Abstract
Extracellular matrix (ECM) interactions regulate both the cell transcriptome and proteome, thereby determining cell fate. Traumatic heterotopic ossification (HO) is a disorder characterized by aberrant mesenchymal lineage (MLin) cell differentiation, forming bone within soft tissues of the musculoskeletal system following traumatic injury. Recent work has shown that HO is influenced by ECM-MLin cell receptor signaling, but how ECM binding affects cellular outcomes remains unclear. Using time course transcriptomic and proteomic analyses, we identified discoidin domain receptor 2 (DDR2), a cell surface receptor for fibrillar collagen, as a key MLin cell regulator in HO formation. Inhibition of DDR2 signaling, through either constitutive or conditional Ddr2 deletion or pharmaceutical inhibition, reduced HO formation in mice. Mechanistically, DDR2 perturbation alters focal adhesion orientation and subsequent matrix organization, modulating Focal Adhesion Kinase (FAK) and Yes1 Associated Transcriptional Regulator and WW Domain Containing Transcription Regulator 1 (YAP/TAZ)–mediated MLin cell signaling. Hence, ECM-DDR2 interactions are critical in driving HO and could serve as a previously unknown therapeutic target for treating this disease process.
Discoidin Domain Receptor 2 promotes extracellular matrix organization and pathogenic bone formation in tendons following trauma.
INTRODUCTION
Mesenchymal lineage (MLin) cells receive mechanical cues from their local environment (1–3) and differentiate along a variety of lineages, including osteoblasts, chondrocytes, and adipocytes. The interactions between MLin cells and the extracellular matrix (ECM) are known to affect cell proliferation (4), migration (5), and cell fate determination (2, 4). Heterotopic ossification (HO) is a pathologic condition that occurs after traumatic injuries where MLin cell–ECM interactions have gone awry. HO is characterized by aberrant osteochondral differentiation of MLin cells, resulting in the formation of ectopic cartilage and bone in extraskeletal soft tissues, including tendon or muscle (6). In a previous study, we discovered that immobilization of the affected limb inhibits trauma-related extremity HO because of changes in the collagen-rich ECM architecture of the injury site (7). However, in the context of musculoskeletal trauma, the mechanisms by which MLin cells interact with the surrounding ECM and how this interaction alters MLin cell transcriptional changes or ECM collagen architecture, thereby leading to aberrant differentiation, remain undefined.
Here, using a series of unbiased transcriptomic and proteomic tools, we have identified the collagen-binding discoidin domain receptor 2 (DDR2) as a required driver of HO formation and progression. DDRs (i.e., DDR1 and DDR2) are a class of collagen-binding, cell surface receptors that differ from integrins because of their intrinsic tyrosine kinase activity and their selective affinity for collagen subtypes (8–10). These receptors exhibit stratification by stem-like potential; DDR1 is primarily expressed in epithelial cells, whereas DDR2 is more specific to MLin cells (11). DDR2 has been previously shown to be critical in bone development (12), cancer metastasis (13), mechanotransductive signaling (14), and ECM remodeling (15). Nevertheless, the role played by DDR2 signaling in MLin cells at sites of musculoskeletal injury on mechanotransductive signaling, cell fate determination, and changes in ECM collagen deposition and alignment has yet to be defined.
On the basis of our identification of MLin cell DDR2 in HO lesions, we launched a series of studies designed to interrogate its role in regulating musculoskeletal MLin cell fate and ECM architecture after injury through single-cell transcriptomics, epigenomics, and in vivo and in vitro modulation of DDR2 signaling. We find that Ddr2 deletion decreases MLin cell osteogenic differentiation, HO formation, and collagen alignment in a focal adhesion kinase (FAK)/YAP/TAZ–dependent pathway. Together, these studies uncover the central role of DDR2 on MLin cell differentiation leading to HO formation and provide a previously unidentified therapeutic target for potential intervention.
RESULTS
Bone matrix–forming MLin cells express Ddr2
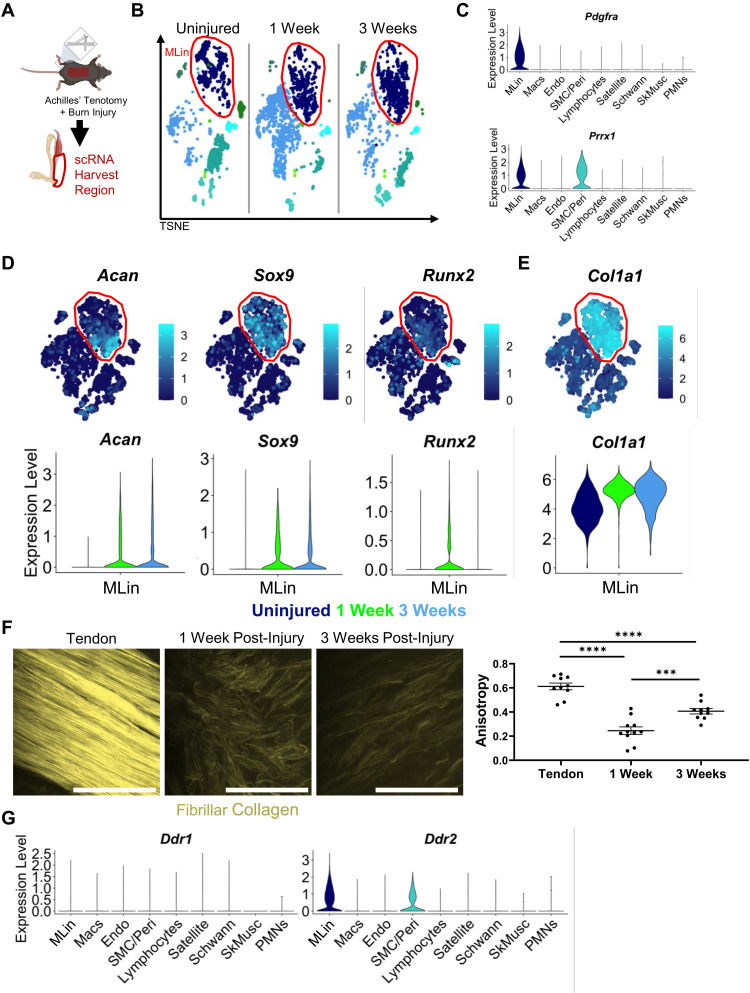
To define transcriptional changes occurring within HO injury sites, mice were injured using a well-characterized burn/tenotomy (BT) injury model of HO, in which mice undergo a 30% total body surface area dorsal burn and full Achilles tendon transection without repair (Fig. 1A) (16, 17). Cells were harvested at three time points (uninjured, 1 week, and 3 weeks after injury) from the tendon injury site for single-cell RNA (scRNA) sequencing analysis (Fig. 1, A and B). Various cell populations were characterized within the harvested cell population by differential gene expression analysis, including MLin cells, macrophages (Macs), vascular endothelial cells (Endos), vascular smooth muscle cells and pericytes (SMCs/Peri), lymphocytes, muscle satellite cells (Satellite), Schwann cells (Schwann), skeletal muscle fibers (SkMusc), and polymorphonuclear leukocytes (PMNs; Fig. 1B and fig. S1, A to I). The identification of these cell populations were consistent with previous scRNA analyses of HO injury sites (18) with MLin cells characterized by their expression of Pdgfrα and Prrx1 (Fig. 1C and fig. S1A). While MLin cells range from multipotent stem cells to more differentiated tenocytes, chondrocytes, etc., reclustering still showed that Pdgfrα and Prrx1 expression was abundant regardless of differentiated state (fig. S2, A to F). Following injury, MLin cells displayed, as expected (17, 19, 20), increased expression of the chondrogenic differentiation markers, Acan (21) and Sox9 (22, 23), as well as the early osteogenic marker, Runx2 (Fig. 1D and fig. S2F) (24). MLin cells also expressed Tppp3, a marker of tendon stem cells (fig. S2E) (25). To determine whether these changes in fate commitment correlated with changes in collagen expression, a hallmark of HO injury sites (19, 26), we next examined fibrillar collagen gene expression and noted a marked increase in Col1a1 expression (Fig. 1E). In addition, Col1a1 and Col1a2 scored second and third, respectively, by log fold change in differentially expressed genes in MLin cells compared to other clusters (table S1). Next, to characterize collagen deposition following injury, we used second harmonic generation (SHG) imaging in uninjured tendons and tendons 1 and 3 weeks following BT (Fig. 1F). Within the injury site 1 week after BT, collagen was highly disorganized (reflected by a low level of fiber anisotropy), with fibrils becoming increasingly aligned at 3 weeks, although still significantly disorganized relative to uninjured controls (Fig. 1F). These findings highlight a progression of MLin cells down an osteochondral lineage with a concomitant increase in collagen deposition and organization following tendon injury.
Fig. 1. MLin cells express fibrillar collagen and collagen binding protein DDR2 after injury.
(A) Schematic of BT injury and single-cell harvest region. (B) TSNE plot of scRNA sequencing of cells harvested from uninjured and 1 and 3 weeks following injury highlighting MLin population in red. (C) Violin plot of relative expression of genes encoding MLin markers Pdgfra and Prrx1. (D) TSNE and violin plots of relative expression of chondrogenic genes (Acan and Sox9), osteogenic gene Runx2, and (E) Col1a1. (F) Second harmonic generation (SHG) imaging of fibrillar collagen in uninjured tendon and at the injury site 1 and 3 weeks following injury. Graph showing anisotropy quantification of collagen ECM (n = 3 mice per group, one to two images per mouse, bars are means ± SEM). Scale bars, 100 μm. (G) Violin plots of relative expression of DDR1 and DDR2 genes Ddr1 and Ddr2. Plots include all time points.
To analyze potential interactions between MLin cells and their surrounding collagen matrix, we also examined expression of DDR proteins 1 and 2 (Ddr1 and Ddr2; Fig. 1G). Ddr1 showed minimal expression before or after injury by scRNA sequencing, while Ddr2 was highly expressed in both MLin and SMC/Peri populations, raising the possibility that it plays an active role in HO.
Collagen receptor DDR2 is present and activated in MLin cells upon tendon injury
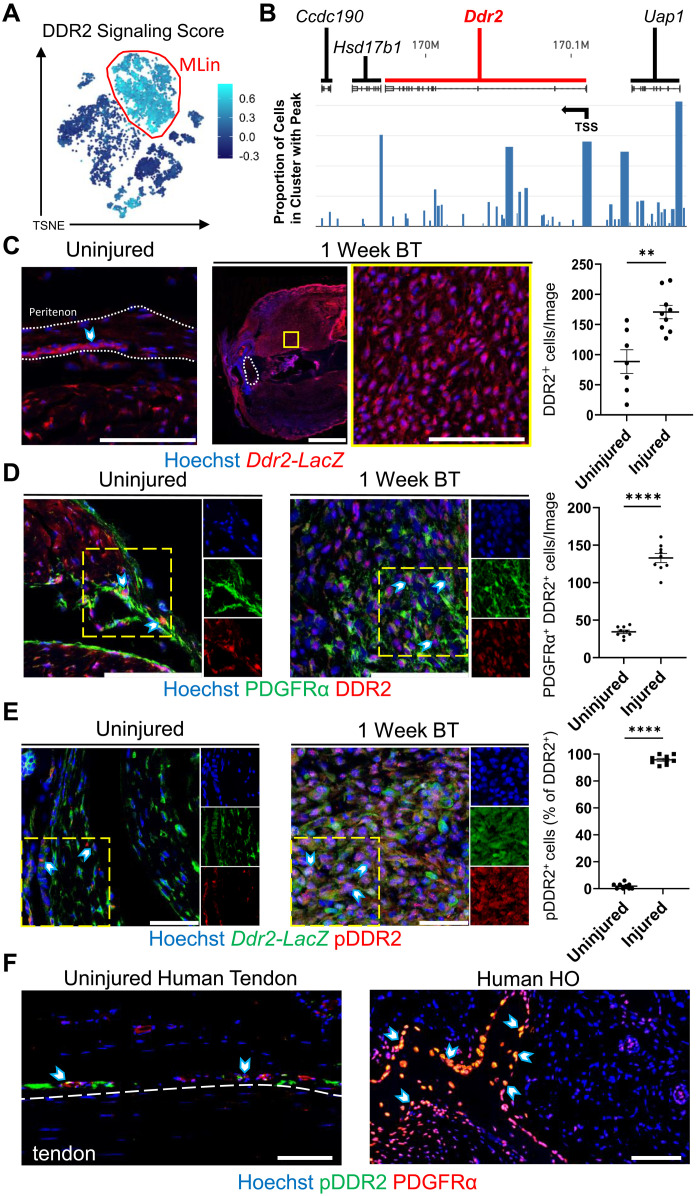
To further elucidate the role of DDR2 in MLin cell differentiation, a list of genes whose expression has been shown to be linked to DDR2 activation was compiled to create a DDR2 signaling score using the AddModuleScore function of Seurat (Fig. 2A and table S2) (13, 27–29). MLin cells showed the highest scores of all cell clusters, suggesting that DDR2 signaling is activated preferentially within this cell population. Single-nucleus assay for transposase-accessible chromatin (snATAC) was next used to view open chromatin regions within the transcriptional initiation and coding regions of Ddr2 and nearby genes (Ccdc190, Hsd17b1, and Uap1) in MLin cells at 1 week after injury. A more open chromatin structure that is easily accessible to transcriptional machinery was detected in MLin cells before and after the Ddr2 transcription start site (TSS), supporting active generation of de novo transcripts (Fig. 2B).
Fig. 2. Ddr2 expression is up-regulated and DDR2 activation increases in MLin cells after injury.
(A) TSNE plot visualizing DDR2 signaling score generated from a list of genes shown to be influenced by Ddr2 activation. (B) snATAC sequencing visualization of Ddr2 and surrounding gene tracks showing the proportion of reads in Pdgfrα-expressing MLin cells 1 week after BT. Arrow labels TSS. (C) Representative confocal images (63×) of uninjured tendon and injury site 1 week after BT in Ddr2-LacZ reporter mice with quantification of the number of β-galactosidase+ cells per image (n = 3 mice per group, n = 2 to 3 images per mouse). The yellow box shows inset region. The region within white outlines is the Achilles tendon. Scale bars on 63× and tile images are 100 and 1000 μm, respectively. (D) Representative confocal images (63×) of uninjured tendon and injury site 1 week after BT in wild-type C57B6 mice with quantification of PDGFRα+ and DDR2+ cells per image (n = 3 mice per group, n = 3 images per mouse). Scale bars, 100 μm. (E) Representative confocal images (63×) of uninjured tendon and injury site 1 week after BT in Ddr2-LacZ mice with quantification of the percentage of β-galactosidase+ cells with pDDR2+ staining per image (n = 3 mice per group, n = 3 images per mouse). Scale bars, 50 μm. (F) Representative confocal images (20×) of uninjured human tendon and mature human HO tissue stained with Hoechst, anti-PDGFRα, and anti-phosphorylated DDR2 (pDDR2). Arrows show PDGFRα+ pDDR2+ cells in peritenon adjacent to uninjured tendon and within and on mature HO tissue. Scale bars, 100 μm. All graphs show means ± SEM. All individual channel images are from the areas within yellow dashes.
To further examine DDR2, we leveraged immunofluorescent imaging in our HO mouse model using a Ddr2-LacZ reporter wherein the LacZ gene is inserted into the Ddr2 gene locus. DDR2-positive cells were found within the tendon, peritenon, and soft tissue areas surrounding the tendon in uninjured and injured mice (Fig. 2C). We confirmed that there was an increase in the number of DDR2-positive MLin cells at the injury site 1 week following injury by costaining MLin cells for platelet-derived growth factor receptor–α (PDGFRα; Fig. 2D). To further examine whether DDR2 is activated following trauma, phosphorylated/ligand-activated DDR2 (pDDR2) was visualized by immunofluorescent labeling (Fig. 2E). In Ddr2-LacZ mice, the percentage of cells positive for pDDR2 increased significantly 1 week following injury relative to uninjured tissue (Fig. 2E). To validate these findings in humans, we next examined patient-derived uninjured tendon and HO tissue. In human samples, we detected high expression of DDR2 by PDGFRα-expressing MLin cells within and along the ectopic bone surface of patients with HO in contrast to uninjured tendon where DDR2 staining was abundant in the peritenon region (fig. S3, A and B). Similarly, human HO tissue samples also showed increased labeling for pDDR2 as compared to cells within uninjured tendon (Fig. 2F). Immunofluorescent labeling , imaging, and analysis of wild-type mice 1 week following BT revealed increased pDDR2 staining in MLin cells closer to the proximal Achilles-gastrocnemius myotendinous junction compared to the distal Achilles-calcaneus enthesis (fig. S4, A and B). Together, these results indicate that DDR2 is expressed and activated in MLin cells following musculoskeletal trauma in both mice and human tissues.
DDR2 regulates HO formation following injury
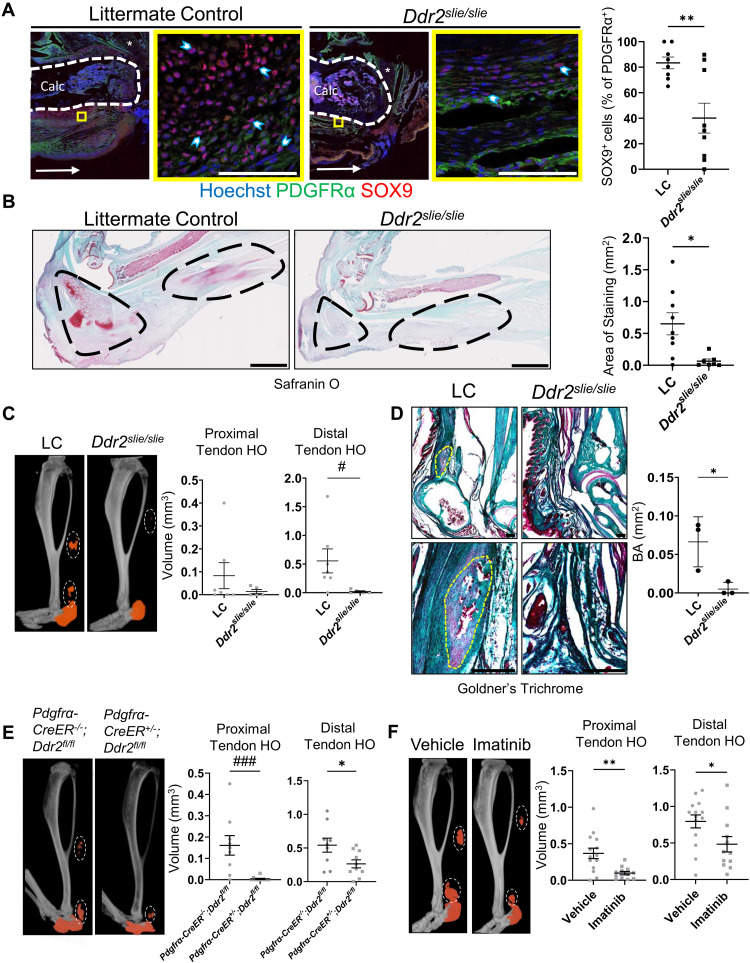
To assess a requirement for DDR2 in HO-induced MLin cell differentiation, mice with a global deletion of Ddr2 (Ddr2slie/slie) underwent BT injury and were compared to littermate controls (LCs). Because HO formation progresses through endochondral ossification, we first assessed ectopic chondrogenesis. Using immunofluorescent imaging, we observed a 51.88% decrease in the average percentage of PDGFRα-expressing MLin cells positively labeled for the chondrogenic marker, SOX9, 1 week following injury in Ddr2slie/slie mice relative to LCs (Fig. 3A). This reduced chondrogenesis was confirmed 3 weeks after injury using Safranin O staining with Ddr2slie/slie mice displaying significant reductions in proteoglycan staining (Fig. 3B). Next, to assess osteogenesis, we leveraged micro–computed tomography (μCT) imaging. Our BT model produces HO in four distinct anatomical regions: calcaneus, tibia, distal residual Achilles tendon, and proximal residual Achilles tendon. We first performed μCT analysis on the injured hindlimbs from Ddr2slie/slie mice and found a significant reduction in HO formation in the distal residual Achilles tendon and a downward trend in the proximal residual tendon 9 weeks following injury (Fig. 3C). We did not find a significant difference in tibial and calcaneal HO (fig. S5A). To confirm our μCT findings of HO formation, we next performed Goldners’ trichrome staining on Ddr2slie/slie and LC mice to visualize the mineralized bone matrix and found that Ddr2slie/slie mice had significantly less bone matrix 9 weeks after injury (Fig. 3D). Given that this model incorporated a germline deletion of Ddr2, we also analyzed tibial length and cortical bone thickness, but these parameters only showed minor differences between Ddr2slie/slie mice and LCs (fig. S5B).
Fig. 3. DDR2 deletion or inhibition prevents osteogenesis.
(A) Representative confocal images (63×) of LCs and Ddr2slie/slie mice 1 week after BT with quantification of the percentage of PDGFRα+ cells expressing SOX9 per image (n = 3 mice per group, n = 2 to 3 images per mouse). Scale bars, 100 μm. Asterisk marks Achilles tendon. Horizontal arrow points toward the dorsal part of the calcaneus. (B) Safranin O staining and quantifications of positive staining in the injured hindlimbs of LCs and Ddr2slie/slie mice 3 weeks after BT (n = 3 mice per group, n = 2 to 3 images per mouse). Areas surrounding calcaneus and Achilles tendon are circled. Scale bars, 2000 μm. (C) μCT imaging of injured hindlimb from LC and Ddr2slie/slie mice (n = 5 to 7 mice per group) quantifying HO bone volume in the proximal and distal residual Achilles tendon 9 weeks after BT. (D) Goldner’s trichome stain for bone area in tendon of LC and Ddr2slie/slie sections 9 weeks after BT (n = 3 mice per group). Scale bars, 200 μm. (E) μCT imaging of injured hindlimb from Pdgfrα-CreER−/−;Ddr2fl/fl and Pdgfrα-CreER+/−;Ddr2fl/fl mice (n = 9 to 11 mice per group) quantifying HO bone volume in the proximal and distal residual Achilles tendon 9 weeks after BT. (F) μCT imaging of injured hindlimb from wild-type mice treated with vehicle or imatinib quantifying HO bone volume in the proximal and distal residual Achilles tendon 9 weeks after BT (n = 12 to 14 mice per group). In μCT images, orange labels ectopic bone and tendon HO is circled in each image. All graphs show means ± SEM.
Nevertheless, to target Ddr2 in an MLin-specific fashion, we crossed mice carrying Ddr2 floxed alleles with animals expressing a tamoxifen-inducible Cre driver Pdgfrα-CreER (Pdgfrα-CreER;Ddr2fl/fl). We confirmed that Pdgfrα MLin cells express chondrogenic (SOX9) and osteogenic (OPN, RUNX2, and OSX) differentiation markers by immunofluorescent analysis in Pdgfrα-CreER;ROSA-TdTomato mice 1 week following injury (fig. S6A). These mice allow for tamoxifen-induced labeling and lineage tracing before injury as observed by a high percentage of TdTomato-expressing MLin cells following cell isolation from the injury site (fig. S6B). We also confirmed effective Ddr2 deletion in MLin cells by quantitative polymerase chain reaction (qPCR) analysis (fig. S6C). As such, we used μCT analysis to quantify HO formation in the Achilles tendon of Ddr2-floxed mice after following injury. Similar to results obtained with the Ddr2slie/slie model, MLin-specific deletion of DDR2 resulted in a significant reduction in both proximal and distal residual tendon HO formation (Fig. 3E) and no difference in tibial and calcaneal HO (fig. S7A) without affecting tibial length or cortical thickness (fig. S7B). Together, these data demonstrate that Ddr2 deletion mitigates aberrant cartilage and bone formation in tendon tissues in vivo following injury.
To determine whether DDR2 targeting could serve as a therapeutic strategy, studies were initiated to inhibit DDR2 activity via pharmacologic intervention immediately following injury. Imatinib is a tyrosine kinase inhibitor that has been demonstrated to inhibit DDR2 phosphorylation and activation (30). In C57B6 mice treated with imatinib daily for 16 days immediately following injury, there was a significant reduction in proximal and distal residual tendon as well as calcaneal HO formation following injury (Fig. 3F and fig. S7C). Under these conditions, neither tibial length nor cortical thickness was affected (fig. S7D). These results indicate that pharmacologic inhibition of DDR2 is an effective strategy for ameliorating HO formation in vivo.
In summary, we observed a reduction in chondrogenesis upon DDR2 deletion. Ectopic bone formation within the proximal and distal residual Achilles tendon—regions that showed increased DDR2 activation (fig. S4, A and B)—was reduced upon deletion or pharmacological inhibition of DDR2.
DDR2 influences ECM alignment
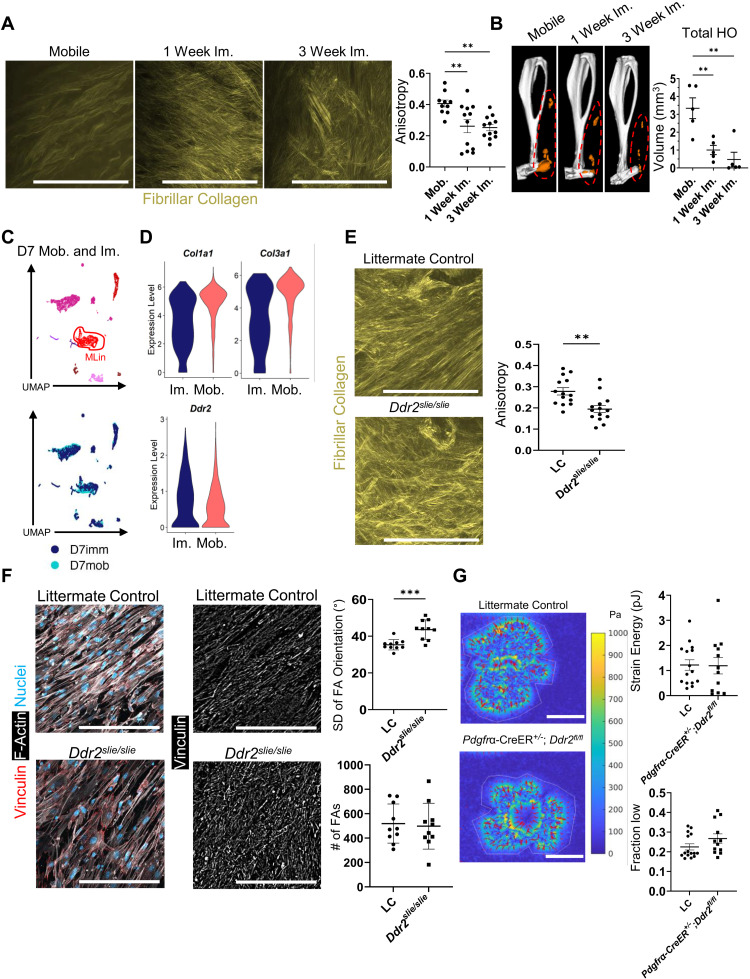
Immobilization of an injured joint has been shown to decrease HO by altering collagen matrix alignment (7). As DDR2 has been reported to play a role in matrix alignment in the tumor microenvironment (14), we sought to determine whether immobilization-dependent alterations in collagen alignment affected DDR2 expression. Mice were subjected to BT injury and either allowed to ambulate freely (mobile) or immobilized for 1 week (1 week im.) and 3 weeks (3 weeks im.) after injury. Tissues were then harvested for histologic analyses 3 weeks after injury. Collagen fibrils of mice following 1 or 3 weeks of immobilization were less anisotropic and more disorganized than those harvested from mobile mice (Fig. 4A). In contrast to that observed in mobilized mice (Fig. 1F), collagen matrix alignment in immobilized mice did not return to baseline and remained disorganized when comparing tissues harvested 1 and 3 weeks after BT injury. One week of immobilization was also sufficient to almost completely mitigate HO (Fig. 4B) without effecting cortical thickness (fig. S8A). Differential gene expression between mobilized and immobilized tissues indicated that MLin cells in mobilized mice expressed significantly higher amounts of Col1a1 and Col3a1 compared to immobilized mice with no differences detected in Ddr2 expression (Fig. 4, C and D). To confirm whether DDR2 could alter matrix alignment independent of joint immobilization, we next performed BT injury in Ddr2slie/slie and LC mice that could ambulate ad libitum, normally creating an aligned matrix (7). Ddr2slie/slie mice (which, at baseline, have matrix alignment greater than LC; fig. S8B) displayed a decrease in ECM alignment 9 weeks following injury despite normal mobilization (Fig. 4E) with matrix misalignment similar to that found in immobilized mice (Fig. 4A).
Fig. 4. DDR2 deletion contributes to a disorganized ECM following injury.
(A) SHG imaging of fibrillar collagen at the injury site of wild-type mice—mobilized or immobilized (1 and 3 weeks)—3 weeks after BT. Anisotropy quantification of collagen ECM (n = 3 mice per group, n = 3 to 5 images per mouse). (B) μCT imaging of mobilized or immobilized (1 and 3 weeks) injured hindlimb from wild-type mice 9 weeks after BT (n = 5 mice per group). Orange labels ectopic bone. (C) UMAP dimensional reduction and (D) violin plots showing relative expression of Col1a1, Col3a1, and Ddr2 from scRNA sequencing of cells harvested from the injury site 1 week after BT with and without limb immobilization (n = 1 mouse per group) [Col1a1: average Log(FC) = 0.63, Padj. = 2.88 × 10−100, Col3a1: average Log(FC) = 0.65, Padj. = 4.94 × 10−107, Ddr2: no significant difference]. (E) SHG imaging of fibrillar collagen at the injury site 1 week after BT in LC and Ddr2slie/slie mice (n = 3 mice per group, n = 4 to 5 images per mouse). (F) Representative confocal images (40×) of MLin cells plated on aligned collagen-coated fibrils from LC and Ddr2slie/slie mice and stained with anti-vinculin, phalloidin for F-actin, and Hoechst. Graphs showing number of FAs and SD of FA orientation (n = 10 images per group). (G) Traction force microscopy heatmaps of LC and Pdgfrα-CreER+/−;Ddr2fl/fl MLin cells on 8-kPa type-I collagen–coated substrates. Graphs quantifying strain energy per cell (n = 12 to 15 cells per group) and fraction of cell footprint with low traction, defined as the fraction of region of interest (ROI) area with traction magnitude less than or equal to half of the mean traction magnitude within the ROI area (n = 12 to 15 cells per group; P = 0.0832 by Mann-Whitney U test). All scale bars, 100 μm. All graphs show means ± SEM.
In considering the mechanisms by which MLin cells might regulate ECM alignment, we focused on focal adhesions (FA), large multicomponent protein complexes that form attachments to ECM components, resulting in mechanotransductive signaling, cytoskeletal rearrangement, and force transmission to the ECM (31, 32). To assess the role of FAs in the context of DDR2-dependent matrix alignment, we isolated MLin cells from Ddr2slie/slie and LC mice 1 week following BT and plated them on matrices of aligned collagen-functionalized fibers that mimic the geometry and mechanics of native collagen fibers (33). While we did not observe a difference in the number of FAs on the MLin cells of Ddr2slie/slie and LC mice (Fig. 4F), a significant increase in the deviation of FA orientation in Ddr2slie/slie cells was noted (Fig. 4F), demonstrating that FAs are more disorganized in the absence of DDR2 signaling. Thus, we hypothesized that MLin cells exert force on their surrounding collagen matrix in an uncoordinated fashion based on their FA orientation, thereby leading to differences in collagen anisotropy.
To evaluate whether DDR2 regulates the degree to which MLin cells exert force on their surrounding matrix, we performed traction force microscopy (TFM) on MLin cells isolated from the injury site of control versus Pdgfrα-CreER+/−;Ddr2fl/fl mice 1 week following injury (Fig. 4G). In TFM, cells are seeded onto an elastic substrate and apply force to a gel with spring-like properties. When the cells apply force to the gel, spring energy is generated and cellular traction forces can be measured by the movement of immunofluorescent beads within the gel based on substrate deformation. Using 8-kPa substrates coated with type I collagen–coated gel, we found that MLin cells recovered from wound sites exert force on the collagen substrate (median strain energy ~1pJ), but Ddr2 deletion did not clearly affect overall contractility (Fig. 4G). Although total force generation was similar, it remained possible that subtle changes in force organization of Pdgfrα-CreER+/−;Ddr2fl/fl MLin cells may explain differences in matrix alignment. Consistent with this proposition, we identified a trend toward an increased fraction of the cell footprint with low contractility in Pdgfrα-CreER+/;Ddr2fl/fl cells compared to LC (Fig. 4G). Hence, while targeting DDR2 did not affect the number of FAs or the cells’ ability to apply pulling forces to the interacting matrix, a lack of coordinated pulling likely contributes to the lack of collagen alignment following injury.
DDR2 regulates YAP/TAZ transcriptional activity
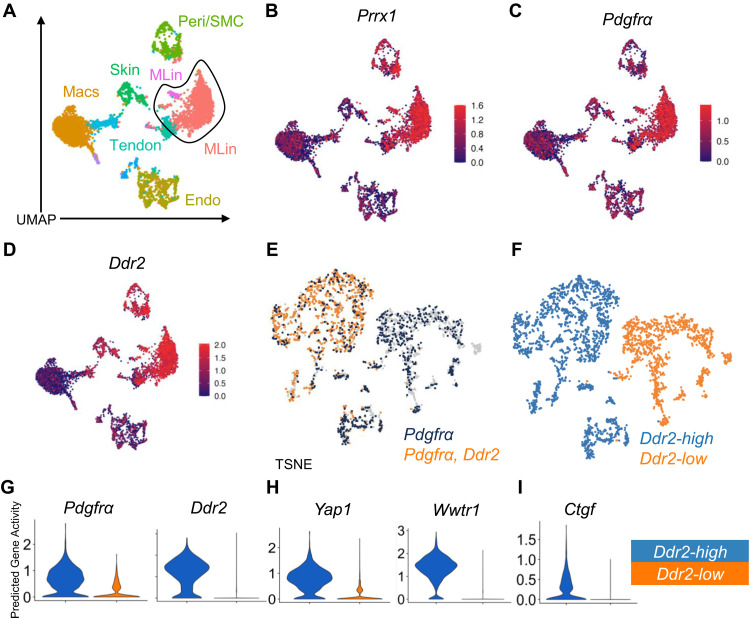
In earlier work, we demonstrated that alterations in collagen alignment and FAs in MLin cells lead to changes in downstream FAK and YAP/TAZ signaling (7). To analyze the downstream mechanisms responsible for the altered collagen alignment found in Ddr2 deleted mice, we queried the status of the FAK/YAP/TAZ axis. First, we used snATAC sequencing of uninjured wild-type mice, 1 week following injury, and 6 weeks following injury to determine predictive genes associated with the activity of the FAK/YAP/TAZ axis (fig. S9, A to L). We identified 11 previously described unique clusters (Fig. 5A) from the injury site (7) and observed that cells in the previously identified MLin clusters, defined by Prrx1 and Pdgfrα predictive gene activity (Fig. 5, B and C), showed increased Ddr2 predictive gene activity (Fig. 5D). This supports our previous findings that Ddr2 is being uniquely transcribed and expressed in MLin cells. Reanalysis of this MLin population from the 1-week post-injury time point revealed two distinct populations of cells with divergent expression of Ddr2 open chromatin reads (Fig. 5E), henceforth referred to as Ddr2-high and Ddr2-low (Fig. 5F). As expected, the Ddr2-high population was confirmed as expressing higher levels of Pdgfrα and Ddr2 gene activity (Fig. 5G). Next, we assessed gene activity for Yap1 (YAP) and Wwtr1 (TAZ) where we found that both genes expressed higher activity in Ddr2-high MLin cells, supporting the potential importance of the YAP/TAZ axis (Fig. 5H). Last, we assessed Ctgf, a downstream target of the YAP/TAZ axis, and confirmed higher activity in the Ddr2-high MLin population (Fig. 5I). Together, these data support a model wherein Ddr2-high MLin cells have actively engaged the YAP/TAZ network.
Fig. 5. snATAC analysis shows that the FAK/YAP/TAZ axis is up-regulated following injury in Ddr2-expressing MLin cells.
(A) UMAP plot showing identified cell clusters from snATAC analysis from cells taken from the injury site taken from days 0, 7, and 42 after BT. (B to D) UMAP plots showing predictive gene activity of Prrx1, Pdgfrα, and Ddr2. (E) TSNE plot visualizing coexpression of at least one open chromatin read for Ddr2 and Pdgfrα (orange) overlaid on cells with Pdgfrα open chromatin (navy blue). (F) TSNE plot showing labeling Ddr2-high and -low clusters. (G to I) Violin plots showing predictive gene activity of Pdgfrα, Ddr2, Yap1, Wwtr1, and Ctgf in Ddr2-high and -low clusters.
While snATAC analysis supports the likelihood of active transcription, we next turned to scRNA analysis to assess transcriptional levels of genes involved in the YAP/TAZ axis. Using an analysis comparing the likelihood that Ptk2, Yap1, or Wwtr1 is coexpressed with Ddr2 in Pdgfrα+ cells relative to two randomly selected genes in 1000 replicate runs, we find that Ddr2 is coexpressed with Ptk2, Wwtr1, and Yap1 in 94.8, 95.6, and 99.5%, respectively, of these iterations (fig. S10).
To examine how Ddr2 targeting changes transcriptional patterning during HO, scRNA analysis was performed on tissues harvested from the injury site 1 week following BT of Ddr2slie/slie and LC mice. We identified 10 unique clusters (fig. S11, A to D) and identified one MLin cluster (cluster 8) based on the expression of Pdgfrα and Prrx1 (fig. S11E) with Ddr2 deletion confirmed in Ddr2slie/slie mice (fig. S11F). The contribution of cluster 8 to HO was confirmed by its high expression of chondrogenic marker genes, Acan and Sox9 (fig. S11G). Focusing on cluster 8, we find a decrease in Ptk2 and Wwrt1 expression in Ddr2-deleted mice (fig. S11H), further supporting the conclusion that Ddr2 regulates the FAK/YAP/TAZ axis at the transcriptional level. Last, to assess downstream signaling in the YAP/TAZ axis, we queried Ctgf and Cyr61 expression and likewise found a decrease in MLin cells harvested from Ddr2-deleted mice (fig. S11I). Together, these data indicate that signaling through the FAK/YAP/TAZ axis is down-regulated following Ddr2 deletion at the transcriptional level.
Deletion of Ddr2 interrupts the FAK/YAP/TAZ axis
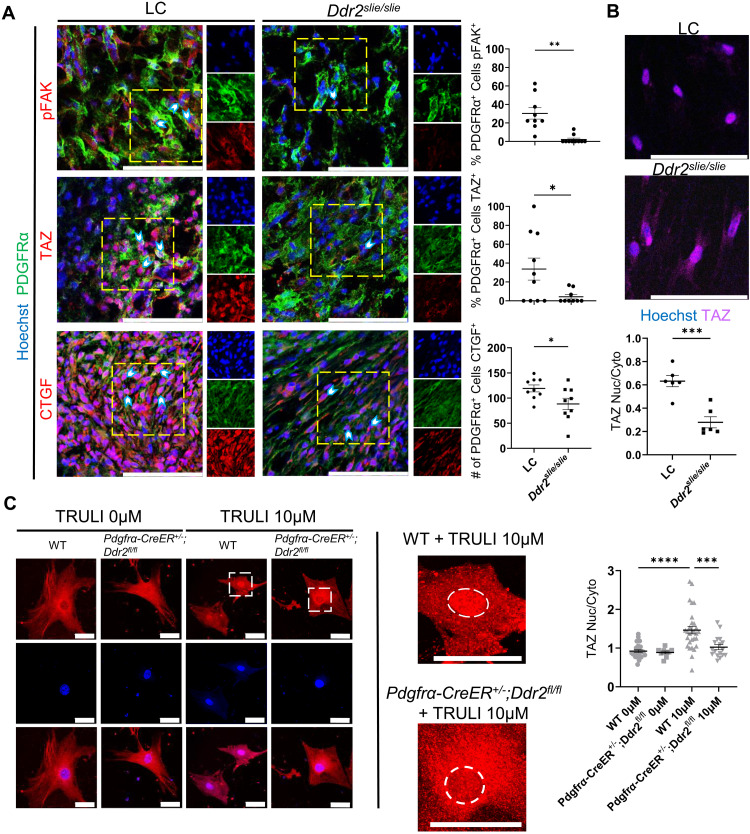
To lastly determine whether Ddr2 deletion affects the FAK/YAP/TAZ axis at posttranscriptional levels, pFAK and TAZ levels were assessed by immunostaining. In Ddr2-deleted mice, pFAK immunostaining and nuclear TAZ levels were significantly decreased relative to LC mice (30.30 versus 2.103% and 33.62 versus 4.302%, respectively; Fig. 6A). Furthermore, the downstream target of TAZ, CTGF, also showed reduced expression in PDGFRα-expressing MLin cells following Ddr2 deletion (119.1 versus 88.33 cells; Fig. 6A). Consistent with these findings, when Ddr2-targeted MLin cells are plated on two-dimensional aligned collagen-functionalized matrices, nuclear TAZ levels are likewise decreased (Fig. 6B).
Fig. 6. DDR2 deletion down-regulates the FAK/YAP/TAZ axis following injury.
(A) Representative confocal microscope images (63×) of injured hindlimb of LC and Ddr2slie/slie mice 1 week after BT, stained with Hoechst, anti-PDGFRα, and anti-phosphorylated FAK (pFAK), anti-TAZ, or anti-CTGF with quantification of the percentage or number of PDGFRα-expressing cells with positive staining of each antigen of interest (n = 3 mice per group, n = 2 to 4 images per mouse). Scale bars, 100 μm. (B) Two-dimensional aligned collagen plates with MLin cells from Ddr2slie/slie uninjured hindlimb and LC uninjured hindlimb and stained with Hoechst and TAZ. Graph shows ratios of positive TAZ staining localized to the nucleus compared to TAZ staining in the cytoplasm of cells from Ddr2slie/slie uninjured hindlimb and LC uninjured hindlimb (n = 2 mice per group, n = 3 images per mouse, bars are means ± SEM). Scale bars, 50 μm. (C) Representative confocal images (40×) of MLin cells isolated from Pdgfrα-CreER+/−;Ddr2fl/fl and wild-type mice 1 week after BT plated on three-dimensional collagen plates treated with 0 or 10 μM TRULI. Cells were stained with Hoechst and anti-TAZ (n = 19 to 29 cells per sample). Scale bars, 100 μm. The region within white dashed lines of high-magnification images is the nucleus. All graphs show means ± SEM.
To further confirm that DDR2 deletion interrupts the YAP/TAZ signaling pathway, we treated cells harvested from Pdgfrα-CreER+/−;Ddr2fl/fl and LC with TRULI in a three-dimensional collagen matrix (34). TRULI is a small-molecule inhibitor of LATS kinases, which stimulates YAP/TAZ signaling (35). Treatment with TRULI resulted in a significant increase in nuclear TAZ in control cells that was significantly abrogated in Pdgfrα-CreER+/−;Ddr2fl/fl cells (Fig. 6C). These data suggest that DDR2 is upstream of, and required for, TAZ activation. Together, our data suggest that DDR2 knockout reduces FAK/YAP/TAZ signaling (fig. S12).
DISCUSSION
Tendon ECM is primarily composed of collagen, specifically type I collagen (36). While the cross-talk between MLin cells and local ECM has been described in various contexts, the molecular influences of MLin cell–receptor interactions on MLin cell fate following musculoskeletal injury have not been clarified previously. The Achilles tendon reflects a unique substrate of dense and intricately aligned collagen fibrils that provide a unique setting in which to investigate the in vivo interactions between type I collagen and MLin cells. As HO lesions often form within the tendon proper following injury, we hypothesized that the ECM composition and organization would alter MLin cell fate. To our surprise, scRNA data revealed that MLin cells undergoing aberrant differentiation into ectopic chondrocytes and osteoblasts up-regulated expression of collagen I. Up-regulation of collagen I is commonly identified following tendon injury (37) and spurred our interest in identifying collagen receptors that might be activated in the damaged tissue. Having queried Ddr1/2 expression, we found Ddr2 to be highly and specifically up-regulated in collagen-expressing HO MLin cells. Our identification of DDR2 as a potential moderator of HO progression aligns, to some degree, with previous findings that DDR2 participates in normal bone development and fracture healing (9, 38, 39). Reduction in tendinous HO in Ddr2slie/slie and Pdgfrα-CreER;Ddr2fl/fl mice and C57B6 mice treated with a DDR2 inhibitor confirms the importance of collagen-DDR2 interactions in driving HO formation within tendons following injury.
The reduced cell-ECM interaction/signal transduction that arises as a consequence of targeting DDR2 further reinforces the importance of mechanical cell-ECM contact in selecting pro-osteogenic programs (40, 41). While there was no difference in FA formation following Ddr2 deletion, the increased deviation in FA orientation implies a decreased linear tension on the surrounding ECM that is required for collagen fiber alignment (42). A previous study of cancer-associated fibroblasts (CAFs) found that Ddr2 deletion resulted in weaker FAs that reduced the tractional force the CAFs generated on ECM collagen (14). By contrast, we found that force generation by Ddr2-deleted MLin cells was similar to control cells. Nevertheless, traction force orientation was disorganized, potentially explaining the differences in anisotropy.
Changes in collagen alignment and FAs have been shown to alter FAK (43) and YAP/TAZ (34, 44, 45) signaling that are known to play important roles in promoting MLin cell osteogenic differentiation and HO progression. Here, we have shown that Ddr2 deletion decreases FAK phosphorylation, TAZ activation and localization, and downstream TAZ signaling in vivo. Furthermore, we validated that DDR2 acts upstream of, and is required for, YAP/TAZ signaling. While we had previously shown that limb immobilization altered matrix alignment, we had not identified the central mechanism by which MLin cell interactions with the surrounding ECM occurred. To our knowledge, this work is the first to link MLin DDR2 activity to fibrillar collagen alignment after musculoskeletal injury in vivo. DDR2 has been previously implicated in matrix reorganization (46) and bone development (9, 12) but has never been implicated in aberrant MLin cell fate determination following injury. DDR2 binds directly to collagen fibrils and is known to trigger downstream signaling cascades that can increase cell proliferation, migration, and differentiation (9, 47, 48). We previously found that organized matrix influences HO formation (7), and in the current study, we found that Ddr2 deletion not only decreased HO formation but also decreased fibrillar collagen alignment without requiring limb immobilization per se. DDR2 appears to be required to form FAs in an organized fashion and then to apply organized mechanical forces to the ECM. While there were no differences in the total force applied to the ECM, we posit that the DDR2-guided orientation of FAs leads to an organized applied force to the matrix, resulting in the alignment of collagen fibers critical to HO progression.
This study has several notable limitations. A major obstacle in studying the role of ECM alignment on MLin cell differentiation is the lack of in vivo methods of altering matrix alignment. Thus, we have used proven in vitro systems with collagen I–based fibrils given that it is the most common collagen found in tendon and HO lesions (7, 34). Still, it is difficult to accurately emulate the in vivo ECM environment and mechanical forces within an in vitro system. In addition, imatinib is regarded as an effective DDR2 inhibitor (30) and has been effective in preventing HO formation in patients living with fibrodysplasia ossificans progressive, a genetic disorder wherein a mutation in the ACVR1 receptor (Alk2 gene) leads to ectopic bone formation both spontaneously and after injury (49, 50). On the basis of our findings, imatinib may prove to be an effective treatment in trauma-induced HO as well. Nevertheless, as a multikinase inhibitor, it may have notable off-target effects and other inhibitors more specific to DDR2 may be more effective in inhibiting DDR2 phosphorylation and activation (51). Previous work has shown that HO formation can be reduced by modulating a variety of factors, such as ECM organization (7), immune cell signaling (52), and progenitor signaling. While both the BT and a tenotomy alone models create a musculoskeletal injury, the BT model triggers a very robust systemic immune response. When the two models are directly compared, the BT model results in more HO formation (26). Previous work has shown that immune cell signaling, such as macrophage transforming growth factor–β (53, 54) or bone morphogenetic protein signaling (55) and mechanically induced netosis from neutrophils (56), can contribute to HO formation. While a tenotomy alone may be sufficient to trigger ECM organization, the BT model ensures that important immune cell populations are at the injury site to facilitate key MLin-immune cell interactions among other factors leading to HO formation. This model has a shortcoming in that we were unable to perform biomechanical testing of tendons following DDR2 knockout or inhibition because we completely transect the Achilles tendon and the tendon does not heal. Future studies will examine the biomechanical properties of tendons in other injury models where the tendon does recover.
Together, this study identifies the role of DDR2 expression in MLin cells on HO formation and ECM alignment following musculoskeletal injury. We have demonstrated a previously unknown DDR2-mediated mechanism by which MLin cell fate determination and collagen fiber alignment are altered. This collagen-binding receptor may be an effective target in preventing aberrant MLin cell differentiation toward osteogenic lineages following injury, expanding the library of potential clinical treatments available in the prevention of debilitating HO.
MATERIALS AND METHODS
Bioinformatics analysis of scRNA sequencing and snATAC sequencing data
Three different previously published datasets were used in the manuscript. Seurat 3.1.1 (29) was used to generate visualizations of scRNA and snATAC analyses. Downstream snATAC sequencing analyses and visualizations were performed using either 10X Loupe Browser or Signac 1.5.0 (57). The first analysis using scRNA in Fig. 1 of uninjured, 1-week, and 3-week post-BT injury was performed on a previously published dataset [GSE126060; (53)]. The second analysis using snATAC in Figs. 2 and 5 of uninjured, 1-week, and 6-week post-BT injury is from a previously published dataset [GSE126118; (7, 18, 53)]. The final analysis using scRNA, shown in Fig. 2, was performed on samples from a recently accepted analysis [GSE150995; (7)] but reanalyzed to target differences between mobilized and immobilized samples 1 week following injury. “Day 7 mobile injury site” and “day 7 immobilize injury site” samples were merged and processed by excluding RNA features [500,6000]. Mitochondrial gene expression was determined and cells with greater than 15% mitochondrial expression were excluded. Data were normalized, scaled, and 2000 variable features were found using the vst method. Principal components analysis considered 100 replicates, and Jackstraw was scored to 20 dimensions. Neighbors were calculated to 20 dimensions, and clusters were determined at a resolution of 0.5. UMAP reductions were calculated and used for visualization.
Ddr2slie/slie (n = 2) and LC (n = 2) male mice underwent BT injury and were harvested 7 days after injury (GSE212281). The contralateral uninjured limb in both Ddr2slie/slie mice and LC mice were also harvested 7 days after injury. Individual cells from each condition (injured Ddr2slie/slie, uninjured Ddr2slie/slie, injured LC, and uninjured LC) were isolated identically to other datasets in this publication. Samples within groups were combined for sequencing. Cells were sequenced using Illumina Nextgen 500 (San Diego, CA, USA). Raw data were processed using 10X genomics protocols. Filtered matrix files were consolidated into a single Seurat object using Seurat 4.1 and further processed excluding cells with RNA features less than 200 and greater than 6000. Mitochondrial gene content was determined, and cells with greater than 30% mitochondrial gene expression were excluded from the dataset. Cells were clustered at a resolution of 0.5. Differential gene expression analysis, enrichR Kyoto Encyclopedia of Genes and Genomes/Gene Ontology pathway analyses, and marker gene expression visualization were used to label clusters. Supplementary analyses were performed to isolate MLin clusters in the BT day 0, 7, and 21 dataset. MLin clusters were isolated, normalized, and integrated using Seurat 4.1 integration (29).
BT injury mouse model
All mice studies were reviewed and approved by The University of Texas Southwestern Medical School Institutional Animal Care and Use Committee (Protocol 2020-102949). Male and female mice between the ages of 8 and 12 weeks old underwent a 30% total body surface area back burn with concurrent Achilles transection as previously described (16). Mice were anesthetized with isoflurane and treated with analgesic buprenorphine sustained-release or hydrochloride. The uninjured right leg served as an uninjured internal control. Pdgfrα-CreER+/+ were crossed with Ddr2fl/fl mice in two rounds to yield Pdgfrα-CreER+/−;Ddr2fl/fl and Pdgfrα-CreER−/−;Ddr2fl/fl, which were used as the LCs. They were given tamoxifen chow (TD. 130860, Envigo, Indiana, USA) starting at adult maturity for 3 weeks and provided normal chow for 1 week to allow tamoxifen to wash out before injury. Pdgfrα-CreER+/−;Rosa-LSL-TdTomato mice were also treated with tamoxifen with the same method. For immobilization studies, mouse limbs were immediately immobilized using 1.5-ml microcentrifuge tubes modified with foam and super glued over the mouse leg, as previously done (7). The surgeon was blinded to genotype/treatment group.
FA organization on aligned fibrous matrices
Dextran (molecular weight, 86 kDa; MP Biomedicals, Santa Ana, CA) functionalized with vinyl sulfone (DexVS) was synthesized as previously described (58). Briefly, pure divinyl sulfone (12.5 ml; Fisher Scientific, Hampton, NH) was added to a sodium hydroxide solution (0.1 M and 250 ml) containing dextran (5 g). This reaction was carried out on a stir plate set to 750 rpm for 3.5 min, after which the reaction was terminated by the addition of 2.5 ml of HCl (12 M). The crude product was dialyzed against Milli-Q water for 3 days before lyophilization. To create heparin methacrylate (HepMA), heparin sodium salt was reacted with methacrylic anhydride following previously described procedures (59). Briefly, heparin sodium salt (500 mg) was dissolved in 50 ml of phosphate-buffered saline (PBS) under vigorous stirring before the addition of methacrylic anhydride (99.3 ml). The reaction was kept under constant stirring at 4°C, and NaOH was added intermittently to maintain a pH of ~8 for 24 hours. The product was dialyzed against Milli-Q water for 3 days and then lyophilized. Final products were characterized by 1H–nuclear magnetic resonance. Electrospun DexVS fibers were generated as described previously (60). Briefly, an electrospinning solution was prepared by dissolving DexVS at 0.60 g/ml in a 1:1 mixture of Milli-Q water and dimethylformamide with a lithium phenyl-2,3,6-trimethylbenzoylphosphinate (LAP; 10 mg/ml) photoinitiator. Electrospinning was performed in an environment-controlled glovebox (21°C and 30 to 35% relative humidity) with a flow rate of 0.15 ml/hour, a gap distance of 8 cm, and a voltage of −7.5 kV. Spinning the grounded collecting mandrel at 1000 rpm resulted in coalignment of deposited fibers, which were collected on coverslips treated with 3-[tris(trimethylsiloxy)silyl]propyl methacrylate. Fiber samples were primarily cross-linked under ultraviolet (UV) light (100 mW/cm2), hydrated in a photoinitiator solution of LAP (1 mg/ml) in Milli-Q water, and then secondarily cross-linked under UV light (100 mW/cm2) for 20 s to stabilize networks. To enable cell adhesion, HepMA was dissolved into secondary cross-linking solutions at 2.5% (w/v) followed by a 30-min incubation in type 1 collagen (100 μg/ml). MLin cells isolated from male mice were trypsinized, counted, and seeded onto substrates at a density of 15,000 cells/cm2. After 2 days of culture, samples were fixed and immunostained for vinculin and F-actin using phalloidin (Invitrogen, A12379). Samples were imaged via confocal fluorescent microscopy, and a custom MATLAB code (https://gist.github.com/chasepagani/ed7e4abacb4450ec3c705f7f153bcd8e) quantified the SD of the orientation of FAs within each field of view.
Histology and immunofluorescent staining
Full hindlimb samples from male and female mice were harvested at 1 week, 3 weeks, and 9 weeks following BT injury. Legs were fixed in 4% paraformaldehyde (PFA) for 24 hours at 4°C, washed with 1× PBS (Gibco, Waltham, MA), and decalcified using 17% EDTA (pH 7.4) for 5 weeks (Sigma-Aldrich, St. Louis, MO). Tissues were then embedded in optimal cutting temperature media (Sakura, Torrance, CA) to prepare either axial or sagittal sections of frozen tissues. Sections were cut at 10 μm for immunofluorescence and Golder’s trichrome staining, 12 μm for Safranin O staining, and 20 μm for SHG imaging on Fisher SuperFrost Microscopy slides (Fisher, Pittsburgh, PA). For immunofluorescence staining, sections were thawed then washed in 1× tris-buffered saline with Tween 20 [TBS-T; 1× TBS (Bio-Rad, Hercules, CA), 0.05% Tween 20 (Sigma-Aldrich, St. Louis, MO)]. Sections were blocked with donkey-serum blocking solution (1% bovine serum albumin, 2% donkey serum, 0.1% cold water fish skin gelatin, 0.05% Triton X-100, 0.05% Tween 20, 300 mM glycine, and 1× TBS, pH 8.4) for 2 hours at room temperature (RT) and then incubated at 4°C overnight with primary antibodies (PDGFRα: R&D Systems AF1062, 1:50; DDR2: LS-B15752, 1:50; pDDR2: MAB25382, 1:50; β-GAL: IC Lab CGAL-45A, 1:50; SOX9: ab185230, 1:50; OPN: AF808, 1:50; SP7: ab22552, 1:50; RUNX2: ab23891, 1:50; pFAK: 44-626G, 1:50; TAZ: NB11058359, 1:50; CTGF: ab6992, 1:100). Slides were washed in 1× TBS-T three times and incubated with fluorescence-conjugated secondary antibodies for 2 hours at RT (Invitrogen donkey anti-goat AF488, Invitrogen donkey anti-goat AF647, Invitrogen donkey anti-rabbit AF488, Invitrogen donkey anti-rabbit AF647, all secondary antibodies at 1:200 dilution, Carlsbad, CA). Then, slides were stained for nucleus with Hoechst 33342 (Thermo Fisher Scientific, Waltham, MA) for 5 min, washed in 1× TBS-T, and mounted with ProLong Glass Antifade Mountant (Invitrogen, Carlsbad, CA) and #1.5 Slip-Rite cover glass (Richard-Allan Scientific, San Diego, CA). For SHG imaging, 20-μm sagittal sections of murine hindlimbs at 1, 3, and 9 weeks were washed in 1× TBST. The slides were mounted with Aqua-Poly/Mount water–based mounting media (Polysciences, Warrington, PA) and #1.5 Slip-Rite cover glass (Richard-Allan Scientific, San Diego, CA) and sealed with nail polish. For Safranin O staining, 12-μm frozen slides were washed three times (3, 3, and 4 min) with deionized water, 1.5 min in 0.02% fast green (Sigma-Aldrich), 30 s in 1% acetic acid, 10 min in 1% Safranin O (Sigma-Aldrich), three times (30 s each) in 95% ethanol, two times (30 s each) in 100% ethanol, three washes of xylene (15 dips each), and mounted using #1.5 Slip-Rite cover glass (Richard-Allan Scientific) and Xylene Substitute Mountant (Epredia). Ten-micrometer slides were stained with Goldner’s trichrome for collagen matrix deposition. The slides were hydrated with deionized water and mordanted in Bouin’s Solution (Sigma-Aldrich, catalog no. HT10132-1L) for 15 min. They were then cooled and washed in running tap water and stained in Working Weigert’s Iron hematoxylin solution (Sigma-Aldrich, catalog no. HT1079-1) for 5 min. The slides were washed in tap water for 5 min, rinsed in deionized water, and placed in Phophotungstic/Phosphomolybdic Acid Solution for 5 min (Sigma-Aldrich, catalog no. HT152; 250 ml, Sigma-Aldrich, catalog no. HT153; 250 ml). Then, they were placed in Light Green solution (SF catalog no. L5382) for 5 min, rinsed in acetic acid 1% for 5 min, dehydrated in 95% alcohol and absolute alcohol with three changes each, cleared in xylene, and mounted. For human samples, the sections were deparaffinized and washed in PBS 3× for 10 min. Normal goat serum (5%) was applied for 30 min and then incubated overnight with primary antibodies at 4°C in a humidified chamber. The next day, slides were washed in PBS and incubated with secondary fluorescent antibodies (Invitrogen anti-rabbit AF488 and goat anti-mouse AF594), for 1 hour at RT, and then nucleus was stained with 4′,6-diamidino-2-phenylindole mounting solution (Vectashield H-1500, Vector Laboratories, Burlingame, CA).
Histology imaging
For immunofluorescent imaging of 10-μm murine sections, 1× and 3.15× computerized zoom images were captured with a 20× oil-immersion lens on a Leica SP8 confocal microscope using Leica’s LAS X software (Wetzlar, Germany), 40× lens on a Zeiss LSM880 inverted confocal microscope (Zeiss, Oberkochen, Germany), or 20× lens on a Leica Stellaris 8 confocal microscope. For SHG imaging, 20-μm slides were imaged using a Zeiss LSM880 inverted confocal microscope (Zeiss, Oberkochen, Germany) with a forward SHG 400 to 480 nm M-2P filter and an 850-nm excitation laser with a transmitted light detector. Images at all three time points of the ECM in the injury site were taken. For Safranin O–stained slides, the entire slide was scanned using Hamamatsu NanoZoomer 2.0-HT (Hamamatsu, Hamamatsu City, Shizuoka, Japan) and 1× tile scans and 5× images used for quantifications were created using NanoZoomer Digital Pathology (NDP.view2) software. Goldner’s trichrome–stained slides were imaged on a Leica DM 6B microscope (Leica Biosystems, Germany). Digital images (20×) of the human sections were captured on a Leica DM6 upright fluorescent microscope (Leica Microsystems Inc., Buffalo Grove, IL).
Histology image analysis
Immunofluorescence images (20×, 40×, and 63×) were analyzed by a blinded, skilled operator for antigen-of-interest count or % of total cell expression quantification. Hoechst 33342–positive cells were first counted to quantify whole cells within the image frame. Then, the number of antigen-of-interest positive cells was counted within the image frame to provide a count or percentage of antigen-of-interest positive cells. Mean and SEM were calculated and plotted in GraphPad Prism 8 (GSL Biotech LLC, Chicago, IL). Multiphoton SHG imaging was used to characterize the collagen organization at the injury site. Collagen fiber alignment was analyzed with Fibril Tool, a validated ImageJ plugin that quantifies anisotropy determined by eigenvectors of the Hessian matrix (61). Four to five regions of interest were selected per image to produce anisotropy values, providing information on the alignment of fibrillar collagen within that region. For Safranin O–stained slides, cartilage content area was quantified using Fiji ImageJ software (62) using the green channel for each image. Thresholding was set using automatic threshold algorithm RenyiEntropy. Goldner’s trichrome stain was quantified using ImageJ software (v1.53c, National Institutes of Health, USA). Bone area of heterotopic bone was measured by intensity thresholding in mouse Achilles tendon.
Human samples
Human HO samples were collected from the left thigh of a 38-year-old male patient and uninjured tendon samples were collected from the right bicep of a 60-year-old male patient. Samples were provided by the Johns Hopkins Bone Pathology Archives and approved by the Johns Hopkins University Institutional Review Board. All human samples were coded to ensure confidentiality and protect personal health information.
Immunocytochemistry
Cells were isolated from male mice and two-dimensional collagen I–coated wells were used for Ddr2slie/slie versus LC experiment. Cells were washed with 1× PBS then fixed in 4% PFA for 15 min at RT. Cells were permeabilized for 5 min in 1× TBS with 0.5% Triton X-100. Cells were then blocked with 10% donkey serum in 1× TBS 0.5% Triton X-100 and then washed twice, and primary antibody was added in blocking solution (2% donkey serum in 1× TBS 1% Triton X-100) and incubated overnight at 4°C (1:200 dilution, rabbit anti-mouse TAZ antibody, Novus Biologicals, Littleton, CO). Cells were then washed three times, and secondary antibody was prepared in blocking solution (1:1000 dilution; donkey anti-rabbit immunoglobulin G Alexa Fluor 647; Thermo Fisher Scientific, Waltham, MA) and incubated for 1 hour at RT. Cells were then nucleus-stained with Hoechst for 15 min, washed, and mounted on glass slides with prolong glass. Cells were imaged on Leica SPX8 Confocal Scanning Microscope. For TRULI treatment experiments and differentiation experiments, cells were isolated from female mice and three-dimensional collagen-coated wells were prepared using collagen I (50 μg/ml). Cells were plated at a final concentration of 4 × 105 cells/ml, and cells were cooled on ice for 3 min. Collagen solution (2 mg/ml; 1,10, pH 7.4) was added and pipetted up and down. A cold tip was used to take 150 to 200 μl of the cell-collagen mixture held for 30 to 60 s and pipetted quickly into wells. Cells were incubated for 1 hour at 37°C and then added to complete medium. The following day, the medium was changed to osteogenic differentiation medium [aMEM with 10% fetal bovine serum (FBS), 10 mM beta-glycerophosphate, and l-ascorbic acid (50 μg/ml)] with 10 μM TRULI (catalog no. Z730688380, Enamine LLC) (35) or vehicle control. After 24 hours, cells were fixed with 4% PFA in 5% sucrose and permeabilized with 0.5% Triton X-100 and then stained with TAZ antibody (catalog no. 3560235, BD Biosciences; 1:50 dilution) (63). After staining was finished, cells were imaged on a Leica Stellaris 8 confocal microscope (Wetzlar, Germany) and quantified using ImageJ.
In vivo DDR2 inhibitor treatments
Male wild-type C57B6 mice were used for all in vivo inhibitor studies. Mice were randomized and treated using intraperitoneal injections with imatinib (50 mg/kg; LC Laboratories) or vehicle (sterile water) daily for 16 days following BT.
Isolation of murine MLin cells
Murine tissue-resident multipotent MLin cells were isolated from tissue collected from the Achilles tendon injury site (soft tissue posterior to the tibia and in between the distal tendon enthesis to the calcaneus and proximal insertion to the gastrocnemius muscle) of mice subjected to BT injury as previously described (7, 17, 64). Briefly, harvested tissues were digested in digestion solution (3 mg/ml collagenase I, 2 mg/ml collagenase II, and 2 mg/ml dispase II), digested tissues were strained through a 40-μm cell strainer, and digestive enzymes were quenched in complete culture media [DMEM-10; Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% FBS (Gibco)] and 1× antibiotics [penicillin (100 U/ml) and streptomycin (100 g/ml)]. Cells were spun down at 300g for 5 min, and the supernatant was discarded. The cell pellet was subsequently expanded in complete media (DMEM-10) and passages with split 1:3 ratio. All cells were used within passages 1 to 4.
𝛍CT imaging and quantification
μCT images were taken at the University of Texas Southwestern Department of Radiology using a MedisoUSA nanoScan PET/CT system (Arlington, VA). The scanning parameters were max zoom with 720 helical projections with an x-ray power of 70 kV at 980 μA and an exposure time of 300 ms. The images were reconstructed using the manufacturer’s program, Nucline nanoScan, with a resulting voxel size of 40 × 40 × 40 μm. Images were quantified using Dragonfly ORS (Montreal, Quebec, Canada) at 800 Hounsfield units (HU). Reconstructions of mouse hindlimbs were created on representative means at 800 HU. Quantifications at 800 HU of ectopic bone formation were performed on four anatomical regions—calcaneus, tibia, distal residual Achilles tendon, and proximal residual Achilles tendon—of the mouse hindlimb by a blinded, skilled operator. Quantifications of total ectopic bone formation included all four of these regions.
Real-time qPCR
Total RNA from LST cells from female mice was extracted with an RNeasy Plus Mini Kit (catalog no. 74136, Qiagen). Two hundred nanograms of total RNA was reverse-transcribed with a High Capacity cDNA Reverse Transcription Kit (catalog no. 4368814, Applied Biosystems). Real-time PCR reactions were performed with an iQ SYBR Green Supermix kit (catalog no. 1708880, Bio-Rad) by the CFX96 touch Real-Time PCR Detection System.
Real-time PCR primers were used: mouse DDR2 forward primer: ATCACAGCCTCAAGTCAGTGG; mouse DDR2 reverse primer: TTCAGGTCATCGGGTTGCAC; mouse glyceraldehyde-3-phosphate dehydrogenase (GAPDH) forward primer: AGGTCGGTGTGAACGGATTTG; mouse GAPDH reverse primer: TGTAGACCATGTAGTTGAGGTCA. Relative mRNA expression by reverse transcription qPCR was calculated according to the equation 2−ΔΔCt.
Statistical analyses
All experiments were performed once and sample sizes were determined on the basis of previously described methods (53). Statistics were performed using GraphPad Prism 9 (San Diego, CA). Tests for normality and heterogeneity of variance were performed to ensure appropriate statistical tests were performed. Grubb’s outlier test was used to identify and omit outliers (fig. S3E, distal tendon HO quantification). When necessary, Welch’s correction was performed on heteroscedastic data and Mann-Whitney U tests were performed for nonparametric data. Statistical significance is displayed on each graph as an asterisk (*) for unpaired two-tailed Student’s t test and a pound sign (#) for Mann-Whitney U test. In experiments with multiple groups or treatments, statistical significance is displayed on each graph as an asterisk (*) for one-way analysis of variance test with Tukey’s post hoc test. All comparisons were performed at α = 0.05. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
TFM and analysis
MLin cells were harvested from the Achilles tendon of LC and Pdgfrα-CreER+/−;Ddr2fl/fl male mice. TFM was carried out on 8-kPa silicone substrates embedded with far-red emitting carboxylate beads as described previously (65). To coat the substrates, each 35-mm-diameter dish was incubated for half an hour at RT with a solution of calcium- and magnesium-free PBS supplemented with 7.5 mg/ml of bovine collagen I and 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride (100 mg/ml). Dishes were rinsed multiple times with PBS and then incubated with a 0.1% weight/volume solution of bovine serum albumin in PBS for an additional half-hour. To evaluate traction forces, 50,000 cells were seeded on the TFM substrates, labeled with 1 μM CellTracker Green CMFDA for 15 min, and imaged 2 hours later with a Nikon CSU-W1 SoRa Spinning Disk Confocal Microscope located in the Quantitative Light Microscopy Core facility at UT Southwestern (NIH 1S100D028630-01). All imaging was performed at 37°C with proper humidity and CO2 control. Z-stacks of the cells and substrates were performed with a 60× numerical aperture 1.4 oil immersion objective at a voxel size of 108 × 108 × 300 nm. Upon evaluation of ~20 cells, a preheated solution of diluted bleach was used to remove the cells from the substrate, thereby allowing the substrate to be imaged in its relaxed, strain-free state.
All image analysis was performed on maximum intensity projections of the data using publicly available software written in MATLAB (65, 66) and accessible on GitHub (https://github.com/DanuserLab/TFM). Substrate displacement was estimated using particle image velocimetry of the substrate images with default package parameters, and the traction field was reconstructed using the Fourier transform traction cytometry method. A regularization parameter of 2 × 10−5 was selected based on L-curve analysis of a subset of images (66) and then held constant for analysis of the full dataset. Strain energy, which represents the mechanical work of the cell on the elastic substrate, was calculated as ½ × (displacement · traction), integrated over the segmented cell area (67).
Acknowledgments
We would like to thank A. Velazquez, S. Harris, and X. Sun from the Department of Radiology at the University of Texas Southwestern for assisting in μCT imaging and K. Padmanabhan at the University of Michigan for assistance in analyzing the snATAC data. We would also like to thank The University of Michigan Bioinformatics Lab and Spencer Barnes for single-cell sequencing assistance. Some of the visualizations in this manuscript were generated on BioRender.com.
Funding: This work was supported by the Department of Defense Congressionally Directed Medical Research Programs W81XWH-20-1-0571 (R.T.F.), Dermatology Foundation (B.A.N.), National Institutes of Health grant R01AR078324 (B.L.), National Institutes of Health grant R01AR071379 (B.L.), National Institutes of Health grant T32AR065969 (B.A.N.), National Institutes of Health grant 1R01DK127589 (K.M.D.), National Institutes of Health grant 1R21HD105189 (K.M.D.), National Institutes of Health grant 5P30CA142543 (K.M.D.), National Institutes of Health grant U54CA26CA268072 (K.M.D.), National Institutes of Health grant R01DE029465 (R.T.F.), Ruth L. Kirschstein Institutional National Research Service Award Postdoctoral Fellowship T32-CA009672 (A.L.S.), and UTSW-UNC Center for Cell Signaling Analysis 1 RM1 GM 145399 (K.M.D.).
Author contributions: Conceptualization: C.A.P., A.C.B., S.J.W., R.T.F., and B.L. Methodology: C.A.P., A.C.B., R.J.T., Y.S., R.N.K., A.L.S., B.A.N., K.M.D., B.M.B., A.W.J., S.J.W., R.T.F., and B.L. Investigation: C.A.P., A.C.B., N.L., Y.S., J.Y.H., R.N.K., A.L.S., J.H.N., J.M.M., N.P., B.A.N., K.M.D., Z.L., and C.G. Visualization: C.A.P., A.C.B., N.L., Y.S., J.Y.H., R.N.K., and A.L.S. Formal analysis: C.A.P., A.C.B., N.L., Y.S., J.Y.H., R.N.K., A.L.S., B.A.N., and K.M.D. Writing—original draft: C.A.P., A.C.B., and B.L. Writing—review and editing: C.A.P., A.C.B., R.J.T., N.L., R.N.K., B.A.N., K.M.D., B.M.B., A.W.J., S.J.W., R.T.F., and B.L.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Data from Ddr2slie/slie and littermate control sequencing experiment are available at NCBI GEO (https://www.ncbi.nlm.nih.gov/geo {GSE212281}), and additional bioinformatics analyses were performed on previously published data.
Supplementary Materials
This PDF file includes:
Figs. S1 to S12
Table S2
Other Supplementary Material for this : manuscript includes the following:
Table S1
REFERENCES AND NOTES
- 1.M. F. Pittenger, A. M. Mackay, S. C. Beck, R. K. Jaiswal, R. Douglas, J. D. Mosca, M. A. Moorman, D. W. Simonetti, S. Craig, D. R. Marshak,Multilineage potential of adult human mesenchymal stem cells. Science 284,143–147 (1999). [DOI] [PubMed] [Google Scholar]
- 2.F. Guilak, D. M. Cohen, B. T. Estes, J. M. Gimble, W. Liedtke, C. S. Chen,Control of stem cell fate by physical interactions with the extracellular matrix. Cell Stem Cell 5,17–26 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.K. Narayanan, K. J. Leck, S. Gao, A. C. Wan,Three-dimensional reconstituted extracellular matrix scaffolds for tissue engineering. Biomaterials 30,4309–4317 (2009). [DOI] [PubMed] [Google Scholar]
- 4.A. J. Steward, D. J. Kelly,Mechanical regulation of mesenchymal stem cell differentiation. J. Anat. 227,717–731 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.W. J. Hadden, J. L. Young, A. W. Holle, M. L. McFetridge, D. Y. Kim, P. Wijesinghe, H. Taylor-Weiner, J. H. Wen, A. R. Lee, K. Bieback, B. N. Vo, D. D. Sampson, B. F. Kennedy, J. P. Spatz, A. J. Engler, Y. S. Choi,Stem cell migration and mechanotransduction on linear stiffness gradient hydrogels. Proc. Natl. Acad. Sci. U.S.A. 114,5647–5652 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.D. Dey, B. M. Wheatley, D. Cholok, S. Agarwal, P. B. Yu, B. Levi, T. A. Davis,The traumatic bone: Trauma-induced heterotopic ossification. Transl. Res. 186,95–111 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.A. K. Huber, N. Patel, C. A. Pagani, S. Marini, K. R. Padmanabhan, D. L. Matera, M. Said, C. Hwang, G. C. Hsu, A. A. Poli, A. L. Strong, N. D. Visser, J. A. Greenstein, R. Nelson, S. Li, M. T. Longaker, Y. Tang, S. J. Weiss, B. M. Baker, A. W. James, B. Levi,Immobilization after injury alters extracellular matrix and stem cell fate. J. Clin. Invest. 130,5444–5460 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.W. Vogel, G. D. Gish, F. Alves, T. Pawson,The discoidin domain receptor tyrosine kinases are activated by collagen. Mol. Cell 1,13–23 (1997). [DOI] [PubMed] [Google Scholar]
- 9.C. Ge, Z. Wang, G. Zhao, B. Li, J. Liao, H. Sun, R. T. Franceschi,Discoidin receptor 2 controls bone formation and marrow adipogenesis. J. Bone Miner. Res. 31,2193–2203 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.R. Khosravi, K. L. Sodek, M. Faibish, P. C. Trackman,Collagen advanced glycation inhibits its discoidin domain receptor 2 (DDR2)-mediated induction of lysyl oxidase in osteoblasts. Bone 58,33–41 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Y. Itoh,Discoidin domain receptors: Microenvironment sensors that promote cellular migration and invasion. Cell Adh. Migr. 12,378–385 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.F. F. Mohamed, C. Ge, A. Binrayes, R. T. Franceschi,The role of discoidin domain receptor 2 in tooth development. J. Dent. Res. 99,214–222 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.M. E. Gonzalez, E. E. Martin, T. Anwar, C. Arellano-Garcia, N. Medhora, A. Lama, Y. C. Chen, K. S. Tanager, E. Yoon, K. M. Kidwell, C. Ge, R. T. Franceschi, C. G. Kleer,Mesenchymal stem cell-induced DDR2 mediates stromal-breast cancer interactions and metastasis growth. Cell Rep. 18,1215–1228 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.S. V. Bayer, W. R. Grither, A. Brenot, P. Y. Hwang, C. E. Barcus, M. Ernst, P. Pence, C. Walter, A. Pathak, G. D. Longmore,DDR2 controls breast tumor stiffness and metastasis by regulating integrin mediated mechanotransduction in CAFs. eLife 8,e45508 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.E. Olaso, H. C. Lin, L. H. Wang, S. L. Friedman,Impaired dermal wound healing in discoidin domain receptor 2-deficient mice associated with defective extracellular matrix remodeling. Fibrogenesis Tissue Repair 4,5 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.J. R. Peterson, S. De La Rosa, H. Sun, O. Eboda, K. E. Cilwa, A. Donneys, M. Morris, S. R. Buchman, P. S. Cederna, P. H. Krebsbach, S. C. Wang, B. Levi,Burn injury enhances bone formation in heterotopic ossification model. Ann. Surg. 259,993–998 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.S. Agarwal, S. Loder, C. Brownley, D. Cholok, L. Mangiavini, J. Li, C. Breuler, H. H. Sung, S. Li, K. Ranganathan, J. Peterson, R. Tompkins, D. Herndon, W. Xiao, D. Jumlongras, B. R. Olsen, T. A. Davis, Y. Mishina, E. Schipani, B. Levi,Inhibition of Hif1α prevents both trauma-induced and genetic heterotopic ossification. Proc. Natl. Acad. Sci. U.S.A. 113,E338–E347 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.C. A. Pagani, A. K. Huber, C. Hwang, S. Marini, K. Padmanabhan, N. Livingston, J. Nunez, Y. Sun, N. Edwards, Y. H. Cheng, N. Visser, P. Yu, N. Patel, J. A. Greenstein, H. Rasheed, R. Nelson, K. Kessel, K. Vasquez, A. L. Strong, G. E. Hespe, J. Y. Song, D. M. Wellik, B. Levi,Novel lineage-tracing system to identify site-specific ectopic bone precursor cells. Stem Cell Rep. 16,626–640 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.L. Lin, Q. Shen, T. Xue, C. Yu,Heterotopic ossification induced by Achilles tenotomy via endochondral bone formation: Expression of bone and cartilage related genes. Bone 46,425–431 (2010). [DOI] [PubMed] [Google Scholar]
- 20.A. Augello, C. De Bari,The regulation of differentiation in mesenchymal stem cells. Hum. Gene Ther. 21,1226–1238 (2010). [DOI] [PubMed] [Google Scholar]
- 21.I. Sekiya, K. Tsuji, P. Koopman, H. Watanabe, Y. Yamada, K. Shinomiya, A. Nifuji, M. Noda,SOX9 enhances aggrecan gene promoter/enhancer activity and is up-regulated by retinoic acid in a cartilage-derived cell line, TC6. J. Biol. Chem. 275,10738–10744 (2000). [DOI] [PubMed] [Google Scholar]
- 22.E. Wright, M. R. Hargrave, J. Christiansen, L. Cooper, J. Kun, T. Evans, U. Gangadharan, A. Greenfield, P. Koopman,The Sry-related gene Sox9 is expressed during chondrogenesis in mouse embryos. Nat. Genet. 9,15–20 (1995). [DOI] [PubMed] [Google Scholar]
- 23.W. Bi, J. M. Deng, Z. Zhang, R. R. Behringer, B. de Crombrugghe,Sox9 is required for cartilage formation. Nat. Genet. 22,85–89 (1999). [DOI] [PubMed] [Google Scholar]
- 24.T. Komori, H. Yagi, S. Nomura, A. Yamaguchi, K. Sasaki, K. Deguchi, Y. Shimizu, R. T. Bronson, Y. H. Gao, M. Inada, M. Sato, R. Okamoto, Y. Kitamura, S. Yoshiki, T. Kishimoto,Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell 89,755–764 (1997). [DOI] [PubMed] [Google Scholar]
- 25.T. Harvey, S. Flamenco, C. M. Fan,A Tppp3+Pdgfra+ tendon stem cell population contributes to regeneration and reveals a shared role for PDGF signalling in regeneration and fibrosis. Nat. Cell Biol. 21,1490–1503 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.J. R. Peterson, P. I. Okagbare, S. De La Rosa, K. E. Cilwa, J. E. Perosky, O. N. Eboda, A. Donneys, G. L. Su, S. R. Buchman, P. S. Cederna, S. C. Wang, K. M. Kozloff, M. D. Morris, B. Levi,Early detection of burn induced heterotopic ossification using transcutaneous Raman spectroscopy. Bone 54,28–34 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.D. Kim, E. You, J. Jeong, P. Ko, J. W. Kim, S. Rhee,DDR2 controls the epithelial-mesenchymal-transition-related gene expression via c-Myb acetylation upon matrix stiffening. Sci. Rep. 7,6847 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.C. A. Corsa, A. Brenot, W. R. Grither, S. Van Hove, A. J. Loza, K. Zhang, S. M. Ponik, Y. Liu, D. G. DeNardo, K. W. Eliceiri, P. J. Keely, G. D. Longmore,The action of discoidin domain receptor 2 in basal tumor cells and stromal cancer-associated fibroblasts is critical for breast cancer metastasis. Cell Rep. 15,2510–2523 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.T. Stuart, A. Butler, P. Hoffman, C. Hafemeister, E. Papalexi, W. M. Mauck III, Y. Hao, M. Stoeckius, P. Smibert, R. Satija,Comprehensive integration of single-cell data. Cell 177,1888–1902.e21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.E. Day, B. Waters, K. Spiegel, T. Alnadaf, P. W. Manley, E. Buchdunger, C. Walker, G. Jarai,Inhibition of collagen-induced discoidin domain receptor 1 and 2 activation by imatinib, nilotinib and dasatinib. Eur. J. Pharmacol. 599,44–53 (2008). [DOI] [PubMed] [Google Scholar]
- 31.J. Seong, N. Wang, Y. Wang,Mechanotransduction at focal adhesions: From physiology to cancer development. J. Cell. Mol. Med. 17,597–604 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.K. Burridge, C. Guilluy,Focal adhesions, stress fibers and mechanical tension. Exp. Cell Res. 343,14–20 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.C. D. Davidson, D. K. P. Jayco, D. L. Matera, S. J. DePalma, H. L. Hiraki, W. Y. Wang, B. M. Baker,Myofibroblast activation in synthetic fibrous matrices composed of dextran vinyl sulfone. Acta Biomater. 105,78–86 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Y. Tang, R. G. Rowe, E. L. Botvinick, A. Kurup, A. J. Putnam, M. Seiki, V. M. Weaver, E. T. Keller, S. Goldstein, J. Dai, D. Begun, T. Saunders, S. J. Weiss,MT1-MMP-dependent control of skeletal stem cell commitment via a β1-integrin/YAP/TAZ signaling axis. Dev. Cell 25,402–416 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.N. Kastan, K. Gnedeva, T. Alisch, A. A. Petelski, D. J. Huggins, J. Chiaravalli, A. Aharanov, A. Shakked, E. Tzahor, A. Nagiel, N. Segil, A. J. Hudspeth,Small-molecule inhibition of Lats kinases may promote Yap-dependent proliferation in postmitotic mammalian tissues. Nat. Commun. 12,3100 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.C. T. Thorpe, H. R. Screen,Tendon structure and composition. Adv. Exp. Med. Biol. 920,3–10 (2016). [DOI] [PubMed] [Google Scholar]
- 37.T. Sakabe, K. Sakai, T. Maeda, A. Sunaga, N. Furuta, R. Schweitzer, T. Sasaki, T. Sakai,Transcription factor scleraxis vitally contributes to progenitor lineage direction in wound healing of adult tendon in mice. J. Biol. Chem. 293,5766–5780 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.F. F. Mohamed, C. Ge, R. T. Cowling, D. Lucas, S. A. Hallett, N. Ono, A. A. Binrayes, B. Greenberg, R. T. Franceschi,The collagen receptor, discoidin domain receptor 2, functions in Gli1-positive skeletal progenitors and chondrocytes to control bone development. Bone Res. 10,11 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.A. Binrayes, C. Ge, F. F. Mohamed, R. T. Franceschi,Role of discoidin domain receptor 2 in craniofacial bone regeneration. J. Dent. Res. 100,1359–1366 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Y. Zhang, J. Su, J. Yu, X. Bu, T. Ren, X. Liu, L. Yao,An essential role of discoidin domain receptor 2 (DDR2) in osteoblast differentiation and chondrocyte maturation via modulation of Runx2 activation. J. Bone Miner. Res. 26,604–617 (2011). [DOI] [PubMed] [Google Scholar]
- 41.K. L. Lin, C. H. Chou, S. C. Hsieh, S. Y. Hwa, M. T. Lee, F. F. Wang,Transcriptional upregulation of DDR2 by ATF4 facilitates osteoblastic differentiation through p38 MAPK-mediated Runx2 activation. J. Bone Miner. Res. 25,2489–2503 (2010). [DOI] [PubMed] [Google Scholar]
- 42.R. J. Klebe, H. Caldwell, S. Milam,Cells transmit spatial information by orienting collagen fibers. Matrix 9,451–458 (1989). [DOI] [PubMed] [Google Scholar]
- 43.R. M. Salasznyk, R. F. Klees, W. A. Williams, A. Boskey, G. E. Plopper,Focal adhesion kinase signaling pathways regulate the osteogenic differentiation of human mesenchymal stem cells. Exp. Cell Res. 313,22–37 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.S. Dupont, L. Morsut, M. Aragona, E. Enzo, S. Giulitti, M. Cordenonsi, F. Zanconato, J. Le Digabel, M. Forcato, S. Bicciato, N. Elvassore, S. Piccolo,Role of YAP/TAZ in mechanotransduction. Nature 474,179–183 (2011). [DOI] [PubMed] [Google Scholar]
- 45.Q. Cong, Y. Liu, T. Zhou, Y. Zhou, R. Xu, C. Cheng, H. S. Chung, M. Yan, H. Zhou, Z. Liao, B. Gao, G. A. Bocobo, T. A. Covington, H. J. Song, P. Su, P. B. Yu, Y. Yang,A self-amplifying loop of YAP and SHH drives formation and expansion of heterotopic ossification. Sci. Transl. Med. 13,eabb2233 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.C. M. Borza, A. Pozzi,Discoidin domain receptors in disease. Matrix Biol. 34,185–192 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.I. Kawai, T. Hisaki, K. Sugiura, K. Naito, K. Kano,Discoidin domain receptor 2 (DDR2) regulates proliferation of endochondral cells in mice. Biochem. Biophys. Res. Commun. 427,611–617 (2012). [DOI] [PubMed] [Google Scholar]
- 48.B. Poudel, Y.-M. Lee, D.-K. Kim,DDR2 inhibition reduces migration and invasion of murine metastatic melanoma cells by suppressing MMP2/9 expression through ERK/NF-κB pathway. Acta Biochim. Biophys. Sin. 47,292–298 (2015). [DOI] [PubMed] [Google Scholar]
- 49.F. S. Kaplan, D. T. Teachey, J. R. Andolina, D. M. Siegel, E. E. Mancilla, E. C. Hsiao, M. Al Mukaddam, D. M. Rocke, R. J. Pignolo,Off-on-off-on use of imatinib in three children with fibrodysplasia ossificans progressiva. Bone 150,116016 (2021). [DOI] [PubMed] [Google Scholar]
- 50.F. S. Kaplan, J. R. Andolina, P. C. Adamson, D. T. Teachey, J. Z. Finklestein, D. H. Ebb, B. Whitehead, B. Jacobs, D. M. Siegel, R. Keen, E. Hsiao, R. J. Pignolo,Early clinical observations on the use of imatinib mesylate in FOP: A report of seven cases. Bone 109,276–280 (2018). [DOI] [PubMed] [Google Scholar]
- 51.W. R. Grither, G. D. Longmore,Inhibition of tumor-microenvironment interaction and tumor invasion by small-molecule allosteric inhibitor of DDR2 extracellular domain. Proc. Natl. Acad. Sci. U.S.A. 115,E7786–E7794 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.M. R. Convente, H. Wang, R. J. Pignolo, F. S. Kaplan, E. M. Shore,The immunological contribution to heterotopic ossification disorders. Curr. Osteoporos. Rep. 13,116–124 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.M. Sorkin, A. K. Huber, C. Hwang, W. F. Carson IV, R. Menon, J. Li, K. Vasquez, C. Pagani, N. Patel, S. Li, N. D. Visser, Y. Niknafs, S. Loder, M. Scola, D. Nycz, K. Gallagher, L. K. McCauley, J. Xu, A. W. James, S. Agarwal, S. Kunkel, Y. Mishina, B. Levi,Regulation of heterotopic ossification by monocytes in a mouse model of aberrant wound healing. Nat. Commun. 11,722 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.X. Wang, F. Li, L. Xie, J. Crane, G. Zhen, Y. Mishina, R. Deng, B. Gao, H. Chen, S. Liu, P. Yang, M. Gao, M. Tu, Y. Wang, M. Wan, C. Fan, X. Cao,Inhibition of overactive TGF-β attenuates progression of heterotopic ossification in mice. Nat. Commun. 9,551 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.F. Wei, Y. Zhou, J. Wang, C. Liu, Y. Xiao,The immunomodulatory role of BMP-2 on macrophages to accelerate osteogenesis. Tissue Eng. Part A 24,584–594 (2018). [DOI] [PubMed] [Google Scholar]
- 56.S. Agarwal, S. J. Loder, D. Cholok, J. Li, G. Bian, S. Yalavarthi, S. Li, W. F. Carson, C. Hwang, S. Marini, C. Pagani, N. Edwards, M. J. Delano, T. J. Standiford, J. S. Knight, S. L. Kunkel, Y. Mishina, P. A. Ward, B. Levi,Disruption of neutrophil extracellular traps (NETs) links mechanical strain to post-traumatic inflammation. Front. Immunol. 10,2148 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.T. Stuart, A. Srivastava, S. Madad, C. A. Lareau, R. Satija,Single-cell chromatin state analysis with Signac. Nat. Methods 18,1333–1341 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Y. Yu, Y. Chau,One-step "click" method for generating vinyl sulfone groups on hydroxyl-containing water-soluble polymers. Biomacromolecules 13,937–942 (2012). [DOI] [PubMed] [Google Scholar]
- 59.G. C. J. Brown, K. S. Lim, B. L. Farrugia, G. J. Hooper, T. B. F. Woodfield,Covalent incorporation of heparin improves chondrogenesis in photocurable gelatin-methacryloyl hydrogels. Macromol. Biosci. 17, (2017). [DOI] [PubMed] [Google Scholar]
- 60.D. L. Matera, W. Y. Wang, M. R. Smith, A. Shikanov, B. M. Baker,Fiber density modulates cell spreading in 3D interstitial matrix mimetics. ACS Biomater Sci. Eng. 5,2965–2975 (2019). [DOI] [PubMed] [Google Scholar]
- 61.A. Boudaoud, A. Burian, D. Borowska-Wykret, M. Uyttewaal, R. Wrzalik, D. Kwiatkowska, O. Hamant,FibrilTool, an ImageJ plug-in to quantify fibrillar structures in raw microscopy images. Nat. Protoc. 9,457–463 (2014). [DOI] [PubMed] [Google Scholar]
- 62.J. Schindelin, I. Arganda-Carreras, E. Frise, V. Kaynig, M. Longair, T. Pietzsch, S. Preibisch, C. Rueden, S. Saalfeld, B. Schmid, J. Y. Tinevez, D. J. White, V. Hartenstein, K. Eliceiri, P. Tomancak, A. Cardona,Fiji: An open-source platform for biological-image analysis. Nat. Methods 9,676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.V. V. Artym, K. Matsumoto,Imaging cells in three-dimensional collagen matrix. Curr. Protoc. Cell Biol. Chapter 10,Unit 10.18.1-20 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.S. Agarwal, S. J. Loder, D. Cholok, J. Peterson, J. Li, C. Breuler, R. Cameron Brownley, H. Hsin Sung, M. T. Chung, N. Kamiya, S. Li, B. Zhao, V. Kaartinen, T. A. Davis, A. T. Qureshi, E. Schipani, Y. Mishina, B. Levi,Scleraxis-lineage cells contribute to ectopic bone formation in muscle and tendon. Stem Cells 35,705–710 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.S. J. Han, E. V. Azarova, A. J. Whitewood, A. Bachir, E. Guttierrez, A. Groisman, A. R. Horwitz, B. T. Goult, K. M. Dean, G. Danuser,Pre-complexation of talin and vinculin without tension is required for efficient nascent adhesion maturation. eLife 10,e66151 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.S. J. Han, Y. Oak, A. Groisman, G. Danuser,Traction microscopy to identify force modulation in subresolution adhesions. Nat. Methods 12,653–656 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.J. P. Butler, I. M. Tolić-Nørrelykke, B. Fabry, J. J. Fredberg,Traction fields, moments, and strain energy that cells exert on their surroundings. Am. J. Physiol. Cell Physiol. 282,C595–C605 (2002). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S12
Table S2
Table S1