Background:
Osteosarcoma (OS) is the most common bone cancer in adolescents, and has a high propensity to metastasize. Ferroptosis is a unique modality of cell death, driving the metastasis of cancer cells. Identifying ferroptosis-related genes (FRGs) as prognostic factors will be critical to predict the outcomes of OS. This study aimed to explore the prognostic value of FRGs in OS and build a prognostic model to indirectly improve OS patients’ outcomes.
Methods:
OS data were downloaded from the TARGET database and 2 Gene Expression Omnibus datasets. Univariate Cox regression was conducted to assess FRGs. A risk score model basing on 5 FRGs was constructed via LASSO-Cox regression. Multivariate Cox regression analysis was used to determine the independent prognostic factors. The Nomogram model was built using independent prognostic factors. The relationship between the risk score and the immune cell infiltration was estimated by CIBERSORT, and the correlation between the risk score and immune checkpoints was also analyzed.
Results:
Based on the prognosis-related FRGs, we built a regression model: Risk score = (−0.01382853 × ACSL4) − (0.05371778 × HMOX1) − (0.02434655 × GPX4) − (0.16432810 × PRNP) − (0.15567120 × ATG7). OS patients with high risk score tended to suffer from poor prognosis, validated in 2 Gene Expression Omnibus datasets. The Nomogram model showed the combination of the risk score and the tumour-node-metastasis stage improved predictive effectiveness. The risk score was also related to immune cell infiltration and immune checkpoint expression.
Conclusion:
The risk score model based on 5 FRGs was a reliable prognostic predictive indicator for OS patients.
Keywords: ferroptosis, LASSO-Cox regression, Nomogram, osteosarcoma, risk score model
1. Introduction
Osteosarcoma (OS) is the most common malignant bone cancer in children and adolescents, peaking at 15 to 19 years old.[1,2] The morbidity is over 3 cases per million per year worldwide. The mortality is higher in males than in females.[3,4] Besides, the 5-year disease-free survival rate of primary cancer patients is 52.9%, and it is even lower in OS patients with metastasis.[5] The tumor mass of OS often found in the distal femur and proximal bones is mainly composed of tumor cells that are related to the production of osteoid tissue or immature bone.[6,7] Current treatments for OS mainly include resection of the primary tumor with or without adjuvant chemotherapy. The development of treatment methods for OS patients has improved the survival rates, but the prognosis of OS patients with metastasis or recurrent disease remains is still poor.[8,9] Grade, size, and location are the 3 most important prognostic parameters in clinical cases, but the significant morphologic overlaps between bone cancer subtypes and the preference for less invasive methods raise great challenges for pathologists.[10,11]
Ferroptosis, a term coined by Dr Brent R Stockwell, is a form of regulated cell death depending on iron,[12] which is caused by the accumulation of reactive oxygen species based on lipid.[13] It has a direct or indirect effect on the glutathione peroxidase through different pathways, leading to a reduction of antioxidant capacity and accumulation of lipid reactive oxygen species, eventually causing oxidative cell death.[14] Previous studies have indicated that the induction of ferroptosis was a potential treatment option, as it is able to cause certain cancer cell death.[15,16] Erastin (a type of ferroptosis inducer) also has anti-cancer effects in certain cancer cells when it is used with or without chemotherapy drugs such as cisplatin.[17] The ferroptosis-related genes (FRGs) have been reported to correlate with the prognosis of hepatocellular carcinoma and glioma.[15,18] Zhu et al[19,20] have shown that FRGs may also be involved in the progression and prognoses of esophageal adenocarcinoma and bladder cancer. Moreover, prognostic models constructed by FRGs have exhibited potential prognostic values and may help predict the prognosis of cancer patients to assist clinical doctors in choosing individual treatments.[21] However, only a few studies about the prognostic values of FRGs for OS patients were reported.[22]
Herein, we systematically analyzed the prognostic values of FRGs in OS patients hoping to construct a reliable FRG-based prognostic model to facilitate OS diagnosis and treatment.
2. Materials and Methods
2.1. Data collection
The mRNA expression profiles of 88 OS patients were downloaded from the Therapeutically Applicable Research to Generate Effective Treatment (TARGET, https://ocg.cancer.gov/programs/target). Eight-four patients had complete survival information (Table 1). We also downloaded GSE16091 (n = 34) and GSE21257 (n = 53) datasets from the Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo/). A gene list including 40 FRGs was obtained from the GSEA database (http://www.gsea-msigdb.org/).
Table 1.
Clinicopathological characteristics of OS patients from TARGET database.
| Characteristics | Patients (N = 84) | ||
|---|---|---|---|
| N | % | ||
| Gender | Female | 37 | 44.05 |
| Male | 47 | 55.95 | |
| Age (median) | ≤14 | 44 | 52.38 |
| >14 | 40 | 47.62 | |
| Grade | I/II | 19 | 22.62 |
| III/IV | 16 | 19.05 | |
| Unknown | 49 | 58.33 | |
| Survival time | Long (>5 yr) | 28 | 33.33 |
| Short (<5 yr) | 56 | 66.67% | |
| OS status | Dead | 27 | 32.14% |
| Alive | 57 | 67.86% | |
OS = osteosarcoma.
2.2. Cluster analysis
Cluster analysis was performed on the 84 OS patients based on the mRNA expression levels of FRGs using the k-means method in R.
2.3. Construction and validation of the risk score model
The 84 OS samples in TARGET database were taken as training set, and the GSE16091 (n = 34) and GSE21257 (n = 53) were merged as an independent validation set, named meta-GEO (n = 87).
The univariate Cox regression analysis was applied to select prognosis-related FRGs, and the filtering criteria was “P value < .05.”
Then, we utilized the “glmnet” package in R[23] to apply LASSO-Cox regression analysis to further optimize the prognosis-related FRGs. The screened genes were used to calculate the Risk Score for each patient using the following formula:
In this formula, Coefi represented the risk coefficient and Xi represented the gene expression value. The “survival,” “survminer” packages in R were used to determine the best cutoff value of risk score. And patients in the training set and validating set were divided into low-risk and high-risk groups based on the best cutoff value.
2.4. Survival analysis
Kaplan–Meier method in “survival” and “survminer” packages was used to estimate the overall survival rates in different groups and the significance of difference was tested by the log-rank test. The Multivariate Cox regression was used to analyze the independent prognostic value of the risk score model for OS patients compared with other clinical features.
2.5. Construction and validation of nomogram model
To predict the prognosis of OS patients in 1, 3, and 5 years, we utilized the “rms” package (https://CRAN.R-project.org/package=rms) in R to construct a nomogram model. Independent factors filtered from the multivariate Cox regression were included to build the nomogram model. Calibrated curves were drawn to test the prognostic power of the nomogram model.
2.6. Calculation of immune cells infiltration proportion
The CIBERSORT was used to calculate the relative proportion of immune cells in each sample.[24] CIBERSORT utilized a 547 barcode gene expression matrix to characterize the composition of immune cells.
2.7. Statistical analysis
The correlations between the immune cells were analyzed by Pearson or Spearman coefficients. The difference in immune checkpoints (ICPs) expression between high-risk and low-risk groups was analyzed by the Wilcoxon rank sum test. All analyses were conducted in R (version 4.0.2, R Core Team, Vienna, Austria).
3. Results
3.1. Cluster of OS patients based on FRGs expression levels
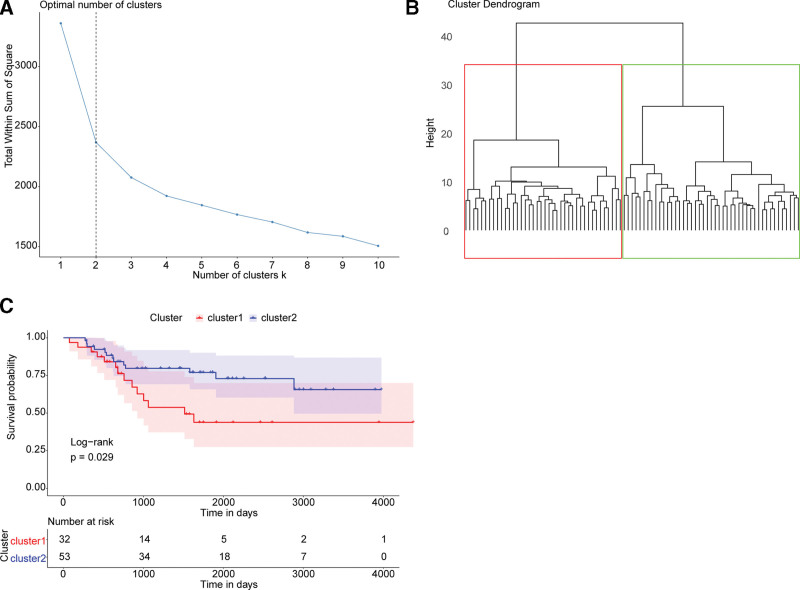
To identify the potential influence of FRGs expression on OS development, we obtained a gene list containing 40 FRGs from GSEA database (Table 2) and performed cluster analysis on the OS samples in the TARGET database based on the FRGs expression levels. According to the sum of the squared errors in the k-means method, the number of cluster k = 2 was chosen (Fig. 1A). OS samples were separated into 2 groups, and they were named the FRGs-high group and the FRGs-low group (Fig. 1B). After conducting the Kaplan–Meier survival analysis, we found a significant difference in overall survival between the 2 groups (P = .029) (Fig. 1C). We speculated that FRGs expression had the potential to predict the outcomes of OS patients.
Table 2.
FERROPTOSIS_Gene.
| MAP1LC3C | SLC39A14 |
| SLC11A2 | ATG5 |
| SLC39A8 | SLC3A2 |
| FTH1 | TFRC |
| SLC40A1 | TP53 |
| NCOA4 | LPCAT3 |
| STEAP3 | PCBP1 |
| HMOX1 | FTMT |
| ACSL6 | PCBP2 |
| CYBB | ACSL4 |
| CP | SLC7A11 |
| MAP1LC3B | ACSL3 |
| TF | FTL |
| SAT1 | ALOX15 |
| VDAC2 | GCLM |
| GCLC | MAP1LC3A |
| VDAC3 | PRNP |
| ACSL5 | SAT2 |
| ATG7 | GSS |
| ACSL1 | GPX4 |
Figure 1.
FRGs expression associated with prognosis of osteosarcoma. (A) Elbow diagram to determine the best number of clusters. The x and y axes are the number of clusters K and the SSE, respectively. The optimal number of clusters K = 2 was chosen. (B) Schematic diagram of samples clustering. The color of the boxes indicates clusters. (C) Kaplan–Meier survival curves of clusters. The x-axis shows time and the y-axis shows survival rate. Color represents the group. The P value is obtained from the log-rank test. FRGs = ferroptosis-related genes, SSE = squared errors.
3.2. Identification and validation of a risk score model for OS
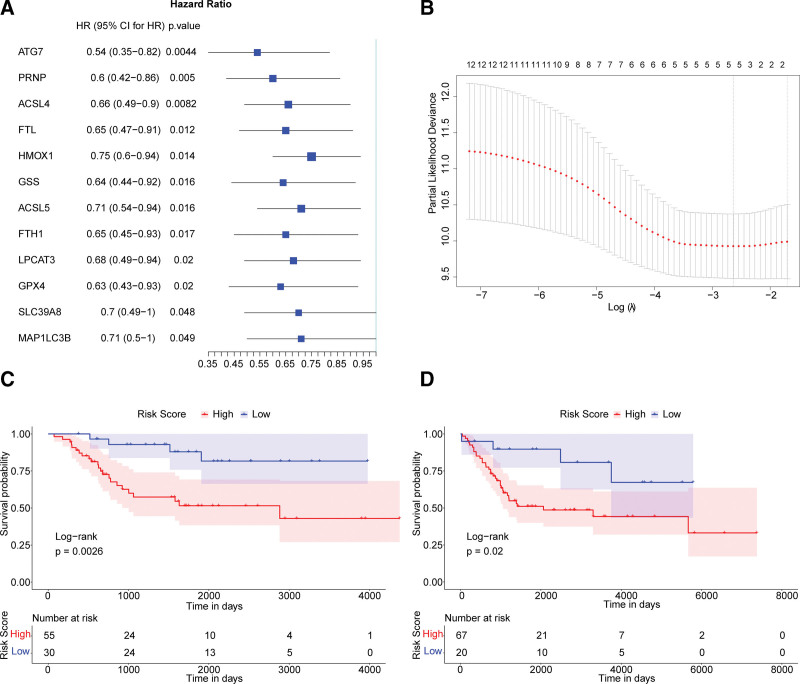
To obtain the independent OS-associated prognostic markers in the 40 FRGs, the univariate Cox regression analysis was conducted. According to the hazard ratio, 12 genes (ATG7, PRNP, ACSL4, FTL, HMOX1, GSS, ACSL5, FTH1, LPCAT3, GPX4, SLC39A8, and MAP1LC3B) were found significantly associated to the outcome of OS patients (Fig. 2A). Then the LASSO-Cox analysis was performed to optimize the 12 independent prognostic markers into 5 FRGs, including Acyl-CoA synthetase long-chain family member 4 (ACSL4), heme oxygenase-1 (HMOX1), glutathione peroxidases 4 (GPX4), prion protein (PRNP) and autophagy related 7 (ATG7) (Fig. 2B). We finally constructed the regression model: risk score = (−0.01382853 × ACSL4) − (0.05371778 × HMOX1) − (0.02434655 × GPX4) − (0.16432810 × PRNP) − (0.15567120 × ATG7).
Figure 2.
Construction and validation of the risk score model. (A) Forest map showing 12 prognosis-associated FRGs analyzed by the univariate Cox analysis. (B) Point plot of the LASSO regression model determining the best tuning parameter lambda. The x and y axes are the value of log (lambda) and the partial likelihood of deviance, respectively. The Kaplan–Meier survival curves of the TARGET dataset (C) and meta-GEO dataset (D). Color represents the group. The P values are calculated by the log-rank test. FRGs = ferroptosis-related genes, GEO = Gene Expression Omnibus.
To validate the predictive performance of the risk score basing on the 5 FRGs, we regrouped OS patients in TARGET (training set) and meta-GEO (combination of 2 GEO datasets, validation set) datasets into high-risk and low-risk groups according to the optimal cutoff point (value = −0.175). The survival analysis showed the overall survival of patients in the high-risk group (score > −0.175) was worse than those in the low-risk group in both the training set and validating set (Fig. 2C and D). It suggested the risk score constructed by ACSL4, HMOX1, GPX4, PRNP, and ATG7 represented a reliable performance to predict the prognosis of OS patients.
3.3. Risk score acted as an independent prognostic marker for OS
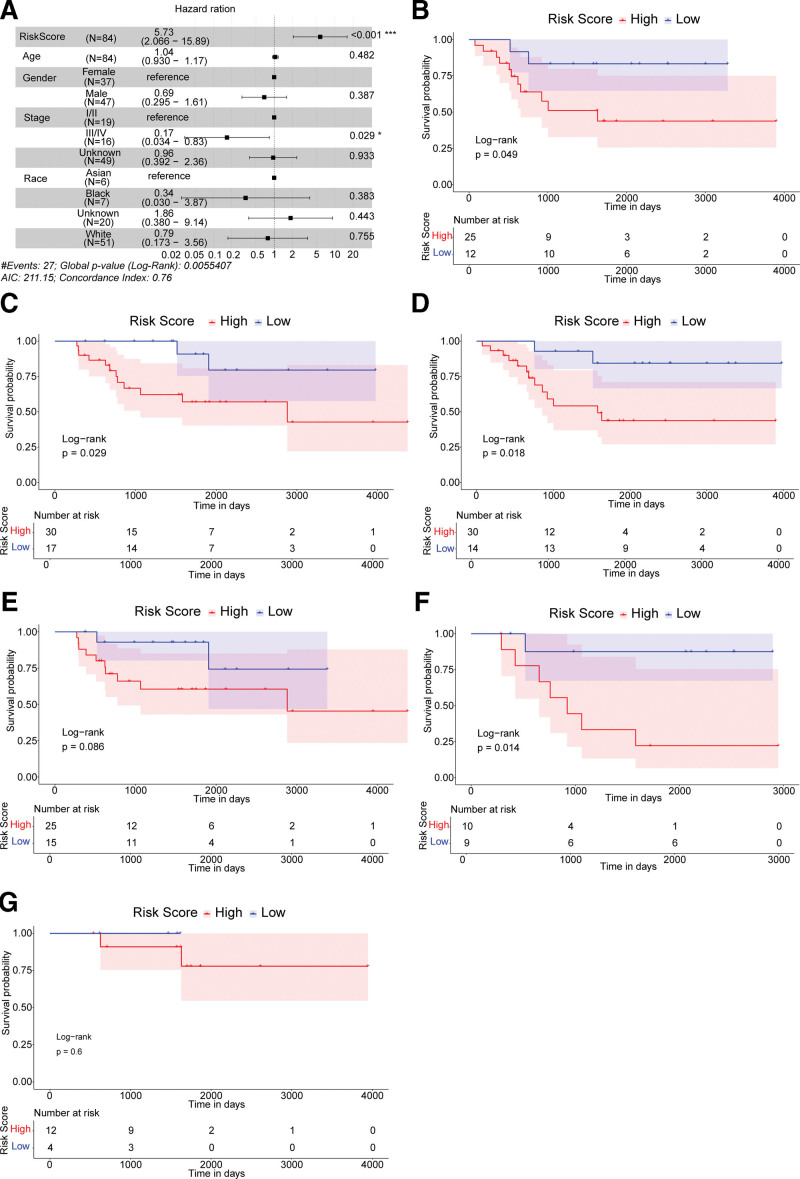
We utilized the multivariate Cox regression analysis to determine whether the risk score could act as an independent prognostic indicator compared with other clinical characteristics: age, sex, race, and the tumour-node-metastasis (TNM) stage. Results indicated that the prognosis of OS was significantly associated with the risk score and the TNM stage. The patients with higher risk score had higher death risk (hazard ratio = 5.73, 95% confidence interval = 2.066–15.89, P < .01) (Fig. 3A).
Figure 3.
The risk score acted as an independent prognostic indicator. (A) Forest map of the multivariate Cox regression analysis. A hazard ratio >1 is considered to indicate a high death risk. (B and C) The Kaplan–Meier survival curves of female subgroups and male subgroups. (D and E) The Kaplan–Meier survival curves of <14 years old subgroups and >14 years old subgroups. (F and G) The Kaplan–Meier survival curves of different TNM stages subgroups, TNM = tumour-node-metastasis.
To further explore the prognostic value of risk score under different situations, OS patients were regrouped for survival analysis according to the pathological factors (including age, sex, and the TNM stage). In the female (Fig. 3B), the male (Fig. 3C), the ≤14 years old (Fig. 3D) and the stage Ⅰ/Ⅱ subgroups (Fig. 3F), the overall survival rates of the high-risk group were significantly lower compared to the low-risk group. The limited sample size might be the reason for the non-significance (P > .05) in the >14 years old (Fig. 3E) and the stage III/IV (Fig. 3G) subgroups. These results suggested the risk score could independently predict the prognosis of OS patients.
3.4. Construction of a nomogram model based on the risk score and the TNM stage
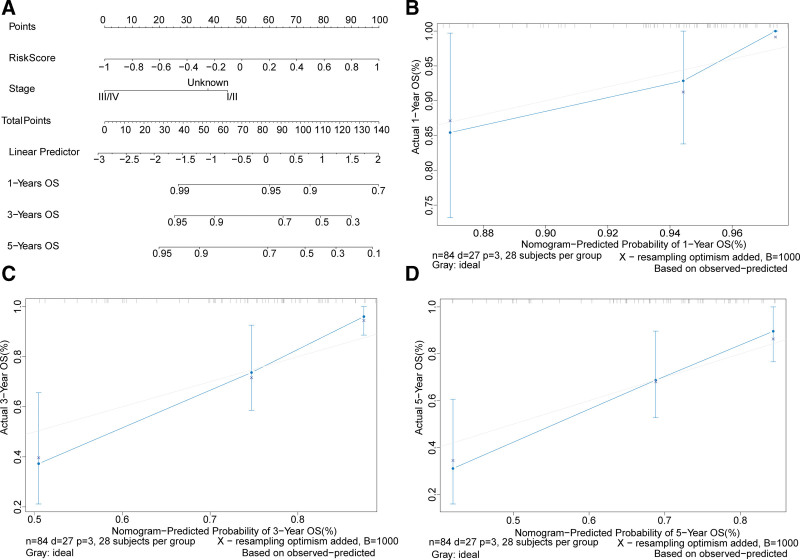
To further evaluate the clinical utility of the risk score, a nomogram model was built based on 2 independent prognostic factors, the risk score and the TNM stage (Fig. 4A). The calibrated diagrams of the 1-year (Fig. 4B), 3-year (Fig. 4C), and 5-year (Fig. 4D) OS showed the nomogram model had the best performance for predicting 1-year OS, suggesting the combination of the risk score and the TNM stage increased the power of predicting prognostic outcomes.
Figure 4.
Construction and validation of a nomogram model to predict the survival probability. (A) A nomogram model to predict the survival probabilities of osteosarcoma patients in 1, 3, and 5 yr. Calibrated Nomogram curves to predict the overall survival of OS patients in 1 yr (B), 3 yr (C), and 5 yr (D). OS = osteosarcoma.
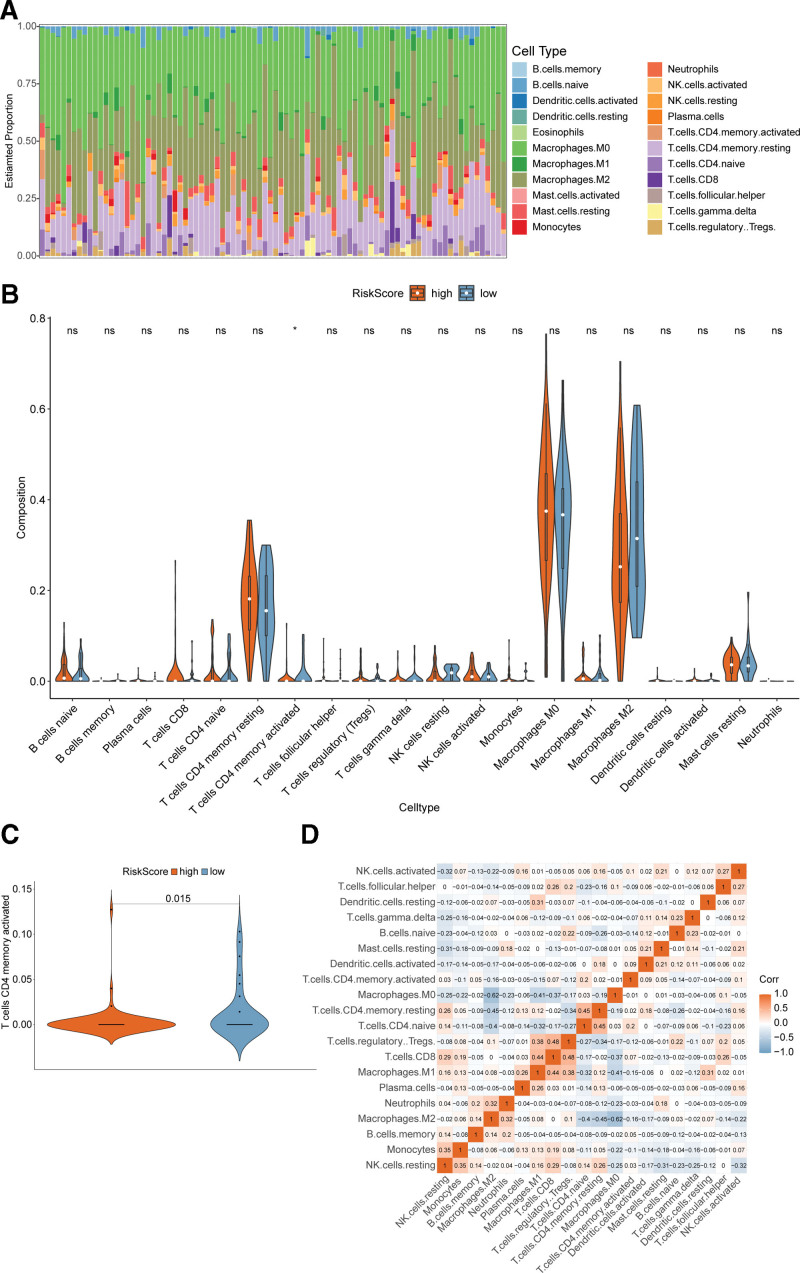
3.5. The immune cells infiltration difference between the high-risk and low-risk groups
To figure out whether the OS patients with high or low risk score were different in the tumor immune microenvironment, the analytical tool CIBERSORT was used to characterize the proportions of immune cell infiltration in the high-risk and the low-risk groups from the TARGET dataset (total number = 84). The overall immune cells distribution in each patient was shown in the stacked percentage barplot, and the ratio changes among patients may represent the intrinsic difference (Fig. 5A). The infiltration ratios of immune cells were different between high or low-risk groups (Fig. 5B), but only the activated CD4 memory T cells proportions were significantly higher in the low-risk group compared with the high-risk group (Fig. 5C). Activation of CD4 memory T cells had been reported to be associated with a low risk of disease relapse in colorectal cancer,[25,26] which agreed with our result that OS patients in the low-risk group had a better prognosis. However, the correlations between different immune cells were weak (Fig. 5D).
Figure 5.
Immune cell infiltration difference between the high-risk and low-risk groups. (A) Stacked percentage barplot showing the relative proportion of immune cells in all patients. (B) Violin plots of immune cells showing infiltration ratio differences between high-risk and low-risk groups. (C) Violin plots of activated CD4 memory T cells. (D) Correlation matrix of immune cells. Orange represents positive correlation and light blue represents negative correlation. The darkness of color indicates the correlation.
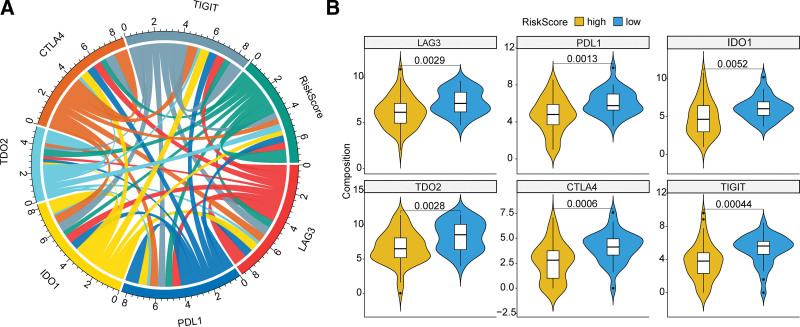
3.6. Relationship between the risk score and the ICPs
ICPs had become popular immunotherapy targets for OS patients. To look into the role of the risk score in immunotherapy, we analyzed the relationship between the risk score and crucial ICPs. Six ICPs (CTLA4, PDL1, LAG3, TIGIT, IDO1, and TDO2) expression levels were all found to be significantly correlated with the risk score (Fig. 6A). And these ICPs were significantly up-regulated in the low-risk group than in the high-risk group (LAG3 P = .0029, PDL1 = 0.0013, IDO1 = 0.0052, TDO2 = 0.0028, CTLA4 = 0.0006, and TIGHT = 0.00044) (Fig. 6B), suggesting the risk score might help selecting the patients who could benefit from immunotherapy. ICPs were associated with immune-related adverse effects and were reported to serve as prognostic biomarkers in stomach adenocarcinoma[27] and renal clear cell carcinoma,[28] which were in accord with our results.
Figure 6.
Correlation between the risk score and crucial ICPs. (A) Chord diagram showing the correlation between the risk score and the expression of 6 ICPs. The width of the line indicates the correlation. (B) Violin plots of ICPs between high-risk and low-risk groups. Yellow represents the high-risk group and blue represents the low-risk group. The P value is calculated by the Wilcoxon rank sum test. ICPs = immune checkpoints.
4. Discussion
Enormous efforts had been put into the prevention, diagnosis, and treatment of OS, however, the outcome has not significantly changed over these years. It is super urging to find more reliable and sensitive markers to improve the prognostic prediction of OS patients.[29,30] Ferroptosis has been reported as an important biological process in many types of cancers,[14] and regulators in the ferroptosis process showed great value in understanding the pathophysiological processes in cancers. But little was known about ferroptosis in OS, leading us to explore the probability of FRGs as biomarkers in OS.
To understand the potential role of the FRGs in the prognosis of OS patients, we collected a 40 FRGs gene list from the GSEA database and downloaded the mRNA expression profiles from the TARGET database. The OS patients could be clustered into 2 groups according to the 40 FRGs expression levels, suggesting FRGs might act as prognosis biomarkers. The univariate Cox regression analysis identified 12 FRGs which were significantly associated with the prognosis of the OS patients. The LASSO-Cox regression optimized the 12 FRGs into 5 FRGs and the risk score model was finally built: risk score = (−0.01382853 × ACSL4) − (0.05371778 × HMOX1) − (0.02434655 × GPX4) − (0.16432810 × PRNP) − (0.15567120 × ATG7).
These 5 FRGs had already been reported participating in tumorigenesis and development. ACSL4 was one of the acyl-CoA synthetase proteins and was a necessary component for lipid peroxidation and ferroptosis metabolism.[31–33] ACSL4 presented great predictive value for the prognosis of hepatocellular carcinoma.[34–36] And the abnormal expression of ACSL4 was correlated with cancer development.[37] ACSL4 also suppressed the proliferation of glioma cells by activating ferroptosis.[31,38] Nevertheless, the detailed role of ACSL4 in OS has never been reported as far as we know, which deserved deepening exploration. The rate-limiting enzyme HMOX1 catalyzed the degeneration of pro-oxidant heme and HMOX1 overexpression was associated with mitophagic cell death of the glioma cells.[39,40] Genetic inhibition of HMOX1 might serve as an anticancer approach in various cancer types.[41] GPX4 could protect cells from ferroptosis by eliminating phospholipid peroxides via presuming on glutathione, and it was a central regulator of ferroptosis.[42,43] Moreover, 1 way to trigger cancer cell death is ferroptosis induction via the inhibition of GPX4 and targeting GPX4 had emerged as a therapeutic strategy for clear-cell carcinomas.[44,45] The prion protein gene encoded a conserved cell surface glycoprotein (PrP) expressed within almost all mammalian cells.[46] The mutation in PRNP could induce dysfunction of PrP, which led to tumorigenesis in many cancers.[47–49] ATG7 encoded an enzyme that was essential for autophagy. Reports had shown that autophagy could cause ferroptosis by degradation of ferritin, and ATG7 low expression could limit erastin-induced ferroptosis.[50] Collectively, previous studies have provided more evidence supporting our present ferroptosis-related prognostic signature in OS.
We further validated the risk score in the meta-GEO datasets and found patients with higher risk score tended to suffer from a worse outcome. Through the multivariate Cox analysis, we found the risk score was an independent prognostic factor. After constructing a nomogram model, we found the combination of the risk score and the TNM stage could increase the prediction performance of 1-year OS. We also found that the activated CD4 memory T cells infiltration ratios were significantly different between the high-risk and low-risk groups. The risk score was significantly associated with expression levels of CTLA4, PDL1, LAG3, TIGIT, IDO1, and TDO2, indicating the risk score may facilitate immunotherapy in OS.
FRGs-based signatures had been identified with prognostic values in colon cancer,[51] lung adenocarcinoma,[52,53] gastric cancer,[54] breast cancer,[55] and bladder cancer.[21,56] And nomograms built by FRGs for predicting survival probabilities had already been used in lung adenocarcinoma and oral squamous cell carcinoma patients.[57,58] Our findings also indicated that the FRGs-based model may be a reliable prognostic marker for OS patients. However, this work needed more big cohorts to validate these results. Other clinical indicators should also be included in the model to improve the prediction power. On the other hand, we have herein preliminarily explored the prognostic effects of the FRG-related signature in OS patients. More details in the exact underlying functional pathways of the 5 key FRGs in OS cannot be clearly concluded at present and should be further investigated in our future work.
5. Conclusions
In summary, we identified a Risk-Score model predicting the outcome of OS patients, risk score = (−0.01382853 × ACSL4) − (0.05371778 × HMOX1) − (0.02434655 × GPX4) − (0.16432810 × PRNP) − (0.15567120 × ATG7) in TARGET dataset via the univariate Cox regression analysis and the LASSO-Cox regression analysis. Herein, we have revealed an independent prognostic signature basing on 5 FRGs for OS for the first time (validated in 2 cohorts). Our findings are promising to give more insights into accurate prognosis prediction and better treatment strategy making of OS patients.
Author contributions
ZG and DS conceived and designed the study, analyzed the data, prepared figures and/or tables, authored or reviewed drafts of the paper, and approved the final draft.
Conceptualization: Zhanyong Ge, Delei Song.
Data curation: Zhanyong Ge, Delei Song.
Formal analysis: Zhanyong Ge, Delei Song.
Funding acquisition: Zhanyong Ge, Delei Song.
Investigation: Zhanyong Ge, Delei Song.
Methodology: Zhanyong Ge, Delei Song.
Project administration: Zhanyong Ge, Delei Song.
Resources: Zhanyong Ge, Delei Song.
Software: Zhanyong Ge, Delei Song.
Supervision: Zhanyong Ge, Delei Song.
Validation: Zhanyong Ge, Delei Song.
Visualization: Zhanyong Ge, Delei Song.
Writing – original draft: Zhanyong Ge, Delei Song.
Writing – review & editing: Zhanyong Ge, Delei Song.
Abbreviations:
- ACSL4 =
- Acyl-CoA synthetase long-chain family member 4
- ATG7 =
- autophagy related 7
- FRGs =
- ferroptosis-related genes
- GEO =
- Gene Expression Omnibus
- GPX4 =
- glutathione peroxidases 4
- HMOX1 =
- heme oxygenase-1
- ICPs =
- immune checkpoints
- OS =
- osteosarcoma
- PRNP =
- prion protein gene
- TARGET =
- therapeutically applicable research to generate effective treatment
- TNM =
- tumour-node-metastasis
Informed consent was obtained from all individual participants included in the study.
The authors have no funding and conflicts of interest to disclose.
The datasets generated during and/or analyzed during the current study are publicly available.
How to cite this article: Ge Z, Song D. A five ferroptosis-related genes risk score for prognostic prediction of osteosarcoma. Medicine 2022;101:50(e32083).
References
- [1].Ferris ITJ, Berbel Tornero O, Ortega Garcia JA, et al. [Risk factors for pediatric malignant bone tumors]. An Pediatr (Barc). 2005;63:537–47. [DOI] [PubMed] [Google Scholar]
- [2].Mirabello L, Troisi RJ, Savage SA. International osteosarcoma incidence patterns in children and adolescents, middle ages and elderly persons. Int J Cancer. 2009;125:229–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Tian H, Guan D, Li J. Identifying osteosarcoma metastasis associated genes by weighted gene co-expression network analysis (WGCNA). Medicine (Baltimore). 2018;97:e10781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Tebbi CK, Gaeta J. Osteosarcoma. Pediatr Ann. 1988;17:285–300. [DOI] [PubMed] [Google Scholar]
- [5].Yasin NF, Abdul Rashid ML, Ajit Singh V. Survival analysis of osteosarcoma patients: a 15-year experience. J Orthop Surg (Hong Kong). 2020;28:2309499019896662. [DOI] [PubMed] [Google Scholar]
- [6].Misaghi A, Goldin A, Awad M, et al. Osteosarcoma: a comprehensive review. Sicot J. 2018;4:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Picci P. Osteosarcoma (osteogenic sarcoma). Orphanet J Rare Dis. 2007;2:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Li Y, Zhao C, Yu Z, et al. Low expression of miR-381 is a favorite prognosis factor and enhances the chemosensitivity of osteosarcoma. Oncotarget. 2016;7:68585–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Whelan JS, Davis LE. Osteosarcoma, chondrosarcoma, and chordoma. J Clin Oncol. 2018;36:188–93. [DOI] [PubMed] [Google Scholar]
- [10].Ducimetiere F, Lurkin A, Ranchere-Vince D, et al. [Incidence rate, epidemiology of sarcoma and molecular biology. Preliminary results from EMS study in the Rhone-Alpes region]. Bull Cancer. 2010;97:629–41. [DOI] [PubMed] [Google Scholar]
- [11].Horvai AE, Roy R, Borys D, et al. Regulators of skeletal development: a cluster analysis of 206 bone tumors reveals diagnostically useful markers. Mod Pathol. 2012;25:1452–61. [DOI] [PubMed] [Google Scholar]
- [12].Dixon SJ, Lemberg KM, Lamprecht MR, et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149:1060–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Hirschhorn T, Stockwell BR. The development of the concept of ferroptosis. Free Radic Biol Med. 2019;133:130–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Li J, Cao F, Yin HL, et al. Ferroptosis: past, present and future. Cell Death Dis. 2020;11:88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Liang JY, Wang DS, Lin HC, et al. A novel ferroptosis-related gene signature for overall survival prediction in patients with hepatocellular carcinoma. Int J Biol Sci. 2020;16:2430–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Liang C, Zhang X, Yang M, et al. Recent progress in ferroptosis inducers for cancer therapy. Adv Mater. 2019;31:e1904197. [DOI] [PubMed] [Google Scholar]
- [17].Xie Y, Hou W, Song X, et al. Ferroptosis: process and function. Cell Death Differ. 2016;23:369–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Zhuo S, Chen Z, Yang Y, et al. Clinical and biological significances of a ferroptosis-related gene signature in glioma. Front Oncol. 2020;10:590861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Zhu L, Yang F, Wang L, et al. Identification the ferroptosis-related gene signature in patients with esophageal adenocarcinoma. Cancer Cell Int. 2021;21:124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Zhu L, Tian Q, Jiang S, et al. A novel ferroptosis-related gene signature for overall survival prediction in patients with breast cancer. Front Cell Dev Biol. 2021;9:670184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Liu J, Ma H, Meng L, et al. Construction and external validation of a ferroptosis-related gene signature of predictive value for the overall survival in bladder cancer. Front Mol Biosci. 2021;8:675651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Lei T, Qian H, Lei P, et al. Ferroptosis-related gene signature associates with immunity and predicts prognosis accurately in patients with osteosarcoma. Cancer Sci. 2021;112:4785–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J Stat Softw. 2010;33:1–22. [PMC free article] [PubMed] [Google Scholar]
- [24].Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12:453–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Gasper DJ, Tejera MM, Suresh M. CD4 T-cell memory generation and maintenance. Crit Rev Immunol. 2014;34:121–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Kamal Y, Dwan D, Hoehn HJ, et al. Tumor immune infiltration estimated from gene expression profiles predicts colorectal cancer relapse. Oncoimmunology. 2021;10:1862529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Shen K, Liu T. Comprehensive analysis of the prognostic value and immune function of immune checkpoints in stomach adenocarcinoma. Int J Gen Med. 2021;14:5807–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Liao G, Wang P, Wang Y. Identification of the prognosis value and potential mechanism of immune checkpoints in renal clear cell carcinoma microenvironment. Front Oncol. 2021;11:720125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Bishop MW, Janeway KA, Gorlick R. Future directions in the treatment of osteosarcoma. Curr Opin Pediatr. 2016;28:26–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Kong C, Hansen MF. Biomarkers in osteosarcoma. Expert Opin Med Diagn. 2009;3:13–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Cheng J, Fan YQ, Liu BH, et al. ACSL4 suppresses glioma cells proliferation via activating ferroptosis. Oncol Rep. 2020;43:147–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Doll S, Proneth B, Tyurina YY, et al. ACSL4 dictates ferroptosis sensitivity by shaping cellular lipid composition. Nat Chem Biol. 2017;13:91–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Lei G, Zhang Y, Koppula P, et al. The role of ferroptosis in ionizing radiation-induced cell death and tumor suppression. Cell Res. 2020;30:146–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Tang B, Zhu J, Li J, et al. The ferroptosis and iron-metabolism signature robustly predicts clinical diagnosis, prognosis and immune microenvironment for hepatocellular carcinoma. Cell Commun Signal. 2020;18:174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Liu Y, Zhang X, Zhang J, et al. Development and validation of a combined ferroptosis and immune prognostic classifier for hepatocellular carcinoma. Front Cell Dev Biol. 2020;8:596679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Du X, Zhang Y. Integrated analysis of immunity- and ferroptosis-related biomarker signatures to improve the prognosis prediction of hepatocellular carcinoma. Front Genet. 2020;11:614888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Adang MJ, Staver MJ, Rocheleau TA, et al. Characterized full-length and truncated plasmid clones of the crystal protein of Bacillus thuringiensis subsp. kurstaki HD-73 and their toxicity to Manduca sexta. Gene. 1985;36:289–300. [DOI] [PubMed] [Google Scholar]
- [38].Yuan H, Li X, Zhang X, et al. Identification of ACSL4 as a biomarker and contributor of ferroptosis. Biochem Biophys Res Commun. 2016;478:1338–43. [DOI] [PubMed] [Google Scholar]
- [39].Meyer N, Zielke S, Michaelis JB, et al. AT 101 induces early mitochondrial dysfunction and HMOX1 (heme oxygenase 1) to trigger mitophagic cell death in glioma cells. Autophagy. 2018;14:1693–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Hull TD, Boddu R, Guo L, et al. Heme oxygenase-1 regulates mitochondrial quality control in the heart. JCI Insight. 2016;1:e85817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Podkalicka P, Mucha O, Jozkowicz A, et al. Heme oxygenase inhibition in cancers: possible tools and targets. Contemp Oncol (Pozn). 2018;22:23–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Stockwell BR, Jiang X, Gu W. Emerging mechanisms and disease relevance of ferroptosis. Trends Cell Biol. 2020;30:478–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Yang WS, SriRamaratnam R, Welsch ME, et al. Regulation of ferroptotic cancer cell death by GPX4. Cell. 2014;156:317–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Bersuker K, Hendricks JM, Li Z, et al. The CoQ oxidoreductase FSP1 acts parallel to GPX4 to inhibit ferroptosis. Nature. 2019;575:688–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Zou Y, Palte MJ, Deik AA, et al. A GPX4-dependent cancer cell state underlies the clear-cell morphology and confers sensitivity to ferroptosis. Nat Commun. 2019;10:1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Ryskalin L, Busceti CL, Biagioni F, et al. Prion protein in glioblastoma multiforme. Int J Mol Sci. 2019;20:5107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Du L, Rao G, Wang H, et al. CD44-positive cancer stem cells expressing cellular prion protein contribute to metastatic capacity in colorectal cancer. Cancer Res. 2013;73:2682–94. [DOI] [PubMed] [Google Scholar]
- [48].Corsaro A, Bajetto A, Thellung S, et al. Cellular prion protein controls stem cell-like properties of human glioblastoma tumor-initiating cells. Oncotarget. 2016;7:38638–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Iglesia RP, Prado MB, Cruz L, et al. Engagement of cellular prion protein with the co-chaperone Hsp70/90 organizing protein regulates the proliferation of glioblastoma stem-like cells. Stem Cell Res Ther. 2017;8:76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Estes JW. Platelet size and function in the heritable disorders of connective tissue. Ann Intern Med. 1968;68:1237–49. [DOI] [PubMed] [Google Scholar]
- [51].Qi X, Wang R, Lin Y, et al. A ferroptosis-related gene signature identified as a novel prognostic biomarker for colon cancer. Front Genet. 2021;12:692426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Zhou J, Wang X, Li Z, et al. Construction and analysis of a novel ferroptosis-related gene signature predicting prognosis in lung adenocarcinoma. FEBS Open Bio. 2021;11:3005–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Ma C, Li F, Luo H. Prognostic and immune implications of a novel ferroptosis-related ten-gene signature in lung adenocarcinoma. Ann Transl Med. 2021;9:1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Liu G, Ma JY, Hu G, et al. Identification and validation of a novel ferroptosis-related gene model for predicting the prognosis of gastric cancer patients. PLoS One. 2021;16:e0254368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Liu Q, Ma JY, Wu G. Identification and validation of a ferroptosis-related gene signature predictive of prognosis in breast cancer. Aging (Albany NY). 2021;13:21385–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Wan RJ, Peng W, Xia QX, et al. Ferroptosis-related gene signature predicts prognosis and immunotherapy in glioma. CNS Neurosci Ther. 2021;27:973–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Wang S, Wu C, Ma D, et al. Identification of a ferroptosis-related gene signature (FRGS) for predicting clinical outcome in lung adenocarcinoma. PeerJ. 2021;9:e11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Li H, Zhang X, Yi C, et al. Ferroptosis-related gene signature predicts the prognosis in Oral squamous cell carcinoma patients. BMC Cancer. 2021;21:835. [DOI] [PMC free article] [PubMed] [Google Scholar]