257.Yarza P, Yilmaz P, Pruesse E, Glöckner FO, Ludwig W, Schleifer K-H, Whitman WB, Euzéby J, Amann R, Rosselló-Móra R. 2014. Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat Rev Microbiol 12:635–645. 10.1038/nrmicro3330. [DOI] [PubMed] [Google Scholar]
- 258.Huggett MJ, Apprill A. 2019. Coral microbiome database: integration of sequences reveals high diversity and relatedness of coral‐associated microbes. Environ Microbiol Rep 11:372–385. 10.1111/1758-2229.12686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Lewis WH, Tahon G, Geesink P, Sousa DZ, Ettema TJG. 2021. Innovations to culturing the uncultured microbial majority. Nat Rev Microbiol 19:225–240. 10.1038/s41579-020-00458-8. [DOI] [PubMed] [Google Scholar]
- 260.Wang F, Li M, Huang L, Zhang X-H. 2021. Cultivation of uncultured marine microorganisms. Mar Life Sci Technol 3:117–120. 10.1007/s42995-021-00093-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Lagier J-C, Armougom F, Million M, Hugon P, Pagnier I, Robert C, Bittar F, Fournous G, Gimenez G, Maraninchi M, Trape J-F, Koonin EV, La Scola B, Raoult D. 2012. Microbial culturomics: paradigm shift in the human gut microbiome study. Clin Microbiol Infect 18:1185–1193. 10.1111/1469-0691.12023. [DOI] [PubMed] [Google Scholar]
- 262.Ma L, Kim J, Hatzenpichler R, Karymov MA, Hubert N, Hanan IM, Chang EB, Ismagilov RF. 2014. Gene-targeted microfluidic cultivation validated by isolation of a gut bacterium listed in Human Microbiome Project’s Most Wanted taxa. Proc Natl Acad Sci USA 111:9768–9773. 10.1073/pnas.1404753111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Watterson WJ, Tanyeri M, Watson AR, Cham CM, Shan Y, Chang EB, Eren AM, Tay S. 2020. Droplet-based high-throughput cultivation for accurate screening of antibiotic resistant gut microbes. Elife 9:1–22. 10.7554/eLife.56998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Salam N, Xian W-D, Asem MD, Xiao M, Li W-J. 2021. From ecophysiology to cultivation methodology: filling the knowledge gap between uncultured and cultured microbes. Mar Life Sci Technol 3:132–147. 10.1007/s42995-020-00064-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Frank U, Rabinowitz C, Rinkevich B. 1994. In vitro establishment of continuous cell cultures and cell lines from ten colonial cnidarians. Mar Biol 120:491–499. 10.1007/BF00680224. [DOI] [Google Scholar]
- 266.Lawson CA, Raina J, Kahlke T, Seymour JR, Suggett DJ. 2018. Defining the core microbiome of the symbiotic dinoflagellate, Symbiodinium. Environ Microbiol Rep 10:7–11. 10.1111/1758-2229.12599. [DOI] [PubMed] [Google Scholar]
- 267.Maire J, Girvan SK, Barkla SE, Perez-Gonzalez A, Suggett DJ, Blackall LL, van Oppen MJH. 2021. Intracellular bacteria are common and taxonomically diverse in cultured and in hospite algal endosymbionts of coral reefs. ISME J 15:2028–2042. 10.1038/s41396-021-00902-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Baldassarre L, Ying H, Reitzel AM, Franzenburg S, Fraune S. 2022. Microbiota mediated plasticity promotes thermal adaptation in the sea anemone Nematostella vectensis. Nat Commun 13:3804. 10.1038/s41467-022-31350-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Wu H-J, Ivanov II, Darce J, Hattori K, Shima T, Umesaki Y, Littman DR, Benoist C, Mathis D. 2010. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity 32:815–827. 10.1016/j.immuni.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Abdel-Gadir A, Stephen-Victor E, Gerber GK, Noval Rivas M, Wang S, Harb H, Wang L, Li N, Crestani E, Spielman S, Secor W, Biehl H, DiBenedetto N, Dong X, Umetsu DT, Bry L, Rachid R, Chatila TA. 2019. Microbiota therapy acts via a regulatory T cell MyD88/RORγt pathway to suppress food allergy. Nat Med 25:1164–1174. 10.1038/s41591-019-0461-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Voolstra CR. 2013. A journey into the wild of the cnidarian model system Aiptasia and its symbionts. Mol Ecol 22:4366–4368. 10.1111/mec.12464. [DOI] [PubMed] [Google Scholar]
- 272.Peixoto RS, Voolstra CR, Sweet M, Duarte CM, Carvalho S, Villela H, Lunshof JE, Gram L, Woodhams DC, Walter J, Roik A, Hentschel U, Thurber RV, Daisley B, Ushijima B, Daffonchio D, Costa R, Keller-Costa T, Bowman JS, Rosado AS, Reid G, Mason CE, Walke JB, Thomas T, Berg G. 2022. Harnessing the microbiome to prevent global biodiversity loss. Nat Microbiol. 10.1038/s41564-022-01173-1. [DOI] [PubMed] [Google Scholar]
- 273.Venn AA, Bernardet C, Chabenat A, Tambutté E, Tambutté S. 2020. Paracellular transport to the coral calcifying medium: effects of environmental parameters. J Exp Biol 223(Pt 17):jeb227074. 10.1242/jeb.227074. [DOI] [PubMed] [Google Scholar]
- 274.Al-Horani FA. 2005. Effects of changing seawater temperature on photosynthesis and calcification in the scleractinian coral Galaxea fascicularis, measured with O2, Ca2+, and pH microsensors. Sci Mar 69:347–354. 10.3989/scimar.2005.69n3347. [DOI] [Google Scholar]
- 275.Evensen NR, Edmunds PJ. 2018. Effect of elevated pCO2 on competition between the scleractinian corals Galaxea fascicularis and Acropora hyacinthus. J Exp Mar Bio Ecol 500:12–17. 10.1016/j.jembe.2017.12.002. [DOI] [Google Scholar]
- 276.Al-Horani FA, Al-Moghrabi SM, de Beer D. 2003. Microsensor study of photosynthesis and calcification in the scleractinian coral, Galaxea fascicularis: active internal carbon cycle. J Exp Mar Bio Ecol 288:1–15. 10.1016/S0022-0981(02)00578-6. [DOI] [Google Scholar]
- 277.Marshall AT, Clode P. 2004. Effects of calcium-free and low-calcium artificial seawater on polyps of a scleractinian coral Galaxea fascicularis. Coral Reefs 23:277–280. 10.1007/s00338-004-0371-4. [DOI] [Google Scholar]
- 278.Kramarsky-Winter E, Loya Y. 1996. Regeneration versus budding in fungiid corals: a trade-off. Mar Ecol Prog Ser 134:179–185. 10.3354/meps134179. [DOI] [Google Scholar]
- 279.Chuang PS, Mitarai S. 2020. Signaling pathways in the coral polyp bail-out response. Coral Reefs 39:1535–1548. 10.1007/s00338-020-01983-x. [DOI] [Google Scholar]
- 280.Conti-Jerpe IE, Thompson PD, Wong CWM, Oliveira NL, Duprey NN, Moynihan MA, Baker DM. 2020. Trophic strategy and bleaching resistance in reef-building corals. Sci Adv 6:eaaz5443. 10.1126/sciadv.aaz5443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Baker AC. 2003. Flexibility and specificity in coral-algal symbiosis: diversity, ecology, and biogeography of Symbiodinium. Annu Rev Ecol Evol Syst 34:661–689. 10.1146/annurev.ecolsys.34.011802.132417. [DOI] [Google Scholar]
- 282.Sunagawa S, Woodley CM, Medina M. 2010. Threatened corals provide underexplored microbial habitats. PLoS One 5:e9554. 10.1371/journal.pone.0009554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Ying H, Cooke I, Sprungala S, Wang W, Hayward DC, Tang Y, Huttley G, Ball EE, Forêt S, Miller DJ. 2018. Comparative genomics reveals the distinct evolutionary trajectories of the robust and complex coral lineages. Genome Biol 19:175. 10.1186/s13059-018-1552-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.van Woesik R, Sakai K, Ganase A, Loya Y. 2011. Revisiting the winners and the losers a decade after coral bleaching. Mar Ecol Prog Ser 434:67–76. 10.3354/meps09203. [DOI] [Google Scholar]
- 285.Reichert J, Arnold AL, Hammer N, Miller IB, Rades M, Schubert P, Ziegler M, Wilke T. 2021. Reef-building corals act as long-term sink for microplastic. Glob Chang Biol 28:33–45. 10.1111/gcb.15920. [DOI] [PubMed] [Google Scholar]
- 286.Baker DM, Andras JP, Jordán-Garza AG, Fogel ML. 2013. Nitrate competition in a coral symbiosis varies with temperature among Symbiodinium clades. ISME J 7:1248–1251. 10.1038/ismej.2013.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Berkelmans R, Van Oppen MJH. 2006. The role of zooxanthellae in the thermal tolerance of corals: a ‘nugget of hope’ for coral reefs in an era of climate change. Proc R Soc B 273:2305–2312. 10.1098/rspb.2006.3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Loya Y, Sakai K, Yamazato K, Nakano Y, Sambali H, van Woesik R. 2001. Coral bleaching: the winners and the losers. Ecol Lett 4:122–131. 10.1046/j.1461-0248.2001.00203.x. [DOI] [Google Scholar]
- 289.Flot J-F, Blanchot J, Charpy L, Cruaud C, Licuanan WY, Nakano Y, Payri C, Tillier S. 2011. Incongruence between morphotypes and genetically delimited species in the coral genus Stylophora: phenotypic plasticity, morphological convergence, morphological stasis or interspecific hybridization? BMC Ecol 11:22. 10.1186/1472-6785-11-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Kenkel CD, Matz MV. 2016. Gene expression plasticity as a mechanism of coral adaptation to a variable environment. Nat Ecol Evol 1:e0014. 10.1038/s41559-016-0014. [DOI] [PubMed] [Google Scholar]
- 291.Donelson JM, Sunday JM, Figueira WF, Gaitán-Espitia JD, Hobday AJ, Johnson CR, Leis JM, Ling SD, Marshall D, Pandolfi JM, Pecl G, Rodgers GG, Booth DJ, Munday PL. 2019. Understanding interactions between plasticity, adaptation and range shifts in response to marine environmental change. Philos Trans R Soc Lond B Biol Sci 374:20180186. 10.1098/rstb.2018.0186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Sharp KH, Distel D, Paul VJ. 2012. Diversity and dynamics of bacterial communities in early life stages of the Caribbean coral Porites astreoides. ISME J 6:790–801. 10.1038/ismej.2011.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Leite DCA, Leão P, Garrido AG, Lins U, Santos HF, Pires DO, Castro CB, van Elsas JD, Zilberberg C, Rosado AS, Peixoto RS. 2017. Broadcast spawning coral Mussismilia hispida can vertically transfer its associated bacterial core. Front Microbiol 8:176. 10.3389/fmicb.2017.00176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Putnam HM, Stat M, Pochon X, Gates RD. 2012. Endosymbiotic flexibility associates with environmental sensitivity in scleractinian corals. Proc Biol Sci 279:4352–4361. 10.1098/rspb.2012.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Davies S, Gamache MH, Howe-Kerr LI, Kriefall NG, Baker AC, Banaszak AT, Bay LK, Bellantuono AJ, Bhattacharya D, Chan CX, Claar DC, Coffroth MA, Cunning R, Davy SK, del Campo J, Diaz-Almeyda EM, Frommlet JC, Fuess LE, Gonzalez-Pech RA, Goulet TL, Hoadley KD, Howells EJ, Hume BCC, Kemp DW, Kenkel CD, Kitchen SA, LaJeunesse TC, Lin S, McIlroy S, McMinds R, Nitschke MR, Oakley CA, Peixoto RS, Prada C, Putnam HM, Quigley K, Reich HG, Reimer JD, Rodriguez-Lanetty M, Rosalas S, Saad OS, Sampayo EM, Santos S, Shoguchi E, Smith EG, Stat M, Stephens TG, Strader ME, Suggett DJ, Swain TD, et al. 2022. Building consensus around the assessment and interpretation of Symbiodiniaceae diversity. Preprints. 10.20944/preprints202206.0284.v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Kovacevic G. 2012. Value of the Hydra model system for studying symbiosis. Int J Dev Biol 56:627–635. 10.1387/ijdb.123510gk. [DOI] [PubMed] [Google Scholar]
- 297.Perez SF, Cook CB, Brooks WR. 2001. The role of symbiotic dinoflagellates in the temperature-induced bleaching response of the subtropical sea anemone Aiptasia pallida. J Exp Mar Bio Ecol 256:1–14. 10.1016/S0022-0981(00)00282-3. [DOI] [PubMed] [Google Scholar]
- 298.Sawyer SJ, Muscatine L. 2001. Cellular mechanisms underlying temperature-induced bleaching in the tropical sea anemone Aiptasia pulchella. J Exp Biol 204:3443–3456. 10.1242/jeb.204.20.3443. [DOI] [PubMed] [Google Scholar]
- 299.Bieri T, Onishi M, Xiang T, Grossman AR, Pringle JR. 2016. Relative contributions of various cellular mechanisms to loss of algae during cnidarian bleaching. PLoS One 11:e0152693. 10.1371/journal.pone.0152693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Mansfield KM, Carter NM, Nguyen L, Cleves PA, Alshanbayeva A, Williams LM, Crowder C, Penvose AR, Finnerty JR, Weis VM, Siggers TW, Gilmore TD. 2017. Transcription factor NF-κB is modulated by symbiotic status in a sea anemone model of cnidarian bleaching. Sci Rep 7:16025. 10.1038/s41598-017-16168-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Detournay O, Schnitzler CE, Poole A, Weis VM. 2012. Regulation of cnidarian-dinoflagellate mutualisms: evidence that activation of a host TGFβ innate immune pathway promotes tolerance of the symbiont. Dev Comp Immunol 38:525–537. 10.1016/j.dci.2012.08.008. [DOI] [PubMed] [Google Scholar]
- 302.Neubauer EF, Poole AZ, Weis VM, Davy SK. 2016. The scavenger receptor repertoire in six cnidarian species and its putative role in cnidarian-dinoflagellate symbiosis. PeerJ 4:e2692. 10.7717/peerj.2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Tivey TR, Parkinson JE, Mandelare PE, Adpressa DA, Peng W, Dong X, Mechref Y, Weis VM, Loesgen S. 2020. N-linked surface glycan biosynthesis, composition, inhibition, and function in cnidarian-dinoflagellate symbiosis. Microb Ecol 80:223–236. 10.1007/s00248-020-01487-9. [DOI] [PubMed] [Google Scholar]
- 304.Tortorelli G, Rautengarten C, Bacic A, Segal G, Ebert B, Davy SK, van Oppen MJH, McFadden GI. 2022. Cell surface carbohydrates of symbiotic dinoflagellates and their role in the establishment of cnidarian-dinoflagellate symbiosis. ISME J 16:190–199. 10.1038/s41396-021-01059-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Weis VM. 1991. The induction of carbonic anhydrase in the symbiotic sea anemone Aiptasia pulchella. Biol Bull 180:496–504. 10.2307/1542351. [DOI] [PubMed] [Google Scholar]
- 306.Cui G, Liew YJ, Li Y, Kharbatia N, Zahran NI, Emwas A-H, Eguiluz VM, Aranda M. 2019. Host-dependent nitrogen recycling as a mechanism of symbiont control in Aiptasia. PLoS Genet 15:e1008189. 10.1371/journal.pgen.1008189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Rädecker N, Chen JE, Pogoreutz C, Herrera M, Aranda M, Voolstra CR. 2019. Nutrient stress arrests tentacle growth in the coral model Aiptasia. Symbiosis 78:61–64. 10.1007/s13199-019-00603-9. [DOI] [Google Scholar]
- 308.Freeman C, Stoner E, Easson C, Matterson K, Baker D. 2016. Symbiont carbon and nitrogen assimilation in the Cassiopea-Symbiodinium mutualism. Mar Ecol Prog Ser 544:281–286. 10.3354/meps11605. [DOI] [Google Scholar]
- 309.Freeman C, Stoner E, Easson C, Matterson K, Baker D. 2017. Variation in δ13C and δ15N values suggests a coupling of host and symbiont metabolism in the Symbiodinium-Cassiopea mutualism. Mar Ecol Prog Ser 571:245–251. 10.3354/meps12138. [DOI] [Google Scholar]
- 310.Mortillaro JM, Pitt KA, Lee SY, Meziane T. 2009. Light intensity influences the production and translocation of fatty acids by zooxanthellae in the jellyfish Cassiopea sp. J Exp Mar Bio Ecol 378:22–30. 10.1016/j.jembe.2009.07.003. [DOI] [Google Scholar]
- 311.Verde E, McCloskey L. 1998. Production, respiration, and photophysiology of the mangrove jellyfish Cassiopea xamachana symbiotic with zooxanthellae: effect of jellyfish size and season. Mar Ecol Prog Ser 168:147–162. 10.3354/meps168147. [DOI] [Google Scholar]
- 312.Holcomb M, Cohen AL, McCorkle DC. 2012. An investigation of the calcification response of the scleractinian coral Astrangia poculata to elevated pCO2 and the effects of nutrients, zooxanthellae and gender. Biogeosciences 9:29–39. 10.5194/bg-9-29-2012. [DOI] [Google Scholar]
- 313.Burmester EM, Breef-Pilz A, Lawrence NF, Kaufman L, Finnerty JR, Rotjan RD. 2018. The impact of autotrophic versus heterotrophic nutritional pathways on colony health and wound recovery in corals. Ecol Evol 8:10805–10816. 10.1002/ece3.4531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Trumbauer W, Grace SP, Rodrigues LJ. 2021. Physiological seasonality in the symbiont and host of the northern star coral, Astrangia poculata. Coral Reefs 40:1155–1166. 10.1007/s00338-021-02119-5. [DOI] [Google Scholar]
- 315.Dimond J, Carrington E. 2008. Symbiosis regulation in a facultatively symbiotic temperate coral: zooxanthellae division and expulsion. Coral Reefs 27:601–604. 10.1007/s00338-008-0363-x. [DOI] [Google Scholar]
- 316.Maire J, Blackall LL, van Oppen MJH. 2021. Microbiome characterization of defensive tissues in the model anemone Exaiptasia diaphana. BMC Microbiol 21:152. 10.1186/s12866-021-02211-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Fleck J, Fitt W. 1999. Degrading mangrove leaves of Rhizophora mangle Linne provide a natural cue for settlement and metamorphosis of the upside down jellyfish Cassiopea xamachana Bigelow. J Exp Mar Bio Ecol 234:83–94. 10.1016/S0022-0981(98)00140-3. [DOI] [Google Scholar]
- 318.Grasis JA, Lachnit T, Anton-Erxleben F, Lim YW, Schmieder R, Fraune S, Franzenburg S, Insua S, Machado G, Haynes M, Little M, Kimble R, Rosenstiel P, Rohwer FL, Bosch TCG. 2014. Species-specific viromes in the ancestral holobiont Hydra. PLoS One 9:e109952. 10.1371/journal.pone.0109952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Brüwer JD, Voolstra CR. 2018. First insight into the viral community of the cnidarian model metaorganism Aiptasia using RNA-Seq data. PeerJ 6:e4449. 10.7717/peerj.4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Bathia J, Schröder K, Fraune S, Lachnit T, Rosenstiel P, Bosch TCG. 2022. Symbiotic algae of Hydra viridissima play a key role in maintaining homeostatic bacterial colonization. Front Microbiol 13:869666. 10.3389/fmicb.2022.869666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Goldsmith DB, Pratte ZS, Kellogg CA, Snader SE, Sharp KH. 2019. Stability of temperate coral Astrangia poculata microbiome is reflected across different sequencing methodologies. AIMS Microbiol 5:62–76. 10.3934/microbiol.2019.1.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Van Etten JL, Meints RH, Kuczmarski D, Burbank DE, Lee K. 1982. Viruses of symbiotic Chlorella-like algae isolated from Paramecium bursaria and Hydra viridis. Proc Natl Acad Sci USA 79:3867–3871. 10.1073/pnas.79.12.3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Li X-Y, Lachnit T, Fraune S, Bosch TCG, Traulsen A, Sieber M. 2017. Temperate phages as self-replicating weapons in bacterial competition. J R Soc Interface 14:20170563. 10.1098/rsif.2017.0563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Lange J, Fraune S, Bosch TCG, Lachnit T. 2019. The neglected part of the microbiome: prophage TJ1 regulates the bacterial community of the metaorganism Hydra. bioRxiv. https://www.biorxiv.org/content/10.1101/607325v1.
- 325.Downs CA, Fauth JE, Downs VD, Ostrander GK. 2010. In vitro cell-toxicity screening as an alternative animal model for coral toxicology: effects of heat stress, sulfide, rotenone, cyanide, and cuprous oxide on cell viability and mitochondrial function. Ecotoxicology 19:171–184. 10.1007/s10646-009-0403-5. [DOI] [PubMed] [Google Scholar]
- 326.Biscéré T, Ferrier-Pagès C, Grover R, Gilbert A, Rottier C, Wright A, Payri C, Houlbrèque F. 2018. Enhancement of coral calcification via the interplay of nickel and urease. Aquat Toxicol 200:247–256. 10.1016/j.aquatox.2018.05.013. [DOI] [PubMed] [Google Scholar]
- 327.Pavia RTB, Estacion JS. 2019. Survival and growth of isolated polyps of Galaxea fascicularis (Linnaeus 1767) on six kinds of culture substrates: implications for mariculture, aquarium culture, and conservation. J World Aquacult Soc 50:219–230. 10.1111/jwas.12538. [DOI] [Google Scholar]
- 328.Puntin G, Craggs J, Hayden R, Engelhardt K, McIlroy S, Sweet M, Baker DM, Ziegler M. 2022. The reef-building coral Galaxea fascicularis: a new model system for coral symbiosis. bioRxiv. https://www.biorxiv.org/content/10.1101/2022.06.02.494472v1.
- 329.McIlroy SE, Coffroth MA. 2017. Coral ontogeny affects early symbiont acquisition in laboratory-reared recruits. Coral Reefs 36:927–932. 10.1007/s00338-017-1584-7. [DOI] [Google Scholar]
- 330.Richmond R. 1985. Reversible metamorphosis in coral planula larvae. Mar Ecol Prog Ser 22:181–185. 10.3354/meps022181. [DOI] [Google Scholar]
- 331.Fordyce AJ, Camp EF, Ainsworth TD. 2017. Polyp bailout in Pocillopora damicornis following thermal stress. F1000Res 6:687. 10.12688/f1000research.11522.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Suwa R, Nakamura M. 2018. A precise comparison of developmental series of oocyte growth and oocyte maturation between real-oocytes and pseudo-oocytes in the coral Galaxea fascicularis. J Coral Reef Stud 20:1–7. 10.3755/galaxea.20.1_1. [DOI] [Google Scholar]
- 333.Miller M, Szmant A. 2006. Lessons learned from experimental key-species restoration, p 219–235. In Precht W (ed), Coral reef restoration handbook. CRC Press, Boca Raton, FL. [Google Scholar]
- 334.Romano SL, Palumbi SR. 1996. Evolution of scleractinian corals inferred from molecular systematics. Science 271:640–642. 10.1126/science.271.5249.640. [DOI] [Google Scholar]
- 335.Voolstra CR, Li Y, Liew YJ, Baumgarten S, Zoccola D, Flot J-F, Tambutté S, Allemand D, Aranda M. 2017. Comparative analysis of the genomes of Stylophora pistillata and Acropora digitifera provides evidence for extensive differences between species of corals. Sci Rep 7:17583. 10.1038/s41598-017-17484-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Kerr AM. 2005. Molecular and morphological supertree of stony corals (Anthozoa: Scleractinia) using matrix representation parsimony. Biol Rev Camb Philos Soc 80:543–558. 10.1017/S1464793105006780. [DOI] [PubMed] [Google Scholar]
- 337.Lajeunesse TC, Bhagooli R, Hidaka M, DeVantier L, Done T, Schmidt G, Fitt W, Hoegh-Guldberg O. 2004. Closely related Symbiodinium spp. differ in relative dominance in coral reef host communities across environmental, latitudinal and biogeographic gradients. Mar Ecol Prog Ser 284:147–161. 10.3354/meps284147. [DOI] [Google Scholar]
- 338.Huang H, Dong Z, Huang L, Yang J, Di B, Li Y, Zhou G, Zhang C. 2011. Latitudinal variation in algal symbionts within the scleractinian coral Galaxea fascicularis in the South China Sea. Mar Biol Res 7:208–211. 10.1080/17451000.2010.489616. [DOI] [Google Scholar]
- 339.Silverstein R, Correa A, LaJeunesse T, Baker A. 2011. Novel algal symbiont (Symbiodinium spp.) diversity in reef corals of Western Australia. Mar Ecol Prog Ser 422:63–75. 10.3354/meps08934. [DOI] [Google Scholar]
- 340.Keshavmurthy S, Meng P-J, Wang J-T, Kuo C-Y, Yang S-Y, Hsu C-M, Gan C-H, Dai C-F, Chen CA. 2014. Can resistant coral-Symbiodinium associations enable coral communities to survive climate change? A study of a site exposed to long-term hot water input. PeerJ 2:e327. 10.7717/peerj.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Lin Z, Chen M, Dong X, Zheng X, Huang H, Xu X, Chen J. 2017. Transcriptome profiling of Galaxea fascicularis and its endosymbiont Symbiodinium reveals chronic eutrophication tolerance pathways and metabolic mutualism between partners. Sci Rep 7:42100. 10.1038/srep42100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Radice VZ, Brett MT, Fry B, Fox MD, Hoegh-Guldberg O, Dove SG. 2019. Evaluating coral trophic strategies using fatty acid composition and indices. PLoS One 14:e0222327. 10.1371/journal.pone.0222327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Mass T, Einbinder S, Brokovich E, Shashar N, Vago R, Erez J, Dubinsky Z. 2007. Photoacclimation of Stylophora pistillata to light extremes: metabolism and calcification. Mar Ecol Prog Ser 334:93–102. 10.3354/meps334093. [DOI] [Google Scholar]
- 344.Muir PR, Wallace CC, Done T, Aguirre JD. 2015. Limited scope for latitudinal extension of reef corals. Science 348:1135–1138. 10.1126/science.1259911. [DOI] [PubMed] [Google Scholar]
- 345.Rinkevich B, Loya Y. 1979. The reproduction of the Red Sea coral Stylophora pistillata. I. Gonads and planulae. Mar Ecol Prog Ser 1:133–144. 10.3354/meps001133. [DOI] [Google Scholar]
- 346.Harrison PL, Babcock RC, Bull GD, Oliver JK, Wallace CC, Willis BL. 1984. Mass spawning in tropical reef corals. Science 223:1186–1189. 10.1126/science.223.4641.1186. [DOI] [PubMed] [Google Scholar]
- 347.Harrison PL. 1988. Pseudo-gynodioecy: an unusual breeding system in the scleractinian coral Galaxea fascicularis, p 699–705. In 6th International Coral Reef Symposium. Executive Committee, Townsville, Australia. [Google Scholar]
- 348.Szmant AM. 1986. Reproductive ecology of Caribbean reef corals. Coral Reefs 5:43–53. 10.1007/BF00302170. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download mmbr.00053-22-s0001.xlsx, XLSX file, 0.01 MB (13.3KB, xlsx)