Abstract
From a mass-excised Staphylococcus aureus λZapII expression library, we cloned an operon encoding a novel ABC transporter with significant homology to bacterial siderophore transporter systems. The operon encodes four genes designated sstA, -B, -C, and -D encoding two putative cytoplasmic membrane proteins (sstA and sstB), an ATPase (sstC), and a membrane-bound 38-kDa lipoprotein (sstD). The sst operon is preceded by two putative Fur boxes, which indicated that expression of the sst operon was likely to be iron dependent. SstD was overexpressed in Escherichia coli, purified by Triton X-114 phase partitioning, and used to generate monospecific antisera in rats. Immunoblotting studies located SstD in the membrane fraction of S. aureus and showed that expression of the lipoprotein was reduced under iron-rich growth conditions. Triton X-114 partitioning studies on isolated membranes provided additional biochemical evidence that SstD in S. aureus is a lipoprotein. Immunoreactive polypeptides of approximately 38 kDa were detected in a wide range of staphylococcal species, but no antigenic homolog was detected in Bacillus subtilis. Expression of SstD in vivo was confirmed by immunoblotting studies with S. aureus recovered from a rat intraperitoneal chamber implant model. To further define the contribution of SstD in promoting growth of S. aureus in vitro and in vivo, we used antisense RNA technology to modulate expression of SstD. Expression of antisense sstD RNA in S. aureus resulted in a decrease in SstD expression under both iron-rich and iron-restricted growth conditions. However, this reduction in SstD levels did not affect the growth of S. aureus in vitro in an iron-limited growth medium or when grown in an intraperitoneal rat chamber implant model in vivo.
Acquisition of nutrients, such as iron, for growth in the host environment is essential for bacterial pathogens to establish an infection. An effective mechanism for scavenging iron involves the production and secretion of low-molecular-weight ferric-iron chelators, siderophores, which scavenge iron from the host and transport it into the cell via specific ABC transporters (27). In comparison to the wealth of information available concerning gram-negative siderophore transport (11, 26), very little is known about ferric-siderophore uptake in staphylococci. It has been demonstrated that Staphylococcus aureus produces at least three siderophores, staphyloferrins A (22) and B (8, 12) and aureochelin (7), and it also utilizes a range of exogenous siderophores, such as the enterobacterial siderophore enterobactin (21). However, to date there are no published reports on staphylococcal genes coding for siderophore biosynthesis and very little is known about siderophore uptake. Three iron-regulated staphylococcal ABC transporters have been identified, but these have only been partially characterized, and in each case the transported solute has not been identified. In S. aureus, the sirABC operon has homology to gram-negative siderophore transporters, in particular, the cbr locus of Erwinia chrysanthemi (14), and the FhuABC transporter is homologous to the Bacillus subtilis ferrichrome transporter (36). A putative iron-manganese transporter, SitABC (6), has been identified in S. epidermidis, and an antigenically related protein is present in S. aureus. As in gram-negative bacteria, these transporters span the cytoplasmic membrane but the proposed ferric-siderphore-binding receptor is the lipoprotein, which is anchored to the cytoplasmic membrane via its N-terminally linked lipid moiety (13).
In gram-negative bacteria, the ferric-uptake regulator protein Fur mediates the iron-dependent transcriptional regulation of genes involved in iron transport (9). Sequence analysis has recently identified three Fur homologues in the S. aureus genome (36; unpublished observations). One homologue, Fur, mediates the negative iron regulation of the sirABC and fhuABC operons (14, 36), but as yet there has been no published functional analysis of the other two Fur homologues. B. subtilis also has three Fur homologues, involved in the regulation of iron (Fur) (4), peroxide stress response (PerR) (4), and zinc uptake (Zur) (10). It is possible that, in addition to Fur, staphylococci have another ferric-iron repressor, SirR (15), which is a homologue of the ferric-iron repressor DtxR (32).
Our previous studies identified a number of iron-regulated S. aureus and S. epidermidis proteins, which are expressed in vivo both during human infection (30, 35) and in an experimental animal model (23, 25). To date, only two of these proteins have been characterized. These are the cytoplasmic membrane-associated 32-kDa SitC lipoprotein (6) and a 42-kDa cell wall transferrin-binding protein (24, 25). This paper describes the molecular cloning and characterization of a 38-kDa lipoprotein, SstD, which is part of a novel ABC transporter from S. aureus, which has strong homology to siderophore transporters of B. subtilis and Campylobacter jejuni. We also describe the use of antisense RNA technology to disrupt the function of the transporter to investigate its contribution to the growth of S. aureus in vitro and in vivo.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
S. aureus W, S. haemolyticus, S. hominis, S. warneri, S. cohni, S. lugdunensis, S. saprophyticus, and S. epidermidis 901 are clinical isolates obtained from the University and City Hospital NHS Trusts, Nottingham, United Kingdom. S. aureus BB (originally isolated from a case of bovine mastitis), RN4220, and RN6390-B and B. subtilis 360 were from our laboratory culture collection. S. carnosus TM300 was provided by F. Gotz.
Staphylococcal strains were cultured aerobically at 37°C in tryptic soy broth (TSB) or on TSB agar, and chloramphenicol (5 μg/ml) was added when required. To maximize iron-regulated protein expression, staphylococci were cultured under iron-deficient conditions in a two-stage protocol. First, staphylococci were grown statically for 18 h at 37°C in 5% CO2 in 10-ml volumes of RPMI 1640 tissue culture medium containing 2 mg of NaHCO3 per ml. Iron-rich conditions were obtained by addition of 20 μM Fe2(SO4)3. Bacteria were pelleted by centrifugation at 3,500 × g for 5 min, and the pellet was resuspended in 1 ml of RPMI medium which had been depleted of iron by overnight batch incubation with 6% (wt/vol) Chelex 100 (Sigma). One hundred microliters of this bacterial suspension was then used to inoculate 10-ml volumes of Chelex-treated RPMI 1640 medium which had been supplemented with 10% (vol/vol) RPMI 1640 medium (CRPMI) to provide essential trace elements for staphylococcal growth. These cultures were incubated as described above for 6 h before harvesting. Where indicated, the medium was supplemented with 20 μM Fe2(SO4)3. Iron-starved B. subtilis bacteria were obtained by growing the organisms statically for 48 h at 37°C in 5% CO2 in RPMI 1640 medium.
E. coli TOPO (Invitrogen) XL1-Blue, or BL21 (Novagen) bacteria were cultured at 37°C in Luria-Bertani (LB) broth (2) or on LB agar containing appropriate antibiotics. Long-term stocks of bacterial strains were stored at −80°C in 10% (vol/vol) glycerol.
Intraperitoneal rat chamber model for in vivo growth of staphylococci.
Staphylococci were grown in intraperitoneal chambers implanted in rats as described by Pike et al. (28). Four animals were used for each strain tested. Inocula for chambers were grown overnight in LB broth containing the appropriate antibiotic and diluted in sterile phosphate-buffered saline (PBS), pH 7.4. Chambers were sampled for viable counting (by dilution and plating on LB agar) at 24 and 48 h postinoculation and after 96 h to assess the phenotype of the in vivo-grown bacteria recovered.
DNA preparation and manipulation.
Genomic DNA was prepared from staphylococci by the cetyltrimethylammonium bromide method described by Ausubel et al. (2). E. coli and staphylococcal plasmid DNAs were extracted using Qiagen mini and maxi kits in accordance with the manufacturer's instructions, except that staphylococcal cell walls were digested with lysostaphin (100 μg/ml; Sigma) in P1 buffer at 37°C for 5 min before the addition of P2 buffer. Restriction endonucleases were purchased from Pharmacia, and DNA manipulation enzymes were from Promega. Restriction-deficient S. aureus strain RN4220 was transformed with E. coli harvested plasmid DNA using an electroporation method described by Kraemer and Iandolo (19), with a few minor modifications. An overnight culture of RN4220 was diluted 1 in 50 in TSB and shaken at 37°C until a culture optical density at 600 nm (OD600) of 0.3 to 0.8 was reached. The cells were collected by centrifugation at 8,000 × g for 5 min and washed once with an equal volume of 500 mM sucrose in water (filter sterilized). After centrifugation, the cell pellet was resuspended in 0.5 volume of 500 mM sucrose and placed on ice for 15 to 30 min. The cells were repelleted and resuspended in 0.1 vol of 500 mM sucrose. Aliquots of this suspension were frozen at −70°C for up to 1 month. For electroporation, 50 μl of cell suspension was mixed with 1 μg of plasmid DNA in a 0.2-cm Bio-Rad Gene Pulser cuvette and incubated at room temperature for 30 min. The cells were given a single pulse with the electroporation apparatus set at 25 mF, 2.5 kV, and 100 Ω. Immediately after the pulse, 0.8 ml of SMMP medium (5) was added to the cells, which were then incubated with shaking at 37°C for 60 min to allow expression of the antibiotic resistance genes. The cells were then plated onto selective agar. Plasmid DNA extracted from RN4220 was used to transform wild-type S. aureus strain RN6390-B by electroporation. The plasmids used in this study are listed in Table 1.
TABLE 1.
Bacterial plasmids used in this study
| Plasmid | Comment | Source or reference |
|---|---|---|
| pBK-CMV | Excised E. coli phagemid | Stratagene |
| pBK-CMV/BB | Excised S. aureus BB phagemid library | This study |
| pJM10 | pBK-CMV containing partial sst operon sequences | This study |
| pSPT18 | E. coli vector for making digoxigenin-labeled RNA probes with SP6 and T7 RNA polymerases | Boehringer Mannheim |
| pSPT18/sstD | pSPT18 with 0.96-kb SmaI/PstI fragment containing sstD | This study |
| pET30a | E. coli expression vector | Novagen |
| pET30a/sstD | pET30a containing sstD coding sequences | This study |
| pS10 | pMK4 E. coli-S. aureus shuttle vector with S. aureus S10 ribosomal gene promoter and T2 transcriptional terminator from E. coli | This study |
| pS1038 | pS10 with 0.96-kb SmaI/PstI fragment containing sstD | This study |
Construction and screening of a genomic DNA library.
A genomic library of MunI fragments of S. aureus BB DNA was constructed in the phage vector λZapII (Stratagene) in accordance with manufacturer's instructions. The phage library was then mass excised into E. coli XLOLR (Stratagene) to generate a pBK-CMV phagemid library. The resultant colonies were plated onto selective LB agar, and colonies expressing staphylococcal antigens were identified by colony immunoblotting. Colonies were lifted onto nitrocellulose filters (Gelman) and then delipidated by incubation in chloroform vapor for 5 min. The filters were air dried, and cell debris was removed by vigorous washing of the filters in PBS. The filters were incubated for 1 h in a blocking solution of 3% (wt/vol) bovine serum albumin–0.1% (vol/vol) Tween 20 in PBS. The filters were then incubated overnight with polyclonal antistaphylococcal rat serum diluted 1/500 (vol/vol) in 0.1% (wt/vol) bovine serum albumin–0.1% (vol/vol) Tween 20 in PBS. Bound antibody was detected by with anti-rat peroxidase-conjugated antibodies, H2O2, and 4-chloro-1-naphthol (7). The plasmid DNA from reactive colonies was purified using Qiagen spin prep mini columns and subjected to restriction analysis. Inserts were sequenced with an ABI automated DNA sequencer. The DNA and predicted protein sequences were analyzed using the BLAST software available at www.ncbi.nlm.nih.gov.
Inverse PCR was used to clone the sequences 5′ and 3′ to the original insert DNA encoding the sst operon. The first round of inverse PCR using SpeI-digested BB genomic DNA and primers 38A rev (5′-GCTGAGAATGTAACTAAATTC-3′) and 38B for (5′-GCGTCCTGTTATTCAGATCAG-3′) resulted in a 700-bp PCR product. The digested genomic DNA was circularized with DNA ligase (Promega) overnight at room temperature before PCR amplification was performed under the following conditions: an initial denaturation at 94°C for 10 min; followed by 30 cycles of 94°C 1 min, 50 to 55°C for 1 min, and 72°C for 3 min; with a final step of elongation at 72°C for 10 min. The resultant PCR products were purified using the Qiagen spin prep column kit and then sequenced. However, the 5′ sequences were still incomplete so a second round of inverse PCR and subsequent sequencing were required. MunI-digested DNA and primers 38C rev (5′-TCATCGTATGAGGGATAGC-3′) and 38D for (5′-ACCGTCTATTGACACCATC-3′) were used to produce an approximately 3,000-bp PCR product, which completed the sequence of the ABC transporter operon.
Southern and Northern blot analyses.
Staphylococcal genomic DNA was digested with MunI, electrophoresed, and transferred to Hybond N+ membrane. The blot was incubated with a digoxigenin-labeled probe (Boehringer Mannheim) obtained by random priming of a 1,000-bp PCR product from BB genomic DNA and primers 38A for (5′-CGAATTTAGTTACATTCTC-3′) and 38B rev (5′-CTGATCTGAATAACAGGACGC-3′). Hybridization was performed overnight at 42°C, and blots were washed sequentially in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.1% (wt/vol) sodium dodecyl sulfate (SDS) at room temperature and in 0.5× SSC–0.1% (wt/vol) SDS at 68°C. Bound probe was visualized using anti-digoxigenin–alkaline phosphatase conjugate and the luminogenic substrate CDP-star (Boehringer Mannheim) in accordance with manufacturer's protocol. The signal was captured by exposure to X-ray film. Staphylococcal RNA was extracted using a Qiagen Rneasy total RNA kit but with lysostaphin at 100 μg/ml added to the initial cell lysis step. RNA samples and RNA markers (Promega) were electrophoresed on 1.5% agarose-formaldehyde gels and then transferred to Hybond N+ membrane as described in the Promega Protocols and Applications Guide, 3rd ed. The Northern blots were incubated overnight at 60°C with digoxigenin-labeled RNA probes. These were prepared by ligation of a 0.96-kb SmaI/PstI DNA fragment encoding the sstD open reading frame into pSPT18 (Boehringer Mannheim). Digoxigenin-labeled RNA probes sense and antisense to sstD were then produced in accordance with the manufacturer's protocol (Boehringer Mannheim). The hybridized filter was washed sequentially in 2× SSC–0.1% (wt/vol) SDS at room temperature for 5 min and in 0.1× SSC–0.1% (wt/vol) SDS at 68°C for 20 min. The bound probe was visualized using CDP-star (Boehringer Mannheim) in accordance with manufacturer's protocol.
Construction of an E. coli-S. aureus shuttle vector for use in antisense RNA studies.
To construct a suitable vector for antisense RNA studies, a strong, constitutive promoter and a transcriptional terminator were added to the E. coli-S. aureus shuttle vector pMK4. The transcriptional terminator from the rRNA (rrnB) operon in E. coli (accession no. AE000471.1) was amplified using primers T2for (5′-GGACCAGGCATGCATCGTAGCGCCGATGGTAG-3′) and T2rev (5′-GCGGCCGTACTGCAGGAGTTTGTAGAAACGCAAAAAG-3′). The resulting 268-bp PCR product was digested with PstI and NsiI, while pMK4 was digested with PstI. The digested insert and plasmid DNAs were purified, ligated together, and transformed into E. coli TOPO by electroporation. Transformants were selected on LB agar plus ampicillin (100 μg/ml) and screened by colony PCR using primers T2for and T2rev, resulting in the isolation of plasmid pMK4/T2. The S10 ribosomal gene promoter was then amplified from S. aureus BB genomic DNA using primers S10 for (5′-CTGAGAATTCCCGTTCTTATGACTA-3′) and S10 rev (5′-CTGACCCGGGCTTATTCGTCTACA-3′). The 320-bp PCR product and pMK4/T2 were digested with EcoRI and SmaI, purified, ligated together, and transformed into E. coli TOPO by electroporation. Transformants selected on LB agar plus ampicillin (100 μg/ml) were screened by colony PCR using the S10 for primer and a T2 for/rev primer. Isolation of the correct combination of S10 promoter and T2 transcriptional terminator in pMK4 was confirmed by sequence analysis, and the isolated plasmid was designated pS10.
Construction of plasmid pS10 containing the antisense sstD gene.
The sstD gene was amplified from S. aureus DNA using primers 38P for (5′-CACCTGCAGGGTAACAATTCTGA-3′) and 38S rev (5′-TAACCCGGGATCAAGTTCCTCAAT-3′), digested with PstI and SmaI, purified, and ligated into SmaI- and PstI-digested and purified pS10. The ligations were transformed into E. coli TOPO by electroporation and screened by colony PCR using S10 for and 39P for primers and restriction analysis. Both parental plasmid pS10 and antisense sstD plasmid pS1039 were transformed first into S. aureus RN4220 and then into RN6390-B by electroporation. Transformants were selected on TSB agar plus chloramphenicol (5 μg/ml).
Overexpression and purification of SstD in E. coli.
sstD was amplified by PCR from BB genomic DNA using primers 38E (5′-GCGGGCTCCATGGAGAAAACAGTCTTATATTTAG-3′) and 38F (5′-CGCCCAGGATCCAAACAATGATTAAGACCTTTAACC-3′). The PCR fragment obtained, which includes the prelipoprotein cleavage site, was digested with NcoI and BamHI and ligated into similarly digested pET30a (Novagen) to generate plasmid pET30a/sstD, which was transformed first into E. coli XL1-Blue and subsequently into E. coli BL21. SstD expression was induced by addition of isopropyl-β-d-thiogalactopyranoside (IPTG) at a final concentration of 1 mM to 3-h LB broth cultures (OD600, 0.3 to 0.4) of BL21 containing pET30a/sstD. Incubation of cultures was continued in the presence of IPTG for a further 3 h at 37°C before bacteria were pelleted by centrifugation at 3,500 × g for 5 min.
SstD was purified from the E. coli cell pellets by Triton X-114 phase partitioning following resuspension in PBS and lysis by sonication (4 × 30 s on ice at 8 mA on an MSE Soniprep sonicator fitted with a 3-mm-diameter probe). Insoluble debris and bacterial membranes were pelleted by centrifugation at 13,000 × g for 10 min, and the soluble fraction was retained and cooled to 4°C. Two hundred microliters of Triton X-114 (10% [vol/vol] in PBS) was added per ml of E. coli soluble fraction, and the mixture was incubated at 4°C overnight. Phase partitioning was achieved by incubation of the mixture at 37°C, centrifugation, and washing as previously described (6). SstD was precipitated from the Triton X-114 phase by overnight incubation with 10 volumes of acetone at −20°C and centrifugation at 13,000 × g for 10 min (3).
Fractionation of bacterial cells.
Total lysostaphin-soluble and membrane fractions of staphylococci were prepared essentially as described by Wilcox et al. (35), except that raffinose was omitted from the incubation mixture. Quantities of bacteria for digestion were standardized on the basis of OD600 measurements. Total soluble and membrane fractions of B. subtilis were prepared in a similar way, except that lysozyme (100 μg/ml in PBS) was used instead of lysostaphin. Triton X-114 phase partitioning was performed as previously described (6).
SDS-PAGE and immunoblotting.
All samples were solubilized by boiling in Laemmli sample buffer (20) for 5 min. Polypeptides were separated by SDS-polyacrylamide gel electrophoresis (PAGE) using a 4% (wt/vol) acrylamide stacking gel and a 10% (wt/vol) resolving gel in a Bio-Rad Mini Protean II gel apparatus as previously described (1). For immunoblotting, polypeptides were transferred to BioTrace NT membrane (Gelman), followed by blocking, incubation with primary antibody (1/500 dilution overnight) and conjugate (1/2,000 dilution of anti-rat peroxidase conjugate for 4 h), and detection of bound antibody as previously described (1).
Polyclonal antibody production.
Polyvalent rat anti-S. aureus BB serum was collected from chamber-implanted (28), BB-inoculated Wistar rats approximately 21 days postinoculation. Antibody to Triton X-114-purified, acetone precipitated, recombinant SstD was produced in female Wistar rats. SstD was resuspended in sterile PBS and administered subcutaneously initially in Freund's complete adjuvant and 2 and 4 weeks later in Freund's incomplete adjuvant. Serum was collected 2 weeks after the third immunization.
Nucleotide sequence accession number.
The DNA sequence of the S. aureus sstD operon is available in the GenBank database under accession no. AJ005352.
RESULTS
Isolation and characterization of a staphylococcal operon encoding an ABC transporter.
Our previous studies (6, 15, 23, 24, 35) have identified several iron-regulated proteins in S. aureus, including a lipoprotein of 32 kDa and two lipoproteins of 36 kDa. To further characterize S. aureus iron-regulated antigenic proteins, we screened an S. aureus BB λZapII genomic expression library with polyvalent anti-BB rat serum. This serum was obtained from chamber-implanted, BB-inoculated rats and potentially contains antibodies against S. aureus antigens expressed under the in vivo growth conditions simulated in this model. The λZapII library was mass excised prior to screening, so that any unstable clones were already eliminated from the library. Consequently, a number of colonies stably expressing staphylococcal antigens were identified by colony immunoblotting and the plasmids were recovered and analyzed. Initial sequencing of one of the DNA inserts identified a clone, pJM10, which encoded an ABC transporter with significant homology to both gram-negative and gram-positive ferric-siderophore ABC transporters. Therefore, pJM10 was analyzed further.
Southern blotting was used to demonstrate that the 3.4-kb MunI insert of pJM10 was a single locus in the S. aureus genome and to identify a homolog in S. epidermidis. Probing with the digoxigenin-labeled insert from pJM10 identified a single locus in MunI-digested S. aureus strain BB, RN4220, and W genomic DNAs (Fig. 1A). Moreover, the probe also identified a single locus in S. epidermidis 901 genomic DNA.
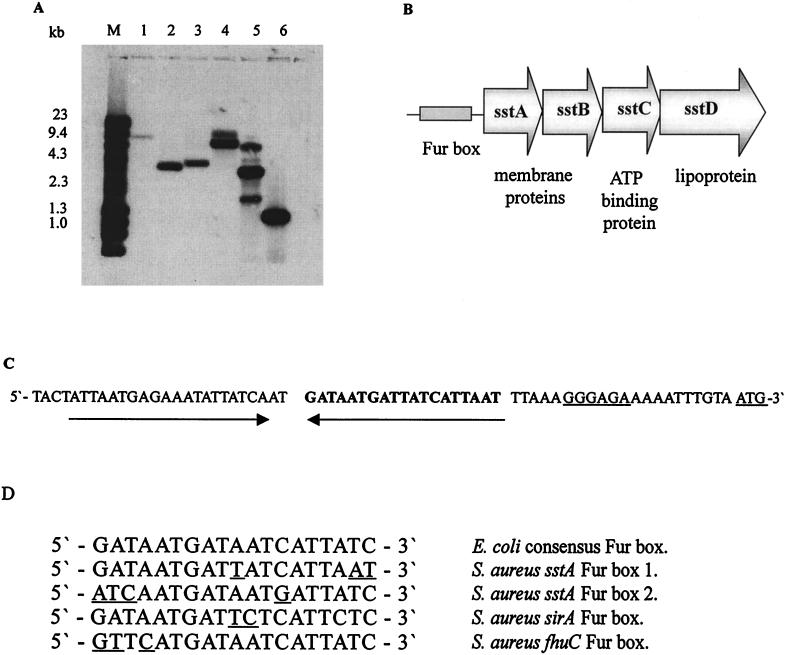
FIG. 1.
(A) Southern blot analysis showing DNA homology among the S. aureus BB sstABCD operon, other S. aureus strains, and S. epidermidis. Plasmid and genomic DNAs were digested with MunI, and the Southern blots were hybridized with a digoxigenin-labeled PCR product amplified from BB genomic DNA using primers 38A for and 38B rev. Lanes: M, λHindIII/phiX174 HaeIII DNA markers; 1, S. epidermidis 901; 2, S. aureus BB; 3, S. aureus W; 4, S. aureus RN4220; 5, plasmid pJM10; 6, diluted PCR product used as the probe. (B) Organization of the S. aureus sstABCD ABC transporter operon. (C) The upstream sequences of the sstABCD operon showing one of the putative Fur box sequences in bold and the position of the potential inverted repeat region. The ribosome-binding site and the translational start site are underlined. (D) Alignment of the sstABCD putative Fur box sequences with the E. coli Fur box consensus sequence (9) and the Fur box sequences from the S. aureus sirA (14) and fhuA (36) genes. The differences are highlighted.
The cloned S. aureus DNA locus shows significant homology to bacterial siderophore ABC transporters.
DNA sequence analysis of the 3.4-kb MunI insert of pJM10 identified the 3′ end of an incomplete open reading frame and two complete open reading frames whose predicted polypeptides showed homology to components of ABC transporters involved in the uptake of ferric siderophores (see below). Inverse PCR was used to clone the missing upstream sequences. Once all of the overlapping sequences were put together, four open reading frames encoding putative polypeptides of 34, 25, 28, and 38 kDa were identified (Fig. 1B). The genes encoding these polypeptides were designated sstA, -B, -C, and -D, respectively (sst stands for staphylococcal siderophore transporter).
The putative SstA and SstB proteins are hydrophobic proteins with significant homology at the peptide level to the cytoplasmic membrane proteins of a number of bacterial siderophore transporters, including the B. subtilis FatD and FatC homologs, C. jejuni CeuB and CeuC, and the Vibrio anguillarum FatD and FatC proteins (Table 2). SstC shows significant homology to several ATP-binding proteins involved in ferric-siderophore transport, including B. subtilis FecE and C. jejuni CeuD. SstC contains two conserved ATP-binding domains (34GPNGAGKSTLLS46 and 136LSGGQRQRAYIAMTIAQDTEYILLDEPLNNLD168), which are found in the same approximate location as in other ATP-binding proteins (27). The SstD polypeptide has significant homology to the lipoproteins of a number of ABC transporters involved in ferric-siderophore uptake. These include the V. anguillarum lipoprotein FatB, which is involved in the transport of the V. anguillarum siderophore anguibactin and the FatB homologue of B. subtilis (Table 2). There is also strong homology with the enterobactin transporters of C. jejuni and Neisseria gonorrhoeae. The SstD lipoprotein has a consensus prelipoprotein signal peptide cleavage sequence (16LAAC19) (13) and partial conservation of an amino acid consensus sequence identified as a signature motif for siderophore-binding proteins (127EVNFDKIAATKPEVI141) (31).
TABLE 2.
Sequence homologies of S. aureus sstABCD transporter proteins
| Polypeptide | Homologous polypeptide | Identity (%) | Similarity (%) | Accession no. |
|---|---|---|---|---|
| SstA | B. subtilis FatD homologue (YclN) | 45 | 68 | BAA09012 |
| C. jejuni CeuB | 38 | 58 | CAB73779 | |
| V. anguillarum FatD | 30 | 54 | P37738 | |
| SstB | B. subtilis FatC homologue (YclO) | 35 | 56 | BAA09013 |
| C. jejuni CeuC | 31 | 53 | CAB73780 | |
| V. anguillarum FatC | 20 | 42 | P37737 | |
| SstC | B. subtilis YclP | 55 | 76 | BAA09014 |
| C. jejuni CeuD | 52 | 71 | CAB73781 | |
| SstD | B. subtilis FatB homologue (YclQ) | 38 | 57 | BAA09015 |
| C. jejuni CeuE | 33 | 55 | CAB73782 | |
| N. gonorrhoeae FetB | 29 | 47 | AAD29611.1 | |
| V. anguillarum FatB | 28 | 52 | P11460 |
All of the putative open reading frames are preceded by potential staphylococcal Shine-Dalgarno sequences 10 to 11 bp upstream of the start codons. Sequence analysis suggests that the genes are transcribed as a single polycistronic mRNA, as there are −35 and −10 signal sequences at the beginning of the operon and a rho-independent transcription terminator downstream of sstD. It is probable that transcription is regulated by iron, since the sstA gene is preceded by a potential stem-loop structure, which contains within it two putative Fur box consensus sequences (Fig. 1C). Fur box 1 (232 to 251) has 16 of the 19 E. coli Fur box consensus nucleotides conserved and is one arm of the stem-loop structure, while Fur box 2 has 15 of 19 conserved nucleotides (Fig. 1D).
The SstD protein is a 38-kDa membrane-bound lipoprotein expressed maximally under iron-deficient conditions in vitro and in vivo.
On the basis of DNA analysis, the SstD protein was predicted to be an iron-regulated 38-kDa lipoprotein which was likely to be associated with the bacterial cytoplasmic membrane. However, initial SDS-PAGE and immunoblotting studies using S. aureus BB grown overnight in RPMI 1640 medium as described in our earlier report (6) failed to identify an iron-regulated membrane-associated protein of 38 kDa (data not shown). Based on this result, we reasoned that our failure to detect the predicted 38-kDa lipoprotein could have been due either to low-level expression of SstD in S. aureus grown overnight in RPMI 1640 medium and/or a low titer of specific antibody to this lipoprotein in the polyvalent rat serum used for immunoblotting. To generate a high-titer, monovalent antiserum to SstD, we therefore first cloned the sstD gene into pET30a and overexpressed the lipoprotein in E. coli under induction with IPTG. To enhance the potential immunogenicity of the antigen, the prelipoprotein cleavage sequence was included to permit lipidation of the preprotein in E. coli after cleavage of the N terminus. That this was achieved was shown both by failure of the recombinant protein to bind to an Ni2+ column (indicating loss of the N-terminal His tag from pET30a) and our ability to purify the recombinant SstD from E. coli lysates using Triton X-114 phase partitioning (data not shown). The purified recombinant lipoprotein, which migrated at a molecular mass of approximately 45 kDa on SDS-PAGE (data not shown), was then used to generate monospecific anti-SstD sera in rats.
In an attempt to enhance expression of SstD in S. aureus BB, we also modified the growth medium and conditions used to grow the bacteria from our original method (6). To overcome potential variation in the iron content of different batches of RPMI 1640 medium and any effect this may have had on levels of iron-regulated protein expression in different experiments, we used Chelex resin to remove iron from the growth medium. Preliminary experiments showed that growth for 6 h in Chelex-treated, supplemented CRPM1 gave reproducible, high-level expression of several previously characterized iron-regulated proteins (data not shown).
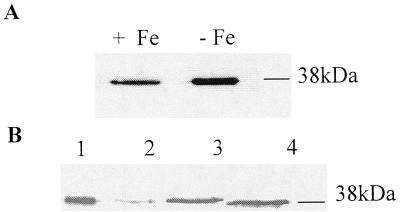
Immunoblots of S. aureus BB membrane preparations prepared from bacteria grown in this way and probed with anti-SstD serum identified a polypeptide of around 38 kDa, which was partially repressed under iron-rich growth conditions (Fig. 2A). This 38-kDa antigen was also extractable with Triton X-114 from membrane preparations of iron-restricted S. aureus BB, supporting the identity of the antigen as a lipoprotein (Fig. 2B). Comparison of lanes 2 and 3 of Fig. 4B shows a reduced level of SstD extractable from bacteria grown for 18 h in RPMI 1640 medium compared to that extracted from those grown in CRPMI for 6 h. A 37-kDa polypeptide which reacted with the anti-SstD serum was also extractable with Triton X-114 from iron-restricted S. epidermidis 901 membrane preparations (Fig. 2B). Thus, the SstD lipoprotein is expressed in vitro and that expression is partially regulated by iron. To determine whether S. aureus grown in vivo expresses the SstD lipoprotein, S. aureus BB was grown in a rat intraperitoneal chamber model. Figure 3 shows that membrane preparations isolated from S. aureus recovered without subculture from the chambers contain the 38-kDa protein, demonstrating that the SstD lipoprotein is expressed in vivo.
FIG. 2.
Immunoblots with rat anti-SstD serum showing membrane association, iron regulation, and Triton X-114 solubility of SstD. (A) Membrane fractions of S. aureus BB grown for 6 h under iron-restricted (lane 1) or iron-rich (lane 2) conditions. The 38-kDa SstD lipoprotein is indicated. (B) Triton X-114 extracts of S. aureus BB grown for 18 h in RPMI 1640 medium (lane 2) and S. aureus BB (lane 3) or S. epidermidis 901 (lane 4) grown for 6 h in CRPMI. Lane 1 shows the membrane fraction of S. aureus BB grown for 6 h in CRPMI. The 38-kDa SstD protein is indicated.
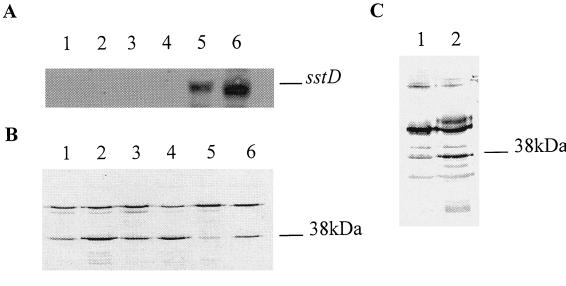
FIG. 4.
(A) Northern blot analysis of antisense sstD transcripts. Total RNAs were isolated from logarithmically growing S. aureus strains grown under iron-rich or iron-restricted conditions in vitro. The Northern blot was hybridized with digoxigenin-labeled sstD RNA. The approximately 1-kb antisense transcript is indicated. (B) Western blot analysis of membrane fractions from S. aureus RN6390-B showing the decrease in SstD protein production in the presence of sstD antisense RNA. Lanes: 1, RN6390-B (with Fe); 2, RN6390-B (without Fe); 3, RN6390-B pS10 (with Fe); 4, RN6390-B pS10 (without Fe); 5, RN6390-B pS1038 (with Fe); 6, RN6390-B pS1038 (without Fe). (C) Immunoblot of membrane preparations from S. aureus recovered without subculture from the chambers and probed with anti-SstD serum. Lanes: 1, RN6390-B S1038; 2, RN6390-B S10.
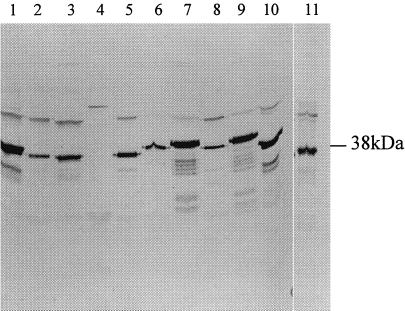
FIG. 3.
Immunoblot of Triton-X114 extracts prepared from S. aureus, coagulase-negative staphylococci, and B. subtilis grown under iron-restricted conditions in vitro and S. aureus BB grown in vivo. The immunoblots were reacted with monospecific rat antiserum to the 38-kDa SstD lipoprotein. Lanes: 1, S. aureus BB; 2, S. haemolyticus; 3, S. carnosus; 4, B. subtilis; 5, S. hominis; 6, S. warnerii; 7, S. cohni, 8, S. lugdunensis; 9, S. saprophyticus; 10, S. epidermidis; 11, S. aureus BB grown in vivo.
The SstD lipoprotein antigen is found in a range of staphylococcal species.
The anti-SstD serum was used in immunoblotting studies to investigate the conservation of this antigen among a range of staphylococcal species and B. subtilis (Fig. 3). Antigenic polypeptides, of approximately 38 kDa were detected in membrane fractions of all nine species of staphylococci grown under iron-deficient conditions. Thus, the SstD lipoprotein antigen is conserved among staphylococci. However, no cross-reacting polypeptide was identified in B. subtilis even though the SstD protein and its B. subtilis homolog are 38% identical at the amino acid level.
sstD antisense RNA downregulates sstD expression in vitro.
To determine the function and importance of the SstABCD transporter for growth of S. aureus, we used antisense RNA technology to disrupt the expression of the Sst transporter. A 960-bp fragment of the sstD gene was cloned in the 3′-to-5′ orientation downstream of a constitutive promoter in an E. coli-S. aureus shuttle vector, pS10. The sstD antisense plasmid (pS1038) and the parental plasmid (pS10) were then transformed separately into S. aureus RN6390-B, resulting in isogenic transformants which vary only in the production of antisense sstD RNA. S. aureus strain RN6390-B was used for these studies because we were unable to genetically manipulate the wild-type S. aureus strain, BB, used previously. RN6390-B was shown to have an sstABCD homologue by Southern hybridization with a BB DNA probe, PCR using BB-specific primers with RN6390-B template DNA, and Western blot analysis using the SstD antibody (data not shown).
Northern blot analysis was used to examine antisense RNA production in wild-type S. aureus strain RN6390-B and the RN6390-B transformants S10 and S1038 when grown under iron-rich or iron-deficient conditions. An antisense transcript was observed in RN6390-B(pS1038) grown in either iron-rich, or iron-deficient medium (Fig. 4A). No antisense transcripts were observed in wild-type RN6390-B or the S10 transformant. Thus, antisense sstD RNA is constitutively produced only in the S1038 transformant.
Immunoblots of membrane preparations from RN6390-B, S10, and S1038 grown under iron-rich or iron-deficient conditions probed with anti-sstD serum showed a reduction in the amount of SstD protein produced by S1038, whereas SstD protein expression was unaffected in the wild type and parental-plasmid-containing transformant S10 (Fig. 4B). No growth differences were observed between the strains (data not shown). Therefore, sstD gene expression is partially disrupted by the production of antisense sstD RNA, resulting in decreased levels of expression of SstD.
Downregulation of sstD expression by antisense RNA does not affect the ability of S. aureus to grow in vivo in a rat chamber implant model.
To determine the effect of the downregulation of the SstD protein on growth in vivo, the isogenic transformants S10 and S1038 were inoculated into separate rat intraperitoneal chambers and growth was monitored over 48 h by sampling and viable counting. There was no major difference in the growth rate or final cell density achieved for S10 or S1038 with mean bacterial counts increasing from approximately 106 bacteria per ml at the time of inoculation to 6 × 108 bacteria per ml for S10 and 9 × 108 bacteria per ml for S1038, respectively, after 48 h. Bacteria isolated at each time point were tested for chloramphenicol resistance to assess plasmid stability in the chamber implant model. All of the bacteria isolated at all time points maintained chloramphenicol resistance, and pS10 and pS1038 were recoverable from the isolated bacteria, confirming plasmid stability. Immunoblots of membrane preparations of the bacteria isolated without subculture from the chambers showed a reduction in the amount of SstD protein produced in vivo due to the presence of antisense sstD RNA (Fig. 4C).
DISCUSSION
Staphylococci are able to utilize both endogenous and exogenous siderophores to acquire iron in iron-deficient environments. Due to their small size, ferric siderophores are likely to be able to freely diffuse through the gram-positive bacterial cell wall due to its porosity but an active transport mechanism is required to facilitate their transport through the cytoplasmic membrane. Although siderophore transport mechanisms and their regulation have been well studied in gram-negative bacteria, only two putative staphylococcal siderophore transporters, SirABC (14) and FhuABC (36), have been identified. We can now report the cloning and characterization of a third genetic locus, sstABCD, which, on the basis of sequence homology, is likely to encode a third staphylococcal siderophore transporter.
SstABCD constitutes a classical ABC transporter, with a ligand-binding lipoprotein, an ATP-binding protein, and two cytoplasmic membrane proteins which potentially act as permeases (27). The SstABCD operon is preceded by a Fur box consensus sequence, suggesting regulation by iron, and our immunoblotting analysis confirms that SstD expression is partially repressed by addition of ferric iron to the growth medium. Moreover, SstD expression is induced in vivo, a highly iron-restricted environment. However, although induced under iron-deficient conditions, SstD expression is still relatively poor. No sstABCD transcripts could be identified by Northern blot analysis, and in contrast to our earlier studies with other iron-regulated staphylococcal lipoproteins (6), the SstD lipoprotein could not be directly identified in staphylococcal membrane fractions following SDS-PAGE. SstD may not be a highly expressed protein, or it is possible that, as found with siderophore transporters in some gram-negative bacteria, expression of the Sst transporter is regulated on at least two levels, a global level of regulation in response to iron starvation mediated by Fur and a local level of control on which the presence of the siderophore molecule itself is required for induced expression of the transporter. This type of regulation has been described for the ferric dicitrate transport system of E. coli, pseudobactin transport in Pseudomonas putida, and pyoverdin, enterobactin, and pyochelin transport in P. aeruginosa (33). Therefore, further enhancement of the expression of SstD may require the presence of the ligand. The apparently constitutive low level of expression of SstD observed in the staphylococcal membrane in the presence of ferric iron may facilitate rapid responses to changes in iron availability and specific ligand concentration. How this induction would be mediated is unclear, since there are no open reading frames encoding identifiable regulators near the sstABCD operon. A second possible explanation for the low level of induction of SstD expression in response to iron limitation is the potential secondary structure associated with the Fur box found upstream of the operon. The formation of a stem-loop structure containing the Fur box could conceivably influence the Fur-mediated regulation of the system or may disrupt RNA polymerase activity. The importance of this region in the promoter of the sstABCD operon requires further investigation.
To begin to address the importance of the SstABCD transporter for staphylococcal growth and virulence, we initially attempted to disrupt its function by mutation using allelic replacement. However, we were unable to isolate sstABCD mutants using this approach. Therefore, we attempted to disrupt sstD gene expression by antisense RNA technology. This method of modification of gene expression has previously been very successfully used with S. aureus to decrease alpha-toxin production, resulting in attenuation of virulence in a murine infection model (17, 18), but our data represents the first use of this approach to investigate iron-regulated protein and ABC transporter functions in staphylococci. In this study, we used the S. aureus S10 ribosomal protein promoter, a strong, constitutive promoter active during the logarithmic stage of growth, to drive sstD antisense RNA production. This ensured that the antisense sstD transcript was produced in large enough quantities and at an appropriate time in the growth cycle to effectively disrupt SstD expression. Northern blot analysis confirmed the presence of the antisense sstD transcript, while immunoblotting confirmed a decrease in SstD protein levels in vitro.
Attempts to identify the specific siderophore transported by the sstABCD operon using parental plasmid pS10 or antisense SstD construct pS1038 in S. aureus RN6390B or RN4220 using plate bioassays in vitro proved inconclusive. No consistent difference in the utilization of a range of siderophores or siderophore precursors was observed in the presence or absence of reduced SstD expression (data not shown). These findings may be due to a number of factors. First, although SstD expression was reduced in the presence of the antisense construct, the residual level of expression may be sufficient to maintain a wild-type phenotype and growth characteristics. Second, other staphylococcal ABC transporters or ligand-binding lipoproteins may compensate for the loss of SstD function. Other bacterial pathogens have been shown to express more than one uptake system for individual siderophores; for example, Salmonella typhimurium has at least two enterobactin transporters (29). Third, although SstD amino acid sequence homology suggests that the transported siderophore is structurally related to either enterobactin or anguibactin, it is conceivable that the ligand is an as yet uncharacterized siderophore produced either by the staphylococci or by any of the diverse range of other bacteria and fungi that staphylococci may come into contact with during colonization or infection of mammalian hosts. Given the conservation of an antigenic and presumably functional homolog of SstD among a range of staphylococcal species, it also seems likely that the ligand is a siderophore commonly encountered by the staphylococci, the identity of which is the subject of our ongoing work.
Further information concerning the contribution of SstD to staphylococcal growth and virulence may also be obtained from additional in vivo studies. Recent studies of S. typhimurium have, however, highlighted the importance of using a range of infection models to identify the contribution of specific iron uptake systems to bacterial virulence (16). sitABCD iron uptake operon mutants showed no loss of virulence when administered intraperitoneally to mice but were attenuated when administered orally, indicating a differential role for the transporter in different tissues. Given these observations, our finding that reduction of SstD expression revealed no effect on the growth of S. aureus in a peritoneal implant model may not be surprising. It is also possible that SstABCD and other staphylococcal iron transporters show tissue-specific expression regulated by the differential availability of iron sources, e.g., siderophores, transferrin, lactoferrin, heme, etc., at different body sites (34). This possibility will be addressed in our future studies. However, in vivo data presented here demonstrates that SstD and the antisense sstD RNA are expressed in vivo. The stability of plasmids pS10 and pS1038 in S. aureus in vivo in the absence of any antibiotic selection also indicates the greater potential of these specific vectors, and antisense technology in general, for studying the roles of potential staphylococcal virulence determinants.
ACKNOWLEDGMENT
This work was supported by program grant G9219778 from the Medical Research Council to P.W.
REFERENCES
- 1.Arbuthnott J P, Arbuthnott E, Arbuthnott A D J, Pike W J, Cockayne A. Investigation of microbial growth in vivo: evaluation of a novel in vivochamber implant system. FEMS Microbiol Lett. 1992;100:75–80. doi: 10.1111/j.1574-6968.1992.tb14022.x. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Greene Publishing Associates and Wiley-Interscience; 1994. [Google Scholar]
- 3.Brusca J S, Radolf J D. Isolation of integral membrane proteins by phase partitioning with Triton X-114. Methods Enzymol. 1994;228:182–193. doi: 10.1016/0076-6879(94)28019-3. [DOI] [PubMed] [Google Scholar]
- 4.Bsat N, Herbig A, Casillas-Martinez L, Setlow P, Helmann J D. Bacillus subtiliscontains multiple Fur homologues: identification of the iron uptake (Fur) and peroxide regulon (PerR) repressors. Mol Microbiol. 1998;29:189–198. doi: 10.1046/j.1365-2958.1998.00921.x. [DOI] [PubMed] [Google Scholar]
- 5.Chang S, Cohen N. High frequency transformation of Bacillus subtilisprotoplasts by plasmid DNA. Mol Gen Genet. 1979;168:111–115. doi: 10.1007/BF00267940. [DOI] [PubMed] [Google Scholar]
- 6.Cockayne A, Hill P J, Powell N B L, Bishop K, Sims C, Williams P. Molecular cloning of a 32-kilodalton lipoprotein component of a novel iron-regulated Staphylococcus epidermidisABC transporter. Infect Immun. 1998;66:3767–3774. doi: 10.1128/iai.66.8.3767-3774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Courcol R J, Trivier D, Bissinger M C, Martin G R, Brown M R W. Siderophore production by Staphylococcus aureusand identification of iron-regulated proteins. Infect Immun. 1997;65:1944–1948. doi: 10.1128/iai.65.5.1944-1948.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Drechsel H, Freund S, Nicholson G, Haag H, Jung O, Zahner H, Jung G. Purification and chemical characterization of staphyloferrin B, a hydrophilic siderophore from staphylococci. Biometals. 1993;6:185–192. doi: 10.1007/BF00205858. [DOI] [PubMed] [Google Scholar]
- 9.Escolar L, Perez-Martin J, de Lorenzo V. Opening the iron box: transcriptional metalloregulation by the Fur protein. J Bacteriol. 1999;181:6223–6229. doi: 10.1128/jb.181.20.6223-6229.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gaballa A, Helmann J D. Identification of a zinc-specific metalloregulatory protein, Zur, controlling zinc transport operons in Bacillus subtilis. J Bacteriol. 1998;180:5815–5821. doi: 10.1128/jb.180.22.5815-5821.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Griffiths E, Williams P. Iron proteins. In: Bullen J J, Griffiths E, editors. Iron and infection: molecular, physiological and clinical aspects. 2nd ed. New York, N.Y: John Wiley & Sons, Inc.; 1999. pp. 27–86. [Google Scholar]
- 12.Haag H, Fiedler H P, Meiwes J, Drechsel H, Jung G, Zahner H. Isolation and biological characterisation of staphyloferrin B, a compound with siderophore activity from staphylococci. FEMS Microbiol Lett. 1994;115:125–130. doi: 10.1111/j.1574-6968.1994.tb06626.x. [DOI] [PubMed] [Google Scholar]
- 13.Hayashi S, Wu H C. Lipoproteins in bacteria. J Bioenerg Biomembr. 1990;22:451–471. doi: 10.1007/BF00763177. [DOI] [PubMed] [Google Scholar]
- 14.Heinrichs J H, Gatlin L E, Kunsch C, Choi G H, Hanson M S. Identification and characterization of SirA, an iron-regulated protein from Staphylococcus aureus. J Bacteriol. 1999;181:1436–1443. doi: 10.1128/jb.181.5.1436-1443.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hill P J, Cockayne A, Landers P, Morrissey J A, Sims C M, Williams P. SirR, a novel iron-dependent repressor in Staphylococcus epidermidis. Infect Immun. 1998;66:4123–4129. doi: 10.1128/iai.66.9.4123-4129.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Janakiraman A, Slauch J M. The putative iron transport system SitABCD encoded on SPI1 is required for full virulence of Salmonella typhimurium. Mol Microbiol. 2000;35:1146–1155. doi: 10.1046/j.1365-2958.2000.01783.x. [DOI] [PubMed] [Google Scholar]
- 17.Ji Y, Marra A, Rosenberg M, Woodnutt G. Regulated antisense RNA eliminates alpha-toxin virulence in Staphylococcus aureusinfection. J Bacteriol. 1999;181:6585–6590. doi: 10.1128/jb.181.21.6585-6590.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kernodle D S, Volardi R K R, Menzies B E, Hager C C, Edwards K M. Expression of an antisense hla fragment in Staphylococcus aureusreduces alpha-toxin production in vitro and attenuates lethal activity in a murine model. Infect Immun. 1997;65:179–184. doi: 10.1128/iai.65.1.179-184.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kraemer G R, Iandolo J J. High-frequency transformation of Staphylococcus aureusby electroporation. Curr Microbiol. 1990;21:372–376. [Google Scholar]
- 20.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 21.Maskell J P. The functional interchangeability of enterobacterial and staphylococcal chelators. Antonie van Leeuwenhoek. 1980;46:343–351. doi: 10.1007/BF00421981. [DOI] [PubMed] [Google Scholar]
- 22.Meiwes J, Fiedler H P, Haag H, Zahner H, Konetschny-Rapp S, Jung G. Isolation and characterisation of staphyloferrin A, a compound with siderophore activity from Staphylococcus hyicusDSM 20459. FEMS Microbiol Lett. 1990;67:201–206. doi: 10.1111/j.1574-6968.1990.tb13863.x. [DOI] [PubMed] [Google Scholar]
- 23.Modun B, Williams P, Pike W J, Cockayne A, Arbuthnott J P, Finch R, Denyer S P. Cell envelope proteins of Staphylococcus epidermidisgrown in vivo in peritoneal chamber implant. Infect Immun. 1992;60:2551–2553. doi: 10.1128/iai.60.6.2551-2553.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Modun B, Kendall D, Williams P. Staphylococci express a receptor for human transferrin: identification of a 42-kilodalton cell wall transferrin-binding protein. Infect Immun. 1994;62:3850–3858. doi: 10.1128/iai.62.9.3850-3858.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Modun B, Cockayne A, Finch R G, Williams P. The Staphylococcus aureus and Staphylococcus epidermidis transferrin-binding proteins are expressed in vivoduring infection. Microbiology. 1998;144:1005–1012. doi: 10.1099/00221287-144-4-1005. [DOI] [PubMed] [Google Scholar]
- 26.Neilands J B. Siderophore systems of bacteria and fungi. In: Beveridge T J, Doyle R J, editors. Metal ions and bacteria. New York, N.Y: John Wiley & Sons, Inc.; 1989. pp. 141–163. [Google Scholar]
- 27.Nikaido H, Hall J A. Overview of bacterial ABC transporters. Methods Enzymol. 1998;292:3–20. doi: 10.1016/s0076-6879(98)92003-1. [DOI] [PubMed] [Google Scholar]
- 28.Pike W J, Cockayne A, Webster C A, Slack R C B, Shelton A P, Arbuthnott J P. Development and design of a novel in vivochamber implant for the analysis of microbial virulence and assessment of antimicrobial chemotherapy. Microb Pathog. 1991;10:443–450. doi: 10.1016/0882-4010(91)90109-n. [DOI] [PubMed] [Google Scholar]
- 29.Rabsch W, Voight W, Reissbrodt R, Tsolis R M, Baumler A J. Salmonella typhimuriumIroN and FepA proteins mediate uptake of enterobactin but differ in their specificity for other siderophores. J Bacteriol. 1999;181:3610–3612. doi: 10.1128/jb.181.11.3610-3612.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith D G E, Wilcox M H, Williams P, Finch R G, Denyer S P. Characterization of the cell envelope proteins of Staphylococcus epidermidiscultured in human peritoneal dialysate. Infect Immun. 1991;59:617–624. doi: 10.1128/iai.59.2.617-624.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tam R, Saier M H J. Structural, functional, and evolutionary relationships among extracellular solute-binding receptors of bacteria. Microbiol Rev. 1993;57:320–346. doi: 10.1128/mr.57.2.320-346.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tao X, Schiering N, Zeng H Y, Ringe D, Murphy J R. Iron, DtxR and the regulation of diphtheria toxin expression. Mol Microbiol. 1994;14:191–197. doi: 10.1111/j.1365-2958.1994.tb01280.x. [DOI] [PubMed] [Google Scholar]
- 33.Venturi V, Weisbeek P, Koster M. Gene regulation of siderophore-mediated iron acquisition in Pseudomonas: not only the Fur repressor. Mol Microbiol. 1995;17:603–610. doi: 10.1111/j.1365-2958.1995.mmi_17040603.x. [DOI] [PubMed] [Google Scholar]
- 34.Weinberg E D. Patho-ecological implications of microbial acquisition of host iron. Rev Med Microbiol. 1998;9:171–178. [Google Scholar]
- 35.Wilcox M H, Williams P, Smith D G E, Finch R G, Denyer S P. Variation in the expression of cell envelope proteins of coagulase-negative staphylococci cultured under iron-restricted conditions in human peritoneal dialysate. J Gen Microbiol. 1991;137:2561–2570. doi: 10.1099/00221287-137-11-2561. [DOI] [PubMed] [Google Scholar]
- 36.Xiong A, Singh V K, Cabrera G, Jayaswal R K. Molecular characterisation of the ferric regulator, Fur, from Staphylococcus aureus. Microbiology. 2000;146:659–668. doi: 10.1099/00221287-146-3-659. [DOI] [PubMed] [Google Scholar]