Abstract
Parasitic nematode infections cause an enormous global burden to both humans and livestock. Resistance to the limited arsenal of anthelmintic drugs used to combat these infections is widespread, including benzimidazole (BZ) compounds. Previous studies using the free-living nematode Caenorhabditis elegans to model parasitic nematode resistance have shown that loss-of-function mutations in the beta-tubulin gene ben-1 confer resistance to BZ drugs. However, the mechanism of resistance and the tissue-specific susceptibility are not well known in any nematode species. To identify in which tissue(s) ben-1 function underlies BZ susceptibility, transgenic strains that express ben-1 in different tissues, including hypodermis, muscles, neurons, intestine, and ubiquitous expression were generated. High-throughput fitness assays were performed to measure and compare the quantitative responses to BZ compounds among different transgenic lines. Significant BZ susceptibility was observed in animals expressing ben-1 in neurons, comparable to expression using the ben-1 promoter. This result suggests that ben-1 function in neurons underlies susceptibility to BZ. Subsetting neuronal expression of ben-1 based on the neurotransmitter system further restricted ben-1 function in cholinergic neurons to cause BZ susceptibility. These results better inform our current understanding of the cellular mode of action of BZs and also suggest additional treatments that might potentiate the effects of BZs in neurons.
Keywords: Benzimidazole resistance, C.elegans, High-throughput assay
Graphical abstract
Highlights
-
•
Expressing wild-type ben-1 only in neurons restores susceptibility to benzimidazoles.
-
•
Expression of ben-1 in cholinergic neurons restores susceptibility to benzimidazoles.
-
•
GABAergic neurons might also play a role in benzimidazole sensitivity.
-
•
Broad implications for molecular mechanisms of benzimidazole mode of action.
1. Introduction
Anthelmintic drugs are crucial to combat parasitic nematode infections, which affect billions of people and livestock each year (Kaplan and Vidyashankar, 2012; Hotez et al., 2014). However, only a limited arsenal of drugs are approved, comprising four major classes: benzimidazoles (BZs), nicotinic acetylcholine receptor agonists (nAChRs), macrocyclic lactones (MLs), and amino-acetonitrile derivatives (AADs). BZs have been used extensively for over 50 years (Roos et al., 1995; Abongwa et al., 2017). Because of the intensity of administration of the few anthelmintics available, resistance was documented in Haemonchus contortus within a few years after its introduction (Theodorides et al., 1970). Resistance to commonly used BZs continues to be widespread today (Kaplan and Vidyashankar, 2012).
Following the establishment of resistance to BZs, mutations in beta-tubulin genes were correlated with resistance in the fungus Aspergillus nidulans and then in the free-living nematode Caenorhabditis elegans (Hastie and Georgopoulos, 1971; Sheir-Neiss et al., 1978; Driscoll et al., 1989). Loss-of-function mutations in the beta-tubulin gene ben-1 were identified in strains resistant to BZs (Driscoll et al., 1989). Redundancy among the six beta-tubulin genes in C. elegans allows strains with non-functional ben-1 to still develop normally (Driscoll et al., 1989). Resistance alleles corresponding to point mutations in ben-1 homologs in parasitic nematode populations continue to be identified (Hahnel et al., 2018; Avramenko et al., 2019; Mohammedsalih et al., 2020; Dilks et al., 2021), and many have been validated to cause resistance using genome editing and highly sensitive C. elegans drug response assays (Hahnel et al., 2018; Dilks et al., 2020, 2021). These techniques are difficult (if not impossible) in parasites, and C. elegans has been proven to be a suitable model to study resistance complementary to parasitic nematodes (Wit et al., 2021a, Wit et al., 2021b).
New resistance alleles continue to be identified in parasite populations, which emphasizes the need to develop compounds that can be used in conjunction with BZs to potentiate their effects. However, the mechanism of action beyond beta-tubulin binding is still unknown. In susceptible animals, BZs binding to beta-tubulin inhibits tubulin polymerization necessary to form microtubules (Ireland et al., 1979; Laclette et al., 1980; Lacey and Prichard, 1986; Lacey, 1990), but it is not well understood in what tissues and specific cells microtubule formation is inhibited. Preliminary studies of BZ susceptibility in H. contortus taken from sheep treated with fenbendazole were found to have gross disintegration of the intestine, suggesting that BZs target beta-tubulin in the intestine of these animals (Jasmer et al., 2000). We can use genetic tools such as transgenesis and high-throughput assays developed for C. elegans to further investigate the mode of action of BZs with the caveat that responses observed in C. elegans are not necessarily representative of the entire phylum.
Here, we re-introduced the wild-type ben-1 gene into a ben-1 knockout strain background using transgenesis (Rieckher and Tavernarakis, 2017), where multi-copy arrays express ben-1 in specific tissues. Plasmids containing the coding sequence of ben-1 fused to tissue-specific promoters, including neurons, hypodermis, muscles, and intestine, as well as endogenous and ubiquitous expression, formed extrachromosomal arrays in transgenic animals. Loss of ben-1 causes BZ resistance (Hahnel et al., 2018), so transgenic addition of wild-type ben-1 can restore BZ sensitivity. We then performed high-throughput fitness assays to quantitatively assess the response to albendazole (ABZ). We found that when ben-1 is expressed in neurons, the wild-type susceptibility phenotype is restored. We then generated transgenic strains that expressed ben-1 in cholinergic, dopaminergic, GABAergic, or glutamatergic neurons to narrow down the neurons where BZs cause lethality. We found that ben-1 expression in cholinergic neurons was sufficient to restore wild-type BZ susceptibility. These results offer insights into the mode of action of BZs and suggest that BZs might have a similar cellular target as other classes of anthelmintics.
2. Materials and methods
2.1. Strains
Strains were maintained at 20 °C on modified nematode growth media plates (NGMA) with 1% agar, 0.7% agarose, and Escherichia coli OP50 bacteria for food (Andersen et al., 2014). To alleviate starvation effects, strains were grown for three generations before each assay (Andersen et al., 2015). Most strains were generated using the ECA882 strain, ben-1(ean64), which has the laboratory-derived reference strain background (N2) and a deletion of ben-1 exons 2 through 4. This strain has been previously shown to be resistant to albendazole (Hahnel et al., 2018). The N2 strain was also used as a background for some control transgenic strains.
2.2. Plasmid construction
All ben-1 plasmids with alternative promoters were constructed by VectorBuilder (Table S1). Tissue-specific promoters included myo-3 (muscles, Fire and Waterston, 1989; Frøkjaer-Jensen et al., 2008), unc-119 (pan-neuronal, Maduro and Pilgrim, 1995; Maduro and Pilgrim, 1996; Johnson et al., 2005), col-19 (hypodermis, Liu et al., 1995; Johnstone, 2000; Thein et al., 2003), and ges-1 (intestines, Kennedy et al., 1993). A plasmid with the eft-3 promoter for ubiquitous expression was designed as well (Frøkjær-Jensen et al., 2012). Neurotransmitter-specific promoters included unc-17 (cholinergic, Serrano-Saiz et al., 2020), eat-4 (glutamatergic, Serrano-Saiz et al., 2020), dat-1 (dopaminergic, Flames and Hobert, 2009), and unc-25 (GABAergic, Serrano-Saiz et al., 2020). The plasmid with the ben-1 endogenous promoter was constructed using the ben-1 cDNA and an amplicon of the ben-1 promoter and assembled using Gibson cloning (Gibson et al., 2009). The co-injection marker plasmid, pBCN27 (myo-2p::GFP::unc-54_3′UTR) was a gift from Ben Lehner (Addgene, plasmid #26347) (Semple et al., 2010).
2.3. C. elegans transgenesis
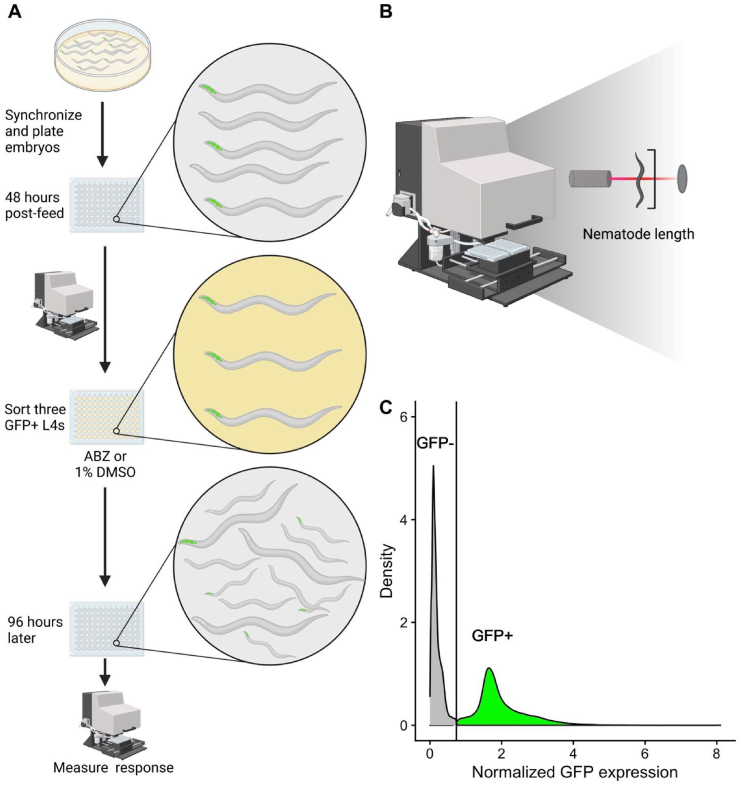
The C. elegans microinjection technique has been previously described (Rieckher and Tavernarakis, 2017). Briefly, a ben-1 expression plasmid was combined with the myo-2p::GFP co-injection marker (5 ng/uL) and 1 kb DNA ladder (Invitrogen #10787018) at the specified concentrations (Table S2, Abrahante et al., 1998; Eastman et al., 1999; Marshall and McGhee, 2001; Carvelli et al., 2004; Frøkjaer-Jensen et al., 2008; Frøkjær-Jensen et al., 2012; Hao et al., 2012; Serrano-Saiz et al., 2013; Kaymak et al., 2016; Muñoz-Jiménez et al., 2017; Thomas et al., 2019). This mixture was injected into the gonads of adult hermaphrodites harboring the ben-1 deletion (Fig. 1). Post-injection, a single adult was placed onto a 6 cm plate. F1 progeny expressing GFP in the pharynx were identified 48–72 h following injection and subsequently singled (Fig. 1). Two independent lines per rescue construct were selected based on the presence of the transgene in the F2 generation (Fig. 1). Wild-type GFP control strains were made by injecting the co-injection marker into the N2 strain. Deletion GFP control strains were constructed using the co-injection marker injected into ECA882.
Fig. 1.
The generation of transgenic strains with specified expression of ben-1 using microinjection. A mixture of ben-1 plasmid fused to a specific promoter and a GFP co-injection marker that expressed GFP in the pharynx were injected into the gonads of adult worms. After 48–72 h, offspring were scored for presence of GFP in the pharynx, which was indicative of successful transgenesis. Strains with approximately 70% transmission were selected for high-throughput fitness assays.
2.4. High-throughput fitness assays
The COPAS BIOSORT high-throughput phenotyping assay has been previously described (Andersen et al., 2015; Zdraljevic et al., 2017; Evans et al., 2018; Hahnel et al., 2018; Brady et al., 2019; Dilks et al., 2020, 2021; Evans and Andersen, 2020). Additional measures were taken at each step of the propagation protocol to select for animals with the transgene. Briefly, a small chunk from a starved 6 cm NGMA plate was placed onto a fresh plate. After 48 h, GFP-positive, gravid hermaphrodites were transferred to a plate with a bleach solution (40 mL NaOCl (Fisher #SS290-1), 10 mL of 10 M NaOH added to 150 mL of distilled water). Approximately 24 h later, GFP-positive L1 larvae were transferred to a fresh 6 cm NGMA plate. After 48 h, five animals at the L4 stage were picked to a new plate. After 72 h to allow for offspring to grow to the L4 stage, five GFP-positive L4s were placed onto a fresh 6 cm NGMA plate and allowed to develop and propagate. After 96 h, strains were washed off of plates using M9 buffer into 15 mL conicals and treated with fresh bleach solution to dissolve gravid adults and obtain a large number of unhatched embryos. Embryo pools were washed three times using M9 and once using K medium (51 mM NaCl, 32 mM KCl, 3 mM CaCl2, and 3 mM MgSO4 in distilled water) (Boyd et al., 2012) before being resuspended in K medium. Clean embryos were diluted to approximately one embryo per μL in K medium and then aliquoted into 96-well plates at approximately 50 embryos in each well (Fig. 2A). After hatching overnight, arrested L1 larvae were fed lyophilized E. coli strain HB101 (Pennsylvania State University Shared Fermentation Facility, State College, PA) at a concentration of 5 mg/mL (García-González et al., 2017). After 48 h, L4 larvae were sorted using the COPAS BIOSORT (Union Biometrica, Holliston MA). The COPAS BIOSORT is able to measure the time-of-flight (TOF) and green fluorescence of each object as it flows through the device (Fig. 2B–C, Andersen et al., 2015; Zdraljevic et al., 2017; Evans et al., 2018; Hahnel et al., 2018; Brady et al., 2019; Dilks et al., 2020; Evans and Andersen, 2020; Dilks et al., 2021). Three GFP-positive L4 larvae were sorted from each well to wells in a new 96-well plate containing HB101 lysate at 10 mg/mL and 12.5 μM albendazole in 1% DMSO or 1% DMSO alone (Fig. 2A). We verified the correct sorting of fluorescent animals by running the COPAS BIOSORT protocol for one row of wild-type animals with the co-injection marker and validating the presence of the GFP marker in all animals sorted. This concentration of albendazole has been previously used with this protocol (Hahnel et al., 2018). Four days after exposure to albendazole, wells were treated with 50 mM sodium azide and scored using the COPAS BIOSORT (Fig. 2A).
Fig. 2.
COPAS BIOSORT high-throughput fitness assay. A) Illustration of the high-throughput fitness assay is shown. Synchronized worms were treated with bleach solution and embryos were distributed into 96-well plates. 48 h post-feeding, three GFP-positive L4 animals were sorted using the COPAS BIOSORT into each well of a 96-well plate containing 12.5 μM albendazole in 1% DMSO or 1% DMSO alone. After 96 h, animals were scored for GFP fluorescence and length using the COPAS BIOSORT. B) Illustration of COPAS BIOSORT is shown. Animals were passed through a flow cell and a laser. The time the detector is interrupted is equated to the length of the nematode. Illustration adapted from (Wit et al., 2021). C) Distribution of GFP fluorescence of animals is shown. The x-axis is the GFP expression of each animal normalized by the length of each animal. The y-axis is the distribution of the population. The vertical line represents the threshold for establishing if animals are GFP-positive (green) or GFP-negative (gray). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
2.5. Data analysis
Raw data from the COPAS BIOSORT were processed using the R package easysorter (Shimko and Andersen, 2014) as previously described (Hahnel et al., 2018; Dilks et al., 2020, 2021). An average value for each phenotypic trait measured in the control condition (1% DMSO) was deducted from the albendazole condition data to normalize the data for each strain. The distribution of green fluorescence values was analyzed to establish a threshold to filter for GFP-positive animals that had the transgene (Fig. 2C). The average mean TOF value was summarized for each well per strain and the distribution of mean TOF values for each transgenic strain was compared to the mean TOF values for the ben-1 deletion strain. Statistical tests and analyses were performed in R using the tukeyHSD function in the Rstatix package. The ANOVA model (phenotype ∼ strain) was used to compare differences in phenotypic responses to BZs between the ben-1 deletion strain and the other strains.
3. Data availability
A list of plasmids used with vendor ID information and a list of C. elegans strains and genotypes used in experiments are included as supplementary information (Tables S1 and S2). The data and code used to process these data are available at https://github.com/AndersenLab/2022_ben1sensitivity_SBG.
4. Results
4.1. ben-1 function in neurons rescues BZ sensitive phenotype
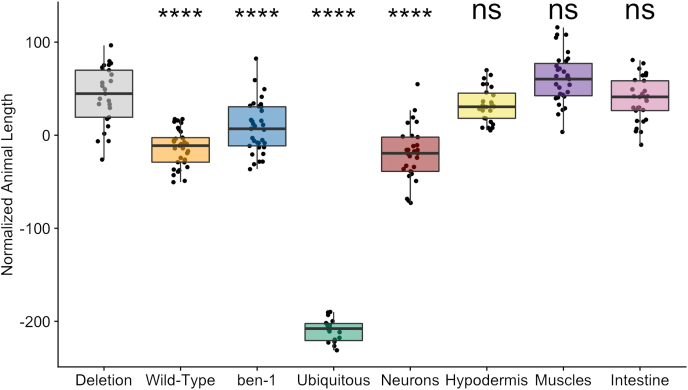
We created transgenic strains where ben-1 was expressed in different tissues, including muscles, neurons, hypodermis, intestines, as well as ubiquitous expression and with the endogenous ben-1 promoter. These different expression constructs were made into transgenes in the ECA882 ben-1 deletion background. For each candidate tissue-specific expression strain, two independent strains were generated and assayed to ensure that the measured effects were caused by the transgenes and not a vagary of the injection process. Assay results with the second strain are available as supplemental information (Figs. S1 and S2). Animals with a ben-1 specific transgene were identifiable by a green fluorescent pharynx caused by the pharyngeal expression of the co-injection transgenesis marker. We then performed high-throughput assays to test if any of the tissue-specific expression strains rescued susceptibility to albendazole. A strain with the resistant ben-1 knockout background and the pharyngeal expression marker and a strain with the susceptible wild-type background and the pharyngeal expression marker were used as controls. We measured growth in control conditions (DMSO) and albendazole (ABZ) conditions in 44 replicates per strain. The high-throughput assay was performed as described (See 2.4, Fig. 2A). Following 48 h of growth, three GFP-positive animals for each strain were sorted into each well of a 96-well plate using the COPAS BIOSORT. These animals gave rise to a population in each well that was then scored 96 h after exposure to ABZ using the COPAS BIOSORT. The green fluorescence measurements were used to filter the data to only include animals that had the transgene (Fig. 2C). We used the length of animals as a measure of developmental rate. Strains with lower average length values, when compared to the ben-1 deletion strain, indicate susceptibility to ABZ. As expected, we measured a significant difference in animal length between the wild-type strain as compared to the ben-1 deletion in response to ABZ (Fig. 3). We found that expression of ben-1 under its endogenous promoter or when highly expressed in all tissues caused susceptibility to ABZ (Fig. 3 and S1). Furthermore, the high level of expression using the eft-3 promoter caused ABZ susceptibility far beyond wild-type levels. When ben-1 expression was driven only in neurons using the unc-119 promoter, the animals were equally susceptible to ABZ as animals with wild-type expression of ben-1 (Fig. 3). These results suggest that neuronal ben-1 might be the endogenous target of BZ compounds. Expression of ben-1 in hypodermis, muscles, and intestine did not restore the ABZ susceptibility phenotype (Fig. 3), suggesting that neurons might be the main target of BZs.
Fig. 3.
ben-1 function in neurons underlies BZ sensitivity. The x-axis denotes the specificity of the ben-1 expression from the transgene or the wild-type or deletion genotype. The y-axis is the normalized length of animals after exposure to 12.5 μM of albendazole. Phenotypic data was normalized by subtracting an average value for each trait from the control data. Each data point is the mean value for the population of GFP-positive worms in a single well. Tukey box plots have horizontal lines at the third quartile on the top, the median in the middle and the first quartile at the bottom. The whiskers are extended within 1.5 range from each quartile. The significant difference between the transgene and the deletion genotype is shown above the transgene (**** = p < 0.0001, one-way ANOVA, Tukey HSD). The strains shown from left to right are ECA2842 (Deletion), ECA2821 (Wild-type), ECA2824 (ben-1, ben-1p), ECA2828 (Ubiquitous, eft-3p), ECA2833 (Neurons, unc-119p), ECA2836 (Hypodermis, col-19p), ECA2846 (Muscles, myo-3p), and ECA2840 (Intestine, ges-1p).
4.2. ben-1 function in cholinergic and GABAergic neurons restores the BZ sensitivity phenotype
Although at least one beta-tubulin gene is expressed in every cell of an organism, ben-1 has been specifically shown to be broadly expressed in neurons (Boulin et al., 2006; Hurd, 2018). We used the C. elegans Neuronal Gene Expression Map and Network (CeNGEN) dataset (Taylor et al., 2021) to further explore neuron-specific expression of ben-1 and found that it is expressed in 97 of the 128 cell types distinguished in the dataset and primarily in cholinergic and glutamatergic neurons (Fig. S3). The thorough characterization of each neuron in the C. elegans nervous system offered the opportunity to generate transgenic animals with ben-1 expression specific to subsets of neurons. We hoped to narrow down which neurons are specifically targeted by BZs by creating transgenic strains with ben-1 expression separated by neurotransmitter system and perform the same high-throughput analyses.
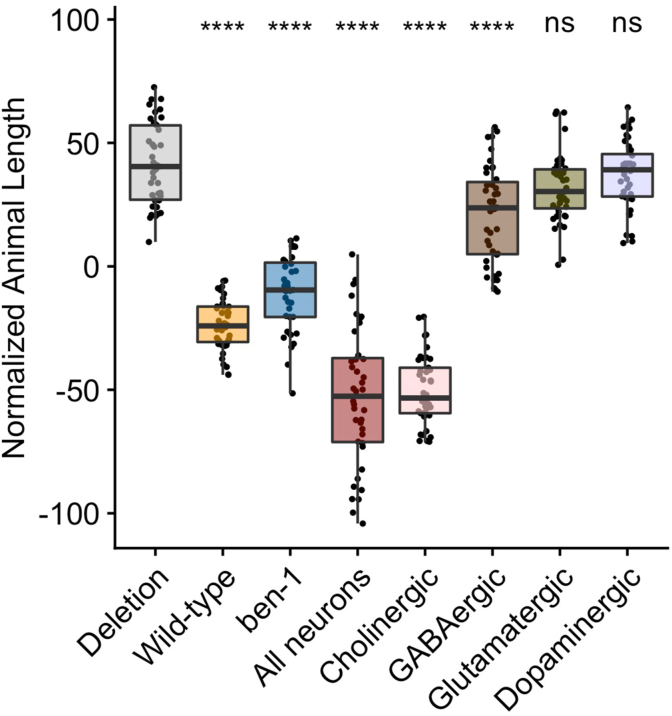
We created transgenic strains where ben-1 expression in neurons was subsetted by the neurotransmitter system, including cholinergic, glutamatergic, dopaminergic, and GABAergic neurons. Two independent lines for each neuronal type were generated in the ECA882 ben-1 deletion background. The same pharyngeal expression marker was used to identify transgenic animals as well as the same wild-type and resistant control strains. We compared the responses of the four neurotransmitter system-specific transgenes as well as the endogenous and pan-neuronal expression strains to the two control strains with 44 replicates per strain. Offspring were scored 96 h after three GFP-positive parents were sorted into each well of plates containing either ABZ or the control DMSO condition. These data were filtered to only compare the lengths of GFP-positive animals. We found that animals expressing ben-1 in cholinergic regions were significantly smaller (e.g., developmentally delayed because of susceptibility to ABZ) than the resistant strain when exposed to ABZ and were comparable in size to animals expressing ben-1 in all neurons (Fig. 4). It is worth noting that a large proportion of the neuron classes that express ben-1 are cholinergic, including a few of the neurons with the highest levels of expression (Fig. S3, Loer and Rand, 2010; Taylor et al., 2021). We observed no difference in animal length between the resistant strain and animals expressing ben-1 in glutamatergic and dopaminergic neurons, suggesting that BZs do not target these neurons (Fig. 4). In animals that expressed ben-1 in GABAergic neurons, we found a less extreme ABZ response (Fig. 4), suggesting that ben-1 function in cholinergic and potentially GABAergic neurons was sufficient to restore ABZ susceptibility.
Fig. 4.
ben-1 function in cholinergic and GABAergic neurons underlies BZ sensitivity. The x-axis denotes the specificity of the ben-1 expression from the transgene or the wild-type or deletion genotype. The y-axis is the normalized lengths of animals after exposure to 12.5 μM of albendazole. Phenotypic data was normalized by subtracting an average value for each trait from the control data. Each data point is the mean value for the population of GFP-positive worms in a single well. Tukey box plots have horizontal lines at the third quartile on the top, the median in the middle and the first quartile at the bottom. The whiskers are extended within 1.5 range from each quartile. The significant difference between the transgene and the deletion genotype is shown above the transgene (**** = p < 0.0001, one-way ANOVA, Tukey HSD). The strains shown from left to right are ECA2842 (Deletion), ECA2821 (Wild-type), ECA2824 (ben-1, ben-1p), ECA2833 (Neurons, unc-119p), ECA3335 (Cholinergic, unc-17p), ECA3356 (GABAergic, unc-25p), ECA3333 (Glutamatergic, eat-4p), and ECA3354 (Dopaminergic, dat-1p).
5. Discussion
5.1. ben-1 function in cholinergic and GABAergic neurons is sufficient to rescue organismal BZ susceptibility
Although it is well understood that reduction- or loss-of-function of ben-1 confers resistance to BZs (Driscoll et al., 1989; Hahnel et al., 2018), little has been done to better characterize BZ sensitivity at the cellular level. Understanding cell-type specific targets of BZs could enable development of co-treatments that potentiate BZ effects. Here, we generated transgenic C. elegans where ben-1 expression was limited to one of four tissue types, all tissues (ubiquitous, high-level), or expression under control of the endogenous ben-1 promoter. The tissue-specific promoters used in these experiments have been well characterized and used extensively (Fire and Waterston, 1989; Kennedy et al., 1993; Liu et al., 1995; Maduro and Pilgrim, 1995, 1996; Johnstone, 2000; Johnson et al., 2005; Frøkjaer-Jensen et al., 2008; Frøkjær-Jensen et al., 2012). The ability to rescue the susceptible phenotype is a strong indication that ben-1 function is targeted in that tissue. Whereas, a negative result where no rescue is observed does not rule out function in that tissue because levels of expression might not be sufficient to cause susceptibility. Each strain's response to the BZ drug albendazole (ABZ) was compared to ABZ responses in the resistant ben-1 deletion strain. When ben-1 is expressed only in neurons, we see that animal development is delayed in ABZ conditions (Fig. 3), suggesting that ben-1 expression in neurons is sufficient to rescue sensitivity to BZs. Although in the replicate experiment we also found a statistically significant difference in animal length when ben-1 was expressed in the hypodermis or the intestine (Fig. S1), these results were not reproducible between experiments so could be attributed to the overexpression of genes by different extrachromosomal arrays. These experiments all rely on the overexpression of ben-1 in a tissue-specific manner. This overexpression could cause an imbalance in endogenous levels of beta-tubulins and could cause a slight deleterious effect on fitness. However, transgenic animal development was not significantly reduced in the control conditions when compared to the wild-type strain (Figs. S4 and S5). The normalized length values for strains expressing ben-1 in the hypodermis and the intestine are also close to zero or positive, unlike the negative normalized values for animals expressing ben-1 in neurons where the difference between the response of the resistant strain is much more pronounced (Fig. 3, S1). Single-cell expression studies show that ben-1 is primarily expressed in neurons (Hurd, 2018; Taylor et al., 2021), so neurons might require this specific form of beta-tubulin. The hypersensitivity to ABZ that is observed when ben-1 is expressed ubiquitously might also suggest that combinatorial expression between multiple tissues could be involved. As mentioned above, the lack of a susceptible phenotype in a transgenic strain does not rule out the role of ben-1 in that tissue. Although C. elegans has proven to be a powerful model of parasitic nematode resistance to anthelmintic compounds, it is important to acknowledge that parasite-specific pathways are not present in C. elegans (Viney, 2018; International Helminth Genomes Consortium, 2019; Hahnel et al., 2020). Because of their significant divergence in evolutionary history, it is possible that the C. elegans responses might not be representative of all parasitic nematodes. The debate of how reliable the C. elegans model is for research into parasites is ongoing (Hahnel et al., 2020). Future experiments can confirm if ben-1 function in neurons is necessary for the BZ susceptibility phenotype across other nematode species.
The detailed understanding of the C. elegans nervous system anatomy and function allowed us to identify specific neurons that are targeted by BZs (White et al., 1986; Taylor et al., 2021). We subsetted the 118 neuron classes by neurotransmitter system to determine which neurons might require ben-1 function to be susceptible to BZ compounds. Transgenic strains were generated with ben-1 fused to cholinergic, dopaminergic, GABAergic, or glutamatergic-specific promoters (Flames and Hobert, 2009; Serrano-Saiz et al., 2020) and assayed using techniques similar to the tissue-specificity experiments. When ben-1 is expressed in cholinergic neurons, the drug-treated animals were significantly smaller than the resistant strain, similar to when ben-1 expression was expressed in all neurons (Fig. 4). Although some glutamatergic and dopaminergic neurons also have higher levels of ben-1 expression, a significant phenotypic effect was not observed in animals with ben-1 expression in these neurons (Fig. 4 and S2). It is possible that BZs also target GABAergic neurons as a less-significant BZ susceptibility phenotype was consistently observed across experiments (Fig. 4 and S2). Neuronal expression data of ben-1 support that cholinergic neurons are more likely to be the target of BZs because they express ben-1 at higher levels than the few GABAergic neurons with ben-1 expression (Loer and Rand, 2010; Taylor et al., 2021).
5.2. Further investigation of ben-1 function in specific tissues is required
C. elegans transgenesis using microinjection of plasmids containing ben-1 is an effective model to suggest where ben-1 function might cause susceptibility to BZs. The re-introduction of ben-1 as an extrachromosomal array causes overexpression, which might not reflect endogenous levels and can cause artefactual conclusions. To definitively show the role of ben-1 in specific neurons, we need experiments that test the requirement of ben-1 in BZ resistance. Conditional knockout (cKO) experiments where the ben-1 gene is removed from wild-type animals in specific tissues and/or cells is achievable using the Cre/loxP system developed for C. elegans (Kage-Nakadai et al., 2014). Genes flanked with loxP sites are removed from the genome using the Cre recombinase (Austin et al., 1981; Kage-Nakadai et al., 2014). Conditionally expressed Cre will only excise ben-1 from specific target tissues and/or cells, and the remaining tissues of the organism will express ben-1 endogenously. We can create strains with ben-1 knocked out in specific tissue types or neurons and perform the same high-throughput assays to assess the relative fitness of each strain when exposed to BZs. If animals with ben-1 knocked out only in neurons are resistant to BZs, ben-1 function in this tissue is necessary for BZ sensitivity.
5.3. Which specific neurons are targeted by BZs?
With the suggestion of ben-1 function in cholinergic and GABA neurons underlying BZ sensitivity, we can further narrow down the neurons targeted by BZs. We can continue to generate transgenic worms using promoters for genes specific to different neuron classes and leverage other tools unique to C. elegans, such as the NeuroPAL strain set (Yemini et al., 2021) and the CRF_ID annotation framework (Chaudhary et al., 2021). NeuroPAL (Neuronal Polychromic Atlas of Landmarks) strains are transgenic animals with a differentiated fluorescence pattern for every neuron in the organism (Yemini et al., 2021). Specialized software was developed for the system that can be used to identify each neuron from images based on fluorescence (Yemini et al., 2021). However, the annotation software is semi-automatic and requires some manual input to label microscopy images (Yemini et al., 2021). An automated annotation framework called CRF_ID has been developed to label cells with an algorithm based on the graphical-model based framework Conditional Random Fields (CRF) (Chaudhary et al., 2021). It has been shown to improve accuracy in labeling whole-brain images from C. elegans and be compatible with labeling color-based NeuroPAL animal images (Chaudhary et al., 2021). This sophisticated imaging and annotation system would allow us to simultaneously assay the entire C. elegans nervous system to identify specific targets of BZs. Neurons affected by BZs would be identified by comparing NeuroPAL animals treated with BZs to ones developing in control conditions. The ability to assay the entire nervous system will be particularly useful as not all of the 118 neuron classes were accounted for in the neurotransmitter system-specific assays as a few neurons expressing ben-1 use other neurotransmitters or their neurotransmitter system is unknown (Fig. S3).
5.4. Benzimidazole neuron targets might be shared with other anthelmintics in C. elegans
Benzimidazole targeting of beta-tubulin in neurons suggests that the specific neurons might be in common with other anthelmintic compounds. Resistance to two other widely used drug classes, macrocyclic lactones (MLs) and nicotinic acetylcholine receptor agonists (nAChRs), has been associated with genes expressed in neurons. Resistance to MLs has been linked to genes coding for the subunits of glutamate-gated chloride channels, including avr-14, avr-15, and glc-1 (Dent et al., 2000), and genes involved in resistance to nAChRs, including lev-1, lev-8, unc-29, unc-38, and unc-63 are known (Lewis et al., 1980; Qian et al., 2008). Overlaps in expression between ML and nAChR resistance genes and ben-1 using the CeNGEN data set (Taylor et al., 2021) could identify neurons potentially targeted by multiple anthelmintic compounds. Comparing responses NeuroPAL animals have to each compound could confirm if some neurons might be targeted and affected by multiple anthelmintic drugs, which might allow for the development of co-treatments compatible with more than one anthelmintic compound. Overall, the development of sophisticated analytical systems for assaying the entire C. elegans nervous system has the potential to improve our understanding of mechanisms of action for multiple anthelmintic classes.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We would like to thank Clay Dilks, Katie Evans, and Nicole Banks for their help with experiments and analysis and Amanda Shaver for her helpful comments on this manuscript. This work was supported by the National Institutes of Health NIAID grant R01AI153088 to ECA. This study used data made available by the C. elegans Neuronal Gene Expression Map and Network (NIH NINDS R01NS100547). Neurotransmitter system data are from WormAtlas neurotransmitter tables by Curtis M. Loer and James B. Rand (Loer and Rand, 2010), compiled from (Pereira et al., 2015; Gendrel et al., 2016; Serrano-Saiz et al., 2017). Some diagrams in figures were created using BioRender.com.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ijpddr.2022.10.004.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Abongwa M., Martin R.J., Robertson A.P. A brief review on the mode of action of ANTINEMATODAL drugs. Acta Vet. 2017;67(2):137–152. doi: 10.1515/acve-2017-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abrahante J.E., Miller E.A., Rougvie A.E. Identification of heterochronic mutants in Caenorhabditis elegans. Temporal misexpression of a collagen::green fluorescent protein fusion gene. Genetics. 1998;149(3):1335–1351. doi: 10.1093/genetics/149.3.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen E.C., Bloom J.S., Gerke J.P., Kruglyak L. A variant in the neuropeptide receptor npr-1 is a major determinant of Caenorhabditis elegans growth and physiology. PLoS Genet. 2014;10(2) doi: 10.1371/journal.pgen.1004156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen E.C., Shimko T.C., Crissman J.R., Ghosh R., Bloom J.S., Seidel H.S., Gerke J.P., Kruglyak L. A powerful new quantitative genetics platform, combining Caenorhabditis elegans high-throughput fitness assays with a large collection of recombinant strains. G3. 2015;5(5):911–920. doi: 10.1534/g3.115.017178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin S., Ziese M., Sternberg N. A novel role for site-specific recombination in maintenance of bacterial replicons. Cell. 1981;25(3):729–736. doi: 10.1016/0092-8674(81)90180-x. [DOI] [PubMed] [Google Scholar]
- Avramenko R.W., Redman E.M., Melville L., Bartley Y., Wit J., Queiroz C., Bartley D.J., Gilleard J.S. Deep amplicon sequencing as a powerful new tool to screen for sequence polymorphisms associated with anthelmintic resistance in parasitic nematode populations. Int. J. Parasitol. 2019;49(1):13–26. doi: 10.1016/j.ijpara.2018.10.005. [DOI] [PubMed] [Google Scholar]
- Boulin T., Etchberger J.F., Hobert O. Reporter gene fusions. WormBook. 2006;1–23 doi: 10.1895/wormbook.1.106.1. Apr 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd W.A., Smith M.V., Freedman J.H. Caenorhabditis elegans as a model in developmental toxicology. Methods Mol. Biol. 2012;889:15–24. doi: 10.1007/978-1-61779-867-2_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady S.C., Zdraljevic S., Bisaga K.W., Tanny R.E., Cook D.E., Lee D., Wang Y., Andersen E.C. A novel gene underlies bleomycin-response variation in Caenorhabditis elegans. Genetics. 2019;212(4):1453–1468. doi: 10.1534/genetics.119.302286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvelli L., McDonald P.W., Blakely R.D., DeFelice L.J. Dopamine transporters depolarize neurons by a channel mechanism. Proc. Natl. Acad. Sci. U. S. A. 2004;101(45):16046–16051. doi: 10.1073/pnas.0403299101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhary S., Lee S.A., Li Y., Patel D.S., Lu H. Graphical-model framework for automated annotation of cell identities in dense cellular images. Elife. 2021;10 doi: 10.7554/eLife.60321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent J.A., Smith M.M., Vassilatis D.K., Avery L. The genetics of ivermectin resistance in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U. S. A. 2000;97(6):2674–2679. doi: 10.1073/pnas.97.6.2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilks C.M., Hahnel S.R., Sheng Q., Long L., McGrath P.T., Andersen E.C. Quantitative benzimidazole resistance and fitness effects of parasitic nematode beta-tubulin alleles. Int. J. Parasitol. Drugs Drug Resist. 2020;14:28–36. doi: 10.1016/j.ijpddr.2020.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilks C.M., Koury E.J., Buchanan C.M., Andersen E.C. Newly identified parasitic nematode beta-tubulin alleles confer resistance to benzimidazoles. Int. J. Parasitol. Drugs Drug Resist. 2021;17:168–175. doi: 10.1016/j.ijpddr.2021.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driscoll M., Dean E., Reilly E., Bergholz E., Chalfie M. Genetic and molecular analysis of a Caenorhabditis elegans beta-tubulin that conveys benzimidazole sensitivity. J. Cell Biol. 1989;109(6 Pt 1):2993–3003. doi: 10.1083/jcb.109.6.2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastman C., Horvitz H.R., Jin Y. Coordinated transcriptional regulation of the unc-25 glutamic acid decarboxylase and the unc-47 GABA vesicular transporter by the Caenorhabditis elegans UNC-30 homeodomain protein. J. Neurosci. 1999;19(15):6225–6234. doi: 10.1523/JNEUROSCI.19-15-06225.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans K.S., Andersen E.C. The gene scb-1 underlies variation in Caenorhabditis elegans chemotherapeutic responses. G3. 2020;10(7):2353–2364. doi: 10.1534/g3.120.401310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans K.S., Brady S.C., Bloom J.S., Tanny R.E., Cook D.E., Giuliani S.E., Hippleheuser S.W., Zamanian M., Andersen E.C. Shared genomic regions underlie natural variation in diverse toxin responses. Genetics. 2018;210(4):1509–1525. doi: 10.1534/genetics.118.301311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A., Waterston R.H. Proper expression of myosin genes in transgenic nematodes. EMBO J. 1989;8(11):3419–3428. doi: 10.1002/j.1460-2075.1989.tb08506.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flames N., Hobert O. Gene regulatory logic of dopamine neuron differentiation. Nature. 2009;458(7240):885–889. doi: 10.1038/nature07929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frøkjær-Jensen C., Davis M.W., Ailion M., Jorgensen E.M. Improved Mos1-mediated transgenesis in C. elegans. Nat. Methods. 2012;9(2):117–118. doi: 10.1038/nmeth.1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frøkjaer-Jensen C., Davis M.W., Hopkins C.E., Newman B.J., Thummel J.M., Olesen S.-P., Grunnet M., Jorgensen E.M. Single-copy insertion of transgenes in Caenorhabditis elegans. Nat. Genet. 2008;40(11):1375–1383. doi: 10.1038/ng.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-González A.P., Ritter A.D., Shrestha S., Andersen E.C., Yilmaz L.S., Walhout A.J.M. Bacterial metabolism affects the C. elegans response to cancer chemotherapeutics. Cell. 2017;169(3):431–441. doi: 10.1016/j.cell.2017.03.046. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendrel M., Emily G Atlas, Hobert O. A cellular and regulatory map of the GABAergic nervous system of C. elegans. Elife. 2016 doi: 10.7554/elife.17686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson D.G., Young L., Chuang R.-Y., Venter J.C., Hutchison C.A., 3rd, Smith H.O. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 2009;6(5):343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- Hahnel S.R., Dilks C.M., Heisler I., Andersen E.C., Kulke D. Caenorhabditis elegans in anthelmintic research – old model, new perspectives. Int. J. Parasitol. Drugs Drug Resist. 2020;14:237–248. doi: 10.1016/j.ijpddr.2020.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahnel S.R., Zdraljevic S., Rodriguez B.C., Zhao Y., McGrath P.T., Andersen E.C. Extreme allelic heterogeneity at a Caenorhabditis elegans beta-tubulin locus explains natural resistance to benzimidazoles. PLoS Pathog. 2018;14(10) doi: 10.1371/journal.ppat.1007226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Y., Hu Z., Sieburth D., Kaplan J.M. RIC-7 promotes neuropeptide secretion. PLoS Genet. 2012;8(1) doi: 10.1371/journal.pgen.1002464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastie A.C., Georgopoulos S.G. Mutational resistance to fungitoxic benzimidazole derivatives in Aspergillus nidulans. J. Gen. Microbiol. 1971;67(3):371–373. doi: 10.1099/00221287-67-3-371. [DOI] [PubMed] [Google Scholar]
- Hotez P.J., Alvarado M., Basáñez M.-G., Bolliger I., Bourne R., Boussinesq M., Brooker S.J., Brown A.S., Buckle G., Budke C.M., et al. The global burden of disease study 2010: interpretation and implications for the neglected tropical diseases. PLoS Neglected Trop. Dis. 2014;8(7) doi: 10.1371/journal.pntd.0002865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurd D.D. Tubulins in C. elegans. WormBook. 2018;1–32 doi: 10.1895/wormbook.1.182.1. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Helminth Genomes Consortium . Vol. 51. Nature Publishing Group; 2019. Comparative Genomics of the Major Parasitic Worms; pp. 163–174. (Nat Genet). 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ireland C.M., Gull K., Gutteridge W.E., Pogson C.I. The interaction of benzimidazole carbamates with mammalian microtobule protein. Biochem. Pharmacol. 1979;28(17):2680–2682. doi: 10.1016/0006-2952(79)90049-2. [DOI] [PubMed] [Google Scholar]
- Jasmer D.P., Yao C., Rehman A., Johnson S. Multiple lethal effects induced by a benzimidazole anthelmintic in the anterior intestine of the nematode Haemonchus contortus. Mol. Biochem. Parasitol. 2000;105(1):81–90. doi: 10.1016/s0166-6851(99)00169-3. [DOI] [PubMed] [Google Scholar]
- Johnson N.M., Behm C.A., Trowell S.C. Heritable and inducible gene knockdown in C. elegans using Wormgate and the ORFeome. Gene. 2005;359:26–34. doi: 10.1016/j.gene.2005.05.034. [DOI] [PubMed] [Google Scholar]
- Johnstone I.L. Cuticle collagen genes. Expression in Caenorhabditis elegans. Trends Genet. 2000;16(1):21–27. doi: 10.1016/s0168-9525(99)01857-0. [DOI] [PubMed] [Google Scholar]
- Kage-Nakadai E., Imae R., Suehiro Y., Yoshina S., Hori S., Mitani S. A conditional knockout toolkit for Caenorhabditis elegans based on the Cre/loxP recombination. PLoS One. 2014;9(12) doi: 10.1371/journal.pone.0114680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan R.M., Vidyashankar A.N. An inconvenient truth: global worming and anthelmintic resistance. Vet. Parasitol. 2012;186(1–2):70–78. doi: 10.1016/j.vetpar.2011.11.048. [DOI] [PubMed] [Google Scholar]
- Kaymak E., Farley B.M., Hay S.A., Li C., Ho S., Hartman D.J., Ryder S.P. Efficient generation of transgenic reporter strains and analysis of expression patterns in Caenorhabditis elegans using library MosSCI. Dev. Dynam. 2016;245(9):925–936. doi: 10.1002/dvdy.24426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy B.P., Aamodt E.J., Allen F.L., Chung M.A., Heschl M.F., McGhee J.D. The gut esterase gene (ges-1) from the nematodes Caenorhabditis elegans and Caenorhabditis briggsae. J. Mol. Biol. 1993;229(4):890–908. doi: 10.1006/jmbi.1993.1094. [DOI] [PubMed] [Google Scholar]
- Lacey E. Mode of action of benzimidazoles. Parasitol. Today. 1990;6(4):112–115. doi: 10.1016/0169-4758(90)90227-u. [DOI] [PubMed] [Google Scholar]
- Lacey E., Prichard R.K. Interactions of benzimidazoles (BZ) with tubulin from BZ-sensitive and BZ-resistant isolates of Haemonchus contortus. Mol. Biochem. Parasitol. 1986;19(2):171–181. doi: 10.1016/0166-6851(86)90122-2. [DOI] [PubMed] [Google Scholar]
- Laclette J.P., Guerra G., Zetina C. Inhibition of tubulin polymerization by mebendazole. Biochem. Biophys. Res. Commun. 1980;92(2):417–423. doi: 10.1016/0006-291x(80)90349-6. [DOI] [PubMed] [Google Scholar]
- Lewis J.A., Wu C.H., Berg H., Levine J.H. The genetics of levamisole resistance in the nematode Caenorhabditis elegans. Genetics. 1980;95(4):905–928. doi: 10.1093/genetics/95.4.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Kirch S., Ambros V. The Caenorhabditis elegans heterochronic gene pathway controls stage-specific transcription of collagen genes. Development. 1995;121(8):2471–2478. doi: 10.1242/dev.121.8.2471. [DOI] [PubMed] [Google Scholar]
- Loer C., Rand J. WormAtlas; 2010. WormAtlas Neurotransmitters Table - the Evidence for Classical Neurotransmitters in Caenorhabditis elegans. (In Press. Albert Einstein College of Medicine). Oct. [DOI] [Google Scholar]
- Maduro M., Pilgrim D. Identification and cloning of unc-119, a gene expressed in the Caenorhabditis elegans nervous system. Genetics. 1995;141(3):977–988. doi: 10.1093/genetics/141.3.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maduro M., Pilgrim D. Conservation of function and expression of unc-119 from two Caenorhabditis species despite divergence of non-coding DNA. Gene. 1996;183(1–2):77–85. doi: 10.1016/s0378-1119(96)00491-x. [DOI] [PubMed] [Google Scholar]
- Marshall S.D., McGhee J.D. Coordination of ges-1 expression between the Caenorhabditis pharynx and intestine. Dev. Biol. 2001;239(2):350–363. doi: 10.1006/dbio.2001.0442. [DOI] [PubMed] [Google Scholar]
- Mohammedsalih K.M., Krücken J., Khalafalla A., Bashar A., Juma F.-R., Abakar A., Abdalmalaik A.A.H., Coles G., von Samson-Himmelstjerna G. New codon 198 β-tubulin polymorphisms in highly benzimidazole resistant Haemonchus contortus from goats in three different states in Sudan. Parasites Vectors. 2020;13(1):114. doi: 10.1186/s13071-020-3978-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz-Jiménez C., Ayuso C., Dobrzynska A., Torres-Mendéz A., Ruiz P. de la C., Askjaer P. An efficient FLP-based toolkit for spatiotemporal control of gene expression in Caenorhabditis elegans. Genetics. 2017;206(4):1763–1778. doi: 10.1534/genetics.117.201012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira L., Kratsios P., Serrano-Saiz E., Sheftel H., Mayo A.E., Hall D.H., White J.G., LeBoeuf B., Garcia L.R., Alon U., et al. Vol. 4. eLife Sciences Publications, Ltd; 2015. A Cellular and Regulatory Map of the Cholinergic Nervous System of C. elegans. (Elife). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian H., Robertson A.P., Powell-Coffman J.A., Martin R.J. Vol. 22. Wiley; 2008. Levamisole Resistance Resolved at the Single-Channel Level in Caenorhabditis elegans; pp. 3247–3254. (FASEB J). 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieckher M., Tavernarakis N. Caenorhabditis elegans microinjection. Bio Protoc. 2017;7(19) doi: 10.21769/BioProtoc.2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roos M.H., Kwa M.S.G., Grant W.N. New genetic and practical implications of selection for anthelmintic resistance in parasitic nematodes. Parasitol. Today. 1995;11(4):148–150. [Google Scholar]
- Semple J.I., Garcia-Verdugo R., Lehner B. Rapid selection of transgenic C. elegans using antibiotic resistance. Nat. Methods. 2010;7(9):725–727. doi: 10.1038/nmeth.1495. [DOI] [PubMed] [Google Scholar]
- Serrano-Saiz E., Gulez B., Pereira L., Gendrel M., Kerk S.Y., Vidal B., Feng W., Wang C., Kratsios P., Rand J.B., et al. Modular organization of cis-regulatory control information of neurotransmitter pathway genes in Caenorhabditis elegans. Genetics. 2020;215(3):665–681. doi: 10.1534/genetics.120.303206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano-Saiz E., Pereira L., Gendrel M., Aghayeva U., Bhattacharya A., Howell K., Garcia L.R., Hobert O. A neurotransmitter Atlas of the Caenorhabditis elegans male nervous system reveals sexually dimorphic neurotransmitter usage. Genetics. 2017;206(3):1251–1269. doi: 10.1534/genetics.117.202127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano-Saiz E., Poole R.J., Felton T., Zhang F., De La Cruz E.D., Hobert O. Modular control of glutamatergic neuronal identity in C. elegans by distinct homeodomain proteins. Cell. 2013;155(3):659–673. doi: 10.1016/j.cell.2013.09.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheir-Neiss G., Lai M.H., Morris N.R. Identification of a gene for beta-tubulin in Aspergillus nidulans. Cell. 1978;15(2):639–647. doi: 10.1016/0092-8674(78)90032-6. [DOI] [PubMed] [Google Scholar]
- Shimko T.C., Andersen E.C. COPASutils: an R package for reading, processing, and visualizing data from COPAS large-particle flow cytometers. PLoS One. 2014;9(10) doi: 10.1371/journal.pone.0111090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor S.R., Santpere G., Weinreb A., Barrett A., Reilly M.B., Xu C., Varol E., Oikonomou P., Glenwinkel L., McWhirter R., et al. Molecular topography of an entire nervous system. Cell. 2021;184(16):4329–4347. doi: 10.1016/j.cell.2021.06.023. e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thein M.C., McCormack G., Winter A.D., Johnstone I.L., Shoemaker C.B., Page A.P. Caenorhabditis elegans exoskeleton collagen COL-19: an adult-specific marker for collagen modification and assembly, and the analysis of organismal morphology. Dev. Dynam. 2003;226(3):523–539. doi: 10.1002/dvdy.10259. [DOI] [PubMed] [Google Scholar]
- Theodorides V.J., Scott G.C., Lademan M.S. Strains of Haemonchus contortus resistant against benzimidazole anthelmintics. Am. J. Vet. Res. 1970;31(5):859–863. [PubMed] [Google Scholar]
- Thomas B.J., Wight I.E., Chou W.Y.Y., Moreno M., Dawson Z., Homayouni A., Huang H., Kim H., Jia H., Buland J.R., et al. CemOrange2 fusions facilitate multifluorophore subcellular imaging in C. elegans. PLoS One. 2019;14(3) doi: 10.1371/journal.pone.0214257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viney M. The genomic basis of nematode parasitism. Brief Funct. Genom. 2018;17(1):8–14. doi: 10.1093/bfgp/elx010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J.G., Southgate E., Thomson J.N., Brenner S. The structure of the nervous system of the nematode Caenorhabditis elegans. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1986;314(1165):1–340. doi: 10.1098/rstb.1986.0056. [DOI] [PubMed] [Google Scholar]
- Wit J., Dilks C.M., Andersen E.C. Complementary approaches with free-living and parasitic nematodes to understanding anthelmintic resistance. Trends Parasitol. 2021;37(3):240–250. doi: 10.1016/j.pt.2020.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wit J., Rodriguez B.C., Andersen E.C. Natural variation in Caenorhabditis elegans responses to the anthelmintic emodepside. Int. J. Parasitol. Drugs Drug Resist. 2021;16:1–8. doi: 10.1016/j.ijpddr.2021.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yemini E., Lin A., Nejatbakhsh A., Varol E., Sun R., Mena G.E., Samuel A.D.T., Paninski L., Venkatachalam V., Hobert O. NeuroPAL: a multicolor Atlas for whole-brain neuronal identification in C. elegans. Cell. 2021;184(1):272–288. doi: 10.1016/j.cell.2020.12.012. e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdraljevic S., Strand C., Seidel H.S., Cook D.E., Doench J.G., Andersen E.C. Natural variation in a single amino acid substitution underlies physiological responses to topoisomerase II poisons. PLoS Genet. 2017;13(7) doi: 10.1371/journal.pgen.1006891. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
A list of plasmids used with vendor ID information and a list of C. elegans strains and genotypes used in experiments are included as supplementary information (Tables S1 and S2). The data and code used to process these data are available at https://github.com/AndersenLab/2022_ben1sensitivity_SBG.