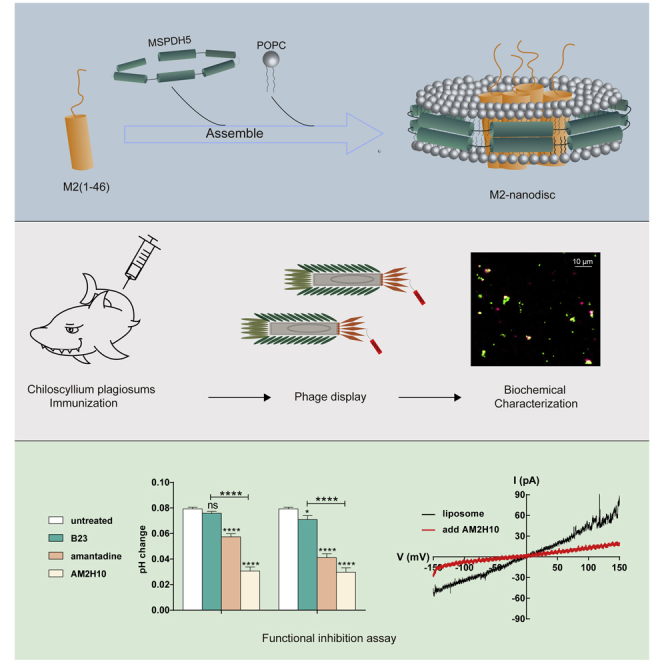
Summary
Influenza A virus poses a constant challenge to human health. The highly conserved influenza matrix-2 (M2) protein is an attractive target for the development of a universal antibody-based drug. However, screening using antigens with subphysiological conformation in a nonmembrane environment significantly reduces the generation of efficient antibodies. Here, M2(1-46) was incorporated into nanodiscs (M2-nanodiscs) with M2 in a membrane-embedded tetrameric conformation, closely resembling its natural physiological state in the influenza viral envelope. M2-nanodisc generation, an antigen, was followed by Chiloscyllium plagiosum immunization. The functional vNARs were selected by phage display panning strategy from the shark immune library. One of the isolated vNARs, AM2H10, could specifically bind to tetrameric M2 instead of monomeric M2e (the ectodomain of M2 protein). Furthermore, AM2H10 blocked ion influx through amantadine-sensitive and resistant M2 channels. Our findings indicated the possibility of developing functional shark nanobodies against various influenza viruses by targeting the M2 protein.
Subject areas: Virology, Nanoparticles
Graphical abstract
Highlights
-
•
M2-nanodisc mimics the natural conformation of tetrameric M2e protein
-
•
AM2H10 binds with tetrameric M2 instead of monomeric M2e
-
•
AM2H10 can inhibit the activities of both amantadine sensitive and resistant M2 channels
Virology; Nanoparticles
Introduction
Influenza A virus causes seasonal influenza infection, resulting in nearly 500,000 annual deaths worldwide.1 The main therapeutic strategies for controlling influenza are vaccines and anti-viral drugs. Currently available clinical vaccines are mainly based on two highly variable envelope proteins, hemagglutinin (HA) and neuraminidase (NA). However, these vaccines do not provide the substantial protection expected in case the annual ‘Flu vaccine’ mismatches with the virus strains causing a pandemic. Unfortunately, the mismatch may contribute to influenza outbreaks, thus worsening the situation.2 Approved anti-influenza drugs, including NA inhibitors, zanamivir and oseltamivir, and matrix-2 protein (M2) inhibitors, amantadine and rimantadine, demonstrate substantial discounted curative effect because of high antigenic variability, drug resistance, and security.3,4,5 Therefore, the development of additional anti-viral therapeutic agents, like antibody-based drugs, is an important research direction.1,6,7,8
Influenza M2 is an important target of antibodies.9,10,11,12,13 As a tetrameric transmembrane protein, M2 functions as a proton channel and participates in virus uncoating in endosomes.14,15,16 Despite numerous epidemics, the ectodomain of M2 (M2e) protein shows a high degree of conservation. Since the isolation of the first batch of influenza strains in 1981, it has essentially remained unchanged,17 implying that M2e peptide possesses a cross-therapeutic potential for various influenza virus infections. Several monoclonal antibodies (mAbs) targeting M2e-only fragments have been developed and their potential therapeutic activities have been demonstrated.18,19,20,21,22,23 However, the current methods for generating anti-M2e antibodies yield poor antibody titers because of the low immunogenicity of M2e.24,25 Almost all previous studies were based on the M2e protein solution without modeling the membrane environment, resulting in the subphysiological conformation of M2e, and consequently, reduced possibility of obtaining efficient antibodies. The predicament may be solved by the nanodisc assembly technique. Nanodisc is an exceptional system for solubilizing membrane proteins and provides a near-native membrane condition. The nanodisc-solubilized membrane protein used in antibody development shows certain enhanced immunogenicity.26,27,28,29
Previous studies suggest that the viral neutralization ability of traditional M2-targeted antibodies is severely limited because of the shielding effect of abundant HA and NA on the virus surface.25,30 Nanobodies, like heavy chain variable domain antibodies (VHH) found in Camelidae or variable new antigen receptors (vNARs) in sharks, possess unique properties including small size, excellent solubility, superior stability, quick clearance from blood, and deep tissue penetration31,32,33,34,35 and may help overcome the above problem.35 In recent years, nanobodies have emerged as a promising tool for diagnosing and treating various diseases.36,37,38,39,40,41 In August 2018, the first nanobody drug, Caplicizumab, was approved for the treatment of a blood clotting disorder.42
vNAR is an unexplored field in the development of therapeutic antibodies against influenza. With a molecular weight of approximately 12 kDa, vNAR is the smallest naturally occurring antigen-binding unit among vertebrates.43 In this study, we assembled M2(1-46)-containing extracellular and transmembrane domains into nanodiscs (Figure 1A). A large immunized library from Chiloscyllium plagiosum was used to identify anti-M2 nanobodies by panning with M2-nanodiscs and counter selecting with empty nanodiscs. After four rounds of subtractive panning, several anti-M2 vNARs were isolated. One of the vNARs, AM2H10, displayed a cross-reactive inhibitory effect on both amantadine-sensitive and resistant M2 channels in the in vitro proton flux and patch clamp assays. Our results demonstrated that AM2H10 could interfere with the ion channel function of M2 including amantadine-sensitive and resistant subtypes in vitro.
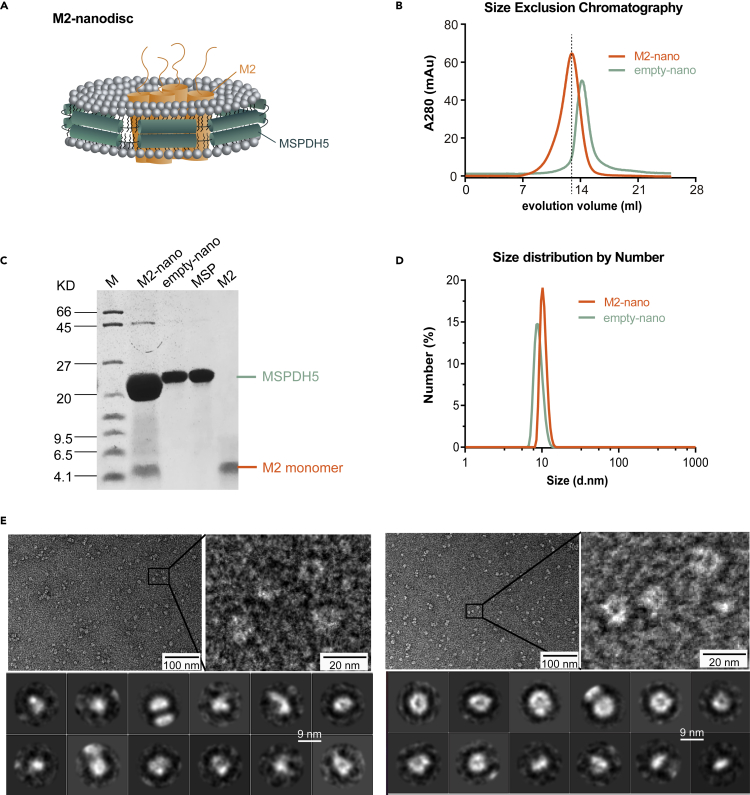
Figure 1.
Assembly and characterization of M2-nanodiscs
(A) The schematic diagram for M2-nanodisc assembly.
(B) The assembled nanodiscs were purified on Superdex 200 column by SEC.
(C) The peak fractions of SEC visualized on SDS-PAGE.
(D) DLS analysis for M2-nanodiscs and empty nanodiscs indicates a uniform distribution of M2-nanodiscs, with an average diameter of 10.29 nm, slightly larger than that of empty nanodiscs (8.964 nm).
(E) TEM data for empty nanodiscs (left) and M2-nanodiscs (right); images (scale bar: 100 nm or 20 nm) are shown on the top and 2D classifications (scale bar: 9 nm) at the bottom.
Results
Assembly and characterization of M2-nanodiscs
For the nanodisc assembly (M2-nanodisc), we synthesized M2 peptides of influenza A/Hong Kong/8/68 (H3N2) containing residues 1 to 46 following the method described previously and the sequence displayed in Table 1.44 The M2-nanodiscs mixture was purified by size-exclusion chromatography (SEC) and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (Figures 1B and 1C). SEC showed a single peak with the peak for M2-nanodiscs elution shifting about 1 mL relative to that of empty nanodiscs. SDS-PAGE analysis showed that the M2-nanodiscs sample had two bands corresponding to M2(1-46) and Membrane Scaffold Protein 1D1 Delta-Helix 5 (MSPDH5), indicating that the former was successfully incorporated into nanodiscs. Dynamic light scattering (DLS) showed that the average diameter of M2-nanodiscs was 10.29 nm, slightly larger than that of empty nanodiscs with a mean of 8.96 nm (Figure 1D). 2D class averages of transmission electron microscopy (TEM) imaging shows that nanodisc samples were homogeneous, approximately 10 nm in size, and M2 insertion did not change the discoidal shape and stability of nanodiscs (Figure 1E), consistent with DLS results. Therefore, M2-nanodiscs showed homogeneous distributions.
Confirmation of tetramer formation of M2-inserted nanodiscs
The formation of homologous tetramers of M2 in a phospholipid membrane environment has been widely reported in the literature.10,45,46 For the M2 nanodisc sample, we conducted a BS3 chemical cross-linking experiment. SDS-PAGE showed a new band between 20 and 27 kDa (Figure 2A, left), corresponding to the molecular weight of the M2 homotetramer. Western blotting with an M2-specific antibody further verified that the new band corresponded to the M2 protein (Figure 2A, right). The upper bands (>35 kDa) may denote the complex of MSPDH5 (∼22 KDa) and tetrameric M2 (∼20 KDa) owing to crosslinking by BS.3
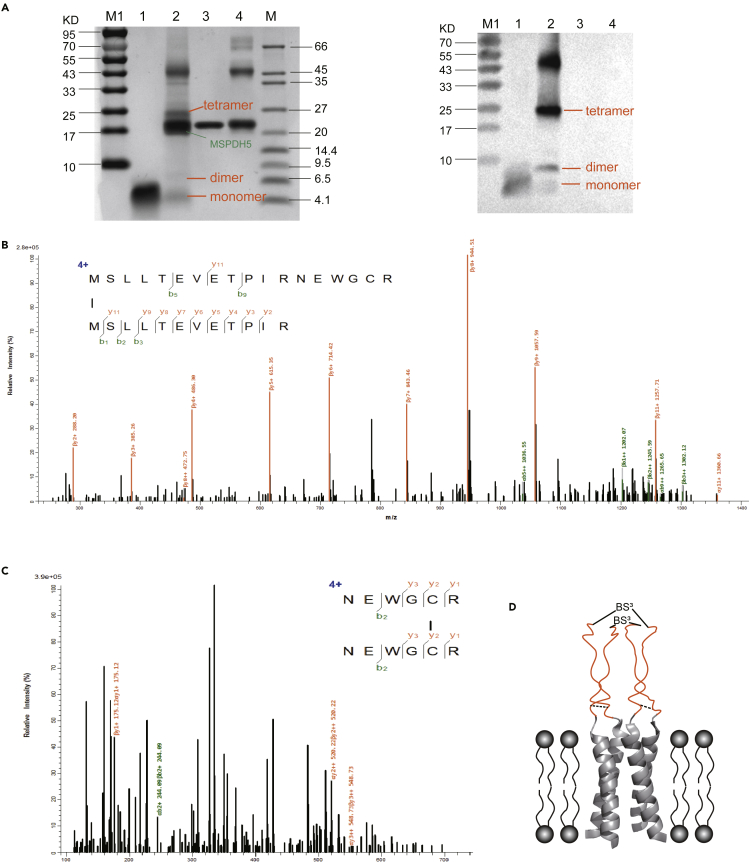
Figure 2.
The confirmation of tetramer formation of M2 inserted nanodiscs
(A) SDS-PAGE (left) and western blotting (right) stained with anti-M2 antibody (14C2) for purified nanodisc samples crosslinked with M2-nanodiscs by BS3. Lanes M1 and M, molecular mass markers; lane 1, M2(1-46); line 2, M2-nanodisc +1 mM BS3; line 3, MSPDH5; line4, MSPDH5 + 1 mM BS3.
(B and C) ESI mass spectra of crosslinked M2-nanodiscs with N terminal cross-linking analysis (B) and disulfide bond analysis (C).
(D) The schematic graph for a tetrameric M2 model based on the NMR structure (PDB: 2RLF).
Ionization mass spectrometry (ESI-MS) analysis for the amino acid sequence of the tetramer band was consistent with the western blotting result (Figures 2B and 2C) for the M2 protein. Secondary mass spectrometry analysis showed the presence of the disulfide bonds between the chains (Figure 2C: 13-NEWGCR-18) and N-terminal chemical crosslinking (Figure 2B: MSLLTEVETPIRNEWGCR(1)-MSLLTEVETPIR(1)). These data indicated that the tetramer was a homotetramer with parallel orientations. To better describe cross-linking data, we obtained a schematic model of a tetrameric M2 in nanodiscs based on the NMR structure (PDB: 2RLF)46 as shown in Figure 2D.
Library construction and selection of vNARs
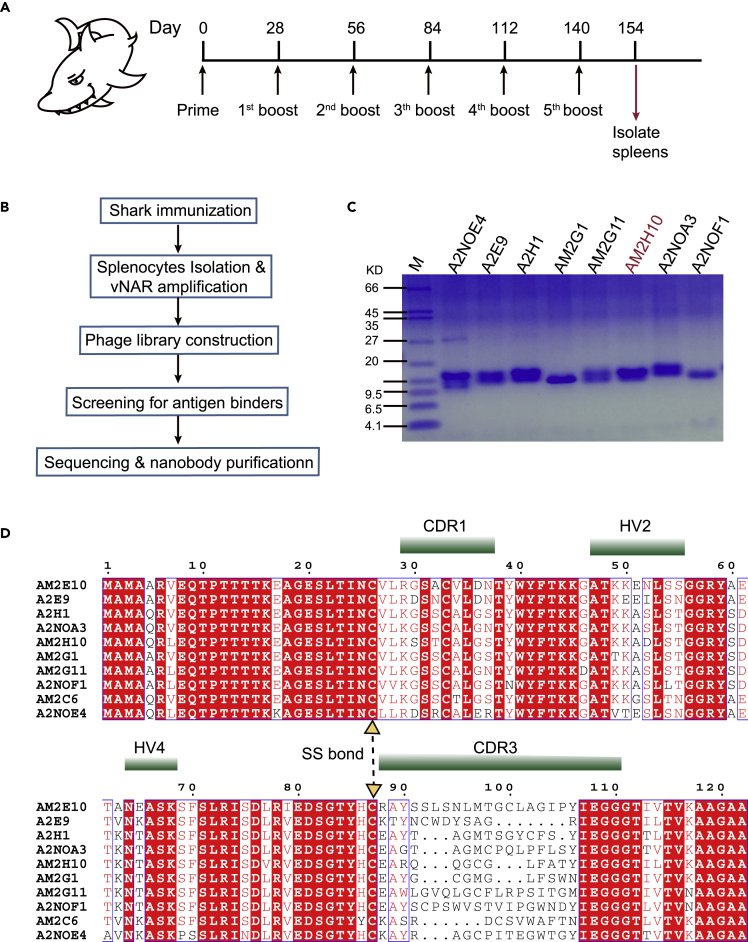
C. plagiosum was subcutaneously vaccinated in the lateral fin using 100 μg of M2-nanodiscs at week 0, followed by booster immunization at weeks 4, 8, 12, 16, and 20. Figure 3A shows the vaccination schedule. To isolate the single domain antibody fragments targeting natural M2 of influenza virus from shark heavy-chain antibodies (vNARs), shark spleens were excised after 2 weeks of vaccination for constructing the phage surface-displayed library and nanobody screening as shown in the workflow in Figure 3B. The vNAR regions were amplified by PCR using primer sequences based on the natural framework repertoire of the bamboo shark.47 The resulting PCR fragments were 350 bp long with sfiI sites at both ends (Figure S1A). The fragments were ligated into a pcomb3XSS48 vector between the two sfiI sites and electroporated into ER2738, resulting in a library of 4×108 (Figure S1B). Four rounds of selection yielded 13 different nanobodies and 3 defective peptides; kinship analysis is shown in Figure S1C. These vNAR genes were recloned into the pcoldII vector and expressed in Shuffle T7. Finally, 10 vNARs were successfully expressed. Figure 3D shows the amino acid sequences and the CDR regions of these 10 nanobodies. Protein quantities expressed per liter are listed in Table S1. Two poorly expressing nanobodies (AM2C6 and AM2E10) were excluded from further investigation, and Figure 3C shows the remaining 8 highly expressing vNARs.
Figure 3.
Library construction and selection of vNARs
(A) Scheme of immunization and sampling.
(B) Workflow for the generation of vNARs against nanodisc-reconstituted M2.
(C) SDS-PAGE for purified vNARs after selection by phage display.
(D) The amino acid sequences and CDR regions of screened vNARs. CDR: complementarity-determining region; HV: hypervariable region; SS bond: disulfide bond.
Binding affinity analysis for vNARs
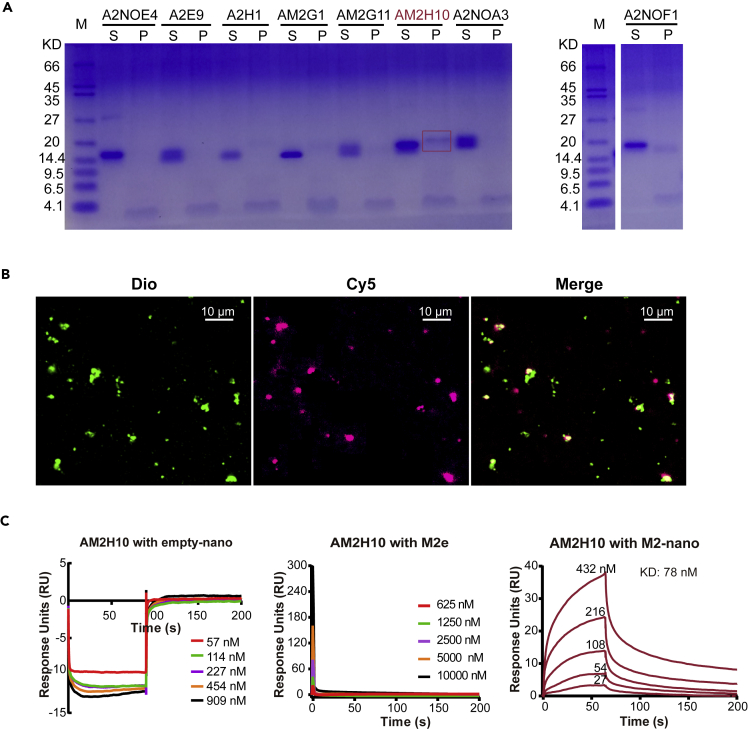
To preliminarily screen the functional nanobody targeting the active ectodomain of M2 (M2e), we performed a pull-down assay. We employed proteoliposome-containing M2 instead of M2-nanodiscs to pull down nanobodies of interest to exclude the influence of MSPDH5 proteins. Synthesized M2(1-60) was incorporated into a lipid vesicle to form a proteoliposome, similar to that in the influenza virus. Larger proteoliposomes settled to the bottom pellet when subjected to ultracentrifugation following incubation with nanobodies. We collected the supernatant and pellets, followed by treatment with ice-cold acetone before SDS-PAGE analysis. Figure 4A shows the AM2H10 pellet sample with a clear band for the nanobody, indicating AM2H10 binding with active M2(1-60).
Figure 4.
Binding affinity analysis for vNARs
(A) Pull-down assay of M2-nanodiscs with vNARs.S: the supernatant of the pull-down; P: the pellet of the pull-down (Irrelevant lanes was eliminated in the right picture).
(B) Fluorescence colocalization for liposomes with M2 and AM2H10, with liposomes stained by Dio (green) and Cy5 tagging AM2H10 (red). Scale bar:10 μm.
(C) The binding abilities of AM2H10 to empty nanodiscs (left), M2e (middle), and M2-nanodiscs (right) were assessed by SPR, indicating that AM2H10 could bind to M2-nanodiscs but rarely to empty nanodiscs and M2e.
To further verify the binding of AM2H10 to M2, fluorescence colocalization experiments were performed (Figure 4B). Reconstituted proteoliposomes including M2 were stained by Dio (3,3′-Dioctadecyloxacarbocyanine perchlorate) and Cy5-SE (Sulfo-Cyanine5 Succinimidyl Ester) marking AM2H10. Similar to the procedure of the pull-down assay, the last resuspended compounds were observed by laser confocal microscopy. In the fluorescence photographs, free proteoliposomes and AM2H10 appeared green and red, respectively. Some yellow signals in the merged images (Figure 4B, right) indicated M2 in proteoliposomes bound to AM2H10.
Subsequently, the binding affinity of AM2H10 for natural M2 protein was measured by SPR on the Biacore T200 instrument. The nanobody was immobilized on a CM5 chip in sodium acetate buffer at pH 4.0. The M2-nanodisc was serially diluted as an analyte. Empty nanodiscs were allowed to flow on another channel of the chip as a control reference. The observed resonance unit (RU) from the sample channel minus the RU from the reference channel was used for calculating the binding affinity. SPR data suggested that the binding of AM2H10 to M2-nanodisc had a KD value of 78 nM (Figure 4C, right), well within the range of values for antibodies in the literature.49 We also measured the binding ability of AM2H10 to empty nanodiscs (Figure 4C, left) and M2e (Figure 4C, middle). AM2H10 was neither bound to empty nanodiscs nor the 24-amino acid M2e (the weak dynamic change in empty nanodiscs may have been caused by the slight nonspecific adsorption of AM2H10 onto the CM5 chip). Thus, the synthesized M2e was different from the conformationally active M2e on the surface of the virus and our vNARs preferentially recognized the physiological M2 tetramer.
In summary, AM2H10 could bind to physiological M2 instead of recombinant M2e alone, with a KD of 78 nM.
Inhibition of the M2 proton channel by AM2H10
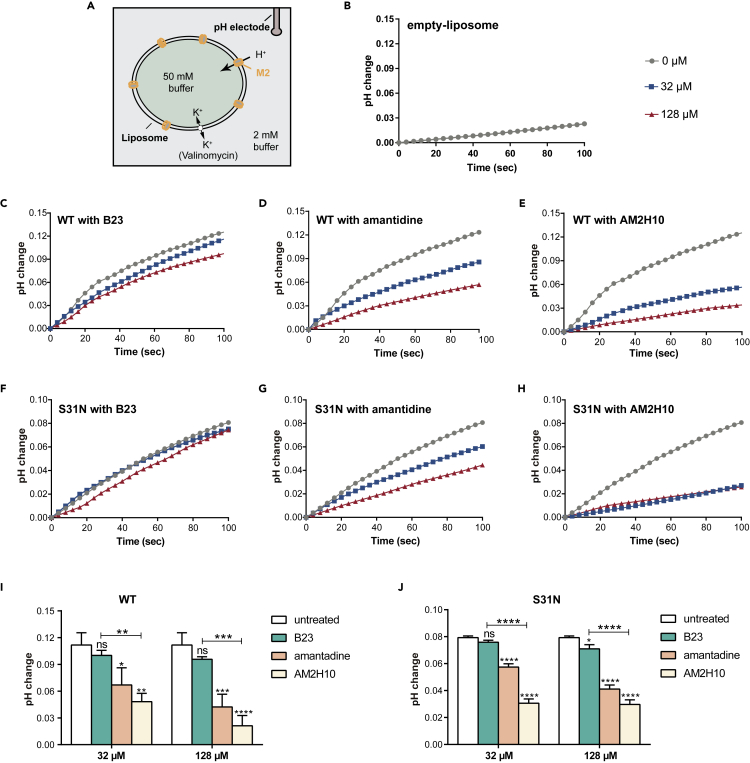
To evaluate the inhibition of AM2H10 on M2 channels, we conducted proton flux (Figure 5) and patch clamp (Figure 6) assays. Figure 5A shows the experimental setup for the proton flux assay, simulating the host cell acidifying the endosomal cavity after the endocytosis of the influenza virus.16 M2 channels of WT (amantadine-sensitive) and S31N mutant (amantadine-resistant) were reconstituted into liposomes, respectively, with strong buffer (50 mM phosphate/50 mM citrate/122 mM KCl/122 mM NaCl/0.01% NaN3, pH 7.7) outside and weak buffer (2 mM phosphate/2 mM citrate/122 mM KCl/122 mM NaCl/0.01% NaN3, pH 7.8) inside. Proton flow was initiated once concentrated hydrochloric acid was added to the exterior, and the external solution increased the pH as H+ migrated down the pH gradient into the vesicle’s interior. As a control, we confirmed very little ion translocation in the absence of proton channels (Figures 5 and 6B). The nanobody had a specific inhibitory effect on WT channels like amantadine compared to the control vNAR B23, which was separated from a shark and screened for anti-hyaluronan synthase (Figure 5C–5E and 5I). The influenza A virus M2 is a crucial target of the adamantane family of anti-influenza drugs, however, serious drug resistance problems have arisen because of drug abuse, and the S31N mutation accounts for the vast majority of all resistant viruses.50 Thus, we tested AM2H10’s effect on M2 S31N mutant channels. AM2H10 displayed significant inhibitory activity against M2 S31N mutant channels, similar to WT channels (Figures 5F–5H and 5J). From the bar graphs for analysis of significant differences, the proton flux of WT and S31N were found to be significantly inhibited by both amantadine and AM2H10 (Figures 5I and 5J). For the control vNAR, B23, sight inhibition to S31N at 128 μM compared to untreated groups was observed. There were considerable differences between AM2H10- and B23-treated groups in both WT and S31N. AM2H10 had a remarkable inhibitory effect on the proton channel function of M2 owing to its specificity for M2e.
Figure 5.
Proton flux experiments for inhibition of the proton conduction of the M2 channel
(A) Experimental setup for the proton flux assay.
(B) The proton conduction traces for empty liposomes with little proton flux.
(C–E) The proton flux of WT M2 channels was inhibited by 2 different concentrations of B23 (C), amantadine (D), and AM2H10 (E).
(F–H) The proton flux of S31N M2 channels was inhibited by 2 different concentrations of B23 (F), amantadine (G), and AM2H10 (H).
(I and J) The bar graphs show significant difference analysis for pH changes in WT (I) and S31N (J) groups subjected to different treatments, n = 3. ∗p< 0.05, ∗∗p< 0.01, ∗∗∗p< 0.001 or ∗∗∗∗p< 0.0001 compared to the untreated group or B23. The error bars indicated the standard deviation.
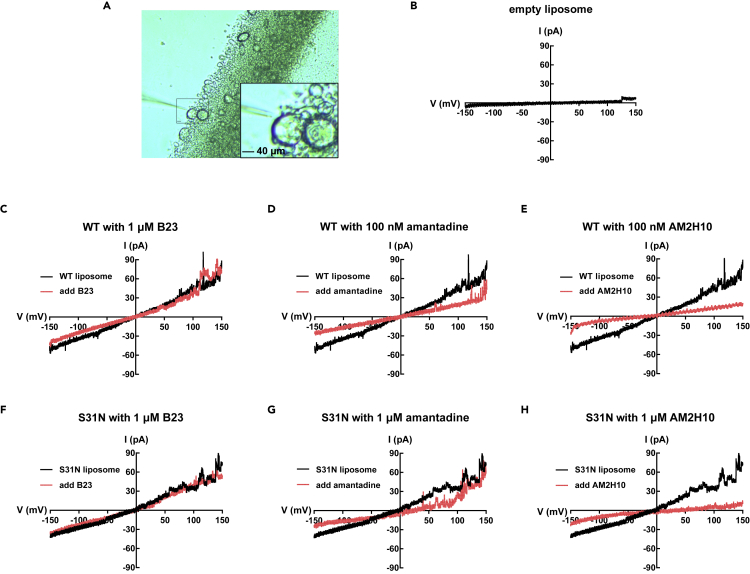
Figure 6.
Patch clamp experiments for inhibition of the ion conduction of M2 channels
(A) Representative image of a giant liposome (120 μm in diameter) under pipette tip.
(B) The current traces for empty liposomes with little electricity flow.
(C–E) The channel currents for WT in the absence or presence of B23 (C), amantadine (D), and ABM2H10 (E).
(F–H) The channel currents for S31N in the absence or presence of B23 (F), amantadine (G), and ABM2H10 (H).
To further verify the inhibitory activity of AM2H10, patch-clamp electrophysiology techniques were used to record single-channel currents of the M2 WT and S31N mutant constructs. Previously, the ion channel activity of M2 protein has been demonstrated by whole cell patch clamp in CV-1 cells and two-electrode voltage clamp in oocytes.15,51 However, ion channels expressed in eukaryotic cells require complex steps and take a long time. In our experiment, we reconstituted the M2 peptide in giant liposomes which have been successfully employed in the research of multiple ion channels.52 Herein, we mainly used giant liposomes with diameters measuring 120 μm and showing spherical, multilamellar appearance to record single-channel currents (Figure 6A). Similar to the results of the proton flux assay, the WT channel activity was inhibited by amantadine and AM2H10 nanobody at 100 nM compared to untreated and vNAR B23 groups (Figures 6C–6E). AM2H10 showed a moderate inhibitory activity for M2 S31N channels insensitive to amantadine (Figures 6F–6H).
In summary, these results revealed that AM2H10 had a specific inhibitory effect on WT M2 channels, comparable to amantadine owing to its specificity for M2e. Of interest, for amantadine-resistant M2 S31N mutant channels, this vNAR showed a modest blocking effect.
Discussion
This study aimed to generate an efficient vNAR targeting the native conformational ectodomain of influenza A M2 protein. The M2 protein forms a homotetramer consisting of two disulfide-linked dimers held together by non covalent interactions.14,45
The development of vaccines and antibodies based on M2 has focused only on the external domain of M2, which lacks tetrameric conformation. The resulting antibodies and protection conferred might not be optimal owing to weak immunogenicity or deviation from native conformation.10,53 In the native configuration, interactions in the transmembrane regions contribute to the tetrameric assembly.49 Hence, using tetrameric M2 in the native conformation as an antigen may be beneficial to produce ideal nanobodies. M2e-nanoparticle vaccines (with tetrameric conformation) have been described previously,54 and this study provides novel insight as we complexed M2e with membranous nanodiscs to create an immunogen presenting M2e in a membrane-embedded context.
M2-nanodiscs were designed in our study not only to increase the immunogenicity of the antigen but also to mimic the natural conformation of the tetrameric ectodomain of the M2 protein.27,28 Unlike other membrane model systems like liposomes, bicelles, and micelles, nanodiscs have the advantages of accessibility to binding epitopes on both faces of the membrane, homogeneous particle size, and stabilization of membrane proteins in their native conformation in a detergent-free environment.55 The membrane proteins reconstituted into nanodiscs support selection and individual characterization of oligomeric states.56,57,58 We successfully assembled M2 channels employing the nanodisc technique, and the cross-linking combined with western blotting and mass spectrometry assays including functional experiments showed that M2 successfully formed an active ion channel homotetramer in nanodiscs with two disulfide-linked dimers, similar to the native M2 protein, and as an antigen, could be used for immunization and antibody screening.
Because of natural characteristics like large size, complex structure, and sensitivity to extreme ambient temperature, traditional mAbs suffer from some limitations in certain applications.59,60 vNARs are emerging as new versatile reagents for the diagnosis and treatment of various diseases, including malaria, Ebola hemorrhagic fever, and toxins.33,37,61,62 Unlike traditional antibodies with flat or concave antigen binding epitopes, vNARs possess a wide variety of loop lengths and structures, making accessibility to more cryptic epitopes like catalytic clefts of enzymes, possible.43,63 Compared to camelids single-domain antibodies (VHHs), vNARs are thus far the smallest naturally occurring antibodies in the animal kingdom owing to the lack of CDR2, with a molecular mass of nearly 12 kDa.63 The inherent solubility and stability of vNARs enable biological activity maintenance in shark blood containing 350 mmol/L urea and 1000 mOsm/kg salt ions.64 Because of reasons including the difficulty of preparation, studies on vNAR are rare worldwide, especially in China; only a few teams have conducted relevant work. Unhesitatingly, we constructed vNAR phage display libraries to screen the vNARs targeting the tetrameric M2 ion channel.
AM2H10, one of the eight vNAR candidates from our phage display screening, showed affinity to the M2-nanodiscs, closely resembling the natural conformation of the tetrameric M2 (Figure 4). AM2H10 not binding to the synthetic M2e peptide confirmed the importance of using an antigen in its native conformation. AM2H10 inhibited ion conductance of both WT A/Hong Kong/8/68 (H3N2) and the S31N mutant (Figures 5 and 6). This is the first vNAR targeting the M2 channel, and in addition, a vNAR capable of binding both WT and amantadine-resistant influenza A viruses M2 channels in vitro. Based on these observations, we proposed a hypothesis that the suppression of the proton influx might be caused by a conformational change of the M2 ion channel transmembrane helices induced by the binding of AM2H10 rather than by AM2H10 directly blocking the tetrameric ectodomain of M2. The binding activity of the vNAR supported this hypothesis and the theory of conformational change of the M2 ion channel has been proposed by James’ group to explain the mechanism underlying amantadine inhibition and resistance.65 We further speculated that the binding epitope of AM2H10 might be conformation-specific as the vNARs generated using M2 reconstituted nanodiscs preferentially recognized native M2 conformation rather than ectodomain alone in the SPR studies.
Although AM2H10 has a moderate binding strength (KD = 78 nM) for M2 protein, we can perform affinity maturation assays based on the CDRs as described previously.66 Furthermore, multivalent AM2H10 also can be engineered via a peptide linker to generate vNAR variants with higher affinity, better avidity, and longer half-life. Guo et al. generated a VHH targeting M2 from a synthetic library of 1012 with an affinity similar to that reported herein.49 Compared to VHHs, although vNARs against multiple targets have been panned, most of them have only focused on specificity, affinity, and stability. Their application in diagnostics or therapeutics is rare. Moreover, fewer than 10 shark species are currently being studied for the development of vNARs, with nurse sharks being the most common. We hope our findings can contribute to paving the path for vNAR application as a clinical drug candidate.
In summary, this study demonstrated the feasibility of the nanodisc system as an immunological reagent and screening of functional nanobodies targeting membrane proteins from the immune library of C. plagiosum. This approach has therapeutic potential and complements other preclinical work aimed at developing M2-specific nanobody vaccines.67 M2-nanodisc, a stable membrane protein system, with the M2 physiological tetramerization, lays the foundation for panning active vNARs targeting the extra-viral part of native M2 channels. AM2H10 showed preferential binding to native M2 tetramer, and significant inhibitory activities for both WT and amantadine-resistant M2 channels, likely by interfering with the ion channel function of M2. Engineering for potential broad-spectrum protection against influenza A viruses is a viable research direction. Although AM2H10 effectively inhibits the activities of M2 WT and S31N channels in liposomes, the structural details of its interaction and anti-viral activity in cells or in vivo have not been studied. Further studies should focus on infection challenge experiments to study the potential anti-viral capacity of engineered AM2H10.
Limitations of the study
Although AM2H10 effectively inhibited the activities of M2 WT and S31N channels in liposomes, the structural details of its interactions were not studied. The absence of structural data limited further engineering for developing a better potential broad-spectrum nanobody against influenza A viruses. The anti-viral activity of AM2H10 was not verified in vitro and in vivo. Further studies should focus on infection challenge experiments to study the potential anti-viral capacity of engineered AM2H10.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Influenza A m2 Antibody (14C2) | Santa Cruz Biotechnology | Cat# sc-32238; RRID: AB_627808 |
| His-Tag (D3I1O) XP® Rabbit mAb | Cell Signaling Technology (CST) | Cat# 12698S |
| Anti-mouse IgG, HRP-linked antibody | CST | Cat# 7076S |
| Anti-rabbit IgG, HRP-linked antibody | CST | Cat# 7074S |
| Anti-M13 (HRP) | HuaBio | Cat# EM1902-18 |
| Bacterial and virus strains | ||
| SHuffle®T7CompetentE. coli | New England Biolabs (NEB) | Cat# C3026J |
| M13KO7 Helper Phage | New England Biolabs (NEB) | Cat# N0315S |
| ER2738 | CRGEN | Cat# PB0421 |
| Chemicals, peptides, and recombinant proteins | ||
| 1-palmitoyl-2-oleoyl-glycero-3-phosphocholine (16:0-18:1 PC (POPC)) | Avanti Polar Lipids | Cat# 850457P |
| M2e | GL Biochem (Shanghai) Ltd | N/A |
| M2(1-46) | GL Biochem (Shanghai) Ltd | N/A |
| M2(1-60) | GL Biochem (Shanghai) Ltd | N/A |
| Molecular weight ladder (M) | BBI life sciences corporation | Cat# C600201 |
| Molecular weight ladder (M1) | Biosharp | Cat# BL712A |
| Freunds Adjuvant, complete | BIOFROXX | Cat# 2203ML010 |
| Freund’s Adjuvant incomplete | BIOFROXX | Cat# 1643ML010 |
| sfiI | NEB | Cat# R0123L |
| T4 DNA ligase | NEB | Cat# M0202T |
| pComb3XSS | Andris-Widhopf et al.,48 | Cat# 63890 (Addgene) |
| pcold II | Sangon | Cat# B540188-0001 |
| 3,3′-Dioctadecyloxacarbocyanine perchlorate (Dio) | MCE | Cat# 34215-57-1 |
| Sulfo-Cyanine5 Succinimidyl Ester(Cy5-SE) | MCE | Cat# 146368-14-1 |
| Deposited data | ||
| NMR structure | Schnell et al.,46 | PDB: 2RLF (https://www.rcsb.org) |
| Oligonucleotides | ||
| 5′-CGTGGCCCAGGCGGCCATGGCC SMACGGSTTGAACAAACACC-3′ |
This study | N/A |
| 5′-GCTGGCCGGCCTGGCCWTTCA CAGTCASARKGGTSCC-3′ |
This study | N/A |
| Software and algorithms | ||
| EMAN2 | Tang et al.,68 | https://cryoem.bcm.edu/cryoem/downloads/view_eman2_versions |
| relion3.1.3 | Scheres et al.,69 | https://relion.readthedocs.io/en/release-3.1/index.html |
| ImageJ | National Institute of Health | RRID:SCR_003070; https://imagej.net/ |
| Graphpad Prism 7.0 | GraphPad Software | https://www.graphpad.com/ |
Resource availability
Lead contact
Information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Hongxin Zhao (zhx@hmfl.ac.cn).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Our study does not use experimental models typical in the life sciences.
Method details
Peptides
All the M2 peptides involved in the experiments were from influenza A/Hong Kong/8/68 (H3N2) and synthesized in GL Biochem (Shanghai) Ltd at a high purity of more than 95%. Table 1 shows the sequences of M2 peptides with different lengths.
Table 1.
The amino acid sequences of M2 in different lengtha
| M2 | Domain | Amino acid sequence |
|---|---|---|
| M2e | Ectodomain | MSLLTEVETPIRNEWGCRCNDSSD |
| M2(1-46) | Ectodomain and transmembrane domain | MSLLTEVETPIRNEWGCRCNDSSD PLVVAASIIGILHLILWILDRL |
| M2(1-60) | Ectodomain, transmembrane domain and intracellular helix | MSLLTEVETPIRNEWGCRCNDSSD PLVVAASIIGILHLILWILDRLFFKC IYRFFEHGLK |
The numbers in parentheses indicate the residue order from N-terminal of M2.
Preparation of empty nanodiscs
Empty nanodiscs were assembled following the published protocols by Sligar lab (https://publish.illinois.edu/sligar-lab/nanodisc-technology) and Dieter Willbold.44 Briefly, POPC (AvantiPolarLipids, Alabaster, AL, USA) was solubilized in chloroform and a film was generated under a nitrogen stream followed by freeze-drying. The lipid/detergent stock was prepared with sodium cholate buffer (20 mM Tris, 100 mM NaCl, pH 7.4, 160 mM sodium cholate) by dissolving lyophilized POPC (final concentration: 80 mM). The hybrid stock was incubated on an orbital shaker at 37 °C to make the solution clear. Nanodiscs were assembled using appropriate volumes of POPC/cholate and purified MSPDH5. E. coliexpressing MSPDH5 and recombinant plasmid were obtained from the work of former labmates of the laboratory, and the procedure of expression and purification was previously described.70 The assembly molar ratio of POPC:sodium cholate:MSPDH5 was 50:100:1 with 0.2 mM MSPDH5. The compound was freeze-thawed twice in liquid nitrogen and 37 °C. Once detergent was removed using BioBeads SM-2 (BioRad, München, Germany), the lipid/cholate and MSPDH5 started self-assembling into nanodiscs. BioBeads were pretreated according to the procedure described previously.44 The suspension was gently shaken at room temperature and BioBeads were added at hours 0, 4, and 8. The nanodiscs were purified by SEC using Superdex 200 column (GE Healthcare, Freiburg, Germany) to remove any lipid aggregates and redundant MSPDH5 molecules, and PBS was the running buffer.
Assembly of M2-nanodiscs
Synthesized M2(1-46) was incorporated into nanodiscs similar to empty nanodiscs. However, the molar ratio of POPC:sodium cholate:MSPDH5 was adjusted. Specifically, M2(1-46) was resuspended in Tris buffer (20 mM Tris, 100 mM NaCl, pH 7.4) and added to the mixture of lipid/cholate and MSPDH5. The molar ratio of POPC:sodium cholate:MSPDH5:M2 was 20:40:1:2 with 0.4 mM M2(1-46). The procedure for sodium cholate elimination from M2-nanodiscs and their purification was the same as that for empty nanodiscs. All peak fractions of SEC were collected for SDS-PAGE analysis to confirm M2 assembly into nanodiscs. M2-rich fractions in the nanodisc peak were pooled and assembled M2 and MSPDH5 were quantified by ELISA.26,27 Briefly, 96-well ELISA plates were coated with various amounts of M2 and MSPDH5 in 0.1 M carbonate buffer (PH9.6) by overnight incubation at 4°C. PBS containing Tween 20 (0.05% v/v) and BSA (3% w/v) was used to block the plates (25°C for 2 h). The plates were further incubated (37°C for 1 h) with anti-M2 antibodies (14C2, 1:50) (mouse IgG; Santa Cruz) or anti-His antibodies (D3I1O, 1:1,000) (rabbit IgG; Cell Signaling Technology). The second antibodies employed anti-mouse (for M2) or anti-rabbit (for MSPDH5) IgG (1:5,000; Cell Signaling Technology). Concentration-dependent standard curves were constructed for M2 and MSPDH5 from the respective OD450 values measured with a microplate reader (Tecan spark) at 450 nm. The M2 and MSPDH5 concentrations of unknown M2-nanodiscs mixtures were then calculated from the corresponding standard curves.
DLS and TEM
DLS (dynamic light scattering) and TEM (transmission electron microscopy) were used to characterize M2-nanodiscs for total protein concentrations of 0.68 mg/mL and 0.0035 mg/mL in PBS respectively. DLS tests were performed on a Malvern Zetasizer Nano ZS at 25°C and the data were analyzed using the Zetasizer Software 7.11 (Malvern Instruments Ltd.). The TEM experiments were conducted on Tecnai G2 F20 using air-dried stained samples. Before imaging, samples loaded on carbon-coated copper grids (Zhongke Microscope) were stained using 2% uranyl acetate, and the excess staining solution was removed with filter paper. For the details of particle diameter distribution and discoidal shape analysis, 2D class averaging was performed by selecting at least 4,000 particles, with EMAN268 picking and relion3.1.3 69 classification.
Identification of tetrameric M2
Chemical cross-linking coupled with mass spectrum was used to verify the tetramer of M2. In the cross-linking assay, bis sulfosuccinimidyl suberate (BS,3 Thermo Fisher Scientific Inc.) was dissolved in PBS buffer at 12.5 mM and added to nanodiscs samples with the final concentration of M2 at 0.4 mg/mL of and BS3 at 1 mM. Samples reacted for 2 h on ice and were subsequently quenched using 20 mM Tris buffer (PH 7.5) for 15 minat room temperature. The reaction products were visualized by SDS-PAGE (Coomassie Blue staining, Tanon 5200 Multi& Tanon 5200 imaging) and western blot with 14C2 (Santa Cruz Biotechnology, Inc.) as M2-specific antibodies. For Electrospray Ionisation mass spectrometry (ESI-MS), the interested gel band of cross-linked M2-nanodiscs was excised and cut into small pieces, followed by TCEP reduction, iodoacetamide alkylation, and trypsin digestion.71,72 The ESI-MS analysis was performed on a Thermo Q-Exactive instrument in a 60-min gradient and the raw data was processed with pLink software.
Immunization of sharks and separation of splenocytes
C. plagiosum was fed according to the protocol reported by Müller.73 Briefly, adult sharks were acclimatized to captivity for a minimum of four weeks prior to immunization. During the period of this study, these animals were maintained at 25°C in filtered natural seawater in small aquariums and kept away from direct light. The seawater kept circulating and was replaced regularly every morning. Marine fish and shrimp were fed to these sharks. Before immunization, animals were submerged in a smaller container with MS-222 dissolved in seawater for anesthetization. Once the desired level of sedation was attained, the animal was placed on the operating table for immunization. For priming, 100 μg of purified M2-nanodiscs in PBS were mixed with an equal volume of Freund’s Complete Adjuvant (FCA). Emulsified antigen was administered subcutaneously to the lateral fin. Four weeks after the initial injection, the shark was boosted with M2-nanodiscs in Freund’s Incomplete Adjuvant. Subsequently, 4 boosters of M2-nanodiscs were administered subcutaneously without adjuvant every 4 weeks. Two weeks after the final immunization, spleens were collected for isolating splenocytes from immunized C. plagiosum using lymphocyte preparation kits, following the manufacturer’s protocol (P5000, Solarbio). All animal experiments were approved by the Ethics Committee of Institute of Oceanology, Chinese Academy of Sciences (No. MB2010-1).
vNAR library construction
Total RNA was extracted from splenocytes using TRIzol following the manufacturer’s instructions (1596026, Invitrogen). For first-strand cDNA synthesis, RT-PCR was performed using the HiScript III 1st strand cDNA Synthesis Kit (Vazyme). After first-strand cDNA synthesis, products were incubated at 95 °C for 7 min to inactivate the reverse transcriptase and denature the template for constructing immune vNAR repertoire. The primer sequences for the vNAR library were designed based on the literature with minor changes.47,74 PCR was performed to amplify vNAR repertoire using forward primer: CGTGGCCCAGGCGGCCATGGCCSMACGGSTTGAACAAACACC (sfiI site) and reverse primer: GCTGGCCGGCCTGGCCWTTCACAGTCASARKGGTSCC (sfiI site). The genes were inserted into the pComb3XSS48 vector between the sfiI sites. The recombinant plasmid was purified using a PCR Cleanup Kit (Axygen) and transformed into competent ER2738 cells by electroporation using Bio-Rad MicroPulserTM (Ec2). After electroporation, the cells were resuspended in a pre-heated 2YT medium and incubated for 1h at 37 °C. Sample aliquots from each transformation were used to prepare 100-fold serial dilutions in 2YT medium for estimating the final size of the library by plating. The remaining bacterial suspensions were plated onto 250 mm square Petri dishes containing 2YT medium, supplemented with ampicillin (100 μg/mL) and tetracycline (25 μg/mL). All plates were incubated overnight at 37 °C. The percentage insert was estimated by colony PCR and the library diversity was assessed by colony sequencing. All bacteria were scraped from the plates and stored in 10 mL of 2YT medium with 20% glycerol at −80°C.
Phage display and selection of vNARs
The preparation of recombinant phage particles has been described previously.75 Briefly, the library stock was grown to log phase (OD600 0.4–0.8) at 37°C by shaking at 220 rpm, and M13KO7 helper phage was added and incubated for 30 minat 37 °C in a 2YT/A/K medium (2YT containing 100 μg/mL ampicillin and 50 μg/mL kanamycin) for viral infection. After incubation, the culture was centrifuged at 4,000 × g at 4 °C for 10 min. The cell pellet was resuspended in 100 mL of 2YT/A/K medium, and the culture was incubated overnight at 30 °C by shaking for library expression. PEG/NaCl (PEG: NaCl = 4:1) precipitation was used to purify and concentrate phages and the phage pellet was dissolved and resuspended in PBS. The purified phage stock can be stored at 4°C for short term, or at −20°C for the longer term with the addition of sterile glycerol to 20% (v/v).
Purified M2-nanodiscs were coated on an enzyme-linked immune sorbent assay (ELISA) plate (Sangon) for panning vNARs against M2 from the phage display library.76 Coated 96-well ELISA plates with varying amounts of M2-nanodiscs (from 10 to 1.25 μg) by overnight incubation at 4°C. Washed the plates twice with PBS before blocking with PBS containing Tween 20 (0.05% v/v) and BSA (3% w/v) for 2 hat 25°C. The phage stock then incubated with the blocked plates containing 10 μg of M2-nanodiscs in the first round of screening. Non-specifically absorbed phages were eliminated by PBST (0.1% Tween 20) washing during selection. The bound phages were collected using 100 mM Glycine-HCl (pH 2.2), immediately neutralized in 1 M Tris-HCl (pH 7.4), and subsequently amplified by infecting with ER2738. Then repeated the incubated-neutralized steps but the contents of M2-nanodiscs reduced to 5, 2.5, 1.25 μg in the following rounds. After four rounds, randomly picked 88 phage-vNAR clones were subjected to subtractive binding to empty nanodiscs by phage-ELISA. Specifically, two ELISA plates were coated with 2 μg of M2-nanodiscs and empty nanodiscs, respectively, overnight at 4 °C with PBS or BSA coating as a negative control. Plates were blocked with 4% nonfat milk in PBS and incubated with randomly picked 88 phage-vNAR clones in 2% BSA for 1 hat 37 °C. HRP-conjugated anti-M13 monoclonal antibody (HuaBio) was used to identify phage-vNAR clones, which could bind against the antigen to the wells containing M2-nanodiscs, with no affinity for wells coated with empty nanodiscs. The positive clones were sent to Sangon for sequencing analysis.
Expression and purification of vNARs
The vNAR genes from the chosen clones were inserted into the pcoldII plasmid and chemically transformed into Shuffle T7 competent E. coli (NEB) using CaCl2. The strains containing recombinant vNARs were grown for protein expression in 2 YT with 100 mM of ampicillin until the OD600 reached nearly 0.8. Protein production was induced with 0.5 mM IPTG by shaking for 20 hat 16 °C. The collected bacteria were sonicated via ultrasound and proteins were initially purified on the Ni-NTA Sepharose column (GE Healthcare) followed by SEC. For proton influx and patch clamp functional assays, the gene was inserted into the pcold-sumo vector to avoid the effect of His-tag. The sumo tag with 6×His was cleaved by sumo enzyme and tag-cleaved vNARs were purified on a second Ni-NTA Sepharose column. The eluate from Ni-column was run on SEC using Superdex 75 column for further purification. The eluted fractions were evaluated by SDS-PAGE for purity and concentrated using concentrators with a molecular mass cut-off of 3 kDa. The concentration was quantified using a Pierce TM BCA Protein Assay Kit (Thermo Fisher Scientific Inc.).
Pull-down assay
M2(1-60) peptides were reconstituted into liposomes for screening nanobodies with binding activities. Specifically, 2 mg of POPC (Avanti Polar Lipids) and 40 nmol of M2(1-60) peptides were dissolved in 1 mL of chloroform and methanol mixture with the molar ratio of 2:1. A thin film of the mixture was obtained under a nitrogen stream, resuspended in 1 mL of chloroform, and blown again. For high-quality and uniform proteoliposome preparation, the doubly-blown films were dried overnight using a lyophilizer, re-dissolved in 1 mL of PBS buffer, and extruded several times through 0.2 μM polycarbonate membranes on a mini-extruder.
The purified nanobodies at 40 nmol were ultracentrifuged at 218,000 g (BECKMAN) for 30 minat 4 °C. Supernatant (150 μL) was incubated with 150 μL of proteoliposomes at 4 °C for 30 min as indicated. The mixtures were ultracentrifuged at 218,000 g for 1 h and the proteoliposomes remained in the precipitate due to their heavier weight, whereas free nanobodies (not bound to M2) were in the supernatant due to their lighter weight. The precipitates were washed twice gently with 300 μL of PBS and resuspended in 200 μL of PBS. Subsequently, 800 μL of pre-chilled acetone was added to precipitate proteins at −20°C for further analysis by SDS-PAGE.
Fluorescence colocalization assay
The process of assembling liposomes was the same as for the proteoliposomes in the pull-down assay but without the extruding step. Reconstituted proteoliposomes including M2 peptides were stained for 1 hat 37°C with 10 μM of DiO (3,3′-Dioctadecyloxacarbocyanine perchlorate, MCE). 90 μM of AM2H10 was tagged with 900 μM of Cy5-SE (Sulfo-Cyanine5 Succinimidyl Ester, MCE) for 2 h by shaking at room temperature, and the reaction was terminated using 90 mM of Tris before excess Cy5-SE was removed on an ion column. The next step was similar to the procedure for the pull-down assay. Briefly, stained proteoliposomes were incubated with ultracentrifuged (retain supernatant) AM2H10-Cy5 for 30 minat 4°C. The compounds were ultracentrifuged and sediments were resuspended in PBS after washing thrice. The resuspended compounds were observed and images were captured using an oil lens in the Olympus microscope (SpinSR10) at 60x.
Surface plasmon resonance (SPR) analysis
The binding kinetics and affinity of AM2H10 to M2-nanodiscs, empty nanodiscs, and synthesized M2e (ectodomain of M2) were measured by SPR on the Biacore T200 instrument (GE Healthcare). Subsequently, 10 μg/mL of AM2H10 was captured onto a CM5 sensor chip in 10 mM sodium acetate buffer at pH 4.0 for 200 sat 10 μL/min. At the end of each cycle, the running buffer flowed for 300 s, followed by 50 mM NaOH for regenerating the surface of the CM5 chip. Biacore data were analyzed using the Biacore T200 Evaluation Software (GE Healthcare Bio-Sciences AB) according to the method described previously.77 The 1:1 Langmuir binding model with mass transfer control was used and data were fed into a global fit to extract the binding characteristics.
Proton flux assay
To verify the inhibition of AM2H10 against M2, we designed a liposomal proton flux assay based on the role of M2 as H+ channels described by James J. Chou’s group.65 This test utilized a proton gradient to drive proton flux through the M2 channel, mimicking the events when the influenza virus invades a cell. Figure 5A illustrates the liposomal proton flux assay system. Briefly, M2(1-60) was reconstituted into the membrane of the liposome with identical pH and ion concentrations inside and outside. However, inside, the solution was strongly buffered, while outside, the solution was weakly buffered. Hydrochloric acid was added by continuous rapid stirring, and proton flux through M2 from the exterior into the interior was initiated. The process was monitored using a pH microelectrode (METTLER TOLEDO) in the external bath.
The wild type (WT) of M2(1-60) from influenza A/Hong Kong/8/68 (H3N2) and S31N mutant constructs were synthesized by GL Biochem (Shanghai) Ltd. M2 reconstitution into liposomes was similar to liposome assembly in the pull-down assay, with minor changes. Specifically, using E.coli polar lipid extract (Avanti Polar Lipids) instead of POPC, the external vesicle buffer differed from the interior, and potassium ionophore valinomycin was introduced in this system for perfectly monitoring the metergasis of M2. Specifically, 20 nmol of M2 peptides (WT or S31N) were mixed with 10 mg of lipid, and 0.2 nmol of valinomycin in a 1.1 mL of chloroform and methanol mixture. The solution was subjected to a cycle of drying-resuspending-drying, and finally dissolved in strongly-buffered internal liposome buffer (50 mM phosphate/50 mM citrate/122 mM KCl/122 mM NaCl/0.01% NaN3, pH 7.7) to form proteoliposomes. After extruding through 0.2 μM polycarbonate membranes, the solution was exchanged for external vesicle buffer (2 mM phosphate/2 mM citrate/122 mM KCl/122 mM NaCl/0.01% NaN3, pH 7.8) using a desalting column (GE Health Sciences). In a 1.5 mL elution, 3 μL of 1 M HCl was added by continuous rapid mixing to drive the conductance of protons from the bath into the liposomes. A negative control sample was analyzed in parallel, with the M2 peptide excluded from the system. vNAR B23 (MAERVEQTPTTTTKEAGESLTVNCVLKGSSCALGNTYWYFTKKGATKKASLSTGGRYSDTKNTASKSFSLRISDLRVEDSGTYHCEAYTCAGGTPWRHYIEGGGTILTVK), separated from shark and screened for anti-hyaluronan synthase, was used as an isotype control. Amantadine and nanobodies were added and incubated for 30 min with the system before initiation of conduction to assess channel inhibition.
Patch clamp assay
The WT and S31N mutant constructs of M2 were reconstituted in a giant liposome in the absence or presence of amantadine and nanobodies for tracking single-channel currents of M2 by patch-clamp electrophysiology techniques. Giant liposomes were prepared based on previously published protocols.52,78,79 Specifically, 60 mg of POPC was dissolved in 4 mL of distilled water and the lipid solution was incubated for 20 min using a vortex. The resulting suspension was sonicated for 10 min under nitrogen, followed by supplementing with 2 mL of 2× dialysis buffer (1× buffer: 140 mM NaCl, 5.3 mM KCl, 0.5 mM MgSO4, 15 mM HEPES, pH 7.2-7.4). Subsequently, 40 ng (0.005 nmol) of WT and S31N peptides in 100 μL of 0.2% (v/v) Triton X-100 were incubated separately in an 800 μL lipid suspension. The detergent was removed using Biobeads or dialysis. The mixture was ultracentrifuged at 160,000 g for 1 h and the precipitate was resuspended in 80 μL of 15 mM HEPES buffer (pH 7.2-7.4) with 5% (v/v) ethylene glycol. The resuspension was dropped onto a glass coverslip in 15 μL aliquots and the sample was dried in a box at 4 °C for 3–6 h with a desiccant. Before the experiment, the dried lipid droplet was rehydrated overnight at 4 °C with 15 μL of dialysis buffer. The sample was placed in a small cylindrical cell containing 500 μL bath solution. Patch-clamp single-channel recording for giant liposome membranes was conducted using patch pipettes filled with dialysis buffer as the intracellular solution at 30–40 MΩ resistance. The experiment was executed on an EPC9 patch clamp amplifier (HEKA Elektronik, Lambrecht/Pfalz, Germany) using the voltage-clamp mode, with the bath solution the same as the intracellular solution. The ramp protocol of depolarization from −150 mV to 150 mVat 1000 ms was implemented using the PatchMaster (HEKA) software. The currents were digitized at every 0.15 ms and filtered at 0.5 kHz. To evaluate the inhibitory effect of AM2H10, the channel currents were recorded and compared before and after amantadine or nanobody application.
Quantification and statistical analysis
Statistically significant differences were analyzed in proton flux data among different by one-way analysis of variance (ANOVA) followed by Bonferroni’s multiple comparisons using GraphPad Prism 7.0. p < 0.05 was considered a statistically significant difference and p < 0.01 indicated a highly significant difference compared to the untreated group.
Acknowledgments
We are grateful to mass spectrometry system at the National Center for Protein Science Shanghai (NCPSS) for instrument support and technical assistance. We thank Guiqin Wang at Nanjing Advanced Academy of Life and Health at data management for help with data management. We thank This work was supported by funds from, Anhui Technologies Major Program (grants: 201903a07020026), President of The Chinese Academy of Sciences (Grant No: YZJJZX202004), Users with Excellence Program of Hefei Science Center, CAS (Grant No. 2020HSC-UE018), National Natural Science Foundation of China (Grant No. 41876168) and the Taishan Young Scholar Program of Shandong Province (tsqn201812108).
Author contributions
J.F.W. and H.X.Z. designed and supervised the project. C.D.Y. executed and participated in all experiments, organized and analyzed the data, and wrote the article. W.D., B.S., and M.S.R. performed the patch clamps experiments. L.S.Z., Y.K.D., Y.C.B., and H.W. participated in shark immunization, phage display, and nanobody screening. M.L., Y.J.W., D.M.Q., Y.Z.W., and Z.H.L. performed and analyzed the TEM experiments. L.N.Z., F.J.C., L.L., and L.X. purified the nanobodies. H.T.X., Y.H.Z., and J.J.L performed fluorescence colocalization assays. W.Q.W. and P.C. conducted the ESI-MS assays. Z.L., B.W., and H.D. designed and analyzed the data for ESI-MS experiments. All authors discussed the results and commented on the manuscript.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: January 20, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.105736.
Contributor Information
Lisha Zha, Email: zhalisha@ahau.edu.cn.
Hongxin Zhao, Email: zhx@hmfl.ac.cn.
Junfeng Wang, Email: junfeng@hmfl.ac.cn.
Supplemental information
Data and code availability
All data reported in this paper will be shared by the lead contact upon request. The displayed NMR structure in Figure 2D was from published PDB data in https://www.rcsb.org and the PDB ID has been listed in the key resources table.
This study did not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contactupon request.
References
- 1.Cerulli R.A., Kritzer J.A. Stringing together a universal influenza antibody. Biochemistry. 2019;58:1943–1944. doi: 10.1021/acs.biochem.9b00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xie H., Wan X.F., Ye Z., Plant E.P., Zhao Y., Xu Y., Li X., Finch C., Zhao N., Kawano T., et al. H3N2 mismatch of 2014-15 northern hemisphere influenza vaccines and head-to-head comparison between human and ferret antisera derived antigenic maps. Sci. Rep. 2015;5:15279. doi: 10.1038/srep15279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fiorentino F., De Angelis M., Menna M., Rovere A., Caccuri A.M., D'Acunzo F., Palamara A.T., Nencioni L., Rotili D., Mai A. Anti-influenza A virus activity and structure-activity relationship of a series of nitrobenzoxadiazole derivatives. J. Enzyme Inhib. Med. Chem. 2021;36:2128–2138. doi: 10.1080/14756366.2021.1982932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vonitzstein M., Wu W.Y., Kok G.B., Pegg M.S., Dyason J.C., Jin B., Van Phan T., Smythe M.L., White H.F., Oliver S.W., et al. Rational design of potent sialidase-based inhibitors of influenza-virus replication. Nature. 1993;363:418–423. doi: 10.1038/363418a0. [DOI] [PubMed] [Google Scholar]
- 5.Kim C.U., Lew W., Williams M.A., Liu H., Zhang L., Swaminathan S., Bischofberger N., Chen M.S., Mendel D.B., Tai C.Y., et al. Influenza neuraminidase inhibitors possessing a novel hydrophobic interaction in the enzyme active site: design, synthesis, and structural analysis of carbocyclic sialic acid analogues with potent anti-influenza activity. J. Am. Chem. Soc. 1997;119:681–690. doi: 10.1021/ja963036t. [DOI] [PubMed] [Google Scholar]
- 6.Sedeyn K., Saelens X. New antibody-based prevention and treatment options for influenza. Antiviral Res. 2019;170:104562. doi: 10.1016/j.antiviral.2019.104562. [DOI] [PubMed] [Google Scholar]
- 7.Nachbagauer R., Krammer F. Universal influenza virus vaccines and therapeutic antibodies. Clin. Microbiol. Infect. 2017;23:222–228. doi: 10.1016/j.cmi.2017.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Naesens L., Stevaert A., Vanderlinden E. Antiviral therapies on the horizon for influenza. Curr. Opin. Pharmacol. 2016;30:106–115. doi: 10.1016/j.coph.2016.08.003. [DOI] [PubMed] [Google Scholar]
- 9.Ebrahimi S.M., Tebianian M. Influenza A viruses: why focusing on M2e-based universal vaccines. Virus Gene. 2011;42:1–8. doi: 10.1007/s11262-010-0547-7. [DOI] [PubMed] [Google Scholar]
- 10.Mezhenskaya D., Isakova-Sivak I., Rudenko L. M2e-based universal influenza vaccines: a historical overview and new approaches to development. J. Biomed. Sci. 2019;26:76. doi: 10.1186/s12929-019-0572-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kolpe A., Schepens B., Ye L., Staeheli P., Saelens X. Passively transferred M2e-specific monoclonal antibody reduces influenza A virus transmission in mice. Antiviral Res. 2018;158:244–254. doi: 10.1016/j.antiviral.2018.08.017. [DOI] [PubMed] [Google Scholar]
- 12.Hikono H., Miyazaki A., Mase M., Inoue M., Hasegawa M., Saito T. Induction of a cross-reactive antibody response to influenza virus M2 antigen in pigs by using a Sendai virus vector. Vet. Immunol. Immunopathol. 2012;146:92–96. doi: 10.1016/j.vetimm.2012.01.017. [DOI] [PubMed] [Google Scholar]
- 13.Muto N.A., Yoshida R., Suzuki T., Kobayashi S., Ozaki H., Fujikura D., Manzoor R., Muramatsu M., Takada A., Kimura T., Sawa H. Inhibitory effects of an M2-specific monoclonal antibody on different strains of influenza A virus. Jpn. J. Vet. Res. 2012;60:71–83. [PubMed] [Google Scholar]
- 14.Sugrue R.J., Hay A.J. Structural characteristics of the M2 protein of influenza A viruses: evidence that it forms a tetrameric channel. Virology. 1991;180:617–624. doi: 10.1016/0042-6822(91)90075-m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pinto L.H., Holsinger L.J., Lamb R.A. Influenza-virus M2 protein has ion channel activity. Cell. 1992;69:517–528. doi: 10.1016/0092-8674(92)90452-I. [DOI] [PubMed] [Google Scholar]
- 16.Helenius A. Unpacking the incoming influenza virus. Cell. 1992;69:577–578. doi: 10.1016/0092-8674(92)90219-3. [DOI] [PubMed] [Google Scholar]
- 17.Lamb R.A., Choppin P.W. Identification of a second protein (M2) encoded by RNA segment 7 of influenza virus. Virology. 1981;112:729–737. doi: 10.1016/0042-6822(81)90317-2. [DOI] [PubMed] [Google Scholar]
- 18.Zebedee S.L., Lamb R.A. Influenza A virus M2 protein: monoclonal antibody restriction of virus growth and detection of M2 in virions. J. Virol. 1988;62:2762–2772. doi: 10.1128/JVI.62.8.2762-2772.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu W., Zou P., Chen Y.H. Monoclonal antibodies recognizing EVETPIRN epitope of influenza A virus M2 protein could protect mice from lethal influenza A virus challenge. Immunol. Lett. 2004;93:131–136. doi: 10.1016/j.imlet.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 20.Fu T.M., Freed D.C., Horton M.S., Fan J., Citron M.P., Joyce J.G., Garsky V.M., Casimiro D.R., Zhao Q., Shiver J.W., Liang X. Characterizations of four monoclonal antibodies against M2 protein ectodomain of influenza A virus. Virology. 2009;385:218–226. doi: 10.1016/j.virol.2008.11.035. [DOI] [PubMed] [Google Scholar]
- 21.Feng M., Yuan Z., Xia W., Huang X., Wang X., Yan Y., Liao M., Zhou J. Monoclonal antibody against the universal M2 epitope of influenza A virus. Appl. Microbiol. Biotechnol. 2018;102:5645–5656. doi: 10.1007/s00253-018-9019-0. [DOI] [PubMed] [Google Scholar]
- 22.Wang R., Song A., Levin J., Dennis D., Zhang N.J., Yoshida H., Koriazova L., Madura L., Shapiro L., Matsumoto A., et al. Therapeutic potential of a fully human monoclonal antibody against influenza A virus M2 protein. Antiviral Res. 2008;80:168–177. doi: 10.1016/j.antiviral.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 23.Grandea A.G., 3rd, Olsen O.A., Cox T.C., Renshaw M., Hammond P.W., Chan-Hui P.Y., Mitcham J.L., Cieplak W., Stewart S.M., Grantham M.L., et al. Human antibodies reveal a protective epitope that is highly conserved among human and nonhuman influenza A viruses. Proc. Natl. Acad. Sci. USA. 2010;107:12658–12663. doi: 10.1073/pnas.0911806107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feng J., Zhang M., Mozdzanowska K., Zharikova D., Hoff H., Wunner W., Couch R.B., Gerhard W. Influenza A virus infection engenders a poor antibody response against the ectodomain of matrix protein 2. Virol. J. 2006;3:102. doi: 10.1186/1743-422X-3-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee Y.N., Kim M.C., Lee Y.T., Kim Y.J., Kang S.M. Mechanisms of cross-protection by influenza virus M2-based vaccines. Immune Netw. 2015;15:213–221. doi: 10.4110/in.2015.15.5.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sheng J.R., Grimme S., Bhattacharya P., Stowell M.H.B., Artinger M., Prabahakar B.S., Meriggioli M.N. In vivo adsorption of autoantibodies in myasthenia gravis using Nanodisc-incorporated acetylcholine receptor. Exp. Neurol. 2010;225:320–327. doi: 10.1016/j.expneurol.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bhattacharya P., Grimme S., Ganesh B., Gopisetty A., Sheng J.R., Martinez O., Jayarama S., Artinger M., Meriggioli M., Prabhakar B.S. Nanodisc-incorporated hemagglutinin provides protective immunity against influenza virus infection. J. Virol. 2010;84:361–371. doi: 10.1128/JVI.01355-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qiang M., Dong X., Zha Z., Zuo X.K., Song X.L., Zhao L., Yuan C., Huang C., Tao P., Hu Q., et al. Selection of an ASIC1a-blocking combinatorial antibody that protects cells from ischemic death. Proc. Natl. Acad. Sci. USA. 2018;115:E7469–E7477. doi: 10.1073/pnas.1807233115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dominik P.K., Borowska M.T., Dalmas O., Kim S.S., Perozo E., Keenan R.J., Kossiakoff A.A. Conformational chaperones for structural studies of membrane proteins using antibody phage display with nanodiscs. Structure. 2016;24:300–309. doi: 10.1016/j.str.2015.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zebedee S.L., Richardson C.D., Lamb R.A. Characterization of the influenza virus M2 integral membrane protein and expression at the infected-cell surface from cloned cDNA. J. Virol. 1985;56:502–511. doi: 10.1128/JVI.56.2.502-511.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dumoulin M., Conrath K., Van Meirhaeghe A., Meersman F., Heremans K., Frenken L.G.J., Muyldermans S., Wyns L., Matagne A. Single-domain antibody fragments with high conformational stability. Protein Sci. 2002;11:500–515. doi: 10.1110/ps.34602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stanfield R.L., Dooley H., Verdino P., Flajnik M.F., Wilson I.A. Maturation of shark single-domain (IgNAR) antibodies: evidence for induced-fit binding. J. Mol. Biol. 2007;367:358–372. doi: 10.1016/j.jmb.2006.12.045. [DOI] [PubMed] [Google Scholar]
- 33.Henderson K.A., Streltsov V.A., Coley A.M., Dolezal O., Hudson P.J., Batchelor A.H., Gupta A., Bai T., Murphy V.J., Anders R.F., et al. Structure of an IgNAR-AMA1 complex: targeting a conserved hydrophobic cleft broadens malarial strain recognition. Structure. 2007;15:1452–1466. doi: 10.1016/j.str.2007.09.011. [DOI] [PubMed] [Google Scholar]
- 34.Kovaleva M., Johnson K., Steven J., Barelle C.J., Porter A. Therapeutic potential of shark anti-ICOSL VNAR domains is exemplified in a murine model of autoimmune non-infectious Uveitis. Front.Immunol. 2017;8:1121. doi: 10.3389/fimmu.2017.01121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gonzalez-Sapienza G., Rossotti M.A., Tabares-da Rosa S. Single-domain antibodies as versatile affinity reagents for analytical and diagnostic applications. Front. Immunol. 2017;8:977. doi: 10.3389/fimmu.2017.00977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Asaadi Y., Jouneghani F.F., Janani S., Rahbarizadeh F. A comprehensive comparison between camelid nanobodies and single chain variable fragments. Biomark. Res. 2021;9:87. doi: 10.1186/s40364-021-00332-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cheong W.S., Leow C.Y., Abdul Majeed A.B., Leow C.H. Diagnostic and therapeutic potential of shark variable new antigen receptor (VNAR) single domain antibody. Int. J. Biol. Macromol. 2020;147:369–375. doi: 10.1016/j.ijbiomac.2020.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schoof M., Faust B., Saunders R.A., Sangwan S., Rezelj V., Hoppe N., Boone M., Billesbølle C.B., Puchades C., Azumaya C.M., et al. An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive Spike. Science. 2020;370:1473–1479. doi: 10.1126/science.abe3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shen Z., Xiang Y., Vergara S., Chen A., Xiao Z., Santiago U., Jin C., Sang Z., Luo J., Chen K., et al. A resource of high-quality and versatile nanobodies for drug delivery. iScience. 2021;24:103014. doi: 10.1016/j.isci.2021.103014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ubah O.C., Lake E.W., Gunaratne G.S., Gallant J.P., Fernie M., Robertson A.J., Marchant J.S., Bold T.D., Langlois R.A., Matchett W.E., et al. Mechanisms of SARS-CoV-2 neutralization by shark variable new antigen receptors elucidated through X-ray crystallography. Nat. Commun. 2021;12:7325. doi: 10.1038/s41467-021-27611-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weinstein J.B., Bates T.A., Leier H.C., McBride S.K., Barklis E., Tafesse F.G. A potent alpaca-derived nanobody that neutralizes SARS-CoV-2 variants. iScience. 2022;25:103960. doi: 10.1016/j.isci.2022.103960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scully M., Cataland S.R., Peyvandi F., Coppo P., Knöbl P., Kremer Hovinga J.A., Metjian A., de la Rubia J., Pavenski K., Callewaert F., et al. Caplacizumab treatment for acquired thrombotic thrombocytopenic purpura. N. Engl. J. Med. 2019;380:335–346. doi: 10.1056/NEJMoa1806311. [DOI] [PubMed] [Google Scholar]
- 43.Streltsov V.A., Carmichael J.A., Nuttall S.D. Structure of a shark IgNAR antibody variable domain and modeling of an early-developmental isotype. Protein Sci. 2005;14:2901–2909. doi: 10.1110/ps.051709505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Glück J.M., Wittlich M., Feuerstein S., Hoffmann S., Willbold D., Koenig B.W. Integral membrane proteins in nanodiscs can Be studied by solution NMR spectroscopy. J. Am. Chem. Soc. 2009;131:12060–12061. doi: 10.1021/ja904897p. [DOI] [PubMed] [Google Scholar]
- 45.Holsinger L.J., Lamb R.A. Influenza virus-M2 integral membrane-protein is a homotetramer stabilized by formation of disulfide bonds. Virology. 1991;183:32–43. doi: 10.1016/0042-6822(91)90115-R. [DOI] [PubMed] [Google Scholar]
- 46.Schnell J.R., Chou J.J. Structure and mechanism of the M2 proton channel of influenza A virus. Nature. 2008;451:591–595. doi: 10.1038/nature06531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grzeschik J., Könning D., Hinz S.C., Krah S., Schröter C., Empting M., Kolmar H., Zielonka S. Generation of semi-synthetic shark IgNAR single-domain antibody libraries. Methods Mol. Biol. 2018;1701:147–167. doi: 10.1007/978-1-4939-7447-4_8. [DOI] [PubMed] [Google Scholar]
- 48.Andris-Widhopf J., Rader C., Steinberger P., Fuller R., Barbas C.F., 3rd Methods for the generation of chicken monoclonal antibody fragments by phage display. J. Immunol. Methods. 2000;242:159–181. doi: 10.1016/s0022-1759(00)00221-0. [DOI] [PubMed] [Google Scholar]
- 49.Wei G., Meng W., Guo H., Pan W., Liu J., Peng T., Chen L., Chen C.Y. Potent neutralization of influenza A virus by a single-domain antibody blocking M2 ion channel protein. PLoS One. 2011;6:e28309. doi: 10.1371/journal.pone.0028309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bright R.A., Shay D.K., Shu B., Cox N.J., Klimov A.I. Adamantane resistance among influenza A viruses isolated early during the 2005-2006 influenza season in the United States. Jama-J Am Med Assoc. 2006;295:891–894. doi: 10.1001/jama.295.8.joc60020. [DOI] [PubMed] [Google Scholar]
- 51.Wang C., Lamb R.A., Pinto L.H. Direct measurement of the influenza A virus M2 protein ion channel activity in mammalian cells. Virology. 1994;205:133–140. doi: 10.1006/viro.1994.1628. [DOI] [PubMed] [Google Scholar]
- 52.Keller B.U., Hedrich R., Vaz W.L., Criado M. Single channel recordings of reconstituted ion channel proteins: an improved technique. Pflugers Arch. 1988;411:94–100. doi: 10.1007/BF00581652. [DOI] [PubMed] [Google Scholar]
- 53.Vemula S.V., Sayedahmed E.E., Sambhara S., Mittal S.K. Vaccine approaches conferring cross-protection against influenza viruses. Expert Rev. Vaccines. 2017;16:1141–1154. doi: 10.1080/14760584.2017.1379396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang Y., Deng L., Gonzalez G.X., Luthra L., Dong C., Ma Y., Zou J., Kang S.M., Wang B.Z. Double-layered m2e-NA protein nanoparticle immunization induces broad cross-protection against different influenza viruses in mice. Adv. Healthc. Mater. 2020;9:e1901176. doi: 10.1002/adhm.201901176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu H., Hill J.J., Michelsen K., Yamane H., Kurzeja R.J.M., Tam T., Isaacs R.J., Shen F., Tagari P. Characterization of the direct interaction between KcsA-Kv1.3 and its inhibitors. Biochim. Biophys. Acta. 2015;1848:1974–1980. doi: 10.1016/j.bbamem.2015.06.011. [DOI] [PubMed] [Google Scholar]
- 56.Banerjee S., Huber T., Sakmar T.P. Rapid incorporation of functional rhodopsin into nanoscale apolipoprotein bound bilayer (NABB) particles. J. Mol. Biol. 2008;377:1067–1081. doi: 10.1016/j.jmb.2008.01.066. [DOI] [PubMed] [Google Scholar]
- 57.Bayburt T.H., Leitz A.J., Xie G., Oprian D.D., Sligar S.G. Transducin activation by nanoscale lipid bilayers containing one and two rhodopsins. J. Biol. Chem. 2007;282:14875–14881. doi: 10.1074/jbc.M701433200. [DOI] [PubMed] [Google Scholar]
- 58.Boldog T., Grimme S., Li M., Sligar S.G., Hazelbauer G.L. Nanodiscs separate chemoreceptor oligomeric states and reveal their signaling properties. Proc. Natl. Acad. Sci. USA. 2006;103:11509–11514. doi: 10.1073/pnas.0604988103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Barelle C., Gill D.S., Charlton K. Shark novel antigen receptors-the next generation of biologic therapeutics? Adv. Exp. Med. Biol. 2009;655:49–62. doi: 10.1007/978-1-4419-1132-2. [DOI] [PubMed] [Google Scholar]
- 60.Zielonka S., Empting M., Grzeschik J., Könning D., Barelle C.J., Kolmar H. Structural insights and biomedical potential of IgNAR scaffolds from sharks. mAbs. 2015;7:15–25. doi: 10.4161/19420862.2015.989032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Anderson G.P., Teichler D.D., Zabetakis D., Shriver-Lake L.C., Liu J.L., Lonsdale S.G., Goodchild S.A., Goldman E.R. Importance of hypervariable region 2 for stability and affinity of a shark single-domain antibody specific for Ebola virus nucleoprotein. PLoS One. 2016;11 doi: 10.1371/journal.pone.0160534. e0160534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu J.L., Anderson G.P., Delehanty J.B., Baumann R., Hayhurst A., Goldman E.R. Selection of cholera toxin specific IgNAR single-domain antibodies from a naive shark library. Mol. Immunol. 2007;44:1775–1783. doi: 10.1016/j.molimm.2006.07.299. [DOI] [PubMed] [Google Scholar]
- 63.Stanfield R.L., Dooley H., Flajnik M.F., Wilson I.A. Crystal structure of a shark single-domain antibody V region in complex with lysozyme. Science. 2004;305:1770–1773. doi: 10.1126/science.1101148. [DOI] [PubMed] [Google Scholar]
- 64.Liu J.L., Zabetakis D., Brown J.C., Anderson G.P., Goldman E.R. Thermal stability and refolding capability of shark derived single domain antibodies. Mol. Immunol. 2014;59:194–199. doi: 10.1016/j.molimm.2014.02.014. [DOI] [PubMed] [Google Scholar]
- 65.Pielak R.M., Schnell J.R., Chou J.J. Mechanism of drug inhibition and drug resistance of influenza A M2 channel (vol 106, pg 7379, 2009) P Natl. Acad. Sci. USA. 2009;106:11425. doi: 10.1073/pnas.0905916106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zielonka S., Weber N., Becker S., Doerner A., Christmann A., Christmann C., Uth C., Fritz J., Schäfer E., Steinmann B., et al. Shark Attack: high affinity binding proteins derived from shark vNAR domains by stepwise in vitro affinity maturation. J. Biotechnol. 2014;191:236–245. doi: 10.1016/j.jbiotec.2014.04.023. [DOI] [PubMed] [Google Scholar]
- 67.Van Hoecke L., Verbeke R., De Vlieger D., Dewitte H., Roose K., Van Nevel S., Krysko O., Bachert C., Schepens B., Lentacker I., Saelens X. mRNA encoding a bispecific single domain antibody construct protects against influenza A virus infection in mice. Mol. Ther. Nucleic Acids. 2020;20:777–787. doi: 10.1016/j.omtn.2020.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tang G., Peng L., Baldwin P.R., Mann D.S., Jiang W., Rees I., Ludtke S.J. EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 69.Scheres S.H.W. A bayesian view on cryo-EM structure determination. J. Mol. Biol. 2012;415:406–418. doi: 10.1016/j.jmb.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wan C., Wu B., Song Z., Zhang J., Chu H., Wang A., Liu Q., Shi Y., Li G., Wang J. Insights into the molecular recognition of the granuphilin C2A domain with PI(4, 5)P2. Chem. Phys. Lipids. 2015;186:61–67. doi: 10.1016/j.chemphyslip.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 71.Yan C., Hang J., Wan R., Huang M., Wong C.C.L., Shi Y. Structure of a yeast spliceosome at 3.6-angstrom resolution. Science. 2015;349:1182–1191. doi: 10.1126/science.aac7629. [DOI] [PubMed] [Google Scholar]
- 72.Han J., Li T., Li Y., Li M., Wang X., Peng C., Su C., Li N., Li Y., Xu Y., Chen Y. The internal interaction in RBBP5 regulates assembly and activity of MLL1 methyltransferase complex. Nucleic Acids Res. 2019;47:10426–10438. doi: 10.1093/nar/gkz819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Müller M.R., O'Dwyer R., Kovaleva M., Rudkin F., Dooley H., Barelle C.J. Generation and isolation of target-specific single-domain antibodies from shark immune repertoires. Methods Mol. Biol. 2012;907:177–194. doi: 10.1007/978-1-61779-974-7_9. [DOI] [PubMed] [Google Scholar]
- 74.Zhang Y., Gao H., Li H., Guo J., Ouyang B., Wang M., Xu Q., Wang J., Lv M., Guo X., et al. The white-spotted bamboo shark genome reveals chromosome rearrangements and fast-evolving immune genes of cartilaginous fish. iScience. 2020;23:101754. doi: 10.1016/j.isci.2020.101754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Flajnik M.F., Dooley H. The generation and selection of single-domain, v region libraries from nurse sharks. Methods Mol. Biol. 2009;562:71–82. doi: 10.1007/978-1-60327-302-2_6. [DOI] [PubMed] [Google Scholar]
- 76.Reichart T.M., Baksh M.M., Rhee J.K., Fiedler J.D., Sligar S.G., Finn M.G., Zwick M.B., Dawson P.E. Trimerization of the HIV transmembrane domain in lipid bilayers modulates broadly neutralizing antibody binding. Angew. Chem. Int. Ed. Engl. 2016;55:2688–2692. doi: 10.1002/anie.201508421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Glück J.M., Koenig B.W., Willbold D. Nanodiscs allow the use of integral membrane proteins as analytes in surface plasmon resonance studies. Anal. Biochem. 2011;408:46–52. doi: 10.1016/j.ab.2010.08.028. [DOI] [PubMed] [Google Scholar]
- 78.Zhang G.P., Shi Y.L., Wang W.P., Liu W.Y. Cation channel formed at lipid bilayer by Cinnamomin, a new type II ribosome-inactivating protein. Toxicon. 1999;37:1313–1322. doi: 10.1016/s0041-0101(99)00078-1. [DOI] [PubMed] [Google Scholar]
- 79.Zhou S., Ruan M., Li Y., Yang J., Bai S., Richter C., Schwalbe H., Xie C., Shen B., Wang J. Solution structure of the voltage-gated Tim23 channel in complex with a mitochondrial presequence peptide. Cell Res. 2021;31:821–824. doi: 10.1038/s41422-020-00452-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data reported in this paper will be shared by the lead contact upon request. The displayed NMR structure in Figure 2D was from published PDB data in https://www.rcsb.org and the PDB ID has been listed in the key resources table.
This study did not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contactupon request.