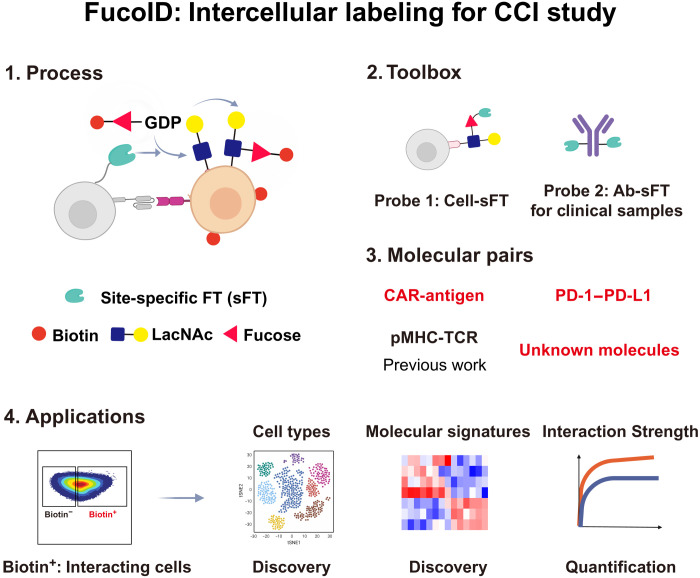
Abstract
FucoID is an intercellular proximity labeling technique for studying cell-cell interactions (CCIs) via fucosyltransferase (FT)–meditated fucosyl-biotinylation, which has been applied to probe antigen-specific dendritic cell (DC)–T cell interactions. In this system, bait cells of interest with cell surface–anchored FT are used to capture the interacting prey cells by transferring a biotin-modified substrate to prey cells. Here, we leveraged FucoID to study CCIs directed by different molecular pairs, e.g., programmed cell death protein-1(PD-1)/programmed cell death protein-ligand-1 (PD-L1), and identify unknown or little studied CCIs, e.g., the interaction of DCs and B cells. To expand the application of FucoID to complex systems, we also synthesized site-specific antibody-based FT conjugate, which substantially improves the ability of FucoID to probe molecular signatures of specific CCI when cells of interest (bait cells) cannot be purified, e.g., in clinical samples. Collectively, these studies demonstrate the general applicability of FucoID to study unknown CCIs in complex systems at a molecular resolution.
FucoID is a platform that can be used to discover and quantify cell-cell interactions mediated by diverse molecular pairs.
INTRODUCTION
Cell-cell interactions (CCIs) initiate intercellular communications to maintain homeostasis and coordinate physiological functions in multicellular organisms (1, 2). In many circumstances, abnormal cellular interactions are the cause of severe disease and disorder (3, 4). As is known for decades, many autoimmune diseases are triggered by the interactions of T cells with normal cells due to aberrant indiscrimination of self-antigens (5–7). On the other hand, understanding how cells interact with each other may channel the development of therapeutic agents that empower the precise manipulation of particular CCIs in human patients (8). Preeminent examples include bispecific antibodies and chimeric antigen receptor (CAR)–T cells (CAR-Ts) (9, 10). Hence, how to precisely identify CCIs for their characterization and quantification has attracted the attention of prospectors aiming at expanding this burgeoning field with the increasing demands from both fundamental research and translational medicine.
A fair number of approaches for studying CCIs have been reported, ranging from cellular imaging tools to transcriptomic data analysis–based bioinformatic techniques (11). Microscopic imaging widely used to visualize CCIs has provided critical information regarding spatial organizations and dynamic movements during an interaction (8, 12). However, imaging techniques alone cannot reveal molecular details of previously unknown CCIs. On the other side, with the advancement of single-cell RNA sequencing (scRNA-seq), databases and algorithms, such as CellPhoneDB, CellChat, iTALK, and NicheNet, have been developed for CCI prediction based on the bioinformatic analysis of single-cell gene expressions (13–16). However, these putative interactions predicted by bioinformatic analysis require further validation and are largely affected by the selection of appropriate parameters, which often can only be used as a reference.
Chemical biology tools based on proximity labeling are the game changers of this field as they not only provide tools to identify unknown CCIs in complex systems but also enable the discovery of molecular signatures involved in such interactions (17–19). Moreover, proximity labeling also provides a means to quantify the strength of CCIs, which is particularly important in understanding the mechanism of the interaction-induced signaling transduction, e.g., T cell receptor (TCR) activation in T cells (20, 21). Although proximity labeling systems of TurboID, APEX2, and horseradish peroxidase have been widely used in different applications, the contact-independent reactive species in this system is limited in probing CCIs (11).
Recently, we developed FucoID as a genetic engineering–free tool for capturing contact-dependent CCIs via the fucosyl-biotinylation of N-acetyl-d-lactosamine (LacNAc), a common cell surface disaccharide, located at the interface of interacting cells (22). In this system, bait cells of interest are chemoenzymatically functionalized with a fucosyltransferase (FT) on the cell surface and used to capture the interacting prey cells in a cell mixture by transferring a biotin-modified FT donor substrate guanosine diphosphate (GDP)–fucose (Fuc)–biotin (GF-Biotin) to the LacNAc acceptor on the glycocalyx of prey cells (23, 24). FucoID enables the facile detection of tumor-specific antigen (TSA)–reactive T cells from tumor-infiltrating lymphocytes (TILs). Its application to a clinical setting may accelerate the pace for the discovery of TSA-reactive TCR and lower the cost and accessibility of personalized TCR relevant therapy (25–27). A distinct advantage of contact-dependent proximity labeling for studying CCI is its small labeling radius, which accounts to the requirement for direct physical contact of the enzyme, donor substrate, and acceptor substrate. This unique property promotes us to further develop and expand the toolbox of FucoID for studying CCIs beyond dendritic cell (DC)–T cell interactions (Fig. 1), in which specific and quantitative tools are less reported.
Fig. 1. Intercellular labeling system for studying CCIs with molecular resolution.
Schematic diagram of the FucoID system for detecting, isolating, and analyzing a broad spectrum of CCIs. The FucoID system contains two probes, cell-sFT (probe 1) and Ab-sFT (probe 2), built on the sFT conjugates. As a specific, versatile, and quantitative proximity labeling system, FucoID enables the quantification of the strength and the discovery of molecular signatures of CCIs through the multiple analyzing methods after labeling and isolation, e.g., flow cytometry and RNA-seq analyses.
Here, we upgraded the FucoID platform by constructing site-specific cell and antibody-conjugate as a next generation of FT probes (cell-sFT and Ab-sFT) to enable FucoID to be applied to a broader setting. We found that not only can cell-sFT be used to probe DC–T cell interactions but this platform can also be applied to probe cancer cell–T cell and DC–B cell interactions. These types of CCIs were governed by chimeric receptor-antigen interaction, programmed cell death protein 1 (PD-1)–programmed death-ligand 1 (PD-L1) interaction or unknown interacting molecular pairs. Furthermore, using the newly developed Ab-sFT reagents, we realized FucoID in human patient samples using the endogenous cancer cells as bait cells without the need of their purification. The prey cells in the unknown CCI system, identified by FucoID, were characterized by RNA-seq or flow cytometry analysis to dissect distinct molecular signatures, opening the avenue to study a broader spectrum of CCIs in complex systems.
RESULTS
Development of site-specific FucoID probes
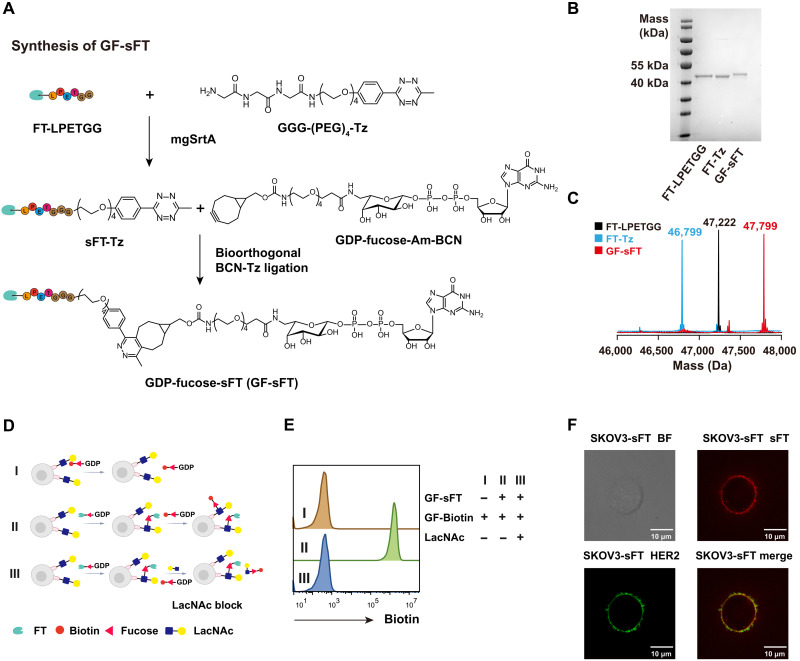
Our previously reported GDP-Fuc-FT (GF-FT) conjugates were constructed using nonspecific N-hydroxysuccinimide ester–based lysine coupling reactions. This approach leads to varying numbers of GDP-Fuc per FT in different batches (fig. S1, A to D), which may raise problems in consistency for translational study. To address this issue, we sought to construct an sFT conjugate using sortase-mediated transpeptidation (sortagging). Briefly, FT with a C-terminal LPETGG tag was labeled by a tetrazine (Tz) group via the sortase-mediated ligation (Fig. 2A and fig. S1F) (18). The resulting FT-Tz conjugate was characterized by SDS–polyacrylamide gel electrophoresis (PAGE) and mass spectrometry as a uniform product without batch effects (Fig. 2, B and C). GDP-Fuc functionalized with the biorthogonal reaction handle bicyclo[6.1.0]nonyne (BCN) (GF-BCN; fig. S2) was then used to react with FT-Tz to form the site-specific GDP-Fuc-sFT (GF-sFT) via the inverse electron demand Diels-Alder (IEDDA) reaction.
Fig. 2. Construction of the site-specific GF-sFT conjugate.
(A) Chemoenzymatic synthesis of GF-sFT. sFT is constructed via the mgSrtA-mediated transpeptidation to install a C-terminal Tz group. The following IEDDA reaction of sFT-Tz and GDP-Fuc-Am-BCN gives the final product of GF-sFT. (B and C) Characterization of FT-LPETGG, FT-Tz, and GF-sFT by SDS-PAGE gel and mass spectrometry. (D and E) Self–fucosyl-biotinylation of NK92 cells by cell surface–anchored FT. NK92 cells were incubated GF-sFT (0.2 mg/ml) to install FT on cell surface, followed by wash and incubation with GF-Biotin or GF-Biotin and LacNAc for blocking labeling. (F) Confocal imaging of sFT and HER2 on SKOV3-sFT. BF, bright field. Scale bars, 10 μm.
As GF-sFT is a self-catalyst that transfers FT onto the cell surface by forming a Fuc–N-acetylglucosamine linkage within the LacNAc unit, we characterized the activity of GF-sFT by incubating NK92 cells with different concentrations of GF-sFT. NK92 cells engineered with FT were then treated with GF-Biotin to trigger the self–fucosyl-biotinylation on the cell surface. The detection of the biotinylation signal was realized by streptavidin-conjugated fluorescent probes. As shown in Fig. 2D, the self-biotinylation signal via the FT-triggered unnatural fucosylation was specifically detected on the cell surface, which could be fully blocked by adding the free glycan acceptor substrate, LacNAc (Fig. 2E). Moreover, the self–fucosyl-biotinylation signal was positively correlated with the GF-sFT concentration used in the first step, ranging from 0.025 to 0.8 mg/ml, which was saturated at 0.2 mg/ml (fig. S1E). We then used GF-sFT (0.2 mg/ml) to label a human epidermal receptor 2–positive (HER2+) ovarian cancer cell line, SKOV3, followed by staining with a polyclonal anti-FT antibody. As revealed by confocal imaging, FT is uniformly distributed on the cell surface of the labeled cells (Fig. 2F).
Characterization of cell-sFT probes in detecting DC–T cell interactions
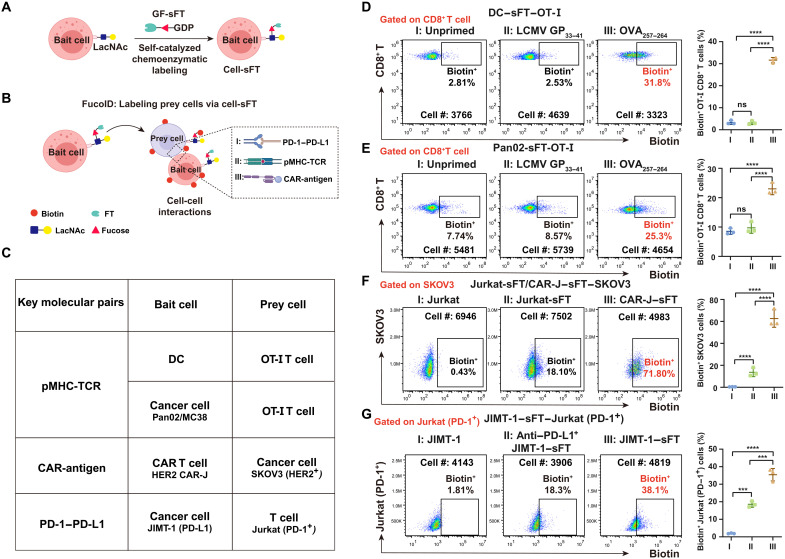
In our FucoID protocol, prey cells from a heterogeneous cell mixture are detected by the externally added bait cells that are engineered to display FT on the cell surface (Fig. 3A). The interaction between prey cells and bait cells leads to prey cell fucosyl-biotinylation catalyzed by FT anchored on bait cells (Fig. 3B). The labeled prey cells can then be isolated for the subsequent analysis. To validate the activity and efficiency of GF-sFT in the FucoID system, we first modified DC with GF-sFT and used DC-sFT to label antigen-specific T cells that participate in antigen-driven interactions (Fig. 3D and fig. S3). We pulsed sFT-labeled DC with the ovalbumin (OVA)257–264 peptide (SIINFEKL) that is recognized by the transgenic TCR of CD8+ OT-I T cells in the context of the major histocompatibility complex (MHC), H-2Kb. We then initiated the interaction by mixing naïve OT-I T cells with the SIINFEKL peptide–primed DC-sFT cells. After coculturing for 2 hours, the addition of GF-Biotin enabled the proximity labeling of the interacting OT-I T cells by DC-sFT within 30 min. Robust fucosyl-biotinylation was found on the interacting CD8+ T cells with a signal-to-background ratio of 32% (Fig. 3D). By contrast, unprimed and DC-sFT cells primed with the irrelevant antigenic peptide GP33–41 derived from lymphocytic choriomeningitis virus only induced the minimum level of labeling on OT-I T cells (~2.5%; Fig. 3D). Notably, the installation of FT on DCs did not alter its property of T cell activation (fig. S3, B and C).
Fig. 3. Expansion of the CCI systems studied by FucoID under the control of different molecular pairs.
(A and B) Schematic illustration of cell-sFT–based FucoID system for detecting of CCIs. (C) A list of the CCIs evaluated. FT was installed onto bait cells. Key molecular pairs designate the molecules directing the interaction. (D and E) DC-sFT and Pan02-sFT cells were primed by OVA257–264 to interact with the cognate OT-I T cells. Flow cytometry–based quantification of antigen-specific Fuc-biotin of CD8+ T cells by DC-sFT (D) or Pan02-sFT (E) primed with OVA257–264 in OT-I splenocytes. n = 3. Interaction time, 2 hours; labeling time, 30 min. The background is defined as the signal produced on OT-I CD8+ T cells when incubated with DC or Pan02 without membrane-anchored sFT. (F) Flow cytometry–based quantification of interaction-dependent fucosyl-biotinylation of HER2+ SKOV3 cells using Jurkat, Jurkat-sFT, and CAR-J–sFT. n = 3. Interaction time, 1 hour; labeling time, 30 min. The background is defined as the signal produced on SKOV3 incubated with Jurkat without membrane-anchored sFT. (G) Flow cytometry–based quantification of interaction-dependent Fuc-biotin of Jurkat–PD-1 cells using JIMT-1–sFT (PD-L1+) with or without PD-L1 blockade treatment. n = 3. Interaction time, 1 hour; labeling time, 30 min. The background is defined as the signal produced on Jurkat incubated with JIMT-1 without membrane-anchored sFT. n, number of biological repeats. ns, P > 0.05; ***P < 0.001; ****P < 0.0001.
Application of cell-sFT in detecting CCIs beyond DCs and T cells
To expand the application of FucoID beyond probing antigen-specific DC–T cell interactions, we sought to explore other types of bait cells for detecting CCIs. Because cancer cells are recognized and attacked by T cells via the antigen-specific peptide MHC (pMHC)–TCR recognition, we investigate the possibility of using cancer cells as bait cells to capture cognate T cells. In this case, cancer cells work as antigen-presenting cell (APC) to present antigens that pMHC on cancer cells then interact with TCR on T cells. We functionalized Pan02, a mouse pancreatic cancer cell line expressing H-2Kb MHC-I, with sFT, pulsed the FT-labeled Pan02 cells with the SIINFEKL peptide, and then initiated the interaction-dependent labeling with naïve CD8+ OT-I T cells. As shown in Fig. 3E, OT-I T cells were strongly labeled by Pan02-sFT primed with OVA257–264 (Biotin+, ~25%), but only very weakly labeled by unprimed Pan02-sFT or Pan02-sFT primed with GP33–41(Biotin+, ~7%). Notably, the fucosyl-biotinylation was also highly correlated with CD69 expression in response to different concentrations of SIINFEKL (fig. S4, B and C). Similar results were observed when using the sFT-functionalized MC38 murine colon carcinoma as cell probes (fig. S4, B and D), demonstrating the general applicability of FucoID in studying antigen-dependent cancer cell–T cell interactions. We also evaluated the influence of FT labeling on the killing ability of T cell. The results demonstrated that FT-labeled T cells showed uninfluenced cytotoxic effect against cancer cells primed with OVA257–264 (fig. S4, E and F).
All FucoID-enabled labeling we explored so far was governed by the interactions of pMHC and TCR (Fig. 3C). As an pMHC-independent therapeutic modality, CAR-Ts combine a tumor antigen recognition domain with intracellular signaling domains to enable the recognition and killing of target tumor cells. To assess whether we could use FucoID to probe CAR-T–target cell interactions, we constructed CAR-Ts by introducing an anti-HER2 CAR into Jurkat, a human T cell line, via lentivirus transduction (fig. S5, A and B). Unmodified Jurkat cells and CAR-Jurkat (CAR-J) cells were then modified with GF-sFT and used to interrogate the interactions with SKOV3 (HER2+). Compared to Jurkat-sFT, CAR-J–sFT induced notably higher fucosyl-biotinylation of SKOV3 cells (72% versus 18%; Fig. 3F and fig. S5, C to E). Besides the molecular pairs of antibody/antigen, we also tested the feasibility of applying the upgraded FucoID system to study CCIs directed by ligand-receptor interactions. As proof of principle, we chose the well-studied interactions of PD-L1 and PD-1. The PD-1/PD-L1 pathway represents an adaptive immune resistance mechanism exploited by tumor cells in response to endogenous T cell–mediated antitumor activity. We constructed a stable cell line on Jurkat cells that express high levels of PD-1 (Jurkat-PD-1; fig. S6). Next, JIMT-1, a human breast carcinoma cell line that expresses high levels of PD-L1, was functionalized with sFT. When JIMT-1–sFT was incubated with Jurkat–PD-1 to trigger FucoID, 38% of Jurkat–PD-1 cells were labeled by fucosyl-biotinylation, whereas only 18% of Jurkat–PD-1 cells were labeled in the presence of a PD-L1 blocking antibody, which is similar to that of untransduced Jurkat cells (Fig. 3G and fig. S6). These results strongly suggest that the PD-1/PD-L1 molecular pairs critically contribute to the interactions between JIMT-1 and Jurkat–PD-1 cells.
Determining the intensity and specificity of the interaction between CAR-J and cancer cells
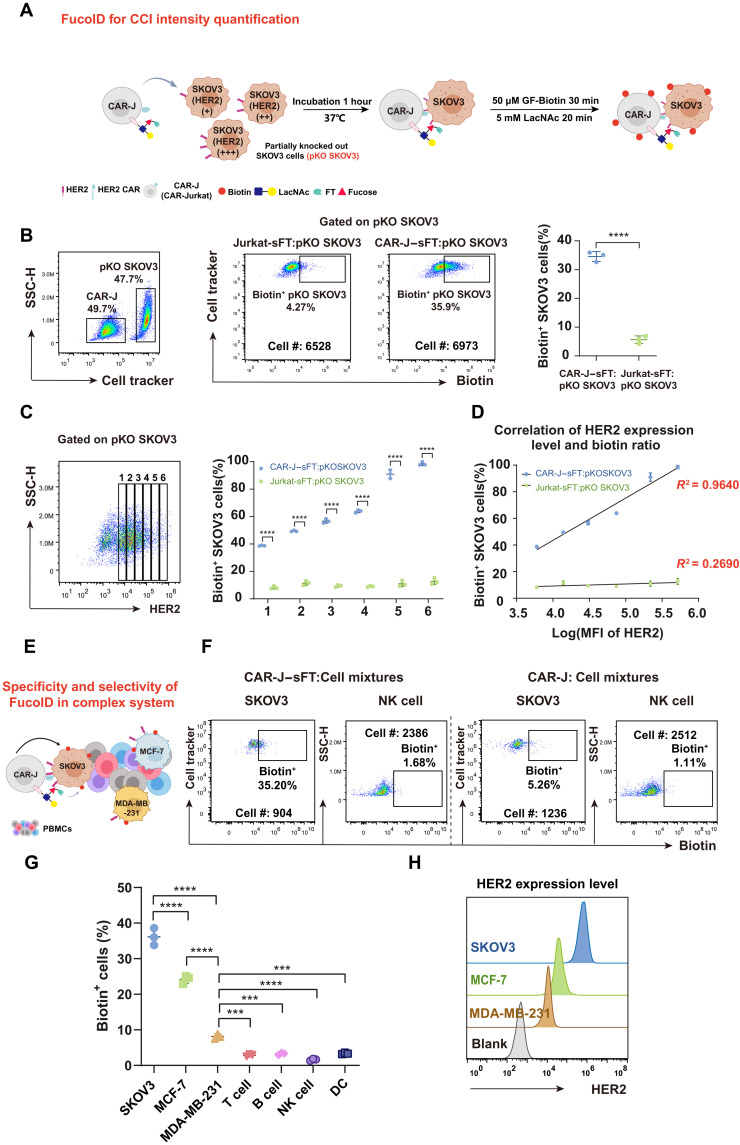
Quantitatively distinguishing a strong interaction from a weak interaction is essential to gain a molecular-level understanding of CCIs. To explore whether FucoID can be used for the quantitative analysis of CCIs, we used a model co-culture system of CAR-J and SKOV3 as the proof of concept (Fig. 4A). First, we established SKOV3 cells with varying HER2 expression levels via transfecting human ERBB2-targeting CRISPR-Cas9 ribonucleoprotein (RNP). Because of the high ERBB2 copy number of SKOV3 and incomplete editing resulting from transient RNP, treated SKOV3 cells showed a broad range of HER2 expression levels (partial HER2 knockout), which was named pKO SKOV3. Next, CAR-J–sFT and Jurkat-sFT cells, respectively, were cocultured with pKO SKOV3 cells to trigger the FucoID labeling. The ratio of fucosyl-biotinylation of pKO SKOV3 induced by CAR-J–sFT was ~36%, whereas the ratio of fucosyl-biotinylation triggered by Jurkat-sFT was less than 10% (Fig. 4B). We then divided the HER2+ SKOV3 cells of pKO SKOV3 into six groups according to the expression level of HER2 and analyzed the ratios of fucosyl-biotinylation in these groups (Fig. 4C and fig. S7A). As expected, the fucosyl-biotinylation ratios induced by CAR-J–sFT were positively correlated with the expression levels of HER2 among these six groups (Fig. 4D), which nicely indicates that the intensity of CCIs could be detected by FucoID quantitatively. Next, to further demonstrate the specificity and selectivity of FucoID in a complex system, we added the cancer cells with different HER2 expression levels into human peripheral blood mononuclear cells (PBMCs), including SKOV3 (HER2, high), MCF-7 (HER2, medium), and MDA-MB-231 (HER2, low; Fig. 4, E and H). Fucosyl-biotinylation was only detected on the cancer cells, while almost all PBMCs were left unlabeled by CAR-J–sFT (Fig.4G and fig. S7C). Moreover, the ratio of fucosyl-biotinylation was consistent with the expression levels of HER2 on cancer cells, in which SKOV3, MCF-7, and MDA-MB-231 showed a signal-to-background labeling ratio of 35, 25, and 7%, respectively (Fig. 4, F and G). Together, the FucoID labeling system displayed good specificity and selectivity for detecting CCIs governed by molecular pairs beyond pMHC-TCR and the potential to quantify CCI intensity in a complex system.
Fig. 4. Characterization of the selectivity and specificity of CAR-J–sFT probe in CAR-J–SKOV3 interaction system.
(A) Workflow of analyzing CCI intensity in the model of CAR-antigen–mediated interactions. (B) Representative flow cytometric plots and statistics showing biotinylation of pKO SKOV3 cells mediated by CAR-J–sFT and Jurkat-sFT. n = 3. The background is defined as the signal produced on pKO SKOV3 incubated with Jurkat without membrane-anchored sFT. (C) Statistical analysis of labeling ratio among six groups of pKO SKOV3 cells with different HER2 expressions. (D) Correlation analysis of biotin ratio and HER2 expression level in the six groups divided in (C). (E) Schematic illustration of CAR-J–sFT as a probe for detecting the specificity and selectivity of FucoID in a complex system. (F and G) Flow cytometry–based quantification of selective and specific labeling of SKOV3, MCF-7, and MDA-MB-231 in cell mixtures by CAR-J–sFT (doped with SKOV3, MCF-7, and MDA-MB-231 in PBMCs at a ratio of 1:1:1:10). n = 3. NK, natural killer. (H) Flow cytometric analysis of expression level of HER2 on SKOV3, MCF-7, and MDA-MB-231. ***P < 0.001; ****P < 0.0001.
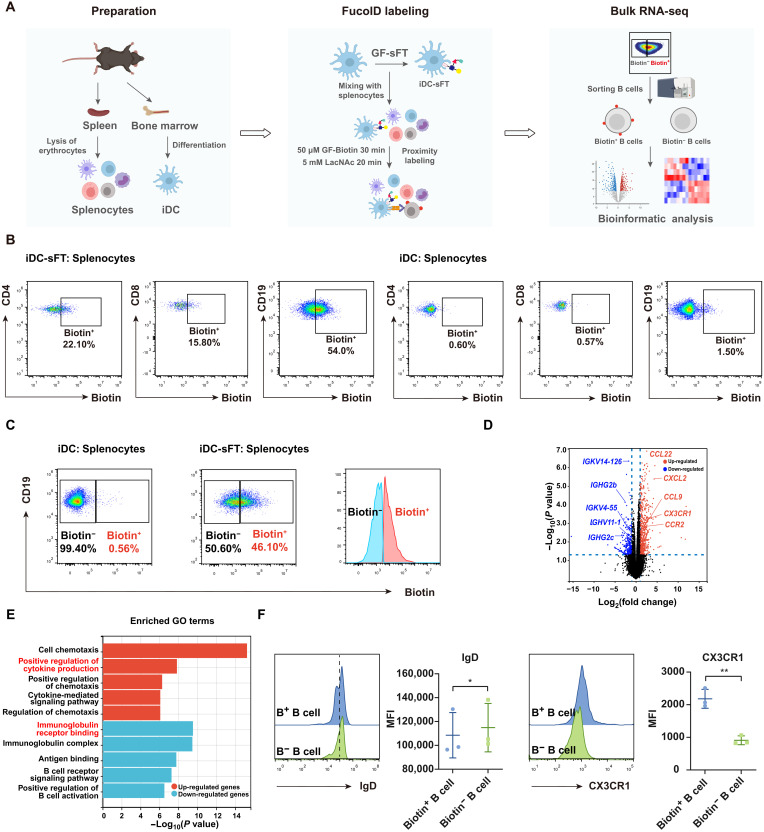
Profiling unknown interactions in splenocytes by FucoID
In the aforementioned experiment, we also observed weak fucosyl-biotinylation in the group that known receptor-mediated interactions did not exist, e.g., Jurkat and SKOV3 cells (Biotin+, ~18%), suggesting that there may exist interactions controlled by unknown molecular pairs, which is worthy of further investigation. Thus, we further explored the utilization of cell-sFT probes to characterize a broader spectrum of CCIs to determine whether unknown or little-studied CCIs could be identified. We chose immature DCs (iDCs) as bait cells to profile their interactions with immune cells in mouse splenocytes due to its rich source of diversified immune cell types (Fig. 5A). Specifically, unprimed iDC-sFT was incubated with splenocytes in the presence of GF-Biotin to capture transient interactions, which is distinct from our published procedure, in which GF-Biotin was introduced upon the formation of long-term, stable cell-cell contacts. To our surprise, after 30 min, B cells exhibited a significantly higher labeling ratio (54%) than both CD4+ and CD8+ T cells (~20%; Fig. 5B). Previously, the DC–B cell interaction as a venue to transfer antigens from DCs to B cells and trigger B cell activation and maturation has not been thoroughly studied (28–30). The preferred labeling of B cells by iDCs promoted us to investigate the possible mechanism governing their interactions. We sorted the interacting (Biotin+) and bystander (Biotin−) B cells. Bulk RNA-seq analysis of these samples allowed us to compare their gene expression profiles (Fig. 5, A and C, and fig. S9). Significantly, compared to Biotin− B cells, the Biotin+ B cell subset was found to expressed lower levels of immunoglobulin receptor related genes, including IGKV14-126, IGHG2b, IGKV4-55, IGHV11-1, IGHG2c, IGHV5-39, IGHV11-2, and IGHG3. By contrast, Biotin+ B cells had higher expression of regulation of cytokine related genes, including CCL22, CXCL2, CCL9, CX3CR1, CCR2, CCR6, and CCL5 (Fig. 5D). These results were also confirmed by pathway and function enrichment analysis of their different gene signatures (fig. S9B). Gene Ontology (GO) enrichment analysis suggested that down-regulated genes in Biotin+ B cells were enriched in immunoglobulin receptor binding and up-regulated genes in Biotin+ B cells were enriched in positive regulation of cytokine production (Fig. 5E and fig. S9C). We then evaluated the protein expression levels of a few significantly changed genes, and confirmed that CX3CR1 was up-regulated and immunoglobulin D (IgD) was down-regulated in Biotin+ B cells (Fig. 5F). These data were consistent with the sequencing results of bulk RNA-seq. As well documented, these genes play critical roles in the maturation of B cells (31). Recently, a study reported that DCs interact with B cells through trogocytosis in splenocytes (32). Accordingly, we also observed a weak trogocytosis taking place through which B cell surface markers could be transferred and detected on DCs but not vice versa (fig. S10). Together, these results strongly suggest that down-regulation of immunoglobulin receptors and up-regulation of cytokine production on B cells are involved in the augmented interactions between iDCs and B cells.
Fig. 5. Use of the iDC-sFT probe to identify DC–B cell interactions in splenocytes.
(A) Schematic workflow for the preparation, labeling and analysis of splenocytes interacting with iDCs. (B) Representative flow cytometric plots showing the fucosyl-biotinylation of CD4+ T cells, CD8+ T cells, and B cells in splenocytes mediated by iDC-sFT and iDCs. iDC-sFT or iDCs were mixed with splenocytes under 50 μM GF-Biotin for 30 min. The background is defined as the signal produced on splenocytes by incubating iDCs with splenocytes. (C) Flow cytometric gating strategy for isolating Biotin+ (interacting) and Biotin− (bystander) B cells via fluorescence-activated cell sorting (FACS). (D) Volcano plots of differentially expressed genes between Biotin+ and Biotin− B cells. n = 4. Membrane protein related genes are highlighted. (E) GO terms enriched by the up-regulated and down-regulated genes between Biotin+ and Biotin− B cells. Bar represents the P value. n = 4. (F) Representative flow cytometric histograms and statistics showing IgD and CX3CR1 expressions on Biotin+ (B+) and Biotin− (B−) B cells. MFI, mean fluorescence intensity. *P < 0.05; **P < 0.05.
Development of Ab-sFT probes for applying FucoID to clinically relevant samples
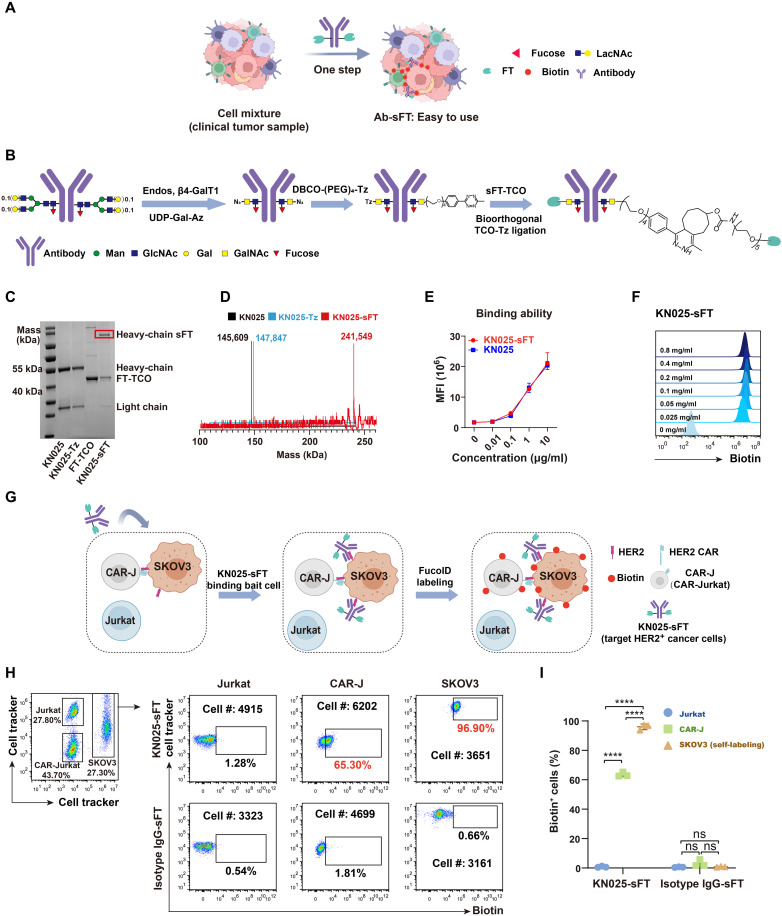
Because the cell-based FT probes require the preinstallation of FT onto isolated bait cells, it is challenging to be applied to systems in which bait cells to be used are primary cells that are hard to purify and isolate (Fig. 6A). To overcome this obstacle, we developed antibody-based probes to expand the toolbox of FucoID. We introduced Ab-sFT as an alternative probe to cell-sFT via constructing site-specific anti–HER2-sFT conjugate as a proof of concept (Fig. 6B). In this design, FT with a C-terminal LPETGG tag was labeled by a trans-cyclooctene (TCO) group via the sortase-mediated ligation [GGG-(PEG)5-TCO as substrate]. An anti-HER2 antibody (KN025) targeting a different epitope of Herceptin on HER2 was site-specially modified by glycoengineering via the cascade enzymatic reaction to introduce an unnatural sugar containing the azide group (N-azidoacetylgalactosamine, GalNAz). The given product of site-specific KN025-(GalNAz)2 was further modified by dibenzocyclooctyne (DBCO)-(PEG)4-Tz via click reaction to obtain the site-specific KN025-(Tz)2. Last, the sFT-TCO was mixed with the site-specific KN025-(Tz)2 conjugate directly to form the site-specific KN025-sFT conjugate via the IEDDA reaction (Fig. 6B and fig. S11). The site specificity and enzyme per antibody number of this antibody-enzyme (FT) conjugate were characterized by the SDS-PAGE and mass spectrometry (Fig. 6, C and D), in which the clear dual modification of FT enzyme on KN025 Fc glycan demonstrated the power of IEDDA reaction (33). Furthermore, sFT-modified KN025 was found to exhibit similar antigen-binding capabilities as compared to the parent antibody (Fig. 6E), and KN025-sFT (0.1 mg/ml) was sufficient to mediate saturating labeling of NK92 cells with GF-Biotin (Fig. 6F).
Fig. 6. Development of Ab-sFT for detecting CCIs.
(A) Schematic illustration of the application of Ab-sFT probes to study clinical samples. (B) Characterization of synthesis process of Ab-sFT. (C and D) Characterization of KN025, KN025-Tz, and KN025-sFT by (C) SDS-PAGE and (D) mass spectrometry. (E) Titration of the HER2 binding ability of KN025-sFT. (F) Characterization of FT activity of KN025-sFT by labeling LacNAc on NK92 cells with GF-Biotin. (G) Schematic overview of probing the interactions between CAR-J/Jurkat-SKOV3 by KN025-sFT. CAR-J, Jurkat, and SKOV3 were mixed as a model of CCI. KN025-sFT was added to the system to label HER2+ cells and their interacting cells. (H) Flow cytometric analysis and (I) summary statistics of the interaction-dependent fucosyl-biotinylation of Jurkat, CAR-J, and SKOV3 in the model system. n = 3. Interaction time, 1 hour; labeling time, 30 min. The background is defined as the signal produced on cells treated with anti-HER2 antibody. ns, P > 0.05; ****P < 0.0001.
To assess whether KN025-sFT can be directly administered to a cell mixture to initiate the interaction-dependent labeling, we mixed SKOV3 with Jurkat and CAR-J (1:1:1) as a model system (Fig. 6G) followed by the addition of KN025-sFT. In this system, the interaction between CAR-J and SKOV3 cells was stronger than that between Jurkat and SKOV3 cells. After removing the unbound KN025-sFT, GF-Biotin was added to trigger FucoID. Flow cytometry analysis revealed that 97% of SKOV3 and 65% of CAR-J were selectively labeled by fucosyl-biotinylation, whereas only 2% unmodified Jurkat cells were labeled (Fig. 6, H and I). Notably, none of the cells in the isotype IgG-sFT administered control group were labeled. We then titrated the concentrations of KN025-sFT and found that KN025-sFT (10 μg/ml) could label nearly all CAR-J cells that interact with SKOV3 but left Jurkat cells untouched (fig. S12).
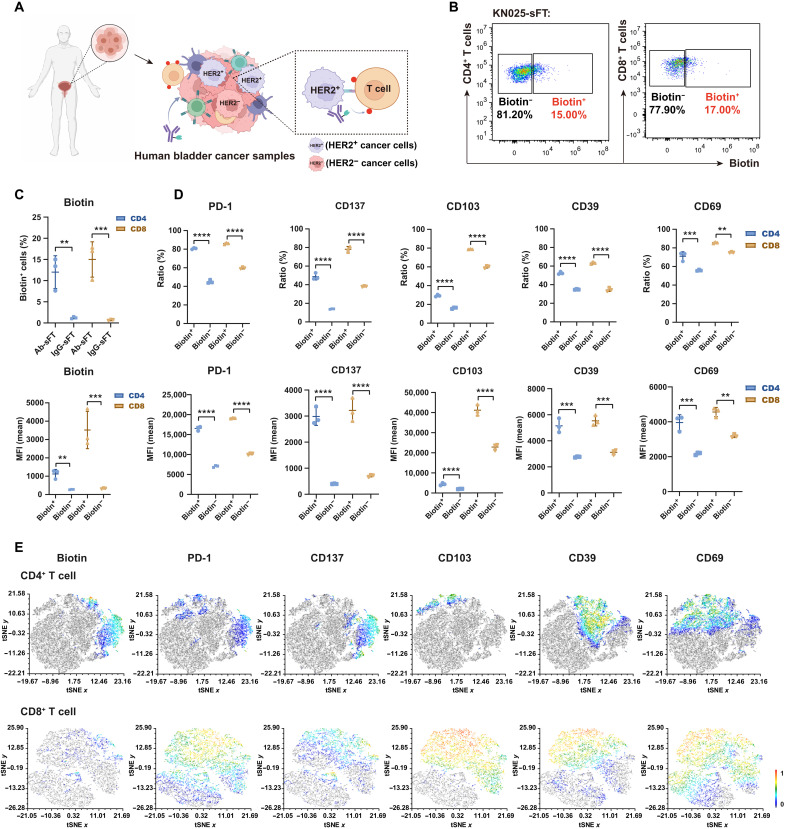
The success of applying KN025-sFT to probe interactions between CAR-J and cancer cells prompted us to interrogate the interactions of primary HER2+ cancer cells with other cells present in human bladder carcinoma (Fig. 7A). This is an excellent test of the scope of FucoID because primary cancer cells are hard to be purified as bait cells. Following the same procedures as described above, we added KN025-sFT into the single-cell suspensions prepared from bladder tumors by transurethral resection. To our delight, 15% of CD4+ T cells and 17% of CD8+ T cells from TILs were labeled by fucosyl-biotinylation (Fig. 7, B and C). By contrast, under the same condition, isotype control IgG-sFT labeled none of these cells (Fig. 7C and fig. S13C). We then characterized the molecular phenotypes of interacting (Biotin+) and bystander (Biotin−) T cells using a panel of T cell activation or exhaustion markers, including CD137, CD103, PD-1, CD39, and CD69 (34–39). We found that the expression of these biomarkers was significantly higher in Biotin+ group than that in Biotin− group (Fig. 7D and fig. S13D), suggesting that T cells interacting with HER2+ cancer cells displaying an activation/dysfunction phenotype. The t-stochastic neighbor embedding (tSNE) plot analysis revealed that compared with the expression of other functional markers, the expression of CD137 had a more pronounced overlap with the signal of fucosyl-biotinylation on CD4+ T cells (Fig. 7E).
Fig. 7. Detection of cells interacting with HER2+ bladder cancer cells in a clinical sample.
(A) Schematic overview of studying the interactions between HER2+ cancer cells and T cells via KN025-sFT in the cell suspensions of human bladder cancer patient samples. (B and C) Flow cytometric analysis and summary statistics of the interaction-dependent fucosyl-biotinylation of CD4+ and CD8+ T cells via KN025-sFT in bladder cancer samples. n = 3. Incubation time, 1 hour; labeling time, 30 min. (D) The expression ratios (top) and intensities (bottom) of functional markers in biotin+/− CD4+ T cells and CD8+ T cells according to flow cytometric analysis. n = 3. (E) Representative tSNE map showing the correlation between functional phenotypes and biotin signal on CD4+ T cells and CD8+ T cells. Human sample from one patient, three biological repeats; **P < 0.05; ***P < 0.001; ****P < 0.0001.
DISCUSSION
In summary, we have upgraded the FucoID platform by introducing two new reagents—GF-sFT and Ab-sFT conjugates. GF-sFT has been further used to construct different cell-based probes, such as CAR-T–sFT and cancer cell–sFT, demonstrating its general applicability, while Ab-sFT allows the application of FucoID to a native context. Featuring easy preparation and ready-to-adopt protocols, FucoID serves as an attractive approach for the precise identification and quantification of CCIs under physiologically relevant conditions by providing resolved molecular information.
Using iDC-sFT, we realized the capture of diverse types of CCIs without previous knowledge of their molecular basis. Combining the iDC-sFT–enabled labeling and bulk RNA-seq analysis, we found that iDC–B cell interactions are regulated by cytokine signaling and B cell receptor on the B cell surface. Furthermore, the invention of Ab-sFT probes has realized the direct characterization of cells interacting with HER2+ cancer cells in clinical samples via the multicolor flow cytometry analysis, which provides an opportunity to resolve the molecular signatures of tumor-specific T cells that could directly recognize tumor cells (22). This approach may be generalized by integrating FT with other high-affinity targeting modalities, e.g., nanobodies, which makes FucoID potentially more convenient to use in tissue samples (fig. S14). Because there were very few in-depth investigations of the molecular signatures of CCIs (11, 17, 40), our results point to a direction for identifying molecular pairs involved in tuning these immune cell interactions. In future studies, the combination of more advanced single-cell omic techniques, e.g., cellular indexing of transcriptomes and epitopes (CITE-seq), with FucoID, could further empower intercellular proximity labeling tools to capture the heterogeneous distributions of molecular determinates that play critical roles in modulating these CCIs (41).
Last, many new drug modalities have emerged in recent years, e.g., checkpoint inhibitors, bispecific antibodies, bispecific T cell engagers, and engineered T cells, that either block or redirect CCIs in human patients for cancer treatment (42–46). In comparison to in vitro drug screens that assess potential perturbations to ligand-receptor interactions using purified recombinant proteins, screens that probe the impact on CCIs may reflect a more native setting. In this regard, we believe FucoID may find wide applications to probe the outcomes of CCIs intervened by these targeting agents in future efforts to search for new drug candidates.
MATERIALS AND METHODS
Mice
OT-I+/− and CD45.1+/− mice were bred and housed under specific pathogen–free conditions. C57BL/6J mice were purchased from GemPharmatech. OT-I+/− or CD45.1+/− mice were generated by cross-breeding OT-I or CD45.1+/+ and C57BL/6J mice, respectively. Both male and female mice of 6 to 12 weeks of age were used for all experiments. All animal experiments were approved by the Animal Ethical and Welfare Committee of Nanjing University (no. IACUC-2012003).
Materials and reagents
All chemical reagents and solvents were obtained from Sigma-Aldrich and used without further purification. PEI (#919012) was purchased from Sigma-Aldrich. NucleoBond Xtra Midi Plus (#740412.10) was purchased from MACHEREY-NAGEL, zombie (#423101), CellTracker CM-DiI Dye (#C7000), CellTracker Green CMFDA (#C7025), eBioscience Cell Proliferation Dye eFluor 450 (#65-0842-85), and eBioscience Cell Proliferation Dye eFluor 670 (#65-0840-85) were purchased from Thermo Fisher Scientific. APC-Cy7 anti-mouse CD45.1 (#110715), APC anti-mouse CD3 (#100236), PB anti-mouse CD4 (#100531), PerCPCy5.5 anti-mouse CD8α (#100734), fluorescein isothiocyanate (FITC) anti-mouse CD19 (#152404), phycoerythrin (PE) anti-mouse CD69 (#104508), PB anti-mouse CD45.1 (#110722), APC streptavidin (#405207), PE streptavidin (#405245), PE-Cy7 streptavidin (#405206), Alexa Fluor 700 anti-human CD3 (#317340), PB anti-human CD19 (#302232), FITC anti-human PD-1 (#329904), APC anti-human CD103 (#350216), APC-Cy7 CD137 (#309830), Brilliant Violet 605 anti-human CD39 (#328236), Brilliant Violet 650 anti-human CD69 antibody (#310934), anti-mouse CD16/32 TruStain FcX PLUS, and Human TruStain FcX (Fc receptor blocking solution, #422301) were purchased from BioLegend. Mouse granulocyte-macrophage colony-stimulating factor (GM-CSF; #TL-655) was purchased from Tongli Haiyuan. Biotin-(PEG)4-alkyne (#TA105) and methyltetrazine-(PEG)4-alkyne (#1013-100) were purchased from Click Chemistry Tools. GGG-(PEG)4-Tz (#BSG-3-200721) and GGG-(PEG)5-TCO were purchased from CONFLUORE. Mouse 1× lymphocyte separation medium (#7211011) was purchased from Dakewe.
Plasmids
The ErbB2-specific single-chain fragment variable (scFv) 4D5 (25) sequences were cloned into the pMDLg/pRRE vector for lentivirus packaging. FT with a C-terminal LPETGG tag was cloned into the pET-28a(+) vector for protein expression. pMD2.G (#12259) and pRSV-Rev (#12253) were purchased from Addgene.
Cell lines
Cell lines were all purchased from American Type Culture Collection or National Infrastructure of Cell line Resource, China. SKOV3, JIMT-1, and Pan02 cell lines were grown in DMEM (Dulbecco’s modified Eagle’s medium; GlutaMAX, Gibco) supplemented with 10% fetal bovine serum (FBS; Omega Scientific Inc.) and 1× penicillin-streptomycin (Gibco). MC38 were grown in RPMI 1640 (DMEM, GlutaMAX, Gibco) supplemented with 10% FBS (Gibco) and 1× penicillin-streptomycin (Gibco). Jurkat and CAR-J were grown in T cell medium (RPMI 1640, GlutaMAX, Gibco) supplemented with 10% FBS (Gibco), 1× penicillin-streptomycin (Gibco), 1× 1 M Hepes (Gibco), 1× NEAA (Gibco), 1× sodium pyruvate (Gibco), and 0.5× β-mercaptoethanol (Gibco). Human embryonic kidney (HEK) 293–GTX were grown in packing virus medium [high-glucose DMEM supplemented with 10% FBS (Gibco) and 1× penicillin-streptomycin (Gibco), 1× 1 M Hepes (Gibco), and 1× NEAA (Gibco)]. All cell cultures were incubated at 37°C under 5% CO2/95% air.
Primary cells
OT-I T cells were isolated from the spleens of OT-I mice. Murine iDCs were induced to differentiate from the bone marrow. Following erythrocyte lysis, the bone marrow cells were resuspended in complete T cell medium with GM-CSF (20 ng/ml). The culture medium was changed every 3 days with fresh complete T cell medium with GM-CSF (20 ng/ml). After 7 days of culturing, nonadherent cells and loosely adherent cells were harvested and gently washed by phosphate-buffered saline (PBS) for downstream labeling experiments. All cells cultures were incubated at 37°C under 5% CO2/95% air.
Human tissue and blood samples
The bladder cancer tissues were obtained from the patient who underwent radical cystectomy or transurethral resection of bladder tumor at the Affiliated Drum Tower Hospital of Nanjing University, School of Medicine (Nanjing, China). The tumor tissues were collected during surgical operation and preserved in the T cell medium to obtain the T cells. Under sterile conditions, tumors were cut into small pieces and digested in RPMI 1640 supplemented with collagenase IV at 1 mg/ml (Sigma-Aldrich) and deoxyribonuclease (DNase) at 30 U/ml (Sigma-Aldrich) at 37°C for 45 min. Digested tissue suspensions were filtered through a 70-μm filter and washed in RPMI 1640 + 5% FBS + DNase (15 U/ml). After the above operations, TILs were obtained and stained analysis by flow cytometry. All TILs were cryopreserved until further analysis. Human bloods samples were collected from a healthy donor who provided paper informed consent. PBMCs were obtained by Ficoll (Ficoll-Paque Plus, GE Healthcare) density gradient centrifugation, were then immediately frozen in liquid nitrogen to maintain the initial cell state, and were thawed before the FucoID labeling experiment. The present study was approved by the Ethics Committee of Nanjing Drum Tower Hospital in accordance with the Declaration of Helsinki (protocol 2021-394-01).
Cloning, expression, and purification of recombinant FT-LPETGG protein
Gene of Helicobacter pylori 26695α1, 3FucT was cloned into PET-28a(+) with a C-terminal LPETGG tag, followed by a His tag for purification. FT-LPETGG plasmids were transformed into BL21 (DE3) Escherichia coli, picking single clone to inoculate into 5 ml of LB medium containing kanamycin (50 mg/ml) and grown at 37°C. Culturing at logarithmic phase (optical density at 600 nm, 0.6) was induced with 0.5 mM isopropylthiogalactoside overnight at 20°C for 20 hours. The bacterial cells were pelleted and lysed via homogenizer in phosphate buffer solution. FT-LPETGG was purified by affinity column. Purified proteins were concentrated in Millipore concentration tubes (<30 kDa) and desalted into PBS using a G25 desalting column.
SDS-PAGE analysis of the molecular weight of protein conjugates
Twenty-microliter protein samples were mixed with 5 μl of 5× SDS-PAGE sample buffer [100 mM tris (pH 6.8), 8% glycerol, bromophenol (0.1 mg/ml), 2% SDS, and 10 mM dithiothreitol] to be analyzed by SDS-PAGE. All samples were boiled at 95°C for 5 min, and a 20-μl aliquot of each sample was electrophoresed in 0.75-mm-thick SDS-PAGE gels containing 12% acrylamide. Gels were run at 80 to 100 V and stained with Coomassie brilliant blue.
Mass spectrometry analysis of the molecular weight of protein conjugates
Protein separations were performed using a Waters ACQUITY UPLC I-Class Plus coupled online to a Xevo G2-XS QTOF MS System (Waters Corporation) equipped with an electrospray ionization source. Solvent A, water with 0.1% formic acid, and solvent B, acetonitrile with 0.1% formic acid, were used as the mobile phase at a flow rate of 0.5 ml/min. Gradients used are as follows: isocratic 95% H2O for 2 min, 95% to 10 H2O in 4 min, 10% H2O for 1 min, 10% to 95% H2O in 1 min, and then 95% H2O for 2 min. Total mass spectra were reconstructed from the ion series using the MaxEnt algorithm preinstalled on MassLynx software (v. 4.1 from Waters) according to the manufacturer’s instructions. To obtain the ion series described, the major peak(s) of the chromatogram were selected for integration and further analysis.
Synthesis procedure of site-specific GF-sFT
Reaction was carried in a 1.5-ml Eppendorf tube. Tz group was first introduced onto FT-LPETGG and then produced site-specific GDP-Fuc-FT conjugates by a click reaction. Briefly, a 50 mM stock of GGG-(PEG)4-Tz in dimethyl sulfoxide (DMSO) was added to solution of FT-LPETGG (8 mg/ml, 200 μl) at a final concentration of 5 mM. The reaction was catalyzed by 50 μM mgSrtA at room temperature for 1 hour. Excess FT-LPETGG in the solution was removed by magnetic beads, and solution was desalted into PBS by G25 desalting column to obtain sFT-Tz. GF-BCN (50 mM) was added to solution at a final concentration of 1.1 mM (approximately 10 equivalent of sFT-Tz) and incubated at room temperature for 5 min to obtain GF-sFT.
General protocols for preparing cell-sFT probe
A total of 1 million live cells were resuspended in 100 μl of Hanks’ balanced salt solution buffer containing 20 mM MgSO4, 3 mM Hepes, and 0.5% FBS. Cells were treated with GF-sFT (0.2 mg/ml). After incubating on ice for 20 min, cell-sFT was washed twice with PBS and ready for further application.
General protocols for preparing Ab-sFT probe
Reaction was carried in a 1.5-ml Eppendorf tube. TCO group was first introduced onto FT-LPETGG and then producing site-specific Ab-sFT conjugates by a click reaction. Briefly, a 50 mM stock of GGG-(PEG)5-TCO in DMSO was added to solution of FT-LPETGG (8 mg/ml, 200 μl) at a final concentration of 5 mM. The reaction was catalyzed by 50 μM mgSrtA at room temperature for 1 hour. Excess FT-LPETGG in the solution was removed by magnetic beads, and solution was desalted into PBS by G25 desalting column to obtain FT-TCO. Anti-HER2 antibody (KN025, targeting different epitopes of trastuzumab, 31 mg/ml, 30 μl) was incubated with endo S (48 mg/ml, 3.5 μl), uridine 5ridiphosphate (UDP)–GalNAz (100 mM, 4.5 μl), and β4-GalT1(Y289L; 3.88 mg/ml, 3.5 μl) in 10 mM MnCl2 and tris-HCl (25 mM) (pH 8.0) for 16 hours at 30°C. Purification with Protein A resins gave KN025(GalNAz)2. A solution of DBCO-(PEG)4-Tz was added KN025(GalNAz)2 and incubated for 5 hours at 30°C (approximately 10 equivalent of Ab-Az). Then, KN025(GalNTz)2 was obtained through a G25 desalting column. FT-TCO (3.5 mg/ml) was added to KN025-(Tz)2 (2.25 mg/ml) and incubated at room temperature for 4 hours (approximately 8 equivalent of KN025-Az), purification with Protein A resins gave KN025-sFT, and site-specific KN025-sFT conjugates were ready to use (33). KN025-sFT on SDS-PAGE was the sample before Protein A purification.
Fluorescence imaging
SKOV3-sFT cells were seeded in a confocal dish, incubated with polyclonal FT antibody (10 μg/ml; made by GenScript, mouse IgG) for 30 min on ice, washed three times with fluorescence-activated cell sorting (FACS) buffer, and then incubated with Alexa Fluor 488 anti-human CD340 (10 μg/ml; erbB2/HER-2) and PE anti-mouse IgG (10 μg/ml) for 30 min on ice. After washing three times with FACS buffer, cell imaging was performed on Olympus SpinSR. PE was excited by 561-nm laser and collected with 560 to 600 nm. Alexa Fluor 488 was excited by 488-nm laser and collected between 490 and 550 nm.
Detection of pMHC-TCR–mediated interactions via cell-sFT
iDC-sFT or cancer cell–sFT was prepared according to the protocol of preparing cell-sFT probe. iDC-sFT or cancer cell–sFT was resuspended in T cell medium and left untreated, primed with 100 nM OVA257–264 or lymphocytic choriomeningitis virus GP33–41 at 37°C for 30 min. After washing twice with PBS, iDC-sFT or cancer cell–sFT was cocultured with OT-I splenocytes in a 96-well plate at 37°C for 2 hours. GF-Biotin (50 μM) was added gently to initiate the FucoID reaction for 30 min. LacNAc (final concentration, 5 mM) was added to quench the reaction. Cell mixtures were washed three times with PBS and stained analysis by flow cytometry (22).
Construction of the HER2-CAR plasmid and CAR-J–expressing Herceptin scFv
The ErbB2-specific scFv 4D5 (25) sequences were synthesized and subcloned into pMDLg/pRRE vector for lentivirus packaging, and hemagglutinin (HA) tag (YPYDVPDYA) was cloned after the CD8 leader. Lentivirus particles were produced using a third-generation system (47–49). Briefly, 6 million HEK293-GTX cells cultured on 10 cm tissue culture–treated dishes were transfected with packaging plasmids pRSV-Rev (4.5 μg), pMD2.G (1.875 μg), and pMDLg/p (4.5 μg), and the transfer plasmid (5.625 μg) in Opti-MEM and PEI. Medium exchange was performed after 8 hours of transfection. Media containing lentivirus particles were collected after 36 to 48 hours and concentrated at 50,000g at 4°C for 3 hours by ultracentrifugation. Jurkat cells (1.2 million) were seeded in six-well plate, followed by the addition of 48 μl of 1 M Hepes, 4.8 μl of protamine sulfate (7.5 mg/ml), and 50 μl of concentrated virus. The six-well plate was centrifuged at 1000g for 90 min at slow acceleration and slow deceleration at 33°C for 1.5 hours. After centrifugation, the six-well plate was placed in an incubator at 37°C for 24 hours before medium exchange. The positive ratio of viral infection was determined by detecting expression level of HA tag of Jurkat cells. After 2 weeks of expansion, Jurkat cells of positive HA were sorted for subsequent experiments.
Detection of CAR-antigen–mediated interactions via cell-sFT
Jurkat-sFT and CAR-J–sFT were prepared according to the protocol of preparing cell-sFT probe. Jurkat-sFT and CAR-J–sFT were resuspended in T cell medium and then cocultured with HER2+ SKOV3 stained with CellTracker CMFDA at 37°C for 1 hour. GF-Biotin (50 μM) was added gently to initiate the FucoID reaction. The reaction was incubated at 37°C for 30 min before quenched with LacNAc (5 mM). Cell mixtures were then stained and analyzed by flow cytometry.
Selectivity
The prepared CAR-J–sFT were added to cell mixtures of SKOV3, MCF-7, MDA-MB-231, and PBMCs (at the ratio of 1:1:1:1:10) and then incubated at 37°C for 1 hour. The following steps are the same as above.
Quantitative labeling
Construction of SKOV3 cells expressing heterogeneous level of HER2
SKOV3-HER2pKO cell line was established through CRISPR-Cas9 RNP–based gene editing. Briefly, Cas9 single-guide RNAs (sgRNAs) targeting human ERBB2 exon 12 (sg1-TGAGTGGGTACCTCGGGCAC) and exon 15 (sg2-ACTTGTGCAGAATTCGTCCC) were chemically synthesized with sgRNA scaffold (GenScript) and suspended in TE buffer at a concentration of 100 μM. For RNP formation, 36 pmol of each sgRNA and 72 pmol of Cas9 protein were mixed in 25 μl of Opti-MEM (Thermo Fisher Scientific) at room temperature for 15 min. For transfection, 3.75 μl of Lipofectamine 3000 (Thermo Fisher Scientific) diluted in 25 μl of Opti-MEM was mixed gently with RNP mixture, incubated at room temperature for 10 min, and added into SKOV3 cells at 60% confluency in 24-well plate. On day 7, cells were collected, stained with anti-HER2 APC to confirm the heterogeneous expression level of HER2.
Quantification of CCI
CAR-J–sFT and Jurkat-sFT were incubated with pKO SKOV3 cells at 37°C for 1 hour, and 50 μM GF-Biotin was added gently to initiate the FucoID reaction. The reaction was incubated at 37°C for 30 min before quenched with LacNAc (5 mM). Cell mixtures were then stained and analyzed by flow cytometry. To quantify CCI, pKO SKOV3 cells were divided into six groups according to expression level of HER2, and biotinylation ratio was determined in each group. Correlation analysis of biotin ratio and HER2 expression level on pKO SKOV3 cells was characterized in GraphPad Prism software (version 7.0) via fitting a linear curve.
Detection of PD-1–PD-L1–mediated interactions via cell-sFT
JIMT-1–sFT was prepared according to the protocol of preparing cell-sFT probe. A portion of JIMT-1–sFT was treated with anti-human PD-L1 on ice for 30 min. JIMT-1–sFT and JIMT-1–sFT pretreated by anti-human PD-L1 were resuspended in T cell medium and then cocultured with Jurkat (PD-1+) stained with CellTracker CMFDA at 37°C for 1 hour. GF-Biotin (50 μM) was added gently to initiate the FucoID reaction. The reaction was incubated at 37°C for 30 min before quenched with LacNAc (5 mM). Cell mixtures were then stained and analyzed by flow cytometry.
Detection of interactions between murine iDCs and immune cells in murine splenocytes
C57BL/6J splenocytes were prepared according to the reported protocol. iDC-sFT was mixed with splenocytes at the ratio of 1:1 immediately. GF-Biotin (50 μM) was added gently to initiate the FucoID reaction. The reaction was incubated at 37°C for 30 min before quenched with LacNAc (5 mM). Cell mixtures were then stained and analyzed by flow cytometry.
Detection of Jurkat/CAR-J–SKOV3 interactions via KN025-sFT
SKOV3 cells were stained with CellTracker CMFDA, while Jurkat cells were stained with eBioscience Cell Proliferation Dye eFluor670. SKOV3, Jurkat, and CAR-J were mixed at a ratio of 1:1:1. A total of 60,000 cells were added to each well of 96-well plate and cultured at 37°C for 30 min. Different concentrations of KN025-sFT (anti–HER2-sFT) or isotype IgG-sFT were added to cell mixtures at 37°C for 30 min. Then, the unbound KN025-sFT or isotype IgG-sFT conjugates were washed twice with PBS. Cell mixtures were cocultured at 37°C for 1 hour, and 50 μM GF-Biotin was added gently to initiate the FucoID reaction. The reaction was incubated at 37°C for 30 min before quenched with LacNAc (5 mM). Cell mixtures were then stained and analyzed by flow cytometry.
Detection of interactions between cancer cells (HER2+) and T cells in human bladder samples via KN025-sFT
The cell suspension from human bladder cancer sample was resuscitated. A total of 50,000 cells were added to each well of 96-well plate and cultured at 37°C for 30 min with Ab-sFT or IgG-sFT. Single-cell suspensions were washed twice with PBS and incubated at 37°C for another 1 hour. GF-Biotin (50 μM) was added gently to initiate the FucoID reaction for 30 min. LacNAc (final concentration, 5 mM) was added to quench the reaction. Cell mixtures were washed with PBS for three times and then stained analysis by flow cytometry.
Flow cytometry and cell sorting
Flow cytometry data analyses were performed using FlowJo software (10.6.2). tSNE analysis was performed using FCS Express 7 from De Novo software. Cells were suspended in T cell medium supplemented with anti-mouse CD16/32 to block Fc receptors before staining with fluorescent antibodies against cell surface epitopes. Samples were stained using the following antibodies: zombie, FITC anti-mouse CD19, PB anti-mouse CD45.1, APC streptavidin, and anti-mouse CD16/32 TruStain FcXPLUS purchased from BioLegend. First, dead cells were excluded through zombie, then splenocytes were distinguished through anti-mouse CD45.1, B cells were isolated through anti-mouse CD19, and last, Biotin+ B cells and Biotin− B cells were sorted through streptavidin. The background was defined as the signal produced on B cells by incubating with iDCs without membrane-anchored sFT.
RNA sequencing
Library preparation for RNA-seq
A total amount of 1 μg of qualified RNA per sample was used as input material for the library preparation. The sequencing libraries were generated using the VAHTS mRNA-seq v2 Library Prep Kit for Illumina (Vazyme, NR601) following the manufacturer’s recommendations. First, mRNA was purified from total RNA using poly-T oligo-attached magnetic beads. Fragmentation was performed using divalent cations under elevated temperature in Vazyme Frag/Prime Buffer. The cleaved RNA fragments were copied into first-strand cDNA using reverse transcriptase and random primers. Second-strand cDNA synthesis was subsequently performed using buffer, dNTPs, DNA polymerase I, and ribonuclease H. Then, the cDNA fragments were end repaired with the addition of a single “A” base at the 3′-end of each strand, ligated with the special sequencing adapters (Vazyme, N803) subsequently. The products were purified, and sizes were selected with VAHTS DNA Clean Beads (Vazyme, N411) to get appropriate size for sequencing. Polymerase chain reaction amplification was performed, and products were purified.
Library examination
Library concentration was measured using Qubit RNA Assay Kit in Qubit 3.0 to preliminary quantify. Insert size was assessed using the Agilent Bioanalyzer 2100 system.
Library clustering and sequencing
The clustering of the index-coded samples was performed on a cBot Cluster Generation System (Illumina, USA) according to the manufacturer’s instructions. After cluster generation, the library preparations were sequenced on an Illumina HiSeq X Ten platform and 150–base pair paired-end module.
Data evaluation
The software package limma was used to detect differentially expressed genes. We applied a threshold of P < 0.05 and a log fold change of >1.0 for up-regulated genes and <−1.0 for down-regulated genes. Volcano plots and heatmaps were performed using the software SangerBox tools, a free online platform foe data analysis (www.sangerbox.com/index.html). The GO term analysis and Kyoto Encyclopedia of Genes and Genomes pathway analysis were performed using cluster Profiler.
Statistical analysis
Statistical analyses were performed by GraphPad Prism software (version 7.0). Comparisons over groups were analyzed using two-way analysis of variance (ANOVA), and comparisons of multiple samples at one group were analyzed using two tailed t test or one-way ANOVA. In all figures with error bars, data are presented as the means ± SD. In all figures, ns, P > 0.05; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Acknowledgments
We thank P. R. Chen for providing the sortase A plasmid.
Funding: J.P.L. acknowledges the support from the National Key R&D Program of China (2019YFA09006600), National Natural Science Foundation of China (21977048 and 92053111), Natural Science Foundation of Jiangsu Province (BK20202004), Beijing National Laboratory for Molecular Sciences (BNLMS202008), Program for Innovative Talents and Entrepreneur in Jiangsu, and the Excellent Research Program of Nanjing University (ZYJH004). P.W. is partially supported by the NIH, USA (R01AI143884).
Author contributions: J.P.L. and P.W. designed the experimental strategies and wrote the manuscript. S.Q., Z.Z., M.W., Q.X., Y.Y., S.O., W.L., L.Z., W.W., and R.Y. performed the experiments. S.Q. and Z.Z. prepared the figures, and all authors analyzed the data and edited the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S15
Table S1
Supplementary Methods
REFERENCES AND NOTES
- 1.Cukierman E., Pankov R., Yamada K. M.,Cell interactions with three-dimensional matrices. Curr. Opin. Cell Biol. 14,633–640 (2002). [DOI] [PubMed] [Google Scholar]
- 2.Armingol E., Officer A., Harismendy O., Lewis N. E.,Deciphering cell-cell interactions and communication from gene expression. Nat. Rev. Genet. 22,71–88 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Batista F. D., Dustin M. L.,Cell:cell interactions in the immune system. Immunol. Rev. 251,7–12 (2013). [DOI] [PubMed] [Google Scholar]
- 4.Huse M.,Mechanical forces in the immune system. Nat. Rev. Immunol. 17,679–690 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cain M. P., Hernandez B. J., Chen J.,Quantitative single-cell interactomes in normal and virus-infected mouse lungs. Dis. Model. Mech. 13,dmm044404 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chua R. L., Lukassen S., Trump S., Hennig B. P., Wendisch D., Pott F., Debnath O., Thürmann L., Kurth F., Völker M. T., Kazmierski J., Timmermann B., Twardziok S., Schneider S., Machleidt F., Müller-Redetzky H., Maier M., Krannich A., Schmidt S., Balzer F., Liebig J., Loske J., Suttorp N., Eils J., Ishaque N., Liebert U. G., von Kalle C., Hocke A., Witzenrath M., Goffinet C., Drosten C., Laudi S., Lehmann I., Conrad C., Sander L. E., Eils R.,COVID-19 severity correlates with airway epithelium-immune cell interactions identified by single-cell analysis. Nat. Biotechnol. 38,970–979 (2020). [DOI] [PubMed] [Google Scholar]
- 7.Noack M., Miossec P.,Importance of lymphocyte–stromal cell interactions in autoimmune and inflammatory rheumatic diseases. Nat. Rev. Rheumatol. 17,550–564 (2021). [DOI] [PubMed] [Google Scholar]
- 8.Vladimer G. I., Snijder B., Krall N., Bigenzahn J. W., Huber K. V. M., Lardeau C. H., Sanjiv K., Ringler A., Berglund U. W., Sabler M., de la Fuente O. L., Knöbl P., Kubicek S., Helleday T., Jäger U., Superti-Furga G.,Global survey of the immunomodulatory potential of common drugs. Nat. Chem. Biol. 13,681–690 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blanco B., Domínguez-Alonso C., Alvarez-Vallina L.,Bispecific immunomodulatory antibodies for cancer immunotherapy. Clin. Cancer Res. 27,5457–5464 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Depil S., Duchateau P., Grupp S. A., Mufti G., Poirot L.,‘Off-the-shelf’ allogeneic CAR T cells: Development and challenges. Nat. Rev. Drug Discov. 19,185–199 (2020). [DOI] [PubMed] [Google Scholar]
- 11.Bechtel T. J., Reyes-Robles T., Fadeyi O. O., Oslund R. C.,Strategies for monitoring cell-cell interactions. Nat. Chem. Biol. 17,641–652 (2021). [DOI] [PubMed] [Google Scholar]
- 12.Wollman A. J., Nudd R., Hedlund E. G., Leake M. C.,From Animaculum to single molecules: 300 years of the light microscope. Open Biol. 5,150019 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Efremova M., Vento-Tormo M., Teichmann S. A., Vento-Tormo R.,CellPhoneDB: Inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes. Nat. Protoc. 15,1484–1506 (2020). [DOI] [PubMed] [Google Scholar]
- 14.Jin S., Guerrero-Juarez C. F., Zhang L., Chang I., Ramos R., Kuan C. H., Myung P., Plikus M. V., Nie Q.,Inference and analysis of cell-cell communication using CellChat. Nat. Commun. 12,1088 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Y. Wang, R. Wang, S. Zhang, S. Song, C. Jiang, G. Han, M. Wang, J. A. Ajani, A. M. Futreal, L. Wang, iTALK: An R package to characterize and illustrate intercellular communication. bioRxiv 10.1101/507871 [Preprint]. 4 January 2019. 10.1101/507871. [DOI]
- 16.Browaeys R., Saelens W., Saeys Y.,NicheNet: Modeling intercellular communication by linking ligands to target genes. Nat. Methods 17,159–162 (2020). [DOI] [PubMed] [Google Scholar]
- 17.Pasqual G., Chudnovskiy A., Tas J. M. J., Agudelo M., Schweitzer L. D., Cui A., Hacohen N., Victora G. D.,Monitoring T cell-dendritic cell interactions in vivo by intercellular enzymatic labelling. Nature 553,496–500 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ge Y., Chen L., Liu S., Zhao J., Zhang H., Chen P. R.,Enzyme-mediated intercellular proximity labeling for detecting cell-cell interactions. J. Am. Chem. Soc. 141,1833–1837 (2019). [DOI] [PubMed] [Google Scholar]
- 19.Liu Q., Zheng J., Sun W., Huo Y., Zhang L., Hao P., Wang H., Zhuang M.,A proximity-tagging system to identify membrane protein-protein interactions. Nat. Methods 15,715–722 (2018). [DOI] [PubMed] [Google Scholar]
- 20.Zhong S., Malecek K., Johnson L. A., Yu Z., Vega-Saenz de Miera E., Darvishian F., McGary K., Huang K., Boyer J., Corse E., Shao Y., Rosenberg S. A., Restifo N. P., Osman I., Krogsgaard M.,T-cell receptor affinity and avidity defines antitumor response and autoimmunity in T-cell immunotherapy. Proc. Natl. Acad. Sci. U.S.A. 110,6973–6978 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li L., Guo X., Shi X., Li C., Wu W., Yan C., Wang H., Li H., Xu C.,Ionic CD3-Lck interaction regulates the initiation of T-cell receptor signaling. Proc. Natl. Acad. Sci. U.S.A. 114,e5891–e5899 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Z., Li J. P., Chen M., Wu M., Shi Y., Li W., Teijaro J. R., Wu P.,Detecting tumor antigen-specific T cells via interaction-dependent fucosyl-biotinylation. Cell 183,1117–1133.e19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parmar S., Liu X., Najjar A., Shah N., Yang H., Yvon E., Rezvani K., McNiece I., Zweidler-McKay P., Miller L., Wolpe S., Blazar B. R., Shpall E. J.,Ex vivo fucosylation of third-party human regulatory T cells enhances anti-graft-versus-host disease potency in vivo. Blood 125,1502–1506 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sackstein R., Merzaban J. S., Cain D. W., Dagia N. M., Spencer J. A., Lin C. P., Wohlgemuth R.,Ex vivo glycan engineering of CD44 programs human multipotent mesenchymal stromal cell trafficking to bone. Nat. Med. 14,181–187 (2008). [DOI] [PubMed] [Google Scholar]
- 25.Stevanović S., Pasetto A., Helman S. R., Gartner J. J., Prickett T. D., Howie B., Robins H. S., Robbins P. F., Klebanoff C. A., Rosenberg S. A., Hinrichs C. S.,Landscape of immunogenic tumor antigens in successful immunotherapy of virally induced epithelial cancer. Science 356,200–205 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tran E., Robbins P. F., Lu Y. C., Prickett T. D., Gartner J. J., Jia L., Pasetto A., Zheng Z., Ray S., Groh E. M., Kriley I. R., Rosenberg S. A.,T-cell transfer therapy targeting mutant KRAS in cancer. N. Engl. J. Med. 375,2255–2262 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zacharakis N., Chinnasamy H., Black M., Xu H., Lu Y. C., Zheng Z., Pasetto A., Langhan M., Shelton T., Prickett T., Gartner J., Jia L., Trebska-McGowan K., Somerville R. P., Robbins P. F., Rosenberg S. A., Goff S. L., Feldman S. A.,Immune recognition of somatic mutations leading to complete durable regression in metastatic breast cancer. Nat. Med. 24,724–730 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heath W. R., Kato Y., Steiner T. M., Caminschi I.,Antigen presentation by dendritic cells for B cell activation. Curr. Opin. Immunol. 58,44–52 (2019). [DOI] [PubMed] [Google Scholar]
- 29.Wykes M., MacPherson G.,Dendritic cell-B-cell interaction: Dendritic cells provide B cells with CD40-independent proliferation signals and CD40-dependent survival signals. Immunology 100,1–3 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qi H., Egen J. G., Huang A. Y., Germain R. N.,Extrafollicular activation of lymph node B cells by antigen-bearing dendritic cells. Science 312,1672–1676 (2006). [DOI] [PubMed] [Google Scholar]
- 31.Rickert R. C., Jellusova J., Miletic A. V.,Signaling by the tumor necrosis factor receptor superfamily in B-cell biology and disease. Immunol. Rev. 244,115–133 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schriek P., Ching A. C., Moily N. S., Moffat J., Beattie L., Steiner T. M., Hosking L. M., Thurman J. M., Holers V. M., Ishido S., Lahoud M. H., Caminschi I., Heath W. R., Mintern J. D., Villadangos J. A.,Marginal zone B cells acquire dendritic cell functions by trogocytosis. Science 375, (2022). [DOI] [PubMed] [Google Scholar]
- 33.van Geel R., Wijdeven M. A., Heesbeen R., Verkade J. M. M., Wasiel A. A., van Berkel S. S., van Delft F. L.,Chemoenzymatic conjugation of toxic payloads to the globally conserved N-glycan of native mAbs provides homogeneous and highly efficacious antibody–drug conjugates. Bioconjug. Chem. 26,2233–2242 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Gros A., Robbins P. F., Yao X., Li Y. F., Turcotte S., Tran E., Wunderlich J. R., Mixon A., Farid S., Dudley M. E., Hanada K., Almeida J. R., Darko S., Douek D. C., Yang J. C., Rosenberg S. A.,PD-1 identifies the patient-specific CD8+ tumor-reactive repertoire infiltrating human tumors. J. Clin. Investig. 124,2246–2259 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gros A., Parkhurst M. R., Tran E., Pasetto A., Robbins P. F., Ilyas S., Prickett T. D., Gartner J. J., Crystal J. S., Roberts I. M., Trebska-McGowan K., Wunderlich J. R., Yang J. C., Rosenberg S. A.,Prospective identification of neoantigen-specific lymphocytes in the peripheral blood of melanoma patients. Nat. Med. 22,433–438 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yossef R., Tran E., Deniger D. C., Gros A., Pasetto A., Parkhurst M. R., Gartner J. J., Prickett T. D., Cafri G., Robbins P. F., Rosenberg S. A.,Enhanced detection of neoantigen-reactive T cells targeting unique and shared oncogenes for personalized cancer immunotherapy. JCI Insight 3,e122467 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Duhen T., Duhen R., Montler R., Moses J., Moudgil T., de Miranda N. F., Goodall C. P., Blair T. C., Fox B. A., McDermott J. E., Chang S. C., Grunkemeier G., Leidner R., Bell R. B., Weinberg A. D.,Co-expression of CD39 and CD103 identifies tumor-reactive CD8 T cells in human solid tumors. Nat. Commun. 9,2724 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scheper W., Kelderman S., Fanchi L. F., Linnemann C., Bendle G., de Rooij M. A. J., Hirt C., Mezzadra R., Slagter M., Dijkstra K., Kluin R. J. C., Snaebjornsson P., Milne K., Nelson B. H., Zijlmans H., Kenter G., Voest E. E., Haanen J., Schumacher T. N.,Low and variable tumor reactivity of the intratumoral TCR repertoire in human cancers. Nat. Med. 25,89–94 (2019). [DOI] [PubMed] [Google Scholar]
- 39.Simoni Y., Becht E., Fehlings M., Loh C. Y., Koo S. L., Teng K. W. W., Yeong J. P. S., Nahar R., Zhang T., Kared H., Duan K., Ang N., Poidinger M., Lee Y. Y., Larbi A., Khng A. J., Tan E., Fu C., Mathew R., Teo M., Lim W. T., Toh C. K., Ong B. H., Koh T., Hillmer A. M., Takano A., Lim T. K. H., Tan E. H., Zhai W., Tan D. S. W., Tan I. B., Newell E. W.,Bystander CD8+ T cells are abundant and phenotypically distinct in human tumour infiltrates. Nature 557,575–579 (2018). [DOI] [PubMed] [Google Scholar]
- 40.Oslund R. C., Reyes-Robles T., White C. H., Tomlinson J. H., Crotty K. A., Bowman E. P., Chang D., Peterson V. M., Li L., Frutos S., Vila-Perelló M., Vlerick D., Cromie K., Perlman D. H., Ingale S., Hara S. D. O., Roberts L. R., Piizzi G., Hett E. C., Hazuda D. J., Fadeyi O. O.,Detection of cell–cell interactions via photocatalytic cell tagging. Nat. Chem. Biol. 18,850–858 (2022). [DOI] [PubMed] [Google Scholar]
- 41.Stoeckius M., Hafemeister C., Stephenson W., Houck-Loomis B., Chattopadhyay P. K., Swerdlow H., Satija R., Smibert P.,Simultaneous epitope and transcriptome measurement in single cells. Nat. Methods 14,865–868 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Houot R., Schultz L. M., Marabelle A., Kohrt H.,T-cell-based immunotherapy: Adoptive cell transfer and checkpoint inhibition. Cancer Immunol. Res. 3,1115–1122 (2015). [DOI] [PubMed] [Google Scholar]
- 43.Duong C. P., Yong C. S., Kershaw M. H., Slaney C. Y., Darcy P. K.,Cancer immunotherapy utilizing gene-modified T cells: From the bench to the clinic. Mol. Immunol. 67,46–57 (2015). [DOI] [PubMed] [Google Scholar]
- 44.Haanen J. B., Robert C.,Immune checkpoint inhibitors. Prog. Tumor Res. 42,55–66 (2015). [DOI] [PubMed] [Google Scholar]
- 45.Krishnamurthy A., Jimeno A.,Bispecific antibodies for cancer therapy: A review. Pharmacol. Ther. 185,122–134 (2018). [DOI] [PubMed] [Google Scholar]
- 46.Einsele H., Borghaei H., Orlowski R. Z., Subklewe M., Roboz G. J., Zugmaier G., Kufer P., Iskander K., Kantarjian H. M.,The BiTE (bispecific T-cell engager) platform: Development and future potential of a targeted immuno-oncology therapy across tumor types. Cancer 126,3192–3201 (2020). [DOI] [PubMed] [Google Scholar]
- 47.Dull T., Zufferey R., Kelly M., Mandel R. J., Nguyen M., Trono D., Naldini L.,A third-generation lentivirus vector with a conditional packaging system. J. Virol. 72,8463–8471 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hammill J. A., Afsahi A., Bramson J. L., Helsen C. W.,Viral engineering of chimeric antigen receptor expression on murine and human T lymphocytes. Methods Mol. Biol. 1458,137–157 (2016). [DOI] [PubMed] [Google Scholar]
- 49.Carter P., Presta L., Gorman C. M., Ridgway J. B., Henner D., Wong W. L., Rowland A. M., Kotts C., Carver M. E., Shepard H. M.,Humanization of an anti-p185HER2 antibody for human cancer therapy. Proc. Natl. Acad. Sci. U.S.A. 89,4285–4289 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S15
Table S1
Supplementary Methods