Abstract
Severe acute respiratory syndrome coronavirus (SARS-CoV-2) is rampant all over the world, and rapid and effective virus detection is the best auxiliary to curb the spread of the epidemic. A diagnosis can only be made if two or more different nucleic acid sequences are confirmed at the same time, and in most of traditional detection technologies, these target sequences have been detected separately. In this work, an electrochemiluminescent (ECL) biosensor employing a single ECL probe as signal output and responding to dual-target simultaneously is proposed for the first time. Taking the two sequences located in ORF 1ab region and N region of SARS-CoV-2 gene sequence as the model target and nitrogen doped carbon quantum dots (CDs) as ECL beacon, supplemented with catalytic hairpin assembly (CHA) reaction for signal amplification, the presented strategy has been successfully applied to the rapid detection of SARS-CoV-2. The developed SARS-CoV-2 biosensor based on the series CHA systems can realize the quantitative determination of SARS-CoV-2 in the range of 50 fM to 200 pM within 40 min. Moreover, the clinical validity of this method has been verified by the high consistency between the detection results of using this method and those using RT-qPCR for seven clinical pharyngeal swab samples.
Keywords: Electrochemiluminescence, Dual-target detection, SARS-CoV-2, Catalytic hairpin assembly, Carbon quantum dots
1. Introduction
Nucleic acid, including DNA and RNA, is an important carrier of genetic information in life activities, and its programmability and Watson-Crick base pairing also make it one of the effective elements for exploring superior sensing systems and improving analytical performance [1]. Electrochemiluminescence (ECL), as a powerful analytical technique, has been widely applied in clinical laboratory diagnosis, environmental analysis, food safety regulation and biosensing due to its high sensitivity, easy operation and simple optical equipment [2], [3], [4], [5]. In the past few years, a great deal of effort has been devoted to construct various ECL nucleic acid sensors and exploring their potential for practical applications. The vast majority of nucleic acid-based ECL sensors that have been developed are aimed at quantitative analysis of a single target [6], [7], [8], [9]. However, in some specific cases, especially for the accurate determination of virus infection, in order to feed back the viral load information more accurately, and to make the sensor have higher specificity to avoid the interference of virus’s nucleic acid from different sources, simultaneously detect sequences located in two or more characteristic regions of the viral gene is usually required [10], [11]. For example, for the detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which is currently raging around the world and causing coronavirus disease 2019 (COVID-19), two or three sequences located in its ORF 1ab, N, and E gene regions are usually selected as targets at the same time [12], [13], [14]. Therefore, it is necessary to establish a non-single target-responsive sensing strategy to meet the needs of the ECL sensing platform in accurately determining viral infection.
Dual-target detection nucleic acid biosensors based on two different beacons to output two distinguishable signals have been developed and applied in practice. For example, in real-time PCR, two pairs of fluorescent groups with different emission wavelengths and their quenching groups are often used to label probes [15]. While in electrochemical sensors, two substances with electroactivity at different potentials are generally selected as beacons [16], [17], [18], [19]. In multiplex analysis, some sensing strategies using different ECL indicator simultaneously have been reported [20], [21], [22], [23], [24]. However, coupling multiple luminophores to biomolecules is not only cumbersome, but also increases the difficulty of the experiment and leads to more uncertainties in the detection results. In addition, there are problems such as crosstalk between different reaction systems due to poor compatibility of co-reactants or quenching mechanisms between probes, as well as compromising the accuracy of quantitative analysis due to the use of the same co-reactant. In order to overcome the cross-reaction between ECL probes, in recent years, several ECL sensing strategies have been developed that employ Boolean logic gates to achieve multiple target detection with a single ECL emitter [25], [26], [27], [28]. However, these ECL strategies can only detect analytes sequentially rather than simultaneously, and thus cannot provide the determination information of coexisting multiple targets. It should be noted that the two targets used in virus detection are located on the same gene and have equal expression levels in the sample. Therefore, the output of a single signal through the joint regulation of two targets is expected to provide a new solution for the construction of ECL sensor that simultaneously detects two sites in the virus gene sequence.
In this study, taking two sequences in the ORF 1ab region and N region of the SARS-CoV-2 gene sequence as model targets, a dual-target responsive ECL biosensor was designed and applied for SARS-CoV-2 detection. Carbon quantum dots (CDs), has rich functional groups on the surface, are favorable for conjugated with nucleic acids, had been chosen as ECL probe [29], [30], [31], [32], [33]. Since the concentrations of targets in the sample are very low, signal amplification is necessary. Catalytic hairpin assembly (CHA), developed according to toehold-mediated strand displacement, is a typical nucleic acid signal amplification technology with constant temperature and does not require the participation of enzymes, which has the advantages of low cost, low requirements on reagent storage conditions [34], [35]. Therefore, CHA had been chosen in this study to amplify the signal. The designed biosensor has simple operation, short reaction time, and excellent specificity for the dual-sites detection of SARS-CoV-2.
2. Materials and methods
2.1. Regents and instruments
Citric acid, ethylenediamine, N-Hydroxysuccinimide (NHS), alumina polishing powder, 6-mercaptohexanol (MCH) and Tris(2-carboxyethyl) phosphine (TCEP) were purchased from Sigma-Aldrich (Shanghai, China). 1-(3-Dimethylaminopropyl)− 3-ethylcarbodiimide hydrochloride (EDC) was purchased from Alfa Aesar China Co. Ltd. (Tianjin, China). The related oligonucleotides are shown in Table S1. The instruments are listed in the Supplementary Material.
2.2. Synthesis of CDs and preparation of the CDs-labeled NH2 (NH2-CDs)
The synthesis steps of CDs and the preparation of the CDs-labeled NH2 (NH2-CDs) was described in the Supplementary Material.
2.3. Preparation of Au electrode for probe connection
The activation steps of Au electrode in this experiment, please referring to the Supplementary Material for details. The thiol modified capture probe (1 µM) pretreated by TCEP was dropped onto the surface of the activated Au electrode and placed at 37 ℃ for 3 h. Through the interaction of Au-thiol bonds, the Au electrode connected with the capture probe was obtained. And then MCH (1.0 mM) was added for 1 h to block the nonspecific active binding sites. Further, 0.4 µM OH2 was continuously added onto the above electrode, the capture probe and OH2 hybridized based on base complementary pairing, and the free end of the capture probe on the electrode surface was blocked by OH2. The prepared electrode was stored at 4 °C for further use.
2.4. Analysis procedure
The obtained electrode was immersed in a mixture of 0.4 µM OH1, 0.4 µM NH1, 5 µL NH2-CDs and a certain concentration of targets (equal concentration of O and N) and reacted at 37 °C for 40 min. The three-electrode system was immersed in 1 × PBS buffer solution containing 10 mM K2S2O8 by using Au electrode as working electrode, platinum wire electrode as counter electrode and Ag/AgCl electrode as reference electrode. The voltage of the photomultiplier tube was set to − 1000 V, conduct cyclic voltammetry scanning at the speed of 100 mV/s in the range of 0 to − 1.3 V, and collect signals by a BPCL ultra-weak luminescence analyzer. Each sample was determined three times and averaged for quantitative analysis.
3. Results and discussion
3.1. The principle of the proposed ECL biosensor
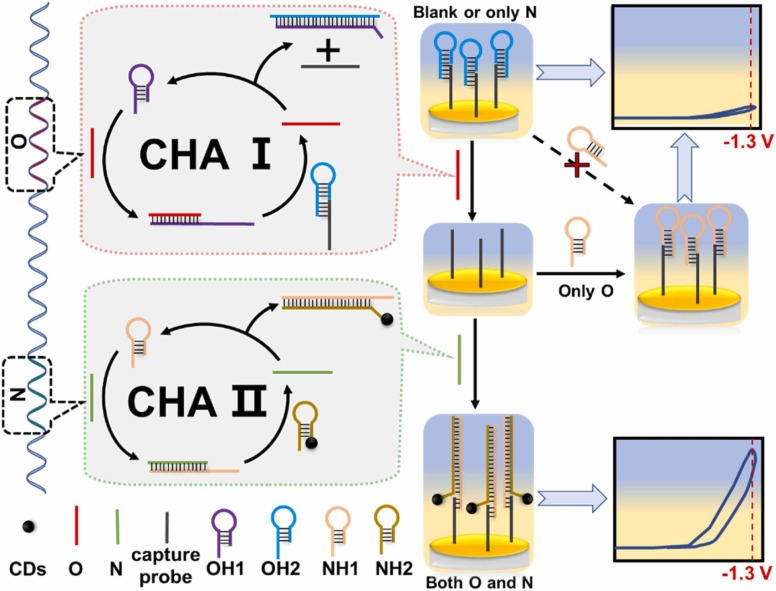
The mechanism regarding the proposed dual-target responsive ECL biosensor based on two sets of CHA signal amplification systems in series is systematically presented in Scheme 1. Taking the two sequences located in ORF 1ab region and N region of SARS-CoV-2 gene sequence as model targets (denoted as O and N, respectively) and CDs as ECL beacons. The thiol-modified capture probe was immobilized on the surface of the Au electrode through Au-thiol bonds, and hybridized with the hairpin probe OH2 by base complementary pairing. Target O and target N were designed as the initiators of the CHA Ⅰ and CHA Ⅱ reactions, respectively. When O is present, O opens the hairpin probe OH1 of the CHA Ⅰ reaction through the toehold nature, forming an O-OH1 double-stranded complex. In this complex, the exposed single-stranded region of OH1 can be hybridized with another hairpin probe OH2 (pre-combined on the electrode surface by hybridization) for the CHA Ⅰ reaction to form an OH1–2 double-stranded complex. At the same time, O is replaced into the next cycle of CHA Ⅰ, the capture probe previously blocked by OH2 is exposed and can hybridized with the hairpin probe NH1 for the reaction of CHA Ⅱ. In the presence of N, the CHA Ⅱ reaction was triggered, and the obtained amplification product NH1–2-CDs double-stranded complex was captured on the electrode surface due to the hybridization of the NH1 segment with the capture probe. With S2O8 2- as co-reactant, CDs shows excellent ECL response at − 1.3 V potential (vs Ag/AgCl), therefore, a strong ECL signal can be measured when O and N are present simultaneously. Conversely, when O is absent (regardless of whether N is present or not), the stability of the capture probe hybridized with OH2 is stronger than that of the capture probe hybridized with NH1. Therefore, the binding site of the capture probe is always blocked by OH2, and CDs cannot be aggregated to the electrode surface, so only weak ECL signal can be obtained. In addition, when O is present, but N is absent, even if the capture probe can bind to NH1, only weak ECL signals can be detected because the CHA Ⅱ reaction is not activated and NH2-CDs cannot bind to the electrode surface. According to the designed strategy, the determination of the presence of two co-existing targets can be accomplished using only a single ECL signaling probe.
Scheme 1.
The schematic diagram of dual-target responsive ECL sensor using single ECL probe based on CHA signal amplification systems in series.
3.2. Feasibility assay
The size and surface groups of the as-prepared CDs were investigated by transmission electron microscopy (TEM), the high-resolution TEM (HRTEM) (Fig. S1A) and FTIR analysis (Fig. S1B), and the ECL properties of the CDs were also investigated (Fig. S2). The migration rate of each band in gel electrophoresis is feedback of the molecular weight of nucleic acid. Sequences with more bases have higher relative molecular mass and slower migration rate in gel electrophoresis. Electrophoretic analysis was carried out using 12% polyacrylamide gel. The results of gel electrophoresis (Fig. S3) confirmed that the probes used were reasonably designed and the two sets of reactions of CHA Ⅰ and CHA Ⅱ were successfully triggered. Detailed information was described in the Supplementary Material.
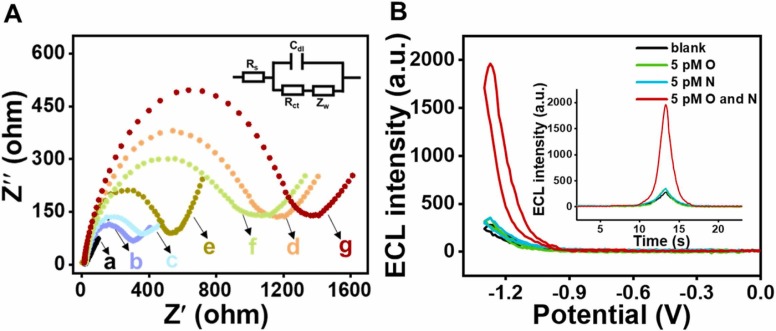
In addition, various forms of the negatively charged nucleic acid structures capable of repelling the negatively charged Fe(CN)6 3- and Fe(CN)6 4- to different extents. As shown in Fig. 1A, electrochemical impedance spectroscopy was used to characterize the electrodes in different states. Compared with the bare Au electrode (curve a), the impedance of the electrode modified with capture probe (curve b, 275 ohm) increased significantly due to the negative charge of the nucleic acid, and further increased after treatment with MCH (curve c, 325 ohm). Curve d (1143 ohm) represents the impedance measured after blocking the capture probe with OH2. When O exists, OH2 separate from the electrode surface due to the triggering of the CHA Ⅰ reaction, and the impedance of the electrode surface reduce (curve e, 514 ohm), while the capture probe previously blocked by OH2 is exposed and could continue to hybridize with NH1 (curve f, 1044 ohm), and the electrochemical impedance increased (compared with curve e). When both O and N are present, the surface resistance of the electrode is further increased (curve g, 1361 ohm) due to the binding of NH1–2-CDs to the electrode via the capture probe.
Fig. 1.
(A) Electrochemical impedance spectroscopy of various probes modified at electrode: (a: bare Au electrode; b: electrode ‘a′ + capture probe; c: electrode ‘b′ + MCH; d: electrode ‘c′ + OH2; e: electrode ‘d′ + CHA Ⅰ-Ⅱ system (but without NH2) + 100 pM O and N; f: electrode ‘d′ + CHA Ⅰ-Ⅱ system + 100 pM O; g: electrode ‘d′ + CHA Ⅰ-Ⅱ system + 100 pM O and N) in 5.0 mM [Fe(CN)6]3-/4- including 100 mM KCl. (B) ECL intensity of the proposed sensing system in the absence of target and in presence of 5 pM O, 5pM N, and 5 pM O and N, respectively.
The changes of the ECL signal caused by the presence of target in the sensor is also investigated. As shown in Fig. 1B, when there is only O or only N, the measured ECL strength is equivalent to that in the absence of both O and N, while the ECL strength of the system is significantly enhanced in the presence of both O and N at the same time. It shows that the sensor only responds to the simultaneous existence of O and N, which proves the feasibility of the designed sensing strategy.
3.3. Optimization of experimental conditions
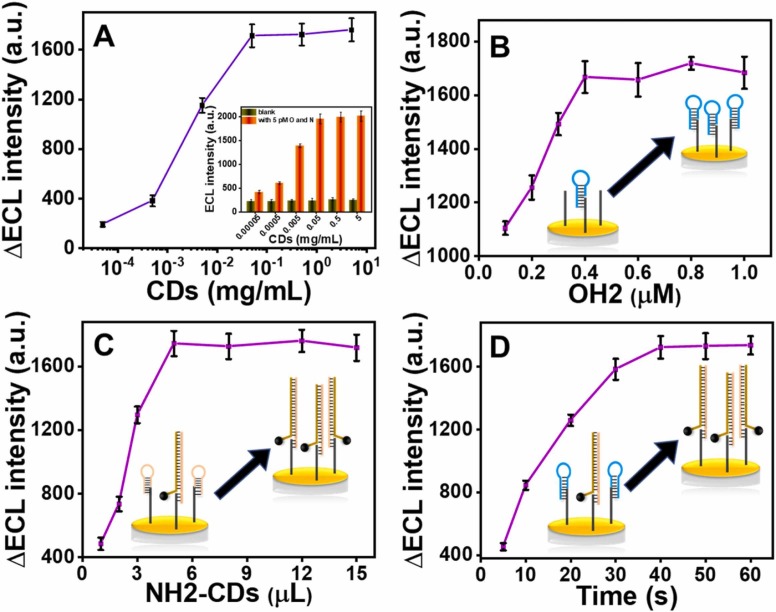
Some important conditions and parameters of the experiments were optimized for the best sensing performance. In order to ensure that CDs can be combined with NH2 as much as possible, the concentration of CDs was firstly optimized. Fig. 2A shows the ΔECL intensity (ΔECL refers to the difference between the measured ECL signal and the signal measured for the blank.) of NH2-CDs prepared with different concentrations of CDs for sensor construction. With the increase of CDs concentration, the ΔECL intensity first increased and reached a plateau at the concentration of 50 µg/mL, indicating that at this concentration, CDs has fully coupled with the added NH2 used to form NH2-CDs. Therefore, 50 µg/mL CDs was selected as the best synthesis condition for NH2-CDs synthesis.
Fig. 2.
The effect of (A) the concentration of CDs, (B) the concentration of OH2, (C) the dosage of NH2-CDs and (D) the reaction time on the change of ECL intensity. The error bars represent the standard deviation of three replicate detections.
Since the signal is generated from CDs, and the amount of CDs enriched on the electrode surface is limited by the hybridization of NH2-CDs with capture probe, the dosage of NH2-CDs and the concentration of OH2 used to block the free end of the capture probe also need to be optimized. The results in Fig. 2B show the relationship between the dosage of synthetic NH2-CDs and the measured ΔECL intensity. With the gradual increase of the dosage of NH2-CDs, the ΔECL intensity also gradually increases and tends to be stable at 5 µL. Therefore, the optimal volume of NH2-CDs is determined as 5 µL. As shown in Fig. 2C, ΔECL intensity increases with the increase of OH2 concentration and reaches a plateau at 0.4 µM, indicating that 0.4 µM OH2 is sufficient to block the free segment of the capture probe to prevent the NH1 probe from interacting with the capture probe. Considering that the hybridization ratio of the two hairpin probes in CHA reaction product is 1:1, and in the designed sensing strategy, the dosage of NH1–2-CDs that can be connected to the capture probe is equivalent to that of the CHA Ⅰ reaction product (OH1–2). Therefore, the concentrations of OH1 and NH1 in the experiment were also determined to be 0.4 µM.
Finally, the effect of reaction time was investigated. As shown in Fig. 2D, with the increase of reaction time, the ΔECL also increased, and when the incubation time exceeded 40 min, the ΔECL was not significantly increased, indicating that 40 min is sufficient to conduct the CHA reaction and complete the enrichment of NH1–2-CDs on the electrode surface.
3.4. Performance of the developed biosensor
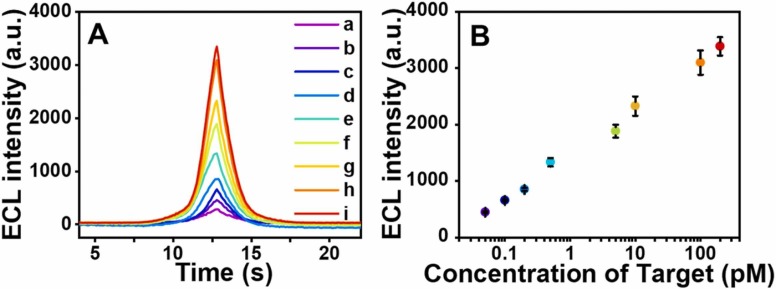
Since the two targets are sequences located in two regions of the same gene, they have the same expression level. Therefore, the quantitative detection performance of the sensor for the two targets at equal concentrations was investigated under the optimal experimental conditions. As depicted in Fig. 3 A, in the concentration range of 50 fM to 200 pM, the ECL intensity increases with the increase of the target concentration. And within this concentration range, the measured ECL intensity and the logarithm of target concentration shows a good linear relationship (Fig. 3B). The correlation equation is shown as follows:
| ECL = 775·6 Lg Ctarget +1434·4 R2 = 0·987 |
where ECL refers to the measured ECL intensity, C target refers to the concentrations of O and N added, and R is the correlation linear coefficient, the detection limit is calculated to be 20 fM (S/N = 3).
Fig. 3.
(A) The ECL signal of the target (equal concentrations of O and N) at various concentrations, a to i: 0, 50 fM, 100 fM, 200 fM, 500 fM, 5 pM, 10 pM, 100 pM, 200 pM. (B) Linear plot of ECL signal vs the concentration of the target. The error bars represent the standard deviation of three replicate detections.
3.5. Selectivity and reproducibility of the ECL biosensor
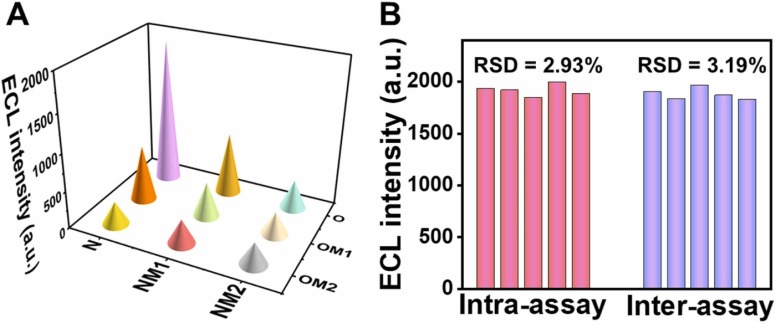
Single base mismatch sequences (OM1 and NM1) and double base mismatch sequences (OM2 and NM2) were designed for two targets (O and N) to study the specificity of the method. The sensing method was applied to the detection of two targets with the same concentration (5 pM) and their mismatched sequence samples. The result of Fig. 4A shows that the ECL signals obtained with the target sample is much higher than those obtained with the mismatched sequence samples, which proves that the method has good specificity for target detection.
Fig. 4.
(A) Selectivity and (B) reproducibility analysis of the proposed ECL biosensor.
To explore the reproducibility of this sensor, NH2-CDs were prepared with the same batch and different batches of CDs, respectively, for the construction of the sensor to detect 5 pM targets. The relative standard deviation (RSD) of ECL intensity of same batch CDs is 2.93%, while that of different batches CDs is 3.19%, indicating that the sensing platform has good reproducibility (as shown in Fig. 4B).
3.6. Application of the proposed biosensor
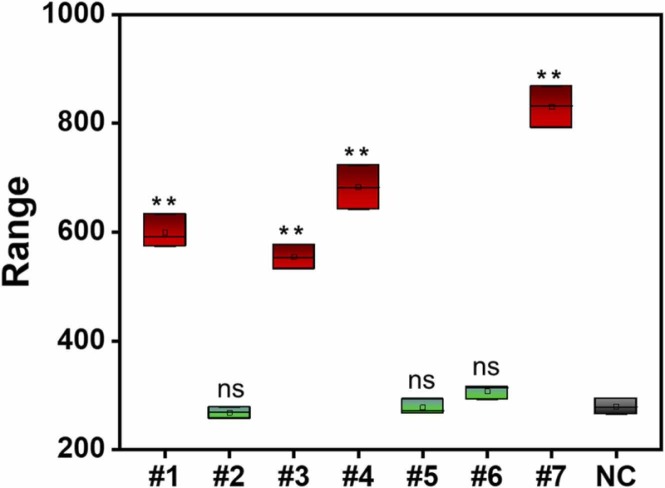
In clinical practice, oropharyngeal swabs are mainly collected to extract viral nucleic acid and gene amplification method is used to determine whether patients are infected with novel coronavirus. To validate the clinical practicability of the proposed assay for the detection of SARS-CoV-2, seven samples were obtained from Affiliated Hospital of Putian University (the samples numbered #2, #5, #6 are uninfected negative samples, and the positive samples numbered #1, #3, #4, #7 are taken from patients with confirmed COVID-19). RNA from these clinical samples was extracted using MagaBio plus Virus DNA/RNA Purification Kit III (Hangzhou Bioer Technology Co., Ltd.), and fragmented by ultrasound to obtain short fragments of RNA. The above work was completed in the laboratory of the Affiliated Hospital of Putian University, and the experiment was conducted in strict accordance with the protocol approved by the Ethics Committee of the Affiliated Hospital of Putian University. As shown in the Fig. 5, compared with the no target control, the samples from the four confirmed cases resulted in a significant statistical difference (P < 0.01), while no statistically significant differences were found between the signals from the three healthy volunteers and the no template controls (P > 0.01). Four SARS-CoV-2 positive clinical samples and three samples of healthy volunteers can be identified by the sensing method proposed in this paper, which were consistent with the test results of the hospital.
Fig. 5.
SARS-CoV-2 detection in clinical throat swab samples. #1, #2, #3, #4, #5, #6 and #7 correspond to clinical samples. NC correspond to the no template control. Values represent the mean of replicate samples (n = 3) ± SD. ** represent statistical differences concerning negative control with a p-value < 0.01, ns: non-significant (p > 0.01).
4. Conclusions
In this study, a highly selective ECL biosensor was developed for the rapid detection of SARS-CoV-2. The most important highlight of this biosensor strategy is that by regulating the connection domain of the capture probe modified on the electrode surface, two sets of the same signal amplification strategies are connected in series, and an ECL nucleic acid sensing platform with a single signal output by the joint regulation of dual-target is successfully established for the first time. The proposed biosensor was successfully applied to the detection of SARS-CoV-2 in clinical throat swab samples, and the detection results of SARS-CoV-2 RNA extracted from clinical samples are 100% consistent with the hospital report using RT-qPCR detection, indicating its promise in clinical use. In addition to this, the approach enables rapid analysis of SARS-CoV-2 in a response time of 40 min, compared to that of 3–5 h in other electrochemical and ECL studies. Although the detection limit is not better than some viral nucleic acid detection methods, and more efforts are still needed to improve the performance of detection sensing platforms, the proposed ECL sensing strategy opens an interesting avenue for virus nucleic acid analysis based on dual-target response.
CRediT authorship contribution statement
Ying Zhang: Methodology, Formal analysis, Data curation, Writing – original draft. Xiaocui Huang: Methodology, Formal analysis, Data curation, Writing – review & editing. Weixin Li: Methodology, Formal analysis, Data curation, Writing – review & editing. Qunfang Xie: Formal analysis, Writing – review & editing. Jie Zhang: Formal analysis, Writing – review & editing. Fang Luo: Formal analysis, Writing – review & editing. Bin Qiu: Funding acquisition, Supervision, Writing – review & editing. Zhonghui Chen: Methodology, Formal analysis, Data curation. Zhenyu Lin: Conceptualization, Funding acquisition, Supervision, Writing – review & editing. Guoyan Xu: Methodology, Conceptualization, Supervision, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
This project was supported by National Sciences Foundation of China (22174018, 21974020), the Natural Sciences Foundation of Fujian Province (2020J05250, 2020J01968).
Biographies
Ying Zhang obtained a Ph.D. in Analytical Chemistry from Fuzhou University and now works in the Central Laboratory of the First Affiliated Hospital of Fujian Medical University. Her research focuses on biosensors for disease diagnosis and synthesis & application of DNA nanomaterials.
Xiaocui Huang is presently pursuing for her Ph.D. in College of Chemistry at Fuzhou University (China). Her research focuses on electrochemiluminescence biosensor for disease forewarning & diagnosis.
Weixin Li is presently pursuing for his M.S. in College of Chemistry at Fuzhou University (China). His research focuses on the design and application of photoelectrochemical biosensors.
Qunfan Xie obtained his master's degree in clinical medicine from Fujian Medical University and now works in Department of General Practice of the First Affiliated Hospital of Fujian Medical University. His research focuses on new technology for diagnosis and treatment of respiratory diseases.
Jie Zhang received her master's degree from Nankai University. She is now working at the Department of Geriatrics in the First Affiliated Hospital of Fujian Medical University.
Fang Luo received her M.Sc. at the Fuzhou University in 2012, and her PhD in 2016 from the University of Newcastle, Australia under the supervision of Zuliang Chen. Currently she is an associate professor in College of Biology Science and Engineering at Fuzhou University (China). Her research focuses on synthesis and application of nanomaterials.
Bin Qiu is currently a professor in College of Chemistry at Fuzhou University (China). He obtained his pH.D. in analytical chemistry from Fuzhou University in 2008. Subsequently, he worked as a postdoctoral fellow in Hong Kong Baptist University. His research focuses on biosensors for food safety analysis.
Zhonghui Chen obtained a Ph.D. from Fuzhou University and now works in the Affiliated Hospital of Putian University. His research focuses on research and development of in vitro diagnostic methods and kits for major diseases.
Zhenyu Lin is currently a full professor in College of Chemistry at Fuzhou University (China). He obtained his B.S. degree in polymer material and engineering from Beijing Institute of Technology (China) and PhD in analytical chemistry from Fuzhou University. He joined the Ministry of Education Key Laboratory of Analysis for Food Safety & Biology at Fuzhou University since 2007. Subsequently, he worked as a postdoctoral fellow in Graduate School of Environmental Studies & School of Engineering, Tohoku University (Japan). He is a 2013 recipient of the National Science Fund for Outstanding Young Scholar and a 2014 recipient of Natural Science Found of Fujian Province for Distinguished Young Scholar. His research focuses on biosensors for food safety analysis and disease forewarning & diagnosis.
Guoyan Xu is an associate professor at Fujian Medical University and the vice director of Department of General Practice of the First Affiliated Hospital of Fujian Medical University. His research focuses on biosensors for disease forewarning & diagnosis.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.snb.2022.133223.
Appendix A. Supplementary material
Supplementary material.
.
Data Availability
Data will be made available on request.
References
- 1.Takahashi S., Sugimoto N. Watson–crick versus hoogsteen base pairs: chemical strategy to encode and express genetic information in life. Acc. Chem. Res. 2021;54:2110–2120. doi: 10.1021/acs.accounts.0c00734. [DOI] [PubMed] [Google Scholar]
- 2.Ding Z.F., Quinn B.M., Haram S.K., Pell L.E., Korgel B.A., Bard A.J. Electrochemistry and electrogenerated chemiluminescence from silicon nanocrystal quantum dots. Science. 2002;296:1293–1297. doi: 10.1126/science.1069336. [DOI] [PubMed] [Google Scholar]
- 3.Richter M.M. Electrochemiluminescence (ECL) Chem. Rev. 2004;104:3003–3036. doi: 10.1021/cr020373d. [DOI] [PubMed] [Google Scholar]
- 4.Tang Y., Li J., Guo Q., Nie G. An ultrasensitive electrochemiluminescence assay for Hg2+ through graphene quantum dots and poly(5-formylindole) nanocomposite. Sens. Actuators B Chem. 2019;282:824–830. [Google Scholar]
- 5.Wang D., Jiang S., Liang Y., Wang X., Zhuang X., Tian C., et al. Selective detection of enrofloxacin in biological and environmental samples using a molecularly imprinted electrochemiluminescence sensor based on functionalized copper nanoclusters. Talanta. 2022;236 doi: 10.1016/j.talanta.2021.122835. [DOI] [PubMed] [Google Scholar]
- 6.Lu J., Wu L., Hu Y., Wang S., Guo Z. Ultrasensitive Faraday cage-type electrochemiluminescence assay for femtomolar miRNA-141 via graphene oxide and hybridization chain reaction-assisted cascade amplification. Biosens. Bioelectron. 2018;109:13–19. doi: 10.1016/j.bios.2018.02.062. [DOI] [PubMed] [Google Scholar]
- 7.Zhang Q., Liu Y., Nie Y., Ma Q. Magnetic-plasmonic yolk-shell nanostructure-based plasmon-enhanced electrochemiluminescence sensor. Sens. Actuators B Chem. 2020;319 [Google Scholar]
- 8.Liu P.-F., Zhao K.-R., Liu Z.-J., Wang L., Ye S.-Y., Liang G.-X. Cas12a-based electrochemiluminescence biosensor for target amplification-free DNA detection. Biosens. Bioelectron. 2021;176 doi: 10.1016/j.bios.2020.112954. [DOI] [PubMed] [Google Scholar]
- 9.Yu L., Zhu L., Yan M., Feng S., Huang J., Yang X. Electrochemiluminescence biosensor based on entropy-driven amplification and a tetrahedral DNA nanostructure for miRNA-133a detection. Anal. Chem. 2021;93:11809–11815. doi: 10.1021/acs.analchem.1c02361. [DOI] [PubMed] [Google Scholar]
- 10.Dronina J., Samukaite-Bubniene U., Ramanavicius A. Advances and insights in the diagnosis of viral infections. J. Nanobiotechnol. 2021;19:348. doi: 10.1186/s12951-021-01081-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nasrollahi F., Haghniaz R., Hosseini V., Davoodi E., Mahmoodi M., Karamikamkar S., et al. Micro and nanoscale technologies for diagnosis of viral infections. Small. 2021;17:2100692. doi: 10.1002/smll.202100692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Navarro A., Gómez L., Sanseverino I., Niegowska M., Roka E., Pedraccini R., et al. SARS-CoV-2 detection in wastewater using multiplex quantitative PCR. Sci. Total Environ. 2021;797 doi: 10.1016/j.scitotenv.2021.148890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yin H., Wu Z., Shi N., Qi Y., Jian X., Zhou L., et al. Ultrafast multiplexed detection of SARS-CoV-2 RNA using a rapid droplet digital PCR system. Biosens. Bioelectron. 2021;188 doi: 10.1016/j.bios.2021.113282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zayani R., Rezig D., Fares W., Marrakchi M., Essafi M., Raouafi N. Multiplexed magnetofluorescent bioplatform for the sensitive detection of SARS-CoV-2 viral RNA without nucleic acid amplification. Anal. Chem. 2021;93:11225–11232. doi: 10.1021/acs.analchem.1c01950. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Y.L., Zhang D.B., Li W.Q., Chen J.Q., Peng Y.F., Cao W. A novel real-time quantitative PCR method using attached universal template probe. Nucleic Acids Res. 2003;31 doi: 10.1093/nar/gng123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu R., Zhang X., Chi K.-N., Yang T., Yang Y.-H. Bifunctional MOFs-based ratiometric electrochemical sensor for multiplex heavy metal ions. ACS Appl. Mater. Interfaces. 2020;12:30770–30778. doi: 10.1021/acsami.0c06291. [DOI] [PubMed] [Google Scholar]
- 17.Zhao M.-h, Shi H.-h, Li C.-c, Luo X., Cui L., Zhang C.-y. Construction of a target-triggered DNAzyme motor for electrochemical detection of multiple DNA glycosylases. Sens. Actuators B Chem. 2022;361 [Google Scholar]
- 18.Derkus B., Acar Bozkurt P., Tulu M., Emregul K.C., Yucesan C., Emregul E. Simultaneous quantification of myelin basic protein and tau proteins in cerebrospinal fluid and serum of multiple sclerosis patients using nanoimmunosensor. Biosens. Bioelectron. 2017;89:781–788. doi: 10.1016/j.bios.2016.10.019. [DOI] [PubMed] [Google Scholar]
- 19.Liu Z., Lei S., Zou L., Li G., Ye B. Grafting homogenous electrochemical biosensing strategy based on reverse proximity ligation and Exo III assisted target circulation for multiplexed communicable disease DNA assay. Biosens. Bioelectron. 2020;167 doi: 10.1016/j.bios.2020.112487. [DOI] [PubMed] [Google Scholar]
- 20.Guo W., Ding H., Gu C., Liu Y., Jiang X., Su B., et al. Potential-resolved multicolor electrochemiluminescence for multiplex immunoassay in a single sample. J. Am. Chem. Soc. 2018;140:15904–15915. doi: 10.1021/jacs.8b09422. [DOI] [PubMed] [Google Scholar]
- 21.Lv Y., Zhou Z., Shen Y., Zhou Q., Ji J., Liu S., et al. Coupled fluorometer-potentiostat system and metal-free monochromatic luminophores for high-resolution wavelength-resolved electrochemiluminescent multiplex bioassay. ACS Sens. 2018;3:1362–1367. doi: 10.1021/acssensors.8b00292. [DOI] [PubMed] [Google Scholar]
- 22.Shao H., Lin H., Lu J., Hu Y., Wang S., Huang Y., et al. Potential-resolved Faraday cage-type electrochemiluminescence biosensor for simultaneous determination of miRNAs using functionalized g-C3N4 and metal organic framework nanosheets. Biosens. Bioelectron. 2018;118:247–252. doi: 10.1016/j.bios.2018.07.064. [DOI] [PubMed] [Google Scholar]
- 23.Cao J.-T., Liu F.-R., Fu X.-L., Ma J.-X., Ren S.-W., Liu Y.-M. A novel electrochemiluminescence resonance energy transfer system for simultaneous determination of two acute myocardial infarction markers using versatile gold nanorods as energy acceptors. Chem. Commun. 2019;55:2829–2832. doi: 10.1039/c9cc00563c. [DOI] [PubMed] [Google Scholar]
- 24.Mo G., He X., Qin D., Meng S., Wu Y., Deng B. Spatially-resolved dual-potential sandwich electrochemiluminescence immunosensor for the simultaneous determination of carbohydrate antigen 19–9 and carbohydrate antigen 24-2. Biosens. Bioelectron. 2021;178 doi: 10.1016/j.bios.2021.113024. [DOI] [PubMed] [Google Scholar]
- 25.Liu X., Liu H., Li M., Qi H., Gao Q., Zhang C. Highly sensitive electrochemiluminescence assay for cardiac Troponin I and adenosine triphosphate by using supersandwich amplification and bifunctional aptamer. Chemelectrochem. 2017;4:1708–1713. [Google Scholar]
- 26.Zhang Y., Li X., Xu Z., Chai Y., Wang H., Yuan R. An ultrasensitive electrochemiluminescence biosensor for multiple detection of microRNAs based on a novel dual circuit catalyzed hairpin assembly. Chem. Commun. 2018;54:10148–10151. doi: 10.1039/c8cc06102e. [DOI] [PubMed] [Google Scholar]
- 27.Feng D., Tan X., Wu Y., Ai C., Luo Y., Chen Q., et al. Electrochemiluminecence nanogears aptasensor based on MIL-53(Fe)@CdS for multiplexed detection of kanamycin and neomycin. Biosens. Bioelectron. 2019;129:100–106. doi: 10.1016/j.bios.2018.12.050. [DOI] [PubMed] [Google Scholar]
- 28.Nie Y., Yuan X., Zhang P., Chai Y.-Q., Yuan R. Versatile and ultrasensitive electrochemiluminescence biosensor for biomarker detection based on nonenzymatic amplification and aptamer-triggered emitter release. Anal. Chem. 2019;91:3452–3458. doi: 10.1021/acs.analchem.8b05001. [DOI] [PubMed] [Google Scholar]
- 29.Yang E., Zhang Y., Shen Y. Quantum dots for electrochemiluminescence bioanalysis - a review. Anal. Chim. Acta. 2021 doi: 10.1016/j.aca.2021.339140. [DOI] [PubMed] [Google Scholar]
- 30.Zheng L., Chi Y., Dong Y., Lin J., Wang B. Electrochemiluminescence of water-soluble carbon nanocrystals released electrochemically from graphite. J. Am. Chem. Soc. 2009;131:4564–4565. doi: 10.1021/ja809073f. [DOI] [PubMed] [Google Scholar]
- 31.Zhang R., Adsetts J.R., Nie Y., Sun X., Ding Z. Electrochemiluminescence of nitrogen- and sulfur-doped graphene quantum dots. Carbon. 2018;129:45–53. [Google Scholar]
- 32.Liu Q., Ma C., Liu X.-P., Wei Y.-P., Mao C.-J., Zhu J.-J. A novel electrochemiluminescence biosensor for the detection of microRNAs based on a DNA functionalized nitrogen doped carbon quantum dots as signal enhancers. Biosens. Bioelectron. 2017;92:273–279. doi: 10.1016/j.bios.2017.02.027. [DOI] [PubMed] [Google Scholar]
- 33.Qin D., Jiang X., Mo G., Feng J., Yu C., Deng B., Novel A. Carbon quantum dots signal amplification strategy coupled with sandwich electrochemiluminescence immunosensor for the detection of CA15-3 in human serum. ACS Sens. 2019;4:504–512. doi: 10.1021/acssensors.8b01607. [DOI] [PubMed] [Google Scholar]
- 34.Liu J., Zhang Y., Xie H., Zhao L., Zheng L., Ye H. Applications of catalytic hairpin assembly reaction in biosensing. Small. 2019;15 doi: 10.1002/smll.201902989. [DOI] [PubMed] [Google Scholar]
- 35.Luo Z., Li Y., Zhang P., He L., Feng Y., Feng Y., et al. Catalytic hairpin assembly as cascade nucleic acid circuits for fluorescent biosensor: design, evolution and application. TrAC, Trends Anal. Chem. 2022;151 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material.
Data Availability Statement
Data will be made available on request.