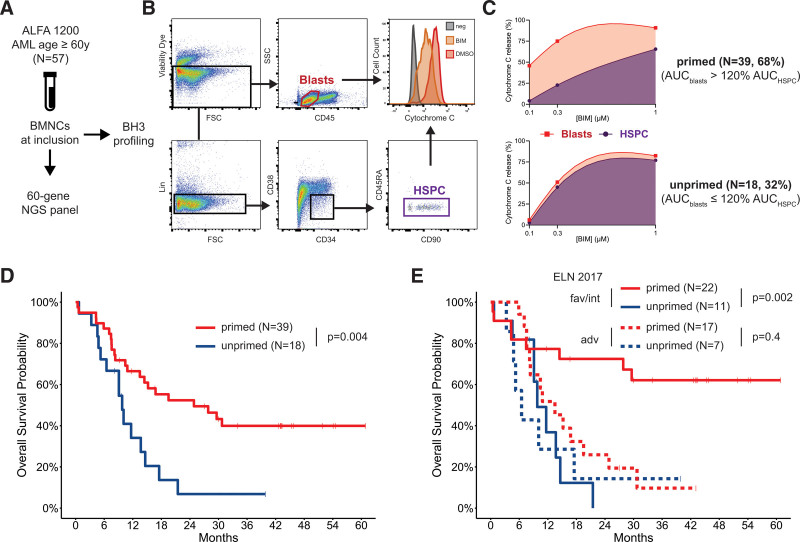
High mitochondrial priming of blasts for apoptosis relative to hematopoietic stem cells, measured by mitochondrial membrane depolarization upon exposure to proapoptotic BH3 peptides, has been suggested to predict longer overall survival (OS) in acute myeloid leukemia (AML).1 Whether its prognostic impact is independent of genetic risk remains unknown. We interrogated the mitochondrial priming of AML cells in patients 60 years or older uniformly treated with intensive chemotherapy in a clinical trial and found that higher blast priming predicted prolonged OS independently of genetic risk.
Intensive chemotherapy remains the standard of care of fit adults older than 60 years with newly diagnosed AML. However, results of 7 + 3 based chemotherapy regimens remain disappointing with long-term survival rates below 30%.2 Identifying patients that benefit from intensive chemotherapy thus remains an important area of investigation. Recent efforts have focused on integrating a greater number of genetic lesions to predict OS of intensively treated older AML patients.3,4 However, the predictive power of risk classifiers solely based on (cyto)genetics remains limited.2–4 In recent years, functional precision medicine has emerged as a promising approach to complement oncogenetics in treatment decision making.5 BH3 profiling is one such functional approach, whereby sensitivity to mitochondrial apoptosis triggering agents, including conventional chemotherapy, is estimated by challenging cells with peptides derived from BH3-only proteins and measuring cytochrome C release or loss of mitochondrial transmembrane potential. In a landmark study, high mitochondrial priming of AML blasts relative to their cognate hematopoietic stem cells was found to predict longer OS in univariable analysis.1 A previous study on 62 patients reported that BH3 profiling could predict remission after various cytarabine-based regimens independently of age and cytogenetics.6 Whether BH3 profiling can predict OS in a homogeneously treated AML population independently of up-to-date oncogenetic risk stratification remains unknown. To address this question, we interrogated the mitochondrial priming of primary AML cells in 57 diagnostic samples from patients 60 years or older treated intensively in the ALFA 1200 clinical trial (NCT01966497).
The ALFA 1200 trial accrued patients 60 years or older with newly diagnosed AML (de novo, secondary to myelodysplastic syndromes or therapy-related), excluding acute promyelocytic leukemia and Ph-positive AML. Patients received a 7 + 3 induction course with idarubicin as anthracycline. Patients failing to achieve complete remission (CR) or CR with incomplete platelet recovery (CRp) could receive an intermediate-dose cytarabine (IDAC) course. Patients in CR/CRp received 2 such IDAC courses as consolidation and could proceed to allogeneic hematopoietic cell transplantation (allo-HCT). Further details on treatment and centralized molecular genetics and cytogenetics procedures have been previously published.3,4 For this study, cryopreserved mononuclear cells from bone marrow samples collected at inclusion in the ALFA 1200 trial in 65 patients were thawed, washed in with MEMα (Life Technologies) and 10% Fetal Bovine Serum (FBS, BioTec) and a second time in Phosphate Buffer Saline (PBS, Eurobio). 5 × 106 cells were stained with Zombie Aqua Fixable Viability Dye (Bio Legend) according to manufacturer’s instructions, then stained for surface markers with anti-CD34 PE-Cy7 (clone 581), anti-CD38 PerCP-Cy5.5 (clone HB7), anti-CD45 APCH7 (clone 2D1), anti-CD45RA APC (clone HI30), anti-CD3 PC5 (clone SP34-2), anti-CD19 PC5 (clone HIB19), anti-CD20 PC5 (clone 2H7), and anti GPA PC5 (clone GA-R2, all from BD Biosciences), Alexa Fluor anti-CD90 (clone 5E10) and anti-CD123 PE (clone 6H6, both Sony Biotechnology). Cells were then washed and resuspended in DTEB Buffer (135 mM trehalose, 50 mM KCl, 20 µM EDTA, 20 µM EGTA, 5 mM succinate, 0.1% Bovine Serum Albumin, 10 mM HEPES-KOH final pH 7.5, all from Sigma-Aldrich). 6 × 105 cells per condition were incubated in the presence of a BIM peptide (Ac-MRPEIWIAQELRRIGDEFNA-NH2, purity >95%, Proteogenix, France) dissolved in DMSO at the indicated concentrations (1, 0.3, 0.1 µM) for 1 hour in the presence of 0.001% digitonin (Sigma-Aldrich) for cell permeabilization. Cells were then fixated with Formaldehyde 8% in PBS (Sigma-Aldrich). Fixation was terminated by N2 buffer solution (1.7 M Tris, 1.25 M Glycine pH 9.1, both from ThermoFisher). Cells were then labeled with anti-human cytochrome C (CytC) Alexa Fluor 488 (BD Biosciences, clone 6H2.B4) in the presence of adequate amounts of Perm/Wash 10× buffer (BD Biosciences) and incubated overnight at 4°C. All experiments included 2 DMSO 2% sample (without BIM peptide), 1 labeled with anti-Cytochrome C and 1 unlabeled as positive and negative controls for mitochondrial Cytochrome C content, respectively. Flow cytometry acquisition was performed on a BD Fortessa analyser (BD Biosciences) on >200,000 events and analyzed with FlowJo v10 (TreeStar). Mitochondrial priming was defined as the percentage of cytochrome C release based on median fluorescence intensities (MFI) according to the following formula: 1 − ((MFIBIM − MFIFMO)/(MFIDMSO − MFIFMO)) ×100 (%) (Figure 1A). We studied the mitochondrial priming of total leukemic blasts (CD45dim/SSClow, mean 1.06 × 105 cells, interquartile range [IQR] 0.50–1.59) and of Hematopoietic Stem and Progenitor Cells (HSPCs, Lin-/CD34+/CD38−/CD45RA−, mean 2.58 × 104 cells, IQR 0.65–2.82) as internal control (Figure 1B).1 Absolute priming of each cell compartment was measured as the area under the curve (spline interpolation method) of Cyt C release across the 3 BIM concentrations (Figure 1C). All analyses were conducted with R 4.1.0 (cran.r-project.org). Of 65 samples, only 57 could be analyzed, due to low cell viability after thawing in 8 samples. In this study population (31 males, 26 females), median age was 71 years (IQR 67–73) and median white blood cell count was 27.1 × 109/L (IQR 9.0–70.2). AML was de novo, secondary to MDS and therapy-related in 46 (81%), 8 (14%), and 3 (5%) cases, respectively. According to European LeukemiaNet (ELN) 2017 guidelines,2 genetic risk was favorable, intermediate, and adverse in 24 (42%), 9 (16%), and 24 (42%) patients, respectively. Thirty-five (61%) patients achieved CR/CRp. As a continuous variable, relative blast priming (ie, AUC of cytochrome C release in blasts normalized to the AUC in the HSPC compartment) was similar in patients achieving CR/CRp after 1 (n = 34) or 2 (n = 1) course and in those with primary induction failure (n = 19) or early death (n = 3) (Mann-Whitney test P = 0.29). With a median follow-up of 43.1 months, median OS in the study population was 14.6 months. Two-year OS was 60.8% (95% confidence interval [CI], 43.6-84.8), 33.3% (95% CI, 13.2-84.0), and 22.4 (95% CI, 10.4–42.8) in patients with favorable, intermediate, and adverse ELN risk, respectively (log-rank test, P = 0.07). In univariable analysis, higher relative blast priming as a continuous variable was associated with a significantly longer OS (univariable Cox model, hazard ratio (HR) = 0.97, 95% CI, 0.95-0.99, P = 0.045). We empirically assigned patients whose blasts had a global priming >120% of their cognate HSPCs into a “primed” category and all others into an “unprimed” group (Figure 1C). Of the 57 patients, 39 (68%) had “primed” blasts. The genetic profile of primed and unprimed samples was comparable (online Supplement). In univariable analysis, 2-year OS of primed patients was 52.4% (95% CI, 38.5-71.2) compared to 6.8% (95% CI, 1.0-44-8) in unprimed patients (log-rank test P = 0.004, Figure 1D). In a multivariable Cox model accounting for ELN risk (reference: favorable risk), primed status was associated with prolonged OS (HR = 0.39, 95% CI, 0.20-0.78, P = 0.008) independently of intermediate (HR = 1.02, 95% CI, 0.37-2.86, P = 0.96) and adverse (HR = 2.08, 95% CI, 0.98-4.39, P = 0.055) ELN risk. Only 4 patients (3 primed, 1 unprimed) underwent allo-HCT in first remission. Censoring at the time of allo-HCT did not abrogate the independent favorable prognostic value of primed status on OS (HR = 0.44, 95% CI, 0.22-0.88, P = 0.02). Specifically, 17 (70.8%), 5 (55.6%), and 17 (70.8%) patients with favorable, intermediate, and adverse ELN risk had primed blasts, respectively. At 2 years from diagnosis, none of the 11 patients with favorable or intermediate risk AML but unprimed blasts were alive, while 2-year OS was 72.4% (95% CI, 55.9-93.9) in the 22 primed patients with favorable or intermediate risk AML (log-rank test P = 0.002). In contrast, 2-year OS was comparable in primed (25.9%, 95% CI, 11.2-59.7) and unprimed (14.3%, 95% CI, 2.3-87.7) patients with adverse risk AML (log-rank P = 0.4, Figure 1E). Among the 33 patients with favorable or intermediate risk AML achieving CR/CRp, 2-year disease-free survival was 80.0% (95% CI, 62.1-100) in the 15 primed patients, whereas none of the 8 unprimed patients were alive in remission 2 years from remission (log-rank P = 0.007). We previously reported a 3-tier genetic classifier to identify patients older than 60 years benefiting from intensive therapy.4 According to this ALFA classifier, 24, 31, and 2 patients were assigned to the “go-go,” “slow-go,” and “no-go” groups, respectively. Eighteen (75%) and 20 (64.5%) of the “go-go” and “slow-go” patients had primed blasts, respectively. In those patients, relative blast priming also predicted longer OS (HR = 0.46, 95% CI, 0.23-0.94, P = 0.03) independently of the ALFA classifier (HR = 2.11, 95% CI, 0.98-4.52 for slow-go [reference: go-go], P = 0.06). Our study is the first to account for genetic risk to investigate the predictive value of BH3 profiling in uniformly treated AML patients. Our results, albeit on a limited cohort skewed toward proliferative cases, suggest that relative mitochondrial priming could complement genetic risk stratification to identify older patients achieving long-term survival with intensive therapy. Functional assays such as BH3 profiling are currently restricted to specialized labs. Their prospective investigation in larger cohorts is warranted to confirm the emerging role of functional precision oncology in AML.
Figure 1.
Relative mitochondrial priming and overall survival in older AML patients treated intensively. (A) Summary of study workflow. (B) Gating strategy for CD45dim/SSClow blasts and live Lin−/CD34+/CD38−/CD45RA− HSPCs. (C) AUC (cubic spline interpolation) of Cytochrome C release in blasts (red) and HSPCs (purple) of 2 exemplary cases, 1 (top) assigned to the “primed” group based on an AUCblasts >120% AUCHSPCs, and 1 (bottom) assigned to the unprimed (AUCblasts ≤120% AUCHSPCs) group. (D) Overall survival probability from study inclusion in patients with primed (n = 39, red curve) or unprimed (n = 18, blue curve) blasts. (E) Overall survival probability from study inclusion in patients with primed (red curve) or unprimed blue curve blasts according to their ELN 2017 risk groups (fav/int favorable or intermediate risk, solid lines, adv adverse risk, dashed lines). All P values from log-rank tests. AML = acute myeloid leukemia; AUC = area under the curve; BMNC = bone marrow mononuclear cell; FSC = forward scatter; HSPC = hematopoietic stem and progenitor cell.
ACKNOWLEDGMENTS
We are indebted to Sophie Duchez, Christelle Doliger and Niclas Setterblad from the Saint-Louis Research Institute Core Flow Facility, and Céline Decroocq from the Lille University Hospital Tumor Bank. This work was supported by Association Laurette Fugain (ALF2020-01) to R. Itzykson and is part of the THEMA program funded by the Agence Nationale pour la Recherche (ANR IHU-B-2018).
AUTHOR CONTRIBUTIONS
RI, RDB, and AP designed the study, performed analyses, and drafted the manuscript. RDB, KP, CC, and JP performed experiments. SM, CR, and AP assisted in flow cytometry analyses. LA, TB, CB, ER, DL, TC, HD, and CG accrued patients. CG was the principal investigator of the trial. KCL and HD manage the ALFA cooperative group. All authors reviewed the manuscript and approved its final version.
DISCLOSURES
The authors have no conflicts of interest to disclose.
SOURCES OF FUNDING
This study was funded by Association Laurette Fugain (ALF2020-01).
Supplementary Material
Footnotes
Supplemental digital content is available for this article.
REFERENCES
- 1.Vo TT, Ryan J, Carrasco R, et al. Relative mitochondrial priming of myeloblasts and normal HSCs determines chemotherapeutic success in AML. Cell. 2012;151:344–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dohner H, Estey E, Grimwade D, et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood. 2017;129:424–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gardin C, Pautas C, Fournier E, et al. Added prognostic value of secondary AML-like gene mutations in ELN intermediate-risk older AML: ALFA-1200 study results. Blood Adv. 2020;4:1942–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Itzykson R, Fournier E, Berthon C, et al. Genetic identification of patients with AML older than 60 years achieving long-term survival with intensive chemotherapy. Blood. 2021;138:507–519. [DOI] [PubMed] [Google Scholar]
- 5.Letai A, Bhola P, Welm AL. Functional precision oncology: testing tumors with drugs to identify vulnerabilities and novel combinations. Cancer Cell. 2022;40:26–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pierceall WE, Kornblau SM, Carlson NE, et al. BH3 profiling discriminates response to cytarabine-based treatment of acute myelogenous leukemia. Mol Cancer Ther. 2013;12:2940–2949. [DOI] [PMC free article] [PubMed] [Google Scholar]