Abstract
Volatile organic compound (VOC) emissions were measured from Chinese Hamster Ovary (CHO) cell and T cell bioreactor gas exhaust lines with the goal of non-invasively metabolically profiling the expansion process. Measurements of cellular ‘breath’ were made directly from the gas exhaust lines using polydimethylsiloxane (PDMS)-coated magnetic stir bars, which underwent subsequent thermal deso- rption-gas chromatography-mass spectrometry (TD–GC–MS) analysis. Baseline VOC profiles were observed from bioreactors filled with only liquid media. After inoculation, unique VOC profiles correlated to cell expansion over the course of 8 d. Partial least squares (PLS) regression models were built to predict cell culture density based on VOC profiles of CHO and T cells (R2 = 0.671 and R2 = 0.769, respectively, based on a validation data set). T cell runs resulted in 47 compounds relevant to expansion while CHO cell runs resulted in 45 compounds; the 20 most relevant compounds of each cell type were putatively identified. On the final experimental days, sorbent-covered stir bars were placed directly into cell-inoculated media and into media controls. Liquid-based measurements from spent media containing cells could be distinguished from media-only controls, indicating soluble VOCs excreted by the cells during expansion. A PLS- discriminate analysis (PLS–DA) was performed, and 96 compounds differed between T cell-inoculated media and media controls with 72 compounds for CHO cells; the 20 most relevant compounds of each cell line were putatively identified. This work demonstrates that the volatilome of cell cultures can be exploited by chemical detectors in bioreactor gas and liquid waste lines to non-invasively monitor cellular health and could possibly be used to optimize cell expansion conditions ‘on-the-fly’ with appropriate control loop systems. Although the basis for statistical models included compounds without certain identification, this work provides a foundation for future research of bioreactor emissions. Future studies must move towards identifying relevant compounds forunderstanding of underlying biochemistry.
Keywords: bioreactors, VOCs, cell expansion, T cells, process analytical technologies, CHO cells, GC–MS
Introduction
In the search for new process analytical technologies (PAT), downstream volatile organic compound (VOC) emissions from cell cultures can be measured by soft sensors for online bioprocess monitoring. A few examples of this have been demonstrated, measuring cellular VOCs with various technologies. An electronic nose uses an array of chemical reactions/binding to show total volatile profiles, tracked with growth of Chinese Hamster Ovary (CHO) cells in a bioreactor [1]. Biomass and growth rates were predicted from VOC profiles of Escherichia coli batch cultivations [2], and the electronic nose also detected VOC changes in animal cell reactor cultures due to microbial and viral contam- inations, including E. coli [3]. However, one major disadvantage of the electronic nose technology is the lack of structural information to confidently identify chemical species, an important step toward assessing the biological relevance of targeted VOCs in any analysis. In addition, those sensors drift over time and must constantly be recalibrated.
Other reports have noted that changes in VOC headspace can be measured from mammalian cells using traditional mass spectrometry, and those chan- ges correlated with single gene expression levels [4]. Mass spectrometry techniques provide additional information for compound identification and have trended towards incorporation as online sensors in reaction monitoring [5] such as in bioreactors. Proton transfer reaction-mass spectrometry (PTR-MS) was incorporated into an E. coli bioreactor and VOC pro- files correlated to culture growth [6]. Traditional mass spectrometry (such as single quadrupole MS) was applied to bioreactors of animal and yeast cells to mea- sure liquid-phase concentrations as well as of gases, such as oxygen and carbon dioxide, and VOCs, such as acetone [7]. Using gas chromatography-mass spectro- metry (GC–MS), murine stem cells and fibroblasts were also observed to have volatilome shifts correlated to growth [8]. GC–MS was also used to demonstrate volatile differences among adipogenically differentiat- ing and nondifferentiating human adipose tissue- derived mesenchymal stromal/step cells [9]. Finally, the volatilome of Mycobacterium avium ssp. para- tuberculosis also correlated with bacterial density [10].
This body of work demonstrates that volatiles emitted by cell cultures relate to metabolism and the cellular ‘breath’ profile is expected to change—even when cells are unperturbed. These results are also important for breath researchers, who investigate volatile signatures in lung cancer lines [11] or rhino- virus-infected human airway cells [12].
Furthermore, there has been an expanding field to miniaturize technologies for VOC analysis and produce ‘lab-on-a-chip’ sensors. Micro-electro-mechanical sys- tems (MEMS) for VOC detection often start with a sample preconcentration step, in which a tiny sorbent- filled chip [13] or other functionalized surface [14] adsorbs VOCs from a gaseous sample. This is poten- tially followed by separation of the chemical species in a micro gas chromatograph [15, 16]. A MEMS-based sensor such as a micro fabricated ion mobility spectro- meter [17], metal oxide semiconductor (MOS) [18], or mass spectrometer, then detects the VOCs [19].
As smaller and more mobile sensors emerge, there are exciting opportunities for bioprocess engineering applications to include VOC-based process analytical controls to provide non-invasive assessments related to the health and proliferation of cell cultures based on cellular ‘breath’. Thus, researchers continue to explore new opportunities for VOC exploitation in bior- eactors, with a drive towards biomarker identification to allow confirmation of the cellular metabolic path- ways that would explain the observed VOC changes.
In this paper, we correlate VOC profiles from bior-eactors over a significant time course of cell expansion for both CHO and T cells. These cell types are impor- tant cell expression models for use in bioprocess engi- neering and cellular immunotherapy workflows. Using headspace sorptive extraction (HSSE) and stir- bar sorptive extraction (SBSE) techniques coupled with GC–MS, we provide critical new information into the types of VOC compounds related to cell growth and cell health. While some work has been performed on CHO cultures previously; to our knowl- edge, we are the first to measure VOC emissions from primary T cell cultures and to describe their VOC profiles.
Identification of all measured compounds is not entirely possible within this scope of work, as putative attempts yield mass spectra matches that are similar for dozens of compounds. For example, it may be pos- sible to know a measured VOC is some type of alde- hyde based on the obtained spectrum, but the exact structure is still unknown. Also, there is a lack of reten- tion indeces in the literature for many potential com- pounds that match our obtained spectrum. This phenomenom has been observed in other cell culture studies [20]. These unknown features are still included in this body of work, as these compounds were con- sistently measured from bioreactors and were found to correlate with cellular expansion. Future studies must make progress to identify these volatile metabolites to elucidate the underlying biochemistry.
Method and materials
Xuri bioreactor setup, cell lines and media Primary T cells were isolated from buffy coats (sourced from Canadian Blood Services) from 2 donors using a Ficoll density gradient and cultured in T flasks for 6 d prior to inoculation in a Xuri Cell Expansion System (CES, GE Healthcare) at ~7 × 105 cells ml−1 in 1 l of T cell culture medium. T cell culture medium was Xuri Expansion Medium (GE Healthcare) with 1% penicil- lin-streptomycin (Hyclone), 5% human AB serum (GemCell), and 350 IU/ml Xuri IL-2. CHO-M cells (courtesy of GE Healthcare, Uppsala, Sweden) were cultured in T flasks in ActiPro (Hyclone) medium with 1% penicillin-streptomycin and 2 mM L-glutamine (Hyclone). CHO cells were inoculated in a Xuri CES at~2 × 105 cells ml−1 in 1 l.
Four, 2 l Xuri Cellbags (working volume of 1 leach) with dissolved oxygen (DO) and pH sensors were connected to Xuri CESs. The 2 l Cellbag was inflated with compressed air and 5% CO2 and then left overnight with 200 ml culture medium to equilibrate the DO/pH sensors. Temperature was set to 37 °C and the platform set to rock at 10 RPM at a 6° angle. For 2 min per hour, the platform rocked at 2 rpm at a 2° angle. Perfusion was initiated using a step-wise proto- col based on a combination of lactate measurements as well as cell density. Below 2 × 106 cells ml−1, no per- fusion was initiated. Above 2 × 106 cells ml−1, med- ium was perfused at 0.5 l/d at VCD between 2 × 106–10 × 106 cells ml−1, at 0.75 l/d for VCD between 10 × 106–15 × 106 cells ml−1, and at 1 l/d for VCD greater than 15 × 106 cells ml−1. A step up in perfusion level was initiated regardless of the VCD in the event of a lactate concentration exceeding 20 mM.
Bioreactor VOC exhaust measurements
A previously developed cell culture HSSE–GC–MS method [21] was adapted for use in immunotherapy cell expansion bioreactors for this study. Cell culture VOC emissions from the gas exhaust line of bioreac- tors were measured using a HSSE technique. An illustration of this experimental setup is provided (figure 1). Bioreactor air exhaust was directed via PTFE tubing through the lid of a capped 250 ml borosilicate jar (Part 1117-S008, Quality Environmental Contain- ers Inc., Beaver, WV). Each bioreactor was connected with a single jar and the same jar was used throughout the course of the entire experiment. Each jar contained four sterile and pre-conditioned HSSE stir bars (‘Twisters®’, Part 011222–001-00, Gerstel US, Linthi- cum Heights, MD), held in place to the side of the jar by magnets, providing four technical replicates per sample. The commercially available HSSE bars were 10 mm in length and coated with a 0.5 mm thick layer of polydimethylsulfide (PDMS) sorbent. Twisters® were left to extract cell culture VOCs in 24 h increments. After this period, the lids were removed from the jars, the four Twisters® were collected and replaced with four fresh HSSE bars, and the lid was screwed back onto the jar.
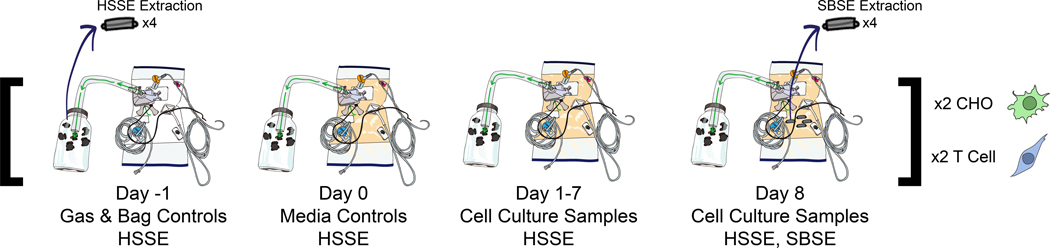
Figure 1.
Experimental design. VOCs from bioreactor gas exhaust were measured via a headspace sorptive extraction (HSSE) method for 10 d (one day of ‘gas and blank controls’, one day of ‘media controls’, eight days of cell culture growth). On the final day 8, stir bar sorptive extraction (SBSE) was performed directly in the cell culture media to extract liquid-phase cellular VOCs.
Liquid-phase in situ VOC measurements
A final time point measurement to examine VOCs dissolved in the liquid media was made using Twisters® in a stir bar sorbtive extraction (SBSE) immersion technique. This was not performed until the end of the experiment to reduce the risk of cell culture contamination. During the final 24 h of the experiment, four sterilized Twisters® (soaked in 70% ethanol for 10 min) were dropped directly into each cell culture via a port on the CellBag bioreactor. Once extraction was complete (24 h), the bioreactor bags were sliced open and the Twisters® were collected. The experiment ended at this point and cells were destroyed via exposure to 10% bleach. For media controls, additional Twisters® were placed directly into 20 ml of cell-free media of each type for 24 h and incubated at the same temperature as the cultures.
Time course explanation
A visual representation of the experimental execution is shown in figure 1. The day prior to media equilibra- tion (day −1), four empty Xuri CellBags were attached to the Xuri units with air flow (compressed air + 5% CO2) and ‘bag and gas controls’ were collected to measure background VOCs. The day of media addi- tion (day 0), two bioreactors had 200 ml T cell media added and two reactors had 200 ml CHO media added; ‘media controls’ were collected (no media perfusion during this day). On the day of cell seeding (day 1), the bioreactors were inoculated with their respective cell lines. HSSE VOC measurements were conducted over 8 d of cell expansion. On day 8, the liquid SBSE measurements and HSSE measurements were concur- rently collected. Every 3–4 d, four unused Twisters® were pulled aside for ‘sorbent controls’ which acted as shipping and handling controls to ensure VOCs of unknown origin did not compromise the experiment.
Twice a day, an aliquot (5–10 ml) from the bior- eactors was collected for measurements of culture attributes/metabolites: viable cell density (VCD), % viability, glutamine, glutamate, glucose, lactate, ammonium, sodium, potassium, calcium, pH and pO2. VCD and percent viability were measured on a Nucleocounter NC-200 (Chemometec, Allerod, Denmark). Metabolite measurements were conducted on a BioProfile FLEX 2 Analyzer (Nova Biomedical, Waltham, MA). Cells were counted by taking an ali- quot of the culture using a syringe through the sam- pling port, pipetting to disrupt aggregates, and the counting on a Nucleocounter NC-200. If cell density was >5 × 106 cells ml−1, then the sample to count was diluted in the same media used to culture the cells in order to obtain a sample in range for measurement on the Nucleocounter.
Twister®-GC–MS analysis
There were two biological replicates (2 Xuri CellBags) for T cells and two biological replicates for CHO cells, with four technical replicates (Twister® sorbent bars) at each time point. All Twisters® were pre-conditioned prior to use, according to manufacturer specifications.
As soon as Twisters® were extracted from the cell culture reactors, they were placed into 2 ml bor- osilicate vials and an aliquot of the first internal stan- dard (1 μl of a 1 ppm naphthalene-D8 in ethanol solution) was pipetted into each vial. Twisters® were kept frozen until analysis. Just prior to analysis, they were transferred into thermal desorption tubes along- side an aliquot of the second internal standard (1 μl of a 0.1 ml l−1 decane-D22 in ethanol).
Individual Twisters® were thermally desorbed using a thermal desorption unit (TDU, Gerstel US) and cooled injection system (CIS, Gerstel US). The TDU was initially set to 30 °C for 0.5 min and heated at 60 °C min−1 until reaching 300 °C and held for 3 min. A flow of helium led desorbed analytes into the CIS, which was held at−80 °C. After desorption, the CIS heated at 12 °C s−1 to 300 °C and was held for 3 min. This process splitlessly injected analytes onto the head of the GC column.
Chromatography occurred on an Agilent 7890A GC (Agilent Technologies Inc., Santa Clara, CA) equipped with a DB-5ms column (30 m × 250 μm × 0.25 μm, Agilent Technologies Inc.). The column was initially at 35 °C for 3 min, then heated at 2 °C min−1 to 200 °C, then heated at 30 °C min−1 to 300 °C and held for5 min. Total runtime was 93.8 min. The GC was operated in constant flow mode (1.5 ml min−1 of helium). Analytes eluted into a 5975C single quadrupole mass spectro- meter (MS, Agilent Technologies Inc.). The MS scanned from 33 to 300 m/z. Its source and quad were set to 230 °C and 150 °C, respectively.
A bake out of the TDU–CIS–GC–MS system was conducted every ~20 injections. After every 30–40 GC–MS injections, a standard mixture of C8-C24 alkanes was analyzed to serve as an external control of the instrument and also to calculate Kovats retention indices of compounds.
GC–MS data processing
GC–MS data files were deconvoluted and aligned using the recursive feature extraction on Profinder (Version B.08.00, Agilent Technologies Inc.). Peak areas were normalized to the first internal standard. Features with siloxane base peaks (73, 147, 207, 221 and 281 m/z) were removed. Statistical analyses were performed using GeneSpring (Version B.14.9, Agilent Technologies Inc.) and PLS_Toolbox (Version 8.6, Eigenvector Research Inc., Manson, WA).A p-value of p < 0.05 was used throughout for significance. Puta- tive peak identification was possible through spectral matching with the NIST 14 mass spec database along with comparison of calculated Kovats Retention Index comparisons to reported literature values.
To model changes in VOC profiles related to cell growth, HSSE data from both CHO cell reactors were pooled together and VOC data from both T cell reac- tors were pooled together, and data were autoscaled. Within each of these two groups, the data were ran- domly separated: 67% for a calibration training set and 33% for a validation set. Partial least squares regres- sion (PLS) was applied to correlate live cell densities (the Y space) to the VOC profiles (the X space) using PLS_Toolbox software (Eigenvector Research Inc., Manson, WA). Cross-validation was performed using the venetian blinds technique, where the calibration data were split into 10 random splits and one sample per split was used to cross-validate the model. To clus- ter compounds of similar changes in intensity, agglomerative hierarchical clustering was applied using the shortest distance algorithm in MATLAB R2017a software (MathWorks, Natick, MA).
SBSE data were divided into the two cell types and their respective controls. A PLS-discriminate analysis (PLS–DA) was performed on each cell type to categori- cally distinguish media controls from cell samples.
Results and discussion
Cell expansion
At the time of media inoculation, the concentrations of CHO cells were 2.2 × 105 and 2.6 × 105 cells ml−1 per reactor respectively, and T cells were 7.0 × 105 and 8.0 × 105 cells ml−1 (supplemental figure 1 is available online at stacks.iop.org/JBR/14/016002/mmedia). By the end of the experiment, the majority of the bioreactors increased cell density by 16–30 times indicating exponential growth over the culture dura- tion in the Xuri CES. On the final day of the experiment, one of the CHO reactors (CHO 2) experienced an unrelated technical issue and lost much of its media, resulting in a sudden spike in cell density for the CHO 2 reactor on day 8. Twister® samples from this reactor from this day were excluded from VOC analysis.
Measured metabolites are also provided in supplemental figure 2 for the duration of culture in the Xuri CES. Monovalent and divalent cations such as K+, Ca2+, and Na+ had fairly stable levels throughout the experiment. As expected, during the initial days of cul- ture in the Xuri CES, pO2, glutamine and glucose concentrations dropped as these metabolites were consumed and lactate and ammonia rose as these byproducts were accumulated. Similarly a con- comitant decrease in pH was observed over the course of the early days of culture corresponding to an increase in lactate. After perfusion was initiated, nearly all metabolites attained steady state levels.
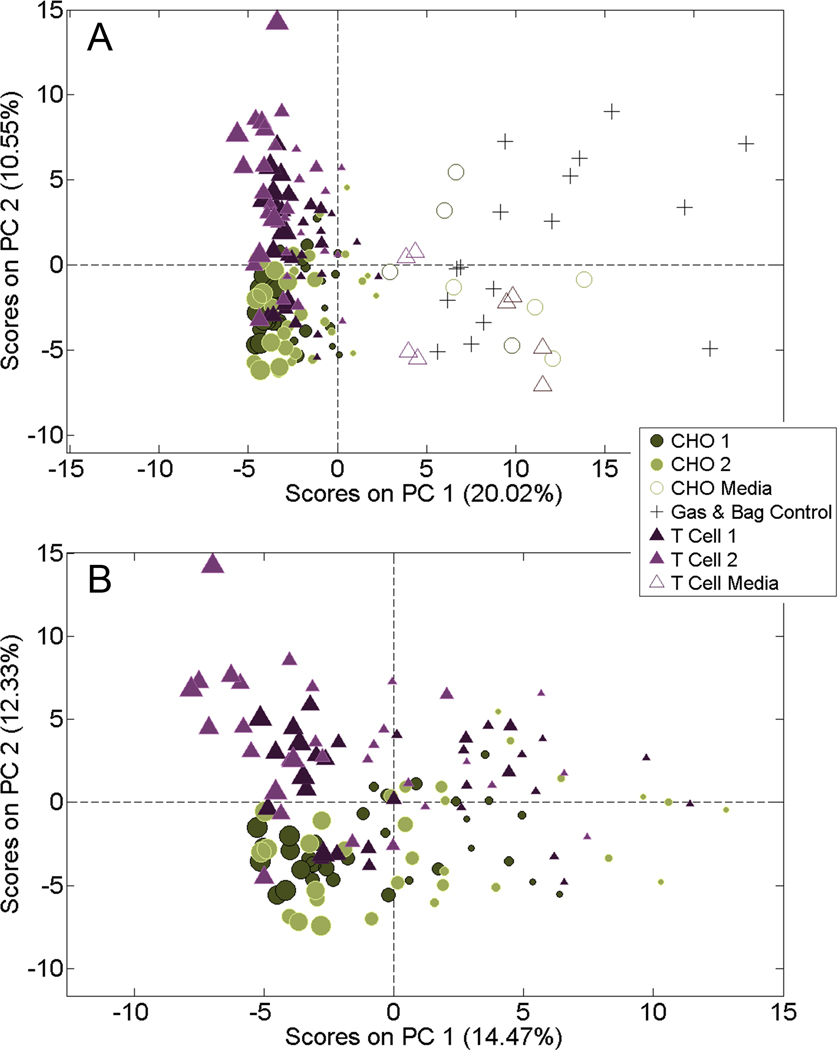
VOC profiles of downstream bioreactor emissions Principal components analysis (PCA) was applied to all HSSE samples (figure 2(A)). VOC profiles of the two control types (media, gas and bag) differed from bioreactors containing cells. Cell samples separated from controls along PC 1, which explained 20.02% of the variance. PCA is an unsupervised method that does not take into account meta-information about the sample (such as sample treatment or type) in its analysis. Instead, PCA only plots the variation between the GC–MS samples. Having controlsamples separate from cell samples along the first principal component suggests that the bioreactors with CHO and T cells exhaust cellular VOCs in levels that make them distinguishable from bioreactors filled with only media.
Figure 2.
PCAs of headspace volatile compound emissions from four bioreactors (two CHO, two T cell cultures). Cell culture samples are sized by day of expansion (smallest: day 1, largest: day 8). (A) Comparison of bioreactor bag and gas controls, media controls and cell culture samples, which separated along PC 1. (B) Cell culture samples during the eight days of expansion exhibited a VOC profile change along PC 1.
In addition to separating from controls, there was a trend for cell types to separate (figure 2(B)). T cell samples had a tendency to separate from CHO cell samples along PC 2, which explained 12.33% of the variance, indicating unique VOC profiles among the cell types.
CHO cells are non-human derived, and are immortalized as a cell line, thus their growth rates and, consequently, metabolic activity would necessarily be different to primary T cells, which would reach senes- cence after a finite number of doublings. Cell lines are engineered to be a cost-effective way to produce a desired outcome—for example output of viral titer or protein, which is one of the primary applications of CHO cells. In contrast, T cells in an autologous setting may be obtained from leukapheresis units or whole blood. Therefore, they may be more sensitive to cul- ture conditions, but may also be more representative of human heterogeneity and disease states. These inherent biological differences will directly impact the metabolism of the cells and the types and amounts of metabolites produced and consumed during bior- eactor expansion. On top of this, both cell types were grown in different media, with likely differing amounts of key media components such as carbon sources (e.g. glucose, L-glutamine), proteins (e.g. albumin) and other components such as trace metals, ions, and growth factors. Since each media is ulti- mately optimized for the growth of its associated cell type, this may further influence their metabolism. Indeed, the different VOC traces measures may offer insights into these metabolic differences, which is one of the key outcomes of this work.
More interesting was the gradual shift of samples that occurred along PC 1 (figure 2(B)), which explained 14.47% of the variance. PC 1 showed strong correlation to experimental day. With the bioreactors controlling all of the conditions of the reactor (gas flow, media perfusion, temperature, etc), the shift along PC 1 is strongly suspected to correlate to viable cell density, which increased with experimental day (supplemental figure 1).
Prior to any statistical analysis, including PCA, samples were normalized to the internal standard. This practice would account for any potential signal drift caused by the GC–MS instrument. Further, visualization of the internal standards results do not suggest an instrument drift occurred (data not shown), confirming that changes in the VOC profile must have related to changes in the bioreactor.
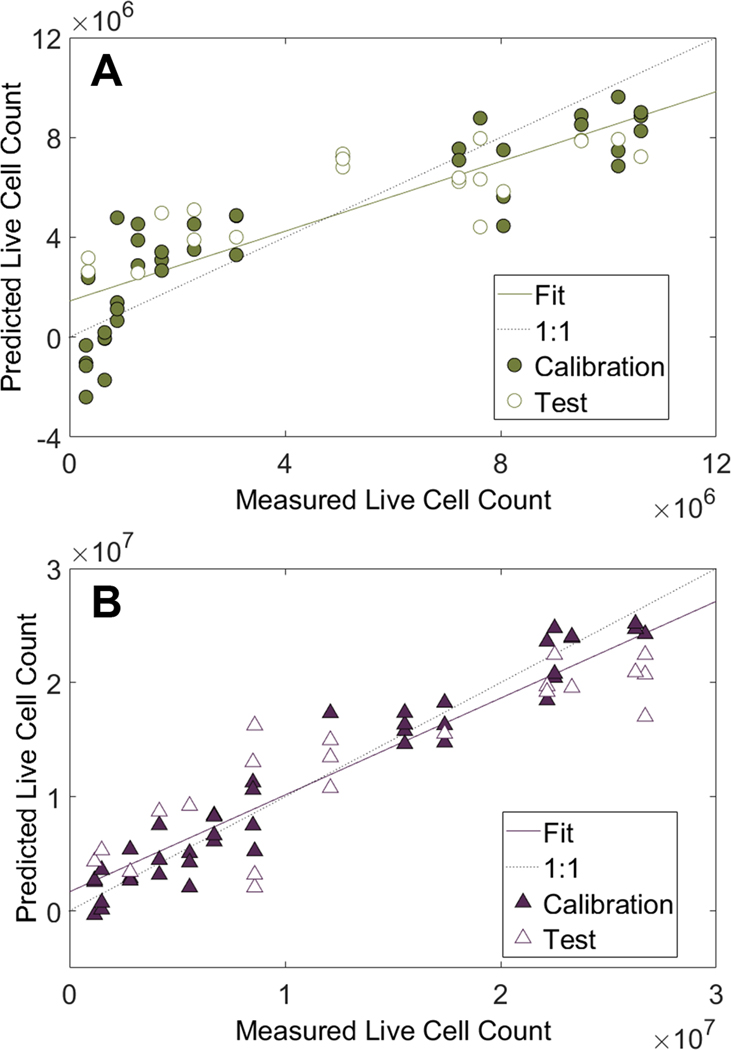
To correlate cell growth to VOC profiles, two PLS regression models were built, one for CHO cells and one for T cells. Within each cell type, 67% of data were used to train and calibrate the PLS model, which was then applied to the remaining 33% as a blinded valida- tion set. Models showed a correlation between the live cell density and the VOC profiles collected using the HSSE–GC–MS extraction technique (figure 3). Based on R2 values, the T cell model had a slightly better lin- ear fit, relative to CHO cells (table 1); although both cell models performed very well with high R2 values. As a measure of accuracy, T cells had slightly higher root-mean-square error (RMSE), even when normal- ized to the range of cell counts (maximum cell count minus minimum). In the validated sets, T cells had more than twice the normalized RMSE than CHO cells, although in general all of these RMSE values are fairly low.
Figure 3.
PLS regression models built from VOC profiles of (A) CHO cells and (B) T cells. Samples were randomly split into 67% calibration and 33% validation (test) sets. Cell counts are reported per ml of media.
Table 1.
Linear correlations (R2), root-mean-square errors (RMSE) and normalized RMSE (NRMSE, normalized to cell count range) from the two PLS models relating VOC profiles to live cell density (figure 3).
| CHO cells | T cells | |
|---|---|---|
|
| ||
| R2 Cross-validation set | 0.724 | 0.842 |
| RMSE Cross-validation set | 2.04 × 106 | 3.47 × 106 |
| NRMSE Cross-validation set | 1.98 × 10−1 | 3.37 × 10−1 |
| R2 Validation set | 0.671 | 0.769 |
| RMSE Validation set | 2.12 × 106 | 4.53 × 106 |
| NRMSE Validation Set | 2.06 × 10−1 | 4.40 × 10−1 |
In a PLS analysis, variable importance in projec- tion (VIP) scores are generated for each variable (in this case, a chemical VOC of interest). Variables with a VIP score greater than one are typically considered relevant to the regression. T cells had 47 compounds with a VIP > 1, and CHO cells had 45 compounds; 26 compounds overlapped between the two cell lines.
Putative identifications were made on the 20 com- pounds with the highest VIP score for the T cell model and the 20 compounds with the highest VIP score for the CHO model (table 2). 27.0% of these compounds were classified as a type of alkane, while 15.4% were esters, 7.7% alcohols, 7.7% oximes, and 23.0% others with 19.2% unknown.
Table 2.
Based on downstream bioreactor VOC emissions. Putative identifications of the 20 compounds with the highest VIP scores for the T cell regression model and the 20 compounds with the highest VIP scores for the CHO cell regression model (figure 3), combined into one table. KI: Kovats index, calculated (Calc) and as reported in the literature (Lit); Major Ions: the top three most abundant ions from respective mass spectra; MS Score: Score of acquired mass spectrum compared to the NIST mass spectral database; Cluster: group applicable to the clusters in figure 4.
| VIP Score (if >1) | Cluster | ||||||||
|---|---|---|---|---|---|---|---|---|---|
|
|
|
||||||||
| Compound | KI (Calc) | KI (Lit) | CAS # | Major Ions (m/z) | MS Score | T cells | CHO | T cell | CHO |
|
| |||||||||
| Undecane | 1100 | 1100 | 1120-21-4 | 57.1, 71.1, 85.1 | 93.71 | 2.57 | 2.69 | 4 | 4 |
| Unknown 1 (alkane) | 1170 | 57.1, 71.1, 85.1 | 2.49 | 2.77 | 4 | 4 | |||
| 2-(2-hydroxyethoxy)ethyl acetate | 1124 | 1000351-92-4 | 87.0, 89.0, 58.1 | 83.52 | 2.46 | 2.70 | 4 | 4 | |
| Unknown 2 (alkane) | 1097 | 71.1, 57.1, 85.1 | 2.37 | 2.65 | 4 | 4 | |||
| 2-ethylhexanal | 952 | 955 | 123-05-7 | 57.1, 72.1, 55.1 | 82.42 | 2.20 | 2.20 | 4 | 4 |
| Docosane | 2206 | 2200 | 629-78-7 | 57.1, 71.1, 85.1 | 89.69 | 2.17 | 2.59 | 1 | 1 |
| Unknown 3 (alkane) | 2220 | 57.1, 71.1, 85.1 | 2.13 | 2.47 | 1 | 1 | |||
| Unknown 4 | 1169 | 57.1, 154.7, 203.0 | 2.12 | 1.57 | 4 | 4 | |||
| 2-ethyl-1-hexanol | 1033 | 1029 | 104-76-7 | 57.1, 55.1, 70.1 | 96.06 | 1.98 | 3 | ||
| Diisobutyl phthalate | 1863 | 1868 | 84-69-5 | 149.0, 150.0, 223.1 | 76.01 | 1.92 | 1.29 | 2 | |
| Unknown 5 | 969 | 193.0, 209.0, 56.1 | 1.84 | 4 | |||||
| Unknown 6 | 1170 | 45.0, 250.0, 125.0 | 1.80 | 4 | |||||
| Unknown 7 (phthalic acid, alkane ester) | 2202 | 149.0, 167.0, 55.1 | 1.74 | 1 | |||||
| 2-methyldecane | 1062 | 1065 | 6975-98-0 | 57.1. 71.1. 85.1 | 84.10 | 1.66 | 1.85 | 4 | 4 |
| Unknown 8 | 1345 | 45.0, 117.0, 237.0 | 1.65 | 1.21 | 1 | ||||
| Decane | 1001 | 1000 | 124-18-5 | 57.1, 71.1, 85.1 | 72.22 | 1.64 | 1.98 | 4 | 4 |
| Benzaldehyde | 955 | 958 | 100-52-7 | 77.0, 106.0, 105.0 | 60.04 | 1.57 | 1.47 | 4 | 2 |
| Unknown 9 (haloalkane) | 950 | 95.0, 69.0, 131.0 | 1.55 | 1.65 | 4 | 4 | |||
| 1-methyl-4-propyl-2-pyrazoline | 1050 | 993 (est) | 33063-77-3 | 85.1, 41.0, 126.1 | 55.06 | 1.54 | 1.99 | 4 | 4 |
| Methoxyphenyloxime | 943 | 1000222-86-6 | 151.0, 133.0, 179.0 | 65.25 | 1.53 | 1.64 | 4 | 4 | |
| Methoxyphenyloxime (2) | 939 | 1000222-86-6 | 151.0, 133.0, 179.0 | 68.71 | 1.08 | 2.53 | 4 | ||
| 1-dodecanol | 1475 | 1469 | 112-53-8 | 55.1. 56.1. 43.1 | 79.75 | 1.04 | 2.01 | 4 | |
| 1,2-dibutoxyethane | 1190 | 1144 | 112-48-1 | 56.1, 57.1, 41.1 | 69.59 | 1.72 | 4 | ||
| Unknown 10 | 1251 | 41.1, 91.0, 65.0 | 1.80 | 3 | |||||
| Unknown 11 (ketone) | 1154 | 43.1, 71.1, 55.0 | 1.81 | 4 | |||||
| 1(3H)-isobenzofuranone | 1335 | 1272 (est) | 87-41-2 | 105.0, 77.0, 134.0 | 87.64 | 1.56 | 4 | ||
By using HSSE–GC–MS, we believe we are the first group to report the identities of VOCs emitted by CHO and T cells in a bioreactor during cell expansion. Without other studies to offer comparison, we com- pare these results to other cell culture experiments and find that the types of VOCs identified in this work are in general agreement with other mammalial cultures. 2-ethyl-1-hexanol was found relevant to viral infec- tions of human laryngeal cancer cells [22]. Benzalde- hyde has been observed in emissions of human fibroblasts (hFB) [11]. Esters have been observed in cultures of human B-lymphoblastoid cells [23]. Alkanes and alcohols have been observed in epithelial cell cultures [21]. Known background compounds were not included in statistical analyses, such as silox- anes from the PDMS sorbent and GC column bleed.
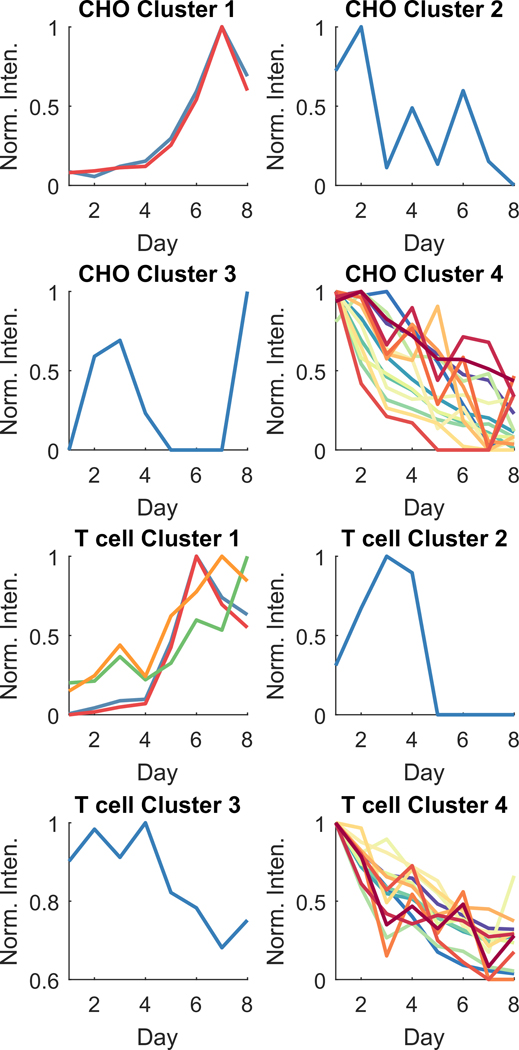
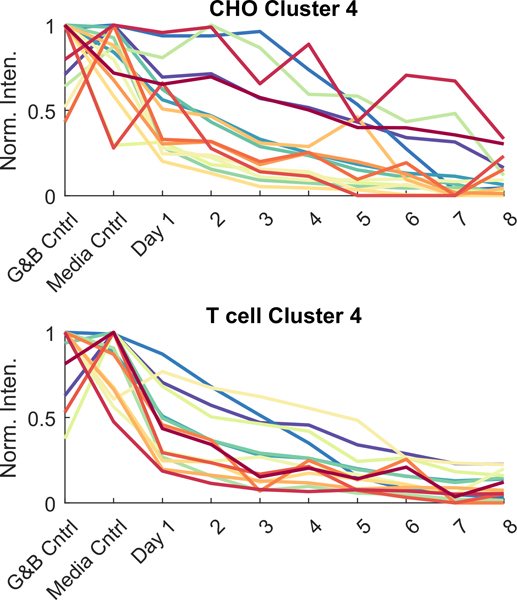
Some compounds increased in intensity with cell expansion while others decreased. To group com- pounds by patterns of change, hierarchical clustering was applied to the top 20 CHO and 20 T cell com- pounds from table 2. Each dendrogram was divided in such a way to yield four clusters of VOCs. Each cluster was plotted to demonstrate the compounds’ inten- sities over the course of the 8 d of cell expansion (figure 4). Both CHO and T cells exhibited com- pounds that increased over the course of cell expan- sion (Cluster 1 compounds). Two compounds increased over time in both cell lines: docosane and an unidentified alkane. Both cell types had a compound that increased until day 3–4, and then suddenly dis- appeared (CHO: Cluster 3, unknown 10; T cell: Clus- ter 2, benzaldehyde).
Figure 4.
How the 20 VOCs most relevant to cell culture expansion changed over 8 d of cell expansion. Compounds were split into 4 clusters via hierarchical clustering. VOCs in each cluster are found in table 2 and are presented as normalized to the maximum intensity within a compound (Norm. Inten.). Each point is the average of n = 8 replicates (four technical replicates × two biological replicates).
The compounds that increased over time are likely direct emissions from the cell cultures. These com- pounds could be directly monitored and exploited in a VOC-based PAT. By measuring downstream VOC emissions, there is no risk to contaminate the cell cultures, as is currently the case with withdrawing 5–10 ml from the reactor to manually measure cell count. VOC-based PAT could provide substantial cost savings with its non-invasive ability to assess cell cul- ture health.
The majority of these most relevant VOCs decreased during cell expansion (Cluster 4 compounds, figure 4). Figure 5 includes the gas and bag controls and media controls with these decreasing compounds. All compounds were present in bior- eactor controls prior to introduction of cells. Thus, it is possible that the cultures are metabolizing these com- pounds during expansion. Although media perfusion is occurring, this rate might not be fast enough to replenish these compounds as quickly as the cells are consuming them. This provides another opportunity for VOC exploitation: in addition to monitoring VOCs emitted by the cell cultures, it is possible to monitor the nutrients found in the media and adjust perfusion rates to provide sufficient growth material for optimal cell growth. Other researchers have also observed VOCs present in lower concentrations in inoculated media relative to pure media, such as stem cell cultures [8, 9].
Figure 5.
VOCs that decreased during cell expansion (from Cluster 4, figure 4), including gas and bag (G&B) controls and media controls. VOCs in each cluster are found in table 2 and are presented as normalized to the maximum intensity within a compound (Norm. Inten.). Each point is the average of n = 8 replicates (four technical replicates × two biological replicates).
As the two T cell cultures were obtained from dif- ferent donors, we had an opportunity to look at donor-based VOC differences. It is possible that expansion of human primary cells could lead to multi- ple heterogeneous functional populations. As noted in the methods, the added mitogens were IL-2 and human AB serum, however mitogens may also have been present in the basal medium, of which the for- mulation is not publicly available. A limited flow cyto- metry analysis was performed one day prior to media inoculation and again on day 8 across two biological replicates. When gating on the CD3 + T cell popula- tion, the CD4 + population appeared to be selected for in one donor (day −1: 65% CD4 + and 32% CD8 +, day 8: 84% CD4 + and 16% CD8+), but not in the other (day −1: 52% CD4 + and 40% CD8+, day 8: 53% CD4 + and 45% CD8+). The sample size of two biological replicates did not allow for a thorough sta- tistical analysis, thus additional replicates are needed to draw conclusions about the impact of mitogens on specific T cell subpopulations.
Although there were differences observed between the donors in regards to the ratio of CD3+/CD4 + and CD3+/CD8+, the viability and over-all cells expansion were similar (data not shown). This indicates that the mitogens or bioreactor expansion conditions did not preferentially trend toward expanding one donor over another, however, once again, a statistical analysis was not possible due to the low power of the experiment.
PCA (figure 2) suggests that the volatile profile of the two T cell lines were similar to each other, as the two cultures overlapped along PC 1 and 2. Overall, we observed a trend that compounds that increased with expansion in one line also increased in the other. This suggests that there may not be strong volatile differ- ences based on donor, although only 2 donors were used in this study. Potential sources of variability in the data, though not specifically noted, tested, or iden- tified in this work, could be donor age, donor health state, operator-to-operator variability during cell expansion, batch-to-batch variability in reagents or consumables used across the study such as human AB serum or IL-2, all of which may be confounding fac- tors to the analysis of VOCs. Future work is required to address these concerns specifically.
Liquid-phase VOC profiles of cell cultures
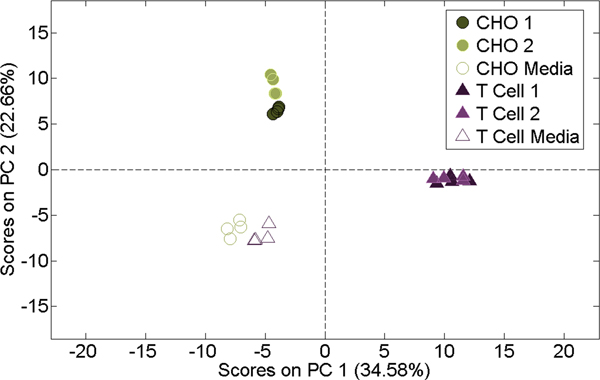
SBSE measurements made directly in bioreactor bags isolated more cellular VOCs from media controls than HSSE measurements of bioreactor gas exhaust. A PCA of these liquid-phase extractions (figure 6) showed clear differences between the two cell types and mediacontrols, which separated between PC 1 and PC 2, explaining a total of 57.24% of the variance.
Figure 6.
PCA of dissolved volatiles in the liquid media from media controls and cell culture inoculated media.
Two PLS–DA analyses were performed that distinguished liquid media controls from respective cell lines. Similar to PLS regression, each variable (in this case, chemical VOC compound) was assigned a VIP score. CHO cells had 72 compounds with a VIP score >1 and T cells had 96 compounds, with 43 over- lapping between cell lines. T cells had 16 compounds with VIP scores >1 in both downstream VOC emis- sion measurements (HSSE) and cell-inoculated liquid measurements (SBSE); there were nine such com- pounds for CHO cells.
The 20 compounds with the highest VIP scores for each cell types were putatively identified (table 3). Not all of these compounds were present in liquid media controls. Compared to HSSE, SBSE extracted more compounds of higher molecular weights. Many com- pounds contained aromatic rings including toluenes, phenols, benzoic acids, benaldehydes, or acet- ophenones. One compound, unknown 10, appears in both tables 2 and 3, having importance only in CHO cells in both HSSE and SBSE measurements.
Table 3.
Based on measurements made directly in cell-inoculated media. Putative identifications of the 20 compounds with the highest VIP scores for the T cell PLS–DA and the 20 compounds with the highest VIP scores for the CHO PLS– DA combined into one table. KI: Kovats index, calculated (Calc) and as reported in the literature (Lit); Major Ions: the top three most abundant ions from respective mass spectra; MS score: score of acquired mass spectrum compared to the NIST mass spectral database.
| VIP score (if >1) | |||||||
|---|---|---|---|---|---|---|---|
|
|
|||||||
| Compound | KI (Calc) | KI (Lit) | CAS # | Major Ions (m/z) | MS score | T cells | CHO |
|
| |||||||
| 2-pentadecanone | 1696 | 1694 | 2345-28-0 | 71.1, 58.1, 113.1 | 77.04 | 1.56 | |
| Unknown 12 | 1553 | 57.1, 179.1, 165.1 | 1.56 | 1.85 | |||
| 3,5-bis(1,1-dimethylethyl)-4-ethyl-1H-pyrazole | 1363 | 125281-21-2 | 193.1, 166.1, 124.0 | 81.65 | 1.56 | 1.85 | |
| Unknown 13 | 2072 | 178.1, 251.1, 223.0 | 1.56 | 1.84 | |||
| 3,5-dimethoxy-4-hydroxytoluene | 1497 | 1447 | 6638-05-7 | 57.1, 153.0, 168.0 | 64.70 | 1.55 | 1.87 |
| Unknown 14 (alkylated phenol) | 1563 | 57.1, 159.1, 191.1 | 1.54 | 1.63 | |||
| 3,4-dimethoxybenzoic acid | 1666 | 1670 | 93-07-2 | 57.1, 182.1, 167.1 | 70.51 | 1.54 | 1.83 |
| Unknown 15 (alcohol) | 1984 | 57.1, 182.1, 167.1 | 1.54 | ||||
| Unknown 16 (ketone) | 2018 | 79.0, 80.0, 150.1 | 1.54 | ||||
| Unknown 17 | 1345 | 45.0, 71.0, 85.1 | 1.54 | 1.85 | |||
| 3,5-bis(1,1-dimethylethyl)-4-methyl-1H-pyrazole | 1586 | 1527 (est) | 18712-47-5 | 179.1, 194.1, 57.0 | 64.21 | 1.54 | 1.74 |
| Unknown 18 | 1858 | 43.0, 79.0, 182.1 | 1.54 | ||||
| Unknown 19 (alcohol) | 1786 | 97.1, 79.0, 43.0 | 1.54 | ||||
| 3,5-di-tert-butyl-4-hydroxybenzaldehyde | 1737 | 1774 | 1620-98-0 | 153.1, 240.1, 149.0 | 78.24 | 1.54 | |
| 1-amino-2-naphthalenol | 1724 | 1764 (est) | 2834-92-6 | 159.0, 130, 201.0 | 69.25 | 1.53 | |
| Butyl citrate | 2111 | 2150 | 77-94-1 | 185.1, 129.0, 259.1 | 97.19 | 1.53 | |
| Undecane | 1100 | 1100 | 1120-21-4 | 57.1, 71.1, 85.1 | 93.71 | 1.53 | |
| γ-dodecalactone | 1674 | 1673 | 2305-05-7 | 85.1, 55.0, 69.1 | 91.82 | 1.53 | 1.84 |
| Unknown 20 (fatty acid derivative) | 2139 | 79.0, 80.0, 91.0 | 1.52 | ||||
| Unknown 21 (benzene dervative) | 1655 | 57.1, 149.1, 164.1 | 1.52 | 1.29 | |||
| 4-methyl-quinazoline | 1329 | 1363 | 700-46-9 | 144.0, 103.0, 129.0 | 87.77 | 1.52 | 1.82 |
| Cholesterol | >2400 | 3075 | 57-88-5 | 93.1, 55.1, 107.1 | 73.74 | 1.51 | 1.75 |
| 3,5-di-tertbutyl-4-hydroxyacetophenone | 1809 | 1903 (est) | 14035-33-7 | 233.1, 248.1, 205.1 | 92.94 | 1.51 | 1.84 |
| Unknown 22 (alkylated ester) | 2091 | 157.0, 112.0, 57.1 | 1.48 | 1.84 | |||
| p-benzoquinone | 1459 | 1458 | 719-22-2 | 177.1, 220.1, 131.0 | 87.31 | 1.33 | 1.77 |
| Sulfurous acid, nonyl 2-propyl ester | 1345 | 1000309-12-0 | 57.0, 71.0, 85.1 | 71.73 | 1.29 | 1.77 | |
| 3,5-di-tertbutyl-4-hydroxybenzaldehyde | 1754 | 1774 | 1620-98-0 | 219.1, 234.1, 191.1 | 78.24 | 1.26 | 1.85 |
| 5-hexyldihydro-2(3H)-furanone | 1463 | 1463 | 706-14-9 | 85.1, 55.0, 69.1 | 94.67 | 1.09 | 1.86 |
| 1-methyl-2(1H)-quinolinone | 1653 | 1669 | 606-43-9 | 130.0, 159.0, 158.0 | 81.69 | 1.86 | |
| Unknown 23 (alkylated acetophenone) | 1624 | 217.1, 232.1, 210.1 | 1.85 | ||||
| Unknown 10 | 1251 | 41.1, 91.0, 65.0 | 1.82 | ||||
| Dihydro-5-pentyl-2(3H)-furanone | 1359 | 1360 | 104-61-0 | 85.0, 43.0, 55.0 | 89.74 | 1.79 | |
| 7,9-di-tert-butyl-1-oxaspiro(4,5)deca-6,9-diene-2,8-dione | 1911 | 1917 | 82304-66-3 | 205.1, 217.1, 57.1 | 96.80 | 1.77 | |
| Methyl ether-β-citronellol | 1588 | 1000333-81-4 | 69.1, 85.1, 67.0 | 70.90 | 1.76 | ||
Some compounds appear related to the mevalo- nate pathway. Important to cell membrane function and steroid synthesis, cholesterol was putatively iden- tified in both CHO and T cell bioreactors. A derivative of citronellol was found in CHO cells, which may be a hydrogenated product of geraniol, a compound involved in cholesterol synthesis pathways [24].
P-benzoquinone could be attributed to exposure to benzene derivatives or as a breakdown product of ubi- quinone. Naphthols such as 1-amino-2-naphthalenol may derive from biomarkers related to exposure to polycyclic aromatic hydrocarbons, such as plasticizers [25]. Heretocyclic compounds such as quinazolines, quinolinones and pyrazoles may have resulted from other steroids.
Measurements of liquid-based VOCs may provide another opportunity for future PAT. Similar to gas exhaust, chemical sensors could be attached to the media waste lines of the bioreactors to monitor target compounds related to cellular health or to perform untargeted analysis to warn users when the waste stream has deviated from a ‘normal’ state. This could help optimize media perfusion rates by monitoring waste and nutrient concentrations within the bioreactor.
Conclusion
We observed a shift in the specific VOC profile of cellular ‘breath’ as cultures expanded over the course of 8 d. These profiles were used to create PLS regression models that could predict cell culture densities. The volatile compounds most relevant to cell culture expansion for CHO and T cells were putatively identified and discussed. Additionally, measurements of VOCs were made directly in cell-inoculated media during the final day of the experiment. Cell-inoculated media samples were rich in VOCs, not present in liquid media controls (no cells present). A PLS–DA analysis revealed the volatile compounds most rele- vant to the cell cultures and were putatively identified and discussed.
Our work supports the idea of using VOC-based detection methods on either gas or liquid waste lines of bioreactors to monitor cell culture health. Methods using VOCs could develop rapid ‘real time’ techniques to optimize cell culture growth, as evidenced by the CHO and T cell lines used in this report. We predict that future work will provide VOC profiles that enable us to rapidly identify early pathogen contamination or nutrient starvation in cell culture systems.
Supplementary Material
Acknowledgments
We are grateful to Canadian Blood Services and donors for providing research samples for the comple- tion of this project. The reporting and interpretation of the research findings are the responsibility of the author(s). The views expressed herein do not necessa- rily represent the views of Canadian Blood Services.
This study was partially supported by: GE Healthcare industry agreement 201700771 (CED, MS, RWH, SEE, NJK); NIH award U01 EB0220003-01 [CED, NJK]; the NIH National Center for Advancing Translational Sciences (NCATS) through grant UL1 TR000002 [CED, NJK]; NIH award 1P30ES023513-01A1 [CED, NJK]; NIH award UG3-OD023365 [CED, NJK]; and NIH award T32 HL007013 [MSY]. The contents of this manuscript are solely the responsibility of the authors and do not necessarily represent the official views of the funding agencies.
Footnotes
Supplementary material for this article is available online
References
- [1].Bachinger T, Riese U, Eriksson R and Mandenius CF 2000. Monitoring cellular state transitions in a production-scale CHO-cell process using an electronic nose J. Biotechnol 76 61–71 [DOI] [PubMed] [Google Scholar]
- [2].Bachinger T, Martensson P and Mandenius CF 1998. Estimation of biomass and specific growth rate in a recombinant Escherichia coli batch cultivation process using a chemical multisensor array J. Biotechnol 60 55–66 [DOI] [PubMed] [Google Scholar]
- [3].Kreij K et al. 2005. On-line detection of microbial contaminations in animal cell reactor cultures using an electronic nose device Cytotechnology 48 41–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Aksenov AA, Gojova A, Zhao W, Morgan JT, Sankaran S, Sandrock CE and Davis CE 2012. Characterization of volatile organic compounds in human leukocyte antigen heterologous expression systems: a cell’s ‘Chemical Odor Fingerprint’ ChemBioChem 13 1053–9 [DOI] [PubMed] [Google Scholar]
- [5].Ray A, Bristow T, Whitmore C and Mosely J 2018. On-line reaction monitoring by mass spectrometry, modern approaches for the analysis of chemical reactions Mass Spectrom. Rev. 37 565–79 [DOI] [PubMed] [Google Scholar]
- [6].Luchner M, Gutmann R, Bayer K, Dunkl J, Hansel A, Herbig J, Singer W, Strobl F, Winkler K and Striedner G 2012. Implementation of proton transfer reaction-mass spectrometry (PTR-MS) for advanced bioprocess monitoring Biotechnol. Bioeng. 109 3059–69 [DOI] [PubMed] [Google Scholar]
- [7].Oeggerli A and Heinzle E 1992. On-line analysis of volatiles in fermenter exhaust gas using mass spectrometry IFAC Symp. Series 1992 295–8 [Google Scholar]
- [8].Bischoff AC, Oertel P, Sukul P, Rimmbach C, David R, Schubert J and Miekisch W 2018. Smell of cells: volatile profiling of stem- and non-stem cell proliferation J. Breath Res12 026014 [DOI] [PubMed] [Google Scholar]
- [9].Klemenz A-C, Meyer J, Ekat K, Bartels J, Traxler S, Schubert JK, Kamp G, Miekisch W and Peters K 2019. Differences in the emission of volatile organic compounds (VOCs) between non-differentiating and adipogenically differentiating mesenchymal stromal/stem cells from human adipose tissue Cells 8 697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Kuntzel A, Fischer S, Bergmann A, Oertel P, Steffens M, Trefz P, Miekisch W, Schubert JK, Reinhold P and Kohler H 2016. Effects of biological and methodological factors on volatile organic compound patterns during cultural growth of Mycobacterium avium ssp paratuberculosis J. Breath Res 10 037103 [DOI] [PubMed] [Google Scholar]
- [11].Filipiak W, Sponring A, Filipiak A, Ager C, Schubert J, Miekisch W, Amann A and Troppmair J 2010. TD-GC-MS analysis of volatile metabolites of human lung cancer and normal cells in vitro Cancer Epidemiology Biomarkers 19 182–95 [DOI] [PubMed] [Google Scholar]
- [12].Schivo M, Aksenov AA, Linderholm AL, McCartney MM, Simmons J, Harper RW and Davis CE 2014. Volatile emanations from in vitro airway cells infected with human rhinovirus J. Breath Res 8 037110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].McCartney MM, Zrodnikov Y, Fung AG, LeVasseur MK, Pedersen JM, Zamuruyev KO, Aksenov AA, Kenyon NJ and Davis CE 2017. An easy to manufacture micro gas preconcentrator for chemical sensing applications ACS Sensors 2 1167–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Strand N, Bhushan A, Schivo M, Kenyon NJ and Davis CE 2010. Chemically polymerized polypyrrole for on-chip concentration of volatile breath metabolites Sensors ActuatorsB 143 516–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Garga A, Akbar M, Vejerano E, Narayanan S, Nazhandali L, Marr LC and Agah M 2015. Zebra GC: a mini gas chromatography system for trace-level determination of hazardous air pollutants Sensors Actuators B 212 145–54 [Google Scholar]
- [16].Terry SC, Jerman JH and Angell JB 1979 Gas-chromatographic air analyzer fabricated on a silicon-wafer IEEE Trans. Electron Devices 26 1880–6 [Google Scholar]
- [17].Miller RA, Nazarov EG, Eiceman GA and King AT 2001. A MEMS radio-frequency ion mobility spectrometer for chemical vapor detection Sensors Actuators A 91 301–12 [Google Scholar]
- [18].Hichwa P and Davis CE 2018. Modern application potential of miniature chemical sensors Sensors for Diagnostics and Monitoring ed Yallup K, Basiricò L and Iniewski K (New York: CRC Press; ) ( 10.1201/9781351250092) [DOI] [Google Scholar]
- [19].Wapelhorst E, Hauschild JP and Muller J 2007. Complex MEMS: a fully integrated TOF micro mass spectrometer Sensors Actuators A 138 22–7 [Google Scholar]
- [20].Purcaro G, Rees CA, Wieland-Alter WF, Schneider MJ, Wang X, Stefanuto PH, Wright PF, Enelow RI and Hill JE 2018. Volatile fingerprinting of human respiratory viruses from cell culture J. Breath Res 12 026015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Yamaguchi MS, McCartney MM, Linderholm AL, Ebeler SE, Schivo M and Davis CE 2018. Headspace sorptive extraction-gas chromatography-mass spectrometry method to measure volatile emissions from human airway cell cultures J. Chromatography B 1090 36–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Giorgia P, Christiaan AR, Wendy FW-A, Mark JS, Xi W, Pierre-Hugues S, Peter FW, Richard IE and Jane EH 2018. Volatile fingerprinting of human respiratory viruses from cell culture J. Breath Res 12 026015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Aksenov AA, Sandrock CE, Zhao WX, Sankaran S, Schivo M, Harper R, Cardona CJ, Xing Z and Davis CE 2014. Cellular scent of influenza virus infection ChemBioChem 15 1040–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Grunler J, Ericsson J and Dallner G 1994. Branch-point reactions in the biosynthesis of cholesterol, dolichol, ubiquinone and prenylated proteins Bba-Lipid Lipid Met. 1212 259–77 [DOI] [PubMed] [Google Scholar]
- [25].Preuss R, Angerer J and Drexler H 2003. Naphthalene—an environmental and occupational toxicant Int. Arch. Occup. Environ. Health 76 556–76 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.